Abstract
Background
Esophageal squamous cell carcinoma (ESCC) is sometimes detected in non‐drinker and non‐smoker females who are considered to have very low risk of ESCC development in daily practice. This study examined the clinicopathological and genomic characteristics of ESCCs in females with no history of drinking and smoking.
Methods
The sample comprised 118 ESCC lesions occurring in 95 female patients who underwent endoscopic submucosal dissection at our department between January 2008 and December 2019. The patients were categorized into two groups: 51 lesions in 49 patients with no history of drinking and smoking (nondrinker/nonsmoker [NDNS] group) and 69 lesions in 45 patients with a history of drinking or smoking (drinker/smoker [DS] group). We analyzed the differences in clinicopathological and cancerous genomic characteristics between the groups. Significant genomic alterations were validated using immunohistochemistry.
Results
Multiple logistic regression revealed that older age, fewer multiple Lugol‐voiding lesions (LVLs), and reflux esophagitis (RE) were independently associated with the occurrence of ESCCs in the NDNS group. ESCC lesions in the NDNS group were predominantly located in the mid‐thoracic esophagus, posterior wall side, with 0‐IIa, the aspect ratio of the lesion >2 (vertical/horizontal), and endoscopic keratinization. Genetic analysis showed that CDKN2A driver alterations were significantly more frequent and KMT2D alterations were significantly less frequent in the NDNS group than in the DS group. KMT2D alterations were strongly correlated with immunostaining.
Conclusion
Older nondrinker, nonsmoker females with RE and fewer multiple LVLs may develop longitudinal 0‐IIa ESCC with keratinization of the posterior wall of the mid‐thoracic esophagus. ESCCs in nondrinker, nonsmoker females had fewer KMT2D alterations and more CDKN2A alterations, which may be a biomarker for treatment.
Keywords: esophageal neoplasms, esophageal squamous cell carcinoma, genomics, nondrinker, nonsmoker females, reflux esophagitis
Our study revealed that older non‐drinker, non‐smoker females with RE and fewer multiple LVLs might develop longitudinal 0‐IIa ESCC with keratinization of the posterior wall of the mid‐thoracic esophagus. These lesions had a low frequency of KMT2D alterations and a high frequency of CDKN2A alterations, with less KMT2D‐positive and more p16‐positive immunostaining.
1. INTRODUCTION
Globally, esophageal cancer is the seventh most common cancer and the sixth most common cause of cancer‐related deaths. 1 It has a poor prognosis, and the 5‐year survival rate ranges from 15% to 25%. 2 However, recent progress in endoscopy has facilitated its detection at an early stage, which has dramatically improved the prognosis. 3 , 4 , 5 Esophageal carcinomas are of two types: squamous cell carcinoma (SCC) and adenocarcinoma. Epidemiologically, over 80% of esophageal cancers in Asian countries such as Japan are SCC, whereas adenocarcinomas predominate in Western countries. 6 Globally, esophageal SCC (ESCC) is more common in males than in females, and in Japan, males are six times more likely to be affected than females. 7 , 8 Consumption of alcohol and tobacco smoking are major risk factors for esophageal cancer, 9 , 10 and their synergistic effects have also been observed in carcinogenesis. 11 They are strongly linked to acetaldehyde metabolism and are mainly regulated by alcohol dehydrogenase 1B (ADH1B) and aldehyde dehydrogenase 2 (ALDH2). 12 Male sex, alcohol consumption, smoking, ADH1B and ALDH2 gene polymorphisms, and multiple Lugol‐voiding lesions (LVLs) significantly affect the incidence of numerous metachronous SCCs. 13 , 14 Therefore, it is crucial to carefully monitor the esophagus in patients with risk factors for ESCC. However, ESCC is sometimes detected in nondrinker and nonsmoker females who are considered to have a very low risk of ESCC development in daily practice. There are few reports on ESCCs in nondrinking and nonsmoking females who have a low risk of ESCC development, and the detailed carcinogenic mechanisms are unclear.
This study aimed to examine the clinicopathological and genomic characteristics of ESCCs in females without a history of smoking and alcohol consumption.
2. METHODS
2.1. Patients
We retrospectively enrolled consecutive patients with superficial ESCCs who underwent endoscopic submucosal dissection at Hiroshima University Hospital between January 2008 and December 2019. Males with ESCC were excluded. The included patients were categorized into two groups based on history of drinking or smoking: the nondrinker, nonsmoker group (NDNS group) and the drinker and/or smoker group (DS group) (Figure 1). We analyzed the differences in clinicopathological and genomic characteristics between the groups.
FIGURE 1.

Study flowchart showing a comparison of clinicopathologic and genomic characteristics between nondrinking, nonsmoking females who underwent ESD for ESCC and those who drink and/or smoke. DS, drinker and/ or smoker; ESCC, esophageal squamous cell carcinoma; ESD, endoscopic submucosal dissection; NDNS, nondrinker nonsmoker.
2.2. Definitions
We defined drinkers as patients with a daily drinking habit and smokers as those with a current or past habit of smoking at least one cigarette daily. Esophageal LVLs, reflux esophagitis (RE), degree of gastric atrophy, longitudinal location, macroscopic type, and circumferential location of the esophagus were previously defined 15 , 16 , 17 and classified with respect to the Japanese Classification of Esophageal Carcinoma. 18 We diagnosed hiatal hernias endoscopically by observing the discordance between the diaphragmatic hiatus and esophagogastric junction. 19 A “longitudinal lesion” was defined as a lesion extending along the long axis of the esophageal lumen with a length more than twice that of the short axis. When white keratinizing epithelial adherence was observed in the lesion, it was described as “endoscopic keratinization.”
2.3. Pathological examination
We evaluated the specimens according to the Japanese guidelines for esophageal carcinoma. 20 The components of intraepithelial carcinoma were classified into two types: the total layer type, in which tumor cells replace the entire epithelial layer, and the basal layer type, in which tumor cells grow mainly in the basal layer and differentiate toward the superficial layer into a more acidophilic cytoplasm. Parakeratosis is characterized by the presence of nuclei in stratum corneum cells, which should not be normally observed. 21
2.4. Tissue collection and DNA extraction
We prepared several 10‐μm‐thick slides using formalin‐fixed paraffin‐embedded (FFPE) specimens. From these specimens, we dissected the pathological tumor tissues (cancer) and nontumor tissues surrounding the tumor using the Laser Capture Microdissection System (Leica LMD 6500). We used the GeneRead DNA FFPE Kit (Qiagen, Valencia, CA, USA) to extract DNA from these tissues and the Qubit 1.0 Fluorometer (Life Technologies, Grand Island, NY, USA) to determine DNA concentrations. Furthermore, we ascertained the quantity and quality of FFPE‐derived DNA samples by calculating the normalized DNA integrity scores (ΔΔCq) via quantitative polymerase chain reaction analysis using the Agilent NGS FFPE QC Kit (Agilent Technologies, Santa Clara, CA, USA).
2.5. Target enrichment and next‐generation sequencing
We developed a sequencing library based on DNA extracted from the tumors and nontumor mucosa using the SureSelect XT HS Kit (Agilent Technologies) after fragmenting into 150–200 bp using the XT Low Input Enzymatic Fragmentation Kit (Agilent Technologies). The amount of DNA was measured using TapeStation D1000 (Agilent Technologies) before hybridization and used if the prepared library was >500 ng. However, three samples were not obtained. To perform target capture, the SureSelect XT Target Enrichment System (Agilent Technologies) was mounted on 468 cancer genes from the MSK‐IMPACT Clinical Sequencing Cohort 22 (Table S1). The resulting pooled libraries were sequenced by paired‐end reads using the HiSeq X platform (Illumina, San Diego, CA, USA) after the quality control check with the High Sensitivity D1000 ScreenTape Assay (Agilent Technologies) (Figure 2).
