Abstract
Elephants have emerged as a model system to study the evolution of body size and cancer resistance because, despite their immense size, they have a very low prevalence of cancer. Previous studies have found that duplication of tumor suppressors at least partly contributes to the evolution of anti-cancer cellular phenotypes in elephants. Still, many other mechanisms must have contributed to their augmented cancer resistance. Here, we use a suite of codon-based maximum-likelihood methods and a dataset of 13,310 protein-coding gene alignments from 261 Eutherian mammals to identify positively selected and rapidly evolving elephant genes. We found 496 genes (3.73% of alignments tested) with statistically significant evidence for positive selection and 660 genes (4.96% of alignments tested) that likely evolved rapidly in elephants. Positively selected and rapidly evolving genes are statistically enriched in gene ontology terms and biological pathways related to regulated cell death mechanisms, DNA damage repair, cell cycle regulation, epidermal growth factor receptor (EGFR) signaling, and immune functions, particularly neutrophil granules and degranulation. All of these biological factors are plausibly related to the evolution of cancer resistance. Thus, these positively selected and rapidly evolving genes are promising candidates for genes contributing to elephant-specific traits, including the evolution of molecular and cellular characteristics that enhance cancer resistance.
Introduction
Large body sizes have repeatedly evolved in many mammalian lineages such as whales (Cetaceans), elephants and their extinct relatives (Proboscideans), extinct hornless rhinos (Paraceratherium, aka ‘Walter’), giant ground sloths (Megatherium), and giant armadillos (Glyptodon); this tendency for lineages to increase in size over evolutionary time is relatively common across the tree of life (Purvis and Orme, 2005). There are many constraints on the evolution of gigantic body sizes, including an increased risk of developing cancer. For example, if all cells in all organisms have a similar risk of malignant transformation and equivalent cancer suppression mechanisms, then organisms with many cells should have a higher prevalence of cancer than organisms with fewer cells, particularly because the size of most cell types is similar across species with different body sizes (Hoogduijn et al., 2013; Savage et al., 2007). While there is a positive correlation between body size and cancer prevalence within species, for example, humans and dogs (Benyi et al., 2019; Dobson, 2013; Green et al., 2011; Zhou et al., 2022), there is no positive correlation between body size and cancer risk between mammalian species (Abegglen et al., 2015; Bulls et al., 2022; Compton et al., 2023); this lack of correlation is called ‘Peto’s Paradox’ (Peto, 2015, 1975). Indeed, within mammals, there is a statistically significant negative correlation between body mass and cancer prevalence (Bulls et al., 2022), indicating that mammals with enormous bodies have evolved particularly effective anticancer mechanisms.
Afrotherian mammals are an excellent model system to study the evolution of cancer resistance because while most lineages such as golden moles (~70g), tenrecs (120g), elephant shrews (~170g), hyraxes (~3kg), aardvarks (~60kg), and sea cows (600–900kg) are small- to large-bodied, gigantic Proboscideans including African Savannah (4,800kg) and Asian elephants (3,200kg) and extinct species such as Deinotherium (~12,000kg), Mammut borsoni (16,000kg), and the straight-tusked elephant (~14,000kg) are nested deeply within the small-bodied species and evolved from a small-bodied ancestor (Elliot and Mooers, 2014; Puttick and Thomas, 2015; Vazquez and Lynch, 2021). Large body masses in Proboscideans evolved stepwise (O’Leary et al., 2013a, 2013b; Puttick and Thomas, 2015; Springer et al., 2013; Vazquez and Lynch, 2021), coincident with dramatic reductions in intrinsic cancer risk (Vazquez and Lynch, 2021) and the evolution of anticancer cellular phenotypes. For example, elephant cells induce apoptosis at very low levels of DNA damage (Abegglen et al., 2015; Sulak et al., 2016a; Vazquez et al., 2018), are resistant to oxidative stress-induced cell death (Gomes et al., 2011), are resistant to experimental immortalization (Fukuda et al., 2016; Gomes et al., 2011), and repair DNA damage more rapidly than cells from smaller bodied species (Francis et al., 1981; Hart and Setlow, 1974; Promislow, 1994).
Many mechanisms must have contributed to the evolution of enhanced cancer resistance. For example, the sensitivity of elephant cells to DNA damage is mediated in part by an increase in the number of tumor suppressor genes in the elephant lineage (Caulin et al., 2015; Doherty and Magalhães, 2016; Sulak et al., 2016b; Tollis et al., 2020; Vazquez et al., 2018; Vazquez and Lynch, 2021) whereas the resistance to oxidative stress-induced cell death may be mediated by a duplicate superoxide dismutase 1 (SOD1) gene (Vazquez and Lynch, 2021). Here, we explore the possibility that the functional divergence of protein-coding genes may also have contributed to anti-cancer traits in the elephant lineage by identifying genes with evidence of positive selection and rapid evolution, an evolutionary signature often associated with the functional divergence of proteins (Slodkowicz and Goldman, 2020; Tennessen, 2008). We used maximum likelihood models to characterize the strength and direction of selection acting on 13,310 orthologous coding gene alignments from the genomes of 261 Eutherian mammals and identified several hundred positively selected and rapidly evolving genes in the elephant lineage. These genes are enriched in gene ontology (GO) and pathway terms related to various kinds of regulated cell death, DNA damage repair, regulation of the cell cycle, EGFR–signaling, and immune functions, particularly neutrophil biology, which have essential roles in cancer biology (Hedrick and Malanchi, 2021; Huang et al., 2020; Mollinedo, 2019; Normanno et al., 2006; Sigismund et al., 2018; Vorobjeva and Chernyak, 2020). Thus, these are good candidates for genes that contribute to elephant-specific traits, including the evolution of cancer resistance.
Results
Hundreds of genes experienced an episode of positive selection or rapid evolution in elephants
Episodic adaptive evolution (“positive selection”) is thought to be associated with functional divergence (Slodkowicz and Goldman, 2020; Tennessen, 2008). Therefore, we used two complementary methods, ABSREL (Smith et al., 2015) and BUSTED (Murrell et al., 2015a), to identify genes with evidence of positive selection in the elephant lineage. We used a dataset of alignments from 13,310 orthologous genes from 261 Eutherian mammals that we previously reported (Bowman et al., 2023b) and a phylogeny with Paenungulates collapsed to a polytomy to reflect that most genes lack significant phylogenetic support for the resolution of this clade (Figure 1A) (Bowman et al., 2023a). The ABSREL and BUSTED models are variants of the branch-sites random effect likelihood (BSREL) model of coding sequence evolution (Pond et al., 2011a; Smith et al., 2015), which allows for variation in dN/dS (ω) rates across lineages and sites; however, while ABSREL selects the optimal number of rate categories for each gene, BUSTED imposes three rate categories such that ω1≤ω2≤ω3 (Figure 1B). Both models also accommodate synonymous rate variation across sites [S] (Pond and Muse, 2005; Wisotsky et al., 2020), double and triple multi-nucleotide mutations per codon [MH] (Lucaci et al., 2021), and both synonymous rate variation and multi-nucleotide mutations [SMH]; failure to account for synonymous rate variation (Dunn et al., 2019; Rahman et al., 2021; Wisotsky et al., 2020) and multi-nucleotide mutations can lead to widespread false positive inferences of positive selection (Dunn et al., 2019; Lucaci et al., 2023a; Nozawa et al., 2009; Suzuki, 2008; Venkat et al., 2018; Yang and Reis, 2011). We used small sample corrected Akaike Information Criterion values (ΔAICc≥10) to select for the best [S], [MH], and [SMH] model and inferred positive selection for a gene when the best fitting model included a class of sites with ω>1 and a likelihood ratio test P≤0.05.
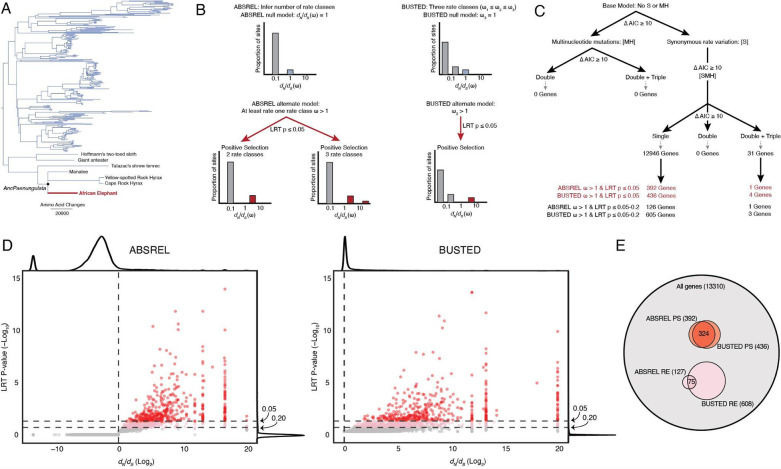
Figure 1. Identification and positively selected and rapidly evolving genes in the elephant lineage.
A. Phylogeny of 261 Eutherian mammals used in selection tests. The Atlantogenatan part of the tree is expanded to show which species of Afrotheria and Xenarthra are included in the analyses. Branch lengths are shown as amino acid changes per lineage. B. Cartoon description of the ABSREL and BUSTED models.
C. Decision tree showing [S], [MH], and [SMH] model selection (inferred with ABSREL) and the number of genes inferred to be positively selected and rapidly evolving.
D. Volcano plot showing the dN/dS rate ratio (Log2) and likelihood ratio test (LRT) P-value (−Log10) for each gene inferred from the best-fitting ABSREL (right) and BUSTED (left) tests in the elephant lineage. Red dots indicate genes with dN/dS>1 and P≤0.05, and dots in pink indicate genes with dN/dS>1 and P=0.051–0.20.
E. Venn diagram showing the number and overlap of genes inferred to be positively selected (PS) or rapidly evolving (RE) by the ABSREL and BUSTED tests.
Including synonymous rate variation across sites improved model fit for all genes; however, only 31 genes had significant evidence for multi-nucleotide mutations (Figure 1C). These data are consistent with previous observations synonymous substitution rate variation is common (Pond and Muse, 2005; Wisotsky et al., 2020) and that shorter branches tend to be more impacted by multiple hits than longer branches (Lucaci et al., 2023b, 2021) such as the elephant lineage. The best-fitting ABSREL model identified 393 genes with a class of sites with ω>1 at P≤0.05 in the elephant lineage (Figure 1C), whereas the best-fitting BUSTED model identified 427 genes with a class of sites with ω>1 at P≤0.05 in the elephant lineage (Figure 1C); 324 genes were identified as positively selected by both methods (Figure 1D). We also defined a set of rapidly evolving genes with a class of sites with ω>1 in either the ABSREL or BUSTED methods, but at P=0.051–0.20; these genes did not meet the statistical significance threshold for positive selection. However, the ABSREL and BUSTED methods are conservative, and they are likely rapidly evolving. ABSREL and BUSTED identified 127 and 608 rapidly evolving genes (Figure 1C), respectively, with 75 genes identified as rapidly evolving by both methods (Figure 1D). Positively selected and rapidly evolving genes were longer than the background gene set (Figure 1 – Figure Supplement 1), consistent with previous observations that ABSREL and BUSTED have more statistical power to detect ω>1 in longer genes (Murrell et al., 2015b; Pond et al., 2011b).
Amino acid substitutions in positively selected genes are likely function-altering
To infer the putative functional consequences of amino acid changes in genes with signatures of positive selection and rapid evolution, we used IQTREE2 (Minh et al., 2020) to reconstruct the ancestral Paenungulate (AncPaenungulata) amino acid sequence for each gene (Figure 1A) and identified amino acid changes in the elephant lineage. Then, we used a fixed effect likelihood (FEL) model (Pond and Frost, 2005) to estimate the strength and direction of selection acting on each of these codon sites across mammals; codons with dN/dS<1 evolve under selective constraints (i.e., selection against amino acid changes). Thus, substitutions at these sites are likely function-altering. In contrast, codons with dN/dS>1 evolve under diversifying selection (i.e., selection favors amino acid changes), and codons with dN/dS≈1 evolve under weak or absent selective constraint. We found that 40.5% (4048/9981) of elephant-specific amino acid changes in positively selected genes occurred at sites with dN/dS<1, whereas 2.5% (251/9981) occurred at sites with dN/dS>1, and 47.4% (4728/9981) occurred at sites with dN/dS≈1 (Figure 2A). Similarly, 38.4% (7868/20366) of elephant-specific amino acid changes in rapidly evolving genes occurred at sites with dN/dS<1, whereas 2.3% (462/20366) occurred at sites with dN/dS>1, and 50.4% 10262/20366) occurred at sites with dN/dS≈1 (Figure 2A).
