Summary
Alternative polyadenylation (APA) is a key post-transcriptional regulatory mechanism; yet, its regulation and impact on human diseases remain understudied. Existing bulk RNA sequencing (RNA-seq)-based APA methods predominantly rely on predefined annotations, severely impacting their ability to decode novel tissue- and disease-specific APA changes. Furthermore, they only account for the most proximal and distal cleavage and polyadenylation sites (C/PASs). Deconvoluting overlapping C/PASs and the inherent noisy 3′ UTR coverage in bulk RNA-seq data pose additional challenges. To overcome these limitations, we introduce PolyAMiner-Bulk, an attention-based deep learning algorithm that accurately recapitulates C/PAS sequence grammar, resolves overlapping C/PASs, captures non-proximal-to-distal APA changes, and generates visualizations to illustrate APA dynamics. Evaluation on multiple datasets strongly evinces the performance merit of PolyAMiner-Bulk, accurately identifying more APA changes compared with other methods. With the growing importance of APA and the abundance of bulk RNA-seq data, PolyAMiner-Bulk establishes a robust paradigm of APA analysis.
Keywords: alternative polyadenylation (APA), post-transcriptional regulation, deep learning, large language model (LLM), bioinformatics, computational biology, gene regulation
Graphical abstract
Highlights
-
•
C/PAS-BERT deep learning model recapitulates the underlying C/PAS grammar
-
•
Softclipped-assisted C/PAS deconvolution aids in tissue-specific C/PAS selection
-
•
PolyAIndex ranking accounts for read density and C/PAS distribution along a gene
-
•
PolyAMiner-Bulk reveals APA dynamics and pathways in scleroderma
Motivation
Alternative polyadenylation (APA) is a pivotal post-transcriptional mechanism that produces multiple mRNA isoforms with diverse 3′ UTR lengths, impacting gene expression. This diversity in mRNA isoforms is not only crucial in cellular processes but also has implications in a range of human diseases, including neurodegeneration and cancer. Understanding and leveraging APA dynamics could unlock new therapeutic avenues. However, current computational methods for detecting cleavage and polyadenylation sites (C/PASs) and analyzing 3′ UTR length variations in bulk RNA-seq data face major hurdles, such as inadequate C/PAS annotations, challenges in disentangling overlapping C/PASs, and difficulties in pinpointing specific APA site changes. These challenges become more pronounced in large-scale cohort studies, such as ROSMAP, TCGA, and Answer ALS, which lack dedicated 3′ UTR sequencing data. This study introduces PolyAMiner-Bulk, a robust bioinformatics tool, to address these limitations. Utilizing an advanced deep learning model, C/PAS-BERT, PolyAMiner-Bulk aims for precise C/PAS identification and comprehensive APA analysis, bridging the gap in APA research using bulk RNA-seq data.
In the era of abundant bulk RNA-seq data, Jonnakuti et al. introduce PolyAMiner-Bulk, a powerful attention-based deep learning algorithm for accurate analysis of alternative polyadenylation (APA) in bulk RNA-seq data. Overcoming limitations of existing methods, PolyAMiner-Bulk captures tissue-specific APA changes, resolves overlapping cleavage and polyadenylation sites, and generates visualizations.
Introduction
Alternative polyadenylation (APA) is a post-transcriptional regulatory mechanism that cleaves a pre-mRNA molecule and appends adenosine residues at one of its potentially several cleavage and polyadenylation sites (C/PASs), ultimately resulting in multiple mRNA isoforms with varying 3′ UTR lengths. By controlling the length of the 3′ UTR, APA allows the differential inclusion of binding sites specific for microRNAs (miRNAs) and RNA-binding proteins.1 As more than half of human genes contain C/PASs and undergo APA, this widespread phenomenon plays critical roles in development, and its misregulation has been implicated in several diseases, including neurodegeneration and cancer.2,3,4 With increasing awareness of its role in human health and disease, researchers have recognized APA as a more critical post-transcriptional mechanism than previously realized. Consequently, the community has developed specialized deep 3′ UTR sequencing protocols such as PAC-Seq, PAS-Seq, and 3′READS to further study this phenomenon in various disease models.5,6,7 These specialized APA-aware datasets represent only a small fraction of all currently available transcriptomic data, which are mostly generated using bulk RNA sequencing protocols.
There is an immediate need for a robust computational model that can leverage existing bulk RNA-seq datasets to decipher APA dynamics accurately and precisely. For example, multiomics data consortiums like the Religious Orders Study/Memory and Aging Project (ROSMAP) contain robust bulk RNA sequencing (RNA-seq of the human frontal cortex for aging and Alzheimer’s disease.8,9,10 However, they are notably devoid of corresponding 3′ UTR sequencing datasets required for the direct study of APA dynamics. Furthermore, resequencing the more than 800 samples from the ROSMAP data consortium is cumbersome and impractical. Other data consortiums, like The Cancer Genome Atlas (TCGA), which contains over 20,000 samples from control and primary cancer disease populations spanning 33 cancer types, and the Answer ALS data portal, which contains over 1,200 samples from control and neurodegenerative disease populations, can similarly benefit from such a tool. 11,12
Current computational approaches for identifying C/PASs and quantifying 3′ UTR length changes from bulk RNA-seq data fail to unravel tissue- and disease-specific APA dynamics (Figure S1). The current generation of bioinformatics tools predominantly relies on (1) a priori C/PAS annotations, (2) transcript reconstruction, (3) poly(A)-capped reads, and (4) read density fluctuations near the 3′ UTR.13 Databases containing predefined a priori C/PAS annotations are incomplete, contain artificial noise, and do not converge with other a priori C/PAS databases.14,15,16,17,18,19,20,21 Methods that try to infer 3′ UTR usage by transcript reconstruction from bulk RNA-seq data are hampered by inherent limitations of transcript assembly. In addition to being computationally demanding when reconstructing lowly expressed transcripts, these tools often ignore isoforms with shorter 3′ UTRs, as they inaccurately assign reads when shorter isoforms are embedded in longer isoforms.22,23 Furthermore, tools that only rely on poly(A)-capped reads or reads that contain unmapped stretches of adenosines suffer from low sensitivity, as these softclipped reads are relatively scarce in standard bulk RNA-seq data due to the inherently reduced read coverage and noise near transcript ends.24 Last, tools whose core APA inference engine centers around detecting read density fluctuations near the 3′ UTR require good coverage of the 3′ UTR.25,26,27 This restriction limits the number of qualified genes in a sample for APA analysis after discarding genes with low read coverage. Furthermore, this class of tools is particularly vulnerable to non-biological variability and read density heterogeneity.
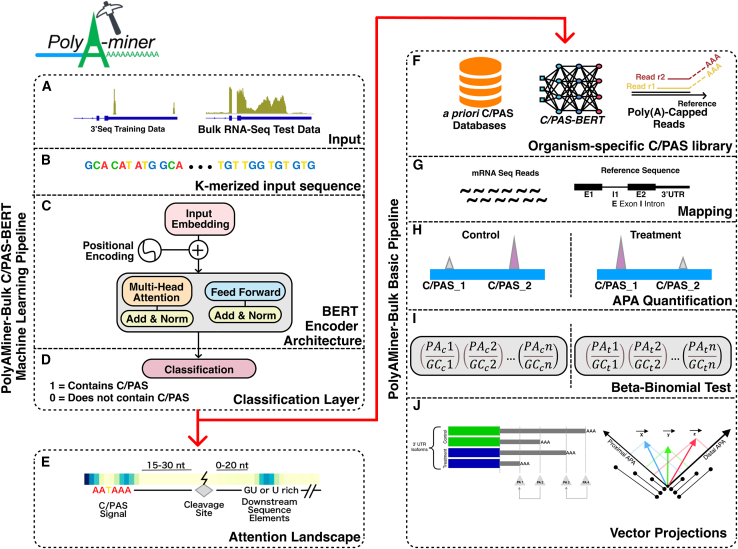
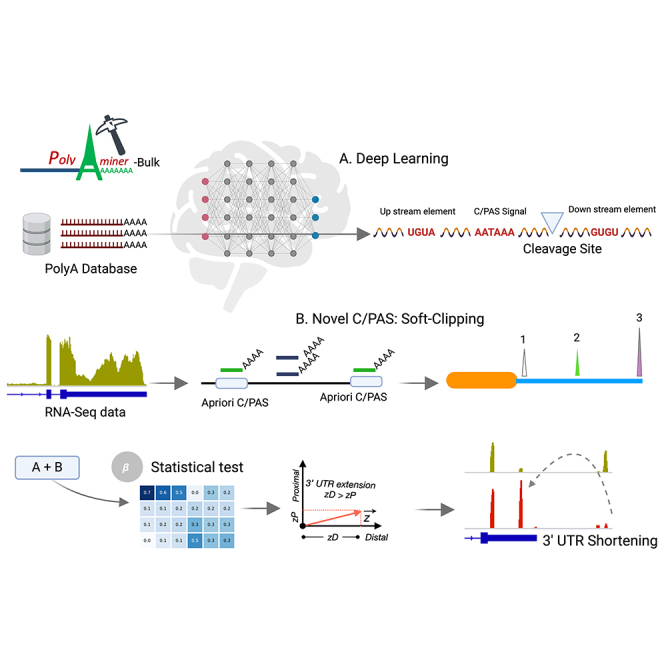
In sum, the current generation of bioinformatics tools for identifying C/PASs and quantifying 3′ UTR length changes from bulk RNA-seq data are limited by poor C/PAS annotations that do not converge with other C/PAS databases, intrinsic limitations of de novo C/PAS detection, failure to deconvolute overlapping C/PASs, and inability to detect intra-distal or intra-proximal APA changes. Recently, an attention-based deep learning model, DNABERT, has been used to detect alternative splice sites from genomic sequences using a directional encoder representation (bidirectional encoder representations from transformers [BERT]) to capture a global understanding of genomic sequences based on neighboring nucleotide contexts.28 This landmark study showcases the power of attention-based models, as they do not rely on motifs’ presence; instead, they model DNA as a language and capture hidden genomic grammar and the semantic dependency between multiple DNA sequence features. However, no deep learning model with a similar attention-based architecture exists to identify C/PASs. The contextual semantic insights garnered by such a model would overcome the limitations of current C/PAS databases by filtering sequence artifacts and retaining true C/PASs. Here, we develop a bioinformatics algorithm and application, PolyAMiner-Bulk, that addresses not only these concerns but also offers an end-to-end paradigm for the complete analysis of APA changes from input bulk RNA-seq data. The methodical flow of PolyAMiner-Bulk is illustrated in Figure 1. In brief, PolyAMiner-Bulk detects de novo C/PASs, merges them with a priori C/PAS databases like PolyA_DB and PolyASite, filters these candidate C/PASs using the C/PAS-BERT deep learning model to create an accurate and comprehensive C/PAS collection, deconvolutes overlapping C/PASs, and employs vector projections to examine APA dynamics throughout the gene body. A detailed description of the proposed approach is given in the STAR Methods, with its key merits illustrated in Figures 2, 3, and 4.
