Abstract
Complete activation of most cyclin-dependent protein kinases (CDKs) requires phosphorylation by the CDK-activating kinase (CAK). In the budding yeast, Saccharomyces cerevisiae, the major CAK is a 44-kDa protein kinase known as Cak1. Cak1 is required for the phosphorylation and activation of Cdc28, a major CDK involved in cell cycle control. We addressed the possibility that Cak1 is also required for the activation of other yeast CDKs, such as Kin28, Pho85, and Srb10. We generated three new temperature-sensitive cak1 mutant strains, which arrested at the restrictive temperature with nonuniform budding morphology. All three cak1 mutants displayed significant synthetic interactions with loss-of-function mutations in CDC28 and KIN28. Loss of Cak1 function reduced the phosphorylation and activity of both Cdc28 and Kin28 but did not affect the activity of Pho85 or Srb10. In the presence of the Kin28 regulatory subunits Ccl1 and Tfb3, Kin28 was phosphorylated and activated when coexpressed with Cak1 in insect cells. We conclude that Cak1 is required for the activating phosphorylation of Kin28 as well as that of Cdc28.
Cyclin-dependent kinases (CDKs) are important regulators of basic cellular processes. Although best known for their role in the control of the eukaryotic cell division cycle, CDKs have also been implicated in signal transduction pathways and transcription (37, 39). The wide range of CDK functions is clearly apparent in the budding yeast, Saccharomyces cerevisiae, which contains at least five CDKs involved in a variety of regulatory pathways (2, 37, 38). The best-understood yeast CDK is Cdc28, an essential regulator of cell cycle progression. When bound to the G1 cyclins Cln1 to Cln3, Cdc28 regulates passage through START; when bound to the B-type cyclins Clb1 to Clb4 or Clb5 and Clb6, Cdc28 controls the onset of mitosis or S phase, respectively (38). Another essential yeast CDK is Kin28, which associates with a single cyclin, Ccl1, in a complex that interacts with the basal transcription factor TFIIH. Kin28 may function to phosphorylate the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (13, 49, 54, 55), and kin28 mutants display broad defects in gene transcription (4, 54). A third yeast CDK is Pho85, which associates with a large family of cyclins involved in a variety of nonessential functions in gene expression and metabolic regulation (2, 21, 30). Srb10 (Ssn3, Ume5, Are1) is another nonessential CDK, which associates with the cyclin-like protein Srb11 (Ssn8, Ume3) in the RNA polymerase II holoenzyme (26, 31, 46, 58). Finally, the CDK-like protein kinase Ctk1, with its cyclin-like partner Ctk2, may also be involved in control of polymerase II-dependent transcription (25, 45).
In addition to cyclin binding, complete activation of most CDKs requires phosphorylation of a conserved threonine residue in a region known as the T-loop; phosphorylation at this site is catalyzed by the CDK-activating kinase (CAK) (20, 37). Mutation of the activating residue in several CDKs, including Cdc28, abolishes kinase activity and function in vivo (8, 15, 18, 33, 44). The major CAK activity in S. cerevisiae is a monomeric protein kinase, Cak1 (Civ1), that is distantly related to CDKs (10, 22, 52). cak1 mutants display defects in the phosphorylation and activity of Cdc28 in vivo (22, 52), and purified Cak1 protein phosphorylates Cdc28 in vitro (10, 22, 52); thus, it seems likely that Cak1 is directly responsible for the phosphorylation and activation of Cdc28. However, cak1 mutants arrest with nonuniform morphologies unlike those observed in cdc28 mutants, raising the possibility that Cak1 contributes to the activation of other CDKs (3, 22, 47, 52). Cells expressing a CDC28 mutant that functions without activating phosphorylation can grow in the absence of CAK1 (5), suggesting that the major essential function of Cak1 is the activation of Cdc28; however, the poor growth of these cells also suggests that Cak1 has other, nonessential functions.
The role of phosphorylation in the activation of Kin28, Pho85, and Srb10 has not been studied in detail. The T-loop of Kin28 contains a threonine (T162), but the role of this site in Kin28 function has not been determined. Studies of the vertebrate Kin28 homologue, Cdk7, may provide insight into the mechanisms governing Kin28 activation. Cdk7 phosphorylation at T170 is required for the activation of Cdk7-cyclin H dimers (9, 14, 15, 35, 51). However, addition of the assembly factor Mat1 allows the formation of an active Cdk7-cyclin H-Mat1 trimer in the absence of phosphorylation (9, 14, 51). Because most Cdk7 exists in the trimeric form in vivo (14, 36), the importance of activating phosphorylation of Cdk7 remains unclear. Similarly, in the budding yeast, Kin28 associates with a cyclin H homologue, Ccl1, and a Mat1 homologue, Tfb3 (Rig2) (11, 12, 49, 56). However, active Kin28-Ccl1 dimers are readily separated from the other components of yeast TFIIH, including Tfb3, raising the possibility that Kin28 is more dependent on activating phosphorylation (13, 48).
Pho85 contains a serine (S166) that aligns with activating-phosphorylation sites in other CDKs. Replacement of this residue with alanine abolishes Pho85 activity and function in vivo (41), but phosphorylation at this site has not been demonstrated in vivo or in vitro. Similarly, Srb10 contains a threonine (T320) in its T-loop region, but the requirement for phosphorylation at this site is unknown.
To address the role of Cak1 in the activation of yeast CDKs other than Cdc28, we generated new cak1 mutants and analyzed their effects on cell morphology and the activity of Kin28, Pho85, and Srb10. Cdc28 and Kin28 phosphorylation and activity were reduced in cak1 strains, but the activities of Pho85 and Srb10 were unaffected. Coexpression of Cak1 and Kin28 in insect cells led to the phosphorylation and activation of Kin28. We therefore conclude that Cak1 plays a role in the activation of Kin28 but not in the activation of Pho85 or Srb10.
MATERIALS AND METHODS
Strains and plasmids.
The wild-type strain used in this work is from a W303 background with the following genotype: MATa ura3-52 leu2-3112 trp1-1 his3-11 ade2-1 can1-100. All the other yeast strains in this work were derived from this strain by one-step replacement or extensive backcrossing (at least four times) into that strain by standard techniques. The parental kin28-3 strain was a kind gift of G. Faye (56). A pRS313-based (42) plasmid containing a 2,320-bp CAK1 insert and flanking sequence was mutagenized in vitro by treatment with hydroxylamine. Mutagenized plasmids were screened for temperature-sensitive rescue of Δcak1::HIS3. cak1 temperature-sensitive strains were generated by transplacement of Δcak1::URA3 as described previously (34).
Cak1-independent cdc28 mutant strains were the kind gift of F. Cross and have been described previously (5). Strains 1834-1B and 1836-1A have their endogenous CDC28 gene deleted and contain plasmids encoding the Cak1-independent Cdc28-4324 or Cdc28-5331 protein, respectively (5). Strains 1834-2A and 1836-5D are identical to 1834-1B and 1836-1A, respectively, except that the CAK1 gene is also deleted.
