ABSTRACT
Clinical genetics is increasingly recognized as an important area within nephrology care. Clinicians require awareness of genetic kidney disease to recognize clinical phenotypes, consider use of genomics to aid diagnosis, and inform treatment decisions. Understanding the broad spectrum of clinical phenotypes and principles of genomic sequencing is becoming increasingly required in clinical nephrology, with nephrologists requiring education and support to achieve meaningful patient outcomes. Establishment of effective clinical resources, multi-disciplinary teams and education is important to increase application of genomics in clinical care, for the benefit of patients and their families. Novel applications of genomics in chronic kidney disease include pharmacogenomics and clinical translation of polygenic risk scores. This review explores established and emerging impacts and utility of genomics in kidney disease.
Keywords: clinical utility, genetic kidney disease, genetic testing, genomics
INTRODUCTION
Integrating genomics into clinical practice enhances clinical diagnosis, identifies potential treatment strategies, and improves awareness of genetic kidney disease (GKD) to progress patient outcomes. Combining clinical nephrology with advances in genomics to improve diagnostics in chronic kidney disease (CKD) has direct impacts on individuals and their families. GKD is variably but substantially prevalent across adult and paediatric populations, with significant clinical heterogeneity across disease phenotypes (Fig. 1) [1–3]. The ability to accurately diagnose affected patients informs prognosis, treatment decisions, reproductive options, and screening of at-risk family members, highlighting the ongoing importance of integrating genomics into everyday clinical care [4]. Advances in genomics have led to improvements in clinical diagnosis that are key to advancing nephrological care. Integrating genomics into clinical nephrology has health economic benefits through improving diagnostic work-up, avoidance of kidney biopsy, rationalizing treatment decisions, and informing family screening [4–6]. The complex interplay of genetic and environmental factors is exemplified in kidney disease, an area where understanding polygenic traits and pharmacogenomics can inform personalized medicine. In synthesizing recent literature (Methods S1, see online supplementary material), this review discusses the impact and utility of genomics in genetic kidney disease, challenges in integrating genomics into clinical medicine, and areas for future research.
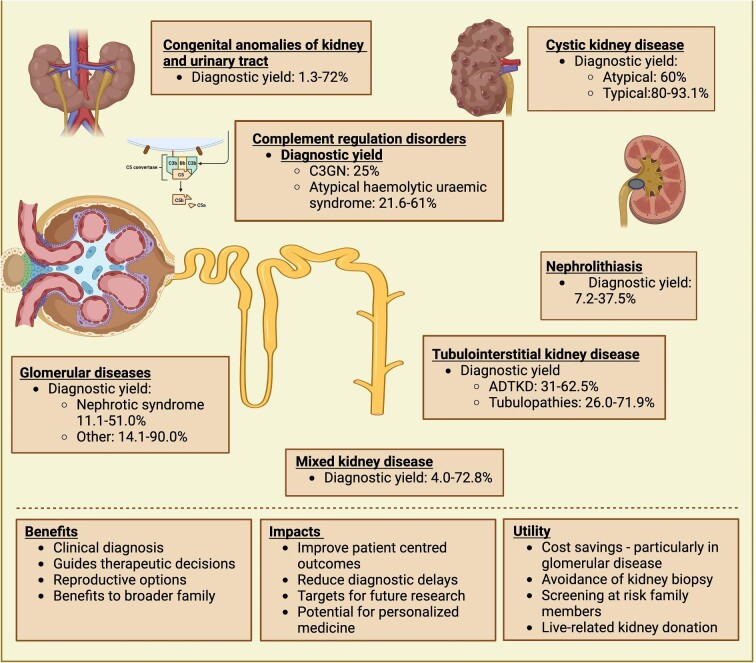
Figure 1:
Clinical applications of genetic testing strategies. The diagnostic yield of genetic diagnosis depends on the clinical phenotype of kidney disease. The benefits of diagnostic testing have direct patient impacts and broader associations. Image made with BioRender.
Genetic kidney disease
Advances in genomics for genetic kidney disease have become an integral component of nephrology with direct patient impacts. GKD are well-described, and understanding their genetic basis has broad clinical applications with critical significance in clinical care and research. Integration of clinical phenotyping and genetic counselling with advances in genomics is high value with direct implications for patients, and requires upskilling of nephrologists to identify suspected cases of GKD [7, 8]. Advances in genomic sequencing have enabled identification of over 400 clinically relevant kidney genes, with ten key genes accounting for nearly 50% of GKD [3, 4]. A paediatric cohort of 188 children with kidney failure identified a genetic cause of kidney disease in 39.9%, impacting management decisions in 34.7% of cases and enabling sequential family testing in 34.7% of families [1]. In another paediatric cohort with kidney disease, whole-exome sequencing (WES) was performed in those with a family history, strong clinical suspicion of genetic kidney disease, or where genetic result would impact management, establishing a genetic diagnosis in 37.1% of patients [2]. The prevalence of GKD in the adult population is markedly more varied depending on the characteristics of the population studied, and certain phenotypes such as ciliopathies, cystic kidney disease (47.7–93.1%), and glomerulopathy (11.1–90.0%) have higher diagnostic yields than congenital anomalies of the kidney and urinary tract (CAKUT; Fig. 1, Table 1) [3, 9].
Table 1:
Overview of diagnostic yield in different cohorts and populations with genetic kidney disease
| Author | Year | PMID | Cohort (n) | Family history | Testing | Panel | Diagnostic yield | Comments | |
|---|---|---|---|---|---|---|---|---|---|
| Groopman [24] | 2019 | 30 586 318 | AURORA (1128) CUMC (2187) |
50–80 years with end stage kidney failure. Adults with CKD 46.7% on dialysis. |
28.7% positive family history | WES | 625 nephropathy genes + 59 medically actionable genes in CUMC cohort | 9.3% diagnostic variants detected | Large population with high incidence of kidney failure, low family history |
| Tanudisastro [79] | 2021 | 33 664 247 | Adult and children referred for genetic testing from 2013–2018 | 552 271 adult 281 paediatric |
Not reported | 2013–2018 TruSight one panel 2018–19 WES |
Disease-specific tests/panels | 35% 32% 38% |
Did not include patients referred for microarray of pKD1/2 genetic testing |
| Elhassan [20] | 2021 | 35 099 770 |
Irish Kidney gene project | 698 adults | 73.9% | Total Panel (416) WES (193) WGS (25) MUC1/UMOD (43) Sanger (49 ADPKD) |
Phenotype directed testing | 51.4% 34% 56% 70% |
Included ADPKD patients, limited to Irish population |
| Jayasinghe [97] | 2021 | 32 939 031 | Patients with suspected GKD | 204 81 paediatric 123 adult |
57.4% | WES (1 microarray) |
Disease specific panels | 39% 47% 34% |
5% consanguinity |
| Connaughton [23] | 2019 | 30 773 290 | Irish patients 2014 to 2017 with suspected GKD | 114 | 73% | WES | Phenotype specific panel then 478 known kidney gene | 39% | Included PKD, predominantly Irish ancestry, high family history |
| Lata [131] | 2018 | 29 204 651 | Adults with CKD unknown OR hypertension from CUMC + 11 from other local institutions | 344 screened with 81 patients with clinical concern for gkd | 58% | WES | 287 Nephropathy genes | 23% | Excluded ADPKD |
| Bullich [29] | 2018 | 29 801 666 | Suspected inherited CKD Cystic Glomerular |
207 98 |
44% 81% |
NGS | 140 kidney gene panel | 78% 62% |
Targeted testing in high clinical likelihood |
| Domingo-Gallego [30] | 2022 | 33 532 864 | Suspected genetic CKD < 30 years | 460 | 49% | NGS + target c.428dupC MUC1 |
316 custom kidney panel | 65% | Included cystic kidney disease |
| Chen [1] | 2021 | 34 031 707 | Kidney failure before 19 years old | 188 | 18.1% | NGS | 2703 targeted genes, proband-WES, TRIO-WES, confirmati on of copy number variants | 39.9% | Genetic diagnosis reclassified diagnosis in 9.3% of cases |
| Rao [132] | 2019 | 31 328 266 | Chinese Children Genetic Kidney Disease Database | 1001 | WES | 2703 gene list | 42.1% | ||
| Saha [25] | 2023 | 37 464 296 | Retrospective cohort study >18 years in India | 86 | 10.5% consanguineous | Sanger, NGS | Disease specific tests/panels | 46% | Clinician initiated testing strategy |
| Mansilla [133] | 2021 | 31 738 409 | Cohort referred for genetic testing | 127 | 8.6% | Targeted panel | Disease-specific tests/panels | 44% | |
| Al-Hamed [134] | 2022 | 36 177 613 | CKD with likely GKD (15–79 years) | 102 | 59% (54% consanguineous) | NGS/Sanger | 336 panel | 42% | Highly consanguineous population |
| Pode-Shakked [135] | 2022 | 34 993 602 | Suspected GKD (aged 0–70 years) | 74 | 48% | NGS | Diseas-specific tests/panel | 57% | 85% Jewish ancestry |
| Doreille [28] | 2023 | Adults with unknown CKD < 45 years, clinical suspicion of GKD or planned for live donor | 538 | 42% | ES | Panel testing including customized Groopman nephropathy genes panel | 24% | Excluded ADPKD, 6% diagnosis were copy number variant |
Given the broad range of genetic causes underpinning clinical disorders, gene panels (engineered or virtual) based on clinical phenotypes have been developed for Alport syndrome, complement mediated disorders, CAKUT, glomerulopathy, cystic kidney disorders, and tubulopathies. The clinical genome resource (ClinGen) has developed a framework to evaluate the evidence supporting a gene-disease association from ‘no reported evidence’, ‘limited’, ‘moderate’, ‘strong’, and ‘definitive’, to guide inclusion of genes on panels [10, 11]. An important aspect of gene panels is regular review to ensure up-to-date, clinically relevant information [12, 13]. Two resources are PanelApp Australia (https://panelapp.agha.umccr.org/) and ClinGen Kidney Disease Clinical Domain Working Group (CDWG; https://clinicalgenome.org/working-groups/clinical-domain/clingen-kidney-disease-clinical-domain-working-group/), which are curated by expert clinicians and scientists [14, 15]. Virtual panel analyses are applied to genomic data from whole exome or genome sequencing, where the coding or entire genome, respectively, is sequenced and variants in the genes of the panel are reviewed. Gene panels are limited as they require regular updates to stay current and are not cost effective in some diseases, where exome sequencing may be more appropriate. The rapid expansion in knowledge requires ongoing review and validation to ensure clinicians have updated, accurate information. The ClinGen Kidney Disease CDWG supported by the National Institutes of Health was established to support aggregation of genomic data, curation of genetic variation, dissemination of tools, standardization, and to evaluate and improve integration of genomics into clinical practice [7, 15–17]. This ensures standardization of clinical genomics, translation into practice, and application of the American College of Medicine and Genetics criteria (ACMG) for variant interpretation [18].
