Abstract
Plant crops serve as essential sources of nutritional sustenance, supplying vital nutrients to human diets. However, their productivity and quality are severely jeopardized by factors such as pests, diseases, and adverse abiotic conditions. Addressing these challenges using innovative biotechnological approaches is imperative for advancing sustainable agriculture. In recent years, genome editing technologies have emerged as pivotal genetic tools, revolutionizing plant molecular biology. Among these, the CRISPR–Cas9 system has gained prominence due to its unparalleled precision, streamlined design, and heightened success rates. This review article highlights the profound impact of CRISPR/Cas9 technology on crop improvement. The article critically examines the breakthroughs, ongoing enhancements, and future prospects associated with this cutting-edge technology. In conclusion, the utilization of CRISPR/Cas9 presents a transformative shift in agricultural biotechnology, holding the potential to mitigate longstanding agricultural challenges.
Keywords: CRISPR/Cas9, Crop improvement, Genome editing, ZFNs, TALENs, MegaN
Introduction
The worldwide populace persists in its rapid expansion [1], yet there remains an unsatisfactory increase in the accessibility of food resources [2]. Projections suggest that the world's population could reach 10 billion by the year 2050. This rapid population growth, combined with the adverse effects of climatic conditions, poses potential challenges to food security. This situation could be exacerbated by diminishing arable land and declining crop yields. Recent findings from the International Rice Research Institute (IRRI) reveal that approximately every 7.7 s, one hectare of fertile land is lost. The impact of this loss could be even more significant if the current acceleration in global temperatures persists [3, 4]. To ensure food security, there is a pressing need to nearly double crop yield capacities and develop cultivars that are highly resilient to various stresses, as emphasized by Jinek et al. [5]. Horticultural practices, including the cultivation of vegetables, fruits, spices, tubers, and medicinal plants, play a pivotal role in the economy. These crops contribute significantly to both food and nutritional security. With the expanding global population, it becomes imperative to enhance agricultural productivity to sustain a consistent food supply. The advancement of next-generation crops is of paramount importance, given that conventional breeding methods have been extensively employed but are time-consuming [6]. An alternative approach is transgenesis, though its adoption depends on public acceptance for commercialization. Recent progress in recombinant DNA technology involving nucleases like ZFNs, TALENs, and CRISPR/Cas9 has demonstrated its effectiveness in precisely modifying targeted genomic locations [7, 8]. This technique is currently extensively utilized in diverse agricultural crops, and its widespread adoption is eagerly anticipated in the coming years.
Genome Editing
Genome editing comprises a collection of sophisticated molecular techniques that enable the precise, efficient, and targeted modification of specific nucleotide sequences. Researchers utilize this technology to delve into the genome's capabilities more profoundly and to develop crops that exhibit resistance to pests, offer improved nutritional content, and can be grown in arid environments [9]. The application of genome editing methods based on site-specific nucleases (SSNs) has demonstrated widespread genetic modification across a diverse range of plant species over the past generation. SSNs function by utilizing endonucleases capable of cleaving DNA within a specific region of the genome. The active region of the SSN is tethered to it through either a DNA-binding domain or an RNA sequence. These SSNs are responsible for inducing double-stranded breaks (DSBs) in the targeted DNA. Repair mechanisms like nonhomologous end joining (NHEJ) and homology-directed recombination (HDR) are employed to repair these DSBs, resulting in insertions/deletions (INDELS) and substitutions in the host locus [5]. Upon targeting the specific nucleotide sequences, cellular DNA repair processes lead to changes in gene expression at the designated sites.
Engineered nucleases used in genome editing encompass several types, including designed Meganuclease (MegaN), Zinc finger nucleases (ZFNs), Transcription activator-like effector nucleases (TALENs) and the Clustered regularly interspaced short palindromic repeat (CRISPR/Cas9) nuclease system. These techniques have facilitated efficient and direct modifications of the genome in a rapid and cost-effective manner.
Engineered Meganuclease (MegaN)
Meganucleases (MegaN) are naturally occurring endonucleases that were first discovered in the late 1980s. These endonucleases possess the ability to identify and cleave extensive nucleotide sequences, ranging from 12 to 40 base pairs, which exhibit substantial variations across diverse genomes. Among exemplary meganucleases are I-SceI, sourced from yeast mitochondria, and I-CreI, derived from algal photosynthetic enzymes. Despite their scarcity in important genomes, meganucleases have been engineered to recognize sequences beyond their original targets. Due to the somewhat longer recognition sites, there is a greater risk of cleavage and subsequently more minor off-target effects. Conversely, the adoption of engineered meganucleases has been comparatively limited when compared to other contemporary nucleases, even though the challenge of adapting meganucleases to recognize new specificities remains [10].
Zinc Finger Nuclease-Based Engineering
Zinc finger nucleases (ZFNs), a class of synthetic nucleases, have brought a revolutionary impact to the realm of programmable nucleases. ZFNs were created by fusing multiple zinc finger DNA-binding domains to the non-specific cleavage property of the restriction endonuclease FokI [7, 9]. This amalgamation allows protein molecules to discern DNA sequences that are only a few nucleotides apart. The paired endonucleases form a dimer, enabling them to cleave double-stranded DNA [11]. Additionally, each motif within the zinc finger array recognizes a distinct 3-nucleotide complementary strand. This offers the flexibility of selecting a variable sequence to suit the desired target. ZFNs were initially applied for sequence-specific mutagenesis in tobacco during the early 2000s, marking one of the earliest instances where engineered endonucleases identified and modified chromosomal DNA [12]. Despite these remarkable achievements, the utilization of ZFNs in agriculture has been constrained, primarily due to factors like the technical intricacies of the production process and the limited availability of target sites compared to more recently developed techniques in functional genomics.