To add hemagglutinin (HA) epitope tags to yeast CDKs, cells were transformed with an integrating plasmid (42) containing a 5′ fragment of the CDK coding region fused to an HA tag (underlined) followed by the ACT1 terminator. YIpFHE83 contains a 744-bp fragment of the CDC28 coding region and alters the C terminus of Cdc28 from QES to QESMAYPYDVPDYASLGPGP. YIpJC01 contains an 866-bp fragment from the KIN28 coding region and alters the C terminus of Kin28 from IRN to IRTMAYPYDVPDYASLGPGP. YIpFHE48 contains a 667-bp fragment of the PHO85 coding region and alters the C terminus of Pho85 from HAS to HASMAYPYDVPDYASLGPGP. YIpFHE101 contains a 1,097-bp fragment from the SRB10 coding region and alters the C terminus of Srb10 from NRR to NRTMAYPYDVPDYASLGPGP. Similar methods were used to construct YIpFHE106, which contains the KIN28 coding sequence fused to a C-terminal Myc epitope tag, resulting in a change of the sequence from IRN to IRAMEQKLISEEDLN. None of the epitope-tagged constructs caused phenotypes associated with loss-of-function alleles of the CDKs.
GAL-driven expression of KIN28 was performed with YIpFHE104, which contains a 918-bp fragment that includes the full-length coding sequence of KIN28 with the intron removed (43), fused to an HA tag (as above) and controlled by the GAL1-10 promoter in the vector pRS306 (42). pFHE104T162A is a version of YIpFHE104 in which the KIN28 coding sequence contains a single nucleotide change that converts codon 162 from ACA to GCA; nucleotide sequencing confirmed that this is the only mutation in the entire Kin28A coding sequence.
Baculoviruses encoding Kin28HA, Ccl1, six-histidine-tagged Tfb3, and six-histidine-tagged Cak1 were constructed and used to infect Sf9 insect cells by conventional methods (8). All Cak1 infections also included coinfection with a virus encoding budding yeast Cdc37 (17), which is required in yeast for Cak1 stability and enhances Cak1 expression in insect cells (10a).
Microscopy.
Samples were fixed for 2 h in 3.7% formaldehyde, washed three times in HBS (10 mM HEPES-NaOH [pH 7.5], 150 mM NaCl), stained with 1 μg of 4′,6-diamidino-2-phenylindole (DAPI) per ml, and washed three more times in HBS. For microscopy, cell clumps were dispersed by sonication for 3 s at 50% power and then mounted on polylysine-treated glass slides. The budding indices of cell populations were assessed at a magnification of ×100 with Nomarski optics or ×60 by phase-contrast microscopy, and nuclear morphology was assessed by DAPI fluorescence microscopy. Small buds were defined as being less than 50% of the size of the mother; large buds were greater than 50% of the size of the mother.
Immunoprecipitations.
For yeast, frozen cell pellets (∼15 optical density units at 600 nm) were resuspended in lysis buffer (25 mM HEPES-NaOH [pH 7.5], 250 mM NaCl, 0.2% Triton X-100, 5 mM β-glycerophosphate, 5 mM NaF, 10% glycerol, 1 mM EDTA, 1 mM dithiothreitol [DTT], 1 μg of leupeptin per ml 1 μg of pepstatin A per ml, 0.1 TIU of aprotinin per ml, 1 mM phenylmethylsulfonyl fluoride [PMSF]) and lysed by mechanical disruption in a Beadbeater (Biospec). For baculovirus-infected Sf9 cells, frozen cell pellets (∼106 cells) were resuspended in hypotonic lysis buffer (10 mM HEPES-NaOH [pH 7.5], 10 mM NaCl, 0.2% Triton X-100, 5 mM β-glycerophosphate, 5 mM NaF, 10% glycerol, 1 mM EDTA, 1 mM DTT, 1 μg of leupeptin per ml, 1 μg of pepstatin A per ml, 0.10 TIU of aprotinin per ml, 1 mM PMSF), allowed to swell for 10 min on ice, vortexed, and adjusted to 250 mM NaCl. Lysates were clarified by centrifugation for 15 min at 14,000 × g, and the protein content was assessed by the Bio-Rad protein assay. A 20-μl volume of protein A-Sepharose (Sigma) and 1 μg of monoclonal antibody 12CA5 (BAbCo) were added to 1 to 8 mg of crude lysate and rotated for 1 h at 4°C. The beads were then washed twice with HBST (10 mM HEPES-NaOH [pH 7.5], 150 mM NaCl, 0.2% Triton X-100) and twice with HBS and divided into two parts for Western blotting and kinase assays. Western blotting was performed with monoclonal antibody 12CA5 as described previously (17). Polyclonal anti-Myc serum (Santa Cruz Biotechnology) was used for immunoprecipitations and Western blotting of Myc-tagged Kin28.
Phosphatase treatment.
Immunoprecipitates were resuspended in phosphatase buffer (50 mM Tris-HCl [pH 7.8], 5 mM DTT, 1 mg of bovine serum albumin per ml, 1 μg of leupeptin per ml, 1 μg of pepstatin A per ml, 0.10 TIU of aprotinin per ml, 1 mM PMSF) to which was added either 2 mM MnCl2, 2 mM MnCl2 plus 100 U of λ-phosphatase, or phosphatase plus 2 mM ZnCl2, 50 mM NaF, and 1 mM Na3VO4. After incubation for 1 h at 30°C, the immunoprecipitates were washed three times with HBS and analyzed by immunoblotting.
Kinase assays.
Immunoprecipitates were resuspended in kinase assay buffer (10 mM HEPES-NaOH [pH 7.5], 150 mM NaCl, 10 mM MgCl2, 100 μM ATP) plus 1 μCi of [γ-32P]ATP and 5 μg of histone H1 for Cdc28 assays, 2.5 μCi of [γ-32P]ATP and 5 μg of glutathione S-transferase–CTD (a gift of Rick Young) for Kin28 and Srb10 assays, and 2 μCi of [γ-32P]ATP and 2 μg of Pho4 for Pho85 assays. The reaction mixtures were incubated for 15 min at room temperature, and the reaction products were analyzed by polyacrylamide gel electrophoresis and autoradiography.
RESULTS
Generation of new cak1 mutants.
We used hydroxylamine mutagenesis of a CAK1-bearing plasmid to generate new temperature-sensitive cak1 mutants that allowed growth in a cak1Δ strain at 24°C but not at 37°C. We obtained three mutants, each containing a single point mutation in the CAK1 coding region (Table 1). The Cak1-23 protein is mutated at the position of an aspartate residue that is highly conserved among all kinases (19), while the mutation in Cak1-95 occurs at a residue that is conserved among CDK-related kinases but not among kinases in general (19). In contrast, the glycine mutated in Cak1-34 is not well conserved (19). We also reconstructed a previously described temperature-sensitive allele, cak1-22, which was originally generated by alanine substitution in a nonconserved region near the carboxy terminus of Cak1 (Table 1) (22, 47).
TABLE 1.
