Abstract
The genus Bifidobacterium widely exists in human gut and has been increasingly used as the adjuvant probiotics for the prevention and treatment of diseases. However, the functional differences of Bifidobacterium genomes from different regions of the world remain unclear. We here describe an extensive study on the genomic characteristics and function annotations of 1512 genomes (clustered to 849 non-redundant genomes) of Bifidobacterium cultured from human gut. The distribution of some carbohydrate-active enzymes varied among different Bifidobacterium species and continents. More than 36% of the genomes of B. pseudocatenulatum harbored biosynthetic gene clusters of lanthipeptide-class-iv. 99.76% of the cultivated genomes of Bifidobacterium harbored genes of bile salt hydrolase. Most genomes of B. adolescentis, and all genomes of B. dentium harbored genes involved in gamma-aminobutyric acid synthesis. B. longum subsp. infantis were characterized harboring most genes related to human milk oligosaccharide utilization. Significant differences between the distribution of antibiotic resistance genes among different species and continents revealed the importance to use antibiotics precisely in the clinical treatment. Phages infecting Bifidobacterium and horizontal gene transfers occurring in genomes of Bifidobacterium were dependent on species and region sources, and might help Bifidobacterium adapt to the environment. In addition, the distribution of Bifidobacterium in human gut was found varied from different regions of the world. This study represents a comprehensive view of characteristics and functions of genomes of cultivated Bifidobacterium from human gut, and enables clinical advances in the future.
Keywords: Bifidobacterium, Gut microbiome, Bile salt hydrolase, Phages, Horizontal gene transfers
1. Introduction
The genus Bifidobacterium, which belongs to Actinomycetota, is one of the dominant members of microbes in the human gut, particularly during the early life [1]. Bifidobacterium makes up 60–70% of infant gut microbes and approximately 10–15% of adult gut microbes [2]. A higher average Bifidobacterium level was observed in Japanese adults than individuals from other countries [3]. Based on the development of culture-based methods and metagenomic analyses, the distribution and function of Bifidobacterium in the human gut have been studied in a lot [4,5] and their relationships with human health have been investigated. Large-scale culture-based collections of human gut microbiota have been established and have provided a large number of bacterial strains and reference genomes of Bifidobacterium which could be used for clinical research [[6], [7], [8], [9]].
Bifidobacterium are closely associated with human health. The phylogenomics of Bifidobacterium revealed relatively less ancestral gene loss occurrences and more gene acquisition events during the evolution development of this genus [10]. Antibiotic resistance genes are often carried by gene acquisitions [11]. The ability to degrade diet- and host-derived carbohydrate solidified the long-term colonization of Bifidobacterium in human intestine [12]. Most strains of Bifidobacterium can consume monosaccharides, disaccharides, and oligosaccharides, but there are still many variations in the metabolism of other carbohydrates among different species [13,14]. Through bile salt hydrolase (BSH), conjugated bile acids are transformed into free bile acids, which is important for lowering cholesterol and preventing cardiovascular diseases (CVD) [15]. The genus Bifidobacterium is considered one of the BSH-active bacteria that could be utilized as a combination of therapeutic probiotics for CVD in the future [16]. Gamma-aminobutyric acid (GABA) is an important inhibitory neurotransmitter with an anti-depressive function [17]. It can regulate Parkinson's disease [18], Huntington's chorea [19] and Alzheimer's disease [20]. Bifidobacterium adolescentis has been confirmed to synthesize GABA and has the potential to modulate the gut-brain axis response [21]. The Bifidobacterium longum subspecies infantis (B. longum subsp. infantis), which harbors genes encoding enzymes involved in human milk oligosaccharides (HMO) degradation, can regulate the immune system of infants in their early lives [22], and can be applied to the treatment of acutely malnourished infants [23]. Other species of Bifidobacterium have also been found to have antibacterial activities, which may prevent the endogenous Escherichia coli infection [24].
Although Bifidobacterium has been utilized in the treatment of various diseases as a probiotic for a long time [25,26], the underlying mechanisms of its probiotic effects still need to be explored. Evidence has revealed that people from different nationalities have different enterotypes of the gut microbiome, and the functions of the gut microbiome also varied [27]. The functional differences of cultivated Bifidobacterium among different nationalities also need extensive exploration. To gain a deeper understanding of Bifidobacterium in the human gut, we collected 1512 Bifidobacterium genomes from three large databases of human gut cultivated microbiota and performed an extensive analysis of their functions, infecting phages, and horizontal gene transfer (HGT) to explore the functional differences of Bifidobacterium strains among different regions. We then analyzed the distribution of these Bifidobacterium species in 12,461 metagenomes of three large cohorts from different regions.
2. Materials and methods
2.1. Collection of cultivated genomes of Bifidobacterium from human gut
We collected 451 Bifidobacterium genomes from the expanded version of Cultivated Genome Reference (CGR2) [6], 851 genomes from the Broad Institute-OpenBiome Microbiome Library (BIO-ML) [9], and 210 genomes from the Unified Human Gastrointestinal Genome (UHGG) [7] (Supplementary Table 1). The genomes of CGR2 can be accessed from the CNGB Sequence Archive (CNSA) [28] of the China National GeneBank DataBase (CNGBdb) [29] with accession number CNP0001833. The genomes of BIO-ML were accessed from https://www.ncbi.nlm.nih.gov/bioproject/PRJNA544527. The genomes of UHGG were downloaded from http://ftp.ebi.ac.uk/pub/databases/metagenomics/mgnify_genomes/human-gut/. Genome quality was evaluated using CheckM (v1.0.12) [30].
2.2. Phylogenetic, and taxonomic determination, and genomes selection
GTDB-TK (v2.1.0) [31] with the database Release 214 [31] was used to perform taxonomic annotation of each genome and construct a maximum-likelihood phylogenetic tree based on 120 conserved single-copy genes. The pairwise alignment average nucleotide identity (ANI) was calculated using fastANI (v1.32) [32]. The phylogenetic tree was visualized using iTOL (v6.5.6, https://itol.embl.de/). Multi-lineage taxonomy was only considered when calculating the proportions of species in metagenomes.
To avoid the bias of Bifidobacterium functions raised by identical genomes, we clustered according to the ANI values using R package hclust (with threshold <99.99%). In each cluster, the genomes with the largest genome sizes were retained. For the genomes of the same cluster but from different sources, the genomes with the largest genome sizes from each continent were retained. Therefore, 849/1512 genomes were retained for functional analysis.
2.3. Functional annotation of genomes
The protein-coding sequences (CDS) of each genome were predicted and annotated using Prokka v1.14.6 [33]. Preliminary functional annotation was performed using eggNOG-mapper v2 [34] (eggNOG database version 5.0.2 [35]). The results of COG, EC number, and KO annotation were extracted from the eggNOG-mapper results and counted by functional category.
The genes encoding CAZymes were predicted using dbCAN database [36] (downloaded from http://bcb.unl.edu/dbCAN2/download/Databases/V11/CAZyDB.08062022.fa). Diamond v0.8.22 [37] (blastp) was used for mapping the CDS to the database, with the identity above 60% and coverage above 50%.
