Abstract
Janus kinase 2 (JAK2) mediates type I/II cytokine receptor signaling, but JAK2 is also activated by somatic mutations that cause hematological malignancies by mechanisms that are still incompletely understood. Quantitative superresolution microscopy (qSMLM) showed that erythropoietin receptor (EpoR) exists as monomers and dimerizes upon Epo stimulation or through the predominant JAK2 pseudokinase domain mutations (V617F, K539L, and R683S). Crystallographic analysis complemented by kinase activity analysis and atomic-level simulations revealed distinct pseudokinase dimer interfaces and activation mechanisms for the mutants: JAK V617F activity is driven by dimerization, K539L involves both increased receptor dimerization and kinase activity, and R683S prevents autoinhibition and increases catalytic activity and drives JAK2 equilibrium toward activation state through a wild-type dimer interface. Artificial intelligence–guided modeling and simulations revealed that the pseudokinase mutations cause differences in the pathogenic full-length JAK2 dimers, particularly in the FERM-SH2 domains. A detailed molecular understanding of mutation-driven JAK2 hyperactivation may enable novel therapeutic approaches to selectively target pathogenic JAK2 signaling.
The molecular and structural basis for JAK2 activation in erythropoietin receptor and pathogenic signaling is revealed.
INTRODUCTION
The Janus kinase (JAK)–signal transducer and activator of transcription (STAT) signaling pathway is used by approximately 60 cytokines and hormones in the regulation of hematopoiesis, metabolism, and orchestration of inflammatory and immune responses (1, 2). Ligand binding to cytokine receptor extracellular domain induces dimerization/oligomerization of the cytoplasmic domains of the receptors, leading to transphosphorylation of cytoplasmic JAKs. Each cytokine receptor interacts and activates a specific set of JAKs [JAK1 to JAK3 and TYK2 (tyrosine kinase 2)] that function as triggering kinases for cellular cytokine signaling. The activated JAKs phosphorylate specific tyrosine residues in the cytoplasmic tails of the receptors recruiting cytoplasmic signaling proteins, such as members of the STAT family of transcription factors to become phosphorylated and activated by JAKs. Substantial advances in our understanding of the receptor activation mechanisms and structural arrangements of JAK activation have been achieved, but an integrated structural understanding of normal and pathogenic JAK2 activation is still lacking.
Activation of cytokine receptor signaling requires pairing of JAKs, which in most cases involves two different JAK isoforms. JAK2 is unique, as it is the only family member also activated by homodimerization in hormone receptors such as erythropoietin receptor (EpoR), thrombopoietin receptor (TpoR), and growth hormone receptor (GHR) (3–5). EpoR signaling has received substantial interest not only as it is responsible for orchestrating the daily development of 200 billion red blood cells (80% of all new cells) but also as a relevant model for physiological and pathogenic cellular signaling. Aberrant activation of EpoR signaling results in its mild form in erythrocytosis due to familial EpoR mutations (6), and in a severe form in polycythemia vera (PV), a form of myeloproliferative neoplasm (MPN) (7, 8). A somatic JAK2 pseudokinase domain [JAK homology 2 (JH2)] mutation V617F at exon 14 was first identified as the molecular basis for PV (>95% of PV cases) and in 60% of the other MPN phenotypes, primary myelofibrosis (PMF) and essential thrombocythemia (ET) (9–12). Subsequently, human pathogenic JAK2 mutations have been found to concentrate in or around the JH2 and the identified >40 JH2 mutations cluster mainly in three regions. In addition to the prevalent V617F in exon 14, pathogenic mutations also occur in exons 12 and 16, which result in different clinical phenotypes; exon 12 mutations lead to PV and exon 16 mutations predominantly in acute lymphocytic leukemia (ALL) (13–15).
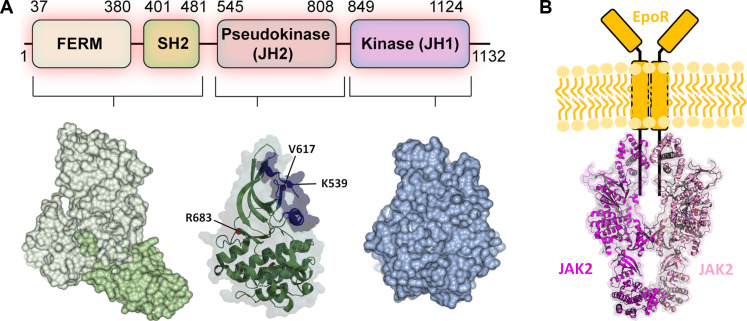
JAK kinases share a unique modular structure consisting of FERM-SH2 domain at the N terminus that mediate receptor interactions, followed by a pseudokinase JH2 domain and the active tyrosine kinase domain (JH1) (Fig. 1). JH2 was originally identified in JAK2 as a key regulator of activation that functions in a dualistic fashion; JH2 maintains JAKs in an inactive state in the absence of cytokine stimulation (autoinhibitory activity), but it is also required for ligand-induced activation (stimulatory activity) (16–18). Structural studies have proven this concept correct and have shown that, in the absence of stimulation, JH2 forms an autoinhibitory interaction with JH1 maintaining the inactive state (19, 20). The C-helix (αC) in JH2 binds to the backside of JH1 stabilizing the inactive state, leading to conformational restriction and inhibition of JH1. The disruption of autoinhibitory interaction as a mechanism for activation was supported by findings that several JAK2 activating mutants map to the JH2-JH1 interface, especially the exon 16 mutations, such as R683G/S, which is observed in approximately 20% of high-risk pediatric ALL patients (14, 19). However, the other main JAK2 JH2 mutations including V617F and K539L are not directly at this interface, suggesting an alternate mechanism for pathogenic activation. The mechanisms of the positive regulatory function of JH2 have been less well defined, but physiological and pathogenic JAK2 activation has now been shown to involve dimerization of JH2 domains that promotes close apposition and activation of the kinase domains (21–23). The first full-length mouse JAK1 V657F cryo–electron microscopy (cryo-EM) structures provided important structural insights for the activated JH2 dimeric interface in JAK homodimers (24, 25). The JAK1 V657F dimer structures, however, included a cross-linked IFN-λ (interferon-λ) receptor peptide (24, 25) and stabilized by a nanobody (24), possibly leading to a nonnative conformation of the dimer. In addition, JAK1 is not known to form homodimers in cells, and the JAK1 V657F is an experimental homolog for the human pathogenic JAK2 V617F mutation, which might complicate the biological insights from the structures (26).
Fig. 1. Domain structure of JAK2 and JAK2-EpoR signaling complex.
(A) Single-domain structures of JAK2. Residues in JAK2 JH2 often mutated in pathogenic signaling are highlighted. The dimerization interface identified from mouse JAK1 (PDB IDs: 7T6F and 8EWY) is shown in lilac. (B) Schematic illustration of JAK2-EpoR complex. JAK2 dimer structure is represented by mouse JAK1 structure (PDB ID: 8EWY). The two monomers are colored in magenta and pink.
RESULTS
Dimerization of monomeric EpoR upon cytokine binding
The superresolution imaging–based quantitative single-molecule localization microscopy (qSMLM) approach (fig. S1) (27) was used to determine the cell surface organization of EpoR in γ2A cells, which lack endogenous EpoR and JAK2. Isogenic clones of γ2A FlpIN-TetOn cells stably expressing inducible EpoR-mEos3.2 and JAK2 [wild type (WT) or mutants] were generated. For the calibration of qSMLM experiments, the monomeric protein cluster of differentiation 86 (CD86) and the covalent dimeric cytotoxic T lymphocyte–associated protein 4 (CTLA-4) were used as controls (fig. S2, A and B) (28, 29). Epo-induced JAK2 activation was verified by immunoblotting (fig. S2C), and in JAK2-expressing cells, EpoR-mEos3.2 showed clear membrane localization in confocal images (fig. S3), confirming that the fusion of mEos3.2 did not affect the localization and signaling activation.
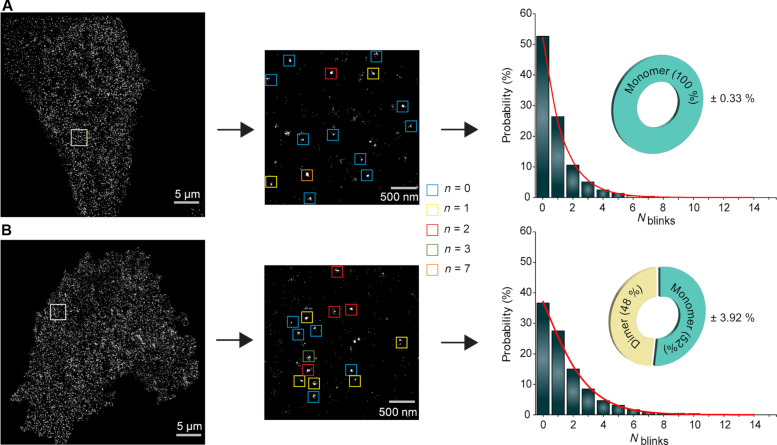
Photo-activated localization microscopy (PALM) images showed individual clusters optimal for qSMLM analysis. The FlpIN-TetOn approach enabled us to obtain physiological cell surface receptor expression levels, which ranged between ~2 and 3.5 receptor complexes per square micrometer (fig. S2D) (21, 30, 31). The qSMLM analysis of EpoR nanoclusters showed a similar blinking histogram as the monomeric CD86-mEos3.2 calibration standard. Fitting the blinking statistics to a mixed population model (27) did not show the presence of any dimers, indicating that the receptors are 100% monomeric on the cell membrane (Fig. 2A). The receptor organization upon Epo stimulation showed 50% dimeric receptors (Fig. 2B). These results show that EpoR exists as a monomeric receptor, and Epo induces dimerization of the receptors.
Fig. 2. qSMLM reveals cytokine-induced EpoR dimerization.
(A) EpoR and (B) EpoR + Epo. The left-hand side images are representative PALM images of mEos3.2-tagged EpoR receptors on a cell surface. The image is representative of 10 cells in three independent experiments. The two middle images are zoomed from the boxed region in the left image. The magnified boxed region (two middle images) showing localization (left) and the colored square boxes (right) illustrate the representation of the selected cluster and the number of blinks from each cluster. The bar graph is generated by single-molecule blinking histogram, and the corresponding fit (in red) shows the oligomeric status of the receptor ± Epo.
Induction of receptor dimerization by pathogenic JAK2 mutants
Next, we sought to study receptor stoichiometry changes in ligand-independent pathogenic JAK2 signaling. For this purpose, we made γ2A FlpIN-TetON-EpoRmEos3.2 cells stably expressing JAK2 V617F or JAK2 R683S mutants. Notably, the pathogenic mutants did not show EpoR membrane localization in normal culture conditions at 37°C. This phenomenon is likely due to continuous receptor internalization, as a similar phenotype was observed in cells grown in the presence of Epo (fig. S3). Receptor internalization was overcome by lowering the temperature to 30°C as described previously for other receptors (28). We also generated cells that lack JAK2 expression to study the role of JAK2 in EpoR organization. These cells did not show any membrane localization even after changing the culture conditions, verifying that JAK2 is essential for EpoR cell surface expression (fig. S3) (32).
