Summary
Method development for mass spectrometry (MS)-based thermal shift proteomic assays have advanced to probe small molecules with known and unknown protein-ligand interaction mechanisms and specificity, which is predominantly used in characterization of drug-protein interactions. In the discovery of target and off-target protein-ligand interactions, a thorough investigation of method development and their impact on the sensitivity and accuracy of protein-small molecule and protein-protein interactions is warranted. In this review, we discuss areas of improvement at each stage of thermal proteome profiling data analysis that includes processing of MS-based data, method development, and their effect on the overall quality of thermal proteome profiles. We also overview the optimization of experimental strategies and prioritization of an increased number of independent biological replicates over the number of evaluated temperatures.
Keywords: thermal proteomic profiling, drug discovery, target engagement, cellular thermal shift assays, protein-ligand interactions
Thermal proteomic profiling explores small molecule and protein interactions, encompassing both known targets and unknown off-targets. In this review, Figueroa-Navedo and Ivanov discuss recent progress in experimental design and mass spectrometry-based data acquisition technologies in thermal proteomic profiling and their impact on the overall quality of thermal proteome profiles.
Introduction
Accurately identifying safe and potent small molecules that affect disease-dependent biology is a critical task for pharmaceutical companies, but it presents significant challenges due to the limitations of currently available approaches.1,2 Despite unpredictable timelines for target deconvolution, phenotypic drug discovery was instrumental in discovering potent compounds that typically require further chemical optimization.1 Phenotypic drug discovery is a hypothesis-free method that begins with a disease and a relevant cellular or animal model to screen for compounds that can modulate the disease phenotype.3 In contrast, target-based drug discovery has shown promise in situations where known and heavily studied targets are involved, such as specific HIV protease inhibitors and drugs that target the renin-angiotensin system for hypertension.1,4 Recent reviews have shown that both approaches are useful in label-based and label-free assays, with target engagement and verification being essential for small-molecule candidates.1 Targeted cellular thermal shift assays (CETSA) were introduced in 2013 using western blot to measure protein-drug interactions and later expanded to proteome-level studies using mass spectrometry (MS)-based techniques.5,6 Afterward, thermal proteome profiling (TPP) enabled a proteome-wide assessment of the effect of small molecules on protein thermal stability.5 The suggested fit for TPP is a sigmoidal fit, which follows thermodynamic theory.7 TPP was further advanced to identify possible targets, off-targets, and the effect of small molecules on disease phenotypes by characterizing the level of protein unfolding and aggregation within cell lysates, cells, physiological fluids, and tissues.1,6,8,9,10,11,12,13,14 CETSA is directly translatable to clinical settings, with no protein or target modifications necessary.4,11,15 One-dimensional (1D) MS-based TPP) assays, relying on the effect of temperature-based protein unfolding and aggregation, have been used to elucidate binding and affinity behavior.5
One of the main parameters proposed by TPP approaches to detect a thermal shift is the melting temperature, or Tm, which is the temperature recorded at half of the initial protein abundance.5 The Tm, which can also be measured at the inflection point of a sigmoid curve, represents the temperature at which 50% of the initial protein abundances at the lowest temperature is recorded.13 Two-dimensional assays have focused on monitoring both temperature- and compound concentration-dependence on protein targets.11 Thermal shift assays can identify compounds that alter the thermal stability of the protein and provide insight into protein-protein interactions (PPI) and protein-ligand/co-factor interactions in various biological matrices, including tissues, intact cells, or cell lysates.11,16 A disadvantage of one-dimensional (2D) thermal shift assays is the filtering criteria based on curve-quality-parameters. Although this filtering step is useful to ensure a reliable measure of Tm and decrease the number of false positives, it can also increase the possibility of having false negatives, although the inclusion of false negatives is noted as a rare occurrence.17,18 Moreover, peptide-level TPP has shown differential profiles from post-translational modifications (PTMs) and proteoforms.19,20,21,22
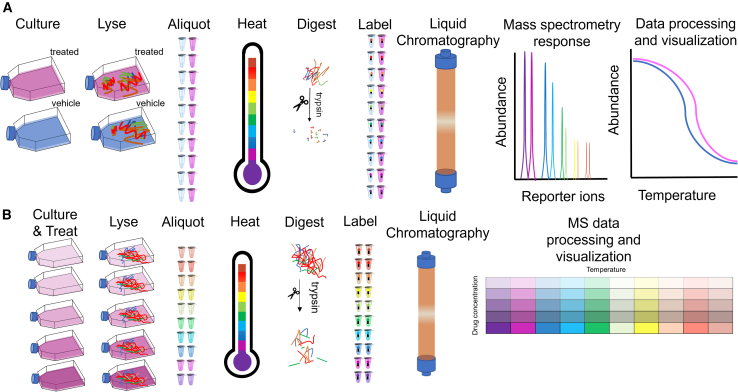
Based on the previous findings, non-parametric analysis of response curves (NPARC) was implemented to compare protein interactions from curve-fitting parameters rather than Tm.2 NPARC provides a statistical summary that implements hypothesis testing using an F-statistic, which is computationally stable (i.e., less model convergence issues).2 Specifically, the implementation of TPP using a specific temperature range does not ensure that all proteins would unfold and aggregate within this temperature window.2,23 Several groups have noted that for up to 20% of the analyzed proteome, Tm values are not determined because they are presumably outside of the typically used temperature ranges.10,24 More recently, a new statistical method based on a hierarchical Gaussian process (GPMelt) was developed to analyze the melting profiles without any fitting, filtering, or reliance on Tm.23 GPMelt relates back to hypothesis testing in a similar way to NPARC and the Bayesian semi-parametric model developed for proteomic profiling.23,25 Figure 1A demonstrates the outline of the 1D thermal shift assay experimental workflow, where the schematic shows cell cultures treated with either a compound of interest or vehicle as the starting biological material, which is harvested, lysed, and aliquoted into eight or more identical pairs of samples each depending on the selected tandem mass tag (TMT)-plex. Each of these ten or more pairs of samples is then subjected to a heating challenge at a specific temperature. Initial publications suggest digestion followed by TMT10-plex (1D- or 2D-assay) stable isotope labeling while allocating one TMT tag per temperature is performed prior to bottom-up liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomic profiling.5,9,26,27,28 With the availability of TMT 16- and 18-plex, more recent work has shown increased throughput with 2D-assays.29 To evaluate protein interactions, data processing, curve fitting, and visualization are typically performed to determine possible targets and off-targets.
