Abstract
Background
Hepatocellular carcinoma (HCC) is a multistep process involving sophisticated genetic, epigenetic, and transcriptional changes. However, studies on microRNA (miRNA)'s regulatory effects of N6-methyladenosine (m6A) modifications on HCC progression are limited.
Methods
Cell Counting Kit-8 (CCK-8), clone formation, and Transwell assays were used to investigate changes in cancer cell proliferation, invasion, and migration. RNA m6A levels were verified using methylated RNA immunoprecipitation. Luciferase reporter assay was used to study the potential binding between miRNAs and mRNA. A mouse tumor transplant model was established to study the changes in tumor progression.
Results
Follistatin-like 5 (FSTL5) was significantly downregulated in HCC and inhibited its further progression. Additionally, methyltransferase-like 3 (METTL3) reduced FSTL5 mRNA stability in an m6A-YTH domain family 2(YTHDF2)-dependent manner. Functional experiments revealed that METTL3 downregulation inhibited HCC progression by upregulating FSTL5 in vitro and in vivo. Luciferase reporter assay verified that miR-186-5p directly targets METTL3. Additionally, miR-186-5p inhibits the proliferation, migration, and invasion of HCC cells by downregulating METTL3 expression.
Conclusions
The miR-186-5p/METTL3/YTHDF2/FSTL5 axis may offer new directions for targeted HCC therapy.
Keywords: microRNA, Hepatocellular carcinoma, METTL3, FSTL5, mRNA stability
Abbreviations
- HCC
Hepatocellular carcinoma
- IHC
Immunohistochemical
- miRNA
microRNA
- m6A
N6-methyladenosine
- FSTL5
Follistatin-like 5
- YTHDF2
YTH domain family 2
- METTL3
methyltransferase-like 3
- TCGA
the cancer genome atlas
- ATCC
American Type Culture Collection
- DMEM
Dulbecco's modified Eagle medium
- FBS
foetal bovine serum
- shRNA
short hairpin RNA
- IGF2BP
insulin-like growth factor 2 mRNA-binding protein 2
- HE
Haematoxylin and eosin
- ECL
Enhanced chemiluminescence
- RIPA
Radio-Immunoprecipitation Assay
- CCK-8
Cell Counting Kit-8
- qRT-PCR
Real-time reverse transcription polymerase chain reaction
- hnRNPA2B1
heterogeneous nuclear ribonucleoprotein A2B1
- METTL14
Methyltransferase-like 14
- ALKBH5
AlkB homologue 5
- FTO
Fat mass and obesity-associated protein
- SOCS2
suppressor of cytokine signalling 2
- YAP
Yes-associated protein
- TAZ
transcriptional coactivator with PDZ-binding motif
- KLF4
Krüppel-like factor 4
- SETD7
SET domain-containing lysine methyltransferase 7
- HBXIP
hepatitis B X-interacting protein
- PBS
phosphate-buffered saline.
1. Introduction
Hepatocellular carcinoma (HCC) is a life-threatening disease that ranks sixth among the most common cancer types globally [2,3]. HCC is induced by nonalcoholic fatty liver disease, type 2 diabetes mellitus, and obesity, thereby increasing its annual incidence [4]. Clinical treatment of HCC is limited due to inconspicuous early symptoms and late diagnosis [5]. Therefore, the molecular mechanisms underlying HCC should be understood to offer a better approach to its diagnosis and treatment.
The follistatin family comprises extracellular matrix glycoproteins that play a role in inflammatory responses, tissue remodeling, and embryonic development [6]. Follistatin-like 5 (FSTL5), a member of the follistatin family, binds directly to activin [7]. Further studies have demonstrated that FSTL5 regulates tumor development. Remke et al. found that FSTL5 is a poor prognostic marker for non-WNT/non-SHH medulloblastoma [8] and may be an attractive therapeutic target and prognostic indicator [9]. However, the role of FSTL5 in HCC requires further investigation.
Several studies have revealed that gene mutations and epigenetic regulation, such as methylation, chromatin modification, and remodeling promote HCC [10]. N6-methyladenosine (m6A) RNA methylation, the most abundant RNA modification, plays a critical role in tumor progression [11,12]. Moreover, m6A modification regulates gene expressions through mRNA stability, splicing, translation, and microRNA maturation [13] and is mediated by m6A “writers,” “erasers,” and “readers” for addition, removal, and recognition, respectively [14]. Methyltransferase-like 3 (METTL3), the major component of m6A “writers,” promotes m6A RNA modification, which is recognized by m6A “readers”, such as YTH domain family 2 (YTHDF), the heterogeneous nuclear ribonucleoprotein A2B1, and insulin-like growth factor 2 mRNA-binding protein 2 [15,16]. m6A “writers,” “erasers,” and “readers” play a vital role in a variety of cancers, including bladder cancer, gastric cancer, colorectal cancer, and HCC [[17], [18], [19], [20]].
MicroRNAs (miRNAs) are endogenous small non-coding RNA 19–22 nucleotides in length that can downregulate gene expression by targeting the 3′-UTR of genes [21]. Several miRNAs negatively modulate METTL3 expression and participate in cancer development and progression [22,23]. For example, miR-600 prevents lung cancer progression by downregulating METTL3 [22]. MiR-4429 prevents gastric cancer progression by targeting METTL3 to inhibit m(6)A-induced SEC62 23. However, it remains obscure whether miRNAs regulate METTL3 expression in HCC. Interestingly, online databases have demonstrated that miR-186-5p binds complementarily to METTL33′-UTR region. Evidence has demonstrated that miR-186-5p inhibits m6A levels by targeting METTL3 in esophageal cancer [24]. Additionally, miR-186-5p plays an important role in HCC [25,26]. Herein, we hypothesized that miR-186-5p is involved in HCC by targeting METTL3, which regulates m6A-mediated stabilization of FSTL5. In this study, we aimed to test this hypothesis and determine the mechanism of action of miR-186-5p in HCC.
2. Methods
2.1. Tissue sample collection
A total of 28 pairs of HCC and adjacent non-tumor tissues were obtained from the First Affiliated Hospital of Bengbu Medical College from January 2020 to December 2020. This study was approved by the Ethics Committee of Bengbu Medical College for the Study of Humans (No. 2017228) and Animals (No. 2017059), and all patients provided written informed consent. None of the patients received preoperative chemotherapy or radiotherapy. Supplementary Table 1 depicts the characteristics of all the participants. The prognosis of the patient cohort was evaluated by using the Cancer Genome Atlas (TCGA) dataset (https://portal.gdc.cancer.gov/).
2.2. Cell culture and transfection
Human HCC cell lines (Huh-7, MHCC97-H, and SMMC-7721) and normal liver cells (HL-7702) were purchased from American Type Culture Collection (ATCC, USA). The Hep3B cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Guangzhou, China). All cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco, USA, Cat#11995065) supplemented with 10% fetal bovine serum (FBS, Gibco, USA, Cat#16140071) at 37 °C in a water-saturated atmosphere with 5% carbon dioxide (CO2). Lipofectamine 2000 (Invitrogen, USA, Cat#11668019) was used to transfect cells with a 50-nm mimic control and miR-186-5p mimic or inhibitor (Ribo bio, Shanghai, China). For FSTL5, METTL3, or YTHDF2 overexpression in the cells, the cDNAs of human FSTL5, METTL3, and YTHDF2 were cloned into the pLV-EF1a-EGFP (2A) Puro vector using GenePharma (Shanghai, China) at the original concentration of 0.5 μg/μL. The dosage was 5, 6, and 5 μL/well, respectively, with 5 μl/well lipofectamine 2000 in the 6-well plate, when the cells encompass approximately 80% of the cell plate. Short hairpin RNA (shRNA) against METTL3 and FSTL5 was obtained from GenePharma (Shanghai, China) at an original concentration of 0.5 μg/μL. The dosage was 10 and 12 μL/well, respectively, with 5 μL/well lipofectamine 2000 in the 6-well plate, when the cells encompass approximately 80% of the cell plate. Supplementary Table 2 depicts the shRNA sequences.
