Abstract
We had previously identified a macrophage surface protein whose expression is highly induced, transient, and specific, as it is restricted to actively fusing macrophages in vitro and in vivo. This protein is recognized by monoclonal antibodies that block macrophage fusion. We have now purified this protein and cloned its corresponding cDNA. This protein belongs to the superfamily of immunoglobulins and is similar to immune antigen receptors such as the T-cell receptor, B-cell receptor, and viral receptors such as CD4. We have therefore named this protein macrophage fusion receptor (MFR). We show that the extracellular domain of MFR prevents fusion of macrophages in vitro and therefore propose that MFR belongs to the fusion machinery of macrophages. MFR is identical to SHPS-1 and BIT and is a homologue of P84, SIRPα, and MyD-1, all of which have been recently cloned and implicated in cell signaling and cell-cell interaction events.
Membrane fusion is a ubiquitous event that occurs in a wide range of cell biological processes. While intracellular membrane fusion mediating interorganelle traffic is well studied, much less is known about cell-cell fusion mediating sperm cell-oocyte, myoblast-myoblast, and macrophage-macrophage fusion. These fusion events are required for fertilization, muscle, and osteoclast development, respectively. In the case of mononuclear phagocytes, fusion is associated not only with the differentiation of osteoclasts, cells which play a key role in the pathogenesis of osteoporosis, but also with the formation of giant cells that are present in chronic inflammatory reactions and in tumors. Despite the biological and pathophysiological importance of intercellular fusion events, the actual molecular mechanisms of cell-cell fusion are still unclear.
Our initial understanding of membrane fusion was gained through the study of viral fusion reactions (17). Studies of viruses, especially influenza virus and human immunodeficiency virus (HIV), have provided strong evidence that viral fusion is mediated by both viral and cellular membrane proteins whose function is to help overcome the repulsive forces that inhibit fusion and/or promote the hydrophobic forces that favor fusion. The influenza virus hemagglutinin protein became the model for all so-called fusion proteins (46). It is possible, if not likely, that common mechanisms exist among all, including cell-cell, fusion events. Indeed, two structurally and functionally similar cell membrane fusion proteins have been identified: the proteins fertilin (PH-30) (3, 34) and meltrin (48) in sperm cells and myoblasts, respectively. However, the extent to which these proteins are involved in the actual fusion event remains unclear.
Increasing evidence suggests that the molecular machinery mediating virus-cell and cell-cell fusion is more complicated than anticipated and involves numerous factors. While it had been thought that HIV needed only CD4 to infect cells, several chemokines have now been demonstrated to slow the growth of HIV in cultures. It has been determined that the chemokine family of G-protein-coupled receptors, most notably CXCR4 and CCR5, are involved in HIV infection (1, 9, 12, 14). To date, at least 10 chemokine receptors have been identified as HIV coreceptors (10). Furthermore, the interaction between the adhesion molecules leukocyte function-associated antigen-1 (LFA-1) and intercellular adhesion molecule-1 (ICAM-1) has been described with respect to both virus-cell and cell-cell fusion events. HIV-induced syncytium formation is blocked by a monoclonal antibody (MAb) directed against the β subunit of LFA-1 (19). In addition, cytokine-induced multinucleate giant cell formation in peripheral blood monocyte cultures (22, 29) as well as osteoclast development (24) is inhibited by antibodies directed against LFA-1 and ICAM-1. Members of the cadherin family of homophilic cell adhesion molecules have also been implicated in cell-cell fusion events. While N-cadherin-mediated adhesion appears necessary for myoblast fusion (28), inhibition of E-cadherin function prevents the fusion of osteoclast precursors to form osteoclasts (27).
Another protein thought to participate in cell-cell fusion is the purinergic receptor P2Z/P2X7, which binds extracellular ATP. While J774 cell clones that express this pore-forming receptor at very high levels as well as HEK293 cells stably transfected with P2X7 cDNA demonstrate some level of multinucleation when grown to confluence (5), oxidized ATP inhibits giant cell formation from concanavalin A- and gamma interferon-stimulated monocytes (13). In such cells, however, multinucleation is accompanied by cell death. Most recently, a set of proteins thought to enhance or induce cell fusion, initially termed FRP-1 and FRP-2 and now known to be CD98 and integrin α3, respectively (18, 30), have been identified in a number of cell lines infected with several different viruses as well as on the surface of monocytes and macrophages. MAbs directed against these proteins stimulate polykaryocyte formation in CD4+ U937 cells transfected with the HIV gp160 gene (32) and in HeLa and FL cells infected with Newcastle disease virus (21). In addition, anti-FRP antibodies inhibit giant cell formation in cultures of peripheral blood monocytes (39). While none of these proteins appear as actual fusion proteins and may not therefore mediate the actual fusion event, together they suggest that the fusion mechanism of viruses and mammalian cells involves both regulatory proteins and adhesion molecules.
Mononuclear phagocytes are unique cells in that they are ubiquitously distributed in tissues and can be programmed, in specific instances, to fuse in order to differentiate into osteoclasts or multinucleated giant cells. This is in contrast to the other fusogenic cells such as sperm cells and myoblasts, which must fuse to allow fertilization and muscle development, respectively. While giant cells differentiate primarily in chronic inflammatory sites and tumors, osteoclasts differentiate in bone, which they eventually resorb. Multinucleation therefore appears to provide cells of the mononuclear phagocyte lineage with an added value to facilitate the resorption of the substrate onto which they differentiate and adhere. We had hypothesized that proteins mediating macrophage-macrophage fusion resemble those used by viruses to infect cells. To test this hypothesis, we previously generated four MAbs that had the ability to block the fusion of macrophages in vitro. Each of these antibodies recognizes a 150-kDa surface protein that is specifically, transiently, and highly expressed in macrophages at the onset of fusion (35). We now report the cDNA of this protein, which we have named macrophage fusion receptor (MFR). This protein structurally resembles immune antigen receptors such as T-cell and B-cell receptors and, like them, belongs to the immunoglobulin (Ig) superfamily of proteins, whose members are well known to play a role in cell surface recognition (47). We show that the soluble extracellular domain of MFR (MFRe) binds macrophages competent for fusion and prevents fusion in vitro. These findings support our hypothesis and further suggest that in order to fuse, macrophages may use a protein machinery similar to that used by viruses. MFR is a potential component of the macrophage fusion machinery, which is likely to include numerous proteins. Our results open avenues to study the molecular mechanism of fusion not only of macrophages but also potentially of sperm cells, oocytes, and myoblasts.
MATERIALS AND METHODS
Cells.
Rat alveolar and peritoneal macrophages were obtained from 12-week-old Fisher rats (Charles River, Kingston, N.Y.) by tracheobronchial and peritoneal lavage, respectively, plated in fusogenic conditions, and cultured in minimal essential medium supplemented with Earle’s salts (MEME)–5% human serum (HS) as previously described (40–43). COS-7 cells were grown as previously described (11).
Chemicals.
N-Glycanase was purchased from Genzyme Corporation (Cambridge, Mass.), and N-glycosidase F was purchased from Boehringer GmbH (Mannheim, Germany); both enzymes were used as specified by the manufacturers. Unless otherwise stated, all chemicals were purchased from Sigma (St. Louis, Mo.).
Antibodies.
MAbs 10C4, 10C5, 10B11, and 12D6 were previously described (35). Mouse anti-rat macrophage CD4 antibody W3/25 and major histocompatibility complex type II (MHCII) antibody RT1B, which are of the IgG1 isotype, were obtained from Serotec (Raleigh, N.C.). Goat anti-mouse IgG–horseradish peroxidase (HRP) conjugate and lissamine rhodamine sulfonyl chloride (LRSC)-conjugated F(ab′)2 goat anti-mouse IgG (heavy plus light [H+L] chains) were obtained from Jackson Immunoresearch Laboratories Inc. (West Grove, Pa.). Goat anti-glutathione S-transferase (GST) was obtained from Pharmacia (Uppsala, Sweden), and rabbit anti-goat IgG conjugated to HRP was obtained from Zymed (South San Francisco, Calif.). Mouse anti-Myc was purchased from Invitrogen (Carlsbad, Calif.), and sheep anti-mouse–HRP conjugate was obtained from Amersham (Arlington Heights, Ill.).
Immunoprecipitation and SDS-PAGE analysis of cellular antigens following metabolic labeling with [35S]methionine.
