Abstract
Activation of c-Raf-1 (referred to as Raf) by Ras is a pivotal step in mitogenic signaling. Raf activation is initiated by binding of Ras to the regulatory N terminus of Raf. While Ras binding to residues 51 to 131 is well understood, the role of the RafC1 cysteine-rich domain comprising residues 139 to 184 has remained elusive. To resolve the function of the RafC1 domain, we have performed an exhaustive surface scanning mutagenesis. In our study, we defined a high-resolution map of multiple distinct functional epitopes within RafC1 that are required for both negative control of the kinase and the positive function of the protein. Activating mutations in three different epitopes enhanced Ras-dependent Raf activation, while only some of these mutations markedly increased Raf basal activity. One contiguous inhibitory epitope consisting of S177, T182, and M183 clearly contributed to Ras-Raf binding energy and represents the putative Ras binding site of the RafC1 domain. The effects of all RafC1 mutations on Ras binding and Raf activation were independent of Ras lipid modification. The inhibitory mutation L160A is localized to a position analogous to the phorbol ester binding site in the protein kinase C C1 domain, suggesting a function in cofactor binding. Complete inhibition of Ras-dependent Raf activation was achieved by combining mutations K144A and L160A, which clearly demonstrates an absolute requirement for correct RafC1 function in Ras-dependent Raf activation.
c-Raf-1 (herein referred to as Raf) is a member of a serine/threonine protein kinase family implicated in the transduction of signals from the cell surface to the nucleus which occurs via activation of a mitogen-activated protein kinase module by a GTPase switch (3, 14, 49). Raf provides an immediate downstream target for Ras and is a pivotal regulator of cell proliferation and differentiation (37, 38, 47, 62–64, 70). Signaling from Ras to Raf is initiated by binding of activated Ras to the Ras binding domain of Raf (RafRBD). Ras recruits Raf to the plasma membrane, and the requirement for Ras in Raf activation can be overcome by fusion of a Ras membrane-targeting motif to the Raf C terminus (40, 60).
The raf oncogene was initially identified as the transforming part of murine sarcoma virus 3611. While the Raf protein kinase consists of an N-terminal noncatalytic region and a C-terminal kinase domain, the N-terminal part is missing in the v-Raf oncoprotein. This leads to a constitutive activity of the kinase domain, indicating that the N-terminal part locks the kinase in an inactive conformation (14, 49). The noncatalytic N terminus of Raf contains two regions that are highly conserved between different members of the Raf family. The first conserved region (CR1) consists of two structural modules that are referred to as RafRBD and a C1-type cysteine-rich domain (RafC1). RafRBD encompasses amino acids 51 to 131 and has the ubiquitin superfold (52, 53). RafC1 (amino acids 139 to 184) is a structural homologue of the protein kinase C (PKC) phorbol ester binding domain (26, 30, 50, 67, 69). RafRBD constitutes an autonomous structural domain sufficient for GTP-dependent binding of Ras (9, 16, 19, 25, 58). Functional analysis of the interaction between Ras and Raf demonstrated that the single Raf-R89L mutation is sufficient to abrogate Ras-dependent Raf activation completely and that the activation of Raf correlates quantitatively with the binding affinity between Ras and RafRBD (4, 17).
Whereas the role of RafRBD in Ras binding is understood in great detail, numerous reports have provided conflicting evidence with regard to the role of RafC1 in Ras-Raf interaction and Raf activation. Initial studies showed a decrease in the binding of Raf fragments to nonfarnesylated Ras in vitro when a zinc binding cysteine was mutated (C168S) (70) or when the RafC1 domain was depleted of the zinc ions that are structurally essential (64). The C168S exchange was also shown to inhibit Ras-dependent Raf activation in vivo, and this mutation was found to abolish the dominant negative effects exerted by noncatalytic Raf fragments (6). These results are in agreement with a report showing that Raf failed to bind to farnesylated Ras with the double mutation C165S C168S (43). In contrast to this, it was recently found that this double mutation had no effect on Ras-dependent membrane targeting of Raf (57). Deletion of the complete RafC1 domain did not reveal an essential role of RafC1 in Ras binding and Ras-dependent Raf activation (8, 56). Some reports have shown RafC1 or parts of this domain to be involved in binding to nonfarnesylated Ras (5, 9, 11, 15, 20), whereas others were unable to confirm these results (22, 27, 31). Upon binding to nonfarnesylated Ras, the RafC1 domain was found to interact preferentially with Ras-GTP with high affinity (5, 11). In contrast to these reports, the RafC1 domain has been reported to be involved in binding to farnesylated Ras exclusively (27, 28, 43). Binding of RafC1 to farnesylated Ras was found to be independent of GTP or GDP loading of Ras and was inhibited by the C168S mutation (27).
In Drosophila Raf (D-Raf), mutations of residues within the D-RafC1 domain (F290I and P308L) have been observed to rescue signaling by a Drosophila D-Raf mutant that is deficient in Ras binding (42). Mutations of the corresponding residues (F163I and P181L) in Raf were shown to induce constitutive activation, whereas Ras binding was inhibited (13). In addition, the RafC1 domain has been reported to play a role in binding to 14-3-3 proteins since either the double mutation C165S C168S or R143E K144E led to disruption of 14-3-3 binding and resulted in activation of the kinase (12, 45). In summary, many reports suggest a role of the RafC1 domain in Ras-dependent Raf activation. Yet, the contribution of RafC1 to Ras binding and to Raf activation has remained elusive.
Most of the previous studies have used mutations that alter structurally important residues within RafC1 and are likely to cause gross conformational changes. This approach precludes detailed analysis of the function of RafC1. Analysis of functional epitopes can be performed by mutational scanning analysis of surface-exposed residues (10, 36, 55, 65, 66). Based on nuclear magnetic resonance (NMR) analysis of the structure of RafC1 (50), we performed extensive surface scanning mutagenesis to investigate the function of RafC1 in Ras binding and Ras-dependent Raf activation. In our study, we defined a high-resolution map of multiple functional epitopes within RafC1 that are required for both negative control of the kinase and the positive function of the protein. The effect of each RafC1 mutation was independent of Ras lipid modification. We were able to assign different functions, such as Ras binding and cofactor interaction, to individual epitopes, which demonstrates an essential and highly complex role of the RafC1 domain in Ras-dependent Raf activation.
MATERIALS AND METHODS
Expression vectors and site-directed mutagenesis.
pSVK3-Ras(G12V) and pcDNA3-Raf plasmids were constructed as previously described (4). The additional plasmids used for the reporter gene assay were E743-tk80-luc and tk80-luc reporter constructs, β-galactosidase expression vector pEQ176 (34), ERK-1 (44), and pSG-ER81 (35). The pcDNA3-Raf(K375W) construct was generated by site-directed mutagenesis. Membrane-targeted Raf kinase was generated by fusion of the sequence coding for the K-Ras membrane-targeting region that consisted of the 17 C-terminal amino acids of K-Ras to the Raf C terminus (60).
E1(Q37I)-Ras(G12V/C181/C184/C186S) [termed QI-Ras(G12V)] was constructed by fusion of the 42-amino-acid transmembrane helix from the E1 glycoprotein to the Ras N terminus, which contains the Q37I mutation (61). A linker of 22 amino acids coding for GSS repeats was inserted between the E1(Q37I) transmembrane helix and the Ras N terminus (23). The QI-Ras(G12V) construct was cloned into either pSVK3 (Pharmacia) or pcDNA3 (Invitrogen). Lipid modification of QI-Ras(G12V) was prevented by Ras mutations C181S, C184S, and C186S [Ras(C181/C184/C186S)] (41).
The two-hybrid Ras constructs were generated by PCR amplification of Ras residues 1 to 166 or of full-length Ras by using the Ras(G12V) template (32). Ras constructs were cloned into the pPC97 DNA binding domain fusion vector (7). Wild-type Raf [Raf(wt)] and mutant forms thereof, that were generated in pcDNA3-Raf, were cloned into the pPC86 GAL4 activation domain fusion vector.
Site-directed mutagenesis was performed by two subsequent PCR amplifications (1). To facilitate PCR mutagenesis, XbaI and MfeI sites were introduced 5′ and 3′ of the RafC1 domain, respectively, into a Raf construct that does not contain an MfeI site in the catalytic domain. As a template, we used Raf in pcDNA3 (Invitrogen) for the first PCR step or Raf in pcDNA3 that was digested with XbaI for the second PCR step. In the first step, the megaprimer was generated by oligonucleotide priming within the RBD region of Raf and the corresponding mutagenesis primer. The megaprimer was used in a second PCR for which XbaI-digested pcDNA3-Raf was used as the template. Mutant RafC1 domains were cloned into the full-length pcDNA3-Raf(wt), -Raf(K375W), or -Raf(CAAX) construct after XbaI/MfeI digestion. All constructs were verified by dideoxy sequencing.
Reporter gene assay.
Rabbit kidney epithelium-like RK13 cells were grown to 25% confluency on 6-cm-diameter dishes and then transfected with a total of 10 μg of DNA by the calcium phosphate coprecipitation method. A 2-μg sample of a reporter construct (E743-tk80-luc or tk80-luc), 0.5 μg of a β-galactosidase expression vector (pEQ176), 1.5 μg of expression vector ERK-1, 1.5 μg of pSG-ER81, and 1.5 μg of expression plasmid pcDNA3 alone or expression plasmid pcDNA3 containing the respective Raf construct were used for each transfection. Where indicated, 80 ng of pSVK3-Ras(G12V) was additionally transfected. At 36 h after transfection, cells were harvested and lysed and luciferase and β-galactosidase activities were determined as previously described (4). Relative luciferase activity was obtained by normalizing luminescence to β-galactosidase activity.
Raf kinase assay.
