Abstract
Background
Therapeutic efficacy studies (TESs) and detection of molecular markers of drug resistance are recommended by the World Health Organization (WHO) to monitor the efficacy of artemisinin-based combination therapy (ACT). This study assessed the trends of molecular markers of artemisinin resistance and/or reduced susceptibility to lumefantrine using samples collected in TES conducted in Mainland Tanzania from 2016 to 2021.
Methods
A total of 2,015 samples were collected during TES of artemether-lumefantrine at eight sentinel sites (in Kigoma, Mbeya, Morogoro, Mtwara, Mwanza, Pwani, Tabora, and Tanga regions) between 2016 and 2021. Photo-induced electron transfer polymerase chain reaction (PET-PCR) was used to confirm presence of malaria parasites before capillary sequencing, which targeted two genes: Plasmodium falciparum kelch 13 propeller domain (k13) and P. falciparum multidrug resistance 1 (pfmdr1).
Results
Sequencing success was ≥ 87.8%, and 1,724/1,769 (97.5%) k13 wild-type samples were detected. Thirty-seven (2.1%) samples had synonymous mutations and only eight (0.4%) had non-synonymous mutations in the k13 gene; seven of these were not validated by the WHO as molecular markers of resistance. One sample from Morogoro in 2020 had a k13 R622I mutation, which is a validated marker of artemisinin partial resistance. For pfmdr1, all except two samples carried N86 (wild-type), while mutations at Y184F increased from 33.9% in 2016 to about 60.5% in 2021, and only four samples (0.2%) had D1246Y mutations. pfmdr1 haplotypes were reported in 1,711 samples, with 985 (57.6%) NYD, 720 (42.1%) NFD, and six (0.4%) carrying minor haplotypes (three with NYY, 0.2%; YFD in two, 0.1%; and NFY in one sample, 0.1%). Between 2016 and 2021, NYD decreased from 66.1% to 45.2%, while NFD increased from 38.5% to 54.7%.
Conclusion
This is the first report of the R622I (k13 validated mutation) in Tanzania. N86 and D1246 were nearly fixed, while increases in Y184F mutations and NFD haplotype were observed between 2016 and 2021. Despite the reports of artemisinin partial resistance in Rwanda and Uganda, this study did not report any other validated mutations in these study sites in Tanzania apart from R622I suggesting that intensified surveillance is urgently needed to monitor trends of drug resistance markers and their impact on the performance of ACT.
Keywords: Malaria, Molecular markers, Therapeutic efficacy studies, Plasmodium falciparum kelch 13 (k13), Plasmodium falciparum multidrug resistance 1 (pfmdr1), Plasmodium falciparum, Tanzania
Background
Anti-malarial drugs, particularly artemisinin-based combinations, are recommended and widely used for effective case management, but drug resistance is a major threat that has impacted their effectiveness for malaria control and elimination. The threat is higher, especially in sub-Saharan African (SSA) countries, which contributed over 95.0% of cases and deaths globally in 2021 [1]. Artemisinin-based combination therapy (ACT) was introduced in the early 2000s in most malaria-endemic countries following the World Health Organization (WHO) recommendations due to widespread resistance to previously used anti-malarials, including chloroquine (CQ) and sulfadoxine-pyrimethamine (SP) [2]. Before and after the adoption of ACT, the efficacy of the artemisinin-based combinations has remained above 90.0% in many countries in SSA, including Tanzania, and this has played a vital role in the reduction of malaria burden between 2000 and 2015 [1]. However, progress has stalled since 2015, and the emergence of artemisinin partial resistance (ART-R) threatens the gains attained over the past two decades and the ongoing elimination efforts [1]. In Africa, the emergence of ART-R has been reported in Rwanda, Uganda, Eritrea and Tanzania [3–7]; and this could potentially impact both malaria case management and elimination strategies. Thus, there is an urgent need to intensify surveillance to monitor the efficacy as well as track the emergence and spread of antimalarial-resistant parasites, particularly in SSA [8].
Following deployment of ACT, the WHO recommended monitoring both the efficacy and safety of ACT to support effective case management strategies [9]. According to the WHO, analysis of molecular markers associated with anti-malarial resistance should also be done within TES to capture the emergence and track the spread of resistant parasites to artemisinins and partner drugs. Polymorphisms in different parasite genes, including Plasmodium falciparum kelch 13 (k13), P. falciparum chloroquine resistance transporter (pfcrt), P. falciparum dihydrofolate reductase (pfdhfr), P. falciparum dihydropteroate synthase (pfdhps), and P. falciparum multidrug resistance 1 (pfmdr1), have been identified and are commonly used as key molecular markers for tracking anti-malarial resistance to the respective drugs [10, 11]. For commonly used artemisinin-based combinations, such as artemether-lumefantrine (AL), mutations in k13 have been linked to ART-R while mutations in pfmdr1 are associated with tolerance or reduced sensitivity to lumefantrine and/or resistance to other drugs such as CQ and amodiaquine (AQ). Currently, the WHO recommends monitoring any of the 13 validated non-synonymous mutations in k13 as markers of ART-R (A675V, R622I, C580Y, P574L, R561H, P553L, I543T, R539T, Y493H, M476I, C469Y, N458Y, and M476I), while eight mutations are considered to be candidate markers (P441L, G449A, C469F, A481V, R515K, P527H, G538V, and V568G) [12]. In recent years, ART-R has been confirmed in multiple African countries, particularly in the Horn of Africa in Eritrea [5] associated with the R622I. Reports have also come from East African countries like: (1) Rwanda, where ART-R was associated with R561H mutation [3]; (2) Tanzania, with R561H and 675 mutations [7, 13] and (3) Uganda, where C469Y and A675V mutations have been reported [4, 6]. Due to the threat of potential spread and impact of ART-R in Africa, monitoring the efficacy of current and future anti-malarials through clinical evaluation and detection of drug resistance markers is necessary and urgently needed.
The commonly detected polymorphisms within the pfmdr1 gene include N86Y, Y184F, S1034C, N1042D, and D1246Y, and the most frequently reported mutations are N86Y, Y184F, and D1246Y [10]. Whereas N86 (wild-type) has been linked to reduced sensitivity to lumefantrine, the 86Y mutation has been associated with reduced sensitivity and/or resistance to AQ and CQ. The presence of the N86, 184F, and D1246 (NFD) haplotypes is linked to reduced sensitivity to lumefantrine, while the 86Y, Y184, and 1246Y (YYY) haplotypes have been reported to cause decreased sensitivity to CQ and AQ [14]. However, the mutations in the pfmdr1 gene have not been associated with clinical failure or resistance to lumefantrine, and there is no recommended marker for this important partner drug.
