ABSTRACT
Streptococcus suis (S. suis) has been increasingly recognized as a porcine zoonotic pathogen that threatens the health of both pigs and humans. Multidrug-resistant Streptococcus suis is becoming increasingly prevalent, and novel strategies to treat bacterial infections caused by these organisms are desperately needed. In the present study, an untargeted metabolomics analysis showed that the significant decrease in methionine content and the methionine biosynthetic pathway were significantly affected by the Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis in drug-resistant S. suis. The addition of L-methionine restored the bactericidal activity of macrolides, doxycycline, and ciprofloxacin on S. suis in vivo and in vitro. Further studies showed that the exogenous addition of methionine affects methionine metabolism by reducing S-adenosylmethionine synthetase activity and the contents of S-adenosylmethionine, S-adenosyl homocysteine, and S-ribose homocysteine. Methionine can decrease the total methylation level and methylesterase activity in multidrug resistant S. suis. The drug transport proteins and efflux pump genes were significantly downregulated in S. suis by exogenous L-methionine. Moreover, the exogenous addition of methionine can reduce the survival of S. suis by affecting oxidative stress and metal starvation in bacteria. Thus, L-methionine may influence the development of resistance in S. suis through methyl metabolism and metal starvation. This study provides a new perspective on the mitigation of drug resistance in S. suis.
IMPORTANCE
Bacterial antibiotic resistance has become a severe threat to human and animal health. Increasing the efficacy of existing antibiotics is a promising strategy against antibiotic resistance. Here, we report that L-methionine enhances the efficacy of macrolides, doxycycline, and ciprofloxacin antibiotics in killing Streptococcus suis, including multidrug-resistant pathogens. We investigated the mechanism of action of exogenous methionine supplementation in restoring macrolides in Streptococcus suis and the role of the methionine cycle pathway on methylation levels, efflux pump genes, oxidative stress, and metal starvation in Streptococcus suis. It provides a theoretical basis for the rational use of macrolides in clinical practice and also identifies a possible target for restoring drug resistance in Streptococcus suis.
KEYWORDS: Streptococcus suis, methionine, methyl metabolism, macrolides, metal starvation
INTRODUCTION
Streptococcus suis (S. suis) is an important bacterial pathogen that can cause meningitis, septicemia, and even sudden death in humans and pigs (1), which has resulted in huge economic losses to the pig farming industry and threatened the public health and safety of the community (2). One of the main causes of persistent S. suis infections is the varying degrees of drug resistance of clinically isolated strains to drugs, including macrolides, fluoroquinolones, and tetracyclines (3–5). Under the pressure of long-term selection of antimicrobial drugs, bacteria become resistant through adaptive resistance, genetic mutation, heterogeneous resistance, and many other ways, resulting in the clinical failure of antimicrobial drug therapy (6). However, the spread of drug-resistant bacteria and the failure to treat some bacterial diseases cannot be fully explained. As the mechanisms of bacterial drug resistance have been investigated, the metabolism of bacteria and the products in their metabolic pathways have been found to be directly related to their drug resistance (7). When bacteria are exposed to drugs, their protein networks are altered, resulting in the corresponding metabolic pathways being altered, which makes the bacteria less susceptible to antimicrobial drugs and promotes the production of drug-resistant and persistent bacteria (8). Moreover, bacterial metabolism and the transmission of signaling molecules are also related to their drug resistance (9).
Amino acids are used as an essential part of protein composition and are the physiological core of all living cells. Amino acids are substrates involved in protein biosynthesis and are also used as an important source of carbon and nitrogen in physiological metabolism (10). Moreover, amino acids are also involved in bacterial cell wall biosynthesis and maintenance of bacterial homeostasis (11). Recent studies have shown that bacteria can maintain their own survival and resist various exogenous stimuli by regulating the dynamic balance of amino acids (12). Among them, sugar metabolism, amino acid metabolism, and components of metabolic pathways being altered can affect the susceptibility of bacteria to drugs (13). Metabolites such as glycine were added exogenously to significantly increase the susceptibility of various bacteria (Escherichia coli, Klebsiella pneumoniae, Staphylococcus aureus, and Pseudomonas aeruginosa) to ampicillin and kanamycin by regulating purine metabolism, TCA cycle, and glycine, serine and threonine metabolic pathways (14).
The methionine (MET) biosynthetic pathway, carried by almost all prokaryotes, has been studied as an adjuvant for a variety of broad-spectrum antibiotics (15). MET is essential for the biosynthesis of bacterial proteins and is required for the initiation and extension phases of translation (16). Meanwhile, MET is a precursor of the important methyl donor S-adenosylmethionine (SAM), which is involved in the activated methyl cycle and is required for DNA methylation, protein methylation, and polyamine biosynthesis (17). In the methyl cycle, SAM is the main methyl donor and forms S-adenosyl homocysteine (SAH) catalyzed by S-adenosylmethionine synthetase (SAMs). Then, SAH is converted into homocysteine (HCY) through a two-step process (18). It has been shown that after the gene expressing DNA cytosine methyltransferase was knocked out in E. coli, the drug tolerance transporter protein showed an overexpressed state and was tolerant to ethidium bromide (19). However, back-complementation of the gene restored the susceptibility of E. coli to ethidium bromide (19), suggesting that altering the methyl cycle affects methylation and has a modulating effect on bacterial drug resistance. However, there are few studies on whether S. suis resistance is associated with changes in physiological metabolism. A recent study by Li et al. investigated how S. suis serine/threonine kinases can affect the virulence and drug resistance of S. suis (20). We found that L-serine can reduce the survival rate of resistant strains of S. suis by increasing the production of reactive oxygen species (ROS) (21). In addition, gallic acid can restore the sulfonamide sensitivity of multidrug-resistant S. suis by regulating the expression of dihydrofolate reductase (22).
At the same time, the oxidative stress and metal starvation encountered by S. suis during the infection process cannot be ignored. The regulation of oxidative stress and metal starvation responses by ferric uptake regulator (Fur)-like proteins is also important for the survival of S. suis in vivo (23). We investigated whether exogenous metabolites have the potential to restore the susceptibility of S. suis to antimicrobial drugs by altering its physiological metabolic profile. Here, we describe in detail the metabolic changes in S. suis following antibiotic resistance and demonstrate that L-methionine (L-met) affects the methyl cycle of S. suis and, in the presence of metabolites, inhibits methyl metabolism and affects perR-regulated oxidative stress, restoring bacterial susceptibility to antibacterial drugs.
RESULTS
Analysis of Streptococcus suis resequencing data
We conducted an analysis of the resequencing data for S. suis strains T-I-8, T-I-64, and T-I-256. In the case of S. suis T-I-8, we identified only one single nucleotide polymorphism (SNP). However, this SNP was located downstream/upstream and appeared to have little functional significance. In the strain S. suis T-I-64, seven SNPs were identified, all of which are nonsynonymous and are located in the coding sequence (CDS) region. We annotated seven candidate genes that play important roles in various biological processes, including ribosomal protein synthesis, fatty acid biosynthesis, magnesium ion transport, ATP binding, antibiotic resistance, and oxidative stress (Table 1). The results showed that nine high-quality SNPs based on whole-genome resequencing were discovered in S. suis T-I-256 (seven nonsynonymous, one stop-gain, and one noncoding), among which eight SNPs were located in the CDS region. Out of the nine SNPs, only three were distinct from those observed in S. suis T-I-64. Two of these SNPs were associated with aquaporin family proteins and ABC transporter ATP-binding proteins, while one had no apparent function. Furthermore, we identified 50 indels (insertions or deletions) in the genome of S. suis T-I-8 when compared to the reference genome (NC_012925.1). Remarkably, these indels were consistent with those found in the genomes of S. suis T-I-64 and T-I-256. In the case of S. suis T-I-8, 24 of these indels were located within the CDS region, spanning across 20 different genes. These indels were associated with various biological processes, including metabolism, membrane protein synthesis, transposons, protein export, and cell wall surface anchor family proteins. However, 52 indels were found in S. suis T-I-64 and T-I-256 genomes. In addition to the same indels as S. suis T-I-8, these two strains displayed two unique indels within the CDS region, primarily linked to the purine salvage pathway and the cysteine pathway (Table 2).
TABLE 1.
| #CHROM | POS | POS | REF | T-I-256 | T-I-64 | T-I-8 | Region | Type | Gene | Function |
|---|---|---|---|---|---|---|---|---|---|---|
| NC_012925.1 | 73309 | 73309 | C | C | T | C | CDS | Nonsynonymous | gene-SSU_RS00435 | 50S ribosomal protein L4 |
| NC_012925.1 | 255014 | 255014 | A | C | A | A | CDS | Nonsynonymous | gene-SSU_RS01380 | MIP/aquaporin family protein |
| NC_012925.1 | 697705 | 697705 | T | T | T | C | Downstream and upstream | – | cds-WP_002935444.1-cds-WP_011922251.1 | – |
| NC_012925.1 | 1123232 | 1123232 | C | A | A | C | CDS | Nonsynonymous | gene-SSU_RS05600 | 50S ribosomal protein L20 |
| NC_012925.1 | 1124391 | 1124391 | T | C | T | T | Upstream and downstream | – | cds-WP_032497583.1-cds-WP_002937055.1 | – |
| NC_012925.1 | 1221394 | 1221394 | G | A | A | G | CDS | Nonsynonymous | gene-SSU_RS06070 | Glycosyltransferase family 4 protein |
| NC_012925.1 | 1614291 | 1614291 | A | C | C | A | CDS | Nonsynonymous | gene-SSU_RS08100 | Acyl carrier protein |
| NC_012925.1 | 1640610 | 1640610 | A | C | C | A | CDS | Nonsynonymous | gene-SSU_RS08200 | Magnesium transporter CorA family protein |
| NC_012925.1 | 1849055 | 1849055 | G | C | G | G | CDS | Stopgain | gene-SSU_RS09190 | ABC transporter ATP-binding protein |
| NC_012925.1 | 1851356 | 1851356 | C | A | A | C | CDS | Nonsynonymous | gene-SSU_RS09195 | ABC transporter ATP-binding protein |
| NC_012925.1 | 1852443 | 1852443 | T | G | G | T | CDS | Nonsynonymous | gene-SSU_RS09200 | MarR family transcriptional regulator |
–, not applicable.
CHROM, chromosome; POS, position; REF, reference strain.
TABLE 2.
