Abstract
The period (per) and timeless (tim) genes encode key components of the circadian oscillator in Drosophila melanogaster. The per gene is thought to encode three transcripts via differential splicing (types A, B, and C) that give rise to three proteins. Since the three per mRNA types were based on the analysis of cDNA clones, we tested whether these mRNA types were present in vivo by RNase protection assays and reverse transcriptase-mediated PCR. The results show that per generates two transcript types that differ only by the presence (type A) or absence (type B′) of an alternative intron in the 3′ untranslated region. Transgenic flies containing transgenes that produce only type B′ transcripts (perB′), type A transcripts (perA), or both transcripts (perG) rescue locomotor activity rhythms with average periods of 24.7, 25.4, and 24.4 h, respectively. Although no appreciable differences in type A and type B′ mRNA cycling were observed, a slower accumulation of PER in flies making only type A transcripts suggests that the intron affects the translation of per mRNA.
The period (per) and timeless (tim) genes encode key components of the circadian oscillator in Drosophila melanogaster. The expression of these genes is required for circadian clock function, and an important aspect of their expression is circadian fluctuations in their mRNA and protein levels (17). These rhythms in per and tim gene products are controlled by a circadian feedback loop in which PER and TIM proteins control the expression of their own mRNAs (17, 25, 26). This feedback is mediated predominantly at the transcriptional level, though posttranscriptional regulation is also involved (5, 16, 29, 31, 32). The role of PER in this process is unknown, but its lack of a known DNA binding domain and inability to bind DNA indicate that it does not regulate transcription directly (17, 25).
Analysis of per cDNA clones uncovered three splice variants that encode three different PER isoforms (3). The most abundant of these transcripts, type A, defined both the structure of the per gene and the prototypical 1,218-amino-acid PER protein. Type B transcripts differ from type A by having two additional introns; one removes 288 nucleotides (nt) from exon 5 of type A transcripts, and the other excises 89 nt from the 3′ untranslated region (3′UTR) of exon 8. After excising the intron from exon 5, type B transcripts produce a protein that is 96 amino acids shorter. The least abundant transcript (only one partial cDNA clone was isolated), type C, differs from type A by retaining introns 5, 6, and 7, thereby producing a transcript whose exon 5 spans exons 5 to 8 in type A transcripts. Due to the inclusion of these additional introns, the last 107 amino acids of the putative type C protein sequence are entirely different from the last 149 amino acids of type A protein sequence. All three per cDNAs are capable of rescuing behavioral rhythms in per01 flies, though the type C construct may mediate behavioral rescue by generating both type A and type B transcripts (3, 4).
Given the critical role that PER plays in controlling the circadian feedback loop in Drosophila, it is important to determine which isoforms contribute to the feedback loop mechanism and what impact this contribution may have on behavioral rhythms. Since the initial characterization of per mRNA splice variants was based on the structure and abundance of partial cDNA clones, we tested whether these per transcripts exist in vivo and function equally to rescue locomotor activity rhythms.
Our studies failed to detect per splice variants that generate different PER isoforms. However, two per transcripts that differ by an alternatively spliced intron within their 3′UTRs were found; type A contains the 89-bp intron, and type B′ lacks this intron. Transgenes that produce type A mRNA, type B′ mRNA, or both mRNA types each rescue robust locomotor activity rhythms, but the period of these rhythms tends to be longer in the transgene that produces only type A transcripts. Type A and type B′ transcripts are indistinguishable with respect to circadian cycling, but the levels of PER derived from the transgene expressing only type A transcripts rise with a later phase than PER derived from transgenes expressing only type B′ or both type A and type B′ transcripts. These results suggest that the alternatively spliced intron alters the translation of per mRNA.
MATERIALS AND METHODS
RNase protection probes.
To make RNase protection probe 1, a sense primer (5′-CCAAGCTTCACTCCACGCAC-3′) starting at 5849 (numbering of exon, intron, and nucleotide sequences follows the type A cDNA structure in reference 3) and an antisense primer (5′-CATGTACTGCAGCGGC-3′) ending at 6053 were used with the per gene as a template to generate a 204-bp per DNA fragment via PCR. After HindIII and PstI digestion, this fragment was cloned into pBluescript KS, forming plasmid BSE5. Plasmid BSE5 was linearized by HindIII and transcribed by T7 RNA polymerase to generate RNase protection probe 1. To make RNase protection probe 2, a PCR product generated from the per gene by using a sense primer (5′-GCGGATCCGTAAGTCAGTG-3′) starting at 6227 and an antisense primer (5′-GGAAGCTTCTGCAAAAGAAC-3′) ending at 6797 was cloned into pBluescript KS after BamHI and HindIII digestion to form plasmid Intron 5-7. Plasmid Intron 5-7 was linearized by BamHI and transcribed by T3 RNA polymerase to generate probe 2. To produce RNase protection probe 3, a PCR DNA fragment was generated from the per gene by using a sense primer (5′-CACGGACGGATCGGAGAGTC-3′) starting at 6678 and an antisense primer (5′-CCGAATTCGCTTCGATGTTCGAACCAC-3′) ending at 7280. The PCR product was blunt ended with the Klenow fragment of DNA polymerase I, digested with EcoRI, and cloned into pBluescript KS, thus forming plasmid Exon 8. Exon 8 was linearized with BamHI and transcribed with T3 RNA polymerase to make probe 3. To generate a plasmid for probe 4, a PCR fragment was synthesized by using the same primers as for probe 3 except that a mutant perA gene was used as the template. This fragment was cloned into pGEM-T Easy vector, linearized with EcoRI, and transcribed with T7 RNA polymerase to make probe 4 (Fig. 1 and 6).
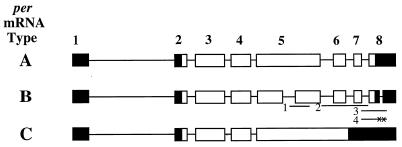
FIG. 1.
Structures of type A, B, and C per transcripts, based on cDNA analysis. Transcript splicing patterns are represented as boxes (exons) and lines (introns), where open boxes denote coding sequences and black boxes denote noncoding sequences. Exons are numbered only in the type A cDNA. In the type B cDNA, RNase protection probes 1 to 4 are shown. Probe 1 spans the 3′ junction of the type B-specific alternative splice event in exon 5, probe 2 extends from the start of intron 5 (with respect to type A) to the end of intron 7, probe 3 extends from the end of exon 7 to 220 bp downstream of the type B-specific intron in the 3′UTR, and probe 4 is the same as probe 3 except that the 5′ and 3′ splice junctions of the type B-specific intron in the 3′UTR are replaced by two EcoRI sites which are indicated by x’s.