Biosynthetic gene clusters (BGCs) were identified using the antiSMASH 6.0 [38]. The parameters --cb-general --cb-knownclusters --cb-subclusters were used to blast the gene sequences to known classification of stimulated metabolism, and --smcogs were used to analyze the family of secondary metabolic genes. The predicted BGCs were mapped against the MiBIG database [39] to characterize the BGCs with >70% identity as known functions.
We used BLAST (Blastn, v2.9.0) [40] to identify the genes encoding bile salt hydrolases (BSH), mapping to the BSH reference sequence provided by Déjean et al. [41], with identity >95% and coverage >75%. We used EC number 3.5.1.24 to identify the cbh gene. And we used EC number 4.1.1.15 and KO number K20265 to identify the gadB and gadC gene, respectively. The method of identifying HMO-related protein sequences were referred to Sakanaka et al. and the reference sequences were downloaded from each accession number listed by the article [42]. Occurrence of the HMO degrading genes were examined using tblastn v2.5.0+ [40], with the identity >60% and query coverage >50%, e value < 1e-5.
2.4. Annotation of ARGs and VFs
The “main” feature with default parameter of Resistance Gene Identifier (RGI) version 5.2.0 and the Comprehensive Antibiotic Resistance Database (CARD [43], version 3.1.2) was used to annotate ARGs. The VFs annotation of all CDS was performed by BLAST v2.2.26 (-evalue 0.01) against the Virulence Factor Database (VFDB [44], setB, 2021-07) with identity higher than 60% and coverage higher than 50%.
2.5. Phage sequences detection and functional annotation
To study the phages infecting Bifidobacterium, the contigs of genomes were scanned using VirSorter2 (v2.2.3) [45] to identify viral-like sequences, and sequences with score >0.7 will be selected. The sequences defined as “not-determined” by CheckV (v0.8.1) [46] were eliminated. The phage proteins were predicted by Prodigal (v2.6.3) [47], and vConTACT2 (v0.11.3) [48] was used to group the viral proteomes into viral clusters (VCs). We then used eggNOG-mapper v2 [34] (eggNOG database version 5.0.2 [35]) for functional annotation of the viral genomes. PFAMs annotation results were extracted from the eggNOG-mapper results.
2.6. Detection of horizontal gene transfer (HGT)
In this study, we focused on horizontal gene transfer occurring between Bifidobacterium and other genera. We performed the detection of HGT as described by Mathieu Groussin et al. [49]. HGTs occurring within Bifidobacterium species were not considered. Diamond v2.0.11.149 [37] (blastp) was used to detect potentially transferred genes. Protein database file “hgtdb_20211121.tar. xz” was downloaded from https://arizonastateu-my.sharepoint.com/:f:/g/personal/qzhu44_asurite_asu_edu/ErLl2qExtFhAiS1J0sCpZqgBEebKHtBilj1IDlaitOVZXg, which included all protein sequences of NCBI RefSeq [50] genomes of bacteria. BLAST hits that were larger than 500 bp, with 100% similarity, 100% coverage, and e-value < 1e-5 were selected. The hit with the highest score for each gene was retained. The COG annotations of transferred genes were extracted from result of eggNOG annotations.
2.7. Calculation of the relative abundance of 15 species in metagenomes
We downloaded 3550 gut metagenomes of a Chinese cohort (part of 4D-SZ) [51] from the CNGB Sequence Archive (CNSA) [28] of the China National GeneBank DataBase (CNGBdb) [29] with accession number CNP0000426; 8244 metagenomes from a cohort established in the Netherlands [52], retrieved from the European Genome-Phenome Archive (EGA, https://ega-archive.org/) [53] under accession number EGAS00001005027; and 667 metagenomes from HMP (Human Microbiome Project [54], https://portal.hmpdacc.org/). Fastp (v0.23.1) [55] was used to filter out low-quality reads and bases with partial parameters ‘--qualified_quality_phred 15 --complexity_threshold 30 --length_required 30’. Bowtie (v2.4.4) [56] was used to remove host contamination by mapping the reads to the human genome (GRCh38). The default parameter (--sensitive) was used, and ‘--un-conc-gz’ was used to output the sequence file after removing host contamination. Considering that there was a multi-lineage species in the taxonomic annotation, we selected 15 genomes of each species with the longest genome sequence, under the premise of highest completeness, as the representative genomes of 15 species. A representative genome was regarded as a bacterial genome reference in the Kraken2 [57] (v 2.1.1) database, and the combination of Kraken2 and Bracken [58] was used to estimate the abundance of representative genomes.
2.8. Statistical analysis
Statistical tests were performed using R v4.1.2. For principal co-ordinates analysis (PCoA), Bray-Curtis dissimilarities were calculated using the vegdist function. The Wilcoxon test was used as the significance test. The packages ggplot2 and pheatmap in R were used for plotting. Venn diagrams were plotted using Venn software (jvenn, http://jvenn.toulouse.inra.fr/app/example.html). Adobe Illustrator CC 2018 was used to adjust the colors and construct figures.
3. Results
3.1. The genome collection of cultivated Bifidobacterium in human gut
In our previous work (CGR2), we obtained 453 Bifidobacterium strains from Chinese individuals using 25 anaerobic culture conditions, including modified peptone-yeast extract-glucose (MPYG) medium and blood-brain heart infusion (BHI) medium [8], proving that Bifidobacterium can be cultured in a variety of media. Two genomes with completeness of less than 95% in CGR2 were excluded. To extensively investigate the genomic characteristics and functional diversity of cultivated Bifidobacterium in the human gut, we further downloaded the Bifidobacterium genomes from the Unified Human Gastrointestinal Genome (UHGG) [7] and the Broad Institute-OpenBiome Microbiome Library (BIO-ML) [9], the two largest databases of human gut-cultivated microbes. Three databases were used to generate an integrated catalog of Bifidobacterium genomes (1,512) of the human gut (Supplementary Table 1). Genomes with >95% completeness and <5% contamination, according to CheckM [30], were retained (Supplementary Fig. 1a).
To further examine the taxonomy of the genomes, we annotated them using the Genome Taxonomy Database [31] (GTDB, https://gtdb.ecogenomic.org/). These 1512 genomes were classified into 14 species (including subspecies) (Fig. 1a, Supplementary Table 1). In addition to genomes with unknown regions, 981 genomes were obtained from 13 countries of four continents. Most of the Bifidobacterium genomes from Asia were contributed by CGR2. Although the number of genomes of Bifidobacterium in UHGG was the lowest, it contributed the greatest diversity of Bifidobacterium species among the three databases (Fig. 1b). The average nucleotide identity (ANI) values were calculated to estimate the similarity among the 1512 genomes. To avoid the bias of Bifidobacterium functions raised by identical genomes, we removed some redundant genomes (ANI >99.99%) and retained 849 representative genomes for further genomic and functional analysis.
Fig. 1.
