Abstract
For proper male sexual differentiation, anti-Müllerian hormone (AMH) must be tightly regulated during embryonic development to promote regression of the Müllerian duct. However, the molecular mechanisms specifying the onset of AMH in male mammals are not yet clearly defined. A DNA-binding element for the steroidogenic factor 1 (SF-1), a member of the orphan nuclear receptor family, located in the AMH proximal promoter has recently been characterized and demonstrated as being essential for AMH gene activation. However, the requirement for a specific promoter environment for SF-1 activation as well as the presence of conserved cis DNA-binding elements in the AMH promoter suggest that SF-1 is a member of a combinatorial protein-protein and protein-DNA complex. In this study, we demonstrate that the canonical SOX-binding site within the human AMH proximal promoter can bind the transcription factor SOX9, a Sertoli cell factor closely associated with Sertoli cell differentiation and AMH expression. Transfection studies with COS-7 cells revealed that SOX9 can cooperate with SF-1 in this activation process. In vitro and in vivo protein-binding studies indicate that SOX9 and SF-1 interact directly via the SOX9 DNA-binding domain and the SF-1 C-terminal region, respectively. We propose that the two transcription factors SOX9 and SF-1 could both be involved in the expression of the AMH gene, in part as a result of their respective binding to the AMH promoter and in part because of their ability to interact with each other. Our work thus identifies SOX9 as an interaction partner of SF-1 that could be involved in the Sertoli cell-specific expression of AMH during embryogenesis.
In mammals, male sex determination starts by activation of the testis-determining factor gene, SRY, within cells of the supporting cell precursor lineage (15, 44). When produced, SRY protein will then trigger differentiation of these embryonic cells into Sertoli cells. After differentiation, the Sertoli cells will export the male determining signal via the production of a member of the transforming growth factor β family, the anti-Müllerian hormone (AMH), also known as Müllerian inhibitory substance (for a review, see reference 25). AMH production promotes the regression of Müllerian ducts, the anlagen of the female reproductive organs, and thus appears to be critical for establishing the male phenotype. Molecular studies have been performed by several groups to reconstruct the pathway of primary male sex determination initiated by SRY and terminated by AMH secretion by Sertoli cells. Since AMH expression will follow Sertoli cell determination initiated by SRY, one attractive hypothesis was the direct control of AMH expression via the SRY gene product, a high-mobility group (HMG) box containing transcription factor (19–21). However, the time lag between SRY expression and AMH expression (18) and the absence of any transactivation domain in the human SRY protein (8, 9) have led many investigators to refute this hypothesis (13, 14, 43). It has been suggested that other genes in the cascade located downstream of SRY have to fulfil this role. The recent description of additional transcription factors involved in the sex-determining pathway and the compilation of conserved DNA-binding sites located in the AMH promoter sequences from diverse mammalian species have opened new tracks for investigating the control of AMH expression during male embryogenesis.
An important initial finding came from deletion analysis of the AMH promoter region that led to the identification of a 180-bp segment required for correct AMH expression in primary Sertoli cells (43). Characterization and analysis of this region in humans, bovines, mice, and rats (43) indicate that it contains at least two highly conserved sequence elements plus a characteristic TATA box (see Fig. 1A). The proximal element that includes a single estrogen receptor half-site, AGGTCA, is known to interact with a protein designated steroidogenic factor 1 (SF-1), the mammalian homologue of the Drosophila orphan nuclear receptor fushi tarazu factor 1 (FTZ-F1), a factor that regulates expression of the fushi tarazu homeobox gene during early development (23, 28, 33). The functional importance of this conserved SF-1-binding site was supported by several observations such as its high conservation among species, its binding in vitro to purified SF-1 protein, a coincident expression profile between SF-1 and AMH, and the ability of a DNA fragment containing this site to drive the transcription of a reporter gene in a Sertoli cell-specific manner as demonstrated in transgenic animals (13, 43). However, the SF-1 binding site failed to activate gene expression from the AMH promoter in heterologous cells such as HeLa cells (43). This activation was shown to require removal of the ligand-binding domain of the SF-1 protein, suggesting that a Sertoli cell-specific ligand or cofactor must be necessary for SF-1 to fulfil its transcriptional activity.
FIG. 1.
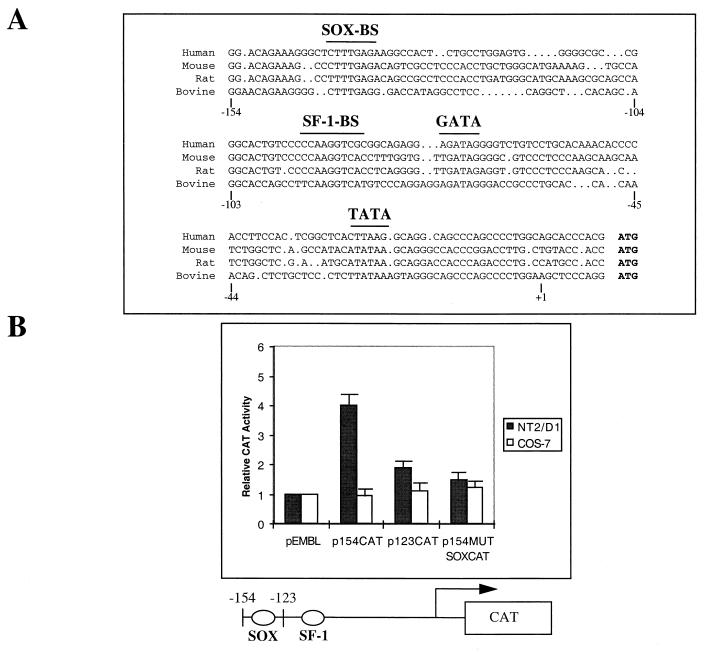
Deletional analysis of the AMH proximal promoter region. (A) Sequence comparison among human, mouse, rat, and bovine AMH proximal promoter regions. The characteristic TATA box, the SF-1-binding site (SF-1-BS), the SOX-binding site (SOX-BS), and a putative GATA site (GATA) that are conserved among the different sequences are indicated. Numbers below the sequences correspond to the human gene. (B) Intrinsic activity of the human AMH proximal promoter in NT2/D1 cells. Various human AMH promoter-CAT reporter gene constructs were transfected in NT2/D1 and COS-7 cells. CAT reporter activities were quantitated after cotransfection of 1 μg of constructs containing either the bp −154 (p154CAT), the bp −123 human AMH promoter (p123CAT), or the bp −154 AMH promoter mutated on the SOX-binding site (p154MUTSOXCAT) and 0.2 μg of pCMV–β-galactosidase. The values shown represent the means of four transfection experiments. Increases in activation are shown relative to pEMBL empty vector. Standard deviations are indicated by bars.
While the existence of a SOX-binding element in the 180-bp minimal promoter led many investigators to postulate the testis-determining factor SRY as a candidate to control AMH expression, rather contradictory results and hypotheses have been produced so far (18, 20, 42, 43, 50). Among the additional sex-determining genes described in recent years that were shown to be expressed concomitant with or shortly after SRY, the SRY-related gene SOX9 is the most attractive candidate to contribute to this control. SOX9 was initially identified by positional cloning as associated with the skeletal malformation syndrome campomelic dysplasia, in which two-thirds of XY individuals show sex reversal (10, 47). The recent detailed analysis of mouse Sox9 expression during gonadal development coincident with Sertoli cell differentiation and its upregulation preceding the onset of AMH expression in mice and chickens are the first arguments to support such a hypothesis (3, 26, 32). Furthermore, unlike human SRY, SOX9 was shown to act as a transcriptional activator during chondrocyte differentiation (2, 30) and to display a high level of protein conservation across vertebrate evolution.
We now address the possibility that the conserved SOX-binding sequence present within the 180-bp proximal promoter region of the human AMH gene is a binding site for SOX9. We demonstrate that SOX9 interacts with this sequence and increases the expression of an AMH promoter/reporter gene construct. We provide evidence for direct protein-protein interaction between SOX9 and SF-1 and for additive activation of the human AMH promoter by these proteins. We also demonstrate that potentiation of SF-1 activity by SOX9 requires both the SOX9 transactivation domain and the SF-1 ligand-binding domain. We conclude that SOX9 and SF-1 have to function in a cooperative manner for the proper control of AMH gene expression during early male gonadal development.
MATERIALS AND METHODS
Cloning and plasmid constructions.
Human SF-1 cDNA was obtained by screening a λZAPII human embryonic cDNA library (24) with a human SF-1-derived genomic 153-bp fragment. This fragment was the result of the reverse transcription-PCR amplification of human embryonic total RNA with the help of primers (forward, 5′-GGTGTCCGGCTACCACTACGG-3′; reverse, 5′-CCAACGCCGACAAGGGACAGC-3′) designed from the human SF-1 partial genomic sequence deposited in GenBank under accession no. U32592. The amplification product was next sequenced to verify its identity to the SF-1 sequence. The SF-1-derived probe was labeled with [α-32P]dCTP by random priming (Megaprime; Amersham) and then used for the screening. SF-1 cDNA-containing phagemids were excised in vivo from the λZAPII vector by coinfection of Escherichia coli XL1-blue cells with VCSM13 helper phage (Stratagene) as specified by the supplier and then fully sequenced. This sequence is deposited in GenBank (accession no. U32591). Human SOX9 cDNA was cloned in pcDNA3 vector (Invitrogen).