Impact on clinical practice
Integrating genomics into mainstream nephrology practice has direct and indirect impacts for patients and families. Genomic testing offers diagnostic clarity for patients and potential to avoid investigations such as diagnostic kidney biopsy, guide screening programs and inform treatment decisions [9, 19]. Genetic testing can result in re-classification of diagnosis in 9.3–13% of cases such as reclassifying dominant to recessive disorders in cystic kidney disease or reclassifying clinical diagnosis such as recognizing FSGS as Alport syndrome, directly impacting patient care [1, 20, 21]. Recent evidence shows genomics is important to help clarify diagnosis in cases where kidney biopsy is inconclusive [22]. Treatment decisions include immunosuppression, renin-angiotensin blockade strategies, and screening for associated manifestations such as hearing involvement [9]. Integrating genomics early into clinical practice and the diagnostic journey will enable better patient outcomes and personalized care.
Genetic testing in CKD in practice
CKD clinics cover a heterogenous population with a variety of disease processes driving kidney impairment, relying on nephrologists to select possible cases of GKD and implement appropriate genetic testing. Nephrologists should consider GKD, particularly in those with a young age, family history, and extra-kidney features such as hearing loss, gout, diabetes, retinitis pigmentosa, and liver disease, though there are many varied presentations where genetic testing can be appropriate [20, 23, 24]. A phenotype-first approach for genetic testing provides a framework to identify potential GKD cases. Current dogma classifies GKD into phenotypes including cystic kidney disease, glomerulopathies, tubulopathies, tubulointerstitial disease, complement disorders, and CKD of unknown aetiology [1, 23, 25]. Clinical assessment and phenotyping enables clinicians to recognize disease patterns associated with forms of GKD and consider genomic testing. Phenotype-directed genetic testing focuses on understanding the nuances of genetic testing in each area category, including likelihood of positive test result, and specific limitations in current sequencing strategies such as limitations in APDKD testing or limitations in MUC1 for autosomal dominant tubulointerstitial kidney disease (ADTKD) [26, 27]. A genomics-first approach for patients with suspected GKD utilizes broader exon-based panel testing, with analysis interpreted within the clinical context. This genomics-first approach broadens genetic testing from phenotype-specific testing and can improve diagnostic yields [24, 28–30]. Genetic variants in Alport-associated genes, COL4A3-5, have been detected in a broad array of clinical phenotypes including glomerular diseases such as FSGS or IgA nephropathy, congenital disorders including CAKUT, vascular diseases such as hypertensive nephropathy and unknown cause of CKD, which may have been missed with a phenotype-first search strategy [23, 24, 28, 31, 32]. Genomics-first strategies carry the risk of detecting additional findings such as APOL1 allele status or cancer risk genes such as BRCA1/2, which require additional counselling to patients [28]. Nephrologists require an understanding of the rationale for different genomic testing strategies in clinical practice.
Cystic kidney disease
In the adult population, autosomal dominant polycystic kidney disease (ADPKD) is a well-known cause of kidney failure, accounting for 6.7–9.8% of patients receiving kidney replacement therapy [33, 34]. Genetic testing has enabled clarification of diagnosis, with the ability to distinguish phenocopies and to provide prognostic information and is becoming part of mainstream clinical care, with diagnostic yields amongst typical cases of 80.0–93.1% [3, 35]. Common genes in which variation is associated with ADPKD are PKD1 and PKD2, with several phenocopy disorders that are important to distinguish including heterozygous variants in GANAB, IFT140, ALG5, ALG8, ALG9, SEC61B, PKHD1, PRKCSH, SEC63, DNAJB11, and HNF1B [36, 37]. DNAJB11-associated disease presents with kidney cysts and overlapping ADTKD, distinguished by later onset of kidney failure [38]. Renal cysts and diabetes (RCAD) is associated with HNF1B mutations causing cystic kidneys with associated diabetes, hypomagnesemia, pancreatic, and liver abnormalities warranting distinct management [39]. Development of a specific disease-modifying therapeutic intervention, tolvaptan, can delay progression of kidney failure in ADPKD [40]. Diagnosis of ADPKD enables opportunity within families for early diagnosis, targeted genetic counselling, clinical prognostication, and genomic diagnosis can facilitate screening of at-risk relatives, living-related kidney donor work-up and reproductive planning [35].
Glomerulopathies
Genetic forms of glomerular diseases have highly variable clinical presentations including haematuria, proteinuria, abnormal glomerular basement membrane, congenital nephrotic syndrome, non-immune glomerulopathy, and more. Studying early-onset CKD found the highest diagnostic yield in glomerular phenotypes, with 80/131 (61%) with a confirmed genetic diagnosis, where 58/131 (73%) had positive family history and 30/80 (36%) had extra-renal manifestations [30]. An adult cohort with biopsy-proven FSGS identified 42.9% with a genetic disorder, particularly in those with absence of nephrotic syndrome, family history, and female gender [41]. In a systematic review, 11.1–51.0% of patients with nephrotic syndrome had a genetic diagnosis and other glomerulopathies had diagnostic yields of 14.1–91.0%, associated with family history, consanguinity, and age of onset [3]. Alport syndrome (COL4A3-5) genes were frequently identified, highlighting the clinical heterogeneity across Alport syndrome. X-linked Alport syndrome is associated with hemizygous COL4A5 mutations in males, but female COL4A5 heterozygous variants may have haematuria, proteinuria, hearing loss, and ocular defects, with risk of kidney failure increasing after age 60 years [42]. Considerations for Alport syndrome testing include clarification of unexplained haematuria and proteinuria, even with negative family history, in patients with focal segmental glomerulosclerosis (FSGS), familial IgA nephropathy, and kidney failure of unknown cause [30, 43]. COL4A3-5 mutations account for over 50% of the genetic diagnoses in FSGS and should especially be considered with extra-renal features and family history [31, 44]. MYH9 gene mutations can mimic Alport syndrome, with CKD, hearing loss, giant platelets and liver dysfunction, and genetic diagnosis can avoid unnecessary treatments including splenectomy or immunosuppression [45]. Understanding GKD inheritance clarifies risk to family members. Recessive disorders such as Finnish Congenital Nephrotic syndrome have NPHS1 antenatal screening in high-risk populations [46].
Tubulopathies
The complexity of kidney pathophysiology is highlighted by mutations in kidney tubular and related transporters in diseases such as Bartter syndrome, Gitelman syndrome, primary hyperoxaluria, cystinosis, cystinuria, and Dent disease [47]. Genetic testing clarifies diagnosis and disease associations, such as hypophosphataemic rickets association with Dent disease [48]. Cystinosis specific treatment can stabilize kidney function, with potential gene therapy [49, 50]. In rarer disorders, the rare stone consortium (https://www.rarekidneystones.org/) compiles cases to coordinate education and research, such as adenine phosphoribosyltransferase deficiency, a rare cause of kidney stones and kidney failure amenable to xanthine oxidase inhibitors [51]. Guidelines for the clinical detection and management of disorders such as Bartter syndrome have been developed to support antenatal and prenatal management, recommendations for genetic testing and management [52]. Understanding tubulopathies has expanded our knowledge of complex kidney physiology, and underpins our understanding of diuretics.
