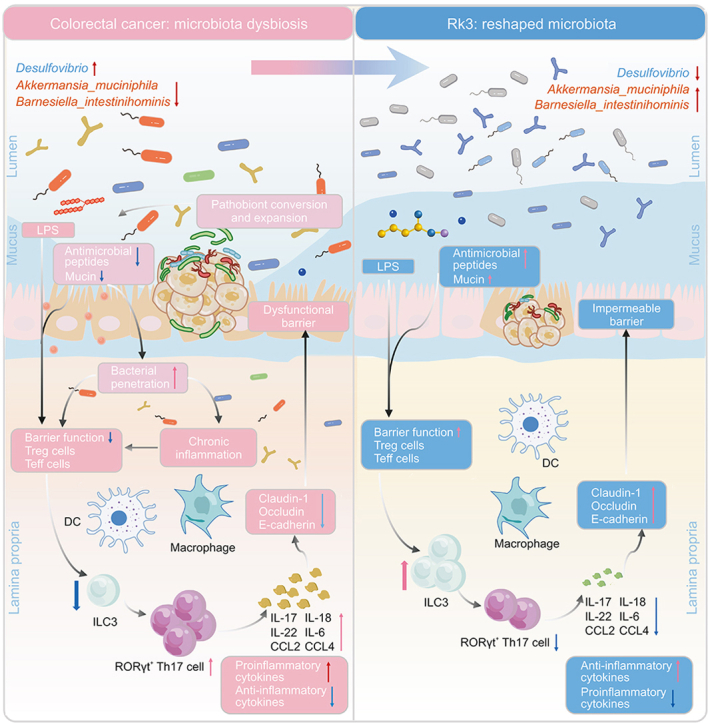
Abstract
The gut microbiota plays a pivotal role in the immunomodulatory and protumorigenic microenvironment of colorectal cancer (CRC). However, the effect of ginsenoside Rk3 (Rk3) on CRC and gut microbiota remains unclear. Therefore, the purpose of this study is to explore the potential effect of Rk3 on CRC from the perspective of gut microbiota and immune regulation. Our results reveal that treatment with Rk3 significantly suppresses the formation of colon tumors, repairs intestinal barrier damage, and regulates the gut microbiota imbalance caused by CRC, including enrichment of probiotics such as Akkermansia muciniphila and Barnesiella intestinihominis, and clearance of pathogenic Desulfovibrio. Subsequent metabolomics data demonstrate that Rk3 can modulate the metabolism of amino acids and bile acids, particularly by upregulating glutamine, which has the potential to regulate the immune response. Furthermore, we elucidate the regulatory effects of Rk3 on chemokines and inflammatory factors associated with group 3 innate lymphoid cells (ILC3s) and T helper 17 (Th17) signaling pathways, which inhibits the hyperactivation of the Janus kinase-signal transducer and activator of transcription 3 (JAK-STAT3) signaling pathway. These results indicate that Rk3 modulates gut microbiota, regulates ILC3s immune response, and inhibits the JAK-STAT3 signaling pathway to suppress the development of colon tumors. More importantly, the results of fecal microbiota transplantation suggest that the inhibitory effect of Rk3 on colon tumors and its regulation of ILC3 immune responses are mediated by the gut microbiota. In summary, these findings emphasize that Rk3 can be utilized as a regulator of the gut microbiota for the prevention and treatment of CRC.
Keywords: Colorectal cancer, Ginsenoside, Immune cells, Gut microbiota
Graphical abstract
Highlights
-
•
Rk3 suppresses colorectal tumorigenesis.
-
•
Rk3 modulates dysbiosis of gut microbiota.
-
•
Rk3 regulates intestinal ILC3 immune response.
-
•
The antitumor effects of Rk3 depend on gut microbiota.
-
•
Rk3-modulated gut microbiota inhibits colorectal tumorigenesis.
1. Introduction
Colorectal cancer (CRC) is a prevalent malignant tumor and the third most common cancer worldwide [1]. Despite various treatment options, such as surgery, chemotherapy, and immune checkpoint inhibitors, the treatment prospects for colon and rectal cancer patients remain discouraging owning to limitations such as drug resistance and toxicity [2,3]. Therefore, there is an urgent need to develop novel strategies for preventing and treating CRC by targeting its causative factors.
The etiology of CRC involves the long-term accumulation of genetic, epigenetic, and environmental factors [4]. Accumulating studies have shown that the gut microbiota is involved in the occurrence, progression, and treatment of CRC [5]. The gut microbiota can exert direct or indirect effects on host health, encompassing alterations in gut microbiota and metabolites, as well as interactions between the microbial community and the immune microenvironment [6,7]. The interaction between the host and bacteria that exacerbate colon tumor progression triggers inflammation and immune responses, which are among the most common mechanisms involved in the development of colon tumors. Several bacterial can elicit the production of chemokines within the host, thereby activating the immune response and fostering the progression of colon tumorigenesis via the secretion of cytokines [8]. Innate lymphoid cells (ILCs), a recently acknowledged family of innate immune cells, play a pivotal role in regulating the interactions between the host and microbiota. They are considered gatekeepers of intestinal barrier integrity and mucosal homeostasis [9]. Notably, group 3 innate lymphoid cells (ILC3s), which serve as innate counterparts to T helper 17 (Th17) cells, exert regulatory control over gut microbiota via the production of cytokines such as interleukin 17 (IL-17) and IL-22. Th17 cells have been demonstrated to directly participate in the development of colon tumors induced by enterotoxigenic Bacteroides fragilis [10]. Furthermore, metabolite production serves as an additional significant mechanism that drives CRC progression. These compositions of these metabolites include lipids, bile acids (BAs), organic acids, and amino acids [11]. Different metabolites exhibit distinct characteristics, such as harmful metabolites such as deoxycholic acid, lithocholic acid, and trimethylamine oxide, which promote CRC development by facilitating tumor cell proliferation, invasion, and migration [12,13]. Beneficial metabolites, such as ursodeoxycholic acid, butyrate, and amino acids, can inhibit the development of CRC by promoting the production of tight junction proteins, suppressing inflammation, and facilitating tumor cell apoptosis [14]. With the increasing comprehension of the interplay between the gut microbiota and the immune system in carcinogenesis, modulating the gut microbiota has emerged as a promising strategy for preventing and treating CRC.
Recent evidence has suggested that natural products can positively affect disease treatment by affecting the gut microbiota. There exists a reciprocal interaction between natural compounds and the gut microbiota. On one hand, the gut microbiota can metabolize natural compounds, influencing their bioavailability and anti-tumor activity [15]. On the other hand, natural compounds can prevent and treat various tumors by modulating the composition and metabolism of the gut microbiota [16]. Furthermore, natural compounds regulate the host immune system by modulating the gut microbiota, thereby affecting the host's response to immunotherapy [17]. For instance, berberine, a plant-derived natural compound, effectively suppresses CRC associated with a high-fat diet by regulating the modulation of gut microbial-mediated lysophosphatidylcholine [18]. Besides, dihydromyricetin has been shown to alleviate colitis in mice by modulating gut microbial dysbiosis and bile acid metabolism [19]. Myrciaria dubia exhibits anti-tumor activity and overcomes anti-programmed cell death protein 1 (anti-PD-1) resistance by modulating the gut microbiota [20]. Ginsenosides, the major active ingredients of the natural product ginseng, have made significant contributions to disease prevention and treatment [21,22]. Previous research has demonstrated that ginsenoside Rk3 (Rk3) regulates autophagy and apoptosis by targeting the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) pathway, thus inhibiting liver cancer cells [23]. Additionally, Rk3 can improve intestinal inflammation by upregulating butyrate content and regulating the Toll-like receptor 4 (TLR4)/nuclear factor-κB (NF-κB) signaling pathway [24,25]. However, the effects of Rk3 on colon tumors and the gut microbiota have not been reported.
In this study, we postulated that Rk3 may exert inhibitory effects on colorectal tumorigenesis by modulating the gut microbiota and influencing the immune system. To validate this hypothesis, the impact of Rk3 on colon tumors and intestinal barrier damage, as well as its effects on gut microbiota and metabolomics, were first appraised in mice treated with CRC induced by azoxymethane (AOM)/dextran sulfate sodium (DSS). Subsequently, we elucidated the potential molecular mechanism of Rk3 in inhibiting CRC, with a particular focus on the impact of Rk3 on the immune response of ILC3 and the tumor-promoting Janus kinase (JAK)-signal transducer and activator of transcription 3 (STAT3) signaling pathway. Importantly, fecal microbiota transplantation (FMT) was performed to investigate whether the inhibitory effect of Rk3 on CRC was mediated by gut microbiota. Our study reveals the mechanism of Rk3 inhibiting CRC by modulating the gut microbiota, which provides a novel strategy for the prevention and treatment of CRC.
2. Materials and methods
2.1. Reagents
Rk3 (purity ≥ 99%) was purchased from the Biomedicine Institute of Northwest University (Xi'an, China). Rk3 was dissolved in a 0.2% carboxymethyl cellulose sodium solution. AOM was purchased from Sigma-Aldrich (St. Louis, MO, USA), and DSS was purchased from MP Biomedicals (Solon, OH, USA). Primary antibodies against Claudin-1, E-cadherin, IL-6, JAK3, and STAT3 were provided by Proteintech Group (Chicago, IL, USA). The antibodies against phospho-JAK3 (p-JAK3) and phospho-STAT3 (p-STAT3) were purchased from Cell Signaling Technology (Danvers, MA, USA), and secondary antibodies were provided by Abcam (Cambridge, UK).
2.2. Animal experiments
Six-week-old male C57BL/6 mice with a body weight of 20–22 g were purchased from Air Force Medical University (Xi'an, China) and were housed for one week under standard conditions with ad libitum access to food and water. This study was approved by the Animal Experiment Ethics Committee of the Northwest University (Approval No.: NWU-AWC-20220903 M).
For the AOM/DSS experiment, mice were randomly divided into three groups (normal, model, and Rk3; n = 12 mice/group) and allowed to adapt for one week under standard conditions, with free access to food and water. The model and Rk3 treatment group mice were intraperitoneally injected with 10 mg/kg AOM. One week after AOM injection, 2.0% DSS was added to the drinking water for five days, followed by two weeks of normal drinking water. This step was repeated three times. The Rk3 group (60 mg/kg Rk3, i.g.) was administered by gavage once a day starting from the first week of the experiment, as shown in Fig. 1A, whereas the normal and model groups mice were administered saline by gavage.
