Abstract
During assembly, bacterial virus phi29 utilizes a motor to insert genomic DNA into a preformed protein shell called the procapsid. The motor contains one twelve-subunit connector with a 3.6 nm central channel for DNA transportation, six viral-encoded RNA (packaging RNA or pRNA) and a protein, gp16, with unknown stoichiometry. Recent DNA-packaging models proposed that the 5-fold procapsid vertexes and 12-fold connector (or the hexameric pRNA ring) represented a symmetry mismatch enabling production of a force to drive a rotation motor to translocate and compress DNA. There was a discrepancy regarding the location of the foothold for the pRNA. One model [C. Chen and P. Guo (1997) J. Virol., 71, 3864–3871] suggested that the foothold for pRNA was the connector and that the pRNA–connector complex was part of the rotor. However, one other model suggested that the foothold for pRNA was the 5-fold vertex of the capsid protein and that pRNA was the stator. To elucidate the mechanism of phi29 DNA packaging, it is critical to confirm whether pRNA binds to the 5-fold vertex of the capsid protein or to the 12-fold symmetrical connector. Here, we used both purified connector and purified procapsid for binding studies with in vitro transcribed pRNA. Specific binding of pRNA to the connector in the procapsid was found by photoaffinity crosslinking. Removal of the N-terminal 14 amino acids of the gp10 protein by proteolytic cleavage resulted in undetectable binding of pRNA to either the connector or the procapsid, as investigated by agarose gel electrophoresis, SDS–PAGE, sucrose gradient sedimentation and N-terminal peptide sequencing. It is therefore concluded that pRNA bound to the 12-fold symmetrical connector to form a pRNA–connector complex and that the foothold for pRNA is the connector but not the capsid protein.
INTRODUCTION
In living systems, components are routinely and actively transported by molecular motors, such as F1-ATPase (1–3), kinesin (4–6), myosin (7–9), helicase (10,11), viral DNA or RNA-packaging motors (12–14). The viral DNA-packaging motor of double-stranded DNA (dsDNA) bacteriophages (e.g. phi29, λ, P22, T4 and T7) can translocate DNA with remarkable velocity into a limited space in a preformed protein shell called a procapsid at the expense of ATP hydrolysis during assembly (15–21). Studies on bacteriophage phi29 and T7 have shown that a motor is also involved in the ejection of DNA from the procapsid during viral infection (22,23).
The procapsid of Bacillus subtilis bacteriophage phi29 consists of the major capsid protein gp8 (235 copies), head fiber protein gp8.5 (55 copies), scaffolding protein gp7 (∼180 copies) and connector protein gp10 (12 copies) (24–27). Through studies involving the bacterial virus phi29 DNA-packaging motor, a molecule known as pRNA that plays a novel and essential role in packaging DNA into procapsids was discovered (28). Six copies of pRNA have been found to form a hexameric ring (29–31) to drive the DNA-packaging motor. The construction pathway for pRNA goes from monomer to dimer and then to hexamer (32).
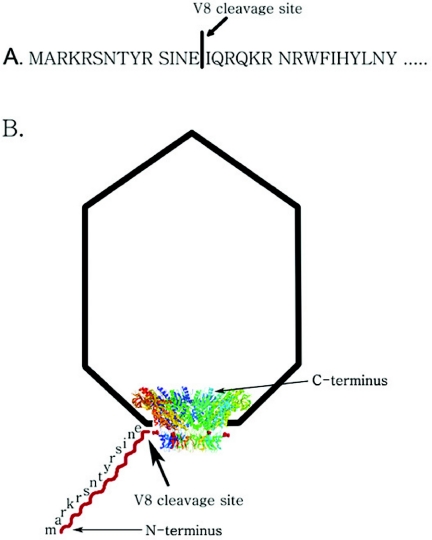
One of the essential components of the phi29 DNA-packaging motor is the connector complex, a dodecameric cylindrical structure with a 36 Å central channel, through which viral DNA is packaged into the capsid and exits during the infection process. The specific structure of the phi29 phage portal protein has been determined at atomic resolution (33–35). The phi29 connector ring consists of 12 alpha-helical subunits, with an inner channel being formed by three long helices of each subunit. The wider end of the connector is located in the procapsid, with the narrow end partially protruding from the procapsid. Owing to the structural flexibility of both the N- and C-terminal regions, the crystal structure of 13 amino acids at the N-terminus and 24 amino acids at the C-terminus were not solved and were missing from PDB file 1H5W (34), while 10 amino acids at the N-terminus and 24 amino acids at the C-terminus were missing from PDB file 1FOU (35). From structural analysis, it is evident that the N-terminus protrudes from the narrow end of the connector and the C-terminus protrudes from the wider end of the connector (Figure 1) (34–36). The C-terminus is buried within the procapsid and the N-terminus is exposed to the solvent.
Figure 1.
Schematic representation of endoproteinase Glu-C (protease V8) cleavage site. (A) V8 cleavage site in the N-terminal sequence of connector protein gp10, which has been verified by N-terminal amino acid sequencing. (B) V8 cleavage site as shown in a procapsid model.
Some DNA-packaging models proposed that the 5-fold procapsid vertexes and 12-fold connector (or the hexameric pRNA ring) produced a force to drive the phi29 rotation motor to stuff DNA (34,35,37,38). Rotation of the connector, which is a 12-subunit complex, within the capsid, a 5-fold symmetrical environment, could constitute a mechanical motor in which the relative motion of the two rings could drive DNA into the procapsid. Although the notion of the generation of physical force from a rotary motor with a symmetry mismatch between two rings is particularly interesting, no one has directly proven this model with experimental data. The published DNA-packaging models all agreed that pRNA bound to one component to execute a consecutive rotation (34,35,38). There were discrepancies regarding the location of the foothold for the pRNA, and the determination of which component is the rotor and which is the stator. To make two rings rotate relatively, at least one additional component is needed to provide a propelling force. The model by Chen and Guo (38) suggested that the foothold for pRNA was the connector and that the pRNA–connector complex was part of the rotor. pRNA could thus be a candidate for this third component. It was demonstrated that pRNA forms a hexamer (29–31). Six copies of pRNA are bound to the connector and work sequentially (38), and pRNA contains two functional domains (Figure 2) (39,40). One domain, composed of the central region of the pRNA, binds to the procapsid. The other domain, which functions as a DNA translocation domain, is located at the 5′/3′ paired ends. In this configuration, it is free for interaction with an additional but as yet unidentified component, such as gp16, the 5-fold symmetrical capsid membrane or genomic DNA–gp3. The interaction of pRNA or of the pRNA/gp16 complex, which is tethered to the connector, with the capsid membrane or DNA lattice may provide the propelling force for connector rotation. However, another model (35) proposed that the foothold for pRNA was the 5-fold vertex of the capsid protein and that pRNA was the stator. To elucidate the mechanism of phi29 DNA packaging, it is critical to confirm whether pRNA binds to the 5-fold vertex of the capsid protein or to the 12-fold symmetrical connector. In this report, we demonstrate that the N-terminal region of gp10 is essential for pRNA binding, and that the foothold of pRNA is not the 5-fold vertex of the capsid protein but instead the connector. Removal of the N-terminus of gp10 from both purified connector and the procapsid resulted in undetectable binding of pRNA to the procapsid.