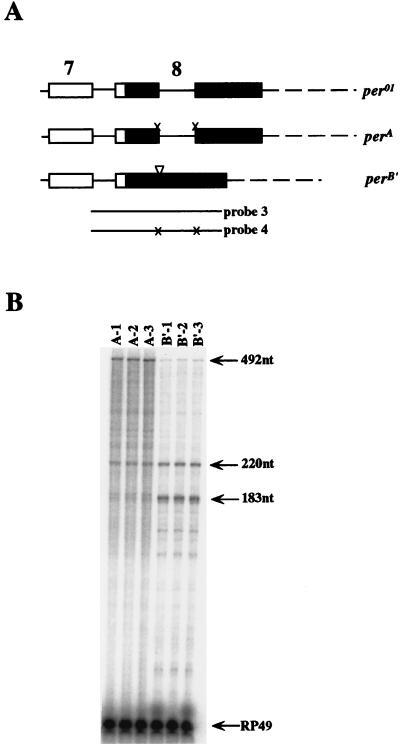
FIG. 6.
perA and perB′ produce the expected per transcript types. (A) Map showing the differences between the perA transgene, the perB′ transgene, and the endogenous per01 gene in the 3′UTR. White boxes, coding sequences; black boxes, noncoding sequences; thin lines, introns; broken lines, per 3′ flanking sequences; x’s in perA, mutagenized splice junctions; inverted triangle in perB′, deleted intron. RNase protection probes 3 and 4 are depicted as thick lines, where probe 4 contains the same mutagenized sequences as the perA transgene. (B) RNase protection assays for three perA and perB′ transgenic lines. perA and perB′ flies were collected in constant light so that RNA levels among lines could be compared. RNase protection probe 3 was used in perB′ transgenic flies, and probe 4 was used in perA transgenic flies. Both probes protect one type A band of 492 nt and two type B′ bands of 220 and 183 nt. In y per01 w;perA transgenic flies, only transcripts from the perA transgene are protected as a type A band of 492 nt; both type A and type B′ transcripts from the endogenous per01 gene are protected as type B′ bands because probe 4 is perA specific. The 183-nt type B′ band in perA lines is a doublet resulting from incomplete digestion of the 183-nt band at the mutagenized end. In y per01w;perB′ transgenic lines, the endogenous per01 gene generates both type A and type B′ transcripts but the perB′ transgene generates only type B′ transcripts.
Transformation plasmids.
Mutant per genes which specifically generate either type A (perA), type B′ (perB′), or both type A and type B′ (perG) transcripts were constructed. To make the perA gene, the 5′ and 3′ splice junctions of the type B-specific intron in the 3′UTR were mutagenized by oligonucleotide-mediated in vitro mutagenesis (19). The 1.6-kb BamHI-to-HindIII per genomic DNA fragment was cloned into a modified M13 phage (pTL) kindly provided by James Carrington. A 5′ splice donor junction primer (5′-CTGGAAACGAATTCGCAATTGCC-3′) from 6880 to 6902 and a 3′ splice acceptor junction primer (5′-CCCTTCGAATTCTTCGAATCAACG-3′) from 7060 to 7083 were used in mutagenesis so that both the 5′ and 3′ splice junctions were mutagenized to EcoRI sites. The mutagenized BamHI-to-HindIII fragment was used to replace the corresponding wild-type fragment in the 13.2-kb per genomic sequence (3) and cloned into pCaSpeR4 to form perA. The perB′ transgene was constructed by replacing the ApaLI-to-HindIII fragment in exon 8 of per with the corresponding region of type B′ cDNA, which is missing the type B′ alternative intron sequence. A 1.3-kb per genomic DNA fragment (from the BamHI site in exon 5 to the ApaLI site in exon 8) and a type B′ per cDNA fragment (from ApaLI to HindIII in exon 8) were ligated into a BamHI- and HindIII-digested pBluescript KS vector. This mutant 1.5-kb BamHI-to-HindIII per fragment was used to replace the corresponding region in the wild-type 13.2-kb per genomic sequence and cloned into pCaSpeR4 to generate perB′. The wild-type 13.2kb per transgene was made by first inserting a 3.7-kb BamHI-to-EcoRI fragment (containing the 3′ part of per and flanking sequences) into the pCaSpeR4 transformation vector, forming clone 22. A 9.5-kb XhoI-to-BamHI fragment (containing the 5′ part of per plus upstream regulatory sequences) was then inserted into clone 22, thereby forming perG.
RNase protection assays.
RNase protection assays on total RNA from heads, male bodies, or other tissues were done as described previously (14). Ten micrograms of DNase-treated total RNA was used for each time point in all the experiments except those represented in Fig. 5; in the latter case, the flies were dried with acetone and dissected under a dissecting microscope as described previously (15, 36), and different amounts of DNase-treated total RNA were used for the different tissues (see the legend to Fig. 5). Quantitation of RNase protection assays was done with a Fujix BAS 2000 phosphorimager using MacBas software. RNase protection probe 1 spans the 3′ splice junction of the type B-specific alternative intron in exon 5 (Fig. 1) and protects 204 nt of type A transcripts and 157 nt of type B transcripts. RNase protection probe 2 spans from the start of intron 5 to the end of intron 7 of per and protects 236 nt of exon 6 and 142 nt of exon 7. Probe 2 also protects splice precursors of various sizes, including the 570-nt one arising from the entire intron 5-to-intron 7 sequence. RNase protection probe 3, which extends from the end of exon 7 to 200 bp downstream of the type B′ alternative intron, can distinguish between type A, type B, and type C transcripts. Probe 3 should protect a 492-nt band corresponding to type A transcripts, 183- and 220-nt bands corresponding to type B transcripts, and a 611-nt band corresponding to type C transcripts.
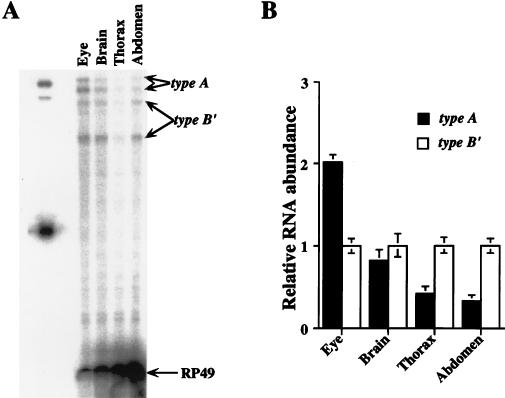
FIG. 5.
Tissue distribution of type A and type B′ transcripts. (A) RNase protection assays were performed with probe 3 (Fig. 1) on total RNA samples prepared from dissected eyes, eyeless heads (brain), thoraxes, and abdomens of CS flies collected at ZT15. The amounts of total RNA used for the samples were 5 μg (eye), 10 μg (brain), 20 μg (thorax), and 30 μg (abdomen). type A, the protected type A per transcript which is split into 254- and 235-nt bands because of a DNA polymorphism in the alternative intron within the per 3′UTR (see Materials and Methods); type B′, the two protected type B′ bands which are 220 and 183 nt; RP49, protected ribosomal protein 49 band. Similar results were obtained from three independent experiments. The lower protected type A band overlaps with a nonspecific band from the probe itself. (B) Ratios of type A and type B′ transcripts in different tissues (see Materials and Methods for transcript quantitation). For a given tissue, the abundance of type A transcript is compared to that of type B′ transcripts, which are normalized to 1.
During this study, a per gene polymorphism in the wild-type (Canton-S [CS]) strain used in this work was determined by both RNase protection assays and cDNA sequencing (data not shown). This polymorphism is due to a deletion of 3 nt (7034 to 7036) in the type B′-specific intron. Due to this DNA polymorphism, probe 3 protects two type A bands of 235 and 254 nt and two type C bands of 354 and 254 nt. The DNA polymorphism does not affect type B protected bands. This probe generates several background bands in RNase protection assays, one of which is ∼235 nt, which masks the lower protected type A band. However, this probe gives much cleaner results on male body RNA samples than head RNA samples of both male and female flies. The perA-specific probe 4, which is the same as probe 3 except that it includes the two mutagenized splice junctions, was used for RNase protections of perA transgenic lines (see Fig. 1 and 6A for depictions of probes).
