Abstract
Methylation at the DNA sequence 5′-CpG is required for mouse development. MeCP2 and MBD1 (formerly PCM1) are two known proteins that bind specifically to methylated DNA via a related amino acid motif and that can repress transcription. We describe here three novel human and mouse proteins (MBD2, MBD3, and MBD4) that contain the methyl-CpG binding domain. MBD2 and MBD4 bind specifically to methylated DNA in vitro. Expression of MBD2 and MBD4 tagged with green fluorescent protein in mouse cells shows that both proteins colocalize with foci of heavily methylated satellite DNA. Localization is disrupted in cells that have greatly reduced levels of CpG methylation. MBD3 does not bind methylated DNA in vivo or in vitro. MBD1, MBD2, MBD3, and MBD4 are expressed in somatic tissues, but MBD1 and MBD2 expression is reduced or absent in embryonic stem cells which are known to be deficient in MeCP1 activity. The data demonstrate that MBD2 and MBD4 bind specifically to methyl-CpG in vitro and in vivo and are therefore likely to be mediators of the biological consequences of the methylation signal.
DNA methylation is the major modification of eukaryote genomes. In vertebrates, this occurs predominantly at position 5 of cytosines when followed by guanosine (CpG). DNA methylation can repress transcription and for this reason has been implicated in stable alterations of gene expression in development (3). Whereas the genomes of certain invertebrates appear to contain “compartments” of either mostly methylated or mostly unmethylated DNA (43), the somatic genomes of vertebrates are globally methylated, with the exception of so-called CpG islands (6). CpG islands are GC-rich regions of DNA, stretching for an average of about 1 kb, which are coincident with the promoters of approximately 60% of human RNA polymerase II-transcribed genes (1). Methylation of CpG islands and subsequent silencing of associated transcription units have been found to occur in genes located on the inactive X chromosome (39), genes silenced by genomic imprinting (36, 38), and genes silenced in transformed cell lines and tumors (2, 8, 16, 18, 40). DNA methylation is known to play an essential role in mammalian development because mice lacking a functional gene encoding the maintenance DNA methyltransferase (DNMT) are developmentally retarded and die at midgestation (29). In contrast to the situation in somatic cells, undifferentiated embryonic stem (ES) cells lacking a functional DNMT gene apparently grow normally despite containing approximately 5% of the wild-type DNA methylation level (25, 35).
One mechanism by which DNA methylation can cause transcriptional repression is by directly interfering with the binding of sequence-specific transcription factors to DNA. Some transcription factors have been shown to be unable to bind to their target sequences when methylated (14, 21). The observations that DNA methylation is capable of repressing transcription at some distance (12, 23) and that repression of transgenes only occurs after chromatin assembly (11) are inconsistent with the direct mechanism and indicate that a more indirect mechanism also exists. Two proteins have been identified, MeCP1 and MeCP2, which bind specifically to methylated DNA in any sequence context (27, 30). Both are capable of inhibiting transcription (9, 32). It is likely that MeCP1 and MeCP2 are important in interpreting the signal that methylation of DNA represents.
MeCP2 consists of a single polypeptide that contains both a methyl-CpG binding domain (MBD) and transcriptional repression domain (TRD) (32, 33). MeCP2 is capable of binding to a single symmetrically methylated CpG pair and was found to bind to chromosomes at sites known to contain methylated DNA (35). On mouse chromosomes, this is visualized as prominent binding to the highly methylated major satellite located just proximal to centromeres, whereas on human or rat chromosomes which do not contain highly methylated satellite DNAs, general chromosomal binding is observed (32). MeCP2 localization is disrupted in ES cells lacking a functional DNMT gene, demonstrating that MeCP2 is a bona fide methyl-CpG binding protein in vivo (35). Like DNMT-deficient ES cells, MeCP2-deficient ES cells appear to grow normally in culture but are not capable of supporting embryonic development (42). In contrast to DNMT deficiency, however, MeCP2 deficiency is compatible with somatic cell viability, and certain imprinted genes known to be misexpressed in the absence of DNMT (5, 28) are not misexpressed in MeCP2-deficient cells (17a). These observations suggest that the effects of DNA methylation are not solely dependent on MeCP2, as the DNA methylation-dependent transcriptional silencing observed at imprinted loci does not appear to rely upon its presence. The known characteristics of MeCP2 point to it being responsible for genomewide transcriptional repression or “transcriptional noise reduction” (7). Its involvement in the repression of specific genes remains unproven.
In contrast to MeCP2, MeCP1 has been shown to require >10 methyl-CpGs to bind DNA (30), making it more likely to be involved in DNA methylation-mediated transcriptional repression of specific genes. Indeed, Boyes and Bird have shown that MeCP1 is capable of repressing transcription of densely methylated promoters (9). Further, MeCP1 was found to be capable of repressing transcription from sparsely methylated promoters, though the interaction is weak and can be overcome by the presence of an enhancer (10). MeCP1 activity is ubiquitous in somatic cells and tissues but is notably absent in ES cells which are also tolerant of DNMT deficiency (30). Thus, MeCP1 is likely to be important in the DNA methylation-mediated repression of genes in somatic cells.
MeCP1 is a large protein complex of 400 to 800 kDa. Cross et al. (15) recently identified a component of MeCP1 by searching the XREF database (4) for sequences homologous to the MBD of MeCP2. A human expressed sequence tag (EST) was identified and found to encode a novel protein containing a MBD-like motif located at its N terminus. This protein, called MBD1 (formerly PCM1), was shown to bind methylated DNA via its MBD-like region and to repress transcription from a methylated promoter in vitro. MBD1 is a component of the MeCP1 protein complex. We now report the identification of three novel MBD-containing proteins. We show that two of these novel proteins specifically bind methylated DNA in vitro and colocalize with methylated sequences in vivo and are thus candidates for mediators of the effects of DNA methylation in mammalian cells.
MATERIALS AND METHODS
cDNA identification and cloning.
Full-length human MeCP2 and the MBD region of MBD1 protein sequences were used to search the XREF database (http://www.ncbi.nlm.nih.gov/XREFdb) or dbEST directly via BLAST (NCBI BLAST Server; http://www.ncbi.nlm.nih.gov/BLAST). The murine homologue of MBD1 was not present in the EST database so degenerate primers designed from the amino acid sequences of the cysteine-rich motifs of the human MBD1 gene (primer G235 [5′-CARACNCARGARGAYTGYGG-3′] and primer D355 [5′-CCNCCRAAYTTRGGYTTRTC-3′]; designed by M. Carr) were used to amplify a portion of the murine cDNA. This PCR product was then used to screen a mouse brain cDNA library (Stratagene), and full-length clones were isolated and sequenced. Approximately 2 × 105 plaques were plated out and lifted onto nitrocellulose filters. Filters were hybridized in Church buffer (7% sodium dodecyl sulfate [SDS], 0.5M sodium phosphate [pH 7.2]) supplemented with 15 μg of denatured salmon sperm DNA per ml at 68°C overnight and washed to a final stringency of 0.1% SDS–0.1 × SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 65°C.