Tubulointerstitial disease
Tubulointerstitial disease is characterized by bland urinary sediment and impaired kidney function, with/without family history. Kidney biopsy reveals non-specific interstitial fibrosis and tubular atrophy, though protein accumulation may be detected with special staining [53]. Common genetic causes of ADTKD include UMOD, MUC1, HNF1B, SEC61A1, DNAJB11, and REN, and possible extra-renal manifestations such as gout in ADTKD-UMOD [3]. Recognizing ADKTD is important due to onset of kidney disease in the second to fourth decades of life [54]. An autosomal recessive cause of kidney failure is nephronophthisis, common in paediatric populations and increasingly recognized amongst adults; in a series of 5606 patients with adult-onset kidney failure, 26 (0.5%) had homozygous deletions in NPHP1, with 88% of them being previously attributed to unknown CKD [55]. Tubulointerstitial diseases can be challenging diagnostically in the absence of specific pathognomonic clinical or histopathologic features, so clinician awareness of the potential diagnosis is paramount, especially given potential novel targeted therapies [56].
Complement disorders
Atypical haemolytic uraemic syndrome (aHUS) and C3 glomerulopathy (C3GN) are two distinct forms of GKD potentially underpinned by genetic mutations in the alternate complement regulatory pathway. Genetic diagnoses in aHUS were initially reported between 51–61% in a French registry population though more recent studies found lower diagnostic yields: 21.6% in a Korean paediatric population; 39% in a global registry; 40.9% in a pregnancy registry; and 46% in a Japanese study [57–62]. C3GN has a genetic diagnosis in up to 20–25% of cases, with mutations in C3, CFB, CFH, CFI, CFHR5 having a clear association [63–65]. Regional variants occur in these genes, such as CFHR5 in familial kidney disease with Cypriot ancestry [64]. These disorders require a ‘second hit’ complement amplifying event such as infection or pregnancy, causing complement pathway activation. Confirming genetic diagnosis enables specific treatment with complement blockade, eculizumab, especially peri-transplant [66]. Penetrance in aHUS family members is variable, between 10–70%, impacted by type of mutation, affected siblings, and age [67, 68].
Unknown kidney disease
Advances in genomic testing have improved diagnosis in CKD of unknown cause. Whole-exome sequencing (WES) in an older population of kidney disease, 28.3% with positive family history, found a genetic diagnosis for unknown CKD in 48/281 (17.1%) of patients [24]. The Irish Kidney Gene project performed WES in unknown CKD, without family history, and identified a causative gene in 29.7% [20]. Genetics may inform diagnosis instead of kidney biopsy [19, 22]. Advances in genomic testing for GKD disorders should be considered to aid diagnosis, even without family history.
Transplantation
Genomic diagnosis has several implications for kidney transplantation. Firstly, a genomic diagnosis can impact management strategies in specific disease states, such as primary hyperoxaluria which previously required consideration of dual liver-kidney transplantation, and intensive dialysis prior to transplantation to prevent recurrence, though recent advances in treatment for primary hyperoxaluria, lumasiran and nedosiran, can be considered [1, 69, 70]. Complement disorders with an underlying mutation direct use of eculizumab to pre-empt recurrence and improve transplantation [71]. In low-resource settings, genetic diagnosis in congenital nephrotic syndrome may provide reassurance disease recurrence is low-risk after kidney transplant [72]. Genomic testing enables screening of potential live-related donors across a broad spectrum of dominant, recessive, and X-linked disorders given potential for variable expression and penetrance [43]. In cystic kidney disease, reclassification to recessive disorders or negative genetic testing can enable relatives to donate [73]. Performing broader genomic testing panels can find unexpected results, such as APOL1 or complement disorder genes, that unknowingly exclude prospective donors [74]. Incorporating genetic diagnosis in kidney transplant can benefit patients and prevent harm to potential donors, requiring additional research and development of specific guidelines for implementation in transplantation practice [75, 76].
Testing strategies
Advances in sequencing technology have improved genetic diagnosis with variable testing strategies used across the spectrum of kidney disease (Fig. 2). As an overarching principle, we primarily espouse a phenotype-driven approach to any genomic testing strategy for a case of suspected GKD. Further, it is important to ensure that a clinical question underpins a genomic testing strategy to maximize patient benefit and optimize outcomes. Informed consent and its documentation is an absolute requirement that founds the principle that genetic counselling is undertaken around a genomic testing strategy. The ordering clinician is encouraged to deliver the eventual result wherever possible, for continuity in patient care in these complex topics.
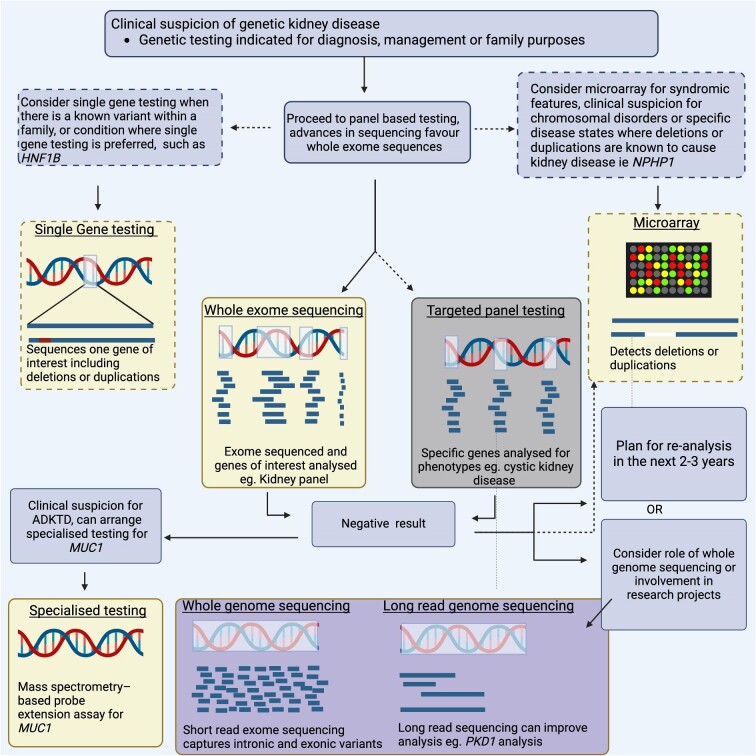
Figure 2:
Comparison of genomic diagnostic techniques and applicability in genetic kidney disease. Image made with BioRender.
Certain GKD are due to missing or additional genetic material, termed copy number variants (deletions/duplications) which are best detected with specific assays (micro-array or multiplex ligand probe amplification) and are important for certain genes, such as HNF1B, NPHP1 and in congenital kidney malformation including CAKUT [3, 39, 55, 77, 78]. Another testing approach is to develop assays for sets of genes of interest for a phenotype of interest (targeted sequence analysis) that has high diagnostic yields of 62–78% in selected patient cohorts with clear clinical phenotypes for CKD, particularly Alport syndrome and ADPKD, and is advantageous in offering targeted, effective testing at a lower cost [29, 30]. WES sequences the coding portion of the genome (∼2%) with analysis restricted to virtual gene panels for CKD. This approach has significant variation in diagnostic yields in CKD depending on the population, from 9.1% in dialysis patients to 51.5% in highly selected patients, and does not cover intronic or structural variants [20, 24, 79]. Whole-genome sequencing (WGS) sequences both the coding and non-coding genome, which may improve genetic diagnosis, including intronic variants, with risk of additional findings at higher costs [80]. The value of this expanded testing to improve diagnostic yield is uncertain in CKD, with further cohort data awaited, including results from the HIDDEN study [81]. Genomic sequencing strategies can determine genetic diagnosis in GKD and are established in clinical practice.
Many cases of clinical GKD that do not have a genomic diagnosis despite WES strategies require further testing strategies. MUC1 is a common form of ADTKD that is difficult to detect on WES due to high regions of CG repeats, and requires dedicated genetic sequencing or mass-spectrometry, though recent advantages in long-read sequencing have potential [27, 82]. In ADPKD, PKD1 is difficult to sequence using WES due to pseudogenes, with only 50% of mutations successfully identified on standard WES, though this is now improving with newer WES approaches [36, 83]. WGS techniques diagnosed 81% of typical ADPKD cases and 60% of atypical ADPKD [26]. Short- and long-read sequencing with RNA studies found a genetic diagnosis in 89% of ADPKD with prior negative genetic testing, including intronic splice site mutations not detected on exome sequencing [84]. Incorporating WGS into testing strategies improves the diagnostic yield in suspected genetic CKD populations to 51.4%, particularly to find genetic diagnosis in rare genetic diseases, though they may find unexpected additional findings [20, 85, 86]. Mitochondrial disorders cause a broad spectrum kidney disease and require clinical suspicion for specific testing, especially considering the unique inheritance modality and requisite genetic counselling [87–89]. Mosaic disorders such as tuberous sclerosis complex have variable levels of gene variants detected in blood, making detection difficult. Improvements in technology with targeted sequencing of the TSC1 and TSC2 genes at greater read depths has improved diagnostic capabilities [90]. Overall, there is no ‘best test’ for diagnosing GKD, and clinicians must understand suitability of each testing type for specific diseases.
