Abstract
Primary human fibroblasts arrest growth in response to the inhibition of mitosis by mitotic spindle-depolymerizing drugs. We show that the mechanism of mitotic arrest is transient and implicates a decrease in the expression of cdc2/cdc28 kinase subunit Homo sapiens 1 (CKsHs1) and a delay in the metabolism of cyclin B. Primary human fibroblasts infected with a retroviral vector that drives the expression of a mutant p53 protein failed to downregulate CKsHs1 expression, degraded cyclin B despite the absence of chromosomal segregation, and underwent DNA endoreduplication. In addition, ectopic expression of CKsHs1 interfered with the control of cyclin B metabolism by the mitotic spindle cell cycle checkpoint and resulted in a higher tendency to undergo DNA endoreduplication. These results demonstrate that an altered regulation of CKsHs1 and cyclin B in cells that carry mutant p53 undermines the mitotic spindle cell cycle checkpoint and facilitates the development of aneuploidy. These data may contribute to the understanding of the origin of heteroploidy in mutant p53 cells.
The onset of each phase of the cell cycle depends on the completion of the previous phase. These events are coordinated by cell cycle checkpoints (19). Cell cycle checkpoints are pathways that ensure the timely progression of the cell cycle, and they play a crucial role in the maintenance of genomic integrity by coordinating the cell cycle regulatory machinery with DNA repair and cell death pathways. At mitosis, the mitotic spindle cell cycle checkpoint prevents the onset of anaphase, the actual segregation of chromosomes, if the integrity of the mitotic spindle is compromised. Recent data have suggested a role for the p53 tumor suppressor gene product in the control of cell ploidy. Fibroblasts isolated from individuals with Li-Fraumeni syndrome have a marked tendency to become heteroploid in culture (5). These individuals are born with heterozygous mutations in the p53 tumor suppressor gene (39, 57). Heteroploidy is also commonly found in p53 null cells in culture (27) and in vivo (13) in p53 knockout mice. Intriguingly, overexpression of a mutant p53 protein on a p53 null background accelerated the appearance of polyploidy in a myelomonocytic cell line (50). Also, the expression of mutant p53 proteins in human colon carcinoma cells and murine cell lines causes karyotypic abnormalities, including an increase in ploidy levels during growth in culture (1). We have seen that Li-Fraumeni syndrome fibroblasts that carry heterozygous structural p53 mutant proteins progress to polyploidy when incubated in the presence of mitotic spindle inhibitors (22). However, normal human fibroblasts, p53 null Li-Fraumeni fibroblasts, and normal human fibroblasts infected with a retrovirus that expresses the human papillomavirus 16 E6, which binds to and promotes the degradation of p53, arrest growth when incubated in the presence of mitotic inhibitors (22). Progression to polyploidy in E6-expressing human fibroblasts, however, has been reported by others (16). In agreement with our previous results, Lanni and Jacks have recently reported that p53 null mouse fibroblasts have a normal mitotic spindle checkpoint (35). However, these fibroblasts may progress to polyploidy due to the inactivation of a p53-dependent postmitotic checkpoint (35). In addition, inactivation of wild-type p53 by the overexpression of a truncated (C terminus) p53 protein in a murine prolymphocytic cell line led to polyploidy (41).
The cell cycle G2/M transition and progression through mitosis is driven by the kinase activity of a complex referred to as maturation- or M-phase-promoting factor (MPF). This complex consists of a catalytic subunit (34-kDa cyclin-dependent kinase, cdc2), a regulatory subunit (cyclin B proteins), and associated proteins (47). Entry into mitosis requires MPF activation, a process that depends upon an increase in cyclin B expression and the dephosphorylation of cdc2. Progression through mitosis and cytokinesis requires the subsequent inactivation of MPF, which depends in part on cyclin B degradation. Experiments with yeast indicate that the mitotic spindle cell cycle checkpoint feeds into the cell cycle regulatory machinery at mitosis by a pathway that delays the degradation of cyclin B and maintains cdc2 kinase activity (3). Thus, cyclin B is degraded and MPF is inactivated only after certain aspects of mitosis related to spindle assembly and disassembly are properly completed. The activity of the p34cdc2-cyclin B complex is thought to be regulated by its interaction with other proteins (47). We have focused our attention on two low-molecular-weight proteins known as cdc2/cdc28 kinase subunit Homo sapiens, CKsHs1 and CKsHs2 (53). These proteins were previously identified as the human homologs of the small cdc28- and cdc2-associated proteins of Saccharomyces cerevisiae, Cks1, and Schizosaccharomyces pombe, Suc1 (53). CKsHs1 and CKsHs2 bind the cyclin B-cdc2 complex (53). In S. pombe, inactivation of suc1 causes cells to arrest in mitosis with high levels of Cdc13 (the S. pombe cyclin B homolog) and high MPF kinase activity (4, 42). Thus, suc1 inactivation mimics the effects of the mitotic spindle cell cycle checkpoint on cyclin B levels and MPF activity. It has been proposed that Cks family members may play a critical role in the mitotic spindle cell cycle checkpoint (54).
We show that primary human fibroblasts arrest growth in response to the inhibition of mitosis by mitotic spindle-depolymerizing drugs. This growth arrest was transient and was accompanied by a delay in the metabolism of cyclin B and a transient decrease in the expression of CKsHs1. Inactivation of p53 by the expression of the human papillomavirus 16 protein E6 did not affect the effect of a mitotic inhibitor on cyclin B metabolism. By contrast, primary human fibroblasts expressing a dominant mutant p53 protein responded very differently to mitotic inhibition: they failed to downregulate CKsHs1 expression, degraded cyclin B despite the absence of chromosomal segregation, and underwent DNA endoreduplication. These results show that in human cells, mutant p53 proteins abrogate the mitotic spindle cell cycle checkpoint by interfering with the regulation of CKsHs1 expression and cyclin B metabolism at mitosis. Consistent with this conclusion, we further show that ectopic expression of CKsHs1 abrogates the control of cyclin B metabolism by the mitotic spindle cell cycle checkpoint pathway and facilitates the development of polyploidy. These data demonstrate that CKsHs1 plays a key role in the control of mitosis in human cells and that altered expression of CKsHs1 in human cells carrying mutant p53 proteins contributes to the development of aneuploidy.
MATERIALS AND METHODS
Plasmids and cell culture.
The expression vectors pBabe CKsHs1 and pBabe CKsHs2 were created by subcloning CKsHs1 and CKsHs2 cDNA fragments into the retroviral vector pBabe, which contains a puromycin selectable marker (42a). CKsHs1 and CKsHs2 cDNA fragments were cloned by reverse transcription PCR (RT-PCR) with primers 5′-AGAGCGATCATGTCGCACAAACAA-3′ and 5′-TCATTTCTTTGGTTTCTTGGGTAG-3′ (CKsHs1) and primers 5′-ACGAGGATGGCCCACAAGCAGATCTACTAC-3′ and 5′-TTTTTGTTGATCTTTTGGAAGAGG-3′ (CKsHs2) and 1 μg of poly(A) mRNA isolated from neonatal human foreskin fibroblasts (NHF). CKsHs cDNA sequences were then subcloned into the pBabe snaB1 site. Plasmid construction was verified by DNA sequencing. The retroviral expression vector pBabe p53 143A was a gift from J. Jacobberger, Case Western Reserve University (Cleveland, Ohio), and was generated by the subcloning of a p53 mutant 143A cDNA fragment formed by BamHI digestion of plasmid CMV-p53 143A into the pBabe BamHI site (33). The murine sarcoma virus long terminal repeat-based p53 expression vectors were a gift from C. Finlay, GlaxoWellcome (Research Triangle Park, N.C.) (21). The retroviral vectors LXSN and LXSN-E6 were a gift from D. A. Galloway, Fred Hutchinson Cancer Research Center (Seattle, Wash.) (26). The CMV-bcl2 plasmid was a gift from M. W. Mayo and A. S. Baldwin, University of North Carolina Lineberger Comprehensive Cancer Center (Chapel Hill, N.C.).
NHF, C2C12, and 10T1/2 cells were incubated in Dulbecco’s modified Eagle’s medium (DMEM) plus 10% dialyzed fetal bovine serum (FBS; GIBCO) supplemented with penicillin (10 U/ml) and streptomycin (10 U/ml). NHF were a gift from C. Muro-Cacho, University of South Florida (Tampa, Fla.). C2C12 and 10T1/2 cells were a gift from K. Guo, Rhone Poulenc Rorer (Philadelphia, Pa.). NHF were incubated in Polybrene-supplemented medium obtained from PA317 cells infected with the retroviral vector pBabe, pBabe-p53 143A, or pBabe-CKsHs1. Selection was carried out by incubation in media supplemented with 3 μg of puromycin (Sigma) per ml. NHF pBabe/LXSN, NHF pBabe/LXSN-E6, and NHF pBabe 143A/LXSN-E6 cells were generated by coinfection of primary NHF with the corresponding pBabe- and LXSN-based vectors followed by double selection in puromycin (3 μg/ml)- and G418 (400 μg/ml)-supplemented media. C2C12-pBabe and C2C12-pBabe 143A cells were generated by transfection of pBabe or pBabe p53 143A followed by selection in puromycin-supplemented medium. C2C12-bcl2/pBabe and C2C12-bcl2/pBabe p53 143A cells were generated by cotransfection of C2C12 cells with CMVneo-bcl2 and pBabe or pBabe p53 143A and double selection in puromycin (3 μg/ml)- and G418 (500 μg/ml)-supplemented media. Control 10T1/2 cells (10T1/2 pBabe) and 10T1/2 cells overexpressing CKsHs1 or CKsHs2 were generated by transfection of 10T1/2 cells with the respective pBabe-based vector followed by selection in puromycin-supplemented medium. Cell populations were analyzed at passages 1 to 3 after drug selection.
Analysis of mitotic spindle cell cycle checkpoint status.