Figure 1.
Schematic representation of workflows
(A) General experimental workflow for one-dimensional thermal proteome profiling and (B) general experimental workflow for two-dimensional thermal proteome profiling.
Figure 1B describes a 2D thermal shift assay experimental workflow, where both temperature and drug concentration vary, allowing for multiplexing, higher throughput, and increased sensitivity.16 Instead of running vehicle and treated conditions in different TMT-plexes, 2D-TPP allows both conditions to be measured within the same TMT-plex.16 In addition, the implementation of different drug concentrations permits calculation of compound affinity. Though 2D-TPP is flexible toward other experimental configurations—such as time points after treatment, a combination of compounds, and other variations—curves are no longer visualized.16
MS-centered method development can improve sensitivity for interacting proteins with thermal shift assays
The reviewed thermal shift assays utilize mMS acquisition settings employing data-dependent acquisition (DDA) approaches. DDA is typically preferred for its capability to detect low-abundance peptides, thus offering improved structural information for the identified peptides.8,30,31 DIA is usually applied in label-free quantitative approaches because of its consistent quantitative performance across replicates and samples, especially for low-abundance peptides and proteins.8,30,31 In a mass spectrometer, DDA involves a full survey single-stage MS scan, where the instrument typically selects higher abundance precursor ions to fragment and perform tandem MS2 scans for further fragmentation. However, DDA approaches are susceptible to precursor ion interference, limited reproducibility due to dynamic exclusion and stochastic ion selection for MS2 fragmentation, and limited accuracy of quantification for peptides with substantially varied intensities. Peptide co-isolation has a significant impact on thermal shift assays due to ratio compression, which may lead to an underestimation of the true fold-change in peptide abundance unless the issue is diminished using substantial fractionation or alternative techniques.8,10,32,33 Two types of ratio compression effects are ion suppression and ion interference, the former being one peptide ion that co-elutes with another peptide ion (or several peptides or other ion species) and, thus, lowering the ion signal and affecting the accuracy of the quantitative attributes (e.g., peak height, peak area) of the peptide.32,34 Interference occurs when co-eluting peptides have overlapping isotopic distributions and potential fragmentation patterns.32,34 With DDA, solutions to minimize co-fragmentation of co-eluting peptides include sample high performance liquid chromatography (HPLC)- or solid phase extraction (SPE)-based fractionation, narrowing isolation windows, and using MS3-based quantitation of TMT-derived ion abundances,.10,32,33,34,35,36,37,38 Recently introduced, SPE tip-based manual fractionation involves the implementation of thermal proteome profiling with some modifications: mainly, the heating and centrifugation occur on the same PCR tube followed by sample processing on a multimode SPE tip and fractionation on a different SPE tip with a C18 membrane.38 MS3-based DDA quantitation methods isolate fragments after MS2 fragmentation, which results in increased quantitation accuracy at the cost of a reduction in proteomic coverage.39 Another option to remediate ratio compression effects is to employ an OnePot approach introduced by Gaetani et al.17 The OnePot approach consists of a physical pooling all temperature-challenged sample aliquots prior to isobaric labeling of the sample.17 DDA quantitation using OnePot 2D over a narrow temperature range has shown greater sensitivity compared to 1D thermal profiling approaches utilizing wider temperature ranges.40
DIA for thermal shift assays has been proposed with modifications of experimental designs to reduce sample analysis times.31,39 In data-independent acquisition (DIA), precursor ions are isolated using pre-defined m/z windows for further fragmentation, which results in accurate label-free proteome quantification, a decreased number of missing values, and an increased throughput for identification of kinase interactors.31,41 DIA mode is more cost-effective and straightforward, suffers less from compression of quantitative ratios, and reportedly improves sensitivity for screening sensitivity over TPP and is comparable with 2D-TPP.31 Among the disadvantages of both DIA and DDA approaches, there is a balance between proteomic coverage and throughput. Hybrid library searches and retention time alignments performed for DIA data are more time-consuming than data processing in DDA workflows for thermal profiling, with a slight increase in the number of identified protein groups for DIA.39 When compared to DDA, DIA resulted in fewer protein group identifications in the less complex samples generated at higher temperatures.39 In addition, data processing time increases due to the large number of fragment ions generated and the need for retention time alignment of precursor and fragment ions.42 In terms of processing times, DIA was shown to reduce instrument time for thermal profiling when only one temperature was used to measure protein interactions.31 The caveat of this approach is the sensitivity to protein interactions, which can occur at a different temperature.31 These findings were analogous to a DDA-based thermal profiling study, in which temperature selection was also relevant to sensitivity.43
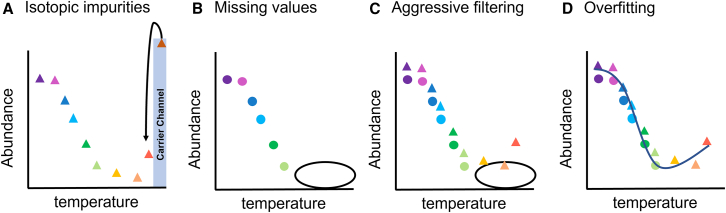
Since MS2 reporter ion abundances in TMT-based TPP assays are often underestimated because of ratio compression, Isobarquant software was implemented for better estimation of the peptide-level fold changes between replicates without having to decrease the throughput or alter instrumental settings.8,32 However, the Isobarquant authors also acknowledge that the vendor or open-source software can mitigate ratio compression at the peptide level, which would remove a step in the data processing pipeline from the user’s side.5 Recent implementations of carrier proteomes to increase the coverage of thermal proteome profiling can result in isotopic impurities that spill into adjacent channels, as shown in Figure 2A.44,45 The spillage in the signal can be reduced by leaving the adjacent channel(s) empty or can be corrected to some degree with software and the reagent manufacturer’s corrective values.44 Other factors that affect TPP curve fitting include missing values at higher temperatures, as shown in Figure 2B, aggressive filtering (Figure 2C), and overfitting (Figure 2D), which will be discussed herein.