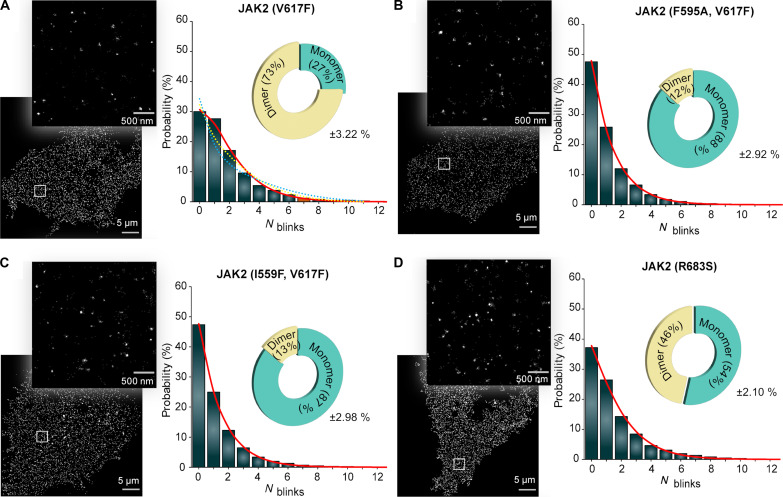
PALM imaging followed by the qSMLM analysis of the EpoR in JAK2 V617F cells revealed that 73% of receptors were dimers, indicating a strong dimerization phenotype for JAK2 V617F mutation (Fig. 3A). Dimerization by JAK2 V617F was also shown before (21–23), and it could provide the basis for pathogenic activation. No trimeric or tetrameric receptors were observed, ruling out the possible formation of higher oligomeric clusters for signaling as shown recently for interleukin-2 (IL-2) cytokine receptor (31).
Fig. 3. Ligand-independent dimerization of pathogenic JAK2 mutants.
PALM image and its magnified boxed region showing localization of cells stably expressing EpoR-mEos3.2 and different JAK2 mutants in the cell membrane: (A) V617F, (B) F595A with V617F, (C) I559F with V617F, and (D) R683S. The image is representative of nine cells taken from three independent experiments. The blinking histogram with fits (red line) and the oligomeric states obtained by the qSMLM analysis is shown on the right. In (A), monomer-trimer and monomer-tetramer fits are shown in yellow and blue dotted lines, respectively, which show that the fits for higher oligomeric states do not converge, indicating the absence of trimers or tetramers.
To gain additional insight into the JAK2 V617F activation mechanism, we used cells expressing JAK2 mutations suppressing the V617F-mediated signaling. Previous studies have identified several mutations in the JAK2 JH2 domain, particularly in the αC-helix and in the adenosine triphosphate (ATP) binding pocket, that suppress pathogenic JAK2 V617F activation (19, 22, 23). Furthermore, αC-helix suppression mutations have been shown to reduce JAK2 V617F–induced dimerization of TpoR in a total internal reflection fluorescence (TIRF)–based co-locomotion analysis (21). The αC-helix and ATP-pocket suppression mutants, JAK2 V617F/F595A and JAK2 V617F/I559F, showed only 12 and 13% dimeric receptors, respectively, indicating that both ATP-blocking and αC-helix suppression mutations revert the dimeric state of JAK2-V617F back to monomeric (Fig. 3, B and C). We then extended our study to another prevailing JAK2 pathogenic mutation R683S, which was observed to be monomeric in the co-locomotion analysis (21). The qSMLM analysis revealed that the JAK2 R683S mutation also induced dimerization of the receptors to a similar level as Epo stimulation in JAK2 WT but to a lesser extent than V617F (Fig. 3D). These results demonstrate that JAK2 pathogenic mutants V617F and R683S induce receptor dimerization but with varying potencies. The results also indicate that both JH2 ATP pocket and αC-helix suppression mutants (I559F and F595A, respectively) inhibit V617F-induced JAK2 and receptor dimerization.
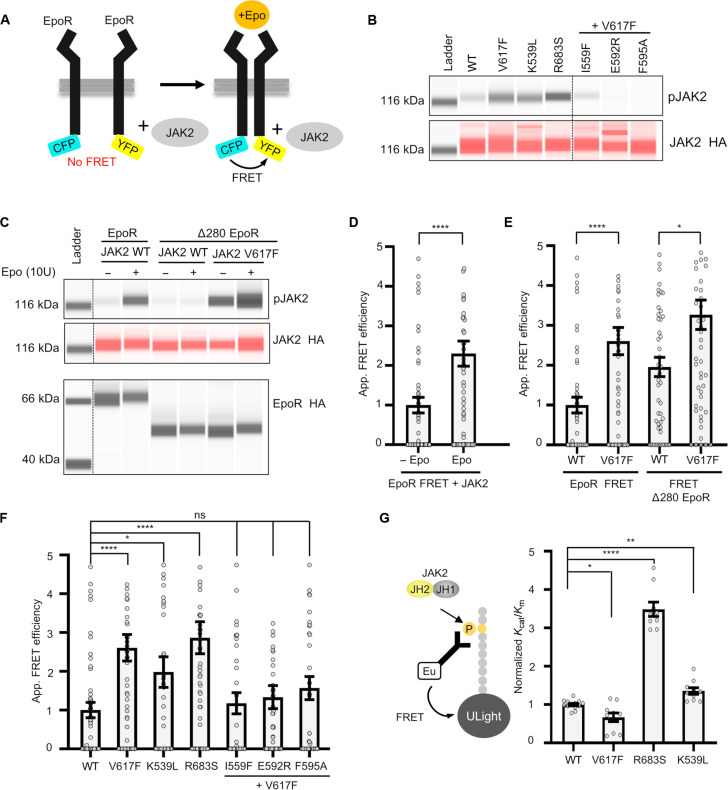
To verify the observations from the superresolution approach, we used EpoR fluorescence resonance energy transfer (FRET) reporters developed to study dynamics of the EpoR cytoplasmic region (Fig. 4A). The functionality of EpoR FRET constructs was verified in γ2A cells by immunoblot analysis after Epo stimulation and with different JAK2 mutants (K539L, V617F, and R683S) (Fig. 4, B and C). The results indicate that the FRET response is due to a change in oligomerization status rather than from conformational changes occurring in predimerized receptor JAK2 complex (fig. S4). Epo stimulation resulted in an increase of FRET, verifying the receptor-induced dimerization observed in the qSMLM analysis (Fig. 4D). Similarly, increased FRET was observed with JAK2 V617F (Fig. 4E). To analyze the role of receptor extracellular domain in pathogenic receptor dimerization, we truncated the EpoR by removing its extracellular domain. JAK2 V617F–induced similar FRET efficiency with WT and truncated EpoR, indicating that the mutation-induced dimerization is independent of receptor extracellular domain (Fig. 4E). In addition, suppressive ATP pocket and αC helix mutations reverted JAK2 V617F–induced FRET (Fig. 4F). These results confirmed that the FRET assay can be used to study EpoR receptor dimerization. Next, we proceeded to analyze other JAK2 mutations: K539L, which is associated with PV (13, 33), and R683S, which is associated with ALL (14, 15). The FRET results showed receptor dimerization in both K539L- and R683S-expressing cells (Fig. 4F).
Fig. 4. Analysis of EpoR dimerization using FRET-based reporters.
(A) Outline of the FRET reporters. (B) Capillary-based immunoblot showing phosphorylation of JAK2 for different JAK2 mutants used in FRET assay. (C) Capillary-based immunoblots showing Epo-induced JAK2 activation [pJAK2 and JAK2 hemagglutinin (HA)] with the EpoR FRET construct and the lack of JAK2 activation in extracellular region deleted EpoR construct (Δ280 EpoR). The extracellular region–independent JAK2 V617F activation is also shown in the blots. The bottom panel shows EpoR levels. (D) FRET efficiency in cells transiently expressing the EpoR FRET construct and JAK2 ± Epo stimulation. (E) Effect of deletion of extracellular domain in EpoR on FRET efficiency in JAK2 V617F–induced dimerization. (F) FRET efficiency in cells expressing V617F, K539L, and R683S and suppressive I559F/V617F, E592R/V617F, and F595A/V617F. (G) Scheme of kinase assay with purified recombinant JAK2 JH2-JH1 and the graph showing kinase activity for JAK2 R683S, K539L, and V617F. For FRET experiments, a minimum of 50 cells were analyzed for each condition from at least two independent experiments, and statistical significance was accessed by Kruskal-Wallis followed by Dunn’s multiple comparison post test. ns, not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P < 0.0001.
To study the activation mechanisms further, we used an in vitro kinase assay using purified JAK2 JH2-JH1 proteins (see Materials and Methods). In this assay, R683S showed increased catalytic activation, and the activity of K539L was also moderately increased, while V617F mutation did not affect the kinase activity (Fig. 4G). The difference observed for R683S was due to increase in Kcat, whereas K539L had lower apparent Km compared to WT (fig. S5). Of note, the cytokine receptor dependency of JAK2-V617F has also been shown before (34). These results support the notion that different mutations use distinct mechanisms for pathogenic activation.
Structural studies on TYK2 and our previous modeling studies with JAK2 have shown that R683 resides at the JH2-JH1 interface (19, 20). The most plausible interpretation of the qSMLM and kinase assay data is that disruption of the autoinhibitory JH2-JH1 interface by R683S allows for JH2 dimerization similarly as in physiological Epo stimulation, but the mutation does not stabilize the dimer as seen in V617F mutation. The weaker dimerization affinity in qSMLM is in line with the lack of dimerization observed for R683G in co-locomotion assay (21). However, these findings cannot explain the difference in K539L activation, which displays dimerization both in FRET assay and in a co-locomotion assay (21), but, unlike V617F, also showed slightly increased kinase activity (Fig. 4G). In summary, our findings demonstrate that the pathogenic activation of human JAK2 relies on JAK2-driven EpoR dimerization and suggest that activation involves unique molecular mechanisms and rearrangements in different mutants.
Structural basis for hyperactivation of JAK2 JH2 mutants
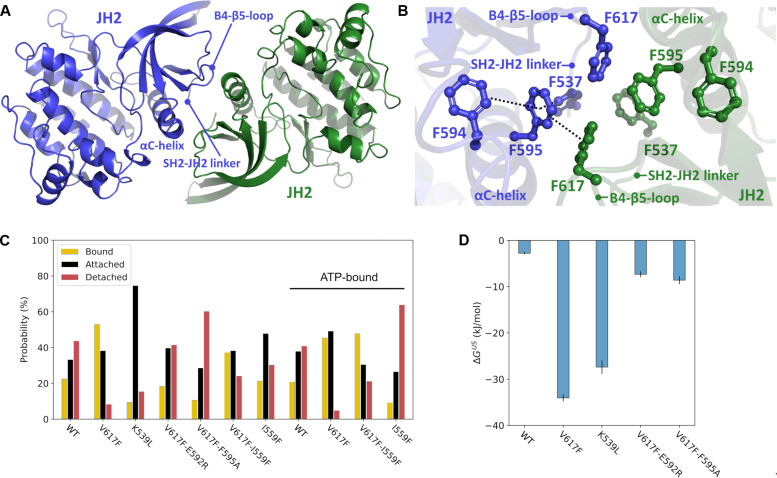
To define the molecular mechanisms for the observed different hyperactivating phenotypes, we proceeded to structural studies on JAK2 JH2 dimers. To study the dimerization interface in atomic detail, we analyzed all available crystal structures of JAK2 JH2 domains from the Protein Data Bank (PDB) database to identify crystallographic dimers that would explain the observed dimerization phenotypes of the JAK2 mutants. We found a JAK2 JH2 V617F structure (PDB ID: 6D2I), where a dimer interface was formed via αC-helices, a β4-β5 loop, and the beginning of an SH2-JH2 linker between two monomers related by crystallographic symmetry (Fig. 5A). We observed an intermolecular π-stacking network across the dimer interface, where stabilizing intermolecular stacking interactions form between F617 and F595 from both monomers. In addition, F537 and F594 stack intramolecularly with F595, further stabilizing the hydrophobic network (Fig. 5B). The JAK2 interface is also stabilized via two hydrogen bonds (N542-E592 and R588-D620) and symmetrical salt bridges (R541-E596) at the edges of the interface (fig. S6A). The dimer interface is highly similar to the full-length mouse JAK1 V657F cryo-EM structure (PDB ID: 7T6F) (fig. S6, B to E). The presence of a biological (cryo-EM–like) JH2-JH2 interface in the crystal structure of JH2 V617F indicates that the mutation can stabilize the biological interface in the crystal. This dimer arrangement explains the dimerizing effect of the V617F mutant based on the qSMLM data and the observed inhibitory effect of F595A and other αC-helix mutants, including charge-reversal E592R and phenylalanine F537A, F594A, and F595A mutations on V617F hyperactivation (21, 23, 35–37).