A summary of 52 indels in S. suis
| #CHROM | POS | POS | REF | Type | T-I-256 | T-I-64 | T-I-8 | Region | Type | Gene | Function |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NC_012925.1 | 13100 | 13100 | – | Insertion | 1/1 | 1/1 | 0/0 | CDS | Nonframeshift | gene-SSU_RS00070 | Hypoxanthine phosphoribosyltransferase |
| NC_012925.1 | 71772 | 71772 | A | Deletion | 1/1 | 1/1 | 1/1 | Downstream and upstream | – | cds-WP_014635810.1-cds-WP_011922546.1 | – |
| NC_012925.1 | 87409 | 87415 | TGCCAAA | Deletion | 1/1 | 1/1 | 1/1 | Downstream | – | cds-WP_012774898.1 | – |
| NC_012925.1 | 87410 | 87413 | GCCA | Deletion | 1/1 | 1/1 | 1/1 | Downstream | – | cds-WP_012774898.1 | – |
| NC_012925.1 | 87420 | 87420 | A | Deletion | 1/1 | 1/1 | 1/1 | Downstream | – | cds-WP_012774898.1 | – |
| NC_012925.1 | 126107 | 126107 | A | Deletion | 1/1 | 1/1 | 1/1 | Downstream and upstream | – | cds-WP_012775422.1-cds-WP_012774908.1 | – |
| NC_012925.1 | 127988 | 127989 | AA | Deletion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS00830 | Folylpolyglutamate synthase/dihydrofolate synthase family protein |
| NC_012925.1 | 129155 | 129164 | AAAATATAGA | Deletion | 1/1 | 1/1 | 1/1 | Upstream | – | cds-WP_011921730.1-cds-WP_012774909.1 | – |
| NC_012925.1 | 146069 | 146069 | A | Deletion | 1/1 | 1/1 | 1/1 | Downstream and upstream | – | cds-WP_011921748.1-cds-WP_011921749.1 | – |
| NC_012925.1 | 163461 | 163469 | CTTCTGAGC | Deletion | 1/1 | 1/1 | 1/1 | CDS | Nonframeshift | gene-SSU_RS00995 | YSIRK-type signal peptide-containing protein |
| NC_012925.1 | 234833 | 234833 | A | Deletion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS01275 | DUF1430 domain-containing protein |
| NC_012925.1 | 259413 | 259424 | GCAAACACCACA | Deletion | 1/1 | 1/1 | 1/1 | CDS | Nonframeshift | gene-SSU_RS01400 | LPXTG cell wall anchor domain-containing protein |
| NC_012925.1 | 332301 | 332301 | T | Deletion | 1/1 | 1/1 | 1/1 | Upstream | – | cds-WP_012774967.1 | – |
| NC_012925.1 | 357506 | 357506 | T | Deletion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS01860 | Cysteine hydrolase family protein |
| NC_012925.1 | 396871 | 396871 | T | Deletion | 1/1 | 1/1 | 1/1 | Downstream and upstream | – | cds-WP_004194171.1-cds-WP_004194167.1 | – |
| NC_012925.1 | 396912 | 396913 | GA | Deletion | 1/1 | 1/1 | 1/1 | Downstream and upstream | – | cds-WP_004194171.1-cds-WP_004194167.1 | – |
| NC_012925.1 | 427004 | 427004 | – | Insertion | 1/1 | 1/1 | 1/1 | Downstream | – | cds-SSU_RS10460 | – |
| NC_012925.1 | 853630 | 853630 | – | Insertion | 1/1 | 1/1 | 1/1 | Downstream and upstream | – | cds-WP_011922397.1-cds-SSU_RS10100 | – |
| NC_012925.1 | 853881 | 853881 | – | Insertion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS10100 | Abi family protein |
| NC_012925.1 | 853947 | 853947 | – | Insertion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS10100 | Abi family protein |
| NC_012925.1 | 879988 | 879993 | CTAGCT | Deletion | 1/1 | 1/1 | 1/1 | CDS | Nonframeshift | gene-SSU_RS04360 | FAD-dependent oxidoreductase |
| NC_012925.1 | 910335 | 910337 | CCC | Deletion | 1/1 | 1/1 | 1/1 | CDS | Nonframeshift | gene-SSU_RS04490 | ZmpA/ZmpB/ZmpC family metallo-endopeptidase |
| NC_012925.1 | 967801 | 967801 | T | Deletion | 1/1 | 1/1 | 1/1 | Upstream and downstream | – | cds-WP_012775157.1-cds-WP_012027071.1 | – |
| NC_012925.1 | 1073309 | 1073309 | T | Deletion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS10585 | Hyaluronate lyase |
| NC_012925.1 | 1073790 | 1073791 | AT | Deletion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS10585 | Hyaluronate lyase |
| NC_012925.1 | 1084376 | 1084381 | CAAAAT | Deletion | 1/1 | 1/1 | 1/1 | CDS | Nonframeshift | gene-SSU_RS05405 | IS110 family transposase |
| NC_012925.1 | 1210965 | 1210968 | GTGT | Deletion | 1/1 | 1/1 | 1/1 | Upstream and downstream | – | cds-WP_012027299.1-cds-WP_012027300.1 | – |
| NC_012925.1 | 1220207 | 1220207 | A | Deletion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS06065 | Hypothetical protein |
| NC_012925.1 | 1246570 | 1246571 | CA | Deletion | 1/1 | 1/1 | 1/1 | Upstream | – | cds-WP_012775226.1-cds-WP_012775227.1 | – |
| NC_012925.1 | 1246634 | 1246634 | – | Insertion | 1/1 | 1/1 | 1/1 | Upstream | – | cds-WP_012775226.1-cds-WP_012775227.1 | – |
| NC_012925.1 | 1246643 | 1246644 | AG | Deletion | 1/1 | 1/1 | 1/1 | Upstream | – | cds-WP_012775226.1-cds-WP_012775227.1 | – |
| NC_012925.1 | 1246648 | 1246648 | – | Insertion | 1/1 | 1/1 | 1/1 | Upstream | – | cds-WP_012775226.1-cds-WP_012775227.1 | – |
| NC_012925.1 | 1246650 | 1246654 | TAGAA | Deletion | 1/1 | 1/1 | 1/1 | Upstream | – | cds-WP_012775226.1-cds-WP_012775227.1 | – |
| NC_012925.1 | 1251294 | 1251294 | T | Deletion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS06175 | Preprotein translocase subunit SecG |
| NC_012925.1 | 1252772 | 1252772 | – | Insertion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS06190 | Dephospho-CoA kinase |
| NC_012925.1 | 1252989 | 1252989 | – | Insertion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS06190 | Dephospho-CoA kinase |
| NC_012925.1 | 1252992 | 1252992 | T | Deletion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS06190 | Dephospho-CoA kinase |
| NC_012925.1 | 1262328 | 1262328 | – | Insertion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS06235 | HAD family hydrolase |
| NC_012925.1 | 1353156 | 1353156 | – | Insertion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS10185 | Hypothetical protein |
| NC_012925.1 | 1356965 | 1356965 | G | Deletion | 1/1 | 1/1 | 1/1 | Upstream and downstream | – | cds-WP_162286914.1-cds-WP_012775258.1 | – |
| NC_012925.1 | 1356972 | 1356972 | – | Insertion | 1/1 | 1/1 | 1/1 | Upstream and downstream | – | cds-WP_162286914.1-cds-WP_012775258.1 | – |
| NC_012925.1 | 1394635 | 1394635 | – | Insertion | 1/1 | 1/1 | 1/1 | Upstream and downstream | – | cds-WP_012027489.1-cds-WP_012027490.1 | – |
| NC_012925.1 | 1396137 | 1396138 | TG | Deletion | 1/1 | 1/1 | 1/1 | Upstream and downstream | – | cds-WP_012027490.1-cds-WP_002937312.1 | – |
| NC_012925.1 | 1396144 | 1396144 | – | Insertion | 1/1 | 1/1 | 1/1 | Upstream and downstream | – | cds-WP_012027490.1-cds-WP_002937312.1 | – |
| NC_012925.1 | 1470643 | 1470643 | – | Insertion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS07350 | IS110 family transposase |
| NC_012925.1 | 1492597 | 1492597 | T | Deletion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS07455 | YSIRK-type signal peptide-containing protein |
| NC_012925.1 | 1534545 | 1534547 | AAT | Deletion | 1/1 | 1/1 | 0/0 | CDS | Nonframeshift | gene-SSU_RS07705 | Endolytic transglycosylase MltG |
| NC_012925.1 | 1602665 | 1602667 | CTT | Deletion | 1/1 | 1/1 | 1/1 | CDS | Nonframeshift | gene-SSU_RS08025 | Restriction endonuclease subunit S |
| NC_012925.1 | 1677032 | 1677032 | – | Insertion | 1/1 | 1/1 | 1/1 | Upstream and downstream | – | cds-WP_002942324.1-cds-WP_012775331.1 | – |
| NC_012925.1 | 1811025 | 1811025 | – | Insertion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS08975 | 3′−5′ exoribonuclease YhaM family protein |
| NC_012925.1 | 1916456 | 1916456 | A | Deletion | 1/1 | 1/1 | 1/1 | CDS | Frameshift | gene-SSU_RS10595 | Pilin N-terminal domain-containing protein |
| NC_012925.1 | 1958662 | 1958662 | – | Insertion | 1/1 | 1/1 | 1/1 | Upstream and downstream | – | cds-WP_012028014.1-cds-WP_012028015.1 | – |
Non-targeted high performance liquid chromatography-MS/MS metabolic profile analysis
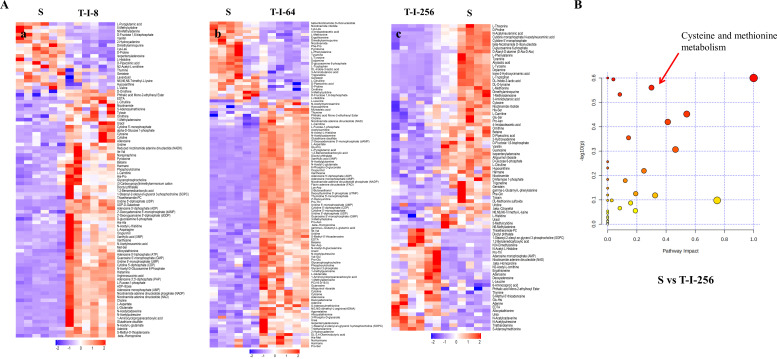
The metabolomic profiles of S and T-I-8, S and T-I-64, and S and T-I-256 were analyzed by liquid chromatography-mass spectrometry. The analytical performance was evaluated by injecting multiple quality control (QC) samples in between the biological samples, and the correlation coefficient of the QCs during the entire MS run was calculated as 0.98, which suggested that our analytical method is robust and reproducible. Each tested bacteria group also contained six biological replicates to ensure biological reproducibility. Unsupervised principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) were used to evaluate the three groups of strains, and the results are shown in Table 3. The scores of R2Y and Q2 were both greater than 0.5, which proved that the model was established reliably and had good stability. To further decipher the metabolic differences between the three comparison groups, a combined comparison of metabolite t-test P-value and fold change (FC) of control samples compared to T-I-8, T-I-64, and T-I-256 samples was conducted, respectively. Differential metabolites were obtained by t-test (P < 0.05), fold change (FC > 1.5 or FC < 0.667), and model variable weight (variable influence of projection, VIP > 1) threshold screening. S and T-I-8, S and T-I-64, and S and T-I-256 groups were detected with 89, 113, and 84 differentially expressed metabolites, respectively (Fig. 1A; Tables S2 to S4). The pictures of differential metabolites are given in detail in the supplemental material (Fig. S1 to S3). Combining the results of the analysis of the three groups, it was found that the FC score of the differential metabolite L-met showed a regular decrease and was the lowest value in the S and T-I-256 groups (Table 4). Meanwhile, as shown in Fig. 1B; Table 5, we found that the metabolism of cysteine and methionine was significantly affected by the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis in the S and T-I-256 groups. These results prompted us to explore whether the phenotype of S. suis changes in the presence of L-met.
