Abstract
Cytotoxic lymphocytes (CLs) induce caspase activation and apoptosis of target cells either through Fas activation or through release of granule cytotoxins, particularly granzyme B. CLs themselves resist granule-mediated apoptosis but are eventually cleared via Fas-mediated apoptosis. Here we show that the CL cytoplasmic serpin proteinase inhibitor 9 (PI-9) can protect transfected cells against apoptosis induced by either purified granzyme B and perforin or intact CLs. A PI-9 P1 mutant (Glu to Asp) is a 100-fold-less-efficient granzyme B inhibitor that no longer protects against granzyme B-mediated apoptosis. PI-9 is highly specific for granzyme B because it does not inhibit eight of the nine caspases tested or protect transfected cells against Fas-mediated apoptosis. In contrast, the P1(Asp) mutant is an effective caspase inhibitor that protects against Fas-mediated apoptosis. We propose that PI-9 shields CLs specifically against misdirected granzyme B to prevent autolysis or fratricide, but it does not interfere with homeostatic deletion via Fas-mediated apoptosis.
Virus-infected and tumor cells are killed on contact by cytotoxic lymphocytes (CLs), which trigger intrinsic cell death programs by using either one of two systems. The first system depends on the ability of perforin to mediate the entry of the serine proteinase granzyme B into the target cell, where it activates cytoplasmic cysteine proteinases known as caspases (reviewed in reference 38). Alternatively, death is triggered by binding of Fas ligand on the CL to Fas/Apo1/CD95 (Fas) on the target cell, resulting in the activation of the intracellular caspase zymogen, procaspase-8. Activation of other caspases follows, leading to the degradation of a variety of nuclear and cytoplasmic substrates and the characteristic biochemical and morphological changes associated with apoptosis (reviewed in reference 24).
Like other activated lymphocytes, CLs die in response to a variety of apoptotic stimuli, including Fas receptor ligation, which is used to remove redundant CLs from the immune system postinfection to preserve long-term tissue homeostasis (reviewed in reference 24). Prior to deletion, functioning CLs are likely to be exposed to multiple cytotoxins as they sequentially engage and destroy target cells, yet they apparently do not commit fratricide or undergo autolysis (13, 17, 25). To forestall premature death, CLs must therefore be able to control misdirected granzyme B and have some means of preventing caspase activation in response to Fas ligand. Studies of viral inhibitors of apoptosis suggest several ways that Fas-induced death can be controlled. For example, herpesvirus and molluscipox virus produce v-FLIPs, which block early events in Fas-mediated apoptosis by preventing the recruitment and activation of caspase-8 at the receptor complex (52), and cellular homologs of the v-FLIPs have been described (15). One of these is produced early in T-cell activation but disappears as the sensitivity of the cells to Fas-induced apoptosis increases, making it a strong candidate as a repressor of Fas-mediated apoptosis in T cells.
Other viruses produce caspase inhibitors, such as the baculovirus p35 protein (4) and the orthopoxvirus cytokine response modifier A (CrmA) (33). CrmA potently inhibits activated caspase-8 and is thought to prevent both Fas- and tumor necrosis factor (TNF)-induced apoptosis (40, 49, 63). It belongs to the serine proteinase inhibitor (serpin) superfamily in both structure and mode of action, but is distinguished from other serpins by its ability to inhibit caspases. CrmA is also a moderately efficient inhibitor of granzyme B that may prevent granzyme B-induced apoptosis under certain conditions (21, 32, 51). Caspases and granzyme B prefer to cleave substrates after Asp (29, 53), and this is reflected in the reactive center loop of CrmA, which has an Asp at the crucial P1 position (33). To date, a cellular homolog of CrmA with a P1 Asp has not been discovered, although the ability of CrmA to inhibit Fas-mediated and (perhaps) granule-mediated apoptosis suggests that endogenous serpins may regulate the apoptotic proteinases.
We have recently described a human intracellular serpin, proteinase inhibitor 9 (PI-9), that efficiently inhibits granzyme B in vitro and is expressed at high levels in the cytoplasm of CLs (45). PI-9 is very similar to CrmA, but surprisingly has Glu rather than Asp at the P1 position. Here we show that PI-9 protects transfected cells against granzyme B-induced but not Fas-induced apoptosis and that the P1 Glu confers specificity for granzyme B and not the caspases. We propose that PI-9 protects CLs (and perhaps bystander cells) against premature death triggered by miscompartmentalized or misdirected granzyme B, but does not interfere with the deletion of cells from the immune system via the Fas pathway.
MATERIALS AND METHODS
Site-directed mutagenesis and plasmid constructions.
Hexahistidine-tagged CrmA was produced by PCR amplification from a plasmid template (kindly provided by D. Pickup) with the primers (5′-TCTGCCATCATGCATCATCATCATCATCATGATATCTTCAGGGAAATC-3′ and 5′-TTAATTAGTTGTTGGAGAGC-3′. The PCR used 20 pmol of each primer and 1 ng of template in Vent polymerase reaction buffer (New England Biolabs) containing 200 μM deoxynucleoside triphosphates (dNTPs) and 1 U of Vent polymerase (New England Biolabs). Thirty cycles of 95°C for 90 s, 57°C for 45 s, and 72°C for 60 s were performed. Amplified fragments were separated by 1% agarose gel electrophoresis, purified from the gels, and cloned into pCRII (Invitrogen) for sequence analysis. A clone with an in-frame fusion of the His tag and no second-site mutations was chosen for the subsequent steps. The modified cDNA was released from pCRII by EcoRI digestion, cloned into the vector pHIL-D2, and expressed as an intracellular protein in Pichia pastoris as previously described (47).
PI-9 cDNA in the P. pastoris vector pHIL-D2 was mutated by the Deng and Nickoloff method (9). One primer was designed to remove a unique XbaI site in the vector (5′-CGGTGAGCATGCAGACCTTCAAC-3′). Other primers were designed to mutate the PI-9 sequence 340Glu to Asp (5′-AGTTGCAGACTGCTGCATG-3′), 340Glu to Ala (5′-AGTTGCAGCGTGCTGCATGG-3′), or 327Thr to Arg (5′-GAAGGCAGGGAGGCAGCG-3′). These specific alterations and the absence of second-site mutations were confirmed by DNA sequencing.
The PI-9 cDNAs were cloned into the EcoRI site of the mammalian expression vector pCMV2 (1) to give pCMV/PI9, pCMV/PI9E340D, and pCMV/PI9T327R and were also cloned into a derivative of pCMV2 containing a neomycin transcriptional unit. This plasmid (pCMVneo) was constructed by digestion of pCMV2 with SmaI and XbaI, followed by T4 DNA polymerase treatment and religation to remove a BamHI site in the polylinker. The resulting plasmid had unique XhoI and BamHI sites downstream of a transcriptional unit comprising the CMV promoter, polylinker, and human growth hormone terminator. It was then digested with XhoI and BamHI and ligated to a 1.8-kb XhoI-BamHI fragment from pPNT (59) containing a neomycin resistance gene under the control of the human phosphoglycerate kinase promoter and terminator. A unique EcoRI site in the polylinker of the resulting plasmid (pCMVneo) was then used to insert EcoRI fragments containing the PI-9 cDNAs or the CrmA cDNA. DNA sequencing was used to confirm the identity of all plasmids.
Production of recombinant serpins.
Hexahistidine-tagged PI-9, PI9E340D, PI9E340A, PI9T327R, and CrmA were produced in the methylotropic yeast species P. pastoris and purified by methods described previously (45, 47).
Inhibition of granzyme B and caspase activity by PI-9 and derivatives.
Purification of active granzyme B from YT cell granules has been described previously (54). For stoichiometric determinations, 10 pmol of granzyme B was incubated with different concentrations of inhibitor at 37°C in a mixture of 20 mM HEPES (pH 7.4), 100 mM NaCl, and 0.05% (wt/vol) Nonidet P-40 (32). Residual enzyme activity was determined after 15 min by a two-stage assay with Boc-Ala-Ala-Asp-S-benzyl and 5,5′-dithiobis(nitrobenzoic acid) (29). The rate of inhibition of granzyme B by each inhibitor was determined by incubation of equimolar enzyme and inhibitor at 37°C and determination of residual activity periodically (2, 47). The second-order rate constant was calculated as described previously (47).
With the exception of caspase-3 (produced in P. pastoris [46]), recombinant caspases were produced in Escherichia coli by using the pET and pGEX expression systems (39). Briefly, 50 ml of exponentially growing bacteria was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 18°C. Cells were collected, resuspended in 1 ml of 20 mM HEPES (pH 8.0)–0.1% CHAPS {3-[(cholamidopropyl)-dimethyl-ammonio]-1-propanesulfonate}, and lysed by sonication. Lysates were cleared by centrifugation at 100,000 × g, and the supernatant was stored at −70°C. Protease activity was assessed with the fluorogenic substrates z-Tyr-Val-Ala-Asp-7-amino-4-trifluoromethyl coumarin (z-YVAD-AFC) and z-Asp-Glu-Val-Asp-7-amino-4-trifluoromethyl coumarin (zDEVD-AFC) from Enzyme Systems Products (Dublin, Calif.). Assays were performed with a mixture of 100 mM HEPES (pH 7.5), 10% sucrose, 0.1% CHAPS, and 10 mM dithiothreitol containing 50 μM substrate at 25°C. Hydrolysis was monitored by measuring the fluorescence (λem = 500 nm and λex = 420 nm).
