Abstract
Vitamin A and its derivatives (retinoids) have profound effects on the proliferation and differentiation of many cell types and are involved in a diverse array of developmental and physiological regulatory processes, including those responsible for the development of the mature nervous system. Retinoid signals are mediated by retinoic acid (RA) receptors (RARs) and retinoid X receptors (RXRs), which show distinct spatio-temporal patterns of expression during development and in adult tissues. We have used SK-N-BE2(c) neuroblastoma cells to study the effects of reciprocal regulation of expression of various RARs. We show that in these cells RARγ1 acts as a repressor of RARβ2 transcription in the absence of an agonist. In the presence of RA, the expression of RARγ1 is reduced and that of RARβ2 is induced. Overexpression of RARγ1 neutralizes the effects of RA on RARβ induction. Expression of an RARγ1-specific antisense construct leads to the constitutive expression of RARβ2. Although both overexpression of RARγ1 and its reduction of expression can result in inhibition of cell proliferation, they induce different morphological changes. Reduction of RARγ1 (and induction of RARβ) leads to increased apoptosis, whereas RARγ1 overexpression leads to differentiation in the absence of apoptosis. Thus, RARγ1 appears to control a differentiation-apoptosis switch in SK-N-BE2(c) neuroblastoma cells.
Retinoids, the natural and synthetic derivatives of vitamin A, are known to regulate a broad range of biological processes, including vertebrate development, growth, and differentiation (24, 40, 56). The common denominator for these various effects is the ability of retinoids to trigger regulatory switches, modifying the repertoire of genes expressed by a given cell (24). The effects of retinoids are mediated by two families of ligand-responsive regulators, i.e., retinoic acid (RA) receptors (RARs) and retinoid X receptors (RXRs), which are members of the nuclear receptor superfamily (8, 22, 23, 30, 31, 42–45, 52). RARs bind and are activated by all-trans-RA and 9-cis-RA, whereas RXRs bind and are activated only by 9-cis-RA (26, 38). Both types of receptors are coded for by three genes (α, β, and γ) from which multiple isoforms can be generated by the use of alternative promoters or differential splicing (29, 37, 71). Differential tissue distribution of the various isoforms suggests that they may possess functional specificity, and given the pleiotropic effects of RA, it has been suggested that a fine tuning of retinoid receptor expression levels might be an essential requirement for correct development (9, 14, 58). RARs and RXRs modulate the expression of their target genes by binding to RA response elements (RAREs) (46, 67). One such RARE is located in the RARβ2 promoter region, and it has been shown that this response element mediates RARβ2 induction in response to RA in several cell lines and tissues (13, 27, 62). This autoregulation of the RARβ gene is thought to play an important role in amplifying the RA response, thereby enhancing the final biological response. This is supported by recent studies which have shown that altered receptor expression can be associated with tumor development: for example, RARβ is not expressed in certain malignant tumors, including lung carcinomas and breast carcinomas (21, 63, 72, 73). A subclone of the RA-responsive murine P19 embryonal carcinoma cells carrying an RARα mutation was found to be RA resistant (55). Similarly, an RA-resistant subclone of HL60 (human myeloid leukemia) cells was found to have a dominant negative RARα (11), while F9 murine teratocarcinoma cells with disrupted RARγ genes exhibit an altered differentiation response (7). Other studies, however, suggest that individual retinoid receptors may not perform unique functions, since mice carrying null mutations for RARα1 or RARγ2 showed no obvious abnormalities (31, 39).
Retinoids have also been implicated in many aspects of neuronal differentiation. Depending on the time of RA administration, teratogenic doses of RA cause defects in neural tube closure (3), and one of the most sensitive teratogenic targets is the neural crest (50).
Neuroblastoma (NB) is the most common extracranial malignant solid tumor of childhood; it arises from neural crest cell derivatives, and retinoids can induce its differentiation in vitro (5, 12, 66), generating a neuronal phenotype and causing a marked reduction in cell proliferation (1, 2, 60, 61). In contrast to the differentiation-promoting activity of RA, the synthetic analog N-(4-hydroxyphenyl)retinamide dramatically suppresses NB cell growth by inducing programmed cell death (54). NB cells therefore appear to represent a suitable model to investigate the mechanisms of neuronal cell death apoptosis and its relation to differentiation. RARα and RARγ have been found to be constitutively expressed in NB cells, while RARβ upregulation depends on the presence of RA (19, 69). RARβ, which could be involved in the ontogenesis of the nervous system (47, 58), is one of the genes known to be induced by RA in cells that have the neuronal phenotype (10). The effects of RA on the differentiation of NB cells do not depend entirely on the induction of RARβ, since in the NB cell line LAN-5, RARβ2 is constitutively expressed but differentiation takes place only in the presence of high concentration of RA, while the LAN-1 cell line can differentiate in the presence of RA even in the absence of RARβ2 (19, 69).
The present study was undertaken to identify the roles that the different RAR subtypes play in the RA response and differentiation pathway. The SK-N-BE2(c) cell line, selected for these studies, expresses RAR and RXR subtypes, and RARβ2 induction has been reported as an early marker of the RA response (19). We show here that RARγ1 controls the expression of the RARβ2 gene and, most interestingly, that the levels of RARγ1, independently of RA addition, are critical for the expression of different cell phenotypes with modified growth rates. In addition, we observed a correlation between the levels of RARγ1 and the induction of differentiation or apoptosis.
MATERIALS AND METHODS
Cell cultures.
The human NB SK-N-BE2(c) cells used in this study were kindly provided by G. Tonini (G. Gaslini Children’s Hospital, Genova, Italy) and grown in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), 1% glutamine, and 1% nonessential amino acids.
All-trans-RA (Sigma) was dissolved in ethanol at the concentration of 5 mM and kept at −80°C. Stock solutions of retinoid antagonists CD2331 and CD2366 (2 mM) were made in a dimethyl sulfoxide-ethanol (1:1) mixture and were maintained at −20°C. Further dilutions were made in culture medium.
RT-PCR.
Reverse transcription-PCR (RT-PCR) was performed under previously described conditions (18, 20) with, as primers, specific oligonucleotides that allow the unequivocal distinction between receptor subtypes and isoforms (20). For each analysis the quantity and quality of RNA were normalized by the coanalysis of β-actin messenger (20).
Plasmids.
Plasmids pECE-RAR (α, β, and γ) and pECE-RXRα have been previously described (35, 53). For stable RARγ1 transfection, the BamHI insert from pSG5 hRARγ1, kindly provided by P. Chambon, was inserted into the BamHI site of the eukaryotic expression vector pHβ Apr-1-neo (25), and the correct sense orientation was determined by restriction analysis. To obtain the RARγ1-specific antisense expression vector, the BalI/BstXI fragment from pSG5 hRARγ1 was made blunt and cloned into the EcoRV site of pBluescript SK. The EcoRI/HindIII insert was subsequently cloned into the EcoRI/HindIII sites of pHβ Apr-1-neo and analyzed for the correct orientation.
Stable transfections.
The recombinant constructs were stably transfected into SK-N-BE2(c) cells by the DOTAB method (Boehringer Mannheim) and screened with 400 μg of G418 (Gibco BRL) per ml. Clones were obtained through serial dilutions. To allow for cell growth, total transfectants and clones were cultured in the presence of the specific antagonist and routinely frozen within 1 week of culture. Experiments utilizing transfected cells were conducted on freshly thawed cells cultured in regular medium. Antagonists were added when needed. The expression of exogenous RARγ1 sense and antisense cDNAs was evaluated by RT-PCR or Northern blotting.
Transient transfection and CAT assay.
Transient transfections were carried out by using a modified calcium phosphate precipitation procedure, as described previously (53), with green monkey kidney cells (CV-1) grown in Dulbecco’s modified Eagle’s medium supplemented with 10% FCS. To measure the transcriptional activation, TREpal-tk linked with the chloramphenicol acetyltransferase (CAT) gene was used as a reporter gene. Briefly, 100 ng of reporter gene, 200 ng of β-galactosidase expression vector (pCH110; Pharmacia), and 50 ng of receptor expression vector were mixed with carrier DNA (pBluescript; Stratagene) to give a total of 1,000 ng of total DNA per well. After the cells were grown in the presence of the various retinoids for 24 h, CAT and β-galactosidase activities were assayed as previously described (53). CAT activity was normalized for transfection efficiency by the corresponding β-galactosidase activity.
Western blotting and immunostaining analyses.
