Abstract
Several structurally divergent proteins associate with molecular chaperones of the 70-kDa heat shock protein (hsp70) family and modulate their activities. We investigated the cofactors Hap46 and Hop/p60 and the effects of their binding to mammalian hsp70 and the cognate form hsc70. Hap46 associates with the amino-terminal ATP binding domain and stimulates ATP binding two- to threefold but inhibits binding of misfolded protein substrate to hsc70 and reactivation of thermally denatured luciferase in an hsc70-dependent refolding system. By contrast, Hop/p60 interacts with a portion of the carboxy-terminal domain of hsp70s, which is distinct from that involved in the binding of misfolded proteins. Thus, Hop/p60 and substrate proteins can form ternary complexes with hsc70. Hop/p60 exerts no effect on ATP and substrate binding but nevertheless interferes with protein refolding. Even though there is no direct interaction between these accessory proteins, Hap46 inhibits the binding of Hop/p60 to hsc70 but Hop/p60 does not inhibit the binding of Hap46 to hsc70. As judged from respective deletions, the amino-terminal portions of Hap46 and Hop/p60 are involved in this interference. These data suggest steric hindrance between Hap46 and Hop/p60 during interaction with distantly located binding sites on hsp70s. Thus, not only do the major domains of hsp70 chaperones communicate with each other, but cofactors interacting with these domains affect each other as well.
In recent years, a growing interest has developed in the mechanisms and pathways by which unfolded polypeptide chains attain their native and functionally active conformations. In this respect, molecular chaperones of approximately 70 kDa are of particular importance (for reviews, see references 4, 17, 22 and 28). In eukaryotes, these proteins are either stress inducible as the heat shock protein hsp70 or constitutively expressed as the cognate counterpart hsc70. The bacterial homologue DnaK is probably the best-studied member of this group of proteins.
Members of the hsp70 family of chaperones are characterized by a distinct structure consisting of two major domains. The 44-kDa ATP binding domain is located at the amino terminus, and its three-dimensional structure shows a centrally located cleft harboring the nucleotide binding site. The substrate binding site lies within the carboxy-terminal domain of about 28 kDa. It binds extended short peptides preferentially if they have hydrophobic characteristics, in addition to binding unfolded or misfolded polypeptides. However, substrate binding does not require the whole of the carboxy-terminal domain (4, 36, 40). It is a major function of hsp70 and hsc70 to bind to exposed hydrophobic stretches of unfolded polypeptides, thus preventing the formation of insoluble aggregates. This is the prerequisite for the ATP-dependent protein folding reaction itself, which is known to require the presence of hsp40 or other members of the DnaJ family of cochaperones (for a review, see reference 8) and, in various instances, chaperonin complexes (4, 28).
There must be some intricate interactions between these major domains of hsp70 and hsc70. This is evident from the fact that the intact heat shock protein has only low basal ATPase activity in the absence of polypeptide substrate but the isolated ATP binding fragment exhibits much stronger hydrolytic activity (6). Moreover, binding of ATP or ADP, as well as polypeptide binding, was found to alter the conformation of both the ATP binding and the carboxy-terminal domains of hsp70s (12, 37). However, the details of molecular interactions between these major domains and the mode of information transfer between them are largely unknown.
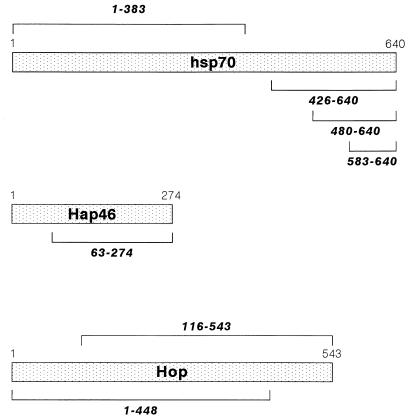
Several structurally unrelated hsp70- and hsc70-interacting proteins which may modulate the chaperoning function either individually or in concert have been discovered in recent years. The “hsp70- and hsc70-associating protein” Hap46 (Fig. 1), which has an apparent molecular weight of 46,000, was originally detected as a factor which associates with nuclear hormone receptors (38). Subsequently we learned that interaction with such receptors and other regulatory proteins occurs via binding to hsp70 or hsc70 (39). The protein Hop/p60 (Fig. 1), with a molecular weight of about 60,000, is known to interact with both hsp70 and hsp90 chaperones and was thus called the “hsp70-hsp90 organizing protein” (7, 21, 24). It plays an important role in the assembly of steroid hormone receptors with heat shock proteins (for a review, see reference 14). Still another factor is the “hsc70-interacting protein” Hip/p48 (18, 31, 32). Hap46 and Hip/p48 interactions occur with the amino-terminal ATP binding domain (13, 18, 39) and consequently compete for binding to hsc70 (13). On the other hand, Hop/p60 binds to the carboxy-terminal domain of hsc70 (9, 13).
FIG. 1.
Linearized structures of human hsp70, Hap46, and Hop/p60. Deletions are indicated by brackets.
Preliminary experiments suggested that the hsp70 and hsc70 accessory proteins affect quite differently the chaperoning activity of hsc70 in a standard protein refolding system (13). We therefore further investigated the interplay between these accessory proteins as they bind to hsp70 and hsc70. Hap46 and Hop/p60 were of major interest to us because of their binding to different parts of the heat shock protein. Nevertheless, we observed a striking interference between Hap46 and Hop/p60 in terms of molecular interactions with hsc70.
MATERIALS AND METHODS
Materials.
Bovine hsc70 and recombinant human hsp70 were purchased from Stressgen. [γ-33P]ATP (10 μCi/μl, 3,000 Ci/mmol) and [35S]methionine (10 μCi/μl, 1,175 Ci/mmol) were from ICN.
ATP binding assay.