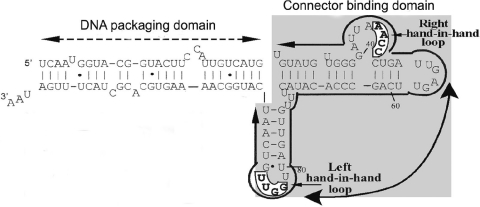
Figure 2.
Primary sequences and secondary structure of wild-type phenotype pRNA (I-i′). Right-hand and left-hand loops involved in hand-in-hand interaction are boxed and in bold. Two functional domains are outlined.
MATERIALS AND METHODS
Connector production
The construction of the plasmid harboring the gene coding for gp10, the over-expression of gp10 and the purification of phi29 connector have been reported recently (36,41). The connector with N-terminal his-tag was over-produced in Escherichia coli and purified by a one-step elution with imidazole from Ni2+-charged His-Bind columns (His•Bind Kits, Novagen).
Synthesis of pRNA
Methods for the synthesis and isolation of wild-type pRNA I-i′ have been described previously (32,42,43).
UV crosslinking of [32P]pRNA to procapsid and connector
[32P]pRNA I-i′ was co-incubated with the connector in TMS buffer (50 mM Tris–HCl pH 7.8, 10 mM MgCl2 and 100 mM NaCl) for 5–10 min at ambient temperature. Procapsid–[32P]pRNA I-i′ complex was produced by dialyzing the pRNA/procapsid mixture against TBE on a 0.025 μm VS filter (Millipore Corp.) at ambient temperature for 15 min, and then dialyzed against TMS for 30 min. Both complexes were then irradiated with short wavelength UV light (254 nm maximum) using a MINERALIGHT® lamp (UVP Model UVGL-58, Inc.) for 20 min at a distance of 5 cm. The crosslinked samples were treated with RNase A and resolved on 10% SDS–PAGE. The gel was then autoradiographed (71).
Binding of pRNA to purified connector
An aliquot of 120 pmol gp10 was mixed with 12 pmol pRNA I-i′ in TMS buffer (50 mM Tris–HCl, pH 7.8, 10 mM MgCl2 and 100 mM NaCl) for 5–10 min at ambient temperature. The complex was then loaded on 0.8% agarose gel in 1× TAE buffer containing 10 mM MgCl2, and the gel was run with constant current at 4°C. The gel was first stained with ethidium bromide to show the presence of RNA bands. The agarose gel was then stained by Coomassie brilliant blue (0.004% Coomassie brilliant blue in 0.5% methanol and 8.5% acetic acid) with gentle agitation at RT overnight to show the protein bands.
Proteolytic treatment of connector or procapsid
The connector or connector–pRNA complex was treated by proteinase V8 (endoproteinase Glu-C from Staphylococcus aureus V8, Roche) with an enzyme:substrate ratio of ∼1:20 (w/w) for 16 h at 25°C (46). Procapsid was treated in the same manner as the connector. The proteolytic products were detected by 10% SDS–PAGE.
N-terminal amino acid sequence analysis of proteolytic connector
The proteinase V8 pretreated connector was separated by 10% SDS–PAGE and then transferred to PVDF membrane and stained by 0.2% Ponceau S. The desired band was cut out and subjected to N-terminal peptide sequencing.
Separation of connector–pRNA complex or procapsid–pRNA complex by sucrose gradient sedimentation
Sucrose gradient sedimentation was performed to separate the connector–[3H]pRNA complex, connector–[3H]pRNA complex treated by V8 before or after adding [3H]pRNA I-i′, procapsid–[3H]pRNA I-i′ complex and procapsid–[3H]pRNA I-i′ complex treated by V8 before or after adding [3H]pRNA I-i′. Procapsid–[3H]pRNA I-i′ complex was prepared as described above. Subsequently, it was treated by V8 in the same manner as the connector–pRNA complex. The above complexes were loaded onto the top of a linear 5–20% sucrose gradient in TMS and spun in a Beckman L-80 ultracentrifuge at 35 000 r.p.m. for 30 min at 20°C in a SW55 rotor. Upon centrifugation, fractions were collected from the bottom of the tube and prepared for scintillation counting.
Electron microscopy of connector–pRNA complex or procapsid–pRNA complex
Carbon-coated transmission electron microscope (TEM) grids were made hydrophilic by glow discharge and floated for 10 min on drops of connector–pRNA complexes, which were purified from sucrose gradient centrifugation. The grids were passed >4 drops of 10 mM MgCl2 for 5 min and stained with 2% uranyl acetate containing 10 mM MgCl2 for 5 min. Electron micrographs were taken on a Philips CM10 TEM. Procapsid–pRNA complexes were made in the same way as the connector.
In vitro phi29 virion assembly assay
The purification of procapsids, gp16, DNA–gp3, the preparation of neck and tail proteins and the assembly of infectious phi29 virion in vitro have been described previously (44). Briefly, 1.5 μl of purified procapsids and V8-cleaved procapsids with or without 100 ng pRNA were dialyzed on a 0.025 μm VS filter against TBE for 15 min at ambient temperature and were then dialyzed against TMS for another 30 min. The pRNA-enriched procapsids were mixed with gp16, DNA–gp3, and reaction buffer to complete the DNA-packaging reaction. After 30 min, neck, tail and morphogenic proteins were added to the DNA-packaging reactions to complete assembly of infectious virions, which were assayed by standard plaque formation.
RESULTS
Photoaffinity crosslinking of pRNA to connector
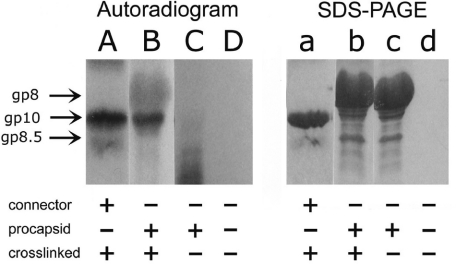
To elucidate the mechanism of pRNA action, it is necessary to know whether pRNA binds to the connector gp10 or to the capsid protein gp8. A UV crosslinking assay was performed to address this question. After binding [32P]labeled pRNA to either the procapsid or the purified connector (Figure 3), the RNA/procapsid or pRNA/connector complex was irradiated with short wavelength UV light. When [32P]pRNA was crosslinked with procapsid, the band in SDS gel corresponding to gp10 was specifically labeled, while gp8 was not, indicating that pRNA selectively bound to gp10 (Figure 3, lanes A and B; lanes a and b).
