Summary
The diagnosis and management of hepatocellular carcinoma (HCC) have improved significantly in recent years. With the introduction of immunotherapy-based combination therapy, there has been a notable expansion in treatment options for patients with unresectable HCC. Simultaneously, innovative molecular tests for early detection and management of HCC are emerging. This progress prompts a key question: as liquid biopsy techniques rise in prominence, will they replace traditional tissue biopsies, or will both techniques remain relevant? Given the ongoing challenges of early HCC detection, including issues with ultrasound sensitivity, accessibility, and patient adherence to surveillance, the evolution of diagnostic techniques is more relevant than ever. Furthermore, the accurate stratification of HCC is limited by the absence of reliable biomarkers which can predict response to therapies. While the advantages of molecular diagnostics are evident, their potential has not yet been fully harnessed, largely because tissue biopsies are not routinely performed for HCC. Liquid biopsies, analysing components such as circulating tumour cells, DNA, and extracellular vesicles, provide a promising alternative, though they are still associated with challenges related to sensitivity, cost, and accessibility. The early results from multi-analyte liquid biopsy panels are promising and suggest they could play a transformative role in HCC detection and management; however, comprehensive clinical validation is still ongoing. In this review, we explore the challenges and potential of both tissue and liquid biopsy, highlighting that these diagnostic methods, while distinct in their approaches, are set to jointly reshape the future of HCC management.
Keywords: HCC, liver cancer, liquid biopsy, cfDNA, extracellular vesicles, tissue biopsy, precision medicine
Introduction
Background
Liver cancer, mainly hepatocellular carcinoma (HCC), is a growing cause of worldwide cancer-related fatalities and is predicted to cause more than 1.3 million annual deaths by 2040.1 HCC typically arises on cirrhotic livers, most commonly in patients with chronic liver diseases including chronic hepatitis B or C (HBV or HCV) infection, alcohol-related liver disease, or the increasingly prevalent metabolic dysfunction-associated steatotic liver disease.1,2 Several new tyrosine kinase inhibitors and immunotherapies have recently gained approval for the treatment of advanced HCC, signalling a promising shift in therapeutic options. Additionally, there are ongoing developments in molecular stratification of HCC. However, HCC continues to be diagnosed only at advanced stages, complicating treatment efforts. Moreover, we lack robust, predictive and prognostic biomarkers to tailor therapeutic strategies, making it difficult to identify which patients would derive the most benefit from specific therapies. This highlights the critical need for access to molecular data on HCC to refine biomarker development for improved detection and management.
The promise of precision medicine in HCC
The advent of molecular diagnostics in oncology has ushered in a new era of precision medicine. By detecting specific genetic alterations within cancer cells, these tests can reveal the molecular underpinnings of the disease, facilitate early diagnosis, and guide the choice of targeted treatments. These molecular tests can enable us to tailor therapy to the patient’s unique genetic profile, thus increasing the likelihood of a positive response. Furthermore, molecular diagnostics can be used to monitor disease progression and detect minimal residual disease, enhancing our ability to prevent recurrence. They may also aid in predicting patient prognosis, providing valuable information for patient counselling and management. In essence, the application of molecular diagnosis can streamline the path from diagnosis to treatment, improving patient outcomes in cancer care. Despite these promising advances, the full potential of molecular stratification and prognostication in HCC is yet to be realised, underscoring the need for continued innovation in this crucial area of research.
To create high-precision tests for HCC, comprehensive tumour molecular data is vital. Yet, such data remains elusive in HCC due to the infrequency of tissue biopsies. Liquid biopsies have emerged as an innovative solution, providing an alternative and minimally invasive means of obtaining critical genetic data on tumours. By enabling analysis of circulating tumour cells (CTCs) and tumour products like DNA in the blood, liquid biopsies serve as a genomic looking glass into the dynamic tumour landscape. They can offer a real-time view of tumour evolution, tracing disease progression and response to treatment. Yet, the use of tissue or liquid biopsies has not yet been integrated into standard clinical practice for HCC. Uncertainties concerning their sensitivity, specificity, and cost-effectiveness, as well as the relative merits of tissue vs. liquid biopsies, are under intense investigation. It remains to be seen whether liquid biopsies can fully obviate the need for tissue biopsies or if each retains a distinct role. In this review, we discuss recent advances in the field of precision medicine for HCC, addressing the role of both tissue and liquid biopsy.
Molecular landscape of HCC
HCC is a complex and heterogeneous disease involving both cancer cell-intrinsic genetic aberrations and changes within the tumour immune microenvironment.3 Comprehensive multiomic profiling has been used to uncover this diversity and complexity, and could identify potential opportunities for individualised therapeutic strategies and biomarkers. Certain common genomic occurrences, such as a mutation in the promoter region of the telomerase reverse transcriptase (TERT) gene, Wnt pathway disruptions, TP53 and AT-rich interactive domain-containing protein (ARID) gene mutations, MYC oncogene activation, and transforming growth factor-β (TGFβ) pathway dysregulation, show promise as potential therapeutic targets or predictive biomarkers in HCC.4–7 For instance, pTERT mutations, which are the most frequent genetic alterations observed in HCC, are evident in early-stage HCC and are associated with a poorer recurrence-free survival rate.8,9 This suggests their potential use as a tool for early detection and risk stratification. The Wnt pathway, crucial in liver development and regeneration,10 is often disrupted in HCC.11 Activation of this pathway has been suspected to be associated with a reduced response to immunotherapy,12,13 however, post hoc analyses of the CheckMate 459 and IMbrave150 trials did not confirm this association.14,15 Further data are awaited. Additionally, recurrent TP53 gene mutations, particularly common in advanced-stage HCC, correlate with an aggressive disease phenotype, poor overall survival, and tumour recurrence.16–19 Mutations in ARID genes, present in 3–5% of HCC cases, are associated with advanced-stage tumours characterised by high vascularity.20 The MYC oncogene, regularly overexpressed in HCC, is a key promoter of tumorigenesis, although targeting it directly remains a challenge.21
Other promising targets include NRF2/KEAP1, a cytoprotective system, and TGFβ signalling, which is activated in a subset of HCC tumours.22,23 Mutations in mechanistic target of rapamycin (mTOR) pathway genes or downstream activation of mTOR signalling in subsets of patients often leads to metabolic dysregulation, and may eventually be targeted using mTOR inhibitors in select patients with mTOR addiction, presenting yet another possible therapeutic approach for HCC, potentially in combination with immunotherapy.24,25 Thus, detection of these genetic events via tissue or liquid biopsy has the potential to change management. Lastly, histological features may also provide insights into genetic alterations and oncogenic pathways, and thus guide patient prognosis. For example, poorly differentiated, proliferative tumours tend to harbour TP53 mutations, FGF19 (fibroblast growth factor 19) amplifications, or activation of TGFβ, RAS/MAPK, PI3K/AKT pathways. On the other hand, well-differentiated, non-proliferative tumours tend to harbour CTNNB1 (encoding β-catenin) mutations, show activation of Wnt and JAK/STAT pathways, and have microtrabecular and pseudoglandular patterns on histology, with less immune infiltration.26,27
Overview of current management of HCC
Treatment options for HCC depend on the stage of the disease.28–30 In early-stage HCC, potential curative approaches include surgical resection, transplantation, or local ablation. Organ scarcity limits the availability of liver transplantation, which offers high survival rates with low recurrence risk by addressing both HCC and underlying cirrhosis. For intermediate-stage HCC, intra-arterial therapies such as transarterial embolisation, transarterial chemoembolisation, and transarterial radioembolisation serve either as first-line treatments or as bridging therapies before transplantation.31 Stereotactic body radiotherapy, with its high local control rates, has emerged as another potential bridging therapy option for patients with HCC awaiting transplantation.32 In advanced-stage HCC, systemic therapies involving targeted therapies and immune checkpoint inhibitors are commonly employed. The groundbreaking IMbrave150 study demonstrated improved overall survival, along with significant enhancements in overall response rate and progression-free survival, by combining the PD1 (programmed death 1) inhibitor atezolizumab and the VEGF (vascular endothelial growth factor) inhibitor bevacizumab, leading to its adoption as the new frontline standard of care for advanced HCC.33 Subsequently, the combination of the anti-CTLA-4 (cytotoxic T lymphocyte antigen-4) antibody tremelimumab and the anti-programmed death ligand 1 (PD-L1) agent durvalumab has also been approved as a first-line treatment option for advanced HCC.34 This consensus is reinforced by international guidelines, consolidating the significance of immune checkpoint inhibitor-based regimens in HCC treatment.30 Thus, recent advances in treatment have vastly extended treatment options for patients with HCC, both for early and advanced stages.
Tissue biopsy for diagnosis and management of HCC
Current non-invasive imaging diagnostic tests for HCC
Screening for early detection of HCC is crucial since the therapeutic window for curative surgical intervention for HCC is often limited to its earliest stages. The American Association for the Study of Liver Diseases and the European Association for the Study of the Liver currently recommend biannual ultrasound surveillance, with or without serum alpha-fetoprotein (AFP) measurement, for high-risk populations.35,36 This includes individuals with cirrhosis of both viral and non-viral aetiology, as well as non-cirrhotic patients with chronic HBV infection. Ultrasound, while central to routine surveillance, is not without its challenges. Despite demonstrating a sensitivity of 58–89% and a specificity of over 90%, adherence to regular ultrasound examinations is often undermined by various factors.37 These factors include knowledge gaps, social disparities impacting healthcare access, and interobserver variability, which can lead to high false-positive rates. Furthermore, ultrasound often falls short when it comes to visualising and differentiating early HCC lesions and dysplastic nodules, although contrast enhancement can potentially boost its sensitivity.38,39
Non-invasive imaging tools such as triphasic CT or MRI have become invaluable in diagnosing and staging HCC. These methods leverage the distinctive vascular changes during hepatic carcinogenesis and the high likelihood of HCC in cirrhotic livers. Diagnostically, HCC often exhibits a radiographic signature consisting of arterial phase hyperenhancement (wash-in) and venous phase hypo-enhancement (wash-out) of contrast agents, owing to their arterial hypervascularity.40,41 For lesions exceeding 2 cm, this radiographic pattern presents over 90% sensitivity and specificity.42 Moreover, tumour necrosis visualised on CT/MRI can define a response to treatment according to RECIST criteria for HCC.43 In summary, while the importance of ultrasound surveillance in early HCC detection cannot be overstated, it is challenged by variability and adherence issues. On the other hand, cross-sectional imaging with contrast-enhanced CT or MRI, the primary methods for diagnosing HCC and assessing treatment responses, also face obstacles, particularly false positivity, and cost. Thus, it is essential to consider these limitations and strive to develop more accessible, cost-effective, and accurate screening and diagnostic tests for HCC.
Current role of tissue biopsy in diagnosis and management of HCC
Even though tissue biopsy is not routinely employed for the diagnosis of HCC, there are specific instances where a biopsy is necessary. For instance, lesions between 1–2 cm that remain indeterminate on dynamic contrast-enhanced CT or MRI can be biopsied to arrive at a definitive diagnosis.36 A biopsy is generally necessary in the non-cirrhotic liver setting, where the specificity of imaging in diagnosing HCC declines.36 In addition, a liver tumour biopsy is indicated in patients with suspected intrahepatic cholangiocarcinoma, mixed HCC-cholangiocarcinoma, or secondary malignancies, as histological markers can differentiate HCC from other tumours.44 Moreover, evaluation of the non-tumoural liver in these biopsies may also be useful to characterise the underlying liver disease and provide insight into HCC prognosis.44 Another major advantage of tissue biopsy is the ability to classify HCC into histological subtypes that correlate with molecular classifications and have prognostic implications, such as the highly differentiated, CTNNB1-mutated and the poorly differentiated, TP53-mutated subtypes.26,45,46 While pathology-based biomarkers have yet to demonstrate clinical efficacy in terms of guiding treatment selection, tissue biopsy may become a more routine diagnostic procedure in the era of precision medicine.