Figure 1.
Illustration of PolyAMiner-Bulk pipeline
(A) Input data.
(B) K-merized input sequence.
(C) BERT encoder architecture.
(D) Classification layer.
(E) Attention landscape.
(F) Amassed and filtered candidate C/PAS library.
(G) Mapping.
(H) APA quantification.
(I) Beta-binomial statistical testing.
(J) Vector projections. In brief, PolyAMiner-Bulk detects de novo C/PASs, merges them with an a priori C/PAS database like PolyA_DB or PolyASite, filters these candidate C/PASs using the C/PAS-BERT machine learning model to create an accurate and comprehensive C/PAS collection, and employs vector projections to examine APA dynamics in all genic regions.
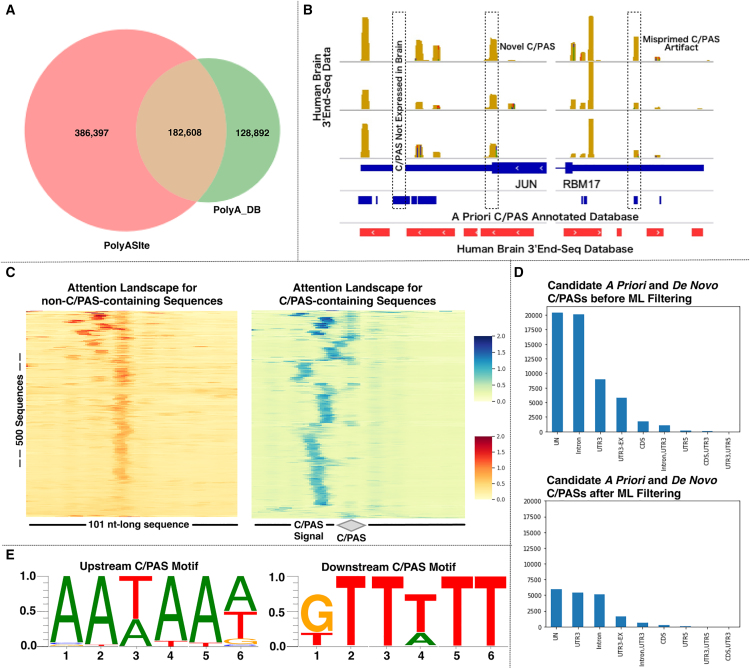
Figure 2.
C/PAS-BERT successfully filters artificial C/PASs and recapitulates the underlying C/PAS grammar
(A) Venn diagram set analysis of PolyASite and PolyA_DB and comparisons of read density visualizations of in-house human brain-specific 3′ UTR-seq data with PolyASite and PolyA_DB. The limitations of current a priori C/PAS databases are the inclusion of C/PASs that are not present in the tissue of interest (brain in this example) but present in other tissues, exclusion of novel C/PASs, and inclusion of misprimed C/PAS artifacts.
(B) Overall performance metrics of the C/PAS-BERT machine learning model.
(C) Attention landscapes for (non-)C/PAS-containing sequences.
(D) Genic C/PAS distribution before and after C/PAS-BERT filtering.
(E) Motif enrichment analysis of the high attention regions of C/PAS-containing sequences.
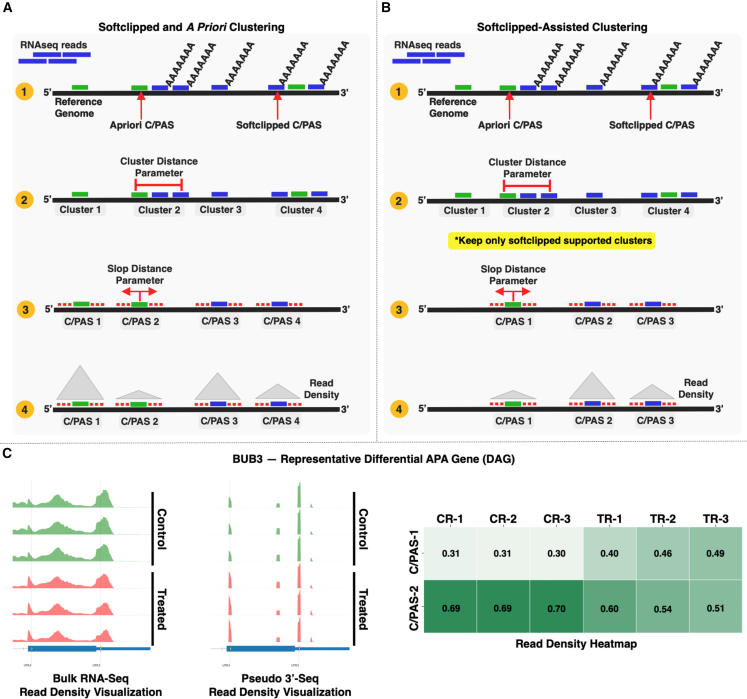
Figure 3.
C/PAS clustering module
(A) Softclipped and a priori clustering mode.
(B) Softclipped-assisted clustering mode. In this mode, PolyAMiner-Bulk only keeps softclipped-supported clusters, allowing for additional specificity in selecting C/PASs supported by the dataset.
(C) Representative differential APA gene identified using the softclipped-assisted clustering mode.
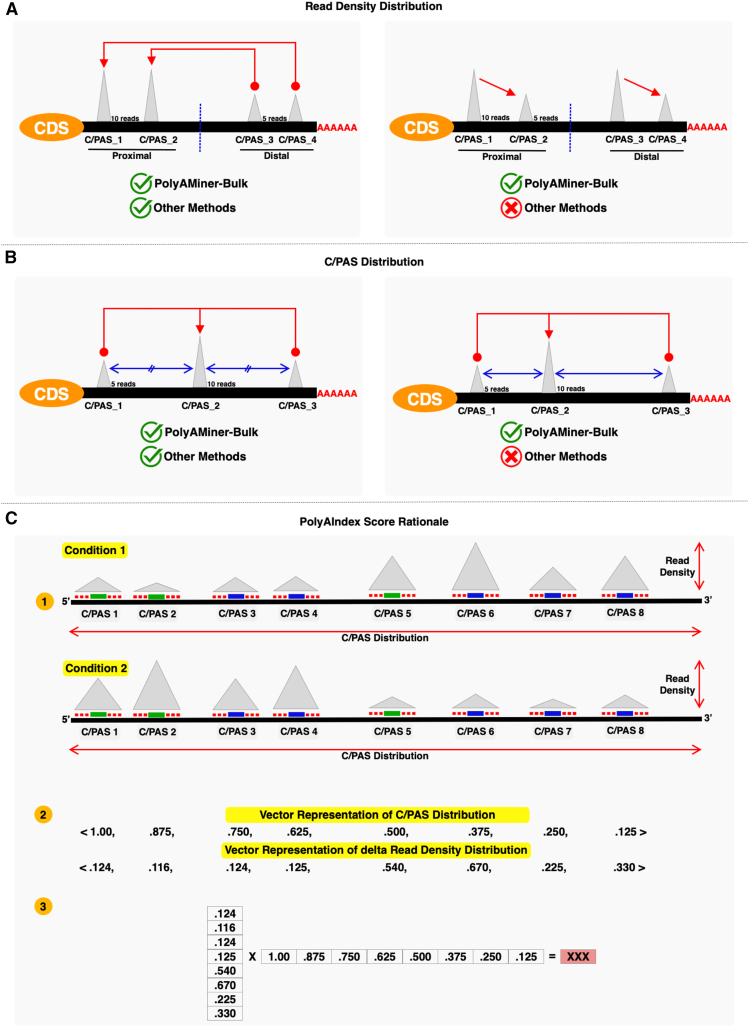
Figure 4.
Vector projection module
(A) Taking read density distribution accounts for all identified APA isoforms, unlike every other methodology that ignores APA changes involving intermediate C/PASs.
(B) Taking C/PAS distribution accounts for non-equidistant 3′ UTR length changes, unlike every other methodology that assumes C/PASs to be uniformly distributed.
(C) Modified vector projection-based engine that takes into account both the distribution of C/PASs along a gene and the read density underlaying each C/PAS.
Results
The attention-based C/PAS-BERT machine learning model successfully filters artificial C/PASs and recapitulates the underlying C/PAS grammar
Filtering artificial C/PASs is highly complex due to the existence of polysemy and distant semantic relationships. Other researchers have previously published a pre-trained bidirectional encoder representation, named DNABERT, that forms global and transferrable understanding of genomic DNA sequences based on up- and downstream nucleotide contexts. In their study, they have convincingly demonstrated that their model, after easy fine-tuning using small task-specific data, can achieve state-of-the-art performance on many sequence prediction tasks and can outperform other deep learning-based architectures like convolutional neural networks (CNNs). For our C/PAS filtering task, we fine-tuned the pre-trained DNABERT model with task-specific data to create C/PAS-BERT. We chose this approach for several reasons. First, BERT is a bidirectional model, allowing it to take the entire context of a genomic sequence into account, both left and right of the target C/PAS. This feature is especially useful for detecting C/PAS motifs, as these signals may appear in different positions within a genomic sequence. Second, DNABERT is pre-trained on large amounts of genomic data, allowing it to learn a broad range of linguistic patterns. This pre-training makes it easier to fine-tune the model on a specific task, such as filtering artificial C/PASs. Third, DNABERT can be fine-tuned on a relatively small amount of labeled data, making it easier to train on C/PAS datasets that may not be comparatively as large. Last, at the heart of C/PAS-BERT is the attention mechanism, which differentially weighs the importance of different parts of the input. This attention mechanism has been effective for a wide variety of natural language processing tasks, and the task of deciphering gene-regulatory code to filter artificial C/PASs out from the candidate C/PAS library can similarly be modeled as a natural language processing task.29,30,31 Just as one may skim through a text corpus and focus on the most important sentences to generate a sense of the main ideas, the attention mechanism in C/PAS-BERT tries to focus on the most relevant parts (or motifs) of the genomic input to filter out artificial C/PASs from the candidate C/PAS library.
