Abstract
The regulation of Werner’s syndrome gene (WRN) expression was studied by characterizing the cis-regulatory elements in the promoter region and the trans-activating factors that bind to them. First, we defined the transcription initiation sites and the sequence of the 5′ upstream region (2.8 kb) of WRN that contains a number of cis-regulatory elements, including 7 Sp1, 9 retinoblastoma control element (RCE), and 14 AP2 motifs. A region consisting of nucleotides −67 to +160 was identified as the principal promoter of WRN by reporter gene assays in HeLa cells, using a series of WRN promoter-luciferase reporter (WRN-Luc) plasmids that contained the 5′-truncated or mutated WRN upstream regions. In particular, two Sp1 elements proximal to the transcription initiation site are indispensable for WRN promoter activity and bind specifically to Sp1 proteins. The RCE enhances WRN promoter activity. Coexpression of the WRN-Luc plasmids with various dosages of plasmids expressing Rb or p53 in Saos2 cells lacking active Rb and p53 proteins showed that the introduced Rb upregulates WRN promoter activity a maximum of 2.5-fold, while p53 downregulates it a maximum of 7-fold, both dose dependently. Consistently, the overexpressed Rb and p53 proteins also affected the endogenous WRN mRNA levels in Saos2 cells, resulting in an increase with Rb and a decrease with p53. These findings suggest that WRN expression, like that of other housekeeping genes, is directed mainly by the Sp1 transcriptional control system but is also further modulated by transcription factors, including Rb and p53, that are implicated in the cell cycle, cell senescence, and genomic instability.
Werner’s syndrome (WS) is a rare autosomal recessive genetic disorder causing symptoms of premature aging, such as gray hair, baldness, cataracts, and osteoporosis (9, 15, 23), accompanied by rare cancers (14). In vitro studies of fibroblast growth characteristics also suggest that WS may be related to normal aging: the life span of WS fibroblasts as expressed by population doubling levels is much shorter than that of normal fibroblasts (10, 32). The hypermutator phenotype, such as represented by genetic instability, also occurs frequently in WS fibroblasts and lymphoid cells (27, 31, 33, 41).
The gene responsible for WS (WRN) has been identified by positional cloning from the 8p11-p12 region (48) and is composed of a total of 35 exons (25, 49) that generate mRNA with 5,189 nucleotide residues. We have recently demonstrated that the gene product of WRN is an active RecQ-type DNA helicase by expressing WRN in insect cells, and we postulated that defective DNA metabolism is involved in a complex process of premature aging in WS patients (40). DNA helicases are enzymes that unwind the energetically stable double-stranded structure of DNA to provide the single-stranded template for important cellular processes such as replication, recombination, and repair (42).
Mutations occurring in more than 100 WS patients have been extensively investigated by us and others. The more than 19 different types of mutations identified to date are distributed over the entire coding region of WRN and exist in individual patients as either homozygous or compound heterozygous mutations (13, 25, 48, 49). We recently found that most of these mutations generate truncated DNA helicase molecules lacking the nuclear localization signals in the C-proximal region, rendering the WRN products unable to be transported to the nucleus, where the DNA helicase is believed to function (26). This finding not only explains why WS patients with different types of mutations manifest similar clinical phenotypes but also suggests that WRN expression in patient cells is totally inhibited at the stage of protein transportation. Thus, to understand the molecular mechanism underlying WS, elucidation of the regulation of WRN transcription, which prevents individuals without WS from premature aging, is imperative.
The WRN mRNA is expressed in many organs but shows some organ-specific features, e.g., high expression in testis, ovary, and pancreas and low expression in lung, brain, kidney, and leukocytes (reference 48 and our unpublished results). Regarding the levels of WRN mRNA in healthy individuals and WS patients, we recently reported that both fibroblasts and Epstein-Barr virus-transformed B-lymphoblastoid cell lines from patients with homozygous mutation 4 or mutation 6 had much less mRNA than normal cells. This reduction in the WRN mRNA level suggested an augmented specific degradation of the WRN mRNA in patient cells, as was shown in other cases in which the nonsense codons affected RNA metabolism in vertebrate cells (47). However, little is known about the various aspects of WRN transcription, for example, the promoter, its cis-acting elements, and trans-activating protein factors, such as Rb, p53, and Sp1, that regulate WRN expression as discussed in this report.
The tumor suppressor proteins Rb and p53 are nuclear phosphoproteins involved in the control of cell proliferation and regulation of the cell cycle. The Rb and p53 proteins have been shown to act positively or negatively in the transcriptional regulation of various cellular genes. Rb interacts with several transcription factors, such as E2F and ATF2, to modulate their activity as was shown for transforming growth factor β2 gene (TGF-β2) expression (5, 20). Rb also regulates Sp1-mediated transcription through the retinoblastoma control element (RCE) motif by interacting directly with Sp1-I (a negative regulator of Sp1) and by liberating the Sp1 transcription factor from Sp1–Sp1-I complexes (6, 44). The RCE motif exists in several growth-related genes, such as c-fos and TGF-β1, and regulates transcription positively or negatively, depending on the cell type (19, 30). By contrast, control by p53 is known to depend on promoter sequence; it activates transcription of the p21/WAF1, GADD45, and mdm2 genes by binding to the p53 response elements in their promoter regions (17). Conversely, p53 represses other genes, such as c-fos and topoisomerase IIα, that lack the p53 response element (21, 45). Transcriptional repression by p53 is mediated by protein-protein interactions, such as with the TATA-binding protein, the CAAT-binding factor, or the Sp1 transcription factor, resulting in the inactivation of their trans-activating abilities (1, 4, 11, 46).
In this study, we elucidated the functional cis-acting elements in the WRN promoter and demonstrated that trans-activating factors Sp1, Rb, and p53 control WRN expression. Knowledge obtained from these studies should contribute to our understanding of the regulation of WRN expression in vivo.
MATERIALS AND METHODS
Cell lines.
HeLa cells (epithelioid carcinoma, cervix) were used for reporter gene assays. Saos2 cells (osteosarcoma) that lacked active Rb and p53 proteins were used for assaying the effects of these proteins on the WRN promoter. Two Saos2 cell lines, SRb-7 and Sp53-3 (29), that carry the tetracycline-inducible expression plasmids for Rb and p53, respectively, were made by one of us (J.Y.) and used to examine the individual effects of Rb and p53 on WRN transcription. All cell lines were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal calf serum. The culture medium for SRb-7 and Sp53-3 contained hygromycin B (0.3 mg/ml), G418 (0.5 mg/ml), and tetracycline (1 μg/ml). The cell lines were cultured at 37°C in an incubator with 5% CO2.
Determination of transcription start sites.
The transcription start sites of WRN mRNA were determined by an oligonucleotide-capping method (36), with slight modification. Briefly, the cap structure of poly(A)+ RNA obtained from HeLa cells was removed by highly purified tobacco acid pyrophosphatase (Nippon Gene), and decapped mRNA was ligated to the RNA linker 5′-CGAAUCGUAACCGUUCGUACGAGAAUCCGCU-3′ by using T4 RNA ligase. The product was then reverse transcribed by using a random hexamer and was amplified by PCR using a combination of the RNA linker-specific primer 5′-ATCGTAACCGTTCGTACGAGAATCGC-3′ and the WRN-specific primer 5′-CCCACCACATCCCCATCTGATAGACTC-3′ (positions 453 to 479), followed by the WRN-specific nested primer 5′-AGGAAAGAGCAATCACTAGCATCG-3′ (positions 411 to 434). The PCR product was cloned into the pGEM-T vector (Promega), and the nucleotide sequence encompassing the capping site of WRN mRNA was determined after sequence analysis by PCR-based cycle sequencing using a Prism sequencing kit (Perkin-Elmer) with 17 independent clones. The S1 nuclease mapping analysis described by Berk and Sharp (3) was performed to confirm the transcription initiation sites obtained by the oligonucleotide-capping method. A 5′-32P-labeled DNA fragment (245 bp, encompassing nucleotide residues −143 to +102) was prepared by EcoO109I and BssHII digestion and by subsequent 5′-end labeling with [γ-32P]ATP and T4 polynucleotide kinase. The labeled duplex DNA fragment (106 cpm) was purified on a Centrisep column (Applied Biosystem), denatured by boiling followed by chilling, and hybridized to the total RNA (25 μg) from human K562 cells. As a control, yeast tRNA (25 μg) was also used in place of K562 cell RNA. Hybridization conditions were heating at 80°C for 10 min and annealing at 50°C for 3 h in hybridization buffer [50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES)–NaOH buffer (pH 6.4), 1 mM EDTA, 80% formamide, 0.4 M NaCl]. The annealed RNA-DNA heteroduplex was digested in S1 digestion buffer containing 30 mM sodium acetate buffer (pH 4.6), 280 mM NaCl, 1 mM ZnSO4, 100 μg of salmon sperm DNA per ml, and 1,000 U of S1 nuclease per ml (TaKaRa) at 37°C for 30 min, and the reaction products were analyzed by 5% polyacrylamide gel electrophoresis.