Plasmid constructions used in this study are described below. Further details as well as plasmid maps are available upon request.
(i) Yeast expression plasmids.
Full-length or partial SF-1 open reading frames, including the sequences encoding amino acids 1 to 461, 1 to 226, and 223 to 461, were obtained by PCR amplification with the appropriate oligonucleotides containing upstream BamHI sites and downstream SalI sites. PCR products were next cloned into pUC18 (SureClone ligation kit; Pharmacia) and checked by sequencing. The different fragments were then subcloned into the BamHI and SalI sites of pGBT11 as a fusion with the GAL4 DNA-binding domain. pGADGH-SOX9 constructs were obtained by inserting the human SOX9 open reading frame or the SOX9 fragment, spanning positions 1 to 304, from the pcDNA3 construct into the BamHI site of pGADGH (Clontech) as a fusion with the GAL4 activating-domain DNA sequence. Again, the authenticity of the constructs and ligation junctions was checked by sequencing.
(ii) Bacterium expression vectors.
Both SF-1 and SOX9 proteins were bacterially expressed as glutathione S-transferase (GST) fusion proteins after PCR amplification of the corresponding cDNA and cloning into the BamHI and EcoRI sites of the pGEX-4T3 expression vector (Pharmacia). The authenticity of the constructs was checked by sequencing.
(iii) Mammalian reporter and expression plasmids.
To construct the reporter gene plasmid, pEMBL8-AMH (16) was used as a template to amplify AMH promoter DNA from positions +10 to −154 by PCR. The two primers contain the recognition site for either SalI or SacI. After SalI-SacI digestion, one copy of the 164-bp PCR product was inserted in the SalI-SacI sites of the pEMBL-CAT vector (Stratagene) upstream of the chloramphenicol acetyltransferase (CAT) coding sequence. This construct is referred to as p154CAT. The same strategy was used to produce the p123CAT construct. Mutagenesis of the SOX-binding site to produce p154MUTSOXCAT was performed by using the QuickChange site-directed mutagenesis kit (Stratagene) with the help of the SOX-MUT oligonucleotide (see below). Full-length SF-1 cDNA was cloned as a BamHI-EcoRI fragment into the pcDNA3 vector, which directs transcription from the cytomegalovirus promoter. pcDNA-SOX9 and pcDNA-SOX9 1–304 were described previously (46).
(iv) Plasmid constructs for in vitro translation.
pcDNA-SOX9 HMG was described previously (31). The SOX9 1–118 deletion mutant was obtained by BssHII digestion of pcDNA-SOX9. For the other two deletions (SOX9 1–208 and SOX9 1–95), pcDNA-SOX9 was digested with BamHI and the released insert was cloned in pBluescript vector. This construct was finally digested with either PstI or HincII and ligated to create SOX9 1–208 and SOX9 1–95 mutants, respectively.
Synthesis of proteins in vitro and preparation of nuclear extracts.
SOX9 protein, SOX9 mutants, and SF-1 protein were synthesized by in vitro transcription-translation with the expression vectors described above and with the TNT system (Promega). Nuclear extracts from NT2/D1 cells were prepared as described previously (41).
Production and purification of bacterially expressed pGEX-SOX9 and pGEX–SF-1 fusion proteins.
After being cloned in the pGEX-4T3 expression vector, the two recombinant proteins were produced in bacterial strain BL21(DE3) after induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 2 h of induction at 30°C, cells were collected by centrifugation, resuspended in lysis buffer (150 mM NaCl, 1 mM dithiothreitol [DTT], 5 mM EDTA, 25% sucrose, 50 mM Tris [pH 7.5]) supplemented with bovine DNase I (Pharmacia) and 0.5 mM Pefabloc-SC-AEBSF (Interchim), and sonicated for 5 min at 4°C. Bacterial debris were removed by centrifugation at 25,000 rpm for 20 min. Lysate was loaded onto glutathione-Sepharose beads and washed three times with buffer I (5 mM EDTA, 250 mM NaCl, 50 mM Tris [pH 7.6]) and three times with buffer II (5 mM EDTA, 120 mM NaCl, 50 mM Tris [pH 7.6]). The purified proteins were checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis and used directly for in vitro binding assay studies or eluted from the matrix with buffer II plus 10 mM reduced glutathione after a 30-min incubation at 4°C.
Production, purification, and characterization of SF-1 and SOX9 antibodies.
Polyclonal human SF-1-specific rabbit serum and rat serum were raised against the GST fusion protein containing amino acids 121 to 232 from the human SF-1 protein. The fusion protein was overexpressed in BL21 (DE3) cells and purified on a glutathione-Sepharose column as described above. Male New Zealand White rabbits and rats were injected with purified protein mixed with complete Freund’s adjuvant (Sigma) and bled 10 days after each injection. For purification, only the rabbit polyclonal antiserum was loaded onto GST beads overnight and affinity purified by blotting overnight onto the SF-1 peptide coupled to an Immobilon membrane (Millipore). After saturation with polyvinylpyrrolidone (Sigma), SF-1 antibody was eluted with 0.2 M glycine (pH 2.5) and dialyzed against 1 M Tris base (pH 7.5). The antibody was finally aliquoted and stored at −80°C.
SF-1 antibody specificity was checked by immunostaining of SF-1-transfected COS-7 cells and also of empty COS-7 cells as control. In each case, nuclear extract proteins were transferred to nitrocellulose and immunostained as described previously (41). In the transfected cells, only an expected 53-kDa protein was detected (data not shown).
Polyclonal human SOX9-specific rat serum was raised against the bacterially expressed SOX9 TA domain (amino acids 408 to 504) fused to GST. SOX9 antibody specificity was checked as described above by using COS-7 cells expressing full-length SOX9 protein and immunostaining.
DNA-binding assays.
Protein binding to AMH promoter DNA probes was assessed by the electrophoretic mobility shift assay (EMSA). For these experiments, double-stranded oligonucleotides were labeled by a fill-in reaction in the presence of [α-32P]dCTP and DNA polymerase I Klenow fragment for 1 h at 37°C. The labeled probe was purified on a 5% nondenaturing acrylamide gel and eluted overnight in 0.8 M ammonium acetate–5 mM EDTA–0.1% SDS buffer at 50°C. For the binding reaction, 5 to 10 μg of nuclear extract or recombinant protein was mixed with 32P-labeled probe (10,000 cpm) and 2 μg of poly(dI-dC) for SF-1 protein or 2 μg of poly(dG-dC) for SOX9 protein in a 20-μl final volume of binding buffer (20 mM HEPES [pH 7.9], 20% glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM DTT). For competition or supershift experiments, unlabeled competitor oligonucleotide or antibody solution was incubated for 15 min at room temperature before the probe was added. The DNA-protein complexes were resolved by electrophoresis on a 5% polyacrylamide gel in Tris-borate-EDTA buffer at 4°C and visualized by autoradiography after being fixed with 10% methanol–10% acetic acid and dried.
The DNA probes used in these assays were complementary double-stranded DNA oligonucleotides including the SOX binding site of the AMH proximal promoter (SOX-BS), a mutated version of this site (SOX-MUT), or the SF-1-binding site from the same promoter (SF-1-BS). The nucleotide sequences of the top strands of the oligonucleotides are as follows: SF-1-BS, 5′-GGCACTGTCCCCCAAGGTCGC-3′; SF-1-MUT, 5′-GGCACTGTCCCCCAATTTCGC-3′; SOX-BS, 5′-GGACAGAAAGGGCTCTTTGAGAAGGCCA-3′; and SOX-MUT, 5′-GGACAGAAAGGGCTCTGGGAGAAGGCCA-3′. The mutated nucleotides are in boldface type.
Cell culture and transfection assays.
The human NT2/D1 cells (N-Tera 2, clone D1, a human pluripotent embryonic carcinoma cell line [ATCC CRL 1973]) were obtained from the American Type Culture Collection (Biovaley, France). NT2/D1 and COS-7 cells were cultured in Dulbecco’s modified Eagle’s medium (Imperial Laboratories, Flobio, France) containing 10% (vol/vol) fetal calf serum (Life Technologies), penicillin/streptomycin, and 2 mM glutamine with a partial pressure of CO2 of 5% at 37°C in humidified air. Plasmids used for transfections were purified with the maxiprep reagent system (Qiagen). COS-7 or NT2/D1 cells at 60% to 80% confluence were washed twice with serum-free medium before undergoing cotransfection with 1 μg of reporter plasmid, 200 ng of pCMV–β-galactosidase plasmid (Stratagene) used as an internal control for transfection efficiency, and different SF-1 and SOX9 expression plasmids with 7 μl of Lipofectamine (Life Technologies) in 200 μl of serum-free medium. After a 6-h incubation, the medium was replaced with 2 ml of medium supplemented with 10% serum and the cells were harvested after 48 h of culture. CAT assays were performed on cell extracts with [3H]acetyl coenzyme A (200 mCi/mmol; Amersham, Little Chalfont, United Kingdom) by a nonchromatographic method as described by Nielsen et al. (37). Promoter activities were expressed as CAT activity units per β-galactosidase unit, and each value represents the mean of the results from four separate wells. Error bars represent the standard errors.
Protein interaction assays.
