Abstract
Hepatocellular carcinoma (HCC) does not respond well to current treatments, even immune checkpoint inhibitors. PD‐L1 (programmed cell death ligand 1 or CD274 molecule)‐mediated immune escape of tumor cells may be a key factor affecting the efficacy of immune checkpoint inhibitor (ICI) therapy. However, the regulatory mechanisms of PD‐L1 expression and immune escape require further exploration. Here, we observed that DDX1 (DEAD‐box helicase 1) was overexpressed in HCC tissues and associated with poor prognosis in patients with HCC. Additionally, DDX1 expression correlated negatively with CD8+ T cell frequency. DDX1 overexpression significantly increased interferon gamma (IFN‐γ)‐mediated PD‐L1 expression in HCC cell lines. DDX1 overexpression decreased IFN‐γ and granzyme B production in CD8+ T cells and inhibited CD8+ T cell cytotoxic function in vitro and in vivo. In conclusion, DDX1 plays an essential role in developing the immune escape microenvironment, rendering it a potential predictor of ICI therapy efficacy in HCC.
Keywords: cytotoxic CD8+ T cell, DEAD‐box helicase 1, hepatocellular carcinoma, immune checkpoint inhibitor
In summary, we report that, as an oncoprotein, DDX1 can increase IFN‐γ‐mediated PD‐L1 expression and induce immune escape. The molecular mechanism may be the interaction between DDX1 and p65 that contributes to increasing p65 phosphorylation and NF‐κB activation. Further studies and clinical cohorts are needed to prove the idea that DDX1 could be a biomarker or biological target for enhancing ICI therapy efficacy.
1. INTRODUCTION
Liver cancer is the second leading cause of cancer‐related death worldwide. 1 In ~854,000 new cases of liver cancer annually, 85%–90% of cases are diagnosed as hepatocellular carcinoma (HCC), making it the sixth most common cancer worldwide. 2 HCC is an epithelial tumor that arises typically in patients with liver disease of varying extents. The 5‐year survival of advanced HCC is <15%. 2 In recent years, immune checkpoint inhibitor (ICI) therapy has shown great potential for treating a variety of cancers, including HCC. 3 , 4 , 5 While that approach is promising, ICI therapy efficacy for HCC ranges from 15% to 23%. 3 Hence, exploring the factors of poor therapeutic efficacy is necessary.
It has been indicated that immune checkpoint protein expression, lymphocyte infiltration, and mutation load are important predictors in ICI therapy. 6 There has been much research on the relationship between immune checkpoint and cancer therapeutic efficacy. 6 PD‐L1 (programmed cell death ligand 1 or CD274 molecule) expression in tumor cells is considered one of the most common factors of immune escape in the tumor microenvironment, inducing T cell anergy and exhaustion. 7 , 8 PD‐L1 levels in tumors are regulated in a highly complex manner, such as via genomic aberrations, transcriptional control, mRNA stability, oncogenic signaling, and protein stability, 9 while the transcription factor NF‐κB (nuclear factor kappa B) plays a central role in such factors. 10 Exploring the regulatory mechanism of PD‐L1 expression and the NF‐κB signaling pathway may be an effective means of improving ICI therapy efficacy.
DEAD‐box (DDX) RNA helicases play an essential role in RNA metabolism. 11 DDX1 is a member of the DDX RNA helicase family and is involved in the processes of cancer development or progression. 12 The first indications of DDX1 involvement in tumor development came from studies that co‐amplified it with the MYCN gene in retinoblastomas and neuroblastomas. 13 Breast cancer and HCC studies have also found that DDX1 expression may be related to patient survival. 14 , 15 DDX1 acts as a coactivator to enhance NF‐κB‐mediated transcriptional activation by binding to the NFKB promoter region via association with NF‐κB p65 (also known as RELA). 16 However, whether DDX1 can mediate PD‐L1 expression through NF‐κB in liver cancer is unknown.
In the present study, analysis of multiple online databases showed that DDX1 was upregulated in HCC tissues, and was related to a significant reduction in disease‐free survival in patients with HCC. Further experiments showed that, under interferon gamma (IFN‐γ) stimulation, DDX1 promoted PD‐L1 expression on tumor cells by binding to NF‐κB p65, thereby inhibiting the antitumor effect of CD8+ T cells. Therefore, our study suggests that DDX1 can be used as a predictor of immunotherapy efficacy and a new therapeutic target for treating HCC.
2. MATERIALS AND METHODS
2.1. Patients and liver specimens
In total, 80 paired liver specimens (Table S1) were collected from Guangdong Second Provincial General Hospital (Guangzhou, China). Patients with human immunodeficiency virus (HIV) infection or concurrent autoimmune disease were excluded. Written informed consent was obtained from the patients and the protocol was approved by the hospital ethical review board. In addition, a tissue array (HLivH180Su15) that contains 90 HCC tumor and para‐tumor specimens was obtained from Shanghai Outdo Biotech Co. Ltd.
2.2. Bioinformatic analysis
HCC RNA sequencing data were obtained from The Cancer Genome Atlas (TCGA), International Cancer Genome Consortium (ICGC), and Gene Expression Omnibus (GEO) databases. DDX1 expression and the correlations with other genes in normal or tumor tissues were conducted using R (4.0.2). Kaplan–Meier survival analysis of patients with HCC with low or high DDX1 expression was conducted using TCGA database and the KmPlot platform. The potential functions of DDX1 were analyzed using Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses, and gene set enrichment analysis (GSEA).
2.3. Cell culture, transfection, and stimulation
Human liver cancer cell lines (Huh7, SMMC‐7721, QGY‐7703, HepG2) and LO2 normal human hepatic cells were obtained from Guangzhou Jennio Biotech Co., Ltd (Guangdong, China) and cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. The cells were transfected with a control vector or DDX1 overexpression plasmids. In some experiments, recombinant human IFN‐γ (100 ng/mL; InvivoGen, CA, USA) and NF‐κB inhibitor (triptolide, 15 μmol/L; Selleck Chemicals, Houston, USA) were added to the culture systems.
2.4. Immunohistochemistry (IHC) and immunofluorescence
Formalin‐fixed, paraffin‐embedded HCC specimens were cut into 4‐μm thick sections. Then, the slides underwent deparaffinization, hydration, antigen retrieval, and blocking. For IHC, the slides were incubated with rabbit polyclonal anti‐DDX1 antibody (1:100, Proteintech, Wuhan, China) or anti‐CD8 antibody (1:200; Bioss, Beijing, China) at 4°C overnight, washed with phosphate‐buffered saline (PBS) five times, and stained with an anti‐rabbit/mouse IHC Secondary Antibody Kit (GK500705; GTVision, Shanghai, China). For immunofluorescence, the slides were stained with a multiplex immunofluorescence staining kit (abs50012; absin, Shanghai, China) according to the manufacturer's instructions. Then, the slides were incubated with antibody against DDX1 (1:100; Proteintech), PD‐L1 (1:200; Proteintech), and NF‐κB p65 (1:300; ZENBIO, Chengdu, China) at room temperature for 1 h. Subsequently, the slides were incubated with anti‐rabbit horseradish peroxidase‐conjugated immunoglobulin G (IgG) at room temperature for 15 min and then with fluorophore‐conjugated tyramine molecules (PPD 520, PPD 570, or PPD 650) for 15 min. Finally, the nuclei were stained with DAPI.
