Abstract
The baker’s yeast Saccharomyces cerevisiae possesses a single gene encoding heat shock transcription factor (HSF), which is required for the activation of genes that participate in stress protection as well as normal growth and viability. Yeast HSF (yHSF) contains two distinct transcriptional activation regions located at the amino and carboxyl termini. Activation of the yeast metallothionein gene, CUP1, depends on a nonconsensus heat shock element (HSE), occurs at higher temperatures than other heat shock-responsive genes, and is highly dependent on the carboxyl-terminal transactivation domain (CTA) of yHSF. The results described here show that the noncanonical (or gapped) spacing of GAA units in the CUP1 HSE (HSE1) functions to limit the magnitude of CUP1 transcriptional activation in response to heat and oxidative stress. The spacing in HSE1 modulates the dependence for transcriptional activation by both stresses on the yHSF CTA. Furthermore, a previously uncharacterized HSE in the CUP1 promoter, HSE2, modulates the magnitude of the transcriptional activation of CUP1, via HSE1, in response to stress. In vitro DNase I footprinting experiments suggest that the occupation of HSE2 by yHSF strongly influences the manner in which yHSF occupies HSE1. Limited proteolysis assays show that HSF adopts a distinct protease-sensitive conformation when bound to the CUP1 HSE1, providing evidence that the HSE influences DNA-bound HSF conformation. Together, these results suggest that CUP1 regulation is distinct from that of other classic heat shock genes through the interaction of yHSF with two nonconsensus HSEs. Consistent with this view, we have identified other gene targets of yHSF containing HSEs with sequence and spacing features similar to those of CUP1 HSE1 and show a correlation between the spacing of the GAA units and the relative dependence on the yHSF CTA.
All organisms possess a highly conserved response to elevated temperatures and to a variety of chemical and physiological stresses commonly designated as the heat shock response (38). In eukaryotic cells this response involves the rapid activation of a transcription factor known as heat shock transcription factor (HSF) (70). Once activated, HSF induces the expression of genes whose products ensure the survival of the cell during stressful conditions by providing defense against general protein damage. These heat shock proteins (Hsps) also play essential roles in the synthesis, transport, translocation, proteolysis, and proper folding of proteins under both normal and stressful conditions (38). Although the heat shock response is conserved among eukaryotes, both the number and overall sequence of HSFs vary widely among different species. Yeasts (Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Kluyveromyces lactis) and Drosophila melanogaster appear to have a single HSF gene, whereas most vertebrates and higher plants possess multiple HSF genes: at least three HSF genes have been isolated from the human, mouse, chicken, and tomato genomes (15, 23, 28, 40, 41, 44, 50, 51, 53, 60, 69). Despite sequence divergence, all members of the HSF family have two highly conserved features: a helix-turn-helix DNA binding domain and coiled-coil hydrophobic repeat domains which mediate the trimerization of HSF (26, 45, 66).
A key step in the induction of heat shock gene transcription is the interaction of HSF with a short, highly conserved cis-acting DNA sequence, the heat shock element (HSE) found in the promoters of HSF-responsive genes. All HSEs contain multiple copies of the repeating 5-bp sequence 5′-nGAAn-3′ (where n is any nucleotide) arranged in alternating orientation (2, 71). The number of pentameric units in an HSE can vary; while a minimum of three is thought to be required for heat-inducible expression, some HSEs harbor eight contiguous inverted repeats (19). Furthermore, the degree of homology of each pentameric unit to the consensus nGAAn motif can vary, as can the nature of the initial pentamer, beginning with either GAA or its complement TTC, with the latter displaying significantly higher levels of biological activity in yeast cells and the capability to bind two HSF trimers instead of one (4). A functional HSE can tolerate a 5-bp insertion between repeating units, provided that the spacing and orientation of the pentameric elements are maintained (2). The binding of HSF to DNA has been shown to be highly cooperative, and deviations from the nGAAn consensus sequence may be tolerated in vivo because multiple HSEs foster cooperative interactions between multiple HSF trimers (4, 65, 72). These variations in the sequence of the binding site can influence the affinity of HSF for the HSE(s) of a particular heat shock gene, thereby influencing the level of transcriptional activation, and ultimately fine-tune the nature of the heat shock gene response.
The existence of multiple HSF species in higher eukaryotes suggests that HSF isoforms may have specialized functions that can be triggered by distinct stimuli or may activate specific target genes. For example, in human K562 erythroleukemia cells, HSF2 responds to hemin treatment and is constitutively active in mouse embryonal carcinoma cells and at the blastocyst stage during embryogenesis and spermatogenesis (46, 55). These observations are consistent with HSF2 functioning as a regulator of heat shock gene expression during development and differentiation, such as its potential regulation of the hsp70.2 gene during spermatogenesis (33, 49). Human HSF1 responds to thermal stress and other stresses at the level of trimerization, phosphorylation and DNA binding to activate transcription of Hsp genes (16, 48, 74). Consistent with the possibility that distinct mammalian HSF isoforms activate different target genes, mouse HSF1 (mHSF1) utilizes a higher degree of cooperativity in DNA binding and demonstrates a preference for HSEs containing four to five pentamers, while mHSF2 has a binding preference for HSEs containing only two to three pentamers (30). This notion is further supported by a recent functional analysis of human HSF1 and HSF2 expressed in yeast, which showed that HSF1 bound with highest affinity to and activated transcription most potently from the SSA3 promoter, which has an extended array of pentameric elements in the HSE (35). On the other hand, HSF2 bound with highest affinity to and activated transcription most potently from the yeast metallothionein gene, CUP1, which has only three pentamers in HSE1 and has a gap between the last two pentamers and an A-to-G substitution (GAG) in the last pentameric unit (35).
Yeast cells utilize the single essential HSF to activate the expression of a wide variety of genes in response to heat and other stresses and to coordinate the expression of genes required for growth under normal physiological conditions. The DNA binding domain of yeast HSF (yHSF) may be more conformationally flexible than HSF1 or HSF2 from higher eukaryotes (20) and allow a wide range of distinct interactions of the DNA binding domain with HSEs. The observation that a single amino acid substitution in the DNA binding domain of yHSF alters the specificity of HSF on different promoters is consistent with this idea (54). A feature distinguishing yHSF from HSFs of higher eukaryotes is the presence of two transactivation domains which respond differentially to heat shock (42, 56). Studies of a synthetic HSE-lacZ reporter gene suggested that the yHSF amino-terminal activation domain mediates a transient response to elevated temperatures, while the carboxyl-terminal activation domain (CTA) is required to regulate both a transient and a sustained response (56). Both activation domains are restrained under normal growth conditions by intramolecular interactions with the DNA binding domain, the trimerization domain, and a short conserved element, denoted CE2 (5, 14, 28, 42, 56). The presence of two activation domains in yHSF may provide additional levels of regulation or selectivity in gene activation. Previous studies have established that the CUP1 gene is transcriptionally activated by yHSF via heat and oxidative stress (36, 63). Interestingly, expression of CUP1 in response to heat shock and oxidative stress exhibits a strong requirement for the CTA of HSF (36, 63). In contrast, this region is largely dispensable for the heat shock activation of the SSA1 and SSA3 genes, encoding members of the Hsp70 family (63). It is interesting that in addition to the differential requirement for the CTA of yHSF, activation of CUP1 by yHSF differs from that of typical heat shock genes in that the robust activation of CUP1 requires a temperature of 39 rather than 37°C (63).
The CUP1 promoter HSE is thought to be atypical in that it contains only one HSE (HSE1) composed of three pentameric units. A compilation of HSEs from many organisms demonstrated that for promoters that contain an HSE composed of three pentameric units, additional flanking HSEs are present (4, 43). Furthermore, HSE1 deviates significantly from consensus HSEs in that there is a gap between the second and third pentamers; however, the gap preserves both the spacing and the orientation between these two repeats. Since yHSF-dependent activation of CUP1 and the SSA genes is distinct, we have carried out a detailed analysis of CUP1 gene expression to understand how yHSF regulates the activation of genes via distinct HSEs and with distinct transactivation domain requirements. We present evidence for a second nonconsensus HSE in the CUP1 promoter, HSE2, which serves to modulate the transcriptional activation of CUP1 in response to both heat and oxidative stress. Furthermore, we demonstrate that the nature of HSE1 plays a crucial role in the dependence on the yHSF CTA for CUP1 activation by heat stress. The expression of two additional yeast genes which contain a gapped HSE is also strongly dependent on the yHSF CTA. Chymotrypsin sensitivity assays show that the arrangement of pentameric units in the CUP1 HSE1 affects the conformation of DNA-bound yHSF and suggests that at least part of the distinct features of CUP1 activation by yHSF may be due to the generation of specific yHSF structures by the HSE. Therefore, this work demonstrates that yeast cells activate and fine-tune the expression of a wide variety of target genes via a single HSF isoform, in part by virtue of the nature of the yHSF binding sites and distinct transactivation domain requirements.
MATERIALS AND METHODS
Strains and growth conditions.
S. cerevisiae MCY1093, a gift from Marian Carlson, was used as the wild-type parental strain throughout this study and is designated DTY123. Strain PS145 (a gift from Hillary Nelson) contains a deletion of the endogenous yHSF gene (60). The HSF(1-583) strain, DTY179, has been previously described (63). Cell culture conditions for inducing CUP1 expression by heat shock and oxidative stress using menadione treatment were as previously described (36, 63). All CUP1-lacZ fusion plasmids are URA3 based, and all strains were grown in synthetic complete medium minus uracil unless otherwise specified. Strain DTY123skn7 is isogenic to DTY123 and carries a hisG-URA3-hisG (1) disrupted SKN7 gene (10). The SKN7 gene was disrupted following previous protocols (11) by transforming DTY123 with an SKN7/hisG-URA3-hisG fragment that was released from plasmid pBS:SKN7:URA3 by XbaI-XhoI digestion. The chromosomal arrangement of this disrupted skn7 allele was confirmed by both PCR and the increased sensitivity of the skn7 disruption strain to tert-butyl hydroperoxide (37).
Plasmids.
