Abstract
Protists encompass a vast widely distributed group of organisms, surpassing the diversity observed in metazoans. Their diverse ecological niches and life forms are intriguing characteristics that render them valuable subjects for in-depth cell biology studies. Throughout history, viruses have played a pivotal role in elucidating complex cellular processes, particularly in the context of cellular responses to viral infections. In this comprehensive review, we provide an overview of the cellular alterations that are triggered in specific hosts following different viral infections and explore intricate biological interactions observed in experimental conditions using different host-pathogen groups.
Subject terms: Cellular microbiology, Virus-host interactions
An in-depth review compiling information about the effects of virus infections in protists and discussing how these interactions are important for cell biology, ecology and biotechnology.
Introduction
The taxon “Protozoa” was formalized by the English biologist, comparative anatomist, and paleontologist Richard Owen in 18581. At the time, the taxon comprised “numerous organisms of minute size retaining the form of nucleated cells, which manifest the common organic characters, but without the distinctive superadditions of true plants or animals”1. Since then, the terms protozoa, protozoans, protista and protists have been used to designate a variety of eukaryotic groups2,3. As this taxon does not encompass well established clades that diverged from it, protists form a paraphyletic group. Similar to other paraphyletic taxa, it proves challenging to identify universal characteristics among all its members2. Nevertheless, protists can be defined as predominantly unicellular eukaryotic organisms, which do not develop their tissues through the process of embryonic stratification4,5. Their cell morphology showcases an astonishing diversity of forms, functions, and survival strategies. They are widespread worldwide, and comprise marine, freshwater, terrestrial, symbiotic, and pathogenic strains2,6,7. However, taxonomists estimate that the known representatives compose only a small portion of the total of protists on Earth today, and that the diversity of this group is greater than that of metazoans6,7.
With such abundance and richness, these eukaryotes are part of communities with a wide diversity of organisms and are especially studied for their interactions with pathogens or regarding their own pathogenicity7. A frequent example in literature are amoebae. Most amoebae are free-living protists of great diversity that have garnered significant attention due to their intricate associations with a plethora of microorganisms from all domains of life8–19. Free-living amoebae occupy different ecological niches and have already been found in a wide range of natural and anthropized environments7. The most notorious feature of amoebae is their ability to alter their cell shape by creating temporary extensions of cytoplasm known as pseudopods, which serves both for feeding and movement7. Due to their foraging behaviors and predatory nature, amoebae are often considered as professional phagocytes, mainly consuming microorganisms to fulfill their nutritional needs9. However, many microorganisms successfully evade the phagocytic pathway and thrive within the amoebae, turning them into transmission vehicles or incubators and enabling various types of ecological relationships to occur, from symbiotic to parasitic9. Besides, their cell biology and active grazing behavior serve as a widely used infection route for giant viruses, although miscellaneous entry mechanisms have already been described for this phylum of viruses10,20,21. Given the substantial size of their viral particles, they can trigger phagocytosis in amoebae, creating entry opportunities11–14. In fact, the efficiency of this route lead other giant viruses that lack the requisite size for phagocytosis to employ a mimicry strategy to use the phagocytic pathway as an entry route15.
The myriad of interactions involving amoebae renders them exceptional experimental models to explore and investigate a diversity of scientific fields16. Over the years, amoebae have contributed to the understanding of a wide array of subjects. These encompass the unraveling of mechanisms related to the resistance and pathogenicity of microorganisms, the intricacies of cell locomotion, the functioning of non-muscle contractile systems, the dynamics of populations and communities, the implications of cell nucleus removal and transplantation, events involving horizontal gene transfer, and the evolution of organelles, that may provide valuable insights into the origin and evolution of eukaryotic cells9,22–29. This shows that the study of protists can help advance different fields of science. However, to gain a more comprehensive understanding, it is important that we delve deeper into the basic aspects of protist cell biology.
Viruses are unique tools to study protists biology
Viruses have been unique tools in helping to comprehend complex biological processes in different host models. As mandatory intracellular parasites, viruses have an intimate relationship with their host cells, often depending on and controlling their cell structure, metabolism, biochemical machinery and behavior30,31. The diversity of structures, genomes, and replication strategies that viruses exhibit reflects thousands-to-billions of years of coevolution with their hosts, some older than the origin of the eukaryotic cell32. This is a key point in biology, as over the centuries, the study of host-virus interactions has led to important discoveries. For instance, in genetics, these findings include the discovery of DNA as the source of heritage, mRNA, mRNA processing, RNA interference, as well as the regulation of gene expression, transcriptional control elements, and transcription factors30,31,33,34. Additionally, these studies have contributed significantly to our comprehension of biochemistry, elucidating tyrosine kinases and signal transduction pathways30,31,33. Furthermore, they have shed light on immunology and many other intrinsic aspects of host cells. These include our understanding of histocompatibility antigen function, interferons, cellular oncogenes, tumor suppressor proteins, and apoptosis in eukaryotic hosts30,31,33,35. They’ve also contributed to our knowledge of the CRISPR/Cas9 mechanism in bacterial and archaeal hosts, which is now widely used for genome editing for many biotechnological purposes36.
Undoubtedly, the field of virology has made substantial contributions to science, extending beyond its significance in diseases. Throughout history, the discovery of viruses has primarily been driven by the concerns of human health, and the health of other animals and plants of economic importance30,33,37. This implies that most of the described viruses have direct connections to humans, whether in the realms of economics, medicine, or biotechnology, resulting in an extremely anthropocentric known virosphere37. However, the viruses known to humankind represent only a small fraction of the viral diversity on Earth. As viruses infect organisms of the three domains of life, and some studies estimate the count of viral particles on Earth to be of the order of 10³¹, there remains a large number of undiscovered viruses35,38. Still, due to the absence of a universal molecular marker for viruses, the true extent of global viral diversity remains largely unknown.
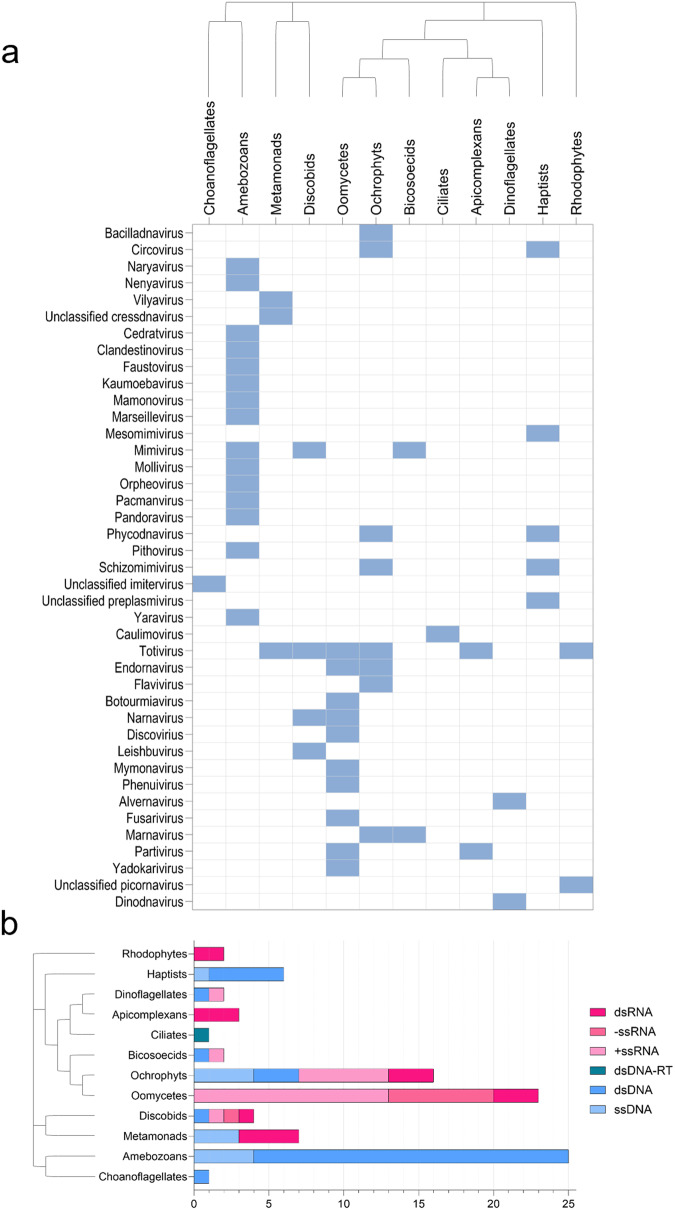
Despite having a small number of identified representatives so far, viruses that infect protists exhibit a remarkable diversity (Fig. 1). One of the earliest mentions of virus-like particles (VLP) in protozoa seems to have been documented in 1960 within Entamoeba histolytica39. Over the years, many taxa of viruses that infect protists have been identified, with representatives containing DNA or RNA genomes of various topologies and sizes, which can exist either in a naked form or be enclosed within capsids, that also exhibit diverse sizes and symmetries40–47. Among these viruses, a group that garners significant attention due to the complexity of their genomes and the size of their particles are the viruses of the phylum Nucleocytoviricota. Regardless of their hypothetical monophyletic origin, the viruses belonging to this phylum display a broad host range within eukaryotes. This group includes the giant viruses (GVs) that infect amoebae, the ones holding the distinction of possessing the largest genomes and viral particles found in the virosphere48. Although these viruses infect amoebae in the laboratory, their natural hosts are still uncertain. Over the past two decades, there has been dedicated research into the exceptional biological traits of these viruses, the enigmatic composition of their genomes and particles, and their ecological significance.
Fig. 1. Overview of the current extent of the known protist virosphere.
a Protist supergroups are shown on the x-axis while viral groups are shown on the y-axis. Highlighted squares on the matrix represent a group of viruses infecting a respective group of protists. The clades Viridiplantae, Fungi and Metazoa were removed from the dendrogram (latest eukaryotic phylogeny was retrieved and adapted from Keeling & Eglit, 2023223. Viral groups at family or genus level (unclassified cressdnavirus and preplasmivirus at phylum level and unclassified imitervirus and picornavirus at order level) are also on the y-axis. b The viral counts at genus level for each virus class (DNA or RNA, single-stranded or double-stranded) were retrieved from VirusHostdb224 and manual curation of the literature. Abbreviatures: double stranded (ds), single stranded (ss), negative sense (−), positive sense (+), double-stranded DNA genome that has an RNA intermediate (dsDNA-RT). Detailed information regarding viruses and respective hosts used to make this figure can be found in supplementary data file (tab 1).
Virology is an integrative science. Host-parasite interactions can reveal valuable information about cellular alterations and host defense mechanisms that can be used to study basic cellular processes30,33. During a viral infection, the virion and host experience modifications of such magnitude that both start to present a different nature, sometimes even shifting the cellular role to solely producing the viral progeny. In consideration of this, the term ‘virocell’ was coined to refer to this specific moment when a virus is actively infecting a cell49. One of the most characteristic changes during a viral infection is the establishment of a viral factory (VF), that can be formed in both the cytoplasm or nucleus of the cell. It is the place where viral progeny is assembled, and often, the genetic material is replicated. Its formation is usually accompanied by extensive cytoplasmic rearrangements50. Therefore, studying host-pathogen interactions can uncover how protists respond to viral infections and highlight the features that were evolutionarily selected to counter viral threats. Moreover, viruses can be engineered for various biotechnological purposes, including gene delivery and genome editing. This technology can help elucidate protist biology for basic research or practical purposes51. Besides, these studies can also provide enlightenment into general cell biology that may apply to a wider range of eukaryotic organisms. In this review, we provide an overview of the effects of virus infection and replication on protists and discuss some prospects that the investigation into viral infections in protists could offer.
Cell machinery and morphology modulation in protist cells during viral infections
Over the past two decades, studies concerning giant viruses have highlighted virus-protist interactions52. Amoeba, particularly the genus Acanthamoeba and the species Vermamoeba vermiformis, are commonly studied hosts that suffer cell machinery and morphological changes after infection53. The ciliate Tetrahymena sp. also shows alterations following GV inoculation. Limited details exist for protists infected with GVs and other viruses, including diatoms, flagellates, dinoflagellates and algae. A list of publications mentioning cellular alterations derived from viral infections in protists is shown in the supplementary data file (tab 2). Below, we discuss these alterations in detail for each main host group.
