Abstract
The mRNA of vascular endothelial growth factor (VEGF), the major angiogenic growth factor, contains an unusually long (1,038 nucleotides) and structured 5′ untranslated region (UTR). According to the classical translation initiation model of ribosome scanning, such a 5′ UTR is expected to be a strong translation inhibitor. In vitro and bicistronic strategies were used to show that the VEGF mRNA translation was cap independent and occurred by an internal ribosome entry process. For the first time, we demonstrate that two independent internal ribosome entry sites (IRESs) are present in this 5′ UTR. IRES A is located within the 300 nucleotides upstream from the AUG start codon. RNA secondary structure prediction and site-directed mutagenesis allowed the identification of a 49-nucleotide structural domain (D4) essential to IRES A activity. UV cross-linking experiments revealed that IRES A activity was correlated with binding of a 100-kDa protein to the D4 domain. IRES B is located in the first half of the 5′ UTR. An element between nucleotides 379 and 483 is required for its activity. Immunoprecipitation experiments demonstrated that a main IRES B-bound protein was the polypyrimidine tract binding protein (PTB), a well-known regulator of picornavirus IRESs. However, we showed that binding of the PTB on IRES B does not seem to be correlated with its activity. Evidence is provided of an original cumulative effect of two IRESs, probably controlled by different factors, to promote an efficient initiation of translation at the same AUG codon.
The vascular endothelial growth factor (VEGF) is a potent endothelial cell mitogen that plays a crucial role in the regulation of both physiologic and pathologic angiogenesis (10, 44). VEGF is involved not only in embryogenic development and differentiation of the vascular system, in wound healing, and in reproductive function but also in pathologic angiogenic processes such as proliferative retinopathies, tumor growth, arthritis, and psoriasis (10).
Numerous studies have been devoted to understanding the expression regulation of this factor, especially at the transcriptional level. A wide range of cytokines or oncogenic proteins, including interleukins 1β (31) and 6 (8), insulin-like growth factor 1 (IGF-1) (14), tumor growth factor β (TGF-β) (5), c-Src (36), v-Raf (15), and Ras (46), and oxygen tension have been shown to regulate VEGF gene transcription (48). VEGF may also be posttranscriptionally regulated. The VEGF pre-mRNA undergoes alternative splicing which generates four polypeptide isoforms of 121, 165, 189, and 206 amino acids (11), the functions of which have not yet been fully defined. VEGF mRNA stability is also influenced by hypoxic conditions or by IGF-1 expression (49, 57). Finally, posttranslational modifications of VEGF isoforms, including plasmin and urokinase proteolysis or glycosylation, have been described (11, 43).
Surprisingly, very little is known about the possible translational control of VEGF messenger except for a stimulatory effect on VEGF translation in CHO cells overexpressing the cap binding protein eukaryotic initiation factor 4E (eIF4E) (25). However, the VEGF mRNA presents unusual features also found in other RNAs of viral origin or transcribed from cellular proliferation regulator genes, such as the fibroblast growth factor 2 (FGF-2) gene, the platelet-derived growth factor (PDGF) gene, or the c-myc proto-oncogene. The 5′ untranslated region (UTR) of the mRNA is unusually long, as the transcription starting point is located 1,038 nucleotides (nt) upstream from the AUG initiation codon and heavily structured due to a high percentage of G and C residues. The 5′ UTR region also contains three noncanonical upstream CUG codons in frame with the initiator AUG codon. All of these elements make the use of a conventional ribosome scanning model for translation initiation very difficult (26).
A cap-independent mechanism involving an internal ribosome entry site (IRES) has been identified in picornavirus messengers, which are uncapped and present a long 5′ UTR (20, 41). The presence of an IRES has also been reported for many viral (cardiovirus, rhinovirus, and aphthovirus) (21) and some cellular human (Bip, FGF-2, IGF-II, eIF4G, PDGF, and c-Myc) (2, 13, 33, 37, 50, 51, 53) messengers and in Drosophila antennapedia and ultrabithorax mRNAs (39, 59). The IRESs discovered so far differ in their primary sequences but show similarities in their secondary structures which appear to be crucial to IRES function (21, 28). In several picornaviruses, the internal entry process has been shown to require cellular factors including the polypyrimidine tract binding protein (PTB) (1), which is also involved in the nuclear splicing regulatory pathway (40, 56).
We show here that the mRNA of the major angiogenic factor is translated by an internal ribosome entry process. Furthermore, we demonstrate that the VEGF 5′ mRNA leader contains two independent IRESs which are able to promote efficient translation at the AUG start codon. The patterns of cellular proteins binding to the two IRESs are clearly different. These data suggest that different factors could control the activities of these IRESs.
MATERIALS AND METHODS
Plasmid constructions.
The VEGF cDNAs and the DNA fragment corresponding to 5′ UTR of the messenger were kindly provided by J. Abraham (52) and cloned into a pKS-derived plasmid. PCR was performed with sense oligonucleotide 5′AAATCTAGAATTCGCGGAGGCTTGGGGCA3′ and antisense oligonucleotide 5′GGTATCGATTGGATGGCAGTAG3′ to construct a translational fusion between VEGF and chloramphenicol acetyltransferase (CAT). The amplified fragment extending from positions 1 to 1205 (corresponding to the 5′ UTR and the 167 nt downstream from the AUG codon) was cloned in a previously described pSCT-derived vector (45) upstream from the CAT coding sequence, under the control of cytomegalovirus (CMV) and T7 promoters. This chimeric construct, called pVC, was expected to encode one VEGF-CAT protein of 32 kDa.
A hairpin (ΔG = −40 kcal/mol) (described in reference 53) was introduced into the pVC construct between the promoters and the 5′ end of VEGF cDNA, leading to the pHVC construct. Bicistronic vectors were constructed from the previously described pSCTCAT plasmid, which contains the CAT gene downstream from the CMV promoter (45). The IVS2β intron was removed from this plasmid, and a synthetic intron, obtained from plasmid pRLCMV (Promega), was added just downstream from the CMV promoter. A second CAT gene was then introduced downstream from the intron. This intermediate vector was called pCC. All or part of the 5′ UTR leader of VEGF plus 167 nt of the coding sequence was then cloned in the intercistronic region between the two CAT genes. The bicistronic vector containing the entire 5′ UTR (nt 1 to 1205), designated pCVC, was expected to encode two CAT proteins of 24 and 32 kDa. The above-described hairpin (ΔG = −40 kcal/mol) was introduced into pCVC between the CMV promoter and the first cistron. This construct was designated pHCVC. Different deletions of the VEGF 5′ UTR were performed. Removal of the first 475 nt in the 5′ UTR (up to the BamHI site) resulted in plasmid pCVC1, removal of the first 654 nt (up to the XmnI site) resulted in pCVC2, removal of the first 745 nt (to Nhe site) yielded pCVC3, removal of the first 846 nt (to the BanII site) resulted in pCVC4, and removal of the first 917 nt (up to the Sma site) gave pCVC5. We then constructed another bicistronic plasmid to determine the 3′ border of the IRES. This plasmid contained the entire VEGF 5′ UTR, in which the AUG codon of VEGF was directly fused to a chimeric CAT gene (fCAT) resulting from translational fusion of part of the nucleolin gene with the CAT gene (pSVNC82) (9). This chimeric fCAT gene was used to discriminate between the products of the two CAT cistrons encoded by this bicistronic vector during Western immunoblotting. This 5′ UTR-fCAT fusion was obtained by PCR amplification using oligonucleotides T7 (matching sequence upstream from the 5′ UTR), and 5′ AAACCTAGGGCCCAAGTTCATGGTTTCGGAG 3′ (matching nt 1029 to 1046) on plasmid pVC. This PCR fragment was digested at the ApaI site (underlined sequence) and fused to fCAT in the pCC vector deleted from the second CAT cistron. This CAT-VEGF 5′ UTR (nt 1 to 1046)-fCAT-containing plasmid was called pCVC0. Several deletions were then made from this plasmid in the VEGF 5′ UTR. The plasmid was called pCVC30 when the first 745 nt were deleted, pCVC60 when nt 654 (XmnI site) to 917 (Sma site) were removed, pCVC70 when nt 554 (NaeI site) to 1013 (NaeI site) were deleted, pCVC90 when nt 1 to 91 (AgeI site) as well as nt 554 to 1013 were removed, and pCVC80 when nt 332 (SacI site) to 1013 (NaeI site) were deleted.
We also constructed a set of bicistronic vectors containing two different reporter cistrons, i.e., the renilla and firefly genes coding for luciferase enzyme, designated LUCr and LUCf, respectively. LUCr was cloned from plasmid pRLCMV (Promega), whereas the LUCf gene was obtained from plasmid pGL3LUCf (Promega). For technical convenience, the XbaI and NarI sites present in the 5′ end of the LUCf cDNA were mutated by a single base pair change using complementary oligonucleotides (5′ CTAGTGGATAGAATGGTGCCGGGCCTTTCTTTATGTTTTTGGCGTCTTCCATACCA 3′ and 5′ AGCTTGGTATGGAAGACGCCAAAAACATAAAGAAAGGCCCGGCACCATTCTATCCA 3′). The backbone of this new bicistronic vector was the same as that of pCVC. The full 5′ UTR (nt 1 to 1046) was cloned between the two LUC genes; this construct was called pRVL1. We also cloned nt 745 to 1046 in the intercistronic region and called the construct pRVL2.
