Abstract
TATA-binding protein-associated factors (TAFIIs) within TFIID control differential gene transcription through interactions with both activators and core promoter elements. In particular, TAFII150 contributes to initiator-dependent transcription through an unknown mechanism. Here, we address whether TAFIIs within TFIID are sufficient, in conjunction with highly purified general transcription factors (GTFs), for differential core promoter-dependent transcription by RNA polymerase II and whether additional cofactors are required. We identify the human homologue of Drosophila TAFII150 through cognate cDNA cloning and show that it is a tightly associated component of human TFIID. More importantly, we demonstrate that the human TAFII150-containing TFIID complex is not sufficient, in the context of all purified GTFs and RNA polymerase II, to mediate transcription synergism between TATA and initiator elements and initiator-directed transcription from a TAFII-dependent TATA-less promoter. Therefore, TAFII-promoter interactions are not sufficient for the productive core promoter-selective functions of TFIID. Consistent with this finding, we have partially purified novel cofactor activities (TICs) that potentiate the TAFII-mediated synergism between TATA and initiator elements (TIC-1) and TAFII-dependent transcription from TATA-less promoters (TIC-2 and -3). Furthermore, we demonstrate an essential function for TFIIA in TIC- and TAFII-dependent basal transcription from a TATA-less promoter. Our results reveal a parallel between the basal transcription activity of TAFIIs through core promoter elements and TAFII-dependent activator function.
The structure of the core promoter region (i.e., DNA sequences flanking the transcription start site and including the TATA, initiator, and downstream elements that interact with the general transcription machinery) of protein-coding genes has an important influence both on the efficiency of basal transcription and on the ability of the core promoter to respond to upstream promoter-bound activators in vivo and in vitro (reviewed in references 33, 38, and 46). Although the general transcription machinery has been well characterized, little is known about the factors and mechanisms that control its activity in a core promoter-specific manner. Whereas various factors like E2F, YY1, TFII-I, and USF may regulate transcription not only through upstream promoter elements but also through interactions with the core promoter regions of certain genes (for reviews, see references 38 and 50), several components of the basal transcription machinery have intrinsic and more general core promoter-selective functions. For instance, the differential affinity of TATA-binding protein (TBP) for different TATA box DNA sequences and conformations can influence the efficiency of transcription initiation (28, 37, 39; for a review, see reference 3). Previous studies have also demonstrated differential requirements for the general transcription factors (GTFs) TFIIE, TFIIF, and TFIIH for basal transcription from different supercoiled promoter templates (27). In addition, RNA polymerase II (pol II), by itself, appears to have weak sequence preferences (4) and is involved in start site selection in yeast in conjunction with TFIIB and TFIIF (29, 40). More recently, TBP-associated factors (TAFIIs) within TFIID have been shown to be essential both for the synergistic function of TATA and initiator elements (14, 19, 45) and for basal transcription from TATA-less promoters through an alternative transcription initiation pathway that is largely independent of TBP-DNA interactions (19, 20). Importantly, the core promoter-selective function of TAFIIs has been further confirmed in studies of yeast cells (24, 35).
How do TAFIIs contribute to core promoter-specific transcription? Depending on the core promoter context, TAFIIs have been shown to either stabilize or destabilize TBP/TFIID-DNA interactions (10, 14, 17, 45). DNase I footprinting and UV cross-linking experiments with highly purified TFIID have provided evidence for several TAFIIs being in close proximity to DNA, both immediately upstream and downstream of the TATA box and over the initiator and downstream region of certain TATA-containing and TATA-less promoters (2, 5, 14, 25, 31, 44, 53). In particular, Drosophila TAFII150 has been reported to interact directly with the adenovirus major late (AdML) and Drosophila Hsp70 core promoters (44, 45). Interestingly, this interaction correlates with increased stability of the TFIID-promoter complex and with the requirement for Drosophila TAFII150, as part of a complete TFIID or in a trimeric complex with TBP and TAFII250, for TATA-dependent initiator function in vitro (13, 45).
Similarly, human TFIID interacts synergistically with the TATA, initiator, and downstream regions of the AdML and model TATA- and initiator-containing synthetic core promoters in a manner that is dependent on functional initiator sequences and strengthened by TFIIA (10). However, because so far a homologue of Drosophila TAFII150 has not been observed within the human TFIID complex, it has been suggested that, in contrast to the situation in Drosophila, human TAFII150 is not involved in initiator recognition by human TFIID (10, 15). Nevertheless, a recently cloned human factor (CIF150) that is similar to Drosophila TAFII150, but apparently not associated with human TFIID/TBP, was reported to be required in addition to TFIID for TATA-dependent initiator activity in a partially purified in vitro transcription system (16).
The studies cited above clearly establish an important core promoter-specific role of TAFIIs and implicate TAFII150 in TATA-dependent functions of the initiator and downstream regions in certain promoters. However, besides TFIID/TAFIIs, the factors and molecular mechanisms involved in differential core promoter-selective transcription, especially for specific transcription initiation at TAFII-dependent TATA-less promoters, remain poorly characterized. This is because earlier studies used either partially purified systems and/or, in the case of TATA-less promoters, core promoter constructs that are not strictly TAFII dependent in vitro (i.e., that can support TBP-mediated transcription through low-affinity but functional TBP binding sites). In a further analysis of the molecular mechanisms involved in TAFII-mediated core promoter selectivity, we have cloned the human homologue of Drosophila TAFII150. We show that it is essentially identical to CIF150, but in contrast to what was reported recently for CIF150 (16), we establish that it is a stably associated component of the human TFIID complex. Furthermore, and more importantly, we have identified at least three novel TAFII- and initiator-dependent cofactors (TICs) that are different from the previously characterized initiator region-binding proteins YY1, TFII-I, and USF and are selectively required for the core promoter-specific functions of the human TAFII150-containing TFIID complex through the initiator region of TATA-containing and TATA-less promoters. In addition, our analysis of the factors required for specific transcription initiation from the TATA-less terminal deoxynucleotidyltransferase (TdT) promoter reveals a novel core promoter-specific basal function for TFIIA that is different from a simple facilitation of TBP-DNA interactions.
MATERIALS AND METHODS
Plasmids.
pTdT(−1300/+59), pTdT(−41/+59), pG5TdT(−41/+59) (G5-TdT), pG5TdT(−41/+33) (G5-TdT+33), pG5TdT(−41TATA/+33) (INR+), pG5TdT(−41TATA/Inr−+33) (INR−), pG5E1bCAT (E1b), pG5hβPol(−41/+58)CAT (β-Pol), pAdML(−257/+33)CAT, and phHsp70(−33/+99)CAT (Hsp) have been described previously (19, 20), as has pMLI(−45/+65)CAT (ML) (8).
Cloning and expression of recombinant human TAFII150.
A GenBank search revealed three nonoverlapping mouse epitope sequence tag (EST) sequences (AA028778, AA103510, and W13567) with 50 to 60% amino acid identity with Drosophila TAFII150. These sequences were used to design degenerate oligonucleotides for PCR-mediated cloning of human TAFII150 cDNA sequences. A 3.8-kb cDNA encoding a full-length human TAFII150 was finally isolated by PCR with Pfu polymerase (Stratagene) from a HeLa Marathon-Ready cDNA library (Clontech) essentially as recommended by the manufacturers. The sequence was confirmed from at least two independent clones, except for one nucleotide position that differs in two independent clones and could extend the N terminus by 10 amino acids (see the legend to Fig. 1). A full-length FLAG-tagged human TAFII150 (f:TAFII150) cDNA was cloned into pT7Blue vector (Novagen) between SalI and EcoRI sites, and the resulting pT7-hFL150 expression vector was used to express recombinant protein in coupled in vitro transcription-translation reactions with the TNT system (Promega). The sequence of the FLAG-tagged N terminus is MDYKDDDDKNRKK (the FLAG sequence is underlined). For baculovirus-mediated expression of human f:TAFII150, the f:TAFII150 cDNA was inserted between the SmaI and NotI sites of pVL1393 (Pharmingen). The resulting pVL1393-f:T150 vector was cotransfected with BacVector 3000 (Novagen) Autographa californica nuclear polyhedrosis virus DNA into Sf9 cells to produce recombinant viruses (essentially as recommended by the manufacturers). Recombinant human f:TAFII150 was purified from recombinant baculovirus-infected Sf9 cells by sonication in BC-300 buffer (20 mM Tris-HCl [pH 7.9], 20% glycerol, 300 mM KCl) containing 0.1% Nonidet P-40 (NP-40), centrifugation at 100,000 × g, loading the supernatant on M2-agarose (Kodak), washing the M2 resin extensively with BC-300, eluting with BC-100 containing FLAG peptide (0.2 mg/ml), loading the eluate onto Ni2+-nitrilotriacetic acid (NTA)-agarose (Qiagen), washing extensively with BC-1000, BC-500, and BC-100, and eluting the purified recombinant f:TAFII150 with 250 mM imidazole in BC-100. For antiserum production, a human TAFII150 cDNA fragment encoding amino acids 539 to 655 cloned into His6T-pET11d was used to express in bacteria a hexahistidine (His6)-tagged protein that was purified on Ni2+-NTA-agarose and used to immunize rabbits by standard protocols (Covance). Western blotting was done by standard protocols with an enhanced chemiluminescence detection kit (Amersham).
FIG. 1.
Predicted amino acid sequence of human TAFII150. Positions identical to those in Drosophila TAFII150 are marked with asterisks (ClustalW alignment program of MacVector with a blosum matrix; gap distance, 8; open gap penalty, 10; extend gap penalty, 0.1). The underlined sequence (amino acids 90 to 100) corresponds to a Glu-C peptide microsequence from the 135-kDa TAFII protein of highly purified eTFIID. A mouse TAFII150 N-terminal EST sequence (GenBank accession no. AA200390) is 100% identical at the amino acid level to the human sequence shown here from position 120 to the first methionine and diverges upstream with no other ATG (data not shown). However, an independent human 5′-end PCR clone has one nucleotide difference, which creates an in-frame upstream but weaker (non-Kozak) initiator ATG and adds the 10 predicted amino acids MPLTGVEPAR at the N terminus. This upstream ATG has also been observed recently in independent CIF150/TAFII150 cDNAs (12a, 16).
Cell lines and purification of GTFs, pol II, and TICs.
