Abstract
Synthetic biology approaches offer potential for large-scale and sustainable production of natural products with bioactive potency, including ginsenosides, providing a means to produce novel compounds with enhanced therapeutic properties. Ginseng, known for its non-toxic and potent qualities in traditional medicine, has been used for various medical needs. Ginseng has shown promise for its antioxidant and neuroprotective properties, and it has been used as a potential agent to boost immunity against various infections when used together with other drugs and vaccines. Given the increasing demand for ginsenosides and the challenges associated with traditional extraction methods, synthetic biology holds promise in the development of therapeutics. In this review, we discuss recent developments in microorganism producer engineering and ginsenoside production in microorganisms using synthetic biology approaches.
Graphical abstract
1. Introduction
A variety of valuable biomaterials exist in nature. Secondary metabolites refer to bioactive compounds and chemicals produced by various life forms that are not directly involved in growth, development, or reproduction. These metabolites serve various functions, such as defense against pests and pathogens, adaptation to environmental stressors, and communication with other organisms [1,2]. Several secondary metabolites include saponins, polysaccharides, flavonoids, and phenolic compounds [3]. Each category has distinct chemical structures and biological activities, contributing to their overall medicinal properties. For example, in ginseng, various secondary metabolites called ginsenosides exist, which are a class of triterpene saponins that have a 4-ring, steroid-like dammarane structure, and can affect the cardiovascular system, central nervous system, and immune system of animals. Ginsenosides have antioxidant, anticancer, antiaging, and neuroprotective properties [4].
Over the past few years, approximately 200 ginsenosides have been isolated and identified from various parts of the Panax species, including leaves, flower buds, roots, and fruits [5,6]. Ginsenosides, the major bioactive constituents of ginseng, possess a wide range of pharmacological activities, including anti-cancer, anti-diabetic, neuroprotective, radioprotective, anti-amnestic, and anti-aging effects [4,5,[7], [8], [9], [10]]. These diverse pharmacological activities are attributed to the physical structures and chemical properties of ginsenosides. Despite their versatile bioactivities, the low abundance of ginsenosides in Panax plants, long cultivation periods (6–7 years for maturity), and complicated extraction-separation processes make direct extraction of ginsenosides from Panax ginseng commercially infeasible. To address these issues, recent advancements in metabolic engineering and synthetic biology have emerged as promising alternatives for the production of microbial ginsenosides. Microbial synthesis of ginsenosides by heterologous hosts such as Saccharomyces cerevisiae and Escherichia coli offers numerous advantages, including rapid growth with high cell density cultivation, simple nutritional requirements, well-established and reliable genetic manipulation methods, and easy extraction and separation [11,12]. Although there are as yet few reports regarding the microbial biosynthesis of ginsenosides compared to studies on the pharmacological effects of ginsenosides, substantial advances have been made to transform microorganisms into cellular factories for ginsenoside overproduction.
In this review, we will focus on and provide a detailed explanation of research aimed to increase the productivity of ginsenosides and non-ribosomal peptides or polyketides using synthetic biology. We highlight below recent developments in the production and engineering of natural products by synthetic biology, focusing on synthetic biology approaches for ginsenosides.
2. Synthetic biology approaches in natural product production
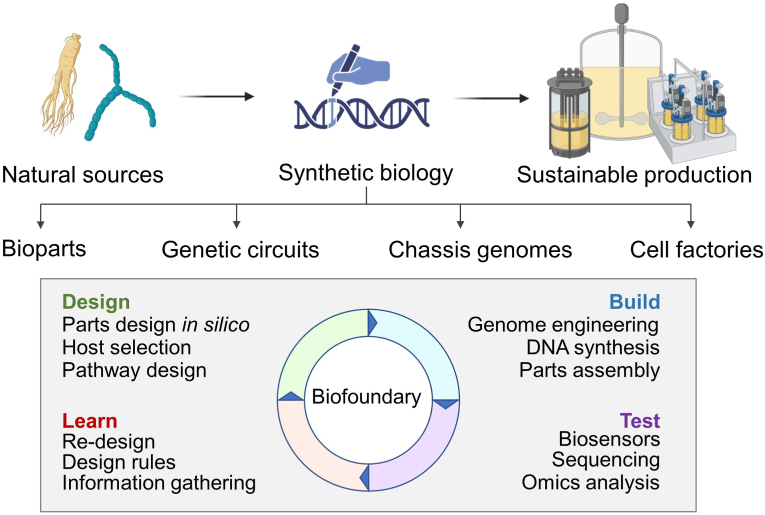
Synthetic biology is a multidisciplinary field that combines principles from biology, engineering, and computer science to design and create new biological systems and organisms with unique functionalities [13]. Regarding secondary metabolites, synthetic biology offers the potential to engineer plants or other organisms to produce higher yields of these valuable compounds (Fig. 1) [14]. By utilizing the Synthetic Biology Open Language to create standard nomenclature for biological terms, databases of genes, proteins, and metabolic pathways were constructed [15]. Using established databases to identify controllable genetic modifications or to optimize metabolic pathways, synthetic biology can overcome the limitations of natural production and enable scalable and cost-effective production of all kinds of metabolites [[16], [17], [18]]. One powerful tool within synthetic biology for improving secondary metabolite production is the collection of databases such as Bioparts, which combines information from the Inventory of Composable Elements (ICE) parts registry, SynBioHub parts registry, iGEM parts registry, NCBI, Addgene, Genescript, and other databases [17,19]. Bioparts are genetic components that can be assembled to create new functionalities within living organisms, and there is an expanding database of DNA, RNA, protein, and metabolic pathways available for this purpose. These bioparts can enhance key enzyme expression in secondary metabolite synthesis to integrate modified metabolic pathways. Researchers can increase the production or potency of specific bioactive compounds by optimizing genetic pathways and introducing synthetic genes into plants in vivo or into bacteria or yeasts in vitro.
Fig. 1.
Overview of synthetic biology to produce natural products. The key technologies and disciplines in each step (design, build, test, and learn) are interconnected for further optimization of the production process.
Genetic circuits, also known as gene regulation networks, are a collection of molecular regulators that interact with each other and other substances in the cell to govern the gene expression levels of mRNA and proteins, which can enhance continuous secondary metabolite production [20]. These circuits include genetic components such as promoters, terminators, and regulatory elements that precisely control gene expression, similar to an electric diagram [21]. By incorporating ideas from electric circuits, NAND, NOR, or XOR gates into the genetic makeup of plants or artificial cells, researchers can continuously optimize metabolic pathways to produce the desired secondary metabolites [22]. Genetic circuits can regulate key enzyme expression levels, ensuring a continuous and steady yield of targeted bioactive compounds.
Two in vitro methods, utilizing cell factories or creating artificial cells, can enhance the continuous production of secondary metabolites. The first method involves the use of cell factories, which are powerful tools in the field of synthetic biology. Researchers can engineer microorganisms, such as bacteria or yeast, to biosynthesize valuable compounds [23,24]. This approach overcomes the limitations of traditional cultivation methods, enabling the optimization of production processes for continuous, high-yield, and efficient production of the desired metabolites. Cell factories also offer scalability and controllability, making them a compelling choice for large-scale continuous production. The second method is the creation of artificial cells, which can be designed and built in the laboratory. These engineered cells, often called protocells, mimic the metabolic processes of natural cells while adding specific functionalities [23]. By introducing genes and genetic circuits into these artificial cells, that may or may not be whole cells but membranes or organelles, researchers can program protocells to continuously produce high levels of metabolites. These artificial cells, which may not be considered “alive” but act in a directed, controlled manner, can be fine-tuned to respond to various environmental conditions, ensuring consistent and efficient production [25,26].