FIGURE 2.
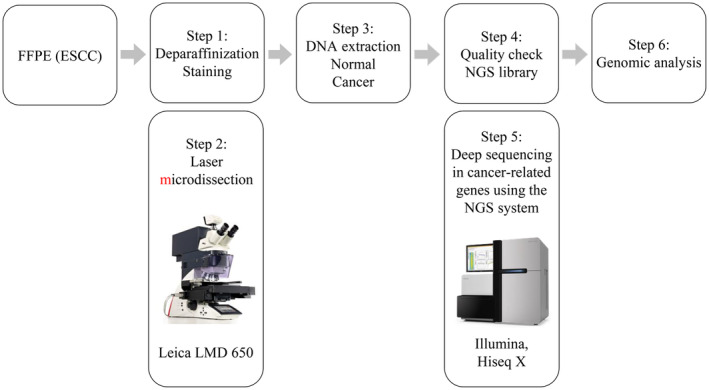
Workflow of multigene panel testing for cancer. DNA, deoxyribonucleic acid; ESCC, esophageal squamous cell carcinoma; FFPE, formalin‐fixed and paraffin‐embedded; LMD, laser microdissection; NGS, next‐generation sequencing.
2.6. Variant detection
Sequencing reads were preprocessed using fastp v0.20 and mapped to hg19 using BWA‐MEM v0.7.17. 23 GATK best practice was used for variant calling. To reduce false positives, somatic mutations were defined as read depths >50 and variant allele frequencies >4%. Copy number analysis was performed using CNVkit v0.9.9 and PureCN v2.0.1. 24 Vcf2maf v1.6.21 (https://zenodo.org/record/1185418#.Y_W6cC_3IUs), oncokb‐annotator v3.2.1 (https://github.com/oncokb/oncokb‐annotator/releases), and InterVar v2.2.2 25 were used for annotation. We defined alterations as mutations, amplifications, or deletions that are classified as oncogenic or likely oncogenic status in OncoKB (https://www.oncokb.org) or pathogenic or likely pathogenic status in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/). R package maftools v2.8.5 (https://bioconductor.org/packages/release/bioc/html/maftools.html) was used for plotting.
2.7. Immunohistochemistry
Sections of paraffin‐embedded human esophageal cancer tissue specimens (2–3‐μm‐thick) were mounted on positively charged slides. Antigen retrieval was performed in Tris‐EDTA buffer (pH 6.0) in a microwave oven at 800 W for 30 min. Subsequently, the tissues were incubated with the primary antibodies p16 and KMT2D at room temperature (20°C–25°C). Bound primary antibodies were detected using the Dako EnVision System (Dako, Copenhagen, Denmark). The slides were then counterstained using hematoxylin after immunostaining. The immunohistochemical evaluations were performed blinded with regard to the histological diagnosis.
2.8. Statistical analyses
Categorical variables were compared using the Chi‐square and Fisher's exact tests. Continuous variables were compared using the Student's t‐test. Multivariate logistic regression analysis was performed with stepwise selection. The Kappa statistic was used to determine the correlation between genomic alterations and KMT2D and CDKN2A immunohistochemistry. All statistical analyses were performed using JMP version 15 (SAS Institute Inc., Cary, North Carolina, USA). p < 0.05 was considered statistically significant.
3. RESULTS
3.1. Patient characteristics
There were 48 and 47 patients in the NDNS and DS groups, respectively. The characteristics of the patients are presented in Table 1. Significant differences were observed between the groups with respect to age (NDNS group: 73.0 ± 8.7 years vs. DS group: 62.8 ± 8.9 years, p < 0.01), body mass index (21.2 ± 3.3 vs. 19.7 ± 2.9 mm, p < 0.05), and height (150.7 ± 5.3 vs. 155.6 ± 6.9 cm, p < 0.01); however, no differences were found with respect to weight. Significant differences were observed regarding the history of cancer (25.0% [12/48] vs. 44.7% [21/47], p < 0.05), history of head and neck cancer (4.2% [2/48] vs. 19.2% [9/47], p < 0.05), and multiple ESCCs (6.3% [3/48] vs. 36.2% [17/47], p < 0.01). Significant differences were found with respect to multiple LVLs (10.4% [5/48] vs. 68.1% [32/47], p < 0.01), RE (64.6% [31/48] vs. 8.5% [4/47], p < 0.01), hiatal hernia (39.6% [19/48] vs. 14.9% [7/47], p < 0.01), Barrett's esophagus (43.8% [21/48] vs. 23.4% [11/47], p < 0.05), symptomatic (37.5% [18/48] vs. 8.5% [4/47], p < 0.01), and Helicobacter pylori eradication (35.4% [17/48] vs. 8.5% [4/47], p < 0.01); however, no significant differences were observed for atrophy of the stomach. Multiple logistic regression revealed that older age, fewer multiple LVLs, and RE were independently associated with the occurrence of ESCCs in the NDNS group (Table 2).
TABLE 1.
Characteristics of female patients who underwent ESD for ESCC.
| NDNS group n = 48 | DS group n = 47 | pvalue | |
|---|---|---|---|
| Age (years, mean ± SD) | 73.0 ± 8.7 | 62.8 ± 8.9 | <0.01 |
| Body mass index (kg/m2) | 21.2 ± 3.3 | 19.7 ± 2.9 | 0.0225 |
| Height (cm, mean ± SD) | 150.7 ± 5.3 | 155.6 ± 6.9 | <0.01 |
| Body weight (kg, mean ± SD) | 48.2 ± 8.4 | 47.7 ± 7.5 | 0.7665 |
| History of cancer, n (%) | 12 (25.0) | 21 (44.7) | 0.0430 |
| History of head and neck cancer, n (%) | 02 (04.2) | 09 (19.2) | 0.0273 |
| Post gastrectomy, n (%) | 01 (02.1) | 04 (08.5) | 0.2038 |
| Family history of ESCC, n (%) | 02 (04.2) | 02 (04.3) | 1.0000 |
| Synchronous/metachronous ESCC, n (%) | 03 (06.3) | 17 a (36.2) | <0.01 |
| Synchronous ESCC, n (%) | 02 (04.2) | 10 (21.3) | 0.0144 |
| Metachronous ESCC, n (%) | 01 (02.1) | 12 (25.5) | <0.01 |
| Multiple Lugol‐voiding lesions | |||
| (−), n (%) | 43 (89.6) | 15 (31.9) | < 0.01 |
| (+), n (%) | 05 (10.4) | 32 (68.1) | |
| Reflux esophagitis | 31 (64.6) | 04 (08.5) | <0.01 |
| LA‐Grade A/B, n (%) | 31 (64.6) | 04 (08.5) | |
| LA‐Grade C/D, n (%) | 00 (00.0) | 00 (00.0) | |
| Regular use of PPI or P‐CAB, n (%) | 24 (50.0) | 04 (08.5) | <0.01 |
| Symptomatic, n (%) | 18 (37.5) | 04 (08.5) | <0.01 |
| Hiatal hernia, n (%) | 19 (39.6) | 07 (14.9) | <0.01 |
| Barrett's esophagus, n (%) | 21 (43.8) | 11 (23.4) | 0.0347 |
| Atrophic gastritis, n (%) | 20 (41.7) | 19 (40.4) | 0.9021 |
| Helicobacter pylori eradication, n (%) | 17 (35.4) | 04 (08.5) | <0.01 |
Note: We divided all 95 patients into two groups: NDNS group (n = 48) and DS group (n = 47).
