Abstract
Roaz, a rat C2H2 zinc finger protein, plays a role in the regulation of olfactory neuronal differentiation through its interaction with the Olf-1/EBF transcription factor family. An additional role for the Roaz/Olf-1/EBF heterodimeric protein is suggested by its ability to regulate gene activation at a distinct promoter lacking Olf-1/EBF-binding sites. Using an in vitro binding-site selection assay (Selex), we demonstrate that Roaz protein binds to novel inverted perfect or imperfect repeats of GCACCC separated by 2 bp. We show that Roaz is capable of binding to a canonical consensus recognition sequence with high affinity (Kd = 3 nM). Analysis of the structural requirement for protein dimerization and DNA binding by Roaz reveals the role of specific zinc finger motifs in the Roaz protein for homodimerization and heterodimerization with the Olf-1/EBF transcription factor. The DNA-binding domain of Roaz is mapped to the N-terminal 277 amino acids, containing the first seven zinc finger motifs, which confers weak monomeric binding to a single half site and a stronger dimeric binding to the inverted repeat in a binding-site-dependent manner. Full-length protein can form dimers on both the inverted repeat and direct repeat but not on a single half site. These findings support the role of the TFIIIA-type Zn fingers in both protein-protein interaction and protein-DNA interaction and suggest distinct functions for specific motifs in proteins with a large number of zinc finger structures.
Gene expression is temporally and spatially regulated in a complex and organized sequence. Higher-order complex formation requires the interplay between specific DNA sequences and DNA-binding transcription factors, as well as additional components that interact with the DNA-protein complexes through protein-protein interaction. Certain structural elements, such as zinc finger motifs of the transcription factor IIIA (TFIIIA) type (4, 25), the basic helix-loop-helix motifs (27), basic leucine zippers (b-zip) (39, 40) and POU/homeodomains (20, 33), are shared by DNA-binding proteins and serve as an interface for DNA recognition. Many of these proteins function in embryonic development and regulate cell proliferation and differentiation. On the other hand, leucine zipper motifs (22), helix-loop-helix domains (1), LIM/double zinc finger domains (13), and PAS domains (21) are implicated in mediating protein-protein interactions that are essential for the function of those transcription factors.
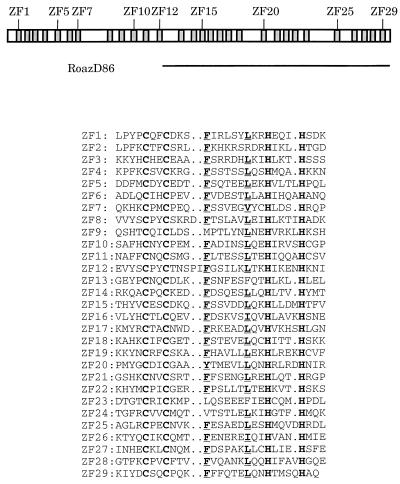
Roaz (rat Olf-1/EBF-associated zinc finger protein) is a 134-kDa protein containing 29 C2H2 zinc finger domains of the TFIIIA type (Fig. 1). The C-terminal half of Roaz (RoazD86), containing 17 zinc finger structures, was identified by its ability to associate with Olf-1/EBF in a yeast two-hybrid screen and subsequently shown to display specific interactions by biochemical assay and in cell line-based expression systems (38). Further studies demonstrated that Roaz functioned as a coregulator of Olf-1/EBF proteins to control olfactory gene expression. Roaz abolished Olf-1/EBF-mediated transactivation of the native promoters upstream of the olfactory marker protein (OMP) and type III adenylyl cyclase (ACIII) by sequestering Olf-1/EBF proteins in a heteromultimeric complex which failed to bind the Olf-1/EBF-binding site. Expression of Roaz mRNA in adult mice was restricted to the lower one-third of the olfactory epithelium consisting of basal cells, neuronal precursors, and immature neurons. Notably, OMP and other mature neuronal markers regulated by Olf-1/EBF proteins were expressed in the middle and upper regions of the epithelium in a complementary manner to Roaz. This pattern of expression, in conjunction with its biochemical properties, suggested that Roaz acts as a switch protein that contributes to the control of transition between the proliferative and differentiated states.
FIG. 1.
Schematic diagram and sequence comparison of the 29 zinc fingers of Roaz. A schematic diagram of Roaz with individual zinc finger structures represented by shaded boxes is depicted at the top. The amino acid alignment of the 29 zinc fingers of Roaz is shown below. The zinc-coordinating Cys and His residues are shown in bold letters, and the conserved aromatic amino acids and branched aliphatic amino acids are underlined. ZF, zinc finger.
In contrast to the negative regulation at Olf-1/EBF-binding sites, coexpression of Roaz and Olf-1/EBF proteins in a cell line-based transactivational assay led to a dramatic activation of reporter expression at a simian virus 40 (SV40) minimal promoter which lacked Olf-1/EBF-binding sites. Further experiments revealed that Roaz protein displayed direct binding to a specific region of this promoter containing the six consecutive SP1 sites (38). In this study, we identified a novel DNA sequence recognized specifically by Roaz protein with high affinity through an in vitro binding-site selection assay (Selex) and characterized the protein structures involved in homomeric and heteromeric protein-protein interaction and DNA-protein interaction. We showed that distinct TFIIIA zinc finger motifs can mediate protein-protein or DNA-protein interaction and that high-affinity DNA-protein association requires the presence of two consensus half sites arranged in a palindromic sequence.
MATERIALS AND METHODS
Plasmid constructs.
pCIS-GSTRoaz was constructed by subcloning a 4.5-kb EcoRV-NotI fragment isolated from pBS-Roaz into the pCMV-GST expression vector (38). pBS-Roaz was the composite cDNA clone reconstituted from three different cDNA clones by using PmlI and BspEI conserved sites between JBOZ2.1 and JBOZ1.3 and between JBOZ1.3 and RoazD86 (38). pCIS-XPRoazD86 and pCIS-XPIC2 plasmid constructs contain the RoazD86 and IC2 inserts (an irrelevant polypeptide), respectively, tagged with an XPress sequence (DLYDDDDK; Invitrogen, San Diego, Calif.); pCIS-GSTRoazD86 contained the RoazD86 insert fused with glutathione S-transferase (GST) in pCMV-GST. pPC97-RoazD86 and pPC86-RoazD86 contain the RoazD86 inserts cloned in the yeast expression vectors containing the GAL4 DNA-binding domain (pPC97) or GAL4 transactivator domain (pPC86) (5).
Terminal truncation constructs were made with the Erase-a-base kit (Promega, Madison, Wis.). N-terminal deletions of pPC86-RoazD86 were made by protecting the SalI site and deleting from the BspEI site. C-terminal deletions of pPC86-RoazD86 were made by protecting the SpeI site and deleting from the HpaI site. All deletion constructs were verified by DNA sequence analysis.