P1/PAC DNA and determination of DNA sequences.
The P1/P1-derived artificial chromosome (PAC) library was screened by a PCR-based strategy, and positive clones were isolated by Genome Systems, Inc. (St. Louis, Mo.). PAC 12339 DNA was isolated by an alkaline lysis procedure essentially described by Smoller et al. (38), with slight modification, and purified by density equilibrium centrifugation with CsCl-ethidium bromide gradients. After digestion with Sau3AI, the PAC 12339 DNA was subcloned into the BamHI site of pBluescriptII KS(+) (Stratagene). To obtain the promoter region, this DNA library was screened by PCR using primers designed from the sequence of the first exon of WRN: 5′-GGGAATAAAGTTTGCTGATTT-3′ (positions 50 to 70) and 5′-CAGTCCAACAGGTCTTCTTCA-3′ (positions 134 to 154). The sequences of nine positive clones were determined. Sequence homology to any other known human genomic DNA was analyzed by Intelligenetics software and the FASTA and TFASTA programs from the Genetics Computer Group database searching software package (2).
Construction of WRN-Luc plasmids for promoter assay.
After the pBS/12339-17 plasmid DNA containing the putative promoter region of WRN was digested with BamHI, the 2,776-bp insert fragment was placed at the BglII site of the pGL3-Basic vector (Promega) upstream of the firefly luciferase gene, which was used as a reporter gene (pGL3/A). To generate a series of 5′-deletion mutants, the WRN promoter region of pGL3/A was digested with SacI/XhoI and then treated with exonuclease III and mung bean nuclease (pGL3/B to -N), or was amplified by PCR with specific primers, and was placed back into the pGL3-Basic plasmid (pGL3/O to -R), yielding what we refer to as the WRN-Luc plasmids. Mutations with base substitutions were made for each Sp1 element and/or RCE motif by using an LA PCR in vitro mutagenesis primer set for pBluescriptII (TaKaRa) according to the manufacturer’s protocol (creating pGL3/S3m, -S2m, -S1m, -S32m, -S321m, -S3m/Rm, -S32m/Rm, -S321m/Rm, and -Rm). To generate the 3′-deleted pGL3/O and pGL3/S321m/Rm mutants missing residues 20 to 160, the plasmids were digested simultaneously at nucleotide position 20 by BglI and upstream of the vector by MluI, and the excised fragment was ligated to the SmaI site of the pGL3-Basic vector (yielding pGL3/OΔ3′ and pGL3/S321m/RmΔ3′). The directions and sizes of the modified promoter region of all constructs were confirmed by DNA sequencing.
DNA transfection and luciferase assay.
HeLa and Saos2 cell lines were transfected by the lipofection method as described by Felgner et al. (12). Briefly, the WRN-Luc plasmid DNAs (1 μg) and a plasmid (pRL-TK; Wako Pure Chemicals, Osaka, Japan) containing the herpes simplex virus thymidine kinase promoter and sea pansy luciferase (0.1 μg) used as an internal control were mixed with Lipofectin reagent (GIBCO BRL) and cotransfected into target cells grown to 60 to 70% confluence in six-well plates. After 5 h of incubation with the Lipofectin-DNA complex, the cells were washed twice in medium containing serum and cultured for 48 h. The cells were then lysed and assayed for firefly and sea pansy luciferase activities separately, using a double-luciferase assay system (Wako Pure Chemicals) and LUMAT LB 9507 luminometer (EG&G Bertholdo). The activity of the WRN promoter evidenced by firefly luciferase activity was evaluated after normalizing for small differences in transfection efficiency by the sea pansy luciferase activity. Expression plasmids that produce Rb (pCMV1-Rb), wild-type p53 (pC53-SN3), and mutant p53 (pC53-249) were kindly provided by Eiji Hara (Kyoto Prefectural Medical School) and Takashi Uchida (Nippon Roche Research Institute). In the Rb or p53 overexpression experiments, the WRN-Luc plasmid DNAs (1 μg), the Rb or p53 expression plasmid (0.03 to 5 μg), and pRL-TK (0.1 μg) were cotransfected and assayed for luciferase activity as described above.
Preparation of nuclear extracts from HeLa cells.
Nuclear extracts were prepared essentially as described by Dignam et al. (8) from confluent cultures of HeLa cells. Approximately 108 cells were washed with ice-cold phosphate-buffered saline, pelleted, and resuspended in 10 mM Tris-HCl (pH 7.9)–1.5 mM MgCl2–10 mM KCl–0.5 mM dithiothreitol (DTT). After incubation on ice for 10 min, the cells were homogenized in a Dounce homogenizer with approximately 20 strokes. The nuclei were then pelleted and resuspended in 20 mM Tris-HCl (pH 7.9) buffer containing 1.5 mM MgCl2, 20% glycerol, 0.5 mM DTT, 0.3 M KCl, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF). After rocking at 4°C for 30 min, the supernatant was dialyzed for 2 h at 4°C against 4 liters of 20 mM Tris-HCl (pH 7.9) buffer containing 0.1 M KCl, 0.2 mM EDTA, 20% glycerol, 0.5 mM DTT, and 0.5 mM PMSF. The extracts were mixed with a cocktail of protease inhibitors to final concentrations of 1 mM PMSF, 1 mM EGTA, 0.021 mM leupeptin, 0.01 mM pepstatin, 0.1 mM Nα-p-tosyl-l-lysine chloromethylketone (TLCK) and 1 mM N-methylmaleimide, aliquoted, and stored at −70°C until used. The protein concentration was measured by using a Pierce bicinchoninic acid protein assay kit.
EMSA.
Electrophoretic mobility shift assays (EMSAs) were done as described by Singh et al. (37), with slight modifications. A HeLa cell nuclear extract (2 to 5 μg) was first incubated in 10 mM Tris-HCl (pH 7.5) buffer containing 50 mM NaCl, 0.5 mM DTT, 0.5 mM EDTA, 1 mM MgCl2, 0.4 mg of bovine serum albumin per ml, 4% glycerol, and 50 to 250 μg of double-stranded poly(dI-dC) (Pharmacia, Piscataway, N.J.) per ml for 20 min at room temperature. The mixture was then incubated for an additional 20 min with approximately 10,000 cpm of double-stranded 32P-labeled oligonucleotides containing Sp1-3 (positions −86 to −57), RCE (positions −70 to −51), Sp1-2 (positions −65 to −36), or Sp1-1 (positions −46 to −17). In the competition experiments, a 50-fold molar excess of specific (Sp1) or nonspecific (AP1) competitor (nonradioactive oligonucleotides; see below) was incubated with the HeLa cell nuclear extract before addition of 32P-labeled oligonucleotides. The competitor oligonucleotides (only sense strands are shown) were 5′-ATTCGATCGGGGCGGGGCGAGC-3′ for Sp1 and 5′-CGCTTGATGAGTCAGCCGGAA-3′ for AP1. For the supershift experiment, the nuclear extract was preincubated with 0.5 mg of Sp1-specific polyclonal antibody or nonimmunized rabbit immunoglobulin G (IgG) fraction (Santa Cruz Biotechnology, Inc.) per ml at 4°C for 1 h. The antibody-treated extract was used for the EMSAs as described above. The reaction mixtures were electrophoresed in a 4% polyacrylamide gel in 0.5× TBE buffer (45 mM Tris-borate, 1 mM EDTA [pH 8.0]) at 100 V for 2.5 h and were analyzed for retardation of labeled oligonucleotides during electrophoretic migration. After drying, the gels were exposed to X-ray film (Kodak) at −70°C with intensifying screens for 24 to 48 h.