Temperature-sensitive cak1 mutants used in this study
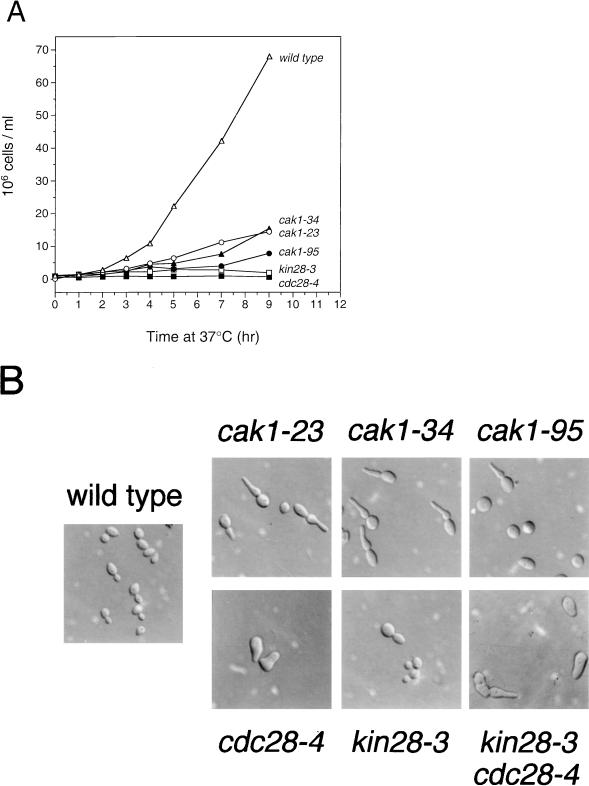
Studies of cak1 phenotypes were performed with yeast strains in which the endogenous CAK1 genes were replaced with the mutant sequences. At 37°C, these strains exhibited a severe growth defect compared to wild-type cells (Fig. 1A) but maintained nearly 100% viability after 24 h (data not shown). At 25°C, cak1 cells were slightly elongated compared to wild-type cells but the budding indices were similar (data not shown). At the restrictive temperature, the majority of cak1-95 cells arrested as unbudded cells but a small fraction arrested as large-budded cells with elongated buds (Fig. 1B; Table 2). In contrast, cak1-23 and cak1-34 cells arrested with nonuniform morphology, and a higher fraction displayed elongated buds (Fig. 1B, Table 2). When cak1 mutants were first arrested in G1 by alpha-factor pheromone treatment at 25°C and then released at 37°C, similar unbudded and elongated budding phenotypes were observed while the number of small-budded cells declined (Table 2). Analysis of cells treated with DAPI revealed a single DNA mass in unbudded cak1-23 and cak1-34 cells, whereas the DNA morphology in large-budded cells was heterogeneous (57 to 70% had one mass, 4 to 7% had a stretched mass, and 18 to 20% had two masses). In cak1-95 cells, the DNA morphology of the large-budded cells was more uniform (97% had a single DNA mass).
FIG. 1.
Growth rates and morphologies of temperature-sensitive cak1, cdc28, and kin28 mutants. (A) Isogenic strains carrying the indicated mutations were grown to mid-log phase at 25°C and switched to 37°C at time zero. At the indicated times, the cells were counted with a hemocytometer. (B) The indicated strains were grown for 3 h at 37°C and analyzed by Nomarski microscopy.
TABLE 2.
Phenotypes of cak1 mutants
| Strain | % of total populationa that was:
|
|||
|---|---|---|---|---|
| Unbuddedb | Small buddedc | Large buddedd | Elongated budding | |
| Log phase at 25°C | ||||
| Wild type | 49 | 23 | 28 | 0 |
| cak1-23 | 47 | 30 | 22 | <1 |
| cak1-34 | 40 | 36 | 23 | <1 |
| cak1-95 | 41 | 35 | 23 | <1 |
| cdc28-4 | 43 | 30 | 27 | 0 |
| kin28-3 | 51 | 22 | 26 | <1 |
| cdc28-4 kin28-3 | 42 | 34 | 23 | <1 |
| 3 h at 37°C | ||||
| Wild type | 48 | 26 | 25 | <1 |
| cak1-23 | 36 | 18 | 4 | 41 |
| cak1-34 | 43 | 13 | 3 | 40 |
| cak1-95 | 70 | 3 | 5 | 14 |
| cdc28-4 | 82 | <1 | 18 | <1 |
| kin28-3 | 46 | 26 | 26 | <1 |
| cdc28-4 kin28-3 | 62 | 7 | 2 | 19 |
| Release from G1 at 37°Ce | ||||
| cak1-23 | 52 | 7 | <1 | 42 |
| cak1-34 | 33 | <1 | <1 | 67 |
| cak1-95 | 65 | <1 | <1 | 34 |
| cdc28-4 | 97 | 3 | <1 | 0 |
| kin28-3 | 15 | 8 | 73 | 0 |
n > 300 cells.
Includes cells with mating projections.
Bud size less than 50% of the size of the mother cell.
Bud size more than 50% of the size of the mother cell.
Mid-log-phase cells were treated with alpha-factor at 25°C until >90% of the cells were arrested in G1, shifted to 37°C for 30 min, and then released from the arrest at 37°C for 3 h.
We compared these phenotypes to those of isogenic cdc28-4, kin28-3, and cdc28-4 kin28-3 mutant cells. As observed previously, cdc28-4 cells at 37°C arrested primarily as large unbudded cells and kin28-3 cells arrested with nonuniform budding morphology (56) (Fig. 1B; Table 2). kin28-3 cells released from a G1 arrest at 37°C arrested primarily with large buds (Table 2) and heterogeneous DNA morphology (40% had one mass, 5% had a stretched mass, and 54% had two masses), and 18% of the large-budded cells rebudded. Interestingly, cdc28-4 kin28-3 double mutants arrested at 37°C with a distribution of phenotypes similar to that of the cak1-95 mutant, suggesting that the phenotype of cak1-95 cells results from simultaneous loss of both Cdc28 and Kin28 function.
Genetic interactions between CAK1 and CDC28 or KIN28.
It might be expected that a reduction in activating phosphorylation by Cak1 would exacerbate the defect in conditional CDK mutants. We therefore tested whether cak1 mutations enhanced the growth defect in temperature-sensitive cdc28 and kin28 mutants.
The cak1-23 cdc28-4 and cak1-34 cdc28-4 double mutants were viable but displayed a growth defect compared to strains with the individual mutations. The maximum permissive temperatures were reduced to 23°C in double mutants (Table 3). The effect of the cak1-95 mutation in a cdc28-4 background was less pronounced. Interactions between CAK1 and KIN28 were more striking: all three of our cak1 mutations were synthetically lethal when present with kin28-3 (Table 3), as observed previously with a different cak1 allele (mca28-782) (54). These results are consistent with the possibility that CAK1 is required for KIN28 function.
TABLE 3.
Synthetic interactions between cak1, cdc28, and kin28 mutants
| Mutation | Growth temp for double mutanta:
|
|||
|---|---|---|---|---|
| cak1-23 (35°C) | cak1-34 (35°C) | cak1-95 (35°C) | cdc28-4 (35°C) | |
| cdc28-4 (35°C) | 23°C | 23°C | 30°C | |
| kin28-3 (35°C) | Lethalb | Lethal | Lethal | 33°C |
The maximum permissive temperature of the single mutations is shown in parentheses. The maximum permissive temperature of the double mutants is indicated in the body of the table.
“Lethal” indicates that the strain did not grow at any temperature tested.
Full Kin28 activity requires phosphorylation at T162.
Our next goal was to determine if Cak1 is required for activating phosphorylation of Kin28. However, it was first necessary to establish that Kin28 is phosphorylated at the putative activating site (T162) in vivo and that this phosphorylation is required for Kin28 activity.
