Abstract
Antimicrobial resistance (AMR) contamination in the environment is one of the most significant worldwide threats of the 21st century. Since sludge is heavily exposed to diverse contaminants, including pharmaceuticals, the inhabitant bacterial population is expected to exhibit resistance to antimicrobial agents. In this study, sewage treatment plant (STP) sludge samples were analyzed to assess the antibiotic-resistant bacterial population, abundance of AMR genes (ermF, qnrS, Sul1, blaGES, blaCTX-M, and blaNDM), and mobile genetic elements (intl1 and IS26). Out of 16, six bacterial isolates exhibited resistance to 13 antibiotics with a high multiple antibiotic resistance index (MARI) (0.93) and high metal tolerance. Quantitative polymerase chain reaction showed the abundance of target genes ranging from 6.6 × 103 to 6.5 × 108 copies g−1 sludge. The overall outcome reveals that STP sludge comprised varied multidrug-resistant bacterial populations. It will give insights into the functions of heavy metals and biofilm development in the selection and spread of AMR genes and the associated bacteria. Therefore, the application of sludge needs proper screening for AMR and metal contamination prior to its countless applications. This study will contribute immensely to the risk analysis of STP effluents on environmental health, including control of AMR transmission.
Keywords: sewage treatment plant, sludge, antimicrobial resistance gene, multi-drug resistance, biofilm, metal resistance
STP sludge study reveals diverse drug-resistant bacteria harboring ARGs, stressing screening to curb AMR and environmental risks.
Introduction
Water is indispensable for the survival of various life forms and for sustaining the environment. The expanding population has led to large-scale production and use of pharmaceuticals. The imprudent and overuse of antibiotics in healthcare, veterinary, and agricultural sectors has increased their quantity in the environment by several folds (Ahmad et al. 2021). As a result, these contaminants are excessively released into the environment, depleting natural water resources, leading to massive wastewater generation and, thereby, water scarcity (Van Vliet et al. 2021). The misuse or abuse of antibiotics has led to unnatural selective pressure on the resistance determinants in clinical and natural environments, expanding the spread of antimicrobial resistance (AMR) and resulting in the emergence of multidrug-resistant (MDR) bacteria or superbug bacteria (Holmes et al. 2006, Xu et al. 2015, Salam et al. 2023). AMR has become one of the principal problems of the 21st century, posing a serious threat to the treatment and prevention of diseases (Davies and Davies 2010, Tang et al. 2023). According to the GRAM (Global Research on AMR) report of 2019, six of the leading resistant pathogens, Staphylococcus aureus, Escherichia coli, Streptococcus pneumoniae, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii were accountable for ∼3·6 million casualties and ∼1 million deaths associated with AMR. By 2050, 10 million people per year could face death due to AMR (O'Neill 2016, Murray et al. 2022).
Due to the depletion of water resources by these contaminants, over 700 million people, or 40% of the world's population, could be affected by severe water scarcity by 2030 (Kölbel et al. 2018, Van Vliet et al. 2021). According to a 2018, NITI Aayog report, 600 million of the Indian population is experiencing severe water stress, and it is predicted that by 2030, water demand may double the available supply, resulting in severe water scarcity and a ∼6% decline in the nation's Gross Domestic Product (GDP). The global production of wastewater is estimated to be roughly 359 billion cubic meters (Jones et al. 2021). India alone produces about 38 354 million liters of wastewater every day (Kaur et al. 2012, Maity et al. 2023). One of the legitimate strategies for wastewater management involves the treatment of wastewater by STPs. STPs handle wastewater from various sources and produce effluent that is suitable for release into the environment or reuse after treatment (Schellenberg et al. 2020). By 2025, sludge production is expected to amount to between 150 and 200 million tonnes (Mohajerani et al. 2017, Vaithyanathan and Cabana 2021). A typical STP forms a byproduct after the removal of the pathogenic bacteria, dissolve solids, organic matter, and nutrients like nitrogen and phosphorus from wastewater. However, several investigations reported that the treatment methods installed in STPs are insufficient to completely remove metal, antibiotic residues, antibiotic-resistant bacteria (ARB), and antibiotic resistance genes (ARGs) (Turolla et al. 2018, Ding et al. 2020, Rumky et al. 2022). Therefore, STP sludge provides a conducive niche where the prevailing bacterial population gets acclimatized to respective environmental conditions and shows high tolerance and resistance against various antibiotics and metals (Sun et al. 2021).
India being a developing country is accelerating its wastewater treatment facilities, as a result, sludge production is rapidly rising in recent times (Kesari et al. 2021). The proper treatment and disposal of huge sludge generated is an expensive and environmentally difficult task. Though a number of ways are available, including incineration and landfilling due to higher costs, reusing sewage sludge turns out to be a potential solution to the problem of sludge waste management (Pedrero et al. 2010, Seleiman et al. 2020). Both developed and developing countries regularly use STP sludge as a source of fertilizer in agriculture as it is rich in nutrients (Seleiman et al. 2020). However, concerns have been raised about the negative effects of the use of untreated sewage sludge in agriculture as it carries various antibiotic residues, ARBs, ARGs, and mobile genetic elements (MGEs) (Manaia et al. 2018, Sorinolu et al. 2021, Zhang et al. 2022). When this sewage sludge enriched in ARBs, ARGs, etc. is used as a fertilizer for field applications, all of these contaminants could infiltrate the soil (da Silva Souza et al. 2020). It poses serious threats to the soil quality and crops on field applications and thus, a potential risk to human health. Several studies reported a huge abundance of tetracycline resistance genes in sludge and effluent water of STPs (Pei et al. 2006, Al-Jassim et al. 2015, Zhang et al. 2020). Extended-spectrum cephalosporin or carbapenem-resistant E. coli isolated from STP effluent, hospital effluent, and other sources have been studied to establish the distribution and connection of AMR determinants (Diwan et al. 2012, Akiba et al. 2016, Guruge et al. 2021, Cho et al. 2023). A higher concern has been raised with the recent exacerbation of the overuse of antibiotics following the COVID-19 pandemic (Mokni-Tlili et al. 2022). Several independent research in India identified bacteria with beta-lactamase genes from sewage water and hospital effluent samples (Beg and Khan 2018, Aggarwal et al. 2020, Paul et al. 2021). A comparison of ARGs diversity and abundance in pharmaceutical effluent contaminated environment was made by Bombaywala et al. (2021). Another cross-sectional pan-India study was performed to evaluate sewage microbiomes by Singh et al. (2023), who reported a high representation of gut microbiomes in sewage. Therefore, the sludge derived from STPs plays a significant role in the environmental transmission of ARGs and ARBs, accelerating the global threat imposed by the phenomena of AMR (Boxall 2004,Wang et al. 2023).
The available literature elucidates that there is a paucity of information regarding the presence of resistance determinants (ARGs and ARBs) in STP sludge. Our previous reports revealed the occurrence and dispersion of ARGs and ARBs in water and sediments of River Ganga (Reddy and Dubey 2019, Reddy and Dubey 2021). Since STPs receive a massive amount of wastewater for the treatment process, there is a prevalence of numerous contaminants in sludge, including antibiotics, ARGs and ARBs, as they remain persistent even after wastewater treatment. The application of this sludge for various purposes further provokes the risk of transmission of ARGs and ARBs in the environment. Monitoring sludge for AMR and ARB benefits public health by detecting early resistance, guiding targeted treatment and safeguarding the environment from resistant bacteria and genes. It aids compliance with regulations, supports research, and aligns with One Health approach for holistic health management. Therefore, the present work aimed to investigate the occurrence of ARGs and ARBs in sludge derived from STPs with the objectives (i) screening and characterization of MDR bacterial population and assessment of AMR genes (ermF, qnrS, Sul1, blaGES, blaCTX-M, and blaNDM) and MGEs (intl1 and IS26), (ii) examine the correlation between tolerance to heavy metals and the prevalence of AMR, (iii) establishment of correlation of ARGs with sludge physico-chemical parameters, and (iv) evaluation of MDR bacteria for biofilm production dynamics in response to different antibiotics.
Materials and methods
Experimental site and sludge sampling
Three different STPs with different range of treatment capacities from two different regions of Varanasi, India, designated as STP1 (Bhagwanpur) (25.27274 N and 83.00519 E), STP2 (Dinapur) (25.34762 N and 83.04844 E), and STP3 (Dinapur) (25.34762 N and 83.04844) with treatment capacity of 8, 80, and 140 million liters per day (MLD), respectively were chosen for study. Three aliquots of sewage sludge were taken from each STP drying tank, where the sludge was gathered for solidification in September 2021. The freshly accumulated sludges were collected in sterile plastic bags and brought to the laboratory. Then, it was sieved through a ∼2 mm sieve to physically remove the debris and kept at 4°C for microbial analysis and metagenomic DNA extraction.
Physico-chemical characteristics of sludge and wastewater
Fresh sludge samples were used for physico-chemical analysis. The pH and electrical conductivity (EC) of the sludge samples were measured in 1:2.5 (soil:water) suspensions (Jackson 1973). EC was measured using a digital conductivity meter (Systronics, India), and the pH by pH meter (CyberScan pH 510, Eutech Instruments Pte Ltd). The bulk density and water holding capacity (WHC) of the sludge were measured by the method suggested by Piper (1945). Walkley and Black (1934), micro-Kjeldhal techniques (Jackson 1958), ascorbic acid method (Olsen et al. 1965), and 1 N ammonium acetate method (Nelson and Heidel 1952) were used to measure the organic carbon, the total nitrogen (N), available phosphorus (P), and available potassium content in sludge samples, respectively. According to Nieuwenhuize et al. (1991), aqua regia (HCl:HNO3; 3:1) was used for the digestion of sludge samples in order to analyze total micronutrients and heavy metals in all sludge samples. The heavy metals (Cd, Cr, Ni, and Pb) and micronutrients (Cu, Mn, Fe, and Zn) were analyzed on an Atomic Absorption Spectrophotometer (AAS) (Agilent FS-240) (Lindsay and Norvell 1978). Physico-chemical characteristics of the influent wastewater and treated water were done by the previously described standard method (Reddy and Dubey 2021, Chourasia et al. 2022).
