Abstract
Posttranscriptional silencing of chalcone synthase (Chs) genes in petunia transformants occurs by introducing T-DNAs that contain a promoter-driven or promoterless Chs transgene. With the constructs we used, silencing occurs only by T-DNA loci which are composed of two or more T-DNA copies that are arranged as inverted repeats (IRs). Since we are interested in the mechanism by which these IR loci induce silencing, we have analyzed different IR loci and nonsilencing single-copy (S) T-DNA loci with respect to the expression and methylation of the transgenes residing in these loci. We show that in an IR locus, the transgenes located proximal to the IR center are much more highly methylated than are the distal genes. A strong silencing locus composed of three inverted T-DNAs bearing promoterless Chs transgenes was methylated across the entire locus. The host Chs genes in untransformed plants were moderately methylated, and no change in methylation was detected when the genes were silenced. Run-on transcription assays showed that promoter-driven transgenes located proximal to the center of a particular IR are transcriptionally more repressed than are the distal genes of the same IR locus. Transcription of the promoterless Chs transgenes could not be detected. In the primary transformant, some of the IR loci were detected together with an unlinked S locus. We observed that the methylation and expression characteristics of the transgenes of these S loci were comparable to those of the partner IR loci, suggesting that there has been cross talk between the two types of loci. Despite the similar features, S loci are unable to induce silencing, indicating that the palindromic arrangement of the Chs transgenes in the IR loci is critical for silencing. Since transcriptionally silenced transgenes in IRs can trigger posttranscriptional silencing of the host genes, our data are most consistent with a model of silencing in which the transgenes physically interact with the homologous host gene(s). The interaction may alter epigenetic features other than methylation, thereby impairing the regular production of mRNA.
Genes that are packaged into heterochromatin or whose promoters are heavily methylated are often transcriptionally repressed. These epigenetic gene inactivation mechanisms are known to be involved in genomic imprinting (2) and X-chromosome inactivation in mammals (78), in the control of homeotic genes in flies (29), and in the repression of silent mating-type loci in yeast (33). Also, the expression of transgenes in genetically modified animals (13–15), plants (4, 12, 37, 42, 64), and lower eukaryotes (52) is often epigenetically controlled. Transgenes usually integrate into the genome at random positions, and their expression is therefore affected by chromosomal position effects. Besides the chromosomal position, the number of transgene copies integrated at a particular chromosomal site affects their expression (15, 42, 64). Tandemly repeated transgenes are often silenced. This repeat-induced gene silencing (RIGS) has also been observed in untransformed plants for tandemly linked homologous host genes (18, 51, 66). RIGS is frequently associated with an increased level of DNA methylation (1, 26, 31, 36, 73). However, despite the lack of DNA methylation in Drosophila (62), RIGS does occur in Drosophila, where it is accompanied by the formation of heterochromatin (15, 16).
RIGS may result from interactions between the linked repeats; however, repeat-induced silenced genes may also interact with homologous sequences that are unlinked. For example, multicopy loci whose transgenes or endogenous genes are silenced appear to be able to transfer the silent state onto an allelic or nonallelic homologue in trans (26, 30, 36, 38, 73). If the silencing locus is methylated, the homologous sequences of the target locus also become methylated, suggesting that the two loci have interacted with each other, presumably by DNA pairing. The most convincing evidence for interactions between homologous sequences comes from studies of repeat-induced point mutation (RIP) in Neurospora crassa (58, 59) and methylation-induced premeiotically (MIP) in Ascobolus immersus (24, 53).
In plants, transgenes can be silenced transcriptionally (transcriptional gene silencing [TGS]) and posttranscriptionally (posttranscriptional gene silencing [PTGS]) (reviewed in references 4, 12, 42, and 64). For PTGS, the transgene-activated RNA degradation machinery attacks any homologous RNA that is present in the cell, irrespective of the origin. Although the mechanism of the sequence-specific RNA degradation is largely unknown, several studies suggest that it can be activated in different ways. One is via excessive production of transgene RNA (10, 11, 19, 22, 34, 50, 60, 75). However, not all cases of PTGS are associated with highly transcribed transgenes (20, 45, 69). It has therefore been proposed that only RNAs that are somehow aberrant and which may comprise a small fraction of the total RNA pool may trigger the RNA degradation machinery (4, 34). Besides possibly being by-product of an excessive RNA synthesis, these aberrant RNAs may also be derived from the transcription of transgenes that are methylated and/or located in repeats (20, 27, 57, 72). If transgenes that are identical or sufficiently homologous to endogenous genes are used, the aberrant RNAs may also be derived from the endogenous gene(s), which somehow would be triggered by the transgenes (4, 63). We are examining this possibility by studying the posttranscriptional silencing of the flower pigmentation gene chalcone synthase (Chs) in petunia. Silencing of Chs expression results in white corollas or corollas bearing white sectors (28, 46, 69, 70). Although Chs silencing can be induced by highly transcribed Chs transgenes (50), it can also be induced by Chs transgenes that are transcriptionally mostly inactive, as was found when promoterless transgenes were used (69). However, when the transgenes are transcriptionally inactive, achieving silencing seems to require that two or more T-DNAs, which carry the transgenes, be inserted at the same chromosomal site where the T-DNAs have to be arranged as inverted repeats (IR) (63). This observation suggests that the palindromic sequence arrangement is important for inducing PTGS, perhaps by facilitating cross talk with the homologous host gene(s). To gain more insight into the properties of IR loci and to obtain evidence that IR loci are indeed able to undergo cross talk with unlinked homologous sequences, we examined different IR loci and, when present in the primary transformants, the accompanying nonsilencing single-copy (S) T-DNA loci. We show that promoter-driven transgenes located proximal to the center of an IR locus that is composed of two T-DNAs are more highly methylated and transcribed at a lower level than the transgenes located distal to the center of the IR. Transcription of the promoterless Chs transgenes could not be detected. The methylation and expression characteristics of the S transgene loci were comparable to those of the unlinked partner IR loci, suggesting that in the primary transformant, the two transgene loci have interacted. We discuss the possibility that an IR locus also interacts with the homologous endogenous genes, thereby triggering their silencing.
MATERIALS AND METHODS
T-DNA constructs and plant material.
The ChsA T-DNA constructs pSE19, pSE6, and pSE21 and the petunia V26 primary transformants PSE6-2, PSE19-3, PSE19-1, PSE21-1, and PSE21-6, which are transgenic for these constructs, have been described by Van Blokland et al. (69). The analyses described in this report were performed on cuttings of progeny plants of PSE6-2 (W7017 and W7016), PSE19-3 (S5055 and V7055), PSE19-1 (W7001), PSE21-6 (W7003), and PSE21-1 (W7002) (63). The T-DNA insertions in these transformants and their zygosity are indicated in the figures. The plants were maintained in a greenhouse under a light regimen of 16 h of light and 8 h of darkness.
GUS enzyme activity.
To examine β-glucuronidase (GUS) expression from the uidA (GUS)-ChsA transgenes, corolla limbs at developmental stage 4, at which corollas become pigmented, and young leaves were collected and ground in liquid N2 in the presence of Dowex-1 (Sigma) to remove flavonoids. GUS enzyme activities in protein extracts were measured as described previously (63).
Nucleic acid purification and blot hybridization conditions.
For the isolation of RNA, DNA, and nuclei, about 50 corollas (stage 4) of each plant were pooled and ground in liquid N2. One-tenth of the material was used for the isolation of RNA and DNA. Nuclei were isolated from the remainder. RNA was obtained by 4 M LiCl precipitation. For Northern blot analysis 10 μg of total RNA was size fractionated on a 1.2% formaldehyde gel. Before loading, 1 μl of ethidium bromide (1 mg/ml) was added to each sample.
Genomic DNA was isolated from the supernatant of the LiCl RNA precipitation by the addition of 2 volumes of ethanol followed by a 15-min centrifugation at 13,000 rpm in an Eppendorf centrifuge 5414. The DNA pellet was resuspended in 300 μl of Tris-EDTA (TE), after which 300 μl of CTAB buffer (0.2 M Tris-HCl [pH 7.5], 50 mM EDTA, 2 mM NaCl, 2% N-cetyl-N,N,N-trimethylammonium bromide) was added, followed by a 15-min incubation at 65°C. After a single chloroform-isoamyl alcohol (24:1) extraction, the DNA was precipitated by the addition of 600 μl of isopropanol. The pellet was washed in 70% ethanol and dissolved in 300 μl of TE. The DNA (2.5 to 5 μg) was digested with the appropriate restriction enzymes for at least 14 h. The DNA fragments were separated on a 1.8% agarose gel overnight at low voltage.
RNA and DNA were blotted onto Hybond-N+ membranes (Amersham) by capillary blotting, followed by alkali fixation essentially as described by the manufacturer. The RNA and DNA blots were hybridized at 65 and 60°C, respectively, for about 18 h in a solution containing 10% dextran sulfate, 1% sodium dodecyl sulfate (SDS), 50 mM Tris (pH 7.5), 1 M NaCl, 0.1 mg of sheared herring sperm DNA/ml, and a double-stranded, 32P-random-primed DNA probe. After the hybridizations, the filters were washed in SSPE buffer (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) with a final 5-min wash in 0.1× SSPE–0.1% SDS at 65°C. The hybridizing fragments were visualized and if necessary quantified with a PhosphorImager (Molecular Dynamics) and ImageQuant software. For rehybridizations, the RNA and DNA filters were washed in 0.1 and 0.5% SDS, respectively, at 100°C for 5 min. The 3′nos, PCaMV, uidA, and ChsA probes have been described by Stam et al. (63). The DfrA probe was a BamHI fragment from VIP178 (70), and the nptII probe was an EcoRI fragment from PCGN1548 (39).