In Tanzania, TES for artemisinin-based combinations has been implemented before and after they were deployed as recommended by the WHO to ensure effective case management [15–18]. After the deployment of ACT in 2006 [19], TES have normally focused on AL, which is the first-line anti-malarial drug for the treatment of uncomplicated falciparum malaria, together with alternative artemisinin-based combinations, such as artesunate-amodiaquine (ASAQ), the first-line drug in Zanzibar since 2003, and dihydroartemisinin-piperaquine (DP), which was deployed in Mainland Tanzania in 2014 [20]. These TESs have been implemented by the Technical Working Group (TWG) of the National Malaria Control Programme (NMCP) and its partners, and the studies have been consistently done since 1997. Together with the studies done in the 2000s, the TES have shown that AL has maintained a cure rate of over 95.0%, while the cure rate of DP has been reported to be > 98.0% [16]. The efficacy of ASAQ was less than 90.0% before the deployment of ACT in 2006, but its performance has increased, reaching 100% in 2017, possibly linked to the withdrawal of CQ in Tanzania [17, 18]
Like in many malaria-endemic countries, routine detection of molecular markers of antimalarial resistance was not consistently performed as part of TES in Tanzania due to limited capacity. In 2016, the capacity to detect markers of resistance to different drugs was established with the support of the Partnership for Antimalarial Resistance Monitoring in Africa (PARMA) network [21], and the analysis has been performed as part of TES except in 2017 which is not covered in this study. The molecular analysis that has been done within TES since 2016 aimed at generating data on mutations in pfmdr1 and k13. In 2021 ART-R was detected in Tanzania, through country-wide surveys of malaria parasites [13] and confirmed using TES [7]. Thus, it is critical to conduct a thorough assessment of markers of drug resistance in Tanzania using retrospective and prospectively collected samples and data to explore the presence of ART-R in the sites covered by TES. This study was undertaken using the data generated through molecular analysis of samples collected in TESs from 2016 to 2021 to assess the trends of markers of resistance to artemisinins (k13) and/or reduced susceptibility to lumefantrine (pfmdr1). The findings provide evidence to NMCP and its partners to support malaria case management guidelines and policy as well as identification of foci of ART-R and planning of targeted malaria molecular surveillance (MMS) with a focus on areas with high prevalence and risk of resistant parasites in Mainland Tanzania.
Methods
Study site
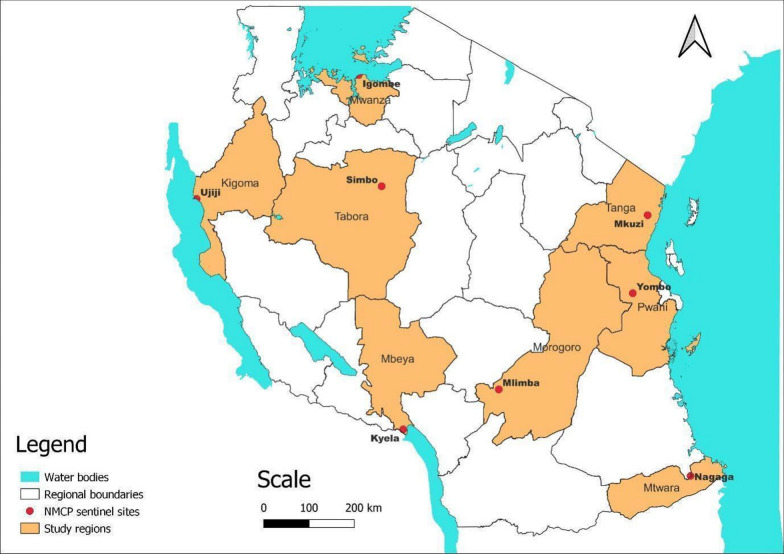
This study was based on the analysis of samples that were collected by the TWG of NMCP during TESs, which were conducted to assess the efficacy and safety of AL. The samples were collected from 2016 to 2021 at eight TES sentinel sites of NMCP, which were: Igombe health centre (Mwanza), Ipinda (Mbeya), Yombo (Pwani), Mkuzi (Tanga), Mlimba (Morogoro), Nagaga (Mtwara), Simbo (Tabora) and Ujiji health centre (Kigoma) (Fig. 1). The sites and details of study design and sample collection have been fully described in previous TES [22] and were based on the WHO protocol of 2009 [9]. According to the TWG’s framework, each sentinel site conducts a TES at least once every two years and thus all the sites were sampled three times during the study period. However, TES 2017 samples were not genotyped using the protocol recommended by the US Centres for Disease Control and Prevention (CDC) and are not included in this study.
Fig. 1.
Map of Tanzania showing the regions and the eight National Malaria Control Programme sentinel sites
Sample collection, processing and molecular analysis
The studies that provided samples for this analysis enrolled malaria patients meeting specific criteria as per the WHO protocol of 2009 [9], and details of enrolment procedures have been fully described in a previous study [22]. In summary, enrolled patients were aged 6 months to 10 years and had uncomplicated malaria with P. falciparum mono-infections and 250–200,000 asexual parasites/µl of blood, as well as fever at presentation or history of fever in the past 24 h. The enrolled patients received AL and were followed up for 28 days as per WHO protocol [9].
Dried blood spot (DBS) samples were collected on Whatman 3-mm filter paper (Whatmann No. 3, GE Healthcare Life Sciences, PA, USA) from enrolled patients on Day 0 (before treatment) and during follow-up visits in the case of recurrent infections, according to the procedures described earlier [22]. Genomic DNA was extracted using QIAamp blood mini-kits (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions and stored at 4 °C before use. Molecular analysis was performed on all samples collected upon enrolment (day 0) and during follow-up in the case of recurrent infections to confirm the presence of malaria parasites before sequencing. This was done at both genus (Plasmodium) and species (P. falciparum) levels using photo-induced electron transfer polymerase chain reaction (PET-PCR), which was performed as previously described [23]. Only samples with positive results for both Plasmodium genus and P. falciparum proceeded to the subsequent step of sequencing.
