Abstract
Background
Lumbar disc degeneration (LDD) is an important pathological basis for the development of degenerative diseases of the lumbar spine. Most clinical patients have low back pain as their main symptom. The deterioration of the biomechanical environment is an important cause of LDD. Although there is a large amount of basic research on LDD, there are fewer reports that correlate biomechanical mechanisms with basic research. Our research aims to identify 304 key genes involved in LDD due to biomechanical deterioration, using a bioinformatics approach. We focus on SMAD3, CAV1, SMAD7, TGFB1 as hub genes, and screen for 30 potential target drugs, offering novel insights into LDD pathology and treatment options.
Methods
The Gene Cards, GenCLip3, OMIM and Drugbank databases were explored to obtain genes associated with biomechanics and LDD, followed by making veen plots to obtain both co-expressed genes. GO enrichment analysis and KEGG pathway analysis of the co-expressed genes were obtained using the DAVID online platform and visualised via a free online website. Protein interaction networks (PPI) were obtained through the STRING platform and visualised through Cytoscape 3.9.0. These genes were predicted for downstream interaction networks using the STITCH platform. Then, the GSE56081 dataset was used to validate the key genes. RT-PCR was used to detect mRNA expression of core genes in the degenerated nucleus pulposus (NP) samples and western bolt was used for protein expression. Lastly, the obtained hub genes were searched in the drug database (DGIdb) to find relevant drug candidates.
Results
From the perspective of biomechanics-induced LDD, we obtained a total of 304 genes, the GO functional enrichment and KEGG pathway enrichment analysis showed that the functions of these genes are mostly related to inflammation and apoptosis. The PPI network was constructed and four Hub genes were obtained through the plug-in of Cytoscape software, namely SMAD3, CAV1, SMAD7 and TGFB1. The analysis of key genes revealed that biomechanical involvement in LDD may be related to the TGF-β signaling pathway. Validation of the GSE56081 dataset revealed that SMAD3 and TGFB1 were highly expressed in degenerating NP samples. RT-PCR results showed that the mRNA expression of SMAD3 and TGFB1 was significantly increased in the severe degeneration group; Western blot results also showed that the protein expression of TGFB1 and P-SMAD3 was significantly increased. In addition, we identified 30 potential drugs.
Conclusion
This study presented a new approach to investigate the correlation between biomechanical mechanisms and LDD. The deterioration of the biomechanical environment may cause LDD through the TGF-β signaling pathway. TGFB1 and SMAD3 are important core targets. The important genes, pathways and drugs obtained in this study provided a new basis and direction for the study, diagnosis and treatment of LDD.
Keywords: Bioinformatics, Lumbar disc degeneration, Biomechanics, Genes, Targeted drugs
1. Introduction
Lumbar disc degeneration (LDD) can contribute to a variety of degenerative diseases of the lumbar spine, such as discogenic back pain, lumbar spinal stenosis and lumbar disc herniation. It is a major cause of low back pain and a serious threat to the physical and mental health of patients [1]. It is estimated that around 20% of young people have mild disc degeneration and 80% have experienced back pain during their lifetime [2]. More research has been done on LDD and the results have shown that it has a complex pathogenesis, mainly including biomechanical deterioration, aging, inflammation, apoptosis, nutritional deficiencies and genetics [3]. A strong link exists between the deterioration of the biomechanical environment and LDD. It has been shown that unstable mechanical loading can cause degradation of the extracellular matrix of the nucleus pulposus(NP), ultimately leading to the development of degenerative lumbar spine disease [4].
The structure of the intervertebral disc supports body movement and withstands sustained external mechanical stress. In normal life, the lumbar disc is under pressure and a healthy disc will undergo some deformation when the force is applied to the lumbar spine, when the force disappears it returns to original state. So it is evident that the disc is in a dynamic biomechanical environment over time. When a disc degenerates, the biomechanics of the lumbar spine changes, ultimately affecting the ability of the lumbar spine to transmit and distribute load forces [5]. Nonetheless, excessive mechanical loading can also cause intervertebral disc damage and degeneration [6]. It is evident that LDD and the deterioration of the biomechanical environment is an inferior cyclic process, so it is extremely necessary to pay attention to the mechanisms of biomechanics in the study of disc degeneration (DD).
With the advent of modern information technology, bioinformatics has emerged as a pivotal discipline at the intersection of biology and informatics, revolutionizing our approach to genetic research. It provides an invaluable framework for researchers to mine vast biological databases for pertinent genes, enhancing our understanding and application of these genetic elements [7]. Traditional research on DD often segregates genetic analysis from biomechanical considerations – an approach that fails to consider the synergistic interplay between these two critical factors. This study aims to transcend this traditional dichotomy, leveraging bioinformatics to integrate molecular genetics with biomechanical insights, thereby offering a comprehensive perspective on the pathogenesis of LDD. We specifically focus on unraveling the elusive genetic mechanisms that link biomechanical changes to LDD, employing an integrative methodology that encompasses both bioinformatics and experimental validation. Our goal is to elucidate the intricate molecular interactions underlying LDD, advancing our understanding of its pathophysiology within the context of biomechanical dynamics.
2. Materials and methods
2.1. Data download and pre-processing
Screening of biomechanically relevant targets for LDD in four commonly used human genetic databases (Gene Cards, GenCLip3, OMIM and Drugbank) [[8], [9], [10], [11]]. The Gene Cards database contains a large number of genes, and in order to improve the screening accuracy, only the target genes with the top 500 score values are selected. The search terms were “lumbar disc degeneration”, “degenerative lumbar disc”, “biomechanics” and “biomechanical”. The obtained target information was combined, de-duplicated and corrected.
2.2. Shared targets screening and venn diagramming
Venn diagrams of biomechanics and LDD targets were plotted through the online network (https://bioinfogp.cnb.csic.es/tools/venny/) to identify common genes.
2.3. Shared targets PPI network construction and modular analysis
Import the obtained shared targets into the STRING platform (https://string-db.org/) to build a target PPI network [12]. The PPI network TSV format was downloaded and imported into the Cytoscape 3.9.0 platform for the next step. The molecular complex detection (MCODE) plug-in was used for clustering and the CytoHubba plug-in was used to find hub genes.
2.4. GO functional enrichment analysis and KEGG pathway enrichment analysis
GO enrichment analysis and KEGG pathway analysis were performed using the DAVID online tool (https://david.ncifcrf.gov/) to clarify the functions and possible pathways, and the content was downloaded in text file format. GO analysis was plotted using the Microbiology website (http://www.bioinformatics.com.cn/), which consisted of three main aspects, biological process (BP), molecular function (MF) and cellular component (CC). The KEGG pathway was enriched for shared targets (P < 0.05) and the results are shown as bubble plots. The possible pathways of the shared genes were screened together with the relevant literature. Not only that, STITCH database(http://stitch.embl.de/) was used to demonstrate the interaction network downstream of key genes.