Wherever the abundances of different transcripts are compared, the protected bands are quantitated and the ratio of signal intensity of protected bands is adjusted to the number of uridines (U’s) in each probe fragment. In Fig. 5 to 7, the protected 254-nt (82-U) type A and the 220-nt (76-U) type B′ bands are used to quantitate the ratio of type A and type B′ transcript abundance. For the results of type A and type B′ transcript cycling in heads and male bodies, the values of the RNase protection signals at all time points were added together for type A (perA) or type B′ (perB′) transcripts, and then the ratio of type A to type B′ transcripts was calculated; the average ratio was calculated from three independent head and male body time courses.
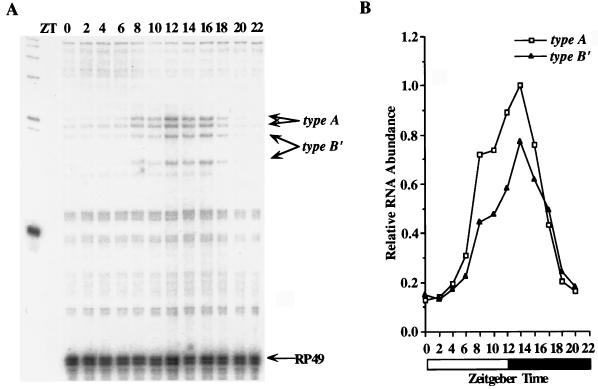
FIG. 7.
Type A and type B′ transcripts cycle in fly heads. (A) RNase protection assays were performed with probe 3 on total RNA samples from CS fly heads collected during LD cycles at the indicated time points. The protected type A, type B′, and RP49 bands are as described for Fig. 5A. (B) Quantitation of the data in panel A. Relative RNA abundance refers to the ratio of per to RP49, where the peak value of type A mRNA was adjusted to 1.0. The white and black boxes represent times when lights were on and off, respectively. This experiment was done in three independent time courses with similar results.
RNase protection assays were performed on the perA and perB′ transgenic flies to determine whether these transgenes were specifically producing type A and type B′ transcripts, respectively. To lock per RNA at a constant overall level in these transgenic lines (i.e., to render them noncycling), the flies were kept in constant light (24). RNase protection probe 4 was used for the perA transgenic flies, and probe 3 was used for the perB′ transgenic flies. Because neither the endogenous per01 gene nor the per transgenes have the DNA polymorphism found in our wild-type (CS) stock, sizes of the protected bands are 492 nt (type A) and 220 nt and 183 nt (type B′). In per01;perA transgenic flies, any transcripts from the endogenous per01 gene are protected as a type B′ band, and transcripts derived from the perA transgene are protected as a 492-nt band because probe 4 is perA specific. In the perB′ transgenic lines, the endogenous per01 gene generates both type A and type B′ transcripts, whereas the perB′ transgene generates only type B′ transcripts that are detected as 220- and 183-nt bands.
RT-mediated PCR (RT-PCR).
Total RNA was prepared from fly heads collected at Zeitgeber time 3 (ZT3), ZT9, ZT15, and ZT21 with Tri-Reagent (Sigma). Poly(A)+ RNA was prepared by passing total RNA over an oligo(dT)-cellulose column (21). First-strand cDNA was synthesized with Moloney murine leukemia virus reverse transcriptase (RT) and oligo(dT)15 primer. The PCR for detection of type B-specific splice event in exon 5 was done with sense primer 1 (5′-GCAAGAAGTCCGCCAATG-3′), which is 208 bp upstream of the 5′ splice junction, and antisense primer 3 (5′-CTCTCGGATGTGCTCATGC-3′), which is at the start of exon 8 (Fig. 4A). The PCR protocol was 5 min at 95°C, 1 min at 60°C, and 3 min at 72°C (99 cycles), followed by 10 min at 72°C; then the reaction product was kept at 4°C. Two PCR primer pairs, 2-3 and 2-4, were used to detect type C transcripts. Primer 2 (5′-ACAACAAGTCGGTGTACACG-3′) is a sense primer at the end of exon 5, and primer 4 (5′-CGGCTTGCATGGGTTCTGGGC-3′) is an antisense primer 113 bp downstream of the 3′ splice junction of the alternative intron in 3′UTR. The PCR protocol for primer pairs 2-3 and 2-4 is the same as that for primer pair 1-3 except that the reaction was cycled 50 times.
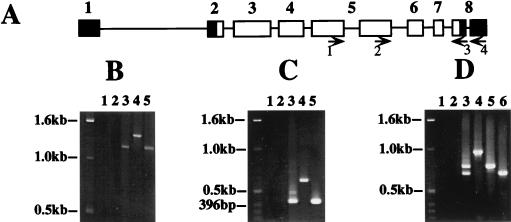
FIG. 4.
Type B-specific alternative splice event in exon 5 and type C transcripts are undetectable by RT-PCR. Approximately equal numbers of CS flies were collected at ZT3, ZT9, ZT15, and ZT21 and mixed. RT-PCR was performed on poly(A)+ RNA isolated from the fly heads. (A) Map of exons in type B per mRNA showing the primers used for PCR analysis. Orientation of the primers is indicated by the arrows. (B) Primers 1 and 3 were used to detect the type B-specific alternative splice event in exon 5. The expected products are 1,220 bp (type A), 932 bp (type B), and 1,412 bp (type C and genomic DNA). Lane 1, PCR with no template; lane 2, PCR with poly(A)+ RNA and no RT; lane 3, RT-PCR; lane 4, PCR with genomic per DNA; lane 5, PCR with type A cDNA. Consistent results were obtained from four experiments with two sets of independently prepared poly(A)+ RNA. (C) Primers 2 and 3 were used to detect type C transcripts. The expected products are 419 bp (types A and B) and 610 bp (type C and genomic DNA). The reactions in lanes 1 to 5 are as described for panel B. Consistent results were obtained from three experiments with two sets of independently prepared poly(A)+ RNA. (D) Primers 2 and 4 were used in PCR to detect type A, B, and C transcripts. The expected PCR products are 782 bp (type A), 693 bp (type B), and 974 bp (type C and genomic DNA). The reactions in lanes 1 to 5 are as described for panel B; the reaction in lane 6 is PCR with type B′ cDNA. Consistent results were obtained from two experiments with one set of poly(A)+ RNA.
Isolation and analysis of per cDNAs.
A size-selected Drosophila head cDNA library having inserts larger than 2 kb was used to screen for per cDNAs (11). This library was made in EXLX (−) vector (22) and kindly provided by Bruce Hamilton. A per cDNA probe spanning from the translation start site in exon 2 to the SalI site in exon 3 was labeled by digoxigenin-dUTP by using a DIG Labeling and Detection kit (Boehringer Mannheim Biochemicals) and used to screen the cDNA library. Hybridization and detection was done according to the manufacturer’s manual. The two full-length per cDNA clones (one type A and one type B′) were analyzed by extensive restriction enzyme digestion and PCR. The type B′ cDNA fragment spanning the alternative intron was sequenced.
Fly stocks and germ line transformation.
All fly strains were raised on cornmeal, sugar, agar yeast, and Tegosept (mold inhibitor) medium at 25°C. P-element-mediated transformation was carried out as described previously (12). Transformant lines with inserts on the second or third chromosome were balanced with In(2LR)Cy0 or In(3LR)TM2, respectively, and crossed into a y per01 w genetic background.
