Abstract
Composting is a natural process of decomposition of organic matter that occurs by the action of microorganisms such as fungi, bacteria, and actinobacteria. The actinobacteria are present throughout the process due to their resistance to different environmental conditions. They are Gram-positive, filamentous bacteria with a high capacity for producing secondary metabolites of biotechnological importance. Thus, the objective of this work was to isolate and characterize actinobacteria from industrial composting soil of oil palm (Elaeis guineensis) in the municipality of Igarapé-Açu, Pará. Ten samples of the material were collected and seeded on soy tryptone agar, Reasoner’s 2A agar, and Columbia agar, using the serial dilution technique. For morphological characterization of the strains, Gram staining and microculture were performed, and for biochemical characterization, the motility, triple sugar iron, Simmons citrate, maltose, phenylalanine, catalase, and DNAse tests were performed. It was observed that compost actinobacteria have a great diversity in morphological and metabolic production, which may be associated with the substrate and cultivation conditions. Therefore, palm oil compost material represents a rich source of bacterial biodiversity, bringing new perspectives for the bioprospecting of actinobacteria of biotechnological importance in little explored environments.
Keywords: Actinobacteria, Composting, Secondary metabolites
Introduction
Composting is a natural process by which organic compounds are degraded by the action of microorganisms, such as fungi, bacteria, and actinobacteria, under aerobic conditions [1]. The organic substrate, the decomposition step, the handling, and the physicochemical conditions of the environment (temperature, pH, humidity, etc.) directly affect the diversity and activity of microorganisms in the site [2, 3]. All the decomposed material is transformed into organic and inorganic products that can be used in soil fertilization and correction [4, 5].
This process occurs in four distinct stages, mesophilic, thermophilic, maturation, and cooling, which ends with the formation of humus [6]. Bacteria are the predominant microorganisms in all phases of composting since they can tolerate the temperature variations of the process [7]. The actinobacteria are a representative group in composting, because they are excellent indicators of organic matter biotransformation, besides having a diverse physiology and metabolic flexibility to produce biocompounds [8, 9].
The actinobacteria are a phylum of Gram-positive bacteria with a genome rich in guanine and cytosine that have fungal-like filamentous structures and reproduce by sporulation [10]. They can be found in various environments, especially in the soil, where they act in the decomposition of residues and in carbon cycling [11]. Their potential for waste degradation is associated with the high production of secondary metabolites and enzymes that not only favor nutrition and survival in the environment, but also have great technological and industrial importance [12].
During maturity, actinobacteria produce different types of reproductive spores, such as arthrospores, conidia, and sporangia, which are fundamental structures for the taxonomy of these bacteria [13]. In culture, they form a network of filaments on the surface and inside the medium, called aerial and vegetative mycelium, respectively [14, 15]. The vegetative mycelium is linked to the nutrition of the bacteria and is the first structure formed after the germination of the spores. When developed, it emerges to the surface giving rise to the aerial mycelium, responsible for spore dispersal and bacterial reproduction [16].
Some genera of actinobacteria, such as Streptomyces, stand out for their ability to produce pigments in the medium [17]. The main ones are the melanoids, which have a dark brown coloration and are produced in response to environmental stress, and the carotenoids, which are produced under different conditions and range from light yellow to reddish orange [18]. The importance of producing pigments from actinobacteria is due to their easy and low-cost growth, their high stability, and their broad biotechnological applicability, encompassing the sectors of agronomy, pharmaceutical, and food industry [19, 20].
Considering the great metabolic and morphological diversity present in actinobacteria, besides their wide industrial and biotechnological applicability, it is essential to explore the properties of this group of bacteria under different conditions in ecosystems little explored. Thus, the objective of this work was to isolate and biochemically and morphologically characterize strains of actinobacteria isolated from industrial oil palm (Elaeis guineensis) composting soils from an oil palm producing company, located in the municipality of Igarapé-Açu, in the interior of Pará.
Material and methods
Study type and location
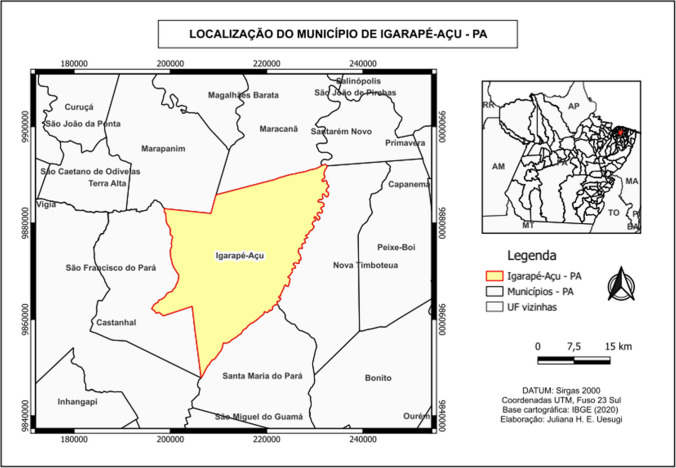
This is an experimental study, with a quantitative and descriptive approach, using dry compost soil samples of oil palm (Elaeis guineensis) provided by the Agroindustrial Palmasa S/A, a company specialized in extracting and refining palm and palm kernel oil, located in the municipality of Igarapé-Açu, in the interior of the state of Pará (Fig. 1).
Fig. 1.
Location of the municipality of Igarapé-Açu, PA
Sample collection
Ten samples of compost soil were collected in ten different points with the aid of spatulas and sterile bags and identified with the number of the collection point, date, time, temperature, latitude, longitude, and composting time. The samples were then transported in an isothermal box to the LABMICRO (CCBS/UEPA) for further analysis. The coordinates (latitude and longitude) of the collection sites are listed below:
Point 1: 1.13587°S; 47.67354°W
Point 2: 1.13592°S; 47.67363°W
Point 3: 1.13532°S; 47.67352°W
Point 4: 1.13546°S; 47.67360°W
Point 5: 1.13771°S; 47.66943°W
Point 6: 1.13767°S; 47.6693°W
Point 7: 1.1365°S; 47.64734°W
Point 8: 1.13713°S; 47.64719°W
Point 9: 1.14473°S; 47.65520°W
Point 10: 1.14457°S; 47.65518°W
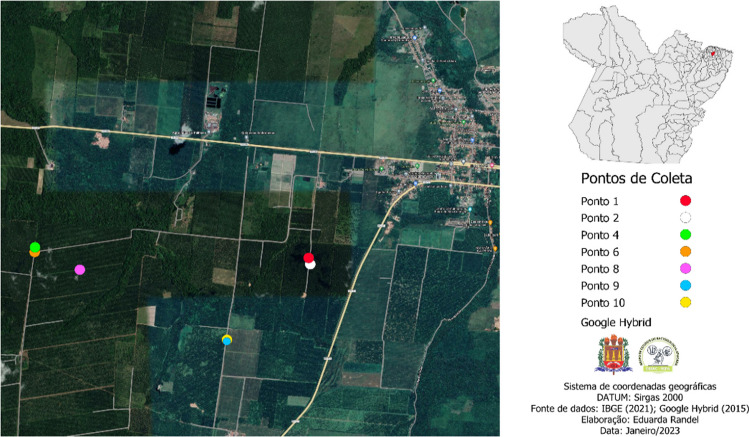
After prospecting the bacteria, points 3, 5, and 7 were discarded, since no strain collected from these locations was selected. Thus, Fig. 2 shows the spatial distribution of the collection points according to the strains used in this study.
Fig. 2.