All plasmids are numbered according to the 5′ and 3′ termini of the CUP1 insert; numbering is relative to the start site of CUP1 transcription. Plasmids containing mutations in the CUP1 HSE that are used for RNA analyses of gene expression are denoted with “m” to distinguish them from plasmids containing mutations in the CUP1 HSE that are used for DNA binding analyses, which are denoted with “M.” For analyzing regions of the CUP1 promoter important for CUP1 activation by HSF, restriction enzyme-generated fragments of the CUP1 promoter containing different 5′ upstream termini but all extending through the 12th codon of CUP1 (BspHI site at +105 from the transcription start site) were ligated into the lacZ fusion vector YEp357R (39). Plasmid pYEpCUP1-807 was generated by using a BspHI-BspHI CUP1 fragment from plasmid pGEXa (63). Plasmids pYEpCUP1-393, pYEpCUP1-241, and pYEpCUP1-167 were generated by using BamHI-, EcoRV-, and XbaI-BspHI CUP1 fragments, respectively, from plasmid pYep336 (12). Mutant CUP1 promoter plasmids pYEpCUP1HSE1P, pYEpCUP1HSE2m, and pYEpCUP1ACEm were generated by using a Chameleon double-stranded site-directed mutagenesis kit (Stratagene, La Jolla, Calif.), plasmid pYEpCUP1-393, and the following oligonucleotides: CUP1HSE1P −131/−172 (5′-CGGAAAAGACGCATCGCTCTGGAAGCTTCTAGAAGAAATGCC-3′), CUP1HSE2m (5′-GCGATGCGTCTTTTTCGCTAAACCGTTTCAGCAAAAAAGACTACC-3′), and CUP1Acem (5′-GCGATGCGTCTTTTCCCGTGAACCGTTCCAGC-3′). By the same procedure, plasmid pYEpCUP1HSE1m and oligonucleotide CUP1HSE2m were used to generate pYEpCUP1HSE1m2m. Plasmids pHSE-WT and pHSE-M were used to prepare identical-sized CUP1 electrophoretic mobility shift assay (EMSA) probes; pHSE-M contains a mutation in CUP1 HSE1, and both plasmids were described previously (63). Plasmids pHSE-2M and pHSE-1M2M contain a CUP1 fragment identical in size to that of pHSE-WT and pHSE-1M, all four plasmids contain CUP1 sequences from −183 to −80 cloned into the EcoRV site of pBluescript SK+. pHSE-2M was constructed by ligating a PCR product derived from plasmid pYEpCUP1HSE2m into the EcoRV site of pBluescript SK+. pHSE-1M2M was constructed by ligating a PCR product derived from plasmid pYEpCUP1HSE1m2m into the EcoRV site of pBluescript SK+. The ability of HSEs from various genes to function as heat-inducible upstream activation sequences (UAS) was tested using the CYC1-lacZ fusion plasmid pCM64, a gift from Charles Moehle.
Plasmid pBS:SKN7:URA3 was constructed as follows. The hisG-URA3-hisG cassette (1) was removed from plasmid pNKY51 by BglII-BamHI digestion, filled in, and ligated into plasmid pBS:SKN7 (a generous gift from Richard Stewart) which had been digested with StyI-MscI. The StyI-MscI digestion removes nucleotides that code for approximately 480 of the 622 amino acids of SKN7 from plasmid pBS:SKN7. Digestion of pBS:SKN7:URA3 with XbaI-XhoI produces a fragment containing the hisG-URA3-hisG cassette with SKN7 sequence flanking each end. To facilitate the purification of full-length yHSF from Escherichia coli, plasmid pET3d-HSF-His6, which contains a six-His tag added to the carboxy terminus of the yHSF open reading frame cloned into pET3d, was constructed. The following plasmids were utilized for making antisense RNA probes by using T7 RNA polymerase and for RNase protection assays. Plasmids pKSACT1 and pKSlacZ, for determining CUP1-lacZ and ACT1 mRNA levels, were described elsewhere (32). Plasmid pKSSSA3 was constructed by ligating a 159-bp EcoRI-HincII fragment from the SSA3 gene into the EcoRI-SmaI sites of pBluescript KS+. Plasmid pSKCUP1 was constructed by inserting a 149-bp EcoRI-BamHI fragment from the CUP1 gene into the same sites of pBluescript SK+. pSKHSC82 was constructed by ligating a PCR product containing a 115-bp fragment of the HSC82 gene to which EcoRI-BamHI sites were introduced into the same sites of pBluescript SK+. Plasmid pSKHSP82 was constructed by ligating a PCR product containing a 109-bp fragment of the HSP82 gene to which EcoRI-BamHI sites were introduced into the same sites of pBluescript SK+. The latter two plasmids were used to generate antisense RNA probes which hybridize specifically to HSC82 or HSP82 mRNA spanning positions +2161 to +2275 and +2196 to +2305, respectively, in the 3′ untranslated regions of both genes (18, 25).
RNA isolation and RNase protection analyses.
RNA from either control, heat shock-treated, or menadione-treated cells was isolated as previously described (36, 63). 32P-labeled antisense CUP1, HSC82, and HSP82 RNAs were produced from BamHI-linearized plasmids. The ACT1 mRNA level was used as a control for normalization for quantitation of RNase protection products throughout this study. RNase protection samples were separated on 6% acrylamide gels; radioactive bands on the dried gels were quantitated by using a PhosphorImager SP and IPLab Gel software (Molecular Dynamics) as described elsewhere (29).
In vitro DNA binding studies.
EMSAs were carried out as described previously (57, 59, 63). Plasmids pHSE-WT, pHSE-M, and pHSE-1M,2M were used to prepare CUP1 HSEWT (wild-type HSE), HSE1M, and HSE1M,2M probes for EMSA by digesting with EcoRI and HindIII and filling in the 103-bp fragments with the Klenow fragment and [α-32P]dATP. Yeast extracts for EMSA were prepared from cells by glass bead disruption in 50 mM Tris-Cl (pH 7.5)–1 mM EDTA–protease inhibitors as previously described (63). Binding reactions were for 30 min at room temperature; the binding buffer was previously described (57, 59). Protein levels in yeast extracts was measured by using the Bio-Rad protein assay (Bio-Rad, Hercules, Calif.). Radiolabeled probes were purified by nondenaturing polyacrylamide gel electrophoresis (PAGE), and probe concentrations were determined spectrophotometrically by UV light absorbance at 260 nm. Quantitative DNA binding studies were carried out, and apparent Kd (Kd,app) and Hill coefficients were determined as described previously (29). Competitive DNA binding assays using CUP1 HSEWT, HSE1M, and HSE1M,2M probes were carried out as previously described (57, 59). Purified yHSF and 0.1 nM probe were used for all Kd,app determinations, and a 4% polyacrylamide gel system with a high cross-linking ratio, 5.6:1, of acrylamide to bisacrylamide was used (62) for all assays with purified yHSF. These gels were run at 4°C and contained 0.5× Tris-borate-EDTA 10% glycerol, and 0.1% Nonidet P-40 (NP-40); the running buffer also contained 0.5× Tris-borate-EDTA and 0.1% NP-40. Following electrophoresis, EMSA gels were fixed (10% acetic, 10% methanol), dried, exposed to X-ray film, and subjected to PhosphorImager analysis.
The DNase I footprinting reactions were carried out as for the EMSA DNA-binding reactions except that after the binding incubation, 1/10 volume of a buffer containing 25 mM MgCl2 and 20 mM CaCl2 and 1 μl of a 1:2,000 dilution of DNase I (10 U/ml; Boehringer Mannheim, Indianapolis, Ind.) were added, and the mixture was incubated for 1 min. DNase I digestion was terminated by the addition of 1/10 volume of 250 mM EDTA and loaded immediately onto an EMSA gel (65). Radioactive bands were excised from the EMSA gel, DNA ethanol precipitated, and fractionated on denaturing polyacrylamide gels (47). The gels were dried and exposed to X-ray film and PhosphorImager screens.
Limited proteolysis of yHSF-HSE complexes.
The proteolytic clipping band shift assay (52) was carried out as previously described (22, 64). Briefly, purified yHSF-DNA complexes formed at room temperature for 30 min were subjected to limited proteolysis with chymotrypsin (amounts of chymotrypsin and lengths of incubation are indicated in figure legends). Binding reactions and the gel system used for these limited proteolysis experiments were identical to those described above used in Kd,app determinations. The chymotrypsin (Worthington Biochemical Corporation) was diluted into water just before use. Chymostatin (Boehringer Mannheim) was used to terminate reactions in the limited proteolytic time course assays. Fixed-time limited proteolysis assays were terminated by direct loading to EMSA gels. Following electrophoresis, EMSA gels were fixed (10% acetic, 10% methanol), dried, exposed to X-ray film, and subjected to PhosphorImager analysis. The amount of yHSF-DNA complexes and free probe remaining after limited proteolysis was quantitated by PhosphorImager analysis of the dried gels.
Expression and purification of yHSF from E. coli.
Full-length yHSF was expressed and purified by standard protocols (3), with minor modifications. Six liters of freshly transformed E. coli BL21(DE3)pLysS cells containing plasmid pET3d-HSF-His6 was grown in Superbroth (Digene Diagnostics, Beltsville, Md.) at 37°C to an A600 of 0.8. Isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.4 mM, and induction was carried out at 25°C for approximately 6 h. Cells were harvested, and cell pellets were frozen in liquid nitrogen and stored at −80°C. All subsequent steps in this purification were carried out at 4°C. Cell pellets were thawed in the presence of protease inhibitor cocktail (3); cells were resuspended in approximately 100 ml of breaking buffer (50 mM sodium phosphate [pH 8], 300 mM NaCl, 10% glycerol) and broken by one passage through a French pressure cell (SLM Aminco, Champaign, Ill.) at 16,000 lb/in2. The cell extract was centrifuged for 30 min at 30,000 × g, the pH of the supernatant was adjusted to approximately 8, and incubation continued for 30 min with gentle mixing at 4°C in batch, using 3 ml of packed Ni-nitrilotriacetic acid (NTA) resin (Qiagen, Chatsworth, Calif.) per 50 ml of extract. The resin was washed twice by centrifugation at 500 rpm in a RT6000B swinging-bucket centrifuge (Sorvall, Wilmington, Del.) at 4°C with wash buffer 1, which was identical to breaking buffer except that it contained 500 mM NaCl and 5 mM imidazole. The resin was then washed once with wash buffer 2, which was identical to wash buffer 1 except that it contained 10 mM imidazole. The resin was pooled, and elution of yHSF was effected by two successive incubations with 6 ml of elution buffer (identical to wash buffer 1 except that it contained 200 mM imidazole) for 15 min with gentle mixing. The eluted sample was aliquoted, frozen by using liquid nitrogen, and stored at −80°C. HSF was further purified by gel filtration chromatography on a Superose 6 HR10/30 column (Pharmacia, Piscataway, N.J.). Procedures for calibration and FPLC (fast protein liquid chromatography) purification using the Superose 6 column (31, 59, 68) and the chromatography buffer (68) have been described elsewhere. Briefly, the Ni-NTA resin eluate was thawed and adjusted to 0.1 mM EDTA–0.1 mM EGTA–0.1% NP-40 immediately prior to injection of 0.2 ml onto the Superose column. The column chromatography buffer was modified by the addition of 0.1% NP-40, the column was run at 0.3 ml/min, and 0.3-ml fractions were taken. After binding of the centrifuged cell lysate to the Ni-NTA resin, the protease inhibitor mix was replaced by the single protease inhibitor Pefabloc (Boehringer Mannheim). Pefabloc was used in all buffers for all remaining steps. HSF eluted at approximately 12 ml (fractions 37 to 45), immediately before the position where the thyroglobulin standard (660 kDa) elutes. The purified yHSF was stable at 0°C for several weeks.
Protein extraction and immunoblotting.