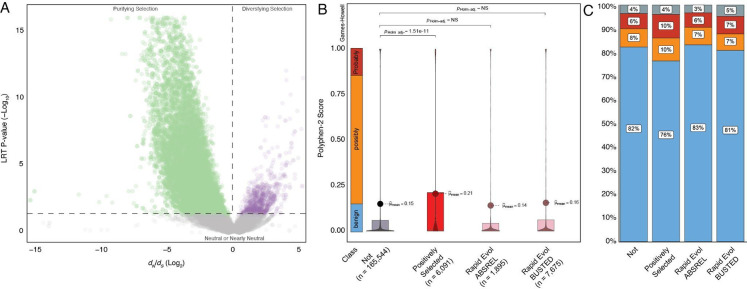
Figure 2. Amino acid substitutions in positively selected genes are likely deleterious.
A. Volcano plot showing the dN/dS (Log2) and likelihood ratio test (LRT) P-value (−Log10) for codons with elephant-specific amino acid changes in positively selected and rapidly evolving genes. dN/dS rates and P-values were estimated with the fixed effect likelihood (FEL) model, the P-value is for the LRT of dN/dS ≠ 1 at each codon. Codons with dN/dS>1 and a P-value≤0.05 are colored purple, those with N/dS<1 and a P-value≤0.05 are colored green, and those with dN/dS ≈ 1 are colored grey.
B. Violin and box plots showing the distribution of PolyPhen-2 scores for elephant-specific amino acid changes. The number of amino acid substitutions is given in parenthesis for each gene set. The range of scores corresponding to benign, possibly damaging, and probably damaging mutations is highlighted. Multiple hypotheses (Holm) corrected p values (pHolm-adj.) are shown for statistically significant comparisons; NS, not significant. Not, amino acid changes in genes with no evidence of positive selection or rapid evolution; Positively selected, amino acid changes in positively selected genes; Rapid Evol ABSREL, amino acid changes in rapidly evolving genes inferred from ABSREL; Rapid Evol BUSTED, amino acid changes in rapidly evolving genes inferred from BUSTED.
C. Bar chart showing the qualitative PolyPhen-2 predictions of amino acid changes. Not, amino acid changes in genes with no evidence of positive selection or rapid evolution; Positively selected, amino acid changes in positively selected genes; Rapid Evol ABSREL, amino acid changes in rapidly evolving genes inferred from ABSREL; Rapid Evol BUSTED, amino acid changes in rapidly evolving genes inferred from BUSTED.
Our observation that many elephant-specific amino acid substitutions in positively selected and rapidly evolving genes occur at sites with an evolutionary signature of purifying selection (Figure 2A) suggests these substitutions are function-altering. To explore this possibility, we used PolyPhen-2 (Adzhubei et al., 2013) to predict the consequences of amino acid substitutions in the elephant lineage; PolyPhen-2 assigns each substitution a score in the range of 0 (benign or unlikely to be function-altering) to 1 (probably deleterious or likely to be function altering). Of 182,333 amino acid changes in the elephant lineage, 175,524 had PolyPhen-2 predictions. We combined genes identified under positive selection by ABSREL and BUSTED into a single gene set because there was considerable overlap between genes (Figure 1C) and found that the mean PolyPhen-2 score of amino acid changes in positively selected genes was 0.21 while the mean score of genes without evidence of selection was 0.15 (Holm adjusted Games-Howell test P=1.91✕10−11); however, there was no difference in mean PolyPhen-2 scores between genes without evidence of selection and rapidly evolving genes identified by ABSREL or BUSTED (Figure 2B). A greater proportion of amino acid substitutions in positively selected genes are also classified as possibly and probably damaging (Figure 2C). These data indicate that many amino acid changes in positively selected genes occurred at sites under purifying selection and are likely function-altering.
Positively selected and rapidly evolving genes are enriched in anti-cancer functions
Elephants have evolved cells with derived anti-cancer phenotypes; for example, they induce apoptosis at low levels of DNA damage (Abegglen et al., 2015; Sulak et al., 2016a; Vazquez et al., 2018), are resistant to oxidative stress-induced cell-death (Gomes et al., 2011), are resistant to experimental immortalization (Fukuda et al., 2016; Gomes et al., 2011), and repair DNA damage faster than smaller-bodied species (Francis et al., 1981; Hart and Setlow, 1974; Promislow, 1994). Therefore, we identified gene ontologies (GO), Reactome (Jassal et al., 2020), Wikipathway (Jassal et al., 2020), and XDeathDB (Gadepalli et al., 2021) terms related to regulated cell death modes, senescence, oxidative stress, DNA damage repair, and the cell cycle, and DisGeNET terms (Piñero et al., 2020) related to neoplasia, malignancy, and metastasis. Next, we used the hypergeometric over-representation (ORA) test in WebGestalt (Liao et al., 2019) to determine if positively selected and rapidly evolving genes were enriched in these terms and pathways. We again combined genes identified under positive selection by both methods into a single gene set and tested rapidly evolving genes identified by ABSREL or BUSTED separately. Positively selected and rapidly evolving genes were statistically enriched in numerous ontologies and pathways (FDR≤0.10) related to multiple cell death modes, regulation of the cell cycle, DNA damage repair, and cancer biology, but only one enriched term was related to oxidative stress and no enriched terms were related to senescence (Figure 3). Next, we tested if positively selected genes were enriched in any GO biological process or molecular function terms, i.e., not using an a priori-defined set of terms as above, and identified four statistically enriched GO terms at an FDR≤0.14 (Figure 4). Similarly, rapidly evolving genes identified by ABSREL were enriched in numerous GO terms at an FDR≤0.10 (Figure 4), most of which were related to immune functions. In contrast, rapidly evolving genes identified by BUSTED were not enriched in any ontology term with an FDR≤0.94. Thus, we conclude that genes related to cell death modes, cancer biology, oxidative stress, senescence, EGFR-signaling, and immune functions, particularly neutrophil biology, have been pervasive targets of positive selection and rapid evolution in the elephant lineage.
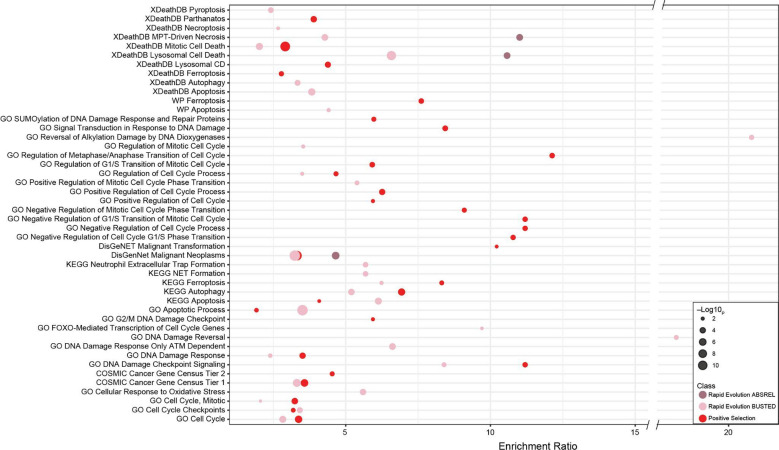
Figure 3. Positively selected and rapidly evolving genes are enriched in cell death pathways and cancer gene sets.
Bubble plot showing the enrichment ratio and hypergeometric P-value of GO, Reactome (R), Wikipathway (WP), and XDeathDB terms related to regulated cell death modes, senescence, oxidative stress, DNA damage repair, and the cell cycle, and DisGeNET terms related to neoplasia, malignancy, and metastasis in which positively selected (red) and rapidly evolving (pink) genes are enriched (FDR≤0.10).
Figure 4. Gene ontology (GO) terms in which positively selected and rapidly evolving genes are enriched.
Bubble plot showing the GO biological process and molecular function name, enrichment ratio, and hypergeometric P-value of terms in which positively selected (red, FDR≤0.14) and rapidly evolving identified by ABSREL (pink, FDR≤0.10) genes are enriched.
Discussion
Elephants have a very low prevalence of cancer given their body size and lifespan (Abegglen et al., 2015; Bulls et al., 2022; Compton et al., 2023; Tollis et al., 2021), indicating that elephants have evolved enhanced cancer prevention mechanisms. Among these anti–cancer phenotypes are cells that induce apoptosis at low levels of DNA damage (Abegglen et al., 2015; Sulak et al., 2016a; Vazquez et al., 2018), are resistant to oxidative stress-induced cell death (Gomes et al., 2011), have faster DNA damage repair rates than smaller-bodied species (Francis et al., 1981; Hart and Setlow, 1974; Promislow, 1994), and resistant to experimental immortalization (Fukuda et al., 2016; Gomes et al., 2011). These cellular traits are at least partly mediated by an increase in the number of tumor suppressors in the elephant lineage (Caulin et al., 2015; Doherty and Magalhães, 2016; Sulak et al., 2016b; Tollis et al., 2020; Vazquez et al., 2018; Vazquez and Lynch, 2021), but many other mechanisms must also have contributed to enhanced cancer resistance including functional divergence of protein-coding genes (Li et al., 2023; Lynch and Wagner, 2008). Here, we used a suite of methods to characterize the strength and direction of selection acting on protein-coding genes in the elephant lineage, focusing on genes with evidence of positive selection and rapid evolution, which is likely associated with functional divergence (Slodkowicz and Goldman, 2020; Tennessen, 2008). We identified 496 genes, 3.73% of alignments tested, with statistically significant evidence for positive selection in the elephant lineage, and 660 genes, 4.96% of alignments tested, that likely evolved rapidly in elephants. These are good candidates for genes that may contribute to elephant-specific traits, such as the evolution of molecular and cellular characteristics that enhance cancer resistance, among many others.
Comparison to previous studies of selection in elephants
Several previous studies have explored the role of positive selection acting on protein-coding genes in the elephant lineage, albeit with smaller numbers of species and alignments than our study or using different methods. Goodman et al. (2009), for example, used the free ratio model in PAML (Yang, 2007) to identify positively selected genes in the elephant lineage using a dataset of 11 species and 5,714 protein-coding genes; they identified 67 genes with dN/dS≥1.01, which were enriched in mitochondrial functions but none of the GO terms we identified (Supplementary dataset 1). There are several important differences between Goodman et al. (2009) and our study: 1) The free ratio model does not test if the dN/dS ratio in a particular lineage is significantly different than one, rather it tests if dN/dS rate variation is different across lineages (Yang, 1998); 2) Unlike ABSREL and BUSTED, the free ratio model does not allow for rate variation across sites, dN/dS>1 is only inferred if the average dN/dS across all sites is greater than 1; and 3) The method implemented in PAML does not account for synonymous rate variation across sites or multiple simultaneous codon substitutions, which can bias inferences of both dS (Dunn et al., 2019; Rahman et al., 2021; Wisotsky et al., 2020) and dN (Dunn et al., 2019; Lucaci et al., 2023a; Nozawa et al., 2009; Suzuki, 2008; Venkat et al., 2018; Yang and Reis, 2011). While we tested 81.6% (4,663) of the genes tested by Goodman et al. (2009), only one (CD3G) was inferred to have been positively selected in our analyses; therefore, it is unsurprising that there was little overlap in our GO enrichment results.
We previously reported the results of a preliminary selection scan using the same dataset of alignments used here and the base ABSREL model, which did not include synonymous rate variation or multiple hits (Raviv et al., 2023). We identified 674 positively selected genes enriched in several Panther pathways (Mi et al., 2013; Mi and Thomas, 2009; Thomas et al., 2022) related to neurotransmitter signaling (Raviv et al., 2023). However, we found in this study that synonymous rate variation across sites improved model fit for all genes, suggesting that at least some of the 674 genes we previously reported as positively selected may be false positives; indeed, only 215 (31.9%) were inferred as positively selected by ABSREL[S]. However, the 393 genes identified as positively selected by ABSREL[SMH] were similarly enriched in Panther pathways related to neurotransmitter signaling (Supplementary dataset 2), particularly serotonin and muscarinic acetylcholine receptor signaling. Thus, the signal for positive selection on genes in these pathways is strong despite the small overlap between genes inferred as positively selected by ABSREL and ABSREL[S].
Li et al. (2023) used BUSTED[S] to test for an episode of positive selection in the stem lineage of African and Asian elephants in a dataset of 12 species and 4,444 protein-coding genes and identified 618 positively selected genes. 492 of these genes were also tested for selection in our dataset, but only 32 (6.45%) were inferred to have been positively selected in our analyses. Not surprisingly, given the low overlap, our datasets had no overlap in GO term or pathway enrichments (Supplementary dataset 3). There are several potential explanations for this low overlap, including differences in our alignments, which can have substantial impacts on inferences of selection (Fletcher and Yang, 2010; Franco et al., 2019; Jordan and Goldman, 2012; Markova-Raina and Petrov, 2011; Spielman et al., 2014) and differences in the number of species included in alignments (261 vs 12), with larger alignments likely having more power and reduced false positive inferences of selection (Pond and Frost, 2005) but also paradoxical effects on inferences of selection, for example, by diluting the signal of dN/dS>1 by the addition of taxa in which positive selection did not occur (Chen and Sun, 2011; Murrell et al., 2012; Smith et al., 2015; Yokoyama et al., 2008); this signal loss may be more likely to occur in cases where the strength of selection is relatively weak or the proportion of sites or lineages under selection relatively small (Yang and Reis, 2011). Thus, we interpret our results as complementary to, rather than in contrast with, Li et al. (2023). Indeed, studies of individual genes should include taxa inclusion sensitivity analyses (such as jackknifing or a priori exclusion of focal taxa) to infer the effects of alignment size and composition on inferences of positive selection.
