Summary
A detailed analysis of the Ewing sarcoma surfacesome has arrived. Robust expression of surface CDH11 and ENPP1 was identified. This “comprehensive catalogue” of the Ewing surfacesome serves as a fresh roadmap to development of new therapeutic approaches, including immunotherapies and multi-modality therapeutic combinations, to target aggressive Ewing tumor subpopulations.
In this issue of Clinical Cancer Research, Mooney and colleagues conduct an extensive, first-in-kind analysis of the Ewing sarcoma cell surface proteome (1). Proteomic approaches, including Tandem Mass Tags-based mass spectrometry, were used to analyze both the surfacesome and global proteome of patient tumor samples from both primary and metastatic Ewing sarcoma. This analysis confirmed the surface expression of known sensitive but not highly specific markers of Ewing sarcoma, such as CD99. The authors generated a prioritization system to identify Ewing surface proteins that may be excellent candidates for development as an immunotherapy target (termed “Group 1” proteins). Group 1 proteins included IL1RAP (2), ADGR2, STEAP1, STEAP2, SLCO5A1, ENPP1, and CHD11. ENPP1 (Ectonucleotide pyrophosphatase/phosphodiesterase 1) was more extensively examined in patient samples of Ewing sarcoma given its rank as a potential immunotherapeutic target and the availability of a suitable commercial antibody. Immunohistochemistry analysis of ENPP1 expression in TMAs of different pediatric solid tumors and normal pediatric tissue revealed ENPP1 is highly expressed in Ewing sarcoma (more so than in other pediatric solid tumors). The authors note that while ENPP1 expression in normal tissues is minimal, low-level expression in the pancreas and liver was noted. Variability in ENPP1 expression intensity was noted across some primary and metastatic Ewing tumor samples. When considering the bench-to-bedside implications of the Mooney et al. (1) Ewing sarcoma surfacesome data, five major themes to consider emerge: 1) tools for next steps, 2) understanding target protein expression heterogeneity, 3) agents that alter surface target protein expression, 4) antibody-drug conjugates, and 5) immunotherapy targets.
When considering tools for next steps, and as Mooney and colleagues (1) note, commercial availability of antibodies is required as an early step in cell surface protein validation. Antibody availability is the reason why the authors chose to proceed with validation of ENPP1 versus CHD11 (no appropriate antibody available) in the current manuscript. To continue to develop ENPP1 and other Ewing sarcoma surface proteins in the preclinical/clinical trial space, antibodies and corresponding standardized, open-access staining protocols would be needed. Ideally, antibodies for both immunohistochemistry (for formalin-fixed paraffin-embedded, FFPE, specimens) and flow cytometry (for fresh single cell tumor suspensions and in vitro cell line specimens) would be worked-up.
In addition to antibodies, other tools for preclinical testing include patient-derived xenografts (PDX) and cell lines (CDX) with ENPP1 (or other surface protein) expression fully characterized. Characterized PDX and CDX Ewing cell surface protein models would ideally be held in a sharable repository to facilitate use by multiple investigators and ease of comparisons across datasets.
A second theme that emerges is understanding target protein expression heterogeneity. Surface protein expression heterogeneity is an incredibly important consideration in the development of tumor-targeting therapies. Mooney and colleagues (1) note that of the 19 patient-derived Ewing sarcoma samples analyzed for ENPP1, 3 primary specimens and 2 metastatic specimens demonstrated lower or negative staining and the average H-score of metastatic cores was lower than that of the primary cores. Many preclinical questions regarding ENPP1 expression heterogeneity exist and need to be addressed as this target continues to be developed. Analysis of paired patient samples (primary, metastatic site and/or relapse) should be conducted to determine persistence in target expression upon disease progression (Fig. 1). Intra-tumoral ENPP1 heterogeneity should be better defined, because if ENPP1-negative cells exist adjacent to ENPP1-positive Ewing cells, targeted therapy efficacy could be reduced. Accordingly, if ENPP1 negative/positive Ewing tumor subsets do exist within a tumor, better defining the characteristics (EWSR1::FLI1 level (3), STAG2 status (4), etc.) of ENPP1 +/− Ewing cell populations could help guide when and how to best utilize therapies targeting ENPP1. In addition to better understanding baseline cell surface protein heterogeneity, a clear understanding of the impact of other treatments on Ewing cell surface protein targets is needed to maximize potential clinical benefit (Fig. 1). For example, do chemotherapies used in the treatment of Ewing sarcoma alter ENPP1 expression? This would be critical to address if an ENPP1 (or other cell surface protein-targeting agent) is being incorporated into an existing Ewing sarcoma treatment backbone. Another question to address is whether there drugs that increase ENPP1 cell surface expression in Ewing sarcoma that could be used to “prime” tumors for ENPP1-targting therapy?
