Abstract
The preprotein translocase of the outer mitochondrial membrane (Tom) is a multisubunit machinery containing receptors and a general import pore (GIP). We have analyzed the molecular architecture of the Tom machinery. The receptor Tom22 stably associates with Tom40, the main component of the GIP, in a complex with a molecular weight of ∼400,000 (∼400K), while the other receptors, Tom20 and Tom70, are more loosely associated with this GIP complex and can be found in distinct subcomplexes. A yeast mutant lacking both Tom20 and Tom70 can still form the GIP complex when sufficient amounts of Tom22 are synthesized. Besides the essential proteins Tom22 and Tom40, the GIP complex contains three small subunits, Tom5, Tom6, and Tom7. In mutant mitochondria lacking Tom6, the interaction between Tom22 and Tom40 is destabilized, leading to the dissociation of Tom22 and the generation of a subcomplex of ∼100K containing Tom40, Tom7, and Tom5. Tom6 is required to promote but not to maintain a stable association between Tom22 and Tom40. The following conclusions are suggested. (i) The GIP complex, containing Tom40, Tom22, and three small Tom proteins, forms the central unit of the outer membrane import machinery. (ii) Tom20 and Tom70 are not essential for the generation of the GIP complex. (iii) Tom6 functions as an assembly factor for Tom22, promoting its stable association with Tom40.
The mitochondrial outer and inner membranes contain multisubunit machineries for the import of nucleus-encoded precursor proteins, termed preprotein translocases of the outer membrane (Tom) and inner membrane (Tim), respectively (36–38, 42, 47). In the past few years, many Tom and Tim proteins have been identified to be involved in the recognition or translocation of preproteins. The Tom and Tim machineries are separate functional entities (20, 41, 51) that can be transiently connected by a preprotein spanning both mitochondrial membranes (9, 19, 49).
The Tom machinery contains import receptors for the initial binding of cytosolically synthesized preproteins and a general import pore (GIP) for membrane translocation of different types of preproteins. Nine different Tom proteins have been found so far, and all are integral proteins of the outer membrane. They have been roughly grouped into two classes according to their function in (i) recognition of preproteins (receptors) or (ii) transport through the GIP.
Tom20, Tom22, and Tom70 function as import receptors for preproteins (4, 6, 17, 18, 26, 32, 48). Tom20 and Tom22 bind preproteins with amino-terminal targeting signals (presequences) (6) and have been proposed to form a complex or a heterodimeric receptor (32). Tom70 shows a preference for preproteins with internal targeting sequences. Tom37 associates with Tom70, and genetic evidence supports a functional interaction, indicating that Tom37 is a subunit of the Tom70 receptor (12). Tom72, a homolog of Tom70, is expressed in only small amounts and loosely associates with the Tom machinery; deletion of its gene does not lead to any significant phenotype, indicating that Tom72 does not play an important role in the import of preproteins (5, 50). The interaction of preproteins with the cytosolic cofactor heat shock protein 70 or the mitochondrial import stimulation factor was reported to influence whether a preprotein is initially recognized by Tom20 or Tom70, respectively (26, 27). In fact, Tom20 and Tom70 show partially overlapping specificities, and preproteins initially recognized by Tom70 are transferred to Tom20 and/or Tom22 before their insertion into the GIP (6, 24, 26).
Tom40 is thought to represent the major component of the GIP (23, 25, 40, 55). The smallest Tom protein, Tom5, functionally links receptors to the GIP and promotes the insertion of preproteins (11). While Tom5 directly interacts with preproteins, two other small Tom proteins, Tom6 and Tom7, do not come into direct contact with preproteins but seem to modulate the stability of the association of Tom components (2, 16, 21). Besides its cytosolic domain, which has receptor function, Tom22 also contains a domain in the intermembrane space (4, 7, 35) that was shown to function as a trans binding site for preproteins with amino-terminal presequences (4, 33). The presence of negatively charged patches in a number of Tom proteins, including Tom20, Tom22 (cytosolic domain and intermembrane space domain), and Tom5, as well as in Tim23 (inner membrane), prompted the hypothesis of an acid chain that directs the import of positively charged presequences (4, 11, 18, 46).
While considerable information has been accumulated about the functions of individual Tom proteins, far less is known about the molecular architecture and organization of the Tom machinery. In the past few years, different views have been suggested—on the one hand, an association of all Tom proteins in one stable complex (24, 25, 34, 54), and on the other hand, the existence of subcomplexes with variable compositions, depending on the study (9–12, 29, 32). Major reasons for this unclear situation are that several methods used to analyze the association of Tom proteins were not quantitative but measured only fractions of the proteins and that no systematic comparison of the various methods was performed. Therefore, for this report we characterized the organization of Tom proteins in complexes by use of distinct biochemical and genetic means, with particular emphasis on a quantitative analysis, including the use of blue native gel electrophoresis. We show that Tom40 and Tom22 are stably associated in a complex with a molecular weight of ∼400,000 (∼400K), henceforth referred to as the GIP complex. This complex also contains all three small Tom proteins. Tom20 and Tom70 are less stably associated with the GIP complex and can be found in distinct subcomplexes. We suggest that Tom22 is predominantly and more stably associated with Tom40 than with Tom20. In fact, the GIP complex can be generated even after the deletion of both the TOM20 and the TOM70 genes. The 400K GIP complex can be dissociated into a subcomplex of ∼100K containing Tom40, Tom7, and Tom5. We demonstrate that Tom6 functions as an assembly factor required for the association of Tom22 with the 100K subcomplex.
MATERIALS AND METHODS
Isolation of mitochondria and immunoprecipitation studies.
The Saccharomyces cerevisiae strains used in this study are listed in Table 1. Mitochondria were isolated by published procedures (8, 14). Immunoprecipitation experiments were performed with digitonin-lysed mitochondria by use of antibodies covalently bound to protein A-Sepharose and obtained from preimmune serum or directed against Tom70, Tom40, Tom22, Tom20, and Tom5 (16). After being washed in digitonin-containing buffer (1), the bound proteins were eluted by the addition of electrophoresis sample buffer (28), separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose, and immunodecorated with antibodies directed against the different Tom proteins.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| YPH499 (wild type) | ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 | 52 |
| MM307 (tom6Δ) | ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tom6::URA3 | 2 |
| AH101 (tom7Δ) | ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tom7::TRP1 | 16 |
| AH610 (tom6Δ tom7Δ) | ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tom6::URA3 tom7::TRP1 | 16 |
| KD56 (tom5Δ) | ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tom5::HIS3 | 11 |
| MM120-C (tom20Δ tom70Δ TOM22↑) | ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tom70::HIS3 tom20::URA3 pG-1(TRP1)-TOM22 | 18 |
Import of preproteins into isolated mitochondria.
Radiolabeled preproteins were obtained by in vitro transcription and translation reactions with rabbit reticulocyte lysate (Amersham) in the presence of [35S]methionine-cysteine (53). Import reactions were performed with bovine serum albumin-containing buffer (3% [wt/vol] fatty-acid-free bovine serum albumin, 80 mM KCl, 5 mM MgCl2 10 mM morpholinepropanesulfonic acid [MOPS]-KOH [pH 7.2]) in the presence of 2 mM ATP and 2 mM NADH. When the membrane potential was dissipated, 8 μM antimycin A, 20 μM oligomycin, and 1 μM valinomycin were added to the import reaction mixture. Radiolabeled preproteins were incubated with mitochondria (25 to 50 μg of protein) at 25°C for various times. Samples were subsequently treated or not treated with proteinase K (50 μg/ml) for 15 min at 4°C. The protease was inactivated by the addition of 1 mM phenylmethylsulfonyl fluoride (PMSF), and samples were incubated for a further 10 min at 4°C. For trypsin treatment, mitochondrial samples in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS-KOH [pH 7.2]) were incubated with trypsin (20 μg/ml) for 20 min on ice. Trypsin was inactivated by the addition of a 30-fold excess of soybean pancreatic trypsin inhibitor, and samples were incubated for an additional 10 min on ice prior to further manipulations. After a washing step with SEM buffer, pelleted mitochondria were lysed in the appropriate detergent-containing buffer and applied to SDS-polyacrylamide or blue native polyacrylamide gels. Radiolabeled proteins were detected by PhosphorImager storage technology (Molecular Dynamics).
