Abstract
Hox proteins control developmental patterns and cell differentiation in vertebrates by acting as positive or negative regulators of still unidentified downstream target genes. The homeodomain and other small accessory sequences encode the DNA-protein and protein-protein interaction functions which ultimately dictate target recognition and functional specificity in vivo. The effector domains responsible for either positive or negative interactions with the cell transcriptional machinery are unknown for most Hox proteins, largely due to a lack of physiological targets on which to carry out functional analysis. We report the identification of the transcriptional activation domains of three human Hox proteins, HOXB1, HOXB3, and HOXD9, which interact in vivo with the autoregulatory and cross-regulatory enhancers of the murine Hoxb-1 and human HOXD9 genes. Activation domains have been defined both in a homologous context, i.e., within a HOX protein binding as a monomer or as a HOX-PBX heterodimer to the specific target, and in a heterologous context, after translocation to the yeast Gal4 DNA-binding domain. Transfection analysis indicates that activation domains can be identified in different regions of the three HOX proteins depending on the context in which they interact with the DNA target. These results suggest that Hox proteins may be multifunctional transcriptional regulators, interacting with different cofactors and/or components of the transcriptional machinery depending on the structure of their target regulatory elements.
Hox genes encode homeodomain-containing transcription factors which control cell fate and developmental patterns in all metazoans, leading to the generation of morphological differences along body axes (reviewed in reference 19). In the Hox gene products (up to 39 in mammalian species), the homeodomain (HD) and flanking amino acids dictate the DNA-binding specificity, characterized by recognition, at least in vitro, of a restricted set of sites containing the core consensus sequence TNAT(G/T)(G/A) (11, 27, 28). Despite this apparently degenerate DNA recognition, Hox proteins act as positive or negative regulators of the transcriptional activity of very specific targets, in cultured cells as well as in embryos (19). The functional specificity of Hox proteins is therefore unlikely to depend on simple DNA-protein interactions and might require the concomitant activity of cofactors determining high-affinity recognition of specific sequences and/or regulating their transcriptional activity (26–28). The HD-containing products of the Drosophila extradenticle (exd) gene and of its vertebrate homologues, Pbx1, Pbx2, and Pbx3, are the first examples of such cofactors, regulating both cooperative DNA-binding and transcriptional activity of Drosophila and mammalian Hox proteins (9, 27, 28). The HD N-terminal and first alpha-helix (21, 44) and a short motif (consensus sequence, YPWM) conserved upstream of the HD in a subset of Hox proteins (7, 18, 31) have been shown to mediate at least some of the protein-protein contacts involved in functional interactions between Hox and other homeodomain-containing proteins (reviewed in reference 28).
For the mammalian system, and more generally for vertebrates, genetic analysis has so far failed to identify the downstream targets of Hox gene function in development or cell differentiation. Analysis of mutant Drosophila embryos showed that in some circumstances, the products of mammalian Hox genes can substitute for the function of the corresponding Drosophila proteins, but it provided no clues about the target genes which are regulated in such a heterologous context (22, 24, 25, 30, 48). Transgenic-mouse analysis, on the other hand, has led to the identification of only a few autoregulatory and cross-regulatory elements within the Hox clusters (14, 23, 32, 33, 43). The lack of “true” target genes and therefore of bona fide Hox-responsive sequences is the single most important factor which has so far limited the analysis of the transcriptional functions of vertebrate Hox proteins. Despite this limitation, a few studies showed the existence of domains with positive or negative transcriptional activity in some Hox proteins, as defined by their ability to regulate the activity of reporter sequences in vitro or in vivo (9, 17, 34, 38, 44, 47). In this respect, Hox proteins thus seem to share the modular structure of most eukaryotic transcription factors, featuring separate DNA-binding domain (DBD) and effector domain.
In this paper, we report the identification of functional domains in the human HOXD9, HOXB1, and HOXB3 proteins, which interact with and activate transcription from the autoregulatory and cross-regulatory elements of the human HOXD9 and murine Hoxb-1 genes (32, 43). Transcriptional activation domains have been identified by deletion analysis in all three proteins and defined by their ability to activate a specific target in a “homologous” context, i.e., within a Hox protein binding either as a monomer to an ATTA-containing sequence or as a Hox-Pbx dimer on a TGAT(T/G)NAT-containing sequence, or in a “heterologous” context, after translocation to the yeast Gal4 DBD. Activation domains have been identified in different regions of the three Hox proteins, depending on the context in which they are brought onto the DNA target. We speculate that Hox proteins may be multifunctional transcriptional regulators, which interact with different cofactors and/or components of the transcriptional machinery depending on the structure of their target regulatory elements.
MATERIALS AND METHODS
Protein expression and reporter plasmids.
All the expression constructs used are derivatives of the simian virus 40 (SV40) promoter-based expression vector pSG5. The HOXD9 expression plasmid and the pTHCR luc, pTUAS luc, pTCBS luc, and Gal-4 reporter plasmids were described previously (43). The D9Δ1–75, D9Δ1–142, and D9Δ1–222 mutants were generated by PCR with 5′ forward primers containing an ATG start codon and subsequently cloned as BamHI fragments into pSG5. D9Δ1–264 was generated by introduction of an ATG codon at a BamHI site corresponding to amino acid (aa) 265 of pSGHOXD9. pSGGal4DBD was obtained by cloning a BamHI-BglII fragment containing the DBD of yeast GAL4 (aa 1 to 147) into pSG5. The HOXD9-Gal4 chimeras were generated by cloning in frame BamHI fragments representing the N-terminal regions of HOXD9 described above. The HOXB1, B1Δ1–155, and PBX1a expressors and the pMLARE reporter plasmids were described previously (9). B1Δ1–38 and B1Δ1–90 were generated by PCR with the pSG5HOXB1 vector (15) as a template, a 5′ forward primer containing an ATG, and a 3′ reverse primer encompassing the XbaI site of pSG5. B1(1–164)-Gal4, B1(38–164)-Gal4, and B1(90–164)-Gal4 were generated by PCR from the corresponding plasmids containing HOXB1 deletions with a 3′ reverse primer terminating at aa 160 of HOXB1, and they were subsequently cloned in frame as EcoRI-SacI fragments into pSGGal4DBD.
