Abstract
The molecular mechanisms which are responsible for restricting skeletal muscle gene expression to specific fiber types, either slow or fast twitch, are unknown. As a first step toward defining the components which direct slow-fiber-specific gene expression, we identified the sequence elements of the human troponin I slow upstream enhancer (USE) that bind muscle nuclear proteins. These include an E-box, a MEF2 element, and two other elements, USE B1 and USE C1. In vivo analysis of a mutation that disrupts USE B1 binding activity suggested that the USE B1 element is essential for high-level expression in slow-twitch muscles. This mutation does not, however, abolish slow-fiber specificity. A similar analysis indicated that the USE C1 element may play only a minor role. We report the cloning of a novel human USE B1 binding protein, MusTRD1 (muscle TFII-I repeat domain-containing protein 1), which is expressed predominantly in skeletal muscle. Significantly, MusTRD1 contains two repeat domains which show remarkable homology to the six repeat domains of the recently cloned transcription factor TFII-I. Furthermore, both TFII-I and MusTRD1 bind to similar but distinct sequences, which happen to conform with the initiator (Inr) consensus sequence. Given the roles of MEF2 and basic helix-loop-helix (bHLH) proteins in muscle gene expression, the similarity of TFII-I and MusTRD1 is intriguing, as TFII-I is believed to coordinate the interaction of MADS-box proteins, bHLH proteins, and the general transcription machinery.
The proteins which make up the contractile apparatus of a striated muscle fiber are the products of multigene families. This large variety of isoforms is derived from distinct genes or alternative splicing of primary transcripts. Throughout development, the functional demands placed upon a particular muscle change. In order to adapt to the changes, different isoforms of various metabolic proteins and proteins of the contractile apparatus are up- or down-regulated, culminating in the appearance of distinct fiber types with distinct functional attributes. The ratio of the various fiber types within a muscle then determines the functional phenotype of that muscle.
A nomenclature based on myosin heavy-chain (MyHC) gene expression is frequently used to define four mature fiber types, one slow and three fast. Slow fibers express MyHC-1, and fast fibers express MyHC-2A, MyHC-2X/D, or MyHC-2B. In adult humans, the other contractile proteins are generally regulated in such a way that expression of the slow and fast isoforms is restricted to slow-twitch and fast-twitch fiber types, respectively. This coordinated pattern of gene expression is not apparent during early fetal development, when embryonic and neonatal isoforms of MyHC are expressed together with various combinations of the other contractile protein isoforms (slow, fast, and cardiac) (36, 37). At the molecular level, nothing is known of the mechanism(s) operating in the establishment of fiber types.
The two best-characterized families of transcription factors with regard to muscle-specific transcription are the myogenic basic helix-loop-helix (bHLH) family (MyoD, Myf5, myogenin, and MRF4) and the MADS-box-containing MEF2 family (22, 26, 31). These appear to be intimately involved in the activation of most muscle-specific genes, whether by binding directly to their respective sequence motifs [bHLH, CANNTG; MEF2, CTA(A/T)4TAG/A] or by binding indirectly through each other’s motifs via a protein-protein interaction (19, 23). Neither of these families has been shown to regulate muscle gene expression in a fiber-specific manner, although myogenin and MyoD transcripts have been shown to be preferentially expressed in slow and fast fibers, respectively (17, 40).
In order to elucidate at least one of the molecular mechanisms responsible for fiber-type-specific gene expression, we have been studying the regulation of the human slow isoform for troponin I (TnIs). Troponin I is the inhibitory subunit of the troponin complex, a heteromeric complex which controls muscle contraction in response to intracellular calcium concentrations. TnIs and the other two isoforms for troponin I, fast (TnIf) and cardiac (TnIc), are each encoded by separate genes. During early fetal development, all three isoforms are coexpressed in both skeletal muscle and cardiac muscle, although TnIs predominates (46). As the coordinated isoform phenotype begins to emerge during late fetal development, TnIs is down-regulated in all muscle fibers except for future slow fibers and is replaced with TnIf in skeletal muscle and TnIc in cardiac muscle (36, 46). In postnatal animals, TnIs expression is restricted to slow fibers and the conductive tissue of heart, while TnIf and TnIc are restricted to fast skeletal muscle fibers and cardiac tissue, respectively. In regenerating rat muscle, slow innervation is required for the induction and maintenance of TnIs expression; in contrast, the expression of TnIf appears to be nerve independent (11). Although the influence of innervation on isoform expression appears to vary with developmental stage, species, and contractile protein (28, 32), there is no doubt that a signaling mechanism exists between the nerve and the nucleus to direct isoform-specific gene expression. The identification of the cis-acting elements necessary for appropriate expression of the TnIs gene will provide a starting point from which to define such a mechanism.
As the characterization of fiber-specific gene expression relies on in vivo models, we injected TnIs-reporter gene plasmids into rat muscle to show that a 157-bp upstream enhancer (USE) is capable of conferring preferential slow-muscle activity upon a heterologous thymidine kinase (TK) minimal promoter (9). Transgenic analysis of the USE linked to the endogenous TnIs −95 minimal promoter confirmed the activity of the enhancer with respect to directing slow-fiber-specific gene expression. This study identifies the nuclear protein binding sites within the USE and correlates these with direct-injection and transgenic data in order to test their functional significance. We describe the isolation of a novel cDNA clone for a protein which interacts with one of the binding sites essential for high-level enhancer activity. This clone encodes a protein that binds to a DNA sequence similar to but distinct from that described for the multifunctional protein TFII-I. Furthermore, this protein bears striking homology to TFII-I in a repeat domain which has yet to be characterized (13, 30, 44). This finding raises the possibility that the two proteins represent the founding members of a new class of transcription factor with a novel repeat domain. Accordingly, we refer to this protein as MusTRD1 (muscle TFII-I repeat domain-containing protein 1).
MATERIALS AND METHODS
Plasmid constructs.
For the direct muscle injection procedure, deletions (5′ or 3′) and point mutations of the USE were generated by PCR such that the resulting fragments had BamHI/BglII ends. These were ligated into the BglII site of pTK81Luc, a pGL3-Basic (Promega)-based luciferase reporter plasmid with the modified TK minimal promoter (−81 to +52) of pT81Luc (25). TnIsUSE-95X1nucZ (9) is a nucleus-targeted lacZ reporter plasmid under the control of the TnIs USE linked to the −95 minimal promoter and exon 1 sequences. TnIsΔB1-USE-95X1nucZ and TnIsΔC1-USE-95X1nucZ are similar plasmids, differing only with respect to the introduction of the USE B1b and USE C1c mutations, respectively.
Muscle nuclear extracts.
All procedures were performed with cold solutions on ice. Soleus (ca. 6 g) and extensor digitorum longus (EDL; ca. 8 g) muscles from 40 to 50 euthanatized male rats were collected into phosphate-buffered saline (calcium and magnesium free). Tendons were removed, and 2-g batches of muscle were processed as follows. Tissue was minced with scissors in a petri dish containing 2 ml of buffer A (14) (300 mM sucrose, 60 mM KCl, 0.15 mM spermine, 0.5 mM spermidine, 0.5 mM EGTA, 2 mM EDTA, 14 mM 2-mercaptoethanol, 10 mg of bovine serum albumin per ml, 15 mM HEPES [pH 7.6]) and then transferred to a 50-ml plastic tube containing a further 28 ml of buffer A. The mixture was homogenized (Kinematica Polytron 10-mW generator) for 90 s at setting 3.6 with aeration by moving the generator in and out of the solution. Speeds and times were determined by monitoring the release of the nuclei from the fibers by 0.2% trypan blue staining so as to enable maximum release with minimum loss due to nuclear rupture. After storage on ice (3 to 5 min), three phases appeared: the top phase contained intact fibers and myofibrils; the middle, clearer phase contained free nuclei and small myofibrils; and the bottom phase contained larger pieces of tissue. The middle phase was collected and stored on ice, and the volume was replaced with buffer A. With a reduction of the homogenization time to 60 s and then 30 s, the homogenization and middle-phase collection steps were repeated until the majority of the nuclei had been recovered (we typically collected a total of 60 to 80 ml per 2 g of muscle).