2.5. Drug-gene interactions
Use the Drug-Gene Interaction Database (DGIdb) (http://www.dgidb.org) [13] to screen for target drugs that may treat the hub gene. Screening criteria were: must be approved by the US Food and Drug Administration. Such drugs are listed and demonstrated.
2.6. Validation of the hub genes
To clarify the expression of hub genes, the specific expression of the obtained pivotal genes was validated by the independent datasets GSE56081 in the GEO datebase(https://www.ncbi.nlm.nih.gov/geo/).
3. Experimental validation
3.1. Subjects
The nucleus pulposus specimens (NPS) were obtained by patients from the Department of Orthopaedics at the Affiliated Hospital of Integrative Medicine, Nanjing University of Traditional Chinese Medicine, who suffered from lumbar disc herniation (LDH) and underwent percutaneous endoscope lumbar discectomy (PELD). Patients were admitted from November 2020 to March 2022. A total of 22 patients were included. Two orthopaedic surgeons scored the degree of DD by the Pfirrmann classification method, and after final selection, NPS from 12 patients were used for the experiment. Six patients with mild degeneration (MD) and six patients with severe degeneration (SD) were included, including eight men and four women.
3.2. Quantitative Real-Time PCR
NPS were removed and quickly frozen at −80 °C. 50 mg of each sample was taken and divided into two EP tubes for the MD and SD groups. Total RNA was extracted using the Trizol method and reverse transcribed into cDNA using the ProtoScript II First Strand cDNA Synthesis Kit (BioLabs). Quantitative Real-Time PCR was then performed using Luna Universal qPCR Master Mix (BioLabs). The relevant expression of key genes was calculated by the 2 - ΔΔCt method. The primer sequence parameters can be found in Table S1. The procedure for cellular experiments is the same as above.
3.3. Western blot
The NPS was ground to a powder with liquid nitrogen and then added to RIPA strong lysis solution, lysed on ice for half an hour and then sonically broken. The supernatant was centrifuged and the protein concentration was measured using the BCA kit, loading buffer was added and the protein was heated at 100 °C for 5 min. The proteins were then separated by electrophoresis in 10% SDS-PAGE and subsequently transferred to PVDF membranes, which were closed with 5% skimmed milk for 1 h. The membranes were cut and incubated with primary antibodies (TGF-β1, Smad3, P-Smad3, GAPDH) overnight on a shaker at 4 °C. Then, the membranes were washed 3 times with TBST and incubated for 1 h at room temperature with secondary antibodies, followed by 3 additional washes with TBST. The membranes were then exposed in a fully automated chemiluminescent image analysis system and the proteins were scanned and analysed. The procedure for cellular experiments is the same as above.