Ten micrograms of DNA-binding proteins (4) was resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis on 10% gels and electroblotted onto a polyvinylidene difluoride (PVDF) membrane. The membrane was reacted with specific anti-RAR and anti-RXR antibodies (Santa Cruz), and protein bands were visualized after addition of enhanced chemiluminescence detection reagent (Amersham) by following the manufacturer’s protocol.
Cytoskeletal proteins were detected by using the 2H3 monoclonal antibody (Developmental Studies Hybridoma Bank) against 165-kDa neurofilaments. Monoclonal antibodies to CD4 receptor were used as a negative control. A positive control for differentiation was obtained by treating SK-N-BE2(c) cells with 10 μM RA for 4 days. Slides were fixed in 4% paraformaldehyde, followed by 10 min at −20°C in ethanol-acetic acid (95:5), and incubated with the diluted antibody. After a second incubation with biotin-conjugated rabbit anti-mouse immunoglobulins (Amersham), the complex was reacted with peroxidase-conjugated streptavidin (Amersham) and visualized with 3-amino-9-ethylcarbazole.
Evaluation of apoptosis and DNA fragmentation detection.
Cells were plated at 15,000/ml on chamber slides and grown as described above. After fixation with cold 2% formaldehyde in phosphate-buffered saline (PBS), the cells were washed with cold PBS and the nuclei were stained with a solution containing 50 μg of propidium iodide per ml, 0.1% Triton X-100, 0.1% Na citrate, and 20 μg of RNase A per ml in PBS for 15 to 20 min at room temperature. Apoptotic nuclei were identified by fluorescence microscopy. DNA fragmentation was measured on floating and adherent cells; 2 × 106 cells for each experiment were lysed and treated as described by Bissonette et al. (6).
Flow cytometric analysis.
Adherent and floating cells were fixed in 70% ethanol, washed twice in PBS, and resuspended in DNA staining solution containing 30 μg of propidium iodide per ml and 0.5 mg of RNase A per ml. DNA flow cytometric measurements were performed on an EPICS Elite instrument (Coulter Corporation, Miami, Fla.), and the Muticycle program (Phoenix Flow Systems, San Diego, Calif.) was used for the analysis of the cell cycle distribution as well as for the evaluation of apoptotic cells.
Cell proliferation assay.
To study anchorage-dependent cell growth, mock-transfected SK-N-BE2(c) cells and sense or antisense transgene cells were seeded at 1,000 to 3,000 cells per well (depending on the time in culture) in 96-well plates and grown in regular medium or treated with various concentrations of retinoid antagonists. Media were changed every 48 h. The number of viable cells was measured by the capacity of cells to reduce nitroblue tetrazolium with a colorimetric cell proliferation kit (MTT assay; Promega) (51).
RESULTS
Induction of RARβ2 correlates with a transient decrease of RARγ1 in SK-N-BE2(c) cells.
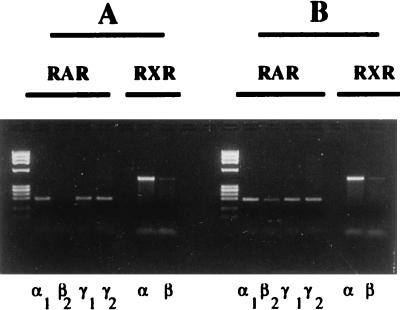
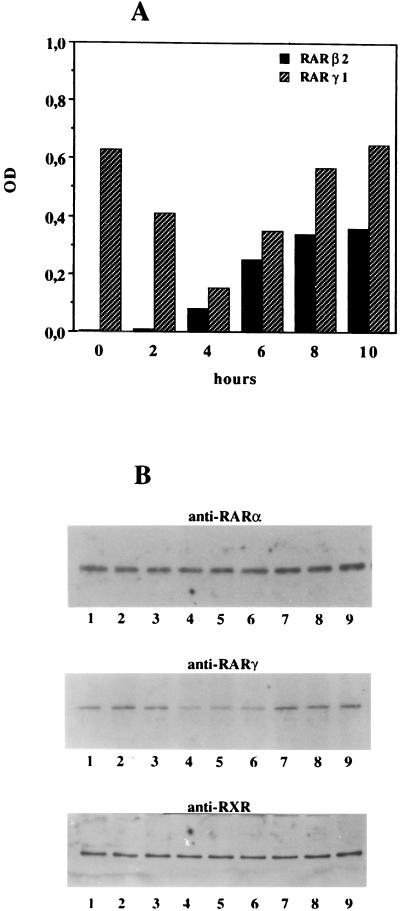
We used an RT-PCR protocol that allows for semiquantitative analysis of RAR and RXR subtype and isoform expression (20). We observed that SK-N-BE2(c) cells constitutively express RARα1, RARγ1, RARγ2, RXRα, and RXRβ (Fig. 1A). Consistent with previous observations (19), exposure to physiological concentrations of RA (10 nM) led to the induction of RARβ2 mRNA, whereas the mRNA levels for the other retinoid receptors were not affected (Fig. 1B). Figure 2A shows that SK-N-BE2(c) cells constitutively express high levels of RARγ1 mRNA; RARβ2 expression is weak, but upon stimulation by RA, it increases strongly in a time-dependent manner. RARβ2 induction correlated with a decrease in RARγ1 mRNA, suggesting that expression of both RARγ1 and RARβ2 is controlled by RA in NB cells. RARα1, RARγ2, and RXRα and -β expression did not change after exposure to RA (data not shown). To assess whether the RARγ1 mRNA decrease was associated with a decrease of its protein, nuclear extracts were analyzed. Western blot analysis of nuclear extracts from control cells and cells exposed to 10 nM RA revealed a reduction in RARγ1 protein at 2 h after RA addition (Fig. 2B). When the same blot was reprobed with anti-RARα and anti-RXR antibodies, no differences between RA-treated and control cells could be detected. Antibodies to RARβ did not recognize specific bands in these analyses.
FIG. 1.
Analysis of RAR and RXR expression in the NB cell line SK-N-BE2(c). One microgram of total cellular RNA was analyzed by RT-PCR with a nested reaction protocol for RAR or RXR subtypes and isoforms as described in Materials and Methods. (A) Control cell cells; (B) cells treated for 24 h with 10 nM RA. The left lane in each panel contains molecular size markers (φX174 RFDNA/HaeIII fragments [GIBCO]).
FIG. 2.
Time course of RA-regulated expression of RARβ2 and RARγ1. (A) SK-N-BE2(c) cells were plated at 106 cells per 25-cm2 tissue culture flask, and after an overnight incubation at 37°C, RA was added (time zero) to a final concentration of 10 nM. At various times after RA addition, total RNA was isolated and 1 μg was analyzed by RT-PCR for RAR or RXR expression as described in the text. Values for RAR and RXR mRNAs were normalized to that for β-actin mRNA used as internal standard for each RNA sample. The degree of amplification was quantitated by scanning densitometry and plotted as a ratio of RAR to β-actin or RXR to β-actin. Only data relative to RARβ2 and RARγ1 are reported, since no modulations were observed for the remaining RARs and RXRs. Five independent experiments with very similar results were conducted. OD, optical density. (B) Ten micrograms of DNA-binding proteins obtained from control cells and cells exposed to 10 nM RA was electrophoresed on SDS-polyacrylamide gels, transferred to PVDF membranes, and probed with antibodies against RARα, RARγ, or RXR. Lanes 1, control cells; lanes 2, cells exposed to RA for 90 min; lanes 3 to lane 9, treated cells collected every 30 min. Prestained molecular size standards were used to identify bands of the correct molecular weight.
RARγ1 limits RA-dependent transactivation of the RARβ2 gene.