The Keasling lab employs yeast cell factories as an exemplary method for producing artemisinic acid, showcasing the potential of cell factories in complex plant secondary metabolite production [27,28]. This approach involves developing genetic circuits in yeast as cell factories to enhance the synthesis of specific bioactive compounds and increase artemisinin production from Artemisia spp. By integrating biosynthetic pathways and regulatory circuits into the genetic makeup of organisms, scientists can finely control gene expression and metabolic processes [29]. This level of precision allows for generating higher quantities and purer forms of the desired metabolites, ultimately enhancing the therapeutic properties and commercial value. Furthermore, using advanced synthetic biology techniques, the Smolke lab has developed methods for enhancing plant phytochemical production. These methods are not limited to plants; they can also be applied to secondary metabolite production [30]. The Smolke lab approach extends to engineering microorganisms, including bacteria and yeast, to produce various compounds [13,31]. By incorporating these techniques into plants, researchers can further boost the production of bioactive compounds. Incorporating synthetic biology involves manipulating the cellular machinery of microorganisms to efficiently produce target molecules, which can subsequently be applied to plants, resulting in increased secondary metabolite production.
Scientists have deployed advanced techniques to expedite synthetic biology research and optimize the design-build-test-learn (DBTL) cycle for secondary metabolite production [32,33]. These methods include high-throughput screening, which swiftly identifies the most promising genetic circuits and biosynthetic pathways [34]. Researchers achieve this by screening vast libraries of genetic components and their variations, pinpointing combinations that yield the highest quantities of target metabolites. Furthermore, machine learning algorithms have been integrated into the design phase [35]. These algorithms predict the performance of genetic circuits and assist in selecting the most optimal designs. This predictive capability enhances the efficiency and precision in secondary metabolite production. Biofoundry is an integrated synthetic biology facility with computers and robots that includes design software, robotic liquid-handling and high-throughput analytical equipment, and data management systems. A global biofoundry alliance has been established to bolster progress, fostering collaboration and cooperation among researchers worldwide. After COVID-19, global research on viruses improved significantly in a short time with the help of biofoundries in analysis of the virus, receptor, and vaccine designs. A global joint development effort coordinating disease treatment and establishing effective partnerships was made [36]. This alliance amplifies output and facilitates collective advancements in synthetic biology, ultimately driving the improvement of secondary metabolite production [37].
3. Modular production of natural products: polyketides and nonribosomal peptides
Natural products including ginsenosides have long served as a source of drugs, mainly including various valuable metabolites such as plant products, polyketides (PKs), and nonribosomal peptides (NRPs) [38,39]. These types of molecules exhibit multifunctional potency against various diseases, including cancer, microbial infections, inflammatory conditions, and metabolism-related diseases. However, chemical diversity and difficulties in isolation have hindered the further development of lead molecules. Recent developments in synthetic biology provide an alternative option for producing natural compounds. We specifically highlight PKs and NRPs because they encompass vast and diverse classes of potent natural products synthesized in various systems, including plants and microorganisms [40,41]. The genetic engineering technologies and producers used for PKs and NRPs have been adapted to produce ginsenosides, which we will discuss in the following chapters.
3.1. Polyketides
Polyketides (PKs) are enzymatically produced through iterative assembly reactions using short-chain acyl units by polyketide synthases (PKS). Due to their high flexibility, the structural diversity of polyketides covers major classes of drugs, including polyenes, polycyclic aromatic compounds, and macrolides [42].
There are three types of PKS: Type I PKSs, mostly found in bacteria and fungi, consist of multi-catalytic domains and are involved in sequential reductive assembly reactions. Type II PKSs, predominantly found in actinomyces, constitute complexes composed of monofunctional enzymes. Type III PKSs, widely found in bacteria, fungi, and plants, are responsible for product chain elongation and termination through ketosynthase [43]. Among these, Type I PKSs have been extensively studied and engineered. The multidomain nature of Type I PKSs enhances polyketide chemical diversity. Type I PKS modules have three key domains that act in the chain elongation process: acetyltransferase (AT), ketosynthase (KS), and acyl carrier protein (ACP) [44]. To incorporate specific acyl building blocks or reduce the polyketide chain, ketoreductase, dehydratase, and enoylreductase are involved, generating a high degree of chemical diversity and stereospecificity. The consecutive polyketide assembly process is completed by releasing the chain from PKS using a thioesterase domain, either in linear or cyclic form, followed by product detachment from ACP through hydrolysis or lactonization (Fig. 2A). The released polyketides can undergo further enzymatic modifications, such as glycosylation and alkylation.
Fig. 2.
Schematic of the modular reaction of polyketide synthase (A) and non-ribosomal peptide synthase (B). ACP, acyl carrier protein; KS, ketosynthase; AT, acyltransferase; ER, enolreductase; DR, dehydratase; TE, thioesterase; A. adenylation domain; C, cyclization domain; PCP, peptidyl carrier proteins.
Recent approaches in the evolution and combinatorial assembly of PKSs have made significant progress, leading to updated PKS modules containing the KS domain and the upstream ACP. This emphasizes the significance of the native connection between PKS domains. Recent studies with the pikromycin and venemycin PKSs have demonstrated successful engineering of hybrid PKSs using updated modules, although challenges remain in the decreased efficiency of hybrid PKSs and the relatively larger size of PKSs, which may hinder further engineering [45]. PKS engineering has recently been applied to the reconstitution of novel biosynthetic routes for tropane alkaloids, such as hyoscyamine. To produce tropinone, an intermediate tropane, a plant-derived Type III PKS was used, achieving a high yield [46].
3.2. Nonribosomal peptides
Nonribosomal peptides (NRPs) are a class of bioactive peptides, such as daptomycin, synthesized by nonribosomal peptide synthetases (NRPSs). Similar to polyketides, NRPSs require adenylation proteins, peptidyl carrier proteins (PCPs), and condensation proteins: adenylation domains function like the AT domain of PKS, PCP domains serve as handles similar to the ACP of PKSs, and condensation domains facilitate the extension of peptide bonds. Finally, a thioester domain releases the peptide product from the PCP of the module. During termination, the peptide product can be cyclized, forming structures such as macrolides, or a linear peptide is liberated. Similar to PKSs, chemical editing, such as intermolecular cyclization and methylation, can be incorporated during the modular production of NRP by additional domains in each module (Fig. 2B) [39]. Recently, NRPS modules have been engineered and designed to increase production yield or develop novel biosynthetic pathways for natural products [47]. In particular, the condensation domain has been newly designed using chimeric domains to allow specificity for exchanging amino acids, employing the concepts of synthetic biology [48].
4. Sustainable production of ginsenosides by synthetic metabolic engineering strategies
Chemically, ginsenosides are a group of glycosylated triterpenes, also known as triterpenoid saponins. They can be classified into the following types based on the backbone and the number of modified glycosides: (1) tetracyclic dammarane-type saponins, including protopanaxadiol (PPD)-type, protopanaxatriol (PPT)-type, and C-17 side-chain isomers; (2) tetracyclic ocotillol (OCT)-type saponins with a side chain containing a furan ring; and (3) oleanolic acid (OA)-type pentacyclic triterpenoids with aglycon OA (Fig. 3 and Table 1) [5,[49], [50], [51]].
