Abstract
Heat shock transcription factor 1 (HSF-1) activates the transcription of heat shock genes in eukaryotes. Under normal physiological growth conditions, HSF-1 is a monomer. Its transcriptional activity is repressed by constitutive phosphorylation. Upon activation, HSF-1 forms trimers, acquires DNA binding activity, increases transcriptional activity, and appears as punctate granules in the nucleus. In this study, using bromouridine incorporation and confocal laser microscopy, we demonstrated that newly synthesized pre-mRNAs colocalize to the HSF-1 punctate granules after heat shock, suggesting that these granules are sites of transcription. We further present evidence that glycogen synthase kinase 3β (GSK-3β) and extracellular signal-regulated kinase mitogen-activated protein kinase (ERK MAPK) participate in the down regulation of HSF-1 transcriptional activity. Transient increases in the expression of GSK-3β facilitate the disappearance of HSF-1 punctate granules and reduce hsp-70 transcription after heat shock. We have also shown that ERK is the priming kinase for GSK-3β. Taken together, these results indicate that GSK-3β and ERK MAPK facilitate the inactivation of activated HSF-1 after heat shock by dispersing HSF-1 from the sites of transcription.
The nuclear translocation, DNA binding, and transcriptional activities of most mammalian transcription factors are regulated by phosphorylation. In many cases, multiple protein kinases can act on a single transcription factor (reviewed in reference 27). Heat shock transcription factor 1 (HSF-1) is subject to complex regulation by phosphorylation. HSF-1 binds to conserved regulatory sequences known as heat shock elements (HSEs) and controls the expression of heat shock proteins in response to chemical, environmental, and physiological stresses (1, 36, 42, 61, 68, 74).
Under normal physiological growth conditions, mammalian HSF-1 exists in a latent, monomeric form (4, 55, 66, 69); is constitutively phosphorylated (4, 10, 43, 55); and is distributed in both the cytoplasm and nucleus. The functional role of phosphorylation in HSF-1 regulation is unclear. Strong evidence suggests that constitutive phosphorylation of HSF-1 negatively regulates HSF-1 activity (9, 31, 32, 43). Upon heat shock, the latent form of HSF-1 is translocated into the nucleus, forms trimers, is hyperphosphorylated, and appears as punctate granules (4, 44, 51, 55). The function of hyperphosphorylation (4, 10, 46, 55) or the role of HSF-1 punctate granules is not known. Punctate granules have been suggested to be important for some activity of HSF-1, perhaps its DNA binding activity (58).
Phosphorylated forms of HSF-1 have been extensively studied by phosphopeptide mapping as well as mutational analysis (30–32, 71). The data suggest that HSF-1 is phosphorylated on multiple serine residues and, perhaps, a threonine residue. Constitutive phosphorylation of serine 307, which is located distal to the transcriptional activation domain, negatively regulates HSF-1 function, since mutation of serine 307 to alanine (31, 71) causes constitutive transcriptional activation of HSF-1.
The protein kinases which phosphorylate HSF-1 have not been identified with certainty in vivo. However, it has been suggested that one or more mitogen-activated protein kinase (MAPK) family members may be involved in the phosphorylation of HSF-1 (9, 30, 32, 43). Recent in vitro studies indicate that the extracellular signal-regulated kinases (ERKs) phosphorylate HSF-1 on serine 307, which then facilitates serine 303 phosphorylation by glycogen synthase kinase 3β (GSK-3β) (9).
The MAPKs respond to diverse stimuli and consist of sequential protein kinase cascades. MAPKs are activated via phosphorylation of specific threonine and tyrosine residues by dual-specificity kinases known as MEK/MKKs. MEK/MKKs are phosphorylated and activated by MEK kinases (MEKKs/MKKKs) (34, 56). There are three well-characterized MAPK pathways: ERK1/ERK2, also known as p42/p44 MAPKs (7); the p38/RK/Mpk2/CSBP protein kinases (22, 37); and the c-Jun amino-terminal kinases/stress-activated protein kinases (JNK/SAPK) (15, 35). Activation of growth factor receptors, G protein-coupled receptors, and some cytokine receptors activates ERKs (56). The p38 protein kinases are activated by proinflammatory cytokines and osmotic shock (22, 28, 52). JNKs are activated by various cellular stresses such as UV, protein synthesis inhibitors, proinflammatory cytokines, G protein-coupled receptors, and growth factor receptors (20, 35, 75). Multiple transcription factors, including ATF2, SAP-1, TCFs/ElK1, MEF2C, CHOP, and c-Jun, are phosphorylated and their activity is regulated by various MAPKs (20, 21, 48, 62, 65, 72).
GSK-3 consists of two isoforms, GSK-3α (51 kDa) and GSK-3β (46 kDa), and was first identified as an activity which phosphorylates and inactivates glycogen synthase (47, 49). A second role of GSK-3 was found when studies showed that inhibition of phosphatase type I activity is relieved when GSK-3 phosphorylates phosphatase inhibitor 2 (25, 38). At least 15 other substrates have been reported to be phosphorylated by GSK-3, including the transcription factors c-Jun, JunD, c-myb, c-myc, L-myc, CREB, and NF-AT, most of which become inactivated when phosphorylated by GSK-3 (5, 18, 47, 54). GSK-3 tends to phosphorylate serine/threonine residues located next to a proline which, in turn, is near another serine residue that has been prephosphorylated by some other protein kinase (referred to as priming kinase) (17, 47). GSK-3 is constitutively active and, as a result, suppresses many of its substrates under normal physiological growth conditions.
In the present study, we examined the characteristic HSF-1 punctate granules which are ubiquitously observed in human cells following heat shock and demonstrate that these HSF-1-containing granules are active transcription complexes. In addition, we investigated the regulation of HSF-1 activity by GSK-3β and MAPK in vivo. Overexpression of GSK-3β causes the rapid disappearance of the HSF-1 punctate granules. This suggests that GSK-3β facilitates HSF-1 inactivation, which is supported by the hsp70-luciferase reporter assay data showing that HSF-1 transcriptional activity after heat shock is decreased by overexpression of GSK-3β. GSK-3β inactivation of HSF-1 requires ERK activity, since treatment of cells with PD98059, a specific MEK inhibitor, before heat shock counteracts the effect of GSK-3β.
MATERIALS AND METHODS
Cell lines and plasmids.
HeLa cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. Expression plasmids HA-MNK1, HA-ERK1, Flag-tagged p38, HA-GSK-3β, and hsp70-luciferase were the generous gifts of T. Hunter, M. Cobb, R. J. Ulevitch, J. P. Woodgett, and R. I. Morimoto, respectively.
Transient-transfection assays.
Transient transfections were performed by electroporation (GenePulser; Bio-Rad). Transfected-DNA mixtures included 5 μg of expression plasmid DNA and, when required, 2 μg of hsp70-luciferase DNA and 0.1 μg of Renilla luciferase DNA with pBluescript carrier DNA added to a total of 20 μg. The DNA mixture was added to 5 × 106 HeLa cells in a 0.4-cm cuvette containing 0.8 ml of serum-free growth medium. Immediately after electroporation (280 V, 950 μF), the cells were fed with 10 ml of growth medium plus 10% fetal calf serum and incubated at 37°C. For immunofluorescence studies, cells were plated in eight-chamber culture slides 24 h after transfection and incubated for an additional 24 h at 37°C before being subjected to further manipulations. For luciferase assays, the cells were plated in 60-mm culture dishes after transfection and left at 37°C for 48 h before undergoing additional treatments. Luciferase assays were performed as specified by the manufacturer (Promega, Madison, Wis.). Renilla luciferase was used as an indicator of transfection frequency.