Abbreviations: DS, drinker and/or smoker; ESCC, esophageal squamous cell carcinoma; ESD, endoscopic submucosal dissection; LA, Los Angeles; NDNS, nondrinker nonsmoker; PPI, proton pump inhibitor; P‐CAB, potassium‐competitive acid blocker; SD, standard deviation.
There are duplicated cases.
TABLE 2.
Multivariate analysis of predictors of ESCC in nondrinking, nonsmoking females.
| Odds ratio | 95% CI | p value | |
|---|---|---|---|
| Age ≥69 years | 12.0 | 2.8–46.5 | <0.01 |
| Multiple Lugol‐voiding lesions (−) | 17.1 | 3.8–77.5 | <0.01 |
| Reflux esophagitis (+) | 12.5 | 2.5–62.0 | <0.01 |
| Regular use of PPI or P‐CAB | 04.3 | 0.9–20.3 | 0.0684 |
Note: We performed multivariate analysis to predict risk factors for ESCC in the NDNS group after selecting factors using the stepwise method.
Abbreviations: CI, confidence interval; ESCC, esophageal squamous cell carcinoma; P‐CAB, potassium‐competitive acid blocker; PPI, proton pump inhibitor.
3.2. Tumor characteristics
There were 50 and 68 lesions in the NDNS and DS groups, respectively. The characteristics of the tumor are presented in Table 3. The mean tumor size and horizontal location ≤1/4 were not significantly different between the groups; however, the mid‐thoracic esophagus in the longitudinal location (72.0% [36/50] vs. 51.5% [35/68], p < 0.05), posterior wall side in the cross‐sectional location (66.0% [33/50] vs. 36.8% [25/68], p < 0.01), macroscopic type (0–IIa 18.0% [9/50], 0–IIc 82.0% [41/50] vs. 4.4% [3/68], 95.6% [65/68], p < 0.05), aspect ratio of the lesion of >2 (vertical/horizontal) (46.0% [23/50] vs. 11.8% [8/68], p < 0.01), and endoscopic keratinization (48.0% [24/50] vs. 11.8% [8/68], p < 0.01) were significantly different between the groups. No differences were found in the pathological tumor depth of invasion. However, significant differences were observed in pathological findings (basal layer type 44.0% [22/50], total layer type 56.0% [28/50] vs. 19.1% [13/68], 80.9% [55/68], p < 0.01), and parakeratosis (42.0% [21/50] vs. 8.8% [6/68], p < 0.01). All the invasive lesions were also of the total layer type.
TABLE 3.
Tumor characteristics of ESCCs in females who underwent ESD.
| NDNS group n = 50 | DS group n = 68 | p value | |
|---|---|---|---|
| Tumor size, mean ± SD, mm | 26.8 ± 17.9 | 25.8 ± 19.3 | 0.7933 |
| Horizontal location ≤1/4, n (%) | 31 (62.0) | 48 (70.6) | 0.3282 |
| Longitudinal location | |||
| Upper esophagus (Ce, Ut), n (%) | 05 (10.0) | 11 (16.2) | 0.4198 |
| Mid‐esophagus (Mt), n (%) | 36 (72.0) | 35 (51.5) | 0.0359 |
| Lower esophagus (Lt. Ae), n (%) | 09 (18.0) | 22 (32.3) | 0.0934 |
| Cross‐sectional location, n (%) | |||
| Anterior 11–2, n (%) | 07 (14.0) | 11 (16.2) | 0.8009 |
| Right 2–5, n (%) | 05 (10.0) | 16 (23.5) | 0.0869 |
| Posterior 5–8, n (%) | 33 (66.0) | 25 (36.8) | <0.01 |
| Left 8–11, n (%) | 03 (06.0) | 12 (17.7) | 0.0918 |
| Whole circumference, n (%) | 02 (04.0) | 04 (05.9) | 1.0000 |
| Macroscopic type | |||
| 0‐IIa, n (%) | 09 (18.0) | 03 (04.4) | 0.0276 |
| 0‐IIc, n (%) | 41 (82.0) | 65 (95.6) | |
| Aspect ratio of the lesion >2 (vertical/horizontal), n (%) | 23 (46.0) | 08 (11.8) | <0.01 |
| Endoscopic keratinization, n (%) | 24 (48.0) | 08 (11.8) | <0.01 |
| En bloc resection, n (%) | 49 (98.0) | 67 (98.5) | 1.0000 |
| Complete en bloc resection, n (%) | 47 (94.0) | 66 (97.1) | 0.4184 |
| Pathological tumor depth of invasion | |||
| EP/LPM, n (%) | 43 (84.0) | 56 (82.4) | 0.4606 |
| MM/SM1, n (%) | 05 (10.0) | 10 (14.7) | 0.4433 |
| SM2, n (%) | 02 (04.0) | 02 (02.9) | 1.0000 |
| Lymphovascular infiltration, n (%) | 04 (08.0) | 05 (07.4) | 1.0000 |
| Pathological findings | |||
| Basal layer type, n (%) | 22 (44.0) | 13 (19.1) | <0.01 |
| Total layer type, n (%) | 28 (56.0) | 55 (80.9) | |
| Parakeratosis, n (%) | 21 (42.0) | 06 (08.8) | <0.01 |
Note: We divided all 118 lesions into two groups: NDNS group (n = 50) and DS group (n = 68).
Abbreviations: Ae, abdominal esophagus; Ce, cervical esophagus; DS, drinker and/or smoker; EP, epithelium; ESCC, esophageal squamous cell carcinoma; ESD, endoscopic submucosal dissection; Lt, lower thoracic esophagus; LPM, lamina propria mucosa; MM, muscularis mucosae; Mt, middle thoracic esophagus; NDNS, nondrinker nonsmoker; SD, standard deviation; SM, submucosa; Ut, upper thoracic esophagus.
3.3. Genomic landscape of female ESCCs
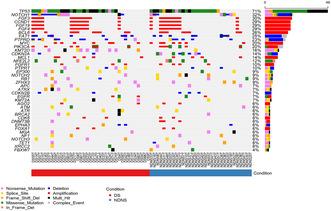
Multigene cancer panel testing of 32 and 37 lesions in the NDNS and DS groups, respectively, was possible. The cancer genome profiles of 69 female ESCC samples are shown in Figures 3, 4 and Tables S2, S3. Overall, pathogenic mutations in 17 genes were detected in >10% of cases, with TP53 having the highest alteration frequency (Figure 3). The frequency of KMT2D alterations was significantly lower (3.1% [1/32] vs. 27.0% [10/37], p < 0.01) and CDKN2A alterations were significantly higher (25.0% [8/32] vs. 5.4% [2/37], p < 0.05) in the NDNS group than in the DS group (Table 4, Figure 4). Of the KMT2D alterations, nonsense mutations were found in the DS group (7/10, 70.0%; Figure 5) rather than in the NDNS group (0/1, 0%; Figure 5). Furthermore, 90% (9/10) of the patients with KMT2D alterations in the DS group were smokers. In contrast, no difference was found in the frequency of CDKN2A deletions between the NDNS and DS groups (9.4% [3/32] vs. 5.4% [2/37], p = 0.5260); however, mutations were found only in the NDNS group (15.6% [5/32] vs. 0% [0/37], p < 0.05; Table 5). Regarding the KMT2D mutations, six cases had a low variant allele frequency (VAF) (<0.1).
FIGURE 3.