3.4. Statistical analysis
All data were analysed by GraphPad Prism 8.0 software and statistics are presented by mean ± standard deviation (‾χ±SD). The experimental data of the two groups were compared using the t-test. When P < 0.05, the difference is indicated as statistically significant.
4. Results
4.1. Search results for total targets
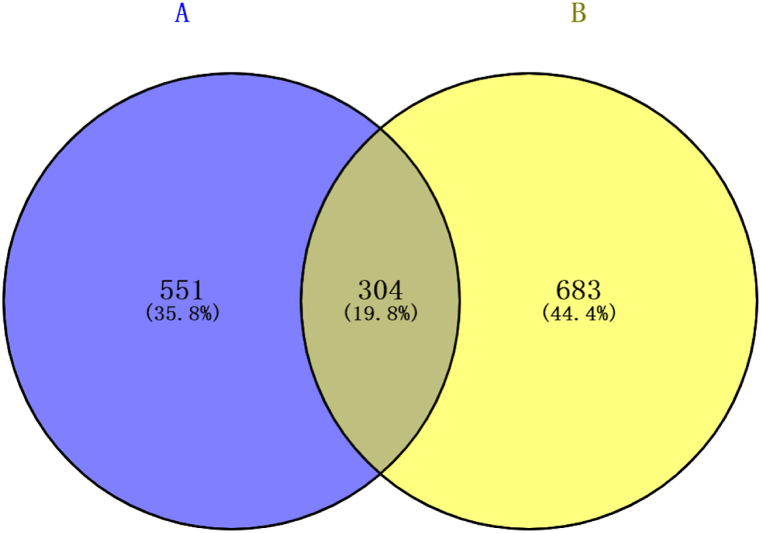
By searching the data for each of the four targets, 855 targets related to LDH and 987 targets related to biomechanics were obtained. A total of 304 targets were found to be associated with LDD and biomechanics by applying venn plots (Fig. 1).
Fig. 1.
Venn map of A (Disc degeneration) and B (Biomechanics) targets. A total of 304 targets are shared between these two groups.
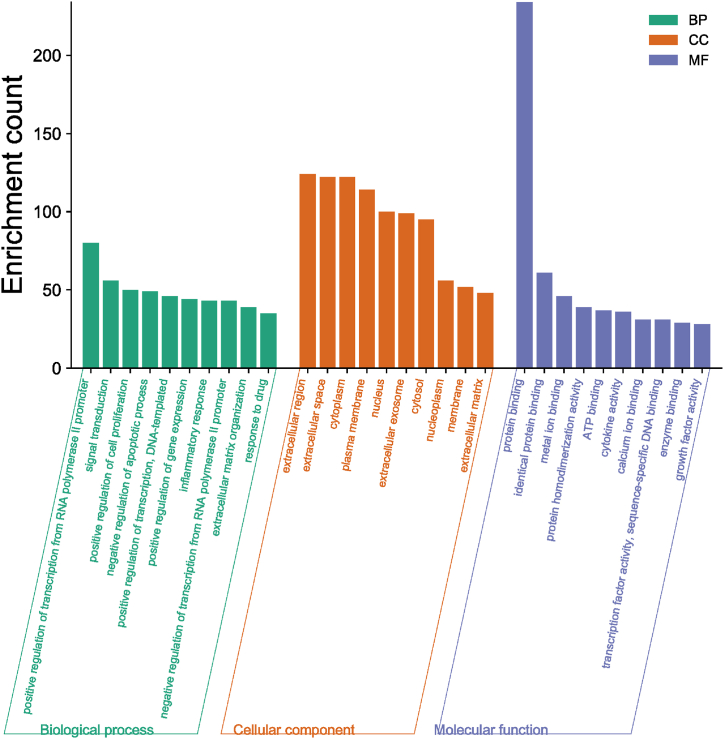
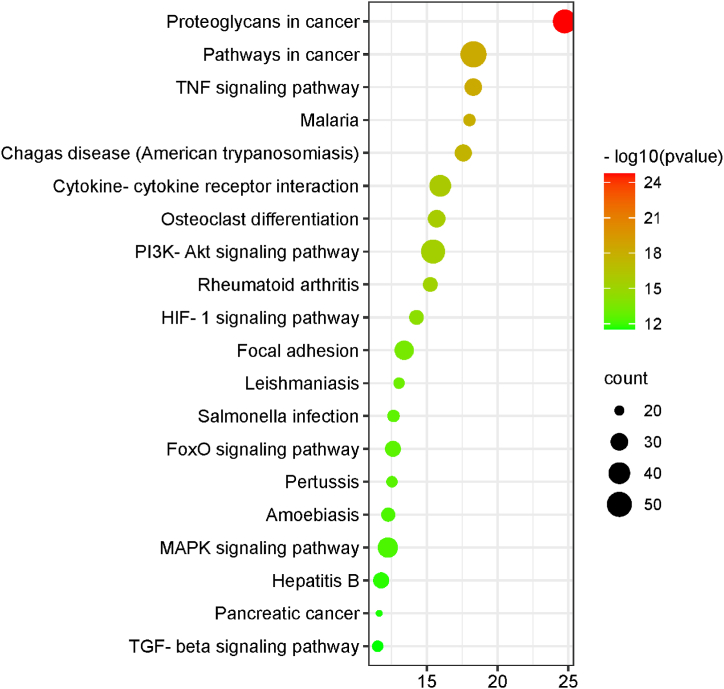
GO enrichment analysis were performed on 304 genes to analyze the functions of genes in regard to BP, CC and MF. In addition, they were enriched for relevant pathways. Visualization of the top 10 GO enrichment results (Fig. 2) showed that in the BP category, the genes were associated with signal transduction, negative regulation of apoptotic process, DNA-templated, positive regulation of gene expression and inflammatory response. In regards to CC, they are mainly associated with extracellular region, extracellular space, cytoplasm, plasma membrane, nucleus, extracellular exosome, cytosol, nucleoplasm and membrane. Concerning MF, this group of genes focuses on protein binding, homologous protein binding, metal ion binding, protein homogenisation activity and ATP binding. In the analysis of the KEGG signaling pathway (Fig. 3), 304 shared genes were associated with TNF signaling pathway, Cytokine-cytokine receptor interaction, Osteoclast differentiation, PI3K-Akt signaling pathway, Rheumatoid arthritis and others.
Fig. 2.
GO Functional enrichment histogram related to Biomechanics and LDD. This represents the top ten GO enrichment results, categorized as BP (Biological Process), CC (Cellular Component), and MF (Molecular Function).
Fig. 3.
Bubble diagram of KEGG pathway enrichment. A total of 304 genes are associated with various pathways including the tumor necrosis factor signaling pathway, cytokine-cytokine receptor interaction, osteoclast differentiation, the PI3K-Akt signaling pathway, and rheumatoid arthritis.
4.2. PPI network and module analysis
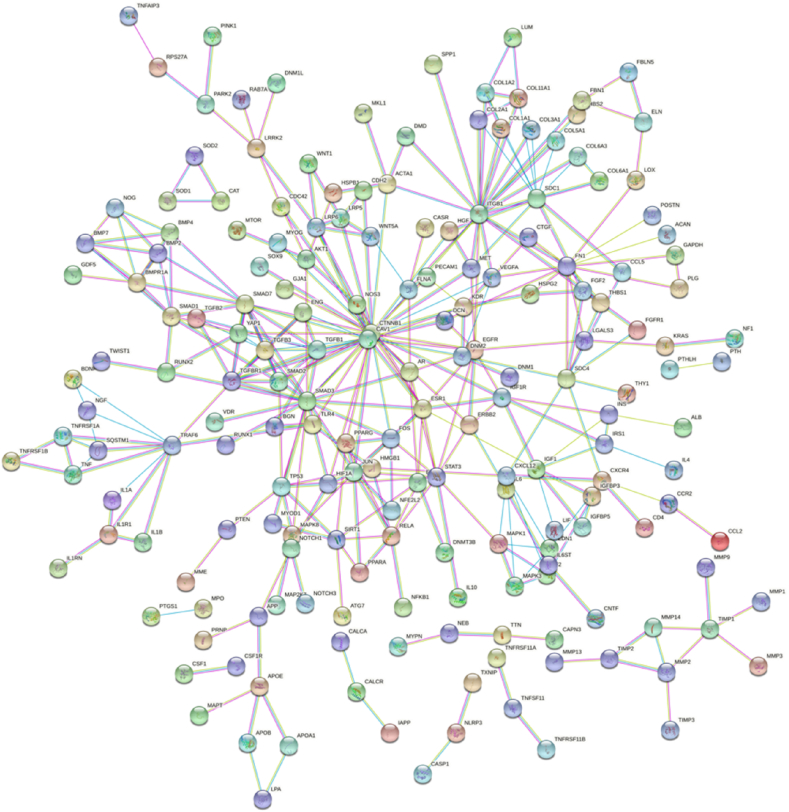
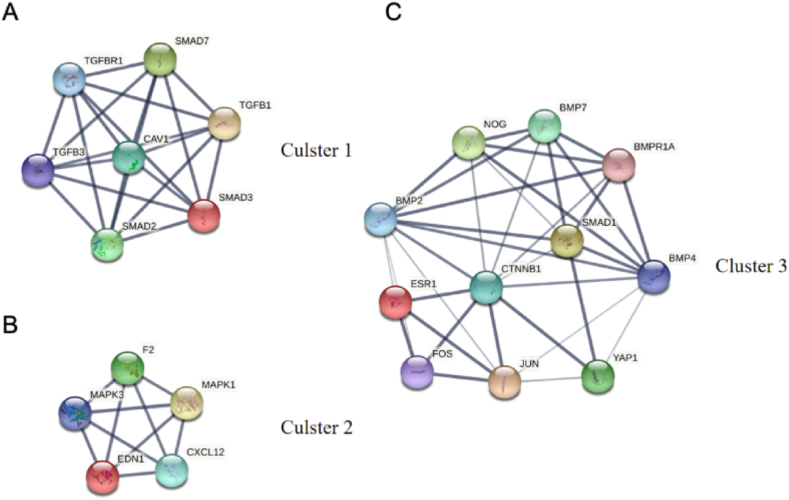
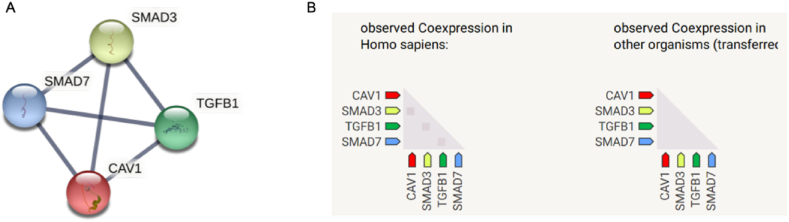
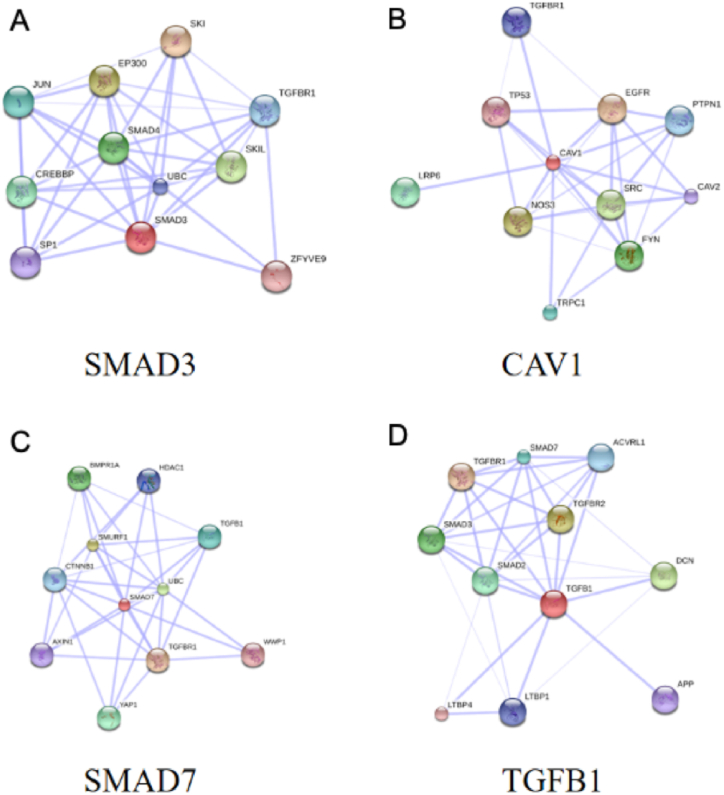