Transcription Activator-Like Effector Nucleases (TALENs)
To enhance the effectiveness, reliability and accessibility of genome editing, the TALEN (transcription activator-like effector nucleases) technology was introduced in 2011. This innovation stemmed from the discovery of transcription activator-like effectors (TALEs) [13]. Much like ZFNs, TALEN constructs synthetic proteins utilizing a flexible arrangement of DNA-binding domains linked to the non-specific cleavage site of FokI. Each unit consists of 33–35 amino acids and recognizes a single nucleotide. The terminal segment, often comprising 20 amino acids, is termed a “half-repeat.” The specific amino acids at positions 12 and 13 determine the nucleotide recognition pattern in DNA (e.g., NI identifies adenine, HD identifies cytosine, NG recognizes thymine, while NN recognizes both guanine and adenine) [14]. These naturally occurring TAL effectors offer segmentation advantages that facilitate genome editing in TALENs. Such repeats are harnessed in TALENs to target distinct genetic regions for expression. In conjunction with TAL effector assemblies, additional TALENs, gene-specific activators and regulatory proteins are employed as gene-targeting agents [15]. Compared to Meganucleases and ZFNs, TALENs provide greater flexibility, hence are more commonly used in plant genome editing. However, the complexity of multiple assays poses challenges in the efficient production and distribution of TALENs within plant tissue.
Clustered Regularly Interspaced Short Palindromic Repeat/CRISPR/Cas9 Nuclease System
Much like TALENs, the CRISPR–Cas system draws inspiration from biological processes. Originally discovered within the adaptive immune systems of bacteria and archaea, CRISPR–Cas nucleases play a role in targeting and cleaving foreign nucleic acids based on instructions encoded by CRISPR-associated (Cas) nucleases. Importantly, this mechanism ensures that prokaryotes employing the CRISPR/Cas system do not target their own genomes, as these representations are not found among externally linked prokaryotes. For a target DNA sequence to be recognized and cleaved, a specific short sequence element within a lateral or segmental context is required. The pioneer of genetic modification tools within this system was the Streptococcus pyogenes CRISPR–Cas9 system (CRISPR–SpCas9), commonly referred to as "CRISPR–Cas9" [16, 17]. However, it's essential to note that CRISPR–Cas9 pertains only to the traits shared by CRISPR–SpCas9 and its orthologs, to avoid confusion. In the proposed CRISPR–Cas9 system, both spacer-containing CRISPR RNA (crRNA) and single guide RNA (sgRNA) contribute to the enhancement of crRNA (tracrRNA). The later is necessary for the development of the functional unit. The sgRNA guides the nuclease complex to a specific DNA region, resulting in the cleavage of the targeted nucleotide sequence. Cas9's structure is intricate, featuring two nuclease domains, RuvC and HNH [9, 18]. The use of the CRISPR–Cas9 system has brought about various advancements in plant genome engineering. Techniques such as cloning, transferring into plant cells and simplified design have contributed to a high success rate in genome editing approaches [19]. Work flow of Genome Editing utilizing the Cas9 System is depicted in Fig. 1. A Comparative Analysis of Genome Editing Tools is given in Table 1.
Fig. 1.
Workflow of Genome Editing utilizing the Cas9 System. The Cas9 gene-editing process entails the formation of a complex between sgRNA and Cas9 protein, DNA unwinding facilitated by sgRNA, gene cleavage by Cas9, utilization of analysis tools, cloning, transformation and more. Notably, this process does not require any foreign elements for successful editing
Table 1.
Comparative analysis of Genome Editing Tools and their properties
| Role | ZFNs | TALENs | MNs | CRISPR/Cas9 | Reference |
|---|---|---|---|---|---|
| Construction | Requires protein engineering for each individual target | Requires protein engineering for each individual target | Utilizes customizable target recognition sequences | Relies on a 20-Nucleotide sequence of sgRNA | [20] |
| Efficacy of target recognition | Higher | Higher | Higher | Higher | [20] |
| Targeting | Relies on protein-DNA interactions, which can be less predictable | Relies on protein-DNA interactions, which can be less predictable | Employs unique DNA binding domains for target specificity | Utilizes DNA-RNA interactions, which are highly predictable | [21] |
| Delivery | Involves the use of two ZFNs positioned around the target sequence | Requires two TALENs positioned around the target sequence | Uses customizable DNA binding proteins along with RNA guides | Employs sgRNA that is complementary to the target sequence along with the Cas9 protein | [22] |
| Feasibility of library construction and transformation for genome-wide screens | Technically demanding and complex | Technically demanding and complex | Enables genome-wide studies with improved accuracy | Highly feasible and practical | [23] |
| Kind of Action | Double-stranded break in target DNA | Double-stranded break in target DNA | Direct conversions in targeted regions | Double-stranded break in target DNA | [21] |
| Mutagenesis | Higher | Middle | Middle | Lower | [20] |
| Multiplexing | Difficult | Difficult | Difficult | Possible | [22] |
| Target range | Unlimited | Unlimited | Unlimited | Limited by PAM | [24] |
| Effects | Lower | Lower | Lower | Lower | [23] |
| Cost | Higher | Higher | Higher | Low | [25] |
| Crop Improvement | Low | Low | Low | Higher | [25] |
| Range | Narrow | Narrow | Narrow | Broad | [25] |
| Dimerization | Required | Not-Required | Not-Required | Not-Required | [23] |
| Types | One | One | One | Many | [23] |
| Future use | Medium | Medium | Medium | High | [25] |
Challenging and Unique CRISPR/Cas9 Variations
CRISPR/Cas9 genome editing, a groundbreaking technique, is re-shaping the landscape of genetic manipulation by inducing small cuts in the double-stranded genome of the target organism. This method harnesses the SpCas9 enzyme derived from Streptococcus pyogenes. Since its inception in 2013, this technology has been widely embraced by researchers who continue to explore its versatile applications in genome editing. Although Cas9 remains remarkably efficient, its utilization in gene editing is not without limitations [26]. For the successful binding and fragmentation of the target genome, Class 1 enzymes rely on multisubunit proteins, which are abundantly present. "Class 2" predominantly encompasses Type II and Type VI effectors. Both classes of effectors facilitate the recognition and cleavage of target nucleic acids, as well as crRNA. Class 2 Type V and Class 2 Type VI utilize distinct domains, while Class 2 Type II employs a single Ruv domain. Similarly, Class 2 Type V employs a combination of Cas9, RuvC, and HNH nuclease domains [27].