Isolation of ARBs
Serial dilution method in 0.9% saline solution and spread plate (10−5 dilution) on Luria Bertani (LB) agar plates supplemented with cycloheximide (50 µg ml−1) used for total bacterial count (TBC) of sewage sludges. For each sample, the colony forming unit (CFU) was counted in triplicate after the plates were incubated at 37°C for 48 h. Pure colonies were obtained by repeated streaking and sub-culturing on LB agar plates incubated for 48 h at 37°C. Single colonies were then streaked on the respective LB agar plates supplemented with 100 µg ml−1 of ampicillin. The obtained bacterial isolates, which were resistant to ampicillin (100 µg ml−1) and with different colony characters, were selected for further studies, and 50% glycerol stocks of isolates were maintained at −80°C for further experiments.
Antibiotic susceptibility testing and multiple antibiotic resistance index
Kirby Bauer disc diffusion method was used for antibiotic susceptibility testing (AST) for 14 antibiotics of different classes. The overnight-grown culture (0.1 ml) broth was picked on a sterile cotton swab and swabbed on Muller Hinton Agar (MHA) plates (Hi-media, PW1184). Discs of ampicillin (A) 10 µg, erythromycin (E) 15 µg, cefoxitin (FOX) 30 µg, chloramphenicol (C) 30 µg, ciprofloxacin (CF) 5 µg, cotrimoxazole (CO) 30 µg, gentamycin (G) 10 µg, kanamycin (K) 30 µg, penicillin-G (P) 10 U, ofloxacin (OFX) 5 µg, rifampicin (RD) 5 µg, streptomycin (S) 10 µg, tetracycline (TE) 30 µg, imipenem (IPM) 10 µg, and azithromycin (AZM) 15 µg, procured from HI Media. The sterile antibiotic discs were aseptically embedded on the swabbed plates, and the negative control consisted of 0.4 mm sterile filter paper immersed in sterile distilled water. After 24 h incubation at 37°C, the diameters corresponding to the zone of inhibition were measured. The sensitivity testing was carried out in accordance with Clinical and Laboratory Standards Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) instructions (Nguyen and Jones 2020), and the isolates were classified as resistant (R), intermediate (I), or susceptible (S). Bacterial isolates that were resistant to a minimum of three classes of antibiotics were categorized as MDR isolates, and the multiple antibiotic resistance index (MARI) was calculated for STPs as well as individual bacterial isolates. The formula for the calculation of sludge is MARI= a/(bxc); where a, b, and c represent the aggregate antibiotic resistant score, the number of antibiotics, and the number of bacterial strains isolated from the sample, respectively; while for each isolate, the MARI = a/b, where a and b are the number of antibiotics to which the isolate was resistant, and the number of antibiotics to which the isolate was exposed, respectively (Krumperman 1983). MARI value < 0.2 indicates a high-risk source of contamination (Christopher et al. 2013).
Identification of the MDR isolates, phylogenetic analysis and BIOLOG assay
Genomic DNA was isolated from an overnight grown culture of selected isolates using MasterPure complete DNA and RNA purification kit using manufacturer's protocol. The isolated genomic DNA was used for the amplification of V1–V9 region of the 16S rRNA gene with the help of the earlier reported standard polymerase chain reaction (PCR) procedure (Frank et al. 2008). PCR was performed using the universal primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGATT-3′) with the steps—initial denaturation at 94°C (5 min), 30 final denaturation cycles at 94°C (30 s), annealing at 52°C (1 min), followed by elongation at 72°C (1 min) and the 16S rRNA gene amplification was examined on 1.2% agarose gel. The amplified PCR product was purified using a MinElute PCR purification kit (Qiagen, Germany). The purified PCR products were subjected to 16S rRNA Sanger sequencing using an Applied Biosystems 3500 Genetic Analyzer (Thermo Fisher Scientific, USA). The Auto-Assembler program was used to edit and assemble raw sequences. The complete consensus sequences (∼1500 bp) of the 16S rRNA gene were compared with existing sequences on the National Center for Biotechnology Information (NCBI) GenBank database using the Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/BLAST) and all the consensus sequences were submitted to GenBank to obtain the accession numbers. A phylogenetic tree was constructed using the neighbor-joining statistical method using MEGA 11.0 software (Tamura et al. 2021).
BIOLOG assay was also carried out for six representative isolates to assess their metabolic potential and diversity test. A volume of 100 µl of Inoculation Fluid (IF) solution of Gen III Microplate BIOLOG sugar utilization assay kits containing culture was applied to GEN III MicroPlateTM (Biolog Inc., Hayward, CA, USA) and incubated at 37°C for 24 h (Mallick et al. 2018). Using IBM SPSS for Windows Version 25.0, the hierarchical cluster analysis method considered the Euclidean distances and produced the dendrogram (SPSS Inc. Chicago, IL, USA).
Metal tolerance, auto-aggregation, co-aggregation, and motility assay of MDR isolates
LB media (4 ml) with various concentrations of different heavy metal salts, including Pb(NO3)2, CuSO4.5H2O, MnSO4.H2O, K2Cr2O7, CdCl2, CoCl2.6H2O, and ZnSO4.7H2O were inoculated with 40 µl overnight grown pure culture of selected 16 different bacterial isolates. The concentrations of each heavy metal used were 0.5, 0.75, 1.0, 1.25, and 1.5 mg ml−1. Tolerance to the heavy metals of each isolate was measured on the basis of their visible growth within 12–36 h, and the absorbance at 600 nm above 0.6 after 36 h incubation at 37°C was regarded as positive. The auto-aggregation assay method suggested by Simões et al. (2008) was utilized. Stationary phase bacterial cells were harvested and centrifuged at 5000 x g (15 min), washed with phosphate buffer saline twice (PBS), pH 7.3 and re-suspended finally in PBS. 2.5 ml of thoroughly mixed bacterial suspensions were put on 24-well Falcon plate and incubated for 0, 2, and 24 h at 28°C. 0.2 ml of the suspension's above portion was transferred to a 96-well plate, and OD600 was measured on an Enzyme-linked immunosorbent assay (ELISA) auto reader (BIORAD 680). The formula for the auto-aggregation used was (1- At)/A0 x 100, where At is the absorbance at time t = 2 and 24 h and A0 at time t = 0. Co-aggregation assays were carried out as previously described for the auto-aggregation assay after overnight cultures had been mixed together (Vlkova et al. 2008). Visual auto-aggregation assay done as suggested by Pandey et al. (2022). For the motility test, semisolid motility test medium (1% tryptone, 0.5% sodium chloride, 0.3% agar, and 50 mg ml−1 Triphenyl tetrazolium chloride dye) was used, and motility halos assessed at 24, 48, and 72 h (Atkinson et al. 2006). All the experiments were performed in triplicate.
Congo Red Agar assay and dynamics of biofilm formation
Congo Red Agar (CRA) method was used as developed by Freeman et al. (1989). The biofilm assay was carried out utilizing the tissue culture plate (TCP) method, as previously described by Risal et al. (2018). LB media without antibiotics was distributed among 96 well plates. Antibiotics were added in the wells in gradually rising concentrations (µg ml−1) of ampicillin (10, 25, 50, and 100), kanamycin (30, 100, 150, and 300), ciprofloxacin (5, 10, 25, and 50), azithromycin (15, 50, 100, and 150), vancomycin (30, 100, 150, and 300), and clarithromycin (15, 50, 100, and 150) in succeeding wells. A volume of 5 µl of overnight grown culture was added in the microtiter plates incubated for 96 h at 37°C. Thoroughly washed the plate with double distilled water and filled the wells with 200 µl of crystal violet solution, incubated at 37°C for an hour. After that, the plate was again rinsed with 90% ethanol, incubated at room temperature (1 h), and run through a micro-ELISA auto reader (BIORAD 680) to determine the OD at 595 nm. Similar experiments were done for a 48 h setup.
Genomic characterization of different ARGs and MGEs in MDR isolates
The criteria for the selection of specific ARGs were based on a comprehensive analysis of the percentages of the resistance exhibited against various antibiotics in different isolates. Isolation of genomic DNA from MDR isolates was done by Master pure complete DNA and RNA purification kit (Lucigen, USA) and Plasmid DNA isolation using the QIAprep Spin Miniprep kit (Qiagen, Germany), both were then subjected to qualitative PCR for the detection of genes widely responsible for resistant to corresponding antibiotics blaTEM (ampicillin), blaGES (ampicillin), blaCTX (ampicillin), tetQ (tetracycline), tetM (tetracycline), tetW (tetracycline), rpoB516 (Rifampicin), rpoB526 (Rifampicin), rpoB531 (Rifampicin), ermF (erythromycin), qnrS (ciprofloxacin), blaNDM (imipenem), and Sul1 (cotrimoxazole and erythromycin) along with 16S rRNA and other MGEs like class 1 integron (intl1) and IS26 gene. PCR using high fidelity Taq Polymerase (Thermo Fisher Scientific, USA) performed as initial denaturation at 94°C (5 min), 35 cycles of denaturation (1 min), annealing (1 min) at the appropriate temperatures listed in Supplementary Table S1, extension (30 sec) at 72°C and a final extension step (7 min) at 72°C. Appropriate band sizes obtained for each target gene were checked on 1.2% agarose gel.
Total sludge metagenomic DNA extraction and qPCR
Metagenomic DNA of all the sludge samples was extracted using the FastDNA spin kit for soil (MP Biomedicals, CA, USA) based on the manufacturer's protocol. DNA concentration and quality were assessed by nanophotometer (Nanodrop 2000, Thermo Fisher Scientific) and confirmed by agarose gel electrophoresis.
The preparation of standard curves involved using plasmid DNA that contained target genes was performed. The number of plasmid DNA copies per microliter is given by the formula: Number of copies = [DNA mass concentration(ng) x 6.022 × 1023]/[Length of DNA (bp) x 660 × 109] (Zhang and Fang 2006). Ten-fold serial dilutions of the plasmid DNA carrying target ARGs were used to create seven-point standard curves for qPCR with copy counts ranging from 102 to 108. A master mix of 20 µl final volume was prepared consisting of 10 µl Maxima SYBR Green/ROX qPCR master mix (2x) (Thermo Fisher Scientific, USA), 1 µl of each primer (10 µM), 0.2 µl of Bovine serum albumin (BSA) (20 mg ml-1), 6.8 µl of nuclease-free water and 1 µl of DNA template. The PCR procedure was set up as follows: 10 min at 95°C, then 40 cycles for 15 s at 95°C and 1 min at 60°C, and finally, a final stage of the melt curve with a ramp from 60°C to 95°C. Each reaction was carried out three times with an additional non-template control, all qPCR tests were carried out in 96-well plates under standard conditions and according to the manufacturer's recommendations (Applied Biosystems). The program (Applied Biosystems 7500 v 2.3) calculated the qPCR efficiencies based on the standard curves. The PCR efficiencies varied from 80 to 110%, and all calibration curves had R2 values above 0.992. For absolute quantitative detection, 6 ARGs with two MGEs (IS26 and intl1) and 16S rRNA were chosen. All of the primers were created by Sigma Aldrich, India, which is listed in Supplementary Table S1, the 16S rRNA gene was also measured so that the gene abundance could be adjusted to the entire bacterial community.