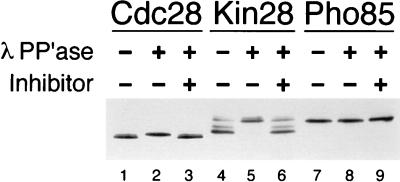
Kin28 is known to migrate as a doublet on polyacrylamide gels (4, 13). The faster-migrating band disappears after phosphatase treatment, indicating that the increase in mobility is dependent on phosphorylation (13). We confirmed this result with a yeast strain in which the KIN28 gene was replaced with a version carrying a carboxy-terminal HA epitope tag; this tag had no discernible effects on KIN28 function. Immunoblotting of Kin28 from these cells revealed a widely spaced doublet that was often accompanied by minor additional bands that may represent degradation products. As in previous work, the lower of the two major bands was consistently more abundant (4, 13) (Fig. 2). This band disappeared upon treatment with phosphatase, demonstrating that the majority of Kin28 is phosphorylated in vivo (Fig. 2).
FIG. 2.
Cdc28 and Kin28 are phosphorylated in vivo. HA epitope-tagged Cdc28, Kin28, and Pho85, under the control of their own promoters, were immunoprecipitated from cell lysates with the 12CA5 antibody and treated with phosphatase buffer alone (lanes 1, 4, and 7), λ-phosphatase (λ PP’ase) (lanes 2, 5, and 8), or λ-phosphatase in the presence of phosphatase inhibitors (lanes 3, 6, and 9). The immunoprecipitates were then subjected to immunoblotting with 12CA5.
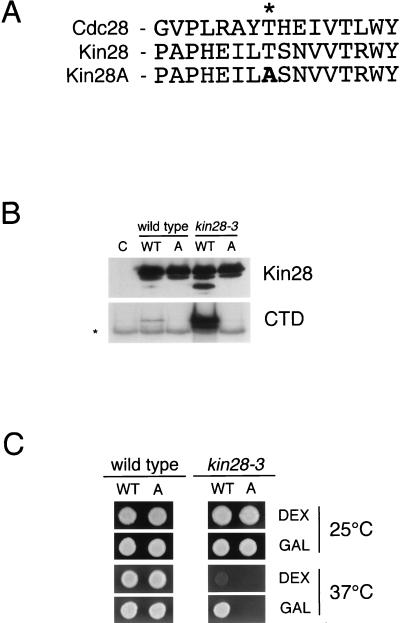
We next mapped the site of phosphorylation in Kin28 by generating a point mutant, Kin28A, in which the putative activating phosphorylation site (T162) is changed to alanine (Fig. 3A). Wild-type and mutant Kin28 proteins were expressed in wild-type or kin28-3 cells at 25°C under the control of the GAL1-10 promoter (Fig. 3B), resulting in Kin28 levels 5- to 10-fold higher than normal (data not shown). The epitope-tagged exogenous Kin28 migrated almost entirely in the low-mobility form, indicating that phosphorylation of the overexpressed protein was less extensive than that of the endogenous protein analyzed in our previous experiments (Fig. 2). Similarly, the CTD kinase activity of the GAL-driven Kin28 was lower than that of endogenous Kin28 (data not shown), particularly when expressed in wild-type cells (Fig. 3B). The low phosphorylation and activity of exogenous Kin28 may be the result of its overexpression at levels higher than those of its activating regulatory subunits Ccl1 and Tfb3, which appear to be required for its phosphorylation and activation (see the description of the insect cell experiments below). Presumably, exogenous Kin28HA is more active when expressed in kin28-3 mutant cells because it more effectively competes with the endogenous mutant protein for limiting regulatory subunits. In any case, analysis of the Kin28A mutant protein revealed that mutation of T162 abolished the high-mobility band on immunoblots and the CTD kinase activity in Kin28 immunoprecipitates. Based on this result and our results with phosphatase-treated Kin28 (Fig. 2), we conclude that activating phosphorylation at T162 causes the mobility shift and is required for Kin28 activity.
FIG. 3.
Mutation of T162 abolishes Kin28 phosphorylation, activity, and function. (A) Alignment of activating loop regions in Cdc28, Kin28, and Kin28A. The activating phosphorylation site (T169) in Cdc28 is indicated by an asterisk. (B) HA epitope-tagged wild-type Kin28 (WT) or Kin28A (A) was expressed under the control of the GAL1-10 promoter in wild-type or kin28-3 cells at 25°C. Anti-HA immunoprecipitates were prepared from cell lysates and subjected to immunoblotting to detect Kin28 (top). Kin28-associated CTD kinase activity was measured in parallel immunoprecipitates (bottom). Lane C contains a control immunoprecipitation from cells lacking epitope-tagged Kin28. The asterisk indicates a background band phosphorylated in CTD kinase assays in the absence of Kin28. (C) Growth of wild-type or kin28-3 strains containing KIN28 (WT) or kin28A (A) under the control of the GAL1-10 promoter at 25°C (top) or 37°C (bottom) on plates containing dextrose (DEX) or galactose (GAL).
GAL-driven expression of wild-type Kin28 or Kin28A had no deleterious effects in wild-type cells (Fig. 3C). Wild-type Kin28 was capable of supporting the growth of kin28-3 cells at 37°C, while the Kin28A mutant was not, arguing that activating phosphorylation of Kin28 is required for full function at high temperature in vivo.
Cak1 is required for Kin28 phosphorylation in vivo.
To assess the role of Cak1 in the activation of Kin28 and other CDKs, we examined the activity and electrophoretic mobility of epitope-tagged CDKs in cak1 mutants. In each case, we generated strains in which endogenous CDK genes were replaced by HA epitope-tagged versions under the control of their own promoters.
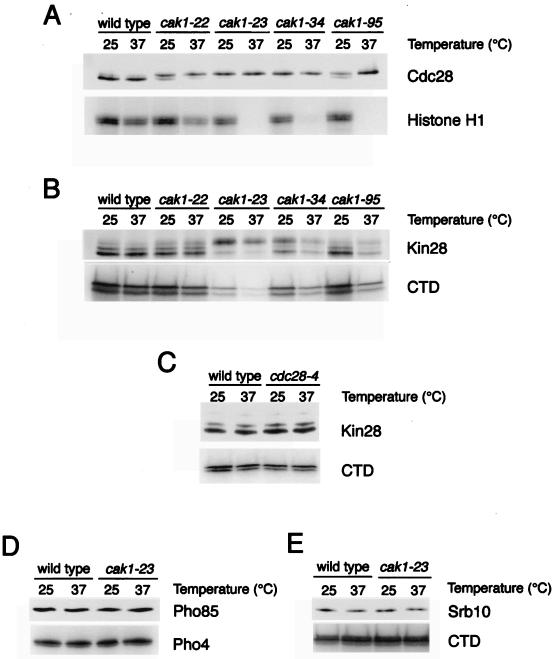
We analyzed the phosphorylation of Cdc28, like that of Kin28, by analyzing mobility shifts on polyacrylamide gels. Cdc28 migrates as a tightly spaced doublet, and the lower band is lost upon phosphatase treatment (Fig. 2). In addition, the lower band is abolished by mutation of the activating-site threonine (T169) in Cdc28 (10a). In a wild-type strain, most Cdc28 was phosphorylated at both the permissive and nonpermissive temperatures (Fig. 4A). The amount of phosphorylated Cdc28 was decreased in all of the cak1 mutants at the permissive temperature (Fig. 4A), and Cdc28 phosphorylation and kinase activity were further reduced in these mutants upon shifting to 37°C (Fig. 4A). Two alleles, cak1-23 and cak1-34, displayed particularly strong phenotypes. These data confirm that Cak1 is required for the phosphorylation and activation of Cdc28.