2.3. Lentiviral construction and transfection
A specific LV-ShMETTL3 (lentiviral LV control carrying luciferase and METTL3 knockdown interference plasmids) was designed by Genepharma (Shanghai, China) and validated by sequencing. The carrier number was GV344 and the original sequence was hU6-MCS-Ubiquitin-firefly-Luciferase-IRES-puromycin. Three lentiviral LV controls, LV-ShMETTL3, were constructed using the original sequence. The lentiviral supernatant was screened for multiple infections (MOI, 50). Cells in the three dishes were transfected with one of the three lentiviruses to screen for the best when the cells encompassed approximately 70% of the cell plate. The stable cells were screened using puromycin [27]. Post 72 h, the cells were harvested for detecting METTL3 mRNA and protein expression using quantitative real-time PCR (qRT-PCR) and western blotting (WB). LV-ShFSTL5 (lentiviral LV controls carrying luciferase and FSTL5 knockdown interference plasmids) and LV-YTHDF2 (lentiviral LV controls carrying luciferase and plasmids expressing YTHDF2), were designed by Genepharma (Shanghai, China), and similar methods were used to transfect cells. Sequences of the METTL3 and FSTL5 interference plasmids are depicted in Supplemental Table 2.
2.4. Real-time reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was separated using the TRIzol reagent (Cat #15596026; Invitrogen, USA). RNA concentration was determined using a NanoDrop spectrophotometer (Thermo, USA, Cat#840–329700). Subsequently, miRNAs and mRNAs were quantified using TaqMan miRNA probes (Cat #4316034; Applied Biosystems) and Synergy Brands (SYBR) Green dye (Cat #4309155; Applied Biosystems), respectively. The RevertAid First Strand cDNA Synthesis Kit (Thermo, USA, Cat#K1622) and Maxima SYBR qPCR mixture (Thermo, USA, Cat#K0251) were used for Reverse transcription in Veriti Dx 96-well Thermal Cycler (Applied Biosystem, USA, Cat#4375305), with 20 μlL total volume. Post product synthesis, amplification is initiated in StepOnePlus Real-Time PCR System (Applied Biosystem, USA, Cat#4376600), with 20 μL total total volume. Relative gene expression was evaluated using the 2−ΔΔCT method. The primers used are enlisted in Supplemental Table 3.
2.5. Western blotting
Total protein was extracted from tumor tissues or cells using a radioimmunoprecipitation assay lysis buffer (20 mM Tris, pH 7.5, 1.0 mM EDTA, 150 mM NaCl, 2.5 mM sodium pyrophosphate, and 1% Triton X-100) supplemented with 1 mM phenylmethylsulfonyl fluoride (Beyotime). The total protein concentration was determined using a bicinchoninic acid detection kit (biosharp, China, BL521A). Proteins (20 μg/well) were separated by electrophoresis in a WIX-miniPRO4 Mini vertical electrophoresis tank (WIX, China), transferred onto polyvinylidene fluoride membranes, and incubated with the appropriate antibodies. Enhanced chemiluminescence (ECL; biosharp, China, BL520A) was used to visualize the membranes using Fusion FX SPECTRA Multifunctional Imaging (Vilber, France). ImageJ software was used to evaluate the protein bands. Antibodies against METTL3 (Cat. #86132) and GAPDH (Cat. #5174) were obtained from Cell Signaling Technology (Danvers, MA, USA). Antibodies against methyltransferase-like 14 (METTL14, Cat#ab220030), AlkB homolog 5 (ALKBH5, Cat#ab234528), fat mass, and obesity-associated protein (FTO, Cat#124892) were purchased from Abcam (Cambridge, USA). Antibodies against FSTL5 (Cat#H00056884–B01P) were purchased from Novus Biologicals (Colorado, CA, USA). Goat anti-rabbit secondary antibodies (Cat#BL003A) were purchased from Biosharp (China). Goat anti-mouse secondary antibodies (Cat#BL001A; Biosharp, China) were also purchased from Biosharp.
2.6. CCK-8 assay
The CCK-8 assay was used to assess cell viability. Huh-7 and Hep3B cells were seeded in 96-well plates at 1 × 104 cells/well density. Subsequently, 10 μL CCK-8 solution (Cat#BS350A, biosharp, China) was incorporated to each well and incubated for 1 h at 37 °C. The cell viability was measured at 450-nm wavelength using a Synergy HT Multi-Mode Microplate Reader (BioTek, USA).
2.7. Clone formation assay
Huh-7 and Hep3B cells were transfected with the corresponding plasmids, shRNAs, or miR-186-5p mimic or inhibitor, and harvested by digestion with 0.25% trypsin (Cat #2520014; Gibco, USA). The cells were resuspended in DMEM, plated in a 6-well plate at 2 × 103 cells/dish density, and incubated at 37 °C for 2 weeks. DMEM was changed every 2 days. Subsequently, the cells were washed with phosphate-buffered saline (PBS, Cat #10010023, Gibco, USA), fixed with 1 mL of 4% paraformaldehyde (Biosharp, China, Cat#BL539A) per well for 15 min, and stained with 1 mL Giemsa (Beyotime, China, Cat#C0121-100 ml) per well for 15 min. The colonies were observed and enumerated under an optical microscope.
2.8. Transwell assay
Transwell assays were used to evaluate Huh-7 and Hep3B cells migration and invasion. For cell migration, 100 μL DMEM without FBS, containing 20,000 cells, was incorporated to the upper chambers (6.5 mm Transwell with 8.0 μm pore polycarbonate membrane insert, Corning, USA, Cat#CLS3422) of the Transwell device. Subsequently, 600-μL DMEM with 10% FBS was incorporated to the lower chamber. For the invasion assay, 100 μL DMEM without FBS was incorporated to the upper chambers to rehydrate Matrigel (Cat #356234; Corning, USA) for 2 h at 37 °C. After removing the supernatant, 40,000 cells containing DMEM without FBS were seeded into the Matrigel-pretreated upper chambers, and 600 μL DMEM with 10% FBS was incorporated to the lower chamber. Following 24-h of incubation, migrating or invading cells were fixed in 4% paraformaldehyde (Biosharp, China, Cat#BL539A) for 20 min and stained with Giemsa (Beyotime, China, Cat#C0121-100 ml). An inverted electron microscope (OLYMPUS IX71, OLYMPUS, Japan) was used to obtain the final images. The number of cells was enumerated.
2.9. Luciferase reporter assay
The wild-type or mutant 3′-UTR of METTL3 was amplified and cloned into the pGL3-basic vector (Promega, USA). The binding site of the 3′-UTR of METTL3 was mutated from UGUACAAUAGCUUUCUUCUUUAU to UGUACAAUAGCUUUCAAGAAAAU for miR-186-5p. Subsequently, Hep3B cells were co-transfected with NC-Luc (0.5 μg/μl, 2 μl/well), wild-type (0.5 μg/μl, 2 μl/well), or mutant luciferase plasmids (0.5 μg/μl, 2 μl/well) and the miR-186-5p mimic (0.5 μg/μl, 6 μl), miR-186-5p inhibitor (0.5 μg/μl, 6 μl/well), or control miRNA (0.5 μg/μl, 6 μl). A dual-luciferase reporter assay system (Promega, WI, USA, Cat#E1960) was used to measure luciferase activity in a multifunctional microplate reader (Synergy II, Bio-Tex, TY2009001641). The relative luciferase activity of each sample was normalized to the Renilla luciferase activity [28].
2.10. m6A RNA methylation quantification
The m6A RNA methylation analysis kit (Cat # p-9005-48; Epigentek, USA) was used to measure the total m6A methylation content. Briefly, 200 ng total RNA was incorporated to each reaction solution after incorporating the antibody solution. The m6A level was quantified based on coloration, and absorbance was measured at 450 nm using a Synergy HT Multi-Mode Microplate Reader (BioTek, USA). To measure FSTL5 m6A enrichment, the total RNA of Hep3B cells was extracted and immunoprecipitation was conducted using a Magna MeRIP m6A kit (Merck Millipore, Germany, Cat#17–10499) with protein A beads and an m6A antibody. Partial RNA was evaluated by qRT-PCR post immunoprecipitation.