Statistical analysis
In the sludge samples, the 16S rRNA gene (denoting total bacterial population) and the chosen target gene (ARGs and MGEs) were denoted as “log transformed gene copy number” per gram of dry sludge weight normalized to the DNA extraction yield. Principal component analysis (PCA) was performed using sludge physico-chemical parameters and metal abundance with the abundance of all six targeted ARGs, two MGEs, and three STPS. The analyses were performed using the OriginPro 2023 software by OriginLab (Massachusetts, USA). All the statistical graphs were generated with the help of GraphPad Prism 8 software (Boston, USA). Average, standard deviations (SD), and standard error of all the data were calculated with Microsoft Excel 2016. One-way analysis of variation (ANOVA), multivariate analysis of variation (MANOVA), and Pearson correlation analysis were carried out by IBM SPSS Statistics 25 software (Chicago, USA). 95% confidence intervals were always used to determine statistical significance (P <.05).
Results
Physico-chemical characteristics of sludge
The physico-chemical properties of the sludge samples of the three STPs are mentioned in Supplementary Table S2. Among these, STP3 sludge had higher bulk density, WHC, available P, and C/N ratio while lower EC and moisture content compared to the other two. In terms of metal composition, it exhibited higher concentrations of Fe, Cu, Zn, Cd, Cr, Ni, and Pb and lower Mn. Mn content was found to be higher in STP1. The heavy metal concentration in the sludge was found to be in the order of STP3>STP2 >STP1, with the exception of Mn, which was higher in STP1 (Supplementary Table S2). Heavy metals Cd, Zn, Fe, Cu, Mn, Cr, Ni, and Pb, were present in a range of (23.39–213.36), (147.86–1173.0), (474.33–778.09), (243.12–436.45), (189.42–257.15), (47.52–83.05), (61.85–90.536), and (42.09–89.006) mg kg−1 sludge, respectively. Zn was found to be most abundant, followed by Fe in sludge samples. The findings for influent wastewater and effluent treated water were also similar (Supplementary Table S3).
Screening and isolation of ARBs
STP sludges were screened for TBC and ampicillin-resistant population. Samples of STP2 contain a higher number of TBC (20.16 × 107 CFU g−1) followed by STP1 (19.98 × 107 CFU g−1) and STP3 (43.48 × 106 CFU g−1) (Supplementary Table S2). In the case of ampicillin-resistant population, this number was dropped to 12.51 × 107, 82.8 × 106, and 78.3 × 107 CFU g−1 for STP1, STP2 and STP3, respectively. Considering colony characteristics, a total of 30 cultivable ampicillin-resistant bacterial colonies were picked up from each STP sludge sample for antibiotic susceptibility test. The culture plate images and colony characteristics for 30 representative isolates from three STPs (10 from each STP) have been shown in Supplementary Fig. S10 and Supplementary Table S7, respectively.
AST and MARI
The antimicrobial susceptibility profiling of the selected 90 bacterial isolates from sludge samples was performed with 14 commonly prescribed antibiotics (including ampicillin); the results are shown in (Fig. 1A, B, C). All the isolates tested positive for ampicillin resistance, and the number of isolates showing resistance to cotrimoxazole, ciprofloxacin, chloramphenicol, streptomycin, rifampicin, gentamycin, ofloxacin, tetracycline, cefoxitin, kanamycin, azithromycin, penicillin G, and imipenem were 67, 74, 56, 56, 64, 44, 70, 60, 85, 55, 59, 81, and 76, respectively.
Figure 1.
Percentage of resistance for selected ampicillin-resistant bacterial isolates for STP sludge samples with 14 different tested antibiotics. (A) percentage of resistant isolates for STP1 sludge, (B) a percentage of resistant isolates for STP2, (C) a percentage of resistant isolates for STP 3, and (D) based on the resistance multiple antibiotic resistance index (MARI) is represented for three STPs.
Based on these findings, a bar diagram (Fig. 1D) was made to illustrate the changes in the MDR level in sludges derived from STPs. The MARI was highest for STP1 (0.79), followed by STP3 (0.76) and STP2 (0.67) (Fig. 1D). MARI for all 90 isolates ranged from 0.5 to 0.93). The percentage resistance to respective antibiotics for bacterial isolates from STP1 sludge were CO-86.67%, CF-100%, C-60%, S-60%, RD-73.33%, G-60%, OFX-93.33%, TE-70%, FOX-100%, K-60%, AZM-63.33%, P-90%, and IPM-90%; STP2 sludge were CO-86.67%, CF-76.67%, C-56.67%, S-96.67%, RD-60%, G-16.67%, OFX-73.33%, TE-50%, FOX-100%, K-56.67%, AZM-56.67%, P-80%, and IPM-86.67%, and STP3 sludge were CO-50%, CF-70%, C-70%, S-80%, RD-80%, G-70%, OFX-83.33%, K-66.66%, AZM-76.66%, P-100%, and IPM-76.66%. The MDR isolates having different colony characteristics, resistant to more than six classes of antibiotics and high MARI (0.64–0.93) were selected for further study. MARI values of 16 selected MDR isolates are given in Table 1.
Table 1.
Multiple antibiotic resistance index (MARI), motility (72 h), auto-aggregation % (24 h), and biofilm-forming ability of selected multi-drug resistance isolates.
| Isolates | MARI | Motility (72 h) (mm) | Auto-aggregation (24 h) (%) | Biofilm-forming ability (24 h) |
|---|---|---|---|---|
| Bacillus sp. BI3 | 0.71 | 90 | 36.67 | Strong |
| Chryseobacterium sp. BI5 | 0.93 | 90 | 24.51 | Moderate |
| Priestia sp. BI6 | 0.93 | 90 | 76.67 | Moderate |
| Microbacterium sp. BI8 | 0.92 | 8 | 23.47 | Strong |
| Staphylococcus sp. BI10 | 0.78 | 90 | 70.41 | Moderate |
| Pantoea sp. BI11 | 0.78 | 90 | 96.89 | Moderate |
| Microbacterium sp. BI13 | 0.64 | 9 | 53.33 | Strong |
| Rhodococcus sp. D1I2 | 0.85 | 3 | 11.76 | Weak |
| Pseudomonas sp. D1I7 | 0.78 | 90 | 10.89 | Strong |
| Glutamicibacter sp. D1I9 | 0.93 | 7 | 14.38 | Moderate |
| Pseudomonas sp. D2I1 | 0.78 | 90 | 71.13 | Strong |
| Enterobacter sp. D2I5 | 0.78 | 90 | 55.18 | Strong |
| Stenotrophomonas sp. D2I6 | 0.86 | 76 | 27.78 | Moderate |
| Sphingobacterium sp. D2I7 | 0.78 | 4 | 77.45 | Weak |
| Bacillus sp. D2I8 | 0.93 | 90 | 66 | Strong |
| Exiguobacterium sp. D2I9 | 0.93 | 90 | 88.02 | Weak |
Characterization of the MDR isolates and phylogenetic analysis
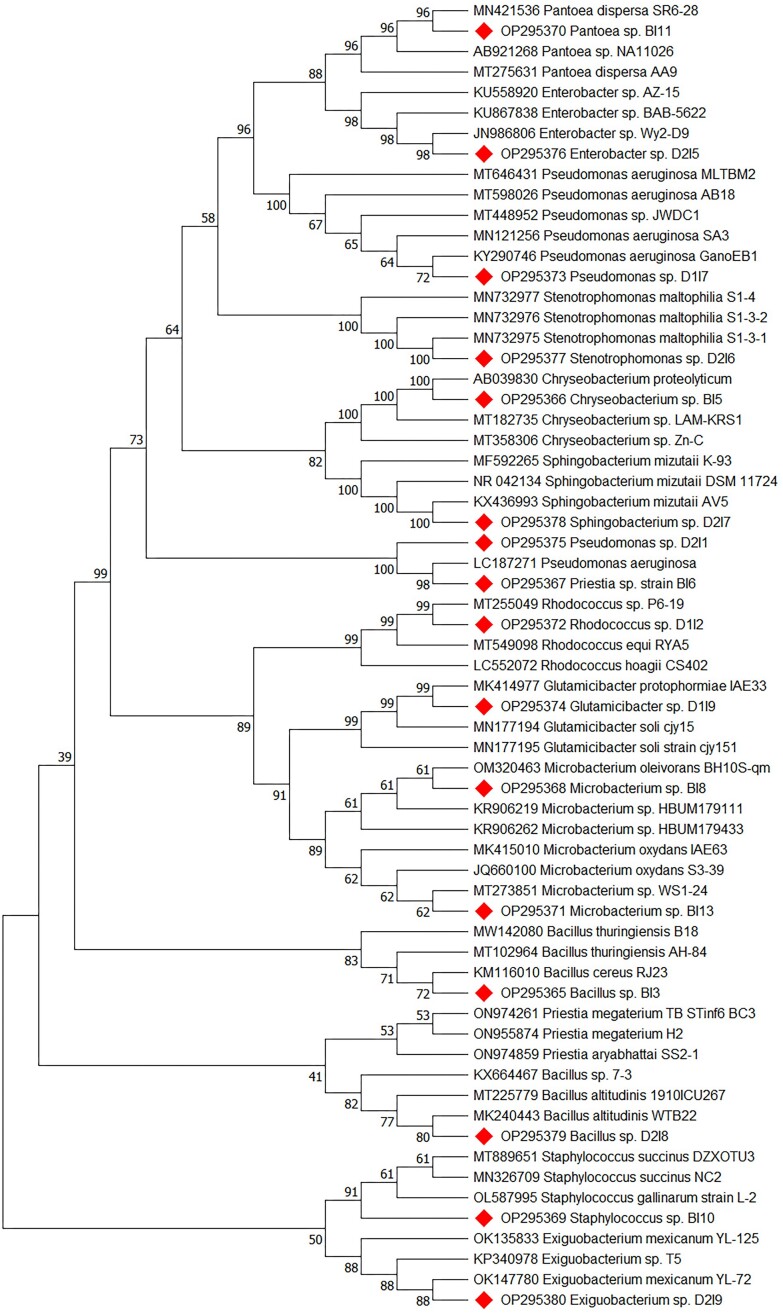
Based on 16S rRNA sequencing, the isolates were identified as Bacillus sp. strain BI3, Chryseobacterium sp. strain BI5, Priestia sp. strain BI6, Microbacterium sp. strain BI8, Staphylococcus sp. strain BI10, Pantoeasp. strain BI11, Microbacterium sp. strain BI13, Rhodococcus sp. strain D1I2, Pseudomonas sp. strain D1I7, Glutamicibacter sp. strain D1I9, Pseudomonas sp. strain D2I1, Enterobacter sp.strain D2I5, Stenotrophomonas sp. strain D2I6, Sphingobacterium sp. strain D2I7, Bacillus sp. strain D2I8, and Exiguobacterium sp. strain D2I9. There were 12 distinct genera altogether and dispersed among nine different orders, which included the actinobacteria and four other phyla of bacteria. The phylum Fermicutes and Proteobacteria shared a dominant number, with 10 isolates accounting for 62.5% of the detected bacteria. Four isolates (25%) were identified as Actinobacteria, while two (12.5%) isolates of the Bacteroidetes were detected. Furthermore, a Biolog assay was applied to assess the metabolic diversity of six representative bacterial isolates (Supplementary Table S4 and Supplementary Fig. S1).