Cells were analyzed by a modification of the technique described previously (22). When both total DNA content and newly synthesized DNA were determined, the cells were labeled with 10 μM bromodeoxyuridine (BrdU) for 4 h, trypsinized, counted, and fixed with 70% ethanol. Fixed cells were centrifuged and treated with 0.08% pepsin for the preparation of nuclei. The nuclear pellet was resuspended in 100 μl of a 1:5 dilution of anti-BrdU fluorescein isothiocyanate-conjugated antibody (Becton Dickinson), incubated for 30 min, washed, stained with 50 μg of propidium iodide (Aldrich Chemical Co.) per ml, and analyzed by flow cytometry for cell cycle distribution of the DNA content. For sorting, cells were stained with Hoechst 33342 (Sigma) at a final concentration of 2.0 μg/ml in culture medium at 37°C for 1 h. Flow cytometry was carried out with a Coulter Elite ESP flow cytometer and analyzed with CellQuest software (Becton Dickinson).
Immunoprecipitations and Western analysis.
Antibodies were purchased from Santa Cruz Biotech, except for anti-β-actin, which was from Sigma. In immunoprecipitation studies, 2 × 107 human fibroblasts were washed twice for 10 min in 10 ml of methionine-free DMEM and incubated for 4 h at 37°C in 5 ml of new medium supplemented with 2.5 mCi of [35S]methionine (1,175 Ci/mmol; NEN). The cells were collected by centrifugation, lysed in 1 ml of immunoprecipitation buffer (phosphate-buffered saline containing 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 1 mM sodium orthovanadate, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride), and subjected to centrifugation at 1,500 × g for 10 min. Cell lysates were incubated in the presence of 1 μg of the indicated antibody at 4°C for 4 h and then subjected to incubation for 1 h with protein A/G-agarose (Santa Cruz). Immunoprecipitates were collected by centrifugation at 5,000 × g for 10 min, washed twice with 250 μl of immunoprecipitation buffer, and resuspended in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer, boiled for 5 min, and subjected to SDS-PAGE (15% polyacrylamide). Gels were exposed to a PhosphorImager screen and analyzed with ImageQuant software (Molecular Dynamics).
For Western analysis, cells were harvested, lysed in 1 ml of lysis buffer (phosphate-buffered saline containing 1% Triton X-100, 0.1% SDS, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride), and subjected to centrifugation at 1,500 × g for 5 min. Equal amounts of proteins were assayed under each set of conditions as determined by the Bradford protein assay (Bio-Rad). SDS-PAGE was carried out at a 30-mA constant current in a 15% polyacrylamide gel (Bio-Rad). Proteins were transferred to Immobilon-P membranes (Millipore) and probed as recommended by the manufacturer. Anti-p53 (DO-1), anti-bcl2, anti-CKsHs1, and anti-β-actin antibodies were used (unless otherwise indicated) at dilutions of 1:500, 1:1,000, 1:100 and 1:10,000, respectively, in phosphate-buffered saline–5% dry milk. The membranes were hybridized overnight at 4°C. For detection, the membranes were incubated for 1 h in a 1:10,000 or 1:5,000 dilution of horseradish peroxidase-linked anti-mouse or anti-rabbit immunoglobulin G, respectively (Santa Cruz). Horseradish peroxidase luminescence reactions were carried out with the ECL kit (Amersham). The membranes were exposed to Hyperfilm (Kodak), and protein bands were detected by autoradiography. Low-exposure autoradiographs were scanned with an LKB densitometer to determine peak areas.
Northern analysis.
RNA was isolated from human fibroblasts, C2C12 cells, or 10T1/2 cells with Trizol reagent (GIBCO). For Northern analysis, 30 μg of total RNA was resolved in a 1.3% agarose–formaldehyde gel, visualized with ethidium bromide, transferred to nitrocellulose filters (Amersham), fixed by UV cross-linking, and baked at 80°C for 1 h. For hybridizations, 106 cpm of a random-primed 32P-labeled (Boehringer Mannheim) CKsHs1 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA fragment per ml was used as a probe. Hybridizations were carried out as described previously (52). The membranes were exposed to Hyperfilm (Kodak), and RNA bands were detected by autoradiography.
RESULTS
Expression of a dominant mutant p53 protein in primary NHF abrogates a transient mitotic spindle cell cycle checkpoint.
To study the molecular mechanisms underlining the effects of mutant p53 on mitosis in human cells, we created cell populations of mutant-p53-expressing primary NHF by the stable expression of a dominant p53 mutant protein in these cells, using a retroviral vector (pBabe p53 143A). Control cells were infected with an insertless retroviral vector (pBabe). Stable expression of the mutant p53 protein in NHF was demonstrated by metabolic labeling and immunoprecipitation with an anti-mutant p53-specific antibody (Fig. 1A).
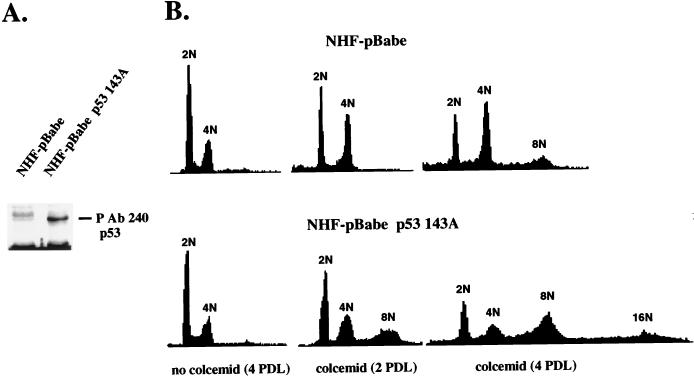
FIG. 1.
(A) Immunoprecipitation of mutant p53 with antibody P Ab 240 in primary NHF infected with vector pBabe (NHF-pBabe) or vector pBabe p53 143A (NHF-pBabe p53 143A). The cells were incubated with radiolabeled methionine, harvested, and lysed, P Ab 240-reactive p53 was immunoprecipitated, and the immunoprecipitates were resolved by SDS-PAGE and exposed to a PhosphorImager screen. (B) Analysis of the cell cycle distribution of the DNA content in NHF-pBabe and NHF-pBabe p53 143A cells. Cells were incubated in the absence or presence of 200 ng of colcemid per ml for two to four population doubling times (PDL). The population doubling times for NHF-pBabe and NHF-pBabe p53 143A cells were 48 and 42 h, respectively. Following incubations, the cells were harvested and processed for flow cytometric determination of DNA content as indicated in Materials and Methods. Polyclonal populations at passage 2 were assayed. The data are representative of at least three independent experiments.
Control and mutant p53-expressing cell populations were analyzed for the integrity of the mitotic spindle cell cycle checkpoint. The cells were incubated in the presence of the mitotic spindle-depolymerizing drug colcemid or nocodazol. Mitotic arrest in the control cells and S-phase reentry and generation of cells with an octaploid DNA (8N) content in the mutant-p53 cell population were observed (Fig. 1B, colcemid 2 PDL). Polyploid DNA content was seen in mutant-p53-expressing cells incubated with a mitotic inhibitor (colcemid or nocodazol) but not with a cell cycle S-phase inhibitor (thymidine or hydroxyurea [results not shown]). When both cultures were incubated with colcemid for up to four population doubling times, a significant level of polyploidy was observed not only in the mutant-p53-expressing cells but also in the control cells (Fig. 1B, NHF pBabe, colcemid 4 PDL, 12% of 8N cells). Nevertheless, the proportion of cells with a polyploid DNA content was always higher in the mutant-p53-expressing cell population (Fig. 1B, NHF pBabe-p53 143A, colcemid 4 PDL, 28% of 8N cells and 4% of 16N cells). The presence of polyploidy in these cells was confirmed by counting chromosomes in metaphase spreads (results not shown). These results extend our previous observations with mutant-p53-expressing fibroblasts (22) and suggest that the activation of the mitotic spindle cell cycle checkpoint in human cells does not result in permanent growth arrest but in transient delay of mitosis. We hypothesized that the cell population carrying a mutant p53 protein accumulated polyploid cells at a higher rate than normal cells did because mutant p53 proteins abrogate a transient mechanism of mitotic arrest.
The results shown in Fig. 1 are consistent with an alternative mechanism. These cells were obtained from biopsy specimens of human skin. Therefore, they are not clonal in origin, and so polyploidy could be originated by the growth of a subpopulation of cells with an inactive mitotic spindle cell cycle checkpoint, namely, mitotic checkpoint-negative cells. In this scenario, the effect of mutant p53 could be but an increase in the fraction of mitotic checkpoint-negative cells rather than the abrogation of the mitotic spindle checkpoint. This hypothesis was discarded on the basis of experiments in which NHF arrested by colcemid at the G2/M boundary (mitotic checkpoint-positive cells) were sorted, grown, and then incubated for four population doubling times in the presence of colcemid. Again, the generation of polyploid cells was observed (data not shown). These experiments support the conclusion that NHF arrest growth in response to an anomalous chromosomal segregation but that this growth arrest is transient and may be abrogated by the expression of a dominant mutant p53 protein.
Unscheduled degradation of cyclin B protein in NHF carrying a mutant p53 protein.
Work by Kung et al. demonstrated that the mitotic spindle cell cycle checkpoint regulates the progression through mitosis in mammalian cells by means of a tight control of cyclin B metabolism (34). Activation of the mitotic spindle cell cycle checkpoint blocks cyclin B degradation and causes sustained levels of cyclin B protein. To elucidate the mechanisms underlying the generation of polyploidy in mutant-p53-expressing NHF, we analyzed the ability of control and p53-expressing NHF to regulate cyclin B turnover in response to mitotic inhibition. Both cell groups were made quiescent and then stimulated to enter the cell cycle synchronously in the presence or absence of colcemid. Cell extracts were prepared at various intervals after stimulation (16 to 152 h) and processed for Western analysis of cyclin B.
