Abstract
Mismatch repair (MMR) proteins actively inhibit recombination between diverged sequences in both prokaryotes and eukaryotes. Although the molecular basis of the antirecombination activity exerted by MMR proteins is unclear, it presumably involves the recognition of mismatches present in heteroduplex recombination intermediates. This recognition could be exerted during the initial stage of strand exchange, during the extension of heteroduplex DNA, or during the resolution of recombination intermediates. We previously used an assay system based on 350-bp inverted-repeat substrates to demonstrate that MMR proteins strongly inhibit mitotic recombination between diverged sequences in Saccharomyces cerevisiae. The assay system detects only those events that reverse the orientation of the region between the recombination substrates, which can occur as a result of either intrachromatid crossover or sister chromatid conversion. In the present study we sequenced the products of mitotic recombination between 94%-identical substrates in order to map gene conversion tracts in wild-type versus MMR-defective yeast strains. The sequence data indicate that (i) most recombination occurs via sister chromatid conversion and (ii) gene conversion tracts in an MMR-defective strain are significantly longer than those in an isogenic wild-type strain. The shortening of conversion tracts observed in a wild-type strain relative to an MMR-defective strain suggests that at least part of the antirecombination activity of MMR proteins derives from the blockage of heteroduplex extension in the presence of mismatches.
Homologous recombination events can be either reciprocal or nonreciprocal in nature. Reciprocal recombination (crossing over) alters the linkage relationships of loci that flank the site of an exchange, whereas nonreciprocal recombination (gene conversion) is defined as the unidirectional transfer of information from one DNA molecule to another. All present models of recombination stipulate the formation of a heteroduplex recombination intermediate, which is formed by complementary base pairing of single strands derived from different duplexes. Correction of a mismatch in heteroduplex DNA can result in a gene conversion event, and such correction is effected by the same mismatch repair (MMR) machinery that corrects errors made during DNA replication. Cocorrection of a series of contiguous mismatches generates a conversion tract, the length of which is considered to be a minimal estimate of the extent of the heteroduplex intermediate formed. The point at which the recombining molecules exchange single strands to form heteroduplex DNA is referred to as a Holliday junction, and cleavage of this junction gives rise to crossovers and noncrossovers at roughly equivalent frequencies.
Homologous recombination usually involves allelic sequences on homologous chromosomes but also can occur between dispersed (ectopic) repeated sequences. Ectopic gene conversion provides a mechanism for homogenizing repeated sequences and hence is important in the concerted evolution of multigene families. In contrast to the relatively benign effects of ectopic gene conversion, ectopic crossing over results in various types of chromosome rearrangements which destabilize overall genome structure. Given the large amount of repetitive DNA present in the genomes of higher eukaryotes and the potentially deleterious nature of ectopic interactions, one would predict that allelic interactions should be highly favored over ectopic interactions. Two features of repeated sequences that might be important in limiting ectopic recombination are the length of the repeats and the degree of sequence identity between the repeats (38).
Studies with bacterial species (1, 22, 28, 29, 39, 46, 51, 57–59), Saccharomyces cerevisiae (4, 7, 12, 13, 20, 24, 30, 32, 33, 35, 36, 40, 43, 44), and higher eukaryotes (14, 15, 52, 56) have uniformly found that sequence divergence acts as a potent barrier to recombination. As first shown in conjugational crosses between Escherichia coli and Salmonella typhimurium (39), much of the recombination barrier associated with sequence divergence in prokaryotes derives from the action of the MMR machinery (1, 16, 22, 28, 29, 51, 57). The MMR system of E. coli (31) is used as a paradigm for eukaryotic MMR, and homologs of the bacterial MutS protein, which binds mismatched base pairs, and of the MutL protein, which interacts with MutS, have been identified in eukaryotes (11). In E. coli, elimination of either MutS or MutL results in a comparable increase in recombination between diverged sequences (1, 39). The antirecombination activity of MMR proteins also has been documented in yeast (7, 12, 13, 24, 32, 43, 44) and mammalian cells (9, 14), indicating that the barrier to recombination imposed by the MMR system is evolutionarily conserved. Interestingly, however, the antirecombination activities of yeast MutS and MutL homologs (Msh2p and Pms1p, respectively) are not equivalent. In yeast, elimination of Msh2p elevates recombination between diverged sequences to a greater extent than does elimination of Pms1p (references 12 and 44 and unpublished data), indicating that the antirecombination effect of Msh2p is not totally dependent on Pms1p. This observation is consistent with in vitro DNA binding studies demonstrating that the yeast MutL homologs increase the mismatch binding efficiency of the MutS homologs (19).
Systematic studies carried out with bacterial cells (28, 51) and yeast (13) have revealed a log-linear relationship between the rate of recombination and the degree of sequence divergence in both MMR-competent and MMR-defective cells. Such a relationship is predicted by models that assume that a minimal length of perfect homology (the “MEPS” [45]) is necessary to initiate recombination; the number of MEPS decreases exponentially with sequence divergence. In the yeast experiments, a single mismatch within a 350-bp substrate was sufficient to reduce recombination fourfold, and this inhibition was completely dependent on the MMR machinery (13). Additional mismatches had a cumulative negative effect on recombination; some of the inhibition was due to MMR-associated antirecombination activity, and some resulted from an MMR-independent process. The MMR-independent inhibition of recombination was attributed to an inability of the recombination machinery to recombine diverged sequences efficiently.
The antirecombination activity of MMR proteins presumably derives from the recognition of mismatches in heteroduplex recombination intermediates, but how this impairs recombination is unclear. Because of the strand-nicking activity associated with the repair of replication errors in E. coli, a heteroduplex destruction model has been considered. According to this model, the attempted repair of multiple mismatches in close proximity would create convergent excision tracts and hence double-strand breaks, which would destroy the recombination intermediate (see reference 6). The efficiencies of transformation of highly mismatched heteroduplex molecules into MMR-competent versus MMR-defective E. coli are similar, however, and this finding is not consistent with the destruction of mismatched heteroduplex DNA (53). Also, there is no evidence of MMR-associated strand nicking in eukaryotes, and such nicking would be a prerequisite for heteroduplex destruction. A second model that has been proposed is the heteroduplex rejection model, which posits that MMR proteins modulate the formation of mismatch-containing heteroduplex DNA (3, 10, 57). One possibility is that MMR-associated helicase activity unwinds mismatch-containing heteroduplex DNA, thus resulting in rejection of the donor strand (10, 57). Alternatively, the MMR machinery might interact directly with the recombination machinery to cause either reverse branch migration or immediate resolution of a recombination intermediate when mismatched heteroduplex is detected (3). A specific prediction of heteroduplex rejection models is that the extent of heteroduplex DNA should be longer in MMR-defective cells than in wild-type cells. Consistent with such a model, Worth et al. (55) have demonstrated that both the rate and the extent of in vitro RecA-catalyzed heteroduplex formation were reduced in the presence of purified MutS and MutL proteins.
One approach to understanding the molecular basis of MMR-associated antirecombination activity is to ask whether recombination products in MMR-competent cells differ significantly from those in MMR-defective cells. Several parameters can be examined in experiments of this sort. One can, for example, examine the ratio of gene conversions to crossovers in order to determine if the MMR machinery biases the resolution of recombination intermediates in a mismatch-dependent manner (see, e.g., reference 5). Alternatively, comparison of gene conversion tracts in MMR-competent and MMR-defective cells will indicate whether the molecular nature of recombination intermediates is impacted by the MMR machinery. The endpoints of the conversion tracts can be examined, or the lengths of the tracts can be determined. A difference in the endpoints of conversion tracts between MMR-competent and MMR-defective cells would imply a role of the MMR machinery in determining preferential sites of recombination initiation and/or resolution; a difference in the lengths of conversion tracts would imply a role for the MMR machinery in monitoring the fidelity of base pairing during heteroduplex formation.
In the present study, we have used an intron-based inverted-repeat (IR) assay system to characterize mitotic gene conversion tracts in wild-type versus MMR-defective yeast strains. With the intron-based system, one selects specifically for events that reverse the orientation of the region between the recombination substrates; reorientation of this region (the invertible segment) can result from either intrachromatid crossover or sister chromatid conversion. Using 94%-identical substrates, we have found an MMR-dependent conversion gradient across the substrates, with an excess of conversion tract endpoints in identity intervals closest to the invertible segment in MMR-competent cells. Such clustering is most easily explained by assuming that recombination occurs predominantly through a sister chromatid conversion mechanism rather than via intrachromatid crossover. Conversion tract length calculations based on an unequal sister chromatid conversion model indicate that conversion tracts are significantly longer in MMR-defective cells than in wild-type cells. The relevance of the MMR-dependent shortening of conversion tracts to the antirecombination activity of MMR proteins is discussed.
MATERIALS AND METHODS
Plasmid constructions.
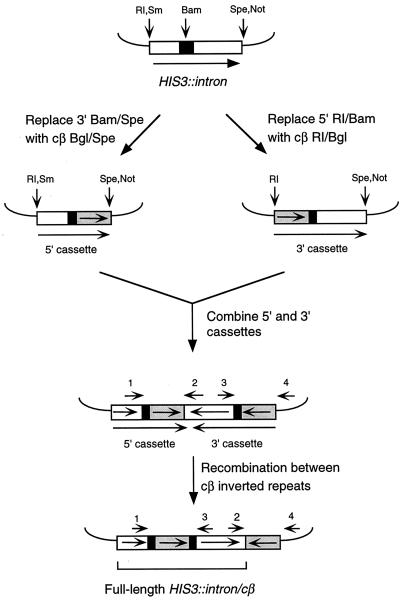
Plasmids with various chicken β-tubulin isoform 2 cDNA sequences (cβ2 sequences) were constructed according to the general scheme outlined in Fig. 1 (see references 12 and 13 for more details). 5′ and 3′ cassettes were derived by replacing sequences in pSR266 (the basic HIS3::intron construct) with PCR-amplified cβ2 sequences. To construct plasmids pSR424, pSR584, and pSR610, a SmaI/SpeI restriction fragment containing the 5′ cassette was inserted into a SpeI/NotI-digested plasmid containing the 3′ cassette (the NotI site was filled in with the Klenow fragment of DNA polymerase). To construct plasmid pSR612, a SmaI/SpeI restriction fragment containing the 5′ cassette was inserted into a NotI-digested plasmid containing the 3′ cassette; the ends of both fragments were filled in with the Klenow fragment of DNA polymerase.
