Abstract
The non-structural protein (nsp) 8 of the porcine epidemic diarrhea virus (PEDV) is highly stable across different PEDV strains and plays an important role in PEDV virulence. In current study, nsp8 prokaryotic expression vectors were constructed based on parental vectors pMAL-c2x-maltose binding protein (MBP) and pET-28a (+). Subsequently, the optimization of expression conditions in Escherichia coli, including induced temperature, time and isopropyl β-D-thiogalactopyranoside concentration were performed to obtain a stable expression of MBP-nsp8 and nsp8. The nsp8 fused with MBP increased the water solubility of the expressed products. Target proteins were further purified from E. coli culture and their immunogenicities were evaluated in vivo by mice. The antibody titers of serum from nsp8 immunized mice were up to 1:7,750,000 when measured by indirect enzyme-linked immunosorbent assay; meanwhile, the mice immunized with MBP-nsp8 gave an antibody titer reaching 1:1,000,000. In all, the expression and purification system of PEDV nsp8 and MBP-nsp8 were successfully established in this work and a strong immune response was elicited in mice by both purified nsp8 and MBP-nsp8, providing a basis for the study of the structure and function of PEDV nsp8.
Key Words: Escherichia coli, Immunogenicity, Non-structural protein 8, Porcine epidemic diarrhea virus
Introduction
The porcine epidemic diarrhea virus (PEDV) is a causative agent of porcine epidemic diarrhea (PED) which is a highly infectious disease for pigs. This porcine intestinal pathogenic coronavirus causes vomiting and dehydration as well as severe diarrhea and high mortality rates in piglets. In this way, significant economic losses are inevitable,1-3 especially, for centuries of swine-farming nations threatened by PEDV infection in Asia, Europe and North America.4-6 The PEDV is an enveloped, positive-sense and single-stranded RNA virus belonging to the genus Alphacoronavirus, subfamily Coronavirinae, family Coronaviridae and order Nidovirales.7 The genome of this virus is approximately 28.00 kb and contains two overlapping non-structural poly-proteins (nsp) open reading frames (ORFs) 1a and b next to 5’ untranslated region; ORF1a produces replicase poly-protein 1 and ORF1b extends replicase poly-protein 1a into poly-protein 1ab. Both poly-proteins are cleaved and mature into nsp1-16 during virus replication.8-10
Since 2010, PED has significantly damaged farming industry of the People of Republic China. Although it is wildly accepted that vaccination is the most effective way to prevent infectious disease, none of the currently available PED vaccines can provide complete immune protection for piglets11-13 and advanced vaccine products and vaccination protocols are long-awaited by the market.14 Despite, the spike protein has received the major attention in the development of coronavirus vaccines and studies showed that specific mutations in some non-structural proteins of coronaviruses significantly attenuated the virus virulence,15-22 indicating an important role of these non-structural proteins in the virulence.
The nsp8, a highly conserved non-structural protein in coronaviruses, functions as an RNA-dependent RNA polymerase (Rd Rp) co-factor, which plays an important role in viral genome replication.23 To complete trans-cription and replication functions of the viral genome, coronaviruses need a large amount of nsps to package into complete enzyme complexes among which the nsp12/ nsp7/nsp8 sub-complex is considered to be the smallest core component mediating coronavirus RNA synthesis.10,24,25 Exposure of the N-terminal amino acid residues of nsp8 is essential for the function of the nsp12/nsp7/nsp8 complex and further stimulating of its RNA polymerase activity.26 Studies of nsp8 have shown that nsp8 functions to bind to 5′-(G/U) CC-3′ on the RNA template to initiate the synthesis of complementary oligonucleotides and it was speculated to have secondary Rd Rp activity.27 By reducing the activity of interferon (IFN) regulatory factor 1 promoters, PEDV nsp8 inhibits type III IFN activity and promotes viral proliferation.28 Considering crucial function and conservative property of nsp8, it may be new outstanding PEDV protein for PED prophylaxis and diagnosis.
In this study, nsp8 was successfully expressed and purified from Escherichia coli BL21 (DE3) and it was capable to induce high antibody titers in immunized mice. The described system in this work would provide potential product for the development of PEDV diagnostic kit.
Materials and Methods
Plasmid, virus and animals. Prokaryotic expression vector pMAL-c2x-maltose binding protein (MBP) and pET-28 a (+) was purchased from EMD Biosciences (Novagen, Beijing, China). The MBP tag protein is 40.00 kDa in size. The MBP fusion protein vector has the advantages of high expression efficiency and easy purification. The competent cells E. coli DH5α and E. coli BL21 (DE3) were purchased from Shanghai Weidi Biotechnology (Weidi, Shanghai, China). The PEDV CV777 attenuated strain (Accession No. KT323979.1) was obtained from Harbin Weike Bio-technology Development Company (Weike, Harbin, China). The 6-week-old female Kunming mice were purchased from Jiangxi University of Traditional Chinese Medicine, Nanchang, China. Jiangxi Agricultural University's Animal Care and User Committee, as well as Laboratory Animal Ethics Committee gave their approval for the mouse research, which was carried out under accordance with their approvals (Reference Number: JXAC20180046).