PPI network of the 304 genes obtained from STRING and Cytoscape software (Fig. 4) and the top 3 key modules were selected from MCODE plug-in (Fig. 5A–C). The STRING database were set up with a confidence score >0.9, excluding disconnected nodes from the network. The specific information statistics for the 3 modules are shown in Table 1. The CytoHubba plug-in from Cytoscape software was used, and top 11 core gene were selected (Table 2). The search for common genes from the top 11 ranked genes with those obtained in the top 3 clusters resulted in four hub genes. They were CAV1, SMAD7, TGFB1 and SMAD3 (Fig. 6 A, B). In addition, a downstream interaction network for SMAD3, CAV1, SMAD7 and TGFB1 were generated using the STITCH database (Fig. 7A–D).
Fig. 4.
PPI network from the 304 genes. PPI network of the 304 genes obtained from STRING and Cytoscape software.
Fig. 5.
Network diagram of the 3 clusters. (A) Cluster1 incluede 7 density and 21 nodes; (B) Cluster2 incluede 5 density and 10 nodes; (C) Cluster3 incluede 11 density and 22 nodes.
Table 1.
Key information on the top 3 modules.
| Clusters | Score | Density | Nodes | Nodes IDs |
|---|---|---|---|---|
| 1 | 7 | 7 | 21 | SMAD7, SMAD3, TGFB3, TGFB1, SMAD2, CAV1, TGFBR1 |
| 2 | 5 | 5 | 10 | MAPK1, EDN1, CXCL12, MAPK3, F2 |
| 3 | 4.4 | 11 | 22 | CTNNB1, SMAD1, ESR1, BMP4, JUN, BMPR1A, BMP2, FOS, YAP1, NOG, BMP7 |
Table 2.
Top 11 core targets.
| Target name | Score | Target name | Score |
|---|---|---|---|
| CTNNB1 | 26 | SDC1 | 12 |
| ITGB1 | 20 | SMAD7 | 12 |
| SMAD3 | 19 | ESR1 | 11 |
| CAV1 | 15 | EGFR | 11 |
| FN1 | 13 | TGFB1 | 11 |
| JUN | 12 | – | – |
Fig. 6.
Ranked display of the top 11 core genes. (A) Construction of network among 4 hub genes; (B) Coexpression analysis of 4 hub genes.
Fig. 7.
Targetable four hub genes (SMAD3, CAV1, SMAD7 and TGFB1) subnetworks. (A) Downstream interaction network for SMAD3; (B) Downstream interaction network for CAV1; (C) Downstream interaction network for SMAD7; (D) Downstream interaction network for TGFB1.
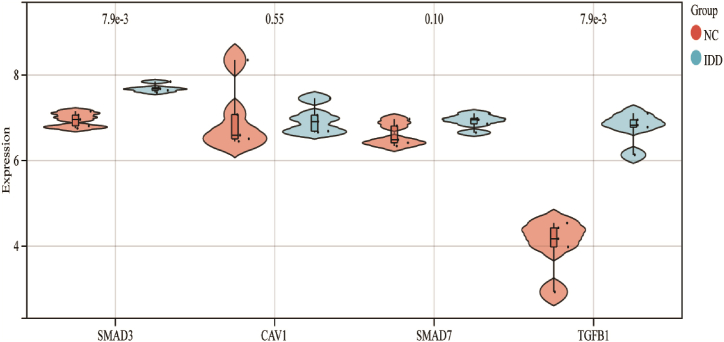
4.3. Verification of the hub genes
To verify the validity of the candidate genes, we searched the GEO database and used the external dataset GSE56081 to verify the differential expression between the two groups. The results revealed that both TGFB1 and SMAD3 were significantly altered in the degenerated group compared to the controls, with statistically significant differences. CAV1 and SMAD7 did not show significant differences. Finally, TGFB1 and SMAD3 were screened as important targets for intervertebral disc degeneration (IDD) (Fig. 8).
Fig. 8.
Validation violin map of hub genes in GSE56081. CAV1 and SMAD7 exhibit no significant differences between the NC (Normal Control) and IDD (Intervertebral Disc Degeneration) groups. TGFB1 and SMAD3 have been identified as crucial targets for IDD.
4.4. Expression of TGFB1 and SMAD3 mRNA in NPS
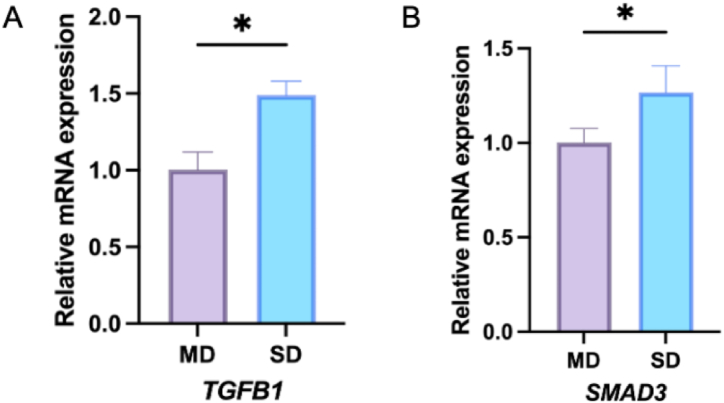
We examined TGFB1 and Smad3 mRNA in clinical NPS and showed that TGFB1 and SMAD3 mRNA was significantly increased in the SD group, with statistically significant differences compared to the MD group (Fig. 9 A, B).