2.5. Western blotting
Tumor cells and HCC tissue from patients were lysed by radioimmunoprecipitation assay buffer and 1% phenylmethylsulfonyl fluoride (PMSF; Beyotime, Shanghai, China). Western blotting was performed as described previously. 17 The primary antibodies were raised against DDX1 (1:1000; Proteintech), PD‐L1 (1:500; Proteintech), phosphorylated (p)‐p65 (1:1000; ZENBIO), and p‐65 (1:1000; ZENBIO). Anti‐GAPDH antibody (1:5000; Proteintech) was used as the control. Semi‐quantitative results were analyzed using ImageJ.
2.6. Immunoprecipitation (IP)
HepG2 cells were transfected with DDX1 overexpression plasmid or control vector by Lipofectamine 2000 (Invitrogen, CA, USA). After 6‐h transfection at 37°C, the cells were treated with IFN‐γ (100 ng/mL; InvivoGen, CA, USA) for 48 h. Then, the cells were lysed in PMSF‐supplemented IP lysis buffer. The cell lysates were incubated with anti‐DDX1 antibody or non‐specific IgG overnight at 4°C. The immunoprecipitated proteins were resolved by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and detected using western blotting.
2.7. Quantitative RT‐PCR (qRT‐PCR)
Total RNA was isolated from HCC cells and patient tissue using TRIzol (TaKaRa, USA). RNA (500 ng) was reverse‐transcribed into complementary DNA. The specific primer sequences were: DDX1 (forward primer, 5′‐TAAAGGAATGGCATGGGTGTAGA‐3′; reverse primer, 5′‐CCCTGCATAACCCTTGGTCAT‐3′), CD274 (PD‐L1, forward primer, 5′‐TGGCATTTGCTGAACGCATTT‐3′; reverse primer, 5′‐TGCAGCCAGGTCTAATTGTTTT‐3′), and GAPDH (forward primer, 5′‐ACAACTTTGGTATCGTGGAAGG‐3′; reverse primer, 5′‐GCCATCACGCCACAGTTTC‐3′). All experiments were performed in duplicate.
2.8. Enzyme‐linked immunosorbent assay (ELISA)
The cytokine concentrations in the cell culture supernatants were quantified using IFN‐γ (Cat# 1110002) and granzyme B (Cat# 1118502) ELISA kits (Dakewe, Beijing, China) following the manufacturer's protocols.
2.9. Tumor cell cytotoxicity detection
CD8+ T cells were isolated from the peripheral blood mononuclear cells of HCC patients. CD8+ T cells were pre‐activated with anti‐CD3 (1 μg/mL; BioLegend, CA, USA) and anti‐CD28 (1 μg/mL; BioLegend) for 1–3 days. The pre‐activated CD8+ T cells were then incubated with HepG2 cells and Huh7 cells that had been pretreated with control plasmid or DDX1 overexpression plasmid at 37°C for 12, 24, and 48 h. The effector:target (E:T) ratio was 1:1, 3:1, and 9:1. The supernatants were collected for detecting CD8+ T cell cytotoxicity using a lactate dehydrogenase (LDH) release assay kit (Promega, Madison, WI, USA). The tumor cells were co‐cultured with the CD8+ T cells, stained with propidium iodide (PI; Beyotime, Shanghai, China), and photographed under a light microscope (Olympus, Tokyo, Japan).
2.10. Animal experiments
Male C57BL/6 mice (6–8 weeks old) were purchased from Guangdong Medical Laboratory Animal Center (Guangzhou, China). All animal experiments were approved by the animal facilities of Guangdong Second Provincial General Hospital. For the subcutaneous tumor model, 1 × 107 Hepa1‐6 cells (transduced with empty lentivirus vector or vector expressing DDX1) were injected into the right groin. Then, anti‐mouse PD‐L1 (150 μg per mouse, clone 10F.9G2; Bio X Cell, Lebanon, NH, USA) or IgG control antibody (clone LTF‐2, Bio X Cell) was injected intraperitoneally two times per week. Tumor growth was monitored every 3 days and tumor size was calculated as 0.5 × width2 × length. For the orthotopic HCC mouse model, 1 × 106 Hepa1‐6 cells were implanted intrahepatically. Intrahepatic and intrasplenic lymphocytes were isolated for further analysis after mice had been killed.
2.11. Intracellular cytokine staining and flow cytometry analysis
T cell functions were detected using intracellular cytokine staining. The mouse spleen and tumor lymphocytes were gently milled and separated using 30%–70% Percoll (GE Healthcare, MA, USA). The white interface layer (lymphocytes) was collected and then stimulated with Hepa1‐6 tumor lysates (100 μg total protein/1 × 106 cells/mL), interleukin‐7 (25 ng/mL), anti‐CD28 (1 μg/mL; BioLegend, CA, USA), anti‐CD49 (1 μg/mL; BioLegend), and brefeldin A (1 μL/mL; BD Biosciences, CA, USA) at 37°C for 6 h. After stimulation, the lymphocytes were stained with surface markers (anti‐CD4 and anti‐CD8). Then, they were fixed and permeabilized (Cytofix/Cytoperm; BD Biosciences). IFN‐γ and granzyme B antibodies were used for intracellular staining. The stained cells were analyzed on a BD FACSCanto II flow cytometer.
3. RESULTS
3.1. DDX1 is an immune‐related gene negatively related to CD8 + T cell frequency in HCC tissue
To obtain the critical genes related to the regulation of the intrahepatic immune microenvironment, differential gene expression, overall survival (OS), progression‐free survival (PFS), and immune‐related gene expression were analyzed using TCGA–LIHC (liver hepatocellular carcinoma) database. After taking the overlap of the above analysis, four genes (ZNF248, SLC25A17, XCR1, DDX1) remained (Figure 1A). Univariate and multivariate Cox regression analysis identified T stage and DDX1 expression as independent prognostic factors in TCGA–LIHC database (Supplementary Table S1). A nomogram was established, showing that DDX1 expression was associated with poor OS in patients with HCC (Figure 1B). The relationship between DDX1 expression and the intrahepatic immune response was explored using correlation analysis and showed that there was a slight positive correlation between the expression of DDX1 and the immune‐related genes (Figure 1C). However, there were negative correlations between DDX1 expression and the frequency of CD8+ T cells, natural killer cells, and cytotoxic cells (Figure 1D–F). Taken together, our analysis suggested that DDX1 might be a key gene that induced immune escape in HCC.
FIGURE 1.