PEDV nsp8 bioinformatics analysis. The hydro-phobicity of PEDV nsp8 was estimated using the Protscale computer program (https://web.expasy.org/protscale/). The predictor of Bepipred was employed to forecast PEDV nsp8 immune epitopes on the Immune Epitope Database and Analysis Resource Website (http://tools.immune- epitope.org/main/). The Swiss-Model (https://swis-smodel.expasy.org/) was used to create the putative three-dimension (3D) of the PEDV nsp8.
Main reagents. An RNA extraction kit was purchased from Vazyme Biotech Co., Ltd (Nanjing, China). The 15 kb DNA Marker, EcoR I, Sal I endonuclease, PrimeScript First Strand cDNA Synthesis Kit, Taq DNA polymerase and T4 DNA ligase were provided from Takara Bio Inc., Kusatsu, Japan. A plasmid extraction kit was obtained from Tiangen Biotech Co. (Beijing, China). The supplier of urea was Xilong Chemical Co., Ltd (Guangzhou, China). The following items were bought from TransGen Biotech (Beijing, China): 14.00 - 100 kDa Protein Marker, 10.00 - 180 kDa Protein Marker and 5.00% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. The Millipore of Sigma-Aldrich Company (St. Louis, USA) supplied the polyvinylidene difluoride (PVDF) membrane. Proteintech Group supplied 6×His-Tag monoclonal antibodies, MBP rabbit polyclonal antibodies, horseradish peroxidase (HRP) and goat anti-mouse immunoglobulin G (IgG). An enzyme-linked immunosorbent assay (ELISA) kit was acquired from Huamei Biological Engineering Co., Ltd (Wuhan, China). Nickel aminotriacetate (Ni-NTA) agarose was purchased from QIAGEN (Hilden, Germany). Thermo Fisher Scientific (Waltham, USA) was the source for the acquisition of Freund's full and incomplete adjuvants.
Plasmid construction . Total RNA of PEDV was extracted from PEDV-infected Vero cells using a total RNA extraction reagent (Vazyme) according to the manufacturer's instructions and then, reverse transcribed into cDNA. The nsp8 gene was amplified with specific nsp8 primers (5’- CCGGA ATTCATGCCGAGCTATGTGATCT-3’ and 5’- ACGCGTCGACT CAGTGGTGGTGGTG-3’) of the PEDV strain CV777 (Accession No. KT323979.1) and subsequently inserted into the pMAL-c2x-MBP and pET-28a (+) vectors. The inserted gene fragments of the plasmid were verified through sequencing for the following operation.
Expression and purification of recombinant protein. Escherichia coli BL21 (DE3) was transformed with the recombinant plasmids pMAL-c2x-MBP-nsp8 and pET-28a (+)-nsp8 separately and induced to express the protein with different induction time points and concentrations of isopropyl β-D-thiogalactopyranoside (IPTG) supplementary under different cultural temperatures. The protein expression levels were measured by 12.00% SDS-PAGE. The cells were then lysed and the supernatant was collected and run through a Ni-NTA affinity column to purify the proteins. The purified proteins were eluted using buffers with varying imidazole concentrations. Aliquots were stored at – 80.00 ˚C for future use.
Immunoblotting. The purified nsp8 was separated by 12.00% SDS-PAGE and then, transferred to a PVDF membrane where it was detected using primary antibody. Afterward, the blocking steps were performed with 5.00% skim milk powder solution to prevent non-specific binding of antibodies to the membrane. The MBP-tag antibody, His-tag antibody and serum from immunized mice (primary antibody) were employed to recognize the target protein, followed by the use of secondary antibody goat anti-mouse IgG to bind to the primary antibody further to produce a signal by the conjugated enzyme HRP that can be visualized with the Clarity Electronics Components Laboratory System (Bio-Rad, Hercules, USA).
Mice immunization. The mice were divided into three groups including phosphate-buffered saline (PBS), nsp8 and MBP-nsp8. Each group was given an intra-peritoneal injection of 40.00 µg of the purified nsp8 being formulated with complete Freund's adjuvant. Two weeks later, a second injection of the purified nsp8 being formulated with incomplete Freund's adjuvant was given followed by a third immunization two weeks later. Blood samples were taken weekly from the experimental mice. The PBS-injected mice were used as a negative control. The mice physiological condition was monitored daily until six weeks after immunization.
Recombinant protein-coated ELISA. Based on the operation instrument provided by the manufacturer, the ELISA plates were prepared by coating them with purified nsp8 in coating buffer overnight at 4.00 ˚C. The plates were then blocked with 5.00% skim milk for an hr at 37.00 ˚C and sera from immunized mice were added to the wells and incubated overnight at 4.00 ˚C. After washing, a goat anti-mouse HRP-conjugated IgG antibody was added and incubated for an hr at 37.00 ˚C, followed by the addition of 3,3',5,5'-tetramethylbenzidine (TMB) and 2.00 M sulfuric acid (Huamei) to terminate the reaction. Finally, the absorbance of each well was measured at 450 nm using the Bio-Rad Microplate Reader.
