Abstract
Templated synthesis of proteins containing non-natural amino acids (nnAAs) promises to vastly expand the chemical space available to biological therapeutics and materials. Existing technologies limit the identity and number of nnAAs than can be incorporated into a given protein. Addressing these bottlenecks requires deeper understanding of the mechanism of messenger RNA (mRNA) templated protein synthesis and how this mechanism is perturbed by nnAAs. Here we examine the impact of both monomer backbone and side chain on formation and ribosome-utilization of the central protein synthesis substate: the ternary complex of native, aminoacylated transfer RNA (aa-tRNA), thermally unstable elongation factor (EF-Tu), and GTP. By performing ensemble and single-molecule fluorescence resonance energy transfer (FRET) measurements, we reveal the dramatic effect of monomer backbone on ternary complex formation and protein synthesis. Both the (R) and (S)-β2 isomers of Phe disrupt ternary complex formation to levels below in vitro detection limits, while (R)- and (S)-β3-Phe reduce ternary complex stability by approximately one order of magnitude. Consistent with these findings, (R)- and (S)-β2-Phe-charged tRNAs were not utilized by the ribosome, while (R)- and (S)-β3-Phe stereoisomers were utilized inefficiently. The reduced affinities of both species for EF-Tu ostensibly bypassed the proofreading stage of mRNA decoding. (R)-β3-Phe but not (S)-β3-Phe also exhibited order of magnitude defects in the rate of substrate translocation after mRNA decoding, in line with defects in peptide bond formation that have been observed for D-α-Phe. We conclude from these findings that non-natural amino acids can negatively impact the translation mechanism on multiple fronts and that the bottlenecks for improvement must include consideration of the efficiency and stability of ternary complex formation.
Graphical Abstract
INTRODUCTION
The mechanism of ribosome-catalyzed polypeptide polymerization offers the opportunity to perform template-driven synthesis of non-protein polymers with benefits for both fundamental research and biomedicine. Over the last several decades, substantial progress has been made towards expanding the genetic code to expand the expressed proteome well beyond the 20 natural α-amino acids. More than 200 different non-natural α-amino acids (nnAA) and a handful of α-hydroxy acids can be incorporated into proteins1–5. There are now several examples of β2- and/or β3-monomers that have been introduced into proteins in cells, either directly6–8 or via rearrangement9. Robust methods of α- and non-α nnAA incorporation into proteins is expected to promote the development of new tools to probe structure-function relationships, discover catalysts, and advance therapeutic approaches.
The most widely adopted nnAA-incorporation strategies require the transplantation of translation components from orthogonal biological systems into model organisms (e.g., Escherichia coli) to promote selective formation of the desired aminoacyl-tRNA10,11. Inefficient suppressor tRNA aminoacylation (charging) initially represented a severe bottleneck, which was largely overcome through the engineering of native aminoacyl tRNA synthases (aaRS)12,13. Such strides have permitted the incorporation of α-amino acids with distinct non-natural side chains into otherwise native proteins11,14. However, the translational machinery did not evolve to support the translation of components with alternative backbones, and significant evidence exists that multiple kinetic bottlenecks likely exist7,15,16. Here we identify kinetic bottlenecks that exist during the delivery and accommodation of acylated tRNA by the ribosome.
During translation, aa-tRNAs are delivered to the ribosome in ternary complex with EF-Tu (eEF1A in eukaryotes) and GTP. The three domains (DI-DIII) of EF-Tu directly engage the amino acid backbone and side chain as well as the TψC and acceptor stems of tRNA to form a high-affinity (ca. 10–100 nM) complex (Fig.1A)17–19. EF-Tu exhibits distinct affinities for each aa-tRNA species 20,21. The prevailing hypothesis is that this variance ensures uniform decoding speeds by balancing the spring-like forces that accumulate in aa-tRNA during initial selection and proofreading steps of tRNA selection on the ribosome that ultimately lead to aa-tRNA dissociation from EF-Tu, allowing the aminoacylated 3’-CCA terminus of the tRNA to enter the ribosome’s peptidyltransferase center22–24. During proofreading selection, which occurs after GTP hydrolysis at the end of initial selection, rate-limiting conformational changes in EF-Tu responsible for triggering its dissociation from aa-tRNA and the ribosome contribute to substrate discrimination by allowing additional time for near- and non-cognate aa-tRNAs to dissociate22,25–30. Together, initial selection and proofreading selection ensure an error rate of approximately one in 1,000–10,000 mRNA codons for natural α-amino acids31–33.
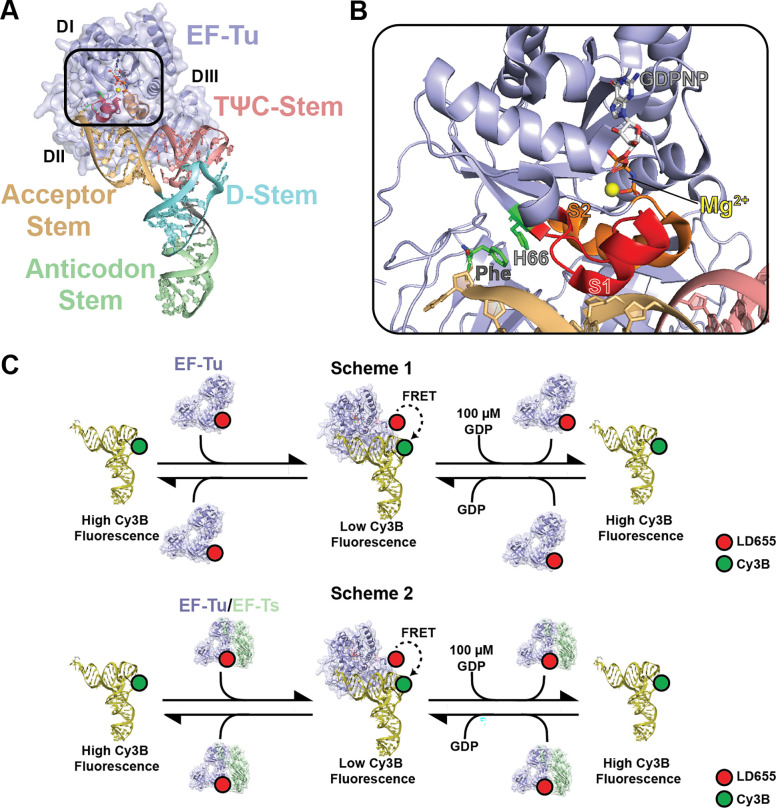
Figure 1. Structures of ternary complex and EF-Tu/Ts.
A) Crystal structure of ternary complex (PDB: 1OB2) highlighting the different structural domains of tRNA (variegated) and EF-Tu (blue). The box signifies the zoom of the amino acid binding pocket of EF-Tu that is shown in panel B. B) Zoomed image of the EF-Tu amino acid binding pocket from the boxed region in panel A highlighting specific functional elements for ternary complex stability. The π-π stacking interaction between phenylalanine (Phe) aminoacylated to the 3’ end of tRNAPhe (yellow) and histidine 66 (H66) in EF-Tu. The switch 1 (S1) and switch 2 (S2) helices which coordinate to the GTP in the nucleotide binding pocket are in red and orange, respectively. The coordinated magnesium (Mg2+) is shown in yellow and the non-hydorlyzable GDPNP analog is shown adjacent to the magnesium in stick form. C) Cartoon schematics of the FRET-based ensemble ternary complex formation assay, with LD655-labeled EF-Tu (blue) without (Scheme 1) or with (Scheme 2) EF-Ts (green) (400 nM) is injected into a cuvette containing 5 nM Cy3B-labeled tRNAPhe (wheat). Formation of ternary complex results in rapid quenching of Cy3B fluorescence via FRET that can be recovered upon dissociation after injection of excess.
The active (GTP-bound) form of EF-Tu harbors a precisely formed binding pocket for the α-amino acid backbone as well as a relatively spacious cavity for the side chains of the 20 naturally occurring amino acids. Upon aa-tRNA engagement, all three EF-Tu domains (DI-III) collapse around the CCA-3’ end of the tRNA acceptor stem to position the constituent components of the amino acid side chain, which concomitantly restructures the switch 1 helix (S1) to engage the γ-phosphate of GTP via Mg2+ coordination to yield the thermodynamically stable ternary complex (Fig. 1B)34,35. This stability strictly depends on tRNA aminoacylation and the presence of a properly positioned γ-phosphate moiety19,36. In line with this exquisite sensitivity, in vitro transcribed tRNAs mis-acylated with native amino acids can exhibit binding affinities for EF-Tu(GTP) that are reduced by up to ~5000-fold20, which affect both the speed and fidelity of tRNA selection on the ribosome37–39. However, the precise contributions of the amino acid backbone to EF-Tu(GTP) affinity have yet to be fully explored.
In bacteria, ternary complex formation is catalyzed by the conserved, EF-Tu-specific guanosine nucleotide exchange factor (GEF), EF-Ts. Under nutrient-rich conditions where GTP concentration far exceeds that of GDP, EF-Ts ensures rapid and abundant ternary complex formation so that ternary complex formation is not rate-limiting to protein synthesis19,36. Under nutrient-poor conditions when GDP concentrations are elevated, EF-Ts instead facilitates ternary complex disassembly, reducing protein synthesis and other energy-intensive cellular processes19,36. The impact of EF-Ts on both ternary complex formation and dissociation indicates that nucleotide exchange and S1 restructuring are dynamic processes that can be influenced by EF-Ts both in the absence or presence of aa-tRNA.
Here we use ensemble and single-molecule FRET-based kinetic assays, together with kinetic simulations, to investigate the effects of both non-natural α-amino acids (specifically, those with a non-natural side chain and a natural α-backbone) and non-natural backbones (specifically, those with a natural side chain and a non-natural β2- or β3-amino acid backbone) on the kinetics and thermodynamics of ternary complex formation and tRNA elongation on the ribosome19,36. Consistent with its widespread use by diverse research labs, our investigations show that the metrics of both ternary complex formation and tRNA selection are virtually identical for tRNAs acylated with α-Phe or the non-natural α-amino acid para-azido-phenylalanine (p-Az-Phe). By contrast, the kinetics of both ternary complex formation and tRNA selection were altered for backbone-modified monomers. The presence of either (R) or (S)-β2 Phe in place of L-α-Phe disrupts ternary complex formation to levels below our in vitro detection limits, whereas (R)- and (S)-β3-Phe reduce ternary complex stability by approximately an order of magnitude. These deficiencies are exacerbated by thermally stable elongation factor EF-Ts, the cellular guanosine nucleotide exchange factor for EF-Tu, and by mutations in both EF-Tu and tRNA previously reported to stabilize ternary complex using in vitro transcribed tRNAs40,41. In line with these observations, ribosomes fail to recognize (R)- and (S)-β2-Phe-charged tRNAs as substrates, while the utilization of (R)- and (S)-β3-Phe stereoisomers was significantly impaired relative to native L-α-Phe. The reduced EF-Tu affinities of tRNAs acylated with either (R)- or (S)-β3-Phe also precipitated defects in the mRNA decoding mechanism, where the proofreading stage of the tRNA selection process immediately after GTP hydrolysis was ostensibly bypassed. Following incorporation into the ribosome, tRNAs charged with (R)- and (S)-β3-Phe stereoisomers were both competent for translocation. However, as predicted using recent metadynamics simulations16, (R)-β3-Phe appeared to exhibit order of magnitude defects in the rate of peptide bond formation, which dramatically reduced the rate of substrate translocation from hybrid-state tRNA positions. We conclude from these findings that the efficiency of ternary complex formation and its thermodynamic stability are key determinants of nnAA incorporation into proteins and that engineering opportunities exist to enable their more efficient utilization.