Figure 2.
Challenges for thermal shift assays for each of the replicates acquired for the same sample type (i.e., either treated or control)
(A) The implementation of carrier proteomes into adjacent channels; (B) the presence of missing values; (C) aggressive filtering due to missing values and channel interference; and (D) overfitting the data with more flexible models.
Colors indicate separate temperature challenges; circles and triangles represent two biological replicates.
Further improvement of data processing and curve-fitting methods can enhance sensitivity for detection of interacting proteins using thermal shift assays
After LC-MS data acquisition, data processing is performed using various vendor software tools, which leads to processing at the protein level or peptide level to generate information to fit curves. The TPP software, introduced in 2015, facilitates proteome-wide measurements of thermal stability by quantifying non-denatured and non-aggregated fractions of cellular proteins as the sample temperature increases.4,8,13 For TPP, a sigmoidal fit was introduced to represent the transition from the native folded state of the protein to the unfolded denatured state leading to aggregation and precipitation as the sample temperature increases.5 The sigmoidal fit is relevant to thermodynamic unfolding, as the melting temperature (Tm) represents the temperature at which half of the proteins have undergone the transition from the natively folded to the unfolded or partially folded state toward aggregation.5 Protein interactions have been measured using ΔTm or the melting temperature difference between treated and control curves.5 Nevertheless, curve fitting has some challenges associated with MS-based thermal profiling. One of these challenges to curve fitting is that the peptides that were mapped to a single gene-formed groups, which could present distinct thermal stability patterns.21 Post-translational modifications, such as phosphorylation, can also cause changes in the thermal stability patterns, where differentially melting phosphoproteins have been compared to unmodified proteins using Tm values.19 Other challenges consist of co-isolated and co-fragmented peptides, which can be affected by ratio compression, as visualized by Phaneuf et al.45
Other groups have focused on the comparison of 3- and 4-parameter log fits for TPP, and found that the 4-parameter log fit increased the determination coefficient R2 and a lower median for Bayesian information criterion (BIC) for the R package Inflect.24 The 4-parameter logistic fit is performed on each condition (vehicle or treated) and on each replicate separately, as the initial TPP version is established. Protein-level differential melting behavior for Inflect was based on the inflection point (Tm) value, which correlated well with the TPP Tm. In addition, the same group detected unique proteins shifts that were not detected by TPP. However, a comparison between TPP and Inflect showed small overlaps between shifting proteins, which is mainly due to the choice of fitting model used for curve fitting. Although Inflect provided an interesting set of results for curve-fitting strategies, the package recommends independent statistical analysis for the results using other software. Recent work has outlined other models that forego the need for sigmoidal assumptions: GPMelt, PSTPP, and I-PISA23,46,47
One of the challenges in performing informative curve fitting is the number of missing values. Missing quantitative values shown in Figure 2B present a challenge to thermal shift assays when reporter ion abundances fall below the instrument’s detection sensitivity threshold.45 For thermostable proteins, when the temperature window is not wide enough to capture detectable changes in protein abundance even at higher temperatures; this presents a challenge to complete the melting profiles of the most heat-resilient proteins.27,48 This directly affects curve-fitting methods that present a value for the plateau and might also affect the efficiency and accuracy in determining Tm value of the protein since the lower plateau may not be well defined. Recent studies for other applications have implemented the addition of a carrier channel to help increase the low-abundance peptides at higher temperatures.49,50 Using carrier channels as a modification to the experimental design promotes more confident identification from a database search and allows for more sensitive quantitation of low-abundance proteins.44,45,49,50 This approach is particularly helpful for peptides heated at higher temperature values, where the reporter ion signal is below the instrument’s limit of detection threshold within an MS1 scan.45 A potential disadvantage to this approach is the amount of carry-over from the carrier channel to adjacent channels, depending on the amount of the added carrier sample, as outlined in Figure 2A.
Quantification faces an additional challenge concerning peptides with shared protein groups, which can impact the estimated protein abundance values.51 Although one approach is to eliminate peptides with shared protein groups, doing so might result in the loss of valuable information from proteins with high homology.51 One study also investigated the effects of filtering peptide-spectrum matches (PSMs) for proteins interacting with a highly specific inhibitor.45 PSM filtering helped remove low-abundance and outlier values, which helped clean up the summarized protein-level curve and improved the accuracy of thermoprofiling.45 PSM filtering provides the advantage of potentially removing low-abundance and noisy profiles, but it comes with the disadvantage of potential loss of low-abundance peptides’ measurements per protein.
For example, at the protein level, TPP has less stringent filters in their most recent implementation of the sigmoidal profile, where the correlation coefficient (R2) is greater than 0.8, and the lower sigmoid plateau parameters must be less than 0.3.2,8 These filters will affect proteins with missing values as well as carrier channel spillage to other TMT channels, as shown in Figure 2C. Though some authors13,17 have recognized that these parameters provide sensitivity constraints, previous publications have kept these or similar parameters because these are implemented in the existing pipelines.9,11,14,18,26,39,43,48,52 Although additional filters—including slope, p value, and Tm value reproducibility—have been suggested originally by TPP and subsequent publications, these filters have not been considered from recent versions of the package.5,53 From a statistical standpoint, stringent filtering underestimates the variance in the models, which can reduce the accuracy of the assay. Overfitting, as shown in Figure 2D, can also reduce the accuracy of the assay if the profile of the curve captures artifacts not linked to the biological or physical properties.