Statistical analysis. The data obtained from the ELISA were analyzed using GraphPad Prism (version 8.0; Graph-Pad Software Inc., San Diego, USA). Statistical significance between two groups was evaluated using the student's t-test. A p-value less than 0.05 was considered statistically significant for each test compared to the control.
Results
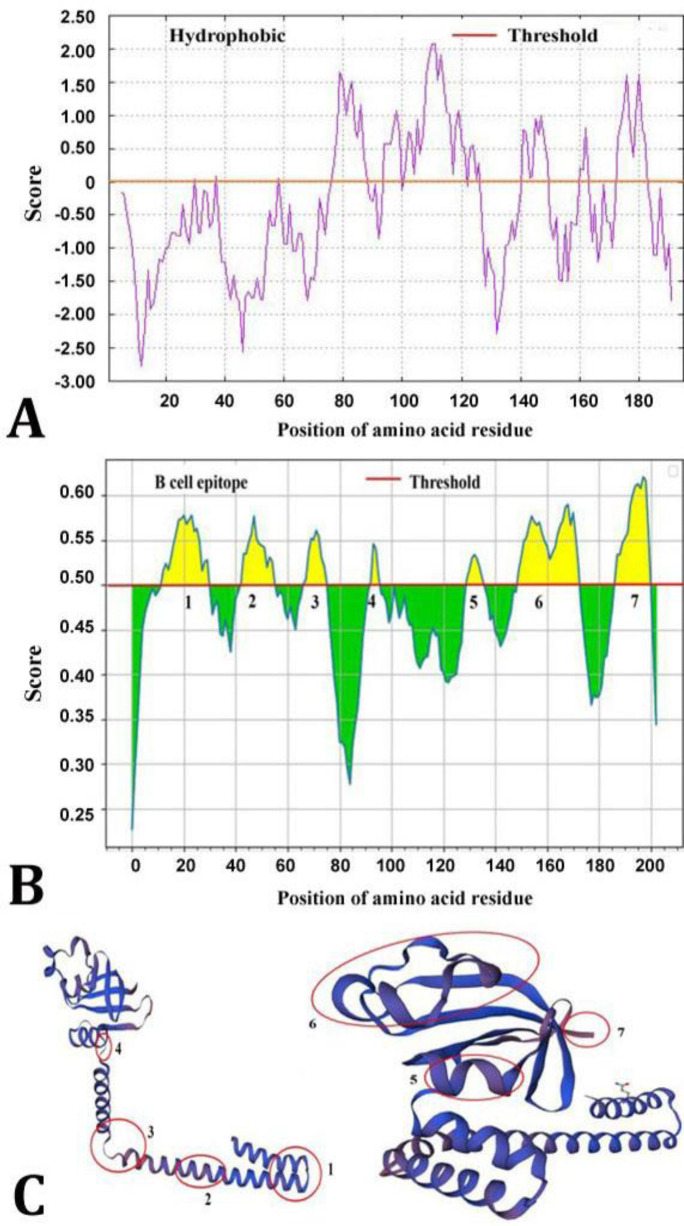
Prediction of hydrophobic, antigenic epitopes and 3D structure of PEDV nsp8. The PEDV nsp8 was analyzed using Protscale, Bepipred and Swiss-Model to predict its linear hydrophobic properties, B cell epitopes and 3D structure, respectively. Protscale analysis revealed the existing of hydrophobic areas at amino acids 80 - 120 (Fig. 1A). Through Bepipred analysis, seven possible B cell epitopes were peculated, being located in amino acid residues 9 to 28, 41 to 54, 65 to 74, 91 to 94, 128 to 134, 148 to 171 and 185 to 192 (Fig. 1B and Table 1). These B cell antigen epitopes were depicted in the PEDV nsp8 3D structure modeled by Swiss researchers (Fig. 1C). These findings collectively suggested that the PEDV nsp8 might be able to elicit host immunological responses.
Fig. 1.
Predictions for the porcine epidemic diarrhea virus non-structural protein (nsp) 8 hydrophobic domains, B cell antigenic epitopes and three-dimension (3D) structure. A) Protscale-based hydrophobic region prediction; B) BepiPred with a threshold value of 0.50 was used to predict the probable antigenic epitopes for B cells; C) The Swiss-model-predicted 3D structure of nsp8 marked with antigenic epitopes. After 90.00-degree rotation of the left panel image, the right panel image was produced
Table 1.