Internal truncations of pPC86-RoazD86 and “broken-finger” mutants were generated by single-stranded DNA mutagenesis. Single-stranded DNA of pCIS-GSTRoazD86 was prepared by cotransfecting CJ236 cells with the plasmid construct and helper phage and annealed with phosphorylated mutagenic oligonucleotides. Extension and ligation reactions were carried out at 37°C for 2 h in the presence of T4 DNA polymerase and T4 DNA ligase. Double-stranded DNA was transformed into HB101 cells. Deletion mutants were first screened by PCR and later confirmed by sequence analysis. The SalI-NotI fragments from individual mutant constructs were subcloned into the yeast expression vector (pPC86) for the yeast two-hybrid interaction assay. The sequences of the mutagenic oligonucleotides are as follows: ind262, 5′-GCGTGAGCCATCGTCCTC|CTCGTTGCTGTGCTTCAC-3′; ind263, 5′-CAGCTGAGCTGCCAGCCAG|TCCGGGCCGGATGTTATG-3′; ind264, 5′-ACACTCATGGTTGATGCC|CTTCTGCATGTGAAAGGTG-3′; bzf275, 5′-GTTCGATGAGGTTACAAAGGAGC-3′; and bzf277, 5′-GCTCATCGTGTTGTTCTGCAAC-3′. Vertical lines in the internal deletion constructs indicate the deletion sites; the mutated nucleotides in the “broken-finger” mutants are underlined.
EMSA.
An electrophoretic mobility shift assay (EMSA) was carried out essentially as described previously (15, 41) with the following modifications. Protein was mixed with specified amounts of probe in a 20-μl binding-reaction mixture and incubated at 20°C for 20 min. The binding-reaction mixture contained 10 mM HEPES (pH7.9), 70 mM KCl, 1 mM dithiothreitol, 4% glycerol, 2.5 mM MgCl2, 100 μM ZnCl2, 1 mM EDTA, 100 μg of poly(dI-dC) per ml, and 20 μg of salmon sperm DNA per ml unless indicated otherwise. The mixture was subjected to electrophoresis on either a 6% polyacrylamide gel (acrylamide/bisacrylamide ratio, 59:1) or a 1.5% agarose gel (SeaKem, Rockland, Maine) in 0.25× Tris-borate-EDTA (TBE) at 4°C. The products were detected by autoradiography of the dried gel.
For the Selex assay and DNA-binding affinity study, PCR products were labeled with [γ-32P]ATP by using T4 DNA polynucleotide kinase in a 20-μl reaction mixture. For the DNA-binding assay of C-terminal truncation, complementary oligonucleotides with a 5′ GATC overhang were annealed in annealing buffer (100 mM KCl, 10 mM Tris [pH 8.0], 1 mM EDTA) at 100°C for 5 min and 68°C for 1 h, and slowly cooled to room temperature. Double-stranded DNA fragments were gel purified, and 75-ng portions of the isolates were labeled with [α-32P]dCTP by using the Klenow fill-in reaction.
Selex.
Optimal DNA sequences for Roaz binding were determined by a PCR-amplified Selex protocol (42, 43). The 76-mer oligonucleotides (5′-CATGAAT TCTCCTATACTGAGT TCATGATN18TGATATCGAACTGTATCGATGAAT TCCAC-3′) including 18 random nucleotides and two PCR primer sequences corresponding to the first (top strand) 20 bases and complementary to the last (bottom) 20 bases were chemically synthesized. A random sequence library was generated by a primer extension reaction carried out with the purified 76-mers as template and the bottom-strand primer in a 20-μl Klenow reaction mixture. Double-stranded DNA fragments were gel purified on a 4% low-melting-point agarose gel and labeled with [γ-32P]ATP by using T4 DNA polynucleotide kinase. Radiolabeled probes (3 × 104 cpm) were mixed with 60 ng of GST-Roaz fusion protein purified through a GST affinity column (38) in EMSA with 20 μl of buffer. The gel was subjected to autoradiography, and the regions corresponding to the shifted bands were cut out and separately eluted in 250 μl of elution buffer (500 mM ammonium acetate, 1 mM EDTA [pH 8.0]) for 4 h at 37°C. A fraction of the eluate (10 μl) was used for subsequent PCR amplification with 10 ng of each primer per μl and 0.2 mM each deoxynucleoside triphosphate in a 50-μl reaction mixture. PCR was programmed as 50°C for 1 min, 72°C for 1 min, and 94°C for 30 s for 35 cycles. The PCR products were then gel purified, radiolabeled, and subjected to the next round of EMSA. Nine rounds of EMSA selection were performed, with PCR amplification after each round. The final products were digested with EcoRI, subcloned into pBlueScript, and analyzed by sequence analysis with T7 primer.
Measurement of the apparent dissociation constant.
The apparent dissociation constant (Kd) of Roaz binding to specific DNA fragments was estimated as the protein concentration at which half of the DNA probe was shifted in an EMSA. Probes were prepared from purified PCR fragments containing different inserts and radiolabeled with [γ-32P]ATP in a kinase reaction. For affinity studies, individual probes (2 pM) were mixed with purified GST-Roaz full-length protein at 1, 2, 4, 8, 16, and 32 nM in the presence of 100 μg of dI-dC per ml and 20 μg of salmon sperm DNA per ml and incubated at room temperature for 30 min to reach equilibrium (11). The intensity of shifted bands was measured with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Affinity chromatography and zinc removal experiment.
RoazD86 or OED5 (a partial sequence of Olf-1/EBF isolated from the original yeast two-hybrid screen [38]) was expressed as a GST fusion protein by transient transfection into HEK-293 cells. Four 100-mm-diameter plates of HEK-293 cells were collected, washed twice with 1× phosphate-buffered saline (PBS), and extracted with 4 ml of extraction buffer (1× PBS, 1% Triton, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, 2 μg of aprotinin per ml, 1 μg of pepstatin A per ml) for 30 min at 4°C. After the extract was precleared by centrifugation at 15,600 × g for 20 min, 300 μl of a 50% slurry of glutathione-bound agarose beads was added to the supernatant and mixed at 4°C for 1 h. The beads were washed once with 1× PBS, once with 1× PBS plus 500 mM NaCl, and three times with 1× PBS. Whole-cell extracts were isolated from HEK-293 cells transfected with pCIS-XPRoazD86 and mixed with GST fusion protein bound to glutathione-agarose beads. After a 2-h mixing at 4°C, the beads were washed once with 1× PBS, twice with 1× PBS plus 500 mM NaCl, and three times with 1× PBS. Sample buffer was added, and a fraction was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% acrylamide). In the zinc removal experiment, whole-cell extract was treated with 1 mM EDTA, 5 mM EDTA, or 1 mM 1,10-phenanthroline and mixed with GST-RoazD86 or GST-OED5 bound to glutathione-agarose beads for the in vitro binding assay. All in vitro binding experiments were carried out essentially as described above unless indicated otherwise in the figure legends.
In vitro transcription-translation of C-terminal truncations of Roaz and Roaz(bzf277).