A HeLa cell line derivative stably expressing f:TAFII100 was established by using the pCIN4 expression vector (32). The cell line stably expressing f:TAFII135 will be described elsewhere (42). Purification of epitope-tagged TFIID (eTFIID) complexes was essentially as described previously (5, 53), with the modifications described in the text. Native TFIIA (Mono Q fraction) was purified as reported elsewhere (21). Native TFIIA (Ni2+-NTA-agarose), f:TFIID, TFIID (Mono S), TFIIE/F/H (Mono S), pol II (Mono Q), and bacterially expressed recombinant (His6)-TFIIB, His6-TBP, Gal4-VP16, and PC4 were purified as described elsewhere (12). Native TFIIH was purified from the phosphocellulose (P11)–0.5 M KCl/DEAE-cellulose–0.12 M KCl (P110.5/DE0.12) fraction, using the BC-buffer system (12; see also above) complemented with 0.2 mM phenylmethylsulfonyl fluoride and either 5 mM dithiothreitol or 10 mM 2-mercaptoethanol and sequential chromatographic steps as follows: Q-Sepharose (Pharmacia), loaded at 0.12 M KCl (BC-120) and step eluted with 0.3 M KCl (BC-300); heparin-Sepharose (Pharmacia), loaded at 0.22 M KCl and step eluted with 0.6 M KCl; S-Sepharose (Pharmacia) flowthrough at 0.22 M KCl; second S-Sepharose, loaded at 0.12 M KCl and developed with a gradient from 0.12 to 0.5 M KCl; and Q-Sepharose (Q2) developed with a gradient from 0.1 to 0.4 M KCl. TFIIH was monitored during purification with antibodies against five of its subunits: ERCC3, p62, CDK7, cyclin H, and MAT-1. In the final (Q2) step, TFIIH eluted at 0.18 M KCl and was free of all the other GTFs and pol II. The pol II DE52 fraction [pol II (DE)] was purified from a resuspended HeLa nuclear pellet fraction (12) by precipitation with 0.09% polyethyleneimine, extraction of the pellet with TGED-250 (250 mM ammonium sulfate in buffer TGED [50 mM Tris-HCl {pH 7.9} at 4°C, 25% glycerol, 0.5 mM EDTA, 0.5 mM EGTA, 2 mM dithiothreitol) supplemented with a cocktail of protease inhibitors (Boehringer Mannheim), and precipitation with 67% saturated ammonium sulfate. The resuspended pellet was loaded onto a DE52 (Whatman) column in TGED-120, washed subsequently with TGED-150, eluted with a step at TGED-600, and then dialyzed to TGED-100. This simple pol II (DE) preparation (0.17 mg/ml; 1.4 mg of total protein for 80 ml of nuclear pellet) is in the A form, 10 to 30% pure by Coomassie blue staining, and free of all GTFs. Recombinant human His6-TFIIE is an equimolar combination of both soluble His6-tagged subunits (34 and 57 kDa) expressed in bacteria and purified essentially as described for His6-TFIIB (12). Recombinant human His6-RAP74 was expressed in bacteria and purified as described above over Ni2+-NTA-agarose but in the presence of 4 M urea. Bacterially expressed recombinant human RAP30 was solubilized from inclusion bodies in 6 M guanidine hydrochloride, renatured by dialysis against BC buffer containing 150 mM KCl (BC-150), loaded onto a heparin-Sepharose column in BC-150, and eluted with BC-500. Recombinant His6-TFIIF was reconstituted by corenaturation of equimolar amounts of RAP30 and His6-RAP74 (mixed in BC-500 with 4 M urea) by step dialysis to reach BC-100. TIC-1 was purified from the P11 0.5/DE 0.12 fraction (with BC buffers) as schematized in Fig. 4F; this included chromatography on Q-Sepharose where TIC-1 activity is found in the 0.12 M KCl flowthrough, followed by chromatography on a heparin-Sepharose column, which was loaded at 0.12 M KCl, washed with 0.25 M KCl, and eluted with 0.5 M KCl. After dialysis against BC-100, TIC-1 was sedimented through a 10 to 40% glycerol gradient (in BC-100), where it sediments between aldolase (163 kDa) and albumin (73 kDa) standards analyzed in parallel on an identical gradient. TIC-1 activity (fraction 8 [Fig. 4D]) was further purified on a Ni2+-NTA-agarose column, where it loses some activity and is found both in the flowthrough and in the bound fractions. The TIC-1 activity in the flowthrough is purer and was used for further characterizations. TIC-2 in the P11 0.5/DE 0.3 fraction was loaded on double-stranded DNA (dsDNA)-cellulose in BC-100 and eluted with BC-600. This fraction was dialyzed to BC-100, loaded onto a heparin-Sepharose column, and step eluted with BC-300 and BC-500. TIC-2 activity was present exclusively in the BC-300 eluate. TIC-1/3 corresponds to the previously described heparin Sepharose USA fraction (12, 21).
FIG. 4.
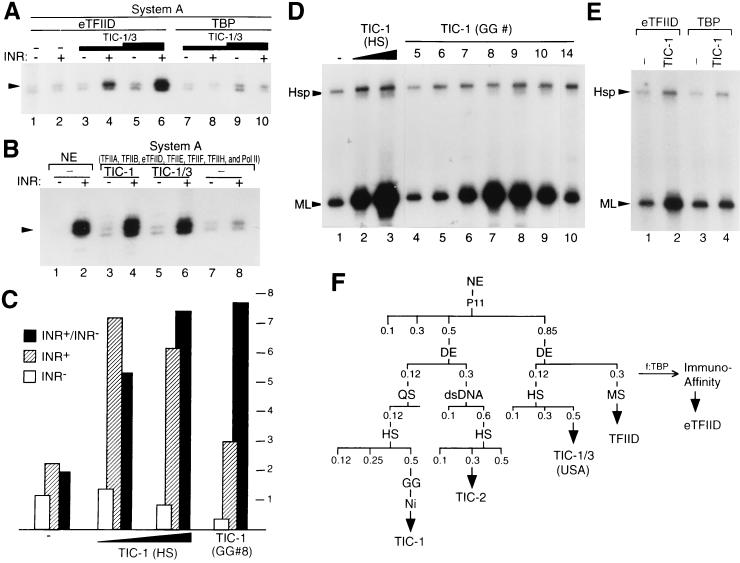
TIC-1 is a core promoter-specific cofactor for TAFII-dependent initiator function. (A) Reconstitution of TAFII-dependent initiator-mediated transcription in the highly purified system with a TIC-1-containing fraction. Transcription from supercoiled INR− and INR+ promoters was analyzed in the complete system A (as for Fig. 3A) but with either 1 μl of eTFIID (lanes 1 to 6) or 10 ng of TBP (lanes 7 to 10) and in the absence (lanes 1 and 2) or presence of 2 μl (lanes 3, 4, 7, and 8) or 4 μl (lanes 5, 6, 9, and 10) of the TIC-1/3 (P11 0.85/DE 0.12) fraction. (B) Two separate fractions, TIC-1 and TIC-1/3, can restore initiator function in the highly purified eTFIID-dependent system. Transcription from linear INR− and INR+ promoters was analyzed as described above in a nuclear extract (NE; lanes 1 and 2) and in the highly purified system A (lanes 3 to 8) either alone (lanes 7 and 8) or with 4 μl of either the P11 0.5/DE 0.12 fraction (TIC-1; lanes 3 and 4) or the P11 0.85/DE 0.12 fraction (TIC-1/3; lanes 5 and 6). (C) Purified TIC-1 fractions with initiator sequence-dependent activity. Transcripts from the linear INR− (open bars) and INR+ (hatched bars) promoters were quantitated in the purified system A alone (−) and in system A complemented with either partially purified TIC-1 (HS) fraction (0.5 and 1.0 μl; black triangle) or the activity peak of glycerol gradient fraction 8 (GG#8; 5 μl; see also below). Closed bars are initiator activities calculated as the ratio of INR+ to INR− (INR+/INR−) transcripts and indicated by the scale on the right side (as fold initiator-mediated stimulation). (D) Core promoter-selective activity of TIC-1 on natural core promoters. Transcription from linear AdML (ML) and human Hsp70 (Hsp) core promoter templates (analyzed by primer extension) was compared in the highly purified system A alone (lane 1) and with either increasing amounts (0.5 and 2.0 μl) of the TIC-1 (HS) fraction (lanes 2 and 3) or with fractions of a subsequent glycerol gradient (5 μl of each; lanes 4 to 10). Peak TIC-1 activity sediments at fraction GG#8 (lane 7). (E) TAFII-dependent core promoter-selective activity of the most purified TIC-1 fraction. Transcription from the Hsp and ML linear templates was performed in system A with either 2 μl of eTFIID (lanes 1 and 2) or 10 ng of TBP (lanes 3 and 4) and in either the absence (lanes 1 and 3) or presence (lanes 2 and 4) of the Ni2+-NTA-agarose TIC-1 fraction. (F) Purification scheme for TICs and TFIID used in the transcription systems. HeLa cell nuclear extracts (NE) were chromatographed as indicated (see also Materials and Methods) on phosphocellulose P11, DE52 (DE), Q Sepharose (QS), heparin-Sepharose (HS), Ni2+-NTA-agarose (Ni), dsDNA-cellulose (dsDNA), Mono S (MS), and anti-FLAG (M2)-agarose (Immuno-Affinity) resins.
Immunodepletions, reconstituted systems, and transcription and primer extension analyses.
Specific immunodepletions of factors from nuclear extracts or derived fractions were performed essentially as previously described (7; see also figure legends) in either BC-100 (TFII-I and TAFII100 depletions) or BC-400 (TFIIA depletion) buffer conditions. System A contains 1 μl of TFIIA (Ni-NTA-agarose), 1 to 2 μl of eTFIID (5 ng of f:TBP/μl), 15 ng of recombinant His6-TFIIB, 20 ng of recombinant His6-TFIIE, 20 ng of recombinant His6-TFIIF, 0.15 μl of TFIIH (Q2), and 0.7 μl of pol II (DE). System B contains 1 μl of TFIIA (Mono Q), 1 μl of eTFIID (5 ng of f:TBP/μl), 15 ng of recombinant His6-TFIIB, 10 ng of recombinant His6-TFIIE, 20 ng of recombinant His6-TFIIF, and 4 μl of P11 0.5/DE 0.3 (TFIIE/F/H/pol II) fraction. System C contains 2 μl of TFIIA (Mono Q), 2 μl of TFIID (Mono S), 30 ng of recombinant His6-TFIIB, 1 μl of TFIIE/F/H (Mono S), and 0.7 μl of pol II (Mono Q). Transcription reactions and primer extension analyses, with primers for TdT (16mer Reverse; New England Biolabs) and chloramphenicol acetyltransferase (CAT) constructs (CAT-30mer) labeled to the same specific activity, were essentially as previously described (19, 20). Quantitations were done on a Molecular Dynamics PhosphorImager.
Nucleotide sequence accession number.
The human TAFII150 cDNA sequence is available under GenBank accession no. AF040701.
RESULTS
Cloning of the human TAFII150 cDNA.