The predicted B cell antigen epitope’s amino acid residues
| No. | Start | End | Peptide | Length |
|---|---|---|---|---|
| 1 | 9 | 28 | NARQQYEDAVNNGSPPQLVK | 20 |
| 2 | 41 | 54 | FDREASTQRKLDRM | 14 |
| 3 | 65 | 74 | EARAVNRKSK | 10 |
| 4 | 91 | 94 | DMSS | 4 |
| 5 | 128 | 134 | DSYNRIQ | 7 |
| 6 | 148 | 171 | IIDIKDNDGKVVHVKEVTAQNAES | 24 |
| 7 | 185 | 192 | KLQNNEIT | 8 |
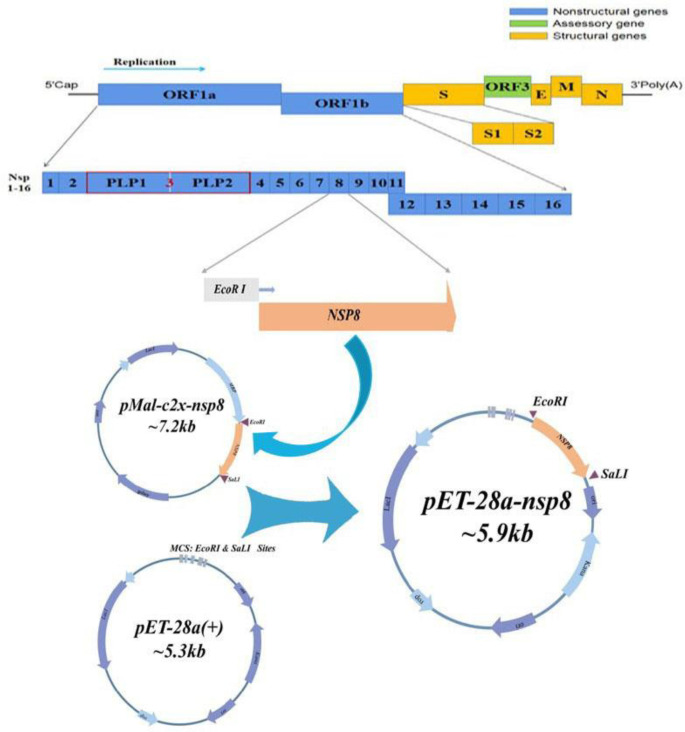
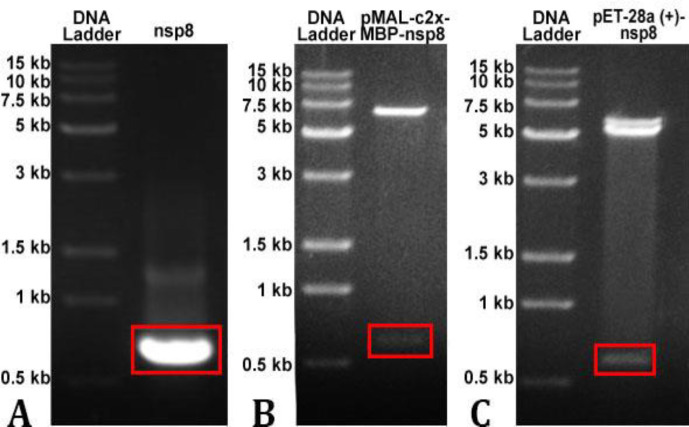
C onstruction of recombinant plasmid pMAL-c2x-MBP-nsp8 and pET-28a (+)-nsp8. As shown in Fig. 2, PEDV cDNA was used as a template to design primers for polymerase chain reaction amplification. The 1.00% gel electrophoresis was used to identify the amplified fragment which showed the expected size of 618 bp (Fig. 3A). The gel-recycling products were ligated to the prokaryotic expression vector pMAL-c2x-MBP after double digestion with EcoR I and Sal I, and the ligation product was transformed into E. coli competent cells DH5α. Double enzyme digestion with EcoR I and Sal I (37.00 ˚C, 1 hr) was used to identify the recombinant plasmids pMAL-c2x-MBP-nsp8 and pET-28a (+)-nsp8 (Figs. 3B and 3C). The identified positive clone was sent for sequencing and verifying that it was indeed PEDV nsp8 sequence as expected. The results mentioned above confirmed that the recombinant expression vectors pMAL-c2x-MBP-nsp8 and pET-28a (+)-nsp8 were successfully constructed.
Fig. 2.
Construction and verification of recombinant plasmid pMAL-c2x-maltose binding protein-non-structural protein (nsp) 8 and pET-28a (+)-nsp8. The position of nsp8 within the porcine epidemic diarrhea virus genome and construction strategy are shown in the schematic diagram
Fig. 3.