A deletion was performed in pRVL2 in which nt 848 (BanII site) to 918 (Sma site) were replaced with complementary oligonucleotides 5′ CATGGACAGGCCCTGGC 3′ and 5′ CCGGGCCAGGGCCTGTCCATGAGCC 3′. This construct was called pRVL2ΔD4. Another deletion plasmid (pRVL3) constructed from pRVL2 consisted of a deletion of nt 917 (Sma site) to 1013 (NaeI site). pRVL4 contained a 5′ UTR fragment limited by nt 801 to 1046 in the intercistronic space. This fragment was obtained by PCR from plasmid pCVCO, using oligonucleotide 5′ TTTGGATCCGAAGGAAGAGGAGAGGGGGC 3′ (matching residues 798 to 817) and a reverse one located in the CAT gene (5′ GCAACTGACTGAAATGCC 3′). This amplified fragment was then restricted at the ApaI (see description of pCVCO above) and BamHI (underlined) sites to obtain only the corresponding VEGF sequence. pRVL6 was obtained after cloning of the 5′ UTR fragment (nt 91 to 554 and 1013 to 1039) contained in plasmid pCVC90 between the two LUC genes. pRVL7, pRVL8, pRVL9, and pRVL10 were constructed from pRVL6 after deletion of nucleotides 483 (PvuI site) to 1013 (NaeI site), 379 (BssHII site) to 1013, 332 (SacI site) to 1013, and 189 (DraI site) to 1013, respectively. pRVL11 contained nt 134 to 483 and 1013 to 1039. This fragment was obtained from plasmid pRVL6 by PCR using oligonucleotide 5′ AAAGAATTCAGATCTTTGATATTCATTGATCCGGG 3′ (matching residues 134 to 155) and a reverse one located in the LUCf gene. This amplified fragment was then restricted by the EcoRI (underlined) and the PvuI sites and recloned in the intercistronic space. pRVL12 was constructed in the same way, using oligonucleotide 5′ AAAGAATTCAGATCTTGAATCGGGCCGACGGCT 3′ (matching residues 241 to 260), and thus contained nt 241 to 483 and 1013 to 1039 between the two LUC genes. pRVL13 consisted of a deletion of nucleotides 379 (BssHII site) to 1013 (NaeI site) from pRVL12.
Monocistronic vectors derived from plasmid pSCTCAT in which VEGF 5′ UTR fragments had been introduced upstream from the CAT gene were constructed for cross-linking experiments. pVC30 contained nt 745 to 1046 upstream from the CAT gene. We also cloned the mutated 5′ UTR fragment contained in pRVL2ΔD4 (nt 745 to 858 and 907 to 1046) upstream from the CAT gene. This plasmid was called pVC30ΔD4. pVC1 was a pVC-derived plasmid in which nt 1 to 475 were deleted. Probes C and D (see Fig. 8A) are derived from plasmids pSKV3 and pKSV4, pKS-derived vectors in which were inserted the VEGF 5′ UTR regions contained in plasmids pRVL11 (nt 134 to 483 and 1013 to 1046) and pRVL12 (nt 241 to 483 to 1013 to 1046), respectively.
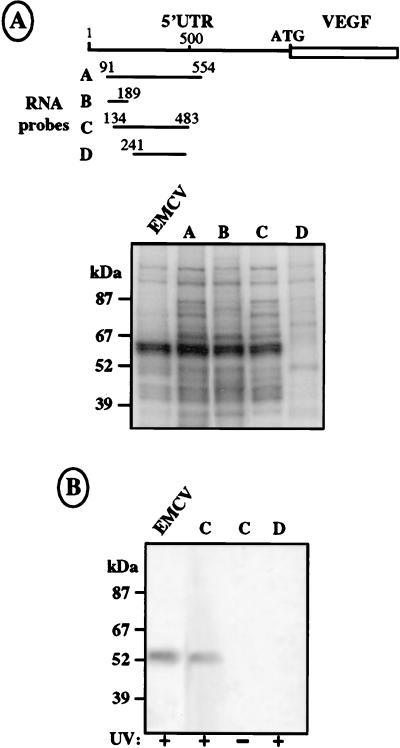
FIG. 8.
UV cross-linking of COS-7 cell proteins on IRES B. (A) Top, drawing of the different 32P-labeled RNA probes, obtained from T7 in vitro transcription and corresponding to the complete or parts of the IRES B sequence. Relative positions of the 5′ and 3′ ends of each probe are indicated. Bottom, UV cross-linking experiments performed with probes A to D and a probe corresponding to the EMCV IRES (first lane). S10 COS-7 cell extracts were incubated with 1.5 × 106 cpm of the different probes followed by UV irradiation and treatment with RNases A and ONE (see Materials and Methods). Size markers are indicated. (B) Immunoprecipitation with an anti-PTB antibody of COS-7 cell protein extract cross-linked to EMCV and VEGF probes C and D (lane 1, 2, and 4). Lane 3 corresponds to a control immunoprecipitation of cell extract and probe C without previous cross-linking. UV cross-linking or absence of cross-linking before immunoprecipitation is indicated by a plus or a minus sign, respectively, below each lane.
Two new plasmids, pVC80 and pVC90, were constructed for in vitro translation by inserting the fragments of the VEGF 5′ UTR contained in plasmids pCVC80 and pCVC90 (nt 1 to 332 and 1013 to 1046 and nt 91 to 554 and 1013 to 1046) into the pSCTCAT vector upstream from the CAT gene. We also constructed vector pVC5, a derivative from pVC in which nt 1 to 917 were deleted.
Probes transcribed from two pKS-derived vectors, called pKSV1 and pKSV2, were used for the RNase protection assays. We inserted nt 1 to 745 (bordered by EcoRI and NheI sites) into plasmid pKS digested at the XbaI and EcoRI sites in pKSV1. In pKSV2, we inserted nt 1 to 1046 (bordered by the XbaI and ApaI sites) into plasmid pKS digested by XbaI and ApaI.
In vitro translation.
Plasmids pVC, pVC90, pVC80, pVC30, and pVC30ΔD4 were linearized downstream from the 3′ end of the CAT coding sequence at the BglII site. pSCT11A (53) was linearized downstream from the 3′ end of the FGF-2 coding sequence at the HindIII site. Capped or uncapped RNAs were generated in vitro, using the T7 mMessage mMachine kit (Ambion) according to the manufacturer’s instructions, with or without adding m7GpppG (0.5 mM) to the transcription reaction. RNA transcripts were quantified by absorbance at 260 nm and ethidium bromide staining on an agarose gel, which also allowed verification of their integrity. In vitro translation in rabbit reticulocyte lysate (RRL; Promega) was performed as previously described (45), in the presence of [35S]methionine (Amersham). Translation products were analyzed by polyacrylamide gel electrophoresis (PAGE) in a sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel (45); the dried gels were scanned with a PhosphorImager apparatus (Molecular Dynamics), and quantification of the bands was performed with Imagequant software.
DNA transfection and Western immunoblotting.
COS-7 monkey cells were transfected with Fugene-6 transfection reagent (Boehringer) according to manufacturer’s instructions or by the DEAE-dextran method as described previously (45). Forty-eight hours after transfection, either (i) the cells were scraped in phosphate-buffered saline (PBS) and the pellets were resuspended in 1% SDS solution and sonicated or (ii) the cells were lysed in 1 ml of Tri-Reagent (Euromedex). Following the latter method and after total RNA extraction (see below), the proteins were precipitated with 1 volume of isopropanol. The protein pellet was then washed five times in 2 ml of a 0.3 M guanidine hydrochloride–95% ethanol buffer and once in 2 ml of ethanol. The protein pellet was then heated to 65°C for 20 min and resuspended in 1% SDS solution. Total proteins were quantified by the bicinchoninic acid assay (Pierce) (absorbance at 562 nm), and 10 μg of proteins from each cell lysate was used for Western immunoblotting. In summary, lysates were heated for 2 min at 95°C in SDS- and dithiothreitol (DTT)-containing sample buffer, separated in a 12.5% polyacrylamide gel, and transferred onto a nitrocellulose membrane. CAT proteins were immunodetected with rabbit polyclonal anti-CAT antibodies prepared in the laboratory (1/10,000 dilution). Antibodies were detected with an enhanced chemiluminescence kit (Amersham).
Cellular RNA purification.
Total cellular RNAs were prepared by the Tri-Reagent method (Euromedex), derived from the guanidinium thiocyanate procedure (7). A total of 5 × 106 transfected cells were scraped, centrifuged, and lysed in 1 ml of Tri-Reagent. RNA was extracted after addition of 0.2 ml of chloroform and precipitated with isopropanol. After an ethanol wash and precipitation, the RNA was quantified by measuring the absorbance at 260 nm and checked for integrity by electrophoresis on an agarose gel and ethidium bromide staining.
RNase mapping.
A complementary-strand RNA probe was generated in vitro by T7 or T3 RNA polymerase (Promega) according to manufacturer’s instructions, using a linearized DNA template and in the presence of 50 μCi of [α-32P]UTP. Probe A was transcribed by using T3 polymerase from plasmid pKSV2 linearized at the XbaI site. Probe C was transcribed by using T3 polymerase from plasmid pKSV2 linearized at the BamHI site. Probe B was transcribed by using T7 polymerase from plasmid pKSV1 linearized at the EcoRI site. 32P-labeled RNA was purified by using the RNaid kit (Bio 101). The RNase protection experiments were performed with an RPAII kit (Ambion) according to the manufacturer’s instructions. A 10-μg aliquot of total cellular RNA and a large excess (2 × 106 cpm) of probe were precipitated with ethanol; 10 μg of yeast RNA can be added to the total RNA before precipitation to increase the size of the RNA pellet. All experiments described were tested with and without addition of RNA. As a control experiment, total RNA samples were incubated 15 min at 37°C in the presence of 10 U of RNase-free DNase 1 prior to the precipitation in order to avoid DNA contamination. The pellet was resuspended in 20 μl of hybridization buffer, heated at 90°C for 4 min, and incubated at 42°C for 14 h. Then 200 μl of RNase digestion buffer containing 20 U of RNase T1 and 1 μg of RNase A were added, and the reaction mixture was incubated for 30 min at 37°C; 300 μl of inhibitor RNase buffer and 10 μg of carrier yeast RNA were then added, and the reaction mixture was precipitated at −20°C for 20 min. The pellet was resuspended in 8 μl of gel loading buffer, denatured, and fractionated on a 5% acrylamide–8 M urea gel at 200 V for 1.5 h. The gel was then fixed, dried, and autoradiographed. Control experiments showed that all probes described, used alone or only with yeast RNA, were totally digested after RNase mix treatment.