The discovery of RNA-dependent RNase enzyme systems from Class 2 Type II (FnCas9) and Class 2 Type VI (C2c2) has paved the way for innovative genome editing approaches. The Leptotrichiashahii bacterium houses the Class 2 Type II C2c2 effector, which is controlled by a single crRNA. This bacterium can be trained to cleave specific ssRNA molecules containing appropriate protospacers. Notably, these effectors exhibit selective cleavage of ssRNAs at varying distances from the crRNA binding site, rather than targeting adenine sequences. Structurally, they consist of two HEPN domains housing catalytic residues. Binding of C2c2 is regulated by a crRNA secondary structure comprising at least one 24-nt stem-loop motif, along with a 22–28-nt complementary sequence to the RNA protospacers. Additionally, a mononucleotide protospacer-flanking site (PFS) consisting of adenine, uracil, or cysteine must be present at the 3' end of the protospacer [27, 28]. In 2013, another RNase-based system was uncovered in the Francisell bacterium [29]. This system, known as FnCas9, targets bacterial mRNA without requiring a protospacer adjacent motif (PAM) and can impact gene expression. Notably, this enzyme demonstrated efficacy in combating hepatitis C virus (HCV) in Huh-7.5 cells through RNA inhibition. By attacking both positive and negative strands of the virus's RNA, FnCas9 disrupts viral RNA translation and replication. FnCas9 exhibits a degree of tolerance for mismatches of up to three to six base pairs at the 3′ or 5′ end; however, mismatches exceeding six base pairs lead to a complete loss of activity. Furthermore, this enzyme can also target DNA [30]. These enzymes can be employed in conjunction with viral vectors to modify plants, imparting desired traits. Viral vectors enable high and transient expression of foreign genes for editing. This was effectively demonstrated in instances such as targeted mutagenesis in Nicotiana benthamiana and Petunia hybrida using the tobacco rattle virus (TRV).
The larger size of SpCas9 (4.2 kb) prevents its expression in plants using the tobacco rattle virus. To address this size constraint, smaller genome editing enzymes from various bacteria—such as SaCas9 (3.2 kb) from Staphylococcus aureus, St1Cas9 (3.4 kb) from Streptococcus thermophilus, and NmCas9 (3.2 kb) from Neisseria meningitidis have been identified as alternatives. These Class 2 Type II immune system enzymes employ the RuvC and HNH domains to cleave double-stranded DNA. Moreover, this group of enzymes specifically targets DNA at a designated site, typically spanning 21–24 nucleotides (nt) in length, located around PAM motifs like 5′-NNGRRT-3′, 5′-NNNRRT-3′, 5′-NNAGAAW-3′, and 5′-NNNNGMTT-3′. In these sequences, N represents any nucleotide, R stands for A or G, M represents A or C, and W represents A or T [31]. Research also indicates that SaCas9 effectively targeted the 5′-NNNGGT-3′ PAM sequence, achieving a notably increased mutation rate (80%) and promoting homologous recombination in the selected lines. For genome editing, these mentioned enzymes primarily focus on a considerably longer PAM sequence [32]. These unique CRISPR/Cas9 variations represent a testament to the dynamic nature of genetic research and the potential for CRISPR technology to revolutionize various fields, from medicine and agriculture to biotechnology and beyond. As scientists continue to refine and expand these variations, the possibilities for precise and targeted genome manipulation are boundless, promising groundbreaking advancements in science and medicine.