Levels of inhibitory activity of the serpins PI-9 and PI9E340D (20 nM) against each caspase (approximately 4 nM) were compared by preincubation of the serpin and protease for 30 min at 37°C prior to addition of the appropriate substrate. The peptide inhibitors Ac-Tyr-Val-Ala-Asp-chloromethylketone (YVAD-cmk) and Ac-Asp-Glu-Val-Asp-aldehyde (DEVD-cho) were purchased from BACHEM and used at 50 μM.
Cells and transfections.
Lp natural killer leukemia cells are a primary culture of cells from a patient with asymptomatic splenomegaly that exhibit potent perforin-dependent cytotoxicity (55). They were maintained in RPMI 1640 containing 10% heat-inactivated fetal bovine serum and 25 U of interleukin 2 (IL-2) per ml. Peripheral blood mononuclear cells were isolated from normal human blood by Isopaque-Ficoll (Pharmacia) centrifugation and incubated for 7 to 21 days in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% heat-inactivated fetal bovine serum and 100 U of IL-2 per ml. FDC-P1 cells were grown in DMEM containing 10% heat-inactivated fetal bovine serum, 10−4 M asparagine, and 50 U of IL-3 per ml. MCF-7 cells were grown in RPMI 1640 medium containing 2 mM glutamine, 5% fetal bovine serum, and 5% newborn bovine serum. Jurkat cells were maintained in RPMI 1640 medium containing 2 mM glutamine, 1 mM pyruvate, 55 μM mercaptoethanol, and 10% heat-inactivated fetal calf serum (GIBCO-BRL).
FDC-P1 and Jurkat cells were transfected with the pCMVneo/PI-9 plasmids, which carry a selectable neomycin resistance marker, or with the control plasmid based on pPNT (59), which carries the same selectable marker. MCF-7 cells were cotransfected with the pCMV/PI9 plasmids and the marker plasmid (in a ratio of 10:1). For FDC-P1 transfections, 107 cells were washed three times in DMEM and then resuspended in 0.25 ml of DMEM. After 10 min on ice and 1 min at 37°C, cells were electroporated with 20 μg of DNA at 240 V and 960 μF with a Gene Pulser (Bio-Rad). Transfectants were selected in the appropriate medium containing 0.4 mg of G418 (GIBCO BRL) per ml. For Jurkat and MCF-7 transfections, 2 × 106 cells in 0.8 ml of HEPES-buffered saline at room temperature were electroporated with 20 μg of DNA at 300 V and 330 μF by using a Cell-Porator (GIBCO-BRL). Transfectants were selected in the appropriate medium containing 0.5 mg of G418 per ml (MCF-7) or 1 mg of G418 per ml (Jurkat).
Cytoxicity and apoptosis assays.
For perforin or granzyme B killing, FDC-P1 cells in 96-well plates (2 × 104 per point) were incubated for 60 min at 37°C in the presence of 100 U of rat perforin per ml (35) and/or 1 μg of granzyme B per ml (54). DNA degradation was assessed by terminal deoxyribonucleotidyl transferase labeling of DNA strands breaks with dUTP (TUNEL) by using a kit from Boehringer Mannheim. Cells were analyzed on a fluorescence-activated cell sorter (FACS) (FACScan; Becton Dickinson).
MCF-7 cells do not exhibit the DNA fragmentation frequently associated with apoptosis (27), so assays dependent on DNA fragmentation (such as TUNEL) are unsuitable for monitoring cell death. However, these cells do exhibit other characteristic morphological changes, such as cytoplasmic vesicularization and shrinkage, as well as nuclear crenation and condensation. Apoptosis can therefore be monitored and quantitated by light microscopy, as previously described for many different cell types (10, 14, 20, 34), including MCF-7 (49, 50). Killing of MCF-7 cells by the Lp cytotoxic cells was assessed as follows. MCF-7 cells were plated at 103 per well on a 12-well microscope slide. The next day, a suspension of Lp cells (5,000, 2,500, or 1,000 cells) was added to each well. At the indicated times, nonadhering Lp cells were washed off, and the remaining cells were fixed and permeabilized with 50% acetone–50% methanol for 2 min at room temperature. Cells were stained with propidium iodide (1 μg/ml in phosphate-buffered saline [PBS]) for 5 min at room temperature, washed in PBS, and mounted in phenylenediamine-buffered glycerol. Slides were examined by phase and fluorescence microscopy in a “blind” procedure (observer unaware of identity of samples) in which those target cells having one or two killer cells attached were counted and scored for apoptosis based on changes in cytoplasmic and nuclear morphology.
Standard 4-h cytotoxicity assays at the effector/target ratios indicated were performed with MCF-7 cells labeled at 37°C for 75 min in RPMI medium supplemented with fetal bovine serum and 51Cr (Amersham) as a marker of cytoplasmic content. All results are expressed as the mean specific release of isotope from triplicate assays ± the standard error of the mean.
Apoptosis of 5 × 104 Jurkat cells per well in microtiter trays was induced by incubation for 20 h with 50 ng of the anti-Fas monoclonal antibody CH-11 per ml (AusPep) (61) or with 1 μM staurosporine (Sigma) in triplicate. Cell viability was then measured by MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay read at 595 nm (16). Apoptotic alterations to cell nuclei were monitored by fluorescence microscopy following staining of whole cells with propidium iodide (100 μg/ml) in PBS and 0.05% Nonidet P-40.
Antibodies.
Rabbit antibodies to recombinant PI-9 have been described previously (45), and a monoclonal antibody (2C5) has been raised against granzyme B (54). Rabbit antisera and 2C5 in ascites fluid were used at 1:2,000 for immunoblotting, and blots were developed with an enhanced chemiluminescence detection kit (Du Pont). The monoclonal antibody to recombinant PI-9 (2E7) was raised by standard procedures and detects an epitope not present on any of the related ovalbumin serpins (14a).
Comparison of PI-9 levels in transfected clones.
Intracellular FACS (31) with the monoclonal antibody 2E7 was used to detect cells expressing PI-9. Analysis of the PI-9-negative parental cells (FDC-P1) and a vector-transfected clone (FDC-neo) showed that 90 to 95% of cells stained below 60 fluorescent units (FU). Transfected cells staining over 60 FU were therefore taken as PI-9 positive. Since every antibody has a detection threshold, and antigen expression in a clonal cell population follows a normal distribution, the proportion of positive cells detected in a clonal population is directly related to the level of PI-9 produced. For example, in low-PI-9 producers, only those cells well to the right of the mode (i.e., those cells in the population producing the most PI-9) will be detected. In contrast, in higher producers, more cells (closer to the mode) will apparently be positive (for example, see Fig. 2A). Therefore, to compare PI-9 levels between clones, FACS analysis was used to determine the percentage of cells in each clone that exhibited staining greater than 60 FU. In each experiment, the percentage of cells over 60 FU in the negative controls (5 to 10%) was subtracted to yield the final value (stated in arbitrary units).
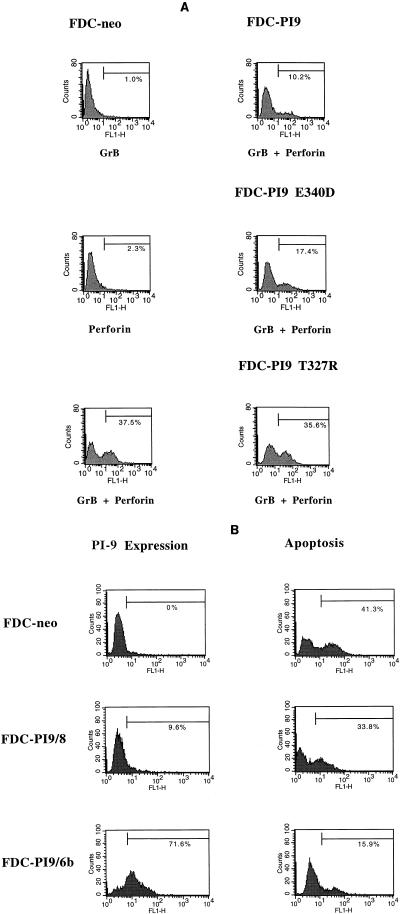
FIG. 2.
Intracellular PI-9 protects FDC-P1 cells from purified granzyme B and perforin. (A) Efficient inhibition of granzyme B-mediated apoptosis by PI-9 requires Glu at the P1 position and mobility of the inhibitory loop. FDC-P1 clones expressing equivalent levels of PI-9, the P1(Asp) mutant (E340D), or the hinge mutant (T327R) were exposed to granzyme B (GrB) and perforin (P) and compared to vector-transfected cells (FDC-neo). Panels show FACS profiles of DNA fragmentation by TUNEL analysis. Cells to the right of the cursor are positive for DNA fragmentation and are shown as a percentage of the whole population. (B) Increased levels of PI-9 correlate with increased protection against apoptosis triggered by granzyme B. Panels to the left show intracellular FACS analysis for PI-9 expression in the FDC-neo control cells, a low-PI-9-expressing clone (FDC-PI9/8), and a higher-PI-9-expressing clone (FDC-PI9/6b). Panels to the right are FACS profiles of the same cells showing DNA fragmentation by TUNEL analysis, following exposure to granzyme B and perforin. All results are representative of three independent experiments.