Genomic diversity of cultivated Bifidobacterium in human gut. a, Phylogenetic tree of 1512 cultivated genomes of Bifidobacterium, based on GTDB annotation. The innermost circle is colored according to the species. The second circle is colored according to the continents from which strains were isolated. The third circle is colored according to the databases from which genomes were obtained. The fourth circle shows the GC content of genomes and the blue line represents a percentage of 62%. The outermost circle represents the genome length, with the red dashed line showing a genome size of 1.9 Mb and the light blue solid line showing a genome size of 2.5 Mb b, The number of genomes of Bifidobacterium collected from three databases, colored according to the species. c, Scatterplot of genomes of 849 non-redundant cultivated Bifidobacterium showing genome size and GC content. The cloud around points represents a simple encirclement of points, using geom_encircle in ggplot2 package of R. d, Principal Co-ordinates Analysis (PCoA) of taxonomic annotation of 849 Bifidobacterium genomes, based on ANI values among the genomes (p = 0.001, R2 = 0.99917, calculated using Adonis test in R). The clouds around the clusters represents a 95% confidence interval. Panel b, c, and d are colored according to the species shown in panel a.
The genome sizes of Bifidobacterium ranged from 1.9 Mb to 2.8 Mb and the GC contents ranged from 54.66% to 63.24%. The genomes of the same species tended to have similar genome sizes and GC contents. B. animalis subsp. animalis and B. catenulatum subsp. catenulatum had relatively smaller genomes, whereas the genome size of B. longum subsp. infantis was larger. B. pseudolongum subsp. globosum and B. bifidum had higher GC contents, whereas B. catenulatum subsp. catenulatum, B. catenulatum subsp. kashiwanohense and B. pseudocatenulatum had lower GC contents (Fig. 1c). The genomes of the same species clustered together according to the ANI (Fig. 1d). B. longum subsp. infantis, a subspecies of B. longum, had the closest relatives to B. longum subsp. longum, but with an ANI <95%. Most of the ANI between B. animalis subsp. animalis and other species were less than 80% (Supplementary Table 2), suggesting that B. animalis subsp. animalis was extremely different from other species. Significantly higher ANI values between genomes within the same continent were observed among B. adolescentis, B. longum subsp. longum, B. pseudocatenulatum, B. catenulatum subsp. catenulatum and B. bifidum, which were not observed in B. breve and B. dentium (Supplementary Fig. 1b).
3.2. The functional variations of Bifidobacterium from human gut among species and continents
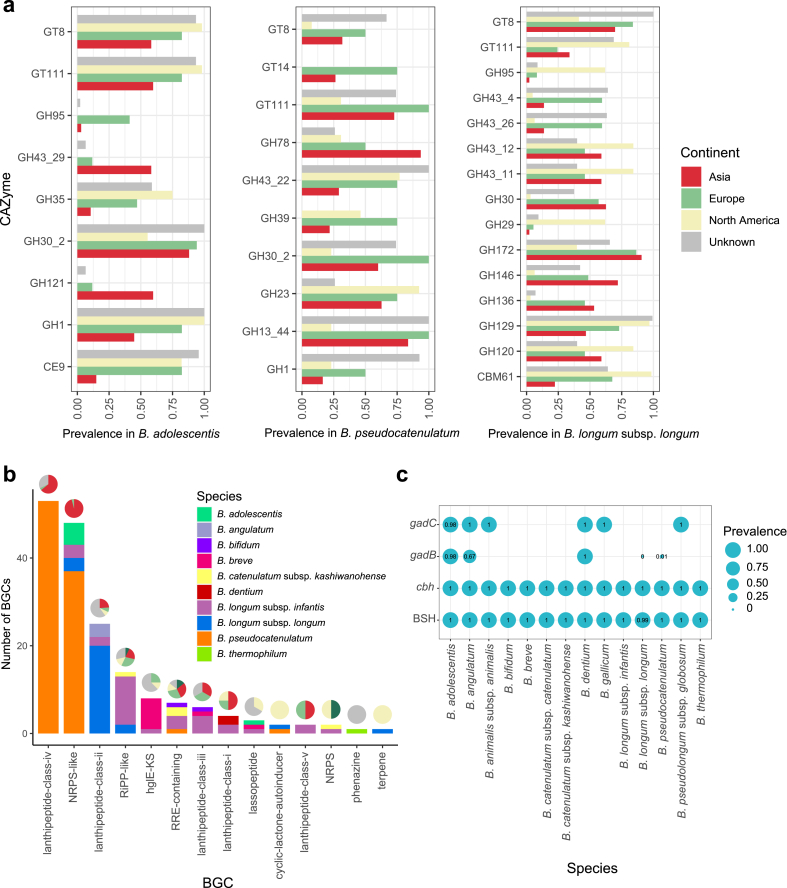
Bifidobacteria were considered able to metabolize the mono-, di- and oligo-saccharides in the gut, but they were unlikely able to metabolize most of the large, complex plant polysaccharides on their own [59]. We then extensively explored the functional diversity of 849 genomes of cultivated Bifidobacterium in the human gut. Regarding the prediction of carbohydrate-active enzyme (CAZyme) genes, genes belonging to GH13_9 (α-1,4-glucan branching enzyme), GH13_11 (amylomaltase/α-glucosidase), GH13_31 (α-transglucosidase/α-amylase), CBM48 (appended to GH13 modules), GT35 (glycogen or starch phosphorylase), GH32 (hexosyltransferases), GH36 (α-galactosidases), GH77 (amylomaltase/4-α-glucanotransferase), GT2 (mannosyltransferase), GT28 (digalactosyldiacylglycerol synthase), GT35 (α-1,4-glucan phosphorylase), GT4 (sucrose synthase) and GT51 (peptidoglycan glycosyltransferase) are widely present in the genomes of Bifidobacterium (Supplementary Fig. 2). Some CAZymes are harbored by specific Bifidobacterium species. For example, GH30_5 (endo-β-1,6-galactanase), GH43_16 (xylan β-1,4-xylosidase), CBM22 (xylan binding) and CBM6 (cellulose-binding) are mostly harbored by B. longum subsp. longum. Nearly all the genomes of B. bifidum harbor genes belonging to GH110 (α-galactosidase), GH84 (hyaluronidase) and GH89 (α-N-acetylglucosaminidase), which are barely present in other species. GH101 (Glycopeptide α-N-acetylgalactosaminidase) was present in most of the genomes of B. bifidum and B. longum subsp. longum, but barely present in genomes of other species. The distribution of CAZymes also varied among the different continents. For B. adolescentis, genes belonging to GH121 (β-L-arabinobiosidase) and GH43_29 (α-1,2-L-arabinofuranosidase) were present in over 50% of genomes from Asia but not a genome from North America. And most of the genomes from Europe and North America harbor GH1 (α-L-arabinopyranosidase) and CE9 (N-acetylglucosamine-6-phosphate deacetylase) while less than 50% of Asian genomes harbor them. For B. pseudocatenulatum, GT111 (β-1,3-galactofuranosyltransferase) and GH13_44 (α-glucosidase) are harbored by most of the genomes from Europe and Asia, while GH43_22 (α-1,3-L-arabinofuranosidase) is harbored by about 75% of the genomes from Europe and North America. For B. longum subsp. longum, GH95 and GH29 (both α-L-fucosidase) are mostly found in genomes from North America, whereas GH43_4 and GH43_26 (both exo-α-1,5-L-arabinofuranosidase) are mostly present in genomes from Europe and unknown region. GH30 (β-1,6-glucosidase), GH146 (β-L-arabinofuranosidase) and GH136 (lacto-N-biosidase) could be found in less genomes from North America (Fig. 2a). We surmised that the differences in CAZymes harbored by Bifidobacterium might result from regional dietary differences (Supplementary Table 3a).