Sequencing to detect mutations in drug-resistance genes
Nested PCR was performed to amplify two genes, pfmdr1 (region 1, codon positions: 86 and 184, and region 2, codon positions: 1034, 1042 and 1246) [24], and the propeller domain of k13 (codon positions: 430 - 726) in separate reactions, as previously described [24]. The amplicons from each reaction were visualized on 2% agarose gel stained with RedsafeTM (Biotium, CA, USA). Capillary sequencing was performed using forward and reverse primers with the BIG dye terminator chemistry v3.1 (Applied Biosystems, UK), according to the protocol adopted from CDC in Atlanta, USA [24].
Downstream analysis was done using Geneious® analysis software version 2022.2.2 (Biomatters, New Zealand; www.geneious.com) as described by others [24]. Raw sequence reads were cleaned using Geneious default setting, and reads with high-quality scores (the percentage of high-quality bases, ≥ 70%) were retained for further analysis. The pfmdr1 and k13 sequences of 3D7 were used as references and single nucleotide polymorphisms (SNPs) detected in one or both strands were considered true SNPs.
Data management
The SNP data was entered into Microsoft Excel 2016 and later exported into R Studio software (version 4.1.3) for validation, cleaning, and analysis. For the pfmrd1 gene, the analysis focused on the three SNPs (N86Y, Y184F, and D1246Y) and their corresponding haplotypes, which have been associated with reduced susceptibility to lumefantrine. The findings were summarized and presented in text, tables, figures, and maps showing the prevalence and spatial as well as temporal changes of different SNPs and/or haplotypes.
Results
A total of 2,015 samples were collected in the TES, which were conducted from 2016 to 2021 from subjects receiving AL for the treatment of uncomplicated falciparum malaria. The study that was conducted in 2016 has been published [22], while the 2018 and 2019 studies are available online [25, 26] and others done from 2020 to 2021 have not yet been published. The 2017 data has been reported elsewhere [27] and thus it is not included in this paper because it was generated using an Illumina platform and with a different analysis pipeline from that of CDC, based on Sanger sequencing. All samples were sequenced for detection of drug resistance mutations in pfmdr1 and k13 genes (Table 1). Sequencing success was over 87.8% in both genes, but the pfmdr1 region 1 had a higher success rate, reaching 94.9% (Table 1).
Table 1.
Number of samples collected through TES from 2016 to 2021 and sequenced for detection of markers of drug resistance
| Year | # Samples sequenced | k13* (%) | pfmdr1-region 1* (%) | pfmdr1-region2* (%) |
|---|---|---|---|---|
| 2016 | 423 | 376 (88.9) | 423 (100.0) | 423 (100.0) |
| 2018 | 426 | 359 (84.0) | 401 (94.0) | 379 (89.0) |
| 2019 | 403 | 353 (88.0) | 371 (91.7) | 372 (92.0) |
| 2020 | 412 | 348 (8.0) | 378 (93.0) | 330 (80.0) |
| 2021 | 351 | 333 (95.0) | 335 (95.0) | 270 (77.0) |
| Total | 2015 | 1769 (87.8) | 1908 (94.7) | 1774 (88.0) |
#Number of samples attempted during sequencing
*number and proportions of samples which passed the sequencing quality score criteria and were therefore used in the analysis
Polymorphisms in k13 gene
Overall, 1,778 (88.2%) samples were successfully sequenced for k13, and the majority (1,724/1,769, 97.5%) had wild-type parasites. Of the successfully sequenced samples, 45 (2.5%) had k13 mutations, with 37 (2.1%) samples carrying synonymous mutations at codons P417P, C469C, R471R, V487V, F505F, G538G, R539R and S624S. Eight samples (0.4%) had non-synonymous mutations with seven SNPs that are not validated by WHO as molecular markers of ART-R (I416V, E433D, R471S, P475S, A578S, and Q613E). Only one sample from Morogoro in 2020 had R622I (a WHO-validated marker of ART-R) in a recurrent infection on day 14 (Table 2).