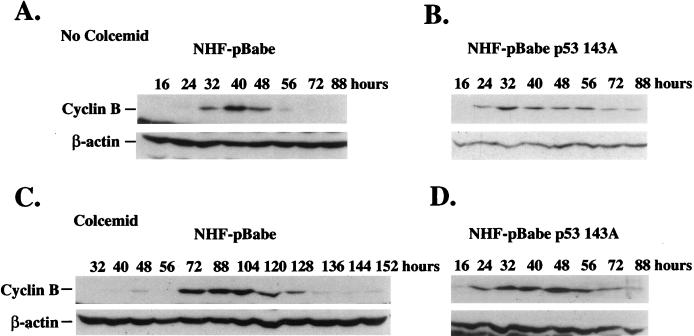
In the absence of colcemid, cyclin B protein levels oscillated similarly in control and mutant-p53-expressing cells (Fig. 2A and B). Cyclin B protein was first detected at 28 h after stimulation; its level reached a maximum at 32 to 40 h and decreased by 56 to 72 h; the level decreased sharply in control cells and at a variable rate in different experiments in mutant-p53-expressing cells (results not shown), suggesting a quick desynchronization of the latter cell group. By contrast, when cells were incubated in the presence of colcemid, a differential ability to regulate cyclin B turnover was observed (Fig. 2C and D). Control fibroblasts accumulated cyclin B protein at 72 h after stimulation, a 40-h delay relative to the situation for control fibroblasts incubated in the absence of the drug. Cyclin B levels remained elevated for up to 128 h and then decreased sharply. Thus, in response to a spindle-depolymerizing agent, NHF delayed cyclin B degradation for approximately 72 h. However, when mutant-p53-expressing cells were incubated in colcemid, the cyclin B levels rose at 32 h and declined by 88 h, an 8-h delay relative to the situation for untreated cells. Similar results were obtained with another two human cell populations infected with the pBabe or pBabe 143A vectors. These results show that mutant-p53-expressing fibroblasts fail to regulate cyclin B levels in response to mitotic spindle depolymerization. These results underscore the role played by the control cyclin B metabolism at the mitotic spindle checkpoint in human cells. Moreover, since cyclin B metabolism eventually proceeded in control cells, these experiments supported the conclusion that the mitotic spindle checkpoint arrest is transient. The accelerated cyclin B degradation in mutant-p53-expressing NHF was accompanied by S-phase reentry as determined by flow cytometry of DNA content (results not shown). Mutant p53 protein levels did not change during the mitotic progression (results not shown).
FIG. 2.
Western analysis of cyclin B and β-actin in NHF carrying the control retroviral vector (NHF-pBabe) or a retroviral vector containing mutant 143A p53 sequences (NHF-pBabe p53 143A). Confluent cell cultures (4 × 104 to 5 × 104 cells/cm2) were synchronized by a 2-day incubation in low-serum medium (0.5% calf serum), incubated at low density (1 × 104 to 2 × 104 cells/cm2) in 10% FBS in the absence (A and B) or presence (C and D) of 200 ng of colcemid per ml, and harvested at the indicated intervals. Colcemid was added at 12 h after cell passage. Western blotting was carried out as indicated in Materials and Methods. Data are representative of three independent experiments.
NHF expressing the human papillomavirus type 16 (HPV16) E6 oncoprotein regulate cyclin B metabolism in response to the inhibition of mitosis.
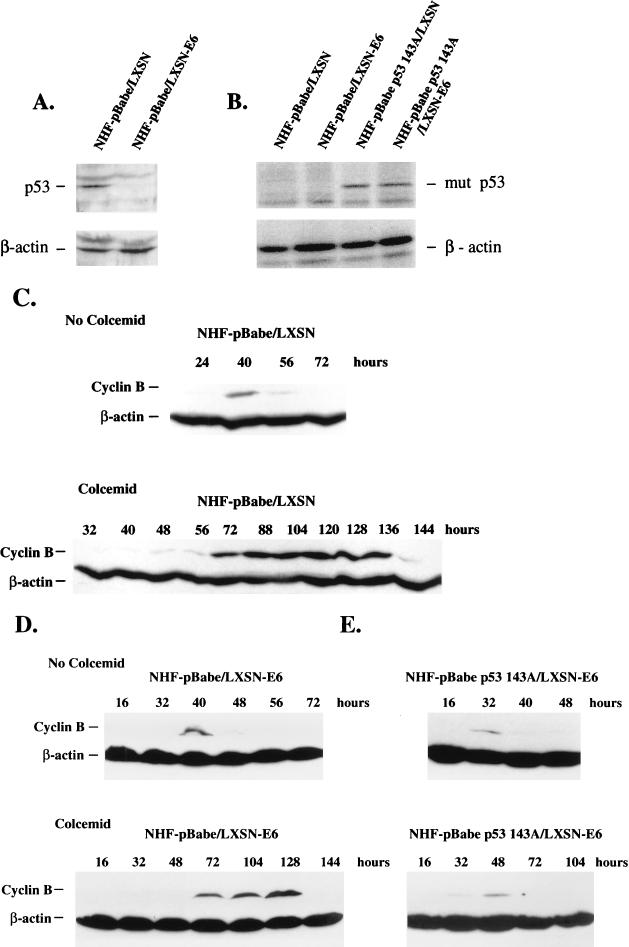
Mutant p53 proteins may exert dominant negative effects on wild-type p53 functions. Alternatively, mutant p53 proteins may exhibit properties that are not originated by the inactivation of wild-type p53 (gain-of-function) (36). Thus, expression of a mutant p53 protein in NHF may interfere with putative wild-type p53-dependent or -independent mechanisms of checkpoint control at mitosis. To distinguish between these two possibilities, we created a functional knockout of wild-type p53 in primary NHF by the expression of the human papillomavirus type 16 E6 oncoprotein in these cells with the retroviral vector LXSN (26). Several experimental controls were introduced in these series of experiments. Primary NHF were coinfected with LXSN (control empty vector) and pBabe (control empty vector) or with LXSN-E6 and pBabe p53 143A. Cells infected with LXSN-E6 were also coinfected with pBabe. The status of p53 in these cell populations is shown in Fig. 3A and B. HPV16 E6 promoted selectively the degradation of wild-type but not mutant p53. Therefore, NHF pBabe p53 143A/LXSN-E6 showed a dramatic decrease in wild-type p53 levels but expressed similar levels of mutant p53 protein to those found in the NHF pBabe p53 143A/LXSN population. Primary NHF coinfected with both control retroviral vectors demonstrated an 80-h delay in cyclin B metabolism in response to colcemid (Fig. 3C). We then compared the ability of the NHF pBabe/LXSN-E6 and NHF pBabe p53 143A/LXSN-E6 to regulate cyclin B metabolism in response to the mitotic inhibitor. E6-expressing NHF delayed cyclin B metabolism in response to colcemid in a similar way to the control population, i.e., by approximately 80 h (Fig. 3C and D). However, the cell population coexpressing E6 and p53 143A showed a much shorter mitotic pause, i.e., 16 h, in response to the mitotic inhibitor (Fig. 3E), similar to what we previously observed in mutant-p53-expressing cells (Fig. 2B and C). A second E6-expressing NHF population also displayed a prolonged delay in cyclin B metabolism when incubated in the presence of colcemid (results not shown). These experiments suggest that expression of a mutant p53 protein, but not wild-type p53 inactivation, abrogates the ability of NHF to regulate the metabolism of cyclin B protein in response to an anomalous chromosomal segregation (see Discussion). This hypothesis is in agreement with recent data by Lanni and Jacks that shows similar mitotic delays in normal and p53 null mouse embryo fibroblasts challenged by a mitotic inhibitor (35). These results do not contradict the fact that p53 inactivation may facilitate an increase in the fraction of polyploid cells in a given cell population by a different mechanism, such as the abrogation of a postmitotic p53-dependent cell death response (35, 41).
FIG. 3.
(A) Western analysis of p53 and β-actin in NHF carrying the retroviral vectors pBabe (puromycin resistance, no insert) and LXSN (neomycin resistance, no insert) or LXSN-E6. For the induction of p53, the cells were incubated for 2 days in 10 μM mycophenolic acid (a GMP biosynthesis inhibitor). (B) Immunoprecipitation of mutant p53 and β-actin in primary NHF following double infection with the retroviral vector pBabe and LXSN or LXSN-E6 or with the vector pBabe p53 143A and LXSN or LXSN-E6. The cells were metabolically labeled and the proteins were analyzed as in Fig. 1. (C to E) Western analysis of cyclin B and β-actin in NHF carrying the retroviral vectors pBabe and LXSN (C), pBabe and LXSN-E6 (D), or pBabe p53 143A and LXSN-E6 (E). The cells were incubated and Western blotting was carried out as in Fig. 2. Data are representative of three independent experiments.
Mutant p53 interferes with the regulation of CKsHs1 expression.
The metabolism of cyclin B is regulated in part by proteins of the Cks family, which interact with the cyclin B-cdc2 complex (42, 49). Two human homologs of these proteins, CKsHs1 and CKsHs2, have been isolated from HeLa cells (53). Using RT-PCR and oligonucleotide primers designed from the reported CKsHs1 and CKsHs2 sequences (53), we cloned the cDNAs encoding these proteins in NHF. Sequence analysis demonstrated no differences from the previously reported HeLa cDNAs (results not shown). Using Northern analysis, we determined the level of expression of CKsHs1 and CKsHs2 transcripts in confluent cultures of control and mutant-p53-expressing cells. Higher levels of CKsHs1 expression were found in mutant-p53-expressing NHF than in their isogenic controls (Fig. 4A). No significant differences in CKsHs2 expression were detected (results not shown). Similar results were obtained by RT-PCR (results not shown).
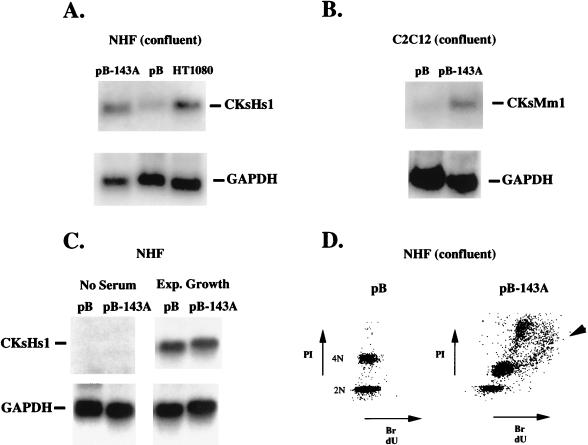
FIG. 4.