Spatial distribution of the collection points of the selected strains
Sample preparation
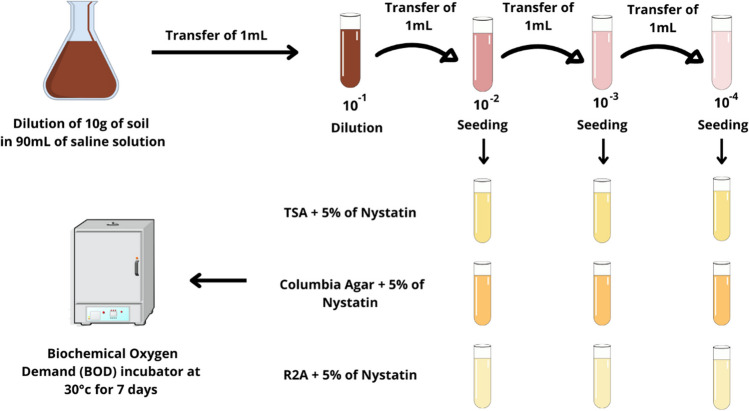
For the sample preparation, the method of Silva [21] and Magron [22] was adapted. In previously sterilized containers, 10 g of each soil sample was diluted in 90 mL of distilled water. This solution was homogenized and left to spontaneously sediment for 30 min. Then, 1 mL of this sample was transferred to another test tube containing 9 mL of distilled water, obtaining a final sample concentration of 10−2 (1:10 dilution). From this first tube, 1 mL of the sample was transferred to a new test tube containing 9 mL of distilled water to obtain a concentration of 10−3 (1:100), and from this, 1 mL was transferred to a third tube containing 9 mL of distilled water, to obtain samples of 10−3 (1:100) and 10−4 (1:1000), respectively.
Actinobacteria isolation
After pre-treatment, the samples were sowed, with the aid of a sterile calibrated loop, in tubes containing Reasoner’s 2A agar, Columbia agar, and soy tryptone agar, all enriched with 5% nystatin to avoid fungal contamination. All dilutions were seeded on the three culture media, totaling nine tubes per sample, in order to obtain isolated colonies and capture as much bacterial diversity as possible. The seeding technique used was depletion and streaking. After seeding, the tubes were incubated in a BOD incubator at 30 °C, in a humid chamber for 10 days (Fig. 3).
Fig. 3.
Primary isolation using the serial dilution technique
The tubes that presented bacterial growth with characteristics of actinobacteria were reisolated in plates containing the culture medium that obtained the highest diversity of colonies in the primary isolation. The following inclusion criteria were considered: Gram-positive bacilli in chains and pigment-producing, spore-forming, aerial and vegetative mycelium. Gram-negative bacteria, bacteria with cocci or cocoid morphology, and bacteria that did not form aerial mycelium were excluded from the study.
The macroscopic characterization of the colonies was performed according to the size, shape, color, density, and consistency of the colonies growing on the culture media used [23–25].
Morphological characterization
Gram staining
Morphological characterization of the isolates will be performed using the Gram staining technique. The Gram staining kit (Kasvi©) was used to perform the technique. The methodology applied will be in accordance with the standard established by the Agência Nacional de Vigilância Sanitária (ANVISA) [26]. The bacteria were characterized based on the morphological classification performed by Barka et al. [10].
Microculture
In order to observe the reproductive structures and carry out a possible identification at the genus level of the bacteria, the microculture technique was employed and performed according to the methodology adapted from Shirling and Gottlieb [27]. A bacterial suspension of concentration similar to the second tube of the MacFarland (3.0 × 108), of each selected colony, was prepared in 0.9% saline solution [28].
From this inoculum, a volume of 50 μL was used to perform the seeding in Petri plates containing the Sabouraud dextrose culture medium, and, with the help of a Drigalski loop, a mat seeding was performed on the medium. Next, three coverslips were inserted in a 45° on the seeding site, and the plates were incubated at 37 °C for 10 days.
Each coverslip was read at intervals of 2, 6, and 10 days, respectively, to visualize the organization of the hyphae and reproductive structures of the bacteria to characterize them at the genus level. To visualize the structures, the removed coverslip was stained with methylene blue dye diluted in distilled water at a 1:1 ratio. The structures were observed under an optical microscope with a 1000 × objective [29].
Biochemical characterization
The biochemical characterization was performed according to Table 1, adapted from Bergey’s Manual of Systematic Bacteriology: Volume 5: The Actinobacteria [30].
Table 1.
Biochemical tests for characterization of the isolated actinobacteria
| Characteristic | Test principle |
|---|---|
| Structural | |
| Motility | Detection of the presence of flagella in a semisolid medium |
| Carbohydrate metabolism | |
| Triple sugar iron | Fermentation of glucose, lactose, sucrose, production of gas (CO2) and H2S |
| Simmons citrate | Use of citrate as the sole carbon source |
| Maltose | Fermentation of maltose |
| Enzyme activity | |
| Phenylalanine | Production of the enzyme phenylalanine deaminase |
| Catalase | Production of the enzyme catalase |
| DNAse | Production of the enzyme DNAse |
Results and discussion
Isolation of actinobacteria
In 10 days of incubation, there was growth in all the seeded tubes, regardless of inoculum concentration, which demonstrated that the increase in dilution does not interfere in bacterial growth. The most expressive growth of the samples was observed in the TSA medium, although the R2A medium showed greater diversity in morphology and coloration of the colonies, which was expected due to the difference in composition of the media.
It was observed that the dilution concentration with the highest growth was 1:1000 in relation to the others. Santos et al. [31] also used the serial dilution method to isolate actinobacteria from composted organic manure, where the highest growth was also in the dilution of lower concentration (10−5). The authors related this result to the nutritional differences for each culture medium and the dilution used.
R2A is a culture medium developed by Reasoner and Geldreich [32] for the purpose of isolating oligotrophic bacteria from treated water sources. It is a low-nutrient agar formulated to simulate the conditions of the environment from which the bacteria were taken. Corroborating the authors Almodovar, Pereira, and Bugno [33] and Raad et al. [34], R2A is considered ideal for the cultivation of soil bacteria because it has a small number of substrates as a source of nutrients for certain groups of bacteria, as was the case of the actinobacteria isolated for this study.
In Hilinski [35]’s work, R2A was used as a substitute for TSA and plate count agar (PCA), as it proved more efficient for the recovery of slow-growing bacteria incubated for periods longer than 7 days. This characteristic is associated with physical changes and metabolic decline in lower nutrient environments, making it ideal for the cultivation of actinobacteria, whose growth is slower compared to other groups of bacteria.
This characteristic was also observed in the work of Park et al. [36], in which a comparative analysis was performed between the use of R2A and TSA media to isolate bacteria from hemodialysis water. In their study, R2A was more sensitive to microbial growth, performing better in detecting pathogens in the material studied.
Nishioka et al. [37] also reported better bacterial growth in R2A compared to TSA. Another characteristic observed by the authors was the production of pigments that, according to Reasoner and Geldreich [32], is higher compared to other media when evaluated over a period of 14 days. These results demonstrate that the lack of nutrients added to the long incubation period leads the bacteria to express their secondary metabolism, which includes the production of pigments in the medium, as was observed among the strains isolated from composting.
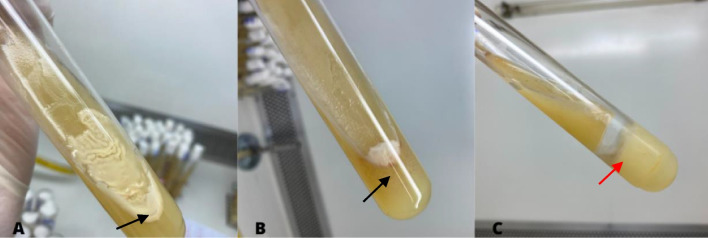
In addition, seeding in test tubes allowed the production of gas by the bacteria to be observed, as well as the formation of aerial mycelia on the sides of the tube (Fig. 4). On the other hand, the use of the tubes caused the colonies to grow overlapping, which made it difficult to isolate some strains.
Fig. 4.
Culture of actinobacteria in a tube, presence of aerial mycelium on the inner wall of the tube (A and B) indicated by the black arrow and gas formation, observed by fragmentation of the medium (C) indicated by the red arrow
No other studies were found in which the primary isolation of soil bacteria was performed in test tubes using a solid culture medium. This method was developed in the laboratory and proved to be a good alternative for cultivating actinobacteria.