Figure 3.
Specific binding of [32P]pRNA I-i′ to connector demonstrated by UV crosslinking assay. Lanes A–D, 10% SDS–PAGE autoradiographed pictures; lanes a–d, the same gel stained by Coomassie blue. Lane A, pRNA crosslinked to connector and then treated by RNase A; lane B, procapsid crosslinked to pRNA and then treated by RNase A; lane C, procapsid–pRNA complex without crosslinking, and then treated by RNase A; lane D, pRNA treated by RNase A. Lanes a, b, c and d correspond to A, B, C and D, respectively.
Binding of pRNA to the N-terminus of the connector investigated by gel shift assay
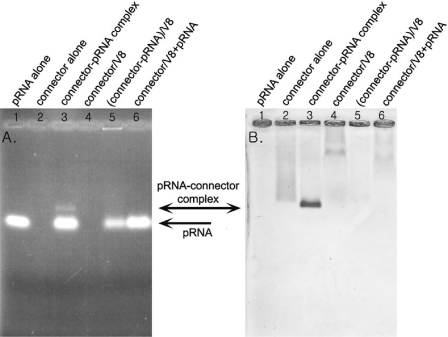
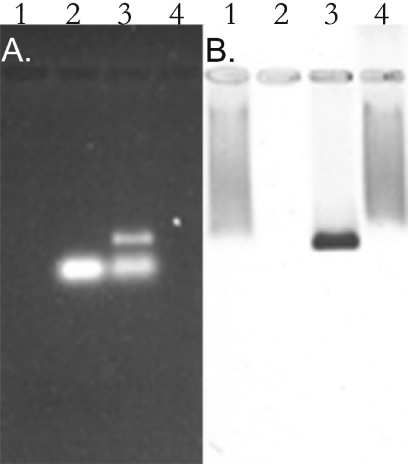
The crystal structure of the connector has been solved (34,35,45). Although the structure and geometrical localization of N-terminal residues of gp10 are not currently known, since their region was missing in the reports published by two separate laboratories (34,35), from the published structure data it can be predicted that the N-terminal residues 1–13 are adjacent to the narrow end of the connector (Figure 1) (36). To identify which part of the connector is responsible for pRNA binding, the connector was purified to homogeneity. The purified connector was incubated with pRNA and then subjected to 0.8% agarose gel electrophoresis. When connector was mixed with pRNA, an extra band with slower migration rate appeared. This nascent band was stainable by both ethidium bromide (Figure 4A, lane 3) and Coomassie brilliant blue (Figure 4B, lane 3). The mixture was also RNase A sensitive (Figure 5A and B, lane 4), suggesting that the band was the connector/pRNA complex.
Figure 4.
Connector–pRNA complex in 0.8% agarose gel. Gel was first stained by ethidium bromide (A) and then by Coomassie brilliant blue (B). Lane 1, 12 pmol pRNA alone; lane 2, 120 pmol gp10 alone (10 pmol connector); lane 3, connector–pRNA complex with a molar ratio of 1:1.2 in the presence of Mg2+; lane 4, 120 pmol gp10 cleaved by endoproteinase Glu-C (protease V8); lane 5, connector–pRNA complex with a molar ratio of 1:1.2 were cleaved by proteaseV8; lane 6, 120 pmol gp10 cleaved by protease V8 then mixed with 12 pmol pRNA.
Figure 5.
Effect of RNase A treatment on connector–pRNA complex. An aliquot of 0.8% agarose gel stained by ethidium bromide showing pRNA (A) and the same gel stained by Coomassie brilliant blue showing the connector protein (B) to evaluate RNase A digestion of the connector–pRNA complex. Lane 1, the purified 120 pmol gp10 alone (10 pmol connector); lane 2, 12 pmol pRNA alone; lane 3, connector–pRNA complex with a molar ratio of 1:1.2 in the presence of Mg2+; lane 4, the same connector–pRNA complex sample as in lane 3 treated with 1 μg/μl RNase A.
When the connector was pre-digested with endoproteinase Glu-C (protease V8) and then mixed with pRNA, the nascent ethidium bromide-sensitive band did not appear (Figure 4A, lane 6) and only the smear Coomassie brilliant blue band presented (Figure 4B, lane 6), similar to the proteolysis profile of free connector treated by V8.
The slow migration rate of the cleaved connector in agarose gel (Figure 4B, lane 4) can be explained by the change in the charge of the connector after V8 cleavage, since the N- terminus of gp10 contained basic charged amino acids protruding from the narrow edge of the connector cylinder (34–36).
N-terminal protein sequence analysis
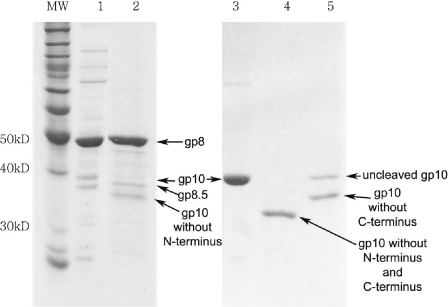
To confirm the specific cleavage site and the removal of the N-terminal residues by V8 proteolysis, a protein band of ∼32 kDa (Figure 6, lane 4) representing the cleaved gp10 product was cut from the SDS–PAGE and subjected to protein sequencing. N-terminal peptide sequencing of the purified V8-treated gp10 revealed that the N-terminal sequence is IQRQKRN, which indicated that the V8 proteolytic site of gp10 was located after the Glu residue. This confirmed that V8 digestion removed the first 14 amino acids from the N-terminal of the gp10, as illustrated in Figure 1. This result also agreed with the previous report with sequence mapping data showing that V8 removes 14 amino acids from the N-terminus and 18 amino acids from the C-terminus of the connector subunit gpl0 (45,46). For the connector–pRNA complex treated by V8, SDS–PAGE yielded two bands (Figure 6, lane5). When the connector was mixed with pRNA, it formed a rosette (Figure 7A). It is evident that the N-terminus of the connector was located within the rosette (36), which protected it from V8 cleavage. Thus, only C-terminus can be cleaved by V8 (Figure 6, lane 5). As noted earlier, the C-terminus of gp10 is buried within the procapsid (Figure 1), and the N-terminus is exposed to the solvent, and thus the N-terminus of gp10 in the procapsid was cleaved by V8 (Figure 6, lane 2). However, since the purified connector is free from the procapsid, both the C- and N-terminal ends are accessible to V8 cleavage (Figure 6, lane 4).