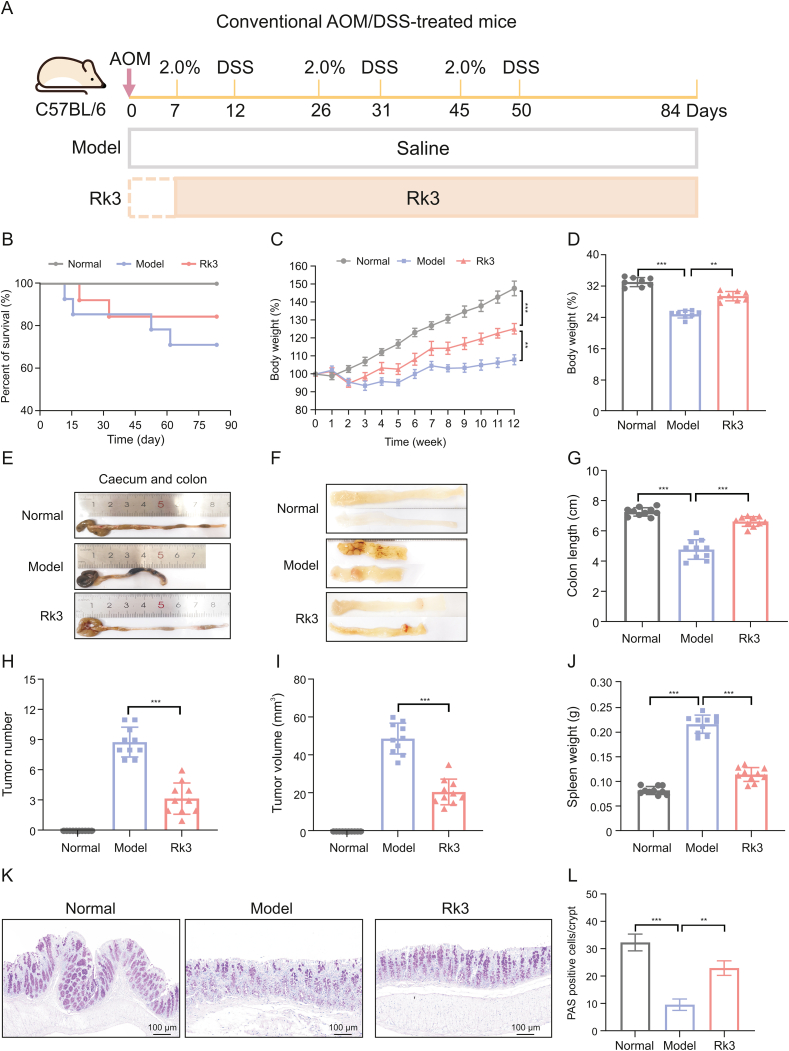
Fig. 1.
Ginsenoside Rk3 (Rk3) inhibits colonic tumorigenesis. (A) Schematic diagram of the animal experiment design. (B) Survival curve of the mice. (C) Rate of change in body weight of the mice during the experiment (n = 10). (D) Final body weight of the mice (n = 10). (E) Colon images of the mice. (F) Medial images of the mice colon. (G) Colon length of the mice (n = 10). (H) Number of colon tumors in the mice (n = 10). (I) Volume of colon tumors in the mice (n = 10). (J) Spleen weight of the mice (n = 10). (K) Periodic acid-schiff (PAS) staining of mice colon. (L) Quantitative analysis of PAS staining (n = 3). ∗∗P < 0.01 and ∗∗∗P < 0.001. AOM: azoxymethane; DSS: dextran sulfate sodium.
For FMT administration, to facilitate proper engraftment of the gut microbiota from the donor group, a two-week antibiotic cocktail treatment (0.2 g/L ampicillin, neomycin, and metronidazole, and 0.1 g/L vancomycin) was administered to the recipient group of mice, thus establishing a germ-free mouse model. Following this, the recipient mice received daily administration of the gut microbiota from the donor group. In summary, fresh fecal samples were from each group, homogenized, and diluted in sterile saline to a final concentration of 100 mg/mL. The composite sample was then centrifuged at 100 g for 3 min, and the resulting supernatant was filtered and used for the FMT treatment of the recipient mice. Concurrently, recipient mice were used to establish an AOM/DSS model according to the aforementioned protocol.
2.3. Histological analysis
Mouse colon tissue samples were fixed with formalin for 48 h. Next, an ethanol gradient dehydration and xylene transparency process were performed and the samples were embedded in paraffin using an embedding machine to create paraffin blocks. Sections (5-mm thick) were cut for hematoxylin-eosin (H&E) and periodic acid-Schiff stain (PAS). For H&E staining, the tissue sections were initially stained with hematoxylin, differentiated in hydrochloric acid, and then immersed in ammonia water to achieve a blue hue. Finally, the sections were stained with eosin. Post-staining, the sections undergo dehydration and transparency procedures and were subsequently embedded in a neutral resin for observation. Alcian blue-PAS staining was performed using a kit following the manufacturer's instructions. The number of goblet cells and capture images was quantified using a microscope (Nikon, Tokyo, Japan).
2.4. Immunofluorescence staining
Tissue slices for immunohistochemistry were first blocked with 2% (V/V) hydrogen peroxide to inhibit endogenous peroxidase and blocked with 3% (V/V) goat serum, and then incubated at room temperature for 2 h. The slices were then incubated with a streptavidin-peroxidase conjugate for 40 min, stained with 3,3'-diaminobenzidine substrate, and counterstained with hematoxylin. Fluorescently labeled antibodies were used for staining, and 4′6-diamidino-2-phenylindole (DAPI) was used for counterstaining for 5 min. Confocal laser scanning microscopy (Nikon) was used to obtain images. The integrated optical density of each marker in each sample was evaluated using ImageJ software.
2.5. Measurement of intestinal permeability
Blood samples were collected from the orbital venous plexus and subsequently centrifuged at 2,500 rpm to obtain serum. The levels of lipopolysaccharides were measured using enzyme-linked immunosorbent assay (ELISA) kits (Shanghai Enzyme-linked Biotechnology Co., Ltd., Shanghai, China) to assess the permeability of the mouse intestines.
2.6. 16S ribosomal RNA (rRNA) analysis of gut microbiota
Fecal samples from mice were collected and promptly frozen in liquid nitrogen before being stored at −80 °C. To investigate the gut microbiota, we performed a 16S rRNA sequencing analysis. Total DNA was extracted from the fecal samples using a commercial kit from Omega Bio-Tek (Norcross, GA, USA). The DNA were assessed using a NanoDrop ND-1,000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA) and agarose gel electrophoresis, respectively. The highly variable V3–V4 region of the bacterial 16S rRNA gene, which is approximately 468 bp long, was selected for sequencing. Polymerase chain reaction (PCR) amplification was performed using specific primers. Sequencing outcomes were examined according to the standardized guidelines for library construction on the Illumina MiSeq platform (San Diego, CA, USA).
2.7. Fecal metabolomics analysis
In this study, we employed the HM400 metabolomics approach, using liquid chromatography-tandem mass spectrometry (LC-MS/MS) for high-throughput targeted metabolomic analysis. This allowed us to simultaneously detect over 400 metabolites, effectively addressing the challenges of untargeted metabolomics validation and the limited coverage of traditional targeted metabolomics. Initially, we conducted metabolite separation and quantitative detection using the Waters ACQUITY UPLC I-Class Plus coupled with a QTRAP6500 plus high-sensitivity mass spectrometer (Milford, MA, USA).
2.7.1. Chromatographic conditions
Samples injected in the bridged ethylene hybrid (BEH) C18 column (Waters Corporation) were eluted for multiple reaction monitoring (MRM) with the following LC gradient: phase ultra pure water (mobile phase A) and acetonitrile (mobile phase B). 0–1 min, 5% mobile phase B; 1–5 min, 5%–30% mobile phase B; 5–9 min, 30%–50% mobile phase B; 9–11 min, 50%–78% mobile phase B; 11–13.5 min, 78%–95% mobile phase B; 13.5–16 min, 95%–100% mobile phase B; and 16.1–18 min, 5% mobile phase B. The flow rate at 0.4 mL/min, the column temperature was set at 40 °C.
2.7.2. Mass spectrum conditions
For the QTRAP 6,500 Plus, equipped with an EST Turbo Ion-Spray interface (SCIEX, Shanghai, China), the source parameters were configured as follows: source temperature, 500 °C and ion spray voltage, 4,500 V (positive mode) or –4,500 V (negative mode). Ion source gas I, gas II, and curtain gas were set at 40 and 20 psi, respectively. The MRM methods were configured in schedule mode for the target metabolites. Quantitative software, Skyline, was employed to generate a data matrix containing information on metabolite identification and quantification results. Subsequently, we imported the results into metaX for downstream bioinformatics analysis. The identified metabolites were then classified and functionally annotated based on their Kyoto Encyclopedia of Genes and Genomes (KEGG) ID, Human Metabolome Database (HMDB) ID, category, and KEGG Pathway information.
2.8. Transcriptomic sequencing analysis
A suitable tissue sample was obtained and homogenize to obtain purified RNA. RNA was denatured at an appropriate temperature to disrupt its secondary structure, and oligo(dT) beads was used to enrich the messenger RNA (mRNA). mRNA was fragmented using a fragmentation reagent at an appropriate temperature and time. A single-stranded synthesis reaction system was prepared and single-stranded complementary DNA (cDNA) was synthesized using a designated reaction program. The product was amplified using a PCR reaction system (Bio-Rad, Hercules, CA, USA), and the reaction program was set. An appropriate detection method was used to inspect the quality of the library. To obtain a single-stranded circular product, the PCR product was denatured into a single-stranded state, a circularization reaction system was prepared, and a reaction program was set up. Linear DNA molecules that fail to circularize and replicate single-stranded circular DNA molecules are digested using rolling circle replication, resulting in a DNA nanoball (DNB) containing multiple copies. The obtained DNBs were loaded onto the network-like pores of the chip using high-density DNA nanochip technology and sequenced using combined probe-anchor synthesis technology.
Gene set enrichment analysis (GSEA) utilizes the GSEA algorithm developed by the Broad Institute (Cambridge, MA, USA) for analysis. It employs predefined gene sets derived from KEGG functional annotations to rank genes based on their differential expression levels between two sample groups. Subsequently, we examined whether the predefined gene sets were enriched at the top or bottom of the ranking table.
2.9. Real-time quantitative PCR (qRT-PCR)
Colon tissue (50 mg) was extracted and pulverized into a homogenate using liquid nitrogen. Total RNA was isolated using the Takara RNA extraction kit (Norcross, GA, USA). RNA was reverse-transcribed into cDNA using a 20 μL of reaction mixture consisting of 50 mmol/L KCl, 10 mmol/L Tris/HCl, 5 mmol/L MgCl2, 1 mmol/L deoxynucleoside triphosphate, 2.5 μmol/L random hexamer, 20 U RNAsin, and 50 U reverse transcriptase. The TaqMan SybrGreen mixture was used for qRT-PCR, and the reaction was performed on the CFX thermal cycling system (Bio-Rad, Hercules, CA, USA).
