Abstract
Filamin B (FLNB) plays an important role in skeletal development. Mutations in FLNB can lead to skeletal malformation such as an abnormal number of ossification centers, indicating that the skeletal segmentation in the embryonic period may be interfered with. We established a mouse model with the pathogenic point mutation FLNB NM_001081427.1: c.4756G > A (p.Gly1586Arg) using CRISPR-Cas9 technology. Micro-CT, HE staining and whole skeletal preparation were performed to examine the skeletal malformation. In situ hybridization of embryos was performed to examine the transcription of HOX genes during embryonic development. The expression of FLNB was downregulated in FLNBG1586R/G1586R and FLNBWT/G1586R mice, compared to FLNBWT/WT mice. Fusions in tarsal bones were found in FLNBG1586R/G1586R and FLNBWT/G1586R mice, indicating that the skeletal segmentation was interfered with. In the embryo of FLNBG1586R/G1586R mice (E12.5), the transcription levels of HOXD10 and HOXB2 were downregulated in the carpal region and cervical spine region, respectively. This study indicated that the loss-of-function mutation G1586R in FLNB may lead to abnormal skeletal segmentation, and the mechanism was possibly associated with the downregulation of HOX gene transcription during the embryonic period.
Keywords: Filamin B, HOX, Skeletal segmentation, CRISPR-Cas9, In situ hybridization of embryo
Highlights
-
•
A mouse model with the pathogenic point mutation G1586R in Filamin B (FLNB) was established using CRISPR-Cas9 technology.
-
•
Fusions in tarsal bones were found in FLNBG1586R/G1586R and FLNBWT/G1586R mice.
-
•
The transcription levels of HOXD10 and HOXB2 were downregulated in the embryo of FLNBG1586R/G1586R mice.
-
•
Disruption of FLNB may lead to abnormal skeletal segmentation by downregulating HOX gene during the embryonic period.
1. Introduction
Filamin B is a cytoplasmic protein encoded by FLNB. Filamin B crosslinks with the actin cytoskeleton, provides a scaffold for many other molecules in the cell, and participates in signaling transduction (Stossel et al., 2001). Mutations in FLNB are associated with a spectrum of distinct skeletal dysplasia, indicating the important role of FLNB in skeletal development (Xu et al., 2017). Gain-of-function mutation in FLNB leads to Larsen syndrome, atelosteogenesis type I, atelosteogenesis type III, and boomerang dysplasia (Maroteaux et al., 1982; Jeon et al., 2014; Kozlowski et al., 1981; Tenconi et al., 1983; Kozlowski et al., 1985). These diseases present short stature and skeletal malformations including supernumerary ossification centers of carpal and tarsal bones, and scoliosis (Larsen et al., 1950; Stanley et al., 1988). Loss-of-function mutation in FLNB leads to spondylocarpotarsal synostosis syndrome (SCT syndrome), presenting skeletal malformations including premature fusions in carpal and tarsal bones, and fusions in vertebrae (Krakow et al., 2004).
These clinical manifestations above indicated that gain/loss-of-function mutations in FLNB might lead to changes in ossification centers, indicating that the skeletal segmentation might be interfered with during embryo development. However, previous researchers only reported that loss-of-function in FLNB inhibited the growth of long bone, which could explain the short stature of such patients (Hu et al., 2014; Lu et al., 2007; Daniel et al., 2012; Sawyer et al., 2009; Zheng et al., 2007). The mechanism of FLNB mutations interfering with the skeletal segmentation process remains unclear.
Skeletal segment identity of vertebral animals is known to be specified by HOX genes in the embryo period (Quinonez and Innis, 2014). Over ten HOX genes have been reported to be associated with skeletal malformation. Interestingly, the skeletal malformation phenotypes were remarkably similar between the HOX-mutated spectrum disorders and FLNB-mutated spectrum disorders, both of which were characterized by alterations in the number of ossification centers (Xu et al., 2017; Quinonez and Innis, 2014).
In this study, we hypothesized that the loss-of-function mutation in FLNB led to skeletal malformation by interfering with skeletal segmentation through the HOX gene during the embryonic period. We established a mouse model with FLNB G1586R loss-of-function mutation using CRISPR-Cas9 technology to examine if the mice acquire skeletal segmentation anomalies and perform in situ hybridization of embryos to detect the transcription of HOX genes in the embryonic period. We aimed to explore the regulation between FLNB and HOX in the embryonic period, and to uncover the pathological mechanism of FLNB-related skeletal disorders.
2. Material and methods
2.1. Generation of mice model with FLNB G1586R mutation
The G1586R mutation in mouse FLNB (NM_001081427.1: c.4756G > A (p.Gly1586Arg)) is homologous to a pathogenic mutation site in human FLNB (NM_001457.3: c.4756G > A (p.Gly1586Arg)) that was previously reported by our group in a patient with severe skeletal malformation (Xu et al., 2018). The sgRNA target was designed around this point mutation site in exon 28 of mouse FLNB (Fig. 1a). The primers were synthesized according to the target sequence, and PCR was performed using the template of the gRNA framework to get the sequence of sgRNA. Then the transcription was performed in vitro using a T7 in vitro transcript kit (#E2050S,NEB, USA), and the sgRNA was purified after ethanol precipitation. The cleavage efficiency of sgRNA was tested using Cas9/gRNA in-vitro activity testing kit (#VK-007, ViewSolid, China), and the target sg1 (5′-tagcgtccggtcttatcaGGG-3′) was screened with the highest activity. The homologous recombination donor sequence was designed based on the sg1 target and point mutation sequence, and the donor ssDNA was synthesized. The transcripts of the sg1, Cas9 mRNA (#R001L, ViewSolid, China), and homologous recombinated ssDNA were mixed, and the mixture was injected into the cytoplasm of the fertilized ovum. The fertilized ovum was transferred into the M2 culture solution to get diploid cells, and transplanted into the ovarian ducts of the recipient mice (ICR) to develop. The genomic DNA was isolated from the tail tips of 10-day-old mice. The point mutation in FLNB was genotyped using the following primers: 1F (5′-CATACAGAGGTGCCAAGTA-3′) and 1R (5′-GACATTAAGTGGATAGAGAAGG-3′) in F0 mice to get FLNBWT/G1586R mice, which were then crossed to get heterozygous mice that can be passaged stably. All animal protocols followed the National Research Council's Guide for the Care and Use of Laboratory Animals, and was approved by the Animal Welfare and Ethics Committee of Peking Union Medical College Hospital (Ethics committee approval code: XHDW-2023-136).