Acanthamoeba castellanii and their broad response to different viruses
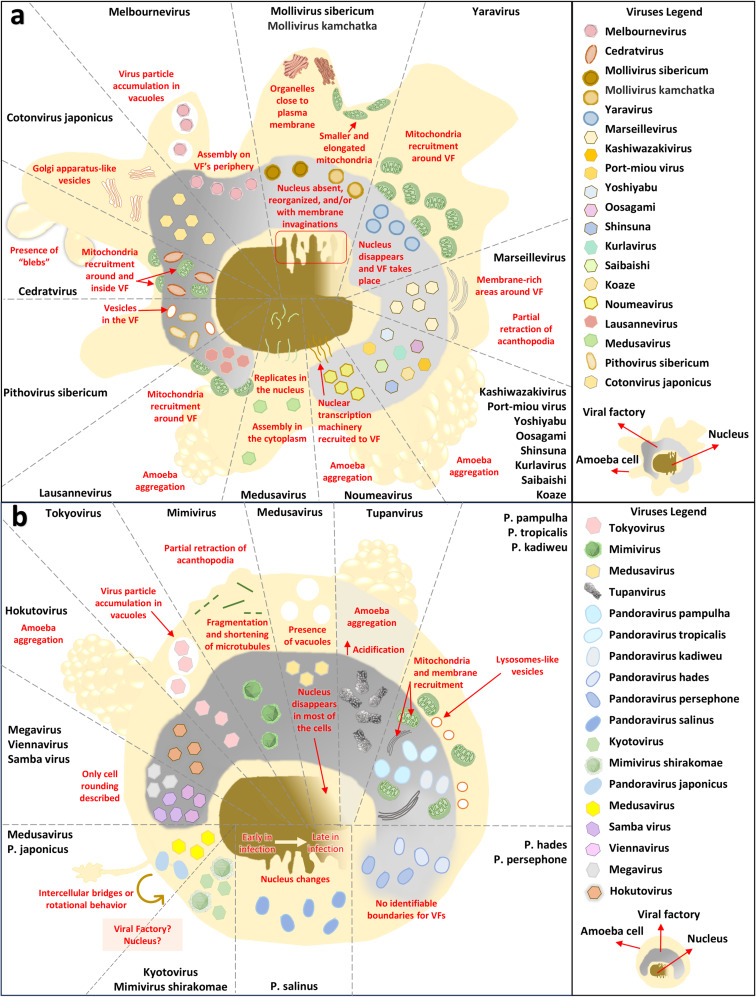
Most known GVs groups were isolated in A. castellanii54. A successful giant virus infection triggers the formation of VFs inside the host cells. However, some studies mention cytopathic effects (CPEs) without evident VFs55–57. Nevertheless, GVs’ DNA replication may also happen in the nucleus, with assembly in cytoplasm58. Noumeavirus, for instance, recruits the host’s nuclear transcription machinery to the cytoplasmatic VF59. An overview of some of the main changes using the A. castellanii model as an example can be seen in Fig. 2.
Fig. 2. Overview of the main changes in the A. castellanii. cell during different giant viruses’ infection.
a “Trophozoite cell shape” to represent viruses that did not interfere in the cell shape structure. b “Rounded cell shape” to represent only the viruses reported to trigger cell rounding. Red text: cell modifications.
Some GVs infections can disorganize and deform the nucleus of A. castellanii, leading to loss of its spherical appearance, membrane invaginations, or degradation55,60–66. The cytoplasm may undergo significant changes including acidification, organelle rearrangement, cytoskeleton modulation with fragmentation and shortening of microtubules, lower quantity of vacuoles, viral particles present inside vacuoles, formation of Golgi apparatus-like vesicles, among others59,61,63,64,67–72 (Fig. 2a).
Viral infection can also modulate the energy of the host, redirecting it to the replication site, often involving the recruitment of mitochondria to VFs60,65,73,74. Mollivirus sibericum infection results in smaller and elongated mitochondria, although host cells did not present changes in cellular adhesion and morphology62. Despite that, cell rounding is one common CPE, observed after GVs infection (Fig. 2b)75–78. Studies also reported loss of cell adhesion decreased motility, and intercellular bridges or rotational behavior55,56,76,79,80. A peculiar CPE is cellular aggregation (“bunches”)77,81,82. This mechanism was suggested to increase viral dissemination during Tupanvirus infection, as mannose-binding protein gene transcripts are significantly increased at earlier times of infection (1, 2 and 4 h post-infection [HPI]) and precedes the formation of bunches (6 HPI) between infected and uninfected cells.
A. polyphaga and V. vermiformis morphology modifications upon giant viruses’ infection
Fewer studies described CPE in A. polyphaga during GVs infection. Acanthamoeba polyphaga mimivirus (APMV) infection leads to nucleus enlargement, prominent VF formation in the cytoplasm, multivesicular bodies and endoplasmic reticulum (ER) membranes appearance around VF, stimulus of ER synthesis, significant structural alteration in microtubules and actin microfilaments resulting in cells becoming smooth, rounded, and losing motility83–85. Marseillevirus infection induces similar VFs to the observed in A. castellanii, with lack of a clear boundary with cytoplasm67,86. Furthermore, cell rounding with formation of cell chains was reported for the first time after a viral infection in amoeba87.
Over the past decade, studies implemented V. vermiformis as a new isolation platform, leading to the discovery of new GVs12,13,88–94. Their infection resulted in cell rounding and loss of adhesion88,94, and cell stretching with increased motility before cell rounding12. The formation of “blebs” and cell bunches seen in V. vermiformis, were previously reported in A. castellanii12,73,91. A distinct response was the encystment of neighbor non-infected cells after the exposure to infected cell-released non-proteic soluble factors, serving as a defense mechanism, as those cysts become unviable trapping the virus inside90. Nuclear alterations of infected V. vermiformis include loss of the rounded shape, decrease in surface area, or nucleus disappearance12,13,90–94. Furthermore, clandestinovirus formed its VF within the nucleus88. V. vermiformis’ mitochondrias were also reported to be recruited around VFs12,13,90,91,93,94. Investigating the unique cellular responses in amoebae to various GVs is essential for establishing correlations in host-virus interaction and comparing potential CPEs to other types of host cells.
Other protists’ morphology and behavior in response to virus inoculation
When exploring protist susceptibility to Tupanvirus beyond amoebae, it was found that the ciliate Tetrahymena sp. experience cytotoxic effects in response to the viral presence. The morphological changes included gradual vacuolization, nuclear degradation, loss of motility, and formation of vesicles, similarly to the observed in A. castellanii. However, the cilliate does not support viral replication61.
In contrast to the extensive use of amoeba to study viral infections, there are relatively fewer reports focusing on interactions between viruses and other protists. Among algae protists, the viral infection of Aureococcus anophagefferens, a component of ocean blooms, can trigger nucleus and organelle degradation (such as chloroplast) in some infected cells, cell lysis, and loss of the outer polysaccharide glycocalyx during viral infection, even in environmental samples95–99. In addition, viral infection of another bloom-forming protist, Heterosigma akashiwo, pointed to nucleus degradation and damaged cell wall100. When infected by HaNIV virus, cell nucleus presented margination of heterochromatin, but no morphological changes101. Conversely, the infection by HaV01 and HaV triggered cell rounding, and loss of cell motility, together with chloroplast degradation for HaV infection102,103. Interestingly, chloroplasts have been reported to remain intact until cell lysis104. Vacuolation, disintegrated cytoplasm and ER swelling were reported during HaRNAV infection in H. akashiwo105. Furthermore, in the haptophyte algae Chrysochromulina parva and Prymnesium parvum the infection by their respective viruses resulted in absence or degradation of cell nucleus106,107.
In the diatoms Chaetoceros sp, Guinardia delicatula and Rhizosolenia setigera the infection by their respective viruses resulted in common CPEs: degradation of chloroplasts and/or photosynthetic pigments98,108–113. Few Chaetoceros species additionally exhibited cytoplasm degradation108–111,114, and nucleus or nucleolus degradation was reported in Chaetoceros salsugineum and Guinardia delicatula98,115. Similarly, chloroplast shrinkage and chlorophyll degradation were suggested during the infection of the phytoplankton coccolithophore E. huxleyi by its lytic virus116,117. Nucleus degradation was identified in infected E. huxleyi cells, as well as cellular aggregation, likely as a defense strategy to sink infected aggregated cells116–118. Furthermore, mitochondrial damage with vacuolar acidification was described during the infection byf EhV99B1119.
Regarding flagellates, the protist Giardia sp. has two life cycle stages, a trophozoite and a cyst form120,121. Only its trophozoite stage is susceptible to viral infection, and few morphological changes have been reported for Giardia canis and Giardia lamblia infected by GCV and GLV, respectively. During GCV infection, the protists presented vacuolization, enlargement of ER, and its cytoplasm became very loose, and during GLV only cell adherence was impaired upon infection122,123. Although, no cell death was reported for both studies, corroborating to the suggestion that giardiaviruses are released from the cell without triggering cell lysis121. G. duodenalis, however, experiences reduction in cell growth without apparent morphological changes124. In the free-living flagellate Bodo saltans infected by the virus BsV, morphological changes included degraded nucleus and intracellular structures, with lipid vesicles migrating through the VF, while kinetoplast remained intact for longer96.
The bloom-forming dinoflagellate Heterocapsa circularisquama, associated with red tides and bivalve mortality, can be infected by RNA and DNA viruses. During the DNA virus infection, a VF is formed with granular or fibrous material inside, in addition to organelles disruption125–127. However, the RNA virus infection triggered organelles, nuclear and chlorophyll-a degradation, and loss of cellular motility128–130. Interestingly, some of the cells were resistant to the RNA virus infection, but not the DNA one, and the non-resistant cells underwent cell lysis129,131. Furthermore, the dsDNA virus infection triggered damage to Symbiodinium cell nucleus initially, and later to organelles132. The dinoflagellate Gymnodinium mikimotoi shows nuclear degradation and swollen chloroplasts upon viral infection133. Nuclear degradation is also reported to the nonphotosynthetic stramenopile from the genus Sicyoidochytrium upon viral infection134. Overall, relating viral responses across different protist organisms and viruses remains challenging, due to the limited number of studies in this field and the scarcity of available isolates for research.
Metabolic changes in protist cells during virus infection
To investigate cell-virus interactions at the molecular level, different approaches, including transcriptomics and proteomics, have been employed, revealing that each virus modulates their host in a unique manner135,136.
Changes in Acanthamoeba sp
Proteomic analysis of A. castellanii infected by Mollivirus sibericum identified a few upregulated proteins related to histones, autophagy, DNA synthesis and packaging; and a few downregulated proteins with no clear functional relationship62. Together with the fact that this virus is released through exocytosis rather than cell lysis, an overall cellular integrity is suggested throughout the replication cycle62. Likewise, few genes were deregulated at the transcriptional level during Marseillevirus infection137. Transcripts expression across early, intermediate, and late time periods showed only 48 genes deregulated among these periods (Table 1), with the upregulation of proteins mainly linked to exosome secretion, transfer RNA biogenesis and genetic information processing; and the downregulation of proteins linked to translation apparatus (rRNA related proteins), tRNA encoding genes, carbohydrate metabolism and lipid biosynthesis137. For Marseillevirus, amoebal and mitochondrial transcript levels decreased after 1 h post-infection (HPI)137.
Table 1.
Overview of the proteomic and transcriptomic studies of different protist organisms infected by their respective viruses
| References | Protist organism | Virus | Technique | Time post infection | FoldChange | Down | Up |
|---|---|---|---|---|---|---|---|
| Rodrigues et al. (2020)137 | A. castellanii | Marseillevirus | RNAseq | 0, 1, 2, 4, 5, 6, 8, 10, and 12HPI | <−1 and >1 | 28 | 19 |
| Legendre et al. (2015)62 | A. castellanii | Mollivirus sibericum | LC-MS/MS | 0 to 6HPI | <−2 and >2 | 38 | 30 |
| Zhang et al. (2021)138 | A. castellanii | Medusavirus | RNAseq | 8 to 16HPI | <−0.445 and >0.364 | 7970 | 2657 |
| Moniruzzaman et al. (2018)160 | A. anophagefferens | Aureococcus anophagefferens virus | RNAseq | 5 min | <−1.5 and >1.5 | 588 | 412 |
| 30 min | ≈ 865 | ≈ 505 | |||||
| 1HPI | 82 | 0 | |||||
| 6HPI | ≈ 1060 | ≈ 690 | |||||
| 12HPI | ≈ 1260 | ≈ 1260 | |||||
| 21HPI | ≈ 1430 | ≈ 1620 | |||||
| Poimala et al. (2022)138 | Phytophthora cactorum | PcBV1 & 2 | RNAseq | -a | <−2 and >2 | 23 | 10 |
| Nanoflow reverse-phase LC-MS | -a | <−1.19 and >1.37 | 17 | 36 | |||
| Provenzano et al. (1997)171 | T. vaginalis | TVV | 2D | - | - | 41 | 47 |
| He et al. (2017)174 | T. vaginalis | TVV | iTRAQ labeling | - | <−1 and >1 | 21 | 29 |
| Rada et al. (2022)175 | T. vaginalis (exossome vesicle) | TVV | LFQ-MS | - | <−2 and >2 | 55 | 20 |
“FoldChange” means threshold utilized by each study to determine when a protein or transcript presents increased expression (upregulated - “up”) or decreased expression (downregulated - “down”). Other protists are not included on this table since information on the number of deregulated proteins or transcripts was not the focus of the publication or this data was not provided.
aThe virus was removed from the protist strain, thus there was no time post-infection.