Fig. 2.
Functional profile of cultivated Bifidobacterium (for 849 non-redundant genomes). a, The prevalence of some CAZyme families in B. adolescentis, B. pseudocatenulatum, and B. longum subsp. longum, colored according to the continent. The CAZymes with obvious differences among different continents in each species are shown. The prevalence represents the number of genomes harboring corresponding CAZyme relative to the total number of corresponding species in this continent. b, The distribution of BGCs in different species. The pie charts show the distribution of continents from which the genomes were obtained. The color of continents is shown in panel a. c, The distribution of genes related to GABA producing and bile salt hydrolysis harbored by each species. The sizes of bubbles and the number shown in the bubbles represent the prevalence of these genes in each species.
An exploration of biosynthetic gene clusters (BGCs) was performed to predict the secondary metabolites produced by Bifidobacterium. Forty-nine non-redundant BGCs were annotated in the 131 genomes (Fig. 2b, Supplementary Fig. 3). Lanthipeptide-class-iv was the most abundant type of BGCs, followed by non-ribosomal peptide synthetase-like (NRPS-like), both of which are mostly produced by B. pseudocatenulatum. Lanthipeptide-class-iv was present in the genomes from Asia and Europe but absent in the genomes from North America. NRPS-like BGCs were mostly present in Asian genomes. This suggests that the production of secondary metabolites also varies among continents. Among these BGCs, B. longum subsp. infantis may have the ability to synthesize the most diverse products, which was consistent with previous research [60]. More than 36% of B. pseudocatenulatum were predicted to harbor gene clusters involved in synthesizing class IV lanthipeptide. The BGCs of different classes of lanthipeptide are harbored by different Bifidobacterium species. Lanthipeptides, also termed lantibiotics, are resistant to pathogens such as Staphylococcus aureus, and have low toxicity to mammals [61]. Owing to the overuse of antibiotics, lantibiotics produced by microbes are much safer and have the potential to replace antibiotics as a new type of antibacterial drug in the future [62]. The lanthipeptides are classified into different classes because of their distinct mechanisms of the dehydration reactions [63], and most of current research has focused on class I and class II lanthipeptides [64,65]. Class IV lanthipeptides are also called as venezuelins, with unclear functions as present [66]. And our study showed that B. pseudocatenulatum has the potential to produce class IV lanthipeptides, which might contribute to clinical advances (Supplementary Table 3b).
We then identified the genes related to BSH and GABA production of cultivated Bifidobacterium in the human gut (Fig. 2c, Supplementary Table 4a). A total of 847 of 849 genomes of Bifidobacterium in the human gut harbored genes encoding BSH, and 848 genomes harbored the cbh gene. Only one genome in our study did not harbor genes encoding any bile salt hydrolases, indicating that most of Bifidobacterium strains in this collection have the potential to be used for the treatment and prevention of CVD. Glutamate decarboxylase (GAD) is the sole enzyme for GABA synthesis [67], which is encoded by the genes gadB and gadC. In our study, 183 of the 186 B. adolescentis genomes harbored these two genes. In addition to B. adolescentis, all genomes of B. dentium also harbored these two genes, which indicated that they might also have the potential to synthesize GABA. In addition, all genomes of B. angulatum, B. animalis subsp. animalis, B. gallicum and B. pseudolongum subsp. globosum harbored only the gene gadC.
B. longum subsp. infantis is a well-known major member of HMO utilization [68]. Here, we analyzed the presence of proteins related to HMO metabolism using tblastn v2.5.0+ [40]. We investigated the distribution of 26 representative amino acid sequences listed by Sakanaka et al. [42]. Genes encoding enzymes involved in extracellular HMO digestion were mostly harbored by B. bifidum. The genes encoding AfcA, AfcB, BbgIII, BbhI, and Siabb2 were conserved in B.bifidum strains, and LnbB was also harbored by most of the B. bifidum and B. breve strains. Strains of B. bifidum, B. breve, B. longum subsp. infantis, and B. longum subsp. longum harbored enzymes involved in uptake of LNB and intracellular LNB digestion. B. longum subsp. infantis harbored the most genes encoding enzymes related to uptake of intact HMOs and intracellular HMO digestion. Genes encoding fucosyllactose transporter solute-binding proteins (FL transporter SBP), lacto-N-tetraose (LNT) beta-1,3-Galactosidase, beta-N-Acetylglucosaminidase, 1,2-alpha-l-Fucosidase, 1,3/4-alpha-l-Fucosidase, and 2,3/6-alpha-Sialidase were harbored by all the B. longum subsp. infantis strains. These results were consistent with the previous research [42]. In addition, the prevalence of HMO-related enzymes might vary among continents. The prevalence of B. bifidum harboring LnbP and LacZ6 from Europe were lower than that from other continents. Blon_2335, Blon_2336 and Blon_2202 were harbored by many B. longum subsp. longum strains of North America while LnbX and LnbY were harbored by the strains of other continents. And the prevalence of B. pseudocatenulatum harboring Blon_0459 or NahA were lower in the strains from North America than that from other continents. (Supplementary Fig. 4, Supplementary Table 4b).
3.3. The safety evaluation of cultivated genomes of Bifidobacterium in human gut
In recent years, a comprehensive understanding of antibiotic resistance is urgently needed to prevent the antibiotic-resistant genes spreading [69]. We identified 57 types of antibiotic resistance genes (ARGs) in 847 genomes with resistance to 24 types of antibiotics. 24 of the 57 types of ARGs were multidrug resistant (Supplementary Fig. 5a, Supplementary Table 5a). Many antibiotics are important antimicrobials listed by the World Health Organization (WHO) [70]. Most of the genomes harbored rifamycin-resistance genes. In addition, almost all the genomes of B. adolescentis, B. catenulatum subsp. catenulatum, B. dentium and B. pseudocatenulatum harbored genes resistant to tetracycline, macrolide, aminocoumarin, and monobactam (Fig. 3a). Among the cultivated Bifidobacterium in the human gut, the genomes of B. catenulatum subsp. kashiwanohense and B. catenulatum subsp. catenulatum harbored the most ARGs. Of note, we found a genome of B. adolescentis (GCA_009094615.1_ASM909461v1_genomic) harboring 37 ARGs (Fig. 3b), which might be threatening and unsuitable for application in clinical treatment. For B. longum subsp. longum, the number of North American genomes with genes resistant to penam, cephalosporin, and cephamycin was higher than that of Asian genomes, while genes resistant to lincosamide, streptogramin, tetracycline, and macrolides were mostly harbored by Asian genomes. This indicated that resistance to antibiotics could differ among the continents of the genomes, even from the same species. Significant differences in the number of ARGs in each genome were observed among Europe, Asia, and North America (Fig. 3c, Supplementary Fig. 5b). Evidence has revealed variations in the consumption rates of antibiotic classes among different regions [71]. This suggests that the composition of ARGs in Bifidobacterium might be affected by the use of different antibiotics on different continents. Therefore, it is essential to use antibiotics distinctly and properly according to the resistance of Bifidobacterium genomes from different continents if they are used as probiotics.