Run-on transcription assay.
The isolation of nuclei and the run-on transcription assays were performed essentially as described by Van Blokland et al. (69). After the nascent RNA hybridization, the final wash was done at 60°C instead of 55°C. The hybridization signals were quantified with a PhosphorImager. For the detection of radiolabelled nascent-sense transcripts, single-stranded M13 recombinant phage DNA was used. The M13-nptII clone for detecting sense nptII RNA was made by ligating a HindIII-EcoRI nptII fragment into M13mp19. The other M13 recombinant phages have been described previously (69).
RESULTS
Position-dependent expression of transgenes located in inverted repeats.
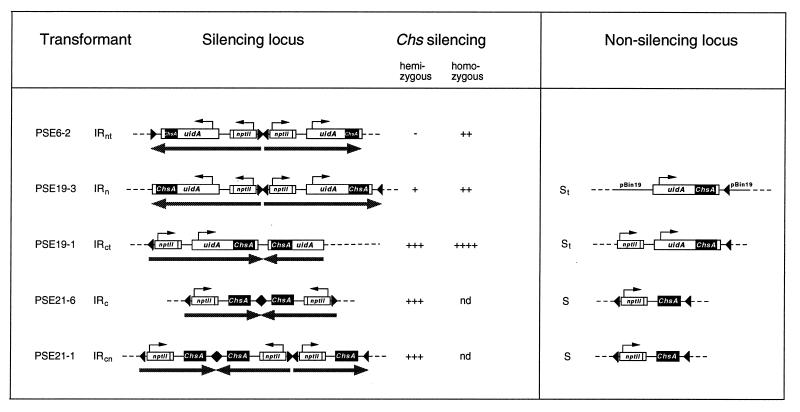
Previously, we generated petunia transformants in which the Chs genes were silenced to various degrees (69). These transformants contained multiple T-DNA loci. By backcrosses, progeny plants which carried single loci were obtained, allowing us to characterize these loci in detail and to identify those that confer silencing (63). This collection of plants was also used for the studies described in this report. Figure 1 shows the physical map of the various T-DNA loci in these plants and their silencing capacity. Three types of IR loci can be distinguished: IR loci with the nptII genes proximal to the center of the IR, indicated by IRn; IR loci with the Chs transgenes near the center, indicated by IRc; and an IR locus which consists of three inverted T-DNAs, indicated by IRcn. The nonsilencing single-copy T-DNA loci are indicated by S. A locus containing a truncated T-DNA is indicated by a subscript t, as the 19-1 IRct locus.
FIG. 1.
Overview of the Chs silencing and nonsilencing T-DNA loci examined in this study. The Chs transformants and the identification of Chs silencing IR loci and the nonsilencing S loci present in these plants have been described previously (63, 69). All T-DNA constructs used in this study contained the neomycin phosphotransferase selectable marker gene (nptII) driven by the nopaline synthase promoter (Nos). The T-DNAs of the PSE6 and PSE19 transformants contained a CaMV-35S promoter-driven chimeric gene composed of the uidA coding region fused to the 5′ half (PSE6) or the full-length (PSE19) ChsA cDNA. The T-DNA of the PSE21 transformants contains the full-length ChsA cDNA. The small arrows in the maps indicate the transcription start sites of the Nos promoter and CaMV-35S promoter. These transgenes are flanked by the 3′nos polyadenylation region, which is indicated by a small open box. The arrowheads flanking the nptII and (uidA-)ChsA transgenes indicate the right and left border of the T-DNAs, respectively. The arrows below the maps of the silencing loci depict the palindromic arrangement of the integrated T-DNAs. The dashed lines indicate flanking plant DNA. Chs silencing by the IR loci in homozygous plants is stronger than in hemizygous plants, as indicated to the right of the physical maps (−, no silencing; +, most corolla limbs contain ≤5% white tissue; ++, about 5 to 50% white tissue; +++, about 50 to 95% white tissue; ++++, >95% white tissue; nd, not determined). pBin19, cointegrated pBin19 vector DNA.
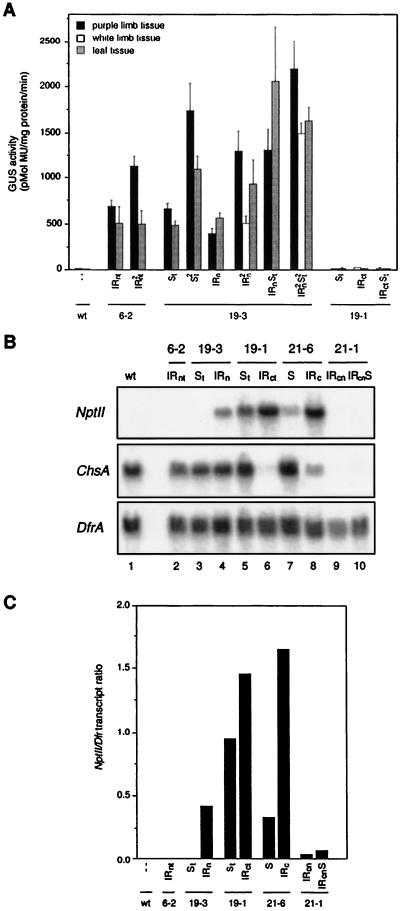
We first examined the expression of the nptII and uidA-ChsA transgenes and of the endogenous ChsA genes in transformants containing either an IRn, IRc, or S locus or combinations of an IR and S locus. Northern analysis showed that in plants containing an IRc or IRcn locus, the ChsA mRNA level in the corollas of these plants was drastically reduced or undetectable, indicative of Chs silencing (Fig. 2B, middle panel, lanes 6 and 8 through 10). The 6-2 IRn and 19-3 IRn loci are weak silencing loci (63) (Fig. 1), as illustrated by the almost wild-type Chs mRNA levels in these transformants (Fig. 2B, middle panel, lanes 2 and 4). In fact, the 6-2 IRn locus induces silencing only in homozygous plants (63) (Fig. 1). The control hybridization with a dihydroflavonol-4-reductase (DfrA) probe, which is also a pigmentation gene, shows that the expression of this gene is not affected by the Chs IR loci (Fig. 2B, bottom panel).
FIG. 2.
Expression of transgenes residing in the S and IR loci. (A) The expression of the uidA-ChsA transgenes in progeny plants of the transformants PSE6-2, PSE19-3, and PSE19-1 was examined by measuring GUS activities. For each genotype, the activities were measured in four different leaves and corolla limbs derived from one to five different plants. If possible, the activities in purple and white corolla sectors were measured separately. This was not possible with the transformants PSE6-2 (IR2nt), PSE19-3 (IRn), and PSE19-3 (IRnSt) because the white sectors were relatively small. GUS activity is expressed as the mean and standard error of the mean (SEM). wt, wild type. (B) Northern blot hybridizations showing expression of the nptII transgenes (top), the ChsA host genes (middle), and the DfrA host genes (bottom), which served as an internal control. The RNAs were isolated from corolla limbs taken from plants that were hemizygous for the T-DNA loci indicated at the top. (C) The intensities of the nptII and DfrA bands in panel B were quantified with a PhosphorImager, and the ratios of nptII to DfrA transcript levels were calculated. −−, no T-DNA present. For further details, see the text.
Expression of the uidA-ChsA transgenes could not be examined by Northern analysis because the chimeric transcripts appeared unstable (69). It was, however, possible to measure the activity of the GUS enzyme, which is encoded by the uidA part of the chimeric transgene transcript. If possible, the GUS activities in the white and purple sectors of the corolla were measured separately. This revealed that the uidA-ChsA genes located distal to the IR center are clearly expressed (Fig. 2A, lanes indicated by 6-2 or by 19-3 IRn), whereas the proximal genes were not (19-1, IRct). This position-dependent expression pattern was also observed for the nptII genes, because the proximal nptII genes of the two IRn loci were not expressed or were expressed at only a low level (Fig. 2B, lanes 2 and 4), while the distal genes of the 19-1 and 21-1 IR locus were highly expressed. It is important to note that each locus will be affected by different chromosomal position effects and that one can therefore compare only the expression levels of transgenes that reside in the same locus. Thus, even though the proximal nptII genes of the 19-3 IRn locus are active, their expression is relatively lower than that of the distal uidA-Chs genes. The difference between the expression of distal and proximal genes was even more pronounced when the transcription rates were measured by run-on assays, which are described below. The three nptII genes of the 21-1 IRcn locus were all silenced since no transcripts were detected in corollas containing this locus (Fig. 2B, top panel, lane 10). Figure 2C shows the quantitation of the nptII Northern data, which are expressed as the ratio of the nptII to DfrA signals.
S transgene loci do not induce silencing (Fig. 2B, lanes 3, 5, and 7), even in homozygous plants (63). Except for the 21-1 S locus, the other S and partner IR loci are on different chromosomes (63), and we expected the transgenes residing in the S and IR loci to be expressed at very different levels. However, as shown in Fig. 2A and B, this was not the case. The expression of the same gene in the S and partner IR locus appeared to be correlated. For example, the nptII genes in both the 19-1 S and IRc locus are expressed at about the same level (lanes 5 and 6), while the nptII genes in the 21-1 S and IRcn locus are all silent. Furthermore, the uidA-ChsA transgenes of both the 19-1 S and IRc loci are not expressed (Fig. 2A), whereas the uidA-ChsA transgenes of the 19-3 S and IRn loci are expressed, with only a twofold difference in expression levels. Only for the 21-6 S and IRc loci did the expression of the nptII genes differ more, and the increase was only slight (Fig. 2B, lanes 7 and 8, and Fig. 2C).