For protein association experiments on glutathione-Sepharose beads, GST–SF-1 fusion proteins were overexpressed in BL21 bacteria and purified as described above. Immobilized SF-1 proteins were then incubated with the different 35S-labeled SOX9-derived proteins obtained by in vitro translation and the diverse SOX9 constructs in pcDNA3 and pBluescript vectors. Incubations were carried out in 300 μl of TBST buffer (10 mM Tris-HCl [pH 7.5], 130 mM NaCl, 0.5% Tween 20)–0.2% bovine serum albumin [BSA]–ethidium bromide (50 μg/ml) at room temperature for 30 min. The Sepharose beads were washed three times with 1 ml of TBST buffer. Bound proteins were eluted by the addition of 5× Laemmli buffer, boiled, and visualized after SDS-PAGE analysis and autoradiography.
Yeast two-hybrid interaction assays.
After the different pGADGH-SOX9 constructs (containing the leucine-selective marker) were obtained, each was transformed into the Matα Y187 yeast strain by standard procedures. On the other hand, the Matα Hf7c yeast strain was transformed with the different pGBT11–SF-1 constructs (containing the tryptophan-selective marker). These two yeast strains both harbor HIS3 and β-galactosidase reporter genes under the control of GAL4-binding sites. Diploids were obtained by mating and were selected on DO-W-L medium without tryptophan and leucine, as reported previously (11). Interaction assays were done for three independent transformants. Histidine assays were conducted on Y187-H7fc diploid strains expressing the designated constructs on DO-W-L-H medium without tryptophan, leucine, and histidine. Quantitative β-galactosidase assays were conducted on the same diploids as the histidine assays, and mean values are given in β-galactosidase units.
In vivo coimmunoprecipitation.
Detection of SF-1/SOX9 complexes was analyzed in vivo in the NT2/D1 cell line. After a 4-h labeling with 100 μCi of [35S]methionine (Amersham; specific activity, >800 Ci/mmol), the cells were washed with phosphate-buffered saline (PBS), collected, lysed for 30 min at 4°C in 1 ml of TBST or TLB (20 mM Tris-HCl [pH 7.6], 140 mM NaCl, 2 mM EDTA, 1% Triton X-100, 25 mM β-glycerophosphate, 10% glycerol, 2 mM sodium pyrophosphate) buffer supplemented with Complete protease inhibitor cocktail (Boehringer Mannheim) with or without 50 μg of ethidium bromide per ml, and vortexed. All subsequent steps were carried out on ice. Cell debris were removed, and the resulting lysate was precleared with protein A-Sepharose beads (Pharmacia). Typically, for each immunoprecipitation, 100 μl of cleared lysate was incubated with anti-SF-1 antibody conjugated to protein A-Sepharose beads at 4°C in a total volume of 1 ml for 1 h with continuous rocking. The beads were pelleted and washed five times in the lysis buffer, and the resultant proteins were diluted in 5× Laemmli buffer and subjected either to SDS-PAGE and autoradiography or to Western blot analysis with the SOX9 antibody (diluted 1/400) revealed with an ECL kit (Amersham).
DNase I footprinting assay.
A double-stranded DNA fragment corresponding to the 164-bp region from the AMH promoter was used for DNase I footprinting analysis. Briefly, a pUC18-AMH 164-bp construct was linearized by digestion with BamHI and labeled with [α-32P]dCTP. The 164-bp fragment was then released by EcoRI. The labeled fragment was gel purified and eluted at 50°C in 0.8 M ammonium acetate–5 mM EDTA–0.1% SDS buffer. For each footprinting reaction, 104 cpm of the probe was added to various amounts of purified SF-1 and SOX9 proteins in 50 μl of mixture containing 10 mM HEPES (pH 7.8), 40 mM KCl, 3 mM MgCl2, 0.5 mM DTT, 5% glycerol, and 100 ng of poly(dI-dC) for SF-1 or 100 ng of poly(dG-dC) for SOX9 and the mixture was incubated at room temperature for 30 min. DNase I digestion was performed by adding 1 U of enzyme diluted in the corresponding buffer (Boehringer-Mannheim) and incubating the mixture for 1 min at 25°C. The reaction was terminated by adding phenol-chloroform. The digested fragments were recovered by ethanol precipitation, resuspended in 3 μl of stop buffer (0.1% xylene cyanol, 0.1% bromophenol blue, 10 mM EDTA, 95% formamide), incubated for 2 min at 90°C, and resolved by electrophoresis on a 6% polyacrylamide–urea gel. A DNA ladder was made by dimethyl sulfide treatment for 5 min of 2 μl of labeled probe and piperidine treatment at 90°C for 30 min. This ladder was used to assign nucleotide positions in the gel.
Immunofluorescence staining of tissue culture cells.
Cells were preincubated in PBS buffer–1% BSA for 30 min at 37°C and then probed with the appropriate antibody, anti-SF-1, anti-SOX9, or anti-AMH diluted 1/100 in PBS/BSA. The cells were washed in PBS, and the primary antibodies were visualized with biotinylated anti-rabbit antibodies (dilution, 1/200) and Texas red-conjugated streptavidin or with Fluorolink Cy2-conjugated anti-rat labeled goat antibodies (dilution, 1/200). In each case, incubations were performed for 30 min under the same conditions described for the primary antibodies. For colocalization analysis, incubation with both appropriate primary antibodies was performed in the same incubation buffer and the same antibody dilutions were used with the secondary antibodies. Cell nuclei were visualized with Hoechst 33286. The cells were washed again and mounted in FluorSave reagent (Calbiochem). Images were collected and processed on a Bio-Rad confocal microscope or on a Zeiss Axiophot.
RESULTS
A consensus and functional SOX-binding site resides within the proximal AMH promoter.
Previous in vitro studies of the AMH promoter indicated that not more than 180 bp is required for proper AMH expression in Sertoli cells (43). Diverse observations including the presence of a conserved SF-1 site (Fig. 1A), mutation analysis, or transgenic experiments have revealed the requirement for this site in AMH gene control (13, 43). However, several results have also pointed out the necessity for a specific Sertoli-related cell environment for activation, indicating that an uncharacterized Sertoli cell factor contributes to this activation (43). We now confirm this requirement by using the COS-7 cell line as the transfection control (Fig. 1B). Pursuing progressive deletions of the minimal human AMH promoter, we show that the nucleotide sequence between −154 and −123 of the AMH promoter is important for promoter function, as demonstrated by transfection assays in the NT2/D1 cell line (Fig. 1B). The NT2/D1 cell line still constitutes one of the rare cell models positive for SRY expression (4, 41). Inspection of the nucleotide sequence confirmed previous analysis by revealing a 7-of-8-bp identity (5′-CCTTGAG, referred to as the SOX site in this study) to a known binding site that represents a potential site for SRY and other related members of the HMG-box class DNA-binding factors such as SOX, TCF-1, or LEF-1 protein (12, 22, 30, 36). Finally, using direct mutagenesis, we also show the necessity for the SOX site in such an additive activation (Fig. 1B).
Purified human SOX9 and SF-1 proteins bind to the AMH promoter in vitro.
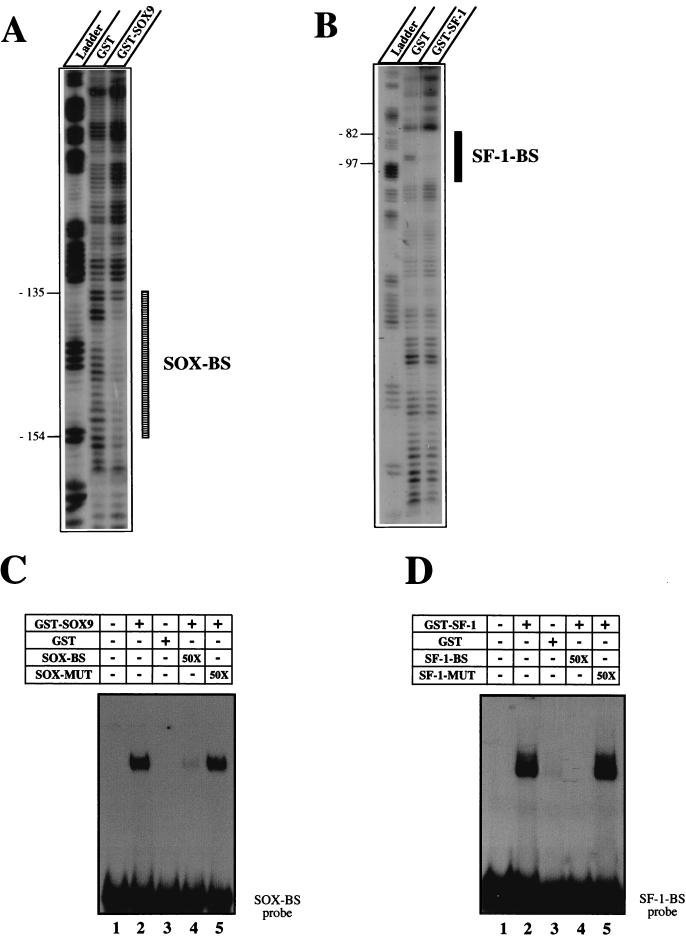
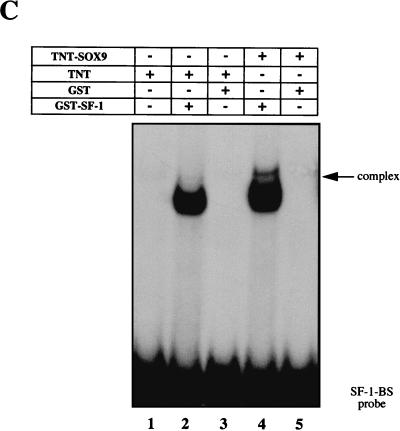
In vitro DNase I footprint analysis showed that purified GST-SOX9 protein could protect the −154 to −135 sequence from the AMH proximal promoter, the sequence spanning the SOX site (Fig. 2A). As a control, by using the same 164-bp labeled oligonucleotide, the same experimental conditions, and the purified GST-SF-1 fusion protein, we also confirmed the protection of the −97 to −82 DNA sequence that includes the SF-1 binding site previously described (43) (Fig. 2B). A labeled 28-bp oligonucleotide probe including the SOX-binding site was next checked in EMSA experiments with GST-SOX9 protein. As shown in Fig. 2C, a single DNA protein complex was evident. Complex formation was blocked by using the same unlabeled oligonucleotide as the competitor. When a double mutation known to abolish SOX protein binding was introduced into the oligonucleotide (SOX-MUT), the competition was abolished. These results indicate that SOX9 binds directly to the DNA in the region of the AMH promoter. On the other hand, similar EMSA experiments with purified GST–SF-1 and the corresponding SF-1 oligonucleotide-binding site designed from the AMH proximal promoter confirm SF-1 protein interaction (Fig. 2D).