Behavioral analysis.
Locomotor activities of wild-type (CS) adult male and heterozygous y per01 w;perG/+, y per01w;perA/+, y per01w;perB′/+ flies were monitored and analyzed as described previously (10). The heterozygous male transgenic flies were obtained by outcrossing y per01 w;perG, y per01 w;perA, and y per01 w;perB′ males to y per01 w females. Flies were entrained in 12-h light/12-h dark (LD) cycles at 25°C for 72 h and then in constant darkness for 7 days. Locomotor activity was monitored from the first day of entrainment, and data collected during constant darkness were analyzed to determine the period and strength of the rhythm. The strength of the rhythms is measured as power, which is the amplitude from the top of the activity peak to the 5% significance line of the χ2 periodogram. Flies with powers greater than 15 and widths (the number of 0.5-h period values within a peak that were statistically significant) greater than 2 in periodogram analyses were designated as rhythmic. One-way analyses of variance (ANOVAs) were used to determine differences among genotypes for period length. Statistical significance of a posterior pairwise comparisons of mean inter- and intragenotypic period values was determined by the Tukey-Kramer method (30).
Western blot analysis.
Heterozygous perA, perB′, and perG transgenic flies were entrained in LD cycles for 3 days and collected at 4-h intervals. Extracts were made from the heads of these flies and used for Western blot analyses as described elsewhere (7), with the following modifications: the primary antibody, a rabbit polyclonal anti-PER antiserum (kindly provided by Ralf Stanewsky) preabsorbed against per01 embryos, was diluted to 1:20,000, and the secondary antibody, an anti-rabbit immunoglobulin G horseradish peroxidase-conjugated antibody (Amersham), was diluted 1:5,000 in blocking solution. X-ray film exposures of Western blots were scanned on an Apple Color Onescanner using OFOTO 2.0.2 software and quantitated with NIH Image 1.6 software. The level of PER at each time point was taken as the PER signal minus the background in each lane. Three independent time courses with different perA, perB′, and perG transgenic lines were analyzed. In each of these time courses, the highest PER signal was set to 1.0 and all other time points were normalized to this value. These values from the three time courses were used to calculate mean values and standard errors of the means for each perA, perB′, and perG time point.
RESULTS
per produces two types of transcripts which encode the same protein.
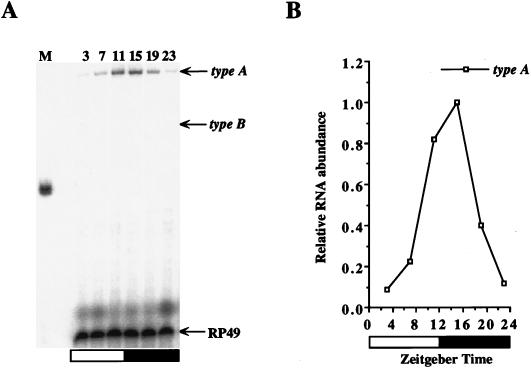
Three per mRNA types have been identified by characterizing per cDNAs isolated from a Drosophila head cDNA library (Fig. 1) (3). To confirm the structure and abundance of these per transcripts in vivo, head RNA was first analyzed by using RNase protection probes that can distinguish between the different per transcript types (Fig. 1). The type B-specific splice event in exon 5 was tested by RNase protection assays using probe 1. A protected fragment of 204 nt, which corresponds to type A transcript, cycles with the same phase and amplitude as that previously reported for per transcripts (Fig. 2) (14, 32). A 157-nt protected band, corresponding to type B transcripts, is not observed in either overnight (Fig. 2) or 10-fold-longer (data not shown) exposures of RNase protection assays. This result indicates that either type B-specific alternative splice events in exon 5 do not exist or type B transcript levels are too low to be detected by RNase protection assays.
FIG. 2.
The type B-specific intron in exon 5 is undetectable by RNase protection assays. (A) RNase protection assays were performed with probe 1 on total head RNA samples from CS flies collected during LD cycles at the indicated time points. M, size markers; type A, the protected 204-nt type A per transcript; type B, the expected 157-nt type B transcript band; RP49, the protected ribosomal protein 49 mRNA band, used as a loading control. (B) Quantitation of the data in panel A. Relative RNA abundance refers to the ratio of type A to RP49, where the peak value of per mRNA was adjusted to 1.0. The white and black boxes represent times when lights were on and off, respectively. Similar results were obtained from six independent time courses.
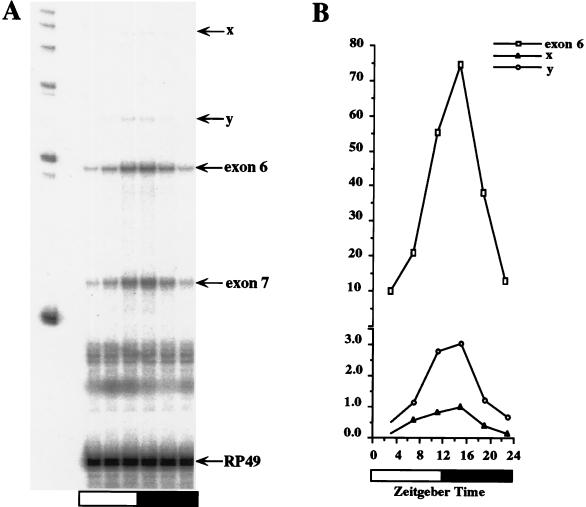
To detect type C transcripts, we performed RNase protection assays with probe 2. In addition to the strong signals expected from exon 6 and exon 7 from type A and type B transcripts, weaker cycling bands were observed (Fig. 3). One of these weak bands (designated band x) corresponds in size (570 nt) to a protected type C transcript. The signal intensity of band x is 1.3% of that of the exon 6 band at the peak time point, ZT15, though the abundance of band x is only 0.48% of that of the exon 6 band when the amount of label in the protected bands (136 U’s in band x and 49 U’s in the exon 6 band) is taken into account. Also detected in the RNase protection assays is a cycling band of ∼300 nt (designated band y), which corresponds in size to exon 6 with intron 5 (306 nt containing 55 U’s) or exon 6 with intron 6 (299 nt containing 57 U’s). This apparent splice precursor is only 2.9% of the abundance of exon 6, which is even more abundant than band x. Since the transcript corresponding to band y is more abundant than the putative type C transcript, band x may simply correspond to a splice precursor instead of representing a true mature type C transcript. This may well be the case, as per pre-mRNA cycles in abundance due to its rhythmic transcription (16, 29).
FIG. 3.
A per transcript corresponding to type C mRNA is low in abundance. (A) RNase protection assays were performed with probe 2 on CS flies collected as for Fig. 2. Exon 6 and exon 7 represent the protected per exon 6 (236-nt) and exon 7 (142-nt) sequences of both type A and type B transcripts. Above the protected exon 6 band are several other protected bands that cycle in abundance, two of which are labeled x and y. The size of band x corresponds to type C transcripts (570 nt [3]). RP49 represents the protected ribosomal protein 49 mRNA. The size of band y corresponds to either exon 6 plus intron 5 (306 nt) or exon 6 plus intron 6 (299 nt). (B) Quantitation of the data in panel A. Relative RNA abundance refers to the ratio of per to RP49, where the peak value of protected band x was adjusted to 1.0. The ordinate is broken, indicating a change in scale. The white and black boxes represent times when lights were on and off, respectively. Similar results were obtained from three independent time courses.