RESULTS
Both the tissue distribution of PI-9 and its efficient in vitro inhibition of granzyme B suggest that this serpin is a physiological regulator of granzyme B. Comparisons with other inhibitory serpins suggest that PI-9 has a mobile C-terminal domain (reactive loop) containing the crucial P1 residue (340Glu) required for inhibitory function (45). Although this putative P1 residue is an acidic amino acid, it does not meet the general expectation that an efficient inhibitor of granzyme B should have a P1 Asp, reflecting the substrate preference of this proteinase (29, 30, 53). For example, the viral serpin CrmA, which closely resembles PI-9, has a P1 Asp and effectively inhibits granzyme B in vitro (32). Before embarking on experiments to test whether PI-9 in transfected cells confers protection against apoptosis induced by granzyme B, we investigated whether 340Glu is required for PI-9 function and whether substitution with Asp improves inhibition of granzyme B.
Substitution of 340Glu with Ala or Asp markedly reduces PI-9 inhibition of granzyme B.
To determine the role of 340Glu, we produced two derivatives having either Ala (PI9E340A) or Asp (PI9E340D) at this position (Table 1). We also produced a third derivative (PI9T327R) that has a 327Thr-to-Arg substitution disrupting the conserved proximal hinge domain required for serpin loop mobility and inhibitory function (42). We found that the rate constant (ka) for complex formation between the P1(Ala) mutant and granzyme B was 300 times less than the ka for the PI-9–granzyme B interaction, indicating that 340Glu is crucial for PI-9 function, as expected for a P1 residue (Table 1). The hinge mutant exhibited even lower inhibitory capacity than the Ala mutant, indicating that inhibition of granzyme B depends on the mobility of the PI-9 reactive loop. Finally, the ka for complex formation between the Asp mutant and granzyme B was 2 orders of magnitude less than the ka for the PI-9–granzyme B interaction and 20-fold less than the CrmA - granzyme B interaction (32). This confirms that the 340Glu is important for PI-9 function and indicates—contrary to dogma—that a P1 Asp is not optimal for inhibition of granzyme B.
TABLE 1.
Properties of serpins used in this study
| Serpin | Sequence of inhibitory regiona | Interaction with granzyme B (ka [M−1 s−1]) |
|---|---|---|
| PI-9 | ‥320VEVNEEGTEAAAASSCFVVAE=CCMESGPRFCADHPFL‥ | (1.7 ± 0.3) × 106 |
| PI9E340D | ‥320VEVNEEGTEAAAASSCFVVAD=CCMESGPRFCADHPFL‥ | (1.4 ± 0.3) × 104 |
| PI9E340A | ‥320VEVNEEGTEAAAASSCFVVAA=CCMESGPRFCADHPFL‥ | (5.8 ± 0.6) × 103 |
| PI9T327R | ‥320VEVNEEGREAAAASSCFVVAE=CCMESGPRFCADHPFL‥ | (8.7 ± 1.0) × 102 |
| CrmA | ‥284IDVNEEYTEAAAAT-CALVAD=CASTVTNEFCADHPFI‥ | 2.9 × 105b |
The sequences of PI-9 and CrmA were taken from references 45 and 33, respectively. Only the portions comprising the hinge and reactive centers of each serpin are shown. A space introduced to optimize sequence alignment is indicated by a single dash. Mutated residues are indicated in boldface. The P1-P1′ bond is indicated by a double dash.
Value taken from reference 32.
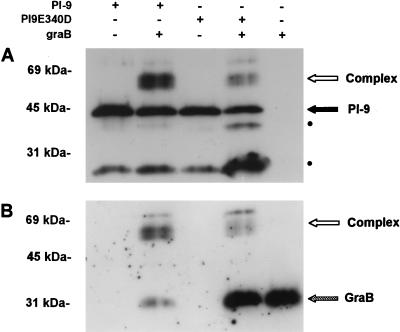
It is accepted that a serpin can act as either an inhibitor or a substrate of a proteinase (30). An efficient inhibitory serpin forms an essentially irreversible complex with the proteinase, and cleavage of the serpin occurs extremely slowly, if at all. In contrast, a substrate-like serpin is rapidly cleaved by the proteinase, resulting in dissociation of the complex and release of active proteinase and inactive serpin. Incubation of the P1(Asp) mutant with granzyme B resulted in the formation of sodium dodecyl sulfate (SDS)-stable complex (Fig. 1). However, much less granzyme B was complexed with the P1(Asp) mutant than with wild-type PI-9. In addition, incubation of the P1(Asp) mutant with granzyme B resulted in the accumulation of 38- and 25-kDa polypeptides that probably reflect serpin cleavage by granzyme B. The appearance of breakdown products and the presence of large amounts of free granzyme B are consistent with the P1 Glu-to-Asp substitution causing a marked shift in the properties of the serpin—from an efficient inhibitor cleaved very poorly by granzyme B to an inefficient inhibitor that is a substrate for the protease. This supports the kinetic data that 340Glu is the P1 residue and that Asp at this position is not required to efficiently inhibit granzyme B.
FIG. 1.
Interaction of PI-9 and the P1(Asp) mutant (PI9E340D) with granzyme B (graB). Equimolar amounts of PI-9 (8 ng) or PI9E340D (8 ng) and granzyme B (5 ng) in 20 mM Tris (pH 7.4)–0.15 M NaCl were incubated for 30 min at 37°C, followed by reduction, boiling, and electrophoresis on an SDS–10% polyacrylamide gel. Protein was transferred to a nitrocellulose membrane and immunoblotted with a rabbit antiserum against PI-9 diluted 1:2,000 (A). The membrane was then stripped and reprobed with a monoclonal antibody (2C5) in ascites fluid against granzyme B diluted 1:2,000 (B). Bound antibody was detected by chemiluminescence. Dots indicate the positions of the 38- and 25-kDa cleavage products.
PI-9 inhibits granzyme B-mediated apoptosis.
We propose that PI-9 protects cells from apoptosis induced by exposure to misdirected or ectopic granzyme B. For example, we envisage that the serpin might protect a CL against autolysis triggered by granzyme B that leaks back into the CL cytoplasm during degranulation and target cell killing. A simple but important prediction of the model is that cytoplasmic PI-9 should protect a transfected cell against apoptosis induced by internalized granzyme B.
Apoptosis of susceptible cells can be induced by exposure to purified granzyme B and perforin or by exposure to a CL which will deliver granzyme B by granule exocytosis. As described below, we used both methods of delivery to test whether PI-9 can protect against granzyme B-induced apoptosis. Given that the PI-9–granzyme B interaction is essentially irreversible and is optimal at a 1:1 stoichiometry (45), the degree of protection observed should depend on the ratio of PI-9 to granzyme B achieved within the cell. This in turn will depend on the amount of serpin present in the cell or the amount of granzyme B it is exposed to. Therefore, we expected that cells expressing higher levels of PI-9 would be more resistant to apoptosis but that resistance might be overcome by higher levels of granzyme B.
To determine whether cytoplasmic PI-9 would prevent apoptosis induced by internalized granzyme B, we produced clones of FDC-P1 cells that express similar amounts of either PI-9, the P1(Asp) mutant, or the hinge mutant, as assessed by immunoblotting and FACS analysis (data not shown). These were then exposed to purified perforin and granzyme B, and apoptosis was monitored by TUNEL assay (Fig. 2A). Compared to mock-transfected cells, those containing PI-9 showed significantly decreased levels of granzyme B-mediated apoptosis. In contrast, cells expressing either the P1(Asp) mutant or the hinge mutant showed limited or no resistance to granzyme B-mediated apoptosis, which is consistent with the kinetic data showing that these mutant serpins are ineffective granzyme B inhibitors. These results indicate that cytoplasmic functional PI-9 can prevent apoptosis induced by granzyme B.
To determine whether the degree of protection conferred by PI-9 is proportional to the amount of serpin present within the cell, we also identified and analyzed several FDC-P1 clones with low to high levels of PI-9 expression (Fig. 2B and Table 2). Levels of PI-9 expression were compared by FACS analysis (for example, see Fig. 2B) and confirmed by immunoblotting (data not shown). We found a good correlation between the amount of PI-9 produced by a particular clone and its resistance to apoptosis (Table 2). That is, the higher the level of PI-9 in a clone, the more resistant it is to granzyme B-mediated apoptosis.
TABLE 2.