Rat alveolar and peritoneal macrophages were collected, plated in six-well plastic dishes at 107 cells/ml, cultured in MEME supplemented with 5% HS for the indicated times, metabolically labeled with [35S]methionine, and subjected to immunoprecipitation as previously described (35). In brief, the postnuclear supernatants were incubated successively for 1 h with either an antifusion MAb (10C4, 10C5, 10B11, or 12D6) or mouse IgG1 (20 μg/ml) and then with goat anti-mouse IgG and Staphylococcus aureus (Zyzorbin; Zymed). The immunoprecipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Recombinant MFRe was immunoprecipitated from COS-7 cells transiently transfected with a plasmid expressing MFR (COS-MFR cells; see below). At 72 h after transfection, the cells were metabolically labeled overnight with [35S]methionine as described above. The postnuclear supernatants (1 ml of each) were incubated with 25 μl of 10C4-conjugated protein A-Sepharose 4-Fast Flow beads (see below) for 2 h at 4°C. The beads were collected and washed twice with immunoprecipitation buffer A (10 mM Tris-HCl, 150 mM NaCl, 0.1% SDS, 0.05% Nonidet P-40 [pH 8.6]) and twice with immunoprecipitation buffer B (0.3 M NaCl, 0.0125 M KH2PO4 [pH 7.4]). The beads were then resuspended in SDS sample buffer, and the proteins were subjected to SDS-PAGE. The dried gels were exposed to X-ray film overnight at −70°C.
Western blot analysis.
Alveolar and peritoneal macrophages were lysed and subjected to electrophoresis on a 10% polyacrylamide gel in nondenaturing, nonreducing conditions. The proteins were transferred to nitrocellulose membranes (Millipore Corp., Bedford, Mass.) by electroblotting overnight at 4°C. The membranes were subsequently incubated in Tris-buffered saline (TBS) supplemented with 5% dry milk for 1 h at room temperature, washed in TBS supplemented with 1% Tween 20, and incubated with MAb 12D6 for 1 h at room temperature, followed by anti-mouse HRP-conjugated IgG (Amersham ECL [enhanced chemiluminescence] kit) for 1 h. Following a quick rinse in TBS, the enzyme reaction was performed according to the manufacturer’s instructions, and the blots were exposed for 30 s on X-ray films.
MFR, CD4, and MHCII ELISA.
To quantitate the levels of MFR, CD4, and MHCII cell surface expression by enzyme-linked immunosorbent assay (ELISA), 5 × 104 alveolar macrophages per well in 96-well dishes were fixed at room temperature in 4% paraformaldehyde for 10 min and then incubated in 100 μl of phosphate-buffered saline (PBS) supplemented with 5% dry milk for 2 h. The cells were subsequently reacted overnight with IgG1, anti-rat MFR, anti-rat CD4 (100 μg/ml), or anti-rat MHCII (10 to 50 μg/ml). Following three washes for 10 min each with PBS, the cells were incubated at room temperature for 2 h with goat anti-mouse IgG-HRP conjugate (1:5,000 dilution). The cells were washed three times for 10 min each with PBS. Surface MFR expression was quantitated by incubating the cells for 5 min in 100 μl of 3,3′,5,5′-tetramethylbenzidine (HRP substrate; Moss Inc., Pasadena, Md.). Optical density at 650 nm (OD650) was measured with an ELISA plate reader (Corning, Corning, N.Y.).
Purification of the antigen recognized by MAb 10C4 and sequencing of proteolytic peptides from purified 10C4 antigen.
Two consecutive affinity purifications, each requiring the isolation and culture of 109 peritoneal macrophages from 200 rats, were performed. The two affinity purifications were performed identically except that a highly purified Coomassie blue stain that facilitated accurate peptide sequencing was used for the second. MAb 10C4 was purified from hybridoma supernatant by fast protein liquid chromatography and conjugated to protein A-Sepharose 4 Fast Flow beads (Pharmacia, Pistcataway, N.J.). Freshly isolated peritoneal macrophages were cultured in fusogenic conditions for 3 days to reach maximum expression of 10C4 antigen. They were then lysed, and the postnuclear supernatants were incubated with 1/10 volumes of 10C4-protein A-Sepharose beads for 3 h at 4°C. The beads were collected in a column and washed with 10 volumes of 10 mM Tris–150 mM NaCl [pH 7.4]. 10C4 antigen was eluted from the 10C4 beads with 10 fractions of 0.5 ml each containing 100 mM glycine at pH 2.8.
Each fraction was supplemented with 5 μl of sodium deoxycholate and 500 μl of 20% trichloroacetic acid and placed on ice for 1 h. Fractions were spun at 12,000 × g for 10 min; the pellets were washed with 500 μl of acetone (−20°C), dried, and analyzed on an SDS–7.5% polyacrylamide gel. The gel was stained with 0.05% Coomassie blue G (Sigma) for 15 min, destained, and soaked in water for 1 h. The 120-kDa band was excised from the gel and subjected to digestion in situ with lysylendopeptidase. The resulting peptides were separated by reverse-phase high-pressure liquid chromatography and sequenced with an automated protein sequencer (Applied Biosystems Inc., Foster City, Calif.). Several of the peptides were found in expressed sequence tag (EST) data banks (Table 1). One EST nucleotide sequence (H31804), originating from a rat PC12 cDNA library, contained one of the peptides (K7), while another EST sequence (R74985), from a mouse brain cDNA library, contained two of the peptides (K13 and K25). The PC12 and mouse brain clones were obtained from Norman Lee (26) and Kevin Brady (unpublished sequence), respectively, and further sequenced. Four of our peptides (K5, K7, K10, and K21) were detected in the rat PC12 clone.
TABLE 1.
Peptides used
| Name | Sequence
|
Identical or homologous peptide | |
|---|---|---|---|
| 1st purification | 2nd purification | ||
| K1 | VLK | ||
| K5 | (S/G)IVEPDTEIK | EST H31804 (PC12) | |
| K6 | XLDPAYYXK | ||
| K7 | ELSHLETTIS(S)(K) | ELSHLETTISSK | EST H31804 (PC12) |
| K9 | P(S)FEYA(S)VQV | ||
| K10 | FTPLYVLAK | SGGGTTLYVLAK | EST H31804 (PC12) |
| K11 | (S)YGF(S)PRITLK | ||
| K13 | KPAPEVPEPNN(H)TEYA | EST R74985 (mouse brain) | |
| XIETXK | |||
| K14 | (F)N(S)YV(D)GVEV | ||
| K15 | MPNMDF(S)I | ||
| K18 | EGQN/ITQIQD(T)N | ||
| K21 | XLVSYGISSTVSVK | EST H31804 (PC12) | |
| K25 | FSTSSTRLHEPPK | EST R74985 (mouse brain) | |
Construction of a fusing rat peritoneal macrophage phage cDNA library.
Freshly isolated rat peritoneal macrophages (9 × 107 cells) were plated under fusing conditions (cells are plated at high density, i.e., 107/ml, in medium supplemented with 5% HS) and cultured for 3 days, a time necessary for newly synthesized 10C4 antigen to reach maximum expression (data not shown). Total RNA was isolated by the guanidinium isothiocyanate procedure (20) followed by centrifugation through a cesium chloride cushion. Approximately 750 μg of total RNA was recovered, and poly(A)+ RNA was prepared by chromatography through oligo(dT)-cellulose. The cDNAs were synthesized by using Moloney murine leukemia virus reverse transcriptase (RT) and inserted into the Lambda ZAP II vector (Stratagene, La Jolla, Calif.). The cDNA library consisted of more than 106 recombinants, and the average size of the inserts was approximately 2.0 kb, as assessed from a pool of approximately 200 recombinants.
Screening of the lambda ZAP II cDNA fusing macrophage library and isolation of positive clones.
A 32P-labeled PCR-generated probe was used to screen approximately 250,000 recombinant plaques from the fusing macrophage cDNA library. The seven clones selected contained inserts ranging in size from 2.0 to 4.3 kb: 2.1, 3.0 kb; 5.1, 2.0 kb; 7.1, 3.7 kb; 7.2, 3.0 kb; 8.1, 3.0 kb; 9.1, 2.0 kb; and 11.2, 3.8 kb. Inserts from five of these clones were subjected to 5′ and 3′ end sequencing and restriction enzyme mapping followed by Southern blot analysis. The longest of these clones (clone 8.1) was subjected to sequencing. DNA was sequenced by using AmpliTaq FS DNA polymerase and fluorescence-labeled dideoxy terminators (Perkin-Elmer, Norwalk, Conn.) in a cycle sequencing method, and the resultant DNA fragments were electrophoresed and analyzed with an automated Applied Biosystems 373A Stretch or 377 DNA sequencer.
Northern blot analysis.