Applications of CRISPR/Cas9-Based Genome Editing System in Crop Improvement
In recent times, CRISPR/Cas9-based genome editing has made substantial advancements in enhancing various attributes of crops. However, the scientific community continues to grapple with emerging challenges while striving to enhance the quality of diverse culinary plants. CRISPR/Cas-based gene editing holds a multitude of applications, including augmenting yield, bolstering disease and pathogen resistance, fortifying herbicide resistance, and boosting stress tolerance. These applications are detailed in Table 2. Gene editing has also been pursued in developmental genes. A host of genes participate in carotenoid production, encompassing Anthocyanin 1 (ANT1), Phytoene Desaturase (SlPDS), Phytochrome Interacting Factor 4 (SlPIF4), and Phytoene Synthetase 1 (PSY1), among others. Notably, under conditions of heat stress, mutant tomato plants with the Slagamous-like 6 (SlAGL6) gene deletion exhibited parthenocarpy fruit development, which would have otherwise impeded fertilization-dependent fruit set. Targeted silencing of the eIF4E gene in melons and tomatoes resulted in resistance to RNA viruses [33, 34]. The CRISPR/Cas9 system was employed to edit the granule-bound starch synthase (GBSS) gene, yielding amylopectin but excluding amylose production [35]. Additionally, the morphology of sweet potatoes (Ipomoea batatas) underwent alteration using the CRISPR/Cas9 system [36]. In the case of chili peppers (Capsicum annuum L.), a mutation induced by CRISPR/Cas9 conferred resistance to anthracnose [37].
Table 2.
Some reports on applications of CRISPR/Cas9 gene editing system for quality and yield enhancement in crops
| Crop | Type of modification | Target gene | Target trait/organism | Result/outcome | Reference |
|---|---|---|---|---|---|
| Grapes | Gene disruption | MLO7 | Powdery mildew | Mutations in target genes; disease resistance not checked | [38] |
| Grapevine | Gene disruption | VvWRKY52 | Botrytis cinerea | Resistance to gray mold disease | [39] |
| Rice | Promoter disruption |
OsSWEET11, OsSWEET14 |
X. oryzae pv. oryzae | Mutations in promoter, resistance not checked | [40] |
| Rice | Gene disruption | OsSWEET14 | X. oryzae pv. oryzae | Resistance to bacterial blight | [40] |
| Rice | Gene disruption | OsERF922 | Magnaporthe oryzae | Resistance to rice blast | [41] |
| Rice | Gene disruption | OsMPK5 | Fungal and bacterial pathogens | Mutations in target, resistance not checked | [41] |
| Rice | Gene disruption | eIF4G | Rice tungro spherical virus | Resistance to rice tungro spherical disease | [41] |
| Tobacco | Gene disruption | Three viral regions (R, CP, and RCR) | Tomato yellow mosaic virus | Significant reduction or attenuation of disease symptoms | [42] |
| Wheat | Gene disruption | TaEDR1 | Powdery mildew | Resistance to Blumeria graminis f.sp. tritici | [43] |
| Apple | Gene disruption | DIPM1, DIPM2, DIPM4 | Fire blight disease | Mutations in target genes; disease resistance not checked | [44] |
| Banana | Viral genome disruption | Viral genes |
Banana streak virus (BSV) |
Inactivation of endogenous banana streak virus integrated in host genome | [45] |
| Citrus | Gene disruption | CsLOB1 | X. citri subsp. citri | Resistance to citrus canker | [46] |
| Citrus | Promoter disruption | CsLOB1 promoter | X. citri subsp. citri | Enhance resistance to citrus canker | [46] |
Furthermore, leveraging CRISPR/Cas9 technology, improvements have been made in rice's PIN5b, GS3, GW2, GW5, and GW2 genes, leading to enhanced yield. Fruit-related genes like CLV and ENO have been successfully modified by scientists to enhance fruit production. Addressing celiac disease triggered by gluten in susceptible individuals, the conserved region has been effectively altered using CRISPR/Cas9 technology, resulting in an impressive 85% reduction in immunoreactivity at wheat loci. Similarly, the potential of CRISPR genome editing to enhance Vitamin A and β-Carotene content in plants has been demonstrated [47].
Genome Editing Safety Guidelines
The advancement of crop resilience against biotic, abiotic, and extreme climatic shifts, as well as the resolution of global policy and governance challenges, heavily rely on plant genome editing techniques. In addition to discussing the current state of agriculture, this narrative delves into reflections on the principled adoption of biotechnology, alongside ethical, social and biological considerations associated with the CRISPR/Cas system. Given the technology's limitations, ethical quandaries surrounding CRISPR have surfaced, prompting the need for both domestic and international attention to navigate questions that serve the broader public interest. Meanwhile, ongoing public discourse revolves around the guidance and regulation of novel methodologies within industrialized nations [47]. The emergence of transgenic plants endowed with nutritional, herbicide-tolerant, and insect-resistant traits has contributed significantly to the surge in genetically modified crop production. A recent estimate indicates that in 2014, 18 million farmers cultivated GMO crops across 181.5 million hectares spanning 28 countries, marking a 3–4 percent increase from 2013 figures [48]. Notable genetically modified crops recently introduced include tomato, corn, soybean, cotton, canola, rice, potato, squash, melon, and papaya. However, soybean, corn, and cotton stand out due to their extensive cultivation and pivotal role in the agricultural economies of numerous nations.
Leading the chart as the world's primary producers and exporters of genetically modified products are the United States, Argentina, and Canada [45]. In India, the management of all GMO-related activities is governed by the Environmental Protection Act (EPA), enacted in 1986. The enforcement of this law falls under the jurisdiction of the Ministry of Environment, Forests, and Climate Change (MoEF & CC) [49, 50].