2.10. Protein extraction and Western blotting (WB)
After homogenizing 20 mg of colon tissue in liquid nitrogen, the sample was lysed in radioimmunoprecipitation assay (RIPA) buffer, containing detergents and protease inhibitors to extract proteins. The samples were incubated on ice to prevent protein degradation and then centrifuged to separate the supernatant containing the proteins from the pellet containing the insoluble materials. The pellet was washed with tris-buffered saline to remove the residual buffer or contaminants. To visualize the protein of interest, the Clarity Western enhanced chemiluminescence (ECL) substrate kit (Waltham, MA, USA) was used, which reacts with horseradish peroxidase to emit light that can be detected using an ECL imaging system (Millipore Corporation, Billerica, MA, USA). This allows for the detection and quantification of specific proteins in a sample.
2.11. Statistical analysis
The data were presented as mean ± standard deviation and analyzed using GraphPad Prism 9.0 program. One-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test was used to compare data between more than two groups. Differences were considered to be statistically significant when the adjusted P value was < 0.05.
3. Results
3.1. Rk3 inhibits the development of colonic tumors in AOM/DSS-induced CRC mice
To investigate the effect of Rk3 on the occurrence and development of CRC, we established a CRC model using AOM/DSS-treated mice supplemented with Rk3 (Fig. 1A). Analogous to human CRC, the AOM/DSS mouse model exhibited CRC clinical features such as low survival rates, weight loss, diarrhea, and rectal bleeding. However, treatment with Rk3 significantly improved survival rate and prevented weight loss in the mice (Figs. 1B−D). Further analysis revealed that supplementing with Rk3 directly alleviated the shortening of the colon (Figs. 1E−G) and reduced the number and volume of colon tumors compared to the model group mice (Figs. 1H and I). Additionally, we observed that the spleens of mice in the model group showed enlargement, indicating abnormal alterations in their immune system, which were mitigated by the administration of Rk3 (Fig. 1J).
Histopathological analysis further confirmed the pathological changes in colon tissue. H&E staining revealed the presence of colon adenomas and adenocarcinomas in the CRC mice, along with abnormal crypt structures, reduced goblet cells, and infiltration of inflammatory cells in the colon tissue. However, treatment with Rk3 markedly ameliorated the histopathological impairment of colon adenomas (Figs. S1A and B). Immunofluorescence staining analysis revealed a significant increase in the number of Ki-67 positive cells in the model group mice, indicating excessive proliferation of colonic epithelial cells. However, Rk3 supplementation partially inhibited colonic cell proliferation (Figs. S1C and D). PAS staining further demonstrated a loss of goblet cells and mucus in the colon of CRC mice, indicating disruption of intestinal mucosal homeostasis, which could be improved through Rk3 treatment (Figs. 1K and L). Our findings highlight the inhibitory effect of Rk3 on colon tumor development in CRC mice, as well as its regulatory role in colonic histopathology and mucosal homeostasis.
3.2. Rk3 regulates the composition of gut microbiota in AOM/DSS-induced CRC mice
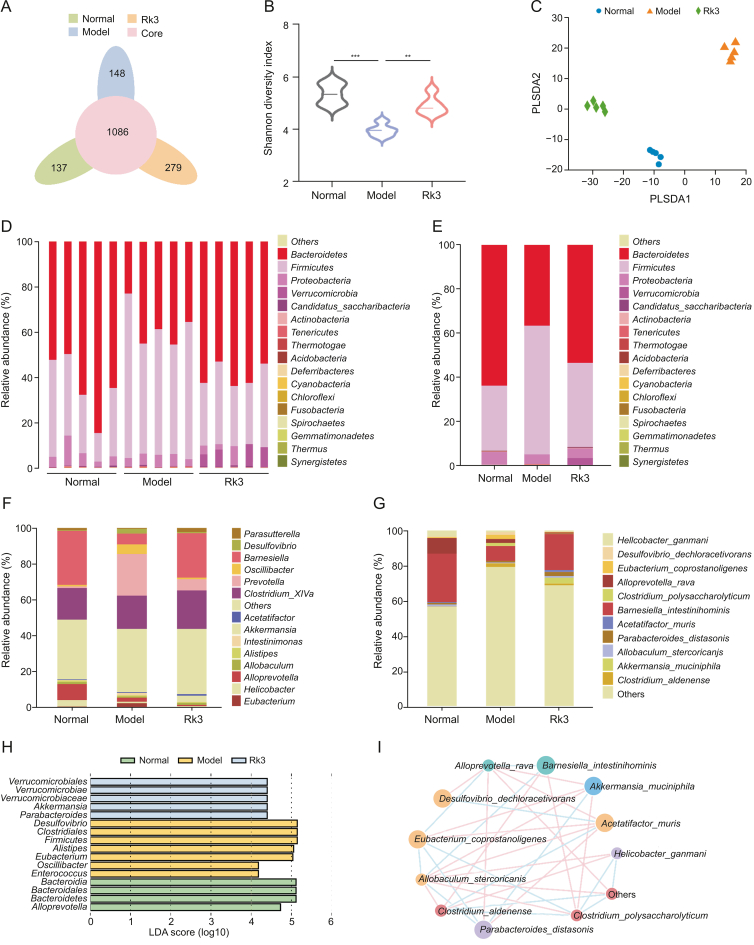
As gut microbiota plays a pivotal role in CRC, we conducted 16S rDNA sequencing analysis of fecal samples to investigate the influence of Rk3 on the gut microbiota of CRC mice. Initially, we assessed the influence of Rk3 on the diversity of gut microbiota in CRC mice. Alpha analysis revealed that the Shannon index was significantly lower in the model group than in the normal and Rk3 groups (Figs. 2A and B). Moreover, the results of partial least squares discriminant analysis (PLS-DA) demonstrated a conspicuous differentiation between the model group and both the normal and Rk3 groups (Fig. 2C). Subsequent investigation was conducted to provide a more in-depth analysis of the gut microbiota composition. Our findings at the phylum level indicated a notable distinction in the abundance of Bacteroidetes and Firmicutes between the model and normal groups. Notably, the administration of Rk3 led to a significant elevation in the abundance of Bacteroidetes and Verrucomicrobia, while concurrently resulting in a decrease in the abundance of Firmicutes (Figs. 2D and E). Subsequent scrutiny at the genus and species level unveiled a substantial decline in beneficial bacteria, including Akkermansia, Barnesiella, and Parasutterella, within the model group. Conversely, there was a notable proliferation of pathogenic bacteria such as Oscillibacter, Desulfovibrio, and Alistipes. However, the administration of Rk3 demonstrated a remarkable capability to counteract this trend and restore a more favorable gut microbiota composition (Fig. 2F). Previous studies have substantiated that Desulfovibrio is the principal producer of hydrogen sulfide (H2S) in the gastrointestinal tract, and H2S is associated with chronic colonic conditions and colonic inflammation. Alistipes demonstrates pathogenicity in CRC, correlating with clinical attributes of CRC patients. The enrichment of beneficial bacteria such as Akkermansia muciniphila, Parabacteroides distasonis, and Barnesiella intestinihominis in the Rk3 group at the species level suggests that Rk3 supplementation may positively modulate the gut microbiota composition. In contrast, the model group showed a large colonization of pathogenic bacteria, particularly Desulfovibrio_dechloracetivorans (Fig. 2G). The dominant bacterial analysis using linear discriminant analysis effect size (LEfSe) further supported these findings, with Verrucomicrobia and Akkermansia being significantly dominant in the Rk3 group, while the model group was dominated by the pathogenic bacteria Desulfovibrio (Fig. 2H). Furthermore, the ecological network analysis of bacterial interactions revealed that the Rk3 group exhibited a significant enrichment of beneficial bacterial interactions, while the model group showed an independent trend of harmful bacterial enrichment (Fig. 2I). Overall, our findings suggest that Rk3 treatment may effectively enrich beneficial bacteria, such as Akkermansia and Barnesiella, while reducing the abundance of pathogenic bacteria, such as Desulfovibrio, in the gut microbiota. The restorative effect of Rk3 on the gut microbiota may ameliorate gut dysbiosis and hinder the advancement of CRC.
Fig. 2.
Ginsenoside Rk3 (Rk3) modulates the composition of gut microbiota. (A) Venn diagram illustrating the overlapping operational taxonomic units (OTUs) in each group (n = 5). (B) Alpha diversity analysis of gut microbiota utilizing the Shannon index (n = 5). (C) Beta diversity analysis of gut microbiota using partial least squares discriminant analysis (PLS-DA) (n = 5). (D) Comprehensive overview of microbial phyla in each sample at the phylum level (n = 5). (E) Comprehensive overview of microbial phyla in each group at the phylum level (n = 5). (F) Overview of microbial genera in each group at the genus level (n = 5). (G) Overview of microbial species in each group at the species level (n = 5). (H) Linear discriminant analysis effect size (LEfSe) analysis to identify differentially abundant bacterial taxa among groups (linear discriminant analysis score > 4; n = 5). (I) Network plot depicting the differentially abundant species between the model and Rk3 groups. ∗∗P < 0.01 and ∗∗∗P < 0.001. LDA: linear discriminant analysis.
