Abstract
Castor (Ricinus communis L.) is an industrially important oil producing crop belongs to Euphorbiaceae family. Castor oil has unique chemical properties make it industrially important crop. It is a member of monotypic genus even though it has ample amount of variability. Using this variability, conventionally many varieties and hybrids have been developed. But, like other crops, the modern and unconventional methods of crop improvement has not fully explored in castor. This article discusses the use of polyploidy induction, distant/wide hybridization and mutation breeding as tools for generating variety. Modern approaches accelerate the speed of crop breeding as an alternative tool. To achieve this goal, molecular markers are employed in breeding to capture the genetic variability through molecular analysis and population structuring. Allele mining is used to trace the evolution of alleles, identify new haplotypes and produce allele specific markers for use in marker aided selection using Genome wide association studies (GWAS) and quantitative trait loci (QTL) mapping. Plant genetic transformation is a rapid and effective mode of castor improvement is also discussed here. The efforts towards developing stable regeneration protocol provide a wide range of utility like embryo rescue in distant crosses, development of somaclonal variation, haploid development using anther culture and callus development for stable genetic transformation has reviewed in this article. Omics has provided intuitions to the molecular mechanisms of (a)biotic stress management in castor along with dissected out the possible genes for improving the yield. Relating genes to traits offers additional scientific inevitability leading to enhancement and sympathetic mechanisms of yield improvement and several stress tolerance.
Keywords: Breeding, Castor, Diversity, Molecular markers, QTL, Tissue culture
Graphical abstract
1. Introduction
Castor (Ricinus communis L., 2n = 2x = 20) is an important non-edible oil crop belongs to the spurge family commonly known as Euphorbiaceae [1]. It is believed to be originated from India or Africa. African origin is most widely accepted because especially the Ethiopian region possesses more castor crop diversity [2]. It is cultivated largely in the world's tropical, subtropical and temperate areas, mainly India, China and Brazil. With a 90% market share, India is the world's largest supplier of castor oil and its byproducts [3]. India produces 15.68 lakh tones of castor bean from the 8.24 lakh hectares growing area with an average productivity of 1902 kg ha−1. In India, Gujarat is a prime castor cultivating state followed by Andhra Pradesh, Telangana, Tamilnadu, Rajasthan and Madhya Pradesh. It has also acquired tiny growing pockets like Orissa, Uttar Pradesh and Karnataka states of India. It is a commercially important crop because of its oil with distinctive chemical properties. Castor oil contains a considerable quantity of the unusual hydroxylated fatty acid “ricinolate” which increases its consumption as a lubricant in power engines [4]. It can readily be dissolved in alcohol and transformed into biodiesel even at low temperatures [5]. It is also used to manufacture soaps, printing inks, linoleum, varnishes and plasticizers. Castor oil is a potent laxative and is also a curing agent for skin problems like sunburn, crinkles and stretch symbols, etc. The by-product of castor oil expeller is the cake which contains vital organic manure with nitrogen (6.4%), P2O5 (2.5%) and K2O (1%) including micronutrients for organic farming. The plant stems are used as firewood and to prepare paper pulp by the paper mills. Beyond this, fresh castor leaves are used to rear silkworms, while desiccated leaves are used as an insecticide in agriculture.
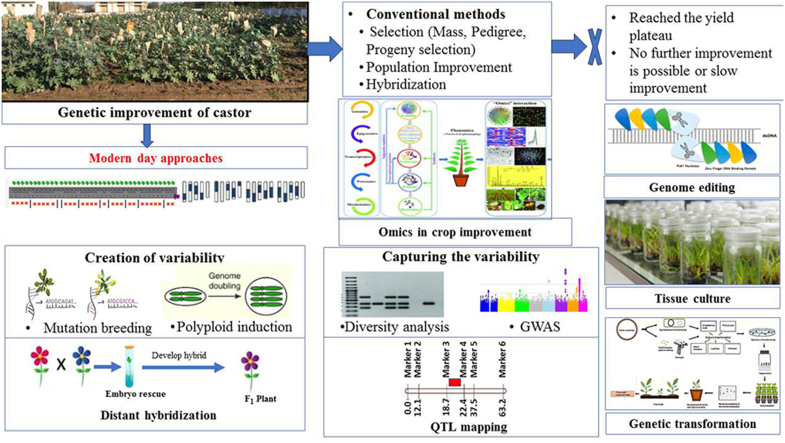
Castor oil is consistently in demand on the international market. But its production is compromised due to several biotic stresses [6]. Semi-looper (Achaea janata), capsule borer (Conogethes punctiferalis), red hairy caterpillar (Amsacta albistriga), jassid (Empoasca flavescens), thrips (Scirtothrips dorsalis) and whitefly (Trialeurodes ricini) are major insects that cause a significant loss to this crop. Similarly, wilt (Fusarium oxysporum f. sp. ricini), root rot (Macrophomina phaseolina), bacterial leaf spot (Xanthomonas campestris pv. Ricinicola), botrytis grey mold (Botrytis ricini) and seedling blight (Phytophthora parasitica) are significant castor diseases. Despite being a monotypic genus, castor exhibits remarkable morphological variability [6]. But the speed of improvement is slow due to less variability available for (a)biotic stresses tolerance and toxic proteins. Castor is a cross-pollinated in nature. Its floral morphology is defined by the arrangement of male and female flowers on the raceme [7]. It is normally monoecious with pistillate flowers on the top portion of the raceme and staminate flowers on the lower. This arrangement is known as regular monoecious. Various categories were proposed by various writers based on the quantity and pattern of male and female flowers arranged on the spike [8]. Castor is a sexually polymorphic species and many sex expression patterns are described [8,9]. It has various sex forms like monoecious, pistillate, interspersed staminate flower (ISF) and sex revertant. Pistillate and ISF sex forms help breeders to produce high-quality hybrid seeds. Until now, the development of various hybrids had resulted in an increase in productivity. Today we have reached to yield plateau in castor and cannot discover new cultivar or recombination which increases the variability regarding stress tolerance of the castor gene pool. Therefore, non-conventional genetic improvement approaches open those doors for creating and selecting elite cultivars through molecular markers, efficient tissue culture protocol, transformation protocols, introgression of genes from extrinsic sources and genome editing. This article reviews the non-conventional approaches to castor improvement and the future scope of research.
2. Genetic improvement through modern day approaches
A significant variation in castor's phenotypic traits has been detected owing to its cross-pollination behavior. Considerable variation has been recorded in quantitative characters in the gene banks of India, Russia and germplasm originated elsewhere. Plant breeders must understand germplasm and its features because germplasm is more than the hundreds of seed bags in the cold room, more than the qualities of the parents of their next variety. But genetic diversity studies of global castor germplasm displayed narrow diversity and demonstrated a lack of geographically grouped genetic populations, irrespective of the molecular marker deployed [10]. The core of castor breeding approaches is selecting better genotype(s) among variants in terms of yield and stress resistance. Generally, castor is improved only through conventional techniques like pedigree method and hybrid development through crossing. But these approaches are mainly restricted due to the monotypic nature of castor as it does not give scope to expand the genetic variability. On the other side, narrow genetic base for various key traits, lack of precision in the selection and time/labour demand limited the scope for improvement through conventional breeding. Moreover, simultaneous improvement for different traits is complex due to the pleiotropic effect and unfavourable linkages between genes.
Various approaches namely mutagenesis, wide hybridization, polyploidization, cis/trans-genesis, genome editing, RNA interference and soma/gameto-clonal variation are now available with plant scientists to create variability in the existing populations of castor. To cope with the problems of time-consumption and genotype by environmental interaction associated with conventional morphological traits/marker-based characterization, different molecular marker systems, especially Simple Sequence Repeats and Single Nucleotide Polymorphism systems are available for castor breeders. DNA marker-based genetic diversity assessment helps to capture an accurate picture of the variability present in global castor germplasm. Association and QTL-mapping help to discover molecular locus controlling the trait of interest. This eventually helps for selecting the superior genotypes within created or existing genetic variability. There are several research priorities in the improvement program designed by market requirements. There is a massive demand for high yielding with low G × E (Genotype × Environment) interactive hybrids which supports mechanical harvesting. Besides, the development of superior genotypes, the focus is required for improving oil quality, ricin free castor, development of horizontal resistance and climate-resilient genotypes. These goals may be achieved by complementing conventional plant breeding practices with modern approaches.
2.1. Creation of variability
2.1.1. Mutation breeding
Mutation breeding is a “modern approach” of crop improvement which provides a way to create different allelic variants of the gene that modulate different phenotypic expressions [11]. Use of mutation breeding started in castor by Kulkarni in 1969 [12] who isolated short duration (matures in <120–150 days) castor mutant through thermal neutron treatment (0.87 × 1013 nh/cm2) with other large numbers of desirable characters. A well-known such mutant is NPH-1-Arun. But systematic mutation breeding program was initiated in 1988 [13] at DOR (Directorate of Oilseed Research, Hyderabad, India) to incorporate wilt resistance in susceptible line VP-1. Lavanya et al. [13] subjected seeds of VP-1 to 45, 55, and 65 Kr gamma radiation at Bhabha Atomic Research Centre, Mumbai and identified a stable pistillate line with wilt resistance in wilt sick plot. They found that progenies of 55 and 65 Kr radiation exhibited marked variation for plant morphology along with wilt resistance. The mutant progenies showed wilt resistance with specific alterations in the plant morphology like short internode, solid stem, slender rachis, cup-shaped leaves, spiny capsules and either with double or triple bloom. In the M13 generation, five pistillate lines namely M 619, M 574, M 584, M 571 and M 568 were identified as a stable pistillate line with long compact spikes. All these mutants were used to develop different hybrids like DCS-591 (M − 574 × DCS 78). It was released in 2006 as an early maturing hybrid having a high yield with resistance to fusarium wilt [14]. Similarly, susceptibility to leaf hopper was also addressed through mutation breeding. This susceptibility had a strong linkage with a monogenic trait i.e. zero bloom [15]. DPC-9 was mutated with gamma rays (550 Gy) and in M5 generation, segregation of bloom was detected from triple (all parts waxy) to zero bloom. Constant selection for bloom in pistillate background fixed nine different mutants had green stem, triple or double bloom, spin on capsules and expression of pistillate till 6th order of spikes. The evaluation for leaf hopper resistance with infester row technique suggested that the mutant genotype IPC-23 was having high resistance for leaf hopper (grade 0) than checks (DCH-177 and DPC-9). This mutant also had high resistance to wilt (8.5%) than check (JI-35; 97%) in a wilt sick plot [15].