The accession numbers of deposited 16S rRNA gene sequences to GenBank were obtained as OP295365–OP295380, and a phylogenetic tree was constructed and presented in Fig. 2.
Figure 2.
Neighbor-joining tree based on distance analysis representing the relationship between the 16S rRNA sequences of 16 bacterial isolates from three different STPs sludge samples and 48 reference sequences (16S rRNA gene) of the related species from NCBI GenBank. Bootstrap values generated from 500 replicates are shown at the nodes.
Metal tolerance assay
To test the co-occurrence of heavy metals and resistance to antibiotics, the metal tolerance ability of the bacterial isolates was investigated. The majority of the bacterial isolates were found to be tolerant to heavy metals than the negative control (E. coli). The results of tolerance assays to seven metals showed that 18.75, 12.5, 18.75, 12.5, 12.5, 12.5, 18.75, 12.5, and 12.5 % of the MDR isolates were tolerant to 500 mg l−1 of Cu2+, 750 mg l−1 of Cu2+, 1500 mg l−1 of Cr3+, 1500 mg l−1 of Pb2+, 750 mg l−1 of Mn2+, 1500 mg l−1 of Mn2+,1500 mg l−1 of Cd2+, 1500 mg l−1 of Co2+, and 1500 mg l−1 of Zn2+, respectively. It was not possible to determine the biocidal concentrations of zinc, cadmium, cobalt, and lead in the present study for > 80% of the strains, same way 68.25% of isolates for Cu and Mn, and 56.25% for Cr. The order of toxicity of the metals to the MDR isolates was found to be Cd>Pb=Co=Zn>Mn>Cu=Cr.
Auto-aggregation, co-aggregation, and motility assay of MDR isolates
The auto-aggregation percentages of the MDR isolates were found to increase with the incubation time (0, 2, and 24 h) (Table 1). The highest auto-aggregation percentage was observed for Pantoea sp. strain BI11 (96.89%) and Exiguobacterium sp. strain D2I9 (88.02%) followed by Sphingobacterium sp. strain D2I7 (77.45%), Priestia sp. strain BI6 (76.67%), Pseudomonas sp. strain D2I1 (71.13%), Staphylococcus sp. strain BI10 (70.41%), Bacillus sp. strain D2I8 (66%), Enterobacter sp. strain D2I5 (55.18%), and Microbacterium sp. strain BI13 (53.33%). The remaining MDR isolates were characterized by low auto-aggregative properties, such as Bacillus sp. strain BI3 (36.67%), Stenotrophomonas sp. strain D2I6 (27.78%), Chryseobacterium sp. strain BI5 (24.51%), Microbacterium sp. strain BI8 (23.47%), Glutamicibacter sp. strain D1I9 (14.38%), Rhodococcus sp. strain D1I2 (11.76%), and Pseudomonas sp. strain D1I7 (10.89%).
The co-aggregation investigation between low aggregative strains revealed that aggregation was more pronounced when Microbacterium sp. strain BI13 was combined with Rhodococcus sp. strain D1I2 and Pantoea sp. strain BI11 with Pseudomonas sp. strain D1I7, with 85.03 and 75.56% of co-aggregation, respectively. In addition, the visual auto-aggregation assay also showed visible flocs for all high auto-aggregative MDR strains and minimal visible flocs for other strains after a given time of incubation. Visible flocs were also observed for the co-aggregation set of low aggregative strains. A considerable number (68.75%) of the MDR isolates were found to be motile and showed a swarming motility pattern on 0.3% agar media plates. (Table 1). In contrast, the non-motile strains included Microbacterium sp. strain BI8, Microbacterium sp. strain BI13, Rhodococcus sp. strain D1I2, Glutamicibacter sp. strain D1I9, and Sphingobacterium sp. strain D2I7.
Biofilm formation dynamics and the produced phenotype
ATCC 35984, a slime-producing/ica-positive strain, and ATCC 12228, a slime-negative/ica-negative strain, were used as the positive and negative controls, respectively. The cut-off value (ODc) for each plate was determined as three SD above the mean optical density (OD) of the negative control: ODc = average OD of the negative control + (3 × SD of the negative control) (Plota et al. 2021). From this investigation, it was found that after 24 h of incubation, 18.75% of the isolates produced a lesser amount of biofilm (0.743 < ODs < 1.486), 37.5% of the isolates formed a moderate amount of biofilm (1.486 < ODs < 2.972), and rest 37.5% of the isolates formed strong biofilm (2.972 < ODs). The ability to form a biofilm of 16 MDR isolates is represented in Table 1. The results of the CRA assay revealed that in contrast to the reference negative bacteria (ATCC 12228), all identified MDR isolates tested positive for the CRA test. Most of the isolates showed black colonies with typical crystalline morphology. The TCP technique also recognized all of the isolates positive for biofilm formation (Supplementary Fig. S6).
Dynamics of biofilm formation by sludge isolates showed that when the antibiotic concentrations of the system were increased, the majority of MDR bacteria followed a Gaussian distribution pattern. The production of biofilm by the isolates was generally enhanced by lowering the concentration of antibiotics (ampicillin, kanamycin, azithromycin, ciprofloxacin, vancomycin, and clarithromycin) (Supplementary Figs. S2, S3, S4, S5, S6, S7). In Set B, when the selected antibiotics were added to the biofilm of the isolates after 48 h of incubation (when the biofilm had already developed), it was found that the bacteria in the biofilm displayed significant resistance to the antibiotics (ampicillin, kanamycin, azithromycin, ciprofloxacin, and vancomycin) (Supplementary Figs. S2, S3, S4, S5, S6, S7). In Set A, when the antibiotic was introduced prior to the bacterial inoculation, a decrease in the biofilm formation was observed at higher antibiotic concentrations by the majority of the isolates. BI8, D2I7, D1I9, D1I2, BI13, and D2I8 did not exhibit any discernible change in the production dynamics of biofilm for most of the antibiotic sets, although in case of isolates D2I1, BI5, and BI8 in the presence of ciprofloxacin, and D2I7 and D2I9 in the presence of ampicillin in Set A revealed that the biofilm was not impacted whereas in case of Set B, the formation of biofilm was gradually and significantly reduced with the increase in antibiotic concentration (Supplementary Figs. S2, S4). The majority of the bacterial isolates from set A, however, displayed greater resistance to clarithromycin (Supplementary Fig. S7). Most interestingly, it was also found that four isolates (BI10, BI11, D2I1, and D2I6) could survive clarithromycin concentrations up to 150 g ml−1 in Set A.
Qualitative assessment of different ARGs in MDR bacteria
In addition to the AST, the molecular genotype of these MDR isolates were revealed by PCR amplification employing primers targeting specific ARGs (blaNDM–100%, qnrS–100%, Sul1–87.5%, blaCTX–12.5%, blaGES–12.5%, Intl1–68.75%, IS26–75%; tetW–31.25%, bla TEM–100%, and rpoB516–75% of the MDR isolates). The amplification of blaTEM genes found in every isolate except D2I5, supported the finding that they had a high level of ampicillin resistance. Several MDR isolates depicted positive results for other genes like blaGES, tetQ, tetW, rpoB516, qnrS, blaNDM, and Sul1, along with 16S rRNA and other MGEs like intl1 and IS26 gene. No PCR amplification was found against tetM, rpoB526, rpoB531, ermF, and blaCTX genes. Among all the 16 MDR isolates, BI5 and BI6 were found to harbor the maximum number of ARGs (nine) in their genomic DNA, whereas D2I5 and D1I7 were found to harbor three and four ARGs, respectively (Table 2). Of 16 isolates 11 have plasmid, but no ARGs were amplified in the plasmid DNA.
Table 2.
Identification and characterization of 16 multi-drug resistance (MDR) isolates. Showing colors for the presence/absence of tested antibiotic resistance genes (ARGs) and mobile genetic elements (MGEs) (derived from PCR experiments) for genotype, also showing colors for resistant/intermediate/sensitive for antibiotic susceptibility test (AST). [Antibiotics denoted as Ampicillin (A), cotrimoxazole (CO), ciprofloxacin (CF), erythromycin (E), chloramphenicol (C), streptomycin (S), rifampicin (RD), gentamycin (G), ofloxacin (OFX), tetracycline (TE), cefoxitin (FOX), kanamycin (K), azithromycin (AZM), penicillin-G (P), and imipenem (IPM)].