UV cross-linking assay and immunoprecipitation.
Cytoplasmic extracts from COS-7 cells were prepared as previously described (53). Confluent cells were scraped in PBS and centrifuged. The cell pellet was resuspended in lysis buffer (10 mM NaCl, 10 mM Tris-HCl [pH 7.4], 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM DTT), and frozen-thawed three times. The extract was centrifuged at 12,000 × g for 10 min, and the supernatant (S10) was brought to 5% (vol/vol) glycerol and frozen in aliquots at −80°C.
Probes A (nt 1 to 1121) and B (nt 1 to 475) (see Fig. 6A) were transcribed by using T7 polymerase from plasmid pVC linearized at the NcoI site and BamHI sites, respectively. Probe C (nt 475 to 1121; Fig. 6A) was transcribed by using T7 polymerase from plasmid pVC1 linearized with NcoI. Probes D (nt 745 to 1046) and E (nt 745 to 858 and 907 to 1046) (Fig. 6A) were transcribed by using T7 polymerase from plasmids pVC30 and pVC30ΔD4 linearized with Bsp120.1. Probes A (nt 91 to 554 and 1013 to 1046) and B (nt 91 to 189) (Fig. 8A) were transcribed from plasmid pVC90 linearized with Bsp120.1 and DraI, respectively. Probes C (nt 134 to 483) and D (nt 241 to 483) (Fig. 8A) were transcribed from plasmids pKSV3 and pKSV4 linearized with NaeI.
FIG. 6.
UV cross-linking of COS-7 cell proteins on the VEGF mRNA 5′ UTR. (A) Drawing of the different 32P-labeled RNA probes, obtained from T7 in vitro transcription and corresponding to the complete or parts of the VEGF 5′ UTR mRNA. Relative positions of the 5′ and 3′ ends of each probe are indicated. (B) UV cross-linking experiments performed with probes A to E. S10 COS-7 cell extracts were incubated with 106 cpm of the different probes followed by UV irradiation and treatment with RNases A and ONE (see Materials and Methods). The control (Ct) lane corresponds to proteinase K treatment of sample loaded in the first lane. Size markers are indicated. (C) 32P-labeled probes corresponding to VEGF probe A (complete 5′ UTR) and to EMCV IRES were cross-linked with proteins extracted from S10 COS-7 cells, and the complex was immunoprecipitated with an anti-PTB antibody. The samples were analyzed before (CL) and after (I) immunoprecipitation. The use of a VEGF or EMCV probe is indicated above the lanes. PTB migration is indicated with an arrow.
32P-labeled RNA probes (1 × 106 to 1.5 × 106 cpm) were incubated with 6 μg of S10 extract, preincubated or not with 2.5 μg of calf liver tRNA (Boehringer Mannheim) for 15 min at 30°C, in buffer containing 5 mM HEPES (pH 7.5), 25 mM KCl, 2 mM MgCl2, 3.8% glycerol, 0.2 mM DTT, and 1.5 mM ATP in a final volume of 10 μl at 30°C for 15 min (34). Samples were transferred to ice and irradiated with a UV Stratalinker (Stratagene) by being fixed 10 cm from the bulbs and routinely irradiated with 400,000 μJ/cm2 at 254 nm; 2.5 μg of calf liver tRNA was then added to calibrate the RNase digestion when no prior incubation of the S10 extract with tRNA had been performed. The samples were then treated with a mix of RNase ONE (5 U; Promega) and 2.5 μg of RNase A at 37°C for 30 min and, when indicated, with proteinase K (Sigma) at 37°C for 20 min at a final concentration of 1 mg/ml. PAGE sample buffer was added, and the samples were heated for 2 min at 95°C and loaded on an SDS–10 or 12.5% polyacrylamide gel. The gel was fixed in 30% methanol–10% acetic acid for 30 min, dried, and autoradiographed.
The cross-linked proteins were immunoprecipitated with Pansorbin as follows. Ten microliters of the cross-linked 32P-labeled sample (see below) was diluted to 150 μl in PBS–Nonidet P-40 (NP-40) buffer (1× PBS, 50 mM NaF, 2 mM EDTA, 2 mM EGTA, 0.05% NP-40) and precleared by incubation with 50 μl of Pansorbin for 10 min at room temperature (RT). The supernatant was incubated for 30 min at RT with 5 μl of anti-PTB antibody (kindly provided by J. G. Patton) (40) and then for 30 min at RT with 50 μl of Pansorbin. After five washes in HEPES–NP-40 buffer (150 mM NaCl, 15 mM HEPES [pH 7.4], 1 mM EDTA [pH 7.4], 0.5% NP-40), the samples were analyzed by PAGE (10 or 12.5% gel) as described above.
Dual luciferase assay.
LUCf and LUCr activities were measured by using the Dual-Luciferase reporter assay system (Promega). Transfected COS-7 cell plates (60 by 15 mm) or 24-well dishes were rinsed twice with PBS, scraped, and homogenized in 400 μl of lysis reagent provided with the kit 24 to 48 h after transfection. The lysate was cleared by a 2-min centrifugation at 4°C. Chemiluminescent signals were measured in a luminometer (Berthold) equipped with automatic injectors. A 20-μl volume of extract was incubated with 100 μl of Luciferase Assay Reagent II (Promega) for 2 s, and LUCf activity was measured for a period of 15 s; 100 μl of Stop and Glo buffer (Promega), stopping the firefly enzymatic reaction and containing the substrate for LUCr enzyme, was then injected. Luminescence corresponding to LUCr activity was measured for 15 s starting 3 s after injection.
RESULTS
Identification of an IRES in the 5′ UTR of the VEGF mRNA.
Two alternative strategies involving hairpin-containing monocistronic vectors or bicistronic vectors were used to study whether VEGF expression was translationally regulated by an internal ribosome entry process. The first approach had been successful for identification of Moloney murine leukemia virus IRES (55), and the second had been used to identify IRESs in several viral and cellular mRNAs (22, 33, 41).
DNA plasmids were designed to contain the 1,038 nt of the VEGF 5′ UTR and the first 167 nt of its coding sequence fused in frame with the CAT coding sequence (Fig. 1A). The predicted size of the chimeric VEGF-CAT protein encoded by this vector was 32 kDa. In the monocistronic plasmid (Fig. 1A, construct A [pVC]), this 5′ UTR-VEGF-CAT sequence was controlled by the CMV and T7 promoters. In a second construct, a stable (ΔG = −40 kcal/mol) hairpin was added downstream from the promoters to impair the cap-dependent ribosome scanning (construct B [pHVC]). This latter construct was expected to encode a 32-kDa VEGF-CAT protein only if there was an IRES in the VEGF mRNA leader.
FIG. 1.
Identification of an IRES in the VEGF mRNA 5′ UTR. (A) Schematic representation of the chimeric constructs used for transfection experiments. pVC (construct A) corresponds to the VEGF-CAT fusion in which nt 1 to 1205 of the VEGF cDNA were fused to the CAT coding sequence (see Materials and Methods). This fusion gives rise to a chimeric VEGF-CAT protein of 32 kDa. pHVC (construct B) is derived from pVC and carries an additional 5′ hairpin (ΔG = −40 kcal/mol) downstream from the CMV promoter. pCVC (construct C) is a bicistronic vector containing the CAT gene as a first cistron upstream from the VEGF-CAT fusion in the pVC construct. pHCVC (construct D), derived from pCVC, contains a 5′ hairpin (ΔG = −40 kcal/mol) upstream from the first CAT cistron. (B) The constructs depicted in panel A were transiently transfected in COS-7 cells, and their expression was analyzed by Western immunoblotting using an anti-CAT antibody. The amount of transfected cell protein extract loaded on each lane was adjusted to the quantity of the mono- and bicistronic mRNAs present in each extract. The control (Ct) lane corresponds to untransfected COS-7 cells. The positions of CAT and VEGF-CAT proteins are indicated by arrows.
Two bicistronic constructs were also derived from the pVC construct by subcloning the CAT coding sequence upstream from the 5′ UTR-VEGF-CAT sequence (Fig. 1A, construct C [pCVC]) and adding a 5′ hairpin structure upstream from the CAT first cistron (construct D [pHCVC]). A 24-kDa CAT protein and a 32-kDa VEGF-CAT protein were expected to be translated from the first and second cistrons, respectively, if the VEGF mRNA 5′ UTR contained an IRES.