Contrastingly, Medusavirus infection in A. castellanii resulted in thousands of differentially expressed host genes, although mitochondrial gene modulation resembled the pattern of Marseillevirus infection. Only 25% of nuclear host genes, mainly related to ribosome and proteosome pathways, increased its expression between 8 to 16 HPI, suggesting the host to be suffering protein degradation, along with increased viral protein synthesis. The remaining 75% experienced decreased expression during that same period. Gene ontology suggested a reduction in transport activity, although at 48HPI many of these genes were downregulated, in addition to an upregulation of encystment-mediated genes138.
Furthermore, Tupanvirus inoculation triggered rRNA shutdown only by the presence of viral particles, unrelated to viral replication61. Genomic analysis of tupanviruses revealed the involvement of the ribonuclease T2, an enzyme related to the reduction of the physiological activity and phagocytosis capacity in protist hosts139.
Bloom-forming and dinoflagellate protists
Infected by lytic and non-lytic viruses, the coccolithophore E. huxleyi, is ecologically crucial for marine carbon flux140,141. The correlation between transcriptomics and metabolomics analysis during viral infection indicated that early during infection (1–4 HPI), there was an upregulation of sphingolipid biosynthesis and glycolysis shuffling energy to fatty acids biosynthesis. Later (24HPI), a shift to the activation of the pentose phosphate pathway to produce nucleotides occurred, while glycolysis and fatty acids became downregulated117. Similar results were found for genes related to sphingolipid metabolism, such as the upregulation of dihydroceramide desaturase (DCD) at 6HPI and its downregulation at 45HPI142. E. huxleyi DCD transcript, together with serine palmitoyl transferase (SPT) transcript (another enzyme from sphingolipid biosynthesis pathway) level results pointed to the same tendency of a decrease in these host transcripts production in the environment143. Other studies also found lipid modulation in E. huxleyi during viral infection, including fatty acids and highly saturated triacylglycerols, and lipid metabolism was suggested to be regulated by modulation of the PI3K-Akt-TOR signaling pathway144–152.
Furthermore, a single cell transcriptomics analysis of E. huxleyi during infection revealed an early shutdown of nuclear transcripts, with mitochondrial and chloroplast transcripts initially higher but gradually declined153. Indeed, there was a decrease in genes involved in photosynthesis at 6, 12 and 24 HPI, although some cells seemed to be intact and photosynthesizing148. Enriched functions associated with E. huxleyi responding to viral infection included modified amino acid, lipid binding, porin activity, calcium channel activity, pore complex, cell outer membrane, bacterial-type flagellum functions, among others154. High expression of glycolysis and nucleotides biosynthesis related genes were reported when studying single protist cells from a coccolithophore bloom151. In addition, there was an upregulation of autophagy related genes, with decreased expression of negative regulatory factors, such as PI3K, and an increase of reactive oxygen species (ROS), which is all related to the programmed cell death (PCD) triggered in E. huxleyi after infection119. In fact, 20 ROS scavenging genes were impacted after viral infection, and ROS-related genes were found to be increased through transcriptomic analysis155,156. Cell death induced by viral presence could occur before viral particles release, which can be associated with cell autophagy as a defense strategy to avoid viral dissemination, although autophagy is also essential for viral propagation117,157. E. huxleyi cell cycle can be affected by virus infection through the modulation of cyclin expression, and host life cycle genes have been found to be impacted during viral takeover of the protist in environment149,158,159. Altogether, these findings suggest substantial cellular changes in E. huxleyi after viral encounter, influencing factors related to cell death, modulation of energy, and specific lipids production, alongside a decline in nuclear activity, potentially playing a role in the modulation of oceans blooms153.
Another bloom forming protist, Aureococcus anophagefferens, exibited transcripts downregulated at six different time points during viral infection. A massive deregulation started as early as 5 min post-infection and continued after 6HPI. Downregulated proteins indicated the suppression of pathways related to host cytoskeleton formation, photosynthesis, fatty acid metabolism, and carbohydrate biosynthesis. The upregulated group of proteins indicated the activation of host cellular respiration, transcription, protein synthesis, polyamine biosynthesis, and RNA processing. Transcripts related to host defense mechanism were likely suppressed. This dramatic cell modulation indicates the virus –host interaction in a time dependent manner until cell lysis occurs, correlating with the virus’s role in brown tide blooms160.
For Phaeocystis globosa, viral infection prevented the accumulation of polyunsaturated fatty acids, decreased protist photosynthetic performance and upregulated mitochondrial respiration, triggered the fragmentation of DNA and activation of caspases, and prevented P. globosa to release star-like structures, which in turn affects host carbon assimilation161–164. The correlation among such changes is speculative, suggesting an association with the lysis mechanism triggered in this organism post-viral infection.
Comparing the transcriptomic responses of the dinoflagellates Symbiodinium tridacnidorum and Symbiodinium C3 under or not UV light exposure (with and without viral replication, respectively), both exhibited upregulated viral transcription and related terms, but only S. tridacnidorum triggered downregulation of transcripts related to host-response to viral infection165.
Protists responsible for causing human or plant disease
After the discovery of Trichomonas vaginalis virus (TVV), few studies investigated Trichomonas vaginalis cell responses to infection166,167. Initial findings described that most clinical isolates had the presence of a dsRNA icosahedral virus, and that the expression of a major immunogen (P270) on cell surface, and the expression of Ig-degrading proteinases, were directly correlated with its presence168–170. An overview of the protein expression pattern of T. vaginalis after TVV infection found at least 47 expressed and 41 suppressed proteins linked to the virus171. Isolates of T. vaginalis might be infected by up to 4 different strains of TVV, and only the cells infected by TVV2 and TVV3 are capable of inducing P270 protein expression172,173. Proteomic studies comparing infected and uninfected cells detected the upregulation of adhesin proteins related to the pathogenicity of T. vaginalis when infecting humans174. Further, metabolic enzymes, ribosomal and heat shock proteins were also differentially impacted175. Extracellular vesicles of T. vaginalis infected by TVV exhibit differences in protein content, such as a protein responsible for increasing adherence, suggesting enhanced exosome binding and pathogenicity175. Overall, TVV plays an important role in T. vaginalis dynamics by modulating different genes that aid the protist to successfully establish in its host.
After the discovery of endosymbiotic viruses in Leishmania’s cytoplasm, most studies focused on the relation Leishmania’s pathogenicity to humans when infected or not by one of its two viruses, LRV and LBV176–178. The metastasizing L. Viannia had a higher amount of LRV1, suggesting that the virus triggers parasitic resistance179. A study looked for transcriptional changes in L. guyanensis and L. major when removing their respective viruses LRV1 and LRV2180. As a result, L. guyanensis did not suffer significant changes, with only 2 differentially expressed genes. On the other hand, proliferation rate of L. major cells decreased, with 67 upregulated and 20 downregulated transcripts after its virus removal. Among the downregulated group, there was a cyclin related to growth kinetics; and the membrane-bound acid phosphatase 2 enzyme, which has been already suggested to affect the biology of Leishmania. The upregulated group presented transcripts related to autophagy, cell response to various stimuli, and nucleosome assembly180.
Oomycetes from the genus Phytophthora are responsible for causing great damage to agriculture181. A metagenomic study of Phytophthora condilina resulted in 15 different putative viruses identified182. Moreover, GVs sequences have been identified in the genome of Phytophthora parasitica183. Phytophthora endornavirus 2 (PEV2) and Phytophthora endornavirus 3 (PEV3), are suggested to stimulate zoosporangium development and inhibit hyphal growth, also reducing the host oomycete sensitivity to the antifungal metalaxyl184. In addition, seven different viruses were able to replicate in P. infestans, and viral infection apparently did not affect the cell host morphology but stimulated the growth of the mycelium mass185. Notably, the infection of P. infestans by PiRV-2 is suggested to increase the protist sporulation, which in turn is associated with a likely hypervirulent factor against its plant host186. However, sporulation of P. cactorum infected by PcBV1 & 2, seemed deeply affected when compared to P. cactorum with PcBV1 & 2 ablation. Its sporangia production and size decreased during viral infection, together with reduced hyphal growth187. This same research used P. cactorum as a model to explore the transcriptomic and proteomic changes after removing PcBV1 & 2 from the protist, and identified 10 up- and 23 downregulated transcripts, as well as 36 up- and 17 downregulated proteins187. The excreted protein elicitin was found upregulated in the infected cells187. This protein is related to the suppression of the plant immune response but can also be recognized by the plant and become a factor that reduces protist pathogenicity188,189. Overall, Phytophthora can be pathogenic or not to its plant host depending on many factors, including the infecting virus type.
Cellular resistance strategies after stress exposure
All living organisms are subjected to the exposure of extrinsic factors that can trigger a stress response at the cellular level. Stress refers to any environmental condition that can be harmful to cells and induce physiological changes that disturb homeostasis190. Biotic stress is caused by interaction with other living beings that will act as stressors, particularly parasitic relationships involving viruses, bacteria, and fungi. Abiotic stress arises from physicochemical factors that can affect cell physiology, such as pH, temperature, radiation, osmolarity and chemical molecules190,191. Despite its disruption of homeostasis, environmental stress is an important factor when it comes to evolution. Cells capable of restoring homeostasis after stress, called acclimatized cells, are favored through natural selection190. Protist cells employ various responses to stressors programmed cell death (PCD). One example is the response of Peridinium gatunense to oxidative stress. At the end of the algal blooms caused by this dinoflagellate, CO2 becomes significantly limited, leading to the production of reactive oxygen species which trigger PCD192. Aureococcus anophagefferens, responds to stress conditions such as low concentrations of inorganic nitrogen and inorganic phosphorus through a transcriptional shift: transcripts related to nitrogen transport and metabolism, and transcripts encoding enzymes that hydrolyze organic phosphorus or alleviate arsenic toxicity are upregulated in these scenarios. In addition, in the context of low light levels, which acts as a stress factor, transcripts encoding enzymes that catabolize organic compounds, restructure lipid membranes, or are involved in the biosynthesis of sulfolipids are upregulated to restructure lipids and renovate the photosynthetic apparatus. The cell undergoes several physiological changes to maintain the survival and ecological success of the species193.
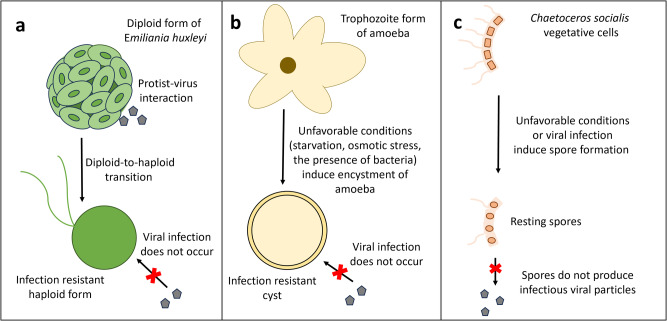
Spore formation as a response to infection
The marine diatom Chaetoceros socialis responds to stress caused by adverse conditions of temperature, light and lack of nutrients through spore formation. Viruses are widely distributed and persistent biological stressors, and some of them are also capable of inducing spore formation194. For instance, CsfrRNAV01 can effectively induce significant spore formation in C. socialis. Viral infection acts as a biotic trigger that induces a substantial formation of heavily silicified spores. Interestingly, their spores do not produce infectious viral particles, making this shift in the life cycle an effective defense strategy against viruses and preventing the loss of a portion of the protist population194.