Table 2.
k13 mutations among samples collected during TES in the eight regions of Tanzania from 2016 to 2021
| Year | Region | #Sequenced | k13* (%) |
k13** (wild-type) (%) |
k13** (mutant) (%) |
k13 ** (SYN mutation) | k13** (NS mutation) |
|---|---|---|---|---|---|---|---|
| 2016 | Kigoma | 118 | 101 (85.6) | 95 (94.1) | 6 (5.9) | 3 (2-R471 & C469) | 3 (2-Q613E &I416V) |
| Morogoro | 104 | 96 (92.3) | 92 (95.8) | 4 (4.2) | 2 (R471 & R539) | 2 (E433D & A578S) | |
| Pwani | 92 | 79 (85.9) | 75 (94.9) | 4 (5.1) | 4 (P417, C469 & 2-V487 | 0 | |
| Tanga | 109 | 100 (91.7) | 95 (95.0) | 5 (5.0) | 4 (P417 & 3-C469) | 1 (R471S) | |
| 2018 | Kigoma | 111 | 96 (86.5) | 96 (100.0) | 0 (0.0) | 0 | 0 |
| Morogoro | 99 | 87 (87.9) | 84 (96.6) | 3 (3.4) | 3 (G538 & 2-R539) | 0 | |
| Pwani | 97 | 79 (81.4) | 76 (96.2) | 3 (3.8) | 3 (S624) | 0 | |
| Tanga | 119 | 97 (81.5) | 97 (100.0) | 0 (0.0) | 0 | 0 | |
| 2019 | Mbeya | 95 | 90 (94.7) | 86 (95.6) | 4 (4.4) | 4 (P417, 2- C469 & R539) | 0 |
| Mtwara | 93 | 88 (94.6) | 84 (95.5) | 4 (4.5) | 3 (C469 & 2-F505) | 1 (P475S) | |
| Tabora | 111 | 96 (86.5) | 94 (98.0) | 2 (2.0) | 2 (P417) | 0 | |
| Mwanza | 104 | 79 (76.0) | 79 (100.0) | 0 (0.0) | 0 | 0 | |
| 2020 | Pwani | 97 | 83 (85.6) | 82 (98.8) | 1 (1.2) | 1 (R539) | 0 |
| Kigoma | 103 | 70 (68.0) | 66 (94.3) | 4 (5.7) | 4 (P417) | 0 | |
| Morogoro | 96 | 95 (99.0) | 91 (95.8) | 4 (4.2) | 3 (P417) | 1 (R622I) | |
| Tanga | 116 | 100 (86.2) | 100 (100.0) | 0 (0.0) | 0 | 0 | |
| 2021 | Mbeya | 48 | 43 (89.6) | 43 (100.0) | 0 (0.0) | 0 | 0 |
| Mtwara | 87 | 87 (100.0) | 87 (100.0) | 0 (0.0) | 0 | 0 | |
| Mwanza | 106 | 96 (90.6) | 96 (100.0) | 0 (0.0) | 0 | 0 | |
| Tabora | 110 | 107 (97.3) | 106 (99.1) | 1 (0.9) | 1 (Y558) | 0 | |
| Total | 2015 | 1769 (87.8) | 1724 (97.5) | 45 (2.5) | 37 (2.1%) | 8 (0.4%) |
k13 = P. falciparum kelch 13, SYN synonymous mutations, and NS non-synonymous mutations
*number of samples that passed the sequencing quality score criteria
**number of samples with either wild-type or mutant genotypes and their corresponding proportions where appropriate
Polymorphisms and haplotypes in the pfmdr1 gene
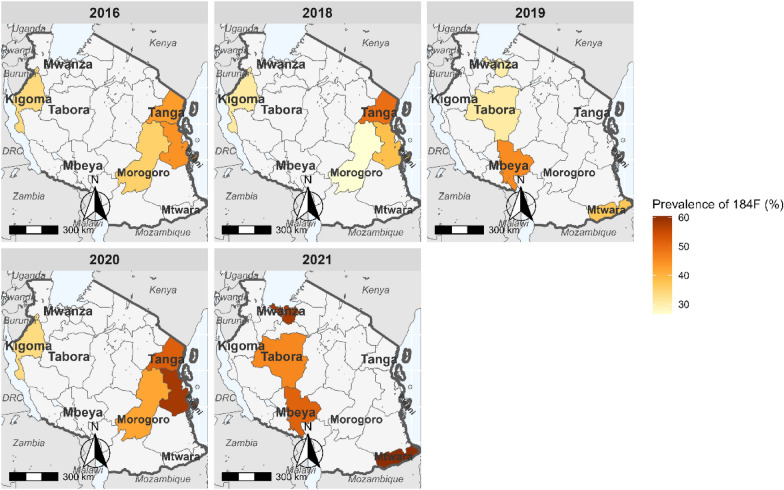
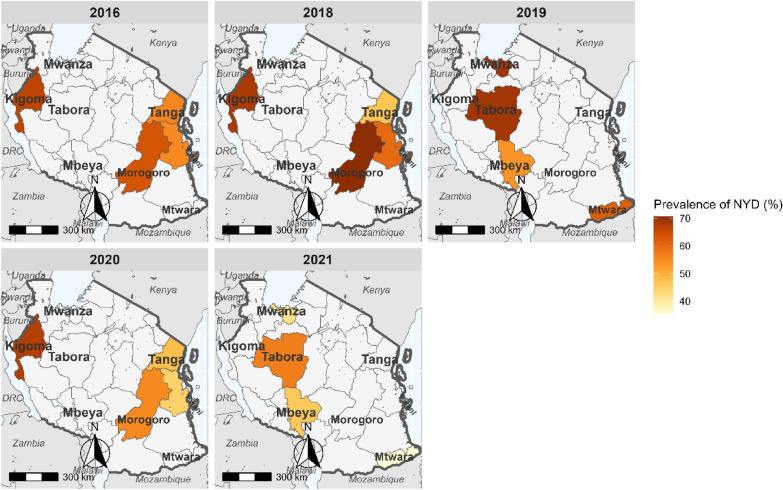
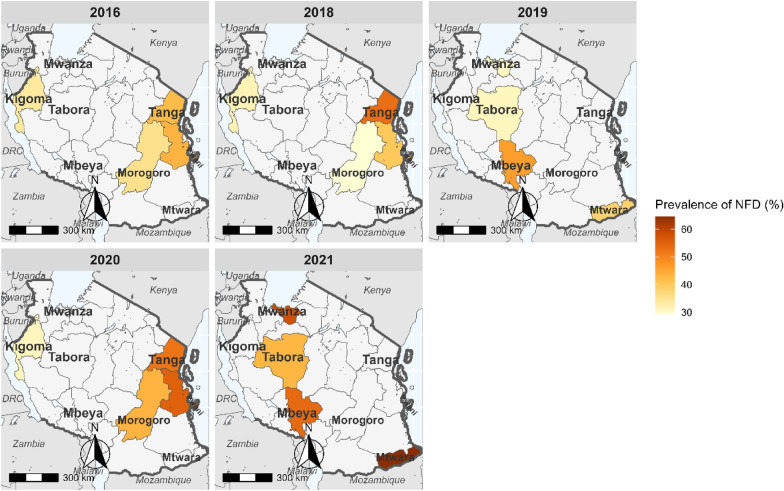
The number of successfully sequenced samples for the pfmdr1 regions covering the three mutations were 1,912 (94.9%) for codons 86 and 184, and 1,774 (88.0%) for codon 1246 (Table 1 and Table 3). For pfmdr1 codon 86, all except two samples (which had 86Y mutations in 2016) carried N86 (wildtype). Mutations at Y184F were more prevalent and increased from 33.9% in 2016 to about 60.5% in 2021 (Fig. 2). D1246Y mutations occurred in four samples (0.2%): two from Morogoro and Tanga in 2016 and two other mutations from Kigoma and Morogoro in 2020. The pfmdr1 haplotypes (N86Y, Y184F, and D1246Y) were constructed with 1,711 (85.0%) samples, and 985 (57.6%) of these had NYD, 720 (42.1%) had NFD, while six samples (0.4%) had minor haplotypes (three with NYY = 0.2%, YFD in two, 0.1%, and NFY in one sample 0.1%). Between 2016 and 2021, pfmdr1 haplotype NYD decreased from 66.1% to 45.2% (Fig. 3) while NFD increased from 38.5% to 54.7% (Fig. 4), but these changes varied among the study regions.