TABLE 3.
Statistical analysis of the metabolomics of the different porcine S. suis in this studya
| Group | PCA | OPLS-DA | |
|---|---|---|---|
| R2X (cum) | R2Y (cum) | Q2 (cum) | |
| T-I-8 vs S | 0.622 | 0.992 | 0.87 |
| T-I-64 vs S | 0.536 | 0.993 | 0.877 |
| T-I-256 vs S | 0.575 | 0.993 | 0.883 |
R2Y means that the square of the percentage of original data is retained in the x-axis direction; Q2 represents the prediction rate of the model.
Fig 1.
Bioinformatics analysis of the metabolomics of S. suis. (A) Overview metabolic profile heat map shows the comparison of control group S with different resistant strains in this study. The six biological replicates of the control group and the six biological replicates of the three resistant strain group are plotted on this heat map. (a) S vs T-I-8 group, (b) S vs T-I-64 group, and (c) S vs T-I-256 group. (B) Results of enrichment analysis of the KEGG pathway in the S and T-I-256 groups. The red arrows mark the cysteine and methionine metabolic pathways.
TABLE 4.
Changes in the metabolism of L-met in S. suis at different levels of drug resistance
| Group | VIP | Fold change | P-value |
|---|---|---|---|
| T-I-256 vs S | 1.789024472 | 0.478270847 | 0.000384802 |
| T-I-64 vs S | 1.591749366 | 0.473527031 | 0.004299033 |
| T-I-8 vs S | 0.52119909 | 0.990764742 | 0.969032166 |
TABLE 5.
Results from pathway analysisa
| KEGG | Total | Expected | Hits | Raw P | −log10(P) | Holm adjust | FDR | Impact | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Valine, leucine, and isoleucine biosynthesis | 6 | 0.97 | 2 | 0.251 | 0.6 | 1 | 1 | 0 |
| 2 | Inositol phosphate metabolism | 6 | 0.97 | 2 | 0.251 | 0.6 | 1 | 1 | 1 |
| 3 | Purine metabolism | 49 | 7.93 | 10 | 0.254 | 0.594 | 1 | 1 | 0.04 |
| 4 | Cysteine and methionine metabolism | 22 | 3.56 | 5 | 0.275 | 0.561 | 1 | 1 | 0.3 |
| 5 | Aminoacyl-tRNA biosynthesis | 45 | 7.28 | 9 | 0.293 | 0.533 | 1 | 1 | 0.08 |
| 6 | Glycerophospholipid metabolism | 13 | 2.1 | 3 | 0.353 | 0.452 | 1 | 1 | 0.54 |
| 7 | Arginine biosynthesis | 8 | 1.29 | 2 | 0.381 | 0.42 | 1 | 1 | 0.41 |
| 8 | Streptomycin biosynthesis | 9 | 1.46 | 2 | 0.442 | 0.355 | 1 | 1 | 0.14 |
| 9 | Methane metabolism | 16 | 2.59 | 3 | 0.494 | 0.307 | 1 | 1 | 0.46 |
| 10 | Valine, leucine, and isoleucine degradation | 11 | 1.78 | 2 | 0.554 | 0.257 | 1 | 1 | 0 |
Total is the total number of compounds in the pathway. Hits refer to the actually matched number from the user-uploaded data. Raw P is the original P value calculated from the enrichment analysis. Holm P is the P value adjusted by Holm-Bonferroni method. FDR P is the P value adjusted using false discovery rate. Impact is the pathway impact value calculated from pathway topology analysis.
Therapeutic effect of L-methionine combined with tylosin in vivo
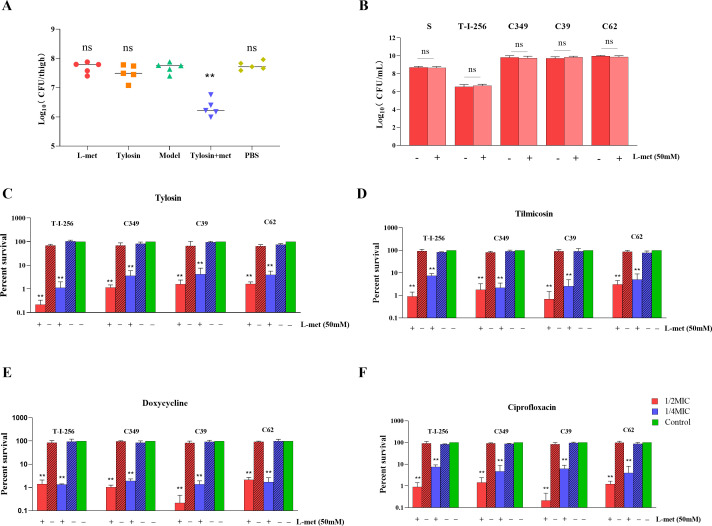
A mouse thigh infection model was established to evaluate the synergetic effect of L-met and tylosin against drug-resistant strains of S. suis in vivo. The change in viable bacterial counts in the mouse thighs was determined by colony counting. The results showed that after the combined treatment with L-met and tylosin, the number of bacteria in the mouse thigh infection model was significantly reduced compared to that seen after treatment with either tylosin or L-met alone (Fig. 2A).
Fig 2.
Effect of L-met addition on the survival of S. suis in in vitro culture and on the treatment of S. suis after host infection in vivo. (A) Therapeutic effects of tylosin and L-met alone and in combination in neutrophilic mice infected with drug-resistant strains. The bacterial load of infected thigh muscle in neutropenic mice (n = 5 per group) was determined by colony counting. Model: the mice with persistent 24 h infection after intramuscular injection of drug-resistant bacteria into the thigh. (B) Effect of adding 50 mM L-met on the survival of S. suis. (C–F) Synergistic effect of 50 mM L-met in combination with tylosin (C), tilmicosin (D), doxycycline (E), and ciprofloxacin. (F) Bacterial survival. Red, antibacterial drug concentration of 1/2 minimum inhibitory concentration (MIC) for strains; blue, antimicrobial drug concentration of 1/4 MIC for strains; green, control group without drugs. All data are the mean ± s.d. P values were determined using an unpaired, two-tailed Student’s t-test. ns, P > 0.05; *P < 0.05; and **P < 0.01.
L-methionine increased the susceptibility of planktonic S. suis cells to different drugs
To verify whether L-met was associated with resistance in S. suis, we used resistant strain T-I-256 obtained by laboratory induction and resistant strain C349, C39, and C62 obtained by clinical isolation, which were resistant to a variety of antimicrobial drugs and showed several times higher MIC (minimum inhibitory concentration) to tylosin, tilmicosin, doxycycline, and ciprofloxacin compared to S. suis S strain (Table 6). We also investigated the toxic effects of L-met on S. suis, and the results showed that the addition of 50 mM of L-met had no significant effect on the growth of either S. suis (Fig. 2B). We observed a significant decrease in bacterial survival after treatment with 1/2 MIC or 1/4 MIC drug levels of tylosin, tilmicosin, doxycycline, and ciprofloxacin in the presence of L-met (Fig. 2C through F). In particular, the combination of an antibacterial drug with 1/2 MIC and L-met achieved a bactericidal effect of more than 90% against resistant strains of S. suis, much higher than the bactericidal effect of the drug acting on S. suis. The time-kill growth curves also showed that the combination of 50 mM L-met with 1/2 MIC or 1/4 MIC concentration of tylosin can significantly inhibit the growth of bacteria (Fig. S4). These data suggest that L-met can enhance the susceptibility of resistant strains of S. suis to antimicrobial drugs.
TABLE 6.
MIC (mg/L) for S. suis in this study
| Strains | Tylosin | Tilmicosin | Ciprofloxacin | Doxycycline |
|---|---|---|---|---|
| S. suis S | 0.125 | 0.25 | 0.0625 | 0.0625 |
| S. suis T-I-256 | 256 | 16 | 2 | 4 |
| S. suis C349 | 640 | 640 | 8 | 32 |
| S. suis C39 | 640 | 640 | 16 | 32 |
| S. suis C62 | 640 | 320 | 8 | 8 |
L-methionine can affect methionine cycle metabolites
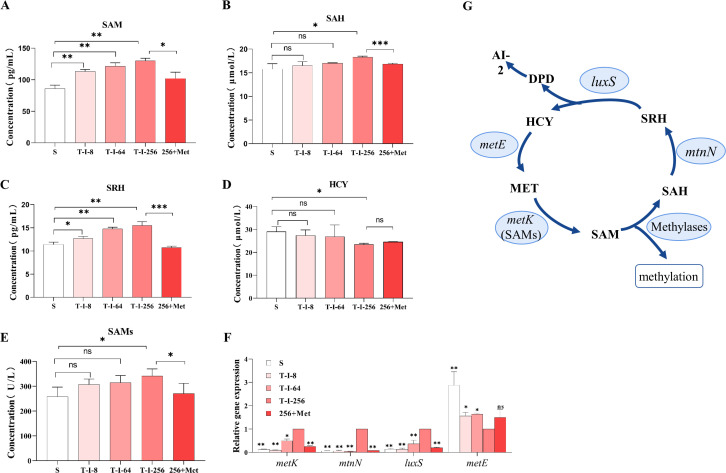
To understand how the methyl cycle is activated by L-met, we further assessed metabolite content, enzyme activity, and gene expression of the MET metabolic pathway. The concentrations of SAM, SAH, and S-ribose homocysteine (SRH) in S. suis were found to increase with the rising level of S. suis resistance, and HCY was found to decrease, and all the tested metabolic contents in the high-level resistant strain T-I-256 changed significantly compared with the sensitive strain S. suis (Fig. 3A through D). A significant increase in the metabolic contents of SAM, SAH, and SRH occurred after the T-I-256 strain was cultured in the medium that was spiked with L-met. SAMs are key enzymes that regulate the production of SAM from methionine. The results showed that the enzymatic activity of SAMs was upregulated with the increase of bacterial resistance (Fig. 3E). However, only the T-I-256 group was significantly different from the sensitive strain S. suis. Moreover, the activity of SAMs was significantly reduced when L-met was added exogenously to the T-I-256 strain. We analyzed the gene expression levels of metK, mtnN, luxS, and metE in the MET metabolic pathway, and the results showed that the gene expression levels of metK, mtnN, and luxS were significantly upregulated with increasing S. suis resistance, while metE was significantly downregulated in T-I-256 strains (Fig. 3F). When L-met was added exogenously to T-I-256, significant downregulation of metK, mtnN, and luxS expression occurred (Fig. 3F). This suggests that exogenous L-met can affect the MET metabolic pathway (Fig. 3G).