RESULTS
Quantifying ternary complex formation.
To directly investigate how various types of non-natural amino acids impact the kinetic features of ternary complex formation, we employed an ensemble approach in which the rate and extent of ternary complex formation was tracked via FRET19,36. In this assay, EF-Tu carrying an LD655 fluorophore at its C-terminus quenches the fluorescence of a Cy3B fluorophore attached to the 3-amino-3-carboxypropyl (acp3) modification at position U47 of native tRNAPhe when the ternary complex forms (Fig. 1C). Reactions were initiated upon stopped-flow addition of 400 nM EF-Tu-LD655 to a solution of 5 nM L-α-Phe-tRNAPhe-Cy3B in a 1.2 mL cuvette with stirring at 25 °C (Methods). Under these conditions, the observed pseudo first-order, apparent rate constant (kobs) for Cy3B fluorescence quenching was 0.12 ± 0.002 s−1, reaching ~65% quenching at equilibrium (Fig. 2A; Table 1). The observed fluorescence quenching amplitude agrees with the close proximity of Cy3B and LD655 fluorophores within the ternary complex (Fig. 1)19. Consistent with the GTP requirement for ternary complex formation, rapid stopped-flow addition of 100 μM GDP to the reaction restored Cy3B fluorescence to ~80% of the initial intensity with an apparent rate constant (koff) of 0.005 ± 0.001 s−1 (Fig. 2A; Table 1). Identical experiments performed using deacylated tRNAPhe-Cy3B in place of L-α-Phe-tRNAPhe-Cy3B resulted in no Cy3B quenching (Fig. 2A), congruous with the selectivity of EF-Tu for aminoacylated tRNA.
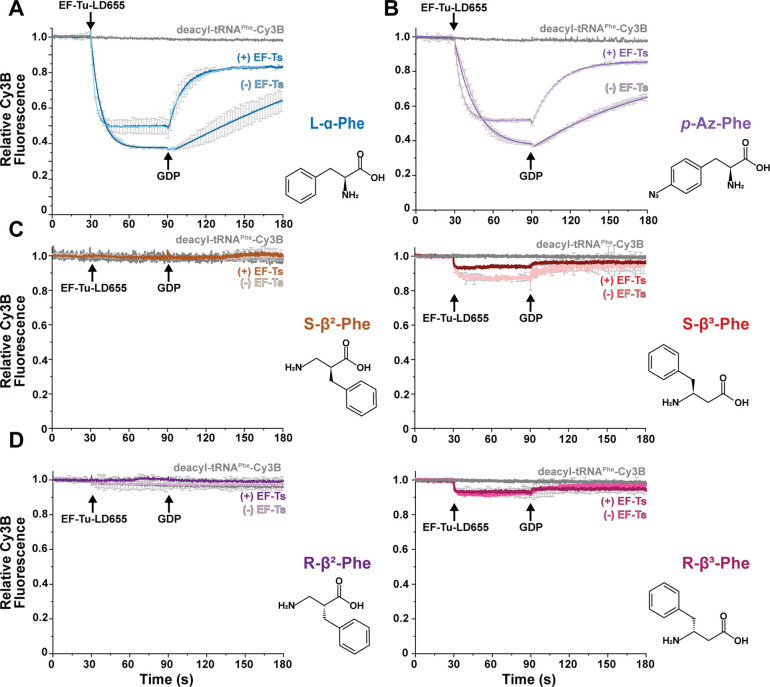
Figure 2. Non-natural amino acids severely disrupt ternary complex stability.
Ensemble ternary complex assays tracking the apparent rates (kobs) of Cy3B relative fluorescence changes upon mixing with LD655-labeled EF-Tu (−/+) EF-Ts (400 nM) in the presence of GTP (100 μM GDP) with Cy3B-labeled tRNAPhe (5 nM) charged with specific amino acid monomers with A) L-α-Phe. B) p-azido-Phe (pAzF). C) S-β2-Phe (left); S-β3-Phe (right). D) R-β2-Phe (left); R-β3-Phe (right). Structures of amino acid monomers are shown to the right of each plot. Error bars represent standard deviation from two experimental replicates.
Table 1.
Apparent reaction rates estimated by single-exponential fitting of the ensemble ternary complex formation experiments shown in Figure 2. Uncertainty estimates represent S.D. from two experimental replicates. Single-exponential fits exhibited R2 values >0.95. The L-α-Phe rates compare well with previously reported results 36.
| Observed Rates | L-α-Phe | p-Az-Phe | ||
|---|---|---|---|---|
|
| ||||
| EF-Ts | − | + | − | + |
|
| ||||
| kobs (S−1) | 0.12 ± 0.002 | 0.31 ± 0.04 | 0.06 ± 0.01 | 0.25 ± 0.02 |
| koff (s−1) | 0.005 ± 0.001 | 0.06 ± 0.007 | 0.01 ± 0.0004 | 0.06 ± 0.01 |
When analogous experiments were performed with the nucleotide exchange factor EF-Ts present in a 1:1 ratio relative to EF-Tu, kobs increased to 0.31 ± 0.04 s−1 and the extent of Cy3B fluorescence quenching at equilibrium was reduced to ~50%. Addition of 100 μM GDP to the same reaction restored the Cy3B fluorescence amplitude with a koff that was approximately 10-fold faster than with EF-Tu alone (0.06 ± 0.007 s−1). As for EF-Tu alone, the fluorescence intensity prior to ternary complex formation was nearly fully restored, although not to the full extent due to residual ternary complex formation under equilibrium conditions (Fig 1C, scheme 2; Fig. 2A). These observations are consistent with previously reported affinities of EF-Tu(GTP) for aa-tRNA (ca. 10–100 nM) and prior conclusions that EF-Ts can engage with EF-Tu in ternary complex to catalyze nucleotide exchange, thereby accelerating ternary complex dissociation rates after GDP addition by ~20-fold19,36.
Ternary complex stability is disrupted by nnAAs.
With this foundation, we next asked how the rates of ternary complex formation and stability were affected when tRNAPhe-Cy3B was aminoacylated with non-natural amino acids such as p-Az-Phe, (R)- and (S)-β2-Phe and (R)- and (S)-β3-Phe. These nnAA were used to acylate native tRNAPhe-Cy3B using flexizyme42–44. Each charged species was purified by FPLC, flash frozen in aliquots, and verified as >95% intact prior to use (Methods; Fig. S1A, B). We confirmed that flexizyme-charged L-α-Phe-tRNAPhe-Cy3B exhibited nearly identical formation/dissociation kinetics and fluorescence quenching/recovery amplitudes as the enzymatically charged species (Fig. S2), validating this experimental tRNA aminoacylation strategy for kinetic assays6,15,43,45.
Using experimental conditions identical to those described above, we next examined the kinetics of ternary complex assembly usingtRNAPhe-Cy3B that was acylated with p-Az-Phe, a nnAA successfully incorporated into proteins by multiple laboratories10,13,46,47. Stopped-flow addition of EF-Tu-LD655 to p-Az-Phe-tRNAPhe-Cy3B resulted in Cy3B fluorescence quenching characterized by a kobs of 0.06 ± 0.01 s−1; the extent of fluorescence quenching reached ~65% at equilibrium (Fig. 2B; Table 1). As observed for the complex of L-α-Phe-tRNAPhe-Cy3B, rapid addition of GDP slowly restored Cy3B fluorescence; in this case, the measured koff was 0.01 ± 0.0004 s−1 (Fig 2B and Table 1). In presence of EF-Ts, ternary complex formation again proceeded more rapidly (kobs = 0.25 ± 0.02 s−1) and the fluorescence quenching reached an amplitude of ~50% at equilibrium (Fig 2B and Table 1). Ternary complex dissociation upon addition of GDP proceeded with a kobs of 0.06 ± 0.02 s−1, restoring Cy3B fluorescence to a ~80% of the initial intensity. Overall, the kinetics parameters measured for assembly and disassembly of ternary complexes containing p-Az-Phe-tRNAPhe-Cy3B were similar to those measured for complexes containing L-α-Phe-tRNAPhe-Cy3B, Thus, the presence of the p-N3 side chain on Phe has, as expected, only a modest impact on ternary complex formation and stability. These results are entirely consistent with the functionalized phenyl sidechain being readily accommodated by EF-Tu.
We next investigated the kinetics of ternary complex assembly and disassembly when tRNAPhe-Cy3B was aminoacylated with the (R)- or (S)- enantiomers of β2- or β3-Phe. Many β2- and β3 -amino acids have been introduced into short peptides using small scale in vitro translation reactions48,49 and a few β3-amino acids have been introduced into proteins in cell lysates50,51. Moreover, one β3-Phe derivative6, three β3-aryl derivatives8, and one β2-hydroxy acid7 have been introduced into proteins in cells. Yet in none of these cases was the level of incorporation especially high, perhaps because of impaired delivery to the ribosome by EF-Tu52,53.
Thus, using experimental conditions identical to those described above, we examined ternary complex formation using tRNAPhe-Cy3B that was acylated with the (R)- and (S)- enantiomers of β2- and β3-Phe (Methods; Fig. 2). Under these conditions we detected no evidence for ternary complex formation in reactions containing (R)- or (S)-β2-Phe-tRNAPhe-Cy3B (Fig. 2C), while reactions containing (R)- or (S)-β3-Phe-tRNAPhe-Cy3B exhibited detectable, yet greatly reduced levels of Cy3B fluorescence quenching (~10–15% vs. ~60–65% with L-α-Phe). Quenching was reversed upon addition of 100 μM GDP, as expected for quenching that resulted from ternary complex formation (Fig. 2C and 2D). Also consistent with ternary complex formation was the observation that addition of EF-Ts to the ternary complex assembly reactions containing (R)- or (S)-β3-Phe-tRNAPhe-Cy3B attenuated Cy3B fluorescence quenching. Due to the impaired signal amplitudes of these reactions, reliable kobs and koff rates could not, however, be estimated.
