Abstract
The translation of specific maternal mRNAs is regulated during early development. For some mRNAs, an increase in translational activity is correlated with cytoplasmic extension of their poly(A) tails; for others, translational inactivation is correlated with removal of their poly(A) tails. Recent results in several systems suggest that events at the 3′ end of the mRNA can affect the state of the 5′ cap structure, m7G(5′)ppp(5′)G. We focus here on the potential role of cap modifications on translation during early development and on the question of whether any such modifications are dependent on cytoplasmic poly(A) addition or removal. To do so, we injected synthetic RNAs into Xenopus oocytes and examined their cap structures and translational activities during meiotic maturation. We draw four main conclusions. First, the activity of a cytoplasmic guanine-7-methyltransferase increases during oocyte maturation and stimulates translation of an injected mRNA bearing a nonmethylated GpppG cap. The importance of the cap for translation in oocytes is corroborated by the sensitivity of protein synthesis to cap analogs and by the inefficient translation of mRNAs bearing nonphysiologically capped 5′ termini. Second, deadenylation during oocyte maturation does not cause decapping, in contrast to deadenylation-triggered decapping in Saccharomyces cerevisiae. Third, the poly(A) tail and the N-7 methyl group of the cap stimulate translation synergistically during oocyte maturation. Fourth, cap ribose methylation of certain mRNAs is very inefficient and is not required for their translational recruitment by poly(A). These results demonstrate that polyadenylation can cause translational recruitment independent of ribose methylation. We propose that polyadenylation enhances translation through at least two mechanisms that are distinguished by their dependence on ribose modification.
The 7-methyl guanosine cap and the poly(A) tail of mRNAs both can stimulate translation. Translation initiation of most mRNAs occurs via recognition of the 5′ cap by initiation factor eIF-4E (reviewed in reference 36), which forms part of the eIF-4F complex (reviewed in reference 58). Yet poly(A), located at the opposite end of the mRNA, also facilitates translational initiation (reviewed in references 18, 23, 46, and 47). Poly(A)’s effects on translation are particularly striking during early development. Elongation of the poly(A) tail of certain maternal mRNAs is correlated with their translational activation, while removal of the tail from others results in their translational inactivation (see, e.g., references 3, 21, 35, 48, 49, 51, and 63). In some cases, the changes in poly(A) tail length are required for the changes in translational activity; exceptions exist, however, and translational repression can be a cause, rather than an effect, of deadenylation (reviewed in reference 18).
The 5′ cap stimulates translation by facilitating recruitment of the 40S ribosomal subunit (reviewed in reference 36). The N-7 methyl group of the cap largely determines translational efficiency in vitro (see, e.g., references 6, 7, and 32), presumably because it enhances the binding of eIF-4E (56, 57). The functional significance of ribose methylation is less clear. Ribose methylation of the first or penultimate nucleotides is common among mRNAs in metazoans, though its abundance varies among species (4). Early experiments suggested that ribose methylation, in contrast to N-7 methylation, confers little if any translational advantage (40, 41). However more recently, ribose methylation of a specific histone-related RNA, B4 (55), was detected during Xenopus oocyte maturation and was suggested to underlie the polyadenylation-dependent increase in that mRNA’s translation (28).
Although the stimulatory effect of poly(A) on translation during early development is striking, the underlying mechanism is not clear. Polyadenylated mRNAs are translated more efficiently than nonadenylated mRNAs when injected into Xenopus laevis oocytes (see, e.g., references 12 and 14), and kinetic studies suggest that poly(A) may facilitate translational reinitiation (14). In Xenopus oocytes, mRNAs compete for translational machinery (31, 45), perhaps accentuating the advantage of a poly(A) tail.
Communication occurs between the cap structure and the poly(A) tail. For example, deadenylation-dependent decapping precedes exonucleolytic degradation of mRNAs in Saccharomyces cerevisiae (5). Information can also flow from the 5′ to the 3′ end: translational repression through binding of a protein to the 5′ untranslated region (5′UTR) can cause deadenylation in somatic cells (39). Communication between the two ends of the mRNA may be mediated, at least in yeast, by a tripartite complex between a protein bound to the poly(A) tail, poly(A) binding protein, and the initiation factors eIF-4G and eIF-4E (60, 61). Thus, poly(A) or PAB may facilitate an interaction between the 5′ and 3′ ends of mRNAs, perhaps resulting in effects on both translation and stability.
Modifications of the 5′ cap structure may regulate translation during early development. For example, in Manduca sexta, N-7 methylation of mRNAs with a GpppG cap occurs following fertilization and may stimulate their translation (24, 25). In the sea urchin Strongylocentrotus purpuratus, N-7 and ribose methylation occurs following fertilization but prior to the two-cell stage. N-7 methylation may be important for the translational activation of histone mRNAs in this species (8). However, translational recruitment of histone mRNAs in the closely related species Lytechinus pictus appears to be independent of cap methylation (53). In somatic cells, translation of insulin mRNA in a pancreatic β-cell tumor may also be controlled by covalent modification of its cap: treatment of rat insulin 2 mRNA with guanylyltransferase and N7 methylase activities specifically increased its translation in vitro (10).
The particularly large stimulatory effect of poly(A) on translation in oocytes and embryos could be explained if translation were relatively insensitive to the 5′ cap structure and entirely dependent on the presence of a long poly(A) tail. In Xenopus oocytes, mRNAs with a cap are translated more efficiently than those without a cap (12), yet translation initiation appears to be insensitive to injection of cap analogs (2) that inhibit translation in vitro (see, e.g., references 1, 17, and 65). Moreover, proteolytic cleavage of eIF-4G, a factor required for cap-dependent initiation, inhibits translation of an injected capped mRNA completely but only modestly decreases translation of endogenous mRNAs. These results suggest that translation of most endogenous mRNAs in oocytes may occur through an eIF-4G- or cap-independent mechanism (26).
In this study, we investigated the role of the cap and poly(A) tail on translation in Xenopus oocytes by injecting synthetic RNAs. By radiolabeling a single phosphate in the cap, we specifically detected cap modifications and assessed their impact on translation. We observed efficient N-7 methylation of an RNA with a GpppG cap during oocyte maturation and demonstrated that this methylation event, in conjunction with polyadenylation, dramatically enhances translation. These findings are consistent with the nearly complete inhibition of translation of an exogenous mRNA by the cap analog m7GpppG. In contrast, ribose methylation of a reporter mRNA bearing a cyclin B1 3′UTR is not required for poly(A)’s stimulation of that mRNA’s translation. We detected neither polyadenylation-dependent modifications of the cap nor deadenylation-dependent decapping. Our findings suggest that the effects on translation of cap modification and cytoplasmic changes in poly(A) length are mechanistically independent.
MATERIALS AND METHODS
Plasmid construction.
All plasmids were named for the sequences that they contain and were created as follows.
(i) Luciferase-cyclin B1 mRNAs.
Those with and without a point mutation in AAUAAA (AAgAAA) were synthesized as previously described (52).
(ii) Luciferase X114 mRNA.
The plasmid pT7-Luc (BglII at stop codon) (15) was cut with BglII and BamHI and religated to produce the plasmid pLUCX114. To generate LUCX114 mRNA, the template was cut with PvuII and transcribed with T7 RNA polymerase (Epicentre Technologies). LUCX114 mRNA contains 1,680 nucleotides of the luciferase gene followed by 114 nucleotides of pBluescript vector (pBSII KS+; Stratagene). The vector sequence does not contain the hexanucleotide AAUAAA.
(iii) Luciferase A100 mRNA.
A BglII-EcoRI fragment containing a 100-nucleotide tract of adenosine residues was isolated from the plasmid pSD5 (16). The isolated fragment was further digested with Sau3AI and cloned into the BglII site of pT7-Luc (BglII at stop codon) (15) to generate the plasmid pLUCA100. To generate LUCA100 mRNA, the plasmid was cut with BglII and transcribed with T7 RNA polymerase. LUCA100 mRNA contains 1,680 nucleotides of the luciferase gene followed by a 100-nucleotide poly(A) tract. The BglII site is immediately adjacent to the poly(A) tract.
Preparation of RNA substrates.
RNAs were prepared in vitro with either T7 or SP6 RNA polymerase (200 U/μl; Epicentre Technologies) and under the suggested reaction conditions (19), including 5 U of inorganic pyrophosphatase (Sigma) per ml (11). Uniformly radiolabeled RNAs (specific activity of approximately 3.8 × 103 dpm/fmol) were prepared in a 20-μl reaction mixture containing the cleaved DNA template, 20 to 80 μCi of [α-32P]UTP (800 Ci/mmol; DuPont), 10 mM cap analog (ApppG, GpppG, or m7GpppG; New England Biolabs), and 250 μM UTP. Transcripts of the appropriate length were eluted from polyacrylamide urea gel slices as previously described (33). The eluate was phenol-chloroform extracted, the resulting aqueous layer was precipitated with ethanol, and the pelleted RNA was washed with 70% ethanol. RNA was redissolved in water and precipitated twice more. The final precipitate was resuspended in water at a concentration of approximately 40 fmol/μl.