Our evaluation showed that C/PAS-BERT performed well, with an accuracy of 0.904, area under the curve of 0.960, F1 score of 0.904, precision of 0.904, and recall of 0.904. Unlike other deep learning models like CNNs, C/PAS-BERT is not entirely a “black box” model because it is based on the transformer architecture, which is a highly interpretable framework. The transformer architecture is designed to allow easy visualization and interpretation of the model’s attention mechanism. Attention weights can be visualized to understand which parts of the input sequence are important for predicting the output. Within the context of our task of filtering out artificial C/PASs from the candidate C/PAS library, we sought to visualize important regions from positively labeled (C/PAS-containing) and negatively labeled (non-C/PAS-containing) sequences as attention landscapes (STAR Methods). Using this methodology, we retrieved two attention landscapes: one for 101-bp non-C/PAS-containing sequences and another for 101-bp C/PAS-containing sequences (where the C/PAS was located directly at the center of the sequence) (Figure 2C). Based on the non-C/PAS-containing sequence attention landscape, we observed that the model placed consistently high attention at the center of the sequence. Based on the C/PAS-containing sequence attention landscape, we can appreciate that the model learned three important genomic features previously validated as necessary for C/PAS detection. Established APA biology suggests that multiple sequence elements are necessary for cleavage and polyadenylation.32 These elements include the C/PAS signal element that is located 15–30 bp upstream of the cleavage site and downstream sequence elements that are located around 20 bp downstream of the cleavage site. Furthermore, the distance between the C/PAS signal element and downstream sequence elements determines the 3′ end formation. These elements themselves and the distance between these elements are variable. As expected, we observed consistently high attention upon (1) the C/PAS, which is located at the center of the sequence, hereafter denoted as position 0; (2) the 15- to 30-nt region upstream of the C/PAS; and (3) the 0- to 20-nt region downstream of the C/PAS.
We performed motif enrichment analysis (STAR Methods) and further characterized these upstream and downstream high-attention regions within our C/PAS-containing genomic sequences to determine the active motifs of the C/PAS-BERT model. Based on established APA biology, we would expect upstream high-attention regions to contain well-conserved C/PAS signal motifs like AATAA and downstream high-attention regions to contain GT- or T-rich sequence elements. The results from our motif enrichment analysis indicate that C/PAS-BERT recapitulates this underlying APA biology. In the 15- to 30-nt region upstream of the C/PAS, we identified AATAAA and its variants (e.g., AAATAA, ATAAAA, ATTAAA, ATAAAT, ATAAAG, CAATAA, TAATAA, ATAAAC, AAAATA, AAAAAA, and AAAAAT) as the upstream C/PAS signal (Figure 2D, left). Moreover, a similar analysis on the nucleotide region downstream of the C/PAS yielded GTTTTT and its variants as the downstream C/PAS signal (Figure 2D, right). Retrieving these well-conserved signals increases our confidence that C/PAS-BERT is actively learning biologically important features to identify C/PASs.
Last, we plotted the distribution of C/PAS locations before and after filtering with C/PAS-BERT (Figure 2E). Before filtering our candidate C/PAS library with C/PAS-BERT, we found that most of the candidate C/PASs were in unannotated or intronic regions (∼20,000 C/PASs each). Most surprisingly, the C/PASs found in either the unannotated or intronic regions outnumber the C/PASs found in the 3′ UTR (∼8,000). This suggests that there may be false-positive C/PASs in these regions and highlights the need for an efficient filtering model. After filtering our candidate C/PAS library with C/PAS-BERT, we observed that the number of C/PASs in the intronic and unannotated genic regions significantly decreased, while the number of C/PASs in the 3′ UTR remained stable. These data suggest that most filtered C/PASs were from unannotated gene and intronic regions, which we expected to contain the highest proportion of artificial C/PASs. This finding further aligns with established APA biology because the 3′ UTR likely contains the lowest proportion of artificial C/PASs, and most C/PASs in the 3′ UTR were preserved.
Figure S2 shows a representative differential APA gene identified using C/PAS-BERT. These findings not only converge with established APA biology but also demonstrate the power of attention-based deep learning models, as they do not rely on the simple presence or absence of motifs. Rather, they employ powerful contextual understanding to simultaneously understand multiple semantic features (like element composition and distance). Taken together, these results support the validity and power of our C/PAS-BERT model, which contains an attention-based architecture to understand distinct DNA sequence semantic relationships around C/PASs.
PolyAMiner-Bulk significantly enhances our ability to decode APA dynamics from bulk RNA-seq data
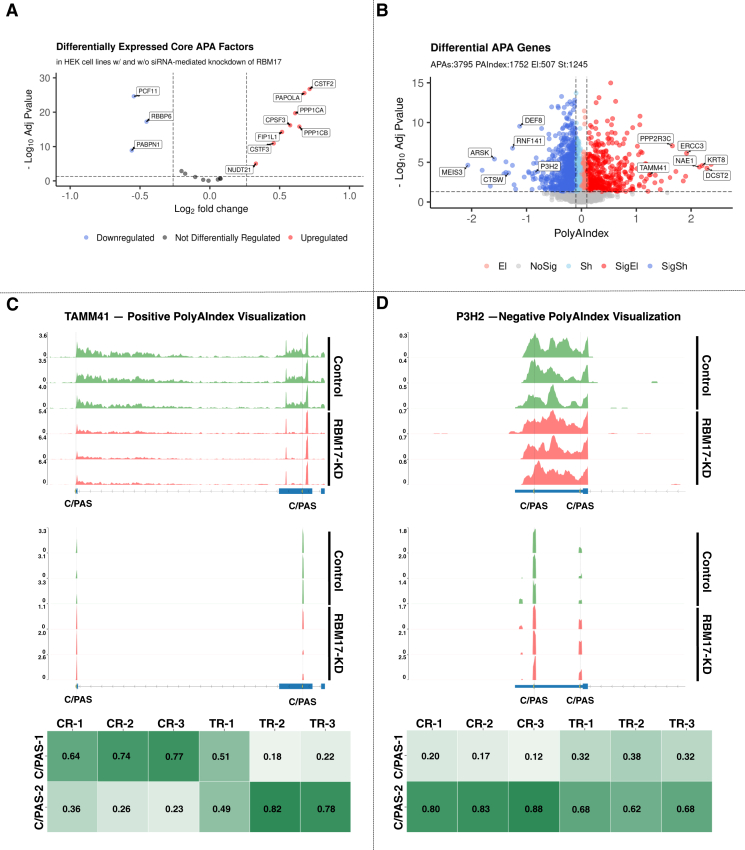
We benchmarked PolyAMiner-Bulk against several of the most common bulk RNA-seq-based APA methods to examine the APA dynamics in a bulk RNA-seq dataset of immortalized human embryonic kidney (HEK293) cells with and without small interfering RNA (siRNA)-mediated knockdown of RNA-binding motif protein 17 (RBM17) (GEO: GSE107648). Previous research has shown this protein to regulate the expression and splicing of RNA-processing proteins.33 Moreover, this RBM17 knockdown and control contrast reveals differential expression of several protein factors that facilitate APA.34 These differential core APA factors include NUDT21, CSTF3, FIP1L1, PPP1CB, CPSF3, PPP1CA, PAPOLA, CSTF2, PABPN1, RBBP6, and PCF11 (Figure 5A). Previously published studies have shown that differential expression of even just one core APA factor like NUDT21 has been shown to substantially perturb APA dynamics.35,36,37,38,39 Since all of these core APA factors aid in the regulation, detection, cleavage, and polyadenylation of a C/PAS, the differential expression of 11 core APA factors strongly suggests that the knockdown of RBM17 substantially perturbs APA dynamics.
Figure 5.
PolyAMiner-Bulk analysis of the bulk RNA-seq benchmarking dataset of HEK293 cells with and without siRNA-mediated knockdown of RBM17
(A) Volcano plot of differentially expressed core APA factors.
(B) Volcano plot of differential APA genes.
(C) Representative differential APA gene with a positive PolyAIndex, suggesting 3′ UTR elongation.
(D) Representative differential APA gene with a negative PolyAIndex, suggesting 3′ UTR shortening.
PolyAMiner-Bulk detected 3,795 significant differential APA genes (DAGs), of which 1,752 genes exhibited a PolyAIndex magnitude greater than 0.1 or less than −0.1. Of these DAGs, 1,245 underwent 3′ UTR shortening, and 507 underwent 3′ UTR lengthening (Figure 5B). These results are in line with expectations and are not surprising.
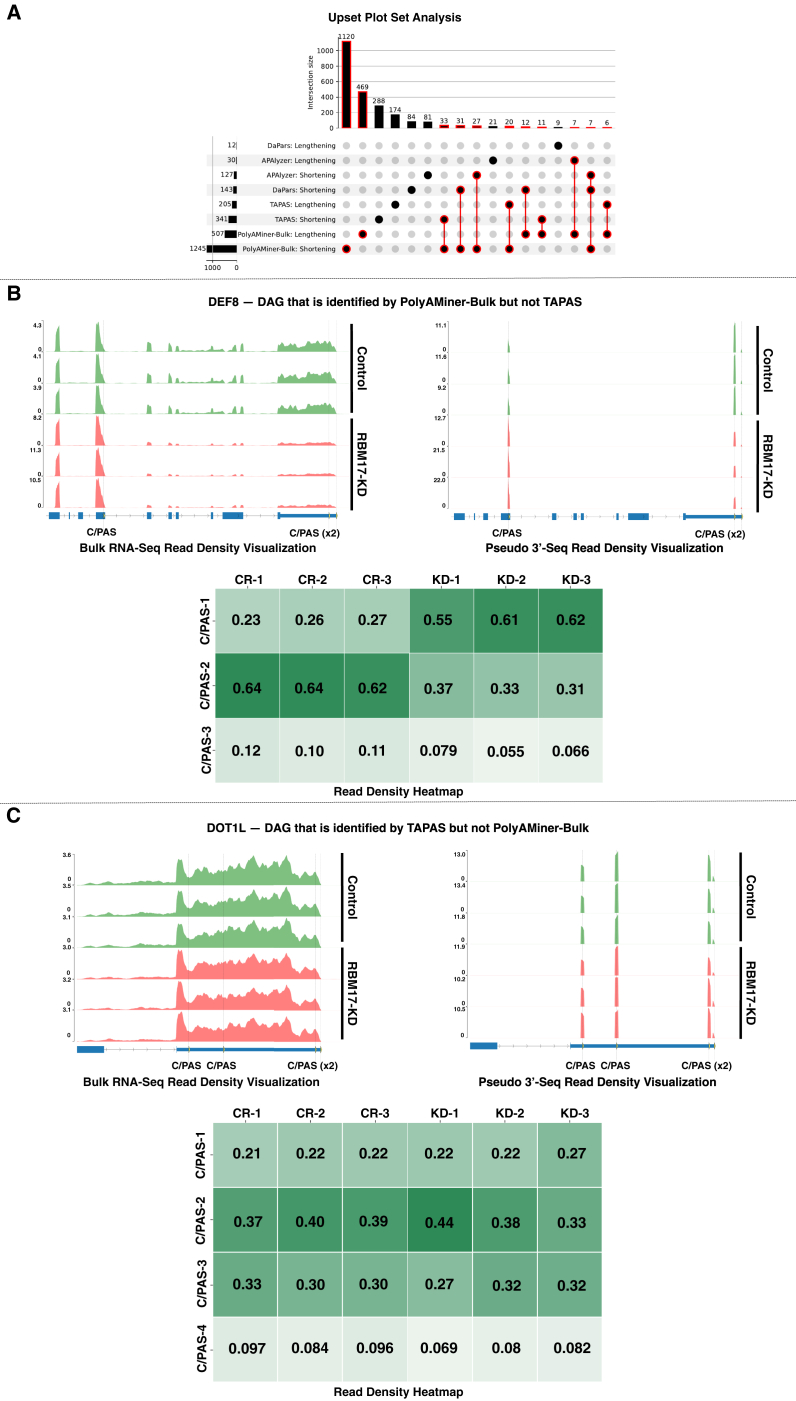
PolyAMiner-Bulk identified 1,120 genes with 3′ UTR shortening and 469 genes with 3′ UTR lengthening that were not detected by other methods (Figure 6A). To validate these predictions, we categorized the genes based on the number of C/PASs and examined changes in read densities at individual C/PASs between the control and RBM17 knockdown conditions using heatmaps (Figure S3). The heatmaps in Figure S3A show an increase in read density at the proximal C/PAS 1 and decreased read density in at the distal C/PAS 2 for 3′ UTR shortening genes with 2 C/PASs and vice versa for elongating genes. Similar results were observed for genes with 3 C/PASs (Figure S3B). The differences in read density observed in these heatmaps strongly support our predictions of 3′ UTR shortening and elongation.