Positive selection and rapid evolution of cancer genes in elephants
We reasoned that genes with roles in cancer biology might have diverged in function in elephants, for example, coincident with the evolution of enhanced tumor suppressor or reduced oncogenic functions. We found that positively selected and rapidly evolving genes were statistically enriched in many GO terms and pathways related to cancer biology, including malignant neoplasms, neoplastic cell transformation, and COSMIC Cancer Gene Census Tier 1 and 2; Tier 1 genes have a documented activity related to cancer and evidence of mutations that promote oncogenic transformation, while Tier 2 genes genes have a strong indication of a role in cancer but with less extensive evidence than Tier 1 genes. Among these genes are many with well-characterized roles in cancer biology, such as APC, a negative regulator of canonical WNT signaling that suppresses invasion and tumor progression, particularly in colorectal cancer (Fodde, 2002; Hankey et al., 2018; Kwong and Dove, 2009), CHEK1 which encodes the checkpoint kinase CHK1 that regulates multiple cell cycle and DNA damage checkpoints (Chen and Sanchez, 2004; Harten et al., 2019; Liu et al., 2000; Patil et al., 2013; Synnes et al., 2002), and POLD1, which encodes the catalytic subunit of the DNA polymerase δ complex that has both DNA polymerizing and a proofreading domain that ensures replication accuracy and genomic stability (Haradhvala et al., 2018); germline mutations in the proofreading domain of POLD1 predispose to colorectal adenomas and carcinomas among others (Palles et al., 2013; Rayner et al., 2016). These data suggest that functional divergence of genes with cancer-protective effects may have contributed to the evolution of cancer resistance in elephants and underlie cellular traits like rapid DNA damage repair.
Positive selection and rapid evolution of cell death pathway genes in elephants
Many mechanisms have likely contributed to the evolution of reduced cancer risk in elephants, including cells that induce apoptosis at low levels of DNA damage (Abegglen et al., 2015; Sulak et al., 2016a; Vazquez et al., 2018) and are resistant to oxidative stress-induced cell death (Gomes et al., 2011). Consistent with these functional studies, we found that positively selected and rapidly evolving genes were statistically enriched in many GO terms and pathways related to cell death, such as apoptosis, ferroptosis, pyroptosis, and parthanatos, and regulated across (Figure 3). These data suggest that positively selected and rapidly evolving genes have contributed to the divergence of cell death pathways in elephants. Among the rapidly evolving genes, for example, is CASP8, the initiator caspase of extrinsic apoptosis, necroptosis, and pyroptosis (Boldin et al., 1996; Fritsch et al., 2019; Muzio et al., 1996), APAF1, which forms the central scaffold of the apoptosome and and thus plays a critical role in apoptosis (Bratton and Salvesen, 2010; Cain et al., 2002), BOK an unusual BCL-2 family protein that directly induces mitochondrial outer membrane permeabilization (MOMP) and apoptosis in response to endoplasmic reticulum (ER) stress (Carpio et al., 2015; Llambi et al., 2016), BCL2L15, another unusual BCL-2 family protein with pro-apoptotic functions that reduces malignant transformation of cells in the gastrointestinal tract (Dempsey et al., 2005; Jang et al., 2022; Özören et al., 2009), BCLAP a tumor suppressor that induces cell cycle arrest at the G1/S and apoptosis through an uncharacterized but p53-independent mechanisms (Fan et al., 2011; Han et al., 2022; Yao et al., 2007; ZHAO et al., 2016), KDM6B, which promotes parthanatos by inhibiting DNA damage-evoked PARP-1 hyperactivation (Yang et al., 2022), and ALKBH7 is required for alkylation and oxidation-induced programmed necrosis (Fu et al., 2013; Jordan et al., 2017; Kulkarni et al., 2020). Remarkably, other cancer-resistant species have also evolved modified cell death pathways. Blind mole rats, for example, evolved a concerted necrotic cell death mechanism (Gorbunova et al., 2012), and naked mole rats evolved cells that induce cell death in response to senescence via the INK4a-RB pathway (Kawamura et al., 2023). These data suggest functional divergence of cell death modes may be shared across cancer-resistant species.
Positive selection and rapid evolution of cell cycle regulatory genes in elephants
The cell cycle and its checkpoints play crucial roles in the development and progression of cancer; loss of checkpoint control, for example, allows cells to proliferate despite DNA damage contributing to genomic instability and resistance to apoptosis (Matthews et al., 2022) Numerous genes with roles in the cell cycle were positively selected or are evolving rapidly in elephants, including PPP2CB, which encodes the catalytic subunit of the master regulator of the cell cycle phosphatase 2A (PP2A) (Wlodarchak and Xing, 2016), CDKAP2, which inhibits cell cycle progression at the G1/S checkpoint by suppressing CDK2 and blocking the interaction between CDK2 and cyclin E and A (Liu et al., 2013), E2F7, which represses the expression of G1/S genes during S-phase progression (Westendorp et al., 2012) and CDC25B, a phosphatase required to activate cyclin/CDK1 complexes at the G2/M checkpoint and induce mitotic progression (Schmidt et al., 2007). Many genes related to the centrosome, which is required for several cell-cycle transitions, including G1 to S-phase, G1 to M, and metaphase to anaphase (Doxsey et al., 2005), were positively selected or evolved rapidly, including CEP57, CEP131, CEP76, CEP350, CEP76, and CEP104. These data suggest that the functional divergence of genes that regulate the cell cycle has likely contributed to the evolution of cancer resistance in elephants.
Positive selection and rapid evolution of DNA damage repair genes in elephants
DNA damage checkpoints are essential to maintain genomic integrity. Perhaps surprisingly, given their critical role, DNA damage responses can dramatically diverge between species. Previous studies have also found that elephant cells repair DNA damage much faster than other smaller-bodied mammals (Francis et al., 1981; Hart and Setlow, 1974; Promislow, 1994), suggesting that DNA damage repair genes may have derived functions in elephants. Among the positively selected and rapidly evolving genes, for example, are RIF1, which cooperates with TP53BP1 to promote DNA repair by non-homologous end joining (NHEJ) in the G1 and S phases of the cell cycle (Escribano-Díaz et al., 2013; Feng et al., 2013; Virgilio et al., 2013), the multifunctional exonuclease EXO1, which is involved in DNA damage checkpoint progression, mismatch repair (MMR), translesion DNA synthesis (TLS), nucleotide excision repair (NER), and limits end resection of double-strand breaks thereby facilitating repair via error-free homologous recombination (HR) rather than error-prone NHEJ (Keijzers et al., 2018; Tomimatsu et al., 2017), MLH1, part of PMS2 MMR complex that generates single-strand breaks near the mismatch and entry points for EXO1 to degrade the strand containing the mismatch (Kadyrov et al., 2006; Kansikas et al., 2011; Sacho et al., 2008), NEK4, which regulates a unique ATM/ATR-independent DNA damage checkpoint and the induction of replicative senescence in response to double-stranded (DSB) DNA damage (Chen et al., 2011; Tomimatsu et al., 2017), KAT5, an acetyltransferase that plays an essential role in DNA damage repair by acetylating and activating ATM and the canonical DSB repair pathway (Sun et al., 2005)and SAMHD1 which promotes DNA end resection to facilitate DSB repair by HR (Daddacha et al., 2017; Kapoor-Vazirani et al., 2022). Similarly, long-lived and cancer-resistant Bowhead whales, which have a maximum lifespan of over 200 years (George et al., 1999), have evolved cells that repair double-strand breaks with high efficiency and accuracy compared to many other mammals (Firsanov et al., 2023), suggesting that the evolution of DNA damage repair genes may be a common route to evolve cancer resistance.
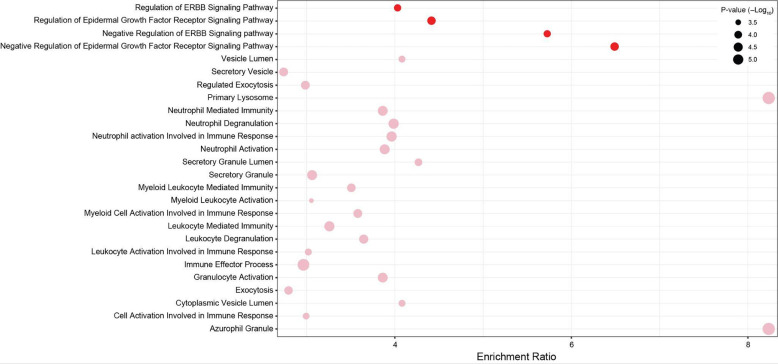
Positive selection of EGFR-signaling genes in elephants
While this study was motivated by the evolution of cancer resistance in the elephant lineage, our genome-wide approach is relatively unbiased with respect to the role of protein-coding genes in cancer, i.e., we tested for selection in all genes rather than genes with previously ascertained roles in cancer biology. Thus, our observation that positively selected genes were only enriched in four GO biological process terms (with an FDR<0.10), and that after redundancy reduction only “regulation of epidermal growth factor receptor signaling pathway” remained is remarkable. Dysregulated epidermal growth factor receptor (EGFR) signaling plays a central role in many cancers, including driving tumor growth and metastasis (Normanno et al., 2006; Sigismund et al., 2018), and EGFR inhibitors are a standard treatment for cancers with oncogenic EGFR mutations (Cooper et al., 2022; Jänne et al., 2015; Levantini et al., 2022). Among the positively selected genes in the EGFR pathway identified by ABSREL and BUSTED is SPRY2, a negative regulator of receptor tyrosine kinases (RTKs) and the Ras/Erk signaling pathway (Guy et al., 2009; Hacohen et al., 1998), that functions as a context-dependent tumor suppressor or oncogene (Holgren et al., 2010; Lo et al., 2006; Saini et al., 2018). For example, epigenetic silencing of SPRY2 is common in prostate cancer (McKie et al., 2005), and it is inactivated in about 18% of primary and over 70% of metastatic tumors (Taylor et al., 2010); SPRY2 cooperates with PTEN and PP2A to act as a tumor suppressor checkpoint in prostate cancer (Patel et al., 2013). In contrast, SPRY2 expression is upregulated in colonic adenocarcinomas, where it promotes cell proliferation, growth of tumor xenografts, metastasis, angiogenesis, and inhibits apoptosis (Holgren et al., 2010). These data suggest a relatively direct connection between positively selected genes in the EGFR–signaling pathway and the evolution of reduced cancer prevalence in elephants.
Derived neutrophil biology in elephants?
Our observation that positively selected and rapidly evolving genes in elephants are enriched in gene ontologies related to neutrophil biology is particularly intriguing. While neutrophils are best known for their role in the innate immune system, they also play an important, if complex, role in cancer biology, where they can both promote and inhibit tumor initiation, growth, and metastasis (Coffelt et al., 2016). Remarkably, many positively selected and rapidly evolving genes are related to neutrophil extracellular trap formation, a form of cell death characterized by the release of decondensed chromatin and granule contents into the extracellular space (Poli and Zanoni, 2023; Remijsen et al., 2011; Vorobjeva and Chernyak, 2020), granules and degranulation, and neutrophil granule proteins have been associated with tumor progression (Mollinedo, 2019). Neutrophils contain (at least) four functionally distinct kinds of granules: 1) primary or azurophilic granules, which store proteins with antimicrobial activity such as myeloperoxidase, elastase, cathepsins, and defensins; 2) secondary or specific granules, which contain lactoferrin and lipocalin, but not matrix metalloprotease-9 (MMP9); 3) tertiary granules, which contain MMP9 but not lactoferrin or lipocalin; and 4) secretory vesicles, which are likely derived from endocytosis of the plasma membrane (Lacy, 2006).
We found that rapidly evolving genes were enriched in GO cellular component terms related to each of these kinds of granules (Figure 4), including granule contents such as CTSS, LYZ, MMP9, and PRDX6, which might be expected to evolve rapidly because they are caught in an arms race with pathogens, but also genes that mediate degranulation including members of the SNAP receptor (SNARE) signaling pathway SNAP23, VAMP2, VAMP7, VAMP8, GCA, STX5, and STXBP2 which might be expected to have conserved functions given their essential roles in conserved biological processes such as membrane trafficking. These data suggest neutrophil phenotypes related to cancer biology may have evolved in the elephant lineage. Indeed, some data indicate that neutrophil biology is different in elephants than in other species. Elephants, for example, have higher concentrations of neutrophils in their blood than expected from their body mass, i.e., neutrophil concentration in blood scales hypermetrically with body size (Downs et al., 2019). Elephant neutrophils are also activated and produce oxygen radicals at much lower stimuli than other species (Smith et al., 1998; Tell et al., 1999). Further studies are needed, however, to determine if these phenotypes are derived in the elephant lineage and to explore if other neutrophil functions such as neutrophil extracellular trap formation (NETosis), a regulated cell death mode characterized by intracellular degranulation and the release of decondensed chromatin and granular contents into the extracellular space (Metzler et al., 2014, 2011; Papayannopoulos et al., 2010; Vorobjeva and Chernyak, 2020), are different in elephants than other species.