In vitro transcription-translation was carried out with a TNT-coupled wheat germ extract system (Promega). The pBS-XPRoaz vector was generated in two steps. First, a 4.5-kb KpnI-NotI fragment from pBSRoaz was subcloned into pEBVHisC (Invitrogen). A 4.9-kb EcoRI-XbaI fragment, containing an in-frame ATG with Kozak consensus sequence, six histidine residues, and an XPress tag at the 5′ end, was isolated and subcloned into pBlueScript vector. pBS-XPRoaz was digested individually with BsgI, HaeII, PmlI, BspEI, and NotI to generate C-terminal truncations (CD1, CD2, CD3, and CD4) and full-length protein, respectively. pBS-XPRoaz(bzf277), encoding a full-length Roaz as described above except for a “broken” 29th finger, was constructed by inserting a BspEI-NotI fragment from pCIS-GSTbzf277 into pBS-XPRoaz. These linearized DNA fragments (2 μg) were used for in vitro transcription-translation with unlabeled methionine as described by the manufacturer.
RESULTS
In vitro selection of the Roaz-binding site.
The ability of Roaz to activate the expression of an SV40 early promoter-regulated reporter gene in the presence of Olf-1/EBF proteins suggested that the Roaz–Olf-1–EBF heterocomplex encoded specific DNA-binding activity. An EMSA with purified Roaz and Olf-1–EBF indicated that the DNA recognition ability of the Roaz–Olf-1–EBF heterocomplex was likely to be conferred by the Roaz protein, and the target sequence was mapped to a region composed mainly of six consecutive SP1 sites (38).
To address the DNA-binding specificity in general and to identify other possible binding sequences, we used a Selex protocol (see Materials and Methods) (42, 43) in which a glutathione-Sepharose-purified GST-Roaz fusion protein (60 ng, 16 nM) was used to select for optimal binding sequences from a DNA library containing a pool of random 18-mers flanked by PCR primer sequences. Shifted DNA-protein complexes of three different mobilities were observed after the first round of selection, with the top and middle bands showing the highest intensities. Two independent Selex procedures were performed individually to select for bound DNA sequences corresponding to either the top or middle band. Eight subsequent rounds of EMSA selection were carried out with purified GST-Roaz fusion protein (60 ng or 16 nM for the first three rounds, and 30 ng or 8 nM for the last five rounds) and radiolabeled PCR products (30,000 cpm) from the previous round of selection.
DNA sequence analysis of 20 isolates from the final round of Selex for the top complex and 14 isolates from the Selex for the middle complex revealed a consensus sequence composed of a perfect or imperfect palindromic sequence [GCACCC(A/ T)(A/T)GGGTGC] in every sequence examined (Fig. 2). The inferred consensus sequence contained a match to the consensus half site GCACCC, a spacer of 2 nucleotides (A or T in either position), and an inverted half site (GGGTGC) or an imperfect inverted half site with a single nucleotide substitution at one of the last two positions (GGGTGA, GGGTGT, GGGTCC). To confirm that the starting library was generated randomly, 25 clones of the original library were sequenced and the average frequency of different nucleotides in each position was estimated to be 24, 28, 24, and 24% for G, A, T, and C, respectively. None of these clones contained a single half site (GCACCC or GGGTGC). The probe derived from the top band or the middle band in each step of the Selex procedure resulted in an increased intensity of all three complexes in the next round of EMSA (data not shown). These observations suggest that these three bands contain similar DNA sequences and are likely to represent monomeric and multimeric complexes of Roaz.
FIG. 2.
Roaz-binding sites from the Selex assay. Sequence alignment of 34 isolates from the final round of PCR amplification is shown. The top 6 sequences were derived from 20 isolates from Selex for the top band; the bottom eight sequences were derived from 14 isolates for the middle band. The conserved sequences are underlined, and the two palindromic repeats are shown in bold letters. In six of the sequences, part of the consensus is contributed by the adjacent primer sequence (parentheses). The number of appearance is given in the right column. Italics indicate non-consensus primer sequence.
Roaz binds a canonical sequence with an apparent Kd of 3 nM.
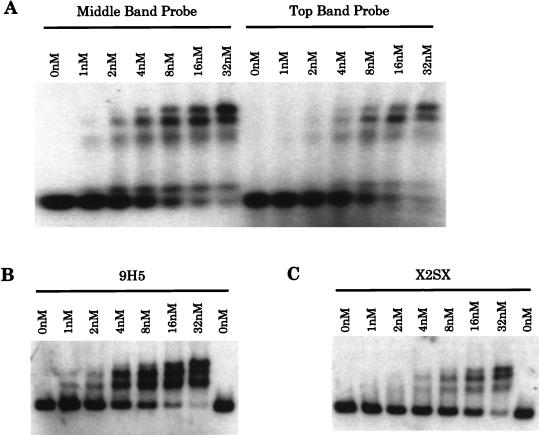
The DNA-binding affinity of Roaz protein was assessed by the protein concentration required to shift half of the DNA probe in an EMSA. In this equilibrium study, less than 2 pM radiolabeled DNA probe was mixed with purified GST-Roaz protein at 1, 2, 4, 8, 16, and 32 nM for 30 min at room temperature, and DNA-protein complexes were resolved on a 1.5% agarose gel. The protein concentration at which half of the DNA probe was shifted was designated the apparent dissociation constant (Kd). The Kd for GST-Roaz binding to the complex DNA probes from the last round of Selex for either the middle or top band was estimated to be around 8 nM (Fig. 3A). The Kd for a canonical palindromic sequence, 9H5 (Fig. 2), was measured in a similar experiment and estimated to be 3 nM (Fig. 3B). To distinguish the orientation and spacing effect from a dosage effect of binding sites, two half sites (GCACCC) were created on a single probe separated by 13 nucleotides and arranged in direct-repeat orientation. The Kd for this probe (X2SX) was estimated to be over 16 nM (Fig. 3C), five- to sixfold higher than that for the inverted repeat.
FIG. 3.
DNA-binding affinity study. (A) A mixture of complex probes (2 pM) from the Selex for the middle band (left) or top band (right) was mixed with purified GST-Roaz, at increasing concentrations as indicated, in the EMSA. (B and C). Parallel experiments to that in panel A, except that a canonical palindromic repeat (9H5) or a direct repeat with 13 bp (X2SX, GCACCCATCGTCGAGATTAGCACCC) was used as a probe in the EMSA, respectively.
Structural analyses of homodimerization and heterodimerization by Roaz.
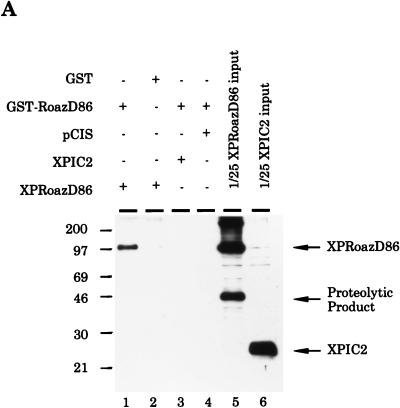
The selection of a large number of independent clones with inverted repeats by Selex (Fig. 2) and the existence of slower-migrating species in the EMSA (Fig. 3) suggested that Roaz might bind DNA as multimeric complexes. The physical interaction was shown by an in vitro binding assay (Fig. 4A). RoazD86 tagged with XPress epitope (XP-RoazD86) could be specifically retained on glutathione-coupled Sepharose beads containing GST-RoazD86 (lane 1) but not on beads containing GST (lane 2). The specificity was confirmed by mixing XPIC2 (an irrelevant polypeptide tagged with XPress epitope) with GST-RoazD86 (lane 3) in a similar experiment.