Mbd2 and Mbd3 were identified by database screening as ESTs (accession no. Z31258 for Mbd2 and accession nos. W81894 and W91000 for Mbd3). The corresponding cDNA clones (MMTEST693, 403236, and 420899, respectively) were obtained and used to screen the mouse brain cDNA library. A number of full-length clones were isolated and sequenced for both Mbd2 and Mbd3. Human MBD2 and MBD3 sequences were identified by subsequent screening of the dbEST database with full-length Mbd2 and Mbd3 sequences. No full-length MBD2 or MBD3 IMAGE clones were identified in database screens so 5′ rapid amplification of cDNA ends (RACE) was used to obtain the 5′ end of MBD3 by the 5′ RACE protocol of Boehringer Mannheim. To obtain the 5′ end of MBD2, a gridded cosmid library (Lawrence Livermore National Laboratories cosmid library LL18NC02; provided by the United Kingdom Human Genome Mapping Project Resource Centre [HGMP], Cambridge, England) was screened with a PCR product corresponding to the 5′ end of MBD2 intron 1 (PCR product of primers 5′-AAAATCTGGGCTAAGTGCTGG-3′ and 5′-ACGCCTTCCACATAAGATGC-3′), using the hybridization conditions described above. Two independent cosmids (clones AD15-O10 and AD17-O6) were identified and subcloned, and the relevant regions were sequenced.
MBD4 was identified as an EST (AA149549). The corresponding clone (588272) was sequenced and found to contain a long open reading frame. No ESTs for Mbd4 were present in the database, so the human cDNA was used as a probe against a mouse 129 genomic lambda library (a gift from A. J. H. Smith). Hybridization conditions were as described above, but filters were washed to a final stringency of 1% SDS–0.5 M NaCl–50 mM Tris (pH 8) at 65°C. Positive clones were identified which, upon subcloning and sequencing, were found to contain the Mbd4 gene. Genomic clones were used to screen a mouse brain cDNA library which resulted in the identification of one partial cDNA containing a short poly(A) tail. This cDNA was used as a probe on a gridded mouse cDNA library (embryonic region cDNA library ER-IV, HGMP; 17), and another partial cDNA clone which ends in a poly(A) at the same location was identified. The full-length Mbd4 sequence was determined by sequencing of cDNAs and by comparison of genomic sequence to that of the human MBD4 cDNA, followed by verification by RT-PCR, 5′ RACE, and sequencing of products.
All IMAGE consortium (LLNL) cDNA clones (26) described here were obtained from the HGMP, except for clones 400458, 403236, and 420899, which were obtained from Research Genetics; clone HIBAA05 (MBD3), which was obtained from the American Type Culture Collection; and clone MMTEST (46), which was obtained from Christer Höög, Stockholm, Sweden. Sequencing was performed with an ABI Automated Sequencer 373A Stretch apparatus. Contig construction, sequence analysis, and comparison was performed by using the Lasergene DNA analysis software package (DNAStar).
Expression analysis.
Total RNA was isolated from tissues of adult mice using acid guanidinium thiocyanate-phenol-chloroform extraction (13). Ten or 20 μg of total RNA was used per lane on Northern blots. Blots were hybridized in Church buffer as described above. Blots were washed to a final stringency of 0.1% SDS–0.1× SSC at 68°C for all probes except Mbd4, which was washed to a final stringency of 1% SDS–0.5 M NaCl–50 mM Tris (pH 8) at 65°C. Signal was detected with a PhosphorImager (Molecular Dynamics). For reverse transcription-PCR (RT-PCR) analysis, 5 μg of total RNA was reverse transcribed at 42°C by Moloney murine leukemia virus reverse transcriptase for 2 h according to the manufacturer’s instructions (Life Technologies). One microliter of the reverse transcriptase reaction mixture was then subjected to 30 cycles of PCR.
Construction of expression vectors and protein expression.
The coding sequences for all four genes were PCR amplified (Mbd1, 5′-AAGCATCCATGGCTGAGTCCTGGC-3′ and 5′-GGGAGGGCAGTAATAAGGCCAGTCA-3′; Mbd2, 5′-GGGAAGACCATGGACTGCCCG-3′ and 5′-GCGGATCCTTACGCCTCATCTCCCTC-3′; Mbd3, 5′-GGCGCCATGGAGCGGAAGAGGTG-3′ and 5′-TCCCCCGGCACTCGCTCTGGCTCCGG-3′; Mbd4, 5′-GCGCCATGGAGAGCCCAAACCTTG-3′ and 5′-GCGGATCCAGGCAGCTTCAAGATAG-3′; MBD4, 5′-CCTGCTCCATGGGCACGACTGGGCTG-3′ and 5′-GCGGGATCCTGAGCTTGAAAGCTGCAG-3′) using Pfu polymerase (Stratagene) or Pwo polymerase (Boehringer Mannheim) according to the manufacturer’s instructions and cloned into the bacterial expression vector pET6H (19). Recombinant proteins were expressed in Escherichia coli BL21(DE3)/pLysS as described previously (15) and purified over a nickel agarose column (Ni2+-NTA-Superflow Agarose [Qiagen]). Extracts were loaded onto the column, washed in a solution containing 60 mM imidazole, 250 mM NaCl, 20 mM Tris (pH 7.9), 10% glycerol, 0.1% Triton X-100, and 10 mM 2-mercaptoethanol, and eluted in elution buffer (wash buffer containing 0.5 M imidazole). Proteins were further purified by fractionation over Fractogel EMD SO3--650(M) (Merck). Proteins partially purified on the nickel-agarose column were loaded onto the Fractogel column, washed in a solution containing 250 mM NaCl, 50 mM HEPES (pH 7), 10% glycerol, 0.1% Triton X-100, and 10 mM 2-mercaptoethanol, and eluted in the same buffer containing 1 M NaCl. All column wash and elution buffers were supplemented with complete protease inhibitor tablets (Boehringer Mannheim).
Constructs producing MBD1-GFP, MBD4-GFP, and MeCP2-GFP were made by PCR amplifying either cloned cDNAs (Mbd1) with Pfu polymerase as described above or reverse-transcribed total ES cell RNA (Mbd4) or murine brain RNA (Mecp2) with Pwo polymerase as described above, digesting the PCR products with HindIII and KpnI or KpnI alone (Mecp2), and ligating into the pCMXGFP mammalian expression vector. The MBD2-GFP construct was made by cloning the coding sequences (NaeI-SspI fragment) of a full-length Mbd2 cDNA into the HindIII site of pCMXGFP. pCMXGFP was constructed by K. Umesono by insertion of the green fluorescent protein (GFP) cDNA (37) into Asp718-BamHI-cut pCMX vector (44). The GFP cDNA is under the control of a cytomegalovirus promoter and both a polyomavirus enhancer and a simian virus 40 enhancer. This version of GFP contains the following amino acid changes: F64L, V163A, S175Q, I167T, and S65T. The GFP-MBD3 construct was made by PCR amplifying an Mbd3 cDNA with Pfu polymerase, digesting with BglII and SpeI, and ligating into the pCMXGFP2 mammalian expression vector. pCMXGFP2 was derived from pCMXGFP by mutating the stop codon for the GFP from TGA to GGG by PCR mutagenesis. Primers used were as follows: for Mbd1, 5′-GCGGAAGCTTATGGCTGAGTCCTGGCAG-3′ and 5′-GCGGTACCCAAAACTTCCTCCTTCAACTGC-3′; for Mbd3, 5′-GCAGATCTATGGAGCGGAAGAGGTGG-3′ and 5′-GCACTAGTACACTCGCTCTGGCTCC-3′; for Mbd4, 5′-GCGAAGCTTATGGAGAGCCCAAACCTTG-3′ and 5′-GCGGTACCAGGCAGCTCCAAGATAGAC-3′; for MeCP2, 5′-GCGGTACCATGGTAGCTGGGATGTTAG-3′ and 5′-GCGGTACCGCTAACTCTCTCGGTCACG-3′.
Band shifts.