India's comprehensive regulatory framework governing genetically modified crops falls under the purview of the Department of Biotechnology, a segment of the Ministry of Science and Technology, as well as the Ministry for Environment and Forestry. This intricate structure comprises six authoritative bodies: the State Biotechnology Coordination Committees (SBCC), the Recombinant DNA Advisory Committee (RDAC), the Genetic Engineering Appraisal Committee (GEAC), the Institutional Biosafety Committees (IBSC), and the District Level Committees (DLC). The Government Environmental Assessment Council (GEAC) in India is tasked with evaluating the environmental impact of GMO-related activities spanning research, industrial production, field applications, and environmental discharge. Crucial regulations essential for the development, environmental release, and marketing of GM crops were established by the Indian Parliament. These regulations encompass the Seeds Act of 1966, the Environment Protection Act of 1986, the Forests and Climate Change (MoEF & CC) Act, and the Seeds (Control) Order overseen by the Ministry of Agriculture. Differing perspectives within the Technical Expert Committee established by the Supreme Court of India for Safety and Guidelines for Genetically Modified Agricultural Research underline the importance of this discourse. Amidst India's infrastructure challenges and a lack of well-defined risk assessment and research protocols for genetically modified crops, the pursuit remains imperative given the nation's pressing needs. As India prepares for future deregulation, continued research, infrastructure development and robust marketing and biosafety regulations are essential and new genetically modified organism (GMO) should be assigned a registration number and date, prominently displayed on a dedicated website or portal to facilitate the commercialization of transgenic products, regardless of the country's approval process [51].
CRISPR-CHOPCHOP: A Beginner's Guide to sgRNA Design
In just a span of three years, CRISPR genome editing has brought about fundamental changes to the field of biology with its applications and acceptance continuing to expand. The need for computational tools that streamline CRISPR targeting remains constant, allowing the integration of these principles to accelerate the process of selecting appropriate targets when new CRISPR mechanisms and target selection criteria emerge. Among these tools, CHOPCHOP (https://chopchop.cbu.uib.no/), a widely recognized web application for genome editing with CRISPR and TALEN, stands as a popular choice. CHOPCHOP provides a user-friendly online platform based on the latest comprehensive research to facilitate target selection, primer generation, and restriction site identification (Fig. 2). This tool offers precise localization of various subsections, including coding regions, UTRs, splice sites, and specific exons, spanning both protein-coding and noncoding genes. It adeptly identifies potential off-target sites for all sgRNAs, automatically generates primers for target sites, and presents all relevant elements through a flexible graphical interface, complete with information pertinent to restriction analysis for further validation [52].
Fig. 2.
Workflow of steps in designing sgRNA in CHOPCHOP tool
Interpretations of Findings
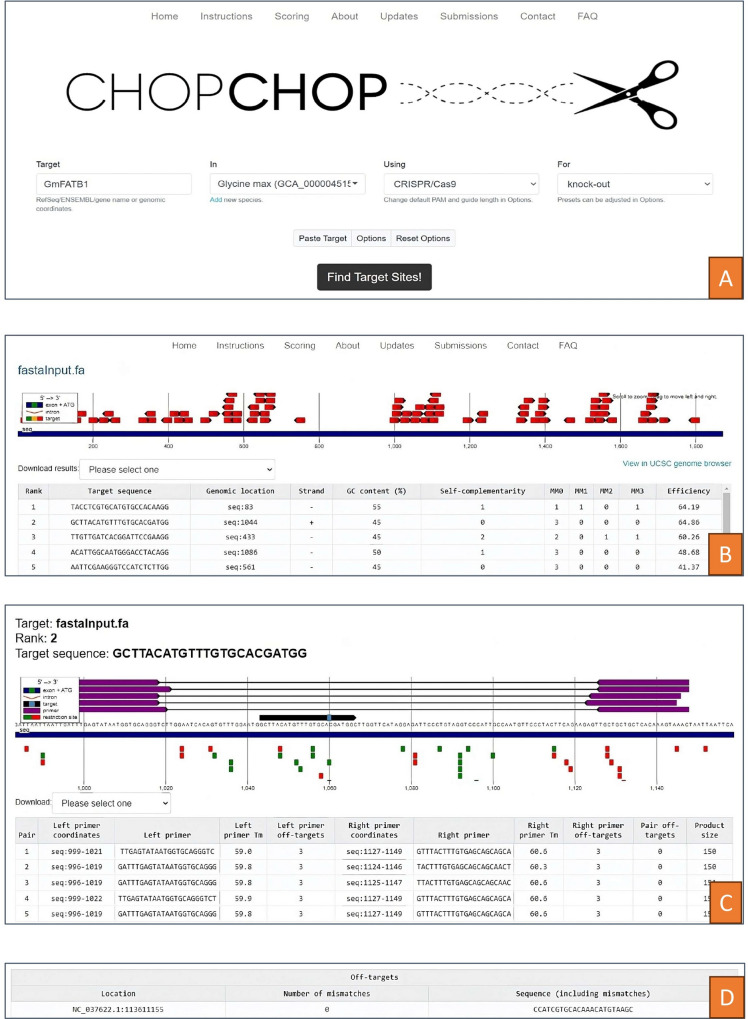
When interpreting the results obtained from the CHOPCHOP tool, the findings are presented through a color-coded system. Within the result window, the color scheme serves as a key indicator: green signifies the absence of off-targets, orange represents a moderate number of off-targets, and red indicates a higher count of off-targets. It's important to note that when generating an appropriate sgRNA, off-targets are not necessary. Clicking on any green symbol in the results window allows users to progress to the next level of target identification. This subsequent stage provides detailed insights into primer sequences, off-target quantities, and the GC content percentage. Primer pairs are highlighted in a distinctive violet color, while corresponding restriction sites are marked by either green or red boxes, with examples like HindIII. An essential consideration is that the PAM sequence must be an integral part of the target sequence. The table presented below the results showcases the genome's location along with potential off-targets, with any mismatches depicted in red. It's worth noting that the phrase "there are no off targets" is used when no off-targets are detected in the analysis. Upon determining the final target, users are advised to copy both the target sequence and the associated primers directly from the CHOPCHOP website, ensuring accurate and reliable replication of the identified genetic information for further research or applications. Designing Guide RNAs Using CHOPCHOP Software is depicted in Fig. 3.
