Abstract
Introduction
Silybum marianum commonly known as milk thistle is one of the most imperative medicinal plants due to its remarkable pharmacological activities. Lately, the antiviral activities of S. marianum extract have been studied and it showed effectiveness against many viruses.
Objective
Although most previous studies were concerned mainly with silymarin content of the fruit, the present study provides comprehensive comparative evaluation of S. marianum different organs’ chemical profiles using UPLC-MS/MS coupled to chemometrics to unravel potentially selective antiviral compounds against human coronavirus (HCoV-229E).
Methodology
UPLC-ESI-TQD-MS/MS analysis was utilized to establish metabolic fingerprints for S. marianum organs namely fruits, roots, stems and seeds. Multivariate analysis, using OPLS-DA and HCA-heat map was applied to explore the main discriminatory phytoconstituents between organs. Selective virucidal activity of organs extracts against coronavirus (HCoV-229E) was evaluated for the first time using cytopathic effect (CPE) inhibition assay. Correlation coefficient analysis was implemented for detection of potential constituents having virucidal activity.
Results
UPLC-MS/MS analysis resulted in 87 identified metabolites belonging to different classes. OPLS-DA revealed in-between class discrimination between milk thistle organs proving their significantly different metabolic profiles. The results of CPE assay showed that all tested organ samples exhibited dose dependent inhibitory activity in nanomolar range. Correlation analysis disclosed that caffeic acid-O-hexoside, gadoleic and linolenic acids were the most potentially selective antiviral phytoconstituents.
Conclusion
This study valorizes the importance of different S. marianum organs as wealthy sources of selective and effective antiviral candidates. This approach can be extended to unravel potentially active constituents from complex plant matrices.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12906-024-04411-7.
Keywords: Milk thistle, Selectivity index, Human coronavirus (HCoV-229E), Metabolomics, LC-MS/MS
Introduction
Silybum marianum (L.) Gaertn is an annual or biennial plant belonging to family Asteraceae [1]. It has many common names, the most widely known one is milk thistle [2]. It is native to the Mediterranean districts of Northern Africa, Southern Europe, and Western Asia, but now it is cultivated throughout the whole world [3], either as vegetable, medicinal or as ornamental plant [4].
Owing to its various beneficial effects, S. marianum is among the most-selling botanical dietary supplements worldwide with an average sale of about US$ 8 billion/annum [5]. Recently, the milk thistle supplements market has globally expanded due to the COVID-19 outbreak, that led to increased need for immunomodulating supplements, in addition to the growing demand of effective anti-inflammatory, anti-aging and skin care natural products. In foods, its leaves and flowers are consumed as a vegetable for salads and a substitute for spinach. Milk thistle seeds can be used in raw form or made into tea. They can be also roasted for use as a coffee substitute [6].
S. marianum has been utilized as a medicinal plant of long ago, mainly for mitigation of liver, kidney, spleen and gall bladder diseases [7, 8]. Nowadays, its extract is sold in the market under many brand names due to its many reported and astonishing pharmacological activities [9]. It has been proved to possess antioxidant [10], hepatoprotective [11], anticancer [12], anti-inflammatory [13], anti-diabetic [14], anti-amnesia [15], antiplatelet [16] and cardioprotective [17] activities. It has also been utilized for alleviation of obsessive-compulsive disorder [18], depression [19], menstrual disorders and varicose veins [20].
Lately, the antiviral activities of S. marianum extract have been studied and it showed effectiveness against many viruses such as the flaviviruses (hepatitis C virus and dengue virus) [21, 22], human immunodeficiency virus [23], togaviruses (Chikungunya virus and Mayaro virus) [24, 25], hepatitis B virus [26] and influenza virus [27]. The remarkable antiviral efficacy of S. marianum extract is attributed to its multi-target activity against host cell. As it showed ability to modulate cell innate immunity [28, 29], inflammation [30], oxidative stress [31] and autophagy [32], which are cellular processes impaired by the viral invasion. In addition to the modulation of the cell environment, S. marianum extract also showed ability to exert direct potent antiviral actions against viral proteins [33]. These findings encouraged the researchers to assess the effectiveness of S. marianum against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19 pandemic. It was computationally found to act as inhibitor of signal transducer and activator of transcription 3 (STAT3), the main modulator of inflammatory and immune response. In addition, it was predicted to inhibit RNA-dependent RNA polymerase (RdRp), the main protein responsible for SARS-CoV-2 replication and transcription [34].
The first human coronavirus (HCoV) strain was found out in 1965. Afterwards, additional 30 strains were recognized, from which HCoV-229E was the prototypic strain that HCoV research focused on until 2002–2003, where severe acute respiratory syndrome coronavirus (SARS-CoV) was flared up. Thereafter, the Middle East respiratory syndrome coronavirus (MERS-CoV) and the 2019 novel coronavirus (SARS-CoV-2) have broken out [35, 36]. Differently from SARS-CoV, MERS-CoV and SARS-CoV-2 that bring about severe respiratory disease, HCoV-229E usually leads to mild to moderate upper-respiratory tract ailment, contributing to about 15–30% of human common cold cases [35]. HCoV-229E is an enveloped, single-stranded RNA virus. It is a member of Alphacoronavirus genus and Duvinacovirus subgenus [37].
S. marianum is rich in diverse secondary metabolites, including silymarin (which is a mixture of flavonolignans), phenolics, fatty acids and other chemical constituents [38]. The majority of previous studies focus only on the phytochemical and biological investigation of the flavonolignans constituents of S. marianum seeds and fruits [39–48], and up to authors’ knowledge there are not previous work on studying the whole metabolome and antiviral activity of all different parts of milk thistle. Therefore, the study in hand aims to investigate the whole chemical profile of different S. marianum organs including fruits, leaves, stems and roots using HPLC-MS/MS and chemometric analysis for the first time and to couple these data with the antiviral activity of these organs aiming at valorizing the unused milk thistle parts. The orthogonal projections to latent structures discriminant analysis (OPLS-DA) was performed to examine the class discrimination between the tested extracts and reveal the chemical markers accountable for such discrimination. Afterwards, the antiviral potentials of the tested extracts against HCoV-229E were determined on African green monkey kidney (Vero E6) cells using cytopathic effect (CPE) inhibition assay. Thereafter, different chemometric models were constructed to identify the biological markers responsible for the bioactive segregation of the studied extracts to exploit them as potential sources of valuable antiviral agents.
Experimental
Collection of the plant material
Five separate samples of the plant material were collected during the flowering-fruiting stage from farms belonging to the Faculty of Agriculture, Alexandria University, Egypt, in July 2022. The plant identity was confirmed via comparison with herbal sample present in the herbarium of the Faculty of Science, Alexandria University, Egypt. A voucher specimen (SM2022) was held at the Department of Pharmacognosy-Faculty of Pharmacy-Alexandria University. The collected plant materials were allowed to dry at room temperature prior to phytochemical analysis.
Preparation of samples
Every plant sample was split up into four organs: fruits, leaves, roots, and stems. Every organ sample (100 g) was extracted individually by ultrasonication in 200 mL of 70% ethanol using an ultrasonic bath 28 kHz/1100 W for 30 min at 45 ºC twice. The filtrates of each organ were collected and evaporated to dryness using a rotary evaporator, under reduced pressure, at 45ºC to get a total of 20 samples.
Profiling the metabolome of S. marianum different organs extracts using UPLC-MS/MS
Samples preparation for UPLC-MS analysis
Methanolic solutions with concentrations of 1 mg ml−1 were prepared for each sample. These solutions were subjected to filtration via membrane filters (0.2 μm) and degassing by sonication before being analyzed via LC-MS. To ensure reproducibility, the above process was performed three times for every sample.
UPLC-ESI- TQD -MS analysis
Chromatographic parameters and conditions
The UPLC system consists of a Waters Acquity QSM pump, an LC-2040 (Waters) autosampler, degasser and Waters Acquity CM detector. 10 µL of each of the previously prepared samples (full loop injection volume) were separately injected into the chromatographic column three times. Chromatographic separation was conducted using a Waters Acquity UPLC BEH C18 column (50 mm × 2.1 mm ID × 1.7 μm particle size) operating at a flow rate of 0.2 mL/min and thermostating at 30 °C.
The analyses were performed using a binary mobile phase consisting of ultrapure water + 0.1% (v/v) formic acid (Phase A) and methanol + 0.1% (v/v) formic acid (Phase B). The mobile phase was prepared by filtration using 0.2 μm membrane disc filter and degassed by sonication before injection. It was pumped at 0.2 mL/min into the UPLC system. The mobile phase gradient elution was programmed as follows: 0.0–2.0 min, 10% B; 2.0–5.0 min, 30% B; 5.0–15.0 min, 70% B; 22.0 min, 90% B; 22.0–25.0 min, 90% B; 26.0 min, 100% B; 26.0–29.0 min, 100% B; 30.0 min, 10% B; followed by 4 min of re-equilibration.
ESI-MS parameters and conditions
For LC/MS analysis, a triple quadrupole mass spectrometer was coupled to the UPLC instrument via an ESI interface. Ultra-high purity helium (He) was used as the collision gas and high purity nitrogen (N2) as the nebulizing gas. The mass spectrometer was monitored in negative ionization mode over 50–1200 m/z mass range. The optimized detection parameters were as follows: temperature 150 °C, cone voltage 30 V, capillary voltage 3 kV, desolvation temperature 440 °C, cone gas flow 50 L/h, and desolvation gas flow 900 L/h. A source fragmentation voltage of 25 V was applied. The mass spectrometer was operated in negative ion mode in order to identify the molecular ions [M-H]− followed by MS/MS product ion experiments to study the fragmentation pattern of the constituents. The analysis process run time lasted for 40 min. Regarding automatic MS/MS fragmentation process of the precursor ions that have been filtered by the first quadrupole (Q1), the mass fragmentation was performed through collision-induced dissociation (CID) energy utilizing Ultra-high purity helium in the second quadrupole (Q2). Eventually, the third quadrupole mass analyzer (Q3) filtered the daughter ions produced from CID that consequently related to the molecular structure of the precursor ions. The collision energy for CID in tandem mass spectrometry analysis, was optimized for each compound, in order to acquire mass spectra with various fragmentation degrees from the precursor ion thus attaining as much structural information as possible. A data-dependent program was utilized for tandem mass spectrometry data acquisition. In this program, molecular ions detected in the negative ion mode were selected for MS2 analysis and the two most abundant fragment ions in the MS2 spectra were then selected for further MS3 fragmentation.
Annotation of UPLC-MS/MS compounds
The raw UPLC–MS data were pre-processed using Mzmine® version 2.8 software that has been utilized for importing data, chromatogram building, peak deconvolution, alignment and annotation. Tentative assignment of metabolites was established via comparing their retention times relative to standards (which were caffeic acid, malic acid, quercetin, coumarin, p-coumaryl alcohol, lanosterol, and linoleic acid that were used as standards to their respective chemical classes), interpreting tandem mass spectra (quasi-molecular ions as well as diagnostic MS/MS fragmentation profiles) combined with our in-house comprehensive database that was set up covering all compounds previously reported in the literature including Dictionary of Natural Products (https://dnp.chemnetbase.com/), PubChem and MassBank (https://massbank.eu/MassBank/) to provide high confidence level of annotation [49, 50].
Semi-quantitation of identified compounds using UPLC-MS/MS
The annotated compounds were semi-quantified in accordance with their chemical class by the use of standard compound solutions. Caffeic acid, malic acid, quercetin, coumarin, p-coumaryl alcohol, lanosterol, and linoleic acid were used as standards for their chemical classes, and they were procured from Sigma–Aldrich (St. Louis, Mo., USA). Stock methanolic solutions, each with concentration of 1 mg ml−1, were prepared for every standard compound. These stock solutions were then diluted to generate working concentrations extending from 0.0125 to 0.625 mg mL−1 using HPLC-grade methanol (Table 1). Each standard solution concentration was analyzed three times under the previously described conditions in UPLC-ESI- TQD -MS analysis section. The standards were analyzed in the same order shown in Table 1: caffeic acid, then malic acid, quercetin, coumarin, p-coumaryl alcohol, lanosterol, then linoleic acid. The calibration curves were constructed by plotting standards peak areas versus their concentrations. For each calibration curve, the equation is y = ax + b, where y is the peak area, x is the concentration of the standard (mg mL−1), a is the intercept, b is the slope and r is the correlation coefficient.
Table 1.
Linearity and sensitivity parameters for caffeic acid, malic acid, quercetin, p-coumaryl alcohol, coumarin, lanosterol and linoleic acid used as S. marianum standards
| Standard compounds | Linearity range (mg mL−1) | Slope (b) | Intercept (a) | r | LOD (mg mL−1) | LOQ (mg mL−1) | |
|---|---|---|---|---|---|---|---|
| 1 | Caffeic acid | 0.0125–0.25 | 32*105 | 35*107 | 0.996 | 0.005 | 0.0125 |
| 2 | Malic acid | 0.025–0.536 | 2.87*108 | -8.35*109 | 0.993 | 0.010 | 0.025 |
| 3 | Quercetin | 0.02–0.525 | 20.51*106 | -1.07*108 | 0.995 | 0.012 | 0.02 |
| 4 | Coumarin | 0.055–0.625 | 5170 | 8.15*108 | 0.994 | 0.003 | 0.0325 |
| 5 | p-Coumaryl alcohol | 0.0325–0.625 | 41.71*106 | -10.39*108 | 0.997 | 0.015 | 0.055 |
| 6 | Lanosterol | 0.0125-0.25 | 1317.3 | 84.58*106 | 0.993 | 0.005 | 0.0125 |
| 7 | Linoleic acid | 0.0135–0.255 | 30.71*102 | 14.99*106 | 0.995 | 0.003 | 0.0135 |
Multivariate statistical analysis
Semiquantitative analysis and biological activity testing were statistically analyzed via ANOVA (one-way analysis of the variance) hiring SPSS 26.0 program (SPSS Inc., Chicago, IL. USA) and Metaboanalyst 4.0 (http://www.metaboanalyst.ca/) which is a web-based tool for processing metabolomics data to construct hierarchical cluster analysis (HCA) heat maps.
In addition, SIMCA v 14 software (Umetrics, Sweden) was applied for the construction of Orthogonal Projections to Latent Structures-Discriminant Analysis model (OPLS-DA) followed by Orthogonal Projections to Latent Structures (OPLS) model that enabled the discrimination of different milk thistle organs extracts based on their chemical profile in addition to antiviral activity. OPLS-DA model enabled the identification of the phytoconstituents that generated such discrimination. Meanwhile, careful examination of the OPLS correlation coefficient plots enabled us to identify the metabolites strongly correlated to the investigated biological activity. Permutations plots were created to validate that the created models were not modelling the noise or over-fitted.
Selective virucidal activity of S. marianum different organs extracts against human coronavirus (HCoV-229E) using cytopathic effect (CPE) inhibition assay
The crystal violet method was used to evaluate antiviral and cytotoxic activities according to Schmidtke et al. (2001) [51]. In brief, Vero E6 cells (Nawah-Scientific, Egypt) were seeded into a 96-well plate at a density of 2 × 104 cells/well one day before infection. Vero E6 cells were cultured in DMEM with 10% fetal bovine serum (FBS) and 0.1% antibiotic/antimycotic solution provided by Gibco BRL (Grand Island, NY, USA). After removing the culture medium the next day, the cells were washed with phosphate-buffered saline. Determination of coronavirus 229E (Nawah-Scientific, Egypt) infectivity was performed using the crystal violet method to monitor CPE and calculate the percentage of cell viability. 0.1 mL of diluted viral suspension of 229E virus with CCID 50 (50% cell culture infective dose of virus stock) was added to mammalian cells to attain the desired CPE after infection. Regarding samples’ treatments, 0.01 mL of desired extract-containing medium was added to the cells. Each test sample’s antiviral activity was estimated by a two-fold diluted concentration range of 0.1–100 µg/mL. The virus controls (virus-infected, non-drug-treated cells) and cell controls (non-infected, non-drug treated cells) were used. For 3 days, culture plates were incubated at 37oC in 5% carbon dioxide. The development of CPE was monitored by light microscopy. Following a PBS wash, fixation then staining of the cell monolayers was done using a 0.03% crystal violet solution in 2% EtOH and 3% formalin. Following washing and drying the optical densities (OD) of individual wells were quantified spectrophotometrically at 540/630 nm. The percentage of antiviral activity of the tested extracts was calculated by Pauwels et al. (1988) method [52], adopting the following equation:
Prior to conducting this assay, we assessed the cytotoxicity on normal cells, cells were seeded at a density of 2 × 104 cells/well in 96-well plate. The next day, the serially diluted extracts- containing culture media were added to the cells then incubated for 48 h then removed and the cells were washed with PBS. The following steps were performed as previously illustrated in the antiviral activity assay. GraphPad PRISM V 8 (San Diego, USA) software was used for determination of 50% cytotoxic concentrations (CC50) and 50% inhibitory concentrations (IC50).
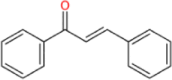
Results and discussion
Characterization of metabolites in S. marianum different organs
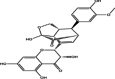
Metabolite profiling of S. marianum tested extracts was accomplished using UPLC-MS-MS. Figure 1 represents the base peak chromatograms of the studied extracts from which 87 metabolites were tentatively annotated by their retention times comparison to references and examining their MS data (Table 2). The details of characterization and fragmentation patterns of the identified metabolites are illustrated below.
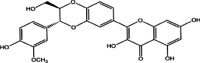
Fig. 1.
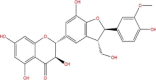
UPLC-ESI- TQD-MS base peak chromatograms of S. marianum extracts in negative ionization mode. Fruit (A), leaves (B), root (C) and stem (D) chromatograms
Table 2.
Metabolites identified in different organs of S. marianum extracts using UPLC-MS in negative ionization mode
| No. | Rt (min.) | [M-H]− | Element composition | MSn ions m/z (-) | Identified compounds | Chemical class | Structure | Reference |
|---|---|---|---|---|---|---|---|---|
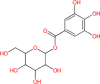
| 1 | 1.36 | 331.25 | C13H16O10 | 169.11, 125.11 | Gallic acid hexoside | Phenolic acid glycoside |
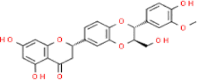
|
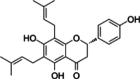
Li et al. 2003 [53] |
| 2 | 1.57 | 327.26 | C14H16O9 | 312.23, 192.13, 164.12 | Bergenin | Phenolic acid glycoside |