A multiple-tissue Northern blot was purchased from Clontech (Palo Alto, Calif.). Each lane contained approximately 2 μg of tissue-derived poly(A)+ mRNA. The blot was hybridized with a 32P-labeled PCR-generated DNA probe according to the manufacturer’s instructions. Alveolar and peritoneal macrophages were isolated, and part of the cells were cultured in a fusogenic milieu for 72 h. Total RNA was isolated by the guanidinium isothiocyanate-cesium chloride method (20); 8 μg was electrophoretically separated in formaldehyde-agarose gels, blotted onto a nylon membrane (GeneScreen Plus; NEN, Boston, Mass.), and hybridized with a 32P-labeled PCR-generated DNA probe corresponding to the full-length MFR cDNA. The signals on the autoradiograms were quantitated by using a Linotype-Hell (Kiel, Germany) scanner with Multi-Analyst software for Macintosh (Bio-Rad Laboratories, Hercules, Calif.).
Transfection of COS-7 cells with MFR cDNA.
Full-length rat MFR cDNA was inserted into the PstI-BamHI site of the pBK-RSV polylinker (Stratagene), generating the pBK-RSV-MFR construct. Fifteen micrograms of pBK-RSV-MFR was mixed with 0.5 ml of 250 mM CaCl2 in a 12- by 75-mm polystyrene tube; 0.5 ml of 2× 2-bromoethanesulfonic acid-buffered saline (pH 6.95) was added, and the DNA was allowed to precipitate for 20 min. The mixture was then added to 80% confluent COS-7 cells cultured in MEME supplemented with 10% fetal calf serum (FCS). The plates were placed in a 3% CO2, 35°C incubator overnight. The following day, the medium was replaced with fresh MEME–10% FCS. After growth for 72 h, cells were subjected to immunofluorescence and immunoprecipitation.
Immunolocalization.
Semiconfluent COS-7 cells were cultured on glass coverslips and transiently transfected with either pBK-RSV-MFR or pBK-RSV as described above. The transiently transfected COS-MFR cells were cultured for 3 days in Dulbecco modified Eagle medium–10% FCS prior to being subjected to immunofluorescence. To investigate whether recombinant MFR was expressed on the surface, the cells were reacted with MAb 10C4 either before or after fixation. The cells were fixed in formaldehyde for 1 h at 4°C, washed for 60 min in PBS-FCS, and incubated overnight in either PBS-FCS alone or PBS-FCS supplemented with MAb 10C4 or mouse IgG1. Following four washes for 15 min each in PBS-FCS, the cells were incubated for an additional hour with LRSC-conjugated F(ab′)2 goat anti-mouse IgG (H+L chains, 1:400 dilution) in the same buffer. For surface expression only, transfected and mock-transfected COS-7 cells were subjected to immunolocalization as described above but fixed after the first antibody incubation. The cells were imaged on either an Olympus microscope equipped with UV light or a Zeiss Axiovert confocal microscope equipped with a confocal Bio-Rad MRC600 system (Bio-Rad, Cambridge, Mass.).
Comparative analysis of 10C4 transcripts in alveolar and peritoneal macrophages by RT-PCR.
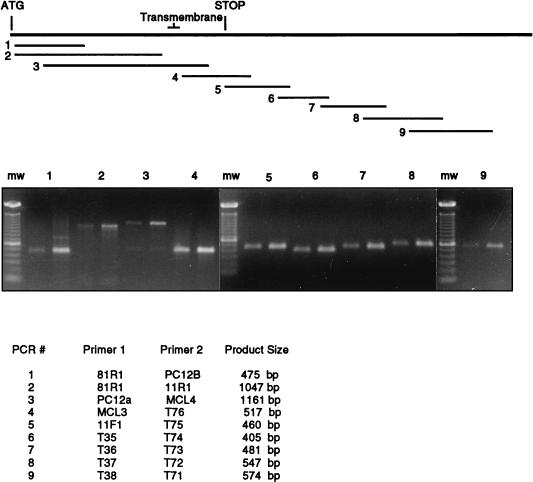
Total cellular RNA was isolated by the guanidinium isothiocyanate-cesium chloride method (20). cDNAs were synthesized by using Moloney murine leukemia virus (GibcoBRL, Gaithersburg, Md.) and amplified with the primers indicated in Fig. 5. The forward and reverse primers were used in PCRs run for 1.5 min at 94°C, 1.5 min at 52°C, and 3 min at 72°C, for a total of 30 cycles. The RT-PCR products generated were analyzed on 1.2% agarose gels.
FIG. 5.
Comparison of 10C4 transcripts in alveolar (left product of each pair) and peritoneal (right product of each pair) macrophages by RT-PCR. PCRs were performed with the primer combinations listed. Primers were designed on the basis of the sequence of cloned MFR cDNA. The corresponding locations of the PCR products with respect to MFR cDNA are shown. PCR primer combinations were chosen such that the products overlap, and the majority of the cDNA sequence was analyzed. PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide. The approximate sizes of the PCR products are indicated. mw, molecular weight markers.
Production of recombinant soluble extracellular domain of MFR (MFRe) and CD4 (CD4e).
MFRe was expressed using two different systems. (i) MFRe was expressed as a GST fusion protein using PGEX-4T (Pharmacia Biotech, Uppsala, Sweden). PCR amplification of MFRe was performed with a sense primer that lacked the MFR leader sequence (5′TGTTTCTGTGCAGAATTCAGCGGGAAAGAACTGAAG; nucleotides [nt] 114 to 150) and an antisense primer (5′ACCCACACCGATGAATTCCTTCCAGTTGTAAGC; nt 1145 to 1181) that did not include the transmembrane domain of MFR. PCR was performed with Pwo polymerase (Boehringer) and full-length MFR cDNA as the template. Both primers were designed to contain EcoRI sites. The amplified PCR fragment was digested with EcoRI and inserted in frame into the EcoRI site of pGEX-4T-1. This construct was used to transform the protease-deficient Escherichia coli strain BL-21. GST-MFRe was isolated from 2 liters of bacterial culture by using the bulk GST purification modules as recommended by the manufacturer. The eluted protein was extensively dialyzed against PBS. A pGEX-calreticulin construct (GST-Cal; courtesy of Ari Helenius, Yale University) was used as the control.
(ii) MFRe was expressed as a fusion protein containing a C-terminal Myc epitope and polyhistidine tag (pcDNA 3.1/Myc-His A, B, and C; Invitrogen). PCR amplification of the extracellular region was performed with the sense primer 5′TCTCCCATCCTTGAATTCCAGCCGCGGCCCATGGAG (nt 5 to 41) and the antisense primer used for the GST fusion protein described above. Full-length MFR cDNA was used as template with Pwo polymerase. The PCR product was digested with EcoRI and ligated into the EcoRI site of pcDNA 3.1 B. This construct was used to transfect COS-7 cells by using Lipofectamine (GibcoBRL) according to the manufacturer’s instructions. Transfected cells were cultured for 72 h in Opti-MEM I reduced serum medium (GibcoBRL). The recombinant protein was purified from culture supernatant by using the Invitrogen Xpress protein purification system according to the manufacturer’s recommendations and extensively dialyzed against PBS.
Recombinant GST-MFRe was quantified by running 5 μl on an SDS–10% polyacrylamide gel and staining the gel with Coomassie brilliant blue. The intensity of the stained band was tested against that of a serial dilution of bovine serum albumin run on the same gel. The concentration of Myc-His-MFRe was then determined by immobilizing serial dilutions of both GST-MFRe and Myc-His-MFRe on nitrocellulose, using a Bio-Dot microfiltration apparatus (Bio-Rad). The immobilized proteins were reacted with MAb 10C4 followed by HRP-linked sheep anti-mouse IgG (Amersham ECL kit). The enzyme reaction was performed according to the manufacturer’s instructions, and the blots were exposed for 30 s on X-ray films. Recombinant Myc-His-MFRe concentration was determined by comparative scanning densitometry of the dots against the concentration of GST-MFRe.
CD4 was expressed as a GST fusion protein, using pGEX-4T. PCR amplification of the extracellular domain of CD4 was performed with a sense primer that lacked the CD4 leader sequence (5′GTTGTCACCCAAGGAGAATTCGTGGTGCTGGGGAAG; nt 120 to 156) and an antisense primer (5′CATTGTCTGGTTCAACCCGAATTCTAAAACCTGGAT; nt 1202 to 1238) (7) that did not include the transmembrane domain of CD4. Macrophage cDNA synthesized from total RNA was used as the template for the PCR. All steps for production of the protein were as described above for MFRe.
Deglycosylation of recombinant Myc-His-MFRe.
Approximately 250 μg of recombinant Myc-His-MFRe was deglycosylated in its native form by incubation with 10 U of N-glycosidase F for 48 h at 37°C in 0.1 M sodium phosphate (pH 7.4). Deglycosylation was analyzed by comparative Western blot analysis of both native and N-glycosidase F-treated Myc-His-MFRe, using anti-Myc antibody (Invitrogen).