Blue native gel electrophoresis.
Blue native PAGE was performed essentially as previously described (9, 43, 45). Briefly, following treatment, mitochondrial pellets (25 to 100 μg of protein) were lysed in 50 μl of ice-cold digitonin buffer (1% [wt/vol] digitonin, 20 mM Tris-HCl [pH 7.4], 0.1 mM EDTA, 50 mM NaCl, 10% [vol/vol] glycerol, 1 mM PMSF) (3) or Triton X-100 buffer (0.5% [vol/vol] Triton X-100, 20 mM Tris-HCl [pH 7.4], 0.1 mM EDTA, 50 mM NaCl, 10% [vol/vol] glycerol, 1 mM PMSF). After a clarifying spin, 5 μl of sample buffer (5% [wt/vol] Coomassie brilliant blue G-250, 100 mM bis-tris [pH 7.0], 500 mM 6-aminocaproic acid) was added, and the samples were electrophoresed through a 6 to 16% polyacrylamide gradient gel (9).
For immunoblotting, the native gel was soaked in blot buffer (20 mM Tris base, 150 mM glycine, 0.08% SDS) prior to transfer to polyvinylidene difluoride (PVDF) membranes (Millipore) by the semidry blotting technique. Immunodecoration was performed by standard procedures, and detection was achieved by the enhanced chemiluminescence method (Amersham). For detection of radiolabeled proteins, the dried gel or PVDF membrane was exposed to PhosphorImager storage cassettes prior to PhosphorImager analysis (Molecular Dynamics).
For two-dimensional gel analysis, individual lanes were excised from the first-dimension native gel and layered on top of the stacking gel of a second-dimension SDS-polyacrylamide gel. Following electrophoresis, proteins were blotted onto nitrocellulose membranes and analyzed by immunodecoration or PhosphorImager analysis.
Quantitation of Tom components.
Purified soluble domains of Tom70, Tom22, and Tom20 (6) and Tom40 protein purified from inclusion bodies after expression in Escherichia coli of known concentrations, along with wild-type mitochondria, were applied in limiting dilutions to an SDS-polyacrylamide gel, which was subsequently immunoblotted. The blot was immunodecorated with antibodies specific for these Tom components, and the signals of the purified proteins and the mitochondrial extracts were directly compared.
Miscellaneous methods.
SDS-PAGE was performed with the Tris-glycine buffer system (28) or the Tris-glycine buffer system (44).
RESULTS
The GIP complex contains Tom40, Tom22, Tom7, Tom6, and Tom5.
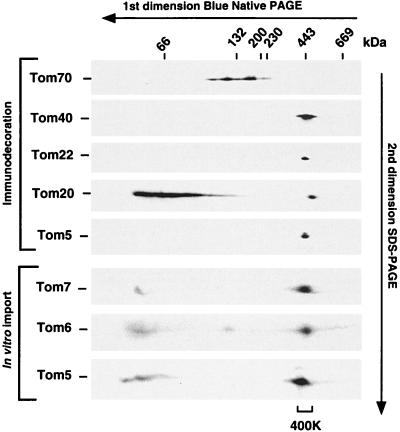
Mitochondria were isolated from the yeast S. cerevisiae, solubilized with digitonin, and separated by blue native gel electrophoresis (10, 45). After electrophoresis on a second-dimension gel under denaturing conditions (9, 11, 43), the presence of distinct Tom proteins was analyzed by immunoblotting with monospecific antisera. Tom40, Tom22, and Tom5 were predominantly present at ∼400K (GIP complex) (Fig. 1). Since specific antibodies against Tom7 and Tom6 were not available, precursors of the small Tom proteins were synthesized in vitro in rabbit reticulocyte lysates in the presence of [35S]methionine-cysteine (1), imported into isolated yeast mitochondria, and subjected to blue native gel electrophoresis and digital autoradiography. All three 35S-labeled small Tom proteins were found at the 400K position (Fig. 1, lower panel). In addition, the small Tom proteins were also observed in the low-molecular-weight range, possibly representing in vitro-imported proteins that were not yet assembled into the 400K complex. With Tom5, this assumption could be proven by a direct comparison between the protein imported in vivo (immunodecoration; Fig. 1, upper panel) and the protein imported in vitro (Fig. 1, lower panel). In vivo-imported Tom5 was present exclusively in the 400K region. Tom5 in the lower-molecular-weight range was observed only with the 35S-labeled protein imported in vitro, indicating that, within the time span of the in vitro import reaction, not all Tom5 molecules could assemble into the 400K complex. We conclude that Tom40, Tom22, and the three small Tom proteins comigrate at the 400K position in blue native gel electrophoresis.
FIG. 1.
Separation of Tom proteins by blue native gel electrophoresis. Isolated S. cerevisiae wild-type mitochondria were lysed in digitonin buffer and subjected to blue native PAGE in the first dimension and SDS-PAGE in the second dimension as described in Materials and Methods. Following electrophoresis, proteins were blotted and then immunodecorated with antibodies specific for various Tom proteins. To analyze the locations of Tom7 and Tom6, these proteins, along with Tom5, which served as a control, were synthesized in the presence of [35S]methionine-cysteine and subsequently imported into mitochondria in vitro. Following two-dimensional gel electrophoresis, radiolabeled Tom proteins were analyzed by PhosphorImager storage technology. The position of the 400K complex is indicated.
The bulk of the receptors Tom20 and Tom70, however, was not found at the 400K position. Tom70 migrated in an area of 100 to 200K, whereas Tom20 migrated in a band of 40 to 100K (Fig. 1). Upon overexposure of the immunoblot, a small amount of Tom20(∼5 to 10%) was found in the higher-molecular-weight range, slightly above the peak of the 400K position (small amounts of Tom40 and other components of the GIP complex were also observed at this slightly higher position) (Fig. 1).
Thus, using blue native gel electrophoresis, we did not detect a stable interaction between Tom70 and the 400K complex. Similarly, most of Tom20 (∼90%) was not stably associated with the 400K complex. A small fraction of Tom20 might be found in association with the 400K complex, leading to a complex with a slightly higher molecular weight.
The GIP complex can be formed in the absence of the receptors Tom20 and Tom70.
We applied two additional approaches to assess the relationship among Tom20, Tom70, and the GIP complex: coimmunoprecipitation of Tom proteins (1, 2, 12, 15, 16, 25, 34, 54) and deletion of the genes TOM20 and TOM70 (18).