Fig. 3.
Annotation of ARGs in the genomes of Bifidobacterium (for 849 non-redundant genomes). a, The distribution of annotated ARGs in Bifidobacterium. The genomes are annotated by their species and continents and the antibiotics are colored according to the importance of drug usage. b, The boxplot of number of ARGs harbored in genomes of different species, colored by their species. Each point represents a genome with annotated ARGs. c, The boxplot of number of ARGs harbored in genomes of different continents, colored by corresponding continent. Only the significant p-values are shown in the figure, calculated by paired wilcoxon test in R.
The virulence of bacteria is another aspect of safety evaluation. To deeply and comprehensively confirm the potential virulence factors carried by Bifidobacterium, we analyzed the virulence factors in the 849 Bifidobacterium genomes. We identified 11 types of virulence factors (VF) in 36 genomes of cultivated Bifidobacterium, annotated with the Virulence Factor Database [72] (VFDB, http://www.mgc.ac.cn/VFs/) (Supplementary Fig. 6, Supplementary Table 5b). Ten out of the eleven VFs were related to evading the host immune system [73], bacterial adhesion, and survival [74], which may not be that threatening to the host. Hyaluronidase, which was harbored by only one genome (GCA_009883615.1_ASM988361v1_genomic), is a pathogenic bacterial spreading factor, cleaves hyaluronan and may also pave the way for other bacterial toxins [75]. This indicated that most of the Bifidobacterium genomes in this study, except those harboring virulence factors, are safe for clinical use.
3.4. Identifying the phages infecting Bifidobacterium
The coevolution of bacteria and phages influences the survival of microbes and evolution of microbial communities [76] and the apparently extensive interactions between Bifidobacterium and phages were identified [77]. Phages are closely associated with human health and have both beneficial and harmful sides [78]. We found 545 non-singleton phages genomes in 361 Bifidobacterium, divided into 38 viral clusters (VCs) (Supplementary Table 6). Among the 38 VCs, 8 could infect all 9 species of this collection, and 34 could infect B. longum subsp. longum. B. longum subsp. longum contributed six unique VCs and B. catenulatum subsp. catenulatum contributed one unique VC (Fig. 4a). This indicated that the VCs might be similar among Bifidobacterium in the human gut. A total of 1275 kinds of protein families (PFAMs) were annotated from the phage genomes, and 461 kinds of PFAMs were shared among phages infecting 9 Bifidobacterium species. The phages infecting B. longum subsp. longum contributed 146 types of unique PFAMs, whereas phages infecting B. adolescentis contributed 61 unique PFAMs (Fig. 4b). GrpE, which participates in the response to hyperosmotic stress and heat shock in association with DnaK, was only found in phages infecting B. longum subsp. longum. PemK_toxin of the type II toxin-antitoxin system was unique in B. longum subsp. longum. ParE_toxin, which is toxic to DNA gyrase, is unique to B. adolescentis. This suggests that in these species, phages might have the potential to help the bacteria cope with toxins or compete with other species, promoting the survival of the bacteria.
Fig. 4.
Taxonomic and functional annotation of phages infecting Bifidobacterium (for 849 non-redundant genomes). a, Venn diagram and histogram showing distribution of VCs infecting Bifidobacterium of different species, colored according to their species. b, Venn diagram and histogram showing distribution of PFAMs of phages, colored according to their species. c-e, Venn diagram showing the distribution of PFAMs encoded by phages infecting B. longum subsp. longum (c), B. pseudocatenulatum (d), and B. adolescentis (e) from different continents, colored according to the continents.
PFAMs of phages infecting Bifidobacterium in the human gut differ among continents. A total of 382 PFAMs were shared among phages infecting B. longum subsp. longum of Asia, Europe, and North America (Fig. 4c). PFAMs of GH129 (α-N-acetylgalactosaminidase) and GH10 (endo-1,4-β-xylanase) were found only in phages infecting B. longum subsp. longum in Asia, suggesting that dietary differences across different continents might influence the functions of phages infecting Bifidobacterium in the human gut. For phages infecting B. pseudocatenulatum, 202 unique kinds of PFAMs were contributed by the genomes from Asia, and 87 types of PFAMs were shared in genomes from the three continents (Fig. 4d). For phages infecting B. adolescentis, genomes from North America contributed 125 unique types of PFAMs, while genomes from Asia contributed 103 unique types of PFAMs. A total of 312 PFAMs types were shared by the genomes from Asia, Europe, and North America (Fig. 4e). This suggests that the PFAMs of phages infecting these three species vary among continents.
3.5. The occurrence of horizontal gene transfer in Bifidobacterium
Horizontal gene transfer (HGT) [79] is widely recognized as an important mechanism for bacterial adaptation [80]. Here, we searched for Bifidobacterium genes that might be transferred from other microbial taxa. In total, 2.45% of the pangenomes of the 849 Bifidobacterium strains were potentially transferred (Supplementary Table 7). Most of the genes were mapped to Terrabacteria and Bacteria. In addition to Actinobacteria, HGTs were also frequently found between Bifidobacterium and Firmicutes. Enterococcus was the genus that was the source of most of the HGT genes detected in Bifidobacterium, followed by Escherichia, Bacteroides and Streptomyces (Fig. 5a). These genera are common in the human gut, reflecting the potentially frequent gene exchange among them in the human gut. Regarding the functions annotated with the Clusters of Orthologous Genes (COG) database, most of the potentially transferred genes were related to information storage and processing (Fig. 5b), even though the genes with metabolic functions accounted for a large proportion of the Bifidobacterium genomes.
Fig. 5.

HGTs occurring in Bifidobacterium genomes (for 849 non-redundant genomes). a, The number of detected HGT genes classified based on the taxa supposed to be the source of the transfer. b, The number of genes derived from HGT events according to the COG annotation, colored by the COG types. c, The percentage of genes derived from HGT events in each genome, classified and colored according to the species. d, The percentage of genes derived from HGT events in each genome of B. adolescentis, B. bifidum, and B. longum subsp. longum, and B. pseudocatenulatum, classified and colored according to continents. P-values are shown in the figure, calculated by paired wilcoxon test in R.
B. longum subsp. longum had the highest proportion of genes acquired by HGT events, with a median of 3.60%, indicating that more gene exchanges with other genera occurred in B. longum subsp. longum than in other Bifidobacterium species (Fig. 5c). In addition, significant differences of the percentage of HGT occurrence in each genome were observed among genomes from different continents in specific species. Among B. adolescentis, HGTs that occurred in the genomes from North America and Asia were significantly higher than the genomes from Europe among B. adolescentis and B. bifidum. And among B. longum subsp. longum, HGTs that occurred in the genomes from North America were significantly higher than those of Europe and Asia (Fig. 5d). This suggests that the occurrence and frequency of HGT events could vary among different region sources, even for the same species, which might result from the difference in life habits and composition of the gut microbiome among different continents. But this difference could not be observed among B. pseudocatenulatum.