Fusion assay using recombinant proteins.
Freshly isolated rat alveolar macrophages were plated at 5 × 106 cells/ml and cultured in a fusogenic milieu supplemented with GST-MFRe, GST-CD4e, GST, Myc-His-MFRe, or deglycosylated Myc-His-MFRe (D-Myc-His-MFRe) at various concentrations. The cells were examined daily for 4 days, and fusion was graded blindly by three investigators on a scale of 1 (absence of fusion) to 5 (fusion greater than 90%).
Binding assay using recombinant fusion proteins.
Freshly isolated rat alveolar macrophages were plated at 5 × 106 cells/ml in triplicate wells, using 96-well dishes (5 × 104 cells per well). The cells were cultured overnight in a fusogenic milieu. At the indicated times, duplicate sets of cells were supplemented with GST-MFRe, GST-CD4e, Myc-His-MFRe, or D-Myc-His-MFRe at the indicated concentrations. Binding proceeded overnight at 4°C. Medium was removed from one set of cells and replaced with fresh medium lacking recombinant protein to allow for dissociation. Dissociation proceeded for 3 h at 4°C. The medium was then removed from all wells, and the cells were fixed for 30 min in 4% paraformaldehyde. Cells were rinsed three times with PBS and blocked for 1 h in 5% milk–PBS. Following three washes for 5 min each in PBS, the cells were incubated with goat anti-GST for 30 min at room temperature. Following three washes for 5 min each in PBS, the cells were incubated with rabbit anti-goat IgG-HRP for 30 min. Following three final washes for 5 min each in PBS, 100 μl of peroxidase substrate (Moss Inc.) was added to each well, and the OD650 with an ELISA plate reader (Corning). Specific binding was determined as the difference in average OD values between the two sets of cells, i.e., between total and nonspecific binding values for each ligand concentration.
Nucleotide sequence accession number.
The sequence shown in Fig. 2 has been assigned GenBank accession no. U62328.
FIG. 2.
Nucleotide and deduced amino acid sequences of rat MFR. The putative transmembrane domain is underlined. Possible N-glycosylation sites (Asn-X-Ser/Thr) are indicated in boldface. Cysteines likely to form disulfide bonds creating the three Ig-like domains (▵) and putative binding sites for SH2 domains of PTPases (–––) are indicated. The putative binding site for SH3 domains is in italics.
RESULTS
Purification of 10C4 antigen.
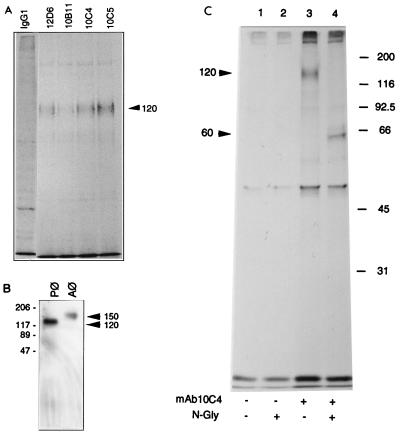
Since the yield of rat alveolar macrophages is relatively low, we explored alternative sources of macrophages in order to secure the purification of sufficient amount of 10C4 antigen for peptide sequencing. Because 10C4 antigen is also expressed by fusing macrophages in vivo (35), we speculated that peritoneal macrophages might be a valuable source of 10C4 antigen. Their yield is 10 times that of alveolar macrophages, and like alveolar cells, they fuse in vitro, although more slowly (10 days to reach 100% fusion instead of 4 days as for alveolar cells) (data not shown). Indirect immunofluorescence performed with all four antifusion MAbs revealed a positive signal in fusing peritoneal macrophages (data not shown). When subjected to metabolic labeling with [35S]methionine followed by immunoprecipitation, all four MAbs precipitated a protein of 120 kDa (Fig. 1A). This molecular mass is lower than that of the protein precipitated from alveolar macrophages (150 kDa [35]). This finding indicated that the antigen expressed by peritoneal cells was also recognized by all four antifusion MAbs and newly synthesized and suggested that the core protein may undergo cell-specific posttranslational modifications. When macrophages were subjected to Western blot analysis in nondenaturing, nonreducing conditions, MAb 10C4 again recognized proteins of 120 and 150 kDa, respectively, suggesting that these proteins were expressed as monomers (Fig. 1B). To further biochemically characterize the 10C4 antigen, peritoneal macrophages were cultured in fusogenic conditions for 24 h and labeled metabolically for 17 h with [35S]methionine, and their MAb 10C4 immunoprecipitates were subjected to N-glycanase digestion. This revealed a 60-kDa protein that was specifically precipitated, indicating that the antigen is heavily glycosylated, possibly in an extended and heterogeneous fashion (Fig. 1C).
FIG. 1.
(A) Antifusion MAbs recognize a 120-kDa protein from fusing rat peritoneal macrophages. Macrophages were cultured for 3 days in fusogenic conditions, metabolically labeled with [35S]methionine for 17 h, and then subjected to immunoprecipitation with either mouse IgG1 or MAb 12D6, 10B11, 10C4, or 10C5. Immunoprecipitates were analyzed by SDS-PAGE. All four MAbs precipitate a 120-kDa protein from peritoneal macrophages. Each lane represents immunoprecipitates from extracts prepared from 5 × 106 plated peritoneal macrophages. In all panels, molecular masses of markers are indicated in kilodaltons. (B) MAb 10C4 recognizes 120- and 150-kDa proteins from fusing peritoneal and alveolar macrophages (Pφ and Aφ), respectively. Macrophages were cultured for 3 days in fusogenic conditions prior to being subjected to Western blot analysis. Total cell lysates were run on a 10% polyacrylamide gel in the absence of both SDS and β-mercaptoethanol and blotted with MAb 12D6 followed by HRP-conjugated goat anti-mouse IgG. The ECL substrate reaction was developed with XAR film. (C) 10C4 antigen is heavily glycosylated. Rat peritoneal macrophages were cultured in fusogenic conditions for 3 days prior to being metabolically labeled with [35S]methionine for 17 h. Immunoprecipitates were subjected to N-glycanase (N-Gly) digestion.
Cloning of MFR, the cDNA coding for 10C4 antigen.
Two successive purifications of 10C4 antigen allowed the isolation and sequencing of several peptides listed in Table 1. Several of these peptides were found in EST data banks (Table 1). Further searches in GenBank revealed that another clone from a human brain cDNA library (CCA53) showed homology with both EST sequences. The degrees of identity at the amino acid level between the PC12 and mouse brain ESTs and the corresponding human brain sequence were 73 and 65%, respectively. Relative to the predicted amino acid sequence of the human brain cDNA, the rat PC12 and mouse brain EST sequences were separated by 166 amino acids, with the PC12 homology being N-terminally located. This information suggested that all six peptides may originate from a single gene.
We designed specific oligonucleotide primers based on the nucleotide sequence of both ESTs and amplified by PCR homologous sequences both from our macrophage cDNA library and from cDNA reverse transcribed from total RNA obtained from fusing alveolar macrophages. The PCR product generated with primers designed according to the mouse brain EST sequence was used to screen our fusing rat macrophage cDNA library. The PCR product generated with primers designed according to the rat PC12 EST was not satisfactory and therefore not used. Several clones were isolated, and the longest, clone 8.1, was subjected to sequence analysis. The nucleotide sequence of clone 8.1 contained one open reading frame encoding a protein of 509 amino acids (Fig. 2) with one predicted transmembrane domain. While 10C4 antigens had apparent molecular masses of 150 and 120 kDa in alveolar and peritoneal macrophages, respectively, the predicted size from the cDNA was 60 kDa, and it contained all of the peptides sequence that we previously obtained. BEAUTY sequence analysis revealed that the protein belonged to the Ig superfamily of proteins. The extracellular domain contained three Ig-related domains. The NH2-terminal Ig domain appeared to be of the V type, while the remaining two more closely resembled the C1 type. The extracellular region also contained 15 putative N-linked glycosylation sites, consistent with our observation that the 10C4 antigen is highly glycosylated. Because the 10C4 antigen resembled the receptors for both HIV (CD4) and coxsackie virus-adenovirus (CAR), we named the 10C4 antigen MFR, for macrophage fusion receptor.