Figure 6.
Comparison of PolyAMiner-Bulk against current-generation tools
(A) UpSet plot analysis.
(B) Representative DAG identified by PolyAMiner-Bulk but not TAPAS, a current-generation tool.
(C) Representative DAG identified by TAPAS but not PolyAMiner-Bulk.
To further validate the PolyAMiner-Bulk predictions, we visualized the C/PAS read density fluctuations of representative DAGs for control and RBM17 knockdown groups (Figures 5C and 5D). For example, TAMM41, involved in mitochondrial translocator assembly and maintenance, is a representative DAG with a positive PolyAIndex metric, suggesting that this gene is undergoing 3′ UTR lengthening in the RBM17 knockdown condition compared with the control condition (Figure 5C).40,41 On the other hand, P3H2, which is involved in collagen chain assembly and stability, is a representative DAG with a negative PolyAIndex metric, suggesting that this gene is undergoing 3′ UTR shortening in the RBM17 knockdown condition compared with the control condition (Figure 5D).42,43 We visualized both genes’ read density as bulk RNA-seq and pseudo-3′ UTR-seq read coverage and plotted their corresponding density proportions as a heatmap. In the control condition, TAMM41 exhibits higher read proportion density in its proximal 3′ UTR C/PAS, whereas TAMM41 shifts a proportion of its read density toward the distal 3′ UTR C/PAS in the RBM17 knockdown condition. By contrast, in the control condition, P3H2 exhibits higher read proportion density in its distal 3′ UTR C/PAS, whereas P3H2 shifts a proportion of its read density toward the proximal 3′ UTR C/PAS in the RBM17 knockdown condition. These data-driven visualizations support PolyAMiner-Bulk predictions.
To assess the performance of PolyAMiner-Bulk, we tested DaPars, APAlyzer, and TAPAS, the currently utilized bulk RNA-seq-based APA methods, on this RBM17 knockdown bulk RNA-seq dataset.19,25,27 Other computational methods identified substantially fewer DAGs, which does not align with one’s expectations of differential APA dynamics in a setting where 11 core APA factors are differentially expressed. DaPars identified 155 DAGs (12 undergoing 3′ UTR lengthening and 143 undergoing 3′ UTR shortening), APAlyzer identified 157 DAGs (30 undergoing 3′ UTR lengthening and 127 undergoing 3′ UTR shortening), and TAPAS identified 546 DAGs (205 undergoing 3′ UTR lengthening and 341 undergoing 3′ UTR shortening). After performing an UpSet plot set analysis, we observed that PolyAMiner-Bulk not only identifies the largest number of unique DAGs but also identifies the highest number of DAGs that were also identified by other methods (Figure 6A). This increased sensitivity in DAG detection can be attributed to (1) our tool’s ability to capture a more comprehensive collection of C/PASs within a feature space and (2) our vector projection-based approach that helps identify significant intra-distal and intra-proximal APA changes that current generation methods would have otherwise ignored. Capturing and quantifying these APA dynamics may be biologically relevant, as a loss (or gain) of these intra-distal and intra-proximal C/PASs can lead to a loss (or gain) of a more significant number of regulatory binding sites for regulatory molecules like RBPs or miRNAs.
We further sought to validate PolyAMiner-Bulk predictions by characterizing the unique genes identified by PolyAMiner-Bulk and no other method as well as the unique genes identified by other methods and not PolyAMiner-Bulk. The DAGs uniquely identified by PolyAMiner-Bulk are well represented by visualizations of read density fluctuations of genes near their respective C/PASs for control and RBM17 knockdown groups. DEF8, involved in cation binding, is a representative gene classified by PolyAMiner-Bulk but not observed in other methods like TAPAS (Figure 6B).44 Compared with the control condition, DEF8 undergoes 3′ UTR shortening in the RBM17 knockdown condition. In the control condition, DEF8 exhibits higher read proportion density in its distal 3′ UTR C/PAS, whereas DEF8 shifts a proportion of its read density toward the proximal 3′ UTR C/PAS in the RBM17 knockdown condition. We also visualized genes uniquely identified as undergoing significant APA changes by methods other than PolyAMiner-Bulk, like TAPAS. DOT1L, involved in methylating lysine 79 of histone H3 in nucleosomes, is one such representative gene (Figure 6C).45 PolyAMiner-Bulk does not classify DOT1L as a DAG since the three samples do not uniformly undergo changes in read density among the four C/PASs between each condition. The corresponding read density and heatmap visualizations further corroborate this result and support the notion that DOT1L was mispredicted as a DAG by other methods. Comparisons between PolyAMiner-Bulk and other methods like APAlyzer and DaPars demonstrate a similar pattern where read density visualizations advocate the merit of PolyAMiner-Bulk (Figure S4).
Revisiting published data using PolyAMiner-Bulk reveals APA dynamics and pathways in scleroderma pathology
Previously published studies have established that NUDT21 (also known as CFlm25), a core APA factor, directs differential APA and that its suppression induces a collection of 3′ UTR shortening events through loss of stimulation of distal C/PASs.35,37,46,47,48 One such study examined the effects of NUDT21 knockdown in normal skin fibroblasts and noted the 3′ UTR shortening of key transforming growth factor β (TGF-β)-regulated fibrotic genes.38 We used PolyAMiner-Bulk to re-analyze this bulk RNA-seq dataset of skin fibroblasts with and without siRNA-mediated knockdown of NUDT21 (GEO: GSE137276) and compared the output with previously published results.
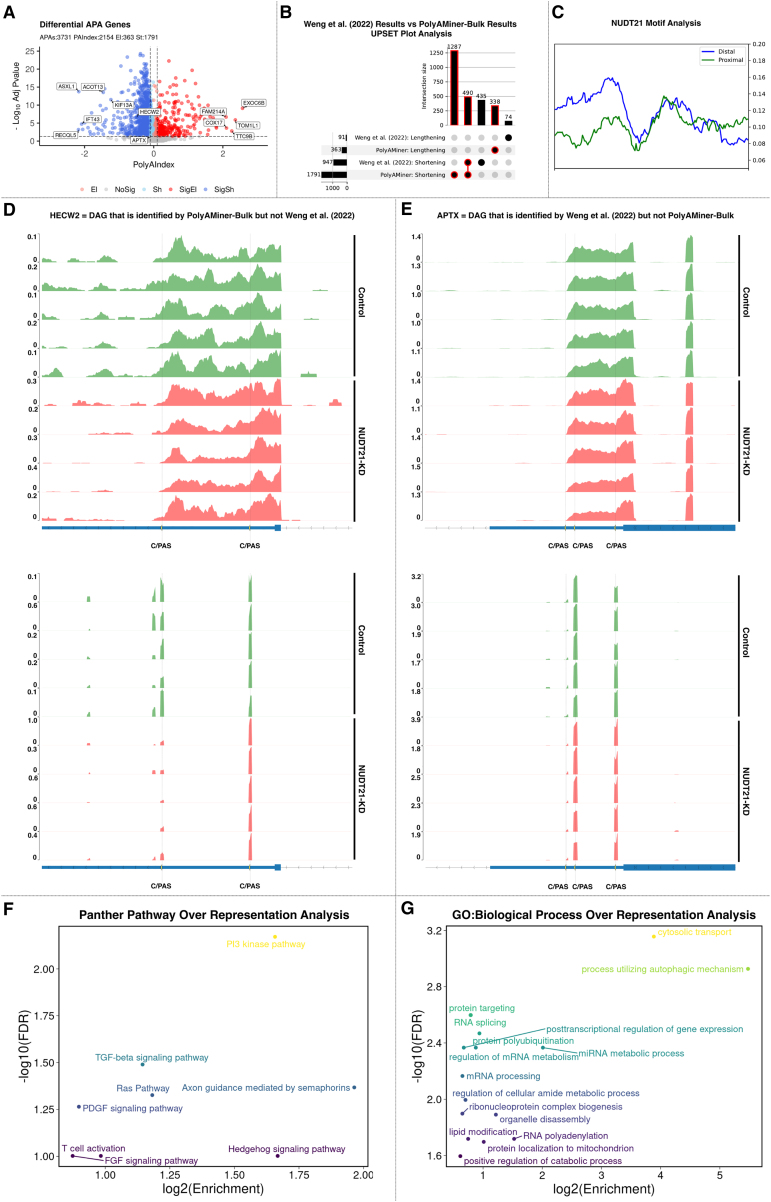
PolyAMiner-Bulk detected 3,731 significant DAGs, of which 2,154 exhibited a PolyAIndex magnitude greater than 0.1. Of these DAGs, 1,791 underwent 3′ UTR shortening, and 363 underwent 3′ UTR lengthening (Figure 7A). In contrast, the previously published study reported only 1,038 DAGs, with 947 undergoing 3′ UTR shortening and 91 undergoing 3′ UTR lengthening (Figure 7B). Of note, PolyAMiner-Bulk not only recapitulated more than 50% of the 3′ UTR shortening DAGs identified by the other study but also identified 1,287 unique 3′ UTR shortening DAGs. As discussed previously, our improved C/PAS identification paradigm and vector projection-based approach underlie PolyAMiner-Bulk’s increased sensitivity.
Figure 7.
PolyAMiner-Bulk analysis of the bulk RNA-seq dataset of skin fibroblasts with and without siRNA-mediated knockdown of NUDT21
(A) Volcano plot of differential APA genes.