Figure 6.
A total of 10% SDS–PAGE to show the connector protein gp10 and procapsid treated before and after V8 cleavage. Lane 1, procapsid alone; lane 2, procapsid cleaved by proteaseV8; lane 3, purified connector protein gp10 alone; lane 4, connector cleaved by V8; lane 5, connector–pRNA complex cleaved by V8. As noted in the text, in the procapsid, the C-terminus of gp10 is located at the wider end of the connector that is buried within the procapsid, while the N-terminus is located at the narrow end of the connector that is exposed to the solvent. Treatment of connector or procapsid with V8 resulted in different sizes of gp10, since V8 can cleave both the N- and C-terminus of gp10 of the free connector, but only the N-terminus of the gp10 that is buried within the procapsid. Gp8 is the capsid protein, while gp8.5 is the fiber protein of the procapsid.
Figure 7.
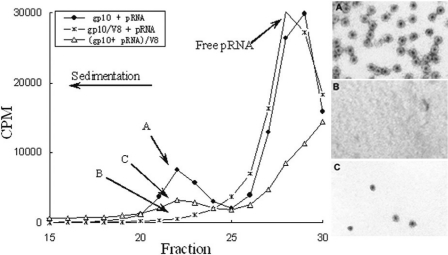
A total of 5–20% sucrose gradient sedimentation of connector–pRNA complex. The closed circle is the connector–pRNA complex (A), while the star is V8 pre-treated connector before adding [3H]labeled pRNA (B). The open triangle is V8-treated connector–pRNA complex (C). Fraction 22 from three samples labeled as A, B and C was collected and subjected to negative stain electron micrograph, as shown on the right in panels A, B and C, respectively. Magnification: ×105 000.
Binding of pRNA to the N-terminus of the connector as demonstrated by sucrose gradient sedimentation
The role of the N-terminus of connector protein gp10 in pRNA binding was further supported by sucrose gradient sedimentation. The purified connector was incubated with [3H]pRNA and then subjected to 5–20% sucrose gradient sedimentation. When connector was mixed with [3H]pRNA, a peak representing the connector–pRNA complex appeared and was centered on fraction 22 (Figure 7) (36). Negative stain electron microscopy revealed that fraction 22 indeed contained pure rosettes, representing the connector–pRNA complexes (Figure 7A). However, this radioactive peak disappeared when the connector was pre-digested with protease V8, indicating that the connector with the N-terminus cleaved could not bind pRNA (Figure 7B). When the connector was pre-incubated with RNA and then digested with V8, a small peak appeared at fraction 22, representing the connector–pRNA complex (Figure 7). Negative stain electron microscopy revealed that this fraction contained only a small amount of connector–pRNA complex (Figure 7C) as compared with fraction 22 in connector–pRNA complex without V8 treatment (Figure 7A). The finding of a small number of connector–pRNA complexes is to be expected, since binding of the connector to pRNA could protect the connector from V8 digestion to a certain degree.
Binding of pRNA to the N-terminus of the connector protein gp10 in procapsid
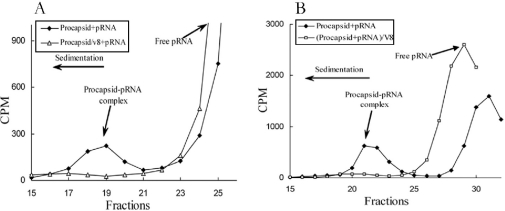
To ascertain whether pRNA can bind to the N-terminus of gp10 assembled in the procapsid, the purified procapsid was incubated with [3H]pRNA and subjected to 5–20% sucrose gradient sedimentation. When the procapsid was mixed with [3H]pRNA, a peak representing the procapsid–pRNA complex appeared and centered on fraction 19–21 (Figure 8A and B). However, when the procapsid was pre-digested with the protease V8, this radioactive peak was again removed (Figure 8A). SDS–PAGE of the cleavage profile of procapsid showed that the band representing gp10 disappeared after V8 digestion, but there was no change in the band representing major capsid protein gp8 and the fiber protein of the procapsid gp8.5 (Figure 6, lane 2). Thus, protease V8 only cleaved the connector protein gp10, but not the capsid protein gp8. This suggests that the procapsid lacking the N-terminal region of connector protein gp10 could not bind pRNA. This result further supports the conclusion that the N-terminus of the connector is essential for pRNA binding. In addition, the procapsid–pRNA complex treated by V8 did not exhibit a peak after sucrose gradient sedimentation (Figure 8B). Furthermore, the procapsid or procapsid–pRNA complexes treated by V8 were inactive in phi29 assembly (Table 1).
Figure 8.
A total of 5–20% sucrose gradient sedimentation of procapsid–pRNA complex treated before and after V8 treatment. The closed rhombus is the procapsid–pRNA complex, while the open triangle is V8 pre-treated procapsid before adding pRNA (A). The open square is V8-treated procapsid–pRNA complex (B).
Table 1.
Effects of V8-treated procapsid or procapsid–pRNA complex on phage assembly
| Packaging system | PFU/ml |
|---|---|
| Procapsid without pRNA | 0 |
| Procapsid–pRNA complex | 5.8 × 107 |
| V8 pre-treated procapsid with pRNA | 0 |
| Procapsid–pRNA complex treated by V8 | 0 |
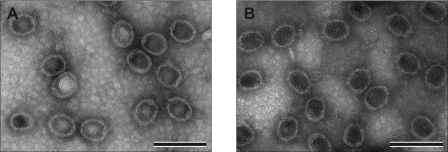
Procapsid with or without treatment by V8 was examined by negative stain electron microscopy. There was no significant change in the structure of procapsid after V8 treatment (Figure 9A and B). In combination with the data from SDS–PAGE showing that the molecular weight of gp8 remains unchanged after V8 digestion, the results indicated that V8 cleavage produced a procapsid with a truncated connector protein gp10 but with unchanged capsid.
Figure 9.
Electron micrographs of negative-stained procapsid–pRNA complex. (A) Procapsid–pRNA complex. (B) V8 pre-treated procapsid plus pRNA. Bar = 200 nm.
DISCUSSION
Phi29 packages its genomic DNA into a procapsid with the aid of the gp16 protein and pRNA molecules. Several DNA-packaging models proposed that 5-fold/6-fold asymmetry produced a force to drive the rotation motor to stuff DNA (34,35,37,38). Although the recent models all agree that pRNA binds to one component to enable connector rotation there were significant discrepancies regarding the location of the foothold for pRNA, and concerning which component is the rotor and which is the stator. The model by Chen and Guo (38) suggested that the foothold for pRNA was the connector and that the pRNA–connector complex was part of the rotor. However, the model by Simpson et al. (35) suggested that the foothold for pRNA was the 5-fold vertex of the capsid protein and that pRNA was the stator. There was also considerable debate concerning whether pRNA was a hexamer (29,31,38,48) or pentamer (35).