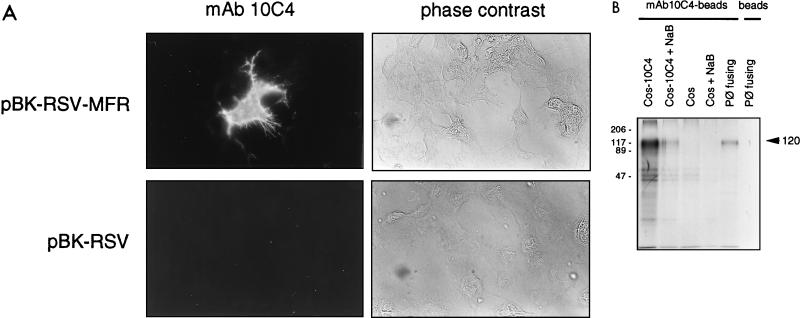
To verify that clone 8.1 cDNA coded for the protein recognized by MAb 10C4, full-length MFR cDNA was inserted into the pBK-RSV vector (creating pBK-RSV-MFR), and COS-7 cells were transiently transfected with either pBK-RSV-MFR or pBK-RSV. COS-MFR cells were reacted unfixed with MAb 10C4. COS cells transfected with pBK-RSV-MFR exhibited a positive signal (Fig. 3A) that was not detected in COS-7 cells transfected with pBK-RSV. COS cells that did not integrate the pBK-RSV-MFR transgene (mock-transfected cells) also failed to react with MAb 10C4 (Fig. 3A, upper panels). This indicated the specificity of the expression and the surface localization. In addition, MAb 10C4 precipitated from radiolabeled COS-7 cells transiently transfected with pBK-RSV-MFR a protein of 120 kDa (Fig. 3B), confirming that clone 8.1 encoded the protein recognized by MAb 10C4.
FIG. 3.
MAb 10C4 recognizes recombinant MFR. (A) COS-7 cells were transiently transfected with either pBK-RSV-MFR or pBK-RSV, and the recombinant protein was detected by incubating unfixed cells with either MAb 10C4 or PBS, followed by LRSC-conjugated F(ab′)2 goat anti-mouse IgG (H+L). Magnification, ×200. (B) COS-7 cells transiently transfected with pBK-RSV-MFR, nontransfected COS-7 cells, and fusing peritoneal macrophages (Pφ) were metabolically labeled with [35S]methionine and subjected to immunoprecipitation with protein A-Sepharose-MAb 10C4-conjugated beads. Peritoneal macrophage lysates were incubated with protein A-Sepharose beads as a control. Immunoprecipitates were analyzed by SDS-PAGE. The mobilities of molecular mass standards are indicated in kilodaltons.
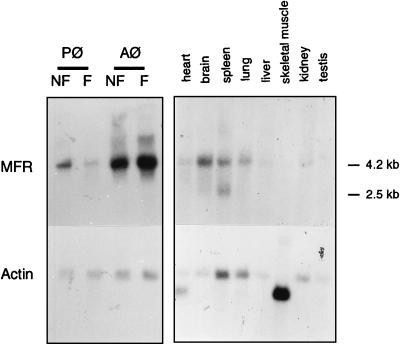
Northern blot analysis revealed a 4.2-kb MFR transcript in both freshly isolated and fusing alveolar and peritoneal macrophages (Fig. 4). MFR transcripts were, however, more abundant in alveolar than peritoneal cells. MFR transcripts were detected in all tissues examined but at low abundance; an additional band of 2.5 kb was detected in the spleen.
FIG. 4.
Northern blot analysis of MFR. Each lane contained approximately 2 μg of mRNA from the tissues indicated and 8 μg of total RNA from freshly isolated nonfusing (NF) and fusing (F) alveolar and peritoneal macrophages (Aφ and Pφ). Hybridization was performed with 32P-labeled PCR-generated mouse brain-based sequence (Table 1) and β-actin probes.
To confirm that alveolar and peritoneal macrophage MFR transcripts were similar, we used comparative RT-PCR analysis. As shown in Fig. 5, such an analysis revealed PCR products of identical size from both cell types, using primer combinations spanning the entire length of clone 8.1. This finding suggested that the two types of macrophages have the same MFR mRNA, and their difference in molecular size was possibly due to posttranslational modifications.
MFR participates in macrophage fusion.
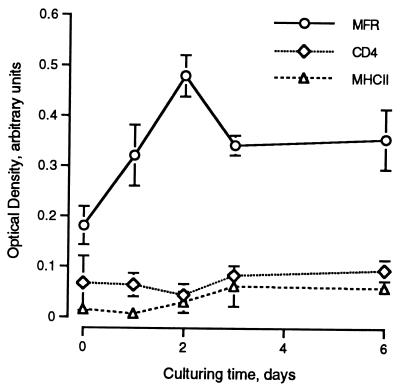
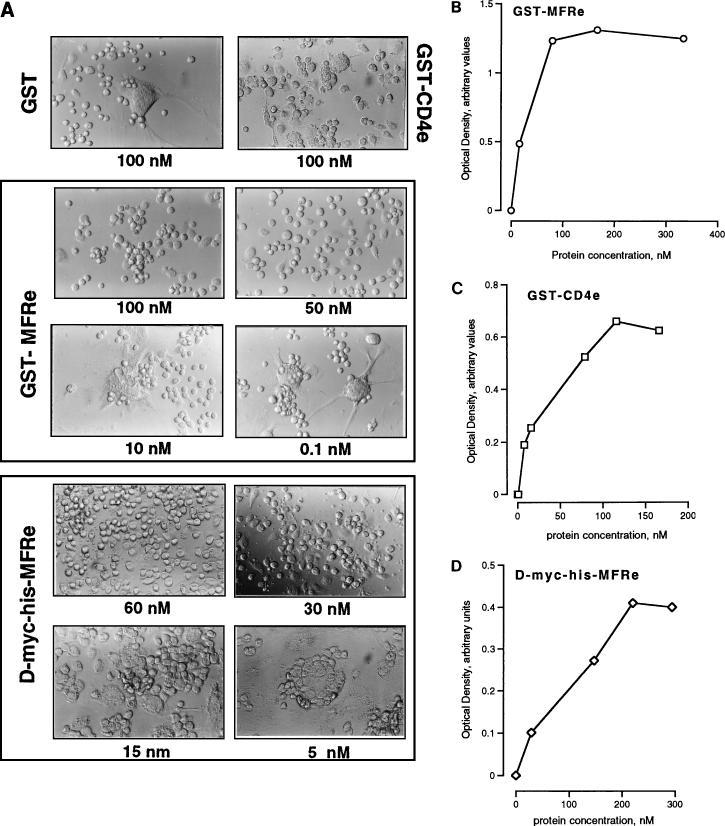
We had previously reported that MFR expression is highly induced in fusing macrophages in vivo and in vitro (35). To investigate the kinetics of MFR expression, levels of expression of cell surface MFR and of two other macrophage surface proteins, CD4 and MHCII, were quantitated by ELISA as a function of time in culture. Figure 6 shows that MFR expression peaked within 2 days after plating of the cells, decreasing and leveling off thereafter. In that experiment, the peak level of MFR expression was approximately 2.5 times greater than that of both CD4 and MHCII, as well as that of MFR 60 min after plating. Neither CD4 nor MHCII expression was altered by fusion. To investigate whether MFR participated in macrophage fusion, we first generated a GST-MFRe fusion protein. Although it was not glycosylated, we were able to immunoprecipitate GST-MFRe with MAb 10C4, indicating that the antibody recognized the core peptide of MFRe (data not shown). SDS-PAGE analysis in reducing and nonreducing conditions indicated that the GST-MFRe fusion protein becomes oxidized, most likely during the purification procedure (data not shown). This was interesting in view of the fact that MAb 10C4 does not recognize the denatured form of its antigen (35). To investigate whether soluble MFRe altered fusion, alveolar macrophages were cultured in the absence or presence of either GST alone, GST-CD4e, or GST-MFRe, and fusion was monitored morphologically and quantitated (Fig. 7A; Table 2). Alveolar macrophages cultured in fusogenic milieu reached 99% fusion within 4 days, as previously shown (35). The addition of 50 nM GST-MFRe completely blocked macrophage aggregation and therefore fusion. However, once the level of GST-MFRe dropped to 10 nM, giant cell formation became very prominent and comparable to that seen in control cultures as well as in cultures supplemented with 100 nM GST or GST-CD4e.
FIG. 6.
Cell surface expression of MFR is transiently induced at the onset of fusion. MFR, CD4, and MHCII cell surface expression was quantitated by ELISA on alveolar macrophages cultured in fusogenic conditions. Expression was recorded at the indicated times.
FIG. 7.
(A) MFRe blocks macrophage fusion in vitro. GST-CD4e, GST-MFRe, and D-Myc-His-MFRe were added at the indicated concentrations to fusing macrophages. Cells were cultured for 4 days prior to being fixed and examined for multinucleation. Magnification, ×100. (B to D) Specific binding of GST-MFRe (B), GST-CD4e (C), and D-Myc-His-MFRe (D) to fusing macrophages in vitro. Fusing macrophages were cultured for 24 h in fusogenic conditions prior to being subjected to binding analysis as described in Materials and Methods. Binding was detected by ELISA.