We acknowledge that tissue biopsy has a few limitations (Fig. 1). Due to sampling bias and tumour heterogeneity, the small tissue cores obtained during biopsy may not be entirely representative. Additionally, liver biopsy is associated with increased risks of pain, bleeding, and very rarely, hypotension, tachycardia, pneumothorax, or biliary peritonitis.47,48 Another possible complication is the potential for “needle tract seeding,” where tumour cells can detach from the primary site and travel along the needle pathway. Although older studies reported a risk of needle tract seeding of up to 3%,49 technical improvements and the performance of a high volume of biopsies are associated with a much lower rate of needle tract seeding of less than 1%.50,51 Based on our own experience, the fear of needle tract seeding appears to be overstated. Thus, despite its limitations, biopsy remains a vital diagnostic tool, and with advances in techniques, particularly in high-volume centres, risks such as needle tract seeding are very low.
Fig. 1. Overview of advantages and disadvantages of tissue vs. liquid biopsy in hepatocellular carcinoma.
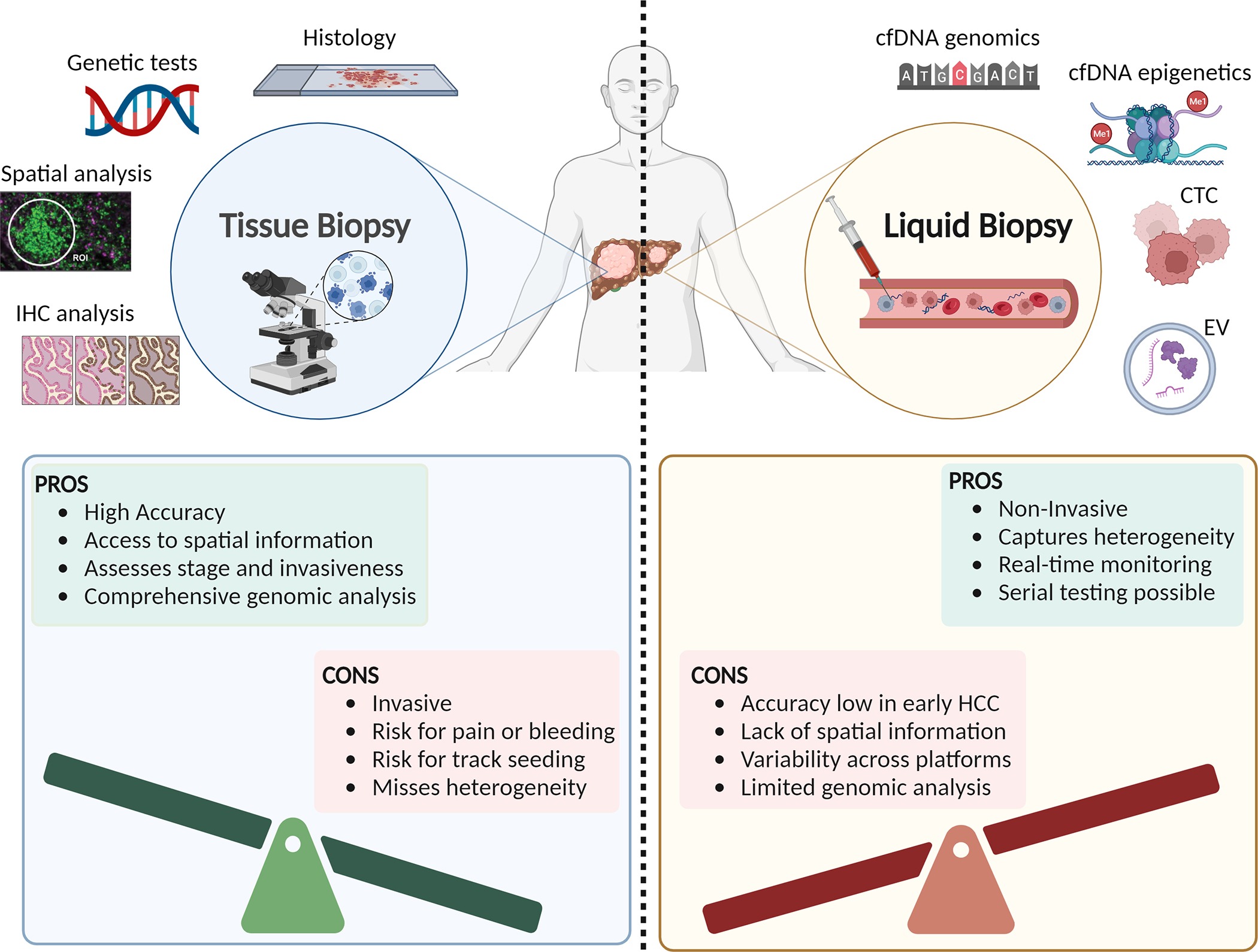
cfDNA, cell-free DNA; CTCs, circulating tumour cells; EVs, extracellular vesicles; TME, tumour microenvironment.
Potential future role of tissue biopsy in management of HCC
As more targeted therapies become available, liver biopsy may play a larger role in guiding target identification and HCC management (Fig. 1). This is based on the premise that, in numerous other cancers, biomarkers derived from biopsy have proven invaluable in identifying cancer subtypes and determining the optimal treatment strategy. Among the 206 distinct oncology therapeutic products approved by the US FDA between 2000 and 2022, an overwhelming majority (97%) are aimed at specific molecular targets, such as PD1, EGFR, BCR-ABL1 fusion, CD19 or HER2.52 In fact, tissue biomarkers have paved the way for site-agnostic approvals, with notable examples being BRAFV600E mutations, NTRK fusions, RET fusions, high tumour mutation burden, microsatellite instability-high, and mismatch repair-deficient status.53 However, it is important to note that these biomarkers may only be applicable to a small proportion (5–10%) of patients with HCC. While biomarker-defined populations accounted for 49% of oncology approvals for targeted therapies by the FDA, liver cancer currently has no biomarker-driven therapy, compared to breast cancer (88% of approvals) or lung cancer (61% of approvals).52 As our understanding of the molecular landscape of HCC continues to improve, the development of targeted therapies and their integration into the treatment armamentarium for HCC becomes more crucial.
Currently, biomarker-driven management is limited, given the lack of evidence supporting the efficacy of targeted therapy in molecular subsets of HCC. However, based on results from the REACH-2 trial, serum AFP has found a place in HCC management. Patients with HCC and AFP >400 are more likely to demonstrate benefit with ramucirumab, based on improved overall survival (7.8 months vs. 4.2 months with placebo) noted in the second-line treatment setting.54 In the future, biomarkers identified from liver biopsy may guide targeted treatment stratification. This is supported by a study which performed targeted exome sequencing of advanced HCC, and found that nearly a quarter of patients had potentially clinically actionable driver mutations involving TSC1/2 (8.5%), FGF19 (6.3%), MET (1.5%), and IDH1 (<1%).13,55 Moreover, the landscape is evolving with ongoing clinical trials for HCC incorporating both all-comer patient enrolment and biomarker-guided approaches. Several tissue-based biomarkers, such as glypican-3 (GPC3: NCT05003895, NCT05103631), epithelial cell adhesion molecule (NCT05028933, NCT03013712), MET (NCT01755767), mucin 1 (NCT02587689), MHC1 (NCT05195294), and TERT (NCT05595473), are being incorporated into these clinical trials. Thus, as our understanding of the biological complexity of HCC deepens, the potential role of biomarkers, and by extension liver biopsy, in guiding treatment choices and improving patient outcomes will become increasingly evident.
Another area of active research is the identification of biomarkers to predict response to immune checkpoint inhibitors. We highlight three such putative predictive biomarkers. First, PD-L1 expression on tissue biopsy may be predictive of response to immunotherapy in patients with HCC. However, the dynamic nature of PD-L1 expression and lack of standardised methods to evaluate its status adds complexity to these assessments. For instance, trials like CheckMate 459 and KEYNOTE-224 observed higher response rates in patients with advanced HCC and PD-L1 positivity when treated with immunotherapy.56,57 In contrast, trials like the CheckMate 040 study reported no significant difference in response between PD-L1-positive and -negative patients treated with nivolumab.58 This discrepancy across trials is not unexpected given the studies were not specifically designed to differentiate responders based on PD-L1 expression. Additionally, PD-L1 expression is generally not a sensitive biomarker across many cancer types, with sensitivities differing based on expression thresholds and regional sampling bias.59,60 Second, high tumour mutational burden or microsatellite instability have been used as biomarkers for response to immunotherapy in other cancers, but less than 1% of HCCs appear to have these features.61 Third, multiple gene signatures which can predict response to immunotherapy have also been reported but require further validation.62–65 These findings underscore the current complexities and highlight the potential for tissue-based biomarkers in the future, which may become instrumental in stratifying patients for immunotherapy in HCC.
Thus, tissue biopsy remains an invaluable tool in cancer diagnosis. By collecting and analysing tumour samples, we can expand our understanding of cancer biology, identify novel biomarkers, and guide the development of new therapies. A retrospective cohort of stored clinical specimens is usually critical to verify biomarkers developed in a preclinical setting, before conducting large, expensive, prospective, randomised-controlled trials. Without biological tissue specimens from a significant number of patients with HCC, especially from those with advanced or metastatic HCC, this pivotal phase in the development of biomarkers for HCC is significantly impeded. Tissue biopsies, when available, can be retrospectively queried in research settings where integrated genomics, digital pathology, and AI can be utilised to derive clinically meaningful biomarkers. The main caveat of existing studies is that tissue biopsies tend to be representative only of early-stage, resected lesions.4–6,66 Nevertheless, investigators have successfully used these tissues to define novel molecular subclasses of patients with HCC for treatment stratification.67 For example, Zeng et al. developed an AI algorithm based on 336 whole-slide histological images integrated with RNA sequencing data to identify immune-related gene signatures potentially correlating with responses to immunotherapy.68 Other studies employed deep learning systems to diagnose HCC, perform histological classification, and provide prognostic indicators through whole-slide imaging analysis.69–71 Thus, it is plausible that deep learning algorithms, fine-tuned on integrated histopathology and spatial transcriptomic data, will enable the diagnosis, tumour classification, and prediction of treatment outcomes for patients with HCC. While the current lack of biomarkers and molecular subtypes may draw into question the ethics of routine biopsies for all patients with HCC, a counter perspective emphasises the urgency of escalating the frequency of tumour biopsies, especially within clinical trial frameworks. Such an approach may hasten the discovery of novel biomarkers and fast-track the evolution of personalised targeted therapies for HCC.
Liquid biopsy for diagnosis and management of HCC
Current serum biomarkers for HCC
With the push towards less invasive techniques in medicine, liquid biopsy provides a supplementary approach to imaging and promises to augment current strategies for the early detection of HCC, the identification of minimal residual disease, treatment selection, and the monitoring of therapeutic responses (Fig. 1). Apart from being able to provide serial samples, liquid biopsy provides a plethora of genomic and transcriptomic information that would otherwise only be accessible via tissue biopsy, as well as enabling the characterisation of intra- and inter-tumoural heterogeneity.72 Intra-tumoural heterogeneity can arise from spatial and temporal clonal differences, while inter-tumoural heterogeneity can arise from differences in the tumour microenvironment (consisting of cancer cells, stroma, endothelial cells, fibroblasts, and immune cells). Through liquid biopsy, serial measurements of blood-based analytes over a treatment window may provide novel insights into biological processes contributing to proteogenomic dynamics, matrix remodelling, metabolic reprogramming, and clonal diversity (Fig. 1).