Band shift assays were performed essentially as described previously (15) except that binding reactions were carried out at room temperature for 30 min and contained 100 ng of sonicated E. coli DNA for MBD1, MBD2, and MBD4 or 200 ng of E. coli DNA for MBD3. Complexes were electrophoresed through 2% agarose gels or 6% polyacrylamide gels in 0.5× TBE (Tris-borate EDTA) at 4°C. The GAM12 and GAC probes have been described previously (27). The double-stranded Sm probe was made by A. Prokhortchouk et al. (37a). The NB probe was made by excising a 125-bp NcoI-BamHI fragment from the MBD1 cDNA clone 222390 (15). Doubly methylated and fully methylated NB were made by methylation with HpaII and HhaI methylases or SssI methylase (New England Biolabs), respectively. The 51 probe was a gift from J. Huntriss and M. Monk (20). The MM1 and MM2 probes were designed by H.-H. Ng. GAM12, GAM5, GAC, MeCG11, and CG11 probes have been described previously (27, 30).
Cell culture, transfection, and fluorescence imaging.
L, HT1080, and CHO cells were grown on coverslips in minimal essential medium alpha (Life Technologies) supplemented with either 10% (L cells) or 5% (HT1080 and CHO cells) bovine calf serum (Hyclone). EFS2 cells were derived from an ES cell line containing a randomly integrated β-geo transgene (42). Embryonic fibroblasts were obtained from this line by injecting ES cells into blastocysts and selecting resistant cells (500 μg of Geneticin per ml and 10% bovine calf serum in minimal essential medium alpha [Life Technologies]) from dissociated embryos in culture (17b). ES cells were grown in Glasgow medium (Life Technologies) supplemented with 10% fetal calf serum (Globepharm), 1× nonessential amino acids (Life Technologies), 1 mM sodium pyruvate, 50 μM 2-mercaptoethanol, and soluble differentiation-inhibitory activity/leukemia-inhibitory factor (41) at 37°C in a 5% CO2 atmosphere. Ten micrograms of each expression construct was transfected at approximately 50% confluence by using Lipofectin or Lipofectamine (Life Technologies) onto 2-cm2 coverslip of cells at 50% confluence according to the manufacturer’s instructions. Approximately 36 h after transfection, cells were fixed in 4% paraformaldehyde in phosphate-buffered saline for 20 min at 37°C and then washed twice in phosphate-buffered saline. Coverslips were mounted in Vectashield (Vector Laboratories) containing 1 μg of 4′,6-diamidino-2-phenylindole (DAPI) per ml and visualized directly. Images were obtained using a Zeiss epifluorescence microscope fitted with a charge-coupled device camera (Photometrics) and controlled by a Macintosh computer running the Quips mFISH software (Vysis Inc.).
Nucleotide sequence accession numbers.
All sequences reported here are available in Genbank under accession nos. AF072240 to AF072252. Human EST contigs for MBD2, MBD3, and MBD4 can be found as entries Hs.25674, Hs.107254, and Hs.35947, respectively, through the UniGene web page: http://www.ncbi.nlm.nih.gov/Schuler/UniGene.
RESULTS
A family of MBD-containing proteins.
We sought to identify novel proteins containing a MBD-like motif by searching the EST database. In addition to Mecp2 and MBD1, three other genes (Mbd2, Mbd3, and Mbd4) were identified in humans and mice encoding proteins containing putative MBDs. Assignment of the initiator AUG codons for each gene was given to the first in-frame ATG in each case. While this location is conserved in the human and murine Mbd1, Mbd2, and Mbd3 genes, it is not conserved in human and murine Mbd4 genes. In both the human and murine Mbd4 genes, the AUG codon at which we propose translation initiates is the only in-frame methionine codon N terminal to the MBD-like region. The first in-frame start codons in the Mbd2 and MBD2 open reading frames is within a CpG island, and the second is located 152 codons downstream, just upstream of the MBD. These two potential initiator codons show 6 of 10 and 5 of 10 matches to the Kozak consensus sequence for initiator codons, respectively (24). Conceptual translation of the region between the first two ATG codons produces a repetitive amino acid sequence due to the high G+C content of the underlying DNA. At this point, we do not known which initiator methionine is used in vivo, though the high degree of conservation between the human and murine genes in this CpG island is consistent with the first methionine being the true initiator methionine.
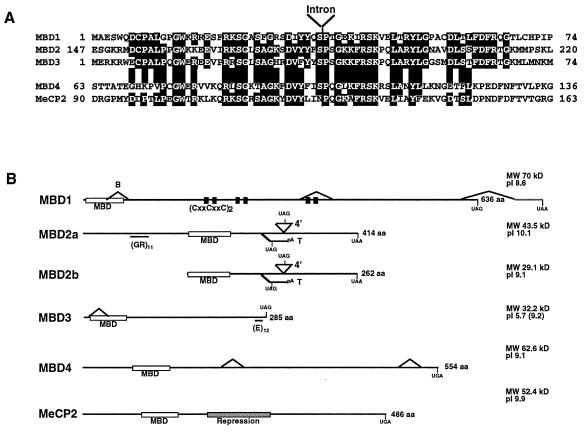
Alignment of the MBD-like regions from the murine MBD1 to MBD4 and MeCP2 proteins is shown in Fig. 1A. The MBD protein family comprises two subgroups based upon the sequences of the putative MBDs (Fig. 1A). The MBD of MBD4 is most similar to that of MeCP2 in primary sequence, while the MBDs of MBD1, MBD2, and MBD3 are more similar to each other than to those of either MBD4 or MeCP2. The MBDs within each protein appear to be related evolutionarily based on the presence of an intron located at a conserved position within all five genes (Fig. 1A).
FIG. 1.
Comparison of MBD family proteins. (A) Amino acid alignment of the MBDs from murine MBD1, MBD2, MBD3, MBD4, and MeCP2 proteins. Amino acids identical in three or more proteins are indicated by shading. The location of an intron present in all five genes is indicated. (B) Comparison of MBD protein structure. Proteins are indicated as horizontal lines, and MBDs are shown as labeled white boxes. Two forms of MBD2 are shown corresponding to initiation of translation at either the first (MBD2a) or second (MBD2b) methionine codons (see text). The cysteine-rich repeats of MBD1 are represented by two black boxes, and the repression domain of MeCP2 is indicated as a stippled box. The glycine-arginine repeat in MBD2a and the C-terminal glutamic acid repeat of MBD3 are indicated as horizontal lines beneath the respective proteins. Alternate splice events are indicated as diagonal lines above or below the line of the protein. Any novel translation termination codons introduced in alternate splice events are indicated. The brain-specific splice of MBD1 and testis-specific splice of MBD2 are labeled B and T, respectively. Predicted molecular masses (in kilodaltons) and isoelectric points of each protein are also listed. The isoelectric point of the MBD3 protein is given both for the full-length protein and, in parentheses, for the protein excluding the glutamic acid repeat. aa, amino acids.
With the exceptions of MBD2 and MBD3, sequence similarity between the proteins is limited to the MBDs themselves. MBD3 is highly similar to MBD2 over most of its length with 71.1% overall amino acid identity (Fig. 2), diverging only at the extreme C terminus where MBD3 has 12 consecutive glutamic acid residues encoded by an imperfect trinucleotide repeat. This characteristic is also conserved in the rat and human MBD3 proteins, although the acidic tail of the latter homologue is slightly longer and contains aspartic acid residues as well as glutamic acid residues. MBD2 and MBD3 show high conservation between human and murine genes (97.6 and 93.8% amino acid identity, respectively), whereas the human and murine homologues of MBD1 and MBD4 are less well conserved (70.9 and 65.5% amino acid identity, respectively [data not shown but all sequences are available in GenBank]). Searches of the protein databases revealed no significant matches for MBD1, MBD2, or MBD3 outside of the MBDs and the CxxCxxC motifs of MBD1 (15). Searches with the MBD4 protein sequence produced a series of low-scoring hits to bacterial DNA repair enzymes (data not shown).