Notably, some potentially transferred genes are related to virulence and resistance. For example, mprF gene acquired from Enterococcus species was found harbored by B. longum subsp. longum. This gene is involved in lysylphosphatidylglycerol (L-PG) production and contributes to bacterial virulence [81], which can help microbes escape from the host immune system. In addition, genes encoding proteins related to the (helix_turn_helix) arsenic (As) resistance operon repressor and (helix_turn_helix) mercury (Hg) resistance were found in the genomes of B. pseudocatenulatum and B. longum subsp. longum respectively, both obtained from North America. These genes are potentially acquired from Firmicutes and Lactobacillaceae. Evidence has shown that both mercury contamination [82] and arsenic contamination [83] exist in North America. This implies that HGT might help Bifidobacterium from North America cope with Hg and As contamination and adapt to the environment.
3.6. Distribution of cultivated genomes of Bifidobacterium from different populations
To explore the distribution of the cultivated Bifidobacterium in the human gut from different regions, we mapped 15 representative genomes of each species (including multi-lineage species B. catenulatum subsp. kashiwanohense_A) collected in this work to 3550 metagenomes from China [51], 8244 metagenomes from the Netherlands [52] and 667 metagenomes from HMP (Human Microbiome Project [54], https://portal.hmpdacc.org/). After normalization, we obtained the proportion of each species in three population cohorts. B. adolescentis and B. longum subsp. longum occupied the most proportion of Bifidobacterium in the Dutch individuals’ gut, accounting for 21.91% and 17.92% respectively (Fig. 6). B. longum subsp. longum and B. pseudocatenulatum occupied the most proportion of Bifidobacterium in the gut of whom participating in Chinese cohort (10.16% and 18.62% respectively) or HMP cohort (12.52% and 12.46% respectively). Significant differences existed in the proportion of each species in three cohorts, indicating that the distribution of Bifidobacterium in gut of people in different area might vary a lot. Given that different Bifidobacterium species could metabolize different kinds of substances, this phenomenon might be caused by the dietary differences among different continents.
Fig. 6.
Distribution of cultivated Bifidobacterium in three different cohorts. Boxplot represents the proportion of each species in the abundance of these 15 Bifidobacterium species in a sample. Each point represents a sample, colored according to the cohorts. Significant differences between the proportion of the same species of different cohorts are marked on the right of the boxplot, calculated by paired wilcoxon test in R (*** represents P < 0.001).
4. Discussion and conclusion
Bifidobacteria, which are common microorganisms in the human gut, are one of the most used probiotics [84]. The application and efficacy of probiotics in the prevention and treatment of intestinal diseases has received great attention. However, even though the same probiotic species are used, different disease types may contribute to the inconsistency of the results [85,86]. Therefore, a deeper understanding of the distribution and function of Bifidobacterium is needed to ensure that these probiotic bacteria can be used properly.
Here, we collected 1512 reference genomes of Bifidobacterium from three large-scale culturable gut microbiota databases and selected 849 non-redundant genomes for further analysis of bacterial functions, infecting phages, and horizontal gene transfer. CAZymes varied among the genomes of different species and continents. Most genomes of B. adolescentis harboring genes encoding α-L-fucosidase (GH95) were from Europe, and most genomes of B. longum subsp. longum harboring GH95 were from North America. GH95, which participates in α-L-fucoside degradation and HMO degradation [87], was also found in most genomes of B. bifidum and B. breve. Notably, all genomes of B. longum subsp. infantis, which is known as an HMO utilizer, also harbored genes encoding GH95. We identified BGCs in Bifidobacterium and found that many B. pseudocatenulatum strains had the potential to synthesize class IV lanthipeptide, which might be a candidate for antibacterial drugs.
Since Bifidobacterium has been widely considered a probiotic, we conducted an in-depth study on probiotic functions such as bile salt hydrolysis, GABA synthesis, and HMO degradation. Almost all genomes of Bifidobacterium in this study harbored genes encoding BSH, indicating that bile salt hydrolysis might be a conserved function in Bifidobacterium. Recently, B. adolescentis was confirmed to synthesize GABA in mouse [21]. However, whether it can be used for the prevention and treatment of neurological diseases in humans requires verification in vivo and in vitro. In addition to B. adolescentis, we found another species (B. dentium) also harbored gadB and gadC, which might provide other choices for probiotic application in the clinical treatment of neurological diseases. Both as a subspecies of B. longum, B. longum subsp. infantis harbors unique and more complete sets of genes for HMO utilization that are not all present in other genomes of B. longum subsp. longum, indicating that B. longum subsp. infantis is more suitable for application in the prevention and treatment of related diseases such as malnutrition [23].
When using Bifidobacterium strains as probiotics, it is necessary to decide which antibiotics could be used according to the species and region source of that strain. In addition, we found a strain of B. adolescentis harboring 37 ARGs and a strain of B. longum subsp. longum harboring the virulent factor hyaluronidase, which might cause more ARGs transfer and harmful effects if used as probiotics.
The phages of 38 VCs were found to infect Bifidobacterium, and their functions varied among different species and continents. This revealed that phages might play an important role in helping probiotic Bifidobacterium compete with other species and adapt to the environment. In our study, genes related to information storage and processing were transferred more frequently between Bifidobacterium and other genera. We also found that Bifidobacterium from North America could cope with local Hg and As contamination and adapt to the environment through HGT. The average proportion of representative genomes of Bifidobacterium in human gut varied among different cohorts, which might be raised by the dietary differences between different countries.
However, there are limitations of our work. Our collection which included three large databases of human gut microbiome covered most of the representatives of Bifidobacterium genomes in the human gut, but some genomes of Bifidobacterium might not be included in our analysis. Even though metagenome-assembled genomes (MAGs) can hardly be used in the experiment, they may represent novel species which can be difficult to isolate. It is worthwhile that we collect more isolated Bifidobacterium genomes and MAGs on a large scale to explore the diversity of Bifidobacterium from different species and countries. In addition, the genetic diversity and different niches adaption of Bifidobacterium isolated from animals, environments, and fermented food remains to be explored. In conclusion, we envisage that our work will serve as a useful summary of the characteristics and functions of genomes of cultivated Bifidobacterium in the human gut and a reference for the clinical application of Bifidobacterium in the future.
Data availability statement
The data that support the findings of this study can be accessed in the Zenodo database, by using the https://doi.org/10.5281/zenodo.7874214.
Ethics declarations
This study was reviewed and approved by the Institutional Review Board on Bioethics and Biosafety of BGI, with the approval number: BGI-IRB 20106-T2. Informed consent was not required for this study because the microbial genomic data in this study were all public.
