Summary
Expansions of CAG trinucleotide repeats cause several rare neurodegenerative diseases. The disease-causing repeats are translated in multiple reading frames, without an identifiable initiation codon. The molecular mechanism of this repeat-associated non-AUG (RAN) translation is not known. We find that expanded CAG repeats create new splice acceptor sites. Splicing of proximal donors to the repeats produces unexpected repeat-containing transcripts. Upon splicing, depending on the sequences surrounding the donor, CAG repeats may become embedded in AUG-initiated open reading frames. Canonical AUG-initiated translation of these aberrant RNAs may account for proteins that have been attributed to RAN translation. Disruption of the relevant splice donors or the in-frame AUG initiation codons is sufficient to abrogate RAN translation. Our findings provide a molecular explanation for the abnormal translation products observed in CAG trinucleotide repeat expansion disorders and add to the repertoire of mechanisms by which repeat expansion mutations disrupt cellular functions.
Graphical Abstract

Blurb
Anderson et al., show that expanded CAG trinucleotide repeats produce 3’ splice sites. Proximal donors splice into the repeats in a repeat-length dependent manner generating unexpected repeat-containing mRNAs. This splicing may place repeats in AUG-initiated frames, potentially explaining aberrant, out-of-frame protein products.
Introduction
Expansions of CAG trinucleotide repeats are associated with at least twelve degenerative disorders, including Huntington’s disease (HD), dentatorubral-pallidoluysian atrophy (DRPLA), spinal and bulbar muscular atrophy (SBMA), and several spinocerebellar ataxias (SCAs)1,2. In each of these diseases, the associated gene is polymorphic for the number of CAG repeats, and disease manifests when the repeat number exceeds a certain threshold1. In most CAG repeat expansion disorders (including HD; SCA types 1, 2, 3, 6, 7, and 17; DRPLA; and SBMA), the CAG repeat tract is located in the protein coding region, and codes for a polyglutamine stretch1. An expanded polyglutamine tract renders proteins aggregation prone, and this repeat-dependent protein aggregation contributes to disease pathology1.
Besides encoding for polyglutamine-containing proteins, CAG repeat expansions can produce cellular dysfunction via at least two additional routes, even when they occur outside of the canonical protein coding regions. First, repeat-containing RNAs can agglomerate in the nucleus as pathogenic foci3,4. RNA foci result from multivalent intermolecular base-pairing interactions templated by the GC-rich repeat tract5. These foci sequester various RNA binding proteins, and cause widespread RNA processing defects6. Second, repeat-containing RNAs undergo translation without requiring an identifiable AUG start codon, in a process known as repeat-associated non-AUG (RAN) translation7-10. RAN translation produces repeat-containing proteins in multiple reading frames, which may form protein aggregates, and are potentially lethal to the cell11. The molecular mechanism of RAN translation is not entirely clear.
In our earlier work, we showed that RNAs that consist primarily of expanded CAG trinucleotide repeats, which are not present in a canonical open reading frame (ORF), accumulate at nuclear foci. These repeat-containing RNAs did not undergo RAN translation even when the repeat number was increased to >400 tandem CAGs5,12. RAN translation is reported to be influenced by the sequences adjacent to the repeat tract10,13. To assess the role of flanking sequences, we previously generated a library of CAG repeat-containing plasmids where various 250 nucleotide long sequences were cloned upstream of 240×CAG repeats12. We found that a subset of these flanking sequences led to RAN translation of the CAG repeat, while others were still retained at nuclear foci12. We were not able to identify sequence motifs in the upstream flanking sequences that differentiated the two classes.
Here, we set out to identify the features of the flanking sequences that determine whether a given CAG repeat-containing RNA will be sequestered at nuclear foci or would undergo RAN translation. We hypothesized that an expanded CAG repeat itself could provide new splicing acceptor site(s), and produce aberrant repeat-containing transcripts. Our hypothesis was guided by three observations. One, live-cell RNA imaging experiments showed that RNAs that undergo RAN translation are first retained at nuclear foci for a prolonged period where they colocalize with the splicing machinery12. Two, mammalian 3’ splice sites have a nearly invariant 'AG' dinucleotide that marks the splice junction (minimal splice acceptor represented as Y6-12NAGNNN, where Y6-12 indicates the upstream pyrimidine rich tract, N is any nucleotide, and the bold bases mark the exon)14. In fact, at least 141,000 (~64%) annotated human splicing acceptor sites harbor a CAG trinucleotide at the splice junction, and >1400 of these sites exhibit two or more (up to nineteen) consecutive CAG trinucleotides (Supp. Fig. 1 A). Three, CAG repeat expansions induce transcriptome-wide splicing changes6,15, and in some cases, are reported to induce mis-splicing of the gene harboring the expanded repeat16-18.
GC-rich repeat-containing sequences are difficult to analyze using standard RNA sequencing approaches, and thus, aberrantly spliced repeat-containing transcripts may have been missed in previous analyses. Thus, we developed an analysis pipeline that specifically captures splicing events at tandem CAG repeats. We find that cognate and near-cognate splicing donors in the surrounding sequences can splice into an expanded CAG repeat tract. This splicing is dependent on the number of CAG repeats, and can place the repeats in unexpected sequence contexts. Interestingly, disease models where expanded CAG repeats are observed to undergo RAN translation, produce RNAs that harbor CAG repeats in AUG-initiated ORFs. Disruption of the relevant splicing donor site or the AUG initiation codon is sufficient to abrogate the production of proteins that are attributed to RAN translation. Our findings provide a cogent mechanistic explanation for how expanded CAG repeats produce abnormal protein products and suggest yet another pathomechanism by which repeat expansions can interfere with RNA processing and cellular functions.
Results
Sequence analysis pipeline for detecting splicing to CAG repeats.
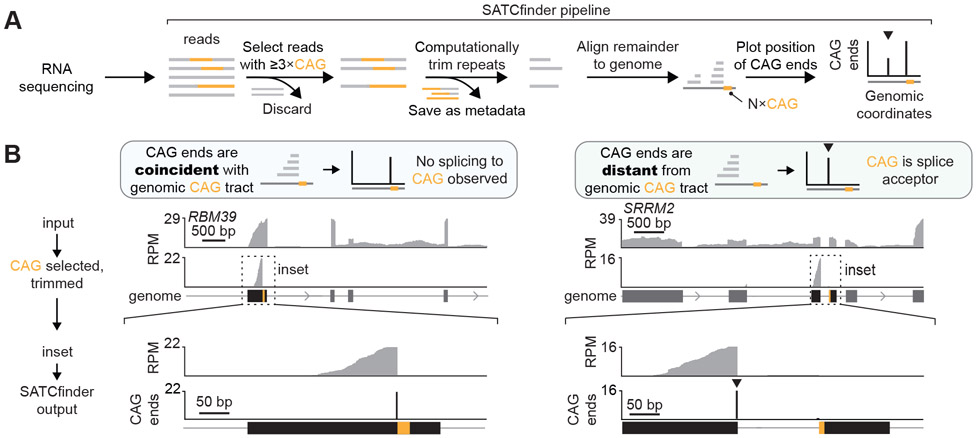
Analysis of repeat-containing RNAs using short-read sequencing technologies is challenging. GC-rich repeats are difficult to reverse transcribe and amplify, and these sequences are substantially under-represented in standard RNA sequencing libraries19,20. The commonly used sequence alignment pipelines also exclude these regions, as repeat-containing reads may map to multiple locations in the genome, or because the reference genome lacks an expanded repeat tract21. To address the first issue, we optimized the library preparation protocol, notably by using a group II intron reverse transcriptase that can reverse transcribe structured22 and GC-rich23,24 RNAs with high fidelity (see STAR Methods). To address the issue of aligning repetitive reads, we developed a pipeline, SATCfinder, to identify and visualize splicing at tandem CAG repeats in RNA sequencing data (Fig. 1A). In brief, SATCfinder’s input is paired end RNA sequencing data. All reads that contain ≥3×CAG/CTG repeats (to capture both sense and antisense transcripts) and at least 15 additional bases that are not a part of the CAG repeat (to allow appropriate alignment and junction identification) are selected. The CAG repeats are computationally trimmed and the remaining non-repetitive portion of the reads, together with the paired read mates, are aligned to the reference genome (see STAR Methods). The mapping coordinate of the CAG end of the trimmed reads (hereafter, CAG end) is tracked. The number of CAG ends coinciding with the genomic coordinate of the CAG repeat versus those aligning farther away to upstream splicing donors allows us to quantitatively assess the extent of splicing to the repeat tract (Fig. 1 A). For example, when CAG repeats are not a part of the splicing acceptor (i.e., they are contained entirely in an exon or an intron), the position of the CAG repeats in mRNA matches the location of the repeat tract in the genome. For a representative gene in this class, RBM39, the CAG ends in RNA identified using SATCfinder coincide with the end of the CAG repeats in the genome (in 99.5% of reads, Fig. 1B left, Supp. Fig. 1B). Likewise, we examined several other genes where CAG repeats are annotated to be in the middle of an exon, and in each case, >99% of CAG ends mapped immediately adjacent to the CAG repeat tract in the genome (Supp. Fig. 1B, D).
Figure 1. Sequence analysis pipeline for detecting splicing to CAG repeats.
A. Schematic for SATCfinder. Reads with >3xCAG/CTG repeats are selected. The CAG repeats are computationally removed and these trimmed reads are aligned to the genome. The genomic coordinates of the base immediately before the repeat (CAG end) in the trimmed and mapped reads is tracked. SATCfinder outputs the number of CAG ends per million mapped reads at a given genomic coordinate. The peak at the repeat reflects reads where CAG repeats are not a part of the splice junction, while a distal upstream peak (at the nearest upstream exon, typically within a few kb) indicates the location of the splicing donor (marked by an arrowhead ▼). B. Representative genes comparing standard RNA sequencing analysis to SATCfinder output.
Sixty-eight human genes are annotated to have ≥3×CAG at splice acceptor sites (Supp. Fig. 1A, Supplemental Table 1). For this class, the non-repetitive sections of CAG-containing reads map to a location upstream of the CAG repeat in the genome (see representative example SRRM2, Fig. 1B, right). A vast majority (~95%) of upstream CAG ends align to the annotated splicing donor site upstream from the tandem CAG acceptor site (Fig. 1B, right, Supp. Fig. 1C, E-F). The remainder (~5%) of CAG ends coincide with the repeat tract in the genome, likely arising from unspliced pre-mRNAs (Fig. 1B, right, Supp. Fig. 1C). Treatment with a splicing inhibitor increased the proportion of reads corresponding to the unspliced CAG repeat-containing RNAs by ~10-fold (from 3% to 33% of CAG ends on treatment with 25 nM pladienolide B, Supp. Fig. 1G). Similar results are observed for other genes in this class (Supp. Fig. 1C-D). Altogether, these results demonstrate that our library preparation protocol coupled with SATCfinder allow us to examine events where CAG repeats form splicing acceptor sites.
CAG repeat expansions result in mis-splicing of repeat-containing RNAs.
We used SATCfinder to examine whether CAG repeat expansions could induce mis-splicing of repeat-containing RNAs. We previously generated a small library of sequences where 240×CAG repeats were cloned downstream of various 250 nucleotide flanking sequences12 (Fig. 2A). In these constructs, multiple stop codons were incorporated immediately upstream of the CAG repeats to eliminate translation readthrough from any upstream ORFs into the repeats. In about half of these constructs, the CAG repeat-containing RNA was retained in the nucleus at foci, while in other cases, the repeat-containing RNA was exported to the cytoplasm, underwent RAN translation, and formed perinuclear aggregates (Fig. 2A). Cytoplasmic localization also coincided with substantial cellular toxicity12.
Figure 2. CAG repeat expansions result in mis-splicing of repeat-containing RNAs.
A. Schematic for constructs with 240xCAG repeats with a variable 250-base flanking sequence12. Some flanking sequences result in retention of the repeat-containing RNA in the nucleus while others induce RAN translation and cell toxicity. B, C, D. Top, SATCfinder output for representative CAG constructs, where the x-axis is the base coordinate within the flanking sequence region and the y-axis indicates the number of CAG ends per million mapped reads. Bottom, Representative fluorescent images of cells expressing the indicated constructs. Micrographs are representative of > 2 independent experiments. Scale bar depicts 10 pm. E. Left, sequence logos for 5’ splice sites annotated in the human genome and those observed in the CAG flanking sequence library, where the x-axis indicates the position within the 9-base donor sequence and the letter height depicts the probability of observing the base. Right, 5’ MaxEnt scores for 220419 annotated human splice donors, all 262144 possible randomly generated 9-mers, and the 20 detected splice donors in the CAG flanking sequence library. F. Quantification of the percentage of cells with cytoplasmic RNA aggregates by fluorescence microscopy. Each data point represents an independent experiment with > 500 cells per experiment, and are summarized as mean ± SD. G. Real-time quantitative PCR quantification of the relative expression of CAGran intron normalized to the expression of the 5’ end of the CAGran transcript. Splicing inhibitor reflects treatment with 25 nM pladienolide B. Data show the mean ± SD for three independent RNA isolations. H. Schematic for CAGran, depicting the transcription initiation site (as a right-facing arrow), flanking sequence, and 240xCAG repeats intervened by stop codons in each frame. The sequence of a representative donor that splices to the CAG tract is shown, with bases in the exon in uppercase. After splicing, the stop codons are removed and the CAG repeat is embedded in an AUG-initiated ORF. I. Immunoblot for cells expressing CAGfoci and CAGran using a polyglutamine antibody. J. Percentage of repeat-containing transcripts where the repeats are observed in AUG-initiated ORFs for constructs that produce RNA foci only or exhibit RAN translation. Each data point is one construct. K. Immunoblot for the indicated samples using a polyglutamine antibody. CAGRAX*donors has point mutations at all splice donor sites; CAGran*ai g has point mutations at two AUGs. Immunoblots are normalized first to tubulin, then to the parent cell line without a repeat-containing construct (mock). Immunoblots and quantification of relative polyglutamine abundance (as mean ± SD) are representative of > 2 independent experiments. *An endogenous protein (TBP) is also detected by this polyglutamine antibody10,65. Significance values in F, G, and J are calculated using Student’s t-test. MS2CP-YFP: bacteriophage MS2 coat protein tagged with YFP.