Preparation of cap-labeled RNA substrates.
RNAs with a radiolabeled cap, G*pppG or m7G*pppG (where “*p” indicates the radiolabeled phosphate), were prepared as previously described (37, 38) with the following modifications. In vitro transcribed RNAs lacking a cap were first incubated at 65°C for 10 min and chilled on ice. Each 20-μl reaction mixture contained the following: RNA (approximately 5 pmol of 5′ triphosphate termini), 1× reaction buffer (50 mM Tris-HCl [pH 7.8], 1.25 mM MgCl2, 0.2 mM EDTA, and 6 mM KCl), 2.5 mM dithiothreitol (DTT), 1 μg of bovine serum albumin, 1 μl of RNasin (40 U/μl; Promega), 0.2 U of inorganic pyrophosphatase (Sigma), 100 μCi of [α-32P]GTP (800 Ci/mmol; DuPont), and approximately 4 U of vaccinia virus guanylyltransferase (GIBCO BRL). RNAs were radiolabeled to a specific activity of approximately 1.8 × 103 dpm/fmol. The reaction contained up to 50 μM S-adenosylmethionine (SAM) when production of a methylated cap structure was desired. Reactions were incubated at 37°C for 1 h. To improve the efficiency of capping, 2 U of guanylyltransferase, 20 U of RNasin, and 0.1 U of pyrophosphatase were added, and the reaction mixture was incubated for an additional hour at 37°C. The reaction was terminated by increasing the volume with water and extracting with phenol-chloroform. The aqueous phase was precipitated in the presence of one-half volume of 7.5 M ammonium acetate and 2 volumes of ethanol to remove unincorporated nucleotides. The pellet was resuspended in water, precipitated with 0.3 M sodium acetate (pH 5.2) and ethanol, washed with 70% ethanol, and resuspended in water at approximately 100 fmol/μl.
Oocyte injections.
Oocyte microinjection and micromanipulation were performed essentially as previously described (66). Briefly, adult females of X. laevis were primed with 50 U of pregnant mare serum (Sigma) 2 to 3 days prior to oocyte isolation. Stage VI oocytes were manually dissected from excised portions of ovary and incubated at 18 to 22°C in Marc’s Modified Ringer’s solution (MMR) (100 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 5 mM HEPES [pH 7.4], and 1 mg of penicillin and streptomycin per ml). Maturation was induced by incubating oocytes in MMR containing 10 μg of progesterone (Sigma) per ml and scored by the appearance of a white spot on the animal pole. Approximately 50 nl of a solution containing 2 to 2.5 fmol of RNA was microinjected into the oocyte cytoplasm on the animal pole side of the midline. In experiments in which cap analogs were injected, the analog was coinjected with the mRNA. Analogs were injected at a concentration of 5 mM to achieve a final in vivo concentration of 250 μM.
Extraction and analysis of RNA.
Five to 10 oocytes were pooled in each sample to decrease any effects due to variation between oocytes. Each sample was homogenized in 400 μl of homogenization buffer (50 mM Tris-HCl [pH 7.9], 5 mM EDTA, 2% sodium dodecyl sulfate, 300 mM NaCl, and 250 μg of proteinase K per ml). The homogenate was extracted with phenol-chloroform, and the aqueous phase was re-extracted prior to precipitation with ethanol and a 70% ethanol wash. When required, extracted RNA was selected by binding to oligo(dT) cellulose (Type-7; Pharmacia) essentially as described previously (50). One-half oocyte equivalent of RNA from each sample was analyzed by electrophoresis through an agarose formaldehyde gel (0.8 to 2%) (50). Radioactivity was detected by autoradiography. Quantitative comparisons were made with a Molecular Dynamics PhosphorImager (ImageQuant Software Version 3.3).
Sucrose gradient analysis.
Oocytes were incubated in MMR containing progesterone for 40 min following GVBD50 (the time at which half the cells in a sample were mature) and for an additional 20 min in MMR containing both progesterone and cycloheximide (20 μg/ml). Each 20-oocyte sample was homogenized in 700 μl of ice-cold gradient homogenization buffer (250 mM KCl, 2 mM MgCl2, 20 mM HEPES [pH 7.4], 0.5% Nonidet P-40, 2.5 mM DTT, 100 U of InhibitAce [5 Prime→3 Prime, Inc.] per ml, and 150 μg of cycloheximide per ml) and incubated on ice for 5 min. Samples were centrifuged for 10 min at 11,750× g, and 500 μl of the clarified cytosol was removed and loaded onto an 11-ml linear sucrose gradient (10 to 50%) containing 250 mM KCl, 2 mM MgCl2, 20 mM HEPES [pH 7.4], 0.5% Nonidet P-40, 2.5 mM DTT, and 0.5 μg of heparin per ml. Gradients were centrifuged at 4°C in a Beckman SW41 rotor at 39,000 rpm for 135 min. Following centrifugation, 11 fractions of approximately 900 μl each were collected from the bottom of the gradient with a Pharmacia P-1 peristaltic pump and a Pharmacia RediFrac fraction collector. Absorption traces were recorded with a Pharmacia UV HR-10 flow cell with an A254 filter. Fifty microliters of each fraction was counted by the Cerenkov method in a Beckman LS 3801 scintillation counter. Fractions numbered 1 to 6 and 7 to 12 were pooled, and RNA was extracted with phenol-chloroform. RNAs were precipitated with 0.3 M sodium acetate and ethanol and were analyzed by nuclease P1 digestion and two-dimensional thin-layer chromatography (2D TLC) as described below, as well as by agarose formaldehyde gel electrophoresis (50).
Analysis of the 5′ terminal cap structure of RNAs.
Nuclease P1 (1 mg/ml; Boehringer Mannheim) was diluted 1:100 in 30 mM ammonium acetate (pH 5.3). The 2.5-μl reaction mixture contained 1 μl of diluted nuclease P1, 0.4 mM zinc sulfate, 30 mM ammonium acetate (pH 5.3), 1 μg of yeast total RNA, and one-half oocyte equivalent of RNA and was incubated at 37°C for 1 h. Cleavage of the cap dinucleotide with tobacco acid pyrophosphatase (Epicentre Technologies) was performed as specified by the manufacturer.
Products of each reaction were separated by 2D TLC on cellulose thin layer plates (Kodak) with a mixture of isobutyric acid, water, and ammonium hydroxide (66:33:1) in the first dimension (bottom to top) (54) and with a mixture of isopropanol, saturated ammonium sulfate, and 1 M sodium acetate (pH 7.0) (2:80:18) in the second dimension (27).
Measurement of luciferase activity.
Protocols and reagents for cell homogenization and initiation of luminescence were obtained from Promega (Madison, Wis.). Luminescence was measured with a Monolight 2010 Luminometer (Analytical Luminescence Laboratories). In each experiment, five oocytes were homogenized for each sample. Luciferase activity was determined by dividing the absolute value for luciferase activity for each sample (relative light units) by the amount of RNA in each sample (determined by quantitating the total radioactivity in each sample). This calculation was used to make comparisons between samples.
Measurement of histone H1 kinase activity.
H1 kinase activity was measured as described previously (42). Briefly, groups of five oocytes were homogenized in 100 μl of buffer A (80 mM β-glycerophosphate, 20 mM EGTA, 15 mM MgCl2, 0.5 mM sodium vanadate, and 10 μg each of chymostatin, leupeptin, and pepstatin [Sigma] per ml). Ten microliters of the homogenate was added to 10 μl of ice cold buffer A and centrifuged at 10,000 × g for 8 min at 4°C. Histone H1 kinase activity was measured in 12 μl of buffer B containing 8 μl of clarified cytosolic extract, 2 μg of histone H1 (Sigma), 300 μM ATP, and 1.5 × 106 cpm of [α-32P]ATP per μl. The reaction mixture was incubated for 15 min at room temperature, and the reaction was stopped by adding 12 μl of 2× sample buffer containing 2.4 μl of β-mercaptoethanol. Samples were analyzed by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (30) and autoradiography.
RESULTS
Cytoplasmic cap methylation is independent of polyadenylation.