The nuclear phase was centrifuged in a Beckman JA14 rotor at 2,500 × g for 5 min. The supernatant was discarded, and the pellet was resuspended in 10 ml of buffer B (same as buffer A but with 0.1 mM EGTA and 0.1 mM EDTA). Larger myofibrils and fibers were removed by filtration through 100-μm mesh. The filtrate was centrifuged in a Beckman JA17 rotor at 2,500 × g for 5 min, and the pellet was resuspended in 7 ml of buffer B. Centrifugation and resuspension were repeated four times, with the final resuspension in 2.5 ml of buffer B. The nuclei and remaining myofibrils were unclumped by three gentle strokes with pestle B in a Wheaton 7-ml hand homogenizer, and the nuclei were counted (typically 7 × 106/g of EDL muscle and 1 × 107 to 2 × 107/g of soleus muscle). The nuclei were pelleted, resuspended in 1 ml of buffer B, and microcentrifuged at 800 × g for 5 min. The nuclei were extracted with four pellet volumes of extraction buffer (20 mM HEPES [pH 7.9], 400 mM KCl, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol [DTT], 2 mM benzamidine, 5 μg of pepstatin per ml, 5 μg of leupeptin per ml, 5 μg of aprotinin per ml, 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) for 45 min on ice. This process also appeared to solubilize the contaminating myofibrils. The nuclei were pelleted in a microcentrifuge (800 × g, 5 min), and the extract (supernatant) was collected. The extract was dialyzed for 50 min with a 10,000-molecular-weight-cutoff Slide-A-Lyzer cassette (Pierce Chemical Company) against dialysis buffer (100 mM KCl, 15 mM HEPES [pH 7.9], 1 mM EDTA, 0.5 mM DTT, 20% glycerol, 0.5 mM PMSF). Precipitates were pelleted in a microcentrifuge at 14,500 × g for 5 min, and the protein concentration of the supernatant was determined (7). We typically recovered 500 μg/g of EDL muscle and 700 μg/g of soleus muscle. Extracts were divided into aliquots and stored at −80°C.
Electrophoretic mobility shift assays.
Probes were prepared by annealing complimentary oligonucleotides (see Fig. 2) with 5′ TCGA overhanging termini and performing a Klenow fill-in reaction with 32P-dCTP. The sequences of the c-fos c-sis/platelet-derived growth factor-inducible element (SIE) and serum response element (SRE) probes were as described previously (13). In a final volume of 29 μl, 25 to 28 μg of nuclear extract (or 4 μl of in vitro translation reaction mixture) was mixed with 0.5 μg of poly(dI-dC) and 3.0 μl of 10× binding buffer (150 mM HEPES [pH 7.9], 50 mM MgCl2, 10 mM EDTA, 5 mM DTT, 20 mM benzamidine, 50 μg of pepstatin per ml, 50 μg of leupeptin per ml, 50 μg of aprotinin per ml, 5 mM PMSF) and adjusted to 0.1% Nonidet P-40–40 mM KCl–5% glycerol. When appropriate, unlabeled annealed oligonucleotides (10 ng) were included as competitors (50-fold molar excess over the probe). This mixture was incubated at room temperature for 10 min prior to the addition of 1 μl of probe (200 pg; 2 × 104 to 9 × 104 cpm). After a further 20 min at room temperature, the reaction mixture was electrophoresed through a native 4% bisacrylamide (bis-acrylamide ratio, 1:29)–2.5% glycerol–0.5× Tris-borate-EDTA (TBE) gel with recirculating 0.5× TBE at 180 V and 4°C for 2.75 h. For supershift analysis, 1 μl of diluted (1:5) anti-FLAG monoclonal antibody (Kodak) was incubated at room temperature for 20 min following the probe incubation.
FIG. 2.
Regions of the USE used as probes and competitors for electrophoretic mobility shift assays. The USE was subdivided into four overlapping regions, A, B, C, and D. These were further subdivided as shown. Sequences similar to those of MEF2 [CTA(A/T)4TA(G/A)], the CCAC-box (CCCACCC), the E-box (CANNTG), the Ets motif [(C/A)(C/A)GGA(A/T)], and overlapping Inr consensus [(T/C)(T/C)AN(T/A)(T/C)(T/C)] binding sites are indicated in bold. The 3-bp mutations introduced to delineate protein binding sites within USE B1 and USE C1 are underlined. Sequence numbering is relative to the transcription initiation site (+1).
Direct muscle injection.
The injection, extraction, and assay of the reporter proteins were performed as described previously (9). Briefly, 100 μg of luciferase reporter plasmid and 140 μg of internal control pUCoriSISCAT plasmid were injected into the soleus and EDL muscles of 6- to 8-week-old Sprague-Dawley rats. After 5 days, the rats were sacrificed, and soleus and EDL muscle extracts were assayed for luciferase and chloramphenicol acetyltransferase activities. For each muscle, variation in the efficiency of DNA uptake was controlled by normalizing the luciferase activity to the chloramphenicol acetyltransferase activity.
Transgenic mouse production, β-galactosidase assays, and histochemistry.
Transgenic mice were generated by standard methods (16) as described previously (21). At the appropriate age, F1-generation mice were sacrificed, and the soleus and EDL muscles were collected. The muscles of one leg were snap frozen in liquid nitrogen for β-galactosidase assays, while the muscles of the other leg were prepared for sectioning by coating with tissue freezing medium (Triangle Biomedical Sciences) prior to freezing. After screening, transgene-positive muscles were powdered on liquid nitrogen with a steel slide ram. Tissue extracts were prepared by lysis in detergent lysis solution (100 mM potassium phosphate [pH 7.8], 1 mM DTT, 0.2% Triton X-100) for 30 min on ice prior to freezing at −80°C. After thawing on ice, cell debris was removed by centrifugation in a microcentrifuge, and the protein concentration was determined (7). β-Galactosidase activity in 20 μg of extracted protein was assayed with a β-galactosidase chemiluminescence detection kit (Clontech) and a Turner Designs 20/20 luminometer. Sections (20 μm) were prepared and stained for β-galactosidase and type 1 MyHC as described previously (9).
cDNA library screening.
The yeast one-hybrid system was used to screen a human quadriceps muscle matchmaker cDNA library (Clontech). A dual-reporter yeast strain (YM4271) was created by stably integrating HIS3 and lacZ reporter genes (derived from pLacZi and pHISi), each with a minimal promoter adjacent to a sequence comprising three tandem repeats of the USE B1 element, AGCCACAGGATTAACATA (see Fig. 2). This strain was transformed with a human quadriceps muscle cDNA library which was constructed in a yeast expression vector (pGAD10). This vector expressed the encoded muscle proteins as fusions with the GAL4 activation domain. Muscle proteins which interacted with the USE B1 element thereby recruited the GAL4 activation domain so as to activate the reporter genes and allow selection with histidine-deficient media and a standard β-galactosidase assay. False-positive clones were identified by their ability to maintain activation of the reporter genes in a dual-reporter yeast strain containing the nonbinding USE B1b sequence, AGCCACAGGATATCCATA (see Fig. 2), as the tandem repeat.
Northern blot analysis.
A multiple-tissue Northern blot (Clontech) of human poly(A)+ RNA (2 μg/lane) was hybridized at 68°C with a randomly primed probe derived from a BamHI fragment (nucleotide positions 31 to 330) of the MusTRD1 cDNA clone.
In vitro translation and epitope tagging.
The coding region of the MusTRD1 cDNA was excised with EcoRI (pGAD10 polylinker) and BstEII (nucleotide position 1564; blunt ended) and subcloned into EcoRI/EcoRV of pcDNA3.1(+) (Invitrogen). Epitope tagging of MusTRD1 was accomplished with a synthetic oligonucleotide carrying the Met-FLAG epitope (MDYKDDDDK) fused to the second codon of MusTRD1. In vitro translations of MusTRD1 and FLAG-tagged MusTRD1 were performed with a TNT T7-coupled rabbit reticulocyte lysate system (Promega).
RESULTS
Deletion analysis of the USE in vivo.