3.3. Rk3 alters the gut microbiota metabolites in AOM/DSS-induced CRC mice
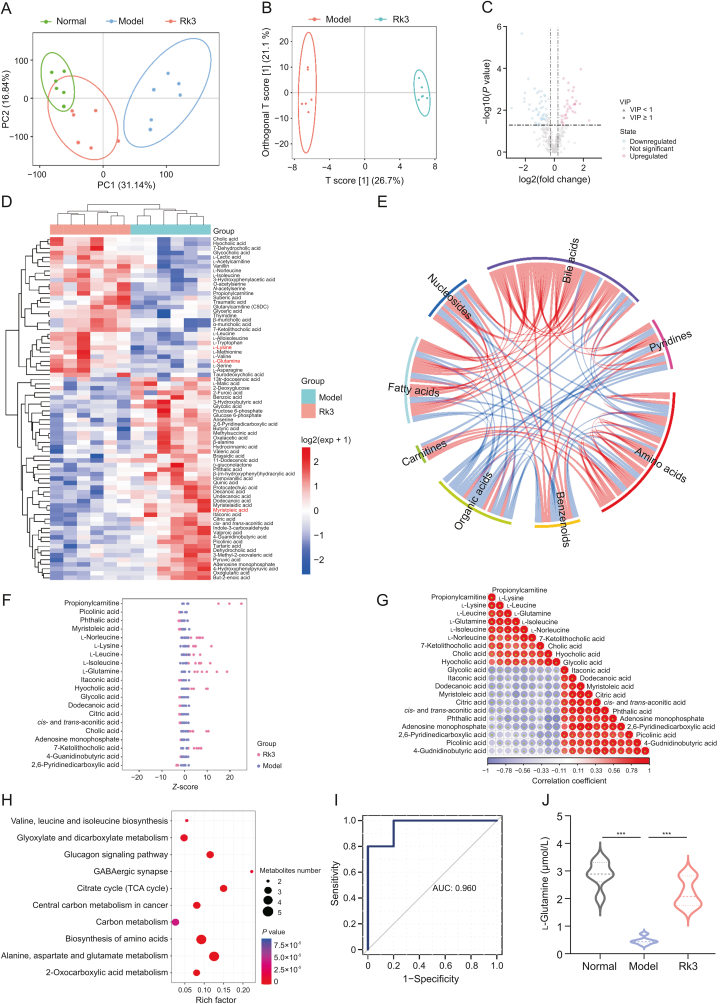
The metabolites produced by the gut microbiota act as an intermediary between the gut microbiota and the host, and they experience modifications in response to changes in the gut microbiota. Hence, to unveil the metabolic changes in different treatment groups of mice, we conducted high-throughput targeted metabolomic analysis on fecal samples using LC-MS/MS technology. Unsupervised principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) score plots demonstrated a distinct differentiation between the metabolite profiles of the three treatment groups (Figs. 3A and B). Compared to the model group, the Rk3 treatment induced alterations in 76 metabolites (Figs. 3C and D). Further categorization revealed that the most significant changes occurred in metabolites associated with amino acids, BAs, organic acids (Fig. 3E). After analyzing the Z-scores of differential metabolites, it was observed that the Rk3 group exhibited a predominance of beneficial metabolites, such as glutamine, leucine, lysine, and cholic acid in comparison to the model group. In contrast, the dominant metabolite cluster in the model group was characterized by the presence of the cytotoxic component myristoleic acid (Fig. 3F). By conducting detailed correlation analysis of differential metabolites, we observed a distinct separation trend between the enriched metabolites in the Rk3 group and the characteristic metabolites in the model group (Fig. 3G). Furthermore, compared to the model group mice, the upregulated metabolites in the Rk3-treated mice were enriched in multiple metabolic signaling pathways. It is noteworthy that alanine, aspartate and glutamate metabolism, and valine, leucine and isoleucine biosynthesis emerged as the major perturbed pathway in the Rk3-treated mice (Fig. 3H). Through receiver operating characteristic (ROC) curve analysis of differential metabolites, we identified glutamine as a crucial indicative metabolite for the Rk3 treatment group (Fig. 3I). Subsequently, we further analyzed the relative abundance of glutamine and found that Rk3 treatment significantly increased its levels (Fig. 3J). As the most abundant amino acid, glutamine is essential for maintaining intestinal mucosal cell metabolism, which plays key roles in protecting the gastrointestinal mucosa, enhancing intestinal cell activity, and improving intestinal immune function. These results indicate that the altered gut microbiota and its metabolites may synergistically contribute to the inhibition of colon tumor development.
Fig. 3.
Ginsenoside Rk3 (Rk3) modulates gut microbiota-associated metabolites. (A) Score plot of principal component analysis (PCA) analysis (n = 6). (B) Score plot of orthogonal partial least squares discriminant analysis (OPLS-DA) between model and Rk3 groups (n = 6). (C) Volcano plot of differential metabolites between model and Rk3 groups (n = 6). (D) Heatmap of hierarchical clustering analysis of differential metabolites expression levels between model and Rk3 groups (n = 6). (E) Correlation and chord diagram of metabolite-metabolite relationship pairs with spearman correlation of P < 0.05. (F) Z-score analysis plot of differential metabolites (n = 6). (G) Correlation analysis of each metabolite based on spearman correlation coefficients between differential metabolites. (H) Metabolic pathway enrichment analysis of differential metabolites based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (n = 6). (I) Receiver operating characteristic (ROC) curve analysis of the glutamine. (J) The quantitative analysis of the glutamine (n = 6). ∗∗∗P < 0.001. VIP: variable importance in projection; PC: principal component; TCA: tricarboxylic acid; AUC: area under the curve.
3.4. Rk3 restores the intestinal barrier function in AOM/DSS-induced CRC mice
The disruption of gut microbiota and its metabolites can lead to an imbalance in intestinal mucosal homeostasis, ultimately causing intestinal barrier dysfunction [26]. Tight junction proteins are critical markers of intestinal barrier integrity [27]. In order to assess the effects of Rk3 on colon barrier function in CRC mice, we first verified through immunofluorescence that the expression of E-cadherin, a cell adhesion molecule, and Claudin-1, a crucial constituent of tight junctions, was significantly decreased in the colon cells of model mice (Fig. S2). Further quantitative analysis through qRT-PCR (Figs. S3A and B) and WB (Figs. 4A−C) experiments revealed that Rk3 treatment significantly increased the expression of colon tight junction proteins. Serum lipopolysaccharide (LPS) levels were measured as indicators of colon cell permeability. Our results demonstrated that the serum LPS concentration was significantly elevated in the model group, yet markedly decreased in mice treated with Rk3 (Fig. S3C). Taken together, these results indicate that Rk3 has the potential to partially restore compromised colon barrier function by modulating the dysregulation of gut microbiota and metabolites.
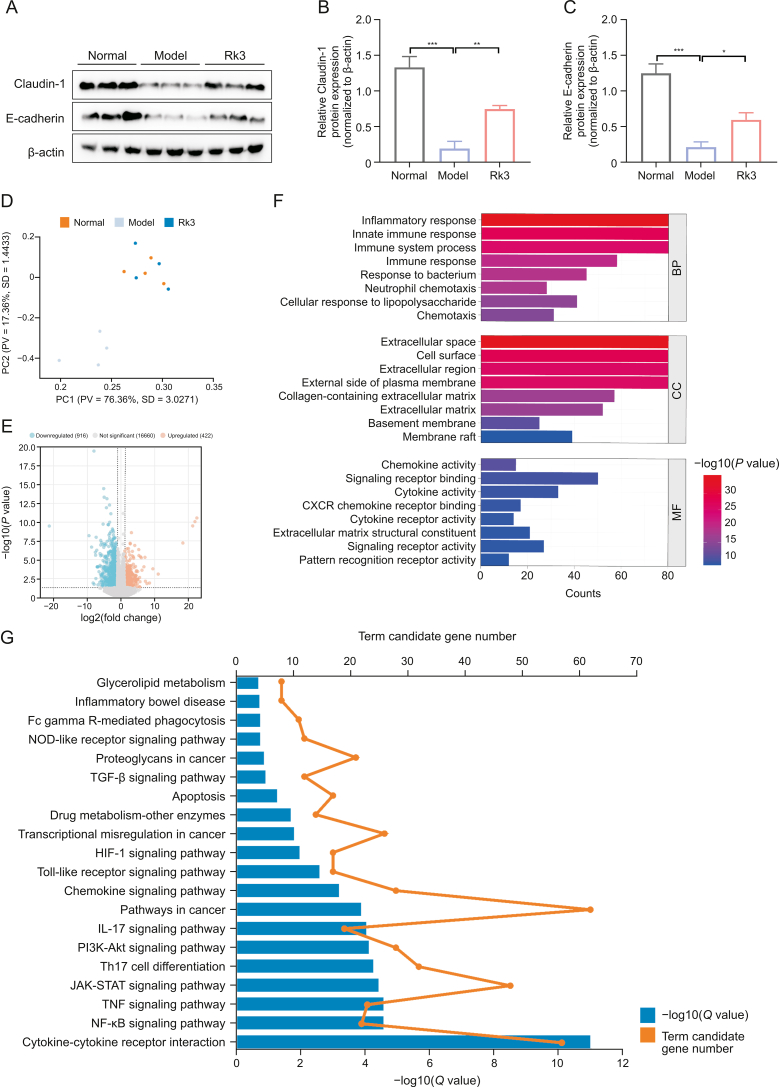
Fig. 4.
Ginsenoside Rk3 (Rk3) restores intestinal barrier function and regulates group 3 innate lymphoid cell (ILC3)-related immune response. (A–C) The expression levels of Claudin-1 and E-cadherin were detected by Western blotting (WB) (n = 3): Western blot analysis of Claudin-1 and E-cadherin (A) and quantitative analysis of Claudin-1 (B) and E-cadherin (C). (D) Principal component analysis (PCA) score plot of transcriptomics analysis (n = 4). (E) Volcano plot of differentially expressed genes between model and Rk3 groups (n = 4). (F) Gene Ontology (GO) enrichment analysis of differentially expressed genes between model and Rk3 groups (n = 4). (G) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of potential molecular signaling pathways and genes regulated by Rk3 in inhibiting colon tumor development (n = 4). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. PC: principal component; PV: proportion of variance; SD: standard deviation; CXCR: C−X−C-chemokine receptor; BP: biological process; CC: cellular component; MF: molecular function; NOD: nucleotide-binding oligomerization domain; TGF: transforming growth factor; HIF-1: hypoxia inducible factor-1; IL: interleukin; PI3K-AKT: phosphatidylinositol-3-kinase (PI3K)-protein kinase B (PKB); Th: T helper; JAK-STAT: Janus kinase-signal transducer and activator of transcription; TNF: tumor necrosis factor; NF-κB: nuclear factor-κB.
3.5. Rk3 modulates the ILC3-associated immune response in AOM/DSS-induced CRC mice
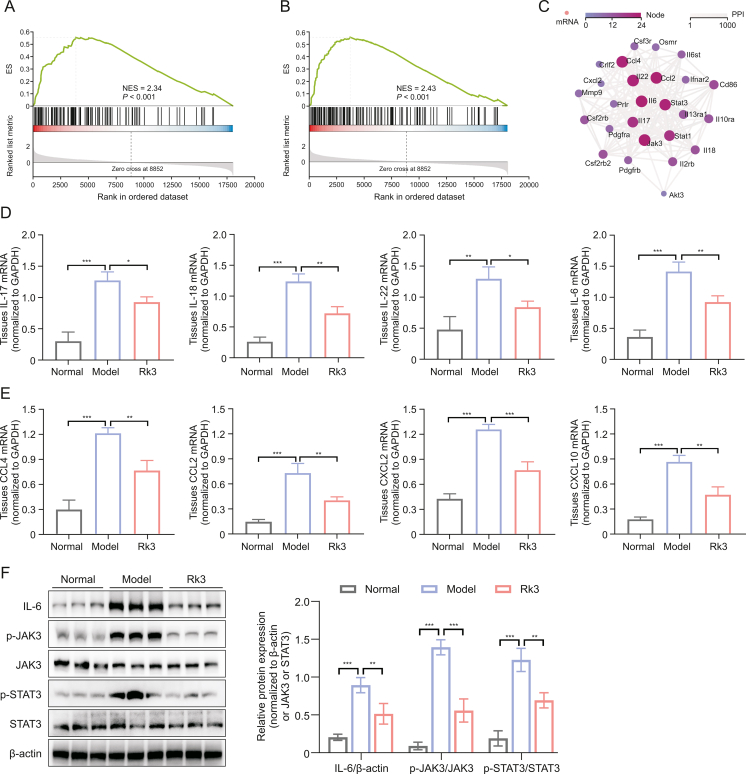
In order to comprehensively elucidate the anticancer effect of Rk3 at the molecular level, we performed transcriptome sequencing of colonic tissues from each group of mice. The results revealed differential expression of genes between the group supplemented with Rk3 and the model group (Figs. 4D and E). To gain insights into the potential biological functions regulated by these genes, further investigations were conducted using the Gene Ontology (GO) database. The findings demonstrated that these genes may be involved in inflammatory response, immune system processes, immune response, cellular response to lipopolysaccharide, and neutrophil chemotaxis (Fig. 4F). Similarly, KEGG enrichment analysis also revealed that Rk3 predominantly regulates genes and signaling pathways related to the immune system, including cytokine-cytokine receptor interaction, Th17 cell differentiation, and JAK-STAT signaling pathway (Fig. 4G). To further investigate the differentially expressed genes, we utilized key driver analysis to focus on key regulatory genes, and predicted their interactions using Cytoscape software. The results demonstrated that in the significantly enriched gene set of Rk3, there are strong interactions among genes associated with cell chemotaxis C–C motif chemokine ligand 2 (CCL2), CCL4, IL-17, IL-22, and cancer pathways (IL-6, JAK3, and STAT3) (Figs. 5A−C). Previous studies have also identified these genes as primarily involved in the ILC3 effector cytokine pathway [28].