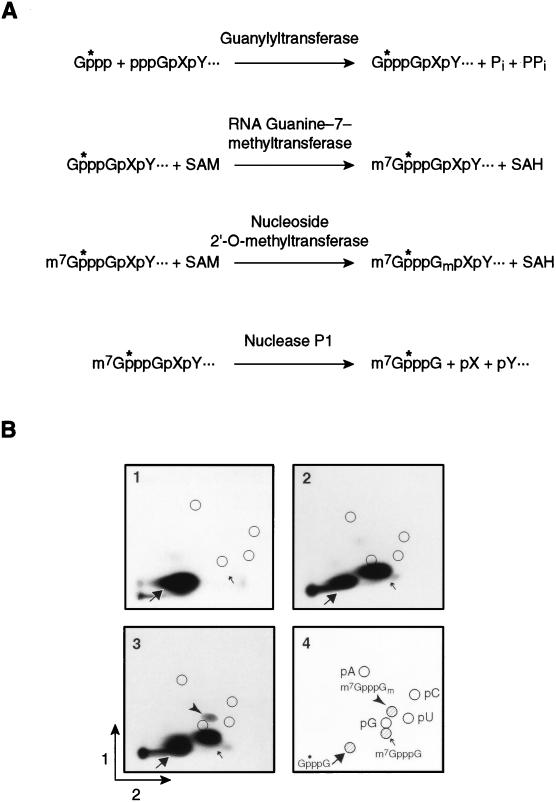
To examine modifications of the 5′ terminal cap structure of RNAs, we used the assay depicted in Fig. 1. To generate RNA molecules with a single 32P residue in the triphosphate linkage of the cap, unlabeled RNAs prepared in vitro were treated with vaccinia virus guanylyltransferase and [α-32P]GTP (Fig. 1A). Inclusion of SAM with the guanylyltransferase yields m7GpppG due to the methylase activity of that enzyme. Further incubation with the vaccinia virus nucleoside 2′-O-methyltransferase yields m7GpppGm. After incubation with RNase P1, which cleaves after any nucleotide to leave a 3′ hydroxyl, dinucleotides containing the cap structure were analyzed by 2D TLC and autoradioagraphy (Fig. 1B). For example, after P1 cleavage, cyclin B1 RNA that had been incubated with the guanylyltransferase and 32P-GTP yielded a spot corresponding to GpppG (Fig. 1B, part 1). The inclusion of SAM in the capping reaction mixture yields a spot corresponding to m7GpppG (Fig. 1B, part 2), while inclusion of SAM plus the 2′-O-methyltransferase yields m7GpppGm (Fig. 1B, part 3). The identities of the different cap species were verified by comigration with chemically prepared GpppG, m7GpppG, and m7GpppGm.
FIG. 1.
In vitro synthesis of radiolabeled cap species. (A) Schematic depicting the in vitro synthesis of the radiolabeled cap species G*pppG, m7G*pppG, and m7G*pppGm and cleavage by nuclease P1. The radiolabeled phosphate is indicated by “*p”. SAH, S-adenosylhomocysteine; Pi and PPi, inorganic phosphate and pyrophosphate, respectively. (B) Characterization of radiolabeled cap dinucleotides by 2D TLC. A portion of the cyclin B1 mRNA 3′UTR was synthesized in vitro and the 5′ cap was added by using vaccinia virus guanylyltransferase. The cap was methylated in the presence of SAM and either guanylyltransferase alone or guanylyltransferase and nucleoside 2′-O-methyltransferase. RNA was digested with nuclease P1, and the products were separated by 2D TLC. Panel 1, guanylyltransferase alone; panel 2, guanylyltransferase and SAM; panel 3, guanylyltransferase, nucleoside 2′-O-methyltransferase, and SAM; panel 4, legend. The positions of nucleoside 5′ monophosphates (circles), GpppG (large arrow, shaded circle), m7GpppG (small arrow, shaded circle), and m7GpppGm (filled arrowhead, shaded circle) are indicated.
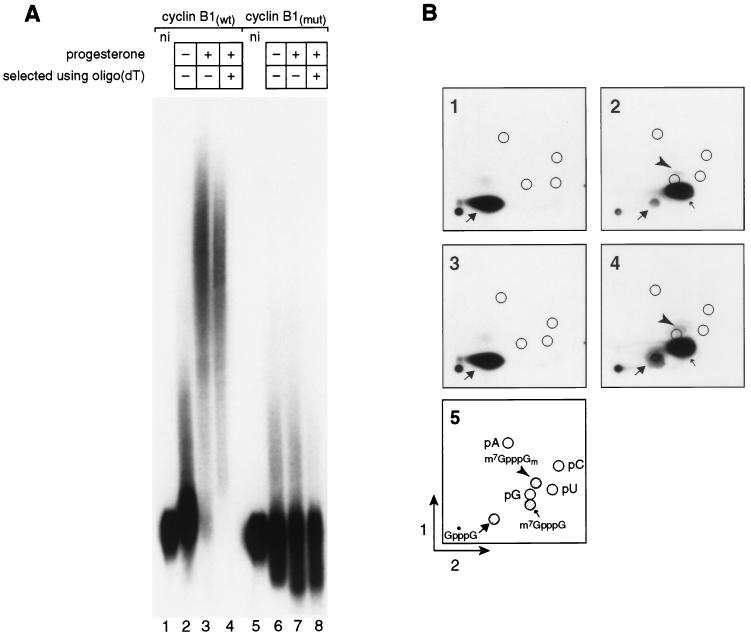
To identify any cap modifications that occur in the Xenopus oocyte cytoplasm, we prepared RNAs corresponding to the 3′UTR of cyclin B1 mRNA (cyclin B1wt). To test whether modifications were dependent on polyadenylation, we prepared an identical RNA with a point mutation in AAUAAA that prevents its polyadenylation (cyclin B1mut) (52). RNAs were cap labeled as described above, yielding a G*pppG 5′ terminus (the radiolabeled phosphate is indicated by “*p”). The identity of the 5′ end was confirmed by the assay described above: both RNAs yielded a single labeled spot that comigrated with chemically and enzymatically prepared cap dinucleotide G*pppG (Fig. 2B, parts 1 and 3). The cap-labeled RNAs were injected into the cytoplasm of stage VI oocytes, and progesterone was added to half of the cells to initiate maturation. Following maturation, RNAs were extracted from the oocytes, and polyadenylated and nonadenylated RNAs were selected by oligo(dT) cellulose chromatography (Fig. 2A, lanes 4 and 8).
FIG. 2.
Cap ribose methylation of synthetic RNA substrates is independent of polyadenylation. (A) Polyadenylation of 3′UTR RNA substrates during oocyte maturation. Samples of five oocytes each were collected and analyzed. Each lane contains RNA equivalent to one-half oocyte. Cyclin B1wt RNA: lane 1, not injected; lane 2, injected, no progesterone; lane 3, injected, progesterone added; lane 4, injected, progesterone added, selection by oligo(dT) cellulose chromatography. Cyclin B1mut RNA: lane 5, not injected; lane 6, injected, no progesterone; lane 7, injected, progesterone added; lane 8, injected, progesterone added, selection by failure to bind oligo(dT) cellulose. (B) Oocyte maturation-dependent cap methylation of RNAs. Samples from panel A were digested with nuclease P1, and the products were separated by 2D TLC. Each chromatogram contains RNA equivalent to one-half oocyte. Chromatograms 1 through 4 contain RNA present in panel A, lanes 1, 4, 5, and 8, respectively. Cyclin B1(wt) RNA: part 1, not injected; part 2, injected, plus progesterone. Cyclin B1(mut) RNA: part 3, not injected; part 4, injected, plus progesterone. Part 5, legend. Nucleoside 5′-monophosphates and cap dinucleotides are labeled as described in the legend to Fig. 1.
Cyclin B1wt RNA was polyadenylated during maturation in a reaction that required AAUAAA, as expected (Fig. 2A). The majority of the polyadenylated RNA was retained on oligo(dT) cellulose (Fig. 2A, lane 4). Virtually none of the mutant transcript was retained; the RNA in lane 8 (and analyzed in Fig. 2B, part 4) is cyclin B1mut RNA that failed to bind to oligo(dT) cellulose. Following oocyte maturation, two new cap structures were present on both polyadenylated wild-type and nonpolyadenylated mutant RNAs. The more prominent structure comigrated with m7G*pppG (Fig. 2B, parts 2 and 4). Further digestion of this material with tobacco acid pyrophosphatase produced the predicted product, m7G*p (data not shown). The second new species (Fig. 2B, parts 2 and 4) comigrated with m7G*pppGm, and was much less abundant.
Although the wild-type and mutant RNAs differed radically in the efficiency with which they received poly(A), the efficiencies with which they were 7-methylated were comparable: over 88% of both RNAs was methylated at the N-7 position of the terminal guanosine (Fig. 2B, parts 2 and 4). Ribose methylation of both RNAs was much less efficient (Fig. 2B, parts 2 and 4): less than 3% of the labeled 5′ ends on polyadenylated cyclin B1wt or nonpolyadenylated cyclin B1mut RNAs comigrated with the ribose-methylated species. The small amount of putative ribose methylation detected was apparently not polyadenylation dependent: for example, ribose-methylated cyclin B1mut RNAs were not depleted by oligo(dT) selection (Fig. 2A, lane 8, and Fig. 2B, part 4). Use of RNAs possessing an m7G*pppG structure at the time of injection did not significantly enhance the efficiency of ribose methylation (data not shown).