We performed RNA sequencing on cells expressing the various 240×CAG repeat-containing RNAs that either form nuclear foci (6 cell lines, with CAGFOCI as a representative of this class, full sequences are provided in Supplemental Table 4) or undergo RAN translation (7 cell lines, with CAGRAN as a representative of this class). Our analysis using SATCfinder revealed that the CAG repeats in RNA were frequently stitched to regions in the upstream flanking sequences, and the intervening region had been removed (see representative examples for CAGFOCI and CAGRAN in Fig. 2B-C, top; and other examples in Supp. Fig. 2A-B).
Several lines of evidence indicate that the junctions identified by SATCfinder arise from splicing to CAG repeats. One, quantitative and end-point PCR on the genomic DNA and RNA confirmed that the desired sequences were integrated into the genome but the intervening sequences were removed from the RNA (Supp. Fig. 2C-E). Two, the readjunctions identified by SATCfinder harbor signatures of splicing donors (Fig. 2E, left,, sequences in Supplemental Table 2). We evaluated these identified donor sites using a computational splice-site prediction algorithm, MaxEntScan25. This algorithm reports a log-odds ratio of observing a motif in true versus decoy splice sites, where a higher MaxEnt score indicates a stronger putative splice site26. Known human splice donors score in the range 0 to 12 (>95% score above 2; median 8.6, Fig. 2E, right). The donors that were identified by SATCfinder to splice to CAG repeats in our library had positive 5’ MaxEnt scores (1.59 – 9.40, median 4.95; Fig. 2E, Supplemental Table 2), indicating that these sequences are canonical splice donors. Three, chemical inhibition of splicing using pladienolide B27 led to a dose-dependent reduction in the frequency of splicing from the upstream flanking region to the CAG repeats (Supp. Fig. 2F, G). Finally, point mutations to the crucial GGU motifs of the donors (GGU→GGA) were sufficient to eliminate splicing to the repeats (Fig. 2D, Supp. Fig. 2H).
Splicing was dependent on the number of CAG repeats, and its frequency progressively increased with the repeat number (Fig. 2G, Supp. Fig. 3 A-G). Similar splicing patterns were observed across various cell lines (Supp. Fig. 21). A variety of donor sequences could be spliced to the CAG repeats, and the consensus sequence of the donors identified in our flanking sequence library closely resembles the consensus sequence for the annotated donor sites in the human genome (Fig. 2E, Supp. Fig. 2J-K). In summary, these results show that splicing from putative donors, occurring by chance in our library of flanking sequences, resulted in unexpected CAG repeat-containing RNAs that differ from the intended sequences that were integrated in the genome.
Even though we engineered stop codons in each frame immediately upstream of the CAG repeats, a subset of these constructs produced polyglutamine-containing proteins (Fig. 2H-I). Splicing to CAG repeats could create mature transcripts where the proximal stop codons are spliced out and the repeats are placed within AUG-initiated ORFs. To test this idea, we assembled the full-length mature mRNAs produced from these constructs using our RNA sequencing data. In all cases where we observed translation of the repeat region, we found AUG-initiated ORFs, generated after splicing, that contain the repeat region (Fig. 2J, Supp. Fig. 4A). On the other hand, sequences that formed nuclear foci either did not exhibit substantial splicing to CAG repeats or, upon splicing, did not produce AUG-initiated ORFs that contain the CAG repeats (Fig. 2J, Supp. Fig. 4A). Across the various cell lines, the abundance of RNAs with repeat-containing AUG-initiated ORFs correlated with the levels of polyglutamine protein produced (Pearson’s correlation coefficient, r = 0.83, between polyglutamine immunofluorescence and RPM of transcripts carrying an AUG-initiated ORF, Supp. Fig. 4B-C).
We chose one representative sequence, CAGRAN, to further examine the role of splicing in the translation of the repeat region. We isolated the polysome fraction from cells expressing CAGRAN and found that a vast majority of CAGRAN transcripts on actively translating ribosomes were spliced (Supp. Fig. 4D-F). Mutation of the identified splice donor sites in CAGRAN, which eliminated splicing to the repeat tract, also abrogated the production of aberrant polyglutamine proteins (Fig. 2K). The mature repeat-containing transcripts produced from CAGRAN contained an ORF with two in-frame AUG codons (Fig. 2H). Disruption of these AUG codons by single-base mutations at each site similarly eliminated the polyglutamine product (Fig. 2K). N-terminal sequencing of the polyglutamine protein produced in CAGRAN confirmed that it is methionine-initiated, and the first six amino acids in this protein (MRFLAT) are as expected for translation initiation from the first AUG in the ORF (Supp. Fig. 4G). Interestingly, pharmacological inhibition of splicing led to the retention of the repeat-containing RNA in the nucleus at foci (Supp. Fig. 4H-I). Likewise, point mutations to the splice donors in CAGRAN were sufficient to convert this sequence to the foci-forming class (Fig. 2D, bottom, and Fig. 2F). Unlike CAGRAN, expression of this sequence with mutated donors (CAGRAN*donors) did not induce cell toxicity (Supp. Fig. 4J), consistent with our earlier report that expression of repeat-containing RNAs that only produce nuclear foci, do not induce overt cell death12. Thus, the aberrant translation products in CAGRAN result from canonical AUG-initiated translation of aberrant transcripts that arise from splicing to expanded CAG repeats, and disruption of splicing or the appropriate AUG start codons is sufficient to eliminate the translation of the repeat tract.
CAG repeats with native disease-associated flanking sequences form splice acceptors.
Besides an acceptor site, splicing requires cis-acting elements, such as a polypyrimidine tract, that facilitate spliceosome assembly. The splicing potential of a putative acceptor site can be evaluated by the 3’ MaxEnt model that takes into account these cis-acting sequences. Most annotated splice sites in the human genome score in the range 0 to 16, with a median score of 8.6 (Supp. Fig. 5A). In our library of flanking sequences, CAG repeats were placed in a synthetic sequence context which likely provided the polypyrimidine tract (3’ MaxEnt score = 2.45; sequence in Supplemental Table 4). We examined whether disease-associated CAG repeats in their native upstream sequence context may also act as splicing acceptors. These loci, in general, have lower 3’ MaxEnt scores than typical human acceptor sites (range −13.3 – 5.9, median = 1.6) (Supp Fig. 5B).
We experimentally examined the prevalence of splicing in cases where expanded CAG repeats are reported to produce aberrant RAN translation products. The first report on RAN translation of CAG repeats utilized a minigene with 107×CAG repeats flanked by sequences native to the ATXN8 gene10, associated with SCA8 (Fig. 3A). Interestingly, Zu et al. also observed splicing from an upstream splice donor to the CAG repeats, but the resulting spliced transcript did not contain ORFs that explained the observed aberrant translation products (Fig. 3A). To investigate whether there were additional donor sites, we transfected HEK293T cells with this ATXN8 minigene, similar to the previous study, and analyzed the resulting RNA using SATCfinder. Our analysis revealed that only about half of the CAG-containing reads were unspliced and mapped to the expected minigene sequence (Fig. 3B). The remaining CAG-containing reads mapped to distant sites on the plasmid, suggesting that additional mature CAG-repeat-containing RNAs are produced in these cells (Fig. 3B). Using a conservative threshold of ≥100 unique reads supporting a putative splice donor, we identified six donor sites (Fig. 3B, Supp. Fig. 5C-D). The donors contain the nearly invariant ‘GGU’ motif found in human splice donors, and have positive 5’ MaxEnt scores (2.7 – 9.82, Fig. 3D). Splicing inhibition using pladienolide B or isoginkgetin28 significantly decreased the number of reads that reflect splicing to the repeat tract (84% and 98% reduction in splicing for 25 nM pladienolide B and 15 μM isoginkgetin treatments, respectively, Fig. 3B). Point mutations to the donor sites completely abolished splicing from the respective donors (Fig. 3C), further validating that these donors are spliced to the CAG repeat tract. Similar results were observed for other CAG-containing mini-genes where the proximal sequences are derived from the genes HTT, JPH3-AS, ATXN3, and DMPK-AS, associated with the diseases HD, Huntington disease-like 2, SCA3, and myotonic dystrophy type 1 respectively (Supp. Fig. 5E).
Figure 3. CAG-repeats with native disease-associated flanking sequences form splice acceptors.
A. Schematic for the ATXN8 mini-gene expressing -100 bp of endogenous A TXN8 sequence directly upstream from 107><CAG repeats10. The ampicillin resistance gene (AS-AmpR) and colEl origin of replication are indicated. B. Left, SATCfinder output for cells transfected with the ATXN8 construct in the presence of splicing inhibitors 25 nM pladienolide B (PB), or 15 μM isoginkgetin (IGG), or 0.1% DMSO (DMSO) as control. Right, quantification of the % of CAG ends that reflect splicing to the repeat. C. Similar to B but for ATXN8 constructs without or with point mutations to identified donor sites. D. Sequences of the identified splicing donors in the ATXN8 construct with corresponding percentage of reads arising from each donor. The sequence logo for the consensus human splice donor is presented for comparison. E. Schematic for the various CAG repeat-containing transcripts produced upon splicing from the ATXN8 construct.
We examined the origin of the splicing donor sequences in these mini-genes, and to our surprise, we found that a significant fraction of the spliced reads arose from a donor located in the ampicillin resistance cassette more than 1 kilobase upstream from the CAG repeats (Fig. 3B). The AmpR gene is encoded in the antisense direction (AS-AmpR) with respect to the CAG repeats. The bacterial AmpR promoter should not initiate transcription in mammalian cells nor would it produce CAG repeat-containing transcripts (see full sequence map in Supp. Fig. 5F). To determine how a donor in the AS-AmpR sequence could splice to the CAG repeats, we examined the raw (non-CAG-selected) read alignments. We noted a region of high coverage beginning within the colE1 E. coli origin of replication and continuing through the AS-AmpR region (Supp. Fig. 5G). This coverage did not result from readthrough of the polyadenylation signal from the adjoining neomycin resistance gene (Supp. Fig. 5G). Transcript assembly using StringTie29 confirmed that the AS-AmpR transcript originated within the colEl region (Supp. Fig. 5H). Reads corresponding to this region were also observed in other plasmids that harbor colEl origin of replication (Supp. Fig. 51), and the transcripts initiating from colEl were ~30% as abundant as those from the SV40 and CMV promoters (Supp. Fig. 5J). This observation is consistent with reports that the colEl origin of replication acts as a cryptic promoter in eukaryotic cells and may result in spurious unintended transcripts in cells transfected with plasmids carrying this bacterial sequence30,31. Thus, transcription from the cryptic colEl promoter and splicing from donor sites in the AS-AmpR region to the repeat tract gives rise to mature transcripts containing CAG repeats embedded in various 5’ sequence contexts (Fig. 3E). Taken together, these results demonstrate that an expanded CAG repeat tract flanked by sequences native to the disease-causing gene can act as splice acceptor sites, and splicing from upstream donors may result in unexpected repeat-containing transcripts.
Canonical translation of aberrantly spliced CAG-repeat-containing RNAs accounts for RAN products.
We then investigated whether the spliced RNAs produced from disease associated mini-genes with repeat-containing ORFs might explain the production of spurious repeat-containing proteins. We examined the ATXN8 mini-gene, where the CAG repeat is immediately preceded by an AUG codon in the polyglutamine frame (Fig. 4A). Transfection of the ATXN8 plasmid in HEK293T cells resulted in the production of multiple polyglutamine-containing proteins (Fig. 4B), as has been previously reported10. Prior work demonstrated that mutation of the in-frame start codon immediately preceding the CAG repeats (construct ATXN8KKQ, Fig. 4A) eliminated one of the polyglutamine products, while other polyglutamine-containing proteins produced from this plasmid are not affected. These products have been attributed to non-canonical RAN translation of the repeat region10.
Figure 4. Canonical translation of aberrantly spliced CAG-repeat-containing RNA results in aberrant protein products.
A. Schematic for ATXN8 mini-gene. Upon splicing, the upstream stop codon is removed and the CAG repeat is embedded in an AUG-initiated ORF. B, C, D. Immunoblots from cells expressing the indicated ATXN8- and ATXN8KKQ- derived constructs that interrupt the predicted ORF by mutating the splice donor (B), mutating the identified in-frame AUG initiation codon (C), or by introducing a stop codon (D). Band intensities are normalized first to NPT (neomycin phosphotransferase), expressed in cis from the plasmid, and then to the endogenous protein (TBP, marked with an asterisk) in the control transfected with a similar vector but encoding for GFP (vector). Tubulin is included to show equivalent loading between conditions, but is not used for normalization due to potential variations in transfection efficiency. Immunoblots and quantification of relative polyglutamine abundance (as mean ± SD) are representative of > 2 independent transfections.