RK13 cells were grown to 25% confluency on 10-cm-diameter dishes and then transfected with a total of 20 μg of DNA by the calcium phosphate coprecipitation method. A 10-μg sample of an empty Raf plasmid or the indicated Raf construct in pcDNA3 was used together with either 100 ng of pcDNA3-Ras(G12V) or 500 ng of the pcDNA3-QI-Ras(G12V) expression construct. At 36 h after transfection, cells were harvested, lysed in Nonidet P-40 (NP-40) lysis buffer (25 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10 mM Na-pyrophosphate, 25 mM Na-glycerophosphate, 2 mM EGTA, 2 mM EDTA, 10% glycerol, 0.5% NP-40), and the lysate was cleared by centrifugation at 12,000 × g for 30 min. For immunoprecipitation, 1 μg of anti-FLAG serum (Santa Cruz) was preabsorbed on protein A-agarose beads (Boehringer Mannheim) and mixed with the lysate for 2 h. Raf kinase assays were performed as previously described, by using recombinant kinase-dead mitogen-activated protein kinase kinase (MEK) as a substrate (18). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted on nitrocellulose. MEK phosphorylation was visualized by autoradiography and analyzed with a Phosphoimager (Fuji). After exposure, membranes were probed with a monoclonal anti-Raf-1 antibody (Transduction Laboratories) and developed by using enhanced chemiluminescence (Amersham).
Western blotting.
Rabbit kidney epithelium-like RK13 cells were grown to 25% confluency on 10-cm-diameter dishes and then transfected with a total of 20 μg of DNA by the calcium phosphate coprecipitation method and harvested 72 h after transfection. A 1.3-μg sample of a β-galactosidase expression vector (pEQ176) and 18.7 μg of expression plasmid pcDNA3 alone or expression plasmid pcDNA3 containing a respective FLAG-Raf construct were used for each transfection. After harvesting, the cells were sonicated and sample loading was normalized according to β-galactosidase activity. Equal proportions of lysate were then used for blotting. Samples were resolved by SDS–9% PAGE and transferred to polyvinylidene difluoride membranes. Western blots were probed with monoclonal M5 anti-FLAG antibody (Eastman Kodak Co.) and developed by using enhanced chemiluminescence.
Comparison of Ras(G12V) and QI-Ras(G12V) expression was performed by transient transfection by using 100 ng of pcDNA3-Ras(G12V) or 500 ng of QI-Ras(G12V), respectively, under conditions identical to those used for Raf kinase assays. After 36 h, cells were harvested and sonicated, and sample loading was normalized according to β-galactosidase activity. Samples were resolved by SDS–15% PAGE and transferred to polyvinylidene difluoride membranes. Western blots were probed with anti-Ras monoclonal antibody Y13-259 (Santa Cruz) and developed by using enhanced chemiluminescence.
For control of expression of pPC86-Raf constructs in the two-hybrid system, yeast cells were grown in selective medium as already described, and equivalent amounts of cells, as determined by optical density at 600 nm (OD600), were lysed in yeast lysis buffer containing 50 mM phosphate (pH 7.4), 1 mM EDTA, 5% glycerol, 1% SDS, and 1 mM phenylmethylsulfonyl fluoride using a glass bead mill. Cell lysate was normalized according to protein concentration for SDS-PAGE. Samples were resolved by SDS–9% PAGE and transferred to polyvinylidene difluoride membranes. Western blots were probed with anti-Raf-C20 serum (Santa Cruz) and developed by using enhanced chemiluminescence.
Yeast two-hybrid assays.
The two-hybrid system employed in this study was developed by Chevray and Nathans (7) and modified here by using yeast strain Y190. For binding studies with Raf and Ras(G12V1-166), competent yeast cells prepared as described by Klebe et al. (37) were cotransformed with 1 μg of each of the two-hybrid vectors and grown on synthetic medium lacking leucine, tryptophan, and histidine and containing 25 mM 3-amino-1,2,3-triazole (Sigma) to monitor interaction between the fusion proteins. Glucose was used as the carbon source. β-Galactosidase activity was detected by filter lifting cells grown on selective medium and staining the yeast colonies by incubation in 0.75-mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) at 37°C for 1 h.
Quantitative two-hybrid assays were performed by using a modified protocol based on the procedure previously described (32). Original transformants were restreaked on selective plates. From these plates, colonies were inoculated in selective medium. The liquid cultures were incubated at 30°C until they reached an OD600 of 0.5 to 1.0. Lysis of yeast cells was performed as described by Bartel and Fields (2) by adding 50 μl of CHCl3 and 50 μl of 0.1% (wt/vol) SDS to 800 μl of resuspended cells. The β-galactosidase activity in this lysate was measured by using the Galacto-Star kit in accordance with the instructions of the manufacturer (Tropix). Relative β-galactosidase activity was obtained by normalizing luminescence to the OD600.
RESULTS
Distinct types of RafC1 surface mutations.
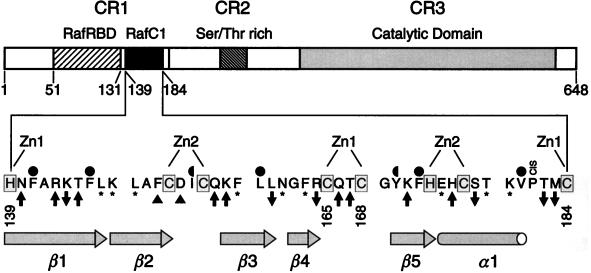
For epitope mapping mutagenesis, we changed 26 amino acids of RafC1 to alanine, since these residues have surface-exposed side chains, according to the structure of the RafC1 domain (50) (Fig. 1). Residues that contribute to the structural integrity of RafC1, such as histidine or cysteine zinc ligands or side chains that constitute the hydrophobic core, were left unchanged. Also, glycine, alanine, and proline residues were not altered. P181 was not mutated, since it was found to constitute a cis proline in the RafC1 structure determined by NMR analysis (50) and a cis proline mutation is likely to result in gross conformational changes due to main chain isomerization.
FIG. 1.
Schematic representation of Raf and the RafC1 domain. Amino acids that represent ligands involved in binding of the structural zinc ions are shaded. Zn1 and Zn2 are the zinc ions to which these ligands are bound. Hydrophobic residues that contribute to the structural core of the domain and which are completely or partially buried in the interior of the domain are indicated by full and half circles, respectively. Residues that did not affect Raf activity and Ras-dependent Raf activation upon mutation are marked with asterisks. Activating and inhibitory mutations are indicated by upward- and downward-pointing arrows, respectively. Mutations leading to a large increase in basal activity are marked by triangles.
To determine the effect of the RafC1 surface mutations on Ras-dependent Raf activation, we used a transient transfection assay with RK13 cells that measures transactivation induced by the Ras/Raf/MEK/ERK pathway (4). This assay utilizes a luciferase reporter gene driven by three E74 binding sites. The E74 binding site is a high-affinity site for Ets transcription factors (33), and Ras/Raf/MEK/ERK-dependent signaling in this assay has been shown to correlate quantitatively with Ras-Raf interaction affinity (4). In addition, we confirmed the results of the reporter gene assay by performing Raf kinase assays (18) with activating and inhibitory mutant forms of RafC1. Six amino acid exchanges decreased Ras/Raf-induced transactivation, whereas mutation of 11 residues to alanine positively affected Ras-dependent Raf activation (Fig. 1; for results, see Fig. 2 and 4). Two of the activating mutations led to a drastic increase in Raf basal activity. The exchange of nine residues had no effect on Raf activation by Ras. This demonstrates that different types of mutations can be distinguished upon surface scanning mutagenesis of the RafC1 domain. In addition, a significant number of surface mutations can be tolerated by the RafC1 domain without any effect on Ras-dependent Raf activation.
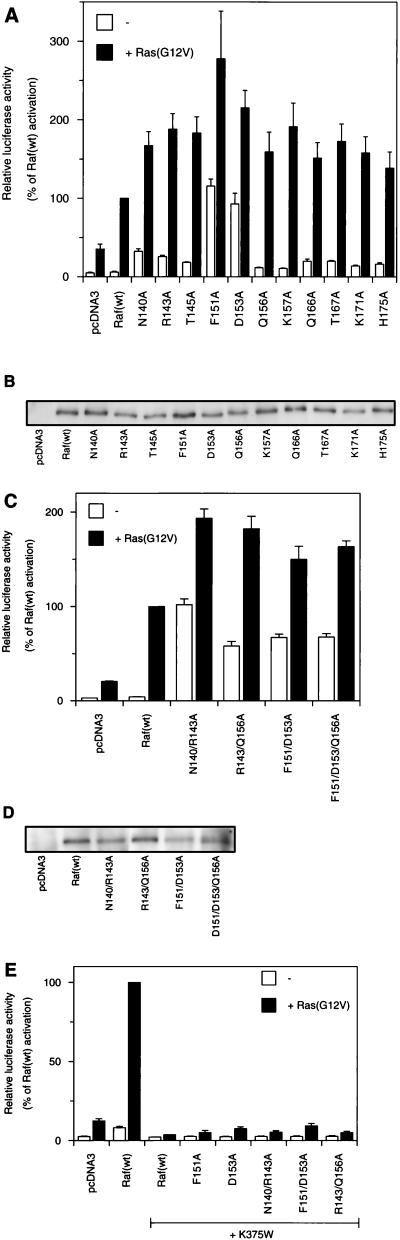
FIG. 2.