Fig 3.
Metabolic pathway analysis. (A–D) SAM (A), SAH (B), SRH (C), and HCY (D) content of the MET metabolic pathway in S, T-I-8, T-I-64, T-I-256, and Met+256 strains. (E) SAMs activity of the MET metabolic pathway in S, T-I-8, T-I-64, T-I-256, and Met+256 strains. (F) The gene levels of the MET metabolic pathways in S, T-I-8, T-I-64, T-I-256, and Met+256 strains were examined by RT-PCR. All data are the mean ± s.d. P values were determined using an unpaired, two-tailed Student’s t-test. ns, P > 0.05; *P < 0.05; and **P < 0.01. (G) Schematic diagram of the MET metabolic pathway.
L-methionine can affect methylation metabolism
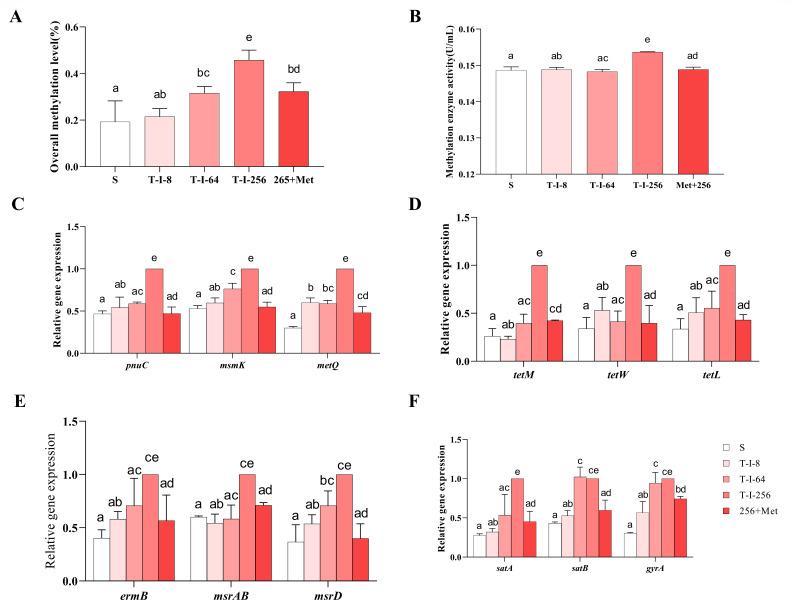
MET is a synthetic precursor of the important methyl donor SAM, while SAM and SAH are important metabolites that affect the methyl cycle. The effect of L-met on SAM and SAH has been demonstrated. To further investigate the effect of L-met on the methyl cycle, we measured total methylation levels and methylation enzymes. The results showed that the total methylation level and methylation enzyme activity of S. suis increased significantly with the increase of drug resistance level (Fig. 4A and B). When L-met was added to T-I-256, the total methylation level and methylesterase activity were significantly decreased.
Fig 4.
Effect of L-met addition on the methyl cycle of S. suis. (A) Total methylation levels of S, T-I-8, T-I-64, T-I-256, and Met+256 strains. (B) Methylesterase activity in S, T-I-8, T-I-64, T-I-256, and Met+256 strains. (C) Gene expression levels of regulatory membrane transporter proteins in S, T-I-8, T-I-64, T-I-256, and Met+256 strains. (D–F) Expression levels of tetracycline (D), macrolide (E), and fluoroquinolone (D) efflux pump genes in S, T-I-8, T-I-64, T-I-256, and Met+256 strains. The data are presented as means ± SEM. The significance of the differences was determined by one-way ANOVA. Bars that do not share the same letters are significantly different (P < 0.05) from each other.
Effect of L-methionine addition on drug transport
To determine whether the drug transport channels induced by the methyl cycle are affected by L-met, we analyzed the expression levels of genes regulating drug transport proteins and efflux pump genes in S. suis by RT-PCR. The expression of pnuC (nicotinamide riboside transporter) (24, 25), msmK (multiple sugar metabolism protein K, ABC transporter) (26, 27), and metQ (L-methionine/D-methionine ABC transporter membrane-anchored binding protein) (28, 29) genes, which regulate drug transport proteins, were significantly upregulated in strain T-I-256 compared with strain S (Fig. 4C). When L-met was exogenously added to strain T-I-256, the expression of pnuC, msmK, and metQ genes was significantly downregulated (Fig. 4C). We also examined the efflux pump genes of different drugs in S. suis. The results showed that the efflux pump genes tetM, tetW, and tetL of tetracyclines (Fig. 4D), ermB, msrAB, and msrD of macrolides (Fig. 4E), and satA, satB, and gyrA of fluoroquinolones (Fig. 4F) were significantly upregulated in strain T-I-256 compared to strain S. Moreover, the exogenous addition of L-met to the T-I-256 strain significantly suppressed the efflux pump genes.
Effect of L-methionine addition on oxidative stress
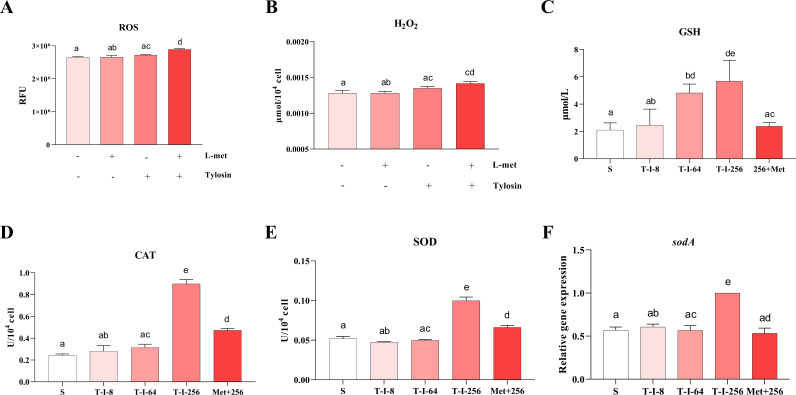
To determine the effects of oxidative stress caused by altered methyl cycle, we measured ROS levels, glutathione (GSH) content, oxidative stress enzyme activity, and sodA gene (superoxide dismutase SodA) expression (30, 31). ROS levels were compared after the addition of macrolides alone or in combination with L-met, using DCFH-DA. The results showed that there was a significant increase in ROS levels within S. suis after the combination of tylosin and L-met (Fig. 5A). It was also found that the H2O2 level in S. suis increased significantly after the combination of tylosin and L-met (Fig. 5B). The level of GSH was also found to increase significantly with increasing levels of S. suis resistance (Fig. 5C). In contrast, the exogenous addition of L-met to strain T-I-256 significantly reduced the level of GSH in the altered strain (Fig. 5C). Also, the scavenging of ROS by superoxide dismutase (SOD) and catalase (CAT) was significantly enhanced in strain T-I-256 (Fig. 5D and E). In contrast, both CAT and SOD activities and sodA gene were significantly decreased when supplemented with L-met (Fig. 5D through F).
Fig 5.
Effect of L-met addition on the level of oxidative stress in S. suis. (A and B) Effect of 50 mM L-met in combination with tylosin on ROS (A) and H2O2 (B) concentration in T-I-256 strains of S. suis. (C) GSH content in S, T-I-8, T-I-64, T-I-256, and Met+256 strains. (D and E) CAT (D) and SOD (E) activities in S, T-I-8, T-I-64, T-I-256, and Met+256 strains. (F) Expression levels of sodA genes in S, T-I-8, T-I-64, T-I-256, and Met+256 strains. The data are presented as means ± SEM. The significance of the differences was determined by one-way ANOVA. Bars that do not share the same letters are significantly different (P < 0.05) from each other.
L-methionine affects metal starvation in S. suis
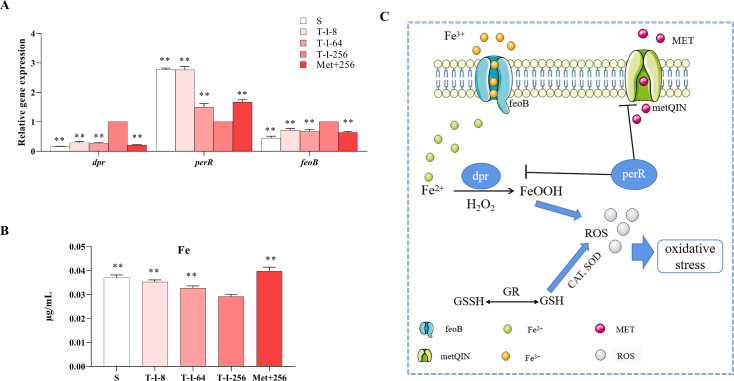
To understand whether the altered survival of S. suis by L-met is related to the modulation of the metal starvation response by fur-like proteins, the fur-like protein regulating gene perR was determined to be significantly downregulated in strain T-I-256, which reduced the repressive effect of perR on dpr (dps-like peroxide resistance protein) and caused a significant upregulation of gene expression of dpr (Fig. 6A). Gene expression of dpr significantly decreased and perR significantly increased after the exogenous addition of L-met in strain T-I-256 (Fig. 6A). The repression of dpr gene contributed to the Fenton response. Therefore, we determined the expression of the feoB gene (ferrous iron transport protein B), which regulates the iron transport protein, and the concentration of iron (32, 33). The results showed that the expression of feoB gene in T-I-256 strain was significantly upregulated, which promoted the efflux of iron and reduced the concentration of iron in the bacteria (Fig. 6A). Moreover, the expression of feoB gene was suppressed after the exogenous addition of L-met in strain T-I-256, which contributed to the accumulation of iron ions and promoted the Fenton reaction (Fig. 6A and B). The above results suggest that the effect of L-met addition on the survival of S. suis may be related to metal starvation (Fig. 6C).