Northern blot analysis.
Two Saos2 cell lines, SRb-7 and Sp53-3 (29), that carry the tetracycline-inducible expression plasmids for Rb and p53, respectively, were cultured in the presence or absence of tetracycline at 37°C for 6 to 12 h. The poly(A)+ RNA was extracted from these cells by the acid guanidinium thiocyanate-phenol-chloroform extraction method (7) and Oligotex-d(T)30 (Takara Co. Ltd., Osaka, Japan). Three micrograms of poly(A)+ RNA was electrophoresed in 1% agarose gels containing formamide and transferred to Hybond-N membranes (Amersham). Hybridization was performed with 32P-radiolabeled probe (2 × 106 cpm/ml) prepared from the C-terminal region, which includes the 3′ untranslated region (nucleotide residues 3199 to 5065) of the WRN gene, at 42°C for 16 h. Then the membranes were washed in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate at 42°C for 30 min and autoradiographed for 5 days. For quantitative analysis, the membranes were stripped and then rehybridized with a 32P-radiolabeled β-actin probe as a control to normalize the density. The relative intensities of individual WRN mRNA bands were estimated with a BAS-1500 Bioimaging Analyzer (Fujifilm).
Nucleotide sequence accession number.
The complete nucleotide sequence of the 2.8-kb WRN fragment was deposited in the GenBank/EMBL Data Bank with accession no. AB003173.
RESULTS
Identification of the transcription start sites of WRN.
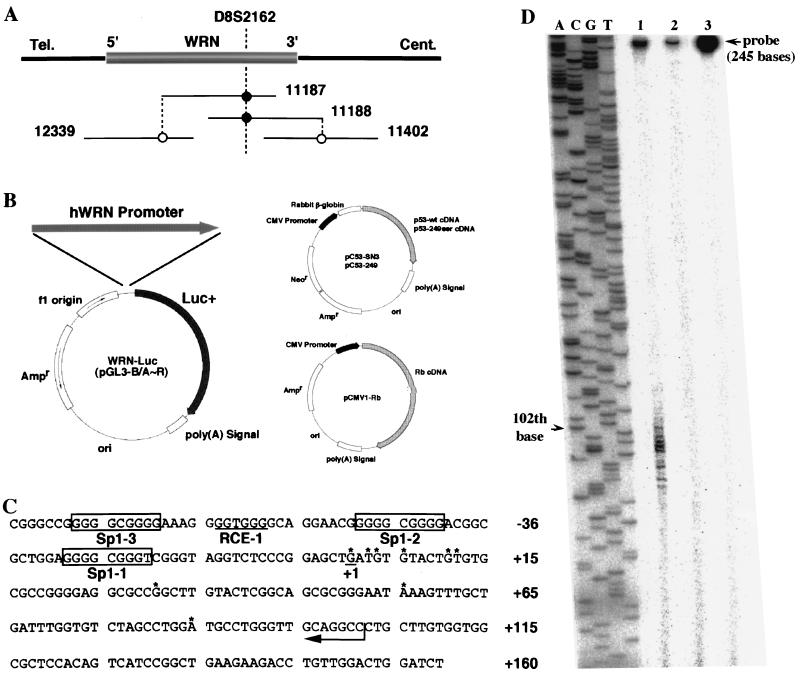
Before identifying the WRN promoter region, we analyzed the transcriptional start sites of WRN by the oligonucleotide-capping method described in Materials and Methods. Primers were designed from the sequences in the fourth and fifth exons of WRN and were used to amplify the 5′ WRN cap site cDNA from the HeLa cell oligonucleotide-capped cDNA library. Several PCR products were obtained, and their nucleotide sequences were determined. As expected, all of the PCR products were found to contain the 5′ untranslated sequence of WRN mRNA. Sequence determination showed that WRN was transcribed from multiple positions, i.e., positions 1, 3, 4, 6, 11, 12, 31, 56, and 85 (Fig. 1C). Positions 1, 4, and 6 starting with G seem to be used most frequently as the initiation start sites of WRN transcription, based both on the results obtained from S1 nuclease mapping analysis (Fig. 1D) and on the number of clones obtained after subcloning of the 5′ WRN cap site cDNA (data not shown). Similarly, multiple transcription initiation sites have recently been found for the human gp130 gene (28). In this report, we assume that the most upstream start site is at position +1.
FIG. 1.
WRN promoter region and structures of plasmids used for promoter assays. (A) Orientation of WRN in the P1/PAC contig map. Solid lines and numbers represent P1/PAC DNAs that form the physical map. The approximate positions of WRN and the STS marker D8S2162 are shown. The transcriptional direction of WRN is indicated by 5′ and 3′. Tel., telomere; Cent., centromere. (B) The 2,776-bp DNA fragment generated from PAC 12339 DNA was placed at the BglII site of the pGL3-Basic vector upstream of the firefly luciferase gene to form pGL3/A, which was used as a reporter gene in the promoter assay. A series of WRN-Luc plasmids containing the sequentially 5′-deleted human WRN (hWRN) promoter regions (pGL3/B to -R) were prepared from pGL3/A. The structures of Rb and p53 expression plasmids (pCMV1-Rb, pC53-SN3, and pC53-249) are shown. CMV, cytomegalovirus. (C) The WRN transcription start sites are indicated by asterisks. The most upstream nucleotide position is assumed to be position +1. The cis-acting Sp1 element binding sites and an RCE motif are boxed and underlined, respectively. (D) Results of S1 nuclease protection assay. The ladder (lanes A, C, G, and T) represents the sequence of pGEM3Zf(+) DNA recognized by the M13 forward primer (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′), and base 102 is shown by an arrowhead. Lane 1, S1 mapping with 106 cpm of 32P-labeled probe DNA for K562 cell RNA that contains WRN mRNA; lane 2, control experiment using yeast tRNA; lane 3, S1 nuclease-undigested 5′ 32P-labeled 245-bp DNA (104 cpm) that encompasses the potential WRN promoter region from −143 to +102, heat denatured prior to electrophoresis. The band remaining in the upper part of the gel indicates the S1 nuclease-resistant duplex DNA formed by annealing of complementary DNAs during hybridization. The band migrating with base 102 (lane 1) corresponds to the most upstream nucleotide from the 32P-labeled 5′ G (from the complementary strand to the 3′-5′ CCG indicated by an arrow in panel C) and is designated the +1 position of the WRN mRNA.
Cloning and nucleotide sequence of the WRN promoter region.
A physical map containing WRN was made with a contig of P1/PAC DNAs, and the precise location of WRN was identified (Fig. 1A). Genomic clones, including the putative promoter region of WRN, were obtained from the PAC 12339 library by PCR using primers designed from the sequence of the first exon of WRN (positions 50 to 154). After the PAC 12339 DNA was digested with Sau3AI, all resulting fragments were subcloned into the BamHI site of pBluescript KS(+) (Stratagene). A PCR-based screening of 100 clones yielded nine independent positive clones containing 1.4- to 4.0-kb inserts. One of them, clone pBS/12339-17, containing an insert of about 2.8 kb, was sequenced, and the analysis confirmed that it contains a 5′-flanking region and the first exon (Fig. 1C). Figure 1C shows part of this 2.8-kb DNA, a region corresponding to −85 to +160 that correlates with WRN promoter activity. The region from positions −85 to −1 contained an extremely high GC content (80%), whereas the overall GC content of the entire 2.8-kb fragment was 58%. In addition, multiple copies of transcription regulatory elements Sp1 (seven), RCE (nine), and AP2 (fourteen) were clustered in the 2.8-kb region. In contrast, no TATA box or CAAT box was evident. These findings collectively indicated that the WRN promoter probably is in this region and has promoter characteristics often associated with housekeeping genes.
Analysis of multiple cis elements in the upstream region of the transcriptional initiation site of WRN.