These results indicate that the transgenes of the S and IR loci have common features. The data shown in Fig. 2A also indicate that the different loci do not affect each others’ expression since they do not induce posttranscriptional silencing. For example, in the purple corolla sectors of plants homozygous for the 6-2 IRn, 19-3 IRn, or S locus, the expression of the uidA-ChsA genes was about twice as high as in hemizygous plants (Fig. 2A, IR2 versus IR and St2 versus St). Also, in plants carrying both the 19-3 IRn and S loci in a hemizygous or homozygous state, the GUS activity was approximately proportional to the number of uidA-ChsA transgenes (Fig. 2A, IRnSt and IRn2St2). A lower GUS activity was observed only in the white corolla sectors of transformants homozygous for the 19-3 IRn locus, where it was 2.5-fold lower than in the pigmented sectors (Fig. 2A, IRn2). This indicates that in the white sectors, the uidA-ChsA transgenes and the endogenous Chs genes are cosuppressed. The residual GUS activity that was measured may come from the L2 cell layer of the corolla in which the uidA-ChsA gene may not be silenced. Silencing may occur only in the upper and lower epidermal L1 cells, where the endogenous Chs genes are highly expressed (35).
Transcriptional silencing of transgenes.
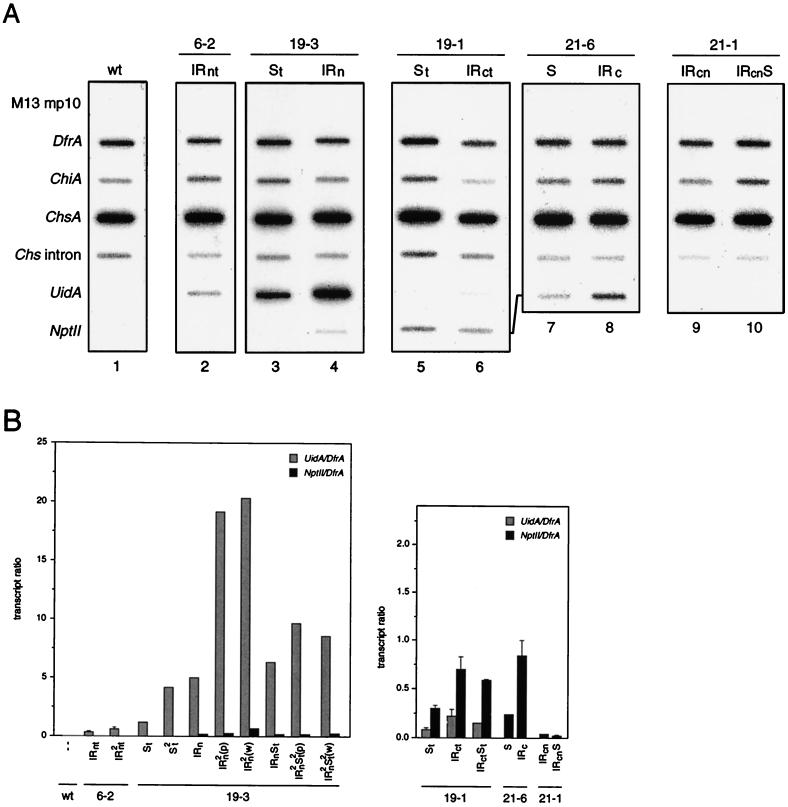
To distinguish between transcriptional and posttranscriptional silencing of the transgenes, we performed run-on transcription assays with corolla nuclei that were obtained from the same plants in which the expression of the transgenes was examined. The nuclear [32P]UTP-labelled nascent RNA was hybridized to filters containing gene and strand-specific single-stranded M13 recombinant phage DNAs. Figure 3A shows a series of representative hybridizations, while Fig. 3B shows the quantitations of all the assays and hybridizations we have done. This revealed that when the transcription rates of the proximal and distal genes of the same IR locus are compared, the proximal genes are transcribed at a lower level than the distal genes, if they are transcribed at all. This is clearly demonstrated for the proximal nptII and distal uidA-ChsA genes of the 6-2 IRn (Fig. 3A, lane 2) and 19-3 IRn (lane 4) loci. The higher transcription of the distal uidA-ChsA transgenes of the 6-2 and 19-3 IRn loci could be explained in part by their stronger 35SCaMV promoter compared to the nos promoter driving the nptII gene. However, the results obtained with the 19-1 IRc locus (lane 6) show that even with this stronger promoter, the proximal uidA-ChsA gene can be transcriptionally silenced whereas the distal nptII gene is not. The nptII genes of the 21-1 IRcn locus (lane 9) and S locus (lane 10) are all transcriptionally silent, since no nascent nptII RNA was detectable. Note that the transcription rates of the nptII and uidA-ChsA transgenes of the S loci (Fig. 3, lanes indicated by S) paralleled the rates of these genes residing in the partner IR loci.
FIG. 3.
Transcriptional activity of transgenes as determined by nuclear run-on assays. (A) Representative examples of nascent RNA hybridizations. Corolla nuclei were obtained from the transformants listed at the top. These plants were hemizygous for the T-DNA loci indicated. Nuclei from untransformed V26 (wild type [wt]) were used as control. The slot blot filters contained gene and strand-specific single-stranded M13 DNAs that were able to hybridize to the RNAs indicated at the left. DfrA and ChiA served as internal controls, and the ChsA intron signal indicates the transcription level of the endogenous Chs genes. M13mp10 was the vector in which the gene sequences were cloned. In addition to the hybridizations shown here, several others were performed with material from transformants carrying the same or other T-DNA loci, as depicted below the bar diagrams. (B) The signals obtained with the uidA, nptII, and DfrA M13 DNAs were quantified with a PhosphorImager, and the ratios of the uidA to DfrA transcripts (grey bars) and nptII to DfrA transcripts (black bars) were calculated. The values are expressed as the mean and SEM when n > 1 (n ranged from 2 to 5).
The run-on data show that transgene expression is transcriptionally controlled; however, the 19-3 IRn transformant is an exception. In the white and pigmented corolla sectors of this transformant, the transcription rates of the uidA-ChsA transgenes were about the same [Fig. 3B, 19-3, IRn2(w) and IRn2(p)] (69), whereas in the white sectors, the GUS activity was lower than in the pigmented sectors (Fig. 2A, 19-3, IRn2). This shows that in the white sectors, the uidA-ChsA transgenes are posttranscriptionally silenced, like the endogenous ChsA genes.
An important question about the mechanism by which promoterless Chs transgene IR loci induce silencing is whether they provide any transcripts that might be implicated in the posttranscriptional silencing process. Although promoterless transgenes are expected to be transcriptionally silent, it is possible that they are transcribed by some readthrough transcription from the upstream nptII gene(s) or from an adjacent plant promoter. For the 21-6 IRc locus, this can occur only by readthrough transcription, because the Chs sequences are juxtaposed and sandwiched between the nptII genes. Detection of possible readthrough Chs transgene transcripts is complicated by the fact that in corollas, the endogenous Chs genes are already highly transcribed. We have previously shown that very little, if any, Chs transgene RNA is produced in the primary PSE21 transformants (69). The data obtained with the progeny of these transformants confirm this. The amount of nascent Chs RNA detected in the transformants in which the Chs genes are silenced is the same as in wild-type corollas (Fig. 3A, lanes 8 to 10; quantitations not shown). This strongly suggests that the promoterless Chs transgenes are indeed transcriptionally inactive.
Methylation of inverted T-DNA repeat loci.