Challenges and future directions
Integrating genomics into clinical practice to provide tailored clinical care enables personalized medicine in nephrology. The nephrologist is pivotal in formulating clinical diagnosis in CKD and to consider genetic causes. The treating clinician is well placed to discuss the merits of genetic testing including impact on management, broader family implications, and reproductive options, and understand how these decisions may change over a patient's lifespan. In ADPKD, for example, while clinical diagnosis can be made based on ultrasound criteria, genetic confirmation can provide prognostic information using the PROPKD score, clarify atypical cases, inform additional screening, and guide reproductive decision making [38, 91]. Each patient will have a different experience and perceived value of the personal benefits of genetic testing and broader implications to their family, with their nephrologist well placed to integrate these into a patient's individualized clinical paradigm. The KDIGO controversies conference on genomics in CKD identified lack of education as a key barrier for nephrologists to implement genetics into practice [92]. The ERA has further identified a lack of genomic literacy, lack of perceived benefit, difficulties with selecting testing strategy, uncertainty around variant interpretation, high cost, and counselling issues [4]. Survey and interview data highlights perceived barriers to implanting genetics in practice including identifying correct patients, selecting the correct test, interpreting results, following up with families, discussing results with patients and families, integrating results into clinical care, ordering the test, and consenting for genetic tests [93, 94]. Strategies to overcome this prioritize education to nephrologists to empower incorporation of clinical genetics and genomics into contemporary clinical practice, increased funding and access to genomic champions [4, 95]. Resources such as genomic decision aids, embedding genetic counsellors into mainstream nephrology clinics, and genetic stewardship programs are all avenues to improve mainstream nephrology access to clinical genomics. Potential avenues to overcome this include development of kidney genetics clinics combining nephrologists, geneticists, genetic counsellors, and patients to streamline and support genetic testing [96].
Health economics
The direct healthcare implications for genetic testing are paramount for patients and families, with broader impacts for health care. In selected patients, particularly children with glomerulopathy, early implementation of genetic testing can determine the diagnosis and prevent invasive testing [19, 97]. Economic analysis shows early integration of WES in children can be cost saving, particularly through avoidance of biopsy [97]. Health economic evaluation of genetic diagnosis in kidney disease identified short- and long-term cost benefit, largely driven by adults and children with glomerular disease. The cost benefit in a genetic diagnosis of atypical cystic kidney disease is through avoiding tolvaptan [5]. Modelling genetic testing to diagnose kidney disease suggests early integration in the work-flow process, in targeted patients, can reduce costs by 20%, with actual cost savings in selected patients reduced by 41% [6]. Genetic testing in adults has led to the discovery of more atypical or unusual presentations of typically paediatric disease, with probably similar benefits in treatment costs and quality of life. Genetic testing can inform diagnosis in diseases such as FSGS and identify cases unlikely to respond to immunosuppression [41]. Cascade testing can identify family members to direct preventative health measures and can avoid screening in negative family members. Kidney failure has disproportionately high health care costs, with annuals costs for home dialysis of  AUD49,137 and
AUD49,137 and  AUD79,072 for hospital dialysis, which are likely more costly than pre-implantation genetic diagnosi,s which substantially reduces the risk of affected children. This is increasingly part of clinical practice for families with established GKD; however, a cost analysis has not been done [98, 99]. With the reducing costs of genomic testing, and expanding knowledge of genomic testing, a ‘genomic first’ approach has the potential to offer cost savings and earlier diagnosis, and may provide clearer diagnosis than kidney biopsy [3, 19].
AUD79,072 for hospital dialysis, which are likely more costly than pre-implantation genetic diagnosi,s which substantially reduces the risk of affected children. This is increasingly part of clinical practice for families with established GKD; however, a cost analysis has not been done [98, 99]. With the reducing costs of genomic testing, and expanding knowledge of genomic testing, a ‘genomic first’ approach has the potential to offer cost savings and earlier diagnosis, and may provide clearer diagnosis than kidney biopsy [3, 19].
The direct impacts of clinical and genetic diagnosis in GKD are unique to each patient, including potential psychosocial impacts. Research in this space is limited, with most studies focusing on clinical results, and more work is needed to prioritize patient outcomes. The SONG-PKD working group noted that ADPKD had a heightened awareness of kidney disease due to seeing the disease across generations, which needs to be appreciated by clinicians [100]. There are also likely non-tangible benefits for genetic testing including alleviating anxiety, reduction in unnecessary testing, and workforce participation. Qualitative analysis focusing on consumer perspectives in patients with ADPKD highlighted the perceived benefit of genetic testing to inform their family members, which is likely to hold true in other forms of GKD [101]. Patients with rarer forms of inherited kidney disease, especially in children, face a long diagnostic odyssey. Genomic testing potentially shortens the time frame to a diagnosis in these patients, though not all will receive an answer [102]. The perceived benefit of genomic testing is highly individualized and perceived risk and benefits need to be evaluated by patients and their clinicians. Genomic testing requires shared decision making between the clinician and patient with a focus on disclosure, clinical benefits, psychosocial impact around guilt, employment impacts, and unexpected secondary findings.
Assessing clinically relevant impacts of genomics in CKD is important to guide integration into clinical practice, but is difficult to standardize given significant heterogeneity across key stakeholders: patients, families, and clinicians. Developing a utility ontology specific for GKD will enable standardization across research to enhance analysis of results, and to establish clear research goals with demonstrable outcomes from integrating genomics in clinical practice [103].
Polygenic risk scores
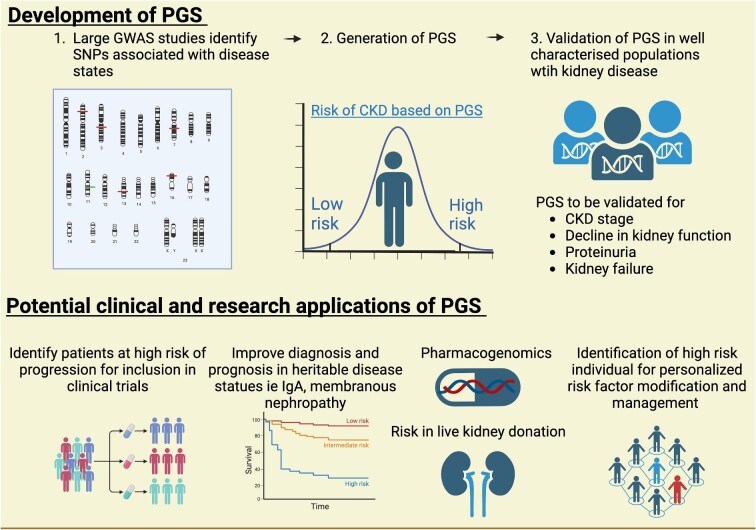
CKD can be a complex trait with both environmental and polygenic determinants, distinct from mendelian forms of GKD. Polygenic risk scores (PGS) capture multiple small genetic factors and aggregate relevant genetic loci for kidney disease from genome-wide association studies (GWAS) into risk scores for CKD traits, such as eGFR, decline in eGFR, CKD stage, and proteinuria (https://www.pgscatalog.org), and then undergo validation (Fig. 3) [104]. PGS can stratify patients into high- and low-risk groups prior to disease development, and evidence shows they correlate well at a population level; however, individual applicability is unclear [105–111]. PGS have the potential to provide early information on CKD risk (distinct from mendelian GKD) where risk can be modified by environment factors, suggesting early intervention in high-risk PGS could prevent, slow, or change development of CKD. Heritability estimates for kidney function from family studies are greater than GWAS/PGS studies, suggesting SNP-derived scores do not capture complexities such as epigenetic factors [112]. Future applications of PGS include risk stratification and prognosis, improved diagnostics, selection of high-risk individuals for tailored nephrological intervention, risk in live kidney donation and informing clinical trial design by selecting those at high risk of CKD progression [105, 113, 114].
Figure 3:
Process of generating and developing polygenic risk scores (PGS) into clinical practice and potential clinical applications. Image made with BioRender.
There are several novel applications of PGS in clinical nephrology. PGS for peritoneal dialysis can provide additional information regarding peritoneal dialysis transporter status [115]. PGS may explain heterogeneity in mendelian GKD, as patients with ADPKD and high-risk PGS had a 54-fold chance of CKD compared to 3-fold risk in the low-risk PGS ADPKD group [116]. PGS for kidney disease with substantial heritability like IgA and membranous nephropathy are being developed and may inform clinical practice for diagnosis, progression risk, and treatment responses [113, 117]. PGS has the potential to provide targeted information on risk for CKD in live-donor work-up, which requires validation in retrospective cohort studies. Post-transplant, PGS may help stratify risk for skin cancer, post-transplant diabetes or risk of CMV in high-risk individuals [118–120]. Further research is necessary prior to PGS integration in clinical care, where potential applications are broad. Future clinical research, refinement, and early implementation may be enabled by greater uptake of genomics, both research and clinical, in nephrology and CKD settings empowered by digital medical record systems.