pSGHOXB3 was described previously (15). B3Δ72–182 was generated by reinserting an EcoRI-PvuII fragment containing the N-terminal 1 to 72 aa of HOXB3 into the EcoRI-SmaI digest of pSGHOXB3. B3Δ1–182 was constructed by ligating a synthetic linker containing an ATG codon to the HOXB3 EcoRI-SmaI fragment. B3Δ273–360 was generated by removing an EclXI-BamHI fragment from the HOXB3 coding region and religating after repairing with Klenow DNA polymerase. B3Δ273–431 was constructed by removing the internal EclXI-AocI fragment and religating after repairing with Klenow DNA polymerase. B3Δ1–182;Δ273–431 was generated as for B3Δ273–431, starting from B3Δ1–182. The B3(1–182)-Gal4 mutant was constructed by cloning an EcoRI-SmaI fragment of HOXB3 in frame into pSGGal4DBD. B3(271–431)-Gal4 was generated by cloning a blunted EclXI-AocI fragment of HOXB3 in frame into pSGGal4DBD. In this mutant, translation starts at an internal Met at position 276 of HOXB3. The Gal4-B3(1–182) and Gal4-B3(273–431) chimeras were generated by using the corresponding HOXB3 fragments cloned in frame into pGal1–147 (35). The HOXB3/B1 mutant was described previously (9). B3/B1Δ72–150 was generated by ligating the EcoRI-PvuII fragment encoding aa 1 to 72 of HOXB3 to the EcoRI-StuI fragment of HOXB3/B1. B3/B1Δ1–123 was PCR generated with a 5′ forward primer containing an ATG and a 3′ reverse primer encompassing the XbaI site of pSG5. The fragment containing the entire coding sequence of B3/B1, starting from aa 123, was then cloned into pSG5 digested with EcoRI and XbaI. B3/B1Δ238–396 and B3/B1Δ238–396 were constructed in the same way as the corresponding B3Δ273–360 and B3Δ273–432, respectively. The same strategy was applied to B3/B1Δ1–123/Δ238–396, starting from B3/B1Δ1–123.
Cell culture and transfection.
HeLa and COS7 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (Gibco), 100 IU of penicillin per ml, and 100 μg of streptomycin per ml. P19 cells were maintained under the same conditions with alpha minimal essential medium. Transfections were carried out by CaHPO4 precipitation. Typically, for a 6-mm-diameter dish, 10 μg of total DNA was added to cells that had reached one-third confluency. A 48 h after transfection, the cells were harvested for luciferase and β-galactosidase activity as described previously (44). Mini nuclear extracts for Western blotting or mobility shift assays were prepared from transfected cells as described previously (12).
Protein production, Western blotting, and DNA-binding assays.
HOX and PBX proteins were produced in vitro from the corresponding pSG5-derived expression vectors by using a T7 polymerase-based transcription and a reticulocyte lysate-based translation system (Promega, Madison, Wis.). A 10-μg portion of transfected nuclear extracts was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted to nitrocellulose membranes, and probed with a monoclonal antibody against the HOXD9 homeodomain. The bound immunocomplexes were then revealed with the ECL peroxidase detection kit (Amersham). A synthetic oligonucleotide containing the 17-bp recognition sequence of the yeast Gal4 protein was end labeled and incubated for 15 min at room temperature with about 4 μg of nuclear extracts from transfected cells in a 20-μl reaction mixture containing 10 mM Tris-HCl (pH 7.4), 50 mM KCl, 1 mM EDTA, 5 mM dithiothreitol (DTT), 50 μg of bovine serum albumin (BSA), 4 μg of poly(dI-dC), and 4% Ficoll. The reaction mixtures were then loaded onto 5% polyacrylamide–0.5× Tris-borate-EDTA (TBE) native gels and electrophoresed for about 2 h. The gels were dried and exposed to a XAR Kodak film with an intensifying screen at −70°C.
To detect Hox protein binding in an electrophoretic mobility shift assay (EMSA), a previously described consensus oligonucleotide (43) was end labeled and incubated in the same binding buffer as described above with 6 μl of reticulocyte lysate and 2 μg of poly(dI-dC). The binding reactions were carried out at 4°C for 30 min, and the reaction mixtures were electrophoresed as described above. The HOX-PBX complex was detected in EMSA as described previously (9), with 4 μl of reticulocyte lysate for each protein.
RESULTS
Identification of activation domains in the N terminus of the HOXD9 protein.
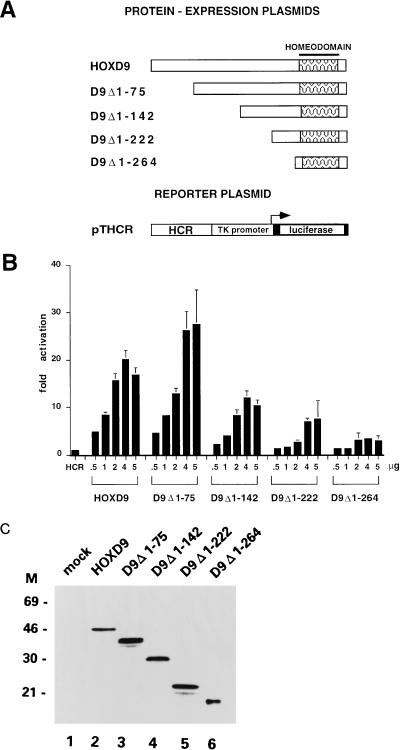
The human HOXD9 gene codes for a 337-aa protein, with 270 aa at the N terminus and 8 aa at the C terminus of the HD (43). We had previously shown in cotransfection experiments that the HOXD9 protein is able to transactivate a luciferase reporter construct (pTHCR) in which the minimal (−81) thymidine kinase (TK) promoter from herpes simplex virus is placed downstream of the Hox control region (HCR), a 90-bp, ATTA-rich autoregulatory element identified in the HOXD9 locus (43, 44). cDNAs encoding the full-length HOXD9 protein and four proteins with N-terminal deletions were cloned under the control of the SV40 promoter-enhancer and cotransfected in 0.5- to 5.0-μg amounts in HeLa cells together with pTHCR. As shown in Fig. 1B, transfection of 4.0 μg of the HOXD9 plasmid led to a 20-fold activation of the pTHCR basal activity. Deletion of the N-terminal 75 aa had no effect on the transcriptional activity of HOXD9 (D9Δ1–75 in Fig. 1A and B), while further deletions up to aa 142, 222, and 264 (D9Δ1–142, D9Δ1–222, and D9Δ1–264, respectively) progressively abolished the activity. All proteins were expressed at comparable levels, as assayed by Western blotting of nuclear extracts obtained from transfected cells with a monoclonal antibody specifically recognizing the HOXD9 HD (Fig. 1C). These experiments indicate that the functional domain necessary for transcriptional activation of the HCR element is spread over a large region of the HOXD9 protein, extending from aa 75 to 222.
FIG. 1.
(A) Schematic representation of the HOXD9 full-length protein and deletion mutants expressed by the pSGHOXD9 series of expression plasmids and of the pTHCR luciferase reporter plasmid. Patterned boxes indicate the HD. (B) Cotransfection assay in HeLa cells. Cells were transfected with 4 μg of reporter plasmid (HCR) and cotransfected with 0.5 to 5 μg of the different expression plasmids. The amount of transfected DNA was kept constant (10 μg) by addition of pSG5 plasmid. Bars represent the luciferase activity of transfected cell extracts (mean ± standard error of the mean [SEM] of at least four independent experiments, each carried out in duplicate), expressed as fold activation over the basal activity of the promoter-only reporter construct. Values were normalized by cotransfection of 0.1 μg of a pCMV–β-gal plasmid as an internal standard. (C) Immunoblot analysis of HeLa cells transfected with 5 μg of the indicated HOXD9 expression plasmids. Nuclear extracts (10 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis blotted onto nitrocellulose filters, and probed with a monoclonal antibody recognizing the HOXD9 homeodomain. M, molecular mass markers (in kilodaltons).
