Abstract
A central unanswered question concerning the initial phases of V(D)J recombination has been at which step the 12/23 rule applies. This rule, which governs which variable (V), diversity (D), and joining (J) segments are able to pair during recombination, could operate at the level of signal sequence synapsis after RAG-HMG1 complex binding, signal nicking, or signal hairpin formation. It has also been unclear whether additional proteins are required to achieve adherence to the 12/23 rule. We developed a novel system for the detailed biochemical analysis of the 12/23 rule by using an oligonucleotide-based substrate that can include two signals. Under physiologic conditions, we found that the complex of RAG1, RAG2, and HMG1 can successfully recapitulate the 12/23 rule with the same specificity as that seen intracellularly and in crude extracts. The cleavage complex can bind and nick 12×12 and 23×23 substrates as well as 12×23 substrates. However, hairpin formation occurs at both of the signals only on 12×23 substrates. Moreover, under physiologic conditions, the presence of a partner 23-bp spacer suppresses single-site hairpin formation at a 12-bp spacer and vice versa. Hence, this study illustrates that synapsis suppresses single-site reactions, thereby explaining the high physiologic ratio of paired versus unpaired V(D)J recombination events in lymphoid cells.
The central feature of specific immunity is its capability to generate an enormous repertoire of antigen receptors by a process called V(D)J recombination. During this process, any one of an array of V (variable) gene segments can be joined to one of many D (diversity) or J (joining) gene segments to generate a new exon encoding the antigen binding domains of immunoglobulins or T-cell receptors. The cis elements mediating the site specificity of the recombination reaction are recombination signal sequences (RSS) flanking the V, D, and J gene segments. They consist of a palindromic heptamer separated from an A+T-rich nonamer by a spacer of either 12 or 23 bp. A single recombination event is directed by two of these joining signals with different spacer lengths, one with a 12-bp spacer (12-signal) and the other with a 23-bp spacer (23-signal). This restriction is referred to as the 12/23 rule (22). The 12/23 rule ensures that the recombining gene segments are joined together correctly.
The recombination reaction can be broadly divided into two phases. The first phase is cleavage at the signal sequences. The second phase is the ligation, or repair, of the gene segment termini to create the new exon. The RAG1 and RAG2 proteins (15, 17, 18) have been found to be necessary and sufficient for cleavage of the recombination signals in vitro (17, 24, 26). After binding and synapsis of the two signals, the reaction occurs by a two-step chemical mechanism during which one DNA strand is nicked at the heptamer of each RSS. The 3′ OH group that is generated attacks the phosphodiester bond of the antiparallel strand in a direct transesterification reaction, leaving hairpinned coding ends and blunt signal ends (25). Recently, a DNA-bending protein, HMG1, was found to markedly enhance the cutting reaction (16, 23). The second phase involves factors that participate in general double-strand break repair. These include the Ku70/86, DNA-PK, XRCC4, and DNA ligase IV (5, 17).
Several studies have indicated that the 12/23 rule is enforced at one of the steps of the cleavage phase of V(D)J recombination rather than at a later step. Such studies include exogenous substrate analysis (19, 21) and assays with crude extract cleavage of linear DNA (3, 16). One of these studies used a cell line that overexpressed RAG proteins to create active crude extracts for the cleavage step of the V(D)J reaction (3). In a series of experiments, the authors examined the ability of these lysates to form nicked or hairpinned products on linearized plasmid-based substrates. However, because these studies utilized plasmid-based substrates of several hundred base pairs, it is difficult to discern the relative rates of nicking and hairpin formation. Furthermore, these studies were performed in crude lysates, which prevent accurate measurements of the contribution of the RAG proteins specifically.
Examination of the role of RAG proteins for establishing the 12/23 rule has been problematic. Specifically, the levels of adherence to the 12/23 rule between RAG proteins overexpressed in crude cell lysates and as purified proteins have been found to differ (16). RAG proteins in crude lysates were shown to have a rate of cutting on 12×23 plasmid substrates at least 25-fold greater than that on non-12×23 substrates (3). Under similar conditions, purified RAG proteins showed only a two to fourfold preference for the 12×23 substrate (16, 26). This important discrepancy has not yet been reconciled.
We generated a novel system that allows the examination of concerted cutting at two recombination signal sequences in a much more detailed manner than was possible with previous systems. Under these conditions, we demonstrated that purified RAG proteins and HMG1 have a strict adherence to the 12/23 rule similar to that of RAG proteins in crude lysates. Having demonstrated that this system contains the essential components to reproduce the 12/23 rule, we went on to study the interaction of the RAG proteins with the 12×23 and alternative 12×12 and 23×23 substrates at each step of the cleavage reaction. We found that binding and nicking by the RAG proteins was no different for the 12×23 and the alternative substrates. However, at the nick to hairpin transition, the RAG proteins were unable to efficiently cleave the 12×12 or 23×23 substrates or prenicked 12×12 or 23×23 intermediates. The only substrates successfully cleaved were those with both the 12-signal and the 23-signal, establishing that the 12/23 rule is enforced at the formation of the double hairpin structure.
MATERIALS AND METHODS
Oligonucleotides.