Whole-cell protein extracts for immunoblotting were prepared exactly as described previously (35) by glass bead extraction using sodium dodecyl sulfate (SDS) harvest buffer (0.5% SDS, 10 mM Tris-HCl [pH 7.4], 1 mM EDTA) containing protease inhibitors. Protein concentration was determined by the Bradford assay (Bio-Rad). Extracts were resolved by SDS-PAGE (10% gel), transferred to nitrocellulose, and immunoblotted under standard conditions. Immunoblotting was carried out with reagents and protocols from Amersham, using anti-yHSF polyclonal antiserum (a gift from P. Sorger), Hsc82/Hsp82p polyclonal antibody (a gift from S. Lindquist), Ssa3/Ssa4p polyclonal antibody (a gift from E. Craig), and monoclonal antibodies against phosphoglycerate kinase (Pgk1p; Molecular Probes, Eugene, Oreg.). Proteins of interest were detected by using the Renaissance chemiluminescence detection system (NEN Life Sciences, Boston, Mass.). Band intensity was estimated using NIH Image version v1.61.
RESULTS
The CUP1 promoter harbors two nonconsensus HSEs.
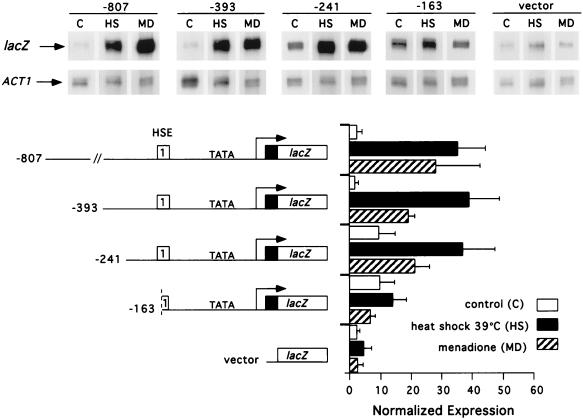
We previously identified a single HSE in the CUP1 promoter required for transcriptional activation in response to heat shock, glucose starvation, and superoxide radical generation (36, 63). Transcription of CUP1 via this HSE is distinct from that of HSP70 genes in its requirement for the yHSF CTA and for heat shock at 39 rather than 37°C (63). To identify other regulatory sites that might function in the transcriptional activation of CUP1 by yHSF, we fused a series of CUP1 promoter deletion mutants to the lacZ gene in YEp357R and analyzed expression from these plasmids by RNase protection experiments (Fig. 1). DNA sequences between −807 and −241 of the CUP1 promoter do not appear to contribute to the magnitude of CUP1 activation in response to either heat shock or menadione treatment (Fig. 1). Deletion of this segment of DNA increases the basal level of CUP1 expression approximately fourfold (Fig. 1). Truncation of the CUP1 promoter to −163, which destroys the first pentameric unit in HSE1, also increased basal transcription fourfold compared to longer promoter fragments but essentially eliminated the activation of CUP1 by HSF in response to both heat shock and oxidative stress (pYEpCUP1-163 [Fig. 1]). Analysis of the 3′ CUP1 promoter region showed that deletions of the CUP1 transcribed region to +9 from the start site of transcription had no significant effect on the magnitude of the transcriptional activation of CUP1 in response to heat shock or oxidative stress (data not shown).
FIG. 1.
Analysis of CUP1 promoter sequences required for heat shock and oxidative stress-inducible transcription. Total RNA was isolated from transformants of strain DTY123 harboring the indicated CUP1-lacZ promoter derivatives, and steady-state mRNA levels for lacZ and ACT1 (indicated with arrows) were analyzed by RNase protection experiments. Values for normalized expression were determined before (control [C]; 27°C) and after heat shock (HS; 39°C) or menadione treatment (MD; 500 μM). Heat shock was carried out for 20 min; cells were exposed to menadione for 1 h. Quantitation was carried out with a PhosphorImager, and in each case the CUP1-lacZ mRNA level was normalized to the respective ACT1 mRNA level. The values represent averages of three separate determinations ± standard deviations. The graph indicates the normalized expression levels of CUP1-lacZ mRNA detected in each lane. Nucleotide numbers refer to positions relative to the start site of transcription of the CUP1 gene; vector represents YEp357R.
Based on the observation that DNA sequences upstream of −241 are not required for heat shock induction of CUP1, we investigated whether the CUP1 HSE1 alone was sufficient to function as a heat-inducible UAS. The HSEs from the SSA1, SSA3, and SSA4 genes were previously shown to be sufficient to function as heat-inducible UASs (6, 73). DNA sequences encompassing the CUP1 HSEs from −168 to −141 or −168 to −116 from the transcription start site were unable to activate heat-induced transcription when fused to the yeast CYC1 basal promoter, while the SSA3 HSE strongly activated heat-induced transcription in this context (data not shown). Longer fragments of the wild-type CUP1 promoter (−393 to −91, −183 to −79, −393 to −1, or −168 to −1 from transcription start) were also unable to activate transcription in this context, implicating a requirement for specific CUP1 basal promoter elements for yHSF-mediated activation of CUP1. Therefore, the CUP1-lacZ fusions used throughout this study contain the CUP1 HSE and basal promoter fused to lacZ.
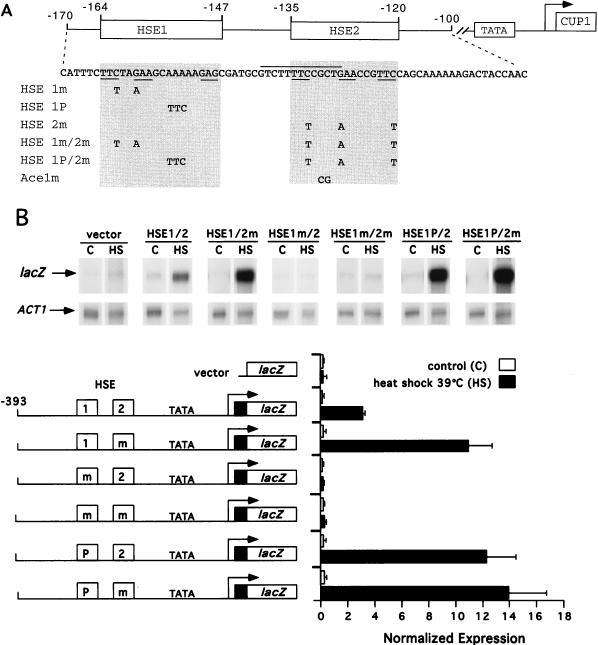
In contrast to the HSEs found within the SSA1 and SSA3 promoters, the CUP1 HSE1 contains only three pentameric units, with a gap between the second and third units (Fig. 2A). However, DNase I footprinting and methylation interference analyses have shown that the HSF trimer interacts with all three pentameric sites (2, 63). Since a 200-fold molar excess of an oligonucleotide containing sequences adjacent to HSE1 (−141 to −107) competes for the binding of yHSF in crude extracts with a probe containing −241 to +37 of the CUP1 promoter (54), we investigated whether a second nonconsensus HSE, HSE2 (Fig. 2A), might function in CUP1 transcriptional activation. The two HSEs are similar in that both contain only three GAA units and both start with TTC.
FIG. 2.
Mutagenesis of HSE1 or HSE2 alters stress-responsive expression from the CUP1 promoter. (A) Diagrammatic representation of the CUP1 promoter, indicating the positions of HSEs and the sequence from −170 to −100 (relative to the transcription initiation site). The nucleotide changes generating mutations in either HSE1, HSE2, or Ace1 sites are shown. Shaded regions delineating the limits of the GAA units of each HSE are numbered, and nucleotide changes for each mutant are displayed in the shaded regions. GAA units are underlined in the CUP1 sequence; the high-affinity Ace1p site (29) which partially overlaps HSE2 is overlined. (B) RNase protection analysis of the transcriptional activation from CUP1-lacZ promoter derivatives containing mutations in HSE1 and HSE2. Heat shock (HS) was carried out at 39°C for 20 min. Controls (C) were not heat shocked. Below the RNase protection data are schematic representations of the CUP1-lacZ promoter derivatives. HSE1 and HSE2 are boxed; mutations in the GAA units of HSE1 or HSE2 (m) and the mutation which fills in the gap in HSE1 (P) are indicated. The histogram shows the normalized expression levels of CUP1-lacZ mRNA detected in each assay.
CUP1 HSE architecture and arrangement modulates transcriptional potency.
To determine whether the gap in the CUP1 HSE1 or the putative HSE2 plays a role in the regulation of CUP1 transcription in response to heat stress, expression from CUP1 promoters with mutationally altered HSE1 and HSE2 elements (summarized in Fig. 2A) was analyzed by RNase protection. Two strategies were adopted. First, the spacing in HSE1 was altered to match the consensus HSEs such as those found in the SSA1 and SSA3 promoters. Second, the putative HSE2 was mutagenized within each pentamer at positions known to be essential for HSF binding to consensus HSEs (19). Conversion of the gapped CUP1 HSE1 to an HSE with four GAA repeats, designated HSE1 “perfect” (HSE1P/2), resulted in 3.9- and 3.6-fold increases in CUP1 activation in response to both heat shock and oxidative stress (data not shown), respectively, compared to the wild type (Fig. 2B). Mutagenesis of HSE2 (HSE1/2m [Fig. 2]) resulted in a 3.5-fold increase in the transcriptional activation of CUP1 in response to heat shock and oxidative stress (data not shown) compared to the wild type (Fig. 2B). This hyperactivation depended on the functional integrity of HSE1, as demonstrated by the inactivity of the double mutant HSE1m/2m (Fig. 2B). Combination of the two mutations (HSE1P/2m) did not result in any significant difference in expression as compared to the CUP1 HSE1P/2 promoter (Fig. 2B). Mutation of HSE1 alone (HSE1m/2 [Fig. 2A]) also resulted in a CUP1-lacZ fusion that was transcriptionally inactive to both heat shock and oxidative stress (Fig. 2B and data not shown), confirming our previous results demonstrating the requirement for HSE1 in the stress induction of CUP1 (36, 63). These results with either double mutation (HSE1P/2m and HSE1m/2m) demonstrate that modulation of the transcriptional activation of CUP1 through HSE2 is highly dependent on the nature of HSE1.
Based on the results obtained with the HSE1P CUP1-lacZ fusion, we synthesized oligonucleotides spanning the HSE1P mutation to determine whether HSE1P could confer heat shock-inducible expression to the CYC1 basal promoter. An oligonucleotide containing the HSE1P mutation and spanning from −168 to −116 or from −168 to −141 of the CUP1 promoter potently activated the CYC1-lacZ reporter in response to heat shock (data not shown). Therefore, the requirement for the basal promoter region in the yHSF-mediated transcriptional activation of CUP1 in response to heat shock can be dispensed with by using a canonical HSE but not the CUP1 HSE1.
The activation of CUP1 by heat shock and oxidative stress has been previously shown to be independent of the Cu ion-dependent transcription factor, Ace1p (54, 63). However, a high-affinity Ace1 binding site overlaps the TTC and partially overlaps the second GAA unit in HSE2 (Fig. 2A), and the HSE2 mutation converts the GAA to AAA, disrupting one nucleotide in this Ace1p site. We therefore analyzed transcriptional activation from a CUP1-lacZ fusion which destroys the high-affinity Ace1p site that overlaps the HSE2 sequence but does not mutate HSE2 (Fig. 2A, Ace1m). Activation of the Ace1m CUP1-lacZ fusion in response to heat stress was indistinguishable from the wild-type promoter (data not shown). Therefore, the increased transcriptional activation observed for the HSE1/2m CUP1-lacZ fusion gene is independent of activation by Ace1p.