FIG. 2.
Amino acid alignment of murine MBD2 and MBD3. Identical residues are indicated with a vertical line between the sequences; homologous residues are indicated with a colon, and similar residues are indicated with a period. Boxed amino acids correspond to the MBD. The glutamic acid repeat of MBD3 is represented by (E12). Amino acid numbering for MBD2 begins from the upstream initiator methionine (see MBD2a in Fig. 1B).
Cloning of the murine Mbd1 gene revealed the presence of three exons with a potential to encode cysteine-rich domains (CxxCxxC [Fig. 1B]), whereas the human gene was reported to encode only two of these domains (15). RT-PCR analysis of various murine tissues detected an alternately spliced RNA form in which the exon encoding the third CxxCxxC motif is excluded from the mature message (Fig. 1B). Further investigation of ESTs present in the databases allowed us to identify human MBD1 ESTs in which an exon encoding a third CxxCxxC motif is present (accession nos. U55972 and R14016). Whereas the 3′ CxxCxxC-encoding exon is alternately spliced in mice, the 5′-most CxxCxxC-encoding exon is alternately spliced in the human gene. Thus, both the human and murine Mbd1 genes can generate proteins containing two or three such motifs depending upon alternate splicing. An additional alternate splicing event was also detected in cDNA derived from mouse brain in which the third exon (encoding the C-terminal half of the MBD) is removed (Fig. 1B, splice labeled B). This splicing event does not maintain the correct reading frame in subsequent exons, and any resulting RNAs would not be expected to encode a functional protein. A third alternate splice would result in the replacement of the C-terminal 29 amino acids with an alternate 44-amino-acid C-terminal tail. This splice variant was found in cDNAs from both brain and embryonic cDNA libraries (this study and IMAGE clone 400458, accession no. W77338) but was not detectable by RT-PCR and is therefore predicted to occur with very low frequency.
A number of cDNAs isolated for the Mbd3 gene contain an in-frame deletion of the first exon which would result in a transcript lacking the coding sequence for the N-terminal half of the MBD (Fig. 1B). This appears to be due to the use of an alternate splice donor site within the first exon. This protein would be only slightly smaller than the full-length protein, but the MBD would be destroyed. Both spliced and unspliced forms of the message are readily detectable in many somatic tissues by RT-PCR, indicating that the shorter message makes up a significant fraction of total Mbd3 message (Fig. 1B and data not shown). The Mbd2 gene is also capable of using alternate splicing to produce potentially nonsense transcripts: a rare, larger form of the transcript includes an alternate fourth exon (4′ [Fig. 1B]) which contains an in-frame stop codon. The testis-specific form seen in Fig. 3B results from the inclusion of an alternate third exon which again results in early termination of the reading frame and truncation of the message (T [Fig. 1B]). This testis-specific exon was found in both the human and murine genes, though the level of sequence conservation in this exon is much lower than that seen for the rest of the coding region. Evidence of alternate splicing was also found for the human and murine Mbd4 transcripts. None of the identified alternately spliced forms affect the coding sequence of the MBD-like region, though one form of the human message found in the EST database would result in a truncated protein lacking the C-terminal 42 amino acids which are completely conserved between the human and murine genes. The significance of these alternately spliced variants is not known but may reflect a common method for the regulation of the MBD protein family members.
FIG. 3.
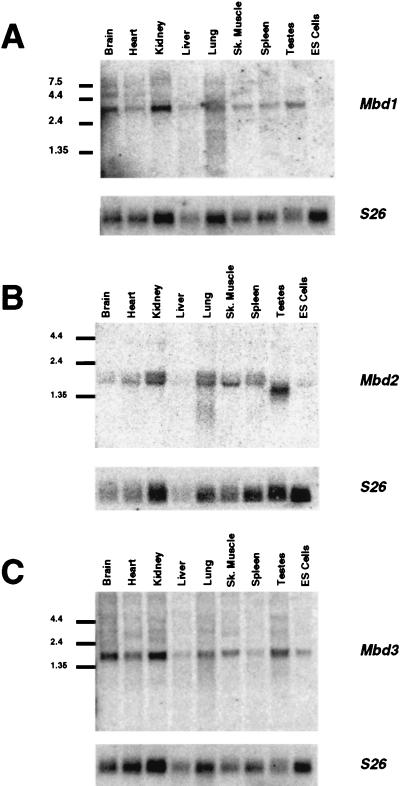
Expression of MBD1 to MBD3. cDNAs corresponding to Mbd1 (A), Mbd2 (B), and Mbd3 (C) were hybridized to Northern blots containing 10 μg of total RNA per lane from various murine tissues and ES cells. Size markers (in kilobases) are shown to the left of each blot. Blots were stripped and rehybridized to a probe corresponding to mouse ribosomal protein 26 cDNA (S26) to act as an RNA loading control. Sk. Muscle, skeletal muscle.
Expression of Mbd genes.
MeCP1 activity has been detected in numerous somatic cell types but is notably absent in ES cells and germ cells (30). In order to determine the expression pattern of the Mbd genes, the corresponding cDNA clones were hybridized to Northern blots containing RNA from various murine tissues including testis and ES cells (Fig. 3). The Mbd genes were expressed in all somatic tissues tested. No Mbd1 transcripts were detectable in ES cells, consistent with the absence of MeCP1 activity in these cells. Similarly, Mbd2 transcript levels were significantly reduced in ES cells, while an alternately spliced transcript of reduced size is detected in testis (see above). DNA methylation is known to be dispensable in ES cells (25) so it would not be surprising for methyl-CpG binding proteins to be reduced or absent in this cell type. Mbd2 expression was detected as a doublet RNA band in somatic tissues, which may reflect alternate polyadenylation site usage. Mbd3 transcripts were detectable in all tissues tested, including testis and ES cells. Mbd4 expression was undetectable on Northern blots except at very low levels in ES cells, though expression was detectable in all tissues by RT-PCR (data not shown). Expression of MBD4 was detectable in RNA from numerous human somatic tissues as well as ovary and testis (data not shown).
MBD proteins bind methylated DNA in vitro.