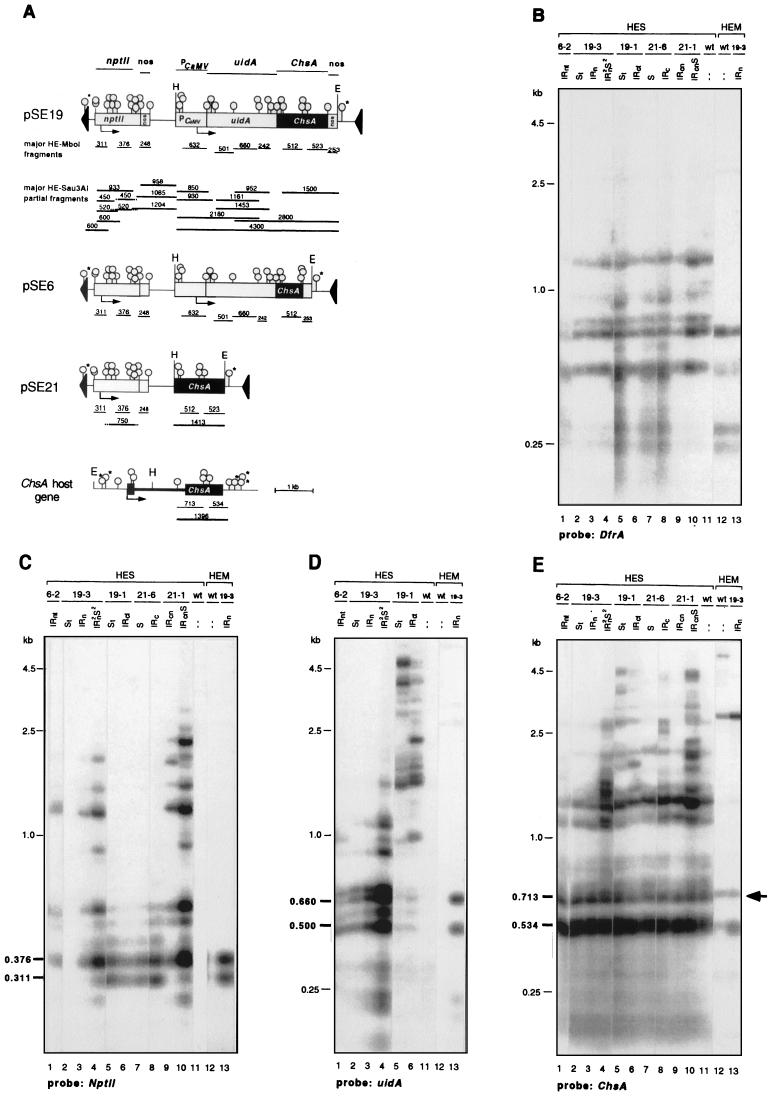
Transcriptional silencing in plants is often associated with DNA methylation (19, 43, 47, 49). Given the differential transcriptional silencing of transgenes in the IR and S loci, it was of interest to see whether this was correlated with a position-dependent methylation of the T-DNA loci. We investigated this by digesting genomic DNA with HindIII and EcoRI and with the methylation-sensitive restriction enzyme Sau3AI (HES digest) or the insensitive isoschizomer MboI (HEM digest). Sau3AI is unable to cleave the sequence GATC when it is GAT5mC/GATC hemimethylated or GAT5mC/GAT5mC bimethylated (65), whereas MboI cleaves these sites. Although some Sau3AI sites are clustered, many are spread along the T-DNAs, which allowed us to get an impression of the methylation status of various parts of the transgene loci. The degree of methylation was estimated on the basis of the size of the partially cleaved fragments in the Sau3AI digests (Fig. 4A), the detection of the same fragments by neighboring probes, and the relative intensities of the bands as measured with a PhosphorImager. The results of this analysis are summarized in Fig. 5, which shows the methylation patterns of the different T-DNA loci. One of the major conclusions is that within a T-DNA IR locus, IRn or IRc, sequences proximal to an IR center are much more highly methylated than are sequences that are more distal to the center and, furthermore, that when a locus is more repetitive, like the 21-1 IRcn locus, all sequences are severely methylated. The data supporting this are described below.
FIG. 4.
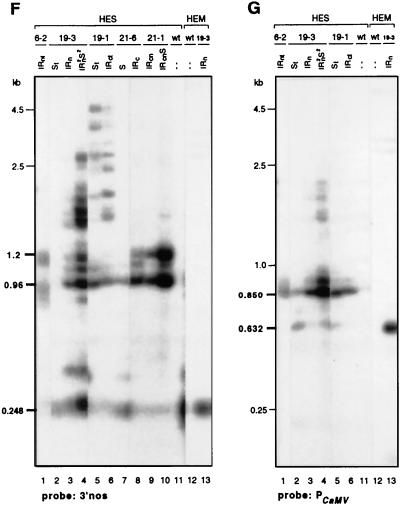
Methylation of T-DNA loci. The extent and level of DNA methylation was determined by digesting genomic DNA from corollas with the methylation-sensitive restriction enzyme Sau3AI. (A) Maps of the T-DNA constructs pSE19, pSE6, pSE21, and the endogenous ChsA host gene. The open circles mark the positions of the Sau3AI and MboI sites, and the EcoRI (E) and HindIII (H) sites are indicated. The probes used for the Southern blot hybridizations are shown at the top. The most prominent restriction fragments after a complete digestion by HindIII-EcoRI-MboI (HE-MboI) are indicated by thin lines beneath the maps. Partially cleaved fragments after digestion with HindIII-EcoRI-Sau3AI (HE-Sau3AI) are indicated by the thick lines. Partial fragments from the pSE6 and PSE21 constructs can be deduced from those indicated below pSE19. The GATC sites indicated by an asterisk (○∗) could not be analyzed because they were out of the reach of the probes. (B to G) Southern blot hybridizations of the transformants indicated at the top of the panels. The T-DNA loci that these transformants contain are also indicated. The blots were hybridized with a DfrA probe (B), nptII probe (C), uidA probe (D), ChsA probe (E), 3′nos probe (F), and PCaMV probe (G). Of the HEM digests, only the data obtained with the untransformed V26 line (wild type [wt]) and the transformant PSE19-3 (IRn) are shown; other transformants carrying the same sequences gave the same results (data not shown). PstI-digested phage lambda DNA was used as the size marker. The sizes in bold type refer to fragments mentioned in the text. The arrow in panel E indicates the position of the 713-bp fragment, which is unique for the endogenous ChsA gene. The intensity of this band is about the same in all lanes, relative to the internal DfrA control, irrespective whether the gene was silenced.
FIG. 5.
(A) Methylation pattern of the various T-DNA loci, where the black boxes indicate the positions of the ChsA sequences. (B) Endogenous ChsA gene, which contains a single intron indicated by the thick line separating the two exons. By using the Southern blot data in Fig. 4, the methylation status of the Sau3AI sites (open and solid circles) and EcoRI sites (E, triangles) was determined. The following criteria were used: the size of the hybridizing partially cleaved fragments; the intensity of the bands, which was quantified with a PhosphorImager; and whether the same fragment was detected by neighboring probes. Sau3AI sites that were not methylated or were barely methylated are indicated by open circles; sites indicated by solid circles are hypermethylated; sites indicated by partially filled circles are moderately methylated; and shaded circles denote sites that are moderately to severely methylated. This classification is also used to indicate the methylation status of the EcoRI sites indicated by the triangles. H, HindIII; E, EcoRI. For details about the maps, see the text and the legends to Fig. 1 and 4.
To be able to compare the hybridization patterns of the T-DNA loci from the different transformants, we first tested whether Sau3AI had digested the DNAs equally by hybridizing the blots with a probe for the endogenous DfrA gene (Fig. 4B). This revealed that the DfrA gene is already partially methylated. In contrast to the MboI digests (lanes 12 and 13), Sau3AI (HES) gives rise to a number of longer fragments, but since the majority of these fragments are present in all DNA samples, the DNAs are digested to the same extent.
The blots were next hybridized to a probe specific for the nptII transgenes. In the Sau3AI digests of the transformants containing the IRc loci (Fig. 4C, 19-1 or 21-6, lanes 6 and 8), two major fragments (of 311 and 376 bp) were detected; these fragments were also seen in the MboI digests of the 19-3 IRn transformant (HEM; lane 13). This shows that the distal nptII genes of these IRc loci are mostly unmethylated. This is different in the transformants containing the IRn loci (Fig. 4C, lanes 1, 3, and 4), in which the nptII genes are proximal to the IR center or the IRcn locus (lanes 9 and 10). In the Sau3AI digests, the intensity of the 311-bp nptII fragment is severely reduced, whereas a number of longer fragments are detected. Thus, the proximal nptII genes of the IRn loci and the three nptII genes of the 21-1 IRcn locus are severely methylated. For the 21-1 IRcn locus, this was confirmed by the 3′nos hybridization. Instead of the 248-bp nos fragment (Fig. 4F, lane 13), Sau3AI generated two partial fragments of about 960 and 1,200 bp (lanes 9 and 10), which indicates that the Sau3AI sites immediately up- and downstream of the nos region are severely methylated. We infer that all T-DNAs of an IR locus are methylated about equally. This is illustrated, for example, by the almost complete absence of the 311-bp nptII fragment in some of the transformants (Fig. 1C, lanes 1, 3, and 9) and the relatively high intensities of the longer partial fragments. These results and the ones described below, in conjunction with the run-on transcription data (Fig. 3), are most consistent with about equally methylated T-DNAs.
Hybridizations with probes for the uidA-ChsA transgenes in the IRn loci and the 19-1 IRc locus resulted in a band pattern for the Sau3AI digests that was opposite to that obtained with an nptII probe. Specifically, in transformants carrying the 6-2 and 19-3 IRn loci (Fig. 4D, lanes 1 and 3), the full-length uidA probe detected predominantly the bands that were also seen when the gene was completely digested with MboI (lane 13). Therefore, the distally located uidA sequences are mostly unmethylated, as opposed to the proximal nptII genes. In contrast, the IR-center-proximal uidA sequences of the 19-1 IRc locus were highly methylated because Sau3AI gave rise to many partially cut fragments (Fig. 4D, lane 6) and the two main uidA MboI fragments of 660 and 500 bp were barely visible (Fig. 4D, lane 13).
The position-dependent methylation pattern of the T-DNAs was confirmed by the ChsA hybridizations. Their interpretation was complicated by the detection of fragments derived from both the transgenes and the endogenous gene, some of which are the same size. However, there is a unique 713-bp fragment which is specific for the endogenous ChsA gene (Fig. 4E). We have taken the intensity of the 713-bp fragment in the Sau3AI digests as a measure of the methylation status of the endogenous gene. The ChsA gene of untransformed V26 petunias already appeared partially methylated, like the DfrA gene, because in addition to the 534- and 713-bp fragments, some longer fragments were detected by the ChsA probe (Fig. 4E, lane 11). In all transformants, irrespective of whether the gene was silenced, the intensity of the 713-bp band was about the same. This shows that the endogenous gene had not undergone a dramatic change in methylation, which was confirmed by hybridizations to five different exon probes (data not shown). Also, for the ChsJ gene, which is 85% identical to ChsA and cosilenced, no methylation changes were detected (data not shown). However, minor changes or an increased methylation of key sites in the Chs genes that were not detected by the Sau3AI digests remains formally possible.