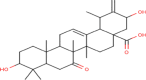
|
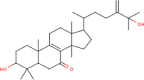
Li et al. 2013 [54] |
| 3 | 1.69 | 341.29 | C15H18O9 | 179.15, 135.15 | Caffeic acid-O-hexoside | Phenolic acid glycoside |

|
Singh et al. 2016 [55] |
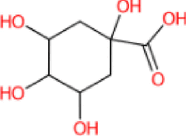
| 4 | 1.81 | 191.16 | C7H12O6 | 147.16 | Quinic acid | Phenolic acid |
|
Haşimi et al. 2017 [56] |
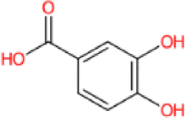
| 5 | 2.01 | 153.11 | C7H6O4 | 109.11 | Protocatechuic acid | Phenolic acid |
|
Haşimi et al. 2017 [56] |
| 6 | 2.32 | 167.14 | C8H8O4 | 152.11, 123.14 | Vanillic acid | Phenolic acid |
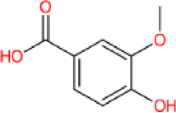
|
Haşimi et al. 2017 [56] |
| 7 | 2.48 | 197.17 | C9H10O5 | 182.14, 153.17 | Syringic acid | Phenolic acid |
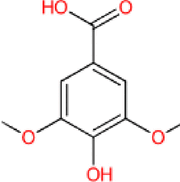
|
Haşimi et al. 2017 [56] |
| 8 | 2.66 | 179.15 | C9H8O4 | 135.15 | Caffeic acida | Phenolic acid |

|
Haşimi et al. 2017 [56] |
| 9 | 7.91 | 163.15 | C9H8O3 | 119.15 | p-Coumaric acid | Phenolic acid |
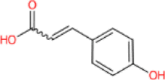
|
Haşimi et al. 2017 [56] |
| 10 | 8.70 | 193.18 | C10H10O4 | 178.15, 149.18 | Ferulic acid | Phenolic acid |
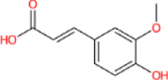
|
Haşimi et al. 2017 [56] |
| 11 | 9.98 | 147.15 | C9H8O2 | 103.15 | Cinnamic acid | Unsaturated carboxylic acid |
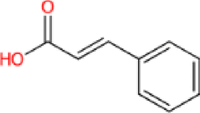
|
Haşimi et al. 2017 [56] |
| 12 | 10.14 | 353.3 | C16H18O9 | 179.15, 191.16 | Chlorogenic acid | Phenolic acid |
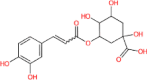
|
Haşimi et al. 2017 [56] |
| 13 | 10.85 | 275.28 | C15H16O5 | 201.25 | Ursinoic acid | Terpenoid |
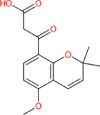
|
Hines et al. 2017 [57] |
| 14 | 11.01 | 133.08 | C4H6O5 |
115.06 71.06 |
Malic acida | Dicarboxylic acid |
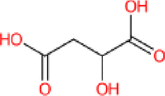
|
Al Kadhi et al. 2017 [58] |
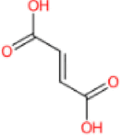
| 15 | 11.12 | 115.06 | C4H4O4 | 71.06 | Fumaric acid | Dicarboxylic acid |
|
Al Kadhi et al. 2017 [58] |
| 16 | 11.74 | 607.49 | C27H28O16 | 313, 295, 285.23, 151.1, 133.12 | Kaempferol 3,7-dihexoside | Flavonoid |

|
Said et al. 2017 [59] |
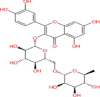
| 17 | 12.18 | 609.51 | C27H30O16 | 301.23, 272.21, 255.2, 151.1 | Rutin | Flavonoid |
|
Kumar et al. 2017 [60] |
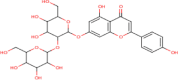
| 18 | 12.22 | 593.5113 | C27H30O15 | 269.23, 151.1, 117.13 | Apigenin 7-dihexoside | Flavonoid |
|
Liu et al. 2010 [61] |
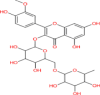
| 19 | 12.31 | 623.54 | C28H32O16 | 315.26, 300.22, 271.2, 151.1 | Isorhamnetin-3-O-dihexoside | Flavonoid |
|
Krasteva and Nikolov, 2008 [62] |
| 20 | 12.39 | 579.53 | C27H32O14 | 271.25, 151.1, 119.14, 107.09 | Naringin | Flavonoid |
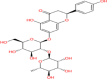
|
Santoro et al. 2020 [63] |
| 21 | 12.45 | 325.29 | C15H18O8 | 163.15, 119.14 | Coumaroyl hexoside | Phenolic acid glycoside |

|
Li et al. 2003 [53] |
| 22 | 12.51 | 491.38 | C22H20O13 | 315.26, 300.22, 271.2, 151.1 | Isorhamnetin-3-O-hexuronide | Flavonoid |

|
Krasteva and Nikolov, 2008 [62] |
| 23 | 12.71 | 447.37 | C21H20O11 | 285.23, 151.1, 133.12 | Luteolin-7-O-hexoside | Flavonoid |
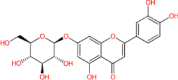
|
Santoro et al. 2020 [63] |
| 24 | 12.80 | 477.4 | C22H22O12 | 325, 315.26, 300.22, 271.2, 151.1 | Isorhamnetin-3-O-hexoside | Flavonoid |
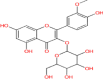
|
Krasteva and Nikolov, 2008 [62] |
| 25 | 12.89 | 431.37 | C21H20O10 | 269.23, 151.1, 117.13 | Apigenin-7-O-hexoside | Flavonoid |
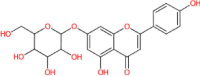
|
Liu et al. 2010 [61] |
| 26 | 13.20 | 432.38 | C21H21O10+ | 286.24, 151.1, 135.1 | Cyanidin-3-O-deoxyhexoside | Flavonoid |
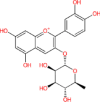
|
Santoro et al. 2020 [63] |
| 27 | 13.47 | 433.39 | C21H22O10 | 271.25, 151.1, 119.14, 107.09 | Naringenin 7-O- hexoside | Flavonoid |

|
Santoro et al. 2020 [63] |
| 28 | 13.53 | 457.36 | C22H18O11 | 125.1, 169.11 | (-)-Epigallocatechin gallate | Flavonoid gallic acid ester |

|
Spáčil et al. 2010 [64] |
| 29 | 13.91 | 461.40 | C22H22O11 | 285.23, 151.1, 133.12 | Rhamnocitrin-O-hexoside | Flavonoid |
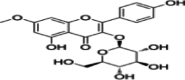
|
Said et al. 2017 [59] |
| 30 | 14.21 | 415.37 | C21H20O9 | 253.23, 135.1, 117.13 | Daidzein-7-O-hexoside | Flavonoid |
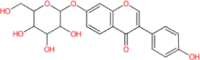
|
Santoro et al. 2020 [63] |
| 31 | 14.44 | 441.37 | C22H18O10 | 125.1, 109.1, 169.11 | Catechin gallate | Flavonoid gallic acid ester |

|
Spáčil et al. 2010 [64] |
| 32 | 14.95 | 445.4 | C22H22O10 | 283.26, 268.23, 224.23 | Trifolirhizin | pterocarpanoid glycoside |
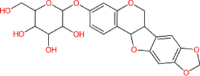
|
Ye et al. 2012 [65] |
| 33 | 15.10 | 473.41 | C23H22O11 | 269.23, 151.1, 117.13 | Apigenin 7- hexuronide, Et ester | Flavonoid |
|
Liu et al. 2010 [61] |
| 34 | 15.33 | 429.4 | C22H22O9 | 267.26, 132.16, 135.1 | Ononin | Flavonoid |