Quantitative assessment of the ARGs and MGEs in STP samples and correlation with sludge physico-chemical parameters
The amplification efficiencies of qPCR for all target genes varied between 80.115% and 114.32% with good linearity (Supplementary Fig. S8). The qPCR results showed a higher copy number of all the target resistance genes than the detection limit. The number of gene target copies per gram of sludge varied greatly on a dry weight basis, from ∼6.6 × 103 (qnrS in STP1) to ∼6.5 × 108 (Sul1 in STP2) (Figs. 3, 4 and Supplementary Table S5). The bacterial population harboring ARG blaCTX gene were ∼0.44%, ∼0.009%, and ∼0.056%; ermF gene were ∼1.26%, ∼1.58%, and ∼2.27%; blaGES were ∼0.95%, ∼0.87%, and ∼0.49%; intl1 were ∼3.83%, ∼11.59%, and ∼7.72%; IS26 were ∼0.52%, ∼11.63%, and ∼24.42%; qnrS were ∼0.0009%, ∼0.0011%, ∼0.0060%, and Sul1 were ∼12.72%, ∼29.42%, and ∼21.94%, in STP1, STP2, and STP3 sludge, respectively. The absolute copies number for all the target genes was normalized to the basal level of 16S rRNA genes in order to minimize the variance caused by differences in the abundance background bacterial population (Figs. 3, 4 and Supplementary Table S5). The absolute abundance of the target ARGs showed significant variation, ranging from 2.0 × 105 to 3.1 × 106 copies g−1 sludge. STP2 sludge had a lower concentration of blaCTX gene (2.0 × 105), one-fold lower than STP1 and STP3 sludge samples. In the case of the ermF gene, the copies number varied from (3.5 × 107 to 8.7 × 106) g−1 sludge and was lower in abundance in STP2 sludge. blaGES gene abundance was higher in STP2 sludge and the copies number ranged from (6.6 × 106 to 1.9 × 107) g−1 sludge. Class 1 integron copies number ranged from 2.6 × 107 to 2.5 × 108 g−1 sludge, higher abundance was seen in STP2 sludge and least abundance in STP1. IS26 copies number varied in 100-folds (3.6 × 106 to 4.6 × 108) g−1 sludge Most abundance of IS26 gene copies number was observed in STP3 and least in STP2. blaNDM copies number ranged from 4.6 × 105 to 1.4 × 106 g−1 sludge and STP1 had shown lower abundance. qnrS was in lower concentration among the other 7 ARGs which ranged from 6.6 × 103 to 1.1 × 105 g−1 sludge. The abundance of qnrS was lower in STP1 sludge and higher in STP3. sul1 gene copies number ranged from 8.9 × 107 to 6.5 × 108 g−1 sludge, STP1 and STP2 had lower and higher abundance of sul1 gene, respectively (Figs. 3, 4 and Supplementary Fig. S9).
Figure 3.
Quantitative real-time polymerase chain reaction (qRT-PCR) for the enumeration of six antibiotic resistance genes (ARGs) (ermF, qnrS, Sul1, blaNDM, blaGES, and blaCTX) and two mobile genetic elements (MGEs) (intl1 and IS26) along with 16S rRNA gene in the three STPs sludge samples. Values (log-transformed) are expressed for the gene copy number per gram of the dry weight of sludge, normalized to the DNA extraction yield.
Figure 4.
The box plot represents the first and third quartile of the six antibiotic resistance genes (ARGs) (A) ermF, (B) qnrS, (C) Sul1, (D) blaGES, (E) blaCTX, (F) blaNDM, and two mobile genetic elements (MGEs) (G) and intl1 (H) IS26 target genes for three STP sludge samples.
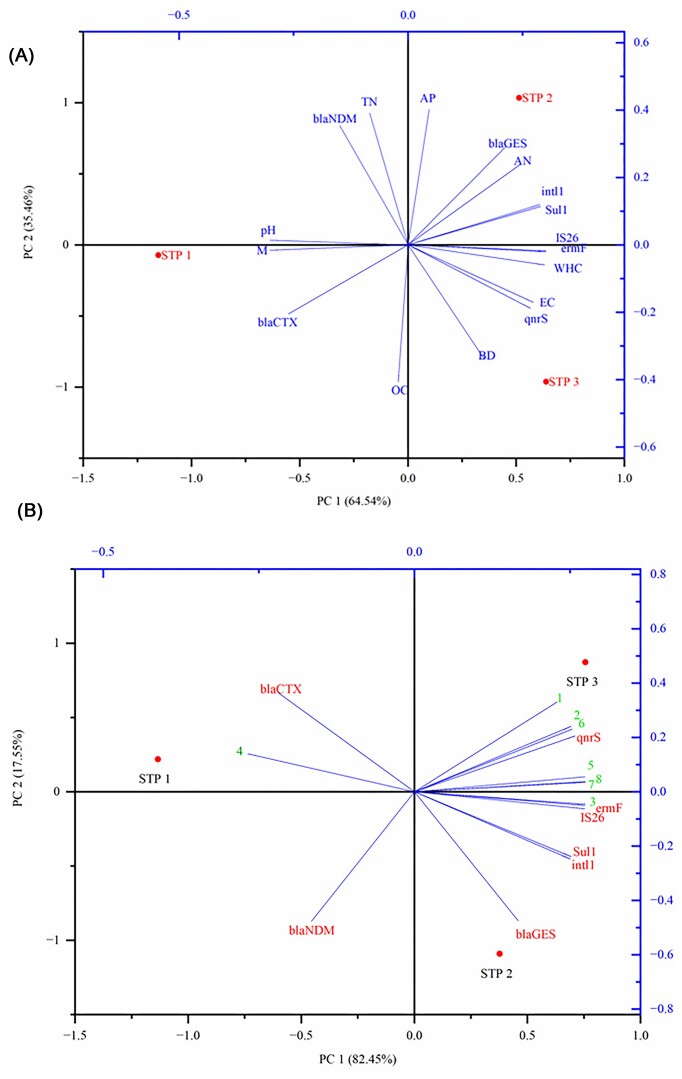
PCA was used considering metal concentration, AMR gene copies number, and different STP sites to evaluate the impact of sludge physico-chemical parameters on the AMR gene abundance and distribution. The abundance of qnrS, ermF, and IS26 genes and the STP3 were positively correlated with the amounts of bulk density, WHC, and electrical conductivity and negatively correlated with moisture and pH (Fig. 5A). On the other hand, blaGES, intl1, sul1, and blaNDM genes have shown positive association with total N, available N, and available P with the STP2 (Fig. 5A). Again, moisture, pH, and organic carbon form a positive link with the blaCTX gene copy numbers with STP1. Similarly, a positive association of metals (Fe, Cu, Zn, Cd, Cr, Ni, and Pb) and qnrS, ermF, IS26, intl1, Sul1, and blaGES genes with STP2 and STP3 (Fig. 5B) and a negative correlation with Mn were found. On the other hand, blaCTX, blaNDM, and Mn form a positive correlation with STP1 (Fig. 5B).
Figure 5.
Principal component analysis (PCA) showing the correlations of ARGs and MGEs gene abundance and different STPs with (A) the physico-chemical parameters denoted as BD = bulk density, WHC = water holding capacity, M = moisture, pH, OC = organic carbon, TN = total nitrogen, AN = available nitrogen, AP = available phosphorus, and EC = electrical conductivity, with antibiotic resistance genes (ARGs) and mobile genetic elements (MGEs) gene abundance, and different STPs (STP1–Bhagwanpur 8 MLD, STP2–Dinapur 80 MLD, and STP3–Dinapur 140 MLD), and (B) Metals denoted as 1 = Fe, 2 = Cu, 3 = Zn, 4 = Mn, 5 = Cd, 6 = Cr, 7 = Ni, and 8 = Pb.
Discussion
Physical parameters such as bulk density, WHC, EC, and moisture and chemical parameters such as heavy metals constituents, organic matter content, nutrients, pH, persistent organic pollutants (POPs), etc. are important determinants of sludge quality. The main sources of heavy metals in sewage sludge are domestic and industrial wastewaters, sewer system corrosion, surface runoff, pharmaceuticals and personal care products, etc. (Rizzardini and Goi 2014, Cantinho et al. 2016). In the present study, the metal concentrations in the sludge were found to be often higher than those in the influent from the STP and the reclaimed wastewater (Supplementary Tables S2, S3). Most of the analyzed metals had concentrations that were within the acceptable range, as per the international regulatory agency, USA Regulation 40 CFR Part 503 503.13. Pb, Zn, and Cd were found to vary greatly, while Ni amounts varied less. Cu and Zn content in STP2 and STP3 were relatively higher. This may be due to the fact that they primarily treat urban wastewater from the main city; the concentration was higher than that of STP1, which primarily treats wastewater from domestic and agricultural sources. Singh and Agrawal (2010a,b,c) also observed similar findings regarding STP2, but contrasting results for STP1 were obtained, a much higher concentration of heavy metals than previous studies (Jatav et al. 2021). The high metal concentration in sludge samples needs careful management and monitoring to guard against potential dangers to human health and the environment (Tytła et al. 2023). A lesser variation in organic C and other parameters was found among the three samples, except P. High levels of organic matter, available P, and N found in sludge may indicate toward higher bacterial counts (Zhang et al. 2017, Turek et al. 2019). The TBC reveals the bacterial population in sludge, and the screening for multi-antibiotic resistance depicts the resistant bacterial population. Primary screening with Ampicillin is used as it is one of the most widely used antibiotics to which resistance has developed over the course of time (Timraz et al. 2017). This study revealed a large population of ampicillin-resistant bacteria in all three sludge samples. Further, screening showed a number of isolates resistant to the 14 most prescribed antibiotics via AST. Based on these findings, the MARI revealed that 16 bacterial isolates were resistant to multiple antibiotics. Almost all the isolates have shown resistance against the β-lactam derivatives like penicillin, cephalosporin, and ampicillin. This may be indicative that the wastewater coming from numerous sources in STPs has antibiotic residues that could not be removed completely even after treatment, and the presence of these antibiotics in sludge in higher amounts exhibits selection pressure on resistant bacterial communities to survive in these environment by developing resistance over time through various mechanisms, including β-lactamase production, efflux pumps, altered penicillin-binding protein, etc. in order to survive in these environment (Worthington and Melander 2013, Giani et al. 2017). Also, STP1 sludge has higher MARI, hence the highest rate of MDR. The MARI for sites and isolates were all greater than 0.2, suggesting that these three STP sites are at high risk of ARGs and ARBs contamination.
The cultivable bacterial population harboring the sludge samples represents a taxonomic diversity. The phylogenetic analysis revealed 12 distinct genera altogether and dispersed among nine different orders, which included the actinobacteria and four other phyla of bacteria. Among these, Fermicutes and Proteobacteria were predominant, collectively representing 62.5% of the detected bacteria, demonstrating a significant microbial diversity. This diversity sets this study apart from many culture-based studies, which often focus on a limited number of genera by exploring a broader range of bacterial strains across various orders and phyla. Nascimento et al. (2018), found Clostridium to be the abundant genus, followed by Treponema, Propionibacterium, Syntrophus, and Desulfobulbus. Acinetobacter baumannii, K. pneumonia, S. aureus, P. aeruginosa, etc. are common examples of bacteria posing resistance to a large class of antibiotics (Akpaka et al. 2017, Vázquez-López et al. 2020, Ahmed 2022, Li et al. 2022).