Fig. 5. Pseudokinase domain–mediated JAK2 dimerization.
(A) Crystallographic dimer of JAK2 JH2-V617F (PDB ID: 6D2I). The structural elements at the dimer interface are labeled. (B) Core aromatic cage of the V617F-induced dimer interface. The intra- and intermolecular π-stacking network is shown in dashed lines. (C) Distribution of the dimer structures observed by unbiased atomistic MD simulations of the WT and JAK2 JH2 mutants categorize the mutants into three different groups describing the stability of the dimer structure (see Materials and Methods): The bound state corresponding to monomers that remain close to the original position during a 1-μs simulation, the attached state referring to monomers that remain together but shift from the original dimer orientation during the simulation, and the detached state in which two monomers detach from the dimer during the simulation (see Materials and Methods). In addition to simulations without ATP (the first seven cases on the left side), selected cases were also considered with ATP, and the results of these MD simulations are marked as “ATP-bound” (see Materials and Methods). (D) Free-energy (ΔGUS) values of JAK2 JH2 dimerization based on umbrella sampling–based simulations (see Materials and Methods).
We proceeded to unveil the behavior of the identified JH2-V617F dimer structure via atomistic molecular dynamics (MD) simulations (see Materials and Methods). We performed multiple independent 1-μs all-atom equilibrium MD simulations and found that the JH2-V617F dimer was highly stable (Fig. 5C). Back mutating F617 to WT valine led to destabilization and breaking of the dimer interface (Fig. 5C). We also included two suppressing mutations, F595A and E592R, key residues in stabilizing the dimer interface and required for V617F-induced EpoR dimerization, to the simulation model. In these simulations, both V617F/F595A and V617F/E592R double mutants led to a WT-like behavior of the dimer, thus agreeing with the observed FRET and qSMLM data. The WT-like behavior of these suppressing mutants was further experimentally confirmed through FRET analysis, demonstrating Epo-induced EpoR dimerization similar to the WT (fig. S2E), and immunoblotting, indicating Epo-induced JAK2 phosphorylation akin to the WT (fig. S2C). To obtain quantitative estimates on the effects of the mutations on dimerization, free-energy calculations were performed (Fig. 5D; see also Materials and Methods). While the V617F interface displayed a high-affinity interaction relative to the WT (~30 kJ/mol more stable), the suppressing V617F double mutants showed WT-like behavior with diminished binding free energies. We also quantitatively analyzed the unbiased MD simulation data with molecular mechanics/generalized Born surface area [MM/GBSA; (38)] approach and, in good agreement with free-energy data, found that V617F dimer is more stable than the WT (fig. S7).
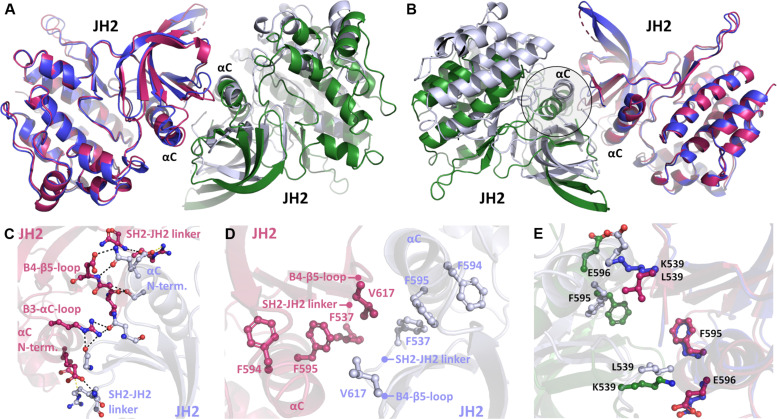
The K539L mutant was also included in equilibrium and free-energy MD simulations (Fig. 5, C and D). While this mutant displayed a highly stabilized dimer (free energy of ~30 kJ/mol more stable than WT) and remained attached in equilibrium simulations, the dimer interface was shifted compared to the starting V617F pose. We hypothesized that this is likely due to an altered dimer interface induced by the K539L mutation. To test this hypothesis, we determined the crystal structure of the JAK2 JH2 K539L mutant. Albeit being partly similar, a nonidentical dimer interface was identified (Fig. 6, A and B). This interface is not stabilized by the π-stacking network observed in the V617F crystal structure (PDB ID: 6D2I) but instead is established by extensive hydrogen bonding with complementing hydrophobic interactions (Fig. 6, C and D). The hydrogen bonding forms at the N-terminal end of the αC-helix and involves residues also from the SH2-JH2 linker and the β3-αC and β4-β5 loops. This unique interface is formed due to the removal of the charged lysine at the 539-position (due to the K539L mutation), which allows the repositioning of the αC-helix toward the SH2-JH2 linker of the dimer pair and further packing of hydrophobic contacts (Fig. 6E). This “hydrophobic collapse” of the interface brings several nonpolar residues to their vicinity, forming a more extensive contact area between the monomers compared to the V617F interface (table S1). Comparison of the K539L mutant simulation (starting from the V617F dimer pose) with the solved crystal structure showed that the structure relaxes toward the K539L crystal structure conformation during the MD simulations. This is observed by analyzing the time-resolved native contacts, which form during the simulations as their number increased by ~30% toward the K539L structure-like contacts with a concomitant ~60% reduction in V617F-like contacts (fig. S6F). This finding is also corroborated by root mean square deviation (RMSD) analysis, which revealed that K539L mutation introduced on the V617F dimer structure relaxes toward the K539L crystal structure (fig. S6G). Overall, both the structural data and atomistic simulations allowed us to identify the differences in the pathologic V617F and K539L interfaces, which could explain the differential signaling caused by these mutations.
Fig. 6. Crystal structure of K539L-induced pathogenic JH2 dimer.
(A and B) Comparison of JAK2 JH2 V617F (blue and green monomers) and K539L (pink and light blue monomers) dimers visualized from both sides of the interface. Blue and pink monomers of V617F and K539L, respectively, have been superposed together. A shift (highlighted) is seen in the position of the superposed αC-helices. (C) Electrostatic interactions across the dimer interface in the K539L mutant. Hydrogen bonds are shown in black, and salt bridges are shown in yellow dotted lines. (D) Aromatic cage region of the K539L dimer. No intermolecular π-stacking is observed unlike with the V617F dimer. (E) Effect of the lysine-leucine mutation at the K539L interface compared to the V617F interface. K539 prevents the collapse of the αC-helix toward the SH2-JH2 linker. Mutation to L539 allows closer packing of the interface.
As we also observed increased dimerization with the hyperactivating R683S mutant, we analyzed whether this mutation induces conformational changes that promote dimerization. Our hypothesis was that the mutation only breaks the JH2-JH1 autoinhibitory interface, thereby facilitating intermolecular JH2 interactions. We determined the crystal structure of JAK2 JH2 R683S and observed a WT-like structure without any mutation-induced structural changes and no V617F-like crystallographic dimer formation, supporting its role in breaking the autoinhibitory interface instead of directly inducing JAK2 dimerization (fig. S8, A and B).
Mutations blocking ATP binding to the JH2 domain have been shown to inhibit JAK activation (23, 37), and the first JH2 ATP competitive drug, TYK2 inhibitor deucravacitinib, has been approved for psoriasis (39). The ATP pocket mutation I559F inhibited V617F-induced EpoR dimerization (Figs. 3 and 4), and hence, we decided to investigate whether this is caused by structural alterations in the JH2 domain. We determined the crystal structure of the JAK2 JH2 I559F. The mutation clearly blocked the ATP binding to the ATP binding site but did not induce any further structural changes in the pocket or near the αC-helix that would explain its inhibitory effect (fig. S8, C to E). Therefore, we hypothesized that ATP binding to JH2 stabilizes the active JAK2 dimer. In previous MD simulations, we have observed the stabilizing effect of ATP on JAK2 JH2 and especially in the αC-helix (40). To investigate this further, we performed unbiased MD simulations of V617F, I559F, and V617F/I559F mutants in ATP-bound and ATP-free states (table S2). In both states, we observed partial destabilization of the V617F dimeric interface by I559F mutation (Fig. 5C and fig. S7), in agreement with the qSMLM analysis (Figs. 2 and 3), suggesting that the ATP binding site mutant V617F/I559F enhances monomerization by destabilizing the active conformation of the JH2-JH1 interface. Furthermore, the observed dimer destabilization was especially pronounced in the ATP-free state of the V617F/I559F mutant (Fig. 5C), highlighting that the effect of the I559F mutation on V617F dimer at least partially arises from the weakened binding of ATP (fig. S7). In summary, our structural and simulation analyses revealed that the major pathogenic JAK2 mutations use distinct JH2 dimerization interfaces and mechanisms for activation.