|
Santoro et al. 2020 [63] |
| 35 | 15.48 | 519.52 | C26H32O11 | 357.38, 339.36, 221.23, 191.2 | Dehydrodiconiferyl alcohol-hexoside | Neolignan |
|
Letourneau and Volmer, 2021 [66] |
| 36 | 15.54 | 161.13 | C9H6O3 | 133.12 | 4-Hydroxycoumarin | Coumarin derivative |
|
Ren et al. 2016 [67] |
| 37 | 15.62 | 145.14 | C9H6O2 | 117.13 | Coumarina | O-hydroxycinnamic acid (Aromatic compound) |

|
Ren et al. 2016 [67] |
| 38 | 15.80 | 149.17 | C9H10O2 | 131.15 | p-Coumaryl alcohola | Monolignol |
|
Liu et al. 2010 [61] |
| 39 | 15.89 | 175.16 | C10H8O3 | 147.15 | 4-Methylumbelliferone | Coumarin derivative |
|
Ren et al. 2016 [67] |
| 40 | 15.99 | 181.21 | C10H14O3 | 163.19 | 2-Hydroxymethyl-5-(2-hydroxypropan-2-yl) phenol | Aromatic compound |

|
Dictionary of Natural Products 30.2 Chemical Search |
| 41 | 16.09 | 181.21 | C10H14O3 | 163.19 | p-Mentha-1,3,5-triene-2,7,8-triol | Aromatic compound |
|
Dictionary of Natural Products 30.2 Chemical Search |
| 42 | 16.13 | 217.2 | C12H10O4 | 175.16 | 4-Methylumbelliferyl acetate | Coumarin derivative |
|
Leonart et al. 2017 [68] |
| 43 | 16.31 | 301.23 | C15H10O7 | 272.21, 255.2, 151.1 | Quercetina | Flavonoid |
|
Kumar et al. 2017 [60] |
| 44 | 16.43 | 303.24 | C15H12O7 | 285.22, 275.23, 151.1, 125, 175 | Taxifolin (Dihydroquercetin) | Flavonoid |
|
Chen et al. 2016 [69] |
| 45 | 16.56 | 305.26 | C15H14O7 | 125.1 | Epigallocatechin | Flavonoid |
|
Spáčil et al. 2010 [64] |
| 46 | 16.61 | 285.23 | C15H10O6 | 151.1, 133.12 | Kaempferol | Flavonoid |
|
Said et al. 2017 [59] |
| 47 | 16.70 | 287.24 | C15H12O6 | 151.1, 135.14 | Dihydrokaempferol (Aromadendrin) | Flavonoid |
|
Said et al. 2017 [59] |
| 48 | 16.77 | 315.26 | C16H12O7 | 300.22, 271.2, 151.1 | Isorhamnetin | Flavonoid |

|
Krasteva and Nikolov, 2008 [62] |
| 49 | 16.89 | 269.23 | C15H10O5 | 151.1, 117.13 | Apigenin | Flavonoid |

|
Liu et al. 2010 [61] |
| 50 | 17.10 | 269.23 | C15H10O5 | 241.22, 225.23 | Genistein | Flavonoid |
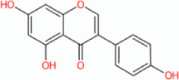
|
Ye et al. 2012 [65] |
| 51 | 18.38 | 271.25 | C15H12O5 | 151.1, 119.14, 107.09 | Naringenin | Flavonoid |
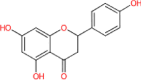
|
Santoro et al. 2020 [63] |
| 52 | 19.27 | 273.26 | C15H14O5 | 151.1, 121.16 | Phloretin | Chalcone derivative |
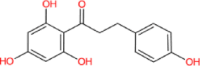
|
Bystrom et al. 2008 [70] |
| 53 | 20.13 | 329.28 | C17H14O7 | 314.25, 299.22, 271.21 | Tricin | Flavonoid |
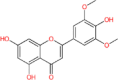
|
Kang et al. 2016 [71] |
| 54 | 20.63 | 283.26 | C16H12O5 | 269.23, 151.1, 117.13 | Genkwanin (7-methoxyapigenin) | Flavonoid |

|
Liu et al. 2010 [61] |
| 55 | 23.55 | 239.25 | C15H12O3 | 130.14, 102.13 | 2’,4’-Dihydroxychalcone | Chalcone derivative |

|
Zhang et al. 2014 [72] |
| 56 | 23.87 | 207.25 | C15H12O | 130.14, 102.13 | Chalcone | α,β-unsaturated ketone |
|
Motiur Rahman et al. 2013 [73] |
| 57 | 24.93 | 497.43 | C25H22O11 | 463, 453, 179, 125, 480, 470, 375, 355 | Silyamandin | Flavonolignan |

|
Csupor et al. 2016 [74] |
| 58 | 24.99 | 479.41 | C25H20O10 | 151, 451.1, 255.03, 466.09 | 2,3-Dehydrosilybin | Flavonolignan |
|
Csupor et al. 2016 [74] |
| 59 | 25.05 | 481.43 | C25H22O10 | 463, 453, 179, 125, 433, 423, 355, 337, 325 | Silychristin | Flavonolignan |
|
Csupor et al. 2016 [74] |
| 60 | 25.23 | 481.43 | C25H22O10 | 463, 453, 179, 125, 409, 151, 301 | Silydianin | Flavonolignan |
|
Csupor et al. 2016 [74] |
| 61 | 25.47 | 481.43 | C25H22O10 | 463, 453, 179, 125, 435, 301, 283, 273, 257, 423 | Silybin A | Flavonolignan |
|
Csupor et al. 2016 [74] |
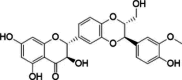
| 62 | 25.69 | 481.43 | C25H22O10 | 463, 453, 179, 125, 435, 301, 283, 273, 257, 423 | Silybin B | Flavonolignan |
|
Csupor et al. 2016 [74] |
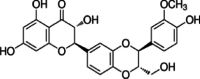
| 63 | 25.81 | 481.43 | C25H22O10 | 463, 453, 179, 125, 435, 301, 283, 273, 257 | Isosilybin A | Flavonolignan |

|
Csupor et al. 2016 [74] |
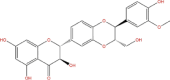
| 64 | 25.99 | 481.43 | C25H22O10 | 463, 453, 179, 125, 435, 301, 283, 273, 257 | Isosilybin B | Flavonolignan |
|
Csupor et al. 2016 [74] |
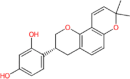
| 65 | 26.05 | 323.36 | C20H20O4 | 201.24 | Glabridin | Flavonoid |
|
Aoki et al. 2005 [75] |
| 66 | 26.15 | 465.43 | C25H22O9 | 151 | Silandrin | Flavonolignan |
|
Csupor et al. 2016 [74] |
| 67 | 26.28 | 407.48 | C25H28O5 | 119.14, 287.33 | 6,8-Diprenylnaringenin | Prenyl flavonoid |
|
Nikolic and van Breemen, 2012 [76] |
| 68 | 26.35 | 633.88 | C37H62O8 | 471.74, 357.55, 339.53 | 24-Methylenelanost-8-ene-3,25,28-triol, 3-O-hexoside | Lanostane triterpenoid |

|
Wu et al. 2011 [77] |
| 69 | 26.41 | 483.66 | C30H44O5 | 465.64, 421.64 | Silymin A | Ursane triterpenoid |
|
Ahmed et al. 2007 [78] |
| 70 | 26.46 | 469.72 | C31H50O3 | 371.53, 353.51 | 3,25-Dihydroxy-24-methylenelanost-8-en-7-one | Lanostane triterpenoid |
|
Wu et al. 2011 [77] |
| 71 | 26.50 | 179.19 | C13H8O | 151.18, 137.15 | 12-Tridecene-4,6,8,10-tetraynal | Unsaturated fatty aldehyde |

|
Suzuki et al. 1980 [79] |
| 72 | 26.58 | 179.19 | C13H8O | 164.16, 149.13 | 1,3-Tridecadiene-5,7,9,11-tetrayne, 1,2-epoxide | Unsaturated fatty epoxide |

|
Suzuki et al. 1980 [79] |
| 73 | 26.63 | 291.41 | C18H28O3 | 273.39, 247.4 | 12-oxo-phytodienoic acid | Unsaturated fatty acid |

|
Brenna, 2013 [80] |
| 74 | 26.70 | 277.42 | C18H30O2 | 259.4, 233.41 | Linolenic acid | Unsaturated fatty acid |
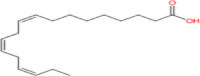
|
Brenna, 2013 [80] |
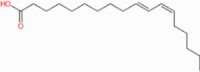
| 75 | 26.74 | 279.44 | C18H32O2 | 261.42, 235.43 | Linoleic acida | Unsaturated fatty acid |
|
Brenna, 2013 [80] |
| 76 | 26.83 | 281.45 | C18H34O2 | 263.43, 237.44 | Oleic acid | Unsaturated fatty acid |
|
Brenna, 2013 [80] |
| 77 | 26.88 | 241.35 | C17H22O | 223.33, 226.32 | 2,9,16-Heptadecatriene-4,6-diyn-8-ol | Unsaturated aliphatic alcohol |
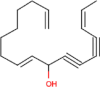
|
El Sayed et al. 2020 [81] |
| 78 | 27.03 | 309.51 | C20H38O2 | 291.49, 265.5 | Gadoleic acid | Unsaturated fatty acid |
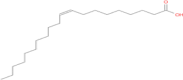
|
Brenna, 2013 [80] |
| 79 | 27.14 | 293.46 | C19H34O2 | 262.43, 221.4, 81.14 | Linoleic acid methyl ester | Unsaturated fatty acid methyl ester |
|
Fagerquist et al. 1999 [82]; Härtig, 2008 [83] |
| 80 | 27.18 | 295.48 | C19H36O2 | 264.45, 221.4, 55.1 | 16-octadecenoic acid methyl ester | Unsaturated fatty acid methyl ester |

|
Fagerquist et al. 1999 [82]; Härtig, 2008 [83] |
| 81 | 27.24 | 227.36 | C14H28O2 | 209.34, 183.35 | Myristic acid | Saturated fatty acid |
|
Brenna, 2013 [80] |
| 82 | 27.32 | 255.42 | C16H32O2 | 237.4, 211.41 | Palmitic acid | Saturated fatty acid |

|
Brenna, 2013 [80] |
| 83 | 27.41 | 283.47 | C18H36O2 | 265.45, 239.46 | Stearic acid | Saturated fatty acid |
|
Brenna, 2013 [80] |
| 84 | 27.51 | 311.52 | C20H40O2 | 293.5, 267.51 | Arachidic acid | Saturated fatty acid |
|
Brenna, 2013 [80] |
| 85 | 27.66 | 339.58 | C22H44O2 | 321.56, 295.57 | Behenic acid | Saturated fatty acid |
|
Brenna, 2013 [80] |
| 86 | 27.88 | 295.52 | C20H40O | 277.5, 71.14 | Phytol | Acyclic diterpene alcohol |