The four constructs were separately transfected into COS-7 cells, and the translation products were analyzed by Western immunoblotting using an anti-CAT antibody (Fig. 1B). The amount of mRNA encoded by the transfected constructs was analyzed in each extract by RNase protection assay, and the signals were quantified with a PhosphorImager (data not shown). The amounts of transfected COS-7 cell protein extracts involved in the Western blotting experiment were adjusted to the quantity of mono- and bicistronic mRNAs present in each extract. A 32-kDa VEGF-CAT protein was detected as a major band with all four constructs, while a minor band band, probably corresponding to a proteolysis product of the 32-kDa protein, was observed at 28 kDa (Fig. 1B, lanes A to D). The protein was expressed from the hairpin-containing monocistronic vector pHVC (lane B) as well as from the bicistronic vector (lanes C and D), even though the level of expression was lower than that obtained from the monocistronic vector pVC. This phenomenon had been observed previously with FGF-2 and Moloney murine leukemia virus mRNAs (53, 55) and can be explained by a contribution of cap-dependent initiation in addition to the internal ribosome entry process in these vectors. Interestingly, the second cistron (VEGF-CAT) was more efficiently expressed than the first cistron (CAT) from the bicistronic pCVC vector (lane C). Furthermore the presence of a 5′ hairpin in the bicistronic construct strongly decreased CAT translation without affecting that of the chimeric VEGF-CAT protein, thereby confirming that expression of the second cistron was not due to a reinitiation event (lane D). These results revealed the presence of a very efficient IRES in the VEGF mRNA leader sequence.
Localization of IRES A in the VEGF mRNA 5′ UTR.
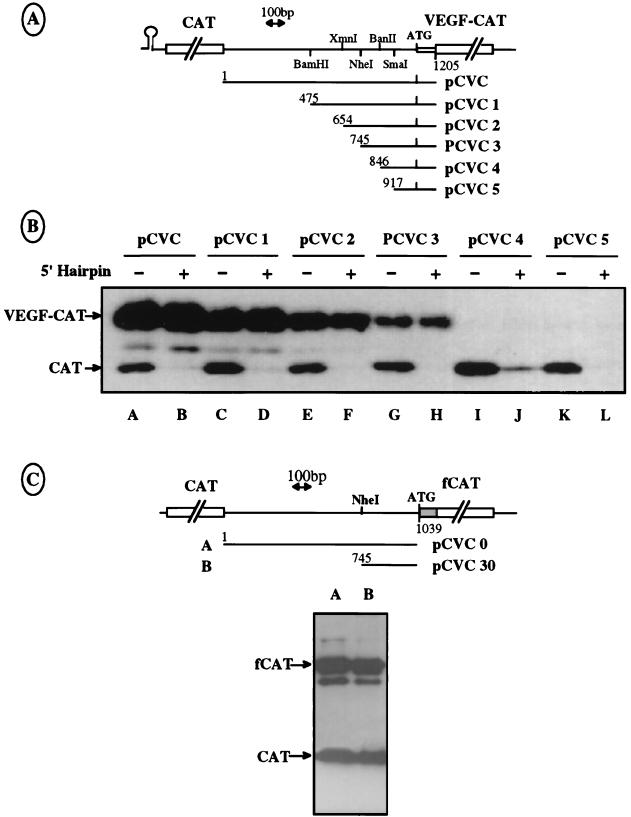
A number of additional bicistronic constructs containing shortened intercistronic UTRs, with or without a 5′ hairpin (Fig. 2A), were designed to more precisely localize the IRES structure in the VEGF mRNA 5′ UTR. Identical amounts of each DNA construct were used to transfect COS-7 cells, and the level of protein synthesis was determined as described above. The same quantity of COS-7 cell protein extracts was loaded in all lanes.
FIG. 2.
Mapping of the IRES by progressive deletions in the VEGF mRNA 5′ UTR. (A) Schematic representation of the different deletions of the 5′ leader performed in the bicistronic vectors pCVC and pHCVC. Only the pHCVC series is represented here. (B) Western immunoblotting was performed as described for Fig. 1B after transfection of COS-7 cells with the constructs detailed in panel A. The presence or absence of a hairpin in the vector, as well as the name of the vector, is indicated above each lane. The same quantity of COS-7 cell protein extract was loaded in all lanes. Positions of the CAT and VEGF-CAT proteins are indicated with arrows. (C) Representation of two bicistronic vectors containing another VEGF-CAT fusion in which nt 1 to 1046 (including the AUG codon) of the VEGF 5′ leader are fused to the chimeric fCAT gene resulting from the translational fusion of part of the nucleolin gene with the CAT gene (9). These two plasmids were transfected in COS-7 cells, and the extracts were analyzed as described for Fig. 1B. The positions of the CAT and fCAT proteins are indicated with arrows.
As expected, the first cistron CAT was expressed in cells transfected with the different constructs lacking the 5′ hairpin (Fig. 2B, lanes A, C, E, G, I, and K), whereas its expression was strongly diminished in cells transfected with plasmids containing the 5′ hairpin (lanes B, D, F, H, J, and L). Expression of the VEGF-CAT second cistron was detected for deletions of the first 475, 654, and 745 nt of the VEGF 5′ UTR (lanes C to H, respectively) and was not affected by the presence of a 5′ hairpin. In contrast, no expression of VEGF-CAT was observed with a shorter intercistronic sequence starting at position 846 or 917 of the leader 5′ end.
These results indicate that the 101-nt-long fragment delimited by positions 745 and 846 (between positions −293 and −192 upstream from the AUG codon) is required for the formation of a functional IRES. Hereafter this fragment will be referred to as the IRES A 5′ region. Although the 293 nt upstream from the AUG codon seem sufficient for IRES function, Fig. 2B shows that the translation efficiency of the VEGF-CAT mRNA decreased with shortening of the 5′ UTR (compare lanes A to H). This finding suggests that sequences located at different positions in the 5′ UTR are required for optimal IRES efficiency.
Two additional constructs were made to determine the involvement of the VEGF coding sequence located downstream from the AUG codon in the internal entry process. They consisted of fusion of the full VEGF 5′ UTR or its 3′ 293 bp up to and including the AUG codon with the chimeric fCAT gene and composed of 560 neutral nucleotides of the nucleolin coding sequence in frame with the CAT gene. This chimeric fCAT reporter gene was used to discriminate the products of the two CAT cistrons in Western immunoblotting. The results showed an efficient expression of the fCAT protein from both constructs (Fig. 2C), revealing that the first 167 coding nucleotides of the VEGF coding region, present in the plasmids used previously (Fig. 2A), were not required for efficient internal entry of the ribosomes on the messenger.
Altogether, these data clearly localize an IRES within the 293 nt upstream from the AUG start codon of VEGF mRNA. This IRES will hereafter be called IRES A.
Verification of the bicistronic mRNA integrity in transfected COS-7 cells.
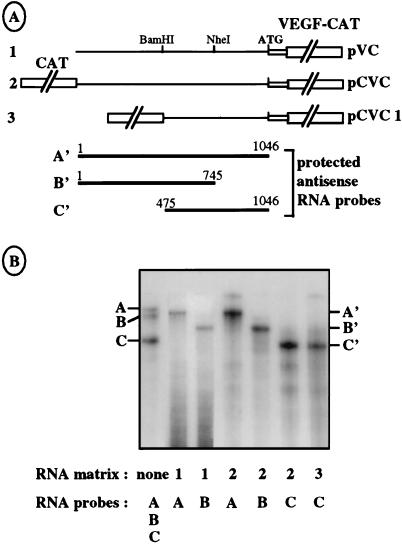
Bicistronic RNA integrity was assessed by RNase protection assays (see Materials and Methods) to rule out the possibility that the observed VEGF-CAT protein could be expressed from unexpected monocistronic mRNA generated by a cleavage or a cryptic promoter located in the VEGF 5′ UTR. The protection experiments were performed with total RNA from COS-7 cells transfected with bicistronic or monocistronic constructs containing either the complete leader sequence, a deletion of the first 475 nt, or a deletion of the last 293 nt (Fig. 3A). Three probes, A, B, and C, with expected sizes of 1054, 748, and 572 nt were used for hybridization with transfected COS-7 total RNA before RNase digestion; they were expected to produce protected fragments of 1,017, 696, and 535 nt, respectively, after RNase digestion. The data presented in Fig. 3B show that the protected fragments are unique and of the expected sizes, thus demonstrating that the intercistronic region is full size in the bicistronic mRNA.
FIG. 3.
Verification of the integrity of the bicistronic mRNA by RNase protection assay. (A) Schematic representation of monocistronic vector pVC (construct 1) and bicistronic vectors pCVC and pCVC1 (constructs 2 and 3) used to generate RNA templates. The regions A′, B′, and C′, protected by the three antisense RNA probes A, B, and C, are indicated. The RNA probes A, B, and C are slightly longer than the protected fragments because of the presence of additional nucleotides in the polylinker regions of the plasmids used as templates for the probes (see Materials and Methods). (B) Vectors shown in panel A were transfected in COS-7 cells. Total mRNAs were purified and analyzed by RNase A and T1 protection (see Materials and Methods), using the RNA probes A (1054 nt), B (748 nt), and C (572 nt), complementary to nt 1 to 1046, 1 to 745, and 475 to 1046, respectively. The first lane corresponds to a mix of the three probes alone, without RNase treatment. The RNA templates and probes used are indicated at the bottom. The fragments protected by the probes A, B, and C are notated as A′ (1,017 nt), B′ (690 nt), and C′ (535 nt), respectively.
Characterization of the structural features of IRES A.