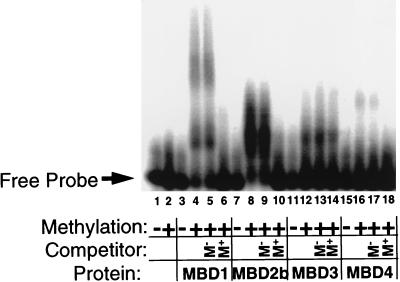
The MBD protein family members were identified due to the presence in each of a MBD-like region. We tested recombinant forms of each protein for the ability to bind to methylated and unmethylated DNA probes in a gel retardation assay. As shown in Fig. 4, MBD1, MBD2, and MBD4 all produce a specific complex with a methylated probe but not with the unmethylated version of the same probe (Fig. 4, compare lanes 3 and 4, 7 and 8, and 15 and 16). The bipartite shift observed with MBD1 is likely due to the presence of a 36-kDa degradation product within the recombinant MBD1 preparation, as was also seen with the human protein (15). The methyl-CpG-specific complexes formed by MBD1, MBD2, and MBD4 are effectively competed away upon addition of 100-fold excess unlabeled probe but not upon addition of the same amount of the unmethylated version of this probe (Fig. 4, compare lanes 5 and 6, 9 and 10, and 17 and 18). While MBD3 forms a complex with the methylated probe and not the unmethylated probe (Fig. 4, lanes 11 and 12), this shift is not competed with either methylated or unmethylated probe (lanes 13 and 14) and thus is not a specific shift. Both MBD1 and MBD2 are capable of binding to a probe containing two symmetrically methylated CpGs located 25 bp apart, though MBD4 failed to bind this probe (NB-HH probe [Table 1]). MBD1, MBD2, and MBD4 all were capable of specifically binding a probe containing one symmetrically methylated CpG (MM2 [Table 1]). Additionally, MBD1 and MBD4 binding can be competed by 100-fold excess hemimethylated oligonucleotide [(GAM)12/(GTC)12] though significantly less well than with a symmetrically methylated version of the same probe (data not shown). The MBD2 shift is not affected by this hemimethylated oligonucleotide. None of these proteins bind single-stranded DNA whether methylated or not, and they do not bind a double-stranded oligonucleotide containing repeats of the sequence TpG (thymine being similar to 5-methylcytosine in that both are pyrimidines with a methyl group at position 5). Thus MBD1, MBD2, and MBD4 proteins specifically bind methylated DNA in vitro and preferably double-stranded, symmetrically methylated DNA, although MBD1 and MBD4 complexes are also competed to a lesser extent by hemimethylated oligonucleotides. The binding of MBD1, -2, and -4 to methyl-CpGs appears to be independent of sequence context in that they bind methylated versions of a number of different oligonucleotide probes (Table 1).
FIG. 4.
Binding of recombinant MBD proteins to methylated DNA in vitro. One hundred picograms of radiolabeled, double-stranded GAC (lanes 1, 3, 7, 11, and 15) or GAM12 (lanes 2, 4 to 6, 8 to 10, 12 to 14, and 16 to 18) DNA (Table 1) was incubated with recombinant MBD proteins in the presence (+) or absence (−) of 200 ng of unlabeled methylated or unmethylated competitor DNA and then electrophoresed through a 2% agarose gel. The location of the free probe is indicated, and protein-DNA interactions are evidenced as slower-migrating DNA. Approximate amounts of recombinant protein used were as follows: MBD1, 20 ng; MBD2b, 10 ng; MBD3, 100 ng; MBD4, 100 ng. All proteins form a slower-migrating complex with methylated probe but not with the unmethylated probe. The shifts of MBD1, MBD2b, and MBD4 are prevented by the presence of excess, unlabeled, methylated DNA during incubation (M+ Competitor), but not by the addition of excess unmethylated DNA (M− Competitor). The full-length MBD2 protein also produces a methyl-specific shift (data not shown). The shift produced by MBD3 is competed by neither methylated nor unmethylated DNA and is therefore considered to be nonspecific.
TABLE 1.
Binding of MBD proteins to methylated DNA in gel shift experiments
| DNA probe | No. of MeCpGs | Sequencea | Bindingb
|
Reference | |||
|---|---|---|---|---|---|---|---|
| MBD1 | MBD2 | MBD3 | MBD4 | ||||
| GAM12 | 12 | GATCMGAMGAMGAMGAMGAMGAMGAMGAMGAMGAMGAMGATC | + | + | (+) | + | 27 |
| GAM5 | 5 | CTAGAAMGAMGTGCMGAAGCTMGACCAGTGMGTGCTAG | + | + | (+) | + | 27 |
| GAC | 0 | GATCCGACGACGACGACGACGACGACGACGACGACGACGATC | − | − | − | − | 27 |
| MeCG11 | 27 | GTACCCAAGMGMGMGCCTGGMGCCMGGGCMGGCTCCAAGMGMGMGCCTGGMGCCMGGCAAGMGMGMGCCTGGMGCCMGGGCMGGGTCCAAGMGMGMGCCTGGMGCCMGGCCMGGGMGCCAGGMGMGMGCTTGGGG | + | + | (+) | ND | 30 |
| CG11 | 0 | GTACCCAAGCGCGCGCCTGGCGCCCGGGCCGGCTCCAAGCGCGCGCCTGGCGCCCGGCAAGCGCGCGCCTGGCGCCCGGGCCGGGTCCAAGCGCGCGCCTGGCGCCCGGCCCGGGCGCCAGGCGCGCGCTTGGGG | − | − | − | ND | 30 |
| NB-Ss | 6 | ATGGCTGAGGACTGGCTGGACTGCCMGGCCCTGGGCCCTGGCTGGAAGMGCMGMGAAGTCTTTMGCAAGTCAGGGGCCACCTGTGGAMGCTCAGACACCTATTACCAGAGCCCCACAGGAGACAG | + | + | − | − | This study |
| NB-HH | 2 | ATGGCTGAGGACTGGCTGGACTGCCMGGCCCTGGGCCCTGGCTGGAAGMGCCGCGAAGTCTTTCGCAAGTCAGGGGCCACCTGTGGACGCTCAGACACCTATTACCAGAGCCCCACAGGAGACAG | + | + | − | − | This study |
| NB | 0 | ATGGCTGAGGACTGGCTGGACTGCCCGGCCCTGGGCCCTGGCTGGAAGCGCCGCGAAGTCTTTCGCAAGTCAGGGGCCACCTGTGGACGCTCAGACACCTATTACCAGAGCCCCACAGGAGACAG | − | − | − | − | This study |
| Me51 | 3 | GCCTTCAGMGCMGMGGATCCAGT | + | + | − | + | 20 |
| 51 | 0 | GCCTTCAGCGCCGCGGATCCAGT | − | − | − | − | 20 |
| MeSm | 3 | AGCAGCMGMGCCAAMGCTGGGAG | + | + | − | − | 37a |
| Sm | 0 | AGCAGCCGCGCCAACGCTGGGAG | − | − | − | − | 37a |
| MM2 | 1 | TCAGATTCGCGCMGGCTGCGATAAGCT | + | + | − | + | This study |
| MM1 | 0 | TCAGATTCGCGCCGGCTGCGATAAGCT | − | − | − | − | This study |
Sequence of one strand of double-stranded DNA probes used in gel shift assays. M, 5 methylcytosine. Methylated CpG dinucleotides are always methylated on both strands.
Binding of the protein to the DNA in band shift assays. Symbols: +, binding; −, no binding; (+), nonspecific binding. ND, not done.
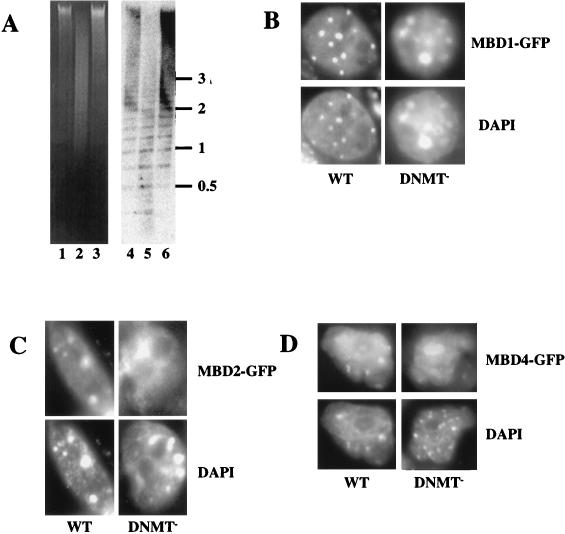
MBD1, -2, and -4 proteins are targeted to methylated DNA in vivo.