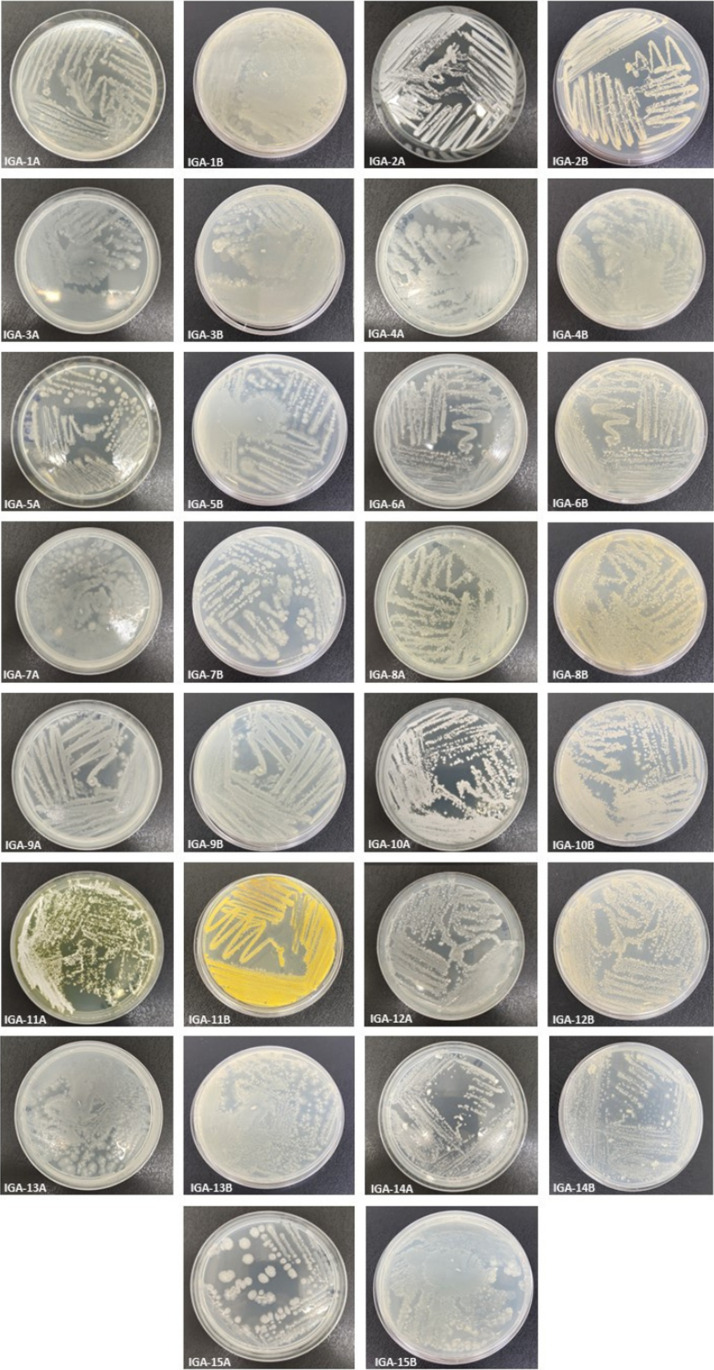
The colonies of actinobacteria are classified according to their morphology, which can vary between radial with furrows, velvety, concentric, umbonate, and convex. They are also classified according to their texture, which can range between powdery, velvety, and cottony. In addition, the colonies can be regular, irregular, or wavy in the medium [23, 24, 38]. These characteristics correspond to the morphologies found in the isolated bacteria, although strains with mucoid and dry texture were also found.
Among the isolated bacteria, 15 strains were selected. Regarding colony morphology, there was a predominance of irregular colonies (26.6%, n = 4), followed by wavy (20%, n = 3), concentric (20%, n = 3), and convex (13.3%, n = 2) morphology. Only one colony showed radial morphology with grooves (6%), and one colony showed regular morphology (6%). There was no growth of any colony with umbonate characteristics. The macroscopic characterization was recorded in Table 2.
Table 2.
Macroscopic characterization of actinobacteria colonies isolated from compost soil
| Strain | Medium | Color | Morphology | Texture | PIG | RM |
|---|---|---|---|---|---|---|
| IGA-1 | R2A | White | Irregular | Mucoid | - | + |
| IGA-2 | R2A | White | Velvety | Cottony | - | - |
| IGA-3 | TSA | White | Irregular | Mucoid | - | + |
| IGA-4 | TSA | White | Wavy | Drought | - | + |
| IGA-5 | COL | White | Convex | Mucoid | - | + |
| IGA-6 | COL | White | Wavy | Mucoid | - | - |
| IGA-7 | TSA | White | Irregular | Mucoid | - | + |
| IGA-8 | R2A | Gray | Concentric | Powdery | Brown | + |
| IGA-9 | R2A | White | Irregular | Mucoid | - | - |
| IGA-10 | R2A | Gray | Concentric | Velvety | - | + |
| IGA-11 | R2A | Gray | Radial with grooves | Powdery | Yellow | - |
| IGA-12 | R2A | Gray | Concentric | Velvety | - | + |
| IGA-13 | TSA | White | Wavy | Drought | - | + |
| IGA-14 | R2A | White | Convex | Drought | - | - |
| IGA-15 | TSA | White | Regular | Mucoid | - | + |
*PIG pigment; RM reverse mycelium; + presence;—absence
In the collected material, the highest temperature found was 49.2 °C in the material of 1 year of decomposition and the lowest was 29 °C, recorded in the material of 4 years of decomposition. According to Feng et al. [39], the increase in soil temperature is due to the increase in microbial metabolism at the site. Therefore, it is evident that microbial activity is higher in materials with less time of decomposition.
As for the colonial diversity of the isolated strains, there was a predominance of the colors: white, gray, and brown (Fig. 5). These colors were also predominant among the bacteria in the work of Silva et al. [24] with soil samples from the Brazilian semiarid region. In the work conducted by Silva et al. [40], on the other hand, there was a predominance of the colors gray, white, and cream among the isolated strains. These results imply that, although collected from different soils, the local microbial population may have similar characteristics.
Fig. 5.
Colonial diversity of the isolated bacteria. *A top view of the colony; B bottom view of the colony
Due to the large number of bacteria and fungi in compost, actinobacteria may produce the pigments in response to competition for nutrients. Chadni et al. [41]; Panwar, Molpa, and Joshi [42]; and Seipke [43] reported that pigment production is tied to the secondary metabolism of actinobacteria. In situations of competition with other microorganisms in the environment, bacteria tend to release products that can act both as chemical signals and defense of their own habitat, as well as possessing antimicrobial, antioxidant, and anticancer action. Such information, added to the findings in this work, demonstrates the biotechnological potential of compost soil bacteria.
According to Salim et al. [44] and Selim, Abdelhamid, and Mohamed [45], pigments can range from blue, violet, pink, red, yellow, green, brown, and black. There is also the presence of pigments in the vegetative mycelium that can be pale yellow, olive green, and brown. These pigments can be diffused in the medium or retained in the mycelium. This information was consistent with the results of the experiment, since the bacteria showed white, gray, yellow, and light brown pigmentation, both present in the mycelium and diffused in the medium.
Another characteristic observed was the presence of distinct colorations between the aerial mycelium and vegetative mycelium of strain IGA-11. This result was also reported by Charousová et al. [46], whose most of the isolated actinobacteria strains showed gray-colored aerial mycelium with yellow-colored vegetative mycelium, they were also absent of melanoid pigmentation, which is indicative of the genus Streptomyces.
According to De Oliveira [47] and Tiwari et al. [48], some actinobacteria can have the same characteristics on the surface, although they present different coloration in their vegetative mycelium. This is probably due to the physiological differences between the hyphae present in the substrate and in the aerial mycelium, a fact that determines the presence of two colorations in the same isolated strain.