TABLE 2.
GST-MFRe and D-Myc-His-MFRe inhibit macrophage fusion in vitro
| Medium | Fusiona |
|---|---|
| Control | 5.0 (0.5) |
| GST, 10 nM | 5.0 (0.5) |
| GST-CD4e, 100 nM | 5.0 (0.5) |
| GST-MFRe (nM) | |
| 100 | 1.0 (0.1) |
| 50 | 1.5 (0.2) |
| 25 | 3.0 (0.5) |
| 10 | 5.0 (0.5) |
| 5 | 5.0 (0.5) |
| D-Myc-His-MFRe (nM) | |
| 60 | 1.0 (0.1) |
| 30 | 1.0 (0.4) |
| 15 | 3.0 (0.4) |
| 5 | 5.0 (0.5) |
5, maximum fusion; 1, absence of fusion (n = 3). Standard deviations are indicated in parentheses.
To ensure that GST-MFRe inhibited fusion by virtue of interacting with specific binding sites on the surface of macrophages, cells were cultured for 24 h in a fusogenic milieu and then incubated overnight at 4°C with increasing concentrations of GST-MFRe or GST-CD4e. Binding was determined by ELISA. Both GST-MFRe and GST-CD4e specifically bound macrophages in a dose-dependent and saturable manner (Fig. 7B and C), while GST did not (data not shown). The specific binding of CD4 to macrophages is very interesting and has not been previously reported. To investigate whether GST-MFRe-specific binding sites were also induced by fusogenic conditions in macrophages, GST-MFRe binding was performed 15 min and 2 days after plating (Table 3). While surface MFR expression had increased nearly 50% by day 2 of culture, recombinant GST-MFRe binding, like GST-CD4e binding, did not change significantly. This may indicate that unlike MFR expression, MFR binding site expression does not increase with induction of fusion.
TABLE 3.
Binding of both GST-MFRe and GST-CD4e to fusing macrophages before and after induction of fusion
| Determination | ODa
|
|
|---|---|---|
| Day 0 | Day 2 | |
| GST-MFRe binding | 0.84 (0.11) | 0.93 (0.10) |
| GST-CD4e binding | 0.91 (0.18) | 0.92 (0.09) |
| MFR expressionb | 0.43 (0.07) | 0.65 (0.19) |
Standard deviations are indicated in parentheses (n = 5).
Cell surface expression of MFR was increased by 50%.
The fact that the nonglycosylated form of MFRe inhibited fusion was suggestive of a role for MFRe core peptide in cell-cell interaction, possibly via binding to a putative MFR ligand. To initiate investigations on the possible role of sugar moieties in MFR-MFR ligand interaction, a glycosylated form of MFRe, Myc-His-MFRe, was expressed in COS cells. Myc-His-MFRe failed to block fusion at all doses tested (data not shown). Indeed, Myc-His-MFRe also failed to bind fusing macrophages in a specific manner (data not shown). However, when Myc-His-MFRe was subjected to enzymatic digestion with N-glycosidase F, leading to over 80% deglycosylation, it recovered the ability to block fusion (Fig. 7A). Although Myc-His-MFRe was partially deglycosylated, it appeared to bind macrophages in a specific and dose-dependent manner (Fig. 7D). These results strongly suggested that the glycosylation imposed by COS-7 cells on MFR was not functional and that the core peptide of MFR played a role in macrophage-macrophage interaction.
DISCUSSION
We had previously identified a macrophage surface protein whose expression is highly induced, transient, and specific, as it is restricted to actively fusing macrophages in vitro and in vivo. We have now purified this protein, called MFR, and cloned its corresponding cDNA. MFR, belongs to the Ig superfamily, which includes proteins well known to play a role in adhesion and cell surface recognition and to serve as immune antigen receptors and receptors for viruses. Given our finding that MFRe prevents fusion of macrophages in vitro by interacting with a specific binding site on macrophages, we propose that MFR in involved in the fusion machinery of macrophages.
In contrast with the other surface proteins proposed to play a role in macrophage multinucleation, MFR surface expression is highly induced by fusogenic conditions in macrophages. This regulated expression may indicate that macrophages reside in tissues as mononucleated cells and can be induced to fuse in specific conditions to differentiate into either osteoclasts or giant cells.
Although multinucleated osteoclasts and giant cells have long been recognized, the mechanism by which their mononucleated precursor cells fuse with each other remains unclear. While a number of adhesion molecules and regulatory proteins have been suggested to participate in the fusion of macrophages, the nature of the assays reported in the literature used to identify these molecules did not allow investigation of the direct role of these molecules in cell-cell adhesion/fusion. These assays used bone marrow cells and monocytes, i.e., cells which are impure and have not yet acquired the status of macrophages. Such cells require additional culturing time to further differentiate and become fusion competent. In contrast, we have used a highly pure and fast fusion assay whereby cells initiate fusion within hours after plating and reach 99% fusion within 3 to 4 days. This assay allows for the investigation of molecules involved in early cell-cell interaction and possibly fusion (41).
MFR is a transmembrane glycoprotein which consists of a core protein of approximately 60 kDa that contains three Ig-like domains in the extracellular region. MFR is highly glycosylated, as suggested by the presence of 15 putative N-linked glycosylation sites on its extracellular domain. Although we have not eliminated the possibility of variants existing as a result of proteolysis, the difference in the apparent molecular weights of MFR between peritoneal and alveolar macrophages is suggestive of tissue-specific differential posttranslational modifications which are common among proteins of the Ig family. Of interest, alveolar and peritoneal macrophages differ not only in their levels of expression of MFR protein and message but also in their fusion rates, as alveolar macrophages fuse about three times faster than peritoneal macrophages (data not shown). It is also possible that glycosylation of MFR, in addition to its expression level, affects the rate of cell-cell recognition or interaction. The role of sugar moieties in MFR function may be implied by our finding that only the deglycosylated 120-kDa form of MFR that is produced by transfected COS-7 cells binds and inhibits the fusion of macrophages, like the bacterially produced form of MFR that is not glycosylated. This observation indicates that modifying the sugar moieties of MFR could affect its function. It is also possible that the sugar moieties added by COS-7 cells mask the core MFR peptide and prevent its binding. Another possibility is that these sugar moieties drive MFR to another, nonspecific binding site.
Of interest, MFR mRNA is as abundant in freshly isolated as in fusing alveolar macrophages, yet MFR protein expression is highly induced by fusogenic conditions in these cells. This finding suggests that either MRF expression is regulated at a posttranslational level or MFR protein is stored intracellularly. Although we were able to detect a significant increase in MFR protein expression as early as 1 h after plating of the cells in several experiments, we did not detect an abundant intracellular signal by immunocytochemistry. This increased expression of MFR contrasts with the abundance of MFR-specific binding sites which appear to remain constant in macrophages, whether freshly isolated or induced to fuse. This could provide a mean to improve the control and regulation of macrophage multinucleation.
MFR also contains four putative tyrosine phosphorylation sites as well as two immunoreceptor tyrosine-based inhibitory motifs and one putative SH3 domain binding site in its cytoplasmic domain (Fig. 2), strongly suggesting the possibility that it is the target of signal transduction cascades. Indeed, MFR is identical to a protein recently cloned from SR-3Y1 cells (v-src-transformed rat fibroblasts) known as SHPS-1 (SHP substrate 1) (15) and a protein cloned from rat brain known as BIT (brain Ig-like molecule with tyrosine-based activation motifs) (36). These proteins are homologous to a protein, known as P84, which was first identified in mouse brain membrane fractions and is widely expressed in the central nervous system (6, 8). Comu et al. (8) also reported the cloning of a smaller version of SHPS-1 that is missing 653 bp corresponding to a single exon flanked by consensus splice acceptor and donor sites, as determined by the analysis of mouse genomic clones. This splicing results in the loss of the two C1-type Ig domains and may correspond to the 2.5-kb transcript seen by Northern blot analysis.