2.1.2. Distant hybridization through embryo rescue technique
Distant hybridization provides a novel way to create new gene combinations and opens multiple doors to widen the genetic background of castor. Euphorbiaceae family consist of several species; Jatropha curcas is one of the potential genera to develop cross with castor, which remain unexploited due to lack of compatibility in diverse castor-jatropha combinations [16]. The reason behind this incompatibility is not well understood but systematic cross-incompatibility of jatropha (as female) with castor (as male) was demonstrated by Reddy et al. [17]. Most inter-specific and inter-generic crosses were unsuccessful because of pollen-pistil incompatibility [[17], [18], [19], [20]]. Laosatit et al. [21] attempted inter-generic crosses between Jatropha spp. and Ricinus for identification of pre- or post-fertilization hindrances. They examined the behavior of pollen tube inside the styles and observed that limited numbers of castor pollens were compatible with the jatropha pistils with normal pollen tube growth. It was also observed within 60 min after pollination where pollen tube touched the style base. Based on this phenomenon, Laosatit et al. [21] concluded a post-fertilization barrier between jatropha and castor. They also suggested that fertilized ovules must be rescued and in vitro culturing should be performed to get the fertilized embryos.
To develop the intergeneric hybrid between castor and jatropha, Premjet et al. [22] treated female parent with a 0.1% calcium-boron solution. Calcium-boron solution stimulates premature pollen germination and enhances the pollen tube growth as such compounds improve cell division during the course of fertilization [23]. This treatment was effective as it displayed the highest seed setting with 100% seedling survival. They excised the young embryo from 40 days old F1 seeds, sterilized and cultured them on regeneration medium (pH 5.7) including MS media [24] augmented with citric acid (30 ppm), 6-benzyl aminopurine (1 ppm), indole-3-butyric acid (0.250 ppm), polyvinyl pyrrolidone (500 ppm), sucrose (3% w/v), kinetin (0.500 ppm) and agar (0.7% w/v). These chemicals supported the conversion of an embryo to a whole plant and the truthfulness of these intergeneric hybrids was validated through RAPD marker OPA-07. OPA-07 amplified specific band of 300 and 400 bp in jatropha. whereas specific band of 300, 400, 800, 900 and 1200 bp in castor. Similarly, their F1 showed clear and specific bands of both the parents. Premjet et al. [22] succeed because J. integerrima and castor are more closely related than other Jatropha species.
2.1.3. Polyploidy induction
Naturally, castor is a diploid (2n = 20) in origin [25], but it is reported to behave as a secondary balanced autopolyploid having basic chromosome number x = 5 [4]. Chromosome segments have undergone significant differentiation and diversification during evolution and leads to a regular bivalent formation [26]. The primary gene pool of castor, consist moderate amount of variability. Polyploid induction through colchicine creates additional variability in castor's primary gene pool [8,9,27]. Colchicine causes chromosomes to double by preventing the production of microtubules during cell division [28]. This artificially produced polyploid expresses different phenotypes like enlarged cells and increased biomass and organ enlargement [29]. Chromosome exchange was observed in colchiploid castor by Narain and Singh [26]. Timko et al. [30] studied the effects of gene dosage on cellular proteins in a euploid series of castor obtained from colchicine treatment of a haploid plant found in the field using haploid, diploid and tetraploid clones. Recently, Baghyalakshmi et al. [31] developed and characterized tetraploid castor plants using different colchicine treatments on 48-1, DCS-107 and AP-41. Colchicine treatment (0.3%) for 24 h was effective to develop a good amount of colchiploid castor. They screened tetraploid plants based on size and number of stomata. Many researchers used this method to confirm increased ploidy level [[32], [33], [34]]. They observed impaired pollen fertility in colchiploid 48-1 and AP-41 plants by 0 % and 15–20%, respectively. Avidov and Lerner [35] utilized the microtubule polymerization inhibitor property of G2/M cell cycle inhibitor under the magnetic field to generate polyploid seeds of castor. This protocol of polyploid generation was patented under US patent act (Patent No. 8,946,513).
2.2. Molecular breeding
2.2.1. Molecular markers for diversity analysis of germplasm
Crop improvement efforts could be advanced with the use of modern breeding methods. Before starting any breeding program, it is necessary to have a basic understanding of genetic diversity and the population structure of breeding material [36]. This information helps in the predicting the amount of inheritance, dissimilarity and degree of heterosis [37]. Genetic variability analysis of global castor germplasm using various molecular markers like Simple Sequence Repeats (SSR), Single Nucleotide Polymorphism (SNP), Expressed Sequence Tag - SSR (EST-SSR), Amplified Fragment Length Polymorphism (AFLP), Restriction Fragment Length Polymorphism (RFLP), Start Codon Targeted Polymorphism (SCoT), Random Amplified Polymorphic DNA (RAPD), Inter-Simple Sequence Repeat (ISSR) and Target Region Amplification Polymorphism (TRAP) have revealed little variability and absence of geographically structured populations [38]. Therefore, it doesn't seem like the great phenotypic variance in castor is a result of a large genetic diversity [39]. This restricts the development of new varieties and hybrids in castor.
The co-dominance and multi-allelic behavior make SSR marker highly reproducible polymorphic and informative. This cost-effective marker is locus-specific and most abundant in any genome after SNP [40]. SSRs have extensively deployed to elucidate the diversity at genetic level in castor [[41], [42], [43], [44], [45], [46]]. Bajay et al. [47] reported 12 polymorphic SSR markers with 38 castor genotypes available with Brazilian Agricultural Research Company (EMBRAPA). Later, Qui et al. [48] reported 118 polymorphic EST-SSR markers from 379 markers in 24 lines of various countries. A total of 350 alleles, with a range of 2–6, were produced by these polymorphic SSRs, with an average of 2.97 alleles per locus. Pranavi et al. [49] exploited expressed sequence tag (EST) data bases to develop novel SSR markers. They used 57895 ESTs and designed a 611 SSR primer. Among them, set of 130 EST-SSRs were used, of which 92 pairs produce robust amplicons. These polymorphic amplicons were used in the genetic purity testing of castor hybrids. SSR markers can be successfully used to identify varieties, assess purity of cultivar and hybrids of castor [49,50].
Since their inception, SRAP (Sequence Related Amplified Polymorphism) markers have only twice been employed to access the genetic diversity of castor [38,51]. Compared to SSR markers which target and amplify both coding and non-coding regions, SRAP markers target only the genome's Open Reading Frames (gene-rich regions). Since SRAP does not require any prior sequence knowledge, it is easier and faster than other marker systems [51]. According to Budak et al. [52], SRAP's genetic diversity potency is greater than that of SSR, ISSR and RAPD and can be employed as cross-species universal primers. Agyenim-Boateng et al. [38] used 361 pairs of SRAP primers, of which 29 were polymorphic. These polymorphic SRAP were used to analyze the diversity of 473 wild castor samples from South China. With more than 80% polymorphism, SRAP marker were reported as useful and effective to detect the genetic diversity of wild castor [38]. Recently, Akhila et al. [3] also characterized 30 castor germplasms with SSR and SRAP and demonstrated that SRAP are more effective than SSR markers as SRAP generated higher mean number of total bands (5.71), polymorphic bands (4.57) and PIC than SSR.
When we work with a large number of samples, genotyping become costly. Homoplasy is become major a concern [53]. Fourth-generation markers like SNPs are a viable option for assessing population diversity. The codominant SNP has its own advantage like it is binary in nature and can quickly measure the heterozygosity available in genotype. The potential of SNP marker does not come from multiple allelism like microsatellites, but it is more potent because it covers a large number of loci. This edge helps us to identify rare nucleotide polymorphism available among genotypes and discriminate the population in several group. The problem of homoplasy is compromised with the help of the identification of evolutionary conserved SNPs [54]. Rapid and efficient genotyping of the huge germplasm is possible due to high throughput and automation of SNP marker system [55]. Foster et al. [56] assess diversity using SNPs in castor. With the use of the Bayesian clustering approach, they exposed five major global groupings and a repeated pattern of mixed genotypes, which demonstrated that few lineages occur and are distributed worldwide.
With a frequency of one SNP per 160 bp, Senthilvel et al. [57] identified 2,179759 SNPs in 14 diverse castor accessions, containing popular cultivars and germplasms. They also developed a genotyping array with 6000 SNPs. In 6K SNP chip, merely 5238 could be incorporated on the Infinium Chips. Recently, Shaw et al. [58] has reported competitive allele specific PCR based SNP (KASP) genotyping assays in castor. A set of 135 KASPs were designed from SNPs list of Senthilvel et al. [57]. These KASPs can be used for genetic diversity and hybrid purity testing. Thus, the accessibility of genomic information opens a new panorama in genetic improvement of castor [50]. Diversity among castor genotypes were captured since last two decades presented in Table 1.
Table 1.
Deployment of DNA markers in castor for diversity analysis and genetic relationships studies.
| Sr. No | Marker system | No. of polymorphic markers | No. of genotypes used | PIC | Reference |
|---|---|---|---|---|---|
| 1 | SNP | 232 | 152 | – | [56] |
| 2 | RAPD | 30 | 22 | 0.82 | [59] |
| ISSR | 5 | 0.88 | |||
| 3 | SSR | 28 | 72 | 0.26 | [41] |
| 4 | SSR | 10 | 60 | 0.82 | [42] |
| 5 | ISSR | 16 | 12 | 0.38 | [60] |
| 6 | RAPD | 13 | 31 | 0.24 | [61] |
| ISSR | 25 | 0.20 | |||
| SCoT | 10 | 0.24 | |||
| 7 | RAPD | 16 | 13 | 0.70 | [62] |
| 8 | SSR | 14 | 27 | 0.43 | [10] |
| 9 | SSR | 45 | 144 | 0.32 | [43] |
| 10 | TRAP | 70 | 40 | 0.24 | [63] |
| 11 | SSR | 14 | 15 | 0.60 | [45] |
| 12 | SSR | 25 | 20 | 0.11 | [46] |
| 13 | SSR | 5 | 20 | 0.68 | [64] |
| 14 | SCoT | 10 | 20 | 0.73 | [65] |
| 15 | SCoT | 37 | 111 | 0.67 | [66] |
| 16 | SSR | 29 | 52 | 0.39 | [67] |
| 17 | SSR | 29 | 5 (species of Euphorbiaceae) | – | [68] |
| 18 | ISSR | 9 | 54 | – | [69] |
| RAPD | 11 | ||||
| 19 | SSR | 9 | 30 | 0.25 | [3] |
| SRAP | 7 | 0.34 | |||
| 20 | SSR | 18 | 36 | 0.27 | [6] |
SNP: Single nucleotide polymorphism, RAPD: Random amplified polymorphic DNA, ISSR: Inter simple sequence repeats, SSR: Simple sequence repeats, SCoT: Start codon targeted polymorphism, TRAP: Target region amplified polymorphism, SRAP: Sequence related amplified polymorphism and PIC: Polymorphism information content.