Emiliania huxleyi life cycle shift as a defense strategy
Another protist that relies on a life cycle shift is E. huxleyi, a photosynthetic organism widely distributed throughout the oceans. It has two independent phases, haploid and diploid, each with distinct morphologies195. The haploid form (N) consists of biflagellated cells covered by thin, organic, non-mineralized scales. In contrast, the diploid (2 N) is nonmotile and contains minute calcite platelets, named coccoliths196. The latter is responsible for large natural algal blooms, an important environmental problem197. It is known that calcium plays an important role during the viral entry process into the host cell in different infection models198–201. One significant difference between the N and 2 N phases of E. huxleyi is the calcium metabolism being considerably greater in the diploid phase than in the haploid phase, affecting virus-host interactions197. Haploid cells are resistant to infection and subsequent lysis by Emiliania huxleyi virus, whereas this infection contributes to the decline of diploid populations during algal blooms197. Evidence suggests that those lytic viruses are responsible for demising the algae population and terminating algae blooms. Thus, Emiliania huxleyi viruses are responsible for regulating the population of this coccolithophore both in abundance of cells in the environment and in composition195,197,202,203. Such protist-virus interaction can directly activate the life cycle transition, since the oxidative stress caused by viral infection can trigger the diploid-to-haploid shift197. Viral infections are known to increase the production of reactive oxygen species in 2 N cells, which induces the activation of metacaspases and, thus PCD is triggered204. However, oxidative stress response leads not only to cell death but also to induction of the aforementioned shift196. Although studies conducted using 1 N E. huxleyi strains found viral RNA and small amounts of EhV glycosphingolipids within the cells146,205, a later study using the host resistant strains have not found any EhV genetic material, which suggests that there is no resistant strain bearing any form of the virus195. Haploid virus-resistant cells are produced as a response to viral infection195. This is referred to as the ‘Cheshire Cat’ strategy, in which the organism escapes the parasite by shifting to a life stage that is infection-resistant197.
Encystment as amoebae’ resistance form against viral infections
The ‘Cheshire Cat’ theory extends to describe a relationship between a giant virus and an amoeba as well. Some amoebae hosts are also capable of shifting to an infection-resistant life form206. Amoebae have two life cycle stages, trophozoites and cysts. The trophozoite stage is the one that predominates when environmental conditions are favorable, such as nutrient supply and temperature, while the cyst is the form of resistance191,206. Under unfavorable conditions such as starvation, or osmotic stress, amoeba trophozoites undergo both cellular and molecular regulatory processes that lead to encystment206–208. Amoeba cysts are more resistant to adverse conditions. For instance, A. castellanii cysts can survive five cycles of freeze–thawing, exposure to high doses of UV or gamma radiation209. Furthermore, giant viruses, such as mimiviruses and marseilleviruses, can only infect amoeba when they are in the trophozoite form. The shift to a non-permissive cell stage is an astounding stratagem for escaping giant viruses206. Some viruses, however, have developed strategies to circumvent this resistance mechanism. Mimivirus isolates and Tupanvirus, can prevent the encystment of amoebae, by reducing the expression of amoebal encystment-mediating subtilisin-like serine proteinase, a proteinase involved in trophozoite encystment. By doing so, the virus can effectively block the formation of the host’s form of resistance and continue its cycle90,206,210.
The tripartite systems
The tripartite systems are an interesting dynamic in which the giant virus-host relationship is accompanied by a further member: a virophage or a bacteria. Such tripartite systems are found, for instance in amoebae. Virophages are viruses capable of infecting giant viruses, they multiply inside the VFs, meaning that they do not replicate if giant viruses are absent. Therefore, the replication of the virophages interferes with the cycle of the giant virus, interrupting it and thus reducing the amoeba mortality related to the viral infection79,211. The first virophage discovered was sputnik, a dsDNA virus associated with mamavirus. Some types of virophages increase the formation of abnormal giant virus particles, reduce infectivity and the ability to lyse their host cell211. The second virophage discovered was the mavirus associated with the Cafeteria roenbergensis virus (CroV). Like the sputnik, this virophage inhibits the cycle of the giant virus. Interestingly, mavirus can integrate its genome into the protist’s genome, but its genes are only expressed when CroV infection occurs. This reactivation is not enough to prevent cell lysis; however, it causes mavirus particles to be released into the environment and protect neighboring cells79,212,213. Despite this, it has been shown that not all virophages negatively affect the cycle of giant viruses; zamilon, for instance, does not affect the ability of the giant virus to lyse the amoeba214. There are also bacterial endosymbionts capable of protecting amoebae from being infected by giant viruses, such as Parachlamydia acanthamoebae, which manages to suppress the viral replication of sympatric Viennavirus, APMV and Tupanvirus by inhibiting the maturation of the VF, which results ultimately in the survival of the infected amoebae215. Such complex systems are also present in other protists, such as Cryptomonas sp. SAG25.80, which is a quadripartite system216. A single cell harbors a phage (MAnkyphage) and two bacterial endosymbionts (Grellia numerosa and Megaira polyxenophila), and a complex community of organelles and selfish elements216. As discussed above, protist organisms have developed various strategies throughout their evolutionary history to survive unfavorable environmental conditions and escape viral infection. A graphical representation of the main strategies discussed is shown by Fig. 3.
Fig. 3. Overview of the strategies employed by different protist life forms to resist stress factors, such as virus infection.
a Upon interaction with viruses, the cocolithophore E. huxleyi shifts to a life form that is resistant to infection195. b Amoebae encyst under unfavorable conditions, and the cysts formed are not able to be infected by a virus206. c Under unfavorable conditions Chaetoceros socialis cells transform into resting spores that are unable to produce infectious viral particles114,194,225. Red text: Cellular resistance strategies after stress exposure.
Summary and concluding remarks
Early eukaryotes are believed to have emerged more than a billion years ago in the Proterozoic oceans217. Despite the ongoing efforts to characterize and explore the diversity of viruses that infect protists, few isolates have been described to date, and the majority of the known diversity was discovered through metagenomic studies. One of the most notable groups of viruses, with the higher number of viral isolates, are the representatives of the phylum Nucleocytoviricota, an ancient component of the eukaryotic virome32,218. Each of these organisms have undergone extensive co-evolution, resulting in their present-day diversity, which encompasses a broad array of eukaryotes as hosts, and different taxa of viruses as parasites48. Even though certain viruses of this phylum possess a highly diverse set of genes involved in different metabolic pathways, their obligated intracellular parasitic nature necessitates an intricate relationship with their hosts.
Viruses often disrupt normal cellular processes to acquire the necessary elements for viral replication, or as a result of dysbiosis originated from the infection process. The intensification of studies on viral-protists host interactions is enhancing our understanding of the molecular mechanisms underlying their interactions, shedding light on viral replication and host defense mechanisms. Following viral infection, host cells often go through significant alterations and begin to exhibit a distinct nature49. Some of the alterations can be observed microscopically and are considered as an indicative of viral infection. These particular alterations, known as cytopathic effects, typically comprise morphological changes that may manifest as noticeable variations in cell shape, size and integrity. In the case of protists, it has been observed that these changes can include increased cellular motility, rounding of cells, reduced adhesion, or increased adhesion to other cells, and cell lysis12,80–83,85–92. Furthermore, morphological modifications are accompanied by intracellular alterations. During a viral infection, intracellular changes in protists cells include structural transformations of the cytoskeleton, degradation of chloroplasts or photosynthetic pigments (if present), recruitment of mitochondria, membranes, vesicles, etc. Even if the VF is exclusively formed in the cytoplasm and the nucleus remains intact throughout viral replication, it can still go through changes, albeit temporary, during the course of the infection12,66,109,117,128,133,219,220. In addition, viral infections induce functional changes in host cells through widespread genetic and metabolic reprogramming. Apart from inducing the expression of viral genes, viruses can elevate the expression of host genes associated with processes such as DNA synthesis, energy generation, and the modification of lipid, protein, and nucleic acid biosynthesis pathways. These alterations collectively contribute to the complex interplay between viruses and host cells during infection62,117,135–138,140,144,153,154,167. Still, deeper exploration of virus-host dynamics is much needed, and can help elucidate host factors influencing susceptibility, pathogenicity, resistance, and protists basic cytology.
As aforementioned, protists are a diverse group of eukaryotic organisms that exhibit a remarkable adaptability when exposed to harsh conditions and invasive encounters. To thrive in ever-changing environments and fend off predators and parasites, protists have evolved a repertoire of sophisticated cellular responses to stress, often including the formation of resistant life forms. Moreover, as they occupy a wide range of ecological niches, protists have a dynamic interplay with other organisms with which they form communities, including viruses. They can act as regulators of population diversity and density, as in the case of Emiliania huxleyi viruses, which are responsible for ending coccolithophorid blooms195,197. Furthermore, the interaction between viruses and protists can also be very complex, as in tripartite systems, in which the virus-host relationship involves another member which may interfere in the replication cycle of the giant virus and protect amoebae from infection79,212,213. Competitive interactions are one of the main driving forces that lead to the diversity and complexity of life on our planet, and the investigation into how viruses influence protists communities, and their ecological interactions can help understand how virus-driven evolution shapes the diversity, dynamics and the impacts in ecosystems, including their role in nutrient cycling and energy flow.
Many are the taxonomic groups of viruses that infect protists, and together, they are present in the majority of Earth’s ecosystems. One outcome of such interactions is horizontal gene transfer. Viruses might act as agents for transferring genetic material across species boundaries, including protists, while certain species are hot spots for horizontal gene transfer (HGT) among viruses, eukaryotes, prokaryotes and mobile elements160,221. This process has the potential to introduce novel genes or traits into protist genomes, and potentially influence their biology, adaptation, and evolution222.Furthermore, it contributes to studies on the evolution of protists and its viruses, providing insights into the coevolutionary dynamics, and as these organisms often have unique genomic features with a high percentage of ORFan genes60, the study of the viral genomes can uncover novel genes and genetic features that may be noteworthy in protist hosts. This can expand our understanding of genetic diversity, the impact of viral genes on protists’ biology, ecology and evolution, as well as potential functions within the protists.
Protists and viruses play a crucial role in Earth’s ecosystems. Understanding how viruses impact protist biology can provide valuable insights into several fields of science, shedding light on evolutionary biology, ecosystem dynamics, nutrient cycling, and biomedical research. However, our knowledge of unicellular organisms and microorganisms known to humankind remains anthropocentric, with little known about the true diversity of these beings. Therefore, it is important that prospecting and characterization studies of these organisms are carried out to better elucidate the complete picture. In summary, viruses can serve as valuable tools and subjects of study in the field of protist biology. Their interactions with protist hosts, unique genomes, and ecological roles can offer new information that enhances our understanding of protist biology and its broader implications for ecosystems and biotechnology.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank our colleagues from Laboratório de Vírus—UFMG for their technical support. We acknowledge financial support from Rede Vírus—Ministério da Ciência, Tecnologia e Inovações (MCTI), Câmara Pox—405249/2022-5. We thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), grant number 88882.348380/2010-1, Fundação de Amparo à Pesquisa do estado de Minas Gerais (FAPEMIG), Programas Institutos Nacionais de Ciência e Tecnologia (INCT), grant number 406441/2022-7, chamada 58/2022, and Pró-Reitorias de Pesquisa e Pós-Graduação of UFMG, and the Centre for New Antibacterial Strategies (CANS) of the Arctic University of Norway (project ID #2520855). R.A.L.R. and J.S.A. are CNPq researchers.
Author contributions
V.F.Q., J.M.T. and B.B.B. performed the literature review and prepared the figures and tables. R.A.L.R., G.M.F.A. and J.S.A. designed the text structure and revised the manuscript. All authors contributed to the writing.
Peer review
Peer review information
Communications Biology thanks Chuan Ku, Guifré Torruella and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Manuel Breuer. A peer review file is available.
Data availability
No datasets were generated or analysed for this review paper. Literature cited is shown in the reference list and in the Supplementary Data File.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Gabriel Magno de Freitas Almeida, Jonatas Santos Abrahao.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06001-2.
References
- 1.Owen, R. Kingdom Protozoain Paleontology. 1–17 (Edinburgh, Colin Macfarquhar & Andrew Bell,1858).
- 2.Cavalier-Smith, T. Kingdom Protozoa and Its 18 Phyla. Microbiol. Rev.57, 953–994 (1993). [DOI] [PMC free article] [PubMed]
- 3.Jahn TL, Votta JJ. Locomotion of protozoa. Annu. Rev. Fluid Mech. 1972;4:93–116. [Google Scholar]
- 4.Adl SM, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryotic Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 5.Caron DA, Worden AZ, Countway PD, Demir E, Heidelberg KB. Protists are microbes too: a perspective. ISME J. 2009;3:4–12. doi: 10.1038/ismej.2008.101. [DOI] [PubMed] [Google Scholar]
- 6.Adl SM, et al. Diversity, nomenclature, and taxonomy of protists. Syst. Biol. 2007;56:684–689. doi: 10.1080/10635150701494127. [DOI] [PubMed] [Google Scholar]
- 7.Brusca, R., Shuster, S. & Moore, W. Invertebrates (2016).