We found that these plasmids produced several spliced RNAs with AUG-initiated ORFs that contain the CAG repeats (Fig. 4A; see other ORFs in Supp. Fig. 5K). One of the major transcripts with an ORF in the polyglutamine frame arises due to splicing from a donor in the AS-AmpR cassette. Point mutations to this donor in AS-AmpR, ~1800 bases upstream from the CAG repeats, that abolished splicing at this site (Fig. 2C), also eliminated the aberrant polyglutamine product in both ATXN8 as well as ATXN8KKQ constructs (Fig. 4B). Splicing from this donor to CAG repeats creates an ORF with an AUG start codon located 135 nucleotides upstream from the donor site. Again, a single mutation to this AUG site, located in AS-AmpR, completely abolished the aberrant polyglutamine protein without affecting the canonical translation product (Fig. 4C). As an additional test for our model, we interrupted the AUG-initiated ORF with a single stop codon. This single-base mutation in the AS-AmpR region to generate a stop codon likewise eliminated the aberrant polyglutamine product (Fig. 4D).
Finally, we deleted a single nucleotide 10 bases upstream from the donor site, which would shift the translation frame for CAG repeats from polyglutamine to polyserine. Consistent with this expectation, the single-base deletion abrogated aberrant polyglutamine production with a concomitant ~500% increase in high-molecular weight polyserine-containing protein (Supp. Fig. 5L). Taken together, these results demonstrate that at least a subset of abnormal translation products arising from these mini-genes can be accounted for by splicing of upstream donors to CAG repeats followed by canonical AUG-initiated translation of the repeat-containing RNA. Even though the splicing donor sequences in these mini-genes used to model disease are non-native, our results suggest that disease-associated expanded CAG repeats, flanked by their native sequences, can act as splicing acceptors and potentially generate aberrant transcripts with repeat-containing ORFs.
Splicing from an endogenous donor in ATXN8 generates AUG-initiated ORFs.
Our observations raise the possibility that a similar mechanism may account for spurious, out of frame protein products observed in disease, where the region upstream of the repeat tract may provide a splice donor. We examined the sequence upstream of CAG repeats in the ATXN8 gene and found several potential splicing donor sites (5’ MaxEnt scores ≥ 1, Supp. Fig. 5M). We cloned 400 nucleotides of this native 5’ flanking sequence and placed it immediately upstream of 47×CAG repeats. Downstream of the repeats, we incorporated distinct epitope tags in each reading frame (Fig. 5A, sequence in Supplemental Table 4). Upon transducing this construct in U-20S cells, we observed one major splice donor that underwent splicing to the CAG repeat (5’ MaxEnt = 8.49). More than 90% of CAG ends reflected splicing to the repeats from this specific site (Fig. 5B). Disruption of this donor by a single-base mutation eliminated splicing from this location (Fig. 5B). Interestingly, when we mutated the original donor site, several new donor sites in the flanking sequence were spliced to the CAG repeats (5’ MaxEnt score for these sites ranging from 1.76 to 5.17 Fig. 5B, Supp. Fig. 5M). This observation is similar to other reports indicating that mutation of a splice donor can result in the activation of cryptic donor sites32, and suggests that the CAG repeat sequence in ATXN8 creates a strong splicing acceptor.
Figure 5. Splicing from an endogenous donor in ATXN8 generates AUG-initiated ORFs.
A. Schematic for design of ATXN8 mini-gene with 400 bases of endogenous ATXN8 sequence fused directly to 47><CAG repeats, followed by epitope tags in each reading frame. B. SATCfinder output for ATXN8 constructs with native upstream sequence, without or with a point mutation to the predicted donor site. C. Schematic for ORF resulting from ATXN8 minigene. Upon splicing, the CAG repeat is embedded in a new AUG-initiated ORF. D. Immunoblot for the indicated samples using an anti-HA antibody. The HA epitope is in the polyalanine frame. Immunoblots are normalized first to tubulin, then to the parent cell line without a repeat-containing construct (mock). Immunoblots and quantification of relative HA abundance (as mean ± SD) are representative of > 2 independent experiments.
Splicing from the identified donor site in ATXN8 creates an AUG-initiated ORF in the polyalanine frame (Fig. 5C). Polyalanine products have been observed in brain tissue from SCA8 patients10. In our synthetic construct, this spliced ORF encodes for an ~7 kD polyalanine containing protein which is detected via an HA-tag encoded downstream of the repeats in the polyalanine frame (Fig. 5D). Point mutations to the splicing donor or to the relevant AUG initiation codon abrogate polyalanine protein production (Fig. 5D, Supp. Fig. 5N-0). Although polyserine products have also been reported in SCA8 patient brains7, we do not observe translation of the repeat in the poly serine frame in this model (Supp. Fig. 5O). The small fragment of the ATXN8 gene that we cloned does not produce RNA with AUG-initiated ORFs in the poly serine frame. Nonetheless, further characterization of the 5’ end of the full-length ATXN8 transcript may reveal splice donors that could account for additional aberrant translation products that are observed in disease.
Discussion
The diseases caused by simple repeat expansions have diverse pathomechanisms that encompass dysfunction at DNA, RNA, and protein levels. Expanded CAG repeats in DNA form secondary structures that induce repeat instability during DNA replication and repair33-36. The repeat-containing RNAs may form inter-molecular base pairs and agglomerate in the nucleus to form foci that sequester essential RNA binding proteins3,4. When translated, the repeats produce aggregation-prone homopolymeric proteins37-39. Our work uncovers yet another route by which the disease-causing CAG repeats may trick the cellular machinery (see model in Fig. 6). Expanded CAG repeats may create new splicing acceptor sites. Nearby donors may splice into the repeats, and thus generate a variety of repeat-containing transcripts. These transcripts may contain AUG-initiated ORFs that encompass the repeat tract and produce unexpected, but canonically translated, polypeptides. This mechanism may account for a subset of abnormal polypeptides that are observed in various CAG repeat expansion disorders40-42.
Figure 6. Model for the sub-cellular localization of RNA with expanded CAG repeats.
In the absence of an expanded repeat, the RNA is normally processed and exported to the cytoplasm. If the RNAs contain expanded CAG repeats outside of an ORF, the RNAs are retained at nuclear foci, where they sequester splicing factors. If the CAG repeats are located downstream of potential splice donors, the donors may be spliced to the CAG repeat in a repeat number dependent manner. Splicing generates new RNA isoforms where the CAG repeat may be present in AUG-initiated ORFs. Translation of these AUG-initiated repeat-containing ORFs produces aberrant homopolymeric proteins that may aggregate and contribute to cellular toxicity.
Mechanistically, how the expanded CAG repeat tract induces mis-splicing of the repeat-containing transcript remains to be investigated. One possibility is that the expanded CAG repeats may provide a battery of alternative acceptor sites. More than 800 human genes contain tandem CAG repeats (≥2×CAG) at the 3’ splice sites43. In several cases, these repeats create alternative splice junctions, with the upstream donor being spliced to either of the two CAGs43. This differential utilization of the tandem splice sites, referred to as NAGNAG splicing, contributes to proteome diversity43,44. To test whether the expanded CAG repeats could be acting as alternative acceptor sites, we analyzed the RNA sequencing reads that span the entire repeat tract for relatively short repeats (10 and 22×CAG) that are accessible via short-read sequencing (Supp. Fig. 3E). At these repeat lengths, we observed that the primary acceptor was the first CAG of the repeat unit, accounting for at least 75% of the splicing events for 10×CAG and 50% of the splicing events for 22×CAG (Supp. Fig. 3F). Similar results were observed for a 47×CAG tract analyzed with long-read sequencing (Supp. Fig. 3G). It is important to note that PCR amplification, such as during sequencing library preparation, can lead to truncation in repeat number45,46. We observe substantial truncations during PCR even when using DNA with a fixed number of CAG repeats (Supp. Fig. 3G). Such truncations increase with repeat number and make it challenging for us to precisely identify the acceptor CAG within the repeat region. Another possibility is that the expanded repeats sequester proteins that alter splice site choice and efficiency. Splicing factors such as MBNL1 and SRSF6 bind RNAs with tandem CAG repeats6,47,48. An expanded repeat tract may recruit multiple copies of these splicing factors and affect splice site utilization. Supporting this model, CAG repeat-dependent RNA processing defects have been documented in multiple diseases6,15,17,18,47. A third possibility is that the repeats may undergo post-transcriptional splicing. CAG-repeat containing RNAs are retained at nuclear foci where they co-localize with numerous splicing factors. This retention at foci increases with repeat number5,49. Increased nuclear retention may augment post-transcriptional splicing, possibly even from moderately strong donor sites. These three models are not mutually exclusive and future work may shed light on the precise mechanisms driving increased splicing to expanded CAG repeats.
To what extent does splicing to tandem CAG repeats occur in disease? To address this question, we examined the publicly available RNA sequencing datasets generated from post-mortem brain and iPSC-derived neural cells from patients affected by HD, the best studied CAG trinucleotide repeat expansion disease50-55. From this composite dataset (~8 billion total reads), we mapped only 396 reads adjacent to the HTTCAG repeat tract (Supp. Fig. 6A), a notoriously under-represented region in sequencing data19 (Supp. Fig. 6B). We did not find statistically significant evidence supporting splicing to the HTT CAG repeat in this dataset beyond the background noise (Supp. Fig. 6A). These observations suggest that splicing to the expanded CAG repeat in HTT is likely rare, and occurs at a frequency of <0.5%, barring potential biases in RNA sequencing library preparation. By necessity, these studies were conducted on regions of the brain that were present in the postmortem tissue and may underrepresent the most affected cells. One potential implication of splicing to repeats would be the production of aberrant RAN translation products. RAN translation is only observed in a subset of cells (up to ~20% cells in the highly affected striatum) in HD postmortem tissue8. It is possible that splicing to the HTT CAG tract is rare and occurs only in a small fraction of cells and is below the detection limit of our current assays (Supp. Fig. 6C). Future studies employing appropriate disease samples and assays with increased coverage in the repeat region may allow us to unequivocally assess the extent of splicing to CAG repeats in disease.
It is also possible that some CAG repeat expansion disorders may not exhibit abnormal splicing to CAG repeats. Most CAG repeat loci in the genome do not serve as splicing acceptors (Supp. Fig 5A, Supp. Fig. IE), and evolutionary pressure may have purged cis-acting splicing-associated features (such as polypyrimidine tract) from these neighboring regions. To assess whether CAG repeat expansions at new genomic locations (i.e., locations that do not endogenously harbor a CAG repeat) could lead to splicing from adjacent donors, we introduced CAG repeats in an intron of ACTB in HEK293T cells (Supp. Fig. 6E). This intron has 8 isolated CAGs, four of which are predicted to be strong splice acceptor sites (3’ MaxEnt 2.76 – 9.47, Supp. Fig. 6F). These native single CAG trinucleotides do not act as splicing acceptors (Supp. Fig. 6G). We incorporated varying numbers of CAG repeats in this intron, isolated monoclonal cells at each repeat number, and characterized the transcribed RNAs using SATCfinder (Supp. Fig. 6H-L). We observed numerous reads where the upstream exon was spliced to the CAG repeats, in a repeat-number dependent manner (Supp. Fig. 6K-L). These reads reflecting splicing to the inserted CAG repeat were ~1% as abundant as the reads corresponding to the correctly spliced ACTB transcript (Supp. Fig. 6M). ACTB is an essential gene and only one locus in our polyploid cells was modified with the repeat insertion (see STAR Methods). Although the fraction of transcripts that are mis-spliced due to the repeat insertion is modest, these results underscore that expansions of CAG trinucleotide repeats in an appropriate context can produce de-novo splicing acceptors that sequester adjacent donor sites and result in aberrantly spliced mRNAs.
Our findings add to various molecular mechanisms that contribute to the aberrant proteins that are attributed to RAN translation40. Besides CAG repeat-expansion disorders, RAN translation has also been observed in several other repeat-expansion diseases. In fragile X tremor ataxia syndrome (FXTAS), caused by CGG repeat expansion in FMR1, the RAN products in the polyglycine frame results from translation initiation at a near-canonical ACG start site56. Likewise, in amyotrophic lateral sclerosis, associated with GGGGCC repeat expansion in c9orf72, RAN translation in the glycine-alanine frame is dependent on the presence of a near-cognate CUG initiation codon in the upstream sequence57. Another possibility is that the secondary structures formed by GC-rich repeat containing RNAs may provide internal ribosome entry sites facilitating cap-independent translation58-60. GC-rich repetitive sequences can also result in ribosomal frameshifting, and generate chimeric proteins57,61-63. We show that in addition to these mechanisms, aberrant splicing of the repeat-containing transcript followed by canonical or potentially near-canonical translation initiation may also contribute to the production of RAN proteins. The location of repeats within a gene is usually annotated based on the wild-type gene without pathogenic expansion. Splicing aberrations may create new isoforms, and modify exon/intron annotations, as has recently been proposed for the c9orf72 repeat expansion64. Advances in long read sequencing technologies in conjunction with methods that allow mapping repetitive regions may reveal the full-length transcripts that are produced in disease, and help elucidate the disease mechanisms.
In conjunction with prior research, our findings help piece together a model for how CAG repeat-expansions affect RNA processing and sub-cellular localization (see model in Fig. 6). CAG repeats potentiate intermolecular RNA-RNA interactions, and an expanded repeat tract results in the sequestration of the repeat-containing RNA at nuclear foci. These foci co-localize with nuclear speckles and sequester various splicing factors, often inducing mis-splicing of other transcripts. If the repeat-containing RNA also harbors an appropriate splice donor, this donor may be spliced to the expanded CAG repeats, facilitating its export to the cytoplasm. Splicing may place the repeats in AUG-initiated ORFs. Canonical translation of such repeat-containing ORFs may produce homo-polymeric peptides that form pathogenic aggregates. Our results provide a new lens to examine the role of cis-acting sequences as modifiers of CAG-repeat induced cell toxicity, and suggest that modulating RNA splicing could be a potential target for the development of therapeutic interventions.