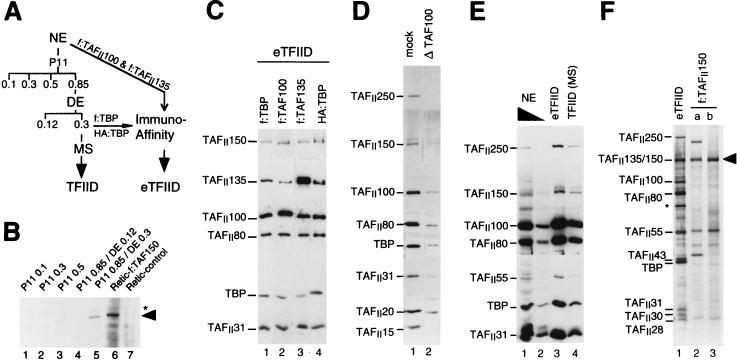
Because of the importance of Drosophila TAFII150 for core promoter recognition and initiator function and the apparent absence of a human homologue in highly purified TFIID preparations (see the introduction), we undertook the cloning of the human TAFII150 cDNA in order to reconstitute, with a complete set of TAFIIs within TFIID, TFIID-mediated core promoter selectivity in a homologous human in vitro transcription system. We used the Drosophila TAFII150 amino acid sequence to search the sequence databases and identified three nonoverlapping mouse EST sequences with 50 to 60% amino acid identity with Drosophila TAFII150. Degenerate oligonucleotides based on the mouse EST sequences were used to obtain (by PCR) corresponding human sequences that further were used to clone (by high-fidelity PCR) a 3.8-kb cDNA encoding a full-length human TAFII150 (Fig. 1; see below and Materials and Methods). Human TAFII150 is overall 46% identical to its Drosophila counterpart (Fig. 1) and clearly is the bona fide human homologue. Northern blot analyses indicate a single 5.7-kb human TAFII150 mRNA species that is expressed comparably in all tissues tested (data not shown and reference 20a). In an immunoblot analysis of HeLa nuclear extracts, polyclonal antibodies raised against a recombinant human TAFII150 protein fragment (amino acids 539 to 655) specifically recognize a ca. 135-kDa protein (see below) that elutes, after chromatography on phosphocellulose P11 and DE52 resins (Fig. 2A), in the P11 0.85/DE 0.3 TFIID-containing fraction (Fig. 2B, lane 5). Note that lower amounts of TAFII150 (and other TAFIIs) are also present in the P11 0.85/DE 0.12 fraction and can be detected after concentration on a subsequent heparin-Sepharose column in the USA fraction (see Fig. 5A, lane 5). In vitro transcription and translation in rabbit reticulocyte lysates of f:TAFII150-encoding cDNA gives rise to a protein with an electrophoretic mobility similar to that of the native human TAFII150 (Fig. 2B, lane 5 versus lane 6). This finding suggests that the cDNA encodes the full (or near-full)-length protein (see also the legend to Fig. 1).
FIG. 2.
Human TAFII150 (hTAFII150) is a tightly associated component of TFIID. (A) Scheme for fractionation and immunopurification of TFIID. Nuclear extracts (NE) of HeLa cells and derived eTFIID-expressing cell lines were chromatographed as indicated on phosphocellulose P11, DE52 (DE), Mono S (MS), and anti-FLAG (M2)-agarose or anti-HA antibody (Immuno-Affinity) resins. Numbers indicate the KCl molarity of elutions. (B) Immunodetection of human TAFII150 in a TFIID-containing fraction with anti-human TAFII150 antiserum and 5 μl of the indicated native fractions (lanes 1 to 5) or of in vitro-translated f:TAFII150 (Retic-f:TAF150; lane 6) and control (Retic-control; lane 7) lysates. The position of native TAFII150 and in vitro-translated f:TAFII150 is indicated by an arrowhead. An asterisk indicates a minor cross-reacting band of unknown origin in lane 5. (C) Stable association of hTAFII150 in immunopurified eTFIID, determined by Western blot analysis of eTFIID complexes from human cell lines expressing f:TBP, HA:TBP, f:TAFII100, or f:TAFII135 and immunopurified directly from nuclear extracts (lanes 2 and 3) or from a two-column-purified TFIID fraction and washed with 0.3 M KCl (lane 1) or with 1.2 M KCl (lane 4; see also text and panel A). The blot first was probed with anti-human TAFII150 antibodies and then was stripped and reprobed with antisera directed against the other TAFIIs and TBP as indicated. The TAFII150 bands on the blot from the first immunoreaction were coincident with the TAFII135 bands in the second immunoreaction (data not shown), and the relevant portion of the first immunoblot is placed above the second blot in the figure. Similar results were obtained in a side-by-side immunoblot analysis of eTFIID and recombinant TAFII135 with TAFII135 and TAFII150 antibodies (data not shown). (D) Human TAFII150 is depleted from a two-column (P11 0.85/DE 0.3) native TFIID fraction with specific anti-TAFII100 antibodies. Lanes 1 and 2 are from an immunoblot analysis of TFIID components present in the supernatant of a control immunodepletion (with protein A resin alone) (mock; lane 1) and in the supernatant after depletion with specific anti-TAFII100 antibodies bound to protein A resin (Δ TAF100; lane 2). (E) The amounts of TAFII150 relative to other TFIID subunits are similar in nuclear extracts and in purified TFIID, as determined by Western blot analysis of TBP and TAFIIs in 5 μl (lane 1) and 1 μl (lane 2) of nuclear extract (NE), 8 μl of immunopurified f:TBP-TFIID (eTFIID; lane 3), and 8 μl of native TFIID (Mono S [MS]) fraction (lane 4). The positions and identities of the different TAFIIs and TBP are indicated. (F) Recombinant human f:TAFII150 has an electrophoretic mobility on SDS-PAGE similar to that of human TAFII135 in eTFIID. Highly purified f:TBP-TFIID (8 μl of eTFIID; lane 1) and two independent preparations of baculovirus-expressed recombinant immunopurified f:TAFII150 (a and b; lanes 2 and 3) were analyzed on an SDS–10% polyacrylamide gel stained with silver. The positions of TAFIIs and f:TBP (TBP) are indicated. An arrowhead indicates the position of recombinant f:TAFII150. An asterisk marks an unspecific (non-TAF) protein band in eTFIID.
FIG. 5.
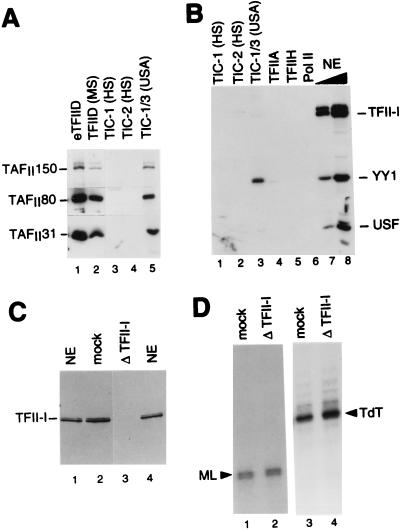
TIC-1 and TIC-2 are different from TFIID, TFII-I, YY1, and USF. (A) Immunoblot analysis of TIC fractions (see the legend to Fig. 4F for definitions) for the presence of TFIID components. Specific antisera against human TAFII150, TAFII80, and TAFII31 were used to probe a Western blot containing 8 μl of eTFIID (lane 1), 8 μl of native TFIID (MS fraction; lane 2), and 5- to 10-fold excess (over amounts used in transcription) of TIC-1 (5-μl HS fraction; lane 3), TIC-2 (5-μl HS fraction; lane 4), and TIC-1/3 (5-μl HS/USA fraction; lane 5). (B) Immunoblot analysis of purified native TIC and GTF fractions for the presence of TFII-I, YY1, and USF. Specific antisera against TFII-I, YY1, and USF1 were used to probe a Western blot containing a 5- to 10-fold excess (over transcription amounts, as above) of the TIC fractions as indicated (5 μl of each; lanes 1 to 3), and of purified native TFIIA (6-μl Ni fraction; lane 4), TFIIH (3-μl Q2 fraction; lane 5), and pol II (4-μl DE fraction; lane 6) used in system A, as well as 1 μl (lane 7) and 5 μl (lane 8) of nuclear extract (NE). (C) Nuclear extracts immunodepleted of TFII-I. Western blot analysis with specific antiserum against recombinant TFII-I (anti-TFII-I/A1) of 9 μg of each of normal nuclear extract (NE; 1.4 μl; lanes 1 and 4), nuclear extract mock depleted twice on protein A-Sepharose (mock; 2.8 μl; lane 2), and nuclear extract depleted twice with affinity-purified anti-TFII-I antibodies (anti-TFII-I/A1) cross-linked to protein A-Sepharose (Δ TFII-I; 2.8 μl; lane 3). By scanning densitometry, about 98% of TFII-I was depleted from the extract. (D) Specific immunodepletion of TFII-I from nuclear extracts does not affect transcription from the AdML and TdT promoters. Transcription from the supercoiled AdML (−257 to +33) (ML; 5 fmol; lanes 1 and 2) and TdT (−1300 to +59) (TdT; 75 fmol; lanes 3 and 4) promoters was analyzed in control mock-depleted (mock; lanes 1 and 3) and TFII-I-immunodepleted (Δ TFII-I; lanes 2 and 4) extracts by primer extension as described in the legend to Fig. 3. Autoradiography was about 10 times shorter for lanes 1 and 2 than for lanes 3 and 4.
Human TAFII150 is stably associated within the TFIID complex.
Our finding of human TAFII150 in P11 0.85-derived TFIID fractions suggested, in contrast to earlier predictions and conclusions of a more recent report (15, 16), that human TAFII150, like other TAFIIs, might be tightly associated with TBP within a TFIID complex. To test this, we immunopurified eTFIID complexes from nuclear extracts of various cell lines expressing a FLAG- or hemagglutinin-tagged version of TBP (f:TBP or HA:TBP), f:TAFII100, or f:TAFII135. Different purification conditions were used to compare the relative stability of association of different TAFIIs within the human TFIID complex (Fig. 2A and C). We found that, independently of the tagged subunit, all highly purified human eTFIID complexes contained essentially the same relative amount of human TAFII150 (Fig. 2C), regardless of whether the immunopurification was carried out at 0.3 M KCl (lanes 1 to 3), at 1.2 M KCl (lane 4), from two column fractions (lanes 1 and 4), or directly from nuclear extracts (lanes 2 and 3). Moreover, TAFII150 is detected mainly in P11 0.85-derived fractions (Fig. 2B; see also below), depletion of TBP and TAFs from a two-column native TFIID fraction with specific anti-TAFII100 antibodies also depletes TAFII150 (Fig. 2D), and the apparent ratios of TAFII150 to other TAFIIs are similar in crude nuclear extracts, partially purified TFIID, and highly purified eTFIID (Fig. 2E). This finding suggests that most of the TAFII150 in nuclear extracts is in TFIID and, potentially, TFIID-related TAFII complexes such as the recently described TBP-free TAFII complex (TFTC [51]) (see Discussion). In addition, native human TAFII150 and TAFII135 comigrate in SDS–7.5% polyacrylamide gels by Western blot analyses (Fig. 2C and legend). Consistent with this, the 135-kDa protein band (TAFII135/TAFII150) in highly purified eTFIID and purified recombinant f:TAFII150 have very similar mobilities, in a side-by-side comparison, on a silver-stained SDS-polyacrylamide gel (Fig. 2F). Furthermore, direct microsequencing of the human 135-kDa protein band in eTFIID revealed a peptide uniquely encoded by TAFII150 (Fig. 1, underlined sequence) as well as peptides encoded by TAFII135, the human homologue of Drosophila TAFII110 (42). This may explain why human TAFII150 was not previously detected in eTFIID. Consistent with the data presented here, independent analyses of eTFIID components by quantitative Western blots with recombinant TAFII standards have established that TAFII150 is present at levels comparable to those of several other TAFIIs (12a).