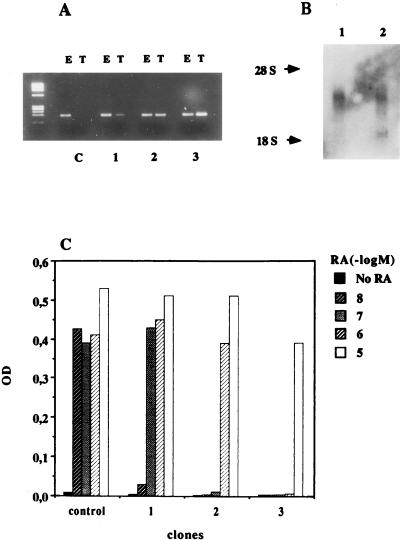
It has been observed that RARγ1 does not act as an RA-dependent activator of the RARβ2 promoter but acts as a transcriptional repressor (27, 28). We investigated whether overexpression of RARγ1 could reduce the RA-induced transcription of RARβ2 in SK-N-BE2(c) cells. The coding region of human RARγ1 was placed under the control of the human β-actin promoter, and stable transfectants were selected. In Fig. 3A, expression levels of endogenous and transfected RARγ1 in three different clones are compared to those for control cells (transfected with the empty vector). To verify the data, RNA levels were also analyzed by Northern blotting, and two bands corresponding to 3.3 and 1.5 kb, as expected, were seen (Fig. 3B). Clones overexpressing RARγ1 were investigated in detail to assess their ability to respond to RA as measured by the activation of the RARβ2 gene. Control clones that contained empty vector and three RARγ1-overexpressing clones were grown in the presence of increasing concentrations of RA for 24 h to achieve maximal induction of RARβ2. An RT-PCR analysis of RNA samples is shown in Fig. 3C. A clear correlation between expression levels of RARγ1 and inducibility of RARβ2 was observed: 10 nM RA was sufficient to induce RARβ2 mRNA in control cells (empty vector clones), while in the RARγ1-transfected clones, increasing the levels of exogenous RARγ1 antagonized the effects of RA. Interestingly, relatively small increases in RARγ1 levels clearly affected RARβ2 expression (the relative amounts of endogenous versus transfected RARγ1 were estimated by scanning densitometry of Fig. 3A). For instance clone 1, where RARγ1 mRNA is augmented 0.5-fold, is resistant to 10 nM RA but still responds to higher concentrations of RA. When the RARγ1 levels are doubled (clone 2), 100 nM RA no longer induces the RARβ2 gene, while the most striking effect is observed in clone 3 (with the highest levels of transfected RARγ1), where the cells have become resistant to even 1 μM RA. Thus, we observed an RARγ1-dependent inhibition of the RARβ2 response to RA in the NB cells, consistent with the previously observed repression of the RARβ2 promoter by RARγ1 (27, 28) in transient-transfection experiments.
FIG. 3.
RARγ1 represses RARβ2 gene induction in a dose-dependent manner. (A) RT-PCR determination of RNA transcripts of endogenous (lanes E) and transfected (lanes T) RARγ1 in three selected clones compared to mock-transfected SK-N-BE2(c) cells (lanes C). The levels of transfected RARγ1 RNA expressed relative to the amount of endogenous RNA, which was taken as 1, were 0.5, 1, and 2 in clones 1, 2, and 3, respectively. (B) Expression of endogenous and transfected RARγ1 determined by Northern blot analysis with total RNA (20 μg) to evaluate their correct sizes. (C) Cells from clones 1, 2, and 3 were treated for 24 h in the presence of increasing RA concentrations or solvent alone. RNA was extracted, and RT-PCR was used to estimate the relative amounts of RARβ2 gene transcripts. RNA transcripts of the β-actin gene were used to normalize the RT-PCR assays. Densitometric scanning of the gel clearly shows that a correlation exists between total RARγ1 levels and the cell response to RA, evaluated as RARβ2 gene induction. OD, optical density.
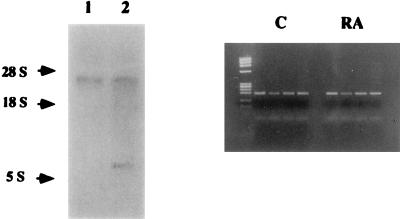
To further analyze the involvement of RARγ1 in the regulation of the RA transduction signals in NB cells, we downregulated its expression by using RARγ1-specific antisense cDNA. RARγ1 differs from RARγ2 in its NH2-terminal region corresponding to the BalI/BstXI fragment of human RARγ1 cDNA (29, 36). As can be seen from Fig. 4, stable transfected cells show detectable levels of RARγ1 antisense expression as determined by Northern blotting. When RAR mRNA expression in cells grown either in FCS-containing medium or in the absence or presence of 10 nM RA was assessed, comparable levels of constitutive RARβ2 expression were observed in both samples (Fig. 4).
FIG. 4.
Analysis of RAR transcripts in RARγ1 antisense transgene-transfected cells. (Left panel) Total RNAs (20 μg) from control (lane 1) and antisense transgene-transfected (lane 2) cells were analyzed by Northern blot hybridization to the BamHI insert of RARγ1 cDNA. Two bands of the correct size (3.3 and 0.167 kb, respectively) can be visualized in transfected cells. (Right panel) RT-PCR for RAR expression in transfected cells grown in regular medium (C) or in the presence of 10 nM RA for 24 h. From left to right are RARα1, -β2 -γ1, and -γ2. Note that RARβ2 mRNA is present independent of RA addition. The left lane contains molecular size markers (φX174 RFDNA/HaeIII fragments [GIBCO]).
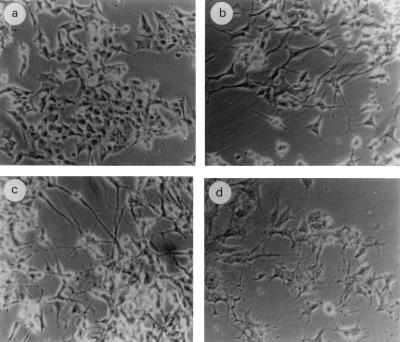
Interestingly, comparing clones expressing RARγ1 sense or antisense transgenes, we noticed clear morphological differences. Two clones, expressing the highest levels of transgenes, were analyzed in more detail. Compared to empty-vector-transfected cells (Fig. 5a), sense transgene-transfected cells (Fig. 5c) showed a more differentiated phenotype, with neurite-like processes resembling those of wild-type cells differentiating in the presence of 10 μM RA (Fig. 5b). Conversely, antisense transgene-transfected cells were relatively round with branched neurites, and a large portion of them became shrunken and eventually detached (Fig. 5d). In both cases the morphological changes were observed in the absence of exogenously added RA.
FIG. 5.
Morphological evaluation of transfected SK-N-BE2(c) cells compared to mock-transfected cells. (a) control cells; (b) cells cultured for 4 days in the presence of 10 μM RA; (c) RARγ1 sense transgene-transfected cells; (d) RARγ1 antisense transgene-transfected cells.
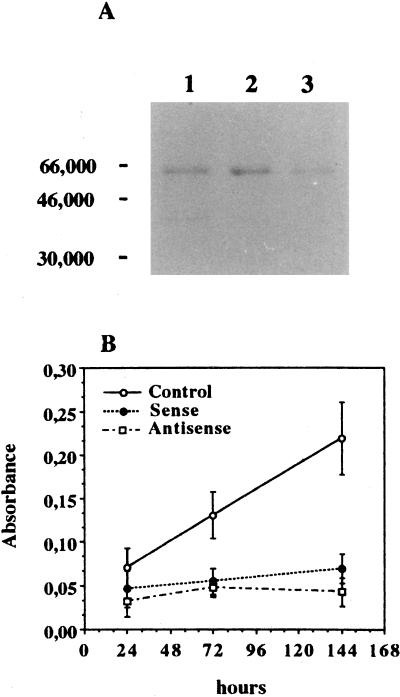
These clones were further analyzed to study the effects of RARγ1 sense or antisense transgenes. The clones showed an appreciable level of transgene expression (data not shown), and Western blot immunostaining revealed an increase of RARγ1 protein in sense transgene-transfected cells and a decrease of the molecule in antisense transgene-transfected cells (Fig. 6A). Comparing their growth rate to that of cells transfected with the empty vector, we observed that both the overexpression of RARγ1 and its reduction (coupled to RARβ2 induction) lead to a strong growth inhibition (Fig. 6B). Cell growth in other clones transfected with either sense or antisense constructs was analyzed, and the levels of transgene expression were proportional to the extent of growth inhibition (data not shown). Thus, very low as well as very high levels of RARγ1 appear to inhibit proliferation of SK-N-BE2(c) cells.
FIG. 6.
Inhibition of cell growth in stable transfected SK-N-BE2(c) cells. (A) Ten micrograms of DNA-binding proteins obtained from mock-transfected SK-N-BE2(c) cells (lane 1), RARγ1 sense transgene-transfected cells (lane 2), and RARγ1 antisense transgene-transfected cells (lane 3) was electrophoresed on an SDS-polyacrylamide gel, transferred to a PVDF membrane, and probed with anti-RARγ antibodies. Numbers on the left are molecular weights in thousands. (B) Recently thawed cells were kept in regular FCS-containing medium for 3 days and then seeded at 1,000 cells per well. Cell growth was evaluated every 48 h. The results were expressed as the A550 of MTT-derived formazan developed by sense and antisense RARγ1 cDNA-transfected cells compared to cells transfected with the empty vector. All data shown are representative of three independent experiments conducted in triplicate. Error bars indicate standard deviations.