Fig 6.
Effect of L-met addition on iron metal starvation in S. suis. (A) Expression levels of iron metal starvation-related genes in S, T-I-8, T-I-64, T-I-256, and Met+256 strains. (B) S, T-I-8, T-I-64, T-I-256, and Met+256 strains for the concentration of iron. The data are presented as means ± SEM. The significance of the differences was determined by one-way ANOVA. Bars that do not share the same letters are significantly different (P < 0.05) from each other. (C) Mechanism of action of MET in porcine S. suis on the effects of iron metal starvation and oxidative stress.
DISCUSSION
S. suis causes systemic infectivity in pigs and is one of the main causes of morbidity and mortality in piglets (34). However, the emergence of multidrug resistant strains has made the prevention, control, and treatment of bacterial diseases more difficult in clinical practice (35). Studies have shown that various endogenous metabolites, such as indole, NO, H2S, and amino acids, are associated with the development of bacterial drug resistance (36–38). These amino acids are the building blocks of proteins and precursors of functional molecules (39, 40). They are indispensable in certain developmental and physiological situations and are essential for normal physiology. Clinically, some essential amino acids are often used as supplements to reduce damage from pathogenic microorganisms and improve growth performance (41). The exogenous addition of citrulline and glutamine can also enhance the susceptibility of Salmonella to the drug in in vitro cultures by modulating the TCA cycle to cause enhanced proton drive, leading to the accumulation of apramycin in the bacteria (42). This demonstrates that the physiological metabolic state of bacteria is altered when they develop drug resistance and that the exogenous addition of metabolites, by modulating metabolic pathways, allows bacterial resistance to be affected.
We conducted a resequencing analysis of S. suis with different levels of resistance induced by tylosin (S. suis T-I-8, T-I-64, and T-I-256). The results indicated that the SNPs in S. suis T-I-64 and T-I-256 were highly identical, and the indels in all three strains were also highly consistent. These findings suggest that the genetic backgrounds of S. suis with varying tylosin resistance levels induced by tylosin are highly identical, especially in S. suis T-I-64 and T-I-256. Furthermore, mutations in genes related to tylosin resistance mechanisms were not observed. Therefore, metabolomics can be employed to further investigate the reasons behind the development of resistance. In order to investigate the mechanism of drug resistance development in S. suis and to find countermeasures, we performed metabolomic assays using S. suis with different levels of drug resistance separately from standard strains to analyze physiological metabolic differences. We found that the inhibition of L-met synthesis by strains T-I-8, T-I-64, and T-I-256 gradually increased, and the L-met content decreased significantly. MET has been reported to exert effects on metabolism, oxidative stress, and disease development. Restriction of MET prevents alterations in transmethylation metabolism, thereby reducing DNA damage and pathogenic processes (43). MET restriction stimulates glutathione production while decreasing oxidative stress (44). At the same time, when MET intake increases, the flux of substrates through the transmethylation pathway decreases and the flux through the transsulfuration pathway increases (45). This explains well the metabolic changes that occur after the development of drug resistance in S. suis. The emergence of drug resistance has led to the limitation of methionine, which implies that the methyl metabolism is disturbed and the resistance of GSH to oxidative stress is enhanced (46). Significant upregulation of gene expression of exocytosis pumps and membrane transport proteins was also detected in drug-resistant strains. This demonstrates that the development of drug resistance reduces the intracellular accumulation of the drug, resulting in weakening of the inhibitory effect of the drug.
Peng et al. (47) found that exogenous alanine or glucose restores the susceptibility of multidrug-resistant Edwardsiella tarda to killing by kanamycin, demonstrating an approach to killing multidrug-resistant bacteria. Therefore, we refer to the method of Peng et al. (47) to restore the metabolic disorders and drug resistance caused by MET restriction by the exogenous addition of MET assay. First, we determined the effect of exogenous L-met addition on the resistance of S. suis. In in vitro tests, the bactericidal efficacy of tylosin, tilmicosin, ciprofloxacin, and doxycycline against resistant strains was significantly enhanced by the addition of exogenous L-met. It was also found in in vivo experiments that the combination of L-met with tylosin significantly improved the therapeutic effect of tylosin in mice infected with S. suis. This demonstrates that drug-resistant strains can take up MET from the medium to recover from the effects of MET restriction.
Our experiments also showed that the addition of L-met led to a decrease in SAM concentration, which caused a disturbance in methyl metabolism and resulted in the suppression of total methylation levels. The metabolite content, enzyme activity, and gene expression were measured in resistant strains after the exogenous addition of L-met to elucidate the relationship between resistance development and MET metabolism in S. suis. The results showed that the concentrations of SAH, SAM, and SRH, the gene expression of metK, luxS, mtnN, and metE, and the activity of SAMs were significantly altered in the resistant strains after the addition of L-met. We, therefore, hypothesize that the decrease in methyl donors due to the reduction in SAM concentration restores the metabolic level of resistant bacteria to a level similar to that of sensitive strains. The inhibition of methylation levels and methylesterase activity also contributed to the recovery of susceptibility of the resistant strains (48). It has been reported that methylation of the methylesterase Cfr, which reduces the ability of antibacterial drugs to bind to bacteria, leads to enhanced resistance of bacteria to chloramphenicol, lincomycin, and some macrolides (49). Methylation of Erm methylesterase can also cause the development of bacterial resistance to macrolides and lincosamides, among others (50). All these research demonstrate the importance of the effect of altered methylation on bacterial drug resistance.
The recovery of susceptibility of S. suis after L-met addition may also be a result of methylation affecting the transmembrane transport of the drug. Methyltransferases can impair transmembrane transport-mediated drug resistance in Escherichia coli by affecting the expression of the sugE gene (19). In our study, we also found that the addition of L-met significantly downregulated the expression of the metQ gene, which regulates membrane transport in drug-resistant strains (28, 29), and the expression of efflux pump genes of several drugs in drug-resistant bacteria was significantly downregulated by the action of L-met, which implies that the addition of L-met may contribute to the accumulation of the drug.
In addition, the effect of MET on oxidative stress and PerR fur-like protein-regulated metal starvation during S. suis infection cannot be ignored (51). The perR gene was found to significantly reduce the susceptibility of S. suis to H2O2 after its deletion (52). We also found that the gene perR was significantly upregulated in drug-resistant strains after L-met addition, whereas the dpr gene was significantly downregulated in drug-resistant strains after L-met addition. The inhibition of dpr allows the Fenton reaction to occur normally (53). Thus, when L-met was added, the level of ROS under the effect of tylosin was significantly increased, contributing to ROS-induced oxidative stress, which allows toxic hydroxyl radicals to be generated (54). The gene sodA can encode protein superoxide dismutase SodA. The main function of SodA is scavenging ROS (30, 31). The detection of diminished SOD and CAT activities and downregulation of sodA gene expression also confirmed the suppression of protective effects against oxidative stress in drug-resistant bacteria (55).
Overall, these results suggest that the MET metabolic pathway is associated with the development of drug resistance in S. suis. L-met may influence the development of resistance in S. suis through methyl metabolism and metal starvation. This study provides a new perspective on the mitigation of drug resistance in S. suis.
MATERIALS AND METHODS
Strains and growth conditions
The S. suis ATCC 700794 used in this study was purchased from the American Type Culture Collection (ATCC) and kept in our laboratory. Unless otherwise stated, bacteria were cultured in Todd-Hewitt Broth (THB) at 37°C under aeration. S. suis strain ATCC 700494 with different levels of resistance to macrolides, named T-I-8 (MIC 8 mg/L), T-I-64 (MIC 64 mg/L), and T-I-256 (MIC 256 mg/L), was obtained using in vitro laboratory induction under long-term selection pressure of tylosin (56). S. suis ATCC 700794 was grown in 96-well plates with increased tylosin concentrations (1/8–4 MIC). The inoculated microplates were incubated at 37℃ for 24 h before being evaluated. The strains surviving in the highest drug concentration were transferred to new 96-well plates with increasing drug concentrations. The bacteria continued to pass through generations until strains with different tylosin resistance levels were induced. In addition, the stability of tylosin resistance was tested by serial passage (10 times) on an antibiotic-free medium. The standard strain ATCC 700494 was described by the abbreviation S. In addition, we also selected three clinically isolated strains of S. suis: C349, C39, and C62, which were used in this study. All experiments were performed using bacteria in the exponential phase. We used the minimum inhibitory concentration to assess the sensitivity to antibiotics.
Antibiotics and chemicals
Tylosin, tilmicosin, doxycycline, ciprofloxacin, and L-methionine (L-met) were purchased from Solarbio (Beijing, China). L-met was prepared with ddH2O amino acid stock solution (500 mM). After filtering the stock solution with a sterile hydrophilic PVDF with a membrane pore size of 0.22 µm (Biosharp, China), it was stored at –20°C. The drugs were diluted to the required concentration.
MIC determination
All isolates were subjected to antimicrobial susceptibility testing via broth microdilution according to the procedure described by the Clinical Laboratory Standards Institute (57). In brief, tylosin, tilmicosin, doxycycline, or ciprofloxacin were mixed with an equal volume of bacterial broth containing 1.5 × 106 CFUs mL−1 and twofold diluted in clear 96-well microtiter plates (Corning, NY, USA). The lowest concentration of the drugs at which no bacterial growth was observed after 24 h of incubation at 37°C was defined as the MIC (58). All samples were tested at least three times for MIC.
Whole-genome resequencing for S. suis strains
Total DNA was extracted from S. suis using the E.Z.N.A. Tissue DNA kit (Omega Bio-Tek) following the Tissue DNA-Spin Protocol. After the DNA samples were delivered, a quality control test was carried out on the specimens, and the qualified DNA (>3 µg; concentration > 30 ng/µL; OD260/OD280 = 1.80–2.00) was used for further study. The genomic DNA library was constructed using the Illumina pair-end sequencing method and selected with an average insert size of ~450 bp using Illumina’s standard genomic DNA library preparation procedure. Sequencing of T-I-8, T-I-64, and T-I-256 was performed with the Illumina NovaSeq 6000 platform (150 bp × 2, Shanghai Biozeron Biotechnology Co., Ltd, Shanghai, China). The public data of reference genome of Streptococcus suis P1/7 were obtained from the NCBI (https://www.ncbi.nlm.nih.gov/nuccore/NC_012925.1/). The gene annotation information (including functional annotation and gene family information) was also downloaded in the corresponding genome database. The clean sequencing reads are aligned to the reference genome sequence using BWA (http://bio-bwa.sourceforge.net/) software (bwa mem -k 32). The valid BAM file was used to detect SNPs and short InDel by GATK (version 4.1.2.0) “HaplotypeCaller” function (http://www.broadinstitute.org/gatk/). When the gene annotation file of reference genome is provided, the annotation of detected variations can be performed by ANNOVAR (http://www.openbioinformatics.org/annovar/), including SNP (synonymous or non-synonymous mutations of SNPs) and InDel. Structure variations were identified by BreakDancer (http://breakdancer.sourceforge.net).