FIG. 4.
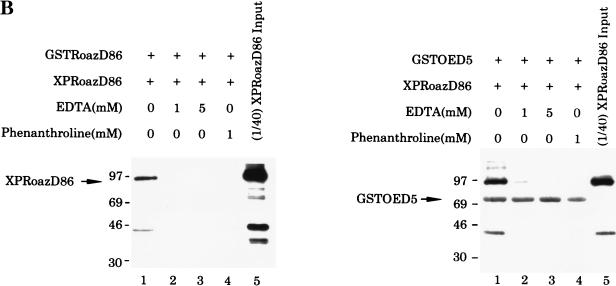
Biochemical characterization of the Roaz protein-protein interaction. (A). Purified proteins (6 to 8 μg of GST or GST-RoazD86 fusion protein bound to 20 μl of a 50% slurry of glutathione-agarose beads) were mixed with whole-cell proteins (400 μg) isolated from HEK293 cells transfected with pCIS, pCIS-XPIC2, or pCIS-XPRoazD86. Bound proteins were extracted with 25 μl of sample buffer, and a portion (8 μl) was fractionated by SDS-PAGE (10% polyacrylamide) and detected by Western blotting with anti-XPress antibody (Invitrogen). A 1/25 portion of the input sample was loaded for comparison. (B) GST fusions of RoazD86 (2 μg) (left) or OED5 (3 μg) (right) were mixed with whole-cell extract (300 μg) isolated from HEK-293 cells transfected with pCIS-XPRoazD86 pretreated with 1 mM EDTA (lane 2), 5 mM EDTA (lane 3), or 1 mM 1,10-phenanthroline (lane 4) for the in vitro binding assay. Half of the bound protein was fractionated by SDS-PAGE (10% acrylamide). A 1/40 portion of the input sample was loaded in lane 5 for comparison.
The only discernible structural motifs present in RoazD86 were 17 C2H2 zinc fingers distributed along 700 amino acids. To examine the possible roles of these zinc finger motifs in mediating the protein-protein interaction, we performed a zinc removal experiment by treating the XPRoazD86-containing extracts with 1 or 5 mM EDTA to remove divalent cations or with 1 mM 1,10-phenanthroline (a zinc chelator). GST-RoazD86 (2 μg) or GST-OED5 (3 μg) was subsequently added to the EDTA- or 1,10-phenanthroline-treated protein extract in a binding assay (Fig. 4B). OED5 encoded residues 221 to 570 of Olf-1–EBF, which were sufficient for Roaz–Olf-1–EBF heterocomplex formation (38). Roaz-Roaz homodimerization (Fig. 4B, left) and Roaz–Olf-1–EBF heterodimerization (right) were both abolished in the presence of EDTA (lanes 2 and 3), or 1 mM 1,10-phenanthroline (lanes 4). The 70-kDa band (Fig. 4B, right) represented the GST-OED5 protein that could cross-react with the anti-XPress antiserum. Coomassie blue staining showed that the binding of GST-RoazD86 and GST-OED5 to glutathione-coupled agarose beads was not affected by the 1 mM 1,10-phenanthroline or 5 mM EDTA treatment (data not shown).
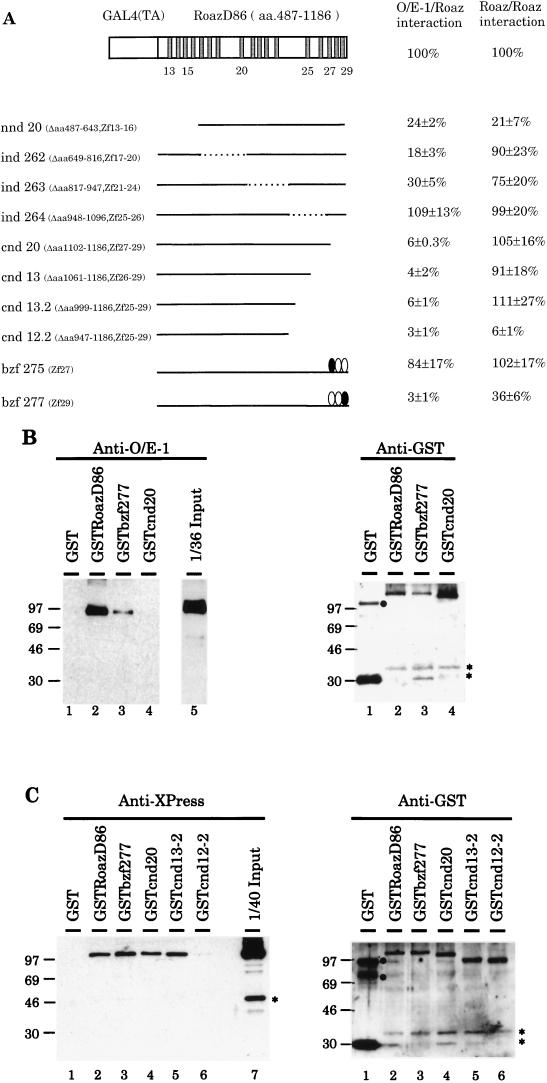
The regions of Roaz involved in homomultimerization and specific intermolecular interactions with Olf-1–EBF were identified in the yeast two-hybrid interaction assay with a series of N-terminal-, C-terminal- and internal-deletion constructs of RoazD86 fusion protein with the Gal4 transactivator domain (Gal4TA-RoazD86). The strength of interaction was determined by a β-galactosidase assay in yeast liquid cultures (2) and expressed as a percentage of the activity observed for interactions of the intact Gal4TA-RoazD86 with the Olf-1–EBF or RoazD86 proteins fused to the Gal4 DNA-binding domain (Fig. 5A). The relative level of protein expression for each construct was assessed by Western blotting with an anti-Gal4TA antibody and found to vary less than twofold, except for bzf277, which was expressed at one-third the level of the intact protein, and nnd20, which was expressed at less than one-third the level (unpublished data). The deletions of RoazD86 revealed that a region corresponding to the last 85 amino acids (amino acids 1102 to 1186) and encoding the last three zinc finger motifs was essential for Roaz–Olf-1–EBF heterodimerization. Loss of this region resulted in a 17-fold reduction in the protein-protein interaction. All constructs which removed this region (cnd20, cnd13, cnd13.2, and cnd12.2) resulted in a dramatic reduction in the Roaz–Olf-1–EBF interaction. Internal deletions of about 150 amino acids across the protein coding region led to activities similar to (ind264) or modestly reduced from (ind262 and ind263) that of the intact protein. The low activity observed in the nnd20 construct can be accounted for by the reduced protein expression level. When the homomultimeric interaction of Roaz with itself was examined with the same set of constructs, normal levels of activity were seen in all cases, with the exception of cnd12.2, where the interaction was reduced 20-fold.
FIG. 5.