RARγ1 levels allow for a switch between neuronal maturation and cell death.
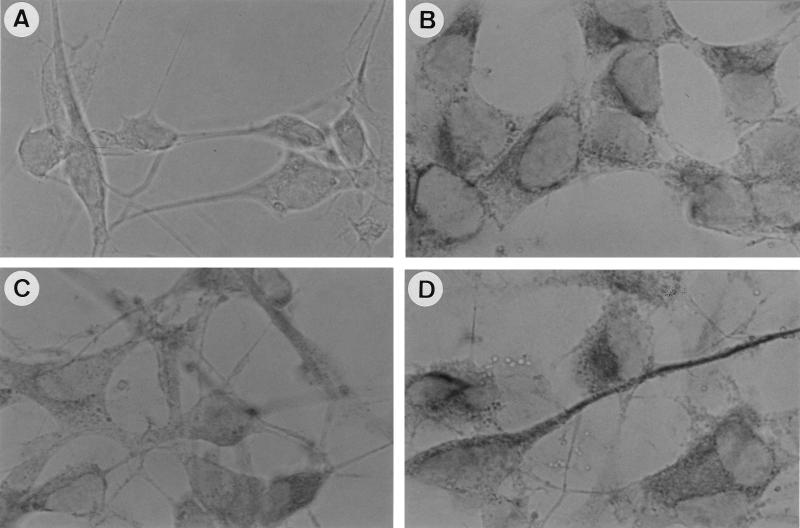
Morphological changes and cytoskeletal protein expression are typical hallmarks of neuronal maturation in NB cell lines, and microscopic inspection of our cell lines showed a similarity between sense transgene-transfected cells (Fig. 5c) and cells exposed for 4 days to 10 μM RA (Fig. 5b). Neurofilaments, specific markers of neurons, were assessed by immunostaining in control, RA-treated, and transgene-containing cells by utilizing the 2H3 monoclonal antibody, which is specific for the 165-kDa neurofilaments. RA treatment (Fig. 7C) caused a shift in the localization of the staining from a diffuse somatic pattern (characteristic of control cells [Fig. 7B]) to an intense perinuclear and neuritic pattern (Fig. 7C), which became more apparent in RARγ1-overexpressing cells (Fig. 7D).
FIG. 7.
Morphological differentiation of sense transgene-transfected SK-N-BE2(c) cells. Effects of RA (10 μM) and RARγ1 overexpression on cytoskeletal proteins were assessed by immunostaining analysis with the 2H3 monoclonal antibody against 165-kDa neurofilaments. (B) Control cells; (C) RA-treated cells; (D) RARγ1-overexpressing cells. As a negative control, RARγ1-overexpressing cells were reacted with anti-CD4 antibodies (A).
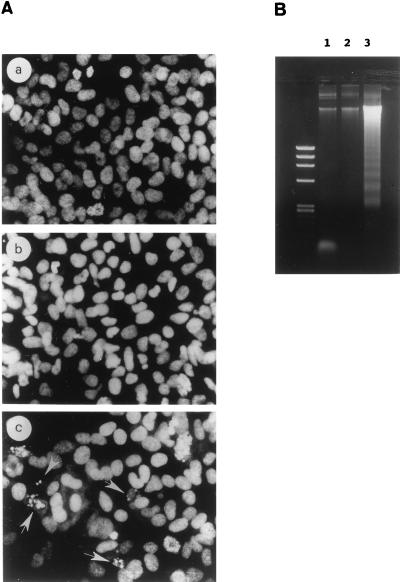
In contrast, the morphology of antisense transgene-transfected cells (Fig. 5d) was consistent with that of cells dying by programmed cell death (17, 70). When nuclei of adherent cells were stained with propidium iodide and examined by fluorescence microscopy (Fig. 8A, panel c), we found that 15% of the cells were smaller and contained condensed and fragmented nuclei with brightly stained chromatin, morphological changes typical of apoptosis. Conversely, sense transgene-transfected cells (Fig. 8A, panel b) showed no alteration in chromatin structure and were similar to control cells in this assay (panel a). During apoptosis, loss of membrane integrity is preceded by chromatin condensation and internucleosomal cleavage of genomic DNA, which produces a characteristic ladder pattern when analyzed by agarose gel electrophoresis (70). When such analyses were performed, we observed strong DNA fragmentation which was absent in control and sense transgene-transfected cells (Fig. 8B). This further confirms that the growth inhibitions observed with sense and antisense transgene-transfected cells result from the induction of different biological programs.
FIG. 8.
Apoptosis in RARγ1 antisense transgene-transfected SK-N-BE2(c) cells. (A) Morphological analysis of propidium iodide-stained nuclei from control cells (a) compared to RARγ1-overexpressing cells (b) and RARγ1 antisense transgene-transfected cells (c). Nuclei with typical morphological features of apoptosis are indicated (arrows). (B) Agarose gel electrophoresis of DNA from mock-transfected SK-N-BE2(c) cells (lane 1), RARγ1-overexpressing cells (lane 2), and RARγ1 antisense transgene-transfected cells (lane 3). Identical numbers of cells from each sample were lysed. DNA was isolated and electrophoresed on a 1.2% agarose gel. The left lane contains molecular size markers (φX174 RFDNA/HaeIII fragments [GIBCO]).
RARβ2- and RARγ1-specific antagonists selectively counteract RA effects.
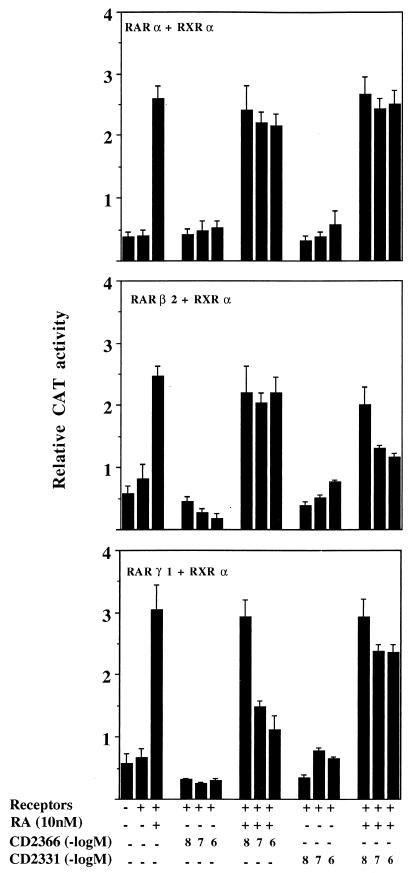
Receptor-selective antagonists can be used as alternative tools to evaluate the roles of individual receptors. We therefore used RARβ- and RARγ-selective antagonists to further evaluate the roles of RARβ and RARγ in the regulation and induction of specific programs in the NB cells. From several compounds reported to possess antagonist activity (34), we selected CD2331 and CD2366. When RARα, -β, and -γ and RXRα expression vectors were cotransfected with a TREpal-tk-CAT reporter gene into CV-1 cells, both CD2331 and CD2366 were unable to activate the reporter gene (Fig. 9). Conversely, in the presence of RA the antagonists caused a dose-dependent reduction of transactivation by RARβ2 and RARγ1, respectively (Fig. 9). At the highest nontoxic antagonist concentration (1 μM), CD2331 could completely suppress the transactivation induced by RARβ2 in the presence of 10 nM RA, while CD2366 inhibited more than 80% of RARγ1-mediated transactivation under the same conditions. The two compounds were ineffective when tested in the presence of RARα (Fig. 9) or 9-cis-RA-activated RXRα (data not shown). These results show that CD2331 antagonizes selectively the transactivation of RARβ2 by RA, while CD2366 antagonizes selectively the transactivation of RARγ1. In both cases a 100-fold excess of the antagonist over RA was required for complete inhibition. If RARγ1 is directly involved in the transduction of the RA signal in SK-N-BE2(c) cells, CD2366 should be able to antagonize this action, and we could expect that the addition of the antagonist to cells exposed to RA prevents RARβ gene induction. In Table 1 the inhibitory activity of CD2366 on RARβ induction by RA is shown. When used at 1 μM, CD2366 completely inhibits induction of RARβ mRNA by 10 nM RA and partially inhibits induction by 100 nM RA. CD2331 was also tested under the same conditions, and no inhibitory effects on RARβ mRNA synthesis were observed (data not shown).