FIG. 4.
Phosphorylation and activity of yeast CDKs in cak1 mutants. The indicated mutant strains were grown to mid-log phase at 25°C and shifted to 37°C for 3 h. Epitope-tagged Cdc28 (A), Kin28 (B and C), Pho85 (D), and Srb10 (E) were immunoprecipitated from cell lysates and analyzed by immunoblotting with anti-HA antibodies (top panels) or kinase activity toward the indicated substrate (bottom panels). Cdc28 phosphorylation does not correlate with kinase activity in panel A, presumably because only a small fraction of Cdc28 is associated with cyclin and is active in asynchronous cells, and kinase activity should be normal even if only this fraction is phosphorylated.
In wild-type cells growing at either the permissive or nonpermissive temperature, the majority of Kin28 migrated in the lower, phosphorylated band on immunoblots. The effects of cak1 mutations on Kin28 mobility and activity varied dramatically among different cak1 alleles and were generally less striking than the effects on Cdc28 (Fig. 4B). In the strongest allele, cak1-23, the phosphorylation and activity of Kin28 were significantly reduced at the nonpermissive temperature and shifting to 37°C resulted in a decrease in Kin28 protein levels and a further drop in kinase activity. Similar but less pronounced defects were observed in the cak1-34 mutant (Fig. 4B). Kin28 phosphorylation in the cak1-95 strain was normal at 25°C; the shift to 37°C caused a decrease in total Kin28 protein levels and a decrease in the relative fraction of phosphorylated protein. Neither the phosphorylation nor the levels of Kin28 changed significantly in the previously described cak1-22 mutant (Fig. 4B) (22, 47). These results suggest that Cak1 is required for the phosphorylation and activation of Kin28 and is also required for normal levels of Kin28 protein production.
The vertebrate homologue of Kin28, Cdk7, can be phosphorylated in vitro by vertebrate Cdc2 or Cdk2 (14, 36), raising the possibility that Cdc28 phosphorylates Kin28 in budding yeast. Thus, the loss of Kin28 phosphorylation in cak1 mutants could be the indirect effect of decreased Cdc28 activity. This possibility seems unlikely, however, since we found that Kin28 phosphorylation was normal in cdc28-4 mutant cells arrested in G1 at 37°C (Fig. 4C). Similar results were obtained with cdc28-13 and cdc28-1N mutants (data not shown).
The activity and electrophoretic mobility of Pho85 (Fig. 4D) and Srb10 (Fig. 4E) were not affected by the cak1-23 mutation. In addition, we analyzed the effects of CAK1 mutations on the expression of PHO5, a gene whose expression increases in the absence of PHO85 function. PHO5 activity is readily assessed by a colorimetric assay of acid phosphatase secretion (53). After 24 h at 37°C, none of our three cak1 mutants displayed a significant increase in PHO5-dependent acid phosphatase secretion (data not shown), in agreement with previous studies of the cak1-22 mutant (47).
Cak1 is required for Kin28 phosphorylation in cells bearing Cak1-independent Cdc28.
Recently, Cross and Levine (5) used an elegant mutagenesis scheme to develop mutant forms of Cdc28 that function in the absence of activating phosphorylation at T169. Cells expressing these versions of Cdc28 are viable in the absence of the CAK1 gene. We obtained two of these mutant CDC28 strains from Cross and colleagues and replaced their Kin28 coding sequences with version carrying a C-terminal Myc epitope tag (HA-tagged Kin28 could not be used for these studies, because the mutant Cdc28 proteins are tagged with the HA epitope). Kin28-Myc displayed the same phosphorylation-dependent mobility shift observed with the HA-tagged protein (Fig. 5, top).
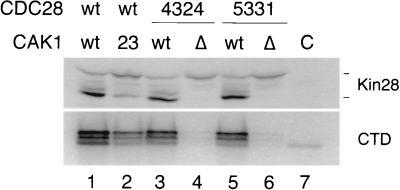
FIG. 5.
Kin28 phosphorylation and activity in wild-type and cak1Δ cells carrying Cak1-independent Cdc28 mutants. Myc-tagged Kin28 mobility on Western blots (top) and CTD kinase activity in immunoprecipitates (bottom) were measured in wild-type (wt) cells (lane 1), cak1-23 cells at room temperature (lane 2), and cells carrying Cak1-independent Cdc28-4324 (lanes 3 and 4) or Cdc28-5331 (lanes 5 and 6), in the presence (lanes 3 and 5) or absence (lanes 4 and 6) of the CAK1 gene. Lane C is a control sample from cells lacking epitope-tagged Kin28.
Kin28-Myc migrated primarily in the phosphorylated form in wild-type cells bearing the Cak1-independent Cdc28 mutants (Fig. 5, top, lanes 3 and 5). However, phosphorylated Kin28 was essentially undetectable in cells lacking the CAK1 gene (lanes 4 and 6). Kin28-associated CTD kinase activity was also greatly reduced in the absence of CAK1 (Fig. 5, bottom). These results further substantiate our conclusion that Cak1 is required for Kin28 phosphorylation and for maximal kinase activation in vivo.
Cells bearing Cak1-independent Cdc28 and lacking CAK1 are thus able to survive, albeit poorly, under conditions where Kin28 phosphorylation and activity are greatly reduced. We therefore conclude that high levels of Kin28 phosphorylation and activity are not required for full function in vivo (see Discussion).
Cak1-dependent phosphorylation of Kin28 in insect cells.
Our results argue that Cak1 is required for the phosphorylation of Kin28 in vivo. We next attempted to demonstrate direct phosphorylation of Kin28 by Cak1 in vitro with purified components expressed in yeast, bacterial, or insect cells. Unfortunately, recombinant Kin28, Ccl1, and Tfb3 were only marginally soluble when expressed separately or together and did not associate when incubated under a variety of conditions (data not shown). Partially soluble Kin28 preparations were not phosphorylated in vitro by Cak1, either alone or in the presence of Ccl1, Tfb3, or crude cell lysates prepared under a variety of conditions (data not shown).
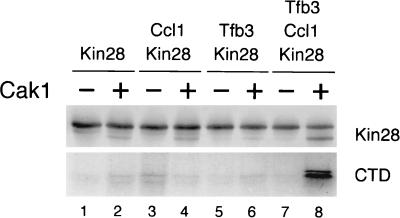
We were able to reconstitute Cak1-dependent Kin28 phosphorylation in insect cells infected with recombinant baculoviruses (Fig. 6). Kin28 was not phosphorylated when expressed in insect cells, but a slight increase in phosphorylation was observed upon coinfection with a virus encoding Cak1. The Kin28 regulatory subunit Ccl1 or Tfb3 had little extra effect when they were added individually, but when added together they caused a striking increase in Cak1-dependent Kin28 phosphorylation and activity. Thus, Cak1 is able to stimulate Kin28 phosphorylation in a heterologous expression system lacking other yeast protein kinases, suggesting that it is capable of catalyzing direct phosphorylation of Kin28.