Indirect immunofluorescence analysis.
Cells were transiently transfected as described above. After 24 h, the cells were trypsinized, plated in eight-chamber tissue culture slides, and incubated at 37°C for an additional 24 h. The cells were treated as described in Results, rinsed with phosphate-buffered saline (PBS), and fixed with 4% paraformaldehyde for 30 min at room temperature. The slides were washed three times with PBS, and the cells were permeabilized for 2 min on ice with a solution containing 0.1% Triton X-100 and 0.1% sodium citrate and rinsed with PBS. To reduce nonspecific binding, the cells were incubated in blocking solution (5% goat serum and 5% bovine serum albumin in PBS) at 37°C for 1 h. They were then incubated in the presence of the primary antibody for 1 h at 37°C, rinsed with PBST (PBS–0.1% Tween 20), and incubated in the presence of secondary antibody (conjugated with fluorescein isothiocyanate [FITC] or Texas red) for an additional 1 h at 37°C. The cells were extensively rinsed with PBST, and the slides were mounted with Pro-Long Antifade (Molecular Probes, Eugene, Oreg.) and examined by fluorescence microscopy.
Antibody specific for HSF-1 was generated in rabbits after multiple injections of a peptide containing amino acids 429 to 454 in the C-terminal region of human HSF-1. Monoclonal antibody 12CA5 specific for hemagglutinin (HA) was obtained from Boehringer Mannheim, Indianapolis, Ind. The anti-Flag M2 was purchased from IBI Flag System, Kodak.
Combined in situ run-on transcription and immunofluorescence.
HeLa cells were plated in chamber slides and incubated at 37°C overnight. The cells were heated at 45°C for 30 min and incubated at 37°C for 1 to 8 h. For the in situ run-on transcription, the procedure of Semmes and Jeang was used (57). Heated cells were washed with Tris-buffered saline (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM MgCl2) and then glycerol buffer (20 mM Tris-HCl [pH 7.4], 5 mM MgCl2, 0.5 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 25% glycerol) for 5 min. The cells were permeabilized for 3 min at room temperature with glycerol buffer containing 0.05% Triton X-100 and then washed with transcription buffer (50 mM Tris-HCl [pH 7.4], 100 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM PMSF, 25% glycerol, 5 U of RNasin/ml). Transcription was performed for 5 min at room temperature with transcription buffer containing 0.1 mM each ATP, CTP, GTP, and UTP-BrUTP (2:1). The reaction was stopped by washing with ice-cold PBS and fixing with 4% paraformaldehyde for 20 min at room temperature. The cells were washed with PBS three times and permeabilized for 2 min on ice with a solution containing 0.1% Triton X-100 and 0.1% sodium citrate. After incubation with a blocking solution (5% goat serum and 5% bovine serum albumin in PBS) for 1 h at 37°C, the cells were incubated with anti-HSF-1 antibody for 1 h at 37°C and then washed three times with PBST. They were then incubated with anti-bromodeoxyuridine (BrdU)-fluorescein antibody (2 μg/ml; Boehringer Mannheim), which also recognizes BrU, and Texas red-conjugated secondary antibody for 1 h at 37°C and washed three times with PBST. Fluorescent signals were examined by laser confocal microscopy (Molecular Dynamics, Sunnyvale, Calif.).
Immunoprecipitation and immune complex kinase assays.
To assess protein kinase activity, the cells were treated as described in Results and lysed in buffer containing 50 mM sodium β-glycerophosphate (pH 7.2), 10 mM MgCl2, 5 mM EGTA, 1 mM EDTA, 10 mM KH2PO4, 1 mM sodium vanadate, and 0.2 mM PMSF (30, 43). To immunoprecipitate and measure GSK-3β activity, lysis buffer containing 10 mM Tris-HCl (pH 7.4), 50 mM NaCl, 2 mM EDTA, 1 mM EGTA, 1% Triton X-100, 1 mM benzamidine, 20 mM sodium pyrophosphate, and 1 mM sodium vanadate was used (73). The lysates were microcentrifuged for 10 min at 4°C, and equal amounts of protein (200 to 300 μg) from each sample were added to 1 μg of the appropriate antibody. After a 1-h incubation at 4°C, 25 μl of a 50% solution of protein A (or G)-Sepharose beads was added and the mixture was incubated at 4°C for an additional 1 h. It was then washed four times with lysis buffer and once with kinase buffer (20 mM β-glycerophosphate [pH 7.3], 5 mM MgCl2, 1 mM EGTA, 10% glycerol, 1 mM dithiothreitol (DTT), 1 mM sodium vanadate, 0.2 mM PMSF). The protein A (or G)-antibody-antigen complex was then incubated for 20 min at 37°C in 10 μl of kinase buffer with 25 μM unlabeled ATP, 20 μCi of [γ-32P]ATP, and appropriate substrates. When HSF-1 peptide containing amino acids 298 to 310 (RKEEPPSPPQSPRV) (15 μg) was used as the substrate, the reactions were stopped by spotting 5 μl of the reaction mixture onto P81 chromatography paper (Whatman) and washing it five times with 1% (wt/vol) phosphoric acid. The filters were dried and quantitated by scintillation counting.
The antibodies used for immunoprecipitation were JNK (C17), ERKs (C16 and C14) (Santa Cruz Biotechnology, Santa Cruz, Calif.), and GSK-3β (Transduction Laboratory, Lexington, Ky.).
Electrophoretic mobility shift assays.
Electrophoretic mobility shift analysis with whole-cell extracts has been described in detail previously (41, 43, 74). Briefly, after each treatment, cells were rinsed with PBS and lysed in 100 μl of extraction buffer (10 mM HEPES [pH 7.9], 0.4 mM NaCl, 0.1 mM EDTA, 0.5 mM DTT, 5% glycerol, 0.5 mM PMSF). The protein concentration of samples was estimated by the bicinchoninic acid method. Equal amounts of protein (15 μg) in extraction buffer (volume not exceeding 15 μl) were added to the reaction mixture, which contained 4 μl of binding buffer (37.5 mM NaCl, 15 mM Tris-HCl [pH 7.4], 0.1 mM EDTA, 0.5 mM DTT, 5% glycerol), 10 μg of yeast tRNA, 1 μg of sheared Escherichia coli DNA, 10 μg of poly(dI-dC), and 1 ng of 32P-labeled HSE oligonucleotide. The mixture was incubated for 15 min at 25°C and resolved on a 4.5% nondenaturing polyacrylamide gel. After electrophoresis, the gels were fixed in 7% (vol/vol) acetic acid for 5 min, rinsed once in distilled water, dried under vacuum, and exposed to X-ray film. The nucleotide sequence used for HSE was 5′-GTCGACGGATCCGAGCGCCTCGAATGTTCTAGAAAAGG-3′ (74). The double-stranded oligonucleotide was labeled with the Klenow fragment of DNA polymerase I, deoxynucleotide triphosphates, and [α-32P]dCTP.