These data corroborate the existence of both cytoplasmic 7-methyltransferase and ribose methylase activities (13, 28, 43). They further demonstrate that base methylation is independent of poly(A) addition (Fig. 2B, parts 2 and 4). Moreover, they demonstrate that ribose methylation of the injected cyclin B1 RNA is very inefficient and appears not to require polyadenylation.
N-7 methylation is independent of polyadenylation and increases at nuclear breakdown.
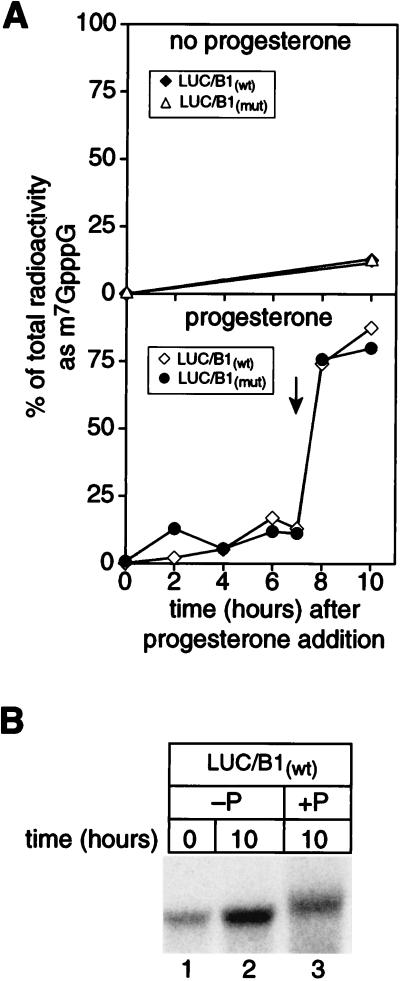
We next determined whether base methylation of the cap was regulated during oocyte maturation. To do so, we injected cap-labeled luciferase mRNA containing a portion of the cyclin B1 3′UTR with and without a point mutation in AAUAAA (LUC/B1mut and LUC/B1wt, respectively). Progesterone was added to induce maturation, and the extent of base methylation was monitored at various times thereafter. Histone H1 kinase activity was monitored to determine when maturation promoting factor was activated. Ribose methylation was too inefficient to be reliably quantitated.
N7-methylation of G*pppG was detectable in the absence of progesterone but was dramatically enhanced during maturation (Fig. 3A). As noted in Fig. 2, its efficiency was unaffected by polyadenylation. Cap guanine 7-methylation activity increased dramatically concomitant with nuclear breakdown and activation of histone H1 kinase (Fig. 3A, bottom). Following maturation, 80% of the cap structures present were N7-methylated; only 12% were methylated in oocytes incubated without progesterone. The injected RNAs are intact after incubation in the oocyte, and LUC/B1wt RNA is polyadenylated during maturation (Fig. 3B). We conclude that cap guanine N-7 methylation activity is regulated during maturation such that it increases dramatically as the nucleus breaks down. Cytoplasmic polyadenylation affects neither its extent nor its timing.
FIG. 3.
N-7 methylation of the cap is independent of polyadenylation and occurs at nuclear breakdown. (A) Cap methylation of mRNAs occurs near the time of nuclear breakdown and is independent of polyadenylation. Cap radiolabeled luciferase-cyclin B1 mRNAs with and without a point mutation in AAUAAA (LUC/B1mut and LUC/B1wt, respectively) were prepared as described in the text. RNAs were injected into oocytes, and progesterone was added. Samples of five oocytes were taken at intervals thereafter, and histone H1 kinase activity was determined for half of each sample. RNA was extracted from the remaining sample and analyzed by polyacrylamide gel electrophoresis (see below), as well as nuclease P1 digestion and 2D TLC. The y axis shows the percentage of total radioactivity in each sample present as m7G*pppG and is plotted against the time after progesterone addition. The time at which histone H1 kinase activity first increased (indicated by an arrow) was concurrent with the onset of nuclear breakdown (approximately 7.5 h). ⧫, LUC/B1wt mRNA, no progesterone; ▵, LUC/B1mut mRNA, no progesterone; ◊, LUC/B1wt mRNA, progesterone added; •, LUC/B1mut mRNA, progesterone added. (B) Polyadenylation of LUC/ B1wt mRNA. The remaining portion of extracted mRNA was analyzed by denaturing gel electrophoresis. Samples were taken immediately following microinjection (lane 1) or after 10 h in the absence (−P; lane 2) or presence (+P; lane 3) of progesterone.
The 5′ cap is not removed following RNA deadenylation in oocytes.
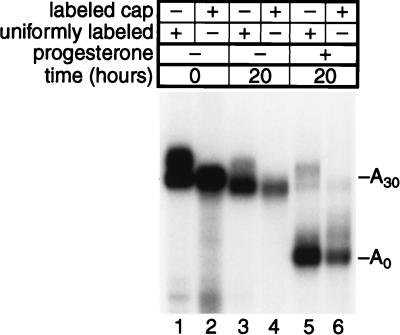
During frog oocyte maturation, specific mRNAs are translationally inactivated due to the removal of their poly(A) tails (18, 62). In yeast, deadenylation of specific mRNAs leads to the removal of their 5′ cap and their subsequent decay (5). Thus, a simple explanation of deadenylation-dependent translational repression observed in oocytes would be that deadenylation causes decapping without triggering mRNA decay. To specifically test the hypothesis that deadenylation results in decapping in oocytes, we prepared RNA containing 3′UTR sequences from the ribosomal protein L1 mRNA followed by a 30-nucleotide poly(A) tail (L1-A30). This RNA lacks sequences required to receive poly(A) during maturation (i.e., a cytoplasmic polyadenylation element [CPE]) and so is deadenylated instead, as is endogenous L1 mRNA (62, 64). RNAs were either uniformly radiolabeled by transcription in the presence of [α-32P]UTP or were cap labeled as shown in Fig. 1. Following injection of RNAs into oocyte cytoplasm, progesterone was added to initiate maturation. RNAs were extracted and analyzed by gel electrophoresis (Fig. 4).
FIG. 4.
Deadenylation during maturation does not result in decapping. L1-A30 RNA was either uniformly labeled by using [α-32P]UTP during transcription in vitro (odd numbered lanes) or labeled exclusively in its cap structure as depicted in Fig. 1 (even numbered lanes). Samples were taken immediately following microinjection (lanes 1 and 2) or after 20 h in the absence (lanes 3 and 4) or presence (lanes 5 and 6) of progesterone.
L1-A30 RNA was efficiently deadenylated during, but not before, maturation (Fig. 4, compare lanes 3 and 5). Retention of the cap on deadenylated L1-A30 RNA was demonstrated by the comparable abundance of uniformly radiolabeled and 5′ cap-labeled L1 RNAs following deadenylation (Fig. 4, lanes 5 and 6).
To examine whether deadenylation caused a cap modification that might inhibit translation, we analyzed the cap structures of L1-A30, L1, and a CPE-containing variant of L1 RNA with and without oocyte maturation; L1-A30 is deadenylated during oocyte maturation, while the CPE-containing variant is polyadenylated. We detected 2′-O-methylation, but this modification was not specific to the presence or absence of a poly(A) tail or to poly(A) addition or removal (data not shown). We conclude that deadenylation during oocyte maturation does not result in decapping and that it is unlikely to cause translational inactivation by cap modification.
Translation in oocytes is sensitive to the N-7 methyl group of the 5′ cap structure.
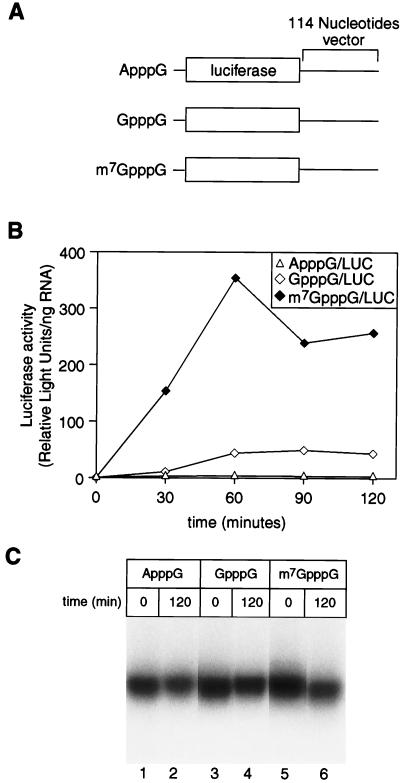
Recent experiments suggest two possible classes of mRNA in oocytes, distinguished by the extent to which their translation is cap dependent (26). To begin to examine the mechanism of cap-dependent translation in oocytes, we first tested whether the N-7 methyl group of the cap was required for translation of an injected mRNA. A set of luciferase mRNAs carrying 5′ termini of ApppG, GpppG, or m7GpppG (Fig. 5A) were injected into oocytes, and translation was assayed by measuring luciferase activity at various times thereafter.