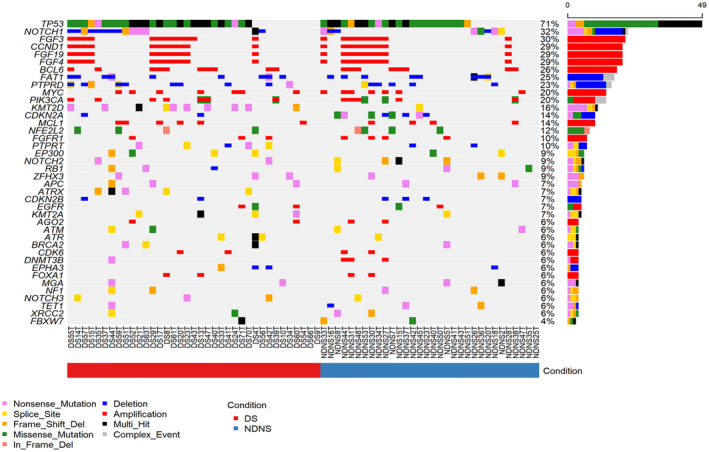
Alteration profiling of 32 lesions in the NDNS group and 37 in the DS group. The main panels contain the mutation patterns of 40 genes with more than 4% somatic mutations from 32 and 37 lesions in the NDNS and DS groups, respectively. Pink, yellow, orange, green, beige, blue, red, black, and gray cells indicate nonsense mutations, splice site mutations, frameshift mutations, missense mutations, in‐frame indels, deletions, amplifications, multiple hits, and complex events, respectively. The bar graph on the right shows the frequency of mutations in each gene. The horizontal bar indicates whether the cases were in the DS (red) or NDNS (blue) group. DS, drinker and/or smoker; NDNS, nondrinker nonsmoker.
FIGURE 4.
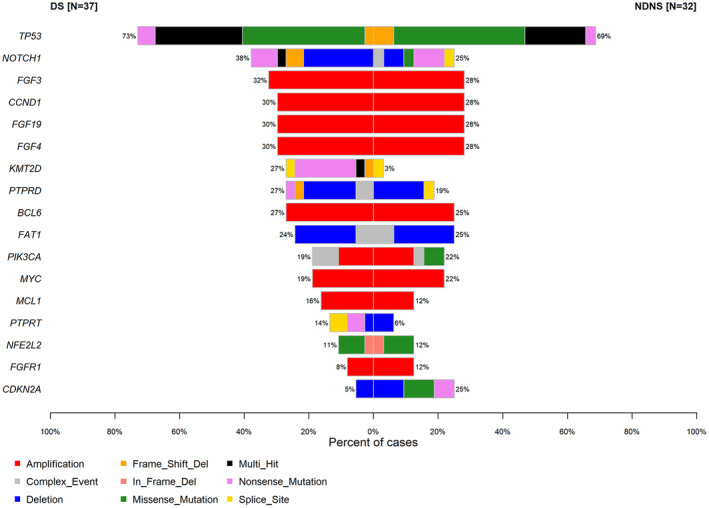
Comparison of genetic alterations between the NDNS and DS groups. Red, gray, blue, orange, green, black, pink, and yellow cells indicate amplification, complex event, deletion, frameshift mutation, missense mutation, multi‐hit, nonsense mutation, and splice site mutation, respectively. DS, drinker and/or smoker; NDNS, nondrinker nonsmoker.
TABLE 4.
Outcome of genetic analysis of ESCCs in females.
| NDNS group n = 32 | DS group n = 37 | p value | |
|---|---|---|---|
| TP53, n (%) | 22 (68.8) | 27 (73.0) | 0.6998 |
| NOTCH1, n (%) | 08 (25.0) | 14 (37.8) | 0.2513 |
| FGF3, n (%) | 09 (28.1) | 12 (32.4) | 0.6978 |
| CCND1, n (%) | 09 (28.1) | 11 (29.7) | 0.8835 |
| FGF19, n (%) | 09 (28.1) | 11 (29.7) | 0.8835 |
| FGF4, n (%) | 09 (28.1) | 11 (29.7) | 0.8835 |
| KMT2D, n (%) | 01 (03.1) | 10 (27.0) | <0.01 |
| PTPRD, n (%) | 06 (18.8) | 10 (27.0) | 0.4141 |
| BCL6, n (%) | 08 (25.0) | 10 (27.0) | 0.8482 |
| FAT1, n (%) | 08 (25.0) | 09 (24.3) | 0.9482 |
| PIK3CA, n (%) | 07 (21.9) | 07 (18.9) | 0.7610 |
| MYC, n (%) | 07 (21.9) | 07 (18.9) | 0.7610 |
| MCL1, n (%) | 04 (12.5) | 06 (16.2) | 0.6607 |
| PTPRT, n (%) | 02 (06.3) | 05 (13.5) | 0.3101 |
| NFE2L2, n (%) | 03 (09.4) | 04 (10.8) | 1 |
| FGFR1, n (%) | 04 (12.5) | 03 (08.1) | 0.6964 |
| CDKN2A, n (%) | 08 (25.0) | 02 (05.4) | <0.05 |
Note: We compared a genetic analysis of TP53, NOTCH1, FGF3, CCND1, FGF19, FGF4, KMT2D, PTPRD, BCL6, FAT1, PIK3CA, MYC, MCL1, ATRX, PTPRT, and CDKN2A between the two groups: 32 lesions in the NDNS group and 37 lesions in the DS group were identified.
Abbreviations: ESCC, esophageal squamous cell carcinoma; DS, drinker and/or smoker; NDNS, nondrinker nonsmoker.
FIGURE 5.
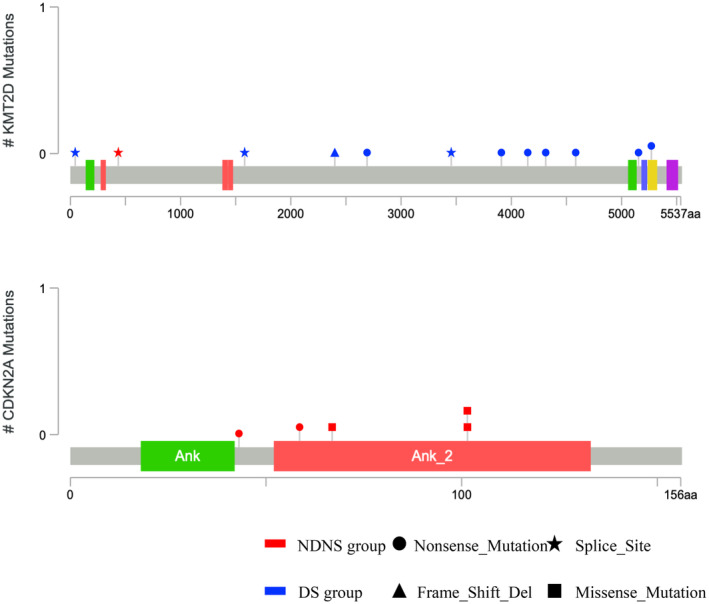
Distribution of KMT2D and CDKN2A somatic mutations. This shows the location of mutated gene positions for ESCCs in the NDNS and DS groups on the KMT2D and CDKN2A structures. DS, drinker and/or smoker; ESCC, esophageal squamous cell carcinoma; NDNS, nondrinker, nonsmoker.
TABLE 5.
Breakdown of KMT2D and CDKN2A alterations in genetic analysis of ESCC in females.
| NDNS group n = 32 | DS group n = 37 | p value | |
|---|---|---|---|
| KMT2D, n (%) | 01 (3.1) | 10 (27.0) | |
| Deletion, n (%) | 0 (0.00) | 0 (0.00) | 1 |
| Mutation, n (%) | 1 (03.1) | 10 (27.0) | <0.01 |
| CDKN2A, n (%) | 08 (25.0) | 02 (05.4) | |
| Deletion, n (%) | 3 (09.4) | 0 2 (05.4) | 0.5260 |
| Mutation, n (%) | 5 (15.6) | 0 0 (0.00) | <0.05 |
Note: We compared the percentage of deletions and mutations in KMT2D and CDKN2A alterations between the two groups. There were 32 lesions in the NDNS group and 37 lesions in the DS group.