2.2.2. Molecular markers for QTL mapping
One of the primary purposes of molecular markers is to map genes and quantitative trait loci (QTLs) [70]. Karl Sax initially introduced the principle of QTL mapping in 1923, when he noticed seed weight segregation related to seed coat colour segregation in Phaseolus vulgaris L. [71]. Genetic linkage mapping is based on the analysis of an organism's genome using DNA markers, determining their relative location on chromosomes and determining their genetic relationship with QTLs [72].
Traditional plant breeding methods are slow and less competent for targeting complex traits like yield, stress resistance, etc. for crop improvement. Conventionally transfer of any quantitative gene from one parent to another parent is time and labor-demanding, especially in the case of unavailability of defined screening approaches and linked molecular markers [73]. Amalgamation of genomic information/techniques with traditional crop improvement tools has been fortunate in some crops. DNA sequencing and DNA marker made it simple to detect marker linkage with the trait of interest through linkage maps and respective QTLs [74].
Developing a mapping population is the most critical aspect in the creation of a linkage map and QTL positioning. Population size is a critical component in the creation of linkage maps and QTLs identification since it can affect the assessment of the number of QTLs, their effects and interactions [75]. Typically, a mapping population with 200–300 lines can identify significant QTLs with considerable effects.
The average yield of castor is around 1250 kg per ha, which is greatly susceptible to three vital fungal diseases viz., charcoal rot (Macrophomina phaseolina), Fusarium wilt (Fusarium oxysporum f. sp. ricini) and grey mold (Botryotinia ricini) as per Liu et al. [76]. Among them, wilt and rot are most prominent in castor growing regions in India [77]. The identification of markers linked with resistance of diseases through linkage maps will be helpful to develop superior varieties. Liu et al. [78] reported first SSR based genetic linkage map in castor. In same year, Tomar et al. [79] developed a mapping population of 190 individuals by crossing parents namely RG 2800 and JC 18 to identify Fusarium wilt resistance QTLs. They screened 786 different molecular markers out of them 141 were polymorphic. The genotyping of segregating population was done with these polymorphic markers. On linkage group 6, they identified one putative QTL (Log of odds: 13.5) controlling wilt resistance near to markers CST 73 and R 83 [79]. Tomar et al. [80] used JI 357 as a resistant and SKI 338 as a susceptible parent for charcoal rot resistance and developed an F2:3 population and find candidate QTL. This population was screened in sick plot for phenotyping and genotyped using molecular markers (RAPD: 520, ISSR: 100, SSR: 300). They developed a map of 1833.4 cM with 10 linkage groups where average inter-markers distance was 6.93 cM. They identified three QTLs for charcoal rot with a R2 of 11.3–71.2%. Among identified QTLs, a QTL with 6.5 LOD value was believed to be a promising QTL for molecular breeding [80].
Of the various types of mapping populations, F2 can be quickly developed and possesses all potential parental allelic combinations [81]. A recombinant inbred line (RIL), also known as an immortal or permanent mapping population can be produced from an F2 generation using the single-seed descent (SSD) approach [82]. The RILs are greatly homozygous, provide precise estimations of genetic components and reveal QTL × environment interactions. The major advantages of RIL population in castor are that it is easy to develop and produces true-breeding lines that are multiplied and reproduced without genetic change. Replicated trials are possible with higher resolution than the F2 and other mortal populations [83].
Senthilvel et al. [57] used SNP genotyping array to minimize the marker interval and developed two high-density linkage maps of castor in recombinant inbred lines (RILs) derived from JC12 × 48-1 and DCS9 × RG1139. The map JC12 × 48-1 had 1090 polymorphic markers (total length: 1139.8 cM) while map of DCS9 × RG1139 (total length: 904.8 cM) was developed with 1978 polymorphic SNPs. The mean inter-marker distance was 1.12 cM for JC12 × 48-1 and 0.81 cM for DCS9 × RG1139. They also developed a consensus map of 995.8 cM using 1978 SNP markers, with one gap of less than 10 cM.
According to Elshire et al. [84], reduced representation strategies like Genotyping by Sequencing (GBS) is one of the reliable and effective technology for SNP identification. This is especially utilizable for the analysis of plants because it possesses a huge genome with high repeats [85]. With GBS, Yu et al. [86] identified 1,270,605 polymorphic SNPs between ZB107 and ZB306. A total of 15,789 SNPs were taken to construct high density linkage map in RIL population (ZB107 × ZB306) consisting 200 individuals. Using 4317 bin markers, a map of 1852.33 cM was constructed. The mean density of the map was 2.33 SNPs per cM. They mapped 16 QTLs (covering 851 candidate genes) associated with seed size and weight, including 4 QTLs for each of seed length (SL), seed width (SW), seed thickness (ST) and single seed weight (SSW). The R2 explained by each QTL was ranged from 4.4% to 20.7%. QTL mapping in plant breeding provides a valuable contribution to find out important traits and disease resistance genes [79]. The linkage map and improved pseudo-chromosome scale genome not only provide opportunities for genomic-assisted breeding of QTL controlling key agronomic traits but also lay foundation for cost-effective molecular breeding of castor [86]. Using a RIL-based (185 F6) linkage map of 1090 SNP maker developed by Shaw et al. [58], a major QTL for Fusarium wilt resistance on linkage group 7 (R2: 44%; LOD: 18.7) was identified.
2.2.3. Genome wide association study
Nowadays, genetic mapping by linkage analysis and GWAS is more common in plant breeding research [[87], [88], [89]]. Both methods take advantage of ability of the recombination to fragment the genome or chromosome into small pieces that can be linked to phenotypic variation. But control of recombination is very different in both strategies [90]. Linkage mapping is close to controlled recombination because it utilizes the bi-parental mapping population to localize the QTL on chromosome [91]. Its limitations include deprived mapping resolution, high cost, lengthy process of developing bi-parental populations and the constraint of only exploring two alleles in a single locus in bi-parental crosses. On the other hand, association mapping has no control over recombination; it exploits a natural population that imitates historical recombination and provides greater resolution compared to linkage mapping [92]. GWAS also referred as a whole genome scanning, enables researchers to control the drawbacks of linkage mapping and even allow researchers to practice cutting-edge genomic techniques to locate loci for desired traits and take advantage of wild populations with higher levels of historical recombination. A limitation of association study is vulnerability to false positives but false positive associations in GWAS can be reduced by information of population structure and kinship in a mixed model [93].
Population structuring is a key step in GWAS which characterizes the domestication history and genetic relatedness among germplasm. Population structuring was done in castor by different molecular markers using many accessions but Foster et al. [56] and Wang et al. [94] found a narrow genetic diversity and low admixture in castor. Agyenim-Boateng et al. [38] found moderate genetic differentiation among South Chinese materials of castor. Betha and Lavanya [44] also recorded the low average distance and Fst (Fst<0.2) value within the main populations and the genotypes are admixed up to 33.3%. Xu et al. [95] found that a relatively low diversity ratio between wild and domesticated populations and suggested a weak domestication bottleneck in castor.
Through GWAS, Fan et al. [96] identified candidate genes for nine important agronomic traits viz., capsule dehiscence, endocarp thickness, hundred-grain weight, panicle height, panicle length, plant height, ratio of male to female flowers, seed length and seed volume. They identified a gene related to glycosyl transferase enzyme which is related to cellulose biosynthesis. It is believed that richness of cellulose and lignin in endocarp is significant factor for dehiscence of capsule. It is an important agronomical trait because it is a cause of major yield loss and harvesting problem. This study provides the basis for breeding and genetic studies in castor. Castor plant has strong anti-oxidant system helps to tolerate cadmium toxicity. This ability of castor makes it useful for phytoremediation of cd-contaminated soils. Yeboah et al. [97] performed GWAS with 175 castor accessions and genotyped with 143 SSR markers to identify valuable alleles and candidate genes controlling tolerance to cadmium. A total of seven traits for Cd tolerance were found associated with 27 significant marker loci and R2 explained by linked markers ranged from 5.63 to 20.47%. Xu et al. [95] identified 17 GWAS signals associated with seed width, single seed weight, seed length, seed thickness and seed area. Earlier GWAS on seed length and seed volume detected the same candidate locus in castor population from China [96]. Colocalization of GWAS marker by Xu et al. [95] and QTLs mapped by Yu et al. [86] which advocated pleiotropism or physical association of genes governing these features of seed size in castor bean. Using 3465 SNPS and 300 inbreds, Shaw et al. [58] detected 69 significant genomic regions linked with wilt resistance and discovered that majority of the linked SNPs were dispersed primarily on linkage group 4, 5 and 8. Summary of GWAS and QTL study is presented in Table 2.
Table 2.
Summary of GWAS and QTL study.
| Sr. No | Population/Parent | Name and number of markers | Identified linked or associated markers | Author |
|---|---|---|---|---|
| Genome Wide Association Study (GWAS) | ||||
| 1 | 300 Individual | 4098 SNPs | 69 SNPs were associated with wilt resistance at 1 × 10−4 probability Level. Among them, 32 32 were significant at FDR threshold q ≤ 0.0.1 | [58] |
| 2 | 280 Individual | 48450 SNPs | 2 SNP for number of nodes, 9 SNP for diameter of the main stem and 2 SNP for plant height above ground. | [95] |
| 3 | 175 Individuals | 143 SSR | 27 SSR were associated with all traits among them 10 for shoot dry weight, 5 for shoot length, 3 for plant height, 3 for shoot fresh weight, 3 for root fresh weight, 2 for root dry weight, 1 for root length | [97] |
| Quantitative Trait Loci (QTL) analysis | ||||
| 4 | 190 F2 segregants developed from the cross RG 2800 × JC 18 | 520 RAPD, 100 ISSR and 166 SSR | 2 QTLs identified which were resistant to Fusarium wilt. | [79] |
| 5 | 190 F2:3 individuals of cross JI 357 × SKI 338 | 520 RAPD, 100 ISSR and 300 SSR | 3 QTLs identified for charcoal rot resistance | [80] |
| 6 | 200 F4 RILs of cross ZB306 × ZB107 | 15,789 SNPs | 16 QTLs identify for seed size and weight which included 4 QTLs for each of seed length, seed width, seed thickness and single seed weight | [86] |
| 7 | 185 RILs of cross JI35 × 48-1 | 1090 SNPs | 1 QTL identified for resistant to Fusarium wilt | [58] |
| 8 | 200 RILs of cross Rc250 × Rc249 | 23413 SNPs | 18 QTLs were identified for seed length, seed width, seed thickness, single seed weight and seed oil content | [95] |
2.3. Tissue culture in castor
Plant tissue culture is a collection of in vitro techniques, procedures and tactics that fall under the aegis of plant biotechnology. Tissue culture has been used to increase the health of planting material (disease free) and create genetic variability [98]. Tissue-culture procedures are available for most of crop species, while several crops including castor, optimized tissue culture protocol is still required. Genetic transformation is widely used to create targeted changes in genome. It is also helpful to create variability in germplasm but success of any transformation completely relays on successful and efficient plant tissue culture protocol which must be fast, consistent and applicable to a large number of genotypes [99]. The reports on in vitro regeneration and genetic transformation protocols in castor are limited. The highly recalcitrance nature of castor makes it less responsive to plant tissue culture. Most of the previous studies are proven inefficient or difficult to reproduce by using vegetative tissue as an ex-plant [100]. The available scanty literature informs about plantlet differentiation in castor and mostly regenerations were achieved in apical meristems and shoot tip callus [17,[101], [102], [103], [104], [105]]. Though, somatic organogenesis in castor was only reported by Ganesh Kumari et al. [106].