Correlation between the levels of PI-9 in transfected FDC-P1 clones and their resistance to granzyme B-or perforin-mediated apoptosis
| Clone | Level of PI-9 expression (arbitrary units)a | % Inhibition of apoptosisb |
|---|---|---|
| FDC-neo | 0 | 0 |
| FDC-PI9/8 | 9.6 | 17.0 |
| FDC-PI9/6a | 38.4 | 43.6 |
| FDC-PI9/11 | 51.6 | 58.3 |
| FDC-PI9/6b | 71.6 | 62.2 |
Levels of PI-9 in the clones were compared by FACS analysis and are expressed in arbitrary units (see Materials and Methods). The vector-transfected cells (FDC-neo) do not express PI-9, as assessed by immunoblotting and RNA analysis. Expression of PI-9 by the transfected clones was confirmed by immunoblotting (data not shown).
Values represent the percent reduction in TUNEL-positive PI-9-expressing cells compared to FDC-neo cells following exposure to purified granzyme B and perforin (as measured by FACS). The results are representative of three experiments in which cells were analyzed by TUNEL and FACS for apoptosis. Under these conditions, 40% of the cells in the FDC-neo control samples routinely became apoptotic.
Fas-negative cells expressing PI-9 resist CL attack.
Numerous studies have implicated granzyme B as the primary proapoptotic granule cytotoxin. To determine whether PI-9 can protect against apoptosis when other granzymes are present, we produced lines of MCF-7 breast cancer cells expressing PI-9 to use as targets for multiple granzyme (granule) delivery by activated CLs. MCF-7 cells produce no endogenous granzyme B or PI-9 and are Fas negative by FACS analysis. They are killed by exposure to purified granzyme B but not by exposure to recombinant soluble Fas ligand (data not shown). CL killing of these cells is therefore mediated by granule cytotoxins. Since MCF-7 cells do not exhibit DNA fragmentation during apoptosis, death can be monitored by monitoring cytoplasmic and nuclear alterations (as described in Materials and Methods).
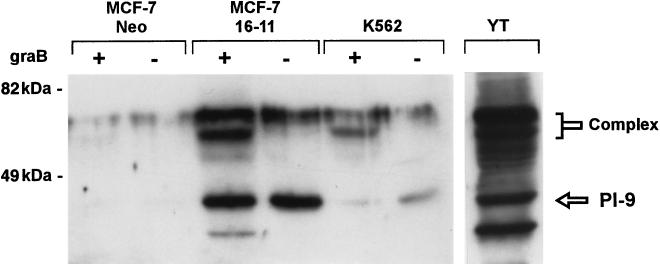
Transfected MCF-7 cells were screened for expression of PI-9 by indirect immunofluorescence and immunoblotting. PI-9 produced in these clones was functional, as indicated by the SDS-resistant complex formed when cytosolic extracts were incubated with granzyme B (Fig. 3). To assess the levels of PI-9 expressed by the clones, they were compared to cell lines that express the serpin constitutively (Fig. 3). The best MCF-7 clone, 16-11, produced about five times more PI-9 than K562 cells (which are sensitive to CL killing), but about five times less than cells of the natural killer leukemia line YT (which resist CL killing). Thus PI-9 expression in the transfectants falls between the levels observed in low and high endogenous producers.
FIG. 3.
Expression level and activity of PI-9 in MCF-7 transfectants compared to those in endogenous PI-9 producers. MCF-7 Neo cells were transfected with the marker plasmid only. Cells were lysed, and 50 μg of protein with (+) or without (−) granzyme B (5 ng) in 20 mM Tris (pH 7.4)–0.15 M NaCl was incubated for 30 min at 37°C. Granzyme B was not added to the YT cell extracts. Samples were reduced and subjected to electrophoresis on an SDS–10% polyacrylamide gel. Immunoblotting with anti-PI-9 antiserum was carried out as described in the legend to Fig. 1.
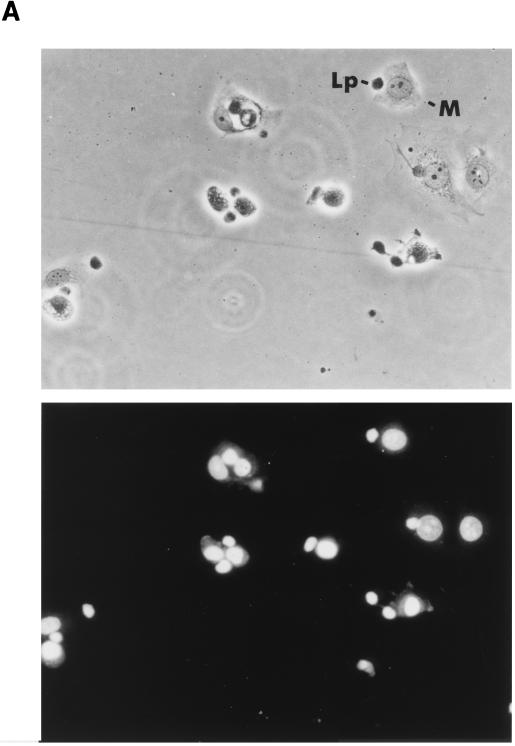
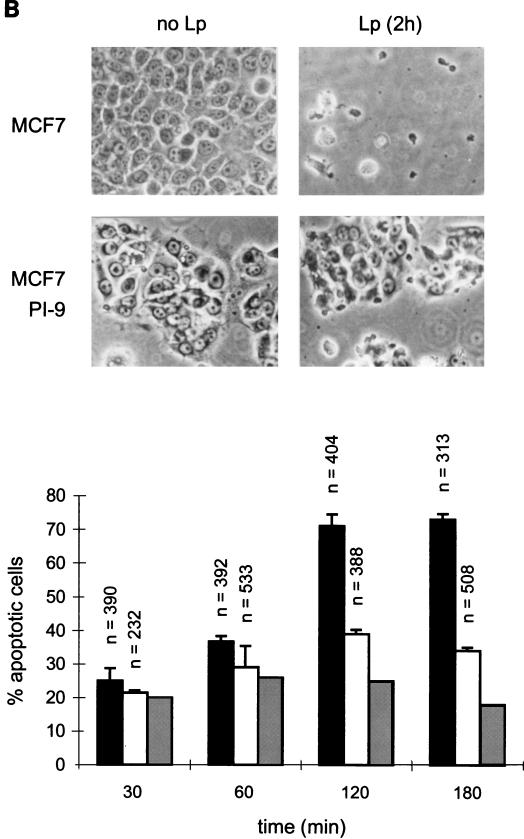
The sensitivity of the MCF-7 clones to CL attack was first tested by incubation with natural killer leukemia cells (Lp [55]). Lp cells are readily distinguished from the much larger MCF-7 cells by phase-contrast microscopy or by fluorescence microscopy following nuclear staining with propidium iodide (Fig. 4A). At an effector/target ratio of 5:1, most of the target cells had multiple Lp cells attached. Untransfected MCF-7 cells were efficiently killed under these conditions, with visible apoptotic alterations occurring within an hour and total destruction of the monolayer after 2 h (Fig. 4B). In contrast, monolayers of cells expressing PI-9 remained essentially intact after 2 h (Fig. 4B).
FIG. 4.
PI-9 protects MCF-7 cells from killing by Lp cytotoxic cells. (A) Phase-contrast (upper panel) and propidium iodide fluorescence (lower panel) microscopy of MCF-7 cells engaged by Lp cells. The cytotoxic (Lp) cells are dark and round, with very little visible cytoplasm, and have small nuclei without visible nucleoli. MCF-7 cells (M) are larger, with prominent cytoplasm and large nuclei containing nucleoli. Several stages of apoptosis of MCF-7 cells induced by attached Lp cells are shown in this field. Cells in early apoptosis show darkened cytoplasm, reduction of cell area, and retraction of pseudopodia. Cells in later stages show nuclear crenation and condensation, cytoplasmic vesicularization, and become refractile. (B) Phase-contrast microscopy of monolayers of untransfected MCF-7 cells and PI-9 transfectants exposed to Lp cells and estimation of the numbers of cells undergoing Lp-induced apoptosis. Only cells having one or two Lp killers attached were counted and assessed for apoptotic changes in a blind assay. Untransfected MCF-7 cells are indicated by solid bars, PI-9 transfectants (clone 16-11) are indicated by open bars, and corrected values to account for negative cells in the latter population are indicated by grey bars.
To quantitate the degree of protection, we examined cells at a 1:1 effector/target ratio, scoring apoptotic changes only in MCF-7 cells conjugated with one or two Lp cells (Fig. 4B). Under these conditions, more than 70% of untransfected MCF-7 cells were apoptotic after 2 h, compared to only 40% of MCF-7 cells expressing PI-9. The degree of protection observed is probably an underestimate, because PI-9 expression in the MCF-7 clones was slightly unstable in long-term culture, leading to the accumulation of PI-9-negative cells in the population (FACS data not shown). When the data were corrected to allow for these negative cells (30%), we estimated that less than 25% of the PI-9-positive cells had been killed. These results show that PI-9 confers protection against apoptosis even when multiple granzymes are delivered, which is consistent with the primary role played by granzyme B in target cell apoptosis. The failure to observe complete protection may reflect the action of one or more granzymes not inhibited by PI-9 or insufficient levels of PI-9 to neutralize all the incoming granzyme B.