Table 3.
pfmdr1 mutations among samples collected in the eight regions from 2016 to 2021
| Year | Region | #Sequenced | pfmdr1* R1, n (%) | N86, n (%) | Y184, n (%) | 184F n (%) | pfmdr1* R2, n (%) | D1246 n (%) | 1246Y n (%) |
|---|---|---|---|---|---|---|---|---|---|
| 2016 | Kigoma | 118 | 118 (100.0) | 118 (100.0) | 78 (66.1) | 40 (33.9) | 118 (100.0) | 118 (100.0) | 0 (0.0) |
| Morogoro | 104 | 104 (100.0) | 104 (100.0) | 67 (64.4) | 37 (35.6) | 104 (100.0) | 103 (99.0) | 1 (1.0) | |
| Pwani | 92 | 92 (100.0) | 92 (100.0) | 51 (55.4) | 41 (44.6) | 92 (100.0) | 92 (100.0) | 0 (0.0) | |
| Tanga | 109 | 109 (100.0) | 109 (100.0) | 62 (56.9) | 47 (43.1) | 109 (100.0) | 108 (99.1) | 1 (0.9) | |
| 2018 | Kigoma | 111 | 110 (99.1) | 110 (99.1) | 76 (69.1) | 34 (30.9) | 104 (93.7) | 104 (100.0) | 0 (0.0) |
| Morogoro | 99 | 94 (94.9) | 94 (94.9) | 69 (73.4) | 25 (26.6) | 86 (86.9) | 86 (100.0) | 0 (0.0) | |
| Pwani | 97 | 86 (88.7) | 86 (88.7) | 53 (61.6) | 33 (38.4) | 87 (89.7) | 87 (100.0) | 0 (0.0) | |
| Tanga | 119 | 111 (93.3) | 111 (93.3) | 56 (50.5) | 55 (49.5) | 102 (85.7) | 102 (100.0) | 0 (0.0) | |
| 2019 | Mbeya | 95 | 88 (92.6) | 88 (92.6) | 48 (54.5) | 40 (45.5) | 84 (88.4) | 84 (100.0) | 0 (0.0) |
| Mtwara | 93 | 89 (95.7) | 89 (95.7) | 56 (62.9) | 33 (37.1) | 89 (95.7) | 89 (100.0) | 0 (0.0) | |
| Tabora | 111 | 98 (88.3) | 98 (88.3) | 68 (69.4) | 30 (30.6) | 97 (87.4) | 97 (100.0) | 0 (0.0) | |
| Mwanza | 104 | 96 (92.3) | 96 (92.3) | 67 (69.8) | 29 (30.2) | 102 (98.1) | 102 (100.0) | 0 (0.0) | |
| 2020 | Pwani | 97 | 90 (92.8) | 90 (92.8) | 38 (42.2) | 53 (58.9) | 79 (81.4) | 79 (100 .0) | 0 (0.0) |
| Kigoma | 103 | 92 (89.3) | 92 (89.3) | 61 (66.3) | 31 (33.7) | 65 (63.1) | 64 (98.5) | 1 (1.5) | |
| Morogoro | 96 | 91 (94.8) | 91 (94.8) | 54 (59.3) | 39 (42.9) | 85 (88.5) | 84 (98.8) | 1 (1.2) | |
| Tanga | 116 | 105 (90.5) | 105 (90.5) | 51 (48.6) | 55 (52.4) | 101 (87.1) | 101 (100.0) | 0 (0.0) | |
| 2021 | Mbeya | 48 | 35 (72.9) | 35 (72.9) | 17 (48.6) | 18 (51.4) | 34 (70.8) | 34 (100.0) | 0 (0.0) |
| Mtwara | 87 | 86 (98.9) | 86 (98.9) | 34 (39.5) | 52 (60.5) | 65 (74.7) | 65 (100.0) | 0 (0.0) | |
| Mwanza | 106 | 105 (99.1) | 105 (99.1) | 44 (41.9) | 61 (58.1) | 94 (88.7) | 94 (100.0) | 0 (0.0) | |
| Tabora | 110 | 109 (99.1) | 109 (99.1) | 59 (54.1) | 50 (45.9) | 77 (70.0) | 77 (100.0) | 0 (0.0) | |
| Total | 2015 | 1908 (94.7) | 1906 (99.9) | 1109 (58.1) | 803 (42.0) | 1774 (88.0) | 1770 (99.0) | 4 (0.2) |
*number of samples which passed the sequencing quality score criteria, n = number of samples. % = percentage of samples analyzed or with a particular genotype
Fig. 2.
Trend of 184F mutation in the eight regions of Mainland Tanzania from 2016 to 2021
Fig. 3.
Trend of pfmdr1 NYD haplotype in the eight regions of Mainland Tanzania from 2016 to 2021
Fig. 4.
Trend of pfmdr1 NFD haplotype in the eight regions of Mainland Tanzania from 2016 to 2021
Discussion
Anti-malarial resistance threatens the effectiveness of current malaria treatments much like it has done for various antimalarials over the past half a century [1]. In recent years, reports of ART-R in Africa [1] have been a growing concern because of the potential emergence and spread of ACTs resistance. Thus, there is a critical need to monitor the effectiveness of these strategies using different approaches, such as TES and MMS. Tracking the emergence and spread of molecular markers of resistance to artemisinin and partner drugs is crucial so as to maintain the effectiveness of malaria treatment, prevent the spread of resistant strains and contribute to global efforts to respond to ART-R as well as control and eliminate malaria. The current study aimed to assess the trends of molecular markers of drug resistance in two genes, k13 and pfmdr1, using the data collected in Mainland Tanzania between 2016 and 2021. Over the study period, WHO-validated mutations in k13, which are associated with ART-R, were only detected in one sample, while significant changes occurred in the pfmdr1 gene.