The human TAFII150-containing TFIID complex cannot support initiator function and TATA-independent transcription with only highly purified GTFs and pol II.
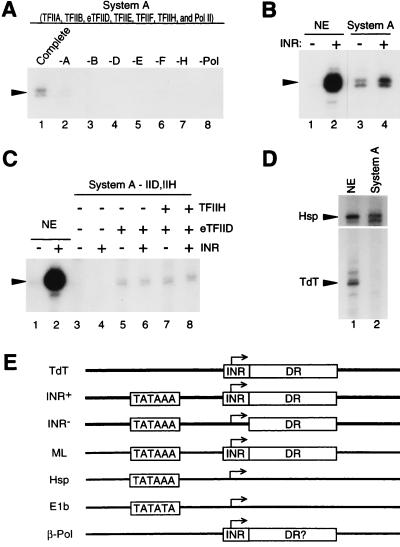
Having established that human TAFII150 is a tightly associated component of a TFIID complex, we tested whether highly purified human eTFIID can direct TAFII-mediated core promoter-specific basal transcription in a homologous assay system (system A; Materials and Methods) reconstituted with highly purified recombinant TFIIB, TFIIE, and TFIIF and native TFIIA, TFIIH, and pol II. With this highly purified assay system and with a linear TATA- and initiator-containing promoter (INR+ [Fig. 3E]), basal transcription is dependent on every GTF and pol II (Fig. 3A) and addition of an excess of TAFII150-containing eTFIID or all other general components does not lead to a significant increase in basal transcription (data not shown). Significantly, under these conditions where GTFs are saturating, the transcription activity of a TATA-containing promoter (Fig. 3E) is similar in the absence (INR−) and presence (INR+) of a functional initiator element on both linear (Fig. 3B, lane 3 versus lane 4) and supercoiled (Fig. 3C, lane 5 versus lane 6 and lane 7 versus lane 8) templates. This is in stark contrast to the dramatic stimulatory effect of the initiator in a crude nuclear extract (Fig. 3B and C, lane 1 versus lane 2) that is, as shown previously, TAFII dependent (14, 19). Interestingly, the activity of the initiator-less promoter is lower in the nuclear extract than in the purified system with both linear (Fig. 3B, lane 1 versus 3) and, to a lesser extent, supercoiled (Fig. 3C, lane 1 versus 5) templates. In contrast, with both supercoiled and linear templates, the activity of the initiator-containing promoter is stronger in the nuclear extract than in the purified system (Fig. 3B, lane 2 versus 4; Fig. 3C, lane 2 versus lanes 6 and 8). Thus, our purified reconstituted system is more efficient than the nuclear extract for basal initiator-independent transcription but apparently lacks components that mediate initiator-specific function. These results also suggest that TAFII-dependent initiator function in the crude nuclear extract may result from both antirepression and positive stimulatory mechanisms. Consistent with the existence of initiator-specific cofactor activities distinct from TFIID and the other GTFs, eTFIID cannot direct basal initiator-mediated transcription from the TAFII-dependent TATA-less TdT core promoter either in the highly purified transcription system described above (Fig. 3D, lane 2) or in less purified systems that also support efficient TATA-dependent transcription (see below). Altogether, these results demonstrate that a complete set of GTFs and TAFIIs (including TAFII150) within the human TFIID complex is insufficient to promote initiator function, either alone or in conjunction with a TATA element in a highly purified system, and therefore that additional activities are required for the TAFII-mediated core promoter-specific (initiator-dependent) functions of TFIID.
FIG. 3.
The human TAFII150-containing TFIID complex is not sufficient for initiator function and TATA-independent transcription in a system reconstituted with highly purified GTFs and pol II. (A) A highly purified transcription system that is dependent on all GTFs and pol II. An arrowhead indicates the position of specific transcripts from the linear INR+ template analyzed by primer extension. The complete system A (lane 1) contains Ni-NTA-agarose-purified native TFIIA (A), recombinant TFIIB (B), eTFIID (D), recombinant TFIIE (E), recombinant TFIIF (F), native TFIIH (H), and pol II (Pol). Lanes 2 to 8, transcription reactions lacking single components as indicated. Note that although TFIIA is required for optimal basal TFIID-dependent transcription from this promoter, its requirement is not absolute since low levels of transcription can be seen in lane 2 on overexposures. (B) The highly purified TFIID-dependent system does not support initiator function from a linear TATA-containing promoter. Transcription from linearized TATA-containing INR− (lanes 1 and 3) and INR+ (lanes 2 and 4) promoters was analyzed in a HeLa cell nuclear extract (NE; lanes 1 and 2) and in the purified system A (lanes 3 and 4) by primer extension using the same 32P-labeled primer. (C) TFIID-mediated transcription from supercoiled templates in a highly purified system is independent of TFIIH and does not support TATA-dependent initiator function. Transcription from the supercoiled TATA-containing INR− and INR+ templates was analyzed as described above in a nuclear extract (lanes 1 and 2) and in either the complete system A (lanes 7 and 8) or in system A lacking TFIIH (lanes 5 and 6) or TFIIH and TFIID (lanes 3 and 4). (D) The highly purified TFIID-dependent system does not support initiator-directed basal transcription from a TAFII-dependent TATA-less promoter. Basal transcription from the supercoiled TATA-containing human Hsp70 core promoter (Hsp) and the TATA-less mouse TdT (−41 to +59) core promoter (TdT) was analyzed as described above in a nuclear extract (lane 1) and in the complete system A (lane 2) by transcribing equimolar amounts of templates in the same reaction. Specific transcripts were analyzed by primer extension using a mixture of two 32P-labeled primers with similar specific activities (see below). (E) Schematic structure of the core promoters used in this study. TdT is the natural murine TdT gene core promoter (positions −41 to +33 or +59 relative to the start site). INR+ is the natural TdT core promoter (−41 to +33) with its −30 DNA sequence converted to a consensus TATAAA element. INR− is the same as INR+ but without a functional initiator element. ML is the natural AdML core promoter (−45 to +65). Hsp is the natural human Hsp70 gene core promoter (−33 to +99). E1b is a construct containing the AdE1b TATA box sequence (as indicated) cloned in a polylinker sequence context upstream of a CAT reporter gene (see Materials and Methods). β-Pol is the human β-polymerase gene core promoter (−41 to +58). A boxed INR represents a functional initiator element. A boxed DR indicates a promoter with TFIID-dependent DNase I footprints extending downstream of the transcription initiation site (bent arrow). For the TdT promoter, this footprint has been observed only in the presence of a TATA box (i.e., in the INR+ construct [20, 26a]). DR? in the β-Pol core promoter indicates that this element has not yet been analyzed. Primer extension analyses of transcripts from the TdT and the INR+ and INR− constructs are performed with the same primer, and transcripts from the ML, Hsp, E1b, and β-Pol CAT gene-containing constructs are analyzed with a CAT gene-specific primer (see Materials and Methods).
TIC-1 potentiates TFIID-mediated initiator function from TATA-containing promoters.
By using the most highly purified set of GTFs and pol II (system A, described above) as a complementation system, we searched for HeLa cell nuclear extract-derived fractions that would restore the TAFII-dependent stimulatory activity of the initiator in the context of TATA-containing core promoters. The activity, which we call TIC-1, was originally identified, at the two-column (P11 and DE52) stage, in two separate fractions (TIC-1 and TIC-1/3 [Fig. 4F]) that, when added independently to the highly purified system, preferentially stimulate the initiator-containing core promoter (Fig. 4A, lanes 1 to 6; Fig. 4B, lanes 3 to 8) in a TAFII-dependent manner (Fig. 4A, lanes 3 to 6 versus lanes 7 to 10). In accord with the results obtained with nuclear extracts (Fig. 3B and C), these TIC-1-containing fractions stimulate initiator-dependent transcription from both supercoiled (Fig. 4A) and linear (Fig. 4B) templates. They also stimulate significantly (about 13-fold) the natural initiator-containing AdML core promoter (ML [Fig. 3E]) but only marginally (about 2.5-fold) the natural initiator-less human Hsp70 core promoter (Fig. 3E, 4D, and 4E, and data not shown). Because the TIC-1/3 fraction is very complex and contains other cofactors (i.e., TIC-3 and both positive and negative USA components [see below]), we have further purified TIC-1 from the P11 0.5/DE 0.12 fraction (summarized in Fig. 4F). TIC-1 activity from the heparin-Sepharose [TIC-1 (HS)] fraction effects a 7- to 8-fold preferential stimulation of the INR+ template (Fig. 4C) and a 13-fold stimulation of the initiator-containing ML template (Fig. 4D, lane 1 versus lane 3) but only a weak (2.5-fold) stimulation of the initiator-less Hsp promoter (Fig. 4D, lane 1 versus lane 3). TIC-1 activity from this heparin-Sepharose fraction sediments on a subsequent glycerol gradient with a native apparent molecular mass in the range of 70 to 160 kDa (Fig. 4D, lanes 4 to 10). TIC-1 from the peak glycerol gradient fraction 8 [Fig. 4C, TIC-1 (GG#8); Fig. 4D, lane 7] was further purified on a Ni-NTA-agarose column, where it splits between the flowthrough and bound fractions and loses part of its activity (see also Materials and Methods). However, TIC-1 activity in the most purified Ni-NTA-agarose flowthrough fraction still retains its TAFII-dependent core promoter-specific activity, as evidenced by a significant (fourfold) stimulation of the ML core promoter with TFIID (Fig. 4E, lane 1 versus lane 2) but not with TBP (lane 3 versus lane 4).