β2-Phe and β3-Phe monomers negatively impact the kinetic features of ternary complex formation
We next employed a micro-volume stopped-flow system (μSFM, Biologic) (Methods) to perform a thorough kinetic study of ternary complex formation for tRNAs carrying (R) or (S)-β3-Phe, which showed the highest fluorescence quenching of the four acylated tRNAs studied above. Using this approach, we measured bimolecular association rate constants for EF-Tu binding to tRNAPhe-Cy3B acylated with either L-α-Phe- or (S)-β3-Phe-charged tRNAPhe by tracking how kobs changed as a function of the concentration of EF-Tu or EF-Tu/Ts. Upon rapid mixing of 5 nM L-α-Phe-charged tRNAPhe-Cy3B with a large excess of EF-Tu (0.2 – 2 μM) and GTP (1 mM), we observed a linear increase in kobs as a function of both [EF-Tu] and [EF-Tu/Ts], consistent with pseudo-first order binding kinetics (Figure 3A and 3B). In line with previous literature19,36, in the absence of EF-Ts, the bimolecular rate constant (kon) was 5.0 ± 0.1 μM−1 s−1 and the dissociation rate constant (koff) was 0.2 ± 0.1 s−1 (Table 2). With equimolar EF-Tu/Ts, kon and koff increased by 1.6- and 4-fold, respectively19,36. Consistent with our initial findings, when L-α-Phe was substituted with (S)-β3-Phe, we observed both a lower kon (1.0 ± 0.1 μM−1 s−1) and a higher koff (1 ± 0.1 s−1) (Table 2). In the presence of EF-Ts, kon and koff increased by 3-fold and 1.8-fold, respectively (Table 2). For (R)-β3-Phe, the fluorescence intensity changes were still too small for reliable estimations of its kinetic parameters (Fig. S3). Additionally, we were unable to rescue the observed defects using tRNA (C49A, G65U) and EF-Tu (N273A) mutations previously reported to stabilize ternary complex formation (Methods; Fig. S3).
Figure 3. Rapid, pre-steady state measurements of ternary complex formation rate constants experiments.
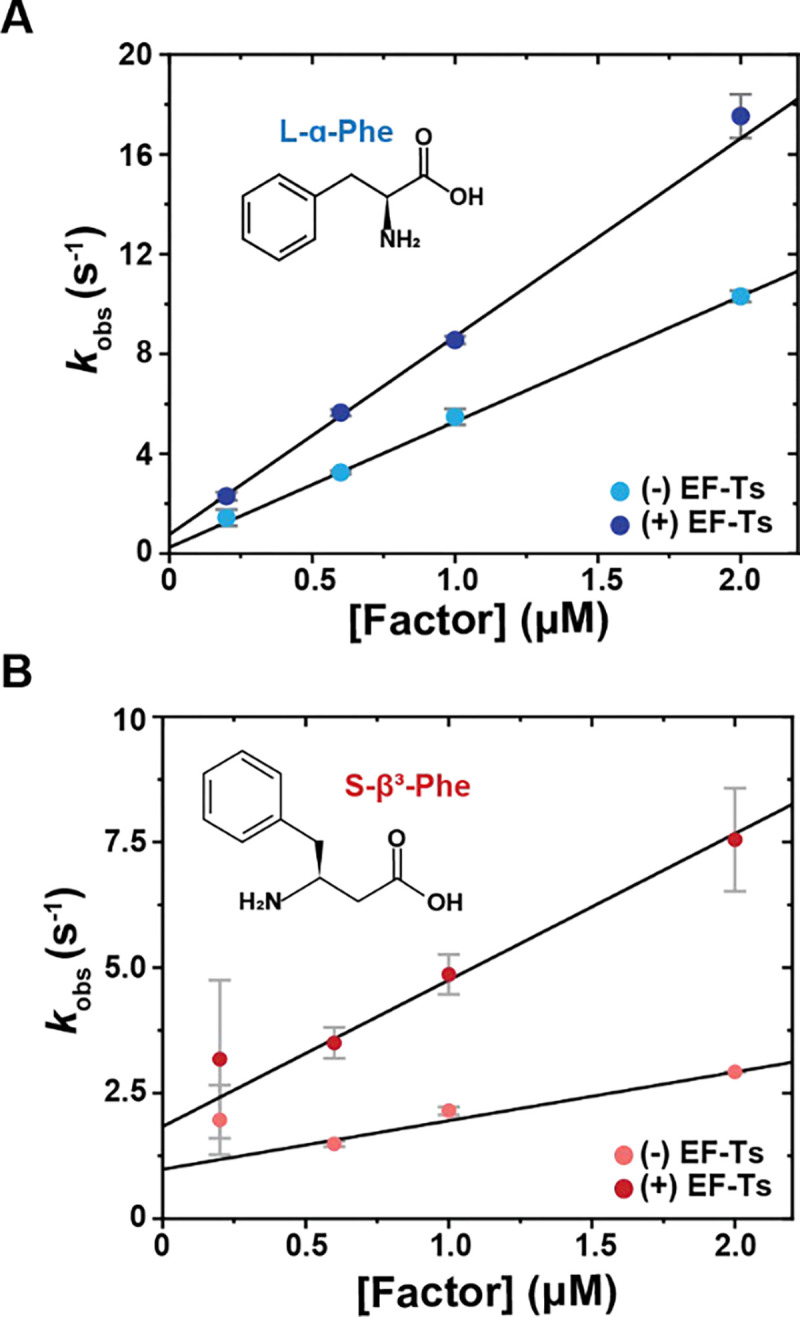
A) Plot showing the apparent rate (kobs) of ternary complex formation for Cy3B-labeled L-α-Phe-tRNAPhe as a function of EF-Tu (sky blue) and equimolar EF-Tu/Ts (navy blue) concentrations performed in the presence of 100 μM GTP. B) Analogous plots of kobs for Cy3B-labeled S-β3-Phe-tRNAPhe in the absence (salmon) and presence (red) of equimolar EF-Ts. C) Kinetic framework for ternary complex formation based on prior literature 19,36,79, indicating ternary complex formation- inactive (grey) and active (black) species of EF-Tu. Error bars represent S.D. from 3–7 experimental replicates.
Table 2.
Kinetic and thermodynamic parameters for aa-tRNAPhe binding to EF-Tu-LD655 in the presence and absence of EF-Ts from the experiments in Figure 3.
From these findings we estimated equilibrium dissociation constants (KD) for the ternary complexes of EF-Tu with L-α -Phe-tRNAPhe-Cy3B of approximately 40 and 100 nM in the absence and presence of EF-Ts, respectively (Table 2). Similar analyses estimated ternary complex KD values of ~1000 and ~600 nM for the analogous complexes containing (S)-β3-Phe, in the absence and presence of EF-Ts, respectively (Table 2). From these KD estimates we calculated ΔG° values of −9 to −11 kcal/mol for complexes containing L-α-Phe, consistent with previous literature20,41,54, while the ΔG° for complexes containing (S)-β3-Phe were only −4.1 to −4.4 kcal/mol. Hence, the stability of the (S)-β3-Phe-containing ternary complex would be outside of the thermodynamic range for ternary complexes that support efficient translation, as both tightly and loosely bound aa-tRNAs to EF-Tu impair translation by slowing peptide bond formation55 or aa-tRNA delivery to the ribosome, respectively54 (Table 2).
These data support the hypothesis that both β2- and β3-Phe monomers interfere with EF-Tu engagement by both reducing the efficiency with which EF-Tu productively engages acyl-tRNA as well as the overall stability of the ternary complex. Despite an approximate 25-fold reduction in affinity for (S)-β3-Phe-tRNAPhe, kinetic simulations based on a simplified framework (Fig. S4A), suggest that (S)-β3-Phe-ternary complexes can be populated significantly in a cellular context, where EF-Tu and EF-Ts are present at ~μM concentrations (Fig. S4B and S4C)56. We speculate that increasing cellular EF-Tu concentrations may only partially alleviate incorporation deficiencies due to complex instability.
β-Phe-charged tRNAs are poorly accommodated on the ribosome and ostensibly bypass proofreading steps.
To examine how ternary complexes formed with nnAAs are incorporated by the ribosome, we employed smFRET imaging methods to directly monitor both the frequency of productive ternary complex binding to the ribosome as well as the process by which an aa-tRNA is released from EF-Tu and incorporated into the ribosomal aminoacyl (A) site22,24,29. This established approach tracks FRET between a donor fluorophore linked to a peptidyl-tRNA bound within the ribosomal P-site and an acceptor fluorophore linked to the incoming aa-tRNA (Fig. 4A).
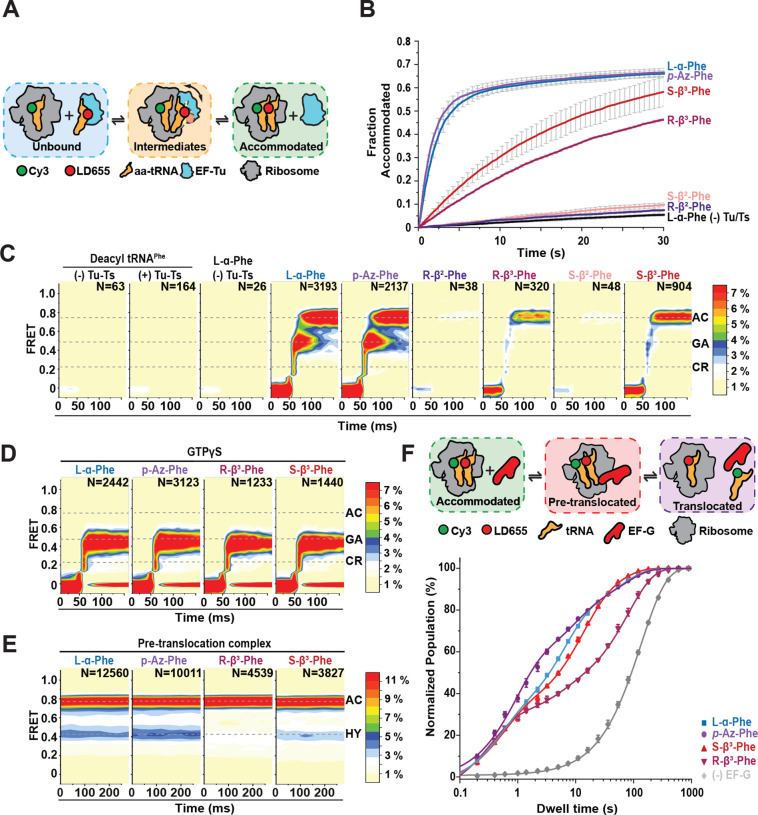
Figure 4. nnAA-tRNA selection on the bacterial ribosome studied by smFRET.
A) Schematic describing the smFRET tRNA selection experimental design, where LD655-labeled aa-tRNAPhe is delivered to surface-tethered ribosomes (blue) bearing Cy3-fMet-tRNAfMet in the P site and tRNA selection intermediates (tan), and the fully accommodated tRNA (green), are distinguished based on their distinct FRET values. B) Fraction of ribosomes bearing a fully accommodated aa-tRNA as a function of time (Methods). Data were collected at 100 ms time resolution; experiments were performed in triplicate. C-D) 2D histograms of smFRET traces containing productive accommodation events with GTP (C) or stalled events with GTPγS (D) (see methods) (N, number selected), post-synchronized to the appearance of FRET above baseline, with the specified amino acids. Contour plots were normalized (scale at right) to the L-α-Phe positive control. Dashed lines indicate FRET efficiency values of tRNA selection intermediates used in kinetic modeling. Data were collected at 10 ms time resolution. E) 2D histograms of smFRET traces of accommodated aa-tRNA after buffer exchange and ~5 min equilibration time, showing both classical (accommodated) and hybrid (P/E, A/P) tRNA conformations. Data were collected at 40 ms time resolution. F) Schematic and fraction of productive elongation cycles over time from tRNA selection and translocation experiments in presence of 10 μM EF-G. Line represent fits to one (L-α-Phe without EF-G) or three (L-α-Phe, p-Az-Phe, (R)- and (S)-β3-Phe) exponential distributions. Three different dwell time regimes are observed: the hybrid state (~0.5 s), the classical state (~8 s) and the “slow” state (~60 s). Data were collected at 200 ms time resolution. For all tRNA selection experiments, LD655-labeled aa-tRNAPhe and EF-Tu/Ts concentrations were 12.5 nM and 125 nM, respectively. For all translocation experiments, LD655-labeled aa-tRNAPhe and EF-Tu/Ts concentrations were 25 nM and 250 nM, respectively.