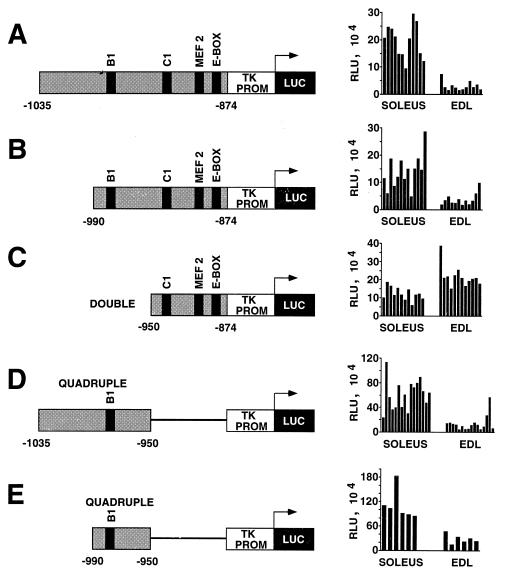
Deletion analysis of the USE by the direct injection assay was used to further dissect the enhancer with the aim of identifying sequence elements involved in slow-fiber expression. We subcloned various portions of the enhancer upstream of a luciferase reporter gene under the control of a TK minimal promoter. The plasmid constructs were injected into two rat muscles: the slow-fiber-rich soleus muscle (77 to 96% slow fibers) and the fast-fiber-rich EDL muscle (92 to 98% fast fibers) (2, 3, 40). Confirming our earlier report (9), the full-length USE (bp −1035 to −874) directed high-level luciferase activity in the soleus muscle as opposed to the EDL muscle (average ratio of luciferase activity in soleus versus EDL muscles, 7.0:1.0), reflecting its slow-fiber specificity (Fig. 1A). As shown in Fig. 1B, preferential slow-fiber expression remained despite the removal of 45 bp from the 5′ end of the USE (soleus/EDL muscle ratio, 3.9:1.0). However, a further 5′ deletion to bp −950 reduced the activity of the reporter construct to background activity (5.9 × 104 ± 0.68 × 104 relative light units [RLU]). Significant activity was restored by duplicating bp −950 to −874 in the reporter construct, but the preferential slow-fiber activity was lost (Fig. 1C). In fact, in this case, slightly higher levels of activity were observed in the EDL muscle than in the soleus muscle (soleus/EDL muscle ratio, 0.6:1.0). As these results suggested that the 5′ region of the USE was important for maintaining high levels of preferential slow-fiber activity, reporter constructs containing either bp −1035 to −950 or bp −990 to −950 were tested (Fig. 1D and E, respectively). Only background activity was obtained with these plasmids (data not shown); however, once again, multimerization (quadruple) restored significant activity. Furthermore, for both constructs, the activity showed a preference for slow fibers (soleus/EDL muscle ratio, >4.0:1.0), suggesting that sequences between −990 and −950 are sufficient to confer slow-fiber specificity. As described below, it is within this region that we have identified an essential protein binding element (USE B1) and a corresponding binding protein (MusTRD1).
FIG. 1.
Deletion analysis of the USE by the direct injection of reporter gene constructs into rat soleus and EDL muscles. Portions of the USE (shaded) were subcloned into a luciferase reporter vector (LUC) with a TK minimal promoter (TK PROM). For C, D, and E, the USE component was ligated in tandem as a doublet or quadruplet. The constructs were injected into rat soleus and EDL muscles, and luciferase activities (RLU) were determined. For each construct, the RLU values represent the normalized values for muscles which had been injected on the same day and assayed 5 days later in parallel. Each bar represents the value obtained from an individual muscle sample. The relative positions of nuclear protein binding sites within the USE are indicated. Sequence numbering is relative to the transcription initiation site (+1).
Identification of protein binding sites within the USE.
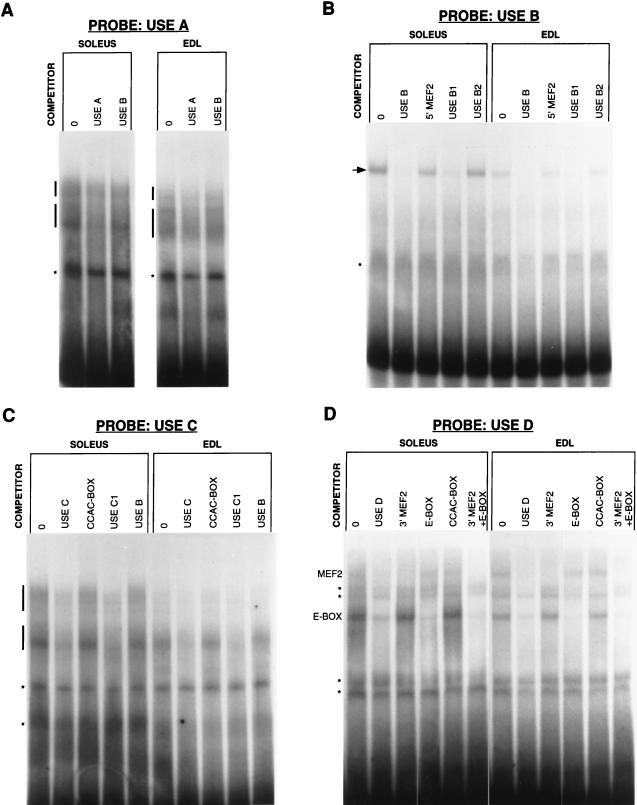
Sequence analysis of the USE identified two consensus-like binding sites for the MEF2 family, an E-box (consensus binding site for the myogenic bHLH proteins), an Ets motif [(C/A)(C/A)GGA(A/T)] (39), an overlapping Inr-like element [(T/C)(T/C)AN(T/A)(T/C)(T/C)] (18), and a CCAC-box (CCCACCC) (Fig. 2). The Ets motif is a binding site for Ets domain proteins, which form ternary complexes with proteins binding to nearby serum response elements (39). The Inr element is a TATA-box analogue that acts as a core transcription initiating element, although other roles have been envisaged with the discovery of novel Inr binding proteins (34). The CCAC-box has been identified as an important element for the transcriptional activation of the slow/cardiac troponin C (27) and myoglobin (4, 5) genes in muscle. To determine whether these and/or other elements within the USE participate in the binding of muscle nuclear proteins, we first divided the USE into four regions, A to D, based on the regions assessed by the direct-injection analysis (Fig. 2). Oligonucleotides corresponding to USE A to USE D were synthesized, annealed, and used for electrophoretic mobility shift assays with nuclear extracts from rat slow (soleus)- and fast (EDL)-fiber-containing muscles. USE A to USE D overlapped each other by 10 bp to minimize the chance of destroying a potential protein binding site. Subfragments of these regions, shown in Fig. 2, were used as competitors and subsequently as probes in electrophoretic mobility shift assays; by a process of elimination, various binding elements were determined.
Despite a high background signal with the USE A probe, two broad complexes were evident in the soleus muscle nuclear extract, while three were detected in the EDL muscle extract (Fig. 3A). Although difficult to resolve, these complexes appeared to be specific for USE A, since their intensity diminished with an excess of unlabeled USE A as a specific competitor compared with USE B as a nonspecific competitor. The lowest complex was unique to EDL muscle. However, given that the direct-injection data indicated that the region corresponding to USE A was dispensable for slow-fiber specificity, we did not investigate this region further.
FIG. 3.
Electrophoretic mobility shift analysis of the USE. USE regions A, B, C, and D (A to D, respectively) were used as probes in binding reactions with nuclear protein extracts derived from rat soleus or EDL muscles. When needed, unlabeled competitors were included at a 50-fold molar excess over the probe. Nonspecific binding is indicated by asterisks, while sequence-specific complexes are indicated by vertical bars and the arrow. MEF2 and E-box complexes are indicated in D. Lanes 0, no competitor.
In contrast to the USE A probe, the USE B probe produced a relatively clean gel shift, revealing a single complex in both soleus and EDL muscles (Fig. 3B). Given that the direct-injection data indicated that this region may contain the sequences conferring slow-fiber specificity, it is noteworthy that this complex was more abundant in soleus muscle nuclear extracts than in EDL muscle nuclear extracts and may reflect a slow-fiber-specific factor. This complex was specifically inhibited by an excess of unlabeled USE B. Within USE B is a sequence resembling a MEF2 consensus binding site. To determine whether the complex binds this sequence or other sequences in USE B, subregions of USE B were used as competitors. 5′ MEF2 and USE B2 were unable to compete for complex formation, while USE B1 was an effective competitor, indicating that USE B1 spans the binding site.