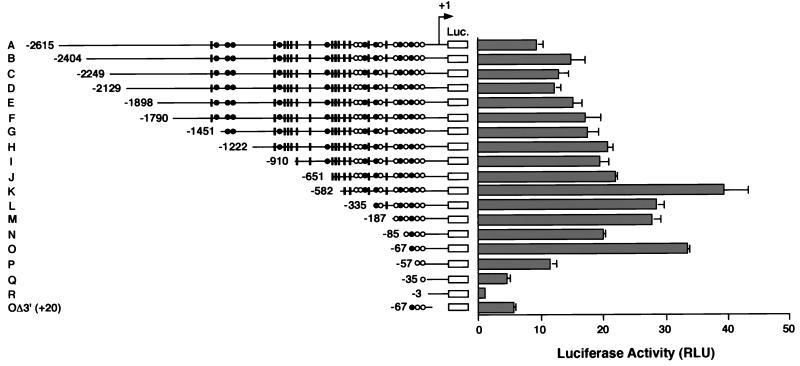
The 2.8-kb DNA fragment was cloned into the pGL3-Basic vector (WRN-Luc plasmid). The resulting construct, pGL3/A (−2615∼+160), was used for promoter assays by a luciferase gene placed downstream of the WRN DNA. To define the region responsible for the WRN promoter, a series of 5′-deletion mutants of pGL3/A was constructed (Fig. 2). These deletion mutants were transfected into HeLa cells, and their promoter activities were monitored by luciferase production after 48 h of cell culture. The majority of about 2.5 kb of sequence upstream of the WRN transcriptional start site was largely dispensable; the short upstream region from positions −67 to +160 was sufficient (Fig. 2, O). This region contains one RCE motif (RCE-1) and two Sp1 elements (Sp1-1 and Sp1-2) that are often indispensable as cis-acting regulatory elements for the constitutive expression of most housekeeping genes. Several AP2 elements in the upstream region of a minimal promoter are clearly of little importance to WRN expression but are required for maximum promoter activity shown by pGL3/K. Truncation at −57 (pGL3/P), which removes the RCE motif, decreased the promoter activity by 66% compared with pGL3/O (Fig. 2, P). Truncation at −35, which left only Sp1-1, largely decreased luciferase expression and resulted in low promoter activity (Fig. 2, Q). These results suggest that Sp1-1 may be nonfunctional by itself but works cooperatively with the other elements at −67 to −36. pGL3/R, which had no Sp1 element or RCE motif, showed very little promoter activity (3% compared to pGL3/O) (Fig. 2, R). The promoter activity of pGL3/OΔ3′, which lacked nucleotides 21 to 160, was about 80% lower than that of pGL3/O, suggesting the presence of another undefined fundamental element that cooperates with the essential Sp1-2, Sp1-1, and RCE sites at −67 to +20. Consequently, the region encompassing positions −67 to +160 seems to represent the minimal WRN promoter region required for full activity in the reporter gene assays.
FIG. 2.
Promoter activities detected in the upstream region of WRN. WRN-Luc plasmids containing various lengths of the WRN upstream region were transiently transfected into HeLa cells as described in Materials and Methods. The diagram on the left shows a map of a series of 5′-truncated WRN promoters in WRN-Luc plasmids. The 5′ ends of the WRN DNA are shown by nucleotide position numbers from the transcription initiation site. Open circles, closed circles, and black boxes indicate Sp1, RCE, and AP2 motifs, respectively. The promoter activities of the 5′-truncated DNA in WRN-Luc plasmids were measured in HeLa cells and are shown as luciferase activity on the right. Each value represents the mean luciferase activity measured in at least three independent experiments. Bars indicate standard deviations of the mean activities, expressed as RLU (relative light units) as specified by the manufacturer (EG&G Bertholdo).
Effect of the substitution mutation in the Sp1 element on WRN promoter function.
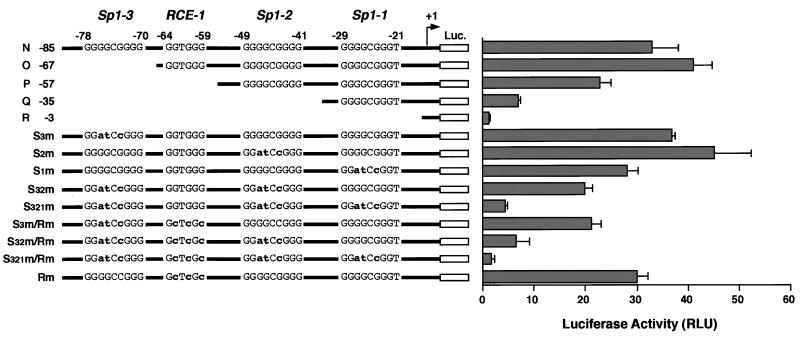
To examine the contribution of each Sp1 element to the expression of WRN, we mutated the Sp1-3, Sp-2, and Sp-1 elements by base substitutions. These changes had been designed to affect the Sp1 elements, and the mutated WRN-Luc plasmids (pGL3/S3m, pGL3/S2m, pGL3/S1m, pGL3/S32m, and pGL3/S321m) were generated from pGL3/N. They were transfected into HeLa cells, and their activities were measured. The single mutation in the Sp1-3, Sp1-2, and Sp1-1 elements had little or no effect on WRN promoter activity (Fig. 3, S3m, S2m, and S1m). A triple mutation of all three Sp1 elements resulted in about 90% reduction of promoter activity, leaving a low level of activity, but 3-fold higher activity than for pGL3/R, which lacked Sp1-3, Sp-2, and Sp-1 (Fig. 3, S321m and R −3). The promoter activity in pGL3/Q was severely reduced, to 20% of that of pGL3/N, upon removal of the Sp1-3 and Sp1-2 elements. By contrast, the double mutations in Sp1-3 and Sp-2 restored 60% of its activity (Fig. 3, Q −35 and S32m). These results strongly suggest that some element(s) other than Sp1 elements in the −85 to −36 region activates the WRN promoter. These findings are consistent with the data obtained for the 5′-deletion experiments and support the hypothesis that the Sp1 elements upregulate WRN transcription. In addition, another response element(s) in the WRN promoter region, such as the RCE motif GGTGGG at −64 to −59, cooperates with the Sp1 elements (see below).
FIG. 3.
Effects of the 5′-deletion and base-substitution mutations in the cis-regulatory Sp1 elements and RCE on WRN promoter activity. WRN-Luc plasmids containing various base-substitution mutations in three Sp1 elements and an RCE motif were generated. Their promoter activities were measured by transfection into HeLa cells and compared with the promoter activity of pGL3/N to -R. The diagram on the left shows the structure of each of the 5′-deletion and base-substitution mutants of the WRN promoter; the substituted bases are represented by lowercase boldface letters. Promoter activities in the modified WRN promoter region are shown as luciferase activity on the right. Each value represents the mean activity of luciferase detected in at least three independent experiments. Bars indicate standard deviations of the mean activities, expressed as RLU (relative light units) as specified by the manufacturer (EG&G Bertholdo).
Effect of the RCE motif on WRN promoter activity.
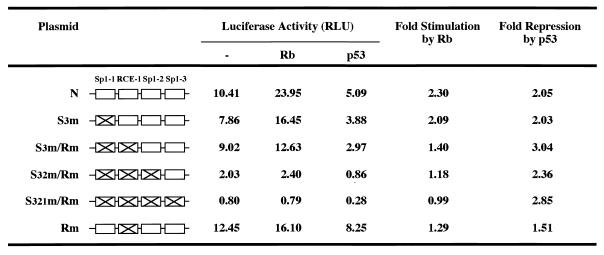
To confirm that the RCE (positions −64 to −59) in the WRN promoter region participates in WRN expression, we constructed a series of WRN-Luc plasmids containing a modified WRN promoter with a substitution mutation in the RCE motif (pGL3/S3m/Rm, pGL3/S32m/Rm, pGL3/S321m/Rm, and pGL3/Rm). In this study, mutation of the RCE motif in pGL3/Rm showed no effect on promoter activity compared to that of pGL3/N (Fig. 3, Rm). However, the activity of pGL3/S3m/Rm was rendered lower than that of pGL3/S3m and was almost the same as that of pGL3/P, which lacked the Sp1-3 element and RCE motif (Fig. 3, S3m/Rm, S3m, and P). The mutation of the RCE motif in pGL3/S32m/Rm reduced its promoter activity to lower than that of pGL3/S32m and to a level equivalent to that of pGL3/Q, which lacked the Sp1-3, Sp1-2, and RCE motifs (Fig. 3, S32m/Rm, S32m, and Q). Very little promoter activity remained in pGL3/S321m/Rm, which has mutations in all four elements, and this level of activity was the same as for pGL3/R, with all four elements deleted (Fig. 3, S321m/Rm and R). These findings suggest that the WRN promoter was upregulated not only by the Sp1 element but also by the RCE motif.