Limitations of the Study
Untargeted RNA sequencing approaches do not provide sufficient coverage in the repeat region and we were unable to detect splicing to CAG repeat tract in RNA-sequencing data in HD patient samples. The contribution of such splicing events to disease thus remains to be determined. Target-enrichment approaches or amplicon sequencing by designing primers based on putative donors in the upstream region may potentially allow one to assess the prevalence of splicing when the pathogenic repeats are expressed from their endogenous loci. The molecular mechanism by which an expanded CAG repeat stimulates splicing has not been addressed by this work and remains to be investigated.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ankur Jain (ajain@wi.mit.edu).
Materials Availability
All cell lines and plasmids generated in this study are listed in the key resources table and are available upon request to the lead contact.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-FLAG | Sigma-Aldrich | Cat# F1804, RRID:AB_262044 |
| Mouse monoclonal anti-HA | BioLegend | Cat# 901501, RRID:AB_2565006 |
| Mouse monoclonal anti-polyglutamine | Millipore | Cat# MAB1574, RRID:AB_94263 |
| Mouse monoclonal anti-Neomycin Phosphotransferase II | Thermo Fisher Scientific | Cat# MA5–15275, RRID:AB_10979669 |
| Rabbit polyclonal anti-β-tubulin | Cell Signaling Technology | Cat# 2146, RRID:AB_2210545 |
| Goat polyclonal anti-rabbit, HRP conjugate | Sigma-Aldrich | Cat# A0545, RRID:AB_257896 |
| Rabbit polyclonal anti-mouse, HRP conjugate | Sigma-Aldrich | Cat# A9044, RRID:AB_258431 |
| Bacterial and virus strains | ||
| Stbl3 E. coli | Invitrogen | Cat# C7373–03 |
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| doxycycline | Sigma Aldrich | CAT# D9891 |
| dimethyl sulfoxide | Sigma Aldrich | CAT# D2650 |
| pladienolide B | Santa Cruz Biotechnology | CAT# SC391691 |
| isoginkgetin | Tocris Bioscience | CAT# 6483 |
| sodium chloride | Invitrogen | CAT# AM9760G |
| NP-40 | Fisher Scientific | CAT# AAJ19628AP |
| sodium deoxycholate | Sigma-Aldrich | CAT# D6750 |
| sodium dodecyl sulfate | Bio-Rad | CAT# 1610302 |
| HALT protease and phosphatase inhibitors | Thermo Scientific | CAT# 78429 |
| Benzonase nuclease | EMD Millipore | CAT# E1014 |
| dithiothreitol | Thermo Scientific | CAT# R0861 |
| methanol | VWR | CAT# EM-MX0475–1 |
| Coomassie Brilliant blue R 250 | Sigma Aldrich | CAT# 1125530025 |
| acetic acid | Sigma Aldrich | CAT# 695092 |
| Hybridase RNase H | Lucigen | CAT# H39500 |
| Turbo DNase | Invitrogen | CAT# AM2238 |
| TGIRT reverse transcriptase | Ingex | CAT# TGIRT |
| Ambion RNase-free buffer kit | Invitrogen | CAT# AM9010 |
| Cycloheximide | Sigma Aldrich | CAT# C1988–1G |
| RNasin Plus | Promega | CAT# N2615 |
| cOmplete protease inhibitor | Roche | CAT# 11836170001 |
| Critical commercial assays | ||
| xGen Broad-Range RNA Library Preparation Kit | IDT | CAT# 10009813 |
| MEGAscript T7 Transcription Kit | Invitrogen | CAT# AMB13345 |
| Deposited data | ||
| Raw RNA sequencing data | This paper | SRA: PRJNA1007766 |
| Raw western blot images | This paper | Mendeley DOI: 10.17632/2r2scm54sn.1 |
| Raw representative fluorescent micrographs | This paper | Mendeley DOI: 10.17632/2r2scm54sn.1 |
| Original code & SATCfinder pipeline | This paper | Zenodo DOI: 10.5281/zenodo.10080617 |
| Huntington’s disease RNA-seq, brain tissue | Labadorf et al.50 | GEO: GSE64810 |
| Huntington’s disease RNA-seq, brain tissue | Lin et al.51 | GEO: GSE79666 |
| Huntington’s disease RNA-seq, iPSC-derived | Mehta et al.52 | GEO: GSE109534 |
| Huntington’s disease RNA-seq, brain tissue | Agus et al.53 | GEO: GSE129473 |
| Huntington’s disease RNA-seq, iPSC-derived | Smith-Geater et al.54 | GEO: GSE144559 |
| Huntington’s disease RNA-seq, iPSC derived | Świtońska et al.55 | GEO: GSE124664 |
| Huntington’s disease RNA-seq, brain tissue | n/a | GEO: GSE159940 |
| Experimental models: Cell lines | ||
| Human: U-2 OS cells | ATCC | Cat# HTB-96; RRID:CVCL_0042 |
| Human: HEK293T cells | ATCC | Cat# CRL-3216; RRID:CVCL_0063 |
| Human: RPE-1 cells | ATCC | Cat# CRL-4000; RRID:CVCL_4388 |
| Mouse: NIH/3T3 cells | ATCC | Cat# CRL-1658, RRID:CVCL_0594 |
| Experimental models: Organisms/strains | ||
| Oligonucleotides | ||
| See Supplemental Table 3 for Oligonucleotides. | ||
| Recombinant DNA | ||
| Unmodified CAGRAN lines | Das et al.12 | Supp. Table 4 |
| Unmodified CAGFOCI lines | Das et al.12 | Supp. Table 4 |
| pHR CAGRAN*donors 240×CAG 12×MS2 WPRE | This paper | Supp. Table 4 |
| pHR CAGRAN*AUGs 240×CAG 12×MS2 WPRE | This paper | Supp. Table 4 |
| pHR CAGRAN5×CAG 5×CAG 12×MS2 WPRE | Das et al.12 | Supp. Table 4 |
| pHR CAGRAN10×CAG 10×CAG 12×MS2 WPRE | This paper | Supp. Table 4 |
| pHR CAGRAN22×CAG 22×CAG 12×MS2 WPRE | Das et al.12 | Supp. Table 4 |
| pHR CAGRAN47×CAG 47×CAG 12×MS2 WPRE | Das et al.12 | Supp. Table 4 |
| pHR CAGRAN-BFP 240×CAG EBFP2 12×MS2 WPRE | Das et al.12 | Supp. Table 4 |
| pcDNA3.1 ATNX8 100×CAG | Zu et al.10 | Supp. Table 4 |
| pcDNA3.1 ATXN8KKQ 100×CAG | Zu et al.10 | Supp. Table 4 |
| pcDNA3.1 HTT 100×CAG | Zu et al.10 | Supp. Table 4 |
| pcDNA3.1 HDL2 100×CAG | Zu et al.10 | Supp. Table 4 |
| pcDNA3.1 SCA3 100×CAG | Zu et al.10 | Supp. Table 4 |
| pcDNA3.1 DMPK-AS 100×CAG | Zu et al.10 | Supp. Table 4 |
| pcDNA3.1 ATXN8KKQ*flanking donor 100×CAG | This paper | Supp. Table 4 |
| pcDNA3.1 ATXN8KMQ*AS-AmpR donor 100×CAG | This paper | Supp. Table 4 |
| pcDNA3.1 ATXN8KMQ*AS-AmpR AUG 100×CAG | This paper | Supp. Table 4 |
| pcDNA3.1 ATXN8KKQ*AS-AmpR donor 100×CAG | This paper | Supp. Table 4 |
| pcDNA3.1 ATXN8KKQ*AS-AmpR AUG 100×CAG | This paper | Supp. Table 4 |
| pcDNA3.1 ATXN8KKQ,AS-AmpR stop 100×CAG | This paper | Supp. Table 4 |
| pcDNA3.1 ATXN8KKQ,AS-AmpR frameshift 100×CAG | This paper | Supp. Table 4 |
| pHR ATNX8 400bp endogenous sequence | This paper | Supp. Table 4 |
| pHR ATXN8*donor 400bp endogenous sequence | This paper | Supp. Table 4 |
| pHR ATXN8*AUG 400bp endogenous sequence | This paper | Supp. Table 4 |
| pcDNA3.1 EGFP, vector control | Xiao et al.66 | Addgene plasmid #129020 |
| pCMV-VSV-G, Lentivirus packaging | Stewart et al.67 | Addgene plasmid #8454 |
| psPAX2, Lentivirus packaging | n/a | Addgene plasmid #12260 |
| pX330-U6-Chimeric_BB-CBh-hSpCas9 | Cong et al.68 | Addgene plasmid #42230 |
| Software and algorithms | ||
| Prism v10.0.3 | GraphPad | RRID:SCR_002798 http://www.graphpad.com/ |
| ImageJ v1.53q | Schneider et al.71 | RRID:SCR_003070 https://imagej.net/ |
| samtools v1.11 | Danecek et al.80 | RRID:SCR_002105 http://www.htslib.org/ |
| BBTools v38.86 | Brian Bushnell | RRID:SCR_016968 https://sourceforge.net/projects/bbmap/ |
| STAR v2.7.1a | Dobin et al.79 | RRID:SCR_004463 https://github.com/alexdobin/STAR |
| pysam v0.16.0.1 | n/a | RRID:SCR_021017 https://github.com/pysam-developers/pysam |
| StringTie v2.2.1 | Pertea et al.29 | RRID:SCR_016323 http://ccb.jhu.edu/software/stringtie/ |
| featureCounts v1.6.2 | Liao et al.81 | RRID:SCR_012919 |
| ggsashimi v1.0.0 | Garrido-Martin et al.84 | https://github.com/guigolab/ggsashimi |
| MaxEntScan | Yeo and Burge.25 | http://hollywood.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html |
| MaxEntPy | n/a | https://github.com/kepbod/maxentpy |
| CHOPCHOP | Labun et al.72 | RRID:SCR_015723 http://chopchop.cbu.uib.no/ |
| Canu v2.1.1 | Koren et al.73 | RRID:SCR_015880 https://github.com/marbl/canu |
| minimap2 v2.24-r1122 | Li.82 | RRID:SCR_018550 https://github.com/lh3/minimap2 |
| LIQA v1.3.0 | Hu et al.83 | https://github.com/WGLab/LIQA |
| Other | ||
| Fetal bovine serum | Gibco | CAT# 26140079 |
| Penicillin-streptomycin-glutamine 100X | Gibco | CAT# 10378016 |
| Dulbecco's Modified Eagle Medium | Gibco | CAT# 11965126 |
| Iscove's Modified Dulbecco's Medium | Gibco | CAT# 12440053 |
| Dulbecco’s Phosphate Buffered Solution | Gibco | CAT# 14190144 |
| Opti-Mem | Gibco | CAT# 31985070 |
| Trypsin-EDTA 0.25% | Gibco | CAT# 25200072 |
| Lipofectamine LTX | Invitrogen | CAT# 15338100 |
| Lipofectamine 3000 | Invitrogen | CAT# L3000001 |
| Polybrene | Millipore Sigma | CAT# TR1003G |
| Tris-HCl pH 7.5 | Invitrogen | CAT# 15567027 |
| PBS, pH=7.2 | Gibco | CAT# 20012–027 |
| Nuclease-free water | Invitrogen | CAT# AM9932 |
| Bovine serum albumin (BSA) | Sigma-Aldrich | CAT# A7906 |
| NuPAGE 4x LDS sample buffer | Invitrogen | CAT# NP0008 |
| 4–12% Bis-tris polyacrylamide gel | Invitrogen | CAT# NW04122 |
| iBlot 2 Transfer Stacks, PVDF | Invitrogen | CAT# IB24002 |
| iBlot 2 Dry Blotting System | Invitrogen | CAT# IB21001 |
| 26-gauge needle | Becton Dickinson | CAT# 305110 |
| 22-gauge needle | Becton Dickinson | CAT# 511055 |
| Skim milk powder | BD Biosciences | CAT# 232100 |
| Tris-buffered saline | Fisher Scientific | CAT# AAJ60764K3 |
| Tween-20 | Fisher Scientific | CAT# BP337 |
| SuperSignal West Femto Maximum Sensitivity Substrate | Thermo Scientific | CAT# 34095 |
| Novex Tris-Glycine SDS Sample Buffer | Fisher Scientific | CAT# LC2676 |
| UltraPure Agarose | Invitrogen | CAT# 16500500 |
| DNA Clean & Concentrate | Zymo | CAT# D4004 |
| PureLink RNA mini kit | Invitrogen | CAT# 12183018A |
| PureLink DNA mini kit | Invitrogen | CAT# K182001 |
| ezDNase | Invitrogen | CAT# 11766050 |
| SuperScript IV VILO Master Mix | Invitrogen | CAT# 11756050 |
| DNA QuickExtract solution | Lucigen | CAT# QE09050 |
| KOD Hot Start 2× Master Mix | Sigma Aldrich | CAT# 71842–3 |
| Advantage GC 2 polymerase | Takara Bio | CAT# 639114 |
| Trypan Blue Stain (0.4%) | Invitrogen | CAT# T10282 |
| RNAClean XP beads | Beckman Coulter | CAT# A63987 |
Data and Code Availability
RNA-seq data have been deposited at the NCBI Sequence Read Archive and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images and fluorescent micrographs were deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. This paper analyzes existing, publicly available data. These accession numbers for these datasets are listed in the key resources table.