Protein mixtures were incubated with ADP and [γ-33P]ATP in buffer A (25 mM HEPES buffer [pH 7.2], 75 mM KCl, 4 mM MgCl2). Free nucleotides were removed by centrifugation through MicroSpin G-50 columns (Pharmacia). Protein-bound [γ-33P]ATP was analyzed by chromatography on polyethylenimine-cellulose as described previously (25). ATP spots were identified by autoradiography and quantified by liquid scintillation counting.
Tagged proteins.
The glutathione S-transferase(GST)-Hap46 fusion protein and His-tagged human hsp40, Hop/p60, and Hip/p48 were as described previously (13, 39). To obtain the amino-terminal deletion (residues 1 to 62) of Hap46 (Fig. 1), the cDNA (38) was digested with BstEII and EcoRI. The fragment encoding residues 63 to 274 was filled in with Klenow fragment to produce blunt ends (1) and ligated in frame into the filled-in EcoRI site of pGEX-2T (Pharmacia) to generate the amino-terminal GST fusion protein. Since this vector introduces a thrombin cleavage site into GST fusion proteins, we obtained untagged versions of Hap46 and Hap46(63-274) by proteolysis (1) with bovine thrombin (Boehringer Mannheim).
The amino-terminally His-tagged ATP binding domain of human hsp70 containing codons 1 to 383 (Fig. 1) was generated by PCR with primers 5′-AATTGGATCCGCATGGCCAAAGCCG-3′ and 5′-AATTAAGCTTGTCCCCCATCAGGAT-3′. The sequence was verified and cloned into BamHI/HindIII sites of pQE-32 (Qiagen), resulting in plasmid pQE-32-hsp70(1-383). Amino-terminally His-tagged fragments of human hsp70 (Fig. 1) were obtained by cutting the cDNA (20) with BglII and HindIII. The fragment containing codons 426 to 640 was converted into blunt ends and ligated into the SmaI site of pQE-30 (Qiagen), resulting in pQE-30-hsp70(426-640). The fragment containing residues 480 to 640 was generated by digesting pQE-30-hsp70(426-640) with ClaI and SacI. DNA was filled in with Klenow fragment and religated, resulting in pQE-30-hsp70(480-640). To obtain the fragment containing residues 583 to 640, we digested pQE-30-hsp70(426-640) with BamHI and BstXI and similarly religated it, resulting in pQE-30-hsp70(583-640).
To obtain amino-terminally His-tagged versions of Hop/p60 with deletions at either the amino or the carboxy terminus (Fig. 1), we used plasmid pET-28a-Hop (13), which was cut either with NcoI or with BglI and XhoI. Fragments containing codons 1 to 448 and 116 to 543 were filled in and cloned into SmaI sites of pQE-30 or pQE-31, respectively, resulting in pQE-30-Hop(1-448) and pQE-31-Hop(116-543).
Expression of proteins was in Escherichia coli JM109 or BL21(DE3), and purifications were carried out as described previously (13, 39).
Substrate binding assay.
[35S]Methionine-labeled human estrogen receptor (ER), obtained by coupled transcription-translation (TnT; Promega) in 50-μl standard reactions, was pretreated with 1 M urea (at 4°C for 1 h) followed by eightfold dilution into buffer A (see above) containing 1 mM ATP. Accessory proteins were added to 150 μl of this dilution (see Fig. 3), and mixtures were incubated with 2.5 μg of the hsc70-specific antiserum K-19 (Santa Cruz) at 4°C for 1 h and then overnight with protein G-Sepharose (Sigma). After extensive washing with saline containing 0.3% Tween 20 (AppliChem), bound proteins were eluted with sodium dodecyl sulfate (SDS) sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) as described in the legend to Fig. 3.
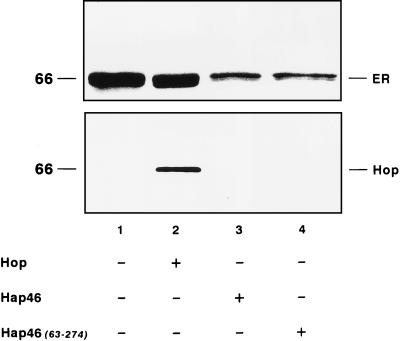
FIG. 3.
Effects of Hap46 and Hop/p60 (Hop) on substrate binding to hsc70. 35S-labeled ER was used as the substrate. It was synthesized in vitro with reticulocyte lysate and subsequently denatured by treatment with urea (see Materials and Methods). Following dilution, the material containing ∼0.3 μM hsc70 (endogenous to reticulocyte lysate) was used without purification. For incubations, Hop/p60 or GST fusion proteins of Hap46 or Hap46(63-274) were added (3 μM each). hsc70-specific immunoprecipitation with K-19 antiserum was done as described in Materials and Methods. Retained material from identical incubations was analyzed either for ER by SDS-PAGE and autoradiography (upper panel) or for Hop/p60 by immunoblotting with antibody F5 (lower panel). In controls without antibody, there were no significant amounts of ER or Hop/p60 (data not shown).
Protein interaction experiments.
Immunoprecipitations were done as described previously (39) in buffer A containing 1 mM ATP with specific antibodies (2.5 to 5 μg each), as described in the figure legends, in a total volume of 150 μl at 4°C for 1 h and subsequently overnight with protein G-Sepharose. After extensive washing with saline containing 0.3% Tween 20, bound proteins were eluted with SDS sample buffer and analyzed by SDS-PAGE and immunoblotting.
Interaction experiments were done with GST fusion proteins of Hap46 or Hap46(63-274) (25 μg each) bound to glutathione-Sepharose (Pharmacia), incubated overnight in buffer A containing 1 mM ATP and 2 mM dithiothreitol, as described previously (38). Incubations were done in a total volume of 200 μl with hsc70 and full-length Hop/p60 or deletions thereof. After extensive washing with saline containing 0.3% Tween 20, bound proteins were eluted with SDS sample buffer and analyzed by SDS-PAGE and immunoblotting.