(B) UpSet plot analysis of PolyAMiner-Bulk results against previously published results.
(C) NUDT21 motif and biding analysis of the unique 3′ UTR shortening genes identified by PolyAMiner-Bulk.
(D) Representative differential APA gene identified by PolyAMiner-Bulk but not the previously published study.
(E) Representative differential APA gene identified by the previously published study but not PolyAMiner-Bulk.
(F) Panther Pathway over-representation analysis of the differential APA genes identified by PolyAMiner-Bulk.
(G) Gene Ontology (GO:Biological Process over-representation analysis of the differential APA genes identified by PolyAMiner-Bulk.
To substantiate our PolyAMiner-Bulk results, we characterized the unique DAGs. NUDT21 loss has been demonstrated to cause widespread 3′ UTR shrinking in many independent studies, including the original study’s authors. PolyAMiner-Bulk predicted 1,287 additional genes with 3′ UTR shortening and 338 additional genes with 3′ UTR elongation compared with the original report. Despite predicting more genes with APA changes, these findings are consistent with the previous observation of predominantly 3′ UTR shrinking, indicating that PolyAMiner-Bulk’s results are biologically and mechanistically valid.
To validate PolyAMiner-Bulk predictions, we investigated the distribution of the NUDT21 binding motif in the genes undergoing 3′ UTR shortening and explored the distribution of the NUDT21 binding motif, UGUA, within their 3′ UTR (Figure 7C). Earlier studies showed that NUDT21 binds to the UGUA motif and reported global 3′ UTR shortening with a significant enrichment of the UGUA motifs near the distal C/PASs compared with the proximal C/PASs in NUDT21 knockdown models.35 Consistent with this hallmark signature and in agreement with previous reports, we found a significant enrichment of the UGUA binding motif frequency upstream of the distal C/PASs compared with the proximal C/PASs within the 3′ UTR of unique DAGs that undergo 3′ UTR following siRNA-mediated knockdown of NUDT21 in skin fibroblasts. This observation supports a model whereby NUDT21 is directed to distal sites to facilitate APA and suggests that the 1,287 unique 3′ UTR shortening DAGs identified by PolyAMiner-Bulk are indeed targets of NUDT21 and actual signals.
Furthermore, PolyAMiner-Bulk results are well supported by the C/PAS read density visualizations of representative DAGs for control and NUDT21 knockdown conditions. For example, HECW2, involved in ubiquitin-protein ligase activity, is a representative DAG identified by PolyAMiner-Bulk and not by the previously published study.49,50,51 This gene underwent 3′ UTR shortening under the NUDT21 knockdown condition compared with the control condition, a finding that is corroborated by read density visualizations (Figure 7D). Under the control condition, HECW2 exhibits higher read proportion density in its distal 3′ UTR C/PAS, whereas HECW2 shifts a proportion of its read density toward the proximal 3′ UTR C/PAS under the NUDT21 knockdown condition. Of significant interest, the authors of this previously published study themselves have independently identified HECW2 as being involved in scleroderma pathogenesis in a separate study.52 We also visualized genes uniquely identified by the previously published study. APTX, involved in single-stranded DNA repair, is one such representative DAG. 53 PolyAMiner-Bulk does not classify APTX as a DAG since the five samples for each condition do not uniformly undergo changes in read density across the three C/PASs. The corresponding read density and heatmap visualizations further corroborate this result and support the merit of PolyAMiner-Bulk in minimizing false positives (Figure 7E).
Last, we performed functional enrichment analyses to compare the biological insights from the DAGs identified by PolyAMiner-Bulk with those identified by the previously published study. We first determined whether any subset within the 3′ UTR shortening DAGs identified by PolyAMiner-Bulk shared more or fewer genes with the “Panther Pathway” database than one would expect by chance. While several pathways, like TGF-β signaling and T cell activation, were enriched by both PolyAMiner-Bulk and the previously published study, other pathways, like phosphatidylinositol 3-kinase (PI3K), Ras, Hedgehog, fibroblast growth factor (FGF), and platelet-derived growth factor (PDGF) signaling pathways were uniquely enriched in the PolyAMiner-Bulk 3′ UTR shortening DAG set (Figure 7F). Furthermore, over-representation functional analysis against the “Gene Ontology: Biological Processes” database reveals significant enrichment for post-transcriptional regulation of gene expression, protein polyubiquitination, mRNA processing, and positive regulation of catabolic processes in the PolyAMiner-Bulk 3′ UTR shortening DAG set (Figure 7G). Taken together, these results demonstrate that identifying these additional DAGs increases our understanding of the underlying biology and reveals previously underappreciated APA dynamics in scleroderma.
Discussion
Limitations of the study
We made every effort within our purview to ensure the rigor and reliability of our computational findings. Although experimental validations were not feasible at this juncture, we are confident in the foundational strength of our computational model. It is important to acknowledge that, as with any methodological approach, there is a potential for false positives. This is particularly pertinent in the context of bulk RNA-seq data, which can have challenges with 3′ UTR coverage, often resulting in variability and noise. We address this by focusing on datasets with high sequencing depth, which significantly mitigates these issues. Nonetheless, we recognize the value of orthogonal approaches to validate the computational predictions of PolyAMiner-Bulk.
Identifying C/PASs is highly complex due to the existence of polysemy and distant semantic relationships. It has been accepted that C/PASs universally contain upstream APA signal motifs and downstream APA signal motifs. However, the simple presence of these established APA signal motifs is not sufficient for C/PAS identification. For example, for a genomic site to be considered a C/PAS, certain upstream and downstream motifs may need to be paired together in a particular order and distance away from the genomic site (much like words in a sentence) for the APA machinery to classify the genomic site as a C/PAS. Due to these reasons, deciphering gene-regulatory code to identify C/PASs can be modeled as a natural language processing (NLP) task.
BERT is a language model that uses a transformer architecture to learn contextual relationships between words in a sentence and has achieved state-of-the-art results on many benchmarking NLP tasks. DNABERT is a pre-trained BERT model that can provide a global and transferrable understanding of genomic DNA sequences based on upstream and downstream nucleotide contexts. Ji et al.28 have demonstrated that easy fine-tuning of DNABERT with small task-specific data can achieve state-of-the-art performance on many sequence prediction tasks, outperforming other deep learning-based architectures such as CNNs. We realized that the task of identifying C/PASs, which has long been a challenge in the APA field, can be addressed by fine-tuning the pre-trained DNABERT model with task-specific data. Thus, we used a fine-tuned variant of DNABERT, called C/PAS-BERT, for our analysis. Compared with other computational approaches, C/PAS-BERT is better equipped to capture the elasticity of the GU-rich and U-rich elements that support C/PAS identification, as the relative position of these elements can vary from one C/PAS to another. Our attention-based learning model can learn and adapt to these variations, as evidenced by the attention heat drawn toward the U/GU-rich and AATAAA regions (Figures 3C and 3E).
CNN models, on the other hand, are commonly used for computer vision tasks, such as image classification and object detection, but can also be used for NLP tasks, such as sentiment analysis and named entity recognition. While both BERT and CNN architectures are powerful deep learning models that can be used for a wide range of NLP tasks, including identifying C/PASs, they are not interchangeable. It is important to choose the right model architecture for the specific task at hand, based on factors such as the nature of the data, the size of the dataset, and the complexity of the task.
We consider BERT to be better suited for our task for several reasons. (1) Bidirectional modeling: BERT is a bidirectional model, meaning it can consider the entire context of a sequence of words, both left and right of the target C/PAS. This is especially useful for detecting C/PAS signals, as these signals may appear in different positions within a genomic sequence. (2) Pre-training: BERT is typically pre-trained on large amounts of text data, allowing it to learn a broad range of linguistic patterns. This pre-training makes it easier to fine-tune the model on a specific task, such as identifying C/PASs. (3) Attention mechanism: BERT uses an attention mechanism to identify which words in a sequence are most important for a given task. This allows it to focus on the most relevant parts of a genomic sequence when identifying C/PASs. (5) Transfer learning: BERT can be fine-tuned on a relatively small amount of labeled data, making it easier to train on C/PAS datasets that may not be very large. Nevertheless, future studies with careful and thorough benchmarking experiments to compare machine learning models with different underlying architectures should be performed.
Conclusion
PolyAMiner-Bulk significantly advances our ability to decode APA dynamics from bulk RNA-seq data (summarized in Table S1). For instance, this is the first tool with an attention-based machine learning architecture to identify C/PASs. Attention-based models do not rely on motifs’ presence; instead, they model DNA as a language and capture the hidden grammar and the semantic dependency between multiple DNA sequence features. The contextual semantic insights garnered by such a model overcome the limitations of current C/PAS databases by separating sequencing artifacts and other noise from the true C/PASs. Furthermore, PolyAMiner-Bulk employs a soft-clipped read filtering module to deconvolute overlapping C/PASs. In addition, using vector projections, PolyAMiner-Bulk accounts for all APA changes, including non-proximal to non-distal changes, and can distinguish the most distal to most proximal changes from most distal to intermediate site changes irrespective of absolute change magnitude. This sensitivity is crucial for estimating the true breadth of 3′ UTR shortening and elongation. In addition, our tool takes raw FASTQ or processed alignment files as input and offers an end-to-end APA analysis paradigm. PolyAMiner-Bulk not only identifies DAGs but also generates (1) read proportion heatmaps and (2) read density visualizations of the corresponding bulk RNA-seq tracks and pseudo-3′ UTR-seq tracks, allowing users to appreciate the differential APA dynamics.
Analysis of bulk RNA-seq datasets of HEK cells with and without siRNA-mediated knockdown of RBM17 and skin fibroblasts with and without siRNA-mediated knockdown of NUDT21 strongly supports the value of PolyAMiner-Bulk, as we demonstrated a substantial increase in the number of dynamic APA events detected. With the emerging importance of APA in understanding development and disease and large-scale availability of bulk RNA-seq data consortia like TCGA, ROSMAP, and the Answer ALS data portal, PolyAMiner-Bulk establishes a paradigm and facilitates a deeper understanding of APA dynamics across various diseases, from cancer to neurodegeneration.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| bulk RNA-seq dataset of immortalized human embryonic kidney (HEK293) cells with and without siRNA-mediated knockdown of RNA-binding motif protein 17 (RBM17) | De Maio et al.33 | GSE107648 |
| bulk RNA-seq dataset of skin fibroblasts with and without siRNA-mediated knockdown of NUDT21 | Weng et al.38 | GSE137276 |
| Software and algorithms | ||
| PolyAMiner-Bulk | This paper |
https://github.com/YalamanchiliLab/PolyAMiner-Bulk.git https://doi.org/10.5281/zenodo.10372661 |
| DaPars | Xia et al.25 | https://github.com/ZhengXia/dapars |
| APAlyzer | Wang and Tian19 | https://bioconductor.org/packages/release/bioc/html/APAlyzer.html |
| TAPAS | Arefeen et al.27 | https://github.com/arefeen/TAPAS |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Hari Krishna Yalamanchili (Hari.Yalamanchili@bcm.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
This study employed existing datasets that are publicly accessible. The specific accession numbers for these datasets are detailed in the key resources table.