To address whether TIC-1 is different from the previously characterized factors implicated in initiator function, i.e., TAFII150, YY1, TFII-I, and USF (reviewed in reference 38; see also the introduction), we performed immunoblot analyses with specific antibodies against these factors and an excess (5- to 10-fold the amount used for transcription) of a highly active TIC-1 (HS) fraction. The results show that the TIC-1 fraction does not contain detectable amounts of TAFII150/TFIID (Fig. 5A, lane 3), YY1, TFII-I, and USF (Fig. 5B, lane 1). Consistent with this, depletion of USF (30) or TFII-I (Fig. 5C) from nuclear extracts does not affect basal transcription from the AdML promoter (30) (Fig. 5D, lane 1 versus lane 2) and YY1 does not mediate initiator function from the AdML promoter (43). Furthermore, in contrast to the YY1 and TFII-I factors that function through TFIIA- and TAFII-independent pathways in a TBP-dependent (i.e., TFII-I) system or in a TBP-independent system requiring a supercoiled promoter (i.e., YY1) (reviewed in references 33, 38, and 43), TIC-1 does not function in conjunction with TBP but, instead, potentiates TFIID/TAFII-mediated initiator-dependent transcription from both linear and supercoiled templates (see above). Thus, TIC-1 is different from TAFII150, YY1, TFII-I, and USF. Altogether, these results demonstrate that a novel initiator-dependent cofactor activity, TIC-1, is required for the TAFII-mediated transcription stimulatory function of human TFIID through the initiator region of TATA-containing promoters.
TIC-2 and TIC-3 are specifically required for TAFII-dependent transcription from TATA-less promoters.
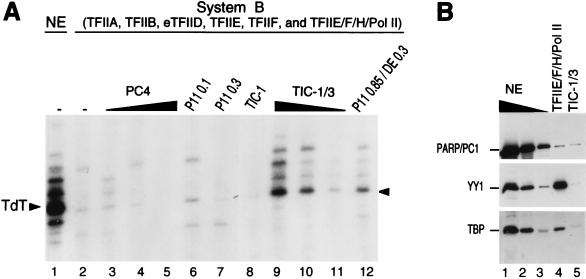
In contrast to what was observed for TATA-containing promoters, the highly purified assay system (system A) described above does not support TATA-independent transcription from the natural TAFII-dependent initiator-containing TdT core promoter (Fig. 3D, lane 2), even in the presence of TIC-1 (see below and data not shown). The only difference between the natural TATA-less TdT core promoter and the TATA-containing INR+ promoter construct used in the above studies is the TATA element (Fig. 3E). This suggests that additional cofactor activities, different from TIC-1, are required specifically for TAFII-mediated TATA-independent transcription. To address this possibility, we tested the TdT core promoter in a less purified assay system (system B), both alone and in combination with various fractions (Fig. 6A). This system (system B [Materials and Methods]) contains highly purified eTFIID, native TFIIA, recombinant TFIIB, recombinant TFIIE, recombinant TFIIF, and a two-column (P11 0.5/DE 0.3) fraction containing TFIIE, TFIIF, TFIIH, and pol II, as well as some endogenous TIC-1 activity (TFIIE/F/H/pol II fraction). This system alone (lane 2) does not support TdT transcription even when supplemented with either the P11 0.1 fraction (lane 6), the P11 0.3 fraction (lane 7), or a two-column-purified (P11 0.5/DE 0.12) TIC-1 fraction (lane 8). In contrast, the two-column (P11 0.85/DE 0.12) TIC-1/3 fraction (lanes 9 to 11) and, to a lesser extent, the P11 0.85/DE 0.3 fraction (lane 12) can restore transcription in this system. This finding suggests that transcription from the TdT core promoter requires an activity that is different from TIC-1 and GTFs, present in the TIC-1/3 fraction and hereafter called TIC-3. TIC-1 and TIC-3 activities present in the two-column TIC-1/3 fraction copurify with the USA coactivator fraction (21) on a subsequent heparin-Sepharose column (Fig. 4F). However, they are different from the major USA component, the coactivator PC4 (11, 18), since recombinant PC4 cannot substitute either for the TIC-1/3 fraction in transcription from the TdT promoter (Fig. 6A, lanes 3 to 5) or for the TIC-1 activity in TAFII-dependent initiator function from a TATA-containing promoter (data not shown). TIC-3 is also different from poly(ADP-ribose) polymerase (PARP)/PC1 (22), YY1, and TFIID since the latter factors are more abundant in the two-column TFIIE/F/H/pol II fraction of system B that lacks TIC-3 activity than in the TIC-1/3 (P11 0.85/DE 0.12) fraction (Fig. 6B, lane 4 versus lane 5). TIC-3 also is different from TFII-I and USF, since these latter factors are absent from the TIC-1/3 (USA) fraction (Fig. 5B, lane 3). Consistent with this finding, immunodepletion of TFII-I from nuclear extracts does not affect basal transcription from the TdT core promoter (Fig. 5D, lane 3 versus lane 4).
FIG. 6.
TIC-1 is not sufficient for initiator-directed transcription from a TATA-less promoter; a novel TIC-3 activity present in the TIC-1/3 fraction is required. (A) TIC-3 activity is different from TIC-1 and PC4 and absent in P11 0.1 and P11 0.3 fractions. Transcription from a supercoiled TdT (−41 to +59) core promoter was analyzed in 5 μl of nuclear extract (lane 1) and in system B (lanes 2 to 12), containing purified native TFIIA (Mono Q fraction), recombinant TFIIB, eTFIID, recombinant TFIIE, recombinant TFIIF, and a two-column (P11 0.5/DE 0.3) TFIIE/F/H/pol II fraction (see text), either alone (lane 2) or complemented with recombinant PC4 (100, 200 and 400 ng; lanes 3 to 5), 500 ng of P11 0.1 (lane 6), P11 0.3 (lane 7), P11 0.5/DE 0.12 (1 μl of TIC-1; lane 8), and P11 0.85/DE 0.3 (lane 12) fractions or with 500 ng (4.5 μl; lane 9), 220 ng (2 μl; lane 10), and 110 ng (1 μl; lane 11) of TIC-1/3 (P11 0.85/DE 0.12) fraction. (B) TIC-3 activity does not correlate with the concentration of PARP/PC1, YY1, and TBP in the TIC-1/3 fraction. Western blot analysis with specific antisera against human PARP/PC1, YY1, and TBP of 5 μl (lane 1), 1 μl (lane 2), and 0.2 μl (lane 3) of nuclear extract (NE) and 4 μl of TFIIE/F/H/pol II (P11 0.5/DE 0.3) fraction (the amount used for system B in panel A; lane 4) and 4.5 μl of TIC-1/3 (P11 0.85/DE 0.12) fraction (the highest amount of TIC-1/3 added to system B in panel A; lane 5).
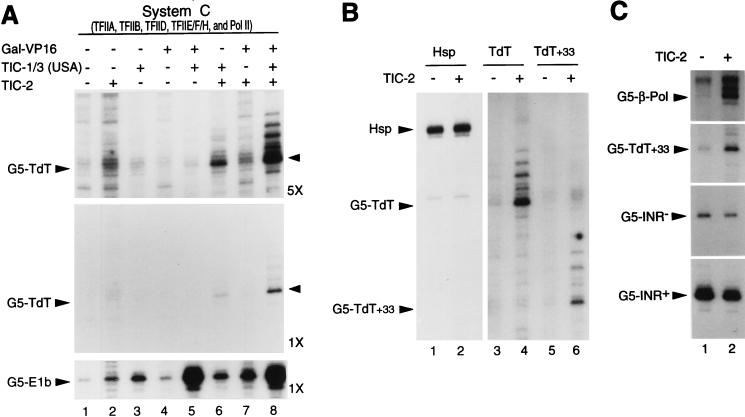
To reconstitute TFIID/TAFII-dependent transcription from the TATA-less TdT promoter in a more purified assay system and to directly compare the factor requirements for both basal and activator-dependent transcription from TATA-dependent and TATA-less promoters, we analyzed transcription from promoter constructs containing either the TATA-less TdT initiator region or the E1b TATA box downstream of five Gal4 binding sites (G5-TdT and G5-E1b) in an assay system (system C [Materials and Methods]) reconstituted with purified native TFIIA, recombinant TFIIB, TAFII150-containing native TFIID (Mono S fraction [Fig. 2E, lane 4; Fig. 5A, lane 2]), pol II, and a four-column-purified (Mono S) fraction containing native TFIIE, TFIIF, and TFIIH (TFIIE/F/H fraction). As expected, and in contrast to the detectable basal transcription from the weak TATA-containing G5-E1B construct, no significant specific basal transcription is observed from the TATA-less G5-TdT promoter (Fig. 7A, lane 1). Interestingly, and in contrast to what was observed in the less purified assay system B (Fig. 6A), the addition of the TIC-1/3 fraction (which contains both TIC-1 and TIC-3 activities) cannot restore basal transcription from the G5-TdT promoter in this more purified system (Fig. 7A, lane 3; note, however, the previously reported basal stimulatory effect of USA components in the TIC-1/3 fraction on the E1b promoter). This finding suggests that another activity is required, in addition to TIC-1/3, for TATA-independent transcription from the TdT initiator region and that this activity is present in a component of system B. Indeed, such an activity, called TIC-2, was found to cofractionate with the two-column (P11 0.5/DE52 0.3) TFIIE/F/H/pol II fraction of system B. TIC-2 from that fraction was further purified on dsDNA-cellulose and heparin-Sepharose (Fig. 4F and Materials and Methods) and analyzed by immunoblotting to show that it lacks detectable TFIID (Fig. 5A, lane 4), TFII-I, YY1, and USF (Fig. 5B, lane 2). The addition of this four-column-purified TIC-2 (HS) fraction (Fig. 4F) alone cannot restore specific basal transcription from the G5-TdT promoter in system C (Fig. 7A, lane 2; note, however, that there is a weak overall increase in nonspecific transcription). However, in conjunction with the TIC-1/3 fraction, TIC-2 specifically promotes basal transcription from the TATA-less G5-TdT promoter, whereas it does not influence TATA-directed transcription from the G5-E1b template (lane 6 versus lanes 2 and 3) or from other TATA-containing promoters that include the natural TATA-containing human Hsp70 core promoter (Fig. 7B, lane 1 versus lane 2; see also below). Importantly, in the presence of the TIC-1/3 fraction, which also contains the USA-derived coactivators PC1 to PC4 (and some TFIID [Fig. 5A, lane 5]), the addition of the strong activator Gal4-VP16 efficiently stimulates transcription from the TATA-containing G5-E1b construct but is without effect on the TATA-less G5-TdT promoter in the absence of TIC-2 (Fig. 7A, lane 3 versus lane 5). The further addition of TIC-2 does not influence the already high level of activated transcription from the G5-E1b construct but dramatically potentiates activated levels of transcription from the G5-TdT promoter (compare lanes 5 and 8). Therefore, basal transcription from the TAFII-dependent TATA-less TdT core promoter in a purified system requires both core promoter-specific cofactors TIC-2 and TIC-3. Moreover, an activator and cognate coactivators of the USA (TIC-1/3) fraction cannot compensate for the absolute requirement for TIC-2. Demonstration of the involvement of TIC-1 in transcription from the TdT promoter, although anticipated from its initiator-specific function on TATA-containing promoters, will require the further purification of TIC-3 from the TIC-1/3 fraction and its separation from TIC-1.