Modeling of full-length activated JAK2
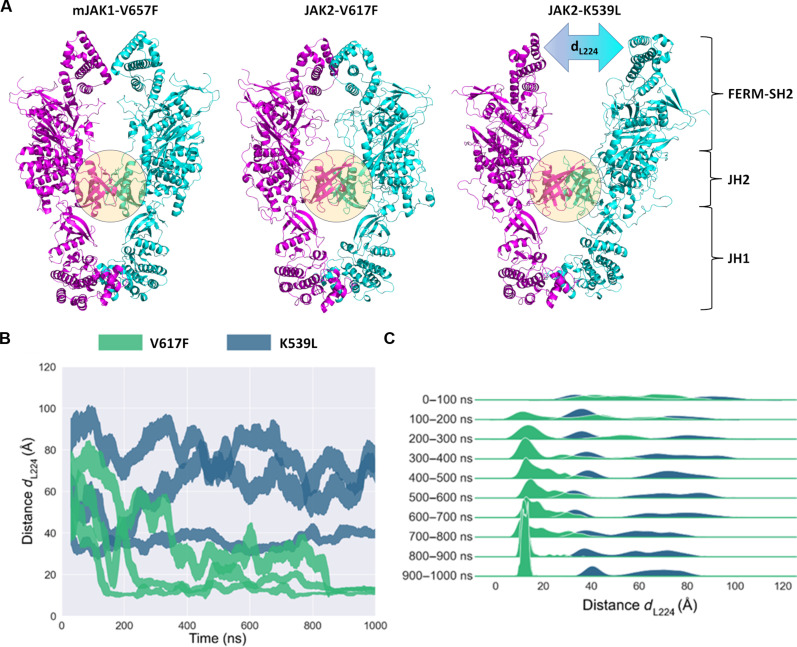
To gain additional structural insights on the effects of the pathogenic mutations on domain organization of JAK2, we superposed the full-length model of JAK2 (from AlphaFold2) to the JAK2 V617F and JAK2 K539L JH2-JH2 dimer structures. Two different monomeric JAK2 structures can be found from the AF2 database: one elongated structure corresponding to the JAK1 V657F cryo-EM monomer (25) and another, compact structure, where JH2-JH1 domain orientations match to the autoinhibited TYK2 JH2-JH1 crystal structure (20). We used the elongated JAK2 structure in constructing the pathogenic full-length JAK2 dimers. The resulting dimers show overall similarity in domain organization to the recent cryo-EM structure of mouse JAK1-V657F dimer (25). The full-length dimers of JAK2 V617F and JAK2 K539L were used as initial structures for microsecond-scale atomistic MD simulations. During the simulations, the conformations of the JH2 dimers observed in the crystal structures remained the same throughout the run. In JAK2 V617F dimers, the FERM domains interact closely with one another across all replicas, converging to a mean distance ∼13 Å between Leu224 residues (dL224) situated at tips of the FERM domains (Fig. 7). In contrast, JAK2 K539L dimers exhibit a separation of 40 to 80 Å between Leu224 residues, and the FERM domains are largely not interacting. Leu224 in JAK2 FERM domain is at the border of the EpoR binding site (box 1 site) (41), making dL224 a good proxy for the distance between EpoR transmembrane domains, which is in agreement with an approximately 50-Å separation observed in active EpoR (42, 43).
Fig. 7. Atomistic MD simulations of full-length AlphaFold2 models for JAK2 V617F and JAK2 K539L dimers.
(A) Comparison of mJAK1 V657F dimer (PDB ID: 8EWY) and snapshots captured at trajectory length of 1000 ns for JAK2 V617F and JAK2 K539L dimers. The distance between L224 residues in K539L dimer is denoted as dL224. Monomers in the dimers are colored in magenta and cyan. The JH2-JH2 dimer interface is highlighted with a sphere. (B) Distance (dL224) between the centers of mass for the L224 residues of JAK2 across all trajectories, with distance lines representing running means ± running SDs. (C) Same information as in (B) but displayed as a ridgeline density plot for specific time subsets of the simulation.
In addition to the differences in the orientation of FERM domains within JAK2 dimers, the JH2 mutations V617F and K539L were found to affect the dynamics of JH1 domains. The initial AF2 model of the JAK2 V617F dimer exhibits direct interactions between JH1 domains (Fig. 7). The JH1 interface is largely formed by αG and αEF helices, the activation loop, and a loop preceding the αG (fig. S9), as also observed by Caveney et al. (25) in JAK1 V657F. In the AF2 model of the JAK2 K539L dimer, the JH1 domains are initially separated, but they converge into dimers across all simulation trajectories exhibiting the in-trans interface similar to that observed in JAK2 V617F (fig. S9). The initial distinct JH1 domain orientations in JAK2 V617F and JAK2 K539L highlight the structural flexibility of JAK2 JH1 domains. These results indicate that the JAK2 V617F and JAK2 K539L mutations lead to different JH2 dimerization interfaces and propagate throughout the JAK2 structure in altered FERM and JH1 domain orientations.
DISCUSSION
The cell surface organization of EpoR, its implications for JAK-STAT signaling, and the mechanisms of pathogenic activation are long-standing questions in the field, especially due to their importance in hematological malignancies (8, 21, 44–47). We undertook a comprehensive analysis consisting of state-of-the-art imaging, biochemical, structural, and artificial intelligence–guided modeling and atomistic simulations of physiological Epo and pathogenic JAK2 signaling using representative mutations of the three major mutational regions (exons 12, 14, and 16) that present distinct clinical phenotypes. Our results show that Epo signaling occurs via ligand-induced dimerization of the monomeric EpoRs and that the three predominant JAK2 mutations use distinct mechanisms for hyperactivation that differ in terms of EpoR dimerization propensity, JH2 dimeric interactions, effect on catalytic activity, and structural organization of full-length JAK2.
Contrary to the traditional view of predimerized EpoRs (48) and in line with the recent findings from Wilmes et al. (21), our data based on qSMLM and FRET analysis prove that human EpoR is organized as a monomer on cell surface and signaling occurs via ligand-induced dimerization of the monomeric receptors. No higher-order oligomers of EpoR were present before or after ligand stimulation. Our results also confirm the crucial role of JAK2 JH2 domain in the dimerization process and for the pathogenic activation of the activating JAK2 disease mutants. Our results show that the somatic mutations V617F and K539L depend on JH2-mediated dimerization but via distinct dimer interfaces. We also found that the R683S mutation has a different mode of activation, but it still depends on JH2 dimerization. Together, our data agree with the JAK activation model, where JAK monomers are kept autoinhibited via intramolecular JH2-JH1 interactions (in cis) and are activated via intermolecular phosphorylation of JH1 in trans (47, 49).
The molecular organization of EpoR was analyzed by the qSMLM approach, which allowed a precise determination of cell surface receptor stoichiometry at physiological levels in cell lines that lacked endogenous expression of EpoR and JAK2 (28, 29, 50). Previous approaches analyzing homomeric cytokine receptor dimerization, including FRET (51), protein complementation (22), co-patching (45), and single-molecule TIRF-based imaging (21), are unable to detect the exact stoichiometry and oligomerization status and are susceptible to interference from endogenous JAK2 expression. The qSMLM approach uses fluorescent proteins that enable stoichiometric labeling and enable accurate determination of receptor stoichiometry and JAK2 interaction. The qSMLM data revealed an exclusive monomeric distribution of EpoR that shifted to a mixed stoichiometry with a dimeric fraction (52% dimers) upon cytokine stimulation. An even higher dimeric EpoR fraction (73%) was observed in V617F mutant cells. Cell-based FRET measurements concurred with these results. In accordance with previous studies, the results also showed that the mutation-induced dimerization is independent of the receptor extracellular domain and the molecular basis for the JAK2 V617F pathogenic activation arises from the mutation-induced dimerization of JAK2 via the JH2 domains (21), but the mutation does not directly increase catalytic activity (Fig. 4E).
The critical role of JH2 in physiological and pathogenic JAK2 signaling is well established in recent studies, but the structural basis has been elusive. We identified a crystallographic dimer of JAK2 JH2 V617F, displaying a nearly identical dimer interface with the JAK1 V657F cryo-EM structure (25). The dimer was stabilized by an aromatic π-stacking network across the monomer-monomer interface mediated by the mutant F617 residue. Atomistic MD simulations confirmed the stable nature of this dimer. In addition, the V617F suppression mutations F595A, I559F, and E592R displayed WT behavior in the MD simulations. This proves and provides a molecular-level explanation for the JAK2 JH2–mediated trans-dimerization model (47), as demonstrated by single-molecule studies (21). The full-length cryo-EM structure of murine JAK1 V657F dimer demonstrated looser packing of the dimer interface compared to the JAK2 V617F structure. This may be due to the potential nonphysiological nature of the cross-linked mini-IFNλR1-JAK1 dimer: JAK1 is not known to form homodimers in cytokine signaling, although it is possible that JAK1 signals as a homodimer, e.g., in IL-6 or IL-11 signaling. The JH2 dimer interface acts as a scaffold in bringing the JH1 domains in close apposition suitable for transphosphorylation and subsequent signaling activation.
V617F and K539L mutations lead to distinct MPN clinical phenotypes, V617F to PV, ET, and PMF, while K539L only to PV (9–13). The mutations also differ in their downstream signaling, and the JAK2 exon 12 mutation has been shown to bind preferentially to EpoR and show increased erythrocytosis but normal leukocyte counts in mouse models (52, 53). The K539L mutation induced EpoR dimerization, and MD simulations also indicated its strong dimerization propensity as also previously reported (21). However, as K539 resides in the SH2-JH2 linker, which is important in stabilizing the autoinhibited conformation, K539L likely influences the autoinhibitory conformation of JAK2, potentially explaining the increased in vitro activity observed for K539L mutant. Structural analysis of K539L revealed a dimer interface with similarity to V617F but exhibiting a more tightly packed interface, leading to extensive hydrophobic interactions and hydrogen bonding between the monomers as opposed to a stabilizing π-stacking network observed in V617F. This finding is in accordance with findings from previous mutational studies demonstrating a distinct activation mechanism for JAK2 mutations, where K539L showed the highest resistance to suppressive mutations compared to V617F>R683S (22, 23). MD simulations of K539L mutant (starting from a V617F dimer pose) also revealed relaxation toward a similar but distinct interface in comparison to V617F. All these points suggest distinct activation mechanisms of K539L and V617F. In addition, the differences in the formation of the interface were found to lead to different JH2 dimer orientations that ultimately affected the conformation of the full-length JAK2. These differences were particularly noticeable in the separation of the FERM-SH2 domains as revealed in MD simulations. The separation of FERM domains in K539L agrees with the distance of activated EpoR transmembrane regions and may correspond to the preference of JAK2 exon 12 mutations for forming dimers when bound to EpoR over the other homomeric receptors (52). The R683S mutation, although predominantly associated with high-risk ALL, can also lead to MPN-like disease (14, 15). R683S showed WT JH2 structure and has also been shown to be able to engage all homomeric receptors (15).
The receptor dimerization analysis was corroborated by suppressive mutations in the αC-helix and ATP binding pocket that were found to inhibit dimerization. The crystal structures provide explanation for the suppressing effect of the αC helix mutations, but the crystal structure of ATP pocket mutant I559F did not provide a direct explanation for its suppressing phenotype. The observed partial destabilization of the dimeric interface in V617F-I559F with the stabilizing effect of ATP on JH2 αC collectively suggests that the ATP binding site mutant destabilizes the active conformation of the JH2-JH1 interface.
R683S mutation was also of particular interest as it was located far from the dimer interface and was shown not to dimerize in co-locomotion analysis (21). The qSMLM results showed dimerization in R683S, although with a smaller dimeric fraction than with V617F, and at the same time, it increased kinase activity in vitro. The crystal structure of R683S did not show any structural changes, suggesting that the mutation acts by destabilization of the autoinhibitory JH2-JH1 interaction, leading to activation as suggested previously by MD simulations (19) and the crystal structure of TYK2 JH2-JH1 (20). The dimeric nature of R683G/S likely stems from a WT-like dimerization but without further stabilization induced by interface mutations. Thus, the higher dimerization propensity resulting from the breaking of the autoinhibitory interface is therefore a result of a more dimerization-prone (nonautoinhibited) conformation of JAK2 that shifts the equilibrium toward the active state. This conclusion is supported by the similar dimeric fraction observed in R683S (54%) and ligand-induced WT JAK2 (52%). Furthermore, as there are no stabilizing interactions at the JH2-JH2 interface, the R683S-mediated dimer will be weaker than, e.g., the V617F dimer explaining why R683G/S is most sensitive to suppressive mutations (23). We assume that the lack of stabilizing interactions at the dimer interface also explains why R683G/S dimerization was not observed in the co-locomotion assay, i.e., the dimer dissociates faster than more stable JAK2 dimers.