RESULTS
HSF-1 punctate granules are sites of transcription.
In most human cell lines, HSF-1 appears as punctate granules throughout the nucleus upon heat shock (11, 29, 41, 55). The appearance and distribution of HSF-1 foci are similar when different fixatives (paraformaldehyde, methanol, or glutaraldehyde), or different antibodies to HSF-1 (11, 29, 41, 55) are used, indicating that HSF-1 granule appearance after heat shock is not an artifact of sample preparation.
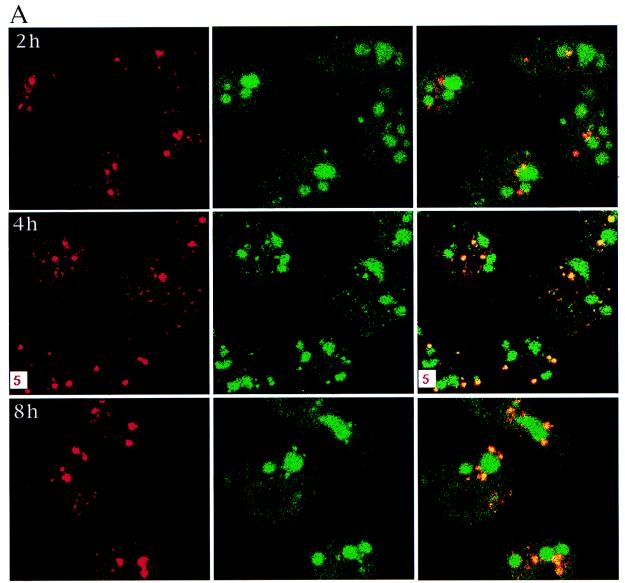
Since it has been shown that the kinetics of HSF-1 granule formation correlate with its transcriptional activity after heat shock (11), we examined whether the punctate granules are sites of transcription by performing an in situ run-on transcription assay. HeLa cells were heated at 45°C for 30 min, which results in 60% cell survival as measured by colony formation analysis and was found to result in the best representative HSF-1 granule appearance (data not shown). The cells were then allowed to recover at 37°C for up to 8 h. Run-on transcription was performed in the presence of BrUTP. The cells were then fixed and incubated with anti-HSF-1 and then with Texas red-conjugated secondary antibody to detect HSF-1 and with fluorescein-conjugated anti-BrdU (which also recognizes incorporated BrU) to detect newly synthesized pre-mRNAs, as described in Materials and Methods. The cells were analyzed by confocal microscopy. Most of the cells with a recovery time of 30 min to 1 h after heat shock showed HSF-1 granules smaller than 0.5 μm, and colocalization of the granules with nascent transcripts was difficult to score due to the small size of the granules (data not shown). The data for longer incubation times (2 to 8 h) are shown in Fig. 1A. The left panels show HSF-1 punctate granules; the middle panels show the detection of newly synthesized pre-mRNAs; and the right panels show the images in the left and middle panels superimposed. At 2 h after heat shock, some granules began to show colocalization with newly synthesized pre-mRNAs (indicated as yellow spots [top right]). At 4 h after heat shock, all the granules showed colocalization (middle right). At 8 h after heat shock, all the granules showed colocalization, with some granules beginning to show reddish edges (bottom right). These reddish edges could suggest a lower level of transcription compared to that at the 4-h time point. Figure 1B (higher magnification than Fig. 1A) further demonstrates the colocalization of HSF-1 punctate granules with nascent transcripts 4 and 8 h after heat shock (left). Vertical sections through a single cell showing the colocalization as well as accumulated nascent transcripts are shown on the right. Taken together, the above results indicate that transcription is occurring in the HSF-1 punctate granules that are characteristic in cells subjected to heat shock.
FIG. 1.
HSF-1 punctate granules are sites of transcription. Combined in situ run-on transcription and immunofluorescence were performed as described in Materials and Methods. Cells were heated at 45°C for 30 min and allowed to recover at 37°C for 2, 4, or 8 h. Run-on transcription involving BrUTP incorporation was performed. The cells were then fixed and stained and examined by confocal microscopy. (A) HSF-1 was detected with anti-HSF-1 antibody and Texas red-conjugated secondary antibody (showing as granular structures in the left panels). Nascent transcripts containing BrU were detected with FITC-conjugated anti-BrdU antibody (showing as patches of green fluorescence in the middle panels). The right panels show left and middle panels superimposed. Image areas of yellow staining indicate colocalization of HSF-1 and newly synthesized pre-mRNAs. The size (in micrometers) is indicated in the corner of the panels. (B) Cells were treated as for panel A but are shown at higher magnifications. The left panels show cells with a 4-h (top) or 8-h (bottom) recovery time. The right panels are vertical sections through a single cell at the same time points as in the left panels, showing colocalization as well as accumulated nascent transcripts.
Rapid inactivation of HSF-1 by GSK-3β after heat stress.
Because GSK-3β has been shown to phosphorylate HSF-1 in vitro (9), we examined the role of GSK-3β in regulating HSF-1 activity in vivo. HeLa cells were transiently transfected with expression constructs containing HA-tagged GSK-3β cDNA. The cells were untreated or heated at 45°C for 30 min and analyzed by indirect immunofluorescence after various recovery times at 37°C. The results show that before heat shock, HSF-1 was distributed in both the cytoplasm and nucleus (Fig. 2A, control, right). Immediately after heat shock, HSF-1 was translocated into the nuclei of all cells and appeared as 8 to 10 intensely stained foci per nucleus (right, 0 h). These granules became larger and more distinct with longer recovery times (right, 4 and 8 h) in untransfected cells whereas HSF-1 appeared as diffuse staining throughout the nucleus, indicating signs of recovery, in cells transfected with GSK-3β. Quantitation of the immunofluorescence analysis indicates that 44% ± 9% of the GSK-3β-transfected cells showed diffuse HSF-1 staining within 2 h of heating and that the number increased to 75% ± 7% by 4 h after heating whereas in untransfected cells a recovery time of 10 h was required for about the same percentage of cells (73% ± 4%) to show the similar HSF-1 staining pattern (Fig. 2B). These results indicate that overexpression of GSK-3β causes rapid dispersion of HSF-1 granules after heat shock.
FIG. 2.
Indirect immunofluorescence analysis of HSF-1 in HeLa cells transiently transfected with GSK-3β. (A) Representative immunofluorescence photographs (magnification, ×1,000) of cells transfected with GSK-3β. HeLa cells were transiently transfected with 5 μg of HA–GSK-3β. At 48 h after transfection, the cells were untreated (control) or heat shocked at 45°C for 30 min and allowed to recover at 37°C for 0, 4, 8, or 24 h. Overexpression of GSK-3β was detected with mouse monoclonal primary antibody to HA and FITC-conjugated secondary antibody. The endogenous HSF-1 was detected with rabbit polyclonal antibody to HSF-1 and Texas red-conjugated secondary antibody. Arrows in the left panels indicate the cells with overexpressed GSK-3β, and arrows in the right panels indicate the same cells stained for HSF-1. (B) Quantitation of the effect of overexpression of GSK-3β on the HSF-1 staining pattern. The bars indicate the percentage of cells showing HSF-1 recovery from punctate granules (i.e., cells with a diffuse staining pattern). More than 100 untransfected or GSK-3β-transfected cells were counted for each time point. (C) Gel mobility shift analysis. HeLa cells were untreated (lane C) or heated at 45°C for 30 min and then incubated at 37°C for 0, 2, 6, 8, 10, or 24 h and analyzed by gel mobility shift assays as described in Materials and Methods. (D) Quantitation of the data from panel C by PhosphorImager analysis.