Proteins were separated by standard SDS-PAGE (10 to 12% acrylamide) and immunoblotted onto Immobilon-P membranes (Millipore) as was done previously (13). His-tagged proteins showed reduced mobilities in SDS gels. Hsc70 was detected by antibody N27F3-4 (Stressgen) or K-19 (Santa Cruz), and His-tagged polypeptides were detected by Penta · His antibody (Qiagen) followed by incubation with peroxidase-conjugated second antibodies and chemiluminescence (ECL; Amersham). Far-Western blotting was done as described previously (13) with Hop-specific antibody F5 (SRA-1500; Stressgen).
RESULTS
Hap46 and Hop/p60 differentially affect ATP binding to hsc70.
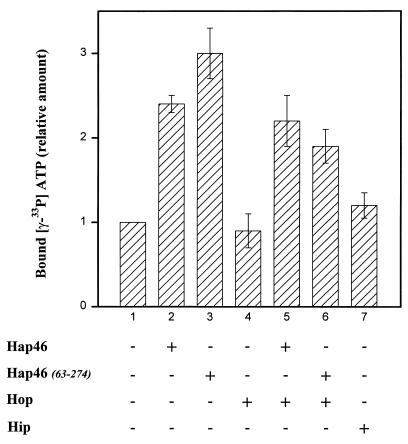
When we measured the steady-state binding of ATP by exposing hsc70, prebound with ADP, to radiolabeled ATP, we found that Hap46 stimulated this exchange two- to threefold (Fig. 2, cf. bars 1 and 2). With the amino-terminal truncation Hap46(63-274) (Fig. 1), which retains full hsp70 and hsc70 binding ability (see below), we obtained roughly the same increase in ATP binding (Fig. 2, cf. bars 1 and 3). By contrast, addition of Hop/p60 had no effect on ATP binding (Fig. 2, bar 4) and did not interfere with the stimulation of ATP binding brought about by Hap46 (Fig. 2, bar 5). This perfectly agrees with the recent observation that Hop/p60 does not alter the ATPase activity of hsp70 and hsc70 (9, 21). All these ATP binding experiments were carried out in the presence of hsp40. When hsp40 was omitted, the stimulation by Hap46 was no more noticeable (data not shown).
FIG. 2.
Effects of Hap46 and Hop/p60 (Hop) on ATP binding to hsc70. hsc70 (1.4 μM) and His-tagged hsp40 (1.2 μM) were preincubated for 10 min at 30°C with accessory proteins (3.4 μM each) and 50 μM ADP. The reaction (total volume, 20 μl) was started by the addition of [γ-33P]ATP (1 μl), and incubation was continued for another 10 min. Samples were cooled on ice, and protein-bound, labeled ATP was quantified as described in Materials and Methods. Hap46 and Hap46(63-274) were added as GST fusion proteins, and Hop/p60 and Hip/p48 (Hip) were added as His-tagged proteins, as indicated. The averages of three separate experiments are given. Error bars show standard deviations. In the control (bar 1), 1.4 to 2.0 pmol of [γ-33P]ATP was bound per μg of hsc70.
We also checked the effect of Hip/p48 in the presence of hsp40 and obtained a marginal increase in ATP binding (Fig. 2, bar 7). This is in accordance with the previous observation that Hip/p48 stabilizes the ADP-bound state of hsc70 (18). However, Hip/p48 had no effect on reactivation of thermally denatured firefly luciferase (13). Since Hap46 and Hop/p60 significantly affected refolding, we further concentrated our efforts on the interactions of these accessory proteins with hsp70 chaperones.
Hap46 and Hop/p60 differentially affect the binding of substrate to hsc70.
In investigating the substrate binding ability of hsc70, we used as the model protein in vitro-synthesized receptor protein ER, which was partially denatured by pretreatment with 1 M urea, followed by dilution. In the presence of ATP, the addition of Hap46 caused a roughly threefold decrease in the amount of ER coprecipitated with hsc70-specific antibody (Fig. 3, upper panel, cf. lanes 1 and 3) (39). Hap46 by itself is known not to be bound by hsc70 as substrate (13, 39). As shown in Fig. 3 (upper panel, lane 4) the amino-terminally truncated version Hap46(63-274) similarly reduced the amount of receptor protein retained on the hsc70-specific matrix.
In similar experiments, we found that Hop/p60 has no effect on the binding of partially denatured receptor protein to hsc70 (Fig. 3, upper panel, cf. lanes 1 and 2), even if used in much larger amounts (roughly 10-fold larger) than hsc70, as in this experiment. Thus, Hop/p60 does not compete for substrate binding. Similarly, Hop/p60 did not interfere with the inhibitory effect of Hap46 on the binding of denatured protein to hsc70 (data not shown). These data show that Hop/p60 does not bind to hsc70 as a substrate, possibly due to denaturation during the course of its preparation, and does not affect substrate binding to hsc70.
Hop/p60 binds to a specific region in the carboxy-terminal domain of hsp70.
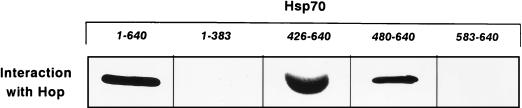
By use of far-Western blotting and live yeast cells, we recently established that Hop/p60 interacts with the carboxy-terminal domain of hsp70 comprising amino acids 384 to 640 (13). To further delineate the binding region, we investigated several deletions in human hsp70 (Fig. 1). Figure 4 shows the results of experiments in which full-length human hsp70 and fragments thereof were incubated with Hop/p60 and interaction was monitored by use of an Hop/p60-specific antibody. Hop/p60 reacted with intact hsp70 but not with the amino-terminal domain (residues 1 to 383) used in these experiments as a negative control. Of the portions originating from the carboxy-terminal domain (residues 384 to 640), fragments 426-640 and 480-640 readily associated with Hop/p60. From these data we conclude that the binding site for Hop/p60 is located between residues 480 and 640, which comprise the α-helical segment of this domain of hsp70 (see Discussion). By contrast, fragment 583-640 did not interact with Hop/p60 (Fig. 4).
FIG. 4.