- 8.da Silva Ferreira M, de Souza Gonçalves D, Medeiros EG, Peralta JM, Guimarães AJ. “Feast-Fit-Fist-Feat”: overview of free-living amoeba interactions with fungi and virulence as a foundation for success in battle. Curr. Trop. Med. Rep. 2021;8:18–31. [Google Scholar]
- 9.Mungroo MR, Siddiqui R, Khan NA. War of the microbial world: Acanthamoeba spp. interactions with microorganisms. Folia Microbiologica. 2021;66:689–699. doi: 10.1007/s12223-021-00889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Queiroz VF, et al. Amoebae: hiding in plain sight: unappreciated hosts for the very large viruses. Annu. Rev. Virol. 2022;9:79–98. doi: 10.1146/annurev-virology-100520-125832. [DOI] [PubMed] [Google Scholar]
- 11.Korn ED, Weisman RA. Phagocytosis of latex beads by Acanthamoeba. II. Electron microscopic study of the initial events. J. Cell Biol. 1967;34:219–227. doi: 10.1083/jcb.34.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Souza F, et al. In-depth analysis of the replication cycle of Orpheovirus. Virol J. 2019;16:158. doi: 10.1186/s12985-019-1268-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva, L. C. F. et al. Microscopic analysis of the tupanvirus cycle in Vermamoeba vermiformis. Front. Microbiol.10, 671 (2019). [DOI] [PMC free article] [PubMed]
- 14.Andrade, A.C.S.P. et al. Filling knowledge gaps for mimivirus entry, uncoating, and morphogenesis. J. Virol.91, e01335-17 (2017). [DOI] [PMC free article] [PubMed]
- 15.Arantes TS, et al. The large marseillevirus explores different entry pathways by forming giant infectious vesicles. J. Virol. 2016;90:5246–5255. doi: 10.1128/JVI.00177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi, Y. et al. The ecology and evolution of amoeba-bacterium interactions. Appl. Environ. Microbiol. 87, e01866-20 (2021). [DOI] [PMC free article] [PubMed]
- 17.Scola BL, et al. A giant virus in amoebae. Science. 2003;299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 18.Horn M. Bacterial endosymbionts of free-living amoebae. J. Eukaryot. Microbiol. 2004;51:509–514. doi: 10.1111/j.1550-7408.2004.tb00278.x. [DOI] [PubMed] [Google Scholar]
- 19.Samba-Louaka A, Delafont V, Rodier MH, Cateau E, Héchard Y. Free-living amoebae and squatters in the wild: ecological and molecular features. FEMS Microbiol. Rev. 2019;43:415–434. doi: 10.1093/femsre/fuz011. [DOI] [PubMed] [Google Scholar]
- 20.de Souza GAP, Queiroz VF, Coelho LFL, Abrahão JS. Alohomora! What the entry mechanisms tell us about the evolution and diversification of giant viruses and their hosts. Curr. Opin. Virol. 2021;47:79–85. doi: 10.1016/j.coviro.2021.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Kao, S., Kao, C.-F., Chang, W. & Ku, C. Widespread distribution and evolution of poxviral entry-fusion complex proteins in giant viruses. Microbiol. Spectr.11, e0494422 (2023). [DOI] [PMC free article] [PubMed]
- 22.De la Fuente IM, et al. The nucleus does not significantly affect the migratory trajectories of amoeba in two-dimensional environments. Sci. Rep. 2019;9:16369. doi: 10.1038/s41598-019-52716-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DuBose, J. G., Robeson, M. S., Hoogshagen, M., Olsen, H. & Haselkorn, T. S. Complexities of inferring symbiont function: paraburkholderia symbiont dynamics in social amoeba populations and their impacts on the amoeba microbiota. Appl. Environ. Microbiol.88, e0128522 (2022). [DOI] [PMC free article] [PubMed]
- 24.De la Fuente IM, López JI. Cell motility and cancer. Cancers. 2020;12:2177. doi: 10.3390/cancers12082177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeon, K. W. Nuclear control of cell movement in amoebae: nuclear transplantation study. Exp. Cell Res.50, 467–471 (1967). [DOI] [PubMed]
- 26.Lhee D, et al. Amoeba genome reveals dominant host contribution to plastid endosymbiosis. Mol. Biol. Evol. 2021;38:344–357. doi: 10.1093/molbev/msaa206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulz F, et al. Life in an unusual intracellular niche: a bacterial symbiont infecting the nucleus of amoebae. ISME J. 2014;8:1634–1644. doi: 10.1038/ismej.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shu L, et al. Symbiont location, host fitness, and possible coadaptation in a symbiosis between social amoebae and bacteria. Elife. 2018;7:1–25. doi: 10.7554/eLife.42660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas V, Greub G. Amoeba/amoebal symbiont genetic transfers: lessons from giant virus neighbours. Intervirology. 2010;53:254–267. doi: 10.1159/000312910. [DOI] [PubMed] [Google Scholar]
- 30.Enquist LW. Virology in the 21st century. J. Virol. 2009;83:5296–5308. doi: 10.1128/JVI.00151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang AL, Wang CC. Viruses of parasitic protozoa. Parasitol. Today. 1991;7:76–80. doi: 10.1016/0169-4758(91)90198-w. [DOI] [PubMed] [Google Scholar]
- 32.Krupovic M, Dolja VV, Koonin EV. The virome of the last eukaryotic common ancestor and eukaryogenesis. Nat. Microbiol. 2023;8:1008–1017. doi: 10.1038/s41564-023-01378-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oldstone MBA, Levine AJ. Virology in the next millennium minireview achievements led to the eradication of viral diseases. Cell. 2000;100:139–142. doi: 10.1016/s0092-8674(00)81690-6. [DOI] [PubMed] [Google Scholar]
- 34.Hershey AD, Chase M. Independent functions of viral protein and nucleic acid in growth of bacteriophage*. J. Gen. Physiol. 1952;36:39–56. doi: 10.1085/jgp.36.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hendrix RW. There 10 31 virus particles on earth, or more, or fewer? Proc. Natl Acad. Sci. USA. 1999;96:2192–2197. [Google Scholar]
- 36.Li T, et al. CRISPR/Cas9 therapeutics: progress and prospects. Sig. Transduct. Target. Ther. 2023;8:36. doi: 10.1038/s41392-023-01309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodrigues, R. A. L. et al. An anthropocentric view of the virosphere-host relationship. Front. Microbiol.8, 1673 (2017). Demonstration that the known virosphere is extremely anthropocentric and that viruses that infect protists are among the least described ones. [DOI] [PMC free article] [PubMed]
- 38.Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 39.Miller JH, Swartzwelder JC. Virus-like particles in an Entamoeba histolytica Trophozoite. Source: J. Parasitol. 1960;46:523. [Google Scholar]
- 40.Attoui H, et al. Micromonas pusilla reovirus: a new member of the family Reoviridae assigned to a novel proposed genus (Mimoreovirus) J. General Virol. 2006;87:1375–1383. doi: 10.1099/vir.0.81584-0. [DOI] [PubMed] [Google Scholar]
- 41.Coy S, Gann E, Pound H, Short S, Wilhelm S. Viruses of eukaryotic algae: diversity, methods for detection, and future directions. Viruses. 2018;10:487. doi: 10.3390/v10090487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diamond LS, Mattern CFT. Protozoal viruses. Adv. Virus Res. 1976;20:87–112. doi: 10.1016/s0065-3527(08)60502-3. [DOI] [PubMed] [Google Scholar]
- 43.Fukuhara T. Endornaviruses: persistent dsRNA viruses with symbiotic properties in diverse eukaryotes. Virus Genes. 2019;55:165–173. doi: 10.1007/s11262-019-01635-5. [DOI] [PubMed] [Google Scholar]
- 44.Khramtsov NV, Upton SJ. Association of RNA polymerase complexes of the parasitic protozoan Cryptosporidium parvum with virus-like particles: heterogeneous system †. J. Virol. 2000;74:5788–5795. doi: 10.1128/jvi.74.13.5788-5795.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malathi VG, Renuka Devi P. ssDNA viruses: key players in global virome. Virus Dis. 2019;30:3–12. doi: 10.1007/s13337-019-00519-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nibert ML, Woods KM, Upton SJ, Ghabrial SA. Cryspovirus: a new genus of protozoan viruses in the family Partitiviridae. Arch. Virol. 2009;154:1959–1965. doi: 10.1007/s00705-009-0513-7. [DOI] [PubMed] [Google Scholar]
- 47.Short SM, Staniewski MA, Chaban YV, Long AM, Wang D. Diversity of viruses infecting eukaryotic algae. Curr. Issues Mol. Biol. 2020;39:29–62. doi: 10.21775/cimb.039.029. [DOI] [PubMed] [Google Scholar]
- 48.Koonin, E. V. et al. Global organization and proposed megataxonomy of the virus world. Microbiol. Mol. Biol. Rev.84, e00061-19 (2020). [DOI] [PMC free article] [PubMed]
- 49.Forterre P. The virocell concept and environmental microbiology are the great virus comeback. ISME J. 2013;7:233–236. doi: 10.1038/ismej.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Castro IF, Volonté L, Risco C. Virus factories: biogenesis and structural design. Cell. Microbiol. 2013;15:24–34. doi: 10.1111/cmi.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bisio H, et al. Evolution of giant pandoravirus revealed by CRISPR/Cas9. Nat Commun. 2023;14:428. doi: 10.1038/s41467-023-36145-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.La Scola B. Looking at protists as a source of pathogenic viruses. Microb. Pathog. 2014;77:131–135. doi: 10.1016/j.micpath.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 53.Diesend, J., Kruse, J., Hagedorn, M. & Hammann, C. Amoebae, giant viruses, and virophages make up a complex, multilayered threesome. Front. Cell. Infect. Microbiol.7, 527 (2018). [DOI] [PMC free article] [PubMed]
- 54.Machado, T. B., de Aquino, I. L. M. & Abrahão, J. S. Isolation of giant viruses of Acanthamoeba castellanii. Curr. Protoc.2, e455 (2022). [DOI] [PubMed]
- 55.Philippe, N. et al. Pandoraviruses: Amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science(1979)341, 281–286 (2013). [DOI] [PubMed]
- 56.Alempic JM, et al. An update on eukaryotic viruses revived from ancient permafrost. Viruses. 2023;15:564. doi: 10.3390/v15020564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akashi M, Takemura M. Co-isolation and characterization of two pandoraviruses and a mimivirus from a riverbank in Japan. Viruses. 2019;11:1123. doi: 10.3390/v11121123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshikawa, G. et al. Medusavirus, a novel large DNA virus discovered from hot spring water. J. Virol.93, e02130-18 (2019). [DOI] [PMC free article] [PubMed]
- 59.Fabre E, et al. Noumeavirus replication relies on a transient remote control of the host nucleus. Nat. Commun. 2017;8:15087. doi: 10.1038/ncomms15087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boratto PVM, et al. Yaravirus: a novel 80-nm virus infecting Acanthamoeba castellanii. PNAS. 2020;117:16579–16586. doi: 10.1073/pnas.2001637117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abrahão J, et al. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 2018;9:749. doi: 10.1038/s41467-018-03168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Legendre M, et al. In-depth study of Mollivirus sibericum, a new 30,000-yold giant virus infecting Acanthamoeba. Proc. Natl Acad. Sci. USA. 2015;112:E5327–E5335. doi: 10.1073/pnas.1510795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quemin, E. R. et al. Complex membrane remodeling during virion assembly of the 30,000-year-old mollivirus sibericum. J. Virol.93, e00388-19 (2019). [DOI] [PMC free article] [PubMed]
- 64.Christo-Foroux, E. et al. Characterization of Mollivirus kamchatka, the first modern representative of the proposed Molliviridae family of giant viruses. J. Virol.94, e01997-19 (2020). [DOI] [PMC free article] [PubMed]
- 65.Pereira Andrade, A.C.S. et al. New isolates of pandoraviruses: contribution to the study of replication cycle steps. J. Virol.93, e01942-18 (2019). [DOI] [PMC free article] [PubMed]
- 66.Fukaya, S., Masuda, L. & Takemura, M. Analysis of morphological changes in the nucleus and vacuoles of Acanthamoeba castellanii following giant virus infection. Microbiol. Spectr.11, e0418222 (2023). Changes on the nucleus, vacuoles and A. castellanii cell shape are triggered during four different giant virus infection. [DOI] [PMC free article] [PubMed]