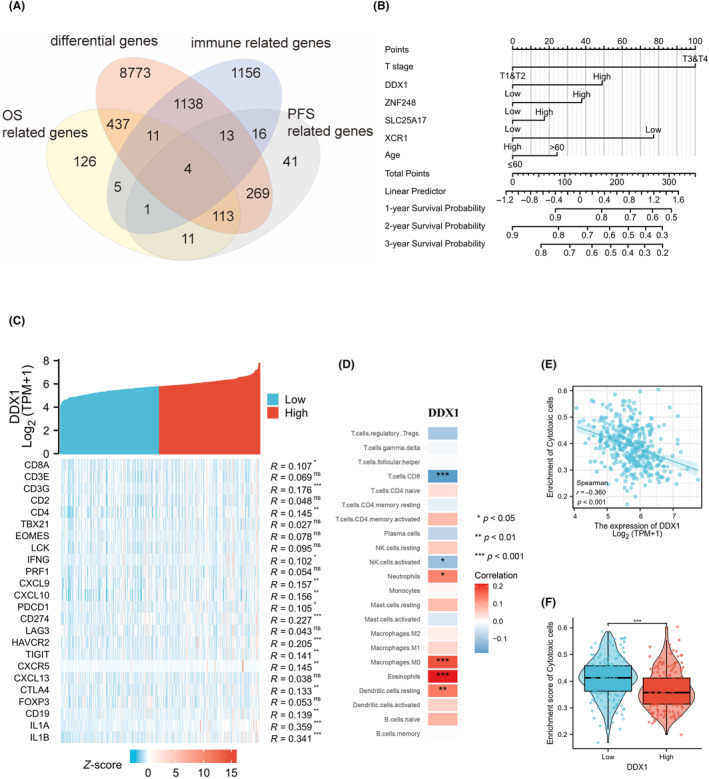
DDX1 is involved in the formation of the tumor immune microenvironment in HCC patients. (A) Venn diagrams showing the genes common to different sets of groups. (B) Development of the nomogram for analyzing the factors associated with OS. (C–E) Correlation between the expression of DDX1 and the immune genes (C), frequency of immune cell clusters (D), and enrichment score of cytotoxic cells (E). (F) Enrichment score of cytotoxic cells in patients with low or high DDX1 expression. *p < 0.05; **p < 0.01; ***p < 0.001.
3.2. DDX1 was upregulated in HCC tissues
We explored DDX1 expression in normal and tumor tissues from patients with HCC. Network database analysis of TCGA, ICGC, and GEO datasets showed that DDX1 was highly expressed in tumor tissues compared with normal tissues (Figure 2A). To confirm the bioinformatic analysis results, we obtained tumor tissue and normal tissue (or para‐tumor tissue) from patients with HCC and performed IHC (Figure 2B), western blotting (Figure 2C), and qRT‐PCR (Figure 2D). DDX1 upregulation was also detected using tissue samples. Patients with a higher clinical stage (T3 and 4 or stage III and IV) had higher DDX1 expression (Figure 2E,F). Receiver operating characteristic (ROC) curve analysis showed that the diagnostic efficiency of DDX1 expression in the diagnosis of HCC was 0.879 (Figure 2G), indicating that DDX1 expression had a high diagnostic efficiency for HCC and had a high sensitivity in distinguishing whether patients had HCC. Overall, these data indicated that DDX1 was overexpressed in HCC tissue.
FIGURE 2.
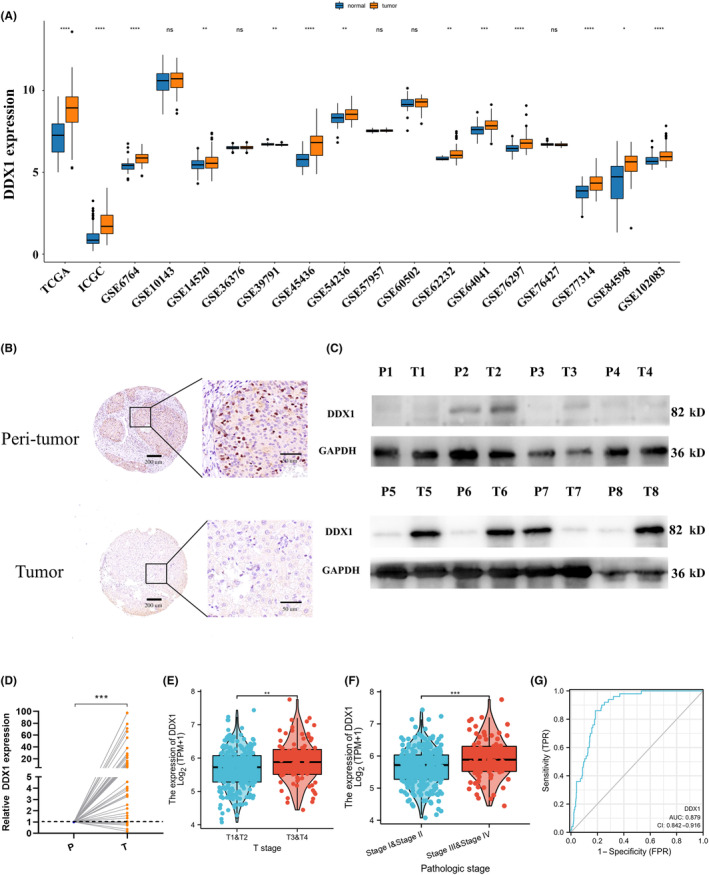
DDX1 was overexpressed in tumor tissue from HCC patients. (A) DDX1 mRNA expression was analyzed using multiple online databases. (B) IHC staining, (C) western blotting (8 pairs), and (D) qRT‐PCR (52 pairs) confirmed DDX1 expression in tumor and peri‐tumor tissues. (E, F) DDX1 expression in different T stages (E) and pathologic stages (F) using TCGA HCC database. (G) ROC curve showing that DDX1 expression was significantly related to the diagnosis of HCC. *p < 0.05; **p < 0.01; ***p < 0.001.
3.3. DDX1 upregulation predicted poor prognosis in patients with HCC
We studied the prognostic value of DDX1. In TCGA cohort, patients with low DDX1 mRNA expression exhibited better OS and PFS than patients with high DDX1 mRNA expression (Figure 3A,B). Moreover, the protein expression of 80 HCC tissue samples at our hospital was analyzed using IHC. Kaplan–Meier survival analysis also indicated that higher DDX1 protein levels in HCC patients were associated with poorer survival rates (Figure 3C). Interestingly, GSEA showed that high DDX1 expression was associated with the poor‐prognosis gene sets of liver and lung cancer (Figure 3D), which suggests that the high expression of DDX1 may be related to the strong proliferation and poor prognosis of liver cancer. These results indicate that high DDX1 expression was an unfavorable prognostic factor for patients with HCC.
FIGURE 3.
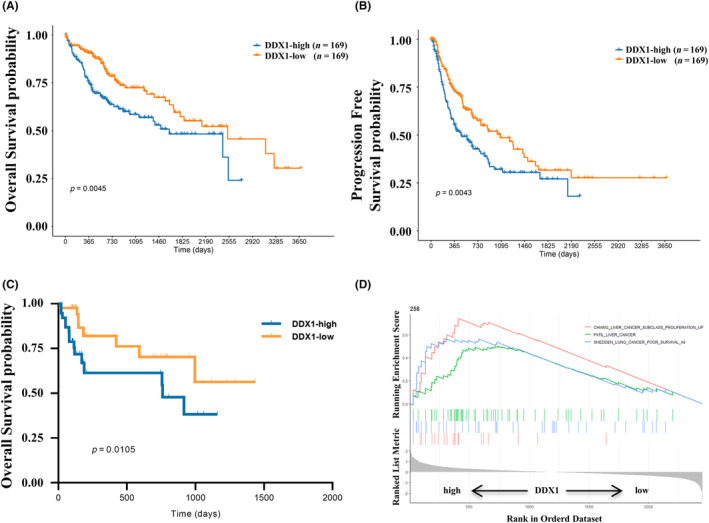
The prognostic effect of DDX1 expression in patients with HCC. (A–D) Comparisons of OS (A) and PFS (B) according to low or high DDX1 expression of patients with HCC in TCGA database. (C) Kaplan–Meier survival analysis of DDX1 expression in patients with HCC from the validation cohort. (D) Gene Set Enrichment Analysis (GSEA) indicating poor survival in HCC and lung cancer patients with high DDX1 expression.