To perform these studies, bacterial ribosome complexes were programmed with a synthetic 5’-biotinylated mRNA that positions Cy3-fMet-tRNAfMet in the P site and a UUC codon in the A site. Initiation complexes were then tethered via a biotin-streptavidin bridge proximal to an optically transparent, polyethylene glycol (PEG)-passivated surface. Single-molecule tRNA selection experiments were subsequently initiated by stopped-flow injection of ternary complexes formed with flexizyme-charged tRNAPhe-LD655, where EF-Tu/Ts (125 nM) is in 10-fold excess over aa-tRNA (12.5 nM) (Methods).
We first assessed the apparent rates and extents of ternary complex utilization by the ribosome by collecting movies at low-time resolution (100 ms per video frame) where photobleaching is minimized. Consistent with our ensemble ternary complex formation studies (Figs. 2, 3), p-Az-Phe-tRNAPhe-LD655 was utilized as efficiently as L-α-Phe-tRNAPhe-LD655 (Fig. 4B; Table 3). Both (R)- and (S)-β2-Phe-LD655 were not utilized by the ribosome, while both (R)- and (S)-β3-Phe-tRNAPhe-LD655 were utilized >10-fold less compared to L-α-Phe-tRNAPhe-LD655, defects that we attribute to defects in ternary complex formation (Fig. 4B; Table 3).
Table 3.
Apparent aa-tRNA accommodation rates on the ribosome calculated from the experiments in Figure 4. Uncertainty estimates represent S.D. from three experimental replicates.
| Amino acid | k1 obs (S−1) | k2 obs (S−1) |
|---|---|---|
|
| ||
| L-α-Phe (−) Tu-Ts | 0.025 ± 0.002 | N/A |
| L-α-Phe | 0.46 ± 0.04 | 0.033 ± 0.001 |
| p-Az-Phe | 0.59 ± 0.03 | 0.036 ± 0.002 |
| R-β2-Phe | 0.024 ± 0.003 | N/A |
| R-β3-Phe | 0.041 ± 0.002 | N/A |
| S-β2-Phe | 0.024 ± 0.001 | N/A |
| S-β3-Phe | 0.056 ± 0.006 | N/A |
To gain more specific insights into the tRNA selection mechanism after ternary complex binding to the ribosome, we performed analogous investigations at 10-fold higher time resolution (10 ms per video frame). As previous smFRET studies of bacterial tRNA selection have shown22,29, experiments of this kind allow direct assessment of codon recognition (CR), GTPase activation (GA), and accommodation (AC) as each reaction endpoint exhibits a distinct FRET efficiency value (~0.21, 0.45, and 0.75, respectively; Methods).
By computationally isolating FRET trajectories reflecting productive ternary complex binding events to individual ribosomes (Methods)22 (i.e. those that stably accommodate) (Fig. 4C), we found that L-α-Phe and p-Az-Phe exhibited similar progression probabilities through tRNA selection, including the rate-limiting progression from the GTPase-activated state (~0.45, GA) to the fully accommodated state (~0.75, AC) during proofreading selection. As expected, tRNA selection events were ostensibly not observed for ternary complexes in which tRNA was acylated with (R)- or (S)-β2-Phe, whereas we observed ~5–10-fold fewer productive tRNA selection events for (R)- and (S)-β3-Phe ternary complexes (Fig. 4C). Strikingly, these rare events of (R)- and (S)-β3-Phe tRNA accommodation exhibited much more rapid passage of the intermediate states of tRNA selection compared to L-α-Phe.
To test whether the tRNA selection process for (R)- and (S)-β3-Phe bypassed GTP hydrolysis, we performed identical smFRET experiments with the non-hydrolyzable GTPγS analog to stall tRNA selection at the GTP hydrolysis step at the end of initial selection36. In the presence of GTPγS, both (R)- and (S)-β3-Phe-tRNAPhe-LD655 were efficiently stalled in the GA state, as were both L-α- and p-Az-Phe-tRNAPhe-LD655 (Fig. 4D). These findings reveal that GTP hydrolysis is indeed required for (R)- and (S)-β3-Phe-tRNAPhe to for GA state passage and the proofreading process. The observation that proofreading is significantly more rapid for (R)- and (S)-β3-Phe-tRNAPhe supports the notion that conformational changes in EF-Tu during proofreading related to 3’-CCA release are rate-limiting to the tRNA selection process22,29 and that this impact can be specifically attributed to the β-Phe monomers. Reduced thermodynamic stability leads to more rapid EF-Tu dissociation from aa-tRNA and likely the ribosome.
Since we were able to observe some β3-Phe accommodation on the ribosome, we next wanted to assess if (R)- and (S)-β3-Phe-tRNAPhe impact later steps of the elongation cycle, including peptide bond formation and EF-G catalyzed translocation. To examine these steps, first we examined the equilibrium between classical hybrid state pre-translocation complex conformations (A/P, P/E and A/A, P/E) that are adopted spontaneously after tRNA selection57–59 (Fig. 4E). L-α-Phe and p-Az-Phe showed a 64:36 (± 2) split ratio between classical and hybrid pre-translocation complexes. Notably, (S)-β3-Phe did not significantly affect the classical-hybrid split ratio (67:33 ± 3), while (R)-β3-Phe preferentially adopted classical conformations (74:26 ± 2; p < 0.05; Methods). These results indicate that, while (S)-β3-Phe behaves similarly to its α-amino acid counterparts, (R)-β3-Phe also perturbs spontaneous transitions to translocation-ready hybrid states23. Hence, incorporation of (R)-β3-Phe into proteins is predicted to increase the frequency of elongation pauses.
We next performed analogous smFRET experiments under cell-like conditions, where productive tRNA selection events are rapidly followed by translocation. To initiate the complete elongation cycle, we stopped-flow injected ternary complexes formed with LD655-labeled tRNAPhe (250nM EF-Tu/Ts, 25nM aa-tRNA) together with EF-G (10 μM) in the presence of 1 mM GTP. In this experiment, the appearance of FRET reports on aa-tRNA incorporating into the ribosome. The loss of FRET reports on dissociation of Cy3-labeled initiator tRNAfMet from the E site during, or after, the process of translocation60 (Fig. 4F). Correspondingly, FRET lifetime in these experiments reports on the total duration of the elongation cycle. We measured complete elongation cycle times using ternary complexes formed with L-α-Phe, p-Az-Phe as well as (R)- and (S)-β3-Phe-tRNAPhe, while including a control study lacking EF-G to estimate the contribution of fluorophore photobleaching (Fig. 4F). Consistent with each complex undergoing EF-G catalyzed translocation, the average FRET lifetimes were substantially shorter (ca. 2.5–8 fold) than photobleaching (165.8 ± 2.8 s). In line with prior investigations of translocation23,60–64, the FRET lifetime distributions for each amino acid type displayed multi-modal behaviors, characterized by a very fast (~0.3 s), fast (~5–10 s) and slow (~30–40 s) kinetic populations (Table 4). We attribute these sub-populations to pre-translocation complexes that undergo rapid translocation from hybrid state conformations, classical pre-translocation complexes that must wait for hybrid state conformations to spontaneously occur and pre-translocation (“slow”) complexes that either exhibit more substantial delays in translocation or retain deacylated tRNA in the E site after translocation is complete. L-α-Phe, p-Az-Phe as well as (R) and (S)-β3-Phe each exhibit rapidly translocating sub-populations, consistent with rapid peptide-bond formation followed by rapid translocation from hybrid state conformations. Sub-populations were also present for all four amino acids that exhibited long-lived FRET lifetimes, consistent with relatively stable, classical ribosome conformations requiring additional time to first transition to hybrid states before being able to productively engage EF-G and then rapidly translocate23,60,62 (Table 4). We note in this context that (R)-β3-Phe, the monomer most dissimilar to L-α-Phe, exhibited a relatively large sub-population of pre-translocation complexes that exhibit “slow” translocation, which may reflect specific deficiencies in peptide bond formation, hybrid state formation or deacylated tRNA release from the E site.
Table 4.
Dwell times for hybrid, classical and “slow” states from the fittings to three exponential distributions (Methods). Photobleaching lifetime is 165.8 ± 0.03 s.
| Hybrid state | Classical state | “Slow” state | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| Amino acid | Dwell time (s) | Population (%) | Dwell time (s) | Population (%) | Dwell time (s) | Population (%) |
|
| ||||||
| L-α-Phe | 0.44 ± 0.11 | 30.1 ± 3.6 | 6.12 ± 0.68 | 48.2 ± 3.1 | 59.17 ± 3.12 | 21.7 ± 1.7 |
| p-Az-Phe | 0.87 ± 0.08 | 53.3 ± 2.2 | 8.45 ± 1.13 | 26.2 ± 1.6 | 67.46 ± 3.35 | 20.5 ± 1.2 |
| S-β3-Phe | 0.53 ± 0.07 | 33.6 ± 2.9 | 11.81 ± 0.83 | 46.7 ± 2.2 | 47.56 ± 3.76 | 19.7 ± 2.5 |
| R-β3-Phe | 0.33 ± 0.03 | 33.9 ± 0.9 | 4.67 ± 0.80 | 12.1 ± 1.2 | 80.79 ± 3.31 | 54.0 ± 1.1 |
DISCUSSION
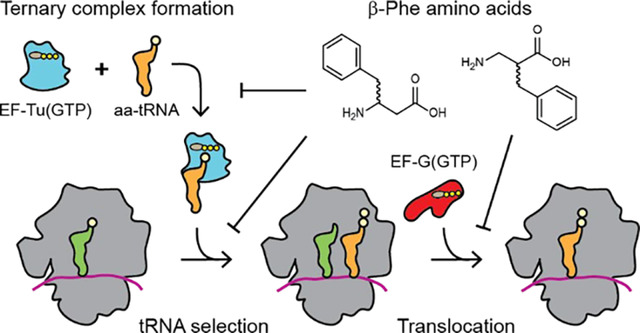
Efficient tRNA selection by the ribosome is paramount to the transfer of genetic information from mRNA to protein65. Harnessing this template-driven platform through genetic code expansion to synthesize previously unknown and useful bio-polymers promises myriad tools to advance research and medicine11. Achieving this goal requires continual technology development to overcome bottlenecks that limit the efficiency of non-α amino acid incorporation. These bottlenecks in principle include, but are not limited to, differences in monomer uptake and cellular stability, aminoacyl-tRNA synthetase activity and fidelity, ternary complex formation of the aa-tRNA with EF-Tu(GTP), constraints present within the ribosome PTC itself, as well as other fidelity mechanisms evolved to reduce erroneous amino acid incorporation by the ribosome13,66–69. Here we employ both ensemble and single-molecule kinetic assays, we show that ternary complex formation and utilization by the ribosome represent significant bottlenecks for the use of β-Phe monomers.
tRNAs acylated with (R)- and (S)-β-Phe monomers are disruptive to ternary complex formation and thus their utilization by the ribosome (Fig. 2C, D; Fig. 4B, C). tRNAs acylated with (R)- and (S)-β2-Phe appeared unable to form ternary complex with EF-Tu under the experimental conditions examined. The incorporation of (R)- and (S)-β2-Phe-tRNAs into the ribosome was below the detection limits of our tRNA selection smFRET assays. By contrast, (R)- and (S)-β3-Phe monomers inefficiently formed ternary complex, reducing ternary complex abundance and thus the number of detectable tRNA selection events per unit time (Fig. 4B, C). Strikingly, the proofreading stage of tRNA selection was accelerated for the events observed, consistent with faster rates of EF-Tu dissociation from both tRNA and the ribosome. We infer from these findings that β3-Phe monomers lower ternary complex stability to an extent that the proofreading stage of tRNA selection after GTP hydrolysis is ostensibly bypassed. As L-α-amino acids misacylated onto non-native tRNAs have been reported to exhibit identical accommodation rates despite evidence of ternary complex formation defects70, further experiments will be needed to discern whether the impact on proofreading is specific for β3-Phe monomers.