Nuclear proteins that bind to the promoter region of WRN.
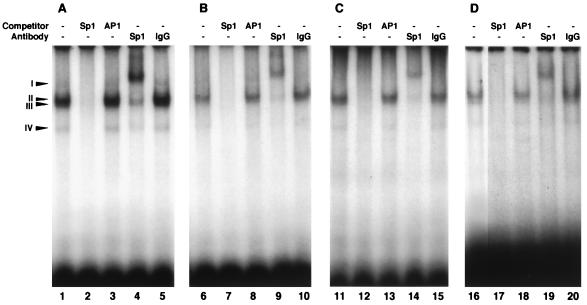
To examine if the Sp1 elements and RCE motif indeed bind to the corresponding nuclear protein factor, we performed EMSAs with HeLa cell nuclear extract. First, EMSAs were done with three double-stranded oligonucleotide probes containing the Sp1-3, Sp1-2, or Sp1-1 element in the WRN promoter region. We observed four DNA-protein complexes (complexes I to IV [Fig. 4A]) when we used Sp1-3 and Sp1-2 as probes (Fig. 4, lanes 1 and 6), while complex I was not detected with Sp1-1 alone (lane 11). Complex formation was inhibited specifically by adding a 50-fold excess of unlabeled Sp1 consensus oligonucleotide (lanes 2, 7, and 12) but not by the AP1 oligonucleotide (lanes 3, 8, and 13), indicating that these DNA-protein complexes were formed by some Sp1-specific binding protein(s) and the Sp1 oligonucleotide. Indeed, in subsequent supershift analysis using a human Sp1-specific polyclonal antibody, antibody addition further decreased the mobility of the majority of these DNA-protein complexes (lanes 4, 9, and 14), while the nonimmunized rabbit IgG added to the binding reaction mixtures as a control did not affect the EMSA profiles (lanes 5, 10, and 15). When the efficiency of complex formation with Sp1 proteins was compared among the probe oligonucleotides, the Sp1 protein showed an increasing order of Sp1-3 > Sp1-2 > Sp1-1, suggesting that the adjacent sequences of Sp1 elements, and perhaps the intervening RCE motif (and included in Sp1-3 and Sp1-2 probes), stimulate complex formation. In any event, these data confirmed that the cellular Sp1 protein can bind specifically to these Sp1 elements, with some differences in binding efficiency. We also noted minor complexes but clearly of another type because their migration profiles were not affected by the Sp1-specific antibody (lanes 4, 9, and 14). These protein complexes were apparently specific in binding to the Sp1 element but may have included proteins other than the known Sp1 protein.
FIG. 4.
Characterization of the nuclear protein that interacts with oligonucleotides containing Sp1 elements and an RCE motif. Nuclear extracts from HeLa cells (2 to 5 μg of protein) were incubated with the 32P-labeled double-stranded oligonucleotide probes that contained WRN Sp1-3 (positions from −86 to −57) (A), Sp1-2 (positions from −65 to −36) (B), Sp1-1 (positions from −46 to −17) (C), and RCE-1 (positions from −70 to −51) (D) in the absence (lanes 1, 6, 11, and 16) or presence (lanes 2, 7, 12, and 17) of 50-fold molar excess amounts of unlabeled Sp1 oligonucleotides. The mixtures were electrophoresed in a 4% polyacrylamide gel as described in Materials and Methods. Similar experiments were also done with 50-fold excess amounts of unlabeled AP1 oligonucleotide (lanes 3, 8, 13, and 18). In the experiments represented by lanes 4, 9, 14, and 19, the Sp1 protein-specific rabbit IgG antibodies (5 μg) were included in the reaction mixtures before incubation; as the control for these experiments, the same reactions were performed with nonimmunized rabbit IgG fraction (5 μg) (lanes 5, 10, 15, and 20).
Udvadia et al. (44) reported that transcriptional regulation of the c-fos gene by RCE is mediated by (i) the Sp1 protein binding to the RCE motif and regulating promoter activity and (ii) the Rb protein activating the RCE-dependent transcription through Sp1. To confirm the binding of the Sp1 protein to the RCE motif in the WRN promoter region, similar EMSAs were performed with a double-stranded oligonucleotide probe containing the RCE motif. Three DNA-protein complexes, II, III, and IV, were detected (Fig. 4, lane 16). The unlabeled Sp1 consensus oligonucleotide, but not AP1 oligonucleotide, inhibited the formation of these DNA-protein complexes (lanes 17 and 18). A human Sp1-specific polyclonal antibody added to the binding reaction mixture decreased the mobility of these DNA-protein complexes in the supershift analysis (lane 19). Addition of nonimmunized rabbit IgG had no effect on the mobility of these complexes (lane 20), indicating that the complex formation is Sp1 specific. The same results were obtained when we performed EMSAs with purified human Sp1 protein (data not shown). These data indicated that the Sp1 protein binds to the RCE motif in the WRN promoter region as was reported by Udvadia et al. (44).
The WRN promoter is upregulated by Rb and downregulated by p53.
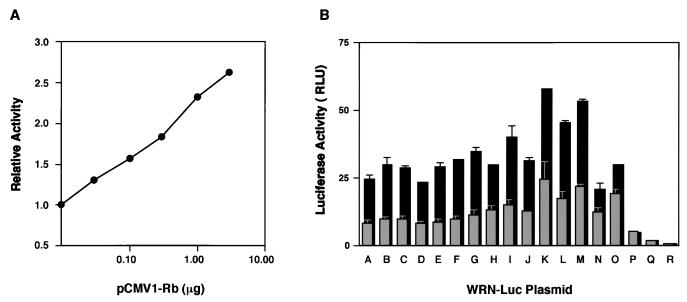
To examine the effects of the Rb protein on WRN promoter activity, we used Saos2 cells lacking the active Rb and p53 proteins (18). First, we cotransfected the Saos2 cells with the WRN-Luc plasmid pGL3/A and increasing amounts of the Rb expression plasmid (pCMV1-Rb) and measured luciferase activities. Strikingly, cotransfection with the Rb expression plasmids resulted in a significant stimulation (maximum of 2.5-fold) of WRN promoter activity in a plasmid dose-dependent manner (Fig. 5A). The production of Rb was confirmed by Western blot analysis using lysates of cells transfected with the Rb expression plasmid (data not shown). To confirm that the RCE motif or Sp1 elements indeed contributed to the Rb-mediated upregulation, we cotransfected WRN-Luc plasmids (pGL3/A to -R) with pCMV1-Rb (1 μg) and measured promoter activities. WRN promoter activities in pGL3/A to -O were increased by the Rb protein about twofold (Fig. 5B), but this stimulatory effect did not occur in pGL3/P to -R, which lacked the RCE motif (Fig. 5B). These results suggest that Rb protein upregulates WRN expression and that the region from −67 to −57 was essential for this effect. Consistent with these results, a series of promoter assays with various WRN-Luc plasmids (pGL3/S3m, -S3m/Rm, -S321m/Rm, and -Rm) containing substitution mutations showed that the stimulation by Rb is pronounced when the RCE motif is intact, while the overall promoter activity appears to depend on the number of intact Sp1 elements in this −78 to −21 region (Table 1).
FIG. 5.
Regulatory effects of Rb protein on WRN promoter activity in Saos2 cells. (A) pGL3/A, a WRN-Luc plasmid containing the full-length WRN promoter region, was cotransfected with increasing amounts of pCMV1-Rb into Saos2 cells, which are known to be deficient in active Rb and p53. The activity of the WRN promoter in the transfected cells was determined and compared with the activity of pGL3/A alone. The values represent relative activities of pGL3/A expressed in the presence of Rb, assuming the luciferase activity with pGL3/A alone to be 1.0. The data were reproducible in two independent experiments. (B) WRN-Luc plasmids (pGL3/A to -R) containing various lengths of the WRN promoter region were cotransfected with Rb-expressing pCMV1-Rb (1 μg) in Saos2 cells, and the site of the WRN promoter that responds to Rb was investigated. Gray columns show WRN promoter activities associated with pGL3/A to -R alone; black columns show WRN promoter activities obtained in the presence of Rb. The data were reproducible in two independent experiments. Luciferase activity is expressed as RLU (relative light units) as specified by the manufacturer (EG&G Bertholdo).