To further investigate the discrepancy, it was critical to confirm whether pRNA binds to the 5-fold vertex of the capsid protein gp8 or to the 12-fold symmetrical connector. UV crosslinking of RNA/procapsid or RNA/connector complex proved that pRNA was specifically bound to the connector (Figure 3), which has 12-fold (12 subunit) symmetry, but not the capsid protein gp8. The nonappearance of pRNA/gp8 (capsid protein) binding in the UV crosslinking experiment does not completely exclude the possibility of transient interaction of pRNA with capsid protein. From the data, it can be concluded that the affinity of pRNA to connector is much stronger than to capsid, and that the connector is the foothold for pRNA. Competition assays revealed that only the pRNA binding to the procapsid is specific (32). The formation of connector–pRNA complex or procapsid–pRNA complex was further demonstrated by agarose gel shift assay (Figures 4 and 5) and/or sucrose gradient sedimentation (Figures 7 and 8). In agarose gel shift assay, the nascent RNase A-sensitive bands indicated that pRNA binds to the connector (Figure 5A and B, lanes 3 and 4). It is difficult to calculate the binding ratio between pRNA and connector, because there were protein smears or traps inside the well of the gel. If the connector or procapsid was pre-digested by V8, N-terminal peptide sequencing data proved that V8 removed 14 amino acids from the N-terminus of the connector protein gp10 (Figure 1). EM imaging and X-ray crystallography have shown that the N-terminus is located at the narrow end of the connector, which exposes outside the procapsid, while the C-terminal of gp10 protein is at the wider end of the connector, which is buried within the procapsid and covered by the capsid protein (34,35). Thus, although V8 can also remove 18 amino acids from the C-terminus of the purified connector protein gp10 (45,46), it is impossible that pRNA bind to the C-terminus of the connector within the procapsid. It was also found that the inhibition of procapsid–pRNA complex formation was not due to the cleavage of procapsid protein gp8 by V8, since SDS–PAGE as well as EM imaging indicated that the capsid protein gp8 was still present while gp10 was cleaved (Figure 6 and Figure 9A and B). Since this gp10-truncated procapsid with intact capsid protein could not bind pRNA, the capsid protein gp8 cannot be the foothold for pRNA. Therefore, the conclusion is that pRNA bound to the 12-fold symmetrical connector to form a pRNA–connector complex and that the foothold for pRNA is the connector. The finding that V8 treatment produced a procapsid that could not bind pRNA argues against the conclusion from a previous publication (35) that five copies of pRNA binds to the capsid protein, which holds a 5-fold symmetrical vertex. Thus, the gp10 N-terminal cleavage accounts for the inhibition of phi29 in vitro virion assembly by V8 (Table 1).
All DNA-packaging motors of the dsDNA virus involve one pair of nonstructural components. In 1987, we classified these components into two categories according to their role in DNA packaging (28). The larger component is involved in procapsid binding, whereas the smaller component is involved in interaction with DNA. Subsequent extensive studies have supported this categorization. Procapsid binding components in the well-studied phages, other than phi29, include gpA in phage lambda (49), gp12 in phi21 (50), gp17 in T4 (51), gp19 in T3 and T7 (52,53). The DNA interacting components include gpNu1 in lambda (54,55), gp1 in phi21 (56), gp16 in T4 (57) and gp18 in T3/T7 (58,59). Emerging information reveals that the DNA-packaging mechanism of cytomegalovirus and adenovirus is very similar to that of phages. Herpes simplex virus contains a pair of DNA packaging proteins ULI5 and UL28 (64). Cytomegalovirus contains a pair of DNA-packaging proteins pUL56 and pUL89 (60), while the pair of DNA-packaging proteins for adenovirus is IVa2 and the L1 52/55 kDa proteins (61–63,65,66). In bacteriophage T3, the interaction of the larger DNA-packaging component gp19 with the connector has also demonstrated. Six molecules of gp19 bound to the prohead at a saturating amount of gp19, but the gp19 did not bind to the prohead lacking the connector (the gp8-defecient prohead) (67). For lambda, it has been found that the target component of gpA binding is the connector protein, since studies with the gpA and connector double-mutant revealed that a connector mutation can suppress a gpA mutation that hinders the gpA binding to procapsid (68,69). In phi29, it has been reported that there were three non-capsid components that were involved in DNA packaging: the terminal protein gp3, the DNA-packaging protein gp16 and pRNA. Gp3 can be excluded from the group of nonstructural DNA-packaging components, since gp3 is a structural component in the mature virion. Consequently, one gp16 and pRNA were considered as the candidate members of the pair of the nonstructural DNA-packaging protein. Accumulated data have positively confirmed that pRNA is the procapsid-binding component (28,32,70,71). In this paper, we have demonstrated that the target component of the procapsid for pRNA binding is the connector, not the procapsid protein gp8. This is in an agreement with the studies of phages T3 and λ, and reveals that pRNA is the counterpart of gpA of λ and gp19 of T3, since pRNA, gpA and gp19 all bind to the connector and contain the ATP-binding motif (72–76).
The conclusion that pRNA binds to the N-terminus of the connector protein gp10 agrees with other findings published previously. Antibody probing and 3D reconstructions of phi29 procapsid indicated that pRNA interacts with the narrow end of the connector (77). Labeling of the narrow end of the connector with monoclonal antibodies hinders the binding of pRNA to the connector (48,77). Comparing the procapsid plus pRNA with the procapsid without pRNA led to the conclusion that pRNA was situated as a ring embracing the narrow end of the connector (48). The docking of connector structure solved by X-ray in the portal region of reconstructed procapsids demonstrated that the pRNA forms a protruding ring that embraces the distal (narrower) domain of the interlaced beta-strands of the apical region of the gp10 dodecamer (34,47,48). The results agree with the 3D computer model of phi29 motor complex, which shows that six pRNAs form a hexameric ring surrounding the narrow end of the connector (78).
Acknowledgments
The authors thank Mary Bower and Doris Terry for their kind assistance on peptide sequencing; Nicola Stonehouse from University of Leeds for providing a plasmid expressing a his-tagged connector; and Jeremy Hall for his assistance in the preparation of this manuscript. Data in Figure 3 were prepared by Kyle Garver. The research was supported by NIH grant R01-EB003730. W.-D.M. is an Erwin-Schroedinger Fellow supported by the Austrian Science Fund FWF J2356. We appreciate Dr Jose Carasscosa for his communications and comments on the manuscript. Funding to pay the Open Access publication charges for this article was provided by R01-EB003730.