|
El Sayed et al. 2020 [81] |
| 87 | 28.30 | 409.63 | C28H42O2 | 394.6, 379.57, 151.19 | (R)-gamma-Tocotrienol | Fat soluble vitamin |
|
Zhao et al. 2010 [84] |
aThe standards used for retention times comparison and semi-quantitation of the annotated compounds were caffeic acid, malic acid, quercetin, coumarin, p-coumaryl alcohol, lanosterol, and linoleic acid
Phenolics
The mass spectra of compounds 1, 3 and 21 exhibited molecular ion peaks at m/z 331.25, 341.29 and 325.29, respectively. They were characterized as galloyl hexoside, caffeic acid-O-hexoside and coumaroyl hexoside respectively, due to the loss of hexoside (-162.14 Da) and CO2 (-44.01 Da) moieties and the presence of characteristic fragments at m/z 169.11 [M-H-hexoside]- and m/z 125.11 [M-H-hexoside-CO2]- for galloyl hexoside; at m/z 179.15 [M-H-hexoside]- and m/z 135.15 [M-H-hexoside-CO2]- for caffeic acid-O-hexoside; and at 163.15 [M-H-hexoside]- and m/z 119.14 [M-H-hexoside-CO2]- for coumaroyl hexoside [53, 55].
Whereas compound 2 having [M-H]- at m/z 327.26 was annotated as bergenin. This annotation was suggested by the daughter peaks in the MS2 spectrum at m/z 312.23 representing the loss of methyl group, and the characteristic Retro Diels-Alder (RDA) fragment at m/z 192.13, which then lost carbonyl group to yield another fragment ion at m/z 164.12 [54].
Furthermore, the MS spectra of compounds 5, 6, 7, 8, 9, 10 and 11 displayed [M–H]- ions at m/z 153.11, 167.14, 197.17, 179.15, 163.15, 193.18 and 147.15, respectively, together with their decarboxylated fragments. For compounds 6, 7 and 10 another fragment generated by methyl group (15.03 Da) loss was noticed in their MS2 spectra at m/z 152.11, 182.14, and 178.15, respectively. Therefore, these compounds were annotated as, protocatechuic acid (compound 5), vanillic acid (compound 6), syringic acid (compound 7), caffeic acid (compound 8), coumaric acid (compound 9), ferulic acid (compound 10) and cinnamic acid (compound 11), respectively [56].
Moreover, the MS spectrum of compound 12 revealed [M–H]− ion at m/z 353.3, in addition to characteristic daughter fragments at m/z 179.15 and 191.16 representing caffeic and quinic acids, respectively. Therefore, compound 12 was recognized as monocaffeoylquinic acid (chlorogenic acid) [56]. Also, quinic acid was present as free acid as shown in compound 4 spectrum which had a base peak [M-H]− ion at m/z 191.16.
Compound 13 displaying a parent ion peak at m/z 275.28 was proposed to be ursinoic acid. This annotation was suggested by its MS/MS fragment ion at m/z 201.25 [M-H-44-30.03]− owing to CO2 and methoxy group elimination [57].
Dicarboxylic acids
Two dicarboxylic acids were recognized. Malic acid (compound 14) was proposed for the parent ion at m/z 133.08 which was then fragmented to yield peaks at m/z 115.06 and 71.06 due to successive loss of H2O and CO2. Whereas fumaric acid (compound 15) was proposed for the parent ion at m/z 115.06 which decarboxylated to give fragment ion at m/z 71.06 [58].
Flavonoids
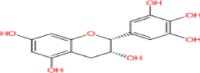
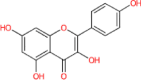
Kaempferol 3,7-dihexoside (compound 16) generated RDA fragments at m/z 313 and 295, indicating that both rings A and B of the flavonoidal structure contained a hexose moiety. It also demonstrated a characteristic ion at m/z 285.23 due to [C12H18O10]− loss. Rhamnocitrin-O-hexoside (compound 29) produced the same fragment ion because of successive loss of methyl and hexose units. These compounds together with kaempferol (compound 46) exhibited fragment ions at m/z 151.1 and 133.12 due to RDA reaction [85]. Compound 47 revealed a parent peak at m/z 287.24 which is 2 Da higher than that of kaempferol. Therefore, it was characterized as dihydrokaempferol (aromadendrin). It gave RDA fragments at m/z 151.1 and 135.14 [59].
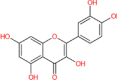
Similarly, rutin (compound 17) showed a distinguishing ion at m/z 301.23 due to [C12H20O9]− loss. This compound together with quercetin (compound 43) generated fragment ions at m/z 272.21 and 255.2 owing to eliminating [CHO]− and [CO + H2O]−, respectively. They also produced RDA fragment at m/z 151.1 [60].
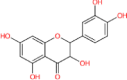
Peak 44 was assigned as taxifolin. It demonstrated its [M − H]− ion at m/z 303.24 and then lost H2O and CO from the C ring to generate fragment ions at m/z 285.22 and 275.23, respectively. There were also the ions at m/z 151.1 and m/z 125 which were generated from RDA reaction in the C ring. Additionally, the fragment ion at m/z 175 was generated from [M − H − H2O]− at m/z 285.22 due to loss of the B ring moiety. By referring to literature [69], peak 44 was recognized as taxifolin.
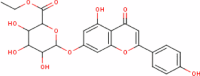
Furthermore, apigenin 7-dihexoside (compound 18), apigenin-7-O-hexoside (compound 25), apigenin 7- hexuronide, ethyl ester (compound 33) and genkwanin (7-methoxyapigenin) (compound 54) exhibited a characteristic fragment ion at m/z 269.23 because of elimination of [C12H20O10]− (dihexose), [C6H10O5]− (monohexose), [C8H12O6]− and [CH2]− units, respectively. These compounds together with apigenin (compound 49) generated fragment ions at m/z 151.1 and 117.13 owing to RDA reaction [61].
Meanwhile, isorhamnetin-3-O-dihexoside (compound 19), isorhamnetin-3-O-hexuronide (compound 22) and isorhamnetin-3-O-hexoside (compound 24) exhibited a distinct fragment ion at m/z 315.26 due to elimination of C12H20O9, C6H8O6 and C6H10O5, respectively. All these compounds and isorhamnetin (compound 48) showed fragment ions at 300.22 and 271.2 due to eliminating [CH3]- and [CH3 + CHO]−, respectively. They also showed the characteristic RDA fragment at m/z 151.1 [62]. Compounds 19, 22, and 24 exhibited their sugar moieties in the second RDA fragment that was at m/z 487, for compound 19; at m/z 339, for compound 22; and at m/z 325, for compound 24, indicating that ring A of these compounds were free of sugar moieties.
In addition, naringin (compound 20), naringenin 7-O- hexoside (compound 27) and naringenin (compounds 51) were fragmented similarly except that naringin and naringenin 7-O- hexoside had extra 308.28 and 162.14 Da corresponding to disaccharide and monosaccharide moieties, respectively. These compounds showed RDA fragments at m/z 151.1 and 119.14, and a fragment at 107.09 corresponding to [151.1-CO2]− [63]. Compound 52 having additional 2 Da to the molecular ion of naringenin was suggested to be dihydronaringenin (phloretin). It was affirmed by its RDA fragments at m/z 151.1 and 121.16 [70]. Whereas compound 67 with [M-H]− at m/z 407.48 and fragment ions at m/z 119.14 and 287.33 was proposed to be 6,8-diprenylnaringenin. It was identified by having extra 136 Da in its molecular ion and RDA fragment than those of naringenin [76].
Compounds 23, 26, 30, 32 and 34 were assigned as flavonoidal glycosides. They displayed their flavonoidal aglycones at m/z 285.23, 286.24, 253.23, 283.26 and 267.26, respectively. These compounds were annotated as luteolin-7-O-hexoside, cyanidin-3-O-deoxyhexoside, daidzein-7-O-hexoside, trifolirhizin and ononin, respectively. Compounds 23, 26, 30 and 34 were subjected to RDA reaction and generated characteristic fragments at m/z 151.1 and 133.12, for compound 23; 151.1 and 135.1, for compound 26; 135.1 and 117.13, for compound 30; and 132.16 and 135.1, for compound 34 [63]. While compound 32 generated fragments ions at m/z 268.23 and 224.23 owing to loss of methyl and CO2 groups, successively [65].
Two isoflavones were identified which are genistein (compound 50) and glabridin (compound 65). They displayed their molecular ions at m/z 269.23 and 323.36, successively. Genistein produced fragment ions in MS2 spectrum at m/z 241.22 and 225.23 corresponding to neutral loss of CO and CO2, respectively [65]. Whereas glabridin lost [C7H6O2]− to yield fragment ion at m/z 201.24 [75]. On the other hand, compound 53 was assigned to be the methoxylated flavone, tricin. It demonstrated its molecular ion at m/z 329.28 which was subjected to fragmentation to generate ions at m/z 314.25, 299.22 and 271.21 that are related to successive loss of two methyl and carbonyl groups [71].
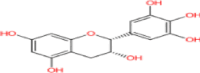
Moreover, compounds 28, 31 and 45 displaying molecular ions at m/z 457.36, 441.37 and 305.26 were characterized as epigallocatechin gallate, catechin gallate and epigallocatechin, respectively. The characterization was relied on the fragment ion at m/z 125.1 for the three compounds corresponding to [C6H5O3]−, which was originated after two bonds cleavage in ring C and it was composed of the phenolic ring A [64]. Catechin gallate was distinguished from epigallocatechin gallate and epigallocatechin by the existence of the fragment ion [C6H5O2]− at m/z 109.1 which was corresponding to ring B of catechin gallate. Epigallocatechin was differentiated from epigallocatechin gallate and catechin gallate by the lack of the fragment ion [C7H5O5]− at m/z 169.11 indicating the lack of gallate moiety attached to 3-OH [64].
Additionally, chalcone (compound 56) was recognized by the parent peak at m/z 207.25 and the daughter ion peaks in the MS2 spectrum at m/z 130.14, corresponding to the fragment ion [C9H7O-H]− which was formed by loss of one phenolic ring, and 102.13, related to cleavage of 1, 2 bond [73]. Compound 55 with additional 32 Da at the molecular ion and fragmented in the same way as chalcone was recognized as 2’,4’-dihydroxychalcone [72].
The most characteristic components of S. marianum is silymarin mixture. It was identified in the mass spectra by seven compounds, which are silyamandin (compound 57), silychristin (compound 59), silydianin (compound 60), silybin A (compound 61), silybin B (compound 62), isosilybin A (compound 63) and isosilybin B (compound 64). Silybin A and B, isosilybin A and B, silydianin and silychristin all had their [M-H]− ions at m/z 481.43, while silyamandin exhibited its [M-H]− ion at m/z 497.43. All seven compounds had similar fragment ions at m/z 463, 453, 179 and 125. Both silybin and isosilybin generated the following fragment ions in common; 435, 301, 283, 273, 257 m/z. However, silybin produced a fragment ion at m/z 423 that was not generated in case of isosilybin. On the other hand, silydianin produced characteristic fragment ions at m/z 409, 151 and 301 m/z. Meanwhile, silychristin produced the following fragment ions at m/z 433, 423, 355, 337 and 325 m/z. Finally, silyamandin exhibited distinguishable peaks at m/z 480, 470, 375 and 355. The fragmentation patterns of these flavonolignans were similar to those explained in literature [74]. Another two compounds were identified which are 2,3-dehydrosilybin (compound 58) and silandrin (compound 66). 2,3-dehydrosilybin was suggested by its molecular ion at m/z 479.41 which is 2 Da lower than that of silybin. Whereas silandrin (isosilybin; 3-deoxy) was proposed by its molecular ion at m/z 465.43 which is 16 Da lower than that of silybin.
Coumarins
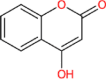
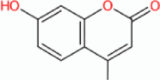
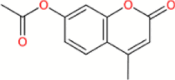
Coumarin (compound 37) was proposed for the parent ion at m/z 145.14 which was further fragmented to yield ion at m/z 117.13 due to loss of carbonyl group [67]. Compound 36 with extra 16 Da in both parent and daughter ions was identified as 4-hydroxycoumarin [67]. Compounds 39 with additional 14 Da than compound 36 was suggested to be 4-Methylumbelliferone. It fragmented in the same way as compounds 36 and 37 by loss of carbonyl group to give fragment ion at m/z 147.15 [67]. Additionally, compound 42 giving parent peak at m/z 217.2 was proposed to be 4-Methylumbelliferyl acetate. It exhibited daughter ion peak at m/z 175.16 related to elimination of acetyl group [M-H-42.04]− [68]. Up to the authors’ knowledge, it is the first report of coumarins in S. marianum.
Alcohols
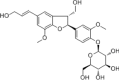