All source code for the SATCfinder pipeline has been deposited at Zenodo and is publicly available as of the date of publication. Other bioinformatic pipelines used in this work are described in STAR Methods.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Cell lines
HEK293T (CRL-3216; RRID:CVCL_0063), RPE-1 (CRL-4000; RRID:CVCL_4388), U-20S (HTB-96; RRID:CVCL_0042), andNIH-3T3 (CRL-1658; RRID:CVCL_0594) were obtained from ATCC and were tested for mycoplasma at least quarterly using MycoAlert (Lonza LT07-318). Cell lines were maintained at 37 °C in 5% CO2 in Dulbecco's Modified Eagle Medium (DMEM; Gibco 11965126) supplemented with 10% fetal bovine serum (FBS; Gibco 26140079) and 1% Penicillin-Streptomycin-Glutamine (PSG; Gibco 10378016). Cells were passaged 3 times per week at 1:10 dilution using Dulbecco’s Phosphate Buffered Solution (DPBS; Gibco 14190144) and trypsin (Gibco 25200072).
Organisms/Strains
Plasmids were propagated in Stbl3 E. coli (Invitrogen C737303) grown at 37°C in Luria-Bertani (LB) medium. AS-AmpR mutants grew poorly on LB/agar plates supplemented with 100 μg/mL carbenicillin without an extended outgrowth period (>1 hour). All plasmids were verified by Oxford Nanopore sequencing (Plasmidsaurus or Quintara Bioscience) or by Sanger sequencing using a modified protocol with added 7-deaza dGTP and betaine to allow sequencing through tandem repeats.
Methods details
Cloning and plasmid generation
Complete sequences for all plasmids used in this study are provided in Supplemental Table 4. Lentiviral transfer plasmids with CAG-repeats and MS2-hairpins were previously described5, as were the plasmids with 240×CAG repeats with various upstream flanking sequences (including CAGRAN and CAGFOCI)12. Plasmids with ~110 CAG repeats and endogenous flanking sequences from ATXN8, JPH3, DMPK, HTT and ATXN3 were a generous gift from Dr. Laura Ranum10. pcDNA3.1(+) EGFP was a gift from Jeremy Wilusz (Addgene plasmid #129020)66. pCMV-VSV-G was a gift from Bob Weinberg (Addgene plasmid #8454)67. psPAX2 was a gift from Didier Trono (Addgene plasmid #12260). pX330-U6-Chimeric_BB-CBh-hSpCas9 was a gift from Feng Zhang (Addgene plasmid #42230)68. Expanded CAG repeats are difficult to amplify using polymerase chain reaction, and make it challenging to incorporate point mutations using site-directed mutagenesis. Instead, mutations were incorporated by using synthetic double stranded DNA fragments that replaced the corresponding fragments in the plasmids. Double stranded DNA fragments with mutations to CAGRAN splice donor and AUG sites (to generate CAGRAN*donors and CAGRAN*AUGs, respectively) were prepared by overlap extension PCR (see oligo sequences in Supplemental Table 3), and inserted between MluI and EcoRI sites in CAGRAN. CAGRAN constructs with varying repeat lengths were prepared by oligo annealing and inserted between the EcoRI and NotI sites. ATXN8 mutants to AS-AmpR were generated with overlap extension PCR, then inserted between BspHI sites in the ATXN8 or ATXN8KKQ constructs. Splice donors adjacent to the CAG repeat tract were mutated by overlap extension PCR, then Gibson assembled into XbaI and SacI-digested ATXN8KKQ. The endogenous 400 bp ATXN8 constructs, and splice donor and AUG mutants, were prepared by overlap extension PCR and inserted between MluI and NotI sites.
Cell culture
U-2OS cells stably expressing a TetOn 3G transactivating protein and MS2-hairpin binding protein fused to YFP were previously described5. RPE-1 and NIH-3T3 cells expressing the same proteins were prepared similarly by sequential lentiviral transduction and selection.
For transient transfections, HEK293T cells were seeded in a 6-well plate at 750,000 cells/well overnight. 2-4 μg of plasmid was mixed with 500 μL Opti-Mem (Gibco 31985070), after which 2.5 μL Plus Reagent and 8 μL Lipofectamine LTX (Invitrogen 15338100) were added. After a 10-minute incubation at room temperature (22 °C), the mixture was added dropwise to wells containing 1 mL Iscove's Modified Dulbecco's Medium (IMDM; Gibco 12440053). After 6 hours, the media was exchanged for DMEM with FBS and PSG.
To prepare lentivirus, HEK293T cells were transfected as described above in 6-well plates with 2 pg of transfer plasmid, 0.5 μg envelope plasmid (Addgene #8454), and 1 μg packaging plasmid (Addgene #12260). 24 hours post transfection, the viral supernatant was collected and spun at 24,000×g for 10 minutes at room temperature. U-20S cells were transduced at varying viral titer in fresh DMEM with FBS and PSG and 10 μg/mL polybrene (Millipore Sigma TR1003G) to increase transduction efficiency.
To induce transgene expression, cells were plated to be 80% confluent at time of use and were treated with 1 μg/mL doxycycline (Sigma Aldrich D9891) for 24 hours prior to use. For splicing inhibition, Tet-inducible cells were induced with 1 μg/mL doxycycline and co-treated with 0.1% DMSO (Sigma Aldrich D2650) or pladienolide B (Santa Cruz Biotechnology, SC391691) at the indicated concentrations (12.5, 25, or 50 nM, depending on the assay) for 24 hours prior to analysis. For splicing inhibition during transient transfection, cells were transfected as above, and after 6 hours, the media was exchanged for DMEM with FBS and PSG, with 25 nM pladienolide B, or 15 μM isoginkgetin (Tocris Bioscience 6483), or 0.1% DMSO as control.
Western blot
Cells were washed with DPBS and lysed with 160 μL RIPA buffer (25 mM Tris-HCl pH 7.5 (Invitrogen 15567027), 150 mMNaCl (Invitrogen AM9760G), 1% (v/v) NP-40 (Fisher Scientific AAJ19628AP), 1% (w/v) sodium deoxycholate (Sigma-Aldrich D6750), 0.1% (w/v) SDS (Bio-Rad 1610302)) supplemented with 1% (v/v) HALT protease and phosphatase inhibitors (Thermo Scientific 78429) and 125 U/mL Benzonase nuclease (EMD Millipore E1014). The cell lysate was homogenized by passing through a 26-gauge needle (Becton Dickinson 305110) 5 times, and incubated on a nutator for 30 minutes at 4 °C. We noticed that polyglutamine-containing proteins were frequently lost in the pellet fraction after centrifugation at 21000×g or even at 3000×g for 5 minutes. Thus, homogenized lysate was used without further clean-up for electrophoresis. Lysates were incubated with NuPAGE 4× LDS buffer (Invitrogen NP0008) with 50 mM dithiothreitol (DTT; Thermo Scientific R0861) at 70 °C for 5 minutes for most assays. When probing polyalanine-containing proteins, we observed substantial aggregation when lysates were heat denatured at or above 70 °C. Instead, when detecting polyalanine proteins, denaturation was performed at 37 °C for 5 minutes, as is recommended for hydrophobic transmembrane proteins69,70, and this denaturation condition substantially reduced polyalanine aggregation (Supp. Fig. 8C). Samples were separated on a Bolt 4-12% Bis-tris polyacrylamide gel (Invitrogen NW04122) run at 200 V for 30 minutes (for 7 kD MW endogenous ATXN8 constructs) or 42 minutes (all other samples). Samples were transferred to PVDF membranes (Invitrogen IB24002) using the iBlot 2 dry blotting system (Invitrogen IB21001). Membranes were blocked in 5% (w/v) skim milk (BD Biosciences 232100) in TBST (tris-buffered saline (Fisher Scientific AAJ60764K3) with 0.1% (v/v) Tween-20 (Fisher Scientific BP337) for one hour at room temperature, then incubated with primary antibodies in 1% (w/v) skim milk in TBST at 4 °C overnight. The membranes were washed 4 times for five minutes each with TBST, then incubated with the appropriate secondary antibody in 1% (w/v) skim milk for one hour at room temperature. After 4 five-minute washes in TBST, chemiluminescence was detected using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific 34095) with a ChemiDoc XRS+ imager (Bio-Rad). Primary antibodies were used at the following dilutions: mouse anti-FLAG (1:1000, Sigma-Aldrich FI804), mouse anti-HA (1:1000, BioLegend 901501), mouse anti-polyQ (1:2000, Sigma-Aldrich MAB1574), mouse anti-NPT (1:2000, Invitrogen MA5–15275), rabbit anti-β-tubulin (Cell Signaling Tech. 2146). Secondary antibodies: goat anti-rabbit HRP conjugate (Sigma Aldrich A0545), rabbit anti-mouse HRP conjugate (Sigma Aldrich A9044). Band intensities were quantified using ImageJ (version 1.53q)71, with background subtraction performed per-lane.
N-terminal protein sequencing of CAGRAN
Four 10-cm dishes expressing CAGRAN fused to BFP at the C-terminus12, downstream of the repeat tract, were induced with doxycycline. After 24 hours, the cells were washed once with DPBS and then lysed with 1 mL of RIP A buffer supplemented with HALT protease and phosphatase inhibitors and DTT. The cell lysate was collected by scraping and homogenized by passing through a 22-gauge needle (Becton Dickinson 511055) 10 times, and incubated on a nutator for 30 minutes at 4 °C. The lysate was loaded onto 70 μL of GFP-Trap beads (Chromotek gtma-20) which were pre-washed once with RIPA wash buffer (as above, but with 0.15% NP-40). The lysates were incubated on a nutator for 90 minutes at 4 °C, after which the beads were sedimented at 2,500×g for 5 minutes at 4 °C, then washed with 500 μL RIPA wash buffer. After four total wash steps, 80 μL of 2× SDS buffer (Fisher Scientific LC2676) and 8 μL of 1 M DTT was added. The beads were boiled at 95 °C for 10 minutes, then the supernatant was separated on a Bolt 4-12% Bis-tris polyacrylamide gel run at 200 V for 32 minutes, and finally transferred to PVDF membrane using the iBlot 2 dry blotting system. The membrane was rinsed with water, then soaked in 100% methanol (VWR EM-MX0475-1) for 10 seconds, and stained with Coomassie blue staining solution (40% (v/v) methanol, lg/L Coomassie R250 (Sigma Aldrich 1125530025), 1% acetic acid (Sigma Aldrich 695092) for one minute. The membrane was destained with 50% (v/v) methanol until the destaining solution remained colorless, rinsed with water, and dried at room temperature. Bands extracted from the PVDF membrane were subjected to N-terminal sequencing by Edman degradation (Molecular Structure Facility, UC Davis).
Generation of ACTBCAG knock-in cell lines
A CRISPR guide was designed using CHOPCHOP72 to select a site which might be able to act as a polypyrimidine tract. We designed a ssDNA repair template with 40×CAG repeats and 40 base homology arms targeting the cut site in intron 3 of ACTB (sequence in Supplemental Table 3, and see Supplemental Fig. 7E-N). This repair template was obtained as a synthetic oligonucleotide (IDT) and transfected in HEK293T cells. In brief, 2 μg of plasmid expressing CRISPR-Cas9 and guide (Addgene #42230) and 40 pmol ssDNA repair template were transfected using Lipofectamine 3000 (Invitrogen L3000001) following manufacturers’ instructions, using 7.5 μL of L3000 reagent. A GFP plasmid (0.2 μg) was co-transfected to facilitate isolation of transfected cells. After 48 hours, cells were dissociated using trypsin, resuspended in DPBS, and sorted by fluorescence-activated cell sorting using an Aria III Cell Sorter (BD Biosciences). The top 25% of GFP positive cells were plated as single cells and expanded to monoclonal populations.
Approximately 50 clones were isolated and examined for the incorporation of CAG repeats at the desired locus using PCR screening as follows. Approximately ~5000 cells were pelleted and after removing the residual media, 50 μL DNA QuickExtract solution (Lucigen QE09050) was added. The mixture was vortexed, heated at 65°C for 10 minutes and 95 °C for 5 minutes, and then diluted to 200 μL with nuclease-free water. The locus with expected insertion was amplified by PCR using 5 μL KOD Hot Start 2× Master Mix (Sigma Aldrich 71842-3), 0.2 μL each primer at 10 μM, 0.5 μL DMSO, 1 μL gDNA, and 3.1 μL water, with denaturation at 95 °C for 20 seconds, annealing at 60 °C for 10 seconds, and extending at 70 °C for8 seconds for 40 cycles. The ~300 bp thus generated amplicons were visualized by 2% agarose gel (Invitrogen 16500500). Unexpectedly, we found wide variability in the amplicon size for 40×CAG reflecting that each clone had a different number of CAG repeats. The number of CAG repeats in each clone were stable and did not measurably vary on our experimental timescales.
We selected 20 clones with variable insertion sizes for further validation using nested PCR. The outer PCR was performed as above, but for 25 cycles with 15 second extension to amplify a ~1300 bp region. Excess primers were degraded by adding 0.5 μL of thermolabile exonuclease I (NEB m0568) at 37 °C for 15 minutes, followed by enzyme inactivation at 80 °C for 5 minutes. 1 μL of this mixture was used as template in a 25 μL reaction for inner PCR for 25 cycles, amplifying an ~800 bp region as above. The final amplicons were subjected to PCR clean up (Zymo D4004) according to the manufacturer’s protocol, and single molecule sequencing using Oxford Nanopore (Plasmidsaurus). Reads entirely spanning the CAG repeat insertion site were selected with sequential runs of bbduk with options "k=20 literal={5’ ACTCTCTTCTCTGACCTGAG or 3’ CTCTCTTCTCTGACCTGAGT} rcomp=t hdist=0 mm=f", then assembled using Canu73 v2.1.1 using options "-p asm useGrid=0 genomeSize=1000 minReadLength=100 minOverlapLength=100 corMinCoverage=50 corMaxEvidenceErate=0.15-nanopore-raw {file.fq}". From 20 clones screened, we found repeats tracts of length 5 – 39×CAG. We speculate that this variability is caused by a hairpin formed by the ssDNA template during repair. The clones we selected for RNA-seq had consistent repeat lengths among the sequencing reads, indicating that a single allele was edited. ~30% of reads spanning the insert region had a CAG repeat, suggesting the ACTB gene is triploid in our HEK293T cells, consistent with prior reports74.