Apical meristems and shoot tip calluses have been used to differentiate plantlets in the majority of reported cases [4]. Effective in vitro regeneration of stem tips was achieved with reduced NO3−/NH4+ lacking FeSO4 in medium supplemented with auxins [107,108]. Embryo axis shoot induction was more successful in medium containing thidiazuron and 300 g/kg of glucose [109]. A technique for inducing shoots and roots from castor seedling's cotyledonary nodes was revealed by Alam et al. [110].
Endosperm is remaining most favorite explant for researchers in castor tissue culture because of its easy culturability, large size and triploid nature. Endosperm is a store of oil and many fatty acids in castor so study of their production path way and understanding its metabolism becomes easy. Triploids are source of trisomics, hence to enable aneuploid breeding and genetic mapping, endosperm culturing or chromosome doubling is more important in castor [[111], [112], [113]]. La Rue [114] first recorded organogenic differentiation in endosperm culture in castor. Mohan Ram and Satsangi [115] demonstrated that mature endosperm can produce cell that has ability to divide further in tissue culture. But organogenesis is still not been achieved in this crop. Mohan Ram and Satsangi [115] were the first to discover that mature endosperm cells can divide and later they established successful mature endosperm cultures. Few researchers have documented regeneration of castor by employing germinated seed and its appendages as an ex-plant in the last 20 years, as indicated in Table 3. This milestone has the potential to facilitate haploid breeding since Liu et al. [125] employed anther as an explant and induce efficient callus by utilizing 8 mg/ml sodium nitroprusside (SoNP). He showed that SoNP may be employed as a NO− donor in tissue culture and that it can interact with auxins to regulate plant cell division, dedifferentiation and redifferentiation.
Table 3.
Tissue culture protocols for culture establishment and micropropagation in castor.
| Sr. No | Explant | Media composition and culture condition | Reference |
|---|---|---|---|
| 1 | Hypocotyl and shoot apical meristem tissues of seed |
Shoot induction: Murashige and Skoog (MS) media having pH 5.7 and supplemented with 30 g l−1 sucrose, 0.5 g l−1 2-(4-morpholino) ethane sulfonic acid (MES). It was further supplemented with either 1 μM thidiazuron (TDZ) or 20 μM 6-benzylaminopurine (BA). Explants were preincubated in the dark up to 7 days and then cultured under the 16h day and 8h night cycle for the rest of the culture period at 100 μ mol m−2 s−1 supplied by cool-white fluorescent lamps (Sylvania, Danvers, MA) at constant 26 °C. Shoot development: Murashige and Skoog (MS) salt having pH 5.7 with 30 g l−1 sucrose as an energy source, 0.5 g l−1 MES supplemented with 2 μM BA |
[100] |
| 2 | Seed-derived cotyledon | MS medium supplemented with 1, 5, or 10 μM thidiazuron (TDZ) with one-week dark reconditioning | [116] |
| 3 | Hypocotyl and shoot apical meristem tissues of mature seeds | Dark pretreatment to explant on media containing MS salt supplemented with 1 μm TDZ | [106] |
| 4 | Embryo axes from mature seeds | Semi-solid Murashige and Skoog's (1962) medium containing 3% sucrose and 0.5 mg l−1 TDZ with adjusted pH of 5.6 ± 0.2 was maintained in 5–7 days in the dark. After this, cultures were maintained at 26 ± 2 °C under a 16/8-h light/dark photoperiod provided by cool white fluorescent lamps at an intensity of 30 μmol m−2 s−1. | [117] |
| 5 | Whole cotyledonary nodes |
Shoot induction: Murashige and Skoog (MS) medium supplemented with 3.0 mg l−1 6-benzylaminopurine (BAP) Shoot elongation: Murashige and Skoog (MS) medium having 1.0 mg l−1 BAP in combination with 0.25 mg l−1 GA3 Root induction: MS medium supplemented with 1.0 mg l−1 NAA. |
[110] |
| 6 | Hypocotyl |
Callus induction: MS salt in the combination of 6-benzyl amino purine 2.0 mg l−1 + 0.5 mg l−1 Naphthalene acetic acid (NAA). Cell culture: MS Salt with the combination of BAP 2.0 mg l−1 and 0.2 mg l−1 NAA. |
[118] |
| 7 | Hypocotyls of germinated seedlings |
In vitro seedlings germination: Growth regulator free MS medium Organogenic callus induction: Murashige and Skoog's (MS) medium supplemented with B5 vitamins and 1.0 mg l−1 BA (80.84%) or 2.0 mg l−1 BA (80.17%) Shoot induction: Murashige and Skoog's (MS) medium with 0.5 mg l−1 KIN + 0.25 mg l−1 BAP (75.00%). Use of 0.2 mg l−1 GA3 in combination with 0.5 mg l−1 KIN and 0.25 mg l−1 BAP induce more shoots. Root induction: IAA, IBA and AgNO3 individually or in blends but root initiation was not attained even after 30 days of culture. |
[119] |
| 8 | Cotyledonary nodes (CN) and Shoot tip |
Multiple shoot proliferation: Modified Murashige and Skoog (mMS) medium supplemented with Thidiazuron (TDZ) 0.3 mg l−1. Shoot Elongation: MS medium supplemented with PF - 68 (0.6 mg l−1) with GA3 (0.3 mg l−1) Rooting and Acclimatization: Full strength MS medium fortified with IBA (1.5 mg l−1) To increase root number: Full strength mMS medium supplemented with 0.6 mg l−1 AgNO3 along with 1.5 mg l−1 IBA |
[120] |
| 9 | Embryo axes isolated from the pre-germinated seeds | Modified 1/2 Murashige and Skoog (MS) basal medium supplemented with 440 mg l−1 Ca2+, 0.2 mg l−1 gibberellic acid and 0.1 mg l−1 TDZ for shoot elongation. Furthermore, 1/2 MS media supplemented with 0.2 mg l−1 1-naphthaleneacetic acid for rooting with one-week dark preconditioning | [121] |
| 10 | Anther | Treatment of anther explant with 15 mg l−1 BA solution for 10 min before being inoculated onto hormone-free Murashige and Skoog (MS) medium containing 8 mg l−1 sodium nitroprusside | [122] |
| 11 | Zygotic embryos of mature seeds | Murashige and Skoog with Gamborg B5 media without growth hormones | [123] |
| 12 | Hypocotyls | MS containing 1 or 2 mg l−1 of zeatin +0.1 mg l−1 of IAA. | [124] |
2.4. Genetic transformation
There is a sustained requirement for new developed cultivar for different growing area with greater tolerance to abiotic as well as biotic factor with yield superiority [126]. Currently, this requirement is achieved by traditional plant breeding but it is not sufficient to cope up today's requirement. It has limitation of transferring new traits from one cultivar to another is very difficult. But now a days, genetic engineering opens these doors of improvement in castor. Contemporary, biotechnology furnishes great assurance in minimizing toxic compounds, enhancing oil content, improving seed quality and enhancing stress tolerance in castor.
The castor cake generated as a byproduct of oil extraction has minimal utility because it includes the cytotoxin ricin as well as ricin agglutinin (RCA120), a potentially hazardous allergen. This is significant since the proper handling and use of castor cake is dependent on the neutralization of both poisonous and allergic components. After detoxification, the meal may be utilized as a feed for animals, giving a source of profit on top of the oil output [127]. Now a days development of ricin free castor becomes a major breeding objective that could be achieved through knocking out or silencing the genes linked to ricin production. Ricin is produced as a precursor pre-proricin (64.1 kDa) which produces both A and B chains. After cleavage of N-terminal signal peptide, pre-proricin converted into proricin (61.6 kDa) during transportation from lumen to endoplasmic reticulum. Pro-peptides are broken down in the vacuoles and ricin gets stored in protein bodies as a mature 58.8 kDa protein [128]. These proteins expressed highly during seed development, but selection of proper promoter and application of gene silencing leads us to suppressing 10,000-fold in seed during seed development and maturing [129].
Published literatures suggested that a significant accumulation of ricin (1.6–32 mg/gram of mature seed) in castor seeds occurs 40 days after pollination [128,130,131]. Sousa et al. [128] exploited the RNAi technique for silencing the genes coding ricin in the endosperm. They designed intron-hairpin vector pRicRNAi containing a 460bp fragment with A-chain of ricin gene cloned directionally in sense and antisense to created siRNA during transcription. Additionally, this cassette contains AHAS imazapyr (herbicide) tolerance gene from Arabidopsis thaliana and screening marker gus gene to screen transformants. They bombarded 270 embryonic axes with vector and isolate four primary transgenic lines with 0.85% transformation efficiency. The gus gene expression helps them to identify true transformed plants and a single line (TB14S-5D) could transfer the trans gene to next generation which was confirmed by PCR analysis of gus, ahas and Δricin transgene. Their findings showed that silencing ricin genes was effective to create a bio-detoxified castor genotype. Castor is adapted to areas facing high temperature and reduced water requirement where farmers don't have many options. This genotype helps to produce animal feed after oil extraction because it is safer to use as an animal feed due to bio-detoxification of ricin.