The following oligonucleotides were used: for the 12×23 substrate, AAAAGAGGAGAGGAAGGAAGGAGGAAAGGAATCCCATGACTGCACAGTGATCATCCTGGAGACAAAAACCTGCAAC (76 nucleotides [nt]), GGAACTGGTTTTTGTCAAGCTTGATTCGGTGAGCAGGTCACTGTGCATGATCTCCCC (57 nt), and CCTTGACCAAAAACAGTTCGAACTAAGCCACTCGTCCAGTGACACGTACTAGAGGGGTTTTCTCCTCTCCTTCCTTCCTCCTTTCCTTAGGGTACTGACGTGTCACTAGTAGGAC CTCTGTTTTTGGACGTTG (133 nt); for the 12×12 substrate, ATCCCATGACTGCACAGTGATCATCCTGGAGACAAAAACCTGCAAC (46 nt), GGAACTGGTTTTTGTGATCGATCGATCCACTGTGCATGATCTCCCC (45 nt), AAAAGAGGAGAGGAAGGAAGGAGGGATCGATCGATCGAAAGGA (44 nt), and CCTTGACCAAAAACACTAGCTAGCTAGGTGACACGTACTAGAGGGGTTTTCTCCTCTCCTTCCTTCCTCCCTAGCTAGC TAGCTTTCCTTAGGGTACTGACGTGTCACTAGTAGGACCTCTGTTTT TGGACGTTG (135 nt); for the 23×23 substrate, GGAACTGGTTTTTGTGATCGAGCGATCGATCGATCGTCCACTGTGCATGATCTCCCC (57 nt), AAAAGAGGAGAGGAAGGAAGGAGGAAAGGAATCCCATGACTGCA CAGTGGATCGATCGATCGATCGTTCGATACAAAAACCTGCAAC (87 nt), and CCTTGACCAAAAACACTAGCTCGCTAGCTAGCTAGCAGGTGACACGTACTAGAGGGGTTTTCTCCTCTCCTTCCTTCCTCCTTTCCTTA GGGTACTGACGTGTCACCTAGCTAGCTAGCTAGCAAGCTATGTTTT TGGACGTTG (144 nt); for the mutant nonamer 12×23 substrate, GGAACACTGCATAGTCAAGCTTGATTCGGTGAGCAGGTCACTGTGCATGATCT CCCC (57 nt), AAAAGAGGAGAGGAAGGAAGGAGGAAAGGAATCCCATGACTGCACAGTGATCATCCTGGAGACAAAAACCTGCAAC (76 nt), and GTTGCAGGTTTTTGTCTCCAGGATGATCACTGTGCAGTCATGGGATTCCTTTCCTCCTTCCTTCCTCTCCTCTTTTGGGGAGATCATGCA CAGTGACCTGCTCACCGAATCAAGCTTGACTATGCAGTGTTCC (133 nt); for the single 12 site substrate, ATCAGGATGTGGTGATGCACAGTGTGATCCCTCCTCACAAAAACCGCAGGTCTTCAGTT (59 nt) and TAGTCCTACACCACTACGTGTCACACTAGGGAGGAGTGTTTTTGGCGTCCA GAAGTCAA (59 nt); for the single 23 site substrate, ATCCCATGACTGCACAGTGGATCGATCGATCGATCGTTCGATACAAAAACCTGCAACGA TC (61 nt) and TAGGGTACTGACGTGTCACCTAGCTAGCTAGCTAGCAAGCTATGTTTTTGGACGTTGCTAG (61 nt); for the nicked 12×23 substrate, GGAACTGGTTTTTGTCAAGCTTGATTCGGTGAGCAGGTCACT GTGCATGATCTCCCCAAAAGAGGAGAGGAAGGAAGGAGGAAAGG AATCCCATGACTG (99 nt), CACAGTGATCATCCTGGAGACAAAAACCTGCAAC (34 nt), CCCTTGACCAAAAACAGTTCGAACTAAGCCACTCGTCCAGTGACAC (45 nt), and GTACTAGAGGGGTTTTCTCCTCTCCTTCCTTCCTCCTTTCCTTAGGGTACTGACGTGTCACTAGTAGGACTCTGTT TTTGGACGTTG (88 nt); for the nicked 12×12 substrate, CACAGTGATCATCCTGGAGACAAAAACCTGCAAC (34 nt), GGAACTGGTTTTTGTGATCGATCGATCCACTGTGCATGATCTCCCCAAAAGAGGAGAGGAAGGA AGGAGGGATCGATCGATCGAAAGGAATCCCATGACTG (101 nt), CCTTGACCAAAAACACTAGCTAGCTAGGTGACAC (34 nt), and GTACTAGAGGGGTTTTCTCCTCTCCTTCCTTCCTCCCTAGCTAGCTAGCTTTCCTTAGGGTACTGACGTGTCACTAGTAGGACCTCTGTTTTTGGACGTTG (101 nt); for the nicked 23×23 substrate, GGAACTGGTTTTTGTGATCGAGCGATCGATCGATCGTCCACTGTG (45 nt), CATGATCTCCCCAAAAGAGGAGAGGAAGGAAGGAGGAAAGGAATCCCATGACTGCACAGTGGA TCGATCGATCGATCGTTCGATACAAAAACCTGCAAC (99 nt), CCTTGACCAAAAACACTAGCTCGCTAGCTAGCTAGCAGGTGACAC (45 nt), and GTACTAGAGGGGTTTTCTCCTCTCCTTCCTTCCTCCTTTCCTTAGGGT ACTGACGTGTCACCTAGCTAGCTAGCTAGCAAGCTATGTTTTTGGACGTTG (99 nt); and for the nonspecific competitor, GGAACTGATCGATCA CAAGCTTGATTCGGTGAGCAGGTGATCGATCATGATCTCCCC (57 nt), AAAAGAGGAGAGGAAGGAAGGAGGAAAGGAATCCCATGACTGGA TCGATATCATCCTGGAGGATCGATGCTGCAAC (76 nt), and GTTGCACGATCGATCCTCCAGGATGATATCGATCCAGTCATGGGATTCCTTTCCTCCTTCCTTCCTCTCCTCTTTTGGGGAGATCATGATCGATCACCTGCTCACCGAATCAAGCTTGTGATCGATCAGTTCC (133 nt).
Expression of RAG and HMG1 proteins.