FIG. 5.
Translation is sensitive to the presence of the N-7 methyl group on the inverted guanosine of the 5′ cap structure. (A) Schematic representation of mRNAs. The coding region of luciferase mRNA was followed by 114 nucleotides of vector (LUCX114). Uniformly radiolabeled mRNAs were synthesized in vitro and differ, as depicted, only in the cap structure which they contain. (B) Translational stimulation of LUCX114 mRNAs is influenced by the cap. mRNAs were injected into oocytes. Luciferase activity (arbitrary units, corrected for the amount of mRNA in each sample) is plotted against the time following injection. ▵, ApppG-capped mRNA; ◊, GpppG-capped mRNA; ⧫, m7GpppG-capped mRNA. (C) mRNAs varying in the 5′ cap structure are equally stable. mRNAs were analyzed by denaturing gel electrophoresis and autoradiography. Lanes 1 and 2, ApppG-capped mRNA; lanes 3 and 4, GpppG-capped mRNA; lanes 5 and 6, m7GpppG-capped mRNA. Samples were taken immediately following microinjection or after a 120-min incubation, as indicated.
Translation of luciferase mRNAs bearing an m7GpppG cap was more efficient than that of mRNAs bearing a GpppG or ApppG terminus (Fig. 5B). Thirty minutes after injection, when the rate of accumulation of luciferase activity was highest, m7GpppG-capped RNA was translated 14-fold more efficiently than GpppG-capped RNA and 48-fold more efficiently than ApppG-capped RNA. All mRNAs were comparably stable, demonstrating that the differences in luciferase activity reflected differences in their translation (Fig. 5C).
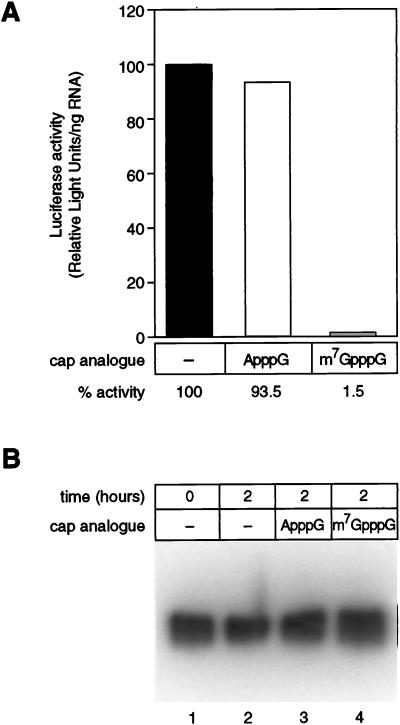
As another means of testing the role of the cap in translation in oocytes, we compared the abilities of various cap analogs to inhibit translation. Previous results suggested that translation of mRNAs in oocytes was insensitive to inhibition by cap analogs, unlike translation in vitro in extracts derived from other cell types (2, 65). An mRNA carrying an m7GpppG 5′ terminus was injected together with the dinucleoside triphosphate A(5′)ppp(5′)G or m7G(5′)ppp(5′)G at concentrations comparable to those used in vitro (1, 17, 59). Following a 2-h incubation, translation was assayed by measuring luciferase activity.
Translation of luciferase mRNA with an m7GpppG cap decreased by greater than 98% in the presence of the m7GpppG cap analog and by less than 7% in the presence of the ApppG cap analog (Fig. 6A). The lack of luciferase synthesis is not due to differential stability of the labeled luciferase mRNA in the presence of different cap analogs, as demonstrated by gel electrophoresis (Fig. 6B). We conclude that translation of injected mRNAs in oocytes is highly sensitive to the cap structure, as evidenced both by the translation of mRNAs bearing different cap structures and by the sensitivity of translation to inhibition by cap analogs.
FIG. 6.
Cap analog inhibition of translation in oocytes. (A) Translation of LUCX114 mRNA is inhibited by the m7G(5′)ppp(5′)G cap analog. Uniformly radiolabeled LUCX114 mRNA was synthesized in vitro and injected into oocytes with the A(5′)ppp(5′)G or m7G(5′)ppp(5′)G cap analog, as indicated. Following a 2-h incubation, three separate five-oocyte samples were collected. Extracts were prepared, and luciferase activity was determined as described in the text. The percentage of luciferase activity (arbitrary units, corrected for the amount of mRNA in each sample) obtained in the absence of a cap analog was arbitrarily set to 100%. Luciferase activity in the other samples is relative to this value. The values presented are means of the three samples. (B) mRNAs are equally stable in the presence of different cap analogs. LUCX114 mRNAs were analyzed by denaturing gel electrophoresis. Lanes 1 and 2, no cap analog, sample taken immediately following microinjection (lane 1) or after a 120-min incubation (lane 2); lane 3, ApppG cap analog, 120-min incubation; lane 4, m7GpppG cap analog, 120-min incubation.
Polyadenylation and cap methylation by endogenous activities stimulate translation synergistically.
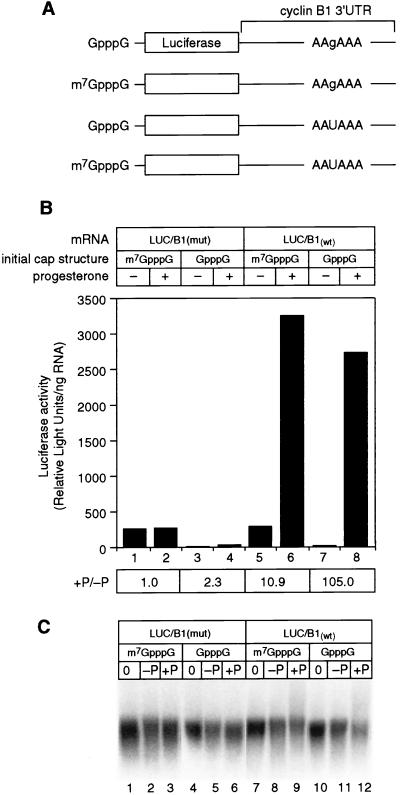
The cap and poly(A) tail interact synergistically to stimulate translation in several systems (15, 47), implying communication between the two ends of the mRNA. To test whether such synergy occurs in oocytes, we relied on endogenous activities to both polyadenylate and methylate RNAs. RNAs were synthesized in vitro with either a GpppG or an m7GpppG cap and with or without a functional AAUAAA polyadenylation signal (Fig. 7A).
FIG. 7.
Polyadenylation and cap methylation stimulate translation synergistically in maturing oocytes. (A) Schematic diagram of mRNAs. LUC/B1wt and LUC/B1mut mRNAs were prepared and manipulated in oocytes as described in the text. LUC/B1wt carries AAUAAA, while LUC/B1mut carries AAgAAA. (B) N-7 methylation of the cap and polyadenylation stimulate translation. The y axis indicates luciferase activity (arbitrary units, corrected for the amount of mRNA injected). Each value is an average of two samples (five oocytes each) which were incubated in the absence or presence of progesterone, as indicated. The ratio of luciferase activity observed in mature versus nonmature oocytes (+P/−P) is indicated. (C) Comparison of mRNA stability. mRNAs were analyzed as described in the text. Samples were taken immediately following injection (lanes 1, 4, 7, and 10), following incubation in the absence of progesterone (lanes 2, 5, 8, and 11), or following incubation in the presence of progesterone (lane 3, 6, 9, and 12).
During maturation, translation was stimulated synergistically by cap N-7 methylation and polyadenylation (Fig. 7B; the ratio of luciferase activity present in mature versus nonmature oocytes [+P/−P] quantifies the fold increase in translation during oocyte maturation). The translational activity of a nonadenylatable mRNA with the cap m7GpppG did not change in the presence and absence of maturation (Fig. 7B, compare lanes 1 and 2). When the initial cap structure was GpppG, such that the cap became N-7 methylated during maturation by the endogenous methylation activity, translation was stimulated twofold (Fig. 7B, compare lanes 3 and 4). Polyadenylation alone stimulated translation 11-fold (Fig. 7B, compare lanes 5 and 6). Interestingly, N-7 methylation and polyadenylation by endogenous activities together stimulated translation over 100-fold (Fig. 7B, compare lanes 7 and 8). In this experiment, GpppG-capped luciferase-cyclin B1 mRNA was somewhat less stable after maturation yet exhibited the largest increase in translational activity (Fig. 7C, compare lanes 8 and 9 to lanes 11 and 12); thus, the level of synergy is a minimum estimate.
Translational recruitment of luciferase-cyclin B1 mRNA is enhanced by polyadenylation and N-7 methylation but does not require ribose methylation.