Real-time quantitative and endpoint PCR
RNA and DNA were isolated from ~106 cells using PureLink RNA (Invitrogen 12183018A) and PureLink Genomic DNA (Invitrogen K182001) mini kits, respectively, according to the manufacturer’s protocols. For reverse transcription, contaminating genomic or plasmid DNA was removed using ezDNase (Invitrogen 11766050), followed by reverse transcription using Superscript IV VILO master mix (Invitrogen 11756050). Briefly, 1 μg total RNA in a 5 μL volume was incubated with ezDNase at 37 °C for 2 minutes. This reaction was diluted with 3 μL nuclease-free water before addition of 2 μL Superscript IV VILO master mix. The mixture was incubated at 25 °C for 10 minutes to anneal primers, 50 °C for 20 minutes for reverse transcription, followed by inactivation at 85 °C for 5 minutes.
For quantitative PCR, 2.5 ng of total RNA, or 5 pg of plasmid DNA, or 10 ng of genomic DNA, were quantified with SYBR Green PCR Master Mix (Applied Biosystems 4309155) using the QuantStudio 3 RT-PCR system (Applied Biosystems A28567). For determination of relative abundance of the CAGRAN intron region, primers targeted the intronic region or the 5’ end of the transcript region. Intron abundance was normalized relative to the 5’ end of the transcript. For CAGRAN*AUG, which has mutations in the 5’ region, the intron was normalized relative to the 3’ end of the repeats. For estimation of transgene copy number, the 5’ end of the transcript was normalized to primers targeting ACTB. See primer sequences in Supplemental Table 3.
For endpoint PCR, 10 ng of total RNA, or 100 ng of genomic DNA was amplified with Advantage GC 2 polymerase (Takara Bio 639114) using 1 M GC Melt and primers spanning the region from upstream of the splice donors to the 3’ end of the repeat, or to the 3’ end of the transcript. The reactions were cycled 25 (cDNA) or 35 (genomic DNA) times, with denaturation at 94 °C for 10 seconds, annealing at 58 °C for 10 seconds, and extension at 68 °C for 25 or 45 seconds. The amplicons were separated on a 3% agarose gel. For sequencing, the PCR reaction was cleaned up with Zymo DNA Clean & Concentrate kit (D4004) and Sanger sequenced (Quintara Biosciences), or by Nanopore long-read sequencing (Plasmidsaurus).
Cell toxicity assays
Cell toxicity assays were performed as previously described 12. In brief, ~10,000 cells were plated into a 6-well plate. The next day, the media was replaced with DMEM with FBS and PSG supplemented with or without doxycycline. After five days, when the cells were approximately 75% confluent, the supernatant containing floating (dead) cells was collected. Cells were washed with DPBS to collect loosely attached cells, and were added to the supernatant. The adherent cells were trypsinized and added to the supernatant. Cells were pelleted at 500×g for 3 minutes, then resuspended in 200 μL DMEM. Dead cells were stained with trypan blue (Invitrogen T10282). The cell count and proportion of dead cells were quantified for ≥ 3 technical replicates using a Countess IIFL Automated Cell Counter (ThermoFisher AMQAflOOO). The cell count for doxycycline induction was normalized to the without-doxycycline condition for each of three biological replicates per condition. The percent of dead cells reflects the number of cells with non-intact membranes, which take up trypan blue.
RNA sequencing
Total RNA was isolated from ~106 (6-well) or ~8x106 (10 cm dish) cells using the PureLink RNA mini kit. RNA quality was verified by denaturing agarose gel or Bioanalyzer. In-house ribodepletion was performed as described previously75. In brief, 2.5 μg of total RNA was mixed with 5 μg of ribodepletion oligos (IDT oligo pool, sequences in Supplemental Table 3) and 2 μL of 5× hybridization buffer (100 mM NaCl, 100 mM Tris-HCl, pH 7.5), to a total volume of 10 μL. The RNA and oligos were hybridized by heating to 95°C for 2 minutes and cooling to 45 °C at −0.1 °C/s. While maintaining samples at 45 °C, 1.5 μL of Hybridase RNase H (Lucigen H39500), 3 μL of 5× RNase H buffer (167 mM NaCl, 50 mM MgCl2), and 0.5 μL of water were added, and the reaction was incubated at 45 °C for 30 minutes. The ribodepleted RNA was cleaned using 2× RNAClean XP beads (Beckman Coulter A63987), eluting with 20 μL DNase digestion reaction mixture (1.5 μL Turbo DNase (Invitrogen AM2238), 2 μL Turbo DNase 10X buffer, and 16.5 μL RNase-free water). The residual DNA oligos were digested at 37 °C for 30 minutes, and the resulting mRNA was cleaned using 2× RNAClean XP beads and eluted in 11.5 μL nuclease-free water.
The ribodepleted RNA (typically ~5% of input, or about 100 ng) was reverse transcribed using a group II intron reverse transcriptase (TGIRT, Ingex) using primers provided by the xGen library preparation kit (IDT 10009813). In brief, 10 μL ribodepleted RNA was mixed with 4 μL 5× TGIRT buffer (2.25 MNaCl, 25 mM MgCl2, 100 mM Tris-HCl, pH 7.5; Ambion RNase-free Buffer Kit AM9010), and 1 μL random hexamer primers (xGen). The mixture was heated to 94 °C for 12 minutes to induce RNA fragmentation. After cooling at 4 °C for 2 minutes to allow primer binding, 1 μL 100 mM DTT (xGen), 1 μL TGIRT enzyme, and 1 μL RNase inhibitor (xGen) was added and incubated at room temperature for 10 minutes to allow TGIRT binding to RNA-primer duplexes. To initiate reverse transcription, 2 μL dNTPs (xGen) were added, followed by sequential incubation for 15 minutes each at 20 °C, 42 °C, 55 °C, and 65 °C for elongation. This step-wise protocol is needed for TGIRT to extend the unstable RNA-DNA duplexes formed by random hexamer primers76. At completion, TGIRT was dissociated from the RNA-DNA duplexes by addition of 1 μL 5 M NaOH (VWR BDH7225-1) with incubation at 95 °C for 3 minutes, then neutralized with 1 μL 5 M HC1 (VWR BDH7419-1). The remainder of the library preparation was performed according to the xGen protocol except that post-ligation, the library was eluted in 30 μL nuclease-free water, followed by a modified protocol for final amplification and incorporation of sequencing adaptors. The final amplification was performed in a 50 μL reaction with 1 μL Advantage GC2 polymerase, 10 μL 5× GC2 buffer, 5 μL (1M) GC melt, 1 μL of 10 mM dNTPs (NEB N0447L), 1 μL GC2 polymerase, 4 μL xGen adapter mix, and 29 μL eluted library. Typically, ~10 cycles of PCR with denaturation at 94 °C for 20 seconds, annealing at 58 °C for 20 seconds, and extension at 68 °C for 45 seconds, for final amplification yielded sufficient material for quality control and sequencing. Libraries were sequenced on a NovaSeq 6000 (Illumina) by the Whitehead Institute Genome Technology Core or by Novogene.
SATCfinder pipeline and RNA-seq analysis
The SATCfinder pipeline and full documentation are available at https://github.com/AnkurJainLab/SATCfmder. In brief, the SATCfinder pipeline first selected reads containing ≥3×CAG or CTG, along with their read mates, if available, using bbduk (BBTools 38.86)77 with options “k=9 hdist=0 mm=f literal=CAGCAGCAG rcomp=t”. At this step, read pairs not containing any repeats were discarded. Low quality read ends and sequencing adapters were removed from the reads using cutadapt (version 3.7)78 with commands “-a AGATCGGAAGAG —error-rate=0.1 —times=1 —overlap=5 — minimum-length=20 --quality-cutoff=20”. Next, the CAG-selected reads were processed using a custom Python script, the SATCfinder trim module. This script converts FASTQ files to unmapped SAM files, during which CAG/CTG repeats (minimum 3, maximum unlimited) are computationally trimmed from the read according to the following rules: (1) CAG repeats and anything downstream of the repeat, is trimmed and saved to a SAM field; (2) CTG repeats, and anything upstream of them, is trimmed and saved to a SAM field. In both cases, the length of the trimmed repeat tract (but not any additional removed sequences) is saved in a SAM field. Repeats will be trimmed from both reads if paired end reads are provided. Reads with at least 15 bases post-trimming and their corresponding read mates are stored in the SAM file. These reads were then aligned to hg38 with GRCh GTF annotation file (38.93) using the short-read alignment tool STAR (version 2.7.1a)79. STAR alignment arguments were outSAMtype BAM SortedByCoordinate “--outSAMunmapped Within -- genomeFastaFiles {construct.fa} --readFilesType SAM (library type}”, where construct.fa refers to a relevant transgene fasta sequence if required, and library type refers to SE or PE for single and paired end libraries respectively.
The aligned BAM files were indexed using samtools (version l.l)80. Trimmed reads were separated from their read mates and stored in a BAM file which only contains reads with trimmed repeats. This file is then processed by a second SATCfinder module, SATCfinder ends. This script takes as input a BAM file, along with genomic coordinates and strand information for a region of interest, and uses pysam (version 0.16.0.1) to generate a .csv file containing the base position and the number of CAG ends at that base for the given region. This csv file can then be used in standard plotting software to generate bar plots as in Fig. 1A-B, which were prepared with GraphPad Prism.
The percentage of CAG ends reflecting splicing to a CAG repeat was calculated using the number of CAG ends more than 10 bases from the repeat divided by the total number of CAG ends. To estimate the percent of AUG-initiated transcripts after splicing, we examined each splice junction supported by more than 100 reads and asked if that junction would generate an AUG-initiated ORF or not. If yes, we counted the % of CAG ends that defined that junction as AUG-initiated. To find the number of CAG removed during splicing, we examined trimmed and mapped reads that aligned to a region upstream of the repeat tract (either at a splice donor, or directly adjacent to the repeat), that also contained at least 10 bases from the 3' end of the repeat in their trimmed sequences.
For standard (non-trimmed) analysis, RNA-seq libraries were processed through cutadapt as previously described and then aligned to hg38 using STAR with arguments “--outSAMtype BAM SortedByCoordinate --outSAMunmapped Within”, along with “--genomeFastaFiles {construct.fa}” if a transgene was expressed. Transcript assembly was performed with StringTie (2.2.1)29 with default options, using all reads mapping to the construct.
Relative expression for SV40, CMV, and the cryptic colEl promoter was calculated by the mean read counts across three replicate libraries for (1) the SV40 transcript, as detected by StringTie; (2) the region between the CMV promoter and CAG repeats, as the 3’ end of the CAG repeat tract could be present on both initiated by CMV and by colEl; and (3) the region of the colE1-initiated transcript detected by StringTie without splicing to the CAG repeat tract.
Reads mapping to genes were quantified by featureCounts (version 1.6.2)81 using arguments “-g gene_name -f −O”. For reads mapping to exons, we first flattened the GTF (flattenGTF), then used featureCounts with additional arguments “--F SAF -t exon -largestOverlap”.
For HD patient RNA-seq analyses, we included datasets with paired end sequencing (≥ 75 bp reads) from HD-affected patient tissue samples and patient-derived cell lines. Accession numbers for these datasets are listed in the key resources table. These studies included unaffected control individuals, however, we included only HD-affected patients in our analysis. Of note, the dataset GSE71191 was released only as a composite dataset of reads mapping to the HTT region from multiple patients, and thus we likely overestimate the prevalence of reads mapping to the HTT exon 1 region. These data were analyzed using SATCfinder as previously described.
For long-read analysis of CAGRAN transcript isoforms, we aligned the reads to the sequence of CAGRAN47×CAG using minimap2 (version 2.24-r1122)82 with option “-x splice”. We used StringTie, as described above, to prepare a GTF file for isoform analysis. We curated the GTF to remove isoform variants in the MS2 loop region (see Supp. Fig. 3B). Next, we quantified the isoform abundance using LIQA (version 1.3.0)83, run with settings “-task quantify -max_distance 10 -f_weight 1”.
Splice site analysis
Sashimi plots without splice junction arcs, as in Fig. 1A-B, were generated with ggsashimi(version 1.1.5)84 with arguments “--out-format pdf --min-coverage 100000 --shrink --fix-y-scale”. To find CAG ends, we used a custom Python script, acessed through SATCfinder ends, as described above.
MaxEnt scores for 5’ and 3’ splice sites were calculated using the MaxEntScan web-server (http://hollywood.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html25). or using maxentpy (https://github.com/kepbod/maxentpy/), a python wrapper for the same model. Genome-wide analysis was performed using a custom python script which calculated the 5’ or 3’ MaxEnt score for every annotated splice donor site or acceptor site in hg38 using the previously described GTF file. No corrections were made for the ~1% of introns which are processed by the minor spliceosome.
To locate repeats in the genome, and to find splice acceptors with tandem CAG repeats, we used a custom python script, the SATCfinder annotate module, which generates a csv file with repeats assigned to genomic regions (gene, intron, exon, or intergene). To find CAG repeat acceptors, we calculated the distance in bases between the annotated exon junctions and the left-most coordinate of CAG repeats within each exon. A distance less than zero meant that the CAG repeats at least partially overlapped with the annotated exon junction and thus defined as the splice acceptor site. For example, ... YYYYCAGCAGCAGCAG... (where Y is the polypyrimidine tract and the exonic bases are indicated in bold) would have a distance of -3, because the CAG repeat begins 3 bases upstream from the exon boundary. A distance greater than zero indicated a small intervening sequence between the annotated acceptor and pure CAG repeat. Acceptors with interruptions between the acceptor ‘CAG’ and pure CAG tract were not included in our analysis, but are included in Supplemental Table 1 as a reference.