Regions essential for hetero- and homomultimerization of Roaz. (A). Yeast strain Y190, harboring the GAL4 DNA-binding domain fusions to Olf-1–EBF or RoazD86, was transformed with constructs encoding C-terminal deletions (cnd), N-terminal deletions (nnd), internal deletions (ind), or broken-finger mutants (bzf) of RoazD86 as GAL4 transactivator domain fusions. Double transformants were assayed for β-galactosidase activity. The positions of the amino acids (aa) which define the deletions in the nnd, cnd, and ind constructs and the zinc finger which is mutated in the bzf constructs are indicated in parentheses. The strength of interaction for an individual mutant is expressed as a percentage relative to that of intact proteins, which is set at 100% and corresponds to 43 ± 7 U for Roaz–Olf-1–EBF and 26 ± 5 U for Roaz-Roaz interactions. All measurements were determined for at least four independent colonies. A schematic diagram of Gal(TA)-RoazD86, with each shaded box representing a zinc finger structure, is shown at the top. Numbers under the boxes refer to the positions of the fingers. Mutated zinc fingers in the bzf constructs are indicated by solid ovals. Zf, zinc finger. (B) Biochemical characterization of heterodimerization between Roaz mutants and Olf-1–EBF. Whole-cell extracts (100 μg) from HEK-293 cells transfected with pCIS–Olf-1–EBF were mixed with 50 ng of individual GST or GST fusions bound to Sepharose beads as indicated. Half of the retained proteins and 2.8 μg of input whole-cell extracts were resolved by SDS-PAGE and Western blotted with either anti-Olf-1–EBF antiserum (left) or anti-GST antiserum (right). (C) Biochemical characterization of homodimerization with Roaz mutants. Whole-cell extracts (400 μg) from HEK-293 cells transfected with pCIS-XPRoazD86 were mixed with 4 μg of individual GST or GST fusions bound to Sepharose beads as indicated. One-third of the retained proteins and 10 μg of input whole-cell extracts were resolved by SDS-PAGE (10% polyacrylamide) and Western blotted with either anti-XPress antiserum (left) or anti-GST antiserum (right). Solid circles indicate the positions of aggregates of GST multimers, and asterisks indicate proteolytic products.
To definitely demonstrate that specific Zn fingers mediate protein-protein interactions, a set of “broken-finger” mutations with histidine-to-asparagine substitutions at the third zinc ligand position (10) was generated at the 27th (bzf275) or 29th (bzf277) zinc finger in a region crucial for heteromultimerization with Olf-1–EBF (Fig. 5A). The strength of the protein-protein interaction was assessed as described above. Roaz with a broken 29th finger preferentially affected its ability to bind Olf-1–EBF (3%) while producing only a modest reduction in interaction with itself (36%). In light of the threefold reduction of fusion protein expression observed with this construct, we inferred that the last finger is specifically involved in Roaz–Olf-1–EBF interaction. In contrast, a broken 27th zinc finger displayed normal interaction with both Olf-1–EBF and Roaz.
The regions of Roaz involved in homo- and heterodimerization were further corroborated by in vitro binding experiments. Three constructs, RoazD86, bzf277, and cnd20 (Fig. 5A), were expressed as GST fusions and assayed for their ability to heterodimerize with Olf-1–EBF. A broken-finger mutation, bzf277, resulted in a significant reduction in the amount of the retained Olf-1–EBF protein (Fig. 5B, left, lane 3) compared with RoazD86 (lane 2), whereas a C-terminal truncation of 85 amino acids (cnd20) exhibited no detectable binding to Olf-1–EBF (lane 4) under these conditions. The use of an anti-GST antibody on a parallel blot (Fig. 5B, right) demonstrated that comparable amounts of the GST fusions were present on the resins used in each sample. The regions of RoazD86 essential for homodimerization were examined in a similar fashion by using RoazD86, bzf277, cnd20, cnd13-2, and cnd12-2 (Fig. 5C). The results confirmed that cnd12-2 had lost most of its ability to interact with RoazD86 (Fig. 5C, left, lane 6), consistent with the observations in the yeast two-hybrid interaction assay. Control experiments for the amount of GST fusions were performed in parallel as described above (Fig. 5C, right). Native GST, a negative control, failed to interact with Olf-1–EBF or RoazD86.
DNA binding of Roaz to a single half site and inverted repeat involves the first seven zinc finger motifs.
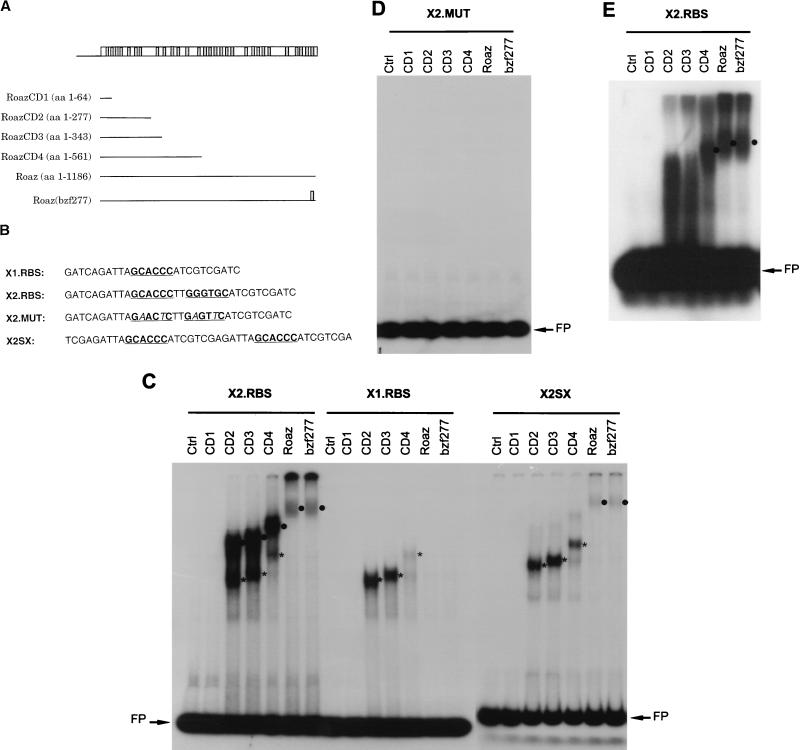
The higher binding affinity of Roaz to the GCACCC inverted repeat than to the direct repeat separated by 13 nucleotides (Fig. 3) and the ability of full-length Roaz to associate with itself (Fig. 4A) suggested that protein dimerization might play an important role in Roaz binding to DNA. Furthermore, Roaz-binding sites identified from the Selex assay with full-length Roaz protein had no affinity for RoazD86, the C-terminal half of Roaz protein, suggesting that DNA binding required the N-terminal half of Roaz (unpublished data). To define the DNA-binding domain in Roaz and to dissect the cooperative binding of the Roaz homodimer from the possible dosage effect, several C-terminally truncated Roaz proteins (RoazCD1, RoazCD2, RoazCD3, and RoazCD4) containing 1 (the first), 7, 8, and 13 zinc finger motifs, respectively, were generated by restriction digestion of the full-length Roaz cDNA and subsequent in vitro transcription-translation (Fig. 6A). These truncated proteins lacked the C-terminal region essential for mediating homodimerization as well as heterodimerization in solution. Roaz(bzf277), encoding a full-length protein with a broken 29th finger, demonstrated little heterodimerization activity, while its ability to homodimerize remained largely intact (Fig. 5). EMSA was performed with an equivalent amount of individual proteins, normalized by measuring the incorporated 35S-labeled methionine, mixed with probes containing an inverted repeat of consensus sites (X2.RBS, GCACCCTTGGGTGC), a single half site (X1.RBS, GCACCC), an inverted repeat with two nucleotide substitutions in the consensus sequence (X2.MUT, GAACTCTTGAGTTC), or a direct repeat of the consensus half site spaced by 13 nucleotides (X2SX) (Fig. 6B). Probes were designed so that the flanking sequences in all probes and spacing sequence in the direct-repeat probe were the same (AGATTA at the 5′ end, ATCGTC at the 3′ end).