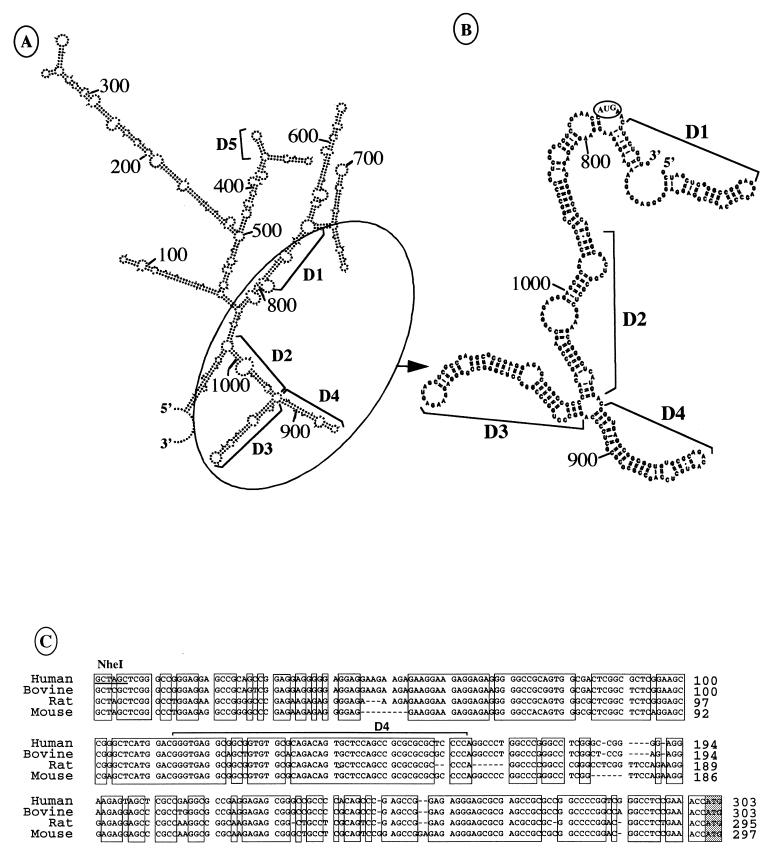
Figure 4 shows the secondary structures, predicted by the Zuker procedure (6), of the entire or 293-bp fragment of the 5′ UTR (nt 745 to 1038) demonstrated to be sufficient for IRES function (Fig. 2B). Extended base pairing is apparent all along this UTR fragment, bringing the AUG codon in the vicinity of the IRES A 5′ region (nt 745 to 846). A stem-loop structure, called D5, bearing an unpaired GNRA (where N is C, G, A, or U and R is G or A; in this case GUGA) sequence is located in the first half of the leader between nt 416 and 434. This motif was shown to be common to picornavirus IRES and implicated in aphthovirus IRES function (32). Involvement of this predicted structure in the VEGF IRES activity is discussed later.
FIG. 4.
IRES A secondary predicted structures and sequence conservation in mammals. (A) Secondary structure of the complete VEGF mRNA 5′ UTR predicted by the ESSA folding program (6). The 5′ and 3′ ends of the sequence corresponding to nt 1 and 1047, respectively, are indicated. Nucleotide positions and the IRES A predicted domains D1 to D4 are also indicated. D5 corresponds to a stem-loop structure bearing an unpaired loop-located GNRA sequence. (B) Secondary predicted structure of the IRES A. Nucleotide positions and the four domains D1 to D4 are indicated. The 5′ and 3′ ends of the region analyzed correspond to nt 745 to 1052, respectively. (C) Alignment of the cDNA sequence of the region corresponding to human IRES A with bovine, rat, and mouse VEGF cDNA sequences. The conserved regions are boxed. The D4 domain is indicated. Relative positions of the nucleotides aligned from the transcription start point of rat, human, and mouse cDNAs are indicated. The complete 5′ UTR of bovine VEGF mRNA is not known.
Interestingly, two predicted structured domains (D3 and D4, from nt 917 to 1013 and 858 to 907, respectively) are present downstream from the IRES A 5′ region (Fig. 4B). Furthermore, a DNA sequence comparison of the human (29), bovine (52), rat (30), and mouse (47) 3′ ends of the VEGF mRNA 5′ UTR revealed several regions highly conserved in these species, particularly the region corresponding to the D4 domain (Fig. 4C). This led us to study the potential role of these evolutionary conserved domains in IRES function.
cis elements involved in IRES A activity.
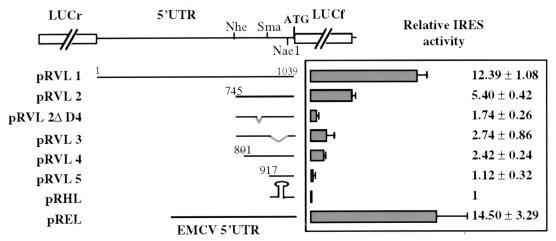
The predicted structural data shown in Fig. 4 were used to design new deletions for further characterization of the VEGF IRES A. A new bicistronic vector was produced in which the LUCr gene was subcloned as the first cistron and the (LUCf) gene was subcloned as the second cistron (Fig. 5) to improve sensitivity and quantification of the assay. The VEGF complete or deleted 5′ UTR (constructs pRVL1, pRVL2, and pRVL5), as well as new deleted fragments corresponding to removal of predicted domains D4 and D3 and the first 56 nt of IRES A (constructs pRVL2ΔD4, pRVL3, and pRVL4, respectively), were introduced into the intercistronic region. Either a hairpin (ΔG = −40 kcal/mol) or the encephalomyocarditis virus (EMCV) IRES (35) was introduced between the LUCr and LUCf genes as controls (construct pRHL or pREL, respectively). These plasmids were transfected in COS-7 cells as described above. The ratio between the activities of the two luciferase enzymes observed in the cell extracts was calculated and compared with the data obtained with the negative control construct pRHL (Fig. 5).
FIG. 5.
Characterization of IRES A cis-acting elements. Schematic drawing of the bicistronic vectors containing the LUCr gene as the first cistron, all or part of the VEGF mRNA leader sequence in the intercistronic region, and the LUCf gene as the second cistron. Construct pREL contains the EMCV IRES in the intercistronic region (positive control); construct pRHL contains a hairpin (ΔG = −40 kcal/mol) in the intercistronic region (negative control). These plasmids were transfected in COS-7 cells, and luciferase activities were measured as described in Materials and Methods. On the right, the histogram and corresponding values represent the ratio between the LUCf/LUCr activities obtained with each construct and that obtained with pRHL. Each value represents the average of at least four independent transfection experiments.
Clearly the full-size VEGF 5′ UTR induced LUCf expression comparable to that induced by the EMCV IRES (pRVL1 and pREL), whereas the 745-nt-deleted 5′ UTR containing the IRES A 5′ region (pRVL2) resulted in a reduced relative activity. These results were in agreement with those obtained in assays using the bicistronic CAT/VEGF-CAT mRNA (Fig. 2B), thereby ruling out any interference of the reporter system with the IRES activity.
Interestingly, removal of the D4 domain strongly affected translation of the LUCf gene (pRVL2ΔD4), while deletion of the D3 domain (pRVL3) resulted in only a 50% decrease in IRES activity compared to that obtained with the pRVL2 construct. In comparison to pRVL2 construct efficiency, deletion of the 5′ 56 nt of IRES A resulted in a 60% decrease of LUCf expression (pRVL4), indicating a very low IRES activity, albeit higher than that obtained with the D4 deletion. This result confirmed the observation made in Fig. 2. These results indicated that the IRES A 5′ end (nt 745 to 801) and the D4 domain (nt 858 to 907) are necessary for the IRES function. Both elements probably form the core of the VEGF IRES, whereas the D3 domain (nt 917 to 1013) seems also to play a role in IRES activity.
Identification of cellular factors bound to VEGF 5′ UTR and to IRES A.
UV cross-linking experiments were performed with COS-7 cell extracts and different 32P-labeled RNA probes corresponding to either the complete 5′ UTR or deleted fragments of the 5′ UTR (Fig. 6A) to identify cellular factors interacting with the VEGF 5′ UTR and particularly with IRES A. As shown in Fig. 6B, the protein pattern differed according to the RNA probe used. The complete leader was able to bind at least nine proteins (Fig. 6B, lane A). Most of these proteins were able to bind to probe B, corresponding to the upstream part of the leader (lane B). In contrast, probes C, D, and E bound only a small number of proteins. Three major proteins migrating at 120, 100, and 85 kDa were detected with probes C and D, corresponding to the downstream part of the leader and both containing the IRES (lanes C and D, bands a, b, and c). Finally probe E, in which the D4 domain was deleted, was able to bind the p85 and the p120 proteins but not p100 (lane E). These results showed a correlation between the cross-linking of p100 to RNA and IRES activity and suggested a potential role of the 100-kDa protein in IRES function.
With regard to the proteins cross-linked to the upstream part of the VEGF 5′ UTR, one of the major bound proteins had an apparent molecular mass of 60 kDa (Fig. 6B, lanes A and B, band P), close to that of PTB, a well-known protein involved in the activities of several picornavirus IRESs. This prompted us to immunoprecipitate the proteins cross-linked to the first half of the leader (probe B) with an anti-PTB antibody (Fig. 6C). An EMCV IRES probe was used as a positive control. As shown in Fig. 6C, PTB could be clearly detected following immunoprecipitation of both VEGF and the EMCV RNA cross-linked proteins with the anti-PTB antibody.
The VEGF mRNA 5′ UTR contains two distinct IRESs.
It is clear from data presented in Fig. 2B and 5 that the complete 5′ UTR behaved as a more efficient IRES than the IRES A fragment containing nt 745 to 1046. Furthermore, deletion of the D4 domain in the full-length 5′ UTR led to a 30% reduced internal entry activity compared to that observed with the corresponding complete leader (data not shown), whereas the same deletion in the IRES A context almost abolished IRES activity. We also showed above that the binding of PTB occurred in the 5′ part of the VEGF mRNA 5′ UTR. These data led us to investigate the presence of a second IRES in the 5′ part of the UTR.