To establish a correlation between the appearance of the punctate granules and DNA binding activity of HSF-1, we used gel mobility shift assays. The time course of DNA binding activity was closely correlated with the appearance and disappearance of the HSF-1 punctate granules observed in the nuclei of heated cells (Fig. 2C and D).
We next wanted to see if the nuclear distribution pattern of the HSF-1 in cells overexpressing GSK-3β could also be observed in cells overexpressing other serine/threonine protein kinases. HeLa cells were transiently transfected with constructs containing cDNA for HA-ERK1, for HA-MNK1, which is located downstream of ERKs in the MAPK signaling pathway (19), or for Flag epitope-tagged p38, which is a stress-activated protein kinase and has been shown to phosphorylate ATF2 transcription factor (22). At 48 h after transfection, the cells were heated at 45°C for 30 min, and after a recovery time of 4 h at 37°C, the HSF-1 staining pattern was analyzed (Fig. 3A). The percentage of cells showing diffuse HSF-1 staining in HA-MNK1- or Flag-p38-transfected cells was about the same as in untransfected (control) cells (Fig. 3B). A similar result was seen when cells were transfected with empty vector (data not shown). Although ERK1 has been shown to be the priming kinase for GSK-3β (9) (see below), transient expression of constructs containing HA-ERK1 cDNA increased the percentage of recovered cells after heat shock to 21% ± 8% (Fig. 3B). However, this number, increased to 46% ± 6% when the cells were cotransfected with c-Ha-Ras, since constitutive active Ras increases the level of activated ERK and in turn increases the effect that ERK exerts on its substrates (43).
FIG. 3.
Indirect immunofluorescence analysis of HSF-1 in HeLa cells transiently transfected with HA-ERK, HA-MNK, or Flag-tagged P38. (A) HeLa cells were transiently transfected with 5 μg of HA-ERK1, HA-MNK1, or Flag-tagged P38 cDNA. At 48 h after transfection, the cells were heat shocked at 45°C for 30 min and allowed to recover at 37°C for 4 h. Overexpression of HA-ERK1 and HA-MNK1 was detected with mouse monoclonal primary antibody to HA. Overexpression of Flag-tagged P38 was detected with monoclonal antibody to the Flag epitope. FITC-conjugated secondary antibody was used to detect the HA and Flag epitope. The endogenous HSF-1 was detected with rabbit polyclonal antibody to HSF-1 and Texas red-conjugated secondary antibody. Arrows in the left panels indicate the cells with overexpressed protein kinases, and arrows in the right panels indicate the same cells stained for HSF-1. Magnification, ×1,000. (B) Quantitation of the effect of overexpression of various protein kinases on the HSF-1 staining pattern. Bars indicate the percentage of cells showing recovery from HSF-1 punctate granules. More than 100 cells of transfected or untransfected cells were counted for each time point.
We demonstrated above that HSF-1- containing granules are sites of active transcription, and overexpression of GSK-3β causes rapid disappearance of these granules, suggesting that GSK-3β overexpression may inactivate HSF-1 after heat shock. To test this, HSF-1 transcriptional activity was assayed in the following experiment. HeLa cells were cotransfected with expression constructs encoding GSK-3β, ERK1, or both GSK-3β and ERK1 along with c-Ha-Ras and hsp70-luciferase reporter constructs (which contain binding sites for HSF-1), as described in Materials and Methods and in the legend to Fig. 4. Cells transfected with hsp70-luciferase only, with no added GSK-3β or ERK1, showed a greater than 30-fold increase in transcriptional activity compared to unheated controls (Fig. 4). However, cells that were cotransfected with GSK-3β showed only a 10-fold increase in transcriptional activity compared to the appropriate unheated controls, indicating that the transcriptional activity of HSF-1 is reduced in cells overexpressing GSK-3β. This result demonstrates that GSK-3β overexpression facilitates HSF-1 inactivation after heat shock. Interestingly, overexpression of ERK1 is as effective as overexpression of GSK-3β in reducing the transcriptional activity of HSF-1 but not as efficient in causing the disappearance of HSF-1 granules, as shown in Fig. 3. This could suggest that ERK1 phosphorylation of HSF-1 in vivo may halt transcription but that this phosphorylation may not be sufficient to disperse HSF-1 from the sites of transcription and thereby cause the disappearance of HSF-1 granules.
FIG. 4.
Overexpression of GSK-3β leads to reduced HSF-1 transcriptional activity. HeLa cells were transiently transfected with constructs encoding c-Ha-Ras and hsp70-luciferase (70-luc) or cotransfected with HA–GSK-3β (+GSK-3 beta), HA-ERK (+ERK), or HA–GSK-3β and HA-ERK together (+GSK-3 beta + ERK). At 24 h after transfection, the cells were serum starved with 0.5% FCS for 24 h. They remained untreated or were heated at 45°C for 30 min and allowed to recover at 37°C for 6 h for accumulation of hsp70-luciferase. The luciferase activity was determined. Data are presented as the fold increase over the unheated control group for each treatment.
Priming by ERK1 is required for GSK-3β inactivation of HSF-1.
High levels of ERK1 activity inhibit HSF-1 transcriptional activity in vivo (9, 43). Recent evidence also indicates that ERK1 can act as a priming kinase for GSK-3β phosphorylation of HSF-1 in vitro (9). To determine if ERK1 can be a priming kinase for GSK-3β in vivo, HeLa cells were transiently transfected with expression constructs encoding HA–GSK-3β. At 48 h after transfection, the cells were pretreated with PD98059, a specific MEK inhibitor which binds to MEK at a site that blocks access to activating enzymes and does not inhibit the activities of other protein kinases such as GSK-3β (2, 13). The cells were then heated at 45°C for 30 min and allowed to recover at 37°C for 4 h. GSK-3β and HSF-1 were detected in cells by indirect immunofluorescence analysis. The results show that after pretreatment with PD98059, HSF-1 in cells overexpressing GSK-3β remained as punctate granules (Fig. 5A), in contrast to cells overexpressing GSK-3β alone (Fig. 2). This result strongly suggests that the activity of ERK1 is required for GSK-3β inactivation of HSF-1.
FIG. 5.