The costs associated with the deregulation of a transgenic trait make the commercial production of a transgenic castor dubious. Moreover, transformation of castor is challenging due to recalcitrant behavior and lack of a commercial regeneration methods [4]. Though, in last two decades numerous attempts have been made to develop transgenics in castor. McKeon and Chen [132], first reported the Agrobacterium mediated genetic transformation of castor exploiting vacuum infiltration of wounded flower buds and recovered 12 true transgenic plants. Since then, an apparently more efficient method using Agrobacterium has been developed [104]. Sujatha and Sailaja [104], used vacuum infiltration for 30 min to transform seed dissected embryos and reported high transformation (0.8%) and survival frequency with poor shoot proliferation. Using above protocol, Malathi et al. [105] developed semilooper resistant transformants (0.42%) by introducing cryIAb into the castor cotyledonary node explants. Later on, through particle gun bombardment, same group reported stable transformed castor plants from germinating embryonic axes [117]. The transformation efficiency with direct gene transfer method was 1.4%. Transgenic castor sheltering cry1EC and cry1AbcF have also been generated through direct and indirect transformation methods [4,133]. Using cry1EC gene, 93 putative transformants were achieved. Through in planta transformation, Kumar et al. [133] introduced cry1AbcF gene against semilooper. To boost the salt stress capacity of castor, Patel et al. [134] transferred vacuolar Na+/H+ antiporter gene SbNHX1 using Agrobacterium. By directly injecting Agrobacterium into endosperm, Sanchez-lvarez et al. [135] described a technique for transient transformation of castor. When injections were performed up to 40 days after pollination, the transgene expression lasted longer for >15 days, thus this approach proved reliable and produced significant amounts of viable altered seeds. The main aim of this study was developing castor as a platform to produce oils of industrial interest. For this, they directly injected Agrobacterium cell suspension into endosperm with a β-ketoacyl-CoA synthase (KCS) from Lesquerella fendleri. Lesquerolic acid (LA) is produced by KCS from ricinoleic acid. Transformed plant exhibited 8 × increments in LA compared to endogenous amount but LA could not get accumulated considerably in mature seeds.
2.5. Use of omics
Omics technology includes genomics, proteomics, transcriptomics, metabolomics and phenomics etc. These all techniques help to identify actual involvement of gene in development of different organs and activation and deactivation of different genes which help to explain functional pathways for rapid improvement. According to Yu et al. [136], seeds are more precious agricultural input for economic and nutritional value. Castor is model plant in seed development studies because of its endosperm remain persistent during whole seed development stage.
Due to their persisting endosperms at maturity, castor seeds are typically endospermic in dicotyledons. Through histology research, Yu et al. [136] showed that cell number instead of cell size in the endosperm of castor-controlled seed size. They used two inbred lines (Large seed: ZB107; small seed: ZB306) for combined and comparative transcriptome and proteomic analysis. According to their investigation, majority of the transcripts playing role in cell division and reservoir accumulation and remain consistence with increased seed size and volume. On the basis of above observation, they concluded that differentially abundant protein species (173) and differentially expressed genes (9545) are crucial to control the seed size in castor. Correlation between transcriptomics and proteomics indicated that numerous genes intricated in cell division, metabolism of protein and carbohydrate and were collectively regulates transcripts and protein levels in seed. These findings help to dissect genetic mechanism in the yield improvement through increased seed volume and size.
Early planting is one of the most important strategies to control disease and pests in castor but in temperate region, low germination percentage found in castor due to lake of cold tolerance in seed during germination period. In Tongbi 5 variety, Wang et al. [137] performed quantitative proteomics in seed during early imbibition and detected differential abundance protein species between cold and control condition. They identify 127 DAPs which were active during carbohydrate and energy metabolism, translation, post translational modification, stress response, lipid transport and signal transduction. On the basis of this study, Wang et al. [137] suggested that cold tolerance genotypes of castor can be created by overexpressing β-ketoacyl-acyl carrier protein synthase I and II. This study helps in understanding molecular mechanism of cold tolerance associated with castor seed germination.
Choubey et al. [7] believed that proteomics is always better than transcriptomics and genomics for quick identification of novel proteins expressed during any biological process. They implemented a comparative proteomics approach to identify proteins released in resistant and susceptible genotypes during infection of Fusarium oxysporum fsp. Ricini. They isolated proteins from the inoculated resistant genotype 48-1 and susceptible genotype JI-35. Isolated proteins were separated through 2D gel-electrophoresis as well as RPLC-MS/MS and concluded that 18 and 8 unique peptides were identified in resistant and susceptible genotypes, respectively. They also perform real time expression study and found that CCR 1, Germin like protein 5-1, RPP8, Laccase 4 and Chitinase like 6 were upregulated during Fusarium infection. They took support of the end-point PCR of c-DNA of three genes Chitinase 6 like, RPP8 and β-glucanase and concluded that these genes may involve in the resistance phenomena in castor while up-regulation of CCR-1 and laccase 4 leads to lignin biosynthesis which gives mechanical strength and prevent the entry of the fungal mycelia.
Many other studies were conducted by various authors like, organelle genome sequencing and global genetic diversity investigation by Rivarola et al. [138]; Isotope labeling-based quantitative proteomics of developing seeds by Nogueira et al. [139]; Genomic understandings about origin, domestication and genetic basis of agronomic traits by Xu et al. [95]; Understanding triacylglycerol lipid biosynthetic pathways by Brown et al. [140] through tissue-specific RNAseq analysis; Complex and interconnected sucrose signaling cascades in developing seeds of castor bean can be revealed by transcriptome analysis [141], Tan et al. [142] used digital gene expression profile and transcriptomics for diverse sex types during development from apical buds to inflorescences. Wu et al. [143] identify ricinolic acid biosynthesis gene using transcriptome analysis of developing castor seed.
3. Conclusion
Castor had an amazing journey from wild species to domesticated species in East African areas 3200 years ago. It became the most significant industrial crop after domestication because of its oil qualities. It is a drought-resistant, high-value industrial crop that is subjected to a variety of biotic and abiotic stressors. Its development is confined to traditional breeding methods. Because it is a monotypic species, it has minimal diversity and room for improvement. To overcome this constraint, contemporary breeding and biotechnology tools must be used. The development of beneficial variations and their use is mostly focused on the enhancement of castor. Molecular marker technology can assist to define diversity within its gene pool as well as enhance castor by cross breeding with other genera such as Jatropha spp. This potential might be opened up by creating a castor synteny map with other crossable species. While intraspecific molecular marker mapping contributes in the improvement of its own gene pool. Molecular markers are extremely useful for capturing genetic diversity that exists naturally or that has been produced via random or controlled mutagenesis as well as molecular marker mapping information can be used to improve castor through marker assisted selection. Castor is particularly recalcitrant to tissue culture response, despite efforts to build a regeneration technique that allows for more targeted variation and focused modification using various gene transformation and genome editing techniques. Omics technology aids in the study of various metabolic pathways and the roles of genes in the development of distinct phenotypes like wilt resistance. This knowledge may open up new avenues for targeted castor improvement in terms of biotic, abiotic and yield improvement.
Data availability statement
No data was used for the research described in the article. Data associated with this study been not deposited into a publicly available repository.
CRediT authorship contribution statement
Rumit Patel: Writing – original draft. Juned Menon: Writing – original draft. Sushil Kumar: Writing – review & editing, Writing – original draft, Supervision, Conceptualization. Márcia Nóbrega: Writing – review & editing. Dipak A. Patel: Supervision. Amar A. Sakure: Writing – original draft. Mahesh B. Vaja: Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Perry B.A. Chromosome number and phylogenetic relationships in the Euphorbiaceae. Am. J. Bot. 1943:527–543. [Google Scholar]
- 2.Memon J., Patel R., Parmar D.J., Kumar S., Patel N.A., Patel B.N., Patel D.A., Katba P. Heliyon; 2023. Deployment of AMMI, GGE-Biplot and MTSI to select elite genotypes of castor (Ricinus communis L.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akhila S.R., Kumar S., Sakure A.A., Patel D.A., Patel M.P. Integration of morpho-physico-biochemical traits with SSR and SRAP markers for characterization of castor genotypes of Indian origin. Oil Crop Science. 2022;7(1):22–30. [Google Scholar]
- 4.Sujatha M., Reddy T.P., Mahasi M.J. Role of biotechnological interventions in the improvement of castor (Ricinus communis L.) and Jatropha curcas L. Biotechnol. Adv. 2008;26(5):424–435. doi: 10.1016/j.biotechadv.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Jeong G.T., Park D.H. Optimization of biodiesel production from castor oil using response surface methodology. Appl. Biochem. Biotechnol. 2009;156(1):1–11. doi: 10.1007/s12010-008-8468-9. [DOI] [PubMed] [Google Scholar]