Fig. 3.
Designing Guide RNAs Using CHOPCHOP Software. The process of designing guide RNAs is illustrated from A to D utilizing the CHOPCHOP bioinformatics tool. A—The home page of CHOPCHOP used for designing sgRNAs. B—Initial stage for identifying off-target regions within the desired genomic sequence. C—Intermediate stage for further identification of off-target regions, displaying %GC content, off-target levels, and primer sequences. D—Final stage depicting the completion of off-target analysis for the desired genomic sequence
Envisioning the Future of CRISPR/Cas9 Technology in Crop Enhancement
In the ever-evolving world of agriculture, the quest for crop improvement and enhancement is unceasing. The demands of a growing global population, coupled with environmental challenges and changing consumer preferences, necessitate innovative solutions to ensure food security and sustainability. Among the transformative technologies on the horizon, CRISPR/Cas9 stands as a beacon of hope in revolutionizing crop enhancement. This revolutionary genome editing technology has the potential to reshape the future of agriculture, ushering in a new era of resilient, nutritious and high-yielding crops.
Moreover, the precision of CRISPR/Cas9 technology minimizes unintended genetic alterations, ensuring that the resulting crops meet safety and regulatory standards. This is in contrast to conventional genetic modification techniques, which often involve the insertion of foreign genes (transgenes) into crops, raising concerns about unintended side effects and potential ecological consequences. CRISPR/Cas9 allows for the introduction of desired traits without the use of transgenes, alleviating these concerns and making the resulting crops more acceptable to consumers and regulatory authorities. As we envision the future of CRISPR/Cas9 technology in crop enhancement, it becomes evident that this technology has the potential to unlock a multitude of possibilities. The ability to precisely edit the genetic makeup of crops offers an unprecedented level of control over their traits, opening up new avenues for crop development. However, it is important to acknowledge that the realization of this vision is not without challenges and ethical considerations. As CRISPR/Cas9 technology advances, questions surrounding intellectual property rights, equitable access, and environmental impacts must be carefully addressed. Ethical discussions about the boundaries of genetic modification and its potential consequences for biodiversity and ecosystems will play a pivotal role in shaping the future of this technology in agriculture.
Conclusion
One existing gap in the current landscape is the need for greater precision and efficiency in crop enhancement. While genome editing, including CRISPR/Cas9 technology, shows immense promise, there is room for improvement in its application. This gap can be bridged by advancing our understanding of crop genetics, optimizing delivery methods, and refining the regulatory frameworks to ensure the responsible use of these technologies. Among the cutting-edge methods for crop improvement, genome editing stands out as a promising avenue. Genome editing enables precise modification of desirable crop traits without the introduction of transgenes, ensuring safety for human health and the environment. Within this realm, CRISPR/Cas9 technology emerges as a powerful tool for enhancing crop attributes. By harnessing CRISPR/Cas9, we can revolutionize crop production, opening new avenues to combat pre- and post-harvest yield losses. This technology not only bolsters future food security but also offers a sustainable path towards reducing crop losses, thus fostering economic growth in the agricultural sector. The potential of CRISPR/Cas9 technology in crop enhancement represents a beacon of hope for more resilient and productive agricultural practices, and with ongoing research and refinement, it holds the promise of closing the existing gap between current agricultural practices and the evolving needs of our expanding nation's economy.
Acknowledgements
The authors are thankful to Director, Research Innovation and Translation, Atmiya University and Head Department of Biotechnology, Atmiya University, Rajkot for providing infrastructure, and facilities.
Author Contributions
AC and RR—Collection of data, literature survey and writing of article; PJ and RST—Conceptualization of idea, critical review and editing of draft.