Our results also show that β3-Phe monomers exhibit significant reductions in the rate of downstream elongation reactions, including EF-G catalyzed translocation (Fig. 4F). (R)-β-Phe monomers, whose stereochemistry mimics that of an unnatural D- α -amino acid, showed significantly greater defects in ternary complex formation, tRNA accommodation during the proofreading stage of tRNA selection and EF-G-catalyzed translocation than their (S)-β-Phe counterparts. These observations are consistent with prior literature indicating that D-α-amino acids ((R)-α-amino acids) result in extended elongation pauses15,45,71. In the cell, such pauses are likely accompanied by futile cycles of EF-Tu catalyzed GTP hydrolysis29 and the induction of rescue pathways72 that may ultimately lead to compromises in cell growth and viability.
We conclude from these findings that ternary complex stability is a significant and perhaps underappreciated bottleneck that limits the incorporation of extended backbone monomers into polypeptides and proteins, both in vitro and in vivo. These observations warrant examination of the extent to which ternary complex stability and EF-Tu catalyzed tRNA selection defects impact the efficiency of nnAA utilization more broadly. Structural data19,36,73,74 suggest that the extended backbones of both β2- and β3-Phe may introduce steric clashes at the interface with EF-Tu that alter its ability to both engage aminoacylated tRNA termini and undergo the rearrangements needed for stable ternary complex formation. Inspection and in silico analysis of the previously reported structure of EF-Tu(GDPNP)-L-α-Phe-tRNAPhe complex (PDB ID: 1OB2) suggested that all four β-Phe monomers would clash with EF-Tu, while p-Az-Phe would not (Fig. S5). Reorientation of domains DI-III of EF-Tu, combined with repositioning of the aminoacylated tRNA termini could, in principle, relieve the observed steric clashes with the β-Phe monomers but this change would likely come at the expense of precise positioning of nucleotide binding pocket elements, including the S1 and S2 regions and therefore ternary complex stability (Fig. 1B). These findings support the hypothesis that engineering ternary complexes to compensate for nnAA-specific perturbations in stability could significantly improve incorporation efficiencies.
Further in-depth analyses of translation kinetics to address the issue of low non-α-amino acid utilization efficiency is warranted. To date, such studies have been performed under two different experimental approaches: in vitro translation48,75 or in cellulo incorporation68. In vitro translation systems allow for optimization over a wide range of experimental conditions and have been successful at incorporating diverse non-α-amino acid monomers in short peptides. However, their reaction yields are seldom reported and only a minor fraction of published studies focus on relative rates or mechanistic bottlenecks16. In cellulo studies that show incorporation of nnAAs outside of α-amino or α-hydroxy acid are far fewer6–8,76. Parallel in vitro and in cellulo investigations are likely to be most informative.
Despite clear evidence that β-Phe monomers cause defects in ternary complex formation and stability, recent experiments show that β3-amino or β2-hydroxy acids can be incorporated into proteins in cells6,7,45. β3-(p-Br)-Phe has also been incorporated into DHFR in E. coli cells expressing a ribosome containing a remodeled PTC6. A β2-hydroxy analogue of Nε-Boc-L-α-Lysine has also been incorporated into sfGFP by a wild-type E. coli strain7, although the yield of protein was far lower based than expected from the in vitro activity of the aaRS. Kinetic simulations of ternary complex formation reveal that this discrepancy is likely explained by the presence of elevated EF-Tu and tRNA concentrations in the cell that help drive ternary complex formation despite significant reductions in stability (Fig. S4). Despite this mass action effect, perturbations to the tRNA selection mechanism, and proofreading specifically, are nonetheless expected to remain.
To achieve incorporation efficiencies approximating native amino acids, engineered EF-Tu and tRNA mutants, and potentially other translation machinery components, will likely be required to compensate for monomer-induced penalties to the system. Such efforts should aid the design of tailored orthogonal ternary complex components to complement existing OTS technologies, increasing nnAA tolerance and desired product yields.
EXPERIMENTAL PROCEDURES
tRNAPhe purification and fluorescent labeling
Native tRNAPhe was expressed and purified as described previously 19,36,77. Briefly, pBS plasmid containing the tRNAPhe V gene was transformed into JM109 cells and incubated overnight at 37 °C with shaking at 250 rpm. Cells pelleted at 10,000 × g for 15 minutes. Cells were lysed by sonication in 20 mM potassium phosphate buffer pH 6.8 with 10 mM Mg(OAc)2 and 10 mM β-mercaptoethanol. Cellular debris was pelleted by high-speed centrifugation at 55,000 × g for 1.5 hours. Supernatant was phenol:chloroform extracted twice followed by a series of precipitation steps consisting of an EtOH precipitation, two isopropanol precipitation steps (added dropwise) and a final EtOH precipitation. Bulk tRNAPhe was then aminoacylated for 10 minutes in charging buffer (50 mM Tris-Cl, pH 8.0; 20 mM KCl; 100 mM NH4Cl; 10 mM MgCl2; 1 mM DTT; 2.5 mM ATP; and 0.5 mM EDTA) with PheRS (crude tRNA in 45-fold molar excess) and a 10-fold molar excess of L-phenylalanine. Phe-tRNAPhe was separated from crude tRNA using a TSK Phenyl-5PW HIC column (Tosoh Bioscience) with a linear gradient starting from buffer A (10 mM NH4OAc, pH 5.8; 1.7 M (NH4)2SO4) to buffer B (10 mM NH4OAc, pH 5.8; 10% MeOH). Elution fractions corresponding to Phe-tRNAPhe were pooled, dialyzed into storage buffer (10 mM KOAc, pH 6.0; 1 mM MgCl2) and concentrated.
Phe-tRNAPhe was fluorescently labeled with Cy™3B Mono NHS ester (Cytiva) using the native aminocarboxypropyluridine (acp3) post-transcriptional modification at the U47 position with previously described procedures 19,36. Briefly, Phe-tRNAPhe was buffer exchanged into 312.5 mM HEPES, pH 8.0. NaCl was added to a final concentration of 1 M. 2 μL of 50 mM Cy™3B in DMSO was added at 15-minute intervals over a 1-hour period. Reaction conditions were such that all the acyl bond is hydrolyzed during labeling yielding deacyl Cy™3B-tRNAPhe. Labeled tRNAPhe was then phenol:chloroform extracted, EtOH precipitated, and purified over the TSK Phenyl-5PW column as described above. Cy3B-tRNAPhe was buffer exchanged into storage buffer, concentrated, aliquoted and stored at −80 °C until further use.
Elongation factor purification and fluorescent labeling
His-tagged versions of elongation factors Tu and Ts were expressed from pPROEX vectors and purified by Ni2+-NTA as described previously 19,36. Fluorescent EF-Tu was labeled on a C-terminal acyl-carrier-protein (ACP) tag with LD655-CoA via ACP synthase. Briefly, 5–10 molar equivalents of LD655-CoA was mixed with EF-Tu-ACP in labeling buffer containing 50 mM HEPES pH 7.5, 10 mM Mg(OAc)2. Labeled EF-Tu-LD655 was separated from ACP-S and free dye via Ni2+-NTA, TEV protease was added to remove the 6X-His tag from EF-Tu-LD655 and run over a second Ni2+-NTA to remove TEV. EF-Tu-LD655/Ts complexes were purified by size exclusion chromatography, concentrated into factor storage buffer containing 10 mM HEPES pH 7.5, 100 mM KCl, 1 mM DTT, and 50% glycerol and stored at −20 °C.
Monomer synthesis
The general procedure for L-α-Phe cyanomethyl ester (CME) monomer synthesis followed previously published procedures with slight modifications 44. To a 5-mL round-bottom flask, N-Boc protected amino acid (0.5 mmol) was dissolved in 1 mL of tetrahydrofuran. Flask was then charged with 315 μL of chloroacetonitrile (5.0 mmol, 10 eq.), followed by addition of 100 μL of N,N-diisopropylethylamine (0.6 mmol, 1.2 eq.). The flask was capped with septa and stirred at room temperature overnight, 16 hours. Solvent was removed via rotary evaporation then the crude material was purified by reverse-phase flash chromatography, 0–100% acetonitrile in water, holding at 60% acetonitrile until product was collected. Solvent removed via rotary evaporation, where the resulting oil was dissolved in 1 mL of tetrahydrofuran for deprotection. To the resulting solution, 1.9 mL of trifluoroacetic acid (25 mmol, 50 eq.) was added, and allowed to stir at room temperature for 2 hours. Upon completion, the solvent was removed followed by purification by reverse-phase flash chromatography utilizing a 2% acetonitrile in water mobile phase. Solvent was removed by lyophilization to yield target materials.
The general procedure for β3-substituted phenylalanine CME monomers was performed as follows. The Boc-protected-Amino Acid-CME (ca 0.5 mmol) was treated with neat formic acid (2 mL). The solution was stirred at rt for 12 h before removing all the formic acid under reduced pressure by azotropic distillation with CHCl3 to afford a pale, yellow oil. The oil was dissolved in minimum amount of THF (ca. 2 mL), triturated with excess MTBE or Et2O until white solid was formed persistently. All the residual solvent was removed under reduced pressure. The white solid was crushed into fine powder, rinsed thoroughly with Et2O (10 mL) and dried over vacuum for overnight. The typical yield over two steps was 50%.