Metabolome assay and data analysis
For targeted metabolomics analysis, the samples were taken out at −80°C, and 1mL of methanol acetonitrile aqueous solution (2:2:1, vol/vol) was added. The vortex was for 60 s, and ultrasound was performed twice at low temperatures for 30 min. The samples were placed at −20°C for 1 h to precipitate the protein and centrifuged at 14,000 rpm at 4°C for 20 min, and the supernatant was freeze-dried for subsequent tests. The samples were analyzed by ultra high performance liquid chromatography (1290 Infinity LC, Agilent Technologies) and coupled quadrupole time-of-flight (AB Sciex TripleTOF6600) equipped with an electrospray ionization source. Separation was achieved by a hydrophilic interaction chromatography on an ACQUITY UPLC BEH Amide column [2.1 × 100 mm, 1.7 µm particle size (Waters, Ireland)] using a gradient of solvent A (25 mM ammonium acetate and 25 mM ammonium hydroxide in water) and solvent B (acetonitrile). The gradient was 95% B for 0.5 min, linearly reduced to 65% in 7 min, and then reduced to 40% in 8 min, kept for 2 min, and then increased to 95% in 9.1 min, with a 4 min re-equilibration period employed. The flow rate was 0.3 mL/min, column temperature was 25°C, autosampler temperature was 4°C, and injection volume was 2 µL. Each sample was detected in both negative and positive ion modes. The raw MS data were converted into the MzXML files using a ProteoWizard MSConvert tool (59) and processed using the XCMS (60) for feature detection, retention time correction, and alignment. The metabolites were identified by accuracy mass (<25 ppm) and MS/MS data, which matched with our standard database. For multivariate statistical analysis, the web-based MetaboAnalyst system (https://www.metaboanalyst.ca/) was used, followed by the Pareto-scaling, PCA, and OPLS-DA. The significantly different metabolites were determined based on the combination of a statistically significant threshold of variable influence of projection value obtained from the OPLS-DA model and two-tailed Student’s t-test (P-value) on the raw data, and the metabolites with VIP values larger than 1.0 and P-values less than 0.05 were considered as significant.
Thigh infection model experiment in mice
A mouse thigh infection model (61) was used to assess the synergistic effect of the combination of L-met and tylosin on S. suis. Six-week-old female specific pathogen-free ICR mice were purchased from the Experimental Animal Center of the Second Affiliated Hospital of Harbin Medical University (Harbin, China).The mice developed neutropenia (neutrophil count < 100 mm3) when cyclophosphamide was administered intraperitoneally at doses of 150 and 100 mg/kg of body weight, respectively, 4 days and 1 day before bacterial inoculation. The immunodeficient mice were divided into five groups for subsequent experiments (n = 5). Mice in the model group were injected intramuscularly with 100 µL of inoculum in each thigh, including 107 log-phase test strains. Two hours after modeling, mice were treated with tylosin (10 mg/kg of body weight) and L-met (0.1 g/kg of body weight) alone or in combination as the tylosin single treatment group, L-met single treatment group, and combination treatment group, respectively. Finally, the mice injected with physiological salt were used as blank controls. Both L-met and tylosin were administered intramuscularly. After 22 h of drug treatment, mice were euthanized. Both posterior thigh muscles of the mice were immediately collected in normal saline, homogenized, and appropriately diluted, and the number of CFUs was determined by planar colony counting. The thigh tissue CFU titer is indicated as log10 CFU/thigh.
Antibiotic survival assay
To obtain exponential and mid-stationary-phase cultures in THB, the 16-h culture was diluted 1:1,000 in fresh medium and grown to the desired turbidity (OD600 = 0.5). The sample was washed twice with PBS, and the culture was collected by centrifugation at 8,000 g for 8 min, and then resuspended in THB medium. Different antibiotics (tylosin, tilmicosin, doxycycline, and ciprofloxacin) with concentrations of 1/2 and 1/4 MIC were added, respectively. For the amino acid supplement experiment, L-met was added to the exponential phase culture at the specified concentration. After incubation, 100 µL aliquots were periodically taken out to perform serial dilutions and then inoculated (10 µL aliquots) on THB agar plates. The agar plates were incubated at 37°C for 24 h, and the number of cfu was calculated. Bacterial survival was calculated as the ratio between the dosing and blank control groups. The results represented the average of three biological replicates, and the error bars represent the standard deviation.
Metabolite concentration detection
MET, SAM, SAH, and HCY concentrations were detected through MET, SAM, SAH, and HCY ELISA Kits, respectively (Sinobestbio, China). GSH level was determined using a content assay kit (Nanjing Jiancheng Bioengineering Institute, China). T-I-256 and L-met were incubated at 37°C for 12 h and named Met+256. The S, Met + 256, T-I-8, T-I-64, and T-I-256 cultures were collected, washed, resuspended in PBS, and adjusted to OD600 of 0.5. After sonicating and centrifuging 1 mL of the sample to break up the cells, the supernatant was collected and analyzed according to the kit to detect the concentration of metabolites in the bacteria. The assays were repeated three times.
Measurement of enzyme activity
The activities of SAMs and methylases were detected by SAMs and methylases ELISA kit (Sinobestbio, China). The S, Met+256, T-I-8, T-I-64, and T-I-256 cultures were collected, washed, resuspended in PBS, and adjusted to OD600 of 0.5. After centrifugation, the supernatant was transferred to a new tube and reacted according to the manufacturer’s instructions, and absorbance at 450 nm was measured using the microplate reader. The activity of the corresponding enzyme was calculated by creating a standard curve and substituting the absorbance value of the sample.
SOD and CAT levels were measured using colorimetric assay kits (Nanjing Jiancheng Bioengineering Institute, China). In brief, bacterial cultures were collected, washed, resuspended in lysis buffer (from the assay kits), and disrupted by sonication. After centrifugation, the supernatant was transferred to a new tube and reacted according to the manufacturer’s instructions, and absorbance at 560 and 240 nm was measured using the microplate reader. SOD and CAT levels were quantified using a commercially available colorimetrical kit (62). The activity of the corresponding enzyme was calculated by creating a standard curve and substituting the absorbance value of the sample.
Quantification of metabolism-related genes by RT-PCR
RT-PCR was used to analyze the relative expression of genes in this study. The genes metK, mtnN, luxS, and metE in the methionine pathway were detected. In order to explore whether exogenous methionine affects the efflux pump of S. suis, the genes pnuC, msmK, and metQ were detected. Drug resistance genes and efflux pump genes tetM, tetW, and tetL of tetracyclines, ermB, msrAB, and msrD of macrolides, satA, satB, and gyrA of fluoroquinolones were detected. Also, the genes sodA (superoxide dismutase SodA), the fur-like protein regulating gene perR, dpr (dps-like peroxide resistance protein), and feoB (ferrous iron transport protein B) associated with oxidative stress response and metal starvation were detected. The primer sequences used are listed in Table S1. The bacterial cultures of S, Met+256, T-I-8, T-I-64, and T-I-256 were collected by centrifugation. Total RNA extraction (Omega, China) and cDNA synthesis (Takara, China) were done according to the manufacturer’s instructions. The relative expression was normalized using the 16S rRNA gene as an endogenous control (56). The reaction conditions were 94°C for 10 min followed by 40 cycles of amplification at 94°C for 15 s and 60°C for 60 s (63). Three replicate analyses were performed for each sample, and expression data were collected from three biological replicate samples. All assays were performed in triplicates.
Total methylation level determination
DNA was extracted from strains S, T-I-8, T-I-64, T-I-256, and T-I-256 treated with L-met using a bacterial genomic DNA extraction kit (A&D Technology Corporation, China). Global DNA methylation was determined with the MethylFlash Methylated DNA 5-mC Quantification kit (Colorimetric) (EpiGentek, Farmingdale, New York, NY, USA). The level of methylation in DNA was then detected using the following colorimetric assay. The extracted DNA was added to the binding solution (provided by the manufacturer) to bond the DNA to the microplate, incubated at 37°C for 90 min, and then eluted with deionized water. A capture antibody for methylated DNA was added, sealed at indoor temperature, and incubated for 30 min before elution. After the addition of the detection antibody as well as the enhancement solution (provided by the manufacturer), it was incubated for 10 min at room temperature and protected from light. At the same time, the color change in the sample wells vs the positive control wells was monitored as sufficiently methylated DNA turned blue. The reaction of the enzyme was terminated by the addition of a fluorogenic developer, and the absorbance was measured and read at 450 nm using a fluorescence analyzer.
Determination of iron concentrations
Cellular iron levels were determined using the colorimetric ferrozine assay method described by Riemer et al. (64). Cultures of S. suis were incubated to OD600 = 0.5, after the addition of L-met, and the cultures were incubated for 12 h and harvested by centrifugation. The cell pellet was washed twice with ice-cold PBS and resuspended in 1 mL of 50 mM NaOH. The lysate was mixed with 10 mM HCl and an iron release reagent and incubated at 60°C for 2 h to quantify the bound iron. After the mixture was cooled to 25°C, 30 mL of the iron detection reagent (6.5 mM ferrozine, 6.5 mM neocuproine, 2.5 M ammonium acetate, and 1M ascorbic acid dissolved in water) was added. After 30 min, the absorbance of the samples was measured at 550 nm.
Measurement of oxidative stress indicators
ROS were detected by the Reactive Oxygen Species Assay Kit based on 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) (Beyotime Biotechnology, China). The samples were incubated with 10 mM DCFH-DA for 20 min. After washing by PBS, cells were treated with tylosin in the presence or absence of 50 mM L-met. Samples were taken after incubated for 20 min and used to monitor fluorescence intensity at an excitation wavelength of 488 nm and an emission wavelength of 525 nm.
GSH and H2O2 (Nanjing Jiancheng Bioengineering Institute, China) levels were determined using content assay kits (Nanjing Jiancheng Institute of Biological Engineering, Nanjing, Jiangsu, China). The resistant strain was incubated in THB medium supplemented with L-met for 12 h at 37°C, and the bacterial cultures were collected, washed, resuspended in PBS, and adjusted to an OD600 of 0.5. After sonicating and centrifuging 1 mL of the sample to break up the cells, the supernatant was collected and analyzed according to the kit instructions to detect the concentration of metabolites in the bacteria. The assays were repeated three times.