Another stress-responsive transcription factor found in S. cerevisiae, Skn7p, possesses significant homology to the DNA binding domain of yHSF (9) and binds to a GAA-containing sequence of the TRX2 promoter (37). Therefore, we investigated whether the increased transcriptional response of the HSE1P/2 and HSE1/2m CUP1-lacZ promoters might be due to Skn7p-mediated activation. The heat shock responses of both wild-type and mutant CUP1-lacZ fusions in a skn7 disruption strain were indistinguishable from that of the wild-type SKN7 strain (data not shown). Taken together, these results demonstrate that the modulation of CUP1 expression in response to heat shock is mediated by HSF, HSE1, and HSE2.
The architecture of CUP1 HSEs imparts specificity to the mode of activation of CUP1.
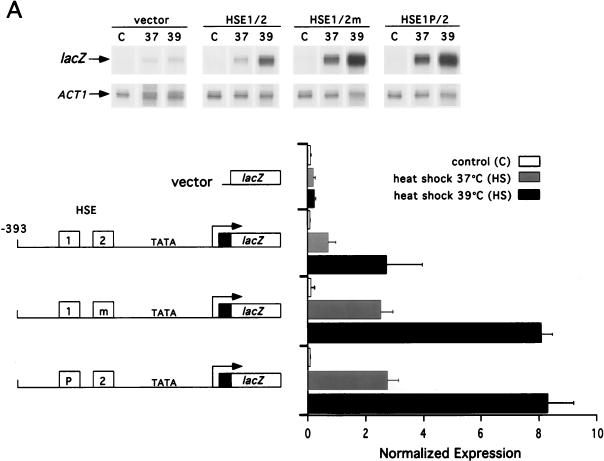
Transcriptional activation of CUP1 by yHSF differs from that of SSA3 in that CUP1 activation requires an optimal heat shock temperature of 39 rather than 37°C (63). Furthermore, CUP1 expression in response to heat shock is highly dependent on the CTA of yHSF, whereas the SSA1 and SSA3 promoters are much less dependent on this domain. To determine if HSE1 and HSE2 are determinants in these features of CUP1 transcriptional activation, we compared expression from the wild-type and mutationally altered CUP1-lacZ fusion genes at 37 and 39°C (Fig. 3A). Consistent with previous results (63), activation of CUP1 at 37°C was only 25% of that observed at 39°C (Fig. 3A). Interestingly, the generation of either HSE1P or HSE2M did not alter the temperature induction profile of CUP1; that is, expression of both derivatives was maximal at 39°C. Both mutations, however, change the efficacy of transcription at 37°C. The HSE1/2m and HSE1P/2 CUP1 derivatives give rise to a level of heat shock-inducible transcription at 37°C that is comparable to that observed for the wild-type fusion at 39°C (Fig. 3A). Therefore, both HSE2 and the gap between pentamers 2 and 3 in HSE1 act to limit the expression of CUP1 at a temperature where many other HSF-responsive genes are near maximal expression.
FIG. 3.
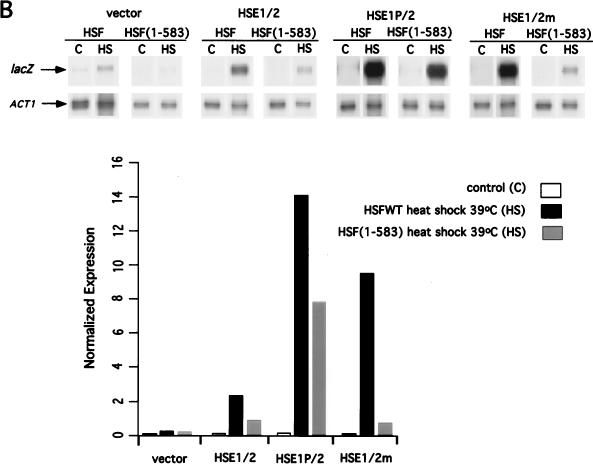
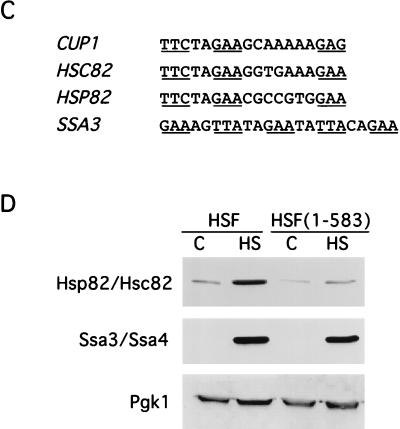
CUP1 promoter HSE mutations alter the temperature response and obviate the requirement for the yHSF CTA. (A) The HSE1P and HSE2m CUP1-lacZ fusions lower the temperature threshold for transcriptional activation of CUP1. The steady-state levels of CUP1-lacZ mRNA from the wild-type, HSE1P, and HSE2m fusions were analyzed before (control [C]; 27°C) and after heat shock (HS) at either 37 or 39°C for 20 min. ACT1 and CUP1-lacZ mRNA levels were assayed and quantitated as described for Fig. 1. Below the data are schematic representations of the CUP1-lacZ promoter derivatives assayed in this RNase protection experiment and normalized expression levels of CUP1-lacZ mRNA. Details of the mutations in the CUP1 HSEs are given in Fig. 2. (B) The HSE1P but not the HSE2m CUP1-lacZ fusion reduces dependency of CUP1 transcriptional activation on the yHSF CTA. Experiments were carried out as described for panel A except that the CUP1-lacZ fusions were also assayed in a yeast strain containing the HSF(1-583) allele. (C) HSC82 and HSP82 possess similar GAA unit arrangements in HSE1, with a gap between units 2 and 3; SSA3 contains a contiguous array of 5-bp units. (D) Deletion of the HSF carboxyl-terminal activation domain results in severe reduction in Hsp82/Hsc82 protein levels, while Ssa3/Ssa4 protein levels are only moderately affected, as determined by Western blot analysis of Hsp82/Hsc82, Ssa3/Ssa4, and Pgk1 protein levels in yeast strains containing either wild-type HSF or HSF(1-583). Yeast cells were heat shocked (HS) at 39°C for 1 h, and extracts were prepared by glass bead disruption as described in Materials and Methods. Samples were subjected to SDS-PAGE and immunoblotted with polyclonal antisera to Hsp82/Hsc82, Ssa3/Ssa4, and Pgk1. Pgk1 levels were used for normalizing sample loads.
Isogenic wild-type HSF and HSF(1-583) cells harboring wild-type and mutant CUP1-lacZ fusions were analyzed to ascertain whether HSE1 or HSE2 plays a role in the dependence of CUP1 transcriptional activation on the yHSF CTA. As shown in Fig. 3B and consistent with previous analyses (36, 63), heat shock activation of the wild-type CUP1-lacZ reporter in the HSF(1-583) strain is greatly reduced (approximately 70%) compared to a strain with wild-type HSF. In contrast, heat shock activation of the HSE1P CUP1-lacZ reporter in the HSF(1-583) strain is reduced only 43% compared to a strain with wild-type HSF. These results suggest that the gapped HSE1 plays a critical role in determining the degree of dependence of CUP1 expression in response to heat shock on the HSF CTA. Transcription from the HSE1P/2 CUP1 promoter derivative was hyperactivated in the HSF(1-583) strain, exhibiting approximately threefold greater activation than the wild-type promoter in the wild-type HSF strain (Fig. 3B). In contrast to expression in a wild-type HSF strain, the HSE1/2m reporter expression is greatly reduced in the HSF(1-583) strain, with an induction approximately equal to that observed for the wild-type reporter in the HSF(1-583) strain (Fig. 3B). This finding suggests that the transcriptional activation observed for the HSE1/2m promoter under stress conditions is dependent on the interaction of yHSF with HSE1. Similar results were obtained in response to oxidative stress using the wild-type, HSE1/2m, and HSE1P/2 reporter plasmids in the HSF(1-583) strain and wild-type HSF strain (data not shown). The data for the HSF(1-583) strain suggest that the HSE1P promoter increases the ability of the amino-terminal activation domain of HSF to activate CUP1 expression. Since the HSF(1-583) protein completely lacks a CTA, the results in Fig. 3B suggest that the gapped HSE1 plays a critical role in determining the contribution of the amino-terminal activation domain of HSF to the magnitude of CUP1 expression in response to both heat shock and oxidative stress.
To ascertain whether the correlation between a gapped HSE architecture and higher dependence on the yHSF CTA is a general phenomenon, other Hsp gene promoters with similarly organized HSEs were examined. Inspection of HSEs found in the promoters of HSP82, HSC82, and CUP1 suggests that nonconsensus HSEs may be commonly used for transcriptional responses to heat shock. Previous analysis of the HSE1 from the HSC82 and HSP82 promoters (8, 18, 24, 25) showed that, like the CUP1 HSE1, they are composed of only three pentameric units containing a gap between pentamers 2 and 3, with all three sites properly oriented and spaced (Fig. 3C). Based on these observations, we measured the heat-induced levels of endogenous CUP1, HSC82, and HSP82 mRNAs to determine if these genes exhibit a strong dependence on the yHSF CTA. Heat shock-induced expression of the CUP1 and HSC82 genes in the HSF(1-583) strain are most affected, with heat shock transcription being only 23 and 24%, respectively, of that observed in an isogenic wild-type HSF strain. Expression of HSP82 in the HSF(1-583) strain is only 37% of that in the wild-type HSF strain, while heat shock expression of SSA3 in the HSF(1-583) strain was least affected by the loss of the yHSF CTA (56% of the level in wild-type strain). This result with SSA3 is identical to that observed with the HSE1P/2 reporter in Fig. 3B, where 56% of the steady-state expression level in the HSF(1-583) background was retained compared to that present in the HSF wild-type strain. To determine whether there are significant physiological consequences for the reduction in the magnitude of transcriptional activation for the HSP82, HSC82, and SSA3 genes in the HSF(1-583) strain, the levels of these proteins in control and heat-shocked cells were determined by immunoblotting. Consistent with the steady-state RNA measurements for the HSP82 and HSC82 genes demonstrating a strong requirement for the HSF CTA in heat-induced expression of these two genes, protein levels of both Hsp90 isoforms were severely diminished (80 to 90%) in HSF(1-583) cells (Fig. 3D). In contrast, the levels of Ssa3/Ssa4 proteins were only slightly reduced (10 to 20%) in HSF(1-583) cells (Fig. 3D). These results strongly suggest that heat shock-induced expression from promoters containing contiguous HSEs is less dependent on the yHSF CTA than expression from promoters with gapped HSEs.
yHSF binding to HSE2 modulates interactions at HSE1.