2.11. RNA immunoprecipitation
Hep3B cells were transfected with a lentivirus expressing YTHDF2. The virus was washed twice with PBS and dissolved in lysine buffer containing a mixture of protease and ribozyme inhibitors for 30 min at 4 °C. The supernatants were collected post centrifugation and incubated with magnetic beads (Sigma, USA, Cat#M8823) for 4 h, which were pre-processed with IgG or YTHDF2 antibodies (Abcam, USA, Cat#ab246514), and then washed five times with wash buffer (50-mM Tris, 0.5-mM DTT, RNase inhibitor, 200-mM sodium chloride, 0.05% NP40, and 2-mM ethylamine tetraethyl). Subsequently, the beads were washed in a solution containing 0.1% sodium dodecyl sulfate and 10-mL of potassium enzyme and incubated for 30 min at 55 °C. The imported nucleic acids and immune precipitates were separated using TRIzol reagent and quantitatively analyzed using PCR and agarose gel electrophoresis in multifunctional horizontal electrophoresis tank (Tanon, HE-120), with 5 μl/well in agarose gel [29].
2.12. RNA stability
Hep3B cells were treated with sh-METTL3, control lentivirus, or YTHDF2 lentivirus for 24 h. Subsequently, 5 mg/mL actinomycin D (Cat #A4262; Sigma) was incorporated to the cells. RNA was isolated using TRIzol reagent at 0, 3, and 6 h and normalized to GAPDH based on qPCR analysis. The t1/2 value of the mRNA was computed.
2.13. Immunohistochemical (IHC) analysis
Tumor tissues were covered with paraffin, waxed, and treated with 3% H2O2 (Cat # 88597; Sigma, USA) to block endogenous peroxidase activity. The tissues were then incubated at 37 °C with 5% bovine serum albumin and with primary antibodies (1:100 diluted, Abcam, USA) against FSTL5, METTL3, or YTHDF2 at 4 °C for 12 h. Following incubation with secondary antibody at 37 °C for 1 h, the tissues were dyed with Diaminobenzidine (DAB, Beyotime, China, Cat#P0203) and representative images were obtained under an optical microscope (Olympus, Japan, Cat#CX23).
2.14. Tumor xenograft model
Approximately 1 × 107 Hep3B cells were suspended in 200-μL PBS and injected into one side of BALB/c nu/nu male mice aged 4–6 weeks (Charles River Lab, Beijing, China). The tumor volume in the mice was recorded every 3 days (volume = [length × width 2]/2). Mice received intratumoral injections of 50 μL METTL3 and FSTL5 knockdown lentivirus (GenePharma, China) at 4 × 107 IU/mL post tumor cell injection. After 15 days, the mice were euthanized as follows: they were kept in a box, and CO2 was injected into the box at a 20% per minute rate. Experiments were conducted in accordance with the ARRIVE guidelines for the care and use of animals.
2.15. Hematoxylin and eosin (HE) staining
Tumor tissues were fixed in 4% paraformaldehyde (biosharp, China, Cat#BL539A) for 48 h and embedded in paraffin. The tissues were sliced and the sections were stained with hematoxylin (Cat #H9627-25 G, Sigma, USA) for 2 min and eosin (Sigma, USA, Cat#212954-25G) for 1 min. Images were viewed using a light microscope (Nikon, Tokyo, Japan).
2.16. Statistical analysis
All cell experiments were conducted in triplicates. Data were evaluated using Prism GraphPad software (version 8.0) and are presented as mean ± standard error of the mean (SEM). A two-tailed Student's t-test was used to compare the two groups. A one-way analysis of variance was used to compare multiple groups. Statistical significance was set at p < 0.05. significance.
3. Results
3.1. FSTL5 inhibited HCC cell proliferation, migration, and invasion
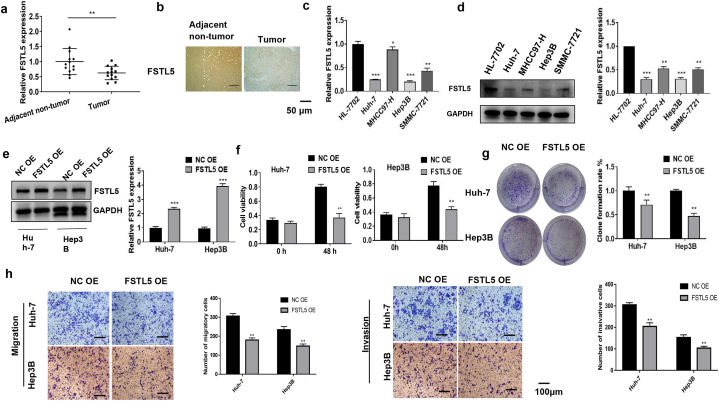
FSTL5 expression was significantly higher in adjacent non-tumor tissues than in HCC tissues (Fig. 1a and b). Similarly, FSTL5 expression was significantly downregulated in four HCC cell lines (Huh-7, MHCC97-H, Hep3B, and SMMC-7721) compared to that in HL-7702 cells (Fig. 1c and d). As the average FSTL5 expression levels in Huh-7 and Hep3B cells were lower than those in the other two HCC cell lines, Huh-7 and Hep3B cells were selected for subsequent experiments. The FSTL5 overexpression plasmid was transfected into Huh-7 and Hep3B cells to overexpress FSTL5 (Fig. 1e). FSTL5 overexpression inhibited the viability, clone formation, migration, and invasion of Huh-7 and Hep3B cells (Fig. 1f–h).
Fig. 1.
FSTL5 inhibits HCC cell proliferation, migration, and invasion (a) qRT-PCR reveals significantly decreased FSTL5 expression in HCC tissues (n = 28). (b) IHC assay indicates lower FSTL5 levels in HCC tissues (bar = 50 μm). (c) qRT-PCR and (d) Western blot analyses indicates lower FSTL5 expression in the HCC cell lines Huh-7, MHCC97-H, Hep3B, and SMMC-7721. (e) WB and qRT-PCR verifies FSTL5 overexpression in cells transfected with FSTL5 OE. (f) Cell viability, (g) clone formation, and (h) migration and invasion inhibited by FSTL5 overexpression (bar = 100 μm). Data represents three separate experiments and are expressed as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, compared with the control group. “NC OE” represents the blank control of “FSTL5 OE.”