FIG. 2.
DNase I footprint analysis of the AMH proximal promoter by using SOX9 and SF-1 recombinant proteins. (A) SOX9 DNase I footprint. The upper strand of the 164-bp AMH promoter was 32P labeled; 104 cpm of the probe was incubated in the presence of 1 μg of either free GST or GST-SOX9. In each case, the reaction mix was resolved on a 6% polyacrylamide sequencing gel. The lane labeled Ladder designates a G+A Maxam-Gilbert sequence ladder obtained with the probe. The protected regions are indicated by a box. (B) SF-1 DNase I footprint. The same radiolabeled probe was incubated with 1 μg of either free GST or GST–SF-1. The lane labeled Ladder represents the G+A Maxam-Gilbert sequence ladder. The protected regions are indicated by a box. (C) Affinity binding of GST-SOX9 to the SOX-binding site (SOX-BS) in the AMH proximal promoter probe. The double-stranded SOX-binding-site oligonucleotide was end labeled and incubated with 10 ng of purified GST-SOX9 protein in the absence (lane 2) or presence (lanes 4 and 5) of a 50-fold molar excess of unlabeled competitors. Free GST was used as the control (lane 3). (D) Affinity binding of GST–SF-1 recombinant protein to the SF-1-binding site (SF-1-BS) in the AMH promoter. The double-stranded SF-1-binding-site oligonucleotide was end labeled and incubated with 10 ng of purified GST-SOX9 protein in the absence (lane 2) or presence (lanes 4 and 5) of a 50-fold molar excess of unlabeled competitors. Free GST was used as control (lane 3).
Transactivation of AMH expression by SOX9 and by SF-1 in COS-7 cells.
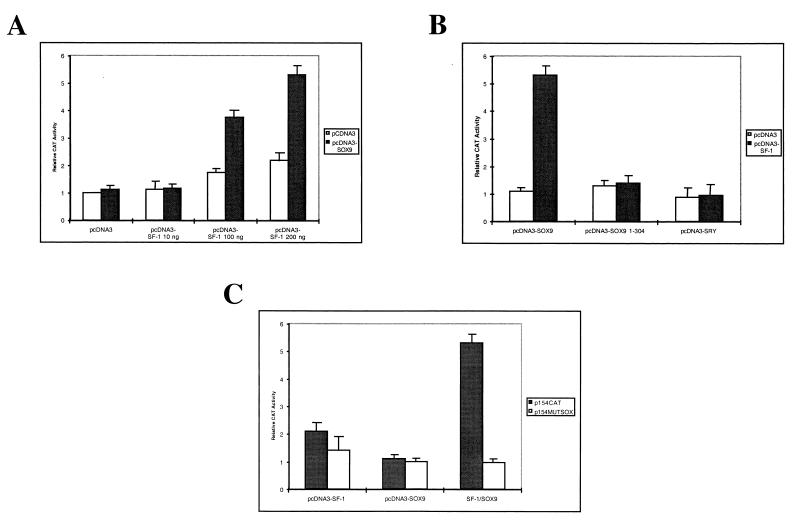
The functional relevance of SOX9 factor binding to the SOX site located in the AMH proximal promoter and the possibility of its cooperation with the SF-1 nuclear receptor in activating the AMH promoter were examined. Cotransfections of COS-7 cells with a reporter plasmid which contains the 164-bp AMH DNA fragment adjacent to the CAT gene and termed p154CAT (see Materials and Methods), together with increasing SF-1 expression plasmid concentrations but with a fixed SOX9 plasmid concentration, were performed (Fig. 3A). The use of 10 ng of pcDNA3-SOX9 along with 200 ng of pcDNA3-SF-1 provides the best activation signal (more than fivefold compared with the empty vector) and was used in the following experiments (Fig. 3A). As a control, under these experimental conditions the specificity of this activation was then tested with a truncated form of SOX9 (deletion of amino acids 305 to 509), a form described as being able to bind the DNA target but unable to activate transcription (30). As shown in Fig. 3B, the SOX9 C-terminal transactivation domain was required for this activation. Similarly, no activation was observed when the effector plasmid expressing SRY, a previous candidate factor for contributing to AMH gene activation, was used (Fig. 3B). While this result confirmed recent data obtained with an artificial promoter (8), it also showed that SRY cannot substitute for SOX9 in the AMH-positive control. If the SOX9 activation domain appears necessary in the activation effect, its DNA-binding capacity is also required, as demonstrated when using a mutated version of the AMH proximal promoter (Fig. 3C).
FIG. 3.
Cooperation between SOX9 and SF-1 proteins in the activation of the AMH minimal promoter in COS-7 cells. (A) Cotransfection assay in COS-7 cells of a reporter plasmid with the CAT gene under the control of an AMH minimal promoter (p154CAT), a constant amount of pcDNA3-SOX9 (10 ng), and increasing amounts of the pcDNA3–SF-1 construct. (B) The specificity of SOX9 activity was tested with a deleted version of pcDNA3-SOX9 (10 ng) or with pcDNA3-SRY (10 ng). (C) Comparison of the SOX9–SF-1 activity on the wild-type AMH proximal promoter (p154CAT) with that on the same promoter mutated on the SOX-binding site p154MUTSOX. The CAT activity of the reporter plasmid alone was set as 1. All values represent the means of three separate transfection experiments (± standard errors).
SOX9 and SF-1 interact directly with each other as shown by two-hybrid analysis and by in vitro assays.
The results just presented and the observation of rather closely spaced SOX- and SF-1-binding sites within the proximal AMH promoter region (Fig. 1B), along with the ability of SOX proteins to bend DNA (12), raised the possibility that the two factors interact with each other, as recently reported for another system involving Sox2 and Oct-3 (1, 52).
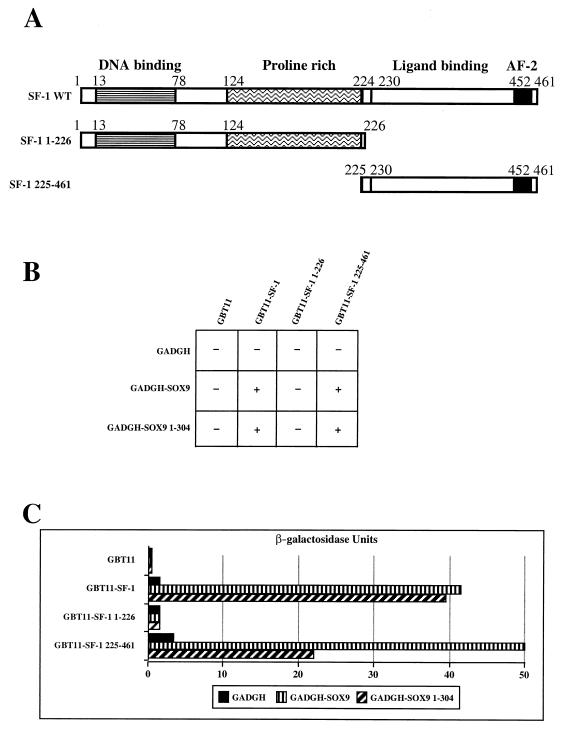
To obtain evidence for such an interaction, we first used the yeast two-hybrid assay and a mating protocol (11). For this, the region encompassing the SF-1 open reading frame was fused to the GAL4 DNA binding domain in the pGBT11 vector and mated with the SOX9 open reading frame fused to the GAL4 activation domain in the pGADGH vector. The yeast diploid transformants were able to grow on selective medium lacking histidine and were also β-galactosidase positive (Fig. 4B and C). This result indicates an interaction between the two proteins. We subsequently tested potential interactions between full-length SOX9 and different regions of SF-1, as depicted in Fig. 4A. We found that the carboxy-terminal region of SF-1 (amino acids 225 to 461) was sufficient for this interaction. Correspondingly, SOX9 did not interact with the N-terminal portion of SF-1 (amino acids 1 to 226). It is worth noting that identical results were obtained when using only the N-terminal part of SOX9 (amino acids 1 to 304) (Fig. 4B). To quantify the strength of the interactions between the different constructs of SF-1 and SOX9 proteins, β-galactosidase activity was measured from the transformant lysates (Fig. 4C). These results confirm that the strongest interaction occurs between the N-terminal region of SOX9 and the C-terminal region of SF-1, including its ligand-binding domain. Although these results demonstrate that SOX9 and SF-1 interact, we cannot exclude the contribution of a posttranslational modification or the involvement of a third partner present in the yeast strain and contributing to the interaction.