Binding of Hop/p60 (Hop) to hsp70 regions. Intact human hsp70 and His-tagged fragments of hsp70 (Fig. 1) were used for interaction experiments as described in Materials and Methods. Far-Western blotting was done with Hop/p60 as the probe and antibody F5 directed against Hop/p60.
With Hop/p60 not competing for the binding of substrate to hsc70 (Fig. 3) and interacting with a distinct region in the carboxy-terminal domain (Fig. 4), we wondered whether both could simultaneously bind to hsc70. This was investigated by immunoblotting for Hop/p60 (Fig. 3, lower panel, lane 2). Radiolabeled ER, pretreated with urea, and Hop/p60 are coprecipitated together with hsc70 and an hsc70-specific antibody. This shows that ternary complexes of substrate-hsc70-Hop/p60 can be formed, further emphasizing that Hop/p60 and substrate binding sites on hsc70 are distinct.
Even though Hop/p60 neither interacted with the ATP binding domain of hsp70 (Fig. 4) nor affected ATP binding to hsc70 (Fig. 2, bar 4), we wondered whether the interaction would be influenced by the presence of nucleotides. In coprecipitation experiments with Hop/p60 and a Hop-specific monoclonal antibody, roughly the same amounts of hsc70 were detected whether or not ATP or ADP (1 mM each) was included (data not shown). This suggests that the interaction between Hop/p60 and hsc70 does not depend on nucleotides.
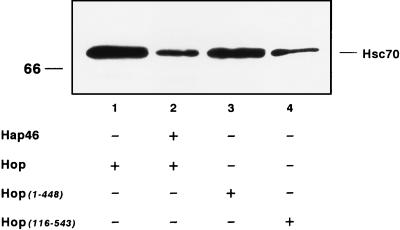
Earlier studies have determined two regions of Hop/p60 through which interaction with hsp70 occurs: roughly amino acid residues 1 to 115 and 225 to 461, both of which regions contain tetratricopeptide repeats (7, 24, 32). We thus constructed two deletion mutants, Hop(116-543) and Hop(1-448), from which either amino- or carboxy-terminal sequences are missing (Fig. 1) but which retain one complete hsp70-hsc70 binding site. Both truncated proteins were used in immunoprecipitation experiments and were found to coprecipitate hsc70, although to different extents (Fig. 5, cf. lane 1 with lanes 3 and 4). Truncated Hop(1-448) and Hop(116-543) reacted with hsc70 with about 75 and 25%, respectively, of the efficiency of full-length Hop/p60. This result agrees with the observation that human and murine Hop/p60s preferentially bind hsp70s through the amino-terminal region (7, 24).
FIG. 5.
Coimmunoprecipitation of hsc70 with Hop/p60 (Hop). hsc70 (0.5 μM) was incubated with Hop/p60 or truncations thereof (0.5 μM each) in either the absence or the presence of GST-Hap46 (2.4 μM). Immunoprecipitations (described in Materials and Methods) were with Penta · His antibody recognizing His-tagged Hop/p60 and fragments thereof. Coprecipitated hsc70 was detected by immunoblotting with antibody N27F3-4.
Hap46 inhibits binding of Hop/p60 to hsc70.
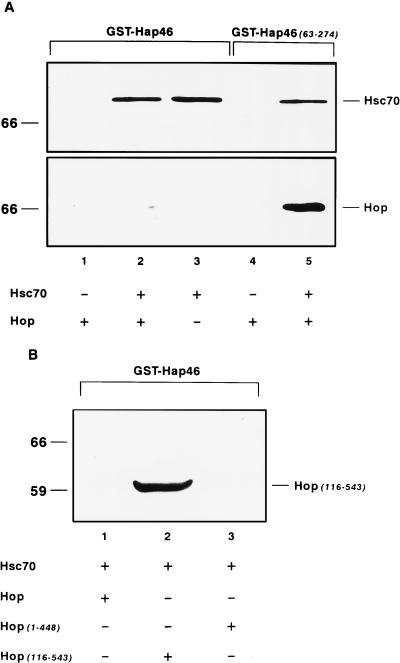
Hap46 fused to GST and attached to glutathione-Sepharose has previously served as affinity matrix which specifically binds hsp70 and hsc70 of mammalian origin (39). Figure 6A (lane 1) shows that Hop/p60 by itself is not retained by this GST-Hap46 matrix but minimal amounts of Hop/p60 are adsorbed in the presence of hsc70 (lane 2). The control immunoblot shown in Fig. 6A (upper panel) shows that the amounts of hsc70 retained on GST-Hap46 are independent of whether Hop/p60 is present (lane 2 versus lane 3), demonstrating that Hop/p60 does not affect the interaction between hsc70 and Hap46.
FIG. 6.
Interaction experiments with GST fusion proteins bound to glutathione-Sepharose. (A) GST fusion proteins with either Hap46 or Hap46(63-274) were used as described in Materials and Methods. Incubations were done in the presence of hsc70 (5 μg) and Hop/p60 (Hop) (10 μg). Identical blots were stained with antibodies F5 against Hop/p60 and N27F3-4 against hsc70. (B) Results of experiments with GST-Hap46, hsc70 (5 μg), and full-length Hop/p60 or deletions thereof (10 μg each). Immunoblotting was done with Penta · His antibody detecting His-tagged Hop/p60 or fragments thereof.
We also tested the above-described Hop/p60 mutations by use of the interaction assay with GST-Hap46. We observed that the carboxy-terminal truncation Hop(1-448) behaves just like full-length Hop/p60 and is unable to bind to hsc70 complexed with Hap46 (Fig. 6B, lane 3). By contrast, amino-terminally abridged Hop(116-543) bound perfectly well to hsc70 in the presence of Hap46 (lane 2), suggesting that Hap46 binding to hsc70 does not interfere with the interaction between hsc70 and the amino-terminal truncation of Hop/p60.