Caveats and limitations
There are several limitations to this study. For example, while there are two major lineages of living elephants, African (Loxodonta africana and Loxodonta cyclotis) and Asian (Elephas maximus ssp.), our alignments only included African savanna elephant (Loxodonta africana). Therefore, our inference of selection combines codon substitutions in the Proboscidean stem lineage with those in the Loxodonta lineage after the Loxodonta–Elephas divergence, which cannot have contributed to the evolution of elephant-specific traits. We do not believe that this adversely affects our results because the length of the Proboscidean stem lineage (~53 million years) is much longer than the divergence between the African and Asian elephant lineages, which is estimated to be 6.6–8.6 MYA (Rohland et al., 2007). Thus, most amino acid changes will have occurred in the Proboscidean stem lineage; codon-based models also have little power to detect recent selection. Perhaps most importantly, the ability of codon-based methods to detect positive selection is sensitive to a range of parameters, including branch length (substitutions per codon), synonymous site saturation, the strength of selection, and the number of selected sites. Tests of positive selection, for example, are conservative and lack power when the strength of selection is weak, limited to one or a few sites, and episodic, i.e., occurs on a single lineage (such as the Proboscidean stem lineage). In these cases, the methods lack the power to reject the null model, and an episode of positive selection will be missed. Thus, our results are not an exhaustive catalog of all genes positively selected in elephants. Future studies could use other methods to detect the signature of protein functional divergence and episodic purifying selection (Gaucher et al., 2002; Gu, 2006, 2001a, 2001b; Gu et al., 2013; Philippe et al., 2003; Pond et al., 2020); these methods, however, would need to include many more Proboscidean species, including living species such as Asian and African forest elephants and extinct species with genomic data such as straight-tusked elephants, mammoths, and mastodons, to have power to detect shifts in amino acid conservation profiles.
Conclusions
Elephants have emerged as a model system to study the evolution of cancer resistance in large and/or long-lived species. While numerous molecular and cellular mechanisms must have contributed to cancer resistance and other traits in the elephant lineage, in this study, we focused on identifying genes that were positively selected or evolved rapidly and may have functionally diverged in elephants. Our results indicate that genes related to many cell death pathways, EGFR–signaling, and neutrophil biology were positively selected or evolved rapidly in the elephant lineage. At least some of the genes likely contributed to the evolution of cancer resistance in Proboscideans and are promising candidates for functional studies. More generally, our results highlight the importance of including variation in synonymous substitution rates and multi-nucleotide mutations in dN/dS-based selection tests; indeed, we found statistically significant evidence for synonymous substitution rate variation in all genes we tested.
Methods
Assembling alignments of orthologous coding genes from 261 Eutherians
We previously reported the assembly of protein-coding gene alignments from the genomes of 261 Eutherian (“Placental”) mammals (Bowman et al., 2023b), which are also used in this study. Briefly, we used a reciprocal best BLAT hit (RBBH) approach to assemble a dataset of orthologous coding gene alignments from the genomes of 261 Eutherian (“Placental”) mammals (Figure 1A; Figure 1 – source data 1). We began by using RBBH with query sequences from human CDSs in the Ensembl v99 human coding sequence dataset and BLAT searching the genomes of 260 other Eutherians, followed by searching the top hit to the human query back to the human genome; we used BLAT matching all possible reading frames, with a minimum identity set at 30% and the “fine” option activated (Kent, 2002), and excluded genes with fewer than 251 best reciprocal hits out of the 261 (human+other mammals) species included in the analysis. Orthologous genes were aligned with Macse v2 (Ranwez et al., 2018), a codon-aware multiple sequence aligner that identifies frameshifts and readjusts reading frames accordingly. Alignments generated by Macse v2 were edited by HMMcleaner with default parameters (Franco et al., 2019) to remove species-specific substitutions that are likely genome sequencing errors and “false exons’’ that might have been introduced during the Blat search. Finally, we excluded incomplete codons and the flanks of indels, which usually have more misaligned substitutions. We thus generated a dataset of 13,310 orthologous coding gene alignments from the genomes of 261 Eutherian mammals; this corresponds to 66.4% of all protein-coding genes annotated in the African savannah elephant genome (Loxafr3.0).
ABSREL and BUSTED selection tests
The multiple sequence alignments used in this study have been described elsewhere (Bowman et al., 2023b). Methods to infer the strength and direction of selection acting on molecular sequences, such as those to identify pervasive and episodic adaptive evolution (“positive selection”), are often based on estimating the ratio of non-synonymous (dN) to synonymous (dS) substitution rates (dN/dS or ω) in an alignment of homologous genes. These methods can be applied to entire genes or regions within genes (Hughes and Nei, 1988), sites (Nielsen and Yang, 1998), branches (Messier and Stewart, 1997; Muse and Gaut, 1994; Yang, 1998), or sites along a specific branch - the latter often called “branch-site” (BSREL) models (Pond et al., 2011a; Smith et al., 2015; Yang and Nielsen, 2002). An advantage of the BSREL models is that they allow for a group of sites to evolve under the action of positive selection, i.e., with ω>1, while the remaining sites can evolve under purifying selection (ω<1) or relatively neutrally (ω=1) in specific branches, and thus can detect episodic positive selection (limited to a subset of branches) and restricted to a subset of sites in a gene. Positive selection is inferred when a class of sites is identified with ω>1, with a LRT P-value ≤ 0.05. We used two related BSREL models implemented in HyPhy (Pond et al., 2005), ABSREL, which infers the number of dN/dS rate categories to be inferred from the alignment, and BUSTED, which has three fixed rate classes. ABSREL and BUSTED can accommodate both synonymous rate variation across sites [S] (Pond and Muse, 2005; Wisotsky et al., 2020) and multi-nucleotide mutations per codon [MH] (Lucaci et al., 2021). We tested for selection using the base ABSREL and BUSTED models, models that account for synonymous rate variation across sites (ABSREL[S] and BUSTED[S]), models that account for multi-nucleotide mutations per codon (ABSREL[MH] and BUSTED[MH]), and models that account for both synonymous rate variation across sites and multi-nucleotide mutations per codon (ABSREL[SMH] and BUSTED[SMH]). HyPhy v2.5.36 was used to run ABSREL v2.3 and BUSTED v3.1, while HyPhy v.5.32 was used to run FEL v2.1. Positive selection is inferred when the best-fitting ABSREL or BUSTED model, determined from a small sample AIC test, includes a class of sites with ω>1 at a likelihood ratio test (LRT) P-value ≤ 0.05, and rapid evolution inferred when the best-fitting ABSREL or BUSTED models inferred a class of sites with ω>1 at a likelihood ratio test (LRT) P-value = 0.051 – 0.20.
Fixed Effects Likelihood (FEL) selection tests
We characterized the strength and direction of selection acting on each codon site in positively selected and rapidly evolving genes in elephant lineage with a fixed effects likelihood model (FEL) that included synonymous rate variation across sites (Pond and Frost, 2005). This model estimates separate dN/dS rates for each site in an alignment and quantifies the strength of selection. We used these per site dN/dS rates to estimate the likely functional consequences of amino acid substitutions at those sites, such that substitutions at sites with dN/dS < 1 are likely to be function-altering and deleterious, substitutions at sites with dN/dS = 1 are likely to be non-function altering, and substitutions at sites with dN/dS > 1 are likely to be function altering and adaptive.
Identification of elephant-specific amino acid changes
We used IQTREE2 (Nguyen et al., 2015) to reconstruct ancestral amino acid sequences of all 13,491 genes to identify elephant-specific amino acid changes. For each gene, we translated nucleotide sequences to amino acid sequences with the -st NT2AA option, imposed the species tree with the Paenungulate lineages as polytomy with the -te option, inferred the best model of amino acid substitution and rate variation across sites with the -m MFP option (Kalyaanamoorthy et al., 2017), and to infer ancestral protein sequences with the --ancestral option; identical protein sequences were maintained in the analyses with the --keep-ident option to ensure consistent internal node numbering across genes. After the reconstructions for each gene were completed, the predicted ancestral Paenungulate (AncPaenungulata), i.e., the last common ancestor of elephant, manatee, and hyraxes, of each gene was compared to the Loxodonta africana sequence to identify amino acid changes in Loxodonta africana. We thus identified 182,333 amino acid changes in 13,310 proteins in the elephant lineage.
PolyPhen-2 amino acid classification
We used PolyPhen-2 (Adzhubei et al., 2013) to classify the functional consequences of amino acid changes in the gorilla lineage using the AncPaenungulata (Figure 1A) reconstructed protein sequences as input and “wildtype” amino acid and amino acid changes in elephant lineage as the “mutant” amino acid. The Classifier model was set to “HumDiv”, and the other parameters to the GRCh37/hg19 genome, canonical transcripts, and missense annotations.
Over Representation Analyses (ORA)
We combined genes identified as positively selected by ABSREL (n=393) and BUSTED (n=427) into a single gene set because 82% (324/393) of genes identified as positively selected by ABSREL were also identified as positively selected by BUSTED, whereas 76% (324/427) of genes identified as positively selected by BUSTED were also identified as positively selected by ABSREL. In contrast, only 59% (75/127) and 12% (75/608) of genes were classified as rapidly evolving by both ABSREL and BUSTED, respectively. Thus, we analyzed these gene sets independently.
We identified gene ontologies (GO), Reactome (Jassal et al., 2020), Wikipathway (Jassal et al., 2020), and XDeathDB (Gadepalli et al., 2021) terms related to regulated cell death modes, senescence, oxidative stress, DNA damage repair, and the cell cycle, and DisGeNET terms (Piñero et al., 2020) related to neoplasia, malignancy, and metastasis directly from each database or with the term search function in Enrichr (Chen et al., 2013; Kuleshov et al., 2016). We used the hypergeometric over-representation (ORA) test in WebGestalt (Liao et al., 2019) to determine if positively selected and rapidly evolving genes were enriched in these terms and pathways. We combined genes identified under positive selection by both methods into a single gene set, tested rapidly evolving genes identified from ABSREL and BUSTED independently, the minimum number of genes for a category was set to 5, we used as the background gene set all genes tested for selection, and controlled false discovery with the Benjamini-Hochberg false discovery rate (Benjamini and Hochberg, 1995).
We also used WebGestalt (Liao et al., 2019) to identify if positively selected and rapidly evolving genes were enriched in any gene ontology (GO) Biological Process and Cellular Component ontologies without a priori selecting candidate gene sets. Because many genes were identified as positively selected or rapidly evolving, we set the minimum number of genes for a category to 30 for tests without a priori identified GO terms and pathways, which excludes GO terms with fewer than 30 annotated genes; this strategy reduces the likelihood of inferring enrichment for ontologies with few annotated genes by chance because our foreground gene sets are large.
Characteristics of positively selected and rapidly evolving genes
We used ShinyGO 0.77 (Ge et al., 2019) to characterize the high-level functions (defined by GO terms) and genomic features of positively selected and rapidly evolving genes; as for the ORA described above, we analyzed positively selected genes as a single group but rapidly evolving gene sets independently. Positively selected and rapidly evolving genes were longer than the background gene set (Figure 1 – figure supplement 1), consistent with previous observations that ABSREL and BUSTED have more statistical power to detect deviation from ω>1 in longer genes (Murrell et al., 2015b; Pond et al., 2011b), positively selected genes were also more GC rich than the background gene set (Figure 1 – figure supplement 1).
Comparison to previous studies of selection in elephants
Comparison to Goodman et al. (2009)
Goodman et al. (2009) used the free ratio model implemented in the codeml program of PAML (Yang, 1997a, 1997b) to identify positively selected genes in the elephant lineage using a dataset of 11 species and 7,768 RefSeq transcripts corresponding to ~6,000 nonredundant protein-coding genes in 2009; note that this is more properly a test of dN/dS rate variation across lineages, and not a statistical test of positive selection because the hypothesis of selection is not directly tested against the null model of dN/dS=1; it is also unclear if the free ratio model was statistically a better fit to the data than a simple one-ratio model, which is the appropriate null model against which the free ratio model is usually tested (Yang, 1998). However, the free ratio model is a branch-based test that does not account for rate variation across sites and is, therefore, a very conservative test for positive selection.