FIG. 1.
Construction of inverted-repeat substrates. The pGAL-HIS3::intron construct contained on plasmid pSR266 is shown at the top. Open boxes correspond to HIS3 sequences, solid boxes to artificial intron sequences, and shaded boxes to cβ2 sequences; boxes are not drawn to scale. The positions of the oligonucleotides used as PCR primers are indicated by arrows numbered as follows: 1, HIS3-702F; 2, HIS3-1751F; 3, HIS3-765R; 4, T3. The left recombinant cβ2 segment was amplified with primers 1 and 3; the right recombinant segment was amplified with primers 2 and 4. Only those restriction sites relevant to constructions are shown. RI, EcoRI; Sm, SmaI; Bam, BamHI; Spe, SpeI; Not, NotI; Bgl, BglII.
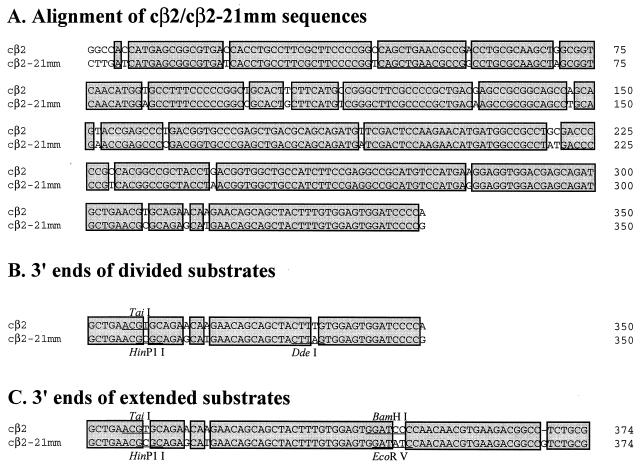
pSR424 contains the 350-bp cβ2 (5′ cassette) and cβ2-21mm (3′ cassette) IR substrates in orientation 1. These substrates have 94% sequence identity (Fig. 2A). pSR612 contains a division construct in which interval 318 to 350 was divided into two equivalent intervals by introducing a base substitution (T334A) (Fig. 2B) into the cβ2-21 mm substrate. The base substitution was introduced by using an appropriate PCR primer. pSR584 contains an extended construct with recombination substrates of 374 bp instead of the standard 350 bp (Fig. 2C). The additional 24 bp extend beyond the 3′ end of the original 350-bp substrates in pSR424 and were introduced as 5′ extensions on reverse PCR primers that anneal to the 3′ ends of cβ2 sequences. The 374-bp substrates in the 5′ and 3′ cassettes were PCR amplified from template plasmids pSR257 (containing cβ2 sequences) and pSR426 (containing cβ2-21mm sequences), respectively.
FIG. 2.
(A) Alignment of recombination substrates. Sequences of perfect identity are boxed and shaded. Although we refer to positions within the aligned sequences using the numbering system shown, it should be noted that the homology between cβ2 and cβ2-21mm does not actually begin until position 7. During the generation of the cβ2-21mm substrate, an unusually high density of PCR errors apparently was introduced at the 5′ end of the product. The homology between cβ2 and cβ2-21mm ends at position 349, for a total of 343 bp of partially homologous (homeologous) sequence. There are 21 mismatches, 2 of which are contiguous, within the 343-bp region of homology. (B and C) Only nucleotides from position 301 are shown; restriction sites used in the restriction site polymorphism analyses are indicated above or below the alignments.
pSR610 contains the orientation 2 construct, in which the orientations of both cβ2 and cβ2-21mm sequences are reversed relative to their orientations in pSR424. The 5′ cassette cβ2 substrate was amplified from pSR257 by using a forward PCR primer with a SpeI site engineered near the 5′ end and a reverse PCR primer with a BglII site engineered near the 5′ end. Ligation of the SpeI/BglII-digested PCR product to SpeI/BamHI-digested pSR266 yielded a 5′ cassette with cβ2 sequences in reverse orientation relative to that in the 5′ cassette of pSR424. To reverse the orientation of the cβ2-21 mm substrate in the 3′ cassette, a 350-bp segment was amplified from pSR426 by using a forward PCR primer with a BglII site engineered near the 5′ end and a reverse PCR primer with an EcoRI site engineered near the 5′ end. Following digestion with BglII and EcoRI, the PCR product was ligated to BamHI/EcoRI-digested pSR266, yielding a 3′ cassette with cβ2-21mm sequences in reverse orientation relative to that in the 3′ cassette of pSR424.
Yeast strain constructions.
All strains used in this study were derived from isogenic strains SJR231 (MATα ade2-101oc his3Δ200 ura3-Nhe), GCY121 (MATα ade2-101oc his3Δ200 ura3-Nhe msh2Δ msh3Δ::hisG), and GCY128 (MATα ade2-101oc his3Δ200 ura3-Nhe pms1Δ) (see reference 12) by lithium acetate transformation (18). pSR424, pSR584, pSR610, or pSR612 was digested with StuI prior to transformation in order to target integration to the URA3 locus. Following selection of Ura+ transformants, single-copy integration of the relevant plasmid was confirmed by PCR or Southern blot analysis.
Generation of independent recombinants.
One-milliliter cultures were grown nonselectively at 30°C for two days in YEP medium (1% yeast extract–2% Bacto Peptone) supplemented with 2% glycerol and 2% ethanol (YEPGE). Cells were harvested, washed once with H2O, and resuspended in 200 μl of H2O. An aliquot of 100 μl was plated on SGGE−His selective medium (synthetic complete medium supplemented with 2% glycerol, 2% galactose, and 2% ethanol but deficient in histidine) to select for His+ recombinants. Only one colony was taken from each culture in order to ensure that all recombinants analyzed were of independent origin.
Molecular analysis of recombinants.
Genomic DNA was extracted by glass bead lysis (21) from each recombinant and used as a template for PCR amplification. The 5′ and 3′ recombination products were amplified by using primers homologous to sequences that flank the recombination substrates (see Fig. 1 for the positions of primers). The 5′ product was amplified by using primers HIS3-702F (5′-GTTTCTGGACCATATG) and HIS3-765R (5′-GCACTCAACGATTAG), and the 3′ recombination product was amplified by using primers HIS3-1751F (5′-GATGGCAAACATGTC) and T3 (5′-TGATGTCGGCGATATAGG). The PCR products were purified by using Qiaquick Spin Columns (Qiagen) and were used as templates for DNA sequencing. Oligonucleotides FAI (5′-ATGGACTAAAGGAGGCT) and T3 were used as sequencing primers for the 5′ and 3′ recombination products, respectively. All sequencing reactions were carried out with ABI Prizm Dye Terminator Cycle Sequencing Ready Reaction Kits and run on an ABI Prizm 377XL DNA Sequencer (Perkin-Elmer Applied Biosystems).
RESULTS
The intron-based IR recombination system.
The relevant features of the intron-based IR recombination system are diagrammed in Fig. 1 (for a more complete description of the system, see references 12 and 13). A galactose-inducible HIS3 gene containing an artificial intron (HIS3::intron) was the starting point for all constructs. 5′ or 3′ recombination cassettes were constructed by replacing sequences downstream of the 5′ intron splice consensus element or upstream of the intron TACTAAC element, respectively, with a fragment derived from cβ2 cDNA. 5′ cassettes contained the 5′ end of HIS3, the 5′ portion of the intron, and a cβ2 recombination substrate, whereas 3′ cassettes were composed of a different cβ2 recombination substrate, the 3′ portion of the intron, and the 3′ end of HIS3. The 5′ and 3′ cassettes were combined in reverse orientation relative to each other, resulting in the 3′ portion of HIS3::intron being flanked by cβ2 IRs. Following integration of the IR construct at the URA3 locus in an appropriate his3Δ strain, cells are phenotypically His−. Recombination between the cβ2 repeats can reverse the orientation of the intervening region (referred to below as the invertible segment), thus placing the 5′ and 3′ parts of the HIS3 gene in the same orientation and reconstituting a functional intron. Such recombinants are phenotypically His+ and can be identified on an appropriate selective medium.
Gene conversion tracts in an MMR-competent strain.