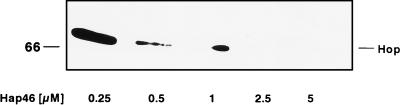
Taken together, these observations clearly show that Hap46 does not directly interact with Hop/p60 but nevertheless affects the binding of Hop/p60 to hsc70. To further substantiate this notion, we carried out a competition experiment in which we incubated equal amounts of hsc70 and Hop/p60 and searched for hsc70 which was coimmunoprecipitated with antibody recognizing Hop/p60. Upon addition of a roughly fivefold molar excess of Hap46, we observed a drastic decrease in the amount of hsc70 retained (Fig. 5, cf. lanes 1 and 2). The concentration dependence of this interference is shown in Fig. 7. In these experiments, we again incubated constant amounts of hsc70 and Hop/p60 but this time assayed for Hop/p60 coimmunoprecipitated with antibody against hsc70. Clearly, increasing concentrations of Hap46 efficiently interfered with the interaction between hsc70 and Hop/p60. Together these data demonstrate that Hap46 and Hop/p60 do not independently interact with hsc70, even though they may bind with different avidities and do recognize very different areas on hsc70. Hap46 interferes with the binding of Hop/p60 to hsc70 but not conversely (cf. Fig. 6A and 7).
FIG. 7.
Coimmunoprecipitation of Hop/p60 (Hop) with hsc70. hsc70 (0.5 μM) was incubated with Hop/p60 (0.5 μM) and increasing amounts of GST-Hap46. Immunoprecipitations (described in Materials and Methods) were done with hsc70-specific antiserum K-19. Coprecipitated Hop/p60 was detected by immunoblotting with antibody F5.
In a previous study, we had not detected the inhibition of Hop/p60 binding to hsc70 by using antibody F5 against Hop/p60 and roughly equal concentrations of Hap46 and Hop/p60 (13). We subsequently turned to immunoprecipitation with a His tag-specific antibody and Hop/p60 with histidine residues added at the carboxy terminus. In this system, interference was observed when we used significantly more Hap46 than Hop/p60 (Fig. 5). This became even more evident when hsc70-specific immunoprecipitation was done: competition was clearly seen at equal concentrations of Hap46 and Hop/p60 (0.5 μM) (Fig. 7, lane 2).
Next, we conducted experiments to find out which part of Hap46 is involved in this interference of Hop/p60 binding to hsc70. Deletion experiments had shown that it is roughly the region of residues 170 to 274 in Hap46 which is involved in binding to hsp70 or hsc70 (data not shown). However, for the interaction experiments of Fig. 6A we used the amino-terminal truncation Hap46(63-274) from which just the region of the conspicuous repetitive Ser-Glu-Glu sequences had been deleted (38). This material was used again as a GST fusion protein. In contrast to intact Hap46, it was found to bind the complex of full-length Hop/p60 and hsc70 perfectly well (Fig. 6A, lane 5). This suggests that it is the amino-terminal region of Hap46 which is involved in inhibiting the binding of Hop/p60, although this part of Hap46 certainly does not by itself contact the heat shock protein. On the other hand, Hap46(63-274) stimulates ATP binding to hsc70 and inhibits the binding of misfolded substrate protein to hsc70 just as Hap46 itself does (Fig. 2 and 3).
Hap46 and Hop/p60 modulate the chaperoning activity.
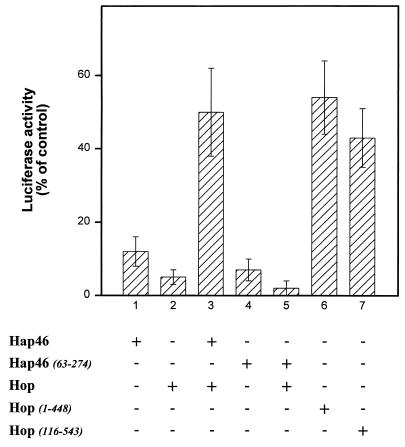
To check the effects of the accessory proteins on the chaperoning activity of hsc70, we used thermally denatured firefly luciferase as a model protein and observed that both Hap46 and Hop/p60 inhibited reactivation while Hip/p48 had no significant effect (13, 39). Intriguingly, Hap46 and Hop/p60 partially compensated each other’s inhibitory activities (13). This prompted us to find out what effects the mutant proteins might exert. We found that the amino-terminal truncation Hap46(63-274) inhibits luciferase refolding just as wild-type Hap46 does (Fig. 8, cf. bars 1 and 4), suggesting that interaction with hsc70 itself causes inhibition of the chaperoning activity. Interestingly, this Hap46 deletion is no longer able to compensate for the inhibitory effect of Hop/p60 on protein refolding (Fig. 8, cf. bars 3 and 5).
FIG. 8.
Effects on refolding of denatured luciferase. Reactivation of thermally denatured firefly luciferase (Sigma) was carried out as described previously (13, 39) with hsc70 (1.4 μM), hsp40 (1.2 μM), and 5% rabbit reticulocyte lysate (Promega). Reconstituted activity in controls without further additions was set at 100%. Other proteins were added (4 μM each) as indicated. Experiments were done in triplicate. The averages of three separate experiments are given. Error bars show standard deviations. The results shown in bars 1 to 3 correspond to previous data (13) and are included for comparison.
In these experiments, we employed GST fusion proteins of Hap46 and the truncation Hap46(63-274). In order to exclude any effect of GST, we also carried out control experiments in which the GST portion was removed by specific proteolysis. We found that protein refolding was affected just as much (data not shown).
For Hop/p60, we observed that inhibition of luciferase reactivation was partially relieved upon deletion of either the amino- or the carboxy-terminal stretches from the primary sequence (Fig. 8, cf. bars 6 and 7 with bar 2). This may be due, at least in part, to decreased binding of Hop(1-448) and Hop(116-543) to hsc70 upon destruction of one hsp70-binding region or the other, as pointed out above (Fig. 5).