Flexizyme charging of native Cy3B-labeled tRNAPhe
All concentrations listed are final. 5 mM of CME monomers were charged onto Cy3B-tRNAPhe with a 5-fold excess of flexizyme. Flexizyme RNA oligo sequence which was developed and described previously, 43,44,78 used in this study is as follows: GGAUCGAAAGAUUUCCGCGGCCCCGAAAGGGGAUUAGCGUUAGGU. were ordered de-protected from IDT, resuspended in ultra-pure water, flash frozen and stored at −80 C. For L-α-Phe and p-Az-Phe, charging reactions were carried out in 100 mM HEPES, pH 6.6, 600 mM MgCl2, and 20% DMSO for 2 hours on ice. For the β2-substituted Phe monomers, reactions were carried out in 50 mM Bicine, pH 9.0, 600 mM MgCl2, and 30% DMSO on ice for 24 hours. For the β3-substituted Phe monomers, reactions were carried out in 50 mM Bicine, pH 9.0, 600 mM MgCl2, and 10% DMSO on ice for 24 hours. All flexizyme charging reactions were quenched with 90 μL of 0.3 M NaOAc, pH 5.3 and EtOH precipitated at −20 °C. Precipitate was centrifuged at 21,000 × g for 10 minutes, EtOH aspirated, precipitate was resuspended in Buffer A and the acylated species was purified from the deacylated species as described above for normal tRNAPhe purification procedures. Purified, charged monomers were dialyzed into storage buffer and concentrated down via Amicon centrifugal filters with a 3k MWCO. Samples were aliquoted, flash frozen, and stored at −80 °C.
Ternary complex assay
Ternary complex assays (Fig. 2 and 3) were performed with a QuantaMaster-400 Spectrofluorometer (Photon Technology International) with 520 nm and 570 nm excitation and emission wavelengths, as previously described or a micro stopped-flow system (μSFM, BioLogic) equipped with a MOS-200/M spectrometer with the excitation monochromator set at 520 nm to monitor changes in Cy3B fluorescence 19,36. In both cases a 532 long bandpass filter was placed in front of the emission PMT to omit noise as a result of excitation light. All concentrations listed are final. Ternary complex formation reactions were carried out in ternary complex reaction buffer containing 100 mM HEPES; pH 7.4, 20 mM KCl, 100 mM NH4Cl, 1 mM DTT, 0.5 mM EDTA, and 2.5 mM Mg(OAc)2. Briefly, ternary complex formation was achieved by stopped-flow injection of 400 nM (unless specified otherwise) EF-Tu-LD655 into a solution containing 5 nM aa-tRNAPhe-Cy3B in ternary complex reaction buffer. Prior to stopped-flow injection, EF-Tu-LD655 was preincubated in ternary complex reaction buffer with 10 μM GTP with or without EF-Ts as indicated. Upon reaction equilibration, ternary complex dissociation was achieved by the stopped-flow injection of 100 μM GDP in ternary complex buffer to the same solution. For the μSFM system, equal volumes of EF-Tu-LD655 with or without 3 μM EF-Ts and aa-tRNAPhe-Cy3B were rapidly mixed together (final volume 24 μL, flow rate 1.2 mL/s) the formation reaction was monitored at 800 V with sampling times of 1 ms for the first 5 seconds and 10 ms for the remaining reaction time. All relative fluorescence values were plotted versus time and fit to:
where n = 1 or 2, as required.
Kinetic simulations
Kinetic analysis of ternary complex formation assays was performed in MATLAB (2021b). kon and koff rates derived from the μstopped-flow data were used in combination with previously reported values to simulate ternary complex levels at physiological concentrations. A system of ordinary differential equations based on the kinetic model in Fig. 3C was solved using the function ode89 in MATLAB at different combinations of aa-tRNA, EF-Tu and EF-Ts concentrations. A proportion of 4*[EF-Tu] = [EF-Ts] was kept constant for each aa-tRNA-EF-Tu pair. Previously reported E. coli cytoplasmic concentrations of GDP (0.69 mM) and GTP (4.9 mM) were used. The equilibrium concentrations of aa-tRNA free and bound were used to determine the ternary complex fraction with the following equation:
Single-molecule FRET experiments
Single-molecule FRET experiments were performed using a custom-built, prism-type TIRF microscope. Bacterial ribosomes programed with Cy3-fMet-tRNAfMet in the P site and a UUC codon displayed in the A site were surface immobilized via streptavidin-biotin interaction to a transparent surface passivated with polyethylene glycol (PEG) polymers doped with biotin-PEG. tRNA selection experiments were initiated by injection of pre-formed ternary complexes (aa-tRNA-LD655, 12.5 nM; EF-Tu/Ts 125 nM; GTP or GTPγS 500 μM) in bacterial polymix buffer (50 mM Tris-OAc; pH 7.5, 100 mM KCl, 5 mM NH4OAc, 0.5 mM Ca(OAc)2, 5 mM Mg(OAc)2, 6 mM 2-mercaptoethanol, 0.1 mM EDTA, 5 mM putrescine and 1 mM spermidine) supplemented with 2 mM PCA/PCD oxygen scavenging system and 1 mM each of cyclooctatetraene (COT), nitrobenzyl alcohol (NBA), and Trolox triplet state quenchers. Translocation experiments were initiated by injection of pre-formed ternary complexes (aa-tRNA-LD655, 25 nM; EF-Tu/Ts 250 nM; GTP 1250 μM) supplemented with 10 μM EF-G in bacterial polymix buffer. Samples were illuminated with a 532 nm diode pumped solid-state laser (Opus, LaserQuantum) at 2.0 and 0.25 kW cm−2 (0.16 kW cm−2 for translocation experiments) with 10 or 100-ms integration times, respectively. Fluorescence emission from donor and acceptor fluorophores was collected using a 60X/1.27 NA super-resolution water-immersion objective (Nikon). Fluorescence was recorded onto two aligned ORCA-Fusion sCMOS cameras (C-14440–20UP, Hamamatsu). Instrument control was performed using custom software written in LabVIEW (National Instruments). Fluorescence intensities were extracted from the recorded videos and FRET efficiency traces were calculated using the SPARTAN software package. FRET traces were selected for further analysis according to the following criteria: 8:1 signal/background-noise ration and 6:1 signal/signal-noise ratio, less than four donor-fluorophore blinking events and a correlation coefficient between donor and acceptor of <0.5. The resulting smFRET traces were further post synchronized to the appearance of FRET and analyzed using the segmental k-means (SKM) algorithm as implemented in the SPARTAN software package v3.8. Data were plotted in OriginPro 2019b (OriginLab, Northhampton, MA). Dwell time curves were fit to:
where n = 1 or 3 as required. Mean and standard deviations of classical-hybrid split ratio were calculated from four independent measurements. Statistical significance (p < 0.05) was assessed by a two-way ANOVA followed by a post-hoc Bonferroni test.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported primarily by the National Science Foundation Center for Genetically Encoded Materials (C-GEM), an NSF Center for Chemical Innovation (NSF CHE-2002182; F.A.C-N., W.C.G., Y-C.C., I.K., A.S., S.J.M., and S.C.B.) C-GEM funding supported protein and tRNA expression, reagent synthesis and purification, chemistry and biochemistry efforts associated with all assays reported, ensemble and single-molecule fluorescence investigations, and manuscript preparation. Additional support was provided by the National Institutes of Health (5R01AI150560; M.M.), primarily for the synthesis of the p-Az-Phe monomer and the fluorophores employed. We thank St. Jude Children’s Research Hospital for their support of J.A., S.K.N. and R.B.A and the Single-Molecule imaging Center, Daniel S. Terry, Zeliha Kilic and Mikael Holm in particular, for early guidance with kinetic simulations, their training and their thoughtful discussions and comments during manuscript preparation.
Footnotes
CONFLICT OF INTEREST
S.C.B and R.B.A. hold equity interests in Lumidyne Technologies
BIBLIOGRAPHY
- (1).Diercks C. S.; Dik D. A.; Schultz P. G. Adding New Chemistries to the Central Dogma of Molecular Biology. Chem 2021. DOI: 10.1016/j.chempr.2021.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Kobayashi T.; Yanagisawa T.; Sakamoto K.; Yokoyama S. Recognition of Non-Alpha-Amino Substrates by Pyrrolysyl-TRNA Synthetase. J. Mol. Biol. 2009, 385 (5), 1352–1360. DOI: 10.1016/j.jmb.2008.11.059. [DOI] [PubMed] [Google Scholar]
- (3).Li Y.-M.; Yang M.-Y.; Huang Y.-C.; Li Y.-T.; Chen P. R.; Liu L. Ligation of Expressed Protein α-Hydrazides via Genetic Incorporation of an α-Hydroxy Acid. ACS Chem. Biol. 2012, 7 (6), 1015–1022. DOI: 10.1021/cb300020s. [DOI] [PubMed] [Google Scholar]
- (4).Fricke R.; Swenson C. V.; Roe L. T.; Hamlish N. X.; Shah B.; Zhang Z.; Ficaretta E.; Ad O.; Smaga S.; Gee C. L.; Chatterjee A.; Schepartz A. Expanding the Substrate Scope of Pyrrolysyl-Transfer RNA Synthetase Enzymes to Include Non-α-Amino Acids in Vitro and in Vivo. Nat. Chem. 2023, 15 (7), 960–971. DOI: 10.1038/s41557-023-01224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Spinck M.; Piedrafita C.; Robertson W. E.; Elliott T. S.; Cervettini D.; de la Torre D.; Chin J. W. Genetically Programmed Cell-Based Synthesis of Non-Natural Peptide and Depsipeptide Macrocycles. Nat. Chem. 2023, 15 (1), 61–69. DOI: 10.1038/s41557-022-01082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Melo Czekster C.; Robertson W. E.; Walker A. S.; Söll D.; Schepartz A. In Vivo Biosynthesis of a β-Amino Acid-Containing Protein. J. Am. Chem. Soc. 2016, 138 (16), 5194–5197. DOI: 10.1021/jacs.6b01023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hamlish N.; Abramyan A.; Schepartz A. Incorporation of Multiple Beta 2-Backbones into a Protein in Vivo Using an Orthogonal Aminoacyl-TRNA Synthetase. BioRxiv 2023. DOI: 10.1101/2023.11.07.565973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Dunkelmann D. L.; Piedrafita C.; Dickson A.; Liu K. C.; Elliott T. S.; Fiedler M.; Bellini D.; Zhou A.; Cervettini D.; Chin J. W. Adding α,α-Disubstituted and β-Linked Monomers to the Genetic Code of an Organism. Nature 2024, 625 (7995), 603–610. DOI: 10.1038/s41586-023-06897-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Roe L. T.; Schissel C. K.; Dover T. L.; Shah B.; Hamlish N. X.; Zheng S.; Dilworth D. A.; Wong N.; Zhang Z.; Chatterjee A.; Francis M. B.; Miller S. J.; Schepartz A. Backbone Extension Acyl Rearrangements Enable Cellular Synthesis of Proteins with Internal β2 -Peptide Linkages. BioRxiv 2023. DOI: 10.1101/2023.10.03.560714. [DOI] [Google Scholar]