Statistical analysis
Data were collected for statistical analysis, and Student’s t-test or one-way ANOVA was conducted using GraphPad Prism 8 (GraphPad Software, Inc., San Diego, CA, USA) and SPSS version 26.0 software (IBM SPSS, Chicago, IL, USA, software version 26.0) (65). Values were reported as the mean ± standard deviation. The different levels of statistical significance were set as ns, P > 0.05, *P < 0.05, and **P < 0.01.
ACKNOWLEDGMENTS
This work was sponsored by grants from the National Natural Science Foundation of China (32102726), the National Key Research and Development Program of China (2023YFD1800100), the Heilongjiang Provincial Natural Science Foundation of China (LH2021C041), and the China Agriculture Research System of MOF and MARA.
Contributor Information
Yan-Hua Li, Email: liyanhua@neau.edu.cn.
Shannon D. Manning, Michigan State University, East Lansing, Michigan, USA
DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding author upon reasonable request. Additionally, raw metabonomic data can be viewed in NODE (https://www.biosino.org/node) under accession no. OEP004815. Resequencing data are available under accession number PRJNA1052407.
ETHICS APPROVAL
All animal experiments conformed to the ethical principles of animal research and were approved by the Institutional Animal Care and Use Committee of the Northeastern Agricultural University (No. NEAUEC20).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.02803-23.
Primers used for the quantitative RT-PCR analysis and differential metabolites.
Fig S1 to S3 (Bioinformatics analysis of the metabolomics of S. suis) and Fig S4 (The time-kill curves of Streptococcus suis treated with tylosin and methionine at different concentrations.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Lun ZR, Wang QP, Chen XG, Li AX, Zhu XQ. 2007. Streptococcus suis: an emerging zoonotic pathogen. Lancet Infect Dis 7:201–209. doi: 10.1016/S1473-3099(07)70001-4 [DOI] [PubMed] [Google Scholar]
- 2. Kerdsin A, Akeda Y, Takeuchi D, Dejsirilert S, Gottschalk M, Oishi K. 2018. Genotypic diversity of Streptococcus suis strains isolated from humans in Thailand. Eur J Clin Microbiol Infect Dis 37:917–925. doi: 10.1007/s10096-018-3208-8 [DOI] [PubMed] [Google Scholar]
- 3. Zhang C, Zhang P, Wang Y, Fu L, Liu L, Xu D, Hou Y, Li Y, Fu M, Wang X, Wang S, Ding S, Shen Z. 2020. Capsular serotypes, antimicrobial susceptibility, and the presence of transferable oxazolidinone resistance genes in Streptococcus suis isolated from healthy pigs in China. Vet Microbiol 247:108750. doi: 10.1016/j.vetmic.2020.108750 [DOI] [PubMed] [Google Scholar]
- 4. Lunha K, Chumpol W, Samngamnim S, Jiemsup S, Assavacheep P, Yongkiettrakul S. 2022. Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs in Thailand, 2018-2020. Antibiotics (Basel) 11:410. doi: 10.3390/antibiotics11030410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cucco L, Paniccià M, Massacci FR, Morelli A, Ancora M, Mangone I, Di Pasquale A, Luppi A, Vio D, Cammà C, Magistrali CF. 2022. New sequence types and antimicrobial drug-resistant strains of Streptococcus suis in diseased pigs, Italy, 2017-2019. Emerg Infect Dis 28:139–147. doi: 10.3201/eid2801.210816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ogawara H. 2019. Comparison of antibiotic resistance mechanisms in antibiotic-producing and pathogenic bacteria. Molecules 24:3430. doi: 10.3390/molecules24193430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duan X, Huang X, Wang X, Yan S, Guo S, Abdalla AE, Huang C, Xie J. 2016. L-serine potentiates fluoroquinolone activity against Escherichia coli by enhancing endogenous reactive oxygen species production. J Antimicrob Chemother 71:2192–2199. doi: 10.1093/jac/dkw114 [DOI] [PubMed] [Google Scholar]
- 8. Zampieri M, Enke T, Chubukov V, Ricci V, Piddock L, Sauer U. 2017. Metabolic constraints on the evolution of antibiotic resistance. Mol Syst Biol 13:917. doi: 10.15252/msb.20167028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zampieri M, Zimmermann M, Claassen M, Sauer U. 2017. Nontargeted metabolomics reveals the multilevel response to antibiotic perturbations. Cell Rep 19:1214–1228. doi: 10.1016/j.celrep.2017.04.002 [DOI] [PubMed] [Google Scholar]
- 10. Shibue R, Sasamoto T, Shimada M, Zhang B, Yamagishi A, Akanuma S. 2018. Comprehensive reduction of amino acid set in a protein suggests the importance of prebiotic amino acids for stable proteins. Sci Rep 8:1227. doi: 10.1038/s41598-018-19561-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miyamoto T, Homma H. 2021. D-amino acid metabolism in bacteria. J Biochem 170:5–13. doi: 10.1093/jb/mvab043 [DOI] [PubMed] [Google Scholar]
- 12. Liu YK, Kuo HC, Lai CH, Chou CC. 2020. Single amino acid utilization for bacterial categorization. Sci Rep 10:12686. doi: 10.1038/s41598-020-69686-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stokes JM, Lopatkin AJ, Lobritz MA, Collins JJ. 2019. Bacterial metabolism and antibiotic efficacy. Cell Metab 30:251–259. doi: 10.1016/j.cmet.2019.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng ZX, Guo C, Chen ZG, Yang TC, Zhang JY, Wang J, Zhu JX, Li D, Zhang TT, Li H, Peng B, Peng XX. 2019. Glycine, serine and threonine metabolism confounds efficacy of complement-mediated killing. Nat Commun 10:3325. doi: 10.1038/s41467-019-11129-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cho DY, Lim DJ, Mackey C, Weeks CG, Peña Garcia JA, Skinner D, Grayson JW, Hill HS, Alexander DK, Zhang S, Woodworth BA. 2018. L-methionine anti-biofilm activity against Pseudomonas aeruginosa is enhanced by the cystic fibrosis transmembrane conductance regulator potentiator, ivacaftor. Int Forum Allergy Rhinol 8:577–583. doi: 10.1002/alr.22088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brewster JL, Pachl P, McKellar JLO, Selmer M, Squire CJ, Patrick WM. 2021. Structures and kinetics of thermotoga maritima mety reveal new insights into the predominant sulfurylation enzyme of bacterial methionine biosynthesis. J Biol Chem 296:100797. doi: 10.1016/j.jbc.2021.100797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jochim A, Shi T, Belikova D, Schwarz S, Peschel A, Heilbronner S, McBain AJ. 2019. Methionine limitation impairs pathogen expansion and biofilm formation capacity. Appl Environ Microbiol 85:e00177-19. doi: 10.1128/AEM.00177-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferla MP, Patrick WM. 2014. Bacterial methionine biosynthesis. Microbiology (Reading) 160:1571–1584. doi: 10.1099/mic.0.077826-0 [DOI] [PubMed] [Google Scholar]
- 19. Militello KT, Mandarano AH, Varechtchouk O, Simon RD. 2014. Cytosine DNA methylation influences drug resistance in Escherichia coli through increased sugE expression. FEMS Microbiol Lett 350:100–106. doi: 10.1111/1574-6968.12299 [DOI] [PubMed] [Google Scholar]
- 20. Li H, Li T, Hu Q, Yao Z, Li L, Huang Q, Zhou R. 2022. Inhibitors targeting the autophosphorylation of serine/threonine kinase of Streptococcus suis show potent antimicrobial activity. Front Microbiol 13:990091. doi: 10.3389/fmicb.2022.990091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu T, Wang X, Dong Y, Xing C, Chen X, Li L, Dong C, Li Y. 2022. Effects of L-serine on macrolide resistance in Streptococcus suis. Microbiol Spectr 10:e0068922. doi: 10.1128/spectrum.00689-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qu Q, Cui W, Huang X, Zhu Z, Dong Y, Yuan Z, Dong C, Zheng Y, Chen X, Yuan S, Li Y. 2023. Gallic acid restores the sulfonamide sensitivity of multidrug-resistant Streptococcus suis via polypharmaceology mechanism. J Agric Food Chem 71:6894–6907. doi: 10.1021/acs.jafc.2c06991 [DOI] [PubMed] [Google Scholar]
- 23. Tram G, Jennings MP, Blackall PJ, Atack JM. 2021. Streptococcus suis pathogenesis-A diverse array of virulence factors for a zoonotic lifestyle. Adv Microb Physiol 78:217–257. doi: 10.1016/bs.ampbs.2020.12.002 [DOI] [PubMed] [Google Scholar]
- 24. Sauer E, Merdanovic M, Mortimer AP, Bringmann G, Reidl J. 2004. PnuC and the utilization of the nicotinamide riboside analog 3-aminopyridine inHaemophilus influenzae. Antimicrob Agents Chemother 48:4532–4541. doi: 10.1128/AAC.48.12.4532-4541.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhu N, Olivera BM, Roth JR. 1989. Genetic characterization of the pnuC gene, which encodes a component of the nicotinamide mononucleotide transport system in Salmonella typhimurium. J Bacteriol 171:4402–4409. doi: 10.1128/jb.171.8.4402-4409.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tan MF, Liu WQ, Zhang CY, Gao T, Zheng LL, Qiu DX, Li L, Zhou R. 2017. The involvement of MsmK in pathogenesis of the Streptococcus suis serotype 2. Microbiologyopen 6:e00433. doi: 10.1002/mbo3.433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tan MF, Hu Q, Hu Z, Zhang CY, Liu WQ, Gao T, Zhang LS, Yao L, Li HQ, Zeng YB, Zhou R. 2021. Streptococcus suis MsmK: novel cell division protein interacting with FtsZ and maintaining cell shape. mSphere 6:e00119-21. doi: 10.1128/mSphere.00119-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nguyen PT, Li QW, Kadaba NS, Lai JY, Yang JG, Rees DC. 2015. The contribution of methionine to the stability of the Escherichia coli MetNIQ ABC transporter-substrate binding protein complex. Biol Chem 396:1127–1134. doi: 10.1515/hsz-2015-0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pei X, Liu J, Liu M, Zhou H, Wang X, Fan H. 2020. Quantitative proteomics revealed modulation of macrophages by MetQ gene of Streptococcus suis serotype 2. AMB Express 10:195. doi: 10.1186/s13568-020-01131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fang L, Shen H, Tang Y, Fang W. 2015. Superoxide dismutase of Streptococcus suis serotype 2 plays a role in anti-autophagic response by scavenging reactive oxygen species in infected macrophages. Vet Microbiol 176:328–336. doi: 10.1016/j.vetmic.2015.02.006 [DOI] [PubMed] [Google Scholar]