The inserted gene fragment verification using 1.00% gel electrophoresis. A) Verification of the size of amplified target fragment of non-structural protein (nsp) 8; B) Double digestion of pMAL-c2x-maltose binding protein-nsp8 using EcoR I and Sal I; C) Double digestion of pET-28a (+)-nsp8 using EcoR I and Sal I. The red rectangles indicate nsp8
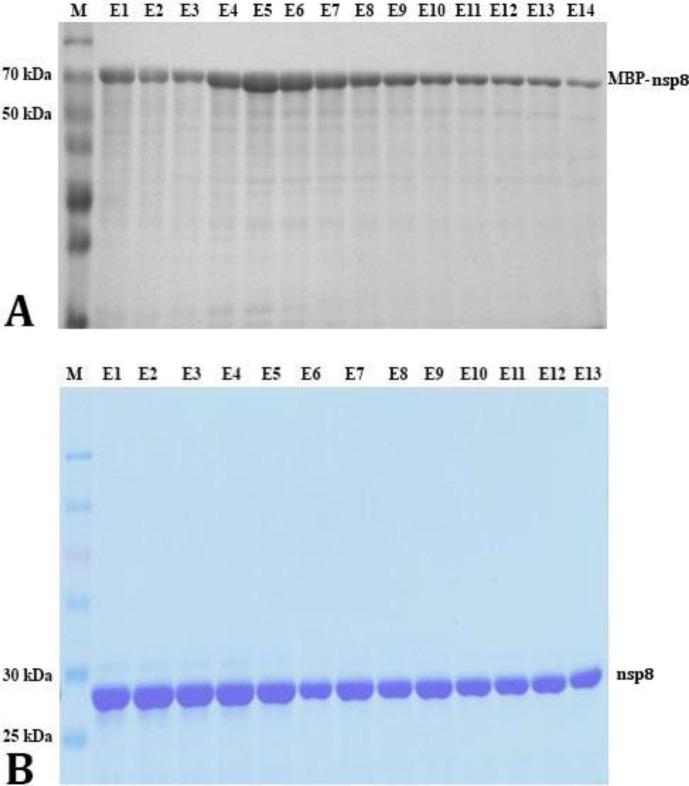
Expression and purification of MBP-nsp8 and nsp8 . For the purpose of protein expression, the recombinant plasmids pMAL-c2x-MBP-nsp8 and pET-28a (+)-nsp8 were transformed into E. coli BL21 (DE3) and then, the IPTG concentration as well as cultivation time and temperature after induction were optimized. The relative molecular weights of MBP-nsp8 and nsp8 were 65.00 and 28.00 kD, respectively. The results showed that the temperature, induction period and IPTG concentration that maximized MBP-nsp8 expression were 37.00 ˚C, 4 hr and 1.00 mM, respectively (Fig. 4A). The optimal condition of nsp8 expression was 4 hr at 37.00 ˚C, being induced with 0.10 mM IPTG supplementary (Figs. 4B and 4C). Results also indicated that MBP-nsp8 was found to be in a pellet and supernatant unlike nsp8 being only found in a pellet (Fig. 4D), showing that nsp8 was fused with MBP and increased the water solubility of the expressed products. Figure 5 shows that the MBP-nsp8 and nsp8 were successfully purified through Ni-column and further eluted into 40.00 mM imidazole. The contents of MBP-nsp8 and nsp8 in the elution fractions were analyzed using 12.00% SDS-PAGE which also indicated the existence of few other proteins. To verify the purified proteins, immunoblotting was performed using commercial MBP-tag or His-tag antibodies (Figs. 6A and 6B). The results showed that two recombinant proteins produced by E. coli were detected with the expected size.
Fig. 4.
Expression and purification of maltose binding protein (MBP)-non-structural protein (nsp) 8 and nsp8. A) Optimization of MBP-nsp8 expression conditions; B) Monitoring the nsp8 induction patterns throughout the course of 4 hr at 37.00 ˚C with various isopropyl β-D-thiogalactopyranoside (IPTG) supplementary; C) Monitoring of nsp8 induction patterns with 0.10 mM IPTG induction at 37.00 ˚C throughout a range of time durations; D) The nsp8 expression in pellet. M: Marker; Pre-I: Pre-induction; Post-I: Post-induction; SN: Supernatant; FT: Flow through
Fig. 5.
A) The maltose binding protein (MBP)-non-structural protein (nsp) 8 content in 40.00 mM imidazole; B) The nsp8 content in 40.00 mM imidazole. M: Marker; E1 - E14: Elution fraction from each number. The protein was eluted gradually at the concentration of 40.00 mmol mL-1 imidazole
Fig 6.