Figure 1. ENPP1 cell surface expression in Ewing sarcoma: better understanding surface expression heterogeneity and persistence.
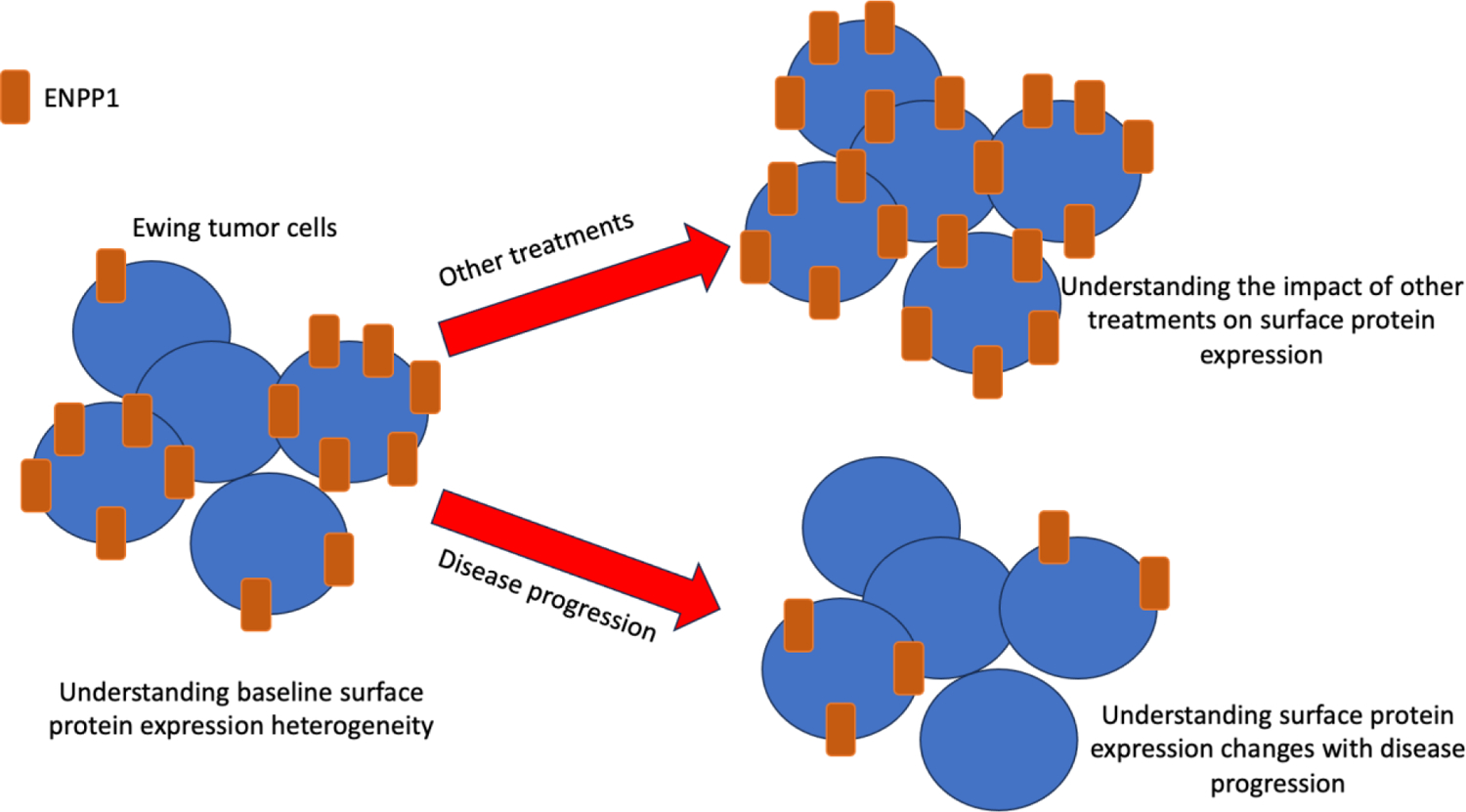
Continued preclinical development of ENPP1 could include better defining baseline intra-tumoral expression heterogeneity, evolution of expression upon disease progression, and understanding the impact of other anti-cancer agents on ENPP1 expression.
Antibody-drug conjugates targeting newly identified Ewing sarcoma cell surface proteins, such as ENPP-1, should be explored. ADCs combine a monoclonal antibody for a target with a cytotoxic “payload” to more specifically direct cellular poisons to tumor cells. Given the low expression of ENPP1 in normal liver and pancreas noted by Mooney et al (1), bispecific ADCs targeting dual tumor-associated surface targets (5) to increase tumor specificity may be needed. For an ADC or bispecific ADC for Ewing sarcoma, payload selection would need to be carefully considered based on what is discovered about the sensitivity of ENPP1 positive/negative Ewing tumor subsets after further preclinical development.