TABLE 1.
Transcriptional activity of mutated WRN promoter cotransfected with Rb and p53 expression plasmids in Saos2 cellsa
WRN-Luc plasmids containing substitution mutations in Sp1-3, Sp1-2, Sp1-1, and the RCE motif of the WRN promoter region were cotransfected with pCMV1-Rb (1 μg) or pC53-SN3 (0.1 μg) in Saos2 cells, and promoter activities were measured. Fold stimulation by Rb and fold repression by p53 were estimated as relative to the activities of WRN-Luc plasmids alone, considered to be 1.0. The data were reproducible in two independent experiments. Luciferase activity is expressed as RLU (relative light units) as specified by the manufacturer (EG&G Bertholdo).
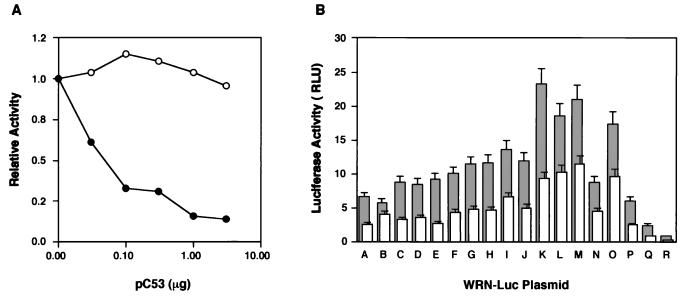
Next, we studied the effect of p53 on WRN promoter activity because p53 protein often participates in transcriptional suppression of the Rb gene and the genes that contain Sp1 elements (4, 35, 46). In cotransfection experiments in which increasing amounts of p53 expression plasmid (pC53-SN3) were expressed in Saos2 cells, WRN promoter activity of pGL3/A was downregulated a maximum of 90% (Fig. 6A, closed circle). By contrast, the inactive p53 mutant protein containing a single amino acid substitution at position 249 (43) failed to show this downregulation (Fig. 6A). Again, the production of p53 in the transfected Saos2 cells was confirmed for two different types of p53 by Western blot analysis using cell lysates (data not shown). To clarify further the cis-regulatory element involved in the p53-mediated downregulation, WRN-Luc plasmids pGL3/A to -R were cotransfected with pC53-SN3 (0.1 μg), and promoter activities were measured (Fig. 6B). Here, the original WRN promoter activities were reduced to about 50% by the presence of p53 protein, regardless of the modifications of WRN promoter elements tested. Unlike the effect of Rb, the suppression of promoter activity by p53 was not affected by alterations in the transcriptional elements in the WRN promoter (Table 1). Downstream deletion at 21 to 160, in addition to mutations in Sp1-3, -2, and -1 and the RCE, however, prevented repression by the overexpressed p53 (data not shown). The mechanism behind this transcriptional suppression may be accounted for by the finding by Borellini and Glazer (4) that the Sp1 protein was prevented from binding to the promoter region by a p53-Sp1 protein complex. Similar types of modulations by Rb or p53 were observed when HeLa cells were cotransfected with WRN-Luc plasmids and the Rb or p53 expression plasmid (data not shown), although the levels of modulation were less pronounced, perhaps due to endogenous Rb and p53 in the HeLa cells. As expected, overexpression of either Rb or p53 did not affect the morphology or proliferation of transfected Saos2 cells, as analyzed by microscopy and cell counting.
FIG. 6.
Regulatory effects of p53 protein on WRN promoter activity in Saos2 cells. (A) pGL3/A, a WRN-Luc plasmid containing the full-length WRN promoter region, was cotransfected with increasing amounts of pC53-SN3 (closed circles) or pC53-249 (open circles) into Saos2 cells, which are known to be deficient in active Rb and p53. The activity of the WRN promoter in the transfected cells was determined and compared with the activity of pGL3/A alone. The values represent relative activities of pGL3/A expressed in the presence of p53, assuming the luciferase activity with pGL3/A alone to be 1.0. The data were reproducible in two independent experiments. (B) WRN-Luc plasmids (pGL3/A to -R) containing various lengths of the WRN promoter region were cotransfected with p53 expressing pC53-SN3 (0.1 μg) in Saos2 cells, and the site of the WRN promoter that responds to p53 was investigated. Gray columns show WRN promoter activities associated with pGL3/A to -R alone; white columns show WRN promoter activities obtained in the presence of p53. The data were reproducible in two independent experiments. Luciferase activity is expressed as RLU (relative light units) as specified by the manufacturer (EG&G Bertholdo).
Effects of Rb and p53 on the endogenous WRN mRNA expression level.
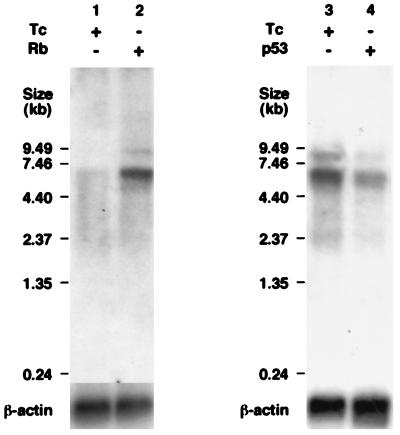
To determine whether the endogenous WRN mRNA level in Saos2 cells is indeed modulated by Rb and p53, we compared the WRN mRNA levels of SRb-7 and Sp53-3 cells by Northern blot analysis in the presence or absence of overexpressed Rb and p53. The level of WRN mRNA in the Rb-overexpressing SRb-7 cells was fourfold higher than that in Rb-nonexpressing cells (Fig. 7, lanes 1 and 2). By contrast, the level of WRN mRNA in the p53-overexpressing Sp53-3 cells was less than 50% of that of p53-nonexpressing cells (lanes 3 and 4). These results indicated that Rb and p53 indeed modulate the WRN gene expression.
FIG. 7.
Northern blot analysis of SRb-7 and Sp53-3 cells carrying the tetracycline-inducible Rb and p53 expression plasmids. SRb-7 (A) or Sp53-3 (B) cells were cultured in the presence or absence of 1 μg of tetracycline (Tc) per ml at 37°C for 6 or 12 h, respectively, poly(A)+ RNA was extracted, and Northern blot analysis was performed with a 32P-radiolabeled probe prepared from the C-terminal region, which includes the 3′ untranslated region of the WRN gene. The relative intensities of individual WRN mRNA bands were estimated with a BAS-1500 Bioimaging Analyzer (Fujifilm).
DISCUSSION
WRN promoter and Sp1-mediated transcription.
Although much progress has been made in determining the genomic structure of WRN, the types of WRN mutations occurring in patients with WS, and the biochemical nature of the DNA helicase gene product, little is known about how WRN expression is regulated and the nature of the cis element(s) and trans-activating factor(s) involved. In this study, we answered some of these questions. First, we analyzed about 2.8 kb of the upstream region of WRN and found that the transcription start site, i.e., the capping site of WRN mRNA, was multiple but that the main sites were the +1, +4, and +6 positions (Fig. 1C). S1 mapping experiments with mRNAs from human K562 cells that express a relatively high level of WRN transcripts supported these findings (Fig. 1D). Proximal to this transcription initiation site cluster, we found two Sp1 elements and one RCE cis-acting motif within the minimal WRN promoter region (positions −67 to +160), required for full activity in a reporter gene assay. Additional elements, including 5 Sp1, 8 RCE, and 14 AP2 motifs, are distributed throughout the upstream 1.4 kb (Fig. 2). Since the Sp1 element is conserved in the promoters of most housekeeping genes, WRN seems to be a housekeeping gene. Consistent with this view, there is neither a TATA box nor a CAAT box proximal to this transcription initiation site. The Sp1 protein that binds to the consensus Sp1 element GGCGGG is also capable of binding to the RCE motif GGTGGG and exerts a stimulatory effect on the transcription (Fig. 4D and reference 44). Perhaps, as reported by Mastrangelo et al. (24), these Sp1 and the Sp1-like elements cooperate in the Sp1-mediated transcription and regulate WRN housekeeping expression.
Upregulation of the WRN promoter by Rb.