Two sets of specific complexes bound USE C (Fig. 3C), with a stronger signal detected for the upper set in soleus muscle nuclear extracts. The CCAC-box and USE B (as a nonspecific competitor) both failed to compete for binding, unlike USE C1 (and USE C), indicating that both sets of complexes bind to sites within USE C1.
The E-box and 3′ MEF2 were both effective competitors for soleus and EDL muscle nuclear extract proteins bound to USE D (Fig. 3D). The faint upper complex (which was more apparent over the background in the EDL muscle) was inhibited by 3′ MEF2 and probably represents binding by MEF2 proteins. The lower complex, which was more pronounced in the soleus muscle, was inhibited by the E-box and most likely reflects binding by the myogenic bHLH proteins.
Point mutations that eliminate protein binding to USE B1 and USE C1.
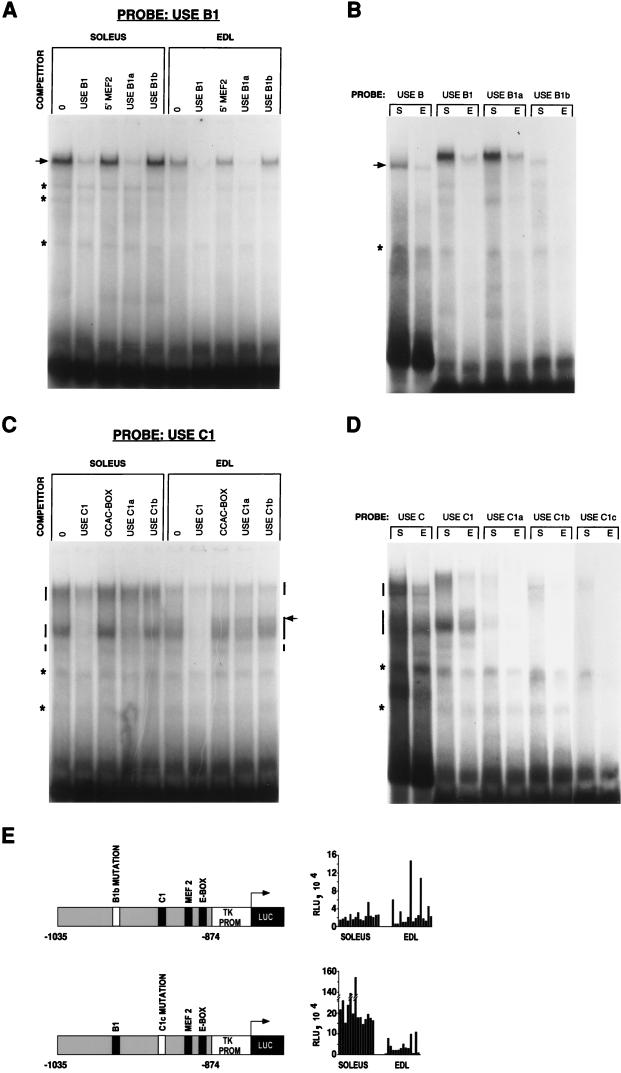
In order to examine the influence of the USE B1 and USE C1 elements in vivo, we first defined specific nucleotides necessary for protein binding for future site-specific mutation analysis. Allowing for the redundancy in most transcription factor binding sites, 3-bp substitutions were introduced into USE B1 (B1a and B1b) and USE C1 (C1a, C1b, and C1c) (Fig. 2). The substitutions in USE B1a disrupted the core [GGA(A/T)] of the Ets motif (38), while the substitutions in USE B1b disrupted the Inr-like element (18).
Confirming our previous results, USE B1 bound an abundant specific complex which was inhibited by unlabeled USE B1 (specific competitor) but not by 5′ MEF2 (nonspecific competitor) (Fig. 4A). USE B1b, unlike USE B1 or USE B1a, was incapable of competing for protein binding, indicating that the 3-bp substitution in USE B1b spanned an important protein binding determinant. This result was confirmed by use of the USE B1 mutations as probes rather than competitors (Fig. 4B). USE B, USE B1, and USE B1a all bound a single abundant complex from both soleus and EDL muscle nuclear extracts. As indicated previously, this complex was more prevalent in soleus muscle than in EDL muscle. Given that binding activity was retained by USE B1a, despite its disrupted Ets motif, it is unlikely that Ets domain proteins can account for USE B1 binding. Significantly, the USE B1b probe possessed negligible binding activity, proving that the 3-bp substitution in this probe would be a viable means by which to assess the influence of this region in vivo. Surprisingly, the complex migrated more slowly with the shorter probes than with the longer USE B probe. A similar phenomenon was observed with the USE C1 probe (see below), and we believe this to be a property of the nondenaturing gels not accurately reflecting the true molecular weight of the complex. For instance, DNA bending is known to influence the migration of DNA-protein complexes in these gels (12).
FIG. 4.
Identification of protein binding sites within USE B and USE C and the influence of nucleotide substitutions in vivo. (A and C) Electrophoretic mobility shift assays were performed with the USE B1 and USE C1 probes and the indicated competitors. Lanes 0, no competitors. (B and D) Binding reactions were performed with soleus (S) and EDL (E) nuclear extracts and the relevant probes in the absence of specific competitors. In A and B, the sequence-specific complex is indicated by an arrow. In C and D, the sequence-specific complexes are indicated by vertical bars. A complex unique to EDL muscle is indicated by an arrow in C. Nonspecific binding is indicated by asterisks. (E) Reporter gene constructs identical to those used in Fig. 1A (see the legend to Fig. 1A for details) but differing only with respect to the introduction of a USE B1b or USE C1c mutation were directly injected into rat soleus and EDL muscles and assayed for luciferase expression (RLU). The introduction of the USE B1b mutation eliminated preferential slow (soleus)-muscle activity.
Confirming that USE C protein binding activity resides within USE C1, two broad specific complexes were evident with the USE C1 probe. A smaller, less abundant specific complex was also resolved with this probe (Fig. 4C). Once again, the upper complex was more abundant in the soleus muscle than in the EDL muscle and may reflect the binding of slow-muscle-specific factors. Interestingly, an additional band in the lower complex was unique to EDL muscle extracts and may represent a fast-muscle-specific transcriptional activator or repressor. Specific binding was determined by competition with unlabeled USE C1, in comparison to the CCAC-box as a nonspecific competitor. The 3-bp mutation in USE C1a spanned a binding determinant for the upper complex, as it was unable to compete for upper-complex binding, although it was still capable of competing for lower-complex binding. The 3-bp mutation in USE C1b eliminated competition for both the upper and the lower complexes, indicating that this 3-bp substitution spanned a binding determinant for both complexes. To verify these findings, mobility shift assays were performed with probes for the USE C1 mutations (Fig. 4D). USE C1a possessed minimal binding activity for the upper complex, as expected, but was also a weak binder of the lower complex, indicating that the 3-bp substitution unique to USE C1a spanned an important binding determinant for both complexes. Binding of both complexes was completely lost with USE C1b; however, some new binding activity became apparent. To eliminate this new binding activity observed with USE C1b, another probe, USE C1c, containing a different 3-bp mutation in the same location as that in USE C1b, was examined and possessed negligible binding activity.
In vivo analysis of USE B1 and USE C1 suggests that only USE B1 plays a significant role in reporter gene activation.
The introduction of the mutations shown to obliterate protein binding in the mobility shift assays revealed the importance of USE B1. Preferential slow-muscle activity was completely lost with a reporter plasmid containing the 3-bp mutation of USE B1b (Fig. 4E). Only low-level activity was detected in the soleus muscle, approximating that in the EDL muscle. This finding is in agreement with our earlier direct-injection data, whereby the 5′ deletion of bp −990 to −950, which includes USE B1, reduced reporter levels to background levels. In contrast, a reporter plasmid containing the 3-bp mutation of USE C1c maintained preferential slow-muscle activity (soleus/EDL muscle ratio, 9.8:1.0) similar to that in the wild type.