Fig. 9.
Relative mRNA expressions of TGFB1 and SMAD3 in NPS. (A) The mRNA expression of TGFB1 in mild degeneration (MD) and severe degeneration (SD); (B) The mRNA expression of SMAD3 in MD and SD. *p < 0.05 (compared with MD).
4.5. Protein expression of the TGF-β signaling pathway in NPS
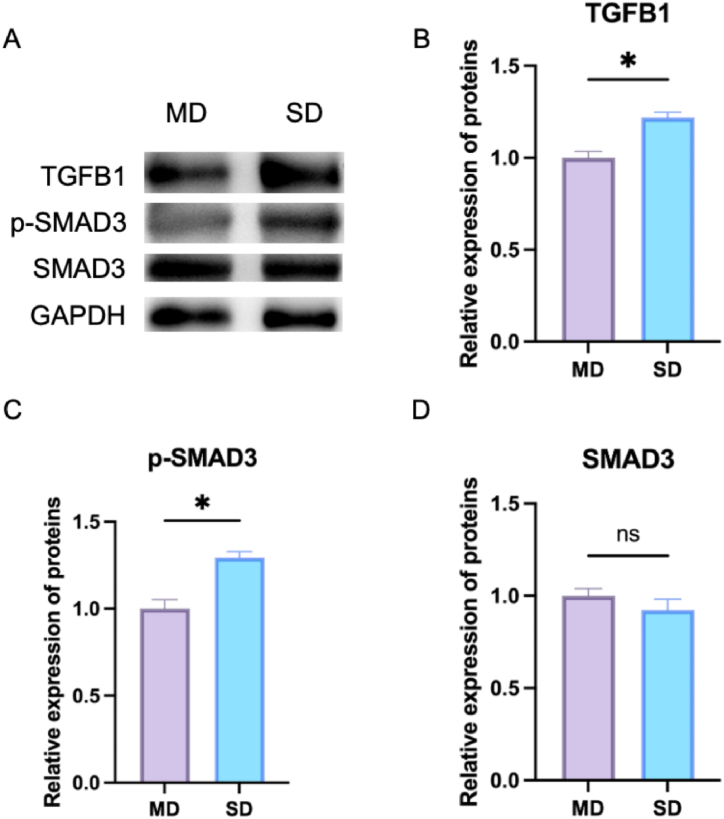
To further validate the importance of the TGFB signaling pathway in IDD, we examined the expression of several important proteins, including TGFB1, SMAD3 and p-SMAD3 (Fig. 10 A). The results revealed that the expression of TGFB1 and p-SMAD3 was significantly higher in the SD NPS compared to the MD group (Fig. 10B–D).
Fig. 10.
Protein expression of TGFB1 and P-SMAD3. (A) Western blot was employed to measure the relative expressions of TGFB1, p-SMAD3 and SMAD3 in MD and SD; (B) The relative expression of TGFB1; (C) The relative expression of p-SMAD3; (D) The relative expression of SMAD3. *p < 0.05 (compared with MD).
4.6. Drug-gene interaction
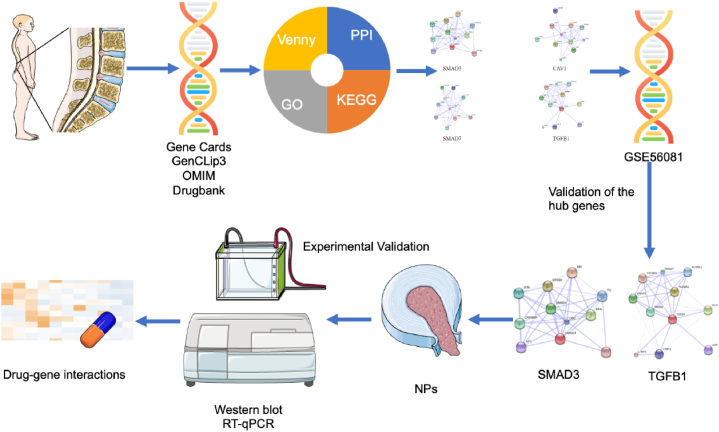
TGFB1 and SMAD3 were screened as possible drug targets and finally a total of 30 available drugs were obtained through the DGIdb database (Table 3). A total of 20 drugs are available for the treatment of TGFB1 target, and last 10 drugs are used for SMAD3 target. Additionally, a schematic diagram of the overall drug screening process is presented (Fig. 11).
Table 3.
List of approved drugs for TGFB1 and SMAD3.
| Drug | Sources | PMIDs | Query Score | Interaction Score |
|---|---|---|---|---|
| RAMIPRIL | NCI | 15716710 | 2.19 | 0.86 |
| PIRFENIDONE | NCI TTD | 12907346 | 1.88 | 0.74 |
| AMIFOSTINE | NCI | 12005544 | 1.75 | 0.69 |
| TOREMIFENE | NCI | 12476040 | 1.25 | 0.49 |
| RITUXIMAB | PharmGKB | 22129793 | 0.63 | 0.25 |
| CLADRIBINE | NCI | 12097998 | 0.58 | 0.23 |
| PIOGLITAZONE | NCI | 17407709 | 0.55 | 0.22 |
| TESTOSTERONE | NCI | 9288184 | 0.49 | 0.1 |
| TRIAMCINOLONE | NCI | 12174062 | 0.38 | 0.15 |
| MELATONIN | NCI | 9649124 | 0.32 | 0.13 |
| ATENOLOL | NCI | 15049387 | 0.26 | 0.1 |
| HYDROCORTISONE | NCI | 7895901 | 0.23 | 0.09 |
| STREPTOZOCIN | NCI | 17968528 | 0.19 | 0.07 |
| NICOTINE | NCI | 10198208 | 0.18 | 0.07 |
| IRINOTECAN | PharmGKB | 27160286 | 0.17 | 0.07 |
| ETOPOSIDE | NCI | 16002781 | 0.17 | 0.07 |
| TAMOXIFEN | NCI | 8019941 | 0.15 | 0.06 |
| ASPIRIN | PharmGKB | 19138248 | 0.12 | 0.05 |
| VERAPAMIL | NCI | None found | 0.11 | 0.04 |
| DOXORUBICIN | NCI | 12956904 | 0.09 | 0.04 |
| CETYLPYRIDINIUM | DTC | None found | 4.38 | 0.28 |
| CHLORQUINALDOL | DTC | None found | 1.1 | 0.07 |
| TRICLOSAN | DTC | None found | 0.73 | 0.05 |
| FLUORESCEIN | TTD | None found | 0.63 | 0.04 |
| EPIRUBICIN HYDROCHLORIDE | DTC | None found | 0.63 | 0.04 |
| TRICLOCARBAN | DTC | None found | 0.63 | 0.04 |
| LANATOSIDE C | DTC | None found | 0.63 | 0.04 |
| LOPINAVIR | DTC | None found | 0.49 | 0.03 |
| OUABAIN | DTC | None found | 0.49 | 0.03 |
| PYRITHIONE | DTC | None found | 0.4 | 0.03 |
Fig. 11.