Over the last several decades, the most widely utilised serum biomarker for HCC screening and diagnosis is AFP. AFP is the foetal analogue of serum albumin, which rises in production during foetal liver development, and diminishes quickly after birth.73 However, various complex mechanisms deregulate this epigenetic silencing and turn on AFP expression in the development of HCC, and thus it has become a widely used screening tool for patients with HCC. Additionally, other prognostic serum proteins identified include fucosylated fraction of AFP (AFP-L3), des-gamma carboxyprothrombin (DCP), and GPC-3.74,75 There are multiple studies investigating AFP as a biomarker for early-stage screening as part of complex panels (e.g., GALAD; gender + age + AFP + DCP + AFP-L3), alongside various methylation markers (e.g., HOXA1, TSPYL5, and B3GALT6),76 or combined with clinicopathologic information.77,78 Moreover, a few groups have assessed AFP as a biomarker for post-resection recurrence or as a biomarker to predict response to treatment with cabozantinib or the combination of atezolizumab + bevacizumab.79,80 Despite the wide acceptance of AFP as a screening tool, limitations include that it is influenced by race/ethnicity, aetiology of HCC (viral vs non-viral), molecular subclasses, and tumour burden, and that it is abundant in other benign liver conditions.81 The rest of this section will detail novel cell-free analytes being tested in the clinic for HCC detection, prognostication, and monitoring of therapeutic responses (Table 1).
Table 1.
Recent Investigations of Liquid Biopsy to Detect and Monitor Treatment Response in HCC
| Study Author, Year | Patient Population | AUC (Sensitivity/ Specificity) | Analyte Detected | Comments |
|---|---|---|---|---|
| Circulating Tumor Cells | ||||
| Wang et al., 2020 | N=344 HCC patients preoperatively | HR=0.46 (TTR) HR=0.32 (OS) |
CTC identified as DAPI+/CD45−/CK+ cells or DAPI+/CD45−/EpCAM+ cells | HCC prognostication of recurrence following TACE |
| Winograd et al., 2020 | N=87 HCC patients | 0.81 (71.1/91.8) early vs metastatic disease HR=3.22 (PD-L1+ CTCs predict shorter OS) 5/5 ICI responders had PD-L1+ CTCs at baseline, while 1/5 ICI nonresponders had PD-L1+ CTCs at baseline |
CTCs identified as DAPI+/CD45−/CK+ cells And PD-L1+ CTCs identified as DAPI+/CD45−/CK+/PD-L1+ cells |
HCC detection and prognostication, prospective study |
| Zhao et al., 2023 | N=270 preoperative HCC patients (N=52 validation set) |
0.94 (NA/NA) CTC number: HR=11.89 CTC clusters: HR=13.67 |
CTCs identified as DAPI+/CD45−/CK+ cells | HCC recurrence post-op prediction |
| Wei et al., 2023 | N=227 HCC patients | 0.84 (NA/NA) CTC number: HR=1.98 |
CanPatrol™ CTC-enrichment technique | HCC extrahepatic recurrence prediction |
| cfDNA Mutations | ||||
| Kaseb et al., 2019 | N=206 HCC patients | N/A 87.9% confirmed alterations |
Blood-derived ctDNA Actionable drivers identified: EGFR, MET, ARID1A, MYC, NF1, BRAF, and ERBB2 Nonactionable drivers identified: TP53, CTNNB1, APC |
Cohort was mix of different stages |
| Qu et al., 2019 | N=331 at risk patients | 0.93 (100.0/94.0) | Blood-derived ctDNA profiling of TERT, TP53, CTNNB1, AXIN1 + HBV integration breakpoint + AFP + DCP + age + gender | Early HCC detection HCCscreen |
| Zhang et al., 2021 | N=571 HCC patients | N/A 77.1% confirmed alterations |
Blood-derived ctDNA Drivers identified: TP53, TERT, CTNNB1, TSC2, RB1, ARID1A, DNMT3A, MLL2, AXIN1, APC |
68% sensitivity early stage detection of ctDNA 86.3% sensitivity late stage detection of ctDNA |
| Nguyen et al., 2023 | N=55 HCC patients | 0.86 (81.0/81.0) | Blood-derived ctDNA genes profiled: APC, ARID1A, AXIN1, BRAF, CDKN2A, CTNNB1, EGFR, KRAS, PIK3CA, PTEN, STK11, TP53, TERT + fragment length profiles | HCC classification compared to healthy participants |
| cfDNA Methylation Signatures | ||||
| Xu et al., 2017 | N=1,098 HCC patients | 0.94 (83.3/90.5) | 401 HCC-specific cfDNA methylation markers | Early HCC detection; prognostic survival |
| Kiesel et al., 2019 | Phase I: N=21 HCC patients Phase II: N=95 HCC patients |
Phase I: 0.91 (86.0/87.0) Phase II: 0.96 (95.0/92.0) |
6 methylated cfDNA markers (HOXA1, EMX1, ECE1, AK055957, PFKP, CLEC11A) normalized by B3GALT6 | Cohort was mix of different stages; phase I tested markers alone or in combination (reported EMX1 and CLEC11A combination here) |
| Luo et al., 2022 | N=120 HCC patients (training set) N=67 HCC patients (validation set) |
0.93 (84.0/96.0) | 2321 methylated cfDNA markers | Early HCC detection |
| Chalasani et al., 2022 | N=156 HCC patients | 0.86 (88.0/82.0) | 3 methylated cfDNA markers (HOXA1, TSPYL5, B3GALT6) + AFP + sex | Early HCC detection |
| Lin et al., 2022 | N=122 HCC patients | 0.94 (75.7/91.2) | 28 methylated cfDNA markers + protein markers (AFP, AFLP-L3, DCP) + clinical variables (age, sex) | Early HCC detection; HelioLiver Test |
| Cheishvilli et al., 2023 | N=504 HCC patients | 0.94 (84.5/95.0) | Differential methylation of CpGs: CHFR, VASH2, CCNJ, GRID2IP, F12 | Early HCC detection; EpiLiver Test |
| ncRNAs Signatures | ||||
| Lin et al., 2015 | N=27 patients in validation set | 0.83 (85.7/91.1) | Seven-miRNA panel (miR-29a, miR-29c, miR-133a, miR-143, miR-145, miR-192, and miR-505) | Early HCC detection from controls at-risk |
| Teufel et al., 2019 | N=349 patients | NA | Nine-miRNA panel (miR-30a, miR-122, miR-125b, miR-200a, miR-374b, miR-15b, miR-107, miR-320, and miR-645) | Prognosticate overall survival after regorafenib (RESORCE trial) |
| Yamamoto et al., 2020 | N=173 patients in validation set | 0.99 (97.7/94.7) | Eight-miRNA panel (miR-320b, miR-6724–5p, miR-6877–5p, miR-4448, miR-4749–5p, miR-663a, miR-4651, and miR-6885–5p) | Early HCC detection from controls at-risk |
| Pratama et al., 2020 | N=86 HCC patients under treatment | 0.84 (72.0/75.0) | Four-miRNA panel (miR-4443, miR-4454, miR-4492, miR-4530) | HCC therapy response and disease-free survival |
| Fu et al., 2022 | N=76 HCC patients in validation set | 0.91 | Three lncRNA panel of AC005332.5, ELF3-AS1 and LINC00665 | HCC detection from healthy controls |
| Ning et al., 2023 | N=171 patients in validation set | 0.94 (84.0/86.0) | 6 cfRNA markers: 1 lncRNA (CYTOR), 1 miRNA (miR-21–5p), 3 cfRNA fragments (WDR74, SNORD89, RN7SL1), and 1 alternative splicing candidate (GGA2) | Early HCC detection HCCMDP Panel |
| Extracellular Vesicles | ||||
| Sun et al., 2020 | N=158 HCC patients | 0.93 (94.4/88.5) | EV-derived 10 mRNA signature (AFP, GPC3, ALB, APOH, FABP1, FGB, FGG, AHSG, RBP4, TF) | Early HCC detection from controls at risk |
| von Felden et al., 2021 | N=209 HCC patients | 0.87 (86.0/91.0) | 3-small RNA cluster | Early HCC detection from controls at risk |
| Rui et al., 2022 | N=124 HCC patients | 0.95 (89.0/92.0) | EV-derived 3 miRNA signature (miR-122–5p, let-7d-5p, and miR-425–5p) | Early HCC detection, classify HCC from non-tumor patients |
| Yang et al., 2022 | N=50 HCC patients | 0.97 (92.0/90.0) | EV-derived 3 miRNA signature (miR-26a, miR-29c, miR-199a) | Early HCC detection from controls at risk |
| Li et al., 2022 | N=60 HCC patients | 0.76 (48.2/93.9) | EV-cfDNA to detect c.747 G > T mutation in TP53 gene | Prediction of HCC and prognosticate microvascular invasion |
| Ye et al., 2022 | N=7 HCC patients | 0.86 (88.0/86.0) | EV-associated proteins (CO9, LBP, SVEP1, VWF) | HCC detection from controls at risk |
| Sun et al., 2023 | N=72 HCC patients (validation cohort) | 0.93 (94.0/81.0) | EpCAM+CD63+, CD147+CD63+, and GPC3+CD63+ EVs | Early HCC detection from controls at-risk |
Circulating tumour cells
Profiling CTCs was one of the initial liquid biopsy approaches in oncology. CTCs migrate from the primary tumour or metastatic site following tumour invasion into nearby vasculature, allowing for entry into systemic circulation where their quantity correlates with tumour burden. Also, profiling CTCs provides a unique window into active cellular processes directly from the tumour. Thus, CTCs may be restricted in their ability to detect early HCC but may be advantageous for monitoring recurrence or treatment response.82 In fact, a recent meta-analysis of 20 studies (1,191 total patients) demonstrated a 95% sensitivity and 60% specificity for CTC-based HCC diagnosis.82 There are many investigations utilising different technologies to isolate, enrich, and profile CTCs for the purpose of monitoring HCC recurrence and treatment response (Table 1). For example, Zhao et al. and Wei et al. profiled CTCs from over 200 patients with HCC preoperatively and were able to predict HCC recurrence with an AUC of 0.95 and 0.84, respectively.83,84 Additionally, Wang et al. demonstrated that transarterial chemoembolisation increased the time to recurrence and death in patients positive for CTCs, but showed no such effect in patients negative for CTCs.85 Additionally, Winograd et al. observed that PD-L1+ CTCs may be useful to predict ICI response.86 Thus, CTCs may hold promise for predicting minimal residual disease, treatment response, and recurrence.
The main challenge associated with using CTCs has been their heterogeneity within patients with HCC. Different investigations have described single-cell RNA sequencing of CTCs and found intra-CTC heterogeneity, likely representing the different origins and various biological factors influencing release under different microenvironmental pressures.87–89 Specifically, Sun et al. utilised single-cell RNA sequencing on CTCs to demonstrate that CCL5 from CTCs recruits regulatory T cells to enable immune evasion and metastasis.89 Thus, improved understanding of CTC biology and their spatial relationships could help to determine which subpopulations provide the most prognostic value.
Cell-free DNA
Cell-free DNA (cfDNA) refers to the large pool of circulating double-stranded DNA fragments associated with nucleosomes, while circulating tumour DNA (ctDNA) refers to smaller (150 bp) fragments transiently present in bodily fluids (<2 h) released by apoptotic, necrotic, or actively proliferating tumour cells. Traditionally, cfDNA has been probed for total quantity, integrity, and copy number alterations,90 while more novel approaches probe for methylation patterns, particularly at CpG islands of tumour suppressor genes, or mutational signatures reflecting the patient’s current “oncologic footprint”. These two cellular processes are closely intertwined in HCC as mutations in chromatin remodelling genes – ARID1A (13%) and ARID2 (7%), which are key constituents of SWI/SNF (SWItch/sucrose non-fermentable) chromatin remodelling components – are frequently observed, along with hypermethylation of CpG islands and hypomethylation in open sea regions.91
Genomic profiling has identified mutations in TERT, CTNNB1, and TP53 as the major drivers in HCC, with various groups attempting to profile these mutations in cfDNA for HCC detection and prognosis (Table 1). Therefore, cfDNA from patients at risk is likely to contain these mutations and could be used to identify early lesions given their truncal role. Seminal investigations have indicated that TERT promoter and TP53 mutations are present in cfDNA samples in >75% of patients with early-stage HCC.92,93 Specifically, for diagnostic detection, Qu et al. developed a panel which profiles the major trunk mutations, HBV integration breakpoints, clinical variables, and protein markers to diagnose early HCC with 100% sensitivity and 94% specificity.94 Additionally, analysing the cfDNA levels of the TERT promoter, NRAS, NFE2L2, and MET mutations may predict long-term treatment outcomes.95,96 However, despite the importance of detecting molecular pathways implicated in disease progression, no precision medicines have been approved for first-line HCC treatment yet.