Given the constant intensity of the 713-bp band of the ChsA gene, differences in the Chs hybridization patterns observed in the transformants must be due to changes in transgene methylation. For example, with DNA containing the 19-1 IRc locus, a number of partially cleaved fragments were seen (Fig. 4E, lane 6), indicating that the ChsA sequences of this locus are highly methylated, like the neighboring uidA sequences (Fig. 4D, lane 6). In contrast, the Chs sequences of the 6-2 and 19-3 IRn loci were not methylated (lanes 1 and 3). Only in plants homozygous for the 19-3 IRn and 19-3 S loci was an elevated methylation of the uidA-Chs transgenes found (Fig. 4D and E, lanes 4).
In the more complex 21-1 IRcn locus, all the nptII genes (Fig. 4B, lane 9) and promoterless ChsA transgenes (Fig. 4E, lane 9) appeared methylated. With probes for both genes, several partial fragments were detected. Some of these fragments were also detected by the neighboring probes, indicating that they encompass a large part of the locus. Even the EcoRI site near the left border of the T-DNAs appeared methylated, because a left-border (LB) probe detected some of these longer fragments as well (results not shown). The same was true for the EcoRI site in the 19-1 IRc locus. Also, the 3′nos hybridization probe indicated that the sequences around the center of the 19-1 IRc are heavily methylated because instead of a band at 250 bp (Fig. 4F, lane 13), Sau3AI generated a number of longer fragments (lane 6) which were also detected by the ChsA probe (Fig. 4E).
Analysis of the CaMV-35S promoter driving the uidA-ChsA transgene in some of the transformants revealed that several of its Sau3AI sites are methylated. Instead of the major 632-bp fragment in the MboI digest (Fig. 4G, lane 13), the CaMV-35S probe detected fragments of 850 bp and larger in the Sau3AI digests (lanes 1, 3, 4, and 6). As assessed by the run-on assays, there was no clear correlation between the methylation of the Sau3AI sites and promoter activity (Fig. 3). Reduced transcription was correlated only with the methylation of the HpaII site, which is about 320 bp upstream of the transcription start site. In the 19-1 IRc and S loci, this site was much more highly methylated than in the other loci (data not shown), which correlates with a lower transcription rate of the uidA-ChsA genes in these loci.
Methylation of single-copy T-DNA loci.
Since the expression and transcription properties of the transgenes of the single copy T-DNA loci were comparable to those of the partner IR loci, it appeared that they also had a common methylation pattern. To test this, DNA from the transformants carrying either the 19-3, 19-1, or 21-6 S locus was digested with Sau3AI and analyzed along the DNAs containing the partner IR loci. This indeed revealed that, overall, the methylation status of the S transgenes was strikingly similar to that of the corresponding IR transgenes. For example, the uidA probe detected in the 19-1 S locus several Sau3AI partial fragments (Fig. 4D, lane 5), as it did for the IRc locus (lane 6), indicating that the uidA genes in both loci are heavily methylated. In contrast, the hybridization with the nptII probe shows that the nptII genes of both loci are predominantly unmethylated (Fig. 4C, lanes 5 and 6). In the 19-3 S locus, the uidA-ChsA transgene is mainly unmethylated, as in the IRn locus (Fig. 4E and D, lanes 2 and 3), and the same holds for the nptII genes of the 21-6 S (Fig. 4C and F, lane 7) and 21-6 IRc loci (lane 8).
For the 21-1 loci, it was not possible to analyze the 21-1 S locus separately from the 21-1 IRcn locus. The two loci are on the same chromosome, and none of the backcross progeny contained only the S locus (63). However, with a plant containing both loci, the overall hybridization pattern with different probes was comparable to that with a plant carrying just the IRcn locus (Fig. 4C and E, lanes 9 and 10). This indicates that the S locus is as highly methylated as the IRcn locus, which is in line with the lack of nptII transcription in both IRcn/S and IRcn plants (Fig. 3A, lanes 9 and 10).
DISCUSSION
In the present study, we have characterized various Chs transgene-containing IR and S loci in transgenic petunias. With the DNA constructs we used, only Chs transgenes residing in IR loci silence the endogenous Chs genes posttranscriptionally, and the IRs with the Chs sequences proximal to the center of the IR confer the strongest silencing (63). We show that these proximal transgenes are transcriptionally repressed and highly methylated compared to the distal transgenes. The level of DNA methylation was examined by using the methylation-sensitive enzyme Sau3AI, for which there are 18 to 31 sites in the T-DNA, depending on the type of T-DNA (Fig. 4A). We found that for an IR locus that consists of two T-DNAs, probes specific for fragments proximal to an IR center detect various partial Sau3AI fragments, whereas probes specific for distal sequences detect mainly the fully digested fragments. One difficulty in defining the T-DNA region confined in a particular partially cleaved Sau3AI fragment involved the clustering of some of the sites. However, since the positions of the sites were known, fragment sizes could be predicted if certain sites were methylated. This allowed us to position most fragments quite precisely. Also, some of the partial fragments were detected by neighboring probes, which indicated that most (clustered) Sau3AI sites separating the probed sequences were modified.
Since an IR locus consists of two or more identical T-DNA copies, it is possible that the partial fragments are derived from just one of the T-DNAs in that one copy is methylated whereas the other(s) is not. However, our data indicate that this is unlikely. For example, for the 19-3 and 6-2 IRn loci, the smallest nptII fragment visible in the MboI digests was not detected in the Sau3AI digests (Fig. 4B) which means that the nptII genes of both T-DNAs are modified. The same holds true for the two proximal uidA-ChsA transgenes of the 19-1 IRc locus (Fig. 4E). Also, the results with the 3′nos probe supports the symmetrical methylation of the nptII genes in the 21-6 IRc locus. If this was not the case, the smallest fragments would be much more intense relative to the larger partial fragments. We therefore infer that an IR, which is composed of two T-DNAs (IRc and IRn), is symmetrically methylated around its center. Although we have examined just one 3-T-DNA IR locus (21-1 IRcn), our results suggest that when there are more T-DNAs arranged as IRs, the entire T-DNA is sensitive to methylation. Small methylation differences between the T-DNA copies, however, cannot be excluded. The run-on data also indicate, although indirectly, that the T-DNA copies are equally modified, since none of the three nptII genes of the 21-1 IRcn locus or the two nptII genes of the 6-2 IRn locus is active (Fig. 3A).
The higher methylation density of DNA around the center of an IR suggests that the palindromic sequence organization itself is somehow responsible for this pattern. Because of the potential for intrastrand base pairing, palindromes can adapt a cruciform structure, which may occur in vivo. The elevated methylation around the IR center may then be explained by the preferential methylation of unusual DNA structures (61), including cruciform structures (6, 32), by DNA methyltransferase. Cruciform structures have been observed in bacteria (48, 81) and mammalian cells (25, 76, 80), and it seems likely that they can also be formed in plants. The reported palindromes range from a few base pairs to about 400 bp, whereas the palindromes we have studied range from 4.5 to about 7.5 kb. It is not known if cruciform structures of this size can ever be formed, but it is conceivable that part of the palindrome transiently adopts a cruciform structure. Another possibility for the distinct IR methylation pattern is a response to the chromatin structure around the IR center. This alternative stems from observations in Drosophila, which lacks C methylation (62) but which nevertheless can epigenetically inactivate tandemly repeated transgenes. Such repeated genes, including IRs, appear to self-associate into silent heterochromatin (16). Whether the IRs we have examined undergo similar interactions remains to be seen, but we have found that the DNA around the center of the IRs is indeed packaged into more highly condensed chromatin than the DNA that is distal to the center (75a). With an IR locus that contains more T-DNAs, like the 21-1 IRcn locus, more interactions between the repeats are possible, which may result in a more highly condensed and silent state (21, 79). Given that repeated genes are sensitive to inactivation via heterochromatinization, the DNA in plants may at some point become methylated, thereby stably maintaining the inactive state.
A third possibility is that the methylation is triggered by RNA (77). However, we consider it unlikely that the distinct methylation pattern of the IRs is mediated by transcripts derived from the transgenes, especially for IR loci containing the promoterless Chs transgenes. We cannot exclude, though, that transcripts from the ChsA host genes are involved and that they may form a triple helix with a putative cruciform structure, thereby triggering methylation. However, the differential methylation of the nptII genes is not readily explained in this way, since there is no endogenous homologue. Thus, based on the data we have obtained thus far, we favor models in which IR methylation is directed by the palindromic sequence arrangement itself.
An important question is how IR loci trigger a posttranscriptional silencing mechanism. There are cases where methylated transgenes are posttranscriptionally silenced (20, 26, 27, 57, 72). Although some of the transgenes we have examined are also methylated, the run-on assays indicate that their expression is mainly transcriptionally controlled (Fig. 3). PTGS of the uidA-Chs transgenes residing in the 19-3 IRn locus occurs only in corollas where the endogenous Chs genes are posttranscriptionally silenced (Fig. 2). How methylated but transcribed transgenes are posttranscriptionally silenced is unknown. In Neurospora and Ascobolus, methylation leads to premature termination of transcription (3, 54). In plants, this has not been observed (27, 72), but it cannot be ruled out that a small number of transcripts terminate prematurely, which would be sufficient to activate the PTGS mechanism. For the methylated Chs transgenes, this possibility cannot formally be excluded, but since the IRs carrying the promoterless transgenes, as well as those carrying the promoter-driven uidA-ChsA transgenes, cause silencing, we consider this possibility unlikely. Moreover, our data indicate that transgene methylation per se is not sufficient to activate PTGS. For example, the uidA-ChsA transgene of the single-copy 19-1 S locus is hypermethylated and transcriptionally weakly active, yet this locus is unable to induce PTGS even in plants homozygous for this S locus (63), in contrast to the 19-1 IRc locus. It seems, therefore, that the palindromic arrangement of the Chs transgenes is a requirement for triggering PTGS of the endogenous homologues.