Overall experiment flowchart. This is a summary of the entire experiment, including a simple flowchart from data collection to analysis to validation.
5. Discussion
LDD is widespread in orthopaedics, especially in spinal surgery, where pain and numbness are the main clinical manifestations, seriously affecting the physical and mental health and quality of life of middle-aged and elderly patients. It is an important pathological basis for the development of lumbar degenerative diseases [14]. Numerous studies have shown that biomechanical deterioration is an important cause of LDD [[15], [16], [17], [18]] [[15], [16], [17], [18]] [[15], [16], [17], [18]]. In normal conditions, the lumbar spine is a dynamic process and biomechanics plays an important role in maintaining normal lumbar spine function, but when changes in the biomechanics of the lumbar spine are caused by various triggers, this state can lead to DD after a period of time. Worse still, the degeneration of the lumbar discs can in turn exacerbate the biomechanical deterioration, and which is an irreversible and vicious process [19].
Despite the extensive literature demonstrating the biomechanical correlation between LDD, the specific underlying mechanisms by which deterioration in the biomechanical environment causes LDD remain unknown. The focus of this research is to explore the common target genes between biomechanics and LDD through a bioinformatics approach and to seek possible therapeutic targets and therapeutic agents. In this article, we used software combined with database to predict the common targets of LDD and biomechanics, construct PPI network diagram of targets, GO enrichment analysis histogram, KEGG pathway enrichment analysis bubble diagram, hub genes downstream network diagram, predict the possible drugs of therapeutic targets through the database, and then systematically refine the specific mechanism of the connection between the both. The results were then validated by means of a dataset from the GEO database. RT-PCR and Western bolt are used to detect mRNA and protein expression of key genes in degenerating NP tissue.
GO functional enrichment analysis of 304 related genes revealed that they have multiple biological functions in terms of BP, MF and CC. BP were mostly enriched for positive regulation of transcription from RNA polymerase II promoter, signal transduction, positive regulation of cell proliferation, negative regulation of apoptotic process, and so on. It can be seen that the deterioration of biomechanics may cause LDD through the regulation of transcription and the promotion of apoptosis. MF is enriched in protein binding, identical protein binding, metal ion binding, protein homodimerization activity, ATP binding, and others. It is clear that from a molecular point of view both may be related to factors, such as proteins, metal ions, and growth factors. In summary, from the point of view of CC, the shared targets are mostly enriched in the extracellular region, extracellular space, cytoplasm, plasma membrane, nucleus. It is apparent that the mechanisms of biomechanically induced LDD are rich and varied, and can act on multiple targets.
A total of 112 pathways were identified by KEGG enrichment analysis of the common targets, and the top 10 pathways were mainly Proteoglycans in cancer, Pathways in cancer 54, TNF signaling pathway, Malaria, Chagas disease (American trypanosomiasis), Cytokine-cytokine receptor interaction, Osteoclast differentiation, PI3K-Akt signaling pathway, Rheumatoid arthritis and HIF-1 signaling pathway. Considering the pathogenesis of LDD and biomechanical mechanisms, combined with the top ten pathways of the disease, the occurrence of biomechanically induced LDD may be related to inflammation, hypoxia, osteoclast differentiation, apoptosis and other factors.
The PPI network construction and network topology resulted in four important gene targets, including SMAD3, CAV1, SMAD7 and TGFB1. We then cited the GSE56081 dataset from the GEO database to validate the obtained genes and found that SMAD3 and TGFB1 were highly expressed in disc degeneration.
SMAD3 is an important target in the study of biomechanical properties [20]. It has been genetically demonstrated that SMAD3 is an important genetic target for the development of degenerative spinal disease and that the differences in SMAD3 gene are statistically significant in normal subjects compared to patients with DD [21]. It can be hypothesized that the deterioration of the biomechanical environment of the lumbar spine may cause LDD by acting on SMAD3 and thus forming a number of responses.
TGFB1 encodes transcriptional growth factor beta, which leads to the recruitment and activation of SMAD family transcription factors, thereby regulating gene expression [22]. This gene also regulates cell proliferation, differentiation and growth, as well as the expression and activation of other growth factors [23]. TGFB1 protein alterations can be detected in the presence of abnormal biomechanical function [24]. TGFB1 has been repeatedly shown to be a central target of IDD using bioinformatics [25,26]. In this study, the two hub genes were found to be enriched in the TGF-β signaling pathway and play an important role in this route. It can be inferred that biomechanically induced LDD may be related to the regulation of the TGF-β signaling pathway.
TGFβ (transforming growth factor beta) is a group of growth factors in the TGF superfamily and is involved in the functional activity of many cells [27]. Studies have shown that TGF-β can repair degenerated discs by promoting matrix synthesis, inhibiting matrix catabolism, suppressing cell loss and inflammatory effects [28]. However, more TGF-β expression is not better, and excessive activation of TGF-β instead accelerates DD. High levels of TGF-β expression can be observed in severe IDD in mice and humans [27,29]. The two important genes and pathways identified in this article are closely related to biomechanics and LDD, and their study will help identify specific functions and explore the mechanisms of biomechanically induced LDD. But the way in which biomechanical factors induce altered TGF-β signaling and then accelerate DD is the question to be discussed. Previous studies have shown that the TGF-β signaling pathway is closely related to the inflammatory response in IDD [30,31]. TGF-β inhibits inflammatory cytokine-induced catabolic processes in IVDD by following down-regulation of MMPs expression [32]. In addition, TGFB1 inhibits TNF-α-induced apoptosis of NPCs, effectively delaying inflammation-mediated DD [31]. DD is a complex process that involves multiple mechanisms. From the results of this study, it can be deduced that deterioration of the biomechanical environment can cause an inflammatory response within the disc via the TGF-β signaling pathway and thus accelerate it.
Our analysis, especially on key genes like SMAD3 and TGFB1, underscores the potential role of the TGF-β signaling pathway in LDD. This finding aligns with studies showing high levels of TGF-β expression in severe intervertebral disc degeneration (IDD). Moreover, the TGF-β signaling pathway has been linked to the inflammatory response in IDD, suggesting a complex interplay between biomechanical stress and inflammatory pathways.
Recent research further supports our findings. A study by Cai [33] demonstrated the biomechanical effects of L4∼L5 lumbar degeneration using numerical simulations, highlighting the impact of biomechanical changes on adjacent segments and overall lumbar spine function. This aligns with our observation that biomechanical deterioration can have far-reaching effects on lumbar spine health, extending beyond the degenerated segment.