This study reported less than 2.5% of non-synonymous mutations in k13 gene, and only one sample had validated k13 mutation (R622I) from Morogoro in 2020. The findings align with most reports in some African countries in which no or a very low prevalence of k13 mutations has been reported [28, 29]. Until recently, R622I has been reported in three countries: Ethiopia, Eritrea, and Sudan [30], and this is the first report of the R622I mutation in Tanzania. In Eritrea, an increase in R622I prevalence from less than 10% in 2016 to 20% in 2019 has been reported [5], while in Ethiopia, the mutation has been reported in several studies, with prevalence ranging from 2.4 to 9.5%. [31, 32]. Similarly, it has been shown that parasites with R622I mutations tend also to carry deletions of histidine-rich protein 2/3 (hrp2/3) gene [33], which is linked to the failure of malaria rapid diagnostic tests that detect the HRP2 antigen to detect P. falciparum infections. In a recent Tanzania-wide surveillance, the R622I was also detected in one sample from Njombe region in 2021, in an area close to Morogoro where the 2020 sample was detected [13]. Njombe is a region that reported more parasites with hrp2/3 single gene deletions in 2021, compared to other regions of Tanzania [34]. More studies will be needed to explore if the parasites with R622I are co-emerging with the gene deletions as reported in Ethiopia and Eritrea.
Despite the reports of ART-R in Rwanda [3] and Uganda [4], this study did not report any other validated mutations in these study sites in Tanzania apart from R622I. However, previous studies in Tanzania reported the presence of R561H mutation in two regions of Pwani in 2020 [35] and Geita [27], and this analysis revealed no evidence for presence of these mutations in these study sites during this period. Recently, the R56IH mutation was detected at high prevalence in some parts of Kagera (reaching over 20%), while few samples with the same mutations were also detected in Tabora, Njombe and Manyara in 2021 [13]. More surveys in Kagera have observed an increase in these mutations in some districts and a spread from three districts in 2021 to five in 2023 (Ishengoma et al., pers.commun.). Another mutation (A675V) has also been reported in Kagera, but at low prevalence compared to R561H, suggesting a potential threat of spreading ART-R in Kagera and other regions (Ishengoma et al., pers.commun.). Therefore, there is an urgent need for other strategies apart from TES for effectively monitoring the spread of ART-R in Tanzania because the presence of these mutations could not be captured by TES in the studies that were done before and after deployment of ACT in 2006.
The polymorphisms in the pfmdr1 gene have been associated with several anti-malarial drugs, including lumefantrine, CQ, and AQ. The N86 and D1246 alleles were observed to be near fixation, and N86, which has been linked to decreased susceptibility to lumefantrine and increased susceptibility to AQ, was only detected in two samples in 2016 (in Pwani and Tanga).
Studies from various parts of Africa have also reported similar results, and this has been linked to AL selecting the N86 allele [36–38]. There was a significant increase in both the 184F and NFD haplotypes over the years, and this might be linked to selection caused by lumefantrine. Another study conducted in Bagamoyo districts in Tanzania using the samples collected from 2006 to 2011 reported similar findings, with a high prevalence of 184F mutations, and an increase from 14.0% in 2006 to 35.0% in 2011 [15]. Several other studies conducted in Africa have also documented an increase of 184F, leading to a rise in NFD haplotype, possibly due to the continued use of AL as the first-line treatment. In Tanzania, the trends of both 184F and NFD haplotype across regions appear to be homogeneous, indicating the drug selection pressure might be similar throughout the country. Similar findings have been reported in other sub-Saharan African countries, where temporal trends of the Y184F mutations and NFD haplotypes have been associated with reduced susceptibility to the lumefantrine component of AL [39, 40]. It is important to note that the changes in pfmdr1 markers were not associated with reduced efficacy of AL, which was > 95.0% for all years [22]. Although these mutations have not been linked to ACT failure, it is essential to monitor their spread and possible association (together with other markers) with ACT resistance in different endemic countries to inform malaria case management strategies.
Conclusions
This is the first report of the R622I (k13 validated mutation) in Tanzania. N86 wildtype, which is associated with decreased susceptibility to lumefantrine and increased susceptibility to AQ and CQ, is near-fixation together with D1246. Changes were observed in pfmdr1, with an increase in Y184F mutations and NFD haplotype reaching over 50% in all regions except Tabora, with over 42% in 2021. Despite the reports of ART-R in Rwanda and Uganda, this study did not report any other validated mutations in these study sites in Tanzania apart from R622I. Following detection of ART-R (561H) in the Kagera region, which was not captured by TES, intensified molecular surveillance is urgently needed to monitor the trends of drug resistance markers and their potential impact on the performance of ACTs.
Disclaimer
The findings and conclusions presented in this report are those of the authors and do not necessarily reflect the official position of the Centers for Disease Control and Prevention and of the US Agency for International Development.
Acknowledgements
The authors are indebted to the children and their parents/guardians for agreeing to take part in the studies and attending the follow-up visits despite the long durations of follow-up. They extend their appreciation to health facilities’ staff, stakeholders, and colleagues from implementing partners and local health authorities for their support during the entire period of implementing the TES and this study. The support provided by the regional and district authorities is greatly acknowledged. Technical and logistic support provided by the CDC, USAID/PARMA, RTI, and NIMR teams is highly appreciated. Permission to publish this paper has been granted by the Director General of NIMR.
Abbreviations
- ACT
Artemisinin-based combination therapy
- AL
Artemether-lumefantrine
- ASA
Artesunate + amodiaquine
- AQ
Amodiaquine
- CDC
US Centers for Disease Control and Prevention
- CQ
Chloroquine
- DBS
Dried blood spot
- DNA
Deoxyribonucleic acid
- DP
Dihydroartemisinin–piperaquine
- MRCC
Medical Research Coordinating Committee of NIMR
- NIMR
National Institute for Medical Research
- NMCP
National Malaria Control Programme
- PARMA
Partnership for Antimalarial Resistance Monitoring in Africa network
- k13
Plasmodium falciparum Kelch 13 gene
- Pfmdr1
Plasmodium falciparum multi-drug resistance 1 gene
- PMI
US President’s Malaria Initiative
- PCR
Polymerase chain reaction
- SNP
Single nucleotide polymorphism
- SP
Sulfadoxine-pyrimethamine
- SSA
Sub-Saharan Africa
- TES
Therapeutic efficacy study
- TWG
Technical Working Group
- WHO
World Health Organization
Author contributions
DSI, CB, RAM, MDS and CIM conceived of the study, designed the experiments and took part in the laboratory analysis of samples with the support of ET, NWL, MV, LM, ESH and UV. DSI, CIM, BN, EK, MA, FF, SB, MC, MKM, RAK and FM implemented TES in the 8 regions with the support of SM, RM, FM, FC, DB, RN, MW, SN, KB, NS, CK and ER. RAM, CB and MDS generated and analyzed the data under the supervision of DSI: CB and DSI wrote the manuscript. All authors reviewed and approved the final manuscript.