Fig. 3.
Ginsenoside biosynthesis pathway, starting with either the mevalonate (MVA) pathway (in yeast) or the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway (in bacteria) and examples of different ginsenoside classes derived from isopentenyl diphosphate (IPP) as the building blocks of ginsenoside. G3P, D-glyceraldehyde-3-phosphate; DXP, 1-deoxy-D-xylulose-5-phosphate; MEP, 2-C-methyl-D-erythritol 4-phosphate; CDP-ME, 4-diphosphocytidyl-2-C-methyl-D-erythritol; CDP-MEP, 4-diphosphocytidyl-2-C-methylerythritol diphosphate; MECPP, methylerythritol cyclodiphosphate; HMBPP, hydroxymethylbutenyl diphosphate; IPP, isoprenyl diphosphate; DMAPP, dimethylallyl diphosphate; FPP, farnesyl pyrophosphate; HMG-CoA, 3-hydroxy-3-methylglutaryl coenzyme A; MVA, mevalonate; MVP, mevalonate-5-phosphate; MVPP, mevalonate-5-diphosphate; SQ, squalene, 2,3-OSQ, 2,3-oxidosqualene; DXS, deoxyxylulose 5-phosphate synthase; DXR, deoxyxylulose 5-phosphate reductoisomerase; ispD, 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase; ispE, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; ispF, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase; ispG, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase; ispH, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; ispA, farnesyl diphosphate synthase; Erg10, acetyl-CoA C-acetyltransferase; Erg13, 3-hydroxy-3-methylglutaryl-CoA; HMG1&2, HMG-CoA reductase; Erg12, mevalonate kinase; Erg8, phosphomevalonate kinase; Erg20, farnesyl pyrophosphate synthase; Erg9, farnesyl-diphosphate farnesyl transferase; Erg1, squalene monooxygenase; DS, dammarenediol-II synthase; PPDS, protopanaxadiol synthase; PPTS, protopanaxatriol synthase; UGT, UDP-glycosyltransferase; URT, UDP-rhamnosyltransferase.
Table 1.
Summary of ginsenoside production in microbial cell factories.
| Classification | Synthetic metabolic engineering strategies | Products | Titer | Cultivation condition | Host strain | Ref |
|---|---|---|---|---|---|---|
| Protopanaxadiol (PPD)-Type |
ERG1, DDS, ATR2-1, PgUGT1, tHMG1, UPC2-1 |
CK | 802.1 μg L−1 | Shake-flask | S. cerevisiae | [69] |
|
ERG20, ERG9, ERG1, PgDDS, PgPPDS-AtCPR1, tHMG1 ERG12, ERG13, MVD1, ERG8, IDI1, ERG10 INO2, UPC2-1 |
PPD | 15.88 g L−1 | Fed-batch | S. cerevisiae | [72] | |
| tHMG1, PgDS, PgUGT74AE2 | 3β-O-Glc-DM | 2.4 g L−1 | Fed-batch | S. cerevisiae | [73] | |
| tHMG1, PgDS, PgUGT1 | 20S–O-Glc-DM | 5.6 g L−1 | Fed-batch | S. cerevisiae | [73] | |
| PgDS, tHMG1, PgPPDS, AtATR2, PgUGT1, UGT74AE2, IDI1, ERG20, ERG9, ERG1, ERG7, HAC1, PGM1, PGM2, UGP1 | F2 | 21.0 mg L−1 | Shake-flask | S. cerevisiae | [74] | |
| PgDS, tHMG1, PgUGT1, IDI1, ERG20, ERG9, ERG1, ERG7, INO2, PGM1, PGM2, UGP1 | 3β,20S-Di-O-Glc-DM | 2.6 g L−1 | Fed-batch | S. cerevisiae | [74] | |
| ERG20, tHMG1, AtCYPOR1, AtCYPOR1, PgDS, PgPPDS, PgCPR5 | PPD | 17.31 mg L−1 | Shake-flask | S. cerevisiae | [78] | |
| PgDS, ERG1, tHMG1, ERG20, ERG9, PgPPDS-AtATR1(1–46) | PPD | 1436.6 mg L−1 | Fed-batch | S. cerevisiae | [79] | |
| PgDDS, ERG1 | DM | 0.10 mg g DCW−1 | Not reported | P. pastoris | [98] | |
| PgDDS, PgPPDS, ATR2.1, tHMG1, ERG20, PgERG1, ERG9, PgUGT45 | Rh2 | 1.45 μmol g DCW−1 | Shake-flask | S. cerevisiae | [67] | |
| PgDDS, PgPPDS, ATR2.1, tHMG1, ERG20, PgERG1, ERG9, PgUGT45, PgUGT29 | Rg3 | 3.49 μmol g DCW−1 | Shake-flask | S. cerevisiae | [67] | |
| PGM2, UGP1, PgUGT1 | CK | 1.7 g L−1 | Fed-batch | S. cerevisiae | [81] | |
| tHMG1, PgCPR1, ERG1, ERG20, ERG9, PgDDS, PgPPDS, ERG12, ERG13, ERG8, ERG19, IDI, ERG10, PgUGT1, UGP1, PGM2, YNK1, ALG5 | CK | 5.74 g L−1 | Fed-batch | S. cerevisiae | [82] | |
| PgCPR1, ERG1, ERG20, ERG9, PgDDS, ERG12, ERG13, ERG8, ERG19, IDI, ERG10, tHMG1, PgPPDS, PnUGT50 | Rh2 | 2.25 g L−1 | Fed-batch | S. cerevisiae | [83] | |
| ScIDI, ispA, ScERG9, McSqE, AtCPR, PgDS | DM | 8.63 mg L−1 | Shake-flask | E. coli | [84] | |
| Microbacterium sp. Gsoil 167 BglG167b | LXXV | 5.7 g | Not reported | E. coli | [85] | |
| PnUGT95 | 20(R)-CK | 0.57 mg L−1 | Not reported | E. coli | [86] | |
| ERG20, tHMG1, AtCPR1, PgDS, PgPPDS, INO2 | PPD | 12.1 mg L−1 | Shake-flask | S. cerevisiae | [88,90] | |
| PEX11, PEX34, ERG9, PgERG1, PgDS, PTS2, tHMG1, PgPPDS, PTS1, PgCPR, ATG36 | PPD | 4 mg L−1 | Fed-batch | S. cerevisiae | [90] | |
| Protopanaxatriol (PPT)-type | PnUGT95 | 20(R)-F1 | 6.48 mg L−1 | Not reported | E. coli | [86] |
| ERG20, PgERG1, ERG9, tHMG1, CYP716A53v2, PgCPR1, PgUGT100 | Rh1 | 98.2 mg L−1 | Shake-flask | S. cerevisiae | [99] | |
| ERG20, PgERG1, ERG9, tHMG1, CYP716A53v2, PgCPR1, PgUGT1 | F1 | 42.1 mg L−1 | Shake-flask | S. cerevisiae | [99] | |
| CYP716A53v2, PgUGT71A54, PgURT94, RHM | Rg2 | 1.3 g L−1 | Fed-batch | S. cerevisiae | [68] | |
| CYP716A53v2, PgUGT71A53, PgUGT71A54, PgURT94, RHM | Re | 3.6 g L−1 | Fed-batch | S. cerevisiae | [68] | |
| Oleanolic acid (OA)-type | GgbAS, MtCPR, MtCYP716A12, tHMG1, ERG9, ERG1 | OA | 606.9 mg L−1 | Fed-batch | S. cerevisiae | [77] |
Pg: Panax ginseng; Sc: Saccharomyces cerevisiae; Pn: Panax notoginseng; Mc: Methylococcus capsulatus; Mt: Medicago truncatula; Gg: Glycyrrhiza glabra.