FIG. 4.
SOX9 interacts with the carboxy-terminal region of the SF-1 protein. The SOX9–SF-1 interaction was scored by a yeast two-hybrid assay (see Materials and Methods). (A) Schematic diagrams of SF-1 wild type, deletion constructs, and functional domains of the human SF-1 protein. (B) Qualitative histidine assays. A positive interaction results in the ability of the Y187-Hf7c diploid expressing the designated constructs to grow (+) or not grow (−) on a medium depleted of tryptophan, leucine, and histidine. Assays were done for three independent diploids. All the SF-1 constructs were tested against the empty pGADGH vector as a negative control, and the SOX9-derived constructs were tested against the empty pGBT11 vector. (C) Quantitative β-galactosidase assays for these interactions were conducted on the same diploids as those used in the histidine assays. Mean values are given in relative β-galactosidase units.
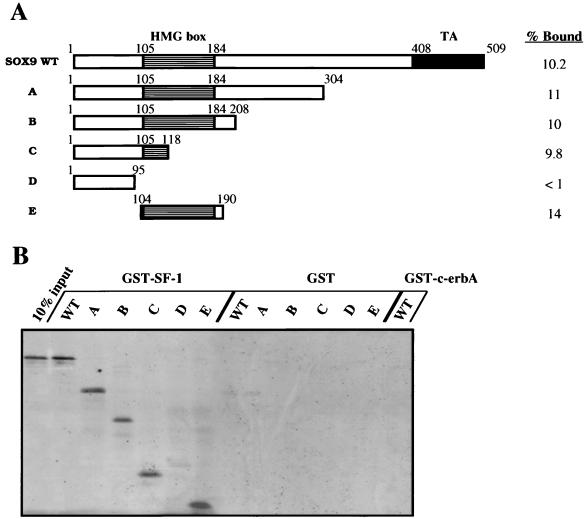
To clarify this point and to better delineate the SOX interaction region, in vitro binding experiments were also performed. For this, SF-1 was expressed as a bacterial GST fusion protein, immobilized on glutathione-Sepharose resin, and subjected to in vitro binding assays by incubation with in vitro-translated radiolabeled SOX9. SOX9 was produced either as full-length protein or as deleted versions (depicted schematically in Fig. 5A). After extensive washing of the resin and analysis by SDS-PAGE, approximately 10% of the input [35S]methionine-labeled SOX9 remained bound on the GST–SF-1 beads (Fig. 5B). These coprecipitation assays show that a GST–SF-1 fusion protein binds specifically to SOX9 in vitro and that the SOX9 interaction domain maps to amino acids 104 to 118, a domain located in the first one-third of the SOX9 HMG box (Fig. 5B). As controls, neither the SOX9-derived proteins bound to GST-Sepharose alone nor the full-length SOX9 protein interacted with an unrelated nuclear receptor such a c-erbA (Fig. 5B). Similar binding was observed after introduction of ethidium bromide, confirming a DNA-independent protein association mechanism (27). Taken together, these results demonstrate a direct protein-protein interaction between SOX9 and SF-1. Finally, this direct protein-protein interaction was confirmed by using band shift experiments including GST–SF-1 purified protein, in vitro-translated SOX9 protein, and the AMH proximal promoter-derived SF-1 binding site (Fig. 5C). At the same time, this data demonstrates the inability of SOX9 protein to interact with the SF-1 DNA-binding site used in this study.
FIG. 5.
SOX9 and SF-1 interact in vitro. (A) The diagram shows the different SOX9 deletion mutants which were used to investigate the physical interaction between SOX9 and SF-1. In each case, the respective percentage of protein bound to the GST–SF-1 phase was determined by phosphorimager analysis. (B) The SOX9 polypeptides depicted in panel A were translated in vitro in the presence of [35S]methionine and analyzed for binding to either GST, GST–SF-1 or GST–c-erbA fusion proteins bound to glutathione-Sepharose. The GST pulldown assay was performed as described in Materials and Methods. The 10% input (left lane) and bound proteins (the other lanes) were separated by SDS-PAGE analysis and then autoradiographed. (C) Binding of the GST–SF-1 fusion protein to the SF-1-binding-site (SF-1-BS) probe (lane 2) is shifted after preincubation with in vitro-translated TNT-SOX9 protein as shown by EMSA (lane 4). The specificity was assessed by the use of either TNT or TNT plus GST alone (lanes 1 and 3). Lane 5 shows the absence of TNT-SOX9 protein binding to the SF-1-binding-site probe in the presence of 2 μg of poly(dI-dC). WT, wild type.
SOX9 and SF-1 interact in vivo.
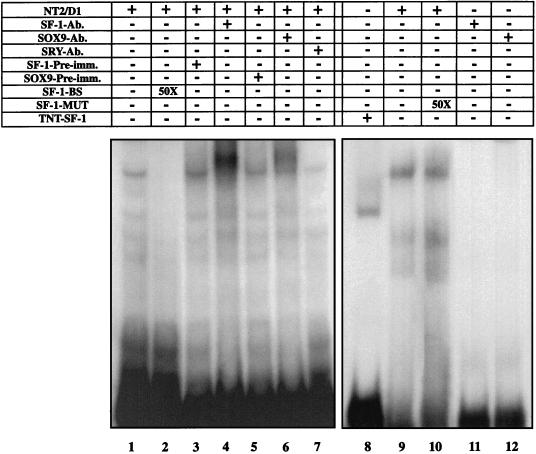
We then examined whether SOX9 and SF-1 form a protein complex in vivo. The ability of SOX9 to interact directly with SF-1 independently of the SOX9 DNA-binding activity was first tested by EMSA in the presence of NT2/D1 cell nuclear extracts and the labeled SF-1 binding-site oligonucleotide whose sequence was derived from the AMH proximal promoter. Nuclear extracts of NT2/D1 cells gave rise to a major protein-DNA complex (Fig. 6, lane 1), a DNA binding that was inhibited by an excess of unlabeled SF-1 oligonucleotide (lane 2) but not by a mutated SF-1 oligonucleotide (lane 10). Evidence for the identity of the retarded major band was obtained in supershift experiments. Preincubation of nuclear extracts in the presence of SOX9 or SF-1 antisera produced a supershift band in both cases (lanes 4 and 6). By contrast, addition of SRY antiserum did not modify the migration of the complex (lane 7), in agreement with previous results (43).
FIG. 6.
SOX9 and SF-1 form a common protein-DNA complex. Nuclear extracts were prepared from NT2/D1 cells. In an EMSA reaction, α-32P-labeled SF-1-binding-site (SF-1-BS) oligonucleotide and 2 μg of nuclear extract were incubated together. The specificity of the retarded bands was controlled by the use of either an excess of cold SF-1-binding-site probe (lane 2) or a mutated form of this oligonucleotide (lane 10). Supershift experiments were performed after preincubation of either 1 μl of SF-1 antibody (SF-1-Ab.) (lane 4) or 1 μl of SOX9 antibody (SOX9-Ab.) (lane 6). The corresponding preimmune serum (SF-1-Pre-imm. and SOX9-Pre-imm.) (lanes 3 and 5) or 1 μl of a specific anti-SRY antibody (SRY-Ab.) (lane 7) was used as the control. The specificity of the observed shifts with the different antibodies was assessed by the use of the corresponding antibody only (lanes 11 and 12). Gel retardation of the TNT–SF-1 protein after binding to the SF-1-binding-site probe was used as a control (lane 8).
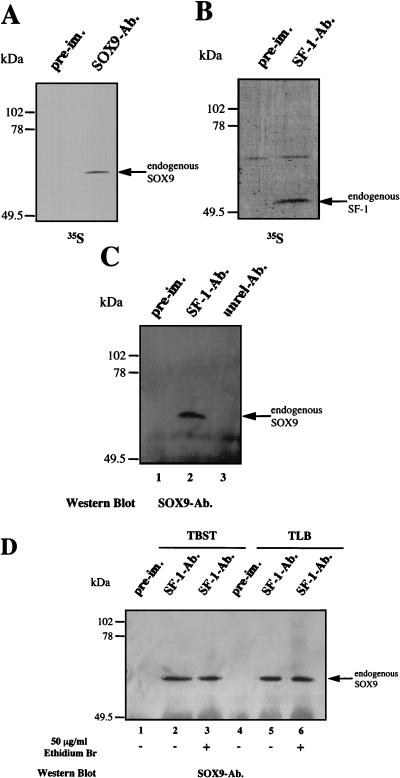
To complete our investigations of the SOX9–SF-1 interaction, we performed coimmunoprecipitation experiments with extracts prepared from [35S]methionine-labeled NT2/D1 cells. Immunoprecipitation was carried out in two steps. First, radiolabeled NT2/D1 cells extracts were immunoprecipitated with SOX9- or SF-1-specific antiserum or with the respective preimmune serum. As shown in Fig. 7A and B, the two antisera precipitated the corresponding proteins with the expected molecular weights, but the preimmune sera did not. In a second step, after immunoprecipitation of NT2/D1 cell extracts with SF-1-specific antiserum, immunoprecipitates were collected on protein A-Sepharose beads and then fractionated by SDS-PAGE. After blotting onto nitrocellulose and probing with the rat SOX9 antibody, a 65-kDa protein with the size expected for SOX9 was detected (Fig. 7C, lane 2), demonstrating the coimmunoprecipitation of SOX9 with SF-1. The 65-kDa protein was not detected when SF-1 preimmune serum was used in the precipitation step (lane 1). As a control, when the unrelated but immunoprecipitating rabbit SIP-1 antibody (40) was used instead of the SF-1 antiserum, no SOX9 protein was detected (lane 3), underlining the specificity of the SOX9 antiserum. Moreover, any potential cross-reactivity of the SF-1 antibody with SOX9 was ruled out by the unsuccessful immunostaining of in vitro-translated SOX9 protein after blotting (data not shown). The specificity of the interaction between SOX9 and SF-1 was confirmed by the use of different detergent conditions and the introduction of ethidium bromide into the coimmunoprecipitation assay (Fig. 7D). The reverse experiment (SOX9 immunoprecipitation followed by SF-1 Western blotting) was unsuccessful because of the similarity between the molecular weights of SF-1 and immunoglobulin light chain.