To ensure a fair comparison between our results, particularly our GO term enrichment results, we downloaded their codeml results from Dryad (https://doi.org/10.5061/dryad.908) and used BioMart (https://www.ensembl.org/info/data/biomart/index.html) to convert ReSeq IDs to HUGO gene IDs based on Ensembl Genes 111 and GRCh38.p14; the redundancy reduced dataset included 5,714 protein-coding genes, of which 67 had dN/dS≥1.01. We again used WebGestalt v. 2019 (Liao et al., 2019) to identify if genes with dN/dS≥1.01 were enriched gene ontology (GO) Biological Process and Cellular Component ontologies using the over-representation analysis (ORA) with the minimum number of genes for a category set to 5 (default) 30 (as in our analyses), the 67 genes with dN/dS≥1.01 as the foreground and the 5,714 genes tested for selection as the background gene set.
The 67 rapidly evolving genes (i.e., those with dN/dS≥1.01) were reported to be enriched in mitochondrial functions (Goodman et al., n.d.), which is similar to previous studies that have reported rapid evolution of mitochondrially encoded protein-coding genes in elephants (Finch et al., 2014; Ngatia et al., 2019), but not in any of the GO terms we identified (Goodman et al., n.d.). Goodman et al. (2009) used an early version of DAVID (Sherman et al., 2007) to identify enriched GO terms, suggesting that differences in our results may arise from differences in our enrichment tests. Therefore, we used WebGestalt to reanalyze GO term enrichment for the 67 rapidly evolving genes identified by Goodman et al. (2009) and found they were enriched (hypergeometric P≤0.05) in many GO biological process and cellular component terms related to the mitochondria (Supplementary dataset 1), but none the terms we identified.
Comparison to Li et al. (2023)
Li et al. (2023) used BUSTED[S] to test for an episode of positive selection in the stem lineage of African and Asian elephants in a dataset of 12 species and 4,444 protein-coding genes and identified 618 positively selected genes. To ensure a fair comparison between our results, mainly GO term enrichment results, Li et al. (2023) provided the list of genes tested for selection and those identified by BUSTED[S] as positively selected. We again used WebGestalt v. 2019 (Liao et al., 2019) to identify if genes with dN/dS≥1.00 were enriched gene ontology (GO) Biological Process and Cellular Component ontologies using the over-representation analysis (ORA) with the minimum number of genes for a category set to 5 (default) 30 (as in our analyses), the 618 genes with dN/dS≥1.00 as the foreground and the 4,444 genes tested for selection as the background gene set. These 618 genes were not enriched in any ontology after FDR correction; however, many terms were enriched with hypergeometric P≤0.05. None of these terms were the same as we identified (Supplementary dataset 3).
Supplementary Material
Figure 1 – source data 1. Genomes used in selection tests.
Figure 1 – source data 2. List of positively selected genes from ABSREL.
Figure 1 – source data 3. List of rapidly evolving genes from ABSREL.
Figure 1 – source data 4. List of positively selected genes from BUSTED.
Figure 1 – source data 5. List of rapidly evolving genes from BUSTED.
Figure 1 – figure supplement 1. Genomic features of positively selected and rapidly evolving genes.
Figure 3 – source data 1. Webgestalt output files for positively selected genes.
Figure 3 – source data 2. Webgestalt output files for ABSREL rapidly evolving genes.
Figure 3 – source data 3. Webgestalt output files for BUSTED rapidly evolving genes.
Figure 4 – source data 1. Webgestalt output files for positively selected genes.
Figure 4 – source data 2. Webgestalt output files for ABSREL rapidly evolving genes.
Figure 4 – source data 3. Webgestalt output files for BUSTED rapidly evolving genes.
Acknowledgments
This study was supported by NIH award 1R56AG071860–01 to V.J.L. Computational support was provided by the Center for Computational Research at the University at Buffalo. We thank S. L. Kosakovsky Pond for advice and help with running HyPhy, X. Zhou for clarifying the details of their analyses and providing IDs for their 4,444 gene dataset, and D. Enard for advice and thoughtful comments on this manuscript.
Data availability
HyPhy json out files from ABSREL, BUSTED, and FEL (http://doi.org/10.5061/dryad.h44j0zpt4), alignments (http://doi.org/10.5061/dryad.5dv41nsbc), and the phylogenetic tree used for selection tests (http://doi.org/10.5061/dryad.j0zpc86n3) are available from Dryad. HyPhy json output files can be visualized with HyPhy Vision in any web browser: http://vision.hyphy.org and https://observablehq.com/@spond/busted-compare.
References
- Abegglen LM, Caulin AF, Chan A, Lee K, Robinson R, Campbell MS, Kiso WK, Schmitt DL, Waddell PJ, Bhaskara S, Jensen ST, Maley CC, Schiffman JD. 2015. Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage in Humans. Jama 314:1850–1860. doi: 10.1001/jama.2015.13134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I, Jordan DM, Sunyaev SR. 2013. Predicting functional effect of human missense mutations using PolyPhen-2. Current protocols in human genetics / editorial board, Jonathan L Haines. [et al. ] Chapter 7:Unit7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57. [Google Scholar]
- Benyi E, Linder M, Adami J, Kieler H, Palme M, Sävendahl L. 2019. Adult height is associated with risk of cancer and mortality in 5.5 million Swedish women and men. J Epidemiol Commun H 73:730. doi: 10.1136/jech-2018-211040 [DOI] [PubMed] [Google Scholar]
- Boldin MP, Goncharov TM, Goltseve YV, Wallach D. 1996. Involvement of MACH, a Novel MORT1/FADD-Interacting Protease, in Fas/APO-1- and TNF Receptor–Induced Cell Death. Cell 85:803–815. doi: 10.1016/s0092-8674(00)81265-9 [DOI] [PubMed] [Google Scholar]
- Bowman J, Enard D, Lynch VJ. 2023a. Phylogenomics reveals an almost perfect polytomy among the almost ungulates (Paenungulata). bioRxiv 2023.12.07.570590. doi: 10.1101/2023.12.07.570590 [DOI] [Google Scholar]
- Bowman J, Silva N, Schüftan E, Almeida JM, Brattig-Correia R, Oliveira RA, Tüttelmann F, Enard D, Navarro-Costa P, Lynch VJ. 2023b. Pervasive relaxed selection on spermatogenesis genes coincident with the evolution of polygyny in gorillas. bioRxiv 2023.10.27.564379. doi: 10.1101/2023.10.27.564379 [DOI] [Google Scholar]
- Bratton SB, Salvesen GS. 2010. Regulation of the Apaf-1–caspase-9 apoptosome. J Cell Sci 123:3209–3214. doi: 10.1242/jcs.073643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulls S, Platner L, Ayub W, Moreno N, Arditi J-P, McCain S, Wagner P, Sonsbeek LGRB- van, Lynch VJ, Claude J, Glaberman, Chiari Y. 2022. Cancer prevalence is remarkably low in turtles and is correlated with life history traits in birds, mammals, and squamates. bioRxiv. doi: 10.1101/2022.07.12.499088 [DOI] [Google Scholar]
- Cain K, Bratton SB, Cohen GM. 2002. The Apaf-1 apoptosome: a large caspase-activating complex. Biochimie 84:203–214. doi: 10.1016/s0300-9084(02)01376-7 [DOI] [PubMed] [Google Scholar]
- Carpio MA, Michaud M, Zhou W, Fisher JK, Walensky LD, Katz SG. 2015. BCL-2 family member BOK promotes apoptosis in response to endoplasmic reticulum stress. Proc Natl Acad Sci 112:7201–7206. doi: 10.1073/pnas.1421063112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulin AF, Graham TA, Wang L-S, Maley CC. 2015. Solutions to Peto’s paradox revealed by mathematical modelling and cross-species cancer gene analysis. Philosophical transactions of the Royal Society of London Series B, Biological sciences 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun Y. 2011. Variation in the Analysis of Positively Selected Sites Using Nonsynonymous/Synonymous Rate Ratios: An Example Using Influenza Virus. PLoS ONE 6:e19996. doi: 10.1371/journal.pone.0019996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chen C-F, Riley DJ, Chen P-L. 2011. Nek1 kinase functions in DNA damage response and checkpoint control through a pathway independent of ATM and ATR. Cell Cycle 10:655–663. doi: 10.4161/cc.10.4.14814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sanchez Y. 2004. Chk1 in the DNA damage response: conserved roles from yeasts to mammals. DNA Repair 3:1025–1032. doi: 10.1016/j.dnarep.2004.03.003 [DOI] [PubMed] [Google Scholar]
- Coffelt SB, Wellenstein MD, Visser KE de. 2016. Neutrophils in cancer: neutral no more. Nat Rev Cancer 16:431–446. doi: 10.1038/nrc.2016.52 [DOI] [PubMed] [Google Scholar]
- Compton Z, Harris V, Mellon W, Rupp S, Mallo D, Kapsetaki SE, Wilmot M, Kennington R, Noble K, Baciu C, Ramirez L, Peraza A, Martins B, Sudhakar S, Aksoy S, Furukawa G, Vincze O, Giraudeau M, Duke EG, Spiro S, Flach E, Davidson H, Zehnder A, Graham TA, Troan B, Harrison TM, Tollis M, Schiffman JD, Aktipis A, Abegglen LM, Maley CC, Boddy AM. 2023. Cancer Prevalence Across Vertebrates. bioRxiv 2023.02.15.527881. doi: 10.1101/2023.02.15.527881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AJ, Sequist LV, Lin JJ. 2022. Third-generation EGFR and ALK inhibitors: mechanisms of resistance and management. Nat Rev Clin Oncol 19:499–514. doi: 10.1038/s41571-022-00639-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daddacha W, Koyen AE, Bastien AJ, Head PE, Dhere VR, Nabeta GN, Connolly EC, Werner E, Madden MZ, Daly MB, Minten EV, Whelan DR, Schlafstein AJ, Zhang H, Anand R, Doronio C, Withers AE, Shepard C, Sundaram RK, Deng X, Dynan WS, Wang Y, Bindra RS, Cejka P, Rothenberg E, Doetsch PW, Kim B, Yu DS. 2017. SAMHD1 Promotes DNA End Resection to Facilitate DNA Repair by Homologous Recombination. Cell Rep 20:1921–1935. doi: 10.1016/j.celrep.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey CE, Dive C, Fletcher DJ, Barnes FA, Lobo A, Bingle CD, Whyte MKB, Renshaw SA. 2005. Expression of pro-apoptotic Bfk isoforms reduces during malignant transformation in the human gastrointestinal tract. FEBS Lett 579:3646–3650. doi: 10.1016/j.febslet.2005.05.050 [DOI] [PubMed] [Google Scholar]
- Dobson JM. 2013. Breed-predispositions to cancer in pedigree dogs. Isrn Vet Sci 2013:941275. doi: 10.1155/2013/941275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty A, Magalhães JP de. 2016. Has gene duplication impacted the evolution of Eutherian longevity? Aging cell 15:978–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs CJ, Dochtermann NA, Ball R, Klasing KC, Martin LB. 2019. The Effects of Body Mass on Immune Cell Concentrations of Mammals. Am Nat 195:107–114. doi: 10.1086/706235 [DOI] [PubMed] [Google Scholar]
- Doxsey S, Zimmerman W, Mikule K. 2005. Centrosome control of the cell cycle. Trends Cell Biol 15:303–311. doi: 10.1016/j.tcb.2005.04.008 [DOI] [PubMed] [Google Scholar]
- Dunn KA, Kenney T, Gu H, Bielawski JP. 2019. Improved inference of site-specific positive selection under a generalized parametric codon model when there are multinucleotide mutations and multiple nonsynonymous rates. BMC Evol Biol 19:22. doi: 10.1186/s12862-018-1326-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot MG, Mooers AØ. 2014. Inferring ancestral states without assuming neutrality or gradualism using a stable model of continuous character evolution. BMC evolutionary biology 14:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Díaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JTF, Tkáč J, Cook MA, Rosebrock AP, Munro M, Canny MD, Xu D, Durocher D. 2013. A Cell Cycle-Dependent Regulatory Circuit Composed of 53BP1-RIF1 and BRCA1-CtIP Controls DNA Repair Pathway Choice. Mol Cell 49:872–883. doi: 10.1016/j.molcel.2013.01.001 [DOI] [PubMed] [Google Scholar]
- Fan D-G, Zhao F, Ding Y, Wu M-M, Fan Q-Y, Shimizu K, Dohjima T, Nozawa S, Wakahara K, Ohno T, Guo Y-S, Ma B-A, Jiang J-L. 2011. BLCAP induces apoptosis in human Ewing’s sarcoma cells. Exp Biol Med 236:1030–1035. doi: 10.1258/ebm.2011.010315 [DOI] [PubMed] [Google Scholar]