RT-PCR was performed to provide maximal sensitivity for detecting type B and type C transcripts. Poly(A)+ RNA was prepared from fly heads collected at ZT3, ZT9, ZT15, and ZT21. First-strand cDNA was synthesized from poly(A)+ RNA and used as the template for PCR. The type B-specific splice event in exon 5 was tested by PCR with sense primer 1, which lies 200 bp upstream of the 5′ splice junction, and antisense primer 3, which lies at the start of exon 8 (Fig. 4A). PCR using these primers should be able to detect the differences between type A, B, and C transcripts, where the expected PCR products are 1,220 bp (type A), 932 bp (type B), and 1,412 bp (type C and genomic DNA). We concentrated on type A and type B transcripts in this experiment because we did not obtain abundant amplification products, which would mitigate against detecting a rare type C transcript. This difficulty is probably related to the fact that cDNA greater than 1,956 bp is required to produce this PCR product, and per cDNAs of this length must be relatively rare. The results show that type B transcripts are not detectable by RT-PCR even though the type A transcripts are easily detected (Fig. 4B, lane 3).
To test whether type C transcripts exist in vivo, RT-PCRs were performed with one sense primer at the end of exon 5 (primer 2) and an antisense primer situated at either the start of exon 8 (primer 3) or 113 bp downstream of the 3′ end of the type B-specific intron in the 3′UTR (Fig. 4A, primer 4). PCR products with primers 2 and 3 can distinguish between type C transcripts and type A (or B) transcripts, where the expected products are 419 bp (type A or B) and 610 bp (type C and genomic DNA). The results in Fig. 4C show that RT-PCR products corresponding to type C transcripts are undetectable even though the product of type A (or B) transcripts is intense, consistent with the RNase protection results. We also detected a weaker band slightly smaller than 500 bp that is the same size as that predicted for precursor transcript equal to type A (or B) plus one of the three introns. PCR with primers 2 and 4 can distinguish between type A, B, and C transcripts, where the expected RT-PCR products are 782 bp (type A), 693 bp (type B), and 974 bp (type C or genomic DNA). Strong type A and type B bands are detected, but a type C band is not detectable (Fig. 4D). Two weaker bands larger than the type A band show up in this experiment, but both are smaller than the expected type C band and may correspond to precursor transcripts containing one or more introns. The RT-PCR products in Fig. 4B to D all hybridized to a per cDNA probe, indicating they are truly amplified per DNA fragments (data not shown).
Type A and type B transcript structure was also analyzed by cDNA cloning. Two full-length (4.5-kb) per cDNA clones were isolated by screening 400,000 PFU of a size-selected Drosophila head cDNA library having inserts larger than 2 kb (11). Restriction enzyme digestion and PCR analysis shows that one of these clones is a type A cDNA having the same structure as described in a previous study (3). Restriction enzyme digestion, PCR, and sequence analysis shows that the other per cDNA clone retains the 288-bp sequence in exon 5 but lacks the type B-specific intron sequence (originally designated a type B-specific intron) in the 3′UTR (data not shown). This result further strengthens the type B transcript structure defined by RNase protection assays and RT-PCR.
Taken together, our data indicate that the per gene generates only two transcript types that differ by an 89-nt intron in the 3′UTR. The per transcript retaining this intron in the 3′UTR is equivalent to the previously characterized type A, and the second per transcript lacking this intron will be referred to as type B′.
Various proportions of type A and type B′ transcripts are found in head and body tissues.
Type A and B′ per transcripts are both present in whole-head RNA. If these transcripts function differently or contribute to different rhythmic processes, then they may be expressed exclusively in certain cell types. To determine the level and distribution of these transcripts, we investigated the tissue distribution of type A and type B′ transcripts by RNase protection assays on dissected fly eyes, eyeless heads (brain), thoraxes, and abdomens using RNase protection probe 3 (Fig. 1). The protected type A band for probe 3 is split into two bands of 254 and 235 nt due to a DNA polymorphism within the 3′UTR of wild-type (CS) flies (see Materials and Methods), and the two protected type B′ bands are 220 and 183 nt. The results show that type A and type B′ transcripts are present in different tissues at various ratios (Fig. 5): 2.0 ± 0.09 in eyes, 0.82 ± 0.14 in the brain, 0.41 ± 0.10 in the thorax, and 0.32 ± 0.09 in the abdomen (Fig. 5B).
These results show that type B′ transcripts predominate in the body, while type A transcripts predominate in the head. No transcript was found to be unique to a particular body part or tissue in this crude analysis; however, dissected brain, thorax, and abdomen contain several per-expressing cell types which may exclusively express type A or B′ mRNAs. The presence of both per transcript types in the compound eye, where per expression is restricted to photoreceptors, argues that at least in this tissue the two per transcripts coexist.
Type A and type B′ transcripts are both capable of rescuing locomotor activity rhythms.
Though both per transcript types are found in heads, per gene function can be measured in only a small group of neurons in the brain (lateral neurons [LNs]) that control locomotor activity rhythms. To determine whether the two per transcript types function equally to rescue locomotor activity, modified per genes which generate only type A transcripts (perA gene), only type B′ transcripts (perB′ gene), or both transcript types (perG gene) were used to transform per01 flies. Differences in behavioral rescue in transgenic lines that produce a single per transcript type would suggest that the 3′UTR alternative intron affects some aspect of per gene expression.
The backbone for the perA, perB′, and perG transgenes is a 13.2-kb per genomic DNA fragment that efficiently rescues locomotor activity rhythms in per01 flies (3, 4). The perA transgene was made by mutagenizing both the 5′ and 3′ splice junctions of the alternative intron in the 3′UTR, the perB′ transgene was constructed by removing the alternative intron in the 3′UTR (see Materials and Methods; Fig. 6A), and the perG transgene (a control that makes both transcript types) consists of an unmodified 13.2-kb per genomic DNA fragment. These three per genomic DNA fragments were inserted into the pCaSpeR transformation vector (33) and used to generate transgenic flies.
All three transgenes rescue locomotor activity rhythms in per01 flies (Table 1). The strengths of these rhythms, as measured by power (i.e., the amplitude from the top of the activity peak to the 5% significance line of the χ2 periodogram), and the penetrance (i.e., percent rhythmic flies) were similar for all transgenic strains, though not necessarily for all lines within a strain. The average periods of these transgenic strains are somewhat different, being 0.7 and 1.0 h longer for perA than for perB′ and perG, respectively (Table 1). When the period differences among these strains were analyzed statistically, however, only the difference between perA and perG flies was significant (ANOVA; a < 0.05). In addition, the periods between different perA transgenic lines and between different perG transgenic lines (i.e., intragenotypical periods) vary significantly (ANOVA; a < 0.05). Much of this perA and perG intragenotypic period variability is due to the lines having the lowest penetrance (i.e., perA-5 and perG-4) and may be due to position effects. Overall, the trend is that strongly rhythmic lines for perA are longer than those for perB′ and perG.
TABLE 1.