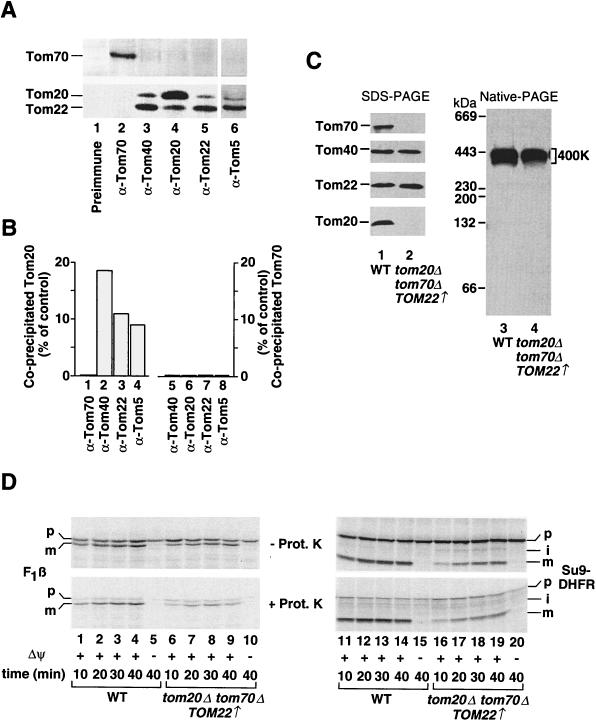
Coimmunoprecipitation of Tom70, Tom20, and Tom22 from digitonin-lysed mitochondria was performed with all available monospecific anti-Tom antibodies: anti-Tom70, anti-Tom20, anti-Tom22, anti-Tom40, and anti-Tom5. Efficient precipitation of Tom70 was possible only with anti-Tom70 (Fig. 2A, upper panel, lane 2). Antibodies directed against Tom40, Tom20, Tom22, or Tom5 precipitated only minute amounts of Tom70 that were close to the background value (Fig. 2A, upper panel, lanes 3 to 6; Fig. 2B, columns 5 to 8). Similarly, only small amounts of Tom20 and Tom22 were coprecipitated with anti-Tom70 (Fig. 2A, lower panel, lane 2; Fig. 2B, column 1). In contrast, Tom22 was coprecipitated with both anti-Tom40 and anti-Tom5 at an efficiency close to that of the direct precipitation of Tom22 with anti-Tom22 (Fig. 2A, lower panel, compare lanes 3 and 6 to lane 5), confirming a stable association of Tom22, Tom40, and Tom5. However, anti-Tom20 precipitated only small amounts of Tom22 (Fig. 2A, lower panel, lane 4). Similarly, Tom20 was efficiently precipitated only by direct precipitation with anti-Tom20 (Fig. 2A, lower panel, lane 4), whereas anti-Tom40, anti-Tom22 and anti-Tom5 coprecipitated ∼10 to 20% of Tom20 (Fig. 2A, lower panel, lanes 3, 5, and 6; Fig. 2B, columns 2 to 4). The coimmunoprecipitation experiments thus support the observations made with blue native gel electrophoresis: a stable association exists among Tom40, Tom22, and Tom5 (GIP complex), while the majority of Tom20 and the majority of Tom70 are less stably associated and can be found separate from each other and the GIP complex. Since Tom40, Tom22, and Tom5 migrate at identical positions on blue native gels (Fig. 1), the efficient coprecipitation demonstrates that they are present in the same 400K complex.
FIG. 2.
Tom20 and Tom70 are not essential for the formation of the 400K Tom complex. (A) Coimmunoprecipitation of Tom proteins. Wild-type mitochondria (250 μg) were lysed in 0.5% digitionin buffer and subjected to coimmunoprecipitation with preimmune serum (lane 1) and with anti-Tom70 (lane 2), anti-Tom40 (lane 3), anti-Tom20 (lane 4), anti-Tom22 (lane 5), and anti-Tom5 (lane 6) antibodies covalently coupled to protein A-Sepharose. The coprecipitated proteins were separated by SDS-PAGE, transferred to nitrocellulose, and immunodecorated with antisera directed against Tom70, Tom22, and Tom20. (B) Quantification of the amounts of coprecipitated Tom20 and Tom70. The experiment was performed as described for panel A. The amounts of Tom20 and Tom70 that were precipitated by their respective antibodies were set to 100% (control). (C) Formation of the 400K Tom complex in mitochondria lacking Tom20 and Tom70. Mitochondria were isolated from the wild type (WT) and a mutant strain lacking Tom20 and Tom70 and expressing Tom22 from a high-copy-number plasmid (tom20Δ tom70Δ TOM22↑) and were subjected to SDS-PAGE and blue native PAGE. Following electrophoresis, proteins were transferred to nitrocellulose and immunodecorated with antibodies directed against Tom70, Tom40, Tom22, and Tom20 (SDS-PAGE) and Tom22 (blue native PAGE). (D) Import of preproteins into mitochondria lacking Tom20 and Tom70. A rabbit reticulocyte lysate containing radiolabeled preproteins (F1-ATPase subunit β [F1β] or a fusion of the presequence of F0-ATPase subunit 9 and dihydrofolate reductase [Su9-DHFR]) was incubated with wild-type mitochondria and tom20Δ tom70Δ TOM22↑ mitochondria in the presence or absence of a membrane potential (Δψ) for the indicated times. When needed, mitochondria were treated with 50 μg of proteinase K (Prot. K) per ml and reisolated. Mitochondrial proteins were separated by SDS-PAGE, and radiolabeled proteins were detected by PhosphorImager storage technology. p, i, and m, precursor intermediate, and mature forms of a preprotein, respectively.
To determine whether Tom20 or Tom70 is needed for the formation of the GIP complex in vivo, we used an S. cerevisiae strain that lacks both Tom20 and Tom70. While deletion of both genes TOM20 and TOM70 is lethal probably because of the involvement of Tom20 and Tom70 in the biogenesis of Tom22 (13, 18, 22, 30, 31, 39), the expression of TOM22 from a high-copy-number plasmid confers viability to the double-deletion strain. The resulting yeast cells show a two- to threefold reduced growth rate on fermentable or nonfermentable medium (18). We isolated mitochondria from a tom20Δ tom70Δ TOM22↑ strain and tested them for Tom protein content. Tom70 and Tom20 were absent, as expected, whereas Tom40 was present in wild-type amounts and the content of Tom22 was slightly increased (Fig. 2C, lane 2). Thus, the expression of TOM22 from a high-copy-number plasmid restores mitochondrial Tom22 content despite the absence of the receptors Tom20 and Tom70 (see Discussion). The mitochondria were then applied to blue native gels and analyzed for the presence of the GIP complex. Lane 4 of Fig. 2C shows that the 400K complex was indeed present, showing that neither Tom20 nor Tom70 is required for the formation of the GIP complex. We wondered what the capability for importing preproteins was when the GIP complex was present but both Tom20 and Tom70 were absent. Mitochondrial precursor proteins were synthesized in rabbit reticulocyte lysates in the presence of [35S]methionine-cysteine. We used two model preproteins that have been used to study the mitochondrial import machinery (1), the β subunit of the F1-ATPase (Fig. 2D, lanes 1 to 10) and a fusion of the presequence of F0-ATPase subunit 9 and dihydrofolate reductase (Fig. 2D, lanes 11 to 20). When the preproteins were incubated with wild-type mitochondria (Fig. 2D, lanes 1 to 4 and 11 to 14) or mutant mitochondria (Fig. 2D, lanes 6 to 9 and 16 to 19) in the presence of a membrane potential, they were proteolytically processed (Fig. 2D, upper panel) and transported to a protease-protected location (Fig. 2D, lower panel). In the absence of a membrane potential, no import of the preproteins was observed with either type of mitochondria (Fig. 2D, lanes 5, 10, 15, and 20). Import into mutant mitochondria thus showed the typical characteristics of mitochondrial protein import, i.e., membrane potential dependence, proteolytic processing, and transport to a protease-protected location. The efficiencies of import of the preproteins into mutant mitochondria represented ∼15 to 25% those into wild-type mitochondria, and the import times were longer (up to 40 min). We conclude that mutant mitochondria lacking both receptors Tom20 and Tom70 are still able to import preproteins, albeit at a significantly reduced efficiency.
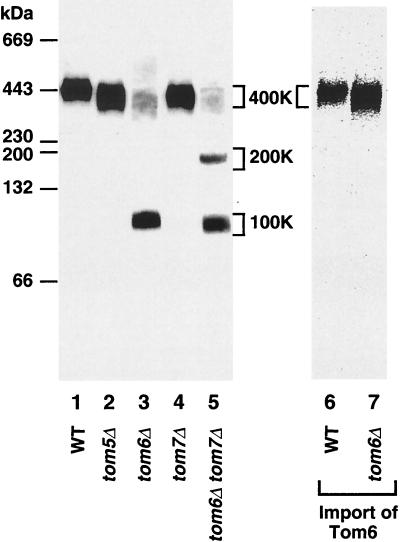
Destabilization of the 400K Tom complex in mitochondria lacking Tom6 leads to the formation of a 100K subcomplex of Tom40, Tom7, and Tom5.