3.4. DDX1 expression correlated positively with PD‐L1 expression
As high DDX1 expression was associated with low frequency of cytotoxic CD8+ T cells (Figure 1D) and poor prognosis in patients with HCC (Figure 3), we analyzed the related mechanisms between expression of DDX1 and immunosuppressive molecule in the HCC tumor microenvironment. High expression of immune checkpoint protein (PD‐L1) or inhibitory receptors (PD‐1, TIM3, CTLA4, TIGIT) was reported to inhibit the T cell antitumor effects significantly. 7 Hence, we conducted a correlation study using TCGA database and found that DDX1 expression correlated with CD274 (PD‐L1) expression in a variety of tumors, including HCC (Figure 4A). Moreover, DDX1 correlated positively with inhibitory receptors, i.e., TIGIT, PDCD1 (PD‐1), HAVCR2 (TIM3), and CTLA4 in patients with HCC (Figure 4A). Using our hospital samples, we verified the positive correlation between DDX1 and CD274 (PD‐L1) in patients with HCC (Figure 4B). In addition, multicolor immunofluorescence revealed that DDX1+ cells co‐stained with PD‐L1 (Figure 4C), which suggested the colocalization of DDX1 and PD‐L1. Furthermore, single‐cell sequencing analysis of the GEO database (LIHC GSE125449) showed that CD274, DDX1, IFNG, and RELA expression was elevated in hepatic cells in patients with HCC treated with anti‐PD‐L1/CTLA4 antibody. Therefore, there was a positive correlation between DDX1 and PD‐L1 expression in patients with HCC.
FIGURE 4.
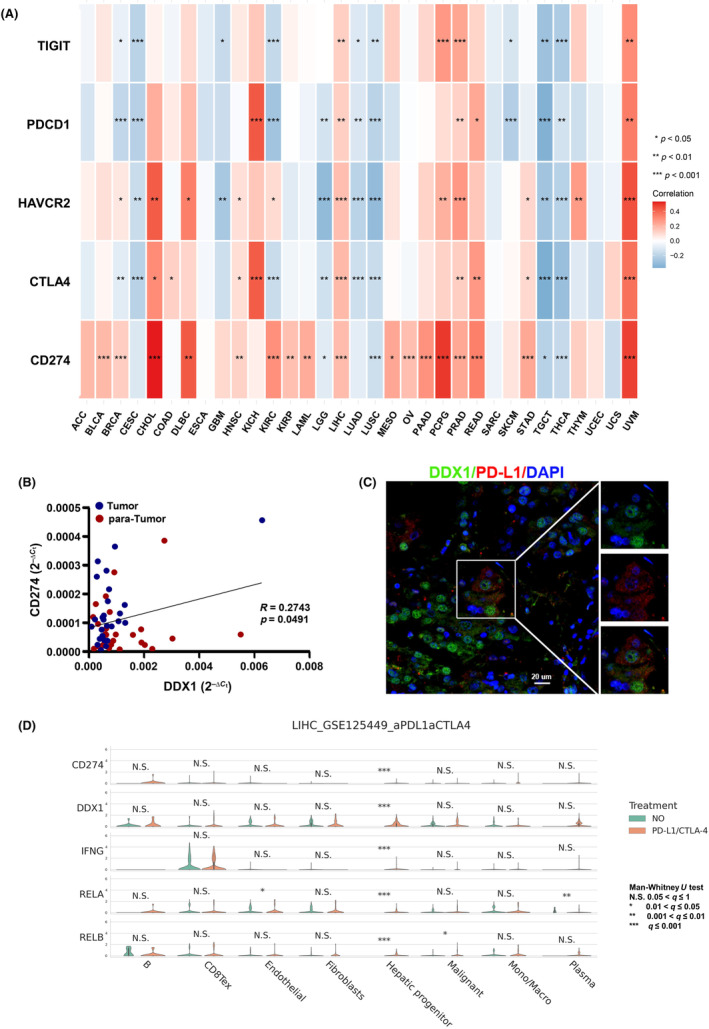
DDX1 expression correlated positively with PD‐L1 expression in HCC patients. (A) Heat map revealing the correlation between DDX1 and immune checkpoints (TIGIT, PDCD1, HAVCR2, CTLA4, CD274) in different cancers. (B) Scatter diagram showing the correlation between DDX1 and PD‐L1 in patients with HCC. (C) Multicolor fluorescence demonstrating the localization relationship between DDX1 and PD‐L1 expression. (D) GEO database (LIHC GSE125449) analysis for determining CD274 (PD‐L1), DDX1, IFNG, RELA (p65) and RELB expression between patients treated with or without antibody against PD‐L1/CTLA4. *p < 0.05; **p < 0.01; ***p < 0.001; N.S., not significant.
3.5. DDX1 enhanced IFN‐γ‐induced PD‐L1 expression on tumor cells
To determine the molecular mechanism between DDX1 and PD‐L1, we first tested DDX1 expression in several hepatoma cell lines and a normal liver cell line (LO2). DDX1 was highly expressed in the hepatoma cell lines SMMC‐7721, QGY‐7703, and HepG2 compared with LO2 cells (Figure 5A). Huh7 and HepG2 cells were selected for further experiments. In these two cell lines, DDX1 overexpression had less effect on PD‐L1 and p65 expression (Figure 5B). IFN‐γ is one of the most studied PD‐L1‐inducers, and IFN‐γ inducible expression of PD‐L1 is related to p50 and p65 activity. 10 Accordingly, we investigated the role of DDX1 in regulating PD‐L1 expression under IFN‐γ stimulation. DDX1 overexpression significantly enhanced PD‐L1 expression and p‐p65 levels (Figure 5C,D). When triptolide inhibited the phosphorylation of the p65 protein, enhanced PD‐L1 expression was weakened both in Huh7 and HepG2 cells (Figure 5C,D). According to a previous report, 16 DDX1 interacts with p65 and enhances NF‐κB‐mediated transcription in 293T cells. Next, the HepG2 cells underwent immunofluorescence staining, which showed that DDX1 and p65 were colocalized (Figure 5E). To confirm the interaction between DDX1 and p65, we performed IP on IFN‐γ‐stimulated and non‐stimulated HepG2 cells. DDX1 was co‐immunoprecipitated efficiently with p65, especially under IFN‐γ stimulation. Therefore, our results indicated that DDX1‐driven IFN‐γ‐mediated PD‐L1 expression was partly dependent on its role in interacting with p65.
FIGURE 5.