Funding
AC acknowledges the Government of Gujarat for the JRF under the SHODH scheme and RR thanks Junagadh Agriculture University for the SRF in a research project funded by the Government of Gujarat.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Singh AK, Singh A, Joshi P. Combined application of chitinolytic bacterium Paenibacillus sp. D1 with low doses of chemical pesticides for better control of Helicoverpa armigera. Int J of Pest Manag. 2016;62:222–227. doi: 10.1080/09670874.2016.1167267. [DOI] [Google Scholar]
- 2.Teraiya S, Nirmal D, Joshi P. Potential scope and prospects of plant growth-promoting microbes (PGPMs) in micropropagation technology. In: Meena M, Swapnil P, editors. Plant-microbe interaction—recent advances in molecular and biochemical approaches. Netherland: Elsevier; 2023. pp. 249–277. [Google Scholar]
- 3.Kumawat A, Yadav D, Srivastava P, et al. Restoration of agroecosystems with conservation agriculture for food security to achieve sustainable development goals. Land Degrad Dev. 2023;34:3079–3097. doi: 10.1002/ldr.4677. [DOI] [Google Scholar]
- 4.Ghadia B, Singh AK, Khatnani T, et al. An improved method of DNA purification from secondary metabolites rich medicinal plants using certain chaotropic agents. Acta Physiol Plant. 2016;38:207. doi: 10.1007/s11738-016-2223-6. [DOI] [Google Scholar]
- 5.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/SCIENCE.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasant G, Bhatt S, Raghav R, Joshi P. Revitalization of PGPR through integrating nanotechnology for sustainable development in agriculture. In: Meena M, Swapnil P, editors. Plant-microbe interaction—recent advances in molecular and biochemical approaches. Netherland: Elsevier; 2023. pp. 227–248. [Google Scholar]
- 7.Wani AK, Akhtar N, Singh R, et al. Genome centric engineering using ZFNs, TALENs and CRISPR-Cas9 systems for trait improvement and disease control in Animals. Vet Res Commun. 2023;47:1–16. doi: 10.1007/s11259-022-09967-8. [DOI] [PubMed] [Google Scholar]
- 8.Becker S, Boch J. TALE and TALEN genome editing technologies. Gene Genome Ed. 2021;2:100007. doi: 10.1016/j.ggedit.2021.100007. [DOI] [Google Scholar]
- 9.Chen K, Gao C. Targeted genome modification technologies and their applications in crop improvements. Plant Cell Rep. 2014;33:575–583. doi: 10.1007/S00299-013-1539-6. [DOI] [PubMed] [Google Scholar]
- 10.Chen K, Wang Y, Zhang R, et al. CRISPR/Cas genome editing and precision plant breeding in agriculture. Ann Rev Plant Biol. 2019;70:667–697. doi: 10.1146/ANNUREV-ARPLANT-050718-100049. [DOI] [PubMed] [Google Scholar]
- 11.Kuang Y, Li S, Ren B, et al. Base-editing-mediated artificial evolution of OsALS1 In planta to develop novel herbicide-tolerant rice germplasms. Mol Plant. 2020;13:565–572. doi: 10.1016/J.MOLP.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 12.Kleinstiver BP, Prew MS, Tsai SQ, et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015;33:1293–1298. doi: 10.1038/NBT.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore R, Chandrahas A, Bleris L. Transcription activator-like effectors: a toolkit for synthetic biology. ACS Synth Biol. 2014;3:708–716. doi: 10.1021/sb400137b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nekrasov V, Wang C, Win J, et al. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep. 2017;7:482. doi: 10.1038/S41598-017-00578-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh D, Kumar A, Sinha N. Targeted genome editing: a new era in molecular biology. In: Mondal S, Singh RL, editors. Advances in animal genomics. Academic press; 2021. pp. 75–89. [Google Scholar]
- 16.Le Rhun A, Escalera-Maurer A, Bratovič M, Charpentier E. CRISPR-Cas in Streptococcus pyogenes. RNA Biol. 2019;16:380–389. doi: 10.1080/15476286.2019.1582974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang T, Zheng Q, Yi X, et al. Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol J. 2018;16:1415–1423. doi: 10.1111/PBI.12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andolfo G, Iovieno P, Frusciante L, Ercolano MR. Genome-editing technologies for enhancing plant disease resistance. Front Plant Sci. 2016;7:1813. doi: 10.3389/FPLS.2016.01813/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rau K, Rentmeister A. CRISPR/Cas9: a new tool for RNA imaging in live cells. ChemBioChem. 2016;17:1682–1684. doi: 10.1002/CBIC.201600342. [DOI] [PubMed] [Google Scholar]
- 21.Khan SH. Genome-editing technologies: concept, pros, and cons of various genome-editing techniques and bioethical concerns for clinical application. Mol Ther Nucleic Acids. 2019;16:326–334. doi: 10.1016/j.omtn.2019.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung CJ, Ménoret S, Brusselle L, et al. Comparative analysis of piggyBac, CRISPR/Cas9 and TALEN mediated BAC transgenesis in the zygote for the generation of humanized SIRPA rats. Sci Rep. 2016;6:31455. doi: 10.1038/srep31455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson RA, Gurevich V, Filler S, Samach A, Levy AA. Comparative assessments of CRISPR-Cas nucleases’ cleavage efficiency in planta. Plant Mol Biol. 2015;87:143–156. doi: 10.1007/s11103-014-0266-x. [DOI] [PubMed] [Google Scholar]
- 24.Takayama K, Igai K, Hagihara Y, et al. Highly efficient biallelic genome editing of human ES/iPS cells using a CRISPR/Cas9 or TALEN system. Nucleic Acids Res. 2017;45:5198–5207. doi: 10.1093/nar/gkx130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahim J, Gulzar S, Zahid R, Rahim K. A systematic review on the comparison of molecular gene editing tools. Int J Innov Sci Res Technol. 2021;6:1–8. [Google Scholar]