FIG. 9.
Antagonistic effects of the synthetic retinoids CD2331 and CD2366 on RA-induced activation of TREpal-tk-CAT and inhibition of specific receptor subtypes. CV-1 cells were transiently transfected with 100 ng of TREpal-tk-CAT reporter together with RARα and RXRα expression plasmids (top panel), RARβ2 and RXRα (middle panel), or RARγ1 and RXRα (bottom panel). Transfected cells were treated with 10 nM RA, with the indicated concentrations of CD2366 and CD2331, or with the combination of RA and antagonists. CAT activity was assayed after 24 h as described in Materials and Methods. The activation obtained in the presence of 10 nM RA alone represents the maximum value. The data shown represent the means from two experiments carried out in duplicate, and the error bars represent standard deviations. The standard errors of the mean values were between 0.02 and 0.5.
TABLE 1.
CD2366 inhibits RARβ2 mRNA induction by RA
| Treatment (concn) | ODa
|
||
|---|---|---|---|
| RARα | RARβ | RARγ | |
| Control | 8.39 | 0 | 0.51 |
| CD2366 (1 μM) | 8.50 | 0 | 0.57 |
| RA (10 nM) | 8.60 | 0.24 | 0.53 |
| RA (100 nM) | 9.10 | 0.33 | 0.56 |
| CD2366 (1 μM) + RA (10 nM) | 8.50 | 0 | 0.54 |
| CD2366 (1 μM) + RA (100 nM) | 9.00 | 0.15 | 0.51 |
Optical density (OD) values were obtained by densitometric scanning of the gel. RNA transcripts of the β-actin gene were used to normalize RT-PCR assays. The experiment was conducted twice with very similar results.
CD2331 and CD2366 partially counteract inhibition of cell proliferation in transfected SK-N-BE2(c) cells.
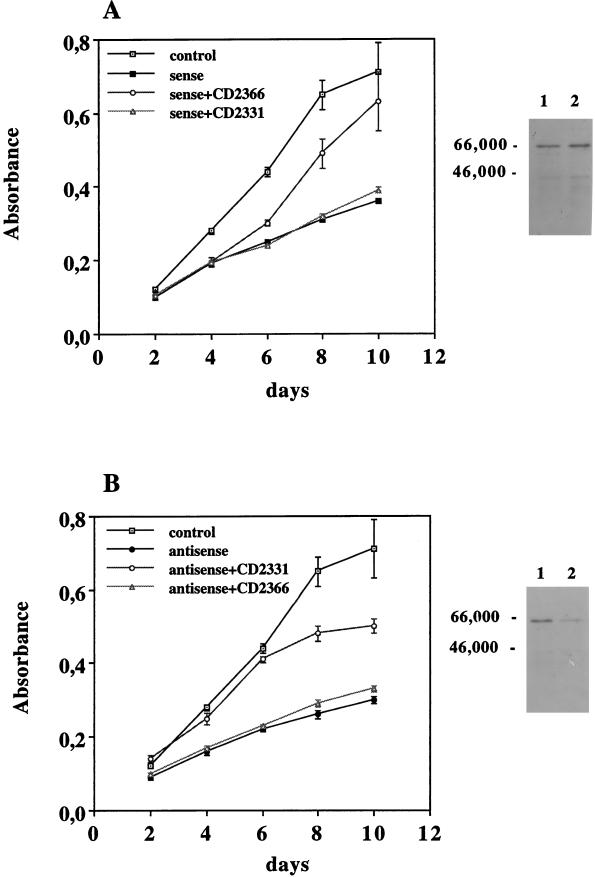
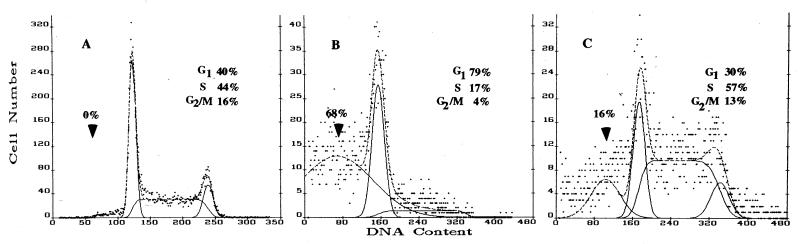
We also assessed the ability of the antagonists to counteract retinoid-induced growth inhibition. To avoid clonal effects due to position-insertion, total transfectant populations were studied. The transfected cultures showed an appreciable level of transgene expression (data not shown), and Western blot immunostaining revealed an increase of RARγ1 protein in sense transgene-transfected cells (Fig. 10A) and a decrease of the molecule in antisense transgene-transfected cells (Fig. 10B). Cells were seeded in the presence of increasing concentration of antagonists to evaluate their effects on cell proliferation. On the basis of the antagonist properties, cells overexpressing RARγ1 were cultured in the presence of CD2366, while CD2331 was utilized to antagonize RARβ2 being constitutively expressed in RARγ1 antisense transgene-transfected cells. Cell proliferation was evaluated by the MTT assay. Growth curves obtained in the presence of 1 μM antagonists are shown in Fig. 10. Lower antagonist concentrations were ineffective in this assay. CD2366 allowed sense transgene-transfected cells (Fig. 10A) to grow faster, with a maximal effect observed at day 8, at which point the growth rate was comparable to that of control cells that lacked the sense transgene. The growth rate of the antisense transgene-transfected cells (Fig. 10B) was partially restored by the addition of CD2331, suggesting that RARβ2 does contribute to cell growth arrest. Indeed, flow cytometric analysis of antisense transgene-transfected cells revealed an apoptotic peak (about 68%) and a decrease in the fraction of S- and G2-plus M-phase cells (Fig. 11B), while antagonist treatment (4 days at 1 μM) decreased the apoptotic peak to 16% and normalized the cell cycle distribution (Fig. 11C).
FIG. 10.
Effect of CD2331 and CD2366 antagonists on SK-N-BE2(c) cell proliferation when transfected with RARγ1 sense and antisense transgenes. Recently thawed cells were kept for 3 days in FCS-containing regular medium and then seeded at 1,000 cells/well in the presence of 1 μM antagonists. Cell growth was evaluated every 48 h by the MTT assay. Three independent experiments were conducted, with very similar results. The data shown represent the means of 10 points from a single experiment. Error bars represent standard deviations. Note that CD2366 can antagonize only RARγ1, while CD2331 is specific for RARβ2. Panels on the right show the relative amount of RARγ1 in transfected cells. Ten micrograms of DNA-binding proteins was electrophoresed on SDS-polyacrylamide gels, transferred to PVDF membranes, and probed with antibodies against RARγ1. Lanes 1, empty vector-transfected cells; lanes 2, RARγ1 sense (A) and RARγ1 antisense (B) transgene-transfected cells. Numbers on the left are molecular weights in thousands.
FIG. 11.
Effect of CD2331 on RARγ1 antisense transgene-transfected cell cycle. Floating and adherent mock-transfected SK-N-BE2(c) cells (A), antisense transgene-transfected cells (B), and antisense transgene-transfected cells cultured for 4 days in the presence of 1 μM CD2331 (C) were analyzed by flow cytometry. Arrowheads point to apoptotic cells.
DISCUSSION
In this study we demonstrate a ligand-sensitive transcriptional cross talk between RARγ1 and RARβ2 in SK-N-BE2(c) cells. Our findings support the existence of a regulatory interplay between members of the retinoid receptor superfamily, consistent with data reported by other groups.