FIG. 6.
Characterization of DNA-binding domains in Roaz. (A) Schematic diagram of C-terminally truncated Roaz proteins (CD1, CD2, CD3, and CD4) and broken-finger mutant Roaz(bzf277). Numbers in parentheses indicate the starting and ending amino acids (aa). (B) DNA sequences of synthetic oligonucleotides containing a consensus half site (X1.RBS), a consensus inverted repeat (X2.RBS), a mutated inverted repeat (X2.MUT) and a direct repeat (X2SX). The consensus sequences recognized by Roaz are indicated by bold letters. (C to E) EMSA with 32P-labeled probes as indicated and truncated forms of recombinant Roaz, full-length protein, single broken-finger mutant (bzf277), and control lysate (Ctrl) as shown in panel A. The positions of dimeric and monomeric forms of recombinant proteins are indicated by circles and asterisks, respectively. FP, free probe. (C and D) 6% polyacrylamide gel. (E) 1.5% agarose gel.
Roaz proteins containing the N-terminal seven zinc finger motifs [RoazCD2, RoazCD3, RoazCD4, Roaz, and Roaz(bzf277)] formed complexes with the inverted repeat (X2.RBS) substrate on a 6% polyacrylamide gel (Fig. 6C, left). Two complexes with different mobilities were observed most clearly with CD2, CD3 and CD4 proteins. The shifted complexes with higher mobility consisted of monomer bound to a single half site, as suggested by a parallel EMSA performed with the single half-site probe (X1.RBS) labeled to the same specific activity (Fig. 6C, middle). With the X1.RBS probe, only the complexes with higher mobility could be seen. In contrast, complexes with lower mobility formed only on the inverted repeat and presumably represented dimer formation on the palindromic sequence. EMSA with full-length Roaz or Roaz(bzf277) showed only dimeric binding on the inverted repeat and no monomeric binding on either the inverted repeat or the single half site. The intensities of the Roaz and Roaz(bzf277) dimers on the inverted repeat were lower than those of RoazCD2, RoazCD3, and RoazC4. One simple explanation was that the majority of radioactivity for full-length Roaz and Roaz(bzf277) was retained in the wells on a 6% polyacrylamide gel due to the size of the multimeric complexes (>260 kDa). To further resolve the larger complexes, the same reaction was run on a 1.5% agarose gel (Fig. 6E). The dimeric protein-DNA complexes of Roaz and Roaz(bzf277) became the predominant species, although a significant amount of radioactivity still remained in the wells.
The formation of dimers on the inverted repeat by RoazCD2, RoazCD3, and RoazCD4 recombinant proteins which lack the essential dimerization domain in solution suggests that two properly oriented and spaced half sites may position these C-terminally truncated proteins in such a way that they can bind cooperatively; alternatively, it may be due to a simple dosage effect or double occupancy on the two half sites. To differentiate these two possibilities, a direct repeat of GCACCC spaced by 13 nucleotides was generated and used in a parallel EMSA experiment (Fig. 6C, right). With the direct-repeat probe, only Roaz and Roaz(bzf277) displayed dimer-DNA complexes whereas the other constructs developed only monomeric binding. The specificity of the consensus site in the EMSA was compared with that of a mutated inverted repeat with the same flanking sequence and two base pair changes in the consensus half site (X2.MUT; Fig. 6B). No shifted DNA-protein complexes could be detected with any of the proteins (Fig. 6D).
DISCUSSION
Roaz binding-site selection.
To demonstrate the specific DNA-binding ability of Roaz and also to identify the candidate target sequences, we used the Selex method to show that Roaz recognizes a novel perfect or imperfect inverted repeat of GCACCC in this assay. The apparent Kd of Roaz to one canonical palindromic sequence, 9H5, was approximately 3 nM. Since this number is 3 orders of magnitude higher than the amount of probe, it is likely to represent the true dissociation constant (Kd) in vitro. The presence of inactive protein generated during the purification process or EMSA and binding of Roaz protein to single-stranded DNA would make the actual Kd even lower than what was measured. In this assay, we limited our findings to sequences less than 18 nucleotides and of relatively high affinity, two limitations intrinsic to the Selex protocol. As a result, important sequences that require a larger neighboring context or are regulated in a dynamic manner will not be identified. Nevertheless, the identification of the Roaz consensus binding sequence allowed us to address its DNA binding mechanism and may provide an important clue to illuminating gene expression controlled by Roaz in vivo. This is particularly significant, given the difficulty and limitation of approaches available for identifying downstream targets of specific transcription factors.
Direct involvement of distinct zinc fingers in protein-protein interaction.
The first zinc-requiring transcription factor, the 5S rRNA gene-specific TFIIIA in Xenopus (17), bears repeated motifs of two Cys and two His residues (4, 25). The DNA-binding ability and crystal structure of C2H2 zinc fingers is demonstrated by EMSA and X-ray crystallography of single-finger or two-finger peptides (28). In the study, each of the C2H2 zinc fingers represents a structurally and functionally independent domain and contributes to the recognition of three consecutive nucleotides in the major groove. A subsequent crystal structure study of a five-finger GL1-DNA complex shows that different fingers can play very different roles in DNA recognition (29). Chemical and biochemical studies of TFIIIA also demonstrate the distinct roles of different finger motifs (6, 7, 10, 18, 19, 32). For proteins with a large number of consecutive zinc fingers, interpretation of the structure and function of individual fingers can be even more problematic. For proteins like Roaz (29 fingers) and Xfin (37 fingers) (34), the contribution of individual finger structures to DNA binding remains largely unknown.
Recent work has revealed an additional role for C2H2 zinc finger motifs in the protein-protein interaction and higher-order complex formation. Such examples include human immunodeficiency virus type 1 integrase (12), SV40 large T antigen (23), GATA-1/erythroid Kruppel-like factor (14), TFIIIA (9), Aiolos/Ikaros (26, 37), YY1/CREB (44), serendipity-δ (30), and aspartate transcarbamoylase (24). Other zinc-requiring transcription factors such as the nuclear receptor of the four-Cys type (31), LIM/double-zinc-finger motifs (13, 35), and modified zinc finger structure (36) have all been shown to participate in protein dimerization. In this study, the direct involvement of zinc finger motifs of Roaz in protein-protein interactions is shown by the zinc removal experiment and by use of a set of broken-finger mutants which abolish the individual zinc finger motif without perturbing the overall protein organization and length. One interesting aspect of Roaz is that the region essential for the Roaz–Olf-1–EBF interaction (amino acids 1102 to 1186) is largely dispensable for Roaz-Roaz homodimerization. A broken finger of the last zinc finger affects predominantly the heterodimerization of Roaz with Olf-1–EBF while preserving its homodimerization activity. In contrast, the lack of interaction of Roaz with cnd12-2 but not with C-terminal truncations that extend to amino acid 999 implies that this region (amino acids 947 to 999) is critical for homomeric interaction. However, internal deletion of this region in ind264 results in normal activity. These observations support a role for individual fingers of Roaz in protein recognition specificity and suggest some degree of redundancy in the regions required for homomeric interactions.