4.1. Ginsenoside biosynthetic pathway
Ginsenosides can be synthesized from 2,3-oxidosqualene by utilizing the universal precursors dimethylallyl diphosphate (DMAPP) and isopentenyl diphosphate (IPP) as their building blocks. Notably, it has been reported that IPP and DMAPP are primarily generated through two different pathways: the mevalonate (MVA) pathway, which is found in most eukaryotes, and the non-MVA pathway, also known as the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway, prevalent in most prokaryotes (Fig. 3) [[52], [53], [54], [55], [56], [57]].
The MVA pathway begins with the condensation of two molecules of acetyl-CoA by acetoacetyl-CoA thiolase (Erg10), forming acetoacetyl-CoA [58,59]. This compound reacts with a third molecule of acetyl-CoA via 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase (Erg13) to produce HMG-CoA. HMG-CoA is subsequently converted to mevalonate by HMG-CoA reductase in two NADPH-dependent reduction steps (Hmg1 and Hmg2). Following the sequential phosphorylation of MVA to MVA-5-diphosphate via MVA kinase (Erg12) and phospho-MVA kinase (Erg8), MVA-5-diphosphate is decarboxylated by an ATP-dependent MVA pyrophosphate decarboxylase (Mvd1) to yield IPP. IPP can then be isomerized to DMAPP via isopentenyl diphosphate:dimethylallyl diphosphate isomerase (Idi1). On the other hand, the MEP pathway initiates with the formation of 1-deoxy-D-xylulose-5-phosphate (DXP) via the condensation reaction between D-glyceraldehyde 3-phosphate (GAP) and pyruvate (PYR) [60]. Subsequently, the reduction reaction of DXP leads to the formation of MEP, followed by several enzymatic steps to convert to (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate (HMBPP). Finally, HMBPP is converted to IPP and DMAPP.
Next, the condensation of IPP and DMAPP is catalyzed by farnesyl diphosphate synthase (FPS/Erg20) to form farnesyl diphosphate (FPP). FPP is transformed into 2,3-oxidosqualene in two enzymatic reactions facilitated by squalene synthase (SqS/Erg9) and squalene epoxidase (SqE/Erg1) [61]. Various kinds of ginsenosides with diverse structures are consequently formed through multiple cyclizations, oxidations, and glycosylations. Specifically, 2,3-oxidosqualene undergoes cyclization by oxidosqualene cyclases (OSCs) as β-amyrin synthase (β-AS) and dammarenediol synthase (DDS) to generate triterpene saponin skeletons [62]. β-AS can cyclize 2,3-oxidosqualene to produce β-amyrin in the biosynthesis of OA-type ginsenosides, while DDS catalyzes the conversion from 2,3-oxidosqualene into dammarenediol-II (DM) in the biosynthesis of dammarane-type ginsenosides. After the formation of the ginsenoside skeletons, various ginsenosides and their derivatives are generated from DM and β-amyrin via hydroxylation by cytochrome P450s (CYP450s) and glycosylation by UDP-glycosyltransferases (UGTs) [8]. These modifications not only enhance the stability and structural diversity of ginsenosides but also influence their physicochemical and physiological properties [[63], [64], [65], [66]].
Among the reported ginseng CYP450 genes, CYP716A47 acts as a protopanaxadiol synthase (PPDS) hydroxylating DM at the C-12 position, yielding PPD. CYP716A53v2 as the protopanaxatriol synthase (PPTS) catalyzes the generation of PPT from PPD for the synthesis of dammarene-type ginsenosides. CYP716A52v2 (OA synthase, OAS) acts as a β-amyrin 28-oxidase modifying β-amyrin into OA for the synthesis of an OA-type saponin. Finally, ginsenosides and analogs are synthesized by UGTs, which is the final step in ginsenoside biosynthesis. UGTs are responsible for synthesizing diverse ginsenosides and their analogs by adding monosaccharides to triterpenoid aglycones, mainly at C-3 and/or C-20 for PPD-type ginsenosides and at C-6 and/or C-20 for PPT-type ginsenosides. This glycosylation modification imparts greater water solubility, stability, and pharmacological properties to these compounds. For example, P. ginseng-derived UGT1 can produce compound K (CK) from PPD and also convert PPT to F1. UGT45 and UGT29 can convert PPD to Rh2 and Rh3, respectively [67]. UGT45 can modify the hydroxyl group at the C3 position of CK to generate F2. URT94, a rhamnosyltransferase, adds another layer of complexity to ginsenoside biosynthesis by transferring rhamnose moieties to specific hydroxyl groups [68].
4.2. Ginsenoside biosynthesis in yeast
Yeast has become a workhorse for ginsenoside synthesis using metabolic engineering and synthetic biology approaches, because of similar intracellular structures to plant cells and because the inherent MVA pathway in yeast enables the reconstruction of ginsenoside pathways into yeast metabolism (Table 1) [[69], [70], [71], [72]]. Typically, the initial steps in ginsenoside production in yeast focus on increasing the flux of the precursors, IPP and DMAPP, in the MVP pathway, while downregulating sterol biosynthesis to improve the availability of 2,3-oxidosqualene. Engineering hubs can be identified at the rate-limiting HMG-CoA reductase, responsible for converting HMG-CoA to mevalonic acid, and the transcriptional activator Upc2, which activates genes in the MVP pathway. In addition, overexpression of IPP isomerase, IDI1, increased production of specific ginsenosides, such as CK and F2 [73]. Overexpression of genes in the MVA pathway, such as ERG20, ERG9, and ERG1, also improved ginsenoside production by enhancing the conversion rate of intermediate molecules [74].
Functional groups are subsequently attached to the ginsenoside skeleton by a variety of enzymes such as OSCs, CYP450s, and UGTs from different sources, primarily from P. ginseng. CYP450s require the delivery of two electrons from NAD(P)H by cytochrome P450 reductase (CPR) to catalyze the oxidation of its substrate. Thus, coexpression of CYP450s along with their redox partner CPR is crucial for converting ginsenosides in yeast [75,76]. For instance, Zhao et al. screened and examined the varying CYP450-CPR pairs for boosting OA production. Among the four redox partners from Arabidopsis thaliana, Lotus japonicas, Glycyrrhiza urelensis, and Medicago truncatula, the combination of CYP716A12 and CPR from M. truncatula exhibited the highest coupling efficiency and enhanced OA production [77].