Several studies have suggested that RARγ1 inhibits the activation of the βRARE by other RARs (28); this function is not due to a general lack of transcriptional enhancer activity of the receptor, since other response elements are efficiently activated (28, 33). A similar conclusion was reached also by Taneja et al. (65), who studied the contribution of RARs and RXRs to the activation of RA target genes by using RAR subtype (α, β, or γ)-specific synthetic retinoids. They observed that even though all three RARs can functionally substitute for each other as activators of RA target genes, one RAR subtype can cell specifically override the activity of the other RAR subtypes, and RARγ can suppress RARβ2 expression in wild-type F9 cells by a mechanism that involves the inhibition of RARα-dependent induction of RARβ2 (65). This inhibitory effect of RARγ1 is likely to be of biological significance for the containment of RA-mediated responses via activation of the βRARE. It is tempting to speculate that the reciprocal tissue expression patterns of RARγ and RARβ might in part be due to such a mechanism. Indeed, unlike RARα, RARβ and RARγ show restricted and mutually exclusive spatio-temporal patterns of expression during embryonic development (15, 58). Ruberte et al. (58) have shown the presence of RARβ transcripts in the closed neural tube, while RARγ transcripts become undetectable at the time of neural tube closure and are absent from the central and peripheral nervous systems throughout development (59). A similar nonoverlapping distribution of RARβ and RARγ transcripts was also seen in the developing limb and in the inner ear region; for the latter region, RARγ transcripts are present only in the otic capsule, whereas RARβ transcripts are found in the mesenchyme surrounding the inner ear epithelium (59). The direct involvement of RARγ in the transduction of the RA signal was also shown in vivo, as RARγ null mutant mice display some of the abnormalities present in animals fed a vitamin A-deficient diet (39) and do not display some of the teratogenic effects caused by maternal RA administration (39).
We have demonstrated that in our system RARγ1 can repress RARβ2 induction and RARβ2 levels determine the inhibition of cell proliferation and induction of apoptosis. A correlation between high levels of RARβ expression and apoptosis has also been observed in vivo in cells in the interdigital regions of the developing limb, in the fusion region of the neural tube, and in the palate (32, 48). Repression of RARβ gene transcription by RARγ1 most likely involves competitive binding between RARγ and RARα-RXR heterodimers to the βRARE. Both RARγ and RARα have been shown to bind to this RARE, but while RARα is an effective activator of this response element, RARγ is not (28). This might explain why even relatively small increases in RARγ can have substantial effects on RARβ induction even in the presence of RA and why high RA concentrations can still induce RARβ2 through RARα upregulation (69). Not unexpectedly, RARγ1 may also be able to substitute for some (but not all) RARβ2 functions (Fig. 5), since overexpression of RARγ1 leads to a phenotype similar to that observed in normal cells in the presence of 10 μM RA. It is well known that RARγ1 has high constitutive activity (28) that may allow it to substitute for the RARγ1 and RARβ2 activity observed in wild-type cells at 10 μM RA. The apparent discrepancy between results obtained when expressing RARγ antisense or inhibiting RARγ by a specific antagonist are most easily explained by the very different modes of action of these two agents. RARγ1 antagonists allow continuous blocking of the βRARE by RARγ-RXR heterodimers. In fact, it is likely that the antagonist represses the activation of RARE-containing genes by constitutively inhibiting RARγ activity (28). Thus, RARγ antagonists can still allow repression of RARβ expression and thereby avoid apoptosis but allow cell proliferation. In contrast, RARγ antisense expression eliminates or reduces RARγ expression, thereby allowing binding of RARα-RXR or RARβ-RXR heterodimers to the βRARE and induction of RARβ (by low concentrations of RA present under the growth conditions) and thus progression towards apoptosis. The orphan receptor nur77 might also function as an activator of the βRARE under those conditions and add to RARβ induction (68). The observation that RARβ antagonists strongly inhibit this pathway indicates that induction of other RARβ-responsive genes is part of this signaling cascade.
Although it has been shown that certain RARγ-selective compounds with retinoid-like activities can induce apoptosis (16, 41), it is generally believed that RARγ can induce cell differentiation. In fact, only RARγ can mediate the RA-induced differentiation of wild-type F9 cells (65), and the overexpression of RARγ directly induces terminal differentiation of human embryonal carcinoma NT2/D1 cells into a neuronal phenotype (49). Conversely, the overexpression of RARα and -β and RXRα does not produce maturation or growth-inhibitory effects (49). In agreement with these findings, we detect a differentiated phenotype in SK-N-BE2(c) cells overexpressing RARγ1; the direct involvement of RARγ in cell growth arrest and differentiation is further demonstrated by the effects of an RARγ-selective antagonist capable of restoring the normal cell growth rate.
A functional redundancy between RARs in vivo and in vitro has been described (31, 57, 64), and the upregulation of the remaining RARs in F9 cells, in which a single RAR has been disrupted, may be sufficient for the maintenance of several functions (64). This is not the case for our antisense transgene-transfected cells, where the loss of RARγ1 and its repressor role cannot be replaced by RARβ2, suggesting that RARγ1 has a particular role in the regulation of genes that control cell growth and differentiation. Thus, our data suggest that both RARγ1 and RARβ2 can control cell growth but that they play distinct roles in determining cell differentiation or apoptosis.
Our study also shows that changes in RAR isoform expression levels can lead to dramatically different effects on the fate of a cell population. Thus, a high level of complexity appears to govern nuclear receptor function in vivo.
ACKNOWLEDGMENTS
We thank B. Shroot and J. M. Bernardon (CIRD Galderma) for supplying us with the antagonists and the Developmental Studies Hybridoma Bank for the 2H3 monoclonal antibody. We also thank Ulrich Pfeffer, Andrea Fanjul, and Javi Piedrafita for helpful discussions and Anna Briata and Francesco Campelli for excellent technical assistance.
This work was supported by grants from PF ACRO-CNR, Associazione Italiana per la Ricerca sul Cancro, Fondazione Italiana per la Ricerca sul Cancro (Luisa Santunione), Ministero della Sanità, CNRS. The work carried out at the Sidney Kimmel Cancer Center was supported by grant CA 55681 to M.P.
REFERENCES
- 1.Abemayor E, Chang B, Sidell N. Effects of retinoic acid on the in vivo growth of human neuroblastoma cells. Cancer Lett. 1990;55:1–5. doi: 10.1016/0304-3835(90)90057-5. [DOI] [PubMed] [Google Scholar]
- 2.Abemayor E, Sidell N. Human neuroblastoma cell lines as models for the in vitro study of neoplastic and neuronal cell differentiation. Environ Health Perspect. 1989;80:3–15. doi: 10.1289/ehp.89803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alles A J, Sulik K K. Retinoic acid-induced spina bifida: evidence for a pathogenetic mechanism. Development. 1990;108:73–81. doi: 10.1242/dev.108.Supplement.73. [DOI] [PubMed] [Google Scholar]
- 4.Andrews N C, Faller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biedler J L, Helson L, Spengler B A. Morphology and growth, tumorigenicity, and cytogenetics of human neuroblastoma cells in continuous culture. Cancer Res. 1973;33:2643–2652. [PubMed] [Google Scholar]
- 6.Bissonnette R P, Echeverri F, Mahboubi A, Green D R. Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature (London) 1992;359:552–554. doi: 10.1038/359552a0. [DOI] [PubMed] [Google Scholar]