- 6.Rajput D., Patel M.P., Kumar S., Patel R., Katba P. Screening of castor germplasm for wilt reaction and morpho-molecular characterization of resistant genotypes. Heliyon. 2023;9(3) doi: 10.1016/j.heliyon.2023.e14598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choubey V.K., Sakure A.A., Kumar S., Vaja M.B., Mistry J.G., Patel D.A. Proteomics profiling and in silico analysis of peptides identified during Fusarium oxysporum infection in castor (Ricinus communis) Phytochemistry. 2023;213(2023) doi: 10.1016/j.phytochem.2023.113776. [DOI] [PubMed] [Google Scholar]
- 8.Kulkarni L.G., Ramanamurthy G.V. Indian Council of Agricultural Research; 1977. Castor. [Google Scholar]
- 9.Moshkin V.A., Dvoryadkina A.G. Castor. Amerind Publi.; New Delhi, India: 1986. Cytology and Genetics of Qualitative Characteristics; pp. 125–132. [Google Scholar]
- 10.Rukhsar S., Patel M.P., Parmar D.J., Kalola A.D., Kumar S. Morphological and molecular diversity patterns in castor germplasm accessions. Ind. Crop. Prod. 2017;97:316–323. [Google Scholar]
- 11.Yali W., Mitiku T. Mutation breeding and its importance in modern plant breeding. J. Plant Sci. 2022;10(2):64–70. [Google Scholar]
- 12.Kulkarni L.G. In Pp 293-299 of Proceedings of the Symposium on Radiation and Radiomimetic Substances in Mutation Breeding. 1969. Bombay Department of Atomic Energy (1969). Indian Agricultural Research Inst.; Hyderabad: 1969. Induction of useful mutations in castor. [Google Scholar]
- 13.Lavanya C., Chakrabarthy S.K., Ramachandran M., Rao C.H., Raoof M.A. Development of wilt resistant pisitillate lines in castor through mutation breeding. J. Oilseeds Res. 2003;20:48–50. [Google Scholar]
- 14.Lavanya C., Varaprasad K.S. Castor hybrids in India: a success story. Seed Technol. 2012;5(4):111–117. [Google Scholar]
- 15.Lavanya C., Duraimurugan P., Lakshmamma P., Manjunatha T., Ramya K.T., Santhalakshmi Prasad M., Vishnuvardhan Reddy A. International Symposium on Plant Mutation Breeding and Biotechnology organized by FAO/IAEA; at Vienna, Austria: 2018. Development of Leafhopper and Wilt Resistant Pistillate Line in castor through Mutation Breeding. [Google Scholar]
- 16.Sujatha M., Prabakaran A.J. New ornamental Jatropha hybrids through interspecific hybridization. Genet. Resour. Crop Evol. 2003;50(1):75–82. [Google Scholar]
- 17.Reddy K.R.K., Swamy N.R., Bir B. Cross-incompatibility between Ricinus and Jatropha. Plant Cell Incompatibility Newsletter. 1987;(19):60–65. [Google Scholar]
- 18.Sathaiah V., Reddy T.P. Seed protein profiles of castor (Ricinus communis L.) and some Jatropha species. Genet. Agrar. 1985;39(1):35–43. [Google Scholar]
- 19.Sujatha M. Osmania University; Hyderabad: 1996. Genetic and Tissue Culture Studies in castor (Ricinus communis L.) and Related Genera (Doctoral Dissertation), Ph. D. Thesis. [Google Scholar]
- 20.Bahadur B., Sujatha M. In: Interspecific Hybridization in the Genus Jatropha. Carels N., editor. Springer; Dordrecht: 2013. Jatropha, challenges for a new energy crop volume 2: genetic improvement and biotechnology; pp. 434–441. [Google Scholar]
- 21.Laosatit K., Mokrong N., Tanya P., Srinives P. Overcoming crossing barriers between jatropha (Jatropha curcas L.) and castor bean (Ricinus communis L.) Crop Breeding and Applied Biotechnology. 2017;17(2):164–167. [Google Scholar]
- 22.Premjet D., Obeng A.K., Kongbangkerd A., Premjet S. Intergeneric hybrid from Jatropha curcas L. And Ricinus communis L.: characterization and polyploid induction. Biology. 2019;8(2):50. doi: 10.3390/biology8020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel R.G., Mankad A.U. In vitro pollen germination-A review. Int. J. Sci. Res. 2014;3(5):304–307. [Google Scholar]
- 24.Murashige T., Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plantarum. 1962;15(3):473–497. [Google Scholar]
- 25.Hagerup C. Uber ployploidie in beziehung zu klima, okologie und phylogenie. Hereditas. 1932;16:19–40. [Google Scholar]
- 26.Narain A., Singh P. Haploid meiosis and its bearing on the constitution of the castor oil plant. J. Hered. 1968;59(5):287–288. [Google Scholar]
- 27.Weiss E.A. second ed. Blackwell Science; 2000. Oilseed Crops. [Google Scholar]
- 28.Elgsti O.J., Dustin P. Colchicine-in agriculture, medicine, biology and chemistry. Colchicine-in agriculture, medicine, biology and chemistry. 1955:50. [Google Scholar]
- 29.Levin D.A. Polyploidy and novelty in flowering plants. Am. Nat. 1983;122(1):1–25. [Google Scholar]
- 30.Timko M.P., Vasconcelos A.C., Fairbrothers D.E. Euploidy in Ricinus. I. Euploidy and gene dosage effects on cellular proteins. Biochem. Genet. 1980;18(1–2):171–183. doi: 10.1007/BF00504367. [DOI] [PubMed] [Google Scholar]
- 31.Baghyalakshmi K., Shaik M., Mohanrao M.D., Shaw R.K., Lavanya C., Manjunatha T., Senthilvel S. Development and characterization of tetraploid castor plants. Plant Genet. Resour. Charact. Util. 2020;1:7. [Google Scholar]
- 32.Speckmann G.J., Post J., Dijkstra H. The length of stomata as an indicator for polyploidy in rye-grasses. Euphytica. 1965;14(3):225–230. [Google Scholar]
- 33.Beck S.L., Dunlop R.W., Fossey A. Stomatal length and frequency as a measure of ploidy level in black wattle, Acacia mearnsii (de Wild) Bot. J. Linn. Soc. 2003;141(2):177–181. [Google Scholar]
- 34.Liu G., Li Z., Bao M. Colchicine-induced chromosome doubling in Platanus acerifolia and its effect on plant morphology. Euphytica. 2007;157(1):145–154. [Google Scholar]
- 35.Avidov A., Lerner A. U.S. Patent and Trademark Office; Washington, DC: 2015. U.S. Patent No. 8,946,513. [Google Scholar]
- 36.Van Inghelandt D., Melchinger A.E., Lebreton C., Stich B. Population structure and genetic diversity in a commercial maize breeding program assessed with SSR and SNP markers. Theor. Appl. Genet. 2010;120(7):1289–1299. doi: 10.1007/s00122-009-1256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi-Lun Y., Ping F., Ke-Cheng K., Guang-Tang P. Genetic diversity based on SSR markers in maize (Zea mays L.) landraces from Wuling mountain region in China. J. Genet. 2008;87(3):287–291. doi: 10.1007/s12041-008-0046-y. [DOI] [PubMed] [Google Scholar]
- 38.Agyenim-Boateng K.G., Lu J., Shi Y., Zhang D., Yin X. SRAP analysis of the genetic diversity of wild castor (Ricinus communis L.) in South China. PLoS One. 2019;14(7) doi: 10.1371/journal.pone.0219667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yi C., Zhang S., Liu X., Bui H., Hong Y. Does epigenetic polymorphism contribute to phenotypic variances in Jatropha curcas L. BMC Plant Biol. 2010;259(10):1–9. doi: 10.1186/1471-2229-10-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaiswal V., Rawoof A., Dubey M., Chhapekar S.S., Sharma V., Ramchiary N. Development and characterization of non-coding RNA based simple sequence repeat markers in Capsicum species. Genomics. 2020;112(2):1554–1564. doi: 10.1016/j.ygeno.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Seo K.I., Lee G.A., Ma K.H., Hyun D.Y., Park Y.J., Jung J.W.…Lee M.C. Isolation and characterization of 28 polymorphic SSR loci from Castor Bean (Ricinus communis L.) Journal of Crop Science and Biotechnology. 2011;14(2):97–103. [Google Scholar]
- 42.Gálová Z., Vivodík M., Balážová Ž., Hlozáková T.K. Identification and differentiation of Ricinus communis L. using SSR markers. Potravinarstvo Slovak Journal of Food Sciences. 2015;9(1):556–561. [Google Scholar]
- 43.Senthilvel S., Shaik M., Anjani K., Shaw R.K., Kumari P., Sarada C., Kiran B.U. Genetic variability and population structure in a collection of inbred lines derived from a core germplasm of castor. J. Plant Biochem. Biotechnol. 2017;26(1):27–34. [Google Scholar]
- 44.Betha U.K., Lavanya C. Molecular diversity and population structure in breeding lines of Castor (Ricinus communis L.) Int.J.Curr.Microbiol.App.Sci. 2019;8(2):2019. [Google Scholar]
- 45.Chaudhari B.A., Patel M.P., Dharajiya D.T., Tiwari K.K. Assessment of genetic diversity in castor (Ricinus communis L.) using microsatellite markers. Biosci Biotechnol Res Asia. 2019;16(1):61–69. [Google Scholar]
- 46.Kachhadiya H.J., Madaria R.B., Bhadani R.V., Nandha A.K., Savaliya N., Antala V. Assessment of molecular diversity in Castor genotypes using SSR markers. Int.J.Curr.Microbiol.App.Sci. 2019;8(7):595–603. [Google Scholar]
- 47.Bajay M.M., Pinheiro J.B., Batista C.E.A., Nobrega M.B.M., Zucchi M.I. Development and characterization of microsatellite markers for castor (Ricinus communis L.), an important oleaginous species for biodiesel production. Conservation Genetics Resources. 2009;1(1):237. [Google Scholar]
- 48.Qiu L., Yang C., Tian B., Yang J.B., Liu A. Exploiting EST databases for the development and characterization of EST-SSR markers in castor bean (Ricinus communis L.) BMC Plant Biol. 2010;10:1–10. doi: 10.1186/1471-2229-10-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pranavi B., Sitaram G., Yamini K.N., Dinesh Kumar V. Development of EST–SSR markers in castor bean (Ricinus communis L.) and their utilization for genetic purity testing of hybrids. Genome. 2011;54(8):684–691. doi: 10.1139/g11-033. [DOI] [PubMed] [Google Scholar]
- 50.Singh A.S., Kumari S., Modi A.R., Gajera B.B., Narayanan S., Kumar N. Role of conventional and biotechnological approaches in genetic improvement of castor (Ricinus communis L.) Ind. Crop. Prod. 2015;74:55–62. [Google Scholar]
- 51.Li G., Quiros C.F. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor. Appl. Genet. 2001;103(2–3):455–461. [Google Scholar]
- 52.Budak H., Shearman R.C., Parmaksiz I., Dweikat I. Comparative analysis of seeded and vegetative biotype buffalograsses based on phylogenetic relationship using ISSRs, SSRs, RAPDs, and SRAPs. Theor. Appl. Genet. 2004;109(2):280–288. doi: 10.1007/s00122-004-1630-z. [DOI] [PubMed] [Google Scholar]
- 53.Estoup A., Angers B. Microsatellites and minisatellites for molecular ecology: theoretical and empirical considerations. Advances in Molecular Ecology Amsterdam: IOS PressCarvalho GR. 1998:55–86. [Google Scholar]
- 54.Brumfield R.T., Beerli P., Nickerson D.A., Edwards S.V. The utility of single nucleotide polymorphisms in inferences of population history. Trends Ecol. Evol. 2003;18(5):249–256. [Google Scholar]