The functional heterologous expression of plant-derived enzymes involved in ginsenoside biosynthesis in microbial hosts poses a significant challenge. To overcome this obstacle, chaperone-assisted expression systems have been extensively employed to improve the expression of heterologous proteins such as CYP450s. Kim et al. demonstrated that the successful pairing of cognate chaperones with CYP450s could facilitate functional assembly of the PPD biosynthesis pathway [78]. When coexpression of cognate chaperones was coupled to the expression of PPD biosynthesis enzymes from P. ginseng, PPD production increased significantly by more than 2.5-fold. Similarly, it was reported that the fusion of P. ginseng PPDS and its redox partner CPR from A. thaliana (truncated ATR1, with 46 amino acids deleted at the N-terminus) had a strong effect on the CYP450 oxidation reaction. The fusion enzyme (PPDS-ATR1) leads to a 71 % increase in PPD production compared to separate expression of the PPDS and ATR1 genes [79]. The results showed a remarkable conversion rate of DM to PPD, reaching 96.8 %, and yielded 1436.6 mg L−1 of PPD during a fed-batch fermentation in a 5 L bioreactor.
In addition, further glycosylation modifications were made to expand the diversity of ginsenosides. Although UGTs have been successfully expressed in yeast, glycosylation catalyzed UGTs remain the key rate-limiting reaction in the ginsenoside synthesis pathway [80]. For instance, an insufficient supply of UDP-glucose as the sugar donor for UGT activity was proved to be the rate-limiting factor. To stimulate UDP-glucose, a PPD-producing yeast strain was engineered by overexpressing PMG2 (phosphoglucose convertase 2) and UGP1 (UTP-glucose-1-phosphate uridyltransferase 1), resulting in a significant increase in the production titer of CK, reaching 1.70 ± 0.16 g L−1 [81]. In addition to improving the UDP-glucose supply, the optimization of P. ginseng-derived UGT1 expression enabled production of 5.74 g L−1 CK in a 1.3 L bioreactor, which is the highest titer achieved to date [82]. In another study, ginsenoside Rh2 was produced by introducing UGT45 from P. ginseng into PPD-producing yeast to glycosylate the hydroxyl group at the C3 position of PPD. However, the low catalytic efficiency of this UGT resulted in a low yield of Rh2, amounting to only 1.45 μmol g DCW−1 [67]. To solve this problem, random mutagenesis of P. ginseng-derived UGT45, coupled with optimization of CYP450 expression levels, enabled the production of 179.3 mg L-1 in a shake flask, which was a 5-fold increase compared to the control [83]. Additionally, the enhanced expression levels and activity of UGT45 enabled the production of 2.25 g L-1 Rh2 in a 10 L bioreactor, which was the first report of gram-scale Rh2 production.
4.3. Ginsenoside biosynthesis in bacteria
In addition to yeast, several other bacterial cells have been exploited to tailor metabolic networks to produce ginsenosides (Table 1). Most plant-derived CYP450s are membrane-anchored to the endomembrane system, such as the endoplasmic reticulum (ER). Accordingly, the main challenge is functional expression of these enzymes in bacteria. Despite the fact that bacteria lack an endomembrane system for attachment of the CYP450s, metabolic engineering assisted by synthetic biology allows bacteria to be an attractive choice for synthesizing ginsenosides [70,71,75].
There have been continued efforts to synthesize ginsenosides by discovering new enzymes that catalyze ginsenoside production while reconstituting and optimizing endogenous metabolic pathways. In E. coli, Li et al. achieved heterologous de novo synthesis of tetracyclic triterpenoid DM, a key precursor of ginsenosides, yielding 8.63 mg L−1 [84]. This was accomplished by reconstructing the synthetic pathway of DM through the coexpression of S. cerevisiae-derived SqS, SqE, and CPR, along with Methylococcus capsulatus-derived SqE and A. thaliana-derived CPR. Furthermore, this approach led to the establishment of two chassis with different SqE/CPR combinations, providing a potential strategy for 2,3-oxidosqualene-derived compound production in E. coli. Interestingly, Cui et al. successfully isolated and characterized a novel ginsenoside-transforming β-glucosidase (BglG167b) derived from ginseng-cultivating soil bacteria [85]. This enzyme can convert various ginsenosides (Rb1, Rb2, Rc, Rg3(S), F2, Re, Rg1 and GypXVII) through selective hydrolysis of the glucose moieties. More recently, Yu et al. found that modified genes related to UGT, originated from Panax notoginseng, could convert 20(R)-PPD and 20(R)-PPT to produce 20(R)-CK and 20(R)-F1, respectively [86].
4.4. Engineering cell factories for the production of ginsenosides using synthetic biology approaches
In parallel, ginsenoside production using cell factories has been significantly advanced through further integration of synthetic biology strategies, allowing more efficient optimization of metabolic fluxes towards target ginsenosides. Recently, successful examples include the use of subcellular organelles and protein scaffolds to colocalize biosynthetic enzymes and pathways, thus increasing enzyme proximity and the availability of substrates [[87], [88], [89], [90], [91], [92]]. Instead of focusing on the assembly of metabolic pathways in the cytosol, utilizing and converting yeast organelles into ginsenoside-producing subcellular compartments provides the ability to enhance the dedicated carbon flux through the isolated pathway (Fig. 4 and Table 1). This organelle compartmentalization helps minimize unwanted metabolic crosstalk and has been recently shown to facilitate the efficiency of metabolic reactions [[93], [94], [95], [96]].
Fig. 4.
Remodeling yeast organelles as subcellular factories for ginsenoside biosynthesis. Remodeling organelle physiology to manipulate inherent activity, morphology, and capacity (e.g., size, number and volume) allows for enhancing cellular synthetic capabilities, thereby facilitating the efficiency of biosynthetic reactions.
In a recent noteworthy example, Kim et al. successfully increased the production titer of a common precursor of PPD-type saponins in yeast by modifying ER biogenesis mediated by overexpression of INO2, an ER biogenesis regulator, to increase the capability and capacity for synthesis and accommodation of ER-localized enzymes [88]. This strategy increased PPD production by 8-fold (from 1.5 to 12 mg L−1) by expanding the size of the ER, which provided more available space for anchoring pivotal ginsenoside pathway enzymes. Thus, the ER expansion strategy is applicable to a versatile yeast framework engineered for biosynthesis of valuable products.
A similar strategy was achieved by Choi et al. through utilization of the yeast peroxisome as a subcellular factory for ginsenoside biosynthesis [90]. Peroxisomes can be used as subcellular compartments for various terpenoids, and their size and number can be dynamically regulated. To expand the storage capacity of the peroxisome membrane, Choi et al. increased the expression of PEX32 and deleted the PEX11 and ATG36 genes, which are involved in peroxisome biogenesis and peroxisome autophagy. This resulted in a 78 % (4 mg L−1) increase in PPD production compared to the control yeast strain.
In addition to the organelle compartmentalization of enzymes or metabolites from other enzymes, a protein scaffold can also be beneficial for optimizing metabolic flux [97]. Through enzyme colocalization by controlled assembly onto a synthetic protein scaffold, substrate channeling can significantly enhance reaction rates while reducing the adverse effects of toxic intermediates on general cell metabolism or byproduct production. In a recent work, Lee et al. demonstrated a multi-enzymatic cascade system using split inteins as a versatile platform for controlling complex bioconversion pathways and increasing the production of ginsenosides in E. coli [91]. Using two types of split inteins, consensus atypical (Cat) and Rma DnaB, three enzymes involved in the CK conversion pathway were tagged to a covalent scaffold, which increased the CK conversion rate and reduced the production time by more than 2-fold. Similarly, in Pichia pastoris, Zhao et al. spatially organized two key enzymes involved in the DM synthesis pathway, encoded by the DDS and ERG1 genes, via protein-protein interaction capable of self-assembly [98]. The assembly of these two enzymes increased DM titer by 2-fold when compared with unassembled system-based production.