We isolated mitochondria from mutant yeast strains with deletions of the genes for one or more of the small Tom proteins in order to determine if small Tom proteins were involved in the formation or stability of the 400K GIP complex. With mitochondria from a tom5Δ strain or a tom7Δ strain, we observed a small mobility shift of the 400K complex (probed with anti-Tom40 antibodies), in agreement with a minor molecular weight change due to the loss of a small subunit (Fig. 3, lanes 2 and 4). With tom6Δ mitochondria, however, a dramatic change occurred. Tom40 was predominantly (∼80% ± 10%) found in the 100K area (Fig. 3, lane 3). A lack of Tom6 thus caused a destabilization of the 400K complex. When in vitro-synthesized Tom6 was imported into tom6Δ mitochondria, the 400K complex was restored (Fig. 3, lane 7), demonstrating that the loss of Tom6 alone, and no indirect effect, was responsible for the dissociation of the 400K complex.
FIG. 3.
Deletion of Tom6 but not Tom7 or Tom5 destabilizes the 400K Tom complex. Wild-type (WT) mitochondria (lane 1) and mitochondria lacking Tom5 (tom5Δ) (lane 2), Tom6 (tom6Δ) (lane 3), Tom7 (tom7Δ) (lane 4), or both Tom6 and Tom7 (tom6Δ tom7Δ) (lane 5) were lysed in digitonin buffer and subjected to blue native PAGE. Proteins were transferred to a PVDF membrane and immunodecorated with antibodies directed against Tom40. For lanes 6 and 7, WT and tom6Δ mitochondria were first incubated with 35S-labeled Tom6 at 25°C for 15 min. Mitochondria were isolated and lysed in digitonin buffer. 35S-labeled Tom6-containing complexes were detected by Phosphorimager storage technology. The Tom40-containing complexes are indicated (400K, 200K, and 100K).
It has been proposed that Tom7 functions in part antagonisticically toward Tom6 (16). Thus, we examined if deletion of Tom7 influenced the Tom complex in tom6Δ mitochondria. Mitochondria were isolated from a tom6Δ tom7Δ double-deletion strain and analyzed by blue native gel electrophoresis. Besides a small mobility shift of the 100K subcomplex (consistent with the loss of a small Tom protein), we observed an additional band at about 200K (Fig. 3, lane 5). The 200K band was present in relatively small amounts and contained Tom40 and probably Tom5 but not Tom22 (data not shown). The lack of Tom7 thus resulted in a partial stabilizing effect on Tom subcomplexes in tom6Δ mitochondria.
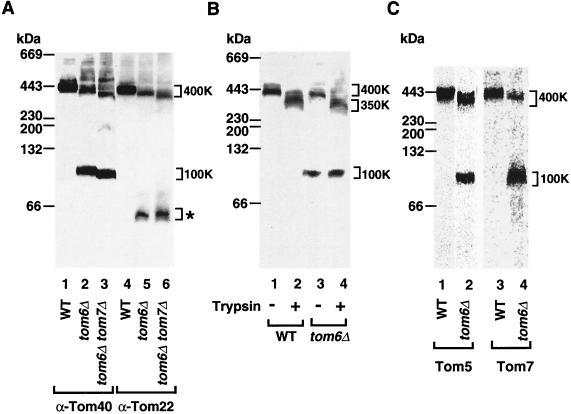
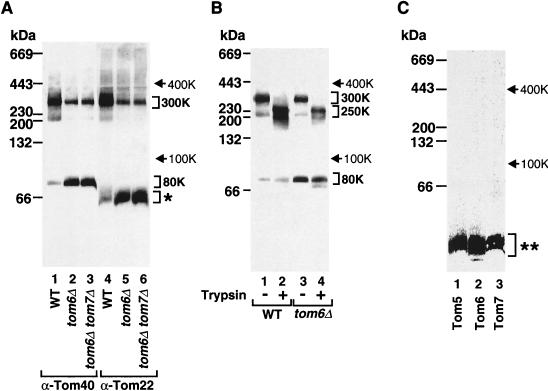
Which Tom proteins are present in the 100K subcomplex? We used anti-Tom40 and anti-Tom22 antibodies in parallel. Only Tom40 was found in the 100K area in both tom6Δ mitochondria and tom6Δ tom7Δ mitochondria (Fig. 4A, lanes 2 and 3), while a considerable amount of Tom22 was observed in a lower-molecular-weight range (Fig. 4A, lanes 5 and 6).
FIG. 4.
The 100K Tom subcomplex contains Tom5 and Tom7 but not Tom22. (A) Tom40 and Tom22 of tom6Δ mitochondria are not tightly associated. Wild-type (WT) mitochondria (lanes 1 and 4) and mitochondria lacking Tom6 (tom6Δ) (lanes 2 and 5) or both Tom6 and Tom7 (tom6Δ tom7Δ) (lanes 3 and 6) were lysed in digitonin buffer and subjected to blue native PAGE and immunodecoration with antibodies against Tom40 (lanes 1 to 3) or Tom22 (lanes 4 to 6). The positions of the 400K and 100K complexes as well as Tom22 found at a low molecular weight (asterisk) are indicated. (B) Trypsin treatment of mitochondria leads to partial degradation of the 400K complex but not of the 100K subcomplex. WT and tom6Δ mitochondria were treated or not treated with 20 μg of trypsin per ml prior to digitonin buffer lysis, blue native PAGE, and immunodecoration with antibodies against Tom40. (C) Tom5 and Tom7 are present in the 100K subcomplex. In vitro-translated 35S-labeled Tom5 (lanes 1 and 2) and 35S-labeled Tom7 (lanes 3 and 4) were incubated with WT or tom6Δ mitochondria at 25°C for 20 min. Mitochondria were isolated, lysed in digitonin buffer, and subjected to blue native PAGE. Radiolabeled complexes were analyzed by PhosphorImager storage technology.
Tom22 exposes to the cytosol a domain that is accessible to trypsin added to mitochondria (22, 24), leading to the removal of ∼10 kDa. When wild-type mitochondria were treated with trypsin prior to lysis and blue native gel electrophoresis, the GIP complex shifted to ∼350K (Fig. 4B, lane 2). As described below, a single GIP complex contains several (three to six) Tom22 molecules; the observed molecular weight shift of the GIP complex is thus in agreement with a loss of the cytosolic domains of the Tom22 molecules. With the 100K subcomplex of tom6Δ mitochondria, however, no mobility shift was observed after treatment of the mitochondria with trypsin (Fig. 4B, compare lanes 3 and 4), confirming the absence of Tom22 from the 100K subcomplex.
We examined whether the other two small Tom proteins, Tom5 and Tom7, were present in the 100K subcomplex of tom6Δ mitochondria. The small mobility shift of the 100K subcomplex between tom6Δ mitochondria and tom6Δ tom7Δ mitochondria (Fig. 4A, compare lanes 2 and 3) may suggest the presence of Tom7. To directly determine their presence, 35S-labeled Tom5 or Tom7 was imported into tom6Δ mitochondria and analyzed by blue native gel electrophoresis. Indeed, major fractions of both small Tom proteins were found in the 100K subcomplex (Fig. 4C, lanes 2 and 4). We conclude that the 100K subcomplex contains Tom40, Tom7, and Tom5.
Release of the three small Tom proteins: Tom6 is not required for maintaining a stable association between Tom40 and Tom22.