Mouse recombinant core RAG1 and RAG2 proteins with an N-terminal maltose binding domain and a C-terminal His tag and myc epitopes (13) were expressed in Hi-5 insect cells. Cell extracts were then purified over a Ni-nitrilotriacetic acid column with imidazole elution. The eluant was further purified with an amylose column and removed with a maltose-containing solution. RAG1 and RAG2 proteins were found to be greater than 95% percent pure by SYPRO orange staining and were present in approximately equimolar amounts. Mouse recombinant C-terminal truncated HMG1 protein was expressed in a bacterial overexpression system as previously described (1).
Cleavage reactions.
The standard cleavage reaction contained 10 nM 32P-oligonucleotide substrate, 50 nM HMG1, 1 μM RAG1 and RAG2, 25 mM MOPS (morpholinepropanesulfonic acid) (pH 7.0), 5 mM Tris (pH 7.5), 30 mM KCl, 2% glycerol, 10 mM MgCl2, and 1 mM dithiothreitol. HMG1 was preincubated with the DNA in the conditions described above on ice for 5 min before the RAG proteins were added. All reactions were incubated at 37°C. Reactions were stopped with a 1% sodium dodecyl sulfate–20 mM EDTA solution. The DNA was then subjected to phenol-chloroform extraction followed by ethanol precipitation. DNA pellets were resuspended in 10 mM Tris (pH 8.0) and 1 mM EDTA (pH 8.0) and mixed with an equal volume of formamide, and half of the total volume was loaded on to a 15% acrylamide urea denaturing gel. Products were then quantified with ImageQuaNT software.
Competition assay.
The cold competitor DNA (100 nM) and the radioactively labeled DNA (10 nM) were mixed together and combined with HMG1 at a ratio of 5 HMG1 molecules to 1 DNA oligonucleotide complex in the standard cleavage reaction conditions described above. The protein and DNA mixture was incubated on ice for 5 min before the RAG proteins were added (at the standard cleavage reaction concentrations). Samples were incubated for 30 to 60 min at 37°C. Samples were then treated the same as the standard cleavage reactions.
RESULTS
Oligonucleotide 12×23 substrates for dissection of the binding, nicking, and hairpin formation steps.
To examine the relationship between nicked and double hairpin products in the context of the 12/23 rule, we developed an oligonucleotide-based substrate that contains both a 12-signal and a 23-signal. An oligonucleotide substrate, which is labeled at a discrete site, allows detailed examination of the phosphodiester breaks induced by the RAG proteins as achieved with the single-site substrates used previously (4, 26). A covalent linkage of the two signals is essential for observing whether hairpin formation at the two sites is coordinated. Therefore, an oligonucleotide must span the entire length of the substrate. It is important to note that previous studies of intersignal length in vivo and in vitro (2, 19) have demonstrated that the distance between the two signals can be less than 60 bp without a reduction in signal joint formation efficiency. Since such a short intersignal distance is still fully active for V(D)J recombination, it appeared possible to design a substrate constructed with oligonucleotides such that the two signals are covalently linked.
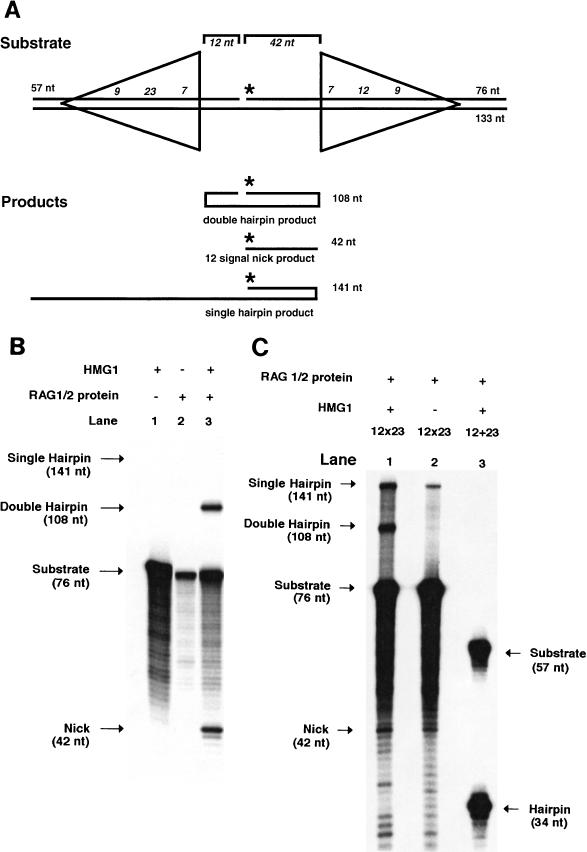
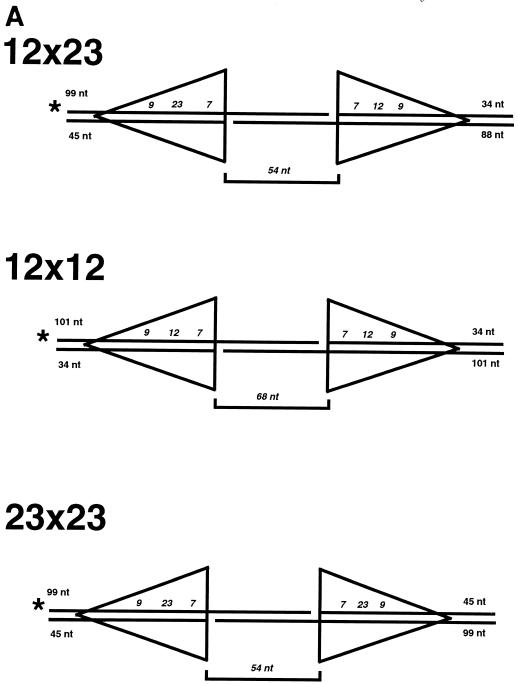
The prototypical substrate used for our study had the recombination signal sequences in the signal joint orientation with an intersignal distance of 54 bp between the cleavage sites. The substrate was constructed with three oligonucleotides (Fig. 1A). The single bottom oligonucleotide spanned the entire 133-bp length of the substrate. The other two top oligonucleotides annealed to create a nick located 12 bp 3′ of the first signal cut site. This allowed the substrate to be labeled in the middle of the two signals, providing high-resolution analysis of both signals. When the substrate underwent hairpin formation at both signal sites, the labeled oligonucleotide included both hairpins, giving it a unique length (108 nucleotides for the 12×23 substrate) that was longer than the input substrate length. The level of nicking at only one of the signal sites could be observed simultaneously, as the nicked product alone has a unique length (42 nucleotides for the 12×23 substrate). Thus, with this configuration, coordinated hairpin formation of the two substrates on the same molecule could be clearly and unambiguously identified concurrently with the level of nicking.