Compound 35 having parent peak at m/z 519.52 was annotated as dehydrodiconiferyl alcohol-hexoside. It was recognized by its daughter ions in the MS/MS spectrum at m/z 357.38 [M-H-162.14]−, corresponding to loss of hexoside moiety, 339.36 [M-H-162.14-18.02]−, due to lack of H2O, 221.23 and 191.2, related to subsequent loss of [C8H8O2]− and methoxy group [66].
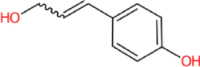
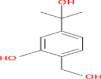
Furthermore, compound 38 giving rise to deprotonated ion at m/z 149.17 was annotated as p-coumaryl alcohol. Upon fragmentation, it generated a daughter ion peak at m/z 131.15 because of water loss. Similarly, compounds 40 and 41 having molecular ions at m/z 181.21 and fragment ions at m/z 163.19 [M-H-H2O]− were identified as 2-hydroxymethyl-5-(2-hydroxypropan-2-yl) phenol and p-mentha-1,3,5-triene-2,7,8-triol, respectively [86].
Triterpenes
Three triterpenoids were identified. One of them was silymin A (compound 69) which is ursane- type triterpene exhibiting its parent peak at m/z 483.66 and having daughter peaks at m/z 465.64 and 421.64 due to successive loss of H2O and CO2 [78]. The other two triterpenoids are 24-methylenelanost-8-ene-3,25,28-triol, 3-O-hexoside (compound 68) and 3,20-dihydroxy-24-methylenelanost-8-en-7-one (compound 70) which belong to lanostane-type triterpenes. Compound 68 was annotated according to its parent peak at m/z 633.88 and daughter peaks at m/z 471.74, 357.55 and 339.53 related to [M-H-C6H10O5]-, [M-H-C6H10O5-C7H14O]- and [M-H-C6H10O5-C7H14O-H2O]-, respectively. Whereas compound 70 demonstrated its parent ion peak at m/z 469.72 and generated daughter peaks at m/z 371.53 and 353.51 related to [M-H-C7H14]- and [M-H-C7H14-H2O]-, successively [77].
Fatty acids
12-Tridecene-4,6,8,10-tetraynal (compound 71) and 1,3-Tridecadiene-5,7,9,11-tetrayne,1,2-epoxide (compound 72) were recognized by their molecular ions at m/z 179.19. They were differentiated by their different fragmentation patterns demonstrated in their MS2 spectra. Compound 71 showed fragment ions at m/z 151.18 and 137.15 due to successive elimination of carbonyl and methyl groups. Whereas compound 72 produced fragment ions at m/z 164.16 and 149.13, owing to sequential loss of two methyl groups [79].
Moreover, the MS spectra of compounds 73, 74, 75, 76, 78, 81, 82, 83, 84 and 85 demonstrated molecular ion peaks at m/z 291.41, 277.42, 279.44, 281.45, 309.51, 227.36, 255.42, 283.47, 311.52 and 339.58, respectively, together with their fragment ions related to neutral loss of H2O and CO2. These compounds were recognized as 12-oxo-phytodienoic acid, linolenic acid, linoleic acid, oleic acid, gadoleic acid, myristic acid, palmitic acid, stearic acid, arachidic acid and behenic acid, respectively [80].
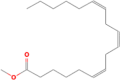
Furthermore, two fatty acids methyl esters were recognized by their deprotonated ions at m/z 293.46 (compound 79) and 295.48 (compound 80). They yield fragment ions in their MS2 spectra at m/z 262.43 (for compound 79) and 264.45 (for compound 80) because of methoxy group loss, and at m/z 221.4 corresponding to lack of McLafferty ion which is characteristic to methyl esters [82]. Compound 79 had a base peak at m/z 81.14 due to lack of the hydrocarbon ion [C6H9]-, while compound 80 demonstrated a base peak at m/z 55.1 owing to lack of the hydrocarbon ion [C4H7]- [83]. Accordingly, these compounds were proposed to be linoleic acid methyl ester (compound 79) and 16-octadecenoic acid methyl ester (compound 80).
In addition, compounds 77 and 86 with parent ions at m/z 241.35 and 295.52 were suggested to be 2,9,16-heptadecatriene-4,6-diyn-8-ol and phytol, respectively. These aliphatic alcohols were assured by their fragment ions corresponding to loss of water at m/z 223.33 (for compound 77) and 277.5 (for compound 86). Moreover, 2,9,16-heptadecatriene-4,6-diyn-8-ol generated another fragment ion at m/z 226.32 due to lack of terminal methyl group. Whereas phytol exhibited a base peak at m/z 71.14 because of the hydrocarbon ion [C5H11]- loss [81].
Finally, (R)-gamma-tocotrienol (compound 87) was identified by its parent ion at m/z 409.63 and fragment ions in the MS/MS spectrum at m/z 394.6 [M-H-CH3]-, 379.57 [M-H-CH3-CH3]- and 151.19 [M-H-C19H30]- [84].
Thereafter, all identified compounds in S. marianum samples were subjected to relative quantitation via the calibration curves illustrated in Semi-quantitation of identified compounds using UPLC-MS/MS section. and their relative contents are presented in Fig. 2 & Table S1.
Fig. 2.
Relative quantitation of the total phenolics, dicarboxylic acids, flavonoids, coumarins, alcohols, triterpenes, and fatty acids in different organs of S. marianum expressed as mg Equivalents (Eq.)/g dry weight
Unsupervised HCA-heat map for chemical profiling of S. marianum different organs
In this section, comparative chemical profiling of S. marianum fruits, leaves, roots and stems was attempted using UPLC-tandem mass analysis combined with multivariable statistical analysis. The semi-quantitative data of characterized compounds in the previous section (Table S1) were implemented to conduct an unsupervised dendritic analysis for the extracts under investigation. As shown in Figs. 1, 2 and 3 considerable variation in chemical profile of milk thistle different organs was observed. Hierarchical clustering analysis (HCA)- heat map (Fig. 3) showed the grouping of the different S. marianum organ extracts into three separate clusters, the first was assigned for the five fruit samples, the second allocated for the root samples while the third one was split into two subclusters; namely the leaves and stem samples indicating relative proximity in their chemical composition and proving the previous findings gained by Javeed et al. that calculated the total phenolic and flavonoid contents in different milk thistle parts and revealed that the leaves and stems extracts were enriched with higher amounts of them [87].
Fig. 3.
Hierarchical analysis heat maps of all identified metabolites in fruit, leaves, roots and stems of S. marianum. Brick red and blue indicate higher and lower abundances, respectively
It was observed that S. marianum fruit samples possessed the highest relative content of flavonolignans such as 2,3-dehydrosilybin, silymin A, silydianin, isosilybin A, silybin A and B, and this finding is in a good agreement with that reported by Korany et al. [39]. Other metabolic classes were diversely distributed among different organ clusters and subclusters as indicated by the dark red color code (Fig. 3). Four compounds were found in all the studied milk thistle organs, namely the phenolic acid “cinnamic acid” which recorded highest accumulation in stems and two flavonoidal aglycones namely naringenin and tricin which were both highly accumulated in fruits followed by seeds, and finally the fatty acid ester “16-octadecenoic acid methyl ester” which was also detected in greater amount in milk thistle fruits. Meanwhile, close observation of Fig. 3. revealed that the two coumarins 4-methylumbelliferone and 4-hydroxycoumarin in addition to 6,8-diprenylnaringenin, 2-hydroxymethyl-5-(2-hydroxypropan-2-yl) phenol and the two flavonolignans silybin A and B were the main constituents in fruit samples. In contrast, milk thistle leaves samples exhibited greater accumulation of the phenolic acid glycoside “caffeic acid-O-hexoside” and the aglycone “quinic acid”. Further, isorhamnetin, gallic acid hexoside, bergenin, caffeic acid-O-hexoside, and apigenin were the major compounds found in roots samples. Finally, cinnamic and syringic acids, genistein, apigenin-7-O-hexoside, taxifolin and phloretin were the main detected constituents in milk thistle stems.
It is worthy to mention that, up to the authors’ knowledge, this is the foremost comprehensive evaluation of S. marianum different organs chemical profiles.
OPLS-DA for supervised multivariate discrimination between different organs
For the sake of inter- and intra-class discrimination of fruits, leaves, roots and stems samples; an OPLS-DA multivariate model was created utilizing the MS data obtained from LC-MS/MS analysis (Fig. 4A and B). Moreover, OPLS-DA was able to unravel the discriminatory markers characteristic for each class chemical profile via coefficient plots of each organ separately (Fig. 4C and F). The first and second latent variables of the constructed model accounted for 46.3% and 30.1% of the variability, respectively. Moreover, the model exhibited high reliability and prediction ability represented by high goodness of fitness (R2 = 0.998) and goodness of prediction (Q2 = 0.996). For validation of the current OPLS-DA model; permutation plots for fruits, leaves, roots and stems (Fig. S1) using 20 permutations for each class were constructed. The blue regression line of Q2 points intersected with vertical axis below the zero, while the green R2 values to the left were lower to the original point to the right which strongly indicated the model validity. ROC curves (Fig. S2) were constructed, and AUC were found to be equal to one for all classes indicating the excellent classification power of the model.
Fig. 4.
OPLS-DA scatter plot (t1 scores vs. t2 scores) (A), Dendrogram derived from the hierarchical cluster analysis, based on the Ward method of fruits, leaves, roots and stems samples of S. marianum (B), Coefficient plots of the OPLS-DA model of S. marianum fruits (C), leaves (D), roots (E), and stems (F) to determine discriminative metabolites
In between class discrimination along the first latent variable (LV1) was observed in the 2D score scatter plot (Fig. 4A) where all the fruit samples where successfully grouped along its positive side, while other organ samples were on the negative side of LV1. Whereas the second latent variable (LV2) successfully separated the root samples on its negative side from the leaves and stem samples on the positive side of the same LV. This classification was in agreement with that observed OPLS-DA dendrogram (Fig. 4B) where it revealed the existence of two principal clusters; one for the fruit samples and the other was additionally sub-clustered into a subcluster for roots and another one comprised of tested stems and leaves samples. The OPLS-DA coefficients plots (Fig. 4C and F) allowed the recognition of phytoconstituents responsible for the segregation of each milk thistle organ samples into separate class. Caffeic acid, naringenin 7-O-hexoside, silydianin, silybin B, isosilybin A and silybin A were the main differentiating markers certainly correlated to fruit samples (Fig. 4C). Meanwhile, daidzein-7-O-hexoside, silandrin, linolenic acid, 1,3-tridecadiene-5,7,9,11-tetrayne 1,2-epoxide, kaempferol 3,7-dihexoside, isorhamnetin-3-O-hexuronide and isosilybin B were found to be the foremost constituents related to leaves samples class (Fig. 4D). In contrast, the flavonoidal aglycone isorhamnetin, coumaroyl hexoside, behenic acid, 12-tridecene-4,6,8,10-tetraynal, p-coumaric acid and ononin were the positively related compounds to root class (Fig. 4E). Moreover, the flavones genistein, dihydrokaempferol and apigenin-7-O-hexoside, the phenolic acids cinnamic, syringic and chlorogenic acid, silychristin, phloretin and linoleic acid were the principal differentiating markers showing positive correlation to stems class (Fig. 4F).
Selective virucidal activity of the tested S. marianum organs extracts against human coronavirus (HCoV-229E) using cytopathic effect (CPE) inhibition assay