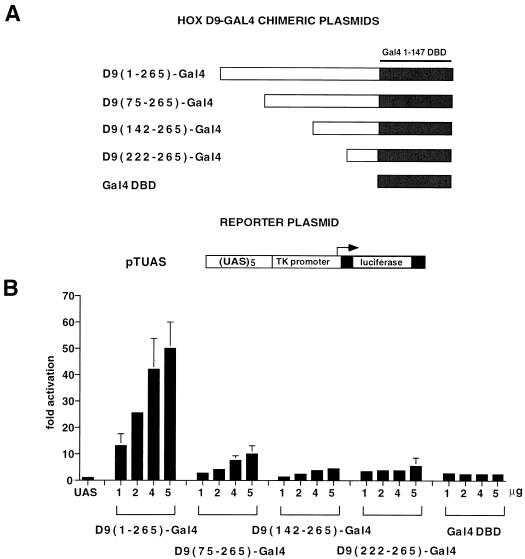
To test the activity of the HOXD9 N-terminal region in a heterologous context, we fused the regions from 1 to 265, 75 to 265, 142 to 265, and 222 to 265 at the N terminus of the DNA-binding domain (aa 1 to 147) of the yeast transcription factor Gal4 (Gal4-DBD). The fusion proteins were cotransfected in HeLa cells with a luciferase reporter gene in which the TK promoter was placed under the control of a 5-mer Gal-4-responsive element (pTUAS). As shown in Fig. 2B, the chimera containing the full-length N-terminal domain of HOXD9 [D9(1–265)-Gal4] was able to activate the pTUAS reporter 40- to 50-fold over the basal level in a dose-dependent fashion. Removal of the first 75 aa of HOXD9 [D9(75–265)-Gal4] caused a 80% reduction in the transcriptional activity of the chimera, while further deletions [D9(142–265)-Gal4 and D9(222–265)-Gal4] showed the same activity of the Gal4 DBD alone on the pTUAS reporter (less than threefold activation, see Fig. 2B). Biosynthesis of the chimeric proteins was tested by EMSA of nuclear extracts obtained from transfected HeLa cells with a 17-bp double-stranded oligonucleotide containing one copy of the Gal4 recognition sequence as the probe, which showed that the D9(1–265)-Gal4, D9(142–265)-Gal4, and D9(222–265)-Gal4 proteins are synthesized at comparable levels, whereas the D9(75–265)-Gal4 protein and the Gal4 DBD are synthesized at >10-fold-higher levels or bind to the probe with a higher affinity (results not shown). After normalization for the DNA-bound protein levels, these experiments indicate that in the context of a protein binding to the Gal4-responsive element, the N-terminal 75 aa of HOXD9 contain a potential transcriptional activator domain, whereas the region containing most of the activity in the context of the native HOXD9 protein (aa 75 to 222) is virtually inactive.
FIG. 2.
(A) Schematic representation of the Gal4 fusion proteins containing the HOXD9 N-terminal domain (positions 1 to 265) or its deletion mutants (positions 75 to 265, 142 to 265, and 222 to 265) and of the pTUAS luciferase reporter plasmid. Solid boxes represent the Gal4 1–147 DBD. (B) Transcriptional activity of HOXD9-Gal4 chimeras in HeLa cells transfected with 1 μg of reporter plasmid (UAS) and cotransfected with 1 to 5 μg of the different expression plasmids. Luciferase activity is expressed as fold activation over the basal activity of the promoter-only reporter construct (see the legend of Fig. 1 for details).
To maintain a HOX-like geometry in the HOXD9-Gal4 chimeras, the Gal4 DBD was placed in the same position of the HD, i.e., at the C terminus of the fusion protein. In the Gal4 protein, however, the DBD is at the N terminus of the protein and the activation domain is at its C terminus. To test the activity of the HOXD9 N-terminal region in a Gal4-like conformation, we constructed a series of chimeric constructs in which the regions from 1 to 298, 75 to 298, 142 to 298, and 222 to 298 were fused at the C terminus of the Gal4 DBD, which were tested by cotransfection with the pTUAS reporter. Although the Gal4-HOXD9 proteins were 50% less active in activating the reporter construct than were their HOXD9-Gal4 counterparts, the region from 1 to 75 contained most of the activity when tested at the C terminus of the DBD (results not shown).
A comparative analysis of all known group 9 human, murine, and Xenopus Hox proteins indicates a modest conservation in the N-terminal regions, with most conserved residues concentrated in the first 130 positions (Fig. 3).
FIG. 3.
Best-fit alignment of the N-terminal regions of group 9 human (all capitals), mouse (m), and Xenopus (x) Hox proteins. Numbers indicate amino acid positions within the HOXD9 protein. Amino acids at the borders of the deletions generated in the HOXD9 N terminus (Fig. 1A) are indicated in boldface type.
Identification of the activation domain of the HOXB1-PBX complex.
The human HOXB1 gene codes for a 296-aa protein with 197 aa at the N terminus and 39 aa at the C terminus of the HD (1). We had previously reported (9) that the HOXB1 protein can cooperatively activate transcription, together with PBX1, from an autoregulatory element directing spatially restricted expression of the murine Hoxb-1 gene (b1-ARE) in the developing hindbrain (32). Selective recognition of the b1-ARE and transcriptional activation are mediated by HOXB1, while DNA-binding and protein-protein interaction functions of both HOXB1 and PBX1 are required for the assembly of a transcriptionally active complex (9). To localize the transcriptional activation domain of the complex, constructs coding for the full-length HOXB1 protein and three proteins with N-terminal deletions were cotransfected with the PBX1a expressor in the murine embryonal carcinoma cell line P19, together with a luciferase reporter construct (pAdMLARE) in which the 148-bp b1-ARE controls the adenovirus major late promoter. Deletion of the first 38 aa of HOXB1 (B1Δ1–38) had no effect on the activity of the HOXB1-PBX1a complex, which was able to transactivate the pAdMLARE reporter 30- to 50-fold over the basal activity (Fig. 4B, column 7). Deletion of the first 90 aa (B1Δ1–90) virtually abolished the activity of the complex (column 8), which showed a residual, eightfold activation level indistinguishable from that of the B1Δ1–90 protein in the absence of PBX1 (column 4). An N-terminal deletion up to aa 155, which does not affect the FDWM domain necessary for cooperative interaction with PBX1 (9), further reduced the activity of the complex, down to three times the reporter basal activity (column 9). All HOXB1 mutants bound cooperatively to the R3 core element (TGATGGATGAG) of the b1-ARE together with PBX1, as checked by EMSA with in vitro-translated proteins (reference 9 and data not shown). These data indicate that the transcriptional activation domain of the HOXB1 protein in the context of the HOXB1-PBX1a complex resides between aa 38 and 90, a Ser-Pro-rich (20%) region only slightly conserved in the N termini of other vertebrate group 1 Hox genes (Fig. 5). The N-terminal region from aa 38 to 90 also contains most of the activating functions when tested in the presence of Prep1, a recently identified PBX1 cofactor forming a HOXB1-PBX1-Prep1 ternary complex on the b1-ARE (5).