- Feng L, Fong K-W, Wang J, Wang W, Chen J. 2013. RIF1 Counteracts BRCA1-mediated End Resection during DNA Repair*. J Biol Chem 288:11135–11143. doi: 10.1074/jbc.m113.457440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch TM, Zhao N, Korkin D, Frederick KH, Eggert LS. 2014. Evidence of Positive Selection in Mitochondrial Complexes I and V of the African Elephant. PLoS ONE 9:e92587. doi: 10.1371/journal.pone.0092587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firsanov D, Zacher M, Tian X, Zhao Y, George JC, Sformo TL, Tombline G, Biashad SA, Gilman A, Hamilton N, Patel A, Straight M, Lee M, Lu JY, Haseljic E, Williams A, Miller N, Gladyshev VN, Zhang Z, Vijg J, Seluanov A, Gorbunova V. 2023. DNA repair and anti-cancer mechanisms in the longest-living mammal: the bowhead whale. doi: 10.1101/2023.05.07.539748 [DOI] [Google Scholar]
- Fletcher W, Yang Z. 2010. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol 27:2257–2267. doi: 10.1093/molbev/msq115 [DOI] [PubMed] [Google Scholar]
- Fodde R. 2002. The APC gene in colorectal cancer. Eur J Cancer 38:867–871. doi: 10.1016/s0959-8049(02)00040-0 [DOI] [PubMed] [Google Scholar]
- Francis AA, Lee WH, Regan JD. 1981. The relationship of DNA excision repair of ultraviolet-induced lesions to the maximum life span of mammals. Mech Ageing Dev 16:181–189. doi: 10.1016/0047-6374(81)90094-4 [DOI] [PubMed] [Google Scholar]
- Franco AD, Poujol R, Baurain D, Philippe H. 2019. Evaluating the usefulness of alignment filtering methods to reduce the impact of errors on evolutionary inferences. Bmc Evol Biol 19:21. doi: 10.1186/s12862-019-1350-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch M, Günther SD, Schwarzer R, Albert M-C, Schorn F, Werthenbach JP, Schiffmann LM, Stair N, Stocks H, Seeger JM, Lamkanfi M, Krönke M, Pasparakis M, Kashkar H. 2019. Caspase-8 is the molecular switch for apoptosis, necroptosis and pyroptosis. Nature 575:683–687. doi: 10.1038/s41586-019-1770-6 [DOI] [PubMed] [Google Scholar]
- Fu D, Jordan JJ, Samson LD. 2013. Human ALKBH7 is required for alkylation and oxidation-induced programmed necrosis. Genes Dev 27:1089–1100. doi: 10.1101/gad.215533.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Iino Y, Onuma M, Gen B, Inoue-Murayama M, Kiyono T. 2016. Expression of human cell cycle regulators in the primary cell line of the African savannah elephant (loxodonta africana) increases proliferation until senescence, but does not induce immortalization. In vitro cellular & developmental biology Animal 52:20–26. [DOI] [PubMed] [Google Scholar]
- Gadepalli VS, Kim H, Liu Y, Han T, Cheng L. 2021. XDeathDB: a visualization platform for cell death molecular interactions. Cell Death Dis 12:1156. doi: 10.1038/s41419-021-04397-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher EA, Gu X, Miyamoto MM, Benner SA. 2002. Predicting functional divergence in protein evolution by site-specific rate shifts. Trends Biochem Sci 27:315–321. doi: 10.1016/s0968-0004(02)02094-7 [DOI] [PubMed] [Google Scholar]
- Ge SX, Jung D, Yao R. 2019. ShinyGO: a graphical enrichment tool for animals and plants. Bioinformatics 36:2628–2629. doi: 10.1093/bioinformatics/btz931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George JC, Bada J, Zeh J, Scott L, Brown SE, O’Hara T, Suydam R. 1999. Age and growth estimates of bowhead whales (Balaena mysticetus) via aspartic acid racemization. Can J Zoöl 77:571–580. doi: 10.1139/z99-015 [DOI] [Google Scholar]
- Gomes NMV, Ryder OA, Houck ML, Charter SJ, Walker W, Forsyth NR, Austad SN, Venditti C, Pagel M, Shay JW, Wright WE. 2011. Comparative biology of mammalian telomeres: hypotheses on ancestral states and the roles of telomeres in longevity determination. Aging cell 10:761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M, Sterner KN, Islam M, Uddin M, Sherwood CC, Hof PR. n.d. Phylogenomic analyses reveal convergent patterns of adaptive evolution in elephant and human ancestries. Proceedings of the National Academy of Sciences 106:20824–20829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Hine C, Tian X, Ablaeva J, Gudkov AV, Nevo E, Seluanov A. 2012. Cancer resistance in the blind mole rat is mediated by concerted necrotic cell death mechanism. Proceedings of the National Academy of Sciences USA 109:19392–19396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Cairns BJ, Casabonne D, Wright FL, Reeves G, Beral V, collaborators MWS. 2011. Height and cancer incidence in the Million Women Study: prospective cohort, and meta-analysis of prospective studies of height and total cancer risk. The Lancet Oncology 12:785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X. 2006. A Simple Statistical Method for Estimating Type-II (Cluster-Specific) Functional Divergence of Protein Sequences. Mol Biol Evol 23:1937–1945. doi: 10.1093/molbev/msl056 [DOI] [PubMed] [Google Scholar]
- Gu X. 2001a. Maximum-Likelihood Approach for Gene Family Evolution Under Functional Divergence. Mol Biol Evol 18:453–464. doi: 10.1093/oxfordjournals.molbev.a003824 [DOI] [PubMed] [Google Scholar]
- Gu X. 2001b. Mathematical Modeling for Functional Divergence after Gene Duplication. J Comput Biol 8:221–234. doi: 10.1089/10665270152530827 [DOI] [PubMed] [Google Scholar]
- Gu X, Zou Y, Su Z, Huang W, Zhou Z, Arendsee Z, Zeng Y. 2013. An Update of DIVERGE Software for Functional Divergence Analysis of Protein Family. Mol Biol Evol 30:1713–1719. doi: 10.1093/molbev/mst069 [DOI] [PubMed] [Google Scholar]
- Guy GR, Jackson RA, Yusoff P, Chow SY. 2009. Sprouty proteins: modified modulators, matchmakers or missing links? J Endocrinol 203:191–202. doi: 10.1677/joe-09-0110 [DOI] [PubMed] [Google Scholar]
- Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. 1998. sprouty Encodes a Novel Antagonist of FGF Signaling that Patterns Apical Branching of the Drosophila Airways. Cell 92:253–263. doi: 10.1016/s0092-8674(00)80919-8 [DOI] [PubMed] [Google Scholar]
- Han F, Hu M, Zhang L, Fan X, Wang J, Lou Z, Wang S, Chen L, Ye Y, Ding Y, Jiao H. 2022. A-to-I RNA editing of BLCAP promotes cell proliferation by losing the inhibitory of Rb1 in colorectal cancer. Exp Cell Res 417:113209. doi: 10.1016/j.yexcr.2022.113209 [DOI] [PubMed] [Google Scholar]
- Hankey W, Frankel WL, Groden J. 2018. Functions of the APC tumor suppressor protein dependent and independent of canonical WNT signaling: implications for therapeutic targeting. Cancer Metastasis Rev 37:159–172. doi: 10.1007/s10555-017-9725-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haradhvala NJ, Kim J, Maruvka YE, Polak P, Rosebrock D, Livitz D, Hess JM, Leshchiner I, Kamburov A, Mouw KW, Lawrence MS, Getz G. 2018. Distinct mutational signatures characterize concurrent loss of polymerase proofreading and mismatch repair. Nat Commun 9:1746. doi: 10.1038/s41467-018-04002-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart RW, Setlow RB. 1974. Correlation Between Deoxyribonucleic Acid Excision-Repair and Life-Span in a Number of Mammalian Species. Proc National Acad Sci 71:2169–2173. doi: 10.1073/pnas.71.6.2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harten AM van, Buijze M, Mast R van der, Rooimans MA, Kemp SRM, Bachas C, Brink A, Walsum MS, Wolthuis RMF, Brakenhoff RH. 2019. Targeting the cell cycle in head and neck cancer by Chk1 inhibition: a novel concept of bimodal cell death. Oncogenesis 8:38. doi: 10.1038/s41389-019-0147-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick CC, Malanchi I. 2021. Neutrophils in cancer: heterogeneous and multifaceted. Nat Rev Immunol 1–15. doi: 10.1038/s41577-021-00571-6 [DOI] [PubMed] [Google Scholar]
- Holgren C, Dougherty U, Edwin F, Cerasi D, Taylor I, Fichera A, Joseph L, Bissonnette M, Khare S. 2010. Sprouty-2 controls c-Met expression and metastatic potential of colon cancer cells: sprouty/c-Met upregulation in human colonic adenocarcinomas. Oncogene 29:5241–5253. doi: 10.1038/onc.2010.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogduijn MJ, Beukel JC van den, Wiersma LCM, Ijzer J. 2013. Morphology and size of stem cells from mouse and whale: observational study. BMJ : Br Méd J 347:f6833. doi: 10.1136/bmj.f6833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Jiang S, Shi Y. 2020. Tyrosine kinase inhibitors for solid tumors in the past 20 years (2001– 2020). J Hematol Oncol 13:143. doi: 10.1186/s13045-020-00977-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Nei M. 1988. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335:167–170. doi: 10.1038/335167a0 [DOI] [PubMed] [Google Scholar]
- Jang DM, Oh EK, Hahn H, Kim HS, Han BW. 2022. Structural insights into apoptotic regulation of human Bfk as a novel Bcl-2 family member. Comput Struct Biotechnol J 20:745–756. doi: 10.1016/j.csbj.2022.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jänne PA, Chih-Hsin YJ, Dong-Wan K, David P, Yuichiro O, S RS, Myung-Ju A, Sang-We K, Wu-Chou S, Leora H, Daniel H, Enriqueta F, Joo-Hang K, Paul F, Mireille C, H BK, A DP, Serban G, Malcolm R. 2015. AZD9291 in EGFR Inhibitor–Resistant Non–Small-Cell Lung Cancer. N Engl J Med 372:1689–1699. doi: 10.1056/nejmoa1411817 [DOI] [PubMed] [Google Scholar]
- Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A. 2020. The reactome pathway knowledgebase. Nucleic Acids Res 48:498–503. doi: 10.1093/nar/gkz1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan G, Goldman N. 2012. The effects of alignment error and alignment filtering on the sitewise detection of positive selection. Mol Biol Evol 29:1125–1139. doi: 10.1093/molbev/msr272 [DOI] [PubMed] [Google Scholar]
- Jordan JJ, Chhim S, Margulies CM, Allocca M, Bronson RT, Klungland A, Samson LD, Fu D. 2017. ALKBH7 drives a tissue and sex-specific necrotic cell death response following alkylation-induced damage. Cell Death Dis 8:e2947–e2947. doi: 10.1038/cddis.2017.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. 2006. Endonucleolytic Function of MutLα in Human Mismatch Repair. Cell 126:297–308. doi: 10.1016/j.cell.2006.05.039 [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, LS Jermiin. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansikas M, Kariola R, Nyström M. 2011. Verification of the three-step model in assessing the pathogenicity of mismatch repair gene variants. Hum Mutat 32:107–115. doi: 10.1002/humu.21409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor-Vazirani P, Rath SK, Liu X, Shu Z, Bowen NE, Chen Y, Haji-Seyed-Javadi R, Daddacha W, Minten EV, Danelia D, Farchi D, Duong DM, Seyfried NT, Deng X, Ortlund EA, Kim B, Yu DS. 2022. SAMHD1 deacetylation by SIRT1 promotes DNA end resection by facilitating DNA binding at double-strand breaks. Nat Commun 13:6707. doi: 10.1038/s41467-022-34578-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Oka K, Semba T, Takamori M, Sugiura Y, Yamasaki R, Suzuki Y, Chujo T, Nagase M, Oiwa Y, Fujioka S, Homma S, Yamamura Y, Miyawaki S, Narita M, Fukuda T, Sakai Y, Ishimoto T, Tomizawa K, Suematsu M, Yamamoto T, Bono H, Okano H, Miura K. 2023. Cellular senescence induction leads to progressive cell death via the INK4a-RB pathway in naked mole-rats. EMBO J 42:e111133. doi: 10.15252/embj.2022111133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijzers G, Bakula D, Petr MA, Madsen NGK, Teklu A, Mkrtchyan G, Osborne B, Scheibye-Knudsen M. 2018. Human Exonuclease 1 (EXO1) Regulatory Functions in DNA Replication with Putative Roles in Cancer. Int J Mol Sci 20:74. doi: 10.3390/ijms20010074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—The BLAST-Like Alignment Tool. Genome Res 12:656–664. doi: 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Research 44:W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni CA, Nadtochiy SM, Kennedy L, Zhang J, Chhim S, Alwaseem H, Murphy E, Fu D, Brookes PS. 2020. ALKBH7 mediates necrosis via rewiring of glyoxal metabolism. eLife 9:e58573. doi: 10.7554/elife.58573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong LN, Dove WF. 2009. APC Proteins. Adv Exp Med Biol 656:85–106. doi: 10.1007/978-1-4419-1145-2_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy P. 2006. Mechanisms of Degranulation in Neutrophils. Allergy, Asthma Clin Immunol 2:98. doi: 10.1186/1710-1492-2-3-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levantini E, Maroni G, Re MD, Tenen DG. 2022. EGFR signaling pathway as therapeutic target in human cancers. Semin Cancer Biol 85:253–275. doi: 10.1016/j.semcancer.2022.04.002 [DOI] [PubMed] [Google Scholar]