Activating effect of mutations in RafC1 on Ras/Raf/MEK/ERK-mediated transactivation in RK13 cells. (A) Transactivation mediated by Raf(wt) and single mutant RafC1 proteins was measured with a luciferase reporter construct driven by three E74 binding sites in transient transfection assays. Where indicated, the Ras(G12V) plasmid was cotransfected. (B) Control of the expression of Raf(wt) and mutant RafC1 proteins by fusion to an N-terminal FLAG epitope. RK13 cells were transfected with the pcDNA3-FLAG-Raf constructs indicated. After harvesting of cells, normalized amounts of cell lysate were resolved by SDS-PAGE and proteins were transferred to polyvinylidene difluoride membranes. Immunoblotting was performed by using a monoclonal anti-FLAG antibody and developed by using enhanced chemiluminescence. (C) Transactivation mediated by multiple RafC1 mutants. (D) Control of expression of multiple RafC1 mutants fused to an N-terminal epitope as described for panel B. (E) Transactivation using mutant RafC1 proteins in combination with the kinase-negative K375W mutation. The data shown are averages of three independent experiments.
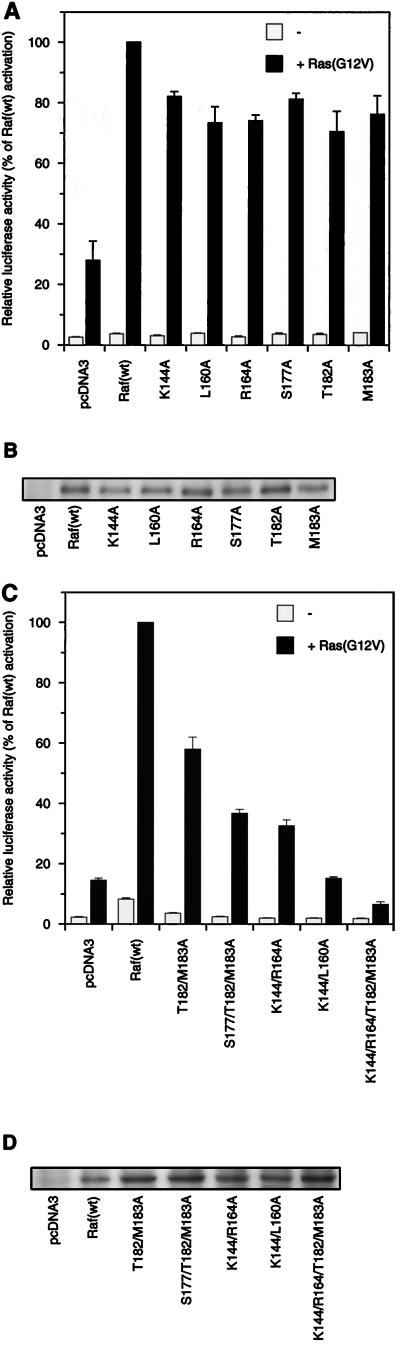
FIG. 4.
Inhibitory effect of RafC1 mutations on Raf-mediated transactivation. (A) Transactivation mediated by Raf(wt) and single mutant RafC1 proteins was measured with an E74 binding site-driven promoter as described in the legend to Fig. 2A. (B) Control of expression of RafC1 mutant proteins fused to an N-terminal epitope as described in the legend to Fig. 2B. (C) Transactivation mediated by multiple mutations of RafC1. The data shown are averages of three independent experiments. (D) Control of the expression of RafC1 multiple mutant proteins fused to an N-terminal FLAG epitope as described in the legend to Fig. 2B.
Activating RafC1 mutations are localized in three distinct epitopes.
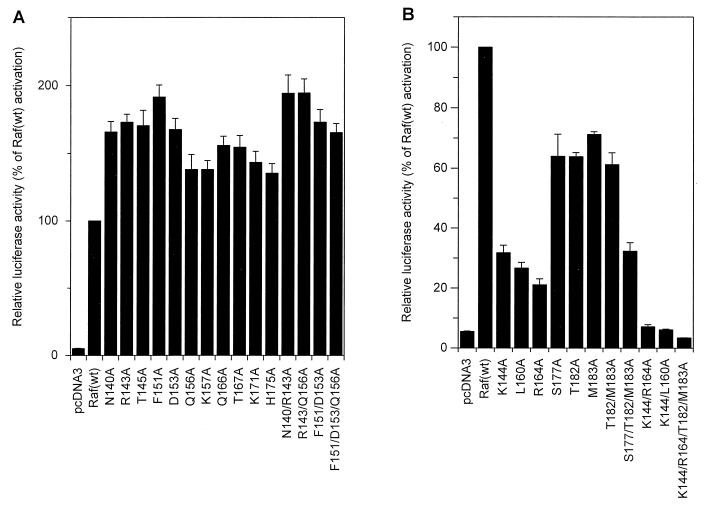
Different types of activating RafC1 mutations could be distinguished by using the E74-driven reporter gene assay. Exchange of T145, Q156, K157, Q166, T167, K171, or H175 for alanine enhanced transactivation by Raf basal activity 1.5- to 2.5-fold and increased Ras(G12V)-induced Raf activation accordingly, to 1.5- to 2-fold over the activity achieved by activation of Raf(wt) by Ras(G12V) (Fig. 2A). N140A and R143A had a slightly stronger effect on basal Raf-induced transactivation and led to a four- to fivefold increase in basal Raf activity. Exchange of F151 or D153 for alanine enhanced the basal Raf-induced transactivation 15- to 20-fold, which is comparable to the level of activation of Raf(wt) by Ras(G12V). Still, these mutant Raf proteins could be further activated by Ras(G12V) about two- to threefold compared to Ras activation of Raf(wt). To test whether the increase in Raf-induced transactivation due to these mutations was caused by enhanced Raf expression, we fused an N-terminal FLAG epitope to these constructs. Control of protein expression showed that mutant RafC1 did not affect protein expression significantly (Fig. 2B). Combination of different activating mutations that elicited only a slight increase in basal activity when exchanged individually, such as N140A R143A or R143A Q156A, resulted in significantly enhanced basal transactivation, whereas Ras(G12V)-dependent Raf activation was not altered compared to that achieved with the single mutant proteins (Fig. 2C). The F151A D153A double mutation also produced increased basal activity. Yet, with this double mutation, Ras(G12V)-induced activation was less efficient than with the single mutation F151A or D153A, which led to only a 1.5-fold increase compared with Raf(wt) activation. This shows that additional activating mutations can even reduce the effect of a single mutation on Ras-induced Raf activation. Adding one additional mutation (F151A D153A Q156A) did not further alter activation compared to that achieved with the double mutation. Multiple amino acid exchanges in the RafC1 domain also did not affect protein expression (Fig. 2D).
To test whether the transactivation induced by RafC1 mutations was due to genuine Raf kinase activity, we combined activating RafC1 mutations with the K375W mutation located in the kinase domain. Raf(K375W) abolishes kinase activity and acts as a dominant negative mutant protein in Ras-induced signaling (6, 24, 39). No kinase activity was observed when RafC1 mutations were combined with the kinase-negative K375W mutation (Fig. 2E). This clearly shows that the enhanced transactivation induced by mutant RafC1 required Raf kinase activity. In addition, the RafC1 mutations, in combination with K375W, appeared to reduce transactivation induced by Ras(G12V) alone (compare to pcDNA3 in Fig. 2E), possibly by sequestration of Ras(G12V) by our inactive mutant Raf kinases. The activation observed in these experiments was specific to the E74 binding site, since no effects were detected with a luciferase reporter gene lacking E74 binding sites (data not shown).
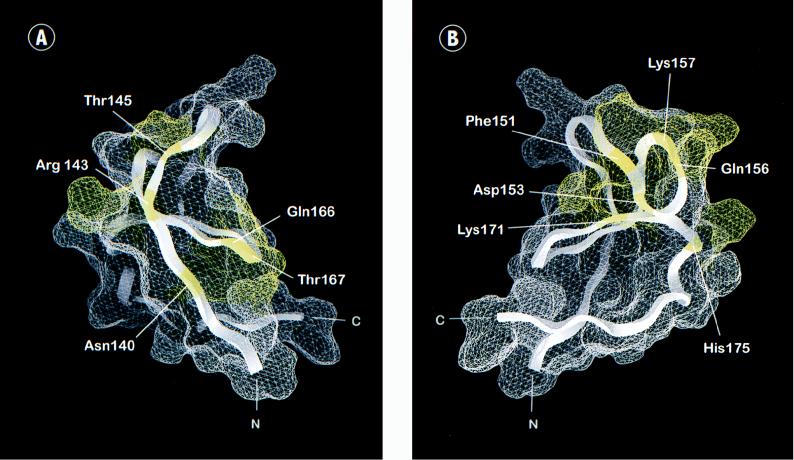
Activating RafC1 mutations can be assigned to three different epitopes. One epitope consists of residues N140, Q166, and T167, which are all immediately adjacent to zinc ligands of the first zinc binding site (Fig. 1 and 3A). A small epitope is located on strand β1 and consists of R143 and T145. A large epitope comprises residues F151, D153, Q156, K157, K171, and H175 from strands β2, β3, and β5 and helix α1, which form a mostly contiguous surface (Fig. 1 and 3B). In summary, RafC1 plays an important role in the negative control of kinase activity by the regulatory N terminus, since mutations within these functional epitopes loosen the control over Raf basal activity and facilitate Ras-induced Raf activation.
FIG. 3.
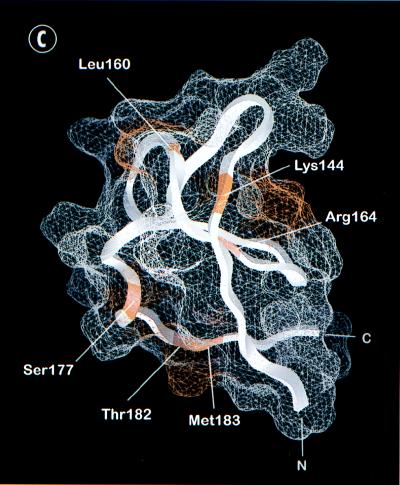
Functional epitopes of the RafC1 domain. The surface was calculated by using GRASP (54) and the RafC1 structure determined by NMR analysis (50). (A) View of the N-terminal part of RafC1 displaying one activating epitope formed by residues that are all immediately adjacent to ligands of the first zinc binding site (N140, Q166, and T167) and one epitope localized on strand β1 (R143 and T145) (in yellow). (B) View of an activating epitope formed by residues localized on strands β2, β3, and β5 and helix α1 (F151, D153, Q156, K157, K171, and H175). (C) Distinct inhibitory epitopes formed by residues located either at the C terminus of RafC1 (S177, T182, and M183), on strands β1 and β4 (K144 and R164), or at a position (L160) analogous to that of the phorbol ester binding site of the PKC C1 domain (in red).