Pharmacogenomics
Pharmacogenomics endeavors to provide personalized medicine, wherein genomic data informs therapeutic decisions, may be beneficial in CKD management. Many medications used in kidney medicine have genetic differences underpinning metabolism, which may guide dosing and drug decisions such as azathioprine, warfarin, and tacrolimus [121]. Metabolism of warfarin varies, and metabolic status has been studied to improve therapeutic outcomes. COAG, a randomized control trial comparing genotype-based warfarin dosing for CYP2C9 and VKORC1 variants with standard-of-care dosing found no difference in time in therapeutic range, suggesting genotypic information didn't translate to clinical benefit [121]. Tacrolimus metabolism is highly varied, with several genetic factors including CYP3A4, CYP3A5, and p-glycoprotein which require further research to understand and guide therapy [122]. Hypertension is a key modifiable risk factors in CKD with hundreds of SNPs associated with hypertension, each exerting only a small influence on blood pressure [123]. Identifying specific genotypic treatments with our current knowledge is challenging. Patients of African ancestry with hypertension respond better to calcium channel blockers and hydrochlorothiazide, which is the recommended first-line treatment in international hypertension guidelines [124]. The rationale behind this is multi-faceted and likely represents both environmental factors from differences in both salt intake and handling to genomic variances in drug metabolism [125]. Differences in the C825T polymorphism of the G protein beta (3) subunit predicts blood pressure response to thiazide treatment, with those with the TT polymorphism, more common in those of African ancestry, having a small but significant improvement in blood pressure, of 1 mmHg [126].
Current clinical translation of pharmacogenomics is underway. The recently published CKD-PGX feasibility trial aimed to optimize hypertension management by screening 11 genetic predictors of pharmacogenomic drug interaction in 335 patients with hypertension. The study noted those with poorer hypertension control were more likely to have a predicted pharmacogenomic drug interaction at the start of the study, such as CYP2C9-reduced metabolism association with poor losartan response and uncontrolled hypertension. Baseline data found 58% of participants with uncontrolled hypertension had an actionable genotype, and 36% of nephrology providers changed treatment based on this data. Over one year of follow-up there was a 4 mmHg systolic blood pressure improvement in the whole group, and a 14.9 mmHg SBP improvement in those with uncontrolled hypertension. In survey data, 96% of patients reported that being provided knowledge of their genotype would prompt them to invest more in their blood pressure control [127, 128]. The GUARDD-US study is currently underway to assess how knowledge of APOL risk alleles impacts on patient blood pressure control [118]. Another smaller study in 61 patients with CKD stage 3–5 found 39% of them had possible actionable changes, with 26 changes made in 20 patients [129]. Integration of the multigene panel into clinical practice can inform therapy selection and reduce adverse drug reactions [130]. Practical realization of pharmacogenomics requires a multi-disciplinary approach with pharmacists and nephrologists supported in genomic upskilling. Genomic predictors of pharmaceutical response may be a feasible tool to optimize blood pressure control, which is an important modifiable risk factor in CKD. An important limitation is the lack of pharmacogenomic studies in non-caucasian populations and to be truly applicable, more research into impacts of genetic loci in global populations is required.
CONCLUSIONS
Genetics is expanding the field of nephrology to enable better diagnostics and treatments. Applications of clinical genomics for CKD have proven diagnostic efficacy and a growing body of evidence for impact and utility. Understanding genomics including differences in sequencing approaches across different phenotypes of GKD is imperative to improve understanding and integration into clinical care. Nephrologists face several challenges in integrating genomics into clinical practice, and require further education and support to improve patient outcomes. Further, genetics has the potential through clinical research to inform future nephrology clinical practice in polygenic disease and pharmacogenomics, with the ultimate goal to improve patient outcomes. Developing clear research priorities in conjunction with key stakeholders is imperative to move the field forwards and enable iterative utility and impact focused implementation.
Supplementary Material
ACKNOWLEDGEMENTS
A.M. is supported by a Queensland Health Advancing Clinical Research Fellowship.
Contributor Information
Julia Jefferis, Genetic Health Queensland, Royal Brisbane and Women's Hospital, Brisbane, Australia; Faculty of Medicine, The University of Queensland, Brisbane, Australia; Kidney Health Service, Royal Brisbane and Women's Hospital, Brisbane, Australia.
Andrew J Mallett, Faculty of Medicine, The University of Queensland, Brisbane, Australia; Institute for Molecular Bioscience, The University of Queensland, Brisbane, Australia; Department of Renal Medicine, Townsville University Hospital, Douglas, Australia; College of Medicine and Dentistry, James Cook University, Douglas, Australia.
DATA AVAILABILITY STATEMENT
No new data were generated or analysed in support of this research.
CONFLICT OF INTEREST STATEMENT
The authors have no relevant disclosures.
REFERENCES
- 1. Chen J, Lin F, Zhai Y et al. Diagnostic and clinical utility of genetic testing in children with kidney failure. Pediatr Nephrol 2021;36:3653–62. 10.1007/s00467-021-05141-5 [DOI] [PubMed] [Google Scholar]
- 2. Vaisitti T, Bracciamà V, Faini AC et al. The role of genetic testing in the diagnostic workflow of pediatric patients with kidney diseases: the experience of a single institution. Hum Genomics 2023;17:10. 10.1186/s40246-023-00456-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Claus LR, Snoek R, Knoers NVAM et al. Review of genetic testing in kidney disease patients: diagnostic yield of single nucleotide variants and copy number variations evaluated across and within kidney phenotype groups. Am J Med Genet C Semin Med Genet 2022;190:358–76. 10.1002/ajmg.c.31995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knoers N, Antignac C, Bergmann C et al. Genetic testing in the diagnosis of chronic kidney disease: recommendations for clinical practice. Nephrol Dial Transplant 2022;37:239–54. 10.1093/ndt/gfab218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu Y, Jayasinghe K, Stark Z et al. Genomic testing for suspected monogenic kidney disease in children and adults: a health economic evaluation. Genet Med 2023;25:100942. 10.1016/j.gim.2023.100942 [DOI] [PubMed] [Google Scholar]
- 6. Becherucci F, Landini S, Palazzo V et al. A clinical workflow for cost-saving high-rate diagnosis of genetic kidney diseases. J Am Soc Nephrol 2023;34:706–20. 10.1681/ASN.0000000000000076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thaxton C, Goldstein J, DiStefano M et al. Lumping versus splitting: how to approach defining a disease to enable accurate genomic curation. Cell Genomics 2022;2:100131. 10.1016/j.xgen.2022.100131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thomas CP, Freese ME, Ounda A et al. Initial experience from a renal genetics clinic demonstrates a distinct role in patient management. Genet Med 2020;22:1025–35. 10.1038/s41436-020-0772-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jayasinghe K, Stark Z, Kerr PG et al. Clinical impact of genomic testing in patients with suspected monogenic kidney disease. Genet Med 2021;23:183–91. 10.1038/s41436-020-00963-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Natasha TS, Erin Rooney R, Adam HB et al. Evaluating the clinical validity of gene-disease associations: an evidence-based framework developed by the Clinical Genome Resource. Biorxiv 2017;111039. https://pubmed.ncbi.nlm.nih.gov/28552198/ [DOI] [PMC free article] [PubMed]
- 11. Wright CF, Fitzgerald TW, Jones WD et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet 2015;385:1305–14. 10.1016/S0140-6736(14)61705-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bean L, Funke B, Carlston CM et al. Diagnostic gene sequencing panels: from design to report—A technical standard of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2020;22:453–61. 10.1038/s41436-019-0666-z [DOI] [PubMed] [Google Scholar]