FIG. 1.
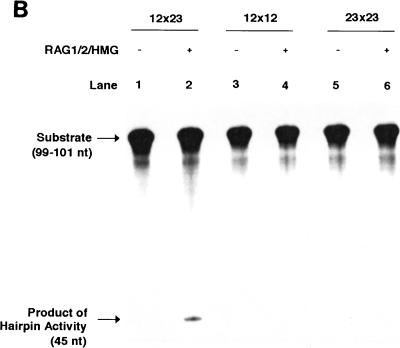
Concerted RAG cutting on an oligonucleotide 12×23 substrate. (A) Structure of the oligonucleotide substrate. The substrate is composed of three oligonucleotides (lines) that form the 12- and 23-signals (shown with triangles). Numbers at the ends of the oligonucleotides indicate the length of the entire adjacent oligonucleotide. Italicized numbers along the DNA strands represent the conserved signal sequences (heptamer, nonamer, and spacer) within the larger oligonucleotides. The structure of this substrate is such that there are 54 bp between the 12- and 23-signals. The brackets above and below the structure give the distances (in italics) between various landmarks, such as the edge of the signal sequence. The location of the radiolabel is denoted with a star. The structures of the nicked and double-hairpinned labeled products are shown, and the lengths of the products are given. (B) RAG1, RAG2, and HMG1 were incubated with the 12×23 substrate, and the products were separated on a 15% denaturing polyacrylamide gel. Lane 1, 12×23 substrate with HMG1 but without RAG1 and RAG2 in Mg2+ conditions; lane 2, 12×23 substrate with RAG1 and RAG2 in Mg2+ conditions; lane 3, 12×23 substrate with HMG1, RAG1, and RAG2 in Mg2+ conditions. (C) RAG1 and RAG2 were incubated with substrates in Mn2+ conditions, and the products were separated on a 15% denaturing polyacrylamide gel. Lane 1, 12×23 substrate with RAG1 and RAG2 with HMG; lane 2, 12×23 substrate with RAG1 and RAG2 alone; lane 3, single site 12 substrate with equimolar cold single site 23 substrate with RAG1 and RAG2 with HMG1.
Concerted cutting is absolutely dependent on the presence of HMG1.
To test the system, the primary substrate with a 12- and a 23-signal site in the signal joint orientation was incubated with purified RAG proteins (Fig. 1B). The cleavage reaction was performed in a Mg2+ buffer similar to that previously described (26). Purified RAG proteins alone failed to lead to hairpin formation (Fig. 1B, lane 2). The RAG protein complex was able to carry out nicking at very low levels on the labeled 12-signal site of a 12×23 substrate. The 42-nucleotide product that illustrates this is not visible in Fig. 1B, lane 2, but is apparent upon longer exposures of this image (data not shown). When the substrate was preincubated with HMG1, coordinated hairpin formation at the two signals occurred (Fig. 1B, lane 3). It is important to note that no products containing one hairpin and one nick were observed. An increase in nicking was also seen with the addition of HMG1 (compare lanes 2 and 3). This increase suggests that HMG1 participates in the initial creation of the complex rather than just at the hairpin stage. This is supported by the finding that the addition of HMG1 after the addition of the RAG proteins greatly decreases the formation of the double hairpin product (data not shown). HMG1 has been reported to augment RAG cutting and stimulate coupled cleavage (16, 23). In our system, HMG1 is essential for double hairpin formation.
Further illustration of the requirement for HMG1 by the 12/23 complex is seen when similar reactions are performed under Mn2+ rather than Mg2+ divalent conditions (Fig. 1C). It has been shown previously that on single-site substrates, RAG proteins cannot create the hairpin product when Mg2+ is the divalent; however, when Mn2+ is the divalent, the hairpin product is formed efficiently (24). With our 12×23 substrate, we found that under Mn2+ conditions, the RAG proteins without HMG1 created only a single hairpin product but no double hairpin products (Fig. 1C, lane 2). However, when HMG1 was present, the RAG protein-HMG1 complex formed both double and single hairpin products (Fig. 1C, lane 1). Interestingly, when RAG proteins were incubated under identical Mn2+-HMG1 conditions with an equimolar concentration of a labeled single 12-bp site substrate plus a cold single 23-bp site substrate with coding ends comparable to those of the 12×23 substrate, significantly more (eightfold) hairpin formation took place than in the 12×23 substrate (Fig. 1C, lane 3). It would appear that under these conditions, the activity of the RAG proteins at either of the sites of the 12×23 substrate was strongly inhibited by the presence of the other site, in contrast to the stimulation of hairpin formation by the second signal in Mg2+-HMG1 conditions (Fig. 1B).
Non-12×23 substrates are not cleaved efficiently.