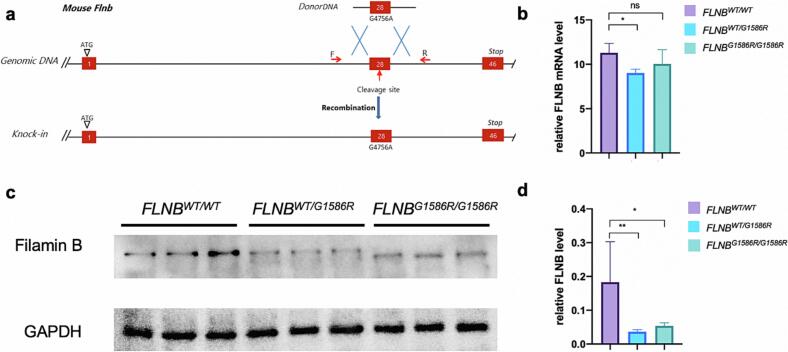
Fig. 1.
Establishment of FLNBG1586R/G1586R mice using CRISPR/Cas9. a Diagrammatic representation demonstrating that FLNB c.4756G>A was introduced into the genomic DNA using CRISPR/Cas9. b qPCR analysis of FLNB transcription in the bone marrow cells of FLNBWT/WT mice, FLNBWT/G1586R mice and FLNBG1586R/G1586R mice (male, 33-week to 34-week, three mice for each group). c-d Western blot analysis of Filamin B and GAPDH expression in the bone marrow cells of FLNBWT/WT mice, FLNBWT/G1586R mice and FLNBG1586R/G1586R mice (male, 33-week to 34-week, three mice for each group). The expression level of Filamin B was significantly down-regulated in the FLNBWT/G1586R mice and FLNBG1586R/G1586R mice, compared to FLNBWT/WT mice.
2.2. Acquisition of bone marrow mononuclear cells and extraction of RNA and protein
FLNBWT/WT, FLNBWT/G1586R, and FLNBG1586R/G1586R mice were euthanized. The femurs and tibias were collected, and the bone marrow was washed out. Red cells were lysed using ammonium chloride cell lysis buffer. The cell mixtures were filtered using 40 μm cell strainers to get the bone marrow cells. Total RNA was extracted with TRNzol universal total RNA extraction kit (cat No. DP424, TIANGEN, China). Total protein was extracted from bone marrow cells using RIPA buffer, and the protein concentration was determined with protein quantification kit (BCA).
2.3. Measurement of the transcription level of FLNB using Real-time PCR
cDNA was amplified from total RNA using PrimeScript™ RT Reagent Kit with gDNA Eraser (cat No. RR047A, Takara, Japan). The reverse-transcribed cDNA was amplified in a real-time fluorescence quantification PCR machine (Quantageneq225, Kubo, China) using Luna Universal qPCR Master Mix (cat No. M3003, NEB, USA). Primers used were as follows: FLNB: 5′-AGGACCCAGCAAGGTGGACATC-3′ (sense) and 5′-CTTCCAGGCACATGCTCGTCAG-3′ (antisense); GAPDH: 5′-ACACTGAGGACCAGGTTGTCTCC-3′ (sense) and 5′-TGTTGCTGTAGCCGTATTCATTGTCA-3′ (antisense). PCR conditions were as follows: 30 s at 95 °C, 45 cycles of 5 s at 95 °C, and 15 s at 60 °C. The amount of FLNB mRNA was normalized to GAPDH mRNA.
2.4. Measurement of the expression of FLNB using Western blotting
Western blotting was performed to measure the expression level of FLNB. The total protein was separated on an 8 % SDS-PAGE gel and transferred onto polyvinylidene difluoride (PVDF) membranes, which were then blocked and incubated with primary antibodies overnight. Primary antibodies used were anti-Filamin B (1:1000; cat No. ab224334, Abcam, UK), and anti-GAPDH (1:5000; cat No. 200306-7E4, Zen-Bio, China). The membranes were then incubated with HRP-conjugated goat anti-rabbit IgG (1:5000; cat No. 511203, Zen-Bio, China) or goat anti-mouse IgG (1: 5000; cat No. 511103, Zen-Bio, China) after a final wash, and developed in enhanced chemiluminescence reagent (Millipore).
2.5. Measurement of serum bone turnover markers
Mice were fasted for 6 h; blood was drawn, allowed to clot for 1 h, and centrifuged at 3000 rpm for 10 min. The serum levels of procollagen type 1 N-terminal propeptide (P1NP), osteocalcin (OCN), C-terminal telopeptides of type 1 collagen (CTX-1), and tartrate-resistant acid phosphatase 5b (TRACP5b) were measured using commercially available enzyme-linked immunosorbent assay (ELISA) kits (MEIMIAN, China) according to the instructions of the manufacturer.