- 67.Goyal, N., Barai, A., Sen, S. & Kondabagil, K. Amoebal tubulin cleavage late during infection is a characteristic feature of mimivirus but not of marseillevirus. Microbiol. Spectr.10, e0275322 (2022). [DOI] [PMC free article] [PubMed]
- 68.Legendre M, et al. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. PNAS. 2014;111:4274–4279. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Legendre M, et al. Diversity and evolution of the emerging Pandoraviridae family. Nat. Commun. 2018;9:2285. doi: 10.1038/s41467-018-04698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takemura M. Morphological and Taxonomic properties of Tokyovirus, the first Marseilleviridae member isolated from Japan. Microbes Environ. 2016;31:442–448. doi: 10.1264/jsme2.ME16107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bertelli C, et al. Cedratvirus lausannensis – digging into Pithoviridae diversity. Environ. Microbiol. 2017;19:4022–4034. doi: 10.1111/1462-2920.13813. [DOI] [PubMed] [Google Scholar]
- 72.Doutre G, Philippe N, Abergel C, Claverie J-M. Genome analysis of the first Marseilleviridae representative from Australia indicates that most of its genes contribute to virus fitness. J. Virol. 2014;88:14340–14349. doi: 10.1128/JVI.02414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silva LKS, et al. Cedratvirus getuliensis replication cycle: an in-depth morphological analysis. Sci. Rep. 2018;8:4000. doi: 10.1038/s41598-018-22398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thomas V, et al. Lausannevirus, a giant amoebal virus encoding histone doublets. Environ. Microbiol. 2011;13:1454–1466. doi: 10.1111/j.1462-2920.2011.02446.x. [DOI] [PubMed] [Google Scholar]
- 75.Campos RK, et al. Samba virus: a novel mimivirus from a giant rain forest, the Brazilian Amazon. Virol. J. 2014;11:95. doi: 10.1186/1743-422X-11-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc. Natl Acad. Sci. USA. 2011;108:17486–17491. doi: 10.1073/pnas.1110889108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aoki K, et al. Marseilleviridae lineage b diversity and bunch formation inhibited by galactose. Microbes Environ. 2021;36:n/a. doi: 10.1264/jsme2.ME20139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scheid PL. New insights into the interaction of free-living amoebae and pandoravirus inopinatum: investigations of the host range and the role of multilamellar bodies. Open Parasitol. J. 2018;6:63–74. [Google Scholar]
- 79.Oliveira G, La Scola B, Abrahão J. Giant virus vs amoeba: fight for supremacy. Virol. J. 2019;16:126. doi: 10.1186/s12985-019-1244-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fukaya, S. & Takemura, M. Kinetic analysis of Acanthamoeba castellanii infected with giant viruses quantitatively revealed process of morphological and behavioral changes in host cells. Microbiol. Spectr.9, e0036821 (2021). Time lapse videos identified cell changes, such as rounding, and behaviour, such as rotation, in A. castellanii after infection of different giant viruses. [DOI] [PMC free article] [PubMed]
- 81.Oliveira G, et al. Tupanvirus-infected amoebas are induced to aggregate with uninfected cells promoting viral dissemination. Sci. Rep. 2019;9:183. doi: 10.1038/s41598-018-36552-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aoki, K. et al. Fifteen marseilleviruses newly isolated from three water samples in Japan reveal local diversity of marseilleviridae. Front. Microbiol.10, 1152 (2019). [DOI] [PMC free article] [PubMed]
- 83.Suzan-Monti M, Scola BL, Barrassi L, Espinosa L, Raoult D. Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus. PLoS One. 2007;2:e328. doi: 10.1371/journal.pone.0000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mutsafi Y, Shimoni E, Shimon A, Minsky A. Membrane assembly during the infection cycle of the giant mimivirus. PLoS Pathog. 2013;9:e1003367. doi: 10.1371/journal.ppat.1003367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yaakov LB, Mutsafi Y, Porat Z, Dadosh T, Minsky A. Kinetics of mimivirus infection stages quantified using image flow cytometry. Cytometry Part A. 2019;95:534–548. doi: 10.1002/cyto.a.23770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boyer M, et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. PNAS. 2009;106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dos Santos RN, et al. A new marseillevirus isolated in Southern Brazil from Limnoperna fortunei. Sci. Rep. 2016;6:35237. doi: 10.1038/srep35237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rolland, C. et al. Clandestinovirus: a giant virus with chromatin proteins and a potential to manipulate the cell cycle of its host Vermamoeba vermiformis. Front. Microbiol.12, 715608 (2021). [DOI] [PMC free article] [PubMed]
- 89.Bajrai L, et al. Kaumoebavirus, a new virus that clusters with Faustoviruses and Asfarviridae. Viruses. 2016;8:278. doi: 10.3390/v8110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Borges, I. et al. Trapping the Enemy: Vermamoeba vermiformis circumvents faustovirus mariensis dissemination by enclosing viral progeny inside cysts. J. Virol.93, e00312-19 (2019). [DOI] [PMC free article] [PubMed]
- 91.Andreani, J. et al. Orpheovirus IHUMI-LCC2: a new virus among the giant viruses. Front. Microbiol.8, 2643 (2018). [DOI] [PMC free article] [PubMed]
- 92.Andreani, J. et al. Morphological and genomic features of the new Klosneuvirinae isolate fadolivirus IHUMI-VV54. Front. Microbiol.12, 719703 (2021). [DOI] [PMC free article] [PubMed]
- 93.Bajrai, L. H. et al. Isolation of Yasminevirus, the first member of Klosneuvirinae isolated in coculture with Vermamoeba vermiformis, demonstrates an extended arsenal of translational apparatus components. J. Virol.94, e01534-19 (2019). [DOI] [PMC free article] [PubMed]
- 94.Reteno DG, et al. Faustovirus, an Asfarvirus-related new lineage of giant viruses infecting amoebae. J. Virol. 2015;89:6585–6594. doi: 10.1128/JVI.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Popovic M, Sarngadharan MG, Read E, Gallo RC. Isolation of virus capable of lysing the brown tide microalga, Aureococcus Anophagefferens. Biophys. Res. Commun. 1984;224:497–500. [Google Scholar]
- 96.Gastrich MD, et al. Ultrastructural analysis of viral infection in the brown-tide alga, Aureococcus Anophagefferens (Pelagophyceae) Phycologia. 1998;37:300–306. [Google Scholar]
- 97.Gastrich MD, et al. Viruses as potential regulators of regional brown tide blooms caused by the alga, Aureococcus anophagefferens. Estuaries. 2004;27:112–119. [Google Scholar]
- 98.Gastrich MD, Anderson OR, Cosper EM. Viral-like particles (VLPS) in the alga, Aureococcus anophagefferens (pelagophyceae), during 1999–2000 brown tide blooms in Little Egg Harbor, New Jersey. Estuarine Res. Federation Estuaries. 2002;25:938–943. [Google Scholar]
- 99.Gobler C, Anderson O, Gastrich M, Mary Downes G, Wilhelm S. Ecological aspects of viral infection and lysis in the harmful brown tide alga Aureococcus anophagefferens. Aquatic Microbial Ecol. 2007;47:25–36. [Google Scholar]
- 100.Nagasaki K, Ando M, Itakura S, Imai I, Ishida Y. Viral mortality in the final stage of Heterosigma Akashiwo (Raphidophyceae) red tide. J. Plankton Res. 1994;16:1595–1599. [Google Scholar]
- 101.Lawrence JE, Chan AM, Suttle CA. A novel virus (HaNIV) causes lysis of the toxic bloom-forming alga Heterosigma akashiwo (Raphidophyceae) 1. J. Phycol. 2001;37:216–222. [Google Scholar]
- 102.Nagasaki K, Tarutani K, Yamaguchi M. Growth characteristics of heterosigma Akashiwo virus and its possible use as a microbiological agent for red tide control. Appl. Environ. Microbiol. 1999;65:898–902. doi: 10.1128/aem.65.3.898-902.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nagasaki K, Yamaguchi M. Isolation of a virus infectious to the harmful bloom causing microalga Heterosigma akashiwo (Raphidophyceae) Aquatic Microbial Ecol. 1997;13:135–140. [Google Scholar]
- 104.Juneau P, Lawrence J, Suttle C, Curtis P, Harrison PJ. Effects of viral infection on photosynthetic processes in the bloom-forming alga Heterosigma akashiwo. Aquatic Microbial Ecol. 2003;31:9–17. [Google Scholar]
- 105.Tai V, et al. Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae) 1. J. Phycol. 2003;39:343–352. [Google Scholar]
- 106.Mirza SF, et al. Isolation and characterization of a virus infecting the freshwater algae Chrysochromulina parva. Virology. 2015;486:105–115. doi: 10.1016/j.virol.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 107.Wagstaff B, et al. Isolation and characterization of a double stranded DNA megavirus infecting the toxin-producing haptophyte Prymnesium parvum. Viruses. 2017;9:40. doi: 10.3390/v9030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tomaru Y, et al. New single-stranded DNA virus with a unique genomic structure that infects marine diatom Chaetoceros setoensis. Sci. Rep. 2013;3:3337. doi: 10.1038/srep03337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shirai Y, et al. Isolation and characterization of a single-stranded RNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus meunier. Appl. Environ. Microbiol. 2008;74:4022–4027. doi: 10.1128/AEM.00509-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kimura K, Tomaru Y. Isolation and characterization of a single-stranded DNA virus infecting the marine diatom Chaetoceros sp. Strain SS628-11 isolated from western Japan. PLoS One. 2013;8:e82013. doi: 10.1371/journal.pone.0082013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kimura K, Tomaru Y. Discovery of two novel viruses expands the diversity of single-stranded DNA and single-stranded RNA viruses infecting a cosmopolitan marine diatom. Appl. Environ. Microbiol. 2015;81:1120–1131. doi: 10.1128/AEM.02380-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nagasaki K, et al. Isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia Setigera. Appl. Environ. Microbiol. 2004;70:704–711. doi: 10.1128/AEM.70.2.704-711.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tomaru Y, et al. Isolation and characterization of a single-stranded RNA virus that infects the marine planktonic diatom Chaetoceros sp. (SS08-C03) Phycolog. Res. 2013;61:27–36. [Google Scholar]
- 114.Tomaru Y, Takao Y, Suzuki H, Nagumo T, Nagasaki K. Isolation and characterization of a single-stranded RNA virus infecting the bloom-forming diatom Chaetoceros socialis. Appl. Environ. Microbiol. 2009;75:2375–2381. doi: 10.1128/AEM.02580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nagasaki K, et al. Previously unknown virus infects marine diatom. Appl. Environ. Microbiol. 2005;71:3528–3535. doi: 10.1128/AEM.71.7.3528-3535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vincent, F., Sheyn, U., Porat, Z., Schatz, D. & Vardi, A. Visualizing active viral infection reveals diverse cell fates in synchronized algal bloom demise. PNAS118, e2021586118 (2021). [DOI] [PMC free article] [PubMed]
- 117.Rosenwasser S, et al. Rewiring host lipid metabolism by large viruses determines the fate of Emiliania huxleyi, a bloom-forming alga in the ocean. Plant Cell. 2014;26:2689–2707. doi: 10.1105/tpc.114.125641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nissimov JI, et al. Dynamics of transparent exopolymer particle production and aggregation during viral infection of the coccolithophore, Emiliania huxleyi. Environ. Microbiol. 2018;20:2880–2897. doi: 10.1111/1462-2920.14261. [DOI] [PubMed] [Google Scholar]
- 119.Yuxuan, X. et al. Virus-induced autophagy in the marine coccolithophorid Emiliania Huxleyi. Haiyang Xuebao45, 143–154 (2023).