FIG. 7.
TIC-2, a TATA-less promoter-specific cofactor required together with TIC-1/3 for both basal and activated transcription from the TdT core promoter. (A) TIC-2 and TIC-1/3 are required for basal and activated transcription from the TATA-less TdT core promoter. Transcription from supercoiled TATA-less G5-TdT and TATA-containing G5-E1b promoters (80 fmol) was analyzed in system C, containing purified native TFIIA (Mono Q fraction), recombinant TFIIB, native TFIID (Mono S fraction), a four-column native TFIIE/F/H fraction, and pol II (see Materials and Methods). System C was used alone (lane 1) or complemented with either 2 μl of TIC-2 (HS) fraction (lane 2), 1 μl of TIC-1/3 (USA) (HS fraction; lane 3), or 3 pmol of Gal4-VP16 (lane 4) independently and in different combinations (lanes 5 to 8) as indicated. The bottom and middle panels are identical exposures (1×) of experiments run on the same gel. The upper panel is a fivefold-longer exposure (5×) of the middle panel. (B) Reduced TIC-2-dependent transcription from the TdT promoter by deletion of natural TdT downstream promoter sequences between +33 and +59. Transcription from the supercoiled Hsp, G5-TdT (−41 to +59), and G5-TdT+33 (−41 to +33) constructs was performed in the reconstituted system C complemented with Gal-VP16 and the TIC-1/3 (USA) fraction, in either the absence (−) or presence (+) of TIC-2 (HS) fraction as for panel A. (C) TIC-2 is a TATA-less promoter-specific cofactor. Transcription from the supercoiled TATA-less G5-β-Pol and G5-TdT+33 constructs and from the TATA-containing G5-INR− and G5-INR+ promoters (see text) was analyzed in the absence (lane 1) and presence (lane 2) of TIC-2 (HS) fraction as for panel B. Positions of specifically initiated transcripts are indicated for each promoter.
To gain more insight into the mechanisms of TIC-2 function, we analyzed, in the reconstituted TIC-2-dependent system (system C complemented with USA–TIC-1/3 and Gal4-VP16), the effect of TIC-2 addition on transcription from different TdT core promoter mutant derivatives. The results presented in Fig. 7B show that a TdT promoter construct lacking the downstream promoter region from +33 to +59 (G5-TdT+33), which is required for optimal basal transcription in crude nuclear extracts (19, 38a), is still responsive to TIC-2 (lane 5 versus lane 6) but has an activity lower than that of the wild-type G5-TdT construct (lane 4 versus lane 6). Since the TIC-2-dependent system contains a TIC-1 activity that is endogenous to the TIC-1/3 fraction and sufficient for initiator function in the context of a TATA-containing promoter (Fig. 4A and B), this result suggests a possible function of TIC-2 through sequences downstream of the initiator (and extending up to the +33/+59 region). Interestingly, derivatives (G5-INR− and G5-INR+) of the G5-TdT+33 construct containing a consensus TATA element and either a mutant (G5-INR−) or a wild-type (G5-INR+) initiator element are not stimulated by TIC-2 in the assay system described above (Fig. 7C). These results indicate that TIC-2 is specifically required in the absence of a functional TATA element and are reminiscent of the observation that the function of the TdT downstream region in crude nuclear extracts is important only in the context of a TATA-less promoter (19). Altogether, these results suggest that TIC-2 may facilitate the recruitment of TFIID in conjunction with specific sequences downstream of the initiator in TATA-less promoters. Consistent with an important role of TIC-2 specifically in TATA-independent transcription, a different initiator-containing TATA-less core promoter, i.e., the human β-polymerase core promoter, also requires TIC-2 for optimal TFIID-mediated transcription in this reconstituted system (Fig. 7C, G5-β-Pol).
TFIIA is essential for basal and activated TIC-mediated transcription from a TATA-less promoter.
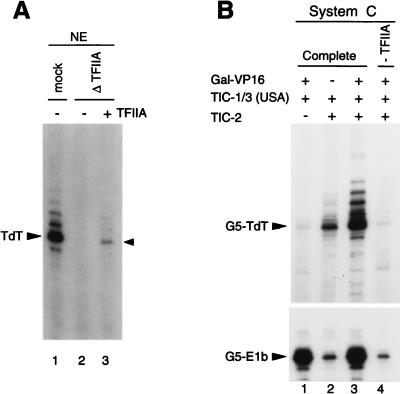
An important function of TFIIA is to facilitate the functional recruitment of TFIID to TATA-containing promoters by competing with specific TBP-interacting inhibitors and by stabilizing TBP-DNA interactions (reviewed in reference 33). However, in contrast to transcription from TATA-containing promoters, transcription from the TATA-less TdT promoter is independent of the affinity of TBP for DNA (20) but requires TICs for functional TFIID recruitment through an alternative TAFII-dependent pathway (see above). Therefore, to better understand the TIC/TAFII-dependent TFIID recruitment pathway at TATA-less promoters, we tested whether TFIIA is needed for specific TATA-independent transcription initiation from the TdT core promoter. Figure 8A shows that specific immunodepletion of TFIIA from nuclear extracts, which inhibits basal transcription from the TATA-containing AdML promoter (7), also inhibits basal transcription from the TdT core promoter (lane 1 versus lane 2) and that addition of TFIIA to the depleted extract stimulates specific TATA-independent transcription initiation at the TdT initiator (lane 2 versus lane 3). We further addressed, in the reconstituted TIC-dependent assay system (system C complemented with TIC-2 and USA–TIC-1/3 fractions), the TFIIA requirement for activator-dependent transcription for both the TATA-containing G5-E1b construct and the TATA-less G5-TdT promoter (Fig. 8B). Whereas TFIIA omission from the system reduces Gal-VP16 activator-mediated transcription from the G5-E1b construct (lane 3 versus lane 4) to a level similar to (or only slightly lower than) the basal level observed in the absence of Gal-VP16 (lane 2 versus 4), it essentially abolishes all specific transcription from the TdT core promoter (G5-TdT [compare lanes 2 and 3 with lane 4]). These results show that TFIIA is crucial for both basal and activated TATA-independent transcription and is even more important for basal transcription from the TATA-less TdT core promoter than for basal transcription from the TATA-containing G5-E1b construct. These results also demonstrate a novel TFIIA role in TIC- and TAFII-dependent transcription from initiator-containing TATA-less promoters that is distinct from a mere stabilization of TBP-DNA interactions.
FIG. 8.
TFIIA is essential for both basal and activator-dependent transcription from a TIC- and TAFII-dependent TATA-less core promoter. (A) Specific immunodepletion of TFIIA from nuclear extracts (NE) inhibits basal transcription from the TATA-less TdT promoter. Transcription from the supercoiled TdT core promoter (−41 to +59) either in a mock-depleted nuclear extract (mock; lane 1) or in a nuclear extract depleted of TFIIA with specific antibodies against TFIIA α/β-p55 subunit (7) (Δ TFIIA; lanes 2 and 3) was analyzed by primer extension; 1.5 μl of TFIIA (Mono Q fraction) was added to the TFIIA-depleted extract in lane 3. (B) TFIIA has a general function in activation and an essential core promoter-selective role in basal transcription from a TIC- and TAFII-dependent TATA-less promoter. Transcription from the supercoiled TATA-less G5-TdT and TATA-containing G5-E1b promoters was analyzed as for panel A, in the complete system C (lanes 1 to 3) or in system C lacking TFIIA (lane 4) and in the presence (+) or absence (−) of Gal-VP16 activator, the TIC-1/3 (USA) fraction, and the TIC-2 (HS) fraction, as indicated.
DISCUSSION
Understanding at the molecular level the gene-specific transcription regulatory role of TAFIIs through core promoter elements requires the reconstitution in vitro of TAFII-dependent core promoter-specific transcription functions with purified components. As a critical first step toward this goal, we have shown that a highly purified human TAFII150-containing TFIID complex is not sufficient for core promoter selectivity in a homologous human system reconstituted with all the other highly purified GTFs and pol II. By the further identification of novel TAFII-dependent core promoter-specific cofactor activities, TICs, we have established the first in vitro transcription systems reconstituted with purified components that support the productive core promoter-specific transcription functions of TFIID through the initiator region of both TATA-containing and TATA-less core promoters. The differential requirement for TBP-DNA interactions, TFIIA, TAFIIs, and TICs as a function of the core promoter structure further suggests the existence of a diversity of regulatory mechanisms for the control of transcription initiation at class II genes and points to potential targets for transcriptional regulators with core promoter-selective functions.
Cloning of human TAFII150, a stably associated component of the TFIID complex.
Human TAFII150 shows 46% identity, at the amino acid level, with its Drosophila homologue and is essentially identical to the recently described CIF150 (16). However, in contrast to the latter report, we clearly show that TAFII150 is stably associated with all human TFIID preparations thus far tested and comigrates on SDS-gels with TAFII135, explaining why it has not been observed previously in silver-stained gels of highly purified eTFIID preparations. The reason for the apparent discrepancy between our results and the recently published results of Kaufmann et al., indicating the absence of TAFII150/CIF150 in their TFIID preparations (16), is not clear but may reflect differences in cells or cell culture conditions, with corresponding differences in TFIID or TAFII150 stability during extract preparation, and/or differences in the sensitivity and specificity of the antibodies used. Our findings have important implications for the conclusions of previous protein-DNA interaction studies that used the human TFIID complex. Indeed, by analogy with Drosophila TFIID, it now seems likely that human TAFII150 also contributes to the extended interactions of TAFIIs over the initiator and downstream regions of initiator-containing promoters and to the resulting increase in stability of the TFIID-TFIIA complex (10). This is consistent with cross-linking studies with highly purified human eTFIID that have shown a TFIIA-induced increase in the cross-linking of 250- and 135-kDa TAFIIs over the AdML initiator region (25). Because a 135-kDa TAFII was also cross-linked upstream of the AdML TATA box and because human TAFII135 interacts directly with TFIIA (42), the 135-kDa TAFII positioned over the AdML initiator region may well be human TAFII150.
TAFII-promoter interactions are not sufficient for the core promoter-selective functions of TFIID.