2.6. Assessment of skeletal structure using micro-CT
For observation of skeletal structures, the distal left forelimbs and distal left hindlimbs were scanned by Inveon MM micro-CT manufactured by Siemens (Berlin, Germany) at a voltage of 70 kV and a current of 400 mA. Three-dimensional reconstruction was performed using the Inveon analysis workstation. For measurement of bone microarchitecture, the left hindlimbs were harvested from the mice above, and the midshaft and proximal metaphysis regions of the femur were scanned. The analysis was performed from 0.5 mm proximal to the growth plate, extending 1 mm towards the diaphysis. A spatial resolution of 35 mm was reconstructed using the Inveon analysis workstation. Trabecular volumetric bone mineral density (Tb.vBMD), cortical volumetric bone mineral density (Ct.vBMD), trabecular thickness (Tb.Th), trabecular separation (Tb.Sp), bone volume fraction (BV/TV), trabecular number (Tb.N), bone surface/volume (BS/BV), and cortical bone thickness (Ct.Th) were computed according to the instruction of the manufacturer. Three mice of each genotype were analyzed.
2.7. Histological observation
For histological observation on the carpal and tarsal bone, the distal right forelimbs and distal right hindlimbs were fixed in 4 % paraformaldehyde for 48 h at 4 °C, decalcified in 30 % methanoic acid for 24 h at 4 °C. Specimens were dehydrated and embedded in paraffin and sectioned at 4 μm, stained with hematoxylin and eosin, and observed using standard light microscope (Olympus, Japan). All images were captured through CaseViewer 2.4 software (3DHISTECH Ltd.). Three mice of each genotype were analyzed.
2.8. Whole skeletal preparation
Mice were dissected, skins were peeled way and all tissues were eviscerated. The remaining tissue was fixed in 95 % ethanol for 72 h, and stained in 0.015 % alcian blue (Sigma-Aldrich, USA) dye for 96 h in dark at 37 °C. The carcasses were destained with 95 % ethanol for 8 h, and treated with 1 % KOH for 72 h until most soft tissue disappeared. After removing the remaining soft tissues with forceps, the mice were stained in 0.005 % alizarin red (Sigma-Aldrich, USA) solution for 96 h in dark. Finally, skeletons were cleared in a 20 % glycerol/1 % KOH solution for 12 h, and stored in 50 % ethanol/50 % glycerol for photography. Two mice of each genotype were analyzed.
2.9. In situ hybridization of embryos
Embryos (E12.5) of FLNBWT/WT, FLNBWT/G1586R, and FLNBG1586R/G1586R mice were collected. Whole-mount in situ hybridization was performed as described (Wilkinson, 1992) with some modifications. Embryos were fixed with 4 % paraformaldehyde at 4 °C overnight. Embryos were then dehydrated and rehydrated through a methanol series. Further permeabilization was achieved by proteinase K treatment, followed by refixation. Digoxygenin-labeled (DIG-labeled) antisense riboprobes were generated from the mouse HOXA4 gene (855-bp region amplified with 5′- CGGGATCCATGACCATGAGCTCGTTTTT-3′ and 5′- CCCAAGCTTTATGGAGGAGGGAATGGGTG-3′), HOXB2 gene (1027-bp region amplified with 5′- CGGGATCCATGAATTTTGAATTTGAGAG-3′ and 5′- CCCAAGCTTTGAAGAAGTCCAGCTCTTC-3′), HOXB4 gene (750-bp region amplified with 5′-CGGGATCCATGGCTATGAGTTCCTTTTT-3′ and 5′-CCCAAGCTTGAGCGCAGGGGGGCCTCCGT-3′), HOXD10 gene(1020-bp region amplified with 5′-CGGGATCCATGTCCTTTCCCAACAGCTC-3′ and 5′-CCCAAGCTTAGAAAAGGTGAGGTTGGC-3′). All 4 gene products above were cloned into the pSPT-18 vector (Roche), and DIG-labeled riboprobes were prepared as previously described (Wilkinson, 1992). Embryos were incubated with DIG-labeled probes in 70 °C humidified chamber overnight, with the unhybridized probes removed by washing, and the non-specific binding sites blocked with sheep serum. The bound probes were detected with an alkaline phosphatase substrate, antibody pre-absorption was omitted, and then incubated directly with the antibody overnight at 4 °C. Before development, embryos were washed with MABTL eight or more times for 1 h each at room temperature, then overnight at 4 °C to achieve lower backgrounds.
2.10. Statistical analysis
Continuous variables were expressed as mean ± standard deviations (SDs); Unpaired t-test was used for comparison between two groups. Two-tailed tests were used for all statistics, and p < 0.05 was defined as statistically significant differences. All figures were performed in GraphPad Prism 8.0 software (GraphPad, La Jolla, USA).
3. Results
3.1. Generation of mice with FLNB G1586R mutation
We have previously reported a patient with severe skeletal malformation including cervical kyphosis, scoliosis, and an increased number of carpal bones, with a missense mutation in FLNB (NM_001457.3, c.4756G > A (p.Gly1586Arg)) (Xu et al., 2018). To understand the role of FLNB in skeletal development, we generated FLNBG1586R/G1586R mice using CRISPR/Cas9 (Fig. 1a). As shown in Fig. 1c and Fig. 1d, the protein level of Filamin B was significantly lowered in the FLNBWT/G1586R mice and FLNBG1586R/G1586R mice, compared to FLNBWT/WT mice. We further performed q-PCR using the same samples. As shown in Fig. 1b, no significant difference in mRNA level was observed between FLNBG1586R/G1586R mice and FLNBWT/WT mice, indicating that the downregulation of FLNB was not a result of reduced transcription of a mutant gene, but was more likely caused by reduced mRNA translation or increased protein degradation.