Free-running behavior of wild-type (CS), y per01 w, and heterozygous per01;perA, per01;perB′, and per01;perG transgenic fliesa
| Genotype | Period (mean ± SEM) | Power (mean ± SEM) | % Rhythmic (nb) |
|---|---|---|---|
| Wild type | 24.0 ± 0.07 | 84.1 ± 10.0 | 85 (26) |
| per01;perA-1 | 25.4 ± 0.12 | 55.6 ± 3.8 | 90 (41) |
| per01;perA-2 | 26.1 ± 0.09 | 44.6 ± 5.8 | 73 (48) |
| per01;perA-3 | 26.0 ± 0.11 | 53.8 ± 4.1 | 83 (46) |
| per01;perA-4 | 24.7 ± 0.28 | 58.6 ± 4.5 | 85 (40) |
| per01;perA-5 | 24.1 ± 0.28 | 39.1 ± 5.9 | 37 (38) |
| Avgc | 25.4 ± 0.09 | 49.8 ± 2.1 | 66 (239) |
| per01;perB′-1 | 24.7 ± 0.27 | 52.6 ± 4.1 | 86 (35) |
| per01;perB′-2 | 24.6 ± 0.10 | 48.8 ± 3.7 | 82 (51) |
| per01;perB′-3 | 24.8 ± 0.11 | 29.4 ± 3.2 | 71 (24) |
| Avg | 24.7 ± 0.10 | 46.4 ± 2.4 | 81 (110) |
| per01;perG-1 | 24.0 ± 0.37 | 47.3 ± 3.7 | 78 (46) |
| per01;perG-2 | 23.9 ± 0.13 | 51.2 ± 4.3 | 88 (33) |
| per01;perG-3 | 24.4 ± 0.07 | 42.0 ± 3.1 | 94 (51) |
| per01;perG-4 | 24.9 ± 0.09 | 45.0 ± 4.6 | 59 (46) |
| per01;perG-5 | 24.7 ± 0.07 | 57.8 ± 4.4 | 91 (46) |
| Avg | 24.4 ± 0.08 | 48.5 ± 1.8 | 82 (222) |
Locomotor activity of young male flies was monitored at 25°C for 7 days in constant darkness following 3 days of entrainment in 12-h light/12-h dark cycles.
Total number of flies tested.
Overall average for all flies containing that transgene.
To ensure that the perA and perB′ transgenes produce the predicted mRNA type, RNase protection assays were used to measure type A and type B′ transcripts in three perA and perB′ transgenic lines (Fig. 6B). Transcripts from perA transgenic lines were measured with probe 4, which protects a 492-nt fragment corresponding to perA transgene-derived transcripts and 220- and 183-nt fragments corresponding to endogenous per01 transcripts. Lanes A-1 to A-3 show that there is a strong transgene-derived type A transcript band and weaker per01-derived transcript bands (Fig. 6B). The presence of the 492-nt fragment shows that the perA transgene indeed makes type A transcripts; however, we cannot exclude the possibility that non-type A transcripts arise from the activation of cryptic splice sites. As expected, protection of three perB′ transgenic strains (lanes B′-1 to B′-3) with probe 3 shows that only type B protected bands are accentuated (Fig. 6B). These experiments show that the perA and perB′ transgenes generate the expected type A and type B′ transcripts, respectively.
Type A and type B′ transcripts cycle in fly heads and bodies.
The perA and perB′ transgenic strains show some difference in the period of locomotor activity rhythms. This difference in behavioral rescue, though small, suggests that the alternative intron in the 3′UTR may affect some aspect of per gene expression. Such an observation is not unprecedented, as 3′UTRs have been shown to regulate gene expression via posttranscriptional mechanisms including mRNA stability and translational efficiency (18). Although per mRNA cycling is controlled primarily at the transcriptional level (12, 16, 29, 32), recent studies indicate that posttranscriptional mechanisms also contribute to per mRNA rhythms (29, 32). To determine whether there are differences in cycling phase and/or amplitude between type A and type B′ transcripts, per transcripts were analyzed in LD cycles via RNase protection assays.
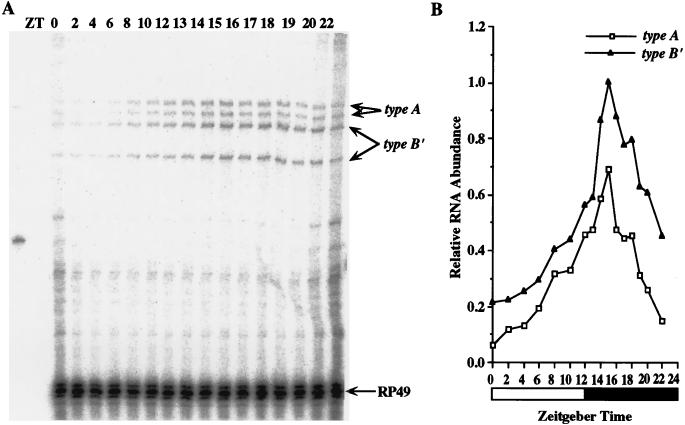
In wild-type fly heads, type A and type B′ transcripts cycle with identical phases but different cycling amplitudes; the average cycling amplitude of type A transcripts is 8.1-fold and that of type B′ transcripts is 5.5-fold in three experiments (Fig. 7). Type A transcript on the average are 1.2-fold more abundant than type B′ transcripts, consistent with stronger type A RT-PCR bands (Fig. 4) and higher type A levels in the eye (Fig. 5). To test whether type A and type B′ transcripts also cycle in fly bodies, we performed RNase protection assays on RNA samples prepared from male bodies since per RNA does not cycle in ovaries (13). Again, type A and type B′ transcripts cycle with identical phases but with different amplitudes: the average cycling amplitude of type A transcripts from three experiments is 7.5-fold, and that of type B′ transcripts is 5.5-fold (Fig. 8). In male bodies, type B′ transcripts are ∼2.0-fold more abundant than type A transcripts, consistent with the levels seen in different body parts (Fig. 5).
FIG. 8.
Both type A and type B′ transcripts cycle in male bodies. (A) RNase protection assays were performed as for Fig. 6A except the total RNA samples were from male fly bodies collected during LD cycles at the indicated time points. (B) Quantitation of data in panel A, performed as described for Fig. 6B. Similar results were obtained from three independent time courses.
PER accumulates to similar levels in lines producing only type A or type B′ transcripts.
The two per transcript types cycle with phases that are indistinguishable in both heads and bodies under LD conditions. The most obvious difference between the two per transcripts is that type B′ has a lower amplitude cycle than type A. This difference in amplitude is unlikely to result from a difference in mRNA stability because changes in mRNA stability affect cycling amplitude and phase in parallel; more stable transcripts will have a lower amplitude and later phase, while less stable transcripts will have a higher amplitude and an earlier phase (29, 32). Since the intron in the 3′UTR does not appear to affect transcript stability, perhaps it functions to regulate the translation of per mRNA. If this were the case, we might expect to see a difference in the level and phase of PER between perA, perB′, and perG transformants.
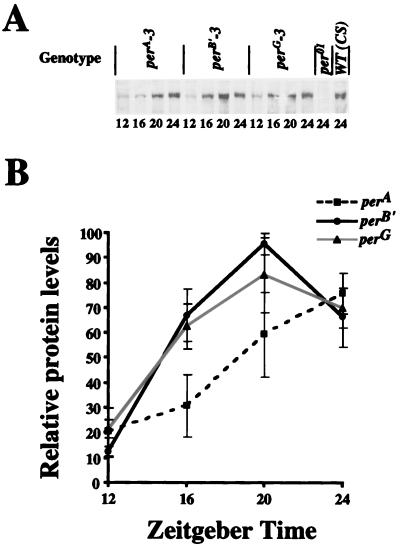
To determine if there are differences in PER levels among these transgenic strains, PER levels were measured by probing Western blots of head extracts with an anti-PER antibody. The overall level of PER in each transgenic strain was similar to that of the wild type at ZT24 (Fig. 9). However, the accumulation of PER in perA flies is delayed compared to that in the other strains, thereby resulting in a later peak (Fig. 9). Since the type A and type B′ mRNAs cycle in phase (Fig. 7 and 8), this result suggests that the intron may affect PER accumulation at the translational level.