FIG. 4.
(A) Schematic representation of the HOXB1 full-length protein and deletion mutants, of the PBX-1a protein, and of the pAdMLARE reporter plasmid. The solid boxes represent the HOXB1 HD and PBX HD, and the hatched boxes represent the conserved PBC-A and PBC-B domains of PBX1. (B) Transcriptional activity of the HOXB1-PBX1a complexes in P19 cells transfected with 4 μg of reporter plasmid (ARE) and cotransfected with 2 μg of the full-length HOXB1 (columns 2 and 6) or the deletion mutants B1Δ1–38 (columns 3 and 7), B1Δ1–90 (columns 4 and 8), or B1Δ1–155 (columns 5 and 9), and 4 μg of PBX1a (columns 1 and 6 to 9). Luciferase activity is expressed as fold activation over the basal activity of the promoter-only reporter construct (see the legend of Fig. 1 for details).
FIG. 5.
Best-fit alignment of the N-terminal regions of group 1 human (all capitals), mouse (m), rat (r), chicken (c) zebra fish (z), and Xenopus (x) Hox proteins. Numbers indicate amino acid positions within the HOXB1 protein. Amino acids at the borders of the deletions generated in the HOXB1 N terminus (Fig. 4A) are indicated in boldface type.
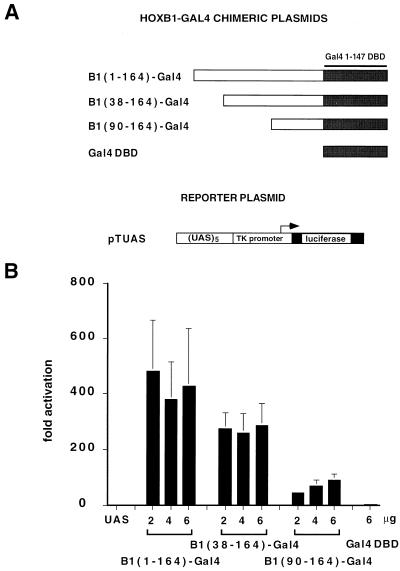
The transcriptional activity of the HOXB1 N-terminal region was also tested as an N-terminal fusion to the Gal4-DBD, by cotransfection in COS7 cells together with the pTUAS reporter. As shown in Fig. 6B, the chimera containing the full-length N-terminal domain of HOXB1 [B1(1–164)-Gal4] was able to activate the pTUAS reporter 300- to 600-fold over the basal level. Removal of the first 38 aa [B1(38–164)-Gal4] or 90 aa [B1(90–164)-Gal4] caused a 40 and 80% reduction, respectively, in transcriptional activity (Fig. 6A). Biosynthesis of the HOXB1-Gal4 chimeric protein in COS7 cells was tested by EMSA as described for the HOXD9-Gal4 chimeras. The full-length N-terminal chimera and the B1(38–164)-Gal4 protein were synthesized at a level comparable to that of the Gal4 DBD, while the B1(90–164)-Gal4 protein accumulated at significantly lower levels in transfected cell nuclei (results not shown). These data indicate that the N terminal of the HOXB1 protein also contains a strong activator domain in the context of a Gal4 DNA-binding protein. After normalization for the DNA-bound protein levels, however, the activator domain appears to be spread over the entire N-terminal region, extending also to the region from aa 90 to 164, which is virtually inactive in the context of the HOXB1-PBX1 complex (Fig. 4).
FIG. 6.
(A) Schematic representation of the Gal4 fusion proteins containing the HOXB1 N-terminal domain (positions 1 to 164) or its deletion mutants (positions 38 to 164 and 90 to 164), and of the pTUAS luciferase reporter plasmid. Solid boxes indicate the Gal4 1 147 DBD. (B) Transcriptional activity of the HOXB1-Gal4 chimeras in COS7 cells transfected with 2 μg of reporter plasmid (UAS) and cotransfected with 2 to 6 μg of B1(1–164)-Gal4, B1(38–164)-Gal4, B1(90–164)-Gal4, and Gal4-DBD. Luciferase activity is expressed as fold activation over the basal activity of the promoter-only reporter construct (see the legend of Fig. 1 for details).
Identification of two activation domains in the N terminus and C terminus of the HOXB3 protein.
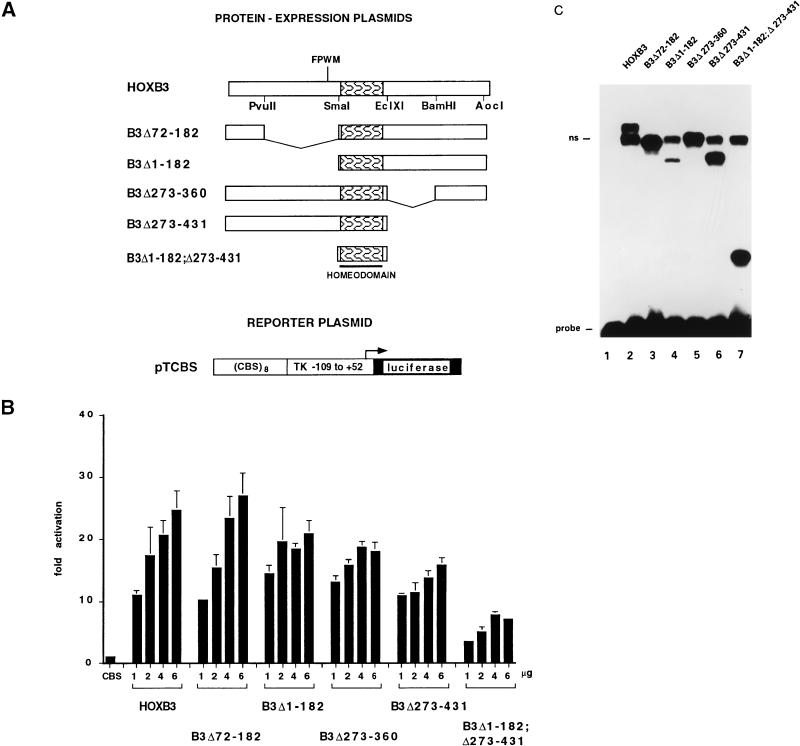
The human HOXB3 gene codes for a 431-aa protein with 187 aa at the N terminus and 184 aa at the C terminus of the HD (1), which is able to transactivate a reporter gene driven by a promoter containing one or more ATTA core sequences (15). Cotransfection of 1 to 6 μg of an expression construct for the full-length HOXB3 protein in COS7 cells led to a 20- to 30-fold, dose-dependent trans-activation of a luciferase reporter construct in which the −109 TK promoter was placed under the control of an ATTA-rich Hox consensus binding site (pTCBS) (44) (Fig. 7A). Partial or total deletion of the N-terminal (B3Δ72–182, B3Δ1–182) or C-terminal (B3Δ273–360, B3Δ273–431) region of HOXB3 had little or no effect on the activity of the protein, while deletion of both regions (B3Δ1–182;Δ273–431) reduced the activity by almost 75% (Fig. 7B). All HOXB3 proteins were synthesized at comparable levels and were able to bind in vitro to the CBS sequence, as shown by EMSA of in vitro-translated proteins (Fig. 7C). These results indicate that both the N terminus and the C terminus of HOXB3 can promote transcriptional activation of an ATTA-containing target element.