To test this hypothesis, four new bicistronic plasmids containing different portions of the upstream half of the VEGF 5′ UTR were designed (Fig. 7A, left) and used to transfect COS-7 cells, and the cell extracts were analyzed by Western immunoblotting with an anti-CAT antibody. It was apparent from these experiments that three VEGF mRNA leader fragments corresponding to nt 1 to 654, 1 to 554, and 91 to 554, respectively, were able to promote translation of the second cistron fCAT (Fig. 7A, right, lanes B, C, and E). fCAT expression was, however, less efficient with these three constructs (lacking the IRES A) than with the complete leader (lane A). In contrast, the fragment extending from positions 1 to 332 exhibited very low IRES activity (Fig. 7A, lane D).
FIG. 7.
Mapping of IRES B. (A) Schematic drawing of the bicistronic constructs containing the complete or truncated VEGF 5′ UTR between the first CAT cistron and the second chimeric fCAT cistron (depicted in Fig. 2C). The 5′ and 3′ boundaries of the deletions are indicated. Western immunoblotting was performed (right) as described for Fig. 1B after transfection of COS-7 cells with plasmids A to E. The CAT and fCAT proteins are shown by arrows. The control (Ct) lane corresponds to untransfected cells. (B) Representation of bicistronic vectors containing a stable hairpin structure upstream from the LUCr gene (first cistron), fragments of the VEGF mRNA leader sequence in the intercistronic region, and the LUCf gene as the second cistron. The 5′ and 3′ boundaries of the deletions are indicated. These plasmids were transfected into COS-7 cells, and luciferase activities measured as described in Materials and Methods. On the right, the histogram and corresponding values represent the LUCf/LUCr activity ratio obtained with each construct and that obtained with pRHL. Each value represents the average of at least four independent transfection experiments.
Taken together, these results suggest that a second IRES is present between positions 91 and 554 of the VEGF 5′ UTR. To obtain a more precise and more quantitative characterization of this IRES, we subcloned a series of segments of the 5′ half of the VEGF UTR into the LUCr-LUCf vector depicted in Fig. 5. COS-7 cell transfection and luciferase activity measurements were performed as described above. With regard to the IRES 3′ border deletions, it was clearly apparent that nt 91 to 483 (pRVL7) retained most of the activity of the reference fragment characterized by the results in Fig. 7A (Fig. 7B, pRVL6, nt 91 to 554) and can be defined as IRES B. In contrast, the fragments containing nt 91 to 379, 91 to 332, and 91 to 189 (pRVL8, -9 and -10) had no significant IRES activity. These results suggest that a 104-bp fragment located between nt 379 and 483 is strictly necessary for IRES B activity since its deletion abolished the internal entry process. Deletions performed in the 5′ border (pRVL11, -12, and -13) showed that the two fragments containing nt 134 to 483 or 241 to 483 retained about 50% of IRES B activity (Fig. 7B, pRVL11 and -12). It was thus apparent that the sequence limited by nt 91 and 134 played a role in IRES B function. These data enabled us to conclude that IRES B was located in a 392-nt-long fragment between nt 91 and 483 and that a 104-nt segment at the 3′ end of this fragment was strictly necessary for IRES activity. It should be noted that this 104-nt segment contains the D5 predicted stem-loop structure bearing a GNRA sequence in the loop (Fig. 4A). We can thus hypothesize a possible role of the GNRA motif in a cellular IRES function.
PTB binding to IRES B is independent of IRES efficiency.
As we had shown that PTB was the main protein bound to the upstream half of the VEGF RNA leader sequence (Fig. 6), we were prompted to see whether PTB interacted with IRES B. As for Fig. 6, this was investigated by UV cross-linking and immunoprecipitation experiments using several fragments of the 5′ part of the leader with or without IRES activity as probes; the EMCV IRES was used as a control (Fig. 8). It was clearly apparent from these experiments that the RNA segments containing nt 91 to 189 and nt 134 to 483 were able to bind PTB with the same efficiency as the segment extending from nt 91 to 554 (Fig. 8, probes and lanes A, B, and C). In contrast, the fragment extending from nt 241 to nt 483 was no longer able to bind this protein (Fig. 8, probe and lane D). PTB binding to probe C was confirmed by immunoprecipitation with anti-PTB antibody (Fig. 8B, lane C).
These results permitted the localization of a PTB binding site between nt 134 and 189, a region which had no influence on IRES activity (Fig. 7B, pRVL11 and -12). Furthermore, PTB bound to fragments B and C but not to fragment D (Fig. 8), while internal entry activity was maintained in fragments C and D but was not detected in fragment B (Fig. 7B). This evidenced an absence of correlation between PTB binding and IRES B activity in our experimental conditions.
Activities of the two IRESs in RRL.
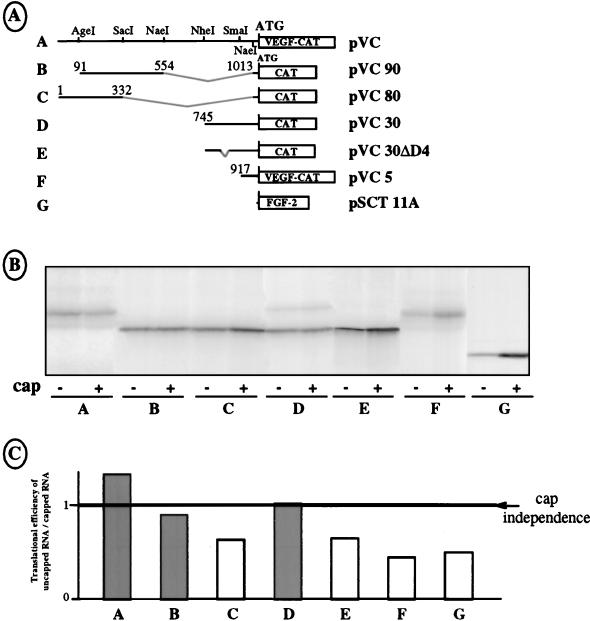
The ability of both IRESs to promote cap-independent translation in vitro was analyzed by using monocistronic RNA containing different fragments of the VEGF 5′ UTR fused to the CAT gene. A cap-dependent control which corresponded to the leader deleted FGF-2 mRNA (53) was included. Equal amounts of uncapped and capped monocistronic mRNAs were transcribed in vitro for each construct and assayed for translation in RRL (Fig. 9B). Cap independence was evaluated by calculating the ratio of CAT expression obtained from uncapped versus capped mRNA (NC/C ratio) (Fig. 9B and C).
FIG. 9.
Activities of IRES A and IRES B in vitro. (A) Schematic drawing of the monocistronic vectors used as T7 polymerase templates. The in vitro transcriptions were performed in the presence or absence of a cap structure. In constructs A and F, the translation product is the chimeric VEGF-CAT protein depicted in Fig. 1A. In constructs B to E, the translation product is the CAT protein. In construct G, the translation product is the FGF-2 protein. (B) Identical quantities of the different capped or noncapped mRNAs were translated in RRL. The presence or absence of a cap structure in the mRNA is indicated by a plus or minus sign, respectively, below each lane, together with the construct used. (C) The lanes in panel B were quantified with PhosphorImager. The histogram indicates the ratio of the translation efficiency observed in uncapped RNA versus capped mRNA. The line corresponds to cap-independent translation initiation (ratio of 1). The experiment reported here is representative of five independent experiments.
Interestingly, the 1,038-nt-long leader showed an NC/C ratio of 1.25, indicating mostly cap independence compared to the cap-dependent control (Fig. 9B and C; compare constructs A and G). Cap independence in the various deleted constructs was conferred by the fragment from nt 91 to 554, with an NC/C ratio of 0.9 (Fig. 8, construct B), whereas translation became cap dependent when the leader contained nt 1 to 332 (Fig. 8, construct C). Furthermore, the segment containing nt 745 to 1038 was also able to confer cap independence to translation, with an NC/C ratio of 1, whereas deletion of the D4 domain led to cap-dependent translation of the CAT gene (Fig. 8, constructs D and E). As expected, translation was cap dependent when the leader was restricted to nt 917 to 1038 (construct F).
These results show that two distinct mRNA segments are able to promote cap-independent translation in vitro. These segments correspond to IRESs A and B characterized in transfected COS-7 cells.
These different approaches enabled us to conclude that the VEGF mRNA leader contains two distinct IRESs which can promote cap-independent translation from AUG 1039 in both RRL and COS-7 transfected cells.
DISCUSSION
In this study, we demonstrate that the synthesis of VEGF, the major angiogenic factor, occurs through an internal ribosome entry site process. This observation, together with the previous discovery of an IRES in the mRNA of another important angiogenic factor, FGF-2 (53), suggests an involvement of IRES-dependent translation in the control of angiogenesis. It is also clearly apparent from our results that two IRESs are present in the 5′ UTR of the VEGF mRNA and that they bind some different factors. This novel and interesting feature of VEGF mRNA suggests that VEGF expression is controlled at the translational level, probably in a specific way, by each of the two IRESs.
IRES A is located in a 293-nt-long fragment just upstream from the AUG codon (nt 745 to 1038 from the mRNA 5′ end [Fig. 2]). Analysis of the cis elements involved in the activity of this IRES revealed a need for at least two elements: the 5′ part of the IRES-containing fragment (IRES A 5′ region, between nt 745 and 846) and the D4 domain (nt 858 to 907 [Fig. 5]). The D4 loop, containing nucleotides AGACA, differs from the sequences of the loop consensus motifs ACCC (21) and GNRA or CAAA (18, 32) conserved in picornavirus IRESs. The data from Fig. 5 also show a detectable but more marginal influence of the D3 domain (nt 917 to 1013) in IRES function. According to its AUG proximal location, this IRES seems to allow ribosome binding and translation initiation without scanning. This is reminiscent of the so-called type II IRESs described in the literature and including the cardiovirus IRESs (21).