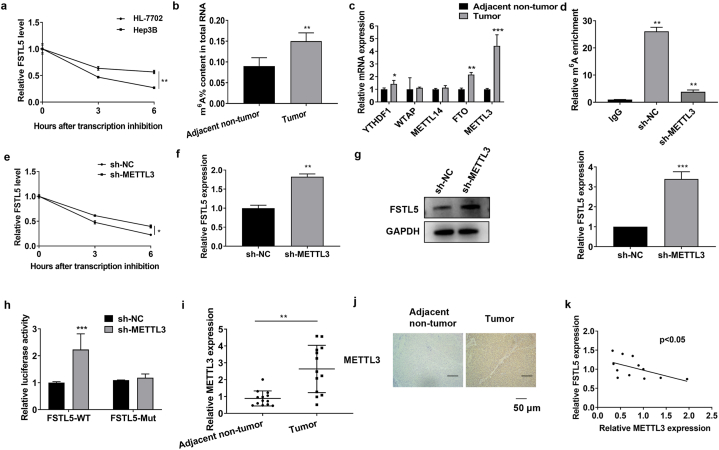
3.2. METTL3 promoted FSTL5 mRNA degradation
We investigated FSTL5 downregulation in HCC. The RNA stability test demonstrated a shortened t1/2 of FSTL5 in HCC cells, suggesting a possible RNA modification (Fig. 2a). m6A modifications regulate mRNA stability to facilitate gene expression in cancer [30,31]. Furthermore, m6A expression was detected in the HCC tissues. Total m6A levels were lower in the adjacent non-tumor than in the tumor tissues (Fig. 2b). m6A modification is mediated by m6A methyltransferases METTL3 and METTL14, which are cleared by fat mass and FTO [32,33]. Remarkably, only METTL3 expression increased in HCC cells (Fig. 2c). METTL3 upregulation is assumed to promote FSTL5 degradation via m6A modification in HCC. MeRIP qPCR evaluation was used to assess the effects of METTL3 on the m6A modification of FSTL5 mRNA. Thus, m6A-specific antibodies were significantly enriched in the FSTL5 gene compared to those in the immunoglobulin group. Additionally, METTL3 inhibition significantly reduced m6A levels in FSTL5 mRNA (Fig. 2d). Furthermore, METTL3 knockdown extended the mRNA t1/2 (Fig. 2e). qRT-PCR and western blotting analyses demonstrated that METTL3 negatively regulated FSTL5 expression in HCC cells (Fig. 2f and g). The luciferase reporter assay revealed that METTL3 inhibition significantly promoted the activity of wild-type luciferase but did not affect the mutant FSTL5 3′-UTR (Fig. 2h). Moreover, qRT-PCR and IHC results indicated that METTL3 expression was increased in HCC tissues (Fig. 2i, j). Pearson's correlation analysis indicated that METTL3 expression negatively correlated with FSTL5 expression in HCC (Fig. 2k). Collectively, these results demonstrated that METTL3-mediated m6A inhibits FSTL5 expression in HCC.
Fig. 2.
METTL3 promotes FSTL5 mRNA degradation (a) RNA stability assay depicts the half-life of FSTL5 mRNA (t1/2) in Hep3B and HL-7702 cells. (b) The total m6A level of the adjacent non-tumor and tumor tissues (n = 28). (c) METTL3, METTL14, FTO, and ALKBH5 levels are detected in HCC tissues using qRT-PCR. (d) The MeRIP qPCR assay is conducted to assess the effects of METTL3 targeting FSTL5 mRNA on m6A modification. (e) The RNA stability assay depicts the half-life of FSTL5 mRNA (t1/2) in Hep3B cells with METTL3 knockdown. (f) qRT-PCR and (g) Western blot analysis depicts that METTL3 negatively regulates FSTL5 expression. (h) Luciferase reporter assay depicts the relationship between METTL3 and FSTL5. (i) qRT-PCR and (j) IHC indicates that METTL3 expression is upregulated in HCC tissues (n = 28; bar = 50 μm). (k) Pearson's correlation analysis depicts the negative correlation between METTL3 and FSTL5 expression in HCC. The data represented three separate experiments and are expressed as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, compared with the control group. “sh-NC″represents the blank control of “sh-METTL3”.
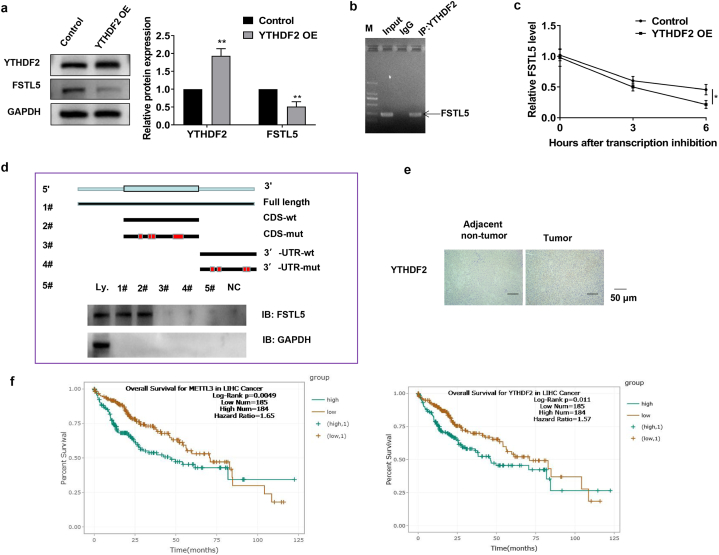
3.3. YTHDF2-bound FSTL5 decreased FSTL5 mRNA stability
YTHDF1/2/3 is a unique m6A reader family that targets thousands of genes via m6A sequencing [34]. Western blotting analysis demonstrated that YTHDF2 overexpression reduced FSTL5 protein expression (Fig. 3a). RIP-RT-PCR revealed that YTHDF2 directly interacts with FSTL5 mRNA, indicating that YTHDF2 can target FSTL5 (Fig. 3b). RNA stability assay demonstrated that the mRNA t1/2 was shortened by YTHDF2 overexpression (Fig. 3c). Only the full-length or wild-type CDS of FSTL5 mRNA reduced YTHDF2 protein expression, indicating that YTHDF2 regulates FSTL5 mRNA stability by binding to the CDS of FSTL5 (Fig. 3d). Moreover, YTHDF2 was upregulated in HCC tissues (Fig. 3e). TCGA dataset analysis revealed that high METTL3 and YTHDF2 expression levels indicated a poor prognosis for HCC (Fig. 3f). Collectively, these results suggested that YTHDF2 shortens the lifespan of FSTL5 mRNA, thereby suppressing FSTL5 protein expression.
Fig. 3.
YTHDF2 can bind FSTL5 and decrease FSTL5 mRNA stability (a) YTHDF2 overexpression inhibits FSTL5 expression. (b) RIP-RT-PCR followed by agarose gel electrophoresis depicts FSTL5 binding to YTHDF2. (c) RNA stability assay depicts the half-life of FSTL5 mRNA (t1/2) in Hep3B cells overexpressing YTHDF2. (d) YTHDF2 expression is reduced in the full-length and wild-type CDS regions of FSTL5 mRNA. (e) IHC depicts YTHDF2 expression in adjacent non-tumor and tumor tissues (bar = 50 μm). (f) Correlation between METTL3 and YTHDF2 expression and prognosis of patients with HCC in TCGA cohort. Data represents three separate experiments and are expressed as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, compared with the control group.
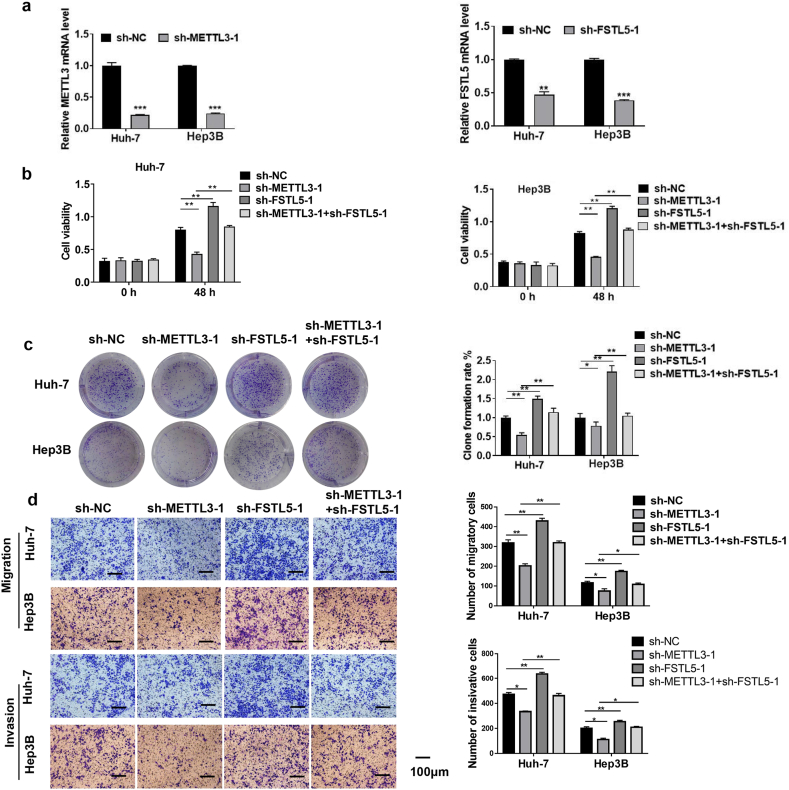
3.4. METTL3 knockdown inhibited HCC cell proliferation, migration, and invasion
To further assess whether METTL3 regulates FSTL5 in HCC, two sequence-specific shRNAs were used to knockdown METTL3 and FSTL5 in Huh-7 and Hep3B cells to exclude off-target effects. The knockdown efficiencies of sh-METTL3-1, sh-METTL3-2, sh-FSTL5-1, and sh-FSTL5-2 in Huh-7 and Hep3B cells were verified using qRT-PCR (Fig. 4a and Supplementary Fig. 1a). METTL3 knockdown significantly inhibits Huh-7 and Hep3B cell proliferation. However, co-transfection with sh-FSTL5 reversed these effects (Fig. 4b–c and Supplementary Figs. 1b–c). Consistently, sh-METTL3 significantly inhibited HCC cell migration and invasion, whereas sh-FSTL5 antagonized these effects of sh-METTL3 (Fig. 4d and Supplementary Fig. 1d).