In conclusion, our study demonstrates a monomeric distribution of EpoR on cell surface and proves that EpoR dimerization is the common denominator for cytokine-induced and pathogenic JAK2 activation. Furthermore, our study identified differences between the dimer interfaces of JAK2 pathogenic mutants and provides a mechanistic model for the activation of full-length JAK2 in pathogenic states. These differences affect receptor phosphorylation patterns and variations in downstream signaling by the JAK2 mutants, as suggested in prior research (52). The mechanistic insights provided in this study offer a valuable background for the divergent clinical outcomes associated with these pathological mutations and may provide opportunities to modulate cytokine receptor signaling and target oncogenic JAK signaling.
MATERIALS AND METHODS
Generation of plasmid constructs
Standard cloning procedures were performed throughout this study for generating plasmids. JAK2 mutants were generated by site-directed mutagenesis of full-length JAK2, which was cloned into Sal I–Not I restriction sites of pCIneo expression vector. This was done using QuikChange (Agilent) according to the manufacturer’s instructions. For generation of constructs for superresolution imaging, the JAK2 WT/mutants were cloned into Asc I–Nhe I site of the bidirectional pBOF vector (54), and the EpoR-mEos3.2 fusion was cloned to the other side of the bidirectional tetracycline-inducible promotor using Bam HI–Mre I site to ensure equal expression of cloned genes. The EpoR-mEos3.2 fusion was made by overlap extension polymerase chain reaction (PCR) of EpoR sequence (23) and mEos3.2 fluorescent protein plasmid (a gift from M. Davidson and T. Xu, Addgene, plasmid #54525). The sequence after amino acid 380 was removed in EpoR before mEos3.2 fusion to reduce internalization of the receptor. A flexible linker sequence RSIAT was also inserted using PCR between EpoR.mEos3.2 to ensure flexibility of the fluorescent protein. qSMLM controls CD86-mEos3.2, and CTLA-4-mEos3.2 was generated by amplifying CD86 and CTLA-4 and fusing the amplified product with amplified mEos3.2 using overlap extension PCR and cloned to Asc I–Nhe I site of pBOF vector. The CD86 and CTLA-4 backbone was from Addgene plasmids #98284 and #98285, respectively. The FRET plasmid used in this study was described previously (23). The plasmid constructs were verified by Sanger sequencing.
Cell culture and generation of stable cell lines
JAK2-deficient γ2A human fibrosarcoma cells (55) maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Lonza, Basel, Switzerland) supplemented with fetal bovine serum (FBS; 10%) (Sigma-Aldrich, St. Louis, MO), l-glutamine (2 mM) (Lonza), and antibiotics (penicillin/streptomycin, 0.5%; Lonza) were used for this study. For imaging studies, the medium was replaced with FluoroBrite DMEM supplemented with FBS (10%), l-glutamine (2 mM), and penicillin/streptomycin (0.5%) at least two passages before seeding cells for imaging. The cells are cultured in normal culture conditions unless otherwise specified. The cells were starved overnight in medium without FBS before Epo stimulation and imaging experiments. All the transfections were performed using FuGENE HD (Promega).
γ2A FlpIN-TetOn cell lines were generated by modifying γ2A cells by two rounds of transfections. In the first round, the γ2A cells were stably transfected with pFRT/lacZeo Vector (Thermo Fisher Scientific) after linearizing with Sca I restriction enzyme. Two days after transfection, cells were selected in zeocin (150 μg/ml) for 12 days. Individual zeocin-resistant colonies were picked manually and expanded, and genomic DNA was isolated from the colonies for screening using the GenElute Mammalian Genomic DNA Miniprep Kit (Sigma-Aldrich). To select the clones that have minimum/single FRT site in the genome, quantitative PCR (qPCR) was performed with primers targeted to FRT site (5′-GGATGCCAAGTTGACCAGTG-3′ and 5′-ATGAACAGGGTCACGTCGTC-3′) and compared with housekeeping genomic α-actinin-1 primers (5′-AGATGGTTTGGGCGGTTCTA-3′ and 5′-AGAACCCCGGGCTAAATTGA-3′). The qPCR was performed using HOT FIREPol EvaGreen qPCR Mix Plus (Solis BioDyne, Tartu, Estonia) in a Bio-Rad CFX-384 Real-Time PCR detection system. The clone that had the lowest FRT site in the genome was selected as γ2A FlpIN cell line and expanded. In the second round of transfection, the γ2A FlpIN cell line was transfected with pUltra-puro-RTTA3 plasmid (a gift from Y. Dogan and K. Kim; Addgene plasmid, #58750), and positive cells were selected by culturing cells in the presence of puromycin (1.5 μg/ml) (Sigma-Aldrich). Individual clones were isolated and screened by analyzing doxycycline-induced activation by transfecting with pBOF vector containing fluorescent protein. The selected γ2A FlpIN-TetOn clone was expanded and maintained in the presence of zeocin (150 μg/ml) and puromycin (1.5 μg/ml).
For generation of stable cells inducibly expressing the EpoR-mEos3.2 and JAK2/mutants, the pBOF vector construct having coding sequence of EpoR-mEos3.2 and JAK2 expressing from the Tet-regulated bidirectional promotor was cotransfected with pOG44 vector (Invitrogen) to γ2A FlpIN-TetOn cells. Two days after transfection, cells were selected in hygromycin B (200 μg/ml) (Thermo Fisher Scientific) and puromycin (1.5 μg/ml) for approximately 20 days until colonies are formed, and then positive clones were pooled and expanded. The stable integrations were verified by PCR from the isolated genomic DNA, followed by sequencing of the amplified product by Sanger sequencing. All the stable cells used in superresolution microscopy experiments were prepared using a similar approach.
Sample preparation and SMLM
Coverslips (0.17 ± 0.005 mm, Carl Roth, Germany) were sonicated for 20 min in a water bath and cleaned extensively by acid/base wash before seeding the cells to reduce background fluorescence. This was done by immersing coverslips in 2 M HCl (overnight), 2 M NaOH (2 hours), and 20% H3PO4 (1 hour) and stored in molecular biology–grade ethanol until use. Wash steps were performed using Milli-Q in between the above steps. Before use, coverslips were coated with fibronectin (50 μg/ml) for 1 hour to facilitate cell adhesion. Cells were seeded in FluoroBrite DMEM medium with supplements, and following overnight attachment, the cells were starved using medium lacking FBS and containing doxycycline (0.1 μg/ml) to induce protein expression. For Epo stimulation, cells were treated with human recombinant Epo (10 U/ml) (NeoRecormon, Roche) for 10 min. Before treatment, the cells are transferred to ice to reduce receptor internalization. The cells are fixed using 4% paraformaldehyde (Electron Microscopy Sciences, USA) and 0.1% glutaraldehyde (Sigma-Aldrich) in phosphate-buffered saline (PBS). After fixation, to reduce background autofluorescence, cells are treated with sodium borohydride (NaBH4) (1 mg/ml; Sigma-Aldrich). Before fixation and between each step, the cells are washed using PBS supplemented with CaCl2 (100 μM), MgCl2 (100 μM), and 4% sucrose. The fixed cells are stored in PBS with CaCl2 and MgCl2 (100 μM each) and imaged in PBS within 1 week. Before imaging, TetraSpeck Microspheres, 0.1 μm (Thermo Fisher Scientific) were added to the sample as fiducial markers for correcting for drifts during SMLM imaging.
SMLM imaging was performed using a Nikon Eclipse Ti2-E (Nikon, Japan) inverted microscope using Nikon NIS Elements 4.11 acquisition software. The objective used was a Nikon 100× Apo TIRF oil immersion objective with 1.49 NA (numerical aperture), and samples were imaged in TIRF mode. First, cells were selected and the imaging plane adjusted using 488-nm laser (Argon, 65 mW) excitation and fluorescence emission filtering using a 525/50-emission filter. For PALM imaging, mEos3.2 was photoconverted using a 405-nm laser (100 mW) starting from 0.1% and gradually increasing up to 10%. Fluorescence excitation of mEos3.2 was conducted with continuous excitation with 561-nm laser (40%, 150 mW) and fluorescence emission filtering using a 605/52 filter. Images were recorded using back-illuminated electron multiplying charge-coupled device camera (Andor iXon DU-897) with a frame integration time of 80 ms and a detector gain of 300 until no further emission events were observed (approximately 20,000 to 30,000 frames). PALM localizations were obtained using the same Nikon NIS software, and localization lists were saved for further analysis.
qSMLM analysis
The qSMLM approach was used to determine the cell surface organization of EpoR receptors. This method uses superresolution localization imaging and reads out the photo-blinking properties of a fluorescent protein. Hence, information on the oligomeric state of proteins within small nanoclusters is accessible, which is beyond the spatial resolution of PALM microscopy (27, 28, 50). In this approach, an SMLM image stack is acquired using a superresolution localization microscope in the TIRF mode from cells expressing the protein of interest tagged with a photoactivable fluorescent protein, and a superresolved localization image is generated. From this images, the number of localizations in each nanocluster are extracted and compiled to yield a histogram, which is fitted with an appropriate model to understand the stoichiometry of proteins in the clusters as described previously (fig. S1) (29, 56, 57). To calibrate the qSMLM assay with the mEos3.2 protein in our cell line and for our imaging setup, we used membrane receptor proteins with known stoichiometry, the monomeric protein CD86, and covalent dimeric CTLA-4. The monomeric and dimeric nature of CD86 and CTLA-4 is well described, and they are widely used as calibration standards in SMLM-based molecular counting experiments (28, 29). Previous studies have demonstrated that the analysis-relevant photophysical parameters of the mEos variants can be transferred to other membrane receptors (27, 29). This makes CD86 and CTLA-4 tagged with mEos3.2 at the cytoplasmic region optimal for stoichiometry detection of EpoR-tagged similarly.
Experimentally, superresolution images generated by SMLM imaging was processed using ThunderSTORM (58) and postprocessed with LAMA (59) and qSMLM tools (https://github.com/SMLMS/qSMLM) (56) by methods developed previously (27, 28, 56). In this approach, the localization coordinate list of images obtained from Nikon SMLM was translated using ChriSTORM (60) to ThunderSTORM format. ThunderSTORM generated image was drift-corrected using the localizations from the fiducial markers, and the background noise localizations were removed using density filter. Spatiotemporal grouping was applied using the merge function of ThunderSTORM to group signals from mEos3.2 within a radius of 90 nm and combining off frames of 65 and with a maximum frame per molecule of 115 frames. This is followed by density-based spatial clustering of applications with noise (DBSCAN)–based clustering and processing in LAMA to obtain histograms from the clusters. Clusters with low brightness, a diameter of >150 nm, or in close vicinity to neighboring clusters were discarded from the analysis. These parameters enabled us to obtain histograms representing monomeric distribution for the monomer control CD86-mEos3.2 and a distinguishably different histogram for the dimer control CTLA-4-mEos3.2–expressing cells (fig. S2, A and B). The histograms were fitted using qSMLM tool with appropriate functions as described by Hummer et al. (56), to obtain the fraction of mEos3.2, which does not blink (P value), and the fraction of undetected fluorophore during the observation time (q value). The P value or the blinking probability value obtained with the monomeric control in our imaging setup was P = 0.55. The q value, which shows the fraction of fluorophore, which does not light up during the observation time was q = 0.31. This translates to the detection efficiency of 69% for mEos3.2 with our cell line and imaging setup. These values were then used to determine the oligomeric status of different EpoR-mEos3.2–expressing cells by fitting the data with all possible combination of fitting functions for monomer, dimer, and trimer using qSMLM tool as described previously (27, 56). To generate the histogram for fitting, the emission events from different cells were combined to a single histogram to increase the statistical strength, and to generate the error values, we regenerated the histograms for (N − 1 cells) and fitted each individual histogram to obtain the mean and error range.