The stability of the 400K complex was tested by lysis of mitochondria with digitonin in the presence of salt or urea. The 400K complex was surprisingly highly stable. Neither up to 0.5 M NaCl nor up to 4 M urea had any influence on the migration of the 400K complex in blue native gel electrophoresis (data not shown). Lysis of mitochondria with Triton X-100, however, had a profound effect on the mobility of the GIP complex (Fig. 5A). When wild-type mitochondria were lysed with Triton X-100, Tom40 and Tom22 migrated mainly at ∼300K (Fig. 5A, lanes 1 and 4). A small amount of Tom40 was also found at ∼80K (Fig. 5A, lane 1), and a small amount of Tom22 was found at an even lower molecular weight (Fig. 5A, lane 4). With tom6Δ mitochondria and tom6Δ tom7Δ mitochondria, the major fraction of Tom40 was found at the 80K position (Fig. 5A, lanes 2 and 3), while Tom22 migrated mainly at a lower position (Fig. 5A, lanes 5 and 6). Trypsin treatment of mitochondria prior to lysis with Triton X-100 caused a shift of the 300K complex to ∼250K (Fig. 5B, lane 2), while the mobility of the 80K subcomplex containing Tom40 was not altered (Fig. 5B, lane 4). This result confirms that Tom22 is not present in the 80K subcomplex.
FIG. 5.
Release of the small Tom proteins leads to a Tom40 core complex (80K). (A) Lysis of mitochondria with Triton X-100 generates 300K and 80K Tom complexes. Wild-type (WT) mitochondria (lanes 1 and 4) and mitochondria lacking Tom6 (tom6Δ) (lanes 2 and 5) or both Tom6 and Tom7 (tom6Δ tom7Δ) (lanes 3 and 6) were lysed in Triton X-100 buffer and subjected to blue native PAGE and immunodecoration with antibodies against Tom40 (lanes 1 to 3) or Tom22 (lanes 4 to 6). The positions of the 300K and 80K Tom40 complexes found in Triton X-100 buffer and Tom22 found at a low molecular weight (asterisk), along with the expected positions of the 400K and 100K complexes (as determined after digitonin buffer lysis; see Fig. 3 and 4), are indicated. (B) Trypsin treatment of mitochondria leads to partial degradation of the 300K complex but not of the 80K complex. WT and tom6Δ mitochondria were treated or not treated with 20 μg of trypsin per ml prior to lysis in Triton X-100 buffer, blue native PAGE, and immunodecoration with antibodies against Tom40. (C) Triton X-100 causes the release of the three small Tom proteins from Tom40. WT mitochondria were incubated with in vitro-translated 35S-labeled Tom5 (lane 1), 35S-labeled Tom6 (lane 2), or 35S-labeled Tom7 (lane 3) for 20 min at 25°C. Mitochondria were isolated and lysed in Triton X-100 buffer prior to analysis by blue native PAGE. Radiolabeled complexes were detected by PhosphorImager storage technology. The expected positions of the 400K and 100K complexes (from digitonin lysis buffer), along with the actual position of the radiolabeled Tom proteins (double asterisks), are indicated.
While it may be argued that the mobility differences of the GIP complex after lysis with digitonin or Triton X-100 can be attributed to an influence of the detergent on the electrophoretic run, we observed a further difference when comparing wild-type mitochondria with tom6Δ or tom6Δ tom7Δ mitochondria. With digitonin-lysed mitochondria, the absence of Tom6 or Tom7 was visible as small mobility shifts of the remaining 400K complexes with both anti-Tom40 and anti-Tom22 antibodies (Fig. 4A). With Triton X-100-lysed mitochondria, however, the mobility of the 300K complexes was not altered by either the presence or the absence of Tom6 or Tom7 (Fig. 5A). Moreover, the subcomplexes of tom6Δ mitochondria containing Tom40 were differentially influenced by the absence of Tom7: after lysis with digitonin, the 100K subcomplex of tom6Δ tom7Δ mitochondria ran faster than that of tom6Δ mitochondria, consistent with the absence of Tom7 (Fig. 4A, lanes 2 and 3); in contrast, after lysis with Triton X-100, the 80K subcomplex of tom6Δ mitochondria (Fig. 5A, lane 2) did not show altered mobility when Tom7 was absent (Fig. 5A, lane 3). These results suggest that small Tom proteins are released from Tom complexes by Triton X-100.
To test this prediction, we imported 35S-labeled Tom5, Tom6, and Tom7 into mitochondria and performed lysis with Triton X-100. None of the small Tom proteins was present in the area of the GIP complex or the 80K subcomplex, but all three, Tom5, Tom6, and Tom7, were found in the very low molecular weight range (Fig. 5C, lanes 1 to 3; data are for wild-type mitochondria; the same results were obtained with tom6Δ mitochondria). We showed above with digitonin lysis that the in vitro-imported small Tom proteins were efficiently assembled into the GIP complex of wild-type mitochondria (Fig. 3, lane 6; Fig. 4C, lanes 1 and 3) or, in case of Tom5 and Tom7, also into the 100K subcomplex of tom6Δ mitochondria (Fig. 4C, lanes 2 and 4). These results demonstrate that Triton X-100 releases the three small Tom proteins from the GIP complex and from Tom40 or Tom22 subcomplexes derived from it.
Since the major fractions of Tom40 and Tom22 from wild-type mitochondria remained associated in Triton X-100 despite the release of Tom6 (Fig. 5A, lanes 1 and 4), we conclude that Tom6 is not essential to maintain the interaction between Tom40 and Tom22. The absence of Tom6 in mitochondria (tom6Δ), however, causes dissociation of large fractions of Tom40 and Tom22 (Fig. 5A, lanes 2 and 5). Thus, Tom6 is required to promote but not to maintain the association of Tom22 with Tom40.
Assessment of the stoichiometry of Tom proteins.
We quantified the mitochondrial amounts of the large Tom proteins Tom20, Tom22, Tom40, and Tom70 by standardized immunoblotting with a direct comparison of expressed and purified Tom protein cytosolic domains and mitochondrial extracts (the antibodies were generated against the expressed proteins) (6, 9). Tom40 was present at 250 to 300 pmol/mg of mitochondrial protein, and Tom22 was present at 200 to 300 pmol/mg (Table 2). Tom20 and Tom70 were found at 60 to 70 pmol/mg. As determined by the accumulation of a two-membrane-spanning preprotein, the amount of translocation contact sites was reported to be 15 pmol/mg, the same amount as that determined for the Tim core complexes of the inner membrane (9). Since only one in three to four GIP complexes contains a two-membrane-spanning preprotein under saturating conditions (9), the amount of GIP complexes is 45 to 60 pmol/mg. We demonstrate here that Tom40 and Tom22 are predominantly present in GIP complexes; therefore, each GIP complex contains about four to six molecules of Tom40 and three to six molecules of Tom22.
TABLE 2.
Assessment of the stoichiometry of Tom proteinsa
| Component or structure | Abundance, pmol/mg of mitochondrial protein (associated with GIP complex) |
|---|---|
| Tom proteins | |
| Tom20 | 60–70 (4–12) |
| Tom22 | 200–300 (200–300) |
| Tom40 | 250–300 (250–300) |
| Tom70 | 60–70 (<3) |
| GIP complexes | 45–60 |
| Translocation contact sites | 15 |
| Tim core complexes | 15–20 |
The abundance of Tom proteins was determined by standardized immunoblotting with expressed and purified protein cytosolic domains (6, 9) and antibodies generated against them. The reactivities of the antibodies with the SDS-denatured proteins were compared for the purified proteins and the mitochondrial extracts. The data for the abundance of GIP complexes, translocation contact sites, and Tim core complexes and the 250-pmol/mg value for Tom40 are derived from reference 9. The association with the GIP complex was determined by blue native gel electrophoresis and coimmunoprecipitation.