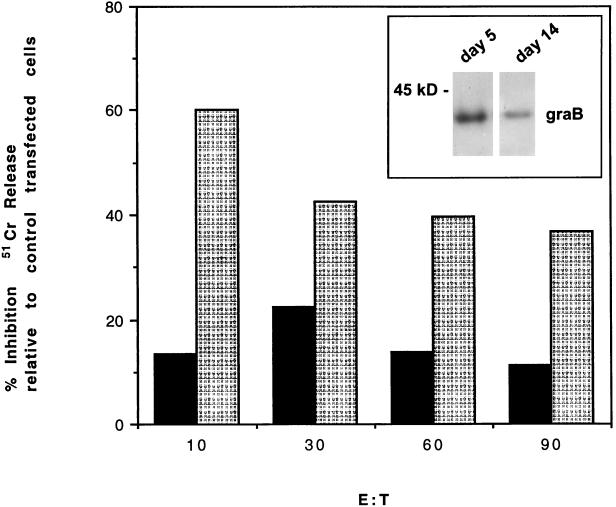
The MCF-7 clones were also used as targets for IL-2-activated killer (LAK) cells in a standard 51Cr-release cytoxicity assay. As shown in Fig. 5B, these LAK cells synthesized granzyme B, with peak production occurring after 5 days in culture. MCF-7 cells expressing PI-9 showed only slight resistance to killing by the day 5 LAK cells, compared to mock-transfected cells (Fig. 5A). However, when MCF-7 cells were exposed to day 14 LAK cells, which were producing significantly less granzyme B, two independent PI-9-expressing clones showed a significant and reproducible resistance to cytolysis (Fig. 5A). This is consistent with the predictions of the model and the results of the FDC-P1 experiments (Fig. 2) that suggest that the degree of protection increases with the ratio of PI-9 to granzyme B. Whereas increased protection correlated with increased expression of PI-9 in the FDC-P1 experiments, in the LAK experiment, increased protection was observed in the context of smaller amounts of granzyme B.
FIG. 5.
PI-9 protects MCF-7 cells from killing by LAK cells. MCF-7 clones expressing PI-9 or cells expressing the marker gene (Neo) only were exposed to LAK cells cultured for 5 or 14 days. Killing was monitored in triplicate by 51Cr release after 4 h at different effector/target ratios (E:T). The results are representative of three independent experiments performed with two independent clones. The inset panel shows granzyme B (graB) levels in the day 5 and day 14 cultured LAK cells. Cells were lysed, and 50 μg of protein was separated by SDS–10% polyacrylamide gel electrophoresis followed by immunoblotting, as described in the legend to Fig. 1. Solid bars, LAK cells cultured for 5 days; stippled bars, LAK cells cultured for 14 days.
PI-9 does not protect T cells from Fas-mediated apoptosis.
CrmA has the unusual ability to inhibit two distinct classes of Aspase: the serine proteinases (granzyme B) and the cysteine proteinases (caspases). The similarity between PI-9 and CrmA suggests that PI-9 might also inhibit caspases, which could enhance the effectiveness of PI-9 as a regulator of granzyme B-induced death. Furthermore, it raises the possibility that PI-9 may also protect cells against Fas-mediated apoptosis.
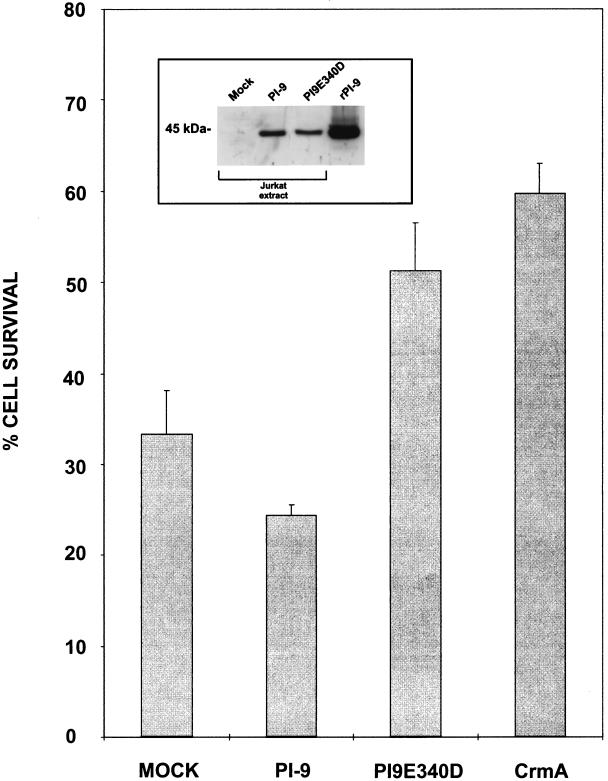
To test whether PI-9 prevents Fas-mediated apoptosis, we expressed it in Fas-sensitive Jurkat cells, which are PI-9 negative (45). In parallel, we produced cells expressing CrmA, which is known to prevent Fas- and TNF-mediated apoptosis (49). Using a monoclonal antibody to cross-link Fas, we were able to trigger apoptosis in pools of transfected and untransfected cells (Fig. 6). Cell viability was measured by MTT assay, which assesses mitochondrial function, and apoptotic cells in the cultures were monitored by the classical alterations to nuclear structure. There was good correlation between the degree of cell death measured by the MTT assay and the proportion of cells showing nuclear disintegration (not shown). As expected, Jurkat cells expressing CrmA were resistant to Fas-induced apoptosis (Fig. 6), and the degree of protection observed was comparable to those in previous studies (36, 49). In contrast, cells expressing PI-9 showed no resistance, indicating that the serpin does not effectively inhibit caspases involved in the Fas pathway.
FIG. 6.
PI-9 does not protect cells from Fas-induced apoptosis. Fas-sensitive Jurkat cells were transfected with expression vectors encoding a neomycin-selectable marker and either PI-9, PI9E340D, or CrmA. G418-resistant pools were then assessed for sensitivity to apoptosis induced by cross-linking Fas with the monoclonal immunoglobulin M antibody CH-11. Cell viability was quantitated by MTT assay. The results shown represent four independent experiments performed in triplicate. The inset panel shows an immunoblot with anti-PI-9 antiserum on total protein (50 μg) extracted from cells expressing the marker plasmid (mock), PI-9, or PI9E340D. rPI-9, purified recombinant PI-9.
The inability of PI-9 to block Fas-mediated apoptosis suggests that its P1 residue (Glu) does not permit interactions with caspases, which cleave preferentially after Asp. If this is correct, it follows that PI-9 with a P1 Asp (thus resembling CrmA more closely) would be more likely to inhibit caspases and therefore block apoptosis. To test this, we also expressed the P1(Asp) mutant in Jurkat cells and assessed the response to Fas ligation. As shown in Fig. 6, these cells produced the same amount of inhibitor as the PI-9-expressing cells, but showed a level of resistance to apoptosis comparable to that of cells expressing CrmA.
PI-9 and the P1(Asp) mutant do not prevent staurosporine-induced apoptosis of T cells.
Apoptosis can be induced by many different agents, and the early steps in the various pathways can sometimes be distinguished. For example, Fas-mediated apoptosis is not inhibited by Bcl-2 but is inhibited by CrmA (44). In contrast, the protein kinase inhibitor staurosporine triggers an apoptosis pathway involving caspase activation that is inhibitable by Bcl-2 but not by CrmA (5, 16). To test if PI-9 or the P1(Asp) mutant behaves differently with CrmA in response to another apoptotic trigger, we treated the transfected Jurkat cells with staurosporine. In our system, 1 μM staurosporine efficiently induced apoptosis of Jurkat cells that was evident by loss of MTT activity and nuclear alterations. Staurosporine-induced death was not prevented by expression of CrmA, PI-9, or the P1(Asp) mutant in Jurkat cells (data not shown).
PI-9 is a poor caspase inhibitor.
The results presented above show that PI-9 can inhibit granzyme B-mediated but not Fas-mediated cell death and suggest that it is incapable of interacting with caspases crucial to the Fas pathway. These results, however, do not exclude the possibility that PI-9 controls caspases downstream of granzyme B that are not important in Fas-mediated death.
Caspases can be divided into three groups (reviewed in reference 26). Caspase-1, -4, and -5 are likely to be involved in cytokine processing rather than apoptosis, are not activated by granzyme B in vitro, and almost certainly do not participate in granule-mediated killing. Caspase-2, -3, -6, and -7 are activated during apoptosis triggered by a wide variety of signals, including Fas, and can be considered to be the main effectors of cell death. Caspase-8, -9, and -10 are upstream activators of apoptosis: caspase-8 is the apical caspase involved in Fas-mediated apoptosis, whereas caspase-9 is the apical caspase in the apoptotic cascade triggered by release of cytochrome c from mitochondria (18). At present, the exact role of caspase-10 is unknown, but it is likely to function in a similar manner to caspase-8.