Approximately 50% of all 5-methylcytosine is concentrated in major satellite in murine cells; subsequently MeCP2 protein has been found to be concentrated at this repetitive sequence (35). Mouse major satellite is organized in foci of constitutive heterochromatin and corresponds to regions of the nucleus that stain brightly with Hoechst 33258 and DAPI (31). The specific localization of MeCP2 is no longer seen in cells with <5% of normal DNA methylation (25), demonstrating that appropriate cellular localization is dependent upon DNA methylation (35). Having shown that the MBD proteins specifically bind methylated DNA in vitro, we wanted to determine whether the cellular localization of the MBD proteins is consistent with them binding methylated DNA in vivo. To do this, we examined their localization in normal murine cells and in cells lacking >95% of normal DNA methylation (Fig. 5A). Constructs designed to express GFP-tagged versions of each of the MBD proteins under the control of a cytomegalovirus promoter were transfected into either wild-type or DNMT-deficient ES cells. As is shown in Fig. 5 and 6, ectopically expressed MBD proteins were predominantly nuclear in murine cells. Nuclear localization was also seen in human (HT1080) and hamster (CHO) cells (data not shown). Overexpressed MBD1-GFP, MBD2-GFP, and MBD4-GFP preferentially localized to regions of the genome known to be highly methylated, as evidenced by colocalization of protein signals with DAPI-bright areas (Fig. 5B to D).
FIG. 5.
Binding of MBD proteins to methylated and hypomethylated genomes in vivo. (A) DNA from wild-type ES cells (lanes 1 and 4), DNMT mutant ES cells (lanes 2 and 5), and murine somatic cells (lanes 3 and 6) was digested with the methylation-sensitive restriction enzyme MaeII (Boehringer Mannheim) and electrophoresed on a 1% agarose gel. Lanes 1 to 3 show the ethidium bromide-stained gel, and lanes 4 to 6 show the gel blotted and probed with the 234-bp major satellite repeat monomer. (B to D) Localization of MBD-GFP fusion proteins in nuclei of ES cells. Either wild-type (WT) or DNMT-deficient (DNMT−) ES cells were transiently transfected with MBD1-GFP (B), MBD2-GFP (C), or MBD4-GFP (D) expression constructs. GFP fluorescence (upper panels) indicates the location of the MBD-fusion protein. DNA is visualized in the same nuclei by DAPI staining (lower panels), and major satellite is visible as intense spots of DAPI fluorescence. All fusion proteins are predominantly nuclear in all cells. MBD1-GFP colocalizes with major satellite in wild-type and mutant cells. MBD2-GFP and MBD4-GFP colocalize with major satellite in wild-type ES cells but fail to localize to major satellite in DNMT-deficient ES cell nuclei. Localization of MBD4-GFP to the nucleolus in DNMT-deficient ES cell nuclei was not observed consistently.
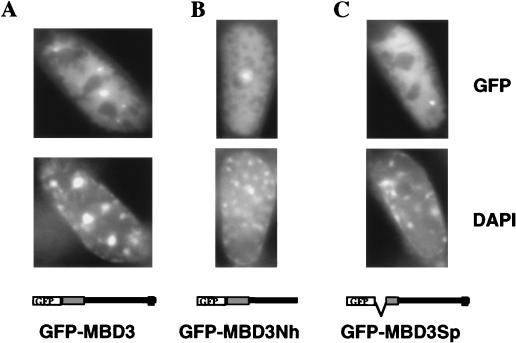
FIG. 6.
Localization of GFP-MBD3 fusion proteins in nuclei of somatic cells. The mouse embryonic fibroblast cell line EFS2 was transiently transfected with GFP-MBD3 fusion protein expression constructs. The top panels show GFP fluorescence, while the lower panels show DAPI staining of the same cells. The constructs used are diagrammed at the bottom, with MBD depicted as a gray box and the glutamic acid repeat depicted as a black box. In all cases, GFP is located at the N terminus. (A) The full-length MBD3 fusion protein localizes to the nucleus and occasionally forms bright foci of GFP fluorescence, but these spots do not correspond to the major satellite. (B) Removal of the glutamic acid repeat (GFP-MBD3Nh) does not alter subnuclear localization, nor does removal of the N-terminal half of the MBD (GFP-MBD3Sp [C]). A fourth construct in which GFP is located at the C terminus of full-length MBD3 showed a localization pattern indistinguishable from that of GFP-MBD3 (data not shown).
MBD1-GFP, MBD2-GFP, and MBD4-GFP all localized to major satellite in transfected wild-type murine cells, consistent with their ability to bind methylated DNA in band shift assays. Whereas MBD1-GFP showed heterochromatic localization in all transfected cells, in cells expressing low levels of MBD2-GFP and MBD4-GFP, unlocalized nuclear staining was often observed. This pattern was seen in somatic murine cells (primary mouse cells and L cells [data not shown]) as well as ES cells. In order to determine whether MBD2-GFP and MBD4-GFP localization was disrupted in DNMT-deficient ES cells, the following screening protocol was used. Coverslips were scanned under ×20 magnification, and cells containing a visible amount of GFP signal at this level of magnification were then analyzed under ×100 magnification and scored for subnuclear localization. Using this selection criterion, 100% of wild-type ES cells analyzed showed localization of MBD2-GFP or MBD4-GFP signal with major satellite. In contrast, DNMT-deficient ES cells expressing MBD2-GFP or MBD4-GFP displayed an overall nuclear localization of the GFP signal (Fig. 5C and D), indicating that the association of MBD2-GFP and MBD4-GFP with major satellite is dependent upon DNA methylation. MBD1-GFP localized to major satellite in all wild-type murine cells analyzed, irrespective of the amount of fluorescence seen. In DNMT-deficient ES cells, MBD1-GFP localized to DAPI-bright regions in most, but not all, nuclei (Fig. 5B). This is in stark contrast with the patterns seen for MBD2-GFP and MBD3-GFP, as well as that reported for MeCP2-LacZ which localized to major satellite in a minority of DNMT-deficient ES cells (35). The possibility of contamination of the DNMT-deficient ES cell cultures by wild-type ES cells was ruled out by verifying the undermethylation of the DNMT-deficient ES cells by digestion of total DNA with the methylation-sensitive restriction enzyme MaeII (Fig. 5A). While normal ES cell DNA and somatic cell DNA is highly resistant to MaeII digestion, the DNMT-deficient ES cell DNA is readily digested with MaeII, which is able to digest the sequence ACGT only when the cytosine is unmethylated (Fig. 5A, compare lane 2 to lanes 1 and 3). Blotting and probing of this gel with major satellite DNA illustrate the high degree of undermethylation of this repetitive sequence in the DNMT-deficient ES cells compared to that of wild-type ES cells or somatic cell DNA (Fig. 5A, compare lane 5 to lanes 4 and 6). This observation may mean that the residual methylation known to exist in the DNMT-deficient ES cells is sufficient to direct MBD1-GFP to the major satellite. An alternate interpretation of these results is that MBD1-GFP is tethered to major satellite via some other protein factor and that this association is independent of DNA methylation.
The MBD3-GFP fusion protein shows diffuse nuclear staining in low-expressing cells and accumulates in many nuclear foci in cells expressing large amounts of the fusion protein. These foci do not coincide with major satellite (Fig. 6). Thus, MBD3 does not prefer to associate with the highly methylated major satellite DNA in mouse cells. To determine whether the failure of MBD3-GFP to localize to methylated DNA in vivo is due to the presence of its acidic C-terminal tail, an MBD3 deletion protein (GFP-MBD3Nh [Fig. 6]), which is truncated at amino acid 248, was expressed in murine cells. This protein also localized to the nucleus in interphase mouse cells but was excluded from the DAPI-bright regions, indicating that it is not the acidic tail which is preventing GFP-MBD3 from associating with methylated DNA in vivo. We next wanted to determine whether the localization of MBD3 was influenced by its MBD-like region. A version of MBD3 (GFP-MBD3Sp [Fig. 6]) lacking amino acids 4 to 36 (the amino-terminal half of the MBD-like region) and corresponding to the common alternately spliced variant of Mbd3 message (Fig. 1B), was expressed as a GFP fusion in murine cells. The localization of this deleted MBD3-GFP protein was indistinguishable from that of the full-length protein, leading us to conclude that the integrity of the MBD-like region is not important for the localization seen in this assay.