ERK1 is a priming kinase for GSK-3β. (A) Indirect immunofluorescence analysis of cells transiently transfected with GSK-3β and pretreated with PD98059 (magnification, ×1,000). The cells were transiently transfected with HA–GSK-3β cDNA. At 48 h after transfection, they were treated for 30 min with 30 μM PD98059, rinsed with PBS, heated at 45°C for 30 min, and allowed to recover at 37°C for 4 h. Overexpressed GSK-3β and the endogenous HSF-1 were detected as described in the text. Arrows in the left panel indicate the cells with overexpressed GSK-3β, and arrows in the right panel indicate the same cells stained for HSF-1. (B) Immune complex kinase assays. ERK1 or JNK1 was immunoprecipitated from HeLa cells which had been heated at 45°C for 30 min. The immunoprecipitated kinases were individually incubated with HSF-1 peptide for 20 min at 30°C in the presence of 25 μM unlabelled ATP. HSF-1 peptide prephosphorylated by ERK1 or JNK or a nonphosphorylated control was then used as the substrate in kinase reactions with purified GSK-3β (100 mU) and [γ-32P]ATP. The reaction mixtures were incubated for 20 min at 30°C, and the products were spotted onto P81 chromatography paper and rinsed five times with 1% phosphoric acid. Radioactivity was determined with a scintillation counter. Experiments were performed in triplicate, and the data are presented as the fold increase in 32P incorporation into HSF-1 peptide prephosphorylated with ERK1 or JNK1 over that in groups phosphorylated with GSK-3β only.
The prerequisite for prior phosphorylation of a substrate by a protein kinase can be efficiently examined by using peptide substrates (47). An HSF-1 peptide containing amino acids 298 to 310 was synthesized to contain serines 303 and 307. ERK1 or JNK1 was immunoprecipitated from heated HeLa cell lysates and used in immune complex kinase reactions to prephosphorylate HSF-1 peptide. These prephosphorylated HSF-1 peptides were subsequently incubated with purified GSK-3β and [γ-32P]ATP, and incorporation of 32P was determined. The results show an increase in 32P incorporation into HSF-1 peptide prephosphorylated with ERK1 but no increase in 32P incorporation into HSF-1 peptide prephosphorylated with JNK1 (Fig. 5B). This indicates that ERK1 can act as a priming kinase for GSK-3β whereas JNK1 cannot. Together with the results presented in Fig. 5A, these data demonstrate that the activity of ERK1 is essential for HSF-1 recovery in cells overexpressing GSK-3β.
Signaling pathways that modulate GSK-3β activity following heat shock.
The activity of GSK-3β is down regulated by 40 to 50% in cells subjected to a variety of stimuli following phosphorylation on serine 9 (13, 47, 49, 63), thereby relieving the repression that GSK-3β exerts on its substrates. Three serine/threonine protein kinases have been shown to phosphorylate and down regulate GSK-3β activity: p70S6K, p90rsk, and PKB/Akt (13, 16). The activity of GSK-3β can also be up regulated following phosphorylation by a tyrosine kinase whose identity is unknown.
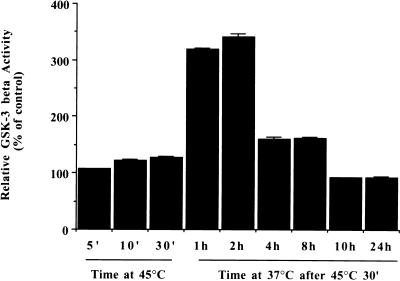
To examine whether the activity of GSK-3β is altered following heat shock, HeLa cells were either heated at 45°C for 5, 10, or 30 min or heated at 45°C for 30 min and allowed to recover at 37°C for up to 24 h. Immune complex kinase assays were performed as described above, with prephosphorylated HSF-1 peptide as the substrate. Figure 6 shows that the activity of GSK-3β was slightly increased over the control level when the cells were heated for 5 or 10 min without a recovery time. There was a 25% increase in GSK-3β activity when the cells were heated at 45°C for 30 min. A dramatic increase in GSK-3β activity, up to 350% of the control level, was seen when the heated cells were allowed to recover at 37°C for 1 to 2 h. The activity of GSK-3β remained elevated for as long as 8 h and returned to control levels by 10 h after heat treatment.
FIG. 6.
Activity of GSK-3β after heat shock. HeLa cells were heated at 45°C for 5, 10, or 30 min or heated at 45°C for 30 min and allowed to recover at 37°C for 1, 2, 4, 8, 10, or 24 h. They were then lysed, and equal amounts of protein were immunoprecipitated with antibody to GSK-3β. Immunoprecipitates were used to phosphorylate 20 μg of HSF-1 peptide prephosphorylated with immunoprecipitated ERK1 as described in Materials and Methods. Phosphorylated HSF-1 peptide was spotted onto P81 chromatography paper and analyzed as described in the legend to Fig. 5. The GSK-3β activity is expressed as percent activity relative to unheated controls.
Although the activities of p70S6K, p90rsk, and PKB/Akt are all induced by heat shock (references 33 and 40 and data not shown), none of these enzymes appeared to phosphorylate and decrease GSK-3β activity during heat shock. Furthermore, blocking p70S6K with rapamycin or blocking phosphoinositide 3-kinase-induced protein kinase B (PKB) activation with wortmannin had no effect on the nuclear appearance of HSF-1 when analyzed by indirect immunofluorescence, on HSF-1 DNA binding activity, or on HSF-1 activation of transcription following heat shock (data not shown).
DISCUSSION
HSF-1 is activated by a variety of environmental stimuli, including heat shock, heavy metals, amino acid analogues, ethanol, hypoxia, and ischemia (39, 45). Many of the signaling pathways that mediate HSF-1 activation are not yet understood. HSF-1 is the target of proline-directed protein kinases such as ERKs in vivo (9, 30, 43, 71) and P38/HOG1 and GSK-3β in vitro (9, 32); phosphorylation of HSF-1 by some of these kinases represses its activity (9, 43). However, the fate of HSF-1 after such phosphorylation is unclear; inactivation may cause HSF-1 to exit the nucleus, as has recently been suggested for NF-AT transcription factors, which are substrates of the proline-directed protein kinases GSK-3β and JNK (5, 8).
In the present study, we have obtained evidence that strongly suggests that GSK-3β and ERK down regulate HSF-1 activity after heat shock in vivo. Overexpression of GSK-3β facilitates the disappearance of HSF-1 punctate granules, which we have shown here to be associated with transcriptional activity; in addition, ERK activity in the cell is essential for this GSK-3β inactivation of HSF-1. Although the approach taken was to overexpress the protein kinase(s) from transfected genes, which could create nonphysiological conditions, such overexpression can be a useful tool in examining the potential role of kinases in vivo. Our data showing that overexpression of GSK-3β and, to a lesser extent, ERK is sufficient to inactivate HSF-1 after heat shock could suggest that elevated levels of GSK-3β and ERK in the cell may play a role in the regulation of HSF-1 activity under normal physiological conditions. This could occur during normal growth, to keep HSF-1 in an inactive form, or during recovery from heat shock, to inactivate HSF-1 so that the cells can resume normal growth. However, it is also possible that under normal physiological conditions and depending on the level or type of stress, phosphorylation of HSF-1 by other protein kinases, such as protein kinase C, JNK, and casein kinase II, and interaction with some regulatory proteins, such as hsp-70 and Hdj 1, will be required to fully inactivate HSF-1 during recovery from heat shock (24, 59, 60, 71).
Overexpression of GSK-3β does not cause HSF-1 to exit the nucleus entirely, a phenomenon which has been described to occur for NF-AT transcription factors (5, 8). It should be noted, however, that during unstimulated growth conditions, NF-AT transcription factors are located primarily in the cytoplasm. In contrast, HSF-1 is normally found in both the cytoplasm and the nucleus (44, 55).