- (10).Chin J. W.; Santoro S. W.; Martin A. B.; King D. S.; Wang L.; Schultz P. G. Addition of P-Azido-L-Phenylalanine to the Genetic Code of Escherichia Coli. J. Am. Chem. Soc. 2002, 124 (31), 9026–9027. DOI: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- (11).Wang L.; Schultz P. G. Expanding the Genetic Code. Chem. Commun. 2002, No. 1, 1–11. DOI: 10.1039/b108185n. [DOI] [PubMed] [Google Scholar]
- (12).Santoro S. W.; Wang L.; Herberich B.; King D. S.; Schultz P. G. An Efficient System for the Evolution of Aminoacyl-TRNA Synthetase Specificity. Nat. Biotechnol. 2002, 20 (10), 1044–1048. DOI: 10.1038/nbt742. [DOI] [PubMed] [Google Scholar]
- (13).Gan R.; Perez J. G.; Carlson E. D.; Ntai I.; Isaacs F. J.; Kelleher N. L.; Jewett M. C. Translation System Engineering in Escherichia Coli Enhances Non-Canonical Amino Acid Incorporation into Proteins. Biotechnol. Bioeng. 2017, 114 (5), 1074–1086. DOI: 10.1002/bit.26239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Brown W.; Liu J.; Deiters A. Genetic Code Expansion in Animals. ACS Chem. Biol. 2018, 13 (9), 2375–2386. DOI: 10.1021/acschembio.8b00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Englander M. T.; Avins J. L.; Fleisher R. C.; Liu B.; Effraim P. R.; Wang J.; Schulten K.; Leyh T. S.; Gonzalez R. L.; Cornish V. W. The Ribosome Can Discriminate the Chirality of Amino Acids within Its Peptidyl-Transferase Center. Proc Natl Acad Sci USA 2015, 112 (19), 6038–6043. DOI: 10.1073/pnas.1424712112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Watson Z. L.; Knudson I. J.; Ward F. R.; Miller S. J.; Cate J. H. D.; Schepartz A.; Abramyan A. M. Atomistic Simulations of the Escherichia Coli Ribosome Provide Selection Criteria for Translationally Active Substrates. Nat. Chem. 2023, 15 (7), 913–921. DOI: 10.1038/s41557-023-01226-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Voorhees R. M.; Ramakrishnan V. Structural Basis of the Translational Elongation Cycle. Annu. Rev. Biochem. 2013, 82, 203–236. DOI: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- (18).Frank J. The Mechanism of Translation. [Version 1; Peer Review: 3 Approved]. F1000Res. 2017, 6, 198. DOI: 10.12688/f1000research.9760.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Burnett B. J.; Altman R. B.; Ferguson A.; Wasserman M. R.; Zhou Z.; Blanchard S. C. Direct Evidence of an Elongation Factor-Tu/Ts·GTP·Aminoacyl-TRNA Quaternary Complex. J. Biol. Chem. 2014, 289 (34), 23917–23927. DOI: 10.1074/jbc.M114.583385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).LaRiviere F. J.; Wolfson A. D.; Uhlenbeck O. C. Uniform Binding of Aminoacyl-TRNAs to Elongation Factor Tu by Thermodynamic Compensation. Science 2001, 294 (5540), 165–168. DOI: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- (21).Dale T.; Sanderson L. E.; Uhlenbeck O. C. The Affinity of Elongation Factor Tu for an Aminoacyl-TRNA Is Modulated by the Esterified Amino Acid. Biochemistry 2004, 43 (20), 6159–6166. DOI: 10.1021/bi036290o. [DOI] [PubMed] [Google Scholar]
- (22).Geggier P.; Dave R.; Feldman M. B.; Terry D. S.; Altman R. B.; Munro J. B.; Blanchard S. C. Conformational Sampling of Aminoacyl-TRNA during Selection on the Bacterial Ribosome. J. Mol. Biol. 2010, 399 (4), 576–595. DOI: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Rundlet E. J.; Holm M.; Schacherl M.; Natchiar S. K.; Altman R. B.; Spahn C. M. T.; Myasnikov A. G.; Blanchard S. C. Structural Basis of Early Translocation Events on the Ribosome. Nature 2021, 595 (7869), 741–745. DOI: 10.1038/s41586-021-03713-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Holm M.; Natchiar S. K.; Rundlet E. J.; Myasnikov A. G.; Watson Z. L.; Altman R. B.; Wang H.-Y.; Taunton J.; Blanchard S. C. MRNA Decoding in Human Is Kinetically and Structurally Distinct from Bacteria. Nature 2023, 617 (7959), 200–207. DOI: 10.1038/s41586-023-05908-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Villa E.; Sengupta J.; Trabuco L. G.; LeBarron J.; Baxter W. T.; Shaikh T. R.; Grassucci R. A.; Nissen P.; Ehrenberg M.; Schulten K.; Frank J. Ribosome-Induced Changes in Elongation Factor Tu Conformation Control GTP Hydrolysis. Proc Natl Acad Sci USA 2009, 106 (4), 1063–1068. DOI: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kavaliauskas D.; Chen C.; Liu W.; Cooperman B. S.; Goldman Y. E.; Knudsen C. R. Structural Dynamics of Translation Elongation Factor Tu during Aa-TRNA Delivery to the Ribosome. Nucleic Acids Res. 2018, 46 (16), 8651–8661. DOI: 10.1093/nar/gky651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Diaconu M.; Kothe U.; Schlünzen F.; Fischer N.; Harms J. M.; Tonevitsky A. G.; Stark H.; Rodnina M. V.; Wahl M. C. Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation. Cell 2005, 121 (7), 991–1004. DOI: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- (28).Liu W.; Chen C.; Kavaliauskas D.; Knudsen C. R.; Goldman Y. E.; Cooperman B. S. EF-Tu Dynamics during Pre-Translocation Complex Formation: EF-Tu·GDP Exits the Ribosome via Two Different Pathways. Nucleic Acids Res. 2015, 43 (19), 9519–9528. DOI: 10.1093/nar/gkv856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Morse J. C.; Girodat D.; Burnett B. J.; Holm M.; Altman R. B.; Sanbonmatsu K. Y.; Wieden H.-J.; Blanchard S. C. Elongation Factor-Tu Can Repetitively Engage Aminoacyl-TRNA within the Ribosome during the Proofreading Stage of TRNA Selection. Proc Natl Acad Sci USA 2020, 117 (7), 3610–3620. DOI: 10.1073/pnas.1904469117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Girodat D.; Blanchard S. C.; Wieden H.-J.; Sanbonmatsu K. Y. Elongation Factor Tu Switch I Element Is a Gate for Aminoacyl-TRNA Selection. J. Mol. Biol. 2020, 432 (9), 3064–3077. DOI: 10.1016/j.jmb.2020.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Parker J. Errors and Alternatives in Reading the Universal Genetic Code. Microbiol. Rev. 1989, 53 (3), 273–298. DOI: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Ninio J. Multiple Stages in Codon-Anticodon Recognition: Double-Trigger Mechanisms and Geometric Constraints. Biochimie 2006, 88 (8), 963–992. DOI: 10.1016/j.biochi.2006.06.002. [DOI] [PubMed] [Google Scholar]
- (33).Johansson M.; Lovmar M.; Ehrenberg M. Rate and Accuracy of Bacterial Protein Synthesis Revisited. Curr. Opin. Microbiol. 2008, 11 (2), 141–147. DOI: 10.1016/j.mib.2008.02.015. [DOI] [PubMed] [Google Scholar]
- (34).Nissen P.; Kjeldgaard M.; Thirup S.; Polekhina G.; Reshetnikova L.; Clark B. F.; Nyborg J. Crystal Structure of the Ternary Complex of Phe-TRNAPhe, EF-Tu, and a GTP Analog. Science 1995, 270 (5241), 1464–1472. DOI: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- (35).Nissen P.; Reshetnikova L.; Siboska G.; Polekhina G.; Thirup S.; Kjeldgaard M.; Clark B. F.; Nyborg J. Purification and Crystallization of the Ternary Complex of Elongation Factor Tu:GTP and Phe-TRNA(Phe). FEBS Lett. 1994, 356 (2–3), 165–168. DOI: 10.1016/0014-5793(94)01254-7. [DOI] [PubMed] [Google Scholar]
- (36).Burnett B. J.; Altman R. B.; Ferrao R.; Alejo J. L.; Kaur N.; Kanji J.; Blanchard S. C. Elongation Factor Ts Directly Facilitates the Formation and Disassembly of the Escherichia Coli Elongation Factor Tu·GTP·aminoacyl-TRNA Ternary Complex. J. Biol. Chem. 2013, 288 (19), 13917–13928. DOI: 10.1074/jbc.M113.460014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Ieong K.-W.; Pavlov M. Y.; Kwiatkowski M.; Ehrenberg M.; Forster A. C. A TRNA Body with High Affinity for EF-Tu Hastens Ribosomal Incorporation of Unnatural Amino Acids. RNA 2014, 20 (5), 632–643. DOI: 10.1261/rna.042234.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Wang J.; Kwiatkowski M.; Forster A. C. Kinetics of Ribosome-Catalyzed Polymerization Using Artificial Aminoacyl-TRNA Substrates Clarifies Inefficiencies and Improvements. ACS Chem. Biol. 2015, 10 (10), 2187–2192. DOI: 10.1021/acschembio.5b00335. [DOI] [PubMed] [Google Scholar]
- (39).Gao R.; Forster A. C. Changeability of Individual Domains of an Aminoacyl-TRNA in Polymerization by the Ribosome. FEBS Lett. 2010, 584 (1), 99–105. DOI: 10.1016/j.febslet.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Achenbach J.; Jahnz M.; Bethge L.; Paal K.; Jung M.; Schuster M.; Albrecht R.; Jarosch F.; Nierhaus K. H.; Klussmann S. Outwitting EF-Tu and the Ribosome: Translation with d-Amino Acids. Nucleic Acids Res. 2015, 43 (12), 5687–5698. DOI: 10.1093/nar/gkv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Schrader J. M.; Chapman S. J.; Uhlenbeck O. C. Understanding the Sequence Specificity of TRNA Binding to Elongation Factor Tu Using TRNA Mutagenesis. J. Mol. Biol. 2009, 386 (5), 1255–1264. DOI: 10.1016/j.jmb.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Bessho Y.; Hodgson D. R. W.; Suga H. A TRNA Aminoacylation System for Non-Natural Amino Acids Based on a Programmable Ribozyme. Nat. Biotechnol. 2002, 20 (7), 723–728. DOI: 10.1038/nbt0702-723. [DOI] [PubMed] [Google Scholar]
- (43).Niwa N.; Yamagishi Y.; Murakami H.; Suga H. A Flexizyme That Selectively Charges Amino Acids Activated by a Water-Friendly Leaving Group. Bioorg. Med. Chem. Lett. 2009, 19 (14), 3892–3894. DOI: 10.1016/j.bmcl.2009.03.114. [DOI] [PubMed] [Google Scholar]