Tm-centered approaches may provide a higher rate of false negatives and lower sensitivity in thermal proteome profiling for proteins that are resistant to unfolding and aggregation
Thermal- or concentration-based shifts have been characterized by sigmoidal models in TPP as well as other dose-response models.8,24,26,54,55 Within each statistical model, a statistical summary defines whether a thermal proteomic shift was detected. One statistical summary that these methods have in common is the inflection point of the curves, where Tm indicates the temperature at half of the initial solubility of the protein by thermal challenge whereas EC50 describes the half maximal effective concentration of the drug required to achieve half of the maximum possible effect. Although Tm was one of the first summary statistics, two advantages include the extrapolation of this value directly from the sigmoid parameter or from half of the maximum initial protein abundance. One of the drawbacks of these fitting methods is not having enough information in the data to extract both the upper and the lower plateau parameters due to the symmetric aspect of the curve, which is also related to the selection of the temperature challenge as well as the provenance of the sample.48 Moreover, statistical conclusions out of one parameter Tm using a t or a z test showed a higher rate of false negatives.14,18 Furthermore, the presence of missing values in the dataset is also a hindrance to sensitivity and accuracy in Tm determination, as discussed in recent reports.17,52,56 For sigmoidal fitting methods, data that do not resemble a sigmoidal fit may lead to long processing times for a proteome-scale complex sample and sometimes to an inability to identify a good fit for possible targets.16,18 More recently, the development of GPMelt would omit the use of curve fitting and ΔTm.23
Another drawback of the ΔTm-centered approaches is related to the verification of the magnitude of the thermal shift for a particular concentration of a small molecule.18,26 This issue has been discussed in several reports and remediated by performing hypothesis testing on the goodness-of-fit18 and the area under the curve (AUC) between treated and vehicle curves,17,56 as well as hypothesis testing using 2D thermal proteome profiling (2D-TPP) by applying nested models. The 2D-TPP assay was introduced because one of the hindrances to performing thermal shift assays with one treatment concentration (i.e., 1D-TPP) was that the 1D-TPP technique could not ensure whether the affinity of the target protein was correlated to the magnitude of the thermal shift ΔTm of the drug.26,57 Whereas the TPP approach mostly depends on one parameter (e.g., ΔTm), hypothesis testing based on goodness-of-fit has increased sensitivity for known targets.13,18
To improve the sensitivity of thermal shift assays for data with missing values, the measured signal difference ΔSm, which is correlated to the AUC, was introduced as a viable option for data analysis in assessing protein-ligand interactions.17,52,56 These methods were fast and translatable because benchmarks on their datasets performed alongside TPP proved that both ΔSm and ΔTm were complementary to obtain known targets.52 In a latter publication, reducing the temperature range for AUC calculation increased the fold changes between vehicle and treated samples.56 Though the experimental and data analysis approaches are more simplified with ΔSm, there is a reported loss of 30% from poorly fitted proteins toward AUC-based assays due to the reliance on high-quality sigmoidal melt curve filters applied based on initial publications.8,52 When contrasting PISA with 2D-TPP, distinctions arise in the methodology and underlying mechanistic principles: PISA involves physical sample pooling of soluble aliquots from the same sample, which is exposed to heat using selected temperatures. In contrast, 2D-TPP involves the sample exposure to a combination of several temperature settings and drug concentration conditions for each sample aliquot.8,17,26 The interpretation of data differs accordingly. In 2D-TPP, alterations in a protein’s thermal stability do not necessarily impact the direct interaction with the treatment molecule.9 Instead, 2D-TPP data may help elucidate whether the observed effect is attributable to protein conformational stabilization, cell state perturbations, such as metabolite levels, PPI, or changes in solubility.9 Disadvantages of 2D-TPP experiments include long analysis time, often leaving the user to conduct one replicate analysis per condition. In the case of PISA, the aliquot pooling before LC-MS analysis may complicate the detection of interactions and the interpretation of the treatment effects on the thermal profile. Therefore, the resulting ΔSm may not offer detailed insights into the specific impact of PTMs, ligand-binding, and PPIs on protein solubility.
Recent improvements to TPP
Thanks to the initial discoveries in data analysis, the field has been able to identify several experimental designs, processing, and curve-fitting modalities to improve TPP.11,14,21,26,27,45,58 Although there is not one tool that would be ideal for all datasets, TPP remains a useful and translational assay. Several papers have noted that poor fit, missing values, the selection of inadequate temperature windows, statistical analysis, overly stringent filters, and implementation of biological and technical variation are factors that must be taken into account when considering thermal shift models.17,18,52,56 For data analysis of thermal shift assays, benchmarking with known, peer-reviewed, and publicly available datasets is instrumental to gauge the translatability of the pipeline. Another suggested change to the experimental design is to allow for an increased number of replicates to enable an informative and thorough assessment of treatment with a compound of interest by implementing TMT 16-plex.59 This study provides the potential to explore more replicates within a TMT-plex with a reduced number of temperatures. Peptide-centric approaches have also been recently developed since the studied proteoforms can exhibit differences in thermal stability, Tm, or thermal profiles.19,20,21,22,60 The functional diversity of proteins is generated through genetic variations, alternative splicing, and post-translational modifications, which lead to altered protein properties, functions, and interaction networks.61
Although curve-fitting models have remained persistent in recent work (and may continue to remain), one group has suggested using one temperature to quantify treatment effects, which was named the isothermal shift assay or iTSA.43 With adequate benchmarking using previously published datasets, relevant ligand-binding proteins were observed by applying one temperature instead of ten.43 However, the selection process for a temperature around the Tm value will vary per the biological system and the protein of interest, as it was shown that rather diverse differentially melting proteins were found.48 Another group suggested pooling the sample temperature aliquots for each condition after heating while labeling using one TMT per replicate, and named this assay the OnePot assay.17 This allows the measurement of five replicates, as shown in (Table 1), employs the same amount of biological material as TPP, and does not rely on curve fitting to measure the difference between vehicle and treated samples.17 However, OnePot as well as other TPP assays, does not distinguish between protein solubility and conformational stability, in contrast to the ion-based PISA approach that interrogates and studies changes in protein solubility.47 One of the challenges related to TPP is that some proteins are insoluble and/or conformationally unstable at increased temperatures.47 The solubility of a protein can be influenced by pH, temperature, ionic strength, detergents, or denaturing agents.47,52,62,63,64 Though the OnePot assay describes a simplified experimental approach to the TPP, the calculation of p values is based on a comparison of the total protein abundance between both conditions. Nevertheless, the obtained results correlate with the findings reported with ΔTm, which would reduce the computational resources involved. Table 1 summarizes the recent trends, where the number of replicates is often favored over the number of temperatures.