A DNA binding assay and a mutational analysis for the WRN promoter region by using a reporter gene assay (Fig. 3 and 4) collectively indicated that WRN expression is directed mainly by the Sp1 elements and cellular Sp1 proteins. The RCE motif in the promoter appeared to have cooperative roles with Sp1 elements, and together they showed maximum promoter activity in Saos2 cells when the Rb protein was overexpressed (Fig. 5 and Table 1). With respect to this stimulatory effect of Rb, Chen et al. (6) reported that the Rb protein upregulates Sp1-mediated gene expression not by directly binding to the Sp1 element or to the Sp1 protein but by binding to the 20-kDa Sp1-I protein, a negative regulator of the Sp1 protein, thus liberating active Sp1 transcription factors from the inactive Sp1-I–Sp1 complex. Thus, the overexpressed Rb protein in Saos2 cells might have activated the WRN promoter by activating the Sp1 proteins that bind to the Sp1 elements and the RCE motifs abundant in the WRN promoter region.
Downregulation of the WRN promoter by p53.
In contrast to the upregulation of the WRN promoter by Rb, the overexpression of wild-type p53, but not of mutant p53, downregulated the WRN promoter activity concentration dependently (Fig. 6A). Wild-type p53 downregulates several genes containing a TATA box by forming a complex with the TATA-binding protein (11). However, this may not be the case for the WRN promoter, which does not contain a TATA box. Rather, this inhibitory effect of the overexpressed p53 is explained by the finding of Borellini et al. (4) that increased levels of p53 resulted in complexes with Sp1 protein, rendering the Sp1 protein inactive for Sp1-mediated transcription. Similarly, the overexpressed p53 in Saos2 cells might have inhibited dose dependently the trans-activating activity of Sp1 by a protein-protein interaction, resulting in negative regulation of the WRN promoter.
Modulation of Sp1-mediated WRN expression by Rb and p53.
The overexpressed Rb and p53 proteins also affected the endogenous WRN mRNA levels in Saos2 cells, consistent with the results obtained for the cotransfection experiments shown in Fig. 7. What is the biological significance of these two ways of regulating WRN expression by Rb and p53? The WRN DNA helicase is a nuclear enzyme nonessential for life and development but important for the suppression of hyperrecombination and genomic instability (16). Its presence prevented normal individuals from the onset of premature aging phenotypes. Although the levels of intact WRN mRNA in the heterozygotes of parents and the relatives of patients carrying a deficient allele are low, they are apparently sufficient (47). Thus, maintaining WRN mRNA over a certain threshold level may be very important for cell homeostasis and for the concentration of WRN helicase during the cell cycle. While the biological function(s) of the WRN helicase remains to be clarified, we speculate that the basal level of WRN expression governed by Sp1-mediated regulation may be further modulated positively by Rb and negatively by p53 in association with various cellular events including the cell cycle, DNA damage, and cell senescence. In this context, Wang et al. (45) reported that p53 inhibits expression of human topoisomerase IIα, another nuclear enzyme involved in DNA metabolism, dose dependently through the inverted CCAAT element in the promoter region. Therefore, a plausible speculation is that the simultaneous downregulation of two important enzymes involved in DNA metabolism by p53 may result in an augmented illegitimate hyperrecombination and/or genomic instability, which are both associated with WS.
WRN expression in WS and natural senescence.
In WS patient cells, overall WRN expression is perturbed completely due to the impaired nuclear import of deficient protein products (26). This perturbation in expression, which would occur even in the case of transcription modulation by Rb and p53, is the primary cause of the generation of premature aging phenotypes in patients at an early stage of life. How, then, does the modulation by Rb and p53 affect WRN expression in the course of natural aging, and what is its relation to the onset of the aging phenotype at a later period of life? This is not clear for Rb. However, Kulju and Lehman (22) reported that the steady-state level of p53 protein increases in the near-senescent human diploid fibroblast cells; also, Sugrue et al. (39) found that the overexpression of wild-type p53 in human EJ cells triggers a rapid onset of G1 and G2/M growth arrest, which is irreversible and results in senescence phenotypes, suggesting a link with p53. These data imply that p53 has important roles in cell senescence, while the role of Rb in the induction of senescence cannot be ruled out (34). Both the defective expression in WS patients and the expression modulated by Rb (upregulation) and by p53 (downregulation) in Saos2 cells prompt us to hypothesize that a potential gradual suppression of WRN expression may occur with natural senescence, accounting for an increased frequency of genetic instability in the senescent cells. Further studies are needed to determine if natural aging, unlike aging of WS patient cells, is associated with a gradual reduction in WRN expression that is regulated by p53 and Rb or by other transcription factors that interact directly or indirectly with the WRN promoter.
ACKNOWLEDGMENTS
This work was supported by the Organization for Drug ADR Relief, R & D Promotion and Product Review, of the Japanese Government.
We thank Masanobu Sugimoto (AGENE Research Institute) for valuable discussion.
REFERENCES
- 1.Agoff S N, Hou J, Linzer D I, Wu B. Regulation of the human hsp70 promoter by p53. Science. 1993;259:84–87. doi: 10.1126/science.8418500. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Berk A J, Sharp P A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977;12:721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- 4.Borellini F, Glazer R I. Induction of Sp1-p53 DNA-binding heterocomplexes during granulocyte/macrophage colony-stimulating factor-dependent proliferation in human erythroleukemia cell line TF-1. J Biol Chem. 1993;268:7923–7928. [PubMed] [Google Scholar]
- 5.Chellappan S P, Hiebert S, Mudryj M, Horowitz J M, Nevins J R. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 6.Chen L I, Nishinaka T, Kwan K, Kitabayashi I, Yokoyama K, Fu Y H, Grunwald S, Chiu R. The retinoblastoma gene product RB stimulates Sp1-mediated transcription by liberating Sp1 from a negative regulator. Mol Cell Biol. 1994;14:4380–4389. doi: 10.1128/mcb.14.7.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Epstein C J, Martin G M, Schultz A L, Motulsky A G. Werner’s syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine (Baltimore) 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Faragher R G, Kill I R, Hunter J A, Pope F M, Tannock C, Shall S. The gene responsible for Werner syndrome may be a cell division “counting” gene. Proc Natl Acad Sci USA. 1993;90:12030–12034. doi: 10.1073/pnas.90.24.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farmer G, Colgan J, Nakatani Y, Manley J L, Prives C. Functional interaction between p53, the TATA-binding protein (TBP), and TBP-associated factors in vivo. Mol Cell Biol. 1996;16:4295–4304. doi: 10.1128/mcb.16.8.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felgner P L, Gadek T R, Holm M, Roman R, Chan H W, Wenz M, Northrop J P, Ringold G M, Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goto M, Imamura O, Kuromitsu J, Matsumoto T, Yamabe Y, Tokutake Y, Suzuki N, Mason B, Drayna D, Sugawara M, Sugimoto M, Furuichi Y. Analysis of helicase gene mutations in Japanese Werner’s syndrome patients. Hum Genet. 1997;99:191–193. doi: 10.1007/s004390050336. [DOI] [PubMed] [Google Scholar]
- 14.Goto M, Miller R W, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- 15.Goto M, Tanimoto K, Horiuchi Y, Sasazuki T. Family analysis of Werner’s syndrome: a survey of 42 Japanese families with a review of the literature. Clin Genet. 1981;19:8–15. doi: 10.1111/j.1399-0004.1981.tb00660.x. [DOI] [PubMed] [Google Scholar]
- 16.Guarente L. Do changes in chromosomes cause aging? Cell. 1996;86:9–12. doi: 10.1016/s0092-8674(00)80072-0. [DOI] [PubMed] [Google Scholar]