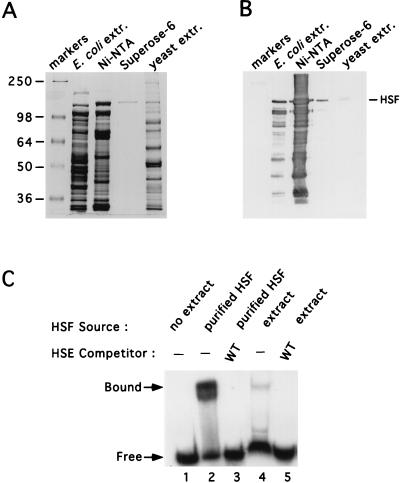
The data described here implicate the presence of a second CUP1 HSE, HSE2, in the modulation of CUP1 transcription that is dependent on both yHSF and the nonconsensus HSE1. To ascertain whether yHSF interacts directly with HSE2 and whether this interaction might modulate the occupancy of HSE1, in vitro DNA binding studies were carried out. Since yeast cells express two proteins (10, 21) bearing homology to the yHSF DNA binding domain that appear to play no role in CUP1 regulation but which may confound in vitro DNA binding studies, full-length yHSF was expressed in and purified from E. coli. To facilitate the purification of yHSF for DNA binding studies, we constructed a yHSF allele in which a His6 tag was placed at the carboxyl terminus of the coding region. This HSF-His6 protein fully complemented the viability defect associated with disruption of the single endogenous yHSF gene at both 30 and 37°C (data not shown). yHSF was obtained after sequential purification on Ni-NTA agarose and FPLC Superose-6 chromatography (31, 59, 68). Purified yHSF migrated on a Coomassie blue-stained SDS-polyacrylamide gel at approximately 150 kDa and comigrated with HSF present in whole-cell yeast extracts from non-heat-shocked cells, as detected by Western blot analysis (Fig. 4A and B). Furthermore, purified yHSF specifically bound to the CUP1 promoter in a manner similar to yHSF present in crude cell extracts from non-heat-shocked cells (Fig. 4C). The amount of yHSF from crude cell extracts binding to CUP1 DNA is lower than that in the recombinant yHSF samples due to the low abundance of endogenous yHSF in the cell extracts used in the binding reaction.
FIG. 4.
Purification of yHSF from E. coli. (A) Coomassie blue-stained SDS–8% polyacrylamide gel of the purified full-length yHSF protein. Molecular mass markers, in kilodaltons, are indicated on the left. Stages in the purification, described in Materials and Methods, are indicated on the top. The right-most lane contains yeast extract which was used for size comparison for Western blotting analysis. (B) yHSF analysis by Western blotting. The same samples used to load the gel shown in panel A were electrophoresed and subjected to immunoblotting with polyclonal anti-yHSF antiserum. Purified yHSF, which comigrates with HSF present in non-heat-shocked yeast cell extract, is indicated by the arrowhead. (C) EMSA of purified yHSF. Lane 1 contains no yHSF protein; lanes 2 and 3 contain purified yHSF; lanes 4 and 5 contain crude yeast extract prepared from non-heat-shocked cells by glass bead disruption as described in Materials and Methods. EMSAs were carried out as described in Materials and Methods. All lanes contain 1 ng of 32P-labeled CUP1 HSEWT probe with or without the CUP1 competitor DNA, at 30-fold molar excess, indicated above the gel.
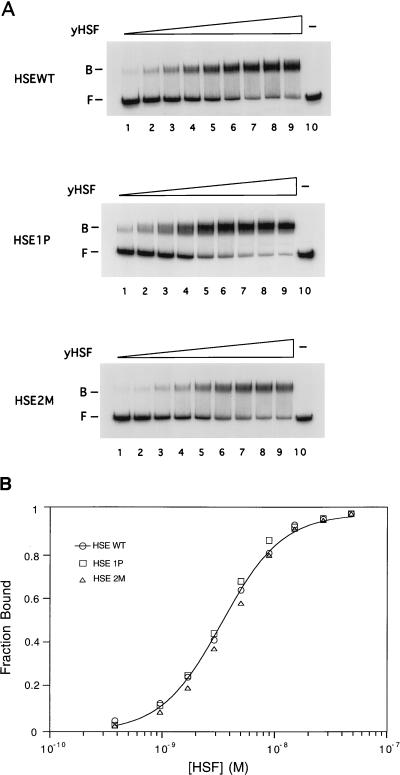
The dissociation constants for yHSF-CUP1 promoter complexes were determined by quantitative EMSAs. As shown in Fig. 5, yHSF interacts with the CUP1 promoter fragment encompassing HSE1 and HSE2 with a Kd,app of (3.7 ± 0.5) × 10−9 M. Since the CUP1 HSE1P/2 derivative gave rise to increased expression in response to stress and this expression was less dependent on the yHSF CTA, we determined whether these effects were due to an increase in binding affinity of yHSF for the CUP1 HSE1P promoter. yHSF bound to the CUP1 HSE1P/2 probe with a Kd,app of (3.4 ± 0.9) × 10−9 M, demonstrating that yHSF does not have a significantly higher affinity for the CUP1 HSE1P compared to the wild-type promoter. The apparent affinity of yHSF for the CUP1 HSE2M probe, (4.0 ± 0.2) × 10−9 M, was not significantly different from that for either the CUP1 HSEWT or CUP1 HSE1P. Furthermore, no difference was observed in the apparent Hill coefficients obtained for the three CUP1 promoter sequences (approximately 1.5), suggesting a lack of differences in yHSF binding cooperativity to these three CUP1 promoter derivatives. This Hill coefficient is highly reproducible, and the intermediate value for the apparent Hill coefficient of between 1 and 2 suggests that one yHSF trimer may bind stably, and a second may bind only weakly or partially, to the CUP1 HSE1. Since the SSA3 and CUP1 promoters also exhibit marked differences in heat shock-inducible gene expression as a function of their HSEs, we explored whether yHSF exhibits different binding affinities for these two promoters. In three independent experiments, differences in neither affinity nor binding cooperativity were observed (data not shown). Therefore, it does not appear that the differences in heat shock-responsive expression between the CUP1 HSE1P, CUP1 HSE2M, and SSA3 promoters and the CUP1 HSEWT promoter are due to differences in the affinity of yHSF for the HSEs or in binding cooperativity. Rather, differences may be due to binding site context-dependent alterations in bound HSF or interactions with other factors.
FIG. 5.
Binding affinity of yHSF for the CUP1 promoter. (A) Results of representative EMSAs of the CUP1 HSEWT (top) CUP1 HSE1P (middle), and CUP1 HSE2M (bottom) promoter fragments titrated with increasing amounts of purified yHSF (lanes 1 through 9). yHSF concentrations used were 5.5 × 10−10, 9.6 × 10−10, 1.6 × 10−9, 2.9 × 10−9, 5 × 10−9, 8.9 × 10−9, 1.5 × 10−8, 2.7 × 10−8, and 4.8 × 10−8 M for lanes 1 through 9, respectively. Lane 10 contains the free probe. The probe concentration in each reaction was 0.1 nM. Positions of the free probe (F) and yHSF-DNA complex (B) are indicated. (B) Graphical representation of the quantitation of the protein titration plots in panel A. The data were quantitated with a PhosphorImager and then plotted and analyzed as described previously (29) and in Materials and Methods. The Kd,app for each probe was derived from at least three independent determinations and is the average ± standard deviation. Kd,apps were (3.7 ± 0.5) × 10−9 M for the CUP1 HSEWT probe (3.4 ± 0.9) × 10−9 M for the CUP1 HSE1P probe, and (4.0 ± 0.2) × 10−9 M for the CUP1 HSE2M probe. Each data point for each probe is taken from the average of at least three independent determinations. The line is drawn only through the data points for the CUP1 HSEWT probe.
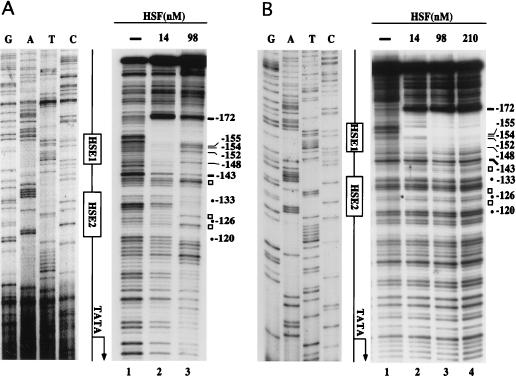
EMSAs using yHSF and CUP1 promoter mutations strongly suggest that yHSF binds to HSE2, albeit with a very low efficiency compared to HSE1 (data not shown). To precisely map the site of interaction of yHSF with the CUP1 HSE2, DNase I footprinting analysis was performed with the CUP1 HSEWT probe (Fig. 6A). At low yHSF concentrations (Fig. 6A, lane 2), strong protection over a region encompassing HSE1, from −172 to −143, was observed. Binding of yHSF to HSE1 is accompanied by DNase I hypersensitivity at several positions upstream of position −172. Additionally, at this concentration of yHSF (14 nM), there is modest protection over the region corresponding to CUP1 HSE2. At a sevenfold-higher concentration of yHSF, however, this DNase I cleavage pattern is altered in several distinct ways (Fig. 6A, lane 3). First, HSE2 is strongly protected from DNase I cleavage by yHSF from positions −134 to −120 on the bottom strand. Second, three sites of DNase I hypersensitivity within or flanking HSE2, at positions −123, −127, and −140, are observed (Fig. 6A). Third, concomitant with more complete occupation of HSE2, there is a marked increase in DNase I cleavage at several positions in the 3′ end of HSE1, including −148, −152, −154, and −155. Therefore, occupation of the lower-affinity HSE2 site by yHSF appears to alter the interaction of yHSF with HSE1.
FIG. 6.
DNase I footprinting analysis of yHSF bound to the CUP1 promoter. The 32P-labeled CUP1 promoter fragments used in DNase I footprinting reactions are described in Materials and Methods, and the sequence of CUP1 encompassing HSE1 and HSE2 is shown in Fig. 2A. The concentration of CUP1 probe in all binding reactions was 0.6 nM, and the probes were labeled at the 5′ end on the bottom strand. Binding reactions were set up, and after 30 min at room temperature, DNase I was added as described in Materials and Methods. The reactions were terminated and loaded directly onto the modified EMSA gels used for Kd determinations. DNA was eluted from the protein-DNA complexes in the EMSA gel, denatured, and loaded on a standard DNA sequencing gel as described in Materials and Methods. (A) DNase I footprinting of the wild-type CUP1 probe. On the left are the reference DNA sequencing reactions showing the sequence of the bottom strand of the CUP1 promoter (for the actual sequence, see Fig. 2A). HSE1 and HSE2 are shown diagrammatically in boxes, the size of each box corresponding to the limits of the three pentameric GAA units in each HSE. The panel on the right shows DNase I footprinting samples: lane 1, DNase I cleavage products generated in the absence of protein; lanes 2 and 3, 0.1 μg (14 nM) and 0.7 μg (98 nM) of yHSF, respectively, added to the binding reactions. Symbols on the right of the DNase I footprinting panel: solid circles labeled −120, −126, and −133 represent the position of the second nucleotide in each pentameric nGAAn unit of HSE2; open squares (not labeled with numbers) represent DNase I-hypersensitive sites; solid dashes labeled −172 and −143 represent the boundary of HSE1. Numbering indicates nucleotide position relative to the start site of transcription. (B) DNase I footprinting of the CUP1 HSE2M mutant probe. Descriptions and symbols are identical to those for panel A except that lane 4 represents 1.5 μg (210 nM) of yHSF added to the binding reaction.