Fig. 4.
METTL3 knockdown inhibits HCC cell proliferation, migration, and invasion (a) Knockdown of METTL3 and FSTL5 in Huh-7 and Hep3B cells. (b) CCK-8 and (c) colony formation assays are used to detect cell proliferation. (d) The Transwell assay is conducted to detect cell migration and invasion (bar = 100 μm). Data are from three separate experiments and are expressed as mean ± SEM. ***p < 0.001, *p < 0.05, **p < 0.01.“sh-NC″represents the blank control of“sh-METTL3-1″or“sh-FSTL5”.
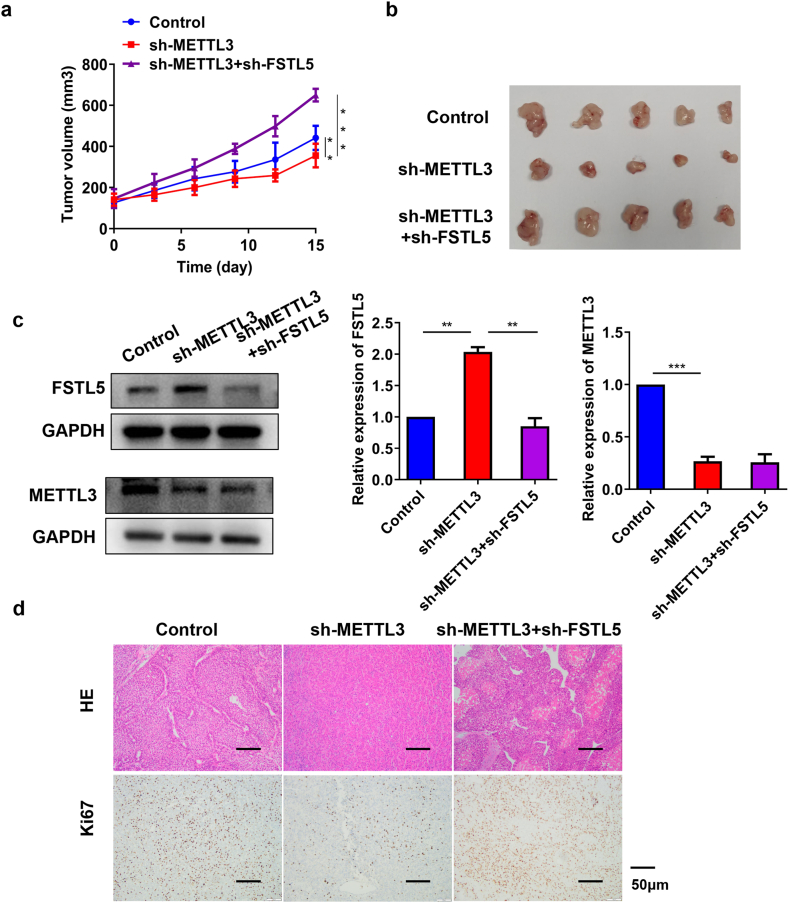
3.5. METTL3 knockdown inhibited HCC tumor growth in vivo
To verify the roles of METTL3 and FSTL5 in HCC, an in vivo xenotransplantation model was established. Hep3B cells were subcutaneously injected into nude mice. METTL3 knockdown inhibited tumor growth and xenograft size, whereas FSTL5 knockdown promoted tumorigenicity (Fig. 5a–b and Supplementary Fig. 2). Furthermore, sh-METTL3 augmented FSTL5 expression, whereas FSTL5 silencing reversed the upregulation of FSTL5 expression by sh-METTL3 (Fig. 5c). Furthermore, HE and IHC revealed that METTL3 knockdown inhibited tumor infiltration and proliferation of Ki67 protein, whereas FSTL5 silencing reversed the effects of sh-METTL3 (Fig. 5d). These results suggest that METTL3 knockdown inhibits HCC tumor growth by upregulating FSTL5 in vivo.
Fig. 5.
METTL3 knockdown inhibits HCC tumor growth in vivo A tumor xenograft model is constructed by subcutaneously injecting 1 × 107 Hep3B cells (a) Images of isolated tumors from each group (n = 5) (b) Tumors of mice in each group (n = 5). (c) METTL3 and FSTL5 expression in tumor tissues (d) HE and IHC analyses of tumor infiltration and Ki67 expression (bar = 50 μm). Data represent three separate experiments and are expressed as mean ± SEM. ***p < 0.001, *p < 0.05, **p < 0.01.
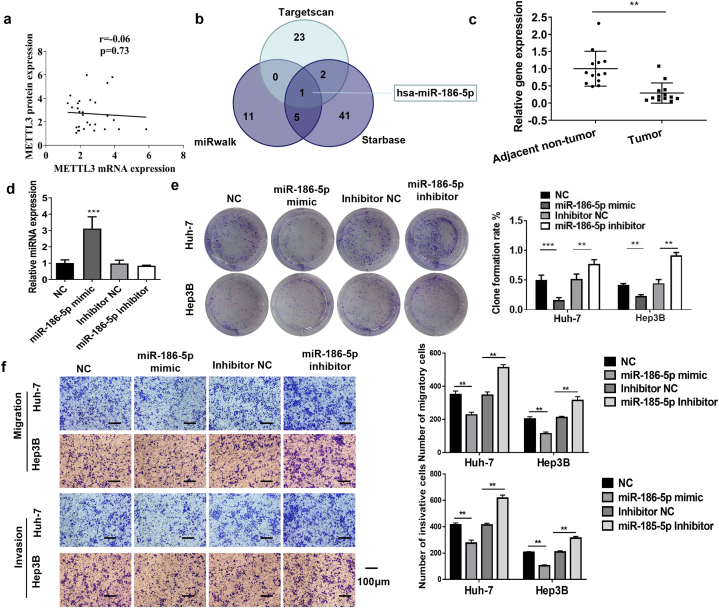
3.6. MiR-186-5p inhibited HCC cell proliferation, migration, and invasion
No association was observed between METTL3 protein and gene expression in most patients in each group (Fig. 6a). The mismatch between METTL3 protein and mRNA expression levels in HCC suggests that METTL3 may be involved in HCC development via post-transcriptional regulation. MiRNAs are vital post-transcriptional regulatory factors that play indispensable roles in HCC [35]. Therefore, we used the online databases TargetScan, miRWalk, and StarBase to predict the possible miRNAs (Fig. 6b). We found that miR-186-5p was a common miRNA in all the three databases (Fig. 6b). RT-qPCR revealed that miR-186-5p expression was downregulated in HCC tissues (Fig. 6c). These data prompted us to investigate the role of miR-186-5p in HCC. The miR-186-5p mimics and inhibitors increased or decreased miR-186-5p expression in cells, respectively (Fig. 6d). The clone formation assay demonstrated that miR-186-5p mimiced or inhibitors inhibited or increased clone formation in Huh-7 and Hep3B cells, respectively (Fig. 6e). Transwell assays demonstrated that the miR-186-5p mimiced or inhibitors inhibited or promoted the migration and invasion of Huh-7 and Hep3B cells, respectively (Fig. 6f).