FIG. 7.
Coimmunoprecipitation of endogenous SOX9 and SF-1 from metabolically 35S-labeled NT2/D1 cells. After labeling, equal amounts of NT2/D1 cell extracts (prepared as described in Materials and Methods) were immunoprecipitated. (A) Immunoprecipitation with SOX9 antibodies (SOX9-Ab.) or preimmune (pre-im.) antibodies. (B) Immunoprecipitation with SF-1 antibodies (SF-1-Ab.) or preimmune (pre-im.) antibodies. (C) Western blot analysis with a SOX9-specific rat antibody of an SF-1 immunoprecipitate (SF-1-Ab.). As a control, an unrelated antibody (unrel-Ab.) was introduced in the precipitation step before revelation with the SOX9 antibody. First-step immunoprecipitation with preimmune (pre-im.) antibodies is also shown. (D) Similar experiments comparing two buffers containing two different detergents, TBST (lanes 1 to 3) or TLB (lanes 4 to 6) (see Materials and Methods), and including or not including ethidium bromide. Lanes 1 and 4 show immunoprecipitation with preimmune (pre-im.) antibodies as controls.
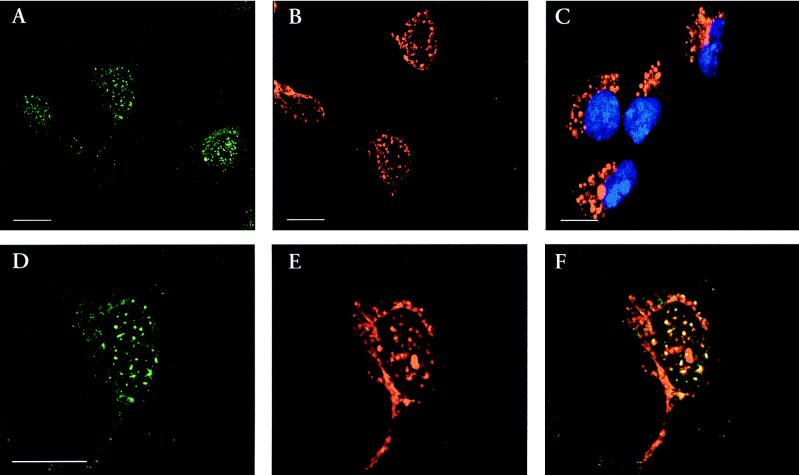
SOX9 and SF-1 colocalize in nuclei of NT2/D1 cells.
We next analyzed the subcellular localization of the SOX9 and SF-1 proteins by performing indirect-immunofluorescence labeling experiments. The use of purified rabbit anti-SF-1 and rat anti-SOX9 antibodies allowed double-labeling experiments on the NT2/D1 cells. Both antibodies revealed a nuclear, punctuate localization for the two proteins, an expression throughout the cell culture (Fig. 8A and B). Cytoplasmic labeling of the AMH protein was observed in the same cells (Fig. 8C). We further analyzed the possibility of subcellular colocalization of SOX9 with SF-1. A good, if incomplete, colocalization between the two proteins was observed after examination by confocal laser-scanning microscopy (Fig. 8F). These results strengthen the notion that SOX9 and SF-1 interact in vivo, possibly as part of a multimeric protein complex.
FIG. 8.
Immunolocalization of SOX9 and SF-1 protein in cultured human NT2/D1 cells. Immunostaining of SF-1 in green (A and D) and of SOX9 in red (B and E) or both (panel F) is shown. As a control, AMH expression was checked by using the corresponding rabbit antibody (in red) along with a counterstaining of cell nuclei with Hoechst 33258 in blue (C). Scale bars, 10 μm.
DISCUSSION
In the developing gonad, when the sex-determining factor Sry is present, the supporting cell precursors differentiate along the Sertoli cell pathway. The first known product of the embryonic Sertoli cell is AMH. Initiation, maintenance, and down-regulation of AMH expression by Sertoli cells still remain a matter of controversy. If AMH has to be tightly regulated during gestation in order to avoid Müllerian duct persistence (17), it also requires a sexually dimorphic regulation because of its necessary absence in females during embryogenesis.
The aim of the work described here was to investigate the functional relationship between cis-acting conserved elements located in the AMH proximal promoter in order to contribute to our understanding of the transcription factors controlling AMH promoter activity and leading to its spatiotemporal regulation during embryogenesis. Sequence analysis showed conserved binding sites for different transcription factors within the proximal and minimal 180-bp AMH promoter (13, 43). In this study, the functional importance of the nuclear receptor SF-1 binding site located in the AMH promoter and previously revealed by studies with primary Sertoli cells (43) was confirmed. However, these results also shed light on the requirement for an appropriate developmental context or cell environment for SF-1 activation of AMH, suggesting that a ligand or a cofactor (or both) for SF-1 is needed (43). As mentioned in this and other reports (18, 43, 50), apart from the now well-described nuclear receptor SF-1 site, a nearly perfect match with the consensus SOX-binding site appears conserved in the promoter of distantly related species. Performing progressive 5′ deletions of the AMH proximal promoter revealed that a deletion of the sequence from −154 to −123 which removes this SOX-binding site caused a strong decrease in the basal activity of the AMH proximal promoter after transfection into the “Sertoli-like” human NT2/D1 cell line. This result was confirmed after mutation of the candidate SOX site. Functional in vitro analysis of this site by either DNase I footprinting or in vitro DNA-binding experiments showed that this putative site was able to specifically bind bacterially expressed SOX9 protein. The choice of this particular SOX protein was dictated by several arguments, including its conservation among vertebrates and its high level of expression closely correlating with Sertoli cell differentiation and subsequent AMH production during testicular development in mice and chickens (32) as well as in humans (unpublished data). Furthermore, the sex reversal phenotype observed in campomelic dysplasia patients carrying an SOX9 mutant is consistent with failure of Sertoli cell differentiation and subsequent absence of AMH production (31). Finally, of the two SOX proteins, SRY and SOX9, that have been identified as being expressed in the male sex-determining pathway, only SOX9 has been shown to act as a potent transcriptional activator in humans via a transactivation domain mapped to its C terminus (30, 46). As suggested by Ambrosetti et al. (1), the requirement for DNA-sequence-specific HMG domain transcription factors such as Sox2 (1, 52), Sox5 (7), and TCF-1 (48) in a particular promoter environment suggests that they are unable to act autonomously but, instead, must collaborate with other transcription factors. Under our experimental conditions with the COS-7 cell line, transient-cotransfection experiments with the SOX9 expression plasmid showed that SOX9 can activate AMH promoter activity to an extent similar to that for SF-1 (data not shown) even when using a single copy of the AMH proximal promoter. Thus, the kidney-derived cell line COS-7 appears to provide a convenient AMH promoter environment to test SF-1 or SOX9 function. Furthermore, these transactivation experiments also confirmed the strict requirement for the C-terminal domain of SOX9, implying that AMH promoter activation by SOX9 is not simply the result of the bending capacity of its HMG domain. In contrast, despite a 71% amino acid similarity to SOX9 in the HMG domain (10), the SOX transcription factor family founder SRY was ineffective in this activation. Because of the ability of both SOX9 and SF-1 to cooperate in AMH proximal promoter activation, it was tempting to investigate if this cooperation involved a protein-protein interaction event. Five different and complementary approaches, namely, two-hybrid analysis, GST pulldown assays, EMSA, coimmunoprecipitation experiments, and immunolocalization studies, all indicate that SOX9 and SF-1 can interact with each other and may be part of a multimeric protein complex. We then performed a preliminary mapping of the amino acid sequence responsible for this interaction. Dissection of the transcription factor SOX9 and subsequent coprecipitation assays with GST-SF-1 showed that the conserved N-terminal region of the SOX9 DNA binding domain is required for its interaction with SF-1. This region of SOX9 contains several amino acid sequence stretches that are conserved between different members of the SOX family and might provide the basis for interaction of SF-1 with other members of the family. Interestingly, another Sox gene product, Sox2, was recently shown to cooperate with the octamer-binding protein Oct-3 in order to synergistically activate the fibroblast growth factor 4 enhancer (1, 52). This activation was dependent both on the protein-protein interaction involving the HMG domain of Sox2 and on the presence of DNA-binding sites for both Sox2 and Oct-3 transcription factors (1). A similar result was obtained with the SOX9/SF-1 couple. We propose that SOX9 could, by its DNA-bending activity (30), induce a local architectural modification of the DNA target, allowing the formation of a transcription complex including SF-1. The SOX9–SF-1 interaction could then stabilize the resulting protein-DNA complex. This hypothesis is now under investigation. On the other hand, the region of SF-1 that was characterized in the two-hybrid experiment described herein as being required for the interaction starts after the proline-rich region of the receptor and extends into the ligand-binding and dimerization domain including both activation domains of SF-1 (6). This region has been previously reported as supporting the cell specificity of SF-1 activation of AMH reporter constructs (43) as well as SF-1 activation by oxysterol (29); it also permits interaction with the nuclear receptor DAX-1 (5). However, despite the possible overlap between the two regions of SF-1 involved in oxysterol activation and SOX9 binding, the SF-1–SOX9 interaction was not modified in the presence of increasing 25-hydroxycholesterol concentrations (data not shown). The ability of this SF-1 sequence region to interact with the coactivator SRC-1 was also recently shown (6). This interaction would allow bridging of the transcription factors such as SOX9 and SF-1 that govern the tissue-specific expression of AMH and the basal transcriptional machinery.