(A) Northern analysis of CKsHs1 and GAPDH expression in NHF-pBabe (pB), NHF-pBabe p53 143A (pB-143A), and HT1080 cells. HT1080 is a fibrosarcoma cell line that carries two mutated p53 alleles. The cells were incubated in DMEM with 10% FBS until confluent (approximately 4 × 104 to 5 × 104 cells/cm2) and processed for Northern analysis. A CKsHs1 sequence obtained by RT-PCR from NHF was subcloned into Bluescribe (Stratagene), sequenced, and used as a probe. RNA integrity was verified by reprobing with a GAPDH sequence (American Type Culture Collection). Other experimental details were as indicated in Materials and Methods. (B) Northern analysis of the expression of a murine homolog of CKsHs1 (CKsMm1) and GAPDH in C2C12-pBabe (pB) and C2C12-pBabe p53 143A (pB-143A) cells. The cells were incubated as in panel A. (C) Northern analysis of CKsHs1 and GAPDH expression in quiescent or exponentially growing NHF-pBabe (pB) and NHF-pBabe p53 143A (pB-143A) cells. The cells were incubated for 2 days at confluence in DMEM with 0.5% calf serum (No Serum) or for 2 days at low density (1 × 104 to 2 × 104 cells/cm2) in DMEM with 10% FBS (Exp. Growth). (D) Flow-cytometric analysis of the cell cycle distribution of the DNA content in NHF-pBabe (pB) and NHF-pBabe p53 143A (pB-143A) cells at confluence. The cells were incubated for 2 days at confluence (4 × 104 to 5 × 104 cells/cm2) in DMEM with 10% FBS, 100 μM BrdU was added, and the cells were incubated for an additional 4 h. The cultures were harvested, fixed, and processed for flow cytometry as indicated in Materials and Methods.
As an additional control, C2C12 myoblasts, which contain only wild-type p53 (56), were stably transfected with plasmids pBabe (no insert) or pBabe p53 143A, the cells were incubated as described above, and the level of the murine homolog of CKsHs1 was determined by Northern analysis. Consistent with our findings in human cells, the expression of the murine CKsHs1 transcript (CKsMm1 [Fig. 4B]) was higher in cells carrying mutant p53.
To further characterize the expression of CKsHs1 in the control and mutant-p53-expressing NHF, we performed Northern analysis of CKsHs1 in these cells incubated under quiescent or exponential growth conditions. Incubation of confluent cell cultures for 2 days in medium containing 0.5% calf serum resulted in no detectable CKsHs1 expression in either cell group (Fig. 4C, No Serum). On the other hand, incubation of control and mutant-p53-expressing cells at low density and with high concentrations of serum resulted in similar levels of CKsHs1 expression in both cells groups (Fig. 4C, Exp. Growth). These experiments indicated that in the absence of cell cycle checkpoint signals, such as contact inhibition, mutant p53 had no effect on the basal expression of CKsHs1. Importantly, confluent cultures of p53-expressing cells incubated in high-serum medium rapidly progressed to a polyploid DNA content (Fig. 4D). Thus, expression of a mutant p53 protein interfered with the downregulation of CKsHs1 expression and cell cycle arrest in confluent cultures of NHF.
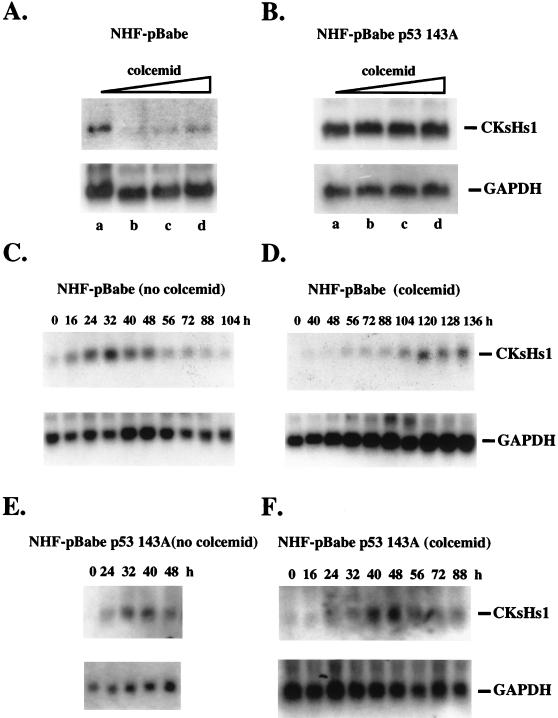
We then investigated whether CKsHs1 expression was under the control of the mitotic spindle cell cycle checkpoint in NHF. For this purpose, we determined the expression of CKsHs1 in control and mutant-p53-expressing cells incubated in the absence or presence of colcemid. Cells were made quiescent and stimulated to enter the cell cycle synchronously for 40 h in the presence of increasing concentrations of colcemid. Colcemid treatment resulted in a drastic decrease in the level of CKsHs1 expression in control fibroblasts (Fig. 5A). However, NHF expressing the mutant p53 143A protein incubated in colcemid at up to 1 μg/ml showed no change in CKsHs1 expression levels (Fig. 5B). Thus, downregulation of CKsHs1 expression in response to mitotic spindle depolymerization was abrogated by expression of mutant p53 in NHF. The decrease in CKsHs1 expression in NHF was not secondary to the accumulation of cells at the G2/M boundary, since maximal levels of CKsHs1 expression were observed at the G2 and M cell cycle phase in control cells incubated in the absence of colcemid (Fig. 5C). In addition, the downregulation of CKsHs1 expression in response to colcemid treatment was transient. CKsHs1 expression recovered partially and peaked at approximately 120 h, an 80-h delay relative to the peak in untreated cells (Fig. 5D). In contrast, a 16-h delay was found in NHF expressing mutant p53 (Fig. 5E).
FIG. 5.
Northern analysis of CKsHs1 expression in NHF-pBabe (A) and NHF-pBabe p53 143A (B) cells incubated in the presence of increasing concentrations of colcemid. The cells were synchronized at G0 by incubation in low-serum medium as in Fig. 2 and then incubated for 40 h in 10% FBS in the presence of 0 (lanes a), 100 (lanes b), 200 (lanes c), or 1,000 (lanes d) ng of colcemid per ml. Colcemid was added 12 h after cell passage. (C and D) Northern analysis of CKsHs1 expression in NHF-pBabe cells incubated in the absence (C) or presence (D) of 200 ng of colcemid per ml. The cells were synchronized and incubated as indicated above. Northern analysis was carried out as in Fig. 4. Data are representative of three independent experiments. (E and F) Northern analysis of CKsHs1 expression in NHF-pBabe 143A cells incubated in the absence (E) or presence (F) of 200 ng of colcemid per ml. The cells were synchronized and incubated as indicated above. Northern analysis was carried out as described in Fig. 4. Data are representative of two independent experiments.
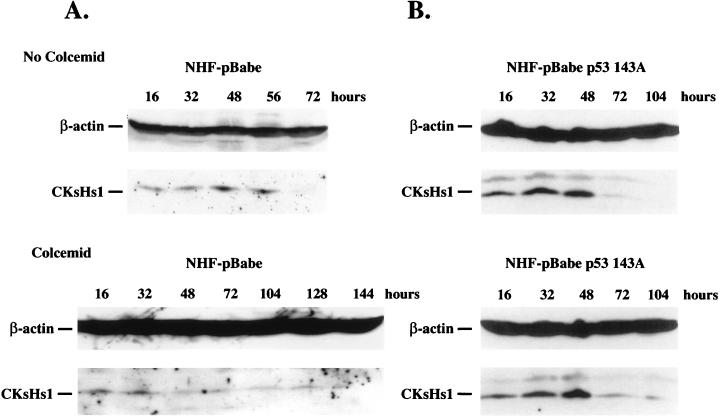
Subsequent experiments investigated the effect of mitotic inhibition and mutant p53 on CKsHs1 protein expression. We performed Western analysis of CKsHs1 protein in control and mutant-p53-expressing NHF treated or not treated with colcemid. The cells were made quiescent by incubation in low-serum medium as indicated above, and CKsHs1 protein levels were determined at different intervals following cell cycle entry. In the control NHF-pBabe cell population, CKsHs1 protein levels were maximal at approximately 48 h following cell passage (Fig. 6A, No Colcemid). Addition of colcemid to the cell culture resulted in a drastic downregulation of CKsHs1 protein levels. The 48-h peak was not detected (Fig. 6A, Colcemid). Instead, CKsHs1 levels increased steadily to reach a moderate peak at 128 h, an 80-h delay with respect to the peak in untreated cells. By contrast, colcemid had a minor effect on the level of CKsHs1 protein in NHF-pBabe p53 143A cells (Fig. 6B). In untreated cells, maximal levels of CKsHs1 protein were found at 32 h, whereas in colcemid-treated cells, CKsHs1 peaked at approximately 48 h. Thus, colcemid treatment caused a marked delay in CKsHs1 protein expression in NHF that was substantially abrogated by mutant p53. Interestingly, peak levels of CKsHs1 protein were detected at the same time as the onset of cyclin B degradation (Fig. 2 and 6). These results are in agreement with genetic and biochemical evidence, obtained with fission yeast and frog eggs, that indicates a requirement for CKsHs1 homologs for cyclin B degradation (4, 42, 49).
FIG. 6.
(A) Western analysis of CKsHs1 and β-actin levels in control NHF (NHF-pBabe) (A) and in NHF expressing mutant p53 proteins (NHF-pBabe p53 143A) (B). The cells were synchronized by a 2-day incubation in low-serum medium (0.5% calf serum) and then incubated in 10% FBS in the absence (No Colcemid) or presence (Colcemid) of 200 ng of colcemid per ml and harvested at the indicated intervals. Colcemid was added 12 h after cell passage. Western blotting was carried out as indicated in Materials and Methods, except for the colcemid-treated NHF-pBabe blot, which was probed with a 1:25 CKsHs1 antibody dilution. Data are representative of three independent experiments.