- 17.Haffner R, Oren M. Biochemical properties and biological effects of p53. Curr Opin Genet Dev. 1995;5:84–90. doi: 10.1016/s0959-437x(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 18.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J-C, Valent A, Minty A, Chalon P, Lelias J-M, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 19.Kim S J, Lee H D, Robbins P D, Busam K, Sporn M B, Roberts A B. Regulation of transforming growth factor beta 1 gene expression by the product of the retinoblastoma-susceptibility gene. Proc Natl Acad Sci USA. 1991;88:3052–3056. doi: 10.1073/pnas.88.8.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S J, Wagner S, Liu F, O’Reilly M A, Robbins P D, Green M R. Retinoblastoma gene product activates expression of the human TGF-beta 2 gene through transcription factor ATF-2. Nature. 1992;358:331–334. doi: 10.1038/358331a0. [DOI] [PubMed] [Google Scholar]
- 21.Kley N, Chung R Y, Fay S, Loeffler J P, Seizinger B R. Repression of the basal c-fos promoter by wild-type p53. Nucleic Acids Res. 1992;20:4083–4087. doi: 10.1093/nar/20.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulju K S, Lehman J M. Increased p53 protein associated with aging in human diploid fibroblasts. Exp Cell Res. 1995;217:336–345. doi: 10.1006/excr.1995.1095. [DOI] [PubMed] [Google Scholar]
- 23.Martin G M. Genetic syndromes in man with potential relevance to the pathobiology of aging. Birth Defects. 1978;14:5–39. [PubMed] [Google Scholar]
- 24.Mastrangelo I A, Courey A J, Wall J S, Jackson S P, Hough P V. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc Natl Acad Sci USA. 1991;88:5670–5674. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumoto T, Imamura O, Yamabe Y, Kuromitsu J, Tokutake Y, Shimamoto A, Suzuki N, Satoh M, Kitao S, Ichikawa K, Kataoka H, Sugawara K, Thomas W, Mason B, Tsuchihashi Z, Drayna D, Sugawara M, Sugimoto M, Furuichi Y, Goto M. Mutation and haplotype analyses of the Werner’s syndrome gene based on its genomic structure: genetic epidemiology in the Japanese population. Hum Genet. 1997;100:123–130. doi: 10.1007/s004390050477. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto T, Shimamoto A, Goto M, Furuichi Y. Impaired nuclear localization of defective DNA helicases in Werner’s syndrome. Nat Genet. 1997;16:335–336. doi: 10.1038/ng0897-335. [DOI] [PubMed] [Google Scholar]
- 27.Monnat R J., Jr Werner syndrome: molecular genetics and mechanistic hypotheses. Exp Gerontol. 1992;27:447–453. doi: 10.1016/0531-5565(92)90080-j. [DOI] [PubMed] [Google Scholar]
- 28.O’Brien C A, Manolagas S C. Isolation and characterization of the human gp130 promoter. Regulation by STATS. J Biol Chem. 1997;272:15003–15010. doi: 10.1074/jbc.272.23.15003. [DOI] [PubMed] [Google Scholar]
- 29.Ookawa K, Tsuchida S, Adachi J, Yokota J. Differentiation induced by RB expression and apoptosis induced by p53 expression in an osteosarcoma cell line. Oncogene. 1997;14:1389–1396. doi: 10.1038/sj.onc.1200976. [DOI] [PubMed] [Google Scholar]
- 30.Robbins P D, Horowitz J M, Mulligan R C. Negative regulation of human c-fos expression by the retinoblastoma gene product. Nature. 1990;346:668–671. doi: 10.1038/346668a0. [DOI] [PubMed] [Google Scholar]
- 31.Runger T M, Bauer C, Dekant B, Moller K, Sobotta P, Czerny C, Poot M, Martin G M. Hypermutable ligation of plasmid DNA ends in cells from patients with Werner syndrome. J Invest Dermatol. 1994;102:45–48. doi: 10.1111/1523-1747.ep12371730. [DOI] [PubMed] [Google Scholar]
- 32.Salk D, Bryant E, Hoehn H, Johnston P, Martin G M. Growth characteristics of Werner syndrome cells in vitro. Adv Exp Med Biol. 1985;190:305–311. doi: 10.1007/978-1-4684-7853-2_14. [DOI] [PubMed] [Google Scholar]
- 33.Schonberg S, Niermeijer M F, Bootsma D, Henderson E, German J. Werner’s syndrome: proliferation in vitro of clones of cells bearing chromosome translocations. Am J Hum Genet. 1984;36:387–397. [PMC free article] [PubMed] [Google Scholar]
- 34.Shay J W, Pereira-Smith O M, Wright W E. A role for both RB and p53 in the regulation of human cellular senescence. Exp Cell Res. 1991;196:33–39. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- 35.Shiio Y, Yamamoto T, Yamaguchi N. Negative regulation of Rb expression by the p53 gene product. Proc Natl Acad Sci USA. 1992;89:5206–5210. doi: 10.1073/pnas.89.12.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimamoto A, Kitao S, Ichikawa K, Suzuki N, Yamabe Y, Imamura O, Tokutake Y, Satoh M, Matsumoto T, Kuromitsu J, Kataoka H, Sugawara K, Sugawara M, Sugimoto M, Goto M, Furuichi Y. A unique human gene that spans over 230 kb in the human chromosome 8p11-12 and codes multiple family proteins sharing RNA-binding motifs. Proc Natl Acad Sci USA. 1996;93:10913–10917. doi: 10.1073/pnas.93.20.10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh H, Sen R, Baltimore D, Sharp P A. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986;319:154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- 38.Smoller D A, Petrov D, Hartl D L. Characterization of bacteriophage P1 library containing inserts of Drosophila DNA of 75-100 kilobase pairs. Chromosoma. 1991;100:487–494. doi: 10.1007/BF00352199. [DOI] [PubMed] [Google Scholar]
- 39.Sugrue M M, Shin D Y, Lee S W, Aaronson S A. Wild-type p53 triggers a rapid senescence program in human tumor cells lacking functional p53. Proc Natl Acad Sci USA. 1997;94:9648–9653. doi: 10.1073/pnas.94.18.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki N, Shimamoto A, Imamura O, Kuromitsu J, Kitao S, Goto M, Furuichi Y. DNA helicase activity in Werner’s syndrome gene product synthesized in a baculovirus system. Nucleic Acids Res. 1997;25:2973–2979. doi: 10.1093/nar/25.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tahara H, Tokutake Y, Maeda S, Kataoka H, Watanabe T, Satoh M, Matsumoto T, Sugawara M, Ide T, Goto M, Furuichi Y, Sugimoto M. Abnormal telomere dynamics of B-lymphoblastoid cell strains from Werner’s syndrome patients transformed by Epstein-Barr virus. Oncogene. 1997;15:1911–1920. doi: 10.1038/sj.onc.1201377. [DOI] [PubMed] [Google Scholar]
- 42.Tuteja N, Tuteja R. DNA helicases: the long unwinding road. Nat Genet. 1996;13:11–12. doi: 10.1038/ng0596-11. [DOI] [PubMed] [Google Scholar]
- 43.Uchida T, Takahashi K, Tatsuno K, Dhingra U, Eliason J F. Inhibition of hepatitis-B-virus core promoter by p53: implications for carcinogenesis in hepatocytes. Int J Cancer. 1996;67:892–897. doi: 10.1002/(SICI)1097-0215(19960917)67:6<892::AID-IJC21>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 44.Udvadia A J, Rogers K T, Higgins P D, Murata Y, Martin K H, Humphrey P A, Horowitz J M. Sp-1 binds promoter elements regulated by the RB protein and Sp-1-mediated transcription is stimulated by RB. Proc Natl Acad Sci USA. 1993;90:3265–3269. doi: 10.1073/pnas.90.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q, Zambetti G P, Suttle D P. Inhibition of DNA topoisomerase II alpha gene expression by the p53 tumor suppressor. Mol Cell Biol. 1997;17:389–397. doi: 10.1128/mcb.17.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webster N J, Resnik J L, Reichart D B, Strauss B, Haas M, Seely B L. Repression of the insulin receptor promoter by the tumor suppressor gene product p53: a possible mechanism for receptor overexpression in breast cancer. Cancer Res. 1996;56:2781–2788. [PubMed] [Google Scholar]
- 47.Yamabe Y, Sugimoto M, Satoh M, Suzuki N, Sugawara M, Goto M, Furuichi Y. Down-regulation of the defective transcripts of the Werner’s syndrome gene in the cells of patients. Biochem Biophys Res Commun. 1997;236:151–154. doi: 10.1006/bbrc.1997.6919. [DOI] [PubMed] [Google Scholar]
- 48.Yu C E, Oshima J, Fu Y H, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin G M, Mulligan J, Schellenberg G D. Positional cloning of the Werner’s syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 49.Yu C E, Oshima J, Wijsman E M, Nakura J, Miki T, Piussan C, Matthews S, Fu Y H, Mulligan J, Martin G M, Schellenberg G D. Mutations in the consensus helicase domains of the Werner syndrome gene. Werner’s Syndrome Collaborative Group. Am J Hum Genet. 1997;60:330–341. [PMC free article] [PubMed] [Google Scholar]