To verify the specificity of yHSF occupation of HSE2 and the effect of yHSF binding to HSE2 on HSE1 binding, DNase I footprinting was carried out with the CUP1 HSE2M probe (Fig. 6B). The complete lack of protection observed over HSE2, even at high yHSF concentrations, demonstrates the specificity of the interaction of yHSF with HSE2 (compare −134 through −120 in Fig. 6A and B). The 5′ boundary of the protected region over HSE1 is identical to that of the HSEWT probe (−172 [Fig. 6]). However, in contrast to the wild-type CUP1 probe, there are no alterations in the protection over HSE1 as more yHSF is added (compare −148 through −155 in Fig. 6A and B). However, the extent of the protected region over HSE1 decreased, and the three hypersensitive sites observed with the wild-type CUP1 probe were abolished (compare Fig. 6A and B). Furthermore, the major cleavage site at −143 (Fig. 6B) and the residues immediately upstream of this site, TCG (bottom strand, −144 to −146) are no longer protected (compare Fig. 6A and B). Therefore, yHSF bound at HSE2 may facilitate the binding of yHSF to the third pentameric unit (GAG) of HSE1.
HSF adopts distinct conformations when bound to consensus and atypical HSEs.
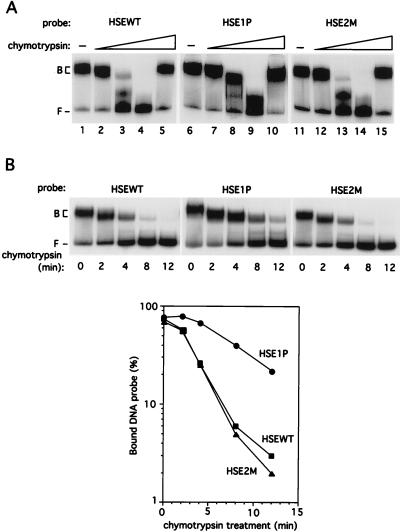
So far, our results show that differences in the transcriptional activity of the CUP1 promoter derivatives cannot be attributed to any changes in either the binding affinity or the cooperativity with which yHSF binds these DNA sequences. To more directly assess whether the differences in transcriptional activation from the CUP1 promoter derivatives are due to changes in the conformation of DNA-bound HSF, protease sensitivity assays were carried out with purified yHSF bound to the CUP1 HSEWT, HSE1P, and HSE2M DNA fragments. The proteolytic clipping band shift assay (52) utilizes limited proteolysis of DNA-bound protein and has been used to probe the structure of transcription factor-DNA complexes (22, 52, 64). HSF-CUP1 HSE complexes were subjected to limited proteolysis by incubation with increasing concentrations of chymotrypsin, and the resulting complexes were separated on EMSA gels (Fig. 7A). The differences in sensitivity to digestion of the HSF-HSEWT and HSF-HSE1P complexes were striking. The HSF-HSE1P complex was routinely more resistant to chymotrypsin treatment than either the HSF-HSEWT or HSF-HSE2M complex (compare lanes 3, 8, and 13 in Fig. 7A). There is approximately an order of magnitude difference in the amount of chymotrypsin required to obtain similar levels of proteolytic sensitivity for the HSEWT and HSE1P. The sensitivities of the HSF-HSEWT and HSF-HSE2M complexes were nearly indistinguishable, suggesting that the major determinant in the chymotrypsin sensitivity of the DNA-bound yHSF is HSE1. Addition of chymostatin to the binding reactions prior to chymotrypsin resulted in complexes which were completely resistant to chymotrypsin (Fig. 7A; compare lanes 5, 10, and 15 with lanes 1, 6, and 11). There were no obvious differences in the pattern of products generated by chymotrypsin digestion of yHSF bound to the HSEWT, HSE1P, or HSE2M (Fig. 7A and data not shown). The data in Fig. 7B shows that the difference in chymotrypsin sensitivity between the HSF-HSE1P and HSF-HSEWT and HSF-HSE2M can also be observed in the rate of proteolysis of these complexes. After 12 min of chymotrypsin digestion, the HSF-HSEWT and HSF-HSE2M complexes are almost completely degraded whereas approximately 25% of the HSF-HSE1P complex remains (Fig. 7B). Thus, the HSF-HSE1P complex adopts a conformation distinct from the HSF-HSEWT and HSF-HSE2M complexes which can be demonstrated as differences in both the concentration and rate of limited digestion with chymotrypsin (Fig. 7).
FIG. 7.
yHSF binds to the CUP1 HSEWT, HSE1P, and HSE2M with distinct conformations. (A) The partial proteolysis with chymotrypsin of the HSF-HSEWT complex is compared with that of the HSF-HSE1P and HSF-HSE2M complexes. Binding reactions containing the indicated probe were carried out exactly as described for Fig. 5 except that all lanes contained 1.5 × 10−8 M HSF. Following the standard binding assay, chymotrypsin was added to all reactions except those in lanes 1, 6, and 11. The HSF-HSE complexes in lanes 2 through 4, 7 through 9, and 12 through 14 were digested with 0.01, 0.1, and 1 ng chymotrypsin, respectively. Binding reaction mixtures were incubated for 10 min following chymotrypsin addition before loading onto EMSA gels. The chymotrypsin inhibitor chymostatin (1 μg) was added to the samples in lanes 5, 10, and 15 immediately prior to chymotrypsin (1 ng) addition. Bound HSF-HSE complexes (B) and free probe (F) are indicated. (B) Graphical representation of the results from time course assays of partial proteolysis of HSF-HSE complexes. Experimental conditions were as for panel A except that chymotrypsin digestion (1 ng) was carried out for the indicated time followed by chymostatin addition to terminate the digestion prior to loading onto EMSA gels. The bound HSF-HSE complexes (B) and free probe (F) were quantitated with a PhosphorImager and plotted. The data represent the averages of two separate experiments.
DISCUSSION
Higher eukaryotic cells possess multiple distinct HSF isoforms, encoded by different genes. This diversity is further increased through differential splicing, responses to distinct stresses, and preferences for binding to distinct arrangements of HSEs (70). In contrast, the S. cerevisiae HSF is encoded by a single, essential gene and binds to some HSEs constitutively, while binding to other HSEs is induced in response to an environmental or pharmacological stimulus (24, 27, 58). Furthermore, yHSF differentially activates gene expression through the use of separate amino- or carboxyl-terminal transactivation domains or by responding to distinct stressors (36, 42, 56, 63). Therefore, yHSF may represent a composite of the functions carried out by individual HSF isoforms in higher eukaryotes. The observation that human HSF isoforms are differentially functional when expressed in yeast cells lacking the endogenous HSF gene further underscores this notion (35).
To explore the mechanisms underlying the differential use of yHSF transactivation domains for target gene activation, HSF-dependent activation of the CUP1 gene was investigated in detail. CUP1 represents a heretofore atypical HSF-dependent gene in that it contains a nonconsensus HSE in its promoter, requires heat shock at 39°C for robust activation, as opposed to 37°C for other HSF target genes, responds to superoxide radical generators for HSF-mediated activation, and exhibits a strong requirement for the yHSF CTA (36, 63). Here, we have demonstrated that the CUP1 promoter harbors two HSEs, neither of which resembles those found in typical HSF-responsive genes, such as SSA1 or SSA3, in their fundamental architecture. Consistent with the known interaction of HSF with HSEs as homotrimeric proteins, both the CUP1 HSE1 and HSE2 harbor three repeats of the pentameric element. Furthermore, the separation of CUP1 HSE1 and HSE2 by one helical turn provides a mechanism for potential interactions between two DNA-bound HSF trimers on the same face of the DNA helix. Although the CUP1 HSE1 contains three pentamers, the distance of one helical turn between pentamers 2 and 3 allows occupancy of major grooves on the DNA with a distinct geometry compared to contiguous pentamers such as those found in SSA1 and SSA3. Indeed, the generation of a CUP1 HSE1 derivative which mimics those found in SSA1 and SSA3 (CUP1 HSE1P) results in stress-responsive transcriptional activation characteristics which more closely resemble these genes in terms of the temperature optimum, their reduced dependence on the yHSF CTA, and ability to activate a heterologous CYC1 basal promoter. One possible mechanism by which the CUP1 HSE1P might enhance CUP1 expression is by affecting the affinity of yHSF for DNA. However, our results suggest no significant difference in the apparent affinity of yHSF for the CUP1 promoter fragment containing HSE1P compared to HSE1WT.
Previous experiments have demonstrated that HSF-dependent activation of CUP1 in response to heat and oxidative stress is absolutely dependent on HSE1 (36, 63). Here, we have shown that although neither HSE1 nor HSE2 functions to activate heat-inducible expression in the context of a fusion to the yeast CYC1 core promoter, HSE2 plays an important role in modulating CUP1 expression through HSE1. The inability of HSE2 alone to function as an activating HSE, even in the context of the CUP1 promoter, may in part be due to its low affinity for yHSF, a consequence of the altered spacing between each of the three pentamers (2, 30, 71). This architecture of HSE2 may also affect structural changes in the CUP1 promoter DNA upon binding of yHSF. DNase I treatment of yHSF-CUP1 promoter DNA complexes results in hypersensitive sites within and adjacent to HSE2, suggesting that the binding of yHSF to the CUP1 DNA induces conformational changes in the DNA. Hypersensitive sites have been observed in DNase I treatment of mHSF1- and mHSF2-HSE complexes (31). Although HSE2 is incapable of autonomously driving yHSF-dependent activation of CUP1 transcription and is bound by yHSF with low affinity, the occupation of HSE2 has dramatic effects on stress-dependent activation of CUP1 transcription and the manner in which HSE1 is bound by yHSF. The generation of a form of HSE2 incapable of binding yHSF renders CUP1 heat inducible transcription hyperactivated at 37 and 39°C, in a manner similar to the conversion of CUP1 HSE1 to HSE1P. Furthermore, consistent with potential interactions between yHSF trimers bound both at HSE1 and HSE2, DNase I footprinting assays demonstrate that the occupation of HSE2 alters the manner in which yHSF is bound at HSE1. Studies of the Drosophila hsp70 promoter have demonstrated the presence of a high- and a low-affinity HSE, the latter of which plays a critical role in the heat-inducible transcriptional response (65). It is interesting that an HSE found in the human prointerleukin 1-β gene, which consists of only two pentameric units, fails to serve as a heat shock-inducible element but restrains expression from the promoter in response to heat shock jointly administered with the inducer, lipopolysaccharide (13). It is thought that this may provide a mechanism to temper the inflammatory response. Furthermore, Westwood et al. have demonstrated the binding of Drosophila HSF to chromosomal loci that far exceed the predicted number of heat shock-inducible genes (67). These observations, taken together with the data described here, suggest that in addition to their role as gene-specific positive transcriptional regulatory elements, HSEs might modulate both HSF activity and the activity of distinct cis-acting promoter elements. Similar context-dependent activation or repression has been observed with the retinoic acid receptor bound to its cognate DNA response element (34).