- (44).Murakami H.; Ohta A.; Ashigai H.; Suga H. A Highly Flexible TRNA Acylation Method for Non-Natural Polypeptide Synthesis. Nat. Methods 2006, 3 (5), 357–359. DOI: 10.1038/nmeth877. [DOI] [PubMed] [Google Scholar]
- (45).Katoh T.; Suga H. Ribosomal Incorporation of Consecutive β-Amino Acids. J. Am. Chem. Soc. 2018, 140 (38), 12159–12167. DOI: 10.1021/jacs.8b07247. [DOI] [PubMed] [Google Scholar]
- (46).Arranz-Gibert P.; Vanderschuren K.; Haimovich A.; Halder A.; Gupta K.; Rinehart J.; Isaacs F. J. Chemoselective Restoration of Para-Azido-Phenylalanine at Multiple Sites in Proteins. Cell Chem. Biol. 2022, 29 (6), 1046–1052.e4. DOI: 10.1016/j.chembiol.2021.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Fladischer P.; Weingartner A.; Blamauer J.; Darnhofer B.; Birner-Gruenberger R.; Kardashliev T.; Ruff A. J.; Schwaneberg U.; Wiltschi B. A Semi-Rationally Engineered Bacterial Pyrrolysyl-TRNA Synthetase Genetically Encodes Phenyl Azide Chemistry. Biotechnol. J. 2019, 14 (3), e1800125. DOI: 10.1002/biot.201800125. [DOI] [PubMed] [Google Scholar]
- (48).Katoh T.; Suga H. In Vitro Genetic Code Reprogramming for the Expansion of Usable Noncanonical Amino Acids. Annu. Rev. Biochem. 2022, 91, 221–243. DOI: 10.1146/annurev-biochem-040320-103817. [DOI] [PubMed] [Google Scholar]
- (49).Cui Z.; Johnston W. A.; Alexandrov K. Cell-Free Approach for Non-Canonical Amino Acids Incorporation Into Polypeptides. Front. Bioeng. Biotechnol. 2020, 8, 1031. DOI: 10.3389/fbioe.2020.01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Daskalova S. M.; Dedkova L. M.; Maini R.; Talukder P.; Bai X.; Chowdhury S. R.; Zhang C.; Nangreave R. C.; Hecht S. M. Elongation Factor P Modulates the Incorporation of Structurally Diverse Noncanonical Amino Acids into Escherichia Coli Dihydrofolate Reductase. J. Am. Chem. Soc. 2023, 145 (43), 23600–23608. DOI: 10.1021/jacs.3c07524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Maini R.; Nguyen D. T.; Chen S.; Dedkova L. M.; Chowdhury S. R.; Alcala-Torano R.; Hecht S. M. Incorporation of β-Amino Acids into Dihydrofolate Reductase by Ribosomes Having Modifications in the Peptidyltransferase Center. Bioorg. Med. Chem. 2013, 21 (5), 1088–1096. DOI: 10.1016/j.bmc.2013.01.002. [DOI] [PubMed] [Google Scholar]
- (52).Katoh T.; Suga H. Ribosomal Incorporation of Negatively Charged D-α- and N-Methyl-l-α-Amino Acids Enhanced by EF-Sep. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2023, 378 (1871), 20220038. DOI: 10.1098/rstb.2022.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Park H.-S.; Hohn M. J.; Umehara T.; Guo L.-T.; Osborne E. M.; Benner J.; Noren C. J.; Rinehart J.; Söll D. Expanding the Genetic Code of Escherichia Coli with Phosphoserine. Science 2011, 333 (6046), 1151–1154. DOI: 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Iwane Y.; Kimura H.; Katoh T.; Suga H. Uniform Affinity-Tuning of N-Methyl-Aminoacyl-TRNAs to EF-Tu Enhances Their Multiple Incorporation. Nucleic Acids Res. 2021, 49 (19), 10807–10817. DOI: 10.1093/nar/gkab288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Schrader J. M.; Chapman S. J.; Uhlenbeck O. C. Tuning the Affinity of Aminoacyl-TRNA to Elongation Factor Tu for Optimal Decoding. Proc Natl Acad Sci USA 2011, 108 (13), 5215–5220. DOI: 10.1073/pnas.1102128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Furano A. V. Content of Elongation Factor Tu in Escherichia Coli. Proc Natl Acad Sci USA 1975, 72 (12), 4780–4784. DOI: 10.1073/pnas.72.12.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Moazed D.; Noller H. F. Intermediate States in the Movement of Transfer RNA in the Ribosome. Nature 1989, 342 (6246), 142–148. DOI: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- (58).Valle M.; Zavialov A.; Sengupta J.; Rawat U.; Ehrenberg M.; Frank J. Locking and Unlocking of Ribosomal Motions. Cell 2003, 114 (1), 123–134. DOI: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- (59).Cornish P. V.; Ermolenko D. N.; Noller H. F.; Ha T. Spontaneous Intersubunit Rotation in Single Ribosomes. Mol. Cell 2008, 30 (5), 578–588. DOI: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Wasserman M. R.; Alejo J. L.; Altman R. B.; Blanchard S. C. Multiperspective SmFRET Reveals Rate-Determining Late Intermediates of Ribosomal Translocation. Nat. Struct. Mol. Biol. 2016, 23 (4), 333–341. DOI: 10.1038/nsmb.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Chen C.; Stevens B.; Kaur J.; Smilansky Z.; Cooperman B. S.; Goldman Y. E. Allosteric vs. Spontaneous Exit-Site (E-Site) TRNA Dissociation Early in Protein Synthesis. Proc Natl Acad Sci USA 2011, 108 (41), 16980–16985. DOI: 10.1073/pnas.1106999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Munro J. B.; Wasserman M. R.; Altman R. B.; Wang L.; Blanchard S. C. Correlated Conformational Events in EF-G and the Ribosome Regulate Translocation. Nat. Struct. Mol. Biol. 2010, 17 (12), 1470–1477. DOI: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Alejo J. L.; Blanchard S. C. Miscoding-Induced Stalling of Substrate Translocation on the Bacterial Ribosome. Proc Natl Acad Sci USA 2017, 114 (41), E8603–E8610. DOI: 10.1073/pnas.1707539114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Wang L.; Altman R. B.; Blanchard S. C. Insights into the Molecular Determinants of EF-G Catalyzed Translocation. RNA 2011, 17 (12), 2189–2200. DOI: 10.1261/rna.029033.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Ogle J. M.; Ramakrishnan V. Structural Insights into Translational Fidelity. Annu. Rev. Biochem. 2005, 74, 129–177. DOI: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- (66).Soye B. J. D.; Patel J. R.; Isaacs F. J.; Jewett M. C. Repurposing the Translation Apparatus for Synthetic Biology. Curr. Opin. Chem. Biol. 2015, 28, 83–90. DOI: 10.1016/j.cbpa.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).DeLey Cox V. E.; Cole M. F.; Gaucher E. A. Incorporation of Modified Amino Acids by Engineered Elongation Factors with Expanded Substrate Capabilities. ACS Synth. Biol. 2019, 8 (2), 287–296. DOI: 10.1021/acssynbio.8b00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Smolskaya S.; Andreev Y. A. Site-Specific Incorporation of Unnatural Amino Acids into Escherichia Coli Recombinant Protein: Methodology Development and Recent Achievement. Biomolecules 2019, 9 (7). DOI: 10.3390/biom9070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Doi Y.; Ohtsuki T.; Shimizu Y.; Ueda T.; Sisido M. Elongation Factor Tu Mutants Expand Amino Acid Tolerance of Protein Biosynthesis System. J. Am. Chem. Soc. 2007, 129 (46), 14458–14462. DOI: 10.1021/ja075557u. [DOI] [PubMed] [Google Scholar]
- (70).Effraim P. R.; Wang J.; Englander M. T.; Avins J.; Leyh T. S.; Gonzalez R. L.; Cornish V. W. Natural Amino Acids Do Not Require Their Native TRNAs for Efficient Selection by the Ribosome. Nat. Chem. Biol. 2009, 5 (12), 947–953. DOI: 10.1038/nchembio.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Melnikov S. V.; Khabibullina N. F.; Mairhofer E.; Vargas-Rodriguez O.; Reynolds N. M.; Micura R.; Söll D.; Polikanov Y. S. Mechanistic Insights into the Slow Peptide Bond Formation with D-Amino Acids in the Ribosomal Active Site. Nucleic Acids Res. 2019, 47 (4), 2089–2100. DOI: 10.1093/nar/gky1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Saito K.; Kratzat H.; Campbell A.; Buschauer R.; Burroughs A. M.; Berninghausen O.; Aravind L.; Green R.; Beckmann R.; Buskirk A. R. Ribosome Collisions Induce MRNA Cleavage and Ribosome Rescue in Bacteria. Nature 2022, 603 (7901), 503–508. DOI: 10.1038/s41586-022-04416-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).De Laurentiis E. I.; Mercier E.; Wieden H.-J. The C-Terminal Helix of Pseudomonas Aeruginosa Elongation Factor Ts Tunes EF-Tu Dynamics to Modulate Nucleotide Exchange. J. Biol. Chem. 2016, 291 (44), 23136–23148. DOI: 10.1074/jbc.M116.740381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Thirup S. S.; Van L. B.; Nielsen T. K.; Knudsen C. R. Structural Outline of the Detailed Mechanism for Elongation Factor Ts-Mediated Guanine Nucleotide Exchange on Elongation Factor Tu. J. Struct. Biol. 2015, 191 (1), 10–21. DOI: 10.1016/j.jsb.2015.06.011. [DOI] [PubMed] [Google Scholar]
- (75).Hecht S. M. Expansion of the Genetic Code through the Use of Modified Bacterial Ribosomes. J. Mol. Biol. 2022, 434 (8), 167211. DOI: 10.1016/j.jmb.2021.167211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Chen S.; Ji X.; Gao M.; Dedkova L. M.; Hecht S. M. In Cellulo Synthesis of Proteins Containing a Fluorescent Oxazole Amino Acid. J. Am. Chem. Soc. 2019, 141 (14), 5597–5601. DOI: 10.1021/jacs.8b12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Dunkle J. A.; Wang L.; Feldman M. B.; Pulk A.; Chen V. B.; Kapral G. J.; Noeske J.; Richardson J. S.; Blanchard S. C.; Cate J. H. D. Structures of the Bacterial Ribosome in Classical and Hybrid States of TRNA Binding. Science 2011, 332 (6032), 981–984. DOI: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Xiao H.; Murakami H.; Suga H.; Ferré-D’Amaré A. R. Structural Basis of Specific TRNA Aminoacylation by a Small in Vitro Selected Ribozyme. Nature 2008, 454 (7202), 358–361. DOI: 10.1038/nature07033. [DOI] [PubMed] [Google Scholar]
- (79).Gromadski K. B.; Wieden H.-J.; Rodnina M. V. Kinetic Mechanism of Elongation Factor Ts-Catalyzed Nucleotide Exchange in Elongation Factor Tu. Biochemistry 2002, 41 (1), 162–169. DOI: 10.1021/bi015712w. [DOI] [PubMed] [Google Scholar]
- (80).Lebedev A. A.; Young P.; Isupov M. N.; Moroz O. V.; Vagin A. A.; Murshudov G. N. JLigand: A Graphical Tool for the CCP4 Template-Restraint Library. Acta Crystallogr. D Biol. Crystallogr. 2012, 68 (Pt 4), 431–440. DOI: 10.1107/S090744491200251X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.