5. Conclusion
Synthetic biology has emerged as a powerful tool for producing secondary metabolites found in plants like ginseng. Significant advancements in ginsenoside production have been achieved through metabolic engineering and synthetic biology approaches. Yeast, due to intracellular structures similar to plant cells and the presence of the MVA pathway, has become an important platform for ginsenoside production. Yeast metabolism has been engineered to enhance the availability of precursors and optimize key enzymes in the ginsenoside biosynthesis pathway. Particularly, chaperone-assisted expression systems have provided a fundamental production platform, overcoming challenges related to the functional expression of plant enzymes in microbial hosts and resulting in significantly improved ginsenoside production yields. Furthermore, the use of subcellular organelles and protein scaffolds in yeast has revolutionized ginsenoside production. By localizing biosynthetic enzymes and pathways within organelles such as peroxisomes, metabolic fluxes have been enhanced, leading to increased yield. Additionally, synthetic biology has enabled the functional expression of plant-derived enzymes in bacterial cells, resulting in successful ginsenoside production. These advancements highlight the potential of synthetic biology in ginsenoside production. The integration of these approaches has paved the way for more efficient and sustainable synthesis methods, addressing the demand for potent natural products.
Declaration of competing interest
The authors declare no competing financial interest.
Acknowledgements
This work was supported by the Basic Science Research Program (NRF-2019R1A2C1090726 to B.S.) and the Bio & Medical Technology Development Program through the National Research Foundation (NRF) grant funded by the Ministry of Science, ICT (MSIT) (NRF-2022M3A9B6082671 to B.S, NRF-2021M3A9I5023254 to J.L.), and by the Ministry of Education (NRF-2022R1A6A1A03054419 to W. L.)
Contributor Information
Bong Hyun Sung, Email: bhsung@kribb.re.kr.
Ju Young Lee, Email: juylee@krict.re.kr.
Wonsik Lee, Email: wonsik.lee@skku.edu.
References
- 1.Pichersky E., Gang D.R. Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci. 2000;5(10):439–445. doi: 10.1016/s1360-1385(00)01741-6. [DOI] [PubMed] [Google Scholar]
- 2.Delgoda R., Murray J. Pharmacognosy. Elsevier; 2017. Evolutionary perspectives on the role of plant secondary metabolites; pp. 93–100. [Google Scholar]
- 3.Abdulhafiz F., et al. Plant cell culture technologies: a promising alternatives to produce high-value secondary metabolites. Arab J Chem. 2022;15(11) [Google Scholar]
- 4.Fan M., et al. Anti-cancer effect and potential microRNAs targets of ginsenosides against breast cancer. Front Pharmacol. 2022;13 doi: 10.3389/fphar.2022.1033017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nag S.A., et al. Ginsenosides as anticancer agents: in vitro and in vivo activities, structure–activity relationships, and molecular mechanisms of action. Front Pharmacol. 2012;3:25. doi: 10.3389/fphar.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye X.W., et al. Saponins of ginseng products: a review of their transformation in processing. Front Pharmacol. 2023;14 doi: 10.3389/fphar.2023.1177819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang Y., Zhao S. Progress in understanding of ginsenoside biosynthesis. Plant Biol. 2008;10(4):415–421. doi: 10.1111/j.1438-8677.2008.00064.x. [DOI] [PubMed] [Google Scholar]
- 8.Kim Y.-J., Zhang D., Yang D.-C. Biosynthesis and biotechnological production of ginsenosides. Biotechnol Adv. 2015;33(6):717–735. doi: 10.1016/j.biotechadv.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Duan L., et al. Panax notoginseng saponins for treating coronary artery disease: a functional and mechanistic overview. Front Pharmacol. 2017;8:702. doi: 10.3389/fphar.2017.00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mancuso C., Santangelo R. Panax ginseng and Panax quinquefolius: from pharmacology to toxicology. Food Chem Toxicol. 2017;107:362–372. doi: 10.1016/j.fct.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J.W., et al. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat Chem Biol. 2012;8(6):536–546. doi: 10.1038/nchembio.970. [DOI] [PubMed] [Google Scholar]
- 12.Krivoruchko A., Nielsen J. Production of natural products through metabolic engineering of Saccharomyces cerevisiae. Curr Opin Biotechnol. 2015;35:7–15. doi: 10.1016/j.copbio.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Church G.M., et al. Realizing the potential of synthetic biology. Nat Rev Mol Cell Biol. 2014;15(4):289–294. doi: 10.1038/nrm3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Latchman D.S. Inhibitory transcription factors. Int J Biochem Cell Biol. 1996;28(9):965–974. doi: 10.1016/1357-2725(96)00039-8. [DOI] [PubMed] [Google Scholar]
- 15.McLaughlin J.A., et al. The Synthetic Biology Open Language (SBOL) version 3: simplified data exchange for bioengineering. Front Bioeng Biotechnol. 2020;8:1009. doi: 10.3389/fbioe.2020.01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villalobos A., et al. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinf. 2006;7:1–8. doi: 10.1186/1471-2105-7-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mante J., et al. Synthetic biology knowledge system. ACS Synth Biol. 2021;10(9):2276–2285. doi: 10.1021/acssynbio.1c00188. [DOI] [PubMed] [Google Scholar]
- 18.Cai P., et al. SynBioTools: a one-stop facility for searching and selecting synthetic biology tools. BMC Bioinf. 2023;24(1):1–11. doi: 10.1186/s12859-023-05281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plahar H.A., et al. BioParts—a biological parts search portal and updates to the ICE parts registry software platform. ACS Synth Biol. 2021;10(10):2649–2660. doi: 10.1021/acssynbio.1c00263. [DOI] [PubMed] [Google Scholar]
- 20.Brophy J.A.N., Voigt C.A. Principles of genetic circuit design. Nat Methods. 2014;11(5):508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson E., Levin M. Gene regulatory networks. Proc Natl Acad Sci USA. 2005;102(14) doi: 10.1073/pnas.0502024102. 4935-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y., et al. Genetic circuit design automation for yeast. Nature Microbiology. 2020;5(11):1349–1360. doi: 10.1038/s41564-020-0757-2. [DOI] [PubMed] [Google Scholar]
- 23.Cho J.S., et al. Designing microbial cell factories for the production of chemicals. JACS Au. 2022;2(8):1781–1799. doi: 10.1021/jacsau.2c00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomes D.G., et al. Current developments in biotechnology and bioengineering. Elsevier; 2023. Bioreactors and engineering of filamentous fungi cultivation; pp. 219–250. [Google Scholar]
- 25.Buddingh’ B.C., Van Hest J.C.M. Artificial cells: synthetic compartments with life-like functionality and adaptivity. Accounts Chem Res. 2017;50(4):769–777. doi: 10.1021/acs.accounts.6b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelletier J.F., et al. Genetic requirements for cell division in a genomically minimal cell. Cell. 2021;184(9):2430–2440. doi: 10.1016/j.cell.2021.03.008. e16. [DOI] [PubMed] [Google Scholar]
- 27.Paddon C.J., et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496(7446):528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 28.Paddon C.J., Keasling J.D. Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat Rev Microbiol. 2014;12(5):355–367. doi: 10.1038/nrmicro3240. [DOI] [PubMed] [Google Scholar]