The organization of the CUP1 HSE1 is very similar to that of the HSE1 in the HSP82 and HSC82 genes. We found that the heat shock induction of these three genes is highly dependent on the yHSF CTA. Like CUP1, HSP82 and HSC82 have multiple HSEs; however, others have shown that only the HSE1 of HSP82 and HSC82 is constitutively occupied in vivo (18). Giardina and Lis have shown that there is a change in the in vivo footprint of the HSP82 HSEs following heat shock (24). The changes in HSF-DNA binding upon heat shock were seen mainly on the low-affinity HSEs, HSE2 and HSE3 of HSP82. The binding of yHSF to these weaker HSEs in the HSP82 promoter was transient, and these sites were largely unoccupied once cells progressed through a recovery stage and into the non-heat-shocked stage. It may be that a similar situation occurs on the CUP1 promoter with HSE1 representing the constitutively occupied HSE with HSE2 occupied only upon stress induction. This is consistent with in vitro DNA binding studies performed in this report showing that HSE2 is a low-affinity site. Our DNase I footprinting assays demonstrate that the occupation of HSE2 alters the manner in which yHSF is bound at HSE1 and perhaps in vivo occupancy of HSE2 following stress induction tempers the transcriptional response of CUP1. The Hill coefficient for the HSE2M was approximately 1.5, suggesting that a second trimer may be only weakly bound to HSE1. The occupancy of HSE2 upon stress induction may also act to stabilize yHSF bound to HSE1. Future in vivo footprinting experiments will address these possibilities.
How do the specific architecture of HSE1 and the presence of HSE2 impart unique HSF-dependent regulatory characteristics to CUP1? One mechanism may be that HSF binds to the CUP1 HSE1 with less cooperativity than for a large contiguous HSE, thereby leading to a tempering of CUP1 expression. mHSF1 and mHSF2 differ in the potential for cooperative interactions with HSEs: mHSF1 binds cooperatively to extended HSEs much like that found in the SSA3 gene, and mHSF2 has a binding preference for HSEs harboring two or three pentamers like that in the CUP1 promoter (30, 31). Since the DNA binding domain of yHSF may be more conformationally flexible than that of mHSF1 or mHSF2 (20), perhaps yHSF extracts binding site context information to influence the level of cooperativity used to bind a given promoter. However, our results suggest that yHSF binds to the CUP1 HSE1WT, HSE1P, and HSE2M with nearly identical levels of apparent cooperativity. It is also possible that when bound to HSE1, HSF adopts a conformation that alters its interactions with the basal transcription machinery in the core promoter of the CUP1 gene. Indeed, our results which demonstrate that a consensus HSE from SSA3, or the CUP1 HSE1P but not the CUP1 HSE1WT element, can confer heat-inducible expression to the CYC1 basal promoter are consistent with a requirement for the adaptation of distinct HSF conformations on the different HSEs. Consistent with this idea, substitution of the Gcn4 leucine zipper for the yHSF trimerization domain has recently demonstrated that the oligomeric state of the DNA-bound HSF-Gcn4 chimeric protein depends on the number and orientation of pentameric units within the HSE (17). Therefore, it is possible that the HSE structure can similarly influence the overall yHSF conformation. The in vitro DNA binding, in vivo gene expression, and protease sensitivity assay results described here are consistent with a change in the conformation of DNA-bound yHSF. The digestion of yHSF-HSE complexes with chymotrypsin shows that a yHSF surface is more readily accessible to the protease in the yHSF-HSEWT complex than it is in the yHSF-HSE1P complex. The ability of DNA to induce conformational changes in transcription factors has been previously proposed for the nuclear hormone receptors (61), the yeast pheromone/receptor transcription factor (64), and nuclear factor NF-κB (p50)2 (22). Therefore, it is possible that HSE structure can similarly influence the conformation of yHSF, and perhaps the extended linker region in yHSF facilitates such changes.
What might be the mechanisms by which HSEs with contiguous pentamers exhibit a reduced dependence on the yHSF carboxyl-terminal activation domain compared to CUP1? Since the CTA is known to harbor an additional coiled-coil domain (14), it is conceivable that this region (HSF584-833) is responsible for intermolecular interactions that serve to stabilize yHSF trimers on the CUP1 promoter or to augment interactions between yHSF trimers bound at HSE1 and HSE2. It is also possible that yHSF receives context information from the HSE that specifies which functional surfaces of yHSF will be presented to the transcriptional machinery or to other, non-DNA binding regulatory factors. The chymotrypsin sensitivity data suggest that a gapped HSE such as the CUP1 HSE1 might induce a conformation of HSF which more efficiently utilizes the C-terminal rather than the N-terminal transactivation domain. The more canonical HSE such as HSE1P might result in more of the HSF N-terminal than C-terminal transactivation domain being presented to the transcriptional machinery. A similar mechanism has been proposed to dictate whether the glucocorticoid receptor functions through its response element as a transcriptional activator or repressor (61). As we show here, the architecture of HSEs in the CUP1, HSC82, HSP82, and HSP70 family of genes plays an important role in the features of the heat shock transcriptional response. Perhaps the sequences of these promoter HSEs have evolved to facilitate differential use of the yHSF transactivation domains and thus impart distinct characteristics to the heat shock transcriptional response. Although HSFs are regulated at many levels in response to stress, these studies demonstrate that promoter context represents a further level for regulation of transcription during the stress response.
ACKNOWLEDGMENTS
We thank David Engelke and members of the Thiele lab for critically reading the manuscript and for valuable suggestions. We thank J. José Bonner and David Gross for excellent advice and suggestions. We gratefully acknowledge gifts of plasmids, yeast strains, and antiserum from Peter Sorger, Hillary Nelson, Charles Moehle, Richard Stewart, Susan Lindquist, and Elizabeth Craig. We thank members of the Thiele lab for valuable discussions and advice during the course of this work and Chen Kuang for helpful technical support.
This work was supported by U.S. Environmental Protection Agency fellowship U 914826-01-2 to Nicholas Santoro. Dennis J. Thiele is a Burroughs Wellcome Toxicology Scholar.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amin J, Ananthan J, Voellmy R. Key features of heat shock regulatory elements. Mol Cell Biol. 1988;8:3761–3769. doi: 10.1128/mcb.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 4.Bonner J J, Ballou C, Fackenthal D L. Interactions between DNA-bound trimers of the yeast heat shock factor. Mol Cell Biol. 1994;14:501–508. doi: 10.1128/mcb.14.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonner J J, Heyward S, Fackenthal D L. Temperature-dependent regulation of a heterologous transcription activation domain fused to yeast heat shock transcription factor. Mol Cell Biol. 1992;12:1021–1030. doi: 10.1128/mcb.12.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boorstein W R, Craig E A. Structure and regulation of the SSA4 HSP70 gene of Saccharomyces cerevisiae. J Biol Chem. 1990;265:18912–18921. [PubMed] [Google Scholar]
- 7.Boorstein W R, Craig E A. Transcriptional regulation of SSA3, an HSP70 gene from Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:3262–3267. doi: 10.1128/mcb.10.6.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borkovich K A, Farrelly F W, Finkelstein D B, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown J L, Bussey H, Stewart R C. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 1994;13:5186–5194. doi: 10.1002/j.1460-2075.1994.tb06849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown J L, North S, Bussey H. SKN7, a yeast multicopy suppressor of a mutation affecting cell wall β-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J Bacteriol. 1993;175:6908–6915. doi: 10.1128/jb.175.21.6908-6915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler G, Thiele D J. ACE2, an activator of yeast metallothionein expression which is homologous to SWI5. Mol Cell Biol. 1991;11:476–485. doi: 10.1128/mcb.11.1.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butt T R, Sternberg E, Herd J, Crooke S T. Cloning and expression of a yeast copper metallothionein gene. Gene. 1984;27:23–33. doi: 10.1016/0378-1119(84)90235-x. [DOI] [PubMed] [Google Scholar]
- 13.Cahill C M, Waterman W R, Xie Y, Auron P E, Calderwood S K. Transcriptional repression of the prointerleukin 1β gene by heat shock factor 1. J Biol Chem. 1996;271:24874–24879. [PubMed] [Google Scholar]
- 14.Chen Y, Barlev N A, Westergaard O, Jakobsen B K. Identification of the C-terminal activator domain in yeast heat shock factor: independent control of transient and sustained transcriptional activity. EMBO J. 1993;12:5007–5018. doi: 10.1002/j.1460-2075.1993.tb06194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clos J, Westwood J T, Becker P B, Wilson S, Lambert K, Wu C. Molecular cloning and expression of a hexameric Drosophila heat shock factor subject to negative regulation. Cell. 1990;63:1085–1097. doi: 10.1016/0092-8674(90)90511-c. [DOI] [PubMed] [Google Scholar]
- 16.Cotto J J, Kline M, Morimoto R I. Activation of heat shock factor 1 DNA binding precedes stress-induced serine phosphorylation. Evidence for a multistep pathway of regulation. J Biol Chem. 1996;271:3355–3358. doi: 10.1074/jbc.271.7.3355. [DOI] [PubMed] [Google Scholar]
- 17.Drees B L, Grotkop E K, Nelson H C M. The GCN4 leucine zipper can functionally substitute for the heat shock transcription factor’s trimerization domain. J Mol Biol. 1997;273:61–74. doi: 10.1006/jmbi.1997.1283. [DOI] [PubMed] [Google Scholar]
- 18.Erkine A M, Adams C C, Gao M, Gross D S. Multiple protein-DNA interactions over the yeast HSC82 heat shock gene promoter. Nucleic Acids Res. 1995;23:1822–1829. doi: 10.1093/nar/23.10.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandes M, Xiao H, Lis J T. Fine structure analyses of the Drosophila and Saccharomyces heat shock factor-heat shock element interactions. Nucleic Acids Res. 1994;22:167–173. doi: 10.1093/nar/22.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flick K E, Gonzalez L, Jr, Harrison C J, Nelson H C. Yeast heat shock transcription factor contains a flexible linker between the DNA-binding and trimerization domains. Implications for DNA binding by trimeric proteins. J Biol Chem. 1994;269:12475–12481. [PubMed] [Google Scholar]
- 21.Fujita A, Kikuchi Y, Kuhara S, Misumi Y, Matsumoto S, Kobayashi H. Domains of the SFL1 protein of yeasts are homologous to Myc oncoproteins of yeast heat-shock transcription factor. Gene. 1989;85:321–328. doi: 10.1016/0378-1119(89)90424-1. [DOI] [PubMed] [Google Scholar]
- 22.Fujita T, Nolan G, Ghosh S, Baltimore D. Independent modes of activation by the p50 and p65 subunits of NF-κB. Genes Dev. 1992;6:775–787. doi: 10.1101/gad.6.5.775. [DOI] [PubMed] [Google Scholar]