Abbreviations: ESCC, esophageal squamous cell carcinoma; DS, drinker and/or smoker; NDNS, nondrinker nonsmoker.
3.4. Immunohistochemistry
The immunohistochemistry results for KMT2D and p16 are shown in Table 6 and Figure 6. Immunohistochemistry for KMT2D and p16 was performed on 50 and 68 lesions in the NDNS and DS groups, respectively. KMT2D was significantly less frequent (2.0% [1/50] vs. 26.5% [18/68], p < 0.01), and p16 was significantly more frequent (62.0% [31/50] vs. 27.1% [19/68], p < 0.01) in the NDNS group than in the DS group. Additionally, KMT2D alterations and positive immunostaining for KMT2D were found in 100% (1/1) of patients in the NDNS group and 90% (9/10) in the DS group. CDKN2A alterations and positive immunostaining for p16 were found in 87.5% (7/8) and 50% (1/2) of patients in the NDNS and DS groups, respectively (Figure 7). Regarding the correlation between alterations and immunostaining, the kappa statistic was 0.8924 and 0.1524 for KMT2D and CDKN2A, respectively (Figure 7). Three of the six cases with KMT2D mutations and low VAF (<0.1) were heterogeneous in immunohistochemistry.
TABLE 6.
Outcome of immunohistochemistry analysis of ESCCs in females.
| NDNS group n = 50 | DS group n = 68 | p value | |
|---|---|---|---|
| KMT2D, n (%) | 01 (02.0) | 16 (23.5) | <0.01 |
| p16, n (%) | 31 (62.0) | 19 (27.1) | <0.01 |
Note: We compared KMT2D and p16 in immunohistochemistry analysis between the two groups: 50 lesions in the NDNS group and 68 lesions in the DS group.
Abbreviations: DS, drinker and/ or smoker; ESCC, esophageal squamous cell carcinoma; NDNS, nondrinker nonsmoker.
FIGURE 6.
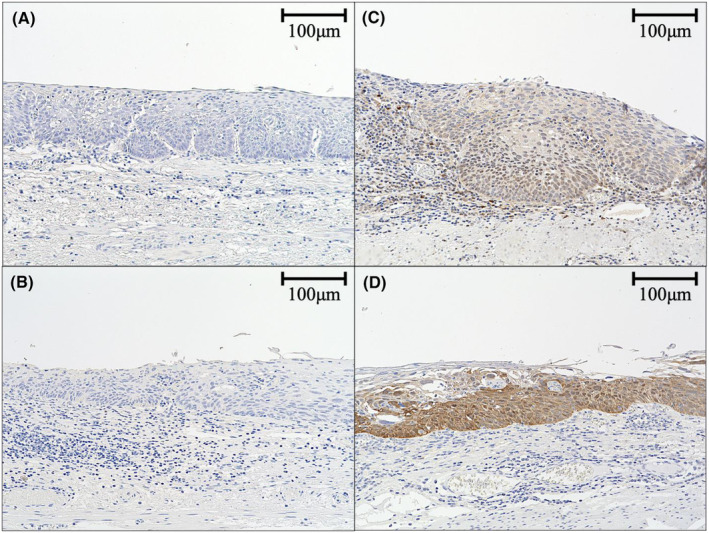
Immunostaining of specimens treated by ESD. Immunostaining of a case negative for KMT2D (A) and p16 (B) immunostaining is shown on the left, while that of a case positive for KMT2D (C) and p16 (D) immunostaining is shown on the right. ESD, endoscopic submucosal dissection.
FIGURE 7.
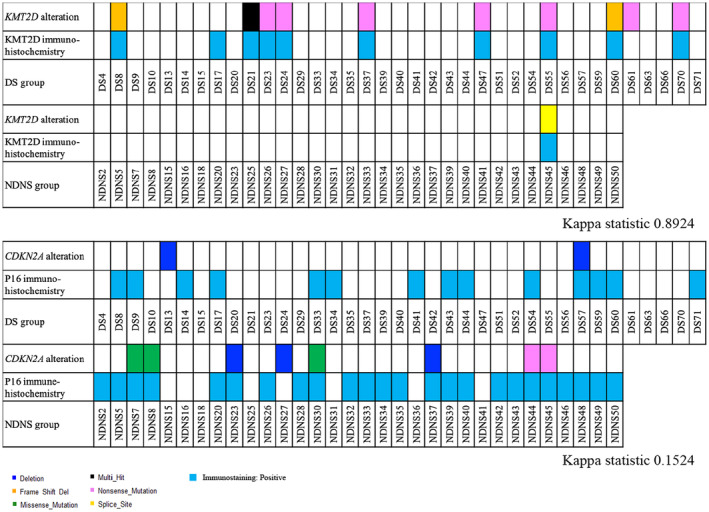
Correlation of KMT2D and CDKN2A alteration status with KMT2D and p16 immunostaining status of ESCCs in the NDNS and DS groups. The KMT2D and p16 immunostaining status was evaluated in 50 and 68 lesions in the NDNS and DS groups, respectively. Pink, orange, black, and yellow cells indicate nonsense mutations, frameshift mutations, multiple hit, and splice sites in KMT2D mutations, respectively. Light blue cells indicate positive staining in KMT2D immunohistochemistry. Blue, green, and pink cells show deletions, missense mutations, and nonsense mutations in CDKN2A mutations, respectively. Light blue cells indicate positive p16 immunohistochemistry. DS, drinker and/or smoker; ESCC, esophageal squamous cell carcinoma; NDNS, nondrinker nonsmoker.
4. DISCUSSION
Our study revealed that older nondrinker, nonsmoker females with RE and fewer multiple LVLs might develop longitudinal 0–IIa ESCC with keratinization of the posterior wall of the mid‐thoracic esophagus. These lesions had a low frequency of KMT2D alterations and a high frequency of CDKN2A alterations, with less KMT2D‐positive and more p16‐positive immunostaining. KMT2D alterations were strongly correlated with immunostaining findings.
Multiple LVLs, which are useful predictors of the risk of metachronous multiple ESCCs, are caused by direct exposure to carcinogens, including alcohol consumption and smoking. Therefore, in females who do not drink or smoke, ESCCs may develop from the background mucosa with a very low risk of ESCC without multiple LVLs, implying that the carcinogenic pathways differ. Chronic RE is typically the predominant causative factor for developing Barrett's esophagus and its progression to esophageal adenocarcinoma. 6 However, some reports have demonstrated a role for gastroesophageal reflux disease in laryngopharyngeal carcinogenesis and identified a relationship between RE and ESCC by reviewing surgical cases of esophageal cancer. 26 , 27 In our study, multivariate analysis revealed that RE was a risk factor for ESCC development in nondrinking, nonsmoking females. In many symptomatic cases, proton pump inhibitors or potassium‐competitive acid blockers, medications, and Barrett's esophagus also reflected the higher incidence of RE in the NDNS group. Shigaki et al. reported that nondrinking, nonsmoking esophageal cancer was more common in older women. 28 Some studies have suggested that exposure to estrogen and progesterone may have protective effects against ESCC development, leading to a lower incidence in women than in men. 29 , 30 , 31 Estrogen suppresses RE by enhancing the defense against damage to the esophageal mucosa. 32 Therefore, it is possible that RE increased in older females with a decline in estrogen levels after menopause and may have contributed to the development of ESCC. In studies of ESCC in nondrinking, nonsmoking women, only this study, which had a larger number of cases than previous reports, 28 , 33 , 34 found significant differences in all clinical findings of older age, RE, and fewer multiple LVLs.