Table 1.
Some of the most notable recent publications on thermal profiling to date
| Year | Authors | Method | Description | Replicates | Temperatures | Reference |
|---|---|---|---|---|---|---|
| 2013 | Molina et al. | CETSA | Western-blot-based thermal profiling | 3 | 12 | Savitski et al.5 |
| 2014 | Savitski et al. | TPP | Mass spectrometry-based thermal profiling | 2 | 10 | Martinez Molina et al.6 |
| 2015 | Franken et al. | TPP | Protocol for mass spectrometry-based thermal profiling | 2 | 10 | Franken et al.8 |
| 2019 | Gaetani et al. | OnePot | Proposed OnePot approach to pool temperatures and increase replicates | 5 | 10-pooled | Gaetani et al.17 |
| 2021 | Ruan et al. | mTSA | Proposed DIA-OnePot approach for thermal profiling using 52°F temperature and 5 concentrations | 5 | 1 | Ruan et al.31 |
| 2022 | Xu et al. | DDA-OnePot | Applied DDA-OnePot approach within 45°F–58°F for improved sensitivity over 1D thermal profiling | 3 | 10-pooled | George et al.39 |
| 2023 | Phaneuf et al. | iMAATSA | Proposed the implementation of CARRIER, field asymetric ion mobility spectrometry (FAIMS), and ΦSDM to increase protein identifications and accuracy of thermal profiling | 3 | 10 | Dwivedi and Rose44 |
Another recent advance in the field of data analysis is the inclusion of bioinformatic tools, which are able to correlate the candidate and known target findings by, for example, implementing STRINGdb, GeneOntology, and CORUM.11,13,55,65,66,67 An implementation of these bioinformatic tools into data analysis for thermal shift assays has proven useful and could provide additional information about possible targets as well as off-targets, when the ligand-protein interaction mechanism and specificity are not well studied.
Concluding remarks
The field of data analysis for thermal profiling assays has been vast and informative in terms of data availability, benchmarking against existing software approaches, curve-fitting approaches, and simulations. Examples of these advances consist of DIA implementations that minimize requirements for thorough sample fractionation and lengthy LC-MS analyses and promote increased identification of proteins and peptide groups.31,39 In terms of the quality of the identifications from different acquisition modes, the complementary identifications from DDA and DIA could provide more detailed insights into protein-ligand interactions.39 There is room for improvement in data analysis tools for thermal shift assays in the fields of curve fitting, sampling time reduction, benchmarking for method development, evaluation of the underlying peptide data, the effect of post-translational modifications on the curves, and time-series measurements. Moreover, the outstanding questions outline possible areas of improvement. With these approaches, the expectation is that the performance be measured on well-studied and highly specific protein-ligand interactions.
Acknowledgments
This work was supported by the National Institutes of Health under the award numbers R01CA218500 (A.R.I.) and R35GM136421 (A.R.I.).
Declaration of interests
The authors declare no competing interests.
References
- 1.Comess K.M., McLoughlin S.M., Oyer J.A., Richardson P.L., Stöckmann H., Vasudevan A., Warder S.E. Emerging Approaches for the Identification of Protein Targets of Small Molecules - A Practitioners’ Perspective. J. Med. Chem. 2018;61:8504–8535. doi: 10.1021/acs.jmedchem.7b01921. [DOI] [PubMed] [Google Scholar]
- 2.Childs D., Bach K., Franken H., Anders S., Kurzawa N., Bantscheff M., Savitski M.M., Huber W. Nonparametric Analysis of Thermal Proteome Profiles Reveals Novel Drug-binding Proteins. Mol. Cell. Proteomics. 2019;18:2506–2515. doi: 10.1074/mcp.TIR119.001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincent F., Nueda A., Lee J., Schenone M., Prunotto M., Mercola M. Phenotypic drug discovery: recent successes, lessons learned and new directions. Nat. Rev. Drug Discov. 2022;21:899–914. doi: 10.1038/s41573-022-00472-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seashore-Ludlow B., Lundbäck T. Early Perspective: Microplate Applications of the Cellular Thermal Shift Assay (CETSA) J. Biomol. Screen. 2016;21:1019–1033. doi: 10.1177/1087057116659256. [DOI] [PubMed] [Google Scholar]
- 5.Savitski M.M., Reinhard F.B.M., Franken H., Werner T., Savitski M.F., Eberhard D., Martinez Molina D., Jafari R., Dovega R.B., Klaeger S., et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science (New York, N.Y.) 2014;346 doi: 10.1126/science.1255784. [DOI] [PubMed] [Google Scholar]
- 6.Martinez Molina D., Jafari R., Ignatushchenko M., Seki T., Larsson E.A., Dan C., Sreekumar L., Cao Y., Nordlund P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science (New York, N.Y.) 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 7.Schellman J.A. The thermodynamics of solvent exchange. Biopolymers. 1994;34:1015–1026. doi: 10.1002/bip.360340805. [DOI] [PubMed] [Google Scholar]
- 8.Franken H., Mathieson T., Childs D., Sweetman G.M.A., Werner T., Tögel I., Doce C., Gade S., Bantscheff M., Drewes G., et al. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat. Protoc. 2015;10:1567–1593. doi: 10.1038/nprot.2015.101. [DOI] [PubMed] [Google Scholar]