3.2. Short stature in FLNBG1586R/G1586R mice
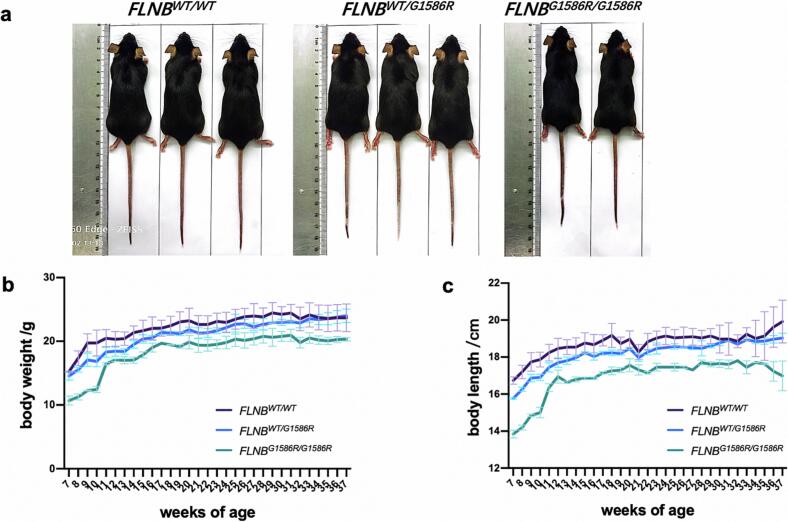
We measured the body weights and body lengths of 3 FLNBWT/WT, 3 FLNBWT/G1586R, and 2 FLNBG1586R/G1586R male mice every week from ages 7 weeks to 37 weeks. As shown in Fig. 2, the body weights of FLNBG1586R/G1586R mice were lower than FLNBWT/G1586R mice and FLNBWT/WT mice, and the body lengths of FLNBG1586R/G1586R mice were shorter than FLNBWT/G1586R mice and FLNBWT/WT mice. These findings suggested that FLNBG1586R/G1586R led to the short stature of mice during development.
Fig. 2.
Body weight and body length of the FLNBWT/WT mice, FLNBWT/G1586R mice and FLNBG1586R/G1586R mice. a Gross pictures of 3 FLNBWT/WT, 3 FLNBWT/G1586R and 2 FLNBG1586R/G1586R mice (male, 37-week). b-c Graphical representation of the body weights and body lengths of 3 FLNBWT/WT, 3 FLNBWT/G1586R and 2 FLNBG1586R/G1586R mice (male, from 7-week to 37-week). The body weights of FLNBG1586R/G1586R mice were lower than FLNBWT/G1586R mice and FLNBWT/WT mice, and the body lengths of FLNBG1586R/G1586R mice were shorter than FLNBWT/G1586R mice, and FLNBWT/WT mice.
3.3. Skeletal malformations in FLNBG1586R/G1586R mice
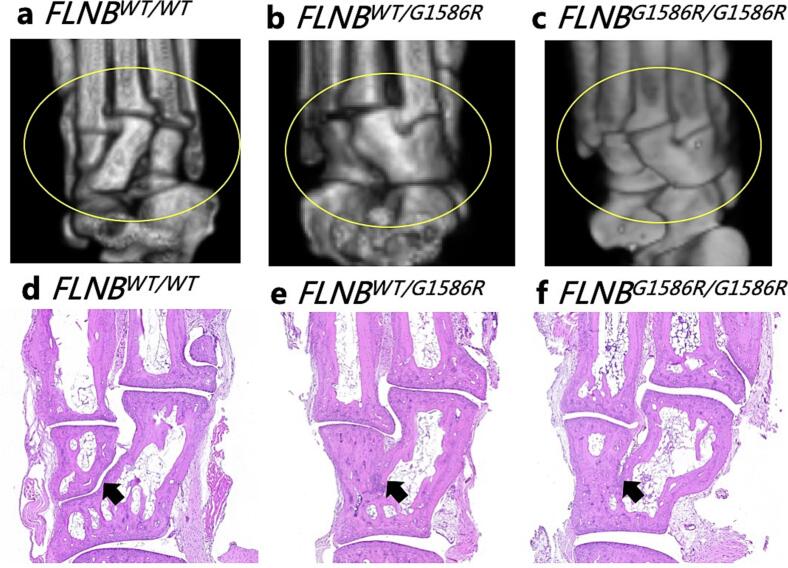
We performed micro-CT, HE staining and whole skeletal preparation in FLNBWT/WT, FLNBWT/G1586R, and FLNBG1586R/G1586R mice. As shown in Fig. 3, tarsal bones were separated in FLNBWT/WT mice; while those tarsal bones were fused in FLNBWT/G1586R mice and FLNBG1586R/G1586R mice. As shown in Figs. S1 and S2, the morphology of carpal bones and vertebrae were not significantly different among FLNBWT/WT mice, FLNBWT/G1586R mice and FLNBG1586R/G1586R mice. These results indicated that FLNBG1586R/G1586R led to the fusion of tarsal bones during development.
Fig. 3.
Micro-CT three dimensional reconstruction of tarsal bones, and hematoxylin and eosin (HE) staining of tarsal bones of the FLNBWT/WT mice, FLNBWT/G1586R mice and FLNBG1586R/G1586R mice (3 male mice in each group, 8-week). In the plantar view of hindlimb, tarsal bones (indicated in oval) were separated in the FLNBWT/WT mice (a, d); while tarsal bones (indicated in oval) were fused in FLNBWT/G1586R mice (b, e) and FLNBG1586R/G1586R mice (c, f);
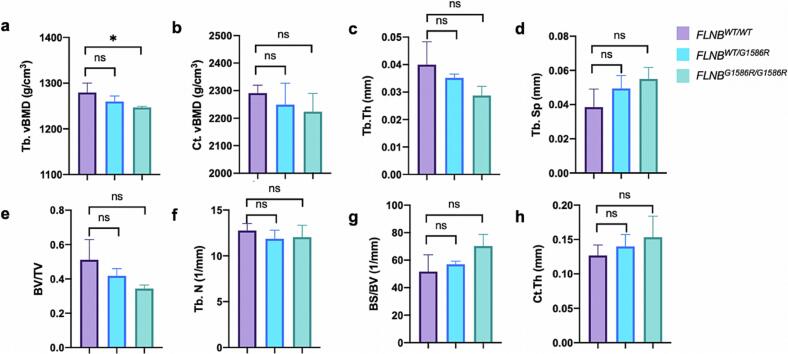
We measured the bone microarchitecture parameters in FLNBWT/WT mice, FLNBWT/G1586R mice and FLNBG1586R/G1586R mice. The trabecular volumetric BMD (Tb.vBMD) was significantly lower in FLNBG1586R/G1586R mice compared to FLNBWT/WT mice. The trabecular thickness (Tb.Th) in the FLNBG1586R/G1586R mice had a trend of downregulation, and the trabecular separation (Tb.Sp) in the FLNBG1586R/G1586R mice had a trend of upregulation, compared to FLNBWT/WT mice, though not statistically significant (Fig. 4). This indicated that the bone microarchitecture had deteriorated in FLNBG1586R/G1586R mice.