The present study also investigated the influence of metal tolerance, auto-aggregation as well as co-aggregation, motility and biofilm production ability of the bacterial isolates in resistance development. By employing both the CRA method and the TCP method, we aimed to ensure a comprehensive and multi-faceted examination of biofilm formation. The tissue culture approach, with its categorization of biofilm strength, provided a detailed and quantitative assessment that complemented the qualitative insights gained from the CRA method. Results elucidated that most MDR isolates have a high tolerance profile to various heavy metals (highest for Cd), good auto-aggregation (highest for Pantoea sp.), as well as co-aggregation (Microbacterium sp. with Rhodococcus sp. and Pantoea sp. with Pseudomonas sp.) properties, and considerable mobility (except four isolates). All five non-motile bacterial strains do not form a strong biofilm. It is well known that non-motile bacterial mutants lack the ability to form biofilms (Basson et al. 2008). Thus, bacterial motility is one of the key elements for the creation of biofilms as it impacts bacterial adhesion to various surfaces (Jałowiecki et al. 2018). Ampicillin, kanamycin, azithromycin, ciprofloxacin, clarithromycin, and vancomycin were among the antibiotics at lower concentrations that helped the MDR sludge isolates build biofilms. This signifies that bacteria sense antibiotics in the sub-MIC concentration and start producing auto-inducer molecules ultimately leading to cell density-dependent signaling pathways such as biofilm induction (Padder et al. 2018). Also, the CRA assay classified all MDR strains to be biofilm-positive. Chryseobacterium sp., P. aeruginosa, S. aureus, and K. pneumonia, Glutamicibacter sp., Exiguobacterium sp., and Bacillus sp. are some of the well-known biofilm producers (Bagge et al. 2004, Chang et al. 2015, Nirwati et al. 2019, Futo et al. 2021, Hoque et al. 2022). Biofilms enhance the emergence resistance phenomena by shielding bacteria against exposure to different antibiotics, promoting genetic exchange (horizontal gene transfer), spreading virulence genes, and boosting stress tolerance (Fisher et al. 2017).
A number of variables, including the physico-chemical characteristics of the sludge, can affect the prevalence of ARGs in sewage sludge. PCA was used to evaluate the impact of sludge physico-chemical parameters on the AMR gene abundance (Fig. 5A) and distribution. This study revealed a strong correlation between the abundance and distribution of the ARGs and MGEs gene and the amounts of metals, and it also indicates the necessity of physico-chemical parameters to regulate AMR gene abundance (Fig. 5A and Supplementary Table S6A). A strong correlation of STP2 and STP3 was obtained with the abundance and distribution of the ARGs and MGEs gene. It may be because of the high amount of contaminated wastewater coming into these two STPs. It represents a relationship between anthropogenic factors and ARG abundance. Also, there is a favorable correlation between the abundance of ARGs in sludge and its bulk density, water retention capacity, and electrical conductivity. The different pH optimums, amount of total organic matter and the availability of nutrients in the sludge may also have an impact on ARG abundance, as these elements can potentially affect bacterial growth and activity.
Qualitative assessment revealed that most of the target genes were present in chromosomal DNA as the plasmid lacked the amplification of these genes. The possible reasons could be genomic DNA mutations taking part in acquired resistance (Van Hoek et al. 2011), loss of a part of the plasmid, etc. (Carroll and Wong 2018). The mechanisms through which resistance genes are acquired, as well as their existence and distribution, can differ amongst bacterial strains. Quantitative assessment through qPCR demonstrated the variance in the number of target gene copies between the various sludge samples, which ranged up to a 100-fold. It may be due to the consideration that STPs with high MLD can process large amounts of wastewater and hence have a high concentration of organic matter and nutrients, providing a selection pressure on the microbial communities to develop resistance toward various antibiotics. This may also create a situation where bacteria can easily exchange mobile genetic components (plasmids, integrons, and transposons) containing ARGs (Tao et al. 2022). Here, intl1 and IS26 showed a substantial positive correlation with most of the targeted ARG families (P ≤ .05) (Supplementary Table S6b), like sulfonamides, β-lactamases, and quinolones; concluding that ARGs linked to MGEs for migration between species in the sludge and also to the respective sink environment. The exact STP design, operational circumstances, and location can all have an impact on the causes for the higher abundance of ARGs in STPs. Therefore, research is required to better understand the link between STPs’ MLD capacity and ARG abundance.
The qualitative and quantitative assessment of resistant determinants in sludge derived from STPs decipher that sludge serves as a prominent factor for the dissemination of ARGs and ARBs. It is the need of the hour to have a critical risk assessment and the management of the sewage sludge and treated water to reduce the spread of MDRs and ARGs to the environmental matrices such as effluent receiving water bodies and sludge-amended agricultural fields.
Conclusions
The present investigation concludes that sludge derived from STPs contained a diverse group of bacterial communities that were resistant to various antibiotics. Chryseobacterium sp. strain BI5, Stenotrophomonas sp. D2I6, Pantoea sp. BI11 strain, Pseudomonas sp. strain D2I7, Enterobacter sp. strain D2I5, and Sphingobacterium sp. strain D2I7 were found to be resistant to multiple drugs. Nearly all the isolates exhibited resistance against the β-lactam derivatives like penicillin, cephalosporin, and ampicillin. AMR genes (blaTEM, blaGES, tetQ, tetW, rpoB516, qnrS, blaNDM, and Sul1), and MGEs (intl1 and IS26) were detected in bacterial genomic DNA significantly. Also, the abundance of various target genes was found to be varied in accordance with sludge from different STPs. A powerful correlation between the abundance and distribution of the ARGs and MGEs gene, the amounts of metals and high biofilm production ability deduce that they aid in antibiotic resistance. Most of the studied isolates could survive in high vancomycin (last resort of antibiotics) and clarithromycin concentrations, indicating the severity of the AMR spreading. The outcomes offer a new window into the complexities of microbial physiology and may offer some guidance in preventing the emergence and subsequent spread of antibiotic resistance. Overall, the research contributes valuable insights into the intricate relationships between sludge characteristics, metal exposure, and antibiotic resistance in bacterial populations, offering a holistic perspective on the environmental implications of antibiotic resistance, by providing peculiar indications of how sludge can act as a key driver of AMR gene/bacteria transmission in the environment. Also, the usage of STP sludge could contaminate the agroecosystems with resistance genes. Therefore, the ingression of ARGs in the environment through STP sludge should be investigated further, and there is a need to formulate potential strategies to combat the spread of AMR in the environment and upgrade the existing STP technologies to remove the contaminants of emerging concern.
Supplementary Material
Acknowledgments
The General Manager, Ganga Pollution and Prevention Unit (Urban), Uttar Pradesh Jal Nigam, Varanasi and the Project Manager, Bhagwanpur Sewage Treatment Plant are gratefully acknowledged for assistance during sludge and wastewater sampling.
Contributor Information
Mrinmoy Patra, Molecular Ecology Laboratory, Department of Botany, Institute of Science, Banaras Hindu University, Varanasi 221005, India.
Bhavana Pandey, Molecular Ecology Laboratory, Department of Botany, Institute of Science, Banaras Hindu University, Varanasi 221005, India.
Suresh Kumar Dubey, Molecular Ecology Laboratory, Department of Botany, Institute of Science, Banaras Hindu University, Varanasi 221005, India.
Author contributions
Mrinmoy Patra (Analysis, Investigation, Writing – original draft), Bhavana Pandey (Analysis, Writing – editing), and Suresh Kumar Dubey (Conceptualization, Funding acquisition, Supervision, Writing – review & editing).
Ethical approval
This study did not contain any data associated with human and animal studies.
Conflict of interest
The authors declared that they have no known competing financial interest or personal relationship that could have appeared to influence the work reported in this paper.
Funding
S.K.D. is thankful to Banaras Hindu University (BHU), Varanasi, India, for financial assistance under the Institution of Eminence (IoE) [Scheme No. 6031] for the completion of this research. B.P. is thankful to DBT, Government of India for financial assistance in the form of a Junior Research Fellowship [DBTHRDPMU/JRF/BET-21/I/2021-22/153].
Data Availability
The 16S rRNA gene sequences for bacterial isolates used in this study are available in NCBI database with accession number OP295365–OP295380.