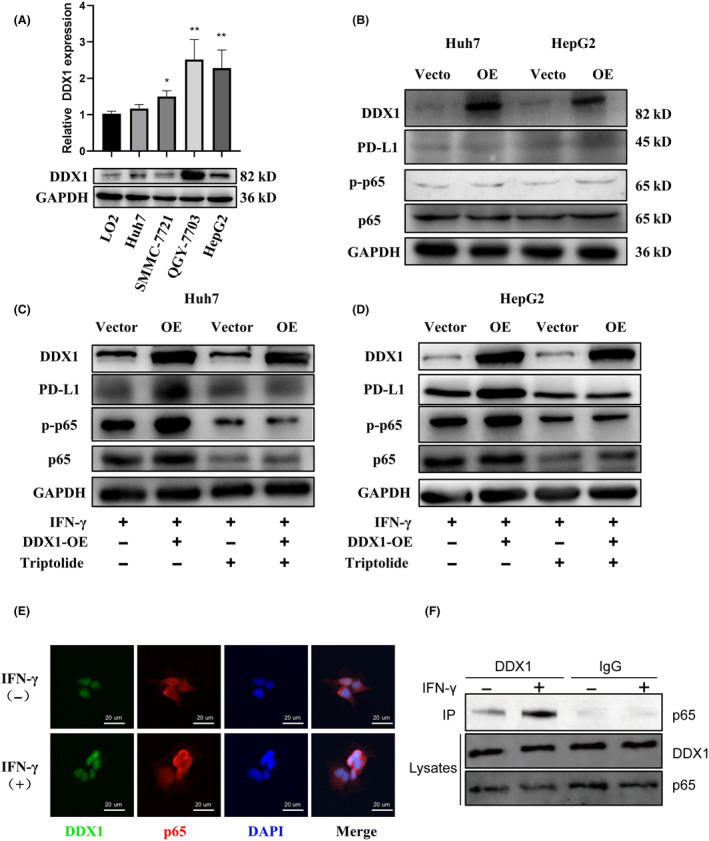
DDX1 expression enhanced IFN‐γ‐induced PD‐L1 expression on tumor cells via p65 phosphorylation. (A) DDX1 mRNA (upper panel) and protein expression (lower panel) levels in the LO2 normal human hepatic cell line and in liver cancer cell lines (Huh7, SMMC‐7721, QGY‐7703, HepG2). (B–D) Western blot analysis of DDX1, PD‐L1, p‐p65, and p65 in Huh7 and HepG2 cells transfected with DDX1 overexpression plasmid (B) and under IFN‐γ stimulation (C, D). (E) DDX1 and RelA localization was determined by multicolor fluorescence with anti‐DDX1 antibody (green), anti‐p65 antibody (red), and the nucleus (blue). (F) Co‐IP of DDX1 and p65. HepG2 cells were treated with or without IFN‐γ (100 ng/mL) for 1 h. The lysates were immunoprecipitated with DDX1 antibody or control IgG. Immunoblotting assay was performed with p65 antibody. *p < 0.05; **p < 0.01.
3.6. DDX1 overexpression in tumor cells attenuated CD8 + T cell antitumor function in vitro
It is well known that CD8+ T cells are key players in antitumor immune responses. 18 Here, DDX1 expression was negatively correlated with CD8+ T cell frequency (Figure 1D). To study the underlying mechanism, isolated CD8+ T cells were co‐cultured with hepatoma cells in vitro. Compared with the control, DDX1 overexpression tumor cells induced significant decreases in the amounts of granzyme B and IFN‐γ from the CD8+ T cells (Figure 6A). Subsequently, the cytotoxicity of CD8+ T cells against tumor cells was assessed by LDH release assay. DDX1 overexpression HepG2 cells significantly inhibited CD8+ T cell cytotoxicity at the E:T ratios of 3:1 and 9:1, while the E:T ratio for DDX1 overexpression Huh7 cells was 9:1 (Figure 6B). Furthermore, CD8+ T cell cytotoxicity was directly detected with PI staining. Fluorescence microscopy (Figure 6C; Figure S1A) and flow cytometry (Figure 6D; Figure S1B) revealed that apoptosis was decreased in the DDX1 overexpression hepatoma cell line co‐culture system, which suggested that DDX1 overexpression attenuated CD8+ T cell cytotoxicity against hepatoma cell lines in vitro. Last, GSEA enrichment analysis of TCGA database showed that DDX1 expression was related positively to cell–cell signaling and negatively regulated the immune response (Figure 6E). Therefore, DDX1 overexpression in the hepatoma cells inhibited the CD8+ T cell antitumor function, namely IFN‐γ and granzyme B secretion and tumor cytotoxicity.
FIGURE 6.
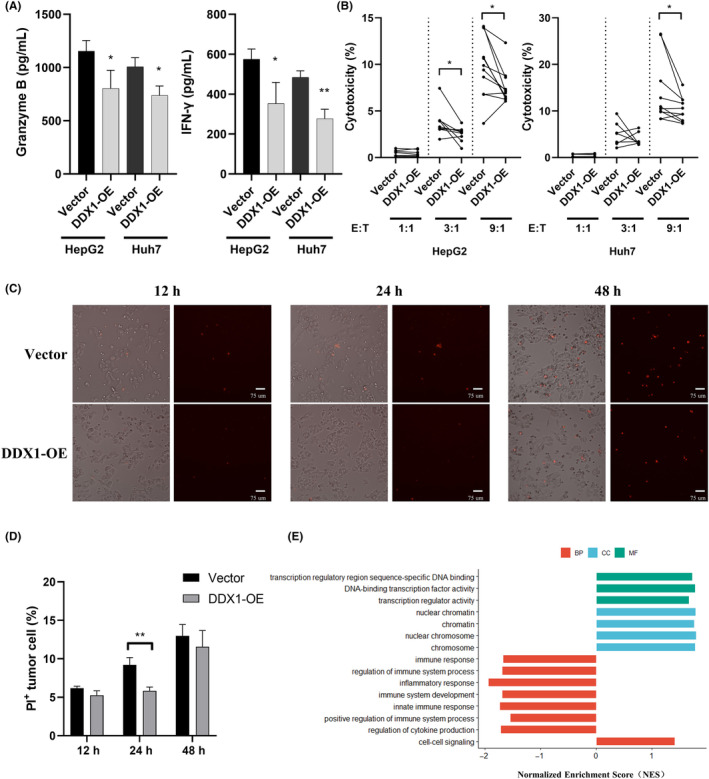
DDX1 suppressed the cytotoxicity of CD8+ T cells against tumor cells in vitro. CD8+ T cells were pre‐activated with anti‐CD3 (1 μg/mL) and anti‐CD28 (1 μg/mL). Vector or DDX1 overexpression (DDX1‐OE) tumor cells were co‐cultured with pre‐activated CD8+ T cells at an indicated effect target ratio (E:T). (A) Granzyme B and IFN‐γ levels in the supernatant from 72 h co‐culture system at a 9:1 effector:target ratio was detected by ELISA. (B) The killing effect of CD8+ T cells incubated with tumor cells at the indicated E:T ratios for 24 h was determined by LDH assay. PI staining of HepG2 tumor cells to reveal dead cells using a light microscope (C) and flow cytometer (D). (E) Signaling pathway were analyzed using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses to compare patients with low or high DDX1 expression from TCGA. *p < 0.05; **p < 0.01.