- 13. Platt CD, Zaman F, Bainter W et al. Efficacy and economics of targeted panel versus whole-exome sequencing in 878 patients with suspected primary immunodeficiency. J Allergy Clin Immunol 2021;147:723–6. 10.1016/j.jaci.2020.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stark Z, Foulger RE, Williams E et al. Scaling national and international improvement in virtual gene panel curation via a collaborative approach to discordance resolution. Am Hum Genet 2021;108:1551–7. 10.1016/j.ajhg.2021.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rehm HL, Berg JS, Brooks LD et al. ClinGen—the clinical genome resource. N Engl J Med 2015;372:2235–42. 10.1056/NEJMsr1406261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pejaver V, Byrne AB, Feng BJ et al. Calibration of computational tools for missense variant pathogenicity classification and ClinGen recommendations for PP3/BP4 criteria. Am J Hum Genet 2022;109:2163–77. 10.1016/j.ajhg.2022.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DiStefano MT, Goehringer S, Babb L et al. The Gene Curation Coalition: a global effort to harmonize gene-disease evidence resources. Genet Med 2022;24:1732–42. 10.1016/j.gim.2022.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Richards S, Aziz N, Bale S et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Robert T, Raymond L, Dancer M et al. Beyond the kidney biopsy: genomic approach to undetermined kidney diseases. Clin Kidney J 2024;17:sfad099. 10.1093/ckj/sfad099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Elhassan EAE, Murray SL, Connaughton DM et al. The utility of a genetic kidney disease clinic employing a broad range of genomic testing platforms: experience of the Irish Kidney Gene Project. J Nephrol 2022;35:1655–65. 10.1007/s40620-021-01236-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yao T, Udwan K, John R et al. Integration of genetic testing and pathology for the diagnosis of adults with FSGS. Clin J Am Soc Nephrol 2019;14:213–23. 10.2215/CJN.08750718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robert T, Greillier S, Torrents J et al. Diagnosis of kidney diseases of unknown etiology through biopsy-genetic analysis. Kidney Int Rep 2023;8:2077–87. 10.1016/j.ekir.2023.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Connaughton DM, Kennedy C, Shril S et al. Monogenic causes of chronic kidney disease in adults. Kidney Int 2019;95:914–28. 10.1016/j.kint.2018.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Groopman EE, Marasa M, Cameron-Christie S et al. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med 2019;380:142–51. 10.1056/NEJMoa1806891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saha A, Kapadia SF, Vala KB et al. Clinical utility of genetic testing in Indian children with kidney diseases. BMC Nephrol 2023;24:212. 10.1186/s12882-023-03240-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mallawaarachchi AC, Lundie B, Hort Y et al. Genomic diagnostics in polycystic kidney disease: an assessment of real-world use of whole-genome sequencing. Eur J Hum Genet 2021;29:760–70. 10.1038/s41431-020-00796-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Okada E, Morisada N, Horinouchi T et al. Detecting MUC1 variants in patients clinicopathologically diagnosed with having autosomal dominant tubulointerstitial kidney disease. Kidney Int Rep 2022;7:857–66. 10.1016/j.ekir.2021.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doreille A, Lombardi Y, Dancer M et al. Exome-first strategy in adult patients with CKD: a cohort study. Kidney Int Rep 2023;8:596–605. 10.1016/j.ekir.2022.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bullich G, Domingo-Gallego A, Vargas I et al. A kidney-disease gene panel allows a comprehensive genetic diagnosis of cystic and glomerular inherited kidney diseases. Kidney Int 2018;94:363–71. 10.1016/j.kint.2018.02.027 [DOI] [PubMed] [Google Scholar]
- 30. Domingo-Gallego A, Pybus M, Bullich G et al. Clinical utility of genetic testing in early-onset kidney disease: seven genes are the main players. Nephrol Dial Transplant 2022;37:687–96. 10.1093/ndt/gfab019 [DOI] [PubMed] [Google Scholar]
- 31. Gast C, Pengelly RJ, Lyon M et al. Collagen (COL4A) mutations are the most frequent mutations underlying adult focal segmental glomerulosclerosis. Nephrol Dial Transplant 2016;31:961–70. 10.1093/ndt/gfv325 [DOI] [PubMed] [Google Scholar]
- 32. Rahimzadeh H, Ajlou S, Nili F et al. Alport syndrome misdiagnosed with IgA nephropathy from familial history: a case report and brief review. BMC Nephrology 2023;24:97. 10.1186/s12882-023-03165-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fernando MR, Dent H, McDonald SP et al. Incidence and survival of end-stage kidney disease due to polycystic kidney disease in Australia and New Zealand (1963–2014). Population Health Metrics 2017;15:7. 10.1186/s12963-017-0123-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Spithoven EM, Kramer A, Meijer E et al. Renal replacement therapy for autosomal dominant polycystic kidney disease (ADPKD) in Europe: prevalence and survival—an analysis of data from the ERA-EDTA Registry. Nephrol Dial Transplant 2014;29 Suppl 4:iv15–25. 10.1093/ndt/gfu017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Elliott MD, James LC, Simms EL et al. Mainstreaming genetic testing for adult patients with autosomal dominant polycystic kidney disease. Can J Kidney Health Dis 2021;8:20543581211055001. 10.1177/20543581211055001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang AR, Moore BS, Luo JZ et al. Exome sequencing of a clinical population for autosomal dominant polycystic kidney disease. JAMA 2022;328:2412–21. 10.1001/jama.2022.22847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lemoine H, Raud L, Foulquier F et al. Monoallelic pathogenic ALG5 variants cause atypical polycystic kidney disease and interstitial fibrosis. Am J Hum Genet 2022;109:1484–99. 10.1016/j.ajhg.2022.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cornec-Le Gall E, Olson RJ, Besse W et al. Monoallelic mutations to DNAJB11 cause atypical autosomal-dominant polycystic kidney disease. Am J Hum Genet 2018;102:832–44. 10.1016/j.ajhg.2018.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heidet L, Decramer S, Pawtowski A et al. Spectrum of HNF1B mutations in a large cohort of patients who harbor renal diseases. Clin J Am Soc Nephrol 2010;5:1079–90. 10.2215/CJN.06810909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Torres VE, Chapman AB, Devuyst O et al. Tolvaptan in patients with autosomal dominant polycystic kidney disease. N Engl J Med 2012;367:2407–18. 10.1056/NEJMoa1205511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miao J, Pinto EVF, Hogan MC et al. Identification of genetic causes of focal segmental glomerulosclerosis increases with proper patient selection. Mayo Clin Proc 2021;96:2342–53. 10.1016/j.mayocp.2021.01.037 [DOI] [PubMed] [Google Scholar]
- 42. Savige J, Colville D, Rheault M et al. Alport Syndrome in Women and girls. Clin J Am Soc Nephrol 2016;11:1713–20. 10.2215/CJN.00580116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Savige J, Lipska-Zietkiewicz BS, Watson E et al. Guidelines for genetic testing and management of Alport syndrome. Clin J Am Soc Nephrol 2022;17:143–54. 10.2215/CJN.04230321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yao Q, Chen Y, Zhou X. The roles of microRNAs in epigenetic regulation. Curr Opin Chem Biol 2019;51:11–17. 10.1016/j.cbpa.2019.01.024 [DOI] [PubMed] [Google Scholar]
- 45. Tabibzadeh N, Fleury D, Labatut D et al. MYH9-related disorders display heterogeneous kidney involvement and outcome. Clin Kidney J 2019;12:494–502. 10.1093/ckj/sfy117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kallinen J, Heinonen S, Ryynänen M et al. Antenatal genetic screening for congenital nephrosis. Prenat Diagn 2001;21:81–84. [DOI] [PubMed] [Google Scholar]
- 47. Downie ML, Lopez Garcia SC, Kleta R et al. Inherited tubulopathies of the kidney: insights from genetics. Clin J Am Soc Nephrol 2021;16:620–30. 10.2215/CJN.14481119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gianesello L, Arroyo J, Del Prete D et al. Genotype phenotype correlation in Dent Disease 2 and review of the literature: OCRL gene pleiotropism or extreme phenotypic variability of Lowe Syndrome? Genes (Basel) 2021;12:1597. 10.3390/genes12101597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kleta R, Bernardini I, Ueda M et al. Long-term follow-up of well-treated nephropathic cystinosis patients. J Pediatr 2004;145:555–60. 10.1016/j.jpeds.2004.03.056 [DOI] [PubMed] [Google Scholar]
- 50. Cherqui S. Hematopoietic stem cell gene therapy for cystinosis: from bench-to-bedside. Cells 2021;10:3273. 10.3390/cells10123273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Runolfsdottir HL, Palsson R, Agustsdottir IMS et al. Kidney transplant outcomes in patients with adenine phosphoribosyltransferase deficiency. Transplantation 2020;104:2120–8. 10.1097/TP.0000000000003088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Konrad M, Nijenhuis T, Ariceta G et al. Diagnosis and management of Bartter syndrome: executive summary of the consensus and recommendations from the European Rare Kidney Disease Reference Network Working Group for Tubular Disorders. Kidney Int 2021;99:324–35. 10.1016/j.kint.2020.10.035 [DOI] [PubMed] [Google Scholar]
- 53. Onoe T, Hara S, Yamada K et al. Significance of kidney biopsy in autosomal dominant tubulointerstitial kidney disease-UMOD: is kidney biopsy truly nonspecific? BMC Nephrol 2021;22:1. 10.1186/s12882-020-02169-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Econimo L, Schaeffer C, Zeni L et al. Autosomal dominant tubulointerstitial kidney disease: an emerging cause of genetic CKD. Kidney Int Rep 2022;7:2332–44. 10.1016/j.ekir.2022.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Snoek R, van Setten J, Keating BJ et al. NPHP1 (Nephrocystin-1) Gene deletions cause adult-onset ESRD. J Am Soc Nephrol 2018;29:1772–9. 10.1681/ASN.2017111200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dvela-Levitt M, Kost-Alimova M, Emani M et al. Small molecule targets TMED9 and promotes lysosomal degradation to reverse proteinopathy. Cell 2019;178:521–535.e523. 10.1016/j.cell.2019.07.002 [DOI] [PubMed] [Google Scholar]