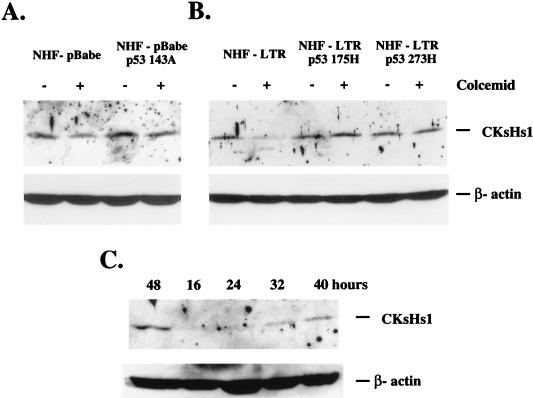
CKsHs1 protein levels decreased by 30 to 70% in exponentially growing control cells but not in mutant-p53-expressing cells following colcemid treatment (Fig. 7A and data not shown). A minor decrease in CKsHs1 protein levels was observed in mutant-p53-expressing cells treated with colcemid, suggesting that mechanisms not affected by mutant p53 may also play a role in the regulation of CKsHs1 levels by the mitotic spindle cell cycle checkpoint. Stable expression of another two structural p53 mutant forms, 175H and 273H, in NHF also blocked the downregulation of CKsHs1 protein levels by colcemid (Fig. 7B). Importantly, these effects of colcemid were caused by its ability to act as a mitotic inhibitor. Addition of this compound to control cells up to 32 h following cell cycle entry significantly decreased CKsHs1 levels (Fig. 7C). Also, colcemid did not affect the progression of cells through S phase; 10 to 15% of control cells were detected in S phase in the absence or presence of colcemid (results not shown).
FIG. 7.
(A) Western analysis of CKsHs1 and β-actin levels in control NHF (NHF-pBabe) and in NHF expressing mutant p53 proteins (NHF-pBabe p53 143A). Exponentially growing cells were incubated in the presence (+) or absence (−) of 200 ng of colcemid per ml for 40 h. (B) Western analysis of CKsHs1 and β-actin in primary NHF stably transfected with murine sarcoma virus long terminal repeat neo-based expression plasmids containing no insert (NHF-LTR), mutant p53 175H (NHF-LTR p53 175H), or mutant p53 273H (NHF-LTR p53 273H) cDNA sequences. The cells were incubated as in panel A. (C) Western analysis of CKsHs1 and β-actin in NHF pBabe cells. Confluent cell cultures (4 × 104 to 5 × 104 cells/cm2) were synchronized by a 2-day incubation in low-serum medium (0.5% calf serum) and then incubated at low density (1 × 104 to 2 × 104 cells/cm2) in 10% FBS. Colcemid (200 ng/ml) was added to the cells at the indicated times, and the cultures were harvested at 48 h. Western analysis was performed as indicated in Materials and Methods. Data are representative of three independent experiments.
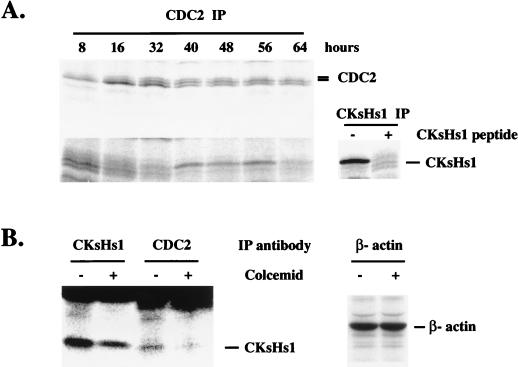
CKsHs1 has been shown to associate with cdc2 (53). Studies with S. pombe and Xenopus extracts suggest that association of CKsHs1 homologs with cdc2 is required for targeting of the cdc2-cyclin B complex to the anaphase-promoting complex and, consequently, for cyclin B proteolysis (4, 42, 49). In addition, it has been argued that members of the Cks1 family are involved in targeting the cdc2 kinase to its mitotic substrates (53). In the absence of these proteins, cdc2 cannot phosphorylate the appropriate substrates, and this results in mitotic arrest. Thus, association of Cks1 proteins with cdc2 seems to be essential for the role of these proteins at mitosis. We investigated the effect of mitotic spindle depolymerization on the association of CKsHs1 with cdc2 in NHF. Using metabolic labeling and immunoprecipitation, we investigated the cell cycle-dependent association of CKsHs1 with cdc2. Figure 8A shows that CKsHs1 associates with cdc2 in the 40- to 56-h period following cell cycle entry. Control NHF were then incubated in the presence or absence of colcemid. Incubation of exponentially growing cells (48 h after synchronization) with colcemid resulted in a 65% decrease in the amount of CKsHs1 protein associated with cdc2 (Fig. 8B).
FIG. 8.
(A) Immunoprecipitation of cdc2-associated CKsHs1. NHF pBabe cells were synchronized and incubated as in Fig. 2 and metabolically labeled for 2 h before being harvested at the indicated times. Preparation of extracts and cdc2 immunoprecipitations (CDC2 IP) were carried out as indicated in Materials and Methods. CKsHs1 was independently immunoprecipitated (CKsHs1 IP) at the 48-h timepoint. As a control, 5 μg of the CKsHs1 peptide epitope was added to half of the CKsHs1 immunoprecipitation extract. Immunoprecipitates were resolved by PAGE (15% polyacrylamide), and the gels were dried and exposed to a PhosphorImager screen. (B) Immunoprecipitation of total and cdc2-associated CKsHs1. Exponentially growing cells (48 h after synchronization) were incubated in the presence (+) or absence (−) of 200 ng of colcemid per ml. Extracts were immunoprecipitated with the indicated antibodies. Other experimental details are as in panel A. Data are representative of three independent experiments.
Ectopic expression of CKsHs1 uncouples cyclin B metabolism from the mitotic spindle cell cycle checkpoint.
Since members of the Cks family are key regulators of progression through mitosis in yeast and frog oocytes (4, 8, 15, 18, 20, 28, 42, 43, 49, 53), we reasoned that CKsHs1 may also play a role in the regulation of mitosis in human cells. Also, CKsHs1 overexpression might interfere with the control of the cell cycle at mitosis in cells with altered p53 status. To test these hypotheses, we attempted to ectopically express CKsHs1 in NHF by using retroviral vectors. However, several attempts to express exogenous CKsHs1 in primary NHF resulted in no viable cells (not shown). In contrast, ectopic expression of CKsHs1 in C2C12 myoblasts resulted in cellular clones. However, when the CKsHs1-expressing C2C12 cells were incubated with colcemid, they suffered extensive cell death (results not shown).
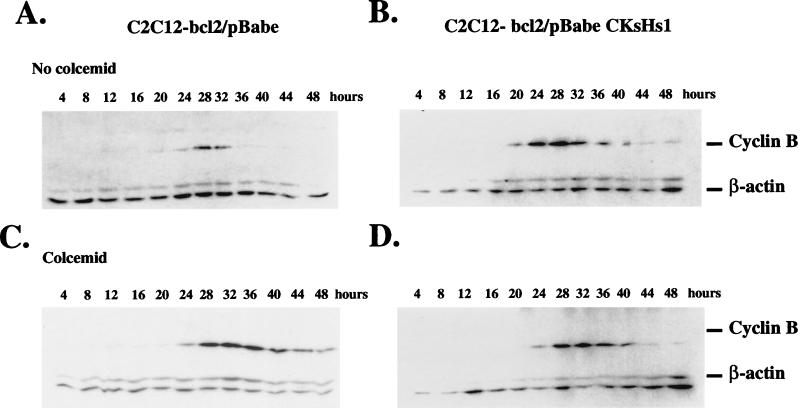
Cells expressing mutant p53 showed high levels of CKsHs1 protein despite their incubation in the presence of colcemid (Fig. 6). However, they did not undergo cell death (Fig. 1). These results suggest that mutant p53 proteins, in addition to abrogating the cell cycle regulatory machinery at mitosis, promote the survival of cells with aberrant DNA content. We reasoned that whereas uncoupling of cyclin B metabolism from the mitotic spindle cell cycle checkpoint could be caused by CKsHs1 overexpression, it was unlikely that the survival effect exerted by mutant p53 proteins was due to higher levels of CKsHs1. Hence, to test the effect of CKsHs1 overexpression while avoiding cell death, we cotransfected C2C12 cells with an expression vector containing cDNA sequences encoding the anti-apoptosis protein bcl2 and the vector pBabe or pBabe CKsHs1. The control cell line C2C12-bcl2/pBabe and the C2C12-bcl2/pBabe CKsHs1 cells were then synchronized and incubated in the presence or absence of colcemid, and the level of CKsHs1 protein was determined by Western analysis. CKsHs1 protein was expressed at lower levels and at later times in control cells when they were incubated in the presence of colcemid (Fig. 9A and B). However, C2C12-bcl2/pBabe CKsHs1 cells demonstrated steady levels of CKsHs1 protein that were not inhibited by colcemid treatment (Fig. 9C). bcl2 expression was also determined by Western analysis. Similar levels were detected in C2C12-bcl2/pBabe and C2C12-bcl2/pBabe CKsHs1 cells (results not shown).
FIG. 9.
Western analysis of CKsHs1 and β-actin in C2C12 cells stably transfected with CMVneo-bcl2 and pBabe or pBabe-CKsHs1 expression vectors. The cells were synchronized as in Fig. 2 and then incubated for the indicated times in 10% FBS in the absence (A, No colcemid) or presence (B and C, Colcemid) of 200 ng of colcemid per ml. Colcemid was added 12 h after cell passage. Western blotting was carried out as indicated in Materials and Methods. Data are representative of two independent experiments.
Control and CKsHs1-overexpressing C2C12 cells were then analyzed for their ability to regulate cyclin B protein levels in response to mitotic inhibition. Control cells delayed the degradation of cyclin B for 8 to 16 h when incubated in the presence of colcemid (Fig. 10A and C and data not shown). However, there was no delay in the onset of cyclin B degradation in cells ectopically expressing CKsHs1 incubated in colcemid (Fig. 10B and D). Thus, overexpression of CKsHs1 abrogated the ability of C2C12 to regulate cyclin B metabolism at mitosis.
FIG. 10.