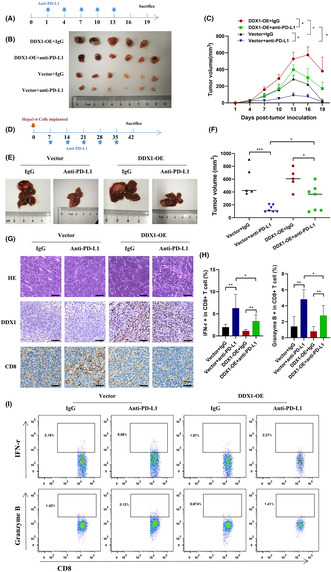
3.7. DDX1 overexpression led to tumor growth and reduced sensitivity to anti‐PD‐L1 therapy
In vivo experiments were conducted to verify the mechanism of DDX1 regulation of the immune response. First, we established a Hepa1‐6 subcutaneous xenograft tumor model using vector (control) or DDX1 overexpression (DDX1‐OE) Hepa1‐6 cells. Compared with control mice, DDX1‐OE mice had markedly increased tumor volume. DDX1‐OE mice were less sensitive to anti‐PD‐L1 therapy (Figure 7A–C). Similar results were observed in the orthotopic HCC mouse model (Figure 7D–F). Immunohistochemistry showed that the DDX1‐OE mice had decreased intrahepatic CD8+ T cells (Figure 7G). Last, we separated intrahepatic lymphocytes from the orthotopic HCC model. Elevated IFN‐γ and granzyme B production from CD8+ T cells was detected when the mice received anti‐PD‐L1 therapy. However, IFN‐γ and granzyme B expression were inhibited in the DDX1‐OE mice (Figure 7H,I).
FIGURE 7.
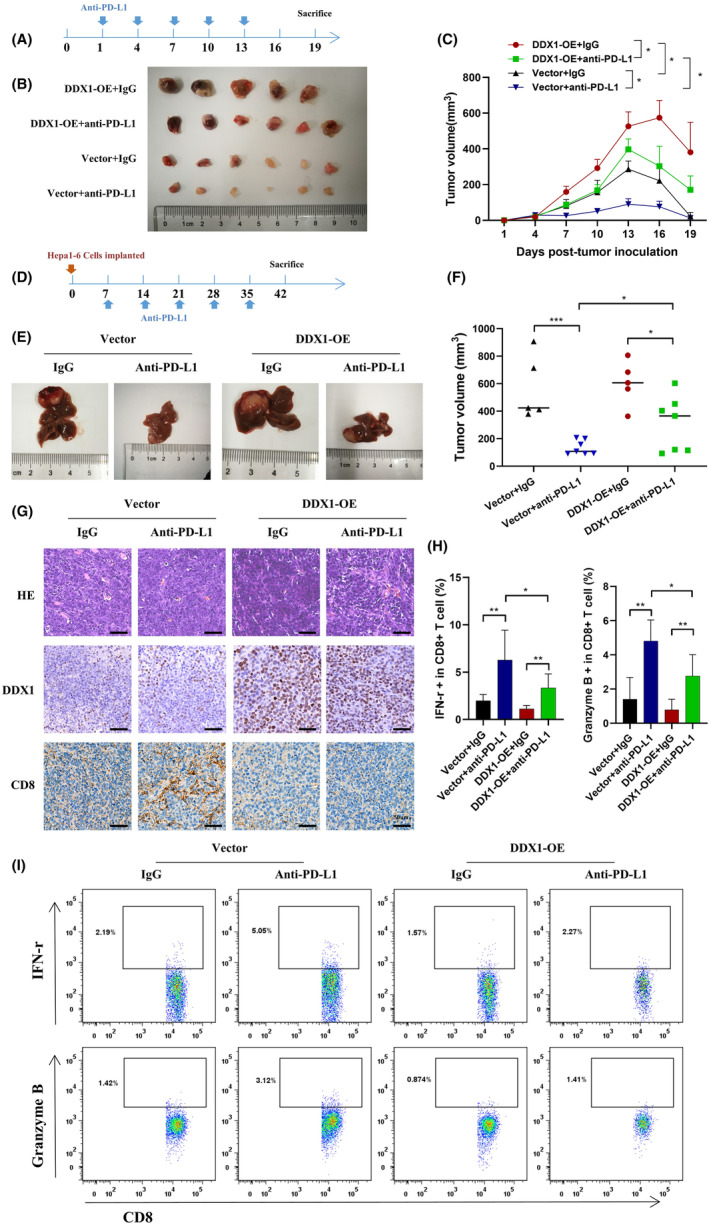
Impact of DDX1 expression on anti‐PD‐L1 treatment in HCC mouse models. (A) Vector control of DDX1 overexpression Hepa1‐6 cells (1 × 107) was inoculated subcutaneously into C57BL/6 mice for 19 days. Anti‐PD‐L1 antibody was given at days 1 and 7 at 150 μg per mouse. (B, C) Tumor volumes were measured every 3 days. (D) Vector control or DDX1 overexpression Hepa1‐6 cells (1 × 106) was inoculated intrahepatically. Anti‐PD‐L1 antibody was given at days 14 and 21 at 150 μg per mouse. (E, F) Tumor volumes were determined at day 42. (G) Representative images of hematoxylin–eosin and IHC staining for DDX1 and CD8 in orthotopic HCC mouse tissue. (H) Intrahepatic lymphocytes were isolated from orthotopic HCC mice. IFN‐γ and granzyme B expression from CD8+ T cells was detected by flow cytometry. (I) The gating strategy of flow cytometry is shown. *p < 0.05; **p < 0.01; ***p < 0.001.
4. DISCUSSION
Strong T cell responses are beneficial for suppressing tumor growth. In contrast, tumor cells elevate the expression of immunosuppressive molecules to inhibit T cell function and promote tumor immune escape. 19 In the present study, the DDX helicase family member DDX1 was upregulated in tumor tissue and correlated negatively with CD8+ T cell frequency and patient survival in HCC. DDX1 enhanced IFN‐γ‐induced PD‐L1 expression in hepatoma cells by interacting with p65. Moreover, DDX1 overexpression depressed CD8+ T cell cytokine secretion and cytotoxicity in vitro and in vivo. Therefore, our results indicate that DDX1 overexpression may account for the unsatisfactory response to anti‐PD‐L1 therapy in HCC.
DDX1 is a highly conserved RNA helicase involved in many cellular functions, including DNA double‐stranded break repair. 20 DDX1 expression has been associated with tumorigenesis in various cancers, including breast cancer, 14 retinoblastoma, 13 neuroblastoma, 13 endometrial carcinomas, and testicular carcinoma. 21 DDX1 plays a key role in microRNA (miRNA) maturation, which induces tumor growth and metastasis. 21 Koenig and colleagues showed that miR‐21 might mediate decreased survival in patients with HCC by targeting DDX1. 15 Although DDX1 participates in activating type I IFN responses in dendritic cells, 22 its role in regulating the HCC microenvironment has not been clearly articulated. In our study, we first systematically analyzed DDX1 expression in patients with HCC using bioinformatic methods and specimens from our hospital. DDX1 was highly expressed in HCC tissues. In addition, patients with high DDX1 expression levels had poorer survival probability than those with low DDX1 expression levels. Furthermore, DDX1 was one of the immune‐related genes that was negatively correlated with CD8+ T cells in the patients. Therefore, we hypothesized that the high DDX1 expression in HCC cells may be related to tumor immune escape.