Our overexpression studies did not directly address the issue of which phosphorylation sites are involved in the inactivation of HSF-1 in vivo by GSK-3β and ERK. However, there is ample in vitro evidence from phosphopeptide mapping as well as mutational analysis indicating that GSK-3β and ERK can phosphorylate HSF-1 on serine 303 and serine 307, respectively (9, 32), and that constitutive phosphorylation of serine 307, and possibly serine 303, plays an important role in the negative regulation of HSF-1 transcriptional activity (9, 31, 32). However, a recent study by Xia et al. (71) provided additional results about the phosphorylation state of serine 303. Their data indicate that HSF-1 is phosphorylated only on serine 307 and not effectively on serine 303 and that mutation of serine 307, but not serine 303, deregulates HSF-1 activity. The discrepancies between different studies may be due to the different chimeric constructs used for phosphopeptide mapping or the different culture conditions used, which may affect the activities of the protein kinases that phosphorylate HSF-1. An alternative explanation may be that phosphorylation of serine 303 is transient, i.e., that it may occur only during recovery from heat shock, while some peptide-mapping experiments were performed immediately after heat shock. More studies are needed to clarify the phosphorylation states of HSF-1 in vivo.
It appears that the activity of HSF-1 can be down regulated by protein kinases which are activated by diverse signal transduction pathways. The ERK MAPK pathway is activated during cell growth and development by multiple signaling pathways (14) that are in turn activated by growth factor receptors, G protein-coupled receptors, ceramide production, and a protein kinase C-dependent pathway (6, 12, 14, 23, 26, 70). Recent evidence from our laboratory suggests that ERK MAPK activation by heat shock may be through ceramide activation of protein kinase Raf-1 (67). The pathways leading to GSK-3β regulation are complex. GSK-3β activity is down regulated by PKB/Akt, p70S6K, or p90rsk as a result of phosphorylation on serine residues (3, 49, 53, 63, 64). Activation of PKB/Akt leads to increased cell survival, as is the case with activation of ERK. The ability of ERK to mediate cell survival is dependent on the activation of transcription factors such as Elk1 and repair of damaged proteins. The ability of PKB/Akt to mediate cell survival is likely to be dependent on downstream effectors such as p70S6K and protein translation, activation of FRAP/TOR, and inhibition of GSK-3β (50). Interestingly, heat shock stimulates the activity of GSK-3β and ERK MAPK. The increase in GSK-3β activity may occur through its phosphorylation on a tyrosine residue by an unknown tyrosine kinase. Thus, it appears that when activated, HSF-1 reduces the expression of most other genes and must be inactivated in a timely manner for cell proliferation to continue. The cell has developed an elegant mechanism for doing this, since some of the enzymes that control cell proliferation are capable of inactivating HSF-1.
ACKNOWLEDGMENTS
We thank Rhea-Beth Markowitz and Demetrius Moskophidis for their critical reading of the manuscript and the Imaging Core Facility at the Institute of Molecular Medicine and Genetics at the Medical College of Georgia for their continuous support and expertise throughout this study.
This work was supported by NIH grant CA62130 from the National Cancer Institute.
REFERENCES
- 1.Abravaya K, Philips B, Morimoto R I. Attenuation of the heat shock response in HeLa cells is mediated by the release of bound heat shock transcription factor and is modulated by changes in growth and in heat shock temperature. Genes Dev. 1991;5:2117–2127. doi: 10.1101/gad.5.11.2117. [DOI] [PubMed] [Google Scholar]
- 2.Alessi D R, Cuenda A, Cohen P, Dudley D T, Saltiel A R. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 3.Azpiazu I, Saltiel A R, Depaoli-Roach A A, Lawrence J C. Regulation of both glycogen synthase and PHAS-I by insulin in rat skeletal muscle involves mitogen activated protein kinase independent and rapamycin sensitive pathways. J Biol Chem. 1996;271:5033–5039. doi: 10.1074/jbc.271.9.5033. [DOI] [PubMed] [Google Scholar]
- 4.Baler R, Dahl G, Voellmy R. Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor HSF-1. Mol Cell Biol. 1993;13:2486–2496. doi: 10.1128/mcb.13.4.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beals C R, Sheridan C M, Turck C W, Gardner P, Crabtree G R. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Nature. 1997;275:1930–1933. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 6.Berra E, Diaz-Meco M T, Lozano J, Frutos S, Municio M M, Sanchez P, Sanz L, Moscat J. Evidence for a role of MEK and MAPK during signal transduction by protein kinase C zeta. EMBO J. 1995;14:6157–6163. doi: 10.1002/j.1460-2075.1995.tb00306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulton T G, Nye S H, Robbins D J, Ip N Y, Radziejewska E, Morgenbesser S D, Depinho R A, Panayotatos N, Cobb M H, Yancopoulos G D. ERKs: a family of protein-serine threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell. 1991;65:663–675. doi: 10.1016/0092-8674(91)90098-j. [DOI] [PubMed] [Google Scholar]
- 8.Chow C-W, Rincon M, Cavanagh J, Dickens M, Davis R J. Nuclear accumulation of NFAT4 opposed by the JNK signal transduction pathway. Science. 1997;278:1638–1641. doi: 10.1126/science.278.5343.1638. [DOI] [PubMed] [Google Scholar]
- 9.Chu B, Soncin F, Price B D, Stevenson M A, Calderwood S K. Sequential phosphorylation by mitogen activated protein kinase and glycogen synthase kinase-3 represses transcriptional activation by heat shock factor-1. J Biol Chem. 1996;271:30847–30857. doi: 10.1074/jbc.271.48.30847. [DOI] [PubMed] [Google Scholar]
- 10.Cotto J J, Kline M, Morimoto R I. Activation of heat shock factor 1 DNA binding precedes stress-induced serine phosphorylation. J Biol Chem. 1996;271:3355–3358. doi: 10.1074/jbc.271.7.3355. [DOI] [PubMed] [Google Scholar]
- 11.Cotto J J, Fox S G, Morimoto R I. HSF-1 granules: a novel stress-induced nuclear compartment of human cells. J Cell Sci. 1997;110:2925–2934. doi: 10.1242/jcs.110.23.2925. [DOI] [PubMed] [Google Scholar]
- 12.Crespo P, Xu N, Simonds W F, Gutkind J S. Ras-dependent activation of MAP kinase pathway mediated by G protein beta gamma subunits. Nature. 1994;369:418–420. doi: 10.1038/369418a0. [DOI] [PubMed] [Google Scholar]