Various groups have defined different epigenetic changes in cfDNA of patients with HCC as a diagnostic tool (Table 1). Detecting methylation signatures may provide improved diagnostic specificity due to the inherent tissue specificity of methylation signatures, along with convenience in quantitating aberrant DNA methylation compared to mutation profiling. Different approaches have been utilised to detect early HCC, including profiling known HCC-associated methylated DNA markers (e.g., HOXA1, EMX1, TSPYL5, SEPT9, ECE1, PFKP, CLEC11A),77,97 alongside clinicodemographic factors (GALAD score),98 or profiling bisulfite-converted cfDNA in an unbiased fashion.99,100 These early studies were limited due to a lack of tissue specificity and high cost. Thus, Cheishvilli et al. utilised publicly available DNA methylation datasets of tumour and normal tissues to define an HCC-specific methylation signature (“epiLiver”) which achieved 84.5% sensitivity and 95% specificity.101
In addition to profiling the mutation and methylation patterns of cfDNA, various groups have begun to integrate cfDNA fragmentation with methylation and mutation signatures.102 cfDNA “fragmentomes” refer to the entire map of circulating cfDNA fragments. Circulating cfDNA is highly fragmented with base pair sizes depending on nucleosome packing. This “fingerprint” provides insights into tissue-specific origins, e.g. patients with HCC show more 4-mer end motifs.103 Early work demonstrated that cfDNA fragmentomes can be used to detect early HCC in at-risk patients (AUC 0.995; 96.8% sensitivity and 98.8% specificity).103,104 This technology has also been shown to detect HBV-related HCC with 87.1% sensitivity and 88.4% specificity.105 More intriguingly, the cfDNA fragmentome can also be probed in an unbiased manner to detect novel CpG methylation patterns which could be used to classify patients with HCC.106 Recently, Wang et al. demonstrated the utility of a multiplex cfDNA mutation + methylation profiling technology, which was validated in a prospective cohort of 311 asymptomatic patients with HBV, demonstrating 80% sensitivity and 94% specificity for the detection of early HCC.107 Lastly, Foda et al. developed an approach to infer genomic alterations driving HCC tumorigenesis from cfDNA and were able to detect HCC with 85% sensitivity and 80% specificity in a large-population of patients at high risk.108
Overall, the main limitation of using cfDNA is that there is a strong correlation between quantity of ctDNA and lesion size, suggesting early-stage lesions are more difficult to detect than advanced disease.109 Therefore, cfDNA may be a better tool for detecting recurrence and/or treatment response and resistance.
Non-coding RNA species
Non-coding RNAs (ncRNAs), such as microRNAs (miRNAs) (25–30 bp) and long non-coding RNAs (>200 bp) are not translated into protein but instead act as important regulatory elements involved in chromatin alterations, DNA transcription, and chromosomal looping. ncRNAs are also involved in diverse cellular processes implicated in cancer pathogenesis, including cellular proliferation, migration, invasion, and cell death. Their stability in the bloodstream has made them attractive candidates for liquid biopsy, along with their tissue-specific expression and ability to distinguish cancer from premalignancy.110 Various groups have defined panels of miRNAs (e.g., miR-21, miR-26a, miR-27a, miR-29a, miR-29c, miR-122, miR-133a, miR-143, miR-145, miR-192, miR-223, miR-505, and miR-801) which have demonstrated the ability to detect early-stage HCC,111–113 predict overall114 and disease-free survival,115 and prognosticate following systemic therapy116 (Table 1). Other groups have utilised long non-coding RNA panels (e.g., AC005332.5, ELF3-AS1 and LINC00665) for diagnosis and prognostication in HBV-related HCC.117 However, the main challenges associated with translating an HCC ncRNA-based test have been the lack of standardised reporting methods, variation in RNA detection techniques, and incomplete data on their contribution to HCC.
Extracellular vesicles
Extracellular vesicles (EVs) are lipid bilayer nanovesicles (50 nm to >1,000 nm in size) spontaneously produced by nearly all mammalian cells and released into the extracellular milieu where they can travel systemically to other organs. The main subclasses of EVs include exosomes, microvesicles, and apoptotic vesicles, with each containing their own distinct nucleic acid species (cfDNA, cfRNA), proteins, and lipids. These cargo molecules participate in diverse cell-to-cell signalling circuits, thereby mediating pathophysiological states.118 EVs have become attractive liquid biopsy analytes given their cargo reflects the genomic and transcriptomic states of their parental cells of origin.119 Various groups have defined EV-associated nucleic acid and proteomic signatures either alone or in combination to detect HCC and monitor treatment responses120,121 (Table 1). For example, EV-derived miRNA panels can classify HCC with >90% sensitivity and specificity.120,122 Also, a meta-analysis of 16 studies determined that exosomal ncRNAs, particularly downregulated miRNAs, can prognosticate disease-free survival in HCC.123 EVs also contain unannotated small RNA clusters which can classify HCC from controls at risk with 86% sensitivity and 91% specificity.124 Additionally, Sun et al. demonstrated that HCC-specific EV subpopulations can be probed to detect early-stage HCC with 94% sensitivity and 84% specificity through profiling epithelial cell adhesion molecule+, CD147+, and GPC3+ EVs.125 Moreover, EV-specific proteins have been shown to detect HCC and predict response to sorafenib +/− radiotherapy.126,127 Lastly, TP53 mutations detected in EV-derived cfDNA have been used to predict microvascular invasion.19
Despite being able to probe for multiple molecular targets in one isolate, there are several limitations associated with utilising circulating EVs for liquid biopsy, including tissue specificity and purity. First, without a method to capture tumour-specific EVs, the cargo molecules isolated from EVs may be derived from other tissue sources, including non-malignant tissues, which spontaneously release EVs into the systemic circulation. Second, best practices for isolating EVs from biofluids are not well defined and can be labour intensive, costly, and result in impurities. However, emerging nanotechnologies may circumvent issues in EV isolation, detection, enumeration, and analyte profiling.128
Update on translation of liquid biopsy to the clinic
Overview of multi-panel analyte-based tests for HCC
In this section, we discuss advanced technologies that employ multi-analyte panels for diagnosing and managing HCC, highlighting their potential and discussing their efficacy. Although these liquid biopsy systems, which identify DNA alterations or hypermethylation and often pair genomic data with protein markers or patient demographics, have not yet become a standard part of clinical practice, a few such platforms have received FDA approval as breakthrough diagnostic tests in recent years (Table 2).
Table 2.
Commercial Liquid Biopsy Platforms for Detection and Treatment Stratification in HCC
| Assay | Analyte(s) Assessed | Intended Purpose | AUC (Sensitivity/Specificity) | Comments and Clinical Studies |
|---|---|---|---|---|
| Elecsys®GALAD | Protein markers: AFP, AFP-L3, DCP; gender; age | Early HCC detection | Overall: 0.947 (85.8/90.8) Early-stage: 0.913 (73.8/90.8) |
FDA Breakthrough Device designation in 2022 |
| Oncoguard® Liver | Methylation markers: HOXA1, TSPYLS, B3GALT6; AFP; sex | Early HCC detection | Overall: 0.94 (88/87) Early-stage: 0.92 (82/87) |
NCT05064553 (ongoing) |
| HelioLiver™ | Methylation markers: 77 CpG sites in 28 genes; protein markers: AFP, AFP-L3, DCP; patient demographics | Early HCC detection | Overall: 0.944 (85.2/91.2) Early-stage: 0.924 (75.7/91.2) |
NCT05059665 (completed); NCT05053412, NCT03694600, NCT04539717 (ongoing) |
| HCCscreen™ | Methylation markers; genomic alterations (SNVs, indels, translocations, viral integration sites) | Early HCC detection and mutation profiling | Overall: 0.93 (90/94) Early-stage: 0.95 (91/95) |
FDA Breakthrough Device designation in 2020 |
| HCCBloodTest | Methylated SEPT9 | Early HCC detection | Overall: 0.93 (85.1/91.07) Early-stage: 0.863 (72.73/86.39) |
|
| Guardant360® | SNVs, indels, CNV, fusion events in 68 genes | Treatment stratification in advanced HCC and mutation profiling | N/A; 79% of patients had potentially actionable alterations | Point mutations: TP53, CTNNB1, PTEN, CDKN2A, ARID1A, MET; amplifications: CDK6, MYC, BRAF, RAF1, FGFR1, CCNE1, PIK3CA, HER2 |
Multi-panel scores based on protein marker analysis
The Elecsys-GALAD score is derived by combining gender and age with an immunoassay that measures serum markers AFP, AFP-L3, and DCP (also known as protein-induced by vitamin K absence-II, or PIVKA-II). A multicentre study comparing the performance of the Elecsys PIVKA-II and Elecsys AFP assays in diagnosing HCC determined that the PIVKA-II assay had an overall sensitivity of 86.9% (95% CI 80.8–91.6) and specificity of 83.7% (77.9–88.4), while the AFP assay achieved an overall sensitivity of 51.8% (44.0–59.5) and specificity of 98.1% (95.1–99.5). When combining both assays, the overall sensitivity for detecting HCC increased to 92% with a specificity of 82%, outperforming the two individual assays.129 A scoring algorithm based on GALAD received a Breakthrough Device designation from the FDA in 2020 for use as an in vitro diagnostic. A large meta-analysis of 15 original studies of 19,021 patients showed that the GALAD score had a pooled sensitivity of 0.82 (95% CI 0.78–0.85) for any-stage HCC and 0.73 (0.66–0.79) for early-stage HCC, easily surpassing the 38% detection rate of AFP alone for early-stage HCC.130 Thus, the robust sensitivity and efficiency of the Elecsys-GALAD score positions it as a promising surveillance tool for HCC. Lastly, the Elecsys-GAAD is another multi-panel diagnostic test for early-stage HCC which incorporates gender and age with immunoassays for just two biomarkers instead of three, AFP and PIVKA-II.131 The performances of Elecsys-GAAD and Elecsys-GALAD are similar, suggesting that the AFP-L3 assay is only a minor contributor towards HCC detection. We await further validation of these results.
Combining insights from cfDNA methylation alterations and protein markers
Other technologies for HCC diagnosis integrate cfDNA methylation state, DNA alterations, and serum marker levels. One such platform, Oncoguard Liver, detects early-stage HCC through a multi-target blood test that measures a panel of methylation sites and serum markers to produce a qualitative result. In a 2021 study of this platform, the panel included four methylation markers (HOXA1, TSPYLS, EMX1, and the reference marker B3GALT6), AFP, and AFP-L3. With these values, the test had an overall sensitivity of 80% (72–86) and early-stage sensitivity of 71% (60–81) at a specificity of 90%, showing greater sensitivity than AFP, AFP-L3, DCP, and GALAD.77 A later study modified the Oncoguard Liver panel by removing EMX1 and AFP-L3 and considering patient sex. In a cohort of 156 patients with HCC and 245 control patients, the overall sensitivity was 88% (82–92) and early-stage sensitivity was 82% (72–89) at a specificity of 87% (82–91). Furthermore, the test maintained its performance across multiple liver aetiologies.77,132 Following the results of this study, the ALTernative to Ultrasound (ALTUS) study (NCT05064553) was initiated to measure the performance of Oncoguard Liver as a diagnostic tool in patients undergoing HCC surveillance with ultrasound or CT/MRI. This study aims to enrol 3,000 patients across 60 sites in the US and will allow for large-scale, longitudinal assessment of the multi-target blood test.133
Another diagnostic platform is HelioLiver, a multi-analyte test designed for monitoring patients at risk of HCC, especially those with cirrhosis, chronic viral hepatitis, and metabolic dysfunction-associated steatotic liver disease.133,134 The test targets 77 methylation sites in 28 target genes using next-generation sequencing libraries and measures AFP, AFP-L3, and DCP from immunoassays. Along with patient demographics, these data are used in a diagnostic algorithm to produce a qualitative test result. The Performance Evaluation of HelioLiver™ Test for Detection of HCC (ENCORE) (NCT05059665), a prospective phase II biomarker study, evaluated HelioLiver in a cohort of 122 patients with HCC and 125 control patients with benign liver disease. The test achieved an overall sensitivity of 85.2% (77.8–90.4) and specificity of 91.2% (84.9–95.0), with an early-stage sensitivity of 76% (59.9–86.7), demonstrating greater sensitivity than AFP, AFP-L3, DCP, and GALAD for both all-stage HCC and early-stage disease.98 Other than ENCORE, three other clinical studies of HelioLiver have been initiated, two of which aim to compare the test results with ultrasound or MRI (NCT05053412, NCT03694600, NCT04539717). Insights from these studies will help decipher the potential role of HelioLiver in clinical applications.