- 29.Ro D.-K., et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440(7086):940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 30.Cravens A., Payne J., Smolke C.D. Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-09848-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayden E.C. Nat News [Internet]; 2016. Biology software promises easier way to program living cells. [Google Scholar]
- 32.Carbonell P., et al. An automated Design-Build-Test-Learn pipeline for enhanced microbial production of fine chemicals. Commun Biol. 2018;1(1) doi: 10.1038/s42003-018-0076-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holowko M.B., et al. Building a biofoundry. Synthetic Biology. 2021;6(1):ysaa026. doi: 10.1093/synbio/ysaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu M.-R., et al. A high-throughput screening and computation platform for identifying synthetic promoters with enhanced cell-state specificity (SPECS) Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-10912-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radivojević T., et al. A machine learning Automated Recommendation Tool for synthetic biology. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-18008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vickers C.E., Freemont P.S. Pandemic preparedness: synthetic biology and publicly funded biofoundries can rapidly accelerate response time. Nat Commun. 2022;13(1) doi: 10.1038/s41467-022-28103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hillson N., et al. Building a global alliance of biofoundries. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-10079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butler M.S., Robertson A.A., Cooper M.A. Natural product and natural product derived drugs in clinical trials. Nat Prod Rep. 2014;31(11):1612–1661. doi: 10.1039/c4np00064a. [DOI] [PubMed] [Google Scholar]
- 39.Sussmuth R.D., Mainz A. Nonribosomal peptide synthesis-principles and prospects. Angew Chem Int Ed Engl. 2017;56(14):3770–3821. doi: 10.1002/anie.201609079. [DOI] [PubMed] [Google Scholar]
- 40.Walsh C.T. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science. 2004;303(5665):1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 41.Sattely E.S., Fischbach M.A., Walsh C.T. Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat Prod Rep. 2008;25(4):757–793. doi: 10.1039/b801747f. [DOI] [PubMed] [Google Scholar]
- 42.Du L., Sanchez C., Shen B. Hybrid peptide-polyketide natural products: biosynthesis and prospects toward engineering novel molecules. Metab Eng. 2001;3(1):78–95. doi: 10.1006/mben.2000.0171. [DOI] [PubMed] [Google Scholar]
- 43.Nivina A., et al. Evolution and diversity of assembly-line polyketide synthases. Chem Rev. 2019;119(24):12524–12547. doi: 10.1021/acs.chemrev.9b00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herbst D.A., Townsend C.A., Maier T. The architectures of iterative type I PKS and FAS. Nat Prod Rep. 2018;35(10):1046–1069. doi: 10.1039/c8np00039e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyazawa T., et al. An in vitro platform for engineering and harnessing modular polyketide synthases. Nat Commun. 2020;11(1):80. doi: 10.1038/s41467-019-13811-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bedewitz M.A., et al. Tropinone synthesis via an atypical polyketide synthase and P450-mediated cyclization. Nat Commun. 2018;9(1):5281. doi: 10.1038/s41467-018-07671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li W., et al. Asperphenamate biosynthesis reveals a novel two-module NRPS system to synthesize amino acid esters in fungi. Chem Sci. 2018;9(9):2589–2594. doi: 10.1039/c7sc02396k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischbach M.A., et al. Directed evolution can rapidly improve the activity of chimeric assembly-line enzymes. Proc Natl Acad Sci U S A. 2007;104(29):11951–11956. doi: 10.1073/pnas.0705348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang W.-z., et al. Saponins in the genus Panax L.(Araliaceae): a systematic review of their chemical diversity. Phytochemistry. 2014;106:7–24. doi: 10.1016/j.phytochem.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 50.Piao X., et al. Advances in saponin diversity of Panax ginseng. Molecules. 2020;25(15):3452. doi: 10.3390/molecules25153452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue-Ni N., et al. Research progress on naturally-occurring and semi-synthetic ocotillol-type ginsenosides in the genus Panax L.(Araliaceae) Chin J Nat Med. 2021;19(9):648–655. doi: 10.1016/S1875-5364(21)60089-4. [DOI] [PubMed] [Google Scholar]
- 52.Rohmer M. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat Prod Rep. 1999;16(5):565–574. doi: 10.1039/a709175c. [DOI] [PubMed] [Google Scholar]
- 53.Boucher Y., Doolittle W.F. The role of lateral gene transfer in the evolution of isoprenoid biosynthesis pathways. Mol Microbiol. 2000;37(4):703–716. doi: 10.1046/j.1365-2958.2000.02004.x. [DOI] [PubMed] [Google Scholar]
- 54.Lange B.M., et al. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc Natl Acad Sci USA. 2000;97(24):13172–13177. doi: 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lombard J., Moreira D. Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Mol Biol Evol. 2010;28(1):87–99. doi: 10.1093/molbev/msq177. [DOI] [PubMed] [Google Scholar]
- 56.Liao P., et al. The potential of the mevalonate pathway for enhanced isoprenoid production. Biotechnol Adv. 2016;34(5):697–713. doi: 10.1016/j.biotechadv.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 57.Xie S.-s., et al. Advances in the metabolic engineering of Escherichia coli for the manufacture of monoterpenes. Catalysts. 2019;9(5):433. [Google Scholar]
- 58.Bochar D.A., et al. Biosynthesis of mevalonic acid from acetyl-CoA. Comprehensive natural products chemistry. 1999;2:15–44. [Google Scholar]
- 59.Kaneda K., et al. An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp. strain CL190. Proc Natl Acad Sci USA. 2001;98(3):932–937. doi: 10.1073/pnas.020472198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Banerjee A., Sharkey T. Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Nat Prod Rep. 2014;31(8):1043–1055. doi: 10.1039/c3np70124g. [DOI] [PubMed] [Google Scholar]
- 61.Lu X., Tang K., Li P. Plant metabolic engineering strategies for the production of pharmaceutical terpenoids. Front Plant Sci. 2016;7 doi: 10.3389/fpls.2016.01647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thimmappa R., et al. Triterpene biosynthesis in plants. Annu Rev Plant Biol. 2014;65(1):225–257. doi: 10.1146/annurev-arplant-050312-120229. [DOI] [PubMed] [Google Scholar]
- 63.Chu L.L., et al. Hydroxylation of diverse flavonoids by CYP450 BM3 variants: biosynthesis of eriodictyol from naringenin in whole cells and its biological activities. Microb Cell Factories. 2016;15(1) doi: 10.1186/s12934-016-0533-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chu L.L., et al. Synthetic analog of anticancer drug daunorubicin from daunorubicinone using one-pot enzymatic UDP-recycling glycosylation. J Mol Catal B Enzym. 2016;124:1–10. [Google Scholar]