- 13.Cross D A E, Alessi D R, Cohen P, Andjelkovich M, Hemming B A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 14.Davis R J. The mitogen activated protein kinase signal transduction pathway. J Biol Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- 15.Derijard B, Hibi M, Wu I-H, Barret T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 16.Eldar-Finkelman H, Seger R, Vandenheede J R, Krebs E G. Inactivation of glycogen synthase-kinase-3 by epidermal growth factor is mediated by mitogen activated protein kinase /p90 ribosomal protein S6 kinase signaling pathway in NIH3T3 cells. J Biol Chem. 1995;270:987–990. doi: 10.1074/jbc.270.3.987. [DOI] [PubMed] [Google Scholar]
- 17.Fiol C J, Mahrenholz A M, Wang Y, Roeske R W, Roach P J. Formation of protein kinase recognition sites by covalent modification of the substrate: molecular mechanism for the synergistic action of casein kinase II and glycogen synthase kinase 3. J Biol Chem. 1987;262:14042–14048. [PubMed] [Google Scholar]
- 18.Fiol C J, Williams J S, Cou C-H, Wang Q M, Roach P J, Andrisani O M. A secondary phosphorylation of CREB 341 at Serine 129 is required for the cAMP mediated control of gene expression. J Biol Chem. 1994;269:32187–32193. [PubMed] [Google Scholar]
- 19.Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta S, Cambell D, Derijard B, Davis R J. Transcription factor ATF2 regulation by the JNK signal transduction. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 21.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 22.Han J, Lee J D, Bibbs L, Ulevitch R J. A MAP kinase targeted by endotoxins and hyperosmolarity in mammalian cells. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 23.Hawes B E, van Biesen T, Koch W J, Luttrell L M, Lefkowitz R J. Distinct pathways of Gi- and Gq-mediated mitogen-activated protein kinase activation. J Biol Chem. 1995;270:17148–17153. doi: 10.1074/jbc.270.29.17148. [DOI] [PubMed] [Google Scholar]
- 24.He B, Meng Y H, Mivechi N F. GSK, ERK MAPK and JNK cooperate to inactivate the heat shock factor 1. 1998. Molecular chaperones and the heat shock response. Unpublished data. [Google Scholar]
- 25.Hemmings B A, Resink T J, Cohen P. Reconstitution of a Mg-ATP dependent protein phosphatase and its activation through a phosphorylation mechanism. FEBS Lett. 1982;150:319–324. doi: 10.1016/0014-5793(82)80760-6. [DOI] [PubMed] [Google Scholar]
- 26.Hii C S, Ferrante A, Edwards Y S, Huang Z H, Hartfield P J, Rathjen D A, Poulos A, Murray A W. Activation of mitogen-activated protein kinase by arachidonic acid in rat liver epithelial WB cells by a protein kinase C-dependent mechanism. J Biol Chem. 1995;270:4201–4204. doi: 10.1074/jbc.270.9.4201. [DOI] [PubMed] [Google Scholar]
- 27.Hunter T, Karin M. The regulation of transcription factor by phosphorylation. Cell. 1993;50:823–829. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- 28.Jiang Y, Chen C, Li Z, Guo W, Gegner J A, Lin S, Han J. Characterization of the structure and function of a new mitogen-activated protein kinase (p38 delta) J Biol Chem. 1996;271:17920–17926. doi: 10.1074/jbc.271.30.17920. [DOI] [PubMed] [Google Scholar]
- 29.Jolly C, Mongelard F, Robert-Nicoud M, Vourch C. Optimization of nuclear transcript detection by FISH and combination with fluorescence immunocytochemical detection of transcription factors. J Histochem Cytochem. 1997;45:1585–1592. doi: 10.1177/002215549704501201. [DOI] [PubMed] [Google Scholar]
- 30.Kim J, Nueda A, Meng Y-H, Dynan W S, Mivechi N F. Analysis of the phosphorylation of human heat shock transcription factor-1 by MAP kinase family members. J Cell Biochem. 1997;67:43–54. doi: 10.1002/(sici)1097-4644(19971001)67:1<43::aid-jcb5>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 31.Kline M P, Morimoto R I. Repression of the heat shock factor 1 transcriptional activation domain is modulated by constitutive phosphorylation. Mol Cell Biol. 1997;17:2107–2115. doi: 10.1128/mcb.17.4.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knauf U, Newton E M, Kyriakis J, Kingston R E. Repression of human heat shock factor 1 activity at control temperature by phosphorylation. Genes Dev. 1996;10:2782–2793. doi: 10.1101/gad.10.21.2782. [DOI] [PubMed] [Google Scholar]
- 33.Konishi H, Matsuzaki H, Tanaka M, Ono Y, Tokunaga C, Kuroda S, Kikkawa U. Activation of Rac-protein kinase by heat shock and hyperosmolarity stress through a pathway independent of phosphatidyl inositol 3-kinase. Proc Natl Acad Sci USA. 1996;93:7639–7643. doi: 10.1073/pnas.93.15.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kyriakis J M, Avruch J. Sounding the alarm: protein kinase cascades activated by stress and inflammation. J Biol Chem. 1996;271:24313–24316. doi: 10.1074/jbc.271.40.24313. [DOI] [PubMed] [Google Scholar]
- 35.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmed M F, Avruch J, Woodgett J R. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 36.Larson J S, Schuetz T J, Kingston R E. Activation in vitro of sequence-specific DNA binding by a human regulatory factor. Nature. 1988;335:372–375. doi: 10.1038/335372a0. [DOI] [PubMed] [Google Scholar]
- 37.Lee J C, Laydon J T, McDonnell P C, Gallagher T F, Kumar S, Green D, McNulty D, Blumenthal M J, Heys J R, Landvatter S W, et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 38.Lim Tung H Y, Reed L J. Purification and characterization of protein phosphatase 1 activating kinase from bovine brain cytosolic and particulate fractions. J Biol Chem. 1989;264:2985–2990. [PubMed] [Google Scholar]
- 39.Lindquist S. The heat shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 40.Matsuzaki H, Konishi H, Tanaka M, Ono Y, Takenawa T, Watanabe Y, Qzaki S, Kuroda S, Kikkawa U. Isolation of the active form of Rac-protein kinase (PKB/Akt) from transfected Cos-7 cells treated with heat shock stress and effects of phosphatidylinositol-3,4,5-triphosphate and phosphatidyl inositol-4,5-biphosphate on its enzyme activity. FEBS Lett. 1996;396:305–308. doi: 10.1016/0014-5793(96)01120-9. [DOI] [PubMed] [Google Scholar]
- 41.Mivechi N F, Murai T, Hahn G M. Inhibitors of tyrosine and serine/threonine phosphatases regulate the heat shock response. J Cell Biochem. 1994;54:186–197. doi: 10.1002/jcb.240540207. [DOI] [PubMed] [Google Scholar]
- 42.Mivechi N F, Ouyang H, Hahn G M. Lower heat shock factor activation and faster rate of HSP-70A mRNA turnover in heat sensitive human leukemias. Cancer Res. 1992;52:6815–6822. [PubMed] [Google Scholar]
- 43.Mivechi N F, Giaccia A J. Mitogen-activated protein kinase acts as a negative regulator of the heat shock response. Cancer Res. 1995;55:5512–5519. [PubMed] [Google Scholar]
- 44.Mivechi N F, Shi X-Y, Hahn G M. Stable overexpression of human HSF-1 in murine cells suggests activation rather than expression of HSF-1 to be the key regulatory step in the heat shock gene expression. J Cell Biochem. 1995;59:266–280. doi: 10.1002/jcb.240590215. [DOI] [PubMed] [Google Scholar]