DISCUSSION
Human Hap46 was originally identified by an interaction screening approach and was found to associate with various members of the nuclear receptor family as well as with a series of completely unrelated proteins (38, 39). This suggested that multiple associations might be indirect and could rather depend on some common adaptor protein. Indeed, we detected members of the hsp70 family as direct interaction partners (39). Furthermore, hsc70 was found to form ternary complexes with Hap46 and several proteins that are structurally very divergent (39). Nevertheless, overexpression of Hap46 was recently found to inhibit glucocorticoid responsiveness in several cell systems (23).
A protein closely related to human Hap46 is the murine “Bcl-2-associated athanogen” BAG-1, which was discovered through its association with the antiapoptotic protein Bcl-2 and certain growth factor receptors (2, 34). Important observations are that both Hap46 and BAG-1 affect protein refolding in hsp70- and hsc70-dependent systems (13, 35, 39) and interact with the amino-terminal ATP binding domain of hsp70 and hsc70 (13, 19, 35, 39). This led us to the view that hsp70 and hsc70 interact through one part, the ATP binding domain, with Hap46 or shorter versions thereof, while a more distal area, the carboxy-terminal domain, is responsible for associations with multiple partners. In this way, Hap46 may affect a variety of proteins involved in very different biological reactions—for example, by regulating their cellular levels, as has been proposed by Zeiner et al. (39). Support for the notion that various indirect associations with Hap46 may take place through the substrate binding domain of hsp70 and hsc70 came from the observation that binding is significantly promoted by pretreatment with 1 to 3 M urea (39), which causes partial unfolding of protein structures. On the other hand, Hap46 might exert pleiotropic effects, some of which could come about independently of the hsp70-hsc70 chaperone system. For example, there is at present no well-characterized function of the amino-terminal area which contains the above-mentioned Ser-Glu-Glu repeats. Moreover, different molecular forms of Hap46 which have recently been described (30) may well have divergent functions.
Even though crystal structures of complete hsp70 molecules have not yet been obtained, those of the major domains were solved in recent years. The ATP binding domain was found to consist of two subdomains between which the nucleotide binding site is located in a central cleft (10, 33). Biochemical studies suggest that Hap46 contacts both these subdomains (13) and stimulates ATP binding to hsc70 (Fig. 2). In the case of the E. coli hsp70 homologue DnaK, the structure of a complex between the ATP binding domain and the nucleotide exchange factor GrpE was recently elucidated (16). These structural data clarify how GrpE interaction induces a distinct rotation of one of these subdomains, promoting dissociation of ADP and hence nucleotide exchange. The eukaryotic cofactor Hap46 may work somewhat differently to produce conformational changes in hsp70s which again affect the ATP and ADP binding sites.
The three-dimensional structure of the carboxy-terminal unit of DnaK is known to consist of a compact β-sandwich structure which is followed towards the carboxy terminus by a region of α-helices (40). Parts of the carboxy-terminal domains of DnaK and mammalian hsc70 are highly conserved, particularly within the region of the β-sandwich structure (3, 27, 40), suggesting that substrate binding occurs within the β-folded region (roughly amino acids 390 to 480) of hsc70. The results of our Hop/p60 binding experiments (Fig. 4), together with the fact that the hsc70-specific peptide antiserum K-19 (recognizing residues 583 to 601) does not interfere with Hop/p60 binding (Fig. 7), suggest that the Hop/p60 interaction region resides between residues 480 and roughly 580. This corresponds to the α-helical portion of the carboxy-terminal domain and is in agreement with recently published mapping data (9). However, our major point is that interactions with substrate and with Hop/p60 are independent and involve different regions. This explains the observation that Hop/p60 and misfolded protein substrate do not compete for binding to hsc70 but can bind simultaneously (Fig. 3). On the other hand, Hop/p60 also does not compete with hsp40 (13), which requires the very carboxy-terminal portion of hsp70 containing the EEVD sequence (9, 11, 13). Thus, Hop/p60 binding to hsp70 and hsc70 does not overlap with either the sites for hsp40 or those for substrate binding.
Perhaps most striking is the observation that Hap46 influences the distally interacting protein Hop/p60. Our data suggest that Hop/p60 binding to hsc70 is strongly inhibited if Hap46 is associated with the amino-terminal domain of hsc70 (Fig. 5 and 6A). In this situation, Hop/p60 may have only one of its binding sites available for interacting with hsc70 due to steric hindrance. We suggest that such interference between hsp70 cofactors could well affect the functions of chaperone proteins by regulatory changes in their relative levels. It is noteworthy in this context that cellular levels of Hap46 vary significantly between cell lines (unpublished results) and can even be externally manipulated (23). On the other hand, Hop/p60 may be growth regulated and affected by stress conditions (the yeast homologue, STI1, is stress inducible [29]).
Interestingly, steric inhibition of Hop/p60 by Hap46, both binding to the same hsc70 molecule, is relieved in some of our deletion mutants. Thus, truncated Hap46(63-274) occupying its site on hsc70 no longer interferes with efficient binding of Hop/p60 (Fig. 6A). Similarly, Hap46 and the amino-terminally deleted variant Hop(116-543) are able to bind simultaneously to hsc70 with high efficiency (Fig. 6B), suggesting that the carboxy-terminal part of Hop/p60 is not involved in such steric inhibition. On the other hand, Hap46 strongly interfered with the binding of the carboxy-terminal deletion Hop(1-448) to hsc70 (Fig. 6B). We thus presume that the amino-terminal portions of Hap46 and Hop/p60 would occupy roughly the same area if both of them were to bind to hsc70 at the same time. By contrast, Hap46 interaction with the ATP-binding domain is not affected by Hop/p60 (Fig. 6A), suggesting that the hsc70 interaction site of Hap46 is not involved in steric hindrance with Hop/p60.
In preliminary experiments, we found that the interaction of Hop/p60 with hsc70 is unaffected by the addition of nucleotides. This is consistent with our far-Western blots (Fig. 4), in which no nucleotides are present. It also agrees with previous results showing that ATP was not required for interaction (7). On the other hand, our data do not exclude the possibility that Hop/p60 prefers to interact with ADP-bound hsp70, as has recently been suggested (21).