- 26.Ortigosa A, Gimenez-Ibanez S, Leonhardt N, Solano R. Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol J. 2019;17:665–673. doi: 10.1111/PBI.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abudayyeh OO, Gootenberg JS, Konermann S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:6299. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sampson TR, Saroj SD, Llewellyn AC, et al. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–257. doi: 10.1038/NATURE12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schunder E, Rydzewski K, Grunow R, Heuner K. First indication for a functional CRISPR/Cas system in Francisella tularensis. Int J Med Microbiol. 2013;303:51–60. doi: 10.1016/j.ijmm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Hirano H, Gootenberg JS, Horii T, et al. Structure and engineering of Francisella novicida Cas9. Cell. 2016;164:950–961. doi: 10.1016/j.cell.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Zhang H, Xu X, et al. Catalytic-state structure and engineering of Streptococcus thermophilus Cas9. Nat Catal. 2020;3:813–823. doi: 10.1038/s41929-020-00506-9. [DOI] [Google Scholar]
- 32.Jia H, Xu J, Orbović V, Zhang Y, Wang N. Editing citrus genome via SaCas9/sgRNA system. Front Plant Sci. 2017;8:2135. doi: 10.3389/fpls.2017.02135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandrasekaran J, Brumin M, Wolf D, et al. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol. 2016;17:1140–1153. doi: 10.1111/mpp.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon YJ, Venkatesh J, Lee JH, et al. Genome editing of eIF4E1 in tomato confers resistance to pepper mottle virus. Front Plant Sci. 2020;11:1098. doi: 10.3389/fpls.2020.01098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toinga-Villafuerte S, Vales MI, Awika JM, Rathore KS. CRISPR/Cas9-mediated mutagenesis of the granule-bound starch synthase gene in the potato variety Yukon Gold to obtain amylose-free starch in tubers. Int J Mol Sci. 2022;23:4640. doi: 10.3390/ijms23094640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ke Q, Kang L, Kim HS, Xie T, et al. Down-regulation of lycopene ε-cyclase expression in transgenic sweet potato plants increases the carotenoid content and tolerance to abiotic stress. Plant Sci. 2019;281:52–60. doi: 10.1016/j.plantsci.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Mishra R, Mohanty JN, Mahanty B, Joshi RK. A single transcript CRISPR/Cas9 mediated mutagenesis of CaERF28 confers anthracnose resistance in chilli pepper (Capsicum annuum L.) Planta. 2021;254:5. doi: 10.1007/s00425-021-03660-x. [DOI] [PubMed] [Google Scholar]
- 38.Malnoy M, Viola R, Jung MH, et al. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front Plant Sci. 2016;7:1904. doi: 10.3389/fpls.2016.01904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fedorina J, Tikhonova N, Ukhatova Y, Ivanov R, Khlestkina E. Grapevine gene systems for resistance to Gray Mold Botrytis cinerea and Powdery Mildew Erysiphe necator. Agronomy. 2022;12:499. doi: 10.3390/agronomy12020499. [DOI] [Google Scholar]
- 40.Zeng X, Luo Y, Vu NT, et al. CRISPR/Cas9-mediated mutation of OsSWEET14 in rice cv. Zhonghua11 confers resistance to Xanthomonas oryzae pv. oryzae without yield penalty. BMC Plant Biol. 2020;20:1–11. doi: 10.1186/s12870-020-02524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nizolli VO, Pegoraro C, Oliveira ACD. Rice blast: strategies and challenges for improving genetic resistance. Crop Breed Appl Biotechnol. 2021 doi: 10.1590/1984-70332021v21Sa22. [DOI] [Google Scholar]
- 42.Roy R, Bagewadi B. CRISPR-Cas system-a promising tool for engineering resistance to plant viruses. In: Awasthi LP, editor. Applied plant virology. Academic Press; 2020. pp. 649–655. [Google Scholar]
- 43.Kang Y, Zhou M, Merry A, Barry K. Mechanisms of powdery mildew resistance of wheat–a review of molecular breeding. Plant Pathol. 2020;69:601–617. doi: 10.1111/ppa.13166. [DOI] [Google Scholar]
- 44.Nishitani C, Osakabe K, Osakabe Y. Genome editing in apple. In: Korban SS, editor. The apple genome. Compendium of plant genomes. Springer; 2021. pp. 213–225. [Google Scholar]
- 45.Tripathi L, Ntui VO, Tripathi JN, Kumar PL. Application of CRISPR/Cas for Diagnosis and management of viral diseases of banana. Front Microbiol. 2021;11:609784. doi: 10.3389/FMICB.2020.609784/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia H, Zhang Y, Orbović V, Xu J, et al. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol J. 2017;15:817–823. doi: 10.1111/PBI.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng X, Kuijer HN, Al-Babili S. Carotenoid biofortification of crops in the CRISPR era. Trends Biotechnol. 2021;39:857–860. doi: 10.1016/j.tibtech.2020.12.003. [DOI] [PubMed] [Google Scholar]
- 48.Lucht JM. Public acceptance of plant biotechnology and GM crops. Viruses. 2015;7:4254–4281. doi: 10.3390/v7082819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahuja V. Regulation of emerging gene technologies in India. BMC Proc. 2018;12:5–11. doi: 10.1186/S12919-018-0106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta S, Kumar A, Patel R, Kumar V. Genetically modified crop regulations: scope and opportunity using the CRISPR-Cas9 genome editing approach. Mol Biol Rep. 2021;48:4851–4863. doi: 10.1007/s11033-021-06477-9. [DOI] [PubMed] [Google Scholar]
- 51.Sprink T, Wilhelm R, Hartung F. Genome editing around the globe: an update on policies and perceptions. Plant Physiol. 2022;190:1579–1587. doi: 10.1093/plphys/kiac359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Labun K, Krause M, Torres Cleuren Y, Valen E. CRISPR genome editing made easy through the CHOPCHOP website. Curr Protoc. 2021;1:e46. doi: 10.1002/cpz1.46. [DOI] [PubMed] [Google Scholar]