MFR/SHPS-1/BIT contains four potential tyrosine phosphorylation sites in its cytoplasmic tail. Cell adhesion and various mitogens were found to induce tyrosine phosphorylation of SHPS-1, possibly by Src family kinases, and subsequent association with the SH2 domain-containing protein tyrosine phosphatase SHP-2. This implies that MFR/SHPS-1/BIT plays an important role with respect to SHP-2-mediated growth factor- and cytokine-induced signal transduction pathways. Similarly, phosphorylated BIT was found to associate with SHP-2 as well as coimmunoprecipitate with Grb2 (31), an adapter molecule known to link growth factor receptors to downstream effector proteins (33). Indeed, MFR contains a potential SH3 domain binding site, suggesting interaction with an SH3 domain-containing signaling molecule such as a Src family kinase or Grb2. It was also shown that BIT-coated substrate could support neurite extension, suggesting that BIT is involved in cell-cell interaction. Recent evidence indicates that MFR/SHPS-1/BIT belongs to a newly recognized family of proteins that seem to regulate signals involved in a number of physiological and pathological processes. Kharitonenkov et al. (23) identified at least 15 members of this gene family, termed SIRPs (signal-regulatory proteins), which seem to inhibit signaling through tyrosine kinase receptors. Indeed, human SIRP-1α is identical to human SHPS-1. Most recently, a member of the SIRPα family has been cloned from cattle monocytes and termed MyD-1 (4). COS-7 cells transfected with MyD-1 cDNA are able to bind CD4+ T cells, while proliferation of resting immune T cells is inhibited by MAbs directed against MyD-1. Hence, MyD-1 may promote the attachment of CD4+ T cells to antigen-presenting cells as well as play a role in signaling and regulation.
While numerous receptors have been implicated in negative regulation of cellular activity because they bear immunoreceptor tyrosine-based inhibitory motifs (25, 44), the biological functions of most inhibitory receptors and the mechanisms by which they, together with activating receptors, regulate cellular activity remain unclear. We now propose that MFR, a member of the newly discovered family of SIRPs, plays a role in macrophage adhesion/fusion. The exact role of MFR in the fusion reaction is yet to be deciphered. One hypothesis is that MFR is one component of a complex fusion machinery. This machinery may consist of numerous proteins, each of which plays a critical role in the adhesion/fusion process. Indeed, one such component may resemble the classical viral fusion protein such as influenza virus HA. It is also very possible that MFR is not directly involved in the fusion reaction per se but instead regulates adhesion/fusion as a part of a specific signaling pathway.
Discerning the role of MFR with respect to the regulation of macrophage fusion may provide insight as to the pathology and treatment of diseases associated with multinucleated macrophages. The question of whether MFR plays a role in osteoclast development remains open. Pure cultures of preosteoclasts have not yet been obtained due to the lack of marker characterizing these cells, thereby complicating the study of osteoclast development. While fusing alveolar macrophages are most likely distinct from fusing preosteoclasts, it is possible that these cells differentiate into either giant cells or osteoclasts, respectively, via similar fusion protein mechanisms. Indeed, we have obtained preliminary immunocytochemical evidence that MFR is present in rat bone marrow cultures (data not shown). We have also identified a number of positive clones isolated from human osteoclastoma and purified normal human osteoclast-like cell cDNA libraries (generously provided by G. David Roodman) which have yet to be extensively analyzed (data not shown).
Whether CD4 plays a role in macrophage-macrophage cross talk is an interesting question. Indeed, the role of CD4 in macrophages remains poorly understood. We had selected CD4 as a control molecule because of its known interaction with HIV. It was therefore interesting to discover that MFR, like CD4, belongs to the Ig superfamily and contains three Ig domains. Unlike MFR, however, CD4 expression is not induced by fusion in macrophages. While the functional consequences of our observation cannot be determined at this time, the fact that CD4 recognizes a binding site on macrophages suggests that it may play a role in cell-cell interaction, possibly via MHCII or another determinant. The identification of the ligand for CD4 will facilitate these investigations.
Since the mechanism by which the molecules previously reported to mediate sperm cell, oocyte, and myoblast fusion has not yet been elucidated (17), our finding opens possibilities to investigate whether MFR homologues participate in both sperm cell, oocyte, and myoblast adhesion/fusion. In this respect, the identification of the ligand for MFR will facilitate studies on the mechanics of cell-cell adhesion/fusion.
ACKNOWLEDGMENTS
We are grateful to G. D. Roodman for providing human osteoclastoma and osteoclast-like cell cDNA libraries and to Norman Lee and Kevin Brady for providing EST clones. We thank Ronald Dirkx, Jr., for help with the transfections and Sarah Whitaker for photographic work. We thank Pietro De Camilli for careful reading of the manuscript.
This work was supported by NIH grant DE12110 (A.V.). H.S. was supported by NIH NRSA AR08395, C.S. was supported by NIH training grant GM07223 and a National Osteoporosis Foundation fellowship, and M.S. was supported by the American Diabetes Association.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C, Feng Y, Kennedy P, Murphy P, Berger E. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Bergelson J, Cunningham J, Droguett G, Kurt-Jones E, Krithivas A, Hong J, Horwitz M, Crowell R, Finberg R. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 3.Blobel C, Wolfsberg T, Turck C, Myles D, Primakoff P, White J. A potential fusion peptide and an integrin ligand domain in a protein active in sperm-egg fusion. Nature. 1992;356:248–252. doi: 10.1038/356248a0. [DOI] [PubMed] [Google Scholar]
- 4.Brooke G, Parsons K, Howard C. Cloning of two members of the SIRPα family of protein tyrosine phosphatase binding proteins in cattle that are expressed on monocytes and a subpopulation of dendritic cells and which mediate binding to CD4 T cells. Eur J Immunol. 1998;28:1–11. doi: 10.1002/(SICI)1521-4141(199801)28:01<1::AID-IMMU1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 5.Chiozzi P, Sanz J, Ferrari D, Falzoni S, Aleotti A, Buell G, Collo G, DiVirgilio F. Spontaneous cell fusion in macrophage cultures expressing high levels of the P2Z/P2X7 receptor. J Cell Biol. 1997;138:697–706. doi: 10.1083/jcb.138.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuang W, Lagenaur C. Central nervous system antigen P84 can serve as a substrate for neurite outgrowth. Dev Biol. 1990;137:219–232. doi: 10.1016/0012-1606(90)90249-i. [DOI] [PubMed] [Google Scholar]
- 7.Clark S, Jeffries W, Barclay A N, Gagnon J, Williams A. Peptide and nucleotide sequence of rat CD4 (W3/25) antigen: evidence for derivation from a structure with four immunoglobulin-related domains. Proc Natl Acad Sci USA. 1987;84:1649–1653. doi: 10.1073/pnas.84.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comu S, Weng W, Olinsky S, Ishwad P, Mi Z, Hempel J, Watkins S, Lagenaur C, Narayanan V. The murine P84 neural adhesion molecule is SHPS-1, a member of the phosphatase-binding protein family. J Neurosci. 1997;17:8702–8710. doi: 10.1523/JNEUROSCI.17-22-08702.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, DiMarzio P, Marmon S, Sutton R, Hill C, Davis C, Peiper S, Schall T, Littman D, Landau N. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 10.Dimitrov D. How do viruses enter cells? The HIV coreceptors teach us a lesson in complexity. Cell. 1997;91:721–730. doi: 10.1016/s0092-8674(00)80460-2. [DOI] [PubMed] [Google Scholar]
- 11.Dirkx R, Thomas A, Li L, Lernmark A, Sherwin R S, DeCamilli P, Solimena M. Targeting of the 67-kDa isoform of glutamic acid decarboxylase to intracellular organelles is mediated by its interaction with the NH2-terminal region of the 65-kDa isoform of glutamic acid decarboxylase. J Biol Chem. 1995;270:2241–2246. doi: 10.1074/jbc.270.5.2241. [DOI] [PubMed] [Google Scholar]