Fig. 5.
Ginsenoside Rk3 (Rk3) regulates Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling pathway. (A) Gene set enrichment analysis (GSEA) plot showing enrichment in T helper 17 (Th17) cell differentiation. (B) GSEA plot showing enrichment in JAK-STAT signaling pathway. (C) Protein-protein interaction (PPI) network analysis of driver genes. (D) Real-time quantitative polymerase chain reaction (qRT-PCR) to detect the expression of inflammatory cytokines including interleukin 17 (IL-17), IL-18, IL-22, and IL-6 in colonic tissue from mice (n = 3). (E) qRT-PCR to detect the expression of chemokines including C–C motif chemokine ligand 2 (CCL2), CCL4, C–X–C chemokine ligand 2 (CXCL2), and CXCL10 in colonic tissue from mice (n = 3). (F) The expression levels of JAK-STAT signaling pathway-related proteins were detected by Western blotting (WB) (n = 3). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. ES: enrichment score; NES: normalized enrichment score; mRNA: messenger RNA; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; p-JAK3: phospho-janus kinase 3; p-STAT3: phospho-signal transducer and activator of transcription 3.
We employed qRT-PCR and WB experiments to perform quantitative analysis of differentially expressed genes. The results revealed that colonic tissues supplemented with Rk3 exhibited decreased expression of inflammatory cytokines, namely IL-22, IL-17, and IL-18, as well as downregulated expression of chemokines CCL4, CCL2, C–X–C chemokine ligand 2 (CXCL2), and CXCL10, when compared to the model group in mice (Figs. 5D and E). It is noteworthy that IL-17 and IL-22 are primarily produced by activated ILC3 immune cells in response to inflammation or infection. IL-22 interacts with its receptor via the JAK-STAT pathway, particularly through the activation of the STAT3 mediated signaling pathway in association with the microbiota [29]. Therefore, we further conducted quantitative analysis of proteins related to the STAT3-mediated signaling pathway, and the results demonstrated that treatment of Rk3 significantly inhibited the phosphorylation of JAK3 and STAT3 (Fig. 5F). In conclusion, we have preliminarily elucidated that the dysregulation of gut microbiota and metabolites caused by CRC leads to massive release of LPS, disrupting the balance between ILC3 and T cells, resulting in the excessive release of inflammatory cytokines and chemokines, as well as activation of the JAK-STAT signaling pathway. However, Rk3 treatment can modulate the dysregulated activation of the gut microbiota-mediated pro-carcinogenic signaling pathway, thus inhibiting the progression of CRC.
3.6. Rk3-modified gut microbiota regulates gut dysbiosis in AOM/DSS-induced CRC mice
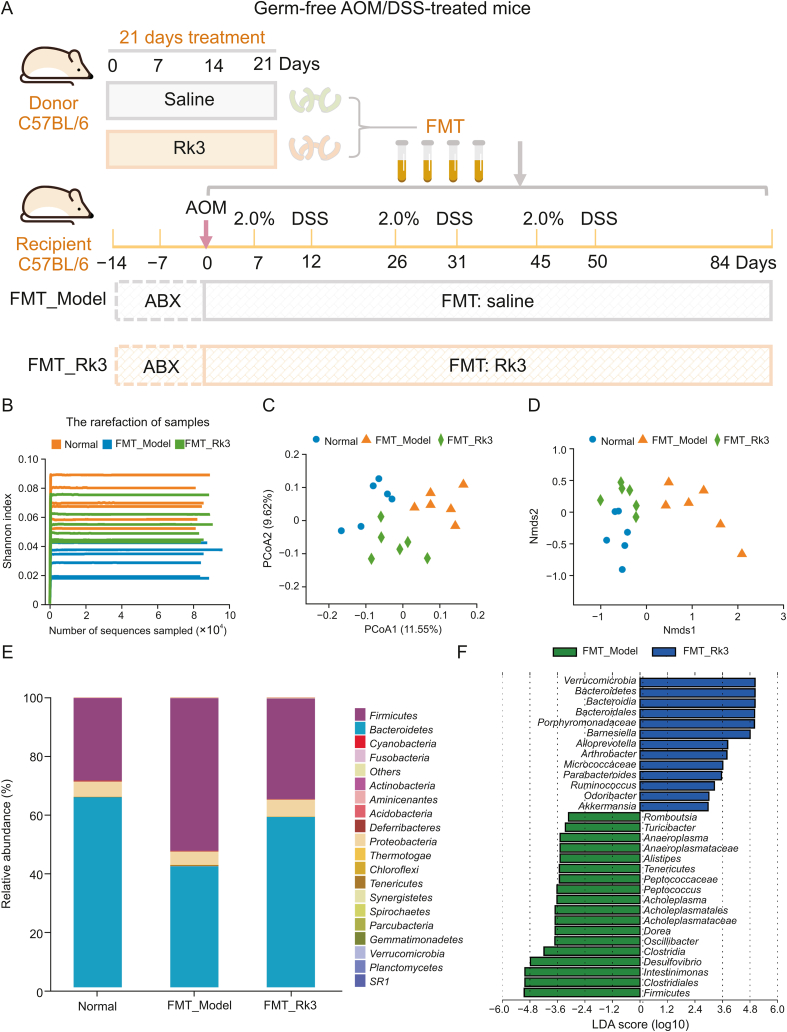
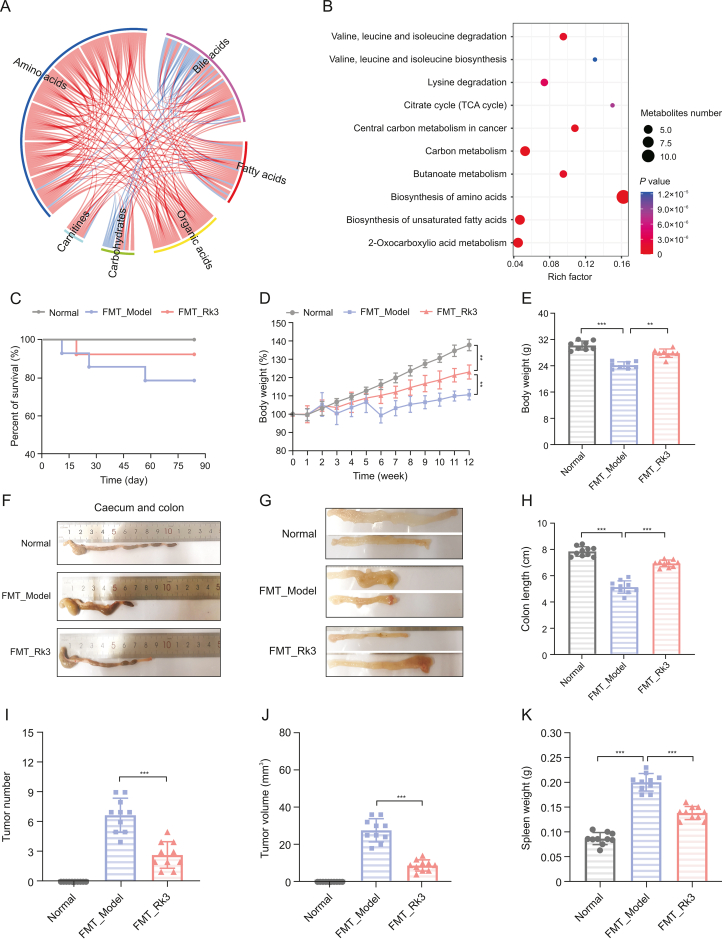
In order to ascertain whether the inhibitory impact of Rk3 on the progression of colorectal tumors is contingent upon the gut microbiota, we conducted an experiment on fecal microbial transplantation in germ-free mice. We administered Rk3-modified or conventional microbiota orally to germ-free mice and triggered a colon cancer model using AOM/DSS (Fig. 6A). Following the animal experiment, we collected fecal samples and conducted 16S rRNA gene sequencing analysis to assess the establishment of the Rk3-modified microbiota. The results were consistent with the outcomes of Rk3 oral administration, as the administration of Rk3-modified microbiota to mice also led to a marked increase in the alpha diversity of the gut microbiota (Fig. 6B). Moreover, beta diversity analysis confirmed significant differentiation of the gut microbiota between the different groups of mice (Figs. 6C and D). Furthermore, we conducted an analysis of the gut microbiota composition at the phylum level, which demonstrated a significant increase in the abundance of the Bacteroidetes phylum in the Rk3-modified gut microbiota (Fig. 6E). Additionally, the LEfSe analysis identified the dominant bacterial taxa in each group, revealing that Verrucomicrobia and Akkermansia still had the highest scores in the group that received Rk3 fecal microbial transplantation, while the pathogenic bacterium Desulfovibrio had the highest score in the model group mice (Fig. 6F). Furthermore, the metabolomics analysis revealed that the Rk3-modified gut microbiota significantly regulated amino acid and bile acid metabolites, as well as the synthesis pathway of amino acids (Figs. 7A and B). More importantly, we emphasize that the results regarding the gut microbiota and metabolites show similar outcomes between conventional Rk3 treatment and Rk3 fecal microbiota transplant therapy. In terms of gut microbiota, both FMT_Rk3 and conventional treatment can regulate the gut microbiota diversity of CRC mice (Figs. S4A and B). Moreover, they both significantly modulated the increase of Firmicutes and the decrease of Bacteroidetes caused by CRC (Figs. S4C and D), and significantly increased the abundance of beneficial genus Akkermansia and the key gut-protective bacteria Barnesiella (Figs. S4E and F), while significantly reducing the abundance of harmful bacteria Desulfovibrio, which can produce toxic substances, and the pro-inflammatory harmful bacteria Alistipes (Figs. S4G and H). In terms of the metabolomic data, it is evident that both FMT_Rk3 group and conventional treatment significantly regulate amino acid and bile acid metabolism (Figs. S5A and B). The KEGG enrichment analysis reveals that both interventions markedly modulate the biosynthesis of amino acids (Figs. S5C and D). Additionally, a detailed analysis of the relative abundance of differential metabolites indicates that both FMT_Rk3 group and conventional treatment significantly decrease the levels of harmful myristic acid while significantly increasing the levels of beneficial glutamine and lysine (Figs. S5E−G). These comparative findings underscore the striking similarity in therapeutic outcomes between FMT_Rk3 group and conventional treatment concerning gut microbiota and metabolomic profiles. These findings further affirm that Rk3-modified gut microbiota effectively colonized the intestines of recipient mice, thereby validating the reliability of FMT.