Besides PTGS of methylated transgenes, PTGS is often associated with multicopy T-DNA loci. In part, this might be related to the preferential methylation of DNA repeats. However, it might also be due to the increased gene dosage and to increased amounts of gene products, since PTGS sometimes occurs only in plants that are homozygous for a particular transgene locus (4, 12, 42, 44, 64). However, there is convincing evidence that PTGS can be activated by single-copy transgenes (19, 28, 74). In these cases, it appears necessary that the transgene be highly transcribed (50, 74) and that the RNA be stable (50). It is unknown how a seemingly excessive production of RNA would trigger cosuppression. It has been proposed that it initiates a negative feedback, ultimately leading to the removal of all homologous RNAs (40). For Chs silencing, it was proposed to be initiated by poly(A)− RNAs produced by aberrant RNA processing due to a localized accumulation of RNA in the nucleus, which allows RNA-RNA pairing followed by endonucleolytic cleavage (41). By cycles of RNA-RNA pairing and cleavage, regular homologous mRNAs would also be targeted for degradation. Another possibility is that the aberrant RNAs serve as template for a host RNA-dependent RNA polymerase by which antisense RNAs can be produced (17, 34, 56).
Que et al. (50) suggested that IR loci trigger silencing by producing aberrant transcripts which are more effective at triggering cosuppression. For IRc loci with the Chs silencing sequences proximal to the center, this may be possible since some readthrough transcription into the Chs transgenes, which potentially can lead to double-stranded RNA, cannot be excluded. However, the run-on assays show that this possibility is unlikely (Fig. 3).
If aberrant RNAs indeed play a key role in triggering silencing, and when it is unlikely that they are not produced by the transgenes, they may come from the endogenous Chs genes. Support for this possibility comes from studies of the natural Chs mutants ‘Red Star’ and ‘Velvet Picotee’, in which the Chs genes in the white corolla sectors are posttranscriptionally silenced (67, 71). In several aspects, this silencing resembles the silencing of Chs expression in transgenic plants (41, 68). Given that endogenous genes themselves can trigger the RNA degradation machinery, it shows that under certain conditions, transgene transcripts are dispensable. It is striking that the natural Chs silencing mutants contain two almost identical Chs genes which are closely linked and organized as a direct repeats (63a). The relevance of this gene organization remains to be determined.
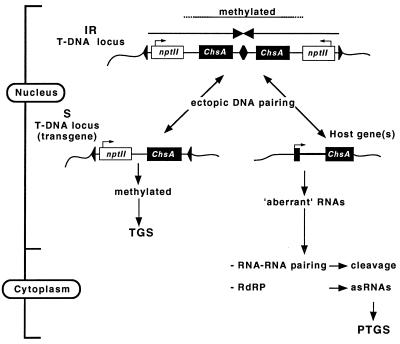
We speculate that in the IR-containing transformants, the host V26 Chs gene(s) is epigenetically modified in a manner similar to that of the two Chs genes in the natural cosuppression mutants, due to an interaction with the IR locus. In this way, the endogenous gene is stimulated to produce aberrant transcripts (the model is shown in Fig. 6). This implies that to induce silencing, the host gene must be transcriptionally active. Two observations support this. First, the strong 19-1 IRc locus can silence an expressed single-copy uidA-Chs transgene posttranscriptionally only in corollas where the host Chs genes are highly transcribed (63). Second, the expression of uidA-ChsA transgenes residing in the 19-3 IRn locus is reduced in the white corolla sectors but not in leaves (Fig. 2A), where the Chs genes are barely active.
FIG. 6.
Proposed model for events that are initiated by an IR T-DNA locus and that lead to the posttranscriptional silencing of the host Chs genes or the transcriptional silencing of genes in a monomeric T-DNA locus (S). When two T-DNAs integrate into the genome as an IR, the sequences proximal to the center are preferentially methylated. If the transgene contains a promoter, this methylation is associated with TGS. A single-copy T-DNA locus (S) is methylated by the IR, and the pattern of methylation will be very similar to that of the IR. This may occur via ectopic DNA-DNA pairing between identical sequences of the IR and the S loci (left). An IR locus may also interact by DNA-DNA pairing with the homologous host gene(s) (right). This interaction does not lead to a dramatic change in DNA methylation but may cause a change in chromatin, which impedes regular transcription of the gene or RNA processing to some degree. In this way, “aberrant” or “unfinished” RNAs are produced, which either may be used as templates for the RNA-dependent RNA polymerase (17, 34) by which antisense RNAs are produced or may initiate a cycle of RNA-RNA pairing events by which they act as a kind of catalytic RNA (41). Both pathways result in the degradation of homologous transcripts and silence the genes posttranscriptionally.
There are indications that IR loci are especially prone to interacting with unlinked homologues. In yeast, for example, IRs create hot spots for mitotic interchromosomal recombination with single-copy sequences (23), indicating that interactions involving IRs are more efficient than those involving single-copy loci. The IR loci we have examined may not be involved in homologous recombination; they are in fact very stable (unpublished results), in contrast to IR loci in mice (7). That IR loci in plants may nevertheless cross talk to homologous sequences can be inferred from the observation that the S and corresponding IR loci (Fig. 1) have a comparable methylation pattern (Fig. 4 and 5A). This was unexpected because the S and the IR loci are at different genomic locations. The S loci can therefore be regarded as independent transgene loci, and there is no reason why they should have a methylation pattern similar to that of the partner IR loci (Fig. 4) unless there has been some sort of cross talk between the IR and S loci in the primary transformant, perhaps via “epigene conversion” (55). There is one other instance of gene methylation in which an IR locus is involved. The four phosphoribosylanthranilate isomerase (PAI) genes in Arabidopsis Wassilewskija are highly methylated, whereas the three PAI genes in Arabidopsis Columbia are not. In strain Wassilewskija, two of the genes are arranged in a tail-to-tail inverted repeat (5), and it has been suggested that the methylated IR locus communicates methylation to the unlinked PAI genes via DNA pairing.
The cross talk underlying methylation of single-copy (trans)-genes is reminiscent of other DNA-DNA interactions believed to take place in plants (36, 49, 73) and in fungi. The latter includes MIP in Ascobolus (24, 53) and RIP in Neurospora (58, 59). Recent studies with Ascobolus indicate that methylation transfer and recombination may be mechanistically related (8), and since methylation transfer between a silencing locus and a target locus appears to occur with high efficiency (36, 73), it was suggested that the process of methylation transfer involves DNA-DNA interactions that are not recombination competent (8).
Although the methylation patterns of the single-copy loci support cross talk between IR loci and unlinked homologous sequences, we have as yet no indication that an IR locus interacts with the endogenous Chs gene(s). The methylation levels of the Sau3AI sites in silenced and unsilenced ChsA (Fig. 4E and 5B) and ChsJ (data not shown) genes were similar. For the ChsA gene, this conclusion is based mainly on the relative intensity of the host gene-specific 713-bp ChsA fragment, which was about the same in all transformants regardless whether the gene was silenced and was similar to that in untransformed plants (Fig. 4E). Whether this indicates that an IR locus is not able to interact with the host ChsA gene is unclear. Since very little is known about the way DNA methylation is transferred to a homologous locus, the absence of increased methylation may be related to a difference in size between T-DNAs and between a T-DNA and a host gene. The size varies from 4.5 to 7.5 kb for the T-DNAs, depending on the construct, and it is 1.4 kb for the genomic ChsA gene, which is even divided into two exons. A smaller size obviously affects DNA-DNA interactions and as a consequence the transfer of methylation (24). The unchanged methylation of the endogenous genes is probably not due to an insensitivity to transgene-induced methylation, because Chs genes in Arabidopsis can be methylated and transcriptionally silenced by a transgene construct containing a 3.9-kb genomic Chs clone (9). In this case, the silent state is meiotically heritable, which is different from Chs silencing in petunia, where it is not meiotically inherited.
Although the methylation of the posttranscriptionally silenced Chs genes was not detectably changed, the elevated level of unspliced transcripts (45a, 68) and the presence of aberrant poly(A)− and shorter poly(A)+ RNAs (41) suggest that the production of mature RNAs is to some extent impaired. How, in our case, IR loci influence RNA processing remains puzzling, but it might be that they are able to physically interact with the host gene and, instead of changing its methylation, change its chromatin structure. This would partially impair the regular synthesis and processing of primary transcripts, which ultimately may give rise to the aberrant RNAs thought to be responsible for activating the RNA degradation pathway.
ACKNOWLEDGMENTS
We thank Susan Kenter for providing technical assistance, Pieter Hoogeveen and Martina Meesters for taking care of the plants, and Rik van Blokland for providing the M13-nptII construct. We are grateful to Vicki Chandler, Titia Sijen, Rik van Blokland, and John Wing for comments on the manuscript and to members of the EU Gene Silencing Network for stimulating and insightful discussions.
This work was financed by Novartis Seeds BV (to M.S.) and by a research fellowship of the Royal Netherlands Academy of Sciences (KNAW) (to J.M.K.).