FIG. 7.
(A) Schematic representation of the HOXB3 full-length protein and deletion mutants and of the pTCBS luciferase reporter plasmid. Patterned boxes indicate the HOXB3 HD. (B) Transcriptional activity of the HOXB3 mutants in COS7 cells transfected with 2 μg of the pTCBS reporter plasmid (CBS) and cotransfected with 1 to 6 μg of the indicated HOXB3 mutant. Luciferase activity is expressed as fold activation over the basal activity of the promoter-only reporter construct (see the legend of Fig. 1 for details). (C) EMSA analysis of the binding of in vitro-synthesized HOXB3 full-length protein and deletion mutants (6 μl of reticulocyte lysate [lanes 2 to 7]) to a labeled double-stranded oligonucleotide containing a HOX consensus binding site. Lane 1, free probe. ns, nonspecific binding.
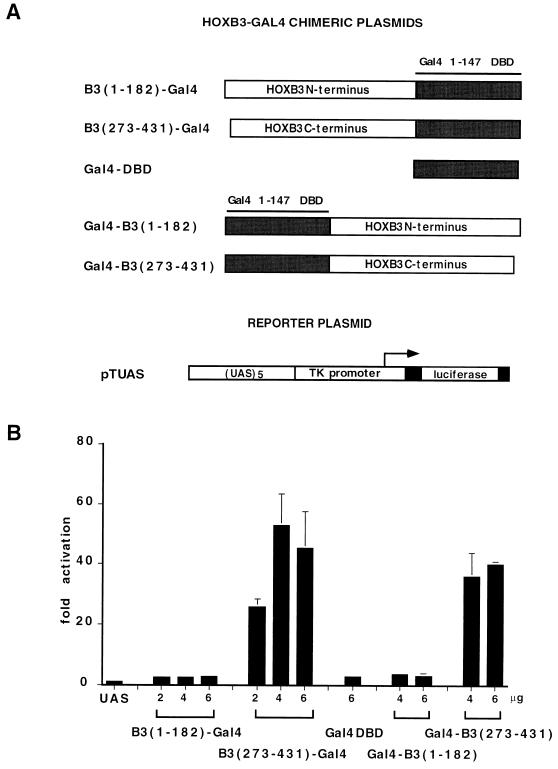
The HOXB3 N terminus and C terminus were also tested as fusions to the Gal4 DBD by cotransfection in COS7 cells together with the pTUAS reporter. Each domain was tested both in HOX-like (i.e., N-terminal to the DBD) and in Gal4-like (i.e., C-terminal) configurations (Fig. 8A). As shown in Fig. 8B, the HOXB3 N terminus was unable to activate transcription of the Gal4-responsive reporter either in the N-terminal [B3(1–182)-Gal4] or in the C-terminal [Gal4-B3(1–182)] configuration, whereas the C terminus led to a 20- to 60-fold activation of the reporter in both orientations. All HOXB3-Gal4 chimeras were able to bind DNA, and they accumulated in COS7 cell nuclei at comparable levels (results not shown). These results indicate that the HOXB3 C terminus contains an activation domain that can be exported on a heterologous DNA-binding protein whereas the N terminus is active only in a HOX protein context.
FIG. 8.
(A) Schematic representation of the fusion proteins between the HOXB3 N terminus or C terminus and the Gal4 1–147 DBD and of the pTUAS reporter plasmid. (B) Transcriptional activity of the HOXB3-Gal4 chimeras in COS7 cells transfected with 2 μg of reporter plasmid (UAS) and cotransfected with 2 to 6 μg of B3(1–182)-Gal4 and B3(273–431)-Gal4, 4 to 6 μg of Gal4-B3(1–182) and Gal4-B3(273–431), and 6 μg of Gal4-DBD expression plasmids. Luciferase activity is expressed as fold activation over the basal activity of the promoter-only reporter construct (see the legend to Fig. 1 for details).
The HOXB3 C terminus is the only activation domain in the context of a heterodimeric complex with PBX1.
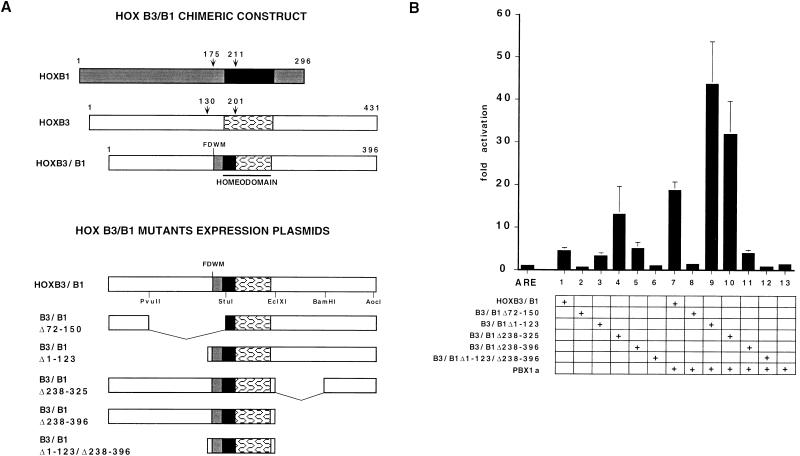
We had previously shown that HOXB3 can cooperatively bind and activate the b1-ARE element together with PBX1 if the N terminus of the HD is replaced with that of HOXB1 (9). To test the activity of the HOXB3 protein in the context of a HOX-PBX heterodimer, expression constructs encoding a full-length HOXB3-HOXB1 chimera (HOXB3/B1, in which the HOXB1 portion encompasses the FDWM motif and the HD N terminus [9]) and five mutants containing a partial N-terminal deletion including the FDWM motif (B3/B1Δ72–150), a complete N-terminal deletion (B3/B1Δ1–123), a partial or a complete C-terminal deletion (B3/B1Δ238–325 and B3/B1Δ238–396), and a combined N-terminal and C-terminal deletion (B3/B1Δ1–123/Δ238–396) were cotransfected in P19 cells together with the PBX1a expression plasmid and the pAdMLARE reporter (Fig. 9A). The full-length HOXB3/B1 chimera induced a PBX-dependent, 20-fold transactivation of the pAdMLARE reporter (Fig. 9B, column 7), as previously reported (9). Deletion of the HOXB3 N-terminal domain from positions 1 to 23 significantly increased the activity of the chimera, resulting in transactivation levels that were >40-fold higher than the basal reporter activity (column 9), while removal of the FDWM HOX-PBX interaction domain (Δ72–150) completely abolished its activity (column 8). Internal deletion of the C-terminal domain from positions 238 to 325 also increased the activity of the chimera. Interestingly, this mutant was able to activate the reporter at significant (>10-fold) levels even in the absence of PBX (column 4). Deletion of the entire C terminus (positions 238 to 396) or of both the N terminus and the C terminus (positions 1 to 123 and positions 238 to 396), completely abolished the activity of the complex (columns 11 and 12). All mutants were translated in vitro and tested for their ability to bind a double-stranded oligonucleotide containing one copy of the HOX-PBX-binding site from the b1-ARE R3 element by EMSA. As shown in Fig. 9C, the control HOXB1, the full-length HOXB3/B1 chimera, and all the mutants were able to bind cooperatively with PBX1 to the R3 element, with the single exception of the Δ72–150 deletion, involving the FDWM motif. These data indicate that in the context of a HOX-PBX heterodimeric complex, the transcriptional activation domain of HOXB3 is contained in the C-terminal 71 residues. The C terminus is more highly conserved than the N terminus in the murine and human group 3 proteins (HOXA3, HOXB3, and HOXD3), with >40% identity throughout the last 71 residues (Fig. 10).