Fig. 6.
The gut microbiota modified by ginsenoside Rk3 (Rk3) were successfully colonized in mice. (A) Animal experimental design for fecal microbiota transplantation (FMT). (B) Analysis of gut microbiome alpha diversity using Shannon index (n = 6). (C) Analysis of gut microbiome beta diversity using principal co-ordinates analysis (PCoA) (n = 6). (D) Analysis of gut microbiome beta diversity using non-metric multidimensional scaling (NMDS) (n = 6). (E) Overview of gut microbial communities at the phylum level for each sample mice (n = 6). (F) The linear discriminant analysis effect size (LEfSe) analysis with linear discriminant analysis (LDA) was used to identify the bacterial communities with differential abundance between the different groups of mice (n = 6). AOM: azoxymethane; DSS: dextran sulfate sodium; ABX: antibiotic cocktail; PCoA: principal co-ordinates analysis; NMDS: non-metric multidimensional scaling.
Fig. 7.
Gut microbiota modified by ginsenoside Rk3 (Rk3) can inhibit colonic tumorigenesis. (A) A chord diagram showing the correlation between metabolite-metabolite relationship pairs with a spearman correlation of P < 0.05. (B) Metabolic pathway enrichment analysis of differential metabolites based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (n = 6). (C) Survival curve of the mice. (D) Rate of change in body weight of the mice during the experiment (n = 10). (E) Final body weight of the mice (n = 10). (F) Colon images of the mice. (G) Medial image of the mice colon. (H) Colon length of the mice (n = 10). (I) Number of colon tumors in mice (n = 10). (J) Volume of colon tumors in mice (n = 10). (K) Spleen weight of the mice (n = 10). ∗∗P < 0.01 and ∗∗∗P < 0.001. TCA: tricarboxylic acid; FMT: fecal microbiota transplantation.
3.7. Rk3-modified gut microbiota inhibits the development of colonic tumors in AOM/DSS-induced CRC mice
After confirming the successful colonization of gut microbiota, we evaluated the inhibitory effect of FMT on colon tumors. Our results showed that FMT_Model mice exhibited symptoms such as weight loss, bloody stool, and shortened colon, while FMT_Rk3 mice showed relief from these symptoms (Figs. 7C−H). Similarly, compared with FMT_Model mice, FMT_Rk3 mice had a decreased number and size of colon tumors (Figs. 7I and J). Additionally, we observed that the spleens of mice in the FMT_Model group showed enlargement. However, after administering FMT_Rk3 treatment, we observed a reduction in splenomegaly (Fig. 7K), indicating potential restoration of the immune system and suggesting the therapeutic effect of FMT_Rk3 on the CRC.
Histological analysis also showed that FMT_Model mice had abnormal crypt structure and abnormal infiltration of inflammatory cells, while supplementing with FMT_Rk3 significantly alleviated histopathological damage to the colon (Figs. S6A and B). FMT_Model mice showed a significant loss of goblet cells and mucus, with a significant increase in epithelial cell proliferation, resulting in a lower PAS index and a higher proportion of Ki-67 positive cells, while FMT_Rk3 improved these symptoms (Figs. S6C−F). These results are consistent with Rk3 conventional treatment group, thus confirming that FMT_Rk3 can inhibit colon cell proliferation and tumor development.
3.8. Rk3-modified gut microbiota restores the intestinal barrier function in AOM/DSS-induced CRC mice
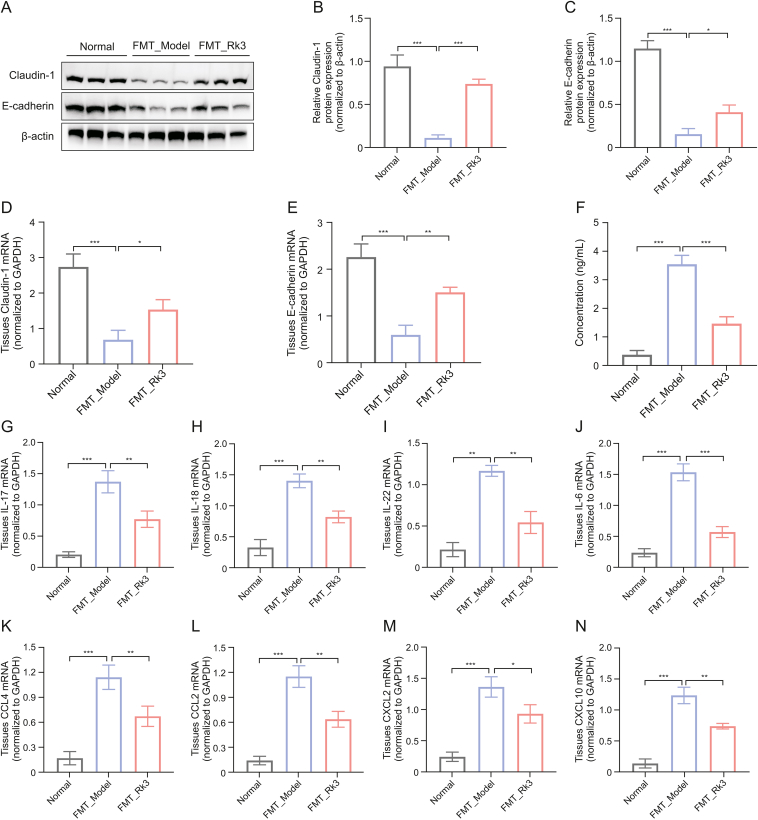
Subsequently, we investigated whether the gut microbiota modified by Rk3 could affect the integrity of the intestinal barrier in germ-free mice. Initially, we observed a lower expression of tight-junction proteins in the colon of FMT_Model mice by immunofluorescence (Fig. S7). However, quantitative analysis revealed significantly higher expression of E-cadherin and Claudin-1 in the FMT_Rk3 group compared to the FMT_Model group (Figs. 8A−E). Furthermore, in contrast to the FMT_Model group, the FMT_Rk3 group exhibited improved intestinal permeability, as evidenced by decreased serum LPS levels (Fig. 8F). Collectively, our findings provide further evidence that the FMT_Rk3 may directly contribute to the repair of impaired intestinal barrier function.
Fig. 8.
Gut microbiota modified by ginsenoside Rk3 (Rk3) can repair intestinal barrier function impairment and regulate group 3 innate lymphoid cell (ILC3)-related immune response. (A–C) The expression levels of Claudin-1 and E-cadherin were detected by Western blotting (WB) (n = 3): Western blot analysis of Claudin-1 and E-cadherin (A) and quantitative analysis of Claudin-1 (B) and E-cadherin (C). (D, E) The expression levels of Claudin-1 (D) and E-cadherin (E) were detected by real-time quantitative polymerase chain reaction (qRT-PCR). (F) Enzyme-linked immunosorbent assay (ELISA) to detect the concentration of lipopolysaccharide (LPS) in mouse serum (n = 5). (G–J) The expression levels of inflammatory factors including interleukin 17 (IL-17) (G), IL-18 (H), IL-22 (I), and IL-6 (J) were detected by qRT-PCR (n = 3). (K–N) The expression levels of chemokines including C–C motif chemokine ligand 4 (CCL4) (K), CCL2 (L), C−X−C chemokine ligand 2 (CXCL2) (M), and CXCL10 (N) were detected by qRT-PCR (n = 3). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. FMT: fecal microbiota transplantation; mRNA: messenger RNA; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
3.9. Rk3-modified gut microbiota regulates the ILC3-associated immune response in AOM/DSS-induced CRC mice
Next, we proceeded to investigate whether Rk3-induced changes in gut microbiota affect inflammatory responses or mucosal immunity. Compared to the FMT_Model mice, the FMT_Rk3 group showed significant downregulation of inflammatory factors IL-17, IL-18, IL-22, and IL-6 related to ILC3 and Th-17 immune regulation (Figs. 8G−J). Similarly, the results of qRT-PCR demonstrated significantly lower mRNA levels of ILC3-associated chemokines, including CCL4, CCL2, CXCL2, and CXCL10 in the FMT_Rk3 group compared to the FMT_Model group (Figs. 8K−N). Furthermore, our additional in vitro experiments have demonstrated that Rk3 can directly enhance the vitality of Akkermansia muciniphila and increase the concentration of glutamine in the Akkermansia muciniphila culture medium (Fig. S8). These results indicate that Rk3-modified gut microbiota can directly regulate ILC3 immune responses, thus confirming that the regulation of immune responses and signaling pathways by Rk3 is mediated by the gut microbiota, which further utilizes metabolites as intermediaries. Beneficial changes in the gut microbiota and metabolites are the main reason why Rk3 inhibits the development of colon tumors.
4. Discussion
CRC is one of the most prevalent digestive tract tumors. However, traditional treatment methods and emerging immunotherapy have proven to be ineffective in treating CRC. In recent years, there has been a growing awareness of the impact of gut microbiota on the occurrence, development, treatment, and prognosis of CRC [30]. Finding effective methods to prevent and treat CRC by regulating the gut microbiota has become a pressing issue to be addressed. In this study, we used genomics, metabolomics, and transcriptomics to elucidate the mechanism by which Rk3 ameliorates gut microbiota imbalance and inhibits the occurrence of CRC at multiple levels. Moreover, the role of the gut microbiota in the antitumor activity of Rk3 against CRC was verified using a mouse model of fecal bacterial transplantation. These findings provide a theoretical basis and offer novel insights into the use of natural compounds to modulate the gut microbiota for the prevention and treatment of CRC.