How do these calculations fit with the relative native sizes of the GIP complex and the various subcomplexes assessed by blue native gel electrophoresis? Treatment of mitochondria with trypsin removes ∼50 kDa from both the digitonin-lysed GIP complex and the Triton X-100-lysed GIP complex, consistent with the removal of the cytosolic domains of three to six molecules of Tom22. The 80K subcomplex in Triton X-100-lysed tom6Δ mitochondria contains neither Tom22 nor the three small Tom proteins. Additionally, since the (already weak) coprecipitation of Tom20 or Tom70 with Tom40 in wild-type mitochondria is further decreased in tom6Δ mitochondria (2), the possibility that Tom20 or Tom70 is quantitatively present in the 80K subcomplex can be excluded. It is therefore likely that the 80K subcomplex consists of Tom40 alone and may represent a dimer. The 100K subcomplex of digitonin-lysed tom6Δ mitochondria contains Tom40, Tom7, and Tom5 but not Tom22. The shift from 80K to 100K agrees with the addition of Tom5 and Tom7 to a Tom40 dimer. The 300K complex of Triton X-100-lysed wild-type mitochondria contains Tom22 and Tom40 but not the small Tom proteins; its size is consistent with the presence of three to six molecules of Tom22 and four to six molecules of Tom40. The size of 400K of the GIP complex of digitonin-lysed wild-type mitochondria agrees with the presence of three to six molecules of Tom22, four to six molecules of Tom40, and the additional presence of small Tom proteins. The absolute amount of small Tom proteins cannot be determined so far due to the lack of expressed and purified proteins (and of monospecific antibodies generated against expressed proteins). The small mobility shifts of the 400K complex in the various mutant mitochondria lacking one or two small Tom proteins (Fig. 3; Fig. 4A) in comparison to the mobility shifts resulting from the removal of the cytosolic domains of Tom22 (Fig. 4B) suggest the presence of a limited number of small Tom proteins (about two to four molecules of each small Tom protein) in the 400K complex. Schägger et al. (43) pointed out that, while blue native gel electrophoresis does not allow an absolute size determination of protein complexes due to the presence of ligands (lipids, detergent, and Coomassie brilliant blue G-250) or the influence of protein shape, the detected molecular weights deviated less than 20% from those determined by other methods. For Tom22 and Tom40, we could compare the assessment by blue native gel electrophoresis with the direct quantification of the protein amounts; we observed good agreement, supporting the value of blue native gel electrophoresis for the assessment of native sizes of membrane protein complexes.
Only 5 to 10% (blue native gel electrophoresis) or 10 to 20% (coimmunoprecipitation) of Tom20 molecules, i.e., ∼4 to 12 pmol/mg, were found in association with the GIP complex (Table 2). This means that only a minority of GIP complexes (60 pmol/mg) have Tom20 stably associated after digitonin lysis. Therefore, only a fraction of Tom22 (present at 200 to 300 pmol/mg) can be observed in association with Tom20. The proposed heterodimer or complex of Tom20 and Tom22 (32) does not seem to represent a major stable form under the conditions used here.
DISCUSSION
This report leads to three main conclusions about the organization of the protein import machinery of the outer mitochondrial membrane: (i) the GIP complex of ∼400K contains the essential subunits Tom40 and Tom22 and the three small Tom proteins; (ii) the receptors Tom20 and Tom70 are not crucial for the formation of the GIP complex; and (iii) Tom6 functions as an assembly factor for Tom22, promoting its stable association with Tom40.
The GIP complex.
We report that a protein complex of 400K represents the central unit of the preprotein translocase of the outer mitochondrial membrane. The complex quantitatively contains the only two essential proteins of the Tom machinery, Tom22 and Tom40. Tom40 is the major constituent of the GIP; thus, the complex is termed the GIP complex. In addition, the complex contains the three small proteins Tom5, Tom6, and Tom7. The GIP complex from digitonin-lysed mitochondria is resistant to treatment with salt and urea (up to 4 M), but the presence of Triton X-100 causes the release of the three small Tom proteins, while Tom22 and Tom40 remain stably associated. This finding suggests that the small Tom proteins may be associated with the Tom40-Tom22 core complex via hydrophobic interactions. By assessment of the relative sizes of complexes by blue native gel electrophoresis and quantification of the amounts of Tom40 and Tom22 in comparison to the number of import sites (Table 2), the GIP complex was found to contain four to six molecules of Tom40 and three to six molecules of Tom22. The three small Tom proteins may be present at two to four copies each.
It may be argued that the exact comigration of Tom40, Tom22, and the three small Tom proteins in blue native gel electrophoresis is fortuitous and does not prove that they are present in the same 400K complex. This possibility can be excluded because these Tom proteins were efficiently coimmunoprecipitated with antibodies directed against Tom40, Tom22, or Tom5 (Fig. 2) (1, 2, 11, 16). Because Tom40, Tom22, Tom5, and (at least) the bulk of Tom6 and Tom7 comigrated at 400K, the efficient coimmunoprecipitation indicated that they were present in the same complex. Observations with mutant mitochondria are supportive of the presence of these Tom proteins in the same complex. The minor mobility shifts of the complexes from tom5Δ or tom7Δ mitochondria in comparison to wild-type mitochondria in blue native gel electrophoresis were in agreement with the loss of the small subunits. In tom6Δ mitochondria, the interaction between Tom40 and Tom22 was destabilized but could be restored by the import of Tom6 into isolated mitochondria. Moreover, the possibility that the association of the Tom proteins occurred after lysis of the mitochondria can be excluded because the 35S-labeled subunits efficiently assembled into the 400K complex when imported into intact mitochondria but did not assemble at all when incubated with lysed mitochondria (data not shown).
Tom20 and Tom70.
Mayer et al. (32) proposed that Tom20 and Tom22 are needed simultaneously in the binding of preproteins, forming a complex that functions as the mitochondrial presequence receptor. We therefore expected that both proteins were present in roughly equimolar amounts in the same complex; however, three lines of evidence suggest a modification of the view.
(i) While Tom22 is stably associated with Tom40 in blue native gel electrophoresis, the majority of Tom20 is found in the low-molecular-weight range. Thus, the vast majority of Tom22 is not present in a stable complex with Tom20 that can be detected by blue native gel electrophoresis (while the associations between the subunits of the GIP complex are highly stable in electrophoresis (Fig. 1), like the associations in the Tim core complex and the associations between a membrane-spanning preprotein and import complexes [9]).
(ii) An additional technique, the coimmunoprecipitation of Tom proteins, led to a conclusion similar to that from blue native gel electrophoresis regarding the loose association between Tom20 and Tom22. Only 4 to 12 pmol of Tom20 per mg (5 to 20%) seems to be stably associated with the 400K Tom complex under the conditions used (where Tom22 is present at 200 to 300 pmol per mg of mitochondrial protein). Both techniques suggest that the bulk of the receptor Tom70 is not stably associated with the GIP complex; <5% (<3 pmol/mg) of total Tom70 seems to be associated with subunits of the GIP complex.
(iii) With both methods, blue native gel electrophoresis and coimmunoprecipitation, it remains formally possible that Tom20 and Tom70 are genuine subunits of the GIP complex and are released from the complex after lysis of the mitochondria. Therefore, we used an in vivo assay with tom20Δ tom70Δ mitochondria. Despite the complete absence of both receptors, the GIP complex was fully stable, and the mitochondria were functional in the import of preproteins, with an efficiency of ∼15 to 25% compared to wild-type mitochondria. The only, but crucial, prerequisite was that Tom22 was present in the mitochondria in wild-type amounts. This requirement could be achieved by expression of TOM22 from a high-copy-number plasmid. The explanation is that Tom20 and Tom70 are involved in the biogenesis of Tom22, and a lack of Tom20 causes a strong reduction in the mitochondrial level of Tom22 (13, 18, 22, 30, 42a). Interestingly, Lithgow et al. (30, 31) isolated a yeast strain with a dominant mutation in an unidentified nuclear gene (termed SUPX). The SUPX mutation apparently caused an increased half-life of Tom22 and thereby suppressed the lethal phenotype caused by a tom20Δ tom70Δ double deletion. Preproteins were shown to be imported into mitochondria isolated from a tom20Δ tom70Δ SUPX strain at a partially reduced efficiency. It is conceivable that the SUPX mutation also stabilizes other components of the mitochondrial import machinery, a suggestion which could explain the relatively mild reduction of protein import into mitochondria isolated from the tom20Δ tom70Δ SUPX strain.