-
•
All original code developed for this study has been deposited in a GitHub repository and can be freely accessed at https://github.com/YalamanchiliLab/PolyAMiner-Bulk.git. In addition, we have archived this code in Zenodo, an open-access repository, available at https://doi.org/10.5281/zenodo.10372661.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Method details
C/PAS-BERT
When we compared two of the field’s most widely used predefined human-specific a priori C/PAS databases, PolyASite and PolyA_DB, we observed that both databases share 182,608 elements that constitute ∼30% of PolyASite and ∼60% of PolyA_DB (Figure 2A). This finding strongly suggests that, although both databases are based on 3′UTR-seq (rather than bulk RNA-seq) technology, they do not capture all C/PASs and most likely contain false-positive C/PAS artifacts. In addition, our in-house human brain-specific 3′UTR-seq data affirms the limitations of current a priori C/PAS databases (Figure 2B). Taken together, these findings showcase the limitations of current a priori C/PAS databases: (i) Inclusion of C/PASs that are not present in the tissue-of-interest (brain in this example) but are present in other tissues, (ii) Exclusion of novel C/PASs, and (iii) Inclusion of misprimed C/PAS artifacts.
To filter false-positive C/PAS artifacts, we extended the pre-trained DNABERT model with task-specific data and developed C/PAS-BERT, an attention-based deep learning model that understands distinct DNA sequence semantic relationships around C/PASs. Candidate C/PASs that both PolyASite and PolyA_DB shared were considered positively labeled C/PASs. Intergenic sites not within 3000 kb upstream and downstream of any annotated gene were considered negatively labeled C/PASs. 6-mer nucleotide sequence representations were first generated by querying for nucleotides that are 50 bp upstream and downstream of the candidate C/PAS and then walking over these 101 nucleotide-long DNA sequences with a 6-nucleotide long sliding window. Breaking DNA sequences into strings of every 6-nucleotide length and using them as vectors allows for sensitive and specific methods for analyzing genomes. The human dataset consisted of 633,786 tuples (6-mer nucleotide sequence representation, C/PAS label). We ensured that this dataset was balanced – the number of positively and negatively labeled tuples was equal. 90% of this overall dataset was used for k-fold cross-validation, while the remaining 10% was used as an independent test set. We employed 12-fold cross-validation to ensures low time complexity for the training process. The resultant C/PAS-BERT model helps to overcome the limitations of current C/PAS databases by filtering sequencing artifacts and better understanding APA dynamics in gene regulation.
PolyAMiner-Bulk pipeline
PolyAMiner-Bulk takes raw FASTQ or processed BAM alignment files as input and offers an end-to-end APA analysis paradigm. We first generate an organism-specific candidate C/PAS annotation library that consists of a priori C/PASs and de novo C/PASs. A priori C/PASs are sourced from pre-existing C/PAS annotation libraries like PolyA_DB and PolyASite, while de novo C/PASs are sourced directly from the dataset itself. C/PAS-BERT subsequently filters artificial C/PASs out from this candidate C/PAS library to retain only high confidence C/PASs. Since alternative polyadenylation is a relatively non-specific process by which cleavage and polyadenylation can occur within a range of a few nucleotides from a C/PAS, we then deconvolute overlapping C/PASs. These high confidence, deconvoluted C/PAS annotations are overlayed across the dataset to create a read density matrix for each C/PAS, for each gene, for each sample. Lastly, we perform vector projection calculations and statistical testing on this read density matrix to collapse these individual C/PAS-level metrics into a singular gene-level PolyAIndex metric that reflects the gene’s dynamic APA usage between conditions. Furthermore, PolyAMiner-Bulk not only identifies differential APA genes but also generates (i) read proportion heatmaps and (ii) read density visualizations of the corresponding bulk RNA-seq tracks and pseudo-3′UTR-seq tracks, allowing users to appreciate the differential APA dynamics.
Step 1: Processing raw reads
PolyAMiner-Bulk can take either the raw read files in fastq format or the mapped alignment files in bam format as input. Raw reads are mapped to the reference genome of origin using STAR and the resulting alignment files (in bam format) are sorted and indexed using samtools. 54,55
Step 2: Extracting de novo C/PASs
PolyAMiner-Bulk amasses a candidate C/PAS collection from two sources: (i) directly from the input data and (ii) indirectly from existing C/PAS databases. In addition to incorporating a priori C/PASs into our candidate C/PAS library for downstream C/PAS-BERT mediated filtering, PolyAMiner-Bulk detects de novo C/PASs using softclipped read detection. The entirety of a read need not be completely aligned to a reference as the read may contain additional bases that are not in the reference or may be missing bases in the reference. This softclipped region phenomenon underscores the de novo C/PAS extraction engine of PolyAMiner-Bulk. Candidate de novo C/PASs are defined as reads from BAM read alignment files whose ends are softclipped regions containing a softclipped length-dependent proportion of adenosines (or thymines, depending on the strandedness of sequencing). For example, a softclipped tail of >12 nucleotides must contain at least 75% adenosines to be classified as a candidate de novo C/PAS. Shorter softclipped tails require a proportionally greater percentage of adenosines. The default settings for this user-adjustable parameter are at least 90% adenosines for a 4 nucleotides long-softclipped tail, at least 85% adenosines for a 4–8 nucleotides long-softclipped tail, at least 80% adenosines for an 8–12 nucleotides long-softclipped tail, and at least 75% adenosines for a >12 nucleotides long-softclipped tail. This loose thresholding approach is crucial as the poly(A) stretch may not necessarily continue until the end of the read because sequencing can continue into primer sequences at the end of fragments, sequencing quality of stretches of the same nucleotide may rapidly deteriorate, and sequencing errors might disrupt the poly(A) stretch.
Step 3: Filtering candidate C/PASs with C/PAS-BERT
These candidate de novo C/PASs from softclipped-based C/PAS detection are merged with our collection of a priori C/PASs from pre-existing C/PAS annotation databases like PolyA_DB and PolyASite to generate our candidate C/PAS library. We subsequently employ C/PAS-BERT to filter artificial C/PASs out from this candidate C/PAS library and to retain only high-confidence C/PASs. Of note, C/PAS-BERT intends to filter artificial C/PAS noise rather than identify de novo C/PASs. Current tools – like methods that rely only poly(A)-capped reads – use a small subset of this candidate C/PAS library, which does not satisfactorily saturate the C/PAS feature space and leads to poor performance. However, simply concatenating all C/PASs identified by each approach into a singular C/PAS library introduces noise and false-positive C/PASs. The C/PAS-BERT filtering module overcomes this issue by reducing the noise and number of false-positive C/PASs from our candidate C/PAS library.
Step 4: Deconvoluting overlapping C/PASs
Since alternative polyadenylation is a relatively non-specific process by which cleavage and polyadenylation can occur within a range of a few nucleotides from a C/PAS, we equipped PolyAMiner-Bulk with two C/PAS deconvolution modes: (i) softclipped and a priori clustering, as well as (ii) softclipped-assisted clustering (Figure 3). De novo and a priori C/PASs are clustered in both modes based on a user-defined cluster distance parameter (default = 30 bp), and PolyAMiner-Bulk selects the most distal C/PAS within a cluster. Notably, in the softclipped-assisted clustering mode, PolyAMiner-Bulk only keeps softclipped-supported clusters (Figures 3A and 3B). This mode allows for additional specificity in selecting C/PASs supported by the dataset. Other parameters are included to refine this specificity even further, such as a parameter for the minimum number of softclipped reads required for a cluster to be kept and another parameter for the minimum number of unique samples that must meet the criteria mentioned above. Figure 3C shows a representative differential APA gene identified using the softclipped-assisted clustering mode.
Step 5: Quantifying APA dynamics using vector projections
We previously deployed a vector projection based PolyAIndex engine to analyze differential APA dynamics from 3’sequencing data.56 In brief, our first-generation PolyAIndex metric ranks genes by the magnitude of APA changes along a genic region. This ranking is critical for any downstream analysis that takes rank as its input, such as Gene Set Enrichment Analysis (GSEA). Furthermore, this vector projection-based approach accounts for ALL identified APA isoforms, unlike other methodologies that ignore APA changes involving intermediate C/PASs (Figure 4A).
We have modified this PolyAIndex engine for PolyAMiner-Bulk, so that our second-generation PolyAIndex metric also accounts for the distribution of C/PASs along a gene (Figures 4A and 4B). Let us take two scenarios to illustrated in Figure 4 the utility of this change: (i) In scenario 1, a gene shifts APA usage between two neighboring C/PASs between two conditions, and (ii) In scenario 2, a gene shifts APA usage between two faraway C/PASs between two conditions (Figure 4B). The revised engine will take the proximity of these C/PASs into account and report the gene in scenario 1 as having a smaller PolyAIndex metric, despite the gene having the same magnitude of read density change in both scenarios. This metric better reflects underlying post-transcriptional biology as a loss (or gain) of C/PASs that are farther away could result in the loss (or gain) of a more significant number of regulatory binding sites for RBPs or miRNAs.
To calculate the PolyAIndex of a gene, PolyAMiner-Bulk first projects the magnitude of C/PAS usage change to a reference C/PAS in an n-dimensional vector space, where n is the number of C/PASs in the gene. Then, it computes the difference in projections of these vectors between conditions and collapses them into a single gene-level magnitude PolyAIndex metric (Figure 4C). A positive PolyAIndex metric suggests overall 3′UTR lengthening, while a negative PolyAIndex metric suggests overall 3′UTR shortening.
Step 6: Statistical testing
A beta-binomial test is used to determine the significance of each PolyAIndex metric. Let J denote the number of C/PAS reads, U denote the total number of reads spanning across all C/PASs, and the set of natural numbers. Assume J is distributed according to a binomial distribution with success probability r , . To capture the variations between biological replicates, we model r through a beta distribution with and , , where is the beta function. For numerical stability, we can parametrize the beta distribution to , where is the expectation of the r and represents the dispersion. The log likelihood of the observed data is given by:
Assuming there are G groups in an experiment, we let be the maximal log likelihood value for group g = 1, …, G. We propose to test the homogeneity of the groups by likelihood ratio test, where the log likelihood ratio statistics S is given by . S is approximately distribution with 2 degrees of freedom. The null hypothesis of this test is that the expectation and dispersion of the different groups are equal. Every gene-level APA change is multiple testing corrected using Benjamini-Hochberg procedure.57 Gene-level APA changes with an adjusted p value <0.05 are predicted as significant APA changes.