- 23.Gallo G J, Prentice H, Kingston R E. Heat shock factor is required for growth at normal temperatures in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol. 1993;13:749–761. doi: 10.1128/mcb.13.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giardina C, Lis J T. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol Cell Biol. 1995;15:2737–2744. doi: 10.1128/mcb.15.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gross D S, Adams C C, Lee S, Stentz B. A critical role for heat shock transcription factor in establishing a nucleosome-free region over the TATA-initiation site of the yeast HSP82 heat shock gene. EMBO J. 1993;12:3931–3945. doi: 10.1002/j.1460-2075.1993.tb06071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison C J, Bohm A A, Nelson H C. Crystal structure of the DNA binding domain of the heat shock transcription factor. Science. 1994;263:224–227. doi: 10.1126/science.8284672. [DOI] [PubMed] [Google Scholar]
- 27.Jakobsen B K, Pelham H R. Constitutive binding of yeast heat shock factor to DNA in vivo. Mol Cell Biol. 1988;8:5040–5042. doi: 10.1128/mcb.8.11.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakobsen B K, Pelham H R B. A conserved heptapeptide restrains the activity of the yeast heat shock transcription factor. EMBO J. 1991;10:369–375. doi: 10.1002/j.1460-2075.1991.tb07958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koch K A, Thiele D J. Autoactivation by a Candida glabrata copper metalloregulatory transcription factor requires critical minor groove interactions. Mol Cell Biol. 1996;16:724–734. doi: 10.1128/mcb.16.2.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kroeger P E, Morimoto R I. Selection of new HSF1 and HSF2 DNA-binding sites reveals difference in trimer cooperativity. Mol Cell Biol. 1994;14:7592–7603. doi: 10.1128/mcb.14.11.7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kroeger P E, Sarge K D, Morimoto R I. Mouse heat shock transcription factors 1 and 2 prefer a trimeric binding site but interact differently with the HSP70 heat shock element. Mol Cell Biol. 1993;13:3370–3383. doi: 10.1128/mcb.13.6.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labbe S, Zhu Z, Thiele D J. Copper-specific transcriptional repression of yeast genes encoding critical components in the copper transport pathway. J Biol Chem. 1997;272:15951–15958. doi: 10.1074/jbc.272.25.15951. [DOI] [PubMed] [Google Scholar]
- 33.Leppa S, Pirkkala L, Saarento H, Sarge K D, Sistonen L. Overexpression of HSF2-β inhibits hemin-induced heat shock gene expression and erythroid differentiation in K562 cells. J Biol Chem. 1997;272:15293–15298. doi: 10.1074/jbc.272.24.15293. [DOI] [PubMed] [Google Scholar]
- 34.Lipkin S M, Nelson C A, Glass C K, Rosenfeld M G. A negative retinoic acid response element in the rat oxytocin promoter restricts transcriptional stimulation by heterologous transactivation domains. Proc Natl Acad Sci USA. 1992;89:1209–1213. doi: 10.1073/pnas.89.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X-D, Liu P C C, Santoro N, Thiele D J. Conservation of a stress response: human heat shock transcription factors functionally substitute for yeast HSF. EMBO J. 1997;16:6466–6477. doi: 10.1093/emboj/16.21.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X-D, Thiele D J. Oxidative stress induces heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev. 1996;10:592–603. doi: 10.1101/gad.10.5.592. [DOI] [PubMed] [Google Scholar]
- 37.Morgan B A, Banks G R, Toone W M, Raitt D, Kuge S, Johnston L H. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 1997;16:1035–1044. doi: 10.1093/emboj/16.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moritomo R I, Tissieres A, Georgopoulos C, editors. The biology of heat shock proteins and chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 39.Myers A M, Tzagoloff A, Kinney D M, Lusty C J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45:299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- 40.Nakai A, Morimoto R I. Characterization of a novel chicken heat shock transcription factor, heat shock factor 3, suggests a new regulatory pathway. Mol Cell Biol. 1993;13:1983–1997. doi: 10.1128/mcb.13.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto R I, Nagata K. HSF4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol Cell Biol. 1997;17:469–481. doi: 10.1128/mcb.17.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nieto-Sotelo J, Wiederrecht G, Okuda A, Parker C S. The yeast heat shock transcription factor contains a transcriptional activation domain whose activity is repressed under nonshock conditions. Cell. 1990;62:807–817. doi: 10.1016/0092-8674(90)90124-w. [DOI] [PubMed] [Google Scholar]
- 43.Nover L. Expression of heat shock genes in homologous and heterologous systems. Enzyme Microb Technol. 1987;9:129–192. [Google Scholar]
- 44.Rabindran S K, Giorgi G, Clos J, Wu C. Molecular cloning and expression of a human heat shock factor, HSF1. Proc Natl Acad Sci USA. 1991;88:6906–6910. doi: 10.1073/pnas.88.16.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rabindran S K, Haroun R I, Clos J, Wisniewski J, Wu C. Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science. 1993;259:230–234. doi: 10.1126/science.8421783. [DOI] [PubMed] [Google Scholar]
- 46.Rallu M, Loones M, Lallemand Y, Morimoto R, Morange M, Mezger V. Function and regulation of heat shock factor 2 during mouse embryogenesis. Proc Natl Acad Sci USA. 1997;94:2392–2397. doi: 10.1073/pnas.94.6.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhodes D. Analysis of sequence-specific DNA binding proteins. In: Creighton T E, editor. Protein function: a practical approach. New York, N.Y: IRL Press; 1989. pp. 177–198. [Google Scholar]
- 48.Sarge K D, Murphy S P, Morimoto R I. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarge K D, Parke-Sarge O K, Kirby J D, Mayo K E, Morimoto R I. Expression of heat shock factor 2 in mouse testis: potential role as a regulator of heat-shock protein gene expression during spermatogenesis. Biol Reprod. 1994;50:1334–1343. doi: 10.1095/biolreprod50.6.1334. [DOI] [PubMed] [Google Scholar]
- 50.Sarge K D, Zimarino V, Holm K, Wu C, Morimoto R I. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 1991;5:1902–1911. doi: 10.1101/gad.5.10.1902. [DOI] [PubMed] [Google Scholar]
- 51.Scharf K D, Rose S, Zott W, Schoffl F, Nover L, Schoff F. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J. 1990;9:4495–4501. doi: 10.1002/j.1460-2075.1990.tb07900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schreiber E, Matthias P, Muller M, Schaffner W. Identification of a novel lymphoid specific octamer binding protein (OTF-2B) by proteolytic clipping bandshift assay (PCBA) EMBO J. 1988;7:4221–4229. doi: 10.1002/j.1460-2075.1988.tb03319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuetz T J, Gallo G J, Sheldon L, Tempst P, Kingston R E. Isolation of a cDNA for HSF2: evidence for two heat shock factor genes in humans. Proc Natl Acad Sci USA. 1991;88:6911–6915. doi: 10.1073/pnas.88.16.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silar P, Butler G, Thiele D J. Heat shock transcription factor activates transcription of the yeast metallothionein gene. Mol Cell Biol. 1991;11:1232–1238. doi: 10.1128/mcb.11.3.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sistonen L, Sarge K D, Phillips B, Abravaya K, Morimoto R I. Activation of heat shock factor 2 during hemin-induced differentiation of human erythroleukemia cells. Mol Cell Biol. 1992;12:4104–4111. doi: 10.1128/mcb.12.9.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sorger P K. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell. 1990;62:793–805. doi: 10.1016/0092-8674(90)90123-v. [DOI] [PubMed] [Google Scholar]
- 57.Sorger P K, Ammerer G, Shore D. Identification and purification of sequence-specific DNA-binding proteins. In: Creighton T E, editor. Protein function: a practical approach. New York, N.Y: IRL Press; 1989. pp. 199–224. [Google Scholar]
- 58.Sorger P K, Nelson H C M. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell. 1989;59:807–813. doi: 10.1016/0092-8674(89)90604-1. [DOI] [PubMed] [Google Scholar]
- 59.Sorger P K, Pelham H R. Purification and characterization of a heat-shock element binding protein from yeast. EMBO J. 1987;6:3035–3041. doi: 10.1002/j.1460-2075.1987.tb02609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sorger P K, Pelham H R B. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988;54:855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- 61.Starr D B, Matsui W, Thomas J R, Yamamoto K R. Intracellular receptors use a common mechanism to interpret signaling information at response elements. Genes Dev. 1996;10:1271–1283. doi: 10.1101/gad.10.10.1271. [DOI] [PubMed] [Google Scholar]
- 62.Stevens S Y, Swanson P C, Glick G D. Application of the gel shift assay to study the affinity and specificity of anti-DNA autoantibodies. J Immunol Methods. 1994;177:185–190. doi: 10.1016/0022-1759(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 63.Tamai K T, Liu X, Silar P, Sosinowski T, Thiele D J. Heat shock transcription factor activates yeast metallothionein gene expression in response to heat and glucose starvation via distinct signalling pathways. Mol Cell Biol. 1994;14:8155–8165. doi: 10.1128/mcb.14.12.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan S, Richmond T J. DNA binding-induced conformational change of the yeast transcriptional activator PRTF. Cell. 1990;62:367–377. doi: 10.1016/0092-8674(90)90373-m. [DOI] [PubMed] [Google Scholar]
- 65.Topol J, Ruden D M, Parker C S. Sequences required for in vitro transcriptional activation of a Drosophila hsp 70 gene. Cell. 1985;42:527–537. doi: 10.1016/0092-8674(85)90110-2. [DOI] [PubMed] [Google Scholar]
- 66.Vuister G W, Kim S J, Orosz A, Marquardt J, Wu C, Bax A. Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor. Nat Struct Biol. 1994;1:605–614. [PubMed] [Google Scholar]
- 67.Westwood J T, Clos J, Wu C. Stress-induced oligomerization and chromosomal relocalization of heat-shock factor. Nature. 1991;353:822–827. doi: 10.1038/353822a0. [DOI] [PubMed] [Google Scholar]
- 68.Westwood J T, Wu C. Activation of Drosophila heat shock factor: conformational change associated with a monomer-to-trimer transition. Mol Cell Biol. 1993;13:3481–3486. doi: 10.1128/mcb.13.6.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wiederrecht G, Seto D, Parker C S. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell. 1988;54:841–853. doi: 10.1016/s0092-8674(88)91197-x. [DOI] [PubMed] [Google Scholar]
- 70.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 71.Xiao H, Lis J T. Germline transformation used to define key features of heat-shock response elements. Science. 1988;239:1139–1142. doi: 10.1126/science.3125608. [DOI] [PubMed] [Google Scholar]
- 72.Xiao H, Perisic O, Lis J T. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell. 1991;64:585–593. doi: 10.1016/0092-8674(91)90242-q. [DOI] [PubMed] [Google Scholar]
- 73.Young M R, Craig E A. Saccharomyces cerevisiae HSP70 heat shock elements are functionally distinct. Mol Cell Biol. 1993;13:5637–5646. doi: 10.1128/mcb.13.9.5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zuo J, Rungger D, Voellmy R. Multiple layers of regulation of human heat shock transcription factor 1. Mol Cell Biol. 1995;15:4319–4330. doi: 10.1128/mcb.15.8.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]