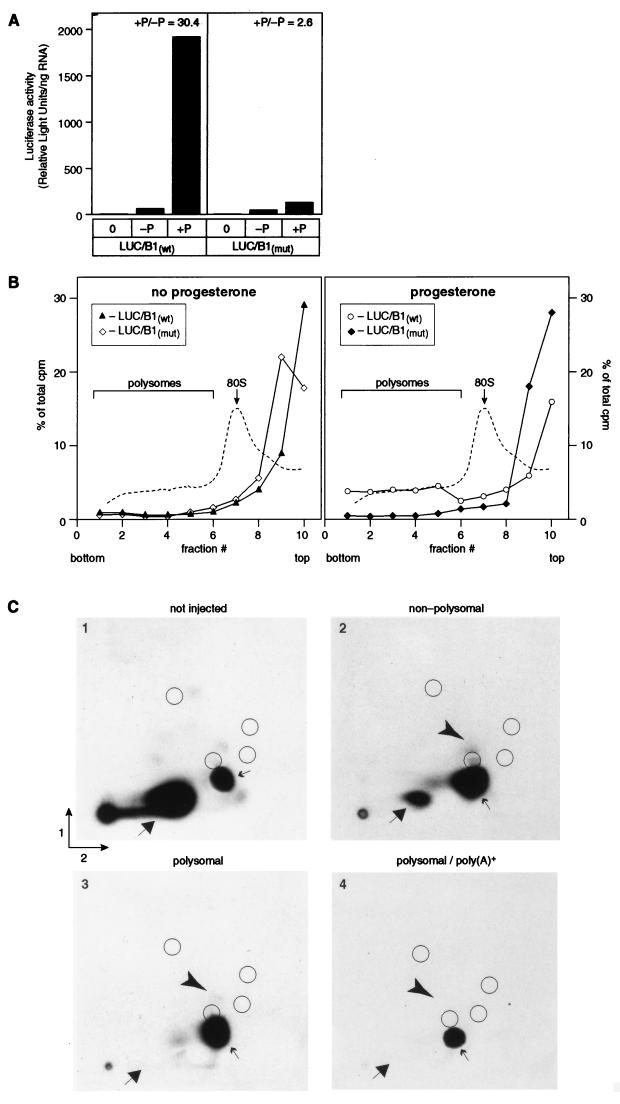
Kuge and Richter (28) have proposed that the stimulation of translation by cytoplasmic polyadenylation is due to cap ribose methylation (reviewed in reference 46). The cyclin B1 3′UTR is very inefficiently ribose methylated in vivo (Fig. 2B), implying that translational recruitment via this 3′UTR might be independent of ribose methylation. To test this hypothesis directly, we examined the translational recruitment and modification state of cap-labeled luciferase mRNAs carrying a cyclin B1 3′UTR. mRNAs containing a mixture of G*pppG and m7G*pppG caps were injected into oocytes and subsequently separated across a linear sucrose gradient into polysome-associated or non-polysome-associated fractions. The cap structure of mRNAs that had been recruited onto polysomes by polyadenylation was determined by 2D TLC, as shown in Fig. 1. In parallel, translational stimulation during maturation was quantitated as the ratio of luciferase activity present in mature versus nonmature oocytes.
Luciferase activity increased dramatically during maturation and required polyadenylation (Fig. 8A). Similarly, over 22% of RNAs containing AAUAAA were polysomally associated, while less than 5% of those bearing AAgAAA were loaded onto polysomes (Fig. 8B). Polyadenylation status was confirmed by gel electrophoresis (data not shown). Less than 5% of either RNA was associated with polysomes in the absence of progesterone. Thus, the recruitment of the luciferase-cyclin B1 chimera is both progesterone and polyadenylation dependent, as predicted.
FIG. 8.
Polyadenylation and N-7 methylation enhance translational recruitment of LUC/B1wt mRNA in the absence of cap ribose methylation. (A) Stimulation of luciferase activity during oocyte maturation. Oocytes were collected within 1 h of maturation, extracts were prepared, and luciferase activity was determined. The data represent the averages of two separate samples (five oocytes each) for each point. Values of luciferase activity (expressed as arbitrary units per amount of mRNA injected) are shown. Samples were collected immediately following oocyte injection (0) or after maturation (approximately 3 h) following incubation in the absence (−P) or presence (+P) of progesterone. The ratio of luciferase activity observed in mature versus nonmature oocytes (+P/−P) is indicated. (B) The fraction of LUC/B1wt mRNA associated with polysomes increases following maturation. Cap-labeled LUC/B1wt and LUC/B1mut mRNAs were synthesized and manipulated as described in the text. Following incubation in the absence or presence of progesterone, oocytes were homogenized, and samples were separated across a 10 to 50% linear sucrose gradient. The percentage of total counts per minute present in each fraction (y axis) is plotted against the fraction number. Only fractions 1 through 10 (of 12 fractions total) are shown. A representative A254 tracing is depicted as a dashed line. ▴, LUC/B1wt mRNA, and ◊, LUC/B1mut, mRNA incubated in the absence of progesterone; ○, LUC/B1wt mRNA, and ⧫, LUC/B1mut, mRNA injected, progesterone added. (C) Polyadenylated LUC/B1wt mRNA becomes polysome associated in the absence of cap ribose methylation. Fractions from the experiment were pooled to reflect polysome-associated (larger than the 80S monosome peak; fractions 1 through 6) and nonpolysome-associated regions of the gradient (fractions 7 through 12). LUC/B1wt mRNA was extracted, digested with nuclease P1, and analyzed by 2D TLC. A portion of the combined fractions 1 through 6 was further selected by binding to oligo(dT) cellulose prior to P1 digestion. Part 1, not injected into oocytes; part 2, progesterone added, nonpolysomal fractions 7 through 12; part 3, progesterone added, polysomal fractions 1 through 6; part 4, progesterone added, oligo(dT)-selected mRNA from polysomal fractions 1 through 6. Nucleoside 5′-monophosphates and cap dinucleotides are as indicated in the legend for Fig. 1. The position of the ribose methylated cap, m7G*pppGm, is indicated by the arrowhead.
If recruitment of luciferase mRNA onto polysomes required ribose methylation, then the fraction of RNAs bearing a 2′-O-methyl group should increase in polysome-associated RNAs, and each mRNA associated with polysomes should be ribose modified. This was not the case: in fact, the population of polyadenylated RNAs loaded onto polysomes was not enriched in ribose methylation (Fig. 8C, part 4). The polysomal fractionation was technically successful, in that the luciferase mRNA associated with polysomes was released upon the addition of EDTA (data not shown), and the polysomal mRNAs were substantially enriched in m7G*pppG versus G*pppG caps relative to either the total (Fig. 8C, part 1) or non-polysome-associated (Fig. 8C, part 2) RNAs. In fact, polysomal, polyadenylated mRNAs appear to contain m7G*pppG caps exclusively. These results corroborate our earlier finding that the presence of an N-7 methyl group enhances translation in the oocyte. We conclude that ribose methylation of luciferase-cyclin B1 mRNA is not required for its poly(A)-mediated translational stimulation.
DISCUSSION
In this report, we demonstrate that RNAs bearing a GpppG cap are methylated during oocyte maturation at both the N-7 position of the terminal guanosine and at the 2′ position of the ribose moiety of the penultimate nucleoside. Cap guanine N-7-methylase activity increases during meiotic maturation and becomes very efficient; in contrast, ribose methylation of the cap on a cyclin B1 3′UTR is inefficient both before and after maturation. Base methylation is independent of polyadenylation; ribose methylation also occurs in the absence of polyadenylation, though its inefficiency makes the possible effects of polyadenylation difficult to quantify. Polysomal recruitment of a luciferase RNA containing the cyclin B1 3′UTR was dependent on polyadenylation and was stimulated substantially by cap N-7 methylation but did not require ribose methylation.
The N-7 methyl group of the cap influences translational efficiency in Xenopus oocytes. However, previous work suggested that cap analogs that are effective translational inhibitors in vitro in extracts prepared from other cell types (see, e.g., references 1, 17, 59, and 65) are ineffective in Xenopus oocytes (2). Using shorter incubation times, we demonstrated efficient translational inhibition by m7G(5′)ppp(5′)G in the oocyte. One possible explanation for these observed differences is that injected cap dinucleotides are labile.
Cytoplasmic guanine-7-methyltransferase activity, first detected in meiotically arrested oocytes (13, 43), is regulated during frog oocyte maturation. The increase in methyltransferase activity is concurrent with nuclear breakdown (Fig. 3). In one simple view, release of a nuclear enzyme to its substrate in the cytoplasm is responsible. However, cap N-7 methylation occurs in the cytoplasm of enucleated oocytes incubated with progesterone and, to a lesser extent, in isolated oocyte nuclei incubated with the RNA substrate in vitro (data not shown). Thus the dramatic increase in N-7 methylation during maturation may require both nuclear and cytoplasmic contributions. Additionally, it seems unlikely that base methylation contributes significantly to translational activation prior to nuclear breakdown as it is only upon GVBD (the time at which cells in a sample are mature) that methyltransferase activity increases dramatically.