To evaluate the inhibitory effect of Rk3 on colon tumors, we established an AOM/DSS-induced CRC mouse model and treated it with Rk3. We confirmed that supplementation with Rk3 reduced the number and size of intestinal tumors in mice, alleviated colon shortening and pathological damage, and inhibited the occurrence of colorectal tumors. These findings demonstrate the antitumor characteristics of Rk3 in colorectal tumors. Based on the crucial role of gut microbiota in CRC development, we investigated the effect of Rk3 on the gut microbiota of CRC mice. Our data demonstrate that treatment with Rk3 can modulate the composition and diversity of the gut microbiota, particularly by upregulating the abundance of beneficial bacteria, such as Akkermansia muciniphila and Barnesiella intestinihominis, thus modulating dysbiosis of the gut microbiota and inhibiting colon tumorigenesis. Previous studies confirmed that Akkermansia_muciniphila promotes the accumulation of M1-like macrophages in vivo and in vitro in a nucleotide-binding oligomerization domain-like receptor thermal protein domain associated protein 3 (NLRP3)-dependent manner, thereby inhibiting colorectal tumorigenesis [31,32]. Similarly, Barnesiella intestinihominis can activate T cells in the spleen, stimulating an anti-cancer immune response and inhibiting colon tumor growth [33]. On the other hand, we observed a significant increase of Desulfovibrio, Oscillibacter, and Alistipes in the model group of mice. Desulfovibrio have been established as exhibiting specificity towards early-stage CRC. Desulfovibrio longreachensis significantly increases in patients with Stage III/IV CRC. Oscillibacter is a typical inflammation-related bacterium, whereas Alistipes is positively associated with colon tumor burden, and specific strains of Alistipes can facilitate the proliferation of CRC cells. Alistipes finegoldii can stimulate CRC formation by activating the STAT3 pathway [[34], [35], [36]]. These data suggest that Oscillibacter and Alistipes are potential pathogens in the development of colon tumors, and supplementation with Rk3 can eliminate these potential pathogenic bacteria. In summary, the reduction of pathogenic bacteria and increase of protective bacteria at least to some extent contribute to the inhibitory effect of Rk3 on the proliferation of colorectal tumors.
Furthermore, gut microbiota not only has a direct impact on CRC, but also produces a large number of metabolites such as short-chain fatty acids, indole, and BAs, which further affect the progression of CRC and the immune system [37]. Therefore, we explored the effects of Rk3 on mouse gut microbiota metabolites using metabolomic analyses. We found that Rk3 treatment significantly modulated amino acid and bile acid metabolite levels. Notably, the glutamine, leucine, and cholic acid levels were significantly enriched. Notably, glutamine, the most abundant amino acid, is absent in the microenvironment of patients with CRC, and glutamine supplementation can slow the growth of melanoma tumors by inhibiting the oncogenic pathway activated by H3K4me3 methylation [38]. In addition, cholic acid has the ability to decrease capillary permeability in inflamed tissues, attenuate inflammatory swelling, and inhibit the production of inflammatory mediators [39,40]. Moreover, the association analysis between gut microbiota and metabolomics further demonstrated the synergistic effect of beneficial bacteria and metabolites. Therefore, our results further emphasize that Rk3 may exert anti-cancer effects by modulating the gut microbiota and their metabolites.
Given that the intestinal mucosal barrier represents the host's foremost defense against harmful pathogenic bacteria and their metabolites, we proceeded to evaluate the effect of Rk3 treatment on the integrity of the mouse intestinal barrier [41]. Our findings indicated that the intestinal barrier function was compromised in CRC mice, with significant reductions in the expression of two pivotal tight junction proteins, namely Claudin-1 and E-cadherin, in the mouse model, coupled with a notable elevation in the toxic metabolite LPS, which is utilized to assess intestinal permeability. The disruption of intestinal tight junctions may heighten colonic permeability, thus facilitating the entry of more harmful bacteria and metabolites into the intercellular crevices of colonic epithelial cells, ultimately expediting disease progression [42]. Nonetheless, the administration of Rk3 has the potential to reverse the damage to the barrier function.
Moreover, in addition to the influence of gut microbiota and its metabolites on CRC, the interaction between gut microbiota and the immune system cannot be ignored. Therefore, we further investigated the changes in the immune system and the potential molecular mechanisms of Rk3 in inhibiting colon tumors. Through transcriptomics analysis, we found that the regulated genes involved changes in many genes related to immune cell trafficking, cytokine-cytokine receptor interaction, chemokines, Th17 cell differentiation, and JAK-STAT signaling pathways. Chemokines represent a distinct subfamily of cytokines responsible for orchestrating immune cell trafficking and contributing to the development of lymphoid tissues [43]. ILC3s expressing CCL4 can promote T cell recruitment to cold tumors and enhance responsiveness to immune checkpoint inhibitor [44]. Surprisingly, our results found that Rk3 supplementation upregulated the expression of multiple chemokines and cytokines related to ILC3 immune cells, including (IL-22, IL-17, CCL2, and CCL4). ILCs are a family of tissue-resident innate lymphoid cells that play a critical role in regulating host-microbe interactions at the surface of mucosal barriers, with ILC3s being the main players [45]. ILC3s are enriched in the intestine and can produce cytokines such as IL-17 and IL-22 to regulate interactions with the microbiota [46]. Additionally, ILC3s can also regulate adaptive immune responses by directly interacting with T cells [47,48]. Recent research has reported a sharp decrease in the number of ILC3 cells and a loss of homeostasis in the interaction between ILC3 and T cells in the tumor microenvironment of CRC patients. This directly exacerbates the malignant progression of colon cancer and leads to resistance to immune therapy. ILC3 is the main producer of IL-22 in the body, which can affect the function of epithelial cells by activating downstream STAT3 signaling [49]. Our results are consistent with these findings. Supplementation with Rk3 significantly reduced the expression of the inflammatory factors IL-17 and IL-22 and inhibited the activation of STAT3 signaling. Reportedly, ILC3 can limit the function of Th17 cells through the major histocompatibility complex-II (MHC-II) and the interaction between MHC-II+ ILC3 and CD4+ T cells is necessary to maintain intestinal immune homeostasis [50]. It is found that the colon of mice with ILC3-specific MHC-II deficiency had a significant reduction in Th1 and CD8 T cells and a significant increase in colon dysplasia score [51,52]. Furthermore, we demonstrate that the dysbiosis of intestinal microbiota and metabolites induced by CRC leads to a significant increase in LPS, disrupting the balance between ILC3 and T cells, promoting the release of pro-inflammatory cytokines and chemokines, and activating the JAK-STAT3 signaling pathway. In contrast, treatment with Rk3 can regulate the gut microbiota to reverse the activation of pro-cancer signaling pathways, thereby inhibiting the progression of CRC.
More importantly, to verify whether the affected gut microbiota and metabolites were the cause or result of CRC alleviation, we constructed a germ-free mouse model using an antibiotic cocktail therapy and transplanted Rk3-modified gut microbiota into germ-free mice. Germ-free mice that received Rk3-modified fecal microbiota exhibited a reduction in tumor numbers, alleviation of colon tissue pathological damage, and reduced cell proliferation, indicating a direct role for Rk3-modified gut microbiota in suppressing colon tumor occurrence. Furthermore, the successful colonization of gut microbiota was confirmed by the similarity in gut microbiota composition between Rk3 mice and FMT_Rk3 mice, with a significant enrichment of Akkermansia muciniphila in both groups. Additionally, Rk3-modified gut microbiota increased the level of glutamine. Notably, FMT_Rk3 was also found to significantly regulate ILC3 immune-related chemokines and inflammatory factors, further elucidating that Rk3-modified gut microbiota can upregulate the abundance of beneficial bacteria Akkermansia muciniphila, as well as the level of beneficial metabolites, further regulating the balance between ILC3 and T cells, inhibiting the activation of pro-inflammatory signals in colon epithelial cells, and thus suppressing the progression of colon tumors. These results confirm that the regulation of the immune responses and signaling pathways by Rk3 is mediated by the gut microbiota and particularly emphasize that beneficial changes in the gut microbiota are the main reason for the inhibitory effect of Rk3 against CRC tumorigenesis.
5. Conclusion
In conclusion, our research highlights the anticancer effects of Rk3 through the regulation of the gut microbiota. Rk3 inhibited the development of colorectal tumors by enriching beneficial bacteria and metabolites, regulating the ILC3 immune response, and suppressing the JAK-STAT3 signaling pathway. Our study elucidates the mechanism by which Rk3 regulates the gut microbiota to inhibit the occurrence and development of CRC. In summary, these findings emphasize that Rk3 can be utilized as a regulator of the gut microbiota for the prevention and treatment of CRC.
CRediT author statement
Xue Bai: Investigation, Methodology, Validation, Project administration, Formal analysis, Data curation, Writing - Original draft preparation, Visualization; Rongzhan Fu: Validation, Methodology, Supervision, Project administration; Writing - Reviewing and Editing, Visualization, Formal analysis, Funding acquisition; Yannan Liu: Methodology, Formal analysis, Supervision, Visualization; Jianjun Deng: Data curation, Investigation, Funding acquisition; Qiang Fei: Investigation, Project administration, Supervision; Zhiguang Duan: Methodology, Formal analysis, Supervision; Chenhui Zhu: Resources, Project administration, Supervision, Writing - Reviewing and Editing, Funding acquisition; Daidi Fan: Resources, Project administration, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the National Key Research and Development Program, China (Grant Nos.: 2021YFC2101500 and 2021YFC2103900), the National Natural Science Foundation of China (Grant Nos.: 22278335 and 21978236), and the Natural Science Basic Research Program of Shaanxi, China (Grant No.: 2023-JC-JQ-17).
Footnotes
Peer review under responsibility of Xi’an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2023.09.010.
Contributor Information
Chenhui Zhu, Email: zch2005@nwu.edu.cn.