- Li X, Wang P, Pan Q, Liu G, Liu W, Omotoso O, Du J, Li Z, Yu Y, Huang Y, Zhu P, Li M, Zhou X. 2023. Chromosome-level Asian elephant genome assembly and comparative genomics of long-lived mammals reveal the common substitutions for cancer resistance. Aging Cell 22:e13917. doi: 10.1111/acel.13917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. 2019. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 47:W199–W205. doi: 10.1093/nar/gkz401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Guntuku S, Cui X-S, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, Donehower LA, Elledge SJ. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev 14:1448–1459. doi: 10.1101/gad.14.12.1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liu X, Gao J, Shi X, Hu X, Wang S, Luo Y. 2013. Overexpression of DOC-1R Inhibits Cell Cycle G1/S Transition by Repressing CDK2 Expression and Activation. Int J Biol Sci 9:541–549. doi: 10.7150/ijbs.5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llambi F, Wang Y-M, Victor B, Yang M, Schneider DM, Gingras S, Parsons MJ, Zheng JH, Brown SA, Pelletier S, Moldoveanu T, Chen T, Green DR. 2016. BOK Is a Non-canonical BCL-2 Family Effector of Apoptosis Regulated by ER-Associated Degradation. Cell 165:421–433. doi: 10.1016/j.cell.2016.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo TL, Fong CW, Yusoff P, Mckie AB, Chua M-S, Leung HY, Guy GR. 2006. Sprouty and cancer: The first terms report. Cancer Lett 242:141–150. doi: 10.1016/j.canlet.2005.12.032 [DOI] [PubMed] [Google Scholar]
- Lucaci AG, Wisotsky SR, Shank SD, Weaver S, Pond SLK. 2021. Extra base hits: Widespread empirical support for instantaneous multiple-nucleotide changes. Plos One 16:e0248337. doi: 10.1371/journal.pone.0248337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucaci AG, Zehr JD, Enard D, Thornton JW, Pond SLK. 2023a. Evolutionary Shortcuts via Multinucleotide Substitutions and Their Impact on Natural Selection Analyses. Mol Biol Evol 40:msad150. doi: 10.1093/molbev/msad150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucaci AG, Zehr JD, Enard D, Thornton JW, Pond SLK. 2023b. Evolutionary shortcuts via multi-nucleotide substitutions and their impact on natural selection analyses. Biorxiv 2022.12.02.518889. doi: 10.1101/2022.12.02.518889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ, Wagner GP. 2008. Resurrecting the role of transcription factor change in developmental evolution. Evolution; international journal of organic evolution 62:2131–2154. [DOI] [PubMed] [Google Scholar]
- Markova-Raina P, Petrov D. 2011. High sensitivity to aligner and high rate of false positives in the estimates of positive selection in the 12 Drosophila genomes. Genome Res 21:863–874. doi: 10.1101/gr.115949.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews HK, Bertoli C, Bruin RAM de. 2022. Cell cycle control in cancer. Nat Rev Mol Cell Biol 23:74–88. doi: 10.1038/s41580-021-00404-3 [DOI] [PubMed] [Google Scholar]
- McKie AB, Douglas DA, Olijslagers S, Graham J, Omar MM, Heer R, Gnanapragasam VJ, Robson CN, Leung HY. 2005. Epigenetic inactivation of the human sprouty2 (hSPRY2) homologue in prostate cancer. Oncogene 24:2166–2174. doi: 10.1038/sj.onc.1208371 [DOI] [PubMed] [Google Scholar]
- Messier W, Stewart CB. 1997. Episodic adaptive evolution of primate lysozymes. Nature [DOI] [PubMed] [Google Scholar]
- Metzler KD, Fuchs TA, Nauseef WM, Reumaux D, Roesler J, Schulze I, Wahn V, Papayannopoulos V, Zychlinsky A. 2011. Myeloperoxidase is required for neutrophil extracellular trap formation: implications for innate immunity. Blood 117:953–959. doi: 10.1182/blood-2010-06-290171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler KD, Goosmann C, Lubojemska A, Zychlinsky A, Papayannopoulos V. 2014. A Myeloperoxidase-Containing Complex Regulates Neutrophil Elastase Release and Actin Dynamics during NETosis. Cell Rep 8:883–896. doi: 10.1016/j.celrep.2014.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD. 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 41:D377–D386. doi: 10.1093/nar/gks1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Thomas P. 2009. Protein Networks and Pathway Analysis. Methods Mol Biol 563:123–140. doi: 10.1007/978-1-60761-175-2_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von HA. 2020. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol 37:1530–1534. doi: 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinedo F. 2019. Neutrophil Degranulation, Plasticity, and Cancer Metastasis. Trends Immunol 40:228–242. doi: 10.1016/j.it.2019.01.006 [DOI] [PubMed] [Google Scholar]
- Murrell B, Weaver S, Smith MD, Wertheim JO, Murrell S, Aylward A, Eren K, Pollner T, Martin DP, Smith DM, Scheffler K, Pond SLK. 2015a. Gene-Wide Identification of Episodic Selection. Mol Biol Evol 32:1365–1371. doi: 10.1093/molbev/msv035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Weaver S, Smith MD, Wertheim JO, Murrell S, Aylward A, Eren K, Pollner T, Martin DP, Smith DM, Scheffler K, Pond SLK. 2015b. Gene-wide identification of episodic selection. Molecular biology and evolution 32:1365–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Pond SLK. 2012. Detecting Individual Sites Subject to Episodic Diversifying Selection. Plos Genet 8:e1002764. doi: 10.1371/journal.pgen.1002764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse SV, Gaut BS. 1994. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol 11:715–724. doi: 10.1093/oxfordjournals.molbev.a040152 [DOI] [PubMed] [Google Scholar]
- Muzio M, Chinnaiyan AM, Kischkel FC, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, Mann M, Krammer PH, Peter ME, Dixit VM. 1996. FLICE, A Novel FADD-Homologous ICE/CED-3–like Protease, Is Recruited to the CD95 (Fas/APO-1) Death-Inducing Signaling Complex. Cell 85:817–827. doi: 10.1016/s0092-8674(00)81266-0 [DOI] [PubMed] [Google Scholar]
- Ngatia JN, Lan TM, Dinh TD, Zhang L, Ahmed AK, Xu YC. 2019. Signals of positive selection in mitochondrial protein-coding genes of woolly mammoth: Adaptation to extreme environments? Ecol Evol 9:6821–6832. doi: 10.1002/ece3.5250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, Haeseler A von, Minh BQ. 2015. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. 1998. Likelihood Models for Detecting Positively Selected Amino Acid Sites and Applications to the HIV-1 Envelope Gene. Genetics 148:929–936. doi: 10.1093/genetics/148.3.929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanno N, Luca AD, Bianco C, Strizzi L, Mancino M, Maiello MR, Carotenuto A, Feo GD, Caponigro F, Salomon DS. 2006. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 366:2–16. doi: 10.1016/j.gene.2005.10.018 [DOI] [PubMed] [Google Scholar]
- Nozawa M, Suzuki Y, Nei M. 2009. Reliabilities of identifying positive selection by the branch-site and the site-prediction methods. Proc Natl Acad Sci 106:6700–6705. doi: 10.1073/pnas.0901855106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary MA, Bloch JI, Flynn JJ, Gaudin TJ, Giallombardo A, Giannini NP. 2013a. Response to comment on “The placental mammal ancestor and the post-K-Pg radiation of placentals”Science. New York, NY. p. 613. doi: 10.1126/science.1238162 [DOI] [PubMed] [Google Scholar]
- O’Leary MA, Bloch JI, Flynn JJ, Gaudin TJ, Giallombardo A, Giannini NP. 2013b. The placental mammal ancestor and the post-K-Pg radiation of placentals. Science 339:662–7. doi: 10.1126/science.1229237 [DOI] [PubMed] [Google Scholar]
- Özören N, Inohara N, Núñez G. 2009. A putative role for human BFK in DNA damage-induced apoptosis. Biotechnol J 4:1046–1054. doi: 10.1002/biot.200900091 [DOI] [PubMed] [Google Scholar]
- Palles C, Cazier J-B, Howarth KM, Domingo E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Salguero I, Sherborne A, Chubb D, Carvajal-Carmona LG, Ma Y, Kaur K, Dobbins S, Barclay E, Gorman M, Martin L, Kovac MB, Humphray S, Thomas HJW, Maher E, Evans G, Lucassen A, Cummings C, Stevens M, Walker L, Halliday D, Armstrong R, Paterson J, Hodgson S, Homfray T, Side L, Izatt L, Donaldson A, Tomkins S, Morrison P, Goodman S, Brewer C, Henderson A, Davidson R, Murday V, Cook J, Haites N, Bishop T, Sheridan E, Green A, Marks C, Carpenter S, Broughton M, Greenhalge L, Suri M, Donnelly P, Bell J, Bentley D, McVean G, Ratcliffe P, Taylor J, Wilkie A, Donnelly P, Broxholme J, Buck D, Cazier J-B, Cornall R, Gregory L, Knight J, Lunter G, McVean G, Taylor J, Tomlinson I, Wilkie A, Buck D, Gregory L, Humphray S, Kingsbury Z, McVean G, Donnelly P, Cazier J-B, Broxholme J, Grocock R, Hatton E, Holmes CC, Hughes L, Humburg P, Kanapin A, Lunter G, Murray L, Rimmer A, Lucassen A, Holmes CC, Bentley D, Donnelly P, Taylor J, Petridis C, Roylance R, Sawyer EJ, Kerr DJ, Clark S, Grimes J, Kearsey SE, Thomas HJW, McVean G, Houlston RS, Tomlinson I. 2013. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 45:136–144. doi: 10.1038/ng.2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. 2010. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol 191:677–691. doi: 10.1083/jcb.201006052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R, Gao M, Ahmad I, Fleming J, Singh LB, Rai TS, McKie AB, Seywright M, Barnetson RJ, Edwards J, Sansom OJ, Leung HY. 2013. Sprouty2, PTEN, and PP2A interact to regulate prostate cancer progression. J Clin Investig 123:1157–1175. doi: 10.1172/jci63672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil M, Pabla N, Dong Z. 2013. Checkpoint kinase 1 in DNA damage response and cell cycle regulation. Cell Mol Life Sci 70:4009–4021. doi: 10.1007/s00018-013-1307-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peto R. 2015. Quantitative implications of the approximate irrelevance of mammalian body size and lifespan to lifelong cancer risk. Philosophical transactions of the Royal Society of London Series B, Biological sciences 370:20150198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peto R. 1975. Cancer and ageing in mice and men. British Journal of Cancer 32:411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Casane D, Gribaldo, Lopez P, Meunier J. 2003. Heterotachy and Functional Shift in Protein Evolution. IUBMB Life 55:257–265. doi: 10.1080/1521654031000123330 [DOI] [PubMed] [Google Scholar]
- Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, Furlong LI. 2020. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res 48:D845–D855. doi: 10.1093/nar/gkz1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli V, Zanoni I. 2023. Neutrophil intrinsic and extrinsic regulation of NETosis in health and disease. Trends Microbiol 31:280–293. doi: 10.1016/j.tim.2022.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond S, Frost S. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Molecular biology and evolution. [DOI] [PubMed] [Google Scholar]
- Pond SK, Muse SV. 2005. Site-to-Site Variation of Synonymous Substitution Rates. Mol Biol Evol 22:2375–2385. doi: 10.1093/molbev/msi232 [DOI] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. [DOI] [PubMed] [Google Scholar]
- Pond SLK, Murrell B, Fourment M, Frost SDW, Delport W, Scheffler K. 2011a. A random effects branch-site model for detecting episodic diversifying selection. Molecular biology and evolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Murrell B, Fourment M, Frost SDW, Delport W, Scheffler K. 2011b. A random effects branch-site model for detecting episodic diversifying selection. Mol Biol Evol. doi: 10.1093/molbev/msr125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Wisotsky SR, Escalante A, Magalis BR, Weaver S. 2020. Contrast-FEL – a test for differences in selective pressures at individual sites among clades and sets of branches. Mol Biol Evol 38:msaa263-. doi: 10.1093/molbev/msaa263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promislow DEL. 1994. DNA Repair and the Evolution of Longevity: A Critical Analysis. J Theor Biol 170:291–300. doi: 10.1006/jtbi.1994.1190 [DOI] [PubMed] [Google Scholar]
- Purvis A, Orme CDL. 2005. Deciphering Growth 1–18. doi: 10.1007/3-540-28902-x_1 [DOI] [Google Scholar]