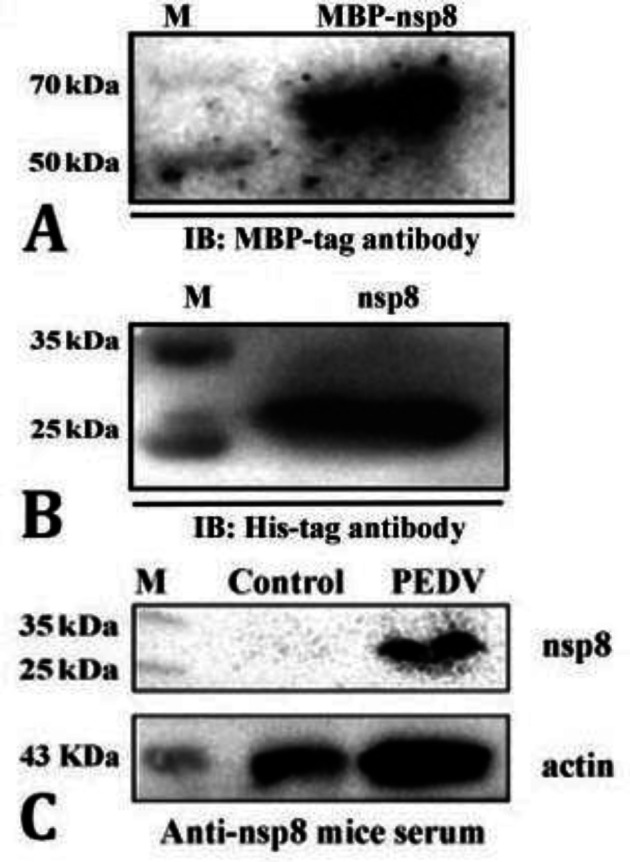
A) The maltose binding protein (MBP)-non-structural protein (nsp) 8 was identified by MBP-tag antibody; B) The nsp8 was identified by His-tag antibody. The electrophoresis of each gel was performed using 12.00% polyacrylamide gels; C) The result of western blots of porcine epidemic diarrhea virus (PEDV)-infected Vero cells with anti-nsp8 mice serum (1:7,000). M: Marker; IB: Immunoblotting
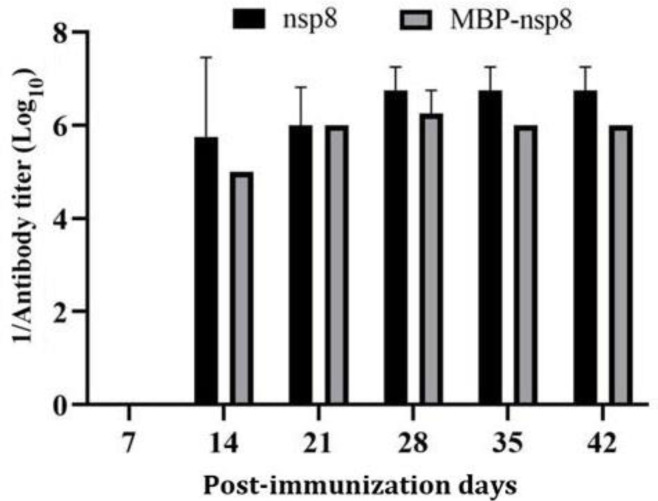
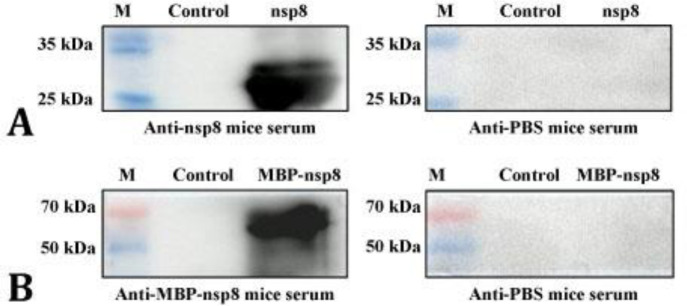
Evaluation of nsp8 immunogenicity. To evaluate the immunogenicity of MBP-nsp8 and nsp8, 6-week-old female Kunming mice received MBP-nsp8 and nsp8 intra-peritoneally. The PBS injection was considered as a negative control. Antibody titers of MBP-nsp8- and nsp8-immunized mice were tested at different time points after immunization. The highest antibody titers (1:7,750,000 for nsp8 and 1:1,000,000 for MBP-nsp8) were observed in immunized mice on day 28 after immunization (Fig. 7). For confirming the specificity of the immune response, the sera were subjected to western blots. The results proved that sera from mice immunized with MBP-nsp8 and nsp8 specifically recognized nsp8; whereas, nsp8 was not recognized in negative control group (Figs. 8A and 8B). The nsp8 in Vero cells infected with PEDV was detected by sera from mice immunized with nsp8 (Fig. 6C). Overall, these results indicated that nsp8 had strong immuno-genicity and could be a potential target for the development of vaccines against PEDV and diagnostic products.
Fig. 7.
The antibody titers of maltose binding protein (MBP)-non-structural protein (nsp) 8 and nsp8 in the serum of immunized mice were tested by enzyme-linked immunosorbent assay at the given time periods by the immunoglobulin G antibodies
Fig. 8.
A) The result of western blots of purified non-structural protein (nsp) 8 with anti-nsp8 mice serum that has been diluted (1:7,000); B) The result of western blots of purified maltose binding protein (MBP)-nsp8 with mice serum that has been diluted (1:7,000). Anti-phosphate-buffered saline (PBS) mice serum was considered as a negative control. M: Marker
Discussion
Partial deletions and substitutions of the S protein as well as mutations in non-structural proteins of PEDV are the main focus of recent research regarding PEDV virulence attenuation. It has been reported that at least 12 PEDV structural and non-structural proteins (nsp1, nsp3, nsp5, nsp7, nsp8, nsp14, nsp15, nsp16, ORF3, E protein, M protein and N protein) inhibit cellular IFN immune responses,28-35 suggesting that mutations or deletions in these proteins potentially reduce PEDV virulence. These studies underlie the importance of exploring the immunogenicity and potential application of PEDV proteins mentioned above.