The appearance of drug-resistant respiratory viral strains to currently used antivirals such as oseltamivir, zanamivir, peramivir, and laninamivir [88] makes the development of natural selective alternatives with diminished toxicity urgently required. In this context, selective virucidal activity of the tested milk thistle organs extracts against human coronavirus (HCoV-229E) using cytopathic effect (CPE) inhibition assay was performed for the first time. The CPE-inhibition assay was used to identify potential antivirals against human coronavirus 229E. The dose-response assay was designed to determine the range of efficacy for the chosen antiviral, i.e. the 50% inhibitory concentration (IC50), as well as the range of cytotoxicity (CC50). This assay is a critical and a well-reputable tool to assess the efficacy of several synthetic and natural agents against many viruses such as metapneumoviruses [89], influenza viruses [90], enteroviruses [91], and herpes simplex virus [92], among others. Selectivity index (SI = cytotoxicity/bioactivity) appeared to be an indispensable parameter to evaluate during the exploring process of novel antiviral candidates rather than focusing only on pharmacological or toxicological parameters separately [93]. As revealed in Fig. 5 all the tested milk thistle organs samples exhibited dose dependent inhibitory activity on HCoV-229E in nanomolar range. The results were compared to the positive control (remdesivir®) (Table 3). Comparison of the IC50 values of organs samples disclosed that fruit samples had the smallest IC50 value among all tested organs of 667.6 ± 0.5 ng/mL indicating its higher activity against the tested HCoV-229E virus while the leaves possessed the largest IC50 value of 2151 ± 0.9 ng/mL. On the contrary, low 50% cytotoxic concentration (CC50) on Vero E6 cells represent an indication of high toxicity of the tested samples on normal cells. Milk thistle fruits possessed the lowest CC50 of 3195 ± 0.3 ng/mL indicating the highest cytotoxicity among other samples (Fig. 5). Meanwhile, the leaves recorded the lowest toxicity with CC50 of 14,598 ± 1.2 ng/mL.
Fig. 5.
50% cytotoxic concentration (CC50) on Vero E6 cells on the left, and 50% inhibitory concentration (IC50) on HCoV-229E virus on the right of different S. marianum organs extracts
Table 3.
Cytotoxicity (pCC50, CC50 in g/L) on Vero E6 cells, HCoV-229E antiviral activity (pIC50, IC50 in g/L), and selectivity indices (SI = pCC50/pIC50) of different S. marianum organs samples
| Organ samples | pCC50 | pIC50 | SI |
|---|---|---|---|
| Fruits | 2.50 | 3.18 | 0.79 |
| Leaves | 1.84 | 2.67 | 0.69 |
| Roots | 2.03 | 2.93 | 0.70 |
| Stems | 2.22 | 3.05 | 0.73 |
| Positive control (remdesivir®) | 2.55 | 2.61 | 0.98 |
Selectivity index (SI) was then calculated by dividing cytotoxicity as pCC50 (-log CC50 in g/L) on HCoV-229E antiviral activity as pIC50 (-log IC50 in g/L) to inspect the samples of high selectivity to virus infected cells without causing toxicity to normal cells (Table 3). The lower the selectivity index the more selective the tested sample. Leaves samples possessing low pCC50 and high pIC50 values, that subsequently yielded low selectivity index, are promising anti- human coronavirus 229E drug-like candidates. Although many other researchers have documented the antiviral efficacy of silymarin and milk thistle supplements [33, 94–96], the present study is the first to compare the antiviral efficacy of different milk thistle organs aiming at valorizing the unused plant parts.
Correlation analysis to selective antiviral activity for unraveling bioactivephytoconstituents from the tested S. marianum organs samples
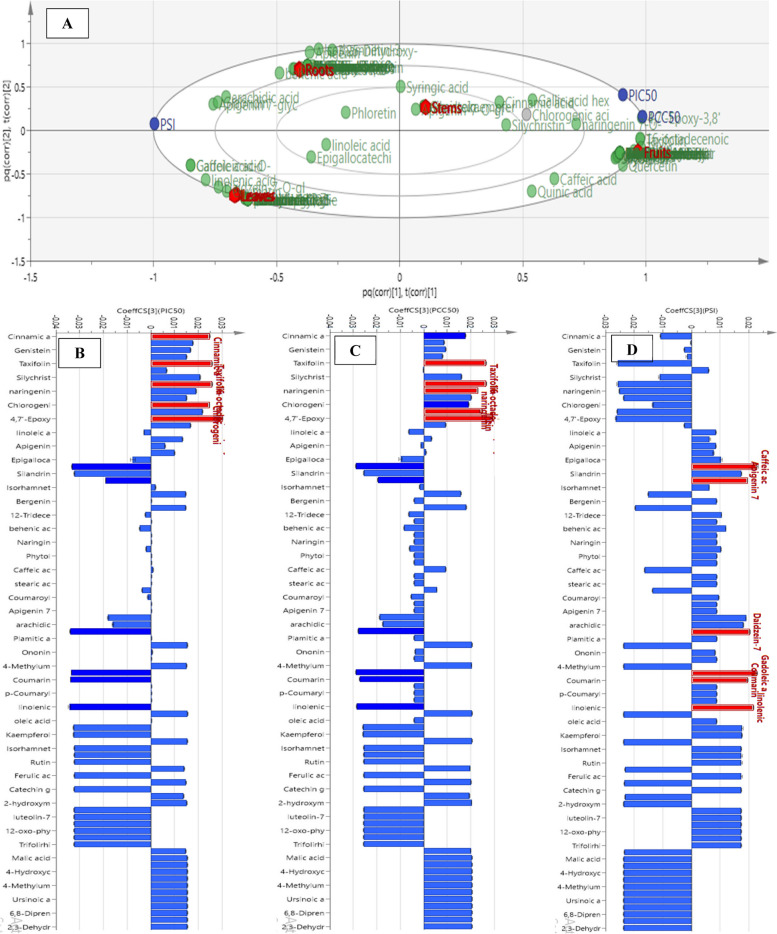
OPLS model and its accompanying correlation coefficient analysis were implemented for detection of significant phytoconstituents having selective virucidal activity against human coronavirus (HCoV-229E) amongst the four milk thistle organ samples studied, as well as evaluating consequent classification of the samples based on bioactivity. The biplot of the constructed OPLS model (Fig. 6A) exposed in-between class discrimination of fruits and stems from roots and leaves samples where the first exhibited spatial relation to cytotoxicity represented as pCC50 and antiviral activity on HCoV-229E as pIC50 while the later classes were in proximity to PSI indicating better selectivity. Further, studying the coefficient plots (Fig. 6B and D) portrayed that 16-octadecenoic acid methyl ester, taxifolin, cinnamic and chlorogenic acids were shown to be the constituents possessing the highest positive correlation to HCoV-229E inhibitory activity (Fig. 6B). While 16-octadecenoic acid methyl ester, taxifolin, tricin and naringenin were the major metabolites positively related to cytotoxic activity on normal cells (Fig. 6C). Finally, Fig. 6D indicated that caffeic acid-O-hexoside, gadoleic and linolenic acids, daidzein-7-O-hexoside, apigenin-7-O-hexoside ethyl ester and coumarin were the most potentially selective anti-human coronavirus 229E phytoconstituents.
Fig. 6.
Orthogonal Projections to Latent Structures (OPLS) biplot of the tested samples in correlation to the bioactive markers (A). Coefficient plots of OPLS model in order to determine biomarkers responsible for the antiviral activity (PIC50) (B), cytotoxicity (PCC50) (C), and selectivity (PIS) (D)
These findings are consistent with previous research that found that octadecenoic acid derivatives can bind to several coronaviruses’ proteins such as RNA-dependent RNA polymerase, main protease, and spike protein S1 to degrees similar to those possessed by the known antiviral drug umifenovir [97]. It also exhibited activity against influenza A and B viruses [98]. In addition, taxifolin showed the ability to inhibit the replication of HCoV-229E in Huh-7 cells at 2.5 µM and this inhibitory activity augmented with increasing its concentration. This activity was explained by its ability to inhibit the viral main protease activity [99]. Moreover, chlorogenic, caffeic, linolenic acids, and daidzein were found to inhibit HCoV S-glycoprotein attachment to host cells. This was illustrated by their ability to impair the function of HSPA5 SDBβ, which is the binding site for viral S-glycoprotein [100]. Furthermore, tricin was found to have antiviral activities against influenza A and B strains by inhibiting viral mRNA synthesis [101]. Besides, naringenin was found to inhibit cytopathic effect in Vero E6 cells infected with SARS-CoV-2 in a time and concentration-dependent manner. This effect was explained by its ability to inhibit endo-lysosomal Two-Pore Channels (TPCs), a pathway facilitating viral entry to host cell [101]. Further, apigenin and coumarins were found to be SARS-CoV-2 main protease inhibitors, thus inhibiting viral replication in the host cell [102, 103].
Conclusion
This study provides the first comparative evaluation of the metabolomes of S. marianum different organs applying UPLC-MS/MS coupled with multivariate analysis. HCA-heat map and OPLS-DA revealed in-between class discrimination between fruits and the other organs samples, in addition to within class discrimination between root samples which were separated from the leaves and stem samples. The OPLS-DA coefficients plots allowed the recognition of phytoconstituents responsible for the segregation of each organ samples into separate class. All studied S. marianum organs extracts were tested for selective virucidal activity against human coronavirus (HCoV-229E), and they all exhibited dose dependent inhibitory activity in nanomolar range with variable degrees of safety, efficacy, and selectivity. OPLS model and its accompanying correlation coefficient analysis were implemented for detection of significant phytoconstituents having effective, safe, and selective antiviral potential amongst the four studied S. marianum organs. The study in hand valorizes the importance of different S. marianum organs as wealthy sources of valuable antiviral agents. The future work will be the isolation of the recognized promising antiviral phytoconstituents from different milk thistle organs, followed by extensive in vitro and in vivo testing of their biological activities to afford more conclusive and comprehensive therapeutic approaches that enable to introduce these drugs to the market.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- ESI
Electrospray ionization
- TQD
Tandem Quadropole Detection
- HCoV-229E
Human coronavirus 229E
- OPLS-DA
Orthogonal Projections to Latent Structures-Discriminant Analysis
- OPLS
Orthogonal Projections to Latent Structures
- HCA
Hierarchical Clustering Analysis
- CPE
Cytopathic Effect
- COVID-19
Coronavirus Disease 2019
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- STAT3
Signal Transducer and Activator of Transcription 3
- RdRp
RNA-dependent RNA polymerase
- HCoV
Human Coronavirus
- MERS-CoV
Middle East Respiratory Syndrome Coronavirus
- Vero E6 cells
African green monkey kidney
- CID
Collision-Induced Dissociation
- ANOVA
One-Way Analysis of the Variance
- DMEM
Dulbecco's Modified Eagle Medium
- FBS
Fetal Bovine Serum
- CCID 50
50 percent cell culture infective dose
- PBS
Phosphate-Buffered Saline
- OD
Optical Density
- CC50
50% Cytotoxic Concentration
- IC50
50% Inhibitory Concentration
- RDA reaction
Retro-Diels–Alder reaction
- R2
Goodness of fitness
- Q2
Goodness of prediction
- ROC curve
Receiver Operating Characteristic curve
- AUC
Area under the curve
- LV
Latent Variable
- SI
Selectivity Index
- HSPA5
Heat shock protein family A member 5
- TPCs
Two-pore channels
Authors’ contributions
A.E-B. and R.S.I.: data acquisition, analysis and interpretation, drafting the work conceptualization of work, experimental design and revision of the manuscript.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). The authors declare that this study was self-funded.
Availability of data and materials
The datasets generated and analyzed during the current study are all mentioned in the presented manuscript.
Declarations
Ethics approval and consent to participate
The plant material was collected during the flowering-fruiting stage from farms belonging to the Faculty of Agriculture, Alexandria University, Egypt, in July 2022 according to relevant guidelines and regulations. The plant identity was confirmed via comparison with herbal sample present in the herbarium of the Faculty of Science, Alexandria University, Egypt. A voucher specimen (SM2022) was held at the Department of Pharmacognosy-Faculty of Pharmacy-Alexandria University.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Abenavoli L, Izzo AA, Milić N, Cicala C, Santini A, Capasso R. Milk thistle (Silybum marianum): a concise overview on its chemistry, pharmacological, and nutraceutical uses in liver diseases. Phytother Res. 2018;32:2202–13. doi: 10.1002/ptr.6171. [DOI] [PubMed] [Google Scholar]
- 2.Porwal O, Sheet M, Ameen M, Anwer ET, Uthirapathy S, Ahamad J, et al. Silybum marianum (milk thistle): review on its chemistry, morphology, ethno medical uses, phytochemistry and pharmacological activities. J Drug Delivery Ther. 2019;9:199–206. doi: 10.22270/jddt.v9i5.3666. [DOI] [Google Scholar]
- 3.Bijak M. Silybin, a major bioactive component of milk thistle (Silybum marianum L. Gaernt.)—Chemistry, Bioavailability, and metabolism. Molecules: J Synth Chem Nat Prod Chem. 2017;22(11):1942. [DOI] [PMC free article] [PubMed]
- 4.Le Q-U, Lay H-L, Wu M-C, Kumar Joshi R, Ming-Chang Wu C. Phytoconstituents and pharmacological activities of Silybum marianum (milk thistle): a critical review. ~ 41 ~ American. J Essent Oils Nat Prod. 2018;6:41–7. [Google Scholar]
- 5.Khan MA, Abbasi BH, Ahmed N, Ali H. Effects of light regimes on in vitro seed germination and silymarin content in Silybum marianum. Ind Crops Prod. 2013;46:105–10. doi: 10.1016/j.indcrop.2012.12.035. [DOI] [Google Scholar]
- 6.Bhattacharya S. Milk thistle (Silybum marianum L: Gaert.) Seeds in Health. Nuts Seeds Health Disease Prev. 2011:759–66. 10.1016/B978-0-12-375688-6.10090-8.
- 7.Flora K, Hahn M, Rosen H, Benner K. Milk thistle (Silybum marianum) for the therapy of liver disease. Am J Gastroenterol. 1998;93:139–43. doi: 10.1111/j.1572-0241.1998.00139.x. [DOI] [PubMed] [Google Scholar]
- 8.Schadewaldt H. The history of silymarin. Contribution to the history of liver therapy. Med Welt. 1969;20:902–14. [PubMed] [Google Scholar]
- 9.Kumar T, Kumar Larokar Y, Kumar Iyer S, Kumar A, Tripathi DK. Phytochemistry and pharmacological activities of Silybum marianum: a review. Int J Pharm Phytopharmacological Res. 2011;1:124–33. [Google Scholar]