Caspases cleaved by granzyme B in vitro that represent potential targets for PI-9 include effector caspase-2, -3, -6, and -7 and apical caspase-8, -9, and -10 (8, 11, 41). Of these, only caspase-3, -6, and -7 are known to be cleaved during granzyme B-induced apoptosis, and at least one (caspase-3) is activated directly by granzyme B (7, 12). It is possible that one or more of the effector or apical caspases are targeted by PI-9 to enhance its control of granzyme B-mediated apoptosis (although our results suggest that PI-9 does not interact efficiently with caspase-8, because it cannot block Fas-mediated apoptosis). To test this, the ability of PI-9 to inhibit 9 of the 10 known human caspases was investigated (Table 3). To reveal even slight PI-9 and caspase interactions, the partially purified recombinant enzymes were incubated with an excess of PI-9. Under these conditions, PI-9 inhibited only caspase-4, and no significant inhibition by PI-9 of the effector or apical caspases was observed. This suggests that PI-9 does not regulate caspases activated by granzyme B or by Fas ligation.
TABLE 3.
Interaction of PI-9 and the PI-9 Asp mutant with recombinant caspasesa
| Caspase type | % Inhibition of enzyme activity
|
||||
|---|---|---|---|---|---|
| PI-9 | PI9E340D | CrmA | DEVD-cho | YVAD-cmk | |
| ICE-like | |||||
| Caspase-1 | 30 | 65 | 100 | NDb | 95 |
| Caspase-4 | 70 | 5 | 40 | ND | 0 |
| Caspase-5 | 30 | 50 | 5 | ND | 10 |
| Effectors | |||||
| Caspase-2 | 5 | 25 | 0 | ND | ND |
| Caspase-3 | 10 | 10 | 5 | 100 | ND |
| Caspase-6 | 10 | 70 | 8 | 100 | ND |
| Caspase-7 | 10 | 60 | 8 | 100 | ND |
| Activators | |||||
| Caspase-8 | 10 | 82 | ND | 95 | ND |
| Caspase-10 | 35 | 90 | ND | 95 | ND |
Recombinant caspases (approximately 4 nM) were incubated with 20 nM PI-9, PI9E340D, or CrmA. The peptide inhibitors were used at 50 μM. Residual enzyme activity was measured and compared to that in an uninhibited reaction. The ICE-like caspases were assayed with the fluorescent substrate z-YVAD-AFC, and the effector and activator caspases were assayed with the fluorescent substrate z-DEVD-AFC (both from Enzyme Systems Products).
ND, not determined.
In contrast, under the same conditions and with equal amounts of inhibitor, the P1(Asp) mutant exhibited a substantially different inhibitory profile against the caspases, which overlapped but was not identical to that observed with CrmA (Table 3). In particular, it was an effective inhibitor of caspase-6, -7, -8, and -10 and, to a lesser extent, caspase-1, -2, and -5. Significantly, the P1(Asp) mutant had essentially no activity against caspase-4, confirming that the P1 substitution has shifted the inhibitory specificity of the serpin. The inhibition of caspase-8, in particular, is consistent with the ability of this mutant to protect against Fas-mediated apoptosis. These results confirm that the P1(Glu) provides PI-9 with only very limited potential to act on caspases, therefore essentially restricting it to inhibition of granzyme B.
DISCUSSION
CLs operate in an environment in which they are exposed to their own potent cytotoxins. As they engage and destroy multiple targets sequentially, they must possess the means to resist self-induced or fraternally induced cytolysis. In this respect, it is known that although extragranular granzyme B can be detected in CLs (56), they resist cytolysis (3, 13, 17) and in fact are more resistant than noncytolytic cells (25). In addition, CLs that produce higher levels of cytotoxins are more resistant to cytolysis than other CLs (19).
Little is known about the protective mechanisms CLs employ to resist their own cytotoxins. Autolysis triggered by exposure to granule cytotoxins is thought to be minimized by the maintenance of granzymes in an inactive state during biosynthesis and packaging into granules (6, 28, 37), by intrinsic differences in the CL membrane compared to other cells (22, 57), and by the short half-life of perforin in serum (58). (Bystander lysis or fratricide would also be minimized by the latter mechanisms.) Although it is important that CLs resist apoptosis induced by their own cytotoxins, it is equally important that this does not interfere with the systems used to delete old or redundant CLs from the immune system. At present, deletion is thought to operate by apoptosis of the activated CL in response to Fas ligand (24) or cytokine depletion (43). It is also possible that TNF-induced apoptosis is involved in clearing CLs (62), although this is likely to represent an ancillary mechanism.
Our recent discovery of an intracellular serpin (PI-9) that is an efficient granzyme B inhibitor suggests an additional means that CLs may employ to prevent autolysis or fratricide. Since PI-9 is found predominantly in lymphocytes (particularly killer cells) or in tissue enriched in immune cells (45), we have postulated that its role is to protect CLs and perhaps antigen-presenting cells (APCs) against death induced by inadvertent exposure to granzyme B. Similarities to the viral serpin CrmA also raise, a priori, the possibility that PI-9 protects cells against exposure to Fas ligand by inhibiting caspases.
In this study, we have investigated whether cytosolic PI-9 can protect a host cell from granzyme B- and Fas-induced apoptosis and have studied its interaction with caspases. We have clearly demonstrated that the presence of PI-9 within transfected cells can inhibit apoptosis induced by exposure to granzyme B, but that it is not sufficient to protect against Fas-mediated apoptosis, most likely because the serpin does not interact significantly with key caspases. Our results also show that the degree of protection against granzyme B depends on the intracellular concentration of the PI-9 and the amount of protease delivered. This is consistent with the properties of the granzyme B–PI-9 interaction, which is optimal at a 1:1 molar ratio and is essentially irreversible (45). The incomplete protection from apoptosis when CLs were used to deliver granzyme B may therefore simply reflect the lack of a transfected clone expressing PI-9 at a high enough level. Alternatively, other granzymes not inhibited by PI-9 may have contributed to target cell death in these systems. The latter possibility is supported by studies of granzyme B knockout mice, which show that target cell apoptosis still occurs (albeit slowly) in the absence of granzyme B.
As a component of the CL and perhaps other immune cells, it is unlikely that PI-9 has evolved to protect against a directed, full-blooded hit delivered by a CL. Rather we envisage that it neutralizes lower levels of misdirected granzyme B that inadvertently threaten the CL or a bystander cell. This is difficult to test directly, so by necessity, our experimental systems have utilized methods of granzyme B delivery that treat the test cells as targets rather than bystanders. It could be argued that this produces a worst-case scenario, because the granzyme B/PI-9 ratio achieved within the transfected test cell is likely to be much greater than that arising from the misdirection of granzyme B into CLs or bystander cells in vivo. Nevertheless, our observation of a significant and reproducible degree of protection under these conditions demonstrates, in principle, that a physiological role of PI-9 is to regulate granzyme B-mediated apoptosis.
Our findings lead to the following model for PI-9 function. Because PI-9 is present in the cytosol of CLs and associated cells, such as APCs (45), we propose that it inactivates granzyme B that either leaks from resting CL granules or enters the cytoplasm of the CL or APCs following target cell recognition and degranulation. Because PI-9 does not inhibit caspases, it cannot interfere with apoptosis induced by Fas ligand, TNF, or cytokine deprivation. We believe that this system has evolved to allow cells to effectively control ectopic granzyme B, yet avoid the consequences of a more general block of apoptosis that would result if PI-9 was also a caspase inhibitor. Deleterious consequences arising from a general block of apoptosis might include the nonclearance of normal cells at the appropriate time and their accumulation in the system (a situation analagous to Bcl-2 overexpression in certain leukemias (for review, see reference 60), or the resistance of malignant or infected cells to Fas-mediated CL attack. The model explains why extragranular granzyme B detected in some CLs is apparently not detrimental and provides an additional or alternative explanation as to why CLs resist cytolysis. It also predicts that the highly cytotoxic CLs that are more resistant to cytolysis will have more PI-9 than other CLs.
One of the original arguments against PI-9 being a physiological granzyme B inhibitor revolved around the presence of Glu at the crucial P1 position in the reactive loop. From the many studies of serpin structure, it was expected that the identity of the P1 residue would reflect the substrate preference of the cognate proteinase and that the best inhibitor of granzyme B should have Asp not Glu at this position. Hence, it could be argued that even though PI-9 is an effective inhibitor of granzyme B, a true granzyme B inhibitor would have a P1 Asp. However, the work described here clearly shows that PI-9 with a P1 Asp is a much less effective granzyme B inhibitor than PI-9 itself, and despite much effort, we have not identified an endogenous serpin with a P1 Asp (45, 48). In addition, it should be noted that CrmA—which has a P1 Asp—is also a much less effective inhibitor of granzyme B than PI-9 is (32). Therefore, an important and unexpected conclusion from this study is that while the P1 residue is crucial for serpin function—as illustrated by the almost total loss of inhibitory capacity of the PI-9 P1 Ala mutant—it does not necessarily reflect the optimal substrate preference of the cognate proteinase.