Morphological characterization
Gram staining
The actinobacteria present themselves in the morphology of Gram-positive bacilli with elongated rods, forming hyphae or filamentous structures and spores. Besides these characteristics, the following can also be observed: spore germination pattern, structure and surface, branching of the mycelium, and formation of aerial and vegetative mycelium [49, 50]. These characteristics were used to select the strains based on bacterial morphology.
Among the isolated strains, most presented Gram-positive bacilli morphology, forming short chains, long chains, pseudohyphae, and spore chains. Gram-negative colonies were also isolated, with isolated bacillary morphology or in chains, as well as bacteria with coccoid morphology, suggestive of Gram-negative cocci, which were excluded from this study.
Microculture
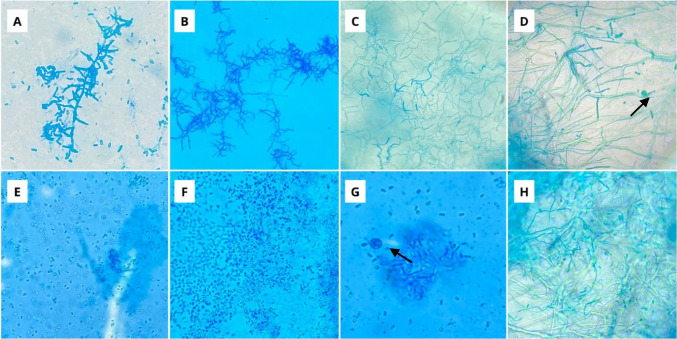
Three consecutive readings of each one of the microcultures were taken in order to observe if there would be a change in the reproductive structures of the bacteria. The first reading was after 2 days of growth, the second was after 6 days, and the last one was after 10 days of growth. In the test, the formation of filaments, hyphae, and pseudohyphae was evaluated, as well as the production of spores, their organization in chains, and reproductive structures (Fig. 6).
Fig. 6.
Reproductive structures of actinobacteria isolated in microculture. *A Monoverticillate without spirals; B biverticillate without spirals; C flexed; D flexed and straight with spore sacs (arrow); E isolated oval; F isolated oval and spherical; G spore sacs (arrow); H straight spores and pseudohyphae
The realization of readings in different periods of time proved to be relevant for the observation of the reproductive structures of bacteria. Some strains, in the first days of incubation, presented only spores and bacillary structures in the microculture, which, as the days passed, acquired filamentous forms with the presence of spore vesicles.
Other strains showed the opposite, presenting pseudohyphae and highly branched structures in the first days of incubation, while in longer incubation periods, they developed the form of bacilli with spore production. Therefore, daily observation of the microculture plates is very important to evaluate the growth of the aerial mycelium and the modifications in the reproductive structures of the bacteria as the days go by.
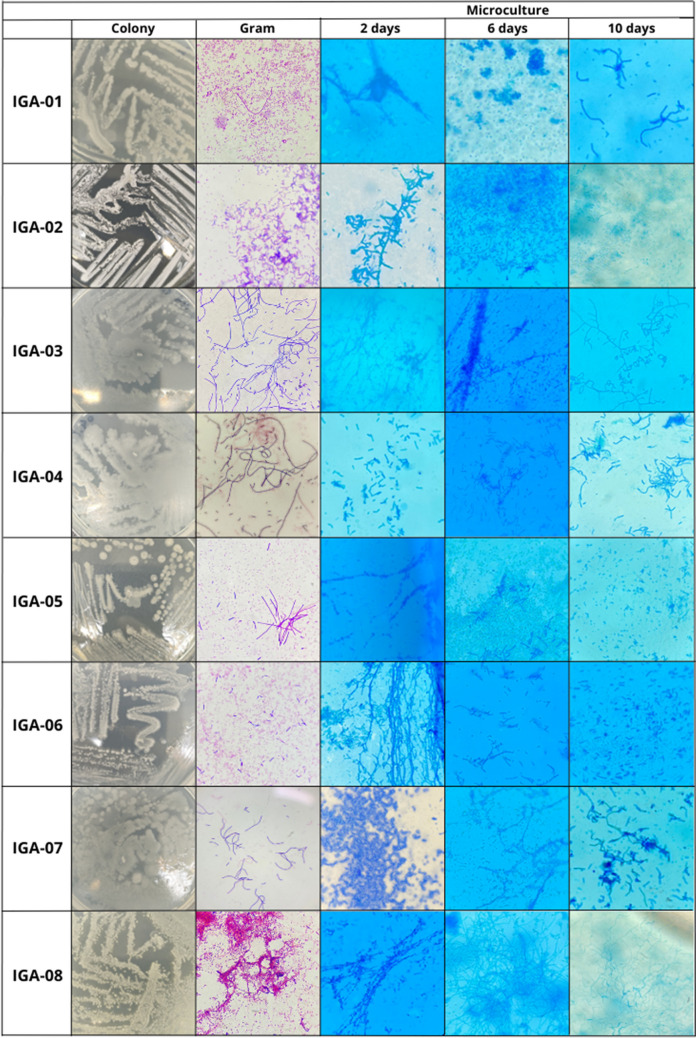
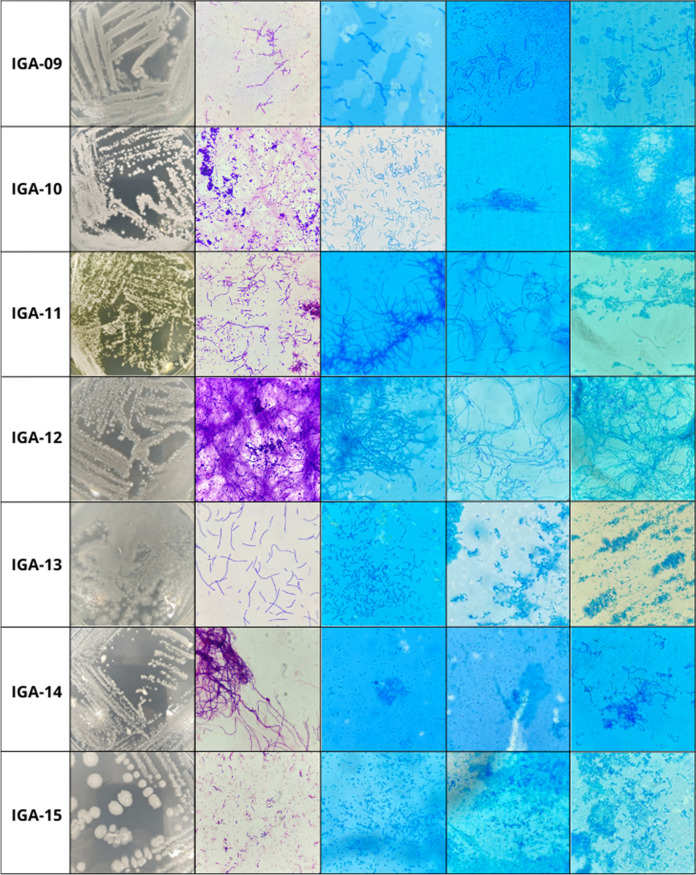
According to Kurtböke [51], the morphological analysis of the structures of the actinobacteria on consecutive days is fundamental, because younger colonies may present a less-developed characteristic compared to older cultures. This characteristic could be observed in this work, as different morphologies were obtained among the microcultures of the isolated actinobacteria in different incubation periods (Fig. 7).
Fig. 7.
Macroscopic and microscopic characterization and microculture of the isolated actinobacterial strains
The organization of reproductive structures is a relevant criterion to be used in the taxonomy of actinobacteria. These structures can be highly branched with spores or spore vesicles, as well as can form short or long chains of spores or vesicles. As for the classification of hyphae and spores, they can be straight, flexed, fasciculated, verticillate, and open and closed spirals [30, 52, 53].