Western analysis of cyclin B levels in C2C12 cells ectopically expressing bcl2 or expressing bcl2 and CKsHs1. The cells were synchronized as in Fig. 2, incubated in 10% FBS in the absence (A and B) or presence (C and D) of 200 ng of colcemid per ml, and harvested at the indicated intervals. Colcemid was added 12 h after cell passage. Western blotting was carried out as indicated in Materials and Methods. Data are representative of two independent experiments.
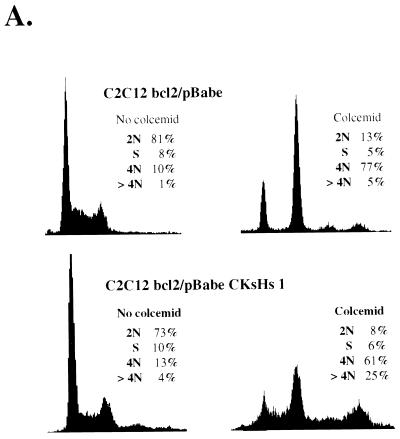
Since CKsHs1 overexpression rendered cells unable to control cyclin B metabolism in response to mitotic inhibition, we reasoned that ectopic expression of CKsHs1 might also allow cell cycle progression in the absence of chromosomal segregation. To test this hypothesis, we performed flow cytometry analysis of the DNA content of the C2C12 cell populations described above, incubated in the presence or absence of colcemid. The results of these experiments are shown in Fig. 11. C2C12-bcl2/pBabe cells incubated for two population doubling times accumulated at the G2/M boundary. However, the C2C12 cells overexpressing CKsHs1 rapidly progressed to an octaploid DNA content (Fig. 11A). Likewise, the bivariate analysis of total DNA content (propidium iodide staining) and newly synthesized DNA (BrdU incorporation) demonstrated that cells overexpressing CKsHs1 had a higher tendency to undergo S-phase reentry at a 4N DNA content than did their isogenic controls (results not shown).
FIG. 11.
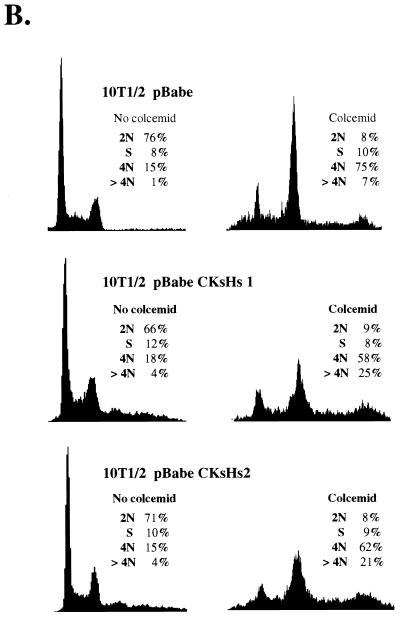
(A) Flow-cytometric analysis of the cell cycle distribution of the DNA content in C2C12 cells ectopically expressing CKsHs1 and/or bcl-2. Cells were incubated in the absence or presence of 200 ng of colcemid per ml for two population doubling times (the population doubling times of C2C12 bcl2/pBabe and C2C12 bcl2/pBabe CKsHs1 cells were 38 and 32 h, respectively). Following incubations, the cells were harvested and processed for flow cytometry of DNA content as indicated in Materials and Methods. Polyclonal populations at passage 2 were assayed. (B) Flow-cytometric analysis of the cell cycle distribution of the DNA content in 10T1/2 cells stably transfected with pBabe, pBabe CKsHs1, or pBabe CKsHs2. The cells were incubated in the absence or presence of 200 ng of colcemid per ml for two population doubling times (72, 66, and 62 h, respectively). Other experimental details are as in panel A. Data are representative of three independent experiments.
In addition, we generated 10T1/2 cell lines that carry wild-type p53 (12), stably expressing CKsHs1 or CKsHs2. Incubation of these cells for two population doubling times in the presence of colcemid also resulted in a marked increase in the number of polyploid cells compared to those for a control cell line stable transfected with an empty pBabe vector (Fig. 11B). In this case, cotransfection with the bcl2 vector was not required to avoid CKsHs toxicity. These experiments confirmed that CKsHs1 overexpression promotes cell cycle progression despite the absence of chromosomal segregation. They also suggested that CKsHs1 and CKsHs2 may have some common functions at mitosis. Overexpression of CKsHs1 and CKsHs2 in these cell lines was determined by Northern analysis of confluent cultures (results not shown).
In conclusion, these experiments demonstrated that an unregulated expression of CKsHs1 leads to abrogation of the mitotic spindle cell cycle checkpoint. Since cells that carry a mutant p53 protein failed to downregulate CKsHs1 in response to mitotic checkpoint signals (Fig. 5 and 6), our results indicate that an altered expression of CKsHs1 mediates, at least in part, the effects of mutant p53 on cyclin B metabolism and the mitotic spindle cell cycle checkpoint.
DISCUSSION
Genetic alterations may occur spontaneously in the general population, but certain individuals are predisposed to their accumulation because of a failure in the mechanisms that correct or eliminate them. We have investigated the activity of the mitotic spindle cell cycle checkpoint, a pathway that contributes to the maintenance of euploidy, in human fibroblasts. Primary human fibroblasts infected with a retroviral vector that drives expression of a mutant p53 protein underwent accelerated DNA endoreduplication in response to spindle-depolymerizing drugs. Normal human fibroblasts arrested growth at mitosis transiently and, eventually, also progressed to mitosis. These results indicated that although the mitotic spindle cell cycle checkpoint delays the cell cycle, mitotic progression eventually takes place. This conclusion is also supported by our cyclin B experiments (see below). The abrogation by a p53 dominant negative mutant (p53 C terminus) of a transient delay in cell cycle progression caused by mitotic spindle-depolymerizing agents has been described previously (41). However, the effect of p53 inactivation appeared to result from the inhibition of the function of this protein at the G1 cell cycle phase that follows a round of anomalous mitosis (without segregation). Our results do not rule out an effect of mutant p53 proteins (working as dominant negatives) on the G1 cell cycle phase following altered mitosis. However, they support the hypothesis that structural p53 mutant proteins have an effect on the control of mitosis.
Cells carrying a mutant p53 protein showed an unscheduled degradation of cyclin B protein. In response to an anomalous chromosomal segregation induced by a mitotic spindle-depolymerizing drug, normal human fibroblasts delayed cyclin B metabolism. However, when the mutant-p53-expressing cells were incubated in colcemid, the onset of cyclin B metabolism was slightly affected (Fig. 2). It has been shown that treatment of HeLa cells with a mitotic inhibitor for 12 to 48 h results in elevated cyclin B levels (34, 51). These data led to the conclusion that prolonged inhibition of the segregation of chromosomes in human cells results in sustained cyclin B levels and mitotic arrest. Our results also underscore the importance of the control of cyclin B metabolism at the mitotic spindle cell cycle checkpoint in human cells. In addition, the use of primary cells and the inclusion of longer incubation periods (up to 152 h) in our studies allowed us to distinguish between normal human cells, in which cyclin B degradation was significantly delayed (48 to 128 h), and human cells carrying a mutant p53 protein, in which altered regulation of cyclin B turnover was observed.
NHF in which wild-type p53 was inactivated by expression of the human papillomavirus type 16 E6 protein demonstrated active control of cyclin B metabolism in response to colcemid treatment. Previous studies have demonstrated that mutant p53 proteins not only inactivate wild-type p53 tumor suppressor function (dominant negative effect) but also have oncogenic properties (gain-of-function). For example, mutated forms of p53 can transactivate certain promoters that do not have wild-type p53 consensus binding sites and can do so in the absence of wild-type p53 protein (11, 14, 17, 23–25, 36, 58). Also, mutant p53 proteins, in addition to blocking wild-type p53-dependent growth arrest, may positively contribute to cell proliferation (9, 10, 17, 31, 32, 44, 59). Using time-lapse videomicroscopy, Lanni and Jacks demonstrated that wild-type p53 does not participate in the mitotic spindle cell cycle checkpoint in mouse fibroblasts (35). Our E6 experiments suggested that p53 is not required for the activity of the mitotic spindle cell cycle checkpoint in NHF. However, we cannot discard the fact that a residual amount of p53 in E6-expressing NHF could be sufficient for the activity of the mitotic spindle checkpoint. p53 null human fibroblast lines have been generated, but they are genetically unstable (38, 62) and strikingly sensitive to colcemid toxicity (22). Therefore, in the absence of primary p53 null human fibroblasts, we can conclude only that expression of a structural mutant p53 protein abrogates the control of cyclin B metabolism by the mitotic spindle checkpoint in NHF.
Our results identified CKsHs1 as a key element in the mitotic spindle cell cycle checkpoint. A role for Cks1 family members at this checkpoint was postulated previously by Rudner and Murray (54). Interestingly, downregulation of CKsHs1 was transient (Fig. 5, 6, and 9), suggesting a phenomenon of adaptation (16, 41) or mitotic slippage (2). Moreover, the regulation of CKsHs1 expression in response to mitotic spindle depolymerization was altered in NHF carrying a mutant p53 protein relative to that in their isogenic controls (Fig. 5 and 6). While control NHF downregulated CKsHs1 expression in response to mitotic inhibition, no change in CKsHs1 expression was observed in mutant-p53-expressing NHF incubated under similar conditions. CKsHs1 may be one of multiple genes targeted by the mitotic spindle checkpoint. A wild-type p53 target, p21cip, has also been implicated in the mitotic spindle cell cycle checkpoint. A wild-type p53 target, p21cip, has also been implicated in the mitotic spindle cell cycle checkpoint. However, p21 expression is controlled by both p53-dependent and p53-independent mechanisms, and recent data indicate that the p21 G2/M growth arrest does not correlate with p53 status (46). p21 may be implicated in a postmitotic growth arrest (35). Transcription of the G2/M cell cycle regulator 14-3-3 has also been shown to be regulated by p53 (29). In addition, recent data show that wild-type p53 negatively regulates the expression of Map4 (45), a microtubule-associated protein that interacts with cyclin B (45, 48).