FIG. 6.
Cak1-dependent activation of Kin28 in insect cells. Sf9 cells were coinfected with recombinant baculoviruses encoding the indicated combinations of Kin28, Ccl1, Tfb3, and Cak1. Kin28 was immunoprecipitated from cell lysates 2 days after infection and analyzed by Western blotting (top) and CTD kinase activity (bottom) assays. All the cells in this experiment were also coinfected with a virus encoding yeast Cdc37 (17), which increases Cak1 expression in insect cells.
DISCUSSION
Considerable genetic and biochemical evidence indicates that Cdc28 is phosphorylated and activated by Cak1. Defects in CAK1 function, as in our cak1 mutants or in the previously described cak1-22 allele, enhance the growth defects of cdc28 mutants (Table 3) and cells deficient in various Cdc28 cyclins (22, 47). Cdc28 phosphorylation and activity are greatly reduced in conditional cak1 mutants (22, 52) (Fig. 4A), and the Cak1 protein is able to catalyze Cdc28 phosphorylation in vitro (10, 22, 52). Thus, it appears likely that Cak1 is directly responsible for the activating phosphorylation of Cdc28 in the cell.
The present work suggests that Cak1 is also required for the phosphorylation and activity of Kin28. Mutations in CAK1 greatly exacerbate the growth defect observed in the kin28-3 mutant (Table 3) (54). Kin28 phosphorylation in vivo is decreased in conditional cak1 mutants and in cells lacking the CAK1 gene, and coexpression of Cak1 and Kin28 in insect cells results in Kin28 phosphorylation and activation. These data are most consistent with a direct role for Cak1 in the phosphorylation of Kin28 in vivo, although clear evidence for direct phosphorylation will require reconstitution of Kin28 phosphorylation by purified Cak1 in vitro.
In addition to causing defects in Kin28 phosphorylation, mutations in CAK1 caused decreased Kin28 protein levels. These decreases occurred gradually at the restrictive temperature (over a period of 3 h [data not shown]). Considering the requirement for KIN28 in mRNA synthesis (4, 13, 54), we suspect that decreases in Kin28 protein levels reflect a decrease in KIN28 transcription that is secondary to the defect in Kin28 activation by phosphorylation. Interestingly, the decrease in Kin28 protein levels in cak1-23 and cak1-34 cells at 37°C was not accompanied by major changes in Kin28 phosphorylation, suggesting that Kin28 phosphorylation was relatively stable under these conditions.
The arrest phenotypes of cak1 mutants can be roughly divided into two classes. Our cak1-95 mutant, like the previously described cak1-4 (civ1-4) mutant, arrests primarily as unbudded G1 cells (52). On the other hand, our cak1-23 and cak1-34 mutants, as well as the previously described cak1-22 mutant and cak1Δ cells (3, 22, 47), arrest with a mixture of phenotypes that includes unbudded cells and cells with elongated buds. The two classes of cak1 arrest phenotypes are not simply the result of differences in the bulk kinase activity of Cdc28 or Kin28; for example, cak1-34 and cak1-95 mutants contain similar levels of these kinase activities but arrest at 37°C with different phenotypes. Instead, variations in cak1 phenotypes may be due in part to differences in the ability of mutant Cak1 proteins to target different CDK-cyclin complexes. For example, the elongated budding phenotype of cak1-23 resembles that seen in cells with compromised Clb-Cdc28 kinase activity (1, 16, 23, 27, 32) while the unbudded phenotype of cak1-95 is reminiscent of the G1 arrest seen in cdc28 mutants or in cells lacking the G1 cyclins Cln1 to Cln3 (6). The complexity of cak1 phenotypes may also reflect differences in the stability of Cak1-dependent CDK activities: we found that shifting cak1 mutant cells to 37°C caused a more rapid loss of Cdc28 activity (10 min) than of Kin28 activity (1 to 2 h). Thus, the major features of the cak1 phenotype may be due in large part to defects in specific Cdc28-cyclin functions but the gradual loss of KIN28-dependent transcriptional activity in these mutants may complicate the phenotype.
Cells carrying Cak1-independent Cdc28 mutants and lacking CAK1 are able to proliferate (at reduced rates) even though the Kin28-associated CTD kinase activity is greatly reduced, indicating that remarkably little Kin28 activity is required for cell viability (assuming that the essential function of Kin28 is dependent on its kinase activity). Furthermore, the absence of detectable Kin28 phosphorylation in these cells indicates that the essential function of Kin28 does not require its phosphorylation. This result seems to contradict our evidence that overexpression of Kin28A does not allow growth of kin28-3 cells at high temperature (Fig. 3C). Perhaps Kin28 in the Cak1-independent cells is phosphorylated by some other kinase at a very low level that is not detectable by our methods but is still sufficient to provide the small amount of Kin28 activity required for cell proliferation. Alternatively, the Kin28A mutant may be defective not only in activating phosphorylation but also in another essential biochemical function.
Our studies with the Kin28A mutant, as well as those with cak1-deficient cells, clearly suggest that phosphorylation of Kin28 at T162 is required for full kinase activity in vivo. Thus, the Kin28-Ccl1 complex appears to be more dependent on phosphorylation than is its vertebrate homologue, Cdk7-cyclin H, whose activation does not require phosphorylation in the presence of the assembly factor Mat1 (9, 14). The S. cerevisiae homologue of Mat1, Tfb3, may not provide the same activating function as Mat1 in the absence of phosphorylation. Indeed, in our insect cell coinfection experiments, we found that coexpression of Kin28, Ccl1, and Tfb3 did not yield an active kinase complex except in the presence of Cak1, suggesting that activating phosphorylation is required even in the presence of Tfb3. Nevertheless, Tfb3 and Ccl1 were required for maximal Kin28 phosphorylation and activity in these experiments. Thus, Cak1 does not appear to promote the phosphorylation of monomeric Kin28, despite its ability to phosphorylate the Cdc28 monomer (10, 22). Further studies, preferably with purified components in vitro, are required to assess the precise role of Tfb3 and Ccl1 in Kin28 assembly, phosphorylation, and substrate targeting.
Our strongest cak1 allele (cak1-23) did not affect the kinase activity, mobility, or levels of Pho85 or Srb10. Similarly, several mutations in CAK1 do not affect the expression of PHO5, a gene whose expression increases in the absence of PHO85 function (reference 47 and data not shown). These results raise the possibility that Pho85 and Srb10 are activated by a different CAK or that their activation does not require phosphorylation. There is little previous data to shed light on these possibilities. For Pho85, mutation of the putative activating site (Ser166) reduces kinase activity and function in vivo, but phosphorylation at this site has not been demonstrated (41). Similarly, nothing is known about the phosphorylation of Srb10; interestingly, its putative human homologue, Cdk8, does not contain a phosphorylatable residue in the T-loop region (29, 40, 50). It is therefore possible that phosphorylation is not necessary for the activation of Pho85 or Srb10. Perhaps nonessential CDKs like these can evolve more easily through the intermediate steps between phosphorylation dependence and independence; these steps may be insurmountable in essential CDKs like Kin28 and Cdc28 (5).