Fig. 6.
MiR-186-5p inhibits HCC cell proliferation, migration, and invasion (a) Correlation between METTL3 protein and mRNA expression in HCC tissues (n = 28). (b) Online databases TargetScan, miRWalk, and Starbase are used to predict possible miRNAs. (c) miR-186-5p levels in HCC tissues are detected using qRT-PCR (n = 28). (d) miR-186-5p mimics or inhibitors increase or decrease miR-186-5p levels. (e) Clone formation in cells treated with miR-186-5p mimic or inhibitor. (f) Invasion and migration of cells treated with miR-186-5p mimic or inhibitor (bar = 100 μm). Data represent three separate experiments and are expressed as mean ± SEM. ***p < 0.001, *p < 0.05, **p < 0.01.“NC”represents the blank control of“miR-186-5p mimic.” “inhibitor NC″represents the blank control of “miR-186-5p inhibitor.”
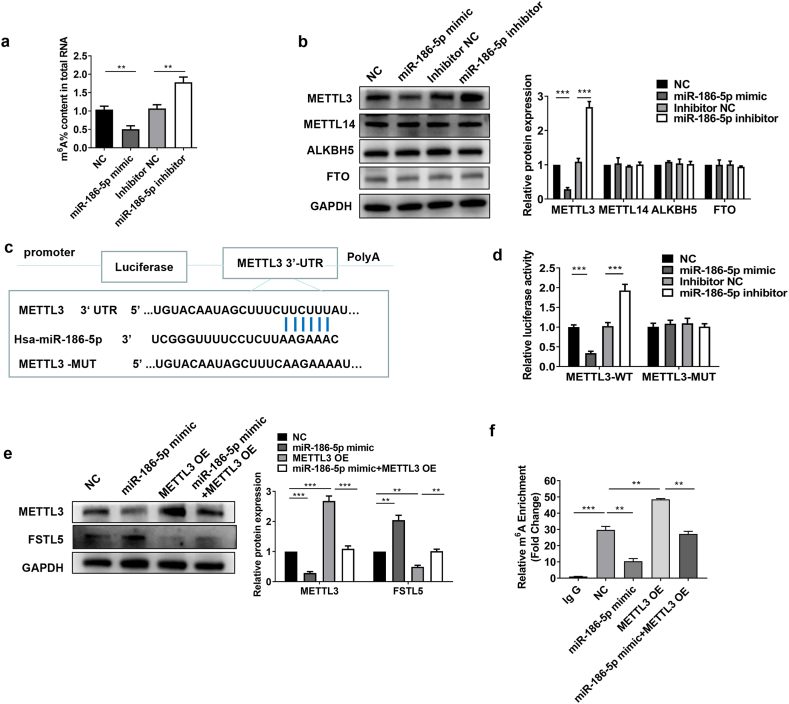
3.7. METTL3 was a direct target of miR-186-5p
In HCC cells, miR-186-5p decreased m6A levels (Fig. 7a). Western blotting analysis demonstrated that miR-186-5p negatively regulated METTL3 expression in HCC cells (Fig. 7b). Potential binding sites were predicted between the miR-186-5p and 3′-UTR of METTL3 (Fig. 7c). The luciferase reporter assay indicated that miR-186-5p significantly reduced the luciferase activity of wild-type METTL3 3′-UTR compared to that of the mutant, indicating that miR-186-5p specificity promoted the combination of 3′-UTR with METTL3 (Fig. 7d). Western blotting analysis demonstrated that the miR-186-5p mimic downregulated METTL3 and upregulated FSTL5 expression, which was reversed by METTL3 overexpression (Fig. 7e). Furthermore, MeRIP qPCR assay revealed that the miR-186-5p mimic significantly reduced m6A levels in FSTL5 mRNA, whereas co-transfection with the METTL3 overexpression plasmid increased m6A levels (Fig. 7f). These results suggested that miR-186-5p regulates FSTL5 mRNA modification mediated by METTL3.
Fig. 7.
METTL3 is a direct miR-186-5p target (a) The total m6A level in Hep3B cells treated with miR-186-5p mimic or inhibitor. (b) METTL3, METTL14, ALKBH5, and FTO protein expression levels are detected using Western blot analysis. (c) The potential binding sites of miR-186-5p and 3′-UTR of METTL3 mRNA. (d) Luciferase reporter assay with reporter plasmid containing the wild-type or mutant METTL3 3′-UTR and NC, miR-186-5p mimic, and miR-186-5p inhibitor. (e) The protein expression of FSTL5 is detected using Western blot analysis. (f) MeRIP qPCR assay is used to evaluate the m6A level of FSTL5 mRNA. Data represent three separate experiments and are expressed as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.“NC”represents the blank control of“miR-186-5p mimic”. “inhibitor NC″represents the blank control of “miR-186-5p inhibitor”.
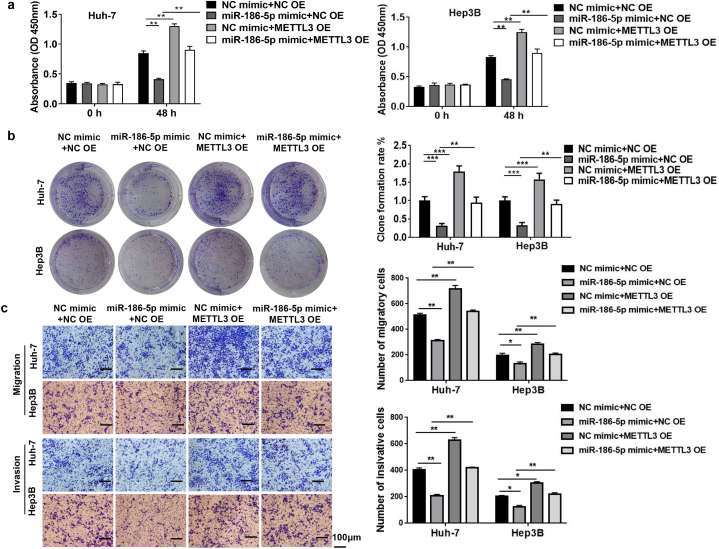
3.8. MiR-186-5p inhibited HCC cell proliferation, migration, and invasion by downregulating METTL3
We verified whether miR-186-5p regulates HCC by targeting METTL3. CCK-8 and colony formation assays demonstrated that miR-186-5p inhibited HCC cell viability and colony formation, whereas METTL3 overexpression reversed these effects of miR-186-5p (Fig. 8a and b). Transwell assays demonstrated that miR-186-5p overexpression inhibited the migration and invasion of HCC cells (Fig. 8c).
Fig. 8.
MiR-186-5p inhibits HCC cell proliferation, migration, and invasion by downregulating METTL3 Huh-7 and Hep3B cells are treated with the miR-186-5p mimic with or without transfection with the METTL3 overexpression lentivirus. (a) Cell viability and (b) clone formation are inhibited by miR-186-5p overexpression, but reversed by METTL3 overexpression. (c) Transwell assay depict that cell invasion and migration are inhibited by miR-186-5p overexpression but are reversed by METTL3 overexpression (bar = 100 μm). Data represent three separate experiments and are expressed as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.“NC mimic”represents the blank control of“miR-186-5p mimic.”“NC OE” represents the blank control of“METTL3 OE”.
4. Discussion
Hepatocellular carcinoma (HCC) is a complex multistep process involving genetic, epigenetic, and transcriptional changes [36]. Herein, we demonstrated that miR-186-5p prevents HCC progression by targeting METTL3 to regulate the m6A-mediated stabilization of FSTL5 (Supplementary Fig. 3).