While our study provides new insights, it also acknowledges limitations, such as the lack of biomechanical degeneration models, the complexity of mechanisms involved in biomechanics-induced LDD, and notably, the constrained number of clinical samples due to the pandemic. This limitation may have impacted our results and represents a crucial area for future research and improvement.
In conclusion, our study offers a novel perspective on the interplay between biomechanics and LDD, highlighting the TGF-β signaling pathway's potential role. This research contributes significantly to understanding LDD's etiology and opens avenues for targeted therapeutic strategies. Addressing the challenges in sample collection and expanding our dataset will be a key focus in our continued efforts to elucidate this complex condition.
6. Conclusion
In conclusion, our research offers novel insights into the biomechanical underpinnings of LDD, a critical concern in spinal health. By elucidating the role of key genes, particularly SMAD3 and TGFB1, within the TGF-β signaling pathway, this study bridges a crucial gap in understanding the molecular basis of biomechanically induced LDD. While providing a foundation for targeted therapeutic development, we acknowledge certain limitations, such as the lack of biomechanical models, underscoring the need for further comprehensive investigations. This work not only advances our understanding of LDD but also proposes a new paradigm for exploring the interplay between biomechanics and gene regulation in spinal pathologies.
Funding
The current research was funded by Jiangsu Provincial Traditional Chinese Medicine Science and Technology Development Plan Project (2020 ZD202008), Natural Science Foundation of Jiangsu Province (BK 20221420), Science and technology projects in Jiangsu Province (2019 BE2019765), Project of National Clinical Research Base of Traditional Chinese Medicine in Jiangsu Province (JD2022 SZXMS07) and Postgraduate Research & Practice Innovation Program of Jiangsu Province (SJCX22_0854).
Ethical approval
This study was approved by the Ethics Committee of the Affiliated Hospital of Integrated Traditional Chinese and Western Medicine, Nanjing University of Chinese Medicine, ethics number 2022-LWKYZ-034.
Data availability statement
We can provide the required data on request. The datasets analysed during the current study are available in the Gene Expression Omnibus (GEO) repository, GSE56081.
CRediT authorship contribution statement
Xiyu Liu: Writing – review & editing, Writing – original draft, Supervision, Software, Resources, Formal analysis, Data curation. Lipeng He: Writing – review & editing, Writing – original draft. Nan Wang: Software, Formal analysis. Lin Xie: Funding acquisition, Formal analysis, Conceptualization. Bin Wu: Project administration, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are particularly grateful to Dr Kang Ran for his guidance and assistance with this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e27016.
Contributor Information
Nan Wang, Email: wangnan082020@163.com.
Lin Xie, Email: xielin@njucm.edu.cn.
Bin Wu, Email: 986213812@qq.com.
Abbreviations
- LDD
Lumbar disc degeneration
- DD
Disc degeneration
- GO
Gene Ontology
- BP
Biological processes
- CC
Cellular components
- MF
Molecular function
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- STRING
Search Tool for the Retrieval of Interacting Genes
- MCODE
Molecular complex detection
- DGIdb
Drug-Gene Interaction Database
- NP
nucleus pulposus
- NPCs
nucleus pulposus cells
- NPS
Nucleus pulposus specimens
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Risbud M.V., Shapiro I.M. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat. Rev. Rheumatol. 2014;10:44–56. doi: 10.1038/nrrheum.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker B.F. The prevalence of low back pain: a systematic review of the literature from 1966 to 1998. J. Spinal Disord. 2000;13:205–217. doi: 10.1097/00002517-200006000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Clouet J., Fusellier M., Camus A., Le Visage C., Guicheux J. Intervertebral disc regeneration: from cell therapy to the development of novel bioinspired endogenous repair strategies. Adv. Drug Deliv. Rev. 2019;146:306–324. doi: 10.1016/j.addr.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 4.Bian Q., Ma L., Jain A., Crane J.L., Kebaish K., Wan M., Zhang Z., Edward Guo X., Sponseller P.D., Séguin C.A., Riley L.H., Wang Y., Cao X. Mechanosignaling activation of TGFβ maintains intervertebral disc homeostasis. Bone Res. 2017;5 doi: 10.1038/boneres.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams M.A., Roughley P.J. What is intervertebral disc degeneration, and what causes it? Spine. 2006;31:2151–2161. doi: 10.1097/01.brs.0000231761.73859.2c. [DOI] [PubMed] [Google Scholar]
- 6.Adams M.A., Lama P., Zehra U., Dolan P. Why do some intervertebral discs degenerate, when others (in the same spine) do not?: why Do Some Intervertebral Discs Degenerate? Clin. Anat. 2015;28:195–204. doi: 10.1002/ca.22404. [DOI] [PubMed] [Google Scholar]
- 7.Sufyan M., Ali Ashfaq U., Ahmad S., Noor F., Hamzah Saleem M., Farhan Aslam M., El-Serehy H.A., Aslam S. Identifying key genes and screening therapeutic agents associated with diabetes mellitus and HCV-related hepatocellular carcinoma by bioinformatics analysis. Saudi J. Biol. Sci. 2021;28:5518–5525. doi: 10.1016/j.sjbs.2021.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y., Kaplan S., Dahary D., Warshawsky D., Guan‐Golan Y., Kohn A., Rappaport N., Safran M., Lancet D. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinforma. 2016;54 doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 9.Wang J.-H., Zhao L.-F., Wang H.-F., Wen Y.-T., Jiang K.-K., Mao X.-M., Zhou Z.-Y., Yao K.-T., Geng Q.-S., Guo D., Huang Z.-X. GenCLiP 3: mining human genes' functions and regulatory networks from PubMed based on co-occurrences and natural language processing. Bioinformatics. 2020;36:1973–1975. doi: 10.1093/bioinformatics/btz807. [DOI] [PubMed] [Google Scholar]