One of the major difficulties inherent in the identification of fiber-type determinants by deletion or mutation analysis of promoters or enhancers is distinguishing true fiber-type-determining elements from general elements necessary for high-level activity only. Attempting to address this problem, we generated transgenic lines of mice with a nuclear lacZ reporter gene under the control of the wild-type USE (TnIsUSE-95X1nucZ), USE B1b (TnIsΔB1-USE-95X1nucZ), and USE C1c (TnIsΔC1-USE-95X1nucZ). These were analyzed not only at the tissue extract level but also at the individual fiber level.
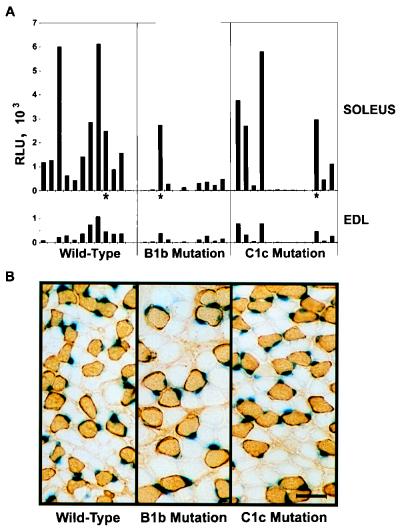
Two transgenic lines for the wild-type construct have been described previously (9) and were shown to express the reporter in a slow-fiber-restricted manner. More transgenic lines were generated, and β-galactosidase expression in the soleus and EDL muscles was assayed. These transgenic lines were compared with similar transgenic lines differing only with respect to the introduction of the USE B1b and USE C1c mutations (Fig. 5A). High-level expression in the soleus muscle compared to the EDL muscle was well established in the 2-week-old wild-type transgenic lines. This result agrees with the establishment of fiber type by this age with respect to endogenous TnIs and TnIf (46). With the Wilcoxin rank-sum test (33, 41), a statistical test for two independent sets of observations which are not normally distributed (appropriate for transgenic lines), expression from the USE B1b-carrying transgene in soleus (P < 0.05) and EDL (P < 0.1) muscles was significantly different from that of the wild type. In contrast, soleus and EDL muscle expression of the USE C1c-carrying transgene was not statistically different from that of the wild type. These results are consistent with our direct-injection data showing that the USE B1b mutation had a profound negative impact on reporter gene expression, unlike the USE C1c mutation. Although not statistically significant, the USE C1c mutation may have had some impact on reporter gene activity, with 6 of the 13 transgenic lines expressing the genes at levels below 100 RLU in soleus muscle.
FIG. 5.
Transgenic line analysis confirms the importance of the region delineated by the USE B1b mutation with respect to high-level expression. (A) Soleus and EDL muscle extracts derived from transgenic lines carrying wild-type USE, USE B1b, or USE C1c were assayed for β-galactosidase activity (RLU). Each bar represents the value obtained from an individual transgenic line, with the EDL muscle value for a given animal being positioned directly below the corresponding value for the soleus muscle. Asterisks indicate the lines sectioned in panel B. (B) Slow-fiber specificity is maintained in transgenic lines containing either the USE B1b or the USE C1c mutation and showing high-level expression. Cross sections of the contralateral soleus muscle for three transgenic animals expressing similar levels of β-galactosidase (asterisks in panel A) were found positive for type 1 (slow) MyHC (immunoperoxidase stain) and were stained for nuclear β-galactosidase activity. Scale bar, 50 μm.
To determine whether either of the mutations had an influence on skeletal muscle-specific expression, tissues (soleus muscle, liver, lung, heart, kidney, and brain) were collected from adult mice of a high-expression line (based upon the results of Fig. 5A) for each of the constructs. Equivalent amounts of tissue extract were assayed for β-galactosidase activity, and for all three transgenic lines, expression in the liver, lung, heart, kidney, and brain was <0.9% that in the soleus muscle, indicating that skeletal muscle-specific expression was being maintained.
Interestingly, for all three transgenic lines, the β-galactosidase levels were higher in soleus muscle than in EDL muscle, even when the expression levels were low (Fig. 5A). While reporter gene expression was undoubtedly compromised, at least by the USE B1b mutation, the preferential expression in the soleus muscle suggested that fiber specificity was still being maintained. To address this issue further, we examined the expression of the reporter gene at the single-fiber level. Although β-galactosidase staining was absent or barely detectable for many of the lines carrying either the USE B1b mutation or the USE C1c mutation (data not shown), fibers which did have β-galactosidase-positive nuclei were almost always of the slow-fiber type (positive for type 1 MyHC). This finding was most obvious in the USE B1b line with the highest expression. As shown in Fig. 5B, this line and a USE C1c line with a similar level of reporter gene activity expressed nuclear β-galactosidase in a slow-fiber-specific manner, similar to the wild type. Thus, while transgenic analysis of the USE B1b mutation within the region from bp −1035 to −874 of the human TnIs USE indicated a severe decrease in expression within the soleus muscle, this mutation did not completely abolish expression in slow, type 1 fibers. These results highlight the importance of testing numerous animals, using sensitive reporter assays, and examining expression at the single-fiber level so as to distinguish whether only general enhancement rather than fiber specificity has been affected.
MusTRD1 is a novel USE B1 binding protein which is expressed predominantly in skeletal muscle.
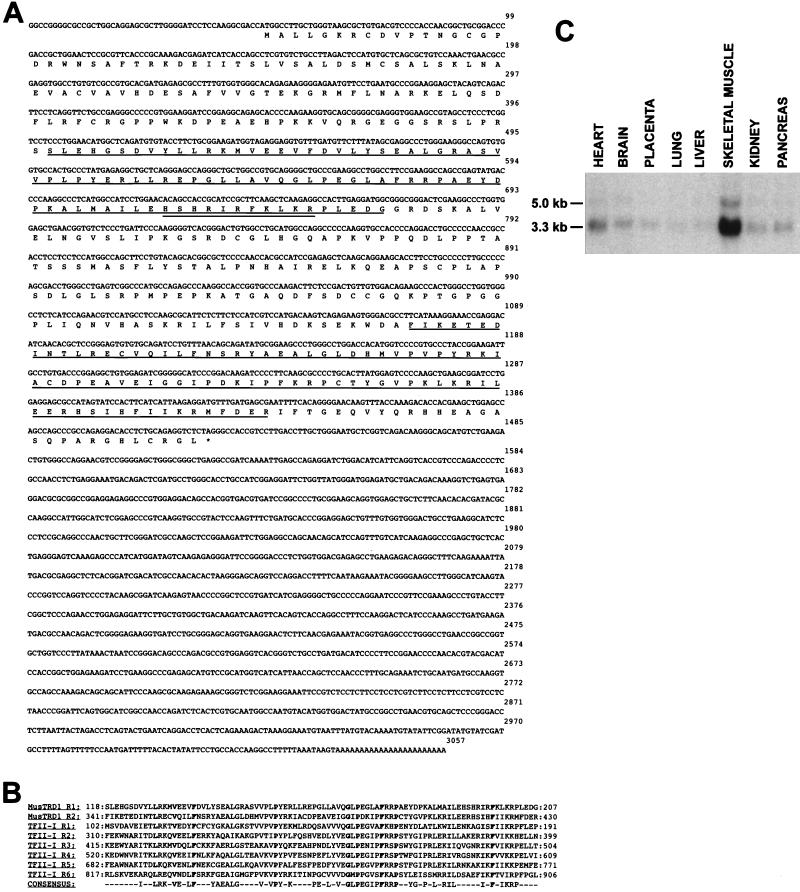
Given the pronounced influence of the USE B1 element on enhancer activity, we used the yeast one-hybrid system (42) to screen a human quadriceps muscle cDNA library for a USE B1 binding protein. Following the transformation of a dual-reporter yeast strain (in which the reporter genes were under the control of a triple repeat of the USE B1 element), 13 positive clones were selected from a screen of approximately 5 × 106 cDNA clones. Of these, two were selected based on their inability to activate reporter genes with three tandem repeats of the USE B1b element, the element with the 3-bp substitution shown to eliminate protein binding. Sequencing revealed them to be identical and novel, with an open reading frame of 458 amino acids encoding a predicted 51-kDa protein (Fig. 6A) which we have termed MusTRD1, based on its homology to TFII-I (see below).
FIG. 6.