Step 7: Visualizing APA changes
After calculating PolyAIndex metrics, PolyAMiner-Bulk can further investigate APA dynamics of individual genes through its visualization module. We implement pyGenomeTracks and Matplotlib to generate gene-level read density coverage plots and corresponding C/PAS usage heatmaps from the bulk RNA-seq input data.58,59,60 Notably, we generate two gene-level read density coverage plots: (i) one showing the entire bulk RNA-seq read density coverage and (ii) the other showing the C/PAS subset of the read density to mimic 3′UTR read density coverage.
Attention landscape
To generate an attention landscape, we scored each nucleotide of the input sequence using the self-attention mechanism. We first extracted the attention of the “entire sequence” on the k-mer subsequences and used it as an importance measure. Then, we converted the attention score from k-mer to the individual nucleotide by averaging the attention scores for all k-mers that contain the nucleotide. Lastly, we plotted attention for individual nucleotides as a heatmap for direct visualization.
Motif enrichment analysis
With an array of attention scores as input, we found contiguous high attention sub-regions from our C/PAS containing sequences. We then extracted fixed, equal length sequences centered at a high-attention motif instance. To obtain a count of instances between input sequences and motif patterns, we used the Aho-Corasick algorithm for efficient multi-pattern matching and subsequently performed a hypergeometric test to find significantly enriched motifs in positive sequences with an adjusted p value threshold of 0.05.61
Quantification and statistical analysis
The statistical analysis in this study involved several steps, including the development and validation of the C/PAS-BERT machine learning model, the application of PolyAMiner-Bulk for APA analysis, and subsequent comparisons with existing tools. The details of the statistical methods, software, and relevant parameters are outlined below.
C/PAS-BERT model development and validation
Dataset composition
The C/PAS-BERT model was trained on a balanced dataset comprising 633,786 tuples of 6-mer nucleotide sequence representations and corresponding C/PAS labels. The dataset was divided into a training set (90%) for 12-fold cross-validation and an independent test set (10%).
Model training
The DNABERT model was extended with task-specific data to create C/PAS-BERT, an attention-based deep learning model. The training process involved 12-fold cross-validation to ensure low time complexity. The model aimed to distinguish positively labeled C/PASs shared by PolyASite and PolyA_DB from negatively labeled intergenic sites.
Performance metrics
The overall performance of the C/PAS-BERT machine learning model was assessed using various metrics, which were detailed in Figure 2B.
PolyAMiner-Bulk APA analysis pipeline
Input data processing
Raw FASTQ or processed BAM alignment files were used as input for PolyAMiner-Bulk. Raw reads were mapped to the reference genome using STAR, and resulting alignment files were sorted and indexed using samtools.
De novo C/PAS detection
PolyAMiner-Bulk detected de novo C/PASs using softclipped read detection, considering softclipped tails with a length-dependent proportion of adenosines. Candidate de novo C/PASs were defined based on softclipped regions in BAM read alignment files.
C/PAS-BERT filtering
The candidate de novo C/PASs were merged with a priori C/PASs from databases like PolyA_DB and PolyASite. C/PAS-BERT was employed to filter artificial C/PASs, ensuring the retention of high-confidence C/PASs.
Clustering and vector projection
Two C/PAS deconvolution modes were implemented: softclipped and a priori clustering, as well as softclipped-assisted clustering. Softclipped-assisted clustering mode retained only softclipped-supported clusters for additional specificity. Vector projection calculations were performed to quantify APA dynamics at the gene level, considering C/PAS distribution and read density.
Statistical testing
A beta-binomial test was used to determine the significance of each PolyAIndex metric. Likelihood ratio tests were employed to assess the homogeneity of groups, and significance was determined based on chi-square distribution. Multiple testing correction using the Benjamini-Hochberg procedure was applied.
Visualization
APA changes were visualized using pyGenomeTracks and Matplotlib, generating gene-level read density coverage plots and C/PAS usage heatmaps. Attention landscapes were generated by scoring each nucleotide using the self-attention mechanism.
Acknowledgments
We are thankful to Dr. Huda Zoghbi, Harini Tirumala, and our other colleagues at the Baylor College of Medicine; Texas Children’s Hospital; and the Jan and Dan Duncan Neurological Research Institute for providing expertise that greatly assisted this research. This work has been supported by the United States Department of Agriculture (USDA/ARS) under Cooperative Agreement no. 58-3092-0-001 and the Duncan NRI Zoghbi Scholar Award (to H.K.Y.), Autism Speaks (to G.C.), and the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health P50 HD103555 IDDRC grant and the National Institute of Mental Health R01MH130356 ( to M.M.S.). V.S.J. is supported by the Gulf Coast Consortia and the National Library of Medicine Training Program in Biomedical Informatics and Data Science (T15 LM0070943).
Author contributions
Conceptualization, V.S.J. and H.K.Y.; methodology, V.S.J. and H.K.Y.; software, V.S.J., validation, V.S.J.; formal analysis, V.S.J. and H.K.Y.; investigation, V.S.J. and H.K.Y.; resources, V.S.J. and H.K.Y.; data curation, V.S.J. and H.K.Y.; writing – original draft, V.S.J.; writing – review & editing, V.S.J., E.J.W., M.M.-S., Z.L., and H.K.Y.; visualization, V.S.J.; supervision, H.K.Y.; project administration, H.K.Y.; funding acquisition, V.S.J., M.M.S., Z.L., and H.K.Y.
Declaration of interests
The authors declare no competing interests.
Published: February 6, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2024.100707.
Supplemental information
References
- 1.Mitschka S., Mayr C. Context-specific regulation and function of mRNA alternative polyadenylation. Nat. Rev. Mol. Cell Biol. 2022;23:779–796. doi: 10.1038/s41580-022-00507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan F., Hankey W., Wagner E.J., Li W., Wang Q. Alternative polyadenylation of mRNA and its role in cancer. Genes Dis. 2021;8:61–72. doi: 10.1016/j.gendis.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel R., Brophy C., Hickling M., Neve J., Furger A. Alternative cleavage and polyadenylation of genes associated with protein turnover and mitochondrial function are deregulated in Parkinson’s, Alzheimer’s and ALS disease. BMC Med. Genom. 2019;12 doi: 10.1186/S12920-019-0509-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agarwal V., Lopez-Darwin S., Kelley D.R., Shendure J. The landscape of alternative polyadenylation in single cells of the developing mouse embryo. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-25388-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Routh A., Ji P., Jaworski E., Xia Z., Li W., Wagner E.J. Poly(A)-ClickSeq: Click-chemistry for next-generation 3’-end sequencing without RNA enrichment or fragmentation. Nucleic Acids Res. 2017;45:e112–e116. doi: 10.1093/nar/gkx286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoque M., Ji Z., Zheng D., Luo W., Li W., You B., Park J.Y., Yehia G., Tian B. Analysis of alternative cleavage and polyadenylation by 3’ region extraction and deep sequencing. Nat. Methods. 2013;10:133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shepard P.J., Choi E.A., Lu J., Flanagan L.A., Hertel K.J., Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett D.A., Schneider J.A., Buchman A.S., Mendes de Leon C., Bienias J.L., Wilson R.S. The Rush Memory and Aging Project: study design and baseline characteristics of the study cohort. Neuroepidemiology. 2005;25:163–175. doi: 10.1159/000087446. [DOI] [PubMed] [Google Scholar]
- 9.Bennett D.A., Schneider J.A., Arvanitakis Z., Wilson R.S. OVERVIEW AND FINDINGS FROM THE RELIGIOUS ORDERS STUDY. Curr. Alzheimer Res. 2012;9:628–645. doi: 10.2174/156720512801322573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett D.A., Schneider J.A., Buchman A.S., Barnes L.L., Boyle P.A., Wilson R.S. Overview and Findings from the Rush Memory and Aging Project. Curr. Alzheimer Res. 2012;9:646–663. doi: 10.2174/156720512801322663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z., Jensen M.A., Zenklusen J.C. A Practical Guide to The Cancer Genome Atlas (TCGA) Methods Mol. Biol. 2016;1418:111–141. doi: 10.1007/978-1-4939-3578-9_6. [DOI] [PubMed] [Google Scholar]
- 12.Baxi E.G., Thompson T., Li J., Kaye J.A., Lim R.G., Wu J., Ramamoorthy D., Lima L., Vaibhav V., Matlock A., et al. Answer ALS, a large-scale resource for sporadic and familial ALS combining clinical and multi-omics data from induced pluripotent cell lines. Nat. Neurosci. 2022;25:226–237. doi: 10.1038/s41593-021-01006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M., Ji G., Fu H., Lin Q., Ye C., Ye W., Su Y., Wu X. A survey on identification and quantification of alternative polyadenylation sites from RNA-seq data. Briefings Bioinf. 2020;21:1261–1276. doi: 10.1093/bib/bbz068. [DOI] [PubMed] [Google Scholar]
- 14.Lee J.Y., Yeh I., Park J.Y., Tian B. PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res. 2007;35:D165–D168. doi: 10.1093/nar/gkl870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X., Zhang Y., Li Q.Q. PlantAPA: A Portal for Visualization and Analysis of Alternative Polyadenylation in Plants. Front. Plant Sci. 2016;7 doi: 10.3389/fpls.2016.00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruber A.J., Schmidt R., Gruber A.R., Martin G., Ghosh S., Belmadani M., Keller W., Zavolan M. A comprehensive analysis of 3’ end sequencing data sets reveals novel polyadenylation signals and the repressive role of heterogeneous ribonucleoprotein C on cleavage and polyadenylation. Genome Res. 2016;26:1145–1159. doi: 10.1101/gr.202432.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R., Nambiar R., Zheng D., Tian B. PolyA_DB 3 catalogs cleavage and polyadenylation sites identified by deep sequencing in multiple genomes. Nucleic Acids Res. 2018;46:D315–D319. doi: 10.1093/nar/gkx1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrmann C.J., Schmidt R., Kanitz A., Artimo P., Gruber A.J., Zavolan M. PolyASite 2.0: a consolidated atlas of polyadenylation sites from 3′ end sequencing. Nucleic Acids Res. 2020;48:D174–D179. doi: 10.1093/nar/gkz918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang R., Tian B. APAlyzer: A bioinformatics package for analysis of alternative polyadenylation isoforms. Bioinformatics. 2020;36:3907–3909. doi: 10.1093/bioinformatics/btaa266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grassi E., Mariella E., Lembo A., Molineris I., Provero P. Roar: Detecting alternative polyadenylation with standard mRNA sequencing libraries. BMC Bioinf. 2016;17:423–429. doi: 10.1186/s12859-016-1254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ha K.C.H., Blencowe B.J., Morris Q. QAPA: A new method for the systematic analysis of alternative polyadenylation from RNA-seq data. Genome Biol. 2018;19 doi: 10.1186/s13059-018-1414-4. 45–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.le Pera L., Mazzapioda M., Tramontano A. 3USS: a web server for detecting alternative 3′UTRs from RNA-seq experiments. Bioinformatics. 2015;31:1845–1847. doi: 10.1093/bioinformatics/btv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Z., Teeling E.C. ExUTR: A novel pipeline for large-scale prediction of 3’-UTR sequences from NGS data. BMC Genom. 2017;18:847. doi: 10.1186/s12864-017-4241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birol I., Raymond A., Chiu R., Nip K.M., Jackman S.D., Kreitzman M., Docking T.R., Ennis C.A., Robertson A.G., Karsan A. KLEAT: CLEAVAGE SITE ANALYSIS OF TRANSCRIPTOMES. Pac Symp Biocomput. 2014;347 [PMC free article] [PubMed] [Google Scholar]