Clearly, the P1 residue of PI-9 limits its specificity with respect to caspase inhibition. Glu at this position does not allow significant inhibition of caspases, whereas Asp allows quite broad caspase inhibition. As a consequence, the P1 Asp mutant is capable of affording protection from Fas-mediated apoptosis. It is likely that the Asp mutant prevents Fas-mediated apoptosis, because it effectively inhibits caspase-8, which is the apical protease of the Fas pathway also thought to be the primary target of CrmA (23, 63). Interestingly, the Asp mutant inhibits a larger range of caspases than CrmA, so the possibility exists that it may interfere with apoptosis triggered by other agents, providing a tool with which to understand caspase activation hierarchies and function during apoptosis.
At present, the physiological role of caspase-4 is unknown, so the significance of its inhibition by PI-9 is unclear and must await more detailed kinetic analysis. On the basis of sequence homology and substrate preference, caspase-4 belongs to the ICE group of caspases, which are proposed to control cytokine processing, rather than participating in apoptosis (reviewed in reference 26). Perhaps PI-9 prevents elaboration of a cytokine or other product by caspase-4 that would be toxic to the host or bystander cells or mark it for attack by other CLs.
In conclusion, we believe that PI-9 is a specific inhibitor of granzyme B that provides effective protection for particular cells against unwarranted apoptosis triggered by ectopic granzyme B-mediated caspase activation. It is clear that the P1 Glu in the PI-9 reactive center provides the precision required to ensure that it inhibits granzyme B and not the apoptotic caspases, thus allowing cells containing PI-9 to die in response to Fas ligation. It remains to be seen if ectopic expression of PI-9 is associated with increased tumor cell resistance to granule-mediated cytotoxicity.
ACKNOWLEDGMENTS
C.H.B., V.R.S., and J.S. contributed equally to this work.
This work was supported by the National Health and Medical Research Council of Australia, Monash University, and the Anti-Cancer Council of Victoria.
We are grateful to D. Pickup (Duke University) for providing the CrmA cDNA, R. Sutherland (Garvan Institute) for the MCF-7 cells, and A. Strasser (Walter and Eliza Hall Institute) for discussions. We thank L. McDonald for technical assistance and F. Scott and M. Chu for help with preparation of figures.
REFERENCES
- 1.Andersson S, Davis D L, Dahlback H, Jornvall H, Russell D W. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 2.Beatty K, Bieth J, Travis J. Kinetics of association of serine proteinases with native and oxidized alpha-1-proteinase inhibitor and alpha-1-antichymotrypsin. J Biol Chem. 1980;255:3931–3934. [PubMed] [Google Scholar]
- 3.Blakely A, Gorman K, Ostergaard H, Svoboda K, Liu C C, Young J D, Clark W R. Resistance of cloned cytotoxic T lymphocytes to cell-mediated cytotoxicity. J Exp Med. 1987;166:1070–1083. doi: 10.1084/jem.166.4.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bump N J, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, Licari P, Mankovich J, Shi L, Greenberg A H, Miller L K, Wong W W. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 5.Cahill M A, Peter M E, Kischkel F C, Chinnaiyan A M, Dixit V M, Krammer P H, Nordheim A. CD95(APO-1/Fas) induces activation of SAP kinases downstream of ICE-like proteases. Oncogene. 1996;13:2087–2096. [PubMed] [Google Scholar]
- 6.Caputo A, Garner R S, Winkler U, Hudig D, Bleackley R C. Activation of recombinant murine cytotoxic cell proteinase-1 requires deletion of an amino-terminal dipeptide. J Biol Chem. 1993;268:17672–17675. [PubMed] [Google Scholar]
- 7.Darmon A J, Ley T J, Nicholson D W, Bleackley R C. Cleavage of CPP32 by granzyme B represents a critical role for granzyme B in the induction of target cell DNA fragmentation. J Biol Chem. 1996;271:21709–21712. doi: 10.1074/jbc.271.36.21709. [DOI] [PubMed] [Google Scholar]
- 8.Darmon A J, Nicholson D W, Bleackley R C. Activation of the apoptotic protease CPP32 by cytotoxic T-cell-derived granzyme B. Nature. 1995;377:446–448. doi: 10.1038/377446a0. [DOI] [PubMed] [Google Scholar]
- 9.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 10.Faleiro L, Kobayashi R, Fearnhead H, Lazebnik Y. Multiple species of CPP32 and Mch2 are the major active caspases present in apoptotic cells. EMBO J. 1997;16:2271–2281. doi: 10.1093/emboj/16.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandes-Alnemri T, Armstrong R C, Krebs J, Srinivasula S M, Wang L, Bullrich F, Fritz L C, Trapani J A, Tomaselli K, Litwack G, Alnemri E S. In vitro activation of CPP32 and Mch3 by Mch4, a novel human apoptotic cysteine protease containing two FADD-like domains. Proc Natl Acad Sci USA. 1996;93:7464–7469. doi: 10.1073/pnas.93.15.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Froelich C J, Orth K, Turbov J, Seth P, Gottleib R, Babior B, Shah G M, Bleackley R C, Dixit V M, Hanna W. New paradigm for lymphocyte granule mediated cytotoxicity: target cells bind and internalize granzyme B but an endosomolytic agent is necessary for cytosolic delivery and subsequent apoptosis. J Biol Chem. 1996;271:29073–29079. doi: 10.1074/jbc.271.46.29073. [DOI] [PubMed] [Google Scholar]
- 13.Golstein P. Sensitivity of cytotoxic T cells to T-cell mediated cytotoxicity. Nature. 1974;252:81–83. doi: 10.1038/252081a0. [DOI] [PubMed] [Google Scholar]
- 14.Grimm S, Stanger B Z, Leder P. RIP and FADD: two death domain-containing proteins can induce apoptosis by convergent, but dissociable pathways. Proc Natl Acad Sci USA. 1997;93:10923–10927. doi: 10.1073/pnas.93.20.10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Hirst, C. E., V. R. Sutton, C. H. Bird, J. A. Trapani, and P. I. Bird. Unpublished observations.
- 15.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer J-L, Schroter M, Burns K, Mattman C, Rimoldi D, French L E, Tschopp J. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 16.Jacobsen M D, Burne J F, Raff M C. Programmed cell death and Bcl-2 protection in the absence of a nucleus. EMBO J. 1994;13:1899–1910. doi: 10.1002/j.1460-2075.1994.tb06459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kranz D M, Eisen H N. Resistance of cytotoxic lymphocytes to lysis by a clone of cytotoxic T lymphocytes. Proc Natl Acad Sci USA. 1987;84:3375–3379. doi: 10.1073/pnas.84.10.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 19.Liu C-C, Jiang S, Persechini P M, Zychlinsky A, Kaufmann Y, Young J D. Resistance of cytolytic lymphocytes to perforin-mediated killing. Induction of resistance correlates with increase in cytotoxicity. J Exp Med. 1989;169:2211–2225. doi: 10.1084/jem.169.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Los M, Van de Craen M, Penning L C, Schenk H, Westendorp M, Baeuerle P A, Droge W, Krammer P H, Fiers W, Schulze-Osthoff K. Requirement of an ICE/CED-3 protease for Fas/Apo-1-mediated apoptosis. Nature. 1995;375:81–83. doi: 10.1038/375081a0. [DOI] [PubMed] [Google Scholar]
- 21.Macen J L, Garner R L, Musy P Y, Brooks M A, Turner P C, Moyer R W, McFadden G, Bleackley R C. Differential induction of the Fas- and granule-mediated cytolysis pathways by the orthopoxvirus cytokine response modifier A/SPI-2 and SPI-1 protein. Proc Natl Acad Sci USA. 1996;93:9108–9113. doi: 10.1073/pnas.93.17.9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller C, Tschopp J. Resistance of CTL to perforin-mediated lysis. Evidence for a lymphocyte membrane protein interacting with perforin. J Immunol. 1994;153:2470–2478. [PubMed] [Google Scholar]
- 23.Muzio M, Salvesen G, Dixit V M. FLICE induced apoptosis in a cell-free system. J Biol Chem. 1997;272:2952–2956. doi: 10.1074/jbc.272.5.2952. [DOI] [PubMed] [Google Scholar]
- 24.Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 25.Nagler-Anderson C, Verret C R, Firmenich A A, Berne M, Eisen H N. Resistance of primary CD8+ cytotoxic T lymphocytes to lysis by cytotoxic granules from cloned T cell lines. J Immunol. 1988;141:3299–3305. [PubMed] [Google Scholar]
- 26.Nicholson D W, Thornberry N A. Caspases: killer proteases. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 27.Pandey S, Walker P R, Sikorska M. Separate pools of endonuclease activity are responsible for internucleosomal and high molecular mass DNA fragmentation during apoptosis. Biochem Cell Biol. 1994;72:625–629. doi: 10.1139/o94-082. [DOI] [PubMed] [Google Scholar]