Conflict of interest statement. None declared.
REFERENCES
- 1.Walker J.E., Saraste M., Gay N.J. E.coli F1-ATPase interacts with a membrane protein component of a proton channel. Nature. 1982;298:867–869. doi: 10.1038/298867a0. [DOI] [PubMed] [Google Scholar]
- 2.Ernster L., Carlsson C., Boyer P.D. Reconstituted mitochondrial oligomycin-sensitive ATPase (F0F1) with intermediate Pi in equilibrium HOH exchange but no Pi in equilibrium ATP exchange activity. FEBS Lett. 1977;84:283–286. doi: 10.1016/0014-5793(77)80707-2. [DOI] [PubMed] [Google Scholar]
- 3.Kinosita K., Jr, Yasuda R., Noji H., Ishiwata S., Yoshida M. F1-ATPase: a rotary motor made of a single molecule. Cell. 1998;93:21–24. doi: 10.1016/s0092-8674(00)81142-3. [DOI] [PubMed] [Google Scholar]
- 4.Svoboda K., Block S.M. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 5.Vale R.D., Funatsu T., Pierce D.W., Romberg L., Harada Y., Yanagida T. Direct observation of single kinesin molecules moving along microtubules. Nature. 1996;380:451–453. doi: 10.1038/380451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sosa H., Peterman E.J., Moerner W.E., Goldstein L.S. ADP-induced rocking of the kinesin motor domain revealed by single-molecule fluorescence polarization microscopy. Nature Struct. Biol. 2001;8:540–544. doi: 10.1038/88611. [DOI] [PubMed] [Google Scholar]
- 7.Rayment I., Rypniewski W.R., Schmidt-Base K., Smith R., Tomchick D.R., Benning M.M., Winkelmann D.A., Wesenberg G., Holden H.M. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 8.Houdusse A., Sweeney H.L. Myosin motors: missing structures and hidden springs. Curr. Opin. Struct. Biol. 2001;11:182–194. doi: 10.1016/s0959-440x(00)00188-3. [DOI] [PubMed] [Google Scholar]
- 9.Inoue A., Saito J., Ikebe R., Ikebe M. Myosin IXb is a single-headed minus-end-directed processive motor. Nature Cell Biol. 2002;4:302–306. doi: 10.1038/ncb774. [DOI] [PubMed] [Google Scholar]
- 10.Nossal N.G., Dudas K.C., Kreuzer K.N. Bacteriophage T4 proteins replicate plasmids with a preformed R loop at the T4 ori(uvsY) replication origin in vitro. Mol. Cell. 2001;7:31–41. doi: 10.1016/s1097-2765(01)00152-6. [DOI] [PubMed] [Google Scholar]
- 11.Ha T., Rasnik I., Cheng W., Babcock H.P., Gauss G.H., Lohman T.M., Chu S. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 12.Doering C., Ermentrout B., Oster G. Rotary DNA motors. Biophys. J. 1995;69:2256–2267. doi: 10.1016/S0006-3495(95)80096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grigoriev D.N., Moll W., Hall J., Guo P. Bionanomotor. In: Nalwa H.S., editor. Encyclopedia of Nanoscience and Nanotechnology. Vol. 1. American Scientific Publishers; 2004. pp. 361–374. [Google Scholar]
- 14.Lisal J., Kainov D.E., Bamford D.H., Thomas G.J., Tuma R. Enzymatic mechanism of RNA translocation in double-stranded RNA Bacteriophages. J. Biol. Chem. 2004;279:1343–1350. doi: 10.1074/jbc.M309587200. [DOI] [PubMed] [Google Scholar]
- 15.Guo P., Peterson C., Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage φ29. J. Mol. Biol. 1987;197:229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- 16.Cue D., Feiss M. Bacteriophage lambda DNA packaging: DNA site requirements for termination and processivity. J. Mol. Biol. 2001;311:233–240. doi: 10.1006/jmbi.2001.4840. [DOI] [PubMed] [Google Scholar]
- 17.Smith D.E., Tans S.J., Smith S.B., Grimes S., Anderson D.L., Bustamnte C. The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 18.Guo P. Introduction: principles, perspectives, and potential applications in viral assembly. Semin. Virol. 1994;5:1–3. [Google Scholar]
- 19.Baumann R.G., Black L.W. Isolation and characterization of T4 bacteriophage gp17 terminase, a large subunit multimer with enhanced ATPase activity. J. Biol. Chem. 2003;278:4618–4627. doi: 10.1074/jbc.M208574200. [DOI] [PubMed] [Google Scholar]
- 20.Catalano C.E. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell. Mol. Life Sci. 2000;57:128–148. doi: 10.1007/s000180050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore S.D., Prevelige P.E., Jr DNA packaging: a new class of molecular motors. Curr. Biol. 2002;12:R96–R98. doi: 10.1016/s0960-9822(02)00670-x. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez-Huici V., Salas M., Hermoso J.M. The push–pull mechanism of bacteriophage Φ29 DNA injection. Mol. Microbiol. 2004;52:529–540. doi: 10.1111/j.1365-2958.2004.03993.x. [DOI] [PubMed] [Google Scholar]
- 23.Molineux I.J. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 2001;40:1–8. doi: 10.1046/j.1365-2958.2001.02357.x. [DOI] [PubMed] [Google Scholar]
- 24.Anderson D.L., Reilly B. Morphogenesis of bacteriophage φ29. In: Sonenshein A.L., Hoch J.A., Losick R., editors. Bacillus subtilis and Other Gram-positive Bacteria: Biochemistry, Physiology, and Molecular Genetics. Washington, DC: American Society for Microbiology; 1993. pp. 859–867. [Google Scholar]
- 25.Peterson C., Simon M., Hodges J., Mertens P., Higgins L., Egelman E., Anderson D. Composition and mass of the bacteriophage phi29 prohead and virion. J. Struct. Biol. 2001;135:18–25. doi: 10.1006/jsbi.2001.4375. [DOI] [PubMed] [Google Scholar]
- 26.Shu D., Huang L., Guo P. A simple mathematical formula for stoichiometry quantitation of viral and nanobiological assemblage using slopes of log/log plot curves. J. Virol. Methods. 2003;115:19–30. doi: 10.1016/j.jviromet.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 27.Wilfried J., Meijer J., Horcajadas J.A., Salas M. Φ29 family of phages. Microbiol. Mol. Biol. Rev. 2001;65:261–287. doi: 10.1128/MMBR.65.2.261-287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo P., Erickson S., Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage φ29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 29.Guo P., Zhang C., Chen C., Trottier M., Garver K. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 30.Trottier M., Guo P. Approaches to determine stoichiometry of viral assembly components. J. Virol. 1997;71:487–494. doi: 10.1128/jvi.71.1.487-494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang F., Lemieux S., Wu X., St.-Arnaud S., McMurray C.T., Major F., Anderson D. Function of hexameric RNA in packaging of bacteriophage phi29 DNA in vitro. Mol. Cell. 1998;2:141–147. doi: 10.1016/s1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]