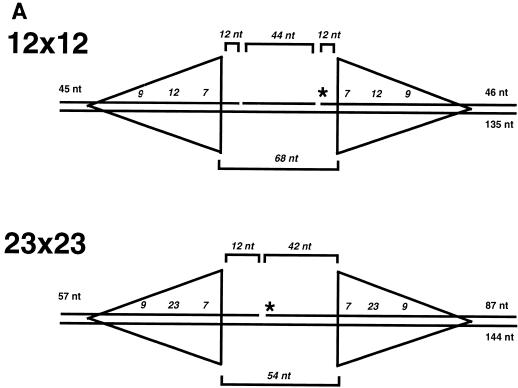
Having demonstrated that HMG1 plus Mg2+ provide for optimal double hairpin formation in a manner comparable to that seen in vivo and in crude extract, we used those conditions in all the remaining studies. Oligonucleotide substrates similar to the 12×23 substrate were created with two 12-RSS sites or two 23-RSS sites (Fig. 2A). The 23×23 substrate contained two nonidentical 23-signal sites with respect to the 23-nucleotide spacers but identical coding end sequences. Likewise, the 12×12 substrate contained nonidentical 12-nucleotide spacers and identical coding end sequences. Both substrates were labeled in a fashion similar to that of the 12×23 substrate with an internal 5′ label (Fig. 2A). The nicking step mediated by the RAG proteins gave a 12-nucleotide product for the 12×12 substrate and a 42-nucleotide product for the 23×23 substrate. Double hairpin formation mediated by the RAG proteins resulted in products of 92 and 108 nucleotides for the 12×12 and 23×23 substrates, respectively.
FIG. 2.
RAG nuclease activity on non-12×23 substrates: double hairpinning is efficient only on 12×23 substrates but nicking can occur on non-12×23 substrates. (A) The 12×12 and 23×23 substrate structures are shown in the same format as in Fig. 1A. (B) RAG1, RAG2, and HMG1 were incubated with the different substrates, and the products were separated on a 15% denaturing polyacrylamide gel. Lane 1, 12×23 substrate with RAG1, RAG2, and HMG1 with a Mg2+ divalent cation; lane 2, same as in lane 1 but with the 23×23 substrate; lane 3, same as in lane 1 but with the 12×12 substrate. The arrowheads mark the positions of the barely detectable single hairpin products, which are more apparent upon darker exposure (data not shown).
These substrates were compared with the 12×23 substrate in a RAG cleavage assay. Under identical conditions, including the presence of HMG1 and Mg2+, a qualitative difference was seen between products of the 12×23 versus the products of the 12×12 and 23×23 substrates (Fig. 2B). No double hairpin products were observed, and single hairpin products were present (Fig. 2B) but were only marginally apparent at this exposure. Significant nicking occurred in all three substrates. As with the 12×12 substrate, barely detectable levels of hairpin product were observed. Thus, there was a significant difference in how the RAG proteins created hairpins in the 12×23 substrate versus substrates that deviated from the 12/23 rule. We interpret the trace levels of hairpin formation in the 12×12 and 23×23 substrates to indicate that a complex similar to that in the 12×23 substrate can occur but that the resulting activity is greatly reduced in accordance with the 12/23 rule.
The RAG complex has a similar affinity for both 12×23 and non-12×23 substrates.
The inability of the RAG proteins to efficiently cleave the 12×12 and 23×23 substrates could be due to a failure at any of the steps along the double hairpin pathway. The RAG proteins may have lower affinity for DNA that cannot form a synaptic complex; that is, a stable complex may not form with the 12×12 and 23×23 substrates. Alternatively, the RAG proteins may fail to efficiently nick the substrates after having bound the DNA. A third possibility is that the RAG proteins may fail to catalyze the formation of the double hairpin.
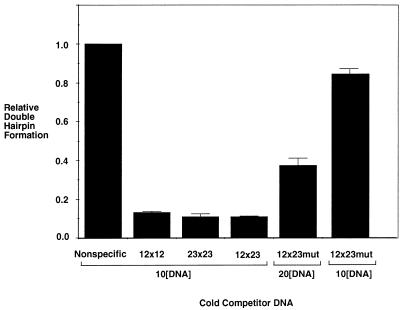
To assess the affinity of the RAG proteins for the different substrates, a competition assay for inhibition of hairpin formation in the 12×23 substrate was performed. Tenfold molar amounts of the cold competitor substrate were mixed with the labeled 12×23 substrate. A fivefold molar excess amount of HMG1 was preincubated with each substrate. This ensured that the levels of HMG1 were consistent per molecule of DNA, as previous studies of HMG1 have shown that excessive amounts of HMG1 can inhibit RAG protein nuclease activity (23). The RAG proteins were then added to the HMG1-DNA complex. Both of the alternative substrates (12×12 and 23×23) had a significant effect on the cutting of the 12×23 substrate compared with the nonspecific negative control (Fig. 3). The presence of the nonspecific competitor DNA had no significant effect on the hairpin formation activity of the RAG proteins. While these substrates competed equally well against the labeled 12×23 substrate, a 12×23 substrate with a mutated nonamer sequence at the 23-signal did not. At twice the molar concentration (which corresponds to an equal concentration of signal sites), this substrate was almost fourfold less effective at competition (Fig. 3). At an equal concentration of that same mutated substrate, the competition was almost 10-fold less effective (Fig. 3). The level of competition by the mutant substrate demonstrates that substrates that have two sites with the capability of synapsis have a higher affinity for the RAG complex than substrates that are much less capable of undergoing synapsis. We conclude that there is an increase in the stability of the complex from synaptic interactions and that these interactions can take place in substrates that do not fulfill the 12/23 rule as well as substrates that do. This indicates that enforcement of the 12/23 rule does not occur at the level of signal binding or synapsis.
FIG. 3.