FIG. 9.
PER levels are similar in perA, perB′, and perG transgenic flies. (A) Flies from the three transgenic strains were entrained in LD cycles for 3 days before collection at the time points indicated. Head protein extracts were made and subjected to Western blotting. The gel strip shows the level of PER immunoreactivity for each sample. Head extracts from wild-type (WT; CS) and per01 flies collected at ZT24 were used as positive and negative controls, respectively, for PER immunoreactivity. Similar results were obtained in three independent time courses. (B) Quantitation of PER cycling in panel A. Data from three independent time courses of perA, perB′, and perG are plotted. For each Western blot, the highest time point was set to 100 and all other time points from the three (perA, perB′, and perG) time courses were set relative to that value. The error bars represent standard errors for the three time courses.
DISCUSSION
In this study, we reexamined the structure and abundance of per transcripts through direct mRNA measurements. Our results show that per generates two types of transcripts, type A and type B′, which differ by an alternatively spliced intron in their 3′UTRs. The intron in the protein coding region of type A, which was thought to arise from an unusual splice event involving a CG splice acceptor site (3), was not detected in either RNase protections or RT-PCR experiments (Fig. 2 and 4). The type C transcripts identified earlier (3) are consistent with a splice precursor because they contain both introns 6 and 7 and are lower in abundance than other transcripts corresponding to per splice precursors (Fig. 3 and 4). From these results, we find no evidence that type B and type C transcripts exist in vivo. It is worth noting, however, that PCR was not available when this earlier study was published (3), making it difficult to easily sort through cloning artifacts.
The abundance of type A and type B′ transcripts varies in different head and body tissues. Since the majority of our dissected body parts contain several per-expressing cell types, we do not know whether a given cell type expresses a single per transcript type or both types. The only dissected tissue (eyes) that contains a single per-expressing cell type (photoreceptors) expresses a 2:1 ratio of type A to type B′ (Fig. 5). This result argues that both RNA types are present in photoreceptors, though we cannot say for certain that both per transcript types are in the same cell because the two inner and six outer photoreceptors are developmentally different and express different opsin genes (6).
Even though various tissues express different proportions of the two per transcript types, these transcripts encode a single PER protein that accounts for all of the known per gene functions. In light of this, it was surprising to find that type A and type B′ transcripts function differently to rescue locomotor activity rhythms; flies expressing only functional type A transcripts (endogenous per01 transcripts are nonfunctional) have average circadian periods of 25.4 h, flies expressing only functional type B′ transcripts have average circadian periods of 24.7 h, and flies that express both type A and type B′ transcripts have average circadian periods of 24.4 h. As expected, the period of perG transformant flies was close to that of wild-type flies, based on previous behavioral rescue experiments using the per 13.2-kb fragment (3, 4, 20, 34, 35). It was surprising to find that the perB′ transgene rescued periods closer to wild type compared to perA since they encode the same protein. This result might suggest that type A transcripts are less important for regulating the circadian period of locomotor activity rhythms. We do not know whether type A transcripts are expressed in the locomotor activity pacemaker cells, the LNs (9, 27, 37), even though type A transcripts are relatively abundant in fly heads (Fig. 5 and 7). Many tissues contain autonomous circadian oscillators in Drosophila, but these tissue oscillators appear to run at slightly different circadian periods which would result in dampening of rhythms under constant dark conditions (2, 8, 9, 13, 23). Perhaps the periods of these tissue autonomous oscillators are controlled, in part, by the ratio of type A and type B′ per transcripts.
Since perA flies tend to have longer period behavioral rhythms than perB′ flies, the molecular events that underlie this difference presumably involve the alternatively spliced intron. This intron, located in the 3′UTR, could affect longer period rhythms by destabilizing per mRNA or by decreasing the translational efficiency of per mRNA. Earlier studies have shown that reducing per gene dosage and/or per mRNA abundance, which presumably results in reduced or delayed PER accumulation, lengthens the period of behavioral rhythms (1, 28). In some genes the 3′UTR is capable of regulating mRNA stability via U-A-rich sequences; more consensus U-A-rich sequences destabilize mRNA (18). There are no consensus U-A-rich sequences in the alternative intron in per (3), and the overall level of transgene-derived per mRNA in perA flies is greater than that of perB′ (data not shown), suggesting that the intron does not destabilize type A transcripts. In addition, a decrease in the stability of type A transcripts would alter their phase compared to that of type B′, but we do not observe any difference in phase between the two per transcripts (Fig. 3, 7, and 8). Another explanation for the long period rescue in perA flies is that the mutant splice junctions may have an effect on overall per RNA abundance or cycling. This possibility is unlikely since the per 3′UTR can be replaced without an effect on per RNA abundance or cycling (32). Since this alternative intron does not appear to destabilize type A transcripts, it may function to alter the period by regulating per mRNA translation.
The 3′UTR has been shown to regulate translation efficiency of some mRNAs by poly(A)-dependent and poly(A)-independent processes (18). If translation efficiency were altered in this case, a difference in the phase of PER cycling or in the overall abundance of PER would be expected. Indeed, PER accumulation is delayed in perA flies (which cannot remove the alternative intron) compared to that of perB′ and perG flies (Fig. 9). This delay is apparent even at low (4-h time point) resolution, suggesting that the magnitude of this difference is large compared to the difference in behavioral periods. Since the LNs that control locomotor activity rhythms represent only a small proportion of PER expression in the head, a relatively high level of PER in the LNs compared to other PER-expressing cells in the head might account for a relatively small change in the period of locomotor activity rhythms.
Timing of the different steps within the Drosophila circadian feedback loop are thought to be important for maintaining a 24-h period. For instance, timing delays between per transcription and per mRNA accumulation, per mRNA accumulation and PER accumulation, and PER accumulation and PER nuclear localization underlie the overall ∼10-h delay between per transcription and PER nuclear localization. This study demonstrates that one factor that effects the delay between per mRNA accumulation and PER accumulation is the per alternative intron, which may act to inhibit the translation of per mRNA. The delay in PER accumulation imposed by this intron, along with delays at other points in the feedback loop, are important for sustaining circadian periodicity.
ACKNOWLEDGMENTS
We thank James Carrington for providing the modified phage vector pTL. We also thank Jerry Houl and Nicole Huynh for collecting samples, Jerry Houl for behavioral analyses, and Nicholas Glossop, Lisa Lyons, and Balaji Krishnan for comments on the manuscript.
This study was supported by NIH grant NS31214.