In this study, the predominant spore morphology was isolated oval, followed by spherical, straight, and flexed. Only one strain demonstrated a characteristic of monoverticillate spores and only one strain had biverticillate spores. There were no strains with spiral morphology (Table 3). The findings were different from the work of Dornelas et al. [54] with actinobacteria from tropical soils, whose predominant morphologies were straight, flexible, spiral, and retinaculum.
Table 3.
Morphological characterization of actinobacteria strains isolated from compost soil
| Strain | Gram | Microculture (spore arrangements) | Suggestive gender | ||
|---|---|---|---|---|---|
| 2 days | 6 days | 10 days | |||
| IGA-1 | GPB in chains with spores | Isolated ovals | Oval and spherical | Isolated ovals | Bacillus sp. |
| IGA-2 | GPB isolates with spore chains | Monoverticillates without spirals | Monoverticillates without spirals | Isolated ovals | Streptomyces sp. |
| IGA-3 | GPB in long chains | Isolated sphericals | Isolated sphericals | Straights | Streptomyces sp. |
| IGA-4 | GPB in long chains | Isolated ovals | - | Isolated ovals | Inconclusive |
| IGA-5 | GPB in chains with spores | Isolated ovals | Isolated ovals | Isolated ovals | Inconclusive |
| IGA-6 | GPB isolates with spore chains | Straights | Isolated ovals | Spore Bags | Bacillus sp. |
| IGA-7 | GPB in short chains | Isolated ovals | Isolated ovals | Straights | Streptomyces sp. |
| IGA-8 | GPB in short chains with spores | Flexed | Flexed | Flexed with spore bags | Streptomyces sp. |
| IGA-9 | GPB in short chains | - | Isolated ovals and straight | Isolated ovals | Bacillus sp. |
| IGA-10 | GPB filamentous with spore chains | Straights | Straights | Straight and flexed with spore bags | Streptomyces sp. |
| IGA-11 | GPB filamentous with spore chains | Biverticullates without spirals | Straight with spore bags | Isolated ovals | Streptomyces sp. |
| IGA-12 | GPB filamentous with spore chains | Straights | Straight and Flexed | Flexed | Streptomyces sp. |
| IGA-13 | GPB in short chains | Isolated spherical | Isolated spherical | Isolated ovals and spherical | Inconclusive |
| IGA-14 | GPB in long chains and pseudohyphae | Spherical isolated and in spore bags | Isolated ovals | Isolated spherical | Streptomyces sp. |
| IGA-15 | GPB isolates with spores | Oval spore bags | Isolated spherical and ovals | Isolated spherical and ovals | Bacillus sp. |
*GPB Gram-positive bacilli
The results were also different from those obtained by Sukmawaty, Sari, and Masri [55] with actinobacteria from pine forests, whose predominant morphologies were flexed and open spirals. From these results, it can be inferred that the spore arrangements for different genera and species of actinobacteria is variable. In addition, the environment in which the bacteria are found may also influence their diversity.
Biochemical characterization
The biochemical characterization was evaluated in three parameters, being structural (presence of flagella), metabolic (utilization of certain nutrient sources), and enzymatic (production of enzymes in the substrate). All tests were read within 24 h, and the results were recorded in Table 4.
Table 4.
Biochemical characterization of actinobacterial strains isolated from compost soil
| Strain | TSI | CIT | MALT | MOT | PHE | CAT | DNASE |
|---|---|---|---|---|---|---|---|
| IGA-1 | AC/AL | + | - | - | - | + | - |
| IGA-2 | AC/AC | - | + | - | - | - | - |
| IGA-3 | AC/AL | - | + | - | - | - | - |
| IGA-4 | AC/AL | - | + | - | - | + | - |
| IGA-5 | AC/AL | - | + | - | - | + | - |
| IGA-6 | AC/AL | - | + | - | - | - | + |
| IGA-7 | AC/AC | - | + | - | - | - | - |
| IGA-8 | AC/AC | - | + | - | - | - | - |
| IGA-9 | AC/AL | + | - | - | - | - | + |
| IGA-10 | AC/AL | - | - | - | - | - | + |
| IGA-11 | AC/AL | - | - | - | - | + | - |
| IGA-12 | AC/AC | - | + | - | - | + | - |
| IGA-13 | AC/AL | - | - | - | - | + | - |
| IGA-14 | AL/AL | - | + | - | - | + | - |
| IGA-15 | AC/AL | - | - | - | - | - | - |
*TSI triple sugar iron agar; AC/AC acid base and acid apex; AC/AL acid base and alkaline apex; AL/AL alkaline base and alkaline apex; CIT Simmons citrate; MALT maltose; MOT motility; PHE phenylalanine; CAT catalase; + positive result;—negative result
Regarding the fermentation of sugars, 93.3% of the strains (n = 14) were able to metabolize glucose alone, while only 26% (n = 4) fermented all three carbohydrates in the medium. In addition, only one of the strains was unable to ferment any of the carbohydrates provided by the medium. There was no production of hydrogen sulfide (H2S) or gas by any of the bacteria.
According to Nurkanto and Agusta [56], the carbon source used by actinobacteria can be from different types of carbohydrates. Fitri et al. [57] pointed out that glucose is the best carbon source for the growth of actinobacteria compared to starch, lactose, sucrose, and fructose. This can be reaffirmed in the present work, since most of the strains tested used glucose as a substrate for growth.
As for the maltose test, 60% of the strains (n = 9) were able to ferment the carbohydrate. The result resembled that of Yun, Roh, and Kim [58], in which 61% of the bacteria fermented maltose. The work of Janardhan et al. [59], on the other hand, demonstrated that among the actinobacteria isolates, glucose and maltose are more favorable carbon sources for the development of the mycelium of actinobacteria compared to sucrose and fructose, as was noticeable in the present work.
As for the citrate test, only 13.3% of the strains (n = 2) were able to use it as a sole carbon source. Similar to the work of Yanti, Setyawati, and Kurniatuhadi [60], in which 16% of the strains were positive for the test. The results were opposed to that obtained by Chaudhary et al. [61], in which 100% of the strains were able to utilize citrate, which may be related to the different types of soils used in their work. The differences between the results are probably related to the different types of soil used in their work.
None of the strains tested presented motility in the medium, indicating the absence of flagella. Most actinobacteria are immobile; generally, when they present motility, it is related to the spores. The motility of spores is associated with their ability to disperse in the environment. Some genera, such as Streptomyces, have immobile spores that make use of the machinery of other soil microorganisms to diffuse in the medium [14, 62, 63].
There was no production of phenylalanine deaminases by any of the strains tested. This result was also obtained by Pathalam et al. [64], Kalyani et al. [65], and Lima et al. [66], in which none of the isolated actinobacteria produced the enzyme. These results suggest that the production of deaminases is not a common feature of this group of bacteria.
Regarding enzyme activity, 46.6% (n = 7) of the strains produced the enzyme catalase. In the work of Díaz-Díaz et al. [67] and Yanti et al. [60], the production of catalase was the majority among bacteria, since the enzyme was produced by 100% of the tested strains. In the research of Almuhayawi et al. [68], the results were similar to those obtained in this study, with 44.4% of the bacteria being positive for the test.
Catalase is an enzyme produced by bacteria in response to oxidative stress produced by reactive oxygen species (ROS), mainly hydrogen peroxide (H2O2). They also act in the development of bacteria and in the production of their secondary metabolites. In environments where there is a large amount of oxygen and nutrients, the production of catalases is very high. In composting, oxygen is essential to obtain a quality compost; therefore, the production of catalases in the strains isolated in this study can be associated with the material used [69–72].
As for DNAse, only three strains (20%) produced the enzyme, different from what was obtained in the work of Kizhakedathil and Subathra [73], in which 80% of the strains produced the enzyme. DNAses contribute to soil functions and are associated with nutrient breakdown, as well as providing an advantage in the survival of bacteria because it makes the environment more favorable within the conditions in which the enzyme acts [74, 75]. Therefore, it is understandable that, for different soil types, the production of DNAses is variable.