- 9.Becher I., Andrés-Pons A., Romanov N., Stein F., Schramm M., Baudin F., Helm D., Kurzawa N., Mateus A., Mackmull M.T., et al. Pervasive Protein Thermal Stability Variation during the Cell Cycle. Cell. 2018;173:1495–1507.e18. doi: 10.1016/j.cell.2018.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai L., Prabhu N., Yu L.Y., Bacanu S., Ramos A.D., Nordlund P. Horizontal cell biology: monitoring global changes of protein interaction states with the proteome-wide cellular thermal shift assay (CETSA) Annu. Rev. Biochem. 2019;88:383–408. doi: 10.1146/annurev-biochem-062917-012837. [DOI] [PubMed] [Google Scholar]
- 11.Perrin J., Werner T., Kurzawa N., Rutkowska A., Childs D.D., Kalxdorf M., Poeckel D., Stonehouse E., Strohmer K., Heller B., et al. Identifying drug targets in tissues and whole blood with thermal-shift profiling. Nat. Biotechnol. 2020;38:303–308. doi: 10.1038/s41587-019-0388-4. [DOI] [PubMed] [Google Scholar]
- 12.Kalxdorf M., Günthner I., Becher I., Kurzawa N., Knecht S., Savitski M.M., Eberl H.C., Bantscheff M. Cell surface thermal proteome profiling tracks perturbations and drug targets on the plasma membrane. Nat. Methods. 2021;18:84–91. doi: 10.1038/s41592-020-01022-1. [DOI] [PubMed] [Google Scholar]
- 13.Kurzawa N., Becher I., Sridharan S., Franken H., Mateus A., Anders S., Bantscheff M., Huber W., Savitski M.M. A computational method for detection of ligand-binding proteins from dose range thermal proteome profiles. Nat. Commun. 2020;11:5783. doi: 10.1038/s41467-020-19529-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leijten N.M., Bakker P., Spaink H.P., den Hertog J., Lemeer S. Thermal proteome profiling in zebrafish reveals effects of napabucasin on retinoic acid metabolism. Mol. Cell. Proteomics. 2021;20:100033. doi: 10.1074/mcp.RA120.002273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seashore-Ludlow B., Axelsson H., Lundbäck T. Perspective on CETSA Literature: Toward More Quantitative Data Interpretation. SLAS Discov. 2020;25:118–126. doi: 10.1177/2472555219884524. [DOI] [PubMed] [Google Scholar]
- 16.Mateus A., Kurzawa N., Perrin J., Bergamini G., Savitski M.M. Drug Target Identification in Tissues by Thermal Proteome Profiling. Annu. Rev. Pharmacol. Toxicol. 2022;62:465–482. doi: 10.1146/annurev-pharmtox-052120-013205. [DOI] [PubMed] [Google Scholar]
- 17.Gaetani M., Sabatier P., Saei A.A., Beusch C.M., Yang Z., Lundström S.L., Zubarev R.A. Proteome Integral Solubility Alteration: A High-Throughput Proteomics Assay for Target Deconvolution. J. Proteome Res. 2019;18:4027–4037. doi: 10.1021/acs.jproteome.9b00500. [DOI] [PubMed] [Google Scholar]
- 18.Childs D., Bach K., Franken H., Anders S., Kurzawa N., Bantscheff M., Savitski M.M., Huber W. Non-parametric analysis of thermal proteome profiles reveals novel drug-binding proteins. Mol. Cell. Proteomics. 2019;18:2506–2515. doi: 10.1074/mcp.TIR119.001481. , mcp.TIR119.001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potel C.M., Kurzawa N., Becher I., Typas A., Mateus A., Savitski M.M. Impact of phosphorylation on thermal stability of proteins. Nat. Methods. 2021;18:757–759. doi: 10.1038/s41592-021-01177-5. [DOI] [PubMed] [Google Scholar]
- 20.Smith I.R., Hess K.N., Bakhtina A.A., Valente A.S., Rodríguez-Mias R.A., Villén J. Identification of phosphosites that alter protein thermal stability. Nat. Methods. 2021;18:760–762. doi: 10.1038/s41592-021-01178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurzawa N., Leo I.R., Stahl M., Kunold E., Becher I., Audrey A., Mermelekas G., Huber W., Mateus A., Savitski M.M., Jafari R. Deep thermal profiling for detection of functional proteoform groups. Nat. Chem. Biol. 2023;19:962–971. doi: 10.1038/s41589-023-01284-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King D.T., Serrano-Negrón J.E., Zhu Y., Moore C.L., Shoulders M.D., Foster L.J., Vocadlo D.J. Thermal Proteome Profiling Reveals the O-GlcNAc-Dependent Meltome. J. Am. Chem. Soc. 2022;144:3833–3842. doi: 10.1021/jacs.1c10621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sueur C.L., Rattray M., Savitski M. Hierarchical Gaussian process models explore the dark meltome of thermal proteome profiling experiments. bioRxiv. 2023 doi: 10.1101/2023.10.26.564129. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCracken N.A., Peck Justice S.A., Wijeratne A.B., Mosley A.L. Inflect: Optimizing Computational Workflows for Thermal Proteome Profiling Data Analysis. J. Proteome Res. 2021;20:1874–1888. doi: 10.1021/acs.jproteome.0c00872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang S., Kirk P.D.W., Bantscheff M., Lilley K.S., Crook O.M. A Bayesian semi-parametric model for thermal proteome profiling. Commun. Biol. 2021;4:810. doi: 10.1038/s42003-021-02306-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becher I., Werner T., Doce C., Zaal E.A., Tögel I., Khan C.A., Rueger A., Muelbaier M., Salzer E., Berkers C.R., et al. Thermal profiling reveals phenylalanine hydroxylase as an off-target of panobinostat. Nat. Chem. Biol. 2016;12:908–910. doi: 10.1038/nchembio.2185. [DOI] [PubMed] [Google Scholar]