In the current study, the characteristic endoscopic finding of ESCC in nondrinking, nonsmoking women was longitudinal morphology with keratinization of the posterior wall of the mid‐thoracic esophagus. The posterior wall of the mid‐thoracic esophagus is where the refluxed stomach acid and contents are retained in the supine position. 35 Therefore, based on morphology and location, it is speculated that retention of refluxed gastric acid and stomach contents in the posterior wall of the mid‐thoracic esophagus led to the occurrence of ESCC in nondrinking, nonsmoking women. Notably, older women with increasing kyphosis due to osteoporosis can develop hiatal hernia of the esophagus because of decreased abdominal cavity volume, and severe and refractory RE is likely to develop due to reflux of gastric contents. 36 Esophageal achalasia induces TP53 mutations through long‐term mechanical and chemical stimuli from food retention, leading to carcinogenesis. 37 , 38 , 39 Additionally, nonacid reflux of duodenal contents may be associated with the development of ESCC in nondrinking, nonsmoking women because carcinogenesis of ESCC due to reflux of duodenal fluid, particularly bile acids, has been demonstrated in rat models of duodenal esophageal reflux. 40 , 41 , 42 There were more cases of 0–IIa ESCC with endoscopic keratinization in the NDNS group. A previous study reported that ESCC in nondrinking, nonsmoking females showed more endoscopic keratinization and more 0–IIa cases and that endoscopic keratinization corresponded to histopathologic parakeratosis. 34 , 43 Endoscopic keratosis, in this case, may have developed through a similar process of repair and regeneration, resulting in changes including mucosal thickening and keratinization in esophagitis related to esophageal achalasia. 37 These findings suggest that the mechanism of longitudinal ESCC with endoscopic keratinization in the posterior wall of the mid‐thoracic esophagus is due to persistent esophageal reflux and retention of refluxed gastric acid, duodenal fluid, and food, leading to chronic hyperplastic esophagitis and eventually to malignant transformation of esophageal epithelial cells via a dysplasia‐carcinoma sequence.
Due to recent advances in next‐generation sequencing (NGS) technology, which has facilitated analysis of the whole genome, exome, and target sequences, a great deal of information on the aberrations of cancer‐related genes causing the development and progression of ESCC has accumulated, leading to the description of the landscape of ESCC‐related somatic aberrations, such as TP53, NOTCH1, PIK3CA, NFE2L2, and CDKN2A. 44 , 45 , 46 Moreover, in our previous study, we performed NGS focusing on early esophageal cancer and reported that somatic alterations such as TP53, NOTCH1, and CDKN2A deletion are more frequent in cancerous mucosa than in noncancerous mucosa. 47 The PI3K/AKT signaling pathway plays an important role in the development of a variety of human carcinomas; PIK3CA mutations in studies of ESCC have been detected in 2.2%–11.8% of analyzed cases. NFE2L2 is a main transcription regulator of the stress response, and its mutations have been detected in 5.0%–11.4% of patients with ESCC. In this study, TP53, NOTCH1, PIK3CA, and NFE2L2 were among the most common genetic mutations, but there was no difference between the NDNS and DS groups. However, KMT2D alterations were less frequent, and CDKN2A alterations were more frequent in the NDNS group than in the DS group. Frequent mutations in the genes involved in histone modifications, including KMT2D, have been found in ESCC. KMT2D mutations occur in 18% of ESCC cases, most of which result in truncated proteins lacking the key methyltransferase domain, indicating a tumor suppressor role for KMT2D in ESCC. 48 , 49 Our results showed that the DS group had a significantly higher frequency of KMT2D alterations and 70% more nonsense mutations than the NDNS group. Mutation location also showed inactivation of all DNA domains after 2409. A study of smoking‐related tumors found that KMT2D alterations were predominantly more common in patients with a smoking history and were considered smoking‐related genetic mutations. 50 In our study, nearly all 15 patients with KMT2D alterations in the DS group had a smoking history, whereas none of the patients in the NDNS group without a smoking history had KMT2D alterations. Among ESCCs, this is the only study to report that KMT2D alterations are more common in patients who smoke. Therefore, KMT2D was rarely found in the NDNS group without a smoking history, suggesting that it is a crucial genetic variant in the DS group, particularly in cases with a smoking history. CDKN2A is a gene involved in the cell cycle regulatory pathway that is mutated in 8% of ESCCs and is the driver gene for ESCC. 51 CDKN2A encodes the cyclin‐dependent kinase inhibitor p16 (Ink4a). p16INK4A is a well‐known surrogate marker for human papillomavirus (HPV) infection. 52 In smokers, HPV infection increases the risk of ESCC development. 53 In our study, CDKN2A alterations were more common in the NDNS group than in the DS group. No significant difference was observed in the frequency of CDKN2A deletions; however, a significant difference was found in the frequency of mutations. The location of the mutation in the NDNS group also suggests the inactivation of Ank_2 in the DNA‐binding domain. Similar to the results of this study, a study of oral floor cancer found no significant difference in the frequency of deletions in the nondrinking, nonsmoking group compared with the drinking and smoking groups, with predominantly more mutations found. 54 Onozato et al. also reported that CDKN2A gene variants were significantly more abundant in the background epithelium of patients with ESCC without risk factors, including alcohol consumption and smoking. 55 This is the first genetic analysis of ESCC in nondrinking, nonsmoking women, where CDKN2A mutations were more common in tumor areas than in nontumor areas. Additionally, p16 positivity, an indirect marker of HPV infection, was more common in the NDNS group, suggesting more HPV‐positive cases among nondrinking, nonsmoking females. The correlation between CDKN2A alterations and p16 was not completely consistent, and many cases were CDKN2A‐negative and p16‐positive. CDKN2A alterations in the presence or absence of HPV infection in head and neck cancer have been reported, with alterations being more common in HPV‐negative cases, 56 indicating that CDKN2A alterations and p16 are not completely consistent. Therefore, there is a distinctive carcinogenic pathway in ESCC in the NDNS group that is not associated with known risk factors, suggesting that CDKN2A plays a vital role in this pathway.
This study has some limitations. First, it was a single‐center, retrospective study. Therefore, a large‐scale, multicenter prospective study should be conducted to confirm our findings. Second, 24‐h multichannel intraluminal impedance‐pH (MII‐pH) monitoring should be conducted to verify that RE was an essential factor in ESCC development in the NDNS group. Third, genetic analysis was challenging to perform in all cases. We attempted to extract DNA by LMD in all cases. However, due to the condition of the specimens and their small diameter, only 69 lesions could be evaluated. Therefore, further genetic analysis is needed to elucidate the origin of the NDNS group tumors. Fourth, we observed cases with low VAF that were relatively homogeneous in immunohistochemistry. The reason for this may be that these were epithelium and lamina propria mucosa cases, and normal tissue may have been included during laser microdissection. This could be a limitation of the laser microdissection technique. Fifth, we observed positive immunostaining in nonsense mutations in KMT2D. One possibility is that the nonsense mutation is located after 2409 with the DNA domains up to 2409 remaining functional, causing positive immunostaining. Functional analysis was needed for accurate evaluation but was unavailable in our study. We consider this a limitation. Sixth, immunostaining for p16 suggested that HPV infection was more common in the NDNS group. We could not determine whether HPV infection was related to carcinogenesis of esophageal cancer in nondrinking, nonsmoking females because we did not perform functional analysis. We plan to investigate this in a future study. Seventh, the findings of a CDKN2A copy number of 0 on NGS analysis were not verified using microarray or real‐time polymerase chain reaction, and complete deletion of the copy number could not be confirmed.