Fig. 4.
Bone microarchitecture parameters in FLNBWT/WT mice, FLNBWT/G1586R mice and FLNBG1586R/G1586R mice (3 male mice in each group, 8-week). Tb.vBMD was significantly lower in FLNBG1586R/G1586R mice compared to FLNBWT/WT mice. Tb.Th in the FLNBG1586R/G1586R mice had a trend of downregulation, and Tb.Sp in the FLNBG1586R/G1586R mice had a trend of upregulation, compared to FLNBWT/WT mice, though not statistically significant. Abbreviations: Trabecular volumetric bone mineral density (Tb.vBMD), cortical volumetric bone mineral density (Ct.vBMD), trabecular thickness (Tb.Th), trabecular separation (Tb.Sp), bone volume fraction (BV/TV), trabecular number (Tb.N), bone surface/volume (BS/BV), and cortical bone thickness (Ct.Th).
We also measured the serum bone turnover markers in FLNBWT/WT mice, FLNBWT/G1586R mice and FLNBG1586R/G1586R mice (Fig. S3). Serum bone formation indexes, P1NP and OCN, presented a trend of downregulation in the FLNBG1586R/G1586R mice compared to FLNBWT/WT mice; and bone resorption indexes, CTX-1 and TRACP5b, presented a trend of upregulation in the FLNBG1586R/G1586R mice compared to FLNBWT/WT mice, though the differences were not statistically significant.
3.4. In situ hybridization of HOX genes in embryos
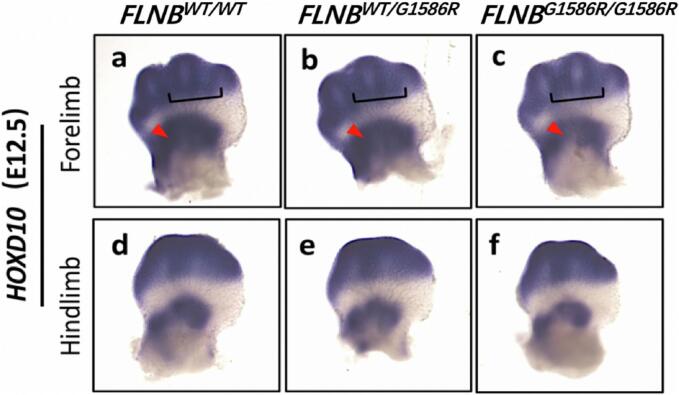
HOXD10 is a key gene regulating the segmentation of the forelimb and hindlimb (Quinonez and Innis, 2014; Wellik and Capecchi, 2003). We performed in situ hybridization of embryos (E12.5) of HOXD10 in FLNBWT/WT, FLNBWT/G1586R, and FLNBG1586R/G1586R mice (Fig. 5). In the metacarpal bones and carpal bones of the forelimb in E12.5, the staining area of HOXD10 was smaller and the staining was weaker in FLNBG1586R/G1586R mice compared with FLNBWT/WT mice and FLNBWT/G1586R. This indicated that the transcription level of HOXD10 was downregulated in FLNBG1586R/G1586R mice during the embryonic development of carpal bone.
Fig. 5.
In situ hybridization of embryo (E12.5) of HOXD10 in FLNBWT/WT mice, FLNBWT/G1586R mice and FLNBG1586R/G1586R mice. (a-c) In the metacarpal bones (pointed by black bracket) and carpal bones (pointed by red arrow) of forelimb, the staining area of HOXD10 was smaller and the staining of HOXD10 was weaker in FLNBG1586R/G1586R mice compared with FLNBWT/WT mice and FLNBWT/G1586R. (d-f) In the metatarsal bones and tarsal bones of hindlimb, the staining of HOXD10 was not significantly different in FLNBG1586R/G1586R mice compared with FLNBWT/WT mice and FLNBWT/G1586R.
HOXB2, HOXA4, and HOXB4 are key genes in the development of cervical spine (Quinonez and Innis, 2014). We performed in situ hybridization of embryos (E12.5) of HOXB2, HOXA4, and HOXB4 in FLNBWT/WT mice, FLNBWT/G1586R mice, and FLNBG1586R/G1586R mice (Fig. S4). In the cervical spine, the staining area of HOXB2 was smaller in FLNBG1586R/G1586R mice compared with FLNBWT/WT mice and FLNBWT/G1586R, while the staining of HOXA4 and HOXB4 was not significantly different in FLNBG1586R/G1586R mice compared with FLNBWT/WT mice and FLNBWT/G1586R. These findings indicated that HOXB2 was downregulated in FLNBG1586R/G1586R mice during the embryonic development of the cervical spine.
4. Discussion
4.1. Summary of the study
In this study, we established a mouse model with a loss-of-function mutation in FLNB using CRISPR/Cas9 technology. The mouse model presented short stature and fusions in tarsal bones, which phenocopied human disease SCT syndrome. We further found that the transcription of different HOX gene subtypes was downregulated in the embryo of mice with loss-of-function FLNB, using in situ hybridization of embryos. The findings above indicated that disruption of FLNB may lead to disorders in skeletal segmentation by regulating the expression of HOX genes.