In addition to ADCs, immunotherapies are another logical class of agents to consider when promising new Ewing cell surface targets emerge. The slow development of immunotherapies for the treatment of Ewing sarcoma has stemmed from the historical lack of detailed understanding of the Ewing tumor immune microenvironment (TIME), historical lack of an immunocompetent in vivo model to study, and the historical lack of a robust cell surface protein target analysis. In the past 5 years, scRNAseq data have helped better define the Ewing TIME (6, 7), immunocompetent zebrafish and humanized mouse models are emerging (8, 9), and Mooney and colleagues have published the current manuscript revealing new surface targets in Ewing tumors (1). These emerging data and tools will help investigators around the world continue to develop new multi-modality treatment strategies, including the incorporation of immunotherapies, for the treatment of aggressive Ewing sarcoma. As discussed above, the status of ENPP1 (or other cell surface protein) expression would need to be carefully considered and re-analyzed when combining ENPP1-targeting immunotherapies with other agents.
In their work in this issue of Clinical Cancer Research “Surface and global proteome analysis identify ENPP1 and other surface proteins as actionable immunotherapeutic targets in Ewing sarcoma” (1), Mooney and colleagues present an exciting new set of cell surface targets in Ewing sarcoma. Here, thoughts on continued, detailed preclinical investigations of these targets to maximize their full therapeutic potential are outlined. Patients with aggressive Ewing sarcoma are in great need of new tumor-targeting agents and a better-defined Ewing cell surfacesome is an exciting next step toward reaching that goal.
Funding Sources
K.M.B. is currently supported by the NCI (1K08CA252178) and Alex’s Lemonade Stand Foundation (Innovator Award). K.M.B. would also like to thank the UPMC Children’s Hospital Foundation.
Footnotes
Conflicts of Interest
The authors declare no potential conflicts of interest.
References
- 1.Mooney B, Negri GL, Shyp T, Delaidelli A, Zhang H-F, Spencer Miko SE, et al. Surface and global proteome analysis identify ENPP1 and other surface proteins as actionable immunotherapeutic targets in Ewing sarcoma. Clin Cancer Res 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H-F, Hughes CS, Li W, He J-Z, Surdez D, El-Nagger AM et al. Proteomic Screens for Suppressors of Anoikis Identify IL1RAP as a Promising Surface Target in Ewing Sarcoma. Cancer Discov. 2021. Nov; 11(11):2884–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franzetti G-A, Laud-Duval K, van der Ent W, Brisac A, Irondelle M, Aubert S, et al. Cell-to-cell heterogeneity of EWSR1-FLI1 activity determines proliferation/migration choices in Ewing sarcoma cells. Oncogene. 2017. June 22;36(25):3505–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shulman DS, Chen S, Hall D, Nag A, Thorner AR, Lessnick SL et al. Adverse prognostic impact of the loss of STAG2 protein expression in patients with newly diagnosed localised Ewing sarcoma: A report from the Children’s Oncology Group. Br J Cancer. 2022. Dec; 127(12):2220–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Shang C, Wang N, An G, Zhang E, Lin Q, et al. Abstract LB214: A first-in-class bispecific antibody-drug conjugate (DM002) targeting HER3 and the juxtamembrane domain of MUC1. Cancer Res (2023) 83 (8_Supplement): LB214. [Google Scholar]
- 6.Cillo AC, Mukherjee E, Bailey NG, Onkar S, Daley J, Salgado C et al. Ewing Sarcoma and Osteosarcoma Have Distinct Immune Signatures and Intercellular Communication Networks. Clin Cancer Res (2022) 28 (22): 4968–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visser LL, Bleijs M, Margaritis T, van de Wetering M, Holstege FCP, Clevers H. Ewing Sarcoma Single-cell Transcriptome Analysis Reveals Functionally Impaired Antigen-presenting Cells. Cancer Res Commun. 2023. Oct 24;3(10):2158–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasileva E, Warren M, Triche TJ, Amatruda JF. Dysregulated heparan sulfate proteoglycan metabolism promotes Ewing sarcoma tumor growth. eLife, 2022. 10.7554/eLife.69734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daley JD, Mukherjee E, Cillo AR, Tufino AC, Bailey NG, Bruno TC et al. Abstract A004: Radiation-induced changes to the immune microenvironment in an immunocompetent mouse model of Ewing sarcoma. Clin Cancer Res (2022) 28 (18_Supplement): A004. [Google Scholar]