HCC has limited treatment options in the advanced and relapsed settings. 2 In recent years, ICI therapy combined with anti‐angiogenic agents has shown great potential for treating advanced liver cancer. 23 , 24 However, patient responses to ICI therapy vary widely. 3 Discovering the factors that affect ICI therapy efficacy has become a hot topic. Emerging evidence has demonstrated that PD‐L1 expression on various types of solid tumors, including HCC, plays a key role in generating an immune escape microenvironment and predicting anti‐PD‐1/PD‐L1 therapy efficacy. 25 In the present study, the worse prognosis for high‐DDX1 expression patients with HCC prompted us to explore the relationship between DDX1 and PD‐L1. We found via TCGA database that DDX1 expression correlated positively with PD‐L1 expression in HCC. Multicolor immunofluorescence assay confirmed that DDX1 and PD‐L1 were coexpressed in some of the tumor cells. Moreover, cell culture experiments in vitro showed that DDX1 overexpression increased PD‐L1 expression significantly in tumor cells after IFN‐γ stimulation. Hence, our study is the first to report DDX1 as an important factor affecting the expression of PD‐L1.
Previous research has suggested that PD‐L1 expression in the tumor microenvironment is regulated in a highly complex manner, 25 such as via genomic aberrations, transcriptional control, mRNA stability, oncogenic signaling, and protein stability. NF‐κB, a central transcription factor of inflammation, is a key positive regulator of PD‐L1 expression. 25 Not only can it directly induce CD274 gene transcription by binding to its promoter, it can also regulate PD‐L1 post‐transcriptionally through indirect pathways. There are at least two mechanisms for activating the NF‐κB pathway directly: binding to p65 26 or triggering IκBα (NFKB inhibitor alpha) phosphorylation by binding to IKKβ‐IKKγ. 27 In the present study, we found that DDX1 overexpression did not directly affect p65 protein phosphorylation and PD‐L1 expression. Tumor necrosis factor alpha (TNF‐α) and IFN‐γ are the most studied PD‐L1 inducers, frequently occurring in the inflammatory tumor microenvironment. We discovered that DDX1 overexpression significantly increased p65 protein phosphorylation after IFN‐γ stimulation. When p65 protein phosphorylation was suppressed, IFN‐γ‐induced PD‐L1 expression was restrained. We also discovered that DDX1 could interact directly with p65, a finding that was consistent with the published literature. Therefore, we identified an uncharacterized role of DDX1 in activating NF‐κB and PD‐L1 transcription by binding to p65.
It is widely known that PD‐L1 engages PD‐1 on T cells and triggers inhibitory signaling downstream of the T cell antigen receptor (TCR). 28 Therefore, PD‐L1 expression on tumor cells plays an important role in inhibiting local antitumor T cell‐mediated responses and generating an immune escape microenvironment in tumors. 28 Previously, we have reported that intrahepatic CD8+ T cells were exhausted following high expression of inhibitory receptors, 29 while anti‐PD‐1 antibodies significantly restored the function of exhausted T cells in vitro. 17 Although ICI therapy is promising in HCC, only 15%–23% of patients with HCC have positive responses and clinical outcomes. 2 Combining other treatments with anti‐PD‐1/PD‐L1 therapy may enhance the curative effect. As we had detected a positive correlation between DDX1 and PD‐L1 expression, we speculated that DDX1 overexpression tumor cells could suppress T cell function, at least in part, via the PD‐L1–PD‐1 pathway. When tumor cells and CD8+ T cells were co‐cultured in vitro, the DDX1 overexpression tumor cells significantly inhibited the IFN‐γ and granzyme B production of CD8+ T cells. The cytotoxicity assay also showed that fewer tumor cells died in the DDX1 overexpression group than in the control group. Furthermore, Hepa1‐6 subcutaneous xenograft and orthotopic HCC models were established. DDX1 overexpression in the tumor cells attenuated the therapeutic effect of anti‐PD‐L1 by reducing CD8+ T cell infiltration and function. Molecular mechanisms are drawn in Figure 8 in order for the reader to understand the mechanisms. Altogether, DDX1 expression may be an indicator for predicting ICI therapy efficacy.
FIGURE 8.
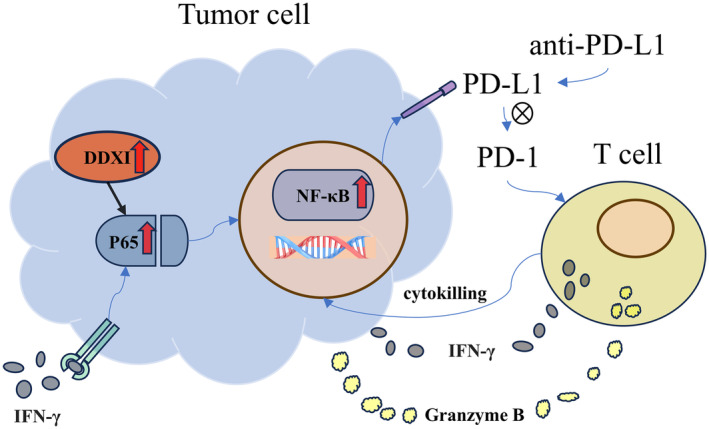
The molecular mechanism diagram of this study.
However, there are still some problems in this study that need to be further investigation. First, the mechanism of DDX1 in promoting tumor growth is complex and it may have the role of directly promoting tumor cell proliferation and differentiation. Additionally, the immune microenvironment plays a huge role in controlling tumors. The effect of DDX1 on the tumor microenvironment is not only in the regulation of PD‐L1 expression, but also on the expression of other immune checkpoints or inhibitory molecules. Moreover, DDX1 is related not only to T cell activation, but also to the functional regulation of other immune cells. The function of DDX1 on other immune cells and its molecular mechanism need to be further investigated.
In summary, we report that, as an oncoprotein, DDX1 can increase IFN‐γ‐mediated PD‐L1 expression and induce immune escape. The molecular mechanism may be the interaction between DDX1 and p65, which contributes to increasing p65 phosphorylation and NF‐κB activation. Further studies and clinical cohorts are needed to confirm the idea that DDX1 could be a biomarker or biological target for enhancing ICI therapy efficacy.