FIG. 9.
(A) Schematic representation of the HOXB1 and HOXB3 proteins, the HOXB3/B1 chimeric protein, and the HOXB3/B1 deletion mutants. Numbers indicate amino acid positions. Shaded and open boxes indicate N- and C-terminal regions from the HOXB1 and HOXB3 proteins, respectively, in the HOXB3/B1 chimeras. Solid and patterned boxes indicate regions from the HOXB1 and HOXB3 HD, respectively. pAdMLARE is represented in Fig. 4A. (B) Transcriptional activity of HOXB3/B1 mutants in P19 cells transfected with 4 μg of reporter plasmid (ARE) and cotransfected with 2 μg of HOXB3/B1 (columns 1 and 7), B3/B1Δ72–150 (columns 2 and 8), B3/B1Δ1–123 (columns 3 and 9), B3/B1Δ238–325 (columns 4 and 10), B3/B1Δ238–396 (columns 5 and 11), B3/B1Δ1–123/Δ238–396 (columns 6 and 12), and 4 μg of PBX1a (columns 7 to 13). Luciferase activity is expressed as fold activation over the basal activity of the promoter-only reporter construct (see the legend of Fig. 1 for details). (C) EMSA analysis of the binding of in vitro-synthesized (4 μl of reticulocyte lysate for each protein) PBX (lane 1), HOXB3-PBX (lane 2) HOXB1-PBX (lane 3), HOXB3/B1-PBX (lane 4), B3/B1Δ72–150-PBX (lane 5), B3/B1Δ1–123-PBX (lane 6), B3/B1Δ238–325-PBX (lane 7), and B3/B1Δ238–396-PBX (lane 8) complexes to a labeled double-stranded oligonucleotide containing the b1-ARE R3 repeat.
FIG. 10.
Best-fit alignment of the N-terminal (A) and C-terminal (B) regions of group 3 human (all capitals) and mouse (m) Hox proteins. Numbers indicate amino acid positions within the HOXB3 protein. Amino acids at the borders of the deletions generated in the HOXB3 N-terminus (Fig. 7A) are indicated in boldface type.
DISCUSSION
Hox proteins are presumed to function as transcriptional regulators of the early steps of vertebrate embryonic development. Although the DNA-binding properties of this family of proteins are characteristically overlapping and nonselective in vitro, the homeodomains are known to mediate functional specificity in vivo (6, 13, 20, 29, 46). Specific target recognition by Hox, or closely related HD proteins such as the Drosophila Ftz, may in fact require the activity of cofactors, regulating both high-affinity DNA-binding and transcriptional activity (9, 16, 26–28, 41). The complexity of the system is further increased by the facts that at least some of the functional specificity of Hox proteins is mediated by protein-protein rather than DNA-protein interactions (2, 8, 10, 36, 37, 39, 44) and that some Hox proteins may work as both activators and repressors of transcription, depending on the context in which their function is tested (36, 38, 44).
While specific functional properties of Hox proteins, such as DNA binding, nuclear localization, and target recognition, have all been assigned to the HD, little is known about the function of the N- and C-terminal regions. Conservation of these regions among vertebrate Hox genes and between these genes and the Drosophila orthologs is minimal and essentially restricted to the YPWM motif and a few N-terminal amino acids. Nevertheless, the regions outside the HDs of two vertebrate Hox proteins, the mouse Hoxa-5 and the chicken Hoxb-1, turned out to be absolutely essential in rather stringent functional tests, such as induction of homeotic transformations (47) or rescue of homeotic mutations (22) in Drosophila embryos. Thus, paradoxically, protein regions that are poorly conserved or not conserved at all during evolution are apparently capable of exerting specific functions across species in vivo, presumably at the level of transcription. Very few studies have addressed the biochemical properties of these “effector” domains and the nature of their interaction with the transcriptional machinery, mainly due to a lack of well-defined target genes and regulatory elements. A significant exception is represented by a few “autoregulatory” enhancers identified by genetic analysis upstream of some Hox promoters, which allowed functional analysis of the transcriptional properties of at least some Hox proteins on bona fide natural target elements (4, 9, 34, 43, 47). In this study, we used two of these elements, the murine Hoxb-1 and the human HOXD9 autoregulatory enhancers, to carry out a functional dissection of the human HOXD9, HOXB1, and HOXB3 proteins.
Hox proteins contain potentially alternative activation domains.
A conventional deletion analysis on the 270-aa N terminus of HOXD9 showed that the first 75 residues contain a potential transcriptional activator when tested in the context of a Gal4 chimeric protein. In contrast, this region is dispensable when the activity of the protein is tested on the HCR, a context in which most of the activating function appears to be located within residues 76 to 264. The regions identified by the two alternative assays share no obvious characteristics with canonical eukaryotic activator domains and are only loosely conserved among different vertebrate species (Fig. 3). In a previous study, we showed that the activation domain of another posteriorly expressed Hox protein, HOXD8, can be localized to a similar sub-N-terminal region (44). HOXD8 and HOXD9 bind the multiple ATTA-containing sites within the HCR as monomers in a noncooperative fashion (42, 43), while Gal4-DBD chimeras bind the Gal4-responsive element (UAS) as a homodimer, a context which could force the HOXD9 N-terminal region to assume a different structural conformation and unmask a potential activating function in the N-terminal 75 residues. For the HOXB1-PBX heterodimer, the analysis carried out on the natural ARE identified a transcriptional activation domain in a Ser-Pro-rich, 52-residue sub-N-terminal region. This region also contained most of the HOXB1 transcriptional activity when tested as a Gal4-DBD chimera, a possible indication that the 52-residue region assumes a similar conformation or activates transcription by a similar mechanism, either in the context of a homodimer or in that of a HOX-PBX heterodimer. The activity of HOXB3 was tested in three different contexts, i.e., upon binding DNA as a monomer to an ATTA-containing element, as a HOX-PBX heterodimer to a bipartite HOX-PBX core element, and as a Gal4-DBD chimeric homodimer to the Gal4-responsive element. Although in the context of a monomer the transcriptional activity was spread over the entire protein sequence, only the C terminus contained a potent activator domain in the context of a Gal4 homodimer or of a PBX heterodimer. The 71-residue C terminus is relatively highly conserved in the mammalian group 3 Hox proteins (Fig. 10).