Fluorescence microscopy
For live-cell imaging with the MS2 system, -10,000 cells were plated on glass bottom 96-well plates (Brooks, MGB096–1-2-LG-L) and induced with doxycycline (and any other treatments, as indicated) the following day. Cells were imaged 24 hours after induction with a Dragonfly 505 spinning-disk confocal microscope (Andor Technologies) equipped with a piezo Z-stage (ASI) and an iXon Ultra 888 EMCCD camera. Pin-hole size was kept at 40 pm. Z-stacks were acquired with a step size of 0.3 to 0.5 pm. Live cells were imaged in a humidified chamber (OKO labs) maintained at 37 °C and 5% (v/v) CO2 using a 100x oil immersion objective NA 1.45 (Nikon MRD01905, pixel size 121 nm x 121 nm). For quantification of cells with aggregates, cells were imaged with a 20x air objective NA 0.75 (Nikon MRD00205). Fixed cells were imaged as above, but at room temperature. DAPI was excited with a 405-nm laser, and fluorescence was collected using a 445/46 bandpass filter. YFP was imaged using a 488-nm laser and corresponding 521/38-nm band pass emission filter. Cy5 and Atto647N-labeled samples were imaged using 640-nm laser line and a 698/77 bandpass emission filter.
RNA FISH
Cells were fixed with a solution of 75% (v/v) methanol and 25% (v/v) acetic acid for 10 min at 4 °C. Fixed cells were washed three times with a wash solution (nuclease-free water with 300 mM NaCl, 30 mM sodium citrate (Calbiochem 567446), 10% (v/v) formamide (Sigma Aldrich 47671), and 0.1% (v/v) NP-40 substitute) at room temperature. Hybridization was performed at 37 °C for 3 hours with 200 nM Atto647N-conjugated probes targeting the CAG repeats. The probes were dissolved in hybridization buffer (100 mg/mL dextran sulfate (Sigma Aldrich D8906), 10% (v/v) formamide, 300 mM NaCl, and 30 mM sodium citrate in nuclease-free water). After hybridization, cells were washed for 30 min with wash solution and counterstained for 30 min with wash solution containing 0.5 pg/mL DAPI (Sigma Aldrich D9542); both this wash step and DAPI staining were performed at 37 °C. Cells were then washed three times with phosphate buffered saline (PBS; Gibco 20012–027), and kept in PBS for imaging.
Immunofluorescence
Cells were fixed with 2% (v/v) formaldehyde (Fisher Scientific 28906) in PBS for 40 min, washed four times with PBS, and then permeabilized with 0.1% (v/v) Triton-X-100 (Sigma Aldrich T8787) in PBS for 10 min at room temperature. Cells were then blocked in 0.45-pm filtered 3% (w/v) bovine serum albumin (BSA; Sigma-Aldrich A7906) in PBS. Primary antibodies targeting poly-glutamine (Sigma-Aldrich MAB1574) were diluted 1:100 in 1% (w/v) BSA in PBS and incubated with cells for 1 h at room temperature. Cells were washed three times with PBS and then incubated with Cy5-conjugated donkey anti-mouse (Jackson Immuno Research Labs 715175151) secondary antibody at 1:2,000 dilution in 1% (w/v) BSA in PBS for 1 h at room temperature. After three washes with PBS, cells were counterstained with DAPI solution (PBS containing 0.5 pg/mL DAPI) for 3 min, washed three times with PBS again, and then kept in PBS for imaging.
Polysome profiling
15 cm dishes of cells at ~75% confluency were induced with 1 pg/mL doxycycline for 12 hours. The cells were washed once with DPBS and scraped in 1 mL lysis buffer (20 mM HEPES pH 7.5, 100 mM KCl, 5 mM MgCl2, 1 mM Triton-X-100, 2 mM DTT, 100 pg/mL cycloheximide [(Sigma Aldrich C1988–1G], 20 μ/mL RNasin Plus [Promega N2615], supplemented with cOmplete protease inhibitor [Roche 11836170001]). Lysates were incubated on ice 10 minutes, homogenized 5x with a 27g needle, and centrifuged at 1500xg for 10 minutes at 4 °C. 300 pL of each supernatant was loaded onto a 10–50% sucrose gradient (20 mM HEPES pH 7.5, 5 mM MgC12, 100 mM KC1, 10–50% (w/v) sucrose, 100 pg/mL cycloheximide, 20 u/mL RNasin Plus, and 2 mM DTT), centrifuged in a Beckman ultracentrifuge (rotor SW41Ti) at 36000 rpm for 2 hours at 4 °C, and then fractioned on a BioComp gradient fractionator. To allow for normalization between fractions, 50 pg of an in-vitro transcribed mCherry RNA (MEGAscript T7 Transcription Kit, Invitrogen AMB13345, prepared as manufacturer’s protocol) was added to 100 pL of each fraction. After pooling the polysome fractions, the free mRNA and pooled polysome fractions were diluted 10x with RNase-free water. 1 pL of this dilution was used without further purification as template for reverse transcription with Superscript IV VILO master mix and analyzed by quantitative PCR, as described above.
Quantification and statistical analyses
Toxicity assays, quantitative PCR, cells with aggregates, and nuclear/cytoplasmic RNA ratios were analyzed by two-tailed unpaired Student’s t-test using GraphPad Prism. The percentage of endogenous transcripts reflecting splicing to annotated CAG acceptors was analyzed by a Fisher’s exact test. Statistical tests are further described in figure legends.
Supplementary Material
Table S3. List of oligonucleotides, related to STAR Methods.
Table S4. List of constructs and full construct sequences, related to STAR Methods.
Table S1. Annotated tandem CAG repeat splice acceptors in hg38, related to Figure 1 and STAR Methods.
Highlights –
Expanded CAG repeats act as splice acceptor sites
Splicing frequency increases with the number of CAG repeats
Splicing may place CAG repeats in RNA in unexpected sequence contexts
AUG-initiated translation of mis-spliced RNAs can account for aberrant proteins
Acknowledgements
We thank David Bartel, Christopher Burge, and members of the Jain lab for helpful discussions. We also thank George Bell of the Whitehead Institute Bioinformatics and Research Computing group for helpful discussions on SATCfinder, and Sumeet Gupta, Amanda Chilaka, Jennifer Love, and Stephen Mraz of the Whitehead Institute Genome Technology Core for supporting our RNA-sequencing projects. Plasmids containing CAG repeats with flanking sequences from ATXN3, ATXN8, JPH3-AS, DMPK-AS, and HTT were a gift from Laura Ranum. This work was supported by grants from the NIH (R00AG053434, R35GM151111), Chan Zuckerberg Initiative, David and Lucile Packard Foundation, and the Smith Family Awards Program. Y.C. was supported by a fellowship from the National Research Foundation of Korea.
Footnotes
Declaration of Interests
The authors declare no competing financial interests. A.J. is a member of the scientific advisory board of Molecular Cell.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Malik I, Kelley CP, Wang ET, and Todd PK (2021). Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat. Rev. Mol. Cell Biol 22, 589–607. 10.1038/s41580-021-00382-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paulson H (2018). Repeat expansion diseases. Handb. Clin. Neurol 147, 105–123. 10.1016/B978-0-444-63233-3.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Mezer M, Wojciechowska M, Napierala M, Sobczak K, and Krzyzosiak WJ (2011). Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 39, 3852–3863. 10.1093/nar/gkql323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wojciechowska M, and Krzyzosiak WJ (2011). Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet 20, 3811–3821. 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain A, and Vale RD (2017). RNA phase transitions in repeat expansion disorders. Nature 546, 243–247. 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mykowska A, Sobczak K, Wojciechowska M, Kozlowski P, and Krzyzosiak WJ (2011). CAG repeats mimic CUG repeats in the misregulation of alternative splicing. Nucleic Acids Res. 39, 8938–8951. 10.1093/nar/gkr608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayhan F, Perez BA, Shorrock HK, Zu T, Banez-Coronel M, Reid T, Furuya H, Clark HB, Troncoso JC, Ross CA, et al. (2018). SCA8 RAN polySer protein preferentially accumulates in white matter regions and is regulated by elF3F. EMBO J. 37. 10.15252/embj.201899023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bafiez-Coronel M, Ayhan F, Tarabochia AD, Zu T, Perez BA, Tusi SK, Pletnikova 0, Borchelt DR, Ross CA, Margolis RL, et al. (2015). RAN Translation in Huntington Disease. Neuron 88, 667–677. 10.1016/j.neuron.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, Chen G, Weatherspoon MR, Clark HB, Ebner TJ, et al. (2006). Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat. Genet 38, 758–769. 10.1038/ngl827. [DOI] [PubMed] [Google Scholar]
- 10.Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram, M.A.C., et al. (2011). Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. U.S.A 108, 260–265. 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen L, Cleary JD, and Ranum LPW (2019). Repeat-Associated Non-ATG Translation: Molecular Mechanisms and Contribution to Neurological Disease. Annu. Rev. Neurosci 42, 227–247. 10.1146/annurev-neuro-070918-050405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das MR, Chang Y, Anderson R, Saunders RA, Zhang N, Tomberlin CP, Vale RD, and Jain A (2023). Repeat-associated non-AUG translation induces cytoplasmic aggregation of CAG repeat-containing RNAs. Proc. Natl. Acad. Sci. U.S.A 120, e2215071120. 10.1073/pnas.2215071120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jazurek-Ciesiolka M, Ciesiolka A, Komur AA, Urbanek-Trzeciak MO, Krzyzosiak WJ, and Fiszer A (2020). RAN translation of the expanded CAG repeats in the SCA3 disease context. J. Mol. Biol 432, 166699. [DOI] [PubMed] [Google Scholar]
- 14.Lee Y, and Rio DC (2015). Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem 84, 291–323. 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sznajder LJ, Thomas JD, Carrell EM, Reid T, McFarland KN, Cleary JD, Oliveira R, Nutter CA, Bhatt K, Sobczak K, et al. (2018). Intron retention induced by microsatellite expansions as a disease biomarker. Proc. Natl. Acad. Sci. U.S.A 115, 4234–4239. 10.1073/pnas.1716617115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorbunova V, Seluanov A, Dion V, Sandor Z, Meservy JL, and Wilson JH (2003). Selectable System for Monitoring the Instability of CTG/CAG Triplet Repeats in Mammalian Cells. Mol. Cell. Biol 23, 4485–4493. 10.1128/MCB.23.13.4485-4493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramani B, Harris GM, Huang R, Seki T, Murphy GG, Costa MDC, Fischer S, Saunders TL, Xia G, McEachin RC, et al. (2015). A knockin mouse model of spinocerebellar ataxia type 3 exhibits prominent aggregate pathology and aberrant splicing of the disease gene transcript. Hum. Mol. Genet 24, 1211–1224. 10.1093/hmg/ddu532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watase K, Barrett CF, Miyazaki T, Ishiguro T, Ishikawa K, Hu Y, Unno T, Sun Y, Kasai S, Watanabe M, et al. (2008). Spinocerebellar ataxia type 6 knockin mice develop a progressive neuronal dysfunction with age-dependent accumulation of mutant Ca v 2.1 channels. Proc. Natl. Acad. Sci. U.S.A 105, 11987–11992. 10.1073/pnas.0804350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gipson TA, Neueder A, Wexler NS, Bates GP, and Housman D (2013). Aberrantly spliced HTT, a new player in Huntington’s disease pathogenesis. RNA Biology 10, 1647–1652. 10.4161/rna.26706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kieleczawa J (2006). Fundamentals of sequencing of difficult templates-an overview. J. Biomol. Tech 17, 207–217. [PMC free article] [PubMed] [Google Scholar]
- 21.Treangen TJ, and Salzberg SL (2012). Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet 13, 36–46. 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, and Pan T (2015). Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837. 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrell ST, Tang Z, Mohr S, Lambowitz AM, and Thornton CA (2018). Detection of expanded RNA repeats using thermostable group II intron reverse transcriptase. Nucleic Acids Res. 46, el. 10.1093/nar/gkx867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nottingham RM, Wu DC, Qin Y, Yao J, Hunicke-Smith S, and Lambowitz AM (2016). RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA 22, 597–613. 10.1261/rna.055558.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeo G, and Burge CB (2004). Maximum Entropy Modeling of Short Sequence Motifs with Applications to RNA Splicing Signals. J. Comput. Biol 11, 2–3. [DOI] [PubMed] [Google Scholar]
- 26.Eng L, Coutinho G, Nahas S, Yeo G, Tanouye R, Babaei M, Dork T, Burge C, and Gatti RA (2004). Nonclassical splicing mutations in the coding and noncoding regions of the ATM Gene: Maximum entropy estimates of splice junction strengths. Hum. Mutat 23, 67–76. 10.1002/humu.10295. [DOI] [PubMed] [Google Scholar]
- 27.Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, and Mizui Y (2007). Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol 3, 570–575. 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 28.O’Brien K, Matlin AJ, Lowell AM, and Moore MJ (2008). The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing. J. Biol. Chem 283, 33147–33154. 10.1074/jbc.M805556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, and Salzberg SL (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol 33, 290–295. 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemp NA, Hiraoka K, Kasahara N, and Logg CR (2012). Cryptic transcripts from a ubiquitous plasmid origin of replication confound tests for cis-regulatory function. Nucleic Acids Res. 40, 7280–7290. 10.1093/nar/gks451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muerdter F, Boryri LM, Woodfin AR, Neumayr C, Rath M, Zabidi MA, Pagani M, Haberle V, Kazmar T, Catarino RR, et al. (2018). Resolving systematic errors in widely used enhancer activity assays in human cells. Nat. Methods 15, 141–149. 10.1038/nmeth.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Habara Y, Takeshima Y, Awano H, Okizuka Y, Zhang Z, Saiki K, Yagi M, and Matsuo M (2009). In vitro splicing analysis showed that availability of a cryptic splice site is not a determinant for alternative splicing patterns caused by +1G->A mutations in introns of the dystrophin gene. J. Med. Genet 46, 542–547. 10.1136/jmg.2008.061259. [DOI] [PubMed] [Google Scholar]