- 12.Dragic T, Litwin V, Allaway G, Martin S, Huang Y, Nagashima K, Cayanan C, Maddon P, Koup R, Moore J, Paxton W. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 13.Falzoni S, Munerati M, Ferrari D, Spisani S, Moretti S, DiVirgilio F. The purinergic P2Z receptor of human macrophage cells. Characterization and possible physiological role. J Clin Invest. 1995;95:1207–1216. doi: 10.1172/JCI117770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Y, Broder C, Kennedy P, Berger E. HIV-1 entry: functional cDNA cloning of a seven transmembrane, G-protein coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 15.Fujioka Y, Matozaki T, Noguchi T, Iwamatsu A, Yamao T, Takahashi N, Tsuda M, Takada Y, Kasuga M. A novel membrane glycoprotein, SHPS-1, that binds the SH2-domain-containing protein tyrosine phosphatase SHP-2 in response to mitogens and cell adhesion. Mol Cell Biol. 1996;16:6887–6899. doi: 10.1128/mcb.16.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harter C, James P, Bachi T, Semenza G, Brunner J. Hydrophobic binding of the ectodomain of influenza hemagglutinin to membrane occurs through the “fusion peptide.”. J Biol Chem. 1989;264:6459–6464. [PubMed] [Google Scholar]
- 17.Hernandez L, Hoffman L, Wolfsberg T, White J. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 18.Higuchi S, Tabata N, Tajima M, Ito M, Tsurudome M, Sudo A, Uchida A, Ito Y. Induction of human osteoclast-like cells by treatment of blood monocytes with anti-fusion regulatory protein-1/CD98 monoclonal antibodies. J Bone Miner Res. 1998;13:44–49. doi: 10.1359/jbmr.1998.13.1.44. [DOI] [PubMed] [Google Scholar]
- 19.Hildreth J, Orentas R. Involvement of a leukocyte adhesion receptor (LFA-1) in HIV-induced syncyium formation. Science. 1989;244:1075–1078. doi: 10.1126/science.2543075. [DOI] [PubMed] [Google Scholar]
- 20.Huynh M. In: DNA cloning: a practical approach. Glover D M, editor. Vol. 1. Oxford, England: IRL Press; 1985. [Google Scholar]
- 21.Ito Y, Komada H, Kusagawa S, Tsurudome M, Matsumura H, Kawano M, Ohta H, Nishio M. Fusion regulation proteins on the cell surface: isolation and characterization of monoclonal antibodies which enhance giant polykaryocyte formation in Newcastle disease virus-infected cell lines of human origin. J Virol. 1992;66:5999–6007. doi: 10.1128/jvi.66.10.5999-6007.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kazazi F, Chang J, Lopez A, Vaas M, Cunningham A. Interleukin 4 and human immunodeficiency virus stimulate LFA-1-ICAM-1-mediate aggregation of monocytes and subsequent giant cell formation. J Gen Virol. 1994;75:2795–2802. doi: 10.1099/0022-1317-75-10-2795. [DOI] [PubMed] [Google Scholar]
- 23.Kharitonenkov A, Chen Z, Sures I, Wang H, Schilling J, Ullrich A. A family of proteins that inhibit signaling through tyrosine kinase receptors. Nature. 1997;386:181–186. doi: 10.1038/386181a0. [DOI] [PubMed] [Google Scholar]
- 24.Kurachi T, Morita I, Murota S. Involvement of adhesion molecules LFA-1 and ICAM-1 in osteoclast development. Biochim Biophys Acta. 1993;1178:259–266. doi: 10.1016/0167-4889(93)90202-z. [DOI] [PubMed] [Google Scholar]
- 25.Lanier L, Phillips J. Inhibitory MHC class I receptors on NK cells and T cells. Immunol Today. 1996;17:86–91. doi: 10.1016/0167-5699(96)80585-8. [DOI] [PubMed] [Google Scholar]
- 26.Lee N, Weinstock K, Kirkness E, Earle-Hughes J, Fuldner R, Marmaras S, Glodek A, Gocayne J, Adams M, Kerlavage A, Fraser C, Venter J. Comparative expressed sequence tag analysis of differential gene expression profiles in PC-12 cells before and after nerve growth factor treatment. Proc Natl Acad Sci USA. 1995;92:8303–8307. doi: 10.1073/pnas.92.18.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mbalaviele G, Chen H, Boyce B, Mundy G, Yoneda T. The role of cadherin in the generation of multinucleated osteoclasts from mononuclear precursors in murine marrow. J Clin Invest. 1995;95:2757–2765. doi: 10.1172/JCI117979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mege R, Goudou D, Diaz C, Nicolet M, Garcia L, Geraud L, Rieger F. N-cadherin and N-CAM in myoblast fusion: compared localisation and effect of blockade by peptides and antibodies. J Cell Sci. 1992;103:897–906. doi: 10.1242/jcs.103.4.897. [DOI] [PubMed] [Google Scholar]
- 29.Most J, Neumayer H, Dierich M. Cytokine induced generation of multinucleated giant cells in vitro requires interferon-γ and expression of LFA-1. Eur J Immunol. 1990;20:1661–1667. doi: 10.1002/eji.1830200807. [DOI] [PubMed] [Google Scholar]
- 30.Ohgimoto S, Tabata N, Suga S, Nishio M, Ohta H, Tsurudome M, Komada H, Kowano M, Watanabe N, Ito Y. Molecular characterization of fusion regulatory protein-1 (FRP-1) that induces multinucleate giant cell formation of monocytes and HIV gp-160 mediated cell fusion. J Immunol. 1995;155:3585–3592. [PubMed] [Google Scholar]
- 31.Ohnishi H, Kubota M, Ohtake A, Sato K, Sano S. Activation of protein-tyrosine phosphatase SH-PTP2 by a tyrosine-based activation motif of a novel brain molecule. J Biol Chem. 1996;271:25569–25574. doi: 10.1074/jbc.271.41.25569. [DOI] [PubMed] [Google Scholar]
- 32.Ohta H, Tsurudome M, Matsumura H, Koga Y, Morikawa S, Kawano M, Kusugawa S, Komada H, Nishio M, Ito Y. Molecular and biological characterization of fusion regulatory proteins (FRPs): anti-FRP mAbs induced HIV mediated cell fusion via an integrin system. EMBO J. 1994;13:2044–2055. doi: 10.1002/j.1460-2075.1994.tb06479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pawson A, Schlessinger J. SH2 and SH3 domains. Curr Biol. 1993;3:434–442. doi: 10.1016/0960-9822(93)90350-w. [DOI] [PubMed] [Google Scholar]
- 34.Primakoff P, Hyatt H, Kline J. Identification and purification of a sperm surface protein with a potential role in sperm-egg membrane fusion. J Cell Biol. 1987;104:141–149. doi: 10.1083/jcb.104.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saginario C, Qian H Y, Vignery A. Identification of an inducible surface molecule specific to fusing macrophages. Proc Natl Acad Sci USA. 1995;92:12210–12214. doi: 10.1073/pnas.92.26.12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sano S, Ohnishi H, Omori A, Hasegawa J, Kubota M. BIT, an immune antigen receptor-like molecule in the brain. FEBS Lett. 1997;411:327–334. doi: 10.1016/s0014-5793(97)00724-2. [DOI] [PubMed] [Google Scholar]
- 37.Scott P, Trinchieri G. The role of natural killer cells in host-parasite interactions. Curr Opin Immunol. 1995;7:34–40. doi: 10.1016/0952-7915(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 38.Stegmann T, White J, Helenius A. Intermediates in influenza induced membrane fusion. EMBO J. 1990;13:4231–4241. doi: 10.1002/j.1460-2075.1990.tb07871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabata N, Ito M, Shimokata K, Suga S, Ohgimoto S, Tsurudome M, Kawano M, Matsumura H, Komada H, Nishio M, Ito Y. Expression of fusion regulatory proteins (FRPs) on human peripheral blood monocytes. J Immunol. 1994;153:3256–3266. [PubMed] [Google Scholar]
- 40.Vignery A. Macrophage multinucleation is accompanied by the expression of new soluble and membrane antigens in mice. Am J Pathol. 1989;135:565–570. [PMC free article] [PubMed] [Google Scholar]
- 41.Vignery A, Niven-Fairchild T, Ingbar D, Caplan M. Polarized distribution of Na+K+-ATPase in giant cells elicited in vivo and in vitro. J Histochem Cytochem. 1989;37:1265–1271. doi: 10.1177/37.8.2546991. [DOI] [PubMed] [Google Scholar]
- 42.Vignery A, Wang F, Qian H-Y, Benz E, Gilmore-Hebert M. Detection of the Na,K-ATPase α3 isoform in multinucleated macrophages. Am J Physiol. 1991;260:F704–F709. doi: 10.1152/ajprenal.1991.260.5.F704. [DOI] [PubMed] [Google Scholar]
- 43.Vignery A, Raymond M, Qian H, Wang F, Rosenzweig S. Multinucleated rat alveolar macrophages express functional receptors for calcitonin. Am J Physiol. 1991;260:1026–1032. doi: 10.1152/ajprenal.1991.261.6.F1026. [DOI] [PubMed] [Google Scholar]
- 44.Vivier E, Daeron M. Immunoreceptor tyrosine based inhibition motifs. Immunol Today. 1997;18:286–292. doi: 10.1016/s0167-5699(97)80025-4. [DOI] [PubMed] [Google Scholar]
- 45.White J, Helenius A, Gething M. Hemagglutinin of influenza virus expressed from a cloned gene promotes membrane fusion. Nature. 1982;300:658–659. doi: 10.1038/300658a0. [DOI] [PubMed] [Google Scholar]
- 46.Wiley D C, Skehel J J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 47.Williams A, Barclay N. The immunoglobulin superfamily-domains for cell surface recognition. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 48.Yagami-Hiromasa T, Sato T, Kurisaki T, Kamijo K, Nabeshima Y, Fujisawa-Sehara A. A metalloprotease-disintegrin participating in myoblast fusion. Nature. 1995;377:652–656. doi: 10.1038/377652a0. [DOI] [PubMed] [Google Scholar]