REFERENCES
- 1.Assaad F F, Tucker K L, Signer E R. Epigenetic repeat-induced gene silencing (RIGS) in Arabidopsis. Plant Mol Biol. 1993;22:1067–1085. doi: 10.1007/BF00028978. [DOI] [PubMed] [Google Scholar]
- 2.Barlow D P. Gametic imprinting in mammals. Science. 1995;270:1610–1613. doi: 10.1126/science.270.5242.1610. [DOI] [PubMed] [Google Scholar]
- 3.Barry C, Faugeron G, Rossignol J L. Methylation induced premeiotically in Ascobolus: coextension with DNA repeat lengths and effect on transcript elongation. Proc Natl Acad Sci USA. 1993;90:4557–4561. doi: 10.1073/pnas.90.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baulcombe D C, English J J. Ectopic pairing of homologous DNA and post-transcriptional silencing in transgenic plants. Curr Opin Biotechnol. 1996;7:173–180. [Google Scholar]
- 5.Bender J, Fink G R. Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell. 1995;83:725–734. doi: 10.1016/0092-8674(95)90185-x. [DOI] [PubMed] [Google Scholar]
- 6.Chen X A, Mariappan S V S, Catasti P, Ratliff R, Moyzis R K, Laayoun A, Smith S S, Bradbury E M, Gupta G. Hairpins are formed by the single DNA strands of the fragile X triplet repeats: structure and biological implications. Proc Natl Acad Sci USA. 1995;92:5199–5203. doi: 10.1073/pnas.92.11.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collick A, Drew J, Penberth J, Bois P, Luckett J, Scaerou F, Jeffreys A, Reik W. Instability of long inverted repeats within mouse transgenes. EMBO J. 1996;15:1163–1171. [PMC free article] [PubMed] [Google Scholar]
- 8.Colot V, Maloisel L, Rossignol J L. Interchromosomal transfer of epigenetic states in Ascobolus: transfer of DNA methylation is mechanistically related to homologous recombination. Cell. 1996;86:855–864. doi: 10.1016/s0092-8674(00)80161-0. [DOI] [PubMed] [Google Scholar]
- 9.Davies G J, Sheih M A, Ratcliffe O J, Couplnad G, Furner I J. Genetics of homology-dependent gene silencing in Arabidopsis: a role for methylation. Plant J. 1997;12:791–804. doi: 10.1046/j.1365-313x.1997.12040791.x. [DOI] [PubMed] [Google Scholar]
- 10.De Carvalho F, Gheysen G, Kushnir S, Van Montagu M, Inzé D, Castresana C. Suppression of β-1,3-glucanase transgene expression in homozygous plants. EMBO J. 1992;11:2595–2602. doi: 10.1002/j.1460-2075.1992.tb05324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Carvalho Niebel F, Frendo P, Van Montagu M, Cornelissen M. Post-transcriptional cosuppression of β-1,3-glucanase genes does not affect accumulation of transgene nuclear mRNA. Plant Cell. 1995;7:347–358. doi: 10.1105/tpc.7.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Depicker A, Van Montagu M. Post-transcriptional gene silencing in plants. Curr Opin Cell Biol. 1997;9:373–382. doi: 10.1016/s0955-0674(97)80010-5. [DOI] [PubMed] [Google Scholar]
- 13.Dobie K, Mehtali M, McClenaghan M, Lathe R. Variegated gene expression in mice. Trends Genet. 1997;13:127–130. doi: 10.1016/s0168-9525(97)01097-4. [DOI] [PubMed] [Google Scholar]
- 14.Dobie K W, Lee M, Fantes J A, Graham E, Clark A J, Springbett A, Lathe R, Mcclenaghan M. Variegated transgene expression in mouse mammary gland is determined by the transgene integration locus. Proc Natl Acad Sci USA. 1996;93:6659–6664. doi: 10.1073/pnas.93.13.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorer D R. Do transgene arrays form heterochromatin in vertebrates? Transgenic Res. 1997;6:3–10. doi: 10.1023/a:1018460413680. [DOI] [PubMed] [Google Scholar]
- 16.Dorer D R, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- 17.Dougherty W G, Parks T D. Transgenes and gene suppression: telling us something new? Curr Opin Cell Biol. 1995;7:399–405. doi: 10.1016/0955-0674(95)80096-4. [DOI] [PubMed] [Google Scholar]
- 18.Eggleston W B, Alleman M, Kermicle J L. Molecular organization and germinal instability of R-stippled maize. Genetics. 1995;141:347–360. doi: 10.1093/genetics/141.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elmayan T, Vaucheret H. A strongly-expressed 35S-driven transgene undergoes post-transcriptional silencing in all tobacco transformants irrespective of the copy number. Plant J. 1996;9:787–797. [Google Scholar]
- 20.English J J, Mueller E, Baulcombe D C. Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell. 1996;8:179–188. doi: 10.1105/tpc.8.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrick D, Fiering S, Martin D I K, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 22.Goodwin J, Chapman K, Swaney S, Parks T D, Wernsman E A, Dougherty W G. Genetic and biochemical dissection of transgenic RNA-mediated virus resistance. Plant Cell. 1996;8:95–105. doi: 10.1105/tpc.8.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordenin D A, Lobachev K S, Degtyareva N P, Malkova A L, Perkins E, Resnick M A. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol Cell Biol. 1993;13:5315–5322. doi: 10.1128/mcb.13.9.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goyon C, Barry C, Gregoire A, Faugeron G, Rossignol J-L. Methylation of DNA repeats of decreasing sizes in Ascobolus. Mol Cell Biol. 1996;16:3054–3065. doi: 10.1128/mcb.16.6.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanke J H, Hambor J E, Kavathas P. Repetitive Alu elements form a cruciform structure that regulates the function of the human CD8α T cell-specific enhancer. J Mol Biol. 1995;246:63–73. doi: 10.1006/jmbi.1994.0066. [DOI] [PubMed] [Google Scholar]
- 26.Hobbs S L A, Warkentin T D, Delong C M O. Transgene copy number can be positively or negatively associated with transgene expression. Plant Mol Biol. 1993;21:17–26. doi: 10.1007/BF00039614. [DOI] [PubMed] [Google Scholar]
- 27.Ingelbrecht I, Van Houdt H, Van Montagu M, Depicker A. Posttranscriptional silencing of reporter transgenes in tobacco correlates with DNA methylation. Proc Natl Acad Sci USA. 1994;91:10502–10506. doi: 10.1073/pnas.91.22.10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jorgensen R, Cluster P, Que Q, English J, Napoli C. Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single copy vs. complex T-DNA sequences. Plant Mol Biol. 1996;31:957–973. doi: 10.1007/BF00040715. [DOI] [PubMed] [Google Scholar]
- 29.Kennison J A. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 30.Kermicle J L, Eggleston W B, Alleman M. Organization of paramutagenicity in R-stippled maize. Genetics. 1995;141:361–372. doi: 10.1093/genetics/141.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kilby N J, Leyser H M O, Furner I J. Promoter methylation and progressive transgene inactivation in Arabidopsis. Plant Mol Biol. 1992;20:103–112. doi: 10.1007/BF00029153. [DOI] [PubMed] [Google Scholar]
- 32.Laayoun A, Smith S S. Methylation of slipped duplexes, snapbacks and cruciforms by human DNA(cytosine-5)methyltransferase. Nucleic Acids Res. 1995;23:1584–1589. doi: 10.1093/nar/23.9.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laurenson P, Rine J. Silencers, silencing, and heritable transcriptional states. Microbiol Rev. 1992;56:543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindbo J A, Silva-Rosales L, Proebsting W M, Dougherty W G. Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell. 1993;5:1749–1759. doi: 10.1105/tpc.5.12.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin C, Gerats T. Control of pigment biosynthesis genes during petal development. Plant Cell. 1993;5:1253–1264. doi: 10.1105/tpc.5.10.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matzke A J M, Neuhuber F, Park Y D, Ambros P F, Matzke M A. Homology-dependent gene silencing in transgenic plants: epistatic silencing loci contain multiple copies of methylated transgenes. Mol Gen Genet. 1994;244:219–229. doi: 10.1007/BF00285449. [DOI] [PubMed] [Google Scholar]
- 37.Matzke M A, Matzke A J M. How and why do plants inactivate homologous (trans)genes? Plant Physiol. 1995;107:679–685. doi: 10.1104/pp.107.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matzke M A, Primig M, Trnovsky J, Matzke A J M. Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J. 1989;8:643–649. doi: 10.1002/j.1460-2075.1989.tb03421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McBride K E, Summerfelt K R. Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol Biol. 1990;14:269–276. doi: 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- 40.Meins F, Jr, Kunz C. Gene silencing in transgenic plants: a heuristic autoregulation model. In: Meyer P, editor. Gene silencing in higher plants and related phenomena in other eukaryotes. Vol. 197. Berlin, Germany: Springer-Verlag KG; 1995. pp. 105–120. [DOI] [PubMed] [Google Scholar]
- 41.Metzlaff M, Odell M, Cluster P D, Flavell R B. RNA-mediated RNA degradation and chalcone synthase A silencing in Petunia. Cell. 1997;88:845–854. doi: 10.1016/s0092-8674(00)81930-3. [DOI] [PubMed] [Google Scholar]
- 42.Meyer P. Repeat-induced gene silencing: common mechanisms in plants and fungi. Biol Chem Hoppe-Seyler. 1996;377:87–95. [PubMed] [Google Scholar]
- 43.Meyer P, Heidmann I, Niedenhof I. Differences in DNA-methylation are associated with a paramutation phenomenon in transgenic petunia. Plant J. 1993;4:89–100. doi: 10.1046/j.1365-313x.1993.04010089.x. [DOI] [PubMed] [Google Scholar]
- 44.Meyer P, Saedler H. Homology-dependent gene silencing in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:23–48. doi: 10.1146/annurev.arplant.47.1.23. [DOI] [PubMed] [Google Scholar]
- 45.Mueller E, Gilbert J, Davenport G, Brigneti G, Baulcombe D C. Homology-dependent resistance: transgenic virus resistance in plants related to homology-dependent gene silencing. Plant J. 1995;7:1001–1013. [Google Scholar]
- 45a.Muskens, M., and J. M. Kooter. Unpublished data.