The nsp8 is a crucial protein in the RNA replication and polymerase activity of PEDV and other coronaviruses. It forms a hexadecameric complex with nsp7, mediating the interaction of nucleic acids with nsp12 and further forming the minimum RNA polymerase complex.36,37 The nsp8 can form complexes with nsp7 and nsp12, separately, and it only exhibits high RNA polymerase activity when mixed with nsp7 and nsp12 at the same temperature.38 It is worth to notice that inhibitors have been developed to target the Rd Rp of coronaviruses including SARS-CoV-2 for the purpose of preventing or reducing viral proliferation.39,40 For instance, the monophosphate of GS-441524 which is a metabolite of remdesivir can bind to SARS-CoV-2 in a binding site made by superimposing two nsp12/nsp7/nsp8 crystal structures that already exist.41 In addition, the deletion of nsp7 or nsp8 regions in the murid herpesvirus produces a deadly phenotype, suggesting that nsp8 may play a significant role in the survival of virus.42 The PEDV nsp8 may also be a potential target for inhibitors to reduce or prevent viral proliferation.43,44
The stable expression of the full-length PEDV nsp8 in the E. coli system paves the way for further PEDV fundamental research as well as development of relative vaccines and diagnostic kit. In our study, by optimizing the expression conditions, we established a favorable prokaryotic expression system which was able to produce enormous amounts of nsp8 and MBP-nsp8. Purified with Ni-column, the nsp8 was specially detected by antibody. The immunogenicity results showed that both nsp8 and MBP-nsp8 were effective immunogens, inducing a strong immunological response in the mice. It was reported that non-structural proteins with mutations in important sites attenuated the virulence of PEDV; but, also the immunogenicity or genetic stability.45,46 In the present study, both MBP-nsp8 and nsp8 produced high antibody titers. This provided a basis for subsequent studies on the effect of mutations at key sites in the nsp8 on PEDV virulence and immunogenicity.
Acknowledgments
This study was funded by research grants from Post-doctoral Research Foundation of Gansu Province (Grant No. 23JRRA550), Natural Science Foundation of Jiangxi Province (Grant No. 20192BAB214001 and 20224B-AB205002), Post-doctoral Research Foundation of China (Grant No. 2022M723442) and National Natural Science Foundation of China (Grant No. 32302859).
Conflict of interest
The authors claim that they have no competing interests.
References
- 1.Pensaert MB, Martelli P. Porcine epidemic diarrhea: a retrospect from Europe and matters of debate. Virus Res. 2016;226:1–6. doi: 10.1016/j.virusres.2016.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun RQ, Cai RJ, Chen YQ, et al. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis. 2012;18(1):161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao X, Zhang L, Jiang X, et al. Porcine epidemic diarrhea: an emerging disease in Tibetan pigs in Tibet, China. Trop Anim Health Prod. 2019;51(2):491–494. doi: 10.1007/s11250-018-1707-3. [DOI] [PubMed] [Google Scholar]
- 4.Lin C-M, Saif LJ, Marthaler D, et al. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016;226:20–39. doi: 10.1016/j.virusres.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun D, Wang X, Wei S, et al. Epidemiology and vaccine of porcine epidemic diarrhea virus in China: a mini-review. J Vet Med Sci. 2016;78(3):355–363. doi: 10.1292/jvms.15-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang E, Guo D, Li C, et al. Molecular characterization of the ORF3 and S1 genes of porcine epidemic diarrhea virus non S-INDEL strains in seven regions of China, 2015. PLoS One. 2016;11(8):e0160561. doi: 10.1371/journal.pone.0160561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King AMQ, Lefkowitz EJ, Mushegian AR, et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2018) Arch Virol. 2018;163(9):2601–2631. doi: 10.1007/s00705-018-3847-1. [DOI] [PubMed] [Google Scholar]
- 8.Kocherhans R, Bridgen A, Ackermann M, et al. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes. 2001;23(2):137–144. doi: 10.1023/A:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song D, Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44(2):167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subissi L, Imbert I, Ferron F, et al. SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets. Antiviral Res. 2014;101:122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo Y, Zhang J, Deng X, et al. Complete genome sequence of a highly prevalent isolate of porcine epidemic diarrhea virus in South China. J Virol. 2012;86(17):9551. doi: 10.1128/JVI.01455-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan Y, Tian X, Li W, et al. Isolation and characterization of a variant porcine epidemic diarrhea virus in China. Virol J. 2012;9:195. doi: 10.1186/1743-422X-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian Y, Yu Z, Cheng K, et al. Molecular characterization and phylogenetic analysis of new variants of the porcine epidemic diarrhea virus in Gansu, China in 2012. Viruses. 2013;5(8):1991–2004. doi: 10.3390/v5081991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung K, Saif LJ, Wang Q. Porcine epidemic diarrhea virus (PEDV): An update on etiology, transmission, pathogenesis, and prevention and control. Virus Res. 2020;286:198045. doi: 10.1016/j.virusres.2020.198045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wathelet MG, Orr M, Frieman MB, et al. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol. 2007;81(21):11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Züst R, Cervantes-Barragán L, Kuri T, et al. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007;3(8):e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham RL, Sims AC, Brockway SM, et al. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J Virol. 