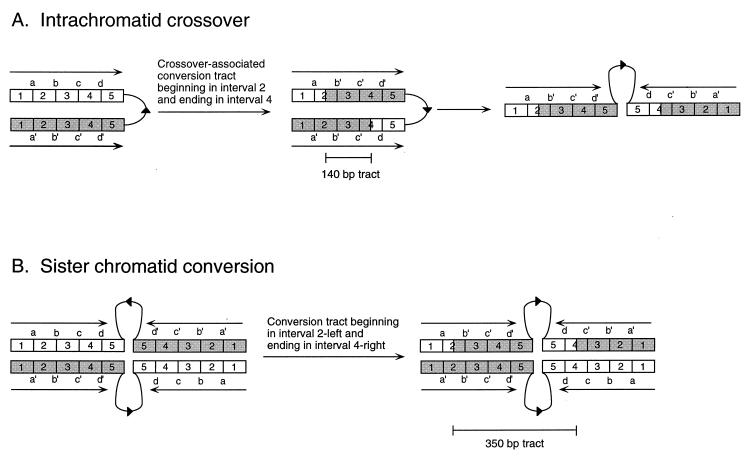
The initial cβ2-derived substrates used to examine gene conversion tracts were 350 bp and 94% identical (cβ2 and cβ2-21mm). cβ2-21mm was generated from a cβ2 plasmid template by low-fidelity PCR. The cβ2–cβ2-21mm pair of substrates was chosen for sequencing studies because the potential mismatches are more randomly distributed than those in naturally diverged sequences. As illustrated in Fig. 2A, the mismatches divide the substrates into 21 intervals of perfect identity ranging in size from 2 to 37 bp. The cβ2 substrates were oriented so that their 3′ ends were proximal and their 5′ ends were distal to the intervening invertible segment (orientation 1 in Fig. 3). Following the isolation of a His+ recombinant, each recombinant cβ2 segment was amplified by PCR using flanking primers (see Fig. 1), and the PCR products were sequenced individually. A given mismatch was considered to have undergone a gene conversion event if the same nucleotide was present in both recombinant cβ2 segments; if the recombinant cβ2 segments still differed at the position of the original mismatch, then the site was considered not to have undergone gene conversion. A gene conversion tract encompasses a series of contiguous mismatches.
FIG. 3.
Orientation of cβ2 substrates relative to the HIS3::intron invertible segment. Arrows indicate the 5′-to-3′ (nucleotides 1 to 350, respectively, as in Fig. 2) orientations of the cβ2 sequences.
Gene conversion tracts were determined for 62 independent His+ recombinants isolated in an MMR-competent strain, and the recombinants were divided into three classes. Most recombinants (50 of 62, or 80%) had a single gene conversion tract with all mismatches converted in the same direction (continuous asymmetric conversions). Six recombinants (10%) had no mismatches converted; the corresponding recombination event was assumed to begin and end in the same identity interval. The final six recombinants (10%) had either noncontiguous conversion tracts or contiguous but bidirectional (symmetric) conversion tracts; these were classified as complex conversions. Because of their complexity, this class was not included in the endpoint distribution analysis (see below) or in the calculations of conversion tract lengths (see Discussion). The total number of recombinants analyzed in terms of conversion tract endpoints and lengths was thus 56, which corresponds to 112 endpoints.
Gene conversion tracts can be analyzed either in terms of their endpoints or in terms of the length of DNA converted. Whereas conversion tract endpoints for a given recombinant can be determined unambiguously from the sequence data, the corresponding conversion tract length calculation varies considerably, depending on whether the event occurs via intrachromatid crossover or via sister chromatid conversion. For this reason, gene conversion tract lengths will not be addressed here but will be considered in detail in the Discussion. An expected distribution of conversion tract endpoints was generated by making the simplifying assumption that the distribution of endpoints is random. The percentage of conversion tract endpoints in a particular identity interval thus should be directly proportional to the length of that interval. For example, because interval 6 to 21 is 14 bp (interval endpoints are defined in terms of the positions of the bordering mismatches) and the sum of all intervals of perfect identity is 322 bp (total homology in the cβ2 repeats [343 bp] minus the 21 mismatched base pairs), one would expect 4.3% of all endpoints to be in this interval. For the experimental data, a conversion tract endpoint was assigned to a given interval if the mismatch defining one side of the interval was converted but the mismatch defining the other side of the interval was not. Each continuous conversion tract had two distinct endpoints, whereas recombinants with no evident conversion tract were assumed to have two endpoints in the same interval. The expected and observed distributions of conversion tract endpoints are presented in Table 1.
TABLE 1.
Distribution of conversion tract endpoints
| Interval | Identity (bp) | Expected endpoints (%) | No. (%) of endpoints in:
|
||||
|---|---|---|---|---|---|---|---|
| WTa orientation 1 | WT orientation 2 | msh2 msh3 orientation 1 | msh2 msh3 orientation 2 | pms1 orientation 1 | |||
| 6–21 | 14 | 4 | 2 (2) | 5 (10) | 9 (9) | 0 (0) | 4 (7) |
| 21–43 | 21 | 7 | 3 (3) | 9 (19) | 8 (8) | 2 (4) | 4 (7) |
| 43–57 | 13 | 4 | 5 (4) | 5 (10) | 2 (2) | 2 (4) | 0 (0) |
| 57–70 | 12 | 4 | 5 (4) | 1 (2) | 3 (3) | 3 (6) | 1 (2) |
| 70–84 | 13 | 4 | 3 (3) | 2 (4) | 5 (5) | 5 (10) | 4 (7) |
| 84–99 | 14 | 4 | 6 (5) | 3 (6) | 15 (14) | 1 (2) | 0 (0) |
| 99–105 | 5 | 2 | 1 (1) | 1 (2) | 1 (1) | 2 (4) | 1 (2) |
| 105–113 | 7 | 2 | 2 (2) | 4 (8) | 0 (0) | 2 (4) | 3 (6) |
| 113–133 | 19 | 6 | 3 (3) | 0 (0) | 2 (2) | 3 (6) | 4 (7) |
| 133–147 | 13 | 4 | 2 (2) | 0 (0) | 3 (3) | 3 (6) | 3 (6) |
| 147–152 | 4 | 1 | 1 (1) | 0 (0) | 1 (1) | 0 (0) | 0 (0) |
| 152–162 | 9 | 3 | 0 (0) | 2 (4) | 1 (1) | 0 (0) | 1 (2) |
| 162–191 | 28 | 9 | 9 (8) | 2 (4) | 13 (13) | 6 (12) | 0 (0) |
| 191–219 | 27 | 8 | 8 (7) | 2 (4) | 14 (13) | 4 (8) | 5 (9) |
| 219–229 | 8 | 2 | 3 (3) | 2 (4) | 1 (1) | 0 (0) | 0 (0) |
| 229–244 | 14 | 4 | 11 (10) | 3 (6) | 3 (3) | 4 (8) | 3 (6) |
| 244–282 | 37 | 11 | 12 (11) | 3 (6) | 7 (7) | 3 (6) | 9 (17) |
| 282–309 | 26 | 8 | 13 (12) | 3 (6) | 11 (11) | 4 (8) | 3 (6) |
| 309–315 | 5 | 2 | 1 (1) | 0 (0) | 2 (2) | 2 (4) | 0 (0) |
| 315–318 | 2 | 1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| 318–350 | 31 | 10 | 22 (20) | 1 (2) | 3 (3) | 4 (8) | 9 (17) |
| Total | 322 | 100 | 112 (100) | 48 (100) | 104 (100) | 50 (100) | 54 (100) |
WT, wild type.
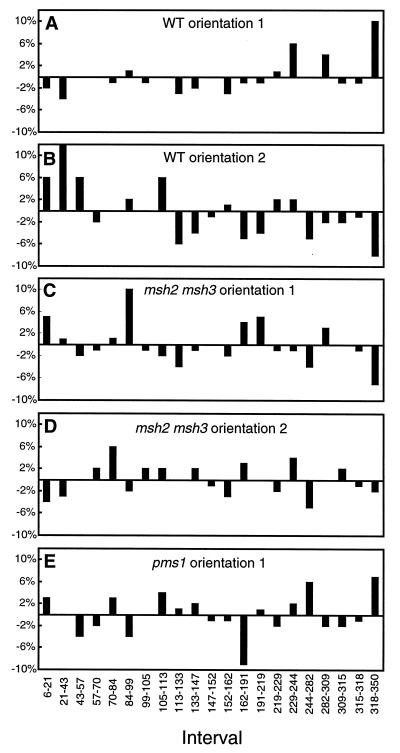
The observed endpoint distribution for the orientation 1 substrates was compared to the expected distribution by subtracting the percentage of endpoints expected in a particular interval from the percentage actually observed in that interval. This method yields positive and negative percentages which, when plotted, indicate intervals containing an excess or deficit of endpoints, respectively. As shown in Fig. 4A, there is a clear excess of conversion tract endpoints in the 3′ half of the cβ2 substrates and a corresponding deficit in the 5′ half. The excess is particularly evident in the 3′-most interval; one would predict that only 10% (31 of 322) of all endpoints should be in interval 318 to 350, yet this interval contained 20% of all endpoints. Because interval 318 to 350 appeared to be particularly “hot” for conversion tract endpoints, this interval was examined in more detail.
FIG. 4.
Conversion tract endpoint distributions. The percentage of endpoints expected in each interval was subtracted from the observed percentage, and the residual value was plotted. Positive or negative percentages indicate excesses or deficits of endpoints, respectively. WT, wild type.
Analysis of conversion tract endpoints in interval 318 to 350 by using a restriction site polymorphism.
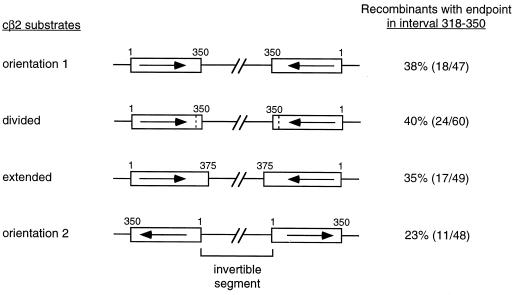
A restriction site polymorphism just upstream of interval 318 to 350 was used to more accurately quantify the number of conversion tracts that end in this interval. A mismatch at position 309 establishes a TaiI site (5′-ACGT-3′) in the cβ2 substrate but a HinP1I site (5′-GCGC-3′) in the cβ2-21 mm substrate. Conversion tracts that either start or end in interval 318 to 350 are assumed to include the mismatch at position 309, and thus both recombination products should have either a TaiI site or a HinP1I site. The observation that only 1 of the 62 recombinants sequenced had an endpoint in interval 309 to 318 validates the use of the polymorphism at position 309 as a marker for assigning endpoints to interval 318 to 350. Forty-seven additional His+ recombinants were isolated and were analyzed for the presence or absence of the restriction site polymorphism. Eighteen (38%) of the recombinants converted the mismatch at position 309. This value agrees well with the DNA sequencing results, where 34% (21 of 62 total recombinants sequenced) of the recombinants had an endpoint(s) in this interval. It should be noted that the restriction site polymorphism studies indicate whether a given recombinant has a conversion tract endpoint in interval 318 to 350 but provide no information as to whether the underlying event is simple or complex. Because of this uncertainty, the percentage of recombinants with an endpoint(s) in interval 318 to 350, instead of the percentage of total endpoints in this interval, was calculated for all constructs.