- 46.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neuhuber F, Park Y D, Matzke A J M, Matzke M A. Susceptibility of transgene loci to homology-dependent gene silencing. Mol Gen Genet. 1994;244:230–241. doi: 10.1007/BF00285450. [DOI] [PubMed] [Google Scholar]
- 48.Noirot P, Bargonetti J, Novick R P. Initiation of rolling-circle replication in pT181 plasmid: initiator protein enhances cruciform extrusion at the origin. Proc Natl Acad Sci USA. 1990;87:8560–8564. doi: 10.1073/pnas.87.21.8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park Y-D, Papp I, Moscone E A, Iglesias V A, Vaucheret H, Matzke A J M, Matzke M A. Gene silencing mediated by promoter homology occurs at the level of transcription and results in meiotically heritable alterations in methylation and gene activity. Plant J. 1996;9:183–194. doi: 10.1046/j.1365-313x.1996.09020183.x. [DOI] [PubMed] [Google Scholar]
- 50.Que Q D, Wang H Y, English J J, Jorgensen R A. The frequency and degree of cosuppression by sense chalcone synthase transgenes are dependent on transgene promoter strength and are reduced by premature nonsense codons in the transgene coding sequence. Plant Cell. 1997;9:1357–1368. doi: 10.1105/tpc.9.8.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ronchi A, Petroni K, Tonelli C. The reduced expression of endogenous duplications (REED) in the maize R gene family is mediated by DNA methylation. EMBO J. 1995;14:5318–5328. doi: 10.1002/j.1460-2075.1995.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rossignol J L, Faugeron G. Gene inactivation triggered by recognition between DNA repeats. Experientia. 1994;50:307–317. doi: 10.1007/BF01924014. [DOI] [PubMed] [Google Scholar]
- 53.Rossignol J L, Faugeron G. MIP: an epigenetic gene silencing process in Ascobolus immersus. Curr Top Microbiol Immunol. 1995;197:179–191. doi: 10.1007/978-3-642-79145-1_12. [DOI] [PubMed] [Google Scholar]
- 54.Rountree M R, Selker E U. DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev. 1997;11:2383–2395. doi: 10.1101/gad.11.18.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabl J F, Laird C D. Epigene conversion: a proposal with implications for gene mapping in humans. Am J Hum Genet. 1992;50:1171–1177. [PMC free article] [PubMed] [Google Scholar]
- 56.Schiebel W, Haas B, Marinkovíc S, Klanner A, Sänger H L. RNA-directed RNA polymerase from tomato leaves. II. Catalytic in vitro properties. J Biol Chem. 1993;268:11858–11867. [PubMed] [Google Scholar]
- 57.Sijen T, Wellink J, Hiriart J B, Van Kammen A. RNA-mediated virus resistance: role of repeated transgenes and delineation of targeted regions. Plant Cell. 1996;8:2277–2294. doi: 10.1105/tpc.8.12.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singer M J, Marcotte B A, Selker E U. DNA methylation associated with repeat-induced point mutation in Neurospora crassa. Mol Cell Biol. 1995;15:5586–5597. doi: 10.1128/mcb.15.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singer M J, Selker E U. Genetic and epigenetic inactivation of repetitive sequences in Neurospora crassa: RIP, DNA methylation, and Quelling. Curr Top Microbiol Immunol. 1995;197:165–178. doi: 10.1007/978-3-642-79145-1_11. [DOI] [PubMed] [Google Scholar]
- 60.Smith H A, Swaney S L, Parks T D, Wernsman E A, Dougherty W G. Transgenic plant virus resistance mediated by untranslatable sense RNAs: expression, regulation, and fate of nonessential RNAs. Plant Cell. 1994;6:1441–1453. doi: 10.1105/tpc.6.10.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith S S, Laayoun A, Lingeman R G, Baker D J, Riley J. Hypermethylation of telomere-like foldbacks at codon 12 of the human c-HA-ras gene and the trinucleotide repeat of the FMR-1 gene of fragile X. J Mol Biol. 1994;243:143–151. doi: 10.1006/jmbi.1994.1640. [DOI] [PubMed] [Google Scholar]
- 62.Smith S S, Thomas C A., Jr The two-dimensional restriction analysis of Drosophila DNAs: males and females. Gene. 1981;13:395–408. doi: 10.1016/0378-1119(81)90019-6. [DOI] [PubMed] [Google Scholar]
- 63.Stam M, De Bruin R, Kenter S, Van der Hoorn R A L, Van Blokland R, Mol J N M, Kooter J M. Post-transcriptional silencing of chalcone synthase in Petunia by inverted transgene repeats. Plant J. 1997;12:63–82. [Google Scholar]
- 63a.Stam, M., and J. M. Kooter. Unpublished results.
- 64.Stam M, Mol J N M, Kooter J M. The silence of genes in transgenic plants. Ann Bot. 1997;79:3–12. [Google Scholar]
- 65.Streeck R E. Single-strand and double-strand cleavage at half-modified and fully modified recognition sites for the restriction nucleases Sau3A and TaqI. Gene. 1980;12:267–275. doi: 10.1016/0378-1119(80)90109-2. [DOI] [PubMed] [Google Scholar]
- 66.Todd J J, Vodkin L O. Duplications that suppress and deletions that restore expression from a chalcone synthase multigene family. Plant Cell. 1996;8:687–699. doi: 10.1105/tpc.8.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Blokland R. Trans-inactivation of flower pigmentation genes in Petunia hybrida. Ph.D. thesis. Amsterdam, The Netherlands: Vrije Universiteit; 1994. [Google Scholar]
- 68.Van Blokland R, Van der Geest N, De Lange P, Stam M, Mol J N M, Kooter J M. Post-transcriptional suppression of chalcone synthase genes in Petunia hybrida and the accumulation of unspliced pre-mRNAs. In: Grierson D, Lycett G W, Tucker G A, editors. Mechanisms and applications of gene silencing. Nottingham, England: Nottingham University Press; 1996. pp. 57–69. [Google Scholar]
- 69.Van Blokland R, Van der Geest N, Mol J N M, Kooter J M. Transgene-mediated suppression of chalcone synthase expression in Petunia hybrida results from an increase in RNA turnover. Plant J. 1994;6:861–877. [Google Scholar]
- 70.Van der Krol A R, Mur L A, Beld M, Mol J N M, Stuitje A R. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990;2:291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van der Meer I M. Ph.D. thesis. Amsterdam, The Netherlands: Vrije Universiteit; 1991. [Google Scholar]
- 72.Van Houdt H, Ingelbrecht I, Van Montagu M, Depicker A. Post-transcriptional silencing of a neomycin phosphotransferase II transgene correlates with the accumulation of unproductive RNAs and with increased cytosine methylation. Plant J. 1997;12:379–392. [Google Scholar]
- 73.Vaucheret H. Identification of a general silencer for 19S and 35S promoters in a transgenic tobacco plant: 90 bp of homology in the promoter sequence are sufficient for trans-inactivation. C R Acad Sci Ser III. 1993;316:1471–1483. [Google Scholar]
- 74.Vaucheret H, Nussaume L, Palauqui J C, Quillere I, Elmayan T. A transcriptionally active state is required for post-transcriptional silencing (cosuppression) of nitrate reductase host genes and transgenes. Plant Cell. 1997;9:1495–1504. doi: 10.1105/tpc.9.8.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vaucheret H, Palauqui J-C, Elmayan T, Moffatt B. Molecular and genetic analysis of nitrite reductase co-suppression in transgenic tobacco plants. Mol Gen Genet. 1995;248:311–317. doi: 10.1007/BF02191598. [DOI] [PubMed] [Google Scholar]
- 75a.Vissers, A., and J. M. Kooter. Unpublished data.
- 76.Ward G K, Shihab-el-Deen A, Zannis-Hadjopoulos M, Price G B. DNA cruciforms and the nuclear supporting structure. Exp Cell Res. 1991;195:92–98. doi: 10.1016/0014-4827(91)90503-m. [DOI] [PubMed] [Google Scholar]
- 77.Wassenegger M, Heimes S, Riedel L, Sanger H L. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 78.Willard H F. X chromosome inactivation, XIST, and pursuit of the X-inactivation center. Cell. 1996;86:5–7. doi: 10.1016/s0092-8674(00)80071-9. [DOI] [PubMed] [Google Scholar]
- 79.Ye F, Signer E R. RIGS (repeat-induced gene silencing) in Arabidopsis is transcriptional and alters chromatin configuration. Proc Natl Acad Sci USA. 1996;93:10881–10886. doi: 10.1073/pnas.93.20.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zannis-Hadjopoulos M, Frappier L, Khoury M, Price G B. Effect of anti-cruciform DNA monoclonal antibodies on DNA replication. EMBO J. 1988;7:1837–1844. doi: 10.1002/j.1460-2075.1988.tb03016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zheng G, Kochel T, Hoepfner R W, Timmons S E, Sinden R R. Torsionally tuned cruciform and Z-DNA probes for measuring unrestrained supercoiling at specific sites in DNA of living cells. J Mol Biol. 1991;221:107–129. doi: 10.1016/0022-2836(91)80208-c. [DOI] [PubMed] [Google Scholar]