Cak1 may also be involved in the activation of kinases other than CDKs. Overexpression of CAK1 suppresses the spore wall defect of cells with mutations in SMK1, a gene encoding a member of the mitogen-activated protein kinase family (24). Cells with defects in CAK1 exhibit a spore wall formation defect that resembles the phenotype of smk1 mutants (57). Like the closely related CDKs, mitogen-activated protein kinases are activated by phosphorylation of residues within the activating loop. Smk1 contains a threonine (T207) in a position that is roughly analogous to the activating site of CDKs (24). Thus, the requirement for Cak1 in spore wall formation may reflect a direct role in the activation of Smk1.
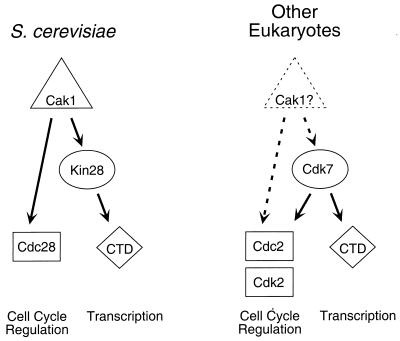
In vertebrates and other higher eukaryotes, the Cdk7-cyclin H-Mat1 complex comprises the major CAK activity in cell lysates, and recent evidence suggests that Drosophila melanogaster cdk7 mutants are defective in the activation of the mitosis-promoting kinase Cdc2 (28). Because of its association with TFIIH, Cdk7 is also thought to contribute to the control of CTD phosphorylation and transcription. Thus, Cdk7 appears to fulfill dual roles in CDK activation and transcription in higher eukaryotes, while its budding yeast homologue Kin28 is involved primarily in transcriptional control. Our results now demonstrate that yeast Cak1 also plays two roles, both in the activation of cell cycle progression through Cdc28 and in the activation of transcription through Kin28 (Fig. 7). This scheme raises the possibility that a higher eukaryotic homologue of Cak1 is responsible for the phosphorylation of Cdk7.
FIG. 7.
Regulatory pathways governing the activities of Kin28 and Cdc28 in S. cerevisiae (left), and homologous pathways in higher eukaryotes (right).
ACKNOWLEDGMENTS
We thank Fred Cross for his generous gift of the Cak1-independent cdc28 mutants and for valuable discussions. We also thank Julia Charles, Rick Young, Erin Peckol, Kayvan Roayaie, and Jean Gabriel Valay for reagents and technical advice.
This work was supported by funding from the National Institute of General Medical Sciences (to D.O.M.), a postgraduate scholarship from the Natural Sciences and Engineering Research Council of Canada (to A.F.), and a Damon Runyon-Walter Winchell Postdoctoral Fellowship (to J.L.N.).
REFERENCES
- 1.Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick—G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- 2.Andrews B, Measday V. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 1998;14:66–72. doi: 10.1016/s0168-9525(97)01322-x. [DOI] [PubMed] [Google Scholar]
- 3.Chun K T, Goebl M G. Mutational analysis of Cak1p, an essential protein kinase that regulates cell cycle progression. Mol Gen Genet. 1997;256:365–375. doi: 10.1007/s004380050580. [DOI] [PubMed] [Google Scholar]
- 4.Cismowski M J, Laff G M, Solomon M J, Reed S I. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cross F, Levine K. Molecular evolution allows bypass of the requirement for activation loop phosphorylation of the Cdc28 cyclin-dependent kinase. Mol Cell Biol. 1998;18:2923–2931. doi: 10.1128/mcb.18.5.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross F R. Cell cycle arrest caused by CLN gene deficiency in Saccharomyces cerevisiae resembles START-I arrest and is independent of the mating-pheromone signalling pathway. Mol Cell Biol. 1990;10:6482–6890. doi: 10.1128/mcb.10.12.6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Bondt H L, Rosenblatt J, Jancarik J, Jones H D, Morgan D O, Kim S-H. Crystal structure of cyclin-dependent kinase 2. Nature. 1993;363:595–602. doi: 10.1038/363595a0. [DOI] [PubMed] [Google Scholar]
- 8.Desai D, Gu Y, Morgan D O. Activation of human cyclin-dependent kinases in vitro. Mol Biol Cell. 1992;3:571–582. doi: 10.1091/mbc.3.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devault A, Martinez A-M, Fesquet D, Labbe J-C, Morin N, Tassan J-P, Nigg E A, Cavadore J-C, Doree M. MAT1 (menage a trois), a new RING finger protein subunit stabilizing cyclin H-cdk7 complexes in starfish and Xenopus CAK. EMBO J. 1995;14:5027–5036. doi: 10.1002/j.1460-2075.1995.tb00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espinoza F H, Farrell A, Erdjument-Bromage H, Tempst P, Morgan D O. A-cyclin-dependent kinase-activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science. 1996;273:1714–1717. doi: 10.1126/science.273.5282.1714. [DOI] [PubMed] [Google Scholar]
- 10a.Farrell, A. Unpublished data.
- 11.Faye G, Simon M, Valay J G, Fesquet D, Facca C. Rig2, a RING finger protein that interacts with the Kin28/Ccl1 CTD kinase in yeast. Mol Gen Genet. 1997;255:460–466. doi: 10.1007/s004380050518. [DOI] [PubMed] [Google Scholar]
- 12.Feaver W J, Henry N L, Wang Z, Wu X, Svejstrup J Q, Bushnell D A, Friedberg E C, Kornberg R D. Genes for Tfb2, Tfb3, and Tfb4 subunits of yeast transcription/repair factor IIH. Homology to human cyclin-dependent kinase activating kinase and IIH subunits. J Biol Chem. 1997;272:19319–19327. doi: 10.1074/jbc.272.31.19319. [DOI] [PubMed] [Google Scholar]
- 13.Feaver W J, Svestrup J Q, Henry N L, Kornberg R D. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 14.Fisher R P, Jin P, Chamberlin H M, Morgan D O. Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell. 1995;83:47–57. doi: 10.1016/0092-8674(95)90233-3. [DOI] [PubMed] [Google Scholar]
- 15.Fisher R P, Morgan D O. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78:713–724. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- 16.Fitch I, Dahmann C, Surana U, Amon A, Nasmyth K, Goetsch L, Byers B, Futcher B. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol Biol Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerber M R, Farrell A, Deshaies R, Herskowitz I, Morgan D O. Cdc37 is required for association of the protein kinase Cdc28 with G1 and mitotic cyclins. Proc Natl Acad Sci USA. 1995;92:4651–4655. doi: 10.1073/pnas.92.10.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gould K L, Moreno S, Owen D J, Sazer S, Nurse P. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 1991;10:3297–3309. doi: 10.1002/j.1460-2075.1991.tb04894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanks S K, Quinn A M. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- 20.Harper J W, Elledge S J. The role of Cdk7 in CAK function, a retro-retrospective. Genes Dev. 1998;12:285–289. doi: 10.1101/gad.12.3.285. [DOI] [PubMed] [Google Scholar]
- 21.Huang D, Moffat J, Wilson W A, Moore L, Cheng C, Roach P J, Andrews B. Cyclin partners determine Pho85 protein kinase substrate specificity in vitro and in vivo: control of glycogen biosynthesis by Pcl8 and Pcl10. Mol Cell Biol. 1998;18:3289–3299. doi: 10.1128/mcb.18.6.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaldis P, Sutton A, Solomon M J. The Cdk-activating kinase (CAK) from budding yeast. Cell. 1996;86:553–564. doi: 10.1016/s0092-8674(00)80129-4. [DOI] [PubMed] [Google Scholar]