- 10.Amberger J.S., Hamosh A. Searching online mendelian inheritance in man (OMIM): a knowledgebase of human genes and genetic phenotypes. Curr. Protoc. Bioinforma. 2017;58 doi: 10.1002/cpbi.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N., Iynkkaran I., Liu Y., Maciejewski A., Gale N., Wilson A., Chin L., Cummings R., Le D., Pon A., Knox C., Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., von Mering C. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner A.H., Coffman A.C., Ainscough B.J., Spies N.C., Skidmore Z.L., Campbell K.M., Krysiak K., Pan D., McMichael J.F., Eldred J.M., Walker J.R., Wilson R.K., Mardis E.R., Griffith M., Griffith O.L. DGIdb 2.0: mining clinically relevant drug–gene interactions. Nucleic Acids Res. 2016;44:D1036–D1044. doi: 10.1093/nar/gkv1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanna R.M., Shetty A.P., Rajasekaran S. Patterns of lumbar disc degeneration are different in degenerative disc disease and disc prolapse magnetic resonance imaging analysis of 224 patients. Spine J. 2014;14:300–307. doi: 10.1016/j.spinee.2013.10.042. [DOI] [PubMed] [Google Scholar]
- 15.Von Forell G.A., Stephens T.K., Samartzis D., Bowden A.E. Low back pain: a biomechanical rationale based on “patterns” of disc degeneration. Spine. 2015;40:1165–1172. doi: 10.1097/BRS.0000000000000982. [DOI] [PubMed] [Google Scholar]
- 16.Sengupta D.K., Fan H. The basis of mechanical instability in degenerative disc disease: a cadaveric study of abnormal motion versus load distribution. Spine. 2014;39:1032–1043. doi: 10.1097/BRS.0000000000000292. [DOI] [PubMed] [Google Scholar]
- 17.Cornaz F., Widmer J., Farshad-Amacker N.A., Spirig J.M., Snedeker J.G., Farshad M. Intervertebral disc degeneration relates to biomechanical changes of spinal ligaments. Spine J. 2021;21:1399–1407. doi: 10.1016/j.spinee.2021.04.016. [DOI] [PubMed] [Google Scholar]
- 18.Di Pauli Von Treuheim T., Torre O.M., Ferreri E.D., Nasser P., Abbondandolo A., Delgado Caceres M., Lin D., Docheva D., Iatridis J.C. Tenomodulin and chondromodulin-1 are both required to maintain biomechanical function and prevent intervertebral disc degeneration. CARTILAGE. 2021;13:604S–614S. doi: 10.1177/19476035211029696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornaz F., Widmer J., Farshad-Amacker N.A., Spirig J.M., Snedeker J.G., Farshad M. Biomechanical contributions of spinal structures with different degrees of disc degeneration. Spine. 2021;46:E869–E877. doi: 10.1097/BRS.0000000000003883. [DOI] [PubMed] [Google Scholar]
- 20.Zhang K., Asai S., Hast M.W., Liu M., Usami Y., Iwamoto M., Soslowsky L.J., Enomoto-Iwamoto M. Tendon mineralization is progressive and associated with deterioration of tendon biomechanical properties, and requires BMP-Smad signaling in the mouse Achilles tendon injury model. Matrix Biol. 2016;52–54:315–324. doi: 10.1016/j.matbio.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liva E., Panagiotou I., Palikyras S., Parpa E., Tsilika E., Paschou P., Mystakidou K. Candidate gene investigation of spinal degenerative osteoarthritis in Greek population. Spine J. 2017;17:1881–1888. doi: 10.1016/j.spinee.2017.06.025. [DOI] [PubMed] [Google Scholar]
- 22.Radwan-Oczko M., Boratyńska M., Ziętek M., Zołędziewska M., Jonkisz A. The relationship of transforming growth factor-β1 gene polymorphism, its plasma level, and gingival overgrowth in renal transplant recipients receiving different immunosuppressive regimens. J. Periodontol. 2006;77:865–873. doi: 10.1902/jop.2006.050086. [DOI] [PubMed] [Google Scholar]
- 23.Liu K., Liu X., Gu S., Sun Q., Wang Y., Meng J., Xu Z. Association between TGFB1 genetic polymorphisms and chronic allograft dysfunction: a systematic review and meta-analysis. Oncotarget. 2017;8:62463–62469. doi: 10.18632/oncotarget.19516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angel P.M., Narmoneva D.A., Sewell-Loftin M.K., Munjal C., Dupuis L., Landis B.J., Jegga A., Kern C.B., Merryman W.D., Baldwin H.S., Bressan G.M., Hinton R.B. Proteomic alterations associated with biomechanical dysfunction are early processes in the Emilin1 deficient mouse model of aortic valve disease. Ann. Biomed. Eng. 2017;45:2548–2562. doi: 10.1007/s10439-017-1899-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu S., Fu Y., Yan B., Shen Z., Lan T. Analysis of key genes and pathways associated with the pathogenesis of intervertebral disc degeneration. J. Orthop. Surg. 2020;15:371. doi: 10.1186/s13018-020-01902-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J., Zhang X., Gao W., Hu H., Wang X., Hao D. lncRNA/circRNA-miRNA-mRNA ceRNA network in lumbar intervertebral disc degeneration. Mol. Med. Rep. 2019 doi: 10.3892/mmr.2019.10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S., Liu S., Ma K., Zhao L., Lin H., Shao Z. TGF-β signaling in intervertebral disc health and disease. Osteoarthritis Cartilage. 2019;27:1109–1117. doi: 10.1016/j.joca.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Yokozeki Y., Uchida K., Kawakubo A., Nakawaki M., Okubo T., Miyagi M., Inoue G., Itakura M., Sekiguchi H., Takaso M. TGF-β regulates nerve growth factor expression in a mouse intervertebral disc injury model. BMC Muscoskel. Disord. 2021;22:634. doi: 10.1186/s12891-021-04509-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakawaki M., Uchida K., Miyagi M., Inoue G., Kawakubo A., Satoh M., Takaso M. Changes in nerve growth factor expression and macrophage phenotype following intervertebral disc injury in mice. J. Orthop. Res. 2019;37:1798–1804. doi: 10.1002/jor.24308. [DOI] [PubMed] [Google Scholar]
- 30.Cui L., Wei H., Li Z.-M., Dong X.-B., Wang P.-Y. TGF-β1 aggravates degenerative nucleus pulposus cells inflammation and fibrosis through the upregulation of angiopoietin-like protein 2 expression. Eur. Rev. Med. Pharmacol. Sci. 2020;24:12025–12033. doi: 10.26355/eurrev_202012_23991. [DOI] [PubMed] [Google Scholar]
- 31.Xie J., Li B., Yao B., Zhang P., Wang L., Lu H., Song X. Transforming growth factor-β1-regulated Fas/FasL pathway activation suppresses nucleus pulposus cell apoptosis in an inflammatory environment. Biosci. Rep. 2020;40 doi: 10.1042/BSR20191726. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Yang H., Gao F., Li X., Wang J., Liu H., Zheng Z. TGF- β 1 antagonizes TNF- α induced up-regulation of matrix metalloproteinase 3 in nucleus pulposus cells: role of the ERK1/2 pathway, Connect. Tissue Res. 2015;56:461–468. doi: 10.3109/03008207.2015.1054030. [DOI] [PubMed] [Google Scholar]
- 33.Cai X., Sun M., Huang Y., Liu Z., Liu C., Du C., Yang Q. Biomechanical effect of l4–l5 intervertebral disc degeneration on the lower lumbar spine: a finite element study. Orthop. Surg. 2020;12:917–930. doi: 10.1111/os.12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We can provide the required data on request. The datasets analysed during the current study are available in the Gene Expression Omnibus (GEO) repository, GSE56081.