- 57. Fujisawa M, Kato H, Yoshida Y et al. Clinical characteristics and genetic backgrounds of Japanese patients with atypical hemolytic uremic syndrome. Clin Exp Nephrol 2018;22:1088–99. 10.1007/s10157-018-1549-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fremeaux-Bacchi V, Fakhouri F, Garnier A et al. Genetics and outcome of atypical hemolytic uremic syndrome: a nationwide French series comparing children and adults. Clin J Am Soc Nephrol 2013;8:554–62. 10.2215/CJN.04760512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Noris M, Caprioli J, Bresin E et al. Relative role of genetic complement abnormalities in sporadic and familial aHUS and their impact on clinical phenotype. Clin J Am Soc Nephrol 2010;5:1844–59. 10.2215/CJN.02210310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schaefer F, Ardissino G, Ariceta G et al. Clinical and genetic predictors of atypical hemolytic uremic syndrome phenotype and outcome. Kidney Int 2018;94:408–18. 10.1016/j.kint.2018.02.029 [DOI] [PubMed] [Google Scholar]
- 61. Huerta A, Arjona E, Portoles J et al. A retrospective study of pregnancy-associated atypical hemolytic uremic syndrome. Kidney Int 2018;93:450–9. 10.1016/j.kint.2017.06.022 [DOI] [PubMed] [Google Scholar]
- 62. Ng MSY, McClymont K, McCallum N et al. CFHR5 Nephropathy in a Greek-Cypriot Australian Family: ancestry-informed precision medicine. Kidney Int Rep 2018;3:1222–8. 10.1016/j.ekir.2018.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Soraru J, Isbel N, Wong G et al. Baseline characteristics of patients with atypical haemolytic uraemic syndrome (aHUS): the Australian cohort in a global aHUS registry. Nephrology (Carlton) 2020;25:683–90. 10.1111/nep.13722 [DOI] [PubMed] [Google Scholar]
- 64. Gale DP, de Jorge EG, Cook HT et al. Identification of a mutation in complement factor H-related protein 5 in patients of Cypriot origin with glomerulonephritis. Lancet 2010;376:794–801. 10.1016/S0140-6736(10)60670-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Iatropoulos P, Noris M, Mele C et al. Complement gene variants determine the risk of immunoglobulin-associated MPGN and C3 glomerulopathy and predict long-term renal outcome. Mol Immunol 2016;71:131–42. 10.1016/j.molimm.2016.01.010 [DOI] [PubMed] [Google Scholar]
- 66. Smith RJH, Appel GB, Blom AM et al. C3 glomerulopathy—understanding a rare complement-driven renal disease. Nat Rev Nephrol 2019;15:129–43. 10.1038/s41581-018-0107-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ardissino G, Longhi S, Porcaro L et al. Risk of Atypical HUS among family members of patients carrying complement regulatory gene abnormality. Kidney Int Rep 2021;6:1614–21. 10.1016/j.ekir.2021.03.885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sullivan M, Rybicki LA, Winter A et al. Age-related penetrance of hereditary atypical hemolytic uremic syndrome. Ann Hum Genet 2011;75:639–47. 10.1111/j.1469-1809.2011.00671.x [DOI] [PubMed] [Google Scholar]
- 69. Baum MA, Langman C, Cochat P et al. PHYOX2: a pivotal randomized study of nedosiran in primary hyperoxaluria type 1 or 2. Kidney Int 2023;103:207–17. 10.1016/j.kint.2022.07.025 [DOI] [PubMed] [Google Scholar]
- 70. Garrelfs SF, Frishberg Y, Hulton SA et al. Lumasiran, an RNAi therapeutic for primary hyperoxaluria type 1. N Engl J Med 2021;384:1216–26. 10.1056/NEJMoa2021712 [DOI] [PubMed] [Google Scholar]
- 71. Regunathan-Shenk R, Avasare RS, Ahn W et al. Kidney transplantation in C3 glomerulopathy: a case series. Am J Kidney Dis 2019;73:316–23. 10.1053/j.ajkd.2018.09.002 [DOI] [PubMed] [Google Scholar]
- 72. Sinha R, Sarkar S, Mandal K et al. Uptake of next-generation sequencing in children with end-stage renal disease secondary to focal segmental glomerulosclerosis and parental decision for kidney transplantation-experience from a low resource setting: a retrospective cohort study. Pediatr Transplant 2021;25:e13960. 10.1111/petr.13960 [DOI] [PubMed] [Google Scholar]
- 73. Thomas CP, Gupta S, Freese ME et al. Sequential genetic testing of living-related donors for inherited renal disease to promote informed choice and enhance safety of living donation. Transpl Int 2021;34:2696–705. 10.1111/tri.14133 [DOI] [PubMed] [Google Scholar]
- 74. Singh G, Gohh R, Clark D et al. Vignette-based reflections to inform genetic testing policies in living kidney donors. Genes (Basel) 2022;13:592. 10.3390/genes13040592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Soraru J, Chakera A, Isbel N et al. The evolving role of diagnostic genomics in kidney transplantation. Kidney Int Rep 2022;7:1758–71. 10.1016/j.ekir.2022.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Thomas CP, Daloul R, Lentine KL et al. Genetic evaluation of living kidney donor candidates: a review and recommendations for best practices. Am J Transplant 2023;23:597–607. 10.1016/j.ajt.2023.02.020 [DOI] [PubMed] [Google Scholar]
- 77. Sanna-Cherchi S, Kiryluk K, Burgess KE et al. Copy-number disorders are a common cause of congenital kidney malformations. Am J Hum Genet 2012;91:987–97. 10.1016/j.ajhg.2012.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sztromwasser P, Michalak A, Małachowska B et al. A cross-sectional study of patients referred for HNF1B-MODY genetic testing due to cystic kidneys and diabetes. Pediatr Diabetes 2020;21:422–30. 10.1111/pedi.12959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tanudisastro HA, Holman K, Ho G et al. Australia and New Zealand renal gene panel testing in routine clinical practice of 542 families. NPJ Genom Med 2021;6:20. 10.1038/s41525-021-00184-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Devarajan P, Chertow GM, Susztak K et al. Emerging role of clinical genetics in CKD. Kidney Med 2022;4:100435. 10.1016/j.xkme.2022.100435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Soraru J, Jahan S, Quinlan C et al. The HIDDEN Protocol: an Australian prospective cohort study to determine the utility of whole genome sequencing in kidney failure of unknown aetiology. Front Med (Lausanne) 2022;9:891223. 10.3389/fmed.2022.891223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kirby A, Gnirke A, Jaffe DB et al. Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing. Nat Genet 2013;45:299–303. 10.1038/ng.2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ali H, Al-Mulla F, Hussain N et al. PKD1 Duplicated regions limit clinical utility of whole exome sequencing for genetic diagnosis of autosomal dominant polycystic kidney disease. Sci Rep 2019;9:4141. 10.1038/s41598-019-40761-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hort Y, Sullivan P, Wedd L et al. Atypical splicing variants in PKD1 explain most undiagnosed typical familial ADPKD. npj Genom Med 2023;8:16. 10.1038/s41525-023-00362-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Orr S, Olinger E, Iosifidou S et al. Molecular genetic diagnosis of kidney ciliopathies: lessons from interpreting genomic sequencing data and the requirement for accurate phenotypic data. Ann Hum Genet 2024;88:76–85. 10.1111/ahg.12508 [DOI] [PubMed] [Google Scholar]
- 86. Stranneheim H, Lagerstedt-Robinson K, Magnusson M et al. Integration of whole genome sequencing into a healthcare setting: high diagnostic rates across multiple clinical entities in 3219 rare disease patients. Genome Med 2021;13:40. 10.1186/s13073-021-00855-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Imasawa T, Hirano D, Nozu K et al. Clinicopathologic features of mitochondrial nephropathy. Kidney Int Rep 2022;7:580–90. 10.1016/j.ekir.2021.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Connor TM, Hoer S, Mallett A et al. Mutations in mitochondrial DNA causing tubulointerstitial kidney disease. PLoS Genet 2017;13:e1006620. 10.1371/journal.pgen.1006620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Viering D, Schlingmann KP, Hureaux M et al. Gitelman-Like syndrome caused by pathogenic variants in mtDNA. J Am Soc Nephrol 2022;33:305–25. 10.1681/ASN.2021050596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. West HD, Nellist M, Brouwer RWW et al. Targeted genomic sequencing of TSC1and TSC2Reveals causal variants in individuals for whom previous genetic testing for tuberous sclerosis complex was normal. Hum Mutat 2023;2023:4899372. 10.1155/2023/4899372 [DOI] [Google Scholar]
- 91. Cornec-Le Gall E, Audrézet MP, Rousseau A et al. The PROPKD score: a new algorithm to predict renal survival in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 2016;27:942–51. 10.1681/ASN.2015010016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Köttgen A, Cornec-Le Gall E, Halbritter J et al. Genetics in chronic kidney disease: conclusions from a kidney disease: improving global outcomes (KDIGO) controversies conference. Kidney Int 2022;101:1126–41. 10.1016/j.kint.2022.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Jayasinghe K, Quinlan C, Mallett AJ et al. Attitudes and practices of Australian nephrologists toward implementation of clinical genomics. Kidney Int Rep 2021;6:272–83. 10.1016/j.ekir.2020.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mrug M, Bloom MS, Seto C et al. Genetic testing for chronic kidney diseases: clinical utility and barriers perceived by nephrologists. Kidney Med 2021;3:1050–6. 10.1016/j.xkme.2021.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kansal A, Quinlan C, Stark Z et al. Theory designed strategies to support implementation of genomics in nephrology. Genes (Basel) 2022;13:1919. 10.3390/genes13101919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mallett A, Fowles LF, McGaughran J et al. A multidisciplinary renal genetics clinic improves patient diagnosis. Med J Aust 2016;204:58–59. 10.5694/mja15.01157 [DOI] [PubMed] [Google Scholar]
- 97. Jayasinghe K, Wu Y, Stark Z et al. Cost-effectiveness of targeted exome analysis as a diagnostic test in glomerular diseases. Kidney Int Rep 2021;6:2850–61. 10.1016/j.ekir.2021.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Cass A., Chadban S, Craig J et al. The Economic Impact of End-Stage Kidney Disease in Australia. Kidney Health Australia, Melbourne, 2006.