- 45.Morimoto R I. Cells in stress: transcriptional activation of heat shock genes. Science. 1993;259:1409–1410. doi: 10.1126/science.8451637. [DOI] [PubMed] [Google Scholar]
- 46.Newton E M, Knauf U, Green M, Kingston R E. The regulatory domain of human heat shock factor 1 is sufficient to sense stress. Mol Cell Biol. 1996;16:839–846. doi: 10.1128/mcb.16.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plyte S E, Hughes K, Nikolakaki E, Pulverer B J, Woodgett J R. Glycogen synthase kinase-3: functions in oncogenesis and development. Biochem Biophys Acta. 1992;1114:147–162. doi: 10.1016/0304-419x(92)90012-n. [DOI] [PubMed] [Google Scholar]
- 48.Price M A, Cruzalegui F H, Treisman R. The p38 and ERK MAP kinase pathways cooperate to activate ternary complex factors and c-Fos transcription in response to UV light. EMBO J. 1996;15:6552–6563. [PMC free article] [PubMed] [Google Scholar]
- 49.Proud C G. Turned on by insulin. Nature. 1994;371:747–748. doi: 10.1038/371747a0. [DOI] [PubMed] [Google Scholar]
- 50.Pullen N, Thomas G. The modular phosphorylation and activation of p70s6k. FEBS Lett. 1997;410:78–82. doi: 10.1016/s0014-5793(97)00323-2. [DOI] [PubMed] [Google Scholar]
- 51.Rabindran S K, Raymond R I, Haroun I, Clos J, Wisniewski J, Wu C. Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science. 1993;259:230–234. doi: 10.1126/science.8421783. [DOI] [PubMed] [Google Scholar]
- 52.Rouse J, Cohen P, Trigon S, Morange M, Alonso-Llamazares A, Zamanillo D, Hunt T, Nebreda A R. A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAP kinase-2 and phosphorylation of the small heat shock proteins. Cell. 1994;78:1027–1037. doi: 10.1016/0092-8674(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 53.Saito Y, Vandenheede J R, Cohen P. The mechanism by which epidermal growth factor inhibits glycogen synthase kinase in A431 cells. Biochem J. 1994;303:27–31. doi: 10.1042/bj3030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saksela K, Makela T P, Hughs K, Woodgett J R, Alitalo K. Activation of protein kinase C increases phosphorylation of the L-myc trans-activator domain at a GSK-3 target site. Oncogene. 1992;7:347–353. [PubMed] [Google Scholar]
- 55.Sarge K D, Murphy S P, Morimoto R I. Activation of heat shock gene transcription by heat shock transcription factor 1 involves oligomerization, acquisition of DNA binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seger R K, Krebs E G. The MAPK signaling cascade. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- 57.Semmes O J, Jeang K T. Localization of human T-cell leukemia virus type 1 Tax to subcellular compartments that overlap with interchromatin speckles. J Virol. 1997;70:6347–6357. doi: 10.1128/jvi.70.9.6347-6357.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheldon L A, Kingston R E. Hydrophobic coiled-coil domains regulate the subcellular localization of human heat shock transcription factor 2. Genes Dev. 1993;7:1549–1558. doi: 10.1101/gad.7.8.1549. [DOI] [PubMed] [Google Scholar]
- 59.Shi Y, Mosser D D, Morimoto R I. Molecular chaperones as HSF-1 specific transcriptional repressors. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soncin F, Zhang X, Chu B, Zhong R D, Stevenson M A, Calderwood S K. HSF-1 regulation by phosphorylation. 1998. Molecular chaperones and heat shock response. Personal communication. [Google Scholar]
- 61.Sorger P K, Pelham H R. Purification and characterization of a heat shock element binding protein from yeast. EMBO J. 1987;6:3035–3041. doi: 10.1002/j.1460-2075.1987.tb02609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strahl T, Gille H, Shaw P E. Selective response of ternary complex factor Sap-1a to different mitogen-activated protein kinase subgroups. Proc Natl Acad Sci USA. 1996;93:11563–11568. doi: 10.1073/pnas.93.21.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sutherland C, Cohen P. The alpha isoform of glycogen synthase kinase-3 from rabbit skeletal muscle is inactivated by p70 S6 kinase or MAP kinase-activated protein kinase-1 in vitro. FEBS Lett. 1994;338:37–42. doi: 10.1016/0014-5793(94)80112-6. [DOI] [PubMed] [Google Scholar]
- 64.Sutherland C, Leighton I A, Cohen P. Inactivation of glycogen synthase-3B by phosphorylation: new kinase connections in insulin and growth factor signaling. Biochem J. 1993;296:15–19. doi: 10.1042/bj2960015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X-Z, Ron D. Stress-induced phosphorylation and activation of the transcription factor CHOP(GADD153) by p38 MAP kinase. Science. 1996;272:1347–1349. doi: 10.1126/science.272.5266.1347. [DOI] [PubMed] [Google Scholar]
- 66.Westwood J T, Wu C. Activation of Drosophila heat shock factor: conformational change associated with a monomer-to-trimer transition. Mol Cell Biol. 1993;13:3481–3486. doi: 10.1128/mcb.13.6.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woessmann, W., Y.-H. Meng, and N. F. Mivechi. An essential role for MAP kinases in preventing heat-induced cell death. Submitted for publication. [DOI] [PubMed]
- 68.Wu C. Activating protein factor binds in vitro to upstream control sequences in heat shock gene chromatin. Nature. 1984;311:81–84. doi: 10.1038/311081a0. [DOI] [PubMed] [Google Scholar]
- 69.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 70.Wu J, Spiegel S, Sturgill T W. Sphingosine-1-phosphate rapidly activates the mitogen activated protein kinase pathway by G protein dependent mechanism. J Biol Chem. 1995;270:11484–11488. doi: 10.1074/jbc.270.19.11484. [DOI] [PubMed] [Google Scholar]
- 71.Xia W, Guo Y, Vilaboa N, Zuo J, Voellmy R. Transcriptional activation of heat shock factor HSF-1 probed by phosphopeptide analysis of factor 32P-labeled in vivo. J Biol Chem. 1998;273:8749–8755. doi: 10.1074/jbc.273.15.8749. [DOI] [PubMed] [Google Scholar]
- 72.Yang D, Tournier C, Wysk M, Lu H-T, Xu J, Davis R J, Flavell R A. Targeted disruption of the MKK4 gene causes embryonic death, inhibition of c-Jun NH2-terminal kinase activation, defects in Ap1 transcriptional activity. Proc Natl Acad Sci USA. 1997;94:3004–3009. doi: 10.1073/pnas.94.7.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang S-D, Lee S-C, Chang H-C. Heat stress induces tyrosine phosphorylation/activation of kinase Fa/GSK-3a (a human carcinoma dedifferentiation modulator) in A431 cells. J Cell Biochem. 1997;65:16–26. [PubMed] [Google Scholar]
- 74.Zimarino V, Wu C. Induction of sequence-specific binding of Drosophila heat shock activator. Nature. 1987;327:727–730. doi: 10.1038/327727a0. [DOI] [PubMed] [Google Scholar]
- 75.Zohn I E, Yu H, Li X, Cox A D, Earp H S. Angiotensin II stimulates calcium dependent activation of c-Jun N-terminal kinase. J Biol Chem. 1995;15:6160–6168. doi: 10.1128/mcb.15.11.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]