Daidi Fan, Email: fandaidi@nwu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Siegel R.L., Miller K.D., Wagle N.S., et al. Cancer statistics, 2023, CA Cancer. J. Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Pan C., Liu H., Robins E., et al. Next-generation immuno-oncology agents: Current momentum shifts in cancer immunotherapy. J. Hematol. Oncol. 2020;13 doi: 10.1186/s13045-020-00862-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weng J., Li S., Zhu Z., et al. Exploring immunotherapy in colorectal cancer. J. Hematol. Oncol. 2022;15 doi: 10.1186/s13045-022-01294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kordahi M.C., Stanaway I.B., Avril M., et al. Genomic and functional characterization of a mucosal symbiont involved in early-stage colorectal cancer. Cell Host Microbe. 2021;29:1589–1598.e6. doi: 10.1016/j.chom.2021.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clay S.L., Fonseca-Pereira D., Garrett W.S. Colorectal cancer: The facts in the case of the microbiota. J. Clin. Invest. 2022;132 doi: 10.1172/JCI155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao R., Wu C., Zhu Y., et al. Integrated analysis of colorectal cancer reveals cross-cohort gut microbial signatures and associated serum metabolites. Gastroenterology. 2022;163:1024–1037.e9. doi: 10.1053/j.gastro.2022.06.069. [DOI] [PubMed] [Google Scholar]
- 7.Karpiński T.M., Ożarowski M., Stasiewicz M. Carcinogenic microbiota and its role in colorectal cancer development. Semin. Cancer Biol. 2022;86:420–430. doi: 10.1016/j.semcancer.2022.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Cremonesi E., Governa V., Garzon J.F.G., et al. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut. 2018;67:1984–1994. doi: 10.1136/gutjnl-2016-313498. [DOI] [PubMed] [Google Scholar]
- 9.Goc J., Lv M., Bessman N.J., et al. Dysregulation of ILC3s unleashes progression and immunotherapy resistance in colon cancer. Cell. 2021;184:5015–5030.e16. doi: 10.1016/j.cell.2021.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Y., Wang Z., Yan Y., et al. Enterotoxigenic Bacteroides fragilis promotes intestinal inflammation and malignancy by inhibiting exosome-packaged miR-149-3p. Gastroenterology. 2021;161:1552–1566.e12. doi: 10.1053/j.gastro.2021.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Dalal N., Jalandra R., Bayal N., et al. Gut microbiota-derived metabolites in CRC progression and causation. J. Cancer Res. Clin. Oncol. 2021;147:3141–3155. doi: 10.1007/s00432-021-03729-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jalandra R., Dalal N., Yadav A.K., et al. Emerging role of trimethylamine-N-oxide (TMAO) in colorectal cancer. Appl. Microbiol. Biotechnol. 2021;105:7651–7660. doi: 10.1007/s00253-021-11582-7. [DOI] [PubMed] [Google Scholar]
- 13.Sinha S.R., Haileselassie Y., Nguyen L.P., et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe. 2020;27:659–670.e5. doi: 10.1016/j.chom.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson T., Caporaso J.G., Yellowhair M., et al. Effects of ursodeoxycholic acid on the gut microbiome and colorectal adenoma development. Cancer Med. 2019;8:617–628. doi: 10.1002/cam4.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang B., Xu Y., Lv H., et al. Intestinal pharmacokinetics of resveratrol and regulatory effects of resveratrol metabolites on gut barrier and gut microbiota. Food Chem. 2021;357 doi: 10.1016/j.foodchem.2021.129532. [DOI] [PubMed] [Google Scholar]
- 16.Chen Z., Zhang Z., Liu J., et al. Gut microbiota: Therapeutic targets of ginseng against multiple disorders and ginsenoside transformation. Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.853981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chrysostomou D., Roberts L.A., Marchesi J.R., et al. Gut microbiota modulation of efficacy and toxicity of cancer chemotherapy and immunotherapy. Gastroenterology. 2023;164:198–213. doi: 10.1053/j.gastro.2022.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Chen H., Ye C., Wu C., et al. Berberine inhibits high fat diet-associated colorectal cancer through modulation of the gut microbiota-mediated lysophosphatidylcholine. Int. J. Biol. Sci. 2023;19:2097–2113. doi: 10.7150/ijbs.81824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong S., Zhu M., Wang K., et al. Dihydromyricetin improves DSS-induced colitis in mice via modulation of fecal-bacteria-related bile acid metabolism. Pharmacol. Res. 2021;171 doi: 10.1016/j.phrs.2021.105767. [DOI] [PubMed] [Google Scholar]
- 20.Messaoudene M., Pidgeon R., Richard C., et al. A natural polyphenol exerts antitumor activity and circumvents anti-PD-1 resistance through effects on the gut microbiota. Cancer Discov. 2022;12:1070–1087. doi: 10.1158/2159-8290.CD-21-0808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chopra P., Chhillar H., Kim Y.J., et al. Phytochemistry of ginsenosides: Recent advancements and emerging roles. Crit. Rev. Food Sci. Nutr. 2023;63:613–640. doi: 10.1080/10408398.2021.1952159. [DOI] [PubMed] [Google Scholar]
- 22.Hou M., Wang R., Zhao S., et al. Ginsenosides in Panax genus and their biosynthesis. Acta Pharm. Sin. B. 2021;11:1813–1834. doi: 10.1016/j.apsb.2020.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qu L., Liu Y., Deng J., et al. Ginsenoside Rk3 is a novel PI3K/AKT-targeting therapeutics agent that regulates autophagy and apoptosis in hepatocellular carcinoma. J. Pharm. Anal. 2023;13:463–482. doi: 10.1016/j.jpha.2023.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai X., Fu R., Duan Z., et al. Ginsenoside Rk3 alleviates gut microbiota dysbiosis and colonic inflammation in antibiotic-treated mice. Food Res. Int. 2021;146 doi: 10.1016/j.foodres.2021.110465. [DOI] [PubMed] [Google Scholar]
- 25.Chen H., Yang H., Deng J., et al. Ginsenoside Rk3 ameliorates obesity-induced colitis by regulating of intestinal flora and the TLR4/NF-κB signaling pathway in C57BL/6 mice. J. Agric. Food Chem. 2021;69:3082–3093. doi: 10.1021/acs.jafc.0c07805. [DOI] [PubMed] [Google Scholar]
- 26.Rogers A.P., Mileto S.J., Lyras D. Impact of enteric bacterial infections at and beyond the epithelial barrier. Nat. Rev. Microbiol. 2023;21:260–274. doi: 10.1038/s41579-022-00794-x. [DOI] [PubMed] [Google Scholar]
- 27.Odenwald M.A., Turner J.R. The intestinal epithelial barrier: A therapeutic target? Nat. Rev. Gastroenterol. Hepatol. 2017;14:9–21. doi: 10.1038/nrgastro.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonçalves P., Di Santo J.P. An intestinal inflammasome – the ILC3-cytokine tango. Trends Mol. Med. 2016;22:269–271. doi: 10.1016/j.molmed.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Chun E., Lavoie S., Fonseca-Pereira D., et al. Metabolite-sensing receptor Ffar2 regulates colonic group 3 innate lymphoid cells and gut immunity. Immunity. 2019;51:871–884.e6. doi: 10.1016/j.immuni.2019.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saeed M., Shoaib A., Kandimalla R., et al. Microbe-based therapies for colorectal cancer: Advantages and limitations. Semin. Cancer Biol. 2022;86:652–665. doi: 10.1016/j.semcancer.2023.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Fan L., Xu C., Ge Q., et al. A. muciniphila suppresses colorectal tumorigenesis by inducing TLR2/NLRP3-mediated M1-like TAMs. Cancer Immunol. Res. 2021;9:1111–1124. doi: 10.1158/2326-6066.CIR-20-1019. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y., Xu Y., Zheng C., et al. Acetyltransferase from Akkermansia muciniphila blunts colorectal tumourigenesis by reprogramming tumour microenvironment. Gut. 2023;72:1308–1318. doi: 10.1136/gutjnl-2022-327853. [DOI] [PubMed] [Google Scholar]
- 33.Daillère R., Vètizou M., Waldschmitt N., et al. Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity. 2016;45:931–943. doi: 10.1016/j.immuni.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 34.Liu X., Tong X., Zou Y., et al. Mendelian randomization analyses support causal relationships between blood metabolites and the gut microbiome. Nat. Genet. 2022;54:52–61. doi: 10.1038/s41588-021-00968-y. [DOI] [PubMed] [Google Scholar]
- 35.Moschen A.R., Gerner R.R., Wang J., et al. Lipocalin 2 protects from inflammation and tumorigenesis associated with gut microbiota alterations. Cell Host Microbe. 2016;19:455–469. doi: 10.1016/j.chom.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Yang J., Wei H., Zhou Y., et al. High-fat diet promotes colorectal tumorigenesis through modulating gut microbiota and metabolites. Gastroenterology. 2022;162:135–149.e2. doi: 10.1053/j.gastro.2021.08.041. [DOI] [PubMed] [Google Scholar]
- 37.Krautkramer K.A., Fan J., Bäckhed F. Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Microbiol. 2021;19:77–94. doi: 10.1038/s41579-020-0438-4. [DOI] [PubMed] [Google Scholar]
- 38.Ishak Gabra M.B., Yang Y., Li H., et al. Dietary glutamine supplementation suppresses epigenetically-activated oncogenic pathways to inhibit melanoma tumour growth. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-17181-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai J., Sun L., Gonzalez F.J. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe. 2022;30:289–300. doi: 10.1016/j.chom.2022.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu J., Xie S., Chi S., et al. Protective effects of taurocholic acid on excessive hepatic lipid accumulation via regulation of bile acid metabolism in grouper. Food Funct. 2022;13:3050–3062. doi: 10.1039/d1fo04085e. [DOI] [PubMed] [Google Scholar]
- 41.Tytgat H.L.P., Nobrega F.L., van der Oost J., et al. Bowel biofilms: Tipping points between a healthy and compromised gut? Trends Microbiol. 2019;27:17–25. doi: 10.1016/j.tim.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 42.Candelli M., Franza L., Pignataro G., et al. Interaction between lipopolysaccharide and gut microbiota in inflammatory bowel diseases. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22126242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vilgelm A.E., Richmond A. Chemokines modulate immune surveillance in tumorigenesis, metastasis, and response to immunotherapy. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruchard M., Geindreau M., Perrichet A., et al. Recruitment and activation of type 3 innate lymphoid cells promote antitumor immune responses. Nat. Immunol. 2022;23:262–274. doi: 10.1038/s41590-021-01120-y. [DOI] [PubMed] [Google Scholar]
- 45.Innate lymphoid cells influence gut microbiota and colorectal cancer. Cancer Discov. 2021;11 doi: 10.1158/2159-8290.CD-RW2021-122. [DOI] [PubMed] [Google Scholar]
- 46.Sano T., Huang W., Hall J.A., et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell. 2015;163:381–393. doi: 10.1016/j.cell.2015.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen L., Ye Y., Sun H., et al. ILC3 plasticity in microbiome-mediated tumor progression and immunotherapy. Cancer Cell. 2021;39:1308–1310. doi: 10.1016/j.ccell.2021.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Serafini N., Jarade A., Surace L., et al. Trained ILC3 responses promote intestinal defense. Science. 2022;375:859–863. doi: 10.1126/science.aaz8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng X., Chen T., Jiang R., et al. Hyocholic acid species improve glucose homeostasis through a distinct TGR5 and FXR signaling mechanism. Cell Metab. 2021;33:791–803.e7. doi: 10.1016/j.cmet.2020.11.017. [DOI] [PubMed] [Google Scholar]
- 50.Castellanos J.G., Longman R.S. Innate lymphoid cells link gut microbes with mucosal T cell immunity. Gut Microbes. 2020;11:231–236. doi: 10.1080/19490976.2019.1638725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hepworth M.R., Monticelli L.A., Fung T.C., et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hepworth M.R., Fung T.C., Masur S.H., et al. Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4+ T cells. Science. 2015;348:1031–1035. doi: 10.1126/science.aaa4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.