- 10.Serçe A, Toptancı BÇ, Tanrıkut SE, Altaş S, Kızıl G, Kızıl S, et al. Assessment of the antioxidant activity of Silybum marianum Extract and its protective effect against DNA oxidation, protein damage and lipid peroxidation. Food Technol Biotechnol. 2016;54(4):455–61. [DOI] [PMC free article] [PubMed]
- 11.Freitag AF, Cardia GFE, da Rocha BA, Aguiar RP, Silva-Comar FMDS, Spironello RA, et al. Hepatoprotective effect of silymarin (silybum marianum) on hepatotoxicity induced by acetaminophen in spontaneously hypertensive rats. Evid-based Complement Altern Med. 2015;2015:538317. [DOI] [PMC free article] [PubMed]
- 12.Ramasamy K, Agarwal R. Multitargeted therapy of cancer by silymarin. Cancer Lett. 2008;269:352–62. doi: 10.1016/j.canlet.2008.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aghazadeh S, Amini R, Yazdanparast R, Ghaffari SH. Anti-apoptotic and anti-inflammatory effects of Silybum marianum in treatment of experimental steatohepatitis. Exp Toxicol Pathol. 2011;63:569–74. doi: 10.1016/j.etp.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Huseini HF, Larijani B, Heshmat R, Fakhrzadeh H, Radjabipour B, Toliat T, et al. The efficacy of Silybum marianum (L.) Gaertn. (silymarin) in the treatment of type II diabetes: a randomized, double-blind, placebo-controlled, clinical trial. Phytother Res. 2006;20:1036–9. doi: 10.1002/ptr.1988. [DOI] [PubMed] [Google Scholar]
- 15.Nazir N, Karim N, Abdel-Halim H, Khan I, Wadood SF, Nisar M. Phytochemical analysis, molecular docking and antiamnesic effects of methanolic extract of Silybum marianum (L.) Gaertn seeds in scopolamine induced memory impairment in mice. J Ethnopharmacol. 2018;210:198–208. doi: 10.1016/j.jep.2017.08.026. [DOI] [PubMed] [Google Scholar]
- 16.Bijak M, Saluk-Bijak J. Flavonolignans inhibit the arachidonic acid pathway in blood platelets. BMC Complement Altern Med. 2017;17:1–8. doi: 10.1186/s12906-017-1897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vilahur G, Casaní L, Peña E, Crespo J, Juan-Babot O, Ben-Aicha S, et al. Silybum marianum provides cardioprotection and limits adverse remodeling post-myocardial infarction by mitigating oxidative stress and reactive fibrosis. Int J Cardiol. 2018;270:28–35. doi: 10.1016/j.ijcard.2018.06.030. [DOI] [PubMed] [Google Scholar]
- 18.Sayyah M, Boostani H, Pakseresht S, Malayeri A. Comparison of Silybum marianum (L.) Gaertn. With fluoxetine in the treatment of obsessive – compulsive disorder. Undefined. 2010;34:362–5. doi: 10.1016/j.pnpbp.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 19.Song X, Liu B, Cui L, Zhou B, Liu W, Xu F, et al. Silibinin ameliorates anxiety/depression-like behaviors in amyloid β-treated rats by upregulating BDNF/TrkB pathway and attenuating autophagy in hippocampus. Physiol Behav. 2017;179:487–93. doi: 10.1016/j.physbeh.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 20.Karimi G, Vahabzadeh M, Lari P, Rashedinia M, Moshiri M. Silymarin, a Promising Pharmacological Agent for Treatment of diseases. Iran J Basic Med Sci. 2011;14:308. [PMC free article] [PubMed] [Google Scholar]
- 21.Wagoner J, Negash A, Kane OJ, Martinez LE, Nahmias Y, Bourne N, et al. Multiple effects of silymarin on the hepatitis C virus lifecycle. Hepatology. 2010;51:1912–21. doi: 10.1002/hep.23587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qaddir I, Rasool N, Hussain W, Mahmood S. Computer-aided analysis of phytochemicals as potential dengue virus inhibitors based on molecular docking, ADMET and DFT studies. J Vector Borne Dis. 2017;54:255–62. doi: 10.4103/0972-9062.217617. [DOI] [PubMed] [Google Scholar]
- 23.McClure J, Lovelace ES, Elahi S, Maurice NJ, Wagoner J, Dragavon J, et al. Silibinin inhibits HIV-1 infection by reducing Cellular activation and proliferation. PLoS ONE. 2012;7:e41832. doi: 10.1371/journal.pone.0041832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lani R, Hassandarvish P, Chiam CW, Moghaddam E, Chu JJH, Rausalu K et al. Antiviral activity of silymarin against chikungunya virus. Scientific Reports. 2015;5:1–10. [DOI] [PMC free article] [PubMed]
- 25.Camini FC, da Silva TF, da Silva Caetano CC, Almeida LT, Ferraz AC, Alves Vitoreti VM, et al. Antiviral activity of silymarin against Mayaro virus and protective effect in virus-induced oxidative stress. Antiviral Res. 2018;158:8–12. doi: 10.1016/j.antiviral.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 26.Wu YF, Fu SL, Kao CH, Yang CW, Lin CH, Hsu MT, et al. Chemopreventive effect of silymarin on liver pathology in HBV X protein transgenic mice. Cancer Res. 2008;68:2033–42. doi: 10.1158/0008-5472.CAN-07-2450. [DOI] [PubMed] [Google Scholar]
- 27.Song JH, Choi HJ. Silymarin efficacy against influenza a virus replication. Phytomedicine. 2011;18:832–5. doi: 10.1016/j.phymed.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 28.Esmaeil N, Anaraki SB, Gharagozloo M, Moayedi B. Silymarin impacts on immune system as an immunomodulator: one key for many locks. Int Immunopharmacol. 2017;50:194–201. doi: 10.1016/j.intimp.2017.06.030. [DOI] [PubMed] [Google Scholar]
- 29.Khairy A, Hammoda HM, Celik I, Zaatout HH, Ibrahim RS. Discovery of potential natural dihydroorotate dehydrogenase inhibitors and their synergism with brequinar via integrated molecular docking, dynamic simulations and in vitro approach. Sci Rep. 2022;12(1):19037. [DOI] [PMC free article] [PubMed]
- 30.Lovelace ES, Wagoner J, MacDonald J, Bammler T, Bruckner J, Brownell J, et al. Silymarin suppresses Cellular inflammation by inducing reparative stress signaling. J Nat Prod. 2015;78:1990–2000. doi: 10.1021/acs.jnatprod.5b00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu SY, Dong Y, Tu J, Zhou Y, Zhou XH, Xu B. Silybum marianum oil attenuates oxidative stress and ameliorates mitochondrial dysfunction in mice treated with D-galactose. Pharmacogn Mag. 2014;10:92–9. doi: 10.4103/0973-1296.127353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang K, Wang W, Jin X, Wang Z, Ji Z, Meng G. Silibinin, a natural flavonoid, induces autophagy via ROS-dependent mitochondrial dysfunction and loss of ATP involving BNIP3 in human MCF7 breast cancer cells. Oncol Rep. 2015;33:2711–8. doi: 10.3892/or.2015.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu CH, Jassey A, Hsu HY, Lin LT. Antiviral Activities of Silymarin and Derivatives. Molecules. 2019;24:1552. [DOI] [PMC free article] [PubMed]
- 34.Bosch-Barrera J, Martin-Castillo B, Buxó M, Brunet J, Encinar JA, Menendez JA. Silibinin and SARS-CoV-2: dual targeting of host cytokine storm and Virus Replication Machinery for Clinical Management of COVID-19 patients. J Clin Med. 2020;9(6):1770. [DOI] [PMC free article] [PubMed]
- 35.Liu DX, Liang JQ, Fung TS. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). Encyclopedia Virol. 2021;428.
- 36.Ibrahim RS, Mahrous RSR, Abu EL-Khair RM, Ross SA, Omar AA, Fathy HM. Biologically guided isolation and ADMET profile of new factor Xa inhibitors from Glycyrrhiza glabra roots using in vitro and in silico approaches. RSC Adv. 2021;11:9995–10001. doi: 10.1039/D1RA00359C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woo PCY, Huang Y, Lau SKP, Yuen KY. Coronavirus Genomics and Bioinformatics Analysis. Viruses. 2010;2:1804. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu F, Han C, Li Y, Zheng M, Xi X, Hu C, et al. The Chemical constituents and pharmacological actions of Silybum Marianum. Curr Nutr Food Sci. 2018;15:430–40. doi: 10.2174/1573401314666180327155745. [DOI] [Google Scholar]
- 39.Korany MA, Moneeb MS, Asaad AM, El-Sebakhy NA, El-Banna AA. The Use of Merbromin fluorescence quenching for the assay of silymarin in its natural source and Pharmaceutical preparations. Anal Chem Lett. 2019;9:152–67. doi: 10.1080/22297928.2019.1613927. [DOI] [Google Scholar]
- 40.Graf TN, Cech NB, Polyak SJ, Oberlies NH. A Validated UHPLC-Tandem Mass Spectrometry Method for Quantitative Analysis of Flavonolignans in milk thistle (Silybum marianum) extracts a validated UHPLC-Tandem Mass Spectrometry Method for Quantitative Analysis of Flavonolignans in milk thistle (Silybum marianum) extracts. J Pharm Biomed Anal. 2016;126:26–33. doi: 10.1016/j.jpba.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mudge E, Paley L, Schieber A, Brown PN. Optimization and single-laboratory validation of a method for the determination of flavonolignans in milk thistle seeds by high-performance liquid chromatography with ultraviolet detection. Anal Bioanal Chem. 2015;407:7657–66. doi: 10.1007/s00216-015-8925-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tayoub G, Alorfi M, Sulaiman H. Quantitative identification of total silymarin in wild Silybum marianum L. by using HPLC. Int J Herb Med. 2018;6:110–4. [Google Scholar]
- 43.Albassam AA, Frye RF, Markowitz JS. The effect of milk thistle (Silybum marianum) and its main flavonolignans on CYP2C8 enzyme activity in human liver microsomes. Chem Biol Interact. 2017;271:24–9. doi: 10.1016/j.cbi.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 44.Nagy M. Identification of Silymarin Constituents: An Improved HPLC–MS Method. Chromatographia. 2012;75:175–80.
- 45.Lee JI, Hsu BH, Wu D, Barrett JS. Separation and characterization of silybin, isosilybin, silydianin and silychristin in milk thistle extract by liquid chromatography–electrospray tandem mass spectrometry. J Chromatogr A. 2006;1116:57–68. doi: 10.1016/j.chroma.2006.03.053. [DOI] [PubMed] [Google Scholar]
- 46.Petrásková L, Káňová K, Biedermann D, Kren V, Valentová K. Simple and rapid HPLC separation and quantification of flavonoid, flavonolignans, and 2,3-dehydroflavonolignans in silymarin. Foods. 2020;9(2):116. [DOI] [PMC free article] [PubMed]
- 47.Wallace S, Carrier DJ, Beitle RR, Clausen EC, Griffis CL. HPLC-UV and LC-MS-MS characterization of silymarin in milk thistle seeds and corresponding products. J Nutraceuticals Funct Med Foods. 2003;4:37–48. doi: 10.1300/J133v04n02_05. [DOI] [Google Scholar]
- 48.Cai X-L, Li D-N, Qiao J-Q, Lian H-Z, Wang S-K. Determination of silymarin flavonoids by HPLC and LC-MS and investigation of extraction rate of silymarin in Silybum marianum fruits by boiling Water. 2009.
- 49.Ibrahim RS, El-Banna AA. Royal jelly fatty acids bioprofiling using TLC-MS and digital image analysis coupled with chemometrics and non-parametric regression for discovering efficient biomarkers against melanoma. RSC Adv. 2021;11:18717–28. doi: 10.1039/D1RA00580D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghallab DS, Mohyeldin MM, Shawky E, Metwally AM, Ibrahim RS. Chemical profiling of Egyptian propolis and determination of its xanthine oxidase inhibitory properties using UPLC–MS/MS and chemometrics. LWT. 2021;136:110298. doi: 10.1016/j.lwt.2020.110298. [DOI] [Google Scholar]
- 51.Schmidtke M, Schnittler U, Jahn B, Dahse HM, Stelzner A. A rapid assay for evaluation of antiviral activity against coxsackie virus B3, influenza virus A, and herpes simplex virus type 1. J Virol Methods. 2001;95:133–43. doi: 10.1016/S0166-0934(01)00305-6. [DOI] [PubMed] [Google Scholar]
- 52.Pauwels R, Balzarini J, Baba M, Snoeck R, Schols D, Herdewijn P, et al. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J Virol Methods. 1988;20:309–21. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- 53.Li W, Sun Y, Liang W, Fitzloff JF, van Breemen RB. Identification of caffeic acid derivatives in Actea racemosa (Cimicifuga racemosa, black cohosh) by liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:978–82. doi: 10.1002/rcm.1008. [DOI] [PubMed] [Google Scholar]
- 54.Li BH, Wu JD, Li XL. LC-MS/MS determination and pharmacokinetic study of bergenin, the main bioactive component of Bergenia purpurascens after oral administration in rats. J Pharm Anal. 2013;3:229–34. doi: 10.1016/j.jpha.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh A, Bajpai V, Kumar S, Sharma R, Kumar B. Profiling of gallic and ellagic acid derivatives in different plant parts of Terminalia arjuna by HPLC-ESI-QTOF-MS/MS. Nat Prod Commun. 2016;11:239–44. [PubMed] [Google Scholar]
- 56.Haşimi N, Ertaş A, Yilmaz M, Boga M, Temel H, Kayıran S, et al. LC-MS/MS and GC-MS analyses of three endemic Astragalus species from Anatolia towards their total phenolic- flavonoid contents and biological activities | semantic Scholar. Biol Divers Conserv. 2017;10:18–30. [Google Scholar]