- 55.Tsuchihashi Z., Dracopoli N.C. Progress in high throughput SNP genotyping methods. Pharmacogenomics J. 2002;2(2):103–110. doi: 10.1038/sj.tpj.6500094. [DOI] [PubMed] [Google Scholar]
- 56.Foster J.T., Allan G.J., Chan A.P., Rabinowicz P.D., Ravel J., Jackson P.J., Keim P. Single nucleotide polymorphisms for assessing genetic diversity in castor bean (Ricinus communis L.) BMC Plant Biol. 2010;10(1):1–11. doi: 10.1186/1471-2229-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senthilvel S., Ghosh A., Shaik M., Shaw R.K., Bagali P.G. Development and validation of an SNP genotyping array and construction of a high-density linkage map in castor. Sci. Rep. 2019;9(1):1–10. doi: 10.1038/s41598-019-39967-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaw R.K., Shaik M., Prasad M.S.L., Prasad R.D., Mohanrao M.D., Senthilvel S. Genomic regions associated with resistance to Fusarium wilt in castor identified through linkage and association mapping approaches. Genome. 2022;65(3):123–136. doi: 10.1139/gen-2020-0048. [DOI] [PubMed] [Google Scholar]
- 59.Gajera B.B., Kumar N., Singh A.S., Punvar B.S., Ravikiran R., Subhash N., Jadeja G.C. Assessment of genetic diversity in castor (Ricinus communis L.) using RAPD and ISSR markers. Ind. Crop. Prod. 2010;32(3):491–498. [Google Scholar]
- 60.Goodarzi F., Darvishzadeh R., Hassani A. Genetic analysis of castor (Ricinus communis L.) using ISSR markers. Journal of Plant Molecular Breeding. 2015;3(1):18–34. [Google Scholar]
- 61.Kallamadi P.R., Nadigatla V.G.R., Mulpuri S. Molecular diversity in castor (Ricinus communis L.) Ind. Crop. Prod. 2015;66:271–281. [Google Scholar]
- 62.Lakhani H.N., Patel S.V., Bodar N.P., Golakiya B.A. RAPD analysis of genetic diversity of castor bean (Ricinus communis L.) Int. J. Curr. Microbiol. App. Sci. 2015;4(1):696–703. [Google Scholar]
- 63.Simões K.S., Silva S.A., Machado E.L., Silva M.S. Genetic divergence in elite castor bean lineages based on TRAP markers. Genet. Mol. Res. 2017;16(3) doi: 10.4238/gmr16039776. [DOI] [PubMed] [Google Scholar]
- 64.Salihu B.Z., Falusi O.A., Adepoju A.O., Arolu I.W., Daudu O.Y., Abejide D.R., Oke C.O. Assessment of genetic diversity of promising castor bean (Ricinus communis L.) genotypes in Nigeria. Not. Sci. Biol. 2019;11(3):467–474. [Google Scholar]
- 65.Vivodík M., Saadaoui E., Balážová Ž., Gálová Z., Petrovičová L. Genetic diversity and population structure in Tunisian castor genotypes (Ricinus communis L.) detected using SCoT markers. Potravinarstvo. 2018;12(1) [Google Scholar]
- 66.Vivodík M., Balážová Ž., Gálová Z., Petrovičová L. Start codon targeted polymorphism for evaluation of functional genetic variation and relationships in cultivated castor (Ricinus communis L.) genotypes. Genetika. 2019;51(1):137–146. [Google Scholar]
- 67.Yang T., Lu J., Yeboah A., Gu S., Li D., Shi Y., Yin X. Construction of castor functional markers fingerprint and analysis of genetic diversity. Biocell. 2020;44(3):381. [Google Scholar]
- 68.Dharajiya D.T., Shah A., Galvadiya B.P., et al. Genome-wide microsatellite markers in castor (Ricinus communis L.): identification, development, characterization, and transferability in Euphorbiaceae. Ind. Crops Prod. 2020;151 [Google Scholar]
- 69.Kim H., Lei P., Wang A., Liu S., Zhao Y., Huang F.…Meng F. Genetic diversity of castor bean (Ricinus communis L.) revealed by ISSR and RAPD markers. Agronomy. 2021;11(3):457. [Google Scholar]
- 70.Jansen R.C. Controlling the type I and type II errors in mapping quantitative trait loci. Genetics. 1994;138:871–881. doi: 10.1093/genetics/138.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sax K. The association of size differences with seed-coat pattern and pigmentation in Phaseolus vulgaris. Genetics. 1923;8:552–560. doi: 10.1093/genetics/8.6.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Ooijen J.W. Accuracy of mapping quantitative trait loci in autogamous species. Theor. Appl. Genet. 1992;84:803–811. doi: 10.1007/BF00227388. [DOI] [PubMed] [Google Scholar]
- 73.Hamdan M.F., Mohd Noor S.N., Abd-Aziz N., Pua T.L., Tan B.C. Green revolution to gene revolution: technological advances in agriculture to feed the world. Plants. 2022;11(10):1297. doi: 10.3390/plants11101297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dhingani R.M., Umrania V.V., Tomar R.S., Parakhia M.V., Golakiya B. Introduction to QTL mapping in plants. Ann Plant Sci. 2015;4(4):1072–1079. [Google Scholar]
- 75.Beavis W.D. In: Molecular Dissection of Complex Traits. Paterson A.H., editor. CRC Press; Boca Raton, FL: 1998. QTL analysis: power, precision, and accuracy; pp. 145–161. [Google Scholar]
- 76.Liu Y., Lu J., Tang J., Guo L., Yin X. Genomic Designing for Biotic Stress Resistant Oilseed Crops. Springer International Publishing; Cham: 2022. Biotic stresses in Castor plant; pp. 289–310. [Google Scholar]
- 77.Rajani V.V., Parakhia A.M. Management of root rot disease (Macrophomina phaseolina) of castor (Ricinus communis L.) with soil amendments and biocontrol agents. J. Mycol. Plant Pathol. 2009;39(2):290. [Google Scholar]
- 78.Liu S., Yin X., Lu J., Liu C., Bi C., Zhu H., Li W. The first genetic linkage map of Ricinus communis L. based on genome-SSR markers. Ind. Crop. Prod. 2016;89:103–108. [Google Scholar]
- 79.Tomar D.S., Kumar S., Singh S.K., Goswami S., Li L. Molecular basis of high viscosity in concentrated antibody solutions: strategies for high concentration drug product development. mAbs. 2016;2(8):216–228. doi: 10.1080/19420862.2015.1128606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tomar R.S., Parakhia M.V., Rathod V.M., Thakkar J.R., Padhiyar S.M., Thummar V.D., Dalal H., Kothari V.V., Kheni J., Dhingani R.M., Sabara P., Golakiya B.A. Molecular mapping and identification of QTLs responsible for charcoal rot resistance in Castor (Ricinus communis L.) Ind. Crop. Prod. 2017;95:184–190. [Google Scholar]
- 81.Lander E.S., Green P., Abrahamson J., Barlow A., Daly M.J., Lincoln S.E., Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 82.Burr B., Burr F.A., Thompson K.H., Albertson M.C., Stuber C.W. Gene mapping with recombinant inbreds in maize. Genetics. 1988;118(3):519–526. doi: 10.1093/genetics/118.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Collard B.C., Jahufer M.Z.Z., Brouwer J.B., Pang E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica. 2005;142(1):169–196. [Google Scholar]
- 84.Elshire R.J., Glaubitz J.C., Sun Q., Poland J.A., Kawamoto K., Buckler E.S., Mitchell S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One. 2011;6(5) doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hamilton J.P., Buell R.C. Advances in plant genome sequencing. Plant J. 2012;70(1):177–190. doi: 10.1111/j.1365-313X.2012.04894.x. [DOI] [PubMed] [Google Scholar]
- 86.Yu A., Li F., Xu W., Wang Z., Sun C., Han B., Wang Y., Wang B., Cheng X., Liu A. Application of a high-resolution genetic map for chromosome-scale genome assembly and fine QTLs mapping of seed size and weight traits in castor bean. Sci. Rep. 2019;9(1):1–11. doi: 10.1038/s41598-019-48492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kump K.L., Bradbury P.J., Wisser R.J., Buckler E.S., Belcher A.R., Oropeza-Rosas M.A., Holland J.B. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat. Genet. 2011;43(2):163. doi: 10.1038/ng.747. [DOI] [PubMed] [Google Scholar]
- 88.Olukolu B.A., Tracy W.F., Wisser R., De Vries B., Balint-Kurti P.J. A genome-wide association study for partial resistance to maize common rust. Phytopathology. 2016;106(7):745–751. doi: 10.1094/PHYTO-11-15-0305-R. [DOI] [PubMed] [Google Scholar]
- 89.Sitonik C., Suresh L.M., Beyene Y., Olsen M.S., Makumbi D., Oliver K., et al. Genetic architecture of maize chlorotic mottle virus and maize lethal necrosis through GWAS, linkage analysis and genomic prediction in tropical maize germplasm. Theor. Appl. Genet. 2019;132(8):2381–2399. doi: 10.1007/s00122-019-03360-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Myles S., Peiffer J., Brown P.J., Ersoz E.S., Zhang Z., Costich D.E., Buckler E.S. Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell. 2009;21(8):2194–2202. doi: 10.1105/tpc.109.068437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Holland J.B. Genetic architecture of complex traits in plants. Curr. Opin. Plant Biol. 2007;10(2):156–161. doi: 10.1016/j.pbi.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 92.Jaiswal V., Gupta S., Gahlaut V., Muthamilarasan M., Bandyopadhyay T., Ramchiary N., Prasad M. Genome-wide association study of major agronomic traits in foxtail millet (Setaria italica L.) using ddRAD sequencing. Sci. Rep. 2019;9(1):1–11. doi: 10.1038/s41598-019-41602-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kang H.M., Zaitlen N.A., Wade C.M., Kirby A., Heckerman D., Daly M.J., Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178(3):1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang M.L., Dzievit M., Chen Z., Morris J.B., Norris J.E., Barkley N.A.…Yu J. Genetic diversity and population structure of castor (Ricinus communis L.) germplasm within the US collection assessed with EST-SSR markers. Genome. 2017;60(3):193–200. doi: 10.1139/gen-2016-0116. [DOI] [PubMed] [Google Scholar]
- 95.Xu W., Wu D., Yang T., Sun C., Wang Z., Han B., Li D.Z. Genomic insights into the origin, domestication and genetic basis of agronomic traits of castor bean. Genome Biol. 2021;22(1):1–27. doi: 10.1186/s13059-021-02333-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fan W., Lu J., Pan C., Tan M., Lin Q., Liu W.…Cui P. Sequencing of Chinese castor lines reveals genetic signatures of selection and yield-associated loci. Nat. Commun. 2019;10(1):1–11. doi: 10.1038/s41467-019-11228-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yeboah A., Lu J., Ting Y., Karikari B., Gu S., Xie Y.…Yin X. Genome-wide association study identifies loci, beneficial alleles, and candidate genes for cadmium tolerance in castor (Ricinus communis L.) Ind. Crop. Prod. 2021;171 [Google Scholar]
- 98.Brown D.C.W., Thorpe T.A. Crop improvement through tissue culture. World J. Microbiol. Biotechnol. 1995;11(4):409–415. doi: 10.1007/BF00364616. [DOI] [PubMed] [Google Scholar]
- 99.Birch R.G. Plant transformation: problems and strategies for practical application. Annu. Rev. Plant Biol. 1997;48(1):297–326. doi: 10.1146/annurev.arplant.48.1.297. [DOI] [PubMed] [Google Scholar]