HCCscreen analyses both cfDNA methylation state and genomic alterations, such as single nucleotide variants, indels (insertions and deletions), translocation, and viral genome integration.135 It uses mutation capsule technology, which simultaneously identifies alterations in both DNA sequence and methylation state through highly sensitive multiplex reactions, resulting in a molecular profile of cancer.94 In 2020, the FDA designated HCCscreen as a Breakthrough Device to accelerate its approval process. A later study by Wang et al. assessed HCCscreen in a cohort of 436 patients with HBV, 148 of whom had HCC. Using a panel of methylation markers and alterations prevalent in HCC tumours, the group constructed an HCC detection algorithm, which demonstrated an overall sensitivity of 90% (0.79–0.96) and specificity of 94% (0.90–0.97). The combined methylation marker and alteration panel performed better than AFP and either the methylation or alteration panels alone in detecting HCC.107 Expanding the clinical application of HCCscreen, an early HCC detection programme has been ongoing in China, where 150,000 tests will be administered in local communities over 3 years.
Some platforms are harnessing the methylation state of cfDNA alone to determine the presence of HCC. One such technology is HCCBloodTest, which assesses hypermethylation of the SEPT9 (Septin 9) gene, a driver of HCC carcinogenesis, in bisulfite-converted DNA derived from cfDNA in plasma. As bisulfite treatment of DNA converts unmethylated cytosine to uracil sulfonate and does not alter 5-methylcytosine, bisulfite-converted DNA can be used to ascertain the degree of methylated SEPT9 after PCR amplification, which is converted to a qualitative result regarding HCC status.136 In an initial observational study, HCCBloodTest achieved a sensitivity of 94.1% (83.8–98.8) and specificity of 94.4 (77.2–90.1), while a subsequent case-control study with age- and gender-matched patients reported similar results, with a sensitivity of 85.1% (71.1–93.8) and specificity of 91.07% (80.4–97.0).137 In both cases, HCCBloodTest had significantly higher accuracy than AFP for detecting HCC. Notably, there was a difference in the specificity of HCCBloodTest between cirrhosis aetiologies, as the specificity ranged from 86.4% for viral hepatitis-associated cirrhosis to 39.4% for metabolic dysfunction-associated steatohepatitis-associated cirrhosis.137 Though the cause for the varying specificity was uncertain, HCCBloodTest should continue to be validated in different patient populations.
Liquid biopsy based on cfDNA-targeted exome sequencing
Although most plasma- or serum-based diagnostics for HCC are intended to aid early detection, this technology can also be used for treatment stratification. Guardant360 CDx is the first FDA-approved comprehensive liquid biopsy for all advanced solid tumours.138 The approved assay utilises next-generation sequencing and profiles single nucleotide variants and indels in 55 genes, copy number variants in two genes, and fusion events in four genes, though alterations in other genes have been identified with the assay. In addition, Guardant360 CDx has been approved as a companion diagnostic for breast cancer and non-small cell lung cancer (NSCLC) to identify whether patients’ mutation profiles are suitable for targeted treatments. In a study by Bauml et al., Guardant360 CDx was used to determine if patients with NSCLC had a KRAS driver mutation that could be targeted with sotorasib, a small molecule inhibitor. Out of 109 patients who were successfully tested, 78 patients were positive for the mutation and began sotorasib therapy, achieving a 36.4% (25.7–48.1) objective response rate.139 The study also assessed agreement between plasma testing with Guardant360 CDx and tissue analysis with PCR, determining that the overall percent agreement was 0.82 (0.76–0.87).139
While targeted therapies for driver mutations in HCC have yet to become the standard of care, this study in patients with NSCLC provides a framework for the potential role of liquid biopsies in stratifying HCC treatment as more precision therapies are developed. In fact, a study by Ikeda et al. evaluated 14 patients with advanced HCC using Guardant360, which features an expanded panel of 68 oncogenes and tumour suppressor genes, to identify actionable alterations. The assay identified somatic alterations in all patients, with 79% of patients having a potentially actionable alteration. Five patients received treatment based on the assay results. One patient with an early-stage HCC began treatment with cabozantinib, a kinase inhibitor, after ctDNA analysis revealed a mutation in the proto-oncogene MET. After 8 weeks, a second ctDNA analysis showed this mutation had disappeared, suggesting a response to therapy.140 As more precision therapies are developed and applied to actionable mutations in HCC, liquid biopsies are expected to play a greater role not only in aiding early diagnosis, but also in selecting treatment, monitoring response, and examining tumour evolution.
There are numerous other multi-cancer detection tests under rigorous evaluation that promise to revolutionise cancer diagnostics. Notable among them are the methylation based MCED test,141 CancerSEEK,142 and FoundationOneCDx (F1CDx),143 to name a few. It is worth noting that, just in 2022, over 200 clinical trials examining a diverse range of liquid biopsy biomarkers were initiated. These continuous advances in liquid biopsy technology, coupled with the sheer volume of ongoing research, signify a promising future for non-invasive cancer detection and management.
Challenges in translating liquid biopsy to the clinic
The rising use of liquid biopsy in liver cancer care showcases its potential, yet several challenges remain, of which we would like to highlight three. First, the majority of liquid biopsy tests still lack robust evidence supporting their clinical validity and utility, relegating their use to research settings. This challenge is likely to be answered by the various ongoing multicentre clinical studies. Second, many of these assays either focus on a single analyte or use a multiparametric approach, but do not have easy scoring criteria. The application of advanced statistical tools employing high-dimensional machine learning techniques can potentially overcome this challenge. The third challenge is the substantial variations in assay results, which complicate clinical interpretations. Homogenising analyte preparatory procedures and standardising reporting can allow for the comparison or integration of results from diverse studies. Collaborative data sharing is equally vital to further investigate integration of liquid biopsy assays into clinical workflows.
Conclusions and future perspectives
Molecular diagnostics, whether through tissue or liquid biopsy, are likely to play a larger role in the management of patients with HCC in the near future. We highlight four specific areas where tissue and/or liquid biopsy can potentially make an impact (Fig. 2). The first is in HCC surveillance. The limitations of imaging surveillance and the challenges with patient adherence to surveillance intervals underscore the appeal of a non-invasive blood-based test that can potentially be employed by primary care physicians and community practices. While liquid biopsy must address current sensitivity, accessibility, and cost issues, it has the potential to complement, if not replace, imaging-based surveillance. Moreover, it can aid in establishing a diagnosis for indeterminate nodules and obviate the need for invasive tissue biopsies. Second, in early-stage HCC where patients undergo surgery or resection, tissue-informed liquid biopsy tests can play a crucial role in detecting minimal residual disease and monitoring for recurrence, thus identifying individuals who may benefit from adjuvant therapies. Third, in patients with intermediate- or advanced-stage HCC receiving targeted or immune-based therapies, biomarker expression analysis from both tissue and liquid biopsy can facilitate the selection of appropriate candidates for specific treatments, leading to improved response rates and reduced side effects. Lasty, the utility of liquid biopsy extends to monitoring the response to therapy at all stages of HCC, making biopsy-based molecular tests a valuable tool in the comprehensive management of this complex disease. In conclusion, the advent of molecular diagnostics through tissue or liquid biopsy heralds a promising era in HCC diagnosis and management, offering sensitive and versatile tools that can be employed across all stages of the disease.
Fig. 2. Potential future applications of liquid or tissue biopsy-based molecular diagnostic tests in HCC.
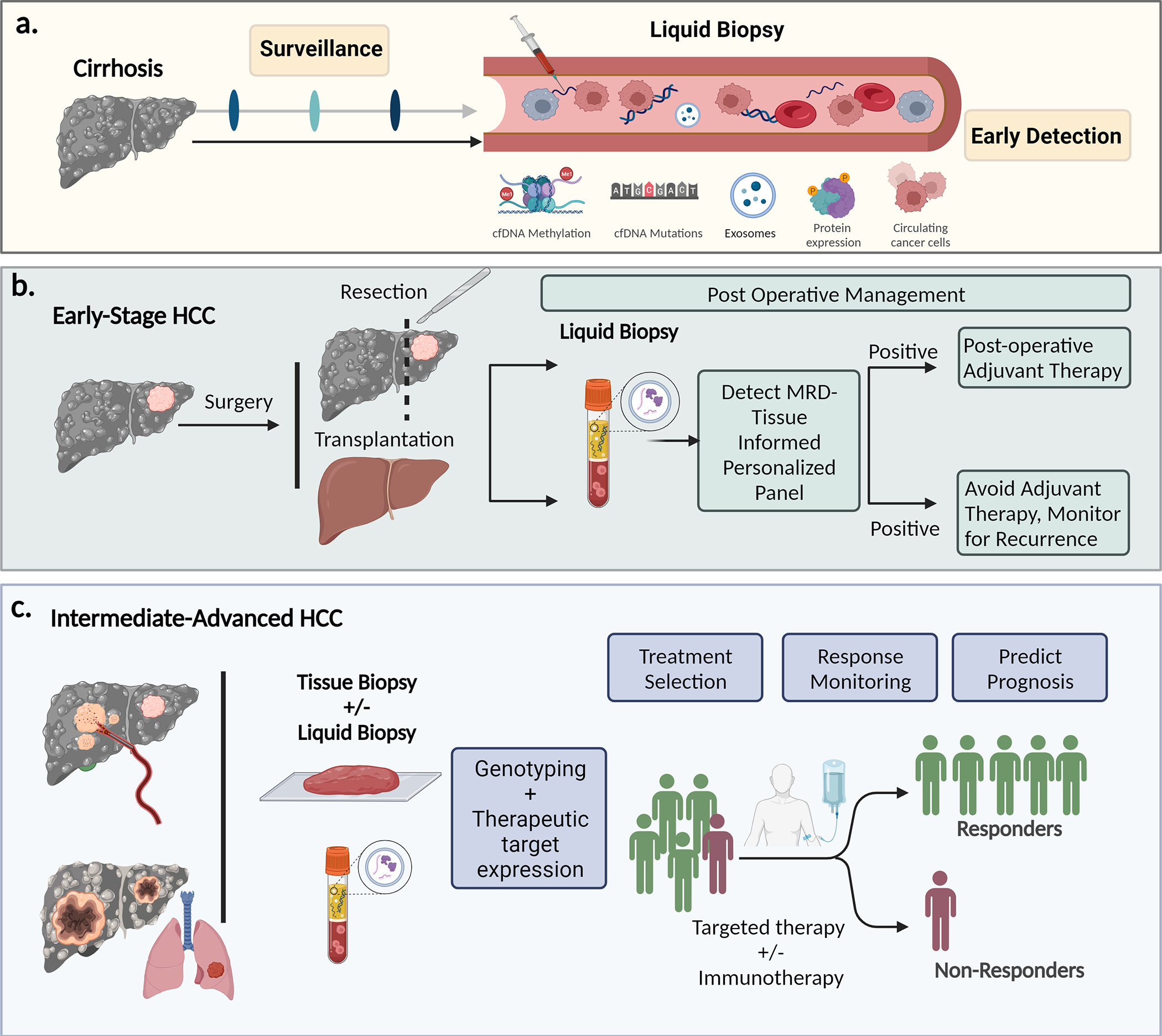
cfDNA, cell-free DNA; CTCs, circulatory tumour cells; EVs, extracellular vesicles; HCC, hepatocellular carcinoma; MRD, minimal residual disease.