N-7 methylation of the cap and cytoplasmic polyadenylation stimulate translation synergistically during oocyte maturation (Fig. 7). This finding raises the possibility that regulated base methylation might contribute to the translational activation of certain maternal mRNAs. Evidence that base modifications may be exploited to regulate mRNAs during early development exists but is inconclusive, in part because definitive analysis of cap structures on endogenous mRNAs is technically difficult (see Introduction).
The guanylyltransferase reaction adds an inverted guanosine very soon after transcription initiation, probably as soon as the nascent RNA chain emerges from the RNA polymerase (9, 20, 34, 44). Base methylation is thought to occur in concert with that reaction. The hypothesis that N-7 methylation is used to regulate specific maternal mRNAs requires either that some mRNAs be produced with an unmethylated cap or that the methyl group be specifically removed from certain mRNAs.
In yeast, deadenylation triggers decapping and mRNA decay (5). In Xenopus, removal of poly(A) during oocyte maturation causes translational inactivation. However, we demonstrate here that RNAs which are fully deadenylated during oocyte maturation retain their caps and that those caps are not specifically modified. Thus, neither decapping nor cap modification is the explanation for deadenylation-dependent translational repression. We note that the same deadenylated RNAs that are stable and translationally repressed during maturation are rapidly degraded after fertilization in a process that requires recognition of the 5′ end of the RNA (unpublished observations). Thus, it is possible that a deadenylation-dependent decapping activity is activated at fertilization and could contribute to translational control in the embryo.
The hypothesis that ribose methylation is a general mechanism by which cytoplasmic polyadenylation enhances translation was prompted by the finding that it appears to be required for poly(A) to stimulate translation of an injected mRNA carrying the B4 3′UTR and occurs only on RNAs that undergo cytoplasmic polyadenylation (28). Moreover, the presence of a 2′-O-methyl group on an injected synthetic mRNA enhances translation in oocytes approximately fourfold (29). Earlier work examining the role of ribose methylation on translation in vitro was inconclusive but suggested only modest effects (40, 41).
The data reported here demonstrate that cytoplasmic polyadenylation can stimulate translation independent of cap methylation. Ribose methylation of the cyclin B1 3′UTR RNA is very inefficient and appears to be independent of polyadenylation (Fig. 2). The translation of chimeric mRNAs bearing the cyclin B1 3′UTR is dramatically enhanced by cytoplasmic polyadenylation, as evidenced both by measurements of luciferase activity and analysis of polysome recruitment. This polyadenylation-dependent translational recruitment occurs despite the lack of detectable ribose methylation, a finding consistent with earlier studies (40, 41). Similarly, the mere presence of a poly(A) tail can enhance translation in a resting oocyte, in which cytoplasmic polyadenylation (and hence any polyadenylation-dependent methylation events) does not occur. In addition, poly(A)-independent N-7 methylation of the cap may by itself be insufficient to fully stimulate the translation of all mRNAs. This is evidenced by the presence of newly N-7-methylated mRNAs in the nonpolysomal fraction (Fig. 8C). Full translational activation may also require an increase in polyadenylation or loss of translational repressors.
Taken together with the findings of others, our data suggest that cytoplasmic polyadenylation can enhance translation during oocyte maturation by at least two mechanisms. One requires ribose methylation and is exemplified by the B4 3′UTR (28). The other is independent of ribose methylation and is exemplified by injected mRNAs bearing the cyclin B1 3′UTR. Perhaps this second mechanism is more analogous to the mechanism by which poly(A) stimulates translation in S. cerevisiae:poly(A) stimulates translation in yeast extracts (22), yet yeast mRNAs are not ribose methylated (4). The existence of two distinct means by which poly(A) can facilitate translation, together with dramatic differences in the efficiencies with which different RNAs are ribose methylated, provides multiple opportunities for complex control during oocyte maturation and early development.
ACKNOWLEDGMENTS
We thank Scott Ballantyne for his helpful comments on the manuscript and other members of the Wickens lab for helpful conversations. Daniel Gallie generously provided the plasmid pT7 LUC (BglII), and Janet Mertz kindly provided the poly(A) tract containing plasmid pSD5. We thank Elsebet Lund and Christian Grimm for their assistance with two-dimensional chromatography. Paul Gershon generously provided the vaccinia virus 2′-O-methyltransferase. We are grateful to Robin Davies, Laura Vander Ploeg, and Adam Steinberg for preparing figures.
This work was supported by an NIH grant to M.W. and a Wellcome International Prize Travelling Research Fellowship to N.K.G.
REFERENCES
- 1.Antony D D, Merrick W C. Analysis of 40S and 80S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J Biol Chem. 1992;267:1554–1562. [PubMed] [Google Scholar]
- 2.Asselbergs F A M, Peters W H M, Van Venrooij W J, Bloemendal H. Cap analogues do not inhibit mRNA translation in Xenopus laevis oocytes. FEBS Lett. 1978;94:195–198. doi: 10.1016/0014-5793(78)80936-3. [DOI] [PubMed] [Google Scholar]
- 3.Bachvarova R, De Leon V, Johnson A, Kaplan G, Paynton B V. Changes in total RNA, polyadenylated RNA, and actin mRNA during meiotic maturation of mouse oocytes. Dev Biol. 1985;108:325–331. doi: 10.1016/0012-1606(85)90036-3. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee A K. 5′-terminal cap structure in eukaryotic messenger ribonucleic acids. Microbiol Rev. 1980;44:175–205. doi: 10.1128/mr.44.2.175-205.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 6.Both G W, Banerjee A K, Shatkin A J. Methylation-dependent translation of viral messenger RNAs in vitro. Proc Natl Acad Sci USA. 1975;72:1189–1193. doi: 10.1073/pnas.72.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Both G W, Furuichi Y, Muthukrishnan S, Shatkin A J. Ribosome binding to reovirus mRNA in protein synthesis requires 5′ terminal 7-methylguanosine. Cell. 1975;6:185–195. doi: 10.1016/0092-8674(75)90009-4. [DOI] [PubMed] [Google Scholar]
- 8.Caldwell D C, Emerson C P., Jr The role of cap methylation in the translational activation of stored maternal histone mRNA in sea urchin embryos. Cell. 1985;42:691–700. doi: 10.1016/0092-8674(85)90126-6. [DOI] [PubMed] [Google Scholar]
- 9.Cho E-J, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordell B, Diamond D, Smith S, Pünter J, Schöne H H, Goodman H M. Disproportionate expression of the two nonallelic rat insulin genes in a pancreatic tumor is due to translational control. Cell. 1982;31:531–542. doi: 10.1016/0092-8674(82)90309-9. [DOI] [PubMed] [Google Scholar]
- 11.Cunningham P R, Ofengand J. Use of inorganic pyrophosphatase to improve the yield of in vitro transcription reactions catalyzed by T7 RNA polymerase. BioTechniques. 1990;9:713–714. [PubMed] [Google Scholar]
- 12.Drummond D R, Armstrong J, Colman A. The effect of capping and polyadenylation of the stability, movement and translation of synthetic messenger RNAs in Xenopus oocytes. Nucleic Acids Res. 1985;13:7375–7394. doi: 10.1093/nar/13.20.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furuichi Y, LaFiandra A, Shatkin A J. 5′-terminal structure and mRNA stability. Nature. 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 14.Galili G, Kawata E, Smith L D, Larkins B A. Role of the 3′-poly(A) sequence in translational regulation of mRNAs in Xenopus laevis oocytes. J Biol Chem. 1988;263:5764–5770. [PubMed] [Google Scholar]
- 15.Gallie D R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 16.Good P J, Welch R C, Barkan A, Somasekhar M B, Mertz J E. Both VP2 and VP3 are synthesized from each of the alternative spliced late 19S RNA species of simian virus 40. J Virol. 1988;62:944–953. doi: 10.1128/jvi.62.3.944-953.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray N K, Hentze M W. Iron regulatory protein prevents binding of the 43S translation pre-initiation complex to ferritin and eALAS mRNAs. EMBO J. 1994;13:3882–3891. doi: 10.1002/j.1460-2075.1994.tb06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray N K, Wickens M P. Control of translation initiation in animal cells. Annu Rev Cell Dev Biol. 1998;14:399–457. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 19.Gurevich V V, Pokrovskaya I D, Obukhova T A, Zozulya S A. Preparative in vitro mRNA synthesis using SP6 and T7 RNA polymerases. Anal Biochem. 1991;195:207–213. doi: 10.1016/0003-2697(91)90318-n. [DOI] [PubMed] [Google Scholar]