- 100.Ahn Y.J., Vang L., McKeon T.A., Chen G.Q. High-frequency plant regeneration through adventitious shoot formation in castor (Ricinus communis L.) Cell Dev. Biol. Plant. 2007;43(1):9–15. [Google Scholar]
- 101.Sangduen N., Pongtongkam P., Ratisoontorn P., Jampatas R., Suputtitada S., et al. Tissue culture and plant regeneration of castor (Ricinus communis L.) SABRAO J Breed Genet. 1987;19:144. [Google Scholar]
- 102.Genyu Z. Callus formation and plant regeneration from young stem segments of Ricinus communis L. Genetic Manipulation in Crops. IRRI, Cassell Tycooly. 1988:393. [Google Scholar]
- 103.Sujatha M., Reddy T.P. Differential cytokinin effects on the stimulation of in vitro shoot proliferation from meristematic explants of castor (Ricinus communis L.) Plant Cell Rep. 1998;17(6):561–566. doi: 10.1007/s002990050442. [DOI] [PubMed] [Google Scholar]
- 104.Sujatha M., Sailaja M. Stable genetic transformation of castor (Ricinus communis L.) via Agrobacterium tumefaciens-mediated gene transfer using embryo axes from mature seeds. Plant Cell Rep. 2005;23(12):803–810. doi: 10.1007/s00299-004-0898-4. [DOI] [PubMed] [Google Scholar]
- 105.Malathi B., Ramesh S., Rao K.V., Reddy V.D. Agrobacterium-mediated genetic transformation and production of semilooper resistant transgenic castor (Ricinus communis L.) Euphytica. 2006;147(3):441–449. [Google Scholar]
- 106.Ganesh Kumari K., Ganesan M., Jayabalan N. Somatic organogenesis and plant regeneration in Ricinus communis. Biol. Plantarum. 2008;52(1):17–25. [Google Scholar]
- 107.Bertozzo F., Machado I.S. Culture media on in vitro development of castor bean (Ricinus communis L.) stem tips. Cienc. E Agrotecnol. 2010;34:1477–1482. [Google Scholar]
- 108.Lan D., Tao W., Wei J.H., Chao G. Study on tissue culture of castor (Ricinus communis L.) Xibei Shifan Daxue Xuebao (Ziran Kexue Ban) 2010;46:79–83. (In Chinese, with English abstract.) doi: CNKI:SUN:XBSF.0.2010-01-019. [Google Scholar]
- 109.Carvalho J.M.F.C., Silva M.M.A., Medeiros M.J.L., Araújo S.S., Milani M. Composition and concentrations of the culture medium in the multiple shoots in vitro of castor bean cultivating BRS Paraguaçu. Portuguese, with English Abstract) Revista Brasileira de Oleaginosas e Fibrosas. 2008;12:119–124. [Google Scholar]
- 110.Alam I., Sharmin S.A., Mondal S.C., Alam J., Khalekuzzaman M., Anisuzzaman M., Alam M.F. In vitro micropropagation through cotyledonary node culture of castor bean (Ricinus communis L.) Aust. J. Crop. Sci. 2010;4(2):81–84. [Google Scholar]
- 111.Gmitter F.G., Ling X.B., Deng X.X. Induction of triploid citrus plants from endosperm calli in vitro. Theor. Appl. Genet. 1990;80(6):785–790. doi: 10.1007/BF00224192. [DOI] [PubMed] [Google Scholar]
- 112.Sikdar A.K., Jolly M.S. Induced polyploidy in mulberry (Morus spp.): induction of tetraploids. Sericologia. 1994;34(1):105–122. [Google Scholar]
- 113.Chaturvedi R., Razdan M.K., Bhojwani S.S. An efficient protocol for the production of triploid plants from endosperm callus of neem, Azadirachta indica A. Juss. J. Plant Physiol. 2003;160(5):557–564. doi: 10.1078/0176-1617-00884. [DOI] [PubMed] [Google Scholar]
- 114.La Rue C.D. Regeneration of endosperm of gymnosperms and angiosperms. Am. J. Bot. 1944;36:798. [Google Scholar]
- 115.Mohan Ram H.M., Satsangi A. Induction of cell divisions in the mature endosperm of Ricinus communis during germination. Curr. Sci. 1963;32(1):28–30. [Google Scholar]
- 116.Ahn Y.J., Chen G.Q. In vitro regeneration of castor (Ricinus communis L.) using cotyledon explants. Hortscience. 2008;43(1):215–219. [Google Scholar]
- 117.Sailaja M., Tarakeswari M., Sujatha M. Stable genetic transformation of castor (Ricinus communis L.) via particle gun-mediated gene transfer using embryo axes from mature seeds. Plant Cell Rep. 2008;27(9):1509–1519. doi: 10.1007/s00299-008-0580-3. [DOI] [PubMed] [Google Scholar]
- 118.Rahman M.A., Bari M.A. Callus induction and cell culture of castor (Ricinus communis L. CV. Shabje) J. Bio. Sci. 2012;20:161–169. [Google Scholar]
- 119.Zalavadiya V.K., Mehta D.R., Javia R.M., Padhiyar S.M., Madariya R.B. Somatic organogenesis and plant regeneration in castor (Ricinus communis L.) Asian J. Bio. Sci. 2014;9:43–52. [Google Scholar]
- 120.Ganesh Kumari K., Jayabalan N. A comparative study of PGRs and Pluronic F68, a surfactant on in vitro regeneration of castor (Ricinus communis L.) using shoot tip and cotyledonary node explant. Int. J. Curr. Biotechnol. 2015;3:1–14. [Google Scholar]
- 121.Zhang J.X., Wang X.Y., Feng Z.Z., Geng X.J., Mu S.M., Huo H.Y., Chen Y.S. In vitro establishment of a highly effective method of castor bean (Ricinus communis L.) regeneration using shoot explants. J. Integr. Agric. 2016;15(6) 1417-142. [Google Scholar]
- 122.Liu Y., Yan S., Yang F., Li D., Tang J., Liu G.…Yang Y. IOP Conference Series: Materials Science and Engineering (Vol. 275, No. 1, P. 012011) IOP Publishing; 2017. High concentration of benzyladenine solution stimulates anthers for inducing callus in Ricinus communis L. December) [Google Scholar]
- 123.Louis G.C., Okafor C.U., Okezie C.E.A. Research article nutrient requirements for in vitro propagation of Ricinus communis L. mature zygotic embryos. Int. J. Agric. Res. 2018;13:19–25. [Google Scholar]
- 124.Alexandrov O.S., Petrov N.R., Varlamova N.V., Khaliluev M.R. An optimized protocol for in vitro indirect shoot organogenesis of impala bronzovaya and zanzibar green Ricinus communis L. Varieties. Horticulturae. 2021;7(5):105. [Google Scholar]
- 125.Liu Y., Yan S., Yang F., Li D., Tang J., Liu G.…Yang Y. IOP Conference Series: Materials Science and Engineering (Vol. 275, No. 1, P. 012011) IOP Publishing; 2017. High concentration of Benzyladenine solution stimulates anthers for inducing callus in Ricinus communis L. December. [Google Scholar]
- 126.Arzani A., Ashraf M. Cultivated ancient wheats (Triticum spp.): a potential source of health‐beneficial food products. Compr. Rev. Food Sci. Food Saf. 2017;16(3):477–488. doi: 10.1111/1541-4337.12262. [DOI] [PubMed] [Google Scholar]
- 127.Endo Y., Mitsui K., Motizuki M., Tsurugi K. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J. Biol. Chem. 1987;262(12):5908–5912. [PubMed] [Google Scholar]
- 128.Sousa N.L., Cabral G.B., Vieira P.M., Baldoni A.B., Aragão F.J. Bio-detoxification of ricin in castor bean (Ricinus communis L.) seeds. Sci. Rep. 2017;7(1):1–9. doi: 10.1038/s41598-017-15636-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen G.Q., He X., Liao L.P., McKeon T.A. 2S albumin gene expression in castor plant (Ricinus communis L.) J. Am. Oil Chem. Soc. 2004;81(9):867–872. [Google Scholar]
- 130.Pinkerton S.D., Rolfe R., Auld D.L., Ghetie V., Lauterbach B.F. Selection of castor for divergent concentrations of ricin and Ricinus communis agglutinin. Crop Sci. 1999;39:353–357. [Google Scholar]
- 131.Baldoni A.B., Carvalho M.H.D., Sousa N.L., Nóbrega M.B.D.M., Milani M., Aragão F.J.L. Variability of ricin content in mature seeds of castor bean. Pesqui. Agropecuária Bras. 2011;46:776–779. [Google Scholar]
- 132.McKeon T.A., Chen G.Q. vol. 6. US Patent; 2003. p. 620. (Transformation of Ricinus communis L., the castor Plant). 986. [Google Scholar]
- 133.Kumar A.M., Sreevathsa R., Reddy K.N., Ganesh P.T., Udayakumar M. Amenability of castor to an Agrobacterium-mediated in planta transformation strategy using a cry1AcF gene for insect tolerance. Journal of Crop Science and Biotechnology. 2011;14:125–132. [Google Scholar]
- 134.Patel M.K., Joshi M., Mishra A., Jha B. Ectopic expression of SbNHX1 gene in transgenic castor (Ricinus communis L.) enhances salt stress by modulating physiological process. Plant Cell Tissue Organ Cult. 2015;122:477–490. [Google Scholar]
- 135.Sánchez-Álvarez A., Ruíz-López N., Moreno-Pérez A.J., Martínez-Force E., Garcés R., Salas J.J. Agrobacterium-mediated transient gene expression in developing Ricinus communis seeds: a first step in making the castor oil plant a chemical biofactory. Front. Plant Sci. 2019;10:1410. doi: 10.3389/fpls.2019.01410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yu A., Li F., Liu A. Comparative proteomic and transcriptomic analyses provide new insight into the formation of seed size in castor bean. BMC Plant Biol. 2020;20(1):1–20. doi: 10.1186/s12870-020-2249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang X., Li M., Liu X., Zhang L., Duan Q., Zhang J. Quantitative proteomic analysis of castor (Ricinus communis L.) seeds during early imbibition provided novel insights into cold stress response. Int. J. Mol. Sci. 2019;20(2):355. doi: 10.3390/ijms20020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rivarola M., Foster J.T., Chan A.P., Williams A.L., Rice D.W., Liu X., Rabinowicz P.D. Castor bean organelle genome sequencing and worldwide genetic diversity analysis. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0021743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nogueira F.C., Palmisano G., Schwammle V., Soares E.L., Soares A.A., Roepstorff P.…Campos F.A. Isotope labeling-based quantitative proteomics of developing seeds of castor oil seed (Ricinus communis L.) J. Proteome Res. 2013;12(11):5012–5024. doi: 10.1021/pr400685z. [DOI] [PubMed] [Google Scholar]
- 140.Brown A.P., Kroon J.T., Swarbreck D., Febrer M., Larson T.R., Graham I.A., Slabas A.R. Tissue-specific whole transcriptome sequencing in castor, directed at understanding triacylglycerol lipid biosynthetic pathways. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang B., Zhang Y., Haque M.E., Xu W., Li F., Liu A. Transcriptomic analyses reveal complex and interconnected sucrose signaling cascades in developing seeds of castor bean. J. Plant Physiol. 2018;221:1–10. doi: 10.1016/j.jplph.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 142.Tan M., Xue J., Wang L., Huang J., Fu C., Yan X. Transcriptomic analysis for different sex types of Ricinus communis L. during development from apical buds to inflorescences by digital gene expression profiling. Front. Plant Sci. 2016;6:1208. doi: 10.3389/fpls.2015.01208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu Z., Yang Q., Xu F., Ouyang Y., Geng X. 2020. Transcriptome analysis of developing castor bean seeds and identification of ricinoleic acid biosynthesis genes, Biol Plantarum 65; pp. 273–282. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article. Data associated with this study been not deposited into a publicly available repository.