4.2. Comparison to previous FLNB-deficient mice models
Previous researchers have generated FLNB-knockout mice models before (Farrington-Rock et al., 2008; Zhou et al., 2007). The model by Farrington-Rock et al. presented fusions in carpal bones and vertebrae, and the model by Zhou et al. presented fusions in vertebrae but no fusions in carpal and tarsal (though reductions in hyaline cartilage in ribs, metacarpals, phalanges, and tarsals were found). In our mouse model, instead of using a gene-trap vector to knock out the FLNB gene, we used CRISPR/Cas9 technology to introduce a known pathogenic point mutation G1586R into FLNB and down-regulated the expression of FLNB. We observed fusions in tarsal bones, which was similar to previous findings in FLNB-deficient mice models, but we did not find obvious fusions in vertebrae in the model. The possible reason may be that in previous models the actin-binding domain (ABD) of FLNB was dysfunctional, while in our model the point mutation G1586R was located in repeat-14 of FLNB, which did not lead to a complete loss-of-function in FLNB, so that the skeletal segmentation was not significantly interfered.
4.3. Known mechanisms of FLNB in causing skeletal malformation
There have been several mechanisms proposed by previous researchers explaining how FLNB regulated skeletal development. ① Loss-of-function mutation in FLNB leads to abnormal growth in the growth plate of long bone. Hu et al. found that in the mouse model with loss-of-function in FLNB, the proliferation zone of the growth plate of the long bone became narrow, while the differentiated hypertrophic zone became broadened, and the osteogenic capacity was reduced (Hu et al., 2014). In addition, Lu et al. found that the apoptosis rate of chondrocytes was significantly increased in the hypertrophic zone of the radius of the mice with loss-of-function in FLNB (Lu et al., 2007). These findings explained why the length and diameter of the long bone are smaller in patients with SCT syndrome. ② Gain-of-function mutation in FLNB leads to the enhancement of binding of FLNB and actin and affects the migration of cell skeleton. Daniel et al. and Sawyer et al. found the missense mutation in the binding domain of FLNB with actin leads to increased binding capacity with actin, which was associated with skeletal malformation (Daniel et al., 2012; Sawyer et al., 2009). Those studies above, though interesting, only explained the phenotype of short stature in FLNB-deficient patients, but did not explain why the number of ossification centers was abnormal (i.e. why the skeletal segmentation was abnormal) in FLNB-related skeletal disorders.
4.4. HOX genes and skeletal malformation
The skeletal segmentation in the embryonic period of vertebral animals is known to be regulated by HOX genes (Quinonez and Innis, 2014). Different HOX genes express in different stages and different segments in the embryonic period and regulate segmentation of the skeleton and other tissues. For example, loss of function in Group 4 to 9 paralogues of HOX genes leads to malformations in cervical, thoracic, and lumbar vertebrae; loss of function in HOXD11 leads to fusion in carpal and fusion in the distal radius and ulna; and loss of function in HOXD12 and HOXD13 leads to fusion in the carpal and metacarpal and shortening of phalanges. Interestingly, the skeletal malformation phenotypes were remarkably similar between the HOX-mutated spectrum disorders and FLNB-mutated spectrum disorders, both of which were characterized by alterations in the number of ossification centers. Therefore, we studied whether loss of function in FLNB interfered with transcription of HOX in the embryonic period.
4.5. Loss of function in FLNB interferes with transcription of the HOX gene
In this study, we found that the transcription level of HOXD10 was downregulated in the metacarpal bones and carpal bones of the FLNBG1586R/G1586R mice during the embryonic development of carpal bone, and the transcription of HOXB2 was downregulated in FLNBG1586R/G1586R mice during the embryonic development of cervical spine (Figs. 5, S4). HOXD10 is a key gene regulating the segmentation of the forelimb and hindlimb, and HOXB2 is a key gene regulating the segmentation of the cervical spine (Quinonez and Innis, 2014; Wellik and Capecchi, 2003). Those findings above suggested that loss of function in FLNB may lead to disorders in the segmentation through HOX genes during the embryonic period, which first established a link between FLNB and HOX gene.
In the metatarsal bones and tarsal bones of the hindlimb, the transcription level of HOXD10 was not significantly different in FLNBG1586R/G1586R mice compared with FLNBWT/WT mice and FLNBWT/G1586R (Fig. 5). The development of the hindlimb is usually earlier than the forelimb by around half a day, so we assume the segmentation of the hindlimb has been finished by E12.5, so the expression of HOXD10 was not different in FLNBG1586R/G1586R mice compared to FLNBWT/WT mice and FLNBWT/G1586R.
4.6. How does FLNB regulate HOX in skeletal development?
Until now, no studies have reported the direct interaction between FLNB and HOX. However, there were some clues indicating that connections between FLNB and HOX might exist through the BMP-Smad signaling pathway. Zheng et al. have reported that FLNB regulated the phosphorylation of Smad. When loss of function occurred in FLNB, the free phosphorylated Smad was increased in the cytoplasm, which increased the transcription activity of Runx2 (Zheng et al., 2007). In addition, Li et al. have reported that HOX proteins were downstream DNA-binding proteins in the BMP signaling cascade and the transcriptional activities of HOX were regulated by Smads (Li et al., 2006). Therefore, we hypothesize that loss of function in FLNB may up-regulate the phosphorylation of Smad, down-regulate the transcription function of HOX gene, and therefore inhibit the skeletal segmentation in the embryonic period and lead to skeletal malformation.
There were also other studies indicating that FLNB may regulate HOX through actin. It is known that FLNB, a scaffold protein, assists in the rearrangement of F-actin in the cytoplasm (Zhao et al., 2016). Ferrai et al. have reported that inhibition of actin polymerization blocked the transcription of HOXB genes (Ferrai et al., 2009). And Miyamoto et al. has found that knock-down of an actin-binding protein resulted in defective HOX gene action and abnormal embryonic development (Miyamoto et al., 2013). Therefore, we hypothesize that loss of function in FLNB may interfere with the polymerization of actin, down-regulate the transcription of HOX gene, and lead to abnormal embryonic skeletal segmentation. However, further experiments need to be performed to test if FLNB truly regulates the activity of HOX gene through either the BMP-Smad signaling cascade or through actin polymerization.