DISCUSSION
Searching the EST database with the amino acid sequence of the MeCP2 MBD has revealed the presence of a family of mammalian MBD-containing proteins. The absence of any other known sequence motifs within MBD2 or MBD3 provides no clues as to the activities of these two proteins. In contrast, homology between MBD4 and bacterial DNA repair enzymes is consistent with a DNA repair activity for MBD4 and may indicate a link between DNA repair and methylation in mammals, but this remains to be proven. Both the sequence and genomic organization of the MBDs within each of the MBD proteins makes it clear that these motifs are related evolutionarily, but the lack of similarity between these proteins outside of the MBD (excluding MBD2 and MBD3) may indicate that each protein carries out a different function within the cell.
Both MeCP2 and MBD1 are capable of transcriptional repression at methylated promoters (15, 32), though there is no sequence in MBD1 (or any of the other MBD proteins) which bears any recognizable sequence homology to the TRD of MeCP2. The recent finding that MeCP2 TRD binds to the Sin3-histone deacetylase complex (22, 34) provides an attractive molecular picture of the potential mechanism of action of MeCP2: MeCP2 binds to methylated DNA via its MBD and attracts the Sin3-histone deacetylase complex via its TRD, resulting in the deacetylation of methylated chromatin and subsequent transcriptional silencing. The absence of any TRD-like sequence in MBD1 may mean that the mechanism by which MBD1 represses transcription is different from that used by MeCP2. Alternately MBD1 may use the same mechanism as MeCP2 but contain a novel Sin3 binding domain or may bind deacetylases directly.
All of the MBD proteins, including MeCP2, are ubiquitously expressed in somatic tissues, but only Mbd3 and Mbd4 transcripts are readily detectable in ES cells. Although ES cells generally have highly methylated genomes, this methylation is not necessary for viability (25). MeCP2 is very weakly expressed in ES cells and is also not needed for viability (42). Thus, DNA methylation and the presence of MeCP2 do not appear to be important in ES cells, and therefore, the finding that Mbd1 and Mbd2 transcript levels are significantly reduced in this cell type is not surprising. Mbd3 and Mbd4 are both well expressed in ES cells, but whether the corresponding proteins are present or important for ES cell viability is not known.
All of the MBD proteins except MBD3 bind to methylated oligonucleotide probes in vitro but do not bind to the unmethylated versions of these oligonucleotides under our conditions. Recently, the chicken homologue of MeCP2 was proposed to be a component of the nuclear matrix and was shown to be capable of binding to a TG-rich “matrix attachment region” sequence in gel shift assays (45). Though low-affinity TG binding has been detected for rat MeCP2 in vitro (35a), we detected no affinity of MBD1, MBD2, MBD3, or MBD4 for TG-containing probes in gel shift assays (data not shown).
We have taken advantage of the fact that approximately half of all 5-methylcytosine in mouse cells is located at the pericentromeric heterochromatin (31) in order to investigate the ability of the MBD proteins to localize to methylated DNA in vivo. Ectopically expressed GFP-fusions of MBD1, MBD2, and MBD4 colocalized with major satellite in mouse cells, but localization of MBD2 and MBD4 was disrupted in cells lacking a functional DNMT gene (Fig. 5). This suggests that MBD2 and MBD4 are capable of binding methylated DNA in vivo as well as in vitro. MBD1 is also capable of associating with methylated DNA in vivo but binds to the same heterochromatic sites in DNMT-deficient ES cells. It is not clear whether MBD1 is binding to low levels of DNA methylation in DNMT-deficient cells or to some other heterochromatin binding protein. Similarly, we cannot formally rule out the possibility that the localization of MBD2 and MBD4 to murine heterochromatin is due to their interaction with some other methylation-sensitive heterochromatin binding protein. It is unlikely that MeCP2 is responsible, as protein activity is undetectable in ES cells (30) and localization of MBD1-, MBD2-, or MBD4-GFP fusion proteins is not disrupted in MeCP2-deficient somatic cells (17a).
MBD3 behaves differently than the other MBD proteins, failing to specifically bind methylated DNA in vitro (Fig. 4C) or colocalize with major satellite in vivo (Fig. 6). The difference in specificity between the highly similar MBD2 and MBD3 proteins cannot be attributable to their divergent C termini, as a truncated version of MBD3 which lacks the C-terminal 37 amino acids, including the glutamic acid repeat, also fails to localize to DAPI-bright regions in mouse nuclei (Fig. 6). Nor can it be attributable to the additional 150 amino acids N-terminal to the MBD of MBD2, since a shorter MBD2-GFP protein which lacks this region also localizes to major satellite in murine cells (data not shown). Thus, the DNA binding specificities are most likely attributable to differences in the MBDs of the two proteins (Fig. 1A and 2). It is possible that the MBD of MBD3 is nonfunctional, since a GFP fusion of an MBD3 variant resulting from the product of the common alternately spliced RNA species (Fig. 1B) which lacks the N-terminal half of the MBD-like sequence shows an in vivo localization pattern indistinguishable from that of the full-length protein (Fig. 6). Despite the high degree of sequence similarity between MBD2 and MBD3, these two proteins display completely different DNA binding activities and in vivo localization patterns. The very high degree of conservation between the murine and human homologues of these two proteins (97.6% amino acid identity for MBD2 and 93.8% for MBD3) indicates that both proteins are functional. One possible explanation for these observations is that Mbd3 arose by gene duplication of Mbd2, but has acquired a novel DNA binding specificity which our ectopic overexpression techniques have not detected. Whether MBD2 and MBD3 perform similar functions despite having different DNA binding abilities remains to be determined.
MeCP2 localizes to major satellite at a very low frequency in DNMT-deficient cells (35). This may be attributable to the nonspecific DNA binding activity of MeCP2 which, in the absence of methyl-CpGs, may allow it to bind to the TG-rich major satellite (27). We have found no in vitro evidence that any of the proteins described here has any sequence specificity other than the absolute requirement for the presence of methylated CpGs. Nevertheless, there are hints of different binding specificities in vivo. Whereas MBD1-GFP location is coincident with DAPI-bright regions in all transfected murine cells, MBD2-GFP and MBD4-GFP fail to bind major satellite in murine cells expressing very small amounts of the fusion protein (data not shown). This observation may indicate a DNA binding preference of MBD2 and MBD4 for methylated euchromatic sites over those found in major satellite. Similarly, the different frequencies with which the MBD proteins are found to localize to major satellite in ES cells lacking >95% of normal methylation levels may reflect the density of methylation required in a given sequence for each protein to bind. For example, MBD1 may be capable of binding to DNA containing low methylation levels, MeCP2 may require a somewhat higher level of methylation, and MBD2 and MBD4 may bind only heavily methylated DNA. This possibility will require quantitative DNA binding assays to be tested rigorously.
DNA methylation is absolutely required for mammalian development (29). The signal that DNA methylation represents is interpreted by methylated DNA binding proteins, and until now only two of these—MeCP2 and MBD1—had been defined molecularly (15, 27). The identification of MBD2 and MBD4 as specific methyl-CpG binding proteins provides two more candidates for proteins that mediate the effects of DNA methylation.