We conclude that Tom20 and Tom70 are not crucial for the formation of the GIP complex but associate with the complex in a more loose manner. It is conceivable that in vivo, all Tom proteins are present in a large dynamic complex that consists of subcomplexes. Tom20 and Tom70 would thereby represent peripheral subunits of such a translocase, while the GIP complex would form the central unit. Moreover, transient interactions between these receptors (15) and the GIP complex may be involved in the transfer of preproteins between the receptors and the GIP complex. The GIP complex contains one subunit with a receptor function (Tom22), such that it is able to mediate the basic import of preproteins by itself. The presence of Tom20 and Tom70 may facilitate the collection of preproteins from all over the mitochondrial surface and thereby increase the rate of import about fivefold.
Tom6 as an assembly factor.
A deletion of Tom6 has a strong effect on the stability of the GIP complex. The interaction between Tom40 and Tom22 is destabilized, and the proteins are preferentially found in the lower-molecular-weight range in blue native gel electrophoresis: Tom40 in an ∼100K subcomplex and Tom22 below 60K. Tom5 and Tom7 remain associated with Tom40 in the 100K subcomplex. When Tom6 is imported into tom6Δ mitochondria, the 400K complex is restored, showing that the lack of Tom6 is solely responsible for the dissociation of Tom22 from Tom40.
Two possibilities for the function of Tom6 are conceivable. (i) Tom6 is a structural component of the complex that must be permanently associated with Tom40 and Tom22, or (ii) Tom6 promotes the assembly of Tom22 with Tom40 but is not required to maintain the interaction between both proteins. The latter possibility seems likely, since most of Tom22 and Tom40 remained stably associated after lysis of wild-type mitochondria with Triton X-100, although all small Tom proteins, including Tom6, were released from the GIP complex. In contrast, when tom6Δ mitochondria were lysed with Triton X-100, a large fraction of Tom22 dissociated from Tom40. Tom6 has to be present in mitochondria in order to promote stable contact between Tom22 and Tom40 but, once established, the interaction is maintained in the absence of Tom6 as well.
We propose that Tom40 forms the core of the GIP complex with which Tom22, Tom7, and Tom5 assemble. While Tom5 and Tom7 associate with Tom40 in a Tom6-independent manner, Tom6 functions as an assembly factor for Tom22.
ACKNOWLEDGMENTS
We are grateful to Klaus Dietmeier for experimental advice.
This work was supported by the Deutsche Forschungsgemeinschaft, the Sonderforschungsbereich 388, and the Fonds der Chemischen Industrie (grant to N.P.) and by long-term fellowships from the Human Frontier Science Program (to P.J.T.D.) and the Alexander-von-Humboldt Foundation (to M.T.R.).
Peter J. T. Dekker and Michael T. Ryan contributed equally to this work.
REFERENCES
- 1.Alconada A, Gärtner F, Hönlinger A, Kübrich M, Pfanner N. Mitochondrial receptor complex from Neurospora crassa and Saccharomyces cerevisiae. Methods Enzymol. 1995;260:263–286. doi: 10.1016/0076-6879(95)60144-9. [DOI] [PubMed] [Google Scholar]
- 2.Alconada A, Kübrich M, Moczko M, Hönlinger A, Pfanner N. The mitochondrial receptor complex: the small subunit Mom8b/Isp6 supports association of receptors with the general insertion pore and transfer of preproteins. Mol Cell Biol. 1995;15:6196–6205. doi: 10.1128/mcb.15.11.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blom J, Dekker P J T, Meijer M. Functional and physical interactions of components of the yeast mitochondrial inner-membrane import machinery (MIM) Eur J Biochem. 1995;232:309–314. doi: 10.1111/j.1432-1033.1995.tb20813.x. [DOI] [PubMed] [Google Scholar]
- 4.Bolliger L, Junne T, Schatz G, Lithgow T. Acidic receptor domains on both sides of the outer membrane mediate translocation of precursor proteins into yeast mitochondria. EMBO J. 1995;14:6318–6326. doi: 10.1002/j.1460-2075.1995.tb00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bömer U, Pfanner N, Dietmeier K. Identification of a third yeast mitochondrial Tom protein with tetratrico peptide repeats. FEBS Lett. 1996;382:153–158. doi: 10.1016/0014-5793(96)00156-1. [DOI] [PubMed] [Google Scholar]
- 6.Brix J, Dietmeier K, Pfanner N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J Biol Chem. 1997;272:20730–20735. doi: 10.1074/jbc.272.33.20730. [DOI] [PubMed] [Google Scholar]
- 7.Court D A, Nargang F E, Steiner H, Hodges R S, Neupert W, Lill R. Role of the intermembrane space domain of the preprotein receptor Tom22 in protein import into mitochondria. Mol Cell Biol. 1996;16:4035–4042. doi: 10.1128/mcb.16.8.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daum G, Böhni P C, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 9.Dekker P J T, Martin F, Maarse A C, Bömer U, Müller H, Guiard B, Meijer M, Rassow J, Pfanner N. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 1997;16:5408–5419. doi: 10.1093/emboj/16.17.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dekker P J T, Müller H, Rassow J, Pfanner N. Characterization of the preprotein translocase of the outer mitochondrial membrane by blue native electrophoresis. Biol Chem. 1996;377:535–538. [PubMed] [Google Scholar]
- 11.Dietmeier K, Hönlinger A, Bömer U, Dekker P J T, Eckerskorn C, Lottspeich F, Kübrich M, Pfanner N. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature. 1997;388:195–200. doi: 10.1038/40663. [DOI] [PubMed] [Google Scholar]
- 12.Gratzer S, Lithgow T, Bauer R E, Lamping E, Paltauf F, Kohlwein S D, Haucke V, Junne T, Schatz G, Horst M. Mas37p, a novel receptor subunit for protein import into mitochondria. J Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harkness T A, Nargang F E, van der Klei I, Neupert W, Lill R. A crucial role of the mitochondrial protein import receptor MOM19 for the biogenesis of mitochondria. J Cell Biol. 1994;124:637–648. doi: 10.1083/jcb.124.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartl F U, Ostermann J, Guiard B, Neupert W. Successive translocation into and out of the mitochondrial matrix: targeting of proteins to the intermembrane space by a bipartite signal peptide. Cell. 1987;51:1027–1037. doi: 10.1016/0092-8674(87)90589-7. [DOI] [PubMed] [Google Scholar]
- 15.Haucke V, Horst M, Schatz G, Lithgow T. The Mas20p and Mas70p subunits of the protein import receptor of yeast mitochondria interact via the tetratricopeptide repeat motif in Mas20p: evidence for a single hetero-oligomeric receptor. EMBO J. 1996;15:1231–1237. [PMC free article] [PubMed] [Google Scholar]
- 16.Hönlinger A, Bömer U, Alconada A, Eckerskorn C, Lottspeich F, Dietmeier K, Pfanner N. Tom7 modulates the dynamics of the mitochondrial outer membrane translocase and plays a pathway-related role in protein import. EMBO J. 1996;15:2125–2137. [PMC free article] [PubMed] [Google Scholar]
- 17.Hönlinger A, Keil P, Nelson R J, Craig E A, Pfanner N. Posttranslational mitochondrial protein import in a homologous yeast in vitro system. Biol Chem. 1995;376:515–519. [PubMed] [Google Scholar]