- 65.Shin J.Y., et al. In vitro single-vessel enzymatic synthesis of novel Resvera-A glucosides. Carbohydr Res. 2016;424:8–14. doi: 10.1016/j.carres.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 66.Chu L.L., et al. Synthesis of umbelliferone derivatives in Escherichia coli and their biological activities. J Biol Eng. 2017;11(1) doi: 10.1186/s13036-017-0056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang P., et al. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab Eng. 2015;29:97–105. doi: 10.1016/j.ymben.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 68.Li C., et al. Pathway elucidation of bioactive rhamnosylated ginsenosides in Panax ginseng and their de novo high-level production by engineered Saccharomyces cerevisiae. Commun Biol. 2022;5(1) doi: 10.1038/s42003-022-03740-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan X., et al. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 2014;24(6):770–773. doi: 10.1038/cr.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ren H., Hu P., Zhao H. A plug‐and‐play pathway refactoring workflow for natural product research in Escherichia coli and Saccharomyces cerevisiae. Biotechnol Bioeng. 2017;114(8):1847–1854. doi: 10.1002/bit.26309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang C., et al. Microbial platform for terpenoid production: Escherichia coli and yeast. Front Microbiol. 2018;9:2460. doi: 10.3389/fmicb.2018.02460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu Y., et al. High-yield production of protopanaxadiol from sugarcane molasses by metabolically engineered Saccharomyces cerevisiae. Microb Cell Factories. 2022;21(1) doi: 10.1186/s12934-022-01949-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu Z.-F., et al. Construction and optimization of microbial cell factories for sustainable production of bioactive dammarenediol-II glucosides. Green Chem. 2019;21(12):3286–3299. [Google Scholar]
- 74.Jiang F., et al. Metabolic engineering of yeasts for green and sustainable production of bioactive ginsenosides F2 and 3β, 20S-Di-O-Glc-DM. Acta Pharm Sin B. 2022;12(7):3167–3176. doi: 10.1016/j.apsb.2022.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pompon D., et al. Methods in enzymology. Elsevier; 1996. [6] Yeast expression of animal and plant P450s in optimized redox environments; pp. 51–64. [DOI] [PubMed] [Google Scholar]
- 76.Dai Z., et al. Identification of a novel cytochrome P450 enzyme that catalyzes the C-2α hydroxylation of pentacyclic triterpenoids and its application in yeast cell factories. Metab Eng. 2019;51:70–78. doi: 10.1016/j.ymben.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 77.Zhao Y., et al. Enhancing oleanolic acid production in engineered Saccharomyces cerevisiae. Bioresour Technol. 2018;257:339–343. doi: 10.1016/j.biortech.2018.02.096. [DOI] [PubMed] [Google Scholar]
- 78.Kim J.E., et al. Pairing of orthogonal chaperones with a cytochrome P450 enhances terpene synthesis in Saccharomyces cerevisiae. Biotechnol J. 2022;17(3) doi: 10.1002/biot.202000452. [DOI] [PubMed] [Google Scholar]
- 79.Zhao F., et al. Optimization of a cytochrome P450 oxidation system for enhancing protopanaxadiol production in Saccharomyces cerevisiae. Biotechnol Bioeng. 2016;113(8):1787–1795. doi: 10.1002/bit.25934. [DOI] [PubMed] [Google Scholar]
- 80.Jung S.-C., et al. Two ginseng UDP-glycosyltransferases synthesize ginsenoside Rg3 and rd. Plant Cell Physiol. 2014;55(12):2177–2188. doi: 10.1093/pcp/pcu147. [DOI] [PubMed] [Google Scholar]
- 81.Nan W., et al. Promotion of compound K production in Saccharomyces cerevisiae by glycerol. Microb Cell Factories. 2020;19(1) doi: 10.1186/s12934-020-01306-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang P., et al. Systematic optimization of the yeast cell factory for sustainable and high efficiency production of bioactive ginsenoside compound K. Synthetic and systems biotechnology. 2021;6(2):69–76. doi: 10.1016/j.synbio.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang P., et al. Synthesizing ginsenoside Rh2 in Saccharomyces cerevisiae cell factory at high-efficiency. Cell Discovery. 2019;5(1) doi: 10.1038/s41421-018-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li D., et al. Heterologous biosynthesis of triterpenoid dammarenediol-II in engineered Escherichia coli. Biotechnol Lett. 2016;38(4):603–609. doi: 10.1007/s10529-015-2032-9. [DOI] [PubMed] [Google Scholar]
- 85.Cui C.-H., et al. Enhanced production of gypenoside LXXV using a novel ginsenoside-transforming β-glucosidase from ginseng-cultivating soil bacteria and its anti-cancer property. Molecules. 2017;22(5):844. doi: 10.3390/molecules22050844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu L., et al. Biosynthesis of rare 20 (R)-protopanaxadiol/protopanaxatriol type ginsenosides through Escherichia coli engineered with uridine diphosphate glycosyltransferase genes. J ginseng research. 2019;43(1):116–124. doi: 10.1016/j.jgr.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deloache W.C., Russ Z.N., Dueber J.E. Towards repurposing the yeast peroxisome for compartmentalizing heterologous metabolic pathways. Nat Commun. 2016;7(1) doi: 10.1038/ncomms11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim J.-E., et al. Tailoring the Saccharomyces cerevisiae endoplasmic reticulum for functional assembly of terpene synthesis pathway. Metab Eng. 2019;56:50–59. doi: 10.1016/j.ymben.2019.08.013. [DOI] [PubMed] [Google Scholar]
- 89.Grewal P.S., et al. Peroxisome compartmentalization of a toxic enzyme improves alkaloid production. Nat Chem Biol. 2021;17(1):96–103. doi: 10.1038/s41589-020-00668-4. [DOI] [PubMed] [Google Scholar]
- 90.Choi B., et al. Organelle engineering in yeast: enhanced production of protopanaxadiol through manipulation of peroxisome proliferation in Saccharomyces cerevisiae. Microorganisms. 2022;10(3):650. doi: 10.3390/microorganisms10030650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee C.-H., et al. Novel split intein-mediated enzymatic channeling accelerates the multimeric bioconversion pathway of ginsenoside. ACS Synth Biol. 2022;11(10):3296–3304. doi: 10.1021/acssynbio.2c00216. [DOI] [PubMed] [Google Scholar]
- 92.Son S.-H., et al. Chain flexibility of medicinal lipids determines their selective partitioning into lipid droplets. Nat Commun. 2022;13(1) doi: 10.1038/s41467-022-31400-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sweetlove L.J., Fernie A.R. The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-04543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cao X., et al. Harnessing sub-organelle metabolism for biosynthesis of isoprenoids in yeast. Synthetic and systems biotechnology. 2020;5(3):179–186. doi: 10.1016/j.synbio.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jiao X., et al. Recent advances in construction and regulation of yeast cell factories. World J Microbiol Biotechnol. 2022;38(4) doi: 10.1007/s11274-022-03241-4. [DOI] [PubMed] [Google Scholar]
- 96.Moon S.Y., et al. Harnessing cellular organelles to bring new functionalities into yeast. Biotechnol Bioproc Eng. 2023:1–13. [Google Scholar]
- 97.Moon S.Y., et al. Designing microbial cell factories for programmable control of cellular metabolism. Curr Opin Struct Biol. 2023 [Google Scholar]
- 98.Zhao C., et al. Enhancing biosynthesis of a ginsenoside precursor by self-assembly of two key enzymes in Pichia pastoris. J Agric Food Chem. 2016;64(17):3380–3385. doi: 10.1021/acs.jafc.6b00650. [DOI] [PubMed] [Google Scholar]
- 99.Wei W., et al. Characterization of Panax ginseng UDP-glycosyltransferases catalyzing protopanaxatriol and biosyntheses of bioactive ginsenosides F1 and Rh1 in metabolically engineered yeasts. Mol Plant. 2015;8(9):1412–1424. doi: 10.1016/j.molp.2015.05.010. [DOI] [PubMed] [Google Scholar]