- 27.Mateus A., Kurzawa N., Becher I., Sridharan S., Helm D., Stein F., Typas A., Savitski M.M. Thermal proteome profiling for interrogating protein interactions. Mol. Syst. Biol. 2020;16 doi: 10.15252/msb.20199232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin J.K., 2nd, Sheehan J.P., Bratton B.P., Moore G.M., Mateus A., Li S.H., Kim H., Rabinowitz J.D., Typas A., Savitski M.M., et al. A Dual-Mechanism Antibiotic Kills Gram-Negative Bacteria and Avoids Drug Resistance. Cell. 2020;181:1518–1532.e1514. doi: 10.1016/j.cell.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z., Kavdia K., Dey K.K., Pagala V.R., Kodali K., Liu D., Lee D.G., Sun H., Chepyala S.R., Cho J.H., et al. High-throughput and Deep-proteome Profiling by 16-plex Tandem Mass Tag Labeling Coupled with Two-dimensional Chromatography and Mass Spectrometry. JoVE. 2020;162:e61684. doi: 10.3791/61684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang T., Choi M., Tzouros M., Golling S., Pandya N.J., Banfai B., Dunkley T., Vitek O. MSstatsTMT: Statistical Detection of Differentially Abundant Proteins in Experiments with Isobaric Labeling and Multiple Mixtures. Mol. Cell. Proteomics. 2020;19:1706–1723. doi: 10.1074/mcp.RA120.002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruan C., Wang Y., Zhang X., Lyu J., Zhang N., Ma Y., Shi C., Qu G., Ye M. Matrix Thermal Shift Assay for Fast Construction of Multidimensional Ligand–Target Space. Anal. Chem. 2022;94:6482–6490. doi: 10.1021/acs.analchem.1c04627. [DOI] [PubMed] [Google Scholar]
- 32.Savitski M.M., Mathieson T., Zinn N., Sweetman G., Doce C., Becher I., Pachl F., Kuster B., Bantscheff M. Measuring and Managing Ratio Compression for Accurate iTRAQ/TMT Quantification. J. Proteome Res. 2013;12:3586–3598. doi: 10.1021/pr400098r. [DOI] [PubMed] [Google Scholar]
- 33.McAlister G.C., Nusinow D.P., Jedrychowski M.P., Wühr M., Huttlin E.L., Erickson B.K., Rad R., Haas W., Gygi S.P. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ting L., Rad R., Gygi S.P., Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roumeliotis T.I., Weisser H., Choudhary J.S. Evaluation of a Dual Isolation Width Acquisition Method for Isobaric Labeling Ratio Decompression. J. Proteome Res. 2019;18:1433–1440. doi: 10.1021/acs.jproteome.8b00870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Searle B.C., Yergey A.L. An efficient solution for resolving iTRAQ and TMT channel cross-talk. J. Mass Spectrom. 2020;55 doi: 10.1002/jms.4354. [DOI] [PubMed] [Google Scholar]
- 37.Niu M., Cho J.H., Kodali K., Pagala V., High A.A., Wang H., Wu Z., Li Y., Bi W., Zhang H., et al. Extensive Peptide Fractionation and y1 Ion-based Interference Detection Enable Accurate Quantification by Isobaric Labeling and Mass Spectrometry. Anal. Chem. 2017;89:2956–2963. doi: 10.1021/acs.analchem.6b04415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu X., Liao B., Sun S., Mao Y., Wu Q., Tian R., Tan C.S.H. Scaled-Down Thermal Profiling and Coaggregation Analysis of the Proteome for Drug Target and Protein Interaction Analysis. Anal. Chem. 2023;95:13844–13854. doi: 10.1021/acs.analchem.3c01941. [DOI] [PubMed] [Google Scholar]
- 39.George A.L., Sidgwick F.R., Watt J.E., Martin M.P., Trost M., Marín-Rubio J.L., Dueñas M.E. Comparison of Quantitative Mass Spectrometric Methods for Drug Target Identification by Thermal Proteome Profiling. J. Proteome Res. 2023;22:2629–2640. doi: 10.1021/acs.jproteome.3c00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y., West G.M., Abdelmessih M., Troutman M.D., Everley R.A. A Comparison of Two Stability Proteomics Methods for Drug Target Identification in OnePot 2D Format. ACS Chem. Biol. 2021;16:1445–1455. doi: 10.1021/acschembio.1c00317. [DOI] [PubMed] [Google Scholar]
- 41.Li J., Smith L.S., Zhu H.J. Data-independent acquisition (DIA): An emerging proteomics technology for analysis of drug-metabolizing enzymes and transporters. Drug Discov. Today Technol. 2021;39:49–56. doi: 10.1016/j.ddtec.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venable J.D., Dong M.Q., Wohlschlegel J., Dillin A., Yates J.R. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 43.Ball K.A., Webb K.J., Coleman S.J., Cozzolino K.A., Jacobsen J., Jones K.R., Stowell M.H.B., Old W.M. An isothermal shift assay for proteome scale drug-target identification. Commun. Biol. 2020;3:75. doi: 10.1038/s42003-020-0795-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dwivedi P., Rose C.M. Understanding the effect of carrier proteomes in single cell proteomic studies-key lessons. Expert Rev. Proteomics. 2022;19:5–15. doi: 10.1080/14789450.2022.2036126. [DOI] [PubMed] [Google Scholar]
- 45.Phaneuf C.G., Aizikov K., Grinfeld D., Kreutzmann A., Mourad D., Lange O., Dai D., Zhang B., Belenky A., Makarov A.A., Ivanov A.R. Experimental strategies to improve drug-target identification in mass spectrometry-based thermal stability assays. Commun. Chem. 2023;6:64. doi: 10.1038/s42004-023-00861-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruan C., Ning W., Liu Z., Zhang X., Fang Z., Li Y., Dang Y., Xue Y., Ye M. Precipitate-Supported Thermal Proteome Profiling Coupled with Deep Learning for Comprehensive Screening of Drug Target Proteins. ACS Chem. Biol. 2022;17:252–262. doi: 10.1021/acschembio.1c00936. [DOI] [PubMed] [Google Scholar]
- 47.Beusch C.M., Sabatier P., Zubarev R.A. Ion-Based Proteome-Integrated Solubility Alteration Assays for Systemwide Profiling of Protein–Molecule Interactions. Anal. Chem. 2022;94:7066–7074. doi: 10.1021/acs.analchem.2c00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jarzab A., Kurzawa N., Hopf T., Moerch M., Zecha J., Leijten N., Bian Y., Musiol E., Maschberger M., Stoehr G., et al. Meltome atlas-thermal proteome stability across the tree of life. Nat. Methods. 2020;17:495–503. doi: 10.1038/s41592-020-0801-4. [DOI] [PubMed] [Google Scholar]