Competition assay with non-12×23 substrates: equivalent competition by 12×23, 12×12, and 23×23 substrates. HMG1 was preincubated at equimolar concentrations with a mixture of radiolabeled 12×23 substrate and a 10-fold concentration of cold competitor substrate. RAG1 and RAG2 were then added under Mg2+ conditions. The products were separated on a 15% denaturing polyacrylamide gel. The intensities of the double hairpin product and the remaining substrate were then quantified with the PhosphorImager system. The double hairpin product value was normalized to the nonspecific DNA sample, which was assigned a relative value of 1. The molar concentration of unlabeled (cold) competitor relative to labeled 12×23 substrate is shown as 10[DNA] or 20[DNA], indicating 10 or 20 times more unlabeled (cold) competitor DNA. 12×23mut designates the substrate that has an ablated nonamer at the 23-signal.
The rate of double hairpin formation is markedly faster for 12×23 substrates relative to 12×12 and 23×23 substrates.
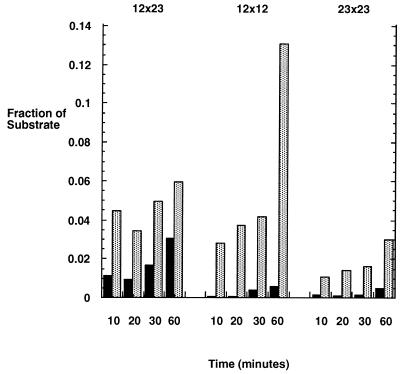
The rate of nicking and double hairpin formation was assessed in a time course assay. After preincubating the substrates with HMG1, RAG incubation was performed for 10, 20, 30, or 60 min (Fig. 4). The 12×23 substrate formed nicks at a rate slightly faster than that of hairpins. The rate of accumulation of the nick product of the 12×12 and 23×23 substrates was 1.6- to 4-fold slower than that of the 12×23 substrate at 10 min, consistent with results of others (3). These differences in nicking were even smaller at subsequent time points. The double hairpin formation was markedly slower for the 12×12 and 23×23 substrates relative to the 12×23 substrate. Double hairpin product reached 50% of total by 10 min with the 12×23 substrate. The hairpin products of the 12×12 and 23×23 substrates were detectable only after 30 min. Quantification of the substrate and double hairpin product showed that the differences in rates between the 12×23 versus the 23×23 and 12×12 substrates were at least 10- and 20-fold, respectively.
FIG. 4.
Time course of the RAG nuclease activity on non-12×23 substrates. RAG1, RAG2, and HMG1 were incubated with the 12×23, 12×12, or 23×23 substrates for 10, 20, 30, and 60 min in Mg2+ conditions. The products were separated on a 15% denaturing polyacrylamide gel. The intensities of the double hairpin and nicked products and the remaining substrate were then quantified with the PhosphorImager system. The ratio of the double hairpin product to the substrate is indicated by the black bars, and the ratio of the nicked product to the substrate is indicated by the stippled bars.
A prenicked substrate does not relieve the 12/23 rule constraint at the double hairpin step.
While it is apparent that the 12/23 rule is applied at some point during the nick to hairpin transition, it is possible that it is the coordinated nicking of a substrate that is the critical step. Though the 12×12 and 23×23 structures create a significant level of nicked products, one could argue that these are the result of an unproductive pathway that branches off from the productive pathway at the nick step. That is, it is possible that the nicks observed in the 12×12 and 23×23 substrates cannot progress to the hairpin step.
The nature of the oligonucleotide system allows intermediate substrates to be examined. New substrates were created to examine whether preexisting nicks were sufficient to induce coordinated hairpin formation in the 12×12 and 23×23 substrates. Substrates were designed so that, adjacent to each heptamer, a nick was positioned as if nicked by the RAG proteins (Fig. 5A). The substrates were labeled at the 5′ end of one of the top strand oligonucleotides. When cleavage occurred in this configuration, we observed the blunt-ended product of the hairpin formation event rather than the hairpin itself. Three nicked substrates were created: 12×23, 12×12, and 23×23. As described above, HMG1 was added prior to incubation with the RAG proteins. As seen in Fig. 5B, only the double-nicked 12×23 substrate was subject to hairpin formation by the RAG proteins. The 12×12 and 23×23 double-nick variations were not cut. Therefore, we can rule out the possibility that coordinated double nicking is the point at which the 12×12 and 23×23 substrates are blocked.
FIG. 5.
RAG activity on prenicked substrates. (A) The prenicked 12×23, 12×12, and 23×23 substrate structures are shown in the same format as in Fig. 1A. (B) RAG1, RAG2, and HMG1 were incubated in Mg2+ conditions with 12×23, 12×12, and 23×23 substrates which had a nick at each signal site. The products were separated on a 15% denaturing polyacrylamide gel. Lanes 1 and 2, 12×23 nicked substrate without and with RAG proteins and HMG1, respectively; lanes 3 and 4, 12×12 nicked substrate without and with RAG proteins and HMG1, respectively; lane 5 and 6, 23×23 nicked substrate without and with RAG proteins and HMG1, respectively.
Having shown that the RAG-HMG1 protein complex can bind and nick at the 12×23 and alternative (12×12 and 23×23) substrates equally well, that double nicked substrates are not able to lead to hairpin formation in the 12×12 and 23×23 substrates, and that hairpins are formed at least 10- to 20-fold faster on the 12×23 substrate, we concluded that the 12/23 rule is enforced at the double hairpin formation stage. The implications of this are discussed below.
DISCUSSION
The 12/23 rule has been examined in the context of experiments in vivo (19, 21) and in cellular extracts (3). However, such studies have not addressed the biochemical steps at which the 12/23 rule applies. Hence, it has been unclear whether this rule applies at the level of the RAG synapsis of the 12- and 23-signals, at their nicking, or at their hairpin formation. It has also been unclear whether the hairpinning occurs sequentially. Here we provide a detailed biochemical examination of the RAG cleavage reaction on double-site (12×23) substrates in the context of purified components. This system uses an oligonucleotide substrate that contains both a 12-bp site and a 23-bp site. An oligonucleotide substrate allows the study of nicking and hairpin formation simultaneously. Using such a system, we examined how the RAG-HMG1 complex interacts with 12×12 and 23×23 substrates as well. We find that the protein complex binds equally well to 12×12 and 23×23 substrates as to 12×23 substrates, but not to substrates that have only one signal. Furthermore, we find that the alternate substrates have a rate of nicking comparable to that of 12×23 substrates. Finally, we find that substrates with preexisting nicks undergo double hairpin formation only if they have the 12×23 configuration. These data demonstrate that the 12/23 rule is enforced at the stage of double hairpin formation and not at earlier stages.