Based on the observations presented in this paper, we conclude that the zinc finger motif in Roaz can serve as an interface for protein-protein interactions as well as playing well-established roles in DNA recognition. An attractive corollary of this hypothesis is that proteins possessing multiple zinc fingers such as Xfin and Roaz may utilize some specific domains for protein-protein interaction, which is important in mediating cooperative DNA binding and recruiting addition cofactors with activational or inhibitory activity. The difference in the functional specificity of individual fingers cannot be resolved at the amino acid sequence level, since sequence comparison of the 29 zinc fingers reveals that in addition to the Cys and His residues that directly coordinate zinc ions, most of them contain a conserved aromatic amino acid, F or Y, and a branched aliphatic amino acid (Fig. 1). Therefore, it is likely that residues which are not conserved in all of the Zn fingers contribute to the distinct functions.
Mechanism of DNA binding by Roaz.
EMSA with a series of C-terminally truncated Roaz protein mapped its DNA-binding domain to the N-terminal seven zinc finger motifs, which display both monomeric binding on a single half site and dimer binding on the inverted repeat. The formation of dimers for CD2, CD3, and CD4 on the inverted repeat but not on the single half site or direct repeat with a 13-bp spacer reveals that DNA recognition of the inverted repeat is contributed by both partners in the homodimer, each interacting with a single half site. Although the first seven zinc fingers fail to form a stable dimer in the solution, they can mediate dimerization in the presence of inverted repeat. A similar property is also described for DNA binding of transcription factors including the N-terminal region of Olf-1–EBF (16). On the other hand, full-length Roaz and Roaz(bzf277) can mediate dimer formation on both the inverted repeat and direct repeat. Under the conditions used, the low binding affinity for monomer binding to a single half site would make a direct repeat with double occupancy almost undetectable. This might explain the lack of noncooperative binding of two monomers of the C-terminal truncations to the direct repeat. These experiments suggest that a majority of Roaz and Roaz(bzf277) assemble multimers in solution and that multimeric-complex formation may interfere with their binding to the single half site. Finally, Roaz(bzf277) demonstrates the same DNA binding pattern as Roaz for all the probes tested. This construct can potentially be used to study the specific function of Roaz in transcriptional regulation.
Possible roles of Roaz-binding sites in vivo.
Roaz-binding sites, GCACCC inverted repeats, identified through the Selex assay are reminiscent of a group of GC-rich sequences recognized by a subset of TFIIIA-type zinc finger protein which includes SP1, Zif268/NGFI-A/ Krox-20,24/Egr1,2/Wilms’ tumor family, and yeast ADR1 (3). Interestingly, the consensus half site bears a close resemblance to the SP1 recognition sequence, CCGCCC, which has been shown to be able to confer Roaz–Olf-1–EBF-mediated reporter activation in a cell-line-based assay (38). To date, we have not been able to mimic this activation by replacing the endogenous SP1 sites in the SV40 early promoter with eight consecutive Roaz half-site (GCACCC) repeats. The arrangements of the sites with respect to other promoter elements may have precluded the activation of transcription from this promoter. A second reporter construct consisting of the eight repeats and a minimal thymidine kinase (TK) promoter was also used. The high background activity of Olf-1–EBF transfection with the minimal TK promoter prohibited us from examining the cotransfection of Roaz–Olf-1–EBF with this reporter. Nevertheless, the lack of activation of Roaz homomultimer on both reporter constructs may suggest that Roaz lacks intrinsic transactivational activity.
A search for Roaz-binding sites in the available proximal promoter region of several olfactory specific genes, including olfactory marker protein (OMP) (8), Golf, type III cyclase (ACIII) olfactory cyclic nucleotide-gated channel, 50.06, and 50.11 (41), reveals no identical sequences to the inverted repeat, although a single half site is found in Golf (position −557) and ACIII (position −114), and a SP1 site in OMP (position −46) and Golf (position −252). Unfortunately, potential target genes for Roaz homodimer or Roaz–Olf-1–EBF heterodimers in the basal-cell population of the olfactory epithelium are unknown. Characterization of the DNA-binding specificity may provide an avenue to identify them.
In summary, Roaz represents a complex transregulatory molecule that utilizes a structural motif, the C2H2 zinc finger, to perform DNA binding and protein dimerization functions. Roaz plays discrete roles in transcriptional regulation. When complexed with the Olf-1–EBF proteins, it inhibits transcriptional activation at the Olf-1–EBF-binding site by sequestering the Olf-1–EBF proteins and preventing the formation of an active DNA-binding Olf-1–EBF homodimer. The heteromeric complex of Olf-1–EBF and Roaz possesses transactivational activity at distinct sites where DNA binding is mediated by the Roaz protein (38). We describe here the direct binding of Roaz to a consensus inverted repeat through the N-terminal seven zinc finger motifs. In vivo, Roaz may activate or repress transcriptional activity by recruiting additional repressor or coactivator through the C-terminal dimerization domain.