- 99. Snoek R, Stokman MF, Lichtenbelt KD et al. Preimplantation genetic testing for monogenic kidney disease. Clin J Am Soc Nephrol 2020;15:1279–86. 10.2215/CJN.03550320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cho Y, Tong A, Craig JC et al. Establishing a core outcome set for autosomal dominant polycystic kidney disease: report of the standardized outcomes in nephrology–Polycystic Kidney disease (SONG-PKD) Consensus Workshop. Am J Kidney Dis 2021;77:255–63. 10.1053/j.ajkd.2020.05.024 [DOI] [PubMed] [Google Scholar]
- 101. Tong A, Tunnicliffe DJ, Lopez-Vargas P et al. Identifying and integrating consumer perspectives in clinical practice guidelines on autosomal-dominant polycystic kidney disease. Nephrology (Carlton) 2016;21:122–32. 10.1111/nep.12579 [DOI] [PubMed] [Google Scholar]
- 102. Schuermans N, Hemelsoet D, Terryn W et al. Shortcutting the diagnostic odyssey: the multidisciplinary Program for Undiagnosed Rare Diseases in adults (UD-PrOZA). Orphanet J Rare Dis 2022;17:210. 10.1186/s13023-022-02365-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Mallett A, Stark Z, Fehlberg Z et al. Determining the utility of diagnostic genomics: a conceptual framework. Hum Genomics 2023;17:75. 10.1186/s40246-023-00524-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Medicine 2020;12:44. 10.1186/s13073-020-00742-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Khan A, Turchin MC, Patki A et al. Genome-wide polygenic score to predict chronic kidney disease across ancestries. Nat Med 2022;28:1412–20. 10.1038/s41591-022-01869-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Steinbrenner I, Yu Z, Jin J et al. A polygenic score for reduced kidney function and adverse outcomes in a cohort with chronic kidney disease. Kidney Int 2023;103:421–4. 10.1016/j.kint.2022.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Yu Z, Jin J, Tin A et al. Polygenic risk scores for kidney function and their associations with circulating proteome, and incident kidney diseases. J Am Soc Nephrol 2021;32:3161–73. 10.1681/ASN.2020111599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wuttke M, Li Y, Li M et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet 2019;51:957–72. 10.1038/s41588-019-0407-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Gorski M, Jung B, Li Y et al. Meta-analysis uncovers genome-wide significant variants for rapid kidney function decline. Kidney Int 2021;99:926–39. 10.1016/j.kint.2020.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Gorski M, Rasheed H, Teumer A et al. Genetic loci and prioritization of genes for kidney function decline derived from a meta-analysis of 62 longitudinal genome-wide association studies. Kidney Int 2022;102:624–39. 10.1016/j.kint.2022.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Bakshi A, Jefferis J, Wolfe R et al. Association of polygenic scores with chronic kidney disease phenotypes in a longitudinal study of older adults. Kidney Int 2023;103:1156–66. 10.1016/j.kint.2023.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Jefferis J, Pelecanos A, Catts V et al. The heritability of kidney function using an older Australian twin population. Kidney Int Rep 2022;7:1819–30. 10.1016/j.ekir.2022.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Sukcharoen K, Sharp SA, Thomas NJ et al. IgA nephropathy genetic risk score to estimate the prevalence of IgA nephropathy in UK Biobank. Kidney Int Rep 2020;5:1643–50. 10.1016/j.ekir.2020.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Xie J, Liu L, Mladkova N et al. The genetic architecture of membranous nephropathy and its potential to improve non-invasive diagnosis. Nat Commun 2020;11:1600. 10.1038/s41467-020-15383-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Mehrotra R, Stanaway IB, Jarvik GP et al. A genome-wide association study suggests correlations of common genetic variants with peritoneal solute transfer rates in patients with kidney failure receiving peritoneal dialysis. Kidney Int 2021;100:1101–11. 10.1016/j.kint.2021.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Khan A, Shang N, Nestor JG et al. Polygenic risk affects the penetrance of monogenic kidney disease. medRxiv 2023. https://pubmed.ncbi.nlm.nih.gov/38097619/ [DOI] [PMC free article] [PubMed]
- 117. Sanchez-Rodriguez E, Southard CT, Kiryluk K. GWAS-based discoveries in IgA nephropathy, membranous nephropathy, and steroid-sensitive nephrotic syndrome. Clin J Am Soc Nephrol 2021;16:458–66. 10.2215/CJN.14031119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Stapleton CP, Birdwell KA, McKnight AJ et al. Polygenic risk score as a determinant of risk of non-melanoma skin cancer in a European-descent renal transplant cohort. Am J Transplant 2019;19:801–10. 10.1111/ajt.15057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Shaked A, Loza BL, Van Loon E et al. Donor and recipient polygenic risk scores influence the risk of post-transplant diabetes. Nat Med 2022;28:999–1005. 10.1038/s41591-022-01758-7 [DOI] [PubMed] [Google Scholar]
- 120. Bodro M, Cervera C, Linares L et al. Polygenic innate immunity score to predict the risk of Cytomegalovirus infection in CMV D+/R- transplant recipients. A prospective multicenter cohort study. Front Immunol 2022;13:897912. 10.3389/fimmu.2022.897912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kimmel SE, French B, Kasner SE et al. A pharmacogenetic versus a clinical algorithm for Warfarin dosing. N Engl J Med 2013;369:2283–93. 10.1056/NEJMoa1310669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Yu M, Liu M, Zhang W et al. Pharmacokinetics, pharmacodynamics and pharmacogenetics of Tacrolimus in kidney transplantation. Curr Drug Metab 2018;19:513–22. 10.2174/1389200219666180129151948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ehret GB, Munroe PB, Rice KM et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011;478:103–9. 10.1038/nature10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Unger T, Borghi C, Charchar F et al. 2020 International Society of Hypertension Global Hypertension Practice Guidelines. Hypertension 2020;75:1334–57. 10.1161/HYPERTENSIONAHA.120.15026 [DOI] [PubMed] [Google Scholar]
- 125. Brewster LM, Seedat YK. Why do hypertensive patients of African ancestry respond better to calcium blockers and diuretics than to ACE inhibitors and β-adrenergic blockers? A systematic review. BMC Med 2013;11:141. 10.1186/1741-7015-11-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Turner ST, Schwartz GL, Chapman AB et al. C825T polymorphism of the G protein beta(3)-subunit and antihypertensive response to a thiazide diuretic. Hypertension 2001;37:739–43. 10.1161/01.HYP.37.2.739 [DOI] [PubMed] [Google Scholar]
- 127. Eadon MT, Cavanaugh KL, Orlando LA et al. Design and rationale of GUARDD-US: a pragmatic, randomized trial of genetic testing for APOL1 and pharmacogenomic predictors of antihypertensive efficacy in patients with hypertension. Contemp Clin Trials 2022;119:106813. 10.1016/j.cct.2022.106813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Eadon MT, Maddatu J, Moe SM et al. Pharmacogenomics of hypertension in CKD: the CKD-PGX study. Kidney360 2022;3:307–16. 10.34067/KID.0005362021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kerskes CHM, van den Eijnde C, Aarnoudse A et al. The effect of genotyping on the number of pharmacotherapeutic gene-drug interventions in chronic kidney disease patients. Pharmacy (Basel) 2023;11:69. 10.3390/pharmacy11020069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Swen JJ, van der Wouden CH, Manson LE et al. A 12-gene pharmacogenetic panel to prevent adverse drug reactions: an open-label, multicentre, controlled, cluster-randomised crossover implementation study. Lancet 2023;401:347–56. 10.1016/S0140-6736(22)01841-4 [DOI] [PubMed] [Google Scholar]
- 131. Lata S, Marasa M, Li Y et al. Whole-exome sequencing in adults with chronic kidney disease: a pilot study. Ann Intern Med 2018;168:100–9. 10.7326/M17-1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Rao J, Liu X, Mao J et al. Genetic spectrum of renal disease for 1001 Chinese children based on a multicenter registration system. Clin Genet 2019;96:402–10. 10.1111/cge.13606 [DOI] [PubMed] [Google Scholar]
- 133. Mansilla MA, Sompallae RR, Nishimura CJ et al. Targeted broad-based genetic testing by next-generation sequencing informs diagnosis and facilitates management in patients with kidney diseases. Nephrol Dial Transplant 2021;36:295–305. 10.1093/ndt/gfz173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Al-Hamed MH, Hussein MH, Shah Y et al. Exome sequencing unravels genetic variants associated with chronic kidney disease in Saudi Arabian patients. Hum Mutat 2022;43:e24–37. 10.1002/humu.24480 [DOI] [PubMed] [Google Scholar]
- 135. Pode-Shakked B, Ben-Moshe Y, Barel O et al. A multidisciplinary nephrogenetic referral clinic for children and adults-diagnostic achievements and insights. Pediatr Nephrol 2022;37:1623–46. 10.1007/s00467-021-05374-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were generated or analysed in support of this research.