The absence of AMH expression before 12.5 days postcoitum in the mouse embryo despite coincident expression of both SF-1 and Sox9 could be questionable (34). This apparent contradiction with our data could be the result either of the absence of an elusive partner or of a missing positive regulation affecting one of the two transcription factors before this developmental stage. Another possibility is a regulation of the subcellular localization of one or more transcription factors contributing to the control of the onset of AMH expression. In this respect, mouse Sox9 expression studies have revealed a predominant cytoplasmic expression in cells of the genital ridge prior to 11.5 days postcoitum, i.e., prior to the sex determination event. At later stages, Sox9 appeared fully nuclear in male embryonic gonads (32). As recently suggested (45), the change in Sox9 localization could be achieved via its interaction with putative protein partner(s) that would mask the Sox9 nuclear localization signal(s). As an alternative, or in parallel, the mechanism used in this cytoplasm-nucleus trafficking could result from the existence of a leucine motif in the HMG domain of Sox9 (10, 47), a motif conserved across SOX9 evolution but also specific for this particular SOX protein (39). This kind of motif has been reported in many cases to act as a nuclear export signal (49). This hypothesis will not require further investigations.
Other gene products, namely, the Wilms’ tumor WT1 and the nuclear receptor DAX-1, are also implicated in mammalian sexual development. Recently, WT1 in its −KTS isoform has been shown to interact and synergize with SF-1 (35). In the same report, DAX-1 was also shown to antagonize this synergy, even when present at low concentration. The inhibitory action of DAX-1 was accompanied by its ability to interact directly with the SF-1 ligand-binding domain. Interestingly, SOX9- and DAX-1-binding sites could overlap with respect to their interaction with SF-1. Furthermore, in the same report, both WT1 and DAX-1 expression levels appear rather similar between male and female mouse embryos 13.5 days postcoitum, i.e., at a time when AMH expression is on (35). Therefore, the only difference between the sexes remains in the expression of the putative transcription factors SRY and SOX9. If, as mentioned above, SRY does not provide an attractive candidate, the high levels of SOX9 observed specifically in male embryos (32) could compete with DAX-1 for its binding to SF-1 and thus could permit WT1 action. This hypothesis would justify the rather low level of AMH stimulation observed in the present data when using SF-1 and SOX9 only.
It is well established that transcriptional regulation of a given gene is the result of combinatorial interactions between multiple proteins forming a higher-order complex based on protein-protein and protein-DNA interactions. We now suggest that AMH gene regulation is no exception to this general rule and that both SF-1 and SOX9 are members of the complex regulating the onset of AMH expression during embryogenesis beyond WT1 and DAX-1. This statement will now require introduction of these four transcription factors in the same assay as well as in vivo characterization. The two proteins SOX9 and SF-1 appear to bind independently to separate DNA sites and could, especially because of the DNA-bending capability of SOX9 (30), facilitate the functional interaction of other regulatory proteins, leading to the formation of the appropriate transcription complex triggering Sertoli-specific AMH expression. Interestingly, another well-conserved binding site in the AMH proximal promoter has homology to the in vitro canonical binding site for the GATA transcription factor family, WGATAR (Fig. 1A). Expression of GATA-1 or GATA-2 factors in Sertoli cells reinforces this hypothesis and makes GATA factors attractive candidates to contribute to the regulation of AMH expression (38, 51), a hypothesis that we are now attempting to test.
ACKNOWLEDGMENTS
We thank V. Laudet for the gift of the c-erbA construction plasmid, R. Rey for the rabbit anti-AMH antibody, J.-Y. Picard for pEMBL8-AMH plasmid, and A. Goldsborough and V. Laudet for comments on and corrections of the manuscript. We are grateful to Catherine Méjean for protein production and purification assistance and to Sandrine Faure for his constant support.
This investigation was supported by Biomed 2 grant BMH4-CT96-0790 from the European Economic Community.
REFERENCES
- 1.Ambrosetti D-C, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol Cell Biol. 1997;17:6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell D M, Leung K K H, Wheatley S C, Ng L J, Zhou S, Ling K W, Sham M H, Koopman P, Tam P P L, Cheah K S E. SOX9 directly regulates the type-II collagen gene. Nat Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- 3.Carré-Eusèbe D, Di Clemente N, Rey R, Pieau C, Vigier B, Josso N, Picard J-Y. Cloning and expression of the chick anti-Müllerian hormone gene. J Biol Chem. 1996;271:4798–4804. doi: 10.1074/jbc.271.9.4798. [DOI] [PubMed] [Google Scholar]
- 4.Clépet C, Schafer A J, Sinclair A H, Palmer M S, Lovell-Badge R, Goodfellow P N. The human SRY transcript. Hum Mol Genet. 1993;2:2007–2012. doi: 10.1093/hmg/2.12.2007. [DOI] [PubMed] [Google Scholar]
- 5.Crawford P A, Dorn C, Sadovsky Y, Milbrandt J. Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1. Mol Cell Biol. 1998;18:2949–2956. doi: 10.1128/mcb.18.5.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford P A, Polish J A, Ganpule G, Sadovski Y. The activation function-2 hexamer is required, but not sufficient, for potentiation by SRC-1. Mol Endocrinol. 1997;11:1626–1635. doi: 10.1210/mend.11.11.9970. [DOI] [PubMed] [Google Scholar]
- 7.Denny P, Swift S, Connor F, Ashworth A. An SRY-related gene expressed during spermatogenesis in the mouse encodes a sequence-specific DNA-binding protein. EMBO J. 1992;11:3705–3712. doi: 10.1002/j.1460-2075.1992.tb05455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desclozeaux M, Poulat F, de Santa Barbara P, Capony J-P, Turowski P, Jay P, Mejean C, Moniot B, Boizet B, Berta P. Phosphorylation of an N-terminal motif enhances DNA-binding activity of the human SRY protein. J Biol Chem. 1997;273:7988–7995. doi: 10.1074/jbc.273.14.7988. [DOI] [PubMed] [Google Scholar]
- 9.Dubin R A, Ostrer H. Sry is a transcriptional activator. Mol Endocrinol. 1994;8:1182–1192. doi: 10.1210/mend.8.9.7838151. [DOI] [PubMed] [Google Scholar]
- 10.Foster J W, Dominguez-Steglich M A, Guioli S, Kwok C, Weller P A, Stevanovic M, Weissenbach J, Mansour S, Young I D, Goodfellow P N, Brook J D, Schafer A J. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 11.Fromont-Racine M, Rain J-C, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 12.Giese K, Cox J, Grosschedl R. The HMG domain of lymphoid enhancer factor-1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 13.Giuili G, Shen W-H, Ingraham H A. The nuclear receptor SF-1 mediates sexually dimorphic expression of Müllerian inhibiting substance, in vivo. Development. 1997;124:1799–1807. doi: 10.1242/dev.124.9.1799. [DOI] [PubMed] [Google Scholar]
- 14.Greenfield A, Koopman P. SRY and mammalian sex determination. Curr Top Dev Biol. 1996;34:1–23. doi: 10.1016/s0070-2153(08)60707-3. [DOI] [PubMed] [Google Scholar]
- 15.Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 16.Guerrier D, Boussin L, Mader S, Josso N, Khan A, Picard J-Y. Expression of the gene for anti-Müllerian hormone. J Reprod Fertil. 1990;88:695–706. doi: 10.1530/jrf.0.0880695. [DOI] [PubMed] [Google Scholar]
- 17.Guerrier D, Tran D, Vanderwinden J M, Hideux S, Van Outryve L, Legeai L, Bouchard M, Van Vliet G, De Laet M H, Picard J Y, Josso N. The persistent Müllerian duct syndrome: a molecular approach. J Clin Endocrinol Metab. 1989;68:46–52. doi: 10.1210/jcem-68-1-46. [DOI] [PubMed] [Google Scholar]
- 18.Hacker A, Capel B, Goodfellow P, Lovell-Badge R. Expression of Sry, the mouse sex determining gene. Development. 1995;121:1603–1614. doi: 10.1242/dev.121.6.1603. [DOI] [PubMed] [Google Scholar]