FRET imaging
For FRET imaging, γ2A cells seeded on 35-mm glass-bottom dish (ibidi) were cotransfected with EpoR FRET plasmid, pLVX-Tet-On Advanced vector (Clontech), and JAK2 construct for 10 hours, and starved for 12 hours in medium supplemented with doxycycline (0.1 mg/ml). Cells were either stimulated with Epo (10 U/ml) (NeoRecormon, Roche) for 30 min or fixed directly with paraformaldehyde (4%) and glutaraldehyde (0.1%) for 15 min in room temperature, washed, and stored in PBS at 4°C before imaging. Imaging was performed with Zeiss LSM 780 laser scanning confocal microscope equipped with Plan Apochromat 63×/1.4 oil immersion objective. The CFP (cyan fluorescent protein) and YFP (yellow fluorescent protein) were excited by using 458- and 514-nm line, respectively, from a multiline argon laser. The fluorescence emission was acquired between 465 and 500 nm for CFP and 525 to 640 nm for YFP with a 32-channel QUASAR GaAsP photomultiplier array detector. Acceptor photobleaching method (61) was used to detect FRET efficiency, and 514-nm laser line (100%) was used for photobleaching the acceptor. The images were processed with Fiji software (62). The cell images were sorted for the membrane expression level using YFP fluorescence before photobleaching, which is independent of FRET. The cells with high levels of FRET construct expression were discarded. The cell membrane region was manually segmented from each image, and apparent FRET efficiency was calculated only from the membrane.
Kinase assay
For in vitro kinase assays, recombinant JAK2 JH2-JH1 (residues 513 to 1132-6xHis) WT, V617F, R683S, and K539L proteins were expressed in High Five insect cells (Thermo Fisher Scientific) by using the Bac-to-Bac expression system (Invitrogen), according to the manufacturer’s instructions. Cells were lysed by means of freeze-thawing and clarified by means of centrifugation, and recombinant proteins were purified with nickel–nitrilotriacetic acid (Ni-NTA) agarose (Qiagen), followed by size exclusion chromatography in a HiLoad 16/600 Superdex 75 pg column (GE Healthcare). Proteins were purified simultaneously to retain comparability. Protein concentrations were measured with the Bradford assay (Bio-Rad Laboratories), and enzymatic activity of recombinant proteins was determined without preactivation by the time-resolved FRET-based Lance Ultra kinase assay (PerkinElmer) as reported earlier (23). The activity parameters of kinase reaction catalysis rate constant (Kcat) and apparent Km for ATP were calculated by fitting reaction velocity versus ATP concentration in GraphPad Prism software. Relative catalytic efficiency for JAK2 mutants was calculated by dividing Kcat/apparent Km with the value for WT. Kinase reactions were performed in triplicate or quadruplicate, and results shown are average of three to four individual experiments.
Immunoblot analysis
The cells (transfected/stably expressing proteins) were washed with ice-cold PBS and lysed in cold Triton X-100 lysis buffer with protease and phosphatase inhibitors [2 mM vanadate, 1 mM phenylmethylsulfonyl fluoride (PMSF), aprotinin (8.3 μg/ml), and pepstatin (4.2 μg/ml)]. The lysates were spun for 20 min at 16,000g at 4°C, and the supernatants were analyzed with Jess (ProteinSimple), automated capillary electrophoresis, and immunodetection system. For protein separation and immunoassay, the manufacturer’s standard protocol for fluorescent (near-infrared) and chemiluminescent detection of proteins with 12- to 230-kDa Jess separation module (SM-W004) was followed. The proteins of interest were detected using primary antibodies against hemagglutinin tag (1:50; Cell Signaling Technology, #3724T and 1:50; Aviva Systems Biology, #OAEA00009) and phosphorylated JAK2 (pJAK2; 1:1000; Y1007/1008, Cell Signaling Technology, #3771). Analysis of capillary Western data was performed using Compass for SW (ProteinSimple) software version 6.1.0.
Protein expression and purification
JAK2 JH2 mutant constructs (residues 536 to 812) were subcloned into pFastBac vector with a C-terminal thrombin-cleavable His6 tag. The proteins were expressed in baculovirus-infected Spodoptera frugiperda-9 cells using the Bac-to-Bac baculovirus expression system (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. After protein expression, the cells were collected by centrifugation and resuspended in lysis buffer {50 mM tris-HCl (pH 8.0), 10% glycerol, 500 mM NaCl, 0.5 mM [tris(2-carboxyethyl)phosphine] (TCEP), and 20 mM imidazole} supplemented with 0.5 mM PMSF, 250 U of benzonase, and 5 mM MgCl2. The cells were lysed by applying four freeze-thaw cycles in liquid nitrogen. The lysates were clarified by centrifugation (30,000g at 4°C for 1 hour) and incubated with Ni-NTA resin at 4°C for 1 hour. The resin was transferred into a gravity-flow column and washed with lysis buffer. The proteins were eluted with 50 mM tris-HCl (pH 8.0), 10% glycerol, 500 mM NaCl, 0.5 mM TCEP, and 250 mM imidazole. The eluted protein was concentrated with Amicon Ultra-15 concentrators (10-kDa molecular weight cutoff) (Merck Millipore Ltd.) and further purified with Superdex 75 16/600 (GE Healthcare) gel filtration column equilibrated with 20 mM tris (pH 8.0), 500 mM NaCl, 10% glycerol, and 0.5 mM TCEP. The purified proteins were concentrated and stored at −80°C in small aliquots.
Protein crystallization
JAK2 JH2 mutants (5 to 10 mg/ml) were crystallized using a sitting-drop vapor-diffusion method. Equal volumes (0.5 μl) of well solution [0.2 M Na acetate or 0.1 M Mg acetate, 0.1 M tris-HCl (pH 7.5 to 8.5), and 16 to 22% polyethylene glycol 3350] and protein solution were mixed and incubated at 4°C. JAK2 JH2-K539L was crystallized in complex of 1 mM ATP and 3 mM MgCl2, and JAK2-R683S with a pyrazolo pyrimidine analog previously identified as JAK2 JH2 binder (63). Before data collection, the crystals were cryo-protected with 20% glycerol in well solution and flash-frozen in liquid nitrogen.
Data collection, structure determination, and refinement
Diffraction data were collected on the beam line I03 at Diamond Light Source, Didcot, UK. Data were processed and scaled with the XDS package (64). Structures were determined by molecular replacement using the JAK2 JH2 structure (PDB ID: 4FVR) (65) as a search model. The refinement was carried out with phenix.refine (66). Coot was used for visualization and manual building of the model (67). Data collection and refinement statistics are shown in table S3.
Atomistic MD simulations
Crystallographic dimer of the JAK2 V617F structure (PDB ID: 6D2I) (68) was used as the initial structure in the atomistic MD simulations. Hydrogens were added, and all amino acids were modeled in their standard protonation states. The system was solvated with water molecules, and a short energy minimization with the steepest descent algorithm was performed (1000 steps). The physiological conditions were introduced by adding salt (KCl, 150 mM) and by adding a sufficient number of counterions to achieve charge neutrality, which was followed by another 1000-step minimization. Subsequently, 1- and 2-ns equilibration runs in NVT and NPT ensembles, respectively, were performed with protein coordinates restrained with a force constant of 400 kJ mol−1 nm−2. Last, production runs were conducted as described in table S2. We used the CHARMM36m force field for all components of the system (69); however, water molecules were described with the TIP3P water model (70). Simulations were conducted using the GROMACS simulation package, version 2020 (71). We note that for the simulations involving the ATP-bound dimer variants (EQ8 to EQ11 in table S2), where each JH2 domain binds one ATP molecule, we sourced the ATP pose from the crystallographic monomer of JAK2 JH2 V617F (PDB ID: 4FVR). This pose was integrated into the dimer structure through protein structure superimposition.
The production runs, along with equilibration, were carried out by the leap-frog integrator with a time step of 2 fs. The LINCS (72) algorithm kept all bonds between hydrogen and heavy atoms constrained. Periodic boundary conditions were applied in all three dimensions. Long-range electrostatics were treated with the particle-mesh Ewald (73) method of the order of 4, applying a cutoff of 1.2 nm between short-range and long-range components. Neighbor search for long-range interactions was carried out every 10 steps. Van der Waals interactions were cut off at 1.2 nm. The Nosé-Hoover thermostat (74, 75) coupled the systems to a heat bath of 310 K, while the Parrinello-Rahman (76) barostat kept the pressure at 1.0 bar, with a compressibility of 4.5 × 10−5 bar−1. At the beginning of each simulation, we assigned random initial velocities using the Boltzmann distribution. The simulation trajectories were saved every 100 ps.
The stability of our JH2 dimer model was assessed by calculating its RMSD values in equilibrium simulations (EQ1 to EQ11 in table S2). Here, we defined three ranges for the RMSD values, such that the domains are either (i) bound in the predicted pose when the RMSD of the dimer falls below 0.8 nm, (ii) attached but in a less-defined pose when RMSD is between 0.8 and 2.0 nm, or (iii) detached when RMSD is higher than 2.0 nm. These intervals were chosen through visual inspection to best reflect the bound state of the dimer; however, we confirmed through additional studies that the classification to these three states was not sensitive to the choices of RMSD values used in the analysis. Figure 5C shows probabilities for all dimer variants to be in each of the defined states. RMSD was calculated for protein heavy atoms every 10 ns.
Free-energy calculations
In the umbrella sampling simulations (systems FE1 to FE5), we pulled the two JH2 subunits apart using a series of umbrella sampling windows (see table S2). In practice, we started from the unbiased MD-relaxed (see above) dimer structure and increased the JH2-JH2 distance by 1.4 Å at every new sampling window using the pull_init option of GROMACS to set a new distance. Each sampling window was simulated for a period of 450 ns, and all the data were used for the analysis of the potential of mean force using the weighted histogram analysis method (WHAM), which is implemented as the gmx wham tool (77, 78) in GROMACS. In the sampling windows, a force constant of 1000 kJ mol−1 nm−2 was used to constrain the distance between the JH2 domains. The other subunit was restrained with a force constant of 1000 kJ mol−1 nm−2 from the heavy atoms of residues 546 to 744. The potential of mean force profiles were shifted so that the free energy reached a level of zero at large JH2-JH2 separation (5.6 nm). The minimum of the potential of mean force (ΔGUS) is reported in Fig. 5D for each variant. Error estimates were calculated by the bootstrap analysis implemented within the gmx wham code.