4.7. Strengths and limitations
There were several strengths in the study. We first established a mouse model with a pathogenic point mutation G1586R in FLNB using CRISPR/Cas9 technology, which presented loss of function in FLNB and skeletal malformations including short stature and fusions in tarsal bones. We first established a link between FLNB and HOX, showing that FLNB may regulate skeletal segmentation through HOX gene. Besides, the point mutation G1586R we introduced to FLNB was located in the repeat-14 of FLNB. Previous studies always focused on the mutations in the actin-binding domain (ABD) of FLNB, but our study indicated that functions in other domains in FLNB were also important and worth further exploration.
There were also limitations in the study. The sample size of experiments in this study was relatively small. This was due to the low survival rate of mice with a homozygous mutation in FLNB, and further experiments should be performed at different stages of the life cycle of the mice with a larger sample size. Besides, since HOXD10 is a key gene regulating the segmentation of the forelimb and hindlimb, we mainly focused on the transcription level of HOXD10 gene in the embryonic development of carpal bones and tarsal bones, but didn't test other HOX gene subtypes which were also responsible for this segmentation process. In addition, we did not further explain how FLNB exactly regulates HOX gene, which needed further experiments to study.
In conclusion, we generated a mouse model with a loss-of-function mutation in FLNB, which showed a shorter stature, and fusions in tarsal bones, and partially phenocopied FLNB-related skeletal disorders in humans. The transcription of HOXD10 was downregulated in the regions of the metacarpal and carpal regions during embryonic development. Loss of function in FLNB may lead to skeletal malformation by interfering with skeletal segmentation through HOX genes.
Declaration of generative AI in scientific writing
We declare that no AI or AI-assisted technologies were used in the writing process of the manuscript.
CRediT authorship contribution statement
Qiming Xu: Writing – review & editing, Supervision, Project administration, Conceptualization. Lijia Cui: Writing – original draft, Funding acquisition, Formal analysis, Data curation. Yude Lin: Data curation. Leigh-Anne Cui: Data curation. Weibo Xia: Writing – review & editing, Supervision, Funding acquisition, Conceptualization.
Declaration of competing interest
Lijia Cui, Yude Lin, Leigh-Anne Cui, Weibo Xia, Qiming Xu declare no conflict of interest.
Acknowledgement
We acknowledge Dr. Yujie Shi from the Scripps Research Institute for her important instruction in the experimental design. This research was supported by the National Natural Science Foundation of China (LC, 82100946; WX, 82270938), Bethune Charitable Foundation (LC, G-X-2020-1107-16), Young Elite Scientists Sponsorship Program by BAST (LC, No.BYESS2023171), CAMS Innovation Fund for Medical Sciences (WX, 2021-I2M-1-002), National Key Research and Development Program of China (WX, 2021YFC2501700), National High Level Hospital Clinical Research Funding (WX, 2022-PUMCH-D-006), and the Non-profit Central Research Institute Fund of the Chinese Academy of Medical Sciences (LC, 2023-PT320-10).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bonr.2024.101746.
Contributor Information
Qiming Xu, Email: xuqiming@jst-hosp.com.cn.
Lijia Cui, Email: cuilijia@pumch.cn.
Yude Lin, Email: linyudesjtu@sjtu.edu.cn.
Leigh-Anne Cui, Email: lcui@scripps.edu.
Weibo Xia, Email: xiawb@pumch.cn.
Appendix A. Supplementary data
Supplementary figures
Data availability
All data are available from the corresponding authors upon reasonable request.
References
- Daniel P.B., Morgan T., Alanay Y., Bijlsma E., Cho T.J., Cole T., Collins F., David A., Devriendt K., Faivre L., Ikegawa S., Jacquemont S., Jesic M., Krakow D., Liebrecht D., Maitz S., Marlin S., Morin G., Nishikubo T., Nishimura G., Prescott T., Scarano G., Shafeghati Y., Skovby F., Tsutsumi S., Whiteford M., Zenker M., Robertson S.P. Disease-associated mutations in the actin-binding domain of filamin B cause cytoplasmic focal accumulations correlating with disease severity. Hum. Mutat. 2012;33(4):665–673. doi: 10.1002/humu.22012. [DOI] [PubMed] [Google Scholar]
- Farrington-Rock C., Kirilova V., Dillard-Telm L., Borowsky A.D., Chalk S., Rock M.J., Cohn D.H., Krakow D. Disruption of the Flnb gene in mice phenocopies the human disease spondylocarpotarsal synostosis syndrome. Hum. Mol. Genet. 2008;17(5):631–641. doi: 10.1093/hmg/ddm188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrai C., Naum-Onganía G., Longobardi E., Palazzolo M., Disanza A., Diaz V.M., Crippa M.P., Scita G., Blasi F. Induction of HoxB transcription by retinoic acid requires actin polymerization. Mol. Biol. Cell. 2009;20(15):3543–3551. doi: 10.1091/mbc.E09-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Lu J., Lian G., Ferland R.J., Dettenhofer M., Sheen V.L. Formin 1 and filamin B physically interact to coordinate chondrocyte proliferation and differentiation in the growth plate. Hum. Mol. Genet. 2014;23(17):4663–4673. doi: 10.1093/hmg/ddu186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon G.W., Lee M.N., Jung J.M., Hong S.Y., Kim Y.N., Sin J.B., Ki C.S. Identification of a de novo heterozygous missense FLNB mutation in lethal atelosteogenesis type I by exome sequencing. Ann. Lab. Med. 2014;34(2):134–138. doi: 10.3343/alm.2014.34.2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski K., Tsuruta T., Kameda Y., Kan A., Leslie G. New forms of neonatal death dwarfism. Report of 3 cases. Pediatr. Radiol. 1981;10(3):155–160. doi: 10.1007/BF00975190. [DOI] [PubMed] [Google Scholar]