Based on its size, interval 318 to 350 would be expected to contain 10% of all endpoints, so approximately 20% of all recombinants should have an endpoint in this interval (each conversion tract has two endpoints). The observation that 34 to 38% of recombinants have an endpoint in interval 318 to 350 clearly indicates that this interval is a hot spot for recombination endpoints (P < 0.01 by the χ2 test for this number of endpoints compared to that expected). There are at least three possible explanations for this observation. First, this 31-bp interval is the second-longest interval of perfect identity in the substrates (only interval 244 to 282 is longer), so the fact that it is a hot spot could simply reflect its length (i.e., longer intervals might contain a disproportional number of endpoints). Second, this interval is at one end of the substrates and so abuts a region of nonhomology, a location that might create a bias in favor of the resolution of recombination events. Finally, the immediate proximity of this interval to the invertible segment may be important; in both substrates the 3′ end of the cβ2 sequence is adjacent to the invertible HIS3::intron segment. In order to test the above hypotheses, interval 318 to 350 was modified as shown in Fig. 3, and the effect of each alteration on the frequency of recombinants with an endpoint(s) in the interval was ascertained by using restriction site polymorphisms.
Division of interval 318 to 350.
A divided construct (Fig. 3) was created in order to determine whether the length of interval 318 to 350 is important for its hot spot activity. The divided construct was derived by introducing a T-to-A transversion at position 334 in cβ2-21mm, which creates an additional mismatch between the recombination substrates and splits interval 318 to 350 into two smaller intervals of equal length (see Fig. 2B). Conversion of the TaiI/HinP1I restriction site polymorphism at position 309 was used to monitor tracts that ended in the 318-to-350 region of the divided construct. Sixty recombinants were examined, and 40% (24 of 60) had an endpoint in interval 318 to 350. This percentage is similar to that obtained with the undivided orientation 1 construct (34 and 38% by DNA sequence and restriction site polymorphism analysis, respectively) and is different from the expected value (P < 0.01 by the χ2 test). Based on the results obtained with the divided construct, we conclude that the excess of endpoints in interval 318 to 350 in the undivided orientation 1 construct is not due to the length of uninterrupted identity in this interval. In addition to dividing interval 318 to 350, the T334A mutation in the cβ2-21mm sequences created a DdeI site (5′-CTNAG-3′). This new polymorphism was used to further refine the positions of endpoints within the two halves of interval 318 to 350. Sixteen of the 24 recombinants had an endpoint in interval 318 to 334, 7 had an endpoint in interval 334 to 350, and 1 had an endpoint in each interval.
Extension of the 3′ ends of the substrates.
An extended construct (Fig. 3) was created in order to test the hypothesis that interval 318 to 350 is a hot spot because it is at the extreme end of the substrate and hence borders a region of complete nonhomology. The original 350-bp substrates were extended at the 3′ end by adding two additional intervals of perfect identity (see Fig. 2C). The first additional interval (interval 347 to 368) in the extended construct was 20 bp, and the second (interval 368 to 375) was 6 bp. A restriction site polymorphism was created by mutations at positions 346 and 347 (C346A and C347T), thus creating an EcoRV site (5′-GATATC-3′) in the extended cβ2-21mm sequences while maintaining a BamHI site (5′-GGATCC-3′) in the extended cβ2 sequences. The two introduced mismatches separate the previous 318 to 350 interval (318 to 346 in the extended construct) from the newly added intervals 347 to 368 and 368 to 375. By monitoring the conversion of the mismatches at positions 309 and 346/347, we were able to determine how many conversion tracts ended either in interval 318 to 346 or in interval 347 to 375. Among 49 recombinants analyzed, 17 (35%) had an endpoint in interval 318 to 346. This percentage is very similar to the proportion of orientation 1 recombinants with endpoints in interval 318 to 350 and is significantly different from the expected value (P = 0.01 by the χ2 test). Interestingly, the two new intervals in the extended construct also contained a disproportional number of endpoints. Because the new intervals contain 7.6% (26 of 344) of the nucleotide identity in the extended substrates, approximately 15% of recombinants would be expected to contain an endpoint in the extended region. Thirteen of the 49 recombinants (27%), however, had an endpoint in the newly added intervals, which is significantly more than expected (P < 0.05 by the χ2 test). Based on the results with the extended substrates, it appears that an interval needs only to be near, rather than at, the 3′ end of the cβ2 substrate in order to contain an excess of conversion tract endpoints. This result is consistent with the general observation (Fig. 4A) that there is an excess of endpoints in the 3′ halves of the substrates and a deficit of endpoints in the 5′ halves. This “gradient” either could be related to the lower density of mismatches in the 3′ halves of the substrates or could reflect the proximity of the 3′ ends of the substrates to the invertible segment. These two possibilities were distinguished by reversing the orientations of the cβ2 substrates relative to the invertible HIS3::intron sequences.
Reorientation of the substrates relative to the invertible segment.
The orientation of each of the cβ2 substrates was reversed relative to the HIS3::intron sequences, thus placing the original 3′ substrate ends (with an excess of endpoints) distal to the invertible segment and the original 5′ ends (with a deficit of endpoints) adjacent to the invertible segment. Substrates in the reverse orientation will be referred to as being in orientation 2 (see Fig. 3). As described above, the proportion of recombinants with a conversion tract endpoint in interval 318 to 350 (note that the same nucleotide receives the same coordinate for orientations 1 and 2) was analyzed by using the TaiI/HinP1I restriction site polymorphism. Only 23% (11 of 48; P > 0.5 by the χ2 test for this number of endpoints compared to that expected) of the orientation 2 recombinants had an endpoint in interval 318 to 350, compared to 34 to 40% of recombinants with an endpoint in this interval with the orientation 1 construct, the divided construct, and the extended construct. This result suggested that altering the orientation of the recombination substrates might have reversed the gradient of conversion tract endpoints evident with the orientation 1 substrates. To examine this further, the first 26 recombinants derived by using the orientation 2 substrates were subjected to DNA sequence analysis in order to determine the precise positions of all conversion tract endpoints. Twenty-two recombinants had continuous conversion tracts, two had complex conversion tracts, and two had no detectable conversion tract. This distribution of recombinant classes was the same as that obtained with the orientation 1 substrates (50 continuous tracts, 6 complex tracts, and 6 simple crossovers; P > 0.90 by the χ2 contingency test). The distribution of conversion tract endpoints for the orientation 2 substrates is given in Table 1 and is compared graphically to the expected distribution in Fig. 4B. Whereas recombinants derived from the orientation 1 cβ2 substrates had an excess of endpoints near the 3′ ends of the substrates (Fig. 4A), the orientation 2 substrates yielded recombinants with a clear excess of endpoints in the 5′ halves of the cβ2 sequences. These data suggest that the distribution of endpoints is determined primarily by the relative proximity of an interval to the invertible HIS3::intron segment located between the substrates rather than by interval size or sequence. In other words, intervals close to the invertible segment are more likely to contain a conversion tract endpoint than are intervals that are farther away from the invertible segment.
Conversion tract endpoints in MMR-defective cells.
The yeast MMR system exerts a strong antirecombination effect on the 94%-identical substrates used in the conversion tract analysis above (13). Therefore, we examined gene conversion tracts in cells defective in mismatch binding activity (an msh2 msh3 mutant) in order to ascertain whether the MMR system impacts the nature of recombination intermediates. Because there is no gene conversion via MMR of heteroduplex DNA in MMR-defective cells, any heteroduplex formed during recombination will be segregated at the next round of DNA replication. Replication of the unrepaired donor DNA thus will produce the equivalent of a gene conversion tract.
Sixty-two His+ recombinants derived from an msh2 msh3 strain containing the orientation 1 substrates were sequenced to estimate the extent of heteroduplex formation. Forty of the 62 recombinants had a single continuous conversion tract, 12 had no mismatches converted, and 10 were classified as complex conversions. This class distribution was the same as that observed in the MMR-competent cells (P > 0.30 by the χ2 contingency test). The distribution of conversion tract endpoints is presented in Table 1, and this distribution is compared to the expected distribution in Fig. 4C. In contrast to the presence of excess endpoints at the 3′ ends of the orientation 1 substrates in wild-type cells, there was no apparent clustering of endpoints in msh2 msh3 cells. A similar loss of endpoint clustering was observed when the orientation 2 substrates were analyzed in an msh2 msh3 background (Fig. 4D). The effect of Pms1p on conversion tract endpoints also was examined, and the data are presented in Table 1 and Fig. 4E. As observed in the msh2 msh3 mutant background, no clear endpoint clustering was evident in the pms1 mutant. These results suggest a causative role for the yeast MMR machinery in establishing the conversion tract endpoint clustering near the invertible segment that was observed with both the orientation 1 and the orientation 2 cβ2 substrates in wild-type cells.