Mutant p53 may directly activate the expression of CKsHs1. Gain-of-function transcriptional properties have been described for some p53 mutants (24, 37, 58), and it has been proposed that mutant p53 proteins mimic the biological functions of a proliferative conformational stage of wild-type p53 (40, 61). The fact that structural mutant p53 forms that carry mutations in their transcriptional domain still can promote polyploidy (22) does not rule out transcription as their mechanism of action, since mutant p53 proteins may work in association with other transcription factors, such as Sp1 (23). Mutant p53 may also regulate CKsHs1 expression indirectly, affecting the levels of a CKsHs1 regulatory factor(s). Interestingly, p53 is itself a target for phosphorylation by cdc2 (6), suggesting a potential mechanism for feedback control of p53 at mitosis. Our data provide one of the first observations of the regulation of the expression of a Cks family member in mammalian cells. In agreement with Richardson et al. (53), we found that CKsHs1 transcript levels oscillate in a cell cycle-dependent manner, with maximal expression at the G2 and M cell cycle phases (Fig. 5). Downregulation of CKsHs1 expression has also been observed in epithelial cells that were growth inhibited by transforming growth factor β (55).
An 8- to 14-h delay in cyclin B metabolism was observed in control murine C2C12 cells treated with colcemid (Fig. 10). This delay was shorter than that observed in NHF (Fig. 2) and similar to that observed in mouse embryo fibroblasts (35). The fact that human cells exhibit a more stringent mitotic spindle cell cycle checkpoint than rodent cells has been reported previously (34). Ectopic expression of CKsHs1 in C2C12 cells caused a failure to regulate cyclin B turnover, S-phase reentry, and the accumulation of cells with a polyploid DNA content (Fig. 9 to 11). These results indicate that CKsHs1 is a positive regulator of the progression through mitosis. A clear understanding of the functions of Cks family members remains elusive. It has been shown that Suc1 is required in fission yeast for cyclin B degradation, cdc2 kinase inactivation, and exit from mitosis (42). Also, conditional mutation of Cks1 in budding yeast decreases the ability of the cells to undergo the G1/S and G2/M transitions (60). Moreover, removal of Xe-p9, the frog Cks homolog, from Xenopus oocyte interphase extracts abolished the activation of the cyclin B-cdc2 complex by tyrosine dephosphorylation (G2/M transition) and the progression through mitosis as a result of a defect in the degradation of cyclin B (49). Interestingly, we have seen that exponentially growing control C2C12 cells have high levels of CKsHs1 protein (Fig. 9, 32 h), similar to the levels observed in C2C12 cells stably transfected with a retroviral vector that drives the expression of CKsHs1 (Fig. 9C). Likewise, exponentially growing control and p53-expressing NHF have similar levels of CKsHs1 mRNA and protein (Fig. 6). Differences in CKsHs1 expression between mutant-p53-expressing cells and control NHF (Fig. 5 and 6) or between C2C12-pBabe and C2C12-pBabe CKsHs1 cells (Fig. 5 and 9) were apparent only following contact inhibition or colcemid treatment. Thus, the phenotype that we observed in cells ectopically expressing CKsHs1 was not the “overexpression” but the “unregulated expression” of CKsHs1. These data indicated that CKsHs1 is normally expressed at maximal levels during exponential growth. These results also suggest that an excess of CKsHs1 may be deleterious to the cells. C2C12 pBabe CKsHs1 cells with excessively high levels of CKsHs1 may suffer negative selection. It has been reported that overexpression of CKsHs1 and CKsHs2 in an S. cerevisiae Cks1 null mutant caused cell enlargement and elongation, suggesting that CKsHs overexpression delays the G2/M transition (53). Likewise, overexpression of Suc1 delays entry into mitosis in fission yeast (28, 30), and addition of Xe-p9 to Xenopus extracts delays rather than promotes cell cycle progression (49). A simple explanation for these potentially contradictory results is that CKsHs1 expression levels may be critical due to its interaction with rate-limiting components of cyclin-cdk complexes. In this regard, it has been proposed that Cks proteins may work as “docking factors” for cdc2 regulators (49). This model is supported by the crystallographic mutational analysis of CKsHs1 complexed with Cdk2 (7).
In conclusion, we show that NHF carrying a p53 protein with a structural mutation fail to regulate the expression of CKsHs1 in response to mitotic spindle depolymerization. Elevated levels of CKsHs1 interfere with the control of cyclin B metabolism by the mitotic spindle cell cycle checkpoint pathway and facilitate the accumulation of cells with a polyploid DNA content. These data provide a key component to our understanding of the origin of heteroploidy in cells carrying mutant p53 proteins.
ACKNOWLEDGMENTS
M. L. Hixon and A. I. Flores contributed equally to this paper.
We thank A. S. Baldwin, C. A. Finlay, D. A. Galloway, K. Guo, J. Jaccoberger, M. W. Mayo, C. Muro-Cacho, G. Nunez, M. I. Rico, P. Ruiz-Lozano, and R. M. Sramkoski for reagents and suggestions. We thank J. Campisi for careful reading of the manuscript and for encouragement.
This work was supported in part by grants NIH AR39750, ACS IRG91022, AHA 9750205N, and CWRU RIG to A.G.
ADDENDUM IN PROOF
While the manuscript was under review, D. Patra and W. G. Dunphy (Genes Dev. 12:2549–2559, 1998) reported that the frog homolog of CKsHs1 is required for the hyperphosphorylation of cdc27 by the cdc2-cyclin B complex and the activation of the cyclin B destruction machinery. These results indicate that Cks proteins regulate substrate recognition by cdc2-cyclinB.
REFERENCES
- 1.Agapova L S, Ilyinskaya G V, Turovets N A, Ivanov A V, Chumakov P M, Kopnin B P. Chromosome changes caused by alterations of p53 expression. Mutat Res. 1996;354:129–138. doi: 10.1016/0027-5107(96)00062-0. [DOI] [PubMed] [Google Scholar]
- 2.Andreassen P R, Margolis R L. Microtubule dependency of p34cdc2 inactivation and mitotic exit in mammalian cells. J Cell Biol. 1994;127:789–802. doi: 10.1083/jcb.127.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basi G, Draetta G. The cdc2 kinase: structure, activation, and its role at mitosis in vertebrate cells. In: Hutchison C, Glover D M, editors. Cell cycle control. Vol. 10. Oxford, United Kingdom: Oxford University Press; 1995. pp. 106–134. [Google Scholar]
- 4.Basi G, Draetta G. p13suc1 of Schizosaccharomyces pombe regulates two distinct forms of the mitotic cdc2 kinase. Mol Cell Biol. 1995;15:2028–2036. doi: 10.1128/mcb.15.4.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischoff F Z, Yim S O, Pathak S, Grant G, Siciliano M J, Giovanella B C, Strong L C, Tainsky M A. Spontaneous abnormalities in normal fibroblasts from patients with Li-Fraumeni cancer syndrome: aneuploidy and immortalization. Cancer Res. 1990;50:7979–7984. [PubMed] [Google Scholar]
- 6.Bischoff J R, Friedman P N, Marshak D R, Prives C, Beach D. Human p53 is phosphorylated by p60-cdc2 and cyclin B-cdc2. Proc Natl Acad Sci USA. 1990;87:4766–4770. doi: 10.1073/pnas.87.12.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourne Y, Watson M H, Hickey M J, Holmes W, Rocque W, Reed S I, Tainer J A. Crystal structure and mutational analysis of the human CDK2 kinase complex with cell cycle-regulatory protein CksHs1. Cell. 1996;84:863–874. doi: 10.1016/s0092-8674(00)81065-x. [DOI] [PubMed] [Google Scholar]
- 8.Brizuela L, Draetta G, Beach D. p13suc1 acts in the fission yeast cell division cycle as a component of the p34cdc2 protein kinase. EMBO J. 1987;6:3507–3514. doi: 10.1002/j.1460-2075.1987.tb02676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen P L, Chen Y M, Bookstein R, Lee W H. Genetic mechanisms of tumor suppression by the human p53 gene. Science. 1990;250:1576–1580. doi: 10.1126/science.2274789. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Chen P L, Lee W H. Hot-spot p53 mutants interact specifically with two cellular proteins during progression of the cell cycle. Mol Cell Biol. 1994;14:6764–6772. doi: 10.1128/mcb.14.10.6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin K V, Ueda K, Pastan I, Gottesman M M. Modulation of activity of the promoter of the human MDR1 gene by Ras and p53. Science. 1992;255:459–462. doi: 10.1126/science.1346476. [DOI] [PubMed] [Google Scholar]
- 12.Coleman W B, Grisham J W, Smith G J. Morphologic transformation of the C3H 10T1/2 cell line is accompanied by altered expression of the p53 tumor suppressor gene. Carcinogenesis. 1994;15:145–152. doi: 10.1093/carcin/15.2.145. [DOI] [PubMed] [Google Scholar]
- 13.Cross S M, Sanchez C A, Morgan C A, Schimke M K, Ramel S, Idzerda R L, Raskind W H, Reid B J. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- 14.Deb S, Jackson C T, Subler M A, Martin D W. Modulation of cellular and viral promoters by mutant human p53 proteins found in tumor cells. J Virol. 1992;66:6164–6170. doi: 10.1128/jvi.66.10.6164-6170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Veylder L, Segers G, Glab N, Casteels P, Van Montagu M, Inze D. The Arabidopsis Cks1At protein binds the cyclin-dependent kinases cdc2aAt and cdc2bAt. FEBS Lett. 1997;412:446–452. doi: 10.1016/s0014-5793(97)00822-3. [DOI] [PubMed] [Google Scholar]
- 16.Di Leonardo A, Khan S H, Linke S P, Greco V, Seidita G, Wahl G M. DNA rereplication in the presence of mitotic spindle inhibitors in human and mouse fibroblasts lacking either p53 or pRb function. Cancer Res. 1997;57:1013–1019. [PubMed] [Google Scholar]