- 20.Hagler J, Shuman S. A freeze-frame view of eukaryotic transcription during elongation and capping of nascent mRNA. Science. 1992;255:983–986. doi: 10.1126/science.1546295. [DOI] [PubMed] [Google Scholar]
- 21.Huarte J, Stutz A, O’Connell M L, Gubler P, Belin D, Darrow A L, Strickland S, Vassalli J-D. Transient translational silencing by reversible mRNA deadenylation. Cell. 1992;69:1021–1030. doi: 10.1016/0092-8674(92)90620-r. [DOI] [PubMed] [Google Scholar]
- 22.Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobson A. Poly(A) metabolism and translation: the closed loop model. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 451–480. [Google Scholar]
- 24.Kastern W H, Berry S J. Non-methylated guanosine as the 5′ terminus of capped mRNA from insect oocytes. Biochem Biophys Res Commun. 1976;71:37–44. doi: 10.1016/0006-291x(76)90246-1. [DOI] [PubMed] [Google Scholar]
- 25.Kastern W H, Swindlehurst M, Aaron C, Hooper J, Berry S J. Control of mRNA translation in oocytes and developing embryos of giant moths. Dev Biol. 1982;89:437–449. doi: 10.1016/0012-1606(82)90332-3. [DOI] [PubMed] [Google Scholar]
- 26.Keiper B D, Rhoads R E. Cap-independent translation initiation in Xenopus oocytes. Nucleic Acids Res. 1997;25:395–402. doi: 10.1093/nar/25.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konarska M M, Padgett R A, Sharp P A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 28.Kuge H, Richter J D. Cytoplasmic 3′ poly(A) addition induces 5′ cap ribose methylation: implications for translational control of maternal mRNA. EMBO J. 1995;14:6301–6310. doi: 10.1002/j.1460-2075.1995.tb00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuge H, Brownlee G G, Gershon P D, Richter J D. Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 1998;26:3208–3214. doi: 10.1093/nar/26.13.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Laskey R A, Mills A D, Gurdon J B, Partington G A. Protein synthesis in oocytes of Xenopus laevis is not regulated by the supply of messenger RNA. Cell. 1977;11:345–351. doi: 10.1016/0092-8674(77)90051-4. [DOI] [PubMed] [Google Scholar]
- 32.Lodish H F, Rose J K. Relative importance of 7-methylguanosine in ribosome binding and translation of vesicular stomatitis virus mRNA in wheat germ and reticulocyte cell-free systems. J Biol Chem. 1977;252:1181–1188. [PubMed] [Google Scholar]
- 33.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 34.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley D L Amgen EST Program. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGrew L L, Dworkin R E, Dworkin M B, Richter J D. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev. 1989;3:803–815. doi: 10.1101/gad.3.6.803. [DOI] [PubMed] [Google Scholar]
- 36.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–69. [Google Scholar]
- 37.Monroy G, Spencer E, Hurwitz J. Characteristics of reactions catalyzed by purified guanylyltransferase from vaccinia virus. J Biol Chem. 1978;253:4490–4498. [PubMed] [Google Scholar]
- 38.Monroy G, Spencer E, Hurwitz J. Purification of mRNA guanylyltransferase from vaccinia virions. J Biol Chem. 1978;253:4481–4489. [PubMed] [Google Scholar]
- 39.Muckenthaler M, Gunkel N, Stripecke R, Hentze M W. Regulated poly(A) shortening in somatic cells mediated by cap-proximal translational repressor proteins and ribosome association. RNA. 1997;3:983–995. [PMC free article] [PubMed] [Google Scholar]
- 40.Muthukrishnan S, Morgan M, Banerjee A K, Shatkin A J. Influence of 5′-terminal m7G and 2′-O-methylated residues on messenger ribonucleic acid binding to ribosomes. Biochemistry. 1976;15:5761–5768. doi: 10.1021/bi00671a012. [DOI] [PubMed] [Google Scholar]
- 41.Muthukrishnan S, Moss B, Cooper J A, Maxwell E S. Influence of 5′-terminal cap structure on the initiation of translation of vaccinia virus mRNA. J Biol Chem. 1978;253:1710–1715. [PubMed] [Google Scholar]
- 42.Nebreda A R, Hunt T. The c-mos proto-oncogene protein kinase turns on and maintains activity of MAP kinase, but not MPF, in cell-free extracts of Xenopus oocytes and eggs. EMBO J. 1993;12:1979–1986. doi: 10.1002/j.1460-2075.1993.tb05847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plessel G, Fischer U, Lührmann R. m3G Cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol Cell Biol. 1994;14:4160–4172. doi: 10.1128/mcb.14.6.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasmussen E B, Lis J T. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci USA. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richter J, Smith L D. Differential capacity for translation and lack of competition between mRNAs that segregate to free and membrane-bound polysomes. Cell. 1981;27:183–191. doi: 10.1016/0092-8674(81)90372-x. [DOI] [PubMed] [Google Scholar]
- 46.Richter J D. Dynamics of poly(A) addition and removal during development. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 481–503. [Google Scholar]
- 47.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 48.Sallés F J, Darrow A L, O’Connell M L, Strickland S. Isolation of novel murine maternal mRNAs regulated by cytoplasmic polyadenylation. Genes Dev. 1992;6:1202–1212. doi: 10.1101/gad.6.7.1202. [DOI] [PubMed] [Google Scholar]
- 49.Sallés F J, Lieberfarb M E, Wreden C, Gergen J P, Strickland S. Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science. 1994;266:1996–1999. doi: 10.1126/science.7801127. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Sheets M, Wu M, Wickens M. Polyadenylation of c-mos mRNA as a control point in Xenopus meiotic maturation. Nature. 1995;374:511–516. doi: 10.1038/374511a0. [DOI] [PubMed] [Google Scholar]
- 52.Sheets M D, Fox C A, Hunt T, Vande Woude G, Wickens M. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- 53.Showman R M, Leaf D S, Anstrom J A, Raff R A. Translation of maternal histone mRNAs in sea urchin embryos: a test of control by 5′ cap methylation. Dev Biol. 1987;121:284–287. doi: 10.1016/0012-1606(87)90161-8. [DOI] [PubMed] [Google Scholar]
- 54.Silberklang M, Gillum A M, RajBhandary U L. Use of in vitro32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- 55.Smith R C, Dworkin-Rastl E, Dworkin M B. Expression of a histone H1-like protein is restricted to early Xenopus development. Genes Dev. 1988;2:1284–1295. doi: 10.1101/gad.2.10.1284. [DOI] [PubMed] [Google Scholar]
- 56.Sonenberg N, Morgan M A, Merrick W C, Shatkin A J. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc Natl Acad Sci USA. 1978;75:4843–4847. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sonenberg N, Rupprecht K M, Hecht S M, Shatkin A J. Eukaryotic mRNA cap binding protein: purification by affinity chromatography on Sepharose-coupled m7GDP. Proc Natl Acad Sci USA. 1979;76:4345–4349. doi: 10.1073/pnas.76.9.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sonenberg N. mRNA 5′ cap-binding protein eIF4E and control of cell growth. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 245–269. [Google Scholar]
- 59.Svitkin Y V, Ovchinnikov L P, Dreyfuss G, Sonenberg N. General RNA binding proteins render translation cap dependent. EMBO J. 1996;15:7147–7155. [PMC free article] [PubMed] [Google Scholar]
- 60.Tarun S Z, Jr, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 61.Tarun S Z, Jr, Wells S E, Deardorff J A, Sachs A B. Translational initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Varnum S M, Wormington W M. Deadenylation of maternal mRNAs during Xenopus oocyte maturation does not require specific cis-sequences: a default mechanism for translational control. Genes Dev. 1990;4:2278–2286. doi: 10.1101/gad.4.12b.2278. [DOI] [PubMed] [Google Scholar]
- 63.Vassalli J-D, Huarte J, Belin D, Gubler P, O’Connell M L, Parton L, Vassalli A, Rickles R J, Strickland S. Regulated polyadenylation controls mRNA translation during meiotic maturation of mouse oocytes. Genes Dev. 1989;3:2163–2171. doi: 10.1101/gad.3.12b.2163. [DOI] [PubMed] [Google Scholar]
- 64.Verrotti A, Thompson S, Wreden C, Strickland S, Wickens M. Evolutionary conservation of sequence elements controlling cytoplasmic polyadenylation. Proc Natl Acad Sci USA. 1996;93:9027–9032. doi: 10.1073/pnas.93.17.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weber L A, Hickey E D, Nuss D L, Baglioni C. 5′-terminal 7-methylguanosine and mRNA function: influence of potassium concentration on translation in vitro. Proc Natl Acad Sci USA. 1977;74:3254–3258. doi: 10.1073/pnas.74.8.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wickens M P, Gurdon J B. Post-transcriptional processing of simian virus 40 late transcripts in injected frog oocytes. J Mol Biol. 1983;163:1–26. doi: 10.1016/0022-2836(83)90027-x. [DOI] [PubMed] [Google Scholar]