Funding
The US President’s Malaria Initiative supported the TES which generated the data for this study.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
All the studies which generated the samples obtained ethical clearance from the Medical Research Coordinating Committee (MRCC) of the National Institute for Medical Research (NIMR—MRCC) in Tanzania.
Consent for publications
Permission to publish the manuscript was sought and provided by the Director General of NIMR.
Competing interests
All authors declare no competing financial interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO. World malaria report 2022. World Health Organization; 2022.
- 2.WHO. Antimalarial Drug Combination Therapy - Report of a WHO Technical Consultation. World Health Organization, Geneva WHO. 2001.
- 3.Uwimana A, Umulisa N, Venkatesan M, Svigel SS, Zhou Z, Munyaneza T, et al. Association of Plasmodium falciparum kelch13 R561H genotypes with delayed parasite clearance in Rwanda: an open-label, single-arm, multicentre, therapeutic efficacy study. Lancet Infect Dis. 2021;21:1120–1128. doi: 10.1016/S1473-3099(21)00142-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balikagala B, Fukuda N, Ikeda M, Katuro OT, Tachibana S-I, Yamauchi M, et al. Evidence of Artemisinin-Resistant Malaria in Africa. N Engl J Med. 2021;385:1163–1171. doi: 10.1056/NEJMoa2101746. [DOI] [PubMed] [Google Scholar]
- 5.Mihreteab S, Platon L, Berhane A, Stokes BH, Warsame M, Campagne P, et al. Increasing Prevalence of Artemisinin-Resistant HRP2-Negative Malaria in Eritrea. N Engl J Med. 2023;389:1191–1202. doi: 10.1056/NEJMoa2210956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conrad MD, Asua V, Garg S, Giesbrecht D, Niaré K, Smith S, et al. Evolution of Partial Resistance to Artemisinins in Malaria Parasites in Uganda. N Engl J Med. 2023;389:722–732. doi: 10.1056/NEJMoa2211803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishengoma DS, Mandara CI, Bakari C, Fola AA, Madebe RA, Seth MD, et al. Evidence of artemisinin partial resistance in North-western Tanzania: clinical and drug resistance markers study. medRxiv. 2024; 10.1101/2024.01.31.24301954.
- 8.Blasco B, Leroy D, Fidock DA. Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat Med. 2017;23:917–928. doi: 10.1038/nm.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO. Methods for surveillance of antimalarial drug efficacy; 2009.
- 10.Ishengoma DS, Saidi Q, Sibley CH, Roper C, Alifrangis M. Deployment and utilization of next-generation sequencing of Plasmodium falciparum to guide anti-malarial drug policy decisions in sub-Saharan Africa: Opportunities and challenges. Malar J. 2019;18:267. doi: 10.1186/s12936-019-2853-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodoameda P. P. falciparum and Its Molecular Markers of Resistance to Antimalarial Drugs. Plasmodium species and drug resistance. IntechOpen; 2021.
- 12.WHO. Malaria: Artemisinin partial resistance. Geneva, World Health Organization [cited 2023 Oct 31]. Available from: https://www.who.int/news-room/questions-and-answers/item/artemisinin-resistance.
- 13.Juliano JJ, Giesbrecht DJ, Simkin A, Fola AA, Lyimo BM, Pereus D, et al. Country wide surveillance reveals prevalent artemisinin partial resistance mutations with evidence for multiple origins and expansion of sulfadoxine-pyrimethamine resistance mutations in northwest Tanzania. bioRxiv. 2023. Available from: https://www.medrxiv.org/content/10.1101/2023.11.07.23298207v1.
- 14.Venkatesan M, Gadalla NB, Stepniewska K, Dahal P, Nsanzabana C, Moriera C, et al. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P. falciparum malaria after artemether-lumefantrine and artesunate-amodiaquine. Am J Trop Med Hyg. 2014;91:833–43. [DOI] [PMC free article] [PubMed]
- 15.Malmberg M, Ngasala B, Ferreira PE, Larsson E, Jovel I, Hjalmarsson A, et al. Temporal trends of molecular markers associated with artemether-lumefantrine tolerance/resistance in Bagamoyo district. Tanzania Malar J. 2013;12:103. doi: 10.1186/1475-2875-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shayo A, Buza J, Ishengoma DS. Monitoring of efficacy and safety of artemisinin-based anti-malarials for treatment of uncomplicated malaria: a review of evidence of implementation of anti-malarial therapeutic efficacy trials in Tanzania. Malar J. 2015;14:135. doi: 10.1186/s12936-015-0649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kakolwa MA, Mahende MK, Ishengoma DS, Mandara CI, Ngasala B, Kamugisha E, et al. Efficacy and safety of artemisinin-based combination therapy, and molecular markers for artemisinin and piperaquine resistance in Mainland Tanzania. Malar J. 2018;17:369. doi: 10.1186/s12936-018-2524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandara CI, Francis F, Chiduo MG, Ngasala B, Mandike R, Mkude S, et al. High cure rates and tolerability of artesunate–amodiaquine and dihydroartemisinin–piperaquine for the treatment of uncomplicated falciparum malaria in Kibaha and Kigoma. Tanzania Malar J. 2019;18:1–12. doi: 10.1186/s12936-019-2740-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ministry of Health. National guidelines for malaria diagnosis and treatment. United Republic of Tanzania Dar es Salaam; 2006.
- 20.Ministry of Health. National guidelines for diagnosis and treatment of malaria. United Republic of Tanzania Dar es Salam; 2014.