- Kozlowski K., Sillence D., Cortis-Jones R., Osborn R. Boomerang dysplasia. Br. J. Radiol. 1985;58(688):369–371. doi: 10.1259/0007-1285-58-688-369. [DOI] [PubMed] [Google Scholar]
- Krakow D., Robertson S.P., King L.M., Morgan T., Sebald E.T., Bertolotto C., Wachsmann-Hogiu S., Acuna D., Shapiro S.S., Takafuta T., Aftimos S., Kim C.A., Firth H., Steiner C.E., Cormier-Daire V., Superti-Furga A., Bonafe L., Graham J.M., Jr., Grix A., Bacino C.A., Allanson J., Bialer M.G., Lachman R.S., Rimoin D.L., Cohn D.H. Mutations in the gene encoding filamin B disrupt vertebral segmentation, joint formation and skeletogenesis. Nat. Genet. 2004;36(4):405–410. doi: 10.1038/ng1319. [DOI] [PubMed] [Google Scholar]
- Larsen L.J., Schottstaedt E.R., Bost F.C. Multiple congenital dislocations associated with characteristic facial abnormality. J. Pediatr. 1950;37(4):574–581. doi: 10.1016/s0022-3476(50)80268-8. [DOI] [PubMed] [Google Scholar]
- Li X., Nie S., Chang C., Qiu T., Cao X. Smads oppose Hox transcriptional activities. Exp. Cell Res. 2006;312(6):854–864. doi: 10.1016/j.yexcr.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Lu J., Lian G., Lenkinski R., De Grand A., Vaid R.R., Bryce T., Stasenko M., Boskey A., Walsh C., Sheen V. Filamin B mutations cause chondrocyte defects in skeletal development. Hum. Mol. Genet. 2007;16(14):1661–1675. doi: 10.1093/hmg/ddm114. [DOI] [PubMed] [Google Scholar]
- Maroteaux P., Spranger J., Stanescu V., Le Marec B., Pfeiffer R.A., Beighton P., Mattei J.F. Atelosteogenesis. Am. J. Med. Genet. 1982;13(1):15–25. doi: 10.1002/ajmg.1320130106. [DOI] [PubMed] [Google Scholar]
- Miyamoto K., Teperek M., Yusa K., Allen G.E., Bradshaw C.R., Gurdon J.B. Nuclear Wave1 is required for reprogramming transcription in oocytes and for normal development. Science. 2013;341(6149):1002–1005. doi: 10.1126/science.1240376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinonez S.C., Innis J.W. Human HOX gene disorders. Mol. Genet. Metab. 2014;111(1):4–15. doi: 10.1016/j.ymgme.2013.10.012. [DOI] [PubMed] [Google Scholar]
- Sawyer G.M., Clark A.R., Robertson S.P., Sutherland-Smith A.J. Disease-associated substitutions in the filamin B actin binding domain confer enhanced actin binding affinity in the absence of major structural disturbance: insights from the crystal structures of filamin B actin binding domains. J. Mol. Biol. 2009;390(5):1030–1047. doi: 10.1016/j.jmb.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Stanley C.S., Thelin J.W., Miles J.H. Mixed hearing loss in Larsen syndrome. Clin. Genet. 1988;33(5):395–398. doi: 10.1111/j.1399-0004.1988.tb03468.x. [DOI] [PubMed] [Google Scholar]
- Stossel T.P., Condeelis J., Cooley L., Hartwig J.H., Noegel A., Schleicher M., Shapiro S.S. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2001;2(2):138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- Tenconi R., Kozlowski K., Largaiolli G. Boomerang dysplasia. A new form of neonatal death dwarfism. Rofo. 1983;138(3):378–380. [PubMed] [Google Scholar]
- Wellik D.M., Capecchi M.R. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science. 2003;301(5631):363–367. doi: 10.1126/science.1085672. [DOI] [PubMed] [Google Scholar]
- Wilkinson D.G. In: In Situ Hybridization: A Practical Approach. Wilkinson D.G., editor. Oxford University Press; New York: 1992. Whole mount in situ hybridization of vertebrate embryos; pp. 89–99. [Google Scholar]
- Xu Q., Wu N., Cui L., Wu Z., Qiu G. Filamin B: the next hotspot in skeletal research? J. Genet. Genomics. 2017;44(7):335–342. doi: 10.1016/j.jgg.2017.04.007. [DOI] [PubMed] [Google Scholar]
- Xu Q., Wu N., Cui L., Lin M., Thirumal Kumar D., Doss C. George Priya, Wu Z., Shen J., Song X., Qiu G. Comparative analysis of the two extremes of FLNB-mutated autosomal dominant disease spectrum: from clinical phenotypes to cellular and molecular findings. Am. J. Transl. Res. 2018;10(5):1400–1412. [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Shapiro S.S., Eto M. F-actin clustering and cell dysmotility induced by the pathological W148R missense mutation of filamin B at the actin-binding domain. Am. J. Phys. Cell Phys. 2016;310(1):C89–C98. doi: 10.1152/ajpcell.00274.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Baek H.J., Karsenty G., Justice M.J. Filamin B represses chondrocyte hypertrophy in a Runx2/Smad3-dependent manner. J. Cell Biol. 2007;178(1):121–128. doi: 10.1083/jcb.200703113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Tian F., Sandzen J., Cao R., Flaberg E., Szekely L., Cao Y., Ohlsson C., Bergo M.O., Boren J., Akyurek L.M. Filamin B deficiency in mice results in skeletal malformations and impaired microvascular development. Proc. Natl. Acad. Sci. USA. 2007;104(10):3919–3924. doi: 10.1073/pnas.0608360104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Data Availability Statement
All data are available from the corresponding authors upon reasonable request.