- 57.Hines KM, Ross DH, Davidson KL, Bush MF, Xu L. Large-scale structural characterization of Drug and Drug-Like compounds by High-Throughput Ion Mobility-Mass Spectrometry. Anal Chem. 2017;89:9023–30. doi: 10.1021/acs.analchem.7b01709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.al Kadhi O, Melchini A, Mithen R, Saha S. Development of a LC-MS/MS Method for the Simultaneous Detection of Tricarboxylic Acid Cycle Intermediates in a Range of Biological Matrices. J Anal Methods Chem. 2017;2017:5391832. [DOI] [PMC free article] [PubMed]
- 59.ben Said R, Hamed AI, Mahalel UA, Al-Ayed AS, Kowalczyk M, Moldoch J et al. Tentative characterization of Polyphenolic compounds in the male flowers of Phoenix dactylifera by Liquid Chromatography coupled with Mass Spectrometry and DFT. Int J Mol Sci. 2017;18(3):512. [DOI] [PMC free article] [PubMed]
- 60.Kumar S, Singh A, Kumar B. Identification and characterization of phenolics and terpenoids from ethanolic extracts of Phyllanthus species by HPLC-ESI-QTOF-MS/MS. J Pharm Anal. 2017;7:214–22. doi: 10.1016/j.jpha.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu W, Kong Y, Zu Y, Fu Y, Luo M, Zhang L, et al. Determination and quantification of active phenolic compounds in pigeon pea leaves and its medicinal product using liquid chromatography-tandem mass spectrometry. J Chromatogr A. 2010;1217:4723–31. doi: 10.1016/j.chroma.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 62.Krasteva I, Nikolov S. Flavonoids in Astragalus Corniculatus. Quim Nova. 2008;31:59–60. doi: 10.1590/S0100-40422008000100012. [DOI] [Google Scholar]
- 63.Santoro V, Parisi V, D’Ambola M, Sinisgalli C, Monné M, Milella L, et al. Chemical profiling of Astragalus Membranaceus roots (Fish.) Bunge Herbal Preparation and evaluation of its Bioactivity. Nat Prod Commun. 2020;15. 10.1177/1934578X20924152.
- 64.Spáčil Z, Nováková L, Solich P. Comparison of positive and negative ion detection of tea catechins using tandem mass spectrometry and ultra high performance liquid chromatography. Food Chem. 2010;123:535–41. doi: 10.1016/j.foodchem.2010.04.048. [DOI] [Google Scholar]
- 65.Ye M, Yang WZ, di Liu K, Qiao X, Li BJ, Cheng J, et al. Characterization of flavonoids in Millettia Nitida var. Hirsutissima by HPLC/DAD/ESI-MSn. J Pharm Anal. 2012;2:35–42. doi: 10.1016/j.jpha.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Letourneau DR, Volmer DA. Mass spectrometry-based methods for the advanced characterization and structural analysis of lignin: a review. Mass Spectrom Rev. 2021. 10.1002/MAS.21716. [DOI] [PubMed]
- 67.Ren Z, Nie B, Liu T, Yuan F, Feng F, Zhang Y et al. Simultaneous Determination of Coumarin and Its Derivatives in Tobacco Products by Liquid Chromatography-Tandem Mass Spectrometry. Molecules. 2016;21:1511. [DOI] [PMC free article] [PubMed]
- 68.Leonart LP, Gasparetto JC, Pontes FLD, Cerqueira LB, de Francisco TMG, Pontarolo R. New Metabolites of Coumarin Detected in Human Urine Using Ultra Performance Liquid Chromatography/Quadrupole-Time-of-Flight Tandem Mass Spectrometry. Molecules. 2017;22:2031. [DOI] [PMC free article] [PubMed]
- 69.Chen G, Li X, Saleri F, Guo M. Analysis of flavonoids in Rhamnus davurica and its antiproliferative activities. Molecules. 2016;21(10):1275. [DOI] [PMC free article] [PubMed]
- 70.Bystrom LM, Lewis BA, Brown DL, Rodriguez E, Obendorf RL. Characterisation of phenolics by LC-UV/Vis, LC-MS/MS and sugars by GC in Melicoccus bijugatus Jacq. Montgomery fruits. Food Chem. 2008;111:1017–24. doi: 10.1016/j.foodchem.2008.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kang J, Price WE, Ashton J, Tapsell LC, Johnson S. Identification and characterization of phenolic compounds in hydromethanolic extracts of sorghum wholegrains by LC-ESI-MSn. Food Chem. 2016;211:215–26. doi: 10.1016/j.foodchem.2016.05.052. [DOI] [PubMed] [Google Scholar]
- 72.Zhang J, Lu J, Zhang Q, Dai L, Liu Y, Tu P, et al. Simultaneous screening and identifying four categories of particular flavonoids in the leaves of murraya exotica L. by HPLC-DAD-ESI-MS-MS. J Chromatogr Sci. 2014;52:103–14. doi: 10.1093/chromsci/bms253. [DOI] [PubMed] [Google Scholar]
- 73.Motiur Rahman AFM, Attwa MW, Ahmad P, Baseeruddin M, Kadi AA. Fragmentation behavior studies of chalcones employing direct analysis in real time (DART) Mass Spectrom Lett. 2013;4:30–3. doi: 10.5478/MSL.2013.4.2.30. [DOI] [Google Scholar]
- 74.Csupor D, Csorba A, Hohmann J. Recent advances in the analysis of flavonolignans of Silybum marianum. J Pharm Biomed Anal. 2016;130:301–17. doi: 10.1016/j.jpba.2016.05.034. [DOI] [PubMed] [Google Scholar]
- 75.Aoki F, Nakagawa K, Tanaka A, Matsuzaki K, Arai N, Mae T. Determination of glabridin in human plasma by solid-phase extraction and LC-MS/MS. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;828:70–4. doi: 10.1016/j.jchromb.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 76.Nikolic D, van Breemen RB. Analytical methods for quantitation of Prenylated flavonoids from Hops. Curr Anal Chem. 2012;9:71–85. doi: 10.2174/1573411011309010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu X, Luo R, He XW, Yu P, Yuan G, Letter Electrospray ionization mass spectral characteristics and fragmentation mechanisms of triterpenoids in Fomes Officinalis. Eur J Mass Spectrom (Chichester) 2011;17:297–304. doi: 10.1255/ejms.1132. [DOI] [PubMed] [Google Scholar]
- 78.Ahmed E, Tun Noor A, Malik A, Ferheen S, Afza N. Structural determination of silymins a and B, new pentacyclic triterpenes from Silybum marianum, by 1D and 2D NMR spectroscopy. Magn Reson Chem. 2007;45:79–81. doi: 10.1002/mrc.1881. [DOI] [PubMed] [Google Scholar]
- 79.Suzuki S, Hori Y, Das RC, Koga O. Chemical ionization Mass Spectrometry of Epoxides. Bull Chem Soc Jpn. 1980;53:1451–2. doi: 10.1246/bcsj.53.1451. [DOI] [Google Scholar]
- 80.Brenna JT. Fatty acid analysis by high resolution gas chromatography and mass spectrometry for clinical and experimental applications. Curr Opin Clin Nutr Metab Care. 2013;16:548–54. doi: 10.1097/MCO.0b013e328363bc0a. [DOI] [PubMed] [Google Scholar]
- 81.El Sayed AM, Basam SM, El-Naggar EMbellah, Marzouk A, El-Hawary HS. S. LC–MS/MS and GC–MS profiling as well as the antimicrobial effect of leaves of selected Yucca species introduced to Egypt. Scientific Reports. 2020;10:1–15. [DOI] [PMC free article] [PubMed]
- 82.Fagerquist CK, Neese RA, Hellerstein MK. Molecular ion fragmentation and its effects on mass isotopomer abundances of fatty acid methyl esters ionized by electron impact. J Am Soc Mass Spectrom. 1999;10:430–9. doi: 10.1016/S1044-0305(99)00003-3. [DOI] [PubMed] [Google Scholar]
- 83.Härtig C. Rapid identification of fatty acid methyl esters using a multidimensional gas chromatography–mass spectrometry database. J Chromatogr A. 2008;1177:159–69. doi: 10.1016/j.chroma.2007.10.089. [DOI] [PubMed] [Google Scholar]
- 84.Zhao Y, Lee MJ, Cheung C, Ju JH, Chen YK, Liu B, et al. Analysis of multiple metabolites of tocopherols and tocotrienols in mice and humans. J Agric Food Chem. 2010;58:4844–52. doi: 10.1021/jf904464u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ćirić A, Prosen H, Jelikić-Stankov M, Durević P. Evaluation of matrix effect in determination of some bioflavonoids in food samples by LC-MS/MS method. Talanta. 2012;99:780–90. doi: 10.1016/j.talanta.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 86.Dictionary of Natural Products. 30.2 Chemical Search. https://dnp.chemnetbase.com/faces/chemical/ChemicalSearch.xhtml. Accessed 9 May 2023.
- 87.Javeed A, Ahmed M, Sajid AR, Sikandar A, Aslam M, Ul Hassan T, et al. Comparative Assessment of Phytoconstituents, antioxidant activity and Chemical Analysis of different parts of milk Thistle Silybum marianum L. Molecules. 2022;27(9):2641. [DOI] [PMC free article] [PubMed]
- 88.Hurt AC, Ison MG, Hayden FG, Hay AJ. Second isirv antiviral group conference: overview. Influenza Other Respir Viruses. 2013;7(SUPPL3):1–7. doi: 10.1111/irv.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kohn LK, Foglio MA, Rodrigues RA, Sousa IM, de O, Martini MC, Padilla MA, et al. In-Vitro antiviral activities of extracts of plants of the Brazilian cerrado against the avian metapneumovirus (aMPV). Revista Brasileira De Ciencia Avicola / Brazilian J Poult Sci. 2015;17:275–80. doi: 10.1590/1516-635X1703275-280. [DOI] [Google Scholar]
- 90.Cho HM, Doan TP, Quy Ha TK, Kim HW, Lee BW, Tung Pham HT, et al. Dereplication by High-Performance Liquid Chromatography (HPLC) with quadrupole-time-of-flight Mass Spectroscopy (qTOF-MS) and antiviral activities of phlorotannins from Ecklonia cava. Mar Drugs. 2019;17(3):149. [DOI] [PMC free article] [PubMed]
- 91.Zhang M, Zhang Y, Wang Y, Lv W, Zhang Y. Automated cell-based luminescence assay for profiling antiviral compound activity against enteroviruses. Sci Rep. 2019;9:6023. [DOI] [PMC free article] [PubMed]
- 92.Cotarelo M, Catalán P, Sánchez-Carrillo C, Menasalvas A, Cercenado E, Tenorio A, et al. Cytopathic effect inhibition assay for determining the in-vitro susceptibility of herpes simplex virus to antiviral agents. J Antimicrob Chemother. 1999;44:705–8. doi: 10.1093/jac/44.5.705. [DOI] [PubMed] [Google Scholar]
- 93.Pereira IV, de Freitas MP. Double focus in the modelling of anti-influenza properties of 2-iminobenzimidazolines: pharmacology and toxicology. SAR QSAR Environ Res. 2021;32:643–54. doi: 10.1080/1062936X.2021.1950832. [DOI] [PubMed] [Google Scholar]
- 94.Anthony K, Subramanya G, Uprichard S, Hammouda F, Saleh M, Antioxidant, Anti-Hepatitis C. Viral Activities of Commercial Milk Thistle Food Supplements. Antioxidants 2013, Vol 2, Pages 23–36. 2013;2:23–36. [DOI] [PMC free article] [PubMed]
- 95.Ferenci P, Scherzer TM, Kerschner H, Rutter K, Beinhardt S, Hofer H, et al. Silibinin is a potent antiviral Agent in patients with chronic Hepatitis C not responding to Pegylated Interferon/Ribavirin Therapy. Gastroenterology. 2008;135:1561–7. doi: 10.1053/j.gastro.2008.07.072. [DOI] [PubMed] [Google Scholar]
- 96.Polyak SJ, Ferenci P, Pawlotsky JM. Hepatoprotective and antiviral functions of Silymarin Components in HCV infection. Hepatology. 2013;57:1262. doi: 10.1002/hep.26179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Elwakil B, Shaaban M, Bekhit A, El-Naggar M, Olama Z. Potential anti-COVID-19 activity of Egyptian propolis using computational modeling. Future Virol. 2021;16:107–16. doi: 10.2217/fvl-2020-0329. [DOI] [Google Scholar]
- 98.El-Shiekh RA, Abdelmohsen UR, Ashour HM, Ashour RM. Novel antiviral and antibacterial activities of hibiscus schizopetalus. Antibiotics. 2020;9:1–16. doi: 10.3390/antibiotics9110756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhu Y, Scholle F, Kisthardt SC, Xie D-Y. Flavonols and dihydroflavonols inhibit the main protease activity of SARS-CoV-2 and the replication of human coronavirus 229E. Virology. 2022;571:21–33. doi: 10.1016/j.virol.2022.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim CH. Anti–SARS-CoV-2 Natural products as potentially therapeutic agents. Front Pharmacol. 2021;12:1015–41. doi: 10.3389/fphar.2021.590509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Khazeei Tabari MA, Iranpanah A, Bahramsoltani R, Rahimi R. Flavonoids as promising antiviral agents against sars-cov‐2 infection: a mechanistic review. Molecules. 2021;26(13):3900. [DOI] [PMC free article] [PubMed]
- 102.Kaul R, Paul P, Kumar S, Büsselberg D, Dwivedi VD, Chaari A. Promising antiviral activities of natural flavonoids against SARS-CoV-2 targets: systematic review. Int J Mol Sci. 2021;22:11069. doi: 10.3390/ijms222011069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Abdizadeh R, Hadizadeh F, Abdizadeh T. In silico analysis and identification of antiviral coumarin derivatives against 3-chymotrypsin-like main protease of the novel coronavirus SARS-CoV-2. Mol Divers. 2021;26:1053–1076. doi: 10.1007/s11030-021-10230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are all mentioned in the presented manuscript.