In conclusion, older nondrinker, nonsmoker females with RE and few multiple LVLs may develop longitudinal 0–IIa ESCC with keratinization of the posterior wall of the mid‐thoracic esophagus and have fewer KMT2D alterations and more CDKN2A alterations. However, even in nondrinkers and nonsmokers at low risk of ESCC development, caution should be exercised, and endoscopy should be performed in older patients with RE and fewer multiple LVLs. Additionally, CDKN2A mutations are common in ESCC in nondrinker, nonsmoker females, which may have implications for drug discovery and selection.
AUTHOR CONTRIBUTIONS
Motomitsu Fukuhara: Writing – original draft (lead). Yuji Urabe: Conceptualization (equal); methodology (equal); writing – original draft (supporting); writing – review and editing (equal). Hikaru Nakahara: Formal analysis (equal); investigation (equal). Akira Ishikawa: Formal analysis (equal); investigation (equal). Kazuki Ishibashi: Formal analysis (equal); investigation (equal). Hirona Konishi: Formal analysis (equal); investigation (equal). Junichi Mizuno: Formal analysis (equal); investigation (equal). Hidenori Tanaka: Formal analysis (equal); investigation (equal). Akiyoshi Tsuboi: Formal analysis (equal); investigation (equal). Ken Yamashita: Formal analysis (equal); investigation (equal). Yuichi Hiyama: Formal analysis (equal); investigation (equal). Hidehiko Takigawa: Formal analysis (equal); investigation (equal). Takahiro Kotachi: Formal analysis (equal); investigation (equal). Ryo Yuge: Formal analysis (equal); investigation (equal). C. Nelson Hayes: Formal analysis (equal); investigation (equal). Shiro Oka: Methodology (equal); supervision (lead); writing – review and editing (equal).
FUNDING INFORMATION
This study was supported by grants from the Japan Society for the Promotion of Science (JSPS KAKENHI, grant number: 21K07963; https://www.jsps.go.jp/j‐grantsinaid/).
CONFLICT OF INTEREST STATEMENT
The authors have no conflict of interest.
ETHICS STATEMENT
The Institutional Review Board and Ethics Committee of Hiroshima University approved this retrospective study (no. E‐1869), and it was performed in accordance with the Declaration of Helsinki and its later amendments.
PATIENT CONSENT STATEMENT
All the participants provided written informed consent.
CLINICAL TRIAL REGISTRATION
N/A.
Supporting information
Table S1. Cancer‐related genes included in MSK‐IMPACT.
Table S2. Raw data of gene mutations identified by genetic analysis.
Table S3. Raw data of copy number variants identified by genetic analysis.
ACKNOWLEDGMENTS
We thank Haru Hashiguchi and Naoyuki Takahashi for their technical support. This work was supported by the Natural Science Center for Basic Research and Development and Program of the Network‐Type Joint Usaga/Research Center for Radiation Disaster Medical Science of Hiroshima University, Nagasaki University, and Fukushima Medical University.
Fukuhara M, Urabe Y, Nakahara H, et al. Clinicopathological and genomic features of superficial esophageal squamous cell carcinomas in nondrinker, nonsmoker females. Cancer Med. 2024;13:e7078. doi: 10.1002/cam4.7078
DATA AVAILABILITY STATEMENT
All data are available in the manuscript or supplementary materials.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381(9864):400‐412. [DOI] [PubMed] [Google Scholar]
- 3. Ishihara R, Tanaka H, Iishi H, et al. Long‐term outcome of esophageal mucosal squamous cell carcinoma without lymphovascular involvement after endoscopic resection. Cancer. 2008;112(10):2166‐2172. [DOI] [PubMed] [Google Scholar]
- 4. Mizumoto T, Hiyama T, Oka S, et al. Diagnosis of superficial esophageal squamous cell carcinoma invasion depth before endoscopic submucosal dissection. Dis Esophagus. 2018;31(7):p.dox142. [DOI] [PubMed] [Google Scholar]
- 5. Mizumoto T, Hiyama T, Quach DT, et al. Magnifying endoscopy with narrow band imaging in estimating the invasion depth of superficial esophageal squamous cell carcinomas. Digestion. 2018;98(4):249‐256. [DOI] [PubMed] [Google Scholar]
- 6. Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64(3):381‐387. [DOI] [PubMed] [Google Scholar]
- 7. Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 2018;154(2):360‐373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tachimori Y, Ozawa S, Numasaki H, et al. Comprehensive registry of esophageal cancer in Japan, 2012. Esophagus. 2019;16:221‐245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oze I, Matsuo K, Ito H, et al. Cigarette smoking and esophageal cancer risk: an evaluation based on a systematic review of epidemiologic evidence among the Japanese population. Jpn J Clin Oncol. 2012;42(1):63‐73. [DOI] [PubMed] [Google Scholar]
- 10. Ishikawa A, Kuriyama S, Tsubono Y, et al. Smoking, alcohol drinking, green tea consumption and the risk of esophageal cancer in Japanese men. J Epidemiol. 2006;16(5):185‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee CH, Wu DC, Lee JM, et al. Carcinogenetic impact of alcohol intake on squamous cell carcinoma risk of the oesophagus in relation to tobacco smoking. Eur J Cancer. 2007;43(7):1188‐1199. [DOI] [PubMed] [Google Scholar]
- 12. Cui R, Kamatani Y, Takahashi A, et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology. 2009;137(5):1768‐1775. [DOI] [PubMed] [Google Scholar]
- 13. Kagemoto K, Urabe Y, Miwata T, et al. ADH1B and ALDH2 are associated with metachronous SCC after endoscopic submucosal dissection of esophageal squamous cell carcinoma. Cancer Med. 2016;5(7):1397‐1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Urabe Y, Kagemoto K, Nakamura K, et al. Construction of a risk model for the development of metachronous squamous cell carcinoma after endoscopic resection of esophageal squamous cell carcinoma. Esophagus. 2019;16:141‐146. [DOI] [PubMed] [Google Scholar]
- 15. Lundell LR, Dent J, Bennett JR, et al. Endoscopic assessment of oesophagitis: clinical and functional correlates and further validation of the Los Angeles classification. Gut. 1999;45(2):172‐180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kimura K, Takemoto T. An endoscopic recognition of the atrophic border and its significance in chronic gastritis. Endoscopy. 1969;1(3):87‐97. [Google Scholar]
- 17. Japan Esophageal Society . Japanese classification of esophageal cancer, 11th edition: part 1. Esophagus. 2017;4(1):1‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oyama T, Momma K. A new classification of magnified endoscopy for superficial esophageal squamous cell carcinoma. Esophagus. 2011;8:247‐251. [Google Scholar]
- 19. Kahrilas PJ, Kim HC, Pandolfino JE. Approaches to the diagnosis and grading of hiatal hernia. Best Pract Res Clin Gastroenterol. 2008;22(4):601‐616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kitagawa Y, Uno T, Oyama T, et al. Esophageal cancer practice guidelines 2017 edited by the Japan esophageal society: part 1. Esophagus. 2019;16:1‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mils SE, ed. Histology for Pathologists. 4th ed. Lippincott Williams & Wilkins; 2012. 7. [Google Scholar]