DISCUSSION
In this study, an intron-based recombination assay system has been used to generate recombinants between a pair of 94%-identical substrates oriented as IRs (see Fig. 1). The IR substrates flank the 3′ end of an intron-containing HIS3 gene, which is in reverse orientation relative to the 5′ end of the gene. Reorientation of the 3′ HIS3::intron segment via recombination involving the flanking IRs reconstitutes a functional HIS3 gene, and such events can be identified as His+ colonies on histidine-deficient medium.
Although reorientation or flipping of a segment of DNA is generally considered to result from intrachromatid crossing over between flanking IRs (Fig. 5A), a sister chromatid gene conversion event also can flip the region between IRs (42). In sister chromatid gene conversion (Fig. 5B), one of the chromatids loops around and pairs with the sister; the substrate upstream of the invertible segment on one chromatid pairs with the downstream substrate on the sister chromatid, and the substrate downstream of the invertible segment likewise pairs with the upstream substrate on the sister. The invertible segment is thus flanked by sister-sister pairings, each of which involves diverged rather than identical substrates. Gene conversion events that initiate in the paired sequences on one side of the invertible segment, extend through the invertible segment, and terminate in the paired sequences on the other side will flip the invertible segment. It should be noted that neither intrachromatid gene conversion nor sister chromatid crossing over will yield His+ recombinants. Intrachromatid gene conversion does not reorient the invertible segment, so recombinants remain His−; sister chromatid crossing over gives rise to acentric and dicentric recombinant chromosomes, resulting in inviable progeny.
FIG. 5.
Reversal of the orientation of a segment of DNA between IRs by either intrachromatid cross over or sister chromatid gene conversion. The open and shaded boxes correspond to the IR substrates that flank the invertible HIS3::intron segment, which is represented by a loop with an arrowhead indicating the orientation. Small open circles represent centromeres.
Distributions of conversion tract endpoints in a wild-type strain.
DNA sequence analysis of both products derived from individual recombination events allowed for the determination of conversion tract endpoints. In the discussion that follows, it is assumed that conversion tracts are an accurate representation of the extent of heteroduplex DNA present in recombination intermediates. We recognize the formal possibility, however, that conversion tract endpoints may not necessarily correspond to positions where recombination events begin and end. At least in MMR-competent cells, endpoints could correspond to repair borders. We also note that the repair of heteroduplex DNA in mitosis is not likely to be 100% efficient (41), so some of the conversion tracts in MMR-competent cells may arise via replicative resolution of heteroduplex intermediates, which is the likely mechanism for generating conversion tracts in MMR-defective cells. Regardless of the mechanism for generating a conversion tract, however, this mechanism presumably operates after the completion of heteroduplex DNA formation and thus should not affect the extent of heteroduplex formed. In determining conversion tract parameters, only the no-conversion and continuous, asymmetric conversion tract classes, which together accounted for 90% of His+ recombinants, were considered. The relative rarity of symmetric or discontinuous conversion tracts is in general agreement with mitotic data obtained in previous studies (2, 49).
The most striking feature of the conversion tracts derived from the orientation 1 cβ2 substrates is an excess of endpoints at the 3′ ends of the substrates (Fig. 4A). Because the 3′ halves of the cβ2 substrates contain fewer mismatches (and hence longer stretches of perfect identity) than the 5′ halves (see Fig. 2A), the biased endpoint distribution could be directly related to mismatch density. This possibility was addressed by reversing the orientations of the recombination substrates relative to the HIS3::intron segments (orientation 2 substrates). If mismatch density is the relevant factor, reversing the orientation of the substrates should not affect the distribution of endpoints; the 3′ half should still contain an excess of endpoints, and the 5′ half should still contain a deficit. We observed, however, that the endpoint clustering was reversed with the orientation 2 substrates, so that the end with the greatest mismatch density contained the largest number of endpoints (Fig. 4B). This result indicates that the endpoint distribution is determined primarily by the orientation of the substrates relative to the intervening invertible segment, such that the identity intervals containing the excess of endpoints are those intervals closest to the invertible segment. As will be discussed in more detail below, we believe that the endpoint distributions are readily explained by a sister chromatid conversion model but are difficult to rationalize if one assumes that most events are the result of intrachromatid crossover.
The distribution of endpoints relative to the lengths of homology blocks also was analyzed in order to determine whether the distribution of conversion tract endpoints is proportional to the length of the homology intervals or whether there is a bias for endpoints to occur in the longer homology blocks. This analysis indicated that endpoints are randomly distributed with respect to the lengths of homology blocks (data not shown). A similar conclusion was reached by Porter et al. (35).
Distributions of conversion tract endpoints in MMR-defective strains.
Conversion tract endpoints were determined in MMR-defective strains in order to ascertain whether endpoint distributions are influenced by the yeast MMR machinery. The clear endpoint clustering evident with the cβ2 substrates in either orientation 1 (Fig. 4A) or orientation 2 (Fig. 4B) was abolished in an msh2 msh3 background (Fig. 4C and D). Similarly, in a pms1 mutant (Fig. 4E) there was no indication of the prominent endpoint clustering that was evident in wild-type cells. The data presented in Fig. 4 thus demonstrate that the biased endpoint distributions evident in wild-type cells are dependent on a functional MMR system. As will be elaborated further below, we speculate that the alteration of conversion tract endpoints by MMR proteins is related to the documented antirecombination activity of these proteins.
Intrachromatid crossover versus sister chromatid conversion.
As shown in Fig. 6, estimates of conversion tract lengths are very different for intrachromatid crossovers and sister chromatid conversion events that have the same endpoints. In principle, an intrachromatid crossover can be distinguished from sister chromatid conversion if the recombination intermediate can be captured and analyzed physically or if the timing of the recombination event in the cell cycle (G1 versus G2) can be determined. Given the low frequency with which our diverged substrates recombine, however, neither approach is feasible at present. As argued below, however, we believe that the majority of the His+ recombinants detected by our assay system occur via sister chromatid conversion rather than via intrachromatid crossover.
FIG. 6.
Estimation of conversion tract lengths. Open and shaded boxes correspond to IR substrates with four hypothetical mismatches (a through d), which divide the 350-bp substrates into five 70-bp intervals of perfect identity (1 through 5). As shown, intrachromatid and sister chromatid events that have identical endpoints (one endpoint in interval 2 and the other in interval 4) correspond to conversion tracts of different lengths: 140 and 350 bp for intrachromatid crossover and sister chromatid conversion events, respectively. If the products are viewed linearly along one chromosome, however, they appear identical.
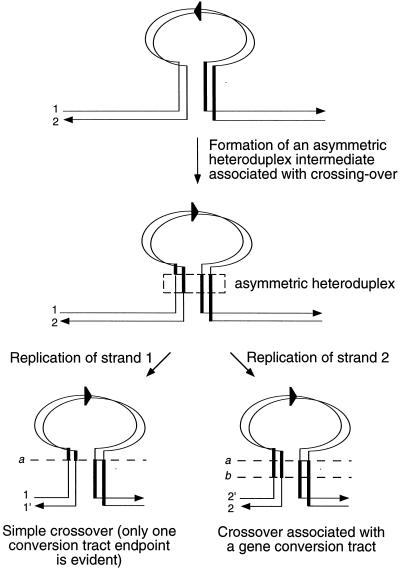
In MMR-defective cells, heteroduplexes formed during mitotic recombination are not repaired and the mismatched strands segregate into different daughter cells after the next round of DNA replication. With an intrachromatid crossover event, it is critical to note that only the flanking IRs are involved in heteroduplex formation; each strand of the heteroduplex DNA intermediate will contain an identical HIS3::intron segment. As shown in Fig. 7, replication of an asymmetric heteroduplex intermediate will give rise to two His+ daughter chromosomes, one of which should contain an apparent conversion tract and the other of which should appear as a simple crossover, with both endpoints in the same identity interval. One would predict, therefore, that at least one-half of all recombinants should appear as simple crossovers. This prediction is not consistent with our experimental data, in which only 20% of msh2 msh3 recombinants were scored as simple crossovers (P < 0.01 by χ2 analysis). Although the data obtained for MMR-defective cells would be consistent with the formation of predominantly symmetric heteroduplex intermediates (both replication products of which should have conversion tracts), symmetric heteroduplexes generally are assumed to be rare relative to asymmetric heteroduplexes in yeast. The extensive analysis of IR recombination carried out by Ahn and Livingston (2) indicates that 90% of mitotic conversion tracts are indeed asymmetric.
FIG. 7.
Replication of an asymmetric heteroduplex intermediate formed during intrachromatid crossing over in a MMR-defective strain. Both strands of the DNA duplexes (1 and 2, 1 and 1′, or 2 and 2′) are shown, with arrows indicating the 3′ ends. Thin and thick vertical lines correspond to the inverted substrates that flank the invertible HIS3::intron segment (represented by a loop with the orientation indicated by an arrowhead). Dashed horizontal lines (a and b) indicate conversion tract endpoints. Resolution of the asymmetric heteroduplex intermediate yields two His+ products, of which one has a conversion tract (right) and the other appears as a simple crossover (left).
In contrast to the high frequency of simple crossovers predicted by the intrachromatid crossover model, the sister chromatid conversion model predicts that the majority of the His+ recombinants isolated from MMR-defective cells should have detectable conversion tracts. In contrast to the exclusion of the invertible segment in the intrachromatid crossover model, the sister chromatid conversion model demands that the invertible segment always be included as part of the heteroduplex intermediate that extends into the flanking IRs. The intact strand of the recipient molecule will have the HIS3::intron segment in its original, His− orientation, whereas the strand donated from the sister will have the HIS3::intron segment in the reverse, His+ orientation. In the absence of MMR, only the strand containing the donated DNA will give rise to a daughter chromosome that carries a His+ allele. Since the donated DNA must contain part of the IRs that flank the invertible HIS3::intron segment, flanking IR segments always will be coconverted along with the invertible segment. In other words, the selection of the donor-derived invertible segment ensures the inheritance of flanking information from the donor as well in all the His+ recombinants. One thus would predict that gene conversion events should be the predominant events, and the data presented in Results are consistent with this prediction. According to the sister chromatid conversion model, recombinants with no detectable gene conversion tract correspond to gene conversion events that have endpoints in the same identity interval on both sides of the invertible segment.