- 32.Chen C., Sheng S., Shao Z., Guo P. A dimer as a building block in assembling RNA. A hexamer that gears bacterial virus phi29 DNA-translocating machinery. J. Biol. Chem. 2000;275:17510–17516. doi: 10.1074/jbc.M909662199. [DOI] [PubMed] [Google Scholar]
- 33.Valpuesta J.M., Fernandez J.J., Carazo J.M., Carrascosa J.L. The three-dimensional structure of a DNA translocating machine at 10 A resolution. Structure Fold. Des. 1999;7:289–296. doi: 10.1016/s0969-2126(99)80039-2. [DOI] [PubMed] [Google Scholar]
- 34.Guasch A., Pous J., Ibarra B., Gomis-Ruth F.X., Valpuesta J.M., Sousa N., Carrascosa J.L., Coll M. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J. Mol. Biol. 2002;315:663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- 35.Simpson A.A., Tao Y., Leiman P.G., Badasso M.O., He Y., Jardine P.J., Olson N.H., Morais M.C., Grimes S., Anderson D.L., et al. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Y., Blocker F., Xiao F., Guo P. Construction and 3-D computer modeling of connector arrays with tetragonal to decagonal transition induced by pRNA of phi29 DNA-packaging motor. J. Nanosci. Nanotechnol. 2005;5:856–863. doi: 10.1166/jnn.2005.143. [DOI] [PubMed] [Google Scholar]
- 37.Hendrix R.W. Symmetry mismatch and DNA packaging in large bacteriophages. Proc. Natl Acad. Sci. USA. 1978;75:4779–4783. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen C., Guo P. Sequential action of six virus-encoded DNA-packaging RNAs during phage phi29 genomic DNA translocation. J. Virol. 1997;71:3864–3871. doi: 10.1128/jvi.71.5.3864-3871.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C.L., Lee C.-S., Guo P. The proximate 5′ and 3′ ends of the 120-base viral RNA (pRNA) are crucial for the packaging of bacteriophage φ29 DNA. Virology. 1994;201:77–85. doi: 10.1006/viro.1994.1267. [DOI] [PubMed] [Google Scholar]
- 40.Reid R.J.D., Bodley J.W., Anderson D. Identification of bacteriophage Φ29 prohead RNA domains necessary for in vitro DNA-gp3 packaging. J. Biol. Chem. 1994;269:9084–9089. [PubMed] [Google Scholar]
- 41.Ibanez C., Garcia J.A., Carrascosa J.L., Salas M. Overproduction and purification of the connector protein of Bacillus subtilis phage φ29. Nucleic Acids Res. 1984;12:2351–2365. doi: 10.1093/nar/12.5.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C.L., Trottier M., Guo P.X. Circularly permuted viral pRNA active and specific in the packaging of bacteriophage φ29 DNA. Virology. 1995;207:442–451. doi: 10.1006/viro.1995.1103. [DOI] [PubMed] [Google Scholar]
- 43.Shu D., Huang L., Hoeprich S., Guo P. Construction of phi29 DNA-packaging RNA (pRNA) monomers, dimers and trimers with variable sizes and shapes as potential parts for nano-devices. J. Nanosci. Nanotech. 2003;3:295–302. doi: 10.1166/jnn.2003.160. [DOI] [PubMed] [Google Scholar]
- 44.Lee C.S., Guo P. A highly sensitive system for the in vitro assembly of bacteriophage φ29 of Bacillus subtilis. Virology. 1994;202:1039–1042. doi: 10.1006/viro.1994.1434. [DOI] [PubMed] [Google Scholar]
- 45.Guasch A., Parraga A., Pous J., Valpuesta J.M., Carrascosa J.L., Coll M. Purification, crystallization and preliminary X-ray diffraction studies of the bacteriophage phi 29 connector particle. FEBS Lett. 1998;430:283–287. doi: 10.1016/s0014-5793(98)00672-3. [DOI] [PubMed] [Google Scholar]
- 46.Donate L.E., Valpuesta J.M., Rocher A., Mendez E., Rojo F., Salas M., Carrascosa J.L. Role of the amino-terminal domain of bacteriophage phi 29 connector in DNA binding and packaging. J. Biol. Chem. 1992;267:10919–10924. [PubMed] [Google Scholar]
- 47.Donate L.E., Valpuesta J.M., Mier C., Rojo F., Carrascosa J.L. Characterization of an RNA-binding domain in the bacteriophage Φ29 connector. J. Biol. Chem. 1993;268:20198–20204. [PubMed] [Google Scholar]
- 48.Ibarra B., Caston J.R., Llorca O., Valle M., Valpuesta J.M., Carrascosa J.L. Topology of the components of the DNA packaging machinery in the phage phi29 prohead. J. Mol. Biol. 2000;298:807–815. doi: 10.1006/jmbi.2000.3712. [DOI] [PubMed] [Google Scholar]
- 49.Catalano C.E., Cue D., Feiss M. Virus DNA packaging: the strategy used by phage lambda. Mol. Microbiol. 1995;16:1075–1086. doi: 10.1111/j.1365-2958.1995.tb02333.x. [DOI] [PubMed] [Google Scholar]
- 50.Frackman S., Siegele D.A., Feiss M. The terminase of bacteriophage λ: functional domains for cosB binding and multimer assembly. J. Mol. Biol. 1985;180:283–300. doi: 10.1016/0022-2836(85)90215-3. [DOI] [PubMed] [Google Scholar]
- 51.Rao V.B., Black L.W. Cloning, overexpression and purification of the terminase proteins gp16 and gp17 of bacteriophage T4: construction of a defined in vitro DNA packaging system using purified terminase proteins. J. Mol. Biol. 1988;200:475–488. doi: 10.1016/0022-2836(88)90537-2. [DOI] [PubMed] [Google Scholar]
- 52.Yamagishi M., Fujisawa H., Minagawa T. Isolation and characterization of bacteriophage T3/T7 hybrids and their use in studies on molecular basis of DNA-packaging specificity. Virology. 1985;144:502–515. doi: 10.1016/0042-6822(85)90290-9. [DOI] [PubMed] [Google Scholar]
- 53.Morita M., Tasaka M., Fujisawa H. DNA packaging ATPase of bacteriophage T3. Virology. 1993;193:748–752. doi: 10.1006/viro.1993.1183. [DOI] [PubMed] [Google Scholar]