FSTL5 is a member of the follistatin gene family that encodes secretory glycogen. FSTL5 inhibited HCC progression by inducing apoptosis and regulating the Wnt/Yes-associated protein (YAP) pathway [37]. The transcriptional coactivator with a PDZ-binding motif (YAP/TAZ) is a transcription-assisted activation factor that interacts with transcription factors to regulate genes involved in HCC. Li et al. have suggested that FSTL5 accelerates HCC cell apoptosis in a caspase-dependent manner [9]. However, the mechanisms underlying FSTL5 downregulation in HCC remain obscure.
Interestingly, our results indicated that the FSTL5 mRNA t1/2 was shortened in HCC cells, suggesting the possibility of RNA modification. Based on these results, we hypothesized that m6A regulates FSTL5 stability in HCC cells. Emerging studies have reported that epigenetic modifications, including miRNA regulation, histone modification, and DNA methylation, are crucial in HCC, with m6A RNA methylation being the most common. Several studies have reported the effects of m6A modification, including its involvement in mRNA stability, which affects HCC development [38,39]. For instance, Chen et al. indicated that METTL3 inhibits cytokine signaling 2 (SOCS2) mRNA expression suppressor by mediating SOCS2 m6A mRNA modification [40]. METTL3 plays a regulatory role via m6A modification-mediated mRNA stability [41] and promotes FSTL5 mRNA degradation. YTHDF2 is a functional m6A-binding protein that regulates mRNA stability [42]. The METTL3/YTHDF2 m6A axis promotes tumor development by decomposing SET domain-containing lysine methyltransferase 7 (SETD7) and Krüppel-like factor 4 mRNAs in bladder cancer [43].
Herein, we found that YTHDF2 overexpression promotes FSTL5 mRNA degradation and inhibits FSTL5 expression by recognizing FSTL5 mRNA CDS region. Moreover, METTL3 abrogates the antitumor effects of FSTL5, which is regulated by METTL3-dependent m6A modifications in HCC. Knockdown of METTL3 can reduce FSTL5 mRNA methylation level, indirectly increase FSTL5 expression, and inhibit the proliferation, migration, and invasion of tumor cells. However, in METTL3 knockdown cells, further FSTL5 knockdown directly decreased FSTL5 expression, which offset the increase in FSTL5 levels caused by METTL3 knockdown, thereby reversing the cell phenotype. MiRNAs play a role in diverse diseases by negatively regulating gene expression post-transcriptional level [44]. The abnormal regulation of miRNA expression is associated with tumor progression. Several miRNAs negatively modulate METTL3 expression via post-transcriptional translation [24,45]. Cai et al. indicated that hepatitis B X-interacting protein promotes breast cancer progression by inhibiting miRNA let-7g to regulate METTL3 and let-7g and downregulate METTL3 expression by targeting the 3′-UTR [45]. Conversely, METTL3 regulates several miRNAs [46,47]. For instance, METTL3-mediated m6A modification of pri-miRNAs promotes their recognition and processing by DGCR8 to produce mature miRNAs [46]. Han et al. found that METTL3 induces colorectal cancer progression through the miR-1246/SPRED2/MAPK signaling pathway [47]. Herein, we found that miR-186-5p regulates METTL3 expression, and that METTL3-dependent m6A modification of FSTL5 stabilizes FSTL5 and inhibits HCC development.
It has been reported that miR-4429 inhibits gastric cancer progression by targeting METTL3 to inhibit m(6)A-induced stabilization of SEC62 23. Herein, we focused on miR-186-5p as it was predicted using three online databases: TargetScan, miRWalk, and StarBase. Diverse miRNAs may regulate METTL3 expression in different cancer types. While miR-4429 may regulate METTL3 expression in gastric cancer, miR-186-5p may regulate METTL3 expression in HCC. qRT-PCR revealed that miR-186-5p was significantly downregulated in HCC tissues. In vitro experiments revealed that miR-186-5p inhibited the proliferation, invasion, and migration of HCC cells. Therefore, we hypothesized that miR-186-5p regulates m6A modification to increase FSTL5 stability or to decrease its degradation in HCC. Furthermore, we detected m6A expression level in miR-186-5p treated HCC cells and found that they decreased upon miR-186-5p overexpression. miR-186-5p is negatively regulated by m6A by directly targeting METTL3 in HCC cells. METTL3 overexpression eliminated the antitumor effects of miR-186-5p, indicating that the effects of miR-186-5p may dependent on METTL3 downregulation.
5. Conclusions
In our study, we first identified abnormally low expression of FSTL5 in HCC tissues and cell lines, and thus speculated that FSTL5 plays a crucial role in HCC. We verified this hypothesis through functional experiments and further explored the upstream and downstream genes that regulate FSTL5 expression in HCC through regression experiments. The results demonstrated that m6A-regulated FSTL5 stability may be mediated by YTHDF2, and Mir-186-5p and METTL3 downregulation can affect HCC progression through FSTL5, which also makes us more confident that the study of the Mir-186-5p/METTL3/YTHDF2/FSTL5 signaling axis will offer promising prospects for molecular targeted therapy of HCC.
Ethics statement
Ethics declaration: This study was reviewed and approved by the Ethics Committee of Bengbu Medical College (approval no. 2017228). All participants (or their proxies or legal guardians) offered informed consent to participate in the study. The study was conducted in accordance with the principles of the Declaration of Helsinki.
Animal Ethics The animal study and euthanasia methods were approved by the Ethics Committee of Bengbu Medical College (No. 2017059). The study was conducted in accordance with the relevant guidelines. Experiments were conducted in accordance with the ARRIVE guidelines for the care and use of animals.
Funding
This study was supported by the Anhui University Natural Science Research Project (KJ2021A0731), the Anhui Province Science Foundation for Youths (1808085QH288), and the Bengbu Medical College Natural Science Key Project(2021byzd129).
Data availability statement
No data associated with this study have been deposited in a publicly available repository. Data are available from the corresponding author upon request.
CRediT authorship contribution statement
Shuoshuo Ma: Formal analysis. Fangfang Chen: Investigation, Formal analysis. Chuanle Lin: Writing – review & editing, Validation, Methodology. Wanliang Sun: Resources, Formal analysis. Dongdong Wang: Resources, Formal analysis. Shuo Zhou: Validation, Formal analysis, Data curation. ShiRu Chang: Funding acquisition, Conceptualization. Zheng Lu: Funding acquisition, Conceptualization. Dengyong Zhang: Writing – review & editing, Supervision, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
“A preprint has previously been published [1].”
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e26767.
Contributor Information
Zheng Lu, Email: zdy527897470@126.com.