- Puttick MN, Thomas GH. 2015. Fossils and living taxa agree on patterns of body mass evolution: a case study with Afrotheria. Proceedings Biological sciences / The Royal Society 282:20152023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Pond SLK, Webb A, Hey J. 2021. Weak selection on synonymous codons substantially inflates dN/dS estimates in bacteria. Proc Natl Acad Sci 118:e2023575118. doi: 10.1073/pnas.2023575118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranwez V, Douzery EJP, Cambon C, Chantret N, Delsuc F. 2018. MACSE v2: Toolkit for the Alignment of Coding Sequences Accounting for Frameshifts and Stop Codons. Mol Biol Evol 35:2582–2584. doi: 10.1093/molbev/msy159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviv L, Jacobson SL, Plotnik JM, Bowman J, Lynch V, Benítez-Burraco A. 2023. Elephants as an animal model for self-domestication. Proc Natl Acad Sci 120:e2208607120. doi: 10.1073/pnas.2208607120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner E, Gool IC van, Palles C, Kearsey SE, Bosse T, Tomlinson I, Church DN. 2016. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer 16:71–81. doi: 10.1038/nrc.2015.12 [DOI] [PubMed] [Google Scholar]
- Remijsen Q, Kuijpers TW, Wirawan E, Lippens S, Vandenabeele P, Berghe TV. 2011. Dying for a cause: NETosis, mechanisms behind an antimicrobial cell death modality. Cell Death Differ 18:581–588. doi: 10.1038/cdd.2011.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Malaspinas A-S, Pollack JL, Slatkin M, Matheus P, Hofreiter M. 2007. Proboscidean mitogenomics: chronology and mode of elephant evolution using mastodon as outgroup. PLoS Biology 5:e207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacho EJ, Kadyrov FA, Modrich P, Kunkel TA, Erie DA. 2008. Direct Visualization of Asymmetric Adenine Nucleotide-Induced Conformational Changes in MutLα. Mol Cell 29:112–121. doi: 10.1016/j.molcel.2007.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini M, Verma A, Mathew SJ. 2018. SPRY2 is a novel MET interactor that regulates metastatic potential and differentiation in rhabdomyosarcoma. Cell Death Dis 9:237. doi: 10.1038/s41419-018-0261-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage VM, Allen AP, Brown JH, Gillooly JF, Herman AB, Woodruff WH. 2007. Scaling of number, size, and metabolic rate of cells with body size in mammals. Proc National Acad Sci 104:4718–4723. doi: 10.1073/pnas.0611235104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Durgan J, Magalhaes A, Hall A. 2007. Rho GTPases regulate PRK2/PKN2 to control entry into mitosis and exit from cytokinesis. EMBO J 26:1624–1636. doi: 10.1038/sj.emboj.7601637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman BT, Huang DW, Tan Q, Guo Y, Bour S, Liu D, Stephens R, Baseler MW, Lane HC, Lempicki RA. 2007. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinform 8:426. doi: 10.1186/1471-2105-8-426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund S, Avanzato D, Lanzetti L. 2018. Emerging functions of the EGFR in cancer. Mol Oncol 12:3–20. doi: 10.1002/1878-0261.12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slodkowicz G, Goldman N. 2020. Integrated structural and evolutionary analysis reveals common mechanisms underlying adaptive evolution in mammals. Proc Natl Acad Sci 117:5977–5986. doi: 10.1073/pnas.1916786117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Pond SLK. 2015. Less Is More: An Adaptive Branch-Site Random Effects Model for Efficient Detection of Episodic Diversifying Selection. Mol Biol Evol 32:1342–1353. doi: 10.1093/molbev/msv022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WL, Tell LA, Kabbur MB, Gage L, Cullor JS. 1998. A rapid isolation of Asian elephant (Elephas maximus) blood heterophils on Percoll density gradients. Comp Haematol Int 8:37–42. doi: 10.1007/bf02628103 [DOI] [Google Scholar]
- Spielman SJ, Dawson ET, Wilke CO. 2014. Limited Utility of Residue Masking for Positive-Selection Inference. Mol Biol Evol 31:2496–2500. doi: 10.1093/molbev/msu183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer MS, Meredith RW, Teeling EC, Murphy WJ. 2013. Technical comment on “The placental mammal ancestor and the post-K-Pg radiation of placentals”. Science 341:613. [DOI] [PubMed] [Google Scholar]
- Sulak M, Fong L, Mika K, Chigurupati S, Yon L, Mongan NP, Emes RD, Lynch VJ. 2016a. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulak M, Fong L, Mika K, Chigurupati S, Yon L, Mongan NP, Emes RD, Lynch VJ. 2016b. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD. 2005. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci 102:13182–13187. doi: 10.1073/pnas.0504211102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y. 2008. False-positive results obtained from the branch-site test of positive selection. Genes Genet Syst 83:331–338. doi: 10.1266/ggs.83.331 [DOI] [PubMed] [Google Scholar]
- Synnes M, Nilssen EA, Boye E, Grallert B. 2002. A novel chk1-dependent G1/M checkpoint in fission yeast. J Cell Sci 115:3609–3618. doi: 10.1242/jcs.00004 [DOI] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. 2010. Integrative Genomic Profiling of Human Prostate Cancer. Cancer Cell 18:11–22. doi: 10.1016/j.ccr.2010.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tell L, Kabbur MB, Smith WL, Gage L, Cullor JS. 1999. Oxygen radical production by Asian elephant (Elephas maximus) heterophils and Holstein cattle (Bos taurus) neutrophils. J Zoo Wildl Medicine Official Publ Am Assoc Zoo Vet 30:402–7. [PubMed] [Google Scholar]
- Tennessen JA. 2008. Positive selection drives a correlation between non-synonymous/synonymous divergence and functional divergence. Bioinformatics 24:1421–1425. doi: 10.1093/bioinformatics/btn205 [DOI] [PubMed] [Google Scholar]
- Thomas PD, Ebert D, Muruganujan A, Mushayahama T, Albou L, Mi H. 2022. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci 31:8–22. doi: 10.1002/pro.4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Ferris E, Campbell MS, Harris VK, Rupp SM, Harrison TM, Kiso WK, Schmitt DL, Garner MM, Aktipis CA, Maley CC, Boddy AM, Yandell M, Gregg C, Schiffman JD, Abegglen LM. 2021. Elephant Genomes Reveal Accelerated Evolution in Mechanisms Underlying Disease Defenses. Mol Biol Evol 38:msab127-. doi: 10.1093/molbev/msab127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Schneider-Utaka AK, Maley CC. 2020. The Evolution of Human Cancer Gene Duplications across Mammals. Mol Biol Evol msaa125-. doi: 10.1093/molbev/msaa125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomimatsu N, Mukherjee B, Harris JL, Boffo FL, Hardebeck MC, Potts PR, Khanna KK, Burma S. 2017. DNA-damage-induced degradation of EXO1 exonuclease limits DNA end resection to ensure accurate DNA repair. J Biol Chem 292:10779–10790. doi: 10.1074/jbc.m116.772475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez JM, Lynch VJ. 2021. Pervasive duplication of tumor suppressors in Afrotherians during the evolution of large bodies and reduced cancer risk. Elife 10:e65041. doi: 10.7554/elife.65041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez JM, Sulak M, Chigurupati S, Lynch VJ. 2018. A Zombie LIF Gene in Elephants Is Upregulated by TP53 to Induce Apoptosis in Response to DNA Damage. Cell reports 24:1765–1776. [DOI] [PubMed] [Google Scholar]
- Venkat A, Hahn MW, Thornton JW. 2018. Multinucleotide mutations cause false inferences of lineage-specific positive selection. Nat Ecol Evol 2:1280–1288. doi: 10.1038/s41559-018-0584-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgilio MD, Callen E, Yamane A, Zhang W, Jankovic M, Gitlin AD, Feldhahn N, Resch W, Oliveira TY, Chait BT, Nussenzweig A, Casellas R, Robbiani DF, Nussenzweig MC. 2013. Rif1 Prevents Resection of DNA Breaks and Promotes Immunoglobulin Class Switching. Science 339:711–715. doi: 10.1126/science.1230624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjeva NV, Chernyak BV. 2020. NETosis: Molecular Mechanisms, Role in Physiology and Pathology. Biochem (Mosc) 85:1178–1190. doi: 10.1134/s0006297920100065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorp B, Mokry M, Koerkamp MJAG, Holstege FCP, Cuppen E, Bruin A de. 2012. E2F7 represses a network of oscillating cell cycle genes to control S-phase progression. Nucleic Acids Res 40:3511–3523. doi: 10.1093/nar/gkr1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisotsky SR, Pond SLK, Shank SD, Muse SV. 2020. Synonymous site-to-site substitution rate variation dramatically inflates false positive rates of selection analyses: ignore at your own peril. Mol Biol Evol 37:2430–2439. doi: 10.1093/molbev/msaa037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarchak N, Xing Y. 2016. PP2A as a master regulator of the cell cycle. Crit Rev Biochem Mol Biol 51:162–184. doi: 10.3109/10409238.2016.1143913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Wang C, Zhou M, Bao L, Yanan Wang, A Kumar, C Xing, W Luo, Yingfei Wang. 2022. KDM6B promotes PARthanatos via suppression of O6-methylguanine DNA methyltransferase repair and sustained checkpoint response. Nucleic Acids Res 50:gkac471-. doi: 10.1093/nar/gkac471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol 24:1586–1591. doi: 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yang Z. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957 [DOI] [PubMed] [Google Scholar]
- Yang Z. 1997a. PAML: a program package for phylogenetic analysis by maximum likelihood. Bioinformatics 13:555–556. doi: 10.1093/bioinformatics/13.5.555 [DOI] [PubMed] [Google Scholar]
- Yang Z. 1997b. PAML: a program package for phylogenetic analysis by maximum likelihood. CABIOS 13:555–556. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. 2002. Codon-Substitution Models for Detecting Molecular Adaptation at Individual Sites Along Specific Lineages. Mol Biol Evol 19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148 [DOI] [PubMed] [Google Scholar]
- Yang Z, dos Reis M. 2011. Statistical Properties of the Branch-Site Test of Positive Selection. Mol Biol Evol 28:1217–1228. doi: 10.1093/molbev/msq303 [DOI] [PubMed] [Google Scholar]
- Yao J, Duan L, Fan M, Yuan J, Wu X. 2007. Overexpression of BLCAP induces S phase arrest and apoptosis independent of p53 and NF-κB in human tongue carcinoma. Mol Cell Biochem 297:81–92. doi: 10.1007/s11010-006-9332-2 [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Tada T, Zhang H, Britt L. 2008. Elucidation of phenotypic adaptations: Molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci 105:13480–13485. doi: 10.1073/pnas.0802426105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- M ZHAO, L ZHANG, X QIU, F ZENG, W CHEN, Y AN, B HU, XUFENG WU, XINXING WU. 2016. BLCAP arrests G1/S checkpoint and induces apoptosis through downregulation of pRb1 in HeLa cells. Oncol Rep 35:3050–3058. doi: 10.3892/or.2016.4686 [DOI] [PubMed] [Google Scholar]
- Zhou E, Wang L, Santiago CN, Nanavati J, Rifkin S, Spence E, Hylind LM, Gills JJ, Luna LL, Kafonek DR, Cromwell DM, Drewes JL, Sears CL, Giardiello FM, Mullin GE, Consortium the BS. 2022. Adult-Attained Height and Colorectal Cancer Risk: A Cohort Study, Systematic Review, and Meta-AnalysisAdult-Attained Height and Colorectal Cancer Risk. Cancer Epidemiology Biomarkers Prev 31:783–792. doi: 10.1158/1055-9965.epi-21-0398 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1 – source data 1. Genomes used in selection tests.
Figure 1 – source data 2. List of positively selected genes from ABSREL.
Figure 1 – source data 3. List of rapidly evolving genes from ABSREL.
Figure 1 – source data 4. List of positively selected genes from BUSTED.
Figure 1 – source data 5. List of rapidly evolving genes from BUSTED.
Figure 1 – figure supplement 1. Genomic features of positively selected and rapidly evolving genes.
Figure 3 – source data 1. Webgestalt output files for positively selected genes.
Figure 3 – source data 2. Webgestalt output files for ABSREL rapidly evolving genes.
Figure 3 – source data 3. Webgestalt output files for BUSTED rapidly evolving genes.
Figure 4 – source data 1. Webgestalt output files for positively selected genes.
Figure 4 – source data 2. Webgestalt output files for ABSREL rapidly evolving genes.
Figure 4 – source data 3. Webgestalt output files for BUSTED rapidly evolving genes.
Data Availability Statement
HyPhy json out files from ABSREL, BUSTED, and FEL (http://doi.org/10.5061/dryad.h44j0zpt4), alignments (http://doi.org/10.5061/dryad.5dv41nsbc), and the phylogenetic tree used for selection tests (http://doi.org/10.5061/dryad.j0zpc86n3) are available from Dryad. HyPhy json output files can be visualized with HyPhy Vision in any web browser: http://vision.hyphy.org and https://observablehq.com/@spond/busted-compare.