Our experiments on the steady-state binding of ATP to hsc70 did not show any stimulation by recombinant Hop/p60 (Fig. 2). Similarly, Johnson et al. (21) found no effect on the ATPase activity or the rate of ADP dissociation from hsp70. This is in contrast to a previous study with Hop/p60 isolated from rabbit reticulocyte lysate (called recycling factor for hsp70 [RF-70]). This material was reported to enhance the ADP-ATP exchange on hsp70 and was proposed to interact with the amino-terminal ATP-binding domain (15). By contrast, our data clearly delineate the area of interaction within the carboxy-terminal domain of hsc70 (Fig. 4). We suppose that these discrepancies are due to different experimental approaches and sources of Hop/p60, as suggested by Johnson et al. (21).
The above structural considerations shed some light on the results of our luciferase reactivation experiments (Fig. 8). Hap46 and Hop/p60 both interfere with efficient refolding but nevertheless partially compensate each other’s effects. While the inhibitory effect of Hap46 is easily explained by reduced binding of denatured substrate to hsc70 (Fig. 3), the mechanism of mutual relief of inhibition by two separately interacting proteins remains enigmatic. The folding assay which we used depends on hsc70, hsp40, ATP, and a minimal amount of rabbit reticulocyte lysate and consistently showed inhibition of luciferase reactivation upon addition of Hop/p60 (Fig. 8, column 2) (13). Using different refolding assays with either undiluted reticulocyte lysate or a mixture of purified chaperone proteins, Johnson et al. (21) recently observed some positive influence of Hop/p60 on luciferase reactivation when they employed roughly 10- to 20-fold-lower concentrations of Hop/p60 than we did. The authors nevertheless state that Hop/p60 is clearly not an essential component of the refolding machinery. Our experiments show that Hop/p60 alone or in combination with Hap46 does not exert any effect on ATP or substrate binding to hsc70 (Fig. 2 and 3). Hop/p60 may rather work in concert with some other components of the folding pathway. For example, the chaperone hsp90 is an alternative interaction partner of Hop/p60 (5, 7, 21, 24). Moreover, chaperonin complexes contained in reticulocyte lysate (26, 28) might well be affected by the accessory proteins Hap46 and Hop/p60. However, the amino-terminally truncated Hap46(63-274), although still inhibitory by itself, no longer counteracts the inhibition by Hop/p60. This goes along with the observation that Hop/p60 can easily associate with hsc70 occupied by Hap46(63-274) but not nearly as efficiently if Hap46 is bound (Fig. 6A). Our data thus suggest that steric hindrance between the amino-terminal portions of Hap46 and Hop/p60 on hsp70 chaperones is the molecular mechanism by which these hsp70 cofactors mutually influence protein refolding.
ACKNOWLEDGMENTS
We thank D. F. Smith and D. O. Toft for interesting and helpful discussions.
This work was supported by the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J-G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley; 1995. [Google Scholar]
- 2.Bardelli A, Longati P, Albero D, Gorupi S, Schneider C, Ponzetto C, Comoglio P M. HGF receptor associates with the anti-apoptotic protein Bag-1 and prevents cell death. EMBO J. 1996;15:6205–6212. [PMC free article] [PubMed] [Google Scholar]
- 3.Boice J A, Hightower L E. A mutational study of the peptide-binding domain of hsc70 guided by secondary structure prediction. J Biol Chem. 1997;272:24825–24831. doi: 10.1074/jbc.272.40.24825. [DOI] [PubMed] [Google Scholar]
- 4.Bukau B, Horwich A L. The hsp70 and hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 5.Chang H-C J, Lindquist S. Conservation of hsp90 macromolecular complexes in Saccharomyces cerevisiae. J Biol Chem. 1994;269:24983–24988. [PubMed] [Google Scholar]
- 6.Chappell T G, Konforti B B, Schmid S L, Rothman J E. The ATPase core of a clathrin uncoating protein. J Biol Chem. 1987;262:746–751. [PubMed] [Google Scholar]
- 7.Chen S, Prapapanich V, Rimerman R A, Honoré B, Smith D F. Interaction of p60, a mediator of progesterone receptor assembly, with heat shock proteins hsp90 and hsp70. Mol Endocrinol. 1996;10:682–693. doi: 10.1210/mend.10.6.8776728. [DOI] [PubMed] [Google Scholar]
- 8.Cyr D M, Langer T, Douglas M G. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 9.Demand J, Lüders J, Höhfeld J. The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol Cell Biol. 1998;18:2023–2028. doi: 10.1128/mcb.18.4.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaherty K M, DeLuca-Flaherty C, McKay D B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 11.Freeman B C, Myers M P, Schumacher R, Morimoto R I. Identification of a regulatory motif in hsp70 that affects ATPase activity, substrate binding and interaction with hdj-1. EMBO J. 1995;14:2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fung K L, Hilgenberg L, Wang N M, Chirico W J. Conformations of the nucleotide and polypeptide binding domains of a cytosolic hsp70 molecular chaperone are coupled. J Biol Chem. 1996;271:21559–21565. doi: 10.1074/jbc.271.35.21559. [DOI] [PubMed] [Google Scholar]
- 13.Gebauer M, Zeiner M, Gehring U. Proteins interacting with the molecular chaperone hsp70/hsc70: physical associations and effects on refolding activity. FEBS Lett. 1997;417:109–113. doi: 10.1016/s0014-5793(97)01267-2. [DOI] [PubMed] [Google Scholar]