Supplementary Material
Keypoints.
HCC is often diagnosed at advanced stages and is associated with poor clinical outcomes, highlighting the critical need for access to molecular insights through either tissue or liquid biopsy.
Utilising this molecular data can pave the way for reliable biomarker development, thereby improving early detection and refining treatment strategies for HCC.
While questions regarding the need for routine HCC biopsies persist, increasing the frequency of tissue biopsies, especially in clinical trials, could expedite the discovery of novel biomarkers and personalised treatments.
Liquid biopsy is transforming cancer care by enabling non-invasive, real-time tumour monitoring and early detection of circulating tumour cells, cell-free DNA, non-coding RNAs, and extracellular vesicles.
Multi-analyte panels for HCC, which combine genomic data with protein markers or patient demographics, are gaining traction, with some recently receiving FDA breakthrough diagnostic test approval.
With the increase in biomarker-stratified trials, tissue biopsies may become pivotal in matching patients to appropriate treatments, and liquid biopsies are likely to play complementary roles in early HCC detection and monitoring of treatment response.
Financial support
This work was supported by NIH grants 1R01CA251155, 1R01CA250227, and Endowed Chair for Experimental Pathology to SPM. This work was also supported in part by NIH grants 1T32EB001026 and 1F30CA284540 to BML. National Institutes of Health (NIH) grant CA222676 from the National Cancer Institute (NCI) (to R.D.).
Abbreviations
- AFP
alpha-fetoprotein
- AFP-L3
fucosylated fraction of AFP
- ARID
AT-rich interactive domain-containing protein
- cfDNA
cell-free DNA
- CTCs
circulating tumour cells
- ctDNA
circulating tumour DNA
- DCP
des-gamma carboxyprothrombin
- EVs
extracellular vesicles
- GPC-3
glypican-3
- HCC
hepatocellular carcinoma
- miRNA/miR
microRNA
- ncRNA
non-coding RNA
- NSCLC
non-small cell lung cancer
- PD-L1
programmed death ligand 1
- TERT
telomerase reverse transcriptase
- TGFβ
transforming growth factor-β
Footnotes
Conflict of interest
The authors of this study declare that they do not have any conflict of interest.
Please refer to the accompanying ICMJE disclosure forms for further details.
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhep.2023.11.030.
References
Author names in bold designate shared co first authorship
- [1].Rumgay H, Arnold M, Ferlay J, et al. Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol 2022;77:1598–1606. 10.1016/j.jhep.2022.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2021;7:6. 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- [3].Dhanasekaran R, Suzuki H, Lemaitre L, et al. Molecular and immune landscape of hepatocellular carcinoma to guide therapeutic decision making. Hepatology 2023. 10.1097/HEP.0000000000000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu, cancer genome atlas research network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017;169:1327–1341.e23. 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012;44:694–698. 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Schulze K, Imbeaud S, Letouzé E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505–511. 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Totoki Y, Tatsuno K, Covington KR, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014;46:1267–1273. 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- [8].Yu JI, Choi C, Ha SY, et al. Clinical importance of TERT overexpression in hepatocellular carcinoma treated with curative surgical resection in HBV endemic area. Sci Rep 2017;7:12258. 10.1038/s41598-017-12469-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li X, Xu W, Kang W, et al. Genomic analysis of liver cancer unveils novel driver genes and distinct prognostic features. Theranostics 2018;8:1740–1751. 10.7150/thno.22010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Russell JO, Monga SP. Wnt/β-Catenin signaling in liver development, homeostasis, and pathobiology. Annu Rev Pathol 2018;13:351–378. 10.1146/annurev-pathol-020117-044010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xu C, Xu Z, Zhang Y, et al. β-Catenin signaling in hepatocellular carcinoma. J Clin Invest 2022:132. 10.1172/JCI154515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ruiz de Galarreta M, Bresnahan E, Molina-Sánchez P, et al. β-Catenin activation promotes immune escape and resistance to anti-PD-1 therapy in hepatocellular carcinoma. Cancer Discov 2019;9:1124–1141. 10.1158/2159-8290.CD-19-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Harding JJ, Nandakumar S, Armenia J, et al. Prospective genotyping of hepatocellular carcinoma: clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clin Cancer Res 2019;25:2116–2126. 10.1158/1078-0432.CCR-18-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Neely J, Yao J, Kudo M, et al. Abstract 2145: genomic and transcriptomic analyses related to the clinical efficacy of first-line nivolumab in advanced hepatocellular carcinoma from the phase 3 CheckMate 459 trial. Cancer Res 2022;82:2145. 10.1158/1538-7445.am2022-2145.2145. [DOI] [Google Scholar]
- [15].Zhu AX, Abbas AR, de Galarreta MR, et al. Molecular correlates of clinical response and resistance to atezolizumab in combination with bevacizumab in advanced hepatocellular carcinoma. Nat Med 2022;28:1599–1611. 10.1038/s41591-022-01868-2. [DOI] [PubMed] [Google Scholar]
- [16].Kancherla V, Abdullazade S, Matter MS, et al. Genomic analysis revealed new oncogenic signatures in -mutant hepatocellular carcinoma. Front Genet 2018;9:2. 10.3389/fgene.2018.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ma Z, Guo D, Wang Q, et al. Lgr5-mediated p53 Repression through PDCD5 leads to doxorubicin resistance in Hepatocellular Carcinoma. Theranostics 2019;9:2967–2983. 10.7150/thno.30562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Villanueva A, Hoshida Y. Depicting the role of TP53 in hepatocellular carcinoma progression. J Hepatol 2011;55:724–725. 10.1016/j.jhep.2011.03.018. [DOI] [PubMed] [Google Scholar]
- [19].Li Y, Wu J, Li E, et al. TP53 mutation detected in circulating exosomal DNA is associated with prognosis of patients with hepatocellular carcinoma. Cancer Biol Ther 2022;23:439–445. 10.1080/15384047.2022.2094666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sun J, Cheng N-S. Comprehensive landscape of ARID family members and their association with prognosis and tumor microenvironment in hepatocellular carcinoma. J Immunol Res 2022;2022:1688460. 10.1155/2022/1688460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dhanasekaran R, Deutzmann A, Mahauad-Fernandez WD, et al. The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol 2022;19:23–36. 10.1038/s41571-021-00549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gonzalez-Sanchez E, Vaquero J, Férnandez-Barrena MG, et al. The TGF-β pathway: a pharmacological target in hepatocellular carcinoma? Cancers 2021;13. 10.3390/cancers13133248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee K, Kim S, Lee Y, et al. The clinicopathological and prognostic significance of Nrf2 and Keap1 expression in hepatocellular carcinoma. Cancers 2020;12. 10.3390/cancers12082128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sun EJ, Wankell M, Palamuthusingam P, et al. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Biomedicines 2021;9. 10.3390/biomedicines9111639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li H, Li X, Liu S, et al. Programmed cell death-1 (PD-1) checkpoint blockade in combination with a mammalian target of rapamycin inhibitor restrains hepatocellular carcinoma growth induced by hepatoma cell-intrinsic PD-1. Hepatology 2017;66:1920–1933. 10.1002/hep.29360. [DOI] [PubMed] [Google Scholar]
- [26].Calderaro J, Ziol M, Paradis V, et al. Molecular and histological correlations in liver cancer. J Hepatol 2019;71:616–630. 10.1016/j.jhep.2019.06.001. [DOI] [PubMed] [Google Scholar]
- [27].Kurebayashi Y, Ojima H, Tsujikawa H, et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology 2018;68:1025–1041. 10.1002/hep.29904. [DOI] [PubMed] [Google Scholar]
- [28].Brown ZJ, Tsilimigras DI, Ruff SM, et al. Management of hepatocellular carcinoma: a review. JAMA Surg 2023;158:410–420. 10.1001/jamasurg.2022.7989. [DOI] [PubMed] [Google Scholar]
- [29].Ducreux M, Abou-Alfa GK, Bekaii-Saab T, et al. The management of hepatocellular carcinoma. In: Current expert opinion and recommendations derived from the 24th ESMO/World Congress on Gastrointestinal Cancer, Barcelona, 2022. ESMO Open; 2023. p. 101567. 10.1016/j.esmoop.2023.101567.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Benson AB, D’Angelica MI, Abbott DE, et al. Hepatobiliary cancers, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2021;19:541–565. 10.6004/jnccn.2021.0022. [DOI] [PubMed] [Google Scholar]
- [31].Llovet JM, De Baere T, Kulik L, et al. Locoregional therapies in the era of molecular and immune treatments for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2021;18:293–313. 10.1038/s41575-020-00395-0. [DOI] [PubMed] [Google Scholar]
- [32].Matsuo Y Stereotactic body radiotherapy for hepatocellular carcinoma: a brief overview. Curr Oncol 2023;30:2493–2500. 10.3390/curroncol30020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med 2020;382:1894–1905. 10.1056/NEJMoa1915745. [DOI] [PubMed] [Google Scholar]
- [34].Abou-Alfa GK, Lau G, Kudo M, et al. Tremelimumab plus durvalumab in unresectable hepatocellular carcinoma. NEJM Evid 2022;1. 10.1056/evidoa2100070. [DOI] [PubMed] [Google Scholar]
- [35].Marrero JA, Kulik LM, Sirlin CB, et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American Association for the Study of Liver Diseases. Hepatology 2018;68:723–750. 10.1002/hep.29913. [DOI] [PubMed] [Google Scholar]
- [36].European Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu, European association for the study of the liver. EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2018;69:182–236. 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- [37].Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: new trends. J Hepatol 2020;72:250–261. 10.1016/j.jhep.2019.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tzartzeva K, Obi J, Rich NE, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology 2018;154:1706–17018.e1. 10.1053/j.gastro.2018.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Meloni MF, Francica G, Chiang J, et al. Use of contrast-enhanced ultrasound in ablation therapy of HCC: planning, guiding, and assessing treatment response. J Ultrasound Med 2021;40:879–894. 10.1002/jum.15471. [DOI] [PubMed] [Google Scholar]
- [40].Lee YJ, Lee JM, Lee JS, et al. Hepatocellular carcinoma: diagnostic performance of multidetector CT and MR imaging-a systematic review and meta-analysis. Radiology 2015;275:97–109. 10.1148/radiol.14140690. [DOI] [PubMed] [Google Scholar]
- [41].Roberts LR, Sirlin CB, Zaiem F, et al. Imaging for the diagnosis of hepatocellular carcinoma: a systematic review and meta-analysis. Hepatology 2018;67:401–421. 10.1002/hep.29487. [DOI] [PubMed] [Google Scholar]
- [42].Terzi E, Ayuso C, Piscaglia F, et al. Liver imaging reporting and data system: review of pros and cons. Semin Liver Dis 2022;42:104–111. 10.1055/s-0041-1732356. [DOI] [PubMed] [Google Scholar]
- [43].Lencioni R, Llovet JM. Modified RECIST (mRECIST) assessment for hepatocellular carcinoma. Semin Liver Dis 2010;30:52–60. 10.1055/s-0030-1247132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Vogel A, Cervantes A, Chau I, et al. Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018;29:iv238–i255. 10.1093/annonc/mdy308. [DOI] [PubMed] [Google Scholar]
- [45].Torbenson MS. Morphologic subtypes of hepatocellular carcinoma. Gastroenterol Clin North Am 2017;46:365–391. 10.1016/j.gtc.2017.01.009. [DOI] [PubMed] [Google Scholar]