We have shown that a complete set of TAFIIs, including TAFII150, within the human TFIID complex cannot support either the transcription synergism between TATA and initiator elements or specific transcription initiation from a TAFII-dependent TATA-less promoter in a highly purified system. This finding indicates that a simple increase in the affinity of TFIID for the core promoter by direct TAFII-initiator and TAFII-downstream region interactions is not sufficient to mediate the productive transcription function(s) of TFIID/TAFIIs through the initiator regions of natural TATA-containing and TATA-less promoters. Consistent with this, and in contrast to synthetic promoter constructs containing only a TATA box and an initiator element (14), more complex natural core promoters like the AdML and the TATA-containing TdT-based promoters (INR+ and INR− [Fig. 3E]) show TAFII interactions over the initiator and natural downstream regions in the absence of a functional initiator element (5, 10, 20, 26a). This is also consistent with the observation that TAFIIs contact specific sequences downstream of the initiator in certain promoters (2, 31). Whatever the role of this core promoter-specific mode of TAFII-DNA interactions (see also below), our data indicate that it is not generally sufficient for productive TAFII-dependent core promoter selectivity with all highly purified GTFs and pol II. In agreement with this observation, we have partially purified novel core promoter-specific cofactors, TICs, that potentiate the core promoter-selective functions of TAFIIs in vitro.
TIC-1, a core promoter-specific cofactor for TAFII-mediated initiator function.
We have identified a human TIC-1 activity that restores the transcription synergism between a TATA box and an initiator element in a highly purified eTFIID-dependent system. This activity was purified from a HeLa nuclear extract-derived two-column (P11 0.5/DE 0.12) fraction similar to the one used previously to functionally identify CIF150 (15, 16). However, the chromatographic properties of TIC-1 on subsequent columns are different from those of CIF150, and TAFII150/CIF150 is not detected by immunoblot analyses in highly active TIC-1 fractions. Furthermore, TAFII150/CIF150, which we found to be stably associated within the human TFIID complex, still requires TIC-1 in a purified system for TATA-dependent initiator function. Therefore, the previously reported effect of ectopic TAFII150/CIF150 in a crude system (15, 16) may have reflected the presence of limiting amounts of TFIID/TAFII150 in the crude fraction used as a source of TFIID and GTFs and is not indicative of the novel TIC-1 cofactor described here. We have shown that TIC-1 is also different from several previously characterized gene-specific transcription factors (YY1, USF, and TFII-I) that have been implicated in initiator function under particular conditions and through TFIIA/TAFII-independent pathways (see Results and references 33 and 38). The TdT initiator element has been shown to stabilize human TFIID (but not TBP) binding to a TATA-containing promoter in a TFIIA-dependent manner (10) but, as shown here, still requires TIC-1 for productive initiator-dependent transcription activity. Therefore, it is possible that TIC-1 functions after TFIID recruitment, perhaps by favoring the recruitment of the other GTFs and pol II. Alternatively, TIC-1 may also function in concert with TFIIA during TFIID recruitment through interactions with DNA and/or TAFIIs either by simply further facilitating or stabilizing TFIIA-mediated TFIID-initiator interactions or by favoring the assembly of a more productive TFIID-promoter conformation. Surprisingly, although TIC-1 supports TAFII-mediated initiator function in the context of TATA-containing promoters, it is not sufficient to promote initiator-directed transcription from a TAFII-dependent TATA-less promoter in a purified system. This finding indicates that additional cofactors are required for TATA-independent initiator-directed transcription that may compensate for the lack of a TATA element (see below).
TIC-2 and TIC-3 are core promoter-specific cofactors for basal transcription from TAFII-dependent TATA-less promoters.
The in vitro study of bona fide TAFII-mediated TATA-independent transcription is complicated by the promiscuous and productive binding of TBP to diverse low-affinity nonconsensus TATA elements. The natural TATA-less initiator-containing TdT promoter is a good model for the in vitro analysis of an alternative TATA-independent transcription initiation pathway because (i) even high concentrations of TBP cannot direct basal TdT transcription, (ii) TAFIIs are absolutely required within a TFIID complex for basal transcription, and (iii) TFIID-mediated transcription from the TdT promoter is largely independent of specific TBP-DNA interactions (19, 20). That such alternate TATA-independent pathways are of significance in vivo is further suggested by transfection experiments with major histocompatibility complex class II gene promoters (reference 1 and references therein). By using the TdT promoter as a model, we have shown that a system reconstituted with a complete set of TAFIIs within TFIID, in conjunction with all the other GTFs and pol II, and even in the presence of TIC-1, cannot support TAFII-mediated TATA-independent transcription. Furthermore, we have isolated and partially purified two novel TATA-less core promoter-specific cofactor activities, called TIC-2 and TIC-3, that are different from YY1, TFII-I, and USF and that, together (in the presence of TIC-1), restore basal initiator-directed transcription from the TdT promoter in a purified system. Because neither TIC-1 nor TIC-2 fractions contain TAFIIs, these factors are also different from the recently described TFTC (51). However, the TIC-1/3 (USA) fraction contains TAFII150, TAFII100, TAFII80, TAFII31, TAFII20, and TAFII15 (Fig. 5A and data not shown) in a complex that does not contain TBP and TAFII250 and is therefore distinct from TFIID (20b) but may be related to TFTC. Therefore, it is formally possible that TFTC contributes to TIC-3 activity in the cruder TAFII-containing TIC-1/3 (USA) fraction. Importantly, in a TFIID-dependent system that is reconstituted with all purified GTFs and pol II and that supports efficient basal and activated transcription from various TATA-containing promoters, a strong activator and cognate coactivators including all other cofactors from the USA fraction cannot alleviate the requirement for TIC-2 in specific initiator-directed transcription from the TATA-less TdT core promoter. Therefore, TIC-2 is also different from all known GTFs and USA coactivators (e.g. PC1 to PC4). Because activators can only function when all factors required for basal transcription (i.e., GTFs) are present in addition to pol II, and since this includes, for the TAFII-dependent TATA-less TdT promoter, the presence of TIC-2 and TIC-3, these cofactors can be viewed as core promoter-specific GTFs or basal transcription factors for the TAFII-dependent TATA-less TdT promoter. Consistent with this, TIC-2 is also required for specific TFIID-mediated transcription from the natural TATA-less core promoter region of the human β-polymerase gene but not from TATA-containing derivatives of the TdT core promoter (INR+ and INR− constructs [Fig. 3E and 7C]). Therefore, TIC-2 may function to compensate, in the context of TFIID (but not TBP), for the lack of stable TBP-DNA interactions on TATA-less promoters, in a manner similar and perhaps related to the function of the TdT promoter downstream region (19). However, TIC-2 is most likely not a high-affinity initiator/downstream sequence-specific DNA-binding protein because preincubation of a TIC-2 (HS) fraction with TdT initiator or downstream DNA regions does not inhibit TIC-2-mediated transcription stimulation from a subsequently added TdT core promoter (data not shown). Thus, TIC-2 may facilitate the functional TAFII-mediated recruitment of TFIID to the initiator and downstream sequences of the TATA-less TdT core promoter in a manner more comparable to the function of TFIIA in the stabilization of TBP-TATA interactions and reorganization of the TFIID-promoter complex on TATA-containing promoters (references 25 and 33 and references therein).
An essential core promoter-specific function for TFIIA in transcription from TAFII-dependent TATA-less promoters.
We have shown that TFIIA plays a crucial role in conjunction with TAFIIs and TICs in basal and activated transcription from the TAFII-dependent TATA-less TdT core promoter. Moreover, our data show that TFIIA is more important for basal (activator-independent) transcription from the initiator-dependent TdT core promoter than from the TATA-containing E1b construct. This finding indicates that TFIIA, like TAFIIs and TICs, has core promoter-specific functions. This observation is consistent with (i) the selective stimulation by TFIIA of TFIID-mediated but not TBP-directed transcription in purified systems that allow efficient TBP-TATA interactions (reviewed in reference 33), (ii) the selective initiator-dependent TFIIA-mediated stabilization of TFIID-DNA interactions on TATA-containing promoters (10), (iii) the TFIIA-dependent rearrangement of TAFIIs over the initiator region of the AdML core promoter (25), and (iv) the requirement for TFIIA in conjunction with TAFIIs in differential transcription from the tandem TATA-containing Drosophila Adh core promoters (13). Moreover, our demonstration of an absolute requirement for TFIIA in addition to TAFIIs and TICs for specific initiator-directed transcription from the TATA-less TdT core promoter indicates a novel role for TFIIA, perhaps related to the function of TFIIA in facilitating TFIID/TAFIIs recognition of the initiator region of TATA-containing promoters described above but independent of TBP-TATA interactions. This provides further evidence for important mechanistic differences, including different rate limiting steps and/or alternative pathways, in the formation of transcription competent preinitiation complexes at class II core promoters.
A parallel between TAFII functions in gene-specific activation and core promoter selectivity.
It is interesting that the requirement for TAFIIs in gene transcription is not general either for activated (24, 26, 35, 47) or for basal (references 33 and 46 and references therein) transcription. It therefore appears that TAFIIs play a critical role through alternative pathways in both gene-specific activation and core promoter-specific functions. Our results further suggest that this parallel can be extended to the mechanisms of TFIID/TAFII function in vitro. Indeed, TAFII-activator and TAFII-core promoter interactions result in an increased recruitment of TFIID and/or stability of the TFIID-promoter complex (33, 46; see also the introduction). However, in contrast to TBP-dependent basal transcription, both of these TAFII-mediated processes require TFIIA and additional cofactors, i.e., coactivators (reviewed in references 5, 12, and 52) or core promoter-specific cofactors (TICs [this report]), for productive transcription stimulation. The mechanisms by which those cofactors potentiate TAFII-mediated functions are still largely unknown. It is possible that both TAFII-dependent activator function and basal TAFII-mediated core promoter-specific transcription stimulation are intimately linked processes at particular genes. This would be consistent with the previously observed core promoter-specific activity of certain activators in vitro and in vivo (6, 9, 23, 36, 49). In accord with a potential role of TAFIIs in selective transcription of a specific group of genes, the levels and activity of TAFIIs are regulated during the cell cycle and TAFIIs play an important role in the regulation of cell growth and cell cycle progression by controlling the transcription of G1 and B-type cyclin genes, many of which have a TATA-less core promoter structure (references 24, 34, 35, 41, 48, and 48a and references therein). The novel TIC- and TAFII-dependent in vitro transcription systems described here will be useful both for the further characterization of TICs and for the analysis of the role that TAFIIs (and TFIIA) play in core promoter selectivity and in specific transcription of cell cycle-regulatory genes.
ACKNOWLEDGMENTS
We thank Z. F. Burton for human RAP30 and RAP74 bacterial expression vectors, M. Guermah for TAFII31 antibodies and for sharing unpublished results, T. C. Gutjahr for anti-TFII-I antibodies and for TFII-I-immunodepleted nuclear extracts, T. Oelgeschläger for an independent f:TFIID preparation, and J. Fu for excellent technical assistance. We also thank T. Oelgeschläger, C. Parada, and M. Teichmann for helpful comments on the manuscript.