Cooperative effects of inhibitory RafC1 mutations.
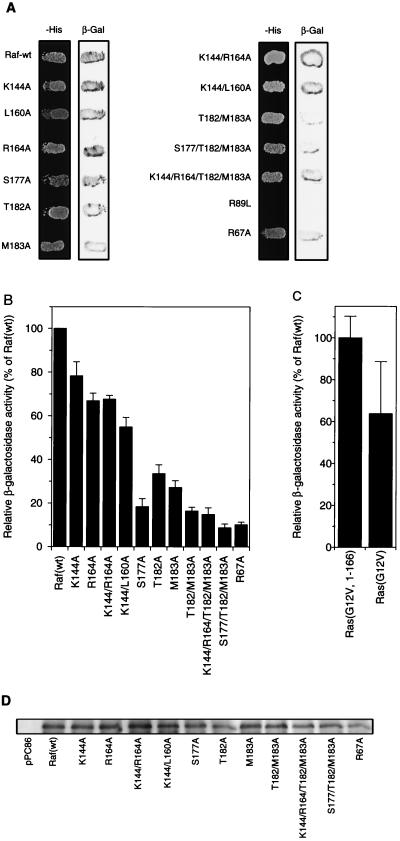
Mutation of K144, L160, R164, S177, T182, or M183 to alanine slightly decreased Ras(G12V)-induced Raf activation to 70 to 80% of the activity of Raf(wt) (Fig. 4A), while expression of FLAG epitope-tagged RafC1 mutant proteins was not altered (Fig. 4B). These inhibitory mutations are also found in three different epitopes (Fig. 3C). One epitope comprises residues S177, T182, and M183. The side chains of K144 and R164 also form a contiguous surface. L160, which is found as a single inhibitory residue, is localized to a position analogous to the phorbol ester binding site in the C1 domain of PKC.
Combining mutations within one epitope reinforced the effect of inhibitory mutations on Ras-dependent Raf activation (Fig. 4C). The mutations T182A M183A and S177A T182A M183A reduced Raf activation to 60 and 35% compared to Raf(wt) activation, respectively. The K144A R164A double mutation affected Raf activation even more drastically, reducing Raf activation to about 30% of the activity of Raf(wt). While inhibition by mutations within a single epitope can be attributed to disruption of a single function of RafC1, this raised the question of whether different functional epitopes may cooperate in Ras-dependent Raf activation. Thus, we combined the L160A mutation, which is localized to a position equivalent to the phorbol ester binding site in PKC, with the K144A mutation. Remarkably, the activation of this double mutant Raf was inhibited to the vector control level. Also, the combination of K144A, R164A, T182A, and M183A led to complete inhibition of Raf activation. Even mutations that completely inhibited Raf activation did not reduce Raf expression (Fig. 4D). Thus, the complete inhibition that was elicited by combination of inhibitory RafC1 mutations from different epitopes clearly demonstrates that RafC1 function is essential in Ras-induced Raf activation.
Raf kinase assays confirm the inhibitory and activating effects of mutant RafC1.
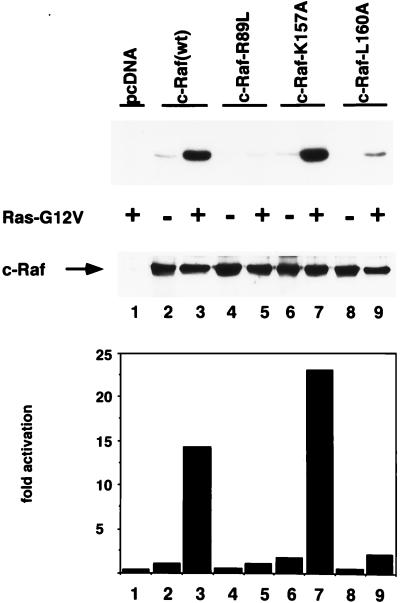
To confirm further that both the activation and inhibition of Raf-dependent transactivation observed in the reporter gene assay were due to changes in Raf activity, we tested inhibitory and activating mutant RafC1 by using a Raf kinase immunoprecipitation assay (18) after transient transfection of RK13 cells. Phosphorylation of kinase-inactive MEK as a substrate was increased 15-fold upon activation of Raf(wt) by Ras(G12V), while Raf(R89L) could not be activated by Ras (Fig. 5, upper and lower panels). The basal activity of K157A mutant RafC1 was enhanced 1.7-fold and corresponded to 23-fold activation of Raf(K157A) by Ras(G12V). Surprisingly, inhibition of Raf activation by the L160A mutation reduced Raf activation to only 15% of the Raf(wt) level. Control of Raf protein levels ruled out the possibility that the strong inhibition by the L160A mutation was due to reduced Raf protein levels (Fig. 5, middle panel). While we cannot explain the stronger inhibitory effect of the mutant Raf(L160A) in the Raf kinase assay compared to the reporter gene assay, these results clearly confirm that the activation and inhibition of Raf-dependent transactivation by mutant RafC1 observed in the reporter gene assay were due to alterations in Raf kinase activity.
FIG. 5.
Effect of mutant RafC1 on Raf kinase activation. MEK phosphorylation by Raf(wt) and by mutant Raf proteins is shown in the upper panel, and quantification of MEK phosphorylation is displayed in the lower panel. Immunodetection of Raf proteins is shown in the middle panel. Activation of Raf(wt) and Raf mutant proteins was measured by using phosphorylation of kinase-inactive MEK by immunoprecipitated FLAG epitope-tagged Raf(wt), Raf(R89L), Raf(K157A), and Raf(160A) proteins that were transiently expressed in RK13 cells. The FLAG antibody was used to immunoprecipitate tagged Raf protein from cells lysed in NP-40 lysis buffer. Proteins were separated by SDS-PAGE and blotted onto nitrocellulose. MEK phosphorylation was visualized by autoradiography and analyzed with a Phosphoimager. After exposure, membranes were probed with a monoclonal anti-Raf antibody and developed by using enhanced chemiluminescence. The data shown are representative of two independent experiments.
RafC1 mutations affect Raf activation independently of Ras lipid modification.
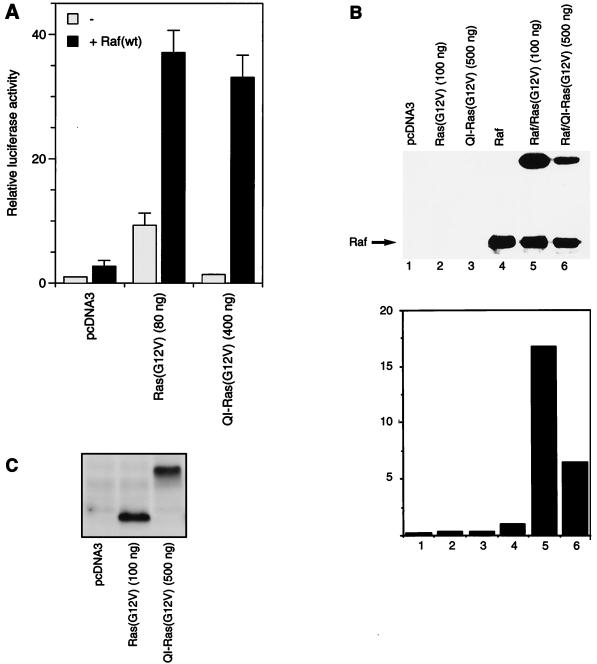
Different studies have suggested that lipid modification of Ras is required for the interaction between Ras and the RafC1 domain and for Raf activation (27, 28, 43). It has been shown that C-terminal lipid modification of Ras is not needed for oncogenic transformation when Ras was targeted to the plasma membrane farnesylation independent by an N-terminally fused transmembrane helix (23). Therefore, we wanted to determine if membrane targeting of Ras is sufficient to activate Raf kinase. We used QI-Ras-(G12V), which cannot be farnesylated (41) and which is targeted to the plasma membrane by the E1(Q37I) transmembrane helix (23, 61). In comparison to Raf activation by farnesylated Ras, the QI-Ras(G12V) construct required a fivefold larger amount of vector to increase Raf-stimulated transactivation to 90% of the level that was achieved with farnesylated Ras(G12V) (Fig. 6A). In the absence of cotransfected Raf, QI-Ras(G12V) increased transcriptional activity only 1.4-fold over the vector control level. We also tested the ability of QI-Ras(G12V) to activate Raf in the direct Raf kinase assay. Activation of Raf by QI-Ras(G12V) was 7-fold, while Ras(G12V) induced a 17-fold increase in Raf activity when a 5-fold greater amount of QI-Ras(G12V) plasmid compared to Ras(G12V) was transfected (Fig. 6B). Since QI-Ras(G12V) is about 10 kDa larger than Ras(G12V), Ras expression could be controlled by direct comparison of overexpressed protein independently of the background of endogenous Ras. Immunoblotting of transfected Ras showed that even when a fivefold excess of the QI-Ras(G12V) plasmid was transfected, expression of this protein was not higher than expression of Ras(G12V) (Fig. 6C). Under these conditions, the endogenous Ras was not detected. It appears that a greater amount of the QI-Ras(G12V) plasmid is necessary to achieve expression of QI-Ras(G12V) similar to that of Ras(G12V). In summary, while the efficiency of Raf activation by QI-Ras(G12V) appears to be reduced, Ras activation of Raf does not require any lipid modification.