- 23.Kellogg D R, Murray A W. NAP1 acts with Clb1 to perform mitotic functions and to suppress polar bud growth in budding yeast. J Cell Biol. 1995;130:675–685. doi: 10.1083/jcb.130.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krisak L, Strich R, Winters R S, Hall J P, Mallory M J, Kreitzer D, Tuan R S, Winter E. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 1994;8:2151–2161. doi: 10.1101/gad.8.18.2151. [DOI] [PubMed] [Google Scholar]
- 25.Kuchin S, Carlson M. Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol Cell Biol. 1998;18:1163–1171. doi: 10.1128/mcb.18.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuchin S, Yeghiayan P, Carlson M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhne C, Linder P. A new pair of B-type cyclins from Saccharomyces cerevisiae that function early in the cell cycle. EMBO J. 1993;12:3437–3447. doi: 10.1002/j.1460-2075.1993.tb06018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larochelle S, Pandur J, Fisher R P, Salz H K, Suter B. Cdk7 is essential for mitosis and for in vivo Cdk-activating kinase activity. Genes Dev. 1998;12:370–381. doi: 10.1101/gad.12.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leclerc V, Tassan J P, O’Farrell P H, Nigg E A, Leopold P. Drosophila Cdk8, a kinase partner of cyclin C that interacts with the large subunit of RNA polymerase II. Mol Biol Cell. 1996;7:505–513. doi: 10.1091/mbc.7.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenburg M E, O’Shea E K. Signaling phosphate starvation. Trends Biochem Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- 31.Liao S-M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 32.Lim H H, Goh P Y, Surana U. Spindle pole body separation in Saccharomyces cerevisiae requires dephosphorylation of the tyrosine 19 residue of Cdc28. Mol Cell Biol. 1996;16:6385–6397. doi: 10.1128/mcb.16.11.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim H H, Loy C J, Zaman S, Surana U. Dephosphorylation of threonine 169 of Cdc28 is not required for exit from mitosis but may be necessary for start in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4573–4583. doi: 10.1128/mcb.16.8.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lundblad V. Manipulation of cloned yeast DNA, Unit 13.10. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1989. [DOI] [PubMed] [Google Scholar]
- 35.Mäkelä T P, Tassan J-P, Nigg E A, Frutiger S, Hughes G J, Weinberg R A. A cyclin associated with the CDK-activating kinase MO15. Nature. 1994;371:254–257. doi: 10.1038/371254a0. [DOI] [PubMed] [Google Scholar]
- 36.Martinez A-M, Afshar M, Martin F, Cavadore J-C, Labbe J-C, Doree M. Dual phosphorylation of the T-loop in cdk7: its role in controlling cyclin H binding and CAK activity. EMBO J. 1997;16:343–354. doi: 10.1093/emboj/16.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgan D O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 38.Nasmyth K. At the heart of the budding yeast cell cycle. Trends Genet. 1996;12:405–412. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 39.Nigg E A. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 40.Rickert P, Seghezzi W, Shanahan F, Cho H, Lees E. Cyclin C/CDK8 is a novel CTD kinase associated with RNA polymerase II. Oncogene. 1996;12:2631–2640. [PubMed] [Google Scholar]
- 41.Santos R, Waters N, Creasy C, Bergman L. Structure-function relationships of the yeast cyclin-dependent kinase Pho85. Mol Cell Biol. 1995;15:5482–5491. doi: 10.1128/mcb.15.10.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon M, Seraphin B, Faye G. KIN28, a yeast split gene coding for a putative protein kinase homologous to CDC28. EMBO J. 1986;5:2697–2701. doi: 10.1002/j.1460-2075.1986.tb04553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solomon M J, Lee T, Kirschner M W. Role of phosphorylation in p34cdc2 activation: identification of an activating kinase. Mol Biol Cell. 1992;3:13–27. doi: 10.1091/mbc.3.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sterner D E, Lee J M, Hardin S E, Greenleaf A L. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol Cell Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Surosky R T, Strich R, Esposito R E. The yeast UME5 gene regulates the stability of meiotic mRNAs in response to glucose. Mol Cell Biol. 1994;14:3446–3458. doi: 10.1128/mcb.14.5.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutton A, Freiman R. The Cak1p protein kinase is required at G1/S and G2/M in the budding yeast cell cycle. Genetics. 1997;147:57–71. doi: 10.1093/genetics/147.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Svejstrup J Q, Feaver W J, Kornberg R D. Subunits of yeast RNA polymerase II transcription factor TFIIH encoded by the CCL1 gene. J Biol Chem. 1996;271:643–645. doi: 10.1074/jbc.271.2.643. [DOI] [PubMed] [Google Scholar]
- 49.Svejstrup J Q, Vichi P, Egly J-M. The multiple roles of transcription/repair factor TFIIH. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 50.Tassan J-P, Jacquenod M, Leopold P, Schultz S J, Nigg E A. Identification of human cyclin-dependent kinase 8, a putative protein kinase partner for cyclin C. Proc Natl Acad Sci USA. 1995;92:8871–8875. doi: 10.1073/pnas.92.19.8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tassan J-P, Jaquenod M, Fry A, Frutiger S, Hughes G J, Nigg E A. In vitro assembly of a functional human CDK7-cyclin H complex requires MAT1, a novel 36 kDa RING finger protein. EMBO J. 1995;14:5608–5617. doi: 10.1002/j.1460-2075.1995.tb00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thuret J-Y, Valay J-G, Faye G, Mann C. Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell. 1996;86:565–576. doi: 10.1016/s0092-8674(00)80130-0. [DOI] [PubMed] [Google Scholar]
- 53.Toh-e A, Oshima Y. Characterization of a dominant, constitutive mutation, PHO0, for the repressible acid phosphatase synthesis in Saccharomyces cerevisiae. J Bacteriol. 1974;120:608–617. doi: 10.1128/jb.120.2.608-617.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valay J-G, Simon M, Dubois M-F, Bensaude O, Facca C, Faye G. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J Mol Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- 55.Valay J G, Dubois M F, Bensaude O, Faye G. Ccl1, a cyclin associated with protein kinase Kin28, controls the phosphorylation of RNA polymerase II largest subunit and mRNA transcription. C R Acad Sci Ser III. 1996;319:183–189. [PubMed] [Google Scholar]
- 56.Valay J G, Simon M, Faye G. The Kin28 protein kinase is associated with a cyclin in Saccharomyces cerevisiae. J Mol Biol. 1993;234:307–310. doi: 10.1006/jmbi.1993.1587. [DOI] [PubMed] [Google Scholar]
- 57.Wagner M, Pierce M, Winter E. The CDK-activating kinase CAK1 can dosage suppress sporulation defects of smk1 MAP kinase mutants and is required for spore wall morphogenesis in Saccharomyces cerevisiae. EMBO J. 1997;16:1305–1317. doi: 10.1093/emboj/16.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wahi M, Johnson A D. Identification of genes required for alpha 2 repression in Saccharomyces cerevisiae. Genetics. 1995;140:79–90. doi: 10.1093/genetics/140.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]