- 33.Lee J-M, Wheeler VC, Chao MJ, Vonsattel JPG, Pinto RM, Lucente D, Abu-Elneel K, Ramos EM, Mysore JS, Gillis T, et al. (2015). Identification of Genetic Factors that Modify Clinical Onset of Huntington’s Disease. Cell 162, 516–526. 10.1016/j.cell.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michlewski G, and Krzyzosiak WJ (2004). Molecular Architecture of CAG Repeats in Human Disease Related Transcripts. J. Mol. Biol 340, 665–679. 10.1016/j.jmb.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 35.Pinto RM, Dragileva E, Kirby A, Lloret A, Lopez E, St Claire J, Panigrahi GB, Hou C, Holloway K, Gillis T, et al. (2013). Mismatch repair genes Mlhl and Mlh3 modify CAG instability in Huntington’s disease mice: genome-wide and candidate approaches. PLoS Genet. 9, el003930. 10.1371/journal.pgen.1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tome S, Manley K, Simard JP, Clark GW, Slean MM, Swami M, Shelbourne PF, Tillier ERM, Monckton DG, Messer A, et al. (2013). MSH3 polymorphisms and protein levels affect CAG repeat instability in Huntington’s disease mice. PLoS Genet. 9, el003280. 10.1371/journal.pgen.1003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies JE, and Rubinsztein DC (2006). Polyalanine and polyserine frameshift products in Huntington’s disease. J. Med. Genet 43, 893–896. 10.1136/jmg.2006.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaspar C (2000). CAG tract of MJD-1 may be prone to frameshifts causing polyalanine accumulation. Hum. Mol. Genet 9, 1957–1966. 10.1093/hmg/9.13.1957. [DOI] [PubMed] [Google Scholar]
- 39.Orr HT, and Zoghbi HY (2007). Trinucleotide Repeat Disorders. Annu. Rev. Neurosci 30, 575–621. 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 40.Gao F-B, Richter JD, and Cleveland DW (2017). Rethinking Unconventional Translation in Neurodegeneration. Cell 171, 994–1000. 10.1016/j.cell.2017.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scoles DR, Ho MHT, Dansithong W, Pflieger LT, Petersen LW, Thai KK, and Pulst SM (2015). Repeat Associated Non-AUG Translation (RAN Translation) Dependent on Sequence Downstream of the ATXN2 CAG Repeat. PLoS ONE 10, e0128769. 10.1371/journal.pone.0128769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soragni E, Petrosyan L, Rinkoski TA, Wieben ED, Baratz KH, Fautsch MP, and Gottesfeld JM (2018). Repeat-Associated Non-ATG (RAN) Translation in Fuchs’ Endothelial Corneal Dystrophy. Invest. Ophthalmol. Vis. Sci 59, 1888. 10.1167/iovs.l7-23265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiller Mv Huse K, Szafranski K, Jahn N, Hampe J, Schreiber S, Backofen R, and Platzer M (2004). Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat. Genet 36, 1255–1257. 10.1038/ngl469. [DOI] [PubMed] [Google Scholar]
- 44.Bradley RK, Merkin J, Lambert NJ, and Burge CB (2012). Alternative Splicing of RNA Triplets Is Often Regulated and Accelerates Proteome Evolution. PLoS Biol. 10, el001229. 10.1371/journal,pbio.1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gymrek M (2017). A genomic view of short tandem repeats. Curr. Opin. Genet. Dev 44, 9–16. 10.1016/j.gde.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 46.Walsh PS, Fildes NJ, and Reynolds R (1996). Sequence Analysis and Characterization of Stutter Products at the Tetranucleotide Repeat Locus VWA. Nucleic Acids Res. 24, 2807–2812. 10.1093/nar/24.14.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sathasivam K, Neueder A, Gipson TA, Landles C, Benjamin AC, Bondulich MK, Smith DL, Faull RLM, Roos RAC, Howland D, et al. (2013). Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl. Acad. Sci. U.S.A 110, 2366–2370. 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun X, Li PP, Zhu S, Cohen R, Marque LO, Ross CA, Pulst SM, Chan HYE, Margolis RL, and Rudnicki DD (2015). Nuclear retention of full-length HTT RNA is mediated by splicing factors MBNL1 and U2AF65. Sci. Rep 5, 12521. 10.1038/srepl2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urbanek MO, Jazurek M, Switonski PM, Figura G, and Krzyzosiak WJ (2016). Nuclear speckles are detention centers for transcripts containing expanded CAG repeats. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1862, 1513–1520. 10.1016/j.bbadis.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 50.Labadorf A, Hoss AG, Lagomarsino V, Latourelle JC, Hadzi TC, Bregu J, MacDonald ME, Gusella JF, Chen J-F, Akbarian S, et al. (2015). RNA Sequence Analysis of Human Huntington Disease Brain Reveals an Extensive Increase in Inflammatory and Developmental Gene Expression. PLoSONE 10, e0143563. 10.1371/journal.pone.0143563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin L, Park JW, Ramachandran S, Zhang Y, Tseng Y-T, Shen S, Waldvogel HJ, Curtis MA, Faull RLM, Troncoso JC, et al. (2016). Transcriptome sequencing reveals aberrant alternative splicing in Huntington’s disease. Hum. Mol. Genet 25, 3454–3466. 10.1093/hmg/ddwl87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mehta SR, Tom CM, Wang Y, Bresee C, Rushton D, Mathkar PP, Tang J, and Mattis VB (2018). Human Huntington’s Disease iPSC-Derived Cortical Neurons Display Altered Transcriptomics, Morphology, and Maturation. Cell Rep 25, 1081–1096.e6. 10.1016/j.celrep.2018.09.076. [DOI] [PubMed] [Google Scholar]
- 53.Agus F, Crespo D, Myers RH, and Labadorf A (2019). The caudate nucleus undergoes dramatic and unique transcriptional changes in human prodromal Huntington’s disease brain. BMC Med Genomics 12, 137. 10.1186/sl2920-019-0581-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith-Geater C, Hernandez SJ, Lim RG, Adam M, Wu J, Stocksdale JT, Wassie BT, Gold MP, Wang KQ, Miramontes R, et al. (2020). Aberrant Development Corrected in Adult-Onset Huntington’s Disease iPSC-Derived Neuronal Cultures via WNT Signaling Modulation. Stem Cell Reports 14, 406–419. 10.1016/j.stemcr.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Switohska K, Szlachcic WJ, Handschuh L, Wojciechowski P, Marczak L, Stelmaszczuk M, Figlerowicz M, and Figiel M (2018). Identification of Altered Developmental Pathways in Human Juvenile HD iPSCWith 71Qand 109Q Using Transcriptome Profiling. Front Cell Neurosci 12, 528. 10.3389/fncel.2018.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kearse MG, Green KM, Krans A, Rodriguez CM, Linsalata AE, Goldstrohm AC, and Todd PK (2016). CGG Repeat-Associated Non-AUG Translation Utilizes a Cap-Dependent Scanning Mechanism of Initiation to Produce Toxic Proteins. Mol. Cell 62, 314–322. 10.1016/j.molcel.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tabet R, Schaeffer L, Freyermuth F, Jambeau M, Workman M, Lee C-Z, Lin C-C, Jiang J, Jansen-West K, Abou-Hamdan H, et al. (2018). CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C90RF72 transcripts. Nat. Commun 9, 152. 10.1038/s41467-017-02643-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng W, Wang S, Mestre AA, Fu C, Makarem A, Xian F, Hayes LR, Lopez-Gonzalez R, Drenner K, Jiang J, et al. (2018). C90RF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through elF2a phosphorylation. Nat. Commun 9, 51. 10.1038/s41467-017-02495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sonobe Y, Ghadge G, Masaki K, Sendoel A, Fuchs E, and Roos RP (2018). Translation of dipeptide repeat proteins from the C90RF72 expanded repeat is associated with cellular stress. Neurobiol. Dis 116, 155–165. 10.1016/j.nbd.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamada SB, Gendron TF, Niccoli T, Genuth NR, Grosely R, Shi Y, Glaria I, Kramer NJ, Nakayama L, Fang S, et al. (2019). RPS25 is required for efficient RAN translation of C9orf72 and other neurodegenerative disease-associated nucleotide repeats. Nat. Neurosci 22, 1383–1388. 10.1038/s41593-019-0455-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McEachin ZT, Gendron TF, Raj N, Garcia-Murias M, Banerjee A, Purcell RH, Ward PJ, Todd TW, Merritt-Garza ME, Jansen-West K, et al. (2020). Chimeric Peptide Species Contribute to Divergent Dipeptide Repeat Pathology in c9ALS/FTD and SCA36. Neuron 107, 292–305.e6. 10.1016/j.neuron.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saffert P, Adamla F, Schieweck R, Atkins JF, and Ignatova Z (2016). An Expanded CAG Repeat in Huntingtin Causes +1 Frameshifting. J. Biol. Chem 291, 18505–18513. 10.1074/jbc.M116.744326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wright SE, Rodriguez CM, Monroe J, Xing J, Krans A, Flores BN, Barsur V, Ivanova MI, Koutmou KS, Barmada SJ, et al. (2022). CGG repeats trigger translational frameshifts that generate aggregation-prone chimeric proteins. Nucleic Acids Res. 50, 8674–8689. 10.1093/nar/gkac626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang S, Wijegunawardana D, Sheth U, Veire AM, Salgado JMS, Agrawal M, Zhou J, Pereira JD, Gendron TF, and Guo JU (2023). Aberrant splicing exonizes C90RF72 repeat expansion in ALS/FTD (Neuroscience) 10.1101/2023.11.13.566896. [DOI] [Google Scholar]
- 65.Lescure A, Lutz Y, Eberhard D, Jacq X, Krol A, Grummt I, Davidson I, Chambon P, and Tora L (1994). The N-terminal domain of the human TATA-binding protein plays a role in transcription from TATA-containing RNA polymerase II and III promoters. EMBO J. 13, 1166–1175. 10.1002/j.1460-2075.1994.tb06366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiao M-S, and Wilusz JE (2019). An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3’ ends. Nucleic Acids Res 47, 8755–8769. 10.1093/nar/gkz576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen ISY, Hahn WC, Sharp PA, et al. (2003). Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501. 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819–823. 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanada K, Yamato I, and Anraku Y (1985). Identification of proline carrier in Escherichia coll K-12. FEBS Lett. 191, 278–282. 10.1016/0014-5793(85)80024-7. [DOI] [PubMed] [Google Scholar]
- 70.Okada N, Yamamoto T, Watanabe M, Yoshimura Y, Obana E, Yamazaki N, Kawazoe K, Shinohara Y, and Minakuchi K (2011). Identification of TMEM45B as a protein clearly showing thermal aggregation in SDS-PAGE gels and dissection of its amino acid sequence responsible for this aggregation. Protein Expr. Purif 77, 118–123. 10.1016/j.pep.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 71.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, and Valen E (2019). CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171–W174. 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, and Phillippy AM (2017). Canu: scalable and accurate long-read assembly via adaptive k -mer weighting and repeat separation. Genome Res. 27, 722–736. 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin Y-C, Boone M, Meuris L, Lemmens I, Van Roy N, Soete A, Reumers J, Moisse M, Plaisance S, Drmanac R, et al. (2014). Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat. Commun 5, 4767. 10.1038/ncomms5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tomezsko P, Swaminathan H, and Rouskin S (2020). Viral RNA structure analysis using DMS-MaPseq. Methods 183, 68–75. 10.1016/j.ymeth.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Diilk S-L, and Rouskin S (2022). Probing RNA Structure with Dimethyl Sulfate Mutational Profiling with Sequencing In Vitro and in Cells. JoVE, 64820. 10.3791/64820. [DOI] [PubMed] [Google Scholar]
- 77.BBMap (2022). SourceForge. https://sourceforge.net/projects/bbmap/. [Google Scholar]
- 78.Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 17, 10. 10.14806/ej.l7.1.200. [DOI] [Google Scholar]
- 79.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience 10, giab008. 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 82.Li H (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. 10.1093/bioinformatics/btyl91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu Y, Fang L, Chen X, Zhong JF, Li M, and Wang K (2021). LIQA: long-read isoform quantification and analysis. Genome Biol 22, 182. 10.1186/sl3059-021-02399-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garrido-Martin D, Palumbo E, Guigo R, and Breschi A (2018). ggsashimi: Sashimi plot revised for browser- and annotation-independent splicing visualization. PLoS Comput. Biol 14, el006360. 10.1371/journal.pcbi.1006360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3. List of oligonucleotides, related to STAR Methods.
Table S4. List of constructs and full construct sequences, related to STAR Methods.
Table S1. Annotated tandem CAG repeat splice acceptors in hg38, related to Figure 1 and STAR Methods.
Data Availability Statement
RNA-seq data have been deposited at the NCBI Sequence Read Archive and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images and fluorescent micrographs were deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. This paper analyzes existing, publicly available data. These accession numbers for these datasets are listed in the key resources table.
All source code for the SATCfinder pipeline has been deposited at Zenodo and is publicly available as of the date of publication. Other bioinformatic pipelines used in this work are described in STAR Methods.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.