We first examined the interaction of RAG1, RAG2, and HMG1 on just the 12×23 substrate (Fig. 1). For efficient and strictly concerted double hairpin formation, all three proteins must be present in Mg2+ conditions (Fig. 1B). Furthermore, in Mn2+ conditions, concerted hairpin formation occurs only in the presence of HMG1 (Fig. 1C), though all the hairpin events are not strictly double hairpins. The comparison of these different conditions further defines the role of HMG1 as an essential component for the synapsis of the two recombination sites regardless of the divalent cation present. It is also clear that HMG1 is involved in the nicking step as well because the presence of HMG1 increases the level of nick formation (Fig. 1B).
Interestingly, we find no products containing a hairpin at one signal and a nick at the other. This strongly suggests that hairpin formation is concerted and tightly coupled with the nicking step. It also indicates that hairpin formation on one signal is very inefficient when the conditions for simultaneous cleavage of the synaptic complex are satisfied.
In Mn2+ conditions, the RAG proteins form a synaptic complex involving contact with both signal sites of the 12×23 substrate (Fig. 1C). However, the efficiency of hairpin formation is significantly different depending on whether the second signal is linked to the first. In Mn2+ conditions, the linkage of both sites causes one site to negatively influence the hairpin formation of the other (Fig. 1C, lanes 2 and 3). In Mg2+ conditions, one site positively influences the hairpin formation of the other (Fig. 1B). However, strictly coordinated hairpin formation cannot proceed without the presence of HMG1 and Mg2+. While either HMG1 or Mg2+ favor synaptic and coordinated cutting, the presence of both is optimal.
The cold competition experiments (Fig. 3) indicate that the RAG complex has an affinity for the 12×12 and 23×23 substrates similar to that for the 12×23 substrate. In these experiments, we chose to present both the competitor DNA and the target DNA to the RAG proteins at the same time rather than to challenge a preformed RAG-HMG1-DNA complex with competitor DNA. The latter design is inferior, as it would examine only the dissociation rate of the RAG proteins on a given substrate rather than both the association and dissociation rates, as in the design we employed. One cannot be certain from these results whether the RAG complex achieves functional synapsis with the 12×12 and 23×23 substrates rather than a nonfunctional binding. There may be several synaptic intermediates that occur before hairpin formation can take place. However, given the equivalent affinity in competition studies, it appears that the RAG complex can achieve a synapsis equivalent to that of the 12×23 substrate and the 12×12 and 23×23 substrates. The degree of inhibition seen with these synaptic substrates contrasts with the significantly weaker inhibition by a substrate with only one functional signal site (but at twice the molar concentration). This substrate had almost fourfold less competition for the 12×23 substrate than the other substrates (Fig. 3). Furthermore, the time course assay (Fig. 4) showed that given a long incubation time, the RAG complex can generate a double hairpin product from 12×12 and 23×23 substrates. This suggests that the RAG complex does achieve synapsis with the 12×12 and 23×23 substrates but that the transition from substrate to hairpin is energetically disfavored relative to the 12×23 substrate. Alternatively, the very low rate of single hairpin formation (Fig. 2B) could lead to sequential cutting (rather than coupled cleavage), resulting in the small amounts of double hairpin product.
The time course assays show that the RAG complex has a high rate of nicking in 12×12 and 23×23 substrates (Fig. 4). Similar results were shown in crude extracts (3). These nicks may go undetected in chromosomal and extrachromosomal studies that examine only the rearranged product. Such events could explain an unusual alternative reaction in V(D)J recombination called open and shut events (8, 11). In such cases, nucleotides are lost or added at the coding-signal border of one V or J element. Such events have been commonly observed in the genome at immunoglobulin and T-cell-receptor loci undergoing recombination (9). It is conceivable that this represents a pathologic activity that may have some significance for the recombining lymphocyte. The high level of nicks potentially generated in a cell where the RAG complex fails to make 12×23 synapsis (and instead makes 12×12 or 23×23 synaptic complexes) could lead to some of the observed chromosomal translocations in lymphoid malignancies (12).
Previous studies have suggested that there may be an additional factor beyond RAG1 and RAG2 that enforces the 12/23 rule (3, 16). These studies have compared cleavage using purified components (where only a two to threefold 12/23 preference was observed) versus that using whole cell extracts (where a 25-fold 12/23 preference was observed). Using similar buffer conditions in the crude and purified systems, these studies describe some level of cutting of 12×12 and 12×23 substrates only in the purified system. However, the previous purified systems utilized substrates with an excess of nonspecific DNA present between the signals. Such plasmid substrates have many cryptic 12- and 23-bp nonconsensus signal sites within them that could provide, for example, many 12-signals to each of the signals in a 23×23 substrate (10). In our system, there is less nonspecific DNA present and also considerably less 12×12 and 23×23 cleavage. Although the intersignal distance is much shorter than that in previous in vitro studies, substrates with very similar intersignal distances are processed by the RAG complex at equivalent efficiencies in vivo (19). The contribution of other DNA binding proteins in the crude extracts may simply be to compete the RAGs away from low-affinity, nonconsensus cryptic sites such that the RAGs ultimately have a higher probability of locking in on synapsis at the correct signals. Our results show that the RAG proteins plus HMG1 can provide what is necessary to achieve adherence to the 12/23 rule to an extent that is equivalent to that seen in crude extracts.