Conclusion
The amount of organic material in composting soils favors the proliferation of a great diversity of microorganisms, especially actinobacteria, which effectively participate in the decomposition process. Their growth in this type of soil is favored due to their high metabolic flexibility and the development of survival mechanisms to the variations of the environment, from the production of pigments and secondary metabolites.
The palm compost is a promising source for the bioprospecting of a great diversity of actinobacteria, although it is a little explored material for bacterial prospection studies. The microculture technique allowed, through the observation of the reproduction structures, to presumptively determine the bacterial genus. The analyses at different incubation periods allowed us to evaluate the morphology of the bacteria at different stages of maturity.
There are few studies in the literature that deal with the diversity of actinobacteria in this type of material. Therefore, the potential of poorly explored environments for the prospecting of new species of actinobacteria is evident. With new studies and through the application of more specific and advanced techniques in the characterization of actinobacteria, it is likely that species will continue to be discovered.
Data Availability
Authors declare that all images and data in this work are their own.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Responsible Editor: Acacio Aparecido Navarrete
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zainudin MHM, Singam JT, Sazili AQ, Shirai Y, Hassan MA. Indigenous cellulolytic aerobic and facultative anaerobic bacterial community enhanced the composting of rice straw and chicken manure with biochar addition. Sci Rep. 2022;12(1):5930. doi: 10.1038/s41598-022-09789-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar-Paredes A, Valdés G, Araneda N, Valdebenito E, Hansen F, Nuti M. Microbial community in the composting process and its positive impact on the soil biota in sustainable agriculture. Agronomy. 2023;13(2):542. doi: 10.3390/agronomy13020542. [DOI] [Google Scholar]
- 3.Shi F, Yu H, Zhang N, Wang Su, Li P, Yu Q, et al. Microbial succession of lignocellulose degrading bacteria during composting of corn stalk. Bioengineered. 2021;12(2):12372–12382. doi: 10.1080/21655979.2021.2002622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayilara MS, Olanrewaju OS, Babalola OO, Odeyemi O. Waste management through composting: challenges and potentials. Sustainability. 2020;12(11):4456. doi: 10.3390/su12114456. [DOI] [Google Scholar]
- 5.Lin C, Cheruiyot NK, Bui XT, Ngo HH. Composting and its application in bioremediation of organic contaminants. Bioengineered. 2022;13(1):1073–1089. doi: 10.1080/21655979.2021.2017624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biyada S, Merzouki M, Dėmčėnko T, et al. Microbial community dynamics in the mesophilic and thermophilic phases of textile waste composting identified through next-generation sequencing. Sci Rep. 2021;11:23624. doi: 10.1038/s41598-021-03191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finore I, Feola A, Russo L, et al. Thermophilic bacteria and their thermozymes in composting processes: a review. Chem Biol Technol Agric. 2023;10:7. doi: 10.1186/s40538-023-00381-z. [DOI] [Google Scholar]
- 8.Moreno J, López-González JA, Arcos-Nievas MA, Suárez-Estrella F, Jurado MM, Estrella-González MJ, López MJ. Revisiting the succession of microbial populations throughout composting: a matter of thermotolerance. Sci Total Environ. 2021;773:145587. doi: 10.1016/j.scitotenv.2021.145587. [DOI] [PubMed] [Google Scholar]
- 9.Al-shaibani MM, Radin Mohamed RMS, Sidik NM, Enshasy HAE, Al-Gheethi A, Noman E, Al-Mekhlafi NA, Zin NM. Biodiversity of secondary metabolites compounds isolated from phylum actinobacteria and its therapeutic applications. Molecules. 2021;26(15):4504. doi: 10.3390/molecules26154504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barka EA, Vatsa P, Sanchez L, et al. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol Mol Biol Rev. 2016;80(1):1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao Y, Dolfing J, Guo Z, Chen R, Wu M, Li Z, Lin X, Feng Y. Important ecophysiological roles of non-dominant Actinobacteria in plant residue decomposition, especially in less fertile soils. Microbiome. 2021;9(1):84. doi: 10.1186/s40168-021-01032-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naligama KN, Weerasinghe KE, Halmillawewa AP. Characterization of bioactive Actinomycetes isolated from Kadolkele Mangrove Sediments, Sri Lanka. Pol J Microbiol. 2022;71(2):191–204. doi: 10.33073/pjm-2022-017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Messaoudi O, Wink J, Bendahou M. Diversity of Actinobacteria isolated from date palms rhizosphere and saline environments: isolation, identification and biological activity evaluation. Microorganisms. 2020;8(12):1853. doi: 10.3390/microorganisms8121853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma M, Dangi P, Choudhary M (2014) Actinomycetes: Source, identification, and their applications. Int J Curr Microbiol Appl Sci 3:801–832. https://www.ijcmas.com/vol-3-2/Mukesh%20Sharma.pdf. Accessed 18 Mar 2023
- 15.Zahr R, Zahr S, Hajj RE, Khalil M (2022) Actinomycetes, promising therapeutic agents: characteristics and active metabolites. J Biol Todays World 11(6):1–8. Retrieved from: https://www.iomcworld.org/articles/actinomycetes-promising-therapeutic-agents-characteristics-and-active-metabolites.pdf. Accessed 10 May 2023
- 16.Neto JM, Bandeira L, Mesquita A, Martins SC, Martins C (2022) biotechnological Potential and enzymes produced by actinobacteria from semi-arid soils. Enciclopedia Biosfera 19(42). https://www.conhecer.org.br/ojs/index.php/biosfera/article/view/5557. Accessed 10 May 2023
- 17.Tran T, Dawrs SN, Norton GJ, Virdi R, Honda JR. Brought to you courtesy of the red, white, and blue pigments of nontuberculous mycobacteria. AIMS Microbiol. 2020;6(4):434–450. doi: 10.3934/microbiol.2020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martins HHS, Caldas DS, Prazeres MCC, Colares TV, Silva JCC, Fernandes CF, Pereira ES, Del-Castillo JCS, Coelho BBF, Bezerra NV (2022) Chromogenic diversity of actinobacteria strains isolated from mangrove soils in the municipality of São Caetano de Odivelas – Pará, Brazil. Int J Dev Res 12(12):60689–60693. https://www.journalijdr.com/sites/default/files/issue-pdf/25867.pdf. Accessed 10 May 2023
- 19.Hemeda NA, Hegazy GE, Abdelgalil SA, Soliman NA, Abdel-Meguid DI, El-Assar SA. Maximization of red pigment production from Streptomyces sp. LS1 structure elucidation and application as antimicrobial/antifouling against human pathogens and marinlucidatees. J Genet Eng Biotechnol. 2022;20(1):1–17. doi: 10.1186/s43141-022-00452-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orlandi VT, Martegani E, Giaroni C, Baj A, Bolognese F. Bacterial pigments: a colorful palette reservoir for biotechnological applications. Biotechnol Appl Biochem. 2022;69(3):981–1001. doi: 10.1002/bab.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silva GR (2013) Bioprospecção de actinobactérias isoladas da rizosfera de Caesalpinia pyramidalis Tul. do bioma Caatinga. Dissertação, Universidade Federal de Pernambuco. https://repositorio.ufpe.br/handle/123456789/13351. Accessed 30 Dec 2022