REFERENCES
- 1.Baylies M K, Bargiello T A, Jackson F R, Young M W. Changes in abundance and structure of the per gene product can alter periodicity of the Drosophila clock. Nature. 1987;328:390–392. doi: 10.1038/326390a0. [DOI] [PubMed] [Google Scholar]
- 2.Cheng Y, Hardin P E. Drosophila photoreceptors contain an autonomous circadian oscillator that can function without period mRNA cycling. J Neurosci. 1998;18:741–750. doi: 10.1523/JNEUROSCI.18-02-00741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Citri Y, Colot H V, Jacquier A C, Yu Q, Hall J C, Baltimore D, Rosbash M. A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature. 1987;326:42–47. doi: 10.1038/326042a0. [DOI] [PubMed] [Google Scholar]
- 4.Cooper M K, Hamblen-Coyle M J, Liu X, Rutila J E, Hall J C. Dosage compensation of the period gene in Drosophila melanogaster. Genetics. 1994;138:721–732. doi: 10.1093/genetics/138.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dembinska M, Stanewsky R, Hall J C, Rosbash M. Circadian cycling of a PERIOD-B-galactosidase fusion protein in Drosophila: evidence for cyclical degradation. J Biol Rhythms. 1997;12:157–172. doi: 10.1177/074873049701200207. [DOI] [PubMed] [Google Scholar]
- 6.Dickson B, Hafen E. Genetic dissection of eye development in Drosophila. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, N.Y: CSHL Press; 1993. pp. 1327–1362. [Google Scholar]
- 7.Edery I, Zwiebel L J, Dembinska M E, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emery I F, Noveral J M, Jamison C F, Siwicki K K. Rhythms of Drosophila period gene expression in culture. Proc Natl Acad Sci USA. 1997;94:4092–4096. doi: 10.1073/pnas.94.8.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frisch B, Hardin P E, Hamblen-Coyle M J, Rosbash M R, Hall J C. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 10.Hamblen M, Zehring W A, Kyriacou C P, Reddy P, Yu Q, Wheeler D A, Zwiebel L J, Konopka R J, Rosbash M, Hall J C. Germ-line transformation involving DNA from the period locus in Drosophila melanogaster: overlapping genomic fragments that restore circadian and ultradian rhythmicity to per0 and per− mutants. J Neurogenet. 1986;3:249–291. doi: 10.3109/01677068609106855. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton B A, Palazollo M J, Meyerowitz E M. Rapid isolation of long cDNA clones from existing libraries. Nucleic Acids Res. 1991;19:1951–1952. doi: 10.1093/nar/19.8.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao H, Allen D L, Hardin P E. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol Cell Biol. 1997;17:3687–3693. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardin P E. Analysis of period mRNA cycling in Drosophila head and body tissues indicates that body oscillators behave differently than head oscillators. Mol Cell Biol. 1994;14:7211–7218. doi: 10.1128/mcb.14.11.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardin P E, Hall J C, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;342:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 15.Hardin P E, Hall J C, Rosbash M. Circadian cycling in the levels of protein and mRNA from Drosophila melanogaster’s period gene. In: Young M W, editor. Molecular genetics of biological rhythms. New York, N.Y: Marcel Dekker; 1992. pp. 155–169. [Google Scholar]
- 16.Hardin P E, Hall J C, Rosbash M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc Natl Acad Sci USA. 1992;89:11711–11715. doi: 10.1073/pnas.89.24.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardin P E, Sehgal A. Molecular components of a model circadian clock: lessons from Drosophila. In: Lydic R, Baghdoyan H, editors. Handbook of behavioral and state control: molecular and physiological mechanisms. Boca Raton, Fla: CRC Press; 1998. pp. 61–74. [Google Scholar]
- 18.Jackson R J. Cytoplasmic regulation of mRNA function: the importance of the 3′ untranslated region. Cell. 1993;74:9–14. doi: 10.1016/0092-8674(93)90290-7. [DOI] [PubMed] [Google Scholar]
- 19.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-directed mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Yu Q, Huang Z, Zwiebel L J, Hall J C, Rosbash M. The strength and periodicity of D. melanogaster circadian rhythms are differentially affected by alterations in period gene expression. Neuron. 1991;6:753–766. doi: 10.1016/0896-6273(91)90172-v. [DOI] [PubMed] [Google Scholar]
- 21.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 22.Palazollo M J, Hamilton B A, Ding D, Martin C H, Mead D A, Mierendorf R C, Raghavan K V, Meyerowitz E M, Lipshitz H D. Phage lambda cDNA cloning vectors for subtractive hybridization, fusion-protein synthesis and Cre-loxP automatic plasmid subcloning. Gene. 1990;88:25–36. doi: 10.1016/0378-1119(90)90056-w. [DOI] [PubMed] [Google Scholar]
- 23.Plautz J D, Kaneko M, Hall J C, Kay S A. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 24.Qiu J, Hardin P E. per mRNA cycling is locked to lights-off under photoperiodic conditions that support circadian feedback loop function. Mol Cell Biol. 1996;16:4182–4188. doi: 10.1128/mcb.16.8.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosbash M, Allada R, Dembinska M, Guo W Q, Le M, Marrus S, Qian Z, Rutila J, Yaglom J, Zeng H. A Drosophila circadian clock. Cold Spring Harbor Symp Quant Biol. 1996;61:265–278. [PubMed] [Google Scholar]
- 26.Sehgal A, Ousley A, Hunter-Ensor M. Control of circadian rhythms by a two component clock. Mol Cell Neurosci. 1996;7:165–172. doi: 10.1006/mcne.1996.0013. [DOI] [PubMed] [Google Scholar]
- 27.Siwicki K K, Eastman C, Petersen G, Rosbash M, Hall J C. Antibodies to the period gene product of Drosophila reveal diverse distribution and rhythmic changes in the visual system. Neuron. 1988;1:141–150. doi: 10.1016/0896-6273(88)90198-5. [DOI] [PubMed] [Google Scholar]
- 28.Smith R F, Konopka R J. Effects of dosage alterations at the per locus on the period of the circadian clock of Drosophila. Mol Gen Genet. 1982;189:30–36. [Google Scholar]
- 29.So V W, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sokal R R, Rohlf F J. Biometry. W. H. New York, N.Y: Freeman; 1995. [Google Scholar]
- 31.Stanewsky R, Frisch B, Brandes C, Hamblen-Coyle M J, Rosbash M, Hall J C. Temporal and spatial expression patterns of transgenes containing increasing amounts of the Drosophila clock gene period and a lacZ reporter: mapping elements of the PER protein involved in circadian cycling. J Neurosci. 1997;15:676–696. doi: 10.1523/JNEUROSCI.17-02-00676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanewsky R, Jamison C F, Plautz J D, Kay S A, Hall J C. Multiple circadian-regulated elements contribute to cycling period gene expression in Drosophila. EMBO J. 1997;16:5006–5018. doi: 10.1093/emboj/16.16.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thummel C S, Pirotta V. New pCaSpeR P-element vectors. Drosoph Inf Serv. 1991;71:150. [Google Scholar]
- 34.Yu Q, Colot H V, Kyriacou C P, Hall J C, Rosbash M. Behavioral modification by in vitro mutagenesis of a variable region within the period gene of Drosophila. Nature. 1987;326:765–769. doi: 10.1038/326765a0. [DOI] [PubMed] [Google Scholar]
- 35.Yu Q, Jacquier A C, Citri Y, Hamblen M, Hall J C, Rosbash M. Molecular mapping of point mutations in the period gene that stop or speed up biological clocks in Drosophila melanogaster. Proc Natl Acad Sci USA. 1987;84:784–788. doi: 10.1073/pnas.84.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng H, Hardin P E, Rosbash M. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO J. 1994;13:3590–3598. doi: 10.1002/j.1460-2075.1994.tb06666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zerr D M, Hall J C, Rosbash M, Siwicki K K. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]