REFERENCES
- 1.Anthony-Cahill S J, Benfield P A, Fairman R, Wasserman Z R, Brenner S L, Stafford W D, Altenbach C, Hubbell W L, DeGrado W F. Molecular characterization of helix-loop-helix peptides. Science. 1992;255:979–983. doi: 10.1126/science.1312255. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 3.Berg J M. Sp1 and the subfamily of zinc finger proteins with guanine-rich binding sites. Proc Natl Acad Sci USA. 1992;89:11109–11110. doi: 10.1073/pnas.89.23.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown R S, Sander C, Argos P. The primary structure of transcription factor TFIIIA has 12 consecutive repeats. FEBS Lett. 1985;186:271–274. doi: 10.1016/0014-5793(85)80723-7. [DOI] [PubMed] [Google Scholar]
- 5.Chevray P M, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Churchill M E, Tullius T D, Klug A. Mode of interaction of the zinc finger protein TFIIIA with a 5S RNA gene of Xenopus. Proc Natl Acad Sci USA. 1990;87:5528–5532. doi: 10.1073/pnas.87.14.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemens K R, Liao X, Wolf V, Wright P E, Gottesfeld J M. Definition of the binding sites of individual zinc fingers in the transcription factor IIIA-5S RNA gene complex. Proc Natl Acad Sci USA. 1992;89:10822–10826. doi: 10.1073/pnas.89.22.10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danciger E, Mettling C, Vidal M, Morris R, Margolis F. Olfactory marker protein gene: its structure and olfactory neuron-specific expression in transgenic mice. Proc Natl Acad Sci USA. 1989;86:8565–8569. doi: 10.1073/pnas.86.21.8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Rio S, Setzer D R. The role of zinc fingers in transcriptional activation by transcription factor IIIA. Proc Natl Acad Sci USA. 1993;90:168–172. doi: 10.1073/pnas.90.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Rio S, Menezes S R, Setzer D R. The function of individual zinc fingers in sequence-specific DNA recognition by transcription factor IIIA. J Mol Biol. 1993;233:567–579. doi: 10.1006/jmbi.1993.1536. [DOI] [PubMed] [Google Scholar]
- 11.Ekker S C, Young K E, von Kessler D P, Beachy P A. Optimal DNA sequence recognition by the Ultrabithorax homeodomain of Drosophila. EMBO J. 1991;10:1179–1186. doi: 10.1002/j.1460-2075.1991.tb08058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellison V, Gerton J, Vincent K A, Brown P O. An essential interaction between distinct domains of HIV-1 integrase mediates assembly of the active multimer. J Biol Chem. 1995;270:3320–3326. doi: 10.1074/jbc.270.7.3320. [DOI] [PubMed] [Google Scholar]
- 13.Feuerstein R, Wang X, Song D, Cooke N E, Liebhaber S A. The LIM/double zinc-finger motif functions as a protein dimerization domain. Proc Natl Acad Sci USA. 1994;91:10655–10659. doi: 10.1073/pnas.91.22.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregory R C, Taxman D J, Seshasayee D, Kensinger M H, Bieker J J, Wojchowski D M. Functional interaction of GATA1 with erythroid Kruppel-like factor and Sp1 at defined erythroid promoters. Blood. 1996;87:1793–1801. [PubMed] [Google Scholar]
- 15.Hagman J, Belanger C, Travis A, Turck C W, Grosschedl R. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 1993;7:760–773. doi: 10.1101/gad.7.5.760. [DOI] [PubMed] [Google Scholar]
- 16.Hagman J, Gutch M J, Lin H, Grosschedl R. EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. EMBO J. 1995;14:2907–2916. doi: 10.1002/j.1460-2075.1995.tb07290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanas J S, Hazuda D J, Bogenhagen D F, Wu F Y, Wu C W. Xenopus transcription factor A requires zinc for binding to the 5 S RNA gene. J Biol Chem. 1983;258:14120–14125. [PubMed] [Google Scholar]
- 18.Hayes J J, Clemens K R. Locations of contacts between individual zinc fingers of Xenopus laevis transcription factor IIIA and the internal control region of a 5S RNA gene. Biochemistry. 1992;31:11600–11605. doi: 10.1021/bi00161a045. [DOI] [PubMed] [Google Scholar]
- 19.Hayes J J, Tullius T D. Structure of the TFIIIA-5 S DNA complex. J Mol Biol. 1992;227:407–417. doi: 10.1016/0022-2836(92)90897-s. [DOI] [PubMed] [Google Scholar]
- 20.Herr W, Sturm R A, Clerc R G, Corcoran L M, Baltimore D, Sharp P A, Ingraham H A, Rosenfeld M G, Finney M, Ruvkun G, et al. The POU domain: a large conserved region in the mammalian pit-1, oct-1, oct-2, and Caenorhabditis elegans unc-86 gene products. Genes Dev. 1988;2:1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- 21.Huang Z J, Edery I, Rosbash M. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature. 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 22.Landschulz W H, Johnson P F, McKnight S L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 23.Loeber G, Stenger J E, Ray S, Parsons R E, Anderson M E, Tegtmeyer P. The zinc finger region of simian virus 40 large T antigen is needed for hexamer assembly and origin melting. J Virol. 1991;65:3167–3174. doi: 10.1128/jvi.65.6.3167-3174.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markby D W, Zhou B B, Schachman H K. A 70-amino acid zinc-binding polypeptide from the regulatory chain of aspartate transcarbamoylase forms a stable complex with the catalytic subunit leading to markedly altered enzyme activity. Proc Natl Acad Sci USA. 1991;88:10568–10572. doi: 10.1073/pnas.88.23.10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J, McLachlan A D, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan B, Sun L, Avitahl N, Andrikopoulos K, Ikeda T, Gonzales E, Wu P, Neben S, Georgopoulos K. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997;16:2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 28.Pavletich N P, Pabo C O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 29.Pavletich N P, Pabo C O. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993;261:1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- 30.Payre F, Buono P, Vanzo N, Vincent A. Two types of zinc fingers are required for dimerization of the serendipity delta transcriptional activator. Mol Cell Biol. 1997;17:3137–3145. doi: 10.1128/mcb.17.6.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rastinejad F, Perlmann T, Evans R M, Sigler P B. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 32.Rollins M B, Del R S, Galey A L, Setzer D R, Andrews M T. Role of TFIIIA zinc fingers in vivo: analysis of single-finger function in developing Xenopus embryos. Mol Cell Biol. 1993;13:4776–4783. doi: 10.1128/mcb.13.8.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenfeld M G. POU-domain transcription factors: pou-er-ful developmental regulators. Genes Dev. 1991;5:897–907. doi: 10.1101/gad.5.6.897. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz i Altaba A, Perry-O’Keefe H, Melton D A. Xfin: an embryonic gene encoding a multifingered protein in Xenopus. EMBO J. 1987;6:3065–3070. doi: 10.1002/j.1460-2075.1987.tb02613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmeichel K L, Beckerle M C. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 36.Scotland P B, Colledge M, Melnikova I, Dai Z, Froehner S C. Clustering of the acetylcholine receptor by the 43-kD protein: involvement of the zinc finger domain. J Cell Biol. 1993;123:719–728. doi: 10.1083/jcb.123.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 1996;15:5358–5369. [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai R Y, Reed R R. Cloning and functional characterization of Roaz, a zinc finger protein that interacts with O/E-1 to regulate gene expression: implications for olfactory neuronal development. J Neurosci. 1997;17:4159–4169. doi: 10.1523/JNEUROSCI.17-11-04159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner R, Tjian R. Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science. 1989;243:1689–1694. doi: 10.1126/science.2494701. [DOI] [PubMed] [Google Scholar]
- 40.Vinson C R, Sigler P B, McKnight S L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246:911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 41.Wang M M, Tsai R Y, Schrader K A, Reed R R. Genes encoding components of the olfactory signal transduction cascade contain a DNA binding site that may direct neuronal expression. Mol Cell Biol. 1993;13:5805–5813. doi: 10.1128/mcb.13.9.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson D, Sheng G, Lecuit T, Dostatni N, Desplan C. Cooperative dimerization of paired class homeo domains on DNA. Genes Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- 43.Xiang M, Zhou L, Macke J P, Yoshioka T, Hendry S H, Eddy R L, Shows T B, Nathans J. The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J Neurosci. 1995;15:4762–4785. doi: 10.1523/JNEUROSCI.15-07-04762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Q, Gedrich R W, Engel D A. Transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB. J Virol. 1995;69:4323–4330. doi: 10.1128/jvi.69.7.4323-4330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]