- 28.Peters P J, Borst J, Oorschot V, Fukuda M, Krahenbuhl O, Tschopp J, Slot J W, Geuze H J. Cytotoxic T lymphocyte granules are secretory lysosomes, containing both perforin and granzymes. J Exp Med. 1991;173:1099–1109. doi: 10.1084/jem.173.5.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poe M, Blake J T, Boulton D A, Gammon N H, Sigal N H, Wu J K, Zweerink H J. Human cytotoxic lymphocyte granzyme B. Its purification from granules and the characterization of substrate and inhibitor specificity. J Biol Chem. 1991;266:98–103. [PubMed] [Google Scholar]
- 30.Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors: structure, function and regulation. J Biol Chem. 1994;269:15957–15960. [PubMed] [Google Scholar]
- 31.Prussin C, Metcalfe D D. Detection of intracytoplasmic cytokine using flow cytometry and directly conjugated anti-cytokine antibodies. J Immunol Methods. 1995;188:117–128. doi: 10.1016/0022-1759(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 32.Quan L T, Caputo A, Bleackley R C, Pickup D J, Salvesen G S. Granzyme B is inhibited by the cowpox virus serpin cytokine response modifier A. J Biol Chem. 1995;270:10377–10379. doi: 10.1074/jbc.270.18.10377. [DOI] [PubMed] [Google Scholar]
- 33.Ray C A, Black R A, Kronheim S R, Greenstreet T A, Sleath P R, Salvesen G S, Pickup D J. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1-beta converting enzyme. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- 34.Sarin A, Najajima H, Henkart P A. A protease-dependent TCR-induced death pathway in mature lymphocytes. J Immunol. 1995;154:5806–5812. [PubMed] [Google Scholar]
- 35.Shi L, Kraut R P, Aebersold R, Greenberg A H. A natural killer cell granule protein that induces DNA fragmentation and apoptosis. J Exp Med. 1992;175:553–556. doi: 10.1084/jem.175.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith K G, Strasser A, Vaux D L. CrmA expression in T lymphocytes of transgenic mice inhibits CD95 (Fas/Apo-1)-transduced apoptosis, but does not cause lymphadenopathy or autoimmune disease. EMBO J. 1996;15:5167–5176. [PMC free article] [PubMed] [Google Scholar]
- 37.Smyth M J, McGuire M J, Thia K Y. Expression of recombinant granzyme B. A processing and activation role for dipeptidyl peptidase I. J Immunol. 1995;154:6299–6305. [PubMed] [Google Scholar]
- 38.Smyth M J, Trapani J A. Granzymes: exogenous proteinases that induce target cell apoptosis. Immunol Today. 1995;16:202–206. doi: 10.1016/0167-5699(95)80122-7. [DOI] [PubMed] [Google Scholar]
- 39.Song Q, Lees-Miller S P, Kumar S, Zhang Z, Chan D W, Smith G C, Jackson S P, Alnemri E S, Litwack G, Khanna K K, Lavin M F. DNA-dependent protein kinase catalytic subunit: a target for an ICE-like protease in apoptosis. EMBO J. 1996;15:3238–3246. [PMC free article] [PubMed] [Google Scholar]
- 40.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Litwack G, Alnemri E S. Molecular ordering of the Fas-apoptotic pathway: the Fas/Apo-1 protease Mch5 is a Crma-inhibitable protease that activates multiple Ced-3/ICE-like cysteine proteases. Proc Natl Acad Sci USA. 1996;93:14486–14491. doi: 10.1073/pnas.93.25.14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srinivasula S M, Fernandes-Alnemri T, Zangrilli J, Robertson N, Armstrong R C, Trapani J A, Tomaselli K J, Litwack G, Alnemri E S. The Ced-3/interleukin 1beta converting enzyme-like homolog Mch6 and the lamin-cleaving enzyme Mch2alpha are substrates for the apoptotic mediator CPP32. J Biol Chem. 1996;271:27099–27106. doi: 10.1074/jbc.271.43.27099. [DOI] [PubMed] [Google Scholar]
- 42.Stein P E, Carrell R W. What do dysfunctional serpins tell us about molecular mobility and disease. Struct Biol. 1995;2:96–113. doi: 10.1038/nsb0295-96. [DOI] [PubMed] [Google Scholar]
- 43.Strasser A. Life and death during lymphocyte development and function: evidence for two distinct mechanisms. Curr Opin Immunol. 1995;7:228–234. doi: 10.1016/0952-7915(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 44.Strasser A, Harris A W, Huang D C S, Krammer P H, Cory S. Bcl-2 and Fas/Apo-1 regulate distinct pathways to lymphocyte apoptosis. EMBO J. 1995;14:6136–6147. doi: 10.1002/j.1460-2075.1995.tb00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun J, Bird C H, Sutton V, McDonald L, Coughlin P B, De Jong T A, Trapani J A, Bird P I. A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes. J Biol Chem. 1996;271:27802–27809. doi: 10.1074/jbc.271.44.27802. [DOI] [PubMed] [Google Scholar]
- 46.Sun J, Bottomley S P, Kumar S, Bird P I. Recombinant caspase-3 expressed in Pichia pastoris is fully activated and kinetically indistinguishable from the native enzyme. Biochem Biophys Res Commun. 1997;238:920–923. doi: 10.1006/bbrc.1997.7370. [DOI] [PubMed] [Google Scholar]
- 47.Sun J, Coughlin P, Salem H, Bird P. Production and characterization of recombinant human proteinase inhibitor 6 expressed in Pichia pastoris. Biochim Biophys Acta. 1995;1252:28–34. doi: 10.1016/0167-4838(95)00108-7. [DOI] [PubMed] [Google Scholar]
- 48.Sun J, Ooms L, Bird C H, Sutton V R, Trapani J A, Bird P I. A new family of 10 murine ovalbumin serpins includes two homologs of proteinase inhibitor 8 and two homologs of the granzyme B inhibitor (proteinase inhibitor 9) J Biol Chem. 1997;272:15434–15441. doi: 10.1074/jbc.272.24.15434. [DOI] [PubMed] [Google Scholar]
- 49.Tewari M, Dixit V M. Fas- and tumor necrosis factor-induced apoptosis is inhibited by the poxvirus crmA gene product. J Biol Chem. 1995;270:3255–3260. doi: 10.1074/jbc.270.7.3255. [DOI] [PubMed] [Google Scholar]
- 50.Tewari M, Quan L T, O’Rourke K, Desnoyers S, Zeng Z, Beidler D R, Poirer G G, Salvesen G, Dixit V M. Yama/CPP32beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 51.Tewari M, Telford W G, Miller R A, Dixit V M. CrmA, a pox-virus-encoded serpin, inhibits cytotoxic T-lymphocyte-mediated apoptosis. J Biol Chem. 1995;270:22707–22708. doi: 10.1074/jbc.270.39.22705. [DOI] [PubMed] [Google Scholar]
- 52.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattman C, Burns K, Bodmer J-L, Schroter M, Scaffidi C, Krammer P, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 53.Thornberry N A, Rano T A, Peterson E P, Rasper D M, Timkey T, Garcia-Calvo M, Houtzager V M, Nordstrom P A, Roy S, Vaillancourt J P, Chapman K T, Nicholson D W. A combinatorial approach defines specificities of members of the caspase family and granzyme B. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 54.Trapani J A, Browne K A, Dawson M J, Smyth M J. Immunopurification of functional Asp-ase (natural killer cell granzyme B) using a monoclonal antibody. Biochem Biophys Res Commun. 1993;195:910–920. doi: 10.1006/bbrc.1993.2131. [DOI] [PubMed] [Google Scholar]
- 55.Trapani J A, Klein J L, White P C, Dupont B. Molecular cloning of an inducible serine esterase gene from human cytotoxic lymphocytes. Proc Natl Acad Sci USA. 1988;85:6924–6928. doi: 10.1073/pnas.85.18.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapani J A, Smyth M J, Apostolidis V A, Dawson M, Browne K. Granule serine proteinases are normal nuclear constituents of natural killer cells. J Biol Chem. 1994;269:18359–18365. [PubMed] [Google Scholar]
- 57.Tschopp J, Masson D. Inhibition of the lytic activity of perforin (cytolysin) and of late complement components by proteoglycans. Mol Immunol. 1987;24:907–913. doi: 10.1016/0161-5890(87)90002-2. [DOI] [PubMed] [Google Scholar]
- 58.Tschopp J, Masson D, Schafer S. Inhibition of the lytic activity of perforin by lipoproteins. J Immunol. 1986;137:1950–1953. [PubMed] [Google Scholar]
- 59.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 60.Yang E, Korsmeyer S J. Molecular thanatopsis: a discourse on the BCL2 family and cell death. Blood. 1996;88:386–401. [PubMed] [Google Scholar]
- 61.Yonehara S, Ishii A, Yonehara M. A cell-killing monoclonal antibody (anti-Fas) to a cell surface antigen co-downregulated with the receptor of tumor necrosis factor. J Exp Med. 1989;169:1747–1756. doi: 10.1084/jem.169.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng L, Fisher G, Miller R E, Peschon J, Lynch D H, Lenardo M J. Induction of apoptosis in mature T cells by tumour necrosis factor. Nature. 1995;377:348–351. doi: 10.1038/377348a0. [DOI] [PubMed] [Google Scholar]
- 63.Zhou Q, Snipas S, Orth K, Muzio M, Dixit V, Salvesen G. Target protease specificity of the viral serpin CrmA. J Biol Chem. 1997;272:7797–7800. doi: 10.1074/jbc.272.12.7797. [DOI] [PubMed] [Google Scholar]