- [46].Torbenson MS. Hepatocellular carcinoma: making sense of morphological heterogeneity, growth patterns, and subtypes. Hum Pathol 2021;112:86–101. 10.1016/j.humpath.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Piccinino F, Sagnelli E, Pasquale G, et al. Complications following percutaneous liver biopsy. A multicentre retrospective study on 68,276 biopsies. J Hepatol 1986;2:165–173. 10.1016/s0168-8278(86)80075-7. [DOI] [PubMed] [Google Scholar]
- [48].Di Tommaso L, Spadaccini M, Donadon M, et al. Role of liver biopsy in hepatocellular carcinoma. World J Gastroenterol 2019;25:6041–6052. 10.3748/wjg.v25.i40.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Silva MA, Hegab B, Hyde C, et al. Needle track seeding following biopsy of liver lesions in the diagnosis of hepatocellular cancer: a systematic review and meta-analysis. Gut 2008;57:1592–1596. 10.1136/gut.2008.149062. [DOI] [PubMed] [Google Scholar]
- [50].Szpakowski J-L, Drasin TE, Lyon LL. Rate of seeding with biopsies and ablations of hepatocellular carcinoma: a retrospective cohort study. Hepatol Commun 2017;1:841–851. 10.1002/hep4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Maturen KE, Nghiem HV, Marrero JA, et al. Lack of tumor seeding of hepatocellular carcinoma after percutaneous needle biopsy using coaxial cutting needle technique. AJR Am J Roentgenol 2006;187:1184–1187. 10.2214/AJR.05.1347. [DOI] [PubMed] [Google Scholar]
- [52].Scott EC, Baines AC, Gong Y, et al. Trends in the approval of cancer therapies by the FDA in the twenty-first century. Nat Rev Drug Discov 2023. 10.1038/s41573-023-00723-4. [DOI] [PubMed] [Google Scholar]
- [53].Thein KZ, Lemery SJ, Kummar S. Tissue-agnostic drug development: a new path to drug approval. Cancer Discov 2021;11:2139–2144. 10.1158/2159-8290.CD-21-0554. [DOI] [PubMed] [Google Scholar]
- [54].Zhu AX, Kang Y-K, Yen C-J, et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased a-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2019;20:282–296. 10.1016/S1470-2045(18)30937-9. [DOI] [PubMed] [Google Scholar]
- [55].Limousin W, Laurent-Puig P, Ziol M, et al. Molecular-based targeted therapies in patients with hepatocellular carcinoma and hepato-cholangiocarcinoma refractory to atezolizumab/bevacizumab. J Hepatol 2023. 10.1016/j.jhep.2023.08.017. [DOI] [PubMed] [Google Scholar]
- [56].Yau T, Park J-W, Finn RS, et al. Nivolumab versus sorafenib in advanced hepatocellular carcinoma (CheckMate 459): a randomised, multicentre, open-label, phase 3 trial. Lancet Oncol 2022;23:77–90. 10.1016/S1470-2045(21)00604-5. [DOI] [PubMed] [Google Scholar]
- [57].Verset G, Borbath I, Karwal M, et al. Pembrolizumab monotherapy for previously untreated advanced hepatocellular carcinoma: data from the open-label, phase II KEYNOTE-224 trial. Clin Cancer Res 2022;28:2547–2554. 10.1158/1078-0432.CCR-21-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].El-Khoueiry AB, Yau T, Kang Y-K, et al. Nivolumab (NIVO) plus ipilimumab (IPI) combination therapy in patients (Pts) with advanced hepatocellular carcinoma (aHCC): long-term results from CheckMate 040. J Clin Oncol 2021;39:269. 10.1200/JCO.2021.39.3_suppl.269.269.33275488 [DOI] [Google Scholar]
- [59].Davis AA, Patel VG. The role of PD-L1 expression as a predictive biomarker: an analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J Immunother Cancer 2019;7:278. 10.1186/s40425-019-0768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lagos GG, Izar B, Rizvi NA. Beyond tumor PD-L1: emerging genomic biomarkers for checkpoint inhibitor immunotherapy. Am Soc Clin Oncol Educ Book 2020;40:1–11. 10.1200/EDBK_289967. [DOI] [PubMed] [Google Scholar]
- [61].Ang C, Klempner SJ, Ali SM, et al. Prevalence of established and emerging biomarkers of immune checkpoint inhibitor response in advanced hepatocellular carcinoma. Oncotarget 2019;10:4018–4025. 10.18632/oncotarget.26998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Dai Y, Qiang W, Lin K, et al. An immune-related gene signature for predicting survival and immunotherapy efficacy in hepatocellular carcinoma. Cancer Immunol Immunother 2021;70:967–979. 10.1007/s00262-020-02743-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lin P, Gao R-Z, Wen R, et al. DNA damage repair profiles alteration characterize a hepatocellular carcinoma subtype with unique molecular and clinicopathologic features. Front Immunol 2021;12:715460. 10.3389/fimmu.2021.715460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sia D, Jiao Y, Martinez-Quetglas I, et al. Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology 2017;153:812–826. 10.1053/j.gastro.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sangro B, Melero I, Wadhawan S, et al. Association of inflammatory biomarkers with clinical outcomes in nivolumab-treated patients with advanced hepatocellular carcinoma. J Hepatol 2020;73:1460–1469. 10.1016/j.jhep.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Letouzé E, Shinde J, Renault V, et al. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat Commun 2017;8:1315. 10.1038/s41467-017-01358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Schmauch B, Romagnoni A, Pronier E, et al. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat Commun 2020;11:3877. 10.1038/s41467-020-17678-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zeng Q, Klein C, Caruso S, et al. Artificial intelligence predicts immune and inflammatory gene signatures directly from hepatocellular carcinoma histology. J Hepatol 2022;77:116–127. 10.1016/j.jhep.2022.01.018. [DOI] [PubMed] [Google Scholar]
- [69].Liao H, Long Y, Han R, et al. Deep learning-based classification and mutation prediction from histopathological images of hepatocellular carcinoma. Clin Transl Med 2020;10:e102. 10.1002/ctm2.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Shi J-Y, Wang X, Ding G-Y, et al. Exploring prognostic indicators in the pathological images of hepatocellular carcinoma based on deep learning. Gut 2021;70:951–961. 10.1136/gutjnl-2020-320930. [DOI] [PubMed] [Google Scholar]
- [71].Cheng N, Ren Y, Zhou J, et al. Deep learning-based classification of hepatocellular nodular lesions on whole-slide histopathologic images. Gastroenterology 2022;162:1948–1961.e7. 10.1053/j.gastro.2022.02.025. [DOI] [PubMed] [Google Scholar]
- [72].Huang A, Zhao X, Yang X-R, et al. Corrigendum to “Circumventing intratumoral heterogeneity to identify potential therapeutic targets in hepatocellular carcinoma” [J Hepatol 67 (2017) 293–301]. J Hepatol 2017;67:1123. 10.1016/j.jhep.2017.08.003. [DOI] [PubMed] [Google Scholar]
- [73].Galle PR, Foerster F, Kudo M, et al. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int 2019;39:2214–2229. 10.1111/liv.14223. [DOI] [PubMed] [Google Scholar]
- [74].Tayob N, Kanwal F, Alsarraj A, et al. The performance of AFP, AFP-3, DCP as biomarkers for detection of hepatocellular carcinoma (HCC): a phase 3 biomarker study in the United States. Clin Gastroenterol Hepatol 2023;21:415–423.e4. 10.1016/j.cgh.2022.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Marrero JA, Feng Z, Wang Y, et al. Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology 2009;137:110–118. 10.1053/j.gastro.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zhu AX, Dayyani F, Yen C-J, et al. Alpha-fetoprotein as a potential surrogate biomarker for atezolizumab + bevacizumab treatment of hepatocellular carcinoma. Clin Cancer Res 2022;28:3537–3545. 10.1158/1078-0432.CCR-21-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Chalasani NP, Porter K, Bhattacharya A, et al. Validation of a novel multi-target blood test shows high sensitivity to detect early stage hepatocellular carcinoma. Clin Gastroenterol Hepatol 2022;20:173–182.e7. 10.1016/j.cgh.2021.08.010. [DOI] [PubMed] [Google Scholar]
- [78].Singal AG, Tayob N, Mehta A, et al. GALAD demonstrates high sensitivity for HCC surveillance in a cohort of patients with cirrhosis. Hepatology 2022;75:541–549. 10.1002/hep.32185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Tsilimigras DI, Moris D, Hyer JM, et al. Serum α-fetoprotein levels at time of recurrence predict post-recurrence outcomes following resection of hepatocellular carcinoma. Ann Surg Oncol 2021;28:7673–7683. 10.1245/s10434-021-09977-x. [DOI] [PubMed] [Google Scholar]
- [80].Kelley RK, Meyer T, Rimassa L, et al. Serum alpha-fetoprotein levels and clinical outcomes in the phase III CELESTIAL study of cabozantinib versus placebo in patients with advanced hepatocellular carcinoma. Clin Cancer Res 2020;26:4795–4804. 10.1158/1078-0432.CCR-19-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Vipani A, Lauzon M, Luu M, et al. Decreasing trend of serum α-fetoprotein level in hepatocellular carcinoma. Clin Gastroenterol Hepatol 2022;20:1177–1179.e4. 10.1016/j.cgh.2021.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Cui K, Ou Y, Shen Y, et al. Clinical value of circulating tumor cells for the diagnosis and prognosis of hepatocellular carcinoma (HCC): a systematic review and meta-analysis. Medicine 2020;99:e22242. 10.1097/MD.0000000000022242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Zhao L, Song J, Sun Y, et al. Tumor-derived proliferative CTCs and CTC clusters predict aggressiveness and early recurrence in hepatocellular carcinoma patients. Cancer Med 2023;12:13912–13927. 10.1002/cam4.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wei H-W, Qin S-L, Xu J-X, et al. Nomograms for postsurgical extrahepatic recurrence prediction of hepatocellular carcinoma based on presurgical circulating tumor cell status and clinicopathological factors. Cancer Med 2023. 10.1002/cam4.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Wang P-X, Sun Y-F, Zhou K-Q, et al. Circulating tumor cells are an indicator for the administration of adjuvant transarterial chemoembolization in hepatocellular carcinoma: a single-center, retrospective, propensity-matched study. Clin Transl Med 2020;10:e137. 10.1002/ctm2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Winograd P, Hou S, Court CM, et al. Hepatocellular carcinoma-circulating tumor cells expressing PD-L1 are prognostic and potentially associated with response to checkpoint inhibitors. Hepatol Commun 2020;4:1527–1540. 10.1002/hep4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sun Y-F, Guo W, Xu Y, et al. Circulating tumor cells from different vascular sites exhibit spatial heterogeneity in epithelial and mesenchymal composition and distinct clinical significance in hepatocellular carcinoma. Clin Cancer Res 2018;24:547–559. 10.1158/1078-0432.CCR-17-1063. [DOI] [PubMed] [Google Scholar]