IRES B is located in a 392-nt segment between nt 91 and 483 from the mRNA 5′ end and exhibits optimal activity in the presence of nt 91 to 134 (Fig. 7). A 104-bp sequence limited by nt 379 and 483 is strictly necessary for IRES B activity. This sequence contains a predicted stem-loop structure bearing a GNRA motif, defined as an element shared by all picornavirus IRES (21, 32). The observation of such a predicted structure also suggests the presence of an IRES in this region. Although involvement of the motif would need to be confirmed, it seems that VEGF mRNA could be an example of cellular IRES the activity of which would require this unpaired GNRA sequence. Strikingly the 3′ border of this IRES was located more than 500 nt from the AUG codon (position 1039). This is reminiscent of the picornavirus type I IRESs, in which the 3′ border is located quite far upstream from the start codon (1). However, in IRES B, the spacer region, which does contain a second IRES, is longer than that of the picornavirus type I IRES, which is usually about 150 nt in length. Insertion of AUG codons in the poliovirus type I IRES, upstream from the authentic start codon, has demonstrated that ribosome scanning occurs from the IRES 3′ border to reach the AUG codon (17). It cannot be excluded that this could be the case for VEGF IRES B, although the length of the spacer region and its predicted stable structure (Fig. 4) would argue against such a hypothesis. Alternatively, two other hypotheses can be proposed. One possibility is that the spacer region allows a jump between the IRES B and the AUG. Such a mechanism has been described for adenovirus (60) and cauliflower mosaic virus (12). In the case of adenovirus, the 5′ part of the mRNA leader allows cap-independent translation to occur, while the 3′ part is required for the jump. This hypothesis cannot be ruled out here. Another possibility is that the 5′ part of the leader, although it contains an independent IRES, can also behave as an enhancer of IRES A. In this case, the two IRESs would together form a single super-IRES structure. Further investigations are necessary before we can choose between these hypotheses.
However, the existence of two independent IRESs is supported by their very different UV cross-linked protein patterns (Fig. 6). Interestingly, very few proteins are UV cross-linked to IRES A (Fig. 6). One, p100, seems to bind to the D4 domain, suggesting a potential role of this protein in IRES function.
Among the proteins which are bound to the 5′ half of the VEGF mRNA leader (Fig. 6), most, including the PTB, are bound on the 5′ part of IRES B (Fig. 8A). These experiments did not permit detection of protein whose binding was correlated to IRES B activity. This one-dimensional analysis was not, however, sufficient for conclusions to be drawn in an absence of IRES-specific protein, as different proteins may have the same electrophoretic mobility. Several proteins were bound to a polypyrimidine-rich sequence located between nt 189 and 241 (Fig. 8A; compare lanes B, C, and D). PTB is the major protein bound to the 5′ part of the RNA leader, and its binding site was mapped to between nt 134 and 189, which suggests that it recognizes the short UUUC sequence present around position 172 of the leader, which corresponds to the PTB consensus binding site described for viral IRESs (19, 42). According to the predicted structural data, this consensus site is located in a paired sequence. However, whereas PTB plays a crucial role in the function of EMCV and foot-and-mouth disease IRESs (4, 23, 38), its binding to IRES B does not seem to be correlated to IRES activity. These data are consistent with several reports showing that PTB is not the universal internal entry factor and that its binding to RNA does not necessarily imply its requirement for IRES function (3, 24).
The occurrence of two IRESs in the VEGF mRNA, probably controlled by different factors, provides interesting possibilities for the regulation of VEGF expression at the translational level. As VEGF plays a central role in angiogenesis, its expression has to be finely regulated. Two independent IRES domains, sensitive to different environmental conditions or cellular contexts, could permit an additional degree of flexibility in the expression of this growth factor. It should also be mentioned that the VEGF 5′ leader presents potential initiation codons located between the two IRESs, in the same open reading frame as the AUG 1039 initiation codon. It could thus be hypothesized that the upstream IRES controls the expression of longer VEGF isoforms, as has been observed for FGF-2 mRNA (53). However, the existence of such alternative VEGF isoforms remains to be demonstrated. It would also be interesting to know whether translation of the various splicing variants of VEGF mRNA is regulated differently by the two IRESs. Indeed, although we have shown that the second IRES does not extend downstream from the AUG codon, we cannot rule out that long-range interactions of IRES A or B with the downstream coding sequence of the messenger could affect or stabilize the IRES structure and influence its ribosome binding efficiency.
The discovery of IRESs in the mRNAs of two major angiogenic growth factors, VEGF and FGF-2, as well as in the mRNA of PDGF, a factor involved in vascular physiology, raises the question as to the possible involvement of this translation initiation mechanism in the control of angiogenesis. We have already shown that FGF-2 expression is translationally activated in response to stress or vascular lesion (8a, 54). Thus, the advantage of IRES-dependent regulation might be to allow an immediate response of the cell to exogenous stimuli. The control of such a process could have important repercussions in cardiovascular disease therapy.
The FGF-2 IRES seems to be constitutively activated in various transformed cell types (54), whereas c-myc is overexpressed in an IRES-dependent manner in Bloom’s syndrome cells (58). The observation of an IRES-dependent translational activation of growth factor or proto-oncogene expression in tumors, added to previous descriptions of eIF4E-dependent translation enhancement (25, 27), favors the hypothesis that translational deregulation of gene expression may play a key role in cell transformation. Furthermore, it has been reported that overexpression of VEGF induces cell transformation in cooperation with FGF-2 (16). Thus, the existence of IRESs in both FGF-2 and VEGF mRNAs suggests that the processes leading to cell transformation and/or tumor neovascularization could involve an IRES-dependent activation of the expression of these angiogenic factors.
ACKNOWLEDGMENTS
We thank B. Michot for predicted secondary structures, F. Bayard and J. Plouet for helpful discussions, and D. Warwick for English proofreading. We thank J. Abraham for the gift of the VEGF cDNA and promoter region and J. G. Patton for sending the anti-PTB antibody.
This work was supported by grants from the Association pour la Recherche contre le Cancer, the Ligue Nationale contre le Cancer, the Conseil Regional de Midi-Pyrenées, the European Community Biotechnology program (subprogram Cell Factory, Actions de Recherches Concertées, contract 94/99-181). I. Huez received fellowships from the Ministère de l’Education Nationale et de la Recherche Scientifique. L. Créancier received a fellowship from the European Community Biotechnology program.
REFERENCES
- 1.Belsham G J, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein J, Sella O, Le S Y, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES) J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 3.Borman A, Howell M T, Patton J G, Jackson R J. The involvement of a spliceosome component in internal initiation of human rhinovirus RNA translation. J Gen Virol. 1993;74:1775–1788. doi: 10.1099/0022-1317-74-9-1775. [DOI] [PubMed] [Google Scholar]
- 4.Borovjagin A, Pestova T, Shatsky I. Pyrimidine tract binding protein strongly stimulates in vitro encephalomyocarditis virus RNA translation at the level of preinitiation complex formation. FEBS Lett. 1994;351:299–302. doi: 10.1016/0014-5793(94)00848-5. [DOI] [PubMed] [Google Scholar]
- 5.Brogi E, Wu T, Namiki A, Isner J M. Indirect angiogenic cytokines upregulate VEGF and bFGF gene expression in vascular smooth muscle cells, whereas hypoxia upregulates VEGF expression only. Circulation. 1994;90:649–652. doi: 10.1161/01.cir.90.2.649. [DOI] [PubMed] [Google Scholar]
- 6.Chetouani F, Monestie P, Thebault P, Gaspin C, Michot B. ESSA: an integrated and interactive computer tool for analysing RNA secondary structure. Nucleic Acids Res. 1997;25:3514–3522. doi: 10.1093/nar/25.17.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Cohen T, Nahari D, Cerem L W, Neufeld G, Levi B Z. Interleukin 6 induces the expression of vascular endothelial growth factor. J Biol Chem. 1996;271:736–741. doi: 10.1074/jbc.271.2.736. [DOI] [PubMed] [Google Scholar]
- 8a.Concina, P., et al. Unpublished results.