MusTRD1 sequence and expression profile. (A) The MusTRD1 cDNA open reading frame of 458 amino acids predicts a 51-kDa protein. The amino acid repeat domains are underlined, and a region rich in basic residues is doubly underlined. (B) Alignment of the two repeat domains of MusTRD1 with the six domains of TFII-I. A consensus based on the conservation of amino acids in at least four of the eight repeats, with a conserved residue in both MusTRD1 and TFII-I proteins, is presented. Amino acids conserved in all eight repeats are shown in bold. (C) Northern blot analysis of human poly(A)+ RNA (2 μg/lane) probed with a MusTRD1 fragment identifies a predominant 3.3-kb skeletal muscle transcript.
Interestingly, MusTRD1 has a repeat domain in its amino- and carboxy-terminal halves which is very similar to a six-repeat domain first described for BAP-135 (Fig. 6B), a target for Bruton’s tyrosine kinase (44). BAP-135 has subsequently been shown to represent the multifunctional DNA binding protein SPIN or TFII-I (13, 30). To the best of our knowledge, MusTRD1 and TFII-I are the first proteins known to share this as-yet-uncharacterized conserved domain. A region rich in basic residues exists within one of the conserved domains of MusTRD1 (amino acids 192 to 202) which could be involved in DNA binding. With a probe outside the conserved domains, Northern blot analysis of eight human tissues revealed that the expression of MusTRD1 was largely restricted to skeletal muscle, unlike the more ubiquitous TFII-I (Fig. 6C). Low-level expression was, however, evident in all tissues upon prolonged exposure, particularly in the heart. The predominant MusTRD1 transcript migrated at a position of approximately 3.3 kb, indicating that our cDNA clone was close to full length. A less abundant, 5-kb transcript was also visible.
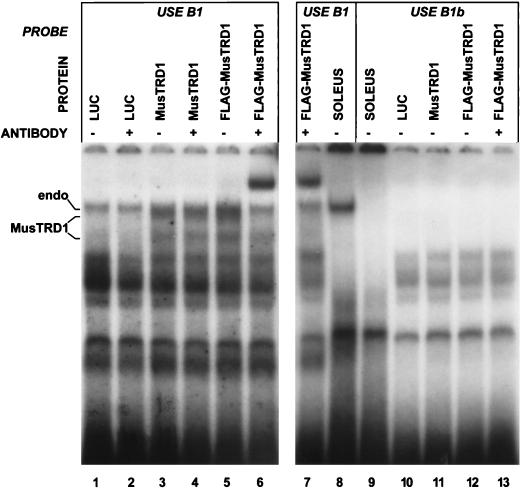
Mobility shift analysis of in vitro-translated MusTRD1 confirmed the DNA binding specificity inherent in the cDNA library screening strategy (Fig. 7). The luciferase in vitro translation control reaction revealed that the rabbit reticulocyte lysate had endogenous USE B1 DNA binding activity (Fig. 7, lanes 1 and 2). Two additional complexes became apparent with in vitro-translated MusTRD1, the uppermost complex migrating at a position similar to that of the upper endogenous complex (lanes 3 and 4). To resolve endogenous reticulocyte binding activity from MusTRD1 binding activity, a FLAG epitope tag was fused to the amino terminus of MusTRD1 (lane 5). Upon the addition of an anti-FLAG monoclonal antibody, only the uppermost complex was supershifted (lane 6). This complex could reflect the interaction of MusTRD1 with itself or other proteins in a manner more favorable for antibody binding. Both the upper endogenous complex and the MusTRD1 complex were dependent upon the integrity of the USE B1 sequence, as neither bound to USE B1b (lanes 10 to 13), displaying specificities and mobilities similar to those of the soleus muscle extract-derived complex (lanes 8 and 9).
FIG. 7.
In vitro-translated MusTRD1 mimics soleus muscle extract binding to USE B1 with respect to specificity and mobility. Electrophoretic mobility shift assays were performed with in vitro-translated firefly luciferase (LUC), MusTRD1, FLAG-tagged MusTRD1 (FLAG-MusTRD1), and soleus muscle nuclear extract. Reactions were performed in the absence (−) or presence (+) of the monoclonal antibody for the FLAG epitope. USE B1-specific binding by MusTRD1 was determined; in comparison, binding with the USE B1b probe was absent. The endogenous rabbit reticulocyte lysate binding complex (endo) which migrates near the MusTRD1 complex (MusTRD1) is indicated. All other complexes appear to represent endogenous reticulocyte lysate binding activities.
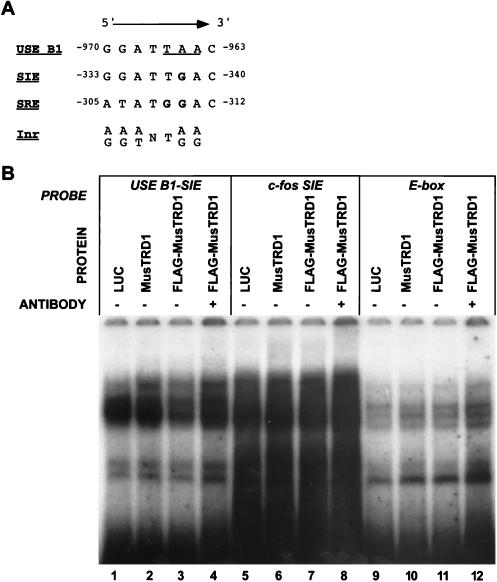
TFII-I has been shown to bind a number of related elements, including SIE and SRE of the c-fos promoter and the Inr element of the adenovirus major late promoter (13, 30). Given the presence of the conserved domain, it seems more than coincidental that MusTRD1 and TFII-I also bind similar DNA elements (Fig. 8A). With the exception of a single nucleotide, USE B1 shows complete identity with the core binding region of c-fos SIE. Interestingly, the differing nucleotide lies within the 3-bp region of USE B1b shown to eliminate binding and corresponds to a guanosine in SIE; in a dimethylsulfate interference assay, this guanosine has been implicated as making direct contact with TFII-I (13). To test whether MusTRD1 exhibits the same DNA binding site requirements as TFII-I, we substituted G for A in USE B1 (nucleotide −965) so as to mimic an SIE. Surprisingly, this single base substitution eliminated all binding of MusTRD1 (Fig. 8B, lanes 2, 3, and 4), as all complexes could be accounted for in the luciferase control reaction (lane 1), and the anti-FLAG antibody failed to produce a supershift (lane 4) like that seen with wild-type USE B1 (Fig. 7, lanes 6 and 7). Similarly, MusTRD1 failed to bind the c-fos SIE probe (Fig. 8B, lanes 6, 7, and 8). TFII-I has also been shown to bind to an upstream E-box of the adenovirus major late promoter (30), an intriguing finding with respect to the regulation of the TnIs gene given the importance of E-boxes in muscle gene regulation. However, examination of the TnIs USE E-box revealed that in vitro-translated MusTRD1 was not bound (Fig. 8B, lanes 10, 11, and 12). Although we cannot rule out the possibility that the native MusTRD1 protein may behave differently, these results indicate that MusTRD1 and TFII-I have similar but distinct DNA binding elements which conform with the Inr consensus sequence.
FIG. 8.
Comparison of the MusTRD1 binding element with TFII-I binding elements. (A) Alignment of the antisense strands for the core binding sites of c-fos SIE, c-fos SRE, and the Inr consensus (TFII-I binding elements) with part of USE B1 (MusTRD1 binding element). The nucleotides substituted in the nonbinding USE B1b sequence are underlined. Nucleotides predicted to contact TFII-I (13) are shown in bold. (B) Electrophoretic mobility shift assays were performed with in vitro-translated firefly luciferase (LUC), MusTRD1, and FLAG-tagged MusTRD1 (FLAG-MusTRD1). Probes included USE B1 with a G-for-A substitution at nucleotide −965 so as to mimic the SIE core sequence (lanes 1 to 4); c-fos SIE (lanes 5 to 8); and the TnIs USE E-box (lanes 9 to 12). Reactions were performed in the absence (−) or presence (+) of the monoclonal antibody for the FLAG epitope. MusTRD1 was unable to bind any of these elements.