While our manuscript was in the final stages of review, another study in which the data were interpreted to indicate that the 12/23 rule is established prior to cleavage (7), specifically at the synapsis step, was published. This necessarily compels us to propose how both sets of data can be interpreted in a unified way. One possibility is that there is a small amount (threefold) of 12/23 rule enforcement at the synapsis step (7), but a larger one (10- to 20-fold) at the double hairpin step (our data). A second possibility is that the use of Ca2+ in the synapsis buffer by Hoim and Gellert (7) resulted in the small (threefold) differences in synapsis seen in their study. Such stable synapsis complexes proceed to cleavage under physiologic Mg2+ conditions. Ca2+ is sufficiently nonphysiologic that it does not permit cleavage. The same distortion that results in failure to cleave in Ca2+ could give synapsis biases that are not seen when one does cold competition studies, as we did, using Mg2+ (Fig. 3).
Despite achieving a very high degree of 12/23 rule adherence in a purified system, we did observe a very low level of 12×12 and 23×23 cleavage. This may correspond to the actual level of physiologic discrimination (6). In the cell, there is a very low level of cleavage at the 12×12 or 23×23 pairing, some of which leads to pathologic results in vivo (9, 20). Such abnormal pairings can be quantitated by using extrachromosomal 23×23 substrates; these were found to recombine at 2% of the rate of 12×23 substrates (6, 11). Examination of in vivo junctions also found violations of the 12/23 rule at a low but readily detectable level (14).
ACKNOWLEDGMENTS
We thank Marco Bianci for providing an HMG1 expression vector. We also thank members of our laboratory, including Ulf Grawunder and Robert Tracy, for reviewing the manuscript.
This research was supported by NIH grants and a Leukemia Society of America Scholar Award to M.R.L. M.R.L. is the Rita and Edward Polusky Basic Cancer Research Professor.
REFERENCES
- 1.Bianchi M E, Falciola L, Ferrari S, Lilley D M. The DNA binding site of HMG1 protein is composed of two similar segments (HMG boxes), both of which have counterparts in other eukaryotic regulatory proteins. EMBO J. 1992;11:1055–1063. doi: 10.1002/j.1460-2075.1992.tb05144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eastman Q M, Leu T M J, Schatz D G. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 3.Eastman Q M, Schatz D G. Nicking is asynchronous and stimulated by synapsis in 12/23 rule-regulated V(D)J cleavage. Nucleic Acids Res. 1997;25:4370–4378. doi: 10.1093/nar/25.21.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grawunder U, Lieber M R. A complex of RAG-1 and RAG-2 persists on the DNA after single-strand cleavage at V(D)J recombination signal sequences. Nucleic Acids Res. 1997;25:1375–1382. doi: 10.1093/nar/25.7.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grawunder U, West R B, Lieber M R. Antigen receptor gene rearrangement. Curr Opin Immunol. 1998;10:172–180. doi: 10.1016/s0952-7915(98)80246-x. [DOI] [PubMed] [Google Scholar]
- 6.Hesse J E, Lieber M R, Mizuuchi K, Gellert M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 1989;3:1053–1067. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 7.Hoim K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 8.Lewis S, Hesse J E, Mizuuchi K, Gellert M. Novel strand exchanges in V(D)J recombination. Cell. 1988;55:1099–1107. doi: 10.1016/0092-8674(88)90254-1. [DOI] [PubMed] [Google Scholar]
- 9.Lewis S M. The mechanism of V(D)J joining: lessons from molecular, immunological and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 10.Lewis S M, Agard E, Suh S, Czyzyk L. Cryptic signals and the fidelity of V(D)J joining. Mol Cell Biol. 1997;17:3125–3136. doi: 10.1128/mcb.17.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis S M, Hesse J E. Cutting and closing without recombination in V(D)J joining. EMBO J. 1991;10:3631–3639. doi: 10.1002/j.1460-2075.1991.tb04929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieber M R. The role of site-directed recombinases in physiologic and pathologic chromosomal rearrangements. In: Kirsch I, editor. The causes and consequences of chromosomal translocations. Boca Raton, Fla: CRC Press; 1992. pp. 239–275. [Google Scholar]
- 13.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 14.Meek K, Hasemann C A, Capra J D. Novel rearrangements at the immunoglobulin D locus. J Exp Med. 1989;170:39–57. doi: 10.1084/jem.170.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oettinger M A, Schatz D G, Gorka C, Baltimore D. Rag-1 and Rag-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 16.Sawchuk D, Weis-Garcia F, Malik S, Besmer E, Bustin M, Nussenzweig M, Cortes P. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J Exp Med. 1997;185:2025–2032. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schatz D G. V(D)J recombination moves in vitro. Semin Immunol. 1997;9:149–160. doi: 10.1006/smim.1997.0068. [DOI] [PubMed] [Google Scholar]
- 18.Schatz D G, Oettinger M A, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 19.Sheehan K M, Lieber M R. V(D)J recombination: signal and coding joint resolution are uncoupled and depend on parallel synapsis of the sites. Mol Cell Biol. 1993;13:1363–1370. doi: 10.1128/mcb.13.3.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steen S, Gomelsky L, Speidel S, Roth D B. Initiation of V(D)J recombination in vivo: role of recombination signal sequences in formation of single and paired double-strand breaks. EMBO J. 1997;16:2656–2664. doi: 10.1093/emboj/16.10.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steen S B, Gomelsky L, Roth D B. The 12/23 rule is enforced at the cleavage step of V(D)J recombination in vivo. Genes Cells. 1996;1:543–553. doi: 10.1046/j.1365-2443.1996.d01-259.x. [DOI] [PubMed] [Google Scholar]
- 22.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 23.van Gent D, Hoim K, Paul T, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 25.van Gent D C, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 26.van Gent D C, Ramsden D A, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]