CRediT authorship contribution statement
Wenxi Li: Writing – original draft, Visualization, Methodology, Investigation, Formal analysis, Conceptualization. Hewei Liang: Methodology, Investigation, Formal analysis. Wenxin He: Visualization, Formal analysis. Xiaowei Gao: Writing – review & editing. Zhinan Wu: Methodology, Formal analysis. Tongyuan Hu: Methodology, Formal analysis. Xiaoqian Lin: Methodology, Formal analysis. Mengmeng Wang: Visualization. Yiyi Zhong: Resources. Haifeng Zhang: Resources. Lan Ge: Resources. Xin Jin: Supervision. Liang Xiao: Project administration. Yuanqiang Zou: Writing – review & editing, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by a grant from the National Natural Science Foundation of China (No. 32100009). This study was supported by Henan Supercomputer Center.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e27270.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Favier C.F., et al. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 2002;68(1):219–226. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arboleya S., et al. Gut bifidobacteria populations in human health and aging. Front. Microbiol. 2016;7:1204. doi: 10.3389/fmicb.2016.01204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishijima S., et al. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016;23(2):125–133. doi: 10.1093/dnares/dsw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta V.K., et al. A predictive index for health status using species-level gut microbiome profiling. Nat. Commun. 2020;11(1):4635. doi: 10.1038/s41467-020-18476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drago L., et al. Cultivable and pyrosequenced fecal microflora in centenarians and young subjects. J. Clin. Gastroenterol. 2012;46(Suppl):S81–S84. doi: 10.1097/MCG.0b013e3182693982. [DOI] [PubMed] [Google Scholar]
- 6.Lin X., et al. The genomic landscape of reference genomes of cultivated human gut bacteria. Nat. Commun. 2023;14(1):1663. doi: 10.1038/s41467-023-37396-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Almeida A., et al. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 2021;39(1):105–114. doi: 10.1038/s41587-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou Y., et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat. Biotechnol. 2019;37(2):179–185. doi: 10.1038/s41587-018-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poyet M., et al. A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat. Med. 2019;25(9):1442–1452. doi: 10.1038/s41591-019-0559-3. [DOI] [PubMed] [Google Scholar]
- 10.Milani C., et al. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the Glycan-Rich gut environment. Appl. Environ. Microbiol. 2016;82(4):980–991. doi: 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mancino W., et al. Mobilome and resistome reconstruction from genomes belonging to members of the Bifidobacterium genus. Microorganisms. 2019;7(12) doi: 10.3390/microorganisms7120638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bottacini F., van Sinderen D., Ventura M. Omics of bifidobacteria: research and insights into their health-promoting activities. Biochem. J. 2017;474(24):4137–4152. doi: 10.1042/BCJ20160756. [DOI] [PubMed] [Google Scholar]
- 13.Derrien M., et al. Insights into endogenous Bifidobacterium species in the human gut microbiota during adulthood. Trends Microbiol. 2022;30(10):940–947. doi: 10.1016/j.tim.2022.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Ryan S.M., Fitzgerald G.F., van Sinderen D. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl. Environ. Microbiol. 2006;72(8):5289–5296. doi: 10.1128/AEM.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ridlon J.M., et al. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microb. 2016;7(1):22–39. doi: 10.1080/19490976.2015.1127483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones M.L., et al. Cholesterol lowering with bile salt hydrolase-active probiotic bacteria, mechanism of action, clinical evidence, and future direction for heart health applications. Expet Opin. Biol. Ther. 2013;13(5):631–642. doi: 10.1517/14712598.2013.758706. [DOI] [PubMed] [Google Scholar]
- 17.Sarasa S.B., et al. A brief review on the non-protein amino acid, gamma-amino Butyric acid (GABA): its production and role in microbes. Curr. Microbiol. 2020;77(4):534–544. doi: 10.1007/s00284-019-01839-w. [DOI] [PubMed] [Google Scholar]
- 18.Muñoz M.D., de la Fuente N., Sánchez-Capelo A. TGF-β/Smad3 signalling modulates GABA Neurotransmission: implications in Parkinson's disease. Int. J. Mol. Sci. 2020;21(2) doi: 10.3390/ijms21020590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manyam B.V., et al. Isoniazid-induced elevation of CSF GABA levels and effects on chorea in Huntington's disease. Ann. Neurol. 1981;10(1):35–37. doi: 10.1002/ana.410100107. [DOI] [PubMed] [Google Scholar]
- 20.Czapski G.A., Strosznajder J.B. Glutamate and GABA in Microglia-Neuron Cross-Talk in Alzheimer's disease. Int. J. Mol. Sci. 2021;22(21) doi: 10.3390/ijms222111677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duranti S., et al. Bifidobacterium adolescentis as a key member of the human gut microbiota in the production of GABA. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-70986-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henrick B.M., et al. Bifidobacteria-mediated immune system imprinting early in life. Cell. 2021;184(15):3884–3898.e11. doi: 10.1016/j.cell.2021.05.030. [DOI] [PubMed] [Google Scholar]
- 23.Barratt M.J., et al. Bifidobacterium infantis treatment promotes weight gain in Bangladeshi infants with severe acute malnutrition. Sci. Transl. Med. 2022;14(640):eabk1107. doi: 10.1126/scitranslmed.abk1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asahara T., et al. Antibacterial effect of fermented milk containing Bifidobacterium breve. Bifidobacterium bifidum and Lactobacillus acidophilus against indigenous Escherichia coli infection in mice. 2001;13(1):16–24. [Google Scholar]
- 25.Ma X., et al. Bifidobacterium infantis strain YLGB-1496 possesses excellent antioxidant and skin barrier-enhancing efficacy in vitro. Exp. Dermatol. 2022;31(7):1089–1094. doi: 10.1111/exd.14583. [DOI] [PubMed] [Google Scholar]
- 26.Kajander K., et al. Clinical trial: multispecies probiotic supplementation alleviates the symptoms of irritable bowel syndrome and stabilizes intestinal microbiota. Aliment. Pharmacol. Ther. 2008;27(1):48–57. doi: 10.1111/j.1365-2036.2007.03542.x. [DOI] [PubMed] [Google Scholar]
- 27.Arumugam M., et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo X., et al. Database; oxford: 2020. CNSA: a Data Repository for Archiving Omics Data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen F.Z., et al. CNGBdb: China national GeneBank DataBase. Yi Chuan. 2020;42(8):799–809. doi: 10.16288/j.yczz.20-080. [DOI] [PubMed] [Google Scholar]
- 30.Parks D.H., et al. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parks D.H., et al. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022;50(D1):D785–d794. doi: 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain C., et al. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018;9(1):5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 34.Cantalapiedra C.P., et al. eggNOG-mapper v2: functional annotation, orthology Assignments, and Domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021;38(12):5825–5829. doi: 10.1093/molbev/msab293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huerta-Cepas J., et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47(D1):D309–d314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng J., et al. dbCAN-seq update: CAZyme gene clusters and substrates in microbiomes. Nucleic Acids Res. 2022;51(D1):D557–D563. doi: 10.1093/nar/gkac1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buchfink B., Reuter K., Drost H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods. 2021;18(4):366–368. doi: 10.1038/s41592-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blin K., et al. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021;49(W1):W29–w35. doi: 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kautsar S.A., et al. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2020;48(D1):D454–d458. doi: 10.1093/nar/gkz882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camacho C., et al. vol. 10. 2009. pp. 1–9. (BLAST+: Architecture and Applications). [Google Scholar]