In addition to the fates of asymmetric heteroduplex DNA intermediates in MMR-defective cells, there are two other observations that are more readily explained by the sister chromatid conversion model than by the intrachromatid crossover model. First, the sister chromatid conversion model can account for the clustering of conversion tract endpoints observed in wild-type cells. Because reorientation via gene conversion of the invertible HIS3::intron sequences located between the IR substrates is selected by the system, each successful recombination event must have one endpoint within substrates on one side of the invertible segment and the other endpoint within substrates on the other side. The closer a mismatch is to the invertible segment, the higher the probability that it will be included in a heteroduplex intermediate. The net result is a conversion gradient that falls off on either side of the selected site, which is the invertible segment in this case. It should be noted that a similar recombination gradient was observed by Willis and Klein (54), using a system in which a kanamycin resistance gene (Kanr) was flanked by 360-bp IRs. By using a small number of restriction site polymorphisms, it was demonstrated that the presumptive crossover events preferentially occurred proximal to the invertible segment. As explained above, a clustering of endpoints close to an invertible segment is exactly what one would predict if the underlying mechanism involves sister chromatid conversion rather than intrachromatid crossover.
A second observation that can be more readily explained by the sister chromatid conversion model concerns the inhibitory effect of MMR proteins on recombination between perfectly identical IR sequences. Using the same intron-based system as that used in this study, we have consistently observed a perplexing two- to threefold stimulation of recombination between identical substrates in msh2 msh3 strains relative to wild-type strains (12, 13). Based on the assumption that recombination occurred exclusively via intrachromatid crossover, we suggested that the MMR machinery might be detecting the extension of heteroduplex DNA into flanking nonhomologous sequences. According to the sister chromatid conversion model, heteroduplex DNA that covers the invertible HIS3::intron region will be at least transiently unpaired (a completely paired inversion loop could form, in principle, when the heteroduplex has extended across the entire region), and this large heterology could be the intermediate recognized by MMR proteins. We suggest that the junction between duplex DNA and unpaired single strands might provide an appropriate target for the Rad1p/Rad10p nuclease, which acts in conjunction with Msh2p and Msh3p to remove nonhomologous ends during recombination (47). Consistent with this possibility, recombination rates between identical sequences are also elevated in a rad1 mutant, but not in an msh6 or pms1 mutant (4a).
One potential problem with the sister chromatid conversion model is that it involves the extension of heteroduplex DNA through a large region of heterology. In the HIS3::intron system used here, the distance between the IR substrates is approximately 1.1 kb. Available evidence indicates that heteroduplex extension through a region this size should not present an obstacle for the yeast recombination machinery. Mitotic gene conversion of lys2 deletions in the 1- to 2-kb range has been reported (8), as well as conversion of a 6-kb Ty element inserted within the URA3 locus (50).
The sister chromatid conversion model applies not only to our system but also to other recombination assay systems that use IRs. The general assumption that the reorientation of the region between IRs is diagnostic of an intrachromatid crossover is likely to be incorrect. The possibility of sister chromatid conversion adds additional complexity to the types of recombination events that can occur between IRs and may alter interpretations of existing data. The majority of recombination within the yeast rDNA array also has been shown to correspond to gene conversion rather than to crossover events (17), so one might argue that crossing over is generally very rare in mitosis. It should be noted, however, that ectopic recombination events involving nonhomologous chromosomes frequently generate reciprocal translocations (25, 27).
Lengths of conversion tracts in wild-type cells.
We believe that our data are most consistent with the sister chromatid conversion model, which involves coconversion of sequences flanking the invertible HIS3::intron segment. The extent of a conversion tract thus becomes the sum of two conversion tracts, one on either side of the invertible segment. Furthermore, those events with no conversion tract would not have two endpoints in the same interval but rather would have an endpoint in each of the relevant identical intervals that flank the invertible segment. For each recombinant examined, DNA sequencing data were used to calculate minimal, maximal, and average tract lengths. Table 2 presents the mathematical mean values for the minimal, maximal, and average tract lengths, as well as the mean number of mismatches included in conversion tracts. With the cβ2 substrates in orientation 1, the mean of the average conversion tract lengths was 275 bp in wild-type cells, and tracts included an average of 14.4 mismatches. Very similar results were obtained with the orientation 2 cβ2 substrates, where the mean of the average conversion tract lengths was 230 bp and tracts included an average of 14.2 mismatches. The mean of the average tract lengths (as well as the means of the minimal and maximal lengths) was slightly larger with the orientation 1 substrates than with the orientation 2 substrates, although the mean number of mismatches converted was the same. This likely reflects the fact that the orientation 1 substrates had the end with the lower mismatch density (the 3′ end) closest to the invertible segment (whose conversion was selected for), whereas the orientation 2 substrates had the end with the higher mismatch density closest to the invertible segment. The relevant parameter for regulating heteroduplex length in MMR-competent cells thus appears to be the number of mismatches traversed. An examination of the distribution of conversion tract endpoints for the orientation 1 and orientation 2 substrates (Fig. 4A and B, respectively) is consistent with this interpretation. As noted previously, there is an excess of endpoints proximal to the invertible segment for both substrate orientations. If 14 total mismatches are included in heteroduplex intermediates, then conversion tracts should include an average of 7 mismatches on each side of the invertible segment and the transition from an excess to a deficit of endpoints should occur approximately 7 mismatches from the invertible segment. As predicted, the position of the shift from an excess of endpoints to a deficit of endpoints occurs seven and eight mismatches away from the invertible segment for the orientation 1 and orientation 2 substrates, respectively.
TABLE 2.
Conversion tract parametersa
| Parameter | Mean value (95% CI) in the following background:
|
||
|---|---|---|---|
| WTb | msh2 msh3 mutantc | pms1 mutantd | |
| Orientation 1 | |||
| Minimal length (bp) | 252 (211, 293) | 364 (321, 407) | 304 (232, 376) |
| Maximal length (bp) | 298 (260, 336) | 406 (365, 447) | 350 (283, 417) |
| Average length (bp) | 275 (235, 315) | 385 (343, 428) | 327 (257, 397) |
| No. of mismatches converted | 14.4 (12, 16.8) | 21.2 (18.5, 23.9) | 17.6 (13.3, 21.9) |
| Orientation 2 | |||
| Minimal length (bp) | 212 (153, 271) | 319 (270, 368) | |
| Maximal length (bp) | 248 (186, 310) | 359 (308, 410) | |
| Average length (bp) | 230 (170, 290) | 339 (289, 389) | |
| No. of mismatches converted | 14.2 (10.3, 18.1) | 21.5 (18.4, 24.6) | |
The minimal, maximal, and average conversion tract lengths, as well as the number of mismatches converted, were determined according to the sister chromatid conversion model. The numbering system for nucleotide positions is that shown in Fig. 2A, where position 1 is the 5′-most nucleotide. The minimal extent of a given conversion tract was calculated as the sum of conversion tracts to the left and right of the invertible segment (see Fig. 6). For the substrates in orientation 1, each of the 5′ ends was distal, and each of the 3′ ends was proximal, to the invertible segment. The left and right conversion tract lengths were calculated individually by subtracting the position of the most distal mismatch converted (i.e., the most distal with respect to the invertible segment) from 350 bp. For the substrates in orientation 2, the 5′ and 3′ ends of the substrates were proximal and distal, respectively, to the invertible segment. The left and right conversion tract lengths were calculated individually by subtracting 6 bp from the position of the most distal mismatch converted. To determine the maximal length of a given conversion tract obtained by using either orientation 1 or orientation 2 substrates, the lengths of the identity intervals that flanked the minimal tract on either side were added to the minimal tract length. The average length of a given conversion tract was calculated as the mean of the minimal and maximal conversion tract lengths. The number of mismatches converted was counted manually for each recombinant according to the sister chromatid conversion model.
WT, wild type. The number of conversion tracts included in the analysis was 56 for orientation 1 substrates and 24 for orientation 2 substrates.
The number of conversion tracts included in the analysis was 52 for orientation 1 substrates and 25 for orientation 2 substrates.
Twenty-seven conversion tracts were included in the analysis.
The mean lengths of conversion tracts in this study are surprisingly similar to ranges reported in other mitotic studies (2, 30, 35, 49), but direct comparisons between these studies are difficult given the very wide variety of assay systems used. Only two studies have estimated conversion tracts for mitotic events involving chromosomal sequences. In one of these studies, Harris et al. (20) sequenced 13 recombinants involving the 85%-identical PMA1 and PMA2 genes, and the mean of the average conversion tract lengths was approximately 250 bp. In the second study, widely spaced, naturally occurring restriction site polymorphisms were used to estimate conversion tracts that encompassed the URA3 locus on chromosome V (26). In contrast to the relatively short conversion tracts (less than 1 kb) observed in other studies, one-half of tracts involving URA3 were at least 4.2 kb in length. The strikingly longer tracts in the study by Judd and Petes (26) could reflect the very high degree of sequence identity between homologous chromosomes and thus would be consistent with the notion that conversion tract lengths are negatively impacted by mismatches in heteroduplex recombination intermediates (see below). Using a transformation-based assay, Supply et al. (48) reached a similar conclusion and suggested that mismatches constitute an effective recombination barrier. In their system, they observed that a high mismatch density near the ends of recombining fragments resulted in short conversion tracts, while a low mismatch density was associated with much longer tracts. Based on available data, one can predict that the lengths of conversion tracts should be inversely proportional to the levels of sequence divergence between the substrates. It should be possible to test this prediction by using the intron-based assay system described here.