- 120.Visvesvara GS. Giardiasis: an overview. IMJ. Illinois Med. J. 1983;164:34–39. [PubMed] [Google Scholar]
- 121.Lagunas-Rangel FA, Kameyama-Kawabe LY, Bermúdez-Cruz RM. Giardiavirus: an update. Parasitol Res. 2021;120:1943–1948. doi: 10.1007/s00436-021-07167-y. [DOI] [PubMed] [Google Scholar]
- 122.Cao L, et al. Giardia canis: ultrastructural analysis of G. canis trophozoites transfected with full length G. canis virus cDNA transcripts. Exp. Parasitol. 2009;123:212–217. doi: 10.1016/j.exppara.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 123.Miller RL, Wang AL, Wang CC. Purification and characterization of the Giardia Lamblia double-stranded RNA virus. Mol. Biochem. Parasitol. 1988;28:189–195. doi: 10.1016/0166-6851(88)90003-5. [DOI] [PubMed] [Google Scholar]
- 124.Marucci G, et al. Re-discovery of giardiavirus: genomic and functional analysis of viruses from Giardia duodenalis isolates. Biomedicines. 2021;9:654. doi: 10.3390/biomedicines9060654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Horiguchi T. Heterocapsa circularisquama sp. nov. (Peridiniales, Dinophyceae): a new marine dinoflagellate causing mass mortality of bivalves in Japan. Phycokogica Res. 1995;43:129–136. [Google Scholar]
- 126.Tarutani K, Nagasaki K, Itakura S, Yamaguchi M. Isolation of a virus infecting the novel shellfish-killing dinoflagellate Heterocapsa circularisquama. Aquatic Microbial Ecol. 2001;23:103–111. [Google Scholar]
- 127.Takano Y, Tomaru Y, Nagasaki K. Visualization of a dinoflagellate-infecting virus HcDNAV and its infection process. Viruses. 2018;10:554. doi: 10.3390/v10100554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Coy SR, et al. Visualization of RNA virus infection in a marine protist with a universal biomarker. Sci. Rep. 2023;13:5813. doi: 10.1038/s41598-023-31507-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tomaru Y, Mizumoto H, Nagasaki K. Virus resistance in the toxic bloom-forming dinoflagellate Heterocapsa circularisquama to single-stranded RNA virus infection. Environ. Microbiol. 2009;11:2915–2923. doi: 10.1111/j.1462-2920.2009.02047.x. [DOI] [PubMed] [Google Scholar]
- 130.Tomaru Y, et al. Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa circularisquama. Aquatic Microbial Ecol. 2004;34:207–218. [Google Scholar]
- 131.Mizumoto H, Tomaru Y, Takao Y, Shirai Y, Nagasaki K. Diverse responses of the bivalve-killing dinoflagellate Heterocapsa circularisquama to infection by a single-stranded RNA virus. Appl. Environ. Microbiol. 2008;74:3105–3111. doi: 10.1128/AEM.02190-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Weynberg KD, et al. Prevalent and persistent viral infection in cultures of the coral algal endosymbiont Symbiodinium. Coral Reefs. 2017;36:773–784. [Google Scholar]
- 133.Nakano S, Suzuki SI, Onji M. Virus-like particles suppress growth of the red-tide-forming marine Dinoflagellate Gymnodinium mikimotoi. Mar. Biotechnol. 2003;5:435–442. doi: 10.1007/s10126-002-0085-y. [DOI] [PubMed] [Google Scholar]
- 134.Takao Y, Nagasaki K, Honda D. Squashed ball-like dsDNA virus infecting a marine fungoid protist Sicyoidochytrium minutum (Thraustochytriaceae, Labyrinthulomycetes) Aquatic Microbial Ecol. 2007;49:101–108. [Google Scholar]
- 135.Westermann AJ, Gorski SA, Vogel J. Dual RNA-seq of pathogen and host. Nat. Rev. Microbiol. 2012;10:618–630. doi: 10.1038/nrmicro2852. [DOI] [PubMed] [Google Scholar]
- 136.Lum KK, Cristea IM. Proteomic approaches to uncovering virus-host protein interactions during the progression of viral infection. Expert Rev. Proteom. 2016;13:325–340. doi: 10.1586/14789450.2016.1147353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rodrigues, R. A. L. et al. Analysis of a Marseillevirus transcriptome reveals temporal gene expression profile and host transcriptional shift. Front. Microbiol.11, 651 (2020). Giant virus infection rapidly triggers the downregulation of genes related to host transcriptional machinery. [DOI] [PMC free article] [PubMed]
- 138.Zhang, R., Endo, H., Takemura, M. & Ogata, H. RNA sequencing of medusavirus suggests remodeling of the host nuclear environment at an early infection stage. Microbiol. Spectr.9, e0006421 (2021). [DOI] [PMC free article] [PubMed]
- 139.Rolland, C., La Scola, B. & Levasseur, A. How tupanvirus degrades the ribosomal RNA of its amoebal host? The ribonuclease T2 track. Front. Microbiol.11, 1691 (2020). Tupanvirus soda lake virus infection triggers rRNA shutdown potentially through RNAse T2 enzyme gene modulation. [DOI] [PMC free article] [PubMed]
- 140.Jones BM, Edwards RJ, Skipp PJ, O’Connor CD, Iglesias-Rodriguez MD. Shotgun proteomic analysis of Emiliania huxleyi, a marine phytoplankton species of major biogeochemical importance. Mar. Biotechnol. 2011;13:496–504. doi: 10.1007/s10126-010-9320-0. [DOI] [PubMed] [Google Scholar]
- 141.Schroeder DC, Oke J, Malin G, Wilson WH. Coccolithovirus (Phycodnaviridae): characterisation of a new large dsDNA algal virus that infects Emiliania huxleyi. Arch. Virol. 2002;147:1685–1698. doi: 10.1007/s00705-002-0841-3. [DOI] [PubMed] [Google Scholar]
- 142.Liu J, et al. Characterization of the sphingolipid profiling of Emiliania huxleyi against virus infection. J. Oceanol. Limnol. 2023;41:1547–1557. [Google Scholar]
- 143.Pagarete A, Allen MJ, Wilson WH, Kimmance SA, De Vargas C. Host-virus shift of the sphingolipid pathway along an Emiliania huxleyi bloom: survival of the fattest. Environ. Microbiol. 2009;11:2840–2848. doi: 10.1111/j.1462-2920.2009.02006.x. [DOI] [PubMed] [Google Scholar]
- 144.Schleyer G, et al. In plaque-mass spectrometry imaging of a bloom-forming alga during viral infection reveals a metabolic shift towards odd-chain fatty acid lipids. Nat. Microbiol. 2019;4:527–538. doi: 10.1038/s41564-018-0336-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Evans C, Pond DW, Wilson WH. Changes in Emiliania huxleyi fatty acid profiles during infection with E. huxleyi virus 86: Physiological and ecological implications. Aquatic Microb. Ecol. 2009;55:219–228. [Google Scholar]
- 146.Hunter, J. E., Frada, M. J., Fredricks, H. F., Vardi, A. & Van Mooy, B. A. S. Targeted and untargeted lipidomics of Emiliania huxleyi viral infection and life cycle phases highlights molecular biomarkers of infection, susceptibility, and ploidy. Front. Mar. Sci. 2, 81 (2015).
- 147.Malitsky S, et al. Viral infection of the marine alga Emiliania huxleyi triggers lipidome remodeling and induces the production of highly saturated triacylglycerol. New Phytol. 2016;210:88–96. doi: 10.1111/nph.13852. [DOI] [PubMed] [Google Scholar]
- 148.Kegel JU, et al. Transcriptional host-virus interaction of Emiliania huxleyi (Haptophyceae) and EhV-86 deduced from combined analysis of expressed sequence tags and microarrays. Eur. J. Phycol. 2010;45:1–12. [Google Scholar]
- 149.Sheyn U, et al. Expression profiling of host and virus during a coccolithophore bloom provides insights into the role of viral infection in promoting carbon export. ISME J. 2018;12:704–713. doi: 10.1038/s41396-017-0004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pagarete A, et al. Unveiling the transcriptional features associated with coccolithovirus infection of natural Emiliania huxleyi blooms. FEMS Microb. Ecol. 2011;78:555–564. doi: 10.1111/j.1574-6941.2011.01191.x. [DOI] [PubMed] [Google Scholar]
- 151.Hevroni G, Vincent F, Sheyn U, Vardi A, Ku C. Daily turnover of active giant virus infection during algal blooms revealed by single-cell transcriptomics. Sci. Adv. 2023;9:41. doi: 10.1126/sciadv.adf7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang E, et al. MicroRNA-mediated regulation of lipid metabolism in virus-infected Emiliania huxleyi. ISME J. 2022;16:2457–2466. doi: 10.1038/s41396-022-01291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ku C, et al. A single-cell view on alga-virus interactions reveals sequential transcriptional programs and infection states. Sci. Adv. 2020;6:1–10. doi: 10.1126/sciadv.aba4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Feldmesser, E., Ben-Dor, S. & Vardi, A. An Emiliania huxleyi pan-transcriptome reveals basal strain specificity in gene expression patterns. Sci. Rep.11, 20795 (2021). [DOI] [PMC free article] [PubMed]
- 155.Xue T, et al. Transcriptome analysis of marine microalga Emiliania huxleyi in response to virus infection[J] Haiyang Xuebao. 2019;41:103–112. [Google Scholar]
- 156.Sheyn U, Rosenwasser S, Ben-Dor S, Porat Z, Vardi A. Modulation of host ROS metabolism is essential for viral infection of a bloom-forming coccolithophore in the ocean. ISME J. 2016;10:1742–1754. doi: 10.1038/ismej.2015.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schatz D, et al. Hijacking of an autophagy-like process is critical for the life cycle of aDNA virus infecting oceanic algal blooms. New Phytol. 2014;204:854–863. doi: 10.1111/nph.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu J, Bratbak G, Zheng T, Thyrhaug R. Effects of virus infection on expression of cell cycle regulatory proteins in the unicellular marine algae Emiliania huxleyi. Acta Oceanol. Sin. 2011;30:89–95. [Google Scholar]
- 159.Jingwen Liu. Virus infection disturbs cyclin expression, leading to cell cycle arrest in the unicellular marine algae Emiliania huxleyi and Chrysochromulina ericina. Afr. J. Microbiol. Res.5, 1801–1807 (2011).