FIG. 6.
Raf activation induced by membrane-targeted Ras(G12V) lacking lipid modification. (A) Transactivation mediated by Raf(wt) that was induced by the indicated amounts of Ras(G12V) or QI-Ras(G12V) was measured with an E74 binding site-driven promoter as described in the legend to Fig. 2. (B) Activation of Raf kinase activity by Ras(G12V) and QI-Ras(G12V). Transient transfection of RK13 cells was performed by using either 100 ng of pcDNA3-Ras(G12V) or 500 ng of QI-Ras(G12V), respectively. The Raf kinase assay was performed as described in the legend to Fig. 5. MEK phosphorylation is shown in the upper panel, and quantification thereof (fold activation) is displayed in the lower panel. Immunodetection of Raf is shown in the middle panel. (C) Detection of Ras constructs was performed after transient transfection of RK13 cells with either 100 ng of Ras(G12V) or 500 ng of QI-Ras(G12V). After harvesting of cells, normalized amounts of cell lysate were resolved by SDS-PAGE and proteins were transferred to polyvinylidene difluoride membranes. Immunoblotting was performed by using anti-Ras monoclonal antibody Y13-259 and developed by using enhanced chemiluminescence.
Transactivation induced by mutant RafC1 was tested by using 400 ng of QI-Ras(G12V) to achieve a Raf(wt)-induced transactivation signal that was suitable for testing of both activating and inhibitory mutant RafC1. When stimulated by QI-Ras(G12V), all of the activating mutant RafC1 proteins displayed 1.5- to 2-fold-increased transactivation compared to the activation of Raf(wt) by QI-Ras(G12V) (Fig. 7A). These results are similar to those obtained upon activation of these mutant proteins by farnesylated Ras(G12V). The inhibitory effect of the RafC1 single mutations S177A, T182A, and M183A was slightly stronger when they were activated by QI-Ras(G12V) than when they were activated by farnesylated Ras(G12V) (Fig. 7B). The combination of the S177A, T182A, and M183A mutations reduced Raf-mediated transactivation to 35% regardless of the Ras construct used for activation (Fig. 4C and 7B). The inhibition by the K144A, L160A, and R164A single mutations was stronger upon stimulation with QI-Ras(G12V), reducing the level of transactivation to 20 to 30%, compared to the 70 to 80% induced by Ras(G12V). In this case, all of the combined mutations completely inhibited QI-Ras(G12V)-induced transactivation. In summary, the inhibitory or activating effects of RafC1 mutations were similar regardless of whether Ras(G12V) or QI-Ras(G12V) was used. Therefore, we conclude that the RafC1 interaction with Ras is independent of Ras farnesylation.
FIG. 7.
Effect of RafC1 mutations on Raf activation induced by membrane-targeted Ras(G12V) lacking lipid modification. (A) Transactivation mediated by activating RafC1 mutants induced by 400 ng of QI-Ras(G12V). Transactivation mediated by Raf(wt) and mutated RafC1 proteins was measured with an E74 binding site-driven promoter as described in the legend to Fig. 2. (B) Transactivation mediated by inhibitory RafC1 mutations activated by QI-Ras(G12V). The data shown are averages of three independent experiments.
Identification of the Ras binding RafC1 epitope.
The fact that Raf activation by Ras could be inhibited by RafC1 surface mutations raised the question of whether this inhibition was due to disruption of the Ras-Raf interaction. To address this question, we used a two-hybrid assay for which we have shown a quantitative correlation between the Ras binding affinities of point mutant RafRBDs measured in vitro and β-galactosidase activity determined with the two-hybrid assay in vivo (32). When tested qualitatively in the context of full-length Raf, all of the mutant RafC1s showed equal growth on selective medium and displayed β-galactosidase activity in combination with Ras(G12V1-166) (Fig. 8A). Even the K144A L160A and K144A R164A T182A M183A mutations, which inhibited Raf activation completely, did not affect Ras-Raf interaction, which is in marked contrast to the RafRBD mutation R89L. In the quantitative assay, the K144A and R164A single mutants and K144A L160A and K144A R164A double mutants retained 60 to 80% of the β-galactosidase activity of Raf(wt) (Fig. 8B). This indicates that these mutations only exert minor effects on Ras-Raf interaction compared to the effect of mutations within RafRBD (32). Mutations T182A and M183A reduced β-galactosidase activity to about 30% of that of Raf(wt). S177A produced the strongest decrease in β-galactosidase activity elicited by a RafC1 single mutation, reducing it to 20% of the wild-type level. The T182A M183A double mutation and the S177A T182A M183A triple mutation further decreased β-galactosidase activity to about 15 and 10% of the wild-type activity, respectively. To compare these mutations to those of RafRBD in the context of full-length Raf, Raf(R67A) was also tested and found to have 10% of the wild-type activity. Thus, the activity of the triple mutation in both Raf activation and Ras binding is equal to that of R67A. Combining the exchanges of K144 and R164 with the T182A M183A mutations did not affect β-galactosidase activity compared to the T182A M183A double mutation.
FIG. 8.
Effect of RafC1 mutations on Ras binding. (A) Qualitative investigation of the effect of RafC1 mutations on Ras binding using the two-hybrid system. Yeast cells were cotransformed with pPC97-Ras(G12V)1-166 and pPC86-Raf constructs containing the mutations indicated in the context of full-length Raf. After 3 days of growth on selective plates, indicated as −His (minus Leu, Trp, and His in the presence of 25 mM 3-amino-1,2,3-triazole), the β-galactosidase (β-Gal) assay was performed as described in Materials and Methods. (B) Quantitative measurement of Ras-Raf binding in vivo using the two-hybrid system. Yeast cells were cotransformed with pPC97-Ras(G12V1-166) and pPC86-Raf constructs containing the mutations indicated in the context of full-length Raf. The assay for β-galactosidase activity was performed as described in Materials and Methods. (C) Quantitative measurement of Ras-Raf binding using full-length pPC97-Ras(G12V) and the pPC86-Raf construct as for panel B. The quantitative data shown are averages of three independent experiments using different clones each time. (D) Control of expression of pPC86-Raf(wt) and mutant pPC86Raf constructs. After 3 days of growth on selective plates, cells were lysed in yeast lysis buffer. Cell lysate was resolved by SDS-PAGE, and proteins were transferred to polyvinylidene difluoride membranes. Immunoblotting was performed by using anti-Raf-C20 serum and developed by using enhanced chemiluminescence.
To test for a possible contribution of the Ras lipid modification to the Ras-Raf interaction in the two-hybrid system, we also employed a full-length Ras(G12V) construct. When full-length Ras(G12V) was used, a reduction of β-galactosidase activity to about 60% of the activity obtained with C-terminally truncated Ras(G12V1-166) was observed (Fig. 8C), indicating that the Ras lipid modification does not contribute to Ras-Raf interaction. Furthermore, the RafC1 mutations S177A, T182A, and M183A strongly decreased β-galactosidase activity; K144A, L160A, and R164A did not (data not shown). Since yeast cells do not contain endogenous Raf, we controlled the expression of mutant Raf by immunoblotting of pPC86-Raf constructs by using an anti-Raf antibody (Fig. 8D). Since Raf expression was not altered by the mutant RafC1, the reduction in β-galactosidase activity clearly reflects a decrease in Ras-Raf binding affinity. The additive effects of the S177A, T182A, and M183A mutations on Ras-Raf interaction strongly suggest that this epitope mediates interaction of the RafC1 domain with Ras, while the other inhibitory epitopes do not contribute to Ras binding.
Different effects of RafC1 mutations on transactivation induced by Raf membrane targeting.
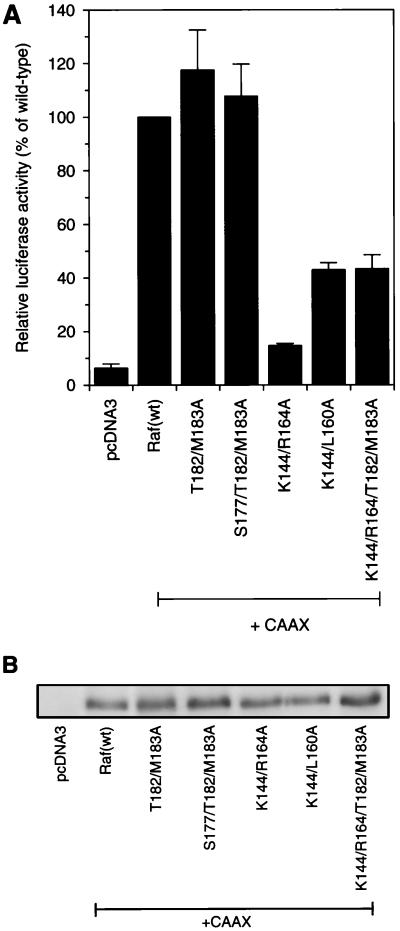
Raf(R89L), which does not bind Ras, can be activated by targeting Raf to the membrane by fusing a CAAX motif to the C terminus of Raf. The level of activation of Raf(R89L-CAAX) was equal to that of activation of Raf(wt-CAAX) (57). For mutant RafC1, the inhibitory effect of the T182A M183A mutations and the S177A T182A M183A mutations was completely overcome by fusing a CAAX motif to the C terminus of these constructs (Fig. 9A). In contrast, the activation of the K144A R164A mutation was strongly reduced, to 15% of Raf(wt-CAAX) activity. The K144A L160A-CAAX and K144A R164A T182A M183A-CAAX mutant retained 40% of Raf(wt-CAAX) activity. Control of the expression of mutant RafC1 proteins showed that the inhibition of Raf(CAAX) activation by these amino acid exchanges was not due to inhibition of protein expression (Fig. 9B). Importantly, the K144A R164A mutation inhibited CAAX-mediated Raf activation much more strongly than did the K144A L160A and K144A R164A T182A M183A mutations, while the latter inhibited Ras(G12V)-mediated activation much more strongly than did the K144A R164A mutation. The different inhibitory potencies of the K144A R164A, K144A L160A, and K144A R164A T182A M183A mutations activated either by CAAX fusion or by Ras reveals that activation by these two stimuli is at least in part mechanistically different. Therefore, these data demonstrate that the inhibitory epitopes in the RafC1 domain have distinct functions in Ras-dependent Raf activation and may even act differently with respect to related stimuli that activate Raf.