In estimating the extent of heteroduplex DNA formed during mitotic recombination in wild-type cells, we have made the simplifying assumptions that gene conversion events correspond to repair tracts and are an accurate reflection of the heteroduplex DNA intermediate. It is possible, however, that the conversion tracts we have measured systematically underestimate the length of the heteroduplex intermediate. This could occur if repair tracts encompass only part of the mismatched heteroduplex intermediate or if repair of an intermediate yields a mixture of both gene conversion and restoration. Although there are no mitotic data that deal specifically with these issues, meiotic studies indicate that repair tracts are generally long and continuous (34).
Lengths of conversion tracts in MMR-defective cells.
Gene conversion tracts presumably arise via repair of heteroduplex DNA and replicative segregation of heteroduplex DNA, respectively, in wild-type and MMR-defective cells. Although they are generated in mechanistically distinct manners, we suggest that conversion tracts generated in the presence or absence of MMR should nevertheless be an accurate reflection of the underlying heteroduplex recombination intermediate. Measuring gene conversion tracts may not be the ideal way to obtain estimates of heteroduplex DNA in mitotically dividing cells, but it is presently the only way to do so.
Elimination of Msh2p and Msh3p had the effect of lengthening gene conversion tracts approximately 50% (Table 2). For the cβ2 repeats in orientation 1, the mean of the average tract lengths was 385 bp and tracts included a mean of 21.2 mismatches (versus 275 bp and 14.4 mismatches in wild-type cells). For the orientation 2 cβ2 repeats, the mean of the average conversion tract lengths was 339 bp and tracts included a mean of 21.5 mismatches (versus 230 bp and 14.2 mismatches in wild-type cells). The differences in tract length between wild-type and MMR-defective cells are highly significant (P < 0.005 by Student’s t test). Data obtained by Negritto et al. (32) also are consistent with conversion tracts being longer in MMR-defective cells than in wild-type cells. Their study utilized the 83%-identical yeast SAM1 and SAM2 genes, and it was noted that coconversion of two restriction site polymorphisms with a selected site was more common in msh2 cells than in wild-type cells.
In recombination assays involving diverged sequences, Msh2p exerts a stronger antirecombination activity than does Pms1p (12, 44). With the 94%-identical substrates used in this study, elimination of MSH2 stimulates mitotic recombination approximately 40-fold, whereas elimination of PMS1 stimulates recombination only 15-fold (29a). The mean of the average conversion tract lengths in a pms1 mutant was 327 bp, which was between the lengths observed in wild-type and msh2 msh3 strains (275 and 385 bp, respectively). Similarly, the mean number of mismatches converted in a pms1 mutant (17.6) was between the numbers observed in wild-type and msh2 msh3 strains (14.4 and 21.2, respectively). Although the differences between pms1Δ strain conversion tracts and those of either the wild-type or msh2Δ strain are not statistically significant (0.14 < P < 0.18 by Student’s t test), we suggest that mismatches have a greater impact on heteroduplex length when Pms1p is present, which is consistent with more-efficient recognition of mismatches by the yeast MutS homologs in the presence of MutL homologs (19).
Relation of conversion tract lengths and endpoints to the antirecombination activity of MMR proteins.
If one assumes that gene conversion tracts are an accurate representation of the extent of heteroduplex DNA formed in the presence of MMR proteins, then our data indicate that MMR proteins regulate the formation of heteroduplex DNA during mitotic recombination. Specifically, heteroduplex tracts are shorter and cover fewer mismatches in wild-type cells than in MMR-defective cells (Table 2). The tract shortening can account for the biased distribution of conversion tract endpoints observed in wild-type cells, where endpoints were clustered at the substrate end closest to the invertible segment (Fig. 4). To our knowledge, no other model could account for such an MMR-dependent clustering of conversion tract endpoints. The idea that MMR proteins might regulate heteroduplex formation when mismatches are present is not a new one (see references 10 and 37), but exactly how this might occur is not clear. MMR proteins could monitor the fidelity of the initial strand exchange reaction, they could regulate the extension of heteroduplex DNA, or they could bias how recombination intermediates are resolved. It also is possible that these proteins could act at more than one step. Recent data from mammalian cells suggest that the initiation, but not the extension, of heteroduplex formation is blocked by mismatches, although the involvement of the MMR machinery was not examined (56). Based on studies of meiotic gene conversion polarity gradients, Alani et al. (3) suggested that MMR proteins specifically block the extension of symmetric heteroduplex intermediates and should, therefore, bias resolution in favor of noncrossovers (see also reference 23). The relevance of the meiotic studies to the data reported here, however, is unclear, since our assay involves recombination between highly mismatched substrates in mitosis.
Regardless of the precise mechanistic explanation, our data indicate that MMR proteins modulate the structures of mitotic heteroduplex recombination intermediates. We suggest that the MMR-associated conversion tract shortening reflects a blockage of heteroduplex extension through mismatched regions, and we think it likely that MMR proteins are associated with the hypothetical yeast recombinasome. A critical question that has not been addressed is how the observed MMR-dependent shortening of cβ2 gene conversion tracts relates to the potent antirecombination activity of MMR proteins reported previously (13). Specifically, can a 50% shortening of conversion tracts by MMR proteins directly account for the observed 50-fold inhibition of recombination? We think not, and we propose the following model to relate the two observations. We suggest that the shortening of conversion tracts by the MMR machinery reflects the role of these proteins in impeding the extension of heteroduplex DNA through mismatched regions. One plausible scenario is that the MMR-associated slowing of heteroduplex extension triggers a helicase-catalyzed reversal of heteroduplex formation, which would be analogous to reverse branch migration. We note that a similar model was proposed by Zahrt and Maloy (57) to explain data obtained in transductions between closely related Salmonella species. According to such a heteroduplex rejection model, there would be competition between extension and removal of heteroduplex DNA, and under normal circumstances, the forward reaction would be highly favored. In the presence of mismatches, however, MMR proteins would impede forward progress, and the reverse reaction would be favored. If MMR proteins are directly associated with the recombination machinery, then it is not difficult to imagine that binding to mismatches in newly formed heteroduplex DNA as the machinery progresses might slow down the forward reaction. The few recombinants detected in MMR-competent cells thus provide a snapshot of the MMR-imposed barrier to heteroduplex extension and represent those few heteroduplexes that survive long enough to produce mature recombinants. The confirmation of this model will likely depend on the further development of in vitro reactions that allow the progress of recombination between mismatched sequences to be directly monitored.
ACKNOWLEDGMENTS
We are indebted to Lorraine Symington for originally pointing out sister chromatid conversion as an alternative to intrachromatid crossover, and we acknowledge the excellent technical assistance of Neal Graber. We thank Gray Crouse and members of the S.J.-R. lab for helpful comments on the manuscript. We also thank Hannah Klein and another, anonymous reviewer for constructive criticisms of the manuscript.
This work was supported by National Institutes of Health (NIH) grant GM38464 (to S.J.-R.). W.C. was supported in part by the Graduate Division of Biological and Biomedical Sciences and by an NIH Medical Scientist Training grant (Emory University).
REFERENCES
- 1.Abdulkarim F, Hughes D. Homologous recombination between the tuf genes of Salmonella typhimurium. J Mol Biol. 1996;260:506–522. doi: 10.1006/jmbi.1996.0418. [DOI] [PubMed] [Google Scholar]
- 2.Ahn B-Y, Livingston D M. Mitotic gene conversion lengths, coconversion patterns, and the incidence of reciprocal recombination in a Saccharomyces cerevisiae plasmid system. Mol Cell Biol. 1986;6:3685–3693. doi: 10.1128/mcb.6.11.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alani E, Reenan R A G, Kolodner R D. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics. 1994;137:19–39. doi: 10.1093/genetics/137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailis A M, Rothstein R. A defect in mismatch repair in Saccharomyces cerevisiae stimulates ectopic recombination between homeologous genes by an excision repair-dependent process. Genetics. 1990;126:535–547. doi: 10.1093/genetics/126.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Bayliss, A., M. Hendrix, J. McDougal, S. Jinks-Robertson, and G. Crouse. Unpublished data.