References
- Aggarwal A, Bhalla M, Fatima KH. Detection of New Delhi metallo-beta-lactamase enzyme gene bla NDM-1 associated with the int-1 gene in gram-negative bacteria collected from the effluent treatment plant of a tuberculosis care hospital in Delhi, India. Access Microbiol. 2020;2. 10.1099/acmi.0.000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad I, Malak HA, Abulreesh HH. Environmental antimicrobial resistance and its drivers: a potential threat to public health. J Glob Antimicrob Resist. 2021;27:101–11. 10.1016/j.jgar.2021.08.001. [DOI] [PubMed] [Google Scholar]
- Ahmed OB. Detection of antibiotic resistance genes in Pseudomonas aeruginosa by whole genome sequencing. Infect Drug Resist. 2022;15:6703–9. 10.2147/IDR.S389959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiba M, Sekizuka T, Yamashita A et al. Distribution and relationships of antimicrobial resistance determinants among extended-spectrum-cephalosporin-resistant or carbapenem-resistant Escherichia coli isolates from rivers and sewage treatment plants in India. Antimicrob Agents Chemother. 2016;60:2972–80. 10.1128/AAC.01950-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akpaka PE, Roberts R, Monecke S. Molecular characterization of antimicrobial resistance genes against Staphylococcus aureus isolates from Trinidad and Tobago. J Infect Public Health. 2017;10:316–23. 10.1016/j.jiph.2016.05.010. [DOI] [PubMed] [Google Scholar]
- Al-Jassim N, Ansari MI, Harb M et al. Removal of bacterial contaminants and antibiotic resistance genes by conventional wastewater treatment processes in Saudi Arabia: is the treated wastewater safe to reuse for agricultural irrigation?. Water Res. 2015;73:277–90. 10.1016/j.watres.2015.01.036. [DOI] [PubMed] [Google Scholar]
- Atkinson S, Chang CY, Sockett RE et al. Quorum sensing in Yersinia enterocolitica controls swimming and swarming motility. J Bacteriol. 2006;188:1451–61. 10.1128/JB.188.4.1451-1461.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagge N, Schuster M, Hentzer M et al. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and β-lactamase and alginate production. Antimicrob Agents Chemother. 2004;48:1175–87. 10.1128/AAC.48.4.1175-1187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson A, Flemming LA, Chenia HY. Evaluation of adherence, hydrophobicity, aggregation, and biofilm development of Flavobacterium johnsoniae-like isolates. Microb Ecol. 2008;55:1–14. 10.1007/s00248-007-9245-y. [DOI] [PubMed] [Google Scholar]
- Beg AZ, Khan AU. Genome analyses of bla NDM-4 carrying ST 315 Escherichia coli isolate from sewage water of one of the Indian hospitals. Gut Pathog. 2018;10:1–6. 10.1186/s13099-018-0247-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombaywala S, Dafale NA, Jha V et al. Study of indiscriminate distribution of restrained antimicrobial resistome of different environmental niches. Environ Sci Pollut Res. 2021;28:10780–90. 10.1007/s11356-020-11318-6. [DOI] [PubMed] [Google Scholar]
- Boxall AB. The environmental side effects of medication: how are human and veterinary medicines in soils and water bodies affecting human and environmental health?. EMBO Rep. 2004;5:1110–6. 10.1038/sj.embor.7400307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantinho P, Matos M, Trancoso MA et al. Behaviour and fate of metals in urban wastewater treatment plants: a review. Int J Environ Sci Technol. 2016;13:359–86. 10.1007/s13762-015-0887-x. [DOI] [Google Scholar]
- Carroll AC, Wong A. Plasmid persistence: costs, benefits, and the plasmid paradox. Can J Microbiol. 2018;64:293–304. 10.1139/cjm-2017-0609. [DOI] [PubMed] [Google Scholar]
- Chang YC, Lo HH, Hsieh HY et al. Identification, epidemiological relatedness, and biofilm formation of clinical chryseobacterium indologenes isolates from central Taiwan. J Microbiol Immunol Infect. 2015;48:559–64. 10.1016/j.jmii.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Cho S, Hiott LM, Read QD et al. Distribution of antibiotic resistance in a mixed-use watershed and the impact of wastewater treatment plants on antibiotic resistance in surface water. Antibiotics. 2023;12:1586. 10.3390/antibiotics12111586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourasia A, Singh AP, Chattopadhyay A et al. Seasonal variation in physico-chemical properties of effluent generated at a sewage treatment plant in Varanasi and its suitability for irrigation. Int J Environ Clim Change. 2022;12:895–907. 10.9734/IJECC/2022/v12i121529. [DOI] [Google Scholar]
- Christopher AF, Hora S, Ali Z. Investigation of plasmid profile, antibiotic susceptibility pattern multiple antibiotic resistance index calculation of Escherichia coli isolates obtained from different human clinical specimens at tertiary care hospital in Bareilly-India. Ann Trop Med Public Health. 2013;6:285. 10.4103/1755-6783.120985. [DOI] [Google Scholar]
- da Silva Souza T, Lacerda D, Aguiar LL et al. Toxic potential of sewage sludge: histopathological effects on soil and aquatic bioindicators. Ecol Indic. 2020;111:105980, 10.1016/j.ecolind.2019.105980. [DOI] [Google Scholar]
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–33. 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Qiao M, Zhong J et al. Characterization of antibiotic resistance genes and bacterial community in selected municipal and industrial sewage treatment plants beside Poyang Lake. Water Res. 2020;174:115603. 10.1016/j.watres.2020.115603. [DOI] [PubMed] [Google Scholar]
- Diwan V, Chandran SP, Tamhankar AJ et al. Identification of extended-spectrum β-lactamase and quinolone resistance genes in Escherichia coli isolated from hospital wastewater from central India. J Antimicrob Chemother. 2012;67:857–9. 10.1093/jac/dkr564. [DOI] [PubMed] [Google Scholar]
- Fisher RA, Gollan B, Helaine S. Persistent bacterial infections and persister cells. Nat Rev Micro. 2017;15:453–64. 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- Frank JA, Reich CI, Sharma S et al. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microb. 2008;74:2461–70. 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman DJ, Falkiner FR, Keane CT. New method for detecting slime production by coagulase negative Staphylococci. J Clin Pathol. 1989;42:872–4. 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futo M, Opašić L, Koska S et al. Embryo-like features in developing Bacillus subtilis biofilms. Mol Biol Evol. 2021;38:31–47. 10.1093/molbev/msaa217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giani T, Antonelli A, Caltagirone M et al. Evolving beta-lactamase epidemiology in Enterobacteriaceae from Italian nationwide surveillance, October 2013: kPC-carbapenemase spreading among outpatients. Euro Surveill. 2017;22:30583. 10.2807/1560-7917.ES.2017.22.31.30583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruge KS, Tamamura YA, Goswami P et al. The association between antimicrobials and the antimicrobial-resistant phenotypes and resistance genes of Escherichia coli isolated from hospital wastewaters and adjacent surface waters in Sri Lanka. Chemosphere. 2021;279:130591. 10.1016/j.chemosphere.2021.130591. [DOI] [PubMed] [Google Scholar]
- Holmes AH, Moore LS, Sundsfjord A et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2006;387:176–87. 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- Hoque MN, Hannan A, Imran S et al. Plant growth-promoting rhizobacteria-mediated adaptive responses of plants under salinity stress. J Plant Growth Regul. 2022;42:1307–26. 10.1007/s00344-022-10633-1. [DOI] [Google Scholar]
- Jackson ML. Soil Chemical Analysis, Vol. 498. Englewood Cliffs, NJ: Prentice Hall Inc, 1958, 183–204. [Google Scholar]
- Jackson ML. Soil Chemical Analysis, Vol. 498. New Delhi: Pentice Hall of India Pvt. Ltd, 1973, 151–4. [Google Scholar]
- Jałowiecki Ł, Żur J, Chojniak J et al. Properties of antibiotic-resistant bacteria isolated from onsite wastewater treatment plant in relation to biofilm formation. Curr Microbiol. 2018;75:639–49. 10.1007/s00284-017-1428-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jatav HS, Singh SK, Jatav SS et al. Sewage sludge quality assessment of sewage treatment plant, Bhagwanpur, Varanasi and its safe utilization in agriculture. J Environ Biol. 2021;42:512–7. 10.22438/jeb/42/2(SI)/SI-271. [DOI] [Google Scholar]
- Jones ER, Van Vliet MT, Qadir M et al. Country-level and gridded estimates of wastewater production, collection, treatment and reuse. Earth Sys Sci Data. 2021;13:237–54. 10.5194/essd-13-237-2021. [DOI] [Google Scholar]
- Kaur R, Wani SP, Singh AK et al. Wastewater production, treatment and use in India. In: National Report Presented at the 2nd Regional Workshop on Safe Use of Wastewater in Agriculture. 2012, 1–13. https://www.ais.unwater.org/ais/pluginfile.php/356/mod_page/content/114/CountryReport_India.pdf. [Google Scholar]
- Kesari KK, Soni R, Jamal QMS et al. Wastewater treatment and reuse: a review of its applications and health implications. Water Air Soil Pollut. 2021;232:1–28. 10.1007/s11270-021-05154-8. [DOI] [Google Scholar]
- Kölbel J, Strong C, Noe C et al. Mapping public water management by harmonizing and sharing corporate water risk information. World Res Institute. 2018;1–20. http://www.wri.org/publication/mapping-public-water. [Google Scholar]
- Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microb. 1983;46:165–70. 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Xin L, Peng C et al. Prevalence and antimicrobial susceptibility profiles of ESBL-producing Klebsiella pneumoniae from broiler chicken farms in Shandong Province, China. Poult Sci. 2022;101:102002. 10.1016/j.psj.2022.102002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay WL, Norvell W. Development of a DTPA soil test for zinc, iron, manganese, and copper. Soil Sci Soc Am J. 1978;42:421–8. 10.2136/sssaj1978.03615995004200030009x. [DOI] [Google Scholar]
- Maity SK, Tyagi U, Sirohi S et al. Fabrication of biocompatible chitosan/graphene based nanocomposite for the competitive adsorption of heavy metal ions from wastewater in binary and ternary systems: scale-up and upgradation studies. J Water Process Eng. 2023;56:104555. 10.1016/j.jwpe.2023.104555. [DOI] [Google Scholar]
- Mallick I, Bhattacharyya C, Mukherji S et al. Effective rhizoinoculation and biofilm formation by arsenic immobilizing halophilic plant growth promoting bacteria (PGPB) isolated from mangrove rhizosphere: a step towards arsenic rhizoremediation. Sci Total Environ. 2018;610:1239–50. 10.1016/j.scitotenv.2017.07.234. [DOI] [PubMed] [Google Scholar]
- Manaia CM, Rocha J, Scaccia N et al. Antibiotic resistance in wastewater treatment plants: tackling the black box. Environ Int. 2018;115:312–24. 10.1016/j.envint.2018.03.044. [DOI] [PubMed] [Google Scholar]
- Mohajerani A, Lound S, Liassos G et al. Physical, mechanical and chemical properties of biosolids and raw brown coal fly ash, and their combination for road structural fill applications. J Clean Prod. 2017;166:1–11. 10.1016/j.jclepro.2017.07.250. [DOI] [Google Scholar]
- Mokni-Tlili S, Hechmi S, Ouzari HI et al. Co-occurrence of antibiotic and metal resistance in long-term sewage sludge-amended soils: influence of application rates and pedo-climatic conditions. Environ Sci Pollut Res. 2022;30:26596–612. 10.1007/s11356-022-23802-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CJ, Ikuta KS, Sharara F et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet North Am Ed. 2022;399;629–55. 10.1016/S140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento AL, Souza AJ, Andrade PAM et al. Sewage sludge microbial structures and relations to their sources, treatments, and chemical attributes. Front Microbiol. 2018;9:1462. 10.3389/fmicb.2018.01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson LB, Heidel H. Soil analysis methods as used in the Iowa state college soil testing laboratory. 1952.
- Nguyen HM, Jones RN. Reanalysis of cefazolin surrogate susceptibility breakpoints utilized as guidances for oral cephalosporin treatments of uncomplicated urinary tract infections: caution concerning application to cefadroxil. Diagn Microbiol Infect Dis. 2020;97:115053. 10.1016/j.diagmicrobio.2020.115053. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuize J, Poley-Vos CH, ávan den Akker AH et al. Comparison of microwave and conventional extraction techniques for the determination of metals in soil, sediment and sludge samples by atomic spectrometry. Analyst. 1991;116:347–51. 10.1039/AN9911600347. [DOI] [Google Scholar]
- Nirwati H, Sinanjung K, Fahrunissa F et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc. 2019;13:1–8. 10.1186/s12919-019-0176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill J. Tackling drug-resistant infections globally: final report and recommendations. 2016. https://apo.org.au/node/63983 (26 June 2023, date last accessed).