FIG. 9.
Effect of RafC1 mutations on transactivation by membrane-targeted Raf. (A) Transactivation induced by 50 ng of pcDNA3-Raf-CAAX containing the mutations indicated was measured with an E74 binding site-driven promoter as described in the legend to Fig. 2. The data shown are averages of three independent experiments. (B) Control of expression of Raf(C1-CAAX) mutant proteins fused to an N-terminal FLAG epitope as described in the legend to Fig. 2B.
DISCUSSION
The RafC1 domain is part of the CR1 region in the regulatory Raf N terminus and is localized immediately adjacent to RafRBD. Although numerous studies have provided evidence regarding a contribution of RafC1 to the Ras-Raf interaction and to Ras-induced Raf activation (5, 6, 8, 11–13, 15, 20, 27, 28, 43, 57, 64, 70), its function has remained elusive. We set out to elucidate the function of the RafC1 domain in detail by performing complete surface alanine scanning mutagenesis, since this approach has been used successfully to define functional epitopes in different protein-ligand interactions (10, 36, 55, 65).
To measure Ras-induced Raf activation, we used an E74-driven reporter gene assay. We have shown previously that the activity measured with this assay correlates quantitatively with Ras-RafRBD binding affinity (4). Results of these assays were supported by results of Raf kinase immunoprecipitation assays (18). In addition, we employed a quantitative two-hybrid assay to measure Ras-Raf interaction by using full-length Raf. We have demonstrated that β-galactosidase reporter gene activity measured in this assay correlates quantitatively with changes in the Ras-Raf binding affinity elicited by RafRBD mutations (32).
Here, we reveal by alanine scanning mutagenesis of surface-exposed residues of RafC1 that multiple distinct functional epitopes are present within this domain. These epitopes are required for both the negative control of the kinase activity and the positive function of the protein. For activating, as well as inhibitory, mutations, three different epitopes were identified. Activating mutations primarily enhanced Ras-dependent activation of Raf, except mutations F151A and D153A, which led to a drastic increase in basal Raf activity. Characteristically, the combination of different activating mutations did not potentiate the effect of the single mutations on Ras-dependent activation but rather resulted in an increase in basal Raf activity. These findings argue in favor of a general role of these residues in the negative control of Raf activity. Nevertheless, some of these residues may also be important for maintaining the activated state of the kinase, since the double mutation F151A D153A even diminished Ras-induced activation compared to the F151A and D153A single mutations. Since the mutations that led to enhanced activation of the kinase are organized in three distinct epitopes, multiple intramolecular interactions, as well as interactions with other regulatory proteins, may be involved in the regulation of kinase activity via these sites.
One possible explanation for activation of Raf via alterations within these epitopes is the dissociation of inhibitory proteins that bind to Raf, which results in activation of the kinase (12, 45). Alternatively, or in addition, these epitopes may play a role in intramolecular interactions. The structures of the protein kinases Src and Hck, which contain different regulatory modular domains in their noncatalytic part, have revealed how their inhibitory regulatory modules act via functional interdomain contacts that retain these kinases in the inactive state (59, 68). In addition, these regulatory modules also provide binding sites for additional protein ligands (48). The weak interactions that are mediated by these negative control domains can be released by competing protein ligands, which leads, in turn, to activation of the kinase (48). Since Raf apparently contains multiple structural modules in the noncatalytic N-terminal part, it is conceivable that these principles also apply to the regulation of Raf kinase activity. In analogy to Src and Hck, the residues in the RafC1 domain that led to enhanced activation of the kinase upon mutation may be part of a complex and interdependent network of intramolecular interactions. Binding of an additional ligand to these sites might release inhibitory intramolecular interactions and activate the kinase. Simultaneously, binding of additional ligands could play a role in correct cellular targeting or stabilize the activated state of the kinase. This model would explain the observation that the activation of the F151A D153A double mutant Raf was lower than the activation of Raf carrying single mutations. The double mutation might weaken the interaction with factors that stabilize the activated state of the kinase further compared to an F151A or D153A single mutation, whereas weak interactions responsible for the negative control of kinase activity by RafC1 may already be disrupted by single mutations. This model is further supported by the recent finding that part of the regulatory domain of the kinase suppressor of Ras stimulates Raf activity in a kinase-independent manner (46).
Inhibitory mutations only had moderate effects when tested as single mutations in the reporter gene assay. In contrast to the mutations that led to activation, the effect of the inhibitory mutations was enhanced when multiple mutations were present. Residues S177, T182, and M183 form a distinct epitope, and mutation of these residues inhibited both Ras-dependent Raf activation and Ras binding. Comparing the decrease of Raf activation with the reduction of Ras binding showed that these two effects correlate closely. The overall inhibition of Ras binding is the same as in the R67A single mutant form of RafRBD. Also, the inhibition of Raf activation caused by the S177A T182A M183A mutation in this study is equal to the effect of the Raf(R67A) mutant described in our previous work (4). Furthermore, inhibition of Raf activation by the S177A T182A M183A mutation (Fig. 9) and by the RafRBD R67A contact surface residue mutation could be overcome completely by fusion of a CAAX motif to the Raf C terminus. This strongly suggests that the epitope consisting of S177, T182, and M183 represents the Ras binding site of RafC1. In comparison with the contact surface of the Ras binding of RafRBD, this epitope is relatively small. In RafRBD, eight amino acid side chains are involved in interaction with Ras, and three of these residues are most important for Ras-dependent Raf activation (4). In RafC1, we identified an interacting epitope of only three side chains that inhibits Ras binding and Raf activation when mutated. Since the decrease of Ras binding energy by mutation of this epitope is equal to the effect of mutation of the single Raf(R67A) side chain, this demonstrates that the contribution of this domain to Ras binding is not equivalent to that of RafRBD.
Assignment of the Ras binding epitope to residues S177, T182, and M183 is in agreement with the strong inhibitory effect of Raf(C165S) and C165S C168S or of the P181L mutation on Ras binding which has been observed in many studies (6, 13, 27, 28, 43, 70). Mutation of C165 and C168 disrupts the first zinc binding site, which will clearly disrupt the conformation of this epitope. These mutations may even lead to steric hindrance of the Ras-RafRBD interaction due to misfolding of parts of the RafC1 interaction surface. Furthermore, mutation of the cis proline at position 181 will cause conformational changes due to cis-trans isomerization and, as a result, will lower Ras-Raf binding. In addition, the C165S C168S mutation will also affect the activating epitope consisting of N140, Q166, and T167, since these residues are localized immediately adjacent to ligands of the first zinc binding site. Thus, the C165S and C168S mutations do affect positive and negative regulatory interactions simultaneously. This strongly supports the notion that it is mandatory for a functional analysis of the RafC1 domain to investigate the effects of mutations within individual regulatory epitopes, thus avoiding gross conformational changes within this domain.
We clearly demonstrate that Raf can be activated by Ras(G12V), even in the absence of lipid modification of Ras, when Ras is targeted to the plasma membrane by fusion to an N-terminal E1(Q37I) transmembrane helix. Thus, Raf activation does not require farnesylated Ras, as membrane targeting of Ras by fusion to an E1(Q37I) transmembrane helix to the Ras N terminus efficiently activates Raf kinase. Furthermore, testing of the Ras-Raf interaction by using full-length Ras in a two-hybrid assay did not provide evidence for a contribution of Ras prenylation to the Ras-Raf interaction. The less efficient activation of Raf by QI-Ras(G12V) could be explained by the insufficient spatial orientation of Ras at the plasma membrane caused by the artificial transmembrane helix and the 22-amino-acid linker. Nearly all RafC1 mutant proteins showed the same effect on Raf activation, regardless of whether Ras(G12V) or QI-Ras(G12V) was used. Inhibition by mutation of K144, L160, and R164 was stronger when QI-Ras(G12V) was used for Raf activation. This indicates that this group of mutations responds systematically differently to membrane targeting of Ras by either farnesylation or fusion of a transmembrane helix. However, if these residues are part of a farnesylation-dependent Ras binding site, the inhibitory effect of these mutations should be abolished when nonfarnesylated QI-Ras(G12V) is used for Raf activation. In summary, we conclude that membrane localization of Ras is required for Raf activation, while farnesylation of Ras is not.
Complete inhibition of Raf activation was caused by combining surface mutations of different epitopes such as K144A L160A or K144A R164A T182A M183A, which demonstrates an essential role of RafC1 in Ras-dependent Raf activation. Mutation K144A R164A or K144A L160A did not interfere with Ras binding significantly but did inhibit Raf activation, even in the presence of a membrane-targeting motif fused to the Raf C terminus. This demonstrates that these residues are essential in the activation of Raf subsequent to membrane targeting of Raf by Ras. Importantly, the potency of the inhibition of Raf activation by these double mutations was interchanged when Raf was activated either by CAAX fusion or by a Ras stimulus. This reveals that Raf activation by CAAX fusion is in part mechanistically different from Ras activation of the kinase. Analogous to the C1 domain of PKC, RafC1 binds to phospholipid vesicles (21). It has been suggested that the C1 domain of PKC is partially inserted into the plasma membrane when bound to phorbol ester (69), and the residues around the phorbol ester binding site have been found to interact with phospholipids (67). The localization of K144 and R164 is in agreement with a possible role of these residues in phospholipid binding, since in the PKC C1 domain, residues localized at these positions were also involved in phospholipid binding (67).