- 54.Becker A., Gold M. Prediction of an ATP reactive center in the small subunit, gpNul of phage lambda terminase enzyme. J. Mol. Biol. 1988;199:219–222. doi: 10.1016/0022-2836(88)90391-9. [DOI] [PubMed] [Google Scholar]
- 55.Duffy C., Feiss M. The large subunit of bacteriophage lambda's terminase plays a role in DNA translocation and packaging termination. J. Mol. Biol. 2002;316:547–561. doi: 10.1006/jmbi.2001.5368. [DOI] [PubMed] [Google Scholar]
- 56.Smith M.P., Feiss M. Sites and gene products involved in lambdoid phage DNA packaging. J. Bacteriol. 1993;175:2393–2399. doi: 10.1128/jb.175.8.2393-2399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin H., Simon M.N., Black L.W. Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA packaging. J. Biol. Chem. 1997;272:3495–3501. doi: 10.1074/jbc.272.6.3495. [DOI] [PubMed] [Google Scholar]
- 58.Sun M., Son M., Serwer P. Formation and cleavage of a DNA network during in vitro bacteriophage T7 DNA packaging: light microscopy of DNA metabolism. Biochemistry. 1997;36:13018–13026. doi: 10.1021/bi971410b. [DOI] [PubMed] [Google Scholar]
- 59.Fujisawa H., Morita M. Phage DNA packaging. Genes Cells. 1997;2:537–545. doi: 10.1046/j.1365-2443.1997.1450343.x. [DOI] [PubMed] [Google Scholar]
- 60.Hwang J.S., Bogner E. ATPase activity of the terminase subunit pUL56 of human cytomegalovirus. J. Biol. Chem. 2002;277:6943–6948. doi: 10.1074/jbc.M108984200. [DOI] [PubMed] [Google Scholar]
- 61.Perez-Romero P., Tyler R.E., Abend J.R., Dus M., Imperiale M.J. Analysis of the interaction of the adenovirus L1 52/55-kilodalton and IVa2 proteins with the packaging sequence in vivo and in vitro. J. Virol. 2005;79:2366–2374. doi: 10.1128/JVI.79.4.2366-2374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang W., Arcos R. Interaction of the adenovirus major core protein precursor, pVII, with the viral DNA packaging machinery. Virology. 2005;334:194–202. doi: 10.1016/j.virol.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 63.Zhang W., Low J.A., Christensen J.B., Imperiale M.J. Role for the adenovirus IVa2 protein in packaging of viral DNA. J. Virol. 2001;75:10446–10454. doi: 10.1128/JVI.75.21.10446-10454.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sheaffer A.K., Newcomb W.W., Gao M., Yu D., Weller S.K., Brown J.C., Tenney D.J. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Viol. 2001;75:687–698. doi: 10.1128/JVI.75.2.687-698.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goding C.R., Russell W.C. Adenovirus cores can function as templates in in vitro DNA replication. EMBO J. 1983;2:339–344. doi: 10.1002/j.1460-2075.1983.tb01428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weber J.M., Khittoo G. The role of phosphorylation and core protein V in adenovirus assembly. J. Gen. Virol. 1983;64:2063–2068. doi: 10.1099/0022-1317-64-9-2063. [DOI] [PubMed] [Google Scholar]
- 67.Fujisawa H., Shibata H., Kato H. Analysis of interactions among factors involved in the bacteriophage T3 DNA packaging reaction in a defined in vitro system. Virology. 1991;185:788–794. doi: 10.1016/0042-6822(91)90550-u. [DOI] [PubMed] [Google Scholar]
- 68.Yeo A., Feiss M. Mutational analysis of the prohead binding domain of the large subunit of terminase, the bacteriophage lambda DNA packaging enzyme. J. Mol. Biol. 1995;245:126–140. [PubMed] [Google Scholar]
- 69.Yeo A., Feiss M. Specific interaction of terminase, the DNA packaging enzyme of bacteriophage lambda, with the portal protein of the prohead. J. Mol. Biol. 1995;245:141–150. doi: 10.1006/jmbi.1994.0013. [DOI] [PubMed] [Google Scholar]
- 70.Guo P., Bailey S., Bodley J.W., Anderson D. Characterization of the small RNA of the bacteriophage φ29 DNA packaging machine. Nucleic Acids Res. 1987;15:7081–7090. doi: 10.1093/nar/15.17.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garver K., Guo P. Boundary of pRNA functional domains and minimum pRNA sequence requirement for specific connector binding and DNA packaging of phage phi29. RNA. 1997;3:1068–1079. [PMC free article] [PubMed] [Google Scholar]
- 72.Shu D., Guo P. A Viral RNA that binds ATP and contains an motif similar to an ATP-binding aptamer from SELEX. J. Biol. Chem. 2003;278:7119–7125. doi: 10.1074/jbc.M209895200. [DOI] [PubMed] [Google Scholar]
- 73.Hang J.Q., Tack B.F., Feiss M. ATPase center of bacteriophage lambda terminase involved in post-cleavage stages of DNA packaging: identification of ATP-interactive amino acids. J. Mol. Biol. 2000;302:777–795. doi: 10.1006/jmbi.2000.4086. [DOI] [PubMed] [Google Scholar]
- 74.Gold M., Becker A. The bacteriophage λ terminase: partial purification and preliminary characterization of properties. J. Biol. Chem. 1983;258:14619–14625. [PubMed] [Google Scholar]
- 75.Morita M., Tasaka M., Fujisawa H. Analysis of the fine structure of the prohead binding domain of the packaging protein of bacteriophage T3 using a hexapeptide, an analog of a prohead binding site. Virology. 1995;211:516–524. doi: 10.1006/viro.1995.1433. [DOI] [PubMed] [Google Scholar]
- 76.Tomka M.A., Catalano C.E. Kinetic characterization of the ATPase activity of the DNA packaging enzyme from bacteriophage lambda. Biochemistry. 1993;32:11992–11997. doi: 10.1021/bi00096a008. [DOI] [PubMed] [Google Scholar]
- 77.Valle M., Kremer L., Martinez A., Roncal F., Valpuesta J.M., Albar J.P., Carrascosa J.L. Domain architecture of the bacteriophage phi29 connector protein. J. Mol. Biol. 1999;288:899–909. doi: 10.1006/jmbi.1999.2731. [DOI] [PubMed] [Google Scholar]
- 78.Hoeprich S., Guo P. Computer modeling of three-dimensional structure of DNA-packaging RNA(pRNA) monomer, dimer and hexamer of Phi29 DNA packaging motor. J. Biol. Chem. 2002;277:20794–20803. doi: 10.1074/jbc.M112061200. [DOI] [PubMed] [Google Scholar]