As a complementary approach, being lighter than the computationally very laborious umbrella sampling free-energy calculations, the binding free energy (ΔGMM/GBSA) between JH2-JH2 variants was also calculated using the molecular MM/GBSA analysis (38). Solvation energy was calculated with the GBOBC2 model (79) at a temperature of 310 K and 0.15 M salt concentration. Frames for MM/GBSA analysis were taken every 10 ns from the last 500 ns of equilibrium runs (see table S2) of four replicas for each variant.
AlphaFold-based structural models
The predicted human full-length JAK2 structure (UniProt code O60674) was acquired from the AlphaFold database (80, 81). Two AlphaFold JAK2 structures were aligned to the JAK2 JH2-V617F and JAK2 JH2-K539L dimers to construct full-length models of activated JAK2 dimers. The dimer models were subjected to MD refinement in a water-ion solution. The simulation setup was prepared using CHARMM-GUI (82) tools. MD simulations were performed (as described above) in three replicas for each model, and each simulation replica covered a timescale of 1 μs.
Acknowledgments
M. Lehtinen and M. Vähä-Jaakkola are acknowledged for technical assistance and the Biocenter Finland Imaging Infrastructure (https://biocenter.fi/) for services. We acknowledge high-performance computing support granted by the CSC-IT Center for Science Ltd., Finland. Protein crystallographic experiments were performed on the I03 beamline at the Diamond Light Source (Didcot, UK). We are grateful to the staff at the beamline for assistance with data collection. Biomedicum Imaging Unit, Helsinki University, and Tampere Imaging Facility (TIF) are acknowledged for microscopy services.
Funding: This work was supported by Academy of Finland grants 287573 (to O.S.), 338176 (to V.S.), 340572 (T.H.), and 331349 and 346135 (to I.V.); Helsinki Institute of Life Science (HiLIFE) Fellow Program (to I.V.); Human Frontier Science Program RGP0059/2019 (to I.V.); Sigrid Juselius Foundation (to O.S., V.S., and I.V.); Finnish Cancer Foundation (to O.S., V.S., and I.V.); Jane and Aatos Erkko Foundation (to O.S. and V.S.); Magnus Ehrnrooth Foundation (to O.S. and V.S.); Tampere Tuberculosis Foundation (to O.S.); Lundbeck Foundation (to I.V.); Competitive Research Funding of the Tampere University Hospital grant 9 V061 (to O.S.); and Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) grants SFB 807 and 1507 (to M.H. and C.K.).
Author contributions: Methodology: B.G.A., T.H., J.V., M.G., A.T.V., V.S., I.V., and O.S. Investigation: B.G.A., T.H., J.V., M.G., A.T.V., and A.K. Provided resources: I.V. Expertise on quantitative analysis of PALM data: M.H. and C.K. Supervision: M.H., V.S., I.V., and O.S. Writing: B.G.A., T.H., J.V., M.G., V.S., I.V., and O.S.
Competing interests: O.S. reports lecture fees from Pfizer, Abbvie, and Novartis and is a member of scientific advisory board of Ajax Therapeutics and chair of scientific advisory board of Finnish hematology registry and biobank. O.S. holds a patent on JAK kinases, US patent no. 8,841,078 (O. Silvennoinen, active, date granted 12 June 2014), and reports royalties or licenses from St. Jude Children’s Research Hospital and stock option for Ajax Therapeutics. B.G.A., T.H., J.V., M.G., A.T.V., A.K., C.K., M.H., V.S., and I.V. declare no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. X-ray data are available at PDB with accession codes 8C08 (JAK2 JH2-K539L), 8C0A (JAK2 JH2-R683S), and 8C09 (JAK2 JH2-I559F).
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1 to S3
Legend for file S1
References
Other Supplementary Material for this manuscript includes the following:
File S1
REFERENCES AND NOTES
- 1.Hammarén H. M., Virtanen A. T., Raivola J., Silvennoinen O., The regulation of JAKs in cytokine signaling and its breakdown in disease. Cytokine 118, 48–63 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Philips R. L., Wang Y., Cheon H., Kanno Y., Gadina M., Sartorelli V., Horvath C. M., Darnell J. E., Stark G. R., O’Shea J. J., The JAK-STAT pathway at 30: Much learned, much more to do. Cell 185, 3857–3876 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witthuhn B. A., Quelle F. W., Silvennoinen O., Yi T., Tang B., Miura O., Ihle J. N., JAK2 associates with the erythropoietin receptor and is tyrosine phosphorylated and activated following stimulation with erythropoietin. Cell 74, 227–236 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Parganas E., Wang D., Stravopodis D., Topham D. J., Marine J.-C., Teglund S., Vanin E. F., Bodner S., Colamonici O. R., van Deursen J. M., Grosveld G., Ihle J. N., Jak2 is essential for signaling through a variety of cytokine receptors. Cell 93, 385–395 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Brown R. J., Adams J. J., Pelekanos R. A., Wan Y., McKinstry W. J., Palethorpe K., Seeber R. M., Monks T. A., Eidne K. A., Parker M. W., Waters M. J., Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat. Struct. Mol. Biol. 12, 814–821 (2005). [DOI] [PubMed] [Google Scholar]
- 6.de la Chapelle A., Träskelin A. L., Juvonen E., Truncated erythropoietin receptor causes dominantly inherited benign human erythrocytosis. Proc. Natl. Acad. Sci. U.S.A. 90, 4495–4499 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watowich S. S., The erythropoietin receptor: Molecular structure and hematopoietic signaling pathways. J. Invest. Med. 59, 1067–1072 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silvennoinen O., Hubbard S. R., Molecular insights into regulation of JAK2 in myeloproliferative neoplasms. Blood 125, 3388–3392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baxter E. J., Scott L. M., Campbell P. J., East C., Fourouclas N., Swanton S., Vassiliou G. S., Bench A. J., Boyd E. M., Curtin N., Scott M. A., Erber W. N., Green A. R., Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365, 1054–1061 (2005). [DOI] [PubMed] [Google Scholar]
- 10.James C., Ugo V., Le Couédic J.-P., Staerk J., Delhommeau F., Lacout C., Garçon L., Raslova H., Berger R., Bennaceur-Griscelli A., Villeval J. L., Constantinescu S. N., Casadevall N., Vainchenker W., A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 434, 1144–1148 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Kralovics R., Passamonti F., Buser A. S., Teo S.-S., Tiedt R., Passweg J. R., Tichelli A., Cazzola M., Skoda R. C., A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 352, 1779–1790 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Levine R. L., Wadleigh M., Cools J., Ebert B. L., Wernig G., Huntly B. J. P., Boggon T. J., Wlodarska I., Clark J. J., Moore S., Adelsperger J., Koo S., Lee J. C., Gabriel S., Mercher T., D’Andrea A., Fröhling S., Döhner K., Marynen P., Vandenberghe P., Mesa R. A., Tefferi A., Griffin J. D., Eck M. J., Sellers W. R., Meyerson M., Golub T. R., Lee S. J., Gilliland D. G., Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 7, 387–397 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Scott L. M., Tong W., Levine R. L., Scott M. A., Beer P. A., Stratton M. R., Futreal P. A., Erber W. N., McMullin M. F., Harrison C. N., Warren A. J., Gilliland D. G., Lodish H. F., Green A. R., JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N. Engl. J. Med. 356, 459–468 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bercovich D., Ganmore I., Scott L. M., Wainreb G., Birger Y., Elimelech A., Shochat C., Cazzaniga G., Biondi A., Basso G., Cario G., Schrappe M., Stanulla M., Strehl S., Haas O. A., Mann G., Binder V., Borkhardt A., Kempski H., Trka J., Bielorei B., Avigad S., Stark B., Smith O., Dastugue N., Bourquin J.-P., Tal N. B., Green A. R., Izraeli S., Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down’s syndrome. Lancet 372, 1484–1492 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Carreño-Tarragona G., Varghese L. N., Sebastián E., Gálvez E., Marín-Sánchez A., López-Muñoz N., Nam-Cha S., Martínez-López J., Constantinescu S. N., Sevilla J., Ayala R., A typical acute lymphoblastic leukemia JAK2 variant, R683G, causes an aggressive form of familial thrombocytosis when germline. Leukemia 35, 3295–3298 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Saharinen P., Takaluoma K., Silvennoinen O., Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol. Cell. Biol. 20, 3387–3395 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saharinen P., Silvennoinen O., The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J. Biol. Chem. 277, 47954–47963 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Saharinen P., Vihinen M., Silvennoinen O., Autoinhibition of Jak2 tyrosine kinase is dependent on specific regions in its pseudokinase domain. Mol. Biol. Cell 14, 1448–1459 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shan Y., Gnanasambandan K., Ungureanu D., Kim E. T., Hammarén H., Yamashita K., Silvennoinen O., Shaw D. E., Hubbard S. R., Molecular basis for pseudokinase-dependent autoinhibition of JAK2 tyrosine kinase. Nat. Struct. Mol. Biol. 21, 579–584 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lupardus P. J., Ultsch M., Wallweber H., Bir Kohli P., Johnson A. R., Eigenbrot C., Structure of the pseudokinase–kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc. Natl. Acad. Sci. U.S.A. 111, 8025–8030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilmes S., Hafer M., Vuorio J., Tucker J. A., Winkelmann H., Löchte S., Stanly T. A., Pulgar Prieto K. D., Poojari C., Sharma V., Richter C. P., Kurre R., Hubbard S. R., Garcia K. C., Moraga I., Vattulainen I., Hitchcock I. S., Piehler J., Mechanism of homodimeric cytokine receptor activation and dysregulation by oncogenic mutations. Science 367, 643–652 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leroy E., Balligand T., Pecquet C., Mouton C., Colau D., Shiau A. K., Dusa A., Constantinescu S. N., Differential effect of inhibitory strategies of the V617 mutant of JAK2 on cytokine receptor signaling. J. Allergy Clin. Immunol. 144, 224–235 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Hammarén H. M., Virtanen A. T., Abraham B. G., Peussa H., Hubbard S. R., Silvennoinen O., Janus kinase 2 activation mechanisms revealed by analysis of suppressing mutations. J. Allergy Clin. Immunol. 143, 1549–1559.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glassman C. R., Tsutsumi N., Saxton R. A., Lupardus P. J., Jude K. M., Garcia K. C., Structure of a Janus kinase cytokine receptor complex reveals the basis for dimeric activation. Science 376, 163–169 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caveney N. A., Saxton R. A., Waghray D., Glassman C. R., Tsutsumi N., Hubbard S. R., Garcia K. C., Structural basis of Janus kinase trans-activation. Cell Rep. 42, 112201 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staerk J., Kallin A., Demoulin J.-B., Vainchenker W., Constantinescu S. N., JAK1 and Tyk2 activation by the Homologous Polycythemia Vera JAK2 V617F mutation: Cross-talk with IGF1 receptor. J. Biol. Chem. 280, 41893–41899 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Karathanasis C., Medler J., Fricke F., Smith S., Malkusch S., Widera D., Fulda S., Wajant H., van Wijk S. J. L., Dikic I., Heilemann M., Single-molecule imaging reveals the oligomeric state of functional TNFα-induced plasma membrane TNFR1 clusters in cells. Sci. Signal. 13, eaax5647 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Fricke F., Beaudouin J., Eils R., Heilemann M., One, two or three? Probing the stoichiometry of membrane proteins by single-molecule localization microscopy. Sci. Rep. 5, 14072 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krüger C. L., Zeuner M.-T., Cottrell G. S., Widera D., Heilemann M., Quantitative single-molecule imaging of TLR4 reveals ligand-specific receptor dimerization. Sci. Signal. 