- 25.Xia Z., Donehower L.A., Cooper T.A., Neilson J.R., Wheeler D.A., Wagner E.J., Li W. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3’2-UTR landscape across seven tumour types. Nat. Commun. 2014;5:5274. doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye C., Long Y., Ji G., Li Q.Q., Wu X. APAtrap: Identification and quantification of alternative polyadenylation sites from RNA-seq data. Bioinformatics. 2018;34:1841–1849. doi: 10.1093/bioinformatics/bty029. [DOI] [PubMed] [Google Scholar]
- 27.Arefeen A., Liu J., Xiao X., Jiang T. TAPAS: tool for alternative polyadenylation site analysis. Bioinformatics. 2018;34:2521–2529. doi: 10.1093/bioinformatics/bty110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji Y., Zhou Z., Liu H., Davuluri R.V. DNABERT: pre-trained Bidirectional Encoder Representations from Transformers model for DNA-language in genome. Bioinformatics. 2021;37:2112–2120. doi: 10.1093/bioinformatics/btab083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J., Yoon W., Kim S., Kim D., Kim S., So C.H., Kang J. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36:1234–1240. doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo R., Sun L., Xia Y., Qin T., Zhang S., Poon H., Liu T.Y. BioGPT: generative pre-trained transformer for biomedical text generation and mining. Briefings Bioinf. 2022;23 doi: 10.1093/bib/bbac409. [DOI] [PubMed] [Google Scholar]
- 31.Wu Y., Liu Z., Wu L., Chen M., Tong W. BERT-Based Natural Language Processing of Drug Labeling Documents: A Case Study for Classifying Drug-Induced Liver Injury Risk. Front. Artif. Intell. 2021;4 doi: 10.3389/frai.2021.729834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magana-Mora A., Kalkatawi M., Bajic V.B. Omni-Polya: A method and tool for accurate recognition of poly(A) signals in human genomic DNA. BMC Genom. 2017;18:620. doi: 10.1186/s12864-017-4033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Maio A., Yalamanchili H.K., Adamski C.J., Gennarino V.A., Liu Z., Qin J., Jung S.Y., Richman R., Orr H., Zoghbi H.Y. RBM17 Interacts with U2SURP and CHERP to Regulate Expression and Splicing of RNA-Processing Proteins. Cell Rep. 2018;25:726–736.e7. doi: 10.1016/j.celrep.2018.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arora A., Goering R., Lo H.Y.G., Lo J., Moffatt C., Taliaferro J.M. The Role of Alternative Polyadenylation in the Regulation of Subcellular RNA Localization. Front. Genet. 2022;12:2791. doi: 10.3389/fgene.2021.818668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brumbaugh J., Di Stefano B., Wang X., Borkent M., Forouzmand E., Clowers K.J., Ji F., Schwarz B.A., Kalocsay M., Elledge S.J., et al. Nudt21 Controls Cell Fate by Connecting Alternative Polyadenylation to Chromatin Signaling. Cell. 2018;172:106–120.e21. doi: 10.1016/j.cell.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu Y., Elrod N., Wang C., Li L., Chen T., Routh A., Xia Z., Li W., Wagner E.J., Ji P. Nudt21 regulates the alternative polyadenylation of Pak1 and is predictive in the prognosis of glioblastoma patients. Oncogene. 2019;38:4154–4168. doi: 10.1038/s41388-019-0714-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masamha C.P., Xia Z., Yang J., Albrecht T.R., Li M., Shyu A.B., Li W., Wagner E.J. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510:412–416. doi: 10.1038/nature13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weng T., Huang J., Wagner E.J., Ko J., Wu M., Wareing N.E., Xiang Y., Chen N.-Y., Ji P., Molina J.G., et al. Downregulation of CFIm25 amplifies dermal fibrosis through alternative polyadenylation. J. Exp. Med. 2020;217 doi: 10.1084/jem.20181384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alcott C., Yalamanchili H.K., Ji P., van der Heijden M., Saltzman A., Leng M., Bhatt B., Hao S., Wang Q., Saliba A., et al. Partial loss of CFIm25 causes aberrant alternative polyadenylation and learning deficits. Elife. 2019;9:1–30. doi: 10.7554/eLife.50895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L., Yan F., Li L., Fu H., Song D., Wu D., Wang X. New focuses on roles of communications between endoplasmic reticulum and mitochondria in identification of biomarkers and targets. Clin. Transl. Med. 2021;11 doi: 10.1002/ctm2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mak H.Y., Ouyang Q., Tumanov S., Xu J., Rong P., Dong F., Lam S.M., Wang X., Lukmantara I., Du X., et al. AGPAT2 interaction with CDP-diacylglycerol synthases promotes the flux of fatty acids through the CDP-diacylglycerol pathway. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-27279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aypek H., Krisp C., Lu S., Liu S., Kylies D., Kretz O., Wu G., Moritz M., Amann K., Benz K., et al. Loss of the collagen IV modifier prolyl 3-hydroxylase 2 causes thin basement membrane nephropathy. J. Clin. Invest. 2022;132 doi: 10.1172/JCI147253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schulten H.J., Al-Adwani F., Saddeq H.A.B., Alkhatabi H., Alganmi N., Karim S., Hussein D., Al-Ghamdi K.B., Jamal A., Al-Maghrabi J., Al-Qahtani M.H. Meta-analysis of whole-genome gene expression datasets assessing the effects of IDH1 and IDH2 mutations in isogenic disease models. Sci. Rep. 2022;12 doi: 10.1038/s41598-021-04214-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujiwara T., Ye S., Castro-Gomes T., Winchell C.G., Andrews N.W., Voth D.E., Varughese K.I., Mackintosh S.G., Feng Y., Pavlos N., et al. PLEKHM1/DEF8/RAB7 complex regulates lysosome positioning and bone homeostasis. JCI Insight. 2016;1 doi: 10.1172/jci.insight.86330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi Y., Ge S. Targeting the histone H3 lysine 79 methyltransferase DOT1L in MLL-rearranged leukemias. J. Hematol. Oncol. 2022;15 doi: 10.1186/s13045-022-01251-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rüegsegger U., Blank D., Keller W. Human Pre-mRNA Cleavage Factor Im Is Related to Spliceosomal SR Proteins and Can Be Reconstituted In Vitro from Recombinant Subunits. Mol. Cell. 1998;1:243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 47.Li W., You B., Hoque M., Zheng D., Luo W., Ji Z., Park J.Y., Gunderson S.I., Kalsotra A., Manley J.L., Tian B. Systematic Profiling of Poly(A)+ Transcripts Modulated by Core 3’ End Processing and Splicing Factors Reveals Regulatory Rules of Alternative Cleavage and Polyadenylation. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin G., Gruber A.R., Keller W., Zavolan M. Genome-wide Analysis of Pre-mRNA 3′ End Processing Reveals a Decisive Role of Human Cleavage Factor I in the Regulation of 3′ UTR Length. Cell Rep. 2012;1:753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 49.Dong Y., Fan X., Wang Z., Zhang L., Guo S. Circ_HECW2 functions as a miR-30e-5p sponge to regulate LPS-induced endothelial-mesenchymal transition by mediating NEGR1 expression. Brain Res. 2020;1748 doi: 10.1016/j.brainres.2020.147114. [DOI] [PubMed] [Google Scholar]
- 50.Krishnamoorthy V., Khanna R., Parnaik V.K. E3 ubiquitin ligase HECW2 mediates the proteasomal degradation of HP1 isoforms. Biochem. Biophys. Res. Commun. 2018;503:2478–2484. doi: 10.1016/j.bbrc.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 51.Krishnamoorthy V., Khanna R., Parnaik V.K. E3 ubiquitin ligase HECW2 targets PCNA and lamin B1. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865:1088–1104. doi: 10.1016/j.bbamcr.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 52.Stern E.P., Guerra S.G., Chinque H., Acquaah V., González-Serna D., Ponticos M., Martin J., Ong V.H., Khan K., Nihtyanova S.I., et al. Analysis of Anti-RNA Polymerase III Antibody-positive Systemic Sclerosis and Altered GPATCH2L and CTNND2 Expression in Scleroderma Renal Crisis. J. Rheumatol. 2020;47:1668–1677. doi: 10.3899/jrheum.190945. [DOI] [PubMed] [Google Scholar]
- 53.Iyama T., Wilson D.M. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair. 2013;12:620–636. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yalamanchili H.K., Alcott C.E., Ji P., Wagner E.J., Zoghbi H.Y., Liu Z. PolyA-miner: Accurate assessment of differential alternative poly-adenylation from 3′Seq data using vector projections and non-negative matrix factorization. Nucleic Acids Res. 2020;48 doi: 10.1093/nar/gkaa398. e69–e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 58.Ramírez F., Bhardwaj V., Arrigoni L., Lam K.C., Grüning B.A., Villaveces J., Habermann B., Akhtar A., Manke T. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 2018;9:189. doi: 10.1038/s41467-017-02525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lopez-Delisle L., Rabbani L., Wolff J., Bhardwaj V., Backofen R., Grüning B., Ramírez F., Manke T. pyGenomeTracks: reproducible plots for multivariate genomic datasets. Bioinformatics. 2021;37:422–423. doi: 10.1093/bioinformatics/btaa692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hunter J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]
- 61.Soroushnia S., Daneshtalab M., Plosila J., Pahikkala T., Liljeberg P. High performance pattern matching on heterogeneous platform. J. Integr. Bioinform. 2014;11:253. doi: 10.2390/biecoll-jib-2014-253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This study employed existing datasets that are publicly accessible. The specific accession numbers for these datasets are detailed in the key resources table.
-
•
All original code developed for this study has been deposited in a GitHub repository and can be freely accessed at https://github.com/YalamanchiliLab/PolyAMiner-Bulk.git. In addition, we have archived this code in Zenodo, an open-access repository, available at https://doi.org/10.5281/zenodo.10372661.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.