- 41.Déjean G., et al. Identifying a novel bile salt hydrolase from the Keystone gut Bacterium Christensenella minuta. Microorganisms. 2021;9(6) doi: 10.3390/microorganisms9061252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakanaka M., et al. Varied pathways of infant gut-associated Bifidobacterium to assimilate human milk oligosaccharides: prevalence of the gene set and its correlation with bifidobacteria-rich microbiota formation. Nutrients. 2019;12(1) doi: 10.3390/nu12010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alcock B.P., et al. Card 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1):D517–d525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen L., et al. Vfdb 2016: hierarchical and refined dataset for big data analysis--10 years on. Nucleic Acids Res. 2016;44(D1):D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo J., et al. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome. 2021;9(1):37. doi: 10.1186/s40168-020-00990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nayfach S., et al. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 2021;39(5):578–585. doi: 10.1038/s41587-020-00774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hyatt D., et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bin Jang H., et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 2019;37(6):632–639. doi: 10.1038/s41587-019-0100-8. [DOI] [PubMed] [Google Scholar]
- 49.Groussin M., et al. Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell. 2021;184(8):2053–2067.e18. doi: 10.1016/j.cell.2021.02.052. [DOI] [PubMed] [Google Scholar]
- 50.O'Leary N.A., et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jie Z., et al. A transomic cohort as a reference point for promoting a healthy human gut microbiome. Medicine in Microecology. 2021;8 [Google Scholar]
- 52.Gacesa R., et al. Environmental factors shaping the gut microbiome in a Dutch population. Nature. 2022;604(7907):732–739. doi: 10.1038/s41586-022-04567-7. [DOI] [PubMed] [Google Scholar]
- 53.Freeberg M.A., et al. The European genome-phenome archive in 2021. Nucleic Acids Res. 2022;50(D1):D980–d987. doi: 10.1093/nar/gkab1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Consortium I.H.i.R.N. The integrative human microbiome Project. Nature. 2019;569(7758):641–648. doi: 10.1038/s41586-019-1238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen S., et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu J., et al. vol. 3. 2017. p. e104. (Bracken: Estimating Species Abundance in Metagenomics Data). [Google Scholar]
- 59.Kelly S.M., Munoz-Munoz J., van Sinderen D. Plant glycan metabolism by bifidobacteria. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.609418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu D., et al. Bifidobacterium longum subsp. infantis as widespread bacteriocin gene clusters carrier stands out among the Bifidobacterium. Appl. Environ. Microbiol. 2023;89(9) doi: 10.1128/aem.00979-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoganathan S., Vederas J.C. Fracturing rings to understand lantibiotics. Chem. Biol. 2008;15(10):999–1001. doi: 10.1016/j.chembiol.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 62.Dischinger J., Basi Chipalu S., Bierbaum G. Lantibiotics: promising candidates for future applications in health care. Int J Med Microbiol. 2014;304(1):51–62. doi: 10.1016/j.ijmm.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 63.Repka L.M., et al. Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem. Rev. 2017;117(8):5457–5520. doi: 10.1021/acs.chemrev.6b00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barbosa J., Caetano T., Mendo S. Class I and class II lanthipeptides produced by Bacillus spp. J. Nat. Prod. 2015;78(11):2850–2866. doi: 10.1021/np500424y. [DOI] [PubMed] [Google Scholar]
- 65.Lee J.H., Li X., O'Sullivan D.J. Transcription analysis of a lantibiotic gene cluster from Bifidobacterium longum DJO10A. Appl. Environ. Microbiol. 2011;77(17):5879–5887. doi: 10.1128/AEM.00571-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Q., et al. Expanded natural product diversity revealed by analysis of lanthipeptide-like gene clusters in actinobacteria. Appl. Environ. Microbiol. 2015;81(13):4339–4350. doi: 10.1128/AEM.00635-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ueno H. Enzymatic and structural aspects on glutamate decarboxylase. J. Mol. Catal. B Enzym. 2000;10(1):67–79. [Google Scholar]
- 68.Sela D.A., et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U. S. A. 2008;105(48):18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bhardwaj S., et al. Antibiotics and antibiotic resistance- flipsides of the same coin. Curr. Pharmaceut. Des. 2022;28(28):2312–2329. doi: 10.2174/1381612828666220608120238. [DOI] [PubMed] [Google Scholar]
- 70.Organization W.H. World Health Organization; 2018. Critically Important Antimicrobials for Human Medicine. 6th revision. [Google Scholar]
- 71.Browne A.J., et al. Global antibiotic consumption and usage in humans, 2000-18: a spatial modelling study. Lancet Planet. Health. 2021;5(12):e893–e904. doi: 10.1016/S2542-5196(21)00280-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu B., et al. Vfdb 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022;50(D1):D912–d917. doi: 10.1093/nar/gkab1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cuccui J., et al. Characterization of the Burkholderia pseudomallei K96243 capsular polysaccharide I coding region. Infect. Immun. 2012;80(3):1209–1221. doi: 10.1128/IAI.05805-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rouquette C., et al. Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol. Microbiol. 1996;21(5):977–987. doi: 10.1046/j.1365-2958.1996.641432.x. [DOI] [PubMed] [Google Scholar]
- 75.Kayaoglu G., Ørstavik D. Virulence factors of Enterococcus faecalis: relationship to endodontic disease. Crit. Rev. Oral Biol. Med. 2004;15(5):308–320. doi: 10.1177/154411130401500506. [DOI] [PubMed] [Google Scholar]
- 76.Koskella B., Brockhurst M.A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 2014;38(5):916–931. doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ventura M., et al. Comparative analyses of prophage-like elements present in bifidobacterial genomes. Appl. Environ. Microbiol. 2009;75(21):6929–6936. doi: 10.1128/AEM.01112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wahida A., Ritter K., Horz H.P. The Janus-Face of Bacteriophages across human Body Habitats. PLoS Pathog. 2016;12(6) doi: 10.1371/journal.ppat.1005634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ochman H., Lawrence J.G., Groisman E.A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 80.Soucy S.M., Huang J., Gogarten J.P. Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 2015;16(8):472–482. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 81.Staubitz P., et al. MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol. Lett. 2004;231(1):67–71. doi: 10.1016/S0378-1097(03)00921-2. [DOI] [PubMed] [Google Scholar]
- 82.Eagles-Smith C.A., et al. Mercury in western North America: a synthesis of environmental contamination, fluxes, bioaccumulation, and risk to fish and wildlife. Sci. Total Environ. 2016;568:1213–1226. doi: 10.1016/j.scitotenv.2016.05.094. [DOI] [PubMed] [Google Scholar]
- 83.McClintock T.R., et al. Arsenic exposure in Latin America: biomarkers, risk assessments and related health effects. Sci. Total Environ. 2012;429:76–91. doi: 10.1016/j.scitotenv.2011.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rivière A., et al. Bifidobacteria and Butyrate-producing Colon bacteria: importance and Strategies for their stimulation in the human gut. Front. Microbiol. 2016;7:979. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bjarnason I., Sission G., Hayee B. A randomised, double-blind, placebo-controlled trial of a multi-strain probiotic in patients with asymptomatic ulcerative colitis and Crohn's disease. Inflammopharmacology. 2019;27(3):465–473. doi: 10.1007/s10787-019-00595-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ritchie M.L., Romanuk T.N. A meta-analysis of probiotic efficacy for gastrointestinal diseases. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0034938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ashida H., et al. Two distinct alpha-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19(9):1010–1017. doi: 10.1093/glycob/cwp082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study can be accessed in the Zenodo database, by using the https://doi.org/10.5281/zenodo.7874214.