- 22.Magron CF (2014) Isolamento, identificação e caracterização de bactérias cultiváveis presentes na compostagem de resíduos orgânicos do zoológico de São Paulo e produtoras de amilases e proteases. Dissertação, Universidade Federal de São Paulo. https://repositorio.unifesp.br/handle/11600/46518. Accessed 30 Dec 2022
- 23.Augustine D, Jacob JC, Ramya K, Philip R. Actinobacteria from sediment samples of Arabian Sea and Bay of Bengal: biochemical and physiological characterization. Int J Res Mar Sci. 2013;2(2):56–63. [Google Scholar]
- 24.Silva MJS, Sousa JB, Martins SCS, Martins CM (2019) Diversidade de cepas de actinobactérias da RPPN “Fazenda Não me Deixes” - QUIXADÁ (CE). Enciclopédia Biosfera 16(29):1857–1869. https://conhecer.org.br/ojs/index.php/biosfera/article/view/312. Accessed 2 Oct 2022
- 25.Bezerra NV, Silva JCC, Uesugi JHE, Fernandes CF, Prazeres MCC, Caldas DS, Pismel JAR, Martins HHS, Júnior JSG. Characterization and evaluation of the antibacterial potential of bacterial microbiota of cultivated soils of Cassava (Manihot esculenta) and Black pepper (Piper nigrum) in the city of Igarapé Açu – Pará, Brazil. Int J Environ Agric Biotechnol. 2021;6(6):106–110. doi: 10.22161/ijeab.66.12. [DOI] [Google Scholar]
- 26.ANVISA (2013) Agência Nacional de Vigilância Sanitária. Microbiologia clínica para o controle de infecção relacionada à assistência à saúde. Módulo 4: Procedimentos Laboratoriais: da Requisição do Exame à Análise Microbiológica e Laudo Final. https://www.saude.go.gov.br/images/imagens_migradas/upload/arquivos/2017-02/modulo-4---procedimentos-laboratoriais---da-requisicao-do-exame-a-analise-microbiologica-e-laudo-final.pdf. Accessed 2 Oct 2022
- 27.Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Evol Microbiol. 1966;16:313–340. [Google Scholar]
- 28.McFarland J. Nephelometer: an instrument for media used for estimating the number of bacteria in suspensions used for calculating the opsonic index and for vaccines. J Am Med Assoc. 1907;49:1176–1178. doi: 10.1001/jama.1907.25320140022001f. [DOI] [Google Scholar]
- 29.Silva WO (2017) Otimização e caracterização parcial de L-asparaginase e L-glutaminase de actinobactéria da rizosfera de Poincianela pyramidalis. Dissertação, Universidade Federal de Pernambuco. https://repositorio.ufpe.br/handle/123456789/30485. Accessed 20 Jan 2023
- 30.Goodfellow M, Kampfer P, Busse H, Trujillo ME, Suzuki KI, Whitman WB (2012) Manual® Bergey’s systematic bacteriology: volume five: Actinobacteria, Part One. Springer, New York
- 31.Santos LS, de Souza GS, da Silva LG, de Araújo LR, da Silva MMR, Lopes EAP. Actinobactérias com potencial biotecnológico agrícola isoladas de adubo orgânico fermentado (Bokashi) Diversitas J. 2021;6(4):3866–3881. doi: 10.48017/dj.v6i4.1946. [DOI] [Google Scholar]
- 32.Reasoner DJ, Geldreich E. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49(1):1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Almódovar AAB, Pereira TC, Bugno A. Eficiência do Agar R2A na contagem de bactérias heterotróficas em água tratada para diálise. Rev Inst Adolfo Lutz. 2009;68(2):232–236. doi: 10.53393/rial.2009.v68.32722. [DOI] [Google Scholar]
- 34.de Raad M, Li Y, Andeer P, Kosina SM, Saichek NR, Golini A et al (2021) A defined medium based on R2A for cultivation and exometabolite profiling of soil bacteria. bioRxiv 2021–05. 10.1101/2021.05.23.445362 [DOI] [PMC free article] [PubMed]
- 35.Hilinski EG (2019) Utilização de método microbiológico rápido para a enumeração de bactérias heterotróficas em água tratada para diálise: técnica de detecção microbiana pelo uso de fluorescência. Dissertação, Universidade de São Paulo. https://www.teses.usp.br/teses/disponiveis/9/9139/tde-26082019-114646/pt-br.php. Accessed 22 Jan 2023
- 36.Park SH, Lee YH, Yeo MH, Lee HR, Kim HS, Chang KS. Comparison of microbial detection of hemodialysis water in Reasoner’s 2A Agar (R2A) and Trypticase Soy Agar (TSA) J Bacteriol Virol. 2021;51(2):79–88. doi: 10.4167/jbv.2021.51.2.79. [DOI] [Google Scholar]
- 37.Nishioka T, Elsharkawy MM, Suga H, Kageyama K, Hyakumachi M, Shimizu M. Development of culture medium for the isolation of Flavobacterium and Chryseobacterium from rhizosphere soil. Microbes Environ. 2016;31(2):104–110. doi: 10.1264/jsme2.ME15144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ayuningrum D, Jati O (2021) Screening of actinobacteria-producing amylolytic enzyme in sediment from Litopenaeus vannamei (Boone, 1931) ponds in Rembang District, Central Java, Indonesia. Biodiversitas J Biol Divers 22(4). 10.13057/biodiv/d220427
- 39.Feng L, Li X, Zhen X, Dong H, Zheng J, Wang Y. Study of enzyme activity changing pattern in livestock manures composting. Appl Ecol Environ Res. 2019;17:6581–93. doi: 10.15666/aeer/1703_65816593. [DOI] [Google Scholar]
- 40.Silva V, Lima JV, Gondim P, Martins C, Martins SC (2015) EFEITO DA IRRIGAÇÃO E DO TIPO DE CULTIVO SOBRE A RIQUEZA E DIVERSIDADE CROMOGÊNICA DE ACTINOBACTÉRIAS DO SOLO DE UMA REGIÃO DO SEMIÁRIDO DO CEARÁ. Enciclopédia Biosfera 11(22). https://www.conhecer.org.br/ojs/index.php/biosfera/article/view/1664. Accessed 30 Jan 2023
- 41.Chadni Z, Rahaman MH, Jerin I, Hoque KMF, Reza MA. Extraction and optimisation of red pigment production as secondary metabolites from Talaromyces verruculosus and its potential use in textile industries. Mycology. 2017;8(1):48–57. doi: 10.1080/21501203.2017.1302013. [DOI] [Google Scholar]
- 42.Panwar AS, Molpa D, Joshi GK. Biotechnological potential of some cold-adapted bacteria isolated from North-Western Himalaya. Microbiology. 2019;88:343–352. doi: 10.1134/S002626171903007X. [DOI] [Google Scholar]
- 43.Seipke RF. Strain-level diversity of secondary metabolism in Streptomyces albus. PLoS ONE. 2015;10(1):e0116457. doi: 10.1371/journal.pone.0116457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salim FM, Sharmili SA, Anbumalarmathi J, Umamaheswari K. Isolation, molecular characterization and identification of antibiotic producing actinomycetes from soil samples. J Appl Pharm Sci. 2017;7(9):069–075. doi: 10.7324/JAPS.2017.70909. [DOI] [Google Scholar]
- 45.Selim MSM, Abdelhamid SA, Mohamed SS. Secondary metabolites and biodiversity of actinomycetes. J Genet Eng Biotechnol. 2021;19(1):72. doi: 10.1186/s43141-021-00156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charousová I, Medo J, Hleba L, Císarová M, Javoreková S (2019) Antimicrobial activity of actinomycetes and characterization of actinomycin-producing strain KRG-1 isolated from Karoo, South Africa. Braz J Pharm Sci 55. 10.1590/s2175-97902019000217249
- 47.Oliveira MPD (2020) Caracterização cultural e micromorfológica de actinobactérias do semiárido nordestino em diferentes níveis de cobertura vegetal. Monografia, Universidade Federal do Ceará. http://www.repositorio.ufc.br/handle/riufc/56882. Accessed 11 Feb 2023