- 21.Halsey ES, Venkatesan M, Plucinski MM, Talundzic E, Lucchi NW, Zhou Z, et al. Capacity Development through the US President’s Malaria Initiative-Supported Antimalarial Resistance Monitoring in Africa Network. Emerg Infect Dis. 2017;23:S53–S56. doi: 10.3201/eid2313.170366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishengoma DS, Mandara CI, Francis F, Talundzic E, Lucchi NW, Ngasala B, et al. Efficacy and safety of artemether-lumefantrine for the treatment of uncomplicated malaria and prevalence of Pfk13 and Pfmdr1 polymorphisms after a decade of using artemisinin-based combination therapy in mainland Tanzania. Malar J. 2019;18:88. doi: 10.1186/s12936-019-2730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucchi NW, Karell MA, Journel I, Rogier E, Goldman I, Ljolje D, et al. PET-PCR method for the molecular detection of malaria parasites in a national malaria surveillance study in Haiti, 2011. Malar J. 2014;13:462–462. doi: 10.1186/1475-2875-13-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talundzic E, Chenet SM, Goldman IF, Patel DS, Nelson JA, Plucinski MM, et al. Genetic analysis and species specific amplification of the artemisinin resistance-associated kelch propeller domain in P. falciparum and P. vivax. PLoS One. 2015;10:e0136099. [DOI] [PMC free article] [PubMed]
- 25.Ngasala B, Chiduo MG, Bushukatale S, Mmbando BP, Makene T, Kamugisha E, et al. Efficacy and safety of artemether-lumefantrine for the treatment of uncomplicated falciparum malaria in mainland Tanzania, 2018. Research Square. 2024[cited 2024 Jan 17]; Available from: https://www.researchsquare.com/article/rs-3520720/v1. [DOI] [PMC free article] [PubMed]
- 26.Ngasala BE, Chiduo MG, Mmbando BP, Francis FT, Bushukatale S, Makene T, et al. Efficacy and safety of artemether-lumefantrine for the treatment of uncomplicated falciparum malaria in mainland Tanzania, 2019. Research Square. 2024[cited 2024 Jan 17]; Available from: https://www.researchsquare.com/article/rs-3786283/v1. [DOI] [PMC free article] [PubMed]
- 27.Moser KA, Madebe RA, Aydemir O, Chiduo MG, Mandara CI, Rumisha SF, et al. Describing the current status of Plasmodium falciparum population structure and drug resistance within mainland Tanzania using molecular inversion probes. Mol Ecol. 2020;mec.15706 – mec.15706. [DOI] [PMC free article] [PubMed]
- 28.Schmedes SE, Patel D, Dhal S, Kelley J, Svigel SS, Dimbu PR, et al. Plasmodium falciparum kelch 13 Mutations, 9 Countries in Africa, 2014–2018. Emerg Infect Dis. 2021;27:1902–1908. doi: 10.3201/eid2707.203230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ndwiga L, Kimenyi KM, Wamae K, Osoti V, Akinyi M, Omedo I, et al. A review of the frequencies of Plasmodium falciparum kelch 13 artemisinin resistance mutations in Africa. Int J Parasitol Drugs Drug Resist. 2021;16:155–161. doi: 10.1016/j.ijpddr.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.WHO. Malaria Policy Advisory Group (MPAG) meeting report, 18–20 April 2023. World Health Organization; 2023 [cited 2023 Dec 7]; Available from: https://www.who.int/publications/i/item/9789240074385.
- 31.Bayih AG, Getnet G, Alemu A, Getie S, Mohon AN, Pillai DR. A Unique Plasmodium falciparum kelch 13 Gene Mutation in Northwest Ethiopia. Am J Trop Med Hyg. 2016;94:132. doi: 10.4269/ajtmh.15-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alemayehu AA, Castaneda-Mogollon D, Tesfa H, Getie S, Mohon AN, Balasingam N, et al. Expansion of the Plasmodium falciparum kelch 13 R622I mutation in Northwest Ethiopia. 2021 [cited 2024 Feb 27]; Available from: https://www.researchsquare.com/article/rs-171038.
- 33.Fola AA, Feleke SM, Mohammed H, Brhane BG, Hennelly CM, Assefa A, et al. Plasmodium falciparum resistant to artemisinin and diagnostics have emerged in Ethiopia. Nat Microbiol. 2023;8:1911–1919. doi: 10.1038/s41564-023-01461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogier E, Battle N, Bakari C, Seth MD, Nace D, Herman C, et al. Plasmodium falciparum pfhrp2 and pfhrp3 gene deletions among patients enrolled at 100 health facilities throughout Tanzania: February to July 2021. medRxiv. 2023 [cited 2023 Dec 7]; Available from: https://www.medrxiv.org/content/10.1101/2023.07.29.23293322. [DOI] [PMC free article] [PubMed]
- 35.Bwire GM, Ngasala B, Mikomangwa WP, Kilonzi M, Kamuhabwa AAR. Detection of mutations associated with artemisinin resistance at k13-propeller gene and a near complete return of chloroquine susceptible falciparum malaria in Southeast of Tanzania. Sci Rep. 2020;10:3500. doi: 10.1038/s41598-020-60549-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adamu A, Jada MS, Haruna HMS, Yakubu BO, Ibrahim MA, Balogun EO, et al. Plasmodium falciparum multidrug resistance gene-1 polymorphisms in Northern Nigeria: implications for the continued use of artemether-lumefantrine in the region. Malar J. 2020;19:439. doi: 10.1186/s12936-020-03506-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kay K, Goodwin J, Ehrlich H, Ou J, Freeman T, Wang K, et al. Impact of drug exposure on resistance selection following artemether-lumefantrine treatment for malaria in children with and without HIV in Uganda. Clin Pharmacol Ther. 2023;113:660–669. doi: 10.1002/cpt.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hailemeskel E, Menberu T, Shumie G, Behaksra S, Chali W, Keffale M, et al. Prevalence of Plasmodium falciparum Pfcrt and Pfmdr1 alleles in settings with different levels of Plasmodium vivax co-endemicity in Ethiopia. Int J Parasitol Drugs Drug Resist. 2019;11:8–12. doi: 10.1016/j.ijpddr.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lobo E, de Sousa B, Rosa S, Figueiredo P, Lobo L, Pateira S, et al. Prevalence of pfmdr1 alleles associated with artemether-lumefantrine tolerance/resistance in Maputo before and after the implementation of artemisinin-based combination therapy. Malar J. 2014;13:300. doi: 10.1186/1475-2875-13-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konaté-Touré A, Gnagne AP, Bedia-Tanoh AV, Menan EIH, Yavo W. Increase of Plasmodium falciparum parasites carrying lumefantrine-tolerance molecular markers and lack of South East Asian pfk13 artemisinin-resistance mutations in samples collected from 2013 to 2016 in Côte d’Ivoire. J Parasit Dis. 2024; Available from: 10.1007/s12639-023-01640-4. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.