Our data indicate that the identification of transcriptionally active regions in Hox proteins is highly dependent on the context in which the activity of the protein is analyzed and possibly depends on the conformation that the different regions of the proteins assume when they are brought onto the DNA targets. It is therefore crucial that functional analysis of these proteins be carried out in the appropriate context, that is, in a native conformation on a natural, HD-binding target, and upon interaction with a natural DNA-binding partner. Previous attempts to identify active domains in the mammalian HOXD4 or Hoxa-7 proteins either gave inconsistent results or identified potential activators and repressors in somewhat unexpected regions of those proteins, such as the HD, when these regions were brought onto DNA via heterologous DBD (34, 38). Interestingly, a Hox protein binding DNA through the HD, for instance HOXB3, has the potential to activate a reporter gene through the activity of alternative regions (N terminus plus C terminus or C terminus only), depending on whether it binds DNA as a monomer or as a Hox-Pbx heterodimer. This would suggest that Hox proteins may be multifunctional transcriptional regulators, interacting with different cofactors and/or components of the transcriptional machinery depending on the context in which they bind DNA and therefore on the nature of the target elements on which they exert their regulatory function.
How do Hox proteins regulate transcription?
The discrepancy between the results obtained by using Hox-binding and Gal4-binding elements as targets may be an indication that Hox proteins exert their function in a way which is intrinsically different from that of classical enhancer-binding transcription factors, such as Gal-4, VP16, or (p65)NF-κB. These factors contain acidic and/or serine/threonine-rich activator domains and may activate transcription by establishing direct contacts with general components of the transcriptional machinery (40, 45). It is conceivable that protein regions acting as activators when tested as chimeras with DBD of this type of factor are only those fitting particular requirements, such as net charge or presence of specific side chains, or those able to assume a restricted set of structural conformations. The effector domains of Hox proteins, on the other hand, might play a different role in gene regulation, such as providing a “positionally” restricted function in the context of regulatory elements, like the Hoxb-1 enhancer used in this study, on which this information is integrated by the interaction with tissue-specific, inducible, or structural factors. These complexes might in turn recruit coactivators or adapter molecules to signal the general transcriptional machinery, as in the case of the homeodomain-containing Oct2 protein (3). In this framework, the function of Hox-containing complexes could be to “open” genes, or sets of genes, to active transcription rather than to directly recruit general transcription factors. The structural constraints for “active” effector domains of transcription factors binding DNA in this type of context may be very different from those required by a classical, Gal4-like type of enhancer and may render the use of specific target sequences, and possibly appropriate cell backgrounds, mandatory in a functional assay.
ACKNOWLEDGMENTS
This work was supported by grants from the Telethon Foundation and the Italian Association for Cancer Research.
REFERENCES
- 1.Acampora D, D’Esposito M, Faiella A, Pannese M, Migliaccio E, Morelli F, Stornaiuolo A, Nigro V, Simeone A, Boncinelli E. The human HOX gene family. Nucleic Acids Res. 1989;17:10385–10402. doi: 10.1093/nar/17.24.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ananthan J, Baler R, Morissey D, Zuo J, Lan Y, Weir M, Voellmy R. Synergistic activation of transcription is mediated by the N-terminal domain of Drosophila fushi tarazu homeoprotein and can occur without DNA binding by the protein. Mol Cell Biol. 1993;13:1599–1609. doi: 10.1128/mcb.13.3.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Annweiler A, Muller-Immergluck M, Wirth T. Oct2 transactivation from a remote enhancer position requires a B-cell-restricted activity. Mol Cell Biol. 1992;12:3107–3116. doi: 10.1128/mcb.12.7.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arcioni L, Simeone A, Guazzi S, Zappavigna V, Boncinelli E, Mavilio F. The upstream region of the human homeobox gene HOX3D is a target for regulation by retinoic acid and HOX homeoproteins. EMBO J. 1992;11:265–277. doi: 10.1002/j.1460-2075.1992.tb05049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthelsen J, Zappavigna V, Ferretti E, Mavilio F, Blasi F. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 1998;17:1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan S K, Mann R S. The segment identity functions of Ultrabithorax are contained within its homeo domain and carboxy-terminal sequences. Genes Dev. 1993;7:796–811. doi: 10.1101/gad.7.5.796. [DOI] [PubMed] [Google Scholar]
- 7.Chang C-P, Shen W-F, Rozenfeld S, Lawrence H J, Largman C, Cleary M. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 8.Copeland J W, Nasiadka A, Dietrich B H, Krause H M. Patterning of the Drosophila embryo by a homeodomain-deleted Ftz polypeptide. Nature. 1996;379:162–165. doi: 10.1038/379162a0. [DOI] [PubMed] [Google Scholar]
- 9.Di Rocco G, Mavilio F, Zappavigna V. Functional dissection of a transcriptionally active, target-specific Hox-Pbx complex. EMBO J. 1997;16:3644–3654. doi: 10.1093/emboj/16.12.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzpatrick V D, Percival-Smith A, Ingles C J, Krause H M. Homeodomain-independent activity of the fushi tarazu polypeptide in Drosophila embryos. Nature. 1992;356:610–612. doi: 10.1038/356610a0. [DOI] [PubMed] [Google Scholar]
- 11.Gehring W J, Qian Y Q, Billeter M, Furukubo-Tokunaga K, Schier A F, Resendez-Perez D, Affolter M, Otting G, Wuthrich K. Homeodomain-DNA recognition. Cell. 1994;78:211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 12.Gerster T, Balmaceda C G, Roeder R G. The cell type-specific octamer transcription factor OTF-2 has two domains required for the activation of transcription. EMBO J. 1990;9:1635–1643. doi: 10.1002/j.1460-2075.1990.tb08283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson G, Schier A, LeMotte P, Gehring W J. The specificities of Sex combs reduced and Antennapedia are defined by a distinct portion of each protein that includes the homeodomain. Cell. 1990;62:1087–1103. doi: 10.1016/0092-8674(90)90386-s. [DOI] [PubMed] [Google Scholar]
- 14.Gould A, Morrison A, Sproat G, White R A, Krumlauf R. Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 1997;11:900–913. doi: 10.1101/gad.11.7.900. [DOI] [PubMed] [Google Scholar]