- 22. Zehir A, Benayed R, Shah R, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703‐713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H, Durbin R. Fast and accurate short read alignment with burrows‐wheeler transform. Bioinformatics. 2009;25(14):1754‐1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Riester M, Singh P, Brannon R, et al. PureCN: copy number calling and SNV classification using targeted short read sequencing. Source Code Biol Med. 2016;11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Q, Wang K. InterVar: clinical interpretation of genetic variants by the 2015 ACMG‐AMP guidelines. Am J Hum Genet. 2017;100(2):267‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang D, Zhou J, Chen B, Zhou L, Tao L. Gastroesophageal reflux and carcinoma of larynx or pharynx: a meta‐analysis. Acta Otolaryngol. 2014;134(10):982‐989. [DOI] [PubMed] [Google Scholar]
- 27. Oda J, Iriguchi Y, Mizutani M, et al. Association of alcohol drinking and smoking with reflux esophagitis in squamous cell carcinoma of the esophagus. Stomach Intestine (Tokyo). 2022;57:1456‐1462. [Google Scholar]
- 28. Shigaki H, Imamura Y, Mine S, et al. Clinicopathological features of esophageal squamous cell carcinoma in never smoker‐never drinkers. Dis Esophagus. 2017;30(5):1‐7. [Google Scholar]
- 29. Freedman ND, Lacey JV Jr, Hollenbeck AR, Leitzmann MF, Schatzkin A, Abnet CC. The 17 association of menstrual and reproductive factors with upper gastrointestinal tract cancers in the NIH‐AARP cohort. Cancer. 2010;116(6):1572‐1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bodelon C, Anderson GL, Rossing MA, Chlebowski RT, Ochs‐Balcom HM, Vaughan TL. Hormonal factors and risks of esophageal squamous cell carcinoma and adenocarcinoma in postmenopausal women. Cancer Prev Res. 2011;4(6):840‐850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu H, Liu G, Zhao P, Zhu L. Hormonal and reproductive factors and risk of esophageal cancer in Chinese postmenopausal women: a case‐control study. Asian Pac J Cancer Prev. 2011;12(8):1953‐1956. [PubMed] [Google Scholar]
- 32. Asanuma K, Iijima K, Shimosegawa T. Gender difference in gastroesophageal reflux diseases. World J Gastroenterol. 2016;22(5):1800‐1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fukuchi T, Hirasawa K, Sato C, et al. Potential roles of gastroesophageal reflux in patients with superficial esophageal squamous cell carcinoma without major causative risk factors. J Gastroenterol. 2021;56(10):891‐902. [DOI] [PubMed] [Google Scholar]
- 34. Inoue T, Ishihara R, Matsuura N, et al. Endoscopic features of superficial esophageal squamous cell carcinoma in patients with very low risk factors (female, non‐drinking, and non‐smoking): a case‐control study. Dig Dis. 2021;39(6):577‐584. [DOI] [PubMed] [Google Scholar]
- 35. Kuwabara H, Chiba H, Tachikawa J, et al. Clinical characteristic of esophageal cancer without lugol‐voiding lesions in the background esophagus. Dig Endosc. 2020;32(4):621‐627. [DOI] [PubMed] [Google Scholar]
- 36. Yamaguchi T, Sugimoto T, Yamauchi M, Matsumori Y, Tsutsumi M, Chihara K. Multiple vertebral fractures are associated with refractory reflux esophagitis in postmenopausal women. J Bone Miner Metab. 2005;23:36‐40. [DOI] [PubMed] [Google Scholar]
- 37. Rake G. Epithelioma of the oesophagus in association with achalasia of the cardia. Lancet. 1931;2(5639):682‐683. [Google Scholar]
- 38. Chino O, Kijima H, Shimada H, et al. Clinicopathological studies of esophageal carcinoma in achalasia: analyses of carcinogenesis using histological and immunohistochemical procedures. Anticancer Res. 2000;20(5C):3717‐3722. [PubMed] [Google Scholar]
- 39. Leeuwenburgh I, Gerrits M, Capello A, et al. Expression of p53 as predictor for the development of esophageal cancer in achalasia patients. Dis Esophagus. 2010;23(6):506‐511. [DOI] [PubMed] [Google Scholar]
- 40. Chen KH, Mukaisho K, Ling ZQ, Shimomura A, Sugihara H, Hattori T. Association between duodenal contents reflux and squamous cell carcinoma–establishment of an esophageal cancer cell line derived from the metastatic tumor in a rat reflux model. Anticancer Res. 2007;27(1A):175‐181. [PubMed] [Google Scholar]
- 41. Mukaisho K, Nakayama T, Yamamoto H, Hagiwara T, Sugihara H. Duodenal contents reflux can induce esophageal squamous cell carcinoma as well as adenocarcinoma lessons from animal experiments. J Cancer Sci Clini Oncol. 2014;1(1):1‐6. [Google Scholar]
- 42. Sato S, Yamamoto H, Mukaisho K, et al. Continuous taurocholic acid exposure promotes esophageal squamous cell carcinoma progression due to reduced cell loss resulting from enhanced vascular development. PloS One. 2014;9(2):e88831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tateishi Y, Fukuchi T, Hirasawa K, et al. Clinicopathological features of superficial esophageal squamous cell carcinoma in patients with no history of smoking and alcohol consumption. Stomach Intestine (Tokyo). 2022;57:1359‐1366. [Google Scholar]
- 44. Gao YB, Chen ZL, Li JG, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46(10):1097‐1102. [DOI] [PubMed] [Google Scholar]
- 45. Lin DC, Hao JJ, Nagata Y, et al. Genomic and molecular characterization of esophageals quamous cell carcinoma. Nat Genet. 2014;46(5):467‐473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sawada G, Niida A, Uchi R, et al. Genomic landscape of esophageal squamous cell carcinoma in a Japanese population. Gastroenterology. 2016;150(5):1171‐1182. [DOI] [PubMed] [Google Scholar]
- 47. Urabe Y, Kagemoto K, Hayes CN, et al. Genomic characterization of early‐stage esophageal squamous cell carcinoma in a Japanese population. Oncotarget. 2019;10(41):4139‐4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang X, Wang Y, Meng L. Comparative genomic analysis of esophageal squamous cell carcinoma and adenocarcinoma: new opportunities towards molecularly targeted therapy. Acta Pharm Sin B. 2022;12(3):1054‐1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sasaki Y, Tamura M, Koyama R, Nakagaki T, Adachi Y, Tokino T. Genomic characterization of esophageal squamous cell carcinoma: insights from next‐generation sequencing. World J Gastroenterol. 2016;22(7):2284‐2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kytola V, Topaloglu U, Miller LD, et al. Mutational landscapes of smoking‐related cancers in Caucasians and African Americans: precision oncology perspectives at wake Forest Baptist Comprehensive Cancer Center. Theranostics. 2017;7(11):2914‐2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang L, Zhou Y, Cheng C, et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet. 2015;96(4):597‐611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. D'Souza G, Kreimer AR, Viscidi R, et al. Case‐control study of human papillomavirus and oropharyngeal cancer. New Engl J Med. 2007;356(19):1944‐1956. [DOI] [PubMed] [Google Scholar]
- 53. Qi Z, Jiang Q, Yang J, et al. Human papillomavirus (HPV) infection and the risk of esophageal squamous cell carcinoma. Dis Esophagus. 2013;26(1):61‐67. [DOI] [PubMed] [Google Scholar]
- 54. Koo K, Mouradov D, Angel CM, et al. Genomic signature of oral squamous cell carcinomas from non‐smoking non‐drinking patients. Cancers (Basel). 2021;13(5):1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Onozato Y, Sasaki Y, Abe Y, et al. Novel genomic alteration in superficial esophageal squamous cell neoplasms in non‐smoker non‐drinker females. Sci Rep. 2021;11(1):20150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. The Cancer Genome Atlas Network . Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576‐582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Cancer‐related genes included in MSK‐IMPACT.
Table S2. Raw data of gene mutations identified by genetic analysis.
Table S3. Raw data of copy number variants identified by genetic analysis.
Data Availability Statement
All data are available in the manuscript or supplementary materials.