- 7.Boylan J F, Lufkin T, Achkar C C, Taneja R, Chambon P, Gudas L J. Targeted disruption of retinoic acid receptor α (RARα) and RARγ results in receptor-specific alterations in retinoic acid-mediated differentiation and retinoic acid metabolism. Mol Cell Biol. 1995;15:843–851. doi: 10.1128/mcb.15.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambon P. The retinoid signaling pathway: molecular and genetic analyses. Semin Cell Biol. 1994;5:115–125. doi: 10.1006/scel.1994.1015. [DOI] [PubMed] [Google Scholar]
- 9.Chambon P, Zelent A, Petkovich M, Mendelsohn C, Leroy P, Krust A, Kastner P, Brand N. The family of retinoic acid nuclear receptors. In: Saurat J H, editor. Retinoids: 10 years on. S. Basel, Switzerland: Karger; 1991. pp. 10–27. [Google Scholar]
- 10.Clagett-Dame M, Verhalen T J, Biedler J L, Repa J J. Identification and characterization of all-trans-retinoic acid receptor transcripts and receptor proteins in human neuroblastoma cells. Arch Biochem Biophys. 1993;300:684–693. doi: 10.1006/abbi.1993.1095. [DOI] [PubMed] [Google Scholar]
- 11.Collins S J, Robertson K A, Mueller L. Retinoic acid-induced granulocytic differentiation of HL-60 myeloid leukemia cells is mediated directly through the retinoic acid receptor (RARα) Mol Cell Biol. 1990;10:2154–2163. doi: 10.1128/mcb.10.5.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Bernardi B, Rogers D, Carli M, Madon E, De Laurentis T, Bagnulo S, Di Tullio M, Paolucci G, Pastore G. Localized neuroblastoma. Cancer. 1987;60:1066–1072. doi: 10.1002/1097-0142(19870901)60:5<1066::aid-cncr2820600523>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 13.de Thé H, Vivanco-Ruiz M M, Tiollais P, Stunnenberg H, Dejean A. Identification of a retinoic acid response element in the retinoic acid receptor β gene. Nature (London) 1990;343:177–180. doi: 10.1038/343177a0. [DOI] [PubMed] [Google Scholar]
- 14.Dollè P, Ruberte E, Kastner P, Petkovich M, Stoner C M, Gudas L J, Chambon P. Differential expression of genes encoding α, β, and γ retinoic acid receptors and CRABP in the developing limbs of the mouse. Nature (London) 1989;342:702–705. doi: 10.1038/342702a0. [DOI] [PubMed] [Google Scholar]
- 15.Dollè P, Ruberte E, Leroy P, Morriss-Kay G, Chambon P. Retinoic acid receptors and cellular binding proteins. I. A systematic study of their differential pattern of transcription during mouse organogenesis. Development. 1990;110:1133–1151. doi: 10.1242/dev.110.4.1133. [DOI] [PubMed] [Google Scholar]
- 16.Fanjul A N, Delia D, Pierotti M A, Rideout D, Qiu J, Pfahl M. 4-Hydroxyphenyl retinamide is a highly selective activator of retinoid receptors. J Biol Chem. 1996;271:22441–22446. doi: 10.1074/jbc.271.37.22441. [DOI] [PubMed] [Google Scholar]
- 17.Fath I, Schweighoffer F, Rey I, Multon M C, Boiziau J, Duchesne M, Tocquè B. Cloning of a Grb2 isoform with apoptotic properties. Science. 1994;264:971–974. doi: 10.1126/science.8178156. [DOI] [PubMed] [Google Scholar]
- 18.Ferrari N, Pfeffer U, Tosetti F, Brigati C, Vidali G. An improved RT-PCR protocol for the quantitation of human retinoic acid receptor RNA. Exp Cell Res. 1994;211:121–126. doi: 10.1006/excr.1994.1067. [DOI] [PubMed] [Google Scholar]
- 19.Ferrari N, Tonini G P, Briata A, Bottini F, Vidali G. Distribution of retinoic acid receptors α, β and γ mRNAs in neuroblastoma-derived cell lines and in fresh tumors. Int J Oncol. 1994;5:1019–1022. doi: 10.3892/ijo.5.5.1019. [DOI] [PubMed] [Google Scholar]
- 20.Ferrari N, Vidali G, Pfeffer U. Use of quantitative PCR to study retinoid receptor expression. Methods Enzymol. 1997;282:48–64. doi: 10.1016/s0076-6879(97)82095-2. [DOI] [PubMed] [Google Scholar]
- 21.Gebert J F, Moghal N, Frangioni J V, Sugarbaker D J, Neel B G. High frequency of retinoic acid receptor β abnormalities in human lung cancer. Oncogene. 1991;6:1859–1868. [PubMed] [Google Scholar]
- 22.Giguère V. Retinoic acid receptors and cellular retinoid binding proteins: complex interplay in retinoid signaling. Endocrinol Rev. 1994;15:61–79. doi: 10.1210/edrv-15-1-61. [DOI] [PubMed] [Google Scholar]
- 23.Glass C K. Differential recognition of target genes by nuclear receptor monomers, dimers, and heterodimers. Endocrinol Rev. 1994;15:391–407. doi: 10.1210/edrv-15-3-391. [DOI] [PubMed] [Google Scholar]
- 24.Gudas L J, Sporn M B, Roberts A B. Cellular biology and biochemistry of the retinoids. In: Sporn M B, Roberts A B, Goodman D S, editors. The retinoids. 2nd ed. New York, N.Y: Raven Press; 1994. pp. 443–520. [Google Scholar]
- 25.Gunning P, Leavitt J, Muscat G, Ng S-Y, Kedes L. A human β-actin expression vector system directs high-level accumulation of antisense transcripts. Proc Natl Acad Sci USA. 1987;84:4831–4835. doi: 10.1073/pnas.84.14.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heyman R A, Mangelsdorf D J, Dyck J A, Stein R B, Eichele G, Evans R M, Thaller C. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann B, Lehmann J M, Zhang X-K, Hermann T, Husmann M, Graupner G, Pfahl M. A retinoic acid-specific element controls the retinoic acid receptor β promoter. Mol Endocrinol. 1990;4:1727–1736. doi: 10.1210/mend-4-11-1727. [DOI] [PubMed] [Google Scholar]
- 28.Husmann M, Lehmann J, Hoffmann B, Hermann T, Tzukerman M, Pfahl M. Antagonism between retinoic acid receptors. Mol Cell Biol. 1991;11:4097–4103. doi: 10.1128/mcb.11.8.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kastner P, Krust A, Mendelsohn C, Garnier J-M, Zelent A, Leroy P, Staub A, Chambon P. Murine isoforms of retinoic acid receptor γ with specific patterns of expression. Proc Natl Acad Sci USA. 1990;87:2700–2704. doi: 10.1073/pnas.87.7.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kastner P, Leid M, Chambon P. The role of nuclear retinoic acid receptors in the regulation of gene expression. In: Blomhoff R, editor. Vitamin A in health and disease. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 189–238. [Google Scholar]
- 31.Kastner P, Mark M, Chambon P. Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 32.Kochhar D M, Jiang H, Harnish D C, Soprano D R. Evidence that retinoic acid-induced apoptosis in the mouse limb bud core mesenchymal cells is gene-mediated. Prog Clin Biol Res. 1993;383B:815–825. [PubMed] [Google Scholar]
- 33.La Vista-Picard N, Hobbs P D, Pfahl M, Dawson M I, Pfahl M. The receptor-DNA complex determines the retinoid response: a mechanism for the diversification of the ligand signal. Mol Cell Biol. 1996;16:4137–4146. doi: 10.1128/mcb.16.8.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee M-O, Dawson M I, Picard N, Hobbs P D, Pfahl M. A novel class of retinoid antagonists and their mechanisms of action. J Biol Chem. 1996;271:11897–11903. doi: 10.1074/jbc.271.20.11897. [DOI] [PubMed] [Google Scholar]
- 35.Lehmann J M, Zhang X-K, Pfahl M. RARγ2 expression is regulated through a retinoic acid response element embedded in Sp1 sites. Mol Cell Biol. 1992;12:2976–2985. doi: 10.1128/mcb.12.7.2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leid M, Kastner P, Chambon P. Multiplicity generates diversity in the retinoic acid signalling pathways. Trends Biochem Sci. 1992;17:427–433. doi: 10.1016/0968-0004(92)90014-z. [DOI] [PubMed] [Google Scholar]
- 37.Leroy P, Krust A, Zelent A, Mendelsohn C, Garnier J-M, Kastner P, Dierich A, Chambon P. Multiple isoforms of the mouse retinoic acid receptor α are generated by alternative splicing and differential induction by retinoic acid. EMBO J. 1991;10:59–69. doi: 10.1002/j.1460-2075.1991.tb07921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levin A A, Sturzenbecker L M, Kazmer S, Busakowski T, Huselton C, Allenby G, Speck J, Kratzeisen C, Rosenberger M, Lovey A, Grippo J F. 9-cis retinoic acid steroisomer binds and activates the nuclear receptor RXRα. Nature (London) 1992;355:359–361. doi: 10.1038/355359a0. [DOI] [PubMed] [Google Scholar]
- 39.Lohnes D, Kastner P, Dierich A, Mark M, LeMeur M, Chambon P. Function of retinoic acid receptor γ in the mouse. Cell. 1993;73:643–658. doi: 10.1016/0092-8674(93)90246-m. [DOI] [PubMed] [Google Scholar]