- 49.Budnik B., Levy E., Harmange G., Slavov N. SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 2018;19:161. doi: 10.1186/s13059-018-1547-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yi L., Tsai C.F., Dirice E., Swensen A.C., Chen J., Shi T., Gritsenko M.A., Chu R.K., Piehowski P.D., Smith R.D., et al. Boosting to Amplify Signal with Isobaric Labeling (BASIL) Strategy for Comprehensive Quantitative Phosphoproteomic Characterization of Small Populations of Cells. Anal. Chem. 2019;91:5794–5801. doi: 10.1021/acs.analchem.9b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saltzman A.B., Leng M., Bhatt B., Singh P., Chan D.W., Dobrolecki L., Chandrasekaran H., Choi J.M., Jain A., Jung S.Y., et al. gpGrouper: A Peptide Grouping Algorithm for Gene-Centric Inference and Quantitation of Bottom-Up Proteomics Data. Mol. Cell. Proteomics. 2018;17:2270–2283. doi: 10.1074/mcp.TIR118.000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Vranken J.G., Li J., Mitchell D.C., Navarrete-Perea J., Gygi S.P. Assessing target engagement using proteome-wide solvent shift assays. Elife. 2021;10 doi: 10.7554/eLife.70784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Childs D., Kurzawa N., Franken H., Doce C., Savitski M., Huber W. TPP: Analyze Thermal Proteome Profiling (TPP) Experiments. R Package Version 3.28.0. 2023 [Google Scholar]
- 54.Di Veroli G.Y., Fornari C., Goldlust I., Mills G., Koh S.B., Bramhall J.L., Richards F.M., Jodrell D.I. An automated fitting procedure and software for dose-response curves with multiphasic features. Sci. Rep. 2015;5 doi: 10.1038/srep14701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feyertag F., Huber K.V.M. TP-MAP - an Integrated Software Package for the Analysis of 1D and 2D Thermal Profiling Data. bioRxiv. 2021 doi: 10.1101/2021.02.22.432361. Preprint at. [DOI] [Google Scholar]
- 56.Li J., Van Vranken J.G., Paulo J.A., Huttlin E.L., Gygi S.P. Selection of Heating Temperatures Improves the Sensitivity of the Proteome Integral Solubility Alteration Assay. J. Proteome Res. 2020;19:2159–2166. doi: 10.1021/acs.jproteome.0c00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mateus A., Määttä T.A., Savitski M.M. Thermal proteome profiling: unbiased assessment of protein state through heat-induced stability changes. Proteome Sci. 2016;15:13. doi: 10.1186/s12953-017-0122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sabatier P., Beusch C.M., Meng Z., Zubarev R.A. System-Wide Profiling by Proteome Integral Solubility Alteration Assay of Drug Residence Times for Target Characterization. Anal. Chem. 2022;94:15772–15780. doi: 10.1021/acs.analchem.2c03506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zinn N., Werner T., Doce C., Mathieson T., Boecker C., Sweetman G., Fufezan C., Bantscheff M. Improved Proteomics-Based Drug Mechanism-of-Action Studies Using 16-Plex Isobaric Mass Tags. J. Proteome Res. 2021;20:1792–1801. doi: 10.1021/acs.jproteome.0c00900. [DOI] [PubMed] [Google Scholar]
- 60.Le Sueur C., Hammarén H.M., Sridharan S., Savitski M.M. Thermal proteome profiling: Insights into protein modifications, associations, and functions. Curr. Opin. Chem. Biol. 2022;71 doi: 10.1016/j.cbpa.2022.102225. [DOI] [PubMed] [Google Scholar]
- 61.Yang X., Coulombe-Huntington J., Kang S., Sheynkman G.M., Hao T., Richardson A., Sun S., Yang F., Shen Y.A., Murray R.R., et al. Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell. 2016;164:805–817. doi: 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meng H., Ma R., Fitzgerald M.C. Chemical Denaturation and Protein Precipitation Approach for Discovery and Quantitation of Protein-Drug Interactions. Anal. Chem. 2018;90:9249–9255. doi: 10.1021/acs.analchem.8b01772. [DOI] [PubMed] [Google Scholar]
- 63.Sridharan S., Hernandez-Armendariz A., Kurzawa N., Potel C.M., Memon D., Beltrao P., Bantscheff M., Huber W., Cuylen-Haering S., Savitski M.M. Systematic discovery of biomolecular condensate-specific protein phosphorylation. Nat. Chem. Biol. 2022;18:1104–1114. doi: 10.1038/s41589-022-01062-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sridharan S., Kurzawa N., Werner T., Günthner I., Helm D., Huber W., Bantscheff M., Savitski M.M. Proteome-wide solubility and thermal stability profiling reveals distinct regulatory roles for ATP. Nat. Commun. 2019;10:1155. doi: 10.1038/s41467-019-09107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miettinen T.P., Peltier J., Härtlova A., Gierliński M., Jansen V.M., Trost M., Björklund M. Thermal proteome profiling of breast cancer cells reveals proteasomal activation by CDK4/6 inhibitor palbociclib. EMBO J. 2018;37 doi: 10.15252/embj.201798359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hashimoto Y., Sheng X., Murray-Nerger L.A., Cristea I.M. Temporal dynamics of protein complex formation and dissociation during human cytomegalovirus infection. Nat. Commun. 2020;11:806. doi: 10.1038/s41467-020-14586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurzawa N., Mateus A., Savitski M.M. Rtpca: an R package for differential thermal proximity coaggregation analysis. Bioinformatics. 2021;37:431–433. doi: 10.1093/bioinformatics/btaa682. [DOI] [PMC free article] [PubMed] [Google Scholar]