- 17.Dittmer D, Pati S, Zambetti G, Chu S, Teresky A K, Moore M, Finlay C, Levine A J. Gain of function mutations in p53. Nat Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 18.Draetta G, Brizuela L, Potashkin J, Beach D. Identification of p34 and p13, human homologs of the cell cycle regulators of fission yeast encoded by cdc2+ and suc1+ Cell. 1987;50:319–325. doi: 10.1016/0092-8674(87)90227-3. [DOI] [PubMed] [Google Scholar]
- 19.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 20.Endicott J A, Nurse P. The cell cycle and suc1: from structure to function? Structure. 1995;3:321–325. doi: 10.1016/s0969-2126(01)00162-9. [DOI] [PubMed] [Google Scholar]
- 21.Finlay C A, Hinds P W, Tan T H, Eliyahu D, Oren M, Levine A J. Activating mutations for transformation by p53 produce a gene product that forms an hsc70-p53 complex with an altered half-life. Mol Cell Biol. 1988;8:531–539. doi: 10.1128/mcb.8.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gualberto A, Aldape K, Kozakiewicz K, Tlsty T D. An oncogenic form of p53 confers a dominant, gain-of-function phenotype that disrupts spindle checkpoint control. Proc Natl Acad Sci USA. 1998;95:5166–5171. doi: 10.1073/pnas.95.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gualberto A, Baldwin A S., Jr p53 and Sp1 interact and cooperate in the tumor necrosis factor-induced transcriptional activation of the HIV-1 long terminal repeat. J Biol Chem. 1995;270:19680–19683. doi: 10.1074/jbc.270.34.19680. [DOI] [PubMed] [Google Scholar]
- 24.Gualberto A, Hixon M L, Finco T S, Perkins N D, Nabel G J, Baldwin A S., Jr A proliferative p53-responsive element mediates tumor necrosis factor alpha induction of the human immunodeficiency virus type 1 long terminal repeat. Mol Cell Biol. 1995;15:3450–3459. doi: 10.1128/mcb.15.6.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gualberto A, Marquez G, Carballo M, Youngblood G L, Hunt S W R, Baldwin A S, Sobrino F. p53 transactivation of the HIV-1 long terminal repeat is blocked by PD 144795, a calcineurin-inhibitor with anti-HIV properties. J Biol Chem. 1998;273:7088–7093. doi: 10.1074/jbc.273.12.7088. [DOI] [PubMed] [Google Scholar]
- 26.Halbert C L, Demers G W, Galloway D A. The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. J Virol. 1992;66:2125–2134. doi: 10.1128/jvi.66.4.2125-2134.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 28.Hayles J, Beach D, Durkacz B, Nurse P. The fission yeast cell cycle control gene cdc2: isolation of a sequence suc1 that suppresses cdc2 mutant function. Mol Gen Genet. 1986;202:291–293. doi: 10.1007/BF00331653. [DOI] [PubMed] [Google Scholar]
- 29.Hermeking H, Lengauer C, Polyak K, He T-C, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. 14-3-3ς is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1998;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 30.Hindley J, Phear G, Stein M, Beach D. Suc1+ encodes a predicted 13-kilodalton protein that is essential for cell viability and is directly involved in the division cycle of Schizosaccharomyces pombe. Mol Cell Biol. 1987;7:504–511. doi: 10.1128/mcb.7.1.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsiao M, Low J, Dorn E, Ku D, Pattengale P, Yeargin J, Haas M. Gain-of-function mutations of the p53 gene induce lymphohematopoietic metastatic potential and tissue invasiveness. Am J Pathol. 1994;145:702–714. [PMC free article] [PubMed] [Google Scholar]
- 32.Iwamoto K S, Mizuno T, Ito T, Tsuyama N, Kyoizumi S, Seyama T. Gain-of-function p53 mutations enhance alteration of the T-cell receptor following X-irradiation, independently of the cell cycle and cell survival. Cancer Res. 1996;56:3862–3865. [PubMed] [Google Scholar]
- 33.Kern S E, Pietenpol J A, Thiagalingam S, Seymour A, Kinzler K W, Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 34.Kung A L, Sherwood S W, Schimke R T. Cell line-specific differences in the control of cell cycle progression in the absence of mitosis. Proc Natl Acad Sci USA. 1990;87:9553–9557. doi: 10.1073/pnas.87.24.9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanni J S, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine A J, Wu M C, Chang A, Silver A, Attiyeh E F, Lin J, Epstein C B. The spectrum of mutations at the p53 locus. Evidence for tissue-specific mutagenesis, selection of mutant alleles, and a “gain of function” phenotype. Ann N Y Acad Sci. 1995;768:111–128. doi: 10.1111/j.1749-6632.1995.tb12115.x. [DOI] [PubMed] [Google Scholar]
- 37.Lin J, Teresky A K, Levine A J. Two critical hydrophobic amino acids in the N-terminal domain of the p53 protein are required for the gain of function phenotypes of human p53 mutants. Oncogene. 1995;10:2387–2390. [PubMed] [Google Scholar]
- 38.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 39.Malkin D, Li F P, Strong L C, Fraumeni J F, Jr, Nelson C E, Kim D H, Kassel J, Gryka M A, Bischoff F Z, Tainsky M A, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 40.Milner J, Watson J V. Addition of fresh medium induces cell cycle and conformation changes in p53, a tumour suppressor protein. Oncogene. 1990;5:1683–1690. [PubMed] [Google Scholar]
- 41.Minn A J, Boise L H, Thompson C B. Expression of Bcl-xL and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev. 1996;10:2621–2631. doi: 10.1101/gad.10.20.2621. [DOI] [PubMed] [Google Scholar]
- 42.Moreno S, Hayles J, Nurse P. Regulation of p34cdc2 protein kinase during mitosis. Cell. 1989;58:361–372. doi: 10.1016/0092-8674(89)90850-7. [DOI] [PubMed] [Google Scholar]
- 42a.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titer retroviral vectors with multiple drug selection markers and complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mottram J C, Grant K M. Leishmania mexicana p12cks1, a homologue of fission yeast p13suc1, associates with a stage-regulated histone H1 kinase. Biochem J. 1996;316:833–839. doi: 10.1042/bj3160833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller B F, Paulsen D, Deppert W. Specific binding of MAR/SAR DNA-elements by mutant p53. Oncogene. 1996;12:1941–1952. [PubMed] [Google Scholar]
- 45.Murphy M, Hinman A, Levine A J. Wild-type p53 negatively regulates the expression of a microtubule-associated protein. Genes Dev. 1996;10:2971–2980. doi: 10.1101/gad.10.23.2971. [DOI] [PubMed] [Google Scholar]
- 46.Niculescu A B, III, Chen X, Smeets M, Hengst L, Prives C, Reed S I. Effects of p21cip1/Waf1 at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol Cell Biol. 1998;18:629–643. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Norbury C, Nurse P. Animal cell cycles and their control. Annu Rev Biochem. 1992;61:441–470. doi: 10.1146/annurev.bi.61.070192.002301. [DOI] [PubMed] [Google Scholar]
- 48.Ookata K, Hisanaga S, Okumura E, Kishimoto T. Association of p34cdc2/cyclin B complex with microtubules in starfish oocytes. J Cell Sci. 1993;105:873–881. doi: 10.1242/jcs.105.4.873. [DOI] [PubMed] [Google Scholar]
- 49.Patra D, Dunphy W G. Xe-p9, a Xenopus Suc1/Cks homolog, has multiple essential roles in cell cycle control. Genes Dev. 1996;10:1503–1515. doi: 10.1101/gad.10.12.1503. [DOI] [PubMed] [Google Scholar]
- 50.Peled A, Schwartz D, Elkind N B, Wolkowicz R, Li R, Rotter V. The role of p53 in the induction of polyploidity of myelomonocytic leukemic M1/2 cells. Oncogene. 1996;13:1677–1685. [PubMed] [Google Scholar]
- 51.Pines J, Hunter T. Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell. 1989;58:833–846. doi: 10.1016/0092-8674(89)90936-7. [DOI] [PubMed] [Google Scholar]
- 52.Reed S I, Ferguson J, Groppe J C. Preliminary characterization of the transcriptional and translational products of the Saccharomyces cerevisiae cell division cycle gene CDC28. Mol Cell Biol. 1982;2:412–425. doi: 10.1128/mcb.2.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richardson H E, Stueland C S, Thomas J, Russell P, Reed S I. Human cDNAs encoding homologs of the small p34cdc28/cdc2-associated protein of Saccharomyces cerevisiae and Schizosaccharomyces pombe. Genes Dev. 1990;4:1332–1344. doi: 10.1101/gad.4.8.1332. [DOI] [PubMed] [Google Scholar]
- 54.Rudner A D, Murray A W. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- 55.Simon K E, Cha H H, Firestone G L. Transforming growth factor beta down-regulation of CKShs1 transcripts in growth-inhibited epithelial cells. Cell Growth Differ. 1995;6:1261–1269. [PubMed] [Google Scholar]
- 56.Soddu S, Blandino G, Scardigli R, Coen S, Marchetti A, Rizzo M G, Bossi G, Cimino L, Crescenzi M, Sacchi A. Interference with p53 protein inhibits hematopoietic and muscle differentiation. J Cell Biol. 1996;134:193–204. doi: 10.1083/jcb.134.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srivastava S, Zou Z Q, Pirollo K, Blattner W, Chang E H. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature. 1990;348:747–749. doi: 10.1038/348747a0. [DOI] [PubMed] [Google Scholar]
- 58.Subler M A, Martin D W, Deb S. Activation of the human immunodeficiency virus type 1 long terminal repeat by transforming mutants of human p53. J Virol. 1994;68:103–110. doi: 10.1128/jvi.68.1.103-110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun Y, Nakamura K, Wendel E, Colburn N. Progression toward tumor cell phenotype is enhanced by overexpression of a mutant p53 tumor-suppressor gene isolated from nasopharyngeal carcinoma. Proc Natl Acad Sci USA. 1993;90:2827–2831. doi: 10.1073/pnas.90.7.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang Y, Reed S I. The Cdk-associated protein Cks1 functions both in G1 and G2 in Saccharomyces cerevisiae. Genes Dev. 1993;7:822–832. doi: 10.1101/gad.7.5.822. [DOI] [PubMed] [Google Scholar]
- 61.Ullrich S J, Anderson C W, Mercer W E, Appella E. The p53 tumor suppressor protein, a modulator of cell proliferation. J Biol Chem. 1992;267:15259–15262. [PubMed] [Google Scholar]
- 62.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]