10, eaan1308 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Kasai R. S., Ito S. V., Awane R. M., Fujiwara T. K., Kusumi A., The class-A GPCR dopamine D2 receptor forms transient dimers stabilized by agonists: Detection by single-molecule tracking. Cell Biochem. Biophys. 76, 29–37 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salavessa L., Lagache T., Malardé V., Grassart A., Olivo-Marin J.-C., Canette A., Trichet M., Sansonetti P. J., Sauvonnet N., Cytokine receptor cluster size impacts its endocytosis and signaling. Proc. Natl. Acad. Sci. U.S.A. 118, e2024893118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang L. J., Constantinescu S. N., Lodish H. F., The N-terminal domain of Janus kinase 2 is required for Golgi processing and cell surface expression of erythropoietin receptor. Mol. Cell 8, 1327–1338 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Passamonti F., Elena C., Schnittger S., Skoda R. C., Green A. R., Girodon F., Kiladjian J.-J., McMullin M. F., Ruggeri M., Besses C., Vannucchi A. M., Lippert E., Gisslinger H., Rumi E., Lehmann T., Ortmann C. A., Pietra D., Pascutto C., Haferlach T., Cazzola M., Molecular and clinical features of the myeloproliferative neoplasm associated with JAK2 exon 12 mutations. Blood 117, 2813–2816 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Andraos R., Qian Z., Bonenfant D., Rubert J., Vangrevelinghe E., Scheufler C., Marque F., Régnier C. H., Pover A. D., Ryckelynck H., Bhagwat N., Koppikar P., Goel A., Wyder L., Tavares G., Baffert F., Pissot-Soldermann C., Manley P. W., Gaul C., Voshol H., Levine R. L., Sellers W. R., Hofmann F., Radimerski T., Modulation of activation-loop phosphorylation by JAK inhibitors is binding mode dependent. Cancer Discov. 2, 512–523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leroy E., Dusa A., Colau D., Motamedi A., Cahu X., Mouton C., Huang L. J., Shiau A. K., Constantinescu S. N., Uncoupling JAK2 V617F activation from cytokine-induced signalling by modulation of JH2 αC helix. Biochem. J. 473, 1579–1591 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dusa A., Mouton C., Pecquet C., Herman M., Constantinescu S. N., JAK2 V617F constitutive activation requires JH2 residue F595: A pseudokinase domain target for specific inhibitors. PLOS ONE 5, e11157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raivola J., Haikarainen T., Silvennoinen O., Characterization of JAK1 pseudokinase domain in cytokine signaling. Cancers 12, 78 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valdés-Tresanco M. S., Valdés-Tresanco M. E., Valiente P. A., Moreno E., gmx_MMPBSA: A new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theory Comput. 17, 6281–6291 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Hoy S. M., Deucravacitinib: First approval. Drugs 82, 1671–1679 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammarén H. M., Ungureanu D., Grisouard J., Skoda R. C., Hubbard S. R., Silvennoinen O., ATP binding to the pseudokinase domain of JAK2 is critical for pathogenic activation. Proc. Natl. Acad. Sci. U.S.A. 112, 4642–4647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrao R. D., Wallweber H. J., Lupardus P. J., Receptor-mediated dimerization of JAK2 FERM domains is required for JAK2 activation. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livnah O., Stura E. A., Johnson D. L., Middleton S. A., Mulcahy L. S., Wrighton N. C., Dower W. J., Jolliffe L. K., Wilson I. A., Functional mimicry of a protein hormone by a peptide agonist: The EPO receptor complex at 2.8 A. Science 273, 464–471 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Syed R. S., Reid S. W., Li C., Cheetham J. C., Aoki K. H., Liu B., Zhan H., Osslund T. D., Chirino A. J., Zhang J., Finer-Moore J., Elliott S., Sitney K., Katz B. A., Matthews D. J., Wendoloski J. J., Egrie J., Stroud R. M., Efficiency of signalling through cytokine receptors depends critically on receptor orientation. Nature 395, 511–516 (1998). [DOI] [PubMed] [Google Scholar]
- 44.Remy I., Wilson I. A., Michnick S. W., Erythropoietin receptor activation by a ligand-induced conformation change. Science 283, 990–993 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Constantinescu S. N., Keren T., Socolovsky M., Nam H., Henis Y. I., Lodish H. F., Ligand-independent oligomerization of cell-surface erythropoietin receptor is mediated by the transmembrane domain. Proc. Natl. Acad. Sci. U.S.A. 98, 4379–4384 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corbett M. S. P., Mark A. E., Poger D., Do all x-ray structures of protein-ligand complexes represent functional states? EPOR, a case study. Biophys. J. 112, 595–604 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hubbard S. R., Mechanistic insights into regulation of JAK2 tyrosine kinase. Front. Endocrinol. 8, 361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atanasova M., Whitty A., Understanding cytokine and growth factor receptor activation mechanisms. Crit. Rev. Biochem. Mol. Biol. 47, 502–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Babon J. J., Lucet I. S., Murphy J. M., Nicola N. A., Varghese L. N., The molecular regulation of Janus kinase (JAK) activation. Biochem. J. 462, 1–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baldering T. N., Bullerjahn J. T., Hummer G., Heilemann M., Malkusch S., Molecule counts in complex oligomers with single-molecule localization microscopy. J. Phys. D Appl. Phys. 52, 474002 (2019). [Google Scholar]
- 51.Brooks A. J., Dai W., O’Mara M. L., Abankwa D., Chhabra Y., Pelekanos R. A., Gardon O., Tunny K. A., Blucher K. M., Morton C. J., Parker M. W., Sierecki E., Gambin Y., Gomez G. A., Alexandrov K., Wilson I. A., Doxastakis M., Mark A. E., Waters M. J., Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science 344, 1249783 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Yao H., Ma Y., Hong Z., Zhao L., Monaghan S. A., Hu M.-C., Huang L. J., Activating JAK2 mutants reveal cytokine receptor coupling differences that impact outcomes in myeloproliferative neoplasm. Leukemia 31, 2122–2131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grisouard J., Li S., Kubovcakova L., Rao T. N., Meyer S. C., Lundberg P., Hao-Shen H., Romanet V., Murakami M., Radimerski T., Dirnhofer S., Skoda R. C., JAK2 exon 12 mutant mice display isolated erythrocytosis and changes in iron metabolism favoring increased erythropoiesis. Blood 128, 839–851 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Haan C., Rolvering C., Raulf F., Kapp M., Drückes P., Thoma G., Behrmann I., Zerwes H.-G., Jak1 has a dominant role over Jak3 in signal transduction through γc-containing cytokine receptors. Chem. Biol. 18, 314–323 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Kohlhuber F., Rogers N. C., Watling D., Feng J., Guschin D., Briscoe J., Witthuhn B. A., Kotenko S. V., Pestka S., Stark G. R., Ihle J. N., Kerr I. M., A JAK1/JAK2 chimera can sustain α and γ interferon responses. Mol. Cell. Biol. 17, 695–706 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hummer G., Fricke F., Heilemann M., Model-independent counting of molecules in single-molecule localization microscopy. Mol. Biol. Cell 27, 3637–3644 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krüger C., Fricke F., Karathanasis C., Dietz M. S., Malkusch S., Hummer G., Heilemann M., Molecular counting of membrane receptor subunits with single-molecule localization microscopy. Proc. SPIE 10071, 100710K (2017). [Google Scholar]
- 58.Ovesný M., Křížek P., Borkovec J., Svindrych Z., Hagen G. M., ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malkusch S., Heilemann M., Extracting quantitative information from single-molecule super-resolution imaging data with LAMA—Localization microscopy analyzer. Sci. Rep. 6, 34486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leterrier C., Potier J., Caillol G., Debarnot C., Rueda Boroni F., Dargent B., Nanoscale architecture of the axon initial segment reveals an organized and robust scaffold. Cell Rep. 13, 2781–2793 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Bastiaens P. I., Majoul I. V., Verveer P. J., Söling H. D., Jovin T. M., Imaging the intracellular trafficking and state of the AB5 quaternary structure of cholera toxin. EMBO J. 15, 4246–4253 (1996). [PMC free article] [PubMed] [Google Scholar]
- 62.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Virtanen A. T., Haikarainen T., Sampathkumar P., Palmroth M., Liukkonen S., Liu J., Nekhotiaeva N., Hubbard S. R., Silvennoinen O., Identification of novel small molecule ligands for JAK2 pseudokinase domain. Pharmaceuticals 16, 75 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kabsch W., XDS. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bandaranayake R. M., Ungureanu D., Shan Y., Shaw D. E., Silvennoinen O., Hubbard S. R., Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat. Struct. Mol. Biol. 19, 754–759 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liebschner D., Afonine P. V., Baker M. L., Bunkóczi G., Chen V. B., Croll T. I., Hintze B., Hung L.-W., Jain S., McCoy A. J., Moriarty N. W., Oeffner R. D., Poon B. K., Prisant M. G., Read R. J., Richardson J. S., Richardson D. C., Sammito M. D., Sobolev O. V., Stockwell D. H., Terwilliger T. C., Urzhumtsev A. G., Videau L. L., Williams C. J., Adams P. D., Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D. Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McNally R., Li Q., Li K., Dekker C., Vangrevelinghe E., Jones M., Chène P., Machauer R., Radimerski T., Eck M. J., Discovery and structural characterization of ATP-site ligands for the wild-type and V617F mutant JAK2 pseudokinase domain. ACS Chem. Biol. 14, 587–593 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B. L., Grubmüller H., MacKerell A. D., CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 71.Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., Lindahl E., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015). [Google Scholar]
- 72.Hess B., Bekker H., Berendsen H. J. C., Fraaije J. G. E. M., LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997). [Google Scholar]
- 73.Darden T., York D., Pedersen L., Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
- 74.Hoover W. G., Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A. Gen. Phys. 31, 1695–1697 (1985). [DOI] [PubMed] [Google Scholar]
- 75.Nosé S., A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 81, 511–519 (1984). [Google Scholar]
- 76.Parrinello M., Rahman A., Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981). [Google Scholar]
- 77.Hub J. S., de Groot B. L., van der Spoel D., g_wham—A free weighted histogram analysis implementation including robust error and autocorrelation estimates. J. Chem. Theory Comput. 6, 3713–3720 (2010). [Google Scholar]
- 78.Kumar S., Rosenberg J. M., Bouzida D., Swendsen R. H., Kollman P. A., The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 13, 1011–1021 (1992). [Google Scholar]
- 79.Onufriev A., Bashford D., Case D. A., Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 55, 383–394 (2004). [DOI] [PubMed] [Google Scholar]
- 80.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A., Žídek A., Green T., Tunyasuvunakool K., Petersen S., Jumper J., Clancy E., Green R., Vora A., Lutfi M., Figurnov M., Cowie A., Hobbs N., Kohli P., Kleywegt G., Birney E., Hassabis D., Velankar S., AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 83.Laskowski R. A., Jabłońska J., Pravda L., Vařeková R. S., Thornton J. M., PDBsum: Structural summaries of PDB entries. Protein Sci. 27, 129–134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 to S3
Legend for file S1
References
File S1