- 31. Yu S, Ma Q, Li Y, Zou J. 2023. Molecular and regulatory mechanisms of oxidative stress adaptation in Streptococcus mutans. Mol Oral Microbiol 38:1–8. doi: 10.1111/omi.12388 [DOI] [PubMed] [Google Scholar]
- 32. Dzyhovskyi V, Stokowa-Sołtys K. 2023. Divalent metal ion binding to Staphylococcus aureus FeoB transporter regions. J Inorg Biochem 244:112203. doi: 10.1016/j.jinorgbio.2023.112203 [DOI] [PubMed] [Google Scholar]
- 33. Huang M, Wang M, Feng Y, Wang M, Gao Q, Zhu D, Jia R, Chen S, Zhao X, Yang Q, Wu Y, Zhang S, Tian B, Huang J, Ou X, Mao S, Sun D, He Y, Wu Z, Cheng A, Liu M, Rogovskyy AS. 2023. Functional characterization of FeoAB in iron acquisition and pathogenicity in riemerella anatipestifer. Microbiol Spectr 11:e0137323. doi: 10.1128/spectrum.01373-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arenas J, Zomer A, Harders-Westerveen J, Bootsma HJ, De Jonge MI, Stockhofe-Zurwieden N, Smith HE, De Greeff A. 2020. Identification of conditionally essential genes for Streptococcus suis infection in pigs. Virulence 11:446–464. doi: 10.1080/21505594.2020.1764173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Holmer I, Salomonsen CM, Jorsal SE, Astrup LB, Jensen VF, Høg BB, Pedersen K. 2019. Antibiotic resistance in porcine pathogenic bacteria and relation to antibiotic usage. BMC Vet Res 15:449. doi: 10.1186/s12917-019-2162-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shatalin K, Shatalina E, Mironov A, Nudler E. 2011. H2S: a universal defense against antibiotics in bacteria. Science 334:986–990. doi: 10.1126/science.1209855 [DOI] [PubMed] [Google Scholar]
- 37. Lee HH, Molla MN, Cantor CR, Collins JJ. 2010. Bacterial charity work leads to population-wide resistance. Nature 467:82–85. doi: 10.1038/nature09354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gusarov I, Shatalin K, Starodubtseva M, Nudler E. 2009. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325:1380–1384. doi: 10.1126/science.1175439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li P, Yin YL, Li D, Kim SW, Wu G. 2007. Amino acids and immune function. Br J Nutr 98:237–252. doi: 10.1017/S000711450769936X [DOI] [PubMed] [Google Scholar]
- 40. Sasabe J, Miyoshi Y, Rakoff-Nahoum S, Zhang T, Mita M, Davis BM, Hamase K, Waldor MK. 2016. Interplay between microbial d-amino acids and host d-amino acid oxidase modifies murine mucosal defence and gut microbiota. Nat Microbiol 1:16125. doi: 10.1038/nmicrobiol.2016.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vieira SL, Lemme A, Goldenberg DB, Brugalli I. 2004. Responses of growing broilers to diets with increased sulfur amino acids to lysine ratios at two dietary protein levels. Poult Sci 83:1307–1313. doi: 10.1093/ps/83.8.1307 [DOI] [PubMed] [Google Scholar]
- 42. Yong Y, Zhou Y, Liu K, Liu G, Wu L, Fang B. 2021. Exogenous citrulline and glutamine contribute to reverse the resistance of Salmonella to apramycin. Front Microbiol 12:759170. doi: 10.3389/fmicb.2021.759170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hosseini SA, Zaghari M, Lotfollahian H, Shivazad M, Moravaj H. 2012. Reevaluation of methionine requirement based on performance and immune responses in broiler breeder hens. J Poult Sci 49:26–33. doi: 10.2141/jpsa.011021 [DOI] [Google Scholar]
- 44. Homma T, Kobayashi S, Fujii J. 2022. Methionine deprivation reveals the pivotal roles of cell cycle progression in ferroptosis that is induced by cysteine starvation. Cells 11:1603. doi: 10.3390/cells11101603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Deobald D, Hanna R, Shahryari S, Layer G, Adrian L. 2020. Identification and characterization of a bacterial core methionine synthase. Sci Rep 10:2100. doi: 10.1038/s41598-020-58873-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martínez Y, Li X, Liu G, Bin P, Yan W, Más D, Valdivié M, Hu C-AA, Ren W, Yin Y. 2017. The role of methionine on metabolism, oxidative stress, and diseases. Amino Acids 49:2091–2098. doi: 10.1007/s00726-017-2494-2 [DOI] [PubMed] [Google Scholar]
- 47. Peng B, Su YB, Li H, Han Y, Guo C, Tian YM, Peng XX. 2015. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab 21:249–262. doi: 10.1016/j.cmet.2015.01.008 [DOI] [PubMed] [Google Scholar]
- 48. Hu X, Wang Y, Gao L, Jiang W, Lin W, Niu C, Yuan K, Ma R, Huang Z. 2018. The impairment of methyl metabolism from luxS mutation of Streptococcus mutans. Front Microbiol 9:404. doi: 10.3389/fmicb.2018.00404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pakula KK, Hansen LH, Vester B. 2017. Combined effect of the Cfr methyltransferase and ribosomal protein L3 mutations on resistance to ribosome-targeting antibiotics. Antimicrob Agents Chemother 61:e00862-17. doi: 10.1128/AAC.00862-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fyfe C, Grossman TH, Kerstein K, Sutcliffe J. 2016. Resistance to macrolide antibiotics in public health pathogens. Cold Spring Harb Perspect Med 6:a025395. doi: 10.1101/cshperspect.a025395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Troxell B, Hassan HM. 2013. Transcriptional regulation by ferric uptake regulator (fur) in pathogenic bacteria. Front Cell Infect Microbiol 3:59. doi: 10.3389/fcimb.2013.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Peng W, Yang X, Wang N, Gao T, Liu Z, Liu W, Zhou D, Yang K, Guo R, Liang W, Chen H, Tian Y, Yuan F, Bei W, Dozois CM. 2022. Perr-regulated manganese import contributes to oxidative stress defense in Streptococcus suis. Appl Environ Microbiol 88:e0008622. doi: 10.1128/aem.00086-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang T, Ding Y, Li T, Wan Y, Li W, Chen H, Zhou R. 2012. A fur-like protein PerR regulates two oxidative stress response related operons dpr and metQIN in Streptococcus suis. BMC Microbiol 12:85. doi: 10.1186/1471-2180-12-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wongsaroj L, Saninjuk K, Romsang A, Duang-Nkern J, Trinachartvanit W, Vattanaviboon P, Mongkolsuk S. 2018. Pseudomonas aeruginosa glutathione biosynthesis genes play multiple roles in stress protection, bacterial virulence and biofilm formation. PLoS One 13:e0205815. doi: 10.1371/journal.pone.0205815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shukla P, Khodade VS, SharathChandra M, Chauhan P, Mishra S, Siddaramappa S, Pradeep BE, Singh A, Chakrapani H. 2017. “On demand” redox buffering by H2S contributes to antibiotic resistance revealed by a bacteria-specific H2S donor. Chem Sci 8:4967–4972. doi: 10.1039/c7sc00873b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Che R-X, Xing X-X, Liu X, Qu Q-W, Chen M, Yu F, Ma J-X, Chen X-R, Zhou Y-H, God’Spower B-O, Liu J-W, Lu Z-X, Xu Y-P, Li Y-H. 2019. Analysis of multidrug resistance in Streptococcus suis ATCC 700794 under tylosin stress. Virulence 10:58–67. doi: 10.1080/21505594.2018.1557505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. 2023. M100: performance standards for antimicrobial susceptibility testing rE. Clinical and laboratory standards institute (CLSI). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Osaki M, Takamatsu D, Tsuji N, Sekizaki T. 2000. Cloning and characterization of the gene encoding O-acetylserine lyase from Streptococcus suis. Curr Microbiol 40:67–71. doi: 10.1007/s002849910013 [DOI] [PubMed] [Google Scholar]
- 59. Gibbons BC, Chambers MC, Monroe ME, Tabb DL, Payne SH. 2015. Correcting systematic bias and instrument measurement drift with mzRefinery. Bioinformatics 31:3838–3840. doi: 10.1093/bioinformatics/btv437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu C, Zhu Z, Du L, Li S, Zhao Q, Wang X. 2023. Urine metabolites and viral pneumonia among children: a case-control study in China. Transl Pediatr 12:1192–1203. doi: 10.21037/tp-23-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen F, Wei MC, Luo YD, Jin Z, Tang YZ. 2020. Synergistic effect of a pleuromutilin derivative with tetracycline against Streptococcus suis in vitro and in the neutropenic thigh infection model. Molecules 25:3522. doi: 10.3390/molecules25153522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xu S, Kang Z, Li K, Li X, Zhang Y, Gao X-J. 2023. Selenium deficiency causes iron death and inflammatory injury through oxidative stress in the mice gastric mucosa. Biol Trace Elem Res. doi: 10.1007/s12011-023-03754-5 [DOI] [PubMed] [Google Scholar]
- 63. Yang YB, Chen JQ, Zhao YL, Bai JW, Ding WY, Zhou YH, Chen XY, Liu D, Li YH. 2016. Sub-MICs of azithromycin decrease biofilm formation of Streptococcus suis and increase capsular polysaccharide content of S. suis. Front Microbiol 7:1659. doi: 10.3389/fmicb.2016.01659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Riemer J, Hoepken HH, Czerwinska H, Robinson SR, Dringen R. 2004. Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells. Anal Biochem 331:370–375. doi: 10.1016/j.ab.2004.03.049 [DOI] [PubMed] [Google Scholar]
- 65. Song M, Liu Y, Huang X, Ding S, Wang Y, Shen J, Zhu K. 2020. A broad-spectrum antibiotic adjuvant reverses multidrug-resistant gram-negative pathogens. Nat Microbiol 5:1040–1050. doi: 10.1038/s41564-020-0723-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for the quantitative RT-PCR analysis and differential metabolites.
Fig S1 to S3 (Bioinformatics analysis of the metabolomics of S. suis) and Fig S4 (The time-kill curves of Streptococcus suis treated with tylosin and methionine at different concentrations.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Additionally, raw metabonomic data can be viewed in NODE (https://www.biosino.org/node) under accession no. OEP004815. Resequencing data are available under accession number PRJNA1052407.