- 9.Creancier L, Prats H, Zanibellato C, Amalric F, Bugler B. Determination of the functional domains involved in nucleolar targeting of nucleolin. Mol Biol Cell. 1993;4:1239–1250. doi: 10.1091/mbc.4.12.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrara N, Davis-Smyth T. The biology of vascular endothelial growth factor. Endocrine Rev. 1997;18:4–25. doi: 10.1210/edrv.18.1.0287. [DOI] [PubMed] [Google Scholar]
- 11.Ferrara N, Keyt B. Vascular endothelial growth factor: basic biology and clinical implications. EXS. 1997;79:209–232. doi: 10.1007/978-3-0348-9006-9_9. [DOI] [PubMed] [Google Scholar]
- 12.Futterer J, Kiss-Laszlo Z, Hohn T. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell. 1993;73:789–802. doi: 10.1016/0092-8674(93)90257-q. [DOI] [PubMed] [Google Scholar]
- 13.Gan W, Rhoads R E. Internal initiation of translation directed by the 5′-untranslated region of the mRNA for eIF4G, a factor involved in the picornavirus-induced switch from cap-dependent to internal initiation. J Biol Chem. 1996;271:623–626. doi: 10.1074/jbc.271.2.623. [DOI] [PubMed] [Google Scholar]
- 14.Goad D L, Rubin J, Wang H, Tashjian A H, Jr, Patterson C. Enhanced expression of vascular endothelial growth factor in human SaOS-2 osteoblast-like cells and murine osteoblasts induced by insulin-like growth factor I. Endocrinology. 1996;137:2262–2268. doi: 10.1210/endo.137.6.8641174. [DOI] [PubMed] [Google Scholar]
- 15.Grugel S, Finkenzeller G, Weindel K, Barleon B, Marme D. Both v-Ha-Ras and v-Raf stimulate expression of the vascular endothelial growth factor in NIH 3T3 cells. J Biol Chem. 1995;270:25915–25919. doi: 10.1074/jbc.270.43.25915. [DOI] [PubMed] [Google Scholar]
- 16.Guerrin M, Scotet E, Malecaze F, Houssaint E, Plouet J. Overexpression of vascular endothelial growth factor induces cell transformation in cooperation with fibroblast growth factor 2. Oncogene. 1997;14:463–471. doi: 10.1038/sj.onc.1200846. [DOI] [PubMed] [Google Scholar]
- 17.Hellen C U, Pestova T V, Wimmer E. Effect of mutations downstream of the internal ribosome entry site on initiation of poliovirus protein synthesis. J Virol. 1994;68:6312–6322. doi: 10.1128/jvi.68.10.6312-6322.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman M A, Palmenberg A C. Mutational analysis of the J-K stem-loop region of the encephalomyocarditis virus IRES. J Virol. 1995;69:4399–4406. doi: 10.1128/jvi.69.7.4399-4406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iizuka N, Yonekawa H, Nomoto A. Nucleotide sequences important for translation initiation of enterovirus RNA. J Virol. 1991;65:4867–4873. doi: 10.1128/jvi.65.9.4867-4873.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson R J. RNA translation. Picornaviruses break the rules. Nature. 1988;334:292–293. doi: 10.1038/334292a0. [DOI] [PubMed] [Google Scholar]
- 21.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 22.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomycarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaminski A, Hunt S L, Patton J G, Jackson R J. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA. 1995;1:924–938. [PMC free article] [PubMed] [Google Scholar]
- 24.Kaminski A, Jackson R J. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA. 1998;4:626–638. doi: 10.1017/s1355838298971898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kevil C G, De Benedetti A, Payne D K, Coe L L, Laroux F S, Alexander J S. Translational regulation of vascular permeability factor by eukaryotic initiation factor 4E: implications for tumor angiogenesis. Int J Cancer. 1996;65:785–790. doi: 10.1002/(SICI)1097-0215(19960315)65:6<785::AID-IJC14>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 26.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazaris-Karatzas A, Montine K S, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 28.Le S Y, Maizel J V., Jr A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Res. 1997;25:362–369. doi: 10.1093/nar/25.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung D W, Cachianes G, Kuang W J, Goeddel D V, Ferrara N. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science. 1989;246:1306–1309. doi: 10.1126/science.2479986. [DOI] [PubMed] [Google Scholar]
- 30.Levy A P, Levy N S, Wegner S, Goldberg M A. Transcriptional regulation of the rat vascular endothelial growth factor gene by hypoxia. J Biol Chem. 1995;270:13333–13340. doi: 10.1074/jbc.270.22.13333. [DOI] [PubMed] [Google Scholar]
- 31.Li J, Perrella M A, Tsai J C, Yet S F, Hsieh C M, Yoshizumi M, Patterson C, Endege W O, Zhou F, Lee M E. Induction of vascular endothelial growth factor gene expression by interleukin-1 beta in rat aortic smooth muscle cells. J Biol Chem. 1995;270:308–312. doi: 10.1074/jbc.270.1.308. [DOI] [PubMed] [Google Scholar]
- 32.Lopez de Quinto S, Martinez-Salas E. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J Virol. 1997;71:4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macejak D J, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 34.Meerovitch K, Pelletier J, Sonenberg N. A cellular protein that binds to the 5′-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 1989;3:1026–1034. doi: 10.1101/gad.3.7.1026. [DOI] [PubMed] [Google Scholar]
- 35.Molla A, Jang S K, Paul A V, Reuer Q, Wimmer E. Cardioviral internal ribosomal entry site is functional in a genetically engineered dicistronic poliovirus. Nature. 1992;356:255–257. doi: 10.1038/356255a0. [DOI] [PubMed] [Google Scholar]
- 36.Mukhopadhyay D, Tsiokas L, Zhou X M, Foster D, Brugge J S, Sukhatme V P. Hypoxic induction of human vascular endothelial growth factor expression through c-Src activation. Nature. 1995;375:577–581. doi: 10.1038/375577a0. [DOI] [PubMed] [Google Scholar]
- 37.Nanbru C, Lafon I, Audigier S, Gensac M C, Vagner S, Huez G, Prats A C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 38.Niepmann M, Petersen A, Meyer K, Beck E. Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J Virol. 1997;71:8330–8339. doi: 10.1128/jvi.71.11.8330-8339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh S-K, Scott M P, Sarnow P. Homeotic gene Antennapedia mRNA contains 5′-noncoding sequences that confer translational initiation by internal ribosome binding. Genes Dev. 1992;6:1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- 40.Patton J G, Mayer S A, Tempst P, Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991;5:1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- 41.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 42.Pestova T V, Hellen C U, Wimmer E. Translation of poliovirus RNA: role of an essential cis-acting oligopyrimidine element within the 5′ nontranslated region and involvement of a cellular 57-kilodalton protein. J Virol. 1991;65:6194–6204. doi: 10.1128/jvi.65.11.6194-6204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plouet J, Moro F, Bertagnolli S, Coldeboeuf N, Mazarguil H, Clamens S, Bayard F. Extracellular cleavage of the vascular endothelial growth factor 189-amino acid form by urokinase is required for its mitogenic effect. J Biol Chem. 1997;272:13390–13396. doi: 10.1074/jbc.272.20.13390. [DOI] [PubMed] [Google Scholar]
- 44.Plouet J, Schilling J, Gospodarowicz D. Isolation and characterization of a newly identified endothelial cell mitogen produced by AtT-20 cells. EMBO J. 1989;8:3801–3806. doi: 10.1002/j.1460-2075.1989.tb08557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prats A C, Vagner S, Prats H, Amalric F. cis-acting elements involved in the alternative translation initiation process of human basic fibroblast growth factor mRNA. Mol Cell Biol. 1992;12:4796–4805. doi: 10.1128/mcb.12.10.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rak J, Filmus J, Finkenzeller G, Grugel S, Marme D, Kerbel R S. Oncogenes as inducers of tumor angiogenesis. Cancer Metastasis Rev. 1995;14:263–277. doi: 10.1007/BF00690598. [DOI] [PubMed] [Google Scholar]
- 47.Shima D T, Kuroki M, Deutsch U, Ng Y S, Adamis A P, D’Amore P A. The mouse gene for vascular endothelial growth factor. Genomic structure, definition of the transcriptional unit, and characterization of transcriptional and post-transcriptional regulatory sequences. J Biol Chem. 1996;271:3877–3883. doi: 10.1074/jbc.271.7.3877. [DOI] [PubMed] [Google Scholar]
- 48.Shweiki D, Itin A, Soffer D, Keshet E. Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature. 1992;359:843–845. doi: 10.1038/359843a0. [DOI] [PubMed] [Google Scholar]
- 49.Stein I, Neeman M, Shweiki D, Itin A, Keshet E. Stabilization of vascular endothelial growth factor mRNA by hypoxia and hypoglycemia and coregulation with other ischemia-induced genes. Mol Cell Biol. 1995;15:5363–5368. doi: 10.1128/mcb.15.10.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoneley M, Paulin F E, Le Quesne J P, Chappell S A, Willis A E. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 51.Teerink H, Voorma H O, Thomas A A. The human insulin-like growth factor II leader 1 contains an internal ribosomal entry site. Biochim Biophys Acta. 1995;1264:403–408. doi: 10.1016/0167-4781(95)00185-9. [DOI] [PubMed] [Google Scholar]
- 52.Tischer E, Mitchell R, Hartman T, Silva M, Gospodarowicz D, Fiddes J C, Abraham J A. The human gene for vascular endothelial growth factor. Multiple protein forms are encoded through alternative exon splicing. J Biol Chem. 1991;266:11947–11954. [PubMed] [Google Scholar]
- 53.Vagner S, Gensac M C, Maret A, Bayard F, Amalric F, Prats H, Prats A C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vagner S, Touriol C, Galy B, Audigier S, Gensac M C, Amalric F, Bayard F, Prats H, Prats A C. Translation of CUG- but not AUG-initiated forms of human fibroblast growth factor 2 is activated in transformed and stressed cells. J Cell Biol. 1996;135:1391–1402. doi: 10.1083/jcb.135.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vagner S, Waysbort A, Marenda M, Gensac M C, Amalric F, Prats A C. Alternative translation initiation of the Moloney murine leukemia virus mRNA controlled by internal ribosome entry involving the p57/PTB splicing factor. J Biol Chem. 1995;270:20376–20383. doi: 10.1074/jbc.270.35.20376. [DOI] [PubMed] [Google Scholar]
- 56.Valcarcel J, Gebauer F. Post-transcriptional regulation: the dawn of PTB. Curr Biol. 1997;7:R705–R708. doi: 10.1016/s0960-9822(06)00361-7. [DOI] [PubMed] [Google Scholar]
- 57.Warren R S, Yuan H, Matli M R, Ferrara N, Donner D B. Induction of vascular endothelial growth factor by insulin-like growth factor 1 in colorectal carcinoma. J Biol Chem. 1996;271:29483–29488. doi: 10.1074/jbc.271.46.29483. [DOI] [PubMed] [Google Scholar]
- 58.West M J, Sullivan N F, Willis A E. Translational upregulation of the c-myc oncogene in Bloom’s syndrome cell lines. Oncogene. 1995;11:2515–2524. [PubMed] [Google Scholar]
- 59.Ye X, Fong P, Iizuka N, Choate D, Cavener D R. Ultrabithorax and antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol Cell Biol. 1997;17:1714–1721. doi: 10.1128/mcb.17.3.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yueh A, Schneider R J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]