DISCUSSION
The identity of the molecular mechanisms responsible for controlling muscle-fiber-type-specific gene expression has proved elusive due, in no small part, to the fact that the experimental approaches rely on in vivo models, usually transgenic lines. Transgenic studies on the regulatory regions of some highly contractile protein isoforms have indicated that fast-fiber-specific gene expression may be quite complex. Characterization of transgenic lines for the rat myosin light chain 1 fast (MLC-1f) (10), the mouse MLC-3f (20), and the quail TnIf (15) genes revealed preferential expression within type 2 fibers in the order MyHC-2B > MyHC-2X > MyHC-2A. It has been postulated that such expression reflects the existence of distinct regulatory mechanisms within fast-fiber subtypes (15). Subsequent work on the expression patterns observed for the MLC-1f transgenic line supports the concept that overall expression patterns are the consequence of cumulative activities directed by discrete sequence elements (29). A recent transgenic study on the human aldolase A pM promoter indicated that a minimal element containing only a MEF3 binding site and an overlapping MEF2-NFI binding site directed reporter gene expression to a subset of fast-twitch muscles (35), unlike the broad fast-muscle activity of larger promoter constructs, lending further support to the idea that expression in all fast fibers results from the combination of discrete fast-fiber-subtype-specific elements (36). Aberrant expression from transgenes in fast fibers could therefore be due to the absence of one or more of these elements in transgenes.
In contrast to the complexities being revealed for fast-fiber-specific gene expression, slow-fiber-specific gene expression may be simpler. In this regard, the TnIs gene is proving to be a convenient model with which to decipher at least one of the mechanisms responsible for directing fiber-specific gene expression. The small sizes of the slow-fiber-specific USE in the human gene (9) and SURE (slow upstream regulatory element) in the rat gene (24) have enabled us to map the protein binding sites by mobility shift assays. The functional importance of these protein binding sites has been assessed by the direct injection of reporter constructs into muscle prior to confirmation in transgenic lines.
Our direct-injection data revealed that the sequence from bp −990 to −874 of the USE is sufficient for slow-muscle activity. We showed that MEF2 and E-box elements within the USE D region bound proteins derived from muscle tissue nuclear extracts, although binding to the MEF2 element was weak. Weak MEF2 binding activity in extracts derived from rat muscle has also been shown for the MEF2 consensus site of the aldolase A pM promoter (36). Interestingly, binding to the E-box was more pronounced in soleus muscle extracts than in EDL muscle extracts. This finding could reflect the differential expression of E-box binding proteins in soleus muscle compared to EDL muscle. Analysis of the muscle creatine kinase gene has implicated different roles for E-boxes in slow- versus fast-muscle-fiber types (33). Furthermore, it has been shown that sequences both within and flanking the consensus E-box can influence its binding or transcriptional activity (1, 6, 43, 45). Therefore, the context of the USE E-box could also favor the binding of soleus muscle (slow)-specific proteins over that of proteins predominant in EDL muscle. In C2 myotubes, a reporter construct in which 20 bp of the USE 3′ terminus had been deleted (a portion which includes the E-box) expressed only 5% activity compared to the full-length construct (8). The role of the E-box element in the context of the USE in vivo remains to be determined. Combinations of factors, including members of the bHLH and MEF2 families, which can bind to this element either directly or indirectly will make it difficult to determine which, if any, of these factors plays a role in determining fiber specificity or simply muscle specificity or enhancement. This determination will be further confounded by their autoregulatory capabilities.
We showed the USE B1 element to be essential for high-level reporter activity in both transgenic and direct-injection experiments. Analysis of the transgenes at the single-fiber level suggested that although reporter gene activity was severely compromised by the introduction of the USE B1b mutation, slow-fiber specificity was still maintained. This result was somewhat surprising given that the direct-injection data suggested that the region from bp −990 to −950 contained the sequence determinants for slow-fiber specificity (Fig. 1E) and that USE B1 accounted for all obvious binding within this region. It could be argued that the 3-bp mutation is not extensive enough to eliminate all USE B1 binding, as some residual binding activity was apparent with the USE B1b probe (Fig. 4B). In the few USE B1b-carrying transgenic lines which maintained appreciable reporter gene activity, such residual binding might be all that is required to maintain slow-fiber specificity. We believe that a more likely explanation is that slow-fiber specificity may not depend on the binding of any single transcription factor but rather on the binding of a combination of factors. One or more of these may be posttranslationally modified in particular fiber types and/or interact with other accessory proteins to direct slow-fiber-specific gene expression. Protein-protein interactions could obviate the absolute requirement for the presence of a high-affinity DNA binding site in order to maintain slow-fiber specificity.
The discovery of MusTRD1 as a USE B1 binding protein is interesting with respect to the above hypothesis of protein-protein interactions, given its homology to TFII-I. TFII-I appears to be quite promiscuous in its choice of both DNA element and protein partner. It has been shown to bind to Inr, Inr-like (SIE and SRE), and E-box elements and to interact with a serum response factor (a MADS box family transcription factor), Phox (a homeodomain protein), and USF1 (a bHLH factor) (13, 30). It is not difficult to envisage that the six 90-amino-acid repeat domains of TFII-I play a significant part in the proposed role of TFII-I as a coordinator of diverse cell signaling responses and the basal transcription machinery. It is reasonable to speculate that the conservation of such domains in MusTRD1 may allow it to interact with other components assembling on the TnIs enhancer and promoter, particularly MEF2 (MADS box family members) and the bHLH myogenic regulatory factors. We have shown that MusTRD1 is expressed predominantly in skeletal muscle and propose that it could play a significant TFII-I-like role in muscle gene regulation. Whether this role includes slow- versus fast-muscle-fiber-specific gene regulation awaits the development of appropriate in vivo models and antibodies.
ACKNOWLEDGMENTS
We thank P. Rowe for encouragement and support. Special thanks are due to X. Badoux, P. Robinson, L. Ferrara, and the animal house team at the Children’s Medical Research Institute for assistance.
This work was supported by grants from the National Health and Medical Research Council of Australia (960775) to J.V.O. and E.C.H. and the National Science Foundation (DCB-9020998), the Muscular Dystrophy Association, and the American Heart Association to R.P.W.