This work was supported by NIH grant CA42567 to R.G.R. E.M. was supported by a fellowship from The Charles H. Revson/Norman and Rosita Winston Foundation.
REFERENCES
- 1.Bellorini M, Dantonel J C, Yoon J-B, Roeder R G, Tora L, Mantovani R. The major histocompatibility complex class II Ea promoter requires TFIID binding to an initiator sequence. Mol Cell Biol. 1996;16:503–512. doi: 10.1128/mcb.16.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burke T W, Kadonaga J T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 4.Carcamo J, Buckbinder L, Reinberg D. The initiator directs the assembly of a transcription factor IID-dependent transcription complex. Proc Natl Acad Sci USA. 1991;88:8052–8056. doi: 10.1073/pnas.88.18.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang C-M, Ge H, Wang Z, Hoffmann A, Roeder R G. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 1993;12:2749–2762. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das G, Hinkley C S, Herr W. Basal promoter elements as a selective determinants of transcriptional activator function. Nature. 1995;374:657–660. doi: 10.1038/374657a0. [DOI] [PubMed] [Google Scholar]
- 7.DeJong J, Roeder R G. A single cDNA, hTFIIA/α, encodes both the p35 and p19 subunits of human TFIIA. Genes Dev. 1993;7:2220–2234. doi: 10.1101/gad.7.11.2220. [DOI] [PubMed] [Google Scholar]
- 8.Du H, Roy A L, Roeder R G. Human transcription factor USF stimulates transcription through the initiator elements of the HIV-1 and the AdML promoters. EMBO J. 1993;12:501–511. doi: 10.1002/j.1460-2075.1993.tb05682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emami K H, Navarre W W, Smale S T. Core promoter specificities of the Sp1 and VP16 transcription activation domains. Mol Cell Biol. 1995;15:5906–5916. doi: 10.1128/mcb.15.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emami K H, Jain A, Smale S T. Mechanism of synergy between TATA and initiator: synergistic binding of TFIID following a putative TFIIA-induced isomerization. Genes Dev. 1997;11:3007–3019. doi: 10.1101/gad.11.22.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge H, Roeder R G. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 12.Ge H, Martinez E, Chiang C-M, Roeder R G. Activator-dependent transcription by mammalian RNA polymerase II: in vitro reconstitution with general transcription factors and cofactors. Methods Enzymol. 1996;274:57–71. doi: 10.1016/s0076-6879(96)74008-9. [DOI] [PubMed] [Google Scholar]
- 12a.Guermah, M., and R. G. Roeder. Unpublished observations.
- 13.Hansen S, Tjian R. TAFs and TFIIA mediate differential utilization of the tandem Adh promoters. Cell. 1995;82:565–575. doi: 10.1016/0092-8674(95)90029-2. [DOI] [PubMed] [Google Scholar]
- 14.Kaufmann J, Smale S T. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev. 1994;8:821–829. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- 15.Kaufmann J, Verrijzer C P, Shao J, Smale S T. CIF, an essential cofactor for TFIID-dependent initiator function. Genes Dev. 1996;10:873–886. doi: 10.1101/gad.10.7.873. [DOI] [PubMed] [Google Scholar]
- 16.Kaufmann J, Ahrens K, Koop R, Smale S T, Müller R. CIF150, a human cofactor for transcription factor IID-dependent initiator function. Mol Cell Biol. 1998;18:233–239. doi: 10.1128/mcb.18.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kokubo T, Gong D-W, Yamashita S, Horikoshi M, Roeder R G, Nakatani Y. Drosophila 230-kD TFIID subunit, a functional homolog of the human cell cycle gene product, negatively regulates DNA binding of the TATA box-binding subunit of TFIID. Genes Dev. 1993;7:1033–1046. doi: 10.1101/gad.7.6.1033. [DOI] [PubMed] [Google Scholar]
- 18.Kretzschmar M, Kaiser K, Lottspeich F, Meisterernst M. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell. 1994;76:525–534. doi: 10.1016/0092-8674(94)90429-4. [DOI] [PubMed] [Google Scholar]
- 19.Martinez E, Chiang C-M, Ge H, Roeder R G. TATA-binding protein-associated factor(s) in TFIID function through the initiator to direct basal transcription from a TATA-less class II promoter. EMBO J. 1994;13:3115–3126. doi: 10.1002/j.1460-2075.1994.tb06610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez E, Zhou Q, L’Etoile N D, Oelgeschläger T, Berk A J, Roeder R G. Core promoter-specific function of a mutant transcription factor TFIID defective in TATA-box binding. Proc Natl Acad Sci USA. 1995;92:11864–11868. doi: 10.1073/pnas.92.25.11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Martinez, E., T. K. Kundu, J. Fu, and R. G. Roeder. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from TFIID. J. Biol. Chem., in press. [DOI] [PubMed]
- 20b.Martinez, E., and R. G. Roeder. Unpublished observations.
- 21.Meisterernst M, Roy A L, Lieu H M, Roeder R G. Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell. 1991;66:981–993. doi: 10.1016/0092-8674(91)90443-3. [DOI] [PubMed] [Google Scholar]
- 22.Meisterernst M, Stelzer G, Roeder R G. Poly(ADP-ribose) polymerase enhances activator-dependent transcription in vitro. Proc Natl Acad Sci USA. 1997;94:2261–2265. doi: 10.1073/pnas.94.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merli C, Bergstrom D E, Cygan J A, Blackman R K. Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev. 1996;10:1260–1270. doi: 10.1101/gad.10.10.1260. [DOI] [PubMed] [Google Scholar]
- 24.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 25.Oelgeschläger T, Chiang C-M, Roeder R G. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 26.Oelgeschläger T, Tao Y, Kang Y K, Roeder R G. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol Cell. 1998;1:925–931. doi: 10.1016/s1097-2765(00)80092-1. [DOI] [PubMed] [Google Scholar]
- 26a.Oelgeschläger, T., E. Martinez, and R. G. Roeder. Unpublished observations.
- 27.Parvin J D, Shykind B M, Meyers R E, Kim J, Sharp P A. Multiple sets of basal factors initiate transcription by RNA polymerase II. J Biol Chem. 1994;269:18414–18421. [PubMed] [Google Scholar]
- 28.Parvin J D, McCormick R J, Sharp P A, Fisher D E. Pre-bending of a promoter sequence enhances affinity for the TATA-binding factor. Nature. 1995;373:724–727. doi: 10.1038/373724a0. [DOI] [PubMed] [Google Scholar]
- 29.Pinto I, Wu W-H, Na J G, Hampsey M. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J Biol Chem. 1994;269:30569–30573. [PubMed] [Google Scholar]
- 30.Pognonec P, Roeder R G. Recombinant 43-kDa USF binds to DNA and activates transcription in a manner indistinguishable from that of natural 43/44-kDa USF. Mol Cell Biol. 1991;11:5125–5136. doi: 10.1128/mcb.11.10.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purnell B A, Emanuel P A, Gilmour D S. TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes Dev. 1994;8:830–842. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- 32.Rees S, Coote J, Stables J, Goodson S, Harris S, Lee M G. Bicistronic vector for the creation of stable mammalian cell lines that predispose all antibiotic-resistant cells to express recombinant protein. BioTechniques. 1996;20:102–110. doi: 10.2144/96201st05. [DOI] [PubMed] [Google Scholar]
- 33.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 34.Segil N, Guermah M, Hoffmann A, Roeder R G, Heintz N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- 35.Shen W-C, Green M R. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 36.Simon M C, Fish T M, Benecke B J, Nevins J R, Heintz N. Definition of multiple, functionally distinct TATA elements, one of which is a target in the Hsp70 promoter for E1A regulation. Cell. 1988;52:723–729. doi: 10.1016/0092-8674(88)90410-2. [DOI] [PubMed] [Google Scholar]
- 37.Singer V, Wobbe C R, Struhl K. A wide variety of DNA sequences can functionally replace a yeast TATA element for transcriptional activation. Genes Dev. 1990;4:636–645. doi: 10.1101/gad.4.4.636. [DOI] [PubMed] [Google Scholar]
- 38.Smale S T. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 38a.Smale S T, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 39.Starr D B, Hoopes B C, Hawley D K. DNA bending is an important component of site-specific recognition by the TATA-binding protein. J Mol Biol. 1995;250:434–446. doi: 10.1006/jmbi.1995.0388. [DOI] [PubMed] [Google Scholar]
- 40.Sun Z W, Tessmer A, Hampsey M. Functional interaction between TFIIB and Rpb9 (Ssu73) subunit of RNA polymerase II in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:2560–2566. doi: 10.1093/nar/24.13.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki-Yagawa Y, Guermah M, Roeder R G. The ts13 mutation in the TAFII250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol Cell Biol. 1997;17:3284–3294. doi: 10.1128/mcb.17.6.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tao, Y., and R. G. Roeder. Unpublished data.
- 43.Usheva A, Shenk T. TATA-binding protein-independent initiation: YY1, TFIIB, and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell. 1994;76:1115–1121. doi: 10.1016/0092-8674(94)90387-5. [DOI] [PubMed] [Google Scholar]
- 44.Verrijzer C P, Yokomori K, Chen J-L, Tjian R. Drosophila TAFII150: similarity to yeast TSM-1 and specific binding to core promoter DNA. Science. 1994;264:933–941. doi: 10.1126/science.8178153. [DOI] [PubMed] [Google Scholar]
- 45.Verrijzer C P, Chen J-L, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 46.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 47.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIs. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 48.Walker S S, Shen W-C, Reese J C, Apone L M, Green M R. Yeast TAFII145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- 48a.Wang E H, Zou S, Tjian R. TAFII250-dependent transcription of cyclin A is directed by ATF activator proteins. Genes Dev. 1997;11:2658–2669. doi: 10.1101/gad.11.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wefald F C, Devlin B H, Williams R S. Functional heterogeneity of mammalian TATA-box sequences revealed by interaction with a cell-specific enhancer. Nature. 1990;344:260–262. doi: 10.1038/344260a0. [DOI] [PubMed] [Google Scholar]
- 50.Weiss L, Reinberg D. Transcription by RNA polymerase II: initiator-directed formation of transcription-competent complexes. FASEB J. 1992;6:3300–3309. doi: 10.1096/fasebj.6.14.1426767. [DOI] [PubMed] [Google Scholar]
- 51.Wieczorek E, Brand M, Jacq X, Tora L. Function of TAFII-containing complex without TBP in transcription by RNA polymerase II. Nature. 1998;393:187–191. doi: 10.1038/30283. [DOI] [PubMed] [Google Scholar]
- 52.Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 53.Zhou Q, Lieberman P M, Boyer T G, Berk A J. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]