- 160.Moniruzzaman, M., Gann, E. R. & Wilhelm, S. W. Infection by a giant virus (AaV) induces widespread physiological reprogramming in Aureococcus anophagefferens CCMP1984-A harmful bloom algae. Front. Microbiol.9, 752 (2018). Giant virus infection triggers fast host transcriptional alterations, such as the supression of transcripts related to photosynthesis, and activation of protein synthesis transcripts. [DOI] [PMC free article] [PubMed]
- 161.Bale NJ, et al. Fatty acid dynamics during viral infection of Phaeocystis globosa. Aquatic Microb. Ecol. 2015;74:85–94. [Google Scholar]
- 162.Chen S, Gao K, Beardall J. Viral attack exacerbates the susceptibility of a bloom-forming alga to ocean acidification. Glob. Chang. Biol. 2015;21:629–636. doi: 10.1111/gcb.12753. [DOI] [PubMed] [Google Scholar]
- 163.Ray JL, et al. Virus infection of Haptolina ericina and Phaeocystis pouchetii implicates evolutionary conservation of programmed cell death induction in marine haptophyte-virus interactions. J. Plankton Res. 2014;36:943–955. doi: 10.1093/plankt/fbu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sheik AR, et al. Viral infection of Phaeocystis globosa impedes release of chitinous star-like structures: quantification using single cell approaches. Environ. Microbiol. 2013;15:1441–1451. doi: 10.1111/j.1462-2920.2012.02838.x. [DOI] [PubMed] [Google Scholar]
- 165.Lawrence SA, Floge SA, Davy JE, Davy SK, Wilson WH. Exploratory analysis of Symbiodinium transcriptomes reveals potential latent infection by large dsDNA viruses. Environ. Microbiol. 2017;19:3909–3919. doi: 10.1111/1462-2920.13782. [DOI] [PubMed] [Google Scholar]
- 166.Wang AL, Wang CC. A linear double-stranded RNA in Trichomonas vaginalis. J. Biol. Chem. 1985;260:3697–3702. [PubMed] [Google Scholar]
- 167.Graves KJ, Ghosh AP, Kissinger PJ, Muzny CA. Trichomonas vaginalis virus: a review of the literature. Int. J. STD AIDS. 2019;30:496–504. doi: 10.1177/0956462418809767. [DOI] [PubMed] [Google Scholar]
- 168.Wang A, Wang CC, Alderete JF. “Trichomonas vaginalis phenotypic variation occurs only among trichomonads infected with the double-stranded RNA virus.”. J. Exp. Med. 1987;166:1. doi: 10.1084/jem.166.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Khoshnan A, Alderete JF. “Trichomonas vaginalis with a double-stranded RNA virus has upregulated levels of phenotypically variable immunogen mRNA.”. J. Virol. 1994;68:6. doi: 10.1128/jvi.68.6.4035-4038.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Provenzano, D. & Alderete, J. F. Analysis of human immunoglobulin-degrading cysteine proteinases of Trichomonas Vaginalis. Infect. Immun.63, 3388–3395 (1995). [DOI] [PMC free article] [PubMed]
- 171.Provenzano D, Khoshnan A, Alderete JF. Involvement of dsRNA virus in the protein compositionand growth kinetics of host Trichomonas vaginalis. Arch. Virol. 1997;142:939–952. doi: 10.1007/s007050050130. [DOI] [PubMed] [Google Scholar]
- 172.Goodman RP, et al. Clinical isolates of Trichomonas vaginalis concurrently infected by strains of up to four trichomonasvirus species (Family Totiviridae) J. Virol. 2011;85:4258–4270. doi: 10.1128/JVI.00220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bessarab IN, Nakajima R, Liu HW, Tai JH. Identification and characterization of a type III Trichomonas vaginalis virus in the protozoan pathogen Trichomonas vaginalis. Arch. Virol. 2011;156:285–294. doi: 10.1007/s00705-010-0858-y. [DOI] [PubMed] [Google Scholar]
- 174.He D, et al. Differential protein expressions in virus-infected and uninfected Trichomonas vaginalis. Korean J. Parasitol. 2017;55:121–128. doi: 10.3347/kjp.2017.55.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Rada, P. et al. Double-stranded RNA viruses are released from Trichomonas vaginalis inside small extracellular vesicles and modulate the exosomal cargo. Front. Microbiol.13, 893692 (2022). [DOI] [PMC free article] [PubMed]
- 176.Tarr, P. I. et al. LR1: a candidate RNA virus of Leishmania (Leishmania Braziliensis Guyanensis/Single-Stranded RNA/Viral Particle/Electron Microscopy). Proc. Natl. Acad. Sci. USA85, 9572–9575 (1988). [DOI] [PMC free article] [PubMed]
- 177.Gupta V, Deep A. An insight into the Leishmania RNA virus. Indian J. Med. Microbiol. 2007;25:7–9. [PubMed] [Google Scholar]
- 178.Cantanhêde, L. M. et al. The maze pathway of coevolution: a critical review over the Leishmania and its endosymbiotic history. Genes12, 657 (2021). [DOI] [PMC free article] [PubMed]
- 179.Ives A, et al. Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Science. 2011;331:775–778. doi: 10.1126/science.1199326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Saura, A. et al. Elimination of LRVs elicits different responses in Leishmania spp. mSphere7, e0033522 (2022). [DOI] [PMC free article] [PubMed]
- 181.Kamoun S, et al. The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 2015;16:413–434. doi: 10.1111/mpp.12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Botella, L. & Jung, T. Multiple viral infections detected in phytophthora condilina by total and small rna sequencing. Viruses13, 620 (2021). [DOI] [PMC free article] [PubMed]
- 183.Hannat, S. et al. Diverse trajectories drive the expression of a giant virus in the oomycete plant pathogen Phytophthora Parasitica. Front. Microbiol.12, 662762 (2021). [DOI] [PMC free article] [PubMed]
- 184.Uchida, K. et al. Two novel endornaviruses co-infecting a phytophthora pathogen of Asparagus officinalis modulate the developmental stages and fungicide sensitivities of the host oomycete. Front. Microbiol.12, 633502 (2021). [DOI] [PMC free article] [PubMed]
- 185.Mascia T, et al. Infection of Colletotrichum acutatum and Phytophthora infestans by taxonomically different plant viruses. Eur. J. Plant Pathol. 2019;153:1001–1017. [Google Scholar]
- 186.Cai, G., Fry, W. E. & Hillman, B. I. PiRV-2 stimulates sporulation in Phytophthora infestans. Virus Res.271, 197674 (2019). [DOI] [PubMed]
- 187.Poimala, A. et al. Bunyaviruses affect growth, sporulation, and Elicitin production in Phytophthora cactorum. Viruses14, 2596 (2022). [DOI] [PMC free article] [PubMed]
- 188.Kamoun S. A catalogue of the effector secretome of plant pathogenic oomycetes. Ann. Rev. Phytopathol. 2006;44:41–60. doi: 10.1146/annurev.phyto.44.070505.143436. [DOI] [PubMed] [Google Scholar]
- 189.Kharel, A., Islam, M. T., Rookes, J. & Cahill, D. How to unravel the key functions of cryptic oomycete elicitin proteins and their role in plant disease. Plants10, 1201 (2021). [DOI] [PMC free article] [PubMed]
- 190.Slaveykova V, Sonntag B, Gutiérrez JC. Stress and protists: no life without stress. Eur. J. Protistol. 2016;55:39–49. doi: 10.1016/j.ejop.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Khan, N. A. Life Cycle and Ecological Significance in Acanthamoeba: biology and pathogenesis. 70-73 (Caister Academic Press, 2009).
- 192.Vardi A, et al. Programmed cell death of the dinoflagellate Peridinium gatunense is mediated by CO2 limitation and oxidative stress. Curr. Biol. 1999;9:1061–1064. doi: 10.1016/s0960-9822(99)80459-x. [DOI] [PubMed] [Google Scholar]
- 193.Frischkorn, K. R., Harke, M. J., Gobler, C. J. & Dyhrman, S. T. De novo assembly of Aureococcus anophagefferens transcriptomes reveals diverse responses to the low nutrient and low light conditions present during blooms. Front. Microbiol.5, 375 (2014). [DOI] [PMC free article] [PubMed]
- 194.Pelusi A, et al. Virus-induced spore formation as a defense mechanism in marine diatoms. New Phytol. 2021;229:2251–2259. doi: 10.1111/nph.16951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Frada, M. J. et al. Morphological switch to a resistant subpopulation in response to viral infection in the bloom-forming coccolithophore Emiliania huxleyi. PLoS Pathog.13, e1006775 (2017). Show that the haploid phase of E. huxleyi is resistant to E. huxleyi viruses, unlike the diploid cell. [DOI] [PMC free article] [PubMed]
- 196.De Vargas, C., Aubry, M. P., Probert, I. A. N., & Young, J. Origin and evolution of coccolithophores: from coastal hunters to oceanic farmers in Evolution of Primary Producers in the Sea. 251–285 (Academic Press, 2007).
- 197.Frada M, Probert I, Allen MJ, Wilson WH, de Vargas C. The ‘Cheshire Cat’ escape strategy of the coccolithophore Emiliania huxleyi in response to viral infection. PNAS. 2008;105:15944–15949. doi: 10.1073/pnas.0807707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zajac, A. J., Amphlett, E. M., Rowlands, D. J. & Sangar, D. V. Parameters influencing the attachment of Hepatitis A virus to a variety of continuous cell lines. J. General Virol. 72,1667–1675 (1991). [DOI] [PubMed]
- 199.Stapleton, J. T., Frederick, J. & Meyer, B. Hepatitis A. Virus attachment to cultured cell lines. J. Infect. Dis. 164, 1098–1103 (1991). [DOI] [PubMed]
- 200.Cheshenko N, Liu W, Satlin LM, Herold BC. Multiple receptor interactions trigger release of membrane and intracellular calcium stores critical for herpes simplex virus entry. Mol. Biol. Cell. 2007;18:3119–3130. doi: 10.1091/mbc.E07-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bishop, N. E. & Anderson, D. A. Early interactions of Hepatitis A virus with cultured cells: viral elution and the effect of PH and calcium ions. Arch Virol.142, 2161–2178 (1997). [DOI] [PubMed]
- 202.Wilson WH, et al. Isolation of viruses responsible for the demise of an Emiliania huxleyi bloom in the English. Channel. J. Mar. Biol. 2002;82:369–377. [Google Scholar]
- 203.Bratbak G, Egge JK, Heldal M. Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar. Ecol. Prog. Ser. 1993;93:39–48. [Google Scholar]
- 204.Evans C, Malin G, Mills GP, Wilson WH. Viral infection of Emiliania huxleyi (Prymnesiophyceae) leads to elevated production of reactive oxygen species. J. Phycol. 2006;42:1040–1047. [Google Scholar]
- 205.Mordecai, G. J., Verret, F., Highfield, A. & Schroeder, D. C. Schrödinger’s cheshire cat: are haploid Emiliania huxleyi cells resistant to viral infection or not? Viruses9, 51 (2017). [DOI] [PMC free article] [PubMed]
- 206.Silva LK, dos S, Boratto PVM, La Scola B, Bonjardim CA, Abrahão JS. Acanthamoeba and mimivirus interactions: the role of amoebal encystment and the expansion of the ‘Cheshire Cat’ theory. Curr. Opin. Microbiol. 2016;31:9–15. doi: 10.1016/j.mib.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 207.Neff’ RJSARWFB, Wilborn M. Induction of synchronous encystment (differentiation) in Acanthamoeba sp. Methods Cell Biol. 1964;1:55–83. [Google Scholar]
- 208.Cordingley JS, Wills RA, Villemez CL. Osmolarity is an independent trigger of Acanthamoeba Castellanli differentiation. J. Cell Biochem. 1996;61:167–171. doi: 10.1002/(SICI)1097-4644(19960501)61:2%3C167::AID-JCB1%3E3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 209.Aksozek A, Mcclellan K, Howard K. Resistance of Acanthamoeba Castellanii cysts to physical, chemical, and radiological conditions. J. Parasitol. 2002;88:621–623. doi: 10.1645/0022-3395(2002)088[0621:ROACCT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 210.Moon EK, Chung D, Il, Hong YC, Kong HH. Characterization of a serine proteinase mediating encystation of Acanthamoeba. Eukaryot Cell. 2008;7:1513–1517. doi: 10.1128/EC.00068-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.La Scola B, et al. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455:100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- 212.Fischer MG, Hackl T. Host genome integration and giant virus-induced reactivation of the virophage mavirus. Nature. 2016;540:288–291. doi: 10.1038/nature20593. [DOI] [PubMed] [Google Scholar]
- 213.Fischer MG, Suttle CA. A virophage at the origin of large DNA transposons. Science. 2011;332:231–234. doi: 10.1126/science.1199412. [DOI] [PubMed] [Google Scholar]
- 214.Gaia, M. et al. Zamilon, a novel virophage with Mimiviridae host specificity. PLoS One9, e94923 (2014). [DOI] [PMC free article] [PubMed]
- 215.Arthofer, P., Delafont, V., Willemsen, A., Panholzl, F. & Horn, M. Defensive symbiosis against giant viruses in amoebae. PNAS119, e2205856119 (2022). [DOI] [PMC free article] [PubMed]
- 216.George EE, et al. A single cryptomonad cell harbors a complex community of organelles, bacteria, a phage, and selfish elements. Curr. Biol. 2023;33:1982–1996.e4. doi: 10.1016/j.cub.2023.04.010. [DOI] [PubMed] [Google Scholar]
- 217.Javaux EJ, Knoll AH, Walter MR. Morphological and ecological complexity in early eukaryotic ecosystems. Nature. 2001;412:66–69. doi: 10.1038/35083562. [DOI] [PubMed] [Google Scholar]
- 218.Guglielmini J, Woo AC, Krupovic M, Forterre P, Gaia M. Diversification of giant and large eukaryotic dsDNA viruses predated the origin of modern eukaryotes. Proc Natl Acad Sci USA. 2019;116:19585–19592. doi: 10.1073/pnas.1912006116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Arsenieff, L. et al. First viruses infecting the marine diatom Guinardia delicatula. Front. Microbiol.10, 3235 (2019). [DOI] [PMC free article] [PubMed]
- 220.Bidle KD, Haramaty L, Barcelos Ramos J, Falkowski P. Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi. PNAS. 2007;104:6049–6054. doi: 10.1073/pnas.0701240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Irwin NAT, Pittis AA, Richards TA, Keeling PJ. Systematic evaluation of horizontal gene transfer between eukaryotes and viruses. Nat. Microbiol. 2022;7:327–336. doi: 10.1038/s41564-021-01026-3. [DOI] [PubMed] [Google Scholar]
- 222.Moliner C, Fournier PE, Raoult D. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol. Rev. 2010;34:281–294. doi: 10.1111/j.1574-6976.2010.00209.x. [DOI] [PubMed] [Google Scholar]
- 223.Keeling, P. J. & Eglit, Y. Openly available illustrations as tools to describe eukaryotic microbial diversity. PLoS Biol.21, e3002395 (2023). [DOI] [PMC free article] [PubMed]
- 224.Mihara, T. et al. Linking virus genomes with host taxonomy. Viruses8, 66 (2016). [DOI] [PMC free article] [PubMed]
- 225.Tomaru Y, Shirai Y, Suzuki H, Nagumo T, Nagasaki K. Isolation and characterization of a new single-stranded DNA virus infecting the cosmopolitan marine diatom Chaetoceros debilis. Aquatic Microbial Ecol. 2008;50:103–112. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed for this review paper. Literature cited is shown in the reference list and in the Supplementary Data File.