- 18.Hönlinger A, Kübrich M, Moczko M, Gärtner F, Mallet L, Bussereau F, Eckerskorn C, Lottspeich F, Dietmeier K, Jacquet M, Pfanner N. The mitochondrial receptor complex: Mom22 is essential for cell viability and directly interacts with preproteins. Mol Cell Biol. 1995;15:3382–3389. doi: 10.1128/mcb.15.6.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horst M, Hilfiker-Rothenfluh S, Oppliger W, Schatz G. Dynamic interaction of the protein translocation systems in the inner and outer membranes of yeast mitochondria. EMBO J. 1995;14:2293–2297. doi: 10.1002/j.1460-2075.1995.tb07223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang S, Jascur T, Vestweber D, Pon L, Schatz G. Disrupted yeast mitochondria can import precursor proteins directly through their inner membrane. J Cell Biol. 1989;109:487–493. doi: 10.1083/jcb.109.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kassenbrock C K, Cao W, Douglas M G. Genetic and biochemical characterization of ISP6, a small mitochondrial outer membrane protein associated with the protein translocation complex. EMBO J. 1993;8:3023–3034. doi: 10.1002/j.1460-2075.1993.tb05971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keil P, Pfanner N. Insertion of MOM22 into the mitochondrial outer membrane strictly depends on surface receptors. FEBS Lett. 1993;321:197–200. doi: 10.1016/0014-5793(93)80107-6. [DOI] [PubMed] [Google Scholar]
- 23.Keil P, Weinzierl A, Kiebler M, Dietmeier K, Söllner T, Pfanner N. Biogenesis of the mitochondrial receptor complex: two receptors are required for binding of MOM38 to the outer membrane surface. J Biol Chem. 1993;268:19177–19180. [PubMed] [Google Scholar]
- 24.Kiebler M, Keil P, Schneider H, van der Klei I J, Pfanner N, Neupert W. The mitochondrial receptor complex: a central role of MOM22 in mediating preprotein transfer from receptors to the general insertion pore. Cell. 1993;74:483–492. doi: 10.1016/0092-8674(93)80050-o. [DOI] [PubMed] [Google Scholar]
- 25.Kiebler M, Pfaller R, Söllner T, Griffiths G, Horstman H, Pfanner N, Neupert W. Identification of a mitochondrial receptor complex required for recognition and membrane insertion of precursor proteins. Nature. 1990;348:610–616. doi: 10.1038/348610a0. [DOI] [PubMed] [Google Scholar]
- 26.Komiya T, Rospert S, Schatz G, Mihara K. Binding of mitochondrial precursor proteins to the cytoplasmic domains of the import receptors Tom70 and Tom20 is determined by cytoplasmic chaperones. EMBO J. 1997;16:4267–4275. doi: 10.1093/emboj/16.14.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komiya T, Sakaguchi M, Mihara K. Cytoplasmic chaperones determine the targeting pathway of precursor proteins to mitochondria. EMBO J. 1996;15:399–407. [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Lithgow T, Glick B S, Schatz G. The protein import receptor of mitochondria. Trends Biochem Sci. 1995;20:98–101. doi: 10.1016/s0968-0004(00)88972-0. [DOI] [PubMed] [Google Scholar]
- 30.Lithgow T, Junne T, Suda K, Gratzer S, Schatz G. The mitochondrial outer membrane protein Mas22p is essential for protein import and viability of yeast. Proc Natl Acad Sci USA. 1994;91:11973–11977. doi: 10.1073/pnas.91.25.11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lithgow T, Junne T, Wachter C, Schatz G. Yeast mitochondria lacking the two import receptors Mas20p and Mas70p can efficiently and specifically import precursor proteins. J Biol Chem. 1994;269:15325–15330. [PubMed] [Google Scholar]
- 32.Mayer A, Nargang F E, Neupert W, Lill R. MOM22 is a receptor for mitochondrial targeting sequences and cooperates with MOM19. EMBO J. 1995;14:4204–4211. doi: 10.1002/j.1460-2075.1995.tb00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moczko M, Bömer U, Kübrich M, Zufall N, Hönlinger A, Pfanner N. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol Cell Biol. 1997;17:6574–6584. doi: 10.1128/mcb.17.11.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moczko M, Dietmeier K, Söllner T, Segui B, Steger H F, Neupert W, Pfanner N. Identification of the mitochondrial receptor complex in Saccharomyces cerevisiae. FEBS Lett. 1992;10:265–268. doi: 10.1016/0014-5793(92)81345-m. [DOI] [PubMed] [Google Scholar]
- 35.Nakai M, Kinoshita K, Endo T. Mitochondrial receptor complex protein. The intermembrane space domain of yeast MAS17 is not essential for its targeting or function. J Biol Chem. 1995;270:30571–30575. doi: 10.1074/jbc.270.51.30571. [DOI] [PubMed] [Google Scholar]
- 36.Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 37.Pfanner N, Craig E A, Hönlinger A. Mitochondrial preprotein translocase. Annu Rev Cell Dev Biol. 1997;13:25–51. doi: 10.1146/annurev.cellbio.13.1.25. [DOI] [PubMed] [Google Scholar]
- 38.Pfanner N, Douglas M G, Endo T, Hoogenraad N J, Jensen R E, Meijer M, Neupert W, Schatz G, Schmitz U K, Shore G C. Uniform nomenclature for the protein transport machinery of the mitochondrial membranes. Trends Biochem Sci. 1996;21:51–52. [PubMed] [Google Scholar]
- 39.Ramage L, Junne T, Hahne K, Lithgow T, Schatz G. Functional cooperation of mitochondrial protein import receptors in yeast. EMBO J. 1993;12:4115–4123. doi: 10.1002/j.1460-2075.1993.tb06095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rapaport D, Neupert W, Lill R. Mitochondrial protein import: Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J Biol Chem. 1997;272:18725–18731. doi: 10.1074/jbc.272.30.18725. [DOI] [PubMed] [Google Scholar]
- 41.Rassow J, Pfanner N. Mitochondrial preproteins en route from the outer membrane to the inner membrane are exposed to the intermembrane space. FEBS Lett. 1991;293:85–88. doi: 10.1016/0014-5793(91)81157-4. [DOI] [PubMed] [Google Scholar]
- 42.Ryan K R, Jensen R E. Protein translocation across mitochondrial membranes: what a long, strange trip it is. Cell. 1995;83:517–519. doi: 10.1016/0092-8674(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 42a.Ryan, M. T., and N. Pfanner. Unpublished data.
- 43.Schägger H, Cramer W A, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 44.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 45.Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 46.Schatz G. Just follow the acid chain. Nature. 1997;388:121–122. doi: 10.1038/40510. [DOI] [PubMed] [Google Scholar]
- 47.Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 48.Schleiff E, Shore G C, Goping I S. Interactions of the human mitochondrial protein import receptor, hTom20, with precursor proteins in vitro reveal pleiotropic specificities and different receptor domain requirements. J Biol Chem. 1997;272:17784–17789. doi: 10.1074/jbc.272.28.17784. [DOI] [PubMed] [Google Scholar]
- 49.Schleyer M, Neupert W. Transport of proteins into mitochondria: translocational intermediates spanning contact sites between outer and inner membranes. Cell. 1985;43:339–350. doi: 10.1016/0092-8674(85)90039-x. [DOI] [PubMed] [Google Scholar]
- 50.Schlossmann J, Lill R, Neupert W, Court D A. Tom71, a novel homologue of the mitochondrial preprotein receptor Tom70. J Biol Chem. 1996;271:17890–17895. doi: 10.1074/jbc.271.30.17890. [DOI] [PubMed] [Google Scholar]
- 51.Segui-Real B, Kispal G, Lill R, Neupert W. Functional independence of the protein translocation machineries in mitochondrial outer and inner membranes: passage of preproteins through the intermembrane space. EMBO J. 1993;12:2211–2218. doi: 10.1002/j.1460-2075.1993.tb05869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Söllner T, Rassow J, Pfanner N. Analysis of mitochondrial protein import using translocation intermediates and specific antibodies. Methods Cell Biol. 1991;3:345–358. doi: 10.1016/s0091-679x(08)61689-1. [DOI] [PubMed] [Google Scholar]
- 54.Söllner T, Rassow J, Wiedmann M, Schlossmann J, Keil P, Neupert W, Pfanner N. Mapping of the protein import machinery in the mitochondrial outer membrane by crosslinking of translocation intermediates. Nature. 1992;355:84–87. doi: 10.1038/355084a0. [DOI] [PubMed] [Google Scholar]
- 55.Vestweber D, Brunner J, Baker A, Schatz G. A 42K outer-membrane protein is a component of the yeast mitochondrial protein import site. Nature. 1989;341:205–209. doi: 10.1038/341205a0. [DOI] [PubMed] [Google Scholar]