REFERENCES
- 1.Apone S, Hauschka S D. Muscle gene E-box control elements. J Biol Chem. 1995;270:21420–21427. doi: 10.1074/jbc.270.36.21420. [DOI] [PubMed] [Google Scholar]
- 2.Ariano M A, Armstrong R B, Edgerton V R. Hindlimb muscle fiber populations of five mammals. J Histochem Cytochem. 1973;21:51–55. doi: 10.1177/21.1.51. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong R B, Phelps R O. Muscle fiber type composition of the rat hindlimb. Am J Anat. 1984;171:259–272. doi: 10.1002/aja.1001710303. [DOI] [PubMed] [Google Scholar]
- 4.Bassel-Duby R, Hernandez M D, Gonzalez M A, Krueger J K, Williams R S. A 40-kilodalton protein binds specifically to an upstream sequence element essential for muscle-specific transcription of the human myoglobin promoter. Mol Cell Biol. 1992;12:5024–5032. doi: 10.1128/mcb.12.11.5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassel-Duby R, Grohe C M, Jessen M E, Parsons W J, Richardson J A, Chao R, Grayson J, Ring W S, Williams R S. Sequence elements required for transcriptional activity of the human myoglobin promoter in intact myocardium. Circ Res. 1993;73:360–366. doi: 10.1161/01.res.73.2.360. [DOI] [PubMed] [Google Scholar]
- 6.Blackwell T K, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Corin S J, Juhasz O, Zhu L, Conley P, Kedes L, Wade R. Structure and expression of the human slow twitch skeletal muscle troponin I gene. J Biol Chem. 1994;269:10651–10659. [PubMed] [Google Scholar]
- 9.Corin S J, Levitt L K, O’Mahoney J V, Joya J E, Hardeman E C, Wade R P. Delineation of a slow twitch myofiber-specific transcriptional element using in vivo somatic gene transfer. Proc Natl Acad Sci USA. 1995;92:6185–6189. doi: 10.1073/pnas.92.13.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donoghue M J, Alvarez J D, Merlie J P, Sanes J R. Fiber type- and position-dependent expression of a myosin light chain-CAT transgene detected with a novel histochemical stain for CAT. J Cell Biol. 1991;115:423–434. doi: 10.1083/jcb.115.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esser K, Gunning P, Hardeman E. Nerve-dependent and -independent patterns of mRNA expression in regenerating skeletal muscle. Dev Biol. 1993;159:173–183. doi: 10.1006/dbio.1993.1231. [DOI] [PubMed] [Google Scholar]
- 12.Garabedian M J, La Baer J, Liu W-H, Thomas J R. Analysis of protein-DNA interactions. In: Hames B D, Higgins S J, editors. Gene transcription: a practical approach. Oxford, England: Oxford University Press; 1993. pp. 243–293. [Google Scholar]
- 13.Grueneberg D A, Henry R W, Brauer A, Novina C D, Cheriyath V, Roy A L, Gilman M. A multifunctional DNA-binding protein that promotes the formation of serum response factor/homeodomain complexes: identity to TFII-I. Genes Dev. 1997;11:2482–2493. doi: 10.1101/gad.11.19.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn C, Covault J. Isolation of transcriptionally active nuclei from striated muscle using Percoll density gradients. Anal Biochem. 1990;190:193–197. doi: 10.1016/0003-2697(90)90180-h. [DOI] [PubMed] [Google Scholar]
- 15.Hallauer P L, Bradshaw H L, Hastings K E M. Complex fiber-type-specific expression of fast skeletal muscle troponin I gene constructs in transgenic mice. Development. 1993;119:691–701. doi: 10.1242/dev.119.3.691. [DOI] [PubMed] [Google Scholar]
- 16.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the mouse embryo: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. Production of transgenic mice; pp. 217–252. [Google Scholar]
- 17.Hughes S M, Taylor J M, Tapscott S J, Gurley C M, Carter W J, Peterson C A. Selective accumulation of MyoD and myogenin mRNAs in fast and slow adult skeletal muscle is controlled by innervation and hormones. Development. 1993;118:1137–1147. doi: 10.1242/dev.118.4.1137. [DOI] [PubMed] [Google Scholar]
- 18.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale S T. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaushal S, Schneider J W, Nadal-Ginard B, Mahdavi V. Activation of the myogenic lineage by MEF2A, a factor that induces and cooperates with MyoD. Science. 1994;266:1236–1240. doi: 10.1126/science.7973707. [DOI] [PubMed] [Google Scholar]
- 20.Kelly R, Alonso S, Tajbakhsh S, Cossu G, Buckingham M. Myosin light chain 3F regulatory sequences confer regionalized cardiac and skeletal muscle expression in transgenic mice. J Cell Biol. 1995;129:383–396. doi: 10.1083/jcb.129.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levitt L K, O’Mahoney J V, Brennan K J, Joya J E, Zhu L, Wade R P, Hardeman E C. The human troponin I slow promoter directs slow fiber specific expression in transgenic mice. DNA Cell Biol. 1995;14:599–607. doi: 10.1089/dna.1995.14.599. [DOI] [PubMed] [Google Scholar]
- 22.Ludolph D C, Konieczny S F. Transcription factor families: muscling in on the myogenic program. FASEB J. 1995;9:1595–1604. doi: 10.1096/fasebj.9.15.8529839. [DOI] [PubMed] [Google Scholar]
- 23.Molkentin J D, Black B L, Martin J F, Olson E N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama M, Stauffer J, Cheng J, Banerjee-Basu S, Wawrousek E, Buonanno A. Common core sequences are found in skeletal muscle slow- and fast-fiber-type-specific regulatory elements. Mol Cell Biol. 1996;16:2408–2417. doi: 10.1128/mcb.16.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nordeen S K. Luciferase reporter gene vectors for analysis of promoters and enhancers. BioTechniques. 1988;6:454–457. [PubMed] [Google Scholar]
- 26.Olson E N, Perry M, Schulz R A. Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev Biol. 1995;172:2–14. doi: 10.1006/dbio.1995.0002. [DOI] [PubMed] [Google Scholar]
- 27.Parmacek M S, Ip H I, Jung F, Shen T, Martin J F, Vora A J, Olson E N, Leiden J M. A novel myogenic regulatory circuit controls slow/cardiac troponin C gene transcription in skeletal muscle. Mol Cell Biol. 1994;14:1870–1885. doi: 10.1128/mcb.14.3.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pette D, Staron R S. Mammalian skeletal muscle fiber type transitions. Int Rev Cytol. 1997;170:143–223. doi: 10.1016/s0074-7696(08)61622-8. [DOI] [PubMed] [Google Scholar]
- 29.Rao M V, Donoghue M J, Merlie J P, Sanes J R. Distinct regulatory elements control muscle-specific, fiber-type-selective, and axially graded expression of a myosin light-chain gene in transgenic mice. Mol Cell Biol. 1996;16:3909–3922. doi: 10.1128/mcb.16.7.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy A L, Du H, Gregor P D, Novina C D, Martinez E, Roeder R G. Cloning of an Inr- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. EMBO J. 1997;16:7091–7104. doi: 10.1093/emboj/16.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudnicki M A, Jaenisch R. The MyoD family of transcription factors and skeletal myogenesis. Bioessays. 1995;17:203–209. doi: 10.1002/bies.950170306. [DOI] [PubMed] [Google Scholar]
- 32.Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev. 1996;76:371–423. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- 33.Shield M A, Haugen H S, Clegg C H, Hauschka S D. E-box sites and a proximal regulatory region of the muscle creatine kinase gene differentially regulate expression in diverse skeletal muscles and cardiac muscle of transgenic mice. Mol Cell Biol. 1996;16:5058–5068. doi: 10.1128/mcb.16.9.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smale S T. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 35.Spitz F, Salminen M, Demignon J, Kahn A, Daegelen D, Maire P. A combination of MEF3 and NFI proteins activates transcription in a subset of fast-twitch muscles. Mol Cell Biol. 1997;17:656–666. doi: 10.1128/mcb.17.2.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutherland C, Elsom V, Gordon M, Dunwoodie S, Hardeman E. Coordination of skeletal muscle gene expression occurs late in mammalian development. Dev Biol. 1991;146:167–178. doi: 10.1016/0012-1606(91)90457-e. [DOI] [PubMed] [Google Scholar]
- 37.Sutherland C L, Esser K A, Elsom V L, Gordon M L, Hardeman E C. Identification of a program of contractile protein gene expression initiated upon skeletal muscle development. Dev Dynamics. 1993;196:25–36. doi: 10.1002/aja.1001960104. [DOI] [PubMed] [Google Scholar]
- 38.Treisman R, Marais R, Wynne J. Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J. 1992;11:4631–4640. doi: 10.1002/j.1460-2075.1992.tb05565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Treisman R. Ternary complex factors: growth factor regulated transcriptional activators. Curr Opin Genet Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 40.Voytik S L, Przyborski M, Badylak S F, Konieczny S F. Differential expression of muscle regulatory factor genes in normal and denervated adult rat hindlimb muscles. Dev Dynamics. 1993;198:214–224. doi: 10.1002/aja.1001980307. [DOI] [PubMed] [Google Scholar]
- 41.Walpole R E, Myers R H. Non parametric statistics. In: Walpole R E, Myers R H, editors. Probability and statistics for engineers and scientists. 4th ed. New York, N.Y: Macmillan Publishing Company; 1990. pp. 615–645. [Google Scholar]
- 42.Wang M M, Reed R R. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature. 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- 43.Wright W E, Binder M, Funk W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol Cell Biol. 1991;11:4104–4110. doi: 10.1128/mcb.11.8.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang W, Desiderio S. BAP-135, a target for Bruton’s tyrosine kinase in response to B cell receptor engagement. Proc Natl Acad Sci USA. 1997;94:604–609. doi: 10.1073/pnas.94.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yutzey K E, Konieczny S F. Different E-box regulatory sequences are functionally distinct when placed within the context of the troponin I enhancer. Nucleic Acids Res. 1992;20:5105–5113. doi: 10.1093/nar/20.19.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu L, Lyons G E, Joya J E, Hardeman E C, Wade R P. Developmental regulation of troponin I isoform genes in striated muscles of transgenic mice. Dev Biol. 1995;169:487–503. doi: 10.1006/dbio.1995.1163. [DOI] [PubMed] [Google Scholar]