Mutation L160A also resulted in decreased Raf-dependent transactivation. Surprisingly, the L160A exchange inhibited Raf activation more potently in the Raf kinase assay. This is especially intriguing because L160 is localized to a position analogous to the phorbol ester binding site in the C1 domain of PKC. The unlikelihood that the small change of leucine to alanine would result in a general decrease in membrane binding, raises the question of whether L160 is involved in the binding of a lipid messenger that is important for Raf activation. Although it is tempting to speculate that ceramide might be a ligand that binds to the RafC1 domain (29), we have been unable to observe a direct effect of ceramide on Raf activation (data not shown), which is in agreement with a recently published study (51). Surprisingly, in RafC1, only the single residue L160 was found to affect Ras-dependent Raf activation. However, in the case of phorbol ester binding to the C1 domain of PKC, most interactions between the C1 domain and the phorbol ester ligand are mediated via main chain interactions (69). If these characteristics also applied to lipid cofactor binding to RafC1 accordingly, our mutational analysis may underestimate the importance of lipid cofactor binding for Raf activation. However, the complete inhibition of Raf activation by the combination of K144A and L160A strongly suggests that L160 is part of a cofactor binding site that plays an essential role in Raf activation.
In summary, the RafC1 domain displays multiple functional features. (i) It appears to be part of a complex intramolecular network of interactions that control the maintenance of the inactive state of the kinase. (ii) It may represent the binding site for inhibitory proteins. (iii) It is involved in mediating Ras binding. (iv) It contains an epitope of charged residues that may be involved in phospholipid binding. (v) It contains a putative binding site for a lipid messenger that is localized to a position analogous to that of the PKC phorbol ester binding site.
This striking complexity of RafC1 function in Raf kinase regulation immediately demonstrates that unravelling of the complete function of RafC1 necessitates the identification of its protein and lipid interaction partners and requires detailed structural and functional characterization of their individual roles in Raf activation.
ACKNOWLEDGMENTS
We thank B. Voss for excellent technical assistance and J. Becker for helpful discussions and critical reading of the manuscript.
This work was supported by DFG grant B1411/1-1 and SFB 394.
REFERENCES
- 1.Barettino D, Feigenbutz M, Valcàrel R, Stunnenberg H G. Improved method for PCR-mediated site-directed mutagenesis. Nucleic Acids Res. 1994;22:541–542. doi: 10.1093/nar/22.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel P L, Fields S. Analyzing protein-protein interactions using the two-hybrid system. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 3.Block C, Wittinghofer A. Switching to Rac and Rho. Structure. 1995;3:1281–1284. doi: 10.1016/s0969-2126(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 4.Block C, Janknecht R, Herrmann C, Nassar N, Wittinghofer A. Quantitative structure-activity analysis correlating Ras/Raf interaction in vitro to Raf activation in vivo. Nat Struct Biol. 1996;3:244–250. doi: 10.1038/nsb0396-244. [DOI] [PubMed] [Google Scholar]
- 5.Brtva T R, Drugan J K, Ghosh S, Terell R S, Campbell-Burk S, Bell R M, Der C J. Two distinct Raf domains mediate interaction with Ras. J Biol Chem. 1995;270:9809–9812. doi: 10.1074/jbc.270.17.9809. [DOI] [PubMed] [Google Scholar]
- 6.Bruder J T, Heidecker G, Rapp U R. Serum-, TPA-, and Ras-induced expression from Ap-1/Ets-driven promoters requires Raf-1 kinase. Genes Dev. 1992;6:545–556. doi: 10.1101/gad.6.4.545. [DOI] [PubMed] [Google Scholar]
- 7.Chevray P M, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of jun. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow Y-H, Pumiglia K, Jun T H, Dent P, Sturgill T W, Jove R. Functional mapping of the N-terminal regulatory domain in the human Raf-1 protein kinase. J Biol Chem. 1995;270:14100–14106. doi: 10.1074/jbc.270.23.14100. [DOI] [PubMed] [Google Scholar]
- 9.Chuang E, Barnard D, Hettich L, Zhang X-F, Avruch J, Marshall M. Critical binding and regulatory interactions between Ras and Raf occur through a small, stable N-terminal domain of Raf and specific Ras effector residues. Mol Cell Biol. 1994;14:5318–5325. doi: 10.1128/mcb.14.8.5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clackson T, Wells J A. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 11.Clark G J, Drugan J K, Terell R S, Bradham C, Der C J, Bell R M, Campbell S. Peptides containing a consensus Ras binding sequence from Raf-1 and the GTPase activating protein NF1 inhibit Ras function. Proc Natl Acad Sci USA. 1996;93:1577–1581. doi: 10.1073/pnas.93.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark G J, Drugan J K, Rossmann K L, Carpenter J W, Rogers-Graham K, Fu H, Der C J, Campbell S L. 14-3-3 ζ negatively regulates Raf-1 activity by interactions with the Raf-1 cysteine-rich domain. J Biol Chem. 1997;272:20990–20993. doi: 10.1074/jbc.272.34.20990. [DOI] [PubMed] [Google Scholar]
- 13.Cutler R E, Morrison D K. Mammalian Raf-1 is activated by mutations that restore Raf signaling in Drosophila. EMBO J. 1997;16:1953–1960. doi: 10.1093/emboj/16.8.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daum G, Eisenmann-Tappe I, Fries H W, Troppmair J, Rapp U. The ins and outs of Raf kinases. Trends Biochem Sci. 1994;19:474–480. doi: 10.1016/0968-0004(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 15.Drugan J K, Khosravi-Far R, White M A, Der C J, Sung Y-J, Hwang Y-W, Campbell S L. Ras interaction with two distinct binding domains in Raf-1 may be required for Ras transformation. J Biol Chem. 1996;271:233–237. doi: 10.1074/jbc.271.1.233. [DOI] [PubMed] [Google Scholar]
- 16.Emerson S D, Waugh D S, Scheffler J E, Tsao K L, Prinzo K M, Fry D C. Chemical shift assignments and folding topology of the Ras-binding domain of human Raf-1 determined by heteronuclear three-dimensional NMR spectroscopy. Biochemistry. 1994;33:7745–7752. doi: 10.1021/bi00191a001. [DOI] [PubMed] [Google Scholar]
- 17.Fabian J R, Vojtek A B, Cooper J A, Morrison D K. A single amino acid change in Raf-1 inhibits Ras binding and alters Raf-1 function. Proc Natl Acad Sci USA. 1994;91:5982–5986. doi: 10.1073/pnas.91.13.5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flory E, Weber C K, Chen P, Hoffmeyer A, Jassoy C, Rapp U R. Plasma membrane-targeted Raf kinase activates NF-κB and human immunodeficiency virus type 1 replication in T lymphocytes. J Virol. 1998;72:2788–2794. doi: 10.1128/jvi.72.4.2788-2794.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fridman M, Tikoo A, Varga M, Murphy A, Nur-E-Kamal M S A, Maruta H. The minimal fragments of c-Raf-1 and NF-1 that can suppress v-Ha-Ras-induced malignant phenotype. J Biol Chem. 1994;269:30105–30108. [PubMed] [Google Scholar]
- 20.Ghosh S, Bell R M. Identification of discrete segments of human Raf-1 kinase critical for high affinity binding to Ha-Ras. J Biol Chem. 1994;269:30785–30788. [PubMed] [Google Scholar]
- 21.Ghosh S, Xie W Q, Quest A F G, Mabrouk G M, Strum J C, Bell R M. The cysteine-rich region of Raf-1 kinase contains zinc, translocates to liposomes, and is adjacent to a segment that binds GTP-Ras. J Biol Chem. 1994;269:10000–10007. [PubMed] [Google Scholar]
- 22.Gorman C, Skinner R H, Skelly J V, Neidle S, Lowe P N. Equilibrium and kinetic measurements reveal rapidly reversible binding of Ras to Raf. J Biol Chem. 1996;271:6713–6719. doi: 10.1074/jbc.271.12.6713. [DOI] [PubMed] [Google Scholar]
- 23.Hart K C, Donoghue D J. Derivatives of activated H-ras lacking C-terminal lipid modifications retain transforming ability if targeted to the correct subcellular location. Oncogene. 1997;14:945–953. doi: 10.1038/sj.onc.1200908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heidecker G, Huleihel M, Cleveland J L, Kolch W, Beck T W, Lloyd P, Pawson T, Rapp U R. Mutational activation of c-raf-1 and definition of the minimal transforming sequence. Mol Cell Biol. 1990;10:2503–2512. doi: 10.1128/mcb.10.6.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrmann C, Martin G A, Wittinghofer A. Quantitative analysis of the complex between p21ras and the Ras-binding domain of the human Raf-1 protein kinase. J Biol Chem. 1995;270:2901–2905. doi: 10.1074/jbc.270.7.2901. [DOI] [PubMed] [Google Scholar]
- 26.Hommel U, Zurini M, Luyten M. Solution structure of a cysteine rich domain of rat protein kinase C. Nat Struct Biol. 1994;1:383–387. doi: 10.1038/nsb0694-383. [DOI] [PubMed] [Google Scholar]
- 27.Hu C-D, Kariya K-I, Tamada M, Akasaka K, Shirouzu K, Shirouzu M, Shigeyuki Y, Kataoka T. Cysteine-rich region of Raf-1 interacts with activator domain of post-translationally modified Ha-Ras. J Biol Chem. 1995;270:30274–30277. doi: 10.1074/jbc.270.51.30274. [DOI] [PubMed] [Google Scholar]