- 19.Haqq C M, King C Y, Donahoe P K, Weiss M A. SRY recognizes conserved DNA sites in sex-specific promoters. Proc Natl Acad Sci USA. 1993;90:1097–1101. doi: 10.1073/pnas.90.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haqq C M, King C-Y, Ukiyama E, Falsafi S, Haqq T N, Donahoe P K, Weiss M A. Molecular basis of mammalian sexual determination: activation of Müllerian inhibiting substance gene expression by SRY. Science. 1994;266:1494–1500. doi: 10.1126/science.7985018. [DOI] [PubMed] [Google Scholar]
- 21.Harley V R, Goodfellow P N. The biochemical role of SRY in sex determination. Mol Reprod Dev. 1994;39:184–193. doi: 10.1002/mrd.1080390211. [DOI] [PubMed] [Google Scholar]
- 22.Harley V R, Jackson D, Hextall P, Hawkins J R, Berkovitz G D, Sockanathan S, Lovell-Badge R, Goodfellow P N. DNA binding activity of recombinant SRY from normal males and XY females. Science. 1992;255:453–456. doi: 10.1126/science.1734522. [DOI] [PubMed] [Google Scholar]
- 23.Honda S, Morohashi K, Nomura M, Takeya H, Kitajima M, Omura T. Ad4BP regulating steroidogenic P-450 gene is a member of steroid hormone receptor superfamily. J Biol Chem. 1993;268:7494–7502. [PubMed] [Google Scholar]
- 24.Jay P, Diriong S, Taviaux S, Roeckel N, Mattéi M-G, Audit M, Bergé-Lefranc J-L, Fontes M, Berta P. Isolation and regional mapping of cDNAs expressed during early human development. Genomics. 1996;39:104–108. doi: 10.1006/geno.1996.4470. [DOI] [PubMed] [Google Scholar]
- 25.Josso N, Cate R L, Picard J-Y, Vigier B, Di Clemente N, Wilson C, Imbeaud S, Pepinsky R B, Guerrier D, Boussin L, Legeai L, Carré-Eusèbe D. Anti-Müllerian hormone the Jost factor. Recent Prog Horm Res. 1993;8:379–418. doi: 10.1016/b978-0-12-571148-7.50005-1. [DOI] [PubMed] [Google Scholar]
- 26.Kent J, Wheatley S C, Andrews J E, Sinclair A H, Koopman P. A male-specific role for SOX9 in vertebrate sex determination. Development. 1996;122:2813–2822. doi: 10.1242/dev.122.9.2813. [DOI] [PubMed] [Google Scholar]
- 27.Lai J-S, Herr W. EtBr provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lala D S, Rice D A, Parker K L. Steroidogenic factor 1, a key regulator of steroidogenic enzyme expression, is the mouse homolog of fushi tarazu-factor 1. Mol Endocrinol. 1992;6:1249–1258. doi: 10.1210/mend.6.8.1406703. [DOI] [PubMed] [Google Scholar]
- 29.Lala D S, Syka P M, Lazarchik S B, Mangelsdorf D J, Parker K L, Heyman R A. Activation of orphan nuclear receptor steroidogenic factor 1 by oxysterols. Proc Natl Acad Sci USA. 1997;94:4895–4900. doi: 10.1073/pnas.94.10.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefebvre V, Huang W, Harley V R, Goodfellow P N, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the proα1(II) collagen gene. Mol Cell Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer J, Südbeck P, Held M, Wagner T, Schmitz M L, Bricarelli F D, Eggermont E, Friedrich U, Hass O A, Kobelt A, Leroy J G, van Maldergem L, Michel E, Mitulla B, Pfeiffer R A, Schinzel A, Schmidt H, Scherer G. Mutational analysis of the SOX9 gene in campomelic dysplasia and autosomal sex reversal: lack of genotype/phenotype correlations. Hum Mol Genet. 1997;6:91–98. doi: 10.1093/hmg/6.1.91. [DOI] [PubMed] [Google Scholar]
- 32.Morais da Silva S, Hacker A, Harley V, Goodfellow P, Swain A, Lovell-Badge R. Sox9 expression during gonadal development implies a conserved role for the gene in testis differentiation in mammals and birds. Nat Genet. 1996;14:62–68. doi: 10.1038/ng0996-62. [DOI] [PubMed] [Google Scholar]
- 33.Morohashi K I, Honda S L, Inamata Y, Handa H, Omura T. A common trans-acting factor, Ad4-binding protein, to the promoters of steroidogenic P-450s. J Biol Chem. 1992;267:17913–17919. [PubMed] [Google Scholar]
- 34.Münsterberg A, Lovell-Badge R. Expression of the mouse anti-Müllerian hormone gene suggests a role in both female and male sexual differentiation. Development. 1991;113:613–624. doi: 10.1242/dev.113.2.613. [DOI] [PubMed] [Google Scholar]
- 35.Nachtigal M W, Hirokawa Y, Enyeart-VanHouten D L, Flanagan J N, Hammer G D, Ingraham H A. Wilms’ tumor 1 and Dax-1 modulate the orphan nuclear receptor SF-1 in sex-specific gene expression. Cell. 1998;93:445–454. doi: 10.1016/s0092-8674(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 36.Nasrin N, Buggs C, Kong X F, Carnazza J, Goebl M, Alexander-Bridges M. DNA-binding properties of the product of the testis-determining gene related protein. Nature. 1991;354:317–320. doi: 10.1038/354317a0. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen D A, Chang T C, Shapiro D J. A highly sensitive, mixed-phase assay for chloramphenicol acetyl transferase activity in transfected cells. Anal Biochem. 1989;179:19–23. doi: 10.1016/0003-2697(89)90193-0. [DOI] [PubMed] [Google Scholar]
- 38.Onodera K, Yomogida K, Suwabe N, Takahashi S, Muraosa Y, Hayashi N, Ito E, Gu L, Rassoulzadegan M, Engel J D, Yamamoto M. Conserved structure, regulatory elements, and transcriptional regulation from the GATA-1 gene testis promoter. J Biochem. 1997;121:251–263. doi: 10.1093/oxfordjournals.jbchem.a021581. [DOI] [PubMed] [Google Scholar]
- 39.Pevny L H, Lovell-Badge R. Sox genes find their feet. Curr Opin Genet Dev. 1997;7:338–344. doi: 10.1016/s0959-437x(97)80147-5. [DOI] [PubMed] [Google Scholar]
- 40.Poulat F, de Santa Barbara P, Desclozeaux M, Soullier S, Moniot B, Bonneaud N, Boizet B, Berta P. The human testis determining factor SRY binds a nuclear factor containing PDZ protein interaction domains. J Biol Chem. 1997;272:7167–7172. doi: 10.1074/jbc.272.11.7167. [DOI] [PubMed] [Google Scholar]
- 41.Poulat F, Girard F, Chevron M-P, Gozé C, Rebillard X, Calas B, Lamb N, Berta P. Nuclear localization of the testis determining gene product SRY. J Cell Biol. 1995;5:737–748. doi: 10.1083/jcb.128.5.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schafer A J, Goodfellow P N. Sex determination in humans. Bioessays. 1996;18:955–963. doi: 10.1002/bies.950181205. [DOI] [PubMed] [Google Scholar]
- 43.Shen W H, Moore C C D, Ikeda Y, Parker K L, Ingraham H A. Nuclear receptor steroidogenic factor 1 regulates the Müllerian inhibiting substance gene: a link to the sex determination cascade. Cell. 1994;77:651–661. doi: 10.1016/0092-8674(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 44.Sinclair A H, Berta P, Palmer M S, Hawkins J R, Griffiths B L, Smith M J, Foster J W, Frischauf A M, Lovell-Badge R, Goodfellow P N. A gene from the human sex-determining region encoding a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 45.Südbeck P, Scherer G. Two independent nuclear localization signals are present in the DNA-binding high mobility group domains of SRY and SOX9. J Biol Chem. 1997;272:27848–27852. doi: 10.1074/jbc.272.44.27848. [DOI] [PubMed] [Google Scholar]
- 46.Südbeck P, Schmitz M L, Baeuerle P A, Scherer G. Sex reversal by loss of the C-terminal transactivation domain of human SOX9. Nat Genet. 1996;13:230–232. doi: 10.1038/ng0696-230. [DOI] [PubMed] [Google Scholar]
- 47.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli F D, Keutel J, Hustert E, Wolf U, Tommerup N, Schempp W, Scherer G. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY related gene SOX9. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 48.Waterman M, Jones K. Purification of TCF-1α, a T-cell-specific transcription factor that activates the T-cell receptor gene enhancer in a context-dependent manner. New Biol. 1990;2:621–636. [PubMed] [Google Scholar]
- 49.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 50.Werner M H, Huth J R, Gronenborg A M, Clore G M. Molecular basis of human 46X,Y sex reversal revealed from the three dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- 51.Yomogida K, Ohtani H, Harigae H, Ito E, Nishume Y, Engel J D, Yamamoto M. Developmental stage- and spermatogenic cycle-specific expression of transcription factor GATA-1 in mouse Sertoli cells. Development. 1994;120:1759–1766. doi: 10.1242/dev.120.7.1759. [DOI] [PubMed] [Google Scholar]
- 52.Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]