- 40.Lotan R. Effect of vitamin A and its analogs (retinoids) on normal and neoplastic cells. Biochim Biophys Acta. 1981;605:33–91. doi: 10.1016/0304-419x(80)90021-9. [DOI] [PubMed] [Google Scholar]
- 41.Lu X-P, Fanjul A, Picard N, Pfahl M, Rungta D, Nared-Hood K, Carter B, Piedrafita J, Tang S, Fabbrizio E, Pfahl M. Novel retinoid-related molecules as apoptosis inducers and effective inhibitors of human lung cancer cells in vivo. Nat Med. 1997;3:686–690. doi: 10.1038/nm0697-686. [DOI] [PubMed] [Google Scholar]
- 42.Mangelsdorf D J, Borgmeyer U, Heyman R A, Zhou J Y, Ong E S, Oro A E, Kakizuka A, Evans R M. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- 43.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 44.Mangelsdorf D J, Ong E S, Dick J A, Evans R M. Nuclear receptors that identifies a novel retinoic acid response pathway. Nature (London) 1990;345:224–229. doi: 10.1038/345224a0. [DOI] [PubMed] [Google Scholar]
- 45.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mangelsdorf D J, Umesono K, Kliewer S A, Borgmeyer U, Ong E S, Evans R M. A direct repeat in the cellular retinol-binding protein type II gene confers differential regulation by RXR and RAR. Cell. 1991;66:555–561. doi: 10.1016/0092-8674(81)90018-0. [DOI] [PubMed] [Google Scholar]
- 47.Mendelsohn C, Ruberte E, LeMeur M, Morriss-Kay G, Chambon P. Developmental analysis of the retinoic acid-inducible RAR-β2 promoter in transgenic animals. Development. 1991;113:723–734. doi: 10.1242/dev.113.3.723. [DOI] [PubMed] [Google Scholar]
- 48.Mendelsohn C, Ruberte E, Chambon P. Retinoid receptors in vertebrate limb development. Dev Biol. 1992;152:50–61. doi: 10.1016/0012-1606(92)90155-a. [DOI] [PubMed] [Google Scholar]
- 49.Moasser M M, Reuter V E, Dmitrovsky E. Overexpression of the retinoic acid receptor γ directly induces terminal differentiation of human embryonal carcinoma cells. Oncogene. 1995;10:1537–1543. [PubMed] [Google Scholar]
- 50.Morriss-Kay G, Murphy P, Hill R E, Davidson D R. Effects of retinoic acid excess on expression of Hox-2.9 and Krox-20 and on morphological segmentation in the hindbrain of mouse embryos. EMBO J. 1991;10:2985–2995. doi: 10.1002/j.1460-2075.1991.tb07849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 52.Pfahl M, Apfel R, Bendik I, Fanjul A, Graupner G, Lee M-O, La Vista N, Lu X-P, Piedrafita J, Ortiz M A, Salbert G, Zhang X-K. Nuclear retinoid receptors and their mechanisms of action. Vitam Horm. 1994;49:327–382. doi: 10.1016/s0083-6729(08)61150-4. [DOI] [PubMed] [Google Scholar]
- 53.Pfahl M, Tzukerman M, Zhang X-K, Lehmann J M, Hermann T, Wills K N, Graupner G. Rapid procedures for retinoic acid receptor cloning and their analysis. Methods Enzymol. 1990;153:256–270. doi: 10.1016/0076-6879(90)89297-u. [DOI] [PubMed] [Google Scholar]
- 54.Ponzoni M, Bocca P, Chiesa V, Decensi A, Pistoia V, Raffaghello L, Rozzo C, Montaldo P G. Differential effects of N-(4-hydroxyphenyl)retinamide and retinoic acid on neuroblastoma cells: apoptosis versus differentiation. Cancer Res. 1995;55:853–861. [PubMed] [Google Scholar]
- 55.Pratt M A, Kralova J, McBurney M W. A dominant negative mutation of the alpha retinoic acid receptor gene in a retinoic acid-nonresponsive embryonal carcinoma cell. Mol Cell Biol. 1990;10:6445–6453. doi: 10.1128/mcb.10.12.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roberts A B, Sporn M B. Cellular biology and biochemistry of the retinoids. In: Sporn M B, Roberts A B, Goodman D S, editors. The retinoids. Vol. 2. Orlando, Fla: Academic Press; 1984. pp. 209–286. [Google Scholar]
- 57.Roy B, Taneja R, Chambon P. Synergistic activation of retinoic acid (RA)-responsive genes and induction of embryonal carcinoma cell differentiation by an RA receptor α (RARα)-, RARβ-, or RARγ-selective ligand in combination with a retinoid X receptor-specific ligand. Mol Cell Biol. 1995;15:6481–6487. doi: 10.1128/mcb.15.12.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruberte E, Dollè P, Chambon P, Morriss-Kay G. Retinoic acid receptors and cellular binding proteins. II. Their differential pattern of transcription during early morphogenesis in mouse embryos. Development. 1991;111:45–60. doi: 10.1242/dev.111.1.45. [DOI] [PubMed] [Google Scholar]
- 59.Ruberte E, Dollè P, Krust A, Zelent A, Morriss-Kay G, Chambon P. Specific spatial and temporal distribution of retinoic acid receptor gamma transcripts during mouse embryogenesis. Development. 1990;108:213–222. doi: 10.1242/dev.108.2.213. [DOI] [PubMed] [Google Scholar]
- 60.Sidell N. Retinoic acid-induced growth inhibition and morphologic differentiation of human neuroblastoma cells in vitro. J Natl Cancer Inst. 1982;68:589–593. [PubMed] [Google Scholar]
- 61.Sidell N, Altman A, Haussler N, Seeger R C. Effects of retinoic acid (RA) on the growth and phenotypic expression of several human neuroblastoma cell lines. Exp Cell Res. 1983;148:21–30. doi: 10.1016/0014-4827(83)90184-2. [DOI] [PubMed] [Google Scholar]
- 62.Sucov H M, Murakami K K, Evans R M. Characterization of an autoregulated response element in the mouse retinoic acid receptor type β gene. Proc Natl Acad Sci USA. 1990;87:5392–5396. doi: 10.1073/pnas.87.14.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swisshelm K, Ryan K, Lee X, Tsou H C, Beacocke M, Sager R. Down-regulation of retinoic acid receptor β in mammary carcinoma cell lines and its up-regulation in senescing normal mammary epithelial cells. Cell Growth Differ. 1994;5:133–141. [PubMed] [Google Scholar]
- 64.Taneja R, Bouillet P, Boylan J F, Gaub M-P, Roy B, Gudas L J, Chambon P. Reexpression of retinoic acid receptor (RAR)γ or overexpression of RARα or RARβ in RARγ-null F9 cells reveals a partial functional redundancy between the three RAR types. Proc Natl Acad Sci USA. 1995;92:7854–7858. doi: 10.1073/pnas.92.17.7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taneja R, Roy B, Plassat J-L, Zusi C F, Ostrowski J, Reczek P R, Chambon P. Cell-type and promoter-context dependent retinoic acid receptor (RAR) redundancies for RARβ2 and Hoxa-1 activation in F9 and P19 cells can be artefactually generated by gene knockouts. Proc Natl Acad Sci USA. 1996;93:6197–6202. doi: 10.1073/pnas.93.12.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsokos M, Scarpa S, Ross R, Triche T J. Differentiation of human neuroblastoma recapitulates neural crest development. Study of morphology, neurotransmitter enzymes, and extracellular matrix protein. Am J Pathol. 1987;128:484–496. [PMC free article] [PubMed] [Google Scholar]
- 67.Umesono K, Murakami K K, Thompson C C, Evans R M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu Q, Li Y, Liu R, Agadir A, Lee M-O, Liu Y, Zhang X K. Modulation of retinoic acid sensitivity in lung cancer cells by a dynamic balance of nur77 and COUP-TF orphan receptor and their heterodimerization. EMBO J. 1997;16:1656–1669. doi: 10.1093/emboj/16.7.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wuarin L, Chang B, Wada R, Sidell N. Retinoic acid up-regulates nuclear retinoic acid receptor-α expression in human neuroblastoma cells. Int J Cancer. 1994;56:840–845. doi: 10.1002/ijc.2910560615. [DOI] [PubMed] [Google Scholar]
- 70.Wyllie A H, Kerr J F R, Currie A R. Cell death: the significance of apoptosis. Int Rev Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- 71.Zelent A, Mendelsohn C, Kastner P, Krust A, Garnier J-M, Ruffenach F, Leroy P, Chambon P. Differentially expressed isoforms of the mouse retinoic acid receptor β are generated by usage of two promoter and alternative splicing. EMBO J. 1991;10:71–81. doi: 10.1002/j.1460-2075.1991.tb07922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang X-K, Liu Y, Lee M-O. Retinoid receptors in human lung cancer and breast cancer. Mutat Res. 1996;350:267–277. doi: 10.1016/0027-5107(95)00102-6. [DOI] [PubMed] [Google Scholar]
- 73.Zhang X-K, Liu Y, Lee M-O, Pfahl M. A specific defect in the retinoic acid response associated with human lung cancer cell lines. Cancer Res. 1994;54:5663–5669. [PubMed] [Google Scholar]