- Olsen SR, Kemper WD, Van Schaik JC. Self-diffusion coefficients of phosphorus in soil measured by transient and steady-state methods. Soil Sci Soc Am J. 1965;29:154–8. 10.2136/sssaj1965.03615995002900020014x. [DOI] [Google Scholar]
- Padder SA, Prasad R, Shah AH. Quorum sensing: a less known mode of communication among fungi. Microbiol Res. 2018;210:51–58. 10.1016/j.micres.2018.03.007. [DOI] [PubMed] [Google Scholar]
- Pandey P, Dubey AP, Mishra S et al. β-lactam resistance in Azospirillum baldaniorum Sp245 is mediated by lytic transglycosylase and β-lactamase and regulated by a cascade of RpoE7→ RpoH3 sigma factors. J Bacteriol. 2022;204:e00010–22. 10.1128/jb.00010-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D, Mazumder NB, Wangkheimayum J et al. Report of a carbapenemase gene blaIMP-4 in multi-drug resistant Escherichia coli from sewage water: a threat on clinical-environmental interphase. Indian J Med Microbiol. 2021;39:556–7. 10.1016/j.ijmmb.2021.04.010. [DOI] [PubMed] [Google Scholar]
- Pedrero F, Kalavrouziotis I, Alarcón JJ et al. Use of treated municipal wastewater in irrigated agriculture—Review of some practices in Spain and Greece. Agric Water Manage. 2010;97:1233–41. 10.1016/j.agwat.2010.03.003. [DOI] [Google Scholar]
- Pei R, Kim SC, Carlson KH et al. Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 2006;40:2427–35. 10.1016/j.watres.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Piper CS. Soil and plant analysis. J Phys Chem. 1945;49:45–45. 10.1021/j150439a012. [DOI] [Google Scholar]
- Plota M, Sazakli E, Giormezis N et al. In Vitro anti-biofilm activity of bacteriophage K (ATCC 19685-B1) and daptomycin against Staphylococci. Microorganisms. 2021;9:1853. 10.3390/microorganisms9091853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy B, Dubey SK. Exploring the allochthonous pollution influence on bacterial community and co-occurrence dynamics of River Ganga water through 16S rRNA-tagged amplicon metagenome. Environ Sci Pollut Res. 2021;28:26990–7005. 10.1007/s11356-021-12342-w. [DOI] [PubMed] [Google Scholar]
- Reddy B, Dubey SK. River ganges water as reservoir of microbes with antibiotic and metal ion resistance genes: high throughput metagenomic approach. Environ Pollut. 2019;246:443–51. 10.1016/j.envpol.2018.12.022. [DOI] [PubMed] [Google Scholar]
- Risal G, Shrestha A, Kunwar S et al. Detection of biofilm formation by Escherichia coli with its antibiogram profile. Int J Community Med Public Health. 2018;5:3771–5. 10.18203/2394-6040.ijcmph20183562. [DOI] [Google Scholar]
- Rizzardini CB, Goi D. Sustainability of domestic sewage sludge disposal. Sustainability. 2014;6:2424–34. 10.3390/su6052424. [DOI] [Google Scholar]
- Rumky J, Kruglova A, Repo E. Fate of antibiotic resistance genes (ARGs) in wastewater treatment plant: preliminary study on identification before and after ultrasonication. Environ Res. 2022;215:114281. 10.1016/j.envres.2022.114281. [DOI] [PubMed] [Google Scholar]
- Salam MA, Al-Amin MY, Salam MT et al. Antimicrobial resistance: a growing serious threat for global public health. Healthcare. 2023;11:1946. 10.3390/healthcare11131946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg T, Subramanian V, Ganeshan G et al. Wastewater discharge standards in the evolving context of urban sustainability–the case of India. Front Environ Sci. 2020;8:30. 10.3389/fenvs.2020.00030. [DOI] [Google Scholar]
- Seleiman MF, Santanen A, Mäkelä PS. Recycling sludge on cropland as fertilizer—advantages and risks. Resour Conserv Recycl. 2020;155:104647. 10.1016/j.resconrec.2019.104647. [DOI] [Google Scholar]
- Simões LC, Simoes M, Vieira MJ. Intergeneric coaggregation among drinking water bacteria: evidence of a role for Acinetobacter calcoaceticus as a bridging bacterium. Appl Environ Microb. 2008;74:1259–63. 10.1128/AEM.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KS, Paul D, Gupta A et al. Indian sewage microbiome has unique community characteristics and potential for population-level disease predictions. Sci Total Environ. 2023;858:160178. 10.1016/j.scitotenv.2022.160178. [DOI] [PubMed] [Google Scholar]
- Singh RP, Agrawal M. Biochemical and physiological responses of rice (Oryza sativa L.) grown on different sewage sludge amendments rates. Bull Environ Contam Toxicol. 2010a;84:606–12. 10.1007/s00128-010-0007-z. [DOI] [PubMed] [Google Scholar]
- Singh RP, Agrawal M. Effect of different sewage sludge applications on growth and yield of Vigna radiata L. field crop: metal uptake by plant. J Ecol Eng. 2010b;36:969–72. 10.1016/j.ecoleng.2010.03.008. [DOI] [Google Scholar]
- Singh RP, Agrawal M. Variations in heavy metal accumulation, growth and yield of rice plants grown at different sewage sludge amendment rates. Ecotoxicol Environ Saf. 2010c;73:632–41. 10.1016/j.ecoenv.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Sorinolu AJ, Tyagi N, Kumar A et al. Antibiotic resistance development and human health risks during wastewater reuse and biosolids application in agriculture. Chemosphere. 2021;265:129032. 10.1016/j.chemosphere.2020.129032. [DOI] [PubMed] [Google Scholar]
- Sun F, Xu Z, Fan L. Response of heavy metal and antibiotic resistance genes and related microorganisms to different heavy metals in activated sludge. J Environ Manage. 2021;300:113754. 10.1016/j.jenvman.2021.113754. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022–7. 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang KWK, Millar BC, Moore JE. Antimicrobial resistance (AMR). Br J Biomed Sci. 2023;80:11387. 10.3389/bjbs.2023.11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao S, Chen H, Li N et al. The spread of antibiotic resistance genes in vivo model. Can J Infect Dis Med Microbiol. 2022;2022:1–11. 10.1155/2022/3348695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timraz K, Xiong Y, Al Qarni H et al. Removal of bacterial cells, antibiotic resistance genes and integrase genes by on-site hospital wastewater treatment plants: surveillance of treated hospital effluent quality. Environ Sci Water Res Technol. 2017;3:293–303. 10.1039/C6EW00322B. [DOI] [Google Scholar]
- Turek A, Wieczorek K, Wolf WM. Digestion procedure and determination of heavy metals in sewage sludge—an analytical problem. Sustainability. 2019;11:1753. 10.3390/su11061753. [DOI] [Google Scholar]
- Turolla A, Cattaneo M, Marazzi F et al. Antibiotic resistant bacteria in urban sewage: role of full-scale wastewater treatment plants on environmental spreading. Chemosphere. 2018;191:761–9. 10.1016/j.chemosphere.2017.10.099. [DOI] [PubMed] [Google Scholar]
- Tytła M, Widziewicz-Rzońca K, Kernert J et al. First comprehensive analysis of potential ecological risk and factors influencing heavy metals binding in sewage sludge from WWTPs using the ultrasonic disintegration process. Water. 2023;15:666. 10.3390/w15040666. [DOI] [Google Scholar]
- Vaithyanathan VK, Cabana H. Integrated biotechnology management of biosolids: sustainable ways to produce value—added products. Front Water. 2021;3:729679. 10.3389/frwa.2021.729679. [DOI] [Google Scholar]
- Van Hoek AH, Mevius D, Guerra B et al. Acquired antibiotic resistance genes: an overview. Front Microbiol. 2011;2:203. https://doi.org/10.3389%2Ffmicb.2011.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vliet MT, Jones ER, Flörke M et al. Global water scarcity including surface water quality and expansions of clean water technologies. Environ Res Lett. 2021;16:024020. 10.1088/1748-9326/abbfc3. [DOI] [Google Scholar]
- Vázquez-López R, Solano-Gálvez SG, Juárez Vignon-Whaley JJ et al. Acinetobacter baumannii resistance: a real challenge for clinicians. Antibiotics. 2020;9:205. 10.3390/antibiotics9040205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlková E, Rada V, Šmehilová M et al. Auto-aggregation and co-aggregation ability in bifidobacteria and clostridia. Folia Microbiol. 2008;53:263–9. 10.1007/s12223-008-0040-z. [DOI] [PubMed] [Google Scholar]
- Walkley A, Black IA. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 1934;37:29–38. [Google Scholar]
- Wang J, Xu S, Zhao K et al. Risk control of antibiotics, antibiotic resistance genes (ARGs) and antibiotic resistant bacteria (ARB) during sewage sludge treatment and disposal: a review. Sci Total Environ. 2023;877:162772. 10.1016/j.scitotenv.2023.162772. [DOI] [PubMed] [Google Scholar]
- Worthington RJ, Melander C. Overcoming resistance to β-lactam antibiotics. J Org Chem. 2013;78:4207–13. 10.1021/jo400236f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xu Y, Wang H et al. Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere. 2015;119:1379–85. 10.1016/j.chemosphere.2014.02.040. [DOI] [PubMed] [Google Scholar]
- Zhang G, Lu S, Wang Y et al. Occurrence of antibiotics and antibiotic resistance genes and their correlations in lower Yangtze River, China. Environ Pollut. 2020;257:113365. 10.1016/j.envpol.2019.113365. [DOI] [PubMed] [Google Scholar]
- Zhang H, Qi HY, Zhang YL et al. Effects of sewage sludge pretreatment methods on its use in agricultural applications. J Hazard Mater. 2022;428:128213. 10.1016/j.jhazmat.2022.128213. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Hu J, Lee DJ et al. Sludge treatment: current research trends. Bioresour Technol. 2017;243:1159–72. 10.1016/j.biortech.2017.07.070. [DOI] [PubMed] [Google Scholar]
- Zhang T, Fang HHP. Applications of real-time polymerase chain reaction for quantification of microorganisms in environmental samples. Appl Microbiol Biotechnol. 2006;70:281–9. 10.1007/s00253-006-0333-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene sequences for bacterial isolates used in this study are available in NCBI database with accession number OP295365–OP295380.