AUTHOR CONTRIBUTIONS
Junhao Liu: Conceptualization; investigation. Ti Yang: Conceptualization; data curation; formal analysis. Zengxin Ma: Data curation; writing – original draft. Yurong Luo: Data curation; formal analysis. Zhitao Yu: Data curation; formal analysis. Lei Zhang: Data curation; formal analysis. Gai Liu: Data curation; investigation. Jianfan Wen: Project administration; supervision. Guankun Lu: Data curation; formal analysis. Guowei Zhang: Data curation; formal analysis. Yujun Zhao: Data curation; formal analysis. Wang Luo: Data curation; formal analysis. Yanan Li: Data curation; formal analysis. Nengjia Yang: Writing – original draft. Jiawei Zhou: Data curation; investigation. Yuhui Lu: Data curation; investigation. Siliang Chen: Conceptualization; funding acquisition; project administration; resources; supervision. Xiancheng Zeng: Conceptualization; funding acquisition; methodology; project administration; resources; supervision; validation; visualization; writing – review and editing.
FUNDING INFORMATION
This study was supported by the Guangdong Natural Science Foundation (No. 2018A0303130184), funding was through Science and Technology Projects in Guangzhou (No. 202201020270), the Hospital Fund of Guangdong Second Provincial General Hospital (3D‐A2020005), the Youth Research Foundation of Guangdong Second Provincial General Hospital (YQ2020‐003), China, and the 3D Printing Project of Guangdong Second Provincial General Hospital (3D‐D2020020), China.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
Approval of the research protocol by an Institutional Review Board: The study protocol was permitted by the Medical Research Ethics Committee of Guangdong Second Provincial General Hospital.
Informed Consent: Informed consents were signed by all patients.
Registry and the Registration No. of the study/trial: N/A.
Animal Studies: Permitted by the Medical Research Ethics Committee of Guangdong Second Provincial General Hospital.
Supporting information
Figure S1
Table S1
ACKNOWLEDGMENTS
Not applicable.
Liu J, Yang T, Luo Y, et al. DEAD‐box helicase 1 inhibited CD8 + T cell antitumor activity by inducing PD‐L1 expression in hepatocellular carcinoma. Cancer Sci. 2024;115:763‐776. doi: 10.1111/cas.16076
Junhao Liu, Ti Yang and Yurong Luo are co‐first author.
Contributor Information
Siliang Chen, Email: chensiliang1991@163.com.
Xiancheng Zeng, Email: llyml2023@163.com.
DATA AVAILABILITY STATEMENT
The data used to support the findings of this study are available from the corresponding author upon request.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380:1450‐1462. [DOI] [PubMed] [Google Scholar]
- 3. Pinter M, Jain RK, Duda DG. The current landscape of immune checkpoint blockade in hepatocellular carcinoma: a review. JAMA Oncol. 2021;7:113‐123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rizvi S, Wang J, El‐Khoueiry AB. Liver cancer immunity. Hepatology. 2021;73(Suppl 1):86‐103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakano S, Eso Y, Okada H, Takai A, Takahashi K, Seno H. Recent advances in immunotherapy for hepatocellular carcinoma. Cancer. 2020;12:775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nishino M, Ramaiya NH, Hatabu H, Hodi FS. Monitoring immune‐checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol. 2017;14:655‐668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zou W, Wolchok JD, Chen L. PD‐L1 (B7‐H1) and PD‐1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8:324r‐328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zheng Y, Fang YC, Li J. PD‐L1 expression levels on tumor cells affect their immunosuppressive activity. Oncol Lett. 2019;18:5399‐5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zerdes I, Matikas A, Bergh J, Rassidakis GZ, Foukakis T. Genetic, transcriptional and post‐translational regulation of the programmed death protein ligand 1 in cancer: biology and clinical correlations. Oncogene. 2018;37:4639‐4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Antonangeli F, Natalini A, Garassino MC, Sica A, Santoni A, Di Rosa F. Regulation of PD‐L1 expression by NF‐kappaB in cancer. Front Immunol. 2020;11:584626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lorsch JR. RNA chaperones exist and DEAD box proteins get a life. Cell. 2002;109:797‐800. [DOI] [PubMed] [Google Scholar]
- 12. Fuller‐Pace FV. DEAD box RNA helicase functions in cancer. RNA Biol. 2013;10:121‐132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Godbout R, Squire J. Amplification of a DEAD box protein gene in retinoblastoma cell lines. Proc Natl Acad Sci USA. 1993;90:7578‐7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Taunk NK, Goyal S, Wu H, Moran MS, Chen S, Haffty BG. DEAD box 1 (DDX1) expression predicts for local control and overall survival in early stage, node‐negative breast cancer. Cancer. 2012;118:888‐898. [DOI] [PubMed] [Google Scholar]
- 15. Koenig A, Barajas J, Guerrero M, Ghoshal K. A comprehensive analysis of argonaute‐CLIP data identifies novel, conserved and species‐specific targets of miR‐21 in human liver and hepatocellular carcinoma. Int J Mol Sci. 2018;19:851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ishaq M, Ma L, Wu X, et al. The DEAD‐box RNA helicase DDX1 interacts with RelA and enhances nuclear factor kappaB‐mediated transcription. J Cell Biochem. 2009;106:296‐305. [DOI] [PubMed] [Google Scholar]
- 17. Li Y, Tang L, Guo L, et al. CXCL13‐mediated recruitment of intrahepatic CXCR5(+)CD8(+) T cells favors viral control in chronic HBV infection. J Hepatol. 2020;72:420‐430. [DOI] [PubMed] [Google Scholar]
- 18. Han J, Khatwani N, Searles TG, Turk MJ, Angeles CV. Memory CD8(+) T cell responses to cancer. Semin Immunol. 2020;49:101435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang X, Wang J, Deng X, et al. Role of the tumor microenvironment in PD‐L1/PD‐1‐mediated tumor immune escape. Mol Cancer. 2019;18:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li L, Monckton EA, Godbout R. A role for DEAD box 1 at DNA double‐strand breaks. Mol Cell Biol. 2008;28:6413‐6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tanaka K, Okamoto S, Ishikawa Y, Tamura H, Hara T. DDX1 is required for testicular tumorigenesis, partially through the transcriptional activation of 12p stem cell genes. Oncogene. 2009;28:2142‐2151. [DOI] [PubMed] [Google Scholar]
- 22. Zhang Z, Kim T, Bao M, et al. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866‐878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med. 2020;382:1894‐1905. [DOI] [PubMed] [Google Scholar]
- 24. Shigeta K, Datta M, Hato T, et al. Dual programmed death receptor‐1 and vascular endothelial growth factor Receptor‐2 blockade promotes vascular normalization and enhances antitumor immune responses in hepatocellular carcinoma. Hepatology. 2020;71:1247‐1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD‐L1 checkpoint. Immunity. 2018;48:434‐452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ahmad R, Raina D, Joshi MD, et al. MUC1‐C oncoprotein functions as a direct activator of the nuclear factor‐kappaB p65 transcription factor. Cancer Res. 2009;69:7013‐7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ahmad R, Raina D, Trivedi V, et al. MUC1 oncoprotein activates the IkappaB kinase beta complex and constitutive NF‐kappaB signalling. Nat Cell Biol. 2007;9:1419‐1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mizuno R, Sugiura D, Shimizu K, et al. PD‐1 primarily targets TCR signal in the inhibition of functional T cell activation. Front Immunol. 2019;10:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zheng X, Zhou Y, Yi X, et al. IL‐21 receptor signaling is essential for control of hepatocellular carcinoma growth and immunological memory for tumor challenge. Onco Targets Ther. 2018;7:e1500673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Table S1
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