- [88].Chen VL, Huang Q, Harouaka R, et al. A dual-filtration system for single-cell sequencing of circulating tumor cells and clusters in HCC . Hepatol Commun 2022;6:1482–1491. 10.1002/hep4.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Sun Y-F, Wu L, Liu S-P, et al. Dissecting spatial heterogeneity and the immune-evasion mechanism of CTCs by single-cell RNA-seq in hepatocellular carcinoma. Nat Commun 2021;12:4091. 10.1038/s41467-021-24386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Tao K, Bian Z, Zhang Q, et al. Machine learning-based genome-wide interrogation of somatic copy number aberrations in circulating tumor DNA for early detection of hepatocellular carcinoma. EBioMedicine 2020;56:102811. 10.1016/j.ebiom.2020.102811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Shen J, Wang S, Zhang Y-J, et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology 2012;55:1799–1808. 10.1002/hep.25569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kaseb AO, Sánchez NS, Sen S, et al. Molecular profiling of hepatocellular carcinoma using circulating cell-free DNA. Clin Cancer Res 2019;25:6107–6118. 10.1158/1078-0432.CCR-18-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Zhang Y, Yao Y, Xu Y, et al. Pan-cancer circulating tumor DNA detection in over 10,000 Chinese patients. Nat Commun 2021;12:11. 10.1038/s41467-020-20162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Qu C, Wang Y, Wang P, et al. Detection of early-stage hepatocellular carcinoma in asymptomatic HBsAg-seropositive individuals by liquid biopsy. Proc Natl Acad Sci U S A 2019;116:6308–6312. 10.1073/pnas.1819799116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Muraoka M, Maekawa S, Katoh R, et al. Usefulness of cell-free human telomerase reverse transcriptase mutant DNA quantification in blood for predicting hepatocellular carcinoma treatment efficacy. Hepatol Commun 2021;5:1927–1938. 10.1002/hep4.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Zhu G-Q, Liu W-R, Tang Z, et al. Serial circulating tumor DNA to predict early recurrence in patients with hepatocellular carcinoma: a prospective study. Mol Oncol 2022;16:549–561. 10.1002/1878-0261.13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Kisiel JB, Dukek BA, Kanipakam RVSR, et al. Hepatocellular carcinoma detection by plasma methylated DNA: discovery, phase I pilot, and phase II clinical validation. Hepatology 2019;69:1180–1192. 10.1002/hep.30244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Lin N, Lin Y, Xu J, et al. A multi-analyte cell-free DNA-based blood test for early detection of hepatocellular carcinoma. Hepatol Commun 2022;6:1753–1763. 10.1002/hep4.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Xu R-H, Wei W, Krawczyk M, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater 2017;16:1155–1161. 10.1038/nmat4997. [DOI] [PubMed] [Google Scholar]
- [100].Luo B, Ma F, Liu H, et al. Cell-free DNA methylation markers for differential diagnosis of hepatocellular carcinoma. BMC Med 2022;20:8. 10.1186/s12916-021-02201-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Cheishvili D, Wong C, Karim MM, et al. A high-throughput test enables specific detection of hepatocellular carcinoma. Nat Commun 2023;14:3306. 10.1038/s41467-023-39055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Nguyen V-C, Nguyen TH, Phan TH, et al. Fragment length profiles of cancer mutations enhance detection of circulating tumor DNA in patients with early-stage hepatocellular carcinoma. BMC Cancer 2023;23:233. 10.1186/s12885-023-10681-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Jiang P, Sun K, Peng W, et al. Plasma DNA end-motif profiling as a fragmentomic marker in cancer, pregnancy, and transplantation. Cancer Discov 2020;10:664–673. 10.1158/2159-8290.CD-19-0622. [DOI] [PubMed] [Google Scholar]
- [104].Zhang X, Wang Z, Tang W, et al. Ultrasensitive and affordable assay for early detection of primary liver cancer using plasma cell-free DNA fragmentomics. Hepatology 2022;76:317–329. 10.1002/hep.32308. [DOI] [PubMed] [Google Scholar]
- [105].Sun X, Feng W, Cui P, et al. Detection and monitoring of HBV-related hepatocellular carcinoma from plasma cfDNA fragmentation profiles. Genomics 2022;114:110502. 10.1016/j.ygeno.2022.110502. [DOI] [PubMed] [Google Scholar]
- [106].Zhou Q, Kang G, Jiang P, et al. Epigenetic analysis of cell-free DNA by fragmentomic profiling. Proc Natl Acad Sci U S A 2022;119:e2209852119. 10.1073/pnas.2209852119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Wang P, Song Q, Ren J, et al. Simultaneous analysis of mutations and methylations in circulating cell-free DNA for hepatocellular carcinoma detection. Sci Transl Med 2022;14:eabp8704. 10.1126/scitranslmed.abp8704. [DOI] [PubMed] [Google Scholar]
- [108].Foda ZH, Annapragada AV, Boyapati K, et al. Detecting liver cancer using cell-free DNA fragmentomes. Cancer Discov 2023;13:616–631. 10.1158/2159-8290.CD-22-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472–484. 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- [110].Roskams-Hieter B, Kim HJ, Anur P, et al. Plasma cell-free RNA profiling distinguishes cancers from pre-malignant conditions in solid and hematologic malignancies. NPJ Precis Oncol 2022;6:28. 10.1038/s41698-022-00270-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Yamamoto Y, Kondo S, Matsuzaki J, et al. Highly sensitive circulating MicroRNA panel for accurate detection of hepatocellular carcinoma in patients with liver disease. Hepatol Commun 2020;4:284–297. 10.1002/hep4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Lin X-J, Chong Y, Guo Z-W, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol 2015;16:804–815. 10.1016/S1470-2045(15)00048-0. [DOI] [PubMed] [Google Scholar]
- [113].Ning C, Cai P, Liu X, et al. A comprehensive evaluation of full-spectrum cell-free RNAs highlights cell-free RNA fragments for early-stage hepatocellular carcinoma detection. EBioMedicine 2023;93:104645. 10.1016/j.ebiom.2023.104645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Pascut D, Pratama MY, Gilardi F, et al. Weighted miRNA co-expression networks analysis identifies circulating miRNA predicting overall survival in hepatocellular carcinoma patients. Sci Rep 2020;10:18967. 10.1038/s41598-020-75945-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Pratama MY, Visintin A, Crocè LS, et al. Circulatory miRNA as a biomarker for therapy response and disease-free survival in hepatocellular carcinoma. Cancers 2020;12. 10.3390/cancers12102810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Teufel M, Seidel H, Köchert K, et al. Biomarkers associated with response to regorafenib in patients with hepatocellular carcinoma. Gastroenterology 2019;156:1731–1741. 10.1053/j.gastro.2019.01.261. [DOI] [PubMed] [Google Scholar]
- [117].Fu P, Gong B, Li H, et al. Combined identification of three lncRNAs in serum as effective diagnostic and prognostic biomarkers for hepatitis B virus-related hepatocellular carcinoma. Int J Cancer 2022;151:1824–1834. 10.1002/ijc.34201. [DOI] [PubMed] [Google Scholar]
- [118].van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018;19:213–228. 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- [119].Luo T, Chen S-Y, Qiu Z-X, et al. Transcriptomic features in a single extrace llular vesicle via single-cell RNA sequencing. Small Methods 2022;6:e2200881. 10.1002/smtd.202200881. [DOI] [PubMed] [Google Scholar]
- [120].Rui T, Zhang X, Guo J, et al. Serum-exosome-derived miRNAs serve as promising biomarkers for HCC diagnosis. Cancers 2022;15. 10.3390/cancers15010205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Sun N, Lee Y-T, Zhang RY, et al. Purification of HCC-specific extracellular vesicles on nanosubstrates for early HCC detection by digital scoring. Nat Commun 2020;11:4489. 10.1038/s41467-020-18311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Yang J, Dong W, Zhang H, et al. Exosomal microRNA panel as a diagnostic biomarker in patients with hepatocellular carcinoma. Front Cell Dev Biol 2022;10:927251. 10.3389/fcell.2022.927251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Nie G, Lian N, Peng D, et al. Prognostic value of exosomal noncoding RNA in hepatocellular carcinoma: a meta-analysis. Carcinogenesis 2022;43:754–765. 10.1093/carcin/bgac066. [DOI] [PubMed] [Google Scholar]
- [124].von Felden J, Garcia-Lezana T, Dogra N, et al. Unannotated small RNA clusters associated with circulating extracellular vesicles detect early stage liver cancer. Gut 2021. 10.1136/gutjnl-2021-325036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Sun N, Zhang C, Lee Y-T, et al. HCC EV ECG score: an extracellular vesicle-based protein assay for detection of early-stage hepatocellular carcinoma. Hepatology 2023;77:774–788. 10.1002/hep.32692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Shuen TWH, Alunni-Fabbroni M, Öcal E, et al. Extracellular vesicles may predict response to radioembolization and sorafenib treatment in advanced hepatocellular carcinoma: an exploratory analysis from the SORAMIC trial. Clin Cancer Res 2022;28:3890–3901. 10.1158/1078-0432.CCR-22-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Ye B, Shen Y, Chen H, et al. Differential proteomic analysis of plasma-derived exosomes as diagnostic biomarkers for chronic HBV-related liver disease. Sci Rep 2022;12:14428. 10.1038/s41598-022-13272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Liang Y, Lehrich BM, Zheng S, et al. Emerging methods in biomarker identification for extracellular vesicle-based liquid biopsy. J Extracell Vesicles 2021;10:e12090. 10.1002/jev2.12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Chan HLY, Vogel A, Berg T, et al. Performance evaluation of the Elecsys PIVKA-II and Elecsys AFP assays for hepatocellular carcinoma diagnosis. JGH Open 2022;6:292–300. 10.1002/jgh3.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Guan M-C, Zhang S-Y, Ding Q, et al. The performance of GALAD score for diagnosing hepatocellular carcinoma in patients with chronic liver diseases: a systematic review and meta-analysis. J Clin Med Res 2023;12. 10.3390/jcm12030949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].A comparative analysis of Elecsys GALAD and Elecsys GAAD score to detect early-stage hepatocellular carcinoma in an international cohort. J Hepatol 2022;77:S937. 10.1016/S0168-8278(22)02154-7. [DOI] [Google Scholar]
- [132].Chalasani NP, Ramasubramanian TS, Bhattacharya A, et al. A novel blood-based panel of methylated DNA and protein markers for detection of early-stage hepatocellular carcinoma. Clin Gastroenterol Hepatol 2021;19:2597–2605.e4. 10.1016/j.cgh.2020.08.065. [DOI] [PubMed] [Google Scholar]
- [133].John B, Porter K, Dahman B, et al. A prospective trial to evaluate the performance of the multitarget hepatocellular carcinoma blood test (mt-HBT) for screening at-risk patients: the Altus study. J Clin Orthod 2022;40:TPS486. 10.1200/JCO.2022.40.4_suppl.TPS486.TPS486. [DOI] [Google Scholar]
- [134].HelioLiver Cancer Test n.d. https://www.helioliver.com/provider/the-helioliver-test (accessed July 22, 2023).
- [135].By P Cell-free technology for detecting cancer n.d. https://www.nature.com/articles/d42473-021-00189-1 (accessed July 22, 2023).
- [136].Oussalah A, Rischer S, Bensenane M, et al. Plasma mSEPT9: a novel circulating cell-free DNA-based epigenetic biomarker to diagnose hepatocellular carcinoma. EBioMedicine 2018;30:138–147. 10.1016/j.ebiom.2018.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Lewin J, Kottwitz D, Aoyama J, et al. Plasma cell free DNA methylation markers for hepatocellular carcinoma surveillance in patients with cirrhosis: a case control study. BMC Gastroenterol 2021;21:136. 10.1186/s12876-021-01714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Website n.d. Guardant360 CDx. Guardant Health, Inc. Accessed July 14, 2023. https://www.guardantcomplete.com/guardant-portfolio/cdx. [Google Scholar]
- [139].Bauml JM, Li BT, Velcheti V, et al. Clinical validation of Guardant360 CDx as a blood-based companion diagnostic for sotorasib. Lung Cancer 2022;166:270–278. 10.1016/j.lungcan.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Ikeda S, Tsigelny IF, Skjevik ÅA, et al. Next-generation sequencing of circulating tumor DNA reveals frequent alterations in advanced hepatocellular carcinoma. Oncologist 2018;23:586–593. 10.1634/theoncologist.2017-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Klein EA, Richards D, Cohn A, et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann Oncol 2021;32:1167–1177. 10.1016/j.annonc.2021.05.806. [DOI] [PubMed] [Google Scholar]
- [142].Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926–930. 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Milbury CA, Creeden J, Yip W-K, et al. Clinical and analytical validation of FoundationOne®CDx, a comprehensive genomic profiling assay for solid tumors. PLoS One 2022;17:e0264138. 10.1371/journal.pone.0264138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