Dengyong Zhang, Email: zhangdengyongdr@bbmc.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhang D., Chen F., Ma S., et al. miR-186-5p prevents hepatocellular carcinoma progression through targeting METTL3 to regulate m6A-mediated stabilization of FSTL5. PREPRINT. 2021 doi: 10.1016/j.heliyon.2024.e26767. (Version 1) available at: Research Square [ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhan L., Cao H., Wang G., et al. Drp1-mediated mitochondrial fission promotes cell proliferation through crosstalk of p53 and NF-κB pathways in hepatocellular carcinoma. Oncotarget. 2016;7(40):65001–65011. doi: 10.18632/oncotarget.11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.Marengo A., Rosso C., Bugianesi E. Liver cancer: connections with obesity, fatty liver, and cirrhosis. Annu. Rev. Med. 2016;67:103–117. doi: 10.1146/annurev-med-090514-013832. [DOI] [PubMed] [Google Scholar]
- 5.Dimitroulis D., Damaskos C., Valsami S., et al. From diagnosis to treatment of hepatocellular carcinoma: an epidemic problem for both developed and developing world. World J. Gastroenterol. 2017;23(29):5282–5294. doi: 10.3748/wjg.v23.i29.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fullerton P.T., Jr., Monsivais D., Kommagani R., Matzuk M.M. Follistatin is critical for mouse uterine receptivity and decidualization. Proc. Natl. Acad. Sci. U. S. A. 2017;114(24):E4772–e4781. doi: 10.1073/pnas.1620903114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kingwell K. FSTL5--a new prognostic biomarker for medulloblastoma. Nat. Rev. Neurol. 2011;7(11):598. doi: 10.1038/nrneurol.2011.156. [DOI] [PubMed] [Google Scholar]
- 8.Remke M., Hielscher T., Korshunov A., et al. FSTL5 is a marker of poor prognosis in non-WNT/non-SHH medulloblastoma. J. Clin. Oncol. 2011;29(29):3852–3861. doi: 10.1200/JCO.2011.36.2798. [DOI] [PubMed] [Google Scholar]
- 9.Li C., Dai L., Zhang J., et al. Follistatin-like protein 5 inhibits hepatocellular carcinoma progression by inducing caspase-dependent apoptosis and regulating Bcl-2 family proteins. J. Cell Mol. Med. 2018;22(12):6190–6201. doi: 10.1111/jcmm.13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson C.L., Mann D.A., Borthwick L.A. Epigenetic reprogramming in liver fibrosis and cancer. Adv. Drug Deliv. Rev. 2017;121:124–132. doi: 10.1016/j.addr.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 1974;71(10):3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun T., Wu R., Ming L. The role of m6A RNA methylation in cancer. Biomed. Pharmacother. 2019;112 doi: 10.1016/j.biopha.2019.108613. [DOI] [PubMed] [Google Scholar]
- 13.Chen J., Fang X., Zhong P., Song Z., Hu X. N6-methyladenosine modifications: interactions with novel RNA-binding proteins and roles in signal transduction. RNA Biol. 2019;16(8):991–1000. doi: 10.1080/15476286.2019.1620060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao W., Qi X., Liu L., Ma S., Liu J., Wu J. Epigenetic regulation of m(6)A modifications in human cancer. Mol. Ther. Nucleic Acids. 2020;19:405–412. doi: 10.1016/j.omtn.2019.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu K., Yang Y., Feng G.H., et al. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017;27(9):1100–1114. doi: 10.1038/cr.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhuang M., Li X., Zhu J., et al. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res. 2019;47(9):4765–4777. doi: 10.1093/nar/gkz157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu D., Shao W., Jiang Y., Wang X., Liu Y., Liu X. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol. Rep. 2017;38(4):2285–2292. doi: 10.3892/or.2017.5904. [DOI] [PubMed] [Google Scholar]
- 18.Nishizawa Y., Konno M., Asai A., et al. Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2018;9(7):7476–7486. doi: 10.18632/oncotarget.23554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J., Wang J.Z., Yang X., et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer. 2019;18(1):110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H., Wang H., Zhao W., et al. SUMO1 modification of methyltransferase-like 3 promotes tumor progression via regulating Snail mRNA homeostasis in hepatocellular carcinoma. Theranostics. 2020;10(13):5671–5686. doi: 10.7150/thno.42539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhaskaran M., Mohan M. MicroRNAs: history, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014;51(4):759–774. doi: 10.1177/0300985813502820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei W., Huo B., Shi X. miR-600 inhibits lung cancer via downregulating the expression of METTL3. Cancer Manag. Res. 2019;11:1177–1187. doi: 10.2147/CMAR.S181058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He H., Wu W., Sun Z., Chai L. MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m(6)A-caused stabilization of SEC62. Biochem. Biophys. Res. Commun. 2019;517(4):581–587. doi: 10.1016/j.bbrc.2019.07.058. [DOI] [PubMed] [Google Scholar]
- 24.Zhang M., Bai M., Wang L., et al. Targeting SNHG3/miR-186-5p reverses the increased m6A level caused by platinum treatment through regulating METTL3 in esophageal cancer. Cancer Cell Int. 2021;21(1):114. doi: 10.1186/s12935-021-01747-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shan Y., Li P. Long intergenic non-protein coding RNA 665 regulates viability, apoptosis, and autophagy via the MiR-186-5p/MAP4K3 Axis in hepatocellular carcinoma. Yonsei Med. J. 2019;60(9):842–853. doi: 10.3349/ymj.2019.60.9.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W., Li Y., Zhong J., Wen G. circ-PRKCI targets miR-1294 and miR-186-5p by downregulating FOXK1 expression to suppress glycolysis in hepatocellular carcinoma. Mol. Med. Rep. 2021;23(6) doi: 10.3892/mmr.2021.12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang M.L., Yang Q., Zhu Y.D., et al. Nobiletin inhibits hypoxia-induced placental damage via modulating P53 signaling pathway. Nutrients. 2022;14(11) doi: 10.3390/nu14112332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Feng Y.C., Gan Y., et al. LncRNA MILIP links YBX1 to translational activation of Snai1 and promotes metastasis in clear cell renal cell carcinoma. J. Exp. Clin. Cancer Res. 2022;41(1):260. doi: 10.1186/s13046-022-02452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y., Wu D., Liu J., et al. Downregulated lncRNA HOXA11-AS affects trophoblast cell proliferation and migration by regulating RND3 and HOXA7 expression in PE. Mol. Ther. Nucleic Acids. 2018;12:195–206. doi: 10.1016/j.omtn.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Ren D., Du Z., Wang H., Zhang H., Jin Y. m(6)A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem. Biophys. Res. Commun. 2018;502(4):456–464. doi: 10.1016/j.bbrc.2018.05.175. [DOI] [PubMed] [Google Scholar]
- 31.Li J., Han Y., Zhang H., et al. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem. Biophys. Res. Commun. 2019;512(3):479–485. doi: 10.1016/j.bbrc.2019.03.093. [DOI] [PubMed] [Google Scholar]
- 32.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 33.Fu Y., Dominissini D., Rechavi G., He C. Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat. Rev. Genet. 2014;15(5):293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 34.Liao S., Sun H., Xu C. YTH domain: a family of N(6)-methyladenosine (m(6)A) readers. Dev. Reprod. Biol. 2018;16(2):99–107. doi: 10.1016/j.gpb.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pu M., Chen J., Tao Z., et al. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. 2019;76(3):441–451. doi: 10.1007/s00018-018-2940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castelli G., Pelosi E., Testa U. Liver cancer: molecular characterization, clonal evolution and cancer stem cells. Cancers. 2017;9(9) doi: 10.3390/cancers9090127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang D.Y., Sun W.L., Ma X., et al. Up-regulated FSTL5 inhibits invasion of hepatocellular carcinoma through the Wnt/β-catenin/YAP pathway. Int. J. Clin. Exp. Pathol. 2017;10(10):10325–10333. [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y., Peng C., Chen J., et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer. 2019;18(1):127. doi: 10.1186/s12943-019-1053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lan T., Li H., Zhang D., et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer. 2019;18(1):186. doi: 10.1186/s12943-019-1106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen M., Wei L., Law C.T., et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 41.Li D., Cai L., Meng R., Feng Z., Xu Q. METTL3 modulates osteoclast differentiation and function by controlling RNA stability and nuclear export. Int. J. Mol. Sci. 2020;21(5) doi: 10.3390/ijms21051660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X., Lu Z., Gomez A., et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie H., Li J., Ying Y., et al. METTL3/YTHDF2 m(6) A axis promotes tumorigenesis by degrading SETD7 and KLF4 mRNAs in bladder cancer. J. Cell Mol. Med. 2020;24(7):4092–4104. doi: 10.1111/jcmm.15063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 45.Cai X., Wang X., Cao C., et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–19. doi: 10.1016/j.canlet.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 46.Alarcón C.R., Lee H., Goodarzi H., Halberg N., Tavazoie S.F. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng W., Li J., Chen R., et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J. Exp. Clin. Cancer Res. 2019;38(1):393. doi: 10.1186/s13046-019-1408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data associated with this study have been deposited in a publicly available repository. Data are available from the corresponding author upon request.