ACKNOWLEDGMENTS
We thank En Li for the DNMT-deficient ES cells, Andrew Smith for the 129 genomic library, Jonathan Pines for the pCMXGFP vector, Christer Höög for the MMTEST clone, John Huntriss and Marilyn Monk for sending oligonucleotide probes, and the HGMP for providing numerous clones and libraries. We also thank members of the Bird lab; Cathy Abbott and Beth Sullivan for advice; Kevin Hardwick for use of the imaging system; Vicky Clark for sequencing; Aileen Greig and Joan Davidson for technical assistance; and Beth Sullivan, Cathy Abbott, Susan Tweedie, Sally Cross, and Huck-Hui Ng for critical reading of the manuscript.
B. H. was the recipient of a Long Term Postdoctoral Fellowship from the Human Frontiers Science Program. The work was also supported by a program grant to A.B. from the Wellcome Trust.
REFERENCES
- 1.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA. 1994;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- 3.Bartolomei M S, Tilghman S M. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- 4.Bassett D E J, Boguski M S, Spencer F, Reeves R, Goebl M, Hieter P. Comparative genomics, genome cross-referencing and XREFdb. Trends Genet. 1995;11:372–373. doi: 10.1016/s0168-9525(00)89109-x. [DOI] [PubMed] [Google Scholar]
- 5.Beard C, Li E, Jaenisch R. Loss of methylation activates Xist in somatic but not embryonic cells. Genes Dev. 1995;9:2325–2334. doi: 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- 6.Bird A P. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 7.Bird A P. Gene number, noise reduction and biological complexity. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- 8.Bird A P. The relationship of DNA methylation to cancer. Cancer Surv. 1996;28:87–101. [PubMed] [Google Scholar]
- 9.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 10.Boyes J, Bird A. Repression of genes by DNA methylation depends upon CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buschhausen G, Wittig B, Graessmann M, Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci USA. 1987;84:1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cedar H. DNA methylation and gene activity. Cell. 1988;53:3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- 13.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 14.Comb M, Goodman H M. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cross S, Meehan R, Nan X, Bird A. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat Genet. 1997;13:256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- 16.de Bustros A, Nelkin B D, Silverman A, Ehrlich G, Poiesz B, Baylin S B. The short arm of chromosome 11 is a “hot spot” for hypermethylation in human neoplasia. Proc Natl Acad Sci USA. 1988;85:5693–5697. doi: 10.1073/pnas.85.15.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison S M, Dunwoodie S L, Arkell R M, Lehrach H, Beddington R S P. Isolation of novel tissue-specific genes from cDNA libraries representing the individual tissue constituents of the gastrulating mouse embryo. Development. 1995;121:2479–2489. doi: 10.1242/dev.121.8.2479. [DOI] [PubMed] [Google Scholar]
- 17a.Hendrich, B. Unpublished results.
- 17b.Hendrich, B., P. Tate, and A. Bird. Unpublished results.
- 18.Herman J G, Merlo A, Mao L, Lapidus R G, Issa J P, Davidson N E, Sidransky D, Baylin S. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci USA. 1995;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu C H, McStay B, Jeong S-W, Reeder R H. xUBF, an RNA polymerase I transcription factor, bind crossover DNA with low sequence specificity. Mol Cell Biol. 1994;14:2871–2882. doi: 10.1128/mcb.14.5.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huntriss J, Lorenzi R, Purewal A, Monk M. A methylation-dependent DNA-binding activity recognising the methylated promoter region of the mouse Xist gene. Biochem Biophys Res Commun. 1997;235:730–738. doi: 10.1006/bbrc.1997.6876. [DOI] [PubMed] [Google Scholar]
- 21.Iguchi-Ariga S M M, Schaffner W. CpG methylation of the cAMP responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 22.Jones P L, Veenstra G J C, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 23.Kass S U, Goddard J P, Adams R L P. Inactive chromatin spreads from a focus of methylation. Mol Cell Biol. 1993;13:7372–7379. doi: 10.1128/mcb.13.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 25.Lei H, Oh S P, Okano M, Jüttermann R, Goss K A, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 26.Lennon G, Auffray C, Polymeropoulos M, Soares M B. The I.M.A.G.E. consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 27.Lewis J D, Meehan R R, Henzel W J, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 28.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 29.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 30.Meehan R R, Lewis J D, McKay S, Kleiner E L, Bird A P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 31.Miller O L, Schnedl W, Allen J, Erlanger B F. Immunofluorescent localization of 5MC. Nature. 1974;251:636–637. doi: 10.1038/251636a0. [DOI] [PubMed] [Google Scholar]
- 32.Nan X, Campoy F, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 33.Nan X, Meehan R R, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nan X, Ng H-H, Johnson C A, Laherty C D, Turner B M, Eisenmann R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 35.Nan X, Tate P, Li E, Bird A. DNA methylation specifies chromosomal localization of MeCP2. Mol Cell Biol. 1996;16:414–421. doi: 10.1128/mcb.16.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Nan, X., S. Tweedie, and A. Bird. Unpublished results.
- 36.Neumann B, Barlow D P. Multiple roles for DNA methylation in gametic imprinting. Curr Opin Genet Dev. 1996;6:159–163. doi: 10.1016/s0959-437x(96)80045-1. [DOI] [PubMed] [Google Scholar]
- 37.Ogawa H, Inouye S, Tsuji F I, Yasuda K, Umesono K. Localization, trafficking, and temperature-dependence of the Aequorea green fluorescent protein in cultured vertebrate cells. Proc Natl Acad Sci USA. 1995;92:11899–11903. doi: 10.1073/pnas.92.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Prokhortchouk, A. V., E. B. Prokhortchouk, and G. P. Georgiev. Unpublished data.
- 38.Razin A, Cedar H. DNA methylation and genomic imprinting. Cell. 1994;77:473–476. doi: 10.1016/0092-8674(94)90208-9. [DOI] [PubMed] [Google Scholar]
- 39.Riggs A D, Pfeifer G P. X-chromosome inactivation and cell memory. Trends Genet. 1992;8:169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- 40.Sakai T, Toguchida J, Ohtani N, Yandell D W, Rapaport J M, Dryja T P. Allele-specific hypermethylation of the retinoblastoma tumor-suppressor gene. Am J Hum Genet. 1991;48:880–888. [PMC free article] [PubMed] [Google Scholar]
- 41.Smith A G, Heath J K, Donaldson D D, Wong G G, Moreau J, Stahl M, Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 42.Tate P, Skarnes W, Bird A. The methyl-CpG binding protein MeCP2 is essential for embryonic development in the mouse. Nat Genet. 1996;12:205–208. doi: 10.1038/ng0296-205. [DOI] [PubMed] [Google Scholar]
- 43.Tweedie S, Charlton J, Clark V, Bird A. Methylation of genomes and genes at the invertebrate-vertebrate boundary. Mol Cell Biol. 1997;17:1469–1475. doi: 10.1128/mcb.17.3.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Umesono K, Murakami K K, Thompson C C, Evans R M. Direct repeats as selective response elements of the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weitzel J M, Buhrmester H, Strätling W H. Chicken MAR-binding protein ARBP is homologous to rat methyl-CpG-binding protein MeCP2. Mol Cell Biol. 1997;17:5656–5665. doi: 10.1128/mcb.17.9.5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan L, Liu J-G, Höög C. Rapid cDNA sequencing in combination with RNA expression studies in mice identifies a large number of male germ cell-specific sequence tags. Biol Reprod. 1995;52:131–138. doi: 10.1095/biolreprod52.1.131. [DOI] [PubMed] [Google Scholar]