- 5.Borts R H, Haber J E. Meiotic recombination in yeast: alteration by multiple heterozygosities. Science. 1987;237:1459–1465. doi: 10.1126/science.2820060. [DOI] [PubMed] [Google Scholar]
- 6.Borts R H, Leung W-Y, Kramer W, Kramer B, Williamson M, Fogel S, Haber J E. Mismatch repair-induced meiotic recombination requires PMS1 gene product. Genetics. 1990;124:573–584. doi: 10.1093/genetics/124.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers S R, Hunter N, Louis E J, Borts R H. The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Mol Cell Biol. 1996;16:6110–6120. doi: 10.1128/mcb.16.11.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chernoff Y O, Kidgotko O V, Demberelijn O, Luchnikova I L, Soldatov S P, Glazer V M, Gordenin D A. Mitotic intragenic recombination in the yeast Saccharomyces: marker-effects on conversion and reciprocity of recombination. Curr Genet. 1984;9:31–374. doi: 10.1007/BF00396201. [DOI] [PubMed] [Google Scholar]
- 9.Ciotta C, Ceccotti S, Aquilina G, Humbert O, Palombo F, Jiricny J, Bignami M. Increased somatic recombination in methylation-tolerant human cells with defective DNA mismatch repair. J Mol Biol. 1998;276:705–719. doi: 10.1006/jmbi.1997.1559. [DOI] [PubMed] [Google Scholar]
- 10.Claverys J-P, Lacks S A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986;50:133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crouse G F. Mismatch repair systems in Saccharomyces cerevisiae. In: Nickoloff J A, Hoekstra M F, editors. DNA damage and repair. 1. DNA repair in prokaryotes and lower eukaryotes. Totowa, N.J: Humana Press; 1998. pp. 411–448. [Google Scholar]
- 12.Datta A, Adjiri A, New L, Crouse G F, Jinks-Robertson S. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1085–1093. doi: 10.1128/mcb.16.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datta A, Hendrix M, Lipsitch M, Jinks-Robertson S. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc Natl Acad Sci USA. 1997;94:9757–9762. doi: 10.1073/pnas.94.18.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 15.Elliott B, Richardson C, Winderbaum J, Nickoloff J A, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinstein S I, Low K B. Hyper-recombining recipient strains in bacterial conjugation. Genetics. 1986;113:13–33. doi: 10.1093/genetics/113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gangloff S, Zou H, Rothstein R. Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 1996;15:1715–1725. [PMC free article] [PubMed] [Google Scholar]
- 18.Geitz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Habraken Y, Sung P, Prakash L, Prakash S. Enhancement of MSH2-MSH3-mediated mismatch recognition by the yeast MLH1-PMS1 complex. Curr Biol. 1997;7:790–793. doi: 10.1016/s0960-9822(06)00337-x. [DOI] [PubMed] [Google Scholar]
- 20.Harris S, Rudnicki K S, Haber J E. Gene conversions and crossing over during homologous and homeologous ectopic recombination in Saccharomyces cerevisiae. Genetics. 1993;135:5–16. doi: 10.1093/genetics/135.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 22.Humbert O, Prudhomme M, Hakenbeck R, Dowson C G, Claverys J-P. Homeologous recombination and mismatch repair during transformation in Streptococcus pneumoniae: saturation of the Hex mismatch repair system. Proc Natl Acad Sci USA. 1995;92:9052–9056. doi: 10.1073/pnas.92.20.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter N, Borts R H. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 1997;11:1573–1582. doi: 10.1101/gad.11.12.1573. [DOI] [PubMed] [Google Scholar]
- 24.Hunter N, Chambers S R, Louis E J, Borts R H. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 1996;15:1726–1733. [PMC free article] [PubMed] [Google Scholar]
- 25.Jinks-Robertson S, Petes T D. Chromosomal translocations generated by high-frequency meiotic recombination between repeated yeast genes. Genetics. 1986;114:731–752. doi: 10.1093/genetics/114.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Judd S R, Petes T D. Physical lengths of meiotic and mitotic gene conversion tracts in Saccharomyces cerevisiae. Genetics. 1988;118:401–410. doi: 10.1093/genetics/118.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lichten M, Haber J E. Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics. 1989;123:261–268. doi: 10.1093/genetics/123.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majewski J, Cohan F M. The effect of mismatch repair and heteroduplex formation on sexual isolation in Bacillus. Genetics. 1998;148:13–18. doi: 10.1093/genetics/148.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matic I, Rayssiguier C, Radman M. Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell. 1995;80:507–515. doi: 10.1016/0092-8674(95)90501-4. [DOI] [PubMed] [Google Scholar]
- 29a.McDougal, J., and S. Jinks-Robertson. Unpublished data.
- 30.Mezard C, Pompon D, Nicolas A. Recombination between similar but not identical DNA sequences during yeast transformation occurs within short stretches of identity. Cell. 1992;70:659–670. doi: 10.1016/0092-8674(92)90434-e. [DOI] [PubMed] [Google Scholar]
- 31.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 32.Negritto M T, Wu X, Kuo T, Chu S, Bailis A M. Influence of DNA sequence identity on efficiency of targeted gene replacement. Mol Cell Biol. 1997;17:278–286. doi: 10.1128/mcb.17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilsson-Tillgren T, Gjermansen C, Holmberg S, Petersen M C L, Kielland-Brandt M C. Analysis of chromosome V and the ILV1 gene from Saccharomyces calsbergensis. Carlsberg Res Commun. 1986;51:309–326. [Google Scholar]
- 34.Petes T D, Malone R E, Symington L S. Recombination in yeast. In: Broach J R, Pringle J R, Jones E W, editors. The molecular biology of the yeast Saccharomyces: genome dynamics, protein synthesis and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 407–521. [Google Scholar]
- 35.Porter G, Westmoreland J, Priebe S, Resnick M A. Homologous and homeologous intermolecular gene conversion are not differentially affected by mutations in the DNA damage or mismatch repair genes RAD1, RAD50, RAD51, RAD52, RAD54, PMS1 and MSH2. Genetics. 1996;143:755–767. doi: 10.1093/genetics/143.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Priebe S D, Westmoreland J, Nilsson-Tillgren T, Resnick M A. Induction of recombination between homologous and diverged DNAs by double-strand gaps and breaks and role of mismatch repair. Mol Cell Biol. 1994;14:4802–4814. doi: 10.1128/mcb.14.7.4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radman M. Mismatch repair and genetic recombination. In: Kucherlapati R, Smith G R, editors. Genetic recombination. Washington, D.C: American Society for Microbiology; 1988. pp. 169–192. [Google Scholar]
- 38.Radman M. Avoidance of inter-repeat recombination by sequence divergence and a mechanism of neutral evolution. Biochimie. 1991;73:357–361. doi: 10.1016/0300-9084(91)90101-6. [DOI] [PubMed] [Google Scholar]
- 39.Rayssiguier C, Thaler D S, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 40.Resnick M A, Zgaga Z, Hieter P, Westmoreland J, Fogel S, Nilsson-Tillgren T. Recombinational repair of diverged DNAs: a study of homeologous chromosomes and mammalian YACs in yeast. Mol Gen Genet. 1992;234:65–73. doi: 10.1007/BF00272346. [DOI] [PubMed] [Google Scholar]
- 41.Ronne H, Rothstein R. Mitotic sectored colonies: evidence of heteroduplex DNA formation during direct repeat recombination. Proc Natl Acad Sci USA. 1988;85:2695–2700. doi: 10.1073/pnas.85.8.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rothstein R, Helms C, Rosenberg N. Concerted deletions and inversions are caused by mitotic recombination between delta sequences in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:1198–1207. doi: 10.1128/mcb.7.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selva E M, Maderazo A B, Lahue R S. Differential effects of the mismatch repair genes MSH2 and MSH3 on homeologous recombination in Saccharomyces cerevisiae. Mol Gen Genet. 1997;257:71–82. doi: 10.1007/pl00008619. [DOI] [PubMed] [Google Scholar]
- 44.Selva E M, New L, Crouse G F, Lahue R S. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen P, Huang H V. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen P, Huang H V. Effect of base pair mismatches on recombination via the RecBCD pathway. Mol Gen Genet. 1989;218:358–360. doi: 10.1007/BF00331291. [DOI] [PubMed] [Google Scholar]
- 47.Sugawara N, Paques F, Colaiacovo M, Haber J E. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Supply P, de Kerchove D’Exaerde A, Roganti T, Goffeau A, Foury F. In-frame recombination between the yeast H+-ATPase isogenes PMA1 and PMA2: insights into the mechanism of recombination initiated by a double-strand break. Mol Cell Biol. 1995;15:5389–5395. doi: 10.1128/mcb.15.10.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sweetser D B, Hough H, Wheldon J, Arbuckle M, Nickoloff J A. Fine-resolution mapping of spontaneous and double-strand break-induced gene conversion tracts in Saccharomyces cerevisiae reveals reversible mitotic conversion polarity. Mol Cell Biol. 1994;14:3863–3875. doi: 10.1128/mcb.14.6.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vincent A, Petes T D. Mitotic and meiotic gene conversion of Ty elements and other insertions in Saccharomyces cerevisiae. Genetics. 1989;122:759–772. doi: 10.1093/genetics/122.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vulic M, Dionisio F, Taddei F, Radman M. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc Natl Acad Sci USA. 1997;94:9763–9767. doi: 10.1073/pnas.94.18.9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waldman A S, Liskay R M. Differential effects of base-pair mismatch on intrachromosomal versus extrachromosomal recombination in mouse cells. Proc Natl Acad Sci USA. 1987;84:5340–5344. doi: 10.1073/pnas.84.15.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westmoreland J, Porter G, Radman M, Resnick M A. Highly mismatched molecules resembling recombination intermediates efficiently transform mismatch repair proficient Escherichia coli. Genetics. 1997;145:29–38. doi: 10.1093/genetics/145.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willis K K, Klein H L. Intrachromosomal recombination in Saccharomyces cerevisiae: reciprocal exchange in an inverted repeat and associated gene conversion. Genetics. 1987;117:633–643. doi: 10.1093/genetics/117.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Worth L, Jr, Clark S, Radman M, Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc Natl Acad Sci USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang D, Waldman A S. Fine-resolution analysis of products of intrachromosomal homeologous recombination in mammalian cells. Mol Cell Biol. 1997;17:3614–3628. doi: 10.1128/mcb.17.7.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zahrt T C, Maloy S. Barriers to recombination between closely related bacteria: MutS and RecBCD inhibit recombination between Salmonella typhimurium and Salmonella typhi. Proc Natl Acad Sci USA. 1997;94:9786–9791. doi: 10.1073/pnas.94.18.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zahrt T C, Mora G C, Maloy S. Inactivation of mismatch repair overcomes the barrier to transduction between Salmonella typhimurium and Salmonella typhi. J Bacteriol. 1994;176:1527–1529. doi: 10.1128/jb.176.5.1527-1529.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zawadzki P, Roberts M S, Cohan F M. The log-linear relationship between sexual isolation and sequence divergence in Bacillus transformation is robust. Genetics. 1995;140:917–932. doi: 10.1093/genetics/140.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]