- 15.Guazzi S, Lonigro R, Pintonello L, Boncinelli E, Di Lauro R, Mavilio F. The thyroid transcription factor-1 gene is a candidate target for regulation by Hox proteins. EMBO J. 1994;13:3339–3347. doi: 10.1002/j.1460-2075.1994.tb06636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guichet A, Copeland J W, Erdelyi M, Hlousek D, Zavorszky P, Ho J, Brown S, Percival-Smith A, Krause H M, Ephrussi A. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature. 1997;385:548–552. doi: 10.1038/385548a0. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi S, Scott M. What determines the specificity of action of Drosophila homeodomain proteins? Cell. 1990;63:883–894. doi: 10.1016/0092-8674(90)90492-w. [DOI] [PubMed] [Google Scholar]
- 18.Knoepfler P, Kamps M. The pentapeptide motif of Hox proteins is required for cooperative DNA binding with Pbx1, physically contacts Pbx1, and enhances DNA binding by Pbx1. Mol Cell Biol. 1995;15:5811–5819. doi: 10.1128/mcb.15.10.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 20.Kuziora M A, McGinnis W. A homeo domain substitution changes the regulatory specificity of the Deformed protein in Drosophila embryos. Cell. 1989;59:563–571. doi: 10.1016/0092-8674(89)90039-1. [DOI] [PubMed] [Google Scholar]
- 21.Lu Q, Kamps M P. Heterodimerization of Hox proteins with Pbx1 and oncoprotein E2a-Pbx1 generates unique DNA-binding specificities at nucleotides predicted to contact the N-terminal arm of the Hox homeodomain—demonstration of Hox-dependent targeting of E2a-Pbx1 in vivo. Oncogene. 1997;14:75–83. doi: 10.1038/sj.onc.1200799. [DOI] [PubMed] [Google Scholar]
- 22.Lutz B, Lu H C, Eichele G, Miller D, Kaufman T C. Rescue of Drosophila labial null mutant by the chicken ortholog Hoxb-1 demonstrates that the function of Hox genes is phylogenetically conserved. Genes Dev. 1996;10:176–184. doi: 10.1101/gad.10.2.176. [DOI] [PubMed] [Google Scholar]
- 23.Maconochie M K, Nonchev S, Studer M, Chan S K, Popperl H, Sham M H, Mann R S, Krumlauf R. Cross-regulation in the mouse HoxB complex: the expression of Hoxb2 in rhombomere 4 is regulated by Hoxb1. Genes Dev. 1997;11:1885–1895. doi: 10.1101/gad.11.14.1885. [DOI] [PubMed] [Google Scholar]
- 24.Malicki J, Bogarad L D, Martin M M, Ruddle F H, McGinnis W. Functional analysis of the mouse homeobox gene HoxB9 in Drosophila development. Mech Dev. 1993;42:139–150. doi: 10.1016/0925-4773(93)90003-g. [DOI] [PubMed] [Google Scholar]
- 25.Malicki J, Schughart K, McGinnis W. Mouse Hox 2.2 specifies thoracic segmental identity in Drosophila embryos and larvae. Cell. 1990;63:961–967. doi: 10.1016/0092-8674(90)90499-5. [DOI] [PubMed] [Google Scholar]
- 26.Manak J R, Scott M P. Able assistants for homeodomain proteins. Curr Biol. 1993;3:318–320. doi: 10.1016/0960-9822(93)90191-p. [DOI] [PubMed] [Google Scholar]
- 27.Mann R. The specificity of homeotic gene function. Bioessays. 1996;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- 28.Mann R S, Chan S-K. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 29.Mann R S, Hogness D S. Functional dissection of Ultrabithorax proteins in D. melanogaster. Cell. 1990;60:597–610. doi: 10.1016/0092-8674(90)90663-y. [DOI] [PubMed] [Google Scholar]
- 30.McGinnis N, Kuziora M A, McGinnis W. Human Hox 4.2 and Drosophila Deformed encode similar regulatory specificities in Drosophila embryos and larvae. Cell. 1990;63:969–976. doi: 10.1016/0092-8674(90)90500-e. [DOI] [PubMed] [Google Scholar]
- 31.Phelan M L, Rambaldi I, Featherstone M S. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol Cell Biol. 1995;15:3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pöpperl H, Bienz M, Studer M, Chang S-K, Aparicio S, Brenner S, Mann R, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/Pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- 33.Pöpperl H, Featherstone M S. An autoregulatory element of the murine Hox-4.2 gene. EMBO J. 1992;11:3673–3680. doi: 10.1002/j.1460-2075.1992.tb05452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rambaldi I, Kovacs E N, Featherstone M S. A proline-rich transcriptional activation domain in murine HOXD-4 (HOX-4.2) Nucleic Acids Res. 1994;22:376–382. doi: 10.1093/nar/22.3.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 36.Saffman E E, Krasnow M A. A differential response element for the homeotics at the Antennapedia P1 promoter of Drosophila. Proc Natl Acad Sci USA. 1994;91:7420–7424. doi: 10.1073/pnas.91.16.7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schier A F, Gehring W J. Functional specificity of the homeodomain protein fushi tarazu: the role of DNA-binding specificity in vivo. Proc Natl Acad Sci USA. 1993;90:1450–1454. doi: 10.1073/pnas.90.4.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnabel C A, Abate-Shen C. Repression of HoxA7 is mediated by the homeodomain and the modulatory action of its N-terminal-arm residues. Mol Cell Biol. 1996;16:2678–2688. doi: 10.1128/mcb.16.6.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sreenath T L, Pollock R A, Bieberich C J. Functional specificity of Hoxa-4 in vertebral patterning lies outside of the homeodomain. Proc Natl Acad Sci USA. 1996;93:9636–9640. doi: 10.1073/pnas.93.18.9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Triezenberg S J. Structure and function of transcriptional activation domains. Curr Opin Genet Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 41.Yu Y, Li W, Su K, Yussa M, Han W, Perrimon N, Pick L. The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein Ftz. Nature. 1997;385:552–555. doi: 10.1038/385552a0. [DOI] [PubMed] [Google Scholar]
- 42.Zappavigna V, Falciola L, Helmer Citterich M, Mavilio F, Bianchi E M. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 1996;15:4981–4991. [PMC free article] [PubMed] [Google Scholar]
- 43.Zappavigna V, Renucci A, Izpisua-Belmonte J C, Urier G, Peschle C, Duboule D. HOX4 genes encode transcription factors with potential auto- and cross-regulatory capacities. EMBO J. 1991;10:4177–4187. doi: 10.1002/j.1460-2075.1991.tb04996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zappavigna V, Sartori D, Mavilio F. Specificity of HOX protein function depends on DNA-protein and protein-protein interactions, both mediated by the homeo domain. Genes Dev. 1994;8:732–744. doi: 10.1101/gad.8.6.732. [DOI] [PubMed] [Google Scholar]
- 45.Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 46.Zeng W, Andrew D J, Mathies L D, Horner M A, Scott M P. Ectopic expression and function of the Antp and Scr homeotic genes: the N-terminus of the homeodomain is critical to functional specificity. Development. 1993;118:339–352. doi: 10.1242/dev.118.2.339. [DOI] [PubMed] [Google Scholar]
- 47.Zhao J J, Lazzarini R A, Pick L. Functional dissection of the mouse Hox-a5 gene. EMBO J. 1996;15:1313–1322. [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao J J, Lazzarini R A, Pick L. The mouse Hox-1.3 gene is functionally equivalent to the Drosophila sex combs reduced gene. Genes Dev. 1993;7:343–354. doi: 10.1101/gad.7.3.343. [DOI] [PubMed] [Google Scholar]