- 48.Tiwari D, Bhati P, Das P, Shouche S (2018) Potential of actinomycetes as bioremediating and biocontrolling agents. Int J Biomed Eng 3(2):25–37. 10.36106/paripex
- 49.Anandan R, Dharumadurai D, Manogaran GP (2016) An introduction to actinobacteria. In Actinobacteria-basics and biotechnological applications. IntechOpen. 10.5772/62329
- 50.Farda B, Djebaili R, Vaccarelli I, Del Gallo M, Pellegrini M. Actinomycetes from caves: an overview of their diversity, biotechnological properties, and insights for their use in soil environments. Microorganisms. 2022;10(2):453. doi: 10.3390/microorganisms10020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurtböke Dİ. Correct interpretation of actinomycete imagery using scanning electron microscopy. Microbiol Aust. 2022;43(1):28–31. doi: 10.1071/MA22009. [DOI] [Google Scholar]
- 52.Fatima A, Aftab U, Shaaban KA, Thorson JS, Sajid I. Spore forming Actinobacterial diversity of Cholistan Desert Pakistan: Polyphasic taxonomy, antimicrobial potential and chemical profiling. BMC Microbiol. 2019;19(1):1–17. doi: 10.1186/s12866-019-1414-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madigan MT, Martinko JM, Bender KS, Buckley DH, Stahl DA (2016) Microbiologia de Brock. 14 Ed. Porto Alegre: Artmed; 2016
- 54.Dornelas JCM, Figueiredo JEF, de Abreu CS, Lana UGP, Oliveira-Paiva CA, Marriel I E (2017) Characterization and phylogenetic affiliation of Actinobacteria from tropical soils with potential uses for agro-industrial processes. http://www.alice.cnptia.embrapa.br/alice/handle/doc/1076062. Accessed 3 Mar 2023 [DOI] [PubMed]
- 55.Sukmawaty E, Sari SR, Masri M. Characterization of soil Actinomycetes from Malino pine forest rhizosphere of South Sulawesi. Elkawnie: J Islam Sci Technol. 2020;6(2):315–328. doi: 10.22373/ekw.v6i2.5383. [DOI] [Google Scholar]
- 56.Nurkanto A, Agusta A. Molecular identification and morpho-physiological characterization of Actinomycetes with antimicrobial properties. J Biologi Indones. 2015;11(2):195–203. [Google Scholar]
- 57.Fitri L, Bessania MA, Septi N, Suhartono S (2021) Isolation and characterization of soil actinobacteria as cellulolytic enzyme producer from Aceh Besar, Indonesia. Biodiversitas J Biol Divers 22(11). 10.13057/biodiv/d221155
- 58.Yun BR, Roh SG, Kim SB. Diversity and physiological properties of soil actinobacteria in Ulleung Island. Korean J Microbiol. 2017;53(4):242–250. doi: 10.7845/kjm.2017.7057. [DOI] [Google Scholar]
- 59.Janardhan A, Kumar AP, Viswanath B, Saigopal DVR, Narasimha G (2014) Production of bioactive compounds by actinomycetes and their antioxidant properties. Biotechnol Res Int 2014. 10.1155/2014/217030 [DOI] [PMC free article] [PubMed]
- 60.Yanti AH, Setyawati TR, Kurniatuhadi R (2020) Composition and Characterization of actinomycetes isolated from Nipah mangrove sediment, gastrointestinal and fecal pellets of Nipah worm (Namalycastis Rhodhocorde). In IOP Conference Series: Earth and Environmental Science (Vol. 550, No. 1, p. 012003). IOP Publishing. 10.1088/1755-1315/550/1/012003
- 61.Chaudhary HS, Yadav J, Shrivastava AR, Singh S, Singh AK, Gopalan N. Antibacterial activity of actinomycetes isolated from different soil samples of Sheopur (A city of central India) J Adv Pharm Technol Res. 2013;4(2):118. doi: 10.4103/2231-4040.111528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muok AR, Claessen D, Briegel A. Microbial hitchhiking: how Streptomyces spores are transported by motile soil bacteria. ISME J. 2021;15(9):2591–2600. doi: 10.1038/s41396-021-00952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Traxler MF, Rozen DE. Ecological drivers of division of labour in Streptomyces. Curr Opin Microbiol. 2022;67:102148. doi: 10.1016/j.mib.2022.102148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pathalam G, Rajendran HAD, Appadurai DR, Gandhi MR, Michael GP, Savarimuthu I, Naif AAD. Isolation and molecular characterization of actinomycetes with antimicrobial and mosquito larvicidal properties. Beni-Suef Univ J Basic Appl Sci. 2017;6(2):209–217. doi: 10.1016/j.bjbas.2017.04.002. [DOI] [Google Scholar]
- 65.Kalyani BS, Krishna PS, Sreenivasulu K. Screening and identification of novel isolate Streptomyces sp., NLKPB45 from Nellore costal region for its biomedical applications. Saudi J Biol Sci. 2019;26(7):1655–1660. doi: 10.1016/j.sjbs.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lima SM, Melo JG, Militão GC, Lima GM, Aguiar JS, Araújo RM, Braz-Filho R, Marchand P, Araújo JM, Silva TG. Characterization of the biochemical, physiological, and medicinal properties of Streptomyces hygroscopicus ACTMS-9H isolated from the Amazon (Brazil) Appl Microbiol Biotechnol. 2017;101:711–723. doi: 10.1007/s00253-016-7886-9. [DOI] [PubMed] [Google Scholar]
- 67.Díaz-Díaz M, Bernal-Cabrera A, Trapero A, Medina-Marrero R, Sifontes-Rodríguez S, Cupull-Santana RD, García-Bernal M, et al. Characterization of actinobacterial strains as potential biocontrol agents against Macrophomina phaseolina and Rhizoctonia solani, the main soil-borne pathogens of Phaseolus vulgaris in Cuba. Plants. 2022;11(5):645. doi: 10.3390/plants11050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Almuhayawi M, Mohamed M, Abdel-Mawgoud M, Selim S, Al Jaouni S, AbdElgawad H. Bioactive potential of several actinobacteria isolated from microbiologically barely explored desert habitat, Saudi Arabia. Biology. 2021;10(3):235. doi: 10.3390/biology10030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abdelrazek SA, Elkony HM. Utilization of oxygen quantity as a compost quality indicator. Zagazig J Agric Res. 2019;46(2):391–401. doi: 10.21608/zjar.2019.33395. [DOI] [Google Scholar]
- 70.Beites T, Pires SD, Santos CL, Osorio H, Moradas-Ferreira P, Mendes MV. Crosstalk between ROS homeostasis and secondary metabolism in S. natalensis ATCC 27448: modulation of pimaricin production by intracellular ROS. PLoS One. 2011;6(11):e27472. doi: 10.1371/journal.pone.0027472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim SY, Park C, Jang HJ, Kim BO, et al. Antibacterial strategies inspired by the oxidative stress and response networks. J Microbiol. 2019;57:203–212. doi: 10.1007/s12275-019-8711-9. [DOI] [PubMed] [Google Scholar]
- 72.Yuan F, Yin S, Xu Y, Xiang L, et al. The richness and diversity of catalases in bacteria. Front Microbiol. 2021;12:645477. doi: 10.3389/fmicb.2021.645477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kizhakedathil MP, Chandrasekaran SD. Screening for extracellular enzymes from actinomycetes isolated from agricultural soils of Kolathur, Tamil Nadu, India. Curr Bioact Compd. 2018;14(4):387–396. doi: 10.2174/1573407213666170615112449. [DOI] [Google Scholar]
- 74.Kamino LN, Gulden RH. The effect of crop species on DNase-producing bacteria in two soils. Ann Microbiol. 2021;71:1–18. doi: 10.1186/s13213-021-01624-w. [DOI] [Google Scholar]
- 75.Sumby P, Barbian KD, Gardner DJ, Whitney AR, Welty DM, Long RD, Bailey JR, Parnell MJ, Hoe NP, Adams GG, Deleo FR, Musser JM. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc Natl Acad Sci. 2005;102(5):1679–1684. doi: 10.1073/pnas.040664110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Authors declare that all images and data in this work are their own.