- 28.Hu C-D, Kariya K-I, Kotani G, Shirouzu M, Yokoyama S, Kataoka T. Coassociation of Rap1A and Ha-Ras with Raf-1 N-terminal region interferes with Ras-dependent activation of Raf-1. J Biol Chem. 1997;272:11702–11705. doi: 10.1074/jbc.272.18.11702. [DOI] [PubMed] [Google Scholar]
- 29.Huwiler A, Brunner J, Hummel R, Vervoodeldonk M, Stabel S, Van den Bosch H, Pfeilschifter J. Ceramide-binding and activation defines protein kinase c-Raf as a ceramide-activated protein kinase. Proc Natl Acad Sci USA. 1996;93:6959–6963. doi: 10.1073/pnas.93.14.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ichikawa S, Hatanaka H, Takeuchi Y, Ohno S, Inagaki F. Solution structure of cysteine-rich domain of protein kinase Cα. J Biochem. 1995;117:566–574. doi: 10.1093/oxfordjournals.jbchem.a124745. [DOI] [PubMed] [Google Scholar]
- 31.Jaitner B K. Ph.D. thesis. Bochum, Germany: Ruhr-Universität-Bochum; 1997. [Google Scholar]
- 32.Jaitner B K, Becker J, Linnemann T, Herrmann C, Wittinghofer A, Block C. Discrimination of amino acids mediating Ras binding from non-interacting residues affecting Raf activation by double mutant analysis. J Biol Chem. 1997;272:29927–29993. doi: 10.1074/jbc.272.47.29927. [DOI] [PubMed] [Google Scholar]
- 33.Janknecht R, Nordheim A. Gene regulation by Ets proteins. Biochim Biophys Acta. 1993;1155:346–356. doi: 10.1016/0304-419x(93)90014-4. [DOI] [PubMed] [Google Scholar]
- 34.Janknecht R, Ernst W H, Pingoud V, Nordheim A. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 1993;12:5097–5104. doi: 10.1002/j.1460-2075.1993.tb06204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janknecht R. Analysis of the ERK-stimulated ETS transcription factor ER81. Mol Cell Biol. 1996;16:1550–1556. doi: 10.1128/mcb.16.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin L, Fendly B M, Wells J A. High resolution functional analysis of antibody-antigen interactions. J Mol Biol. 1992;226:851–865. doi: 10.1016/0022-2836(92)90636-x. [DOI] [PubMed] [Google Scholar]
- 37.Klebe R J, Harriss J V, Sharp Z D, Douglas M D. A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene. 1983;25:333–341. doi: 10.1016/0378-1119(83)90238-x. [DOI] [PubMed] [Google Scholar]
- 38.Koide H, Satoh T, Nakafuku M, Kaziro Y. GTP-dependent association of Raf-1 with H-Ras: identification of Raf as a target downstream of Ras in mammalian cells. Proc Natl Acad Sci USA. 1993;90:8683–8686. doi: 10.1073/pnas.90.18.8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolch W, Heidecker G, Lloyd P, Rapp U R. Raf-1 protein kinase is required for growth of induced NIH/3T3 cells. Nature. 1991;349:426–428. doi: 10.1038/349426a0. [DOI] [PubMed] [Google Scholar]
- 40.Leevers S J, Paterson H F, Marshall C J. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature. 1994;369:411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- 41.Lowy D R, Willumsen B M. Function and regulation of Ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 42.Lu X, Melnick M B, Hsu J-C, Perrimon N. Genetic and molecular analyses of mutations involved in Drosophila raf signal transduction. EMBO J. 1994;13:2592–2599. doi: 10.1002/j.1460-2075.1994.tb06549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo Z, Diaz B, Marshall M S, Avruch J. An intact Raf zinc finger is required for optimal binding to processed Ras and for Ras-dependent activation in situ. Mol Cell Biol. 1997;17:46–53. doi: 10.1128/mcb.17.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meloche S, Pagès G, Pouysségur J. Functional expression and growth factor activation of an epitope-tagged p44 mitogen activated protein kinase, p44mapk. Mol Biol Cell. 1992;3:63–71. doi: 10.1091/mbc.3.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michaud N R, Fabian J R, Mathes K D, Morrison D K. 14-3-3 is not essential for Raf-1 function: identification of Raf proteins that are biologically activated in a 14-3-3- and Ras-independent manner. Mol Cell Biol. 1995;15:3390–3397. doi: 10.1128/mcb.15.6.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michaud N R, Therrien M, Cacace A, Edsall L C, Spiegel S, Rubin G M, Morrison D K. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc Natl Acad Sci USA. 1997;94:12792–12796. doi: 10.1073/pnas.94.24.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moodie S A, Willumsen B M, Weber M J, Wolfman A. Complexes of Ras-GTP with Raf-1 and mitogen activated protein kinase kinase. Science. 1993;260:1658–1661. doi: 10.1126/science.8503013. [DOI] [PubMed] [Google Scholar]
- 48.Moraefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee C-H, Kuriyan J, Miller W T. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 49.Morrison D K, Cutler R E., Jr The complexity of Raf-1 regulation. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 50.Mott H R, Carpenter J W, Zhong S, Ghosh S, Bell R M, Campbell S L. The solution structure of the Raf-1 cysteine-rich domain: a novel Ras and phospholipid binding site. Proc Natl Acad Sci USA. 1996;93:8312–8317. doi: 10.1073/pnas.93.16.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Müller G, Storz P, Bourteele S, Döppler H, Pfizenmaier K, Mischak H, Philipp A, Kaiser K, Kolch W. Regulation of Raf-1 kinase by TNF via its second messenger ceramide and crosstalk with mitogenic signalling. EMBO J. 1998;17:732–742. doi: 10.1093/emboj/17.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nassar N, Horn G, Herrmann C, Scherer A, McCormick F, Wittinghofer A. The 2.2Å crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature. 1995;375:554–560. doi: 10.1038/375554a0. [DOI] [PubMed] [Google Scholar]
- 53.Nassar N, Horn G, Herrmann C, Block C, Janknecht R, Wittinghofer A. Ras/Rap effector specificity determined by charge reversal. Nat Struct Biol. 1996;3:723–729. doi: 10.1038/nsb0896-723. [DOI] [PubMed] [Google Scholar]
- 54.Nicholls A, Sharp K A, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 55.Onrust R, Herzmark P, Chi P, Garcia P D, Lichtarge O, Kingsley C, Bourne H R. Receptor and βγ binding sites in the α subunit of the retinal G protein transducin. Science. 1997;275:381–384. doi: 10.1126/science.275.5298.381. [DOI] [PubMed] [Google Scholar]
- 56.Pumiglia K, Chow Y-H, Fabian J, Morrison D, Decker S, Jove R. Raf-1 N-terminal sequences necessary for Ras-Raf interaction and signal transduction. Mol Cell Biol. 1995;15:398–406. doi: 10.1128/mcb.15.1.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roy S, Lane A, Yan J, McPherson R, Hancock J F. Activity of plasma membrane recruited Raf-1 is regulated by Ras via the Raf zinc finger. J Biol Chem. 1997;272:20139–20145. doi: 10.1074/jbc.272.32.20139. [DOI] [PubMed] [Google Scholar]
- 58.Scheffler J E, Waugh D S, Bekesi E, Kiefer S E, LoSardo J E, Neri A, Prinzo K M, Tsao K-L, Wegrzynski B, Emerson S D, Fry D C. Characterization of a 78-residue fragment of c-Raf-1 that comprises a minimal binding domain for the interaction with Ras-GTP. Biochemistry. 1994;35:22340–22346. [PubMed] [Google Scholar]
- 59.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 60.Stokoe D, Macdonald S G, Cadwallader K, Symons M, Hancock J F. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 61.Swift A M, Machamer C E. A Golgi retention signal in a membrane-spanning domain of coronavirus E1 protein. J Cell Biol. 1991;115:19–30. doi: 10.1083/jcb.115.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Aelst L, Barr M, Marcus S, Polverino A, Wigler M. Complex formation between Ras and Raf and other protein kinases. Proc Natl Acad Sci USA. 1993;90:6213–6217. doi: 10.1073/pnas.90.13.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voijtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 64.Warne P H, Rodriguez Viciana P, Downward J. Direct interaction of Ras and the amino-terminal region of Raf-1 in vitro. Nature. 1993;364:352–355. doi: 10.1038/364352a0. [DOI] [PubMed] [Google Scholar]
- 65.Wells J A. Systematic mutational analysis of protein-protein interfaces. Methods Enzymol. 1991;202:390–410. doi: 10.1016/0076-6879(91)02020-a. [DOI] [PubMed] [Google Scholar]
- 66.Wells J A. Structural and functional epitopes in the growth hormone receptor complex. Bio/Technology. 1995;13:647–651. doi: 10.1038/nbt0795-647. [DOI] [PubMed] [Google Scholar]
- 67.Xu R X, Pawelczyk T, Xia T-H, Brown S C. NMR structure of a protein kinase C-γ phorbol-binding domain and study of protein-lipid micelle interactions. Biochemistry. 1997;36:10709–10717. doi: 10.1021/bi970833a. [DOI] [PubMed] [Google Scholar]
- 68.Xu W, Harrison S C, Eck M J. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 69.Zhang G, Kazanietz M G, Blumberg P M, Hurley J H. Crystal structure of the Cys2 activator-binding domain of protein kinase Cδ in complex with phorbol ester. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 70.Zhang X-F, Settleman J, Kyriakis J M, Takeuchi-Suzuki E, Elledge S J, Marshall M S, Bruder J T, Rapp U R, Avruch J. Normal and oncogenic p21ras proteins bind to the amino-terminal regulatory domain of c-Raf-1. Nature. 1993;364:308–313. doi: 10.1038/364308a0. [DOI] [PubMed] [Google Scholar]