2005;79(21):13399–13411. doi: 10.1128/JVI.79.21.13399-13411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mielech AM, Deng X, Chen Y, et al. Murine coronavirus ubiquitin-like domain is important for papain-like protease stability and viral pathogenesis. J Virol. 2015;89(9):4907–4917. doi: 10.1128/JVI.00338-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fehr AR, Athmer J, Channappanavar R, et al. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J Virol. 2015;89(3):1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng X, StJohn SE, Osswald HL, et al. Coronaviruses resistant to a 3C-like protease inhibitor are attenuated for replication and pathogenesis, revealing a low genetic barrier but high fitness cost of resistance. J Virol. 2014;88(20):11886–11898. doi: 10.1128/JVI.01528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fehr AR, Channappanavar R, Jankevicius G, et al. The Conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio. 2016;7(6):e01721–16. doi: 10.1128/mBio.01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jimenez-Guardeño JM, Nieto-Torres JL, DeDiego ML, et al. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. 2014;10(8):e1004320. doi: 10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clementz MA, Kanjanahaluethai A, O'Brien TE, et al. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology. 2008;375(1):118–129. doi: 10.1016/j.virol.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subissi L, Posthuma CC, Collet A, et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci U S A. 2014;111(37):E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.te Velthuis AJW, van den Worm SHE, Snijder EJ. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40(4):1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorkhali R, Koirala P, Rijal S, et al. Structure and function of major SARS-CoV-2 and SARS-CoV proteins. Bioinform Biol Insights. 2021;15:11779322 211025876. doi: 10.1177/11779322211025876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, Ke H, Blikslager A, et al. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J Virol. 2018;92(4):e01677–17. doi: 10.1128/JVI.01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q, Ma J, Yoo D. Inhibition of NF-κB activity by the porcine epidemic diarrhea virus nonstructural protein 1 for innate immune evasion. Virology. 2017;510:111–126. doi: 10.1016/j.virol.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xing Y, Chen J, Tu J, et al. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J Gen Virol. 2013;94(Pt 7):1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, Fang L, Shi Y, et al. Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J Virol. 2015;90(4):2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q, Shi K, Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Ma Z, Li Y, et al. Porcine epidemic diarrhea virus: molecular mechanisms of attenuation and vaccines. Microb Pathog. 2020;149:104553. doi: 10.1016/j.micpath.2020.104553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y, Zhang H, Shi Z, et al. Porcine epidemic diarrhea virus nsp15 antagonizes interferon signaling by RNA degradation of TBK1 and IRF3. Viruses. 2020;12(6):599. doi: 10.3390/v12060599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi P, Su Y, Li R, et al. PEDV nsp16 negatively regulates innate immunity to promote viral proliferation. Virus Res. 2019;265:57–66. doi: 10.1016/j.virusres.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hillen HS, Kokic G, Farnung L, et al. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584(7819):154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Wu J, Wang H, et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182(2):417–428. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bouvet M, Imbert I, Subissi L, et al. RNA 3'-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc Natl Acad Sci U S A. 2012;109(24):9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krichel B, Falke S, Hilgenfeld R, et al. Processing of the SARS-CoV pp1a/ab nsp7-10 region. Biochem J. 2020;477(5):1009–1019. doi: 10.1042/BCJ20200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin R, Li J, Parvangada A, et al. Genetic conservation of SARS-CoV-2 RNA replication complex in globally circulating isolates and recently emerged variants from humans and minks suggests minimal pre-existing resistance to remdesivir. Antiviral Res. 2021;188:105033. doi: 10.1016/j.antiviral.2021.105033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung LS, Gund TM, Narayan M. Comparison of binding site of remdesivir and its metabolites with NSP12-NSP7-NSP8, and NSP3 of SARS CoV-2 virus and alternative potential drugs for COVID-19 treatment. Protein J. 2020;39(6):619–630. doi: 10.1007/s10930-020-09942-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imbert I, Guillemot J-C, Bourhis J-M, et al. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25(20):4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Brunn A, Teepe C, Simpson JC, et al. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One. 2007;2(5):e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang Y, Tong K, Yao R, et al. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV-2 replication. Cell Biosci. 2021;11(1):140. doi: 10.1186/s13578-021-00644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niu X, Kong F, Hou YJ, et al. Crucial mutation in the exoribonuclease domain of nsp14 of PEDV leads to high genetic instability during viral replication. Cell Biosci. 2021;11(1):106. doi: 10.1186/s13578-021-00598-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng X, van Geelen A, Buckley AC, et al. Coronavirus endoribonuclease activity in porcine epidemic diarrhea virus suppresses type I and type III interferon responses. J Virol. 2019;93(8):e02000–e2018. doi: 10.1128/JVI.02000-18. [DOI] [PMC free article] [PubMed] [Google Scholar]