- 14.Gehring U. Steroid hormone receptors and heat shock proteins. In: Litwack G, editor. Vitamins and hormones. Advances in research and applications. Vol. 54. San Diego, Calif: Academic Press; 1998. pp. 167–205. [DOI] [PubMed] [Google Scholar]
- 15.Gross M, Hessefort S. Purification and characterization of a 66-kDa protein from rabbit reticulocyte lysate which promotes the recycling of hsp70. J Biol Chem. 1996;271:16833–16841. doi: 10.1074/jbc.271.28.16833. [DOI] [PubMed] [Google Scholar]
- 16.Harrison C J, Hayer-Hartl M, Di Liberto M, Hartl F-U, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 17.Hartman D, Gething M-J. Normal protein folding machinery. In: Feige U, Morimoto R I, Yahara I, Polla B S, editors. Stress-inducible cellular responses. Basel, Switzerland: Birkhäuser; 1996. pp. 3–24. [Google Scholar]
- 18.Höhfeld J, Minami Y, Hartl F-U. Hip, a novel cochaperone involved in the eukaryotic hsc70/hsp40 reaction cycle. Cell. 1995;83:589–598. doi: 10.1016/0092-8674(95)90099-3. [DOI] [PubMed] [Google Scholar]
- 19.Höhfeld J, Jentsch S. GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunt C, Morimoto R I. Conserved features of eukaryotic hsp70 genes revealed by comparison with the nucleotide sequence of human hsp70. Proc Natl Acad Sci USA. 1985;82:6455–6459. doi: 10.1073/pnas.82.19.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson B D, Schumacher R J, Ross E D, Toft D O. Hop modulates hsp70/hsp90 interactions in protein folding. J Biol Chem. 1998;273:3679–3686. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- 22.Johnson J L, Craig E A. Protein folding in vivo: unraveling complex pathways. Cell. 1997;90:201–204. doi: 10.1016/s0092-8674(00)80327-x. [DOI] [PubMed] [Google Scholar]
- 23.Kullmann M, Schneikert J, Moll J, Heck S, Zeiner M, Gehring U, Cato A C B. RAP46 is a negative regulator of glucocorticoid receptor action and hormone induced apoptosis. J Biol Chem. 1998;273:14620–14625. doi: 10.1074/jbc.273.23.14620. [DOI] [PubMed] [Google Scholar]
- 24.Lässle M, Blatch G L, Kundra V, Takatori T, Zetter B R. Stress-inducible, murine protein mSTI1. J Biol Chem. 1997;272:1876–1884. doi: 10.1074/jbc.272.3.1876. [DOI] [PubMed] [Google Scholar]
- 25.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melki R, Batelier G, Soulié S, Williams R C., Jr Cytoplasmic chaperonin containing TCP-1: structural and functional characterization. Biochemistry. 1997;36:5817–5826. doi: 10.1021/bi962830o. [DOI] [PubMed] [Google Scholar]
- 27.Morshauser R C, Wang H, Flynn G C, Zuiderweg E R P. The peptide-binding domain of the chaperone protein hsc70 has an unusual secondary structure topology. Biochemistry. 1995;34:6261–6266. doi: 10.1021/bi00019a001. [DOI] [PubMed] [Google Scholar]
- 28.Netzer W J, Hartl F U. Protein folding in the cytosol: chaperonin-dependent and -independent mechanisms. Trends Biochem Sci. 1998;23:68–73. doi: 10.1016/s0968-0004(97)01171-7. [DOI] [PubMed] [Google Scholar]
- 29.Nicolet C M, Craig E A. Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3638–3646. doi: 10.1128/mcb.9.9.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Packham G, Brimmel M, Cleveland J L. Mammalian cells express two differently localized Bag-1 isoforms generated by alternative translation initiation. Biochem J. 1997;328:807–813. doi: 10.1042/bj3280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prapapanich V, Chen S, Nair S C, Rimerman R A, Smith D F. Molecular cloning of human p48, a transient component of progesterone receptor complexes and an hsp70-binding protein. Mol Endocrinol. 1996;10:420–431. doi: 10.1210/mend.10.4.8721986. [DOI] [PubMed] [Google Scholar]
- 32.Smith D F. Sequence motifs shared between chaperone components participating in the assembly of progesterone receptor complexes. Biol Chem. 1998;379:283–288. doi: 10.1515/bchm.1998.379.3.283. [DOI] [PubMed] [Google Scholar]
- 33.Sriram M, Osipiuk J, Freeman B C, Morimoto R I, Joachimiak A. Human hsp70 molecular chaperone binds two calcium ions within the ATPase domain. Structure. 1997;5:403–414. doi: 10.1016/s0969-2126(97)00197-4. [DOI] [PubMed] [Google Scholar]
- 34.Takayama S, Sato T, Krajewski S, Kochel K, Irie S, Millan J A, Reed J C. Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cell. 1995;80:279–284. doi: 10.1016/0092-8674(95)90410-7. [DOI] [PubMed] [Google Scholar]
- 35.Takayama S, Bimston D N, Matsuzawa S, Freeman B C, Aime-Sempe C, Xie Z, Morimoto R I, Reed J C. BAG-1 modulates the chaperone activity of hsp70/hsc70. EMBO J. 1997;16:4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T-F, Chang J, Wang C. Identification of the peptide binding domain of hsc70. J Biol Chem. 1993;268:26049–26051. [PubMed] [Google Scholar]
- 37.Wilbanks S M, Chen L, Tsuruta H, Hodgson K O, McKay D B. Solution small-angle X-ray scattering study of the molecular chaperone hsc70 and its subfragments. Biochemistry. 1995;34:12095–12106. doi: 10.1021/bi00038a002. [DOI] [PubMed] [Google Scholar]
- 38.Zeiner M, Gehring U. A protein that interacts with members of the nuclear hormone receptor family: identification and cDNA cloning. Proc Natl Acad Sci USA. 1995;92:11465–11469. doi: 10.1073/pnas.92.25.11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeiner M, Gebauer M, Gehring U. Mammalian protein RAP46: an interaction partner and modulator of 70 kDa heat shock proteins. EMBO J. 1997;16:5483–5490. doi: 10.1093/emboj/16.18.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu X, Zhao X, Burkholder W F, Gragerov A, Ogata C M, Gottesman M E, Hendrickson W A. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]